Abstract
Oxylipins, including jasmonic acid (JA) and volatiles, are important for signaling in plants, and these are formed by the lipoxygenase (LOX) enzyme family. There is a large gap in understanding of the underlying molecular basis of their roles in tea plants. Here, we identified 11 CsLOX genes from the tea plant (Camellia sinensis), and characterized their phylogeny, gene structure and protein features into three subclasses. We then examined their enzymatic activities, LOX expression and alternative splicing of transcripts during development and in response to abiotic or biotic stresses in tea plants. In vitro expressed protein assays showed that the CsLOX2, 3 and 9 enzymatically function to produce 9/13-HPOT, 13-HPOT and 9-HPOT, respectively. CsLOX2 and CsLOX9 green fluorescent protein (GFP) fusion proteins localized to chloroplasts and the cytoplasm, respectively. RNA sequencing, quantitative reverse transcription–PCR and Northern blot analysis suggested that CsLOX5, 6 and 9 were predominantly expressed in seeds, flowers and roots, respectively. CsLOX2, 3, 4, 6 and 7 were up-regulated after attack by the insect Ectropis oblique, while CsLOX1 was induced after infection with the pathogen Glomerella cingulata. CsLOX3, 7 and 10 were up-regulated by JA but not ABA or salicylic acid. Long-term cold stress down-regulated CsLOX expression while a short duration of cold induced the expression of CsLOX1, 6 and 7. Alternatively spliced transcripts of six CsLOX genes were dynamically regulated through time and varied in relative abundances under the investigated stresses; we propose a mechanism of competing or compensating regulation between isoforms. This study improves our understanding of evolution of LOXs and regulation of their diverse functions in plants.
Keywords: Alternative splicing, Biotic and abiotic stresses, Camellia sinensis, Expression patterns, LOX gene family, Phylogenetic analysis
Introduction
Perennial plants are in general more resistant to environmental stresses than annual plants, which suggests that they possess more elaborate mechanisms for stress adaptation (Rennenberg and Schmidt 2010, Munne-Bosch 2014). The perennial tea tree (Camellia sinensis) is an important economic woody crop planted within tropical to temperate regions (Yu 2011). The leaves of the tree are used for making tea, which is a popular beverage due to its attractive aroma, good taste and health-promoting effects (Zaveri 2006). The quality of tea leaves and thr abundance of natural flavor molecules are affected by the environment. To date, it is known that various abiotic stresses such as cold (Ban et�al. 2017), nitrogen (Li et�al. 2017b) and drought (Liu et�al. 2016), and biotic stresses such as insect attack (Wang et�al. 2016b) and pathogen infection (Wang et�al. 2016a) can induce a series of adaptive responses.
Oxylipins are oxygenated derivatives of fatty acids, and play roles in signaling responses to environmental stresses. In plants, oxylipins include the phytohormone jasmonic acid (JA), hydroxy-, oxo- or keto-fatty acids or volatile aldehydes (Mosblech et�al. 2009). Lipoxygenases (LOXs; EC 1.13.11.12) constitute a family of iron-containing enzymes which catalyze the oxygenation of polyunsaturated fatty acids (PUFAs) to initiate oxylipin biosynthesis in plants (Feussner and Wasternack 2002). During the reaction, LOXs catalyze the insertion of molecular oxygen into position C-9 or position C-13 of the PUFAs to produce 9S-HPOT and 13-HPOT hydroperoxides, respectively. Based on the positional specificity of substrates, LOXs can be classified into 9- and 13-LOXs (Feussner and Wasternack 2002). 13-LOXs can be further classified into type I and type II based on sequence similarity (Liavonchanka and Feussner 2006).
These hydroperoxides can be substrates for the allene oxide synthase (AOS) and hydroperoxide lyase (HPL) pathways. JA, as the product of the AOS pathway, is biosynthesized by a series of enzymatic reactions catalyzed by plastid-localized 13-LOX, AOS, allene oxide cyclase (AOC; Li et al.) and 12-oxo-phytodienoic acid reductase (OPR), and the final product is synthesized by three consecutive β-oxidation steps (Wasternack and Hause 2013). Synthesis of LOX-derived six carbon (C6) volatile compounds, such as C6 aldehydes, is catalyzed by HPL, isomerization factor, alcohol dehydrogenase (ADH) and alcohol acyltransferase (AAT) (Liavonchanka and Feussner 2006, Andreou and Feussner 2009). Both JA and C6 aldehydes commonly play roles in triggering and regulating defense mechanisms against insects, pathogens and environmental stresses in plants, which are mediated by the AOS and HPL pathways via molecular cross-talk and by competition for substrate (Halitschke and Baldwin 2003, Halitschke et�al. 2004, Duan et�al. 2005, Chehab et�al. 2008, Pauwels et�al. 2009, Wasternack and Hause 2013). Therefore, the characterization of LOX as a critical mediator in the AOS and HPL pathways will help us understand these plant defense mechanisms under stresses.
Many LOX-related studies have examined the functions of 9-LOX, 13-LOX or 9/13-LOX in plant development, and abiotic and biotic stresses; several enzymes have been described in several plant species such as Vitis vinifera and Cucumis sativus (Andreou and Feussner 2009, Andriy et�al. 2010, Yang et�al. 2012). In addition, functional characterization studies of LOXs in some model plants have been reported (Bannenberg et�al. 2009, Feng et�al. 2010, Fournier et al. 2010). However, functional differences among the entire LOX gene family in a plant species have remained unclear. Moreover, it is not well known which types of LOXs regulate the AOS and HPL pathways in response to various stresses.
RNA alternative splicing (AS) is a post-transcriptional gene regulation at the mRNA level, resulting in diverse transcripts encoded by the same gene (Filichkin et�al. 2015). AS affects transcript export, localization, mRNA stability, translation, protein stability and degradation (Reddy et�al. 2013). In plants, AS occurs during many stages of growth and development, circadian rhythms and environmental stresses (Crosetto et�al. 2013, Cui and Xiong 2015). For example, photoperiod, heat, low temperature and high salinity stresses induce clock genes such as CCA1, TOC1 and ZTL to undergo AS (Kwon et�al. 2014). However, previous studies on AS were mainly focused on regulatory genes (Kriechbaumer et�al. 2012, Wang et�al. 2015). To the best of our knowledge, AS of LOX genes has not been reported.
This study aims to determine the functions of CsLOX genes in the tea plant. We initially identified 11 CsLOX genes, examined their phylogenetic relationships and compared them with those of other plants. We predicted and then experimentally characterized the functions of CsLOX2, CsLOX3 and CsLOX9 via heterologous expression. We examined the expression patterns of full-length CsLOX transcripts and their alternatively spliced transcripts in developmental tissues, under stresses of cold, insect attack, pathogen infection and phytohormone exposure. Our results improve the understanding of the diverse functions of LOXs in tea plant, and could help to validate further the functions of CsLOX AS.
Results
Identification of tea CsLOX genes and phylogenetic analysis in plant species
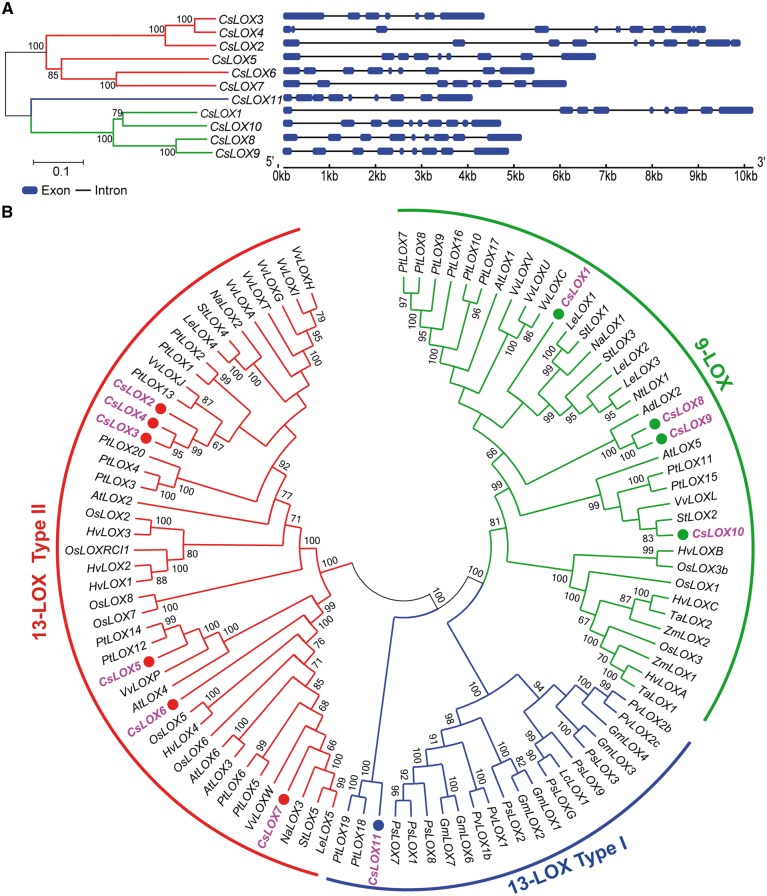
To identify the CsLOX genes in tea, we used the LOX amino acid sequences from four closely related species as a reference, and also used the LOX HMM model in Pfam to detect the CsLOX genes in the tea plant genome sequence (Finn et�al. 2016). Eleven CsLOX genes were identified after a comprehensive and restrictive filtering. Characteristics of these CsLOX proteins were then predicted, and the results are summarized in Supplementary Table S1. These characteristics include coding sequence (CDS) length, amino acid length, molecular weight, isoelectric point and subcellular localization. Ten CsLOX genes were identified as full-length sequences from the tea plant genome, and the remaining full-length sequence, named CsLOX11, was obtained using the 5' rapid amplication of cDNA ends (RACE) experimental technique. The predicted gene structures, which are further supported by our RNA sequencing (RNA-seq) data, are described in Fig.�1A. Most of the CsLOX genes harbor 8–9 introns, whereas CsLOX4 contains 11 introns.
Fig. 1.
Gene structure and phylogenetic tree of CsLOX genes in tea plant. (A) Full-length gene structure of CsLOX genes. The blue blocks indicate exons, and the lines between exons represent introns; lengths of blocks and lines represent relative sequence lengths. (B) Phylogenetic analysis of CsLOX genes. An unrooted Neighbor–Joining phylogenetic tree was constructed from 102 LOX proteins from 17 plant species. A bootstrap test was set to 1,000 replicates to validate tree classification confidence. The bootstrap values of the confidence levels are shown as percentages at branch nodes. The CsLOX genes are highlighted in pink.
