Abstract
Mathematics rules the world of science. Innovative technologies based on mathematics have paved the way for implementation of novel strategies in assisted reproduction. Ascertaining efficient embryo selection in order to secure optimal pregnancy rates remains the focus of the in vitro fertilization scientific community and the strongest driver behind innovative approaches. This scoping review aims to describe and analyze complex models based on mathematics for embryo selection, devices, and software most widely employed in the IVF laboratory and algorithms in the service of the cutting-edge technology of artificial intelligence. Despite their promising nature, the practicing embryologist is the one ultimately responsible for the success of the IVF laboratory and thus the one to approve embracing pioneering technologies in routine practice. Applied mathematics and computational biology have already provided significant insight into the selection of the most competent preimplantation embryo. This review describes the leap of evolution from basic mathematics to bioinformatics and investigates the possibility that computational applications may be the means to foretell a promising future for the IVF clinical practice.
Keywords: Prediction models IVF, Bayesian models IVF, Time-lapse IVF, Devices IVF, Artificial intelligence IVF
Introduction
The success rate during the first few years of in vitro fertilization (IVF) was 6% per cycle [1], which is less than a fifth of today’s success rates. Nowadays, trends have shifted towards improving implantation rates of the embryo and pregnancy rates, as well as better obstetrics and perinatal results along with better pediatric follow-up.
In order to achieve this, successfully selecting the top-quality embryos is of outmost importance. Embryo selection has always been based on a mathematical-algorithmic approach, considering that even the simplest grading systems are based on mathematics. Grading systems initially depended solely on morphological criteria as they were assessed through light microscopy [2]. These early embryo selection systems in reality were “decision trees”—a term commonly encountered in biostatistical and computer models. However, their empirical employment did not include a direct mathematical approach. The natural evolution of this pattern was the creation of mathematical equations—algorithms that could provide a better prediction of the embryo’s potential [3, 4].
Entering the “Computer Era,” it became clear that the mathematical approach would evolve to computer-based prediction models. The invention of devices and more complex software systems followed [5], enhancing the embryologists’ decision-making ability. Computational applications have fully entered the IVF laboratories to the point that the phrase “there is an app for that” is close to becoming a reality in the embryo selection models. More complicated mathematical and statistical tools are now employed, with the assistance of computer software in order to produce more accurate prediction models. The emergence and introduction of various devices in the laboratory have offered the embryologists the opportunity to closely monitor the embryos’ development. Programs created with artificial intelligence are able to detect embryo characteristics that could not be assessed by humans [6]. Nonetheless, prior to accepting novel approaches, we should evaluate when new technology should enter the clinical setting of an IVF laboratory and define what establishes approval and promotes safe practice [7]. The aim of this scoping review is to examine and report on the potential of bioinformatic-based approaches—including both devices and prediction algorithms—that may be part of an IVF laboratory’s routine. It should be noted that the term “bioinformatics” has been defined differently numerous times. Employment of bioinformatics in clinical embryology has a clear identity. For the present review, it is defined as the field that includes “the application of computers to the collection, organization, analysis, manipulation, presentation, and sharing of biologic data” [8]. The outline of the analysis performed in this study is presented in Fig. 1.
Fig. 1.
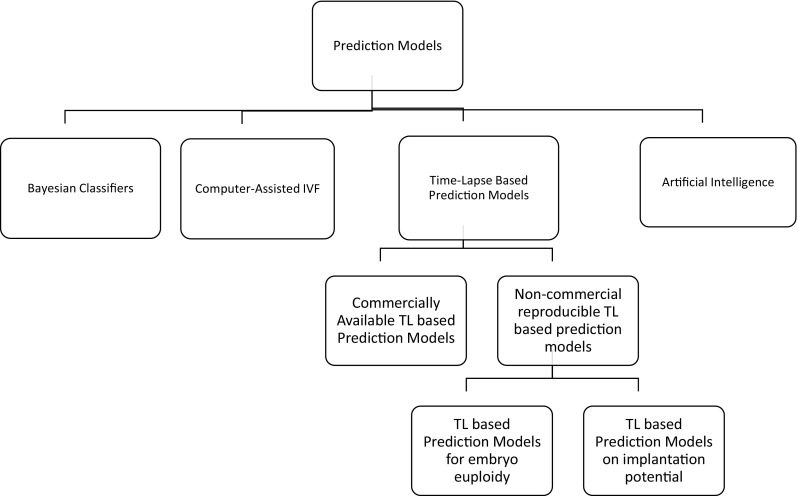
Outline of sophisticated models and software employed in the IVF laboratory and analyzed in this review
Methodology
This scoping review relied on researching the international published literature on the subject of bioinformatics in the service of predicting the IVF outcome. The studies presented in this review article were extracted from Scopus and PubMed. The keywords selected were “IVF prediction algorithms,” “time-lapse IVF,” “bioinformatics IVF,” and “artificial intelligence IVF.” At Scopus, the results were limited only to “medicine.” The overall search yielded 325 articles. Titles and abstracts of the search results were screened and evaluated. Articles that did not fit the concept of our study or that were not in English were excluded on that account by the authors. In addition to forward reference searching, citation mining was conducted by a backward reference search in high-impact articles. Emphasis was given to articles published by research groups specializing on the relevant field, without excluding all relevant contributions. The articles were categorized according to the structure decided for this study and critical analysis on them was performed.
Bayesian classification models: mathematics in the service of embryo selection
Bayesian classifiers are a classification type based on Bayes’ theorem. Bayes’ theorem allows the calculation of an event by using observed data even if the calculation depends on uncertain or missing parameters [9]. The first approach towards Bayesian classification models was reported by Dukic and Hogan [10]. Their model expanded a previous model that relied on applied mathematics, named the EU model [11], to a Bayesian hierarchical model. The EU model’s concept is that for a pregnancy to be achieved, the uterus must be receptive and the embryo must be viable, based solely on maternal age and embryo cell number. The parameters included in their algorithm were hydrosalpinx status, maternal age, oocyte age, and number of embryos which were not transferred. It was suggested that the higher the number of the embryos that were not transferred, the better the quality of the transferred embryos. The interaction between the uterus and the embryo with respect to implantation potential is perhaps considered a “blackbox” in the search for the factors involved in successful implantation. Although the algorithm’s prediction ability was confirmed by the observed pregnancies, the fact that it could be employed following embryo transfer was a serious limitation.
Adding more morphological characteristics of the embryo, Morales and colleagues conducted two studies developing Bayesian classification models [12, 13]. In the first study, different types of Bayesian classifiers were applied in order to select the embryos with the highest implantation potential. It was reported that the semi-naïve Bayesian classifier held the highest positive predictive value (50%), whereas all classifiers had a similar negative predictive value. A serious limitation in this study was the assessment of implantation potential in embryo batches (each batch consisted of 3 embryos) and the assumption that embryos with the highest score were the ones that implanted from each batch [12]. In the second study, seven different types of Bayesian classifiers were examined using a dataset of digital images of 249 embryos with known implantation data taken 40–50 h following fertilization. The embryos were graded according to Mill’s scoring system (1992). The aim was to predict the possibility of successful embryo implantation. The classifiers were successful in their prediction demonstrating an accuracy of at least 78%. The Bayesian type of “TAN with wrapper feature selection” reached 91% whereas logistic regression reached 82.33%. Bayesian classifiers proved to be able to perform successfully towards predicting embryo implantation. The authors concluded that a possible increase in the number of variables used for embryo morphological grading may result to a higher accuracy for the prognostic algorithm [13].
