Abstract
The Set4 protein in the yeast Saccharomyces cerevisiae contains both a PHD finger and a SET domain, a common signature of chromatin-associated proteins, and shares sequence homology with the yeast protein Set3, the fly protein UpSET, and the human protein mixed-lineage leukemia 5 (MLL5). However, the biological role for Set4 and its potential function in chromatin regulation has not been well defined. Here, we analyzed yeast cell phenotypes associated with loss of Set4 or its overexpression, which revealed that Set4 protects against oxidative stress induced by hydrogen peroxide. Gene expression analysis indicated that Set4 promotes the activation of stress response genes in the presence of oxidative insults. Using ChIP analysis and other biochemical assays, we also found that Set4 interacts with chromatin and directly localizes to stress response genes upon oxidative stress. However, recombinant Set4 did not show detectable methyltransferase activity on histones. Our findings also suggest that Set4 abundance in the cell is balanced under normal and stress conditions to promote survival. Overall, these results suggest a model in which Set4 is a stress-responsive, chromatin-associated protein that activates gene expression programs required for cellular protection against oxidative stress. This work advances our understanding of mechanisms that protect cells during oxidative stress and further defines the role of the Set3–Set4 subfamily of SET domain–containing proteins in controlling gene expression in response to adverse environmental conditions.
Keywords: chromatin, chromatin regulation, gene expression, oxidative stress, stress response, yeast, gene regulation, SET domain, Set4
Introduction
Cell survival in the presence of challenging environmental conditions is critically dependent on rapid changes in gene expression. Stress-responsive gene expression programs are controlled by a suite of transcription factors and chromatin proteins that fine-tune expression in response to diverse signals (1–4). These include chromatin-modifying enzymes, such as histone lysine methyltransferases and acetyltransferases, as well as chromatin effector proteins that bind modified histone residues and often stabilize enzymatic activities at chromatin. In Saccharomyces cerevisiae, the roles of numerous transcription factors and chromatin modifiers have been delineated in response to different stresses, including osmotic stress (5, 6), nutrient stress (7, 8), and oxidative stress (9, 10). However, our understanding of the mechanisms controlling stress response gene expression programs is still incomplete, suggesting that the roles of additional chromatin regulators remain to be determined.
A family of methyltransferases that are important for the control of gene expression and linked to stress responses is the SET (Su(var)3–9, Enhancer-of-zeste, Trithorax) domain family, which frequently catalyze methylation of lysine residues. There are 12 SET domain proteins in S. cerevisiae, of which six are known or predicted chromatin regulators, and the other six are nonhistone protein methyltransferases (4, 11). The H3K42 methyltransferase Set1; the H3K36 methytransferase Set2; and the H4K5, H4K8, and H4K12 methyltransferase Set5 have each been linked to cell survival in the presence of different environmental stresses (6, 8, 12, 13). Additionally, the protein Set3, which is not known to be an active methyltransferase, inhibits cryptic transcription and represses gene expression in response to changing environmental conditions, such as carbon source shifts (14). Set3 is a component of a histone deacetylase (HDAC) complex and contains a PHD finger, which interacts with methylated H3K4 and stabilizes the HDAC complex at chromatin, particularly in the 5′-transcribed regions of genes (15, 16).
Set3 shares sequence similarity with the mammalian protein MLL5 (16, 17) and the Drosophila protein UpSET (18, 19), each of which contain a PHD finger and a SET domain. Yeast Set3, mammalian MLL5, and the fly protein UpSET contain divergent SET domains that are predicted to lack catalytic activity based on amino acid substitutions in SAM binding sites and the lack of a tyrosine in a stereotypical position important for both SAM and target lysine binding (17, 20). Whereas mammalian MLL5 had been postulated to catalyze H3K4 methylation, this activity has not been conclusively demonstrated, and the existence of catalytic activity for MLL5 altogether is unlikely based on biochemical and structural analyses (17, 21–23). Additionally, catalytic activity has not been detected for the fly protein UpSET (18). However, each of these proteins play critical roles in gene expression and chromatin organization through physical interactions with methylated histones and chromatin modifiers (18, 19, 24).
The S. cerevisiae genome also contains a paralog to Set3, known as Set4, which possesses a PHD finger and a divergent SET domain that contains similar amino acid substitutions as the SET domains of Set3, fly UpSET, and mammalian MLL5 (Fig. 1A). Based on its similarity to this subfamily of SET domain proteins and the presence of a PHD finger, Set4 is also likely to be a chromatin regulator. Interestingly, genome-wide studies have shown that Set4 is expressed at low levels under normal growth conditions (25), although it appears to be up-regulated during stress, such as anoxia (26), and during stationary phase (27). These observations suggest the possibility that Set4 may be a stress-regulated chromatin protein; however, its biological function has yet to be well-characterized.
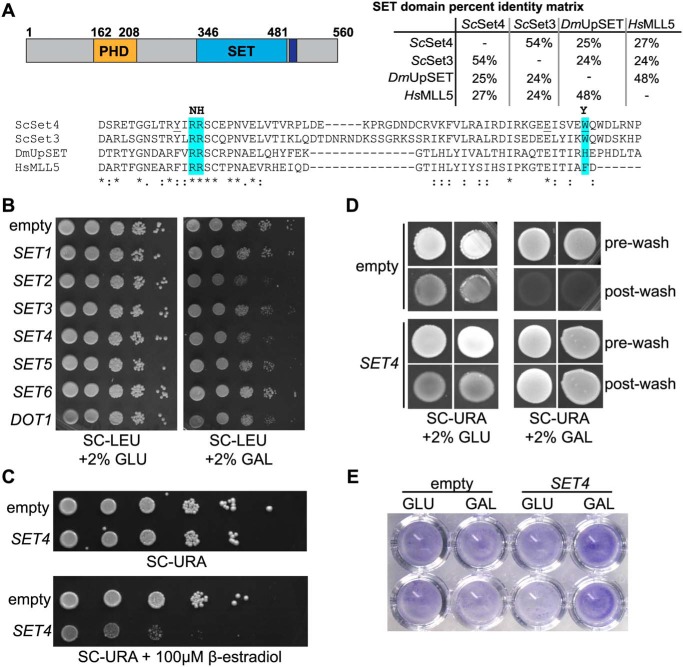
Figure 1.
Overexpression of SET4 is deleterious and promotes cell adhesion. A, the domain structure of Set4 (left), indicating the position of the PHD finger (orange), the SET domain (blue), and the post-SET domain (purple). A percentage identity matrix (right) compares the SET domains of S. cerevisiae Set4 (ScSet4), Set3 (ScSet3), D. melanogaster UpSET (DmUpSET), and H. sapiens MLL5 (HsMLL5). Percentage identity was determined using Clustal Omega. Shown is a sequence alignment of the most conserved region of the SET domain among ScSet4, ScSet3, DmUpSET, and HsMLL5. Amino acids highlighted in blue represent differences from canonical SET domains in regions required for SAM and lysine binding (20). Amino acids predominantly found in other SET domains are indicated in boldface type at the top of the alignment. Underlined amino acids within the Set4 sequence indicate mutations used in Fig. 8C. B, WT yeast (yEG001) carrying plasmids expressing known and putative histone methyltransferases under the control of the GAL1 promoter. 10-Fold serial dilutions were spotted on either SC-LEU with 2% glucose (GLU) or 2% galactose (GAL) and grown at 30 °C for 3 days. C, strains harboring a β-estradiol–inducible artificial transcription factor and either the empty pMN3 vector or pMN3-SET4 were grown in SC-URA, and 10-fold serial dilutions were spotted on SC-URA or SC-URA with 100 μm β-estradiol and grown at 30 °C for 3 days. D, the Σ1278b strain with pRS316-GAL1p-SET4 or an empty vector was grown on SC-URA with either 2% glucose or 2% galactose for 10 days to promote adhesion to the agar and invasion. Images were obtained before washing (pre-wash) and after gentle washing with water to remove nonadherent cells (post-wash). E, crystal violet staining of Σ1278b cells containing an empty vector or pRS316-GAL1p-SET4 grown in either 2% glucose (GLU) or 2% galactose (GAL) and adhered to 96-well polystyrene plates, as described (46).
In this study, we identified a protective role for Set4 during oxidative stress and determined that the regulation of genes linked to the response to oxidative stress depends on Set4. We find that Set4 is a nuclear protein that associates with chromatin. The localization of Set4 to chromatin appears to be tightly controlled by the cell, which provides for careful calibration of stress defense pathways by Set4. This work therefore identifies Set4 as a regulated chromatin factor required for controlling gene expression in response to stress and sheds light on potential additional roles for metazoan orthologs, such as fly UpSET and mammalian MLL5.
Results
Set4 promotes survival in the presence of oxidative stress
In a screen for phenotypes associated with overexpression of known and candidate histone lysine (K) methyltransferases (KMTs), we identified that overexpression of Set4 was deleterious to cells. Observation of yeast strains carrying plasmids expressing KMTs under the control of a galactose-inducible promoter showed that ectopic expression of SET4, SET2, and DOT1 limited cell growth (Fig. 1B). Of these, relatively little is known about the biological functions of Set4; therefore, we focused on this protein for further analysis. To verify our findings in a system independent of the potential confounding effects caused by carbon source shifts, we overexpressed SET4 using the β-estradiol–inducible artificial transcription factor Z3EV, which has been shown to limit off-target effects associated with inducible promoters (28). In this system, we also observed inhibited cell growth when SET4 was highly expressed (Fig. 1C). Our findings are consistent with a previous genome-wide study that reported defective growth upon overexpression of SET4 (29).
Ectopic expression of SET4 had also been linked to increased cell adhesion and haploid invasive growth, a stress response initiated by haploid yeast under nutrient-limiting conditions (30). We transformed the galactose-inducible SET4 overexpression vector and an empty vector into the Σ1278b strain background, which is permissive to adhesion and invasive growth. We performed a standard plate washing assay in which cell adherence to the agar is monitored following vigorous washing of the plate with water (31). In the presence of galactose, cells overexpressing SET4 exhibited increased adherence to the agar after washing, relative to the empty vector control or under glucose conditions (Fig. 1D). We note that cells grown in glucose consistently showed higher background adherence to the agar than those grown in galactose; however, no difference was found in the amount of cells adhered to the plate between empty vector or SET4 vector cells in the uninduced, glucose conditions. In addition, we also observed increased adherence to polystyrene plates using crystal violet staining of Σ1278b cells overexpressing SET4 (Fig. 1E). Together, these data show that ectopic expression of Set4 increases adhesive properties of yeast independent of environmental conditions, suggesting possible inappropriate activation of stress response pathways when Set4 levels are increased.
