Abstract
Current research efforts on neurological diseases are focused on identifying novel disease biomarkers to aid in diagnosis, provide accurate prognostic information and monitor disease progression. With advances in detection and quantification methods in genomics, proteomics and metabolomics, saliva has emerged as a good source of samples for detection of disease biomarkers. Obtaining a sample of saliva offers multiple advantages over the currently tested biological fluids as it is a non-invasive, painless and simple procedure that does not require expert training or harbour undesirable side effects for the patients. Here, we review the existing literature on salivary biomarkers and examine their validity in diagnosing and monitoring neurodegenerative and neuropsychiatric disorders such as autism and Alzheimer's, Parkinson's and Huntington's disease. Based on the available research, amyloid beta peptide, tau protein, lactoferrin, alpha-synuclein, DJ-1 protein, chromogranin A, huntingtin protein, DNA methylation disruptions, and micro-RNA profiles display a reliable degree of consistency and validity as disease biomarkers.
Keywords: Neurological diseases, Salivary biomarkers, Diagnosis, Dementia, Neurodegeneration
Neurological disorders are a family of diseases affecting both the central and peripheral nervous system, ranging from neurodegenerative to neurodevelopmental and psychiatric. Many of these diseases are multifaceted and there is no consensus about their actual cause, although many studies suggest the involvement of multiple combined factors [1], [2]. One aspect of neurological disorders is neurodegeneration, which is caused by a progressive loss of certain classes of neurons that affect either motor skill or memory and cognition. The degeneration of these neurons is usually due to several molecular mechanisms that promote cell death including excitotoxicity, mitochondrial dysfunction, and intracellular inclusions or extracellular aggregation of toxic molecules [3]. Early disease detection is critical in assigning proper treatment therapy to affected patients [4]. Yet diagnosis remains a challenge concerning diseases affecting the central nervous system (CNS) due to multiple delays in the diagnostic procedure and treatment initiation, which ultimately diminishes treatment effectiveness. Usually the tests performed for the diagnosis of neurological conditions are either blood tests or lumbar puncture. Their invasive nature, especially for the lumbar puncture, usually results in discomfort, pain and disagreeable side effects for patients, which necessitates the search for accurate, more advanced and less invasive testing methods [5]. Recent investigations have begun to examine the possible use of urine as a less invasive source of biomarkers, but have not yet been put into clinical use pending further assessment [6], [7], [8], [9].
Saliva is a physiological fluid composed of mucous and serous secretions containing mucin, alpha-amylase and different ions [10]. It fulfils a range of functions including: digestion of nutrients and protection of teeth and oral tissues; through enzymatic action, lubrication and antibacterial properties [11]. Saliva is secreted in the mouth by the principal salivary glands: the sublingual, submandibular and the parotid, which are under direct parasympathetic innervation of the cranial nerves VII (glossopharyngeal) and IX (facial). The facial nerve innervates both the sublingual and submandibular glands through the submandibular ganglion, while the parotid gland is under glossopharyngeal innervation via the otic ganglion [12]. Therefore this close relation between the salivary glands and the nervous system could render these glands' secretions as a useful pool of biomarkers that represent various normal and pathological physiologies of the nervous system [13]. Saliva offers a new and easily accessible physiological fluid that can be collected in a non-invasive manner and assessed using different analytical assays [14]. Although many diseases have confirmed salivary biomarkers [15]; diseases affecting the nervous system have few markers available in saliva which are still being investigated. This narrative review is concerned with the salivary biomarkers of the most known neurological diseases i.e. Alzheimer's, Parkinson's Huntington's, Amyotrophic lateral sclerosis, Multiple sclerosis, Autism spectrum disorders and finally neuropsychiatric disorders, to establish a general idea about the advances made in this field in hopes of providing guidelines for the development of methods to monitor and assess nervous system health using salivary biomarkers.
Alzheimer's disease
Alzheimer's disease (AD) is a chronic neurodegenerative disorder of the CNS. It is mainly manifested by dementia, confusion and cognitive impairment due to the loss of neurons in the hippocampus, basal forebrain and other cortical areas of the brain [16], [17], [18]. AD etiology can be attributed to two molecules, the amyloid beta peptide and tau protein. Amyloid beta peptide is proposed to have normal physiological roles in memory formation, lipid homeostasis, regulation of neuron activity and neurite growth [19], [20], [21], [22], [23], [24]; on the other hand, tau protein is a member of the microtubule associated proteins that maintain proper neuronal structure and intracellular transport [25], [26], [27]. Studies on AD pathophysiology suggest that extracellular accumulation of amyloid beta peptides (Aβ) in amyloid plaques and intracellular tau protein neurofibrillary tangles are the major factors that contribute to neuron cell death [28], [29], [30], [31], [32], [33]. Based on different research efforts, the cause of neurodegeneration cannot be attributed to only one of the aforementioned key players in disease pathophysiology rather than a combinatory effect of both amyloid beta and tau protein pathologies on neuronal cell death.
Amyloid beta peptides
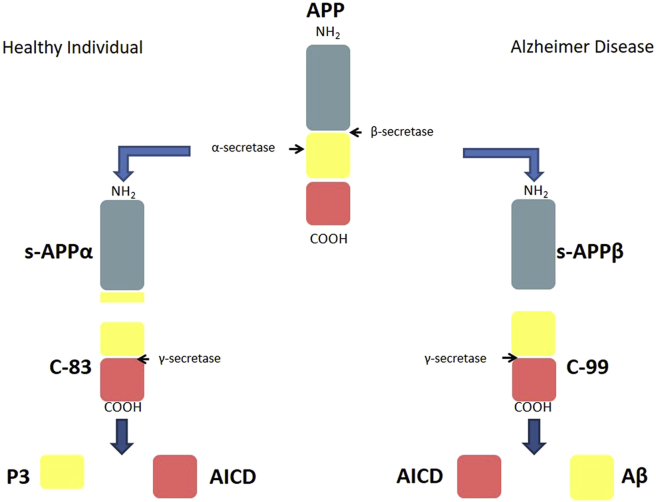
Using animal models and neural cell lines, functions of the soluble APP alpha isoform have been described which encompass neurogenesis, neurite outgrowth, neurite guidance, axonal transport, learning, memory formation and synaptogenesis [34], [35] [Table 1]. In non-pathologic conditions, amyloid precursor protein is cleaved first by α-secretase a member of the ADAM family followed by a subsequent cleavage by γ-secretase. These two proteases are involved in the normal processing and recycling of transmembrane amyloid precursor protein. Normally, APP processing by α-secretase cuts within the amyloid beta sequence and results in the formation of three main peptides: soluble APP alpha (s-APPα), which is released into the extracellular space, and an 83 AA fragment (C-83) embedded in the membrane. C83 is finally cleaved by γ-secretase releasing the p3 peptide and the amyloid precursor protein intracellular domain (AICD) [36], [37] [Table 1] [Fig. 1].
Table 1.
Normal physiological and neurotoxic functions of the derivatives of APP processing. The following table lists the functions that are known to be performed by the derivatives of APP processing. It is worth noting that sAPPβ and p3 are missing due to lack of information regarding the function of these two derivatives.
| sAPPα |
Aβ |
AICD |
|
|---|---|---|---|
| Normal physiological function | Normal physiological function | Neurotoxicity | Neurotoxicity |
Fig. 1.
The normal and AD pathogenic pathway of APP processing into its multiple derivatives. In a healthy individual the primary processing of APP by α-secretase within the Aβ sequence produces the s-APPα which is released to the extracellular domain. This cleavage of the Aβ domain inactivates its amyloidogenic potential. The cleavage of the produced C-83 peptide by γ-secretase produces the P3 peptide which is released to the extracellular space and the AICD which remains inside the cell. The difference between the normal APP processing pathway and that of AD is the initial cleavage of APP by β-secretase in AD, producing an s-APPβ and conserving the Aβ sequence. Aβ is then released after C-99 processing by γ-secretase. Once secreted to the extracellular space, Aβ is free to aggregate and form amyloid plaques.
In AD, α-secretase is substituted by β-secretase known as BACE-1 which does not cleave APP within the beta amyloid structure, thus maintaining its integrity and function. This altered sequence in enzymatic cleavage results in the production of (i) a soluble amyloid precursor protein beta (s-APPβ), and (ii) a 99 AA fragment (C-99) that is subsequently cleaved by γ-secretase to produce a 38–43 amino acid residue amyloid-β peptide and the intracellular AICD domain [38], [39]. The amyloid beta 42 (Aβ42) isoform which is known to be more amyloidogenic and neurotoxic than the other isoforms, accumulates into plaques causing adverse cytopathic effects [40], [41], [42]. This cleavage can also produce the Aβ40 isoform, which has been found to enhance and stabilize Aβ42 oligomer. The synergistic effects of the two Aβ isoforms instigate severe neurite damage along with neuronal toxicity [43], [44], [45], [46] [Fig. 1] [Table 1].
Applying the enzyme-linked immunosorbent assay (ELISA) to saliva samples, Bermejo Pareja et al. [47] compared amyloid beta 40 and 42 levels between AD patients and two sets of controls: healthy controls and Parkinson patients. It was evident that Aβ42 levels increased in patients suffering from mild and moderate AD, but Aβ42 levels were comparable to the healthy controls in severe AD. This reported variation in Aβ42 concentrations with disease progression, mimics previously established CSF findings [48], [49], [50]. Aβ40 levels remained unchanged using this technique for all participants. A single study reported the detection of Aβ42 as a urinary biomarker for both AD diagnosis and monitoring, with patients suffering from severe dementia exhibiting a complete absence of Aβ42 [9].
In contrast to these findings, Kim et al. [51] used antibody-based magnetic nanoparticle immunoassay to show that Aβ42 secretion increases with disease progression from mild cognitive impairment to severe AD; an increase not evidenced in CSF, but in the brain as deposition of intracerebral Aβ42 in amyloid plaques [49]. Aβ40 levels also exhibited an increasing trend similar to that of Aβ42 as opposed to being unchanged in the previous study but remained without statistical significance. Kim et al. have also conducted an ELISA on the saliva samples and the results were in accordance with their findings using the immunoassay. There exist two scenarios that might explain the variability of Aβ42 concentration between the two studies. The first being that, excessive loss of intracerebral neurons in advanced AD contributes to the decrease of released Aβ42 which explains the results obtained by Bermejo-Pareja et al. [47] The second being that, AD induced reduction in submandibular salivary flow might contribute to the increase of concentration of Aβ42 due to decreased sample volume which explains the results obtained by Kim et al. [47], [51]. Yet there is still no consensus or precise explanation for the observed contradiction.
Recent work performed by Lee et al. [52] proved that stabilizing salivary levels of Aβ42 peptides by adding thioflavin S to prevent its aggregation, and inhibiting bacterial growth by using sodium azide greatly enhanced the results obtained by ELISA. These compounds act as preservatives and prevent sample degradation and thus enhance sample quality and Aβ42 detection. Aβ42 detection by using this method accurately differentiated between controls and individuals at risk or affected by AD. The concentration of Aβ42 in healthy controls was around 20 pg/ml and in individuals affected by or at risk of developing AD was above 40 pg/ml. Lee et al. did not report any differences in Aβ42 concentrations for different disease stages (mild-moderate-severe) nor did they attempt the detection of Aβ40 with their method. To test the efficiency of the methods used by Bermejo Pareja et al., Lee et al. performed an ELISA using the same Invitrogen kit used by Bermejo Pareja et al. and found that it was only capable of detecting 25% of Aβ42 in the sample when compared to their method.
Using Parkinson patients as controls is essential to prove the validity and exclusive nature of this diagnostic tool in identifying amyloid beta exclusively in AD patients. Results had shown that controls suffering from Parkinson's disease studied by both Bermejo Pareja et al. and Lee et al. showed no differences in Aβ42 concentrations in comparison to healthy controls and thus proving the specificity of salivary Aβ42 as a salivary biomarker for AD patients. This comes as sound evidence with the findings of amyloid beta pathology in PD patients [53], [54], [55].
Molecular findings have shown that multiple genes have been identified as key players in AD diagnosis and prognosis. The four main genetic mutations in AD target PSEN1, PSEN2, ADAM10 and APOE. PSEN1 and PSEN2 are both subunits of the γ-secretase complex and have been associated with the familial cases of AD [56]. ADAM10 is an α-secretase and APOE4 is a member of the apolipoprotein family which plays a role in lipid metabolism and is speculated to intervene in amyloid beta plaque formation. Both ADAM 10 and APOE4 mutations have been associated with late onset AD [57], [58], [59].
Lee et al. [52] reported three non-AD subjects who showed Aβ42 concentrations higher than other controls two of whom had an extensive family history of AD and one with a PSEN1 mutation. Complimentary findings were provided by Bermejo Pareja et al. [47] who reported that Aβ42 concentrations in the saliva of AD are independent of APOE4 genotype which is linked to late onset AD. Given the above presented data about the implication of PSEN1 mutations in familial AD, it could be concluded that salivary Aβ42 is probably more reflective of familial AD genotype rather than sporadic AD.
Aβ40 concentrations did not show an identical uniform variation between the two stated studies. Aβ40 levels showed no change between the saliva of patients and of controls with the ELISA as shown by Lee et al. [52] and Bermejo-Pareja et al. [47]. On the other hand Aβ40 levels followed an increasing pattern in AD patient saliva that did not reach significance as demonstrated by Kim et al. [51] using the immunoassay.
Tau protein
Tau protein, known as the Microtubule Associated Protein T (MAPT), is a member of the microtubule associated protein family which plays a role in microtubule stabilization and flexibility by binding to tubulin [26], [27]. Normally, tau phosphorylation promotes its disassembly from microtubules and initiates its destabilization and elimination [25], [60]. Mutations in the tau protein sequence altering its phosphorylation site have been reported to induce tau hyper phosphorylation [61]. Phosphorylated tau (p-tau) can aggregate and form intracellular neurofibrillary tangles and work synergistically with Aβ to enhance its cytotoxic functions [28], [61], [62]. Following Mass Spectrometry analysis of patient saliva, Min Shi et al. [13] were the only group who examined p-tau as a salivary biomarker for AD. Analysis of tau protein species, which are the phosphorylated tau (p-tau) and the total tau (t-tau), conveyed that the ratio p-tau/t-tau per individual exhibited a significant increase in affected individuals as compared to healthy controls. This indicates that the increase of phosphorylated tau with respect to the total tau concentration could be used as a potential biomarker for assessment of AD status. It is worth noting that both CSF p-tau and t-tau exhibit an increase in AD patients [63].
Acetylcholinesterase activity
Acetylcholinesterase inhibitors (AChE-I) are the primary medications for AD symptom management and disease control [64], [65]. Acetylcholinesterase is the enzyme required to degrade Acetylcholine (AChE) neurotransmitter released into the synaptic cleft to halt its postsynaptic effects. The Nucleus Basalis of Meynert is a major cholinergic nucleus of the basal forebrain which is a principal target of neurodegeneration in AD [66]. AChE has been demonstrated to accumulate in amyloid plaques and neurofibrillary tangles of AD brains; furthermore many findings have implicated AChE in Aβ pathology and enhancing neurotoxicity [67], [68], [69]. Assessment of AChE catalytic activity in AD patients by conventional methods such as positron emission tomography showed a significant decrease in this activity in the affected brain regions [70], [71]. Sayer et al. [72] were the first to report AChE salivary activity by using the Ellman colorimetric method. They reported that, AChE activity decreases with age among healthy individuals, and was significantly decreased in patients with AD who did not respond to AChE-I treatment. While those who respond to AChE-I treatment did not show any variation when compared to controls.
Further studies conducted by Bakhtiari et al. [73] and Boston et al. [74] also using the Ellman colorimetric method, found a decreasing trend in AChE catalytic activity in AD patients as compared to controls but these results did not reach significance. They both attributed the decrease in catalytic activity observed by Sayer et al. to long term adaptive changes in the production of AChE because of the administered AD medication.
Lactoferrin
Current research continues to supply evidence implicating the immune system as a major player in the course of AD [75], [76]. Recent work presented by Carro et al. names lactoferrin as an impressive new candidate to be one of the first salivary biomarkers for AD early detection and diagnosis [77]. Lactoferrin is an antimicrobial peptide which targets bacteria, viruses, fungi, yeasts and protozoa with a known Aβ-binding ability. It functions in the modulation of immune reactions and inflammation [78], [79], [80], [81], [82], [83], [84], [85], [86], [87]. The fact that there exists concomitant evidence implicating systemic and brain infections with AD [88], [89], [90], [91], further emphasizes the validity of lactoferrin as a probable biomarker for AD. Using Mass spectrometry and ELISA, Carro et al. [77] demonstrated that salivary lactoferrin concentration was significantly reduced in AD patients when compared to healthy controls, and controls suffering from PD. They were also able to prove the value of lactoferrin in early disease detection as an early disease biomarker. 14 out of 18 controls who presented with reduced salivary lactoferrin concentrations comparable to AD associated concentrations developed either mild cognitive impairment or AD over the course of the study; while, none of the controls who presented with normal or high lactoferrin salivary concentrations developed any form of cognitive impairment. Finally, to establish the validity of this marker and prove that it is truly representative of AD pathology, significant correlations were established between lactoferrin levels in saliva and APOE4 allele status, Mini Mental State Examination (MMSE) score, CSF Aβ42 and CSF total tau.
Protein carbonyl levels
There is much evidence concerning oxidative stress being implicated in AD, with reports indicating a wide array of processes involved in, or resulting from free radicals, such as lipid peroxidation, DNA, RNA and protein oxidation [92]. Protein carbonyls originate from protein oxidation. In the process of carbonylation of proteins, carbonyl side chains are added to proteins as they undergo oxidation reactions [93]. Protein carbonyls were found to be elevated in multiple brain regions in AD subjects such as the hippocampus, parahippocampal gyrus, inferior parietal lobule, superior and middle temporal gyri [94], [95].
Haixiang Su et al. [96] were able to quantify protein carbonyl levels by ELISA in saliva samples of AD patients and found that there is no significant difference between AD and controls, though they were able to identify a diurnal variation in carbonyl levels that peak at 2 pm. This intriguing peak was maintained in both AD patients and control, with dampened level in APOE 4 genotype patients but this dampening remained non-significant.
Parkinson's disease
Parkinson disease (PD) is a progressive neurodegenerative disorder resulting in multiple motor and cognitive deficits. The symptoms of this disorder have been classified into two groups, motor and non-motor [97] [98]. In most cases the specific cause of PD remains unknown and the disease has been termed idiopathic; although, familial forms and environmental risk factors have been identified [99], [100]. PD mainly causes loss of two classes of neurons across the brain: the dopaminergic and the serotoninergic. This neuron loss triggered by the aggregation of alpha-synuclein proteins within neurons in structures known as Lewy bodies, the hallmark of PD, leads to the multiple motor and non-motor deficits associated with Parkinson [101], [102], [103]. PD is classified into three main types based on the dominant symptom: tremor dominant type, akinetic-rigid dominant type and the mixed type [104].
Alpha-synuclein
Alpha-synuclein (α-syn) is a 140 amino acid protein and member of the synuclein family. It is abundant and ubiquitously found in multiple brain regions such as the striatum, hippocampus, olfactory bulb, neocortex, thalamus and cerebellum [105]. It is located in the presynaptic terminals of neurons and has shown affinity for SNARE complex proteins such as synaptobrevin-2, synapsin III and rab3A. α-syn has been found to affect synapsin III expression and its cellular distribution in dopaminergic neurons, the caudate and putamen of PD patients [106], [107], [108], [109]. This results in shifts in neurotransmitter vesicle cluster arrangement and dopamine release. Alpha-synuclein is known to exist in and cycle between two forms: (i) the soluble cytosolic form which has no known function [110], and (ii) the membrane bound helical form which functions in membrane fusion in the cascade of synaptic events [111]. Alpha-synuclein pathology in familial PD has been linked to its increased concentrations in neurons, which can be attributed to increased α-syn gene (SNCA) expression due to promoter polymorphisms as well as gene copy number duplication and triplication [112], [113], [114], [115]. Along with gene overexpression, three point mutations in the SNCA gene sequence have been identified in familial PD that affect α-syn aggregation [116], [117], [118], [119], [120], [121]. There are two forms of α-syn aggregation in PD: (i) oligomeric amorphous aggregates which are linked to multiple organelle dysfunctions as well as defects in the axonal transport system [122], [123], [124], [125], [126], [127], [128], [129], [130], and (ii) fibrillar insoluble aggregates that contribute to the formation of Lewy Body pathology [131], [132], [133].
In the quest to establish α-syn as a prominent salivary biomarker of PD diagnosis, Devic et al. [134] used western blotting and spectrometry to establish the presence of α-syn in human saliva. Using Luminex assay they showed that α-syn concentrations significantly decrease in the saliva of PD patients as compared to healthy controls. These findings were replicated by Al-Nimer et al. [135] and validated by ELISA assay. Vivacqua et al. [136] were able to measure, using ELISA, the concentrations of α-syn oligomer (α-synolig) and total α-syn (syntotal) which is constituted of α-syn monomers and to a lesser degree α-syn oligomers in saliva. They detected a significant increase in α-synolig and α-syn olig/α-syntotal ratio in the saliva of PD patients as compared to healthy controls, while α-syntotal exhibited a significant decrease in concentration in saliva of PD patients when compared to healthy controls. These results observed in saliva are mirrored in CSF [63]. Vivacqua et al. suggested that the difference in concentrations between α-syntotal and α-synolig was due to the oligomerization of free monomeric α-syn in saliva, which lead to the reduction of the recorded α-syntotal concentration.
