Abstract
Streptococcus mutans is a Gram-positive human pathogen that is one of the major contributors to dental caries, a condition with an economic cost of over $100 billion per year in the United States. S. mutans secretes a 21-amino-acid peptide termed the competence stimulating peptide (21-CSP) to assess its population density in a process termed quorum sensing (QS) and to initiate a variety of phenotypes such as biofilm formation and bacteriocin production. 21-CSP is processed by a membrane bound protease SepM into active 18-CSP, which then binds to the ComD receptor. This study seeks to determine the molecular mechanism that ties 21-CSP:SepM recognition and 18-CSP:ComD receptor binding, and to identify QS modulators with distinct activity profiles. To this end, we conducted systematic replacement of the amino acid residues in both 21-CSP and 18-CSP and assessed the ability of the mutated analogs to modulate QS. We identified residues that are important to SepM recognition and ComD receptor binding. Our results shed light on the S. mutans competence QS pathway at the molecular level. Moreover, our structural insights of the CSP signal can be used to design QS-based anti-infective therapeutics against S. mutans.
Keywords: Streptococcus mutans, Competence stimulating peptide (CSP), Quorum sensing
Graphical Abstract
Quorum sensing (QS) is a primary bacterial communication system used to assess local population densities and to coordinate group behaviors.1–3 QS bacteria use pheromones to regulate the expression of various genes when the bacterial cell density reaches a certain threshold concentration.4, 5 Gram-negative bacteria generally utilize small-molecule signals, whereas Gram-positive bacteria use autoinducing peptides as the chemical signal to activate QS.6, 7 In both Gram-negative and Gram-positive bacteria QS has been shown to be involved in the regulation of symbiotic group phenotypes such as root nodulation and bioluminescence, as well as pathogenic behaviors including competence, biofilm formation, swarming, and virulence factor production.4, 8–10 Specifically, pathogenic bacteria utilize QS to launch synchronized attacks on their host.11, 12 Thus, QS has received considerable attention as a novel anti-infective target against a variety of prevalent human pathogens (e.g., Streptococcus pneumoniae,13, 14 Staphylococcus aureus,10, 15–18 Vibrio cholera,4, 19 Enterococcus faecalis20, 21 and Pseudomonas aeruginosa22–25). In contrast to antibiotics, interception of QS circuits’ signal:receptor interactions would lead to a reduction in resistance development as this approach does not directly apply selective pressure.26, 27
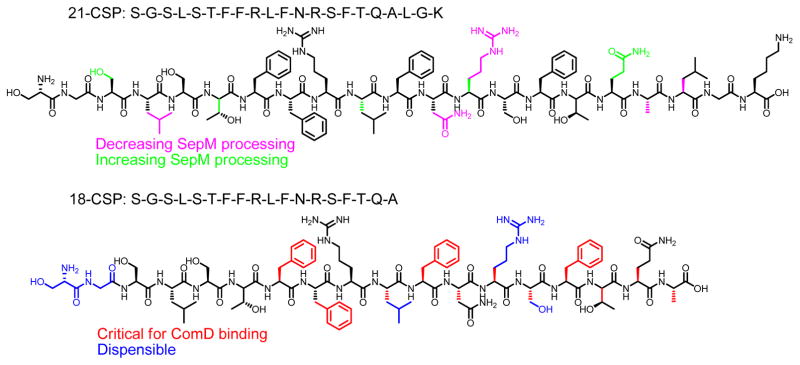
Streptococcus mutans is a human pathogen that is considered to be one of the primary etiological agents of dental caries,28 a condition bearing costs around $100 billion per year in the USA alone.29 In order to thrive in the oral cavity, S. mutans forms robust biofilms on tooth surfaces. This process requires a highly coordinated effort by the entire bacterial population. Thus, S. mutans utilizes a QS system called the competence regulon to govern biofilm formation. This QS circuitry is also required for the regulation of stress responses, the development of genetic competence and the expression of bacteriocin-encoding genes. The competence regulon in S. mutans is centered on a peptide pheromone termed the competence stimulating peptide (CSP). The CSP signal is encoded by the comC gene as a propeptide, ComC, which is processed and secreted by a dedicated ABC transporter complex (ComAB) to generate a 21-residue peptide (21-CSP). 21-CSP is further cleaved at the C-terminal end by a membrane-localized protease, SepM, to form the active signal, 18-CSP, which then interacts with the cognate transmembrane histidine-kinase receptor ComD.30, 31 18-CSP:ComD binding triggers the phosphorylation of ComE, a cytoplasmic response regulator, and stimulates the expression of genes involved in biofilm formation, bacteriocin production and competence development, in addition to upregulating the competence regulon genes comABCDE (Figure 1).30 Another QS system in S. mutans, encoded by comR-comS is also linked to competence development. ComS, a 17-amino acid polypeptide, is secreted and processed to its mature form, XIP (sigma X-inducing peptide), which is then imported back into the cell to activate the ComR regulator. Activated ComR then upregulates the expression of ComS and ComX.32, 33 Development of competence either by CSP or XIP depends on the growth media. In rich media, transformation frequency is dependent on CSP, whereas in a chemically defined medium (CDM), devoid of any exogenous peptides, transformation depends on XIP.34–36
Figure 1.
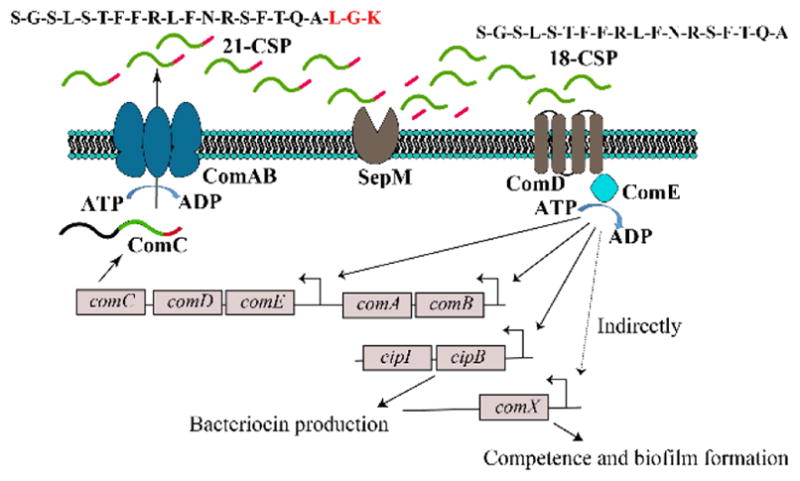
CSP-mediated QS circuit of S. mutans. The CSP pro-peptide, ComC, is processed and secreted by the dedicated ABC transporter (ComAB) as 21-CSP, which is further cleaved by a membrane protease, SepM, to form the active 18-CSP signal. 18-CSP interacts with the cognate transmembrane histidine-kinase receptor ComD, leading to phosphorylation of ComE, a response regulator, and results in autoinduction of the QS circuitry, the production of bacteriocins, and expression of comX, which codes for additional QS-regulated phenotypes.
