Plastid NAD-dependent malate dehydrogenase is essential for chloroplast development, not because of its enzymatic activity, but because it interacts with FtsH proteases at the inner envelope membrane.
Abstract
Malate dehydrogenases (MDHs) convert malate to oxaloacetate using NAD(H) or NADP(H) as a cofactor. Arabidopsis thaliana mutants lacking plastidial NAD-dependent MDH (pdnad-mdh) are embryo-lethal, and constitutive silencing (miR-mdh-1) causes a pale, dwarfed phenotype. The reason for these severe phenotypes is unknown. Here, we rescued the embryo lethality of pdnad-mdh via embryo-specific expression of pdNAD-MDH. Rescued seedlings developed white leaves with aberrant chloroplasts and failed to reproduce. Inducible silencing of pdNAD-MDH at the rosette stage also resulted in white newly emerging leaves. These data suggest that pdNAD-MDH is important for early plastid development, which is consistent with the reductions in major plastidial galactolipid, carotenoid, and protochlorophyllide levels in miR-mdh-1 seedlings. Surprisingly, the targeting of other NAD-dependent MDH isoforms to the plastid did not complement the embryo lethality of pdnad-mdh, while expression of enzymatically inactive pdNAD-MDH did. These complemented plants grew indistinguishably from the wild type. Both active and inactive forms of pdNAD-MDH interact with a heteromeric AAA-ATPase complex at the inner membrane of the chloroplast envelope. Silencing the expression of FtsH12, a key member of this complex, resulted in a phenotype that strongly resembles miR-mdh-1. We propose that pdNAD-MDH is essential for chloroplast development due to its moonlighting role in stabilizing FtsH12, distinct from its enzymatic function.
INTRODUCTION
Malate dehydrogenases (l-malate-NAD-oxidoreductase [MDH]; EC 1.1.1.37) catalyze the reversible interconversion of malate and oxaloacetate, using NAD(H) or NADP(H) as a cofactor. Plant MDHs form a family of enzymes, with isoforms that are present in several compartments within the cell. The Arabidopsis thaliana genome encodes nine isoforms of MDH: two plastidial, two peroxisomal, two mitochondrial, and three that are assumed to be cytosolic, as they have no predicted localization sequence. All of these isoforms use NAD+ as a cofactor, with the exception of one plastidial isoform that is NADP dependent (NADP-MDH). Although it is well established that MDHs play an important role in central metabolism in plants, the exact role(s) of many of the individual isoforms in Arabidopsis remains unclear, particularly for the plastidial and cytosolic isoforms.
MDHs play a critical role in the tricarboxylic acid cycle in the mitochondria, where they generate NADH by oxidizing malate to oxaloacetate. They can also reduce oxaloacetate back to malate in order to generate NAD+ for the decarboxylation reaction that occurs during the conversion of glycine to serine (Journet et al., 1981). An Arabidopsis double mutant lacking both mitochondrial MDH isoforms is viable but has defects in seed germination, stunted growth, and altered leaf respiration and photorespiration rates (Tomaz et al., 2010; Sew et al., 2016). Mitochondrial MDHs are also involved in the carbon-concentrating mechanism in plants that conduct the NAD-malic enzyme type of C4 photosynthesis (Hatch and Osmond, 1976). The two peroxisomal MDH isoforms generate NAD+, which is mainly required for the β-oxidation of fatty acids. An Arabidopsis mutant deficient in both of these isoforms does not efficiently mobilize triacylglycerols and requires the exogenous supply of sugars for seedling establishment (Pracharoenwattana et al., 2007, 2010).
In the plastids, NADP-MDH was proposed to be the key enzyme in the malate valve, a mechanism by which reducing equivalents can be indirectly transported across membranes of organelles (Heber, 1974; Scheibe, 2004; Taniguchi and Miyake, 2012). NADP-MDH is redox-regulated by the ferredoxin-thioredoxin system and is therefore thought to be active only in the light (Scheibe, 1987). According to the malate valve hypothesis, NADPH is generated in the chloroplast through the electron transport chain during the day but does not readily diffuse out of the chloroplast. NADP-MDH reduces oxaloacetate to malate by oxidizing NADPH to NADP, and the malate is shuttled to the cytosol in exchange for oxaloacetate via the dicarboxylate transporter, AtpOMT1. In the cytosol, malate is oxidized back to oxaloacetate by the cytosolic MDHs, regenerating a reducing equivalent in the form of NADH in that compartment (Kinoshita et al., 2011). However, the importance of the malate valve is unclear. Previous studies of the nadp-mdh knockout mutant have reported either no reduction in the growth of mutants compared with the wild type (Hebbelmann et al., 2012) or a slight reduction (Heyno et al., 2014). The latter study also reported that the mutant had higher H2O2 levels under high light conditions, probably because it could not reversibly inactivate catalase activity.
Unlike NADP-MDH, the activity of the plastidial NAD-dependent malate dehydrogenase (pdNAD-MDH) is not redox sensitive (Berkemeyer et al., 1998). pdNAD-MDH was thus proposed to play an important role in the malate valve in dark and non-green plastids. In contrast to the nadp-mdh mutant, the homozygous pdnad-mdh knockout mutant is embryo-lethal (Beeler et al., 2014; Selinski et al., 2014), a phenotype that was not reported for mutants of any other MDH isoform so far. An artificial microRNA silencing construct was used to generate the miR-mdh-1 line, with constitutively reduced pdNAD-MDH levels using the 35S promoter (Beeler et al., 2014). This line was viable but had pale leaves with disordered chloroplast ultrastructure and was severely compromised in growth. Levels of malate, starch, and glutathione during the night were higher in miR-mdh-1 compared with the wild type. The nighttime respiration rate was also lower in the silencing line. These findings are consistent with a role for pdNAD-MDH in metabolism in the dark, but it is difficult to pinpoint a specific role for the enzyme due to the highly pleiotropic nature of the miR-mdh-1 phenotype. In addition, Selinski et al. (2014) showed that pollen tube growth is affected in pdnad-mdh in vitro, but not in vivo, and proposed that the maternal tissue is able to supply substrates for an enzyme generating NAD+, allowing proper tube elongation in vivo. Together, these data suggest that pdNAD-MDH is important for embryogenesis and subsequent growth and that its loss cannot be compensated by the presence of the NADP-MDH in the plastid or of NAD-MDH in other compartments. However, the link between the activity of the enzyme and the phenotypes resulting from its loss remain unclear. Here, we aimed to investigate the role of pdNAD-MDH in both embryo development and postembryonic growth, focusing on chloroplast development, and to test the importance of NAD-MDH activity in these processes.
RESULTS
Embryo-Specific Expression of pdNAD-MDH Complements the Embryo Lethality of pdnad-mdh
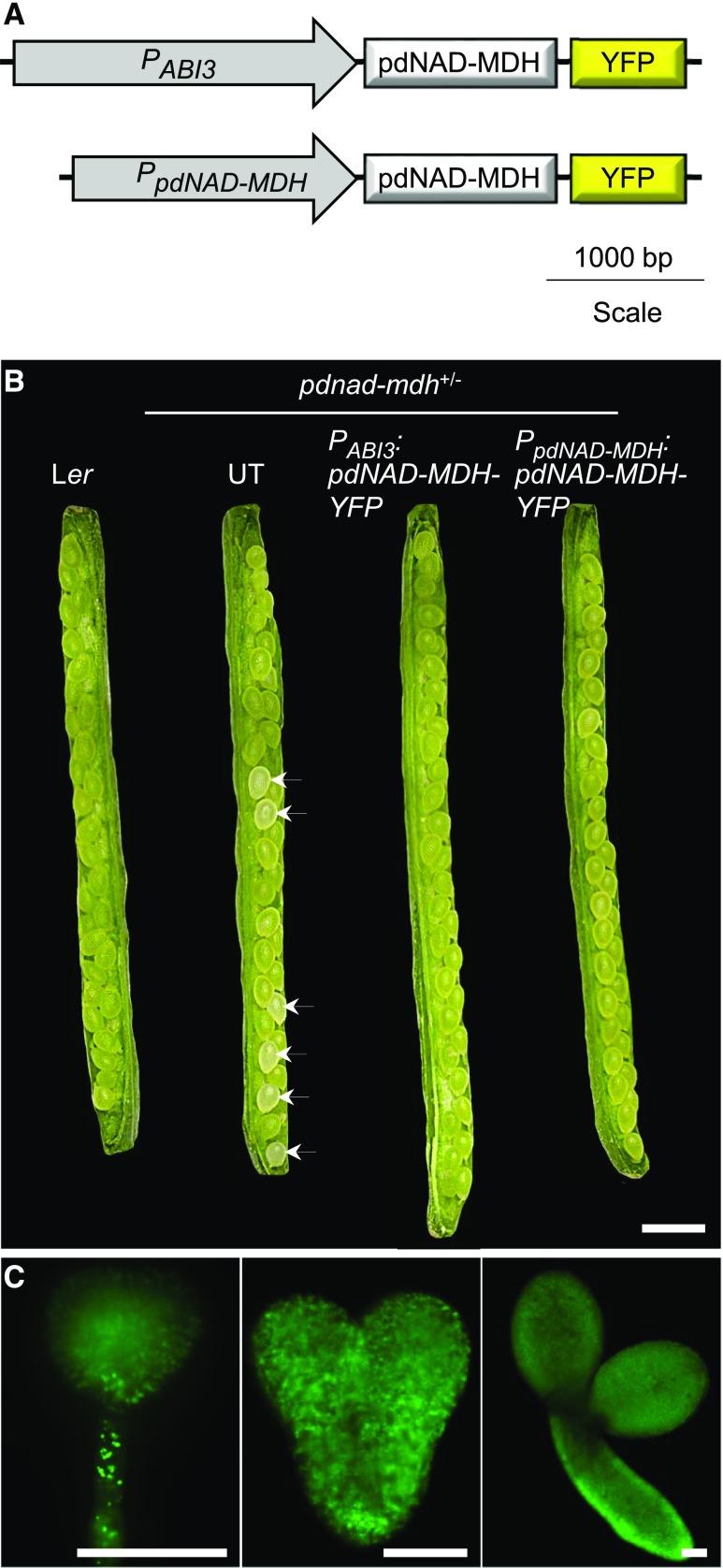
We previously reported that the Arabidopsis pdnad-mdh knockout mutant has an embryo-lethal phenotype and that the silencing line with reduced pdNAD-MDH expression, miR-mdh-1 (carrying a 35S promoter-driven artificial microRNA construct), has a pale, dwarfed phenotype with aberrant chloroplast ultrastructure (Beeler et al., 2014). To investigate how the complete loss of pdNAD-MDH affects postembryogenic growth, we expressed pdNAD-MDH specifically in the embryos of pdnad-mdh. For this, we cloned a PABI3:pdNAD-MDH-YFP construct encoding the pdNAD-MDH protein with a C-terminal YFP tag, driven by the embryo-specific ABI3 promoter (Figure 1A). Previous studies have effectively used the ABI3 promoter to rescue the embryo lethality of various mutants (Despres et al., 2001; Gómez et al., 2010; Candela et al., 2011; Bodi et al., 2012). We transformed heterozygous pdnad-mdh plants with the construct and selected transformed T1 plants using a BASTA resistance marker. The genotype of these T1 plants were determined by PCR amplification of the pdnad-mdh T-DNA insertion, as previously described (Beeler et al., 2014). Four independent lines were selected for further analysis. Plants homozygous for the PABI3:pdNAD-MDH-YFP transgene and heterozygous for pdnad-mdh were identified in the T3 generation. When we opened the siliques of these plants, we found that all seeds within the silique were green (Figure 1B). All seeds were also green in siliques of wild-type Ler plants, and of heterozygous pdnad-mdh plants that were complemented with pdNAD-MDH-YFP expressed under its native promoter (homozygous for the PpdNAD-MDH:pdNAD-MDH-YFP construct; described in Beeler et al., 2014). In contrast, approximately one-quarter of the seeds contained in siliques of untransformed heterozygous pdnad-mdh plants were white. Expression of pdNAD-MDH-YFP under the control of the ABI3 promoter was visualized in isolated embryos using fluorescence microscopy, where we observed YFP signal in embryos at various stages of embryogenesis (globular, heart, and torpedo stage; Figure 1C). These findings suggest that the embryo-specific expression of pdNAD-MDH-YFP can overcome the embryo-lethal phenotype of pdnad-mdh.
