Abstract
Paraoxonase 1 (PON1) is a serum enzyme associated with high density lipoprotein (HDL) regulation through its paraoxonase and arylesterase activity. PON1 inhibits the oxidation of HDL and low density lipoprotein (LDL), and is involved in the pathogenesis of a variety of diseases including atherosclerosis. Conversely, mutations in the low density lipoprotein receptor (LDLR) result in failure of receptor mediated endocytosis of LDL leading to its elevated plasma levels and onset of familial hypercholesterolemia (FH). In the current study we investigated the role of PON1 polymorphisms rs662; c.575A > G (p.Gln192Arg) and rs854560; c.163T > A (p.Leu55Met) in a large family having FH patients harboring a functional mutation in LDLR. Genotypes were revealed by RFLP, followed by confirmation through Sanger sequencing. PON1 activity was measure by spectrophotometry. Our results show significantly reduced serum paraoxonase and arylesterase activities in FH patients compared with the healthy individuals of the family (p < 0.05). PON1 QQ192 genotype showed a significantly higher association with FH (p=0.0002). PON1 Q192 isoform was associated with reduced serum paraoxonase activity by in silico analysis and PON1 R192 exhibited higher serum paraoxonase and arylesterase activity than the other polymorphs. Our results highlight that the combination of LDLR mutations and PON1 MMQQ genotypes may lead to severe cardiac events.
Keywords: Paraoxonase-1, hypercholesteremia, arylesterase, LDLR mutation
Introduction
The human paraoxonase (PON) gene family located on the long arm of chromosome 17 consists of three members, each of which coding for three different calcium dependent esterases: PON1, PON2, and PON3 (La Du et al., 1993; Li et al., 2003; Mackness and Mackness, 2015). PON1 and PON3 are plasma HDL-associated enzymes with antioxidant activities, albeit with differences (Mackness et al., 1993). PON1 serum concentration is affected by inflammation and serum levels of oxidized-LDL (Ceron et al., 2014; Vakili et al., 2014; Mackness and Mackness, 2015). PON3 on the other hand is far less expressed and is not influenced by inflammation or oxidized lipids (Reddy et al., 2001). PON2 is an intracellular enzyme with ubiquitous expression and is thought to protect against oxidative stress (Ng et al., 2001).
In the current study we investigated PON1 in familial hypercholesteremia due to its role in diverse physiological and pathophysiological functions, including atherosclerosis and inflammatory diseases (La Du, 1996; Li et al., 2003). PON1 is associated with high density lipoprotein (HDL) in human serum and prevents oxidation of both low density lipoprotein (LDL) and HDL (Aviram et al., 1998; Durrington et al., 2001; Mackness et al., 1996). The inhibition of LDL and HDL oxidation may protect against various pathologies including cardiovascular diseases (CVD); PON1, therefore, is also considered the gene of longevity (Lescai et al., 2009, Martinelli et al., 2013). A relationship between paraoxonase 1 (PON1) genotype status, anti-oxidant, and anti-atherogenic capacity of the enzyme has been suggested previously (Mackness et al., 1993, 2002; Rosenblat et al., 2006). In addition, PON1 arylesterase/paraoxonase activities have been shown to be inversely correlated to the risk of coronary heart diseases and hypercholesterolemia (Humbert et al., 1993; Garin et al., 1997; Bryk et al., 2005).
Functional mutations in LDLR gene cause the monogenic form of familial hypercholesterolemia (FH) (Diakou et al., 2011; Ahmed et al., 2012). Recently, it was also shown that increased serum paraoxonase activity in LDLR (-/-) mice significantly inhibits progression of atherosclerosis (Leckey et al., 2010). However, the role of PON1 activity has not been studied previously in individuals with mutated LDLR. A large Pakistani family with LDLR associated FH was investigated in this study to understand the role of PON1 in the protection against atherosclerosis (Ajmal et al., 2010). In this study, we report on the role of PON1 coding sequences of single nucleotide polymorphisms (SNPs) rs662 (c.575A > G; p.Gln192Arg) and rs854560 (c.163T > A (p.Leu55Met) in relation to resultant paraoxonase and arylesterase activity in hypercholesterolemia patients with mutated LDLR. The structural and functional aspects of these SNPs have also been studied to explore how different allozymes affect and mediate the paraoxonase and arylesterase activities of the enzyme.
Subjects and Methods
Subjects
A large consanguineous Pakistani family (n=34) with LDLR mutation (c. 2416_2417InsG) presenting clinical FH was identified, including 10 patients suffering from FH and 24 healthy individuals (Ajmal et al., 2010). All subjects were screened for the presence/absence of CHD, diabetes, hypertension, malignant tumors, and acute or chronic infectious diseases. The study was approved by the Ethics Committee and Institutional Review Board of the Department of Biosciences, COMSATS Institute of Information Technology, Islamabad, Pakistan. All patients and participating healthy members of the family were informed about the study in their local language and written consent was obtained from them prior to inclusion in the study.