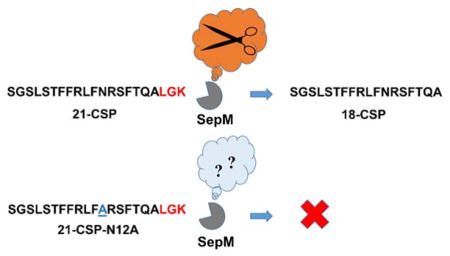
Previous study has shown that 21-CSP failed to induce QS in a ΔsepM mutant of S. mutans.30 This study established that processing of 21-CSP by SepM is critical for forming active 18-CSP, however, it is not known which interactions govern the recognition of 21-CSP by SepM. To better understand the molecular mechanism that drives the S. mutans QS circuitry, especially to identify the structural motifs necessary for SepM binding and processing as well as ComD binding and activation, we set out to study the structure-activity relationships (SARs) of both 21-CSP and 18-CSP. To this end, full alanine and D-amino acid scans of both CSPs were performed and the ability of the resulting analogs to modulate the activation of the ComD receptor was determined. In addition, the overall structural features of the CSP analogs were analyzed using circular dichroism (CD) spectroscopy to investigate the correlation between the CSP analogs’ secondary structures and biological activities. Lastly, we designed a second-generation library of analogs based on our initial SAR analysis and identified potent simplified 18-CSP analogs. These analogs allowed us to determine the minimal structural requirements for effective ComD receptor activation.
Results and Discussion
To identify the key side chains and stereocenters that are involved in CSP:SepM and CSP:ComD interactions, we started our study with systematic evaluations of each residue of 21-CSP and 18-CSP by performing full alanine and D-amino acid scans. Overall, we synthesized 94 CSP analogs using Fmoc/tBu solid-phase peptide synthesis procedures on 4-benzyloxybenzyl alcohol (Wang) resin (see Materials and Methods) and purified them to homogeneity by semipreparative RP-HPLC to afford the purified CSP analogs in acceptable yields (10–20%; see the Supporting information for full characterization details).
Next we tested the ability of these analogs to modulate the competence QS circuitry in S. mutans using two reporter systems: a reporter strain harboring a pCipB-lacZ construct integrated to S. mutans chromosome (SAB249, representing the expression of early-stage bacteriocin genes; see Materials and Methods for strain information), and a ΔcomC reporter strain harboring a pcomX-lacZ plasmid (SMCOM2, representing the expression of late-stage competence genes; see Materials and Methods for strain information). In these reporter strains, the cipB or comX promoter is fused to the lacZ gene, thus upon activation of the QS circuitry through exogenous addition of CSP, lacZ will be transcribe, allowing for quantification of QS activation by measuring β-galactosidase activity.
Alanine scanning of 21-CSP and 18-CSP against ComD
Since 18-CSP is the active signal that binds ComD, any observed activity change of 18-CSP analogs can be attributed to alteration in CSP:ComD binding. To the contrary, changes in activity of 21-CSP analogs can either be the result of modulation in SepM processing or ComD binding. Thus, correlation of the activity trends of both 18-CSP and 21-CSP would allow for investigating changes in CSP:SepM interactions. Starting with 18-CSP, the alanine scan revealed that changes at the hydrophobic residues resulted in dramatic reduction in potency while changes of the hydrophilic residues were more tolerable (compare modifications in Leu4, Phe7, Phe8, Phe11 and Phe15 with modifications in Ser1, Ser3, Ser5, Thr6, Arg9, Asn12, Arg13, Ser14 and Gln17 in Table 1). This observation emphasizes the importance of the hydrophobic central region of 18-CSP in ComD receptor binding. Overall, our analysis of the central region of 18-CSP is in agreement with previous work by Syvitski et al. that showed that the hydrophobic residues in S. mutans CSP are involved in ComD receptor binding.37
Table 1.
EC50 values of the alanine scan of 21-CSP and 18-CSP against S. mutans SAB249a
|
|
|
||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Name | EC50 (nM)b | 95% CIc | Fold changed | Name | EC50 (nM)b | 95% CIc | Fold changed |
| 21-CSP | 240 | 200 – 290 | 18-CSP | 6.2 | 3.7 – 10.0 | ||
| 21-CSP-S1A | 90 | 43 – 190 | 0.38 | 18-CSP-S1A | 2.6 | 1.1 – 6.5 | 0.42 |
| 21-CSP-G2A | 140 | 60 – 330 | 0.58 | 18-CSP-G2A | 9.7 | 4.2 – 22 | 1.6 |
| 21-CSP-S3A | 150 | 100 – 230 | 0.63 | 18-CSP-S3A | 13 | 7.1 – 24 | 2.1 |
| 21-CSP-L4A | 6500 | 3300 – 13000 | 27 | 18-CSP-L4A | 57 | 25 – 130 | 9.2 |
| 21-CSP-S5A | 330 | 140 – 760 | 1.4 | 18-CSP-S5A | 11 | 6.8 – 17 | 1.8 |
| 21-CSP-T6A | 280 | 240 – 320 | 1.2 | 18-CSP-T6A | 6.0 | 3.3 – 11 | 0.97 |
| 21-CSP-F7A | --e | -- | -- | 18-CSP-F7A | 220 | 130 – 370 | 35 |
| 21-CSP-F8A | --e | -- | -- | 18-CSP-F8A | >1,000 | -- | -- |
| 21-CSP-R9A | 1100 | 760 – 1500 | 4.6 | 18-CSP-R9A | 27 | 14 – 51 | 4.4 |
| 21-CSP-L10A | 370 | 200 – 680 | 1.5 | 18-CSP-L10A | 6.5 | 3.5 – 12 | 1.1 |
| 21-CSP-F11A | --e | -- | -- | 18-CSP-F11A | 410 | 300 – 560 | 66 |
| 21-CSP-N12A | --e | -- | -- | 18-CSP-N12A | 33 | 18 – 60 | 5.3 |
| 21-CSP-R13A | 480 | 390 –580 | 2.0 | 18-CSP-R13A | 2.7 | 1.3 – 5.6 | 0.44 |
| 21-CSP-S14A | 160 | 69 – 370 | 0.67 | 18-CSP-S14A | 4.5 | 2.8 – 7.3 | 0.73 |
| 21-CSP-F15A | --e | -- | -- | 18-CSP-F15A | --e | -- | -- |
| 21-CSP-T16A | 1400 | 980 – 2100 | 5.8 | 18-CSP-T16A | 64 | 49 – 83 | 10 |
| 21-CSP-Q17A | 94 | 61 – 140 | 0.39 | 18-CSP-Q17A | 19 | 11 – 31 | 3.1 |
| 21-CSP-L19A | 190 | 85 – 420 | 0.79 | ||||
| 21-CSP-G20A | 190 | 79 – 430 | 0.79 | ||||
| 21-CSP-K21A | 290 | 220 – 390 | 1.2 | ||||
See experimental section for details on reporter strain and methods. See supporting information for primary screening assay results and plots of agonism dose response curves. All assays were performed in triplicate.