Figure 1.
Embryo-Specific Expression of pdNAD-MDH to Rescue the Embryo Lethality of pdnad-mdh Plants.
(A) Schematic diagram of the PABI3:pdNAD-MDH-YFP and PpdNAD-MDH:pdNAD-MDH-YFP constructs. For the former construct, the ABI3 promoter (PABI3) was placed upstream of the pdNAD-MDH coding sequence fused at its C terminus to YFP. The latter construct was described by Beeler et al. (2014).
(B) Opened siliques of pdNAD-MDH+/− plants transformed with the PABI3:pdNAD-MDH-YFP construct. Siliques from the wild type (Ler), untransformed (UT) pdNAD-MDH+/− plants, and pdNAD-MDH+/− transformed with the PpdNAD-MDH:pdNAD-MDH-YFP construct are shown for comparison. White seeds are indicated with an arrow. Bar = 1 mm.
(C) Expression of the PABI3:pdNAD-MDH-YFP construct in Arabidopsis embryos after transformation of pdNAD-MDH+/− plants. Expression was detected by fluorescence microscopy on embryos at three different developmental stages (globular, heart, and torpedo). Bar = 50 µm.
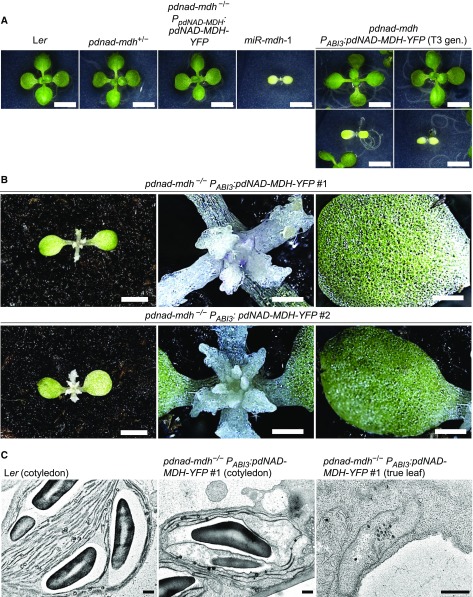
We germinated the progeny from plants that were heterozygous for pdnad-mdh and homozygous for the PABI3:pdNAD-MDH-YFP transgene. At the seedling stage, most of these plants resembled the wild-type (Ler) or heterozygous pdnad-mdh plants, but approximately one-quarter had pale cotyledons resembling those observed in miR-mdh-1 (Figure 2A). These pale seedlings grew very slowly, and once enough material could be harvested for genotyping, they were confirmed to be homozygous for pdnad-mdh. These findings confirmed that the PABI3:pdNAD-MDH-YFP construct could rescue the embryo-lethal phenotype of pdnad-mdh but show that after embryogenesis when the ABI3 promoter is no longer active, these plants become greatly compromised. Aside from the pale cotyledons, plants homozygous for pdnad-mdh initiated albino “true leaves” that failed to undergo proper organogenesis, remaining as small white primordia-like stubs on the meristem (Figure 2B). These plants died 2 to 4 weeks after germination and failed to reach the reproductive stage. The addition of sucrose in the growth medium did not prevent the seedling lethality of these plants, nor did it improve growth (Supplemental Figure 1). By contrast, homozygous pdnad-mdh plants that were complemented with pdNAD-MDH-YFP expressed under its native promoter were indistinguishable from the wild type (Figure 2A).
Figure 2.
Phenotype of pdnad-mdh Mutants Rescued by the Embryo-Specific Expression of pdNAD-MDH.
(A) Photographs of 14-d-old seedlings of pdnad-mdh plants expressing the PABI3:pdNAD-MDH-YFP construct, grown on 0.5× strength MS agar plates under a 12-h-light/12-h-dark regime. The PABI3:pdNAD-MDH-YFP construct was transformed into pdNAD-MDH+/− plants, and a T3 population that was segregating for the pdnad-mdh T-DNA insertion but not for the PABI3:pdNAD-MDH-YFP construct was obtained. Two examples of pale plants that resembled the constitutive silencing line, miR-mdh-1, are shown, while the other plants resembled the wild-type Ler or pdnad-mdh+/−. A pdnad-mdh−/− plant complemented with the PpdNAD-MDH:pdNAD-MDH-YFP construct is also shown. Bars = 500 µm.
(B) Photographs of 4-week-old pdnad-mdh−/− seedlings rescued with the PABI3:pdNAD-MDH-YFP construct. Close-up images of their meristematic zones and their cotyledons are shown. Bars in the left-hand panels = 500 µm, and bars in the middle and right-hand panels = 100 µm.
(C) Transmission electron micrographs of plastids in pdnad-mdh−/− seedlings rescued with the PABI3:pdNAD-MDH-YFP construct. Plastids from cotyledons are shown for the wild type (Ler) and pdnad-mdh−/−, and a plastid in a true leaf of pdnad-mdh−/− is shown on the right. Bars = 500 nm.
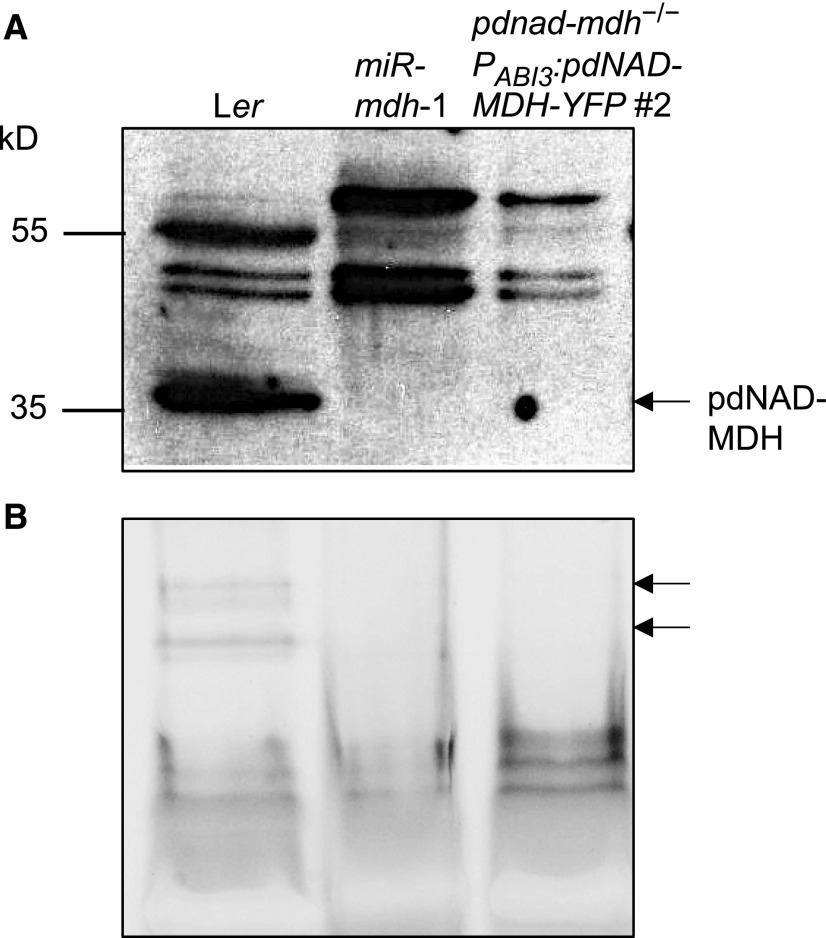
We previously observed that the young leaves of miR-mdh-1 had compromised chloroplast ultrastructure, with fewer thylakoid membranes and starch granules (Beeler et al., 2014). We therefore investigated chloroplast ultrastructure in pdnad-mdh PABI3:pdNAD-MDH-YFP seedlings using transmission electron microscopy. We fixed and embedded cotyledon and true leaf samples from 3- to 4-week-old Ler and pdnad-mdh plants rescued with PABI3:pdNAD-MDH-YFP. The chloroplasts in cotyledons of the rescued pdnad-mdh seedlings contained thylakoid membranes, but they were not as structured as those in wild-type cotyledons (Figure 2C). However, in the white true leaves, no mature chloroplast structures were observed, only proplastid-like structures. We suspect that the chloroplasts in the cotyledons developed further than those in the true leaves due to residual pdNAD-MDH derived from its PABI3-driven expression in cotyledons during embryogenesis (Figure 1C). To confirm that homozygous pdnad-mdh PABI3:pdNAD-MDH-YFP plants do not express pdNAD-MDH protein after embryogenesis, we extracted proteins from these seedlings and performed immunoblots with the pdNAD-MDH antibody, as well as native-PAGE gels followed by MDH activity staining. No bands corresponding to pdNAD-MDH or pdNAD-MDH-YFP were detected in these extracts on the immunoblot (Figure 3A). We observed several unspecific bands on the blot, but our previous cross-reactivity test of this antibody suggests that they are unlikely to be other MDH isoforms (Beeler et al., 2014). These bands were visible in all lines but variable in abundance, possibly due to variation in the amounts of chloroplast proteins such as Rubisco between the lines (gels were loaded on an equal protein basis). On native-PAGE gels, pdNAD-MDH runs as three distinct activity bands, where the lower band corresponds to the free dimer, while the upper bands correspond to pdNAD-MDH in protein-protein interactions. The lower band is difficult to resolve from other NAD-MDH isoforms (Beeler et al., 2014). Thus, it should be noted that only part of the total pdNAD-MDH in the extract can be observed with this technique. The two upper pdNAD-MDH activity bands were clearly observed in extracts of Ler, but not in miR-mdh-1 and pdnad-mdh PABI3:pdNAD-MDH-YFP (Figure 3B). These data confirm that PABI3:pdNAD-MDH-YFP was only active during embryogenesis. Any residual protein from the embryonic tissues was either inactive by this time or below the level of detection.
