Characterization of the venom of an ant reveals a diverse and multifunctional hymenopteran toxin superfamily.
Abstract
Ants (Hymenoptera: Formicidae) are diverse and ubiquitous, and their ability to sting is familiar to many of us. However, their venoms remain largely unstudied. We provide the first comprehensive characterization of a polypeptidic ant venom, that of the giant red bull ant, Myrmecia gulosa. We reveal a suite of novel peptides with a range of posttranslational modifications, including disulfide bond formation, dimerization, and glycosylation. One venom peptide has sequence features consistent with an epidermal growth factor fold, while the remaining peptides have features suggestive of a capacity to form amphipathic helices. We show that these peptides are derived from what appears to be a single, pharmacologically diverse, gene superfamily (aculeatoxins) that includes most venom peptides previously reported from the aculeate Hymenoptera. Two aculeatoxins purified from the venom were found to be capable of activating mammalian sensory neurons, consistent with the capacity to produce pain but via distinct mechanisms of action. Further investigation of the major venom peptide MIITX1-Mg1a revealed that it can also incapacitate arthropods, indicative of dual utility in both defense and predation. MIITX1-Mg1a accomplishes these functions by generating a leak in membrane ion conductance, which alters membrane potential and triggers neuronal depolarization. Our results provide the first insights into the evolution of the major toxin gene superfamily of the aculeate Hymenoptera and provide a new paradigm in the functional evolution of toxins from animal venoms.
INTRODUCTION
Hymenopteran venoms have been the subject of scientific investigation for centuries (1). Detailed analyses of hymenopteran venom composition and the characterization of individual venom components began in the 1950s (2). However, research to date has largely been limited to studies on the venoms of bees and wasps of the families Apidae and Vespidae, respectively, with the major active venom components shown to be peptides. The venoms of other hymenopteran families, including the Formicidae (ants), have remained largely unstudied. In the case of ants, this is probably owing not only to their relatively small size (and venom yield) but also to the widespread misconception that ants have very simple, primarily formic acid (FA)–containing venoms. FA is indeed the major and primary active component of the venoms of ants of the subfamily Formicinae (3). Moreover, the venoms of fire ants (subfamily Myrmicinae) are relatively simple and composed primarily of a suite of small alkaloids (4). However, there is accumulating evidence that these simple, largely nonpeptidic ant venoms are the exception rather than the rule.
In 1964, Cavill et al. (5) revealed for the first time that, like bees and wasps, the venoms of some ants were primarily polypeptidic. The subject of this pivotal study was the ant Myrmecia gulosa (the giant red bull ant) from the subfamily Myrmeciinae (not to be confused with Myrmicinae). Cavill et al. showed that (i) the venom of M. gulosa is primarily polypeptidic [with a small amount of histamine (~2%)], (ii) at least eight distinct polypeptide fractions could be separated by electrophoresis, and (iii) multiple distinct biological activities are associated with some of these polypeptide fractions, namely, hyaluronidase, kinin-like, and hemolytic activity. Further studies have since revealed that the venoms of other species of Myrmeciinae are similarly polypeptidic (6–12), as are those of ants of at least seven other subfamilies, namely, Amblyoponerinae (13), Dorylinae (14), Ectatomminae (15), Myrmicinae (16), Paraponerinae (17), Ponerinae (18), and Pseudomyrmecinae (19).
Until recently, however, little work had been done beyond simply establishing the polypeptidic nature of these ant venoms. For several species, individual venom peptides have now been isolated and characterized [for a review, see (20)]. Venom gland transcriptomes have also been generated for three species (21–23), but in the absence of corresponding proteomic data, their value in defining the genuine venom composition of these species is limited (24–26). In short, a comprehensive characterization of the venom repertoire of any species of ant (Formicidae) has been lacking.
Here, we used a combination of transcriptomics and mass spectrometry (MS)–based proteomics to provide the first comprehensive characterization of the polypeptide repertoire of an ant venom—that of the giant red bull ant, M. gulosa (Fig. 1A). Analysis of the encoding transcripts reveals that all but one of the venom peptides are derived from what appears to be a structurally and functionally hyperdiverse toxin gene superfamily that includes almost all venom peptides previously reported from aculeate Hymenoptera. Pharmacological experiments demonstrated that members of this toxin gene family account for multiple mechanisms of action within the venom of M. gulosa and that the major venom peptide has a mechanism of action that gives it dual utility in both defensive and predatory interactions.
Fig. 1. The polypeptidic venom composition of M. gulosa.
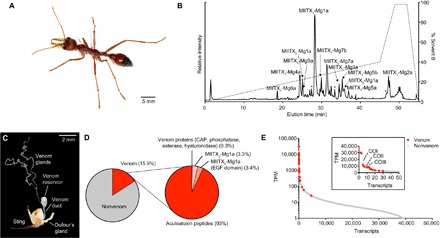
(A) M. gulosa. (B) Total ion chromatogram of M. gulosa venom. Venom (2 μg) was separated by C18 ultrahigh-performance liquid chromatography (uHPLC), using a gradient of 1 to 40% solvent B [90% acetonitrile (ACN) and 0.1% FA] over 45 min and analyzed using MS/MS. Peaks corresponding to identified venom peptides are labeled. Peaks identified as corresponding to partial fragments of larger peptides are labeled with an asterisk. (C) Venom apparatus of M. gulosa. (D) Venom component-encoding transcripts (that is, those encoding peptides detected in the venom itself) comprised 15.9% of total expression. Of these, transcripts encoding aculeatoxin peptides, MIITX2-Mg1a, MIITX3-Mg1a, and venom proteins comprised 93.0, 3.4, 3.3, and 0.3%, respectively, of venom component expression. EGF, epidermal growth factor. (E) Venom component-encoding transcripts (highlighted in red) are found exclusively in the highly expressed portion of the venom apparatus transcriptome, where they constitute most of the most highly expressed transcripts. Inset: Zoom showing the top 50 transcripts (with a linear scale on the y axis). The most highly expressed nonvenom component-encoding transcripts are labeled [cytochrome c oxidase subunits 1 (COI), 2 (COII), and 3 (COIII)].
RESULTS
The venom composition of M. gulosa
To generate a full portrait of the polypeptidic venom composition of M. gulosa, we used a combined transcriptomics and MS-based proteomics strategy. A venom apparatus transcriptome of M. gulosa was generated as follows: 47,042,454 and 45,167,480 demultiplexed raw reads generated by RNA sequencing of M. gulosa venom apparatus [venom gland filaments, venom reservoir (including the convoluted gland), venom duct, and Dufour’s gland; Fig. 1C] and remaining whole body, respectively, were assembled de novo using Trinity (27). Assembly yielded a total of 42,828 transcripts with E90 values (the number of transcripts that are supported by 90% of the expression data) of 14,307 (venom apparatus) and 17,513 (remaining whole body).
Annotation of the M. gulosa assembly using a blastx (28) alignment against a UniProtKB database identified numerous transcripts as “venom”- or “toxin”-associated. These included several of the most highly expressed transcripts in the venom apparatus data set, which were identified as similar to “pilosulins,” a group of peptides previously detected in the venom of Myrmecia pilosula (6, 8, 12). However, many of the venom- or toxin-associated identifications were not supported by our proteomic data as genuine venom components (table S1), highlighting the high rate of false positives associated with similarity-based venom component annotation, as previously noted (24–26).
To identify transcripts encoding bona fide venom components, we collected injected venom from M. gulosa (movie S1) and analyzed it by MS-based proteomics. Bottom-up sequencing, searching liquid chromatography–tandem mass spectrometry (LC-MS/MS) data of reduced, alkylated, and trypsin-digested venom against the translated transcriptome assembly, resulted in identification of 13 unique peptides and six proteins (Table 1). These were named according to the rational nomenclature proposed for other venomous arthropod lineages (29, 30), including ants (20). Two additional peptides (MIITX1-Mg4b and MIITX1-Mg7c) that were not detected in our proteomic data were included as probable venom components based on high-sequence similarity and similarly high-expression levels to identified venom peptides. The six proteins identified included a 19.4-kDa protein [MIITX3-Mg1a, a CAP (cysteine-rich secretory protein, insect venom allergen antigen 5, and pathogenesis-related 1 protein)], a venom dipeptidyl peptidase IV (DPP-4), a phosphatase, an esterase, and a hyaluronidase. Of these, DPP-4 is probably not a functional venom component but instead is responsible for the maturation of venom peptides (31). Phospholipases have been reported in most other hymenopteran venoms studied to date (1), but we did not detect any tryptic peptides derived from phospholipases, suggesting that this class of enzyme is either absent or of very low abundance in the venom of M. gulosa.