Upon analysis of the results presented by Vivacqua et al. for α-syntotal salivary levels, it was evident that α-syntotal is pertinent in more than simply identifying PD patients when compared to controls, as its levels on a smaller scale within the PD population exhibited correlations with disease severity, progression, stages and cognitive impairments. α-syntotal was shown to positively correlate with H&Y scores, MDS-UPDRS total score and LEDD scores. Indicating that it can provide a tool for prediction of disease progression as lower concentrations reflect early disease stages, while higher concentrations reflect late disease stages. Negative correlations were established between α-syntotal and both FAB and MOCA scores, indicating reduced cognitive abilities in PD patients. The increase in α-syntotal with the advancement of disease, was attributed by Vivacqua et al. to disease progression causing advanced synaptic and cellular damage that result in α-syn monomer release into the extracellular medium. Unlike α-syn total, α-syn olig was not demonstrated to have any correlation with disease stages and progression. A urinalysis study conducted on a Korean sample demonstrated that α-syn cannot be detected in urine of PD patients and healthy individuals.
DJ-1
DJ-1 is a 189 Amino Acid protein; its mutation has been associated with rare early onset familial autosomal recessive PD. DJ-1 is speculated to be a pleiotropic neuro-protective protein that functions as an antioxidant and against mitochondrial dysfunction [137], [138], [139], [140], [141]. Normally DJ-1 is located mainly in the cytoplasm and to a lesser extent in mitochondria and nuclei of dopaminergic neurons. Under oxidative stress DJ-1 monomers dimerize and favour mitochondrial localization to finally translocate to the cell nucleus. Recent evidence suggests that DJ-1 recruitment to plasma membrane and mitochondria is efficient in neuro-protection only when cells are under low to moderate oxidative stress [142]. In the face of oxidative stress, DJ-1 has been shown to have the ability to reduce hydrogen peroxide species, stabilize Nrf2 transcription factor which regulates the expression of antioxidant proteins and reduce oxidative stress sustained by neurons upon calcium entry via L-type channels in pacemaker potentials [138], [142], [143]. DJ-1 also plays a role in preventing mitochondrial dysfunction through regulation of SLC25A14 and SLC25A27 which are mitochondrial uncoupling proteins in dopaminergic neurons of the substantia nigra pars compacta [142]. DJ-1 interacts with PINK1 which is a serine threonine kinase that protects cells from stress induced mitochondrial dysfunction [139], [141]. There is evidence that implicates DJ-1 in regulation of astrocyte inflammatory responses as well as astrocytic and neuronal lipid rafts formation [144], [145]. Finally, DJ-1 plays the role of a molecular chaperone to inhibit the formation of α-synuclein fibrils, an essential step in the formation of α-syn oligomers which constitute a key part in PD pathology [146]. Devic et al. [134] examined DJ-1 concentrations in saliva of PD patients and their respective healthy controls by Luminex assay and showed that DJ-1 levels did not correlate with clinical test scores for the UPDRS motor test, yet have a tendency to increase in PD patient saliva as compared to controls. Using the same technique, Kang et al. [147] were able to identify a significant increase in DJ-1 concentration in patients who are classified as PD stage 4 according to the H&Y score as compared to those who were classified as stages 1–3. A significant decrease in DJ-1 concentration was realized in patients with mixed type PD as compared to tremor dominant and akinetic-rigid dominant type suggesting different mechanisms of disease progression in different disease subtypes. Kang et al. reported no correlation between DJ-1 salivary concentration and UPDRS scores in accordance with the findings of Devic et al. In a recent study conducted by Masters et al. [148] and by performing quantitative immunoblotting, DJ-1 showed a significant increase in concentration and a positive correlation with UPDRS motor score and thus reflects motor disability. In turn, total salivary protein concentrations were significantly elevated in saliva of patients with respect to controls and differentiated between them. This elevation was attributed to autonomic dysfunction in PD patients. After adjusting DJ-1 levels with respect to total protein concentration, there was no difference between PD patients and controls. Similar to these studies, results concerning DJ-1 concentrations in CSF remain inconclusive [63]. A urinalysis study conducted on a Korean sample has shown that DJ-1 concentrations significantly increase in the urine of PD affected males when compared to non-PD males, while results for females remained insignificant [8].
Acetylcholinesterase activity
Dopaminergic neuron loss in PD is accompanied by a heterogeneous loss of cholinergic neurons affecting various brain regions. Cholinergic deficit with cholinergic neuron terminal reduction has been recorded in PD patients, being more severe with patients affected by PD associated dementia [149], [150], [151]. It has been established that, the decrease in gait speed associated with PD is attributed to cholinergic degeneration [152]. Recently, AChE inhibitors were shown to be effective in alleviating some PD symptoms [153]. Fedorova et al. [154] analysed AChE in saliva samples obtained from PD affected individuals and their relative controls. They were able to uncover an increase in enzymatic activity in the saliva of PD patients when compared to controls. This increase was representative of disease progression as the increase in AChE catalytic activity mirrored stage progression in the H&Y score. Federova et al. analysed total salivary protein concentration which they found to be significantly increased in PD patient saliva as a result of hyposiallorhea. The ratio AChE activity/Total protein concentration was calculated to emphasize that the increased catalytic activity of AChE can only be attributed to an alteration in enzymatic function and not enzymatic concentration. Despite their significant differences, AChE concentrations and AChE/Total protein ratio displayed an overlap between controls and PD patients as few patients presented with values that would classify them as healthy individuals and vice versa.
Huntington's disease
Huntington's disease (HD) is an autosomal dominant neurodegenerative disorder caused by an expansion of N-terminal (CAG)n trinucleotide repeat of the huntingtin gene (Htt) with locus 4p16.3. Polymorphism in the number of repeats of this trinucleotide is common among the population where repeats ranging from (CAG)9–36 are considered normal, while repeats above (CAG)37 are considered pathological [155], [156], [157]. The length of the CAG repeat has been positively correlated with early disease onset and rate of disease progression [158]. The altered CAG trinucleotide repeat codes for repeats of glutamine in the translated 3142 AA huntingtin protein. Normal huntingtin protein contributes to multiple physiological functions including embryonic development, cell survival, tissue maintenance, and cell morphology [159]. In HD, The mutant huntingtin protein is processed by proteases to release the N-terminal poly-glutamine sequence, these released fragments can interfere with transcription and trigger neurodegeneration. Also resulting from this cleavage is the C-terminal sequence of huntingtin, which mediates cytotoxicity by interfering with dynamin1 function and endoplasmic reticulum homeostasis [160]. Mutant huntingtin proteins have been shown to form cytoplasmic aggregates, whose function is still debated, as some studies find them as neuroprotective while others advocate their role in toxicity [161], [162], [163]. Neurodegeneration in HD affects multiple neuron classes in the neocortex, striatum, cerebellum, hippocampus, substantia nigra, and brainstem nuclei [164]. Usually, genetic testing for diagnostic confirmation of HD aims at identifying Htt gene mutations. Here we present Htt protein as a promising candidate for the diagnosis of HD.
Huntingtin protein
Knowing that testing CSF and blood for Htt protein for diagnostic confirmation of HD can be performed, it is still an impractical measure due to the invasiveness of the testing procedure and because of low Htt concentrations in the acquired samples. Blood and CSF Htt protein levels remain fairly low, with a minimum requirement of a 50 ml blood sample to successfully detect Htt protein. In addition to variations in Htt protein concentrations with different blood cell types [165], [166], [167]. In a study conducted by Bloom et al. [168] Htt protein was successfully detected, using ELISA, in saliva of HD patients and healthy controls. There was a significant increase in total Htt protein concentration in saliva samples obtained from HD patients when compared to controls. Additionally, salivary concentration of mutant Htt was significantly increased in pre-manifest HD patients when compared to healthy controls. Thus, salivary Htt can serve as an early detection biomarker for HD. Given that currently a non-invasive measure of Htt CNS concentration doesn't exist. This method could facilitate diagnostic procedures or even replace existing tests once properly established.
Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS) is a motor neuron degenerative disorder known to affect men and women in unequal ratios, approximately 2:1 [169]. The initial presentation of ALS is muscle weakness followed by progressive paralysis during the course of the disease, ultimately leading to death. ALS can also be associated with cognitive manifestations such as frontotemporal dementia [170], [171]. Histologically, cellular inclusions are not an uncommon feature of neurodegenerative diseases as in Alzheimer and Parkinson. As part of its neuropathology ALS has two different types of immunohistochemical inclusions, Bunina bodies, and Ubiquitin-positive TDP-43 inclusions. The exact mode of toxicity employed by these inclusion bodies remains unknown, along with their contribution to disease pathology and progression.
ALS usually presents with upper motor neuron degeneration symptoms such as spasticity, hyperreflexia and the Hoffman sign in combination with lower motor neuron degeneration symptoms such as fasciculations, muscle cramps, and muscle atrophy [172]. These manifestations are mimicked by several other motor neuron degenerative diseases which affect diagnostic efficiency and duration of the diagnostic period. Familial ALS has been reported with genetic mutations affecting the following genes C9orf72, SOD1, TDP-43 and FUS/TLS, whereas sporadic ALS has shown mutations in SOD1, ANG, TDP-43 and TUBA4A genes. These mutations are rarely tested for, except if there exist a family history of the disease [173], [174], [175], [176]. Because of the strenuous and unspecific nature of diagnosis, the period between disease onset, proper diagnosis and treatment initiation is prolonged, and has been established at 12–14 months [177], [178], [179]. Treatment should begin as early as possible in order to be effective, which makes early diagnosis a crucial and pressing need for ALS patients [180], [181].
Chromogranin A
Chromogranin A (CgA) is a neuroendocrine secretory protein that is among the constituents of large dense core vesicles of neurons and endocrine cells containing neuropeptides and hormones respectively. Normally, CgA performs multiple physiological functions including regulation of calcium, vasoconstriction, regulation of glucose metabolism and storage, antimicrobial, antifungal and a major modulator of the neuroendocrine system [182]. Multiple studies have linked CgA to pathological features of ALS. For instance, CgA was shown to interact in a chaperone-like manner with ALS mutant SOD1 and mediate its secretion, there was a significant loss of CgA expressing neurons accompanied by decreased CgA density in the neuropil and an accumulation of CgA in the remaining neurons [183], [184], [185].
In the study performed by Obayashi et al. [186] using a YK070 chromogranin A EIA kit, salivary CgA levels were found to be only significantly elevated in patients with terminal ALS in comparison with healthy controls, moderate ALS patients and patients suffering from vascular dementia. Finally, CgA concentration positively correlated with the El Escorial score of emotional functioning rather than that of physical mobility, communication and alimentation. Thus according to these findings CgA is reflective of disease severity and the affective state of ALS patients.
Multiple sclerosis
Multiple sclerosis (MS) is a neurodegenerative, inflammatory, demyelinating disorder of the CNS with women being at higher risk than men (2:1) [187], [188], [189], [190]. MS is believed to be an autoimmune disorder in which lymphocyte T cells target the myelin sheaths of CNS neurons [191], [192]. The destruction of myelin and the breach in the BBB result in the appearance of white matter plaques, which are the hallmark of MS. The main symptoms of the disease can be sensory, motor and cognitive [193] [194], [195], [196], [197] [198]. Psychiatric symptoms do exist in patients with MS and are manifested commonly by depression, which is a major cause of mortality in patients by increasing suicide risk [199], [200], [201]. Diagnosing MS usually relies on clinical examination, MRI scans, evoked potential testing and analysis of CSF. Because many demyelinating syndromes mimic MS in its presentation and symptoms, current MS diagnosis is performed by ruling-out these illnesses based on multiple tests and differential diagnosis techniques to confirm a diagnosis with MS [202]. This necessitates the establishment of easily accessible early diagnostic markers indicative of MS.
Soluble human leukocyte antigen, class II
The Human Leukocyte Antigen (HLA) can be grouped into two major classes, HLA class I and HLA class II both of which are coded by the Major Histocompatibility Complex (MHC) gene cluster and play an important role in antigen presentation and immune regulation which are key in eliciting an immune response [203]. Antigen presentation in MS is carried out by antigen presenting cells (APCs) which include macrophages, dendritic cells, microglia and astrocytes. These APCs endocytose myelin proteins, process them as autoantigens via the endolysosomal pathway and the resulting peptides are bound to MHC class II molecules (i.e. HLA class II). Following antigen presentation, immune cells are activated and recruited to launch an immune response targeting the myelin sheath [204], [205], [206]. Interestingly, recent evidence has revealed a correlation between HLA mutations and levels of HLA class II molecules with MS [207], [208], [209], [210], [211].
The HLA classes exist in two forms, membrane bound and soluble. The membrane bound form functions in antigen presentation while the soluble form is considered as immunomodulatory and has been detected in serum, CSF, sweat, synovial fluid and saliva. The exact process by which these soluble HLA are produced or released from the membranes is still unknown. Some studies suggest that soluble HLA class I are produced by the liver and play a role in transplant tolerance; while, soluble HLA class II remain uncharacterized [212], [213], [214], [215].
In the search for novel diagnostic salivary biomarkers for MS, Adamashvili et al. [216] determined, using ELISA, that the concentration of HLA class II is significantly elevated in the saliva of patients with Relapsing Remitting MS (RRMS) compared to healthy controls. HLA class I were not detected by the used technique, even though they are found in low concentrations in saliva. Adamashvili et al. attributed this to the low sensitivity of ELISA to low concentrations of soluble HLA class I. These results conformed to CSF measurements of both HLA classes. In accordance with Adamashvili, Minagar et al. [217] reported, using ELISA, an increase in soluble HLA class II in the saliva of patients suffering from RRMS as compared to healthy controls and undetectable salivary soluble HLA class I concentrations. They also monitored soluble HLA class II salivary levels in response to interferon β1-a, which is a medication commonly used to manage MS. Soluble HLA class II concentrations exhibited an increase in response to interferon β1-a treatment with decline in MRI contrast enhancing lesions and a stable disease course.
Oxidative stress
The release of free radicals such as reactive oxygen or nitrogen species (ROS, RNS) is normally performed during an immune response and inflammation. Cells are said to be under oxidative stress, when the released reactive species overwhelm their antioxidant defences and cause damage to cell structures that could ultimately lead to degeneration [218]. With inflammatory and autoimmune reactions being a major part in MS pathology, oxidative stress is sure to manifest in an MS affected CNS. In MS, free radicals are generated by activated macrophages, microglia and mitochondrial dysfunction [219]. Oxidative damage is known to target lipid membranes, proteins, myelin, macrophages, astrocytes, oligodendrocytes and inhibit gene expression essential for myelination. ROS also contribute to the pathological plaque formation in MS [219], [220], [221]. In the quest for establishing saliva as a reliable pool of oxidative stress markers for disease monitoring, Karlik et al. [222] performed four analytical tests on saliva and blood samples obtained from MS patients. They dosed Advanced Oxidation Protein Products (AOPP) a marker of protein oxidation, Thiobarbituric Acid Reacting Substances (TBARS) a marker of lipoperoxidation and finally advanced glycation end products (AGEs) and fructosamine which are markers of carbonyl stress. Of the four dosed oxidative stress markers, TBARS and AGEs were significantly elevated in saliva of MS patients as compared to healthy controls, while AOPP remained unchanged. All four of the dosed markers exhibited a significant increase in plasma. Karlik et al. also assessed Total Antioxidant Capacity (TAC) and Ferric Ion Reducing Ability of saliva/plasma (FRAS/P), both of which represent saliva's and blood's antioxidant power. They found FRAS to be significantly lower in MS patient saliva samples as compared to healthy controls, while TAC levels were lower without reaching significance. The authors attributed this difference to the fact that FRAS only measures the antioxidative effects of non-protein molecules, but as the name suggests, TAC provides a total measure of antioxidative capacity in a certain fluid and is therefore affected by multiple variables. Meanwhile, plasma TAC levels decreased significantly between MS patients and controls, and FRAP remained unchanged. With the exception of one study reporting no change in CSF oxidative stress markers [223], studies conducted on CSF revealed that TBARS are increased in CSF of MS patients when compared to controls, with a decrease in TAC [221], [224], [225].
Autism spectrum disorders
Autism spectrum disorder (ASD) is a neurodevelopmental disorder presenting with impairments in social communication, interaction and restricted repetitive patterns of behaviour. Autistic disorder, Asperger's disorder, Rett's disorder, Childhood disintegrative disorder, Pervasive developmental disorder-Not otherwise specified-are the group of disorders that make up the ASD [226]. Despite genetic mutations being linked to the disorder and strong evidence supporting the genetic roots of ASD, there is no consensus over the specific etiology of ASD [227], [228], [229]. There exists evidence of both neurodegeneration and neuroinflammation in ASD, manifested by microglial activation, proinflammatory cytokine release and neuron loss [230], [231]. Alterations in the histology and anatomy of the frontal lobe, amygdala and cerebellum have been discussed as part of the neuropathology of ASD with over-all developmental time-course of the brain being affected [232], [233]. Diagnosis of ASD is based on clinical examination to identify the main symptoms of the disorder. Due to the genetic polymorphism of ASD, genetic testing is impractical to establish a diagnosis. Of the existing immunological, biochemical and hormonal biomarkers in CSF and blood for ASD, analysing blood serotonin level is the most established [234], [235]. Functional MRI neuroimaging is also employed to detect differences in neurofunctional and neurophysiological activity of the ASD brain [235]. Given that early detection of ASD is important and that usually a diagnosis is not established before the age of 2 years [236], [237], [238], [239], the search for new and easily accessible biomarkers is required that might precede the morphological and physiological changes of the disorder or help monitor the patients.
Micro-RNAs
With the ongoing search for biomarkers and causes for ASD, after identification of the genetic diversity of the disorder [227], [228], [229], research has turned to epigenetics. Epigenetics plays an important role regulating gene products without altering the nucleotide structure of the gene itself. MicroRNAs (miRNA) are short single stranded RNA sequences that interfere with gene expression by interacting with target mRNA and regulating its translation into protein. Multiple miRNA expression profiles have been elucidated in ASD most of which are involved in nervous system development and function [240], [241], [242]. In efforts to establish new diagnostic biomarkers for ASD, salivary miRNA profiling was performed by Hick's et al. [243] who was able to discover 14 differentially expressed miRNAs in ASD patient saliva when compared to healthy individuals. From these 14 miRNAs 4 were downregulated in saliva samples of ASD patients (miR-23a-3p, miR-27a-3p, miR-30e-5p and miR-32-5p) and the remaining ten were upregulated (miR-140-3p, miR2467-5p, miR-218-5p, miR-28-5p, miR-335-3p, miR-628-5p, miR-7-5p, miR-191-5p, miR-127-3p and miR-3529-3p). All miRNA levels were predictive of the Vineland Adaptive Behaviour score especially for neurodevelopmental scores except for miR-140-3p. Knowing that the Vineland Adaptive Behaviour scale is a psychometric assessment test for psychological and psychiatric disorders and that a low Vineland score indicates an impairment [244], it was interesting to find that the upregulated miRNAs negatively correlated with the Vinland score while downregulated miRNAs positively correlated with the score. Further, investigation of possible target genes for the identified miRNA revealed a vast number of enriched neurodevelopment linked and ASD linked genes. This indicates that creating a miRNA salivary profile for ASD can help properly diagnose infants with the disorder before prominent behavioural manifestations appear.
Salivary proteome
With the immune system suspected to play an important role in ASD pathology, proteins implicated in immune reactions are expected to be deregulated. Studies regarding the ASD related neuroimmune disturbances as well as correlation of ASD with parental autoimmune disorders further verifies that the deregulation of the immune system is either etiological or consequential to ASD [231], [245], [246], [247], [248], [249]. In the quest of establishing new ASD salivary biomarkers, Ngounou Wetie et al. [250] conducted a pilot study in which they examined discrepancies in salivary protein levels and signatures between ASD patients and healthy controls. By employing Mass Spectrometry-based proteomics, they were able to identify a variability in salivary proteomic levels for ASD with 12 proteins having an elevated concentration and 4 proteins with reduced concentration in ASD patient saliva when compared to healthy controls. The list of proteins with elevated and reduced concentrations in saliva of ASD patients along with their physiological functions are summarized in Table 2, Table 3. It is evident that all the mentioned proteins are involved in immune reactions. Further analysis of protein–protein interactions revealed that LTF and PIP both interact with prolactin. A similar analysis revealed that submaxillary gland androgen-regulated protein 3B, statherin and histatin all interact with one another. This observation led Ngounou Wetie et al. to the conclusion that protein complex formation might be compromised in ASD. In the same context, Castagnola et al. conducted a proteomic study aimed at identifying changes in the post-translational modifications of salivary proteins. They were able to detect salivary protein hypophosphorylation for ASD patients when compared to healthy controls. The four studied hypophosphorylated proteins were statherin, histatin and proline rich proteins 1 and 3. These findings support the above data indicating that molecular changes in proteins might affect their physiological functions. Castagnola et al. [251] stated that hypophosphorylation serves more as an explanation for failed protein mechanisms rather than a biomarker and therefore could explain the essence of protein–protein interaction deficits. We can thus conclude that biochemical and physiological changes affect salivary proteins in ASD.
