The winter-dormant shoot apical meristem of the tree species Populus keeps an epigenetic memory of environmental variations that arose during the preceding vegetative period.
Keywords: Differentially methylated regions, DNA methylation, drought, environment, epigenetics, field grown, poplar, shoot apical meristem, stress memory
Abstract
Trees have a long lifespan and must continually adapt to environmental pressures, notably in the context of climate change. Epigenetic mechanisms are doubtless involved in phenotypic plasticity and in stress memory; however, little evidence of the role of epigenetic processes is available for trees growing in fields. Here, we analyzed the possible involvement of epigenetic mechanisms in the winter-dormant shoot apical meristem of Populus × euramericana clones in memory of the growing conditions faced during the vegetative period. We aimed to estimate the range of genetic and environmentally induced variations in global DNA methylation and to evaluate their correlation with changes in biomass production, identify differentially methylated regions (DMRs), and characterize common DMRs between experiments. We showed that the variations in global DNA methylation between conditions were genotype dependent and correlated with biomass production capacity. Microarray chip analysis allowed detection of DMRs 6 months after the stressful summer period. The 161 DMRs identified as common to three independent experiments most notably targeted abiotic stress and developmental response genes. Results are consistent with a winter-dormant shoot apical meristem epigenetic memory of stressful environmental conditions that occurred during the preceding summer period. This memory may facilitate tree acclimation.
Introduction
Trees are sessile perennial organisms, which therefore respond continuously to environmental pressures over many years. Understanding the mechanisms underlying phenotypic plasticity and stress memory in trees is of utmost importance in the context of rapid climate change (Gagliano et al., 2014). Phenotypic plasticity is the ability of a genotype to rapidly generate and display different phenotypes under distinct environmental conditions (Sultan, 2000; Nicotra et al., 2010). While several studies have been conducted for annual plants (Bossdorf et al., 2010; Kooke et al., 2015; Hébrard et al., 2016), DNA methylation under stress conditions has been studied for only a limited number of trees at the genome level to investigate its putative role in phenotypic plasticity and ecophysiological traits (Conde et al., 2017; Lafon-Placette et al., 2018). Indeed, chromatin marks such as DNA methylation (Niederhuth and Schmitz, 2017) provide strong plasticity and modulate the development, morphology, and physiology of plants by controlling gene expression and the mobility of transposable elements (TEs) (Jablonka et al., 1995; Pál and Miklós, 1999; Angers et al., 2010; Mirouze and Paszkowski, 2011; Kooke et al., 2015; Ong-Abdullah et al., 2015; Yakovlev et al., 2016; Richards et al., 2017).
Recent studies have highlighted how DNA methylation helps trees adapt to changing environments and favors the functional diversity associated with productivity and population stability (Choi and Sano, 2007; Bräutigam et al., 2013; Latzel et al., 2013; Rico et al., 2014; Kawakatsu et al., 2016; Richards et al., 2017). Methylation-induced modifications may be either reversible or retained during cell division (an epigenetic phenomenon) and passed on to daughter cells, as occurs during vernalization (intragenerational transmission) or to the next generation, as shown in EpiRIL (EPIgenetic Recombinant Inbred Lines) populations for inter-generational transmission (Johannes et al., 2009; Reinders et al., 2009; Baulcombe and Dean, 2014; Kooke et al., 2015; Meyer, 2015; Kawakatsu et al., 2016). Another well-described footprint or ‘memory’ system in plants is defense priming, which controls responses to pathogen or herbivore attacks (Pastor et al., 2013; Espinas et al., 2016; Mauch-Mani et al., 2017). The concept of priming has also been applied to responses to abiotic stresses, such as water deficit (Sultan et al., 2009; Ding et al., 2012; Fleta-Soriano and Munné-Bosch, 2016; Mauch-Mani et al., 2017). The priming event is followed by a period of stress memory, which involves storing information about the priming stress, potentially through an epigenetic phenomenon, and results in a modified response upon recurring exposure to a stress or a sustained response after the priming stress (Lämke and Bäurle, 2017). This memory may last from several days to weeks for somatic stress memory, and in some cases may even extend to offspring (Lämke and Bäurle, 2017). For instance, the response to repeated drought stress in Arabidopsis thaliana is mediated by transcriptional memory; that is, an increase in the transcription levels of stress response genes occurs (Ding et al., 2012; Avramova, 2015). Hyperosmotic stress memory in A. thaliana is also associated with distinct regions of the Arabidopsis genome that are susceptible to DNA (de)methylation. Furthermore, this memory is transmitted to the immediate progeny through the female lineage, due to widespread DNA glycosylase activity in the male germline (Wibowo et al., 2016). Interestingly, genes related to stress memory have been found to be partially conserved between A. thaliana and Zea mays when the plants are exposed to the same constraints, suggesting that memory could be an evolutionarily conserved response to repeated abiotic stress (Ding et al., 2014). This memory is particularly crucial for perennial organisms such as trees, since episodes of drought are predicted to increase in both frequency and intensity with ongoing global climate change (Allen et al., 2010; IPCC, 2014). Evidence of an epigenetic memory in trees has been described in Norway spruce: different temperatures during embryogenesis induced changes in adult tree traits such as bud phenology or frost tolerance, suggesting an epigenetic memory transmitted from the embryo to the adult plant (Yakovlev et al., 2012). This temperature-related epigenetic memory modifies the expression of bud burst-related genes in epitypes and affects the timing of bud burst in the progeny (Carneros et al., 2017).
Poplar (Populus spp.) is a model tree owing to its widely available genome, the large number of genetic and phenotypic variations it exhibits, and its fast growth associated with large water requirements. This makes poplar ideal for dissecting the relationship between the ecophysiological and molecular determinants of water deficit tolerance (Tuskan et al., 2006; Jansson and Douglas, 2007). In this context, a better characterization of the epigenetic component of tree phenotypic plasticity and stress memory seems crucial (Bräutigam et al., 2013; Plomion et al., 2016). Previous studies on poplar have shown that global DNA methylation varies across hybrids and is correlated with biomass productivity and water deficit (Gourcilleau et al., 2010; Raj et al., 2011). Site-dependent hemimethylation statuses and potential responses to environmental and edaphic conditions have also been reported (Guarino et al., 2015). In addition, gene-body DNA methylation in poplar, as compared with Arabidopsis, is more extensive in the open chromatin state; it is linked to structural gene characteristics and is correlated with tissue-specific gene expression or stress (Vining et al., 2012; Bräutigam et al., 2013; Lafon-Placette et al., 2013; Liang et al., 2014; Lafon-Placette et al., 2018). The importance of poplar’s epigenetic component has recently been underlined in site-dependent growth performance (Schönberger et al., 2016) and in the developmental phenotypic plasticity of shoot apical meristem (SAM) in response to environmental bud-break conditions (Conde et al., 2017) and water availability (Lafon-Placette et al., 2018).
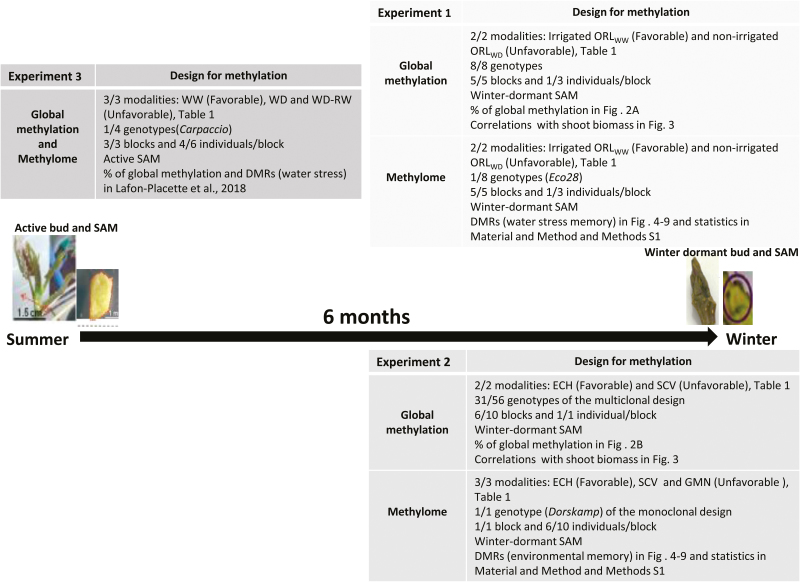
In the present study, we provide new insights into the epigenetic memory of environmental stress in poplar SAM. We analyzed global variations in DNA methylation levels in winter-dormant SAM of Populus deltoides × Populus nigra genotypes that had faced contrasted growing conditions the previous summer, making use of both a field controlled-drought trial (Experiment 1; Fichot et al., 2010, 2011; Table 1, Fig. 1) and pedoclimatic gradient among three sites in France (Experiment 2; Toillon et al., 2013a, b; Table 1, Fig. 1). For global DNA methylation, we used high-performance liquid chromatography (HPLC) to investigate variations in DNA methylation (see Causevic et al., 2005; Gourcilleau et al., 2010; Trap-Gentil et al., 2011; Zhu et al., 2013) and the methylated DNA immunoprecipitation microarray (MeDIP-chip) approach to analyze epigenomic profiles (Hébrard et al., 2016; Lafon-Placette et al., 2018).
Table 1.
Description of the three experiments used for the comparative approach
| Experiment 1 | Experiment 2 | Experiment 3 | |||
|---|---|---|---|---|---|
| Environmental conditions | Type | Outdoor plantations | Outdoor plantations | Greenhouse | |
| Location | Orléans (ORL), France | Echigey (ECH) and Saint-Cyr-en-Val (SCV), France | Echigey (ECH), Saint- Cyr-en-Val (SCV), and Guémené (GMN), France | Nancy, France | |
| Treatments studied | Water availability | Pedoclimatic conditions (mainly water availability and soil fertility) | Pedoclimatic conditions (mainly water availability and soil fertility) | Water availability | |
| Number of treatment conditions | 2 | 2 | 3 | 3 | |
| Treatment conditions | Irrigated (ORLWW, favorable) versus non-irrigated (ORLWD, unfavorable) | ECH (favorable) versus SCV (unfavorable) | ECH (favorable) versus SCV or GMN (unfavorable) | Well-watered (WW, favorable) versus water deficit (WD, unfavorable) or rewatered (WD- RW, unfavorable) | |
| General characterization | ORLWW: Pre‐dawn leaf water potential above –0.20 MPa | ECH: Summer soil VWC >40% (fertile site) | ECH: Summer soil VWC >40% (fertile site) | WW: Soil REW maintained close to 100% | |
| ORLWD: Pre‐dawn leaf water potential of –0.75 MPa at the drought peak | SCV: Summer soil VWC <20% (poor site) | SCV: Summer soil VWC <20% (poor site) | WD: Soil REW maintained at 20% during 2 weeks | ||
| GMN: Summer soil VWC <20% (poor site) | WD-RW: Soil REW maintained at 20% during 8 days followed by rewatering at field capacity for 6 days | ||||
| Plantation design | Culture | Coppice | Coppice | Coppice | Potted cuttings |
| Species | P. deltoides × P. nigra | P. deltoides × P. nigra | P. deltoides × P. nigra and P. trichocarpa | ||
| Number of genotypes | 8 | 56 | 1 | 4 | |
| Design | Randomized multiclonal blocks | Randomized multiclonal blocks | Monoclonal blocks | Randomized multiclonal blocks | |
| Number of blocks | 5 | 10 | 1 | 3 | |
| Number of individuals per genotype per block | 3 | 1 | 10 | 6 | |
| Plantation year | 2006 | 2009 (ECH), 2010 (SCV) | 2009 (ECH, GMN), 2010 (SCV) | 2008 | |
| Coppice year | 2007, 2008 | 2010 (ECH), 2011 (SCV) | 2010 (ECH, GMN), 2011 (SCV) | ||
| Sampling year | 2008 | 2010 (ECH), 2011 (SCV) | 2010 (ECH, GMN), 2011 (SCV) | 2008 | |
| Age at sampling time | 3 years | 2 years | 2 years | 3 months | |
| Previously highlighted effects | Main shoot annual dry mass | G×E ns, G*, E** | G×E***, G***, E*** | ||
| Relative growth rate in height | G×E ***, G*, E** | G×E ns, G***, E*** | |||
| References | Fichot et al., 2010, 2011 | Toillon et al., 2013a | Toillon et al., 2013b | Bizet et al., 2015; Lafon- Placette et al., 2018 | |
REW, Relative extractible water; SAM, shoot apical meristem; VWC, volumetric water content. Effect of growing environments was evaluated with an ANOVA test (G, genotype effect; E, environment effect for growth conditions; G×E, interaction between genotype and environment). Levels of significance: *P<0.05, **P<0.01, ***P<0.001; ns, non-significant. The corresponding references for these data are indicated.
