Summary
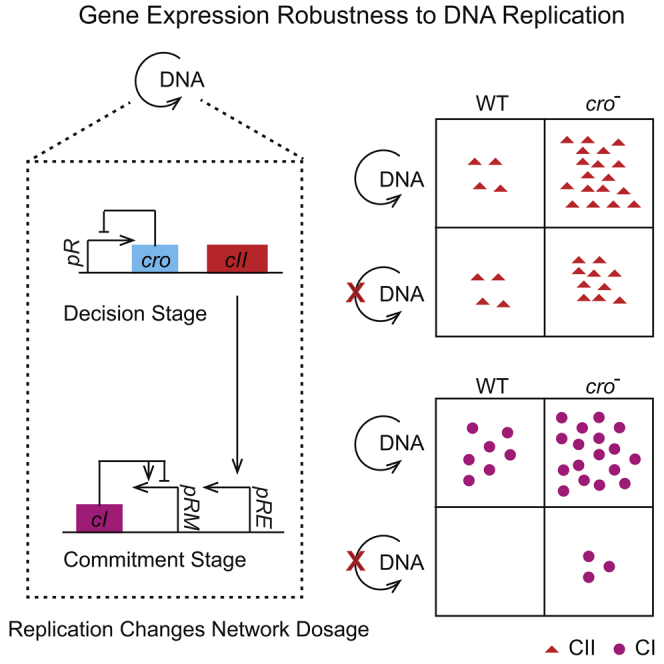
Cellular decision-making arises from the expression of genes along a regulatory cascade, which leads to a choice between distinct phenotypic states. DNA dosage variations, often introduced by replication, can significantly affect gene expression to ultimately bias decision outcomes. The bacteriophage lambda system has long served as a paradigm for cell-fate determination, yet the effect of DNA replication remains largely unknown. Here, through single-cell studies and mathematical modeling we show that DNA replication drastically boosts cI expression to allow lysogenic commitment by providing more templates. Conversely, expression of CII, the upstream regulator of cI, is surprisingly robust to DNA replication due to the negative autoregulation of the Cro repressor. Our study exemplifies how living organisms can not only utilize DNA replication for gene expression control but also implement mechanisms such as negative feedback to allow the expression of certain genes to be robust to dosage changes resulting from DNA replication.
Subject Areas: Gene Network, Microbial Genomics, Bioinformatics, Mathematical Biosciences
Graphical Abstract
Highlights
-
•
One single DNA is not able to commit to either lytic or lysogenic decision
-
•
DNA replication increases lysogenization frequency by boosting cI expression
-
•
CII expression is robust to DNA copy number changes resulting from DNA replication
-
•
Robustness of CII expression is due to Cro negative feedback
Gene Network; Microbial Genomics; Bioinformatics; Mathematical Biosciences
Introduction
Cellular decision-making often relies on the output of its gene regulatory networks, which are sensitive to intra- and extracellular signals (Balazsi et al., 2011, Perkins and Swain, 2009). In some cases, the gene network senses and responds to these signals by up- or downregulating the expression levels of certain genes to optimize the organism's survival (Kramer, 2010). In other cases, however, it is preferable for the cell's phenotype to remain insensitive (robust) to perturbations (Alon et al., 1999, Stelling et al., 2004, von Dassow et al., 2000). There are a great variety of perturbations that gene networks must mitigate, such as changes in physical conditions (e.g., temperature), the abundance of environmental toxins or nutrients, and the fluctuation of intracellular protein and toxin levels. In particular, one such perturbation to gene networks is DNA copy number fluctuation (e.g., by DNA replication), which virtually all living organisms must experience. Gene copy number changes can potentially affect gene expression to cause significant phenotypic changes (Baumgart et al., 2017, Kemkemer et al., 2002, Pollack et al., 2002, Rancati et al., 2008, Seidman and Seidman, 2002). For example, highly amplified genes in cancer cells often show elevated expression (Pollack et al., 2002), whereas inactivation of a single allele in diploid organisms can reduce expression and lead to diseases (Seidman and Seidman, 2002). Theoretical modeling also reveals that a number of commonly observed gene regulatory subnetworks, or network motifs, are sensitive to gene copy number changes due to interactions via a common pool of transcription factors in the duplicated networks (Mileyko et al., 2008).
Organisms can also develop strategies to deal with gene dosage changes and their cognate effects. A recent study reveals that in budding yeast, Saccharomyces cerevisiae, the expression of a large fraction of genes is significantly reduced when the gene dosage is halved, whereas some genes have unaltered expression (Springer et al., 2010). This suggests that there may exist mechanisms and control structures keeping a small number of genes robust to gene dosage variations. Indeed, some studies have reported network structures allowing gene expression insensitivity to DNA dosage changes (Acar et al., 2010, Song et al., 2014). However, those observations are most often based on gene deletions or insertions, and the effects of replication-associated temporal DNA copy number changes remain largely unknown. Given the prevalence of DNA replication and the fluctuation of DNA levels during cell growth, it is natural to ask how gene networks can cope with or take advantage of DNA replication to choose the optimal cell fate.
To understand the effect of DNA copy number fluctuations on gene expression and network level outputs, we use phage lambda as a model system to study how ongoing viral replication affects the lysis-lysogeny decision. The genetic components involved in this lysis-lysogeny decision have been well-characterized (Hendrix, 1983, Oppenheim et al., 2005, Ptashne, 2004). The default lytic pathway for lambda infection is executed by the transcription and translation of the lysis and phage morphological genes, which lead to the bursting of the cell and release of hundreds of phage progeny. These events are triggered when the anti-terminator protein, Q, reaches a threshold, allowing transcription from promoter pR′ to bypass the downstream terminator, tR′ (Cortes et al., 2017, Kobiler et al., 2005, Roberts et al., 1998). The alternative lysogenic pathway culminates in the integration of phage DNA into the E. coli chromosome, and the inhibition of gene transcription from the two major promoters pR and pL by repressor CI (Oppenheim et al., 2005). The master viral regulator, CII, plays a central role in controlling CI production by activating the pRE promoter. The choice between lytic and lysogenic development is therefore shaped by the cascade of regulatory genes expressed early in the infection process. After a decision is made, it is enforced by CI to establish the lysogenic pathway, or by cell destruction through lysis to complete the lytic pathway. Phage DNA replication starts shortly after the infection, causing a radical change in DNA copy number concordant with the expression of lysis-lysogeny decision-making genes. Here, we examine the effect of gene dosage change on the expression of two highly important decision regulators, CII and CI.