Samples
Blood (5 mL) was drawn after 12–14 h fasting and collected in separate tubes for DNA extraction by organic method (Helms et al., 1985) and serum separation for the determination of lipid profile and enzyme activities. For DNA extraction, blood was collected in acid citrate dextrose (ACD) vacutainer (Becton–Dickinson, Franklin Lakes, NJ) and for serum separation, blood was collected in Z Serum Sep Clot Activator vacutainer tubes (Greiner bio-one, Munich, Germany). Serum was separated from clotted blood by centrifuging the vacutainers at 3000 rpm for 10 min, at 4 °C.
Determination of PON1 SNPs
Genomic DNA was extracted from peripheral blood leukocytes using standard procedures (Helms et al., 1985). Two sets of primers were used for genotyping the polymorphisms of codon 192 and codon 55 in the PON1 gene as described by Motti et al. (2001). The primer sequences for PON1 Q192R (rs662) and PON1 L55M (rs854560) were as follows: PON1 Q192R-forward primer TTG AAT GAT ATT GTT GCT GTG GGA CCT GAG and PON1 Q192R-reverse primer CGA CCA CGC TAA ACC CAA ATA CAT CTC CCA GaA; PON1 L55M -forward primer GAG TGATGT ATA GCC CCA GTT TC and PON1 L55M-reverse primer AGT CCATTA GGC AGTATC TCC g. The reverse primers contained mismatched nucleotides as indicated by lower case letters in italics. This allowed a restriction site for HinfI (G/ANTC) to be introduced into the DNA amplification products in the presence of the polymorphisms arginine-PON1-192 or leucine-PON1-55. PON1 gene segments were amplified by PCR using 0.3 mM deoxyribonucleotide triphosphates (dNTPs), 1x PCR buffer (10 mM Tris–HCl pH 9.0, 50 mM KCl), 2.0 mM MgCl2, 0.5 mM of each primer (forward and reverse), 1.5 U of Taq Polymerase, and 50 ng of genomic DNA. The thermal cycling consisted of an initial denaturation at 95 °C for 4 min, followed by 35 cycles of amplification consisting of denaturing at 95 °C for 45 s, primer annealing at 55 °C for 1 min and chain extension at 72 °C for 1 min. A final extension step was performed at 72 °C for 10 min.
Screening of the amplified products
PCR products were purified using a DNA extraction kit (Fermentas Life Sciences, Burlington, Ontario, Canada) and subjected to restriction with Hinf1 enzyme to screen for the type of SNP in the target sequence. Digested fragments were resolved on 8% polyacrylamide gel to reveal the genotypes (Figure S1 (123.7KB, pdf) ). The results were analyzed to compare the prevalence of PON1 L55M and PON1 Q192R SNPs. To confirm the DNA sequence, amplified products were also subjected to bidirectional sequencing by the Sanger sequencing method to reveal the genotypes of all the individuals.
Measurement of PON1 paraoxanase activity
Paraoxonase activity was determined by measuring the increase in absorbance at 412 nm (Thermo Scientific GENESYS 10 UV Scanning UV/Visible Spectrophotometer) due to the formation of 4-nitrophenol (using paraoxon as substrate) (Zehra et al., 2009). The assay mixture contained 1.0 mM paraoxon and 1.0 mM CaCl2 in 50 mM glycine/NaOH buffer (pH 10.5). The amount of 4-nitrophenol liberated was calculated from the molar coefficient 18,290/Mcm.
Measurement of PON arylesterase activity
Arylesterase activity was determined using an assay mixture containing 1.0 mM phenylacetate and 0.9 mM CaCl2 in 20 mM Tris-HCl buffer (pH 8.0). The rate of hydrolysis was monitored at 270 nm. The results were calculated using extinction coefficient 1310/Mcm.
Other paramenters
The lipid profile, including total cholesterol (TC), triglycerides (TG), LDL-cholesterol (LDL-C) and HDL cholesterol (HDL-C), of all the subjects was obtained using a Roche/Hitachi automated system with commercial kits for CHOL (Cholesterol CHOD-PAP), LDL-C plus 2nd generation (LDL Cholesterol), HDL-C plus 3rd generation (HDL-Cholesterol) and TG (Triglyceride GPO-PAP) (all from Roche Diagnostics, Germany).
Statistical analysis
Results for continuous variables are reported as mean ± SD. Differences among patients and healthy family members were assessed using Chi-square (χ2) and Fisher’s Exact test. P-values less than 0.05 were considered statistically significant. Continuous variables were analyzed using ANOVA whereas categorical variables were compared by Chi-squared and Fisher’s Exact test.
Modeling the structure of PON allozymes
Structures of the four different PON1 allozymes, L55, M55, Q192 and R192, were modeled at Modeller (Eswar et al., 2006) upon an already determined structure of human PON1 at 2.2 Å resolution with an R-factor 0.217 (Harel et al., 2004a,b). All four models were energy minimized and validated for various quality factors, which included Ramachandran plot for the backbone dihedral ψ and φ, bad angles, bad bonds, steric clashes, and Z score of the model.
Assessing paraoxonase and arylesterase activities in silico
To assess the paraoxonase and arylesterase activities of PON1 allozymes Q192 and R192, in silico molecular docking was done at Autodock, developed against Paraoxon and Phenyl Acetate and retrieved from Zinc database (Irwin and Shoichet, 2005; Morris et al., 2009). The docked conformations of the ligands were ranked on the basis of binding affinity, analyzed, and assessed at Lig Plot+ and UCSF Chimera (Pettersen et al., 2004; Laskowski and Swindells, 2011). The allozymes L55 and M55 were also analyzed to explore why PON1 L55 has been reported to show high seral concentration than M55 polymorph (Bryk et al., 2005).