Fig. 1.
Summary of the three experiments used for the methylome comparative approach. Plant materials, water regimes, samples, and the previously published ecophysiological or epigenomic data used for the analyses in this paper are cited. For experiment 1, the field trial was located near Orléans, France, under two different water regimes (ORLWW and ORLWD); WW, Well watered (considered favorable for growth); WD, water deficit (considered unfavorable for growth). For experiment 2, field trials were located in Echigey (a site with favorable growing conditions) and Saint-Cyr-en-Val or Guémené, France (both sites with unfavorable growing conditions). For experiment 3, poplar trees were grown in a greenhouse under three water regimes: WW, WD, and water deficit followed by rewatering (WD-RW), also considered unfavorable for growth. SAM, Shoot apical meristem.
The main aims of the study were (i) to assess the range of genetic and environmentally induced variations in global DNA methylation in winter-dormant SAMs and evaluate the relationship with biomass production, (ii) to identify differentially methylated regions (DMRs) in winter-dormant SAMs of trees that had been exposed to different environmental conditions during their vegetative period for each experiment separately, and (iii) to identify conserved DMRs between the two experiments and the corresponding overlapped genes potentially serving as a signature of water stress or unfavorable growth conditions. Overall, we found that global DNA methylation in winter-dormant SAMs varied in response to environmental conditions and among genotypes, and was correlated with biomass production capacity. Winter-dormant SAMs of trees grown in different conditions during the vegetative period exhibited DMRs between favorable and unfavorable vegetative growth periods inside independent experiments, consistent with an epigenetic memory of environmental conditions mediated by DNA methylation that lasted for at least 6 months. In our study, epigenetic memory corresponded to the DMRs that were stable through mitosis in SAM from the time when the induction was performed (summer, active SAM) to the time when we performed our analyses (winter, dormant SAM). In addition, we compared the DMRs reported here with DMRs found for active SAMs in greenhouse-grown trees (Lafon-Placette et al., 2018; Table 1, Fig. 1). We detected conserved DMRs among these three independent experiments that mostly overlapped genes related to abiotic stress or developmental responses. Altogether, our data clearly point to an environmentally induced epigenetic memory in the SAM in field-grown poplar trees.
Materials and methods
Plant materials, experimental designs, growth performance, and samplings
Analyses were performed on samples collected in three independent experiments conducted in different environments with distinct genotypes. These experiments had initially been set up to characterize the ecophysiological response to water availability and pedoclimatic conditions at the genotype level. A comparative analysis was therefore carried out by merging the ecophysiological data already published (Table 1; Fichot et al., 2010, 2011; Toillon et al., 2013a, b; Bizet et al., 2015; Lafon-Placette et al., 2018) and the epigenomic analyses performed in the framework of the present study (Fig. 1).
Experiment 1 consisted of a field drought experiment conducted in a common garden. The experiment was installed in spring 2006 at the INRA Research Station of Forest Genetics in Orléans in central France (Loiret, 47°46ʹN, 1°52ʹE, 110 m above sea level) on poor loamy sand. Hardwood cuttings (25 cm in length) of eight unrelated hybrid genotypes of P. deltoides × P. nigra were planted in the field (Table 1; Fichot et al., 2010, 2011). The eight genotypes were initially selected for their contrasting growth performance and water-use efficiency (Monclus et al., 2005; Fichot et al., 2009). Two twin plots were established 15 m apart on a 250 m2 surface area. Each plot was divided into five complete randomized blocks with three ramets of each genotype per block. All the plants were cut back at the end of 2006 and 2007, thereby creating a coppice system the following years. All the plants were irrigated during the 2007 growing season to ensure successful installation. In 2008, irrigation was maintained on one plot (control plot, ORLWW) and withheld from the other plot (water deficit plot, ORLWD) from June 18 to the end of September. The unirrigated plot received water only from rainfall. There was a summer drought period peaking on July 24, which resulted in a 25% average decrease in annual above-ground biomass production (Fichot et al., 2010). Additional information on the experimental design, growth conditions, and genotype ecophysiological characteristics can be found in Fichot et al. (2010, 2011).
Experiment 2 consisted of a network of site-specific field trials initially established to evaluate genotype × environment interactions in poplars cultivated under short-rotation coppice. Multiclonal field trials were composed of 56 unrelated P. deltoides × P. nigra genotypes and were replicated on two sites in France: at Echigey (ECH; Côte d’Or, 47°10ʹN 5°11ʹE, 197 m above sea level) and Saint-Cyr-en-Val (SCV; Loiret, 49°81ʹN 1°98ʹE, 100 m above sea level) (Toillon et al., 2013a; Table 1; see also Supplementary Table S1 at JXB online). Each field trial was composed of 10 complete randomized blocks with one ramet of each genotype per block. Monoclonal trials of the same genotype (P. deltoides × P. nigra cv. Dorskamp) were also established at the same two sites (ECH and SCV) plus one additional third site: Guémené-Penfao (GMN; Loire-Atlantique, 47°37ʹN 1°49ʹW, 43 m above sea level). The sites were planted in spring 2009 at ECH and GMN, and 2010 at SCV, with 25 cm long woody-stemmed cuttings. Each plantation was managed as an extensive short-rotation coppice using a double-row planting scheme with alternating distances of 0.75 m and 2 m between the rows and 1 m between trees within the rows (~7272 trees ha−1). The ECH site was characterized by fertile, silty-clay soil and high soil water content during the growing season (>40% soil volumetric water content at 20 cm) (Toillon et al., 2013a). The SCV site was characterized by a poor, sandy-loam soil type with low soil water content (<20%). The GMN site was located on a silty-clay soil with a low soil water content (<20%) (Toillon et al., 2013b). ECH was the most productive site, followed by SCV and then GMN, which had, respectively, 32.5% and 50% less annual average above-ground biomass production than ECH (Toillon et al., 2013a, b). Growth conditions were considered ‘favorable’ at the ECH site and ‘unfavorable’ at both the GMN and SCV sites. Additional information on the experimental design, growth conditions, and genotype ecophysiological characterization can be found in Toillon et al. (2013a, b).
Experiment 3 consisted of plants of four commercial hybrids, P. deltoides × P. trichocarpa (cv. Beaupré) and P. deltoides × P. nigra (cv. Carpaccio, Dorskamp, and Soligo), grown in 10 liter pots in Nancy, France in April 2008 (Table 1; Bizet et al., 2015, Lafon-Placette et al., 2018). The plants were exposed to natural daylight (400–900 μmol m−2 s−1). Air temperature and humidity were maintained within the ranges of 19–26 °C and 50–75%, respectively. After 5 weeks at field capacity, the plants were organized in three randomized blocks with six ramets for each genotype per block and then exposed to three distinct water treatments: (i) well watered for 10 days, where evaporative demand was compensated for by watering to field capacity, (ii) water deficit for 10 days, where the relative extractable water content was maintained within the range of 17–23%, or (iii) rewatering, where water deficit was applied for 8 days, followed by rewatering to field capacity for 6 days. Additional information on the experimental design, growth conditions, and genotype ecophysiological characterization can be found in Bizet et al. (2015) and Lafon-Placette et al. (2018).
Winter-dormant buds from P. deltoides × P. nigra genotypes were collected for epigenomic analysis from experimental systems 1 and 2 (Fig. 1). For experiment 1, dormant SAMs were collected for all eight genotypes in winter 2008, 6 months after the summer drought (Table 1, Fig. 1). For experiment 2, dormant SAMs were collected in winter 2010 (at the ECH and GMN sites) and winter 2011 (at the SCV site) 6 months after the summer period for both the multiclonal and monoclonal trials (Table 1, Fig. 1). For the multiclonal trials, SAMs were picked from a subset of 31 genotypes representative of the genetic diversity among the 56 genotypes available. Next, the shoot apices were cut in half with a scalpel and all visible differentiated tissues were discarded under a binocular magnifying glass in order to extract, as much as possible, the SAM, as previously reported (Lafon-Placette et al., 2013). Samples were frozen in liquid nitrogen and stored at –80 °C.
For methylome analysis, we selected for experiment 1 the winter-dormant SAM of the Eco 28 genotype, which showed the highest relative growth rate for the main shoot under well-watered conditions and the most significant effect of drought on DNA methylation (Fichot et al., 2010; Fig. 1); for experiment 2 the winter-dormant SAM of the Dorskamp genotype owing to its positive and well-reported physiological response to moderate drought (Marron et al., 2002, 2003; Toillon et al., 2013a; Fig. 1); and in experiment 3 the active SAM of the Carpaccio genotype owing to its positive and well-reported physiological response to moderate drought (Fig. 1). For global DNA methylation, methylome, and transcriptomic analyses, the corresponding data have been reported in Lafon-Placette et al. (2018).
DNA extraction and determination of global DNA methylation percentage by HPLC
Following Lafon-Placette et al. (2013, 2018), the winter-dormant SAMs from experiments 1 (eight genotypes) and 2 (31 genotypes) were cleared of all visible differentiated tissues to maximize the isolation of the meristematic cells, immersed in liquid nitrogen, and ground to a fine powder in an automatic ball mill (MM 200 Retsch, Germany). Genomic DNA was extracted following the CTAB protocol (Doyle and Doyle, 1987; Lafon-Placette et al., 2018) and stored at –80 °C. The quantity and quality of DNA were estimated with a NanoDrop spectrometer (NanoDrop Instrument, France).
Genomic DNA was enzymatically hydrolyzed into nucleosides and analyzed by HPLC with a GeminiTM column (150 × 4.6 mm, 5 µm; Phenomenex, Le Pecq, France) and an isocratic mobile phase consisting of 0.5% methanol (v/v) and 5 mM acetic acid in water, as described by Zhu et al. (2013). Controls for this procedure included co-migration with commercial standards (Sigma-Aldrich), confirmation by enzyme restriction analysis, and tests for RNA contamination based on the HPLC detection of ribonucleosides. Global DNA methylcytosine percentages (% mC) were calculated as follows:
where C is the 2ʹ-deoxycytidine content and mC is the 5-methyl-2ʹ-deoxycytidine content. For each set of conditions, we analyzed SAMs from four to six individuals per genotype (depending on the possible degradation of SAM for some individuals in the field), with two hydrolysis replicates and two HPLC runs for each.
