Abstract
Background
Automation in microbiology laboratories impacts management, workflow, productivity and quality. Further improvements will be driven by the development of intelligent image analysis allowing automated detection of microbial growth, release of sterile samples, identification and quantification of bacterial colonies and reading of AST disk diffusion assays. We investigated the potential benefit of intelligent imaging analysis by developing algorithms allowing automated detection, semi-quantification and identification of bacterial colonies.
Methods
Defined monomicrobial and clinical urine samples were inoculated by the BD Kiestra™ InoqulA™ BT module. Image acquisition of plates was performed with the BD Kiestra™ ImagA BT digital imaging module using the BD Kiestra™ Optis™ imaging software. The algorithms were developed and trained using defined data sets and their performance evaluated on both defined and clinical samples.
Results
The detection algorithms exhibited 97.1% sensitivity and 93.6% specificity for microbial growth detection. Moreover, quantification accuracy of 80.2% and of 98.6% when accepting a 1 log tolerance was obtained with both defined monomicrobial and clinical urine samples, despite the presence of multiple species in the clinical samples. Automated identification accuracy of microbial colonies growing on chromogenic agar from defined isolates or clinical urine samples ranged from 98.3% to 99.7%, depending on the bacterial species tested.
Conclusion
The development of intelligent algorithm represents a major innovation that has the potential to significantly increase laboratory quality and productivity while reducing turn-around-times. Further development and validation with larger numbers of defined and clinical samples should be performed before transferring intelligent imaging analysis into diagnostic laboratories.
Keywords: Automation, Diagnostic, Bacteriology, Imaging, Growth, Expert
At a glance commentary
Scientific background on the subject
The introduction of laboratory automation has revolutionized conventional clinical bacteriology from samples inoculation to plates incubation and reading. With this new technology, the reading of plates is performed on digital images by technicians that can select microbial colonies for subsequent follow-up work such as identification and antibiotic susceptibility testing.
What this study adds to the field
The study shows as a proof of concept that artificial intelligence may represent a driving innovation in diagnostic bacteriology. Intelligent algorithms for plates analysis linked to expert systems may provide a fully automated approach for microbial growth reading and interpretation that could eventually replace and/or support human-based decisions.
For many years, diagnostic microbiology was not considered as being adapted for laboratory automation due to the variability of the specimen types, the complexity of the various analytical processes and a relatively low level of analytical samples volume compared to other diagnostic units such as chemistry and molecular biology. However, the gradual increase in samples number, limited budget, personal shortage and quality issues as well as laboratories consolidation and liquid-based transport devices have triggered the development and the introduction by different manufacturers of laboratory automation solutions into diagnostic bacteriology laboratories [1], [2], [3], [4]. Several peer-reviewed publications have demonstrated that laboratory automation have the potential to greatly improve the diagnostic processes in bacteriology by increasing the productivity, the quality and the throughput but also by decreasing the time-to results and laboratory cost [1], [5], [6], [7], [8], [9], [10], [11], [12], [13]. Even though the indirect impact of lab automation on patient management remained to be demonstrated in objective, comparative and prospective clinical studies performed by independent laboratories, the shortening of time-to results observed after implementation of laboratory automation strongly suggests that automation will positively improve the clinical management of patients suffering from infectious diseases.
The partial automation available in bacteriology covers four main laboratory processes: inoculation, plate management, incubation and digital imaging [1]. However, a significant part of diagnostic microbiology such as samples pre-processing, microscopy, reading and follow-up work such as identification (ID) and antibiotic susceptibility testing (AST) of isolated colonies remain to be automatized to reach a true total lab automation. Several manufacturers are working on additional hardware solutions to further increase the level of automation in bacteriology such as (1) sample input track, (2) automated colony picking modules including automated deposition of the samples on MALDI-TOF plates as well as automated preparation of bacterial suspension for automated or disk diffusion AST, (3) automated disk dispensing modules and (4) broth incubators [1].
Even though these new technologies will further improve laboratory automation with increased productivity, the next revolution with a major impact on diagnostic microbiology will likely arise from the development of intelligent algorithms and applications linked to expert systems that may in the future monitor several laboratory processes from inoculation to ID/AST without human intervention. To reach such a level of intelligent automation, several algorithmic and application tools need to be developed and validated before being used by intelligent expert systems for the monitoring of laboratory processes. Thus, further improvements will be driven by the development of intelligent image analysis algorithms allowing earlier detection of microbial growth, automated detection and auto-release of sterile samples, automated identification and quantification of bacterial colonies as well as automated reading of AST disk diffusion assays.
We thus investigated the potential benefit of intelligent imaging analysis by developing several algorithms and applications allowing automated detection, identification and semi-quantification of bacterial colonies from both defined and clinical urine samples.
Material and methods
Strains, media, and bacterial suspensions
Most bacterial and yeast strains used in this study (Tables A.1–A.3) were selected according to the most prevalent strains isolated in clinical urine samples in 2014 at the University Hospital of Lausanne (CHUV), Switzerland. The strains were grown on Columbia agar with 5% sheep blood (Columbia III agar; BD, Franklin Lakes, NJ, USA) at 37 °C in normal atmosphere or in 5% CO2 atmosphere incubators. Colonies of each bacterial species were utilized to prepare a bacterial suspension in saline solution adjusted to a 0.5 McFarland turbidity measured with a DensiCheck densitometer instrument (bioMérieux, Marcy l'Etoile, France). The exact bacterial concentration corresponding to a 0.5 McFarland were assessed for each bacterial and yeast species and for each experimental run by measuring the colony forming units (CFU) on Columbia agar with 5% sheep blood (Table A.1). Different concentrations of monomicrobial suspensions were prepared by doing serial 10-fold dilutions in saline solutions, ranging from 1 to 10−5.
Clinical urine sample collection and processing
A total of 218 clinical urine samples were collected with UriSwab™ tubes (Copan, Brescia, Italy) during a 2-months period from outpatients and hospitalized patients at the CHUV, without selection criteria. All clinical urine samples were deidentified prior to testing. Selected urinary samples were immediately processed or stored for maximum 8 h at 4 °C until inoculation. UriSwab™ contains preservative substances (boric acid and sodium formate) that both preserve the microbial viability and prevent microbial growth for up to 48 h.
Inoculation and incubation
For each bacterial strain, 6 different bacterial concentrations, ranging from 1 to 10−5 dilutions of the starting inoculums (Table A.1), were inoculated in duplicate on 3 different media (BD CHROMagar™ Orientation (CHROM), BD Blood agar (COL) and BD MacConkey agar (MAC)) for Gram negative bacteria and on 2 different media (BD CHROMagar™ Orientation, BD Blood agar) for Gram positive bacteria and yeasts. The clinical urine samples were inoculated once on 3 different media (CHROMagar™ Orientation, BD Blood agar, BD MacConkey agar). All the samples were inoculated with the BD Kiestra InoqulA BT module (BD Kiestra, Drachten, Netherlands) using the #17 zig-zag streaking pattern and incubated at 37 °C in normal atmosphere (BD CHROMagar™ Orientation, BD MacConkey agar) or in 5% CO2 atmosphere incubators (BD Blood agar). All the samples processed in the BD laboratory (BD Sparks, Baltimore, USA) for the training of the semi-quantification and identification classifiers were inoculated with the BD Kiestra InoqulA using the #4 zig-zag streaking pattern.
Imaging time
The image acquisition was performed with the ImagA BT using the OPTIS™ software (BD Kiestra, Drachten, Netherlands). The 26 bacterial strains and 99 clinical urine samples were imaged every 2 h from time 0 to time 24 h post inoculation. Each defined and clinical urine samples were processed in 2 runs, 0–12 h and 12–24 h. The first run was inoculated at 8 h in the morning and imaged at times 0–2–4–6–8–10–12–24 h. The second run was inoculated at 20 h in the evening and imaged at times 0–12–14–16–18–20–22–24 h. In addition, 119 clinical urine samples were imaged in 1 run at times 0 h, 16 h, 20 h, and 24 h post inoculation. For the algorithmic analysis, time 0 was chosen as a reference of no growth or no detectable growth. Time 12 h was chosen as an intermediate time point (since most microbial colonies are already detectable) allowing an increase in algorithmic performance for growth prediction. Time 24 h was chosen as the imaging endpoint coinciding with most routine laboratory first plate reading. Other time points were imaged to provide the potential to develop additional applications or to improve the applications presented in this study. Practically, for a time series to be considered as usable by the algorithm, an image must have been captured for each of the subsequent temporal windows: [0–4 h], [10–14 h], [23–25 h]. If multiples time points were matching a temporal window, the time point closest to the central time of the temporal window was kept, others were discarded. Moreover, all samples not captured at time 0 were discarded from the analysis since a reference image at time 0 was needed for algorithmic analysis.