Table 2.
List of the upregulated salivary proteins obtained from ASD patients and their respective functions.
| Protein | Length | Function | References |
|---|---|---|---|
| Prolactin-inducible protein (PIP) | 146 AA |
|
[336] |
| Lactoferrin/Lactotransferrin (LTF) | 711 AA |
|
[78], [79], [82], [85], [86] |
| Ig kappa chain C region (IGKC) IgG gamma-1 chain C region (IGHG1) |
107 AA 330 AA |
|
|
| Annexin A1 (ANXA1) | 346 AA |
|
[337], [338], [339], [340], [341], [342] |
| Neutrophil-defensin 1 (DEFA1/DEFA1B) |
94 AA |
|
[343], [344] |
| Neutrophil elastase (ELANE) |
267 AA |
|
[345], [346], [347], [348], [349], [350] |
| Lactoperoxidase (LPO) | 712 AA |
|
[351] |
| Lipocalin 1 (LCN1) | 176 AA |
|
[352], [353] |
| Polymeric immunoglobulin receptor (PIGR) |
764 AA |
|
[354], [355], [356] |
| Deleted in malignant brain tumours 1 protein (DMBT1) |
2413 AA |
|
[357], [358], [359] |
| Myeloperoxidase (MPO) | 745 AA |
|
[360], [361], [362], [363], [364], [365], [366] |
Table 3.
List of the downregulated salivary proteins obtained from ASD patients and their respective functions.
| Protein | Length | Function | References |
|---|---|---|---|
| Salivary acidic proline rich phosphoprotein (PRH1/2) | 166 AA |
|
[367], [368], [369], [370] |
| Submaxillary gland androgen-regulated protein 3B (SMR3B) | 79 AA |
|
|
| Statherin (STATH) | 62 AA |
|
[371], [372], [373], [374] |
| Histatin-1 (HTN1) | 57 AA |
|
[375], [376], [377] |
Oxytocin
Oxytocin is a neuropeptide produced by the hypothalamus and secreted by the posterior pituitary. It is known to function in the brain in neuromodulation, regulating the mother and infant bond, sexual behaviour and social recognition [252], [253], [254]. Studies have associated oxytocin dysfunction with multiple aspects of ASD and its administration has been found to alleviate social impairment symptoms [255], [256], [257], [258]. In the light of the affiliation present between oxytocin and ASD two studies were performed to dose oxytocin saliva concentrations in ASD patients and compare them with healthy controls. By implementing an ELISA, Feldman et al. [259] proved that detection of salivary oxytocin was possible during early disease stages and salivary oxytocin levels in ASD patients to be significantly lower than those of normal controls. Fujisawa et al., [260] were able to dose oxytocin in saliva of both healthy controls and ASD patients by an ELISA as well. Their assay was directed at associating oxytocin salivary levels with visual attention for social signals which resulted in inconclusive evidence. Finally, a meta-analysis performed by Grazia et al. [261] combined the data of the two studies obtaining a 152 individual sample and concluded that there is no significant difference in salivary oxytocin levels between ASD patients and healthy controls. In addition to saliva, Grazia et al. conducted a meta-analysis on data obtained on oxytocin levels in serum and CSF which yielded the same results obtained from the data on saliva. Not enough data was retrieved on urine to properly conduct a meta-analysis and obtain significant results [261].
Neuropsychiatric disorders
Schizophrenia, bipolar disorder and attention deficit hyperactivity disorder (ADHD) all fall under the title of neuropsychiatric disorders, which present with multiple behavioural symptoms. Symptoms of schizophrenia are grouped into: (i) Positive – delusions and hallucinations; (ii) Negative – reduced motivation, social impairments, reduced pleasure and blunted affect; and finally (iii) Disorganized speech and behaviour [262], [263], [264]. Bipolar disorder is classified as a major mood disorder with symptoms of varying degrees of mania and depression [265]. Aside from the multitude of social impairments that affect patients, a high suicide risk has been correlated with both schizophrenia and bipolar disorder [266], [267]. Of the three mentioned disorders only ADHD is classified as a childhood disorder. ADHD is characterized with severe persistent debilitating hyperactivity that leads to functional and social impairments [265]. A diagnosis is usually established for these disorders after proper clinical examination and observation based on the aforementioned symptoms and other DSM-V criteria [226]. Genetic causes have been associated with all three diseases, but the specific etiology remains to be elucidated [268], [269], [270], [271], [272], [273]. Given that diagnosis of the Neuropsychiatric disorders relies on clinical examination of the patient without the establishment of specific disease biomarkers, exploration of physiological fluids for easily accessible and accurate biomarkers seems essential in efforts to provide innovative and faster diagnostic processes.
DNA Methylation
Multiple genes can be named as players in causing the aforementioned neuropsychiatric diseases, as there is no single culprit capable of explaining the various symptoms and types of the disorders [268], [269], [270], [271], [272], [273]. Because of this, research has turned its attention towards the possible implication of aberrant epigenetics in causing the pathological phenotypes associated with neuropsychiatric disorders [274], [275], [276], [277], [278], [279]. On this basis, investigation of methylation profiles of salivary DNA of schizophrenic, bipolar and ADHD patients allowed the identification of four genes HTR2A, DTNBP1, MB-COMT and VIPR2 [Table 4] with epigenetic alterations when compared to normal controls [280], [281], [282], [283]. Cumulative research efforts are providing mounting evidence for the involvement of HTR2A, DTNBP1 and MB-COMT in both schizophrenia and bipolar disorder [284], [285]. Deregulation of both serotoninergic and dopaminergic systems have been shown, with these genes known as major players in their pathways [286], [287], [288], [289]. Pathological genetic variants of HTR2A, DTNBP1 and MB-COMT were proven to affect memory [290], [291], [292], hallucinations [293], glutamate signalling [291], and symptom severity [294], [295]. Reports of correlations between VIPR2 methylation and ADHD may be premature and thus the function of this gene in ADHD requires further studies [296], [297]. In fact, the study conducted by Wilmot et al. [283], who detected VIPR2 CpG methylation in saliva, was the first of its kind performing a genome wide DNA methylation analysis for ADHD. Aside from the downregulation of DTNBP1, the study conducted by Abdolmaleky et al. [280] characterized the level of gene methylation as an indicator to treatment responsiveness for both schizophrenic and bipolar subjects. Results indicated that patients undergoing and responding to treatment had lower levels of DTNBP1 promoter methylation as compared to untreated patients. The disease-associated hypermethylation was detected in early schizophrenic patients and in healthy first degree relatives, with higher levels of methylation associated with early disease onset. This data supports the merit of DTNBP1 in disease prediction and treatment management. Similar to DTNBP1, HTR2A hypomethylation was also detected in first degree relatives, suggesting the heritability of the epigenetic anomaly [281]. In addition, by comparing salivary samples to post-mortem brain samples, the salivary methylation status of MB-COMT, DTNBP1 and HTR2A was reported to mirror the brain's methylation status for these genes.
Table 4.
Results of salivary epigenetic methylation profile of patients suffering from schizophrenia, bipolar disorder and ADHD. The table below provides information concerning the target region of the epigenetic modification. The chromosomal localization of the gene. The function of the coded protein. As well as the methylation status of the gene for the respective neuropsychiatric disorder.
| Gene Name | Target Region | Locus | Function | Disorder | Status |
|---|---|---|---|---|---|
| 5-hydroxytryptamine receptor 2A (HTR2A) | T102C polymorphic site | 13q14.2 | Serotonin receptor | Schizophrenia/Bipolar disorder | Hypomethylated |
| Dystrobrevin Binding Protein 1 (DTNBP1) | Promoter | 6p22.3 | Organelle Biogenesis | Schizophrenia | Hypermethylated/Down-regulated |
| Membrane-Bound Catechol-O-Methyltransferase (MB-COMT) | Promoter | 22q11.21 | Catecholamine Neurotransmitter degradation | Schizophrenia/Bipolar disorder | Hypomethylated/Up-regulated |
| Vasoactive Intestinal Peptide Receptor 2 (VIPR2) | CpG | 7q36.3 | G-protein coupled receptor for vasoactive intestinal peptide | ADHD | Hypomethylated |
Salivary proteome
Similar to the discussed Neurodegenerative diseases, immune system deregulations have been put into evidence for bipolar disorder and schizophrenia [298], [299], [300], [301]. Thus it is only logical that levels of proteins involved in eliciting an immune response ought to be altered in schizophrenic and bipolar patients when compared to healthy controls. From this standpoint, Iavarone et al. [302] conducted a salivary proteome analysis and found eight immune system-related proteins to be elevated in the saliva of schizophrenic and bipolar patients when compared to healthy controls. The identified proteins were α-defensins 1–4, S100A12, cystatin A and the S-derivatives of cystatin B (glutathionylated and cysteinylated). All were significantly increased in both disorders except for α-defensin 3 in bipolar disorder, where it exhibited a slight non-significant decrease with respect to the controls. All of the mentioned proteins are involved in innate immunity and therefore indicate an immunologic imbalance in schizophrenic and bipolar patients [303], [304], [305], [306].
Discussion
Early detection of neuro-degenerative and neuropsychiatric disorders is imperative for better disease prognosis and the initiation of early treatment [4]. The two main physiological fluids that are used for the detection of such markers are CSF and blood. Acquiring samples of these two fluids has proven to cause a certain level of discomfort and pain to the patient [5]; therefore, it is essential to establish a substitute that is less invasive but remains representative of the body's physiological changes. A good candidate offering promise as a biomarker pool for neurological disease diagnosis and monitoring is saliva. It is an easily accessible biological fluid and its collection is non-invasive, painless and cost effective [14]. In addition to its ease of collection, saliva is generally safer than blood and CSF, and its collection does not expose the healthcare provider to needles, thus reducing the risk of pathogen transmission from patients suffering from chronic infection. Similar to saliva, urine seems to provide a novel and non-invasive source of biomarkers for neurological disorders.
Following the genetic findings provided by Lee et al. [52] and Bermejo Pareja et al. [47], who reported associations between Aβ42 concentration in saliva with AD patient medical history and genetic data, it could be concluded that salivary Aβ42 is probably more reflective of familial AD genotype rather than sporadic AD, and can be used as a tool for disease prediction but requires further investigation. Based on the differential detection of Aβ42 between groups, where it has been found to be secreted in high concentrations in the saliva of individuals suffering from or at risk of developing AD, as compared to healthy controls and controls suffering from PD, Aβ42 can be considered as a prominent candidate as salivary biomarker for AD; however, a gold standard test must be developed for its reproducible and accurate detection in saliva. Further investigation must be carried out in larger population samples to identify the exact diagnostic concentration ranges for salivary Aβ42. Moreover, there is a crucial need to validate the differential detection of Aβ42 in different disease stages. Unlike Aβ42, analysis of Aβ40 has produced conflicting and insignificant findings [47], [52]. This lack of evidence renders it less reliable as a salivary biomarker for AD diagnosis or disease staging without further evaluation. Similar to Aβ42, tau pathology has been reported in Parkinson's disease [63]. Therefore, further investigation with different detection techniques and the inclusion of Parkinson controls must be performed to confirm the validity and potential use of p-tau/t-tau as a biomarker for AD. As described by Carro et al. [77], lactoferrin is highly representative of AD pathophysiology, cognitive impairment and phenotypes and therefore qualifies as a promising salivary biomarker for disease diagnosis. Further longitudinal studies and comparative studies enrolling individuals suffering from multiple cognitive impairments are required to establish the true validity and diagnostic merit of lactoferrin for AD. The weakness of the study conducted by Haixiang Su et al. [96] on protein carbonyls as biomarkers for AD lies in the age difference between controls (mean = 69.20) and AD patients (mean = 82.4) which necessitates a new study with age adjustment among the compared groups for more accurate findings.
Based on the reports on α-syn and DJ-1 as PD biomarkers [134], [135], [136], [147], [148], it could be inferred that the two molecular forms of α-syn, oligomeric and monomeric, can be considered in the search for salivary biomarkers for PD. With α-syn olig serving as a biomarker for disease diagnosis only, due to the lack of data that links it to disease stage and prognosis, and α-syn total serving as a staging biomarker to track disease progression. DJ-1 could be considered as a salivary biomarker for diagnosing the disease and identifying the type of Parkinsonism.
The observations reported on salivary AChE activity in both AD and PD patients [72], [73], [74], [154], indicate that at present salivary AChE cannot be used as a diagnostic marker. In light of present research, AChE could be considered only for monitoring disease pathophysiology and parasympathetic denervation in PD. Properly designed and multifaceted studies must be conducted in order to confirm the potential use of salivary AChE in AD and PD diagnosis. These studies should take into account the various disease stages (mild, moderate, and severe) and treatment programs of every patient, as well as the development of a proper technique for sample collection and storage for maintaining optimum enzyme function.
Due to their involvement in immune system reactions, peptides, oxidative stress markers and soluble HLA II remain inconclusive as predictive biomarkers, pending further investigation. It is crucial to conduct comparative studies between neurological conditions that trigger the immune system and other immunological conditions, in order to establish whether these markers are exclusively indicative of neurological conditions or can be detected in and affected by the presence of other immunological deregulations.
Investigations on salivary biomarkers are still inconclusive because there is not enough evidence to support or negate the actual pertinence of these molecules in saliva. Most of the existing studies were based on relatively small samples that lacked sufficient power, in addition to concluding with few conflicting results. Based on the results presented by our review, we realize that there are some biomarkers which are more prominent than others, especially those proven to be causal for disease phenotypes, while those associated with other physiological processes like immunity and oxidative stress remain somewhat unspecific unless their presence in saliva is proven to be strictly representative of the diseases in question [Fig. 2]. The presence of the reviewed salivary biomarkers detected in neurological conditions was also assessed in different commonly tested physiological fluids: CSF, blood and urine [Table 5], along with a comparison of each fluid's advantages and disadvantages [Table 6].
Fig. 2.
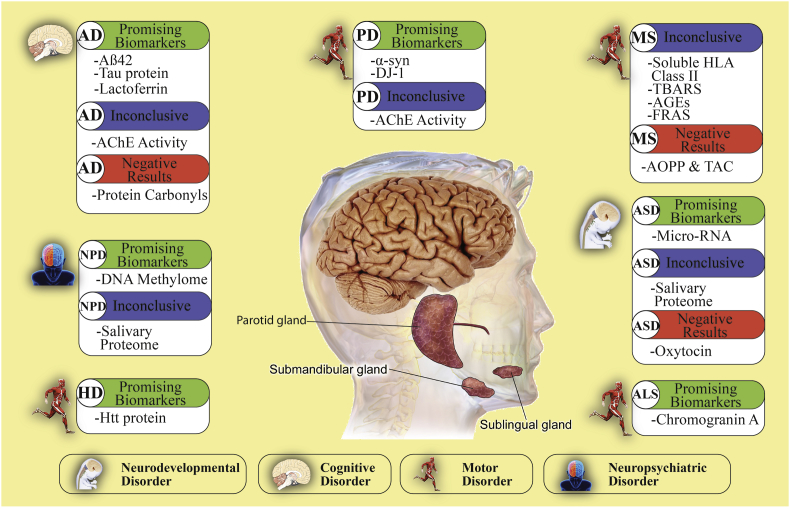
Summary of salivary biomarkers and their current state of validity. This figure classifies the neurological disorders into four different types: neurodevelopmental, cognitive, motor and neuropsychiatric disorders. For each one of these disorders, a categorical system classifies the biomarkers as: promising, inconclusive and negative results based upon the findings of our work. Abbreviations used: AD: Alzheimer's disease; PD: Parkinson's disease; MS: Multiple Sclerosis; ASD: Autism Spectrum Disorder; ALS: Amyotrophic Lateral Sclerosis; HD: Huntington's disease; and NPD: Neuropsychiatric Disorders.
Table 5.
Summary of available reports on the levels of disease biomarkers detected in saliva, CSF, blood and urine and their respective variations.
| Saliva | CSF | Blood | Urine | ||
|---|---|---|---|---|---|
| AD | AB42 | ↑ in Moderate AD [47] ↓ in Severe AD [47] ↑ [51], [52] |
↓ in Severe AD [48], [49], [50], [378], [379] |
↑ [380], [381] ↓ [382], [383] |
Detected [9] |
| AB40 | ↑ [51] No change [47] |
No change [378], [379] | ↑ [378], [380], [384] ↓ [385] |
N/A | |
| Tau | ↑ [13] | ↑ [63], [386], [387] | No change [384] | N/A | |
| Lactoferrin | ↑ [77] | N/A | No change [388], [389] | N/A | |
| AChE activity | ↓ [72], [73], [74] | ↑ in AD with APOE4 mutation [390] No change [391] ↓ [391] |
↓ Erythrocyte AChE activity [392] No change [391] |
N/A | |
| Protein Carbonyls | No change [96] | No change [393] | ↑ [394], [395], [396], [397] | N/A | |
| PD | α-syntotal | ↓ [136], [398] ↑ [134], [135] |
↓ [63] No change [63] |
↓ [399] ↑ [400], [401] |
Not detected [8] |
| α-synolig | ↑ [136] | ↑ [63] | N/A | N/A | |
| α-synolig/α-syntotal | ↑ [136] | ↑ [63] | N/A | N/A | |
| DJ-1 | ↑ [134], [147], [148] | ↓ [398] ↑ [402], [403] |
↑ [404] No change [405], [406] |
↑ [8] | |
| AChE activity | ↑ [154] | No change [391] | ↑ [407] | N/A | |
| HD | mHtt protein | ↑ [168] | ↑ [167] | ↑ [165], [166] | N/A |
| ALS | CgA | ↑ [186] | ↑ [408] | N/A | N/A |
| MS | s-HLA Class II | ↑ [216] | ↑ [216] Detected [409] |
↑ [210] ↓ [409] |
N/A |
| TBARS | ↑ [222] | ↑ [221], [224], [225] No change [223] |
↑ [221], [222], [223], [224] No change [225] |
N/A | |
| AOPP | No change [222] | N/A | ↑ [222] | N/A | |
| AGEs | ↑ [222] | N/A | No change [222] | N/A | |
| TAC | No change [222] | ↓ [221], [224] | ↓ [222] | N/A | |
| FRA | ↓ [222] | N/A | No change [222] | N/A | |
| Fructosamine | N/A | N/A | ↑ [222] | N/A | |
| ASD | Oxytocin | No change [261] | No change [261] | No change [261] | N/A |
All variations reported are in comparison to healthy controls, Legend: ↑: increasing, ↓: decreasing, N/A: no studies found, Detected: no information concerning the concentration only that the biomarker is detected, If biomarker variation is specific to disease stage, phenotype or cell type, then it is clearly stated.
Table 6.
Advantages and disadvantages of sampling saliva, CSF, blood and urine.
| Saliva | Urine | Blood | CSF | |
|---|---|---|---|---|
| Advantages | - Non-invasive | - Highly representative of internal physiology | ||
| - Painless | ||||
| - Cost effective | ||||
| Disadvantages | - Diurnal variation | - Moderately invasive | - Invasive | |
| - Variable volume | - Drowsiness | - Pain & Discomfort | ||
| - Variable biomarker concentration | - Discomfort | - Possible allergic reaction to the anaesthetic | ||
| - Possible blood contamination | - Infection | |||
| - Oral healthrowhead | - Urinary tract infection | - Headache | ||
| - Dizziness | ||||
| - Vomiting | ||||
That being said, we cannot depend on one biomarker to ultimately achieve an accurate diagnosis. Instead, a definitive biomarker profile must be created to be used for future diagnostic purposes. The source of these biomarkers remains to be properly identified. They could be released into saliva from plasma microfiltration through the gums or expressed in salivary glands. It is also probable that they are transported directly from the CNS via the axons of the facial and glossopharyngeal nerves to the salivary glands to ultimately be vacated along with saliva secretion.
Because different fundamental and prominent factors may affect sample quality, saliva collection methods and processing must be refined. In the study conducted by Lee et al., stabilizing salivary levels of Aβ42 peptides by adding thioflavin S to prevent aggregation and using sodium azide to inhibit bacterial growth, enhanced Aβ42 detection in the sample [52]. Furthermore, the available literature indicates that patients suffering from neurological disorders have lower oral health and are more prone to oral disease than healthy individuals [307], [308], [309], [310], [311], [312], [313]. We thus speculate the possible interference of reduced oral health and successful biomarker detection. This raises the question if proper extraction, handling and treatment of the saliva sample with agents and compounds known to stabilize the measured molecule will result in better sample quality and therefore better biomarker detection.
Another aspect of saliva that might affect proper biomarker detection is the fact that patients suffering from some of the neurological diseases listed in this review suffer from sialorrhea or hyposialorrhea which are excess and diminished salivation respectively. The changes in saliva secretion is usually brought on as a side effect of the illness [314], [315], [316], [317], [318] or caused by medication [317], [319], [320]. This indicates that there is a possibility of shifts in biomarker concentrations among healthy controls and patients due to different secreted concentrations of total salivary proteins. Consequently, it is recommended to normalise future measurements of salivary biomarkers against total salivary proteins to account for shifts in concentrations of biomarkers brought about by irregular salivation. Aside from pathological changes in salivary composition and secretion, it is established that there exist circadian rhythms that normally govern saliva production, composition and flow rate [321]. Thus it is imperative that samples from patients and controls be collected during the same time frame in order to avoid normal shifts in saliva protein concentrations from confounding the evidence.
