Abstract
Ubiquitin (Ub) has important roles in a wide range of intracellular signalling pathways. In the conventional view, ubiquitin alters the signalling activity of the target protein through covalent modification, but accumulating evidence points to the emerging role of non-covalent interaction between ubiquitin and the target. In the innate immune signalling pathway of a viral RNA sensor, RIG-I, both covalent and non-covalent interactions with K63-linked ubiquitin chains (K63-Ubn) were shown to occur in its signalling domain, a tandem caspase activation and recruitment domain (hereafter referred to as 2CARD)1,2. Non-covalent binding of K63-Ubn to 2CARD induces its tetramer formation, a requirement for downstream signal activation3. Here we report the crystal structure of the tetramer of human RIG-I 2CARD bound by three chains of K63-Ub2. 2CARD assembles into a helical tetramer resembling a ‘lock-washer’, in which the tetrameric surface serves as a signalling platform for recruitment and activation of the downstream signalling molecule, MAVS. Ubiquitin chains are bound along the outer rim of the helical trajectory, bridging adjacent subunits of 2CARD and stabilizing the 2CARD tetramer. The combination of structural and functional analyses reveals that binding avidity dictates the K63-linkage and chain-length specificity of 2CARD, and that covalent ubiquitin conjugation of 2CARD further stabilizes the Ub–2CARD interaction and thus the 2CARD tetramer. Our work provides unique insights into the novel types of ubiquitin-mediated signal-activation mechanism, and previously unexpected synergism between the covalent and non-covalent ubiquitin interaction modes.
Effective immune defence against viral infection is dependent upon efficient detection of pathogens by innate immune receptors. One such receptor is RIG-I, which recognizes RNAs from a broad range of viruses and activates the type I interferon (IFN) signalling pathway4. RIG-I consists of an amino (N)-terminal 2CARD, which is important for signal activation, and a central DExH/C motif helicase domain and carboxy (C)-terminal domain (CTD), which together function as a viral RNA recognition unit (Fig. 1a). The crystal structure of RNA-free, full-length RIG-I5 showed that 2CARD forms an intramolecular interaction with the helicase domain. Upon RNA binding, 2CARD was proposed to be released from the helicase domain5–7, which would allow its interaction with the N-terminal CARD of the downstream signalling adaptor molecule, MAVS8–10. Upon interaction with RIG-I 2CARD, MAVS CARD forms a self-perpetuating filament, which then recruits TRAF2,3 and 6 to activate downstream IFN-α/β signalling pathways11.
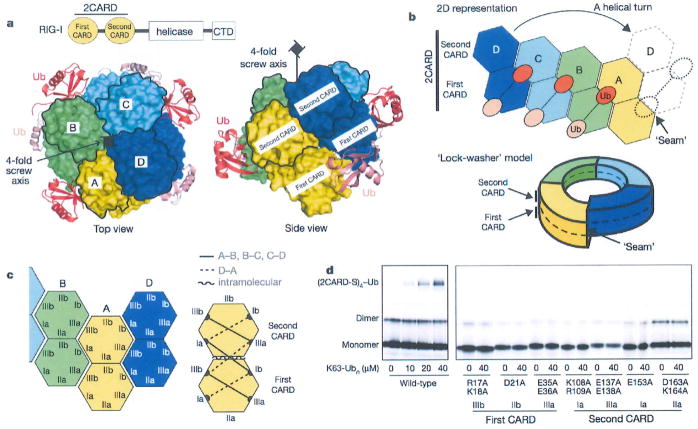
Figure 1. RIG-I 2CARD assembles into a helical tetramer bridged by three copies of K63-Ub2.
a, Domain architecture of RIG-I and top and side views of the 2CARD tetramer (chains A–D, surface representation) bound by Ub chains (cartoon representation). The second CARDs are demarcated by black lines, b, Two-dimensional (2D) representation and ‘lock-washer model of the RIG-I 2CARD tetramer, using the same colour code as for a. c, Definition of the surface types in the first and second CARDs. Six types of intermolecular interactions were observed; three in the A–B, B–C and C–D interactions (solid lines on the right), and another three in the D–A interaction (dotted lines). A single type of intramolecular interaction was observed between IIb of the first CARD and IIa of the second CARD (curved line). See Extended Data Fig. 4. d, Tetramer formation of wild-type and mutant 2CARD fused to an Alexa647-labelled SNAP tag (2CARD-S) with and without K63-Ubn (n > 8) as measured by EMSA. Mutations are on the indicated surface as defined in c.
Accumulating evidence suggests that a simple release of 2CARD is insufficient for the signal activation by RIG-I. It was shown that modification of RIG-I 2CARD with K63-Ubn by the E3 ligase TRIM25 is important for signalling activity of isolated 2CARD2. Subsequent studies showed that unanchored K63-Ubn can non-covalently bind 2CARD to promote its tetramerization and concomitant signal activation1,3. Recently, we and others have shown that RIG-I forms a filament along double-stranded RNA (dsRNA) using its helicase and CTD12,13, and that the resultant proximity can promote 2CARD oligomerization and stimulate MAVS in the absence of K63-Ubn12. This differs from isolated 2CARD, which strictly requires K63-Ubn for oligomerization and signal activation1,3 (Extended Data Fig. 1b). These studies suggest the presence of at least three mechanisms, Ub-conjugation, Ub-binding and filament formation, and more complex signal activation process for RIG-I.
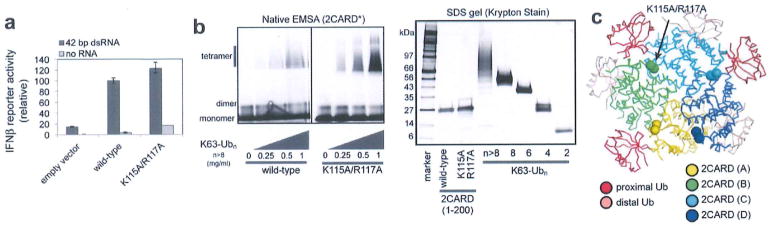
To understand more precisely the roles of covalent and non-covalent interactions with Ub, we have crystallized human RIG-I 2CARD in complex with K63-Ub2, a minimal chain length required for tetramerization of 2CARD1. Our initial crystallization trials included wild-type and 2CARD surface mutants, but crystals were obtained only with the double mutant, K115A/R117A, which retains the wild type’s ability to tetramerize with K63-Ubn and activate the IFN-β pathway (Extended Data Fig. 1a, b). The structure was determined by molecular replacement and was refined to 3.7Å with Rwork of 22.2% and Rfree of 28.5% (Extended Data Table 1). The structure revealed the tetrameric architecture of 2CARD bound with six Ub molecules in the asymmetric unit (Fig. 1a, b and Extended Data Fig. 2a). Despite poor electron density for the K63-linkage, SDS–polyacrylamide gel electrophoresis (SDS–PAGE) analysis of the crystal revealed that K63-Ub2 was intact (Extended Data Fig. 2e). Analysis of the distance between the C terminus and K63 of adjacent Ubs unambiguously indicated the three Ub pairs connected through the K63 linkage (Extended Data Fig. 2f).
Extended Data Table 1.
Data collection and refinement statistics
| Crystal 1 | |
|---|---|
| Data collection | |
| Space group | P1 21 1 |
| Cell dimensions | |
| a, b, c(Å) | 83.5, 101.9, 88.2 |
| α, β, γ (°) | 90.0, 106.9, 90.0 |
| Resolution (Å) | 50-3.7 (3 .93-3 .70)* |
| Rsvm or Rmerge | 0.165 (0 .621) |
| I / σI | 5.92 (1.55) |
| Completeness(%) | 98.1 (96.7) |
| Redundancy | 4.0 (4.0) |
| Refinement | |
| Resolution (Å) | 46.12-3.70 (3 .82-3.70) |
| No. reflections | 29885 (2697) |
| Rwork / Rfree | 0.222 (0.333) / 0.285 (0.401) |
| No. atoms | |
| Protein | 19179 |
| Ligand/ion | |
| Water | |
| B-factors | |
| Protein | 92.3 |
| Ligand/ion | |
| Water | |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.6 |
Values in parentheses are for highest-resolution shell.
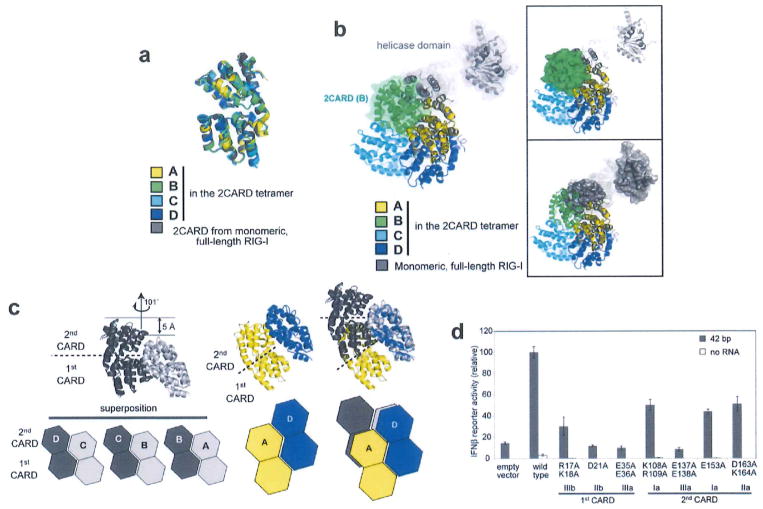
The RIG-I 2CARD tetramer is formed by a rigid-body docking of monomeric 2CARD. The four 2CARD subunits (A–D) display a conformation that is indistinguishable from 2CARD in the monomeric RIG-I5 (Extended Data Fig. 3a) in terms of both individual CARD structures and the relative orientation of the first and second CARDs. Superposition of the full-length RIGT onto one of the 2CARD subunits in the tetramer showed that a part of the 2CARD–2CARD interface is directly masked by the helicase domain in the full-length RIG-I (Extended Data Fig. 3b), indicating that blockade of the tetramerization surface mediates auto-repression of RNA-free RIG-I.