Visual semi-quantification
The visual semi-quantification of the total growth on plate images by technicians was performed using a reading template composed of images of Escherichia coli inoculated at concentrations ranging from 102 to 108 CFU/ml (Fig. C.1). The visual semi-quantification was thus estimated according to the most probable semi-quantification based on growth pattern similarities. Visual semi-quantification was performed in two rounds to establish the “final truth” (gold standard). A first reading was performed and compared to algorithmic prediction. All the images of isolates and clinical urine samples exhibiting discordant results between visual inspection and algorithmic prediction were read a second time to check for possible major reading mistakes, which may introduce a bias in the true performance of algorithmic prediction. The second reading was performed in a blinded manner to avoid the introduction of an important bias in the study.
Then the algorithmic performance was established by comparing algorithmic prediction to the human-based “final truth”.
Identification of microbial colonies in clinical urine samples
Each distinct colony exhibiting a unique phenotypic, morphological and color signature was identified by matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS). These colonies were then manually annotated on images of clinical urine samples allowing the training and the testing of classifiers for colony identification. The characteristics (semi-quantification and identification by MALDI-TOF) of these clinical urines on COL, CHROM and MAC agar plates are summarized in Tables D.1 and D.2.
Image acquisition and image analysis
The major steps of sample image analysis are presented in Fig. A.1. The system is described in the context of an automated sample streaking module (InoqulA BT) using a magnetic rolling bead to inoculate samples on agar plates along a predefined streaking pattern (InoqulA BT pattern #17 in this study).
To overcome classical SNR imaging limitation and to optimize contrast, the BD Kiestra™ OPTIS™ image acquisition software was used in this study to analyze imaged plates which is designed to adjust and optimize in real time the contrast and the SNR for each pixel in each red/green/blue (RGB) channels by performing an acquisition session of multiple frames (22 images acquisition per plate) using several illumination conditions and exposure times in a calibrated and standardized manner. In addition, images were captured using different incident lights (top, side and bottom) on black or white backgrounds to maximize the contrast of the colony to its background and thus the amount of information used to discriminate the different microorganisms growing on a plate. The plates were analyzed in real time to evaluate the best exposure times required to capture all pixel related information matching a given illumination and background condition. Depending on media and growth, the OPTIS™ analysis engine selected the set of exposure times leading to optimized contrast dynamic range and SNR.
In order to maximize the information, a contrast gathering algorithm was used to generate an optimal contrasted image of the plate to detect microbial colonies. Furthermore, images of the plate were captured at different time points during incubation time (time series) to measure the growth kinetic and to image the evolving phenotypic pattern of growing colonies.
Isolated colonies were counted using time series images of plates during the incubation process. For each detected colony at a given time point (e.g. at incubation time = 24 h), the program analyzed earlier time points (e.g. at incubation time = 12 h) to verify if the analyzed colony was previously only a single CFU or if this area was previously composed of multiple CFUs that became confluent at the analyzed time point due to increased size of the colonies.
At a given time point, objects with a sufficient strong spatial or temporal contrast such as colonies, dust, artifacts, air bubbles were detected. A microbial object classifier was developed to allow segregation of microbial and non-microbial objects. Then, microbial objects were analyzed to estimate the microbial load contained in the sample either by directly counting colonies when the number of colonies on a plate was ≤100 CFU or with the bead model count when the number of CFUs was ≥100 (see supplementary material and method A.1).
The bead model or direct count, together with overall contrast measures over the entire media and derivative features relating changes as a function of time serve as input to the semi-quantification classifier, which ultimately predicts the actual CFUs/ml buckets (<102 (no growth), 102–103, 103–104, 104–105 and ≥105) for sample on a given media.
Semi-quantification and identification classifiers
Random forest classifiers were used at three different stages to perform: (1) microbial vs non microbial objects classification, (2) semi-quantification and (3) presumptive identification. The parameterization of each classifier including number of trees, number of nodes and number of features used at each split was chosen to minimize and level off out-of-bag error and to avoid over fitting [14]. The features used as input to the random forest algorithm were chosen as the 20 most important features detected using the Boruta algorithm (R package Boruta) and confirmed with VSURF R package as providing a good balance between out-of-bag error, classification performance, computation speed and classifier object storage size [15], [16]. For each classifier, algorithm update test performance was assessed and compared to baseline performance in a regression testing to ensure that unanticipated effects do not appear. Only algorithm updates showing performance improvement were selected. Training of the classifiers was performed on an entirely separate dataset ensuring that the performance reported here corresponds to the test performance of a cross-validation process. The training of the semi-quantification classifiers was performed on a dataset generated at the BD laboratory (BD Sparks, Baltimore, USA) composed of serial 10-fold dilutions ranging from 10−2 to 10−7 of a 0.5 McFarland bacterial suspension of 5 defined microbial strains for each of 11 different selected bacterial species, of 20 mixed samples composed of 2–3 defined microbial strains and of 200 clinical urines (Table A.2). The semi-quantification classifiers were tested on the CHUV dataset composed of 26 microbial strains and 138 clinical urines (Tables A.1 and A.3). For the identification classifiers, a microbial objects dataset was generated from 8 defined microbial strains of the BD laboratory (Table A.2), 15 defined microbial strains and 41 clinical urines of the CHUV laboratory (Table A.3).
One semi-quantification classifier was developed per media, each providing a probability for each plate to be within ordered count buckets <102 (no growth), 102–103, 103–104, 104–105 and ≥105. Growth detection was performed when comparing the <102 category to the four other count categories. Classification performance was assessed with agreement, agreement with 1 logarithmic tolerance, sensitivity (growth detection only) and specificity (growth detection only). Two-sided 95% confidence intervals were computed by bootstrapped percentile method [17].
Two inputs were used to train and test presumptive identification. The first input was the manually annotated images of clinical urine samples. The second input was monomicrobial strains samples where all objects classified as microbial objects were considered as microbial objects corresponding to inoculated microbial isolate (Tables A.2 and A.3). For the identification classifiers, 582 plates were used. Out of these 582 plates, 1.5 × 106 microbial objects were identified in the whole incubation imaging window [0–25 h], but only 212,682 microbial objects belonged to the [23–25 h] window, which is the time frame used for algorithmic classification. After randomization, the complete set of 212,682 objects was divided into 2 populations: 80% were used for training and 20% for testing. For each object, color and morphometric features were extracted to train and test the identification classifiers.
Multi-reader and imaging algorithm comparisons
Readings from 10 independent technicians and prediction from the algorithm were compared over a set of 150 images randomly selected from the data set to assess the agreements percentage. The algorithm was considered as one independent reader and agreements were evaluated in two conditions, with or without algorithm as part of the reader's dataset. Global agreement between readers was calculated by permutation obtained among 2 to 10 readers, with and without algorithm results, with 0, 1 and 2 logarithmic tolerances. Two-sided 95% confidence intervals were obtained by bootstrapped percentile method performed on reviewed plates during the reader permutation process [17].
Statistical software
Statistical analyses were performed with R 3.3.1 [18].