EC50 values determined by testing peptides over a range of concentrations.
95% confidence interval.
Ratio where each analog’s EC50 is divided by native CSP EC50; a value <1 indicates a better activator than the parent CSP.
EC50 not determined due to the analog’s low induction in primary agonism screening assay.
Green color indicates modifications that improve processing of 21-CSP by SepM, purple color indicates modifications that decrease processing of 21-CSP by SepM.
Red color indicates modifications critical for 18-CSP:ComD binding.
We then evaluated the activity trend of the 21-CSP analogs in comparison to the 18-CSP analogs to identify positions that are important for SepM recognition and processing. First, the EC50 value of 21-CSP is 240 nM, whereas the EC50 of 18-CSP is 6.2 nM. Since 21-CSP is being processed to 18-CSP, the ~40-fold decrease in potency for 21-CSP implies that only a small fraction of 21-CSP is being processed by SepM, thus a significantly higher concentration of 21-CSP is required to afford the effective concentration of the active 18-CSP. When comparing the alanine analogs, starting with the 21-CSP, the G2A substitution resulted in an increased activity of ~2-fold (fold change 0.58) whereas the S3A modification resulted in an increased activity of ~2-fold (fold change 0.63; Table 1). An opposite trend was observed for these modifications in 18-CSP: G2A (~1.5-fold reduction in potency) and S3A (~2-fold reduction in potency). A similar discrepancy was also observed for position Gln17: this modification in 21-CSP resulted in an analog with increased activity of ~3-fold (fold change 0.39), while in 18-CSP this modification resulted in a decrease in potency (~3-fold reduction). These results suggest that alanine replacement in Gly2, Ser3 and Gln17 positions lead to an increased SepM recognition and processing, affording a higher effective concentration of the corresponding 18-CSP analog. Interestingly, alanine replacement of Leu4 or Asn12 resulted in a significant decreased activity for 21-CSP (27-fold reduction in potency and EC50 >10,000 nM, respectively), whereas the same mutations lead to only a moderate reduction in potency for 18-CSP (9-fold and 5-fold reduction in potency, respectively). A similar, although not as profound, trend was observed for Arg13: this modification in 21-CSP resulted in an analog with decreased potency of 2-fold, while in 18-CSP this modification resulted in an increase of ~2-fold in potency (fold change 0.44). The reduced activity of the 21-CSP analogs implies that the Leu4, Asn12 and Arg13 residues are important for SepM recognition and processing, and thus that the modifications in these positions resulted in the inability of SepM to process the 21-CSP analogs to their active 18-CSP counterparts.
With regards to the 21-CSP C-terminal residues that are being removed by SepM (Leu19, Gly20 and Lys21), alanine mutation in all three positions resulted in analogs with similar activities to the parent 21-CSP, suggesting that the identity of these residues is not critical for SepM recognition and processing (Table 1). These results are in contrast to Biswas et al. who reported that the 21-CSP-L19A analog was not processed by SepM.31 The discrepancies in the results may be attributed to the differences in the assays used to evaluate SepM processing: Our assay included synthetic 21-CSP analogs while Biswas’s assay involved the incorporation of plasmids containing mutated comC genes. Thus, the lack of reporter activity observed by Biswas could be a result of the inability of the ABC transporter to process or export ComC as the mature 21-CSP, rather than the inability of SepM to process 21-CSP to 18-CSP.
D-amino acid scanning of 21-CSP and 18-CSP against ComD
Starting with 18-CSP, D-amino acid modifications at the N-terminal region were found to be more tolerant compared to the central and C-terminal regions (Table 2). This trend implies that the majority of the binding contacts between 18-CSP and ComD are located in the central and C-terminal regions of CSP. Moreover, the stringent stereochemistry requirements suggest that 18-CSP binds to a relatively tight binding pocket in ComD. This observation further explains the importance of removing the last three residues in 21-CSP. That is, these three residues are likely causing steric clashes with the ComD binding pocket and thereby preventing 21-CSP from binding and activating the ComD receptor. It is clear from these results, along with the alanine-scanning of 18-CSP, that the N-terminus of CSP only plays a secondary role in ComD binding.
Table 2.
EC50 values of the D-amino acid scan of 21-CSP and 18-CSP against S. mutans SAB249a
|
|
|
||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Name | EC50 (nM)b | 95% CIc | Fold changed | Name | EC50 (nM)b | 95% CIc | Fold changed |
| 21-CSP | 240 | 200 – 290 | 18-CSP | 6.2 | 3.7 – 10.0 | ||
| 21-CSP-s1 | 140 | 100 – 190 | 0.58 | 18-CSP-s1 | 6.4 | 2.6 – 16 | 1.0 |
| 21-CSP-s3 | 160 | 71 – 370 | 0.67 | 18-CSP-s3 | 9.4 | 4.2 – 21 | 1.5 |
| 21-CSP-l4 | 390 | 160 – 920 | 1.6 | 18-CSP-l4 | 4.5 | 2.3 – 8.7 | 0.72 |
| 21-CSP-s5 | 640 | 410 – 1000 | 2.7 | 18-CSP-s5 | 17 | 6.4 – 45 | 2.7 |
| 21-CSP-t6 | 1600 | 1200 – 2100 | 6.7 | 18-CSP-t6 | 300 | 180 – 510 | 48 |
| 21-CSP-f7 | --e | -- | -- | 18-CSP-f7 | >1,000 | -- | -- |
| 21-CSP-f8 | 840 | 440 – 1600 | 3.5 | 18-CSP-f8 | 42 | 33 – 52 | 6.8 |
| 21-CSP-r9 | 1500 | 860 – 2600 | 6.3 | 18-CSP-r9 | 76 | 32 – 180 | 12 |
| 21-CSP-l10 | 760 | 270 – 2200 | 3.2 | 18-CSP-l10 | 670 | 420 – 1100 | 110 |
| 21-CSP-f11 | --e | -- | -- | 18-CSP-f11 | --e | -- | -- |
| 21-CSP-n12 | --e | -- | -- | 18-CSP-n12 | --e | -- | -- |
| 21-CSP-r13 | 2100 | 1600 – 2700 | 8.8 | 18-CSP-r13 | 480 | 380 – 610 | 77 |
| 21-CSP-s14 | --e | -- | -- | 18-CSP-s14 | >1,000 | -- | -- |
| 21-CSP-f15 | --e | -- | -- | 18-CSP-f15 | --e | -- | -- |
| 21-CSP-t16 | --e | -- | -- | 18-CSP-t16 | >1,000 | -- | -- |
| 21-CSP-q17 | --e | -- | -- | 18-CSP-q17 | 130 | 90 – 200 | 21 |
| 21-CSP-a18 | --e | -- | -- | 18-CSP-a18 | >1,000 | -- | -- |
| 21-CSP-l19 | --e | -- | -- | ||||
| 21-CSP-k21 | 330 | 180 – 610 | 1.4 | ||||
See experimental section for details on the reporter strain and methods. See supporting information for primary screening assay results and plots of agonism dose response curves. All assays performed in triplicate.
