This study revealed that FZP functions in grain size and sterile lemma identity, and supported the hypothesis that the lemma, rudimentary glume, and sterile lemma are homologous organs.
Keywords: CRISPR-Cas9, dehydration-responsive element (DRE), FZP, GCC-box, grain size, rice (Oryza sativa)
Abstract
In grass, the spikelet is a unique inflorescence structure that directly determines grain yield. Despite a great deal of research, the molecular mechanisms behind spikelet development are not fully understood. In the study, FZP encodes an ERF domain protein, and functions in grain size and sterile lemma identity. Mutation of FZP causes smaller grains and degenerated sterile lemmas. The small fzp-12 grains were caused by a reduction in cell number and size in the hulls. Interestingly, the sterile lemma underwent a homeotic transformation into a rudimentary glume in the fzp-12 and fzp-13 mutants, whereas the sterile lemma underwent a homeotic transformation into a lemma in FZP over-expressing plants, suggesting that FZP specifically determines the sterile lemma identity. We confirmed the function of FZP by complementation, CRISPR-Cas9 gene editing, and cytological and molecular tests. Additionally, FZP interacts specifically with the GCC-box and DRE motifs, and may be involved in regulation of the downstream genes. Our results revealed that FZP plays a vital role in the regulation of grain size, and first provides clear evidence in support of the hypothesis that the lemma, rudimentary glume, and sterile lemma are homologous organs.
Introduction
Rice (Oryza sativa L.) is the important and staple food. Increasing rice grain yield provides the food demands and security of a rapidly growing population. Grain size and shape in rice (Oryza sativa) is specified by grain length, width, and thickness, which determine overall yield and quality. Multiple genetic factors control grain weight (Hu et al., 2015, Huang et al., 2017), but the underlying regulatory mechanisms that determine grain size and shape are still largely unknown.
The spikelet hull restricts grain growth and determines the final grain size in rice (Li and Li, 2016). Most of the identified genes and quantitative trait loci (QTLs) that influence grain size act through regulation of the proliferation and expansion of the cells in the spikelet hull. For example, GRAIN SIZE 3 (GS3), GRAIN LENGTH 3 (qGL3/GL3.1), and BIG GRAIN 1 (BG1) determine grain size by controlling cell number. GS3 encodes a putative G protein γ subunit, and its loss of function induces long grains as a result of increased cell number (Mao et al., 2010, Fan et al., 2006). qGL3/GL3.1 encodes a phosphatase kelch protein and dephosphorylates its substrate cyclin-T1;3. Loss of function of qGL3/GL3.1 produces longer grains by increasing cell number (Zhang et al., 2012; Qi et al., 2012). BG1 encodes a novel membrane-localized protein and positively regulates auxin response and transport. Over-expression of BG1 results in increased grain size and its knockdown leads to smaller grains by regulating cell number (Liu et al., 2015). Several other factors that control grain size by influencing cell size have also been cloned, including GRAIN LENGTH 2/GRAIN SIZE 2 (GL2/GS2), Grain Length on Chromosome 7 (GL7), POSITIVE REGULATOR OF GRAINLENGTH 1 (PGL1) and PGL2. GL2/GS2 encodes the transcription factor GROWTHREGULATING FACTOR 4 (OsGRF4) and is regulated by the microRNA OsmiR396. Mutation in GL2/GS2 disturbs OsmiR396-directed regulation of GL2/GS2, resulting in large grains predominantly by influencing cell size (Che et al., 2016; Hu et al., 2015). GL7 is homologous to Arabidopsis thaliana LONGIFOLIA, and upregulation of GL7 expression produces slender grains as a result of longitudinally-increased cell length and transversely-decreased cell width (Wang et al., 2015a, 2015b). Atypical helix-loop-helix proteins encoded by PGL1 and PGL2 affect grain length by regulating cell length (Heang and Sassa, 2012a, 2012b). The above studies revealed that cell proliferation and cell expansion play key roles in coordinately controlling grain size in rice.
In rice, the spikelet comprises one terminal fertile floret and unique glumes: sterile lemmas and rudimentary glumes, which is different from those of other grasses (Ren et al., 2018). Despite a great deal of research, the origins of glumes are still debated. Recently, several genes have been cloned, which helped to elucidate the origins of these organs in rice. LONG STERILE LEMMA (G1) encodes a plant-specific gene family protein with an unknown domain. Mutation of G1 causes the sterile lemma to develop lemma-like characteristics (Yoshida et al., 2009). OsMADS34 is an important regulator of the identity of sterile lemma (Kobayashi et al., 2010; Gao et al., 2010). The osmads34 mutant shows enlarged glumes (sterile lemmas and rudimentary glumes) with lemma-like cellular structure. Similarly, the dg1 mutant also displays elongated rudimentary glumes and sterile lemmas (Yu et al., 2017). Furthermore, DROOPING LEAF (DL) signal was found in these abnormal glumes, indicating that both the sterile lemmas and rudimentary glumes had the lemma identity (Lin et al., 2014; Ren et al., 2016; Yu et al., 2017). ABERRANT SPIKELET AND PANICLE 1 (ASP1) and EXTRA GLUME 1 (EG1) determine the identity of sterile lemma (Li et al., 2009; Yoshida et al., 2012). In the asp1 and eg1 mutants, the elongated sterile lemma is caused by homeotic transformation of the sterile lemma into the lemma, and the asp1 mutant also displayed elongated rudimentary glumes, resembling the sterile lemma in size and structure. OsMADS1 plays a role in the identity of the lemma and palea, and its ectopic expression in sterile lemmas induced the lemma-like sterile lemmas (Jeon et al., 2000; Wang et al., 2017). Additionally, another class of genes, including FRIZZY PANICLE (FZP), MULTI-FLORET SPIKELET1 (MFS1), OsINDETERMINATE SPIKELET1 (OsIDS1), and SUPERNUMERARY BRACT (SNB) are also required to regulate the fate of glumes. Among these, loss-of-function of SNB and FZP induced extra rudimentary glumes with a loss of sterile lemmas on the corresponding rachilla position (Lee et al., 2007; Komatsu et al., 2003; Yi et al., 2005; Bai et al., 2017). Mutated MFS1 and OsIDS1 caused the transformation of sterile lemmas into rudimentary glumes (Ren et al., 2013; Lee and An, 2012). These findings suggest that different genes are involved in the regulation of the fate of glumes, and the lemma, rudimentary glume, and sterile lemma are homologous organs. However, the identities and origins of glumes, and the relationship between the rudimentary glume, lemma, and sterile lemma are not yet fully understood in grass.
In the study, we identified a new weak mutant allele of FZP, a mutant we named fzp-12. In this mutant, fzp-12 is mutated in a different genetic background and at a different site compared with the reported fzp mutants. In the previous studies, the fzp mutant failed to produce normal spikelets and grains and instead produced supernumerary rudimentary glumes (Bai et al., 2016; Komatsu et al., 2003). Interestingly, the fzp-12 mutant showed different phenotypes, including small grains and degraded sterile lemmas. Therefore, we particularly investigated the functions of FZP in grain size and sterile lemma identity. Our results revealed that FZP regulates grain size by mediating cell proliferation and expansion. Our findings, along with data on the expression of DL and G1 in sterile lemmas, also strongly support the hypothesis that the lemma, rudimentary glume, and sterile lemma are homologous organs. As such, our study provides a new function of FZP.
Materials and methods
Plant materials
The fzp-12 mutant was obtained from an M2 population from the japonica cultivar Zhonghua 11 (ZH11) with ethylmethane sulfonate treatment. ZH11 was regarded as the wild type in this study. We crossed the fzp-12 mutant with the cultivar Nan Jing 6 (NJ6, indica) to generate the F2 segregation population for gene mapping. Genetic analysis revealed that the fzp-12 mutant trait was determined by a recessive single gene (Supplemental Table 1). All plants used for morphological and genetic analysis were cultivated in the paddies under natural growing conditions at the China National Rice Research Institute (Hangzhou, Zhejiang Province) and (Lingshui, Hainan Island), China.
Microscopy analysis
For paraffin section observation, fresh spikelets from the wild type, fzp-12 mutant, and FZP over-expressing plants were placed in 70% formalin-acetic acid-alcohol (FAA) mixed solution, then samples were treated and stained as described by Ren et al. (2016). Next, we observed the stained sections with a microscope (NIKON ECLIPSE 90i). Meantime, for scanning electron microscopy (SEM) analysis, young and fresh spikelets and panicles were examined using a scanning electron microscope with a −30 °C cooled stage (HITACHI S-3500).