Table 1. Polypeptide venom components of M. gulosa.
TPM, transcripts per million.
| Mass (Da) | TPM | Sequence | Features | |
| MIITX1-Mg1a | 3144.85 | 30120 | GLGRLIGKIAKKGAKIAAEAAANAAAEKAAEAL* | |
| MIITX1-Mg2a | 8482.78 | 29318 | LLSKDQALKHVWGVLKKLGKAAMEYVIQQICAKYNKK | 1 Cys (homodimer) |
| MIITX1-Mg3a | 4325.32 | 16248† | KNEETMEEALKGLNELKERLKKQGIDTAALNLDEKLLT | |
| MIITX-Mg4a | 9220.85 | 11836† | SLVGCPRPDFLPSWNRCKSCVCKNNKLKCPKILKGSLLKTAA | 5 Cys (homodimer) |
| MIITX-Mg4b | 8876.52‡ | 2518 | SLVGCPRPNFLPSWNRCKCICKNNKPMCRKLPNLLKTTA | 5 Cys (homodimer?) |
| MIITX1-Mg5a | 2516.40 | 9654† | SINVKNLMDMIREQITSRLKK | |
| MIITX1-Mg5b | 2515.42 | 3556 | SINVKNLMNMIREQITSRLKK | |
| MIITX1-Mg6a | 1509.80 | 9484 | FRGPCLKIKGYKC | 2 Cys |
| MIITX1-Mg7a | 7110.84 | 8100 | KRSKSSSKTKPKKPKKPKKKIKIPDWVKSGGKMVGEAVAGAVADAAVSAVMDAAVGTTAEPEQ | 3 HexNAc |
| MIITX1-Mg7b | 5596.04 | 7808 | KRRRGLKKIIGKVIKGTGKVAGEAAASAVADAAVSAAIDAVVGTTEEPEQ | 3 HexNAc |
| MIITX1-Mg7c | 5736.17‡ | 4343 | KRRRRLRKIIRKVIKGTGKVAGEAAASAVAGAAVSAAIDAAVGTTEEPEQ | 3 HexNAc? |
| MIITX1-Mg8a | 7632.29‡ | 1027 | WLGALFSFIRFIAPYVIRAVRVLIQVVSKVVKPAKVAMKYAKKIATNVAKDVAKDMATDIAIDTITGGDE | |
| MIITX1-Mg8b | 7373.97‡ | 773 | WLGALFSFIRFIAPYVIKAIRFVVQVASKLVKPAKAAIKFAKNIAKDIAEDMAMDFAMDVITGGDDE | |
| MIITX1-Mg9a | 3246.96 | 861 | NIKWSKYAKKVGKVIVKHGIPLAASIALSQ* | |
| MIITX2-Mg1a | 5787.37 | 5354 | DISDYGDPCSDDLKDYCIHGDCFFLKELNQPACRCYTGYYGSRCEHIDHN | 6 Cys |
| MIITX1-Mg10a | ? | 11791 | ||
| MIITX3-Mg1a | ~19,400‡ | 5208 | ||
| CAP | ~24,200‡ | 378 | ||
| Phosphatase | ~43,700‡ | 90 | ||
| Esterase | ~61,800‡ | 59 | ||
| Hyaluronidase | ~38,700‡ | 26 | ||
| DPP-4 | ~86,500‡ | 233 |
*C-terminal amidation.
†The TPM value is the sum of more than one assembled transcript encoding the same peptide.
‡Predicted mass (complete primary structure not confirmed by MS/MS).
To determine the complete primary structure of each of the identified venom peptides, we used top-down sequencing of nonderivatized and nondigested venom (Fig. 1B). MIITX1-Mg1a, Mg3a, Mg5a, Mg5b, Mg7a, Mg7b, Mg7c, Mg8a, Mg8b, and Mg9a are present in the venom as linear peptides of lengths ranging from 21 to 70 residues. Of these, MIITX1-Mg1a and Mg9a were each detected with an amidated C terminus, while MIITX1-Mg7a and Mg7b were detected each with three N-acetylhexosamine groups. MIITX1-Mg6a and MIITX2-Mg1a were found to contain a single or three intrachain disulfides, respectively, while MIITX1-Mg2a was detected in the venom as a homodimer linked by a single interchain disulfide bond. MIITX1-Mg4a was also detected as a homodimer.
Transcripts encoding venom components corresponded to 15.9% (158,784 TPM) of all venom apparatus–derived reads (Fig. 1D) and were confined to the high-expression portion of the venom apparatus transcriptome where they constituted most of the most highly expressed transcripts (Fig. 1E). TPM values for identified venom peptides (as opposed to proteins) ranged from 773 to 30,120 and accounted for 96.4% of venom component expression (Fig. 1D and Table 1). For the six proteins, with the exception of MIITX3-Mg1a (TPM value of 5208), transcripts were expressed at much lower levels, with TPM values ranging from 26 to 378, all lower than the TPM values of the identified peptidic venom components and together accounting for a mere 0.3% of venom component expression (Fig. 1D and Table 1).
All but one of the identified venom peptides share a set of distinct biophysical features regardless of the presence or absence of cysteine residues: All are rich in lysine residues, all but MIITX1-Mg3 have a net positive charge, and all have sequence characteristics suggestive of a capacity to form amphipathic helices. The only exception is MIITX2-Mg1a, which shares the same six cysteine–containing structural domain as EGF. EGF domain peptides appear to be rare in animal venoms and, to our knowledge, have only been reported in the venoms of some sea anemones (25, 32, 33). Thus, the venom of M. gulosa appears to contain only four general polypeptide toxin types: 13 cationic and likely amphipathic helical peptides; one EGF-domain peptide; three enzymes, each represented by a single isoform; and two proteins of unknown function (CAP and MIITX3-Mg1a).
The aculeatoxin gene superfamily
Analysis of the precursor structures of each of the identified venom peptides revealed that all but MIITX2-Mg1a have a similar signal peptide sequence, and almost all of them have a similar repetitive and highly anionic propeptide sequence (MIITX1-Mg4a and Mg4b lack a propeptide). These data are suggestive of a shared gene family for these venom peptides. In other words, the venom repertoire of M. gulosa appears to be largely, although not exclusively, dominated by peptides derived from a single gene family.
Comparison of the M. gulosa venom peptide precursor sequences (except MIITX1-Mg4a, MIITX1-Mg4b, and MIITX2-Mg1a, which lack propeptide regions) with those previously reported for other experimentally validated venom peptides from Formicidae revealed that all share a similar signal and propeptide sequence (Fig. 2). These include the pilosulins from M. pilosula (Myrmeciinae) (8, 34–36), dinoponeratoxins from Dinoponera quadriceps (Ponerinae) (22, 37), pilosulin-like peptides from Odontomachus monticola (Ponerinae) (21), and bicarinaline and related peptides from Tetramorium bicarinatum (Myrmicinae) (23, 38). These data suggest that, despite the apparent variation in their mature peptide domains (for example, MIITX1-Mg9a versus pilosulin 3a; Fig. 2), these peptides may represent a single gene family. Furthermore, available evidence suggests that this peptide family is not merely present in these other ant species but in fact also dominates their venom repertoire.
Fig. 2. Venom peptides of the aculeate Hymenoptera.
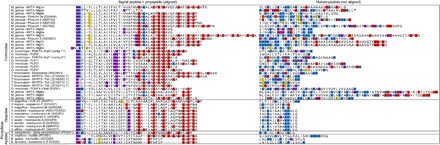
Precursor sequences of aculeate Hymenoptera venom peptides (encoding experimentally validated venom peptides) are aligned on the basis of signal and propeptide regions. Mature peptide regions are diverse in sequence (and have not been aligned). Methionine, lysine/arginine, aspartate/glutamate, and cysteine residues are highlighted in purple, blue, red, and yellow, respectively. Posttranslational modifications are not shown. aTorres et al. (22); bKazuma et al. (21); cBouzid et al. (23).