EC50 values determined by testing peptides over a range of concentrations.
95% confidence interval.
Ratio where each analog’s EC50 is divided by native CSP EC50; a value <1 indicates a better activator than the parent CSP.
EC50 not determined due to the analog’s low induction in primary agonism screening assay.
Green color indicates modifications that improve processing of 21-CSP by SepM, purple color indicates modifications that decrease processing of 21-CSP by SepM.
Red color indicates modifications critical for 18-CSP:ComD binding.
Moving to 21-CSP, with the exception of Thr6, Leu10 and Arg13, the same trend was observed for 18-CSP and 21-CSP D-amino acid scans, either due to the significantly reduced potencies of the D-amino acid-containing analogs, or as a result of the inability of SepM to effectively process the D-amino acid-containing analogs (Table 2). In the case of Thr6, Leu10 and Arg13, D-amino acid modifications to 21-CSP were relatively tolerable (6.7, 3.2 and 8.8 fold decrease in potency, respectively), while the same modifications in 18-CSP resulted in a significant decrease in potency (48, 110 and 77 fold decrease in potency, respectively). These results imply that inverted chirality in these positions allows for better SepM recognition and processing of 21-CSP. Lastly, when looking at the three C-terminal residues of 21-CSP, chirality change of Leu19 was less tolerable than side chain modification (L19A). Since SepM cleaves between Ala18 and Leu19, it is not surprising that it has a strict orientation requirement for both of these residues in order for it to recognize the cleavage site and be able to process the peptide. To the contrary, the Lys21 position appears to be highly modifiable, as both alanine and D-amino acid substitutions resulted in analogs with similar activities.
Second-generation CSP analogs
To further characterize the activity trends revealed through the alanine and D-amino acid substitutions, and to identify CSP-based QS modulators with enhanced activities, we designed a second-generation library of CSP analogs. First, our analysis suggests that the N-terminal residues of both 21-CSP and 18-CSP are dispensable. Moreover, our analysis showed that the identity of the C-terminal residues of 21-CSP is not important. To assess the importance of the N- and C-terminal residues and identify the minimal sequence required for effective ComD binding, we decided to conduct sequential truncation of both 21-CSP and 18-CSP from both ends. The truncated library consisted of sequential truncation of either the five N-terminal residues of 21-CSP and 18-CSP, or truncation of the C-terminal residues of both native signals (Table 3). Our analysis of the N-terminus revealed that for 18-CSP, the first two residues (Ser1 and Gly2) are dispensable, while for 21-CSP removal of these residues is beneficial for SepM processing (Table 3). With the exception of Ser3 for 21-CSP, removal of additional residues from the N-terminus resulted in a significant decrease in activity for both 18-CSP and 21-CSP, suggesting that these residues are required for effective ComD binding. These results are in agreement with the previous work by Syvitski et al. who also conducted sequential truncation of the 21-CSP scaffold.37 As for the C-terminus, removal of Lys21 was tolerated, while removal of Gly20 resulted in a significant decrease in potency, suggesting that two residues are needed beyond the cleavage site for effective SepM processing (Table 3). C-terminal truncation of 18-CSP resulted in complete loss of activity (Table 3) further emphasizing the importance of the C-terminus in 18-CSP:ComD receptor binding. These results are in contrast to the results published by Syvitski et al. who reported that truncation of three residues from the C-terminus of 21-CSP-S3T to afford 18-CSP-S3T resulted in an analog with inhibitory activity.37
Table 3.
EC50 Values of the 2nd generation Analogs of 21-CSP and 18-CSP against the S. mutans SAB249a
| Name | EC50 (nM)b | 95% CIc | Fold changed | Name | EC50 (nM)b | 95% CIc | Fold changed |
|---|---|---|---|---|---|---|---|
| 21-CSP | 240 | 200 – 290 | 18-CSP | 6.2 | 3.7 – 10.0 | ||
| 21-CSP-des-S1 | 23 | 19 – 28 | 0.10 | 18-CSP-des-S1 | 4.8 | 2.3 – 10.0 | 0.77 |
| 21-CSP-des-S1G2 | 120 | 49 – 310 | 0.50 | 18-CSP-des-S1G2 | 7.9 | 4.1 – 15 | 1.3 |
| 21-CSP-des-S1G2S3 | 530 | 340 – 810 | 2.2 | 18-CSP-des-S1G2S3 | 85 | 46 – 160 | 14 |
| 21-CSP-des-S1G2S3L4 | 3600 | 1500 – 8900 | 15 | 18-CSP-des-S1G2S3L4 | 570 | 270 – 1200 | 92 |
| 21-CSP-des-S1G2S3L4S5 | 3300 | 1600 – 6900 | 14 | 18-CSP-des-S1G2S3L4S5 | 950 | 460 – 2000 | 150 |
| 21-CSP-des-K21 | 98 | 52 – 180 | 0.41 | 18-CSP-des-A18 | --e | -- | -- |
| 21-CSP-des-G20K21 | 3600 | 2100 – 6100 | 15 | 18-CSP-des-Q17A18 | --e | -- | -- |
| 21-CSP-S3T | 94 | 76 – 120 | 0.39 | 18-CSP-S3T | 2.1 | 1.0 – 4.2 | 0.34 |
| 18-CSP-s1s3l4 | 17 | 11 – 26 | 2.7 | ||||
| 18-CSP-s1s3l4L10AR13AS14A | 3.4 | 2.9 – 4.00 | 0.55 | ||||
| 18-CSP-S1AL10AR13AS14A | 1.7 | 0.90 – 3.3 | 0.27 |
See experimental section for details on the reporter strain and methods. See supporting information for primary screening assay results and plots of agonism dose response curves. All assays performed in triplicate.
EC50 values determined by testing peptides over a range of concentrations.
95% confidence interval.
Ratio where each analog’s EC50 is divided by native CSP EC50; a value <1 indicates a better activator than the parent CSP.
EC50 not determined due to the analog’s low induction in primary agonism screening assay.
As mentioned above, Syvitski et al. reported that an 18-CSP analog bearing a Ser3 to Thr mutation was capable of competitively inhibiting the competence regulon in S. mutans.37 In an attempt to develop potent QS inhibitors, we synthesized both the 18-CSP and 21-CSP versions of this proposed mutant (18-CSP-S3T and 21-CSP-S3T) and investigated their activity trend. In our hands, both analogs were found to induce lacZ expression, rather than inhibit it, with slightly higher potencies compared to the native CSPs (Table 3). Although we currently do not have an explanation to the apparent discrepancy, we point out to the fact that Biswas et al. also observed the same discrepancy with the Syvitski results as we did, suggesting that our observed activity trend is accurate.31
Lastly, our alanine scan analysis of 18-CSP revealed that alanine replacements of Ser1, Leu10, Arg13 and Ser14 resulted in analogs with similar activities to the native 18-CSP (Table 1). Similarly, the D-amino acid scan revealed that replacements of Ser1, Ser3 and Leu4 with their enantiomers resulted in analogs with similar activities compared to native 18-CSP (Table 2). We therefore hypothesized that combining these mutations together would result in 18-CSP analogs that maintain the binding affinity to ComD but may not be able to activate the receptor, leading to competitive inhibitors. Thus, we synthesized three multiple mutated 18-CSP analogs (18-CSP-s1s3l4, 18-CSP-s1s3l4L10AR13AS14A and 18-CSP-S1AL10AR13AS14A). Analysis of these three analogs revealed that, as expected, they maintained their binding affinity to ComD (Table 3). However, contrary to our expectations, these analogs were still capable of activating the ComD receptor and thus cannot be used as competitive inhibitors. Moreover, the activity trend of these analogs suggest that multiple alanine mutations are more tolerant than multiple D-amino acid mutations. Importantly, 18-CSP-S1AL10AR13AS14A was found to be 4-fold more active than the native 18-CSP, making it the most potent S. mutans QS activator reported to date.