The evolution towards a Bayesian network (BN) was realized and tested against decision tree-based algorithms [14] and other Bayesian classification models [15]. Both studies were conducted by the same group, thus the BN featured the same parameters. Although embryo grading was performed daily and according to Istanbul Consensus Criteria (2011) [16], the grade was then converted to binary and more specifically categorized into “top” and “non-top.” Interestingly, parameters also included maternal age, number of IVF attempts, and the procedure employed for fertilization, adding another level of specificity to the approach. The BN presented similar accuracy to the decision tree but a higher area under curve (AUC), meaning that it was able to perform more reliable estimations [14]. According to the results of the second study, the BN proved to be of greater accuracy than the TAN Bayesian classifier, in contrast to the AODE Bayesian classifier which can be perceived as an ensemble of TANs. The authors interestingly propose a “decision support transfer.” DST suggests that prediction of embryo’s implantation is linked to parents, and an informed decision on embryo’s identity and number is deduced involving the patient, the physician, and the embryology laboratory. Specifically, according to the embryo viability possibility, it is proposed that clinicians and parents should decide if a selective single, a double, or a triple embryo transfer should be performed and what will confer optimization to the treatment—always abiding by the relevant legislative frame. It is a valid hypothesis that integration of a Bayesian network with time-lapse microscopy system will increase the algorithm’s predictive value and become a preferred approach [15].
On another note, the Bayesian hidden Markov model has been successfully proposed as means to enable aneuploidy screening, adding another element towards enriching prediction [17].
The development from Bayes’ theorem to a Bayesian network is challenging and interesting to follow. The study by Corani and colleagues suggesting that the model may enable the patients’ involvement into the embryo-transfer decision is in line with contemporary trends and strengthening the informed decision scenario [15].
The accommodation of Bayesian models may not strike the clinical IVF laboratory routine as straightforward and user friendly. Bayesian classifiers in ART were initially developed as statistical tools to prove mathematically that for a successful IVF, both patient and embryo characteristics are equal protagonists—what now is considered common knowledge. Their evolution towards prediction models encompassing more characteristics and a higher level of complexity appears to have been acknowledged. Bayesian prediction models report relatively high prediction power and equally high goodness-of-fit, as presented in Table 1. Nonetheless, implementation of a software application to enable employment of a Bayesian model from an IVF laboratory could potentially lead to better practice, and it is a direction worth exploring as it would require minimal processing power and would be able to accommodate a large number of samples.
Table 1.
Accuracy and AUC of Bayesian prediction models
| Study | Sample size | Single/multicenter | AUC | Accuracy | Externally validated |
|---|---|---|---|---|---|
| Morales et al. [12] | 249 embryos | S | 0.99 | 88% | N |
| Morales et al. [13]a | 63 cycles | S | – | 71.43% | N |
| Gianaroli et al. [14]a | 388 cycles | S | 0.72 | 81.5% | N |
| Corani et al. [15] | 600 embryos | S | 0.83 | – | N |
aResults correspond to embryo batches transferred and not to single embryos
Computer-assisted IVF
The quest for a better embryo-development evaluation carries a constant effort to improve our understanding of embryo morphology. One of the leading morphological features is blastomeres’ fragmentation. A computer-assisted method was employed including segmentation of digital embryo photographs and ×400 zoom in order to analyze embryo fragmentation and multinucleation. The morphological results were evaluated by DNA staining. The results demonstrated that fragmentation percentage and multinucleation were better evaluated through a computer as the human eye cannot perceive with absolute integrity of the above aspects. More specifically, it was identified that occasionally, large fragments could be mistaken as blastomeres by embryologists and the study proposed a cut-off value for the minimum size of the blastomere [18, 19]. Further to that, a plug-in was created, able to assess 24 morphological characteristics of the zygote and compared to the standard zygote scoring using six characteristics that were part of the laboratory routine [20]. Conclusively, the plug-in created was found to be more successful in accurately categorizing embryos into three groups in comparison to the embryologist.
On a slightly different and more embryo-orientated perspective, Paternot and colleagues conducted two studies [21, 22] in an attempt to create a complete embryo scoring system based on computer models. In the first study, the characteristics of an embryo were evaluated through computer analysis of digital embryo images, in order to create a complete embryo scoring system. The computer embryo evaluation was tested against the laboratory’s standard scoring system (SSS). The computer-assisted scoring system was reported as superior in terms of predicting live birth rates than the SSS [21]. For the 2013 study, evaluation of the optimal cytoplasmic volume was performed and associated with pregnancy outcome. The evaluation of cytoplasmic volume was based on a semi-automated system. It should be noted that in both studies, the computer system was not entirely autonomous as the embryologist contributed to the definition of every cell’s limit [22]. This drawback of the computer system could be resolved by a software that could automatically analyze blastomeres and define their boundaries. A recent approach accomplished this by creating a software that could automatically identify blastomeres and their irregularity on day 2 embryos with promising application [23]. Integration of the developed image-analysis software may automate the process of evaluating the embryo grading, thus creating a more robust entirely computer-based embryo grading system.
The rapid advancement of technology over the past decades has led to the establishment of informatics in every aspect of the human life and the IVF laboratory is no exception. Computer-assisted systems have been employed in the IVF laboratories for a wide range of applications. Aspects of the laboratories’ routine that have been automated include segmentation of digital photographs of the embryo, precise embryo measurements, and decision support algorithms. Since IVF outcome is defined principally by the gametes’ and embryo’s molecular characteristics, their contribution in prediction, in combination with the clinical factors of infertility, and embryology data could potentially be more enlightening. This merge might lead the way forward in computer-assisted IVF.
Time-lapse microscopy
The most widely employed devices introduced to the ART world in 2009 refer to time-lapse microscopes providing the embryologist with a complete view of the morphokinetics of the embryo. Employment of embryo morphokinetics has been related to higher rates of blastocyst formation and pregnancy [24]. Time-lapse systems have gained a fair market share of 17% of IVF laboratories in the USA [25]; however, there is still controversy regarding the clinical efficiency of these systems [24, 26–28].
It has been almost a decade since the emergence of the—still considered to be novel—approach of time-lapse. There have been studies supporting its efficiency and suggest its employment in IVF laboratories. Other studies represent the view that data on the benefits related to time-lapse service are still not convincing enough to justify the corresponding cost of the device and the consumables or the considerable changes that should be adopted regarding the IVF routine work. According to a meta-analysis, comparing time-lapse microscopy-based embryo grading versus conventional grading methods, the former failed to provide a statistically significant difference in improving clinical pregnancy or ongoing pregnancy rates [27]. To add to the complexity, the ability of time-lapse microscopy to predict embryo euploidy has also been examined. A recent systematic review concluded that on the grounds of various contradicting results obtained from literature, time-lapse microscopy may not offer information regarding euploidy in the same scope as PGS conveys—especially in clinical practice. Possible standardization of culture conditions may enable the adjustment of parameters that will in turn allow a possible association between embryo morphokinetics and euploidy [29].
Time-lapse microscopy has not yet earned the trust of the majority of embryologists, despite its availability for almost a decade. Numerous conflicting studies led several laboratories to regard time-lapse systems more as a “gadget” than a “necessity.” Further multicenter and perhaps multicountry, studies are required to solidify its effectiveness.
Commercially available prediction algorithms based on time-lapse systems
Since time-lapse microscopy entered the embryology laboratory, several attempts have been reported in order to generate an algorithm that would surpass the standard scoring system and achieve higher pregnancy rates.