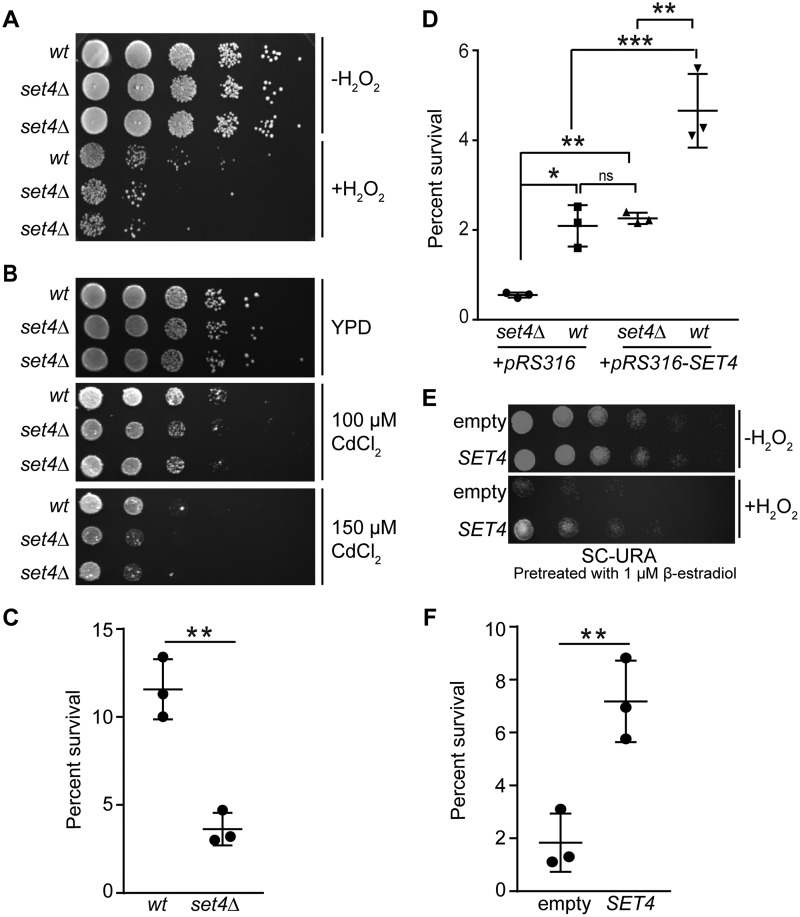
Our data investigating phenotypes associated with overexpression of SET4 suggest that it may play a role in cellular stress responses. To address this possibility, we subjected cells lacking Set4 to ∼25 different environmental stress conditions, including multiple carbon sources, temperature variation, nutrient stress, and genotoxic agents, among others. Of the conditions tested, set4Δ cells showed a specific sensitivity to oxidative insults. Two independent set4Δ isolates showed decreased survival following exposure to H2O2 (Fig. 2A) and cadmium chloride (Fig. 2B). Quantitative analysis of cfu from WT and set4Δ cells following treatment with H2O2 also showed a >50% reduction in survival for set4Δ cells compared with WT (Fig. 2C). To determine whether sensitivity to oxidative stress was specifically due to loss of Set4, we tested survival following H2O2 treatment in WT and set4Δ cells containing either an empty vector or a low-copy (ARS/CEN) vector expressing SET4 from its endogenous promoter. Whereas set4Δ cells with the empty vector showed reduced survival following H2O2 treatment, set4Δ cells harboring the SET4-expressing vector survived to a similar extent as WT cells with the empty vector (Fig. 2D), indicating that the set4Δ phenotype can be rescued by adding back SET4. Interestingly, WT cells containing the SET4-expressing plasmid showed a statistically significant increase in survival relative to those with just the empty vector. This observation suggests that a modest increase in SET4 levels, due to one or two additional copies of the SET4 gene from this ARS/CEN vector, is sufficient to increase cell survival in the presence of oxidative damage.
Figure 2.
Set4 promotes survival during oxidative stress. A, 10-fold serial dilutions of WT (yEG001) and set4Δ (yEG322 and yEG325) strains grown to mid-log phase in YPD or YPD with 4 mm H2O2 for 30 min. B, 10-fold serial dilutions of WT (yEG001) and set4Δ (yEG322 and yEG325) strains spotted on YPD, YPD with 100 μm CdCl2, or YPD with with 150 μm CdCl2. C, percentage survival calculated from cfu of WT (yEG001) and set4Δ (yEG325) strains following treatment with 4 mm H2O2 for 30 min. The scatter plot shows the mean and S.D. (error bars) from three biological replicates. An unpaired t test was used to determine statistical significance. D, percentage survival of WT and set4Δ cells (yEG638, yEG639, yEG640, and yEG641) carrying either the empty pRS316 vector or pRS316-SET4, with SET4 under the control of its endogenous promoter, calculated from cfu on SC-URA following treatment with 20 mm H2O2 for 30 min. One-way analysis of variance was performed, followed by Turkey's post hoc test to determine statistical significance. E, 10-fold serial dilutions of strains carrying either pMN3 (empty) or pMN3-SET4 (yEG372 and yEG375, respectively) that were pretreated with 1 μm β-estradiol for 4–5 h, followed by exposure to 20 mm H2O2 for 30 min. F, percentage survival calculated from cfu of cells grown as in E, except that 50 nm β-estradiol was used to induce SET4, and plated on SC-URA. An unpaired t test was used to determine statistical significance. For all panels, asterisks represent p values as follows: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. ns, not significant.
To further investigate the possibility that increased expression of SET4 promotes survival during oxidative stress, we used the β-estradiol–inducible promoter system to overexpress Set4, as described above. We induced SET4 expression using a low amount of β-estradiol, which minimized the toxic effect of SET4 overexpression, as observed in Fig. 1C. Indeed, cells overexpressing SET4 showed increased survival following exposure to H2O2 both by using a semiquantitative plating assay (Fig. 2E) and quantifying percentage survival of cfu following treatment with H2O2 (Fig. 2F). These data further support our finding that higher protein levels of Set4 in the cell promote survival during oxidative stress.
An alteration in the cell's ability to manage intracellular reactive oxygen species (ROS) levels may result in differing survival rates in the presence of exogenous oxidative stress. Therefore, we next investigated whether cells lacking or overexpressing SET4 had different levels of ROS compared with WT cells grown under the same conditions. To monitor intracellular ROS, flow cytometry was performed using the ROS indicator dye 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA) (32). Following exposure to H2O2, set4Δ cells showed an increase in intracellular ROS relative to WT cells (Fig. 3A), indicating that cells lacking Set4 may not efficiently eliminate ROS, potentially causing decreased survival. Furthermore, even in the absence of exogenous H2O2, WT yeast overexpressing SET4 showed a clear decrease in the amount of intracellular ROS compared with the control strain carrying an empty vector (Fig. 3B), suggesting that cells with high levels of Set4 maintain low levels of endogenous ROS. Overall, our phenotypic analyses show that higher levels of Set4 correlate with increased survival during oxidative stress, whereas loss of Set4 leads to sensitivity in oxidative stress. This suggests that there is a protective role for Set4 in the presence of oxidative stress and raises the possibility that the levels of Set4 in the cell are calibrated to promote survival under normal and stress conditions.
Figure 3.
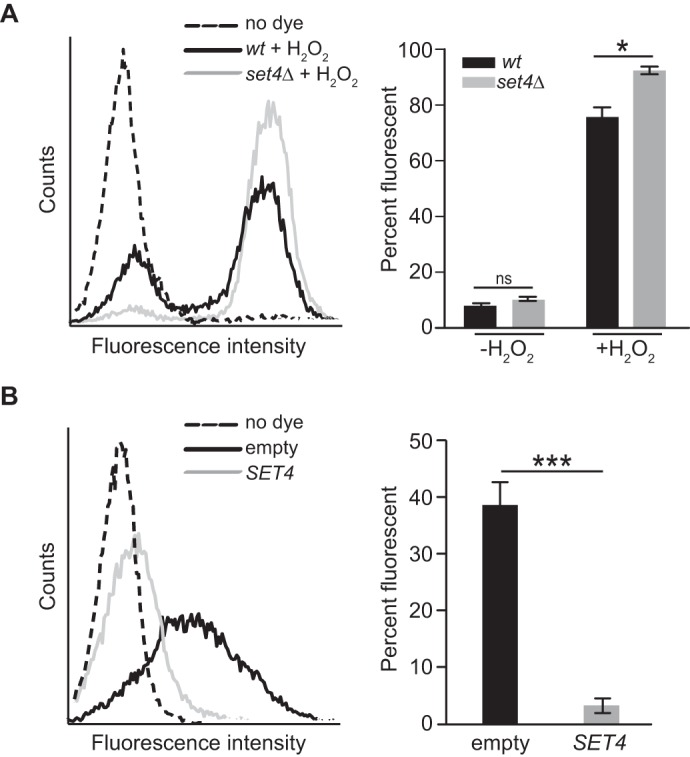
Intracellular ROS levels are altered in set4Δ and SET4-overexpressing cells. Shown are representative flow cytometric histograms of intracellular ROS levels detected using DCFH-DA (left) and percentage fluorescent cells for each strain (right). A, WT (yEG001) and set4Δ (yEG325) strains were grown to mid-log phase in YPD or YPD with 4 mm H2O2 for 30 min. B, yeast carrying either pMN3 (empty) or pMN3-SET4 (yEG372 and yEG375) were grown to early-log phase in SC-URA and treated with 25 μm β-estradiol for 3 h. Error bars, S.E. from three biological replicates. Asterisks, p values from unpaired t tests (*, p ≤ 0.05; ***, p ≤ 0.001).