Map-based cloning and genetic transformation
To identify the mutated gene, the fzp-12 mutant was crossed with the NJ6, and 1189 F2 recessive plants with the mutant trait were used for mapping. The mutated site of the candidate gene was identified by sequencing analysis. To test whether LOC_Os07g47330/FZP was responsible for the mutant phenotypes, we constructed complementation vectors. A genomic DNA fragment covering a 2025-bp promoter and a 1251-bp downstream sequences was cloned into the binary vector pCAMBIA1301 (FZP-COM). The 957-bp full-length coding region of FZP was inserted into the expression vector 35S-1301 with the 35S promoter (Ren et al., 2016) to generate the 35S-FZP over-expression construct. Then, the FZP-COM and 35S-FZP plasmids were introduced into Agrobacterium tumefaciens and used to infect fzp-12 calli. The 35S-FZP vectors was introduced into A. tumefaciens and used to infect ZH11 calli. The primer sequences in Supplemental Table 2.
CRISPR-Cas9 targeting of FZP
For the CRISPR-Cas9 assay, we selected the specific candidate site in FZP. The CRISPR/Cas9 vector was constructed using the target sequence (GAGATACGCGACCCGACCACCA) to create the Cas9-FZP construct. The construct was introduced into A. tumefaciens and used to infect ZH11 calli.
RNA isolation and quantitative reverse transcription PCR test
Total RNA was extracted and further purified following the related kit instructions (Axygen). First strand cDNA was synthesized using the ReverTra Ace quantitative PCR RT Master Mix Kit (Toyobo) using 2 µg purified RNA in a 20 µl reaction volume. The quantitative qPCR was performed as described by Ren et al. (2016). Three biological replicates were performed and rice Actin was used as an endogenous control. Primers used for qPCR analysis in Supplemental Table 2.
In situ hybridization
Fresh young panicles from the wild type, fzp-12 mutant, and FZP over-expressing plants were placed in 70% FAA mixed solution (RNase-free), dehydrated through ethanol and xylene, and then embedded in paraffin (Sigma). Hybridization and immunological treatment of the sections were performed according to the methods in Sang et al., 2012. All probes were prepared using the same method and labeled with a RNA DIG Label (Roche). The primer sequences in Supplemental Table 2.
Promoter activity analysis
The 2033-bp promoter being located upstream of the coding frame was fused to the GUS (ß-glucuronidase) reporter gene and inserted into pCAMBIA1391Z (proFZP-GUS). The proFZP-GUS plasmid was introduced into EHA105 and used to infect ZH11. GUS activity in the transgenic plants was detected as described by Rao et al. (2015). Primers in Supplemental Table 2.
Subcellular localization of FZP protein
The coding frame of FZP was amplified from ZH11 and cloned into the 35S-GFP (S65T)-NOS (pCA1301) and 35S-YFP (S65T)-NOS (pCA1301) vectors to create the FZP-GFP and FZP-YFP recombinant vectors according to the In-Fusion Cloning Kit instructions (Takara). Then, the vectors of GFP and FZP-GFP, and the vectors of YFP and FZP-YFP were transformed into tobacco (Nicotiana tabacum) and rice protoplasts, and expressed, respectively (Ren et al., 2016). The samples were observed using a confocal microscope (OLYMPUS IX71). The primer sequences in Supplemental Table 2.
Yeast assay
The coding frame of FZP was amplified from ZH11. The coding frame of GS2 was amplified from ZH11 and used as the positive control (Hu et al., 2015). The two target fragments were inserted into the pGBKT7 vector to fuse with the GAL4 DNA-binding domain. All plasmids were transformed into yeast strain AH109 and the clones were selected on SD/–Trp plates. Then, the activation ability was tested on selective medium plates without histidine, tryptophan, and adenine (Ren et al., 2016).
Transcriptional activity analysis
Using the dual luciferase reporter assay system, we analyzed the transcriptional activity of FZP in rice protoplasts, and the DLR assay system was used with a GloMax 20-20 luminometer (Promega) to measure the relative luciferase activity (Wu et al., 2013). The primers in Supplemental Table 2.
Electrophoretic mobility shift assay
To produce the recombinant protein of HIS-FZP, the coding region of FZP was cloned into the pET-SUMO vector and expressed in the E. coli DE3 strain. The HIS-FZP recombinant proteins were purified as reported previously (Wang et al., 2013). All probes containing three repeat motifs were synthesized by TSING KE (Hangzhou) and labeled with biotin at the 5’ end for the electrophoretic mobility shift assay (EMSA). One µg of probes and different concentrations of FZP recombinant protein were mixed with binding buffer in a total volume of 15 µl. EMSAs were conducted according to the instructions of the LightShift Chemiluminescent EMSA Kit (Thermo). The primers and probe sequences in Supplemental Table 2.
Results
The fzp-12 mutant displays defects in the sterile lemma identity
The rice spikelet comprises one terminal floret and two pairs of vestigial glumes (sterile lemmas and rudimentary glumes) (Fig. 1A); these develop from the spikelet meristem. The sterile lemmas are present on the rachilla in an alternate phyllotaxy and are formed between the terminal floret and the rudimentary glumes which occur below the sterile lemmas (Fig. 1A, B).
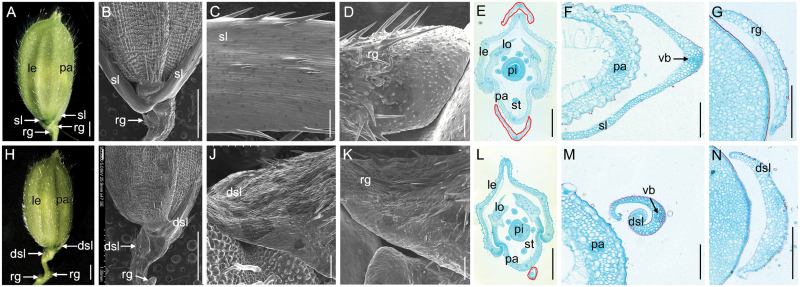
Fig. 1.
Phenotypes observation of spikelets in the wild type and the fzp-12 mutant. A, wild type spikelet. B-D, scanning electronic microscope analysis of wild type spikelet. C, epidermal surface of the sterile lemma. D, epidermal surface of the rudimentary glume. E-G, histological analysis of wild type spikelet. F, histological analysis of the sterile lemma. G, histological analysis of the rudimentary glume. H, fzp-12 spikelet. I-K, scanning electronic microscope analysis of fzp-12 spikelet. J, epidermal surface of the sterile lemma. K, epidermal surface of the rudimentary glume. L-N, histological analysis of fzp-12 spikelet. M, histological analysis of the sterile lemma. N, histological analysis of the sterile lemma. le, lemma; pa, palea; lo, lodicule; st, stamen; pi, pistil; sl, sterile lemma; dsl, degenerated sterile lemma; rg, rudimentary glume; vb, vascular bundle. Regions surrounded by red lines indicate the sterile lemma in E. Region surrounded by red line indicates the degenerated sterile lemma in L. Bars=1000 um in A, B, G, H; 100 um in C-F and I-L.
In wild-type plants, the sterile lemma of the spikelet differs from the rudimentary glume in size and structure (Fig. 1A–G). The sterile lemmas are larger and about 2 mm long, and the rudimentary glumes are about 0.5 mm long. Close examination revealed that in the fzp-12 mutant, the reduced sterile lemma formed at the expense of the normal sterile lemma in the corresponding position (Fig. 1A, B, H, I). The fzp-12 mutant produced smaller sterile lemmas that showed no visible differences from the rudimentary glumes of the fzp-12 mutant or the wild type (Fig. 1A, B, H–K). To address the effect of the fzp-12 mutation in detail, we investigated the cellular morphology of the sterile lemmas between the fzp-12 mutant and the wild type. We found one vascular bundle in the wild-type sterile lemma, and the wild-type epidermis was smooth with regularly arranged cells with rare trichomes on the abaxial surface (Fig. 1C–F). No vascular bundles were found in the wild-type rudimentary glumes, and the epidermis was rough and developed irregularly arranged cells and had lots of trichomes and small protrusions on its surface (Fig. 1D, G). In contrast, the fzp-12 mutant sterile lemmas were reduced to various degrees and even resembled the rudimentary glumes of the wild type or fzp-12 mutant in size (Fig. 1H–N). Abundant trichomes and protrusions were observed on the epidermis of the reduced sterile lemmas (Fig. 1J), which were highly similar to the rudimentary glume (Fig. 1D). Also, the regularly arranged and smooth cells were found in the distal and marginal regions in the sterile lemmas of the fzp-12 mutant (Fig. 1J), which were similar to those of the sterile lemma of the wild type. No homeotic transformations were observed in other spikelet organs in the fzp-12 mutant (Fig. 1E, L). These results suggested that the fzp-12 mutation specifically influences the sterile lemma, and the reduced sterile lemmas in the fzp-12 mutant were converted to rudimentary glume-like organs that had the identities of both normal rudimentary glumes and sterile lemmas.
