Abstract
Objective: In this study, we aimed to expand current knowledge of head and neck squamous cell carcinoma (HNSCC)-associated long noncoding RNAs (lncRNAs), and to discover potential lncRNA prognostic biomarkers for HNSCC based on next-generation RNA-seq. Methods: RNA-seq data of 546 samples from patients with HNSCC were downloaded from The Cancer Genome Atlas (TCGA), including 43 paired samples of tumor tissue and adjacent normal tissue. An integrated analysis incorporating differential expression, weighted gene co-expression networks, functional enrichment, clinical parameters, and survival analysis was conducted to discover HNSCC-associated lncRNAs. The function of CYTOR was verified by cell-based experiments. To further identify lncRNAs with prognostic significance, a multivariate Cox proportional hazard regression analysis was performed. The identified lncRNAs were validated with an independent cohort using clinical feature relevance analysis and multivariate Cox regression analysis. Results: We identified nine HNSCC-relevant lncRNAs likely to play pivotal roles in HNSCC onset and development. By functional enrichment analysis, we revealed that CYTOR might participate in the multistep pathological processes of cancer, such as ribosome biogenesis and maintenance of genomic stability. CYTOR was identified to be positively correlated with lymph node metastasis, and significantly negatively correlated with overall survival (OS) and disease free survival (DFS) of HNSCC patients. Moreover, CYTOR inhibited cell apoptosis following treatment with the chemotherapeutic drug diamminedichloroplatinum (DDP). HCG22, the most dramatically down-regulated lncRNA in tumor tissue, may function in epidermis differentiation. It was also significantly associated with several clinical features of patients with HNSCC, and positively correlated with patient survival. CYTOR and HCG22 maintained their prognostic values independent of several clinical features in multivariate Cox hazards analysis. Notably, validation either based on an independent HNSCC cohort or by laboratory experiments confirmed these findings. Conclusions: Our transcriptomic analysis suggested that dysregulation of these HNSCC-associated lncRNAs might be involved in HNSCC oncogenesis and progression. Moreover, CYTOR and HCG22 were confirmed as two independent prognostic factors for HNSCC patient survival, providing new insights into the roles of these lncRNAs in HNSCC as well as clinical applications.
Keywords: Head and neck squamous cell carcinoma, Long noncoding RNA (lncRNA), Weighted gene co-expression network analysis (WGCNA), Clinicopathological feature, Multivariate Cox regression model
1. Introduction
Head and neck squamous cell carcinoma (HNSCC), the sixth leading cancer worldwide, is characterized by phenotypic, aetiological, biological, and clinical heterogeneity (Leemans et al., 2011). Several factors and clinical features have been implicated to be predictive for HNSCC development, including age, sex, smoking, alcohol, tumor stage, nodal status, and human papillomavirus (HPV) status. However, the prognosis of HNSCC patients is still a formidable challenge on account of distant metastasis, therapeutic resistance, or invasive recurrence, which frequently results in a lethal malignancy (Seiwert et al., 2007; Gold et al., 2009). A thorough understanding of HNSCC pathogenesis and the identification of new disease markers would be extremely useful for selecting suitable therapeutic regimens and improving the prognosis of HNSCC patients (Pritzker, 2015).
Long noncoding RNAs (lncRNAs), a class of noncoding RNAs with a length greater than 200 nucleotides, generally exert a regulatory function at the RNA level. Accumulating evidence suggests that lncRNAs play critical roles in almost every aspect of cell biology, including carcinogenesis (Cheetham et al., 2013; Huarte, 2015; Bartonicek et al., 2016; Schmitt and Chang, 2016; Schmitt et al., 2016). This makes lncRNAs a hot target for translational medicine (Salyakina and Tsinoremas, 2016) in addition to microRNAs and transcription factors (Salazar et al., 2014; Miller et al., 2015; Yan et al., 2016). Taking HOX transcript antisense RNA (HOTAIR) as an example, the best known regulatory mechanism of this lncRNA is to act as a scaffold molecule and form a complex with Polycomb repressive complex 2 (PRC2) to modulate the H3K27me3 level of downstream target genes (Tsai et al., 2010). HOTAIR is highly overexpressed in almost all tested cancer types, and functions as an oncogene in cultured cells (Geng et al., 2011; Kim et al., 2013; Li DD et al., 2013; Li X et al., 2013; Hajjari and Salavaty, 2015). Recently, we conducted a meta-analysis, and clarified that HOTAIR is positively correlated with tumor development and negatively correlated with clinical outcomes (Min et al., 2017). However, the number of lncRNAs is vast. Currently, the GENCODE project has collected about 16 000 lncRNAs, and this number is still climbing due to the development of more robust and sensitive techniques. HOTAIR and other functionally characterized lncRNAs represent only a very small proportion of the total number of annotated lncRNAs (Quek et al., 2015; Signal et al., 2016). Therefore, our knowledge of the majority of lncRNAs remains elusive.
The advancement of RNA sequencing and computational techniques over the past decade has facilitated the generation of rich and diverse information about lncRNAs. Previous transcriptome analyses identified numerous dysregulated lncRNAs which play regulatory roles in multiple cellular processes such as genomic mutation and DNA methylation. However, these studies were either based on microarray analysis focusing on a particular type of HNSCC such as oral squamous cell carcinoma (OSCC) (Zhang et al., 2015) and tongue squamous cell carcinoma (TSCC) (Yu et al., 2017), or based on a relatively small number of HNSCC samples (426 RNA-seq) and lncRNA molecules (500 lncRNAs) (de Lena et al., 2017). In this work, we focused on a more comprehensive dataset with a 28% larger sample size and 2787 lncRNAs. We subsequently performed an integrated transcriptomic analysis incorporating differential expression, weighted gene co-expression networks, functional enrichment, clinical parameters, survival analysis, and a multivariate Cox regression model on 546 RNA-seq datasets, aiming to uncover potential HNSCC-associated lncRNAs and demonstrate their functional roles, and reveal their prognostic signature for patient survival, followed by validation using an independent cohort.
2. Materials and methods
2.1. Data source
The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov) is a comprehensive and coordinated effort to accelerate our understanding of the molecular basis of cancer through the application of genome analysis technologies. Here, we downloaded level 3 RNA-seq data and clinical data of corresponding patients from the TCGA via Genomic Data Commons (GDC; https://portal.gdc.cancer.gov). In total, there were 546 RNA-seq datasets from 501 patients, including data from 43 paired samples from tumor tissue and adjacent normal tissue. From one patient, only a normal tissue sample was available. Two types of expression data were used in this study: upper quartile normalized fragments per kilobase per million mapped reads (FPKM-UQ), and raw counts calculated with HTSeq-count (Anders et al., 2015). Genes in the expression profile table had been annotated with Ensembl gene ID (Aken et al., 2016; Yates et al., 2016) by TCGA. All genes were annotated using BioMart (Kinsella et al., 2011), and lncRNAs were then confirmed with GENCODE annotation (Derrien et al., 2012; Harrow et al., 2012; https://www.gencodegenes.org, Version 26).