Results
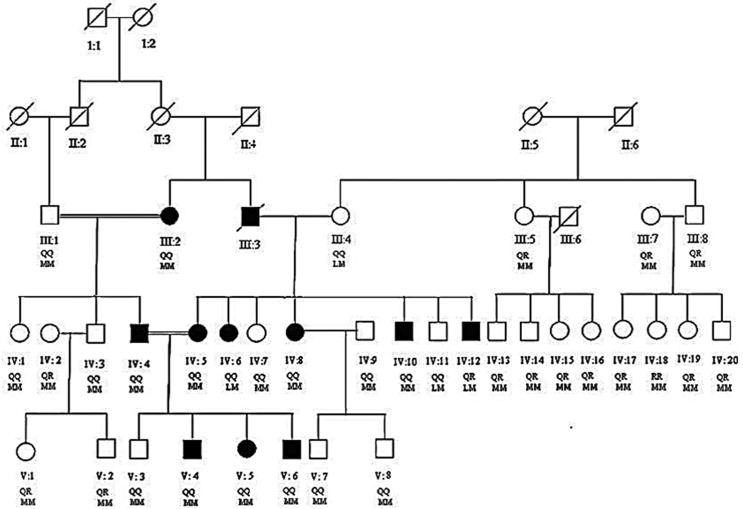
All the living individuals of the family (Figure 1) were sampled and their PON1 genotypes and phenotypes were determined. Demographic characteristics, lipid profiles, and /insparaoxonase and arylesterase activities in hypercholesterolemia patients and healthy individuals are summarized in Table 1.
Figure 1. Pedigree of hypercholesterolemia family. Filled boxes and circles represent male and female patients respectively. Empty boxes and circles represent male and female carriers respectively. Modified from Ajmal et al. (2010).
Table 1. Blood chemistry and clinical data of the affected and normal individuals of the studied family.
| Characteristics | Patients (n = 10) | Control (n = 24) | P-value |
|---|---|---|---|
| Age (Years) | 22.5 ± 16.8 | 26.5 ± 14.6 | NS |
| Male: Female | 5:5 | 12:12 | 1 |
| BMI (kg/m2) | 20 ± 3.2 | 21 ± 3.6 | NS |
| TC (mg/dL) | 422.2 ± 181.5 | 184.9 ± 32.5 | < 0.0001 |
| TG (mg/dL) | 167.6 ± 67.1 | 135.7 ± 80.1 | 0.28 |
| LDL-C (mg/dL) | 323.6 ± 149.6 | 108.5 ± 26.8 | < 0.0001 |
| HDL-C (mg/dL) | 38 ± 8.4 | 45.0 ± 11.0 | 0.084 |
| Paraoxonase activity (U/L) | 116.5 ± 40.3 | 172.5 ± 61.6 | 0.001 |
| Arylesterase activity (kU/L) | 168.3 ± 24.8 | 210.7 ± 37.9 | 0.002 |
| Xanthomas | 1 (10%) | 0 (0%) | — |
| CHD | 1 (10%) | 0 (0%) | — |
Values are given as means ± SD (p < 0.05), continuous variables were analyzed using ANOVA whereas categorical variables were compared by Chi-square. NS: not statistically significant; BMI: body mass index; TC: serum total cholesterol; TG: serum total triglyceride; HDL-C: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol; CHD: coronary heart disease risk.
Clinical data
There was no significant difference between patients and healthy individuals with respect to mean ± SD values of age, gender and body mass index (BMI). Routine laboratory findings of lipid profile indicated significant differences between the patients and healthy individuals for TC (p < 0.0001) and LDL-C (p < 0.0001) while differences between the TG (p = 0.28) and HDL-C (p = 0.084) were not significant. Paraoxonase (p = 0.001) and arylesterase activities (p = 0.002) were significantly lower in patients compared to healthy individuals of the family.
PON1 polymporphisms genotyping data
PCR amplification results and restriction fragment length polymorphism (RFLP) results of amplified products were in accordance with the specific band sizes reported previously (Motti et al., 2001). Sequencing results confirmed the PCR-RFLP based identification of SNPs. The prevalence of various genotypes (PON1 SNPs and combinations) is given in Table 2. PON1 M55 and Q192 allele prevalence was highly correlated with individuals with FH. Similarly, 80% of patients showed M55M55 genotype whereas 90% showed Q192Q192 genotype. PON1 L55 and R192 alleles were deficient in patients compared to healthy individuals of the family. Conversely, PON1 M55 and Q192 alleles were found significantly higher in patients. Similarly, the haplotype MM/QQ was prevalent in 80% of the FH patients whereas 10% of the FH patients exhibited LM/QQ and LM/QR haplotypes.