Comparison of the early spikelet development between the wild type and fzp-12 mutant
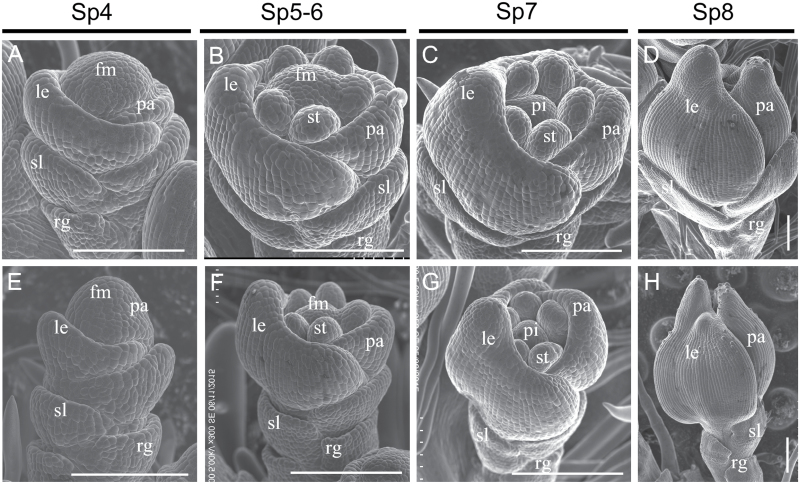
To elucidate how defective spikelet development progressed in the fzp-12 mutant, we monitored early developmental stages of young panicles by SEM (Fig. 2A–H; Fig. S1A–C). At the spikelet 4 stage (Sp4) of the wild type, the palea and lemma primordia were developing, and the wild-type sterile lemma was longer than the rudimentary glume (Fig. 2A). In the fzp-12 mutant, the sterile lemma was similar to the rudimentary glume in morphology and size (Fig. 2E). In the wild type during Sp5-Sp6 stages, we observed stamen primordia, cessation of growth of the rudimentary glume, and continued growth of the sterile lemma (Fig. 2B). In the fzp-12 mutant, the morphologies of the rudimentary glume and sterile lemma were not distinguishable from each other (Fig. 2F). During the Sp7 to Sp8 stage (when the pistil primordia are forming), the wild-type sterile lemma was much larger than the wild-type rudimentary glume (Fig. 2C, D, Fig. S1A–C). At these stages in the fzp-12 mutant, the sterile lemma remained its original size, resembling the rudimentary glume (Fig. 2G–H, Fig. S1A–C).
Fig. 2.
Developmental patterns of early spikelets in the wild type and the fzp-12 mutant. A-D, wild type spikelet. E-H, fzp-12 spikelet. fm, floral meristem; le, lemma; pa, palea; st, stamen; pi, pistil; sl, sterile lemma; rg, rudimentary glume; fm, floral meristem. Bars=100 μm.
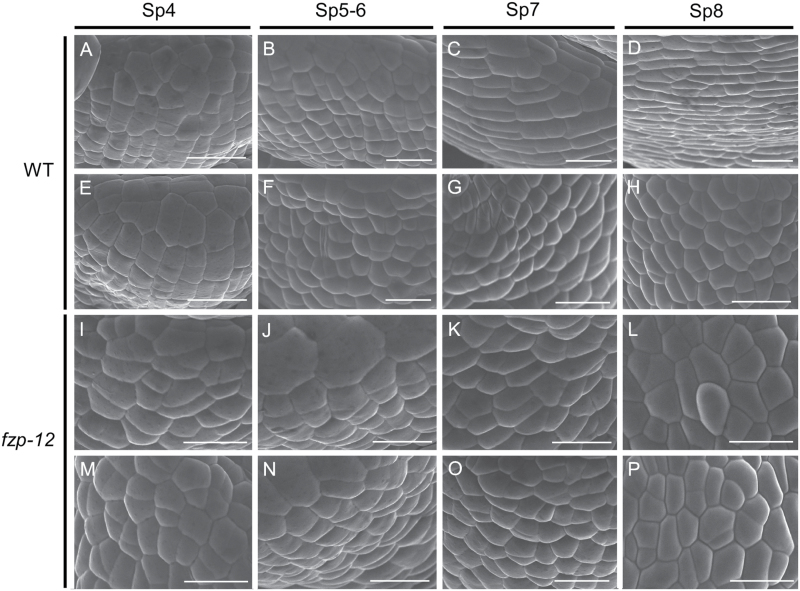
Further observations of the base of the spikelet showed that the identity of epidermal cells of the sterile lemma in the fzp-12 mutant is determined at the early stages (Fig. 3A–P). The surface of the sterile lemmas of the fzp-12 mutant and the wild type were not distinguishable from each other before the stamen initiation (Fig. 3A, B; I, J). In the wild type after the stamen primordia were formed, the epidermal cells in the sterile lemmas started to elongate and formed an orderly arrangement (Fig. 3C, D). Moreover, the epidermal cells of rudimentary glume maintained their size in the wild type (Fig. 3E–H), whereas the epidermal cells in the reduced sterile lemmas of the fzp-12 mutant became more similar to the cells of the rudimentary glume than the cells of the normal sterile lemma (Fig. 3K–P). These results implied that the identity of sterile lemma was altered in the fzp-12 mutant, and the abnormal sterile lemma showed a similar developmental pattern with that of the wild-type rudimentary glume.
Fig. 3.
Development of sterile lemma and rudimentary glume in the wild type and fzp-12 spikelets at early stages. A-D, epidermal surface of sterile lemma in the wild type. E-H, epidermal surface of rudimentary glume in the wild type. I-L, epidermal surface of sterile lemma in the fzp-12 mutant. M-P, epidermal surface of rudimentary glume in the fzp-12 mutant. Bars=10 μm.
The fzp-12 mutant produces small grains
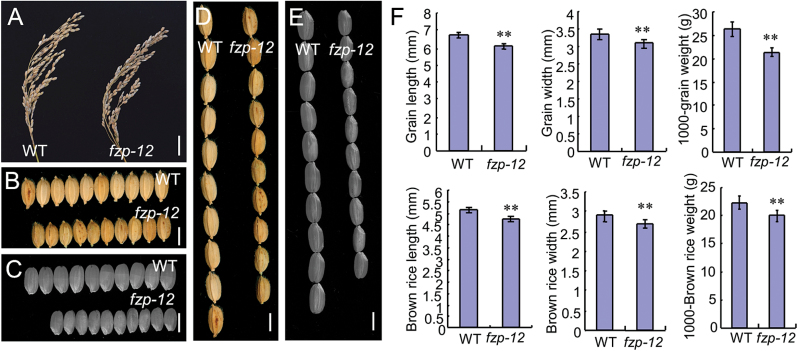
The fzp-12 mutant had obviously smaller grains than the wild type, and no obvious differences of grain number per panicle and seed setting rate were found in the wild type and fzp-12 mutant (Fig. 4A–F, Fig. S1D). Observations of early spikelet development showed that these spikelets were compressed and became dwarf throughout the early spikelet development stages (Fig. 2A–H), consistent with the smaller fzp-12 grains. At maturity, the widths of the grains and brown rice from the fzp-12 mutant were reduced compared with those of the wild type (Fig. 4B, C, F). The average widths of the grains and brown rice from the wild type were 3.3 and 3.0 mm, respectively (Fig. 4F), while the average widths of the grains and brown rice from the fzp-12 mutant were 2.9 and 2.7 mm, respectively (Fig. 4F). Also, the fzp-12 mutant grains and brown rice were shorter in comparison with those of the wild type (Fig. 4D–F). The average lengths of the grains and brown rice were 6.7 and 5.2 mm in the wild type, respectively (Fig. 4F), while the grains and brown rice from the fzp-12 mutant averaged 6.1 and 4.7 mm long, respectively (Fig. 4F). Additionally, the weights of 1000-grain and 1000 brown rice in the fzp-12 mutant were considerably decreased compared to the wild type (Fig. 4F). These results led to significantly decreased grain yield per plant in the fzp-12 mutant compared with the wild type (Fig. S1D). Together, these findings revealed that FZP affects grain length and width as well as grain yield.
Fig. 4.