To assess the differences between early-stage bacteriocin genes (directly under ComE control) and late-stage competence genes (indirectly affected by ComE in complex media), we tested all the 21-CSP and 18-CSP analogs using a second reporter system (pComX:lacZ; SMCOM2 strain). Although the exact EC50 values varied a bit, the overall activity trends observed using the SAB249 strain were also observed for the SMCOM2 strain (Tables S-5 – S-7 and Figures S-13 – S-15). These results reaffirm our initial observations and suggest that once the competence regulon is turned on (ComD is being activated) both early-stage and late-stage genes are being modulated at the same manner.
Structural analysis of 21-CSP and 18-CSP analogs using Circular Dichroism (CD)
In a previous work focused on the competence regulon in S. pneumoniae, we observed a strong correlation between CSP activity and an α-helix conformation.14 Moreover, Syvitski et al. have shown that 21-CSP adopts an α-helix conformation in membrane mimicking conditions.37 Lastly, evaluation of the activity trend of the 18-CSP analogs revealed a periodic sharp reduction in activity every 3–4 residues along the peptide sequence, consistent with an α-helix conformation (Figure 2). We therefore hypothesized that an α-helix conformation is required for effective SepM and ComD binding.
Figure 2.
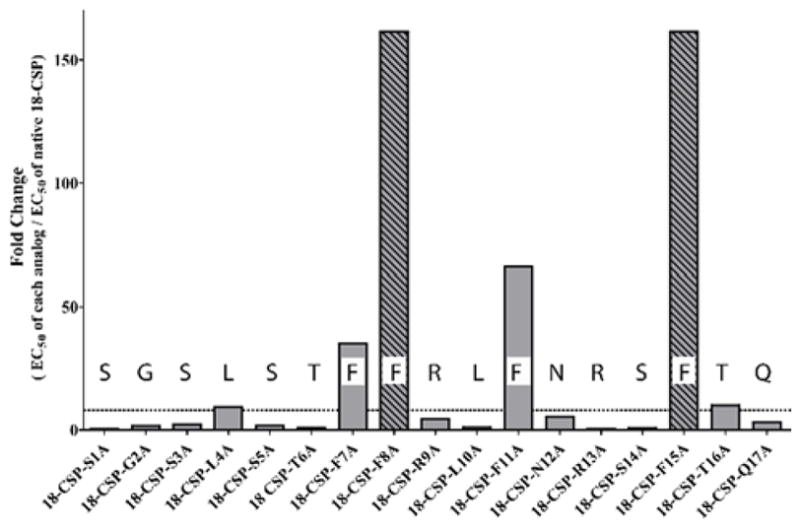
Plot of the ratio of 18-CSP alanine analogs EC50 values divided by the EC50 value of 18-CSP in the SAB249 strain. The plot reveals a periodic sharp reduction in activity every 3–4 residues along the peptide sequence, consistent with an α-helix conformation. Striped bars represent analogs with EC50 values >1,000 nM, thus the EC50 ratio is >160. >8-fold decrease in potency was considered significant (dashed line).
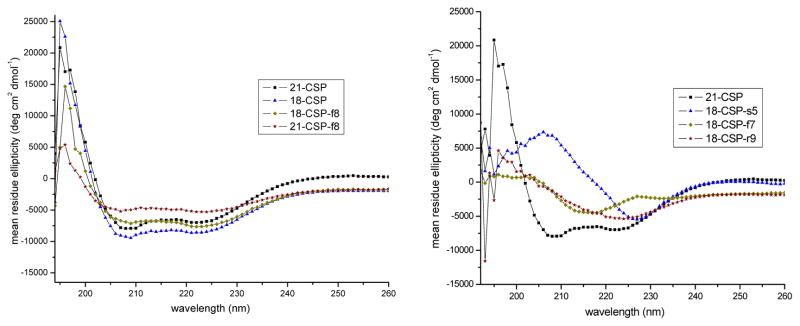
To test our hypothesis and to gain additional structural understanding of the molecular mechanism of 21-CSP:SepM binding and 18-CSP:ComD binding, we evaluated the overall structural motifs of all the CSP analogs using CD spectroscopy. Our analysis included testing all the peptides in both aqueous (PBS buffer, pH 7.4) and membrane mimicking conditions (20% TFE in PBS, pH 7.4). Starting with 21-CSP and 18-CSP, these two peptides were unfolded in aqueous solution (Figures S-16 and S-24) but adopted a helix conformation in membrane mimicking conditions (Figures 3, S-17 and S-25). As expected, most of the 21-CSP and 18-CSP analogs exhibited a random coil pattern in aqueous solution, with a few analogs exhibiting varying degrees of α-helix, β-sheet, or β-turn patterns (Figures S-16, S-18, S-20, S-22, S-24 and S-26). Evaluation of the analogs of both 21-CSP and 18-CSP in membrane mimicking conditions revealed five types of secondary structures: α-helix, distorted α-helix, β-sheet, distorted β-sheet, and β-turn patterns.38 Importantly, all the biologically active peptides maintained either an α-helix or a distorted α-helix secondary structure (Figures S-17, S-19, S-21, S-23, S-25 and S-27), whereas peptides that exhibited β-sheet or β-turn structures were largely inactive (or exhibited significantly reduced potencies; see Tables S-8 – S-15). Interestingly, all the alanine scan and truncated analogs adopted either an α-helix or a distorted α-helix pattern (with the exception of 21-CSP-R9A that exhibited a distorted β-sheet pattern), while D-amino acid mutations at the central regions of both 18-CSP and 21-CSP resulted in either β-sheet (21-CSP-t6, 21-CSP-f7, 21-CSP-r9, 18-CSP-f7 and 18-CSP-r9) or β-turn (21-CSP-l10, 21-CSP-r13 and 18-CSP-s5) patterns (Figures S-19 and S-23). In our previous SAR study of the S. pneumoniae CSPs, D-amino acid substitutions resulted in disruption of β-sheet aggregates and lead to adoption of α-helices,14 while in this study an opposite trend was observed where D-amino acid mutations disrupted the α-helix and resulted in formation of β-sheet or β-turn conformations. Together, these results indicate that D-amino acid substitutions have a stronger effect on peptide conformations than alanine mutations and highlight their potential utility in modifying secondary structures.