The first attempt was reported in 2010 by Wong and colleagues [30] who created an algorithm able to predict blastocyst formation at the four-cell stage prior to embryonic genome activation. Wong’s algorithm employed Sequential Monte Carlo methods in order to create a prediction model by featuring 5-min time frames from time-lapse system. Three features were highly correlated with blastocyst formation, the duration of the first cytokinesis, and time between the 2nd and 3rd-cell stage and between the 3rd and 4th-cell stage. The proposed algorithm was patented and later became commercially available as the Eeva system. Based on these data, a decision tree was created, serving as the Eeva classification algorithm, and the system was completed with the addition of an image analysis software. Two studies followed aiming to examine its effectiveness [31, 32]. In Conaghan’s study, embryologists using the Eeva software, with a two-category system, were able to predict good blastocyst formation rate on day 3 embryos. This was supported with an accuracy rate of more than 66%, resulting to a statistically significant improvement in comparison to the accuracy corresponding to the same embryologists employing respective standard scoring method. These results validated and supported Eeva’s use in clinical practice [31]. In VerMilyea’s study, the Eeva software was further improved to provide a three-category system (high, medium, and low-quality embryos). The study proved the three-category system to be of added benefit in regard to the medium quality embryos. The Eeva software has been implemented in several IVF programs as a user-friendly approach enriching and strengthening the embryologists’ choice on the embryo selection [32]. However, its success on improving the implantation rate in comparison to the ALPHA/ESHRE’s morphology grading-based decision is not solidified [33]. The Eeva system appears to perform more efficiently when compared to morphology-based selection according to ASEBIR criteria [34]. The Eeva system has updated its software adding patient characteristics and morphological parameters thus creating a five-category output. This new update is commercially known as the Eeva Xtend algorithm. The combination of this new algorithm with traditional morphology grading obtained by the embryologist could efficiently promote eSET [35].
An additional time-lapse decision tree-based algorithm is the commercially available KIDScore [33]. KIDScore algorithm, also known as known implantation data score, includes morphokinetic parameters along with cell number following 66 h. The equations were calculated using recursive partitioning of the initial parameters. This work relied on the largest sample encountered namely 3275 embryos ensuring robustness coupled by significant validation on 11,218 embryos. KIDScore algorithm presented a statistically significant higher predictive capability for blastocyst formation and quality than the ALPHA/ESHRE scoring system [33]. It should be noted that an update of the KIDScore includes the addition of a day 5 decision support tool (KIDScore D5) that can be employed only in culture systems of certain oxygen conditions, featuring the above algorithm along with inner cell mass and trophectoderm characteristic evaluation. However, consensus on investigating the results and the corresponding benefits has not yet been reported. The KIDScore has not been evaluated to the same extent as Eeva, possibly due to its more recent emergence and availability counting less than 2 years.
Reproducible prediction algorithms based on time-lapse systems accessible on a non-commercial basis for embryo euploidy
Since 1995, when the first pregnancies following PGS were reported [36], embryo euploidy has been in the center of attention when selecting the embryos to be transferred. Two decades later and the juries are still out for the (cost-)effectiveness of preimplantation genetic test for aneuploidy (PGT-A) [37]. A recent ASRM committee opinion concluded that the effectiveness of PGT-A cannot yet be determined although it is universally employed as a screening test for embryo euploidy [38].
In an effort to develop an algorithm that would be able to accurately predict embryo euploidy, morphokinetical characteristics obtained through time-lapse microscopy were associated with the risk of aneuploidy in human embryos [39]. A decision tree-based algorithm was proposed dividing embryos in three categories (low, medium, high) in regard to aneuploidy risk in association to the time required from insemination to reaching the blastocyst stage (before expansion) and the time from insemination to blastulation. This algorithm demonstrated a high prediction ability of 97% regarding embryos at high risk of being aneuploid, whereas low risk embryos presented a 37% probability of aneuploidy. The algorithm was examined in a following study by the same group, including fetal heart beat and live birth rate data [40]. Robust results were provided with the low risk for aneuploidy group of embryos being associated with improved outcomes. However, it should be noted that the sample in both studies featuring less than 100 embryos each was considerably smaller in comparison to the work of other groups, highlighting the limitation of this work. In the same direction on predicting aneuploidy, a larger study proposed another algorithm based on completely different morphokinetics, evaluating earlier stages of embryo development [41]. The proposed algorithm presented to be statistically more efficient in terms of predicting embryo euploidy. Employment of Basile’s algorithm presents the advantage of enabling embryo transfer on day 3 [41]. However, the statistical significance of the parameters featured has been argued [40]. A study comparing three of the above algorithms (Campbell, Basile 2014, and Cruz) along with three other studies proposing selection/deselection criteria but not scoring algorithms was conducted [42]. Campbell’s algorithm proved to hold the highest positive predictive value and the second highest negative predictive value regarding euploidy status. Basile’s algorithm was externally evaluated resulting to 96% of embryos under the “A” category being euploid whereas only 3% of category “D” embryos were euploid [43]. However, the issue of mosaicism should be considered as a serious limitation regarding PGD studies. It has been reported that up to 24% of embryos that are diagnosed by PGD may be mosaic [44]. The prediction models for embryo euploidy presented in Table 2 are hitherto limited, and according to the reported results, significant efforts on their development are still required towards their implementation in clinical routine. Due to their high negative prediction value, perhaps a possible means of employment for these prediction models could be to assist in the decision of which embryos PGT-A should be performed on.
Table 2.
Positive and negative prediction values of prediction models employing TL regarding embryo euploidy
Reproducible prediction algorithms based on time-lapse systems accessible on a non-commercial basis featuring implantation potential
The first attempt to create a decision tree algorithm applied on time-lapse systems associating embryo morphokinetics to implantation rates was created by Meseguer [45]. The decision tree led to a total of 10 categories ranging from A+ to F. This could provide significant assistance to the embryologist in selecting the top-quality embryo and achieving better implantation rates. Further assessing this algorithm in two subsequent studies was added to validation of results [46, 47]. Meseguer’s algorithm proved to be robust considering the blastocyst formation rate [46]. Furthermore, Meseguer’s algorithm was compared to Wong’s and according to Cruz, it was associated to a greater implantation potential. A serious limitation was that Wong’s algorithm employed a different time-lapse system [46]. Basile and colleagues employed a greater number of embryos (1122) with known implantation data on Meseguer’s algorithm and found the decision tree to be significantly altered, resulting to a lower implantation rate for the top category embryos (A+). In particular, it recorded a major decrease from 66 to 32% [47]. This important work performed by this group and depicted in consecutive publications has successfully proved the scenario of improving the original concept through thorough validations.
An algorithm supporting that embryo cleavage synchronicity is a more reliable marker than absolute time points was developed by Cetinkaya and colleagues in 2015 [48]. Three different equations were developed based on results of previous literature and were respectively evaluated. The equation that included intervals from two to three cells, from four to five cells, and from two to eight cells demonstrated higher top-quality blastocyst formation potential with almost 80% of good and top-quality blastocysts being classified in the top quadrille and 86% of arrested embryos and poor-quality blastocysts being classified in the bottom quadrille. The authors suggested the integration of their algorithm with Campbell’s in order to be able to predict embryo euploidy as well but no study evaluating a possible integration has been performed.