Set4 localizes to chromatin, and its association is enhanced during oxidative stress
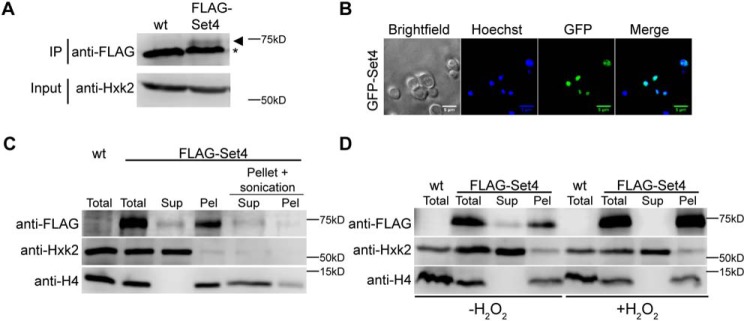
Although commonly associated with chromatin, the localization of SET domain–containing proteins is variable, and some members of this protein family show both nuclear and cytoplasmic distribution. The subcellular localization of Set4 has not been characterized, likely because the expression level of Set4 is very low under normal growth conditions (25). To further investigate the protein levels of Set4, we performed immunoprecipitations (IPs) of N-terminally 3xFLAG-tagged Set4 expressed from the SET4 locus under control of its endogenous promoter. Although FLAG-Set4 was not detectable by immunoblotting of whole-cell lysates (data not shown), we were able to detect FLAG-Set4 by IP from the equivalent of 2 liters of mid-log phase yeast cells grown in rich medium (Fig. 4A). This shows that Set4 is expressed in cells under normal growth conditions, albeit at low abundance.
Figure 4.
Set4 localizes to the nucleus and associates with chromatin. A, immunoblot of anti-FLAG IP from either WT (yEG001) cells or cells expressing FLAG-Set4 from its endogenous locus (yEG513) grown to mid-log phase in YPD. An anti-FLAG blot is shown for the IP eluates, and anti-Hxk2 (hexokinase) is shown for the input as a protein loading control (FLAG-Set4 is not detectable in the input samples). Arrowhead, FLAG-Set4; *, a nonspecific protein that binds to the anti-FLAG beads. B, fluorescence microscopy of cells carrying pGAL1-GFP-SET4 (yEG735), induced in 2% galactose. DNA is stained with Hoechst. Scale bar, 5 μm. C, anti-FLAG immunoblots from chromatin fractionation of extracts from WT cells or cells with FLAG-Set4 (yEG626) induced with 10 μm β-estradiol for 3 h. The soluble supernatant (Sup) and insoluble, chromatin pellet (Pel) fractions are shown. Pellet + sonication, samples that come from sonication of the resuspended chromatin pellet and subsequent centrifugation to separate soluble (Sup) and insoluble (Pel) material. Anti-H4 and anti-Hxk2 blots are shown as controls for chromatin and soluble proteins, respectively. D, immunoblots from chromatin fractionation performed as in C except that cells induced to express FLAG-Set4 were treated with 20 mm H2O2 for 60 min.
The low level of Set4 protein expression precludes the use of common immunological and fluorescence-based assays for determining its localization. Therefore, we analyzed the localization of ectopically expressed Set4 using two different assays. Live-cell imaging of GFP-Set4 expressed from a galactose-inducible promoter showed GFP-Set4 predominantly localized to the nucleus (Fig. 4B). To determine whether or not ectopically expressed Set4 is specifically associated with chromatin, we used a subcellular fractionation assay that separates soluble cytoplasmic and nuclear material from insoluble material associated with chromatin (33, 34). FLAG-Set4 was expressed from a β-estradiol–inducible promoter (as in Fig. 1C) and was observed predominantly in the chromatin fraction, with a small pool of protein in the soluble fraction (Fig. 4C). To verify that FLAG-Set4 was indeed chromatin-associated and not simply insoluble, we used sonication to shear the DNA and solubilize nucleosomes within the pellet containing the chromatin fraction. The pellet was resuspended, sonicated, and subjected to another round of centrifugation. Immunoblot analysis of the supernatant and pellet at this stage showed that the majority of the detectable FLAG-Set4 was released into the supernatant (Fig. 4C), indicating that it is likely to be directly associating with nucleosomes and not simply insoluble. Using this assay, we also tested whether or not the localization of Set4 is altered during oxidative stress. In the presence of H2O2, we observed that all of the FLAG-Set4 is associated with chromatin in the pellet fraction, and there is no FLAG-Set4 in the soluble fraction (Fig. 4D). As further investigated below, this finding suggests the possibility that association of Set4 with chromatin may be regulated during oxidative stress.
Set4 regulates stress response gene expression
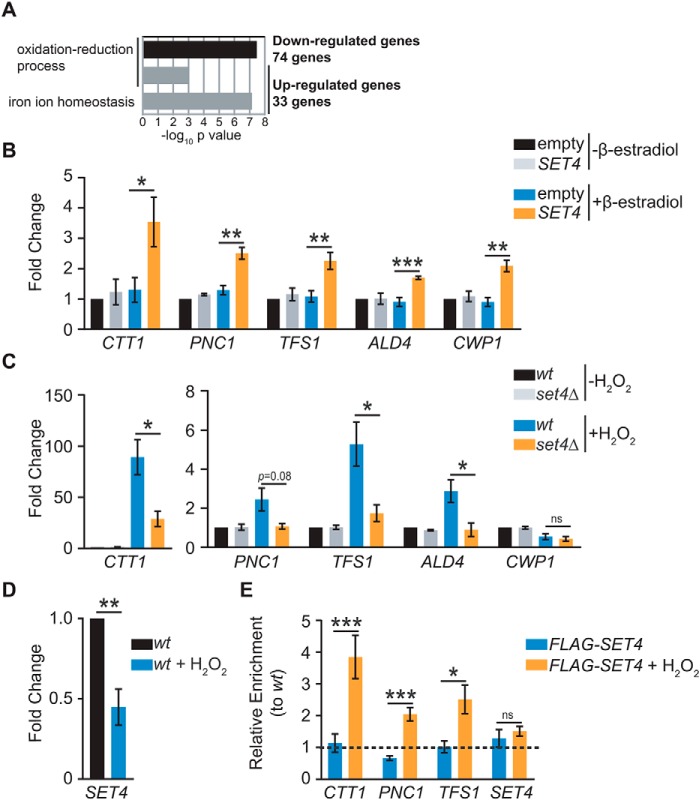
Based on the chromatin localization of Set4, we hypothesized that it may act to regulate stress-responsive genes, potentially as a means to promote survival during oxidative stress. A previously published microarray analysis of set4Δ cells grown under normal conditions identified ∼120 significantly differentially expressed genes compared with WT, with a majority down-regulated in the absence of Set4 (35). Gene ontology (GO) analysis of the 74 down-regulated genes revealed enrichment in oxidation-reduction processes (Fig. 5A), consistent with the phenotypes we have linked to loss of Set4. The 33 up-regulated genes also showed enrichment for the GO categories oxidation–reduction process and iron ion homeostasis. Given the larger quantity of genes down-regulated compared with up-regulated in set4Δ cells, we focused on down-regulated genes for further targeted analysis.
Figure 5.
Set4 promotes activation of stress response genes. A, GO analysis of significantly differentially expressed genes identified by microarray analysis of set4Δ cells from Kemmeren et al. (35). The −log10 p value of the enriched GO biological process categories are shown for both down-regulated and up-regulated genes. B, RT-qPCR of stress response genes identified by Kemmeren et al. (35) from yeast with empty pMN3 or pMN3-SET4 in the presence or absence of β-estradiol. Expression levels were normalized to SCR1. -Fold change relative to empty vector strain without β-estradiol treatment is shown. Error bars, S.E. from at least three biological replicates. C, RT-qPCR of stress response genes from WT (yEG001) and set4Δ (yEG322) strains grown in YPD and treated with 0.4 mm H2O2 for 30 min. Expression levels were normalized to SCR1. -Fold change relative to the WT strain without H2O2 treatment is shown. Error bars, S.E. from at least three biological replicates. PNC1 consistently showed lower expression in set4Δ cells treated with H2O2, although this did not reach the threshold for statistical significance due to variability in its expression levels in H2O2. D, -fold change, as in C, of SET4 mRNA in WT cells without or with 0.4 mm H2O2 for 30 min. E, chIP of FLAG-Set4 (expressed from its endogenous promoter; yEG513) from cells grown to mid-log phase in YPD or YPD with 0.4 mm H2O2 for 30 min. Percentage of input was calculated for each IP and primer set, and enrichment is shown for FLAG-Set4 relative to the WT (untagged) strain either with or without H2O2 treatment. The primers detect the promoter sequences for each of the indicated genes. Error bars, S.E. from three biological replicates. For all panels, asterisks represent p values from unpaired t tests (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ns, not significant), except for E, in which a paired t test was used.
Quantitative RT-PCR was performed to monitor expression of genes that appeared dependent on Set4 in previous work (35) and were linked to the response to oxidative stress. We first analyzed their expression in cells with ectopically expressed Set4 grown under normal conditions without H2O2 treatment. Expression of these genes (CTT1, PNC1, TFS1, ALD4, and CWP1) increased in the presence of excess Set4 (Fig. 5B), suggesting that a higher level of Set4 may be sufficient to promote activation of some stress response genes. We next investigated the expression of these genes in WT and set4Δ cells with or without H2O2 treatment. We observed increased expression of CTT1, PNC1, TFS1, and ALD4, although not CWP1, in the presence of H2O2 in WT cells (Fig. 5C). However, their expression was attenuated in set4Δ mutants under stress, suggesting that Set4 is important for full activation of these stress response genes following H2O2 exposure. Additionally, we monitored the expression of SET4 itself in response to H2O2 treatment of WT cells. Unexpectedly, we observed a decrease in SET4 mRNA expression in H2O2 (Fig. 5D). This suggests negative regulation of the SET4 transcript in oxidative stress, although we postulate that other types of regulatory mechanisms offset this down-regulation of the transcript during stress, as further outlined under “Discussion.”
To determine whether Set4 directly regulates genes induced during oxidative stress, ChIP of cells expressing FLAG-Set4 from its endogenous promoter or untagged control cells was performed either with or without H2O2 treatment. We probed promoters of genes shown to be dependent on Set4 during oxidative stress, including CTT1, PNC1, and TFS1, as well as the promoter of SET4. We were unable to detect substantial association of FLAG-Set4 with any chromatin regions in the absence of H2O2; however, specific enrichment of FLAG-Set4 was observed over the CTT1, PNC1, and TFS1 promoters following treatment with H2O2 (Fig. 5E). We did not see any enrichment at the SET4 promoter, which served as a negative control region. This is consistent with the observation that Set4 localization may be regulated during oxidative stress (Fig. 4D) and indicates that these genes may be direct targets of Set4 during stress.