The RIG-I 2CARD tetramer exhibits a helical assembly, in which the adjacent 2CARD subunits (A–B, B–C and C–D) are related by ~101° rotation and ~5Å rise along the central screw axis (Extended Data Fig. 3c). The interaction between subunits D and A at the helical ‘seam’ (Fig. 1b) differs from the A–B, B–C and C–D interactions by displacement of subunit A by a single CARD (Extended Data Fig. 3c). Consequently, the 2CARD tetramer can be viewed as a ‘lock washer’ with the two ends displaced by half the thickness of the ring (Fig. 1b). Alternatively, it can be viewed as two continuous helical turns of a single CARD, first with four copies of the first CARD followed by four copies of the second CARD. The helical assembly of 2CARD cannot extend beyond the tetramer as the assembly unit is tandem CARD domains, in which the first and second CARDs are rigidly joined through tight intramolecular interactions.
CARD belongs to the death domain (DD) superfamily, members of which share little sequence similarity, but have a common three-dimensional fold14. The helical assembly of 2CARD is reminiscent of those of DD oligomers, such as the Myddosome15, PIDDosome16 and FAS–FADD complex17. Assembly of DD oligomers is commonly mediated by six surface areas (Ia/b, IIa/b and IIIa/b) forming three types of intermolecular interactions (Ia–Ib, IIa–IIb and IIIa–IIIb)14. Similarly, RIG-I 2CARD uses the analogous surface areas of both the first and second CARDs for the intramolecular (IIa–IIb) and intermolecular interactions (Ia–Ib and IIIa–IIIb) in the tetramer (Fig. 1c). The six types of Ia–Ib and IIIa–IIIb interactions were observed; three (solid lines in Fig. 1c) between adjacent subunits along the helical trajectory (A–B, B–C and C–D), and the other three (dotted lines) at the helical seam between subunits D and A. Mutation of residues involved in the inter- or intramolecular interactions impaired the ability of 2CARD to tetramerize with K63-Ubn (as measured by electrophoretic mobility shift assay (EMSA) using 2CARD fused to a fluorescent tag, SNAP12, Fig. 1d), and to activate the IFN-β signalling pathway in cells (Extended Data Fig. 3d). This result suggests the importance of inter-CARD interactions in formation of the signalling competent RIG-I 2CARD tetramer.
We next asked how this tetrameric, but not monomeric, 2CARD can activate downstream signalling pathway, that is, MAVS CARD filament formation. In the case of Myddosome, PIDDosome, and FAS–FADD complex, the helical oligomeric structure of upstream signalling molecules serves as a scaffold to assemble the downstream DD oligomers through helical extension14. We speculated that RIG-I 2CARD may use a similar mechanism to assemble the MAVS CARD filament (Fig. 2a). According to this model, the bottom surface of the first CARD or the top of the second CARD would serve as a platform to recruit MAVS CARD, and the resultant MAVS CARD filament would use the same type of molecular interactions (Ia–Ib, IIa–IIb and IIIa–IIIIb) as in the 2CARD tetramer (Fig. 2a). We found that mutations of surface IIb (top) of the second CARD, but not IIa (bottom) of the first CARD, significantly reduced the signalling activity of RIG-I (Fig. 2c), suggesting that the top of the second CARD may be involved in MAVS recruitment. Consistently, mutations in IIb of the second CARD completely abrogated its ability to stimulate MAVS CARD filament formation without affecting its tetramer formation of 2CARD (Fig. 2d, e), suggesting that the top surface of the second CARD serves as the primary site for MAVS recruitment. In addition, both MAVS CARD filament assembly and cellular signalling activity was significantly affected by mutations in the equivalent surface areas, Ia/b–IIIa/b (Fig. 2f, g), further supporting our ‘helix-extension’ model of MAVS activation by the RIG-I 2CARD tetramer. Although the helical symmetry of RIG-I 2CARD (that is, a pitch in the screw equivalent to the height of a single CARD) prevents helical assembly of RIG-I tandem CARDs beyond the tetramer, it is compatible with filament extension of MAVS as it only has a single CARD.
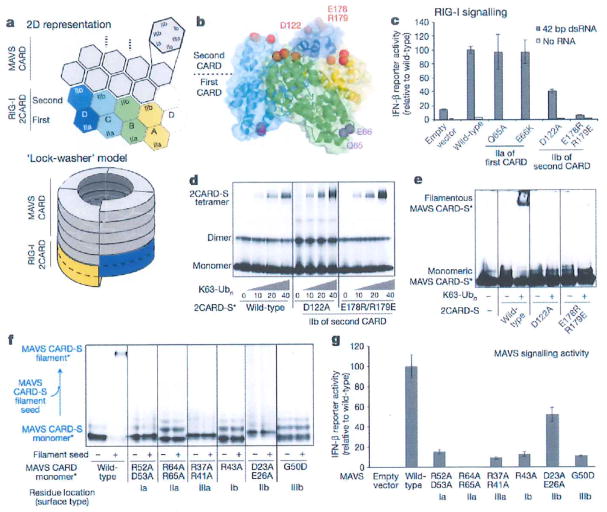
Figure 2. The helical tetramer of RIG-I 2CARD uses the top surface of the second CARD to interact with MAVS CARD and promote MAVS filament formation.
a, A ‘helical extension’ model for how the RIG-I 2CARD tetramer stimulates the MAVS CARD (grey) filament formation. b, D122 and E178/R179 are located at the top of the 2CARD tetramer (surface IIb of the second CARD), whereas Q65 and E66 are at the bottom of the 2CARD tetramer (surface IIa of the first CARD), c, IFN-β signalling activity of mutants with altered surface IIa or IIb of the first and second CARDs (mean ± s.d., n = 3). d, e, 2CARD tetramerization (d) and MAVS filament stimulation (e) of wild-type RIG-I 2CARD and its mutants. The SNAP-fusion allowed fluorescent labelling (indicated with an asterisk) of RIG-I 2CARD and MAVS CARD (2CARD-S and CARD-S, respectively), f, g, Filament formation (f) and IFN-β signalling activity (g) (mean ± s.d., n = 3) of wild-type MAVS and its mutants with altered surface I–IIIa/b.
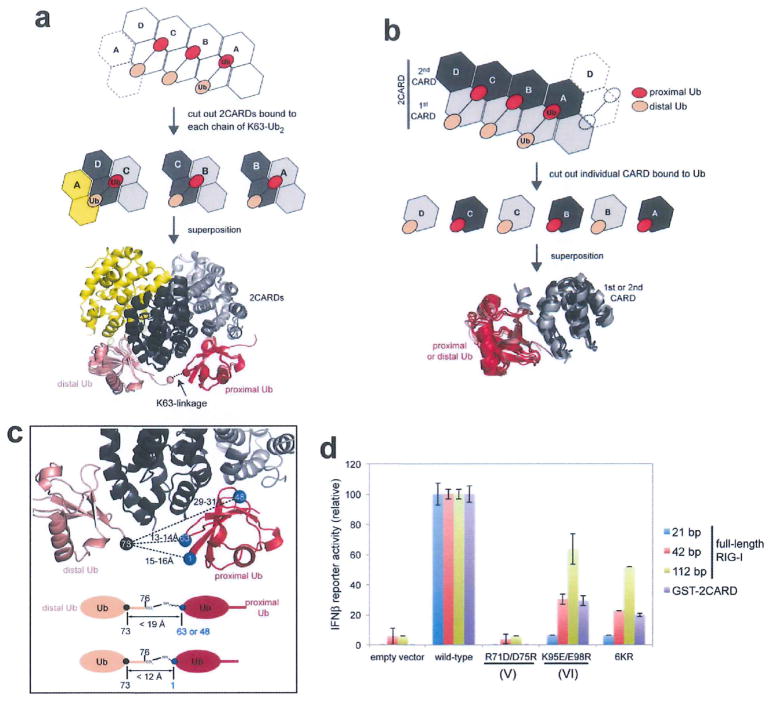
Unlike the DD oligomers, tetramer formation of isolated 2CARD requires K63-Ubn. In our structure, three chains of K63-Ub2 were bound along the outer rim of the helical trajectory (Fig. 3a). Each chain interacts with two adjacent 2CARDs in the same manner (Extended Data Fig. 5a). There are two types of interactions between Ub and 2CARD; the ‘proximal’ Ub fits into the groove between the adjacent 2CARDs, forming composite interactions using two distinct surface patches of Ub (L8/I44/V70 and F45/A46/N60/Q62) (Fig. 3a, b). These two patches of proximal Ub contact surface IV and V of 2CARD, respectively. By contrast, the ‘distal’ Ub interacts with only one 2CARD (surface VI) using the L8/I44/V70 surface patch (Fig. 3a, b). One exception is the distal Ub bound at the helical seam, which interacts with both subunits D (surface VI) and A (surface V). Note that the location of the surface IV in the second CARD is equivalent to that of VI in the first CARD (Extended Data Fig. 5b). Mutations in any one of the three surface areas (IV, V and VI) in 2CARD impaired its ability to tetramerize upon addition of free K63-Ubn (Fig. 3c). However, the cellular signalling activity of RIG-I 2CARD (fused to glutathione S-transferase (GST)) was impaired by mutations of surface V and VI, but not IV (Fig. 3d), suggesting that the 2CARD(IV)–Ub interaction is compensated by other factors in cells, such as covalent conjugation of Ub near IV (explained further in Fig. 4).