Timing of cell divisions is of paramount importance and was implemented in an algorithm featuring timing of the first division all the way through to the five-cell stage, along with the duration of the second-cell cycle and that of the three-cell stage [49]. The feature weight for each parameter was indicated through a multivariate logistic regression analysis. The embryos that developed to blastocysts presented a statistically significant higher score than embryos that failed to reach the blastocyst stage. This one along with the study by Cetinkaya et al. [48] was the sole study not relying on a decision tree for embryo grading. Instead, Milewski’s group created a purely mathematical reproducible algorithm, albeit relying on a relatively small sample. This algorithm was re-evaluated by the same group, on a larger sample, targeting implantation rate instead of blastocyst formation potential [50]. Due to the shortage of the previous algorithm to present a statistically important prediction ability, a new algorithm was created by employing principal component analysis. The new algorithm added two new parameters, blastomere fragmentation at the three-cell stage and maternal age. The proposed algorithm resulted in negative values with close to zero scores being associated with higher implantation rates. By dividing the embryo in four quadrilles according to their scores, the study presented a statistically important correlation between embryo score and implantation potential. Expansion and reexamination of this approach merit further investigation on a multicenter level to cement its contribution towards embryo selection.
A two-part decision tree algorithm was proposed employing morphokinetical parameters in order to successfully predict blastocyst formation rate as well as implantation [51]. For the first part, time required to reach morula stage and time required for transition from the five to eight-cell stage were featured. For the second part, the latter parameter was featured again, however presenting with a different cut-off value, along with time required to reach the blastocyst stage. Following its development, the algorithm was tested using a different set of embryos. The algorithm provided successful associations between the top category embryos which proved to correspond to a higher blastocyst formation rate and higher implantation potential. The authors highlighted that all embryo-transfer selections were performed according to a combination of day 5 morphological criteria assessment along with Meseguer’s algorithm.
Liu and colleagues [52] proceeded with a new time-lapse based prediction algorithm equally employing a decision tree. The variables employed in this decision tree were poor conventional morphology, abnormal cleavage, less than eight cells after 68 h, duration of three-cell stage, and time from pronuclear fading to the five-cell stage. The embryos were categorized into seven categories and the implantation rate ranged from 52.9% for the top category to 0% for the worst category. Liu and colleagues [52] proposed that each laboratory should create its own cut-off values, slightly changing the parameters of the decision tree, since culture variations may alternate the embryo growth. This approach might have to offer the benefit of customization in the sense that each IVF laboratory will define their own algorithm parameters on embryo categorization by adjusting their cut-off values employing the same decision tree. Such an approach deserves to be expanded and adopted in the era of personalized medicine extending significantly in the ART set-up.
A recent retrospective study by Carrasco and colleagues proposed an algorithm where the decision tree included morphokinetics along with the novelty of introducing the parameter of morphological features [53]. The first step in this decision tree algorithm is the assessment of embryo morphology. Embryos on day 3 with less than 6% or an asymmetric evaluation of blastomeres, fragmentation of more than 20%, or multinucleation were categorized as poor. All event timings were statistically analyzed and the time to reach the four-cell stage was determined to be more important followed by the time to reach the eight-cell stage. The poor group presented an implantation rate of less than 10%, the fair group of 17%, and the good quality group corresponded to an implantation rate of 29%.
The current review presents a palette of studies on prediction models relying on algorithms employing mainly the concept of the decision tree based on morphokinetics. The majority of these present more similarities than differences. Similarities include recruitment of decision trees relying on similar parameters referring to time intervals from one-cell stage or cell division to another. Principal differences were identified regarding the size of the sample and embryo pool that the results relied on. This may consequently strengthen some proposed models over others. In addition to that, the prioritization and hierarchy of the decision trees reported to be different along with diverse cut-off values. This fact reflects yet again the discrepancies regarding the considerations on the high or low priority and influence of certain parameters over others. Commercially available updated algorithms include morphological criteria. It should be distinguished that the study by Carrasco was the only—accessible on a non-commercial basis—reproducible algorithm not relying solely on morphokinetics, but employing specific morphological features not taken into account by the other models. All algorithms can be reproduced and the relevant prediction models can be adopted by a standard IVF laboratory featuring time-lapse technology. Regarding commercialization of the findings and software development based on findings, the studies by Wong and Petersen evolved into a commercially available software that can be attached to and employed by any IVF laboratory. The studies by Meseguer, Basile, Milewski, and Liu and colleagues provide reproducible, accessible algorithms.
Petersen and colleagues compared their algorithm to some of the abovementioned ones except from Carrasco’s—which was created afterwards. According to this study, Petersen and Liu’s algorithms were associated with a statistically significant higher blastocyst formation rate and quality, whereas Conaghan’s algorithm resulted to a statistically significant lower blastocyst formation rate and quality. Liu also conducted a study comparing their own algorithm with the KIDScore, along with Meseguer’s algorithms, both the original and Basile’s improved version [54]. According to this study, all algorithms presented reduced predictive value when only single embryo transfer was performed, which further reduced when maternal age and embryo morphology were accounted for. Liu’s algorithm was demonstrated as the more robust. The authors concluded that in order to develop algorithms with higher predictive value, morphological and clinical features should be included. Another recent study compared some of the aforementioned algorithms assessing both interalgorithm comparison and comparison with the embryologists’ embryo selection procedure [55]. The same study highlighted that it is Petersen’s suggested model (KIDScore) that compares well to the practitioner embryologists’ selection criteria. The key conclusion focused on the lack of agreement when comparing the predictability between the algorithms.
The question raised is which of these algorithms carries the highest prognostic value. Most of the time-lapse based prediction models have been validated. Their reports on prediction power and goodness-of-fit (referring to the AUC) are presented on Table 3. However, the validation studies report different prediction capabilities, thus creating a conflict in literature. It is essential that external validation on a large sample prior to reproduction, adoption, and horizontal employment of the suggested models is performed in order to indicate strengths and weakness of the proposed models. It is imperative that the IVF scientific community reaches a consensus on the algorithm use and value as a prognostic tool in the IVF laboratory. Premature adoption of models may be associated with risk in clinical practice. The underlying reason behind the risks involved in wide-ranging application is the fact that different IVF laboratories follow different practices, employ different criteria, and select embryos on different grounds, using different cut-off values. This study highlights the differences—sometimes minor and other times critical—regarding the importance of associated factors and their respective weight and role in the development of each algorithm. Conclusively, each model is different to the next. This observation may stem from the fact that different IVF laboratories rely on divergent selection criteria. It is therefore imperative to take into account this diversity and perhaps strengthen the scenario that different practices should promote and rely on the use of “in-house” algorithms. On the other hand, the use of a universal commonly accepted more evidence-based efficient algorithm remains the goal. This issue highlights yet again the need for a common universal protocol on grading and evaluating an embryo and its corresponding implantation potential. Since 2011 and the Istanbul consensus [16], the field of clinical embryology has made strides in relation to embryo selection. However, horizontal adoption of a consensus on embryo grading has yet to be achieved to a satisfactory extent commonly accepted and practiced.
Table 3.