It was noted previously that the peptide precursor of pilosulin 1 from M. pilosula has sequence similarity to that of the peptide melittin, the major venom component of the European honeybee Apis mellifera, suggestive of a shared gene family (39). While there is limited similarity in the signal peptide sequence, melittin does share a similar repetitive and highly anionic propeptide sequence with the ant venom peptide precursors (Fig. 2). In prepromelittin, this characteristic propeptide sequence represents the recognition site for DPP-4, the enzyme responsible for liberating the mature melittin peptide (31). Given that a DPP-4 enzyme was also detected in the venom of M. gulosa, a shared processing mechanism seems likely for the ant venom peptides.
Broadening the search to venom peptide sequences from other hymenopterans revealed that the precursor sequences of the mastoparans, vespa-chemotactic peptides and vespakinins of Vespidae venoms, the bombolitins of Apidae venoms, and pompilidotoxin of Pompillidae venom also share marked sequence similarity (Fig. 2). As with melittin, there is limited similarity in the signal peptide region but very marked similarity in the propeptide region. Again, these peptides are the dominant bioactive components of their respective venoms (1). No related sequences were detected outside of the infraorder Aculeata. Thus, almost all experimentally validated aculeate hymenopteran venom peptides (for which a corresponding precursor sequence has been determined) including the myrmeciitoxins described herein (except MIITX2-Mg1a), all other ant venoms peptides, melittin, the mastoparans, the vespakinins, the bombolitins, and the pompilidotoxins share a characteristic precursor sequence, suggestive of a shared gene superfamily. For practical reasons, we have tentatively referred to this family of peptides collectively as the “aculeatoxin” gene superfamily.
Investigation of the defensive utility of some M. gulosa venom peptides
M. gulosa uses its venom defensively, and stings from Myrmeciinae species are notoriously painful. We therefore investigated the defensive function of the venom by assaying for depolarization of vertebrate sensory neurons isolated from the mouse dorsal root ganglion (DRG); among which are the neurons responsible for sensing noxious (painful) stimuli. Whole venom (at ~1:1000 dilution) caused an immediate sharp increase in intracellular calcium concentration ([Ca2+]i) in mouse DRG cells (Fig. 3A). In most cells, this initial increase was followed by a drop in [Ca2+]i, accompanied by a corresponding increase in fluorescence of the extracellular medium that indicated leakage of the calcium-sensitive dye from the cells, and, thus, cytolytic activity. This activation of sensory neurons is consistent with cellular depolarization and therefore with the painful effects associated with envenomation.
Fig. 3. Action of M. gulosa venom peptides on vertebrate sensory neurons.
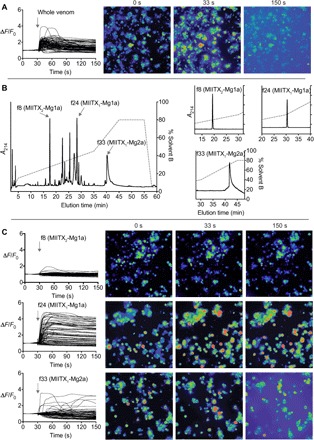
(A) Application of whole M. gulosa venom to DRG cells produced a rapid, non–cell-specific increase in [Ca2+]i followed by release of fluorescent dye into the medium. Each trace represents a single cell in the field of view. Snapshots shown of the recording are at 0 s (baseline), 33 s (3 s after addition of venom/fraction), and 150 s. (B) Three different peptides [fractions 8, 24, and 33 (f8, f24, and f33, respectively)] were purified from the whole venom using a single RP-HPLC step. Inset: Reanalysis of purified fractions by RP-HPLC. (C) Fraction 8 (MIITX2-Mg1a) had no effect on [Ca2+]i in sensory neurons, fraction 24 (MIITX1-Mg1a) caused an increase in [Ca2+]i in all sensory neurons, while fraction 33 (MIITX1-Mg2a) induced nonspecific permeabilization of cell membranes.
We then investigated the potential defensive role of individual M. gulosa toxins by examining the algogenic potential of three peptides purified from the venom (MIITX1-Mg1a, MIITX1-Mg2a, and MIITX2-Mg1a; Fig. 3B). We identified the homodimeric peptide MIITX1-Mg2a as a late-eluting series of broad peaks in a reversed-phase (RP)–HPLC chromatogram (Fig. 3B). Reanalysis by RP-HPLC of individual peaks revealed the same late-eluting series of broad peaks, indicating that the unusual RP-HPLC profile reflects slow conformational exchange of the peptide rather than multiple variants. This peptide caused a drop in [Ca2+]i in all cells and a corresponding increase in fluorescence of the extracellular medium (Fig. 3C), which was preceded, in some cells, by a sharp increase of [Ca2+]i. These data implicate MIITX1-Mg2a as the venom component responsible, at least in part, for the dye leakage (cytolysis) observed with the whole venom. A second major peak purified from the whole venom corresponded to the EGF-like peptide MIITX2-Mg1a (Fig. 3B). This peptide did not cause a change in [Ca2+]i in DRG cells (Fig. 3C). Finally, MIITX1-Mg1a, which accounts for the major peak in the venom RP-HPLC chromatogram (Fig. 3B), produced an immediate sharp increase of [Ca2+]i in all DRG cells. Like MIITX2-Mg1a but unlike MIITX1-Mg2a, however, this influx in Ca2+ plateaued without resulting in any observable dye leakage (Fig. 3C). Together, these data indicate that two of these three peptides are likely major contributors to the pain associated with envenomation but use distinct mechanisms of action to achieve this.
To further investigate its defensive utility, the major venom peptide of M. gulosa, MIITX1-Mg1a, was produced by solid-phase peptide synthesis (SPPS). The synthetically produced peptide coeluted on RP-HPLC with MIITX1-Mg1a purified from the venom (fig. S2) and produced the same effect on sensory neurons (at a concentration of 10 μM) as the purified venom peptide.
Consistent with the observed in vitro effects, intraplantar injection of MIITX1-Mg1a in C57/BL6/J mice produced spontaneous nocifensive behavior (that is, licking, flinching, or shaking of the paw), which lasted 5 min (10 μM) and 7 min (100 μM; Fig. 4A). The minimum effective concentration tested was 10 μM. We observed Mechanical and heat hypoalgesia (reduced pain response to normally painful stimuli) at 20 and 25 min after injection at the highest dose tested (100 μM; fig. S3).
Fig. 4. Defensive and predatory function and mechanism of action of MIITX1-Mg1a.
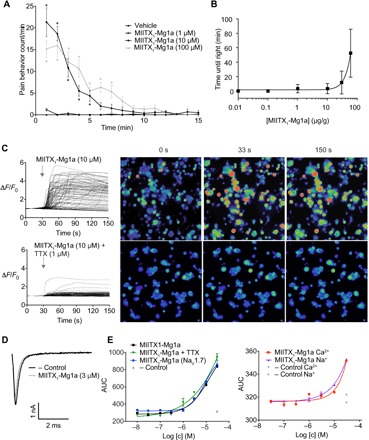
(A) Intraplantar injection of MIITX1-Mg1a causes spontaneous dose-dependent nocifensive behavior in mice, lasting for up to 7 min (n = 6 per group). Data are expressed as means ± SEM. Statistical significance compared to vehicle control was determined using two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test. *P < 0.05. (B) Intra-abdominal injection of MIITX1-Mg1a in crickets (A. domesticus) caused dose-dependent, reversible, and nonlethal incapacitation (inability to right after being placed on back). Effects were observed at doses ≥1 μg/g (0.3 nmol/g), and at the maximum dose tested (60 μg/g, 19 nmol/g), incapacitation lasted an average of 53 min. (C) MIITX1-Mg1a caused an increase in [Ca2+]i in DRG cells, which was mostly blocked in the presence of TTX. (D) Representative sodium currents obtained in HEK293 cells heterologously expressing hNaV1.7, in the presence and absence of 3 μM MIITX1-Mg1a. Currents were obtained by a 50-ms voltage pulse to −20 mV from a holding potential of −90 mV. Compared with buffer control, in hNaV1.7-expressing cells, MIITX1-Mg1a did not change peak current or activation/inactivation kinetics, and it did induce persistent currents. (E) In HEK293 cells [independent of hNaV1.7 expression or the presence of TTX (1 μM)], MIITX1-Mg1a alters membrane potential and causes Na+ and Ca2+ influx in a concentration-dependent manner. Concentration-response curves were evaluated by fitting the data with a four-parameter Hill equation with variable slope. FLIPR data are expressed as the means ± SEM and are representative of at least three independent experiments. AUC, area under the curve.