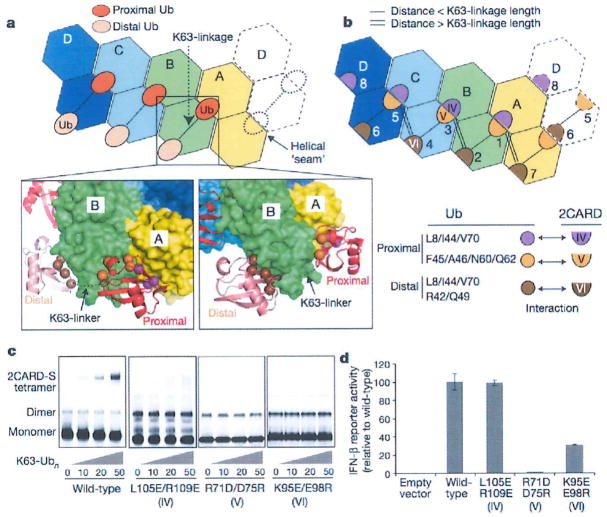
Figure 3. Ub binds to the 2CARD tetramer by decorating the outer rim of the helical trajectory.
a, Two types of Ub–2CARD interactions. Proximal Ub forms a composite interaction with two adjacent 2CARDs using two surface areas (purple and orange spheres). Distal Ub interacts with only one 2CARD using one of the two surface areas (brown spheres), with the exception of Ub bound to D. Dotted lines represent disordered K63-linkers between proximal and distal Ubs. See Extended Data Fig. 6. b, Three types of 2CARD surface areas (IV, V and VI) are involved in Ub binding. Surface IV and VI interact with L8/I44/V70 in proximal and distal Ubs, respectively, whereas surface V interacts with F45/A46/N60/N62 of proximal Ub. Mapping of IV–VI onto the 2D representation of the 2CARD tetramer revealed 7 bona fide Ub binding sites, six of which (sites 1–6) are occupied in our structure, and one potential binding site (site 8, see Extended Data Fig. 7c, d for additional discussion). Single and double lines indicate respectively the distance between adjacent Ubs that can be directly connected through a K63-linkage and those that cannot (thus requiring a spacer Ub). c, EMSA analysis of the tetramer formation by wild-type RIG-I 2CARD-S and its mutants with altered surfaces IV, V and IV, upon incubation with K63-Ubn (n > 8). d, IFN-β reporter activity of wild-type and mutant RIG-I 2CARD in GST fusion (mean ± s.d., n = 3).
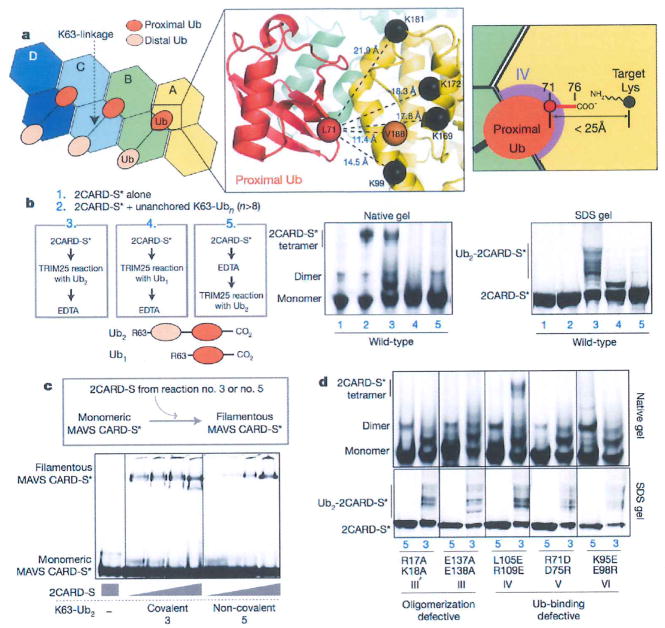
Figure 4. Covalent conjugation and non-covalent binding of Ub synergize to stabilize the 2CARD tetramer.
a, The C-terminal residue of the proximal Ub is within the covalent-linkage distance from the target Lys. As residues 72–76 of proximal Ub were disordered in the structure, distance was measured between Cαs of L71 in proximal Ub (red sphere) and target Lys (K99, K169, K172 andK181, black spheres) in 2CARD. V188 was used in place of K190 and K193, which are in the disordered C-terminal tail of 2CARD. The distance requirement for covalent conjugation (<25Å) is shown on the right, b, EMSA analysis of 2CARD tetramerization stimulated by covalently conjugated or unanchored K63-Ub2 or Ub1. Fluorescently labelled (indicated with an asterisk) 2CARD-S was subject to Ub conjugation (TRIM25 reaction) before or after EDTA quenching. Samples 3 and 5 enable direct comparison between covalently conjugated and unanchored Ub2. As elongation of Ub chains by TRIM25 is prevented by K63R mutation within Ub2 and Ub1, the presence of several bands in SDS–PAGE analysis reflects conjugation of Ub2 to 2CARD at multiple Lys targets, c, Comparison of the MAVS stimulatory activity of 2CARD covalently conjugated with K63-Ub2 (sample 3 from b) or 2CARD with unanchored K63-Ub2 (sample 5 from b). Increasing concentrations of 2CARD-S (47 or 187 or 750 or 3000 nM) were incubated with fluorescently labelled monomeric MAVS CARD-S and MAVS filament formation was monitored by BMSA. d, EMSA analysis of tetramerization of mutant 2CARD by covalently conjugated (sample 3 from (b)) or unanchored K63-Ub2 (sample 5 from (b)).
The observed K63-linkage specificity of 2CARD3 appears to be dictated by avid binding of K63-Ubn chains to the proximal and distal Ub binding sites, as observed with other Ub receptors18. That is, the relative distance and orientation of the proximal and distal Ub binding sites are compatible with K63-linked, but not K48-linked or linear Ub chains (Extended Data Fig. 5c). The poor electron density for the K63-linker, an indicator of conformational flexibility, also argues against a mechanism in which the K63-linkage is directly recognized by 2CARD, although other factors could have also contributed to conformational heterogeneity of the linker (Extended Data Fig. 2g).
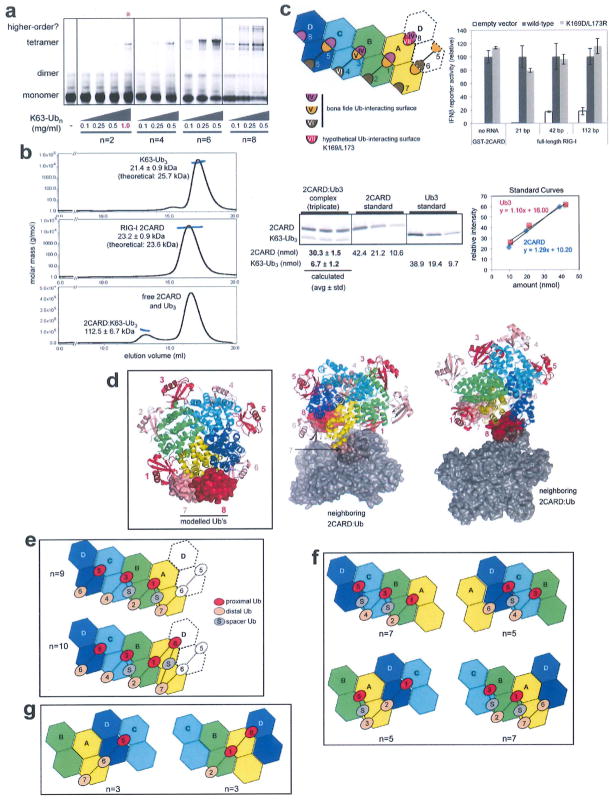
The structure predicts that longer Ub chains could wrap around the 2CARD tetramer at 1:4 or 2:4 molar ratios, thereby increasing the binding avidity and efficiency to promote 2CARD tetramerization. We observed that the efficiency of 2CARD tetramer assembly consistently increases with Ub chain length and that a single chain of Ub3 can stabilize the 2CARD tetramer (Extended Data Fig. 7a, b, g). To understand how K63-Ubn with n > 2 interacts with the 2CARD tetramer, we identified possible Ub-binding sites by mapping the IV, V and VI sites onto the 2CARD tetramer (Fig. 3b). Besides the six sites occupied in our crystal structure (sites 1–6 in Fig. 3b), we located two additional Ub binding sites (site 7, and possibly 8) (Extended Data Fig. 7c, d). As the distance between sites 2 and 3;4 and 5; and 7 and 1 is too great to be directly connected via the K63 linkage (Fig. 3b), a ‘spacer’ Ub would be required between these sites (Extended Data Fig. 7e). Accordingly, the chain length of n = 5–10 would be required to bridge the four 2CARD subunits (Extended Data Fig. 7e, f), which coincides with the chain length of K63-Ubn co-purified with RIG-I 2CARD from cells1.