Regulation of ergosterol biosynthetic genes by Set4 depends on growth conditions and is not a primary influence during the oxidative stress response
Previous studies have shown that cell survival in the presence of H2O2 is directly linked to the permeability of the plasma membrane (36). Ergosterol biosynthetic genes are largely down-regulated in the presence of oxidative stress (37), likely as a means to protect cells by altering membrane permeability. A recent report showed that Set4 maintains repression of ergosterol biosynthetic (ERG) genes during hypoxia (38). To test whether ERG genes are regulated by Set4 during oxidative stress, potentially as a mechanism to promote cell survival, we analyzed the expression of a subset of ERG genes in WT and set4Δ cells following H2O2 treatment. As expected, we observed down-regulation of ERG11, ERG3, ERG2, and ERG6 in WT cells treated with H2O2 (Fig. 6A). set4Δ cells, however, showed reduced expression of these genes in rich medium (YPD) alone, without H2O2 treatment, and they were expressed at similar levels in WT and set4Δ cells that were treated with H2O2. This indicates that ERG genes depend on Set4 for full activation under normal, aerobic growth conditions in rich medium, although Set4 does not play a role in their repression during oxidative stress. When a similar experiment was performed in synthetic complete (SC) medium, no difference in ERG gene expression was observed in set4Δ cells compared with WT (Fig. 6B), suggesting that whereas regulation of ERG genes by Set4 occurs in unstressed cells, its role in gene expression appears to depend on the composition of the growth medium. As elaborated under “Discussion,” this suggests a context-dependent role for Set4 in repression of ergosterol biosynthetic genes (38).
Figure 6.
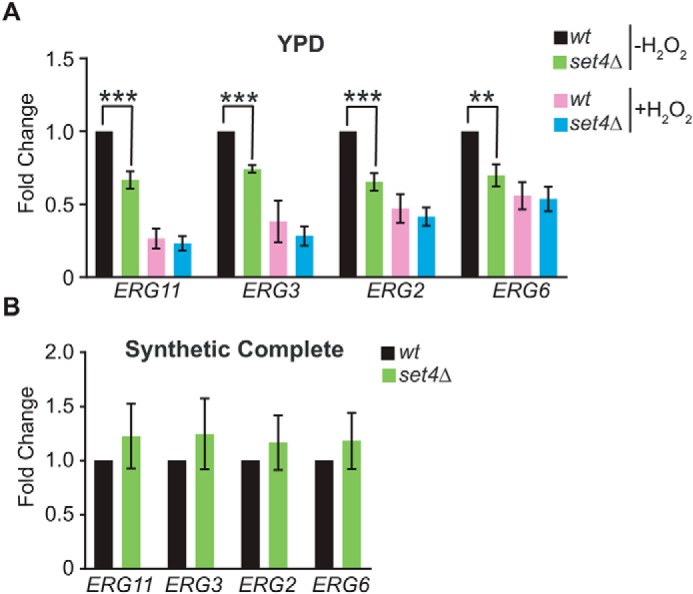
Set4-dependent regulation of ERG gene expression. A, RT-qPCR of ERG genes from WT (yEG001) and set4Δ (yEG322) strains grown in YPD and treated with 0.4 mm H2O2 for 30 min. Expression levels were normalized to SCR1. -Fold change relative to the WT strain without H2O2 treatment is shown. Error bars, S.E. from three biological replicates. B, RT-qPCR as in A except that cells were grown in SC medium, and H2O2 treatment was not performed. Asterisks, p values from unpaired t tests (**, p ≤ 0.01; ***, p ≤ 0.001).
Set4 and Set3 each promote survival during oxidative stress, although they have distinct molecular roles
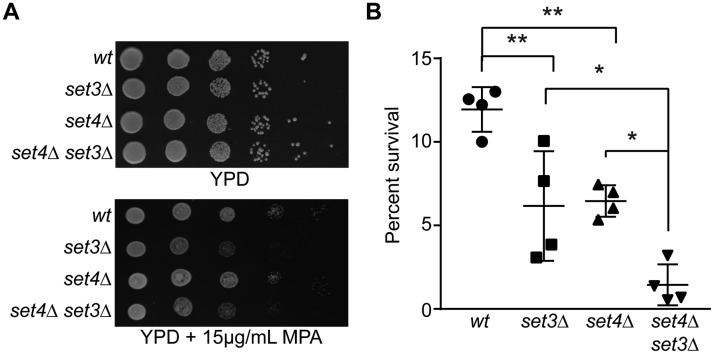
Set4 is a paralog to Set3, although it is unknown whether or not these proteins share functional redundancy. Notably, overexpression of SET3 does not impair cell growth, whereas overexpression of SET4 does (Fig. 1B), and gene expression profiles of set3Δ and set4Δ cells grown under the same conditions are distinct (35). We investigated the extent of genetic redundancy between the two factors by generating double set4Δ set3Δ mutants. These mutants do not show any growth defects in rich medium (Fig. 7A). set3Δ mutants are sensitive to mycophenolic acid (MPA), which interferes with transcription elongation (16); however, set4Δ grew similarly to WT in the presence of MPA (Fig. 7A). The double mutant grew similar to the set3Δ mutant, suggesting no genetic redundancy for this phenotype. We also analyzed growth of the single and double mutants in the 25 different environmental conditions used to screen set4Δ mutants for phenotypes. We did not observe any difference in growth for the set4Δ set3Δ double mutant compared with the single mutants for any of these conditions (data not shown). However, following treatment with H2O2, we observed that set3Δ cells are sensitive to oxidative stress, and the double set4Δ set3Δ mutant shows enhanced sensitivity (Fig. 7B). These data suggest that there is some functional overlap between the two genes in that they each contribute to cell survival during oxidative stress. However, the lack of a shared phenotype in other contexts and their distinct biochemical roles (see “Discussion”) suggest that they have primarily independent molecular functions.
Figure 7.
Partial functional overlap between SET3 and SET4. A, 10-fold serial dilutions of WT (yEG001), set3Δ (yEG330), set4Δ (yEG325), and set4Δ set3Δ (yEG347) strains spotted on YPD or YPD with 15 μg/ml MPA. B, percentage survival of the same strains as in A following growth in YPD and treatment with 4 mm H2O2 for 30 min. The scatter plot shows the mean and S.D. from four biological replicates. One-way analysis of variance was performed, followed by Turkey's post hoc test to determine statistical significance. Asterisks represent p values as follows: *, p ≤ 0.05; **, p ≤ 0.01.
Set4 does not show methylation activity toward histones in vitro
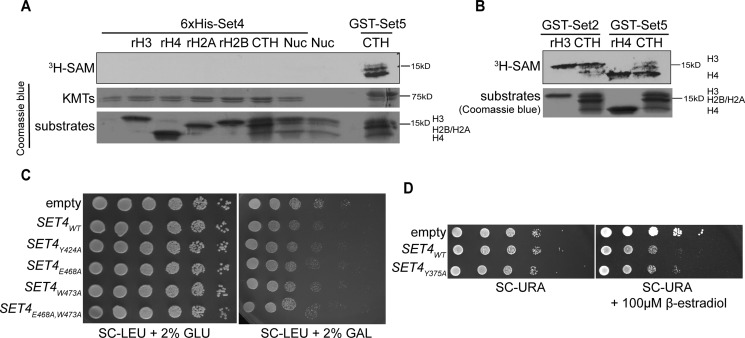
Our data suggest that Set4 is a chromatin-associated protein that plays a role in gene expression regulation. Whereas the SET domain of Set4 is similar to other proteins predicted to lack catalytic activity (Fig. 1A), the potential methyltransferase activity of Set4 has not been reported. We therefore investigated the catalytic activity of Set4 using in vitro methylation assays and multiple histone substrates. Recombinant His6-Set4 was expressed and purified from Escherichia coli and incubated with individual, recombinant histones, histones purified from calf thymus (which contain pre-existing post-translational modifications), and nucleosomes, in the presence of radiolabeled SAM ([3H]SAM) (Fig. 8A). We did not observe a signal on any of these potential substrates, although the positive controls Set2 and Set5 were able to methylate histone substrates (Fig. 8B).
Figure 8.
Recombinant Set4 does not methylate histones in vitro. A, autoradiograph and Coomassie Blue staining of SDS-PAGE with in vitro methylation assays using purified His6-Set4 and histone substrates, including recombinant H3 (rH3), rH4, rH2A, rH2B, calf thymus histones (CTH), and nucleosomes purified from Hela cells (Nuc). A reaction with GST-Set5 and CTH as substrate was run on the same gel as a positive control. Coomassie Blue of KMTs shows both His6-Set4 and GST-Set5 used in the reactions. B, autoradiograph and Coomassie Blue staining of SDS-PAGE with in vitro methylation assays using purified GST-Set2 and GST-Set5 on multiple substrates. C, 10-fold serial dilutions of WT yeast (yEG001) carrying WT and mutant SET4 under the control of the GAL1 promoter spotted on either SC-LEU with 2% glucose (GLU) or 2% galactose (GAL) and grown at 30 °C for 2–3 days. D, serial dilutions of WT strains (yEG315) with either WT or Y375A mutant pMN3-SET4 spotted on SC-URA or SC-URA with 100 μm β-estradiol and grown at 30 °C for 3 days.
We also performed genetic tests to investigate the requirement for the SET domain in Set4 function. We found that complete deletion of the SET domain partially destabilized the protein, precluding use of this mutation in genetic assays (data not shown). We therefore generated point mutations within the SET domain, as indicated in the sequence in Fig. 1A. These mutations include a tyrosine conserved with Set3 (Tyr-424), a conserved glutamic acid within a region implicated in SAM binding in other SET domains (Glu-468), and a tryptophan that replaces a tyrosine found in other SET domains implicated in SAM and target lysine binding or catalysis (Trp-473). In addition, we also mutated a tyrosine conserved with other SET domains, which has been shown to act as a base for catalysis in some enzymes (Tyr-375; not shown in the sequence in Fig. 1A (20)). Using the galactose-induced overexpression system, expression of the Y424A, E468A, and W473A mutants conferred similar growth defects to cells overexpressing WT SET4 (Fig. 8C). Cells overexpressing the Y375A mutant using the β-estradiol–inducible promoter also showed growth very similar to that of cells with WT SET4 overexpression (Fig. 8D), indicating that Set4 function is largely intact in the presence of this mutation. Although there may be additional residues required for a function associated with the SET domain, these data indicate that some of the conserved residues, and those substituted compared with canonical SET domains, are not required for Set4 function.