To investigate the evolutionary relationship of CsLOX genes among plant species, a total of 102 LOX gene sequences from 17 plant species were retrieved as candidate genes to construct a phylogenetic tree (Fig.�1B). The 102 LOX genes were divided into two main subfamilies (9-LOX and 13-LOX) based on their sequence similarities and positional specificities of substrate oxygenation. Seven and four CsLOX genes were grouped into the 9-LOX and 13-LOX categories, respectively. We further classified the genes in the 13-LOX category into type I and II according to the presence or absence of an N-terminal transit peptide. As shown in Fig.�1A, six and one CsLOX genes were grouped into types II and I, respectively.
To reveal the internal relationship of the tea CsLOX family of genes, conserved motifs were predicted and annotated by submitting 11 CsLOX amino acid sequences to the MEME website (Supplementary Fig. S1A). Most of the closely related members have common motif compositions. Of 20 identified motifs, 14 identical motifs were shared among all LOX proteins (Supplementary Fig. S1A). Motif 2 consists of 38 amino acid residues, and includes five highly conserved histidine residues [His-(X)4-His-(X)4-His-(X)17-His-(X)8-His] (Supplementary Fig. S1B). The three-dimensional structures of 11 CsLOX proteins were predicted by homologous modeling (Supplementary Fig. S2) and their secondary structural compositions were analyzed (Supplementary Table S2). Our prediction shows that all CsLOX proteins contain two domains consisting of one N-terminal domain-I forming a β-barrel, and a C-terminal domain-II forming a bundle of helices. To examine further features of CsLOX members, multiple alignments of the 11 CsLOX amino acid sequences were performed, and all CsLOXs were found to harbor the five conserved amino acid residues (His, His, His, Asn and Ile), which were essential for binding of iron (Supplementary Fig. S3).
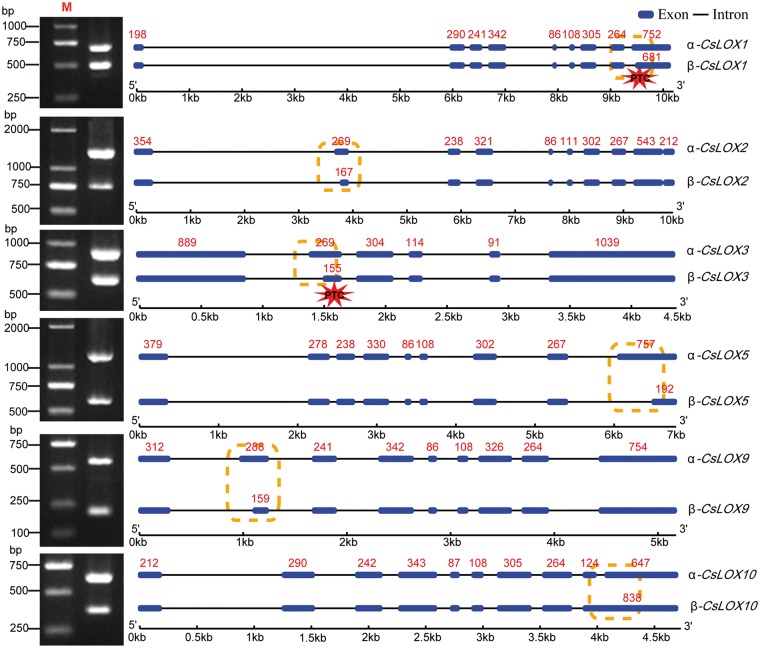
Detection and validation of alternative splicing of CsLOX transcripts
Using the sequence of genomic DNA and RNA-seq data, we identified six CsLOX genes which had undergone alternative splicing where each gene produced two isoforms. The primary type of transcript (α-transcript) retains all the exons with no AS, while the other type (β-transcript) represents an AS variant. The existence of α- and β-transcripts of the six CsLOX genes was validated by reverse transcription–PCR (RT–PCR) (Fig.�2). The β-CsLOX10 isoform retained the ninth intron, while the remaining CsLOX isoforms produced 3' splicing variants. Notably, a premature stop codon (PTC) was found in β-transcripts of CsLOX1, CsLOX2 and CsLOX3, which may result in degradation. However, no PTC was found in those of CsLOX5, CsLOX9 and CsLOX10, indicating that these genes may produce potentially truncated proteins. Therefore, the abnormal isoforms may have distinct functions compared with their respective full-length isoforms.
Fig. 2.
Alternative spliced isoforms of CsLOX genes in tea plant. The dotted box and asterisk indicate the positions of alternative splicing and locations of PTCs, respectively. The red numbers indicate exon length.
Activity of CsLOX2, 3 and 9 expressed in Escherichia coli, and subcellular localization
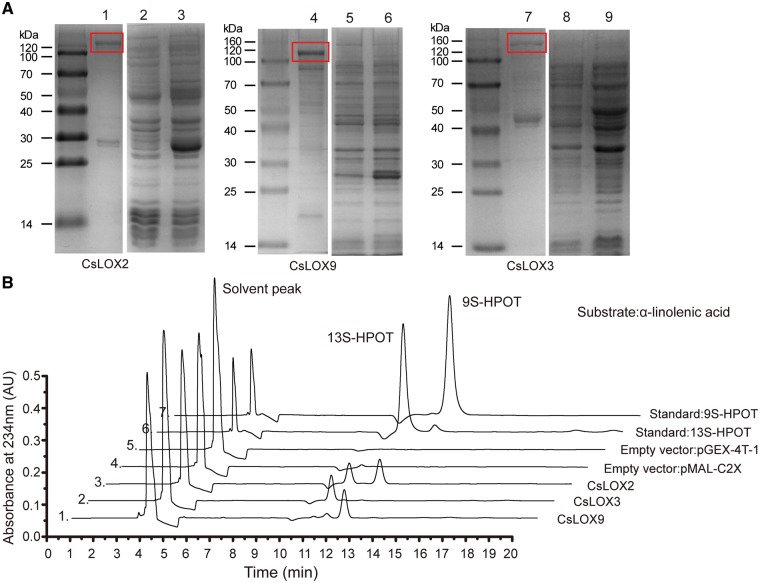
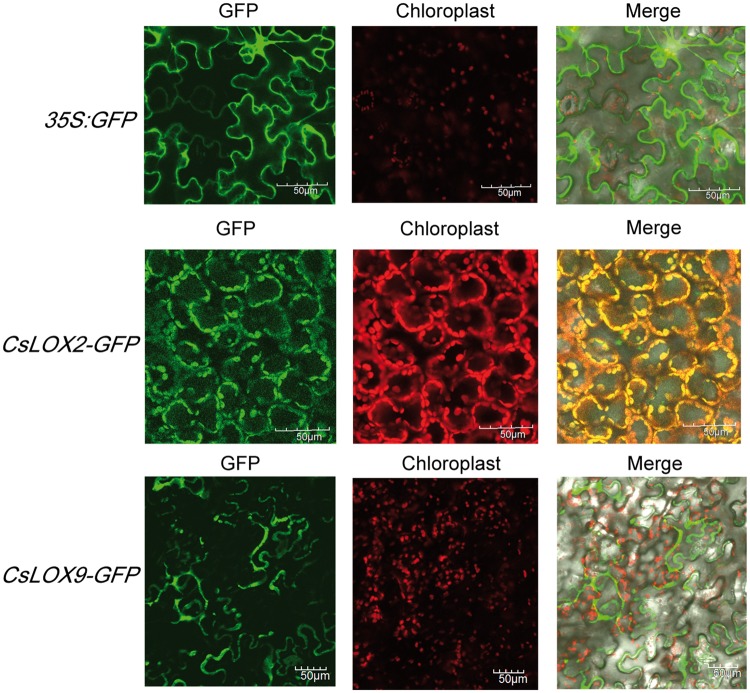
In consideration of the close and distinct phylogenetic relationship between LOX2, 3 and 9, they may have different enzymatic properties and differ in their expression patterns in response to stresses. Heterologous expression was necessary to determine whether CsLOX2, CsLOX3 and CsLOX9 proteins are functional LOXs; to determine this, we cloned the protein-coding regions to examine the heterologous expression of each gene in E. coli under the control of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG). To improve the efficiency of the enzymatic reaction, recombinant proteins were purified and characterized. SDS–PAGE analysis of protein size demonstrated that the expressed proteins were of the expected sizes: CsLOX2, 128 kDa; CsLOX3, 142 kDa; and CsLOX9, 130 kDa (Fig.�3A; Supplementary Fig. S8). To investigate the positional specificity of CsLOX proteins, we performed HPLC analysis to test the products resulting from CsLOX enzymatic reactions. Based on their retention times relative to the references 9S-HPOT and 13S-HPOT, we concluded that the products catalyzed by CsLOX3, CsLOX9 and CsLOX2 were 13-HPOT, 9S-HPOT and 9/13S-HPOT, respectively; the target products were not detected in the vector control pMAL-C2X or pGEX-4T-1 (Fig.�3B). Further quantitative determination indicated that the respective specific enzymatic activities of recombinant CsLOX2, CsLOX3 and CsLOX9 proteins were 8.54, 5.37 and 6.41 U mg–1 when α-linolenic acid was supplied as the substrate. In light of the different predictions for subcellular localizations of LOX2 and 9, we constructed CsLOX2-GFP, CsLOX9-GFP and 35S-GFP fusion protein expression vectors to validate their subcellular localizations. Transient expression in Nicotiana benthamiana leaf showed that CsLOX2–green fluorescent protein (GFP) was localized in the chloroplast, while CsLOX9–GFP protein was diffusely localized in the cytoplasm rather than the chloroplast (Fig.�4).
Fig. 3.
