Abstract
Soil salt-alkalization is a dramatic challenging factor for plant growth. Wild soybean (Glycine soja) exhibits a favorable trait of superior tolerance to salt-alkali stress, and recent discoveries show that response regulator family genes are involved in diverse abiotic stresses. Genomic and transcriptomic analyses of all response regulator genes in wild soybean will provide insight into their function in plant stress response. In this study, we identified and characterized a total of 56 Glycine soja response regulator (GsRR) genes. Phylogenetic analysis suggested that GsRR genes could be classified into five subclasses (A1, A2, B1, B2, and C). We further investigated the chromosome locations, gene duplications and conserved domains of the GsRRs. Furthermore, the clustering analysis of GsRR transcript profiles revealed five different expression patterns under alkali stress. The A1 and A2 subclasses display significantly higher transcriptional levels than the B subclass. In addition, quantitative real-time PCR results verified that the GsRR genes were also significantly influenced by salt stress. Notably, GsRR2a in the A1 subclass showed opposite expression patterns under salt stress comparing with alkali stress. Moreover, overexpression of GsRR2a in Arabidopsis significantly improved the tolerance to alkali stress, but not salt stress. These results suggest the important roles of GsRR genes in response to salt and alkaline stresses, and also provide valuable clues for further functional characterization of GsRR family genes.
Keywords: Glycine soja, alkali stress, salt stress, response regulator, GsRR2a
Introduction
Saline-alkali soil is a major factor limiting crop growth, development, and yields. Salt stress in the soil generally causes osmotic stress and ion injury (Zhu, 2003). Alkali stress in the soil is usually characterized by low availability of nutrients, high concentrations of HCO3- (bicarbonate) and CO32- (carbonate), and high pH (Yang C.W. et al., 2008; An et al., 2016; Song T. et al., 2017). Owing to hydrolyzation of HCO3- and CO32-, plants growing on such soils suffer not only sodium toxicity, but also the precipitation Ca2+, Mg2+, and H2PO4- (Islam et al., 2011), inhibition of ion uptake (Yang et al., 2007) and disruption of cytoplasmic ion homeostasis (An et al., 2016). Some studies have demonstrated that alkali stress imposes much severer effects than salt stress on plants (Sadras et al., 2003; Shi and Sheng, 2005; Yang et al., 2007), and recent researches also point out a great difference in the physiological adaptive mechanisms of plants responding to alkali stress and salt stress (Borsani et al., 2005; Miller et al., 2010; Rouphael et al., 2017).
With the recent advances in high-throughput sequencing technologies, genes associated with high salinity and alkaline tolerance have been identified on a large scale at a genome-wide level (Jin et al., 2008; Sun et al., 2013; Zhang et al., 2016). The current knowledge of salt-alkali stress transcriptome mainly focuses on salt stress, whereas only limited information concerning alkali stress is available. Wild soybean (Glycine soja) exhibits very high adaptability in extreme environments. Our previous studies showed that the wild soybean (G07256) could germinate and set seed even in sodic soil of pH 9.02, and displayed much superior tolerance to 50 mM NaHCO3 treatment (Ge et al., 2010), demonstrating that it has developed molecular and physiological mechanisms to adapt itself to this severe condition. Additional, we have identified 3,380 alkaline-responsive genes using RNA sequencing, and also characterized some functional genes under alkaline stress, such as GsCHX19.3 (Jia et al., 2017), GsJ11 (Song X. et al., 2017), and GsTIFY10 (Zhu et al., 2011). Therefore, it is a suitable model organism for studying the molecular mechanisms of plant stress tolerance and a valuable source for characterizing alkali stress responsive genes.
Cytokinins (CKs) are regulators of plant growth and development, and have been shown to control plant responses to salt stress (Tran et al., 2007; Wang et al., 2015). The early response to CKs in Arabidopsis involves a multi-step signaling network, in which ARRs (Arabidopsis Response Regulators) play central roles (Jeon and Kim, 2013). The ARRs are divided into three types (type A, B, and C). The type-A ARRs (ARR3-9, ARR15-17, and ARR23) are small proteins with a short receiver domain which contains the phosphorylatable aspartate residue. CK-inducible type-A ARRs act mainly as redundant negative regulators in CK signaling (To et al., 2007). The type-B ARRs (ARR1, ARR2, ARR10-14, and ARR18-21) contain a receiver domain and a large C-terminal region harboring a Myb-like DNA-binding domain for transcriptional activation (Yokoyama et al., 2007). The type-B ARRs are not inducible by CKs, but activate transcription factors that induce transcription of type-A ARRs under CK treatment. Type-C ARRs (ARR22 and ARR24) resemble type-A ARRs, but their expression does not depend on CKs (Horak et al., 2008).
In Arabidopsis, the function of ARRs has been well suggested to be involved in plant development and signal transduction. ARR2 is a downstream genes of ETR1 in ethylene signal transduction (Hass et al., 2004). ARR3 and ARR4 play important roles in the circadian control through the CK-independent pathway (Salome et al., 2006). ARR4 also modulates red light signaling by interacting with phytochrome B (Sweere et al., 2001). Furthermore, studies have demonstrated that ARRs play regulatory roles in abiotic stresses. The type-A, -B, and -C ARRs are reported to differentially respond to salt stress (Nishiyama et al., 2012). ARR1 and ARR12 regulate sodium accumulation in the shoots by controlling the expression of HKT1 in Arabidopsis (Mason et al., 2010). Overexpression of ARR5, ARR7, and ARR15 promoted freezing tolerance (Shi et al., 2012). The CK-deficient Arabidopsis mutants displayed enhanced drought and salt tolerance, as well as increased ABA sensitivity (Nishiyama et al., 2011). In addition, type-A ARRs can act as negative regulators in cold stress signaling through the inhibition of the ABA-dependent pathway (Jeon et al., 2010). However, until now, little is known about the RR family genes in response to salt and alkali stresses.
In this study, we identified 56 genes encoding RR proteins in G. soja genome. By using phylogenetics to characterize the variations within the GsRR family, we found expression of GsRR family genes were differentially affected by alkali and salt stresses. We further suggested that one of them, GsRR2a played a positive role in response to alkali stress.
Materials and Methods
Identification and Characteristics of Response Regulator Family Genes in the G. soja Genome
To identify all putative RR family genes in wild soybean, we obtained the G. soja genome and proteome sequences, respectively (Jeon et al., 2010; Qi et al., 2014). Because of the limited sequence information for G. soja, G. max database is used to identify the predicted genes and secondary structure (Zeng et al., 2012). Local BLAST search against G. soja proteome was carried out by using the HMM profile (build 2.3.2) of the response regulator domain as query. The HMM profile of receiver domain (ID PF00072) was downloaded from the Pfam database (Punta et al., 2012). The molecular weight and isoelectric point of GsRR proteins were predicted using online software Compute pI/Mw1.