Discussion
In this study, we investigated the biological functions of the uncharacterized SET domain–containing protein Set4 in budding yeast. Although its yeast paralog, Set3, is well characterized as a regulator of gene expression (14, 16), little has been uncovered about the function of Set4. We identified an important role for Set4 in protecting cells when challenged with oxidative insults, primarily H2O2. Interestingly, high levels of Set4 expression were deleterious to cells under normal growth conditions; however, increased expression of Set4 appeared to promote survival during oxidative stress. Our results also indicated that Set4 promotes proper expression of stress response genes, likely through direct binding to chromatin. The association of Set4 with chromatin also appears regulated, with increased association when cells are treated with H2O2. Overall, our results support a model in which Set4 localization to chromatin is regulated by the cell to fine-tune stress response gene expression, which promotes cell survival following oxidative insults.
Our phenotypic analysis of SET4-overexpressing and set4Δ cells uncovered a protective role for Set4 in the presence of oxidative stress. Based on its domain structure, nuclear localization, and association with chromatin, we hypothesized that Set4 may protect cells by contributing to the cascade of gene expression changes that underlie the response to oxidative stress. Analysis of previously published genome-wide data (35) indicated that Set4 might regulate genes associated with defending against oxidative damage and that it appeared to promote activation of a higher number of genes than it repressed. Additional investigation of Set4-dependent changes in gene expression showed that overexpression of Set4 resulted in increased expression of stress response genes, and the loss of Set4 attenuated induction of these genes following H2O2 treatment, suggesting a role for Set4 in gene activation. We note that we did not see down-regulation of these genes in unstressed conditions, as observed by Kemmeren et al. (35). However, we suspect this difference is due to our low-throughput growth conditions, which have maximum shaking and culture aeration, whereas standard high-throughput growth conditions may induce mild stress on cells. Moreover, we saw a clear defect in induction of these genes in set4Δ cells once the cells were stressed by H2O2.
Interestingly, we observed increased ROS in set4Δ cells upon H2O2 treatment compared with WT and also observed substantially decreased ROS under normal conditions when Set4 is overexpressed (Fig. 3). One possibility is that the Set4-dependent changes in intracellular ROS are due to gene expression differences that contribute to ROS-scavenging activities, most likely cytosolic catalase, encoded by CTT1. In SET4-overexpressing cells, CTT1 is highly expressed, whereas it is not fully induced in set4Δ mutants treated with H2O2. These changes in CTT1 expression correlate with the respective ROS levels in the tested genotypes and are also likely linked to the ability of cells to survive challenges by exogenous H2O2. Whereas there may be gene expression-independent functions of Set4 that contribute to its role in ROS regulation and cell defense pathways in oxidative stress, further experiments are required to address these possibilities. It is also possible that alterations in ROS levels are due to indirect effects associated with SET4 overexpression or loss. Of note, previous work has shown that mouse hematopoietic stem and progenitor cells (HSPCs) lacking MLL5 have increased ROS, which has been linked to defective HSPC function (39). This highlights potential functional similarity between yeast Set4 and mammalian MLL5 and a possible conserved role of this SET domain subfamily in ROS regulation.
Given that increased membrane permeability sensitizes cells to exogenous H2O2 (36), another plausible mechanism for Set4-mediated protection in the presence of H2O2 could be the regulation of ergosterol biosynthetic genes, which control the levels of the plasma membrane sterol, ergosterol, in yeast. A recent report found that Set4 represses ERG genes during hypoxia, in a manner dependent on the transcription factor Hap1 (38). Our data suggest that proper expression of ERG genes depends on Set4 in cells grown in rich medium even under unstressed conditions, although their expression is Set4-independent either with H2O2 treatment or in a more depleted medium, such as synthetic complete medium (Fig. 6). This finding suggests that changes to ERG gene expression in set4Δ cells are not likely to be the primary mechanism driving their sensitivity to H2O2, given that set4Δ cells are sensitive to H2O2 in both rich and synthetic medium (Fig. 2). In addition, these data indicate that the regulation of ERG gene expression by Set4 is not limited to hypoxia (38), and ERG genes show differential expression in set4Δ even in aerobic conditions before hypoxic treatment when cells are grown in rich medium.
Although our observation that Set4 promotes full activation of ERG genes in aerobic conditions in rich medium appears contradictory to the finding that Set4 promotes repression of ERG genes during chronic hypoxia in rich medium (38), this discrepancy may be explained by condition-specific functions of Set4 and may underscore potentially different roles in acute stress (short H2O2 treatment) versus chronic stress. Set4 may be able to both activate and repress genes, particularly in a condition-specific manner. Similar behavior has been characterized for a number of stress-responsive transcription factors, including Hap1, Mot3, and Rox1 (40, 41).
Unlike the most well-studied SET domain chromatin regulators in yeast, including Set1, Set2, and Set3, Set4 has been reported to be expressed at very low levels under normal growth conditions (29). Based on our observations that Set4 was required for full ERG gene expression under normal conditions, as well as survival and gene expression regulation following relatively short treatments with H2O2, we predicted that Set4 is present and functioning at low abundance in cells without stress. Indeed, endogenous levels of FLAG-Set4 are detectable by IP from a high quantity of cells grown in normal conditions (Fig. 4A), indicating that Set4 may function in specific pathways in unstressed cells, even at low abundance. Based on the deleterious effect of Set4 overexpression (Fig. 1), it appears important to maintain low protein levels of Set4 to promote survival in the absence of stress.
In the presence of H2O2, we also observed increased association of Set4 with chromatin. ChIP experiments showed a specific increase in Set4 binding at promoters of genes dependent on Set4 for expression in H2O2 (Fig. 5E), which is also supported by our finding that when Set4 is overexpressed in cells treated with H2O2, it is all associated with chromatin (Fig. 4D). These data indicate the existence of post-translational regulatory mechanisms that promote Set4 binding to chromatin during oxidative stress and suggest that Set4 is highly responsive to environmental conditions. Although increased transcription of SET4 has been reported when cells are grown under anaerobic (26) or hypoxic (38) conditions, our data suggest that acute stress with H2O2 results in lower steady-state levels of the SET4 transcript (Fig. 5D). This raises the possibility that SET4 expression may be subject to negative feedback regulation, at least during acute oxidative stress, which appears to differ from the positive transcriptional regulation of its expression during longer-term exposure to low or no oxygen (38). It is plausible that the negative regulation of SET4 transcript levels during oxidative stress is counteracted by the increased association of Set4 with chromatin and potentially other means of translational or post-translational control. Although the mechanisms that regulate Set4 function are not yet clear, these observations suggest that there are likely multiple modes of regulation acting on Set4, including at the level of its expression and localization. This implies a specialized, regulated role for Set4 activity at chromatin.
Based on the sequence of its SET domain, Set4 belongs to a subfamily of SET domain–containing proteins that includes the yeast paralog Set3 as well as the fly protein UpSET and mammalian MLL5. Biochemical and structural analyses of the SET domains within these proteins have indicated that they lack intrinsic methyltransferase activity (17, 20), although a biochemical test of Set4 activity had not been previously reported. Based on its localization to chromatin and its role in gene expression, we postulated that the most likely substrates for Set4 are histones. However, our in vitro methylation assays with recombinant Set4 and multiple histone substrates did not detect any methylation activity (Fig. 8A). Although it remains formally possible that Set4 requires interacting partners or post-translational modifications for its activity or that it targets nonhistone substrates, this finding is consistent with previous reports regarding Set3, UpSET, and MLL5 (15, 17, 18, 23). Additionally, we generated a number of mutations within the SET domain that did not appear to alter Set4 function (Fig. 8, C and D). It remains possible that there are functional roles for the SET domain independent of histone methylation or that the domain may catalyze methylation through an alternate mechanism; however, there is not yet clear evidence that it is a primary contributor to the biological function of Set4.
The yeast protein Set3 is also thought to lack catalytic activity, although it has clear roles in gene expression regulation by forming a complex with HDACs and other associated proteins (14–16). The full extent of Set4's physical interactions in the cell remain to be determined, but it may be that, similar to Set3, a primary function for Set4 is to regulate the activity of other chromatin modifiers or transcriptional machinery.
Although Set3 and Set4 are paralogs, previous work suggests that they are not likely to have overlapping molecular functions. Set3 binds methylated H3K4 via its PHD finger (16, 42), although the PHD finger of Set4 is not able to bind H3K4me (or H3K36me) in vitro (42, 43). The abundance of Set3 is much higher than Set4 under normal growth conditions, and gene expression profiles of cells lacking either Set3 or Set4 are quite distinct under normal growth conditions (35). Analysis of set4Δ set3Δ mutants suggests that they share partial genetic redundancy in promoting survival during oxidative stress (Fig. 7B). We did not observe any redundancy in the response of the mutants to MPA (Fig. 7A) or other stresses; nor do SET3 and SET4 show similar phenotypes when overexpressed (Fig. 1B). Combined with previous observations regarding molecular functions of these proteins, we postulate that they are likely to each contribute to cell defense during oxidative stress through distinct molecular mechanisms. Further investigation will be required to reveal the molecular details underlying the role of Set3 in oxidative stress and mechanisms driving the stress-responsive regulation of gene expression by Set4.
This study provides insight into the biological functions of Set4 and uncovers a key role for Set4 in protecting cells during oxidative stress, likely through the direct control of gene expression. Set4 localization to chromatin appears to be regulated by the cell, indicating that it is very responsive to changes in environmental conditions. Our results suggest that Set4 functions in a stress defense pathway that allows the cell to carefully calibrate gene expression in response to environmental stress, and we expect that further study will shed light on the mechanisms by which Set4 is regulated and acts at chromatin.
Experimental procedures
Yeast strains, plasmids, and growth conditions
Yeast were grown in standard media, including YPD (1% yeast extract, 2% peptone, 2% dextrose) and synthetic complete (SC) or dropout media (US Biological). Agar plates were prepared with the same medium conditions, and, where necessary, the carbon source was substituted with 2% galactose, or additional reagents were added to the medium following autoclaving, including β-estradiol (Sigma-Aldrich), cadmium chloride, or MPA (MP Biomedicals).
All yeast strains used in this study are listed in Table 1. Yeast gene deletions were constructed using insertion of targeted PCR cassettes amplified from the pFA6a vector series (44). The N-terminal 3xFLAG-tagged SET4 was generated using PCR amplification from plasmid pZM467 (kindly provided by K. Struhl) and loopout of the URA3 marker from transformed cells by selection on plates containing 5-fluoroorotic acid (45). Double set4Δ set3Δ strains were generated by crossing parental strains yEG325 and yEG330 and performing tetrad dissection. All newly constructed strains were confirmed by growth on selective media and colony PCR using primers targeting individual gene deletions or epitope tag insertions.