Table 2. Paraoxonase-1 allele, genotype, and haplotype distributions of L55M and Q192R polymorphisms in patients and healthy individuals of the hypercholesterolemia family.
| Characteristics (Allele/Genotypes) | Patients (n = 10) | Controls (n = 24) | p | |
|---|---|---|---|---|
| Alleles | L55 | 2 (10%) | 2 (4.2%) | 0.336 |
| M55 | 18 (90%) | 46 (95.8%) | 0.927 | |
| Q192 | 19 (95%) | 33 (68.7%) | 0.0164 | |
| R192 | 1 (5%) | 15 (62%) | 0.998 | |
| Genotypes | L55L55 | 0 (0%) | 0 (0%) | 1 |
| L55M55 | 2 (20%) | 2 (8.3%) | 0.334 | |
| M55M55 | 8 (80%) | 22 (91.7%) | 0.933 | |
| Q192Q192 | 9 (90%) | 10 (41.7%) | 0.0113 | |
| Q192R192 | 1 (10%) | 13 (54.2%) | 0.999 | |
| R192R192 | 0 (0%) | 1 (4.2%) | 1 | |
| Haplotypes | M55M55/Q192Q192 | 8 (80%) | 8 (33.3%) | 0.017 |
| L55M55/Q192Q192 | 1 (10%) | 2 (8.3%) | 0.661 | |
| L55M55/Q192R192 | 1 (10%) | 0 (0%) | 0.294 | |
| M55M55/Q192R192 | 0 (0%) | 13 (54.2%) | 1 | |
| M55M55/R192R192 | 0 (0%) | 1 (4.2%) | 1 |
p < 0.05 was considered statistically significant; data were analyzed using Fisher’s Exact Test.
Paraoxonase and arylesterase activities in silico
Both paraoxon and phenyl acetate were docked against allozymes of the PON1, and showed a higher binding affinity to PON1 R192 than Q192. The binding affinity of paraoxon to PON1 Q192 was observed to be -5.6 kcal/mol whereas for PON1 R192 the biding affinity has been significantly higher, i.e., 6.1 kcal/mol. In case of phenyl acetate (PA), the biding affinity for the allozyme R192 was observed to be 6.3 kcal/mol; however, it was 5.7 kcal/mol for Q192 (Table 3).
Table 3. Binding affinities of PON1 Q192 and R192 against paraoxon and phenyl acetate.
| PON1Allozyme | Ligand | Binding Energy (kcal/mol) |
|---|---|---|
| R192 | Paraoxon | -6.10 |
| Phenyl acetate | -6.30 | |
| Q192 | Paraoxon | -5.60 |
| Phenyl acetate | -6.00 |
The binding pocket of PON1 is perfectly designed to spatially accommodate a lactone or arylester, with its distal end distributed by highly basic residues, N168 and N224, to interact with NO2 of paraoxon or OH of phenylacetate through hydrogen bonds. This arrangement helps positioning the acyl bond in proximity to the catalytic dyad of histidine residues H115 and H134, which has been reported to deprotonate a water molecule to generate a hydroxyl ion, thereby hydroxylating the lactones (paraoxon) and arylesters (phenylacetate) (Khersonsky and Tawfik, 2006; Ben-David et al., 2012).
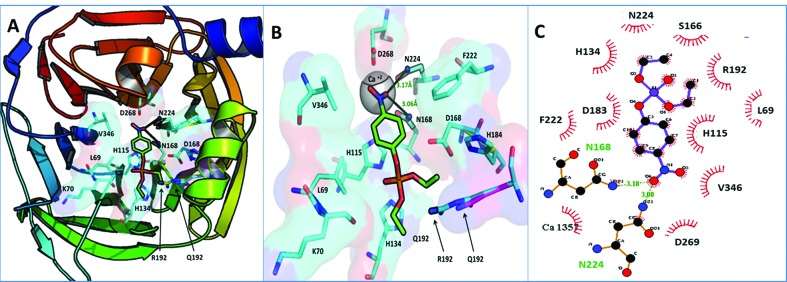
Our results suggest that position 192 lies near the inner lining of the PON1 binding pocket opening. This position is critical because of its sidechain lying closer to the oxygen atoms of acyl group, which is a positively charged basic residue and has been selected by PON1 fold for this position over the course of time. A long sidechain of R192 with highly positively charged terminalguanidinium (HNC(NH2)2) mediates with acyl oxygen atoms then a small sized lysine sidechain with just one –NH3 +, R192 thus helps in positioning the acyl bond of the substrate to the catalytic dyad of H115 and H134, thus the higher arylesterase and paraoxonase activity of R192 allozyme (Figure 2).
Figure 2. Docked conformations of paraoxon in PON1 binding pocket. (A) Paraoxon bound in PON1 binding pocket. (B) Zoomed in binding pocket of PON1, paraoxon terminally making two hydrogen bonds with N168 and N224. Notice that R192 adopts a conformation more proximal to the binding pocket opening than that of Q192, steering the ligand to adopt the most suitable conformation for paraoxonase activity by H 115 and H134 dyad. (C) 2D illustration of paraoxon binding in PON1 binding pocket; paraoxon forms two hydrogen bonds with Asn 224 (3.00 Å) and Asn 168 (3.18 Å) shown in green while a number of hydrophobic interactions with the residues configure the binding pocket.