Phylogenetic Tree Construction and Sequence Analysis
To investigate the phylogenetic relationships among GsRR proteins in plants, Clustal X program (Larkin et al., 2007) was used to perform the multiple sequence alignments of all 56 GsRRs from wild soybean and 24 ARRs from Arabidopsis. The phylogenetic trees were generated and displayed by using software MEGA 5.0 with the NJ (neighbor-joining) method (Kumar et al., 2008). The MEME2 was used to discover conserved motifs of GsRR family proteins. Gene structure maps were generated using GSDS (Gene Structure Display Server)3 (Hu et al., 2015). We defined the gene duplication according to the reported standards (Yang S. et al., 2008).
Plant Materials, Growth Conditions, and Stress Treatments
Seedlings of wild soybean (G07256) were grown in a culture room with the following settings: 60–80% relative humidity, 24–28°C and a light regime of 16 h light/8 h dark. Before sowing, seeds were treated with 98% sulfuric acid for 10–15 min and washed three times with sterile water. Nineteen days after sowing, seedlings were transferred into 1/4 strength Hoagland’s solution with 50 mM NaHCO3 or 200 mM NaCl for alkali or salt stress. Equal amounts of leaves and roots were sampled as three biological replicates at 0, 1, 3, 6 h time points after treatments.
Transcript Level Analysis
In order to analyze the expression profiles of GsRR family genes under alkali stress, hierarchical clustering tree based on the transcript data of GsRR genes was created with TM4: MeV 4.9 software (Saeed et al., 2003). The transcript data of GsRRs in G. soja roots subjected to alkali stress was previously obtained in 1 KP project by using transcriptome sequencing, and the data has been deposited in 1KP project4.
The expression profiles of GsRRs under salt stress were performed by using qRT-PCR (quantitative real-time PCR). The GAPDH in G. soja was used to normalize all values. Primer sequences of GsRRs and GADPH are listed in Supplementary Table S1. To enable statistical analysis, three fully independent biological replicates were obtained and subjected to qRT-PCR runs in triplicate. Expression levels for all candidate genes were calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Transformation of Arabidopsis
The CDS region of GsRR2a was cloned into the pCAM230035S vector under the control of CaMV35S promoter (primer pairs: 5′-CGGGATCCATGGACACGGACA GCT TCG-3′ and 5′-GCGTCGACTCAATCGGTGCTGGTCA-3′). The pCAM230035S:GsRR2a construct was introduced into Agrobacterium tumefactions strain LBA4404 for transformation through floral-dip method (Clough and Bent, 1998). The transformed seeds were selected on 1/2MS medium containing 50 mg L-1 kanamycin, and the T3 generation overexpression lines were randomly chosen for further studies.
Phenotypic Analysis Under Alkali and Salt Stresses
The Arabidopsis seeds were sterilized as described (Sun et al., 2014). During the early seedling growth stage, the WT and overexpression seeds were sown on 1/2 agar medium supplemented with 0, 7, or 8 mM NaHCO3, respectively. The numbers of seedlings with opening and greening leaves were recorded after 12 days. At the adult stage, the 20-day-old WT and overexpression plants grown in nursery pots were irrigated with water or 100 mM NaHCO3 every 3 days. Photos were taken after 21 days. The chlorophyll content was detected using the 80% (v/v) acetone extract (Lewinsohn and Gressel, 1983). The malondialdehyde (MDA) content was determined by using a thiobarbituric acid method (Peever and Higgins, 1989). For salt treatment, the WT and overexpression seeds were sown on 1/2 agar medium supplemented with 0 or 150 mM NaCl, respectively. The germination rates were recorded and photos were taken after 6 days.
All experiments were repeated at least three times and the data was subjected to statistical analyses using the SPSS software by Student’s t-test.
Results
Identification of Response Regulator Genes in G. soja
In order to identify GsRR family genes, we used the amino acid sequences of the RR receiver domains (Pfam: PF00072) as queries for BLASTP searches. Sixty-two putative GsRR genes were acquired. Then we performed a proteome-wide screen for all putative GsRR by using the Pfam database, four genes were discarded due to the incomplete RR receiver domains and two genes were discarded because of redundancy. Consequently, 56 non-redundant GsRR genes were identified, including 19 type-A, 30 type-B, and 7 type-C GsRRs. The characteristics of the GsRR family genes, including the full CDS length, protein length, molecular weight and pI values are presented in Table 1.
Table 1.
Basic information of the GsRR family genes of G. soja.
| Gene name | Full CDS length (bp) | Protein length (aa) | Molecular weight (Da) | pI | Domain | Similarity with Arabidopsis | |
|---|---|---|---|---|---|---|---|