Table 1.
Yeast strains used in this study
| Strain | Background | Genotype | Source |
|---|---|---|---|
| yEG001 | BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | YKO |
| yEG322 | BY4741 | MATa set4Δ::HIS3MX | This study |
| yEG325 | BY4741 | MATa set4Δ::HIS3MX | This study |
| yEG638 | BY4741 | MATa yEG001 + p045 (pRS316) | This study |
| yEG639 | BY4741 | MATa yEG322 + p045 (pRS316) | This study |
| yEG640 | BY4741 | MATa yEG001 + p188 (pRS316-SET4) | This study |
| yEG641 | BY4741 | MATa yEG322 + p188 (pRS316-SET4) | This study |
| yEG513 | BY4741 | MATa SET4::FLAG-SET4 | This study |
| yEG735 | BY4741 | MATa set4Δ::HIS3MX + p171 (pRS315-GAL1p-GFP-SET4) | This study |
| yEG330 | BY4741 | MATα set3Δ::NATMX | This study |
| yEG347 | BY4741 | MATa set4Δ::HIS3MX set3Δ::NATMX | This study |
| yEG315 | DBY12394 | MATa ura3Δ leu2Δ0::ACT1pr-Z3EV-NATMX | Ref. 28 |
| yEG372 | DBY12394 | MATa yEG315 + p183 (pMN3) | This study |
| yEG375 | DBY12394 | MATa yEG315 + p180 (pMN3-SET4) | This study |
| yEG626 | DBY12394 | MATa ura3Δ leu2Δ0::ACT1pr-Z3EV-NATMX set4Δ::HIS3MX + p302 (pMN3-FLAG-SET4) | This study |
| yEG282 | Sigma1278b | MATα ura3-52 | From P. Farabaugh |
| yEG443 | Sigma1278b | MATα yEG282 + p041 (pRS316-GAL1p) | This study |
| yEG444 | Sigma1278b | MATα yEG282 + p175 (pRS316-GAL1p-SET4) | This study |
All plasmids used in this study are listed in Table 2. Plasmids for GAL1 promoter–driven overexpression were constructed by insertion of open reading frames for each gene into the pRS315-GAL1p vector using ApaI and SalI restriction sites. Similarly, GFP-SET4 was amplified from a yeast strain containing an integrated N-terminal GFP tag and cloned into the pRS315-GAL1p vector to generate pRS315-GAL1p-GFP-SET4. The pRS316-SET4 expression vector was generated by cloning a PCR fragment that amplified the SET4 locus, including its promoter and 3′-UTR, into pRS316. SET4 and FLAG-SET4 were amplified from yeast strains yEG001 and yEG513, respectively, and cloned into the pMN3 vector (kindly provided by Scott McIsaac, Botstein laboratory) downstream of the modified GAL1 promoter recognized by the artificial transcription factor Z3EV (28). Mutations in the SET domain were generated in either pRS315-GAL1p-SET4 or pMN3-modGAL1p-SET4 using a QuikChange site-directed mutagenesis protocol. Full-length Set4 was cloned into pRSET-B using the SacI and NcoI restriction sites for expression of His6-Set4 in E. coli.
Table 2.
Plasmids used in this study
| Number | Description |
|---|---|
| p040 | pRS315-GAL1p |
| p185 | pRS315-GAL1p-SET1 |
| p186 | pRS315-GAL1p-SET2 |
| p157 | pRS315-GAL1p-SET3 |
| p132 | pRS315-GAL1p-SET4 |
| p133 | pRS315-GAL1p-SET5 |
| p134 | pRS315-GAL1p-SET6 |
| p187 | pRS315-GAL1p-DOT1 |
| p041 | pRS316-GAL1p |
| p175 | pRS316-GAL1p-SET4 |
| p171 | pRS315-GAL1p-GFP-SET4 |
| p183 | pMN3-modGAL1p (from Ref. 30) |
| p180 | pMN3-modGAL1p-SET4 |
| p302 | pMN3-modGAL1p-FLAG-SET4 |
| p045 | pRS316 |
| p188 | pRS316-SET4 |
| p325 | pRSET-His6-SET4 |
| p008 | pGEX-GST-SET5 |
| p018 | pGEX-GST-SET2 |
| p154 | pRS315-GAL1p-SET4Y424A |
| p155 | pRS315-GAL1p-SET4E468A |
| p156 | pRS315-GAL1p-SET4W473A |
| p161 | pRS315-GAL1p-SET4E468A,W473A |
| p286 | pMN3-modGAL1p-SET4Y375A |
Adhesion assays
To monitor adhesion to agar, the Σ1278b strain (kindly provided by P. Farabaugh) carrying either pRS316-GAL1p or pRS316GAL1p-SET4 was grown to mid-log phase in SC-URA with 2% glucose. 50 μl of each culture was spotted on SC-URA containing either 2% glucose or 2% galactose and incubated at 30 °C for 10 days. The plates were washed with sterile water using a shaking platform, as described (31), and imaged. To monitor adhesion to polystyrene, the strains were grown in SC-URA with 2% glucose or 2% galactose for 7 days at 30 °C. Cell densities were adjusted to an A600 = 2.0, and 100 μl of each culture was added to a well in a 96-well plate. The cells were allowed to settle and were stained with crystal violet, as described (46).
Hydrogen peroxide survival assays
WT (yEG001) and set4Δ (yEG322) cultures were diluted 1:10,000 in YPD and grown to mid-log phase (A600 ∼0.6–0.7). The cultures were normalized such that all strains had the same A600 and then divided in half. One culture received 4 mm H2O2, and both treated and untreated cells were incubated for an additional 30 min, shaking at 30 °C. The cells were serially diluted 10-fold and spotted onto YPD plates. To count cfu, equivalent volumes of cultures before H2O2 treatment were serially diluted and plated on YPD to determine total cfu/culture. Following H2O2 treatment, equivalent volumes of cultures were plated directly on YPD to determine surviving cfu. Percent survival was determined based on surviving cfu after H2O2 treatment compared with total cfu before treatment. For experiments in which the pRS316-SET4 rescue plasmid was used, cells were grown in or plated on SC-URA medium, and a 20 mm H2O2 treatment was used. The higher concentration of H2O2 was used to account for the increased inherent resistance to H2O2 of yeast grown in synthetic medium. To test the role of SET4 overexpression, yeast strain yEG375, carrying the pMN3-SET4 vector, and yEG372, carrying the empty vector control, were grown overnight in SC-URA with shaking at 30 °C. The following day, two cultures diluted back to an A600 ∼0.2 were prepared, and one culture was treated with either 1 μm (plate assay) or 50 nm (cfu assay) β-estradiol. The cells were grown to mid-log phase, and the culture volumes were normalized based on A600. One set of cultures was treated with 20 mm H2O2 for 30 min, and survival was monitored by spotting serial dilutions or counting cfu as described above.
Intracellular ROS detection
Detection of intracellular ROS was performed using DCFH-DA (Sigma) as described previously (32). Briefly, 10 ml of mid-log phase yeast cells (strains yEG001 and yEG322) grown in YPD at 30 °C were treated with 4 mm H2O2 for 30 min. 10 μm DCFH-DA was added to the culture medium for 1 h. For experiments with ectopic Set4 expression, yeast strains yEG372 and yEG375 were grown to early log phase in SC-URA at 30 °C and treated with 25 μm β-estradiol. DCFH-DA (10 μm) was added to the culture for 1 h with shaking. After treatment with DCFH-DA, cells were washed twice in PBS, resuspended in PBS, and analyzed using a Beckman Coulter CyAn ADP flow cytometer. Data were further analyzed using Summit software. WT cells not treated with DCFH-DA were used as a negative control, and cells treated with 6 mm H2O2 and DCFH-DA served as a positive control for dye staining. Gating was performed to calculate the percentage of stained cells per culture by excluding all cells overlapping with the unstained control.
Immunoprecipitation of FLAG-Set4
WT (yEG001) and SET4::FLAG-SET4 (yEG513) strains were grown in 4 liters of YPD to mid-log phase. Cells were harvested, resuspended in lysis buffer (100 mm HEPES pH 8.0, 20 mm MgOAc, 10% glycerol, 10 mm EGTA, 0.1 mm EDTA, 1 mm PMSF, yeast protease inhibitor mixture (Sigma)), and frozen dropwise in liquid nitrogen. Cell pellets were ground in a coffee grinder with dry ice, the dry ice was allowed to evaporate, and lysates were clarified by centrifugation at 7000 rpm for 15 min. IPs were set up with equal protein concentrations of lysate and anti-FLAG M2 magnetic beads (Sigma-Aldrich) and rotated for 3 h at 4 °C. Proteins were eluted with SDS loading buffer, and 50% of the protein eluted from IPs was run on SDS-PAGE and subjected to immunoblotting with mouse anti-FLAG (Sigma-Aldrich; catalogue no. F1804). 0.2% of input protein was also run as a loading control and probed with anti-hexokinase (Hxk2)-HRP (Novus Biologicals; catalogue no. NB120-20547).
GFP-Set4 live cell microscopy
A set4Δ strain carrying pRS315-GAL1-GFP-SET4 (yEG735) was grown overnight at 30 °C in SC-LEU plus 2% glucose, transferred to SC-LEU plus 2% raffinose, and induced with 2% galactose until mid-log phase. The cells were washed with PBS, stained with Hoechst (2 μg/ml; Life Technologies, Inc.), harvested, and mounted on a glass slide in 5% low temperature melting agarose dissolved in SC-LEU plus 2% galactose medium. Images were obtained using a Leica SP5 confocal microscope, and image processing was performed using Image J.
Chromatin fractionation
Yeast strains yEG372 and yEG626, carrying an empty vector or pMN3-FLAG-SET4, were grown to mid-log phase in 25 ml of SC-URA with shaking at 30 °C. FLAG-Set4 expression was induced by treating cells with 10 μm β-estradiol for 3 h, followed by treatment with 20 mm H2O2 for 1 h, harvested, and frozen in liquid nitrogen. Cell pellets were processed for chromatin fractionation as described previously (33, 34, 47). For experiments in which the pellet fraction was subjected to sonication to shear chromatin, the fractions were sonicated for eight cycles of 20 s each at 15% output in a Branson sonifier. Collected fractions were boiled in SDS loading buffer, and equivalent volumes were subjected to SDS-PAGE. Immunoblots were probed with mouse anti-FLAG, mouse anti-H4 (Abcam; catalogue no. ab31830), and anti-Hxk2-HRP. Blot imaging was performed on a LI-COR C-DiGit chemiluminescent Western blotting scanner.