Investigation of the predatory utility of MIITX1-Mg1
Although M. gulosa (and other species of Myrmeciinae) are best known for their defensive stings, they also use their venom for predation. Adult workers not only are nectar feeders but also hunt other arthropods that they incapacitate by stinging, facilitating transport to the colony where they are fed to the carnivorous larvae. We therefore investigated a role in predatory envenomation for MIITX1-Mg1a by assaying for incapacitation of house crickets (Acheta domesticus; Fig. 4B).
Crickets injected with phosphate-buffered saline (PBS; negative control) and placed on their backs were able to immediately right themselves. Injection of MIITX1-Mg1a at doses ≥1 μg/g caused immediate incapacitation (inability to right after being placed on back). Incapacitation was dose-dependent, reversible, and nonlethal and, at the highest dose tested (60 μg/g), lasted an average of 53 min. The biological effects of this peptide are therefore consistent with it having a dual function in defense and predation.
Analysis of venom apparatus by matrix-assisted laser desorption/ionization MS
Some venomous animals are reportedly able to produce and store functionally distinct venom secretions in distinct glands (40), gland regions (41), or cell types (42) and use these different secretions in different contexts (that is, defense or predation). Because M. gulosa uses venom in both predatory and defensive interactions, we examined the contents of the various regions of the venom apparatus for indications of whether topological variations in toxin production might allow the ant to regulate the composition of its secreted venom. Matrix-assisted laser desorption/ionization (MALDI)–MS analysis of the “injected” venom painted a very similar picture of the venom composition to that generated by electrospray ionization (ESI)–MS (fig. S1). Extracts from four regions along the venom gland filaments, the venom reservoir, and venom duct were similar in peptide composition to the venom (fig. S1), with no evidence for regiospecific expression of venom peptides. For example, MIITX1-Mg1a was detected along the entire venom gland, as well as in the venom reservoir and venom duct and in the venom. No venom peptides were detected in the Dufour’s gland (fig. S1), which is consistent with previous reports of this being an aliphatic hydrocarbon-producing gland in M. gulosa (and our observations of it releasing an “oily” substance during dissection) (43). Thus, these data suggest that there is no morphological distinction between any functionally distinct venom components in the venom of M. gulosa and do not support behavioral modulation of venom composition. Further, these data are consistent with our observations of dual utility in defense and predation for the major venom peptide MIITX1-Mg1a.
Mechanism of action of MIITX1-Mg1
Although initial experiments indicated that MIITX1-Mg1a produces an immediate sharp increase in [Ca2+]i in DRG cells (Figs. 3C and 4C), this effect was not cell-specific: All DRG neurons and nonneuronal cells were affected, suggesting that the peptide affects a common cellular target such as the lipid bilayer. However, we did not observe any dye leakage from the cells, which shows that the effect was also not simply cytolytic. Consistent with this observation, the peptide was not hemolytic to human red blood cells, although it did display weak cytotoxicity [inhibition of growth of human embryonic kidney (HEK) 293 cells] and some antimicrobial activity (table S2).
The initial data suggested a nonspecific membrane-mediated mechanism of action for MIITX1-Mg1a. However, we noted that the increase in [Ca2+]i was greater in excitable cells (neurons) than in the nonneuronal cells. Furthermore, in DRG neurons, the increase in [Ca2+]i induced by MIITX1-Mg1a was substantially reduced in the presence of 1 μM tetrodotoxin (TTX), a blocker of some subtypes of voltage-gated sodium (NaV) channels (Fig. 4C). This implied that, in DRG neurons, a large proportion of the increase in [Ca2+]i caused by the peptide was dependent on the presence of functional NaV channels (and possibly other additional cellular components that contribute to cellular excitability). We therefore examined whether the massive increase in [Ca2+]i in excitable cells resulted from an indirect (upstream) action on NaV channels or whether the peptide had the additional ability to act directly on NaV channels.
Whole-cell voltage-clamp electrophysiology was used to examine the effect of MIITX1-Mg1a on human NaV1.7 (the major TTX-sensitive NaV subtype in DRG neurons) heterologously expressed in HEK293 cells. When compared to buffer control, MIITX1-Mg1a, at concentrations <3 μM, did not change peak current or activation and inactivation kinetics and did not induce persistent currents (Fig. 4D). Where concentrations exceeded 3 μM, recording was not possible because of unstable seal from large leak currents, which is consistent with an action of the peptide on the lipid bilayer.
Next, using a fluorescence imaging plate reader (FLIPR) assay, the peptide’s ability to alter membrane potential or to allow Ca2+ and Na+ influx was assessed. In normal HEK293 cells (not expressing any NaV channels), MIITX1-Mg1a altered membrane potential [unaffected by the presence of TTX (1 μM)] and induced Na+ and Ca2+ influx in a concentration-dependent manner (Fig. 4E) with effects observed at concentrations ≥1 μM. The effect on membrane potential was no different in HEK293 cells heterologously expressing hNaV1.7 channels (Fig. 4E). These data again pointed to a membrane-mediated mechanism of action of the peptide.
On the basis of these data, we propose the following model for the mechanism of action of MIITX1-Mg1a: MIITX1-Mg1a interacts with cell membranes, producing a concentration-dependent leak in ion conductance, possibly via multimeric pore formation as is the case with melittin (44). In excitable cells, such as mammalian sensory neurons, this leak in ion conductance is sufficient to shift the membrane potential to threshold, initiating neuronal depolarization (and opening of NaV channels) and subsequent additional (and much larger) Ca2+ influx through voltage-gated calcium channels. This action, on nociceptors, would result in immediate pain. Similarly, the temporary paralysis observed when the peptide is injected into insects could result from the same action on cell membranes of other excitable tissues such as muscle or peripheral motor neurons.
DISCUSSION
Here, we performed the first comprehensive characterization of the polypeptidic venom composition of an ant. Consistent with previous studies of other aculeate hymenopteran venoms such as those of wasps and bees (1), but in contrast to other arthropod venoms, such as those of most spiders, scorpions, or centipedes, the venom composition of M. gulosa is relatively simple. Most M. gulosa venom peptides appear to be derived from a single gene superfamily. This gene superfamily includes almost all peptides identified in the venoms of the aculeate Hymenoptera, and hence, we have named it the aculeatoxin superfamily. Almost all aculeatoxins share certain general structural features, including a net cationic charge. Where examined, they also all interact with lipid bilayers, and in such an environment, they adopt an amphipathic helical structure (1), even where the peptides have multiple cysteine residues (45). However, there is noticeable sequence diversity in the encoded mature peptides (Fig. 2), and this diversity in mature peptide sequence is reflected in a diversity in biological activity (1). For example, melittin, the mastoparans, and the bombolitins all target cell membranes in which they form ion-permeable channels (44, 46), vespakinins probably exert their effect through bradykinin G protein–coupled receptors, while pompilidotoxins act on NaV channels (47). Pharmacological diversity is also present in M. gulosa venom, where we demonstrated that multiple mechanisms of action are encoded in this gene family: The homodimeric peptide MIITX1-Mg2a produces effects in DRG cells that indicate relatively nonspecific membrane permeation (as is the case for the dimeric peptide MIITX-Mp1 (previously described) (45). On the other hand, MIITX1-Mg1a alters membrane potential presumably by forming ion-permeable pores. The observed sequence diversity in the remaining peptides of M. gulosa and other ant venoms suggests that these peptides almost certainly encode further mechanistic diversity.