2.2. Differential expression analysis
The R platform (https://www.r-project.org, Version 3.32) was adopted for data manipulation and statistical analysis (R Development Core Team, 2011). Differential expression analysis was performed using R package edgeR (Version 3.16.5) (Robinson et al., 2010; McCarthy et al., 2012) based on the 43 paired samples (43 tumor tissue samples and 43 adjacent normal tissue samples). Genes with a count per million reads (CPM) (Law et al., 2014) higher than 1 in more than 10 samples were selected for the following analyses. Briefly, Ensembl gene IDs were annotated to gene symbol and Entrez gene IDs to facilitate the following functional profiling. The Benjamini-Hochberg (BH) method was used to correct the P value to a false discovery rate (FDR) for multiple comparisons (Benjamini and Hochberg, 1995). Genes with log2 fold change (log2FC) ≥1 and FDR≤0.05 were considered as significantly up-regulated, while those with log2FC≤−1 and FDR≤0.05 were counted as significantly down-regulated.
2.3. Co-expression network analysis
WGCNA (Langfelder and Horvath, 2008) is a widely used R package for performing weighted gene co-expression network analysis. It transforms gene expression profiles into functional co-expressed gene modules to provide deep insight into the genetic network (Liu R et al., 2015). In our study, all 546 RNA-seq samples and the top 20 000 most abundant genes that accounted for 99.6% of total reads in terms of FPKM-UQ were used in the co-expression network analysis. Low-abundance genes were eliminated as their abundance tends to be highly biased. To construct a more robust network, we applied the bi-weight mid-correlation (bicor) method which is insensitive to outliers for computing the correlation (Song et al., 2012). For a given gene, the soft degree is defined as the sum of connection weights with other network genes. In network analysis, hub genes are the highly connected genes. By scale-free topology fitting of the network, six was determined to be the best soft threshold power (Fig. S1). Finally, Cytoscape software was employed for visualizing and analyzing the co-expression network (http://www.cytoscape.org, Version 3.4.0; Kohl et al., 2011).
2.4. Geneset enrichment analysis
R package clusterProfiler (Yu et al., 2012) was used to perform the enrichment analysis of cancer hallmark gene ontology (GO), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) for the co-expression network gene modules and hub lncRNA-connected genes (weight ≥0.01). The “gmt” files of cancer hallmark gene sets were obtained from the Molecular Signatures Database (MSigDB) on the Gene Set Enrichment Analysis (GSEA) website (http://software.broadinstitute.org/gsea/msigdb ; Subramanian et al., 2005). Reduce visualize gene ontology (REVIGO) was adopted to summarize the enriched GO terms in line with their semantic similarity (http://revigo.irb.hr; Supek et al., 2011). All gene set enrichment analyses were carried out using Fisher’s exact test based on a hypergeometric distribution, and P values were corrected using the BH method for multiple comparisons. Gene sets or functions with FDR≤0.05 were considered significantly enriched.
2.5. Clinical pathological parameters and survival analysis
The association between the expression of each candidate lncRNA and the clinicopathological features of the HNSCC patients was analyzed by calculating the odds ratio (OR) value in terms of a 95% confidence interval (CI) (http://www.medcalc.org/ calc/odds_ratio.php; Parshall, 2013). The analyzed clinicopathological parameters included sex (male versus female), age (<65 versus ≥65 years old), alcohol habit, smoking history, clinical stage (I/II versus III/IV), tumor size (T1/T2 versus T3/T4), the absence or presence of regional lymph node metastasis, and the absence or presence of distant metastasis (M0 versus M1). Patients with clinical stage IV A, B, or C tumors were classified as “Stage IV”. Patients with tumor size stage T4, T4a, or T4b were classified as “T4”. For smoking history, patients categorized as “current smoker” or “current reformed smoker for ≤15 years” were classified as “recent smokers”, while those categorized as “current reformed smoker for >15 years” were classified as “reformed smokers”. Patients with no information available for a given characteristic were filtered from the corresponding analysis.
The R package survival (Therneau, 2017) was used for survival analysis, and multivariate Cox proportional hazards for regression analysis. The Kaplan Meier (KM) curve was visualized with R package survminer (Kassambara and Kosinski, 2017). Multivariate Cox proportional hazards analysis was conducted to evaluate the independence of the prognostic signature of the hub lncRNAs. The hazard ratio (HR), corresponding 95% CI, and P value were calculated. In total, 500 patients with RNA-seq data for cancer tissue (marked as 01 in the sample ID) were used in survival analysis. The FPKM-UQ value of genes was used to divide the patients into a low expression group and a high expression group based on the lower and upper quartiles, respectively. The difference in the survival distributions of the two groups was tested using a log-rank test with a significance level of 0.05. For the multivariate Cox regression model, the gene expression group and other covariates including sex, age, alcohol habit, smoking history, tumor size, node metastasis, and primary tumor site were included. Covariates with P≤0.05 were marked as statistically significant in relation to patient survival. The HPV data were not included in the analysis above because most patients did not have this information in TCGA database ((16.48±1.86)% of total upper and lower quartile samples for HPV status, and (20.39±2.41)% for HPV p16).
2.6. Validation using an external dataset
An independent dataset (Rickman et al., 2008) with 81 HNSCC patients from the Oncomine database was used to validate the clinical relevance of the potential HNSCC-associated lncRNAs discovered based on TCGA dataset. Oncomine (https://www. oncomine.org/resource/main.html) collects biological data from a large number of microarrays and allows the users to investigate using clinical-based and pathology-based analyses (Rhodes et al., 2004). Clinical parameters, overall survival (OS), and multivariate Cox regression analysis adjusting for sex, age, pathological stage (I/II versus III/IV), tumor size, node metastasis, and differentiation (poor, moderate, and well) were considered in this study to validate our findings. Due to the small sample size of this dataset, the median of the expression value was used to divide patients into a high expression group and a low expression group. The chi-squared test was used in the analysis of the association between gene expression group and differentiation. DFS information was not available for this dataset and hence was not analyzed. In total, 79 patients were used for survival analysis because the vital status of two patients was unavailable.
2.7. Cell culture and DDP treatment
TSCC15 cells were purchased from the American Type Culture Collection (ATCC) and cultured following the official instructions. A final concentration of 5 µg/ml cisplatin (diamminedichloroplatinum (DDP), Qilu Pharmaceutical, China) was added to the TSCC15 cells for DNA damage induction.
2.8. RNA interference and transfection
Small interfering RNAs (siRNAs) targeting human CYTOR transcript were purchased from RiboBio Co., Ltd. (Guangzhou, China). The siRNA sequences were as follows: siCYTOR-1 (5'-GAAACAGGAA GCTCTATGA-3'), siCYTOR-2 (5'-CACACTTGAT CGAATATGA-3'), and siCYTOR-3 (5'-CCAGTC TCTATGTGTCTTA-3'). Cells were pre-seeded on 6-well plates at a density of 5×105 per dish the day before transfection. The negative control or 5 µl of siRNAs were transfected into TSCC15 cells using lipofectamine 3000 (Invitrogen, USA) following the manufacturer’s instructions.