Results
Detection
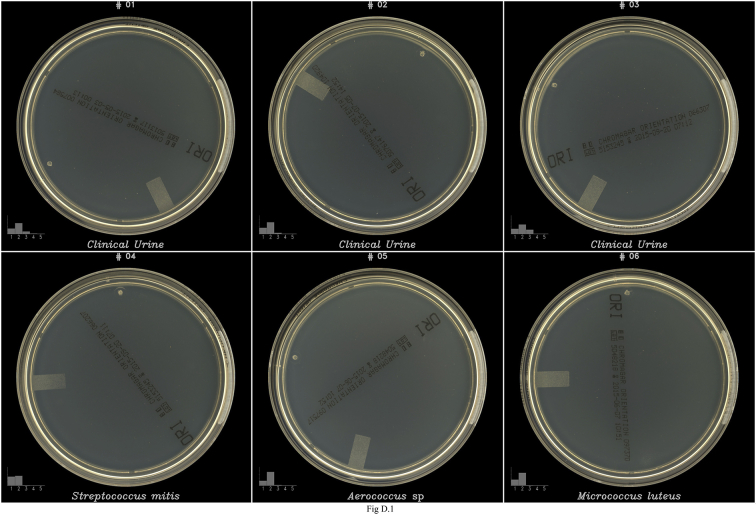
The algorithmic detection of growth was assessed at 24 h post inoculation. The algorithmic performance was compared to human visual inspection of the plates, which was defined as the final truth. The sensitivity (Se), specificity (Sp), positive predictive value (PPV), negative predictive value (NPV) and the ROC curve (Se/1-Sp) of the algorithmic detection of both defined isolates and clinical urine samples were thus calculated compared to the final truth (Table 1 and Fig. B.1). An overall 97.1% Se, 93.6% Sp, 99.0% PPV and 82.6% NPV were measured for all samples types on all tested media plates. No significant difference in Se was observed between microbial isolates and clinical urine samples with a Se ranging from 98.8% to 100% on the BD Blood agar (COL) and BD MacConkey agar (MAC) media, respectively, and a lower Se ranging from 92.5% to 93.9% on BD CHROMagar™ Orientation agar (CHROM). A Sp ranging from 90% to 100% was observed on CHROM and MAC but a lower Sp of 80–84.2% was obtained on COL agar with the two sample types. The PPV was superior to 97% for both microbial isolates and clinical urine samples on all media types except for clinical urine samples, which exhibited a 90% PPV on MAC. On all media types, a better NPV was observed with clinical urine samples than with microbial isolates with an average of 93.2% (77.1–100%) and of 72.2% (36.4–85.7%), respectively. Moreover, on overall, a NPV superior to 74.8% was obtained except with microbial isolates on COL plates which exhibited a low NPV performance of 36.4%. The ROC curve showed that the overall accuracy of the algorithmic detection is slightly higher with bacterial isolates than with clinical urine samples, especially on CHROM agar (Fig. B.1). Finally, 0.8% (15/1796) false positives (FP) and 2.6% (46/1796) false negatives (FN) were obtained following algorithmic detection prediction compared to visual inspection of plate images (Table 2 and Figs. D.1–I.1). Overall, 80.4% (37/46) of FNs were observed on CHROM agar plates, 8/46 (17.4%) on COL agar and 1/46 (2.2%) on MAC agar (Table 2). For both microbial isolates and clinical urine samples, a visual inspection of the FNs plate images showed that FNs could be classified into different categories: (1) microbial colonies not detected by the algorithms, (2) detected microbial colonies wrongly classified as non microbial objects, (3) microbial colonies located on the extreme edges of the plate, which is a zone not included in image analysis and (4) possible false positive reported by the technician (Table 2 and Figs. G.1–I1).
Table 1.
Evaluation of the Algorithm to report positive and negative plates with a detection threshold of 100 CFU/ml.
| Sample type | Media | N Se | Se | CI (se) | N Sp | Sp | CI (Sp) | N PPV | PPV | CI (PPV) | N NPV | NPV | CI (NPV) | N Prev | Prev | CI (Prev) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | All | 1562 | 97.1 | (96.2, 97.8) | 234 | 93.6 | (90.1, 96.5) | 1531 | 99.0 | (98.4, 99.5) | 265 | 82.6 | (77.9, 87) | 1796 | 87.0 | (85.4, 88.6) |
| All | CHROM | 582 | 93.6 | (91.7, 95.6) | 119 | 95.0 | (90.7, 98.4) | 551 | 98.9 | (98, 99.6) | 150 | 75.3 | (67.8, 82.2) | 701 | 83.0 | (80.2, 85.6) |
| All | COL | 703 | 98.9 | (98, 99.6) | 24 | 83.3 | (67.9, 96.6) | 699 | 99.4 | (98.7, 99.9) | 28 | 71.4 | (54.2, 87.5) | 727 | 96.7 | (95.3, 97.9) |
| All | MAC | 277 | 99.6 | (98.6, 100) | 91 | 94.5 | (89.5, 98.9) | 281 | 98.2 | (96.6, 99.6) | 87 | 98.9 | (96.1, 100) | 368 | 75.3 | (70.9, 79.9) |
| Isolates | All | 1292 | 97.1 | (96.2, 98) | 100 | 96.0 | (91.4, 99.1) | 1259 | 99.7 | (99.4, 99.9) | 133 | 72.2 | (64.7, 79.6) | 1392 | 92.8 | (91.5, 94.1) |
| Isolates | CHROM | 476 | 93.9 | (91.5, 96) | 89 | 96.6 | (92.3, 100) | 450 | 99.3 | (98.4, 100) | 115 | 74.8 | (66.4, 82.2) | 565 | 84.2 | (81.2, 87.1) |
| Isolates | COL | 584 | 98.8 | (97.8, 99.7) | 5 | 80.0 | (33.3, 100) | 578 | 99.8 | (99.5, 100) | 11 | 36.4 | (7.7, 66.7) | 589 | 99.2 | (98.3, 99.8) |
| Isolates | MAC | 232 | 99.6 | (98.7, 100) | 6 | 100.0 | (61, 100) | 231 | 100.0 | (98.4, 100) | 7 | 85.7 | (50, 100) | 238 | 97.5 | (95.4, 99.2) |
| Urines | All | 270 | 96.7 | (94.5, 98.5) | 134 | 91.8 | (86.6, 96) | 272 | 96.0 | (93.2, 98.1) | 132 | 93.2 | (88.7, 97) | 404 | 66.8 | (62.4, 71.8) |
| Urines | CHROM | 106 | 92.5 | (86.9, 97.2) | 30 | 90.0 | (77.8, 100) | 101 | 97.0 | (93.2, 100) | 35 | 77.1 | (63.2, 90.3) | 136 | 77.9 | (70.6, 84.6) |
| Urines | COL | 119 | 99.2 | (97.3, 100) | 19 | 84.2 | (65.2, 100) | 121 | 97.5 | (94.3, 100) | 17 | 94.1 | (81.2, 100) | 138 | 86.2 | (80.4, 91.3) |
| Urines | MAC | 45 | 100.0 | (92.1, 100) | 85 | 94.1 | (88.1, 98.7) | 50 | 90.0 | (80.8, 97.9) | 80 | 100.0 | (95.4, 100) | 130 | 34.6 | (26.9, 43.1) |
Abbreviations: All: all sample types or all media types; CHROM: BD BBL CHROMagar Orientation; COL: BD Columbia Agar with 5% Sheep Blood; MAC: BD MacConkey II Agar; N Se: Number of sample analyzed for sensitivity; Se: Sensitivity; CI (Se): 95% confidence interval for Se; N Sp: Number of sample analyzed for specificity; Sp: Specificity; CI (Sp): 95% confidence interval for Sp; N PPV: Number of sample analyzed for PPV; PPV: positive predictive value; CI (PPV): 95% confidence interval for PPV; N NPV: Number of sample analyzed for NPV; NPV: Negative predictive value; CI (NPV): 95% confidence interval for NPV; N Prev: Number of sample analyzed for Prev (i.e. total number of samples per condition); Prev: Prevalence; CI (Prev): 95% confidence interval for Prev.
Table 2.
Discordant results: False positives and false negatives.
| Sample type | Media | Number | Comments |
|---|---|---|---|
| False positives | |||
| Urines | CHROM | 3 |
|
| Urines | COL | 3 | |
| Urines | MAC | 5 | |
| Isolates | CHROM | 3 | |
| Isolates | COL | 1 | |
| Total | 15 | ||
| False negatives | |||
| Urines | CHROM | 8 | Plates containing between 1 and 3 colonies were not detected by the algorithms (see Figs. G.1 and H.1).
|
| Urines | COL | 1 | |
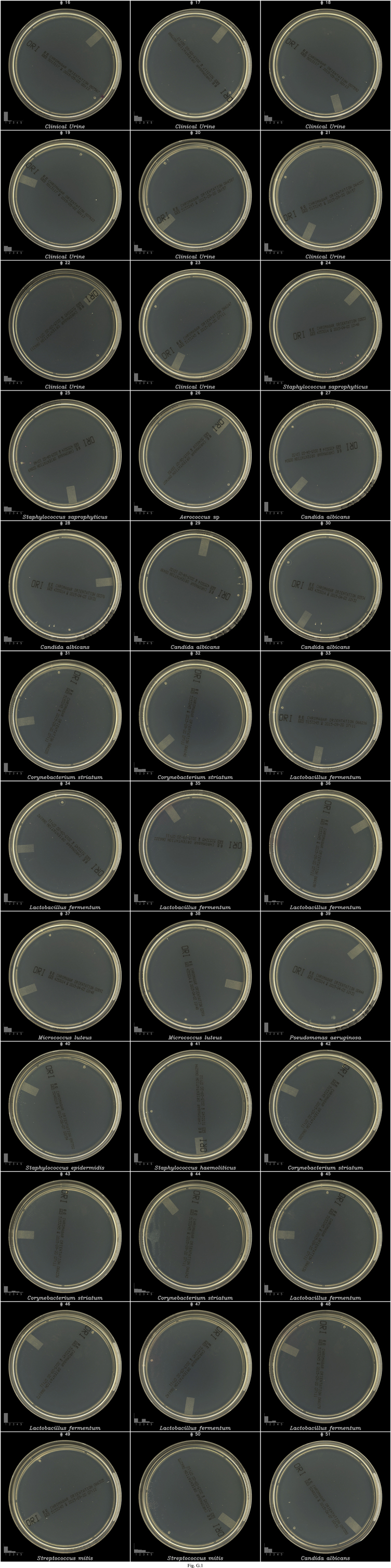
| Isolates | CHROM | 29 | Some plates containing Lactobacillus fermentum (8), Candida albicans (6),Corynebacterium striatum (5), Micrococcus luteus (2), Streptococcus mitis group (2), Staphylococcus saprophyticus (2), Staphylococcus haemolyticus (1), Staphylococcus epidermidis (1), Aerococcus urinae (1) and Pseudomonas aeruginosa (1) were not detected by the algorithms (see Fig. G.1).