Figure 3.
Immunoblot and Native-PAGE Detection of pdNAD-MDH in Ler, miR-mdh-1, and pdnad-mdh−/− Seedlings Rescued with the PABI3:pdNAD-MDH-YFP Construct.
(A) Immunoblots were conducted with the pdNAD-MDH antibody. Equal amounts of soluble protein (5 µg) were loaded. The migration of molecular weight markers is indicated (left).
(B) In-gel activity assay of the NAD-MDH. The two bands corresponding to pdNAD-MDH activity are indicated (arrows, right). Equal amounts of protein (15 µg) were loaded.
Inducible Silencing of pdNAD-MDH at the Rosette Stage Results in the Formation of White Leaves from the Meristem
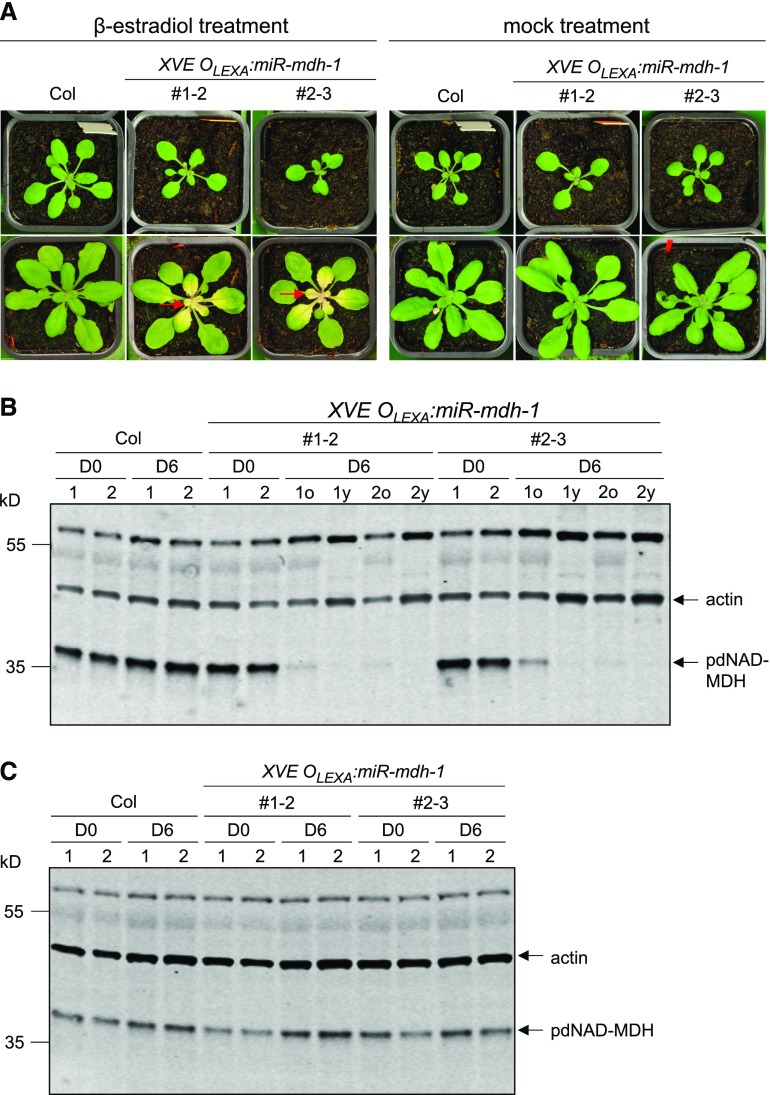
To further investigate the postembryonic role of pdNAD-MDH, we generated Arabidopsis lines where pdNAD-MDH silencing could be induced at later growth stages. The identical microRNA silencing cassette used in miR-mdh-1 was cloned downstream of the XVE/OlexA β-estradiol-inducible promoter system. Wild-type plants were transformed with the construct, and transformed T1 seedlings were selected using the hygromycin resistance marker. Prior to β-estradiol treatment, 3-week-old resistant T2 plants were phenotypically indistinguishable from the wild type. A β-estradiol solution was then sprayed onto the entire rosettes, and this treatment was repeated every 2 d. Six days into the treatment, newly emerging leaves were noticeably pale, whereas the old leaves stayed green (Figure 4A). Paleness was not observed in any leaves of wild-type plants treated with β-estradiol. Immunoblot analysis of proteins extracted from these rosettes confirmed that pdNAD-MDH protein levels were undetectable in the young white leaves of the transgenic line, and only residual amounts of protein were present in the old green leaves (Figures 4B and 4C). The phenotype of the newly emerging leaves suggests that pdNAD-MDH deficiency particularly affects tissues in which chloroplasts are developing. The severe reduction in pdNAD-MDH levels did not have a visible effect on the older leaves, which contain mature chloroplasts.
Figure 4.
β-Estradiol-Inducible Silencing of pdNAD-MDH in Mature Plant Rosettes.
XVE OLEXA:miR-mdh-1 plants photographed before and after a 6-d treatment with β-estradiol or a mock treatment.
(A) Photographs in the top row were taken before β-estradiol treatment (D0) and those in the bottom row after 6 d (D6). β-Estradiol solution (20 µM) was sprayed every second day onto the entire rosette. β-Estradiol was applied to wild-type (Col) plants as a control. Mock-treated samples were sprayed with water containing the same amount of DMSO as the treatment solution (used to dissolve the β-estradiol). Red arrows indicate examples of white, newly emerging leaves.
(B) Immunoblot detection of pdNAD-MDH in total protein extracts of wild-type and XVE OLEXA:miR-mdh-1 plants before (D0) and 6 d into β-estradiol treatment (D6). For the treated XVE OLEXA:miR-mdh-1 plants, proteins were extracted from the old, green leaves (D6o) and young, white leaves (D6y) separately. Gels were loaded on an equal leaf area basis. The migration of molecular mass markers is indicated on the left. pdNAD-MDH and actin (as a loading control) were detected concurrently on the same membrane using secondary antibodies conjugated to different infrared fluorescence dyes (800CW for pdNAD-MDH and 680RD for actin).
(C) As for (B), but with old leaves from mock-treated samples.
pdNAD-MDH Is Important for Early Etioplast and Chloroplast Development
To investigate the role of pdNAD-MDH in chloroplast development in more detail, we studied the deetiolation process in dark-grown miR-mdh-1 seedlings. During deetiolation, plastids undergo a rapid conversion from partially developed chloroplasts (etioplast) to photosynthetic chloroplasts upon illumination (Solymosi and Aronsson, 2013). The prolamellar body (PLB), a crystalline lattice structure within etioplasts, contains structural building blocks for the photosynthetic apparatus, including protochlorophyllide, the enzyme protochlorophyllide oxireductase A (PORA), carotenoids, and fragments of membranes that will form the thylakoids (prothylakoids) (Bahl et al., 1976; Ryberg and Sundqvist, 1982, 1988; Park et al., 2002).