Methyl DNA immunoprecipitation and microarray analysis
Custom microarray probes designed with eArray software (https://earray.chem.agilent.com/earray/; Agilent Technologies, Massy, France) using Populus trichocarpa genome v2 (Tuskan et al., 2006; http://www.phytozome.net/poplar.php) have been described by Lafon-Placette et al. (2018) (GEO accession number GSE46624). For each poplar gene model (over 42000 models), we designed five probes: one probe bound to the 0.5 kb upstream from the start codon (annotated ‘PROMOTER’), one probe bound to the 0.5 kb downstream from the stop codon, and three probes bound within the body of the gene (located between the transcription start site and transcription termination site, and annotated ‘BODY’,). Probes were annotated ‘TE’ if they were identified in a transposable element (TE) described by the RepeatMasker annotation of the P. trichocarpa v3 genome (http://www.phytozome.net/poplar.php). If the probe was present in either a TE inserted into a gene or in a promoter, it was annotated as ‘BODY+TE’ or ‘PROM+TE’, respectively. Otherwise, the probe was annotated as ‘INTERGENIC’. Additional probes covering genes and intergenic regions were also selected based on a previous study (Lafon-Placette et al., 2013). These probes spanned from 2 kb upstream to 2 kb downstream from target loci (either genes or intergenic regions), with a distance between probes of 140 bp for genes and 780 bp for intergenic regions. The microarray also contained 50 probes for reproducibility controls and internal Agilent control probes. Following the release of the v3 P. trichocarpa genome sequence, probe information was updated by BLAST searches of probe sequences against the v3 genome sequence. See Hébrard et al. (2016) and Lafon-Placette et al. (2018) for a description of the procedure for MeDIP-chip, including controls and data analysis under the supervision of IMAXIO (Clermont-Ferrand, France), in accordance with the instructions issued by Agilent Technologies.
For experiments 1 and 2, the set of winter-dormant SAM samples used for methyl DNA immunoprecipitation was composed of four to six individuals for one genotype (Eco28 for experiment 1 and Dorskamp for experiment 2) for each condition (Fig. 1) depending on the possible degradation of the SAM for some individuals in the field.
DMR identification and bioinformatics analyses
DNA methylation data obtained with the MeDIP approach were used to detect DMRs between favorable and unfavorable conditions for each experiment. For experiment 1, we compared the Eco 28 genotype in favorable well-watered (ORLWW) and unfavorable water-deficit (ORLWD) conditions. For experiment 2, we compared the Dorskamp genotype in favorable growing conditions at the ECH site and in unfavorable conditions at SCV or GMN. Identifying DMRs within each experiment allowed us to compare favorable and unfavorable conditions for each experiment without taking into account differences among experiments for the growing design.
To identify DMRs, we applied a mixed model to normalized and filtered signal probes mapped on to the poplar reference genome (ftp://plantgenie.org/Data/PopGenIE/Populus_trichocarpa/v3.0) and estimated the false discovery rate (FDR) for each set of biological conditions, following Lafon-Placette et al. (2018) (see Supplementary Method S1). Briefly, we first estimated the reference mean from 20% of the probes selected at random, for each chromosome under each growth condition and for each experiment. This became our reference mean, or base level of methylation, for a given chromosome in a given biological sample. We used mixed models based on the bimodal empirical distributions for this estimated reference mean (McLachlan and Peel, 2000). We used the R statistical software mixtool package (Benaglia et al., 2009) for subsequent analyses. We first constructed 50 kb analysis windows (~7550 windows) based on the remaining 80% of the probes, following Lafon-Placette et al. (2018). Each defined window was characterized by its specific probe composition and gene model positions. These windows were used to assess whether the mean methylation signal per window differed from the previously determined reference mean. Of the windows, 56% had fewer than 30 probes for experiments 1 and 71% had fewer than 30 probes for experiment 2. We therefore performed a one-sample Wilcoxon signed-rank non-parametric test with a symmetric distribution of the data around the reference mean as the null hypothesis. This test was performed to compare each window mean with the corresponding reference mean to obtain a P-value that would allow us to accept or reject the null hypothesis. An FDR was then used to control for multiple testing (Benjamini and Hochberg, 1995) with α=5%, to control for the global risk of error in the windows selected by the multiple Wilcoxon signed-rank tests. This FDR was estimated with the R package fdrtool (Strimmer, 2008). The results were ranked according to decision rule scores based on the FDR. We defined three levels: not significantly different from the mean reference level of methylation (0), significantly lower levels of methylation than the reference mean (–1), and significantly higher levels of methylation than the reference mean (+1). DMR was defined as a window for which different decision rule scores were obtained for favorable and unfavorable growth conditions for each experiment (0 and +1, 0 and –1, or +1 and –1).
To assess Gene Ontology (GO) term enrichment for the genes inside DMRs, we used the corresponding best BLAST hit for each v3.0 poplar model gene from the best v3.0 BLAST hits with Arabidopsis TAIR10. AgriGO (http://bioinfo.cau.edu.cn/agriGO/) and then REVIGO (http://revigo.irb.hr/) software, set to default parameters, were used for enrichment analysis. A treemap was created with rectangle size adjusted to reflect the absolute log10P-value of the GO term.
Statistical analyses
Statistical analyses were performed with Rstudio statistical software (R Core Team, 2015; http://www.rstudio.com/). Means are expressed with their standard error. Differences between groups were evaluated by ANOVA (one-way ANOVA test, general linear model procedure, two-way ANOVA test, genotype and treatment effect). Tukey rank tests were also performed post hoc, considering only the effect of genotype in a given set of conditions. Relationships between global DNA methylation and ecophysiological parameters were analyzed for normality by carrying out Shapiro–Wilk tests. Linear correlation analyses were then carried out to determine Pearson’s coefficient (r) and Spearman’s coefficient (rho). Significant effects of the methylation pattern on the distribution of DMRs were evaluated with the chi-squared (χ2) test. Finally, a hypergeometric distribution was used to describe the probability of having more common DMRs than the number identified without replacement. The n value indicated in the figure legends corresponds for the ANOVA test and the Pearson correlation to the lowest number of biological replicates for a given condition in an experiment (Fig. 1). Some SAMs could not be harvested owing to the field conditions (e.g. insect or pathogen attack).
Accession numbers
Microarray design and methylome data are available from the GEO database (accession numbers GSE46624 and GSE97144, respectively).
Results
Global DNA methylation in winter-dormant SAMs depends on genotype and environment and correlates with biomass production
Global DNA methylation was measured by HPLC on winter-dormant SAMs of the different poplar genotypes from the two independent experiments 1 and 2, comparing in each case favorable and unfavorable growing conditions (Fig. 1, Table 1).
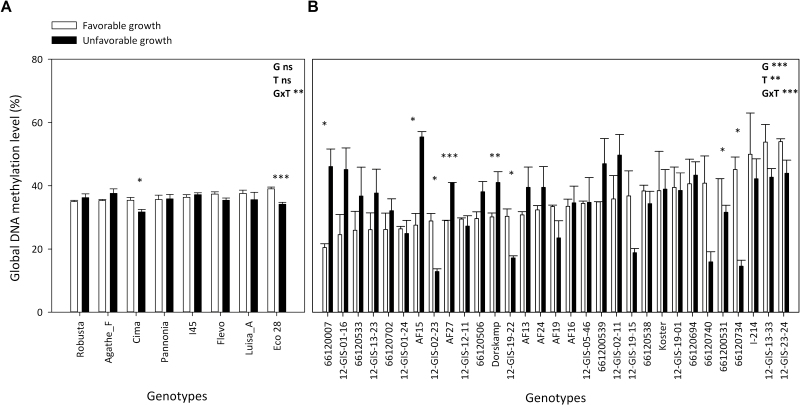
In experiment 1, global DNA methylation varied between 34% and 39% across the eight unrelated poplar genotypes (Agathe_F, Cima, Eco28, Flevo, I45-51, Luisa_Avanzo, Pannonia, and Robusta) and the two water regimes (ORLWW versus ORLWD; Fig. 2A). A significant genotype×treatment interaction was detected; two genotypes (Cima and Eco 28) displayed significant differences between well-watered and water deficit conditions, and in both cases a decrease in the level of DNA methylation was observed in water deficit conditions (Fig. 2A). These two genotypes are among the three most productive of the eight analyzed genotypes in terms of relative growth rates of the main shoot under well-watered conditions and exhibiting a significant decrease of their relative growth rate in response to water deficit (Fichot et al., 2010).
Fig. 2.
Genotypic variation and environmental effect for global DNA methylation on different Populus deltoides × P. nigra hybrids grown in experiment 1 (A) or experiment 2 (B). White and black bars correspond respectively to well-watered (ORLWW, favorable) and water deficit (ORLWD, unfavorable) conditions for experiment 1 (Fichot et al., 2010, 2011), and to favorable (ECH) and unfavorable (SCV) growth conditions for experiment 2 (Toillon et al., 2013a). Values are genotype means ±SE (n=4). Global genotype variations and the effect of growing conditions were evaluated by ANOVA. G, genotype effect; T, treatment effect for growth conditions; G×T: interaction between genotype and treatment. *P<0.05, **P<0.01, ***P<0.001; ns, non-significant.
A larger range of variation was found in experiment 2, where global DNA methylation varied between 20% and 54% across the 31 unrelated genotypes and the two locations (ECH, favorable, versus. SCV, unfavorable; Fig. 2B). A significant genotype×treatment interaction was detected (Fig. 2B). Significant differences across sites were detected for 8 out of 31 genotypes. Four genotypes displayed higher DNA methylation under favorable conditions, while the other four showed lower DNA methylation under favorable conditions (Fig. 2B). No relationship between genetic proximity and variations in global DNA methylation was found (Supplementary Table S1). For example, clones 12-GIS-13–23 and 12-GIS-13–33 came from the same genetic background but showed opposite patterns of variation in their global DNA methylation levels (Supplementary Table S1; Fig. 2B).
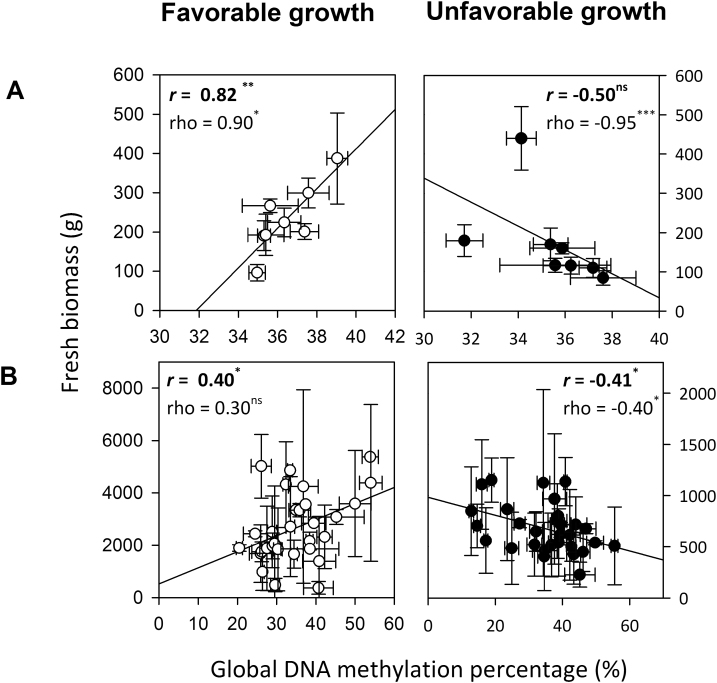
Significant positive correlations were detected between global DNA methylation and shoot biomass under favorable growing conditions for both experiments (Fig. 3), while under unfavorable growing conditions these correlations were negative (Fig. 3).