Figure 3.
CD spectra of select CSP analogs in membrane mimicking conditions. Both 21-CSP and 18-CSP adopt an α-helix conformation. Similarly, 18-CSP-f8 and 21-CSP-f8 also adopt an α-helix conformation. To the contrary, 18-CSP-s5 adopts a β-turn, while 18-CSP-f7 and 18-CSP-r9 adopt a β-sheet conformation.
Contrary to the other D-amino acid mutations at the central region of 18-CSP, 18-CSP-f8 was found to adopt an α-helix conformation. Since 18-CSP-f8 was one of the few central region analogs that maintained a relatively high potency (Table 2), this observation provides additional support to the hypothesis that an α-helix is required for effective ComD binding. To further evaluate the role of an α-helix conformation in bioactivity, we quantified the percent helicity of all the CSP analogs that exhibited an α-helix pattern using the mean residue ellipticity at 222 nm (Tables S-8–S-10) and the BeStSel method (Tables S-11–S-15). This analysis revealed only a weak correlation between percent helicity and bioactivity, suggesting that the degree of helicity is not as critical as assuming an overall helical conformation. Importantly, the two methods provided similar results, with only a few exceptions. Interestingly, the BeStSel method also revealed a correlation between activity and low percent of parallel β-sheet pattern (Tables S-11–S-15). Overall, our structural analysis of the S. mutans CSP signals highlights the importance of an α-helix pattern for bioactivity and is in line with that of the S. pneumoniae CSPs.14
Summary and Conclusions
In conclusion, S. mutans uses the CSP-mediated QS circuit to outcompete other bacterial species, establish and sustain robust infections and thrive in the oral cavity. Studying the molecular mechanism that drives this QS circuitry can provide fundamental understanding of this important pathogen and its behavior, and may lead to the development of therapeutic agents capable of attenuating S. mutans pathogenicity. In the current study, we report the most in-depth analysis of the SARs of S. mutans 21-CSP and 18-CSP and their interactions with SepM and the ComD receptor. Specifically, our analysis revealed specific residues in 21-CSP that can be modified to improve SepM processing (Gly2, Ser3, Thr6, Leu10, Arg13 and Gln17), as well as residues that are important for effective SepM recognition and processing (Leu4 and Asn12) (Figure 4).
Figure 4.
Minimal structural requirements for 21-CSP processing by SepM and 18-CSP binding to the ComD receptor.
Moreover, our structural analysis of the CSP analogs revealed a positive correlation between helicity and bioactivity and a negative correlation between bioactivity and parallel β-sheet pattern. Furthermore, our analysis highlighted the ability of single D-amino acid substitutions to modify the entire secondary structure of peptides and suggest that this type of modification should be applied when trying to identify and fine-tune structural features.
Lastly, our comprehensive SAR analysis of 18-CSP highlighted the importance of the hydrophobic residues located at the central region in ComD binding (Figure 4). Our analysis further uncovered that the C-terminus region of 18-CSP is vital for activity (removal of one amino acid from that region resulted in complete loss of activity) while the N-terminus region is dispensable (the first two residues can be removed without affecting activity). Combined, our analysis revealed the minimal sequence required for effective ComD binding and activation (S-L-S-T-F-F-R-A-F-N-A-A-F-T-Q-A). This minimal structure can be used as a template to design peptidomimetic QS modulators with enhanced pharmacological properties that could be applied to attenuate S. mutans infectivity. Such studies are ongoing in our lab and will be reported in due course.
Materials and methods
Chemical Reagents and Instrumentation
All chemical reagents and solvents were purchased from Sigma-Aldrich and used without further purification. Water (18 MΩ) was purified using a Millipore Analyzer Feed System. Solid-phase resins were purchased from Advanced ChemTech and Chem-Impex International.
Reversed-phase high-performance liquid chromatography (RP-HPLC) was performed using a Shimadzu system equipped with a CBM-20A communications bus module, two LC-20AT pumps, an SIL-20A auto sampler, an SPD-20A UV/vis detector, a CTO-20A column oven, and an FRC-10A fraction collector. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) data were obtained on a Bruker Microflex spectrometer equipped with a 60 Hz nitrogen laser and a reflectron. In positive ion mode, the acceleration voltage on Ion Source 1 was 19.01 kV. Exact mass (EM) data were obtained on an Agilent Technologies 6230 TOF LC/MS spectrometer. The samples were sprayed with a capillary voltage of 3500 V, and the electrospray ionization (ESI) source parameters were as follows: gas temperature of 325 °C at a drying gas flow rate of 8 L/ min at a pressure of 35 psi.
Solid Phase Peptide Synthesis
All the CSP analogs were synthesized using standard Fmoc-based solid-phase peptide synthesis (SPPS) procedures on 4-benzyloxybenzyl alcohol (Wang) resin. Preloaded Fmoc-L-Lys(Boc) Wang resin (0.59 mmol/g) was used for peptides that required a lysine at the C-terminus, and preloaded Fmoc-L-Ala Wang resin (0.8 mmol/g) was used for peptides that required an alanine at the C-terminus. For peptides that have amino acids other than L-lysine or L-alanine at the C-terminus, loading of the first amino acid to the Wang resin linker was done by using the symmetrical anhydride procedure as previously described (for the full procedure, see supporting information).39
Peptide Purification
Crude peptides were purified with RP-HPLC. A semipreparative Phenomenex Kinetex C18 column (5 μm, 10 mm × 250 mm, 110 Å) was used for preparative RP-HPLC work, while an analytical Phenomenex Kinetex C18 column (5 μm, 4.6 mm ×250 mm, 110 Å) was used for analytical RP-HPLC work. Standard RP-HPLC conditions were as follows: flow rates = 5 mL min−1 for semipreparative separations and 1 mL min−1 for analytical separations; mobile phase A = 18 MΩ water + 0.1% TFA; mobile phase B = ACN + 0.1% TFA. Purities were determined by integration of peaks with UV detection at 220 nm. Preparative HPLC methods were used to separate the crude peptide mixture to different chemical components using a linear gradient (first prep 5% B → 45% B over 40 min and second prep 20% B → 30% B over 30 min). Then, an analytical HPLC method was used to quantify the purity of the desired product using a linear gradient (5% B → 95% B over 27 min). Only peptide fractions that were purified to homogeneity (>95%) were used for the biological assays. TOF-MS was used to validate the presence of synthesized peptides. The observed mass-to-charge (m/z) ratio of the peptide was compared to the expected m/z ratio for each peptide (see Tables S-1–S-4).
Biological Reagents and Strain Information
All standard biological reagents were purchased from Sigma-Aldrich and used according to enclosed instructions.