2.9. RT-PCR and quantitative RT-PCR
After twice washing with pre-chilled phosphate-buffered saline (PBS), cultured cells were lysed with TRIzol reagent (Ambion, USA). Total RNA was then extracted according to the manufacturer’s instructions. The first-strand complementary DNA (cDNA) was synthesized using a HiScript II 1st Strand cDNA synthesis kit (Vazyme, China), with RNA treated with DNase I (Thermo Scientific, USA) as a template. For quantitative polymerase chain reaction (qPCR), the cDNA was analyzed on a LightCycler® 96 (Roche, Switzerland) using a SYBR Green qPCR kit (Vazyme, China). The relative expression level of the desired lncRNA was calculated by the 2−ΔΔ C T method normalized to 18S. Primer sequences were as follows: CYTOR (F: 5'-AAAATCACGACTCAGCCCCC-3'; R: 5'-AATGGGAAACCGACCAGACC-3'), 18S (F: 5'-GTAACCCGTTGAACCCCATT-3'; R: 5'-CCAT CCAATCGGTAGTAGCG-3'). The expression level was denoted as the mean±standard error of the mean (SEM) of three independent experiments. Differences in expression levels were tested using a two-tailed Student’s t-test with GraphPad Prism (Version 6.00, La Jolla, California, USA). P≤0.05 was considered significant.
2.10. Apoptosis analysis and EdU proliferation assay
TSCC15 cells were transfected with small interfering CYTORs (siCYTORs) or siRNA control for 24 h. Cells were then treated with indicated doses of DDP (5 µg/ml). TSCC15 cells were harvested by trypsinization and washed with PBS. The 1×105 cells were harvested and an FITC Annexin-V apoptosis detection kit (BD Biosciences, USA) was used according to the manufacturer’s instructions. A Cellometer Vision image cytometer (Nexcelom Bioscience, USA) was used for apoptosis detection. Data were analyzed using FCS4 Express Cytometry software (De Novo Software, USA). Cell proliferation was assessed using a Cell-Light™ EdU Apollo567 in vitro kit (RiboBio, Guangzhou, China), according to the manufacturer’s instructions.
2.11. Cell migration assay and scratch wound healing assay
First, CYTOR in TSCC15 cells was knocked down for 24 h. For the cell migration assay, an 8 μm-pore polycarbonate membrane was used to divide transwell chambers into upper and lower chambers. The 1×105 TSCC15 cells re-suspended with 200 µl serum-free medium were added to the upper chamber, and the medium containing 10% fetal bovine serum (FBS) was added to the lower chamber before both chambers were incubated at 37 °C in 5% CO2 for 24 h. After removing non-migrated cells on the upper side of the membrane using a cotton swab, the membrane was fixed with 95% ethanol for 30 min and stained with 0.25% (2.5 g/L) crystal violet (Sangon, China). The migrated cells were counted using light microscopy (4×). For the wound healing assay, cells were cultured in 6-well plates and incubated until 100% confluence. Uniform scratches were made using a 200-µl pipette tip, then cells were washed using 1×PBS, which finally was replaced with serum-free medium. The cells from a consistent field in each well were photographed at 0, 24, and 48 h. The migration distance was assessed using Image-pro Plus Version 6.0 (Media Cybernetics, USA).
3. Results
3.1. Overview of this study
Firstly, we obtained clinical data and level 3 RNA-seq data from TCGA. A hierarchical clustering based on the gene expression profile suggested that there was no significant batch effect between samples under different batch operating conditions including BatchID, PlateID, Ship Date, and Tissue Source Site (Fig. S2). Subsequently, differential expression analysis was conducted, followed by the construction of a co-expression network which identified nine HNSCC-related lncRNAs. We then performed functional enrichment analyses on these nine lncRNAs to discover their potential functions and interpret their roles in HNSCC. Six of these nine lncRNAs were shown to be significantly associated with clinical parameters in terms of the OR value. Survival analysis showed that two of these six lncRNAs were significantly associated with the survival of HNSCC patients. Multivariate Cox regression models were used to assess the contribution of these lncRNAs to OS and DFS. The prognostic signature and biological roles of these lncRNAs were validated either on an independent cohort or by laboratory experiments. The workflow of this study is illustrated in Fig. S3.
3.2. Transcriptional profile in tumor tissue
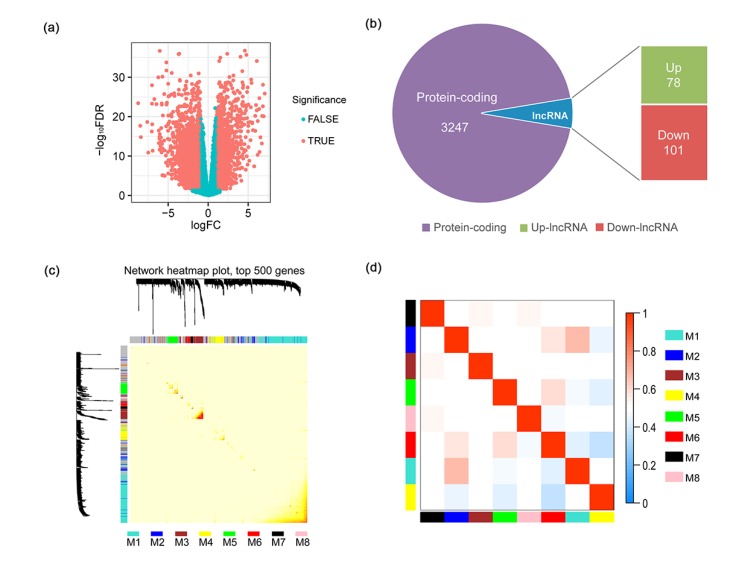
To unearth the gene expression shifts and functional alterations between cancer tissue and its adjacent normal tissue, we performed a differential expression analysis. In total, among 15 851 genes with a CPM past the cutoff described in the methods section, 1385 (8.74%) were identified as significantly up-regulated, and 1956 (12.34%) were down-regulated (Fig. 1a, Table S1). Among those, 78 lncRNAs were up-regulated, and 101 lncRNAs were down-regulated (Fig. 1b, Table S1). The top 10 most differentially expressed lncRNAs are listed in Table S1. HCG22 was the most down-regulated lncRNA (log2FC=−5.96, FDR=1.98×10−15), implicating that it may be highly associated with HNSCC.
Fig. 1.