The relationship between the covalent conjugation and non-covalent binding of K63-Ubn in RIG-I signalling has been controversial1,2. These two mechanisms have been proposed to use the same six Lys residues (K99, K169, K172, K181, K190 and K193)1,2. Our structure showed that none of these residues are involved in K63-Ubn binding or in the 2CARD–2CARD interaction, suggesting that the observed negative impact of mutating the target lysines (6KR)2 reflects the importance of covalent Ub conjugation in the signalling activity of isolated 2CARD (Extended Data Fig. 5d). To understand the interplay of covalent and non-covalent binding of Ub, we examined the distance between the target lysine and the C-terminal tail of proximal Ub. Although 2CARD residues 189–200 containing K190 and K193 are disordered in the crystal structure, the remaining four target lysines and the most C-terminal residue, V188, are within the covalent linkage distance from the C-terminal tail of proximal Ub (Fig. 4a). This observation suggests that covalent conjugation of K63-Ubn to the six lysines is compatible with its non-covalent binding to 2CARD, and may further stabilize the 2CARD tetramer.
To test this hypothesis, we compared the tetramerization efficiency of 2CARD conjugated with K63-Ub2 and 2CARD with free K63-Ub2. We observed the mobility shift of 2CARD when K63-Ub2 was covalently conjugated by TRIM25 (sample 3 in Fig. 4b) similar to the shift observed with unanchored K63-Ubn (n > 8) (sample 2). No shift was observed when mono-Ub was conjugated (sample 4) or Ub2-conjugation was inhibited by EDTA (sample 5), suggesting that the mobility shift was caused by covalent conjugation with Ub2, not by free Ub2 (which requires higher concentrations to induce 2CARD tetramerization, Extended Data Fig. 7a). Accordingly, Ub2-conjugated 2CARD stimulated MAVS filament formation more efficiently than 2CARD with free Ub2 (Fig. 4c). In addition, mutations in the 2CARD–2CARD and 2CARD–Ub interfaces (except for the mutation in IV) abrogated tetramerization of 2CARD conjugated with Ub2 (Fig. 4d). These results suggest that 2CARD tetramerization mediated by covalently conjugated Ut2 requires the same type of non-covalent 2CARD–Ub and 2CARD–2CARD interactions as with that mediated by unanchored K63-Ubn. The loss of tetramerization of Ub2-conjugated 2CARD upon VI mutation is consistent with the observation that conjugation with mono-Ub (and its bridging of IV and V) is insufficient to induce 2CARD tetramerization (Fig. 4b). As for the IV mutation, covalent conjugation of Ub2 compensated for inefficient Ub binding (Figs 3c and 4d), which provides a rationale for its robust signalling activity in cells (Fig. 3d). This result further supports the notion that covalent Ub conjugation near its binding site enhances the stability of the 2CARD tetramer.
We have previously shown that induced proximity of 2CARD within RIG-I filaments formed on long (~60–100 base pair (bp)) dsRNA could contribute to 2CARD oligomerization independtly of Ub12. To better understand how filament formation interplays with Ub-mediated 2CARD oligomerization, we investigated the dependence of the signalling activity of Ub conjugation-defective (6KR) or binding-defective (K95E/E98R) mutants on RNA length. Both mutations significantly reduced the signalling activity of full-length RIG-I with 21 bp dsRNA (Extended Data Fig. 5d), a dsRNA length that does not support filament formation. However, their negative effects were progressively alleviated by increasing the length of dsRNA (Extended Data Fig. 5d). This result suggests that the requirement for Ub-conjugation and Ub-binding can be partially compensated by filament formation12, and that the three mechanisms, Ub-binding, conjugation and filament formation, act synergistically for optimal signal activation.
Ub-dependent signal activation typically involves covalent conjugation of a target protein with Ub upon signal activation, and this ‘Ub signal’ is decoded by a downstream protein which binds to the conjugated Ub in linkage- and length-specific manners18. In the RIG-I signalling mechanism, 2CARD is both an Ub-conjugation target and a receptor, with little structural similarity to previously characterized Ub-binding domains. Our current work shows how these dual functions of RIG-I 2CARD synergize and lead to efficient formation of the signalling competent tetramer and provides unique insights into a novel type of Ub-mediated signal activation mechanism. Furthermore, the resultant helical architecture of the RIG-I 2CARD tetramer offers a previously unexpected signal activation mechanism for RIG-I, in which the 2CARD tetramer is used as a signalling ‘platform’ to recruit and assemble the MAVS filament. Whether there is potential for generalizing our findings to MDA5 or other CARD domains will require future investigations (Extended Data Figs 8 and 9).
METHODS
Plasmid constructs
Wild type and variants of RIG-I 2CARD (residue 1–200) were subcloned between XmaI and HindIII restriction sites in pET47b (Novagen). Site-directed mutagenesis was performed using the KAPA HiFi PCR kits (KAPA biosystems). For the 2CARD-SNAP (2CARD-S) construct, 2CARD was subcloned between the XmaI and HindIII, and SNAP between the XhoI and AvrII restriction sites in pET47b. For MAVS CARD fused to SNAP (CARD-S), CARD and SNAP were sequentially inserted between the BamHI and EcoRI, and between the EcoRI and XhoI restriction sites in pET47b, respectively. The bacterial expression construct for mouse E1 ubiquitin activating enzyme (pET28-mE1) was purchased from Addgene. Human TRIM25 was cloned between KpnI and Sail restriction sites of pET50b. Human UbcH5, Ubc13 and Uev1a were inserted between the XmaI and XhoI restriction sites in pET47b, and human ubiquitin was inserted between the XmaI and HindIII restriction sites in pET47b. For expression of RIG-I in HEK293T cells, wild-type RIG-I and its mutants were inserted between the HindIII and KpnI restriction sites in pFLAG-CMV4 (Sigma). GST fusion constructs for RIG-I 2CARD (in pEBG) and its Lys variant (6KR) were gifts from M. U. Gack (Harvard Medical School).
Material preparation
RIG-I 2CARD and 2CARD-S were expressed in BL21 (DE3) at 20 °C for 16–20 h following induction with 0.4 mM IPTG. The proteins were purified by Ni-NTA affinity chromatography. The 6XHis tag was removed by HRV 3C protease, and 2CARD was further purified by additional Ni-NTA affinity chromatography (which removes uncleaved protein and protease) and SEC in buffer A (20 mM HEPES pH 7.5, 150 mM NaCl). For N-terminal fluorescent labelling of RIG-I 2CARD, the protein (~2 mg ml−1) was incubated with 0.5 mM peptide (LPETGG) conjugated with fluorescein (Anaspec) and 0.3 mM S. aureus sortase A (a gift from Hidde Ploegh, MIT)23 at room temperature (RT) for 2–3 h, followed by Ni-NTA affinity purification to remove sortase and SEC.
MAVS CARD was expressed as a fusion protein with the SNAP tag (CARD-S) in BL21 (DE3) at 20 °C for 16–20 h following induction with 0.4 mM IPTG. MAVS CARD-S was purified using Ni-NTA affinity chromatography and SEC. Purified CARD-S exists in the form of short filaments and was denatured in 6 M guanidinium hydrochloride for 30 min at 37 °C, followed by dialysis against 20 mM HEPES, pH 7.5, 500 mM NaCl, 0.5 mM EDTA and 10 mM BME at 4 °C for 1 h. Refolded CARD-S was labelled with Alexa488-benzylguanine (NEB) according to the manufacturer’s instruction, further purified by SEC in buffer B (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM EDTA) to remove unincorporated dye, and was immediately used for filament formation assays.
Mouse E1, human UbcH5, Ubc13, Uev1a and ubiquitin were prepared as previously reported12. NusA fusion ofTRIM25 was expressed from BL21 (DE3) at 20 °C for 16–20 h following induction with 0.4 mM IPTG and purified by Ni-NTA affinity and SEC.
The 42 and 112 bp dsRNAs were synthesized as before (ref. 12). See ref. 12 for the sequence.
Synthesis of unanchored and 2CARD-conjugated polyubiquitin chains
Lys63-linked ubiquitin chains (K63-Ubn) were generated from a reaction containing 0.4 mM ubiquitin, 4 μM mE1, 20 μM Ubc13 and 20 μM Uev1a in buffer (10 mM ATP, 50 mM Tris pH 7.5, 10 mM MgCl2, 0.6 mM DTT). The K63-Ubn synthesis reaction was performed overnight at 37 °C. Synthesized K63-Ubn chains were purified as described previously24. Briefly, ubiquitin chains were diluted fivefold into 50 mM ammonium acetate, pH 4.5, 0.1 M NaCl and separated over a 45 ml 0.1–0.6 M NaCl gradient in 50 mM ammonium acetate, pH 4.5 using a Hi-Trap SP FF column (GE Healthcare). High molecular weight fractions were applied to an S200 10/300 column equilibrated in Buffer A.