Fig. 3.
Relationships between global DNA methylation percentage and above-ground fresh shoot biomass for experiment 1 (A) and experiment 2 (B). White circles are for favorable (ORLWW, ECH) growing conditions and black circles for unfavorable (ORLWD, SCV) conditions. Values are genotype means (n=4) and the line represents the linear regression correlation among individuals. Shapiro tests were used to confirm values given by a homogeneities distribution test. Linear parametric correlation analyses were then carried out to determine Pearson’s coefficient (r) and Spearman’s coefficient (rho).
Winter-dormant SAM displays hyper- and hypomethylated regions in response to environmental variations from the preceding vegetative period
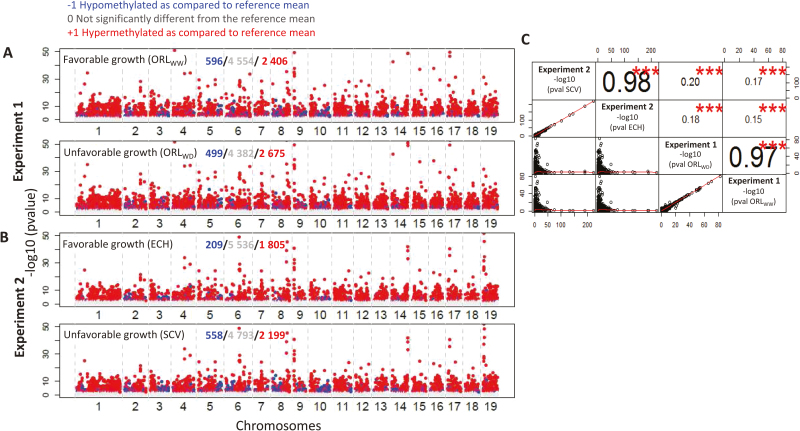
The methylome of winter-dormant SAMs from trees grown in experiments 1 and 2 was assessed by MeDIP-chip analysis on one genotype from each experiment (Fig. 1). P. deltoides × P. nigra clones of genotype Eco28 were used for experiment 1 and of genotype Dorskamp for experiment 2. A custom-made poplar array (1 million probes) was generated as described by Hébrard et al. (2016) and Lafon-Placette et al. (2018). In order to identify DMRs, consecutive 50 kb genomic windows (~7550 windows; see Supplementary Method S1) were first calculated within each biological condition, as described by Lafon-Placette et al. (2018). Manhattan plots were generated (Figs 4A, B; Supplementary Fig. S1A) to represent DNA methylation variations along the 19 chromosomes as –log10P-values for all the consecutive 50 kb genomic windows, ranging from 0 to ~50 (Fig. 4A). One dot represents one 50 kb genomic window and DNA methylation status is color-coded as follows: not significantly different from the average level of methylation (designated 0; grey dots in Fig. 4A), significantly lower (hypomethylated, designated −1; blue dots in Fig. 4A), and significantly higher (hypermethylated, designated +1; red dots in Fig. 4A; see Materials and methods and Supplementary Method S1).
Fig. 4.
Genomic features of DNA methylation changes in poplar shoot apical meristem in response to environmental variations. (A) Experiment 1: Favorable (ORLWW) and unfavorable (ORLWD, unfavorable) growing conditions. (B) Experiment 2: Favorable ( ECH) and unfavorable (SCV) growing conditions. Graphs are based on Manhattan plots from a significant false discovery rate of 5%. The plots show –log10P-values on the y axis, and the location of the different 50 kb windows in the genome with a gap at chromosome locations on the x axis. Blue dots correspond to hypomethylated windows compared with the reference mean, red dots to hypermethylated windows compared with the reference mean, and grey dots to non-significant windows. (C) Relationships (linear correlations) between –log10 of experiment 1 and 2 windows for all growth conditions (n=7550). Lines represent linear regression correlations; numbers represent Pearson’s coefficient (r). *P<0.05, **P<0.01, ***P<0.001; ns, non-significant.
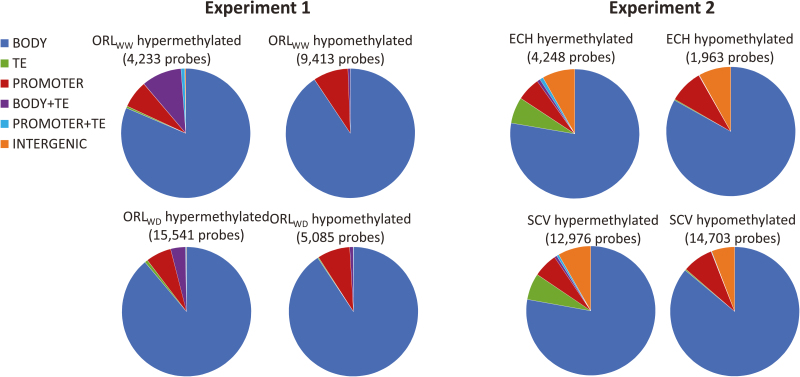
The distribution of hypo- and hypermethylation status over the chromosomes differed significantly between experiments (r values ranging from 0.15 to 0.20 at P<0.001; Fig. 4C). In contrast, within an experiment, similar distributions of DNA methylation on chromosomes were observed between favorable and non-favorable conditions in the 50 kb genomic windows tested, as indicated by r values ranging from 0.97 to 0.98 (at P<0.001; Fig. 4C). Despite this stability, differences between favorable and unfavorable conditions for a given experiment were detected as changes in the number of hyper -and hypomethylated loci. Indeed, 596 hypomethylated and 2406 hypermethylated loci were identified in the genome of trees growing in the favorable condition of experiment 1, while under unfavorable growing conditions, fewer hypomethylated (499) and more hypermethylated (2675) loci were detected within the analyzed windows (Fig. 4A). In experiment 2, both hypomethylated and hypermethylated loci increased between favorable and unfavorable conditions: from 209 to 558 for hypomethylated loci, and from 1805 to 2199 for hypermethylated loci (Fig. 4B; Supplementary Fig. S1). We also determined the association between DNA methylation status and specific DNA sequences, including genic regions and transposons. Variations in DNA methylation status observed in the tested windows were mostly located in the genic region between the transcription start site and transcription termination site, thereafter defined as the gene body (Fig 5; Supplementary Fig. S1B; Supplementary Table S2). This could explain in part the differences observed between variations of methylation for a given genotype for global DNA methylation performed by HPLC (mostly affected by non-genic loci such as repeated sequences) and the methylome by MeDIP-chip (mostly affected by genes) (Figs 2 and 4).
Fig. 5.
Characterization of loci overlapped by DNA methylation changes (DMRs) in response to different environmental conditions. DMRs were classified as hypo- or hypermethylated for each experimental design. BODY, gene body; PROMOTER, 1 kb upstream region; TE, transposable element; BODY+TE, TE inserted into a gene body; PROM+TE, TE inserted into a promoter; INTERGENIC, any other locus.
The identification of hypo- or hypermethylated regions also confirmed that TEs were mainly hypermethylated, while promoters showed similar proportions of hypo- and hypermethylated regions (Fig. 5; Supplementary Fig. S1B; Supplementary Table S2). In each experiment, no impact of the growing conditions was observed on the distribution of methylation status, while TEs and intergenic loci revealed differences between experiments 1 and 2 (Fig. 5 and Supplementary Fig. S1B). Functional annotation of the genes covered by methylation status variation was performed by using GO enrichment analysis. This analysis was based on the annotations for the homologous genes of Arabidopsis in TAIR10 (https://www.arabidopsis.org/). The analysis revealed that these genes were rich in biological processes (Supplementary Table S5; Supplementary Figs S1C and S2). In the ORLWD condition, for example, some of the main categories were associated with genes involved in multicellular organism development, including shoot and meristem development (37%; P=2.7 × 10−53), the abiotic stress response (25%; P=3.9 × 10−56), and the negative regulation of biological processes (13%; P=8.8 × 10−29) (Supplementary Fig. S2, Supplementary Table S5).
Winter-dormant SAM maintains DMRs from the preceding vegetative period
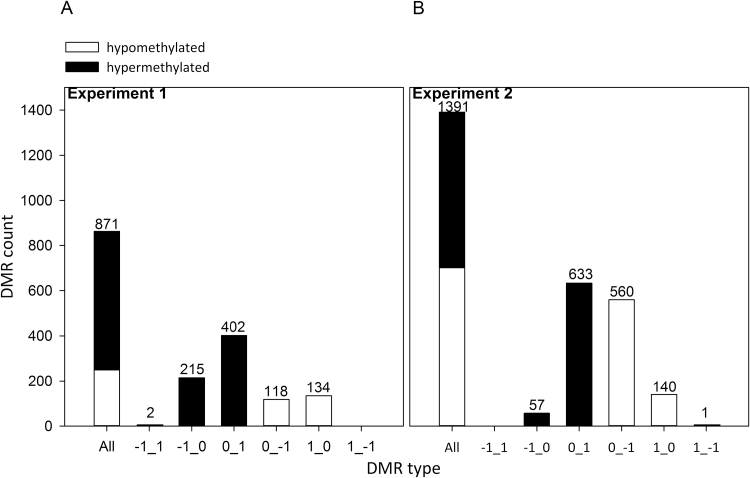
Using the FDR controlling procedure for multiple testing (α=5%; see Supplementary Method S1), 871 DMRs (11.5%) between favorable (ORLWW) and unfavorable (ORLWD) growing conditions were identified in experiment 1. In experiment 2, 1391 DMRs (18.4%) were detected between favorable (ECH) and unfavorable (SCV) sites (Fig. 6A; Supplementary Table S3). In both experiments, ~70% of the DMRs were hypermethylated under unfavorable compared with favorable conditions (Fig. 6A, B). In all cases, nearly all DMRs showed weak DNA methylation variations (–1 to 0 or 0 to +1 scores; Fig. 6).
Fig. 6.
Classification of DNA methylation changes (DMRs) for the intensity of variations for (A) experiment 1 and (B) experiment 2. DMRs were split between hypermethylated (black bars) and hypomethylated (white bars) regions. DMR categories were defined by the size of DNA methylation changes between favorable and unfavorable growing conditions (indicated on the x axis as favorable_unfavorable): mild changes from –1 (hypomethylation) to 0 (average methylation) and from 0 to +1 (hypermethylation), or strong changes from –1 (hypomethylation) to +1 (hypermethylation).