Functional gene modules detected by co-expression network analysis
(a) The volcano plot shows the magnitude of differential expression between cancer tissue and its adjacent normal tissue. Each dot represents one gene, and genes in read are significantly altered. FDR: false discovery rate; logFC: log2 fold change. (b) Pie chart shows the proportion of dysregulated lncRNAs in all differentially expressed genes. (c) Topological overlap heatmap of the gene co-expression network. Each row and column represents a gene. Light color indicates low topological overlap, and on the contrary dark color denotes high topological overlap. The different side colors indicate different modules. The dendrogram suggests the clustering of these genes based on the similarity of their gene expression profiles. (d) The correlation between each module demonstrated based on eigengene. Blue represents a negative correlation, while red represents a positive correlation (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
3.3. Eight distinct functional gene modules detected by co-expression network analysis
By co-expression network construction and analysis using WGCNA, we obtained eight distinct modules, namely M1 (turquoise), M2 (blue), M3 (brown), M4 (yellow), M5 (green), M6 (red), M7 (black), and M8 (pink), in terms of the WGCNA conventions (Zhang and Horvath, 2005). There were 7301 genes that could not be assigned to any module. A module represents a cluster of highly interconnected genes (Zhang and Horvath, 2005). M1 was the largest module, consisting of 5624 genes (Table S2). As illustrated in the topological overlap matrix (TOM) plot of 500 genes with the high soft degree across all modules (Fig. 1c), M3 had the highest cohesion within the module, but the lowest coupling to other modules. Moreover, correlations between eigengenes of each module showed that M1 and M2 were highly related (Fig. 1d). Among these eight modules, M4 contained the most differentially regulated lncRNAs (Table S2).
3.4. Functional analysis of the modules identified by WGCNA
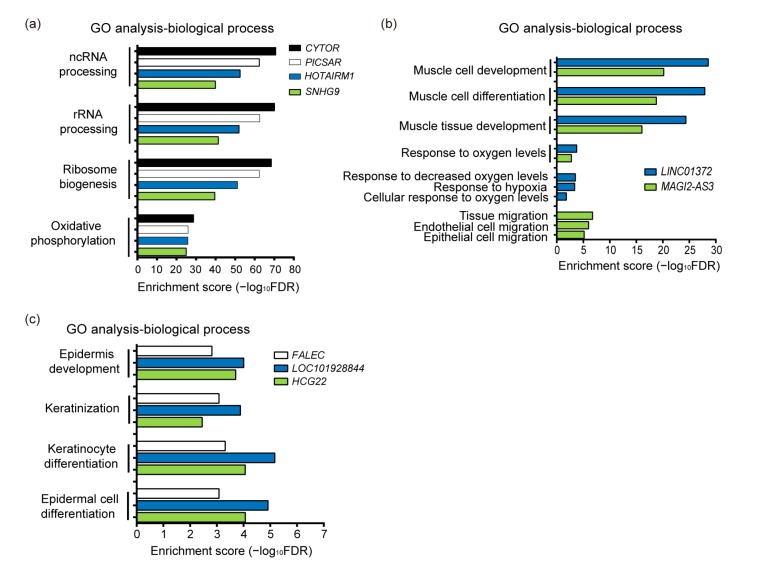
To gain further biological insights into these modules, we performed GO term enrichment analysis. Genes from M1 are enriched primarily in ncRNA processing, ribosome biogenesis, mitochondrial translation, DNA replication, apoptotic signaling pathways, and DNA damage response (Fig. S4a, Table S2). Genes in M3 are involved mainly in muscle cell development, response to oxygen levels, response to hypoxia, and response to decreased oxygen levels (Fig. S4a, TableS2). The most significantly represented biological processes for M4 genes are keratinocyte differentiation, epidermis development, skin development, and epidermal cell differentiation (Fig.S4c, Table S2). These related biological processes have been widely reported to be associated with tumorigenesis or cancer development (Bartkova et al., 2005; Zaidi et al., 2011; Liu BD et al., 2015).
3.5. Potential cancer-associated lncRNAs revealed by co-expression network analysis
Subsequently, we discovered nine HNSCC-associated hub lncRNAs which were differentially regulated in terms of differential expression analysis (|log2FC|≥1, log2CPM>0, and FDR≤0.05) with a soft degree above 10, namely LINC01372, CYTOR, SNHG9, MAGI2-AS3, PICSAR, HOTAIRM1, FALEC, LOC101928844, and HCG22 (Table S2). To investigate the potential functions of these hub lncRNAs, and gain a deeper understanding of their roles in HNSCC, we performed gene set enrichment analyses including cancer hallmarks, GO terms, and KEGG pathways based on the genes connected to each lncRNA. Cancer hallmarks enrichment analysis revealed that CYTOR, PICSAR, HOTAIRM1, and SNHG9 are significantly involved in oxidative phosphorylation, DNA repair, and MYC targets v1 (v2) (Table 1, Table S3). Notably, SNHG9 belongs to the M2 module which is highly connected to M1, implying they have similar biological functions. Among these four lncRNAs, some have been demonstrated to be related to tumor progression (Gabay et al., 2014). For example, CYTOR, which was up-regulated in our data, has also been reported to be overexpressed in gastric cancer and to affect DNA damage, cell cycle arrest, and tumor metastasis (Zhao et al., 2015; Chen et al., 2016). LINC01372 and MAGI2-AS3 are associated mainly with cancer hallmarks of myogenesis (Table 1, Table S3), which play essential roles in tumorigenesis (Lamouille et al., 2014; LeBleu et al., 2014). LOC101928844 and FALEC are especially associated with estrogen response (Table 1, Table S3).
Table 1.
Cancer hallmarks enriched in HNSCC-associated hub lncRNAs
| Gene symbol | ENSG number | Module | Cancer hallmarks |
| CYTOR | ENSG00000222041 | M1 (turquoise) | Oxidative phosphorylation, DNA repair, MYC targets v1 (v2) |
| PICSAR | ENSG00000275874 | ||
| HOTAIRM1 | ENSG00000233429 | ||
| SNHG9 | ENSG00000255198 | M2 (blue) | |
| LINC01372 | ENSG00000235475 | M3 (brown) | Myogenesis |
| MAGI2-AS3 | ENSG00000234456 | ||
| LOC101928844 | ENSG00000267709 | M4 (yellow) | Estrogen response early |
| FALEC | ENSG00000228126 | ||
| HCG22 | ENSG00000228789 | N/A |
ENSG: the Ensign Group, Inc.; N/A: not applicable
GO term enrichment analyses indicated that CYTOR, PICSAR, HOTAIRM1, and SNHG9 are associated mainly with ncRNA processing, ribosomal RNA (rRNA) processing, ribosome biogenesis, or oxidative phosphorylation (Fig. 2a, Table S4). A number of apoptosis and DNA damage related GO terms were also enriched in CYTOR-connected genes, suggesting that CYTOR plays a role in these functions (Table 2). LINC01372 and MAGI2-AS3 are particularly associated with muscle development and response to oxygen levels (Fig. 2b, Table S4). HCG22, LOC101928844, and FALEC are involved mainly in epidermis development, keratinocyte differentiation, epidermal cell differentiation, and keratinization (Fig. 2c, Table S4). Most of these lncRNAs have not been characterized or well investigated in HNSCC, and some have not previously been reported in cancer studies. Hence, these new findings provide a deeper insight into the roles of these uncharacterized lncRNAs in HNSCC, and importantly provide more knowledge regarding the pathogenesis of HNSCC.
Fig. 2.