Heterologous expression of CsLOX in E. coli and determination of positional specificity. (A) SDS–PAGE image shows the purified CsLOX2, CsLOX9 and CsLOX3 recombinant protein: 1, purified protein of CsLOX2; 2, protein extracted from empty pGEX-4T-1 before induction; 3, proteins extracted from empty pGEX-4T-1 after induction; 4, purified protein of CsLOX9; 5, proteins extracted from empty pGEX-4T-1 before induction; 6, proteins extracted from empty pGEX-4T-1 after induction; 7, purified protein of CsLOX3; 8, proteins extracted from empty pMAL-C2X before induction; 9, proteins extracted from empty pMAL-C2X after induction. The solid box shows the expected positions of proteins. (B) HPLC analysis of enzymatic reaction products after incubation of recombinant CsLOX proteins with α-linolenic acid. Lines 1, 2, 3, 4, 5, 6 and 7 represent the product from reactions which used different recombinant proteins (CsLOX9, CsLOX3, CsLOX2, pMAL-C2X and pGEX-4T-1) and two standards (13S-HPOT and 9S-HPOT), respectively.
Fig. 4.
Subcellular localization of CsLOX2 and CsLOX9. The 35:GFP, 35:CsLOX2-GFP and 35:CsLOX9-GFP constructs were each transiently expressed in tobacco leaves. Rows 1, 2 and 3 show confocal imaging for the empty vector fused to a GFP tag, CsLOX2 fused to a GFP tag and CsLOX9 fused to a GFP, respectively. Scale bar = 50 μm.
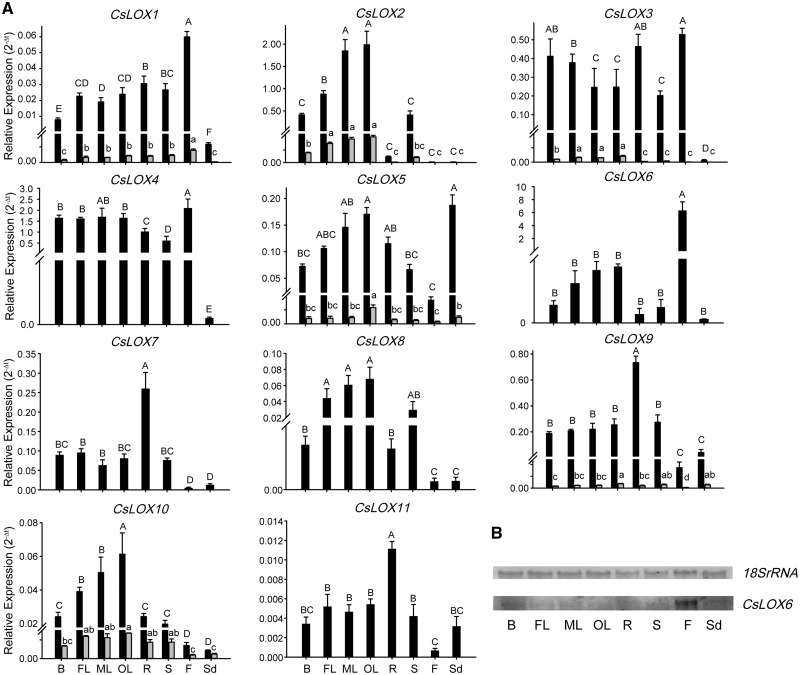
Expression patterns of CsLOX genes in different tissues
We examined the expression patterns of CsLOX genes from RNA-seq data in eight tea plant tissues (Supplementary Fig. S4A), and qRT–PCR was further conducted to verify these expression patterns and gain insight into potential functions of CsLOX genes in bud, first leaf, mature leaf in summer, old leaf in winter, root, stem, flower and seed (Fig.�5A). The results from RNA-seq data were partially consistent with those from qRT–PCR, especially for some tissue-specific CsLOX genes. The expression patterns of CsLOX genes in different tissues were distinct, suggesting their diverse roles during plant development. Most CsLOX genes exhibited high expression levels in mature leaves, but were lower in tender buds; a notable exception was CsLOX3, which had the opposite expression pattern. Moreover, CsLOX genes were barely detectable in seeds except for CsLOX5, which showed the highest expression level in seeds. Notably, some CsLOX genes exhibited prominent tissue specificity, such as CsLOX1 and CsLOX6, which were highly expressed in flowers. Transcripts of CsLOX6 were barely detectable in other tissues, while its expression in flowers was about 600-fold that in the first leaves; this result was further verified by Northern blot analysis (Fig.�5B). The expression pattern of CsLOX6 suggests that it may play a pivotal role in flower development (Fig.�5B). Expression of CsLOX9 was relatively high in roots, but was barely detectable in flowers. CsLOX7 and CsLOX11 followed the expression pattern of CsLOX9, but transcript abundances were slightly lower. Remarkably, all β-CsLOX transcripts showed much lower expression levels than the full-length transcripts in eight tissues, but similar expression patterns were observed between α- and β-transcripts.
Fig. 5.
Expression patterns of CsLOX genes in different tissues of tea plant. (A) qRT–PCR analysis of CsLOX genes across different tissues (B, bud; FL, first leaf; ML, mature leaf in summer; OL, old leaf in winter; R, root; S, stem; F, flower; Sd, seed). Black and gray bars represent the full-length and alternatively spliced transcripts, respectively. Bars show the means � SD (n = 3) of three biological replicates. Different letters above the bars denote significant differences at P < 0.05. (B) Northern blot analysis of CsLOX6.
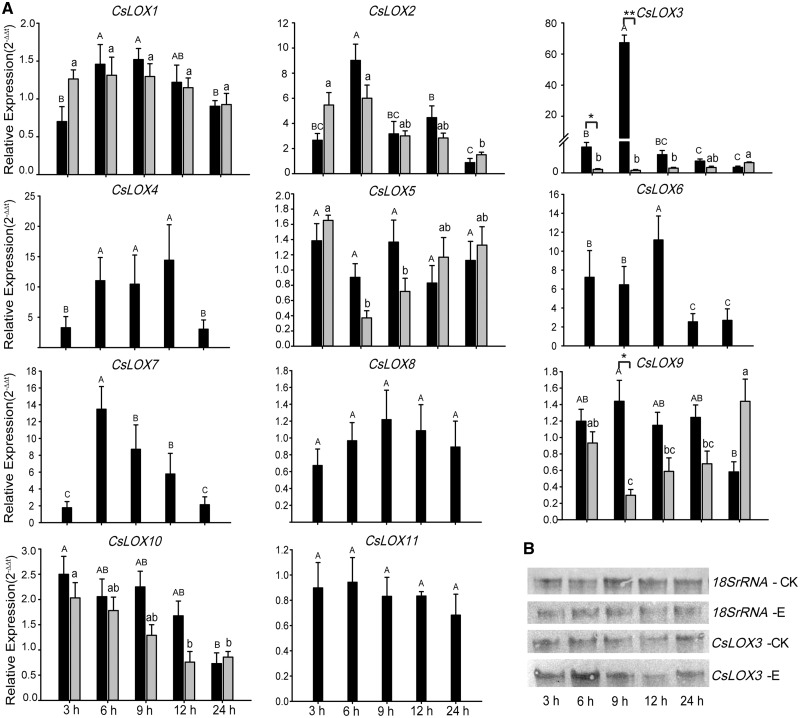
Expression patterns of CsLOX genes in response to biotic stresses
Expression patterns of CsLOX genes in response to feeding by tea geometrids and Glomerella cingulata infection were studied using qRT–PCR and transcriptome analysis. We observed that CsLOX genes were differentially expressed under these two treatments. Under E. oblique attack, almost all CsLOX genes were up-regulated; transcript levels of most CsLOX genes started to increase at 3 h, attained peak levels between 6 and 9 h, and finally decreased to normal transcript levels at 24 h (Fig.�6A; Supplementary Fig. S4B). The 13-LOX type II subfamily members CsLOX2, CsLOX3, CsLOX4, CsLOX6 and CsLOX7 showed strong up-regulation, although the times needed to attain the highest levels differed among them. Compared with other CsLOX genes, the transcript level of CsLOX3 increased 67.3-fold at 6 h, which was followed by a rapid decline. This fact was further supported by Northern blot analysis, which confirmed the significant mRNA accumulation at 6 h (Fig.�6B). Overall, similar expression patterns were observed between α-transcripts and β-transcripts, but higher levels of α-transcripts were detected than those of β-transcripts. For example, the alternatively spliced transcript levels of β-CsLOX3 were very low during the treatment relative to full-length α-CsLOX3 (Fig.�6A).
Fig. 6.
Expression pattern of CsLOX genes under tea geometrid feeding treatments at different time points. (A) qRT–PCR analysis of CsLOX genes under insect feeding treatments (CK, control leaves; E, leaves partially consumed by Ectropis oblique). Black and gray bars represent the full-length and alternatively spliced transcripts, respectively. The bar shows the means � SD (n = 3) of three biological replicates. Different letters above the bars represent significant differences at P < 0.05. The asterisks indicate the significant level (*P < 0.05, ** P < 0.01) based on a Tukey’s honestly significant difference test. (B) Northern blot analysis of the CsLOX3 gene.
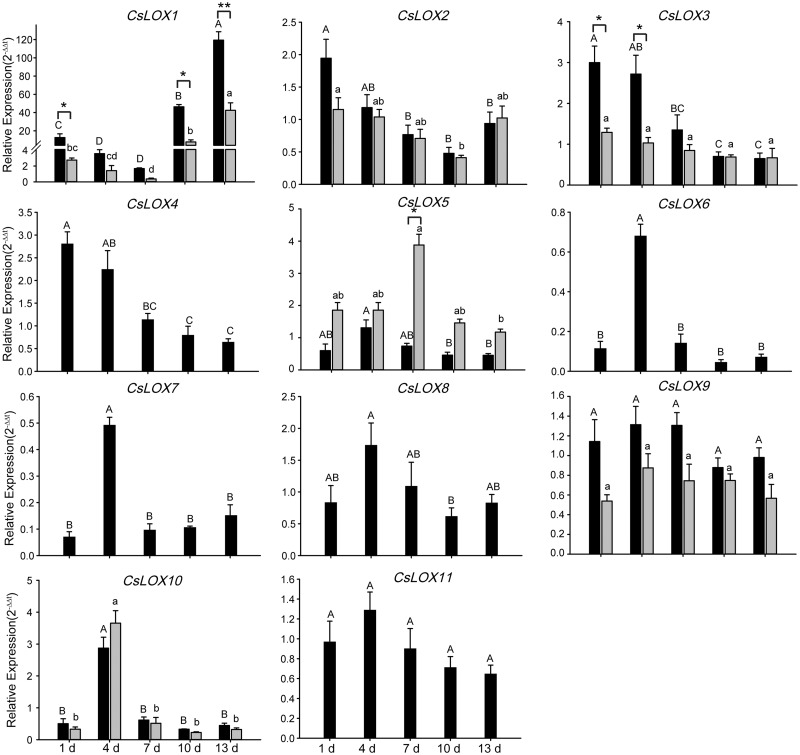
To reveal the pathogen’s effects on the expression of CsLOX genes, G. cingulata were inoculated onto healthy leaves. Results showed that CsLOX1 expression was strongly induced by G. cingulata infection, and reached a peak of up to 119-fold that of the blank control leaves at 13 d post-inoculation (Fig.�7). We also found a down-regulation (1.7-fold) of CsLOX1 expression between days 1 and 7, followed by up-regulation between days 10 and 13 relative to the control, indicating a two-stage regulation of CsLOX1 expression in response to pathogen infection. In addition, the expression patterns of CsLOX2, CsLOX3 and CsLOX4 can be clustered into a group, while those of CsLOX6 and CsLOX7 fell into another group. Under infection, the levels of CsLOX β-transcripts were lower than those of the α-transcripts. However, the β-CsLOX1 transcript level was significantly induced, but remained lower than that of α-CsLOX1. The abundances of α- and β-CsLOX1 reached a maximum at day 13 post-inoculation, and followed a two-stage pattern as mentioned previously. Interestingly, a stronger induction of CsLOX5 β-transcription was observed compared with the corresponding α-transcript. Together, the full-length α-transcripts of most CsLOX genes should function dominantly while spliced β-transcripts may act complementarily or competitively in response to pathogen infection.