FZP affects grain size. A, panicle of the wild type and fzp-12 mutant. B, mature grains of the wild type and fzp-12 mutant. C, brown rice of the wild type and fzp-12 mutant. D, mature grains of the wild type and fzp-12 mutant. E, brown rice of the wild type and fzp-12 mutant. F, length, width, and weight of mature grains and brown rice in the wild type and fzp-12 mutant. Bars=2 cm in A; 5 mm in B-E. **Significant difference at P<0.01 compared with the wild type by Student’s t-test.
Molecular cloning of FZP
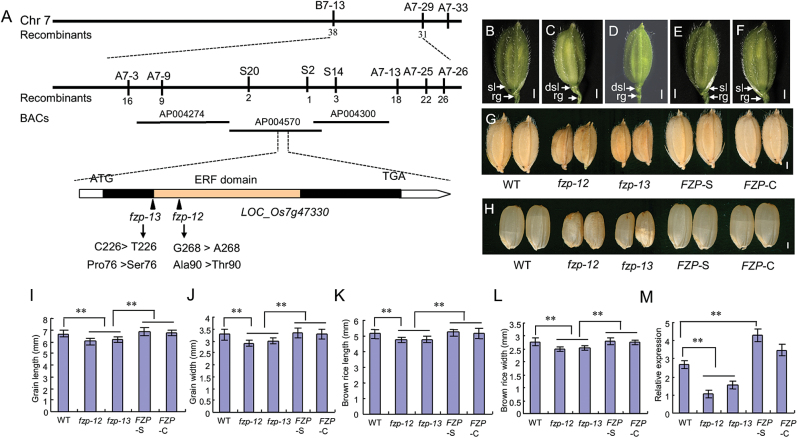
To isolate the mutated gene that determined the mutant phenotypes, we performed a cross between the fzp-12 mutant and NJ6, and all F1 plants showed the wild-type phenotype. In the F2 population, 3612 individuals exhibited the wild-type phenotype and 1189 plants exhibited the fzp-12 mutant phenotype, which fits the Mendelian segregation ratio (3:1) of a single recessive nuclear gene (Supplemental Table 1). Sixty individuals showing the fzp-12 phenotype were selected from the F2 population, and the mutation was preliminarily located between the simple-sequence repeat (SSR) markers B7-13 and A7-29 on chromosome 7 (Fig. 5A). Next, we used 36 SSR/SNP markers and 1129 recessive plants for fine mapping. The fzp-12 locus was delimited between the markers S2 and S20, a 66-kb region, which contained 7 open reading frames (ORFs) (Fig. 5A). By sequencing all predicted genes within this region, we found a single-nucleotide mutation from G to A within an ERF transcription factor (LOC_Os07g47330/FZP), which triggered an amino acid substitution of Ala-90 to Thr-90 in the fzp-12 mutant (Fig. 5A; Fig. S2).
Fig. 5.
Map-based cloning of the mutated gene. A, map position of fzp-12 locus. B, wild type spikelet. C, fzp-12 spikelet. D, fzp-13 spikelet. E, defective spikelets of fzp-12 mutant were rescued in the 35S::FZP–GFP complementation plants. F, defective spikelets of fzp-12 mutant were rescued in the genomic complementation plants. G, mature grains. fzp-13 mutant produced smaller grains, resembling those of fzp-12 mutant. And small grains of fzp-12 mutant were rescued in 35S::FZP–GFP complementation plants and genomic complementation plants. H, brown rice. fzp-13 mutant produced smaller brown rice grains, resembling those of fzp-12 mutant. And small brown rice grains of fzp-12 mutant were rescued in 35S::FZP–GFP complementation plants and genomic complementation plants. I, grain length. J, grain width. K, brown rice length. L, brown rice width. M, expression levels of FZP in the wild type, fzp-12 mutant, fzp-13 mutant, 35S::FZP–GFP complementation plants and genomic complementation plants. fzp-13 indicates the Cas9 knock-out plant. FZP-S indicates the 35S::FZP–GFP complementation plants. FZP-C indicates the genomic complementation plants. dsl, degenerated sterile lemma; sl, sterile lemma; rg, rudimentary glume. Bars=1 mm in B-H. Error bars indicate SD. **Significant difference at P<0.01 compared with the wild type by Student’s t-test.
To confirm whether LOC_Os07g47330 is causally linked to the fzp-12 mutant phenotypes, two plasmids, one contained the LOC_Os07g47330 wild-type genomic fragment and the other containing the wild-type cDNA driven by a 35S promoter (35S::FZP–GFP), were introduced into the fzp-12 calli. We obtained 28 and 20 transgenic lines from each transformation, respectively. Among which, 16 and 12 lines, respectively, harbored the transgene, and were resistant to hygromycin, and showed the wild-type phenotypes, which revealed that the fzp-12 phenotypes were completely rescued (Fig. 5B, C; E–M). Furthermore, we used the CRISPR/Cas9 gene editing to knock out FZP in the ZH11 wild-type plants. Sequencing analysis exhibited that the homozygous knocked out fzp-13 mutant had one single base mutation from C to T, resulting in an amino acid substitution of Pro-76 to Ser-76 (Figs 5A; S2). In the fzp-13 mutant, the expression of FZP was greatly reduced and this plant produced smaller grains and degenerated sterile lemmas (Fig. 5B–D; G–M), resembling the fzp-12 phenotypes. Additionally, we also found the knocked out fzp-14 mutant in the transgenic plants, and this mutant produced many branches that replaced the spikelets, resembling the phenotypes of previous reported fzp mutants. Sequencing analysis showed that the fzp-14 mutant showed one single base C deletion, resulting in a premature translation stop (Fig. S3). Together, these data confirmed that LOC_Os07g47330/FZP is the target gene, the mutated FZP is responsible for the mutant phenotypes.
FZP expression pattern and subcellular localization of its encoding protein
To determine the FZP expression pattern, we detected its expression in the fzp-12 mutant and wild type. The qPCR analysis showed that FZP was mainly expressed in young panicles, with higher levels in panicles shorter than 2 cm (Fig. S4A). No notable expression was found in the other tissues examined including roots, stems, leaves, and sheaths (Fig. S4A). However, FZP showed lower transcript levels in the fzp-12 mutant compared to the wild type (Fig. S4A). We fused the reporter gene (GUS gene) with the FZP promoter (proFZP::GUS), and transformed the recombinant plasmid into ZH11. GUS activity from proFZP-GUS was only found in the spikelets (Fig. S4B–F). Furthermore, Cross-section of the GUS stained spikelets displayed that the FZP promoter was specifically active in the outer spikelet organs including the sterile lemma, lemma and palea (Fig. S4G–I). And, no obvious GUS activity was observed in inner floral organs including the lodicule, stamen, and pistil (Fig. S4I). The expression pattern of FZP was different from the reported in situ hybridization expression pattern (Komatsu et al., 2003), and was consistent with its function in influencing spikelet development and grain size.
The FZPORF-GFP fusion protein and GFP alone, and the FZPORF-YFP fusion protein and YFP alone were transiently expressed in rice protoplasts and tobacco (Nicotiana tabacum) epidermal cells, respectively (Fig. S5). In cells expressing GFP or YFP only, the green and yellow fluorescence signals were visible throughout the cell (Fig. S5A–C; G–I). And, green fluorescence and yellow fluorescence were observed in the nuclei of rice protoplasts and tobacco epidermal cells containing the FZPORF-GFP and FZPORF-YFP fusion proteins (Fig. S5D–F; J–L). These results indicated that FZP encodes a nuclear-localized protein that may function as a transcription factor.
Transcriptional activity analysis of FZP
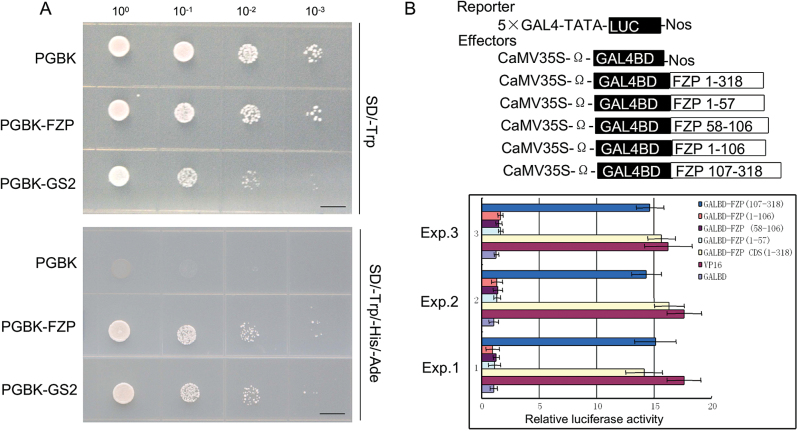
To explore the transcriptional activity of FZP, we fused the coding regions of FZP and the known transcriptional activator GS2 to the DNA-binding domain (BD) of yeast GAL4. The BD-GS2 construct and empty pGBKT7 vector were regarded as positive and negative controls, respectively (Hu et al., 2015). The transformed yeast cells harboring BD-FZP and BD-GS2 grew on the medium without histidine, tryptophan, and adenine, whereas the yeast cells harboring the pGBKT7 empty vector did not survive on the medium without histidine, tryptophan, and adenine (Fig. 6A), suggesting that FZP has transcriptional activation activity.