GO enrichment analysis was used to characterize the genes associated with DMRs (and by the filtered MeDIP probes) as previously mentioned (Supplementary Figs S3–S5; Supplementary Table S5). Similar functional categories were associated with DMRs in both experiments (Supplementary Fig. S3; Supplementary Table S5), of which the most abundant were cellular protein modification process (with RNA metabolism; 18% of genes, P=5.2 × 10−13 in experiment 1; Supplementary Table S5; Supplementary Fig. S3A), response to abiotic stress (14% of genes, P=5.9 × 10−14 in experiment 1), and cellular ketone metabolism (cell cycle, cell division, small molecule metabolism; 10% of genes, P=3.1 × 10−7 in experiment 1). The GO category corresponding to genes involved in the response to abiotic stress was associated with both hyper- and hypomethylated DMRs (Supplementary Figs S4 and S5). Additional GO categories such as phytohormone signaling/metabolism pathways that were found for jasmonic acid, abscisic acid, and salicylic acid were specifically associated with hypomethylated DMRs, whereas RNA processing and small molecule metabolism were associated with hypermethylated DMRs in experiment 1 (Fig. S4). In experiment 2, a similar observation was made, with GO categories containing salicylic acid metabolism being associated with hypomethylated DMRs and anatomical structure development and macromolecule catabolism categories with hypermethylated DMRs.
In order to determine possible correlations between gene expression and change in methylation levels, we used a candidate approach to characterize genes covered by DMRs using an in silico analysis with available transcriptomic data describing differentially expressed genes (DEGs) in the active SAM between the well-watered and the water deficit and rewatering conditions for experiment 3 (Lafon-Placette et al., 2018; Table 1; Fig. 1). This overlap analysis identified 123 genes between DEGs (Lafon-Placette et al., 2018) and DMRs for experiment 1, and 96 genes for experiment 2 (Supplementary Table S4). For experiments 1 and 2, identified genes were enriched in response to stress (29%, P=2.3 × 10−5 and 33%, P=2.2 × 10−11, respectively) (Supplementary Table S5; Supplementary Fig. S6). Specific GO categories were also detected, such as hormones (experiment 1: 11%, P=5.3 × 10−4), regulation of gene expression and epigenetic (experiment 2: 4%, P=8.2 × 10−2), and shoot development (experiment 2: 3%, P=8.2 × 10−2) (Supplementary Table S5; Supplementary Fig. S6). The distribution of DEGs in the DMRs was analyzed. The data showed an over-representation, relative to a random distribution, of hypermethylation for down-regulated (P<0.01) and up-regulated (P<0.001) genes in experiment 1, while hypergeometric tests revealed that DEGs were equally distributed between hypo- and hypermethylated DMRs in experiment 2 (Supplementary Fig. S7).
Finally, we used hypergeometric tests to determine whether the distribution of DEGs over DMRs occurred by chance or not (Supplementary Fig. S7). This revealed that down-regulated DEGs (P<0.01) were over-represented in the hypermethylated DMRs found in experiment 1, whereas up-regulated DEGs (P<0.001) were equally distributed between hypo- and hypermethylated DMRs in experiment 2.
SAM displays common environmentally induced epigenetic signatures
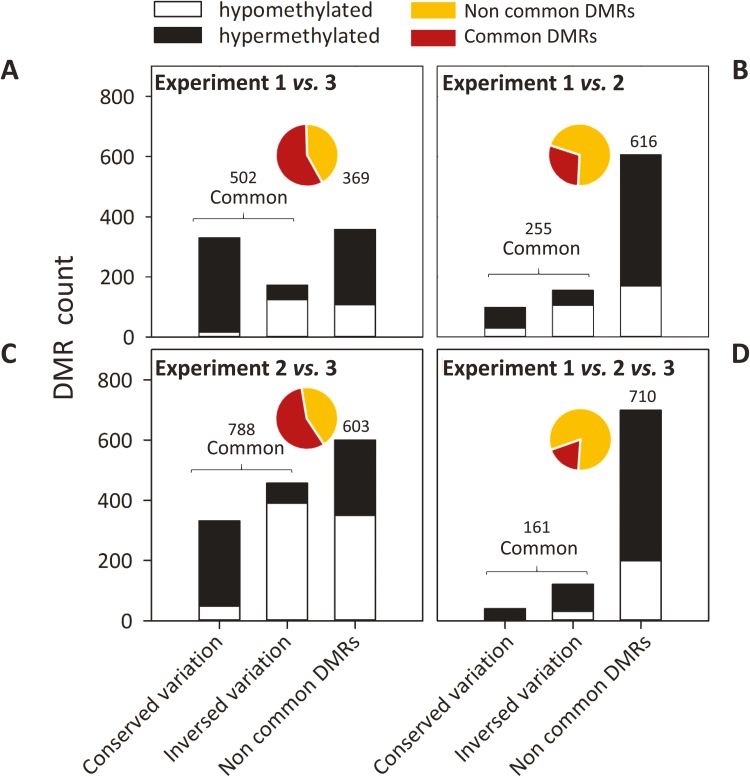
In order to identify the loci systematically and durably targeted in SAM by DNA methylation variations (DMRs called ‘epigenetic signatures’) depending on site growth performances (favorable versus unfavorable conditions), we compared the DMRs identified in the winter-dormant SAMs of the field experiments 1 and 2 among themselves and with the recently reported DMRs in active SAM of experiment 3 (Lafon-Placette et al., 2018; Figs 1 and 7). More than half of the DMRs identified in winter-dormant SAMs (502 out of 871 DMRs, or 57.6%, in experiment 1; 788 out of 1391 DMRs, or 56.6%, in experiment 2) were common to those found in active SAMs of experiment 3 (Fig. 7A, C). Fewer DMRs (255 out of 871, or 29.2%; Fig. 7B) were common to experiments 1 and 2, and 161 DMRs (18.4 %) were common to all three experiments (Fig. 7D). Using hypergeometric tests on common DMR distribution among the experiments, we identified significant over-representations (P<0.001) among all the compared experiments.
Fig. 7.
Comparison of DMRs among (A) experiments 1 and 3, (B) experiments 1 and 2, (C) experiments 2 and 3, and (D) experiments 1, 2, and 3. DMRs inside an experiment correspond to a genomic region showing a significant variation in DNA methylation between favorable and unfavorable growing conditions. ‘Common’ DMRs correspond to the same DMRs found in at least two experiments (if not, DMRs are labeled ‘non-common’). For the common DMRs, the direction of methylation variation (hyper- or hypomethylation) between favorable and unfavorable growing conditions can be maintained (conserved DMRs) or reversed (inversed DMRs). Values indicate DMR counts for common (red) and non-common (yellow) categories and are represented as pie charts. Black bars correspond to hypermethylated DMRs and white bars to hypomethylated DMRs.
Two categories of DMRs common between pairwise comparisons of all the experiments were distinguishable: (i) DMRs with the same direction of methylation changes (i.e. hyper- or hypomethylation) between favorable and unfavorable growth conditions (hereafter called ‘conserved’ DMRs; Fig. 7) and (ii) DMRs with the opposite direction in methylation changes (hereafter called ‘inversed’ DMRs; Fig. 7). Globally, the proportion of common conserved DMRs was lower than of inversed DMRs (Fig. 7B–D). However, notably, experiments 1 and 3 showed more conserved than inversed common DMRs (Fig. 7A). Hypergeometric tests revealed a significant over-representation of hypermethylated DMRs among conserved DMRs relative to a random distribution (P<0.001) among all compared experiments (Fig. 7).
GO enrichment analyses were performed on the genes associated with the common DMRs, either globally or separately for the conserved and inversed DMRs (Fig. 8; Supplementary Figs S8–S11; Supplementary Table S5). In all cases, one of the most significant enrichment category corresponded to genes involved in the response to (abiotic) stresses (~15% of the genes; see Supplementary Table S5). GO enrichment also revealed differences between hyper- or hypomethylated DMRs for experiments (1 versus 2; 1 versus 3; 2 versus 3) of common and conserved or inversed DMRs. These data suggest the possibility of coordinated control of different biological functions, for example, by hyper- or hypomethylation. Indeed, ‘water stress’ was a major GO category for hypomethylated experiment 1 versus experiment 2 DMRs (8% of the genes, P=3.4 × 10−5), while ‘tissue development’ was a major GO category for hypermethylated DMRs (9% of the genes, P=2.0 × 10−6) (Supplementary Fig. S10B, C,; Supplementary Table S5).
Fig. 8.
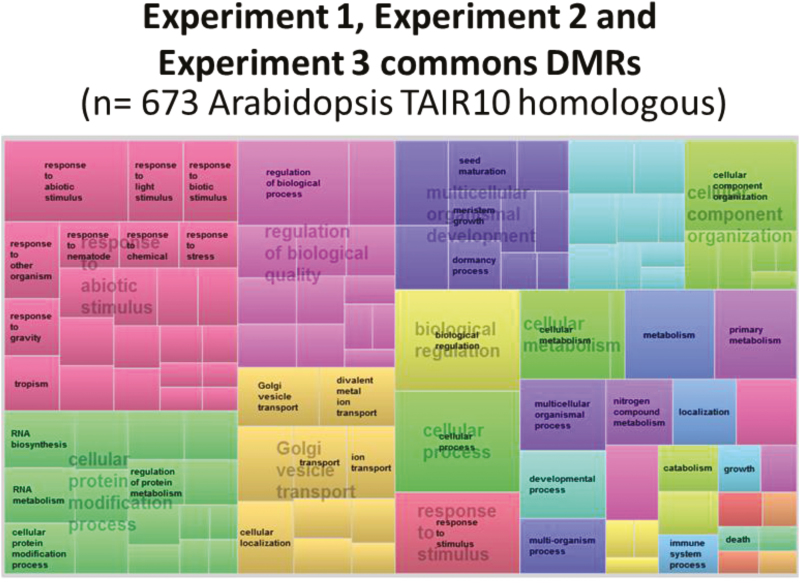
Treemap view of REVIGO for biological process on DMRs common to experiments 1, 2 and 3 (n=673 annotations of homologous Arabidopsis genes). Each rectangle is a single cluster representative of TAIR10 corresponding to poplar v3.0 annotations. The representatives are combined into superclusters of loosely related terms, visualized with different colors. Rectangle size is adjusted to reflect the abs log10P-value of the GO term in the underlying Gene Ontology Annotation database.
Finally, we considered the mapped genes in the 161 DMRs that were common among the three experiments as epigenetic signatures (Fig. 7D). The major identified GO category was ‘response to abiotic stress’ (16% of the genes, P=1.70 × 10−7; Fig. 8, Supplementary Table S5). We then compared these genes in common DMRs with the DEGs reported for experiment 3 (Lafon-Placette et al., 2018; Fig. 1). Eleven DEGs (Supplementary Table S4) were identified, mostly in hypermethylated regions (Supplementary Fig. S7). Among these genes, STRUWWELPETER and SPINDLY are already known to be involve in stress and hormone signaling (Autran et al., 2002; Qin et al., 2011; Sarnowska et al., 2013; Wang et al., 2015; Steiner et al., 2016).
To conclude, abiotic-responsive genes (including some hormone-responsive genes) and shoot-development genes showed epigenetic signatures by preferential DNA methylation changes in response to environmental changes. Variations of gene methylation were conserved from active to winter-dormant SAM. The conservation of methylation marks suggests a memory of epigenetic signatures in the SAM that could contribute to stress memory and adaptation of trees. This could suggest a memory of these epigenetic signatures in the SAM contributing to stress memory and adaptation in trees, as proposed in a model shown in Supplementary Fig. S12.