Ub2 was generated from a reaction containing0.2 mM Ub(D77), 0.2 mM Ub(K63R), 4 μM mE1, 20 μM Ubc13 and 20 μM Uev1a in buffer (10 mM ATP, 50 mM Tris pH 7.5, 10 mM MgCl2, 0.6 mM DTT). Ub4, Ub6 and Ub8 were synthesized as previously reported (ref. 1).
For synthesis of Ub2 covalently conjugated to RIG-I 2CARD, 10 μM wild-type and mutant RIG-I 2CARD-S constructs were incubated with 350 μM Ub2, 4 μM mE1, 2 μM TRIM-25, 150 μM UbcH5 in 50 mM Tris, pH 7.5, 10 mM ATP, 10 mM MgCl2, 2 mM DTT for 2 h at 37 °C.
Crystallization and structure determination
The RIG-I 2CARD–K63-Ub2 complex was assembled by mixing RIG-I 2CARD (residues 1–200, K115A/R117A) (~10.0 mg ml−1) and K63-Ub2 at a 1:2 molar ratio, and was concentrated to ~10 mg ml−1 in buffer A and 2 mM DTT and the resultant complex was purified by SEC. Crystals were initially obtained as needle clusters using the hanging-drop vapour-diffusion method from a 1:1 mixture of sample and reservoir buffer that contained 0.2 M tri-lithium citrate, 20% PEG3350. Crystals were optimized using reservoir buffer containing 0.2 M tri-lithium citrate, 22–24% PEG300, 3% ethylene glycol. Diffraction data were collected at beamline 8.3.1 at the Advanced Light Source and beamline 24ID-C at the Advanced Photon Source and processed using the program XDS19.
The structure was determined by molecular replacement using Phaser20 with duck 2CARD and monoubiquitin (from PDB: 4A2W and 1UBQ, respectively). We first identified the molecular replacement solution for four copies of 2CARD in the asymmetric unit, which was then fixed in the subsequent rounds of molecular replacement to locate six copies of Ub. The solutions for four copies of 2CARD in the tetramer (chains A through D) were unambiguously identified with the translational function Z-scores (TFZ) between 13 and 24. The solutions for Ub chains E, F, G, H and J were also unambiguous with TFZ between 11 and 23. Two competing solutions for the Ub chain I (Ia and Ib in Extended Data Fig. 2b) were found during earlier stages of molecular replacement with TFZs of 15 and 19, respectively. However, once the rest of the complex (that is, four 2CARDs and Ub chains E–H and J) was fixed as a known solution, only Ia was identified. In addition, only Ia, but not Ib, is compatible with the simulated annealing omit map density (Extended Data Fig. 2b). These results suggest that Ia is the correct solution and thus was selected in the final model. The structure was refined by an iterative process of manual model building usingCoot21 and simulated annealing, individual site, TLS refinement with torsion NCS and bulk solvent scaling using Phenix Refine22. Individual CARDs (first and second) and Ub moieties were used as independent TLS groups. The quality of the final model was examined using MolProbity25 and simulated annealing omit maps (Extended Data Fig. 2a, b) calculated with Phenix. Ramachandran plot indicated 94.3% and 0.9% of the protein residues are in favoured and outlier conformations, respectively. Unless mentioned otherwise, molecular graphics figures were generated using PyMOL (Schrodinger). A summary of data collection and structure refinement statistics is provided in Extended Data Table 1.
2CARD tetramerization assay (EMSA)
Unless stated otherwise, EMSA was performed by Alexa647 labelled, purified RIG-I 2CARD-S (10 μM or indicated amount) in buffer A (20 mM HEPES, pH 7.5, 150 mM NaCl, and 2 mM DTT) in the presence and absence of indicated amount of K63-Ubn (see Extended Data Fig. 1b) for 15 min at room temperature, and the complex was analysed on Bis-Tris native PAGE (Life). Fluorescent gel images were recorded using an FLA9000 scanner (Fuji) and analysed with Multigauge (Fuji).
MAVS filament formation assay
The MAVS filament formation assay was performed as previously reported26. Refolded, Alexa647 labelled monomer of CARD-S was prepared as previously described26. In the absence of external stimuli or seed filaments, refolded MAVS CARD remains stably as a monomer over 24 h, after which it spontaneously forms prion-like filaments over the course of days. Thus, all assays involving MAVS filament formation were performed within 6 h after refolding. To monitor stimulation of MAVS filament formation by RIG-I 2CARD, labelled, monomeric MAVS CARD-S (CARD-S*, 10 μM) was incubated with 10 μM 2CARD-S and 10 μM Ubn (10 μM monomeric Ub concentration) for 15 min at room temperature before analysis by Bis-Tris native PAGE (Life) or by electron microscopy. For MAVS filament extension assay in Fig. 2f, monomeric CARD-S* was mixed with unlabelled, wild-type MAVS CARD-S filament seeds, and filament extension by the monomeric CARD-S* was monitored by EMSA. Fluorescent gel images were recorded using an FLA9000 scanner (Fuji). Some level of divergence observed between in vitro MAVS filament formation assay and cellular IFN-β reporter assay may reflect differences in the sensitivity of the assays and signal readout mechanisms.
Interferon reporter assay
293T cells were plated in 48-well plates in Dulbecco’s modified Eagle medium (Cellgro) supplemented with 10% heat-inactivated fetal calf serum (Gibco) and 1 % penicillin/streptomycin. At ~ 95% confluence, cells were co-transfected with pFLAG-CMV4 plasmids encoding RIG-I or mutants (20 ng), IFN-β promoter driven firefly luciferase reporter plasmid (100 ng) and a constitutively expressed Renilla luciferase reporter plasmid (pRL-TK, 10 ng) by using at 1:2 ratio of DNA:lipofectamine2000 (Life) according to the manufacturer’s protocol. The medium was changed 4–6 h post-transfection and cells were subsequently transfected with in vitro transcribed dsRNA (0.5 μg) using 1:1 ratio of lipofectamine2000. For MAVS and GST–2CARD, cells were transfected with plasmids expressing full-length MAVS (MAVS-pcDNA3, 20ng) or GST-2CARD (2CARD-pEBG, 30 ng) with luciferase reporter plasmids (as above) without additional stimulation with dsRNA. Cells were lysed 24 h post-stimulation and IFN-β promoter activity was measured using the Dual Luciferase Reporter assay (Promega) and a Synergy2 plate reader (BioTek). Firefly luciferase activity was normalized against Renilla luciferase activity. Error bars represent standard deviation of three to five independent experiments.
Multi-angle light scattering (MALS)
The molecular masses of wild-type RIG-I 2CARD (residues 1–200), K63-Ub3 and that of their complex were determined by MALS using a Superose 200 10/300 column (GE) attached to a miniDAWN TRI-STAR detector (Wyatt Technology) in 20 mM HEPES, pH 7.5, 150 mM NaCl. The complex of 2CARD and K63-Ub3 was formed by mixing at 1:2 molar ratio of 2CARD–K63-Ub3 before loading on a gel filtration column.
Extended Data
Extended Data Figure 1. RIG-I 2CARD (K115A/R117A) forms the signalling-competent 2CARD tetramer.
a, IFN-β reporter activity of wild-type RIG-I and the K115A/R117A mutant with and without 42 bp dsRNA stimulation (mean ± s.d., n = 3). b, Left, EMSA analysis of tetramerization of wild-type and K115A/R117A RIG-I 2CARD (residues 1–200) with K63-Ubn (n > 8). 2CARD was N-terminally labelled with fluorescein using sortase (see Methods). Right, SDS analysis of wild-type and mutant 2CARD, and K63-Ubn chains used in this study. Unless mentioned otherwise, K63-Ubn indicates the chain length n > 8 throughout the manuscript, c, Mapping of K115 and R117 onto the crystal structure. Although K115 and R117 are located at the edge of the interface, K115A/R117A has little effect on the cellular signalling activity of RIG-I (a) and tetramerization of RIG-I 2CARD (b), indicating that these residues are not critical for mediating inter-domain contacts.