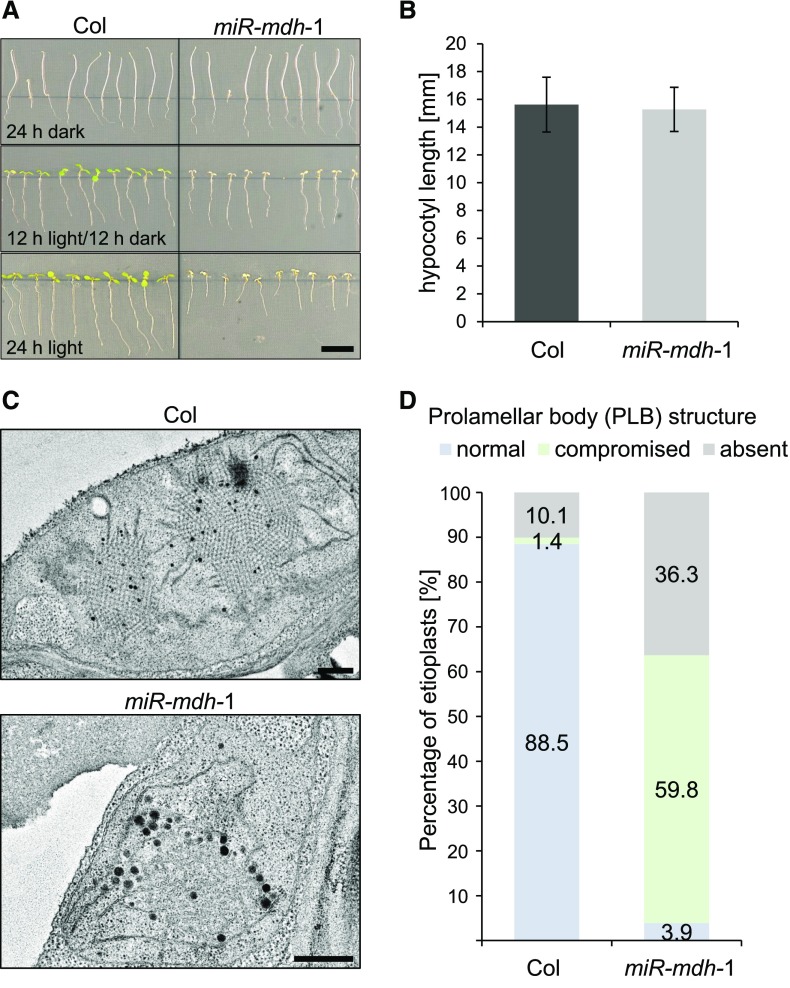
Surprisingly, when grown in the dark, the etiolated miR-mdh-1 seedlings were indistinguishable from the wild type, indicating that skotomorphogenic growth is unaffected by pdNAD-MDH deficiency (Figure 5A). Quantification of the hypocotyl length showed no significant difference between miR-mdh-1 and the wild type (Figure 5B). However, growth of the seedlings in the light was affected in the miR-mdh-1 seedlings. When grown under a 12-h light/12-h dark regime, miR-mdh-1 seedlings had cotyledons that were paler and smaller than those of the wild type (Figure 5A). When grown under continuous light, root growth was further compromised in miR-mdh-1.
Figure 5.
Etiolated Growth of miR-mdh-1 and Etioplast Structure.
(A) Wild-type (Col) and miR-mdh-1 seedlings were grown on 0.5× strength MS agar plates in different diel light conditions (24 h dark, 12 h light/12 h dark, 24 h light). Bar = 1 cm.
(B) Quantification of the hypocotyl length of etiolated wild-type and miR-mdh-1 seedlings. Values represent the mean ± se of measurements conducted on n = 89 and n = 86 seedlings for the wild type and miR-mdh-1, respectively.
(C) Etioplast ultrastructure in cotyledons of etiolated wild-type and miR-mdh-1 seedlings observed by transmission electron microscopy. Bars = 500 nm
(D) Proportion of etioplasts showing normal, compromised, or absent prolamellar body structures in cotyledons of the wild type and miR-mdh-1. The first 139 and 256 etioplasts, observed in wild-type and miR-mdh-1 cotyledons, respectively, were categorized based on their prolamellar body structure.
We then examined etioplast structure in cotyledons of etiolated wild-type and miR-mdh-1 seedlings using transmission electron microscopy. Etioplasts were imaged from sections produced from three different 6-d-old seedlings for both miR-mdh-1 and the wild type. The observed etioplasts were categorized according to their PLB structure—either normal, compromised (with a less ordered structure), or absent (no internal structure observed within the etioplast). Examples of wild-type and miR-mdh-1 etioplasts within each category are shown in Supplemental Figure 2. In wild-type cotyledons, we observed that the vast majority of etioplasts (88.5%, example shown in Figure 5C) had normal PLB structure, and only 1.4% had compromised structure (Figure 5D). PLBs were absent in 10.1% of etioplasts, but this is likely because they were not visible within that thin section of the etioplast. However, in miR-mdh-1, most etioplasts had compromised PLB morphology (59.8%, example shown in Figure 5C) and a substantial proportion had no visible PLB (36.3%).
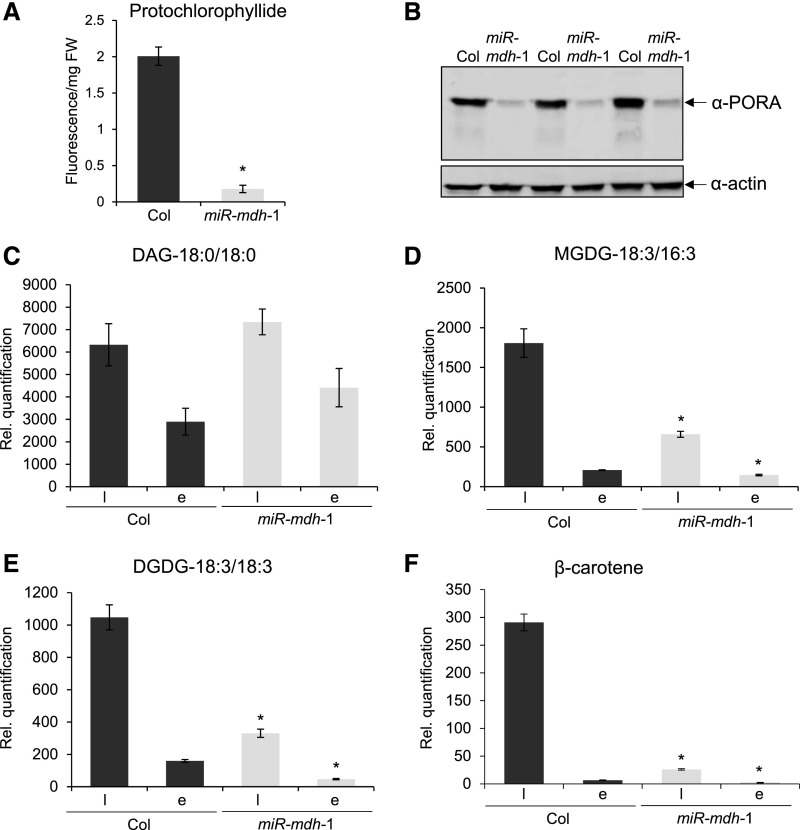
We then quantified the abundance of major prolamellar body components in etiolated miR-mdh-1 seedlings. First, we quantified protochlorophyllide and its binding protein, PORA, using a fluorescence-based assay (Cheminant et al., 2011) and immunoblots, respectively. The levels of both components were greatly reduced in 5-d-old etiolated miR-mdh-1 seedlings compared with the wild type (Figures 6A and B). We also quantified major galactolipids and carotenoids in 6-d-old etiolated seedlings, and, since these molecules are also major components of mature chloroplasts, we simultaneously analyzed extracts of photomorphogenically grown seedlings. Several monogalactosyl- and digalactosyl-diacylglycerol (MGDG and DGDG) species were detected in the extracts. The most abundant MGDG species was MGDG-18:3/16:3, while DGDG-18:3/18:3 was the most abundant DGDG. Interestingly, the levels of MGDG-18:3/16:3 and DGDG-18:3/18:3 were greatly reduced in miR-mdh-1 in both dark-grown and light-grown seedlings, even though there was no difference in the levels of diacylglycerol precursor (Figures 6C to 6E). Similar trends were observed for most other detectable MGDG and DGDG species (Supplemental Table 1). Levels of major carotenoids, such as β-carotene, were also reduced in miR-mdh-1 etiolated seedlings, as well as light-grown seedlings, relative to the wild type (Figure 6F). Similar trends were observed for lutein and viola/zeaxanthin (Supplemental Table 1). In summary, miR-mdh-1 seedlings were deficient in major lipids and carotenoids that are normally present in etioplasts or chloroplasts. Etioplast development is greatly affected by pdNAD-MDH deficiency, even though etiolated growth is not affected. These data further suggest a critical role of pdNAD-MDH in the early stages of chloroplast development.
Figure 6.
Protochlorophylide, Galactolipid, and β-Carotene Contents of miR-mdh-1 Seedlings.
(A) Protochlorophyllide levels in 5-d-old wild-type (Col) and miR-mdh-1 etiolated seedlings, measured by fluorescence after excitation at 440 nm. Values are the means ± se of three biological replicates, each measured with four technical replicates. Each biological replicate is a pool of 100 to 200 individual seedlings.
(B) Immunoblot detection of PORA in total protein extracts from 6-d-old etiolated wild-type and miR-mdh-1 seedlings (upper panel). Immunoblots for actin was performed as a loading control (lower panel). Gels were loaded on an equal fresh weight basis. Three biological replicates are shown. Each biological replicate is a pool of 50 to 100 individual seedlings.
(C) to (F) Lipids and carotenoids were extracted from 6-d-old wild-type and miR-mdh-1 seedlings, either etiolated (E) or light grown (l; grown under a 12-h-light/12-h-dark regime). Levels of DAG (C), MGDG 18:3/16:3 (D) DGDG 18:3/18:3 (E), and β-carotene (F) are shown. For the lipids, only the most common species (in terms of fatty acid chain composition) is shown; similar trends were seen in other detected species (Supplemental Table 1). All lipids and carotenoids were quantified relative to an internal standard, and values were corrected for differences in fresh weight (see Methods for details). Values are the mean ± se from four biological replicates. Each biological replicate is a pool of 50 to 100 individual seedlings. Significant differences (P < 0.05) within the respective light-grown and etiolated samples of miR-mdh-1 and the wild type, determined using a two-tailed t test, are indicated with an asterisk.