Fig. 6.
Transcriptional activity analysis of FZP protein. A, transcriptional activation assay in yeast cell. B, relative luciferase activities of rice protoplasts. PGBK, PGBK-FZP, and PGBK-GS2 represent the empty PGBK vector, the pGBKT7 vector fused with the coding frame of FZP, and the pGBKT7 vector fused with the coding frame of GS2. Bars=1 cm.
Next, we used a dual luciferase (LUC) reporter to further investigate the transcriptional activity, and examined the LUC activity in rice protoplasts. As in the yeast assays described above, the coding frame of the FZP cDNA was fused to the GAL4 DNA-binding domain (BD), but it was driven by the constitutive 35S promoter from Cauliflower mosaic virus (BD-FZP). The LUC gene that has five copies of binding sites was used for GAL4. VP16, a transcriptional activator, was used as a positive control and GAL4-BD was regarded as a negative control (Ren et al., 2016). The VP16, BD-FZP, and GAL4-BD effectors were transiently expressed in rice protoplasts. FZP and VP16 showed higher luciferase activity than that of GAL4-BD (Fig. 6B). These findings further demonstrated that FZP acts as a transcriptional activator and has transcriptional activation activity, implying that FZP may be involved in transcriptional regulation.
Further, we investigated the transcriptional activation activity of a set of truncated FZP proteins. The result revealed that the N-terminal regions including 1–57, 58–106 and 1–106 had no transcriptional activation activity, whereas the C-terminal region 107–318 conferred the same transactivation activity as FZP, suggesting that the C terminal region (107–318) is indispensable for the transcriptional activation activity (Fig. 6B).
Expression patterns of marker genes in the spikelet
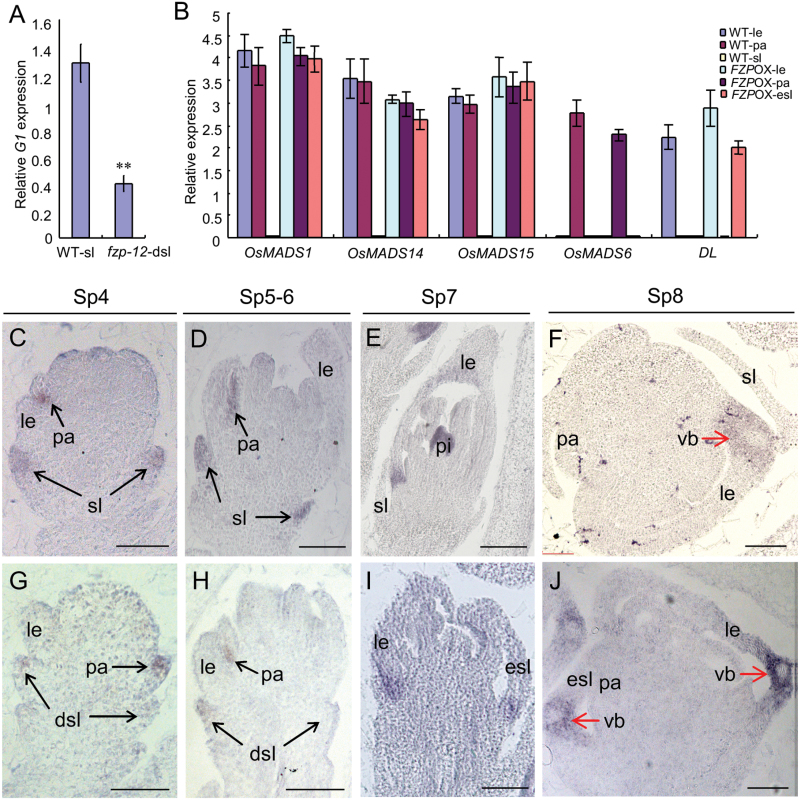
We used qRT-PCR to examine the expression of several genes that specify the sterile lemma or hull (lemma and palea). The expression of the sterile lemma identity gene, G1, was significantly decreased in the fzp-12 reduced sterile lemmas compared with that of the wild type (Fig. 7A), suggesting that the sterile lemma identity was affected in the fzp-12 mutant. In the wild type, the transcripts of the hull marker genes OsMADS15, OsMADS14, and OsMADS1, were detected in the palea and lemma, and the palea identity gene, OsMADS6, and the lemma identity gene, DL, were primarily expressed in the palea and lemma, respectively (Fig. 7B). In the FZP over-expressing plants, the sterile lemma was enlarged and had similar vascular bundles, cell layers, inward hook-like structures, and epidermal cells, including trichomes and protrusions, resembling those of the lemma in the wild type (Fig. S6A–H). Abundant transcripts of DL, OsMADS15, OsMADS14, and OsMADS1 were found, but no OsMADS6 transcript was observed in the elongated sterile lemmas (Fig. 7B). Meantime, the expressions of DL, OsMADS6, OsMADS15, OsMADS14, and OsMADS1 were not changed in the fzp-12 mutant (Fig. S7).
Fig. 7.
Expression of floral organ identity genes in the spikelets of the wild type, fzp-12 mutant and FZP over-expressing plant. A, qPCR analysis of G1 gene in the wild type and fzp-12 mutant. B, qPCR analysis of DL, OsMADS15, OsMADS14, OsMADS6, and OsMADS1 genes in the wild type and FZP over-expressing plant. C-D, G1 expression in the wild-type. E-F, DL expression in the wild-type. G-H, G1 expression in the fzp-12 mutant. I-J, DL expression in the FZP over-expressing plant. dsl, degenerated sterile lemma; esl, enlarged sterile lemma; sl, sterile lemma; le, lemma; pa, palea; pi, pistil; vb, vascular bundle. Bars=50 μm. Error bars indicate SD. **Significant difference at P<0.01 compared with the wild type by Student’s t-test.
Next, we investigated the expression of G1 and DL by in situ hybridization (Fig. 7C–J). Strong G1 expression was detected in sterile lemmas at Sp4-Sp6 stages in the wild type (Fig. 7C, D), but G1 signals were faint in the sterile lemma primordia at these stages in the fzp-12 mutant (Fig. 7G, H), suggesting that the degenerated sterile lemmas were derived from the normal sterile lemmas and partly retained the sterile lemma identity. In wild-type flowers, a strong DL signal was visible in the lemma at the Sp7-Sp8 stages (Fig. 7E, F), whereas in the FZP over-expressing plants, DL signals were observed in the elongated sterile lemmas and the original lemma (Fig. 7I, J), indicating that FZP over-expression induced a homologous change of the sterile lemma to the lemma. Thus, these results revealed that FZP indeed regulated the sterile lemma identity.