| GsRR1a | 735 | 244 | 26489.9 | 5.15 | RR | ARR3 | AT1G59940.1 |
| GsRR2a | 723 | 240 | 26531 | 4.99 | RR | ARR3 | AT1G59940.1 |
| GsRR3a | 747 | 248 | 28265.8 | 5.61 | RR | ARR9 | AT3G57040.1 |
| GsRR4a | 519 | 172 | 19577.6 | 6.44 | RR | ARR9 | AT3G57040.1 |
| GsRR5a | 615 | 204 | 22219.8 | 8.49 | RR | ARR6 | AT5G62920.1 |
| GsRR6a | 441 | 146 | 16118.8 | 8.35 | RR | ARR17 | AT3G56380.1 |
| GsRR7a | 708 | 235 | 26476.7 | 5.32 | RR | ARR9 | AT3G57040.1 |
| GsRR8a | 636 | 211 | 23187.9 | 8.23 | RR | ARR5 | AT3G48100.1 |
| GsRR9a | 699 | 232 | 26149.4 | 5.2 | RR | ARR9 | AT3G57040.1 |
| GsRR10a | 441 | 146 | 16003.6 | 8.34 | RR | ARR17 | AT3G56380.1 |
| GsRR11a | 615 | 204 | 22139.5 | 7.63 | RR | ARR6 | AT5G62920.1 |
| GsRR12a | 672 | 223 | 24610.3 | 5.27 | RR | ARR3 | AT1G59940.1 |
| GsRR13a | 564 | 187 | 21226.3 | 5.62 | RR | ARR9 | AT3G57040.1 |
| GsRR14a | 636 | 211 | 23722 | 5.38 | RR | ARR9 | AT3G57040.1 |
| GsRR15a | 540 | 179 | 20420.5 | 6.53 | RR | APRR7 | AT5G02810.1 |
| GsRR16a | 540 | 179 | 20439.5 | 5.98 | RR | ARR9 | AT3G57040.1 |
| GsRR17a | 624 | 207 | 22844.7 | 7.59 | RR | ARR8 | AT2G41310.1 |
| GsRR18a | 669 | 222 | 24661.3 | 5.26 | RR | ARR3 | AT1G59940.1 |
| GsRR19a | 741 | 246 | 27901.5 | 5.5 | RR | ARR9 | AT3G57040.1 |
| GsRR1b | 1902 | 633 | 69759.6 | 6.27 | RR/Myb-like | ARR2 | AT4G16110.1 |
| GsRR2b | 1971 | 656 | 72493.4 | 6.36 | RR/Myb-like | ARR12 | AT2G25180.1 |
| GsRR3b | 2076 | 691 | 76424.3 | 6.43 | RR/CCT motif | APRR5 | AT5G24470.1 |
| GsRR4b | 1908 | 635 | 71711.8 | 5.35 | RR/Myb-like | ARR11 | AT1G67710.1 |
| GsRR5b | 1233 | 411 | 45938.8 | 8.14 | RR/Myb-like | ARR1 | AT3G16857.1 |
| GsRR6b | 1488 | 495 | 55935.2 | 6.38 | RR/Myb-like | ARR1 | AT3G16857.1 |
| GsRR7b | 2091 | 696 | 76774.2 | 6.51 | RR/Myb-like | ARR12 | AT2G25180.1 |
| GsRR8b | 2103 | 700 | 77092.2 | 6.67 | RR/CCT motif | APRR5 | AT5G24470.1 |
| GsRR9b | 1092 | 633 | 69792.6 | 6.17 | RR/Myb-like | ARR2 | AT4G16110.1 |
| GsRR10b | 2040 | 679 | 74250.5 | 5.72 | RR/Myb-like | ARR2 | AT4G16110.1 |
| GsRR11b | 1206 | 401 | 45439.9 | 5.63 | RR/Myb-like | ARR14 | AT2G01760.1 |
| GsRR12b | 1479 | 492 | 54810.5 | 7.26 | RR/Myb-like | ARR2 | AT4G16110.1 |
| GsRR13b | 1782 | 593 | 66977.2 | 5.23 | RR/Myb-like | ARR11 | AT1G67710.1 |
| GsRR14b | 2022 | 673 | 73845 | 6.1 | RR/Myb-like | ARR2 | AT4G16110.1 |
| GsRR15b | 2097 | 698 | 76222 | 5.57 | RR/Myb-like | ARR12 | AT2G25180.1 |
| GsRR16b | 2298 | 765 | 83325.3 | 5.85 | RR/CCT motif | APRR7 | AT5G02810.1 |
| GsRR17b | 1815 | 604 | 68074.7 | 5.42 | RR/Myb-like | ARR11 | AT1G67710.1 |
| GsRR18b | 1881 | 626 | 68442 | 6.04 | RR/CCT motif | APRR7 | AT5G02810.1 |
| GsRR19b | 2043 | 680 | 75246.4 | 5.94 | RR/Myb-like | ARR12 | AT2G25180.1 |
| GsRR20b | 1998 | 665 | 73548.4 | 5.84 | RR/Myb-like | ARR12 | AT2G25180.1 |
| GsRR21b | 2019 | 672 | 73650.7 | 5.94 | RR/Myb-like | ARR1 | AT3G16857.1 |
| GsRR22b | 2094 | 697 | 76394.6 | 5.83 | RR/Myb-like | ARR12 | AT2G25180.1 |
| GsRR23b | 2010 | 669 | 73975.8 | 8.08 | RR/Myb-like | ARR2 | AT4G16110.1 |
| GsRR24b | 2034 | 677 | 73855.1 | 5.81 | RR/Myb-like | ARR1 | AT3G16857.1 |
| GsRR25b | 2046 | 681 | 75164.1 | 5.31 | RR/Myb-like | ARR12 | AT2G25180.1 |
| GsRR26b | 2004 | 667 | 73691.4 | 5.9 | RR/Myb-like | ARR12 | AT2G25180.1 |
| GsRR27b | 1368 | 455 | 51483.7 | 6.23 | RR/Myb-like | ARR11 | AT1G67710.1 |
| GsRR28b | 1008 | 335 | 38653.4 | 7.04 | RR/Myb-like | ARR2 | AT4G16110.1 |
| GsRR29b | 1107 | 369 | 41812.9 | 6.95 | RR/Myb-like | ARR2 | AT4G16110.1 |
| GsRR30b | 948 | 315 | 35909.7 | 5.39 | RR/Myb-like | ARR2 | AT4G16110.1 |
| GsRR1c | 399 | 132 | 14767 | 6.51 | RR | ARR24 | AT5G26594.1 |
| GsRR2c | 342 | 113 | 12627.8 | 9.05 | RR | ARR24 | AT5G26594.1 |
| GsRR3c | 345 | 114 | 12572.2 | 5.32 | RR | ARR24 | AT5G26594.1 |
| GsRR4c | 426 | 141 | 16341.8 | 8.7 | RR | ARR24 | AT5G26594.1 |
| GsRR5c | 351 | 116 | 12977.9 | 5.31 | RR | ARR24 | AT5G26594.1 |
| GsRR6c | 327 | 108 | 11984.9 | 5.08 | RR | ARR24 | AT5G26594.1 |
| GsRR7c | 453 | 150 | 16958.4 | 5.56 | RR | ARR24 | AT5G26594.1 |
Phylogenetic Analysis of GsRR Proteins
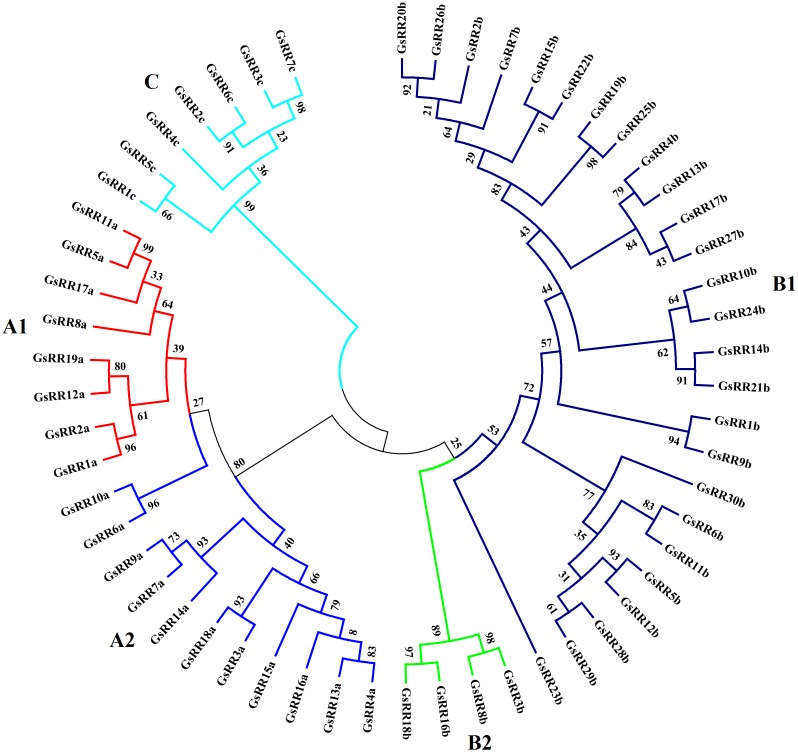
To investigate the evolutionary relationship of GsRRs and homologous ARR proteins, we constructed a NJ tree using MEGA 5.0 (Supplementary Figure S1). Based on the topology and clade robust bootstrap values, the GsRR proteins were classified into three major classes: type-A, type-B, and type-C. Nineteen GsRRs (GsRR1a to GsRR19a), thirty (GsRR1b to GsRR30b) and seven (GsRR1c to GsRR7c) were clustered into type-A, type-B, and type-C, respectively (Table 1). Furthermore, as shown in Figure 1, type-A was further divided into two subclasses, designated as A1and as A2. In addition, type-B was also divided into two subclasses (B1 and B2). Most of type-B GsRR proteins belonged to the B1 subclass, only GsRR3b, GsRR8b, GsRR16b, and GsRR18b were clustered into the B2 subclass.