An alternative source of DNA copy number variation in the lambda infection system is the different MOI (Multiplicity of Infection, or number of infecting phages per cell), which determines the initial concentration of intracellular phage DNA. Current experimental evidence and theoretical models of phage lambda suggest that lysogenization is preferred at a higher MOI, i.e., higher initial DNA concentration (Cortes et al., 2017, Joh and Weitz, 2011, Kourilsky, 1973, Weitz et al., 2008, Zeng et al., 2010). A deterministic model of the CI/Cro bistable switch predicts that a high CI, low Cro state becomes the dominant attractor state when viral concentration is high (Weitz et al., 2008) because higher MOI leads to a higher transient spike in CII levels, causing an overshoot of CI via CII activation of the pRE promoter. This allows the CI/Cro bistable switch to flip to the direction of high CI and low Cro levels even if Cro was initially high, consistent with the lysogenic development (Kobiler et al., 2005). A quasi-stochastic version of this model indicated that this trend holds true even if lytic and lysogenic decisions are determined by threshold crossing of Q and CI, respectively, as opposed to steady state attractors (Joh and Weitz, 2011). Notably, these two models kept the viral DNA level constant throughout the dynamics by leaving out DNA replication, and just varied the infection MOI. Later, DNA replication was introduced in other models, which suggests that DNA replication promotes lysogeny by increasing DNA concentration similar to higher MOI (Cortes et al., 2017, Robb and Shahrezaei, 2014). However, thorough experimental interrogation of the regulation of decision-making by DNA replication is still lacking.
In this work, we experimentally characterize the role of DNA replication in the lysis-lysogeny decision of lambda phage and discover that CII levels remain robust to DNA replication, especially during the early infection period. Combined with mathematical modeling, our work suggests that negative feedback by Cro plays an important role in keeping CII expression robust to DNA replication. On the contrary, expression of cI, the downstream target gene of cII, is extremely sensitive to DNA copy number variations, thereby affecting the lysis-lysogeny decisions. Overall, we show that different elements of a gene network respond distinctly to copy number variations and demonstrate the potential of negative feedback regulation to encode phenotypic robustness against extremely variable DNA copy numbers.
Results
Lack of DNA Replication Leads to Failure in Lytic and Lysogenic Development
To understand how DNA replication may affect the lysis-lysogeny decision, we studied phage infection outcomes in the absence of replication. For this, we investigated how the DNA replication-defective λP- mutant (see Transparent Methods and Tables S1–S4 for more strain information) differs in its ability to make decisions compared with the wild-type (WT) laboratory strain λWT. The lysogenization frequency of λP- has been reported (Kourilsky, 1973) to be lower than that of λWT at low APIs (average phage input, calculated as plaque-forming unit [PFU]/colony-forming unit [CFU]). We confirmed this earlier finding (Figure S1A). In addition, the lysogenic response (percentage of lysogeny) of λP- phage to API follows a Poisson distribution of n ≥ 3, indicating that lysogenization requires 3 or more λP- phages on average, compared with n ≥ 2 for λWT (Figure S1B). This suggests that lysogenic decisions are possible in the absence of DNA replication, but more initial phage inputs are required to cause the same level of lysogenization.
To quantitatively detect the differences in decision-making behaviors in the presence and absence of phage DNA replication, we utilized our established lytic-lysogenic reporter systems (Trinh et al., 2017) to study the decision-making of λP- phage at the single-cell/single-phage level. Briefly, a fluorescent protein (mKO2) is inserted downstream of cI on the phage genome to report cI transcription activity, corresponding to lysogenic events by λWT infections (Figures 1A and S2). Another fluorescent protein (mTurquoise2) was fused to the C terminus of the phage capsid decoration protein, gpD. Thus, mTurquoise2 fluorescence reports lytic development up until host cell lysis for λWT infections (Figure 1A). This method also allows the quantification of MOI for each infection (Figure 1A, blue dot at 0 min). Overall, the λP- phage lysogenized less frequently than λWT (Figure 1C), as predicted by bulk experiments (Figure S1). Remarkably, λP- phage infections showed no lysogenic events at MOI = 1 (Figure 1C, 0 of 35 cells). In the lysogenic cells of MOI > 1, the cI reporter signal was lower than in λWT infections (Figures 1A and 1B), suggesting that cI transcription levels are lower in the absence of DNA replication. In addition, DNA replication is also required for cell lysis, as we only observed very low levels of the lytic reporter expression (Figure 1B). Accordingly, lysis did not occur within the time window of our time-lapse movies (4 hr) as opposed to λWT, where cells lysed at 114 ± 16 min (mean ± SD, N = 243). Overall, our data suggest that the decision-making network outputs, CI and the lysis genes, are severely compromised in the absence of DNA replication. The expression of these genes is regulated by their corresponding transcription factors, CII and Q, respectively. Therefore, we next sought to quantify how DNA replication affects the expression of these transcription factors from the pR promoter using single-molecule fluorescence in situ hybridization (smFISH).
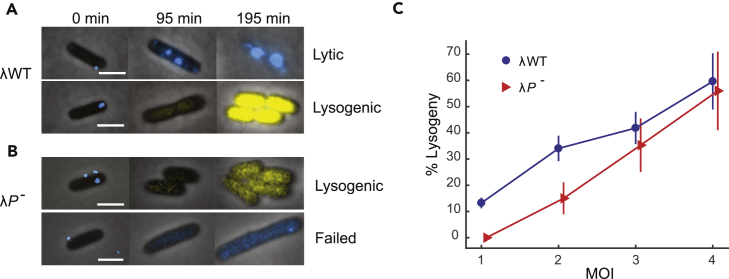
Figure 1.
More than One Copy of Phage DNA Is Required for Lysogenic Establishment
(A) Representative images showing lytic and lysogenic events by λWT. Top: a cell is apparently infected by one λWT phage (blue dot at 0 min), and subsequently gpD-mTurquoise2 expression (blue) is observed. Cell lysis is observed at 195 min. Bottom: a cell is apparently infected by one λWT phage (blue dot at 0 min). cI reporter expression (yellow) and cell division are observed, indicating a successful lysogenization event.
(B) Representative images of lysogenic and failed infection by λP-. Top: a cell is apparently infected by 3 λP- phages (blue dots at 0 min). The cell divides, and expression of the cI reporter (yellow) is observed, indicating lysogenization. Bottom: a cell is apparently infected by one λP- phage (blue dot at 0 min). The cell does not divide, and only minimal expression of gpD-mTurquoise2 is detected, indicating that the phage failed to reach either the lytic or lysogenic decision.