The pdNAD-MDH Protein Is Required for Proper Embryo and Chloroplast Development, but Its Enzymatic Activity Is Not
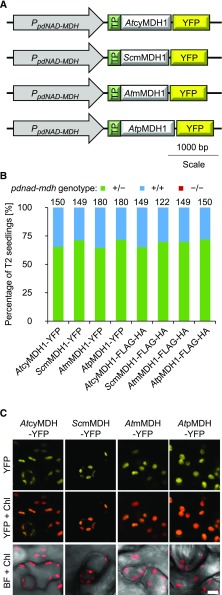
We tested whether we could complement the pdnad-mdh mutant by introducing other NAD-MDH isoforms into the plastid, thereby restoring NAD-MDH activity. The Arabidopsis NAD-MDH isoforms in the mitochondria, peroxisome, and the cytosol have amino acid sequences that are similar to pdNAD-MDH (Supplemental Figure 3). We therefore chose one isoform from each compartment (AtcyMDH1, AtmMDH1, and AtpMDH1) and a more distantly related mitochondrial MDH from yeast (Saccharomyces cerevisiae; ScmMDH1). The coding sequences of the different MDH isoforms were fused directly downstream of the plastidial transit peptide from the Rubisco small subunit (RbcS; amino acids 1–80). For the mitochondrial isoforms, the mitochondrial transit sequence was first predicted using the TargetP 1.1 program (Nielsen et al., 1997; Emanuelsson et al., 2000), and this sequence was replaced with the plastidial one. The constructs were driven by the native pdNAD-MDH promoter and encoded a YFP tag fused to the C-terminal end of each MDH protein (Figure 7A). Constructs were also generated with the smaller Flag-HA peptide tag in place of the YFP. Heterozygous pdnad-mdh plants were transformed with these constructs, and T1 seedlings were selected via BASTA resistance. Expression of the transgene was verified by immunoblotting. All resistant T1 plants were genotyped (5–15 plants per line) for the parental pdnad-mdh mutation, but no homozygous mutant plants were found. We therefore selected T1 plants that were both expressing the transgene and heterozygous for pdnad-mdh, and genotyped the BASTA-resistant T2 progeny (120–180 plants per construct from 5 independent T1 lines [30–36 T2 plants each]). However, we were not able to isolate any plants that were homozygous for pdnad-mdh (Figure 7B). We verified that all of the fusion proteins were correctly targeted to the chloroplast using confocal microscopy on heterozygous pdnad-mdh plants that were transformed with the YFP-tagged constructs. In all lines, YFP signal was detected exclusively in the chloroplasts (Figure 7C). We also verified that the constructs encoded active MDH isoforms via native-PAGE with activity staining. Additional activity bands were observed in extracts from plants expressing AtmMDH1 or ScmMDH1, suggesting that these proteins were indeed active (Supplemental Figure 4). The other isoforms are also potentially active, but their activity on the native-PAGE gel might be masked by the activity bands of the endogenous MDH isoforms. In summary, restoring plastidial NAD-MDH activity alone in pdnad-mdh could not complement the embryo-lethal phenotype.
Figure 7.
Complementation Test with Various NAD-MDH Isoforms Expressed in pdnad-mdh.
(A) Constructs encoding NAD-MDH isoforms under the control of the pdNAD-MDH promoter. The plastidial transit peptide from Rubisco small subunit was fused to the N terminus of each isoform. The constructs shown are tagged at the C terminus with YFP. Similar constructs were cloned with the Flag-HA tag in place of YFP.
(B) Genotyping results of the T2 progeny from T1 plants heterozygous for the pdnad-mdh mutation and expressing the different NAD-MDH isoforms. Numbers above the bars indicate the number of BASTA-resistant T2 plants that were genotyped.
(C) Chloroplast localization of the NAD-MDH isoforms. Similar constructs to those shown in (A), except with the 35S promoter in place of the native promoter, were transiently expressed in Nicotiana benthamiana leaves and imaged using confocal microscopy. Bar = 5 µm.
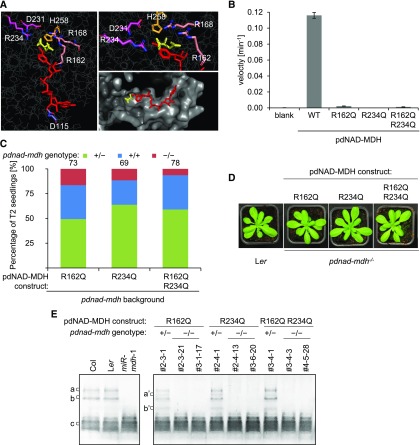
Given these findings, we questioned whether the enzymatic activity of pdNAD-MDH is required at all. Therefore, we generated inactive versions of the pdNAD-MDH protein using site-directed mutagenesis to test whether they could complement the pdnad-mdh mutant. The catalytic site of the malate dehydrogenases is highly conserved among all homologs and includes a histidine, two aspartate, and three arginine residues (Birktoft and Banaszak, 1983; Musrati et al., 1998; Minárik et al., 2002) (Figure 8A). These amino acids are required to coordinate both the substrate and nicotinamide ring of the cofactor within the catalytic pocket. The MDH reaction is initiated by cofactor binding, which then facilitates substrate binding (Silverstein and Sulebele, 1969). When the ternary complex is formed, an external loop closes over the substrate and the residues involved in catalysis (Nicholls et al., 1992; Goward and Nicholls, 1994). Additionally, two conserved arginines on a flexible loop are brought into close proximity to the substrate (Grau et al., 1981; Clarke et al., 1986; Wigley et al., 1992). We mutated the arginines at positions 162 (on the flexible loop) and 234 (in the active site) to glutamines, resulting in three different pdNAD-MDH variants: two single amino acid mutations and one containing both mutations. Both Arg-162 and Arg-234 interact directly with malate, and substitution with glutamine, which has a smaller side chain and lacks a positive charge, should destabilize the substrate binding site. We tested the effect of these mutations on enzyme activity in vitro. Wild-type pdNAD-MDH protein and proteins containing the mutations were expressed in and purified from Escherichia coli and incubated with oxaloacetate and NADH. The reduction of NADH was monitored spectrophometrically at 340 nm. No activity was detected for any of the three mutated proteins (Figure 8B). We then created plant expression constructs encoding these enzymatically inactive pdNAD-MDH proteins fused to a Flag-HA tag at the C-terminal end and driven by the native pdNAD-MDH promoter. Heterozygous pdnad-mdh plants were transformed with the constructs, and transformed T1 plants were selected via BASTA resistance. As described above for the experiments shown in Figure 7B, we selected T1 individuals that were both expressing the transgene and heterozygous for pdnad-mdh and genotyped their T2 progeny (69–78 plants per construct from 3 independent lines [23–26 plants each]). Surprisingly, we identified individuals that were homozygous for pdnad-mdh in the T2 generation for all catalytically inactive pdNAD-MDH constructs, indicating that each of them could complement the embryo-lethal phenotype (Figure 8C). The complemented pdnad-mdh plants grew like the wild type, showing that the enzymatically inactive pdNAD-MDH variants also complemented the growth phenotypes of pdnad-mdh (Figure 8D). We confirmed (using native-PAGE) that no activity bands corresponding to pdNAD-MDH were detected in extracts from complemented pdnad-mdh plants (Figure 8E). Thus, the embryo-lethal phenotype of pdnad-mdh, as well as the pale dwarfed phenotype of miR-mdh-1, are caused primarily by the lack of pdNAD-MDH protein itself, rather than the deficiency of NAD-MDH activity.
Figure 8.
Enzymatically Inactive pdNAD-MDH Proteins Can Complement the Embryo Lethality and Growth Defects of the pdnad-mdh Mutant.
(A) Catalytic center of the Arabidopsis pdNAD-MDH structure. The pdNAD-MDH protein sequence (without cTP) was modeled using the human malate- and NADH-bound MMDH2 crystal structure (PDB: 2DFD) as a template. Left panel shows NADH (red) and malate (yellow), and the relative position of the conserved catalytic amino acid residues of pdNAD-MDH. A detailed view of the malate binding site is shown in the upper right panel. The surface of the catalytic pocket is shown in the lower right panel.
(B) An in vitro activity assay was performed using purified recombinant proteins. The proteins (10 µg) were incubated with an excess of cofactor (NADH) at 22°C. The reaction was started by the addition of excess substrate (oxaloacetate). The velocity was determined by measuring the decrease in absorbance at 340 nm resulting from the conversion of NADH to NAD+. Error bars indicate mean ± se (n = 3).
(C) Genotyping results of T2 progeny from T1 plants heterozygous for pdnad-mdh and expressing the enzymatically inactive pdNAD-MDH proteins. Numbers above the bars indicate the number of BASTA-resistant T2 plants that were genotyped.
(D) Photographs showing 3-week-old rosettes of homozygous pdnad-mdh plants complemented with enzymatically-inactive pdNAD-MDH mutants. For comparison, wild-type (Ler) plants are shown. Plants were grown under long days (16 h light/8 h dark).
(E) NAD-MDH activity observed by native-PAGE. Equal amounts of protein (15 µg) were loaded per lane, and all lanes were run on the same gel. pdNAD-MDH runs in distinct activity bands (a, b, and c). While the fastest migrating band (c) corresponding to the free dimer is masked by other NAD-MDH activity, the two slower migrating bands that correspond to NAD-MDH in protein complexes can be easily observed (a and b). Additional activity bands (a’ and b’) are observed for plants heterozygous for the pdnad-mdh T-DNA insertion expressing the catalytic-inactive pdNAD-MDH variants, possibly due to dimer formation between an inactive pdNAD-MDH with an endogenous form of pdNAD-MDH protein.
pdNAD-MDH Interacts with the FtsH12-FtsHi Complex, Which Is Involved in Chloroplast Development
Since the pdNAD-MDH protein itself is indispensable for chloroplast and embryo development, we investigated whether it interacts with other plastidial proteins. We generated stable Arabidopsis transgenic lines overexpressing YFP-tagged pdNAD-MDH under the control of the constitutive 35S promoter and extracted protein from these lines at three different developmental stages: etiolated and light-grown seedlings, as well as rosette leaves. We then conducted immunoprecipitation (IP) experiments with these extracts using beads that specifically bind YFP. Proteins in the IP were digested with trypsin, and the resulting peptides were analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS). The identified peptides (Table 1; Supplemental Data Set 1) were searched against the TAIR10 genome annotation database. As a control, we also analyzed IPs conducted on extracts from wild-type plants.