Positive and negative prediction values of prediction models employing time-lapse microscopy
| Study | Sample size (embryos) | Single/multicenter | Blastocyst formation | Implantation | AUC | Externally validated | Commercially available | ||
|---|---|---|---|---|---|---|---|---|---|
| PPV | NPV | PPV | NPV | ||||||
| Meseguer et al. [45] | 247 | S | – | – | 66% (52%)b | 92% | – | Y | N |
| Conaghan et al. [31] | 941 | M | 54.7% | 73.7% | – | – | – | Y | Y |
| VerMilyea et al. [32] | 375 | M | – | – | 37% | 85% | – | Y | Y |
| Basile et al. [41] | 1620 | M | – | – | 32% | 83% | – | Y | N |
| Cetinkaya et al. [48] | 3354 | S | 79.2% | 86.2% | – | – | – | N | N |
| Milewski et al. [49] | 432 | S | 90.7% | 73.3% | – | – | 0.813 | Y | N |
| Behr et al. [35] | 216 | M | – | – | 51% | 100% | – | N | Y |
| Milewski et al. [50] | 410 | S | – | – | 46% | 87.9% | 0.703 | N | N |
| Motato et al. [51] | 257[832]c | S | – | – | 90.9% | 58.2% | 0.596 | N | N |
| Liu et al. [52] | 36[270]c | S | – | – | 50% | 100% | 0.783 | Y | N |
| Petersen et al. [33] | 3275 | M | – | – | 36.17% | 94.8% | 0.745 | Y | Y |
| Carrasco et al. [53] | 800 | S | – | – | 29% | 90.5% | – | N | N |
bThe number in brackets represents the PPV of the five-category model whereas the number outside brackets represents the nine-category model
cThe number in square brackets refers to the sample for the development of the algorithm, whereas the number outside square brackets refers to the sample of the test phase
Artificial intelligence—the next big step
Artificial intelligence (AI) is defined as the machine’s ability to learn and exert intelligent behavior. The early steps towards AI in medicine were recorded in the 1960s. First, the use of the naïve Bayesian classifier was introduced, followed by neural networks, symbolic learning (using decision trees), machine learning [56], and nowadays the accomplishment of deep-learning AI. Deep learning allows the computer to discover a structure in a large dataset using a back-propagation algorithm and to conduct small changes in its parameters in order to achieve the algorithm with the optimal representation of the dataset [57].
In the field of IVF, the first attempt to employ AI was in 1997 [58]. An artificial neural network (ANN) was created employing the variables of maternal age, number of oocytes retrieved, and number of embryos transferred and whether embryo cryopreservation was performed. The ANN managed to accurately predict pregnancy in 59% percent of the cases ascertaining a decent performance efficiency. An AI program capable of predicting the outcome of IVF/ICSI employing surgically removed spermatozoa was also described [59]. The variables included in this program were maternal age, sperm type (fresh or frozen), etiology of male factor infertility, and sperm retrieval technique. Only maternal age and sperm type were found to be of statistical significance using reverse regression analysis. This program was only able to successfully predict failed attempts with an accuracy of 82%. The lack of accuracy concerning successful attempts hinders the algorithm’s clinical usefulness.
Focusing on embryo selection as well as clinical characteristics, an algorithm based on support vector machine (SVM) was developed employing both female characteristics and embryo morphology for the prediction of the IVF outcome [60]. Both numerical and categorical features were included. In order to transform their categorical values, the algorithm was tested based on three different techniques: The first was binary encoding including eight categories. The second technique involved frequency-based encoding which transformed the categorical value into numerical defined as the difference between the occurrence of each categorical value in positive and negative outcomes. The third approach referred to expert judgment which required the input of a number (one to four) for each categorical value by embryologists. Frequency-based encoding had the best prediction accuracy, suggesting that machine learning may be less biased than experts and more sensitive than simple binary encoding. This lead to the conclusion that machine learning may offer better predictive capabilities. Although stratified validation was performed, all data were obtained from a single center. The lack of external validation may hinder the algorithm’s robustness.
The creation of an ANN for prediction of IVF outcomes was proposed by selecting at least 10 characteristics from a pool including treatment, embryo, female, and male characteristics. The aim was to define the most important characteristic, removing both the statistically insignificant and those that were highly correlated between them [61]. However, the results of their promising study have not yet been presented.
Machine learning has been assessed against purely statistical methods [62]. Based on clinical characteristics, an algorithm was created via multivariate logistic regression analysis. An artificial network was developed by running the algorithm 30,000 times with random startup parameters each time. The ANN with the highest sensitivity (69%) and specificity (60.3%) was selected, achieving an (AUC) of 0.7026. When compared to the algorithm developed through statistics, the ANN proved to have a strong statistically significant better prediction ability. These results demonstrate the higher capability of ANNs in developing prediction models compared to traditional statistical methods.
The development of a pregnancy prediction model that included both embryo and clinical characteristics ensued [63]. Employing the boosted tree method, the created model named IVF-BT (IVF-BoostedTree), assessed both male and female clinical characteristics, ovarian simulation protocols, embryo grading, and preimplantation genetic screening results and—as anticipated—proved to be more accurate than age alone. Evolving the IVF-BT, a model predicting success of a second attempt cycle based on the first cycle characteristics was proposed and evaluated. The study concluded that information from a first failed cycle could hold prognostic power and prove to be beneficial in designing and performing a second cycle [63].
Choi and colleagues [64] implemented the use of more than 20 clinical characteristics for the creation of their model. This approach slightly altered the scope towards the use of clinical characteristics of the female and the male referring to pre-stimulation factors and prediction of live birth by means of a boosted regression tree approach. Their model, named PreIVF-D (PreIVF-Diversity), was built on a multi-step process using data from different clinics and a total of 13,076 cases. It was later trained for adjusting specific parameters by 1021 other cases and tested on 1058 different cases. The aim of this model was to personalize IVF prediction rates prior to embarking on treatment by thoroughly informing patients regarding their chances of success. The PreIVF-D’s receiver operating characteristic (ROC) analysis was slightly improved in comparison to the control model. It should be noted that the model predicts the success probability per oocyte retrieval and not per fresh ET alone. Additionally, the model may be subjected to slight alterations in case of different clinical protocols, provided that training set is in place, thus making the model widely applicable. The authors propose the employment of personalized prediction models in order to reassure patients. Patients that may be of good prognosis may particularly benefit from such an approach as the stress related to low success rates that may trouble them could be alleviated [64].
An entirely different approach relying entirely on oocyte and embryo data presented novelty [6]. This approach recommended the capture of digital photographs through an inverted microscope and a further magnification of ×20, followed by segmentation of these photographs and a subsequent computer analysis. Following this, two test-points were introduced and performed: the first, in order to evaluate different classifiers for an ANN and the second, to evaluate the proposed classifier (random subspace ensemble of neural networks, RSNN), using two different semi-supervised learning methods. The first learning process included the use of certain instances of the training set to classify the other training patterns. The second learning method consisted of a 10-fold cross validation of the training set, by randomly dividing the dataset into 10 different subsets, repeated 150 times. Nine of the above sets were used for training and the 10th for testing. The features incorporated in this method were not all perceived by the human eye and they were analyzed by artificial intelligence. This fact alone highlights the benefits and ascertains the bioinformatics place in the IVF laboratory. As the study suggests, an integration of this system with time-lapse microscopy could further enhance the prediction of the IVF outcome employing successful selection of an oocyte or indicating the embryo implantation potential by relying on pattern recognition and AI methodologies [6].
Although decent attempts have been committed towards creating an AI program capable of predicting a pregnancy outcome and/or selecting the embryo with the highest implantation potential, none of these are yet suitable and qualified for the IVF laboratory routine. Against anticipated beliefs, the prediction models created with AI do not appear to be of superior effectiveness or credibility in comparison to other prediction models, as initial reported results demonstrate (Table 4). A possible reason behind this is the lack of a comprehensive software that should be all inclusive relying on: male and female clinical characteristics, morphological and morphokinetical embryo characteristics, as well as culture conditions. The creation of such a program would require a learning set consisting of a vast number of IVF cycles and an equally large testing dataset with known implantation data and pregnancy outcome that is yet to be realized. It should be noted that impressive strides in this field are being made and the rapid development and evolution of AI is remarkable. Therefore, further progress is reflected in the potential of AI and its successful implementation in the IVF laboratory is certainly anticipated.