The structural and pharmacological diversity of aculeatoxins provides a marked contrast to the “classic” patterns of functional evolution of other, primarily ion channel–interacting, cysteine-rich peptide toxin gene families of the venoms of sea anemones, cone snails, spiders, scorpions, centipedes, and snakes (48). Unlike the toxins in these other venoms, the nonglobular folds of aculeatoxins make their evolution less likely to be constrained by structural residues that direct folding. Thus, functional, rather than structural, factors probably drive the evolution of aculeate venom peptides to a much greater extent than in most other peptide toxin gene families, as indicated by the structural diversity and lack of “anchor points” available for accurate sequence alignment and molecular evolutionary analyses of the mature aculeatoxins. The interaction with metabolic products (cell membranes) as opposed to gene products (proteins) also presents a rarely considered scenario for toxin-target coevolution. This scenario is relevant not only for the aculeatoxins (which represent the major venom peptide class of the aculeate Hymenoptera) but also other membrane-interacting venom peptides, such as the “antimicrobial peptides” of some spider and scorpion venoms (49, 50). The evolution of membrane-interacting toxin gene superfamilies thus represents a significant challenge to understanding the processes that govern the functional diversification of venom peptides.
One question that arises from our data is why there is more than one pain-producing peptide in the venom of M. gulosa. For the European honeybee, A. mellifera, the peptide melittin is the sole pain-producing component of the venom (51). Another question is why, in the venom of M. gulosa, there are (at least) two different peptides (MIITX1-Mg1a and MIITX1-Mg2a) that act by permeabilizing cell membranes. Both peptides produce an effect that would ultimately lead to neuronal depolarization and pain, and presumably, one of these alone would be sufficient to meet the defensive needs of the ant. One hypothesis, based on the broad range of natural predators and prey that M. gulosa has, is that different venom peptides may have distinct tissue or cell-type selectivity profiles for different membrane compositions. Under this scenario, peptides are likely to act synergistically to produce an effect, either systemically or locally on cell membranes, that is stronger than each peptide alone. If this were the case, then the venoms of not only the Formicidae but also those of the aculeate Hymenoptera as a whole might represent a vast library of membrane-selective peptides with potential use as research tools or even antimicrobial or anticancer drugs. The exploration of the venoms of the aculeate Hymenoptera in this context is an exciting prospect.
MATERIALS AND METHODS
Venom collection
Adult workers of M. gulosa were collected from a single colony near Kooringal, Moreton Island, Queensland, Australia. Venom was collected from three individuals through a combination of defensive stinging (movie S1) followed by electrostimulation of the abdomen (in an effort to deplete the venom reservoir and maximize the expression of toxin genes before RNA extraction for transcriptome sequencing). Defensive stings were collected by holding the ant with a pair of forceps while allowing it to sting through a layer of parafilm covering the opening of a 1.5-ml tube. Each ant aggressively and repeatedly stung the parafilm layer presented to it, depositing, on most occasions, a small drop of clear colorless venom. When no more defensive venom was secreted, the ant was allowed to rest for approximately 10 min. The procedure was then repeated while applying a mild electric current (15 V, ~1 mA) to the abdomen as soon as the stinger was inserted into the parafilm membrane, resulting in additional venom being expelled. Venom was collected from the bottom of the parafilm layer and the sides of the tube by rinsing with 10 μl of ultrapure water. Venom was then stored at −80°C until further analysis. We found no observable differences by LC-MS/MS of native venom (see below) between voluntary secreted venom and that obtained with electrostimulation, and pooled venom drops were used for subsequent analyses.
Transcriptome sequencing and assembly
Venom apparatuses (venom gland filaments, venom reservoir, venom duct, and Dufour’s gland) were dissected from the ants 3 days after venom collection (using the same three individuals from which venom was collected). Using TRIzol (Life Technologies), total RNA was extracted from the complete venom apparatus (venom gland filaments, venom reservoir, venom duct, and Dufour’s gland) and the remaining whole body (yielding 26.8 and 49.2 μg of RNA, respectively).
Complementary DNA library preparation and sequencing was performed by the Institute for Molecular Bioscience Sequencing Facility, The University of Queensland. A dual-indexed library was constructed with the TruSeq-3 Stranded mRNA Sample Prep Kit (Illumina) with oligo (dT) selection and an average insert size of 180 base pairs. Samples were pooled in a batch of 15 samples, and 150-cycle paired-end sequencing was performed on an Illumina NextSeq 500 instrument.
Adapter trimming of demultiplexed raw reads was performed using fqtrim (52), followed by quality trimming and filtering using prinseq-lite (53). Error correction was performed using the BBnorm ecc tool, which is a part of the BBtools package. Trimmed and error-corrected reads were assembled using Trinity (version 2.2.1) (27) with a k-mer length of 31 and a minimum k-mer coverage of 10. Assembled transcripts were annotated using a blastx (28) search (E value setting of 1 × 10−3) against a database derived from UniProtKB. TPM counts were generated using the Trinity RSEM (54) plugin (align_and_estimate_abundance). Toxin transcripts were manually examined using the Map-to-Reference tool of Geneious version 10.2.3 (55). Toxin precursor sequences have been deposited with the transcriptome shotgun assembly database as GGFG01000001–GGFG01000022.
Proteomic analysis of venom
Bottom-up proteomics was used as a first step to identify venom components. Ten micrograms of crude venom was dried by vacuum centrifugation and resuspended in 4 M urea, 10% ACN, and 100 mM ammonium bicarbonate (pH 8). Cystines were reduced by incubation with 5 mM dithiothreitol at 60°C for 5 min and then alkylated with 10 mM iodoacetamide at 37°C for 60 min. Reduced and alkylated venom was then digested by incubating with trypsin (20 ng/μl) overnight at 37°C in 2 M urea, 10% ACN, and 100 mM ammonium bicarbonate (pH 8) at a final substrate-to-enzyme ratio of 50:1. The digested sample was desalted using a C18 ZipTip (Thermo Fisher Scientific) and dried by vacuum centrifugation before LC-ESI-MS/MS analysis.
Digested venom was reconstituted in 0.5% FA, and 2 μg was separated on a Shimadzu (Japan) Nexera uHPLC with an Agilent Zorbax stable-bond C18 column (2.1 mm × 100 mm; particle size, 1.8 μm; pore size, 300 Å), using a flow rate of 180 μl/min and a gradient of 1 to 40% solvent B (90% ACN and 0.1% FA) in 0.1% FA over 45 min and analyzed on an AB Sciex 5600 TripleTOF mass spectrometer equipped with a Turbo-V source heated to 550°C. MS survey scans were acquired at 300 to 1800 mass/charge ratio (m/z) over 250 ms, and the 20 most intense ions with a charge of +2 to +5 and an intensity of at least 120 counts were selected for MS/MS. The unit mass precursor ion inclusion window mass within ±0.7 Da and isotopes within ±2 Da were excluded from MS/MS, with scans acquired at 80 to 1400 m/z over 100 ms and optimized for high resolution. Using ProteinPilot version 5.0 (ABSciex), MS/MS spectra were searched against the venom apparatus transcriptome [translated and filtered to transcripts containing an open-reading frame of >40 amino acids and a signal peptide sequence (signalP D value of >0.3)]. Peptides identified by ProteinPilot were validated by comparison of experimentally derived peaks against a theoretical peak list generated using MS-Product in ProteinProspector 5.19.4 (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msproduct).
Top-down proteomics was used to determine the complete primary structure of identified venom components. Native venom (2 μg) was analyzed by LC-MS/MS, as described above. ProteinPilot was used to match transcripts to major peaks in the total ion chromatogram. All peptides identified by ProteinPilot were validated by comparison of experimentally derived MS/MS peaks against a theoretical peak list generated using MS-Product in ProteinProspector version 5.19.4 (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msproduct). Major peaks remaining unassigned following the ProteinPilot search were sequenced de novo to identify the corresponding transcript in the transcriptome. Venom components were named following the rational nomenclature established by King et al. (29), with the minor modification of a subscript number indicating the gene family to which a venom peptide belongs (30).
Matrix-assisted laser desorption/ionization mass spectromery
The venom apparatus of one “nonmilked” M. gulosa specimen was divided into four gland sections, the venom reservoir, and the venom duct. Each sample was mixed and then centrifuged to pellet tissue. The venom gland samples were concentrated using a C18 ZipTip (Thermo Fisher Scientific): 20 μl was loaded onto the ZipTip, washed with 10 μl of 0.1% trifluoroacetic acid (TFA), and eluted in approximately 2 μl of ACN. Venom reservoir, venom duct, and Dufour’s gland samples and an aliquot of injected venom were diluted 1:10. One microliter of each sample was mixed with 2 μl of diluted α-cyano-4-hydroxycinnamic acid solution [stored as an acetone-saturated solution and diluted 1 in 10 with a solution of ethanol/acetone/0.1% TFA (6:3:1) as a working solution] and spotted onto a polished steel target. MALDI spectra were acquired using a Bruker Autoflex Speed MALDI-TOF/TOF system (Bruker Daltonics Inc.) in linear positive mode at 2000 Hz, with an m/z range from 1000 to 15,000. Ten spectra of 500 shots each were saved. The group of 10 spectra were loaded into ClinProt Tools version 3.0 (Bruker Daltonics Inc.) as a separate group and observed as a gel view, with the averaged spectrum for the entire data set produced after recalibration of the entire loaded sample cohort.