(C) Lysogenization frequency of λWT and λP-. For both phages, the lysogenization frequency increases with MOI. λP- has lower lysogenization frequencies at MOI ≤ 3, and reaches a similar level at MOI = 4. At MOI = 1, no lysogenization events (0 of 35 infections) are observed for λP-. Error bars denote SEM.
Scale bars denote 2 μm. See also Figures S1 and S2.
Single-Molecule Characterization of pR Transcription Activity after Phage Infection
Most of the key lysis-lysogeny-determining genes, including cII and Q, are located on the pR transcript (Figure 2A). Therefore, to determine the overall expression of cellular decision-controlling regulators, we quantified the level of pR transcription at the single-cell level using smFISH (see Transparent Methods and Table S5 for more details), by targeting the cII gene and its neighboring region, as an initial step to uncover the molecular mechanism of the decision-making process. In these experiments, we controlled the MOI by infecting with an API of 0.1–0.2. As the distribution of the number of phage particles per cell follows a Poisson distribution (Zeng et al., 2010), the estimated percentages of infected cells (cells with ≥1 infecting phages) at an API of 0.1 and 0.2 were 9.5% and 18.1%, respectively. Correspondingly, the estimated percentages of MOI = 1 infections within infected cells were 95.1% and 90.3%. Indeed, we observed that the percentage of cells showing cII signals ranges from 10.8% to 15.2% in multiple experiments (Figure 2B), indicating that the infection API is within the range of 0.1–0.2. Under these experimental conditions, most infections are at an MOI of 1. This minimized the effect of MOI, an important factor affecting the lysogenization frequency (Kourilsky, 1973), and therefore allowed us to focus on the role of DNA replication for cell-fate decisions by one single infecting phage. At 0 min after λWT phage infection, a small fraction of cells displayed one cII focus (Figure 2C), which likely corresponds to one single mRNA or a few mRNAs clustering together. At later time points, i.e., 6 min as shown in Figure 2D, the cII mRNA clusters become larger and brighter, indicating that cII mRNA level increases in cells over time. The percentage of cells showing cII transcription quickly reaches a plateau within the first 2 min of infection, indicating that gene expression closely follows phage infection (Figure 2B). To validate our mRNA detection method, we quantified the mRNA numbers from smFISH (see details in Transparent Methods and Figure S3) and then compared the average expression levels with data obtained by qRT-PCR. The data obtained from the two methods were in good agreement (Figure S4). Overall, the average cII mRNA level quickly peaked at around 6–12 min after infection, and subsequently dropped (Figure 2E), reflecting the repression of pR promoter by either CI or Cro (Kobiler et al., 2005, Oppenheim et al., 2005). Moreover, cII levels in different cells showed a wide population distribution (Figure 2F). For example, at 6 min, the number of cII molecules per cell has a mean of 38.8 and a coefficient of variation of 0.47.
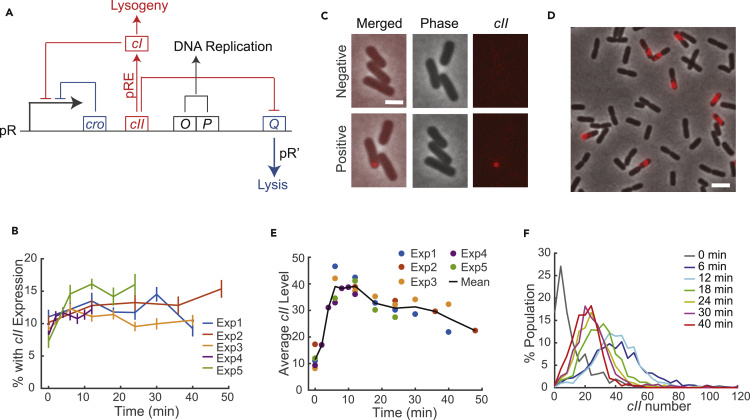
Figure 2.
Schematic of Lambda Lysis-Lysogeny Decision-Making and the Characterization of cII mRNA Expression
(A) The pR transcript includes the cro, cII, O, P, and Q genes. Cro and CI both repress the pR promoter. O and P are required for phage DNA replication. CII activates the expression of CI from the pRE promoter, whereas it represses Q through paQ. Q allows transcription of the lysis and morphogenesis genes from pR′.
(B) Percentage of cells showing cII expression after infection. Data from multiple experiments were shown. A plateau is reached after 2 min of infection, when samples were taken every 2 min (Exp4). Overall, between 10.8% (Exp3, averaged over 6–40 min) and 15.2% (Exp5, averaged over 6–24 min) of the cells show cII expression in multiple experiments, consistent with an API of 0.1–0.2. Error bars denote SEM.
(C) Representative images showing cells from the negative and positive samples. Top: cells without phage infection. None of the cells show cII signal. Bottom: cells with λWT infection at API = 0.2. Images were taken at 0 min. One cell shows a distinct focus (red). The other two cells do not show foci either because they have not started the mRNA expression, or they are not infected.
(D) Representative images showing cII mRNA expression at 6 min. cII mRNA appeared as clusters instead of punctate foci.
(E) Average cII levels (calculated as the average of all cells pooled from all 5 experiments) over time after λWT infection. Data from multiple experiments (dots) and the mean (black line) are shown. Only infected cells with cII expression were included in the calculation. cII expression reaches a peak at around 6–12 min after infection and subsequently drops.
(F) Distribution of cII mRNA levels at different time points. Combined data from Exp1, Exp3, and Exp5 (as in panels D and E) are shown. The cII mRNA distributions at 6 and 12 min are similar and gradually shift to the lower end after 18 min.
Scale bars denote 2 μm. See also Figures S3 and S4.
cII Expression Is Robust to Gene Dosage Changes Arising from DNA Replication
Having validated the smFISH method under our experimental settings, we proceeded to investigate the effect of DNA replication on the expression of transcription factor cII. Unexpectedly, λWT and λP- displayed similar levels of cII mRNA expression on average, especially through the first 18 min (Figure 3A). As these smFISH experiments were performed in lysogeny broth (LB) medium, where cell lysis typically occurs at ∼60 min and lysogenic decisions are reached within the first 20 min, the data suggest that cII expression is not affected by DNA replication in the time window when lysogenic decisions are processed. After 24 min, the cII level for λWT remains higher, whereas λP- seems to drop more rapidly and reach a lower level (Figure 3A). The reasons for the difference in cII level at the late time points can be complicated as different decisions are reached and different feedback regulations are in place. We therefore focused on the early time points (0–18 min) and asked if cII expression dynamics are altered in the absence of DNA replication to result in the decreased lysogenic frequency that we observed through live-cell movies (Figure 1C) and bulk lysogenic assays (Figure S1A). Surprisingly, we did not observe significant differences in the distribution of cII mRNA between λWT and λP- either (Figure 3B).