Table 4.
Prediction power and AUC for prediction models developed with AI
| Study | Sample size | Single/multicenter | Implantation | Clinical pregnancy | AUC | Externally validated | ||
|---|---|---|---|---|---|---|---|---|
| PPV | NPV | PPV | NPV | |||||
| Kauffman et al. [58] | 455 cycles | S | – | – | 39% | 82% | – | N |
| Wald et al. [59] | 113 cycles | S | – | – | – | – | 0.783 | N |
| Uyar et al. [60] | 2429 embryos | S | 65.6% | 67.5% | – | – | 0.712 | N |
| Banerjee et al. [63] | 1676 cycles | S | – | – | – | – | 0.8 | N |
| Milewski et al. [62] | 1995 patients | S | – | – | – | – | 0.642 | N |
| Choi et al. [64] | 1058 cycles | M | – | – | 59.4% | 94.9% | – | N |
| Manna et al. [6] | 269 embryos | S | – | – | – | – | 0.83 | N |
Conclusion
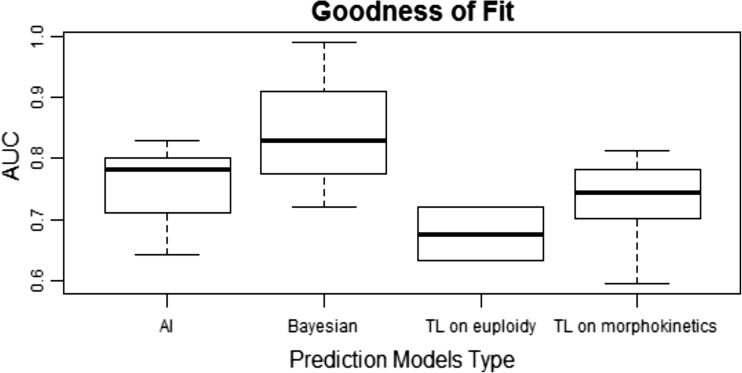
All of the prediction models presented in this review have proved their robustness—to a different extent—through the goodness-of-fit. For all prediction models that tested their goodness-of-fit, the evaluation was made by employing the AUC. A graphic representation of their goodness-of-fit is presented in Fig. 2. The data is there; still the decision regarding the identity of the most efficient prediction model should be made by the respective IVF laboratory. Such a decision should rely on criteria that are custom made and are identified on the rounds of best serving the needs of each clinical embryology laboratory irrespectively of the given prediction model’s efficiency.
Fig. 2.
Goodness-of-fit as measured by area under the curve (AUC) for the four prediction models types that include two or more algorithms: artificial intelligence, Bayesian classifiers, time-lapse (TL) on euploidy, and time-lapse (TL) on morphokinetics
In an ideal scenario and in the best interest of securing optimal embryo culture, the integration of the newly discovered devices and systems is implied to be the key. However, the plausibility of this scenario is questionable. Besides the apparent encountered obstacles, applying this integration by successfully recruiting all important contributions from the world of computational applications into the IVF practice raises several issues. What remains to be seen in the near future is what the IVF world’s verdict will be on adoption of such practices employing algorithms, software, and devices. A horizontal application dictates the way forward and the proposed changes to be manageable and feasible without majorly disrupting the IVF laboratory’s function that strictly relies on morphological criteria for embryo selection. Could it be that the traditional approach of light microscopy evaluation prior to the ET will be totally abandoned or could we simply combine both options and hence leave the final decision on the embryologist taking into account the software ranking on embryo quality? Is our goal to eliminate the human element and its subjective nature from the equation and rely on the objective analysis promised by the automation that computational applications offer? These are all valid considerations and deserve to be addressed. Ultimately, what would be the clear advantage of creating a fully automated laboratory and to which extent should the human element be present and controlling the final choices? An important factor to acknowledge here is the non-invasive approach and practice of the tool of computational applications in the IVF laboratory. The future certainly holds the answers on such considerations and will provide improved models supported by robust data along with conclusions. Further research is required on delineating the practical aspects on accommodating this approach and how the embryologists’ role within the IVF laboratory will be shaped implementing and including computational applications in its routine. Complexity in the era of personalized medicine is a given. Embryology and bioinformatics are two current strikingly developing fields. Perhaps the next step towards implementing bioinformatics in the IVF laboratory lies in bridging the chasm between understanding both intricate fields and explaining the underlying motives behind this merge. It is of essence to impart the principles of embryo development and the basis of an embryology laboratory routine to bioinformatic connoisseurs ascertaining development of more user-friendly means. Concurrently, there is undoubtful benefit in acquainting and familiarizing embryologists with the basics of computational biology. This will allow more control and understanding in decision making and working towards trusting novel approaches on our way to improving IVF practice.
Authors’ contribution and agreement
M. Simopoulou and K. Sfakianoudis conceived and designed the study. M. Simopoulou, E. Maziotis, N. Antoniou, and A. Rapani performed the literature search and contributed to drafting the manuscript. P. Bakas, S. Bolaris, and A. Pantou contributed to the structure of the manuscript. M. Simopoulou and G. Anifandis edited the manuscript. M. Simopoulou, K. Pantos and, M. Koutsilieris revised the manuscript. All authors approved the final draft.