Fig. 7.
Expression pattern of CsLOX genes subjected to strains of Glomerella cingulata infection. qRT–PCR analysis of CsLOX genes at different time points. Black and gray bars indicate the full-length and alternatively spliced transcripts, respectively. Data represent the means � SD (n = 3) of three biological replicates. Different letters above the bars denote significant differences at P < 0.05. The asterisks indicate the significant level (*P < 0.05, **P < 0.01) based on a Tukey’s honestly significant difference test.
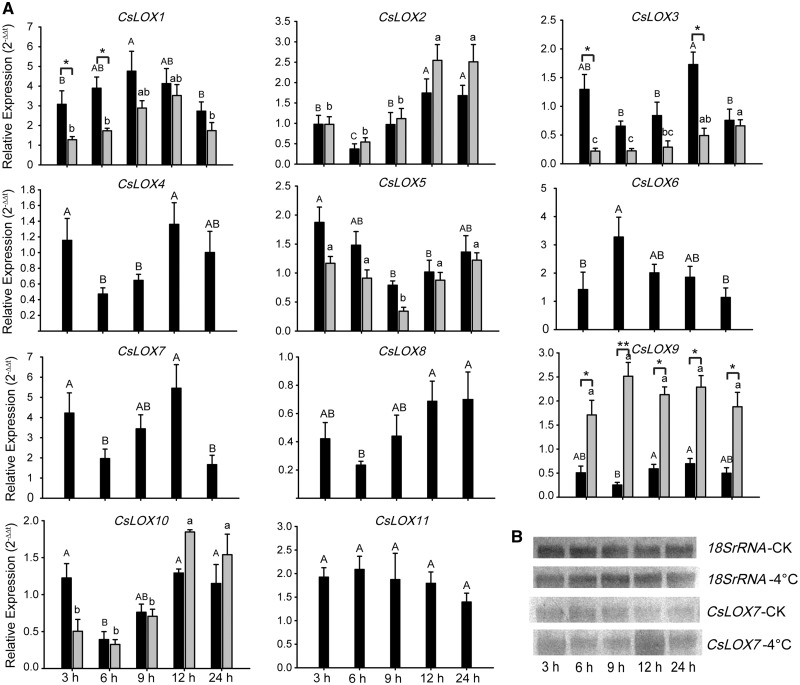
Expression patterns of CsLOX genes in response to low temperature
To further study the in vivo function of CsLOX genes under cold treatment, joint analysis of bioinformatics and qRT–PCR was employed to observe the expression patterns of CsLOX genes. After long-term cold (10, 4 and 0�C) stress treatments, the expression levels of CsLOX genes were down-regulated compared with the control before cold treatment (Supplementary Fig. S5). In addition, another 4�C cold treatment was conducted to study the early expression patterns at up to five time points within 24 h. The results of both RNA-seq and qRT–PCR indicated that CsLOX1, CsLOX6 and CsLOX7 transcripts were induced to high accumulation levels in response to 4�C cold stress, and their expression levels were highest at 9, 6 and 12 h, respectively. As Fig.�8A illustrates, the level of CsLOX7 transcripts increased at 3 h, decreased at 6 h and then increased to a peak of 5.5-fold the initial level at 12 h compared with the control; finally, the expression recovered to a normal level at 24 h. The results of Northern blot analysis were consistent with qRT–PCR results (Fig.�8B). Similarly, a lower level of alternatively spliced transcripts was observed than of full-length transcripts under 4�C treatment. Interestingly, the timing of peak expression differed between α-CsLOX1 and β-CsLOX1, suggesting the dynamic regulation of splicing under cold stress.
Fig. 8.
Expression patterns of CsLOX genes under low-temperature treatments. (A) qRT–PCR analysis of CsLOX genes under 4�C treatments at different time points. Black and gray bars represent the full-length and alternatively spliced transcripts, respectively. Bars indicate the means � SD (n = 3) of three biological replicates. Different letters above the bars denote significant differences at P < 0.05. The asterisks indicate the significant level (*P < 0.05, **P < 0.01) based on a Tukey’s honestly significant difference test. (B) Northern blot analysis of the CsLOX7 gene under 4�C treatments.
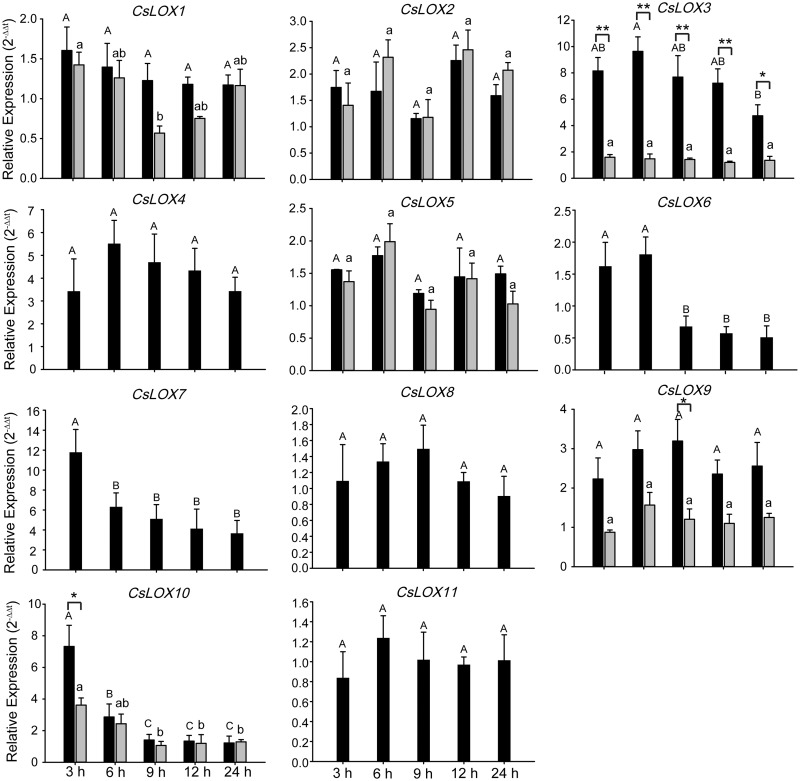
Expression patterns of CsLOX genes under phytohormone treatments
CsLOX genes have been reported to respond to various defense-related hormones, including ABA (Kishimoto et�al. 2005), salicylic acid (SA) and methyl jasmonate (MeJA) (Yang et�al. 2012). Although we found various expression patterns among CsLOX genes, no significant inductions occurred among any CsLOX genes after ABA or SA treatment, suggesting that CsLOX genes were insensitive in response to ABA and SA (Supplementary Figs. S6, S7). However, strong induction of CsLOX3, CsLOX7 and CsLOX10 transcripts was observed in response to MeJA; the highest transcript levels of CsLOX7 and CsLOX10 were at 3 h, while that of CsLOX3 occurred at 6 h (Fig.�9). The significantly up-regulated expression of CsLOX3, CsLOX4 and CsLOX7 was detected in both MeJA treatments and geometrid feeding, which could be caused by wounding from geometirds inducing endogenous accumulation of JA. Interestingly, differences between α-CsLOX3 and β-CsLOX3 induction were observed not only after tea geometrid feeding but also after MeJA treatment.
Fig. 9.
Expression patterns of CsLOX genes under MeJA phytohormone treatments. qRT–PCR analysis of CsLOX genes at different time points. Black and gray bars represent the full-length and alternatively spliced transcripts, respectively. Bars indicate the means � SD (n = 3) of three biological replicates. Different letters above the bars denote significant differences at P < 0.05. The asterisks indicate the significant level (*P < 0.05, **P < 0.01) based on a Tukey’s honestly significant difference test.
Discussion
The LOX gene family (9-LOX, 13-LOX and 9/13 LOX) is known to be involved in lipid catabolism for oxylipin synthesis, which functions in defense, i.e. via the AOS and HPL pathways to synthesize JA and C6 aldehydes (Liavonchanka and Feussner 2006). However, there remains a large gap between understanding LOX gene sequences and their functions in plants. In this study, we identified and characterized 11 LOX genes in the tea plant C. sinensis, including their phylogenetic relationship, their evolution in plant species, and gene and protein structures. We predicted and experimentally verified cellular localization of CsLOXs. We noted differential expression patterns and transcript splicing during tea plant developmental stages, in different tissues, and in response to stresses of insects, pathogens, phytohormones and cold. We concluded that CsLOX genes share sequence and functional similarity but have different roles with tissue specificity during development and plant defense, which could be regulated via alterative transcript splicing. Thus, the study highlights the important contribution and efficacy of AS and provides comprehensive understanding of the functional roles of CsLOX genes.