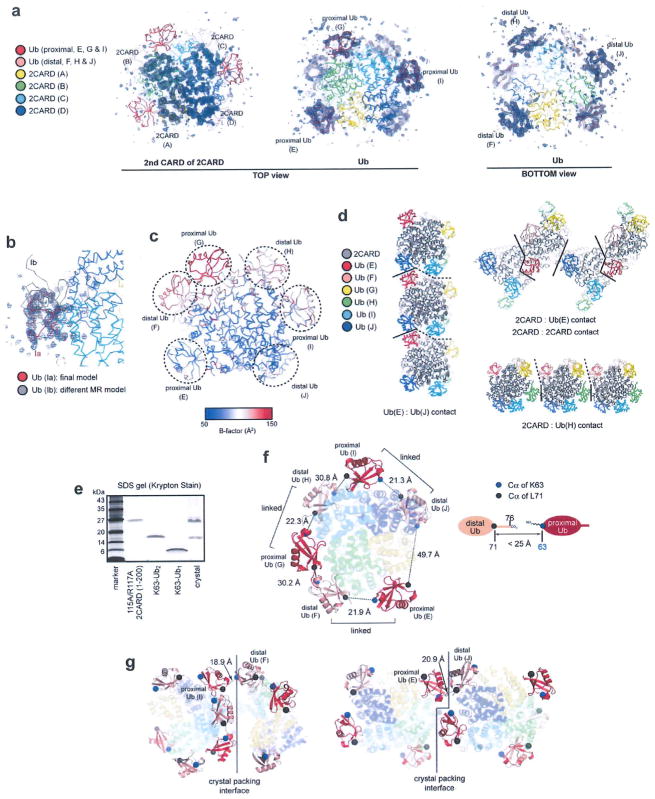
Extended Data Figure 2. Analysis of the crystal structure of the 2CARD-Ub complex.
a, Simulated annealing omit maps (sigma-A weighted Fo-Fc, contoured at σ = 2.5). Four and six maps were separately calculated using models with individual second CARDs (left) and Ubs (middle and right) omitted, respectively, and were overlaid on the crystal structure. The omit maps for Ubs are shown from both the top (middle) and bottom (right) views for better visualization of both proximal and distal Ubs. Although the omit maps for Ub chains F, G, H and I are less well-defined than those for chains E and J, the overall density matches well with individual Ub structures, and thus supports our model. b, Two competing molecular replacement (MR) solutions for Ub chain I. During the early stage of molecular replacement, solutions Ib (grey) as well as Ia (red, final solution) received high Z-scores, but only Ia was confirmed once the rest of the complex (all four 2CARDs and Ub chains E–H and J) was fixed as a known solution (see Methods). In addition, the simulated annealing omit map from (a) matches Ia, not Ib. Based on these results, Ia was selected for our final model. c, B-factor representation of the 2CARD–Ub complex. Note that Ub chains F–I have high B-factors, indicative of a high degree of flexibility. See (d) for an explanation. d, Three types of crystallographic packing interactions. They are mediated by Ub(E)–Ub(J) contacts (left); 2CARD–Ub(E) and 2CARD–2CARD contacts (middle); and 2CARD–Ub(H) contacts (right). Solid lines indicate crystallographic contacts, whereas dotted lines indicate boundaries between adjacent 2CARD tetramers without direct contacts. Note that 2CARD–Ub(H) contacts are less intimate than other contacts. The extensive contacts with Ub(E) and Ub(J) explain lower B-factors observed with these two Ub chains in c. e, SDS–PAGE analysis of the crystals shows that K63-Ub2 in the crystal is intact. f, Analysis of distance between the C terminus and K63 of adjacent Ub chains. As residues 72–76 of several Ubs were disordered in the structure, we measured the distance between Cα of L71 (black sphere) and Cα of K63 (blue sphere). The distance requirement for covalent conjugation (<25Å) is shown on the right, assuming 3.5Å spacing per residue in the missing C-terminal tail, 2Å for the C-terminal carboxylate, 6Å for Lys side chains. This condition is only satisfied with three pairs of Ubs, which enabled us to unambiguously identify pairs of Ubs that are covalently connected through the K63-linkage. g, Two crystallographic packing arrangements, which can potentially allow sharing of a single chain of Ub2 by two neighbouring 2CARD tetramers. In these arrangements, K63 of Ub bound to one 2CARD tetramer is within the covalent conjugation distance (Cα distance <25Å) from L71 of Ub bound to a neighbouring 2CARD tetramer. Such Ub crossover would increase the heterogeneity in the Ub connectivity, and could have contributed to the poor electron density map corresponding to the K63-linker. The same colour code was used as in f.
Extended Data Figure 3. Assembly of the 2CARD tetramer is mediated by rigid-body docking along the helical trajectory with the pitch in the screw equivalent to a single CARD.
a, Superposition of RIG-I 2CARD in the tetramer (subunits A–D) and 2CARD from full-length duck RIG-I (PDB: 4A2W). b, Superposition of full-length RIG-I (grey) onto the 2CARD tetramer by aligning 2CARD in full-length RIG-I with 2CARD subunit A (yellow) in the tetramer. The same colour code was used as in a. The superposition shows that the helicase domain in full-length RIG-I masks the 2CARD(A)–2CARD(B) interface and sterically blocks subunit B (green) from interacting with A. On the right, surface representation was separately shown for 2CARD(B) in the tetramer (top) and the helicase domain in full-length RIG-I (bottom) to further demonstrate the steric clash between the helicase domain and 2CARD(B). c, Geometric relationship between adjacent 2CARDs. Superposition of the ‘cut-out’ dimers of A–B, B–C and C–D (left), showing repetition of intermolecular interactions along the helical trajectory. The D–A interaction (middle) in the helical ‘seam’ (as defined in Fig. 1b) differs from the A–B, B–C and C–D interactions by a relative dislocation of A by a single CARD (right), d, IFN-β reporter activity of wild-type RIG-I and tetramer interface mutants (in Fig. 1d) with and without 42 bp dsRNA stimulation in 293T cells (mean ± s.d., n = 3).
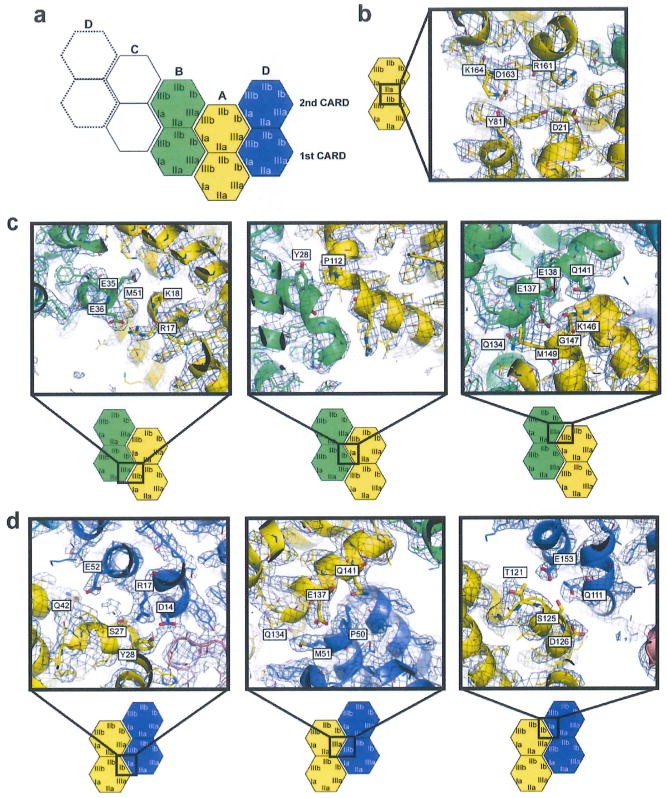
Extended Data Figure 4. Detailed analysis of the 2CARD tetramerization interface.
Electron density map (2Fo-Fc) was contoured at σ = 0.9. A few residues at the interface were displayed as stick models with labels, a, Definition of the interaction surface type (same as in Fig. 1c). b, Intramolecular interaction between surface IIb and IIa of the first and second CARDs, respectively. c, Intermolecular interactions that repeat along the helical trajectory (A–B, B–C and C–D). Shown are the interactions between 2CARD(A) and (B), which are identical to those between (B) and (C), and between (C) and (D) (Extended Data Fig. 3c). These interactions involve three interfaces (shown in the left, middle and right panels). Each of these interfaces consists of a few (< 3–4) residues on each side of the molecules, suggesting that cooperativity of all three interactions might be important for the tetramer stability. The IIIa–IIIb interactions in the first and second CARDs (left and right panels) are more extensive than the Ia–Ib interaction (middle), and are in general electrostatic. Detailed interactions among the interface residues could not be unambiguously determined due to the limited resolution of the structure. d, Intermolecular interactions at the helical seam, which occurs only between 2CARD(D) and 2CARD(A). As with the interaction between 2CARD(A) and (B) (or between B and C, and between C and D along the helical trajectory), each of the three interfaces consists of a few (< 3–4) residues on each side of the molecules, and the IIIa–IIIb interaction is more extensive than either of the two Ia–Ib interactions. Note that IIIa of the second CARD interacts with IIIb of the first CARD at the helical seam, whereas it interacts with IIIb of the second CARD along the helical trajectory (c). This is despite the low level of conservation of IIIb between the first and second CARDs, and thus suggests plasticity in molecular interactions. Similarly, Ia of the second CARD also has two distinct interaction partners, that is, Ib of the second CARD at the helical seam and Ib of the first CARD along the helical trajectory (c). But in this case, Ia utilizes different residues (albeit in the same local area) to accommodate different interaction partners.