Footnotes
M. Simopoulou and K. Sfakianoudis are the first co-authors in this study.
K. Pantos and M. Koutsilieris are the last co-authors in this study.
References
- 1.Steptoe PC, Edwards RG, Purdy JM. Clinical aspects of pregnancies established with cleaving embryos grown in vitro. Br J Obstet Gynaecol. 1980;87:757–768. doi: 10.1111/j.1471-0528.1980.tb04611.x. [DOI] [PubMed] [Google Scholar]
- 2.Rienzi L, Vajta G, Ubaldi F. New culture devices in ART. Placenta. 2011;32(Suppl 3):S248–S251. doi: 10.1016/j.placenta.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 3.Lazzaroni-Tealdi E, Barad DH, Albertini DF, Yu Y, Kushnir VA, Russell H, Wu YG, Gleicher N. Oocyte scoring enhances embryo-scoring in predicting pregnancy chances with IVF where it counts most. PLoS One. 2015;10:e0143632. doi: 10.1371/journal.pone.0143632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holte J, Berglund L, Milton K, Garello C, Gennarelli G, Revelli A, Bergh T. Construction of an evidence-based integrated morphology cleavage embryo score for implantation potential of embryos scored and transferred on day 2 after oocyte retrieval. Hum Reprod. 2007;22:548–557. doi: 10.1093/humrep/del403. [DOI] [PubMed] [Google Scholar]
- 5.Gardner DK, Meseguer M, Rubio C, Treff NR. Diagnosis of human preimplantation embryo viability. Hum Reprod Update. 2015;21:727–747. doi: 10.1093/humupd/dmu064. [DOI] [PubMed] [Google Scholar]
- 6.Manna C, Nanni L, Lumini A, Pappalardo S. Artificial intelligence techniques for embryo and oocyte classification. Reprod BioMed Online. 2013;26:42–49. doi: 10.1016/j.rbmo.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Harper J, Magli MC, Lundin K, Barratt CL, Brison D. When and how should new technology be introduced into the IVF laboratory? Hum Reprod. 2012;27:303–313. doi: 10.1093/humrep/der414. [DOI] [PubMed] [Google Scholar]
- 8.National Research Council (US) Board on Biology. Bioinformatics: Converting Data to Knowledge: Workshop Summary [Internet]. Pool R, Esnayra J, editors. Washington (DC): National Academies Press (US); 2000 [cited 2018 Jun 1]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK44939/ [PubMed]
- 9.Eddy SR. What is Bayesian statistics? Nat Biotechnol. 2004;22:1177–1178. doi: 10.1038/nbt0904-1177. [DOI] [PubMed] [Google Scholar]
- 10.Dukic V, Hogan JW. A hierarchical Bayesian approach to modeling embryo implantation following in vitro fertilization. Biostatistics. 2002;3:361–377. doi: 10.1093/biostatistics/3.3.361. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H, Weinberg CR. Evaluating effects of exposures on embryo viability and uterine receptivity in in vitro fertilization. Stat Med. 1998;17:1601–1612. doi: 10.1002/(SICI)1097-0258(19980730)17:14<1601::AID-SIM870>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Morales DA, Bengoetxea E, Larranaga P. Selection of human embryos for transfer by Bayesian classifiers. Comput Biol Med. 2008;38:1177–1186. doi: 10.1016/j.compbiomed.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Morales DA, Bengoetxea E, Larranaga P, Garcia M, Franco Y, Fresnada M, et al. Bayesian classification for the selection of in vitro human embryos using morphological and clinical data. Comput Methods Prog Biomed. 2008;90:104–116. doi: 10.1016/j.cmpb.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 14.Gianaroli L, Magli MC, Gambardella L, Giusti A, Grugnetti C, Corani G. Objective way to support embryo transfer: a probabilistic decision. Hum Reprod. 2013;28:1210–1220. doi: 10.1093/humrep/det030. [DOI] [PubMed] [Google Scholar]
- 15.Corani G, Magli C, Giusti A, Gianaroli L, Gambardella LM. A Bayesian network model for predicting pregnancy after in vitro fertilization. Comput Biol Med. 2013;43:1783–1792. doi: 10.1016/j.compbiomed.2013.07.035. [DOI] [PubMed] [Google Scholar]
- 16.Alpha Scientists in Reproductive M, Embryology ESIG of. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011; 26:1270–83. [DOI] [PubMed]
- 17.Woo I, Arrach N, Rhodes-Long K, Paco C, Paulson R, Chung K, et al. Detection of mosaicism using Bayesian model. Fertil Steril. 107:e22.
- 18.Hnida C, Engenheiro E, Ziebe S. Computer-controlled, multilevel, morphometric analysis of blastomere size as biomarker of fragmentation and multinuclearity in human embryos. Hum Reprod. 2004;19:288–293. doi: 10.1093/humrep/deh070. [DOI] [PubMed] [Google Scholar]
- 19.Hnida C, Agerholm I, Ziebe S. Traditional detection versus computer-controlled multilevel analysis of nuclear structures from donated human embryos. Hum Reprod. 2005;20:665–671. doi: 10.1093/humrep/deh639. [DOI] [PubMed] [Google Scholar]
- 20.Beuchat A, Thevenaz P, Unser M, Ebner T, Senn A, Urner F, Germond M, Sorzano COS. Quantitative morphometrical characterization of human pronuclear zygotes. Hum Reprod. 2008;23:1983–1992. doi: 10.1093/humrep/den206. [DOI] [PubMed] [Google Scholar]
- 21.Paternot G, Debrock S, D’Hooghe T, Spiessens C. Computer-assisted embryo selection: a benefit in the evaluation of embryo quality? Reprod BioMed Online. 2011;23:347–354. doi: 10.1016/j.rbmo.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Paternot G, Debrock S, De Neubourg D, D’Hooghe TM, Spiessens C. Semi-automated morphometric analysis of human embryos can reveal correlations between total embryo volume and clinical pregnancy. Hum Reprod. 2013;28:627–633. doi: 10.1093/humrep/des427. [DOI] [PubMed] [Google Scholar]
- 23.Strouthopoulos C, Anifandis G. An automated blastomere identification method for the evaluation of day 2 embryos during IVF/ICSI treatments. Comput Methods Prog Biomed. 2018;156:53–59. doi: 10.1016/j.cmpb.2017.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Kirkegaard K, Campbell A, Agerholm I, Bentin-Ley U, Gabrielsen A, Kirk J, Sayed S, Ingerslev HJ. Limitations of a time-lapse blastocyst prediction model: a large multicentre outcome analysis. Reprod BioMed Online. 2014;29:156–158. doi: 10.1016/j.rbmo.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Dolinko AV, Farland LV, Kaser DJ, Missmer SA, Racowsky C. National survey on use of time-lapse imaging systems in IVF laboratories. J Assist Reprod Genet. 2017;34:1167–1172. doi: 10.1007/s10815-017-0964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Racowsky C, Kovacs P, Martins WP. A critical appraisal of time-lapse imaging for embryo selection: where are we and where do we need to go? J Assist Reprod Genet. 2015;32:1025–1030. doi: 10.1007/s10815-015-0510-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M, Wei S, Hu J, Yuan J, Liu F. Does time-lapse imaging have favorable results for embryo incubation and selection compared with conventional methods in clinical in vitro fertilization? A meta-analysis and systematic review of randomized controlled trials. PLoS One. 2017;12:e0178720. doi: 10.1371/journal.pone.0178720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovacs P. Embryo selection: the role of time-lapse monitoring. Reprod Biol Endocrinol. 2014;12:124. doi: 10.1186/1477-7827-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reignier A, Lammers J, Barriere P, Freour T. Can time-lapse parameters predict embryo ploidy? A systematic review. Reprod Biomed Online [Internet]. 2018; Available from: http://www.ncbi.nlm.nih.gov/pubmed/29398421. [DOI] [PubMed]
- 30.Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- 31.Conaghan J, Chen AA, Willman SP, Ivani K, Chenette PE, Boostanfar R, Baker VL, Adamson GD, Abusief ME, Gvakharia M, Loewke KE, Shen S. Improving embryo selection using a computer-automated time-lapse image analysis test plus day 3 morphology: results from a prospective multicenter trial. Fertil Steril. 2013;100:412–419. doi: 10.1016/j.fertnstert.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 32.VerMilyea MD, Tan L, Anthony JT, Conaghan J, Ivani K, Gvakharia M, et al. Computer-automated time-lapse analysis results correlate with embryo implantation and clinical pregnancy: a blinded, multi-centre study. Reprod BioMed Online. 2014;29:729–736. doi: 10.1016/j.rbmo.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen BM, Boel M, Montag M, Gardner DK. Development of a generally applicable morphokinetic algorithm capable of predicting the implantation potential of embryos transferred on day 3. Hum Reprod. 2016;31:2231–2244. doi: 10.1093/humrep/dew188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aparicio-Ruiz B, Basile N, Perez Albala S, Bronet F, Remohi J, Meseguer M. Automatic time-lapse instrument is superior to single-point morphology observation for selecting viable embryos: retrospective study in oocyte donation. Fertil Steril. 2016;106:1379–1385. doi: 10.1016/j.fertnstert.2016.07.1117. [DOI] [PubMed] [Google Scholar]
- 35.Behr B, Tan L, Conaghan J, Liebermann J, Bartolucci A, Chen AA. Non-invasive technology combining time-lapse imaging and statistical modeling: bringing automation into the lab to improve blastocyst selection. Fertil Steril. 104:e152.