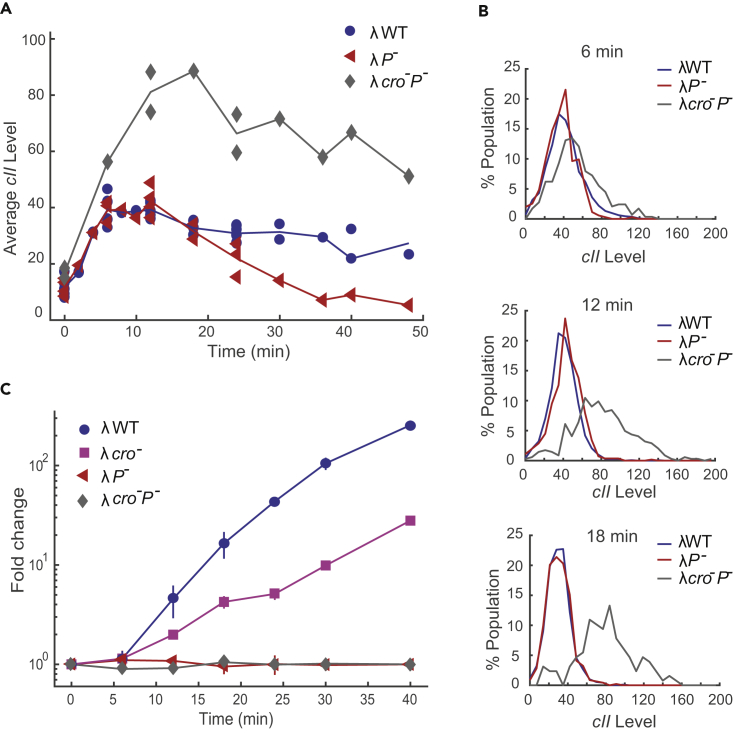
Figure 3.
Early cII Expression Is Not Affected by DNA Replication
(A) Average cII levels comparing λWT, λP-, and λcro-P- infections. Data from multiple experiments are shown, and solid lines represent the mean of all cells pooled from different experiments. In the first 18 min, λWT and λP- have similar cII levels. After 24 min of infection, average λWT cII level is higher compared with λP-. Average cII expression for λcro-P- infection is higher throughout the infection.
(B) Distribution of cII mRNA levels at 6, 12, and 18 min after infection. λWT and λP- have similar cII distributions, whereas λcro-P- infection shows higher cII expression levels.
(C) Average DNA level after infection. Fold change is calculated as the ratio of phage DNA to E. coli DNA normalized to time 0 and further normalized to the mean of λP- and λcro-P- data at the corresponding time point. For λWT and λcro- infection, phage DNA level increases by 256 ± 7-fold and 29 ± 5-fold, respectively, after 40 min of infection. Error bars denote SEM.
See also Figure S5.
To rule out the possibility that phage DNA replication simply does not occur until after 18 min, we quantified the phage DNA level using qPCR. As shown in Figure 3C (see Transparent Methods and Figure S5 for more details), the relative number of phage DNA to E. coli genome increases by 16.5 ± 5 (mean ± SEM) fold at 18 min after λWT infection, indicating that phages undergo a substantial amount of DNA replication within this time frame. On the contrary, λP- infection had no detectable replication, as expected (Figure 3C). Altogether, our results suggest that cII expression is robust to the gene dosage variations resulting from DNA replication during the early infection time window.
Having shown that λP- infection leads to the same levels of cII mRNA expression during the decision-making time window, we next wanted to confirm that the CII protein concentration in the λP- strain is also sufficient for the activation of the pRE promoter. We used a multi-copy plasmid, pRE-mCherry, to report the activation of pRE promoter by CII (Kobiler et al., 2005, Zeng et al., 2010). This system artificially increases the copy number of pRE promoter without affecting the decision-making of λWT phage (Zeng et al., 2010). We used EYFP-labeled fluorescent phages (green dots in Figures 4A and 4B) to quantify the MOI. We found that at MOI = 1, 62.9% (N = 835) of λP- and 50.5% (N = 101) of λWT infections were able to activate this reporter (Figure 4), further confirming that a single λP- DNA is capable of producing a sufficient amount of CII protein to activate the pRE promoter, an essential step in establishing lysogeny. In fact, the percentage activation by λP- was slightly higher than that of λWT infections, which might be due to additional effects of DNA replication on the expression of other genes that also affect the decision outcomes.
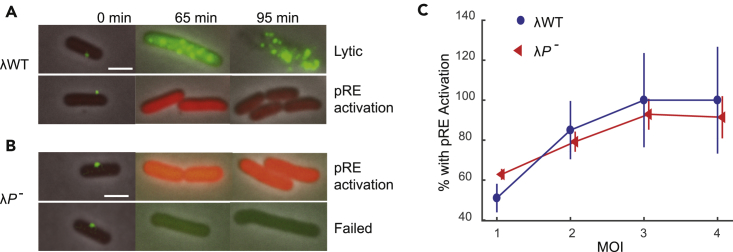
Figure 4.
Normal pRE Activation without DNA Replication
(A) Representative images of lytic and lysogenic events by λWT infections at MOI = 1. Top: expression of gpD-EYFP (green) is observed and the cell lyses at 95 min. Bottom: increase of mCherry (red) expression and normal cell division is observed, indicating a successful lysogenic event.
(B) Representative images showing λP- infections at MOI = 1. Top: increase in mCherry expression is observed, indicating the activation of pRE promoter. Low levels of gpD-EYFP expression are also detected in the cell. Bottom: only a very low level of gpD-EYFP expression is observed. Division is inhibited, and the cell keeps growing longer without lysing. Same contrast was applied to the images in (A) and (B) at 65 and 95 min. At 0 min, a different contrast if applied to see the green phage particles.
(C) Percentages of cells with pRE activation at different MOIs. Both phages show similar levels of pRE activation at different MOIs. Error bars denote SEM.
Scale bars denote 2 μm.
cI Expression Responds Strongly to Gene Dosage Changes Arising from DNA Replication
So far, our data suggest that the main effect of DNA replication during phage lysogenization is on downstream processes of cII expression. To understand why the lack of DNA replication decreases the lysogenic frequency, we next compared cI expression in the presence and absence of DNA replication. By simultaneously detecting cII and cI mRNA, we found that whereas 33.4 ± 1.7% (averaged over time points 24, 36, and 48 min, with N = 152, 126, and 152, respectively) λWT-infected cells show cI expression, very few λP--infected cells (0, 1, 2, 0, and 0 out of N = 62, 157, 111, 102, and 61 cells at time points 0, 12, 24, 36, and 48 min, respectively) express cI mRNA at API ≤ 0.2 (Figure 5A). Moreover, in those rarely observed cI-expressing cells infected by λP-, the expression level is very low (≤2 cI mRNA per cell), confirming a severe deficiency in cI expression in the absence of DNA replication.