Purification of venom peptides by RP-HPLC
Approximately 500 μg of venom was separated on a Phenomenex Gemini NX-C18 column (250 × 4.6 mm; particle size, 3 μm; pore size, 110 Å) using a gradient of 15 to 45% solvent B (90% ACN and 0.05% TFA) over 30 min at a flow rate of 1 ml min−1. Fractions were collected on the basis of absorbance at 214 nm. MALDI-MS was used to confirm the identity of the peptide/s present in each fraction, and purity was assessed by rerunning an aliquot of each under the same conditions. Purified fractions containing MIITX1-Mg1a, MIITX1-Mg2a, and MIITX2-Mg1a were dried by vacuum concentration and resuspended in 80 μl of pure water from which 7 μl-aliquots were used for calcium imaging experiments.
Peptide synthesis
MIITX1-Mg1a was prepared at a 100-μmol scale using Fmoc (9-fluorenyl methoxycarbonyl) SPPS on Protide Rink Amide resin (CEM Corporation). The peptide was elongated on a CEM Liberty Blue automated microwave peptide synthesizer [4-min coupling cycle: 2-min coupling (90°C), 1-min deprotection (90°C), 1-min associated washes and liquid handling] using a fivefold excess of Fmoc amino acid, ethyl cyanohydroxyiminoacetate (Oxyma), and diisopropylcarbodiimide. Fifty micromoles of peptide was cleaved from the resin and deprotected with a TFA-based cleavage solution [TFA/triisopropylsilane/H2O (95:2.5:2.5)] for 2 hours at room temperature and then worked up with diethyl ether and lyophilized. MIITX1-Mg1a was purified from the crude material [to ≥95% with a yield of 11.2 mg (7.1%)] using a 20 to 40% solvent B (99.9% ACN and 0.1% TFA) gradient over 40 min (15 ml/min−1) on a Waters Sunfire C18 OBD preparative column (pore size, 5 μm; 19 mm × 150 mm). The identity of the peptide was confirmed by MS using a Shimadzu UPLC-ESI-MS 2020 instrument (Theoretical [M+2H]2+, 1574.4; observed [M+2H]2+, 1574.4), and purity was determined on a Waters Acquity UPLC system (Acquity C18 BEH; pore size, 1.7 μm; column, 2.1 mm × 50 mm) at 0 to 70% solvent B (99.9% ACN and 0.1% TFA) over 5 min at 0.6 ml min−1.
Calcium imaging assay of vertebrate sensory neurons
DRG from 4- to 8-week-old male C57BL/6 mice were dissociated and plated in Dulbecco’s modified Eagle’s medium (Gibco) containing 10% fetal bovine serum (FBS; Assaymatrix) and penicillin/streptomycin (Gibco) on a 96-well poly-d-lysine–coated culture plate (Corning) and maintained overnight. Cells were loaded with Fluo-4 AM calcium indicator, according to the manufacturer’s instructions (Thermo Fisher Scientific). After loading (1 hour), the dye-containing solution was replaced with assay solution (1× Hanks’ balanced salt solution and 20 mM Hepes). Fluorescence corresponding to [Ca2+]i of typically 100 to 150 DRG cells per experiment was monitored in parallel using a Nikon Ti-E Deconvolution inverted microscope, equipped with a Lumencor Spectra LED Lightsource. Images were acquired at a 20× objective at 1 frame/s (excitation, 485 nm; emission, 521 nm). For each experiment, baseline fluorescence was monitored for 20 s, and at 30 s, the assay solution was replaced with an assay solution containing individual peptides [1:10 dilution of resuspended fraction (described above)] or whole venom (1:1000 dilution). Experiments involving the use of mouse tissue were approved by The University of Queensland animal ethics committee.
Pain behavior experiments
Male adult (12 to 15 weeks old) C57BL/6J mice were used for behavioral experiments. MIITX1-Mg1a (1, 10, or 100 μM) diluted in saline containing 0.1% bovine serum albumin (BSA) was administered in a volume of 20 μl into the hind paw by shallow intraplantar injection. Following injection, spontaneous nocifensive behavior events were counted, from video recordings, by a blinded observer. Control animals were injected with saline containing 0.1% BSA.
Mechanical paw withdrawal thresholds after intraplantar injection of MIITX1-Mg1a (1, 10, or 100 μM) were measured using an electronic von Frey apparatus (MouseMet Electronic von Frey, Topcat Metrology Ltd.). Briefly, the von Frey filament was placed against the plantar surface of the injected hind paw, and the force was gradually increased until the mice responded with the withdrawal of the paw. The average of three measurements per mouse was used as one biological replicate.
Thermal paw withdrawal thresholds after intraplantar injection of MIITX1-Mg1a (1, 10, or 100 μM) were measured using a MouseMet Thermal apparatus (Topcat Metrology Ltd.). Briefly, the preheated (37°C) thermal probe tip was applied to the plantar surface of the injected paw, which automatically induced the gradual heating (+2.5°C/s) of the probe until the mice responded with the withdrawal of the paw. The average of three measurements per mouse was used as one biological replicate.
All mice were acclimatized before experiments, and all measurements were performed by a blinded observer. Experiments involving animals were approved by The University of Queensland animal ethics committee.
Insect incapacitation assay
House crickets (A. domesticus; average mass, 150 mg; Pisces Live Food) were injected intra-abdominally with different doses of MIITX1-Mg1a (in 2 μl of PBS) and placed on their back, and the time taken to “right” was measured and compared with that of negative control crickets injected with 2 μl of PBS. Lethality was assessed at 24 hours.
Whole-cell patch-clamp electrophysiology
Whole-cell patch-clamp experiments were performed with HEK293 cells heterologously expressing human NaV1.7 (SB Drug Discovery) using a QPatch 16X automated electrophysiology platform (Sophion Bioscience). The extracellular solution contained the following: 70 mM NaCl, 70 mM choline chloride, 4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, and 10 mM glucose (pH 7.4; osmolarity, 305 mosmol). The intracellular solution contained the following: 140 mM CsF, 1 mM:5 mM EGTA/CsOH, 10 mM Hepes, and 10 mM NaCl (pH 7.3 with CsOH; osmolarity, 320 mosmol). MIITX1-Mg1a was diluted in extracellular solution with 0.05% BSA at a concentration of 3 μM. MIITX1-Mg1a effects were compared to pretoxin control conditions on the same cell with incubation times for buffer and MIITX1-Mg1a of 5 min each. NaV1.7 currents were induced with a holding potential of −90 mV followed by a 50-ms test pulse to −20 mV and a 0.05-Hz pulse frequency.
FLIPR assays
HEK293 cells and HEK293 cells heterologously expressing human NaV1.7 channels (SB Drug Discovery) were cultured in Eagle’s minimal essential medium (Sigma-Aldrich) containing 10% FBS (Assay Matrix Pty Ltd.), 2 mM l-glutamine, and selection antibiotics [blasticidin (4 μg/ml) and geneticin (0.6 mg/ml); hNaV1.7-expressing cells only). The cells were grown in a humid incubator at 37°C with 5% CO2 and passaged every 3 to 4 days using TrypLE Express (Thermo Fisher Scientific). Dissociated cells were plated into 384-well clear-bottom black-walled imaging plates (Corning) at a density of 10,000 to 15,000 cells per well. After 48 hours, the growth medium was removed from the wells, and the cells were loaded with red membrane potential dye (Molecular Devices), Calcium 4 No-Wash dye (Molecular Devices), or Asante NaTRIUM Green-2 (ANG-2 sodium dye) (Abcam) diluted in physiological salt solution (PSS; composition: 140 mM NaCl, 11.5 mM glucose, 5.9 mM KCl, 1.4 mM MgCl2, 1.2 mM Na2H2PO4, 5 mM NaHCO3, 1.8 mM CaCl2, 10 mM Hepes (pH 7.4)] for 30 min at 37°C, with 5% CO2. MIITX1-Mg1a was diluted in PSS/0.1% BSA and added to HEK293 cells and NaV1.7-expressing HEK293 cells using the FLIPRTETRA. Changes in membrane potential, [Ca2+]i, or intracellular [Na+] were monitored with a cooled charge-coupled device camera (excitation: 515 to 545 nm, 470 to 495 nm, and 470 to 495 nm; emission: 565 to 625 nm, 515 to 575 nm, and 515 to 575 nm, respectively) with reads taken every 1 s for 10 s before (baseline values) and 310 s after peptide addition. PSS/0.1% BSA was used as a negative control.