- 36.Verlinsky Y, Cieslak J, Freidine M, Ivakhnenko V, Wolf G, Kovalinskaya L, White M, Lifchez A, Kaplan B, Moise J, J.Valle, Ginsberg N, Strom C, Kuliev A. Pregnancies following pre-conception diagnosis of common aneuploidies by fluorescent in-situ hybridization. Hum Reprod. 1995;10:1923–1927. doi: 10.1093/oxfordjournals.humrep.a136207. [DOI] [PubMed] [Google Scholar]
- 37.Mastenbroek S, Repping S. Preimplantation genetic screening: back to the future. Hum Reprod. 2014;29:1846–1850. doi: 10.1093/humrep/deu163. [DOI] [PubMed] [Google Scholar]
- 38.Practice Committees of the American Society for Reproductive Medicine and the Society for Assisted Reproductive Technology. Electronic address: ASRM@asrm.org, Practice Committees of the American Society for Reproductive Medicine and the Society for Assisted Reproductive Technology. The use of preimplantation genetic testing for aneuploidy (PGT-A): a committee opinion. Fertil Steril. 2018;109:429–36. [DOI] [PubMed]
- 39.Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CF. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod BioMed Online. 2013;26:477–485. doi: 10.1016/j.rbmo.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Thornton S. Retrospective analysis of outcomes after IVF using an aneuploidy risk model derived from time-lapse imaging without PGS. Reprod BioMed Online. 2013;27:140–146. doi: 10.1016/j.rbmo.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Basile N, Nogales Mdel C, Bronet F, Florensa M, Riqueiros M, Rodrigo L, et al. Increasing the probability of selecting chromosomally normal embryos by time-lapse morphokinetics analysis. Fertil Steril. 2014;101:699–704. doi: 10.1016/j.fertnstert.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Barrie A, Homburg R, McDowell G, Brown J, Kingsland C, Troup S. Examining the efficacy of six published time-lapse imaging embryo selection algorithms to predict implantation to demonstrate the need for the development of specific, in-house morphokinetic selection algorithms. Fertil Steril. 2017;107:613–621. doi: 10.1016/j.fertnstert.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 43.Patel DV, Shah PB, Kotdawala AP, Herrero J, Rubio I, Banker MR. Morphokinetic behavior of euploid and aneuploid embryos analyzed by time-lapse in embryoscope. J Hum Reprod Sci. 2016;9:112–118. doi: 10.4103/0974-1208.183511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastenbroek S, Twisk M, van der Veen F, Repping S. Preimplantation genetic screening: a systematic review and meta-analysis of RCTs. Hum Reprod Update. 2011;17:454–466. doi: 10.1093/humupd/dmr003. [DOI] [PubMed] [Google Scholar]
- 45.Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26:2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 46.Cruz M, Garrido N, Herrero J, Perez-Cano I, Munoz M, Meseguer M. Timing of cell division in human cleavage-stage embryos is linked with blastocyst formation and quality. Reprod BioMed Online. 2012;25:371–381. doi: 10.1016/j.rbmo.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 47.Basile N, Vime P, Florensa M, Aparicio Ruiz B, Garcia Velasco JA, Remohi J, et al. The use of morphokinetics as a predictor of implantation: a multicentric study to define and validate an algorithm for embryo selection. Hum Reprod. 2015;30:276–283. doi: 10.1093/humrep/deu331. [DOI] [PubMed] [Google Scholar]
- 48.Cetinkaya M, Pirkevi C, Yelke H, Colakoglu YK, Atayurt Z, Kahraman S. Relative kinetic expressions defining cleavage synchronicity are better predictors of blastocyst formation and quality than absolute time points. J Assist Reprod Genet. 2015;32:27–35. doi: 10.1007/s10815-014-0341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milewski R, Kuc P, Kuczynska A, Stankiewicz B, Lukaszuk K, Kuczynski W. A predictive model for blastocyst formation based on morphokinetic parameters in time-lapse monitoring of embryo development. J Assist Reprod Genet. 2015;32:571–579. doi: 10.1007/s10815-015-0440-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milewski R, Milewska AJ, Kuczynska A, Stankiewicz B, Kuczynski W. Do morphokinetic data sets inform pregnancy potential? J Assist Reprod Genet. 2016;33:357–365. doi: 10.1007/s10815-016-0649-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motato Y, de los Santos MJ, Escriba MJ, Ruiz BA, Remohi J, Meseguer M. Morphokinetic analysis and embryonic prediction for blastocyst formation through an integrated time-lapse system. Fertil Steril. 2016;105:376–384. doi: 10.1016/j.fertnstert.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Chapple V, Feenan K, Roberts P, Matson P. Time-lapse deselection model for human day 3 in vitro fertilization embryos: the combination of qualitative and quantitative measures of embryo growth. Fertil Steril. 2016;105:656–662. doi: 10.1016/j.fertnstert.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Carrasco B, Arroyo G, Gil Y, Gomez MJ, Rodriguez I, Barri PN, et al. Selecting embryos with the highest implantation potential using data mining and decision tree based on classical embryo morphology and morphokinetics. J Assist Reprod Genet. 2017;34:983–990. doi: 10.1007/s10815-017-0955-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Feenan K, Chapple V, Matson P. Assessing efficacy of day 3 embryo time-lapse algorithms retrospectively: impacts of dataset type and confounding factors. Hum Fertil (Camb) 2018; 1–9. [DOI] [PubMed]
- 55.Storr A, Venetis C, Cooke S, Kilani S, Ledger W. Time-lapse algorithms and morphological selection of day-5 embryos for transfer: a preclinical validation study. Fertil Steril. 2018;109:276–283. doi: 10.1016/j.fertnstert.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 56.Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med. 2001;23:89–109. doi: 10.1016/S0933-3657(01)00077-X. [DOI] [PubMed] [Google Scholar]
- 57.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 58.Kaufmann SJ, Eastaugh JL, Snowden S, Smye SW, Sharma V. The application of neural networks in predicting the outcome of in-vitro fertilization. Hum Reprod. 1997;12:1454–1457. doi: 10.1093/humrep/12.7.1454. [DOI] [PubMed] [Google Scholar]
- 59.Wald M, Sparks A, Sandlow J, Van-Voorhis B, Syrop CH, Niederberger CS. Computational models for prediction of IVF/ICSI outcomes with surgically retrieved spermatozoa. Reprod BioMed Online. 2005;11:325–331. doi: 10.1016/S1472-6483(10)60840-1. [DOI] [PubMed] [Google Scholar]
- 60.Uyar A, Bener A, Ciray H, Bahceci M. A frequency based encoding technique for transformation of categorical variables in mixed IVF dataset. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:6214–6217. doi: 10.1109/IEMBS.2009.5334548. [DOI] [PubMed] [Google Scholar]
- 61.Siristatidis C, Pouliakis A, Chrelias C, Kassanos D. Artificial intelligence in IVF: a need. Syst Biol Reprod Med. 2011;57:179–185. doi: 10.3109/19396368.2011.558607. [DOI] [PubMed] [Google Scholar]
- 62.Milewski R, Milewska Anna J, Więsak T, Morgan A. Comparison of artificial neural networks and logistic regression analysis in pregnancy prediction using the in vitro fertilization treatment. slgr. 2013;35:39. [Google Scholar]
- 63.Banerjee P, Choi B, Shahine LK, Jun SH, O’Leary K, Lathi RB, et al. Deep phenotyping to predict live birth outcomes in in vitro fertilization. Proc Natl Acad Sci U S A. 2010;107:13570–13575. doi: 10.1073/pnas.1002296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi B, Bosch E, Lannon BM, Leveille MC, Wong WH, Leader A, et al. Personalized prediction of first-cycle in vitro fertilization success. Fertil Steril. 2013;99:1905–1911. doi: 10.1016/j.fertnstert.2013.02.016. [DOI] [PubMed] [Google Scholar]