Extended Data Figure 5. Interaction between the 2CARD tetramer and K63-Ub2.
a, Superposition of RIG-I 2CARD in the tetramer (subunits A–D) bound with K63-Ub2. The three Ub2 chains bound to A–B, B–C and C–D–A were superposed by aligning chain A, B and C, respectively, b, Superposition of the first (dark grey) and second (light grey) CARDs from RIG-I 2CARD bound by proximal (red) or distal (salmon) Ubs. The good alignment suggests that the proximal Ub-binding site, IV, in the second CARD is equivalent to the distal Ub-binding site, VI, in the first CARD (see Fig. 3b for the definitions of IV and VI). c, Distance analysis between the C-terminal residue of the distal Ub and K63, K48 and M1 of the proximal Ub. The C-terminal tail (residues 74–76) of the distal Ub is disordered in the structure, and thus the distance was measured between Cα of residue 73 and Cα of the target Lys. Below is the distance requirement for covalent conjugation, assuming 3.5Å spacing per residue in the missing C-terminal tail, 2Å for the C-terminal carboxylate, 6Å for Lys side chains and 1.5Å for N-terminal amine. These distance requirements are satisfied only with K63, thus rationalizing the observed specificity of 2CARD for the K63-linkage. d, IFN-β reporter activity of full-length RIG-I stimulated with 21 bp, 42 bp and 112 bp dsRNAs or isolated 2CARD fused to GST (GST–2CARD) without RNA. Activities were compared between wild type and mutants defective in Ub binding (R71D/D75R and K95E/E98R) or conjugation (6KR), and were normalized against the wild-type values (mean ± s.d., n = 3). 6KR indicates Arg mutation of six Lys (K99, K169, K172, K181, K190 and K193) that are known to be conjugated with K63-Ubn. The signalling activity of full-length RIG-I with 21 bp dsRNA was completely abrogated by R71D/D75R, K95E/E98R or 6KR, suggesting the importance of both Ub-binding and conjugation. The negative impacts of K95E/E98R and 6KR were progressively alleviated by stimulation with increasing length of dsRNA (42 and 112 bp). This restoration of the signalling activity by longer dsRNAs is consistent with our previous report that filament formation of RIG-I on long dsRNA (>60 bp) promotes 2CARD tetramerization by the ‘proximity-induced’ mechanism12. Note that 21 bp, 42 bp and 112 bp dsRNA can accommodate 1–2, 3, 8 RIG-I molecules, respectively. The negative impact of R71D/D75R could not be alleviated by stimulation with longer dsRNAs, which is somewhat at odds with the result with another Ub-binding deficient mutant, K95E/E98R. It is possible that R71D/D75R has more severe defects (in Ub binding or perhaps in 2CARD structure), which could not be overcome by Ub-conjugation or filament formation. For comparison, we also used GST-2CARD, which has been widely used in previous studies1–3. Despite the fact that GST-2CARD cannot form filaments, its sensitivity to the mutations was equivalent to full-length RIG-I with 42 bp, rather than 21 bp dsRNA. This likely reflects the effect of the fusion partner, GST, which forms a constitutive dimer. Isolated 2CARD without GST is a very poor stimulant of IFN-β, thus could not be used in this study.
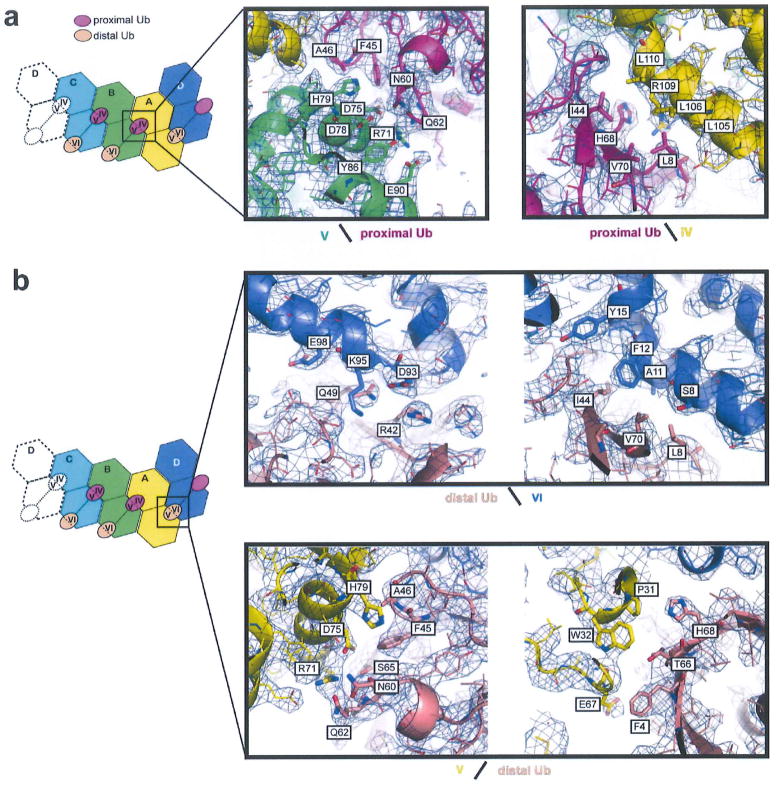
Extended Data Figure 6. Detailed analysis of the 2CARD-Ub interface.
Electron density map (2Fo-Fc) was contoured at σ = 0.9. Surface types were defined in the 2D representation on the left (as in Fig. 3b). a, Proximal Ub occupies surfaces IV and V on 2CARD. Surface IV interacts with the hydrophobic patch (L8/I44/V70) of proximal Ub, whereas V interacts with a combination of hydrophilic (N60/Q62) and hydrophobic (F45/A46) residues of proximal Ub. b, The distal Ub bound to 2CARD(D) simultaneously occupies both surfaces VI and V, whereas distal Ub’s bound to 2CARD(B) and (C) occupy surface VI alone (due to the lack of adjacent V). The interaction between distal Ub and VI is identical no matter whether the same Ub forms an additional interaction with V. Thus, only the distal Ub–2CARD(D) interaction is shown. Surface VI forms a combination of hydrophilic and hydrophobic interactions with Q49/R42 and L8/I44/V70 on distal Ub, respectively. Surface V interacts with distal Ub in the same manner as with proximal Ub, forming contacts with F45/A46/N60/N62 of Ub. Additional interactions involving surface V were seen with F4/T66/H68 of both distal and proximal Ubs (not shown in a), but they appear less intimate than those involving F45/A46/N60/N62.
Extended Data Figure 7. High avidity interaction is required for efficient formation of the 2CARD tetramer by K63-Ubn (n > 2).
a, EMSA analysis of the 2CARD tetramer formation using fluorescently labelled 2CARD-S (50 μM) with increasing concentrations of K63-Ubn (n = 2, 4, 6, 8). Note that an additional higher concentration (1 mg ml−1) was included only for K63-Ub2 (red asterisk), due to its low efficiency to stimulate 2CARD tetramerization. With K63-Ub8, additional bands appeared above the tetramer band, possibly reflecting two or more 2CARD tetramers bridged by a single Ub chains. b, Molecular mass analysis of 2CARD in complex with K63-Ub3i using multi-angle light scattering (MALS) coupled to size exclusion chromatography (SEC). Molecular mass estimated for the complex is 112.5 kDa (± 6.7 kDa), which is consistent with a tetrameric 2CARD (92.8 kDa, 23.2 ± 0.9 kDa as a monomer) bound by a single chain of K63-Ub3 (21.4 ± kDa). This 4:1 binding ratio of 2CARD to Ub3 is further supported by the SDS–PAGE intensity analysis of the complex (purified from MALS-SEC above) using Krypton fluorescence stain (right) (mean ± s.d., n = 3), which suggests the molar ratio of 4.5:1 for 2CARD–Ub3. This result suggests the sufficiency of a single chain of K63-Ubn (n > = 3) for stabilizing the 2CARD tetramer, although it does not exclude potential binding of additional Ubs at saturating concentrations3. Note that previous study3 suggesting 4:4 binding of 2CARD to Ubn (n = 3–6) was performed in a buffer lacking salt (20 mM Tris-HCl (pH 7.5) and 1 mM DTT), whereas the current study was performed with 150 mM NaCl (20 mM HEPES (pH 7.5) and 150 mM NaCl), which could be responsible for the divergent results. The sufficiency of the single chain of Ub3 for stabilizing the 2CARD tetramer suggests that multiple stoichiometries and Ubn binding configurations are possible, depending on the concentrations of 2CARD and Ubn as well as buffer compositions. See g for how a single chain of K63-Ub3 could stabilize the 2CARD tetramer. c, Six Ub binding sites (1–6) were occupied in the crystal structure, with a potential for binding of up to eight total Ub molecules in the 2CARD tetramer. Site 7 is equivalent to sites 2, 4 and 6, and thus is a bona fide Ub-binding site. Site 8 is a hypothetical Ub-binding site, as its interaction with Ub would simultaneously utilize surfaces IV and VII, instead of surfaces IV and V as in sites 1, 3 and 5. Mutations of VII (K169D/L173R) did not affect the signalling activity of RIG-I in cells based on the IFNβ reporter assay (right) (mean ± s.d., n = 3). Although this result suggests that surface VII (which only affects site 8, not 1–7) is not important, it does not exclude the possibility of site 8 serving as another Ub-binding site, as the loss of one out of 8 sites may not have a significant effect on the signalling outcome, d, A model of the 2CARD tetramer with all 8 potential Ub binding sites occupied. Ub bound to sites 7 and 8 (surface representation) were modelled by superposing 2CARDs bound to distal and proximal Ub onto 2CARD(A) and (D), respectively. Ub binding sites are numbered according to the 2D representation in c. Crystallographic packing prevents Ub occupancy of sites 7 and 8. Neighbouring molecules, which occlude the sites 7 (left) and 8 (right), are shown in grey surface. e, Two configurations to occupy 7 or 8 potential Ub-binding sites in the 2CARD tetramer using a single K63-Ubn chain. Ub-binding sites are numbered as in b. Ub labelled ‘S’ stands for the unbound Ub that serves as a spacer. The presence of spacer Ub is consistent with the observed activity of Ub4 with K63- and K48- mixed linkage in stimulating RIG-I 2CARD3, as there is no geometric restriction to impose the linkage specificity for the spacer Ub. f, Minimal length of K63-Ubn chain that allows bridging of four 2CARDs by a single Ub chains. Four examples were shown, in which Ub chains start with 2CARD subunit A, B, C or D. Ub-binding sites are numbered as in b. Ub labelled ‘S’ stands for the unbound Ub that serves as a spacer, g, Two configurations that a single chain of K63-Ub3 can bridge three 2CARDs (without involvement of a spacer Ub).