Discussion
Global DNA methylation in winter-dormant SAM is a biomarker of genotype performances
Variations in global DNA methylation have been reported in different plant species (Lambé et al., 1997; Alonso et al., 2015; Plomion et al., 2016), within the population (Vaughn et al., 2007; Schmitz et al., 2013) and the offspring of the same parents (Becker et al., 2011; Schmitz et al., 2011; Eichten et al., 2012). Variations in global DNA methylation may also depend on environmental constraints or developmental processes (Causevic et al., 2005; Teyssier et al., 2008; Gourcilleau et al., 2010; Trap-Gentil et al., 2011; Teyssier et al., 2014; Conde et al., 2017). In poplar, variations in global DNA methylation have been reported in response to limited water availability, water or soil salinity, competition, and pathogens, or according to geographic origin, in a limited number of genotypes (Gourcilleau et al., 2010; Raj et al., 2011; Latzel et al., 2013; Garg et al., 2015; Song et al., 2016; Lafon-Placette et al., 2018).
Here, we analyzed global DNA methylation variations in winter-dormant SAMs for two collections of poplar genotypes grown in open fields. The levels of methylation reported here for 39 poplar genotypes were in agreement with previously reported data (Gourcilleau et al., 2010; Raj et al., 2011; Lafon-Placette et al., 2018). The range of variation in our study (~34%) was similar to that reported for Dianthus broteri (29–35%; Alonso et al., 2016), a collection of angiosperms (5–40%; Alonso et al., 2015), or for vernalized sugar beet genotypes (20–80%; Trap-Gentil et al., 2011). These results show that DNA sequence variation is not the only determinant of genotypic and probably phenotypic variations; epigenetic variations are also key players (Latzel et al., 2013; Cortijo et al., 2014; Kooke et al., 2015; Kawakatsu et al., 2016; Richards et al., 2017).
In the present study, significant genotype×environment interaction for the global methylation in winter-dormant SAM was identified. In each experiment, correlations of global DNA methylation were established with shoot biomass production. This is in agreement with several studies proposing that variation in DNA methylation has a genetic basis and is a sign of local adaptation (Dubin et al., 2015; Foust et al., 2016).The Arabidopsis 1001 Epigenomes project also provides evidence that methylation is correlated with geography and climate of origin (Kawakatsu et al., 2016). In addition, positive correlations between shoot biomass and DNA methylation have already been reported in a study on active SAM of P. deltoides × P. nigra genotypes under well-watered conditions (Gourcilleau et al., 2010). A recent study also highlights site-specific growth performance related to methylation patterns in P. trichocarpa clones coming from two distinct sites with different phosphorus nutrition; trees growing with adequate phosphorus nutrition had higher dry shoot biomass and global DNA methylation levels (Schönberger et al., 2016). In our study, under favorable growth conditions we identified a positive correlation between fresh shoot biomass and global DNA methylation, while the opposite correlation was detected under unfavorable growth conditions. These results suggest that DNA methylation could be a biomarker of productivity in poplar, at least in the sites analyzed in the present study.
Winter-dormant SAM keeps the epigenetic memory of environmental variations
DNA methylation plays a role in the ability of plants to acclimate and adapt to changing environmental conditions (Boyko and Kovalchuk, 2008; Bräutigam et al., 2013; Rico et al., 2014; Baulcombe and Dean, 2014; Kawakatsu et al., 2016; Xu et al., 2018; Richards et al., 2017), more specifically, through stress memory (Ding et al., 2014; Kinoshita and Seki, 2014; Meyer, 2015; Yong-Villalobos et al., 2015; Espinas et al., 2016; Fleta-Soriano and Munné-Bosch, 2016; Schönberger et al., 2016; Mauch-Mani et al., 2017). Trees are long-lived organisms that are subjected to repeated environmental constraints; therefore, further studies are needed to better understand environmental epigenetic memory (Allen et al., 2010; Raj et al., 2011; Bräutigam et al., 2013; IPCC, 2014; Schönberger et al., 2016). The best-known example concerns the studies of environmental epigenetic memory in Norway spruce somatic embryos. These studies highlight that environmental epigenetic memory regulates bud phenology and acclimation to the cold (Johnsen et al., 2009; Yakovlev et al., 2010; 2011; 2016; Yakovlev and Fossdal, 2017). Furthermore, an epigenetic memory of temperature during embryogenesis was first observed as important differences in bud phenology in epitypes (Carneros et al., 2017).
Recently, the role of DNA demethylation in poplar SAM was demonstrated for bud burst (Conde et al., 2017). We identified that when short-term variations in water availability occur, poplar active SAM integrates hormonal signals through epigenomic and transcriptomic imprints. These imprints modulate shoot growth and morphogenesis (Lafon-Placette et al., 2018). In this study, we showed that trees grown in favorable or unfavorable open field conditions in two independent experiments still exhibited DMRs in their winter-dormant SAMs 6 months after the summer period. Several reports have shown that the epigenetic machinery in meristematic cells is specific and plays a major role in cell fate (Baubec et al., 2014; Birnbaum and Roudier, 2017). In accordance with these findings, we previously showed that the methylated non-condensed chromatin fraction of active SAM in poplar covered 1.9% of the poplar genome. The non-condensed chromatin fraction is a region where variations in DNA methylation could influence the developmental trajectory. This fraction displayed 74% of the poplar gene models, which were mostly exons (Lafon-Placette et al., 2013). Our results also suggest that the dormant SAM kept the DNA methylation changes that had arisen during the active vegetative period and which correspond to stable epigenetic modifications. Such a phenomenon suggests epigenetic memory. We defined this phenomenon as an epigenetic stress memory that transmits from active SAM in the dividing meristem cells to the dormant cell. The transmission of epigenetic memory has been reported for vernalization in Arabidopsis (Baulcombe and Dean, 2014).
We found that most of the DMRs in the winter-dormant SAM were located in genes (body and promoter). Furthermore, although to a lesser extent, these DMRs were also found in TEs inactivated by hypermethylation; this echoes the findings of Secco et al. (2015). Two approaches were then used to characterize the genes covered by DMRs: (i) without any a priori considerations, where all the genes in the DMRs were analyzed for GO enrichment, and (ii) with a candidate approach using the available DEGs analysis on poplar active SAMs exposed to different water regimes and being involved in the abiotic stress response (Lafon-Placette et al., 2018). Other transcriptomic data were not used because we have recently shown that transcriptomics in water-stressed SAM are different from those in the leaves or roots (Lafon-Placette et al., 2018). Globally, we found genes that are mostly involved in RNA metabolism, the abiotic stress response, the cell cycle and division, and phytohormone pathways (jasmonic acid, abscisic acid, and salicylic acid). These results explain well our biological context of SAM in changing environments, and are in agreement with a recent study in active SAM showing that DNA methylation preferentially affects phytohormone pathways in response to water availability (Lafon-Placette et al., 2018). Altogether, our data, combined with the study of Lafon-Placette et al. (2018), suggest that active SAM integrates environmental variations as DNA methylation in epigenetic footprints. Epigenetic footprints could partially be transmitted mitotically, for several months, up until the winter-dormant stage (Supplementary Fig. S12).
SAM displays epigenetic signatures in response to environmental variations
Although epigenetics plays a role in stress memory, the ecological significance in terms of acclimation and adaptation requires more evidence. In this context, more field experiments and studies at the population level are necessary to confirm the role of epigenomics in stress memory (Richards et al., 2017). To our knowledge, previous studies on ecophysiological traits and epigenomics related to SAM of poplar clones have been performed in one ecological condition. This study is the first to propose a comparative analysis of ecophysiological traits and epigenomics related to poplar SAM among three ecological conditions (Fig. 1). Our comparative analysis allowed us to identify the loci that were systematically targeted by DNA methylation variations related to site growth performances (called epigenetic signatures) transmitted from active to winter-dormant SAM.
Interestingly, the common DMRs of distinct clones overlapped with certain genes and DEGs (Lafon-Placette et al., 2018). Those genes are involved in abiotic stress response, phytohormone signaling, and shoot meristem development, which can all contribute to short-term acclimation of trees or their resistance to repeated stress (Zhang et al., 2013; Yona et al., 2015; Yamamuro et al., 2016; Richards et al., 2017; Lafon-Placette et al., 2018; Supplementary Fig. S12). In the common DMRs, it was also possible to detect two known poplar water-stress DEGs involved in stress acclimation, growth regulation, cell differentiation and proliferation, and the regulation of hormone signaling. STRUWWELPETER encodes a positive regulator of transcription involved in defense signaling crosstalk (salicylic acid and jasmonic acid/ethylene) and SPINDLY encodes a regulator of gibberellin and cytokinin signaling that interacts with the chromatin remodeling complex (Autran et al., 2002; Qin et al., 2011; Sarnowska et al., 2013; Wang et al., 2015; Steiner et al., 2016). Our results propose that DNA methylation preferentially affects specific responsive genes and conserves these signatures in the winter-dormant SAM, although several studies have demonstrated the existence of extensive remodeling of DNA methylation in response to stresses. Therefore, a putative selective variation of DNA methylation on stress-responsive genes has been proposed (Colaneri and Jones, 2013; Liang et al., 2014; Garg et al., 2015; Secco et al., 2015; Yong-Villalobos et al., 2015; Wibowo et al., 2016). Our data are also in agreement with two recently published studies on the active SAM of poplar subjected to environmental variations (Conde et al., 2017; Lafon-Placette et al., 2018).
Future studies could use functional analysis to confirm the biological function of these gene categories, which we identified through a GO enrichment approach. In addition, although our comparative analysis was performed on distinct clones and systems of production (see Table 1, Fig. 1, and the Materials and methods for details), a significant number of conserved DMRs were detected. In order to improve comparative analysis and to test the potential ‘priming’ effect (Lämke and Bäurle, 2017) in future investigations, it would be relevant to follow the epigenetic memory and corresponding level of tolerance to abiotic stress. This type of comparative approach has to be managed on a defined set of poplar clones grown under similar management at distinct pedoclimatic sites and analyzed over successive years of growth and in distinct seasons. Another challenge will be to validate the role of the epigenetic signature through a reverse genetic approach with hyper- or hypomethylated RNAi lines (Zhu et al., 2013; Conde et al., 2017; A-L. Le Gac and S. Maury, unpublished results) or through CRISPR/Cas9 technology (Liu et al., 2017, (Fan et al., 2015). It would be relevant to use CRISPR/Cas9 for epigenetic editing for DNA methylation on common DMRs and evaluate the impact on poplar stress memory. Finally, histone modifications, small and long non-coding RNAs, and nucleosome occupancy and chromatin structure should also be investigated to complete our understanding of the epigenetic mechanisms involved in stress memory in trees (Lämke and Bäurle, 2017).
Supplementary data
Supplementary data are available at JXB online.
Table S1. Description of poplar clones used in experiment 2.
Table S2. Gene v3 annotation counts associated with their respective probe category subset.
Table S3. Differentially methylated regions (DMRs) for experiments 1 and 2 and for commonly conserved DMRs.
Table S4. Overlap between genes localized in DMRs and DEGs.
Table S5. Summary of AGRIGO statistical results with the number of genes, P-values and FDRs for major Gene Ontology categories of REVIGO treemap analyses.
Fig. S1. Methylome description for an additional unfavorable site in experiment 2, located at Guémené (GMN).
Fig. S2. REVIGO treemap of biological process GO clustering based on the abs log10P-values of hypo- and hypermethylated probes in DMRs located in the gene body for each condition and experiment.
Fig. S3. REVIGO treemap of biological process GO clustering based on the abs log10P-value of genes in DMRs in experiment 1 and experiment 2.