We propose that future research efforts be directed towards establishing salivary epigenetic signatures based on micro-RNA analysis. Given that micro-RNA can be successfully detected in human saliva [322] and that micro-RNA disruptions do exist in neurological disease [323], [324], it is possible to create disease specific micro-RNA expression profiles. These profiles will allow detection of a pathological fingerprint unique to the disease in question and therefore allow for a better, more accurate diagnosis.
Conclusions
Neurological disorders are generally debilitating for patients and they impose a socioeconomic burden on these patients and their caregivers. This highlights the urgent requirement for the development of easily accessible, non-invasive and cost effective diagnostic tests that aim at early identification of neurological diseases. Saliva is a physiological fluid that shows promise in developing non-invasive testing for disease biomarkers, but the utility and applicability of salivary biomarkers in diagnosis of neurological disorders remains in question. The current research remains inefficient in discerning the reliability of salivary biomarkers in neurological disease detection and monitoring. Due to the lack of data and conflicting results of the existing studies, there is an incessant need for increased research efforts to conduct well-structured clinical trials if a non-invasive screening technique based on salivary biomarkers is to be realized. Proper saliva collection and processing protocols must be standardized in order to decrease biases and allow an accurate identification of salivary biomarkers. Furthermore, we believe that salivary biomarkers, in their initial stages, will not be able to solely detect neurological pathologies, but instead these biomarkers will serve as means aiding in diagnosis, or simply replacing other invasive tests. From this standpoint and based on the reviewed literature, we were able to categorise the available biomarkers [Fig. 2], in hope of guiding future research endeavours along a targeted path to ascertain the validity of the most promising biomarkers available.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by funds from the Lebanese University and the University of the Pacific.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bj.2018.03.004.
Contributor Information
David M. Ojcius, Email: dojcius@pacific.edu.
Najwane Said Sadier, Email: najwane_said@yahoo.fr.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Bertram L., Tanzi R.E. The genetic epidemiology of neurodegenerative disease. J Clin Invest. 2005;115:1449–1457. doi: 10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landrigan P.J., Sonawane B., Butler R.N., Trasande L., Callan R., Droller D. Early environmental origins of neurodegenerative disease in later life. Environ Health Perspect. 2005;113:1230–1233. doi: 10.1289/ehp.7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bossy-Wetzel E., Schwarzenbacher R., Lipton S.A. Molecular pathways to neurodegeneration. Nat Med. 2004;10:S2–S9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- 4.DeKosky S.T., Marek K. Looking backward to move forward: early detection of neurodegenerative disorders. Science. 2003;302:830–834. doi: 10.1126/science.1090349. [DOI] [PubMed] [Google Scholar]
- 5.Evans R.W. Complications of lumbar puncture. Neurol Clin. 1998;16:83–105. doi: 10.1016/s0733-8619(05)70368-6. [DOI] [PubMed] [Google Scholar]
- 6.An M., Gao Y. Urinary biomarkers of brain diseases. Genomics Proteom Bioinform. 2015;13:345–354. doi: 10.1016/j.gpb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan J.-Z., Guan W.-P., Maeda T., Guoqing X., GuangZhi W., Makino N. Patients with multiple sclerosis show increased oxidative stress markers and somatic telomere length shortening. Mol Cell Biochem. 2015;400:183–187. doi: 10.1007/s11010-014-2274-1. [DOI] [PubMed] [Google Scholar]
- 8.Ho D.H., Yi S., Seo H., Son I., Seol W. Increased DJ-1 in urine exosome of Korean males with Parkinson's disease. BioMed Res Int. 2014 doi: 10.1155/2014/704678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takata M., Nakashima M., Takehara T., Baba H., Machida K., Akitake Y. Detection of amyloid β protein in the urine of Alzheimer's disease patients and healthy individuals. Neurosci Lett. 2008;435:126–130. doi: 10.1016/j.neulet.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Schenkels L.C., Veerman E.C., Nieuw Amerongen A.V. Biochemical composition of human saliva in relation to other mucosal fluids. Crit Rev Oral Biol Med. 1995;6:161–175. doi: 10.1177/10454411950060020501. [DOI] [PubMed] [Google Scholar]
- 11.Mandel I.D. The functions of saliva. J Dent Res. 1987;66:623–627. doi: 10.1177/00220345870660S203. [DOI] [PubMed] [Google Scholar]
- 12.Moore Keith L., Dalley A.F., Agur Anne M.R. Lippincott Williams & Wilkins; Philadelphia: 2010. Clinically oriented anatomy. [Google Scholar]
- 13.Shi M., Sui Y.T., Peskind E.R., Li G., Hwang H., Devic I. Salivary tau species are potential biomarkers of Alzheimer's disease. J Alzheim Dis. 2011;27:299–305. doi: 10.3233/JAD-2011-110731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabak L.A. A revolution in biomedical assessment: the development of salivary diagnostics. J Dent Educ. 2001;65:1335–1339. [PubMed] [Google Scholar]
- 15.Streckfus C., Bigler L. Saliva as a diagnostic fluid. Oral Dis. 2002;8:69–76. doi: 10.1034/j.1601-0825.2002.1o834.x. [DOI] [PubMed] [Google Scholar]
- 16.O'Banion M.K., Coleman P.D., Callahan L.M. Elsevier; 1994. Regional neuronal loss in aging and Alzheimer's disease: a brief review; pp. 307–314. Seminars in Neuroscience. 6. [Google Scholar]
- 17.Padurariu M., Ciobica A., Mavroudis I., Fotiou D., Baloyannis S. Hippocampal neuronal loss in the CA1 and CA3 areas of Alzheimer's disease patients. Psychiatr Danub. 2012;24:152–158. [PubMed] [Google Scholar]
- 18.Zarow C., Zaias B., Lyness S., Chui H. Cerebral amyloid angiopathy in Alzheimer disease is associated with apolipoprotein E4 and cortical neuron loss. Alzheimer Dis Assoc Disord. 1999;13:1–8. doi: 10.1097/00002093-199903000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Osta A., Alberini C.M. Amyloid beta mediates memory formation. Learn Mem. 2009;16:267–272. doi: 10.1101/lm.1310209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm M.O., Grimm H.S., Hartmann T. Amyloid beta as a regulator of lipid homeostasis. Trends Mol Med. 2007;13:337–344. doi: 10.1016/j.molmed.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Kontush A., Berndt C., Weber W., Akopyan V., Arlt S., Schippling S. Amyloid-β is an antioxidant for lipoproteins in cerebrospinal fluid and plasma. Free Radic Biol Med. 2001;30:119–128. doi: 10.1016/s0891-5849(00)00458-5. [DOI] [PubMed] [Google Scholar]
- 22.Koo E.H., Park L., Selkoe D.J. Amyloid beta-protein as a substrate interacts with extracellular matrix to promote neurite outgrowth. Proc Natl Acad Sci. 1993;90:4748–4752. doi: 10.1073/pnas.90.10.4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morley J.E., Farr S.A., Banks W.A., Johnson S.N., Yamada K.A., Xu L. A physiological role for amyloid-β protein: enhancement of learning and memory. J Alzheim Dis. 2010;19:441–449. doi: 10.3233/JAD-2009-1230. [DOI] [PubMed] [Google Scholar]
- 24.Nicolas M., Hassan B.A. Amyloid precursor protein and neural development. Development. 2014;141:2543–2548. doi: 10.1242/dev.108712. [DOI] [PubMed] [Google Scholar]
- 25.Drechsel D.N., Hyman A., Cobb M.H., Kirschner M. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol Biol Cell. 1992;3:1141–1154. doi: 10.1091/mbc.3.10.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drubin D.G., Kirschner M.W. Tau protein function in living cells. J Cell Biol. 1986;103:2739–2746. doi: 10.1083/jcb.103.6.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebneth A., Godemann R., Stamer K., Illenberger S., Trinczek B., Mandelkow E.-M. Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer's disease. J Cell Biol. 1998;143:777–794. doi: 10.1083/jcb.143.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bloom G.S. Amyloid-β and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 2014;71:505–508. doi: 10.1001/jamaneurol.2013.5847. [DOI] [PubMed] [Google Scholar]
- 29.Ittner L.M., Götz J. Amyloid-[beta] and tau–a toxic pas de deux in Alzheimer's disease. Nat Rev Neurosci. 2011;12:65–72. doi: 10.1038/nrn2967. [DOI] [PubMed] [Google Scholar]
- 30.Mudher A., Lovestone S. Alzheimer's disease–do tauists and baptists finally shake hands? Trends Neurosci. 2002;25:22–26. doi: 10.1016/s0166-2236(00)02031-2. [DOI] [PubMed] [Google Scholar]
- 31.Ballatore C., Lee V.M.-Y., Trojanowski J.Q. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 32.Buée L., Bussière T., Buée-Scherrer V., Delacourte A., Hof P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 33.Selkoe D.J. The cell biology of β-amyloid precursor protein and presenilin in Alzheimer's disease. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 34.Chasseigneaux S., Allinquant B. Functions of Aβ, sAPPα and sAPPβ: similarities and differences. J Neurochem. 2012;120:99–108. doi: 10.1111/j.1471-4159.2011.07584.x. [DOI] [PubMed] [Google Scholar]
- 35.Mattson M.P. Cellular actions of beta-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol Rev. 1997;77:1081–1132. doi: 10.1152/physrev.1997.77.4.1081. [DOI] [PubMed] [Google Scholar]
- 36.Jorissen E., Prox J., Bernreuther C., Weber S., Schwanbeck R., Serneels L. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci. 2010;30:4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kojro E., Fahrenholz F. Springer; 2005. The non-amyloidogenic pathway: structure and function of α-secretases; pp. 105–127. Alzheimer's Disease. [DOI] [PubMed] [Google Scholar]
- 38.Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P. β-secretase cleavage of Alzheimer9s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 39.Yates D., McLoughlin D.M. The molecular pathology of Alzheimer's disease. Psychiatry. 2008;7:1–5. [Google Scholar]
- 40.Bernstein S.L., Wyttenbach T., Baumketner A., Shea J.-E., Bitan G., Teplow D.B. Amyloid β-protein: monomer structure and early aggregation states of Aβ42 and its Pro19 alloform. J Am Chem Soc. 2005;127:2075–2084. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 41.Kim W., Hecht M.H. Sequence determinants of enhanced amyloidogenicity of Alzheimer Aβ42 peptide relative to Aβ40. J Biol Chem. 2005;280:35069–35076. doi: 10.1074/jbc.M505763200. [DOI] [PubMed] [Google Scholar]
- 42.Allan Butterfield D. Amyloid β-peptide (1-42)-induced oxidative stress and neurotoxicity: implications for neurodegeneration in Alzheimer's disease brain. A review. Free Radic Res. 2002;36:1307–1313. doi: 10.1080/1071576021000049890. [DOI] [PubMed] [Google Scholar]
- 43.Jan A., Gokce O., Luthi-Carter R., Lashuel H.A. The ratio of monomeric to aggregated forms of Aβ40 and Aβ42 is an important determinant of amyloid-β aggregation, fibrillogenesis, and toxicity. J Biol Chem. 2008;283:28176–28189. doi: 10.1074/jbc.M803159200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein A.M., Kowall N.W., Ferrante R.J. Neurotoxicity and oxidative damage of beta amyloid 1–42 versus beta amyloid 1–40 in the mouse cerebral cortex. Ann N Y Acad Sci. 1999;893:314–320. doi: 10.1111/j.1749-6632.1999.tb07845.x. [DOI] [PubMed] [Google Scholar]
- 45.Kuperstein I., Broersen K., Benilova I., Rozenski J., Jonckheere W., Debulpaep M. Neurotoxicity of Alzheimer9s disease Aβ peptides is induced by small changes in the Aβ 42 to Aβ 40 ratio. EMBO J. 2010;29:3408–3420. doi: 10.1038/emboj.2010.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattson M.P., Cheng B., Davis D., Bryant K., Lieberburg I., Rydel R.E. beta-Amyloid peptides destabilize calcium homeostasis and render human cortical neurons vulnerable to excitotoxicity. J Neurosci. 1992;12:376–389. doi: 10.1523/JNEUROSCI.12-02-00376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bermejo-Pareja F., Antequera D., Vargas T., Molina J.A., Carro E. Saliva levels of Abeta1-42 as potential biomarker of Alzheimer's disease: a pilot study. BMC Neurol. 2010;10:108. doi: 10.1186/1471-2377-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andreasen N., Hesse C., Davidsson P., Minthon L., Wallin A., Winblad B. Cerebrospinal fluid β-amyloid (1-42) in Alzheimer disease: differences between early-and late-onset Alzheimer disease and stability during the course of disease. Arch Neurol. 1999;56:673–680. doi: 10.1001/archneur.56.6.673. [DOI] [PubMed] [Google Scholar]
- 49.Grimmer T., Riemenschneider M., Förstl H., Henriksen G., Klunk W.E., Mathis C.A. Beta amyloid in Alzheimer's disease: increased deposition in brain is reflected in reduced concentration in cerebrospinal fluid. Biol Psychiatry. 2009;65:927–934. doi: 10.1016/j.biopsych.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sunderland T., Linker G., Mirza N., Putnam K.T., Friedman D.L., Kimmel L.H. Decreased β-amyloid1-42 and increased tau levels in cerebrospinal fluid of patients with Alzheimer disease. JAMA. 2003;289:2094–2103. doi: 10.1001/jama.289.16.2094. [DOI] [PubMed] [Google Scholar]
- 51.Kim C.B., Choi Y.Y., Song W.K., Song K.B. Antibody-based magnetic nanoparticle immunoassay for quantification of Alzheimer's disease pathogenic factor. J Biomed Optic. 2014;19 doi: 10.1117/1.JBO.19.5.051205. 051205. [DOI] [PubMed] [Google Scholar]
- 52.Lee M., Guo J.P., Kennedy K., McGeer E.G., McGeer P.L. A method for diagnosing Alzheimer's disease based on salivary amyloid-β protein 42 levels. J Alzheim Dis. 2017;55:1175–1182. doi: 10.3233/JAD-160748. [DOI] [PubMed] [Google Scholar]
- 53.Irwin D.J., Lee V.M.Y., Trojanowski J.Q. Parkinson's disease dementia: convergence of [alpha]-synuclein, tau and amyloid-[beta] pathologies. Nat Rev Neurosci. 2013;14:626–636. doi: 10.1038/nrn3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrou M., Dwamena B.A., Foerster B.R., MacEachern M.P., Bohnen N.I., Müller M.L. Amyloid deposition in Parkinson's disease and cognitive impairment: a systematic review. Mov Disord. 2015;30:928–935. doi: 10.1002/mds.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J., Mattison H.A., Liu C., Ginghina C., Auinger P., McDermott M.P. Longitudinal assessment of tau and amyloid beta in cerebrospinal fluid of Parkinson disease. Acta Neuropathol. 2013;126:671–682. doi: 10.1007/s00401-013-1121-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanzi R.E., Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 57.Kim M., Suh J., Romano D., Truong M.H., Mullin K., Hooli B. Potential late-onset Alzheimer's disease-associated mutations in the ADAM10 gene attenuate α-secretase activity. Hum Mol Genet. 2009;18:3987–3996. doi: 10.1093/hmg/ddp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saunders A.M., Strittmatter W.J., Schmechel D., George-Hyslop P.S., Pericak-Vance M., Joo S. Association of apolipoprotein E allele ε4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 59.Suh J., Choi S.H., Romano D.M., Gannon M.A., Lesinski A.N., Kim D.Y. ADAM10 missense mutations potentiate β-amyloid accumulation by impairing prodomain chaperone function. Neuron. 2013;80:385–401. doi: 10.1016/j.neuron.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Billingsley M.L., Kincaid R.L. Regulated phosphorylation and dephosphorylation of tau protein: effects on microtubule interaction, intracellular trafficking and neurodegeneration. Biochem J. 1997;323:577–591. doi: 10.1042/bj3230577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hyman B.T., Augustinack J.C., Ingelsson M. Transcriptional and conformational changes of the tau molecule in Alzheimer's disease. Biochim Biophys Acta (BBA) Mol Basis Dis. 2005;1739:150–157. doi: 10.1016/j.bbadis.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 62.Amadoro G., Corsetti V., Ciotti M., Florenzano F., Capsoni S., Amato G. Endogenous Aβ causes cell death via early tau hyperphosphorylation. Neurobiol Aging. 2011;32:969–990. doi: 10.1016/j.neurobiolaging.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 63.Parnetti L., Castrioto A., Chiasserini D., Persichetti E., Tambasco N., El-Agnaf O. Cerebrospinal fluid biomarkers in Parkinson disease. Nat Rev Neurol. 2013;9:131–140. doi: 10.1038/nrneurol.2013.10. [DOI] [PubMed] [Google Scholar]
- 64.Jann M.W. Rivastigmine, a new-generation cholinesterase inhibitor for the treatment of Alzheimer's disease. Pharmacother: J Hum Pharmacol Drug Ther. 2000;20:1–12. doi: 10.1592/phco.20.1.1.34664. [DOI] [PubMed] [Google Scholar]
- 65.Rogers S., Friedhoff L. Long-term efficacy and safety of donepezil in the treatment of Alzheimer's disease: an interim analysis of the results of a US multicentre open label extension study. Eur Neuropsychopharmacol. 1998;8:67–75. doi: 10.1016/s0924-977x(97)00079-5. [DOI] [PubMed] [Google Scholar]
- 66.Whitehouse P.J., Price D.L., Clark A.W., Coyle J.T., DeLong M.R. Alzheimer disease: evidence for selective loss of cholinergic neurons in the nucleus basalis. Ann Neurol. 1981;10:122–126. doi: 10.1002/ana.410100203. [DOI] [PubMed] [Google Scholar]
- 67.García-Ayllón M.-S., Small D.H., Avila J., Sáez-Valero J. Revisiting the role of acetylcholinesterase in Alzheimer's disease: cross-talk with P-tau and β-amyloid. Front Mol Neurosci. 2011;4 doi: 10.3389/fnmol.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inestrosa N.C., Alvarez A., Perez C.A., Moreno R.D., Vicente M., Linker C. Acetylcholinesterase accelerates assembly of amyloid-β-peptides into Alzheimer's fibrils: possible role of the peripheral site of the enzyme. Neuron. 1996;16:881–891. doi: 10.1016/s0896-6273(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 69.Rees T., Hammond P., Soreq H., Younkin S., Brimijoin S. Acetylcholinesterase promotes beta-amyloid plaques in cerebral cortex. Neurobiol Aging. 2003;24:777–787. doi: 10.1016/s0197-4580(02)00230-0. [DOI] [PubMed] [Google Scholar]
- 70.Rinne J.O., Kaasinen V., Järvenpää T., Någren K., Roivainen A., Yu M. Brain acetylcholinesterase activity in mild cognitive impairment and early Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2003;74:113–115. doi: 10.1136/jnnp.74.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shinotoh H., Namba H., Fukushi K., Nagatsuka S.I., Tanaka N., Aotsuka A. Progressive loss of cortical acetylcholinesterase activity in association with cognitive decline in Alzheimer's disease: a positron emission tomography study. Ann Neurol. 2000;48:194–200. [PubMed] [Google Scholar]
- 72.Sayer R., Law E., Connelly P.J., Breen K.C. Association of a salivary acetylcholinesterase with Alzheimer's disease and response to cholinesterase inhibitors. Clin Biochem. 2004;37:98–104. doi: 10.1016/j.clinbiochem.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 73.Bakhtiari S., Moghadam N.B., Ehsani M., Mortazavi H., Sabour S., Bakhshi M. Can salivary acetylcholinesterase be a diagnostic biomarker for alzheimer? J Clin Diagn Res: JCDR. 2017;11:ZC58. doi: 10.7860/JCDR/2017/21715.9192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boston P.F., Gopalkaje K., Manning L., Middleton L., Loxley M. Developing a simple laboratory test for Alzheimer's disease: measuring acetylcholinesterase in saliva-a pilot study. Int J Geriatr Psychiatry. 2008;23:439–440. doi: 10.1002/gps.1882. [DOI] [PubMed] [Google Scholar]
- 75.McGeer P., Akiyama H., Itagaki S., McGeer E. Immune system response in Alzheimer's disease. Can J Neurol Sci. 1989;16:516–527. doi: 10.1017/s0317167100029863. [DOI] [PubMed] [Google Scholar]