Evolutionarily, the divergence in the gene phylogenetic tree may indicate a divergence in functional roles, while genes within the same cluster may share common functions. LOX genes were proposed to have multiple functions in several plant species such as Populus (Andriy et�al. 2010, Chen et�al. 2015). In this study, the classified subfamilies of 13-LOXs and 9-LOXs, which comprise our 11 identified full-length CsLOX genes, were predicted and proven to have different functions for 9- or 13-LOX specificity. The primary conserved motifs of R/TH, R/TF, R/SF or R/CF for 13-LOX and R/TV for 9-LOX in the substrate-binding region may determine the 9- or 13-LOX specificity (Feussner and Wasternack 2002). Among the tested CsLOX2, 3 and 9, CsLOX3 and CsLOX9 exhibited 13-LOX and 9-LOX catalytic activities as expected, respectively; however, CsLOX2 exhibited dual-positional specificity. Previously, CsLOX1 was initially classified as a 9-LOX, but also has dual-positional specificity (Liu and Han 2010). Similar functions of LOXs have been detected in rice, pea and maize (Kim et�al. 2003, Veronico et�al. 2006, Wang et�al. 2008). Positional specificity may be imparted by amino acids other than those within the conserved motif. Therefore, we propose that additional amino acids should also contribute to the detailed positional specificity of LOXs. In addition, the dual-positional LOX may enrich the potential biological function via simultaneously producing two different hydroperoxides.
There is increasing evidence that LOX gene family members play fundamental roles in lipid and fatty acid catabolism in seed germination, plant development and senescence, formation of signaling molecules, stress responses and plant defenses (Liavonchanka and Feussner 2006, Andreou and Feussner 2009, Yang et�al. 2012). Our analysis of CsLOX expression patterns in different tissues showed relatively high transcript abundances in reproductive organs and roots. Among them, CsLOX7, CsLOX9 and CsLOX11 showed significantly higher expression in roots than in leaves. This may explain a reported observation that strong 9-LOX catalysis of 9S-HPOT synthesis and its derivatives (i.e. 9-KOT, 9-KOD and 9-HOT) may modulate lateral root formation and decrease primary root elongation (Vellosillo et�al. 2007). Therefore, we hypothesize that the most likely biological function of CsLOX9 is to act as a vital regulator via producing 9S-HPOT. Flower-specific expression of LOX was observed in Alstroemeria peruviana during senescence in both sepals and petals, and Arabidopsis lox3 lox4 double mutants were male sterile and defective in global proliferative arrest (Leverentz et�al. 2002, Caldelari et�al. 2011). Previous results showed that expression of CsLOX6 in flowers is 105-fold higher relative to that of CsLOX1 (Liu and Han 2010); considering our findings, we think that CsLOX6, as a 13-LOX member, may be a predominant regulator during tea flower development. Considering the vital and diverse functions of JA during flower development such as anther elongation, proliferative arrest and male inflorescence growth (Acosta et�al. 2009, Brioudes et�al. 2009, Caldelari et�al. 2011), it is reasonable to speculate that CsLOX6 may be the pre-eminent CsLOX gene activating the AOS pathway to synthesize JA, and that production of JA is directly involved in tea plant flower development.
LOX members are involved in plant responses to herbivore attack by initiating either the AOS or HPL pathway (Halitschke et�al. 2004). It has been reported that the 13-LOXs provide the main function in responses to both wounding and herbivore feeding, which subsequently results in up-regulated levels of JA or green leaf volatiles (GLVs) in wounded plant tissues (Halitschke et�al. 2004). In our study, five 13-LOX genes (CsLOX2, 3, 4, 6 and 7) were strongly induced under tea geometrid treatment (Fig.�6; Supplementary Fig. S4B). The AtLOX2 in Arabidopsis, StLOX4 in potato and OsLOXORC1 in rice are homologous with CsLOX2, CsLOX3 and CsLOX4, respectively; these do not contribute to JA biosynthesis and are involved in the HPL pathway by converting the linolenic acid substrate into C6 volatiles in wounded leaves (Schaffrath et�al. 2000, Le�n et�al. 2002, Kishimoto et�al. 2005). Therefore, these three CsLOXs (2, 3 and 4) may function in the HPL-mediated production of C6 aldehydes and alcohols. On the other hand, another two close homologs of CsLOX6 and CsLOX7 (potato StLOX5 and tobacco NaLOX3, respectively) are responsible for wound-induced JA synthesis (Royo et�al. 1996, Halitschke and Baldwin 2003). However, the differences in expression patterns of CsLOX6 and CsLOX7 suggest that the temporal specificity is possibly critical for secondary activation of JA biosynthesis in response to insect feeding.
LOXs are involved in defense response against pathogen infection by formation of hydroperoxides, 13-LOX-derived oxylipin or JA, or by activating autoxidative lipid peroxidation processes to program cell death (G�bel et�al. 2002, Montillet et�al. 2002, G�bel et�al. 2003, Glazebrook 2005, Goossens et�al. 2016). Our results revealed that CsLOX1 was up-regulated 119-fold, and may play a vital role in activating defense pathways under G. cingulata treatment. CsLOX1 was sensitive to JA but not to SA, and this was in accordance with previous findings for maize LOX1 (Cho et�al. 2012). This could be due to an antagonistic interaction between JA and SA in defense reactions (Chen et�al. 1995, Glazebrook 2005, Guo and Stotz 2007, Loake and Grant 2007). The absence of an SA-specific TCA-element in the CsLOX1 promoter (Supplementary Table S3) could also be a factor in SA insensitivity.
Oxylipin metabolism plays roles in low-temperature stress (Laura et al. 2009). Under low temperature, JA can alleviate the oxidative damage through delaying the increases in the O2– production rate and H2O2 content, regulating the ascorbate–glutathione (AsA–GSH) cycle and inducing some transcription factors to activate downstream defense genes such as MYC2, AP2/ERF or WRKY (Blokhina et�al. 2003, Li et�al. 2004, Pr� et�al. 2008, Fernandez-Calvo et�al. 2011, Zhao et�al. 2013, Li et�al. 2017a). Previous studies demonstrated that LOX is strongly induced by low-temperature exposure with the production of JA (He et�al. 2002, Zhao et�al. 2012). Our results showed that the CsLOX6 and CsLOX7 were induced, and these two 13-LOXs were expressed in response to pathogen infection. Therefore, they may be involved in JA production. However, CsLOX1 was surprisingly up-regulated although it is not in the 13-LOX cluster. It is possible that the dual-positional specificity of CsLOX1 functions as 13-LOX enzymatic activity for JA synthesis. In contrast to previous studies (Yamaguchi-Shinozaki and Shinozaki 2005), our results showed that ABA did not induce the expression of CsLOX1, CsLOX6 and CsLOX7, which may result from the absence of ABA-responsive elements (ABREs) in promoters of these genes.
Studies have shown a linkage between environmental stress and AS events (Cui and Xiong 2015). For instance, the clock genes are regulated by extensive AS, and are influenced by various adversities (James et�al. 2012, Kwon et�al. 2014). Plant LOX genes are regulated by circadian rhythms (Nemchenko et�al. 2006). Interestingly, we found similar or opposite expression patterns between CsLOX full-length and alternatively spliced transcripts under diverse stresses (Figs.�6–8), and similar patterns were observed in MtJMJC5 expression profiles in response to cold (Shen et�al. 2016). For example, the splicing variant β-CsLOX3 was significantly inhibited by insect feeding, while the α-CsLOX3 transcript level reached a peak. Previous studies on protein LSm5 and SKIP suggested that alternatively spliced transcripts generally decrease the levels of the corresponding full-length transcripts (Cui et�al. 2014, Feng et�al. 2015). Therefore, LOX function should depend on not only full-length transcripts but also alternatively spliced transcripts via compensation or competition. Moreover, sequence analysis of alternatively spliced CsLOX indicated that the PTC was in β-CsLOX1 and β-CsLOX3 sequences (Fig.�2). The alternatively spliced RNA variants with PTCs were degraded through the nonsense-mediated decay (NMD) pathway (Brogna and Wen 2009, Crosetto et�al. 2013). Therefore, it is possible that alternatively spliced transcripts of CsLOX1 and CsLOX3 are degraded by NMD, and they meanwhile regulate the abundance of corresponding full-length transcripts via competition for splicing machinery. On the other hand, we proposed that the PTC lacking alternatively spliced variants of CsLOX2, CsLOX5, CsLOX9 and CsLOX10 may encode the truncated proteins. In comparison, Arabidopsis β-AtHAB1 and β-AtJAZ2 are truncated proteins and still function at a lower activity than full-length transcripts (Chung et�al. 2010, Wang et�al. 2015); hence, the truncated CsLOX proteins may still have activity. Recent reports indicated the different function of alternatively spliced transcripts in plant (Kriechbaumer et�al. 2012, Liu et�al. 2018). In our study, we found that several alternatively spliced transcripts were significantly induced under diverse stresses; for example, β-CsLOX1 is highly expressed in response to G. cingulate. Therefore, we speculate and emphasize that the basic LOX functionality may not only depend on the constitutive transcripts but may also be augmented or modulated to act on stress induction pathways in an AS-dependent manner. The functional validation of alternatively spliced CsLOX1 could be of interest in our future research.
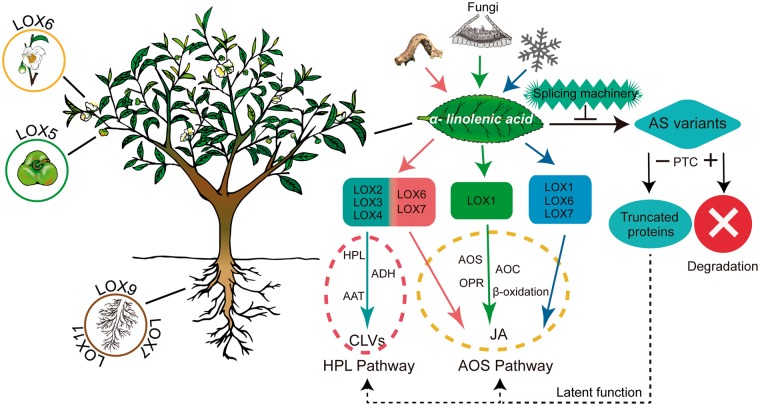
In conclusion, we identified 11 CsLOX genes from the tea plant (Camellia sinensis) genome sequence, classified them into three subfamilies and characterized these CsLOXs by bioinformatic and experimental methods. The regulation of CsLOX genes showed differential spatiotemporal expression, and alternative transcript splicing in plant development and in response to biotic and abiotic stresses (Fig.�10). The AS in CsLOX genes may play a role through compensation or competition with corresponding full-length transcripts. Therefore, this study improves our understanding of evolution of the LOX gene family and their diverse functions in plant.