|
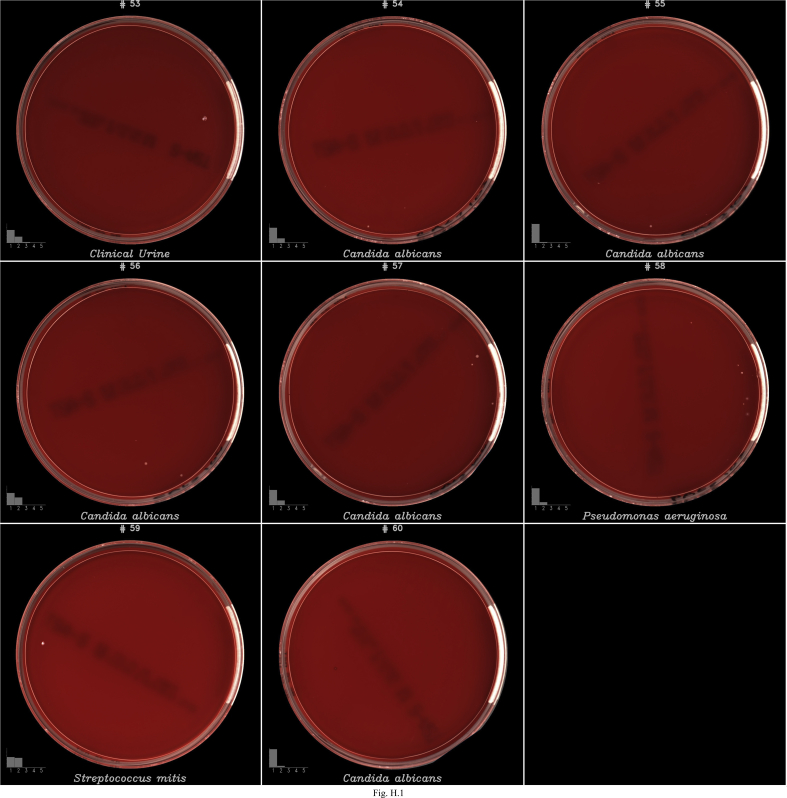
| Isolates | COL | 7 | Some plates containing between 1 to 4 colonies of Candida albicans (5), Streptococcus mitis group (1) and Pseudomonas aeruginosa (1) were not detected by the algorithms (see Fig. H.1).
|
| Isolates | MAC | 1 | A plate containing one colony of Pseudomonas aeruginosa was not detected by the algorithm. The colony was located on the edge of the plate, which is a zone not included in image analysis (see Fig. I.1). |
| Total | 46 | ||
Semi-quantification
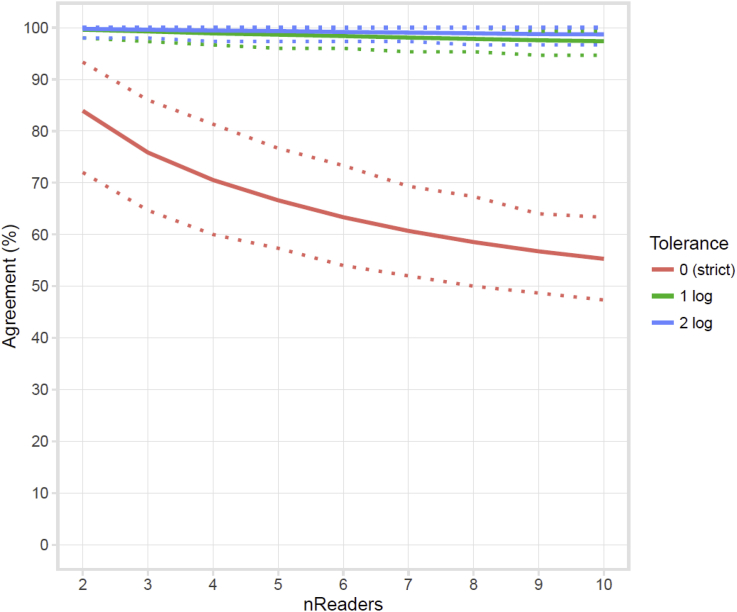
The accuracy of algorithmic semi-quantification was determined by comparing algorithmic to visual semi-quantification performed by laboratory technicians based on a reading template (Table 3 and B.1). The global accuracy of the exact match was equal to 80.2% for all sample types on all media types with an accuracy ranging from 72.2% to 93.8% depending on the sample and media types. However, the global accuracy was significantly improved to 98.6%, ranging from 96.1% to 100% depending on the sample and media types, by allowing a 1 log tolerance to the results reported by the algorithms. On a total of 1796 analyzed images, 1440 (80.2%), 330 (18.4%), 19 (1.0%), 5 (0.3%) and 2 (0.1%) plate images were quantified by the algorithm with a difference of 0 log (no difference), 1 log, 2 log, 3 log and 4 log, respectively, compared to visual quantification on plate images. All plate images except 2 cases with a difference ≥2 log compared to visual semi-quantification were underestimated by the algorithm. A semi-quantification of ≥100,000 was predicted by the algorithm on one MAC plate with no growth but with agar artefacts that were classified as microbial object and quantified by the algorithms. To estimate the accuracy of human visual semi-quantification used to determine the final truth, the analytical variation of visual semi-quantification by laboratory technicians was assessed by submitting 150 plate images to 10 different laboratory technicians and to the algorithms. The 150 plate images were composed of 10 images for each of the 5 concentrations ranging from <102 to ≥105 CFU/ml on each COL, MAC and CHROM media plates. The reading accuracy of the 10 technicians and of the algorithms was assessed by comparing the results to the final truth determined by visual reading of plate images (Table 4). A similar performance was observed between human and algorithmic semi-quantification compared to the final truth with an accuracy ranging from 77.3 to 84.7% and an accuracy ranging from 98.0% to 99.3% with a 1 log tolerance. Moreover, the global agreement between 2 and 10 readers was calculated with 0, 1 and 2 logarithmic tolerances (Fig. 1). The global agreement between human readers ranged from 83.7%, 99.6%, 99.7% (2 Readers) to 56.9%, 97.6% and 98.7% (10 Readers) with logarithmic tolerances of 0, 1 and 2, respectively.
Table 3.
Evaluation of the Algorithm to semi-quantify microbial growth (<102 to ≥105).
| Sample type | Media | N | Accuracy | CI | Accuracy.pm1 | CI pm1 |
|---|---|---|---|---|---|---|
| All | All | 1796 | 80.2 | (78.3, 82) | 98.6 | (97.9, 99.1) |
| All | CHROM | 701 | 78.9 | (75.6, 81.9) | 96.9 | (95.6, 98.1) |
| All | COL | 727 | 75.9 | (72.8, 78.8) | 99.7 | (99.3, 100) |
| All | MAC | 368 | 91 | (88, 93.8) | 99.5 | (98.6, 100) |
| Isolates | All | 1392 | 76.9 | (74.6, 79.2) | 98.3 | (97.6, 98.9) |
| Isolates | CHROM | 565 | 76.5 | (72.7, 80) | 96.1 | (94.5, 97.7) |
| Isolates | COL | 589 | 72.2 | (68.6, 75.7) | 99.7 | (99.2, 100) |
| Isolates | MAC | 238 | 89.5 | (85.7, 92.9) | 100 | (98.4, 100) |
| Urines | All | 404 | 91.6 | (88.4, 94.3) | 99.5 | (98.8, 100) |
| Urines | CHROM | 136 | 89 | (83.8, 94.1) | 100 | (97.3, 100) |
| Urines | COL | 138 | 92 | (87, 96.4) | 100 | (97.3, 100) |
| Urines | MAC | 130 | 93.8 | (89.2, 97.7) | 98.5 | (96.2, 100) |
Abbreviations: All: all sample types or all media types; CHROM: BD BBL CHROMagar Orientation; COL: BD Columbia Agar with 5% Sheep Blood; MAC: BD MacConkey II Agar; N: Number of analyzed samples; CI: 95% confidence interval; Accuracy.pm1: accuracy with a plus or minus 1 log difference tolerance; CI.pm1: 95% confidence interval for the accuracy.pm1.
Table 4.