GO terms enriched in HNSCC-associated hub lncRNAs of co-expression network
Enrichment of GO terms of biological process for: (a) CYTOR, PICSAR, HOTAIRM1, and SNHG9; (b) LINC01372 and MAGI2-AS3; (c) FALEC, LOC101928844, and HCG22. The terms with FDR≤0.05 are considered significantly enriched. The negative log of FDR (base 10) is plotted on the X-axis
Table 2.
List of enriched DNA damage or apoptosis related GO terms for CYTOR
| GO ID | GO term | Count | Background | FDR |
| GO:0042769 | DNA damage response, detection of DNA damage | 9 | 36 | 4.44×10−4 |
| GO:0008637 | Apoptotic mitochondrial changes | 16 | 123 | 2.02×10−3 |
| GO:2001244 | Positive regulation of intrinsic apoptotic signaling pathway | 8 | 51 | 2.49×10−2 |
| GO:2001235 | Positive regulation of apoptotic signaling pathway | 17 | 180 | 3.77×10−2 |
| GO:0043281 | Regulation of cysteine-type endopeptidase activity involved in apoptotic process | 19 | 216 | 4.75×10−2 |
| GO:2001242 | Regulation of intrinsic apoptotic signaling pathway | 15 | 155 | 4.99×10−2 |
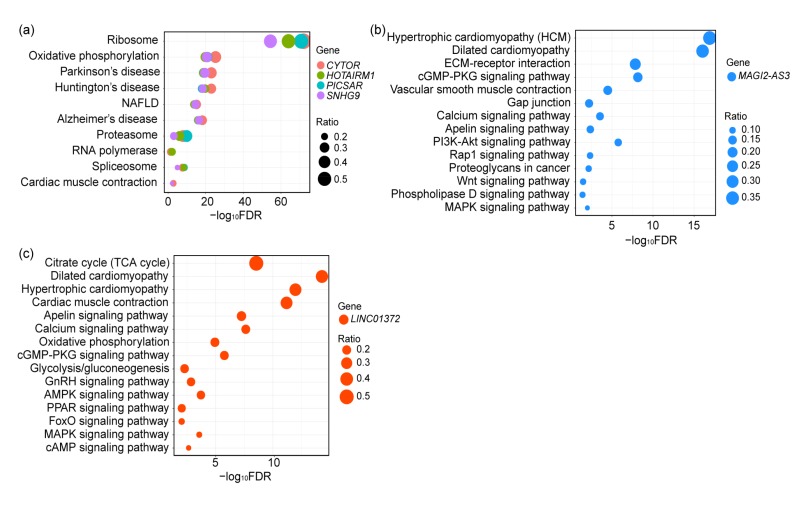
To comprehensively mine the biological roles of these hub lncRNAs, KEGG pathway enrichment analysis was performed using R package clusterProfiler. Fig. 3 and Table S5 list all significantly enriched KEGG pathways with FDR≤0.05 and gene counts ≥5. We discovered that CYTOR, HOTAIRM1, PICSAR, and SNHG9 are involved primarily in ribosome biogenesis and maintenance of genomic stability, and that their dysregulation is connected with a predisposition to multiple diseases. The significantly enriched pathways of MAGI2-AS3 are several cancer-associated signaling pathways such as, cell migration and cell adhesion pathways (extracellular matrix (ECM)-receptor interaction, Gap junction, and Rap1 signaling pathways), angiogenesis signaling pathways (vascular smooth muscle contraction and apelin signaling pathways), and the phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathway. Notably, the functions of enriched pathways are consistent with the observations from GO terms and cancer hallmarks. LINC01372 appears to be relevant to multiple signaling pathways as well, such as metabolism pathways (glycolysis/gluconeogenesis and oxidative phosphorylation), the hypoxia signaling pathway (FoxO signaling pathway), and some oncogenic signaling pathways (calcium, mitogen-activated protein kinase (MAPK), and peroxisome proliferator-activated receptor (PPAR) signaling pathways). However, FALEC, LOC101928844, and HCG22 were not found to be involved in any pathway because there were too few connected genes for enrichment analysis. In summary, the pathway analyses above showed that most of these hub lncRNAs may play key roles in malignancies and disease pathogenesis. Compared to previous observations (Nötzold et al., 2017), some new functions of lncRNAs like LINC01372 and MAGI2-AS3 were revealed in this study. These findings for uncharacterized lncRNAs expand our knowledge of the possible roles of lncRNAs in HNSCC.
Fig. 3.
Significantly enriched KEGG signaling pathways in CYTOR, HOTAIRM1, PICSAR, and SNHG9 (a), MAGI2-AS3 (b), and LINC01372 (c)
The different colors of the dots indicate different lncRNA-correlated genes, and the size of the dot indicates the ratio of number of given genes to total number of genes in each pathway. The X-axis denotes negative log10FDR. NAFLD: 1-non-alcoholic fatty liver disease; ECM: extracellular matrix; cGMP: cyclic guanosine monophosphate; PKG: protein kinase G; PI3K: phosphatidylinositol 3-kinase; MAPK: mitogen-activated protein kinase; TCA: tricarboxylic acid cycle; GnRH: gonadotropin-releasing hormone; AMPK: adenosine monophosphate (AMP)-activated protein kinase; PPAR: peroxisome proliferator-activated receptor; cAMP: cyclic adenosine monophosphate (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
3.6. Correlations between aberrant expression of hub lncRNAs and HNSCC clinical pathological parameters
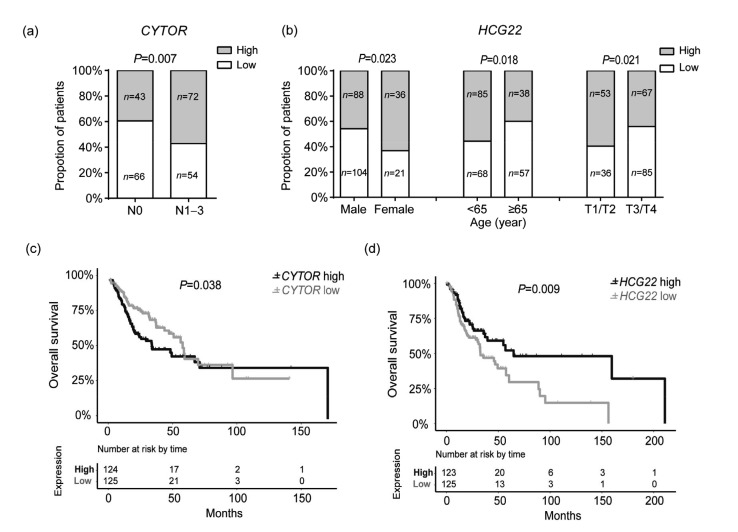
To gain deeper insight into the associations between the hub lncRNAs and pathogenesis of cancer, we examined the correlation between the expression levels of these lncRNAs and the patients’ clinical features in HNSCC. The complete results are shown in Table S6. In total, six lncRNAs were found to be related to one or several features. For example, recent smokers are prone to have a low PICSAR expression level compared to reformed smokers. Patients with a low level of PICSAR and a high level of LINC01372 tend to have a smaller tumor size. Moreover, a high expression of PICSAR is negatively correlated with node metastasis. Male patients have high SNHG9 and HOTAIRM1 levels. CYTOR expression was positively correlated with lymph node metastasis (OR=0.49, 95% CI=0.29–0.82, P=0.007; Fig. 4a). High HCG22 expression was negatively related with tumor size (OR=1.87, 95% CI=1.10–3.18, P=0.021), and more common among female patients (OR=0.49, 95% CI=0.27–0.91, P=0.023) and patients younger than 65 years old (OR=1.88, 95% CI=1.12–3.15, P=0.018), as shown in Fig. 4b. The co-expression network of these six lncRNAs was visualized with Cytoscape based on the top 50 most connected genes for each lncRNA (Fig. S5, Table S7).