Table 1. Proteins Identified in Anti-YFP Immunoprecipitates from Plants Expressing pdNAD-MDH-YFP.
| Accession No. | Identified Proteins (226/242) | P35S:MDH-YFP | ||
|---|---|---|---|---|
| Etiolated | Seedlings | Rosette | ||
| AT3G47520 | pdNAD-MDH | 210 | 102 | 163 |
| ATCG00860 | Chloroplastic Ycf2 | 65 | 14 | 68 |
| AT3G04340 | FtsH extracellular protease family, FtsHi 5, Embryo defective 2458 | 55 | 9 | 59 |
| AT3G01510 | Like SEX4 1 | 9 | 11 | 52 |
| AT1G79560 | FtsH protease 12, Embryo defective 1047, Embryo defective 156, Embryo defective 36 | 38 | 14 | 46 |
| AT5G64580 | FtsH extracellular protease family, FtsHi 4, Embryo defective 3144 | 29 | 8 | 43 |
| AT3G16290 | FtsH extracellular protease family, FtsHi 2, Embryo defective 2083 | 34 | 13 | 41 |
| AT4G23940 | FtsH extracellular protease family, ARC1, FtsHi1 | 21 | 8 | 41 |
| AT4G13670 | Plastid transcriptionally active 5 | 5 | 16 | 30 |
| AT3G23920 | Beta-amylase 1 | 0 | 0 | 12 |
| AT5G53860 | Embryo defective 2737 | 5 | 3 | 10 |
| AT2G40300 | Ferritin 4 | 10 | 3 | 9 |
| AT1G48460 | tRNA-processing ribonuclease BN | 5 | 2 | 7 |
| AT3G56090 | Ferritin 3 | 5 | 0 | 5 |
| AT5G22640 | Embryo defective 1211, Tic100 | 0 | 0 | 5 |
| AT5G63040 | Transmembrane protein | 3 | 2 | 4 |
| AT4G17090 | Beta-amylase 3 | 0 | 0 | 4 |
| AT4G28210 | Embryo defective 1923 | 2 | 2 | 4 |
| AT5G01600 | Ferritin 1 | 8 | 0 | 3 |
| AT5G59500 | Protein C-terminal S-isoprenylcysteine carboxyl O-methyltransferase | 3 | 2 | 3 |
| AT3G61780 | Embryo defective 1703 | 0 | 0 | 3 |
| AT1G31330 | Photosystem I subunit F | 0 | 0 | 2 |
| ATCG00900 | Chloroplast ribosomal protein S7 | 0 | 0 | 2 |
| ATCG00830 | Ribosomal protein L2 | 0 | 0 | 2 |
| AT5G14320 | Embryo defective 3137 | 0 | 0 | 2 |
| ATCG00190 | RNA polymerase subunit beta | 2 | 0 | 0 |
| AT5G16130 | Ribosomal protein S7e family protein | 2 | 0 | 0 |
| AT3G18420 | Slow green 1 | 3 | 0 | 0 |
IPs were conducted with anti-YFP beads on extracts of material harvested at three different developmental stages, and the coeluting proteins were identified using LC-MS/MS. Values represent the total spectrum count of peptides matching each protein. Proteins found in the control samples (IPs from wild-type plant extracts) were assumed to be contaminants and excluded. The full data set can be found in Supplemental Data Set 1.
Within the IP, the largest number of peptides, apart from pdNAD-MDH itself, matched hypothetical chloroplast open reading frame 2 (Ycf2), suggesting that it was present in the highest abundance, although it should be noted that our analysis is semiquantitative, as peptide counts are not strictly correlated with protein abundance. Ycf2 is encoded in the chloroplast genome. It is an essential protein in tobacco (Nicotiana tabacum) and green algae (Chlamydomonas reinhardtii) (Drescher et al., 2000; Nickelsen, 2005). Strikingly, many peptides also matched the FtsH (filamentous temperature sensitive) proteases, chloroplast isoforms of which are also essential (Wagner et al., 2012). The second largest functional group of proteins identified in the IP experiment consists of the proteins involved in starch degradation (including Like SEX4 1, BETA-AMYLASE1 (BAM1), and BAM3). These proteins were detected particularly at the rosette stage. Although these proteins are involved in starch turnover in the leaf mesophyll and stomatal guard cells, no defects in chloroplast structure were reported for mutants deficient in these proteins (Fulton et al., 2008; Comparot-Moss et al., 2010; Horrer et al., 2016). Given the essential nature of the identified FtsH12-FtsHi complex subunits, we focused on the interaction between pdNAD-MDH and these proteins. The possible role of pdNAD-MDH in starch turnover is being pursued in a separate study.
FtsH proteins are a family of membrane-bound proteases containing an ATPase associated with various cellular activities (AAA-ATPase) domain and a zinc binding metalloprotease domain. FtsH proteins are restricted to the mitochondria and chloroplasts in eukaryotes (Wagner et al., 2012). The Arabidopsis genome encodes 17 FtsH genes: 12 predicted proteolytic isoforms and 5 predicted nonproteolytic FtsH isoforms that lack the zinc binding motif for proteolytic activity (FtsHi1-5). FtsH12, along with FtsHi1, FtsHi2, FtsHi3, FtsHi4, and FtsHi5, is located at the inner membrane of the chloroplast envelope, and these subunits are proposed to form a hetero-hexameric complex, as deduced from coexpression analysis and proteomics data (Ferro et al., 2010). FtsH12 and its associated FtsHi subunits (with the exception of FtsHi3) were consistently detected in the IP with pdNAD-MDH at all three of the developmental stages tested.
For further analysis, we focused on FtsH12, as it was the only FtsH protein identified in the IP that has an intact zinc binding motif and, therefore, the potential for proteolytic activity. To confirm the interaction with pdNAD-MDH, we conducted a reciprocal immunoprecipitation with tagged FtsH12 protein. The FtsH12 coding sequence was cloned downstream of the UBIQUITIN10 promoter, and in frame with a YFP tag on the C-terminal end (UBI10:FtsH12-YFP). Wild-type plants were transformed with the construct, and the IP was performed as described above on T1 plants overexpressing FtsH12-YFP. Within the immunoprecipitate, we found peptides matching pdNAD-MDH, as well as those matching all of the FtsHi subunits that were identified in the IP with pdNAD-MDH-YFP and Ycf2 (Table 2). The correct plastidial localization of the FtsH12-YFP protein was confirmed via confocal microscopy (Supplemental Figure 5A).
Table 2. Proteins Identified in Anti-YFP Immunoprecipitates from Two Independent Lines Overexpressing FtsH12-YFP.
| Accession No. | Identified Proteins (236/247) | PUBQ10:FtsH12-YFP | |
|---|---|---|---|
| 1 | 2 | ||
| AT1G79560 | FtsH protease 12, Embryo defective 1047, Embryo defective 156, Embryo defective 36 | 386 | 371 |
| ATCG00860 | Chloroplast Ycf2 | 181 | 237 |
| AT3G04340 | FtsH extracellular protease family, FtsHi 5, Embryo defective2458 | 247 | 179 |
| ATCG01130 | Chloroplast Ycf1, TIC214 | 70 | 111 |
| AT3G16290 | FtsH extracellular protease family, FtsHi 2, Embryo defective 2083 | 46 | 86 |
| AT5G64580 | FtsH extracellular protease family, FtsHi 4, Embryo defective 3144 | 66 | 83 |
| AT4G23940 | FtsH extracellular protease family, ARC1, FtsHi1 | 69 | 52 |
| AT1G01320 | Tetratricopeptide repeat-like superfamily protein | 6 | 32 |
| AT4G02510 | Translocon at the outer envelope membrane of chloroplasts 159 | 37 | 31 |
| AT5G53860 | Embryo defective 2737 | 13 | 26 |
| ATCG00480 | ATP synthase subunit beta | 8 | 23 |
| AT3G47520 | pdNAD-MDH | 5 | 22 |
| AT1G07920 | GTP binding Elongation factor Tu family protein | 15 | 20 |
| AT1G43170 | Ribosomal protein 1 | 5 | 20 |
| ATCG00120 | ATP synthase subunit alpha | 42 | 19 |
| ATCG00490 | Ribulose-bisphosphate carboxylase | 48 | 16 |
| AT5G01590 | Histone-lysine N-methyltransferase ATXR3-like protein | 14 | 16 |
| AT2G41840 | Ribosomal protein S5 family protein | 6 | 16 |
The IPs were conducted with anti-YFP beads on extracts of material harvested from rosette leaves, and the coeluting proteins were identified using LC-MS/MS. Values represent the total spectrum count of peptides matching each protein. Proteins found in the control sample (IPs from wild-type plants) were assumed to be contaminants and excluded. The full data set can be found in Supplemental Data Set 1.
To investigate the function of FtsH12, four independent Arabidopsis mutants harboring T-DNA insertions in the FtsH12 gene were obtained. We confirmed that ftsh12 mutants are embryo-lethal, as previously described (Patton et al., 1991, 1998; Franzmann et al., 1995). Like pdnad-mdh, the ftsh12 mutants arrested near the globular-to-heart transition stage. However, the exact stage at which the embryo arrested varied between lines, likely due to the position of the T-DNA insertions having different effects on transcript and protein accumulation (Supplemental Figures 5B to 5F).