Statistical analyses
Raw FLIPR fluorescence values were converted to response over baseline values, and a negative control correction was performed using the FLIPRTETRA software ScreenWorks 3.2.0.14 (Molecular Devices). The AUC values were exported, and data were plotted and analyzed using GraphPad Prism version 7.00. To calculate concentration-response curves, a four-parameter Hill equation with variable Hill slope was fitted to the data. All data were expressed as the means ± SEM and are representative of at least three independent experiments. Statistical significance was defined as P < 0.05 and was calculated using an unpaired Student’s t tests. For pain behavior experiments, the data were expressed as means ± SEM. Statistical significance compared to vehicle controls was determined in GraphPad Prism version 7.00 using two-way ANOVA with Sidak’s multiple comparison test (spontaneous pain measurements) or one-way ANOVA with Dunnett’s multiple comparison test (mechanical and heat sensitivity measurements). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Material
Acknowledgments
Antimicrobial screening was performed by the Community for Antimicrobial Drug Discovery (CO-ADD). Funding: The authors would like to acknowledge financial support from the Australian Research Council (DECRA Fellowship DE160101142 to E.A.B.U., Discovery Grant DP160104025 to E.A.B.U. and G.F.K., and ARC Linkage Grant LP140100832 to B.R.H. and G.F.K.), an Australian Research Council Future Fellowship (FT130150100 to R.J.P.), and the Australian National Health and Medical Research Council (Principal Research Fellowship APP1044414 to G.F.K.). A.M. was supported by an Australian Government Research Training Program Scholarship. CO-ADD was funded by the Wellcome Trust (UK) and The University of Queensland (Australia). Author contributions: S.D.R. and E.A.B.U. designed the study. R.J.P., I.V., G.F.K., and E.A.B.U. provided reagents and additional input on experimental designs. S.D.R., A.M., D.C., B.R.H., H.S., and E.A.B.U. conducted experiments and analyzed the data. S.D.R. and E.A.B.U. wrote the manuscript with input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Venom component precursor sequences from this Transcriptome Shotgun Assembly project have been deposited at DDBJ/EMBL/GenBank (accession no. GGFG00000000). The version described in this paper is the first version (GGFG01000000). Raw sequencing data have been deposited in the National Center for Biotechnology Information sequence read archive (SRA accession no. SRR6466797). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaau4640/DC1
Supplementary Materials and Methods
Results
Fig. S1. MALDI-MS analysis of M. gulosa venom apparatus.
Fig. S2. Coelution of native (purified from venom) and synthetic MIITX1-Mg1a.
Fig. S3. MIITX1-Mg1a–induced changes in paw withdrawal.
Table S1. Venom- or toxin-associated annotation of nonvenom component transcripts.
Table S2. Assessment of antimicrobial, cytotoxic, and hemolytic activity of MIITX1-Mg1a.
Movie S1. Collection of venom from M. gulosa.
REFERENCES AND NOTES
- 1.T. Piek, Venoms of the Hymenoptera: Biochemical, Pharmacological, and Behavioural aspects (Academic Press, 1986). [Google Scholar]
- 2.Neumann W., Habermann E., Beiträge zur charakterisierung der wirkstoffe des bienengiftes. N.-S. Arch. Ex. Path. Ph. 222, 367–387 (1954). [PubMed] [Google Scholar]
- 3.Osman M. F. H., Brander J., Weitere Beiträge zur Kenntnis der chemischen Zusammensetzung des Giftes von Ameisen aus der Gattung Formica. Zeitschrift für Naturforschung B 16, 749 (1961). [Google Scholar]
- 4.MacConnell J. G., Blum M. S., Fales H. M., The chemistry of fire ant venom. Tetrahedron 27, 1129–1139 (1971). [Google Scholar]
- 5.Cavill G. W. K., Robertson P. L., Whitfield F. B., Venom and venom apparatus of the bull ant, Myrmecia gulosa (Fabr.). Science 146, 79–80 (1964). [DOI] [PubMed] [Google Scholar]
- 6.Davies N. W., Wiese M. D., Brown S. G. A., Characterisation of major peptides in ‘jack jumper’ ant venom by mass spectrometry. Toxicon 43, 173–183 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Ewen L. M., Ilse D., An inhibitor of mitochondrial respiration in venom of the Australian bull dog ant, Myrmecia gulosa. J. Insect Physiol. 16, 1531–1542 (1970). [DOI] [PubMed] [Google Scholar]
- 8.Inagaki H., Akagi M., Imai H. T., Taylor R. W., Wiese M. D., Davies N. W., Kubo T., Pilosulin 5, a novel histamine-releasing peptide of the Australian ant, Myrmecia pilosula (Jack Jumper Ant). Arch. Biochem. Biophys. 477, 411–416 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Lewis J. C., de la Lande I. S., Pharmacological and enzymic constituents of the venom of an Australian ‘bulldog’ and Myrmecia pyriformis. Toxicon 4, 225–234 (1967). [DOI] [PubMed] [Google Scholar]
- 10.Matuszek M. A., Hodgson W. C., Sutherland S. K., King R. G., Pharmacological studies of jumper ant (Myrmecia pilosula) venom: Evidence for the presence of histamine, and haemolytic and eicosanoid-releasing factors. Toxicon 30, 1081–1091 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Matuszek M. A., Hodgson W. C., Sutherland S. K., King R. G., Pharmacological studies of the venom of an Australian bulldog ant (Myrmecia pyriformis). Nat. Toxins 2, 36–43 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Wiese M. D., Chataway T. K., Davies N. W., Milne R. W., Brown S. G. A., Gai W.-P., Heddle R. J., Proteomic analysis of Myrmecia pilosula (jack jumper) ant venom. Toxicon 47, 208–217 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Touchard A., Koh J. M. S., Aili S. R., Dejean A., Nicholson G. M., Orivel J., Escoubas P., The complexity and structural diversity of ant venom peptidomes is revealed by mass spectrometry profiling. Rapid Commun. Mass Spectrom. 29, 385–396 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Hermann H. R. Jr, Blum M. S., The morphology and histology of the hymenopterous poison apparatus. III. Eciton hamatum (Formicidae)1. Ann. Entomol. Soc. Am. 60, 1282–1291 (1967). [Google Scholar]
- 15.Pluzhinikov K. A., Nol’de D. E., Tertyshnikova S. M., Sukhanov S. V., Sobol A. G., Torgov M., Filippov A. K., Arsen’ev A. S., Grishin E. V., [Structure-activity study of the basic toxic component of venom from the ant Ectatomma tuberculatum]. Bioorg. Khim. 20, 857–871 (1994). [PubMed] [Google Scholar]
- 16.Hermann H. R. Jr, Blum M. S., The morphology and histology of the hymenopterous poison apparatus. II. Pogonomyrmex badius (Formicidae)1. Ann. Entomol. Soc. Am. 60, 661–668 (1967). [Google Scholar]
- 17.Hermann H. R. Jr, Blum M. S., The morphology and histology of the hymenopterous poison apparatus. I. Paraponera clavata (Formicidae). Ann. Entomol. Soc. Am. 59, 397–409 (1966). [Google Scholar]
- 18.Orivel J., Redeker V., Le Caer J.-P., Krier F., Revol-Junelles A.-M., Longeon A., Chaffotte A., Dejean A., Rossier J., Ponericins, new antibacterial and insecticidal peptides from the venom of the ant Pachycondyla goeldii. J. Biol. Chem. 276, 17823–17829 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Blum M. S., Callahan P. S., The venom and poison glands of pseudomyrmex pallidus (F. Smith). Psyche 70, 69–74 (1963). [Google Scholar]