- 76.Rogers J., Luber-Narod J., Styren S.D., Civin W.H. Expression of immune system-associated antigens by cells of the human central nervous system: relationship to the pathology of Alzheimer's disease. Neurobiol Aging. 1988;9:339–349. doi: 10.1016/s0197-4580(88)80079-4. [DOI] [PubMed] [Google Scholar]
- 77.Carro E., Bartolomé F., Bermejo-Pareja F., Villarejo-Galende A., Molina J.A., Ortiz P. Early diagnosis of mild cognitive impairment and Alzheimer's disease based on salivary lactoferrin. Alzheimer's Dement: Diagnosis Assess Dis Monit. 2017;8:131–138. doi: 10.1016/j.dadm.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beljaars L., van der Strate B.W., Bakker H.I., Reker-Smit C., Wiegmans F.C., Harmsen M.C. Inhibition of cytomegalovirus infection by lactoferrin in vitro and in vivo. Antivir Res. 2004;63:197–208. doi: 10.1016/j.antiviral.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 79.Berlutti F., Schippa S., Morea C., Sarli S., Perfetto B., Donnarumma G. Lactoferrin downregulates pro-inflammatory cytokines upexpressed in intestinal epithelial cells infected with invasive or noninvasive Escherichia coli strains. Biochem Cell Biol. 2006;84:351–357. doi: 10.1139/o06-039. [DOI] [PubMed] [Google Scholar]
- 80.Gifford J.L., Hunter H.N., Vogel H. Lactoferricin. Cell Mol Life Sci. 2005;62:2588–2598. doi: 10.1007/s00018-005-5373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ito S., Ohtsuki S., Kamiie J., Nezu Y., Terasaki T. Cerebral clearance of human amyloid-β peptide (1–40) across the blood-brain barrier is reduced by complex formation of activated α2-macroglobulin. Neurosci Res. 2007;58:S116. [Google Scholar]
- 82.Orsi N. The antimicrobial activity of lactoferrin: current status and perspectives. Biometals. 2004;17:189–196. doi: 10.1023/b:biom.0000027691.86757.e2. [DOI] [PubMed] [Google Scholar]
- 83.Pietrzik C.U., Jaeger S. Functional role of lipoprotein receptors in Alzheimer's disease. Curr Alzheimer Res. 2008;5:15–25. doi: 10.2174/156720508783884675. [DOI] [PubMed] [Google Scholar]
- 84.Qiu Z., Strickland D.K., Hyman B.T., Rebeck G.W. α2-Macroglobulin enhances the clearance of endogenous soluble β-amyloid peptide via low-density lipoprotein receptor-related protein in cortical neuron. J Neurochem. 1999;73:1393–1398. doi: 10.1046/j.1471-4159.1999.0731393.x. [DOI] [PubMed] [Google Scholar]
- 85.Valenti P., Catizone A., Pantanella F., Frioni A., Natalizi T., Tendini M. Lactoferrin decreases inflammatory response by cystic fibrosis bronchial cells invaded with Burkholderia cenocepacia iron-modulated biofilm. Int J Immunopathol Pharmacol. 2011;24:1057–1068. doi: 10.1177/039463201102400423. [DOI] [PubMed] [Google Scholar]
- 86.Van der Strate B., Beljaars L., Molema G., Harmsen M., Meijer D. Antiviral activities of lactoferrin. Antivir Res. 2001;52:225–239. doi: 10.1016/s0166-3542(01)00195-4. [DOI] [PubMed] [Google Scholar]
- 87.Wang L., Sato H., Zhao S., Tooyama I. Deposition of lactoferrin in fibrillar-type senile plaques in the brains of transgenic mouse models of Alzheimer's disease. Neurosci Lett. 2010;481:164–167. doi: 10.1016/j.neulet.2010.06.079. [DOI] [PubMed] [Google Scholar]
- 88.Hill J.M., Clement C., Pogue A.I., Bhattacharjee S., Zhao Y., Lukiw W.J. Pathogenic microbes, the microbiome, and Alzheimer's disease (AD) Front Aging Neurosci. 2014;6:127. doi: 10.3389/fnagi.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holmes C., Cunningham C., Zotova E., Woolford J., Dean C., Kerr Su. Systemic inflammation and disease progression in Alzheimer disease. Neurology. 2009;73:768–774. doi: 10.1212/WNL.0b013e3181b6bb95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iwashyna T.J., Ely E.W., Smith D.M., Langa K.M. Long-term cognitive impairment and functional disability among survivors of severe sepsis. JAMA. 2010;304:1787–1794. doi: 10.1001/jama.2010.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kamer A.R., Craig R.G., Dasanayake A.P., Brys M., Glodzik-Sobanska L., de Leon M.J. Inflammation and Alzheimer's disease: possible role of periodontal diseases. Alzheimer's Dement. 2008;4:242–250. doi: 10.1016/j.jalz.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 92.Wang X., Wang W., Li L., Perry G., Lee H.G., Zhu X. Oxidative stress and mitochondrial dysfunction in Alzheimer's disease. Biochim Biophys Acta (BBA) Mol Basis Dis. 2014;1842:1240–1247. doi: 10.1016/j.bbadis.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suzuki Y.J., Carini M., Butterfield D.A. Protein carbonylation. Antioxidants Redox Signal. 2010;12:323–325. doi: 10.1089/ars.2009.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aksenov M., Aksenova M., Butterfield D., Geddes J., Markesbery W. Protein oxidation in the brain in Alzheimer's disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 95.Hensley K., Hall N., Subramaniam R., Cole P., Harris M., Aksenov M. Brain regional correspondence between Alzheimer's disease histopathology and biomarkers of protein oxidation. J Neurochem. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- 96.Su H., Gornitsky M., Geng G., Velly A.M., Chertkow H., Schipper H.M. Diurnal variations in salivary protein carbonyl levels in normal and cognitively impaired human subjects. Age. 2008;30:1–9. doi: 10.1007/s11357-007-9042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ruiz P.J.G., Catalan M., Carril J.F. Initial motor symptoms of Parkinson disease. Neurol. 2011;17:S18–S20. doi: 10.1097/NRL.0b013e31823966b4. [DOI] [PubMed] [Google Scholar]
- 98.Chaudhuri K.R., Healy D.G., Schapira A.H. Non-motor symptoms of Parkinson's disease: diagnosis and management. Lancet Neurol. 2006;5:235–245. doi: 10.1016/S1474-4422(06)70373-8. [DOI] [PubMed] [Google Scholar]
- 99.Klein C., Westenberger A. Genetics of Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a008888. a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Priyadarshi A., Khuder S.A., Schaub E.A., Priyadarshi S.S. Environmental risk factors and Parkinson's disease: a metaanalysis. Environ Res. 2001;86:122–127. doi: 10.1006/enrs.2001.4264. [DOI] [PubMed] [Google Scholar]
- 101.Masliah E., Rockenstein E., Veinbergs I., Mallory M., Hashimoto M., Takeda A. Dopaminergic loss and inclusion body formation in α-synuclein mice: implications for neurodegenerative disorders. Science. 2000;287:1265–1269. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- 102.Schulz-Schaeffer W.J. The synaptic pathology of α-synuclein aggregation in dementia with Lewy bodies, Parkinson's disease and Parkinson's disease dementia. Acta Neuropathol. 2010;120:131–143. doi: 10.1007/s00401-010-0711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spillantini M.G., Schmidt M.L., Lee V.M.-Y., Trojanowski J.Q., Jakes R., Goedert M. [alpha]-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 104.Thenganatt M.A., Jankovic J. Parkinson disease subtypes. JAMA Neurol. 2014;71:499–504. doi: 10.1001/jamaneurol.2013.6233. [DOI] [PubMed] [Google Scholar]
- 105.Iwai A., Masliah E., Yoshimoto M., Ge N., Flanagan L., De Silva H.R. The precursor protein of non-Aβ component of Alzheimer's disease amyloid is a presynaptic protein of the central nervous system. Neuron. 1995;14:467–475. doi: 10.1016/0896-6273(95)90302-x. [DOI] [PubMed] [Google Scholar]
- 106.Burré J., Sharma M., Tsetsenis T., Buchman V., Etherton M.R., Südhof T.C. α-Synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen R.H., Wislet-Gendebien S., Samuel F., Visanji N.P., Zhang G., Marsilio D. α-Synuclein membrane association is regulated by the Rab3a recycling machinery and presynaptic activity. J Biol Chem. 2013;288:7438–7449. doi: 10.1074/jbc.M112.439497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maroteaux L., Campanelli J.T., Scheller R.H. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8:2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zaltieri M., Grigoletto J., Longhena F., Navarria L., Favero G., Castrezzati S. α-synuclein and synapsin III cooperatively regulate synaptic function in dopamine neurons. J Cell Sci. 2015;128:2231–2243. doi: 10.1242/jcs.157867. [DOI] [PubMed] [Google Scholar]
- 110.Jensen P.H., Nielsen M.S., Jakes R., Dotti C.G., Goedert M. Binding of α-synuclein to brain vesicles is abolished by familial Parkinson's disease mutation. J Biol Chem. 1998;273:26292–26294. doi: 10.1074/jbc.273.41.26292. [DOI] [PubMed] [Google Scholar]
- 111.Lee H.J., Choi C., Lee S.J. Membrane-bound α-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form. J Biol Chem. 2002;277:671–678. doi: 10.1074/jbc.M107045200. [DOI] [PubMed] [Google Scholar]
- 112.Chartier-Harlin M.C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S. α-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 113.Maraganore D.M., De Andrade M., Elbaz A., Farrer M.J., Ioannidis J.P., Krüger R. Collaborative analysis of α-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 114.Pals P., Lincoln S., Manning J., Heckman M., Skipper L., Hulihan M. α-Synuclein promoter confers susceptibility to Parkinson's disease. Ann Neurol. 2004;56:591–595. doi: 10.1002/ana.20268. [DOI] [PubMed] [Google Scholar]
- 115.Singleton A., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J. α-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 116.Conway K., Lee S.J., Rochet J.C., Ding T., Harper J., Williamson R. Accelerated oligomerization by Parkinson's disease linked α-synuclein mutants. Ann N Y Acad Sci. 2000;920:42–45. doi: 10.1111/j.1749-6632.2000.tb06903.x. [DOI] [PubMed] [Google Scholar]
- 117.Greenbaum E.A., Graves C.L., Mishizen-Eberz A.J., Lupoli M.A., Lynch D.R., Englander S.W. The E46K mutation in α-synuclein increases amyloid fibril formation. J Biol Chem. 2005;280:7800–7807. doi: 10.1074/jbc.M411638200. [DOI] [PubMed] [Google Scholar]
- 118.Li J., Uversky V.N., Fink A.L. Effect of familial Parkinson's disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human α-synuclein. Biochemistry. 2001;40:11604–11613. doi: 10.1021/bi010616g. [DOI] [PubMed] [Google Scholar]
- 119.Narhi L., Wood S.J., Steavenson S., Jiang Y., Wu G.M., Anafi D. Both familial Parkinson's disease mutations accelerate α-synuclein aggregation. J Biol Chem. 1999;274:9843–9846. doi: 10.1074/jbc.274.14.9843. [DOI] [PubMed] [Google Scholar]
- 120.Pandey N., Schmidt R.E., Galvin J.E. The alpha-synuclein mutation E46K promotes aggregation in cultured cells. Exp Neurol. 2006;197:515–520. doi: 10.1016/j.expneurol.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 121.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 122.Chandra S., Gallardo G., Fernández-Chacón R., Schlüter O.M., Südhof T.C. α-Synuclein cooperates with CSPα in preventing neurodegeneration. Cell. 2005;123:383–396. doi: 10.1016/j.cell.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 123.Cooper A.A., Gitler A.D., Cashikar A., Haynes C.M., Hill K.J., Bhullar B. α-Synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dalfo E., Barrachina M., Rosa J., Ambrosio S., Ferrer I. Abnormal α-synuclein interactions with rab3a and rabphilin in diffuse Lewy body disease. Neurobiol Dis. 2004;16:92–97. doi: 10.1016/j.nbd.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 125.Esteves A.R., Arduíno D.M., Swerdlow R.H., Oliveira C.R., Cardoso S.M. Oxidative stress involvement in á-synuclein oligomerization in Parkinson's disease cybrids. Antioxidants Redox Signal. 2009;11:439–448. doi: 10.1089/ars.2008.2247. [DOI] [PubMed] [Google Scholar]
- 126.Gosavi N., Lee H.J., Lee J.S., Patel S., Lee S.J. Golgi fragmentation occurs in the cells with prefibrillar α-synuclein aggregates and precedes the formation of fibrillar inclusion. J Biol Chem. 2002;277:48984–48992. doi: 10.1074/jbc.M208194200. [DOI] [PubMed] [Google Scholar]
- 127.Hsu L.J., Sagara Y., Arroyo A., Rockenstein E., Sisk A., Mallory M. α-Synuclein promotes mitochondrial deficit and oxidative stress. Am J Pathol. 2000;157:401–410. doi: 10.1016/s0002-9440(10)64553-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Outeiro T.F., Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Smith W.W., Jiang H., Pei Z., Tanaka Y., Morita H., Sawa A. Endoplasmic reticulum stress and mitochondrial cell death pathways mediate A53T mutant alpha-synuclein-induced toxicity. Hum Mol Genet. 2005;14:3801–3811. doi: 10.1093/hmg/ddi396. [DOI] [PubMed] [Google Scholar]
- 130.Snyder H., Mensah K., Theisler C., Lee J., Matouschek A., Wolozin B. Aggregated and monomeric α-synuclein bind to the S6′ proteasomal protein and inhibit proteasomal function. J Biol Chem. 2003;278:11753–11759. doi: 10.1074/jbc.M208641200. [DOI] [PubMed] [Google Scholar]
- 131.Baba M., Nakajo S., Tu P.-H., Tomita T., Nakaya K., Lee V. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am J Pathol. 1998;152:879–884. [PMC free article] [PubMed] [Google Scholar]
- 132.Engelender S., Kaminsky Z., Guo X., Sharp A.H., Amaravi R.K., Kleiderlein J.J. Synphilin-1 associates with α-synuclein and promotes the formation of cytosolic inclusions. Nat Genet. 1999;22:110–114. doi: 10.1038/8820. [DOI] [PubMed] [Google Scholar]
- 133.Spillantini M.G., Crowther R.A., Jakes R., Hasegawa M., Goedert M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with Lewy bodies. Proc Natl Acad Sci. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Devic I., Hwang H., Edgar J.S., Izutsu K., Presland R., Pan C. Salivary α-synuclein and DJ-1: potential biomarkers for Parkinson's disease. Brain. 2011;134 doi: 10.1093/brain/awr015. e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Al-Nimer M.S., Mshatat S.F., Abdulla H.I. Saliva α-synuclein and a high extinction coefficient protein: a novel approach in assessment biomarkers of Parkinson's disease. N Am J Med Sci. 2014;6:633–637. doi: 10.4103/1947-2714.147980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vivacqua G., Latorre A., Suppa A., Nardi M., Pietracupa S., Mancinelli R. Abnormal salivary total and oligomeric alpha-synuclein in Parkinson's disease. PLoS One. 2016;11 doi: 10.1371/journal.pone.0151156. e0151156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen J., Li L., Chin L.S. Parkinson disease protein DJ-1 converts from a zymogen to a protease by carboxyl-terminal cleavage. Hum Mol Genet. 2010;19:2395–2408. doi: 10.1093/hmg/ddq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Clements C.M., McNally R.S., Conti B.J., Mak T.W., Ting J.P.Y. DJ-1, a cancer-and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tang B., Xiong H., Sun P., Zhang Y., Wang D., Hu Z. Association of PINK1 and DJ-1 confers digenic inheritance of early-onset Parkinson's disease. Hum Mol Genet. 2006;15:1816–1825. doi: 10.1093/hmg/ddl104. [DOI] [PubMed] [Google Scholar]
- 140.Tanti G.K., Goswami S.K. SG2NA recruits DJ-1 and Akt into the mitochondria and membrane to protect cells from oxidative damage. Free Radic Biol Med. 2014;75:1–13. doi: 10.1016/j.freeradbiomed.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 141.Xiong H., Wang D., Chen L., Choo Y.S., Ma H., Tang C. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119:650. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Junn E., Jang W.H., Zhao X., Jeong B.S., Mouradian M.M. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J Neurosci Res. 2009;87:123–129. doi: 10.1002/jnr.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sekito A., Koide-Yoshida S., Niki T., Taira T., Iguchi-Ariga S.M., Ariga H. DJ-1 interacts with HIPK1 and affects H2O2-induced cell death. Free Radic Res. 2006;40:155–165. doi: 10.1080/10715760500456847. [DOI] [PubMed] [Google Scholar]
- 144.Kim K.S., Kim J.S., Park J.Y., Suh Y.H., Jou I., Joe E.H. DJ-1 associates with lipid rafts by palmitoylation and regulates lipid rafts-dependent endocytosis in astrocytes. Hum Mol Genet. 2013;22:4805–4817. doi: 10.1093/hmg/ddt332. [DOI] [PubMed] [Google Scholar]
- 145.Waak J., Weber S.S., Waldenmaier A., Görner K., Alunni-Fabbroni M., Schell H. Regulation of astrocyte inflammatory responses by the Parkinson's disease-associated gene DJ-1. FASEB J. 2009;23:2478–2489. doi: 10.1096/fj.08-125153. [DOI] [PubMed] [Google Scholar]
- 146.Shendelman S., Jonason A., Martinat C., Leete T., Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits α-synuclein aggregate formation. PLoS Biol. 2004;2:e362. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kang W.Y., Yang Q., Jiang X.F., Chen W., Zhang L.Y., Wang X.Y. Salivary DJ-1 could be an indicator of Parkinson's disease progression. Front Aging Neurosci. 2014;6:102. doi: 10.3389/fnagi.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Masters J.M., Noyce A.J., Warner T.T., Giovannoni G., Proctor G.B. Elevated salivary protein in Parkinson's disease and salivary DJ-1 as a potential marker of disease severity. Park Relat Disord. 2015;21:1251–1255. doi: 10.1016/j.parkreldis.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 149.Bohnen N.I., Müller M.L., Kotagal V., Koeppe R.A., Kilbourn M.R., Gilman S. Heterogeneity of cholinergic denervation in Parkinson's disease without dementia. J Cerebr Blood Flow Metab. 2012;32:1609–1617. doi: 10.1038/jcbfm.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kuhl D.E., Minoshima S., Fessler J.A., Ficaro E., Wieland D., Koeppe R.A. In vivo mapping of cholinergic terminals in normal aging, Alzheimer's disease, and Parkinson's disease. Ann Neurol. 1996;40:399–410. doi: 10.1002/ana.410400309. [DOI] [PubMed] [Google Scholar]
- 151.Müller M.L., Bohnen N.I. Cholinergic dysfunction in Parkinson's disease. Curr Neurol Neurosci Rep. 2013;13:377. doi: 10.1007/s11910-013-0377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bohnen N.I., Frey K.A., Studenski S., Kotagal V., Koeppe R.A., Scott P.J. Gait speed in Parkinson disease correlates with cholinergic degeneration. Neurology. 2013;81:1611–1616. doi: 10.1212/WNL.0b013e3182a9f558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Moreau C., Devos D., Defebvre L. Acetylcholinesterase inhibitors and gait: a steadying hand? Lancet Neurol. 2016;15:232–233. doi: 10.1016/S1474-4422(16)00003-X. [DOI] [PubMed] [Google Scholar]
- 154.Fedorova T., Knudsen C.S., Mouridsen K., Nexo E., Borghammer P. Salivary acetylcholinesterase activity is increased in Parkinson's disease: a potential marker of parasympathetic dysfunction. Parkinson's Dis. 2015;2015 doi: 10.1155/2015/156479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gusella J.F., Wexler N.S., Conneally P.M., Naylor S.L., Anderson M.A., Tanzi R.E. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306:234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 156.MacDonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 157.Ross C.A., Tabrizi S.J. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 158.Tabrizi S.J., Langbehn D.R., Leavitt B.R., Roos R.A., Durr A., Craufurd D. Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8:791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Saudou F., Humbert S. The biology of huntingtin. Neuron. 2016;89:910–926. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 160.El-Daher M.T., Hangen E., Bruyère J., Poizat G., Al-Ramahi I., Pardo R. Huntingtin proteolysis releases non-polyQ fragments that cause toxicity through dynamin 1 dysregulation. EMBO J. 2015;34:2255–2271. doi: 10.15252/embj.201490808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Davies S.W., Turmaine M., Cozens B.A., DiFiglia M., Sharp A.H., Ross C.A. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 162.Miller J., Arrasate M., Shaby B.A., Mitra S., Masliah E., Finkbeiner S. Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into huntington's disease molecular pathogenesis. J Neurosci. 2010;30:10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Saudou F., Finkbeiner S., Devys D., Greenberg M.E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 164.Sapp E., Schwarz C., Chase K., Bhide P., Young A., Penney J. Huntingtin localization in brains of normal and Huntington's disease patients. Ann Neurol. 1997;42:604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 165.Massai L., Petricca L., Magnoni L., Rovetini L., Haider S., Andre R. Development of an ELISA assay for the quantification of soluble huntingtin in human blood cells. BMC Biochem. 2013;14:34. doi: 10.1186/1471-2091-14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Weiss A., Träger U., Wild E.J., Grueninger S., Farmer R., Landles C. Mutant huntingtin fragmentation in immune cells tracks Huntington's disease progression. J Clin Invest. 2012;122:3731–3736. doi: 10.1172/JCI64565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wild E.J., Boggio R., Langbehn D., Robertson N., Haider S., Miller J.R. Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington's disease patients. J Clin Invest. 2015;125:1979–1986. doi: 10.1172/JCI80743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Corey-Bloom J., Aikin A., Garza M., Haque A., Park S., Herndon A. Salivary huntington protein as a peripheral biomarker for Huntington's disease (P1.053) Neurology. 2016;86 [Google Scholar]