Percentage agreement of semi-quantification by 10 technicians and by the algorithms compared to the final truth determined by manual reading of 150 plate images.
| nBoot | Reader | Accuracy | CI | Accuracy.pm1 | CI pm1 |
|---|---|---|---|---|---|
| 10,000 | Algorithms | 78.0 | (71.3, 84.6) | 98.7 | (96.7, 100.0) |
| 10,000 | Reader 1 | 84.7 | (78.7, 90.0) | 98.7 | (96.7, 100.0) |
| 10,000 | Reader 2 | 80.0 | (73.3, 86.0) | 98.7 | (96.7, 100.0) |
| 10,000 | Reader 3 | 81.3 | (74.7, 87.3) | 98.7 | (96.7, 100.0) |
| 10,000 | Reader 4 | 83.3 | (77.3, 88.7) | 98.7 | (96.7, 100.0) |
| 10,000 | Reader 5 | 79.3 | (72.7, 86.0) | 98.0 | (95.3, 100.0) |
| 10,000 | Reader 6 | 82.7 | (76.0, 88.7) | 98.7 | (96.7, 100.0) |
| 10,000 | Reader 7 | 78.0 | (71.3, 84.7) | 99.3 | (98.0, 100.0) |
| 10,000 | Reader 8 | 82.0 | (76.0, 88.0) | 98.0 | (95.3, 100.0) |
| 10,000 | Reader 9 | 77.3 | (70.7, 84.0) | 98.7 | (96.7, 100.0) |
| 10,000 | Reader 10 | 82.0 | (76.0, 88.0) | 98.7 | (96.7, 100.0) |
Abbreviaitons: nBoot: number of bootstrap samples to compute the 95% CI; CI: Confidence interval; Accuracy.pm1: accuracy with a plus or minus 1 log difference tolerance; CI.pm1: 95% confidence interval for the accuracy.pm1.
Fig. 1.
Global agreement between human readers to report growth semi-quantification. The global agreement to report semi-quantification was calculated with a tolerance of 0 (strict quantification), 1 log or 2 log difference as compared to the “final truth”. Solid line: estimated value, dashed lines: 95% CI.
Identification
The performance of algorithmic identification was assessed on both defined microbial strains (Tables A.1–A.3) and manually annotated clinical urine samples. The identification algorithm ability to correctly classify bacterial species and groups (Table C.1 and Fig. J.1) listed in the product information of the CHROMagar™ Orientation medium as well as Staphylococcus aureus was assessed on 212,682 microbial objects derived from microbial strains and clinical urine samples. Among the 212,682 microbial objects, 80% (170,145) were used for training of the algorithms and 20% (42,537) for testing.
A correct classification superior or equal to 96.4% was obtained for E. coli, Staphylococcus saprophyticus, group Proteus spp./Morganella spp./Providencia spp. (PMP), group Klebsiella spp./Enterobacter spp./Serratia spp./Citrobacter spp. (KESC) and S. aureus (Table 5). A 93.8% correct classification was obtained for Enterococcus spp., with 5.7% incorrect classification of them in the group KESC. Similarly, the Streptococcus agalactiae exhibited a correct classification of 94.8% with 4.7% incorrect classification in the group KESC. Interestingly, the Enterococcus spp., S. agalactiae and the group KESC all exhibit a chromatic phenotype located in the blue color space (Fig. J.1). E. coli and the group Enterococcus spp were identified by the algorithm with a 96.4% and 93.8% Se and a 99.9% and 99.8% Sp, respectively (Table 6). The product information of the BD CHROMagar™ Orientation medium states that E. coli and enterococci can be identified without confirmatory testing based on colony color and morphology with a sensitivity and specificity of 97.0% and 99.0%, respectively, for E. coli, and a sensitivity and specificity of 99.0% and 97.0%, respectively, for Enterococcus spp. Thus the performance of the algorithmic identification for E. coli was in line with the performance that can be expected by visual inspection of BD CHROMagar™ Orientation medium and was slightly reduced compared to an expected visual identification for the enterococci group. The other microbial groups were correctly identified by the algorithm with a Se ≥ 94.8%, a Sp ≥ 97.9%, a PPV ≥95.4% and a NPV ≥99.2%, which is an excellent performance for bacterial species and groups that can only be presumptively identified by visual inspection on BD CHROMagar™ Orientation medium before confirmatory tests such as MALDI-TOF ID.
Table 5.
Performance of the algorithmic identification in percentage correct classification (manual reference and percentage computed per column).
| ESCCOL | Group Enterococcus | STASAP | STRAGA | Group KESC | Group PMP | STAAUE | |
|---|---|---|---|---|---|---|---|
| ESCCOL | 96.4% | 0.0% | 1.3% | 0.0% | 0.0% | 0.0% | 0.0% |
| Group Enterococcus | 0.0% | 93.8% | 0.0% | 0.0% | 0.5% | 0.0% | 0.0% |
| STASAP | 2.5% | 0.1% | 97.6% | 0.1% | 0.1% | 0.4% | 0.1% |
| STRAGA | 0.0% | 0.2% | 0.0% | 94.8% | 0.1% | 0.0% | 0.0% |
| Group KESC | 0.5% | 5.7% | 0.6% | 4.7% | 98.9% | 1.1% | 0.4% |
| Group PMP | 0.7% | 0.1% | 0.3% | 0.3% | 0.4% | 97.6% | 0.3% |
| STAAUE | 0.0% | 0.1% | 0.2% | 0.0% | 0.0% | 0.9% | 99.2% |
Abbreviations: ESCCOL: Escherichia coli; KESC: Klebsiella spp./Enterobacter spp./Serratia spp./Citrobacter spp.; PMP: Proteus spp./Morganella spp./Providencia spp.; STAAUE: Staphylococcus aureus; STASAP: Staphylococcus saprophyticus; STRAGA: Streptococcus agalactiae.
Table 6.
Evaluation of the performance of the algorithms to correctly classify bacterial species or groups.
| Group | N Acc | Acc | CI (Acc) | N Se | Se | CI (Se) | N Sp | Sp | CI (Sp) | N PPV | PPV | CI (PPV) | N NPV | NPV | CI (NPV) | N Prev | Prev | CI (Prev) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ESCCOL | 42,611 | 99.6 | (99.6, 99.7) | 3288 | 96.4 | (95.6, 97.0) | 39,323 | 99.9 | (99.9, 99.9) | 3209 | 98.8 | (98.3, 99.1) | 39,402 | 99.7 | (99.6, 99.7) | 42,611 | 7.7 | (7.5, 8.0) |
| Group Enter | 42,611 | 99.1 | (99.0, 99.2) | 4744 | 93.8 | (93.1, 94.4) | 37,867 | 99.8 | (99.7, 99.8) | 4530 | 98.2 | (97.9, 98.6) | 38,081 | 99.2 | (99.1, 99.3) | 42,611 | 11.1 | (10.8, 11.5) |
| Group KESC | 42,611 | 98.3 | (98.2, 98.4) | 17,685 | 98.9 | (98.8, 99.1) | 24,926 | 97.9 | (97.7, 98.1) | 18,028 | 97.1 | (96.8, 97.3) | 24,583 | 99.2 | (99.1, 99.3) | 42,611 | 41.5 | (41.0, 42.1) |
| Group PMP | 42,611 | 99.3 | (99.2, 99.4) | 7470 | 97.6 | (97.3, 98.0) | 35,141 | 99.7 | (99.6, 99.7) | 7416 | 98.4 | (98.0, 98.6) | 35,195 | 99.5 | (99.4, 99.6) | 42,611 | 17.5 | (17.1, 17.9) |
| STAAUE | 42,611 | 99.7 | (99.7, 99.8) | 3965 | 99.2 | (99.0, 99.5) | 38,646 | 99.8 | (99.7, 99.8) | 4014 | 98.0 | (97.6, 98.5) | 38,597 | 99.9 | (99.9, 99.9) | 42,611 | 9.3 | (9.0, 9.6) |
| STASAP | 42,611 | 99.5 | (99.5, 99.6) | 2799 | 97.6 | (97.1, 98.1) | 39,812 | 99.7 | (99.6, 99.7) | 2864 | 95.4 | (94.8, 96.2) | 39,747 | 99.8 | (99.8, 99.9) | 42,611 | 6.6 | (6.3, 6.8) |
| STRAGA | 42,611 | 99.6 | (99.5, 99.7) | 2660 | 94.8 | (93.6, 95.7) | 39,951 | 99.9 | (99.9, 100.0) | 2550 | 98.9 | (98.3, 99.3) | 40,061 | 99.7 | (99.6, 99.7) | 42,611 | 6.2 | (6.0, 6.5) |
Abbreviations: Group: Bacterial species or groups; N Acc: Number of sample analyzed for accuracy; Acc: Accuracy; CI (Acc): 95% confidence interval for Acc; N Se: Number of sample analyzed for sensitivity; Se: Sensitivity; CI (Se): 95% confidence interval for Se; N Sp: Number of sample analyzed for specificity; Sp: Specificity; CI (Sp): 95% confidence interval for Sp; N PPV: Number of sample analyzed for PPV; PPV: positive predictive value; CI (PPV): 95% confidence interval for PPV; N NPV: Number of sample analyzed for NPV; NPV: Negative predictive value; CI (NPV): 95% confidence interval for NPV; N Prev: Number of sample analyzed for Prev; Prev: Prevalence; CI (Prev): 95% confidence interval for Prev; ESCCOL: Escherichia coli; Group Enter: Group Enterococcus spp.; Group KESC: Group Klebsiella spp./Enterobacter spp./Serratia spp./Citrobacter spp.; Group PMP: Group Proteus spp./Morganella spp./Providencia spp.; STAAUE: Staphylococcus aureus; STASAP: Staphylococcus saprophyticus; STRAGA: Streptococcus agalactiae.