Figure 5.
Low cI Expression without DNA Replication
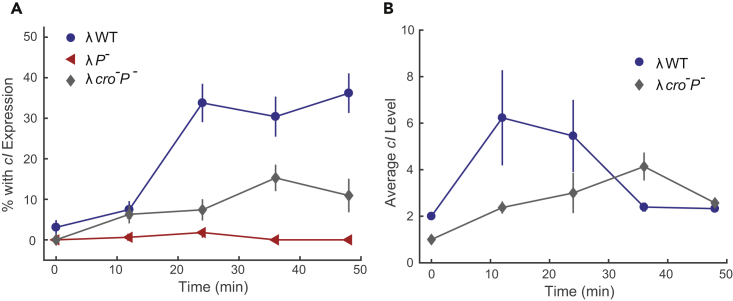
(A) Percentage of infected cells with cI expression. After 24 min, 33.4 ± 1.7% (averaged over 24, 36 and 48 min) of λWT infection leads to cI expression, which is higher than λcro-P-. Frequency of cI expression in λP- infection is very low. Error bars denote SEM.
(B) Comparison of average cI expression level. Only cells with cI expression are included for calculation. cI level for λWT peaks at around 12–24 min. λcro-P- has delayed cI peak time at 40 min with a lower peak level. Error bars denote SEM.
See also Figure S1.
To further dissect the mechanism of lysogenic establishment, we tested a λcro-P- double mutant. Due to the absence of the repressor Cro, this double mutant has much higher cII mRNA levels compared with λWT (Figures 3A and 3B). With this mutant, we characterized how a single phage DNA responds to elevated cII levels. We found that cells infected by this mutant phage showed cI mRNA expression less frequently, despite having higher cII levels than the λWT infections (Figure 5A). The average cI expression level for λcro-P- is also lower (Figure 5B), suggesting that this phage cannot effectively carry out the lysogenic decision despite having ample expression of cII. Our bulk lysogenization assay also showed that λcro-P- does not lysogenize as frequently as λWT (Figure S1C). Altogether, the data suggest that CII is not the only key factor regulating cI expression and lysogenic decisions, and that a single phage DNA is generally incompetent at consummating lysogeny. An increase in DNA copy number is important to enable the production of additional cI transcripts to boost CI levels.
Cro Negative Feedback Enables cII Expression Robustness against Replication-Associated Gene Dosage Variations
To systematically understand how the viral gene network encodes cII robustness to viral DNA replication, we computationally simulated the network behavior in the infection process by adapting a published model (Cortes et al., 2017). This model includes the key components of the decision-making network, namely, Cro, CI, and CII and their interactions, as well as DNA replication (Figures 2A and S6, Tables S6 and S7). The goal of the model is to capture the important behaviors of the true network, not to account for all of its known features. This model simulates transcription of pR mRNA, which is then translated into CII and Cro proteins. CII activates the pRE promoter, driving the transcription and translation of the cI gene. We phenomenologically modeled the repression of the pR promoter by CI and Cro using Hill functions (Cortes et al., 2017, Joh and Weitz, 2011, Robb and Shahrezaei, 2014, Weitz et al., 2008). Likewise, we used Hill functions to model the CI activation and Cro repression of pRM promoter. As we focused on the early decision-making phase of infection when CI is not highly expressed, we did not include the CI repression on pRM promoter for simplicity. We also modeled viral DNA replication and its effects on CI, CII, and Cro expression. Lastly, we phenomenologically modeled the repression of CI on DNA replication using a Hill function. The cII expression level predicted by this model agreed closely with the experimental data (Figure 6A), indicating that the interactions in the model were sufficient to capture the robustness of CII levels to DNA replication. We have also built a more complex model where the binding and unbinding reactions of transcription factors to their cognate promoters are simulated in detail. This model has similar performance to the simplified model that we presented above (see Transparent Methods, Figure S7, Tables S8 and S9 and Data S1 for more details), therefore, only the results of the simplified model will be further discussed here.
Figure 6.
Cro Is Important for the Robustness of cII Expression to DNA Replication
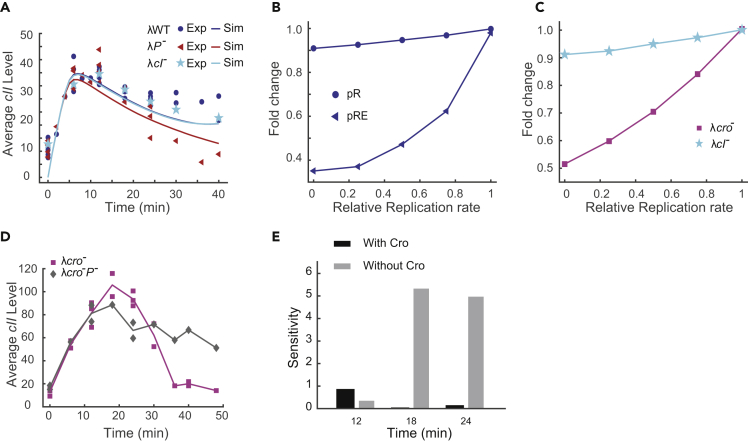
(A) The model prediction of the mean cII level in comparison with the experimental data. The model captures the average cII expression for λWT, λP-, and λcI-. Both experimental quantification and model prediction show that λcI- mutant has similar cII expression levels to λWT and λP- at the early infection period (0–20 min).
(B) Model prediction of the relative pR and pRE mRNA levels under different replication rates. Under the model assumptions, pR transcription level shows minimal changes as the replication rate varies between 0- and 1-fold of the original replication rate, whereas the pRE level varies greatly, agreeing with experimental observations.
(C) Model prediction of the sensitivity of pR mRNA levels for λcro- and λcI- mutants. Removing Cro from the model gives rise to pR transcription sensitivity to DNA replication, whereas removing CI does not cause significant changes.
(D) Experimental validation of the role of Cro in cII expression robustness to DNA replication. In the cro- background, removing DNA replication (λcro-P-) causes a decrease of cII level at 18 and 24 min.
(E) cII expression sensitivity to DNA replication. The sensitivity is calculated as the absolute difference in cII level between λWT and λP- (with Cro) or λcro- and λcro-P- (without Cro) at each time point divided by the DNA number difference within each group. In the absence of Cro, the cII expression sensitivity to DNA replication greatly increases, as shown at 18 and 24 min.