- 169.Mehta P., Kaye W., Bryan L., Larson T.C., Copeland T., Wu J. Prevalence of amyotrophic lateral sclerosis—United States, 2012–2013. MMWR Surveill Summ. 2016;65:1–12. doi: 10.15585/mmwr.ss6508a1. [DOI] [PubMed] [Google Scholar]
- 170.Lomen-Hoerth C., Murphy J., Langmore S., Kramer J., Olney R., Miller B. Are amyotrophic lateral sclerosis patients cognitively normal? Neurology. 2003;60:1094–1097. doi: 10.1212/01.wnl.0000055861.95202.8d. [DOI] [PubMed] [Google Scholar]
- 171.Phukan J., Pender N.P., Hardiman O. Cognitive impairment in amyotrophic lateral sclerosis. Lancet Neurol. 2007;6:994–1003. doi: 10.1016/S1474-4422(07)70265-X. [DOI] [PubMed] [Google Scholar]
- 172.Brooks B.R., Miller R.G., Swash M., Munsat T.L. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Mot Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 173.Hayward C., Swingler R.J., Simpson S.A., Brock D. A specific superoxide dismutase mutation is on the same genetic background in sporadic and familial cases of amyotrophic lateral sclerosis. Am J Hum Genet. 1996;59:1165–1167. [PMC free article] [PubMed] [Google Scholar]
- 174.Pensato V., Tiloca C., Corrado L., Bertolin C., Sardone V., Del Bo R. TUBA4A gene analysis in sporadic amyotrophic lateral sclerosis: identification of novel mutations. J Neurol. 2015;262:1376–1378. doi: 10.1007/s00415-015-7739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Saberi S., Stauffer J.E., Schulte D.J., Ravits J. Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol Clin. 2015;33:855–876. doi: 10.1016/j.ncl.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cellura E., Spataro R., Taiello A.C., La Bella V. Factors affecting the diagnostic delay in amyotrophic lateral sclerosis. Clin Neurol Neurosurg. 2012;114:550–554. doi: 10.1016/j.clineuro.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 178.Paganoni S., Macklin E.A., Lee A., Murphy A., Chang J., Zipf A. Diagnostic timelines and delays in diagnosing amyotrophic lateral sclerosis (ALS) Amyotroph Lateral Scler Frontotemporal Degener. 2014;15:453–456. doi: 10.3109/21678421.2014.903974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Williams J.R., Fitzhenry D., Grant L., Martyn D., Kerr D.A. Diagnosis pathway for patients with amyotrophic lateral sclerosis: retrospective analysis of the US Medicare longitudinal claims database. BMC Neurol. 2013;13:160. doi: 10.1186/1471-2377-13-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Riviere M., Meininger V., Zeisser P., Munsat T. An analysis of extended survival in patients with amyotrophic lateral sclerosis treated with riluzole. Arch Neurol. 1998;55:526–528. doi: 10.1001/archneur.55.4.526. [DOI] [PubMed] [Google Scholar]
- 181.Yorkston K. Early intervention in amyotrophic lateral sclerosis: a case presentation. Augment Altern Commun. 1989;5:67–70. [Google Scholar]
- 182.Taupenot L., Harper K.L., O'connor D.T. The chromogranin–secretogranin family. N Engl J Med. 2003;348:1134–1149. doi: 10.1056/NEJMra021405. [DOI] [PubMed] [Google Scholar]
- 183.Ezzi S.A., Larivière R., Urushitani M., Julien J.P. Neuronal over-expression of chromogranin A accelerates disease onset in a mouse model of ALS. J Neurochem. 2010;115:1102–1111. doi: 10.1111/j.1471-4159.2010.06979.x. [DOI] [PubMed] [Google Scholar]
- 184.Schrott-Fischer A., Bitsche M., Humpel C., Walcher C., Maier H., Jellinger K. Chromogranin peptides in amyotrophic lateral sclerosis. Regul Pept. 2009;152:13–21. doi: 10.1016/j.regpep.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 185.Urushitani M., Sik A., Sakurai T., Nukina N., Takahashi R., Julien J.-P. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- 186.Obayashi K., Sato K., Shimazaki R., Ishikawa T., Goto K., Ueyama H. Salivary chromogranin A: useful and quantitative biochemical marker of affective state in patients with amyotrophic lateral sclerosis. Intern Med. 2008;47:1875–1879. doi: 10.2169/internalmedicine.47.1278. [DOI] [PubMed] [Google Scholar]
- 187.Ahlgren C., Odén A., Lycke J. High nationwide prevalence of multiple sclerosis in Sweden. Multiple Scler J. 2011;17:901–908. doi: 10.1177/1352458511403794. [DOI] [PubMed] [Google Scholar]
- 188.Compston A., Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 189.Orton S.-M., Ramagopalan S.V., Brocklebank D., Herrera B.M., Dyment D.A., Yee I.M. Effect of immigration on multiple sclerosis sex ratio in Canada: the Canadian collaborative study. J Neurol Neurosurg Psychiatry. 2010;81:31–36. doi: 10.1136/jnnp.2008.162784. [DOI] [PubMed] [Google Scholar]
- 190.Wallin M.T., Culpepper W.J., Coffman P., Pulaski S., Maloni H., Mahan C.M. The Gulf War era multiple sclerosis cohort: age and incidence rates by race, sex and service. Brain. 2012;135:1778–1785. doi: 10.1093/brain/aws099. [DOI] [PubMed] [Google Scholar]
- 191.Lassmann H., Brück W., Lucchinetti C.F. The immunopathology of multiple sclerosis: an overview. Brain Pathol. 2007;17:210–218. doi: 10.1111/j.1750-3639.2007.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Steinman L. Multiple sclerosis: a coordinated immunological attack against myelin in the central nervous system. Cell. 1996;85:299–302. doi: 10.1016/s0092-8674(00)81107-1. [DOI] [PubMed] [Google Scholar]
- 193.Rae-Grant A.D., Eckert N.J., Bartz S., Reed J.F. Sensory symptoms of multiple sclerosis: a hidden reservoir of morbidity. Multiple Scler J. 1999;5:179–183. doi: 10.1177/135245859900500307. [DOI] [PubMed] [Google Scholar]
- 194.Betts C.D., D'Mellow M., Fowler C.J. Urinary symptoms and the neurological features of bladder dysfunction in multiple sclerosis. J Neurol Neurosurg Psychiatry. 1993;56:245–250. doi: 10.1136/jnnp.56.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Krupp L.B., Alvarez L.A., LaRocca N.G., Scheinberg L.C. Fatigue in multiple sclerosis. Arch Neurol. 1988;45:435–437. doi: 10.1001/archneur.1988.00520280085020. [DOI] [PubMed] [Google Scholar]
- 196.Rizzo M., Hadjimichael O., Preiningerova J., Vollmer T. Prevalence and treatment of spasticity reported by multiple sclerosis patients. Multiple Scler J. 2004;10:589–595. doi: 10.1191/1352458504ms1085oa. [DOI] [PubMed] [Google Scholar]
- 197.Stevens V., Goodman K., Rough K., Kraft G.H. Gait impairment and optimizing mobility in multiple sclerosis. Phys Med Rehabil Clin. 2013;24:573–592. doi: 10.1016/j.pmr.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 198.Chiaravalloti N.D., DeLuca J. Cognitive impairment in multiple sclerosis. Lancet Neurol. 2008;7:1139–1151. doi: 10.1016/S1474-4422(08)70259-X. [DOI] [PubMed] [Google Scholar]
- 199.Chwastiak L., Ehde D.M., Gibbons L.E., Sullivan M., Bowen J.D., Kraft G.H. Depressive symptoms and severity of illness in multiple sclerosis: epidemiologic study of a large community sample. Am J Psychiatry. 2002;159:1862–1868. doi: 10.1176/appi.ajp.159.11.1862. [DOI] [PubMed] [Google Scholar]
- 200.Sadovnick A., Eisen K., Ebers G.C., Paty D.W. Cause of death in patients attending multiple sclerosis clinics. Neurology. 1991;41:1193–1196. doi: 10.1212/wnl.41.8.1193. [DOI] [PubMed] [Google Scholar]
- 201.Stenager E.N., Stenager E., Koch-Henriksen N., Brønnum-Hansen H., Hyllested K., Jensen K. Suicide and multiple sclerosis: an epidemiological investigation. J Neurol Neurosurg Psychiatry. 1992;55:542–545. doi: 10.1136/jnnp.55.7.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Miller D., Weinshenker B.G., Filippi M., Banwell B., Cohen J., Freedman M. Differential diagnosis of suspected multiple sclerosis: a consensus approach. Multiple Scler J. 2008;14:1157–1174. doi: 10.1177/1352458508096878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Janeway C.A., Jr., Travers P., Walport M., Shlomchik M.J. Garland Science; New York: 2001. The major histocompatibility complex and its functionshttps. Immunobiology: the immune system in health and disease. [Google Scholar]
- 204.Chastain E.M., D'Anne S.D., Rodgers J.M., Miller S.D. The role of antigen presenting cells in multiple sclerosis. Biochim Biophys Acta (BBA) Mol Basis Dis. 2011;1812:265–274. doi: 10.1016/j.bbadis.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Riedhammer C., Weissert R. Antigen presentation, autoantigens, and immune regulation in multiple sclerosis and other autoimmune diseases. Front Immunol. 2015;6:322. doi: 10.3389/fimmu.2015.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Stoeckle C., Tolosa E. Antigen processing and presentation in multiple sclerosis. Results Probl Cell Differ. 2010;51:149–172. doi: 10.1007/400_2009_22. [DOI] [PubMed] [Google Scholar]
- 207.Hillert J., Olerup O. Multiple sclerosis is associated with genes within or close to the HLA-DR-DQ subregion on a normal DR15, DQ6, Dw2 haplotype. Neurology. 1993;43:163–168. doi: 10.1212/wnl.43.1_part_1.163. [DOI] [PubMed] [Google Scholar]
- 208.Martin R., Howell M.D., Jaraquemada D., Flerlage M., Richert J., Brostoff S. A myelin basic protein peptide is recognized by cytotoxic T cells in the context of four HLA-DR types associated with multiple sclerosis. J Exp Med. 1991;173:19–24. doi: 10.1084/jem.173.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Oksenberg J.R., Barcellos L.F., Cree B.A., Baranzini S.E., Bugawan T.L., Khan O. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet. 2004;74:160–167. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ott M., Stecker K., Seidl C., Seifried E., Westhoff U., Grosse-Wilde H. Soluble HLA class I and class II antigens in patients with multiple sclerosis. HLA. 1998;51:301–304. doi: 10.1111/j.1399-0039.1998.tb03106.x. [DOI] [PubMed] [Google Scholar]
- 211.Olerup O., Hillert J. HLA class II-associated genetic susceptibility in multiple sclerosis: a critical evaluation. HLA. 1991;38:1–15. doi: 10.1111/j.1399-0039.1991.tb02029.x. [DOI] [PubMed] [Google Scholar]
- 212.Adamashvili I., Kelley R.E., Pressly T., McDonald J.C., Soluble H.L.A. Patterns of expression in normal subjects, autoimmune diseases, and transplant recipients. Rheumatol Int. 2005;25:491–500. doi: 10.1007/s00296-005-0585-y. [DOI] [PubMed] [Google Scholar]
- 213.Charlton R.K., Zmijewski C.M. Soluble HL-A7 antigen: localization in the β-lipoprotein fraction of human serum. Science. 1970;170:636–637. doi: 10.1126/science.170.3958.636. [DOI] [PubMed] [Google Scholar]
- 214.Güssow D., Ploegh H. Soluble class I antigens: a conundrum with no solution? Immunol today. 1987;8:220–222. doi: 10.1016/0167-5699(87)90170-8. [DOI] [PubMed] [Google Scholar]
- 215.Jankowska-Gan E., Rhein T., Haynes L.D., Geissler F., Mulder A., Kalayoglu M. Human liver allograft acceptance and the “tolerance assay”: II. donor HLA-A,-B but not DR antigens are able to trigger regulation of DTH. Hum Immunol. 2002;63:862–870. doi: 10.1016/s0198-8859(02)00450-0. [DOI] [PubMed] [Google Scholar]
- 216.Adamashvili I., Minagar A., Gonzalez-Toledo E., Featherston L., Kelley R.E. Soluble HLA measurement in saliva and cerebrospinal fluid in Caucasian patients with multiple sclerosis: a preliminary study. J Neuroinflammation. 2005;2:13. doi: 10.1186/1742-2094-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Minagar A., Adamashvili I., Kelley R.E., Gonzalez-Toledo E., McLarty J., Smith S.J. Saliva soluble HLA as a potential marker of response to interferon-β1a in multiple sclerosis: a preliminary study. J Neuroinflammation. 2007;4:16. doi: 10.1186/1742-2094-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sies H. Hydroperoxides and thiol oxidants in the study of oxidative stress in intact cells and organs. Oxid Stress. 1985:73–90. [Google Scholar]
- 219.Van Horssen J., Witte M.E., Schreibelt G., De Vries H.E. Radical changes in multiple sclerosis pathogenesis. Biochim Biophys Acta (BBA) Mol Basis Dis. 2011;1812:141–150. doi: 10.1016/j.bbadis.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 220.Ortiz G.G., Pacheco-Moisés F.P., Bitzer-Quintero O.K., Ramírez-Anguiano A.C., Flores-Alvarado L.J., Ramírez-Ramírez V. Immunology and oxidative stress in multiple sclerosis: clinical and basic approach. Clin Dev Immunol. 2013;2013 doi: 10.1155/2013/708659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang P., Xie K., Wang C., Bi J. Oxidative stress induced by lipid peroxidation is related with inflammation of demyelination and neurodegeneration in multiple sclerosis. Eur Neurol. 2014;72:249–254. doi: 10.1159/000363515. [DOI] [PubMed] [Google Scholar]
- 222.Karlík M., Valkovič P., Hančinová V., Krížová L., Tóthová Ľ., Celec P. Markers of oxidative stress in plasma and saliva in patients with multiple sclerosis. Clin Biochem. 2015;48:24–28. doi: 10.1016/j.clinbiochem.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 223.Naidoo R., Knapp M.L. Studies of lipid peroxidation products in cerebrospinal fluid and serum in multiple sclerosis and other conditions. Clin Chem. 1992;38:2449–2454. [PubMed] [Google Scholar]
- 224.Ghabaee M., Jabedari B., Al-E-Eshagh N., Ghaffarpour M., Asadi F. Serum and cerebrospinal fluid antioxidant activity and lipid peroxidation in Guillain–Barre syndrome and multiple sclerosis patients. Int J Neurosci. 2010;120:301–304. doi: 10.3109/00207451003695690. [DOI] [PubMed] [Google Scholar]
- 225.Hunter M., Nlemadim B., Davidson D. Lipid peroxidation products and antioxidant proteins in plasma and cerebrospinal fluid from multiple sclerosis patients. Neurochem Res. 1985;10:1645–1652. doi: 10.1007/BF00988606. [DOI] [PubMed] [Google Scholar]
- 226.Association A.P. Auteur; Washington, DC: 2013. Diagnostic and statistical manual of mental disorders-DSM-5. [DOI] [PubMed] [Google Scholar]
- 227.Bailey A., Le Couteur A., Gottesman I., Bolton P., Simonoff E., Yuzda E. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 228.De Rubeis S., Buxbaum J.D. Genetics and genomics of autism spectrum disorder: embracing complexity. Hum Mol Genet. 2015;24:R24–R31. doi: 10.1093/hmg/ddv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Xu L.-M., Li J.-R., Huang Y., Zhao M., Tang X., Wei L. AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res. 2011;40:D1016–D1022. doi: 10.1093/nar/gkr1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kern J.K., Geier D.A., Sykes L.K., Geier M.R. Evidence of neurodegeneration in autism spectrum disorder. Transl Neurodegener. 2013;2:17. doi: 10.1186/2047-9158-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Pardo C.A., Vargas D.L., Zimmerman A.W. Immunity, neuroglia and neuroinflammation in autism. Int Rev Psychiatr. 2005;17:485–495. doi: 10.1080/02646830500381930. [DOI] [PubMed] [Google Scholar]
- 232.Akshoomoff N., Pierce K., Courchesne E. The neurobiological basis of autism from a developmental perspective. Dev Psychopathol. 2002;14:613–634. doi: 10.1017/s0954579402003115. [DOI] [PubMed] [Google Scholar]
- 233.Amaral D.G., Schumann C.M., Nordahl C.W. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 234.Muller C.L., Anacker A.M., Veenstra-VanderWeele J. The serotonin system in autism spectrum disorder: from biomarker to animal models. Neuroscience. 2016;321:24–41. doi: 10.1016/j.neuroscience.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ruggeri B., Sarkans U., Schumann G., Persico A.M. Biomarkers in autism spectrum disorder: the old and the new. Psychopharmacology. 2014;231:1201–1216. doi: 10.1007/s00213-013-3290-7. [DOI] [PubMed] [Google Scholar]
- 236.Zwaigenbaum L., Bauman M.L., Fein D., Pierce K., Buie T., Davis P.A. Early screening of autism spectrum disorder: recommendations for practice and research. Pediatrics. 2015;136:S41–S59. doi: 10.1542/peds.2014-3667D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zwaigenbaum L., Bauman M.L., Stone W.L., Yirmiya N., Estes A., Hansen R.L. Early identification of autism spectrum disorder: recommendations for practice and research. Pediatrics. 2015;136:S10–S40. doi: 10.1542/peds.2014-3667C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Osterling J.A., Dawson G., Munson J.A. Early recognition of 1-year-old infants with autism spectrum disorder versus mental retardation. Dev Psychopathol. 2002;14:239–251. doi: 10.1017/s0954579402002031. [DOI] [PubMed] [Google Scholar]
- 239.Eikeseth S., Klintwall L., Jahr E., Karlsson P. Outcome for children with autism receiving early and intensive behavioral intervention in mainstream preschool and kindergarten settings. Res Autism Spectr Disord. 2012;6:829–835. [Google Scholar]
- 240.Flashner B.M., Russo M.E., Boileau J.E., Leong D.W., Gallicano G.I. Epigenetic factors and autism spectrum disorders. Neuromolecular Med. 2013;15:339–350. doi: 10.1007/s12017-013-8222-5. [DOI] [PubMed] [Google Scholar]
- 241.Mellios N., Sur M. The emerging role of microRNAs in schizophrenia and autism spectrum disorders. Front Psychiatry. 2012;3:39. doi: 10.3389/fpsyt.2012.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wink L.K., Plawecki M.H., Erickson C.A., Stigler K.A., McDougle C.J. Emerging drugs for the treatment of symptoms associated with autism spectrum disorders. Expet Opin Emerg Drugs. 2010;15:481–494. doi: 10.1517/14728214.2010.487860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hicks S.D., Ignacio C., Gentile K., Middleton F.A. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016;16:52. doi: 10.1186/s12887-016-0586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sparrow S.S., Balla D.A., Cicchetti D.V., Harrison P.L., Doll E.A. American Guidance Service; Circle Pines, MN: 1984. Vineland adaptive behavior scales. [Google Scholar]
- 245.Ashwood P., Krakowiak P., Hertz-Picciotto I., Hansen R., Pessah I., Van de Water J. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav Immun. 2011;25:40–45. doi: 10.1016/j.bbi.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Estes M.L., McAllister A.K. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci. 2015;16:469–486. doi: 10.1038/nrn3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Gottfried C., Bambini-Junior V., Francis F., Riesgo R., Savino W. The impact of neuroimmune alterations in autism spectrum disorder. Front Psychiatry. 2015;6:121. doi: 10.3389/fpsyt.2015.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Keil A., Daniels J.L., Forssen U., Hultman C., Cnattingius S., Söderberg K.C. Parental autoimmune diseases associated with autism spectrum disorders in offspring. Epidemiology. 2010;21:805–808. doi: 10.1097/EDE.0b013e3181f26e3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Samsam M., Ahangari R., Naser S.A. Pathophysiology of autism spectrum disorders: revisiting gastrointestinal involvement and immune imbalance. World J Gastroenterol: WJG. 2014;20:9942–9951. doi: 10.3748/wjg.v20.i29.9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wetie N., Armand G., Wormwood K.L., Russell S., Ryan J.P., Darie C.C. A pilot proteomic analysis of salivary biomarkers in autism spectrum disorder. Autism Res. 2015;8:338–350. doi: 10.1002/aur.1450. [DOI] [PubMed] [Google Scholar]