Overall, a higher identification performance was obtained by algorithmic classification than by visual inspection of bacteria growing on BD CHROMagar™ Orientation medium.
Discussion
This study is demonstrating, as a proof of concept, that automated image analysis has the potential to further improve laboratory automation in bacteriology by introducing software-based analysis of growth detection, growth semi-quantification and colony identification based on microbial phenotype, morphology and color. The overall performance of the algorithm prediction for all samples and all media types was robust with (1) a detection Se of 97.1%, a Sp of 93.6%, a PPV of 99.0% and a NPV of 82.6%, (2) an exact semi-quantification accuracy ≥ 80.0% and a semi-quantification accuracy ≥ 98% with a 1 log tolerance, and (3) a correct identification at species or group level ≥ 93.0% of bacterial colonies growing on CHROM agar.
Image acquisition and image analysis was designed (1) to detect microbial growth, (2) to determine the microbial load in clinical samples such as urine samples by an automated method based on colony distribution analysis on plates and (3) to identify microbial species or group of species based on morphology and color features.
Counting colonies on plates using streaking patterns designed to generate isolated colonies regardless of the initial microbial load is challenging. Depending on the level of growth, the semi-quantification of the microbial load in the sample is based on the counting of colonies, or on the analysis of the distribution of isolated colonies using a model of the CFU releasing process linked to the streaking system (see rolling bead model), or from pattern similarities with known growth pattern references (Fig. C.1). When microbial growth cannot be partially or entirely distinguished into single CFUs, the exact counting is impossible and growth has to be estimated (semi-quantification) using plate global features (e.g. growth region area and intensity related to the dispensing origin) and/or physical models of growth when reproducible automated inoculation systems are used (see rolling bead model in material and methods section 2 and supplementary material and methods A.1).
Colony detection is strongly dependent on the contrast over the background [contrast = (signal – background)/(signal + background)] which is increasing with the size of the colonies. An object can thus be detected in an image if it is significantly different in brightness, color and/or texture from its surroundings (i.e. the background). In addition, colony detection in an image is mainly dependent on the area covered by a colony in the sensor space and not by the number of pixels used to image the colony. Moreover, the quality and the confidence of a measurement are characterized by the signal-to-noise ratio (SNR) of the measurement. Thus, colonies detection limitations are strongly linked to colony size and contrast (the smaller the colonies and the lower the contrast) which are dependent of the background, and thus of both the morphological traits of colonies and media plates. The BD Kiestra™ OPTIS™ image acquisition software was used to maximize SNR and contrast for each pixel in each RGB channel of the plate image in order to collect multi-sources standardized data information for subsequent image analysis. Due to the internal calibration, correction and normalization, images acquired on different systems (ImageA BT and ReadA Compact) by the OPTIS™ image acquisition software are similar.
The plates streaking process with the InoqulA BT generates a non random uniform pattern characterized by a CFU density gradient along the streaking pathway to produce isolated colonies regardless of the original concentration of CFUs in the sample. The rolling magnetic bead spread the inoculated sample generating thus a print out of what was loaded on the bead. The bead model count (supplementary material and methods A.1) relies on the analysis of the CFU distribution along the streaking pattern allowing an estimation of the microbial load on the bead, which in case of a high microbial load leading to CFUs confluence, represented the best estimation of the microbial load carried on the magnetic bead and thus of the bacterial load contained in defined or clinical samples.
The detection prediction performance was characterized by an overall Se and PPV greater than Sp and NPV for both microbial isolates and clinical urine samples. These results may have been impacted by an inclusion bias since a high prevalence of positive samples was observed in this study, which directly favored the PPVs over the NPVs. In addition, the number of samples analyzed for Sp and for NPV was very low for some sample types inoculated on some media types, which resulted in large 95% CI (Table 1). The high prevalence of positive samples with microbial isolates is originating from the experimental setup which was designed to obtain a growth on all media types inoculated with microbial loads ranging from 108 to 103 CFU/ml. Thus, for instance, only a 36.4% NPV was obtained on COL agar with microbial isolates exhibiting a 98.8% Se and 80.0% Sp, but with a prevalence of 99.2% (Table 1). Similarly, positive clinical urine samples were less prevalent than positive microbial isolates, exhibiting thus a better NPV (Table 1).
The CHROM and MAC media plates have a high degree of transparency compared to COL which enhances the contrast of dust and agar artefacts detected by the algorithms and in some cases wrongly classified as microbial objects, resulting thus in 15 FPs (0.8%) (Table 2) and in decreased PPVs (see Figs. D.1–F.1). Overall, 46/1796 (2.6%) FN samples were observed. A subsequent visual inspection of the images on a data screen did not allow to discriminate reliably between microbial and non microbial objects for some of the FN plates (see images #23, 27, 28, 38, and 42 in Fig. G.1), thus indicating that some plates may have been wrongly considered as positive in the “final truth”, which was determined based on visual inspection of plate images. Moreover, the growth of some bacterial species on CHROM agar produced lawns of tiny and faint colonies after 24 h incubation in normal atmosphere that were not detected, or detected but not considered as microbial object by the algorithms, and thus reported as negative (see Corynebacterium striatum and Lactobacillus fermentum in Fig. G.1), representing 45% (13/29) FN cases on CHROM agar. This result was probably caused by an absence of these growth patterns in the database and/or in the training set of the algorithms, underlying how important it will be to enrich and train the algorithms with the most exhaustive possible set of strains presenting unique growth patterns. Thanks to cloud-based informatics technology, a solution for continuous improvement of detection Se and Sp over time could be to automatically enrich the training databases with samples identified as wrongly assessed by the algorithms during manual reviews of routine cases. Finally, dust, agar artefacts and fingerprints generated by manual plate handling and image acquisition with the ImagA BT created objects interfering in the training and testing of microbial and non-microbial objects classifiers that likely decreased the performance of algorithmic detection of true microbial objects, affecting thus negatively the detection Se and Sp. Thus, a greater sensitivity and specificity with a reduced percentage of FPs and FNs are expected with automated systems.
A performance of semi-quantification with 85.9% exact accuracy and 98.7% accuracy with a 1 log tolerance was obtained in this study. The final truth semi-quantification was determined by visual inspection of the plate based on growth pattern similarities with a reading template containing inoculation of samples ranging from 102 to 108 microorganisms/ml (thus 0 to 106 inoculated microorganisms in 10 μl) (Fig. C.1). This method of semi-quantification is greatly dependent on the visual interpretation of the reading template by technicians, especially at microbial concentration ≥104 CFU/ml where the total number of colonies cannot be counted but may only be estimated. In addition, the visual semi-quantification based on a reading template is dependent on colonies traits such as morphology, size, color and phenotypes, which can significantly influence both human perceptions of growth patterns but also the growth gradient pattern per se. The reading accuracy of 10 laboratory technicians was similar to the performance of algorithmic prediction (Table 4). Moreover, the global agreements between human readers indicated that the reported visual semi-quantification is inaccurate below a 1 logarithmic tolerance [Fig. 1]. Together, these results suggest that the final truth determined following human visual semi-quantification is probably correct in a 1 logarithmic range around the true value, suggesting that the performance of algorithmic semi-quantification prediction is likely comprised between the exact accuracy (80.2%) and the accuracy obtained with a 1 log tolerance (98.6%) (Table 3). Regarding classifiers development and prediction, these results indicate that human limitations in providing the true quantification value have to be considered by introducing a certain degree of uncertainty during the algorithmic training and validation tests processes.