To examine the ability of the synthetic CSP analogs to modulate the ComD receptor, and thus the competence QS circuit in S. mutans, beta-galactosidase assays were performed using two reporter strains: a S. mutans reporter strain harboring a pCipB::lacZ construct integrated to its chromosome (SAB249; representing the expression of early-stage bacteriocin genes),40 and a ΔcomC S. mutans reporter strain harboring a pcomX::lacZ plasmid (SMCOM2; representing the expression of late-stage competence genes).41
Bacterial Growth Conditions
Freezer stocks were created from 1.5 mL aliquots of overnight cultures in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) and 0.5 mL glycerol and stored at −80 °C. For experiments, bacteria from the freezer stocks were streaked onto a THY agar plate containing 250 μg/mL kanamycin (SAB249) or 5 μg/mL erythromycin (SMCOM2). The plate was incubated for 22–24 h in a CO2 incubator (37 °C with 5% CO2). A fresh single colony was transferred to 5 mL of THY broth supplemented with a final concentration of 250 μg/mL kanamycin (SAB249) or 5 μg/mL erythromycin (SMCOM2) and the culture was incubated in a CO2 incubator overnight (15 h). Overnight culture was then diluted (1:100 for SAB249 or 1:25 for SMCOM2) with THY, and the resulting solution was incubated in a CO2 incubator for 3h (SAB249) or 2 h (SMCOM2), until the bacteria reached early exponential stage (0.18–0.2 OD 600nm for SAB249 or 0.20–0.25 OD 600nm for SMCOM2) as determined by using a plate reader.
Beta-Galactosidase Activation Assays
The ability of synthetic CSP analogs to activate the expression of cipB or comX was determined using the corresponding reporter strains grown in THY (pH 7.3). An initial activation screening was performed at a high concentration (10 μM) for all CSP analogs. A total of 2 μL of 1 mM solution of CSP analogs in dimethyl sulfoxide (DMSO) was added in triplicate to a clear 96-well microtiter plate. This concentration was chosen to afford full activation of the QS circuit, as determined from the dose-dependent curves created for the native 21-CSP. A total of 2 μL of DMSO was added in triplicate and served as the negative control. Then, 198 μL of bacterial culture was added to each well containing CSP and analogs. The plate was incubated at 37 °C for 1 h (SAB249) or 2 h (SMCOM2), and the OD 600nm was measured. In order to measure the beta-galactosidase activity in the culture, the cells were lysed by incubating the culture for 30 min at 37 °C with 20 μL of 0.1% Triton X-100. In a new plate, 100 μL of Z-buffer solution (60.2 mM Na2HPO4, 45.8 mM NaH2PO4, 10 mM KCl, and 1.0 mM MgSO4 in 18 MΩ H2O; pH was adjusted to 7.0, and the buffer was sterilized before use) containing 2-nitrophenyl-beta-D-galactopyranoside (ONPG) at a final concentration of 0.4 mg mL−1 was added, followed by 100 μL of lysate, and the plate was incubated for 30 min at 37 °C. The reaction was stopped by adding 50 μL of 1 M sodium carbonate solution, and the OD 420nm and OD 550nm were measured using a plate reader. The final results were reported as percent activation, which is the ratio between the Miller units of the analog and of the positive control. For calculation of Miller units, please see data analysis section below. Analogs that exhibited high activity in the initial screening (>75% activation compared to the native signal) (see Figures S-1–S-5 and S-7–S-11) were further evaluated using a dose-dependent assay in which peptide stock solutions were diluted with DMSO in serial dilutions (either 1:2, 1:3, or 1:5) and assayed as described above. GraphPad Prism 5 was used to calculate the EC50 values, which are the concentration of a drug that gives half-maximal response.
Beta-Galactosidase Inhibition Assays
Analogs that exhibited low cipB or comX activation in the initial screening (see Figures S-1–S-5 and S-7–S-11) were evaluated for competitive inhibition (see Figure S-6 and S-12). The ability of synthetic CSP analogs to inhibit the expression of either cipB or comX by outcompeting 21-CSP for the receptor binding site was evaluated using the same assay conditions as described above, except that in the initial inhibition screening, the native CSP was added to every well in a set concentration (1000 nM 21-CSP for SAB249 and 350 nM 21-CSP for SMCOM2) that was chosen to afford full activation of the QS circuit, as determined from the dose-dependent curves created for the native 21-CSP. A total of 2 μL of native 21-CSP and 2 μL of 1 mM solution of CSP analogs were added to the same well in triplicate in a clear 96-well microtiter plate. A total of 2 μL of native 21-CSP and 2 μL of DMSO were added to the same well in triplicate and served as the positive control. A total of 4 μL of DMSO was added in triplicate and served as the negative control. Then, 196 μL of bacterial culture was added to the wells, and the plate was incubated at 37 °C for 1 h (SAB249) or 2 h (SMCOM2). The procedure for lysis, incubation with ONPG, and all the measurements were as described in the activation assay.
Analysis of Activation/Inhibition Data
Miller units were calculated using equation 1:
| Equation 1 |
Abs420 is the absorbance of o-nitrophenol (ONP). Abs550 is the scatter from cell debris, which, when multiplied by 1.75, approximates the scatter observed at 420 nm. t is the duration of incubation with ONPG in minutes, v is volume of lysate in milliliters, and Abs600 reflects cell density.
Circular Dichroism (CD) Spectroscopy
CD spectra were recorded with an Aviv Biomedical CD spectrometer (model 202–01). All the measurements were performed with a peptide concentration of 200 μM in PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4; pH was adjusted to 7.4) with 0% or 20% trifluoroethanol (TFE). Measurements were performed at 25 °C with a quartz cuvette (science outlet) with a path length of 0.1 cm. Samples were scanned one time at 3 nm min−1 with a bandwidth of 1 nm and a response time of 20 s over a wavelength range 190 to 260 nm. The spectra were analyzed using two methods:
Percent helicity (fH) was calculated for peptides that exhibited a significant helical pattern using equation 2:
| Equation 2 |
[θ]222 is the mean residue ellipticity of the sample peptide at 222 nm, [θ∞]222 is the mean residue ellipticity of an ideal peptide with 100% helicity (−44 000 deg cm2 dmol−1),42 n is the number of residues in the potential helical region, and x is an empirical correction for end effects (2.5).42 Secondary structure contents were also calculated using the BeStSel (Beta Structure Selection) method (http://bestsel.elte.hu/).38
Supplementary Material
Acknowledgments
This work was supported by the Nevada INBRE through a grant from the National Institute of General Medical Sciences (GM103440) from the National Institutes of Health, and the National Science Foundation (CHE-1808370). The S. mutans SAB249 (pCipB::lacZ) reporter strain was generous gift from R. A. Burne (University of Florida). The S. mutans SMCOM2 (ΔcomC, pcomX::lacZ) reporter strain was generous gift from D. G. Cvitkovitch (University of Toronto). We would also like to thank M. J. Tucker (University of Nevada, Reno) for the use of the CD spectrometer.