- 251.Castagnola M., Messana I., Inzitari R., Fanali C., Cabras T., Morelli A. Hypo-phosphorylation of salivary peptidome as a clue to the molecular pathogenesis of autism spectrum disorders. J Proteome Res. 2008;7:5327–5332. doi: 10.1021/pr8004088. [DOI] [PubMed] [Google Scholar]
- 252.Galbally M., Lewis A.J., IJzendoorn M.V., Permezel M. The role of oxytocin in mother-infant relations: a systematic review of human studies. Harv Rev Psychiatry. 2011;19:1–14. doi: 10.3109/10673229.2011.549771. [DOI] [PubMed] [Google Scholar]
- 253.Heinrichs M., von Dawans B., Domes G. Oxytocin, vasopressin, and human social behavior. Front Neuroendocrinol. 2009;30:548–557. doi: 10.1016/j.yfrne.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 254.Stoop R. Neuromodulation by oxytocin and vasopressin. Neuron. 2012;76:142–159. doi: 10.1016/j.neuron.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 255.Guastella A.J., Einfeld S.L., Gray K.M., Rinehart N.J., Tonge B.J., Lambert T.J. Intranasal oxytocin improves emotion recognition for youth with autism spectrum disorders. Biol Psychiatry. 2010;67:692–694. doi: 10.1016/j.biopsych.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 256.Hollander E., Bartz J., Chaplin W., Phillips A., Sumner J., Soorya L. Oxytocin increases retention of social cognition in autism. Biol Psychiatry. 2007;61:498–503. doi: 10.1016/j.biopsych.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 257.Hollander E., Novotny S., Hanratty M., Yaffe R., DeCaria C.M., Aronowitz B.R. Oxytocin infusion reduces repetitive behaviors in adults with autistic and Asperger's disorders. Neuropsychopharmacology. 2003;28:193–198. doi: 10.1038/sj.npp.1300021. [DOI] [PubMed] [Google Scholar]
- 258.Yrigollen C.M., Han S.S., Kochetkova A., Babitz T., Chang J.T., Volkmar F.R. Genes controlling affiliative behavior as candidate genes for autism. Biol Psychiatr. 2008;63:911–916. doi: 10.1016/j.biopsych.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Feldman R., Golan O., Hirschler-Guttenberg Y., Ostfeld-Etzion S., Zagoory-Sharon O. Parent-child interaction and oxytocin production in pre-schoolers with autism spectrum disorder. Br J Psychiatry. 2014;205:107–112. doi: 10.1192/bjp.bp.113.137513. [DOI] [PubMed] [Google Scholar]
- 260.Fujisawa T.X., Tanaka S., Saito D.N., Kosaka H., Tomoda A. Visual attention for social information and salivary oxytocin levels in preschool children with autism spectrum disorders: an eye-tracking study. Front Neurosci. 2014;8:295. doi: 10.3389/fnins.2014.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Rutigliano G., Rocchetti M., Paloyelis Y., Gilleen J., Sardella A., Cappucciati M. Peripheral oxytocin and vasopressin: biomarkers of psychiatric disorders? A comprehensive systematic review and preliminary meta-analysis. Psychiatry Res. 2016;241:207–220. doi: 10.1016/j.psychres.2016.04.117. [DOI] [PubMed] [Google Scholar]
- 262.Crow T.J. Molecular pathology of schizophrenia: more than one disease process? Br Med J. 1980;280:66–68. doi: 10.1136/bmj.280.6207.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lenzenweger M.F., Dworkin R.H., Wethington E. Examining the underlying structure of schizophrenic phenomenology: evidence for a three-process model. Schizophr Bull. 1991;17:515–524. doi: 10.1093/schbul/17.3.515. [DOI] [PubMed] [Google Scholar]
- 264.Strauss J.S., Carpenter W.T., Bartko J.J. Speculations on the processes that underlie schizophrenic symptoms and signs: III. Schizophr Bull. 1974;1:61–69. doi: 10.1093/schbul/1.11.61. [DOI] [PubMed] [Google Scholar]
- 265.Kring A.M., Johnson S.L., Davison G.C., Neale J.M. 13th Ed. Wiley; 2015. Abnormal psychology. [Google Scholar]
- 266.Hawton K., Sutton L., Haw C., Sinclair J., Harriss L. Suicide and attempted suicide in bipolar disorder: a systematic review of risk factors. J Clin Psychiatry. 2005;66:693–704. doi: 10.4088/jcp.v66n0604. [DOI] [PubMed] [Google Scholar]
- 267.Hor K., Taylor M. Suicide and schizophrenia: a systematic review of rates and risk factors. J Psychopharmacol. 2010;24:81–90. doi: 10.1177/1359786810385490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Consortium C-DGotPG Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Consortium IS Common polygenic variation contributes to risk of schizophrenia that overlaps with bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Consortium SPG-WAS Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Group PGCBDW Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lasky-Su J., Neale B.M., Franke B., Anney R.J., Zhou K., Maller J.B. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet Part B: Neuropsychiatric Genet. 2008;147:1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 273.Craddock N., O’donovan M., Owen M. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet. 2005;42:193–204. doi: 10.1136/jmg.2005.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Dempster E.L., Pidsley R., Schalkwyk L.C., Owens S., Georgiades A., Kane F. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Guidotti A., Auta J., Chen Y., Davis J., Dong E., Gavin D. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology. 2011;60:1007–1016. doi: 10.1016/j.neuropharm.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Mill J., Petronis A. Pre-and peri-natal environmental risks for attention-deficit hyperactivity disorder (ADHD): the potential role of epigenetic processes in mediating susceptibility. J Child Psychol Psychiatry. 2008;49:1020–1030. doi: 10.1111/j.1469-7610.2008.01909.x. [DOI] [PubMed] [Google Scholar]
- 277.Petronis A. The origin of schizophrenia: genetic thesis, epigenetic antithesis, and resolving synthesis. Biol Psychiatry. 2004;55:965–970. doi: 10.1016/j.biopsych.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 278.Rao J., Keleshian V., Klein S., Rapoport S. Epigenetic modifications in frontal cortex from Alzheimer's disease and bipolar disorder patients. Transl Psychiatry. 2012;2:e132. doi: 10.1038/tp.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Roth T.L., Lubin F.D., Sodhi M., Kleinman J.E. Epigenetic mechanisms in schizophrenia. Biochim Biophys Acta (BBA) Gen Subj. 2009;1790:869–877. doi: 10.1016/j.bbagen.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Abdolmaleky H.M., Pajouhanfar S., Faghankhani M., Joghataei M.T., Mostafavi A., Thiagalingam S. Antipsychotic drugs attenuate aberrant DNA methylation of DTNBP1 (dysbindin) promoter in saliva and post-mortem brain of patients with schizophrenia and Psychotic bipolar disorder. Am J Med Genet Part B: Neuropsychiatric Genet. 2015;168:687–696. doi: 10.1002/ajmg.b.32361. [DOI] [PubMed] [Google Scholar]
- 281.Ghadirivasfi M., Nohesara S., Ahmadkhaniha H.R., Eskandari M.R., Mostafavi S., Thiagalingam S. Hypomethylation of the serotonin receptor type-2A Gene (HTR2A) at T102C polymorphic site in DNA derived from the saliva of patients with schizophrenia and bipolar disorder. Am J Med Genet Part B: Neuropsychiatric Genet. 2011;156:536–545. doi: 10.1002/ajmg.b.31192. [DOI] [PubMed] [Google Scholar]
- 282.Nohesara S., Ghadirivasfi M., Mostafavi S., Eskandari M.-R., Ahmadkhaniha H., Thiagalingam S. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. J Psychiatry Res. 2011;45:1432–1438. doi: 10.1016/j.jpsychires.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 283.Wilmot B., Fry R., Smeester L., Musser E.D., Mill J., Nigg J.T. Methylomic analysis of salivary DNA in childhood ADHD identifies altered DNA methylation in VIPR2. J Child Psychol Psychiatry. 2016;57:152–160. doi: 10.1111/jcpp.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Craddock N., O'donovan M.C., Owen M.J. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32:9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.López-Figueroa A.L., Norton C.S., López-Figueroa M.O., Armellini-Dodel D., Burke S., Akil H. Serotonin 5-HT 1A, 5-HT 1B, and 5-HT 2A receptor mRNA expression in subjects with major depression, bipolar disorder, and schizophrenia. Biol Psychiatry. 2004;55:225–233. doi: 10.1016/j.biopsych.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 286.Bleich A., Brown S.-L., Kahn R., van Praag H.M. The role of serotonin in schizophrenia. Schizophr Bull. 1988;14:297–315. doi: 10.1093/schbul/14.2.297. [DOI] [PubMed] [Google Scholar]
- 287.Cousins D.A., Butts K., Young A.H. The role of dopamine in bipolar disorder. Bipolar Disord. 2009;11:787–806. doi: 10.1111/j.1399-5618.2009.00760.x. [DOI] [PubMed] [Google Scholar]
- 288.Davis K.L., Kahn R.S. Dopamine in schizophrenia: a review and reconceptualization. Am J Psychiatry. 1991;148:1474–1486. doi: 10.1176/ajp.148.11.1474. [DOI] [PubMed] [Google Scholar]
- 289.Mahmood T., Silverstone T. Serotonin and bipolar disorder. J Affect Disord. 2001;66:1–11. doi: 10.1016/s0165-0327(00)00226-3. [DOI] [PubMed] [Google Scholar]
- 290.Alfimova M., Monakhov M., Abramova L., Golubev S., Golimbet V. Polymorphism of serotonin receptor genes (5-HTR2A) and Dysbindin (DTNBP1) and individual components of short-term verbal memory processes in schizophrenia. Neurosci Behav Physiol. 2010;40:934–940. doi: 10.1007/s11055-010-9348-7. [DOI] [PubMed] [Google Scholar]
- 291.Karlsgodt K.H., Robleto K., Trantham-Davidson H., Jairl C., Cannon T.D., Lavin A. Reduced dysbindin expression mediates N-methyl-D-aspartate receptor hypofunction and impaired working memory performance. Biol Psychiatry. 2011;69:28–34. doi: 10.1016/j.biopsych.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Walton E., Liu J., Hass J., White T., Scholz M., Roessner V. MB-COMT promoter DNA methylation is associated with working-memory processing in schizophrenia patients and healthy controls. Epigenetics. 2014;9:1101–1107. doi: 10.4161/epi.29223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Cheah S.-Y., Lawford B.R., Young R., Morris C.P., Voisey J. Dysbindin (DTNBP1) variants are associated with hallucinations in schizophrenia. Eur Psychiatry. 2015;30:486–491. doi: 10.1016/j.eurpsy.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 294.Wright G.E., Niehaus D.J., van der Merwe L., Koen L., Korkie L.J., Kinnear C.J. Association of MB-COMT polymorphisms with schizophrenia-susceptibility and symptom severity in an African cohort. Prog Neuro Psychopharmacol Biol Psychiatry. 2012;39:163–169. doi: 10.1016/j.pnpbp.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 295.Goghari V.M., Sponheim S.R. Differential association of the COMT Val158Met polymorphism with clinical phenotypes in schizophrenia and bipolar disorder. Schizophr Res. 2008;103:186–191. doi: 10.1016/j.schres.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Peter C.J., Fischer L.K., Kundakovic M., Garg P., Jakovcevski M., Dincer A. DNA methylation signatures of early childhood malnutrition associated with impairments in attention and cognition. Biol Psychiatry. 2016;80:765–774. doi: 10.1016/j.biopsych.2016.03.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hamza M., Halayem S., Bourgou S., Daoud M., Charfi F., Belhadj A. Epigenetics and ADHD: Toward An integrative approach of the disorder pathogenesis. J Atten Disord. 2017 doi: 10.1177/1087054717696769. 1087054717696769. [DOI] [PubMed] [Google Scholar]
- 298.Kim Y.K., Jung H.G., Myint A.M., Kim H., Park S.H. Imbalance between pro-inflammatory and anti-inflammatory cytokines in bipolar disorder. J Affect Disord. 2007;104:91–95. doi: 10.1016/j.jad.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 299.Müller N., Riedel M., Gruber R., Ackenheil M., Schwarz M.J. The immune system and schizophrenia: an integrative view. Ann N Y Acad Sci. 2000;917:456–467. doi: 10.1111/j.1749-6632.2000.tb05410.x. [DOI] [PubMed] [Google Scholar]
- 300.Ortiz-Domínguez A., Hernández M.E., Berlanga C., Gutiérrez-Mora D., Moreno J., Heinze G. Immune variations in bipolar disorder: phasic differences. Bipolar Disord. 2007;9:596–602. doi: 10.1111/j.1399-5618.2007.00493.x. [DOI] [PubMed] [Google Scholar]
- 301.Strous R.D., Shoenfeld Y. Schizophrenia, autoimmunity and immune system dysregulation: a comprehensive model updated and revisited. J Autoimmun. 2006;27:71–80. doi: 10.1016/j.jaut.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 302.Iavarone F., Melis M., Platania G., Cabras T., Manconi B., Petruzzelli R. Characterization of salivary proteins of schizophrenic and bipolar disorder patients by top-down proteomics. J Proteom. 2014;103:15–22. doi: 10.1016/j.jprot.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 303.Foell D., Wittkowski H., Vogl T., Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 304.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 305.Magister Š., Kos J. Cystatins in immune system. J Cancer. 2013;4:45–56. doi: 10.7150/jca.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Pietzsch J., Hoppmann S. Human S100A12: a novel key player in inflammation? Amino Acids. 2009;36:381–389. doi: 10.1007/s00726-008-0097-7. [DOI] [PubMed] [Google Scholar]
- 307.Arnaiz A., Zumárraga M., Díez-Altuna I., Uriarte J.J., Moro J., Pérez-Ansorena M.A. Oral health and the symptoms of schizophrenia. Psychiatry Res. 2011;188:24–28. doi: 10.1016/j.psychres.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 308.Bejerot S., Dahllöf G. A cross-sectional study on oral health and dental care in intellectually able adults with autism spectrum disorder. BMC Oral Health. 2015;15:81. doi: 10.1186/s12903-015-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Clark D.B. Dental care for the patient with bipolar disorder. J Can Dent Assoc. 2003;69:20–24. [PubMed] [Google Scholar]
- 310.Mancini M., Grappasonni I., Scuri S., Amenta F. Oral health in Alzheimer's disease: a review. Curr Alzheimer Res. 2010;7:368–373. doi: 10.2174/156720510791162359. [DOI] [PubMed] [Google Scholar]
- 311.Müller T., Palluch R. Caries and periodontal disease in patients with Parkinson's disease. Spec Care Dent. 2011;31:178–181. doi: 10.1111/j.1754-4505.2011.00205.x. [DOI] [PubMed] [Google Scholar]
- 312.Saft C., Andrich J.E., Müller T., Becker J., Jackowski J. Oral and dental health in Huntington‘s disease-an observational study. BMC Neurol. 2013;13:114. doi: 10.1186/1471-2377-13-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Sheu J.J., Lin H.C. Association between multiple sclerosis and chronic periodontitis: a population-based pilot study. Eur J Neurol. 2013;20:1053–1059. doi: 10.1111/ene.12103. [DOI] [PubMed] [Google Scholar]
- 314.Aziz N., Anguelova G., Marinus J., Van Dijk J., Roos R. Autonomic symptoms in patients and pre-manifest mutation carriers of Huntington's disease. Eur J Neurol. 2010;17:1068–1074. doi: 10.1111/j.1468-1331.2010.02973.x. [DOI] [PubMed] [Google Scholar]
- 315.Proulx M., De Courval F.P., Wiseman M.A., Panisset M. Salivary production in Parkinson's disease. Mov Disord. 2005;20:204–207. doi: 10.1002/mds.20189. [DOI] [PubMed] [Google Scholar]
- 316.Ship J.A., Decarli C., Friedland R.P., Baum B.J. Diminished submandibular salivary flow in dementia of the Alzheimer type. J Gerontol. 1990;45:M61–M66. doi: 10.1093/geronj/45.2.m61. [DOI] [PubMed] [Google Scholar]
- 317.Tumilasci O.R., Cersosimo M., Belforte J.E., Micheli F.E., Benarroch E.E., Pazo J.H. Quantitative study of salivary secretion in Parkinson's disease. Mov Disord. 2006;21:660–667. doi: 10.1002/mds.20784. [DOI] [PubMed] [Google Scholar]
- 318.Young C.A., Ellis C., Johnson J., Sathasivam S., Pih N. Treatment for sialorrhea (excessive saliva) in people with motor neuron disease/amyotrophic lateral sclerosis. Cochrane Database Syst Rev. 2011;11 doi: 10.1002/14651858.CD006981.pub2. [DOI] [PubMed] [Google Scholar]
- 319.Donaldson S. Sialorrhea as a side effect of lithium: a case report. Am J Psychiatry. 1982;139:1350–1351. doi: 10.1176/ajp.139.10.1350. [DOI] [PubMed] [Google Scholar]
- 320.Praharaj S.K., Arora M., Gandotra S. Clozapine-induced sialorrhea: pathophysiology and management strategies. Psychopharmacology. 2006;185:265–273. doi: 10.1007/s00213-005-0248-4. [DOI] [PubMed] [Google Scholar]
- 321.Dawes C. Circadian rhythms in human salivary flow rate and composition. J Physiol. 1972;220:529–545. doi: 10.1113/jphysiol.1972.sp009721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Bahn J.H., Zhang Q., Li F., Chan T.-M., Lin X., Kim Y. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Bian S., Sun T. Functions of noncoding RNAs in neural development and neurological diseases. Mol Neurobiol. 2011;44:359–373. doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Keller A., Leidinger P., Lange J., Borries A., Schroers H., Scheffler M. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4:e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Obregon D., Hou H., Deng J., Giunta B., Tian J., Darlington D. sAPP-α modulates β-secretase activity and amyloid-β generation. Nat Commun. 2012;3:777. doi: 10.1038/ncomms1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Plant L.D., Boyle J.P., Smith I.F., Peers C., Pearson H.A. The production of amyloid β peptide is a critical requirement for the viability of central neurons. J Neurosci. 2003;23:5531–5535. doi: 10.1523/JNEUROSCI.23-13-05531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Esteban J.A. Living with the enemy: a physiological role for the β-amyloid peptide. Trends Neurosci. 2004;27:1–3. doi: 10.1016/j.tins.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 328.Li S., Jin M., Koeglsperger T., Shepardson N.E., Shankar G.M., Selkoe D.J. Soluble Aβ oligomers inhibit long-term potentiation through a mechanism involving excessive activation of extrasynaptic NR2B-containing NMDA receptors. J Neurosci. 2011;31:6627–6638. doi: 10.1523/JNEUROSCI.0203-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Tu S., Okamoto S-i, Lipton S.A., Xu H. Oligomeric Aβ-induced synaptic dysfunction in Alzheimer's disease. Mol Neurodegener. 2014;9:48. doi: 10.1186/1750-1326-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.De Felice F.G., Velasco P.T., Lambert M.P., Viola K., Fernandez S.J., Ferreira S.T. Aβ oligomers induce neuronal oxidative stress through an N-methyl-D-aspartate receptor-dependent mechanism that is blocked by the Alzheimer drug memantine. J Biol Chem. 2007;282:11590–11601. doi: 10.1074/jbc.M607483200. [DOI] [PubMed] [Google Scholar]
- 331.Green K.N., LaFerla F.M. Linking calcium to Aβ and Alzheimer's disease. Neuron. 2008;59:190–194. doi: 10.1016/j.neuron.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 332.Manczak M., Anekonda T.S., Henson E., Park B.S., Quinn J., Reddy P.H. Mitochondria are a direct site of Aβ accumulation in Alzheimer's disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum Mol Genet. 2006;15:1437–1449. doi: 10.1093/hmg/ddl066. [DOI] [PubMed] [Google Scholar]
- 333.Sutton E., Thomas T., Bryant M., Landon C., Newton C., Rhodin J. Amyloid-beta peptide induced inflammatory reaction is mediated by the cytokines tumor necrosis factor and interleukin-1. J Submicr Cytol Pathol. 1999;31:313–323. [PubMed] [Google Scholar]
- 334.Pardossi-Piquard R., Checler F. The physiology of the β-amyloid precursor protein intracellular domain AICD. J Neurochem. 2012;120:109–124. doi: 10.1111/j.1471-4159.2011.07475.x. [DOI] [PubMed] [Google Scholar]
- 335.Chang K.A., Suh Y.H. Possible roles of amyloid intracellular domain of amyloid precursor protein. BMB Rep. 2010;43:656–663. doi: 10.5483/BMBRep.2010.43.10.656. [DOI] [PubMed] [Google Scholar]