FIGURE 1.
Phylogenetic trees of GsRR family genes in G. soja. The phylogenetic tree was inferred by MEGA 5.0 with the NJ (neighbor-joining) method after the alignment of protein sequences of the GsRR family. The numbers beside the branches represent bootstrap values based on 1,000 replications. GsRR family genes were divided into five subclasses and marked by different colors.
Physical Locations and Gene Duplications of GsRRs
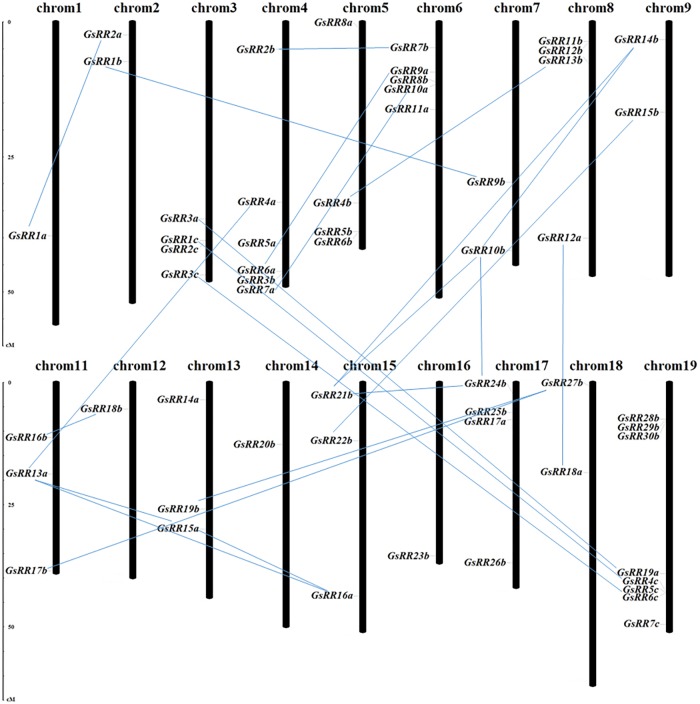
The potential mechanisms driving the evolution of the GsRR family were elucidated by analyzing the gene duplication events. In this study, 56 GsRR genes were distributed among 18 chromosomes, with the exception of chromosome 10 and 20 (Figure 2). The number of GsRR genes in each chromosome differed considerably. For example, 8 GsRRs were located on chromosome 19, which chromosomes 1, 12, 14, and 16 only contain one gene, respectively. Using G. soja genome duplication information, thirty duplicated gene pairs were identified among 56 GsRRs, including three segmental duplication events between chromosomes.
FIGURE 2.
Chromosomal locations and duplications of GsRR genes. The scales represent megabases (Mb). The black bars represent the chromosomes. GsRR genes distribute on the 18 chromosomes. The paralogous genes are connected by lines.
Conserved Domains and Motifs of GsRR Family Genes
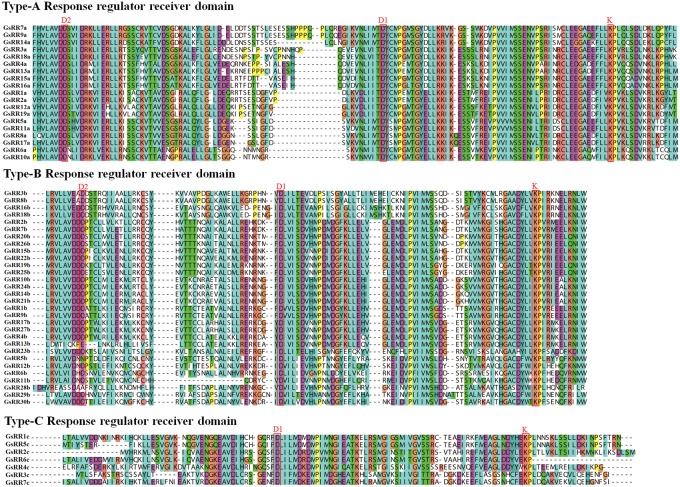
The modular structure of ARRs has been studied thoroughly in Arabidopsis (D’Agostino et al., 2000), which enables us to analyze domain architecture for GsRRs. We identified three conserved domains: a RR receiver domain (PF06200), a Myb-like DNA-binding domain (PF00249) and a CCT motif (PF06203). The RR receiver domain was variable among three types of GsRRs (Figure 3). The RR receiver domain of type-B GsRRs contained approximately 120 amino acids with three exclusively conserved phosph-accepting amino acids: an invariant D1 site in the center, a D2 site at the N-terminus and a K site at the C-terminus. Compared with type-B, each type-A GsRR has a variable short insertion in the receiver domain and a short C-terminal extension. Type-C GsRRs lost the conserved D2 site in the N-terminus. Remarkably, besides the RR receiver domain, all type-B1 GsRRs contained a C-terminal conserved domain designated as Myb-likes DNA binding domain, which functions importantly in CK responses. In addition, four type-B2 GsRRs contained a CCT motif in the C-terminus. In general, the classification of GsRRs based on their domain composition well supported the phylogenetic results described above.
FIGURE 3.
Multiple alignment of GsRR family proteins. Multiple sequence alignment shows the RR receiver domains of type-A, -B, and -C. Multiple alignment was performed with Clustal X. The conserved amino acids sites D1, D2, and K are marked.
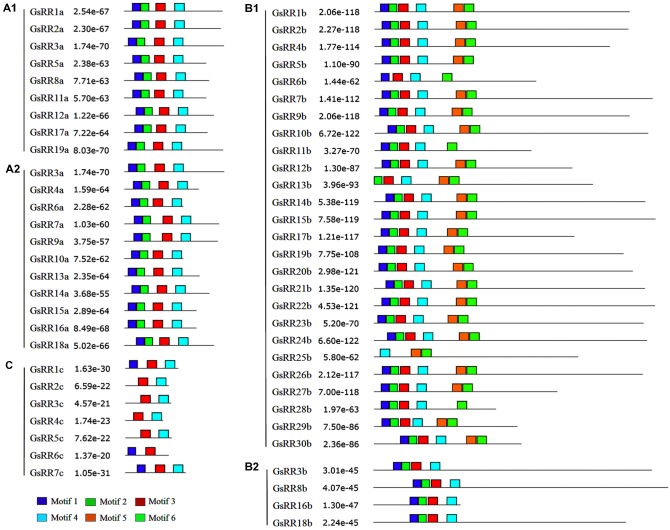
To verify the results of domain prediction, the conserved motifs were discovered using MEME on-line tool (Bailey et al., 2009). As shown in Figure 4 and Supplementary Figure S2, when specifying the RR receiver domain, motifs 1, 2, 3, and 4 were found in most type-A and type-B GsRRs. The type-C GsRRs possessed an incomplete RR receiver domain. The Myb-like DNA binding domain, motifs 5 and 6 were distinctively detected in type-B1 members, except GsRR6b, GsRR11b and GsRR28b only included motif 6.
FIGURE 4.
Distribution of conserved motifs in the GsRR family members. All motifs were identified by MEME using the full-length amino acid sequences of GsRR genes. The p-values are showed. Different conserved motifs are indicated by different colors.