Fig. 10.
Hypothetical model for functions of CsLOX genes in the tea plant. Several CsLOX genes exhibit tissue specificity during tea plant growth. Biotic and abiotic stresses (tea geometrid feeding, infection with strains of Glomerella cingulata and low temperature) initiate the transcription of key CsLOX genes and subsequently activate the AOS or HPL pathway, respectively. Environmental stresses trigger the aberrant splicing machinery and generate the AS of CsLOX genes. The CsLOX genes which harbor PTCs can be degraded by the NMD machinery. In contrast, CsLOX genes without a PTC may possibly be translated into truncated proteins which, as latent functional proteins, can perform their normal functions.
Materials and Methods
Plant materials and treatments
Two-year-old tea plants (C. sinensis cv. Shuchazao) were obtained from the Dechang tea plantation (Shucheng, latitude 31.3N, longitude 117.2E, Anhui, China). Tea plants were grown in an experimental plot with 120 cm between rows and 33 cm between plants within a row. Plants were maintained under a controlled culture environment at 23 � 3�C with 65 � 5% room humidity and a 16/8 h (day/night) photoperiod. All tea plants were fertilized and watered under the same regime. Plants were chosen for our experiments that had uniform heights and canopy width, and without signs of disease and insects. All treatments in this study were performed with these tea plant materials.
Tea geometrids were captured in fields at the Dechang tea plantation (Shucheng, Anhui, China) and were identified as Ectropis oblique. A population of tea geometrids was maintained on a tea plant (C. sinensis cv. Shuchazao) grown in a growth chamber at 25�C with 65 � 5% room humidity and a 14/10 h (day/night) photoperiod. For the insect feeding treatment, tea geometrids at the third larval stage were placed on tea plants (20 geometrids per individual tea plant). After a third of each leaf was consumed, all the tea geometrids were removed and consumed leaves were collected at 3, 6, 9, 12 and 24 h. The samples in the control group without tea geometrid feeding were collected at the same time points.
The Glomerella cingulata strain was isolated from diseased tissues of tea leaves (C. sinensis cv. Shuchazao) showing anthracnose symptoms from the Dechang tea plantation (Shucheng). Conidia were harvested and adjusted to 106 c.f.u. ml–1 with sterile water. Leaves were surface-sterilized with 75% ethanol and washed with sterile distilled water. Five leaves per tea plant were pricked with a sterile needle, and 20 μl of conidial suspension was applied to the wound (Chen et�al. 2017). Inoculated leaves were covered with plastic wrap, and plants were immediately placed under a plastic cover and were incubated in the greenhouse at 28 � 3�C with 85 � 5% room humidity and a 16/8 h (day/night) photoperiod. The control group consisted of plants where healthy leaves were inoculated with sterile water. Infected leaves were collected on days 1, 4, 7, 10 and 13.
Two kinds of cold treatments were used in the study. The first consisted of a cold acclimation treatment; for this, healthy tea plants were placed in a controlled chamber at 25�C/20�C for day and night cycles, respectively. The plants were cold treated at 10�C for 6 h (recorded as CA1–6h) and were subsequently cold treated at 4�C for 7 d (CA1–7d), followed by a freezing treatment at 0�C for 7 d (CA2). Finally, the plants were treated at 25�C/20�C for a 7 d recovery period (DA). A group of tea plants maintained at 25�C/20�C served as the control. The second cold treatment was performed by switching plants from 25�C/20�C directly to 4�C without cold acclimation, with the control plants kept at 25�C/20�C. The second leaves of each plant were collected at 3, 6, 9, 12 and 24 h for both the treatment and control groups.
Healthy tea plants were sprayed with either 1 mM SA, 100 μM ABA or 1 mM MeJA (containing 0.05% Tween-20) until the leaves were completely wet. Leaves treated with distilled water served as controls for SA and ABA treatments, and leaves treated with 0.05% Tween-20 served as the control for MeJA. The second leaf at the same position of each branch was collected at 3, 6, 9, 12 and 24 h following treatment.
The apical bud, first leaf, mature leaf in summer, old leaf in winter, stem, root, flower and seed were collected from identical tea plants. All treatments in this study were performed with three biological replicates, and all collected samples were immediately frozen in liquid nitrogen and stored at –80�C until use.
Identification and collection of CsLOX genes
‘Genwise v2.2.0’ software (Birney et�al. 2004) was used to search the LOX sequence from the tea plant genome sequence by using amino acid sequences of previously reported LOX-encoded proteins from four species having close phylogenetic relationships with the tea plant (C. sinensis) (Xia et�al. 2017): Arabidopsis thaliana (Bannenberg et�al. 2009), Populus trichocarpa (Chen et�al. 2015), Vitis vinifera (Andriy et�al. 2010) and Actinidia deliciosa (Zhang et�al. 2009) (Supplementary Table S4). We simultaneously used Hidden Markov Model (HMM) to search the tea genome sequence with a Pfam sequence (PF00305) containing a typical LOX domain from the Pfam HMM library (http://pfam.jouy.inra.fr/, v.31.0). After filtering the redundant sequences, the remaining sequences were translated into amino acid sequences and manually confirmed for the existence and integrity of the LOX domain and PLAT/LH2 domain using the Simple Modular Architecture Research Tool (SMART) (Schultz et�al. 1998).
Phylogenetic tree construction
Multiple sequences alignment analysis was carried out using the ClustalW program in the software MEGA 6.0. A phylogenetic tree was constructed for 102 LOX CDS protein sequences using the NJ (Neighbor–Joining) method in the software MEGA with the following parameters: bootstrap method (1,000 replicates), Poisson model, Uniform rates, Complete deletion. The accession information of all LOX genes from 17 species is listed in Supplementary Table S4, and some sequences are referenced to published articles (Chen et�al. 2015).
Analyses of conserved motifs, gene structure, promoters and protein characteristics
Motif analysis was conducted on the MEME website (http://meme-suite.org/tools/meme). The parameters were set as 20 motifs; minimum and maximum widths, 6–200. The intron and exon were determined based on genome data and depicting the intron/exon structure of LOX by the online website Gene Structure Display Server (GSDS 2.0, http://gsds.cbi.pku.edu.cn/index.php). Cis-acting regulatory elements were annotated using an online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Basic characteristic data about LOX proteins were calculated using the ProtParam tool (http://web.expasy.org/protparam/). Secondary structures of proteins were analyzed using Scratch Protein Predictor (http://scratch.proteomics.ics.uci.edu/). Homologous modeling was conducted to construct 3-dimensional structures of proteins using an online website (https://swissmodel.expasy.org/interactive). The CsLOX proteins were rebuilt in 3-dimensional models with ribbons by SPDBV software 4.10 (Johansson et�al. 2012). Heat maps were depicted using R language. Subcellular localization was predicted through the following online websites: TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/) and MultiLoc2 (http://abi.inf.uni-tuebingen.de/Services/MultiLoc2).
Transcriptomic data analysis
The 90 bp paired-end RNA-seq reads from different tissues of tea plants (bud, first leaf, mature leaf in summer, old leaf in winter, stem, root, flower and seed) (unpublished) were generated with an Illumina Hiseq™2000 platform. The 90 bp paired-end RNA-seq reads from leaf feeding by the tea geometrid (accession, SRX1543038) were also generated with an Illumina Hiseq™2000 platform. We used Bowtie v2.0.6 to build an index of the tea plant (C. sinensis) reference genome (Langmead and Salzberg 2012). Furthermore, clean paired-end reads were mapped to the reference genome using TopHat v2.0.9 and mapped reads were assembled with Cufflinks v.2.1.1 to generate a genome-based Cufflinks assembly (Trapnell et�al. 2009). We calculated the fragments per kilobase of the exon model per million mapped fragments (FPKM) for all CsLOX genes. Next, ASTALAVISTA (v.3.0) was carried out to identify AS. Intron retention (IR), exon skipping (ES), alternative acceptor (AA) and alternative donor (AD) were defaulted as four main types of AS events, and were analyzed as reported previously (Foissac and Sammeth 2007). All AS events of CsLOX genes were validated for their authentic existence in vivo according to a reported study using P19–P36 primers (Xu et�al. 2017).
RNA extraction and qRT–PCR analysis
Total RNA was extracted from plant materials using an RNAprep Pure Plant Kit (cat. No. DP441, Tiangen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from total RNA using a PrimeScript RT Reagent Kit (cat. No. 6110A, TAKARA) referring to the manufacturer’s instructions. The program and reaction system of qRT–PCR was as per the previous reported protocol from our team (Wang et�al. 2016b). The CsGAPDH gene was selected as the internal control (Wang et�al. 2016b, Yu et�al. 2017). All specific primers (P37–P72) are listed in Supplementary Table S5. The amplification efficiencies of all genes used in this study ranged from 95% to 105% (Kubista et�al. 2006). The relative gene expressions were quantified using the 2−ΔCt and 2−ΔΔCt method in different cases. Statistical analysis of qRT–PCR was performed using DPS software (Tang and Zhang 2013).
RACE and cloning of full-length LOX genes
A SMART RACE Kit (cat 6107, Clontech, Dalian) was used to obtain the full-length sequences. The primers used were P1–P6 (Supplementary Table S5). The amplification of each full-length LOX gene sequence used one pair of specific primers (P7–P10). PCR products were purified and ligated with pEAXY-Blunt vector (cat. No. CB101, Transgen), subsequently transformed into E. coli Trans1-T1 Phage Resistant Chemically Competent Cells (cat. No. CD501, Transgen) and screened to verify clones positive for the gene insert.
Heterologous expression and purification of CsLOX proteins
The open reading frames (ORFs) of CsLOX2, CsLOX3 and CsLOX9 were amplified by PCR with FastPfu polymerase (cat. No. AP221, Transgen) using gene-specific primers (P11–P16) and cDNA template, which was generated from total RNA of tea plant leaves. The PCR products of CsLOX2 and CsLOX9 were purified and directly ligated with the pGEX-4T-1 vector which was digested with restriction enzymes BamHI (TAKARA, 1010A) and XhoI (TAKARA, 1094A) using a ClonExpressII One Step Cloning Kit (cat. No. C112, Vazyme) according to the manufacturer’s instructions. In addition, the CsLOX3 PCR product was similarly processed, but was ligated into pMAL-C2X with BamHI (TAKARA, 1010A) and SalI (TAKARA, 1080A). After confirming the cloned fragments by sequencing, pGEX-LOX2, pGEX-LOX9 and pMAL-LOX3 were transformed into E. coli BL21 (DE3) Chemically Competent Cell (cat. No. CD601, Transgen). Glutathione S-transferase (GST)-tagged recombinant proteins were purified from disrupted cells by affinity chromatography with GST resin (cat. No. DP201, Transgen), and maltose-binding protein (MBP)-tagged recombinant proteins were affinity purified with amylose resin (cat. No. E8021S, New England Biolabs). For SDS–PAGE analysis, 5 μg of each recombinant protein was boiled with loading buffer and subjected to 10% SDS–PAGE.