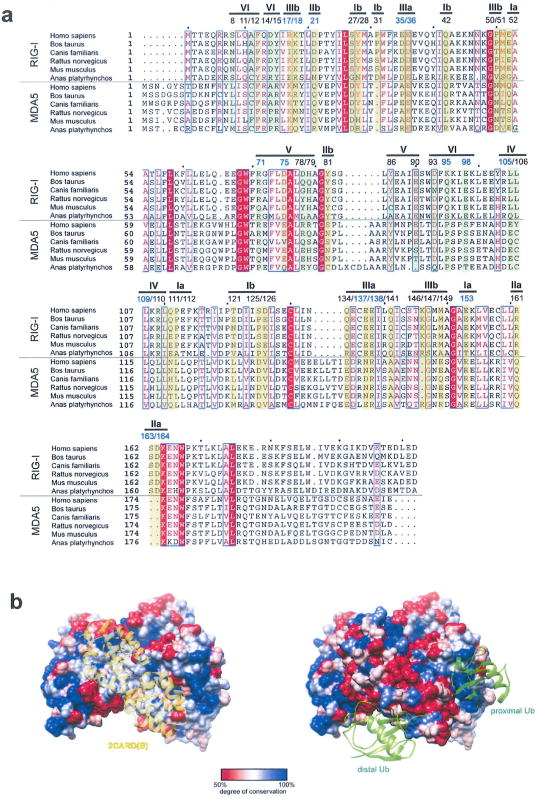
Extended Data Figure 8. Sequence conservation analysis of the 2CARD–2CARD and 2CARD–Ub interface.
a, Sequence alignment of RIG-I and MDA5 2CARD (using the program ClustalOmega27). Residues in RIG-I involved in the 2CARD–2CARD interactions (surface Ia/b–IIIa/b in Extended Data Fig. 4) and Ub binding (surface IV, V and VI in Extended Data Fig. 6), and their equivalent residues in MDA5 are highlighted (yellow and green for 2CARD–2CARD and 2CARD–Ub interfaces, respectively). Residue numbers right above highlights are according to human RIG-I. Residues tested by mutagenesis in this study are indicated by blue colour. Residues involved in the 2CARD–2CARD interactions show, on average, a moderate level of conservation in comparison to other residues on the surface of RIG-I 2CARD (see surface representation in b). Poor conservation of the tetramerization interface is consistent with previous observations that protein-protein interfaces often display evolutionary versatility due to the plasticity of the interaction and co-evolution of the interacting surfaces28,29. The Ub binding surface of RIG-I is more conserved than the 2CARD–2CARD interface (see surface representation in b), possibly reflecting the conserved nature of Ub. When the comparison is made between RIG-I and MDA5, we found that only four residues are well-conserved (F12 and L110 in the Ub-binding surface, and E36 and K164 in tetramerization surface), assuming interchange of residues within each group of F/Y, L/V/I or D/E as well-conserved. This is insufficient to support structural conservation of the 2CARD tetramer or 2CARD–Ub complex between RIG-I and MDA5. The structure of the MDA5 oligomers and/or MDA5–Ub complex would be required to compare 2CARD oligomerization mechanism between RIG-I and MDA5. b, Degree of sequence conservation (within RIG-I based on a) mapped onto the RIG-I 2CARD tetramer structure (generated using the program Chimera). Consistent with the analysis above, the 2CARD–2CARD interfaces show a moderate level of conservation, whereas Ub-binding sites show a higher degree of conservation. Other conserved surface areas may be involved in interactions with other molecules, such as TRIM25 or MAVS.
Extended Data Figure 9. Sequence analysis of the Ub-binding surface in the CARD family.
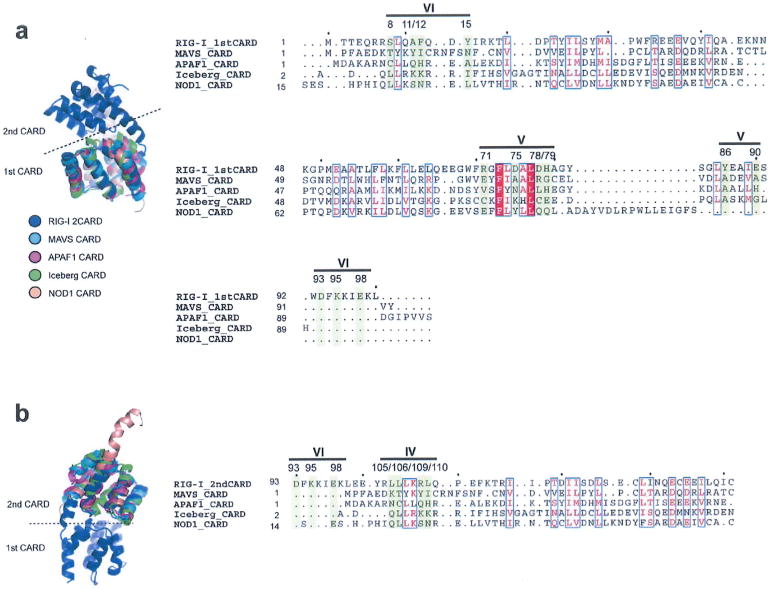
Structure-based sequence alignment (using the program SALGIN30) of various CARD domains. In our effort to further analyse potential generality of 2CARD–Ub interaction observed with RIG-I in our structure, we aligned other CARDs with RIG-I 2CARD. We performed structure-based sequence alignment, as many members of the CARD family share little sequence similarity. Three dimensional protein structure, which is more conserved than the primary sequence, allows more accurate sequence comparison. The first CARD (a) and second CARD (b) of human RIG-I was aligned with other CARDs from MAVS, APAF1, Iceberg and NOD1 (PDB code: 2VGQ, 3YGS, 1DGN and 4E9M, respectively). None of the Ub-binding residues in RIG-I 2CARD are conserved in these CARDs.
Acknowledgments
We thank G. Meigs at ALS BL 8.3.1 for data collection, and H. Wu and M. Eck for comments on the manuscript. This work was supported by GSK fellowship (B.W.), The Welch Foundation (I-1389; Z.J.C.), NIH (R01-GM63692; Z.J.C). Pew Scholarship (S.H.) and Career Development Award from BCH (S.H.).
Footnotes
Online Content Any additional Methods, Extended Data display items and Source Data are available in the online version of the paper; references unique to these sections appear only in the online paper.
Author Contributions S.H. initiated the project idea. A.P., B.W. and S.H. designed the experiments. A.P. and B.W. performed the experiments. H.X. and Z.J.C. provided K63-Ub3–8. A.P. and S.H. interpreted the data and wrote the manuscript
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession number 4NQK.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 3.Jiang X, et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 5.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 6.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seth RB, Sun L, Ea CK, Chen Z. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-β Signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 11.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Patel JR, et al. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrao R, Wu H. Helical assembly in the death domain superfamily. Curr Opin Struct Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park HH, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, et al. The Fas–FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nature Struct Mol Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 19.Kabsch W. XDS. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antos JM, et al. Site-specific N- and C-terminal labeling of a single polypeptide using sortase of different specificity. J Am Chem Soc. 2009;131:10800–10801. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong KC, et al. Preparation of distinct ubiquitin chain reagents of high purity and yield. Structure. 2011;19:1053–1063. doi: 10.1016/j.str.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu B, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 27.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caffrey DR, Somaroo S, Huges JD, Minstseris J, Huang E. Are protein-protein interfaces more conserved sequence than the rest of the protein surface? Protein Sci. 2004;13:190–202. doi: 10.1110/ps.03323604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreani J, Faure G, Guerois R. Versatility and invariance in the evolution of homologous heteromeric interfaces. PLOS Comput Biol. 2012;8:e1002677. doi: 10.1371/journal.pcbi.1002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braberg H, et al. SALIGN: A Webserver for alignment of multiple protein sequences and structures. Bioinformatics. 2012;28:2072–2073. doi: 10.1093/bioinformatics/bts302. [DOI] [PMC free article] [PubMed] [Google Scholar]