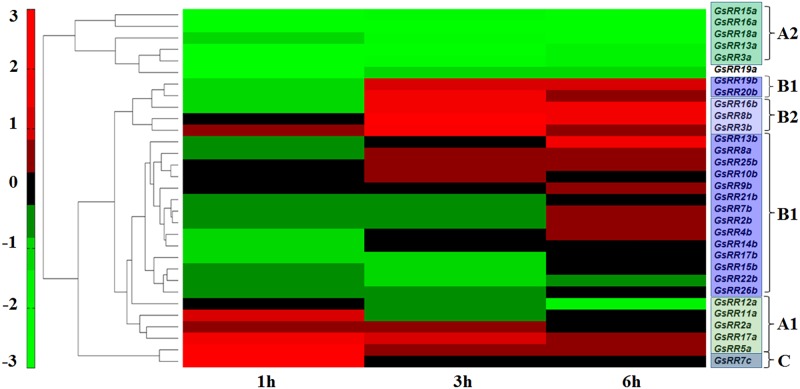
Expression Patterns of GsRRs Genes Under Alkali Stress
The RR family genes are known to be involved in abiotic stress response (Jeon et al., 2010; Mason et al., 2010). The wild soybean G07256 exhibits a much greater tolerance to alkali stress than other plants. Therefore, based on our previous transcriptome data of wild soybean roots under alkali stress (Ge et al., 2010; DuanMu et al., 2015), we performed the expression profiles of GsRR family genes using Pearson correlation Hierarchical Clustering with TM4: MeV 4.9 software. The results showed that 31 GsRRs were responsive to alkali stress, with distinctive induction dynamics (Figure 5). In general, five major expression patterns were unraveled. Five type-A2 GsRRs (15a, 16a, 18a, 13a, 3a) and GsRR19a formed the first cluster, with significant down-regulation from 1 h to 6 h after alkali stress. Six type-B1 GsRRs (25b, 10b, 9b, 21b, 7b, 2b) and GsRR8a showed no obvious change during the treatment. In contrast, type-B GsRRs (19b, 20b, 16b, 8b and 3b) in the third cluster were dramatically up-regulated at 3 h and kept the up-regulated trend in varying degrees until 6 h. The transcript levels of other six GsRR genes (4b, 14b, 17b, 15b, 22b, and 26b) in the fourth cluster were down-regulated and then recovered to the basal levels. It is worth to notice that on the basis of their expression patterns, type-A GsRRs were basically separated into two groups, similar with the classification of subclass A1 and A2. The transcript levels of subclass A1 GsRRs (11a, 2a, 17a, and 5a) were up-regulated at 1 h and then down-regulated at 6 h, which is opposite to subclass A2. These results indicated that GsRRs might have different roles in regulating alkali stress response.
FIGURE 5.
Expression profile of GsRR family genes under alkali stress. Expression profiles of GsRRs are shown according to the RNA-seq data of wild soybean treated with 50 mM NaHCO3. The expression profiles were conducted using Pearson correlation Hierarchical Clustering with TM4: MeV 4.9 software.
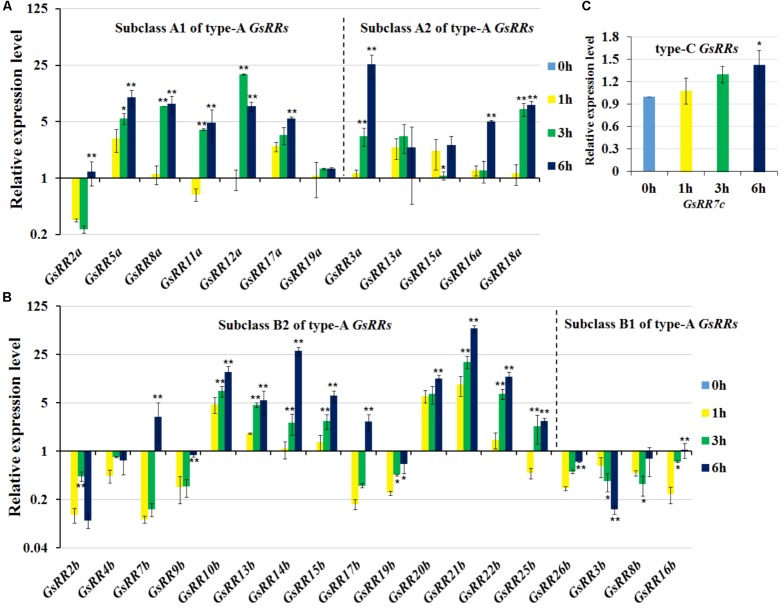
Expression Patterns of GsRRs Under Salt Treatment
To provide insight into the regulatory mechanisms of GsRRs in salt stress, we further analyzed their transcript levels under salt stress using the qRT-PCR analysis. As shown in Figure 6A, most type-A GsRRs were significantly up-regulated from 1 to 6 h under salt stress. Compared with subclass A2, subclass A1 GsRRs responded to salt stress faster and last longer. Unlike alkali stress, among 12 type-A GsRRs, only two were down-regulated under salt stress, indicating they responded to salt and alkali stresses in different pathways. For type-B GsRRs, three subclass B2 members GsRR3b, GsRR8b, and GsRR16b were down-regulated; eight type-B1 GsRRs (10b, 13b, 14b, 15b, 20b, 21b, and 22b) were up-regulated from 1 to 6 h, seven type-B1 genes were down-regulated at 1, 3, or 6 h (Figure 6B). For type-C GsRRs, only GsRR7c slightly responded to salt stress (less than twofold) (Figure 6C).
FIGURE 6.
Expression profile of GsRR family genes under salt stress. (A–C) The expression patterns of GsRRs were measured by qRT-PCR analysis with G. soja seedlings treated with 200 mM NaCl. Values represent the means of three fully independent biological replicates, and three technology replicates for each. ∗P < 0.05, ∗∗P < 0.01 by Student’s t-test.
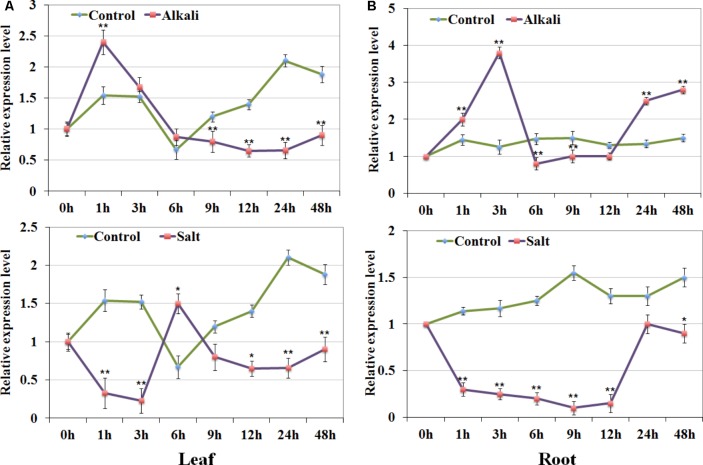
QRT-PCR Validation of GsRR2a Under Salt and Alkali Stresses
According to the expression analysis under salt and alkali stress, we focused on one of the type-A1 genes GsRR2a, whose expression was strongly induced by alkali stresses, but reduced by salt stress. To confirm this finding, we further detected its expression levels in both roots and leaves of G. soja seedlings under 200 mM NaCl or 50 mM NaHCO3 by using qRT-PCR analysis. As shown in Figure 7, under alkali treatment, GsRR2a showed similar tendencies in leaves and roots. The relative transcript abundance of GsRR2a rapidly increased at 1 or 3 h, respectively. Under salt treatment, the transcript abundance of GsRR2a was slightly decreased in roots and leaves. These results suggested that GsRR2a expression indeed differently responded to alkali and salt stresses.