Gene expression analysis by quantitative RT-PCR
To test gene expression changes in the presence of SET4 overexpression, strains yEG372 and yEG375 were grown in SC-URA overnight at 30 °C, diluted back to an A600 of ∼0.2, and treated with 1 μm β-estradiol until cells reached mid-log phase (∼6 h). Cells were harvested and frozen in liquid nitrogen before RNA isolation. To test expression in WT (yEG001) and set4Δ (yEG322), cultures were prepared as described for the H2O2 survival assays, except that 0.4 mm H2O2 treatment for 30 min was performed to provide acute stress but not dramatically decrease survival, as previously performed for measurement of gene expression changes following H2O2 treatment in yeast (9, 48, 49). RNA isolation, removal of genomic DNA contamination, cDNA synthesis, and quantitative RT-PCR were performed as described previously (50, 51). Gene-specific primers for the qPCR are listed in Table 3. Gene expression was normalized to SCR1, which was observed to have stable expression under the growth conditions tested.
Table 3.
Primers used in this study
| Gene | Analysis | Sequence |
|---|---|---|
| CTT1 | RT-qPCR | 5′-AGACCAGACGGCCCTATCTT-3′ |
| 5′-TACACGCTCCGGAACTCTTT-3′ | ||
| TFS1 | RT-qPCR | 5′-CCCAATTCCAGTTCACGTTC-3′ |
| 5′-TGAACAAATCGTCGTCTTGG-3′ | ||
| ALD4 | RT-qPCR | 5′-GCACAACAGACCTCACCAGA-3′ |
| 5′-TTGGAGCTGGGTGGTAAATC-3′ | ||
| PNC1 | RT-qPCR | 5′-GACCACTGTCCTGCTGGATT-3′ |
| 5′-TTGTGGGCCTTCAACTCTTC-3′ | ||
| CWP1 | RT-qPCR | 5′-GAAGGTTCTGAGAGCGATGC-3′ |
| 5′-GACCCGTCCTTGATAGCGTA-3′ | ||
| SET4 | RT-qPCR | 5′-CATTATACCGCCCCAACAAC-3′ |
| 5′-TTGAATTCCGTGTCATTGGA-3′ | ||
| SCR1 | RT-qPCR | 5′-GGGAGTTTTATCCAGCGTCA-3′ |
| 5′-GAAGCGATCAACTTGCACAAT-3′ | ||
| ERG11 | RT-qPCR | 5′-CCTCTTATTCCGTCGGTGAA-3′ |
| 5′-TGTGTCTACCACCACCGAAA-3′ | ||
| ERG3 | RT-qPCR | 5′-GTCGATTTTCAACCATCCTCGT-3′ |
| 5′-TCGACATCCATGGGATAGCAC-3′ | ||
| ERG2 | RT-qPCR | 5′-AGACGCACTTGCCTCTCATT-3′ |
| 5′-GGATACGGAAGCGTGTAGGA-3′ | ||
| ERG6 | RT-qPCR | 5′-GTCTAGCCCCAGAAGGTTCC-3′ |
| 5′-TTCCTAGCGACGAAAAGCAT-3′ | ||
| CTT1 promoter | ChIP | 5′-ATTCGACGTAGCCTGGACAC-3′ |
| 5′-TGGAATAGAGGTAAAGCAACGA-3′ | ||
| PNC1 promoter | ChIP | 5′-TTCAAGGGGCAGGGGTTT-3′ |
| 5′-TATTAGCACATCATAATCGTATCTGGA-3′ | ||
| TFS1 promoter | ChIP | 5′-AAGGGATAGAGGGGCTGTTC-3′ |
| 5′-AGGAGCACTGCAATTCGTTT-3′ | ||
| SET4 promoter | ChIP | 5′-TGAAAAGGAAAGGAGGGAAA-3′ |
| 5′-GCGTTCCACGCTGAAATTAT-3′ |
ChIP
WT (yEG001) and SET4::FLAG-SET4 (yEG513) were grown as described for gene expression analysis, including treatment with 0.4 mm H2O2 for 30 min in 100 ml of YPD. Cells were fixed with 1% formaldehyde for 45 min, and ChIP procedures were performed as described previously (51, 52). Mouse anti-FLAG (Sigma; catalogue no. F1804) was prebound to protein A/G magnetic beads (Pierce) and then incubated with ChIP lysates for 3 h at 4 °C. qPCR was performed on eluted DNA, as described (51, 52), with primers listed in Table 3. The percentage input was calculated relative to 10% input, and the signal for FLAG-Set4 was determined relative to WT (untagged) either with or without H2O2 treatment.
Recombinant protein purification
His6-Set4 was expressed and purified from plasmid p325 using E. coli BL21 (DE3). Protein expression was induced using 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 4 h, and cell lysates were prepared in 20 mm sodium phosphate, pH 7.4, 150 mm NaCl, 10 mm imidazole, 0.5% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitors (ThermoScientific Pierce protease inhibitor tablet). His6-Set4 was bound to nickel–nitrilotriacetic acid resin for 1 h at 4 °C. The resin was washed with lysis buffer, including 300 mm NaCl and 25 mm imidazole. His6-Set4 was eluted with 250 mm imidazole, and subsequently dialyzed into 20 mm Tris-HCl, pH 8.0, 20 mm KCl, 5 mm MgCl2, 5% glycerol before performing methylation assays. Purification of GST-Set5 and GST-Set2 was performed as described previously (13).
In vitro methylation assays
Methylation assays were performed as described previously (13). Briefly, 30 μg of His6-Set4 was incubated with 1 μg of substrate in 50 mm Tris-HCl (pH 8.0), 10% glycerol, 20 mm KCl, 5 mm MgCl2 plus 2 μCi of [3H]SAM (PerkinElmer Life Sciences). Reactions were incubated at 30 °C overnight, and proteins were resolved by SDS-PAGE, followed by autoradiography. For reactions with GST-Set5 and GST-Set2, 5 μg of enzyme was used. Substrates include recombinant histones (New England Biolabs), histones purified from calf thymus (Worthington), and mononucleosomes purified from HeLa cells (Epicypher).
Author contributions
K. T. and E. M. G. conceptualization; K. T. and E. M. G. formal analysis; K. T., Y. J., and D. J. investigation; K. T., Y. J., and D. J. methodology; K. T., Y. J., D. J., and E. M. G. writing-original draft; K. T., Y. J., and E. M. G. writing-review and editing; E. M. G. supervision; E. M. G. funding acquisition.
Acknowledgments
We thank members of the Green laboratory for additional technical support, helpful discussions, and comments on the manuscript. We also thank Leila Bahmani Kazerooni, Pushya Pradeep, and Julie Wolf from the Applied Molecular Biology program at UMBC for technical assistance; Sue Ostrand-Rosenberg, members of the Rosenberg laboratory, and Gregory Szeto for help with flow cytometry; and Tagide deCarvalho of the Keith R. Porter Imaging Facility (funded by National Science Foundation Grant DBI0722569) for assistance with microscopy. We also acknowledge Philip Farabaugh, Lasse Lindahl, Kevin Struhl, and Scott McIsaac for yeast strains and plasmids.
This work was supported in part by National Institutes of Health Grant R01GM124342 (to E. M. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- H3 and H4
- histone H3 and H4, respectively
- me
- methylation
- HDAC
- histone deacetylase
- SAM
- S-adenosylmethionine
- KMT
- lysine methyltransferase
- ROS
- reactive oxygen species
- DCFH-DA
- 2,7-dichlorodihydrofluorescein diacetate
- IP
- immunoprecipitation
- GO
- gene ontology
- SC
- synthetic complete
- MPA
- mycophenolic acid
- qPCR
- quantitative PCR
- HSPC
- hematopoietic stem and progenitor cell
- HRP
- horseradish peroxidase.