Discussion
This is the first study of PON1 activity and region polymorphisms coding in a family with FH with LDLR mutation, although in a couple of recent studies, the role of rs662 was assessed in relation to serum lipid levels and coronary artery disease (Liang, 2016; Chen et al., 2017). PON1 genotype and activity are not being monitored in routine clinical practice for the management of hypercholesterolemia. Understanding a possibly protective role of PON1 activity in patients susceptible to atherosclerosis can help in taking early preventive measures for improvement of life span of hypercholesterolemia patients.
The contribution of PON1 in CVD is minor in healthy populations but it is known that genotypes with minor effects in general population may have more pronounced effects in patients, for example in FH cases (Leus et al., 2001; Wiegman et al., 2004). The beneficial effects of PON1 on the inhibition of atherosclerosis might be more pronounced in FH patients because they are more prone to develop atherosclerosis than the general population (van Himbergen et al., 2005).
Low PON1 activity has been reported in previous studies as one of the leading factors causing atherosclerosis and myocardial infarction (Ayub et al., 1999; James et al., 2000; Mackness et al., 2002; van Himbergen et al., 2005; Bryk et al., 2005; Gur et al., 2006). Numerous clinical studies have shown an association of low PON1 activity with atherosclerosis and cardiovascular diseases (Jarvik et al., 2000; Mackness et al., 2003; Gur et al., 2006; Rosenblat et al., 2006; Soran et al., 2008; Sun et al., 2016; Verit et al., 2008). Thus, significantly decreased paraoxonase and arylesterase activities in patients compared to the healthy individuals in the present study, indicate an increased risk of atherosclerosis in patients. One of the patients (IV-4) in the current family had a history of CVD in addition to FH. He had premature coronary artery disease and had suffered from myocardial infarction at an early age, which may have been due to decreased paraoxonase and arylesterase activity. The lipid profile of this 37-year-old male was not considerably elevated compared to other patients in this family.
Patient V-6 was identified with xanthomas in addition to FH, but without any history of CVD. However, his levels of TC (917 mg/dL) and LDLC (728 mg/dL) were markedly high and HDL-C (22 mg/dL) was lower compared to other FH patients in the family. The presence of tendon xanthomas is high risk factor of CVD among patients with FH, which along with decreased paraoxonase and arylesterase activities may lead to atherosclerosis (Jarvik et al., 2000; Mackness et al., 2003; Gur et al., 2006; Rosenblat et al., 2006; Soran et al., 2008; Sun et al., 2016; Verit et al., 2008).
The low levels of serum paraoxonase and arylesterase activities of patients in this study may be due to their genetic makeup (PON1 coding region SNPs). The L55M polymorphism affects the enzyme concentration (plasma PON1 protein levels indicated by serum arylesterase activity), whereas the Q192R polymorphism affects the catalytic efficiency (serum paraoxonase activity), but not the concentration (Bryk et al., 2005). The PON1 M55 is associated with low plasma PON1 level (Blatter et al., 1993; Mackness et al., 1998a,b). The PON1 R192 allozyme hydrolyzes paraoxon more readily than Q192 (Costa et al., 2005). In our study, the high prevalence of M55 allele in the patients may have been responsible for the low levels of PON1 activity (Aviram et al., 2000). The frequency of the low-activity allele (Q192) and unstable form (M55 isoform, which is sensitive to proteolysis) was collectively higher in patients of this family indicated by high prevalence of MMQQ genotype (8, 80%), which may significantly reduce the function of PON1 for protection against atherosclerosis. The generally low frequency of the M55 is such that in some studies the MM genotype is not observed at all (Santos et al., 2005). M is not favored in human populations, but it is kept in populations in heterozygous individuals. The L55M mutation may considerably affect PON1’s stability and thereby account for the lower enzymatic activity because M55 isoform is an unstable form (sensitive to proteolysis) (Harel et al., 2004a,b; Mackness et al., 1998a).
Indeed, PON1 phenotype (paraoxonase and arylesterase activities) has been shown previously to be a better predictor of vascular disease than PON1 Q192R or PON1 L55M genotypes due to possible effects of other genetic and non-genetic factors (Costa et al., 2005; Jarvik et al., 2000; Rosenblat et al., 2011). Therefore, upregulation of PON1 levels by non-genetic factors (Rosenblat et al., 2011) can have a potential advantage in such cases for better and achievable protection against atherosclerosis. Thus, our results combined with previously published data indicate the need of regular monitoring and upregulation of serum paraoxonase activity in patients with hypercholesterolemia for prevention of atherosclerosis. Our results highlight the importance of exploring PON1 as a therapeutic agent to accommodate the lower level of plasma PON1 in patients susceptible to atherosclerosis.
In the current study, the LDLR mutated patients had low level of PON1, which may indicate some gene-regulation action of LDLR and PON1, thus needing further investigations. With the current data, it is difficult to speculate how LDLR and PON1 are involved in regulating each other.
Both the hypercholesterolemia and PON1 deficiency are independent risk factors for the development of atherosclerosis. In addition to controlling high levels of cholesterol, there is a need to regularly monitor PON1 status of hypercholesterolemia patients and normal family members. A better understanding of factors upregulating PON1 status in humans will have a significant public health impact by saving patients who are otherwise susceptible to atherosclerosis due to their deficient PON1 status. In the current study, controls from the general population were not screened, and this is a limitation of the study. However, based on the current findings, future studies can be designed to screen the population for PON1 and further investigate its role in cardiovascular diseases.