In addition, the semi-quantification classifier was developed using a slightly different zig-zag streaking pattern (#4) than the one used in this study (#17). Even though some features could be normalized to be pattern independent, other features such as overall plate growth are dependent of the streaking pattern and accounted for some decreased level of performance. The future imaging applications will therefore be pattern dependant. The rolling bead model requires the detection of single colonies to generate a semi-quantification prediction. When only a bacterial lawn with no detectable single colonies was observed such as growth of Lactobacillus spp., Corynebacterium spp. and Aerococcus spp. on CHROM agar, the algorithmic prediction of semi-quantification has to rely exclusively on overall plate growth derived features contained in the training dataset. Thus, in this case, inaccurate semi-quantification prediction was generated in absence of similar growth patterns in the database. Finally, the algorithmic semi-quantification prediction is based on a classification probability of the detected growth into 5 “buckets”, <102 (no growth), 102–103, 103–104, 104–105 and ≥105 (Fig. D.1–I.1). Whereas a clear classification with a high probability confidence for one bucket was obtained in most cases, several samples generated close probability classification between two buckets or more. Even though the bucket with the highest probability was selected by the algorithms, the degree of confidence of these quantitative results was significantly reduced. Moreover, the pattern of predicted class probabilities generated following random forest classification may provide a more valuable representation of the true value than the bucket with the highest probability. Thus, the future integration in routine diagnostic of automated detection and semi-quantification algorithmic solutions should be implemented with confidence level indicators to estimate the robustness of the prediction to properly interpret and validate the results. The uncertainty of the true value as illustrated in the reading accuracy between the 10 technicians further indicates the need to report confidence level indicators and/or classification probability distributions.
A correct identification classification ≥93.8% with an accuracy ≥98.3% was obtained following algorithmic identification of bacterial species and group growing on CHROM agar (Table 5, Table 6). The smallest performance was observed with the group Enterococcus spp. and S. agalactiae, which exhibited a 5.7% and 4.7% misclassification, respectively, of the colonies in the KESC Group. The KESC and enterococci groups as well as S. agalactiae are located in the blue color space, which likely explained that some colonies of the group Enterococcus spp. and of S. agalactiae were incorrectly classified by the algorithm in the KESC group. The identification classifiers included multiple features to optimize the identification performance including color intensity, morphology (shape, circularity, colony area, perimeter, …) and contextual information on local density (number of objects, distance from other colonies, distance along the streaking pattern, local temporal and spatial contrast). However, the features could be enriched with additional information to further improve the classification performance on the bacterial species and groups listed in this study but also to investigate the possibility to identify bacterial species within the KESC, PMP and Enterococcus groups as well as other microbial species that may have distinct phenotypic signatures on CHROM agar or on other media types such as COL and MAC.
The identification features used by the classifiers were extracted from microbial isolates colonies but also from manually annotated clinical urine samples plates for which each distinct phenotypic sister colonies were identified by MALDI-TOF. The manual annotation from clinical urine samples allowed thus to enrich the database with microbial intraspecies morphology and color variations and with features belonging to bacterial species not included in the microbial isolates used in this study.
Finally, the performance of the object classification into microbial and non-microbial object affected the identification performance since each non-microbial object such as dust considered as microbial object were analyzed by the identification algorithms, which generated wrong identification prediction based on the features used by the classifiers.
This study demonstrates that intelligent imaging applied to diagnostic bacteriology has the potential to significantly improve laboratory workflow by automating software-based decision making processes that are nowadays human-dependent. Imaging applications may provide the possibility to reach full automation in bacteriology by automating and standardizing samples preparations, management and analysis from sample reception to analytical results. These algorithms can easily be adapted to develop applications as surveillance (MRSA, VRE) [19], [20], alert of positivity for normally sterile samples such as cerebrospinal fluids, pleural fluids, joints, prosthetic material and deep wounds, but also clinical urine applications associated to expert systems that may be used in the future in conjunction with automated colony picking and AST modules to deliver more efficiently and earlier complete identification and AST results. In routine practice, plate management and handling by the robotic systems and not image acquisition and algorithmic analysis will be the limiting factor regarding imaging throughput. In addition, the algorithms presented in this study are requiring images taken at 3 incubation time windows ([0–4 h], [10–14 h], [23–25 h]), which is compatible with the throughput performance of modern automated incubators which can image their entire plates content in about 4 h.
The manual process of data collection has proven to generate more artefacts than usually observed on automated systems, which introduced an additional level of complexity in image analysis and likely a reduced performance of algorithmic prediction. Thus, these algorithms will be further optimized to operate on very standardized fully automated sample processes including inoculation, incubation and imaging. Moreover, this study demonstrated the importance to develop these algorithms on specific media plates from defined manufacturers in order to guarantee the robustness and reliability of algorithmic detection, semi-quantification and identification, despite obvious interest to later adapt these algorithms to in-house made media plates or to media from other manufacturers. We also consider important that imaging application solutions should be delivered with a high degree of flexibility to provide the possibility to the user for adjusting the performance of Se and Sp of the algorithms, for instance according to the sample types, with confidence level indicators to facilitate the technical validation of the results. In addition, imaging applications should be linked to expert rules that could be defined and adjusted by the users to modulate the use and the settings of such applications to different samples types and/or microbial species identification and to help the users for the interpretation of the analytical results. Finally, the number of defined and clinical samples that should be used before transferring intelligent imaging analysis into diagnostic laboratories should be larger than the number used to train the algorithms in this study, indicating that extensive validation studies remained to be performed by the manufacturers.
In conclusion, the development of intelligent algorithms represents a driving innovation that will likely further increase laboratory quality and productivity while significantly reducing turn-around-times. Indeed, these algorithms are required to reach a real level of full automation in bacteriology.
Conflicts of interest
Becton-Dickinson supported this study by funding the salary of one laboratory technician during 6 months and reagents. Three co-authors, Raphaël Marcelpoil, Cédrick Orny and Didier Morel are employed by Becton-Dickinson.
Acknowledgements
We thank Jérôme Blanc for technical assistance and Jean-Marc Volle for assistance and support on computer imaging. Becton-Dickinson supported this study by funding the salary of one laboratory technician during 6 months and reagents.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bj.2017.09.001.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Diagram illustrating the major steps from sample processing to colony forming units' semi-quantification in an automated system. Once sequential images have been acquired with the Optis™ technology, the analysis includes three main steps: (1) plate analysis preparation, (2) semi-quantification feature extraction and (3) semi-quantification classification. PPM: Prepared Plated Media.
Se/1-Sp (ROC curve) of the algorithmic detection of both defined microbial isolates and clinical urine samples compared to the “final truth” (gold standard).
Semi-quantification reading template (E. coli). A reading template consisting of 7 bacterial concentrations of E. coli ranging from 102 CFU/ml to 108 CFU/ml. The technicians used this template to estimate bacterial concentration in microbial isolates and clinical urine samples.
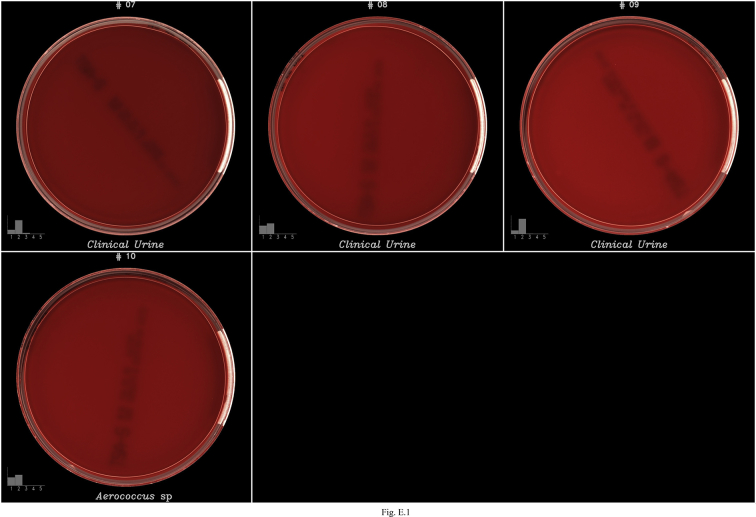
Images of false positives (FP) on CHROM agar. Each image contains at the bottom left a bar graph representing the classification probabilities generated by the algorithms to classify the growth into 5 buckets: (1) <102 (no growth), (2) 102–103, (3) 103–104, (4) 104–105 and (5) ≥105. The algorithms selected the bucket with the highest probability to report growth semi-quantification.
Fig. E.1.