Enzyme activity assay
A total reaction system of 1 ml consists of 0.2 M sodium acetate (pH 5.0), 1.6 mM α-linolenic acid and 10 μg of recombinant protein. The temperature of reaction was set to 25�C and the reaction time was 30 min. The reaction was stopped by adding 0.1 M hydrochloric acid solution until pH 3.0 was reached. The reaction mixture was then added to a 1 ml volume of a chloroform:methanol mixture at a 1:3 ratio, and subjected to 10 min ultrasonic oscillations. We removed the aqueous phase, and completely dried the reaction mixture with a flow of nitrogen gas. Finally, the reaction mixture was dissolved in a 0.5 ml volume of chloroform and methanol mixture at a 1:1 ratio. Product analysis of 9S-HPOT and 13S-HPOT was carried out using a Waters series HPLC system with a silica LC column [Luna Silica (2) 100 �, 250�4.6 mm, 5 μm particle size]. The eluent was composed of n-hexane:2-propanol:acetic acid (100:5:0.1, by vol.) at 1 ml min–1 and the effluent was monitored at 234 nm (Liu and Han 2010). Standards of 9- and 13-HPOT were purchased from Cayman Chemical (cat. Nos. 45120and 45210).
Northern blot analysis
The DIG-High Prime DNA Labeling and Detection Starter Kit II (cat. No. 11745832910, Roche) was used to synthesize the probes of tested genes according to the manufacturer’s instructions. The probe primers were P73–P80, and at least one gene-specific primer was generated in the untranslated region (UTR). To verify successful labeling of probes with DIG, PCR products generated from labeled probes were detected by agarose gel electrophoresis. PCR primers from labeled probes have a slower migration rate than those from non-labeled probes. Northern blot was carried out as previously reported (Santos et�al. 2011).
Subcellular localization of GFP fusion proteins
ORFs of CsLOX2 and CsLOX9 were amplified without a stop codon by RT–PCR (P81–P84) and ligated with pGWB5 vector by gateway technology to construct LOX2–GFP and LOX9–GFP fusion proteins under control of the 35S promoter. The plasmids of pGWB5-LOX2 and pGWB5-LOX9 were then transformed into EHA105 Competent Cells. A positive colony was selected for infiltration of N. benthamiana according to a reported protocol (Shen et�al. 2012). After 48 h of infection, leaves were examined using an Olympus FV1000 confocal microscope.
Funding
This work was supported by the National Natural Science Foundation of China [31171608]; the Special Innovative Province Construction in Anhui Province [15czs08032]; the Special Project for Central Guiding Science and Technology Innovation of Region in Anhui Province [2016080503B024]; and the Program for Changjiang Scholars and Innovative Research Team in University [IRT1101].
Supplementary Material
Acknowledgment
We thank Dr. Liang Zhang for assistance with the HPLC analysis.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- AOS
allene oxide synthase
- AS
alternative splicing
- CDS
coding sequence
- GFP
green fluorescent protein
- HPL
hydroperoxide lyase
- 9S-HPOT
9S-hydroperoxy-10E,12Z,15Z-octadecatrienoic acid
- 13S-HPOT
13S-hydroperoxy-6Z,9Z,11E-octadecatrienoic acid
- JA
jasmonic acid
- LOX
lipoxygenase
- MeJA
jasmonic acid methyl ester
- NMD
nonsense-mediated decay
- ORF
open reading frame
- PTC
premature stop codon
- PUFA
polyunsaturated fatty acid
- qRT–PCR
quantitative reverse transcription–PCR
- RACE
rapid amplification of cDNA ends
- RNA-seq
RNA sequencing; RT–PCR, reverse transcription–PCR
- SA
salicylic acid
Glossary
Footnote
- The nucleotide sequences from this study were submitted to NCBI GenBank under the following accession numbers: CsLOX2 (FJ418174.1), CsLOX3 (FJ794853.1), CsLOX4 (MG708225), CsLOX5 (MG708226), CsLOX6 (MG708227), CsLOX7 (MG708228), CsLOX8 (MG708229), CsLOX9 (MG708230), CsLOX10 (MG708231) and CsLOX11 (MG708232)
References
- Acosta I.F., Laparra H., Romero S.P., Schmelz E., Hamberg M., Mottinger J.P. (2009) tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323: 262–265. [DOI] [PubMed] [Google Scholar]
- Andreou A., Feussner I. (2009) Lipoxygenases—structure and reaction mechanism. Phytochemistry 70: 1504–1510. [DOI] [PubMed] [Google Scholar]
- Andriy P., Jackie W., Brian J., Chris W. (2010) Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Functional Plant Biol. 37: 767–784. [Google Scholar]
- Ban Q., Wang X., Pan C., Wang Y., Kong L., Jiang H., et al. (2017) Comparative analysis of the response and gene regulation in cold resistant and susceptible tea plants. PLoS One 12: e0188514.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannenberg G., Martinez M., Hamberg M., Castresana C. (2009) Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids 44: 85–95. [DOI] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R. (2004) GeneWise and Genomewise. Genome Res. 14: 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhina O., Virolainen E., Fagerstedt K.V. (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot. 91: 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brioudes F., Joly C., Szecsi J., Varaud E., Leroux J., Bellvert F., et al. (2009) Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J 60: 1070–1080. [DOI] [PubMed] [Google Scholar]
- Brogna S., Wen J. (2009) Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 16: 107–113. [DOI] [PubMed] [Google Scholar]
- Caldelari D., Wang G., Farmer E.E., Dong X. (2011) Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 75: 25–33. [DOI] [PubMed] [Google Scholar]
- Chehab E.W., Kaspi R., Savchenko T., Rowe H., Negre-Zakharov F., Kliebenstein D., et al. (2008) Distinct roles of jasmonates and aldehydes in plant-defense responses. PLoS One 3: e1904.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Qiao W., Zeng L., Shen D., Liu Z., Wang X., et al. (2017) Characterization, pathogenicity, and phylogenetic analyses of colletotrichum species associated with brown blight disease on Camellia sinensis in China. Plant Dis. 101: 1022–1028. [DOI] [PubMed] [Google Scholar]
- Chen Z., Chen X., Yan H., Li W., Li Y., Cai R., et al. (2015) The lipoxygenase gene family in poplar: identification, classification, and expression in response to MeJA treatment. PLoS One 10: e0125526.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Malamy J., Henning J., Conrath U., S�nchez-Casas P., Silva H., et al. (1995) Induction, modification, and transduction of the salicylic acid signal in plant defense responses. Proc. Natl. Acad. Sci. USA 92: 4134–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K., Kim Y.C., Woo J.C., Rakwal R., Agrawal G.K., Yoeun S., et al. (2012) Transgenic expression of dual positional maize lipoxygenase-1 leads to the regulation of defense-related signaling molecules and activation of the antioxidative enzyme system in rice. Plant Sci. 185-186: 238–186. [DOI] [PubMed] [Google Scholar]
- Chung H.S., Cooke T.F., Depew C.L., Patel L.C., Ogawa N., Kobayashi Y., et al. (2010) Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto P., Deparis S., Fourestey G., Quarteroni A. (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P., Xiong L. (2015) Environmental stress and pre-mRNA splicing. Mol. Plant 8: 1302–1303. [DOI] [PubMed] [Google Scholar]
- Cui P., Zhang S., Ding F., Ali S., Xiong L. (2014) Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 15: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H., Huang M.Y., Palacio K., Schuler M.A. (2005) Variations in (hydroperoxide lyase) gene expression differentially affect hexenal signaling in the Columbia and Landsberg ecotypes of Arabidopsis. Plant Physiol. 139: 1529–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Dong Z., Xu Z., An X., Qin H., Wu N., et al. (2010) Molecular analysis of lipoxygenase (LOX) genes in common wheat and phylogenetic investigation of LOX proteins from model and crop plants. J. Cereal Sci. 52: 387–394. [Google Scholar]
- Feng J., Li J., Gao Z., Lu Y., Yu J., Zheng Q., et al. (2015) SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant 8: 1038–1052. [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvo P., Chini A., Fernandez-Barbero G., Chico J.M., Gimenez-Ibanez S., Geerinck J., et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I., Wasternack C. (2002) The lipoxygenase pathway. Annu. Rev. Plant Biol. 53: 275–297. [DOI] [PubMed] [Google Scholar]
- Filichkin S., Priest H.D., Megraw M., Mockler T.C. (2015) Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol. 24: 125–135. [DOI] [PubMed] [Google Scholar]
- Finn R.D., Penelope C., Eberhardt R.Y., Eddy S.R., Jaina M., Mitchell A.L., et al. (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44: D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac S., Sammeth M. (2007) ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 35: W297–W299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier J., Pou�nat M.L., Rickauer M., Rabinovltch-Chable H., Rigaud M., Esquerr�-Tugay� M.T. (2010) Purification and characterization of elicitor-induced lipoxygenase in tobacco cells. Plant J. 3: 63–70. [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- G�bel C., Feussner I., Hamberg M., Rosahl S. (2002) Oxylipin profiling in pathogen-infected potato leaves. Biochim. Biophys. Acta 1584: 55–64. [DOI] [PubMed] [Google Scholar]
- G�bel C., Feussner I., Rosahl S. (2003) Lipid peroxidation during the hypersensitive response in potato in the absence of 9-lipoxygenases. J. Biol. Chem. 278: 52834–52840. [DOI] [PubMed] [Google Scholar]
- Goossens J., Fernandez-Calvo P., Schweizer F., Goossens A. (2016) Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91: 673–689. [DOI] [PubMed] [Google Scholar]
- Guo X., Stotz H.U. (2007) Defense against Sclerotinia sclerotiorum in Arabidopsis Is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol. Plant Microbe Interact. 20: 1384. [DOI] [PubMed] [Google Scholar]
- Halitschke R., Baldwin I.T. (2003) Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 36: 794–807. [DOI] [PubMed] [Google Scholar]
- Halitschke R., Ziegler J., Kein�nen M., Baldwin I.T. (2004) Silencing of hydroperoxide lyase and allene oxide synthase reveals substrate and defense signaling crosstalk in Nicotiana attenuata. Plant J. 40: 35–46. [DOI] [PubMed] [Google Scholar]