FIGURE 7.
Expression validation of GsRR2a in G. soja. (A,B) Expression levels of GsRR2a were detected in root and leaves under salt and alkali stresses using qRT-PCR analysis. Nineteen-day-old of G. soja seedlings were submerged into 1/4 Hoagland solution with 50 mM NaHCO3 or 200 mM NaCl, respectively. The untreated plants were used as controls. Values represent the means of three fully independent biological replicates, and three technology replicates for each. ∗P < 0.05, ∗∗P < 0.01 by Student’s t-test.
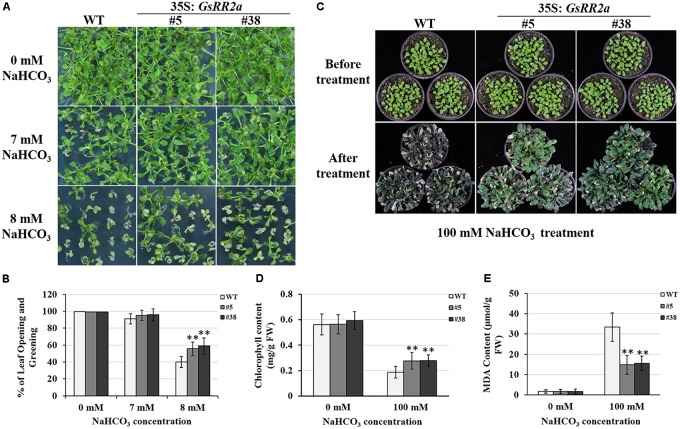
Overexpression of GsRR2a Improved Tolerance to Alkali Stress in Arabidopsis
Considering the responsive expression of GsRR2a under salt and alkali stresses, we further analyzed the effect of GsRR2a overexpression on alkali and salt tolerance. The transgenic lines (#5 and #38) were generated by overexpressing GsRR2a in Arabidopsis. We firstly performed the early seedling growth assays to determine the tolerance of WT (wide-type) and overexpression lines. Under normal conditions, GsRR2a overexpression does not affect plant growth under normal conditions. However, under NaHCO3 stress treatment, GsRR2a overexpression lines exhibited more seedlings with open and green leaves than WT (Figures 8A,B). Furthermore, to evaluate the alkali tolerance at the adult stage, the WT and GsRR2a overexpression lines were irrigated with 150 mM NaHCO3. After 16 days, the overexpression lines appeared much greener and healthier than WT (Figure 8C). In addition, statistical analysis revealed that overexpression lines exhibited higher chlorophyll contents but lower MDA contents than WT (Figures 8D,E). In contrast with alkali stress, no significant difference was observed between WT and the overexpression lines in the presence of 150 mM NaCl (Supplementary Figure S3). These results suggested that overexpression of GsRR2a in Arabidopsis could significantly improve the tolerance to alkali stress, but not to salt stress.
FIGURE 8.
Overexpression of GsRR2a in Arabidopsis enhances the tolerance to alkali stress. (A) The growth performance of WT and overexpression lines on 1/2MS medium containing 0, 7, or 8 mM NaHCO3 at early seeding growth stage. (B) The numbers of seedlings with opening and greening leaves of WT and overexpression lines. (C) The growth performance of WT and overexpression lines before alkali treatment or treated with 100 mM NaHCO3 for 16 days. (D) The chlorophyll content of WT and overexpression lines. (E) The malondialdehyde (MDA) content of WT and overexpression lines. ∗P < 0.05, ∗∗P < 0.01 by Student’s t-test.
Discussion
Recent studies have reported that the RR family genes regulate plant environmental stress responses through two-component systems (Tran et al., 2010). However, there is limited information about the functions of RR genes in soybean. This study identified all RR family genes in G. soja and systematically analyzed their sequences and their responses to salt and alkali stresses. This information may provide useful clues for functional characterization of GsRRs, especially concerning their role in stress tolerance.
In the current study, a total of 56 GsRRs were identified in wild soybean genome. These GsRRs were classified into five subclasses according to their phylogeny, which is consistent with previous reports in Arabidopsis and rice (D’Agostino et al., 2000; Jain et al., 2006). Interestingly, there were more GsRRs containing Myb-like DNA domain in type-B than type-A, which may attribute to gene duplication events. The Arabidopsis genome contains almost the same number of type-A and type-B ARRs. By contrast, the maize genome contains more type-A ZmRRs (Chu et al., 2011). These indicated that type-B RRs containing the Myb-like DNA binding domain might play more important roles in dicots. Different from Arabidopsis, type-A GsRRs are further divided into two subclasses (8 members in subclass A1 and 11 in subclass A2), which suggests possible divergence of their functions during evolution. Moreover, four type-B GsRRs (3b, 8b, 16b, and 18b) were designed as subclass B2. Subclass B2 members are also called the pseudo-response regulators, which are the circadian clock component proteins in Arabidopsis. They contain a receiver-like domain lacking the conserved phosphoacceptor aspartic acid residue, and a CCT motif responsible for transcriptional repression (Chu et al., 2011; Wang et al., 2013).
The motif distribution analyzed by MEME was basically consistent with the phylogenetic analysis. GsRRs in each individual subclass usually shared subclass-specific motifs. Besides, different types of GsRRs contained different numbers of exons (Supplementary Figure S4). For example, type-A GsRRs contained five exons, whereas type-B GsRRs contained four to nine exons. The different numbers of exons possibly shared evolutionary and structural differences.
Roots are the first point perceiving the underground environment stress. To explore the possible functions of RRs under alkali stress, we investigated the transcript levels of GsRRs in wild soybean roots. From their expression profiles, we observed five type-A2 GsRRs (15a, 16a, 18a, 13a, 3a) showed the same expression pattern under alkali stress, where they were significantly and continuously down-regulated upon the NaHCO3 treatment. This result suggested these co-expressed type-A2 GsRRs might function negatively in alkali stress responses. Interestingly, other five type-A1 genes GsRR12a, GsRR11a, GsRR2a, GsRR17a, and GsRR5a were also closely clustered and showed co-expression in roots. This further implies the functional redundancy among GsRRs, and functional divergence between type-A1 and type-A2 in plant tolerance to alkali stress. Moreover, GsRR14b, GsRR15b, GsRR17b GsRR21b, GsRR22b, and GsRR26b in subclass B1 were strongly down-regulated at 1 h or 3 h, while other subclass B1 GsRRs were significantly up-regulated at 3 h or 6 h. The difference among subclass B1 members in alkali stress responses may be resulted from different upstream or downstream regulatory elements or factors, which indicated diversified functions within the same subclass.