References
- 1. Feil R., and Fraga M. F. (2012) Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109 10.1038/nrg3142 [DOI] [PubMed] [Google Scholar]
- 2. Badeaux A. I., and Shi Y. (2013) Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 14, 211–224 10.1038/nrm3545 [DOI] [PubMed] [Google Scholar]
- 3. Suganuma T., and Workman J. L. (2011) Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 80, 473–499 10.1146/annurev-biochem-061809-175347 [DOI] [PubMed] [Google Scholar]
- 4. Jaiswal D., Turniansky R., and Green E. M. (2017) Choose your own adventure: the role of histone modifications in yeast cell fate. J. Mol. Biol. 429, 1946–1957 10.1016/j.jmb.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Nadal E., Zapater M., Alepuz P. M., Sumoy L., Mas G., and Posas F. (2004) The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427, 370–374 10.1038/nature02258 [DOI] [PubMed] [Google Scholar]
- 6. Nadal-Ribelles M., Mas G., Millán-Zambrano G., Solé C., Ammerer G., Chávez S., Posas F., and de Nadal E. (2015) H3K4 monomethylation dictates nucleosome dynamics and chromatin remodeling at stress-responsive genes. Nucleic Acids Res. 43, 4937–4949 10.1093/nar/gkv220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Law M. J., and Ciccaglione K. (2015) Fine-tuning of histone H3 Lys4 methylation during pseudohyphal differentiation by the CDK submodule of RNA polymerase II. Genetics 199, 435–453 10.1534/genetics.114.172841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McDaniel S. L., Hepperla A. J., Huang J., Dronamraju R., Adams A. T., Kulkarni V. G., Davis I. J., and Strahl B. D. (2017) H3K36 methylation regulates nutrient stress response in Saccharomyces cerevisiae by enforcing transcriptional fidelity. Cell Rep. 19, 2371–2382 10.1016/j.celrep.2017.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baker L. A., Ueberheide B. M., Dewell S., Chait B. T., Zheng D., and Allis C. D. (2013) The yeast Snt2 protein coordinates the transcriptional response to hydrogen peroxide-mediated oxidative stress. Mol. Cell Biol. 33, 3735–3748 10.1128/MCB.00025-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morano K. A., Grant C. M., and Moye-Rowley W. S. (2012) The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195 10.1534/genetics.111.128033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarke S. G. (2013) Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem. Sci. 38, 243–252 10.1016/j.tibs.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weiner A., Chen H. V., Liu C. L., Rahat A., Klien A., Soares L., Gudipati M., Pfeffner J., Regev A., Buratowski S., Pleiss J. A., Friedman N., and Rando O. J. (2012) Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 10, e1001369 10.1371/journal.pbio.1001369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green E. M., Mas G., Young N. L., Garcia B. A., and Gozani O. (2012) Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat. Struct. Mol. Biol. 19, 361–363 10.1038/nsmb.2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim T., Xu Z., Clauder-Münster S., Steinmetz L. M., and Buratowski S. (2012) Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150, 1158–1169 10.1016/j.cell.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pijnappel W. W., Schaft D., Roguev A., Shevchenko A., Tekotte H., Wilm M., Rigaut G., Séraphin B., Aasland R., and Stewart A. F. (2001) The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15, 2991–3004 10.1101/gad.207401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim T., and Buratowski S. (2009) Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137, 259–272 10.1016/j.cell.2009.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Madan V., Madan B., Brykczynska U., Zilbermann F., Hogeveen K., Döhner K., Döhner H., Weber O., Blum C., Rodewald H. R., Sassone-Corsi P., Peters A. H., and Fehling H. J. (2009) Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood 113, 1444–1454 [DOI] [PubMed] [Google Scholar]
- 18. Rincon-Arano H., Halow J., Delrow J. J., Parkhurst S. M., and Groudine M. (2012) UpSET recruits HDAC complexes and restricts chromatin accessibility and acetylation at promoter regions. Cell 151, 1214–1228 10.1016/j.cell.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McElroy K. A., Jung Y. L., Zee B. M., Wang C. I., Park P. J., and Kuroda M. I. (2017) upSET, the Drosophila homologue of SET3, is required for viability and the proper balance of active and repressive chromatin marks. G3 (Bethesda) 7, 625–635 10.1534/g3.116.037788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dillon S. C., Zhang X., Trievel R. C., and Cheng X. (2005) The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6, 227 10.1186/gb-2005-6-8-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nin D. S., Yew C. W., Tay S. K., and Deng L. W. (2014) Targeted silencing of MLL5β inhibits tumor growth and promotes γ-irradiation sensitization in HPV16/18-associated cervical cancers. Mol. Cancer Ther. 13, 2572–2582 10.1158/1535-7163.MCT-14-0019 [DOI] [PubMed] [Google Scholar]
- 22. Sebastian S., Sreenivas P., Sambasivan R., Cheedipudi S., Kandalla P., Pavlath G. K., and Dhawan J. (2009) MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc. Natl. Acad. Sci. U.S.A. 106, 4719–4724 10.1073/pnas.0807136106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mas-Y-Mas S., Barbon M., Teyssier C., Déméné H., Carvalho J. E., Bird L. E., Lebedev A., Fattori J., Schubert M., Dumas C., Bourguet W., and le Maire A. (2016) The human mixed lineage leukemia 5 (MLL5), a sequentially and structurally divergent SET domain-containing protein with no intrinsic catalytic activity. PLoS One 11, e0165139 10.1371/journal.pone.0165139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ali M., Rincón-Arano H., Zhao W., Rothbart S. B., Tong Q., Parkhurst S. M., Strahl B. D., Deng L. W., Groudine M., and Kutateladze T. G. (2013) Molecular basis for chromatin binding and regulation of MLL5. Proc. Natl. Acad. Sci. U.S.A. 110, 11296–11301 10.1073/pnas.1310156110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., and Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- 26. Lai L. C., Kosorukoff A. L., Burke P. V., and Kwast K. E. (2006) Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot. Cell 5, 1468–1489 10.1128/EC.00107-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aragon A. D., Rodriguez A. L., Meirelles O., Roy S., Davidson G. S., Tapia P. H., Allen C., Joe R., Benn D., and Werner-Washburne M. (2008) Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol. Biol. Cell 19, 1271–1280 10.1091/mbc.e07-07-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McIsaac R. S., Gibney P. A., Chandran S. S., Benjamin K. R., and Botstein D. (2014) Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res. 42, e48 10.1093/nar/gkt1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S. G., Cyert M., Hughes T. R., Boone C., and Andrews B. (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21, 319–330 10.1016/j.molcel.2005.12.011 [DOI] [PubMed] [Google Scholar]
- 30. Cullen P. J., and Sprague G. F. Jr. (2012) The regulation of filamentous growth in yeast. Genetics 190, 23–49 10.1534/genetics.111.127456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shively C. A., Eckwahl M. J., Dobry C. J., Mellacheruvu D., Nesvizhskii A., and Kumar A. (2013) Genetic networks inducing invasive growth in Saccharomyces cerevisiae identified through systematic genome-wide overexpression. Genetics 193, 1297–1310 10.1534/genetics.112.147876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Azad G. K., Singh V., Mandal P., Singh P., Golla U., Baranwal S., Chauhan S., and Tomar R. S. (2014) Ebselen induces reactive oxygen species (ROS)-mediated cytotoxicity in Saccharomyces cerevisiae with inhibition of glutamate dehydrogenase being a target. FEBS Open Bio. 4, 77–89 10.1016/j.fob.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang C., and Stillman B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11, 3375–3386 10.1101/gad.11.24.3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Donovan S., Harwood J., Drury L. S., and Diffley J. F. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 94, 5611–5616 10.1073/pnas.94.11.5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kemmeren P., Sameith K., van de Pasch L. A., Benschop J. J., Lenstra T. L., Margaritis T., O'Duibhir E., Apweiler E., van Wageningen S., Ko C. W., van Heesch S., Kashani M. M., Ampatziadis-Michailidis G., Brok M. O., Brabers N. A., Miles A. J., Bouwmeester D., van Hooff S. R., van Bakel H., Sluiters E., Bakker L. V., Snel B., Lijnzaad P., van Leenen D., Groot Koerkamp M. J., and Holstege F. C. (2014) Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell 157, 740–752 10.1016/j.cell.2014.02.054 [DOI] [PubMed] [Google Scholar]
- 36. Branco M. R., Marinho H. S., Cyrne L., and Antunes F. (2004) Decrease of H2O2 plasma membrane permeability during adaptation to H2O2in Saccharomyces cerevisiae. J. Biol. Chem. 279, 6501–6506 10.1074/jbc.M311818200 [DOI] [PubMed] [Google Scholar]
- 37. Kelley R., and Ideker T. (2009) Genome-wide fitness and expression profiling implicate Mga2 in adaptation to hydrogen peroxide. PLoS Genet. 5, e1000488 10.1371/journal.pgen.1000488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Serratore N. D., Baker K. M., Macadlo L. A., Gress A. R., Powers B. L., Atallah N., Westerhouse K. M., Hall M. C., Weake V. M., and Briggs S. D. (2018) A novel sterol-signaling pathway governs azole antifungal drug resistance and hypoxic gene repression in Saccharomyces cerevisiae. Genetics 208, 1037–1055 10.1534/genetics.117.300554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tasdogan A., Kumar S., Allies G., Bausinger J., Beckel F., Hofemeister H., Mulaw M., Madan V., Scharfetter-Kochanek K., Feuring-Buske M., Doehner K., Speit G., Stewart A. F., and Fehling H. J. (2016) DNA damage-induced HSPC malfunction depends on ROS accumulation downstream of IFN-1 signaling and Bid mobilization. Cell Stem Cell 19, 752–767 10.1016/j.stem.2016.08.007 [DOI] [PubMed] [Google Scholar]
- 40. Hickman M. J., and Winston F. (2007) Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol. Cell Biol. 27, 7414–7424 10.1128/MCB.00887-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martínez-Montañés F., Rienzo A., Poveda-Huertes D., Pascual-Ahuir A., and Proft M. (2013) Activator and repressor functions of the Mot3 transcription factor in the osmostress response of Saccharomyces cerevisiae. Eukaryot. Cell 12, 636–647 10.1128/EC.00037-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gatchalian J., Ali M., Andrews F. H., Zhang Y., Barrett A. S., and Kutateladze T. G. (2017) Structural insight into recognition of methylated histone H3K4 by Set3. J. Mol. Biol. 429, 2066–2074 10.1016/j.jmb.2016.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shi X., Kachirskaia I., Walter K. L., Kuo J. H., Lake A., Davrazou F., Chan S. M., Martin D. G., Fingerman I. M., Briggs S. D., Howe L., Utz P. J., Kutateladze T. G., Lugovskoy A. A., Bedford M. T., and Gozani O. (2007) Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282, 2450–2455 10.1074/jbc.C600286200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 10.1002/(SICI)1097-0061(199807)14:10%3C953::AID-YEA293%3E3.0.CO%3B2-U [DOI] [PubMed] [Google Scholar]
- 45. Moqtaderi Z., and Struhl K. (2008) Expanding the repertoire of plasmids for PCR-mediated epitope tagging in yeast. Yeast 25, 287–292 10.1002/yea.1581 [DOI] [PubMed] [Google Scholar]
- 46. Cullen P. J. (2015) Biofilm/Mat assays for budding yeast. Cold Spring Harb. Protoc. 2015, 172–175 10.1101/pdb.prot085076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lopes da Rosa J., Holik J., Green E. M., Rando O. J., and Kaufman P. D. (2011) Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics 187, 9–19 10.1534/genetics.110.123117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., and Brown P. O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tsang C. K., Liu Y., Thomas J., Zhang Y., and Zheng X. F. (2014) Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 5, 3446 10.1038/ncomms4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jezek M., Gast A., Choi G., Kulkarni R., Quijote J., Graham-Yooll A., Park D., and Green E. M. (2017) The histone methyltransferases Set5 and Set1 have overlapping functions in gene silencing and telomere maintenance. Epigenetics 12, 93–104 10.1080/15592294.2016.1265712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jaiswal D., Jezek M., Quijote J., Lum J., Choi G., Kulkarni R., Park D., and Green E. M. (2017) Repression of middle sporulation genes in Saccharomyces cerevisiae by the Sum1-Rfm1-Hst1 complex is maintained by Set1 and H3K4 methylation. G3 (Bethesda) 7, 3971–3982 10.1534/g3.117.300150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jezek M., Jacques A., Jaiswal D., and Green E. M. (2017) Chromatin immunoprecipitation (ChIP) of histone modifications from Saccharomyces cerevisiae. J. Vis. Exp. 130, e57080 10.3791/57080 [DOI] [PMC free article] [PubMed] [Google Scholar]