Fig. 4.
Clinical feature correlation and survival analysis of potential HNSCC-associated lncRNAs
(a) Correlation of CYTOR expression with lymph node metastasis in HNSCC patients. (b) Correlation of HCG22 expression with gender, age, and tumor size in HNSCC patients. (c, d) Kaplan-Meier analysis of overall survival in HNSCC patients with CYTOR (c) and HCG22 (d) expression. HNSCC patients with higher HCG22 and lower CYTOR expression have a better overall survival. P-values were calculated by log-rank test
3.7. HCG22 and CYTOR identified as independent prognostic markers of HNSCC
Cancer biomarker genes usually play important roles in the carcinogenesis of certain cancer types, and contribute to the choice of multiple therapeutic alternatives currently available (Bhatt et al., 2010; Mäbert et al., 2014). We applied Kaplan-Meier analysis to discover candidate lncRNA biomarkers, and a log-rank test to identify the lncRNAs, which led to a significant difference in survival outcome between high and low expression patients. The results indicated that patients with a high HCG22 expression level (P=0.009) and a low CYTOR expression level (P=0.038) had significantly better OS time (P=0.016; Figs. 4c and 4d) and disease free survival (DFS) (P=0.001; Figs. S6a and S6b) in a cohort of 500 HNSCC patients. The other four lncRNAs PICSAR, HOTAIRM1, SNHG9, and LINC01372 did not exhibit a prognostic ability for HNSCC. Multivariate Cox regression analyses were performed on these two lncRNAs to ensure that their association with patient survival was independent of prognostic factors. As expected, we discovered that the low expression of CYTOR could be an independent protective factor for DFS with a hazards ratio of 0.33 and P-value of 0.005. HCG22 was identified as a prognostic factor for both OS (HR=1.68, 95% CI=1.08–2.61, P=0.022) and DFS (HR=2.14, 95% CI=1.05–4.38, P=0.037), independent of clinical features. The Cox regression analyses of CYTOR and HCG22 are summarized in Table S6. More recently, overexpression of CYTOR was reported to be positively correlated with tumor invasion depth, progression, and poor survival in gastric cancer (Chen et al., 2016) and TSCC (Yu et al., 2017), which supports the credibility of our findings. In summary, these findings imply that CYTOR might function as an oncogenic lncRNA and HCG22 likely acts as a tumor suppressor in HNSCC.
3.8. Validation of the HNSCC lncRNA signature using an independent cohort
To confirm the effectiveness of these nine hub lncRNAs for HNSCC prognosis, we validated their prognostic signatures (only PICSAR and HCG22 were available) on an independent cohort of 81 HNSCC patients from the Oncomine database. We performed clinicopathological parameter analysis, survival analysis, and multivariate Cox regression analysis for these two genes using this dataset. Kaplan-Meier survival analysis indicated that the low expression group of PICSAR and HCG22 was related to poor survival, although P-values were larger than 0.05. HCG22 showed a negative correlation with tumor size (OR=2.89, 95% CI=1.17–7.12, P=0.021), and maintained its prognostic value according to multivariate Cox regression analysis in the validation cohort (HR=1.97, 95% CI=1.06–3.64, P=0.032). We also analyzed PICSAR, but the results were not statistically significant using the analyses mentioned above. The clinical characteristics and Cox regression results for this cohort are listed in Table S8.
3.9. Effect of CYTOR knockdown on apoptosis in TSCC15 cells
Our results showed that CYTOR was correlated with DNA damage or apoptosis according to GO term enrichment analysis (Table 2). To test the effect of CYTOR on tumorigenesis, we performed in vitro apoptosis, EdU, wound healing, and transwell assays on TSCC15 cells. After introducing three CYTOR RNA interference (RNAi) products individually into TSCC15 cells, the expression of CYTOR was detected by qRT-PCR, which demonstrated a significant decrease in CYTOR expression (Fig. S7a). Cisplatin (also named DDP), a platinum-based chemotherapy drug, is one of the most effective drugs used in the treatment of HNSCC (Posner et al., 2007) and dramatically induces cell death. Annexin V+propidium iodide (PI) apoptosis analysis was carried out to measure the amount of apoptotic cells. Our data revealed that the apoptosis rate increased in the presence of 5 μg/ml DDP compared with the control, after CYTOR was knocked down (Fig. S7b). EdU, wound healing, and transwell assays were implemented to assess the effect of CYTOR on cell proliferation, mobility, and migration. However, we did not observe a significant difference between the presence of control and indicated CYTOR siRNAs in TSCC15 (Figs. S7c–S7e).
4. Discussion
LncRNAs have been increasingly viewed as key factors in various human cancers. Recent studies have established their relationships with gene regulation (Engreitz et al., 2016), epigenetic regulation (Holoch and Moazed, 2015), and cell signaling pathways (Peng et al., 2017). However, the functions of most lncRNAs in cancer have not yet been well characterized. This study focused on identifying HNSCC-associated lncRNAs, and systematically investigating their functions and clinical relevance in HNSCC. Using an integrated approach incorporating differential expression analysis, co-expression network, and gene set enrichment analysis, we discovered nine HNSCC-related lncRNAs. Among them, FALEC, HCG22, and LOC101928844 were strongly differentially regulated between HNSCC tumor and adjacent normal tissue, implying that their dysregulation may play a carcinogenic role in cancer development. GO term enrichment analysis based on co-expression network showed that these three lncRNAs were significantly involved in epidermis development and epidermal cell differentiation. Previous studies have shown that HCG22 expression was reduced in OSCC, and was positively associated with patient survival, but there has been a lack of further functional investigation (Nohata et al., 2016; Feng et al., 2017). In this study, we found that HCG22 was the most down-regulated lncRNA in HNSCC, and might function in epidermis differentiation. Co-expression network showed that HCG22 is closely related to EVPL and PPL (Fig.S5). EVPL and PPL are two important proteins in the plakin family, which play a pivotal role in the maintenance of tissue integrity. An earlier study confirmed that EVPL and PPL were down-regulated in HNSCC tissues, which fits with our findings (Chi et al., 2009). We observed that the expression of HCG22 was negatively correlated with tumor size, and positively correlated with patient OS and DFS. In particular, HCG22 maintained prognostic significance independent of clinical features according to multivariate Cox analysis, which indicated its potential to be an independent prognostic factor in patients with HNSCC. Validation of HCG22 on an independent cohort showed a similar clinical and prognostic performance, which confirmed our findings.