See also Figures S6 and S7.
We next sought to understand which network features or components give rise to the cII expression robustness to DNA replication. Examining this network, we found two network features regulating pR mRNA transcription: the Cro negative feedback regulation and CI negative feedback regulation (Figure 2A). Negative feedback is a network motif that has been shown to possess properties such as reducing gene expression noise and linearizing gene expression level to the input signal (Becskei and Serrano, 2000, Nevozhay et al., 2009). To determine whether any of these two negative feedback regulatory links alone is responsible for cII mRNA expression robustness, we systematically removed the effect of CI and Cro on pR transcription and DNA replication to generate computational mutants with compromised feedback regulation. To examine the sensitivity of mRNA expression to different rates of DNA replication for the various computational mutants, we first calculated for each time point t from 6 to 24 min the average fold change, F (ɛ, t), defined as the ratio of mRNA levels at replication rate ɛ⋅r (for 0 ≤ ɛ ≤ 1) versus mRNA levels at the WT replication rate r. Then, we calculated the time-average of this quantity, F(ε), to get the sensitivity over all time points. In the WT background, the model successfully predicted the cII expression robustness to DNA replication (Figure 6B). Moreover, CI removal did not compromise the robustness of cII expression to a wide range of replication rates (Figure 6C). On the contrary, removing Cro significantly increases the sensitivity of cII expression to DNA replication (Figure 6C), indicating the importance of Cro for the robustness of cII expression to DNA replication.
To validate these theoretical predictions, we then experimentally probed the expression of cII in different mutant backgrounds. As expected, cII expression was similar among the λcI-, λP-, and λWT infections (Figure 6A), suggesting that CI is not essential for cII expression robustness under our experimental settings. We further tested the role of Cro negative feedback by comparing cII expression level between λcro- and λcro-P-. As shown in Figure 6D, λcro-P- infection leads to lower cII levels at 18 and 24 min compared with λcro-. As the replication rate of λcro- phage is lower compared with λWT, we then calculated the cII expression sensitivity as the mean cII level change per extra DNA (Figure 6E), which indicated robustness. Altogether, our data suggest that Cro can overcome the variations of gene dosages to result in cII expression robustness. To understand the difference between the contributions of CI and Cro to the cII expression robustness, we examined the timing and expression of CI versus Cro. CI is only expressed in cells entering the lysogenic state (33.4% for λWT at MOI = 1, Figure 5A), whereas Cro is present in all cells. Moreover, by the time DNA replication starts (∼6 min), there is most likely a substantial amount of Cro present already, since Cro is one of the first two genes to be expressed during infection. On the contrary, cI expression only starts when CII protein reaches a certain threshold. It is therefore possible that the different contributions of CI and Cro to cII expression robustness are due to their difference in the timing and magnitude of expression, yet further experimentation is needed to provide more support.
Discussion
The lysis-lysogeny decision-making of bacteriophage lambda has long served as a paradigm for studying stochastic cell-fate selection, due to the well-established genetic networks involved (Oppenheim et al., 2005). Following decades of studies, researchers have characterized the effects of most genetic components (Hendrix, 1983) and built models to understand this process systematically (Arkin et al., 1998, Joh and Weitz, 2011, Weitz et al., 2008). However, due to the limit of resolution in previous experimental approaches, the effect of DNA copy number changes resulting from DNA replication on the decision-making process has been largely neglected, which may have obscured important aspects of this process. Here, we provided more quantitative measurements of their effects on gene expression, decision-making, and the enforcement of the cell-fate decisions.
Gene expression and DNA replication are both partly stochastic processes (Elowitz et al., 2002, Kaern et al., 2005, Raj and van Oudenaarden, 2008), and in the lambda network, DNA replication and the expression of genes, such as Cro, can affect each other. O and P, proteins required for phage DNA replication, are under the control of pR promoter, and their expression is affected by Cro and CI. In addition, replication initiation seems to require active pR transcription (Dove et al., 1969, Mensa-Wilmot et al., 1989), whereas Cro and CI both repress pR promoter activities. On the other hand, pR transcription goes in the opposite direction of DNA replication, originating from λori located at the O gene region. As head-on collisions between transcription and replication have been shown to slow replication (Merrikh et al., 2012, Pomerantz and O'Donnell, 2010, Soultanas, 2011), pR transcription and phage DNA replication might be constantly affecting each other as well. This dynamic interplay may have interesting impacts on the level of gene expression and DNA replication. Due to the limit of our experimental approaches, we cannot follow the copy number changes of phage DNA simultaneously with the expression of genes. As a result, the exact cellular concentrations of phage DNA and cII and cI mRNAs at the time of lysogenic establishment are unknown. Future experiments with higher resolutions are needed to allow the examination of the correlations of DNA copy number with cell-fate selection.
Increasing the copy number of promoters can titrate transcription factors and lead to complex dosage response (Lee and Maheshri, 2012, Rydenfelt et al., 2014). Using a simple repression regulatory architecture, researchers have found that at a high transcription factor:promoter ratio, the gene expression response is similar to that of a single isolated copy of the gene (Brewster et al., 2014). Networks invariant to gene dosage have also been reported. In Saccharomyces cerevisiae, the activity of the galactose signaling network (GAL network) was invariant to the network dosage changes when the gene copy number was halved (Acar et al., 2010). The finding was further generalized by mathematical simulations to conclude that for any N-component network, containing a 2-component subnetwork with an activator, an inhibitor, as well as a 1:1 stoichiometry interaction between the activator and inhibitor is both necessary and sufficient for the network to be dosage compensated (Acar et al., 2010, Song et al., 2014). An alternative mechanism that provides robustness to DNA copy number changes is an incoherent feedforward loop with DNA copy number as its source node (Segall-Shapiro et al., 2018).
Notably, in these studies, the copy number changes are usually introduced through deletion and insertion of genes, or changing the plasmid copy numbers, and it is possible that the effects of dosage variations resulting from DNA replication can be different. In the lambda system, both MOI and DNA replication affect decision-making by affecting the DNA concentration, although the response of cII expression to these two factors seems to be different. Experimentation and theoretical modeling have suggested that the difference might be due to Cro. Specifically, increasing the MOI leads to the introduction of more copies of DNA into a cell in which Cro is not present yet. On the contrary, by the time DNA replication starts, Cro has been expressed to a certain level such that negative feedback by Cro counteracts the effect of increasing template number on cII expression.