Footnotes
Full details of peptide synthesis and characterization, initial screening results, dose response curves for CSP analogs, and CD spectra of all the CSP analogs. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 3.Bassler BL. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 4.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Bodman SB, Willey JM, Diggle SP. Cell-cell communication in bacteria: united we stand. J Bacteriol. 2008;190:4377–4391. doi: 10.1128/JB.00486-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welsh MA, Blackwell HE. Chemical probes of quorum sensing: from compound development to biological discovery. FEMS Microbiol Rev. 2016;40:774–794. doi: 10.1093/femsre/fuw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2010;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsek MR, Greenberg E. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Palmer AG, Senechal AC, Mukherjee A, Ané JM, Blackwell HE. Plant responses to bacterial N-acyl L-homoserine lactones are dependent on enzymatic degradation to L-homoserine. ACS Chem Biol. 2014;9:1834–1845. doi: 10.1021/cb500191a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Muir TW. Regulation of virulence in Staphylococcus aureus: molecular mechanisms and remaining puzzles. Cell Chem Biol. 2016;23:214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relationships. Infect Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu L, Lin J, Kuang Z, Vidal JE, Lau GW. Deletion analysis of Streptococcus pneumoniae late competence genes distinguishes virulence determinants that are dependent or independent of competence induction. Mol Microbiol. 2015;97:151–165. doi: 10.1111/mmi.13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Koirala B, Sanchez LA, Phillips NR, Hamry SR, Tal-Gan Y. Structure–Activity Relationships of the Competence Stimulating Peptides (CSPs) in Streptococcus pneumoniae Reveal Motifs Critical for Intra-group and Cross-group ComD Receptor Activation. ACS Chem Biol. 2017;12:1141–1151. doi: 10.1021/acschembio.7b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J Am Chem Soc. 2013;135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- 16.Tal-Gan Y, Ivancic M, Cornilescu G, Yang T, Blackwell HE. Highly Stable, Amide-Bridged Autoinducing Peptide Analogues that Strongly Inhibit the AgrC Quorum Sensing Receptor in Staphylococcus aureus. Angew Chem Int Ed Engl. 2016;55:8913–8917. doi: 10.1002/anie.201602974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci U S A. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon CP, Williams P, Chan WC. Attenuating Staphylococcus aureus virulence gene regulation: a medicinal chemistry perspective. J Med Chem. 2013;56:1389–1404. doi: 10.1021/jm3014635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng WL, Perez L, Cong J, Semmelhack MF, Bassler BL. Broad spectrum pro-quorum-sensing molecules as inhibitors of virulence in vibrios. PLoS Pathog. 2012;8:e1002767. doi: 10.1371/journal.ppat.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama J, Yokohata R, Sato M, Suzuki T, Matsufuji T, Nishiguchi K, Kawai T, Yamanaka Y, Nagata K, Tanokura M, Sonomoto K. Development of a peptide antagonist against fsr quorum sensing of Enterococcus faecalis. ACS Chem Biol. 2013;8:804–811. doi: 10.1021/cb300717f. [DOI] [PubMed] [Google Scholar]
- 21.McBrayer DN, Gantman BK, Cameron CD, Tal-Gan Y. An Entirely Solid Phase Peptide Synthesis-Based Strategy for Synthesis of Gelatinase Biosynthesis-Activating Pheromone (GBAP) Analogue Libraries: Investigating the Structure-Activity Relationships of the Enterococcus faecalis Quorum Sensing Signal. Org Lett. 2017;19:3295–3298. doi: 10.1021/acs.orglett.7b01444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amara N, Gregor R, Rayo J, Dandela R, Daniel E, Liubin N, Willems HM, Ben-Zvi A, Krom BP, Meijler MM. Fine-Tuning Covalent Inhibition of Bacterial Quorum Sensing. Chembiochem. 2016;17:825–835. doi: 10.1002/cbic.201500676. [DOI] [PubMed] [Google Scholar]
- 23.Moore JD, Rossi FM, Welsh MA, Nyffeler KE, Blackwell HE. A Comparative Analysis of Synthetic Quorum Sensing Modulators in Pseudomonas aeruginosa: New Insights into Mechanism, Active Efflux Susceptibility, Phenotypic Response, and Next-Generation Ligand Design. J Am Chem Soc. 2015;137:14626–14639. doi: 10.1021/jacs.5b06728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welsh MA, Eibergen NR, Moore JD, Blackwell HE. Small molecule disruption of quorum sensing cross-regulation in pseudomonas aeruginosa causes major and unexpected alterations to virulence phenotypes. J Am Chem Soc. 2015;137:1510–1519. doi: 10.1021/ja5110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci U S A. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 27.Swem LR, Swem DL, O’Loughlin CT, Gatmaitan R, Zhao B, Ulrich SM, Bassler BL. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol Cell. 2009;35:143–153. doi: 10.1016/j.molcel.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pepperney A, Chikindas ML. Antibacterial peptides: opportunities for the prevention and treatment of dental caries. Probiotics Antimicro Prot. 2011;3:68. doi: 10.1007/s12602-011-9076-5. [DOI] [PubMed] [Google Scholar]
- 30.Hossain MS, Biswas I. An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J Bacteriol. 2012;194:5886–5896. doi: 10.1128/JB.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biswas S, Cao L, Kim A, Biswas I. SepM, a streptococcal protease involved in quorum sensing, displays strict substrate specificity. J Bacteriol. 2016;198:436–447. doi: 10.1128/JB.00708-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reck M, Tomasch J, Wagner-Döbler I. The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet. 2015;11:e1005353. doi: 10.1371/journal.pgen.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol. 2012;86:258–272. doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan R, Rukke HV, Høvik H, Åmdal HA, Chen T, Morrison DA, Petersen FC. Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core Streptococcal competence genes. mSystems. 2016;1:e00038–00015. doi: 10.1128/mSystems.00038-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagen SJ, Son M. Origins of heterogeneity in Streptococcus mutans competence: interpreting an environment-sensitive signaling pathway. Phys Biol. 2017;14:015001. doi: 10.1088/1478-3975/aa546c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Syvitski RT, Tian X-L, Sampara K, Salman A, Lee SF, Jakeman DL, Li Y-H. Structure-activity analysis of quorum-sensing signaling peptides from Streptococcus mutans. J Bacteriol. 2007;189:1441–1450. doi: 10.1128/JB.00832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Micsonai A, Wien F, Kernya L, Lee YH, Goto Y, Réfrégiers M, Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci U S A. 2015;112:E3095–3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan WC, White PD. Fmoc solid phase peptide synthesis. Oxford University Press; 2000. [Google Scholar]
- 40.Huang X, Palmer SR, Ahn SJ, Richards VP, Williams ML, Nascimento MM, Burne RA. A Highly Arginolytic Streptococcus Species That Potently Antagonizes Streptococcus mutans. Appl Environ Microbiol. 2016;82:2187–2201. doi: 10.1128/AEM.03887-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aspiras MB, Ellen RP, Cvitkovitch DG. ComX activity of Streptococcus mutans growing in biofilms. FEMS Microbiol Lett. 2004;238:167–174. doi: 10.1016/j.femsle.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 42.Luo P, Baldwin RL. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.