The great difference in expression patterns of GsRRs to alkali and salt stresses bring us to consider there might be other regulatory mechanism and signal pathway in alkali stress. As we know, that salt stress involves osmotic stress and ion injury, and salinity tolerance in plants largely contributed by Na+ exclusion (Yamaguchi et al., 2013). Actually, it has been pointed out that high HCO3- can diminish leaf area and length, decrease shoot biomass, and reduce the photosynthetic rate. However, the molecular mechanism of plant response to alkali stress is rarely known. Considering the important roles of RR proteins in CK signaling, the induction of GsRRs expression by salt and alkali stress provides a molecular link between stress and CK signaling. Moreover, GsRR2a, the homologous gene of ARR3, could enhance plant tolerance to alkali stress, but not to salt stress. One possible reason is that GsRR2a was up-regulated under alkali stress which indicated that this gene may be as a positive regulator of plant tolerance to alkali stress. However, GsRR2a exhibited the opposite expression pattern to salt and alkali stresses, which implied that GsRR2a may participate in different signaling pathways under alkali and salt stresses. In total, these results support the different mechanisms for alkali and salt stresses, and also provide a foundation for future work to elucidate the function of GsRR family genes.
Conclusion
In summary, we identified 56 GsRR genes, which could be classified into three types (five subclasses). GsRR were distributed among 18 chromosomes with gene duplications. Moreover, GsRR genes exhibited different expression patterns under alkali and salt stresses. Furthermore, overexpression of GsRR2a in Arabidopsis significantly improved the tolerance to alkali stress. In total, our results showed that GsRRs play crucial roles in plants responses to alkali and salt stresses. These results provided a foundation for further functional characterization of GsRR family genes.
Author Contributions
CC, AL, HR, YY, HD, and XD performed the experiments and analyzed data. CC and AL wrote the manuscript. BL interpreted data and revised the manuscript. DZ, XS, and YZ provided ideas and designed the research. All authors have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the Heilongjiang Provincial Higher School Science and Technology Innovation Team Building Program (2011TD005), Major Special Projects for New Varieties of Genetically Modified Organisms (2018ZX0800912B), BL is supported by the Swedish Cancer Society (CAN 2017/643 and CAN 2015/406), the Swedish Natural Research Council (VR 2015-04984), the National Natural Science Foundation of China (31771692), and National Natural Science Foundation of China (31671596 and 31500204 to XS).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01306/full#supplementary-material
Phylogenetic trees of GsRR family of G. soja and Arabidopsis. Neighbor-joining phylogenetic tree of the response regulator members in G. soja and Arabidopsis. The tree was inferred by MEGA 5.0 with the neighbor-joining method after the alignment of the full-length amino acid sequences of the 56 G. soja genes and 24 Arabidopsis genes. The numbers beside the branches represent bootstrap values based on 1,000 replications. The scale bar corresponds to 0.1 estimated amino acid substitutions per site.
Distribution of conserved motifs. All motifs were identified by MEME using the complete amino acid sequences of GsRR genes.
Overexpression of GsRR2a in Arabidopsis did not affect the tolerance to salt stress. The WT and overexpression lines are grown on medium containing 0 or 150 mM NaCl. The germination rates were recorded and photos were taken after 6 days.
Structure analysis of GsRR genes using GSDS online tools. The UTRs (upstream/downstream sequences), exons and introns are shown with light blue boxes, yellow boxes, and black lines, respectively.
Gene-specific primers of GsRR family used for q-RT PCR assays.
References
- An Y. M., Song L. L., Liu Y. R., Shu Y. J., Guo C. H. (2016). De novo transcriptional analysis of alfalfa in response to saline-alkaline stress. Front. Plant Sci. 7:931 10.3389/fpls.2016.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37 W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J. H., Verslues P. E., Sunkar R., Zhu J. K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291. 10.1016/j.cell.2005.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z. X., Ma Q., Lin Y. X., Tang X. L., Zhou Y. Q., Zhu S. W., et al. (2011). Genome-wide identification, classification, and analysis of two-component signal system genes in maize. Genet. Mol. Res. 10 3316–3330. 10.4238/2011.December.8.3 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- D’Agostino I. B., Deruere J., Kieber J. J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124 1706–1717. 10.1104/pp.124.4.1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuanMu H., Wang Y., Bai X., Cheng S., Deyholos M., Wong G.-S., et al. (2015). Wild soybean roots depend on specific transcription factors and oxidation reduction related genesin response to alkaline stress. Funct. Integr. Genomics 15 651–660. 10.1007/s10142-015-0439-y [DOI] [PubMed] [Google Scholar]
- Ge Y., Li Y., Zhu Y.-M., Bai X., Lv D.-K., Guo D., et al. (2010). Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol. 10:153 10.1186/1471-2229-10-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C., Lohrmann J., Albrecht V., Sweere U., Hummel F., Yoo S. D., et al. (2004). The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 23 3290–3302. 10.1038/sj.emboj.7600337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak J., Grefen C., Berendzen K. W., Hahn A., Stierhof Y. D., Stadelhofer B., et al. (2008). The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol. 8:77 10.1186/1471-2229-8-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A. Y., Zhang H., Luo J., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. S., Akhter M., El Sabagh A., Liu L. Y., Nguyen N. T., Ueda A., et al. (2011). Comparative studies on growth and physiological responses to saline and alkaline stresses of Foxtail millet (Setaria italica L.) and Proso millet (Panicum miliaceum L.). Aust. J. Crop Sci. 5 1269–1277. [Google Scholar]
- Jain M., Tyagi A. K., Khurana J. P. (2006). Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol. 6:1 10.1186/1471-2229-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Kim J. (2013). Arabidopsis response regulator1 and Arabidopsis histidine phosphotransfer protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 161 408–424. 10.1104/pp.112.207621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Kim N. Y., Kim S., Kang N. Y., Novak O., Ku S.-J., et al. (2010). A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 285 23369–23384. 10.1074/jbc.M109.096644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B., Sun M., Duanmu H., Ding X., Liu B., Zhu Y., et al. (2017). GsCHX19.3, a member of cation/H(+) exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Sci. Rep. 7:9423 10.1038/s41598-017-09772-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Kim H. R., Plaha P., Liu S. K., Park J. Y., Piao Y. Z., et al. (2008). Expression profiling of the genes induced by Na(2)CO(3) and NaCl stresses in leaves and roots of Leymus chinensis. Plant Sci. 175 784–792. 10.1016/j.plantsci.2008.07.016 [DOI] [Google Scholar]
- Kumar S., Dudley J., Nei M., Tamura K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9 299–306. 10.1093/bib/bbn017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., Mcwilliam H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lewinsohn E., Gressel J. (1983). The determination of chlorophylls a and b together with 14CO2 fixation in the same plant tissue samples. Anal. Biochem. 135 438–442. 10.1016/0003-2697(83)90708-X [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mason M. G., Jha D., Salt D. E., Tester M., Hill K., Kieber J. J., et al. (2010). Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J. 64 753–763. 10.1111/j.1365-313X.2010.04366.x [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33 453–467. 10.1111/j.1365-3040.2009.02041.x [DOI] [PubMed] [Google Scholar]