Based on cancer hallmark, GO term, and KEGG pathway enrichment analysis, MAGI2-AS3 is relevant mainly to angiogenesis, cell migration, and several cancer-associated signaling pathways, indicating that it might affect the multistep process in tumor progression. LINC01372 is involved primarily in response to oxygen level, cell development, and hypoxia-related signaling pathways. A large number of findings imply that a local hypoxic microenvironment is one of the most important characteristics of solid tumors (Wilson and Hay, 2011; Eales et al., 2016), suggesting that LINC01372 may contribute to tumorigenesis. LINC01372 also showed a positive correlation with tumor size in this study.
Enrichment analysis showed that the connected genes CYTOR, HOTAIRM1, SNHG9, and PICSAR are potential MYC targets, and are involved in ribosome biogenesis and maintenance of genomic stability. Previous studies have demonstrated that elevated MYC expression might lead to genome destabilization in human cancers via coordinating ribosome biogenesis (Wade and Wahl, 2006; Barna et al., 2008). Compelling evidence suggested that MYC directly regulates ribosome biogenesis through a multitude of mechanisms (van Riggelen et al., 2010). Dysregulation of ribosome biogenesis is closely associated with alterations in cellular proliferation and apoptosis, and contributes to increasing susceptibility to cancer (Ruggero and Pandolfi, 2003). These findings suggest that CYTOR, HOTAIRM1, SNHG9, and PICSAR are closely associated with tumorigenesis. Our in vitro experiments demonstrated that down-regulation of CYTOR contributes to TSCC15 cell apoptosis, which is concordant with our co-expression network and functional enrichment analysis. Remarkably, CYTOR has a high positive correlation with node metastasis, and a negative correlation with HNSCC patient OS and DFS. Multivariate Cox analysis confirmed that CYTOR is an independent predictor of DFS in patients with HNSCC.
In this study, we comprehensively identified HNSCC-associated lncRNAs, and elucidated their biological functions and roles in the oncogenesis of HNSCC. Their prognostic value for patient survival outcome was determined followed by validation on an independent cohort. However, there were some limitations. For example, the tumorigenesis function of the identified lncRNA was tested using cell-based experiments. Further in vivo investigations would be very helpful to fully understand the roles of these lncRNA in TSCC initiation and development. Nevertheless, we are convinced that these findings, based on this large cohort and verified on an independent cohort or by experiments, are valuable not only for fundamental research, but also for potential clinical applications.
5. Conclusions
CYTOR might function in ribosome biogenesis and the maintenance of genomic stability, and had a high positive correlation with node metastasis and a negative correlation with patient OS and DFS. CYTOR inhibited cell apoptosis following treatment with the chemotherapeutic drug DDP. HCG22 might play a role in epidermis differentiation, and was negatively correlated with tumor size and positively correlated with patient OS and DFS. CYTOR and HCG22 were independent prognostic biomarkers for patient outcome. The potential of HCG22 to be a prognostic biomarker independent of clinical features was validated and confirmed using an external dataset.
List of electronic supplementary materials
Determination of parameter β of the adjacency function in the WGCNA algorithm
Hierarchical clustering of samples based on the gene expression profile
Overview of the workflow of the study design
Functional profiling of modules identified by WGCNAFunctional profiling of modules identified by WGCNA
Co-expression of six hub lncRNAs visualized using Cytoscape software based on the top 50 most connected genes of each given lncRNA
Kaplan-Meier analysis of disease free survival in HNSCC patients
Effects of CYTOR knockdown on cell apoptosis, proliferation, and migration in TSCC15 cells
Results of differential expression analysis
GO term enrichment analysis for WGCNA modules and HNSCC-associated lncRNAs
Cancer hallmarks enrichment analysis for HNSCCassociated lncRNAs
GO term enrichment analysis for HNSCC-associated lncRNAs
KEGG pathway enrichment analysis for HNSCCassociated lncRNAs
OR analysis and multivariate Cox regression analysis in HNSCC
WGCNA network of six lncRNAs
OR and multivariate Cox regression analysis based on Oncomine dataset
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 31471226 and 91440108) and the Fundamental Research Funds for the Central Universities (Nos. WK2070000044 and WK2070000034), China
Electronic supplementary materials: The online version of this article (https://doi.org/10.1631/jzus.B1700319) contains supplementary materials, which are available to authorized users
Compliance with ethics guidelines: Yu-zhu GUO, Hui-hui SUN, Xiang-ting WANG, and Mei-ting WANG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Aken BL, Ayling S, Barrell D, et al. The Ensembl gene annotation system. Database, 2016:baw093. 2016 doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders S, Pyl PT, Huber W. HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barna M, Pusic A, Zollo O, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456(7224):971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartkova J, Hořejší Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 5.Bartonicek N, Maag JLV, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15(1):43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- 7.Bhatt AN, Mathur R, Farooque A, et al. Cancer biomarkers–current perspectives. Indian J Med Res. 2010;132:129–149. [PubMed] [Google Scholar]
- 8.Cheetham SW, Gruhl F, Mattick JS, et al. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen WM, Huang MD, Sun DP, et al. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7(9):9773–9787. doi: 10.18632/oncotarget.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi LM, Lee CW, Chang KP, et al. Enhanced interferon signaling pathway in oral cancer revealed by quantitative proteome analysis of microdissected specimens using 16O/18O labeling and integrated two-dimensional LC-ESI-MALDI tandem MS. Mol Cell Proteomics. 2009;8(7):1453–1474. doi: 10.1074/mcp.M800460-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lena PG, Paz-Gallardo A, Paramio JM, et al. Clusterization in head and neck squamous carcinomas based on lncRNA expression: molecular and clinical correlates. Clin Epigenetics, 9:36. 2017 doi: 10.1186/s13148-017-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eales KL, Hollinshead KER, Tennant DA. Hypoxia and metabolic adaptation of cancer cells. Oncogenesis. 2016;5(1):e190. doi: 10.1038/oncsis.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17(12):756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 15.Feng L, Houck JR, Lohavanichbutr P, et al. Transcriptome analysis reveals differentially expressed lncRNAs between oral squamous cell carcinoma and healthy oral mucosa. Oncotarget. 2017;8(19):31521–31531. doi: 10.18632/oncotarget.16358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabay M, Li YL, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4(6):a014241. doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng YJ, Xie SL, Li Q, et al. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39(6):2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 18.Gold KA, Lee HY, Kim ES. Targeted therapies in squamous cell carcinoma of the head and neck. Cancer. 2009;115(5):922–935. doi: 10.1002/cncr.24123. [DOI] [PubMed] [Google Scholar]
- 19.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12(1):1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for the encode project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16(2):71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 23.Kassambara A, Kosinski M. Survminer: Drawing Survival Curves Using ‘ggplot2’. R Package Version 0.4.0. https://CRAN.R-project.org/package=survminer [Accessed on June 10, 2017].2017. [Google Scholar]