Conclusion
This study aimed at exploring the implications of PON1 polymorphism in the individuals affected by familial hypercholesterolemia (FH). The role of PON1 and its various polymorphs has not been studied previously in FH subjects. This work sought to investigate paraoxonase and arylesterase activity of various PON1 SNPs in individuals with mutated LDLR, thereby explore their role in the development of FH. The results suggest that most of the hypercholesterolemia patients with LDLR mutation have homozygous M55 and Q192 PON1 genotype, thus combination of MMQQ PON1 genotype and LDLR mutation might lead to a more severe disease outcome in the form of a fatal heart failure. Further studies are needed to explore the role of LDLR and PON1 pathways in the onset of hypercholesterolemia and atherosclerosis.
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Mara H. Hutz
References
- Ahmed W, Ajmal M, Sadeque A, Whittall RA, Rafiq S, Putt W, Khawaja A, Imtiaz F, Ahmed N, Azam M, et al. Novel and recurrent LDLR gene mutations in Pakistani hypercholesterolemia patients. Mol Biol Rep. 2012;39:7365–7372. doi: 10.1007/s11033-012-1568-1. [DOI] [PubMed] [Google Scholar]
- Ajmal M, Ahmed W, Sadeque A, Ali SH, Bokhari SH, Ahmed N, Qamar R. Identification of a recurrent insertion mutation in the LDLR gene in a Pakistani family with autosomal dominant hypercholesterolemia. Mol Biol Rep. 2010;37:3869–3875. doi: 10.1007/s11033-010-0043-0. [DOI] [PubMed] [Google Scholar]
- Aviram M, Rosenblat M, Bisgaier CL, Newton RS, Primo-Parmo SL, La Du BN. Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. J Clin Invest. 1998;101:1581–1590. doi: 10.1172/JCI1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviram M, Hardak E, Vaya J, Mahmood S, Milo S, Hoffman A, Billicke S, Draganov D, Rosenblat M. Human serum paraoxonases (PON1) Q and R selectively decrease lipid peroxides in human coronary and carotid atherosclerotic lesions: PON1 esterase and peroxidase-like activities. Circulation. 2000;101:2510–2517. doi: 10.1161/01.cir.101.21.2510. [DOI] [PubMed] [Google Scholar]
- Ayub A, Mackness MI, Arrol S, Mackness B, Patel J, Durrington PN. Serum paraoxonase after myocardial infarction. Arterioscler Thromb Vasc Biol. 1999;19:330–335. doi: 10.1161/01.atv.19.2.330. [DOI] [PubMed] [Google Scholar]
- Ben-David M, Elias M, Filippi JJ, Dunach E, Silman I, Sussman JL, Tawfik DS. Catalytic versatility and backups in enzyme active sites: The case of serum paraoxonase 1. J Mol Biol. 2012;418:181–196. doi: 10.1016/j.jmb.2012.02.042. [DOI] [PubMed] [Google Scholar]
- Blatter MC, James RW, Messmer S, Barja F, Pometta D. Identification of a distinct human high-density lipoprotein subspecies defined by a lipoprotein-associated protein, K-45. Identity of K-45 with paraoxonase. Eur J Biochem. 1993;211:871–879. doi: 10.1111/j.1432-1033.1993.tb17620.x. [DOI] [PubMed] [Google Scholar]
- Bryk B, BenMoyal-Segal L, Podoly E, Livnah O, Eisenkraft A, Luria S, Cohen A, Yehezkelli Y, Hourvitz A, Soreq H. Inherited and acquired interactions between ACHE and PON1 polymorphisms modulate plasma acetylcholinesterase and paraoxonase activities. J Neurochem. 2005;92:1216–1227. doi: 10.1111/j.1471-4159.2004.02959.x. [DOI] [PubMed] [Google Scholar]
- Ceron JJ, Tecles F, Tvarijonaviciute A. Serum paraoxonase 1 (PON1) measurement: An update. BMC Vet Res. 2014;10:74. doi: 10.1186/1746-6148-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ding S, Zhou M, Wu X, Liu X, Liu J, Wu Y, Liu D. PON1 L55M and Q192R gene polymorphisms and CAD risks in patients with hyperlipidemia. Herz. 2017 doi: 10.1007/s00059-017-4611-0. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Costa LG, Cole TB, Furlong CE. Paraoxonase (PON1): From toxicology to cardiovascular medicine. Acta Biomed. 2005;76:50–57. [PubMed] [Google Scholar]
- Diakou M, Miltiadous G, Xenophontos SL, Manoli P, Cariolou MA, Elisaf M. Spectrum of LDLR gene mutations, including a novel mutation causing familial hypercholesterolaemia, in North-western Greece. Eur J Intern Med. 2011;22:e55–59. doi: 10.1016/j.ejim.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Durrington PN, Mackness B, Mackness MI. Paraoxonase and atherosclerosis. Arterioscler Thromb Vasc Biol. 2001;21:473–480. doi: 10.1161/01.atv.21.4.473. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M-Y, Pieper U, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2006;5:5.6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin MC, James RW, Dussoix P, Blanche H, Passa P, Froguel P, Ruiz J. Paraoxonase polymorphism Met-Leu54 is associated with modified serum concentrations of the enzyme. A possible link between the paraoxonase gene and increased risk of cardiovascular disease in diabetes. J Clin Invest. 1997;99:62–66. doi: 10.1172/JCI119134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur M, Aslan M, Yildiz A, Demirbag R, Yilmaz R, Selek S, Erel O, Ozdogru I. Paraoxonase and arylesterase activities in coronary artery disease. Eur J Clin Invest. 2006;36:779–787. doi: 10.1111/j.1365-2362.2006.01727.x. [DOI] [PubMed] [Google Scholar]
- Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RB, McCarthy A, Toker L, et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004a;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RBG, McCarthy A, Toker L, et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol, 2004b;11:1253–1253. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- Helms C, Graham MY, Dutchik JE, Olson MV. A new method for purifying lambda DNA from phage lysates. DNA. 1985;4:39–49. doi: 10.1089/dna.1985.4.39. [DOI] [PubMed] [Google Scholar]
- Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet. 1993;3:73–76. doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK. ZINC - A free database of commercially available compounds for virtual screening. J Chem Inf Model, 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RW, Leviev I, Righetti A. Smoking is associated with reduced serum paraoxonase activity and concentration in patients with coronary artery disease. Circulation. 2000;101:2252–2257. doi: 10.1161/01.cir.101.19.2252. [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Rozek LS, Brophy VH, Hatsukami TS, Richter RJ, Schellenberg GD, Furlong CE. Paraoxonase (PON1) phenotype is a better predictor of vascular disease than is PON1(192) or PON1(55) genotype. Arterioscler Thromb Vasc Biol. 2000;20:2441–2447. doi: 10.1161/01.atv.20.11.2441. [DOI] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS. The histidine 115-histidine 134 dyad mediates the lactonase activity of mammalian serum paraoxonases. J Biol Chem. 2006;281:7649–7656. doi: 10.1074/jbc.M512594200. [DOI] [PubMed] [Google Scholar]
- La Du BN. Structural and functional diversity of paraoxonases. Nat Med. 1996;2:1186–1187. doi: 10.1038/nm1196-1186. [DOI] [PubMed] [Google Scholar]
- La Du BN, Adkins S, Kuo CL, Lipsig D. Studies on human serum paraoxonase/arylesterase. Chemico-Biol interact. 1993;87:25–34. doi: 10.1016/0009-2797(93)90022-q. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Swindells MB. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inform Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Leckey LC, Garige M, Varatharajalu R, Gong M, Nagata T, Spurney CF, Lakshman RM. Quercetin and ethanol attenuate the progression of atherosclerotic plaques with concomitant up regulation of paraoxonase1 (PON1) gene expression and PON1 activity in LDLR-/- mice. Alcohol Clin Exp Res. 2010;34:1535–1542. doi: 10.1111/j.1530-0277.2010.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescai F, Marchegiani F, Franceschi C. PON1 is a longevity gene: Results of a meta-analysis. Ageing Res Rev. 2009;8:277–284. doi: 10.1016/j.arr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Leus FR, Zwart M, Kastelein JJ, Voorbij HA. PON2 gene variants are associated with clinical manifestations of cardiovascular disease in familial hypercholesterolemia patients. Atherosclerosis, 2001;154:641–649. doi: 10.1016/s0021-9150(00)00440-8. [DOI] [PubMed] [Google Scholar]
- Li HL, Liu DP, Liang CC. Paraoxonase gene polymorphisms, oxidative stress, and diseases. J Mol Med. 2003;81:766–779. doi: 10.1007/s00109-003-0481-4. [DOI] [PubMed] [Google Scholar]
- Liang BG. Paraoxonase-1 (PON1) rs662 polymorphism and its association with serum lipid levels and longevity in the Bama Zhuang population. Med Sci Monit. 2016;22:5154–5162. doi: 10.12659/MSM.898231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackness M, Mackness B. Human paraoxonase-1 (PON1): Gene structure and expression, promiscuous activities and multiple physiological roles. Gene. 2015;567:12–21. doi: 10.1016/j.gene.2015.04.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackness MI, Arrol S, Abbott C, Durrington PN. Protection of low-density lipoprotein against oxidative modification by high-density lipoprotein associated paraoxonase. Atherosclerosis. 1993;104:129–135. doi: 10.1016/0021-9150(93)90183-u. [DOI] [PubMed] [Google Scholar]
- Mackness MI, Mackness B, Durrington PN, Connelly PW, Hegele RA. Paraoxonase: Biochemistry, genetics and relationship to plasma lipoproteins. Curr Opin Lipidol. 1996;7:69–76. doi: 10.1097/00041433-199604000-00004. [DOI] [PubMed] [Google Scholar]