Fig. S4. REVIGO treemap of biological process GO clustering based on the abs log10P-value of genes in DMRs in experiment 1: hypomethylated DMRs and hypermethylated DMRs.
Fig. S5. REVIGO treemap of biological process GO clustering based on the abs log10P-value of genes in DMRs in experiment 2: hypomethylated DMRs and hypermethylated DMRs.
Fig. S6. REVIGO treemap of biological process GO clustering based on the abs log10P-value of DEGs in DMRs in experiment 1 or 2.
Fig. S7. DEG counts relative to DMRs: down-regulated DEGs and up-regulated DEGs between favorable and unfavorable growth conditions.
Fig. S8. REVIGO treemap of biological process GO clustering based on the abs log10P-value of genes in DMRs common to experiments 1 and 3: experiments 1 and 3 (n=2029), hypomethylated DMRs, and hypermethylated DMRs between favorable and unfavorable growth conditions.
Fig. S9. REVIGO treemap of biological process GO clustering based on the abs log10P-value of genes in DMRs common to experiments 2 and 3: experiments 2 and 3 (n=2900), hypomethylated DMRs, and hypermethylated DMRs between favorable and unfavorable growth conditions.
Fig. S10. REVIGO treemap of biological process GO clustering based on the abs log10P-value of genes in DMRs common to experiments 1 and 2: experiments 1 and 2 (n=998), hypomethylated DMRs, and hypermethylated DMRs between favorable and unfavorable growth conditions.
Fig. S11. REVIGO treemap of biological process GO clustering based on the abs log10P-value of genes in DMRs common to experiments 1 and 2: Common conserved variation DMRs and common inversed variation DMRs between favorable and unfavorable growth conditions.
Fig S12. Model for epigenetic environmental memory in the poplar shoot apical meristem.
Method S1. False discovery rate estimation for experiments 1 and 2.
Supplementary Material
Acknowledgements
CLP and ALLG received PhD grants from the Région Centre and the Ministère de la Recherche et Enseignement Supérieur. This work was funded by the ANR France, within the project BIOENERGIE SYLVABIOM (ANR-08-BIOE-0006). We acknowledge Prof. Phillipe Gallusci (Université INRA Bordeaux, France) and Dr Farshad Roodbarkelari (Institute for Biology III, University of Freiburg, Germany) for their help in improving the final version of the manuscript.
Author contributions
SM designed the study; ALLG, CLP, AD, ILJ, RF, AB, NM, GB, JCB, FB, and SM performed the study; ALLG, CLP, DC, VS, FB, and SM analyzed the data; ALLG, CLP, FB, and SM wrote the manuscript.
References
- Allen CD, Macalady AK, Chenchouni H, et al. 2010. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. Forest Ecology and Management 259, 660–684. [Google Scholar]
- Alonso C, Balao F, Bazaga P, Pérez R. 2016. Epigenetic contribution to successful polyploidizations: variation in global cytosine methylation along an extensive ploidy series in Dianthus broteri (Caryophyllaceae). New Phytologist 212, 571–576. [DOI] [PubMed] [Google Scholar]
- Alonso C, Pérez R, Bazaga P, Herrera CM. 2015. Global DNA cytosine methylation as an evolving trait: phylogenetic signal and correlated evolution with genome size in angiosperms. Frontiers in Genetics 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers B, Castonguay E, Massicotte R. 2010. Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Molecular Ecology 19, 1283–1295. [DOI] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J. 2002. Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. The EMBO Journal 21, 6036–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova Z. 2015. Transcriptional ‘memory’ of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes. The Plant Journal 83, 149–159. [DOI] [PubMed] [Google Scholar]
- Baubec T, Finke A, Mittelsten Scheid O, Pecinka A. 2014. Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Reports 15, 446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC, Dean C. 2014. Epigenetic regulation in plant responses to the environment. Cold Spring Harbor Perspectives in Biology 6, a019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D. 2011. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480, 245–249. [DOI] [PubMed] [Google Scholar]
- Benaglia T, Chauveau D, Hunter D, Young D. 2009. mixtools: An R package for analyzing finite mixture models. Journal of Statistical Software 32, 1–29. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300. [Google Scholar]
- Birnbaum KD, Roudier F. 2017. Epigenetic memory and cell fate reprogramming in plants. Regeneration 4, 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizet F, Bogeat-Triboulot MB, Montpied P, Christophe A, Ningre N, Cohen D, Hummel I. 2015. Phenotypic plasticity toward water regime: response of leaf growth and underlying candidate genes in Populus. Physiologia Plantarum 154, 39–53. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Arcuri D, Richards CL, Pigliucci M. 2010. Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana. Evolutionary Ecology 24, 541–553. [Google Scholar]
- Boyko A, Kovalchuk I. 2008. Epigenetic control of plant stress response. Environmental and Molecular Mutagenesis 49, 61–72. [DOI] [PubMed] [Google Scholar]
- Bräutigam K, Vining KJ, Lafon-Placette C, et al. 2013. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecology and Evolution 3, 399–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneros E, Yakovlev I, Viejo M, Olsen JE, Fossdal CG. 2017. The epigenetic memory of temperature during embryogenesis modifies the expression of bud burst-related genes in Norway spruce epitypes. Planta 246, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causevic A, Delaunay A, Ounnar S, Righezza M, Delmotte F, Brignolas F, Hagège D, Maury S. 2005. DNA methylating and demethylating treatments modify phenotype and cell wall differentiation state in sugarbeet cell lines. Plant Physiology and Biochemistry 43, 681–691. [DOI] [PubMed] [Google Scholar]
- Choi CS, Sano H. 2007. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Molecular Genetics and Genomics 277, 589–600. [DOI] [PubMed] [Google Scholar]
- Colaneri AC, Jones AM. 2013. Genome-wide quantitative identification of DNA differentially methylated sites in Arabidopsis seedlings growing at different water potential. PLoS One 8, e59878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde D, Le Gac AL, Perales M, Dervinis C, Kirst M, Maury S, González-Melendi P, Allona I. 2017. Chilling-responsive DEMETER-LIKE DNA demethylase mediates in poplar bud break. Plant, Cell & Environment 40, 2236–2249. [DOI] [PubMed] [Google Scholar]
- Cortijo S, Wardenaar R, Colomé-Tatché M, et al. 2014. Mapping the epigenetic basis of complex traits. Science 343, 1145–1148. [DOI] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z. 2012. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nature Communications 3, 740. [DOI] [PubMed] [Google Scholar]
- Ding Y, Virlouvet L, Liu N, Riethoven J-J, Fromm M, Avramova Z. 2014. Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biology 14, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19, 11–15. [Google Scholar]
- Dubin MJ, Zhang P, Meng D, et al. 2015. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 4, e05255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Ellis NA, Makarevitch I, et al. 2012. Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genetics 8, e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinas NA, Saze H, Saijo Y. 2016. Epigenetic control of defense signaling and priming in plants. Frontiers in Plant Science 7, 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K. 2015. Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Scientific Reports 5, 12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichot R, Barigah TS, Chamaillard S, LE Thiec D, Laurans F, Cochard H, Brignolas F. 2010. Common trade-offs between xylem resistance to cavitation and other physiological traits do not hold among unrelated Populus deltoides × Populus nigra hybrids. Plant, Cell & Environment 33, 1553–1568. [DOI] [PubMed] [Google Scholar]
- Fichot R, Chamaillard S, Depardieu C, Le Thiec D, Cochard H, Barigah TS, Brignolas F. 2011. Hydraulic efficiency and coordination with xylem resistance to cavitation, leaf function, and growth performance among eight unrelated Populus deltoides × Populus nigra hybrids. Journal of Experimental Botany 62, 2093–2106. [DOI] [PubMed] [Google Scholar]
- Fichot R, Laurans F, Monclus R, Moreau A, Pilate G, Brignolas F. 2009. Xylem anatomy correlates with gas exchange, water-use efficiency and growth performance under contrasting water regimes: evidence from Populus deltoides × Populus nigra hybrids. Tree Physiology 29, 1537–1549. [DOI] [PubMed] [Google Scholar]
- Fleta-Soriano E, Munné-Bosch S. 2016. Stress memory and the inevitable effects of drought: a physiological perspective. Frontiers in Plant Science 7, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust CM, Preite V, Schrey AW, Alvarez M, Robertson MH, Verhoeven KJ, Richards CL. 2016. Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Molecular Ecology 25, 1639–1652. [DOI] [PubMed] [Google Scholar]
- Gagliano M, Renton M, Depczynski M, Mancuso S. 2014. Experience teaches plants to learn faster and forget slower in environments where it matters. Oecologia 175, 63–72. [DOI] [PubMed] [Google Scholar]
- Garg R, Narayana Chevala V, Shankar R, Jain M. 2015. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Scientific Reports 5, 14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino F, Cicatelli A, Brundu G, Heinze B, Castiglione S. 2015. Epigenetic diversity of clonal white poplar (Populus alba L.) populations: could methylation support the success of vegetative reproduction strategy?PLoS One 10, e0131480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourcilleau D, Bogeat-Triboulot M-B, Thiec D, Lafon-Placette C, Delaunay A, El-Soud WA, Brignolas F, Maury S. 2010. DNA methylation and histone acetylation: genotypic variations in hybrid poplars, impact of water deficit and relationships with productivity. Annals of Forest Science 67, 208. [Google Scholar]
- Hébrard C, Peterson DG, Willems G, Delaunay A, Jesson B, Lefèbvre M, Barnes S, Maury S. 2016. Epigenomics and bolting tolerance in sugar beet genotypes. Journal of Experimental Botany 67, 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC 2014. Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part A: Global and Sectoral Aspects. Working Group II contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Field CB, Barros VR, Dokken DJ. et al, eds. New York: Cambridge University Press. [Google Scholar]
- Jablonka E, Oborny B, Molnar I, Kisdi E, Hofbauer J, Czaran T. 1995. The adaptive advantage of phenotypic memory in changing environments. Philosophical Transactions of the Royal Society B: Biological Sciences 350, 133–141. [DOI] [PubMed] [Google Scholar]
- Jansson S, Douglas CJ. 2007. Populus: a model system for plant biology. Annual Review of Plant Biology 58, 435–458. [DOI] [PubMed] [Google Scholar]
- Johannes F, Porcher E, Teixeira FK, et al. 2009. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genetics 5, e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen Ø, Kvaalen H, Yakovlev IA, Dæhlen OG, Fossdal CG, Skrøppa T. 2009. An epigenetic memory from time of embryo development affects climatic adaptation in Norway spruce. In: Gusta LV, Wisniewski ME, Tanino KK, eds. Plant cold hardiness. From the laboratory to the field. Wallingford: CABI, 99–107. [Google Scholar]
- Kawakatsu T, Huang SC, Jupe F, et al. ; 1001 Genomes Consortium. 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166, 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Seki M. 2014. Epigenetic memory for stress response and adaptation in plants. Plant & Cell Physiology 55, 1859–1863. [DOI] [PubMed] [Google Scholar]