Strategies such as negative feedback are commonly utilized by gene regulatory networks to increase gene expression stability (Becskei and Serrano, 2000), or to linearize the input-output response of the genes (Nevozhay et al., 2009). By reducing the gene expression noise, networks can achieve more ordered, “deterministic” outcomes, as reliability is important for many cellular processes. For phage lambda, as a repressor, Cro has been shown to perform multiple important roles in the development of the lytic pathway (Court et al., 2007, Johnson et al., 1978, Ptashne et al., 1980, Svenningsen et al., 2005). Interestingly, by artificially inserting Cro onto the E. coli chromosome and placing Cro under the control of pR with one single operator binding site, oR1 (as opposed to three: oR1, oR2, and oR3, in the native lambda genome), researchers observed oscillations of Cro expression, synchronized to the cell cycle and the associated gene copy changes (Hensel and Marquez-Lago, 2015). However, the period of these oscillations were several fold longer than the time frame of lambda phage decision-making right after infection. Here, our work suggests that Cro-mediated negative feedback can stabilize the activity of the pR promoter against DNA copy number variations resulting from DNA replication, thus creating conditions wherein the early decision-making process is not affected by DNA replication. Since Cro plays two major roles in the network, both to repress pR promoter activity and to modulate the rate of DNA replication, these two functions may seem convoluted in providing cII expression robustness. However, it is hard to decouple the two roles of Cro both experimentally and theoretically due to the complex interplay between Cro and DNA replication as discussed. Future work to elucidate the mechanism of regulation on DNA replication by Cro as well as the development of mutants to separate the two functions of Cro will be of great interest.
Whether or not transcription factor expression robustness to network dosage changes confers any evolutionary advantages to the lambda decision-making circuit remains unknown. Due to the different physiological state or growth phase of the host cell, the levels of DNA replication may be different in different cells. The resource level, i.e., the available DNA polymerase, may fluctuate in response to different environmental stimuli to result in changes in the rate of DNA replication. To address these potential changes, lambda seems to adapt by allowing its effector (cI) expression to respond to the DNA copy number changes rather than altering the actual decision-making (cII expression) behavior. Recent studies showed that dosage-compensating networks can act as a noise reduction module to reduce the effects of extrinsic noise on the network output (Peng et al., 2016). In the lambda system, the behavior of the decision-making circuit in response to fluctuations in different cellular factors remain unknown.
Overall, our study has shown that the lambda decision-making process is composed of an intricate network where both MOI changes and replicating DNA can significantly affect the outcomes. However, the lambda network is far more complicated than what we described here. CIII is also an important factor for lysogenic establishment by promoting CII stability (Herman et al., 1997, Hoyt et al., 1982), and its expression might also be affected by DNA replication, yet it is neglected in our models for simplicity. The anti-terminator N is critical for phage development by allowing the transcription to go beyond N and Cro production at the immediately early stage, and possibly regulates the temporal progression of gene expression as well as decision-making (Oppenheim et al., 2005). On the other hand, the fate-determining genes on the pR transcript, although promoted from the same promoter, are in fact separated by several terminators, tR1–4 (Casjens and Hendrix, 2015, Oppenheim et al., 2005). Although N can allow transcription to go past those terminators, the efficiencies may vary (Gusarov and Nudler, 2001). This adds another layer of regulation, and a more systematic examination of gene expression is required to fully understand the lambda decision-making network.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We are grateful to Ryland Young and Chenxi Qiu for commenting on the earlier versions of the manuscript. We thank Ryland Young and Donald Court for gifting strains, and Ido Golding, Samuel O Skinner, Leonardo A. Sepulveda, and Mengyu Wang for helpful suggestions on the smFISH experiments and data analysis. This research was supported by NIH-NIGMS grant R01GM107597 and Administrative Supplement 3R01GM107597-02S1 to L.Z. and G.B., as well as by NIH-NIGMS grant R35GM122561 and by a Laufer Center for Physical and Quantitative Biology endowment to G.B.
Author Contributions
Q.S., J.T.T., and L.Z. designed the experiments. Q.S., J.T.T., and J.G. conducted the experiments. Q.S. and J.T.T. analyzed the data. M.G.C., Q.S., and G.B. built the mathematical models. Q.S., M.G.C., J.T.T., G.B., and L.Z. wrote the paper. G.B. and L.Z. supervised the project.
Declaration of Interests
The authors declare no competing interests.
Published: August 31, 2018
Footnotes
Supplemental Information includes Transparent Methods, seven figures, nine tables, and one data file and can be found with this article online at https://doi.org/10.1016/j.isci.2018.07.006.
Contributor Information
Gábor Balázsi, Email: gabor.balazsi@stonybrook.edu.
Lanying Zeng, Email: lzeng@tamu.edu.
Supplemental Information
References
- Acar M., Pando B.F., Arnold F.H., Elowitz M.B., van Oudenaarden A. A general mechanism for network-dosage compensation in gene circuits. Science. 2010;329:1656–1660. doi: 10.1126/science.1190544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U., Surette M.G., Barkai N., Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- Arkin A., Ross J., McAdams H.H. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazsi G., van Oudenaarden A., Collins J.J. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart L., Mather W., Hasty J. Synchronized DNA cycling across a bacterial population. Nat. Genet. 2017;49:1282–1285. doi: 10.1038/ng.3915. [DOI] [PubMed] [Google Scholar]
- Becskei A., Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Brewster R.C., Weinert F.M., Garcia H.G., Song D., Rydenfelt M., Phillips R. The transcription factor titration effect dictates level of gene expression. Cell. 2014;156:1312–1323. doi: 10.1016/j.cell.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S.R., Hendrix R.W. Bacteriophage lambda: early pioneer and still relevant. Virology. 2015;479-480:310–330. doi: 10.1016/j.virol.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes M.G., Trinh J.T., Zeng L., Balazsi G. Late-arriving signals contribute less to cell-fate decisions. Biophys. J. 2017;113:2110–2120. doi: 10.1016/j.bpj.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D.L., Oppenheim A.B., Adhya S.L. A new look at bacteriophage lambda genetic networks. J. Bacteriol. 2007;189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove W.F., Hargrove E., Ohashi M., Haugli F., Guha A. Replicator activation in lambda. Jpn. J. Genet. 1969;44:11–22. [Google Scholar]
- Elowitz M.B., Levine A.J., Siggia E.D., Swain P.S. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Gusarov I., Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- Hendrix R.W. Cold Spring Harbor Laboratory; 1983. Lambda II. [Google Scholar]
- Hensel, Z. and Marquez-Lago, T.T. (2015). Cell-cycle-synchronized, oscillatory expression of a negatively autoregulated gene in E. coli. q-bioQM, arXiv:1506.08596v1.