- 336.Hassan M.I., Waheed A., Yadav S., Singh T., Ahmad F. Prolactin inducible protein in cancer, fertility and immunoregulation: structure, function and its clinical implications. Cell Mol Life Sci. 2009;66:447–459. doi: 10.1007/s00018-008-8463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Blume K.E., Soeroes S., Keppeler H., Stevanovic S., Kretschmer D., Rautenberg M. Cleavage of annexin A1 by ADAM10 during secondary necrosis generates a monocytic “find-me” signal. J Immunol. 2012;188:135–145. doi: 10.4049/jimmunol.1004073. [DOI] [PubMed] [Google Scholar]
- 338.Weyd H., Abeler-Dörner L., Linke B., Mahr A., Jahndel V., Pfrang S. Annexin A1 on the surface of early apoptotic cells suppresses CD8+ T cell immunity. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062449. e62449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ernst S., Lange C., Wilbers A., Goebeler V., Gerke V., Rescher U. An annexin 1 N-terminal peptide activates leukocytes by triggering different members of the formyl peptide receptor family. J Immunol. 2004;172:7669–7676. doi: 10.4049/jimmunol.172.12.7669. [DOI] [PubMed] [Google Scholar]
- 340.Walther A., Riehemann K., Gerke V. A novel ligand of the formyl peptide receptor: annexin I regulates neutrophil extravasation by interacting with the FPR. Mol Cell. 2000;5:831–840. doi: 10.1016/s1097-2765(00)80323-8. [DOI] [PubMed] [Google Scholar]
- 341.Perretti M., D'acquisto F. Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat Rev Immunol. 2009;9:62–70. doi: 10.1038/nri2470. [DOI] [PubMed] [Google Scholar]
- 342.Leoni G., Alam A., Neumann P.-A., Lambeth J.D., Cheng G., McCoy J. Annexin A1, formyl peptide receptor, and NOX1 orchestrate epithelial repair. J Clin Invest. 2013;123:443–454. doi: 10.1172/JCI65831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Szyk A., Wu Z., Tucker K., Yang D., Lu W., Lubkowski J. Crystal structures of human α-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749–2760. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Yamaguchi Y., Ouchi Y. Antimicrobial peptide defensin: identification of novel isoforms and the characterization of their physiological roles and their significance in the pathogenesis of diseases. Proc Jpn Acad Ser B. 2012;88:152–166. doi: 10.2183/pjab.88.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Tralau T., Meyer-Hoffert U., Schröder J.M., Wiedow O. Human leukocyte elastase and cathepsin G are specific inhibitors of C5a-dependent neutrophil enzyme release and chemotaxis. Exp Dermatol. 2004;13:316–325. doi: 10.1111/j.0906-6705.2004.00145.x. [DOI] [PubMed] [Google Scholar]
- 346.Eggers C.T., Murray I.A., Valerie A., CRAIK C.S. The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase. Biochem J. 2004;379:107–118. doi: 10.1042/BJ20031790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Papareddy P., Rydengård V., Pasupuleti M., Walse B., Mörgelin M., Chalupka A. Proteolysis of human thrombin generates novel host defense peptides. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000857. e1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Salipante S.J., Rojas M.E., Korkmaz B., Duan Z., Wechsler J., Benson K.F. Contributions to neutropenia from PFAAP5 (N4BP2L2), a novel protein mediating transcriptional repressor cooperation between Gfi1 and neutrophil elastase. Mol Cell Biol. 2009;29:4394–4405. doi: 10.1128/MCB.00596-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Chen H.-C., Lin H.-C., Liu C.-Y., Wang C.-H., Hwang T., Huang T.-T. Neutrophil elastase induces IL-8 synthesis by lung epithelial cells via the mitogen-activated protein kinase pathway. J Biomed Sci. 2004;11:49–58. doi: 10.1007/BF02256548. [DOI] [PubMed] [Google Scholar]
- 350.Nemoto E., Tada H., Shimauchi H. Disruption of CD40/CD40 ligand interaction with cleavage of CD40 on human gingival fibroblasts by human leukocyte elastase resulting in down-regulation of chemokine production. J Leukoc Biol. 2002;72:538–545. [PubMed] [Google Scholar]
- 351.Wijkstrom-Frei C., El-Chemaly S., Ali-Rachedi R., Gerson C., Cobas M.A., Forteza R. Lactoperoxidase and human airway host defense. Am J Respir Cell Mol Biol. 2003;29:206–212. doi: 10.1165/rcmb.2002-0152OC. [DOI] [PubMed] [Google Scholar]
- 352.Pieragostino D., Agnifili L., Fasanella V., D'Aguanno S., Mastropasqua R., Di Ilio C. Shotgun proteomics reveals specific modulated protein patterns in tears of patients with primary open angle glaucoma naive to therapy. Mol Biosyst. 2013;9:1108–1116. doi: 10.1039/c3mb25463a. [DOI] [PubMed] [Google Scholar]
- 353.van't Hof W., Blankenvoorde M.F., Veerman E.C., Amerongen A.V.N. The salivary lipocalin von Ebner's gland protein is a cysteine proteinase inhibitor. J Biol Chem. 1997;272:1837–1841. doi: 10.1074/jbc.272.3.1837. [DOI] [PubMed] [Google Scholar]
- 354.Mostov K.E. Transepithelial transport of immunoglobulins. Annu Rev Immunol. 1994;12:63–84. doi: 10.1146/annurev.iy.12.040194.000431. [DOI] [PubMed] [Google Scholar]
- 355.Bomsel M., Heyman M., Hocini H., Lagaye S., Belec L., Dupont C. Intracellular neutralization of HIV transcytosis across tight epithelial barriers by anti-HIV envelope protein dIgA or IgM. Immunity. 1998;9:277–287. doi: 10.1016/s1074-7613(00)80610-x. [DOI] [PubMed] [Google Scholar]
- 356.Mazanec M.B., Coudret C.L., Fletcher D.R. Intracellular neutralization of influenza virus by immunoglobulin A anti-hemagglutinin monoclonal antibodies. J Virol. 1995;69:1339–1343. doi: 10.1128/jvi.69.2.1339-1343.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Mori M., Shiraishi T., Tanaka S., Yamagata M., Mafune K., Tanaka Y. Lack of DMBT1 expression in oesophageal, gastric and colon cancers. Br J Cancer. 1999;79:211–213. doi: 10.1038/sj.bjc.6690035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Wu W., Kemp B.L., Proctor M.L., Gazdar A.F., Minna J.D., Hong W.K. Expression of DMBT1, a candidate tumor suppressor gene, is frequently lost in lung cancer. Cancer Res. 1999;59:1846–1851. [PubMed] [Google Scholar]
- 359.Somerville R., Shoshan Y., Eng C., Barnett G., Miller D., Cowell J.K. Molecular analysis of two putative tumour suppressor genes, PTEN and DMBT, which have been implicated in glioblastoma multiforme disease progression. Oncogene. 1998;17:1755–1757. doi: 10.1038/sj.onc.1202066. [DOI] [PubMed] [Google Scholar]
- 360.Klebanoff S.J., Kettle A.J., Rosen H., Winterbourn C.C., Nauseef W.M. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J Leukoc Biol. 2013;93:185–198. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Metzler K.D., Fuchs T.A., Nauseef W.M., Reumaux D., Roesler J., Schulze I. Myeloperoxidase is required for neutrophil extracellular trap formation: implications for innate immunity. Blood. 2011;117:953–959. doi: 10.1182/blood-2010-06-290171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Stamp L.K., Khalilova I., Tarr J.M., Senthilmohan R., Turner R., Haigh R.C. Myeloperoxidase and oxidative stress in rheumatoid arthritis. Rheumatology. 2012;51:1796–1803. doi: 10.1093/rheumatology/kes193. [DOI] [PubMed] [Google Scholar]
- 363.Wagner B.A., Buettner G.R., Oberley L.W., Darby C.J., Burns C.P. Myeloperoxidase is involved in H2O2-induced apoptosis of HL-60 human leukemia cells. J Biol Chem. 2000;275:22461–22469. doi: 10.1074/jbc.M001434200. [DOI] [PubMed] [Google Scholar]
- 364.Klinke A., Nussbaum C., Kubala L., Friedrichs K., Rudolph T.K., Rudolph V. Myeloperoxidase attracts neutrophils by physical forces. Blood. 2011;117:1350–1358. doi: 10.1182/blood-2010-05-284513. [DOI] [PubMed] [Google Scholar]
- 365.Klebanoff S.J. Myeloperoxidase: contribution to the microbicidal activity of intact leukocytes. Science. 1970;169:1095–1097. doi: 10.1126/science.169.3950.1095. [DOI] [PubMed] [Google Scholar]
- 366.Odobasic D., Kitching A.R., Yang Y., O'Sullivan K.M., Muljadi R.C., Edgtton K.L. Neutrophil myeloperoxidase regulates T-cell− driven tissue inflammation in mice by inhibiting dendritic cell function. Blood. 2013;121:4195–4204. doi: 10.1182/blood-2012-09-456483. [DOI] [PubMed] [Google Scholar]
- 367.Schlesinger D.H., Hay D.I. Complete covalent structure of a proline-rich phosphoprotein, PRP-2, an inhibitor of calcium phosphate crystal growth from human parotid saliva. Chem Biol Drug Des. 1986;27:373–379. doi: 10.1111/j.1399-3011.1986.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 368.Saitoh E., Isemura S., Sanada K. Inhibition of calcium-carbonate precipitation by human salivary proline-rich phosphoproteins. Arch Oral Biol. 1985;30:641–643. doi: 10.1016/0003-9969(85)90086-x. [DOI] [PubMed] [Google Scholar]
- 369.Bennick A., McLaughlin A., Grey A., Madapallimattam G. The location and nature of calcium-binding sites in salivary acidic proline-rich phosphoproteins. J Biol Chem. 1981;256:4741–4746. [PubMed] [Google Scholar]
- 370.Vitorino R., de Morais Guedes S., Ferreira R., Lobo M.J.C., Duarte J., Ferrer-Correia A.J. Two-dimensional electrophoresis study of in vitro pellicle formation and dental caries susceptibility. Eur J Oral Sci. 2006;114:147–153. doi: 10.1111/j.1600-0722.2006.00328.x. [DOI] [PubMed] [Google Scholar]
- 371.Johnsson M., Richardson C., Bergey E., Levine M., Nancollas G. The effects of human salivary cystatins and statherin on hydroxyapatite crystallization. Arch Oral Biol. 1991;36:631–636. doi: 10.1016/0003-9969(91)90014-l. [DOI] [PubMed] [Google Scholar]
- 372.Gibbons R., Hay D. Human salivary acidic proline-rich proteins and statherin promote the attachment of Actinomyces viscosus LY7 to apatitic surfaces. Infect Immun. 1988;56:439–445. doi: 10.1128/iai.56.2.439-445.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Raj P., Johnsson M., Levine M.J., Nancollas G. Salivary statherin. Dependence on sequence, charge, hydrogen bonding potency, and helical conformation for adsorption to hydroxyapatite and inhibition of mineralization. J Biol Chem. 1992;267:5968–5976. [PubMed] [Google Scholar]
- 374.Douglas W.H., Reeh E.S., Ramasubbu N., Raj P.A., Bhandary K.K., Levine M.J. Statherin: a major boundary lubricant of human saliva. Biochem Biophys Res Commun. 1991;180:91–97. doi: 10.1016/s0006-291x(05)81259-8. [DOI] [PubMed] [Google Scholar]
- 375.Oudhoff M.J., Bolscher J.G., Nazmi K., Kalay H., van't Hof W., Amerongen A.V.N. Histatins are the major wound-closure stimulating factors in human saliva as identified in a cell culture assay. FASEB J. 2008;22:3805–3812. doi: 10.1096/fj.08-112003. [DOI] [PubMed] [Google Scholar]
- 376.Tsai H., Bobek L. Human salivary histatins: promising anti-fungal therapeutic agents. Crit Rev Oral Biol Med. 1998;9:480–497. doi: 10.1177/10454411980090040601. [DOI] [PubMed] [Google Scholar]
- 377.Oppenheim F., Yang Y.-C., Diamond R., Hyslop D., Offner G., Troxler R. The primary structure and functional characterization of the neutral histidine-rich polypeptide from human parotid secretion. J Biol Chem. 1986;261:1177–1182. [PubMed] [Google Scholar]
- 378.Mehta P.D., Pirttilä T., Mehta S.P., Sersen E.A., Aisen P.S., Wisniewski H.M. Plasma and cerebrospinal fluid levels of amyloid β proteins 1-40 and 1-42 in Alzheimer disease. Arch Neurol. 2000;57:100–105. doi: 10.1001/archneur.57.1.100. [DOI] [PubMed] [Google Scholar]
- 379.Spies P., Slats D., Sjogren J., Kremer B., Verhey F., Olde Rikkert M. The cerebrospinal fluid amyloid β42/40 ratio in the differentiation of alzheimer's disease from non-alzheimer's dementia. Curr Alzheimer Res. 2010;7:470–476. doi: 10.2174/156720510791383796. [DOI] [PubMed] [Google Scholar]
- 380.Mayeux R., Honig L., Tang M.-X., Manly J., Stern Y., Schupf N. Plasma Aβ40 and Aβ42 and Alzheimer's disease Relation to age, mortality, and risk. Neurology. 2003;61:1185–1190. doi: 10.1212/01.wnl.0000091890.32140.8f. [DOI] [PubMed] [Google Scholar]
- 381.Schupf N., Tang M.X., Fukuyama H., Manly J., Andrews H., Mehta P. Peripheral Aβ subspecies as risk biomarkers of Alzheimer's disease. Proc Natl Acad Sci. 2008;105:14052–14057. doi: 10.1073/pnas.0805902105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Lui J.K., Laws S.M., Li Q.-X., Villemagne V.L., Ames D., Brown B. Plasma amyloid-β as a biomarker in Alzheimer's disease: the AIBL study of aging. J Alzheim Dis. 2010;20:1233–1242. doi: 10.3233/JAD-2010-090249. [DOI] [PubMed] [Google Scholar]
- 383.Lewczuk P., Kornhuber J., Vanmechelen E., Peters O., Heuser I., Maier W. Amyloid β peptides in plasma in early diagnosis of Alzheimer's disease: a multicenter study with multiplexing. Exp Neurol. 2010;223:366–370. doi: 10.1016/j.expneurol.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 384.Wang T., Xiao S., Liu Y., Lin Z., Su N., Li X. The efficacy of plasma biomarkers in early diagnosis of Alzheimer's disease. Int J Geriatr Psychiatry. 2014;29:713–719. doi: 10.1002/gps.4053. [DOI] [PubMed] [Google Scholar]
- 385.Sundelöf J., Giedraitis V., Irizarry M.C., Sundström J., Ingelsson E., Rönnemaa E. Plasma β amyloid and the risk of Alzheimer disease and dementia in elderly men: a prospective, population-based cohort study. Arch Neurol. 2008;65:256–263. doi: 10.1001/archneurol.2007.57. [DOI] [PubMed] [Google Scholar]
- 386.Andreasen N., Vanmechelen E., Van de Voorde A., Davidsson P., Hesse C., Tarvonen S. Cerebrospinal fluid tau protein as a biochemical marker for Alzheimer's disease: a community based follow up study. J Neurol Neurosurg Psychiatry. 1998;64:298–305. doi: 10.1136/jnnp.64.3.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Vigo-Pelfrey C., Seubert P.P., Barbour R., Blomquist C., Lee M., Lee D. Elevation of microtubule-associated protein tau in the cerebrospinal fluid of patients with Alzheimer's disease. Neurology. 1995;45:788–793. doi: 10.1212/wnl.45.4.788. [DOI] [PubMed] [Google Scholar]
- 388.Thome J., Gsell W., Rösier M., Kornhuber J., Frölich L., Hashimoto E. Oxidative-stress associated parameters (lactoferrin, superoxide dismutases) in serum of patients with Alzheimer's disease. Life Sci. 1996;60:13–19. doi: 10.1016/s0024-3205(96)00583-8. [DOI] [PubMed] [Google Scholar]
- 389.Thome J., Münch G., Müller R., Schinzel R., Kornhuber J., Blum-Degen D. Advanced glycation endproducts—associated parameters in the peripheral blood of patients with Alzheimer's disease. Life Sci. 1996;59:679–685. doi: 10.1016/0024-3205(96)00349-9. [DOI] [PubMed] [Google Scholar]
- 390.Soininen H., Lehtovirta M., Helisalmi S., Linnaranta K., Heinonen O., Riekkinen S.P. Increased acetylcholinesterase activity in the CSF of Alzheimer patients carrying apolipoprotein epsilon4 allele. Neuroreport. 1995;6:2518–2520. doi: 10.1097/00001756-199512150-00017. [DOI] [PubMed] [Google Scholar]
- 391.Sirviö J., Kutvonen R., Soininen H., Hartikainen P., Riekkinen P. Cholinesterases in the cerebrospinal fluid, plasma, and erythrocytes of patients with Alzheimer's disease. J Neural Transm. 1989;75:119–127. doi: 10.1007/BF01677425. [DOI] [PubMed] [Google Scholar]
- 392.Smith R.C., Ho B.T., Hsu L., Vroulis G., Claghorn J., Schoolar J. Cholinesterase enzymes in the blood of patients with Alzheimer's disease. Life Sci. 1982;30:543–546. doi: 10.1016/0024-3205(82)90267-3. [DOI] [PubMed] [Google Scholar]
- 393.Korolainen M., Pirttilä T. Cerebrospinal fluid, serum and plasma protein oxidation in Alzheimer's disease. Acta Neurol Scand. 2009;119:32–38. doi: 10.1111/j.1600-0404.2008.01057.x. [DOI] [PubMed] [Google Scholar]
- 394.Conrad C.C., Marshall P.L., Talent J.M., Malakowsky C.A., Choi J., Gracy R.W. Oxidized proteins in Alzheimer's plasma. Biochem Biophys Res Commun. 2000;275:678–681. doi: 10.1006/bbrc.2000.3356. [DOI] [PubMed] [Google Scholar]
- 395.Greilberger J., Fuchs D., Leblhuber F., Greilberger M., Wintersteiger R., Tafeit E. Carbonyl proteins as a clinical marker in Alzheimer's disease and its relation to tryptophan degradation and immune activation. Clin Lab J Clin Lab Lab Relat. 2010;56:441. [PubMed] [Google Scholar]
- 396.Greilberger J., Koidl C., Greilberger M., Lamprecht M., Schroecksnadel K., Leblhuber F. Malondialdehyde, carbonyl proteins and albumin-disulphide as useful oxidative markers in mild cognitive impairment and Alzheimer's disease. Free Radic Res. 2008;42:633–638. doi: 10.1080/10715760802255764. [DOI] [PubMed] [Google Scholar]
- 397.Puertas M., Martinez-Martos J., Cobo M., Carrera M., Mayas M., Ramirez-Exposito M. Plasma oxidative stress parameters in men and women with early stage Alzheimer type dementia. Exp Gerontol. 2012;47:625–630. doi: 10.1016/j.exger.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 398.Hong Z., Shi M., Chung K.A., Quinn J.F., Peskind E.R., Galasko D. DJ-1 and α-synuclein in human cerebrospinal fluid as biomarkers of Parkinson's disease. Brain. 2010;133:713–726. doi: 10.1093/brain/awq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Li Q.-X., San Mok S., Laughton K.M., McLean C.A., Cappai R., Masters C.L. Plasma α-synuclein is decreased in subjects with Parkinson's disease. Exp Neurol. 2007;204:583–588. doi: 10.1016/j.expneurol.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 400.Duran R., Barrero F.J., Morales B., Luna J.D., Ramirez M., Vives F. Plasma α-synuclein in patients with Parkinson's disease with and without treatment. Mov Disord. 2010;25:489–493. doi: 10.1002/mds.22928. [DOI] [PubMed] [Google Scholar]
- 401.Lee P., Lee G., Park H., Bang O., Joo I., Huh K. The plasma alpha-synuclein levels in patients with Parkinson's disease and multiple system atrophy. J Neural Transm. 2006;113:1435–1439. doi: 10.1007/s00702-005-0427-9. [DOI] [PubMed] [Google Scholar]
- 402.Waragai M., Wei J., Fujita M., Nakai M., Ho G.J., Masliah E. Increased level of DJ-1 in the cerebrospinal fluids of sporadic Parkinson's disease. Biochem Biophys Res Commun. 2006;345:967–972. doi: 10.1016/j.bbrc.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 403.Shi M., Bradner J., Hancock A.M., Chung K.A., Quinn J.F., Peskind E.R. Cerebrospinal fluid biomarkers for Parkinson disease diagnosis and progression. Ann Neurol. 2011;69:570–580. doi: 10.1002/ana.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Waragai M., Nakai M., Wei J., Fujita M., Mizuno H., Ho G. Plasma levels of DJ-1 as a possible marker for progression of sporadic Parkinson's disease. Neurosci Lett. 2007;425:18–22. doi: 10.1016/j.neulet.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 405.Maita C., Tsuji S., Yabe I., Hamada S., Ogata A., Maita H. Secretion of DJ-1 into the serum of patients with Parkinson's disease. Neurosci Lett. 2008;431:86–89. doi: 10.1016/j.neulet.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 406.Shi M., Zabetian C.P., Hancock A.M., Ginghina C., Hong Z., Yearout D. Significance and confounders of peripheral DJ-1 and alpha-synuclein in Parkinson's disease. Neurosci Lett. 2010;480:78–82. doi: 10.1016/j.neulet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Coelho H., Azevedo M., Proença C., de Silva J.M., Manso C. Plasma dopamine-beta-hydroxylase and erythrocyte acetylcholinesterase in a group of patients with Parkinson disease. J Neural Transm. 1978;42:163–166. doi: 10.1007/BF01675356. [DOI] [PubMed] [Google Scholar]
- 408.Kaiserova M., Grambalova Z., Otruba P., Stejskal D., Prikrylova Vranova H., Mares J. Cerebrospinal fluid levels of chromogranin A and phosphorylated neurofilament heavy chain are elevated in amyotrophic lateral sclerosis. Acta Neurol Scand. 2017;136:360–364. doi: 10.1111/ane.12735. [DOI] [PubMed] [Google Scholar]
- 409.Filaci G., Contini P., Brenci S., Gazzola P., Lanza L., Scudeletti M. Soluble HLA class I and class II molecule levels in serum and cerebrospinal fluid of multiple sclerosis patients. Hum Immunol. 1997;54:54–62. doi: 10.1016/S0198-8859(97)00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.