FZP influences cell proliferation and expansion in spikelet hulls
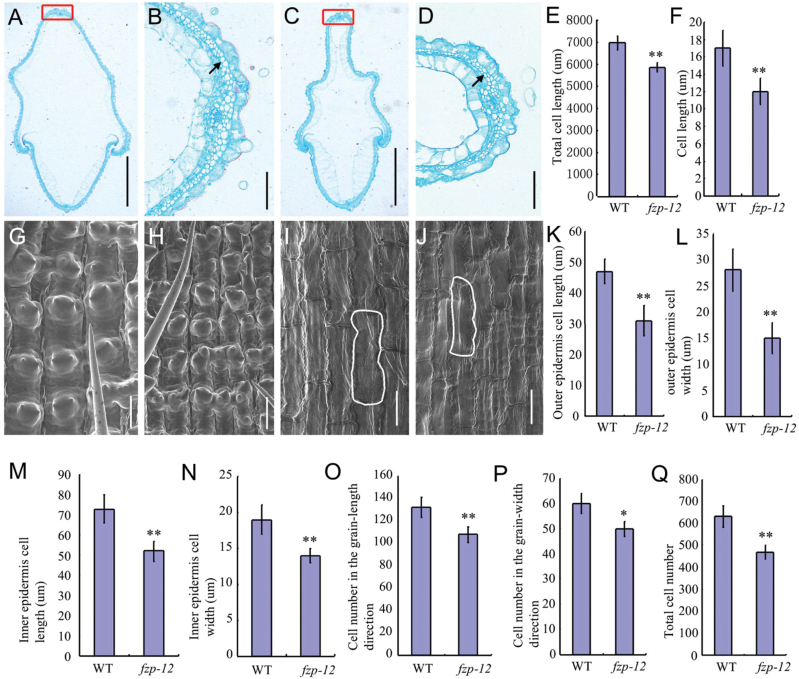
In rice, the spikelet hull restricts grain growth, thereby determining grain size (Duan et al., 2016). During organogenesis, cell proliferation and expansion coordinately regulate the organ growth (Jiang et al., 2012). To reveal the cellular basis underlying the change in grain size, we investigated cell size and number in the hulls of the wild type and the fzp-12 spikelet by SEM and paraffin sectioning (Fig. 8A–Q). Cross-sections of central region of the spikelet hull revealed that the parenchymal cell layer in the fzp-12 mutant hull was shorter and the hull contained smaller and fewer cells than the same cell layer in the wild type (Fig. 8A–D; E, F; Q). Similarly, there were fewer outer epidermal cells in the grain-width direction than in the wild type (Fig. 8P). The fzp-12 spikelet hulls had narrower and shorter cells in the outer epidermis than those of the wild-type spikelet hulls (Fig. 8G, H; K, L). Similarly, the fzp-12 mutant spikelet hulls had short, narrow cells in the inner epidermis compared to those of the wild type (Fig. 8I, J; M, N). We measured the number of outer epidermal cells along the longitudinal grain-length direction in the wild type and the fzp-12 mutant. The total cell number along the longitudinal grain-length direction was significantly reduced in the fzp-12 mutant compared with that of the wild type (Fig. 8O). These results suggested that the small-grain phenotype of the fzp-12 mutant mainly results from changes in cell size and number in the spikelet hull.
Fig. 8.
FZP influences cell proliferation and expansion in spikelet hulls. A, cross-sections of spikelet hulls in the wild type. B, magnified views of the box in A. C, cross-sections of spikelet hulls in the fzp-12 mutant. D, magnified views of the box in C. E, total cell length in the outer parenchymal cell layers of spikelet hulls. F, cell length in the outer parenchymal cell layers of spikelet hulls. G, SEM analysis of the outer surface in the wild type. H, SEM analysis of the outer surface in the fzp-12 mutant. I, SEM analysis of the inner surface in the wild type. J, SEM analysis of the inner surface in the fzp-12 mutant. K, outer epidermis cell length. L, outer epidermis cell width. M, inner epidermis cell length. N, inner epidermis cell width. O, cell number in the grain-length direction. P, cell number in the grain-width direction. Q, total cell number of outer parenchymal cell layers of spikelet hulls. Arrows indicate the parenchymal cell. Bars=1000 um in A and C; 100 um in B and D; 50 um in G and J. **Significant difference at P<0.01 compared with the wild type by Student’s t-test; *Significant difference at P<0.05 compared with the wild type by Student’s t-test.
Several grain-size genes regulate cell proliferation and expansion. SPL13/GWL7, GL7/GW7/SLG7, GS2, SRS5, PGL1, PGL2, and APG influence cell expansion, and GS3, GL3, GS5, GW2, and GW8 are involved in regulation of cell proliferation. To reveal how FZP regulates cell proliferation and expansion in spikelet hulls, we detected their expression levels in the wild-type and fzp-12 mutant panicles. Compared with the wild type, the expression levels of GWL7, SRS5, GL7, PGL1, PGL2, GS3, GS5, and GW8 were significantly reduced in fzp-12 panicles (Fig. S8A), and the expression levels of GS2, GL3, GW2, APG in fzp-12 panicles were similar between the fzp-12 mutant and the wild type (Fig. S8A).
Furthermore, we investigated the expression of several genes that regulate cell cycle and cell expansion. Compared with the wild type, ten cell cycle-related genes, CYCT1, CYCA2-1, CYKA2, CYCD3, CYCD20, H1, E2Fa, MAD2, MCM3, and MCM5, showed significantly lower expression in the fzp-12 mutant than in wild type (Fig. S8B). The expression levels of five cell expansion-related genes, OsEXPA32, OsEXPA2, OsEXPA1, OsEXPB11, and OsEXPB9, were also much lower in the fzp-12 mutant compared with that of the wild type (Fig. S8B). Together, these results supported the role of FZP in the regulation of cell proliferation and expansion in spikelet hulls, and thereby in grain size.
FZP interacts specifically with the GCC-box and DRE motifs
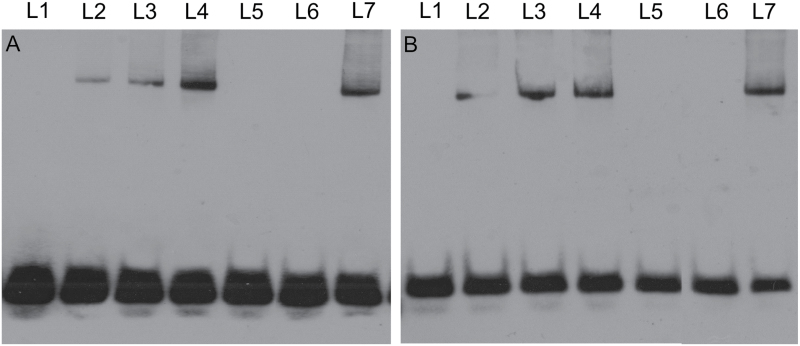
The GCC-box and dehydration-responsive element (DRE) motifs are recognized by ERF subfamily proteins, which belong to the AP2/ERF superfamily (Rong et al., 2014). Therefore, ERF proteins can directly or indirectly activate the expression of downstream genes with the GCC-box or DRE motifs, thereby affecting plant development. In this study, FZP also encodes an ERF domain protein and belongs to the AP2/ERF superfamily. We investigated whether FZP binds to the GCC-box and DRE motifs, as well as to GCC-box and DRE motifs containing two point mutations, by an electrophoretic mobility shift assay (EMSA). The EMSA results showed that FZP could directly combine with the normal GCC-box and DRE DNA fragments, but it failed to combine with the mutated GCC-box and DRE DNA fragments (Fig. 9A, B). Additionally, different concentrations of FZP exhibited different binding abilities with the normal GCC-box and DRE DNA fragments (Fig. 9A, B). These results indicated that FZP interacts specifically with the GCC-box and DRE motifs in the promoters of related downstream genes, and may thereby directly regulate their expression.
Fig. 9.
FZP protein interacts with the GCC-box and DRE motifs. A, FZP protein could bind to the normal GCC-box and failed to interact with mutated GCC-box (mGCC-box). B, FZP protein could bind to the normal DRE and failed to interact with mutated DRE (mDRE). L1, L2, L3 L4 L5, L6 and L7 in A indicate GCCbox, GCCbox+0.5 ug protein, GCCbox+1 ug protein, GCCbox+1.5 ug protein, mGCCbox+1.5 ug protein, GCCbox+1.5 ug protein+100×specific competitor, and GCCbox+1.5 ug protein+100×non-specific competitor, respectively. L1, L2, L3 L4 L5, L6 and L7 in in B indicate DRE, DRE+0.5 ug protein, DRE+1 ug protein, DRE+1.5 ug protein, mDRE+1.5 ug protein, DRE+1.5 ug protein+100×specific competitor, and DRE+1.5 ug protein+100×non-specific competitor, respectively. L, lane.
Discussion
FZP plays an important role in determining grain size
Grain size is coordinately determined by grain length, width and thickness, which are controlled by several genetic factors in rice. However, our knowledge of the complicated genetic mechanisms by which rice grain size is controlled is still limited.
FZP encodes an ERF protein with transcriptional activation activity, and the fzp-12 loss-of-function mutant showed short and narrow grains. Early spikelets of fzp-12 mutant became dwarf, consistent with the smaller fzp-12 grains. The knocked out fzp-13 mutant also produced small grains that resembled those of the fzp-12 mutant. These results suggest that FZP is an important regulator of grain size and shape in rice. Cell proliferation and cell expansion determine organ size (Jiang et al., 2012). Our results revealed that FZP is required for cell proliferation and expansion in rice grains. The spikelet hulls of the fzp-12 mutant had smaller and fewer cells than the wild-type spikelet hulls, and the transcript levels of several genes involved in cell proliferation and expansion were significantly reduced in the fzp-12 mutant. These findings suggested that FZP influences grain size by modifying cell proliferation and expansion.