- Kooke R, Johannes F, Wardenaar R, Becker F, Etcheverry M, Colot V, Vreugdenhil D, Keurentjes JJ. 2015. Epigenetic basis of morphological variation and phenotypic plasticity in Arabidopsis thaliana. The Plant Cell 27, 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Placette C, Faivre-Rampant P, Delaunay A, Street N, Brignolas F, Maury S. 2013. Methylome of DNase I sensitive chromatin in Populus trichocarpa shoot apical meristematic cells: a simplified approach revealing characteristics of gene-body DNA methylation in open chromatin state. New Phytologist 197, 416–430. [DOI] [PubMed] [Google Scholar]
- Lafon-Placette C, Le Gac A-L, Chauveau D, et al. 2018. Water availability footprint on epigenome and transcriptome targets hormone pathways in the poplar shoot apical meristem. Journal of Experimental Botany 69, 537–551. [DOI] [PubMed] [Google Scholar]
- Lambé P, Mutambel HSN, Fouché J-G, Deltour R, Foidart J-M, Gaspar T. 1997. DNA methylation as a key process in regulation of organogenic totipotency and plant neoplastic progression?In Vitro Cellular & Developmental Biology - Plant 33, 155–162. [Google Scholar]
- Lämke J, Bäurle I. 2017. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biology 18, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latzel V, Allan E, Bortolini Silveira A, Colot V, Fischer M, Bossdorf O. 2013. Epigenetic diversity increases the productivity and stability of plant populations. Nature Communications 4, 2875. [DOI] [PubMed] [Google Scholar]
- Liang D, Zhang Z, Wu H, et al. 2014. Single-base-resolution methylomes of Populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genetics 15(Suppl 1), S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Xie C, Si H, Yang J. 2017. CRISPR/Cas9-mediated genome editing in plants. Methods 15, 121–122. [DOI] [PubMed] [Google Scholar]
- Marron N, Delay D, Petit JM, Dreyer E, Kahlem G, Delmotte FM, Brignolas F. 2002. Physiological traits of two Populus × euramericana clones, Luisa Avanzo and Dorskamp, during a water stress and re-watering cycle. Tree Physiology 22, 849–858. [DOI] [PubMed] [Google Scholar]
- Marron N, Dreyer E, Boudouresque E, Delay D, Petit JM, Delmotte FM, Brignolas F. 2003. Impact of successive drought and re-watering cycles on growth and specific leaf area of two Populus × canadensis (Moench) clones, ‘Dorskamp’ and ‘Luisa_Avanzo’. Tree Physiology 23, 1225–1235. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Baccelli I, Luna E, Flors V. 2017. Defense priming: an adaptive part of induced resistance. Annual Review of Plant Biology 68, 485–512. [DOI] [PubMed] [Google Scholar]
- McLachlan G, Peel D. 2000. Finite mixture models. New York: Wiley. [Google Scholar]
- Meyer P. 2015. Epigenetic variation and environmental change. Journal of Experimental Botany 66, 3541–3548. [DOI] [PubMed] [Google Scholar]
- Mirouze M, Paszkowski J. 2011. Epigenetic contribution to stress adaptation in plants. Current Opinion in Plant Biology 14, 267–274. [DOI] [PubMed] [Google Scholar]
- Monclus R, Dreyer E, Delmotte FM, Villar M, Delay D, Boudouresque E, Petit JM, Marron N, Bréchet C, Brignolas F. 2005. Productivity, leaf traits and carbon isotope discrimination in 29 Populus deltoides × P. nigra clones. New Phytologist 167, 53–62. [DOI] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, et al. 2010. Plant phenotypic plasticity in a changing climate. Trends in Plant Science 15, 684–692. [DOI] [PubMed] [Google Scholar]
- Niederhuth CE, Schmitz RJ. 2017. Putting DNA methylation in context: from genomes to gene expression in plants. Biochimica et Biophysica Acta 1860, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong-Abdullah M, Ordway JM, Jiang N, et al. 2015. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525, 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Miklós I. 1999. Epigenetic inheritance, genetic assimilation and speciation. Journal of Theoretical Biology 200, 19–37. [DOI] [PubMed] [Google Scholar]
- Pastor V, Luna E, Mauch-Mani B, Ton J, Flors V. 2013. Primed plants do not forget. Environmental and Experimental Botany 94, 46–56. [Google Scholar]
- Plomion C, Bastien C, Bogeat-Triboulot M-B, et al. 2016. Forest tree genomics: 10 achievements from the past 10 years and future prospects. Annals of Forest Science 73, 77–103. [Google Scholar]
- Qin F, Kodaira KS, Maruyama K, Mizoi J, Tran LS, Fujita Y, Morimoto K, Shinozaki K, Yamaguchi-Shinozaki K. 2011. SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiology 157, 1900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2015. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Raj S, Bräutigam K, Hamanishi ET, Wilkins O, Thomas BR, Schroeder W, Mansfield SD, Plant AL, Campbell MM. 2011. Clone history shapes Populus drought responses. Proceedings of the National Academy of Sciences, USA 108, 12521–12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wulff BB, Mirouze M, Marí-Ordóñez A, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J. 2009. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes & Development 23, 939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CL, Alonso C, Becker C, et al. 2017. Ecological plant epigenetics: evidence from model and non-model species, and the way forward. Ecology Letters 20, 1576–1590. [DOI] [PubMed] [Google Scholar]
- Rico L, Ogaya R, Barbeta A, Peñuelas J. 2014. Changes in DNA methylation fingerprint of Quercus ilex trees in response to experimental field drought simulating projected climate change. Plant Biology 16, 419–427. [DOI] [PubMed] [Google Scholar]
- Sarnowska EA, Rolicka AT, Bucior E, et al. 2013. DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiology 163, 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. 2011. Transgenerational epigenetic instability is a source of novel methylation variants. Science 334, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, et al. 2013. Patterns of population epigenomic diversity. Nature 495, 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönberger B, Chen X, Mager S, Ludewig U. 2016. Site-dependent differences in DNA methylation and their impact on plant establishment and phosphorus nutrition in Populus trichocarpa. PLoS One 11, e0168623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Wang C, Shou H, Schultz MD, Chiarenza S, Nussaume L, Ecker JR, Whelan J, Lister R. 2015. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4, e09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Ci D, Tian M, Zhang D. 2016. Stable methylation of a non-coding RNA gene regulates gene expression in response to abiotic stress in Populus simonii. Journal of Experimental Botany 67, 1477–1492. [DOI] [PubMed] [Google Scholar]
- Steiner E, Livne S, Kobinson-Katz T, Tal L, Pri-Tal O, Mosquna A, Tarkowská D, Muller B, Tarkowski P, Weiss D. 2016. SPINDLY inhibits class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiology 171, 1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K. 2008. A unified approach to false discovery rate estimation. BMC Bioinformatics 9, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SE. 2000. Phenotypic plasticity for plant development, function and life history. Trends in Plant Science 5, 537–542. [DOI] [PubMed] [Google Scholar]
- Sultan SE, Barton K, Wilczek AM. 2009. Contrasting patterns of transgenerational plasticity in ecologically distinct congeners. Ecology 90, 1831–1839. [DOI] [PubMed] [Google Scholar]
- Teyssier E, Bernacchia G, Maury S, How Kit A, Stammitti-Bert L, Rolin D, Gallusci P. 2008. Tissue dependent variations of DNA methylation and endoreduplication levels during tomato fruit development and ripening. Planta 228, 391–399. [DOI] [PubMed] [Google Scholar]
- Teyssier C, Maury S, Beaufour M, Grondin C, Delaunay A, Le Metté C, Ader K, Cadene M, Label P, Lelu-Walter MA. 2014. In search of markers for somatic embryo maturation in hybrid larch (Larix×eurolepis): global DNA methylation and proteomic analyses. Physiologia Plantarum 150, 271–291. [DOI] [PubMed] [Google Scholar]
- Toillon J, Fichot R, Dallé E, Berthelot A, Brignolas F, Marron N. 2013a. Planting density affects growth and water-use efficiency depending on site in Populus deltoides × P. nigra. Forest Ecology and Management 304, 345–354. [Google Scholar]
- Toillon J, Rollin B, Dallé E, Feinard-Duranceau M, Bastien JC, Brignolas F, Marron N. 2013b. Variability and plasticity of productivity, water-use efficiency, and nitrogen exportation rate in Salix short rotation coppice. Biomass and Bioenergy 56, 392–404. [Google Scholar]
- Trap-Gentil MV, Hébrard C, Lafon-Placette C, Delaunay A, Hagège D, Joseph C, Brignolas F, Lefebvre M, Barnes S, Maury S. 2011. Time course and amplitude of DNA methylation in the shoot apical meristem are critical points for bolting induction in sugar beet and bolting tolerance between genotypes. Journal of Experimental Botany 62, 2585–2597. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Tanurdzić M, Lippman Z, et al. 2007. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biology 5, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining KJ, Pomraning KR, Wilhelm LJ, Priest HD, Pellegrini M, Mockler TC, Freitag M, Strauss SH. 2012. Dynamic DNA cytosine methylation in the Populus trichocarpa genome: tissue-level variation and relationship to gene expression. BMC Genomics 13, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yao J, Du X, Zhang Y, Sun Y, Rollins JA, Mou Z. 2015. The Arabidopsis mediator complex Subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiology 169, 856–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibowo A, Becker C, Marconi G, et al. 2016. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 5, e13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhou S, Gong X, Song Y, van Nocker S, Ma F, Guan Q. 2018. Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnology Journal 16, 672–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev IA, Asante DK, Fossdal CG, Junttila O, Johnsen Ø. 2011. Differential gene expression related to an epigenetic memory affecting climatic adaptation in Norway spruce. Plant Science 180, 132–139. [DOI] [PubMed] [Google Scholar]
- Yakovlev IA, Carneros E, Lee Y, Olsen JE, Fossdal CG. 2016. Transcriptional profiling of epigenetic regulators in somatic embryos during temperature induced formation of an epigenetic memory in Norway spruce. Planta 243, 1237–1249. [DOI] [PubMed] [Google Scholar]
- Yakovlev IA, Fossdal CG. 2017. In silico analysis of small RNAs suggest roles for novel and conserved miRNAs in the formation of epigenetic memory in somatic embryos of Norway spruce. Frontiers in Physiology 8, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev IA, Fossdal CG, Johnsen Ø. 2010. MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytologist 187, 1154–1169. [DOI] [PubMed] [Google Scholar]
- Yakovlev I, Fossdal CG, Skrøppa T, Olsen JE, Jahren AH, Johnsen Ø. 2012. An adaptive epigenetic memory in conifers with important implications for seed production. Seed Science Research 22, 63–76. [Google Scholar]
- Yamamuro C, Zhu JK, Yang Z. 2016. Epigenetic modifications and plant hormone action. Molecular Plant 9, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona AH, Frumkin I, Pilpel Y. 2015. A relay race on the evolutionary adaptation spectrum. Cell 163, 549–559. [DOI] [PubMed] [Google Scholar]
- Yong-Villalobos L, González-Morales SI, Wrobel K, Gutiérrez-Alanis D, Cervantes-Peréz SA, Hayano-Kanashiro C, Oropeza-Aburto A, Cruz-Ramírez A, Martínez O, Herrera-Estrella L. 2015. Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proceedings of the National Academy of Sciences, USA 112, E7293–E7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Fischer M, Colot V, Bossdorf O. 2013. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytologist 197, 314–322. [DOI] [PubMed] [Google Scholar]
- Zhu R, Shevchenko O, Ma C, Maury S, Freitag M, Strauss SH. 2013. Poplars with a PtDDM1-RNAi transgene have reduced DNA methylation and show aberrant post-dormancy morphology. Planta 237, 1483–1493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.