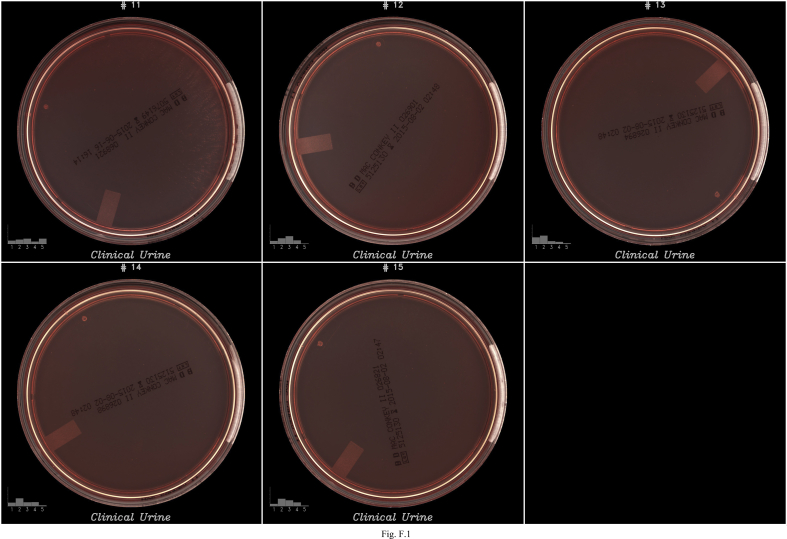
Images of false positives (FP) on COL agar. Each image contains at the bottom left a bar graph representing the classification probabilities generated by the algorithms to classify the growth into 5 buckets: (1) <102 (no growth), (2) 102–103, (3) 103–104, (4)104–105 and (5) ≥105. The algorithms selected the bucket with the highest probability to report growth semi-quantification.
Fig. F.1.
Images of false positives (FP) on MAC agar. Each image contains at the bottom left a bar graph representing the classification probabilities generated by the algorithms to classify the growth into 5 buckets: (1) <102 (no growth), (2) 102–103, (3) 103–104, (4) 104–105 and (5) ≥105. The algorithms selected the bucket with the highest probability to report growth semi-quantification.
Fig. G.1.
Images of false negatives (FN) on CHROM agar. Each image contains at the bottom left a bar graph representing the classification probabilities generated by the algorithms to classify the growth into 5 buckets: (1) <102 (no growth), (2) 102–103, (3) 103–104, (4) 104–105 and (5) ≥105. The algorithms selected the bucket with the highest probability to report growth semi-quantification.
Fig. H.1.
Images of false negatives (FN) on COL agar. Each image contains at the bottom left a bar graph representing the classification probabilities generated by the algorithms to classify the growth into 5 buckets: (1) <102 (no growth), (2) 102–103, (3) 103–104, (4) 104–105 and (5) ≥105. The algorithms selected the bucket with the highest probability to report growth semi-quantification.
Fig. I.1.
. Images of false negatives (FN) on MAC agar. Each image contains at the bottom left a bar graph representing the classification probabilities generated by the algorithms to classify the growth into 5 buckets: (1) <102 (no growth), (2) 102–103, (3) 103–104, (4) 104–105 and (5) ≥105. The algorithms selected the bucket with the highest probability to report growth semi-quantification.
Fig. J.1.
Classification of microbial species and groups that can be definitely or presumptively identified on CHROM agar according to the product information of the BD CHROMagar™ Orientation medium. Esc col: Escherichia coli, Sta sap: Staphylococcus saprophyticus, Stc aga: Streptococcus agalactiae, Sta aur: Staphylococcus aureus, KPN: Klebsiella pneumonia, CKO: Citrobacter koseri, CFR: Citrobacter freundii, ECL: Enterobacter cloacae group, EAE: Enterobacter aerogenes, SMA: Serratia marcescens, MMO: Morganella morganii, PMI: Proteus mirabilis, PVU: Proteus vulgaris.
References
- 1.Croxatto A., Prod'hom G., Faverjon F., Rochais Y., Greub G. Laboratory automation in clinical bacteriology: what system to choose? Clin Microbiol Infect. 2016;22:217–235. doi: 10.1016/j.cmi.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 2.Bourbeau P.P., Ledeboer N.A. Automation in clinical microbiology. J Clin Microbiol. 2013;51:1658–1665. doi: 10.1128/JCM.00301-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greub G., Prod'hom G. Automation in clinical bacteriology: what system to choose? Clin Microbiol Infect. 2011;17:655–660. doi: 10.1111/j.1469-0691.2011.03513.x. [DOI] [PubMed] [Google Scholar]
- 4.Lina G., Greub G. Automation in bacteriology: a changing way to perform clinical diagnosis in infectious diseases. Clin Microbiol Infect. 2016;22:215–216. doi: 10.1016/j.cmi.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Hombach M., Jetter M., Blochliger N., Kolesnik-Goldmann N., Bottger E.C. Fully automated disc diffusion for rapid antibiotic susceptibility test results: a proof-of-principle study. J Antimicrob Chemother. 2017;72:1659–1668. doi: 10.1093/jac/dkx026. [DOI] [PubMed] [Google Scholar]
- 6.Croxatto A., Dijkstra K., Prod'hom G., Greub G. Comparison of inoculation with the InoqulA and WASP automated systems with manual inoculation. J Clin Microbiol. 2015;53:2298–2307. doi: 10.1128/JCM.03076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bielli A., Lacchini C., Vismara C., Lombardi G., Sironi M.C., Gesu G. 25th European Congress of Clinical Microbiology and Infectious Disease. 2015. WASPLab urine validation study: comparison between 16 and 24 hours of incubation. Abstract EVO535. [Google Scholar]
- 8.Mutters N.T., Hodiamont C.J., de Jong M.D., Overmeijer H.P., van den Boogaard M., Visser C.E. Performance of Kiestra total laboratory automation combined with MS in clinical microbiology practice. Ann Lab Med. 2014;34:111–117. doi: 10.3343/alm.2014.34.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Froment P., Marchandin H., Vande Perre P., Lamy B. Automated versus manual sample inoculations in routine clinical microbiology: a performance evaluation of the fully automated InoqulA instrument. J Clin Microbiol. 2014;52:796–802. doi: 10.1128/JCM.02341-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mischnik A., Mieth M., Busch C.J., Hofer S., Zimmermann S. First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach. J Clin Microbiol. 2012;50:2732–2736. doi: 10.1128/JCM.05501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphrey G., Malone C., Gough H., Awadel-Kariem F.M. 21st European Congress of Clinical Microbiology and Infectious Disease. 2011. Experience with KIESTRA's Total Lab Automation solution to meet the challenge of universal MRSA screening for Lister Hospital, a large UK district general hospital. Abstract P-1793. [Google Scholar]
- 12.Bentley N., Farrington M., Doughton R., Pearce D. 21st European Congress of Clinical Microbiology and Infectious Disease. 2011. Automating the bacteriology laboratory. Abstract P-1792. [Google Scholar]
- 13.Dauwalder O., Landrieve L., Laurent F., de Montclos M., Vandenesch F., Lina G. Does bacteriology laboratory automation reduce time to results and increase quality management? Clin Microbiol Infect. 2016;22:236–243. doi: 10.1016/j.cmi.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 14.James G., Witten D., Hastie T., Tibshirani R. 2013. An introduction to statistical learning with applications in R. [Google Scholar]
- 15.Kursa M.B., Rudnicki W.R. Feature selection with the Boruta package. J Stat Softw. 2010;36 [Google Scholar]
- 16.Genuer R., Poggi J.-M., Tuleau-Malot C. VSURF: an R package for variable selection using random forests. R J. 2015;7:19–33. [Google Scholar]
- 17.Davison A.C., Hinkley D.V. Cambridge University Press; Cambridge: 1997. Bootstrap methods and their application. [Google Scholar]
- 18.Team R.C. 2016. R: A language and environment for statistical computing.https://www.R-project.org/ [Google Scholar]
- 19.Faron M.L., Buchan B.W., Vismara C., Lacchini C., Bielli A., Gesu G. Automated scoring of chromogenic media for detection of methicillin-resistant Staphylococcus aureus by use of WASPLab image analysis software. J Clin Microbiol. 2016;54:620–624. doi: 10.1128/JCM.02778-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faron M.L., Buchan B.W., Coon C., Liebregts T., van Bree A., Jansz A.R. Automatic digital analysis of chromogenic media for vancomycin-resistant-Enterococcus screens using copan WASPLab. J Clin Microbiol. 2016;54:2464–2469. doi: 10.1128/JCM.01040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagram illustrating the major steps from sample processing to colony forming units' semi-quantification in an automated system. Once sequential images have been acquired with the Optis™ technology, the analysis includes three main steps: (1) plate analysis preparation, (2) semi-quantification feature extraction and (3) semi-quantification classification. PPM: Prepared Plated Media.
Se/1-Sp (ROC curve) of the algorithmic detection of both defined microbial isolates and clinical urine samples compared to the “final truth” (gold standard).
Semi-quantification reading template (E. coli). A reading template consisting of 7 bacterial concentrations of E. coli ranging from 102 CFU/ml to 108 CFU/ml. The technicians used this template to estimate bacterial concentration in microbial isolates and clinical urine samples.
Images of false positives (FP) on CHROM agar. Each image contains at the bottom left a bar graph representing the classification probabilities generated by the algorithms to classify the growth into 5 buckets: (1) <102 (no growth), (2) 102–103, (3) 103–104, (4) 104–105 and (5) ≥105. The algorithms selected the bucket with the highest probability to report growth semi-quantification.