- Mackness B, Mackness MI, Arrol S, Turkie W, Julier K, Abuasha B, Miller JE, Boulton AJ, Durrington PN. Serum paraoxonase (PON1) 55 and 192 polymorphism and paraoxonase activity and concentration in non-insulin dependent diabetes mellitus. Atherosclerosis. 1998a;139:341–349. doi: 10.1016/s0021-9150(98)00095-1. [DOI] [PubMed] [Google Scholar]
- Mackness MI, Mackness B, Durrington PN, Fogelman AM, Berliner J, Lusis AJ, Navab M, Shih D, Fonarow GC. Paraoxonase and coronary heart disease. Curr Opin Lipidol. 1998b;9:319–324. doi: 10.1097/00041433-199808000-00006. [DOI] [PubMed] [Google Scholar]
- Mackness B, Durrington PN, Boulton AJ, Hine D, Mackness MI. Serum paraoxonase activity in patients with type 1 diabetes compared to healthy controls. Eur J Clin Invest. 2002;32:259–264. doi: 10.1046/j.1365-2362.2002.00977.x. [DOI] [PubMed] [Google Scholar]
- Mackness B, Durrington P, McElduff P, Yarnell J, Azam N, Watt M, Mackness M. Low paraoxonase activity predicts coronary events in the Caerphilly Prospective Study. Circulation. 2003;107:2775–2779. doi: 10.1161/01.CIR.0000070954.00271.13. [DOI] [PubMed] [Google Scholar]
- Martinelli N, Consoli L, Girelli D, Grison E, Corrocher R, Olivieri O. Paraoxonases: Ancient substrate hunters and their evolving role in ischemic heart disease. Adv Clin Chem. 2013;59:65–100. doi: 10.1016/b978-0-12-405211-6.00003-6. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motti C, Dessi M, Gnasso A, Irace C, Indigeno P, Angelucci CB, Bernardini S, Fucci G, Federici G, Cortese C. A multiplex PCR-based DNA assay for the detection of paraoxonase gene cluster polymorphisms. Atherosclerosis. 2001;158:35–40. doi: 10.1016/s0021-9150(00)00765-6. [DOI] [PubMed] [Google Scholar]
- Ng CJ, Wadleigh DJ, Gangopadhyay A, Hama S, Grijalva VR, Navab M, Fogelman AM, Reddy ST. Paraoxonase-2 is a ubiquitously expressed protein with antioxidant properties and is capable of preventing cell-mediated oxidative modification of low density lipoprotein. J Biol Chem. 2001;276:44444–44449. doi: 10.1074/jbc.M105660200. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Reddy ST, Wadleigh DJ, Grijalva V, Ng C, Hama S, Gangopadhyay A, Shih DM, Lusis AJ, Navab M, Fogelman AM. Human paraoxonase-3 is an HDL-associated enzyme with biological activity similar to paraoxonase-1 protein but is not regulated by oxidized lipids. Arterioscler Thromb Vasc Biol. 2001;21:542–547. doi: 10.1161/01.atv.21.4.542. [DOI] [PubMed] [Google Scholar]
- Rosenblat M, Karry R, Aviram M. Paraoxonase 1 (PON1) is a more potent antioxidant and stimulant of macrophage cholesterol efflux, when present in HDL than in lipoprotein-deficient serum: relevance to diabetes. Atherosclerosis. 2006;187:74–81. doi: 10.1016/j.atherosclerosis.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Rosenblat M, Volkova N, Aviram M. Injection of paraoxonase 1 (PON1) to mice stimulates their HDL and macrophage antiatherogenicity. BioFactors. 2011;37:462–467. doi: 10.1002/biof.188. [DOI] [PubMed] [Google Scholar]
- Soran N, Altindag O, Cakir H, Celik H, Demirkol A, Aksoy N. Assessment of paraoxonase activities in patients with knee osteoarthritis. Redox Rep. 2008;13:194–198. doi: 10.1179/135100008X308911. [DOI] [PubMed] [Google Scholar]
- Sun T, Hu J, Yin Z, Xu Z, Zhang L, Fan L, Zhuo Y, Wang C. Low serum paraoxonase1 activity levels predict coronary artery disease severity. Oncotarget. 2016;8:19553–19454. doi: 10.18632/oncotarget.14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakili L, Navab KD, Shabihkhani M, Pourtabatabaei N, Vazirian S, Barseghian Z, Seyedali S, Hough G. Systemic inflammation, intestine, and paraoxonase-1. Adv Exp Med Biol. 2014;824:83–88. doi: 10.1007/978-3-319-07320-0_8. [DOI] [PubMed] [Google Scholar]
- van Himbergen TM, Roest M, de Graaf J, Jansen EH, Hattori H, Kastelein JJ, Voorbij HA, Stalenhoef AF, van Tits LJ. Indications that paraoxonase-1 contributes to plasma high density lipoprotein levels in familial hypercholesterolemia. J Lipid Res. 2005;46:445–451. doi: 10.1194/jlr.M400052-JLR200. [DOI] [PubMed] [Google Scholar]
- Verit FF, Erel O, Celik H. Paraoxonase-1 activity in patients with hyperemesis gravidarum. Redox Rep. 2008;13:134–138. doi: 10.1179/135100008X259259. [DOI] [PubMed] [Google Scholar]
- Wiegman A, de Groot E, Hutten BA, Rodenburg J, Gort J, Bakker HD, Sijbrands EJ, Kastelein JJ. Arterial intima-media thickness in children heterozygous for familial hypercholesterolaemia. Lancet. 2004;363:369–370. doi: 10.1016/S0140-6736(04)15467-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.