FZP determines sterile lemma fate in rice
The sterile lemma and rudimentary glume of grasses are unique organs (Li et al., 2009). In previous studies, the fzp mutants only produced supernumerary bract-like rudimentary glumes, and lacked normal florets and sterile lemmas (Komatsu et al., 2003; Yi et al., 2005). Rarely, the bract-like organs partly harbored the identity of the sterile lemma in the fzp mutant (Komatsu et al., 2003). In our study, the fzp-12 and knocked out fzp-13 mutants produced the spikelets with one pair of reduced sterile lemmas, which resembled the rudimentary glume of the wild type and the bract-like organs of the reported fzp mutants in size. The sterile lemmas of the fzp-12 and fzp-13 mutants, and the bract-like organs of the reported fzp mutants exhibited similar protrusions and trichomes to those of the rudimentary glume of the wild type (Fig. 1; Komatsu et al., 2003). These results indicated that these reduced glume-like organs or bract-like rudimentary glumes may be derived from the sterile lemmas. Next, we found no expression of DL, OsMADS15, OsMADS14, OsMADS6, and OsMADS1 in sterile lemmas of the wild type and fzp-12 mutant, suggesting that the sterile lemma of the fzp-12 mutant does not have the lemma/palea identity. Compared with the wild type, G1 signals were significantly decreased in the sterile lemmas of the fzp-12 mutant, indicating that the sterile lemma identity is altered in the fzp-12 mutant. The sterile lemma in the FZP over-expressing plants was enlarged and had a similar cellular structure, resembling the wild-type lemma. We detected abundant transcripts of DL, OsMADS15, OsMADS14, and OsMADS1, but no OsMADS6 signals were found in the sterile lemmas of the FZP over-expressing plants. These findings indicated that the sterile lemma is converted to the lemma-like organ and had the lemma identity in these plants. Cross-section of the spikelets showed that GUS activity was obviously visible in the sterile lemma, which was consistent with our phenotypic observations (i.e. reduced sterile lemmas). Together with the strong expression of FZP in the sterile lemma, our results showed that FZP controls the identity of the sterile lemma, and is vital in the development of sterile lemma by repressing the development of the rudimentary glume.
Two hypotheses have been proposed on the evolution and origin of the sterile lemma (Ren et al., 2013; Takeoka et al., 1993). One hypothesis suggests that the spikelet of a putative ancestor of Oryza contained three florets: a terminal floret and two lower lateral florets. In time, the lateral florets lost all inner floral organs and subsequently degenerated into the lemma (Kellogg, 2009). The sterile lemma was regarded as the remnant of the morphologically modified lemma (Yoshida et al., 2009). An alternative hypothesis proposes that the spikelet of a putative ancestor of Oryza only produced one floret, and the rudimentary glume and sterile lemma were reduced, bract-like organs. In the dg1, g1, eg1, asp1, and osmads34 mutants, the sterile lemmas were elongated and transformed into lemma-like organs, which supports the first idea that the lemma and sterile lemma were homologous organs. In the osids1 and mfs1 mutants, the sterile lemma was not fully formed and acquired the identity of the rudimentary glume, thereby supporting the second hypothesis and the view that the rudimentary glume and sterile lemma were homologous organs. In our study, the sterile lemma of the fzp-12 mutant was reduced and resembled the rudimentary glume, whereas the sterile lemma was elongated and converted to the lemma-like structure in the FZP over-expressing plants. In addition, the rudimentary glumes were also enlarged and had the identity of the sterile lemma or lemma in the asp1, dg1, and osmads34 mutants (Yoshida et al., 2012; Ren et al., 2016; Yu et al., 2017). The spikelet of most grass species contains florets and glume-like organs that are equivalent to the rudimentary glumes of Oryza spp., but lacks sterile lemma-like organs (Hong et al., 2010; Yoshida et al., 2009). In some grass species, the bract-like glume organ is similar in structure and size to the lemma (Yoshida et al., 2009; Ren et al., 2013), whereas in Oryza spp., this organ is severely reduced (Bommert et al., 2005; Li et al., 2009; Ren et al., 2013). Together with these hypotheses and previous studies, our results strongly support the view that the lemma, rudimentary glume, and sterile lemma were homologous structures.
A conserved function of AP2/ERF genes
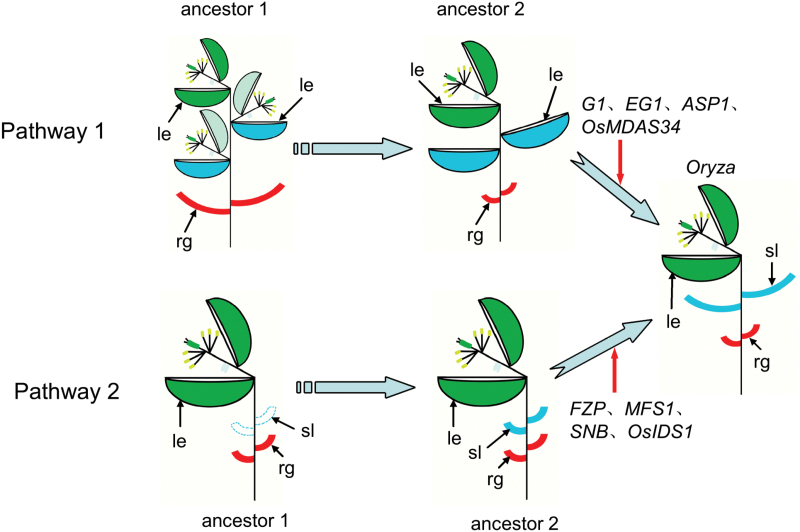
The plant-specific AP2/ERF gene family includes four subfamilies: ERF, AP2, DREB, and RAV (Sharoni et al., 2011). Previous studies showed that SNB and OsIDS1 encode an AP2 domain protein, and play a role in the specification of sterile lemma fate. FZP and MFS1 belong to the ERF subfamilies and also determine the sterile lemma identity. The plants with mutations in SNB produced extra rudimentary glumes; however, no corresponding sterile lemmas were observed in the original position. Loss of function of FZP, MFS1, and OsIDS1 caused the transformation of the sterile lemmas into rudimentary glumes. These AP2/ERF genes have a unique AP2 domain (Fig. S9), showed similar expression patterns and were strongly expressed in the sterile lemma. The other class of genes including EG1, ASP1, G1/ELE1, and OsMADS34, encode non-AP2/ERF domain proteins, and also affect the development of the sterile lemma. The sterile lemmas were elongated and had the lemma identity in eg1, asp1, g1 and osmads34 mutants. These findings suggest that the development of the sterile lemma is controlled by two different regulatory pathways. EG1, ASP1, G1/ELE, and OsMADS34 determine the sterile lemma identity by preventing the homologous transformation of the sterile lemma into the lemma, whereas, the functions of the AP2/ERF genes FZP, MFS1, OsIDS1, and SNB are conserved in the regulation of the sterile lemma identity by mainly preventing the degeneration of the sterile lemma into the rudimentary glume. Interestingly, O. grandiglumis has a large sterile lemma that resembles the lemma, whereas the genus Leersia belongs to Oryzeae and lacks the sterile lemma. These results support the above-mentioned hypotheses, and suggest the existence of four putative ancestors of Oryza spikelet that lead to the different types of spikelets and two evolutionary pathways that determine the origin of sterile lemma (Fig. 10). In hypothesized pathway 1, the putative ancestor of the spikelet of genus Oryza contained three florets: two lateral florets and one terminal floret. Subsequently, the lateral florets degenerated into the lemmas, consistent with the previous idea (Yoshida et al., 2009; Kellogg, 2009; Ren et al., 2013), suggesting that EG1, ASP1, G1/ELE, and OsMADS34 may be recruited to specify the sterile lemma by repressing the lemma identity during evolution (Fig. 10). In hypothesized pathway 2, the putative ancestor of the spikelet of genus Oryza had a terminal floret and a pair of rudimentary glumes, similar with the genus Leersia. During evolution, the spikelet developed the rudimentary glume-like sterile lemma, which subsequently were elongated, suggesting that the AP2/ERF genes (FZP, MFS1, OsIDS1, and SNB) may be recruited to specify the sterile lemma by suppressing the rudimentary glume identity (Fig. 10).
Fig. 10.