- He Y., Fukushige H., Hildebrand D.F., Gan S. (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128: 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A.B., Syed N.H., Bordage S., Marshall J., Nimmo G.A., Jenkins G.I., et al. (2012) Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24: 961–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.U., Zoete V., Michielin O., Guex N. (2012) Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics 13: 173.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.S., Choi E., Kim Y., Cho K., Lee A., Shim J., et al. (2003) Dual positional specificity and expression of non-traditional lipoxygenase induced by wounding and methyl jasmonate in maize seedlings. Plant Mol. Biol. 52: 1203–1213. [DOI] [PubMed] [Google Scholar]
- Kishimoto K., Matsui K., Ozawa R., Takabayashi J. (2005) Volatile C6-aldehydes and Allo-ocimene activate defense genes and induce resistance against Botrytis cinerea in Arabidopsis thaliana. Plant Cell Physiol. 46: 1093–1102. [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V., Wang P., Hawes C., Abell B.M. (2012) Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 70: 292.. [DOI] [PubMed] [Google Scholar]
- Kubista M., Andrade J.M., Bengtsson M., Forootan A., Jon�k J., Lind K., et al. (2006) The real-time polymerase chain reaction. Mol. Aspects Med. 27: 95–125. [DOI] [PubMed] [Google Scholar]
- Kwon Y.J., Park M.J., Kim S.G., Baldwin I.T., Park C.M. (2014) Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 14: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laura S., Mar�aT S., Felix R., FranciscoB F. (2009) Physiological, hormonal and molecular mechanisms regulating chilling injury in horticultural species. Postharvest technologies applied to reduce its impact. J. Sci. Food Agric. 89: 555–573. [Google Scholar]
- Le�n J., Royo J., Vancanneyt G., Sanz C., Silkowski H., Griffiths G., et al. (2002) Lipoxygenase H1 gene silencing reveals a specific role in supplying fatty acid hydroperoxides for aliphatic aldehyde production. J. Biol. Chem. 277: 416–423. [DOI] [PubMed] [Google Scholar]
- Leverentz M.K., Wagstaff C., Rogers H.J., Stead A.D., Chanasut U., Silkowski H., et al. (2002) Characterization of a novel lipoxygenase-independent senescence mechanism in Alstroemeria peruviana floral tissue. Plant Physiol. 130: 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E.T. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Lu X., Ma H., Lyu D. (2017) Jasmonic acid regulates the ascorbate–glutathione cycle in Malus baccata Borkh. roots under low root-zone temperature. Acta Physiol. Plant. 39: 174. [Google Scholar]
- Li W., Xiang F., Zhong M., Zhou L., Liu H., Li S., et al. (2017) Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis). Sci. Rep. 7: 1693.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liavonchanka A., Feussner I. (2006) Lipoxygenases: occurrence, functions and catalysis. J. Plant Physiol. 163: 348–357. [DOI] [PubMed] [Google Scholar]
- Liu G.F., Liu J.J., He Z.R., Wang F.M., Yang H., Yan Y.F., et al. (2018) Implementation of CsLIS/NES in linalool biosynthesis involves transcript splicing regulation in Camellia sinensis. Plant. Cell Environ. 41: 176–186. [DOI] [PubMed] [Google Scholar]
- Liu S., Han B. (2010) Differential expression pattern of an acidic 9/13-lipoxygenase in flower opening and senescence and in leaf response to phloem feeders in the tea plant. BMC Plant Biol. 10: 228.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-C., Jin J.-Q., Ma J.-Q., Yao M.-Z., Ma C.-L., Li C.-F., et al. (2016) Transcriptomic analysis of tea plant responding to drought stress and recovery. PLoS One 11: e0147306.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loake G., Grant M. (2007) Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 10: 466–472. [DOI] [PubMed] [Google Scholar]
- Montillet J.L., Agnel J.P., Ponchet M., Vailleau F., Roby D., Triantaphylid�s C. (2002) Lipoxygenase-mediated production of fatty acid hydroperoxides is a specific signature of the hypersensitive reaction in plants. Plant Physiol. Biochem. 40: 633–639. [Google Scholar]
- Mosblech A., Feussner I., Heilmann I. (2009) Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 47: 511–517. [DOI] [PubMed] [Google Scholar]
- Munne-Bosch S. (2014) Perennial roots to immortality. Plant Physiol. 166: 720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemchenko A., Kunze S., Feussner I., Kolomiets M. (2006) Duplicate maize 13-lipoxygenase genes are differentially regulated by circadian rhythm, cold stress, wounding, pathogen infection, and hormonal treatments. J. Exp. Bot. 57: 3767–3779. [DOI] [PubMed] [Google Scholar]
- Pauwels L., Inze D., Goossens A. (2009) Jasmonate-inducible gene: what does it mean? Trends Plant Sci. 14: 87–91. [DOI] [PubMed] [Google Scholar]
- Pr� M., Atallah M., Champion A., De Vos M., Pieterse C.M.J., Memelink J., et al. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.S.N., Marquez Y., Kalyna M., Barta A. (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennenberg H., Schmidt S. (2010) Perennial lifestyle—an adaptation to nutrient limitation? Tree Physiol. 30: 1047–1049. [DOI] [PubMed] [Google Scholar]
- Royo J., Vancanneyt G., P�rez A.G., Sanz C., St�rmann K., Rosahl S., et al. (1996) Characterization of three potato lipoxygenases with distinct enzymatic activities and different organ-specific and wound-regulated expression patterns. J. Biol. Chem. 271: 21012–21019. [DOI] [PubMed] [Google Scholar]
- Santos T.B.D., Budzinski I.G.F., Marur C.J., Petkowicz C.L.O., Pereira L.F.P., Vieira L.G.E. (2011) Expression of three galactinol synthase isoforms in Coffea arabica L. and accumulation of raffinose and stachyose in response to abiotic stresses. Plant Physiol. Biochem. 49: 441. [DOI] [PubMed] [Google Scholar]
- Schaffrath U., Zabbai F., Dudler R. (2000) Characterization of RCI-1, a chloroplastic rice lipoxygenase whose synthesis is induced by chemical plant resistance activators. Eur. J. Biochem. 267: 5935–5942. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., He X.Z., Poovaiah C.R., Wuddineh W.A., Ma J.Y., Mann D.G.J., et al. (2012) Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193: 121–136. [DOI] [PubMed] [Google Scholar]
- Shen Y., Wu X., Liu D., Song S., Liu D., Wang H. (2016) Cold-dependent alternative splicing of a Jumonji C domain-containing gene MtJMJC5 in Medicago truncatula. Biochem. Biophys. Res. Commun. 474: 271–276. [DOI] [PubMed] [Google Scholar]
- Tang Q.Y., Zhang C.X. (2013) Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Entomologia Sin. 20: 254–260. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellosillo T., Mart�nez M., L�pez M.A., Vicente J., Casc�n T., Dolan L., et al. (2007) Oxylipins produced by the 9-lipoxygenase pathway in arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 19: 831–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronico P., Giannino D., Melillo M.T., Leone A., Reyes A., Kennedy M.W., et al. (2006) A novel lipoxygenase in pea roots. Its function in wounding and biotic stress. Plant Physiol. 141: 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Y., Cao H., Hao X., Zeng J., Yang Y., et al. (2016) Transcriptome analysis of an anthracnose-resistant tea plant cultivar reveals genes associated with resistance to Colletotrichum camelliae. PLoS One 11: e0148535.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Shen W., Liu L., Jiang L., Liu Y., Su N., et al. (2008) A novel lipoxygenase gene from developing rice seeds confers dual position specificity and responds to wounding and insect attack. Plant Mol. Biol. 66: 401–414. [DOI] [PubMed] [Google Scholar]
- Wang Y.N., Tang L., Hou Y., Wang P., Yang H., Wei C.L. (2016) Differential transcriptome analysis of leaves of tea plant (Camellia sinensis) provides comprehensive insights into the defense responses to Ectropis oblique attack using RNA-Seq. Funct. Integr. Genomics 16: 383–398. [DOI] [PubMed] [Google Scholar]
- Wang Z., Ji H., Yuan B., Wang S., Su C., Yao B., et al. (2015) ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 6: 8138.. [DOI] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia E.H., Zhang H.B., Sheng J., Li K., Zhang Q.J., Kim C., et al. (2017) The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 10: 866–877. [DOI] [PubMed] [Google Scholar]
- Xu Q., Zhu J., Zhao S., Hou Y., Li F., Tai Y., et al. (2017) Transcriptome profiling using single-molecule direct RNA sequencing approach for in-depth understanding of genes in secondary metabolism pathways of Camellia sinensis. Front. Plant Sci. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10: 88–94. [DOI] [PubMed] [Google Scholar]
- Yang X.Y., Jiang W.J., Yu H.J. (2012) The expression profiling of the lipoxygenase (LOX) family genes during fruit development, abiotic stress and hormonal treatments in cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 13: 2481–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-Y., Yang H., Wei C.-L., Yu O., Zhang Z.-Z., Jiang C.-J., et al. (2011) Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 12: 131.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu O. (2011) Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC genomics 12: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaveri N.T. (2006) Green tea and its polyphenolic catechins: medicinal uses in cancer and noncancer applications. Life Sci. 78: 2073–2080. [DOI] [PubMed] [Google Scholar]
- Zhang B., Yin X.R., Li X., Yang S.L., Ferguson I.B., Chen K.S. (2009) Lipoxygenase gene expression in ripening kiwifruit in relation to ethylene and aroma production. J. Agric. Food Chem. 57: 2875–2881. [DOI] [PubMed] [Google Scholar]
- Zhao J., Li S., Jiang T., Liu Z., Zhang W., Jian G., et al. (2012) Chilling stress—the key predisposing factor for causing Alternaria alternata infection and leading to cotton (Gossypium hirsutum L.) leaf senescence. PLoS One 7: e36126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.L., Wang J.N., Shan W., Fan J.G., Kuang J.F., Wu K.Q., et al. (2013) Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 36: 30–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.