- Herman C., Thevenet D., D'Ari R., Bouloc P. The HflB protease of Escherichia coli degrades its inhibitor lambda cIII. J. Bacteriol. 1997;179:358–363. doi: 10.1128/jb.179.2.358-363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Knight D.M., Das A., Miller H.I., Echols H. Control of phage lambda development by stability and synthesis of cII protein: role of the viral cIII and host hflA, himA and himD genes. Cell. 1982;31:565–573. doi: 10.1016/0092-8674(82)90312-9. [DOI] [PubMed] [Google Scholar]
- Joh R.I., Weitz J.S. To lyse or not to lyse: transient-mediated stochastic fate determination in cells infected by bacteriophages. PLoS Comput. Biol. 2011;7:e1002006. doi: 10.1371/journal.pcbi.1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A., Meyer B.J., Ptashne M. Mechanism of action of the cro protein of bacteriophage lambda. Proc. Natl. Acad. Sci. USA. 1978;75:1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M., Elston T.C., Blake W.J., Collins J.J. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Kemkemer R., Schrank S., Vogel W., Gruler H., Kaufmann D. Increased noise as an effect of haploinsufficiency of the tumor-suppressor gene neurofibromatosis type 1 in vitro. Proc. Natl. Acad. Sci. USA. 2002;99:13783–13788. doi: 10.1073/pnas.212386999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiler O., Rokney A., Friedman N., Court D.L., Stavans J., Oppenheim A.B. Quantitative kinetic analysis of the bacteriophage lambda genetic network. Proc. Natl. Acad. Sci. USA. 2005;102:4470–4475. doi: 10.1073/pnas.0500670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourilsky P. Lysogenization by bacteriophage lambda. I. Multiple infection and the lysogenic response. Mol. Gen. Genet. 1973;122:183–195. doi: 10.1007/BF00435190. [DOI] [PubMed] [Google Scholar]
- Kramer R. Bacterial stimulus perception and signal transduction: response to osmotic stress. Chem. Rec. 2010;10:217–229. doi: 10.1002/tcr.201000005. [DOI] [PubMed] [Google Scholar]
- Lee T.H., Maheshri N. A regulatory role for repeated decoy transcription factor binding sites in target gene expression. Mol. Syst. Biol. 2012;8:576. doi: 10.1038/msb.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensa-Wilmot K., Carroll K., McMacken R. Transcriptional activation of bacteriophage lambda DNA replication in vitro: regulatory role of histone-like protein HU of Escherichia coli. EMBO J. 1989;8:2393–2402. doi: 10.1002/j.1460-2075.1989.tb08369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H., Zhang Y., Grossman A.D., Wang J.D. Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol. 2012;10:449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileyko Y., Joh R.I., Weitz J.S. Small-scale copy number variation and large-scale changes in gene expression. Proc. Natl. Acad. Sci. USA. 2008;105:16659–16664. doi: 10.1073/pnas.0806239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevozhay D., Adams R.M., Murphy K.F., Josic K., Balazsi G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:5123–5128. doi: 10.1073/pnas.0809901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim A.B., Kobiler O., Stavans J., Court D.L., Adhya S. Switches in bacteriophage lambda development. Annu. Rev. Genet. 2005;39:409–429. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- Peng W., Song R., Acar M. Noise reduction facilitated by dosage compensation in gene networks. Nat. Commun. 2016;7:12959. doi: 10.1038/ncomms12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins T.J., Swain P.S. Strategies for cellular decision-making. Mol. Syst. Biol. 2009;5:326. doi: 10.1038/msb.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack J.R., Sorlie T., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz R.T., O'Donnell M. What happens when replication and transcription complexes collide? Cell Cycle. 2010;9:2537–2543. doi: 10.4161/cc.9.13.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Cold Spring Harbor Laboratory Press; 2004. Genetic Switch: Phage Lambda Revisited. [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A.D., Maurer R., Meyer B.J., Pabo C.O., Roberts T.M., Sauer R.T. How the lambda repressor and cro work. Cell. 1980;19:1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Raj A., van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G., Pavelka N., Fleharty B., Noll A., Trimble R., Walton K., Perera A., Staehling-Hampton K., Seidel C.W., Li R. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb M.L., Shahrezaei V. Stochastic cellular fate decision making by multiple infecting lambda phage. PLoS One. 2014;9:e103636. doi: 10.1371/journal.pone.0103636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.W., Yarnell W., Bartlett E., Guo J., Marr M., Ko D.C., Sun H., Roberts C.W. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb. Symp. Quant. Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Rydenfelt M., Cox R.S., 3rd, Garcia H., Phillips R. Statistical mechanical model of coupled transcription from multiple promoters due to transcription factor titration. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2014;89:012702. doi: 10.1103/PhysRevE.89.012702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall-Shapiro T.H., Sontag E.D., Voigt C.A. Engineered promoters enable constant gene expression at any copy number in bacteria. Nat. Biotechnol. 2018;36:352–358. doi: 10.1038/nbt.4111. [DOI] [PubMed] [Google Scholar]
- Seidman J.G., Seidman C. Transcription factor haploinsufficiency: when half a loaf is not enough. J. Clin. Invest. 2002;109:451–455. doi: 10.1172/JCI15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R., Liu P., Acar M. Network-dosage compensation topologies as recurrent network motifs in natural gene networks. BMC Syst. Biol. 2014;8:69. doi: 10.1186/1752-0509-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P. The replication-transcription conflict. Transcription. 2011;2:140–144. doi: 10.4161/trns.2.3.15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Weissman J.S., Kirschner M.W. A general lack of compensation for gene dosage in yeast. Mol. Syst. Biol. 2010;6:368. doi: 10.1038/msb.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J., Sauer U., Szallasi Z., Doyle F.J., Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Svenningsen S.L., Costantino N., Court D.L., Adhya S. On the role of Cro in lambda prophage induction. Proc. Natl. Acad. Sci. USA. 2005;102:4465–4469. doi: 10.1073/pnas.0409839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh J.T., Szekely T., Shao Q., Balazsi G., Zeng L. Cell fate decisions emerge as phages cooperate or compete inside their host. Nat. Commun. 2017;8:14341. doi: 10.1038/ncomms14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G., Meir E., Munro E.M., Odell G.M. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Weitz J.S., Mileyko Y., Joh R.I., Voit E.O. Collective decision making in bacterial viruses. Biophys. J. 2008;95:2673–2680. doi: 10.1529/biophysj.108.133694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Skinner S.O., Zong C., Sippy J., Feiss M., Golding I. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell. 2010;141:682–691. doi: 10.1016/j.cell.2010.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.