- 20.Touchard A., Aili S. R., Fox E. G., Escoubas P., Orivel J., Nicholson G. M., Dejean A., The biochemical toxin arsenal from ant venoms. Toxins 8, E30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazuma K., Masuko K., Konno K., Inagaki H., Combined venom gland transcriptomic and venom peptidomic analysis of the predatory ant Odontomachus monticola. Toxins 9, E323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres A. F. C., Huang C., Chong C.-M., Leung S. W., Prieto-da-Silva Á. R. B., Havt A., Quinet Y. P., Martins A. M. C., Lee S. M. Y., Rádis-Baptista G., Transcriptome analysis in venom gland of the predatory giant ant Dinoponera quadriceps: Insights into the polypeptide toxin arsenal of hymenopterans. PLOS ONE 9, e87556 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouzid W., Klopp C., Verdenaud M., Ducancel F., Vétillard A., Profiling the venom gland transcriptome of Tetramorium bicarinatum (Hymenoptera: Formicidae): The first transcriptome analysis of an ant species. Toxicon 70, 70–81 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Robinson S. D., Undheim E. A. B., Ueberheide B., King G. F., Venom peptides as therapeutics: Advances, challenges and the future of venom-peptide discovery. Expert Rev. Proteomics 14, 931–939 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Madio B., Undheim E. A. B., King G. F., Revisiting venom of the sea anemone Stichodactyla haddoni: Omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteomics 166, 83–92 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Smith J. J., Undheim E. A. B., True Lies: Using proteomics to assess the accuracy of transcriptome-based venomics in centipedes uncovers false positives and reveals startling intraspecific variation in Scolopendra subspinipes. Toxins 10, E96 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., Macmanes M. D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C. N., Henschel R., LeDuc R. D., Friedman N., Regev A., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 29.King G. F., Gentz M. C., Escoubas P., Nicholson G. M., A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 52, 264–276 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Undheim E. A. B., Jones A., Clauser K. R., Holland J. W., Pineda S. S., King G. F., Fry B. G., Clawing through evolution: Toxin diversification and convergence in the ancient lineage chilopoda (Centipedes). Mol. Biol. Evol. 31, 2124–2148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreil G., Haiml L., Suchanek G., Stepwise cleavage of the pro part of promelittin by dipeptidylpeptidase IV. Eur. J. Biochem. 111, 49–58 (1980). [DOI] [PubMed] [Google Scholar]
- 32.Honma T., Kawahata S., Ishida M., Nagai H., Nagashima Y., Shiomi K., Novel peptide toxins from the sea anemone Stichodactyla haddoni. Peptides 29, 536–544 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Shiomi K., Honma T., Ide M., Nagashima Y., Ishida M., Chino M., An epidermal growth factor-like toxin and two sodium channel toxins from the sea anemone Stichodactyla gigantea. Toxicon 41, 229–236 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Donovan G. R., Baldo B. A., Sutherland S., Molecular cloning and characterization of a major allergen (Myr p I) from the venom of the Australian jumper ant, Myrmecia pilosula. Biochim. Biophys. Acta 1171, 272–280 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Street M. D., Donovan G. R., Baldo B. A., Molecular cloning and characterization of the major allergen Myr p II from the venom of the jumper ant Myrmecia pilosula: Myr p I and Myr p II share a common protein leader sequence. Biochim. Biophys. Acta 1305, 87–97 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Inagaki H., Akagi M., Imai H. T., Taylor R. W., Kubo T., Molecular cloning and biological characterization of novel antimicrobial peptides, pilosulin 3 and pilosulin 4, from a species of the Australian ant genus Myrmecia. Arch. Biochem. Biophys. 428, 170–178 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Cologna C. T., Cardoso Jdos S., Jourdan E., Degueldre M., Upert G., Gilles N., Uetanabaro A. P. T., Costa Neto E. M., Thonart P., de Pauw E., Quinton L., Peptidomic comparison and characterization of the major components of the venom of the giant ant Dinoponera quadriceps collected in four different areas of Brazil. J. Proteomics 94, 413–422 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Rifflet A., Gavalda S., Téné N., Orivel J., Leprince J., Guilhaudis L., Génin E., Vétillard A., Treilhou M., Identification and characterization of a novel antimicrobial peptide from the venom of the ant Tetramorium bicarinatum. Peptides 38, 363–370 (2012). [DOI] [PubMed] [Google Scholar]
- 39.D. R. Hoffman, in Natural Toxins 2: Structure, Mechanism of Action, and Detection, B. R. Singh, A. T. Tu, Eds. (Springer, 1996), pp. 169–186. [Google Scholar]
- 40.Walker A. A., Mayhew M. L., Jin J., Herzig V., Undheim E. A. B., Sombke A., Fry B. G., Meritt D. J., King G. F., The assassin bug Pristhesancus plagipennis produces two distinct venoms in separate gland lumens. Nat. Commun. 9, 755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dutertre S., Jin A.-H., Vetter I., Hamilton B., Sunagar K., Lavergne V., Dutertre V., Fry B. G., Antunes A., Venter D. J., Alewood P. F., Lewis R. J., Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 5, 3521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran Y., Genikhovich G., Gordon D., Wienkoop S., Zenkert C., Özbek S., Technau U., Gurevitz M., Neurotoxin localization to ectodermal gland cells uncovers an alternative mechanism of venom delivery in sea anemones. Proc. R. Soc. Lond. B Biol. Sci. 279, 1351–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavill G. W. K., Williams P. J., Constituents of Dufour’s gland in Myrmecia gulosa. J. Insect Physiol. 13, 1097–1103 (1967). [Google Scholar]
- 44.Tosteson M. T., Tosteson D. C., The sting. Melittin forms channels in lipid bilayers. Biophys. J. 36, 109–116 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dekan Z., Headey S. J., Scanlon M., Baldo B. A., Lee T.-H., Aguilar M. I., Deuis J. R., Vetter I., Elliott A. G., Amado M., Cooper M. A., Alewood D., Alewood P. F., Δ-Myrtoxin-Mp1a is a helical heterodimer from the venom of the jack jumper ant that has antimicrobial, membrane-disrupting, and nociceptive activities. Angew. Chem. Int. Ed. Engl. 56, 8495–8499 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Okumura K., Inui K.-I., Hirai Y., Nakajima T., The effect of mastoparan on ion movement in black lipid membrane. Biomed. Res. 2, 450–452 (1981). [Google Scholar]
- 47.Sahara Y., Gotoh M., Konno K., Miwa A., Tsubokawa H., Robinson H. P. C., Kawai N., A new class of neurotoxin from wasp venom slows inactivation of sodium current. Eur. J. Neurosci. 12, 1961–1970 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Undheim E. A. B., Mobli M., King G. F., Toxin structures as evolutionary tools: Using conserved 3D folds to study the evolution of rapidly evolving peptides. Bioessays 38, 539–548 (2016). [DOI] [PubMed] [Google Scholar]
- 49.D. M. Santos, P. V. Reis, A. M. C. Pimenta, in Spider Venoms, P. Gopalakrishnakone, G. A. Corzo, M. E. de Lima, E. Diego-García, Eds. (Springer, 2016), pp. 361–377. [Google Scholar]
- 50.Harrison P. L., Abdel-Rahman M. A., Miller K., Strong P. N., Antimicrobial peptides from scorpion venoms. Toxicon 88, 115–137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prince R. C., Gunson D. E., Scarpa A., Sting like a bee—The ionophoric properties of melittin. Trends Biochem. Sci. 10, 99 (1985). [Google Scholar]
- 52.G. Pertea, Fqtrim: v0. 9.4 release. 10.5281/zenodo.20552 (2015).
- 53.Schmieder R., Edwards R., Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A., Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaau4640/DC1
Supplementary Materials and Methods
Results
Fig. S1. MALDI-MS analysis of M. gulosa venom apparatus.
Fig. S2. Coelution of native (purified from venom) and synthetic MIITX1-Mg1a.
Fig. S3. MIITX1-Mg1a–induced changes in paw withdrawal.
Table S1. Venom- or toxin-associated annotation of nonvenom component transcripts.
Table S2. Assessment of antimicrobial, cytotoxic, and hemolytic activity of MIITX1-Mg1a.
Movie S1. Collection of venom from M. gulosa.