- 24.Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinsella RJ, Kähäri A, Haider S, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database, 2011:bar030. 2011 doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. In: Hamacher M, Eisenacher M, Stephan C (Eds.), Data Mining in Proteomics. Humana Press; 2011. pp. 291–303. [DOI] [PubMed] [Google Scholar]
- 27.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform, 9:559. 2008 doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law CW, Chen Y, Shi W, et al. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeBleu VS, O'Connell JT, Gonzalez Herrera KN, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 32.Li DD, Feng JP, Wu TY, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182(1):64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Wu Z, Mei Q, et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer. 2013;109(8):2266–2278. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu BD, Sun LJ, Liu Q, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Liu R, Cheng Y, Yu J, et al. Identification and validation of gene module associated with lung cancer through coexpression network analysis. Gene. 2015;563(1):56–62. doi: 10.1016/j.gene.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 36.Mäbert K, Cojoc M, Peitzsch C, et al. Cancer biomarker discovery: current status and future perspectives. Int J Radiat Biol. 2014;90(8):659–677. doi: 10.3109/09553002.2014.892229. [DOI] [PubMed] [Google Scholar]
- 37.McCarthy DJ, Chen YS, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller DL, Davis JW, Taylor KH, et al. Identification of a human papillomavirus-associated oncogenic miRNA panel in human oropharyngeal squamous cell carcinoma validated by bioinformatics analysis of the cancer genome atlas. Am J Pathol. 2015;185(3):679–692. doi: 10.1016/j.ajpath.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min SN, Wei T, Wang XT, et al. Clinicopathological and prognostic significance of homeobox transcript antisense RNA expression in various cancers: a meta-analysis. Medicine (Baltimore) 2017;96(23):e7084. doi: 10.1097/MD.0000000000007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nohata N, Abba MC, Gutkind JS. Unraveling the oral cancer lncRNAome: identification of novel lncRNAs associated with malignant progression and HPV infection. Oral Oncol. 2016;59:58–66. doi: 10.1016/j.oraloncology.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nötzold L, Frank L, Gandhi M, et al. The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells. Sci Rep, 7:2265. 2017 doi: 10.1038/s41598-017-02357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parshall MB. Unpacking the 2×2 table. Hear Lung J Acute Crit Care. 2013;42(3):221–226. doi: 10.1016/j.hrtlng.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posner MR, Hershock DM, Blajman CR, et al. Cisplatin and fluorouracil alone or with docetaxel in head and neck cancer. New Engl J Med. 2007;357(17):1705–1715. doi: 10.1056/nejmoa070956. [DOI] [PubMed] [Google Scholar]
- 45.Pritzker KPH. Predictive and prognostic cancer biomarkers revisited. Expert Rev Mol Diagn. 2015;15(8):971–974. doi: 10.1586/14737159.2015.1063421. [DOI] [PubMed] [Google Scholar]
- 46.Quek XC, Thomson DW, Maag JLV, et al. LncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43(D1):D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Development Core Team R. R: A Language and Environment for Statistical Computing. 2011 [Google Scholar]
- 48.Rhodes DR, Yu JJ, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rickman DS, Millon R, de Reynies A, et al. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27(51):6607–6622. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3(3):179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 52.Salazar C, Calvopiña D, Punyadeera C. miRNAs in human papilloma virus associated oral and oropharyngeal squamous cell carcinomas. Expert Rev Mol Diagn. 2014;14(8):1033–1040. doi: 10.1586/14737159.2014.960519. [DOI] [PubMed] [Google Scholar]
- 53.Salyakina D, Tsinoremas NF. Non-coding RNAs profiling in head and neck cancers. NPJ Genomic Med, 1:15004. 2016 doi: 10.1038/npjgenmed.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmitt AM, Garcia JT, Hung T, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet. 2016;48(11):1370–1376. doi: 10.1038/ng.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seiwert TY, Salama JK, Vokes EE. The chemoradiation paradigm in head and neck cancer. Nat Clin Pract Oncol. 2007;4(3):156–171. doi: 10.1038/ncponc0750. [DOI] [PubMed] [Google Scholar]
- 57.Signal B, Gloss BS, Dinger ME. Computational approaches for functional prediction and characterisation of long noncoding RNAs. Trends Genet. 2016;32(10):620–637. doi: 10.1016/j.tig.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Song L, Langfelder P, Horvath S. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinformatics, 13:328. 2012 doi: 10.1186/1471-2105-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Supek F, Bošnjak M, Škunca N, et al. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Therneau T. A Package for Survival Analysis in S. R Package Version 2.41-2. https://CRAN.R-project.org/package=survival [Accessed on Mar. 20, 2017].2017. [Google Scholar]
- 62.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 64.Wade M, Wahl GM. c-Myc, genome instability, and tumorigenesis: the devil is in the details. Curr Top Microbiol Immunol. 2006;302:169–203. doi: 10.1007/3-540-32952-8_7. [DOI] [PubMed] [Google Scholar]
- 65.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11(6):393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 66.Yan L, Zhan C, Wu JH, et al. Expression profile analysis of head and neck squamous cell carcinomas using data from The Cancer Genome Atlas. Mol Med Rep. 2016;13(5):4259–4265. doi: 10.3892/mmr.2016.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yates A, Akanni W, Amode MR, et al. Ensembl 2016. Nucleic Acids Res. 2016;44(D1):D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu GC, Wang LG, Han YY, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J Integr Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu JJ, Liu Y, Guo C, et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8(4):523–530. doi: 10.7150/jca.17510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaidi MR, Davis S, Noonan FP, et al. Interferon-γ links ultraviolet radiation to melanomagenesis in mice. Nature. 2011;469(7331):548–553. doi: 10.1038/nature09666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(1):Article 17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 72.Zhang SC, Tian LL, Ma PH, et al. Potential role of differentially expressed lncRNAs in the pathogenesis of oral squamous cell carcinoma. Arch Oral Biol. 2015;60(10):1581–1587. doi: 10.1016/j.archoralbio.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 73.Zhao J, Liu YC, Zhang WH, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14(19):3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Determination of parameter β of the adjacency function in the WGCNA algorithm
Hierarchical clustering of samples based on the gene expression profile
Overview of the workflow of the study design
Functional profiling of modules identified by WGCNAFunctional profiling of modules identified by WGCNA
Co-expression of six hub lncRNAs visualized using Cytoscape software based on the top 50 most connected genes of each given lncRNA
Kaplan-Meier analysis of disease free survival in HNSCC patients
Effects of CYTOR knockdown on cell apoptosis, proliferation, and migration in TSCC15 cells
Results of differential expression analysis
GO term enrichment analysis for WGCNA modules and HNSCC-associated lncRNAs
Cancer hallmarks enrichment analysis for HNSCCassociated lncRNAs
GO term enrichment analysis for HNSCC-associated lncRNAs
KEGG pathway enrichment analysis for HNSCCassociated lncRNAs
OR analysis and multivariate Cox regression analysis in HNSCC
WGCNA network of six lncRNAs
OR and multivariate Cox regression analysis based on Oncomine dataset