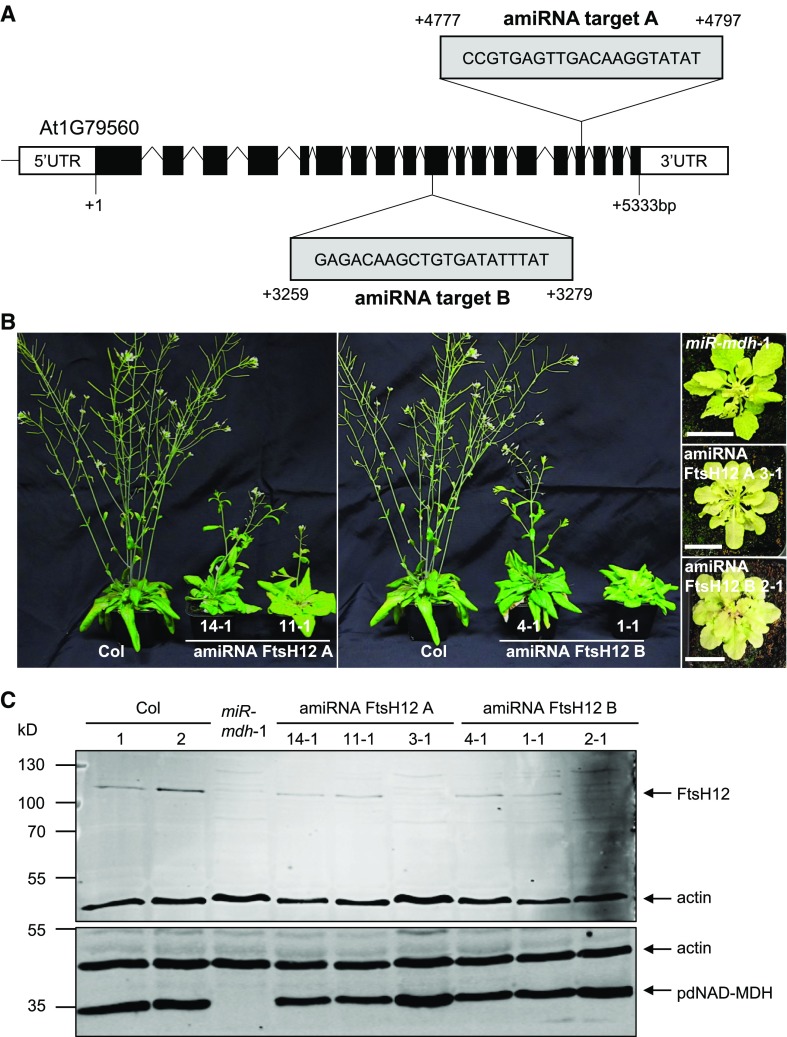
To further study the effect of FtsH12 deficiency, we generated lines constitutively expressing artificial microRNAs. We designed microRNA silencing cassettes targeting regions 3259 to 3279 (amiRNA target B) and 4777 to 4797 (amiRNA target A) of the FtsH12 coding sequence, respectively (Figure 9A) (Ossowski et al., 2008). Both cassettes were cloned downstream of the constitutive 35S promoter. Wild-type plants were transformed with the constructs, and T1 seedlings were selected via kanamycin resistance. For both silencing constructs, the T1 plants had varying degrees of paleness, from wild-type-like plants to very pale plants that were stunted in growth (Figure 9B). Notably, the pale plants strongly resembled miR-mdh-1 plants. Immunoblots were performed on protein extracts from the leaves of these silencing lines, using antibodies against FtsH12 and pdNAD-MDH to test the impact of FtsH12 silencing on these proteins. The molecular mass of FtsH12 is 115 kD, including the chloroplast transit peptide (49 amino acids), and the mature peptide is predicted to be 110 kD. No FtsH12 protein was detectable in the T1 plants with the strongest pale phenotype (amiRNA FtsH12 A 3-1 and amiRNA FtsH12 B 2-1), and those with intermediate levels of paleness had reduced amounts of FtsH12 protein (Figure 9C). Surprisingly, there was also no detectable FtsH12 protein in the miR-mdh-1 plants. Quantitative immunoblot analysis showed a 7-fold reduction in FtsH12 protein abundance in miR-mdh-1 leaves relative to the wild type (Supplemental Figure 6). However, pdNAD-MDH protein levels were unaffected by FtsH12 silencing.
Figure 9.
Constitutive and Inducible Silencing of the FtsH12 Protein.
(A) In Arabidopsis, the FtsH12 gene consists of 19 exons and 18 introns. The sequence and position of the target for artificial microRNA silencing are indicated. Numbers represent nucleotide positions relative to the translational start site +1.
(B) Constitutive silencing of FtsH12 resulted in plants with varying degrees of paleness and delayed growth phenotype in the T1 generation. Plants were grown under a 12-h-light/12-h-dark regime for 6 weeks. Both amiRNA constructs produced plants with comparable phenotypes. The identical wild type (Col) plant was used for both the left and right panels. Bar = 2 cm.
(C) Immunoblot analysis of total protein extracts from the amiRNA FtsH12 lines, using the FtsH12 antibody (upper panel), pdNAD-MDH antibody (lower panel), and actin antibody as a loading control (both panels). FtsH12 and actin, as well as pdNAD-MDH and actin, were detected concurrently on the same membrane using secondary antibodies conjugated to different infrared fluorescence dyes (800CW for FtsH12 and pdNAD-MDH, and 680RD for actin). The migration of molecular mass markers is indicated (left). The gel was loaded on an equal protein (15 µg) basis.
pdNAD-MDH Activity Is Not Required for Its Interaction with the FtsH12-FtsHi Complex
We tested whether the FtsH12-FtsHi complex could also interact with the NAD-MDH isoforms from other cellular compartments that could not complement the embryo lethality of pdnad-mdh when targeted to chloroplasts. We extracted proteins from 4-week-old rosettes of heterozygous pdnad-mdh plants expressing the YFP-tagged NAD-MDH isoforms (Figure 7A) and performed anti-YFP IPs for analysis by LC-MS/MS. IPs were also performed with extracts from pdnad-mdh PpdNAD-MDH:pdNAD-MDH-YFP plants (as a positive control) and from wild-type Ler plants (as a negative control). We did not detect peptides matching Ycf2, FtsH12, or the FtsHi proteins in the IPs with most NAD-MDH-YFP isoforms. One exception was the IP with AtmMDH1-YFP, where four peptide hits were found matching Ycf2 and four matching FtsH12, but none matching FtsHi subunits. These peptide counts were very low compared with those found in the pdNAD-MDH-YFP IP, which yielded 193 and 155 peptides matching Ycf2 and FtsH12, respectively, and many peptides matching FtsHi subunits (Table 3). Thus, none of these NAD-MDH isoforms readily associated with the FtsH12-FtsHi complex. However, in the IPs of the NAD-MDH isoforms, peptides matching pdNAD-MDH were found. This suggests that the NAD-MDH isoforms may dimerize with the endogenous pdNAD-MDH protein, since MDHs are known to form dimers (Minárik et al., 2002).
Table 3. Proteins Identified in IPs of YFP-Tagged NAD-MDH Proteins (AtpdNAD-MDH, AtcyMDH1, ScmMDH1, AtmMDH1, and AtpMDH) Expressed in Heterozygous pdnad-mdh Plants.
| Accession No. | Identified Proteins (102/115) | pdnad-mdh+/− | ||||
|---|---|---|---|---|---|---|
| At pdNAD-MDH | At cyMDH1 | Sc mMDH1 | At mMDH1 | At pMDH1 | ||
| GFP_AEQVI | YFP-tag | 108 | 7 | 3 | 66 | 162 |
| AT3G47520 | pdNAD-MDH | 142 | 3 | 0 | 18 | 6 |
| AT1G04410 | cyMDH1 | 0 | 8 | 5 | 0 | 0 |
| AT1G53240 | mMDH1 | 4 | 0 | 0 | 96 | 0 |
| AT2G22780 | pMDH1 | 0 | 2 | 2 | 0 | 312 |
| ATCG00860 | Chloroplast Ycf2 | 193 | 0 | 0 | 4 | 0 |
| AT3G04340 | FtsH extracellular protease family, FtsHi 5, Embryo defective2458 | 192 | 0 | 0 | 0 | 0 |
| AT1G79560 | FtsH protease 12, Embryo defective 1047, Embryo defective 156, Embryo defective 36 | 155 | 0 | 0 | 4 | 0 |
| AT5G64580 | FtsH extracellular protease family, FtsHi 4, Embryo defective 3144 | 102 | 0 | 0 | 0 | 0 |
| AT3G01510 | Like SEX4 1 | 82 | 0 | 0 | 0 | 0 |
| AT4G23940 | FtsH extracellular protease family, ARC1, FtsHi1 | 69 | 0 | 0 | 0 | 0 |
| ATCG01130 | Chloroplast Ycf1 protein | 59 | 0 | 0 | 0 | 0 |
| AT3G16290 | FtsH extracellular protease family, FtsHi 2, Embryo defective 2083 | 54 | 0 | 0 | 0 | 0 |
| AT5G53860 | Embryo defective 2737 | 29 | 0 | 0 | 0 | 0 |
| AT4G02510 | Translocon at the outer envelope membrane of chloroplasts 159 | 29 | 0 | 0 | 0 | 0 |
IPs were conducted with anti-YFP beads on extracts of rosette leaves, and the coeluting proteins were identified using LC-MS/MS. Values represent the total spectrum count of peptides matching each protein. Proteins also identified in the control sample (Ler) were assumed to be contaminants and excluded. The full data set can be found in Supplemental Data Set 1.
We conducted a similar experiment with the enzymatically inactive pdNAD-MDH-FlagHA proteins, which complemented the embryo lethality of homozygous pdnad-mdh plants. Instead of beads conjugated to an anti-YFP antibody, we used beads conjugated to an anti-HA antibody, since the inactive pdNAD-MDH proteins were tagged with a Flag-HA tag. Again, pdnad-mdh plants complemented with PpdNAD-MDH:pdNAD-MDH-Flag-HA and wild-type plants served as positive and negative controls, respectively. Peptides matching the Flag-HA-tagged pdNAD-MDH proteins were present with the highest abundance within each IP, indicating that each of the fusion proteins was effectively enriched. All of the components of the FtsH12-FtsHi complex copurified with the different pdNAD-MDH catalytic mutants, with peptide counts that were similar to those in the IPs with wild-type protein (Table 4). Thus, the point mutations in the catalytic center did not affect the protein-protein interaction between pdNAD-MDH with the FtsH12-FtsHi complex.