- Nishiyama R., Le D. T., Watanabe Y., Matsui A., Tanaka M., Seki M., et al. (2012). Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One 7:e32124 10.1371/journal.pone.0032124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R., Watanabe Y., Fujita Y., Le D. T., Kojima M., Werner T., et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 23 2169–2183. 10.1105/tpc.111.087395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peever T. L., Higgins V. J. (1989). Electrolyte leakage, lipoxygenase, and lipid peroxidation induced in tomato leaf tissue by specific and nonspecific elicitors from Cladosporium fulvum. Plant Physiol. 90 867–875. 10.1104/pp.90.3.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40 D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Li M. W., Xie M., Liu X., Ni M., Shao G., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 5:4340 10.1038/ncomms5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouphael Y., Cardarelli M., Bonini P., Colla G. (2017). Synergistic action of a microbial-based biostimulant and a plant derived-protein hydrolysate enhances lettuce tolerance to alkalinity and salinity. Front. Plant Sci. 8:131 10.3389/fpls.2017.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadras V., Baldock J., Roget D., Rodriguez D. (2003). Measuring and modelling yield and water budget components of wheat crops in coarse-textured soils with chemical constraints. Field Crops Res. 84 241–260. 10.1016/S0378-4290(03)00093-5 [DOI] [Google Scholar]
- Saeed A., Sharov V., White J., Li J., Liang W., Bhagabati N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34 374–378. [DOI] [PubMed] [Google Scholar]
- Salome P. A., To J. P. C., Kieber J. J., Mcclung C. R. (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18 55–69. 10.1105/tpc.105.037994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D. C., Sheng Y. M. (2005). Effect of various salt-alkaline mixed stress conditions on sunflower seedlings and analysis of their stress factors. Environ. Exp. Bot. 54 8–21. 10.1016/j.envexpbot.2004.05.003 [DOI] [Google Scholar]
- Shi Y., Tian S., Hou L., Huang X., Zhang X., Guo H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and Type-A ARR genes in Arabidopsis. Plant Cell 24 2578–2595. 10.1105/tpc.112.098640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T., Xu H., Sun N., Jiang L., Tian P., Yong Y., et al. (2017). Metabolomic analysis of alfalfa (Medicago sativa L.) root-symbiotic rhizobia responses under alkali stress. Front. Plant Sci. 8:1208 10.3389/fpls.2017.01208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Duanmu H., Yu Y., Chen C., Sun X., Zhu P., et al. (2017). GsJ11, identified by genome-wide analysis, facilitates alkaline tolerance in transgenic plants. Plant Cell Tissue Organ Cult. 129 411–430. 10.1007/s11240-017-1188-5 [DOI] [Google Scholar]
- Sun X., Luo X., Sun M., Chen C., Ding X., Wang X., et al. (2014). A Glycine soja 14-3-3 protein GsGF14o participates in stomatal and root hair development and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 55 99–118. 10.1093/pcp/pct161 [DOI] [PubMed] [Google Scholar]
- Sun Y. P., Wang F. W., Wang N., Dong Y. Y., Liu Q., Zhao L., et al. (2013). Transcriptome exploration in Leymus chinensis under saline-alkaline treatment using 454 pyrosequencing. PLoS One 8:e53632 10.1371/journal.pone.0053632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweere U., Eichenberg K., Lohrmann J., Mira-Rodado V., Baurle I., Kudla J., et al. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294 1108–1111. 10.1126/science.1065022 [DOI] [PubMed] [Google Scholar]
- To J. P. C., Deruere J., Maxwell B. B., Morris V. F., Hutchison C. E., Ferreira F. J., et al. (2007). Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19 3901–3914. 10.1105/tpc.107.052662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.-S. P., Shinozaki K., Yamaguchi-Shinozaki K. (2010). Role of cytokinin responsive two-component system in ABA and osmotic stress signalings. Plant Signal. Behav. 5 148–150. 10.4161/psb.5.2.10411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L. S. P., Urao T., Qin F., Maruyama K., Kakimoto T., Shinozaki K., et al. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104 20623–20628. 10.1073/pnas.0706547105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kim J., Somers D. E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 110 761–766. 10.1073/pnas.1215010110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shen W., Chan Z., Wu Y. (2015). Endogenous cytokinin overproduction modulates ROS homeostasis and decreases salt stress resistance in Arabidopsis thaliana. Front. Plant Sci. 6:1004 10.3389/fpls.2015.01004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Hamamoto S., Uozumi N. (2013). Sodium transport system in plant cells. Front. Plant Sci. 4:410 10.3389/fpls.2013.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Chong J., Li C., Kim C., Shi D., Wang D. (2007). Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant Soil 294 263–276. 10.1007/s11104-007-9251-3 [DOI] [Google Scholar]
- Yang C. W., Shi D. C., Wang D. L. (2008). Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca (Bge.). Plant Growth Regul. 56 179–190. 10.1007/s10725-008-9299-y [DOI] [Google Scholar]
- Yang S., Zhang X., Yue J.-X., Tian D., Chen J.-Q. (2008). Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genomics 280 187–198. 10.1007/s00438-008-0355-0 [DOI] [PubMed] [Google Scholar]
- Yokoyama A., Yamashino T., Amano Y. I., Tajima Y., Imamura A., Sakakibara H., et al. (2007). Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 48 84–96. 10.1093/pcp/pcl040 [DOI] [PubMed] [Google Scholar]
- Zeng Q.-Y., Yang C.-Y., Ma Q.-B., Li X.-P., Dong W.-W., Nian H. (2012). Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 12:182 10.1186/1471-2229-12-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang J., Jiang W., Liu J., Yang S., Gai J., et al. (2016). Identification and analysis of NaHCO3 stress responsive genes in wild soybean (Glycine soja) roots by RNA-seq. Front. Plant Sci. 7:1842 10.3389/fpls.2016.01842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Bai X., Chen C., Chen Q., Cai H., Li Y., et al. (2011). GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol. Biol. 77 285–297. 10.1007/s11103-011-9810-0 [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6 441–445. 10.1016/S1369-5266(03)00085-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic trees of GsRR family of G. soja and Arabidopsis. Neighbor-joining phylogenetic tree of the response regulator members in G. soja and Arabidopsis. The tree was inferred by MEGA 5.0 with the neighbor-joining method after the alignment of the full-length amino acid sequences of the 56 G. soja genes and 24 Arabidopsis genes. The numbers beside the branches represent bootstrap values based on 1,000 replications. The scale bar corresponds to 0.1 estimated amino acid substitutions per site.
Distribution of conserved motifs. All motifs were identified by MEME using the complete amino acid sequences of GsRR genes.
Overexpression of GsRR2a in Arabidopsis did not affect the tolerance to salt stress. The WT and overexpression lines are grown on medium containing 0 or 150 mM NaCl. The germination rates were recorded and photos were taken after 6 days.
Structure analysis of GsRR genes using GSDS online tools. The UTRs (upstream/downstream sequences), exons and introns are shown with light blue boxes, yellow boxes, and black lines, respectively.
Gene-specific primers of GsRR family used for q-RT PCR assays.