Hypothesized evolutionary pathway in spikelet architecture of Oryza (Yoshida et al., 2009) and a conserved function of AP2/ERF genes. rg, rudimentary glume; sl, sterile lemma; le, lemma. The pathway 1 indicates the evolutionary process in which the sterile lemma is gradually reduced, and this hypothesized process has been shown (Yoshida et al., 2009). The pathway 2 indicates the evolutionary process in which the sterile lemma starts to appear and elongate. The putative ancestor 1 of genus Oryza had three florets (one terminal floret and two lower lateral florets) and a pair of rudimentary glumes in a single spikelet in the pathway 1. The putative ancestor 2 of genus Oryza had one terminal floret, two lower lemma-like organs, and a pair of rudimentary glume in a single spikelet in the pathway 1. The putative ancestor 1 of genus Oryza had one terminal floret and a pair of rudimentary glume, and lacked the sterile lemma-like organ in a single spikelet in the pathway 2. The putative ancestor 2 of genus Oryza had one terminal floret and two rudimentary glume-like organs in a single spikelet in the pathway 2.
Supplementary data
Supplementary data are available at JXB online.
Supplemental Figure 1. Investigation of early spikelets and agronomic traits in the wild type and fzp-12 mutant.
Supplemental Figure 2. Protein sequence alignment of FZP, fzp-12, and fzp-13.
Supplemental Figure 3. fzp-14 mutant and protein sequence alignment of FZP and fzp-14.
Supplemental Figure 4. Expression pattern of FZP.
Supplemental Figure 5. Subcellular localization of the FZP protein.
Supplemental Figure 6. Spikelets and FZP expression in the wild type and FZP over-expressing plants.
Supplemental Figure 7. Expression of floral organ identity genes in the wild type and fzp-12 mutant.
Supplemental Figure 8. Expression levels of related genes in the wild type and fzp-12 mutant.
Supplemental Figure 9. Protein sequence alignment of AP2/ERF genes.
Supplemental Table 1. Genetic analysis of the mutant phenotypes of fzp-12.
Supplemental Table 2. Primers used in the study.
Supplementary Material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31521064 and 91735304), the National Science and Technology Major Project (2016ZX08009003-003-008), the Zhejiang Natural Science Foundation (LY18C130007), the National Natural Science Foundation of China (31671666), and the National GMO New Variety Breeding Program of PRC (2016ZX08011-001). The authors declare that they have no competing interests.
Author contributions
Q.Q., and R. D. designed the research. R.D., H.J., X.Q., C.Y. and X.D. performed the experiments. Z.D., Z.G., G.Z., Z.L. and G.L. analyzed the data. Z.Y., R.Y., S.L., and C.G., conducted the fieldwork. R.D., H.J., X.Q., and Q.Q. wrote the manuscript.
References
- Bai X, Huang Y, Mao D, Wen M, Zhang L, Xing Y. 2016. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Scientific Reports 6, 19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Huang Y, Hu Y, Liu H, Zhang B, Smaczniak C, Hu G, Han Z, Xing Y. 2017. Duplication of an upstream silencer of FZP increases grain yield in rice. Nature Plants 3, 885–893. [DOI] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY. 2005. Genetics and evolution of inflorescence and flower development in grasses. Plant & Cell Physiology 46, 69–78. [DOI] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, et al. 2016. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nature Plants 2. [DOI] [PubMed] [Google Scholar]
- Duan P, Ni S, Wang J, Zhang B, Xu R, Wang Y, Chen H, Zhu X, Li Y. 2016. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nature Plants 2, 15203. [DOI] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. 2006. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theoretical and Applied Genetics 112, 1164–1171. [DOI] [PubMed] [Google Scholar]
- Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D. 2010. The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiology 153, 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D, Sassa H. 2012a. An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breeding Science 62, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D, Sassa H. 2012b. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One 7, e31325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Qian Q, Zhu K, Tang D, Huang Z, Gao L, Li M, Gu M, Cheng Z. 2010. ELE restrains empty glumes from developing into lemmas. Journal of Genetics and Genomics 37, 101–115. [DOI] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, et al. 2015. A rare allele of GS2 enhances grain size and grain yield in rice. Molecular Plant 8, 1455–1465. [DOI] [PubMed] [Google Scholar]
- Huang K, Wang D, Duan P, Zhang B, Xu R, Li N, Li Y. 2017. WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. The Plant Journal 91, 849–860. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, et al. 2000. Leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. The Plant Cell 12, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Bao L, Jeong SY, Kim SK, Xu C, Li X, Zhang Q. 2012. XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. The Plant Journal 70, 398–408. [DOI] [PubMed] [Google Scholar]
- Kellogg E. 2009. The evolutionary history of Ehrhartoideae, Oryzeae, and Oryza. Rice 2, 1–14. [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. 2010. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant & Cell Physiology 51, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J. 2003. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841–3850. [DOI] [PubMed] [Google Scholar]
- Lee DY, An G. 2012. Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. The Plant Journal 69, 445–461. [DOI] [PubMed] [Google Scholar]
- Lee DY, Lee J, Moon S, Park SY, An G. 2007. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. The Plant Journal 49, 64–78. [DOI] [PubMed] [Google Scholar]
- Li H, Xue D, Gao Z, Yan M, Xu W, Xing Z, Huang D, Qian Q, Xue Y. 2009. A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. The Plant Journal 57, 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Li Y. 2016. Signaling pathways of seed size control in plants. Current Opinion in Plant Biology 33, 23–32. [DOI] [PubMed] [Google Scholar]
- Lin X, Wu F, Du X, Shi X, Liu Y, Liu S, Hu Y, Theißen G, Meng Z. 2014. The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytologist 202, 689–702. [DOI] [PubMed] [Google Scholar]
- Liu L, Tong H, Xiao Y, Che R, Xu F, Hu B, Liang C, Chu J, Li J, Chu C. 2015. Activation of Big Grain 1 significantly improves grain size by regulating auxin transport in rice. Proceedings of the National Academy of Sciences, USA 112, 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q. 2010. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proceedings of the National Academy of Sciences, USA 107, 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang L, Gao JP, Lin HX. 2012. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Research 22, 1666–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Yang Y, Xu J, et al. 2015. EARLY SENESCENCE1 Encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiology 169, 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Li Y, Zhao F, et al. 2013. MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiology 162, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Rao Y, Leng Y, et al. 2016. Regulatory role of OsMADS34 in the determination of glumes fate, grain yield, and quality in rice. Frontiers in Plant Science 7, 1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yu H, Rao Y, et al. 2018. ‘Two-floret spikelet’ as a novel resource has the potential to increase rice yield. Plant Biotechnology Journal 16, 351–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong W, Qi L, Wang A, Ye X, Du L, Liang H, Xin Z, Zhang Z. 2014. The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnology Journal 12, 468–479. [DOI] [PubMed] [Google Scholar]
- Sang X, Li Y, Luo Z, et al. 2012. CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiology 160, 788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharoni AM, Nuruzzaman M, Satoh K, Shimizu T, Kondoh H, Sasaya T, Choi IR, Omura T, Kikuchi S. 2011. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant & Cell Physiology 52, 344–360. [DOI] [PubMed] [Google Scholar]
- Takeoka Y, Shimizu M, Wada T. 1993. Panicles. In: Matsuo T, Hoshikawa K, eds. Science of the Rice Plant, Vol I Tokyo: Nobunkyo, 295–326. [Google Scholar]
- Wang P, Du Y, Zhao X, Miao Y, Song CP. 2013. The MPK6-ERF6-ROS-responsive cis-acting Element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiology 161, 1392–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zeng X, Zhuang H, Shen Y, Chen H, Wang Z, Long J, Ling Y, He G, Li Y. 2017. Ectopic expression of OsMADS1 caused dwarfism and spikelet alteration in rice. Plant Growth Regulation 81, 433–442. [Google Scholar]
- Wang S, Li S, Liu Q, et al. 2015a. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nature Genetics 47, 949–954. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiong G, Hu J, et al. 2015b. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nature Genetics 47, 944–948. [DOI] [PubMed] [Google Scholar]
- Wu X, Tang D, Li M, Wang K, Cheng Z. 2013. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiology 161, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G, Choi JH, Jeong EG, Chon NS, Jena KK, Ku YC, Kim DH, Eun MY, Jeon JS, Nam MH. 2005. Morphological and molecular characterization of a new frizzy panicle mutant, “fzp-9(t)”, in rice (Oryza sativa L.). Hereditas 142, 92–97. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Suzaki T, Tanaka W, Hirano H. 2009. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proceedings of the National Academy of Sciences, USA 106, 20103–20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano HY. 2012. ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. The Plant Journal 70, 327–339. [DOI] [PubMed] [Google Scholar]
- Yu H, Ruan B, Wang Z, et al. 2017. Fine mapping of a novel defective glume 1 (dg1) mutant, which affects vegetative and spikelet development in rice. Frontiers in Plant Science 8, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang J, Huang J, et al. 2012. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proceedings of the National Academy of Sciences, USA 109, 21534–21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.