Table 4. Proteins Identified in Anti-HA Immunoprecipitates from Homozygous pdnad-mdh Plants Expressing Noncatalytic Versions of pdNAD-MDH.
| Accession No. | Identified Proteins (917/929) | pdnad-mdh−/− | |||
|---|---|---|---|---|---|
| pdNAD-MDH WT | pdNAD-MDH R162Q | pdNAD-MDH R234Q | pdNAD-MDH R163Q R234Q | ||
| AT3G47520 | pdNAD-MDH | 96 | 49 | 31 | 22 |
| ATCG00860 | Chloroplast Ycf2 | 283 | 457 | 332 | 405 |
| AT3G04340 | FtsH extracellular protease family, FtsHi 5, Embryo defective2458 | 408 | 330 | 210 | 362 |
| AT1G79560 | FtsH protease 12, Embryo defective 1047, Embryo defective 156, Embryo defective 36 | 373 | 309 | 212 | 317 |
| AT5G64580 | FtsH extracellular protease family, FtsHi 4, Embryo defective 3144 | 221 | 207 | 164 | 159 |
| AT3G16290 | FtsH extracellular protease family, FtsHi 2, Embryo defective 2083 | 125 | 228 | 158 | 179 |
| ATCG01130 | Chloroplast Ycf1 | 70 | 166 | 174 | 223 |
| AT4G23940 | FtsH extracellular protease family, ARC1, FtsHi1 | 153 | 141 | 153 | 109 |
| AT4G02510 | Translocon at the outer envelope membrane of chloroplasts 159 | 25 | 95 | 113 | 133 |
| AT3G01510 | Like SEX4 1 | 159 | 58 | 105 | 23 |
| AT5G53860 | Embryo defective 2737 | 30 | 54 | 55 | 54 |
The IPs were conducted with anti-HA beads on extracts of rosette leaves, and the coeluting proteins were identified using LC-MS/MS. Values represent the total spectrum count of peptides matching each protein. Proteins found in the control sample (IPs from wild-type [Ler] plant extracts) were assumed to be contaminants and excluded. The full data set can be found in Supplemental Data Set 1.
Mutated Forms of pdNAD-MDH That Cannot Bind NADH Still Complement the Knockout Phenotype
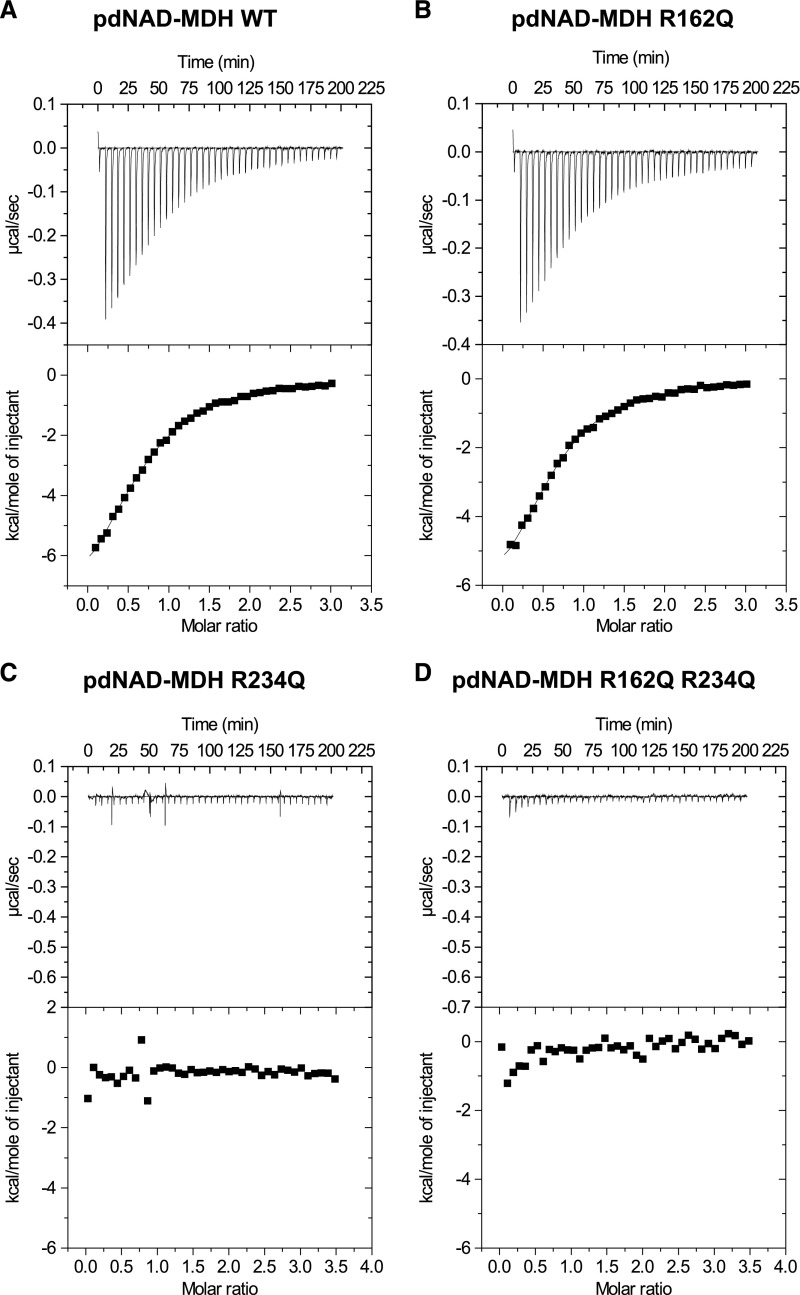
Our data show that the enzymatic activity of pdNAD-MDH is dispensable, yet the protein has a critical role within the FtsH12-FtsHi complex. Our strategy to abolish the enzymatic activity of pdNAD-MDH disrupted the malate/oxaloacetate binding site. However, it is still possible that the mutants can bind their cofactor NADH and might function as a redox sensor for the AAA-ATPase complex. To investigate this, we performed isothermal titration calorimetry (ITC) with the enzymatically inactive pdNAD-MDH proteins to test if they were still capable of binding NADH. ITC measures the heat released or taken up as the titrated cofactor (NADH) binds the protein (pdNAD-MDH), depending on whether it is an exothermic or endothermic binding event. We titrated NADH into the wild-type pdNAD-MDH recombinant protein in four independent experiments, each using freshly purified recombinant protein, and observed binding with a mean dissociation constant (Kd) of 7.54 ± 0.53 µM (Figure 10A). Titration of NADH into the R162Q variant resulted in binding with a similar mean Kd (6.75 ± 0.15 µM, from two independent experiments; Figure 10B), suggesting that this mutation does not abolish cofactor binding. However, no NADH binding could be detected for the R234Q variant or for the R162Q R234Q variant (Figures 10C and 10D). Since these two variants could also complement pdnad-mdh, we can rule out the possibility that cofactor binding of the pdNAD-MDH protein plays a significant role in the FtsH12-FtsHi complex.
Figure 10.
ITC Thermograms of NADH Titrated into the pdNAD-MDH Wild-Type and Inactive Proteins.
(A) ITC profile of NADH injected into a solution of recombinant His-pdNAD-MDH wild-type protein at 25°C. The upper panel shows the raw calorimetric data. The plot below shows the integrated enthalpy as a function of the NADH/pdNAD-MDH molar ratio. Reactions are exothermic.
(B) to (D) Experiments were conducted as described for (A) with the His-pdNAD-MDH R162Q protein (B), the His-pdNAD-MDH R234Q protein (C), and the His-pdNAD-MDH R162Q R234Q protein (D).
DISCUSSION
pdNAD-MDH Is Required for Chloroplast Development
Our results demonstrate that the pdNAD-MDH protein plays a vital role in plastid development, both during and after embryogenesis in Arabidopsis. During embryogenesis, plastids start to differentiate into chloroplasts in cotyledons at the late globular stage. They expand during the early heart stage, as thylakoids and grana stacks begin to develop (Mansfield and Briarty, 1991). Embryos of the pdnad-mdh knockout mutant arrest in the globular-to-heart transition stage (Beeler et al., 2014), like many other mutants that are defective in genes essential for chloroplast biogenesis (Yu et al., 2004; Ruppel and Hangarter, 2007; Feng et al., 2014; Lu et al., 2014).
Previously, we studied the viable constitutive silencing line, miR-mdh-1, where pdNAD-MDH levels were reduced, but not abolished, at most or all stages of plant growth. Using the ABI3 promoter to drive embryo-specific expression of pdNAD-MDH enabled us to complement the embryo lethality of homozygous pdnad-mdh plants and determine the role of pdNAD-MDH in vegetative growth (Figure 1). These plants were seedling-lethal, with albino leaves containing highly aberrant plastids (Figures 2 and 3). This confirms the notion that pdNAD-MDH plays a vital role in postembryogenesis chloroplast development. The phenotype of seedlings resulting from the embryo-specific complementation of pdnad-mdh is more severe than, but consistent with, those observed in miR-mdh-1 plants, which also have pale leaves with aberrant chloroplast ultrastructure. This pale phenotype of miR-mdh-1 results from MDH deficiency during leaf development rather than from defective embryogenesis. This is further supported by the finding that inducible silencing of pdNAD-MDH at the rosette stage resulted in white/pale newly emerging leaves (Figure 4).
We propose that pdNAD-MDH is required for the early stages of plastid differentiation. This is most obviously reflected by the absence of internal membrane structure in leaf plastids of embryo-complemented pdnad-mdh and by the reduced thylakoid structure in miR-mdh-1 chloroplasts. However, the aberrant formation of internal structure was even observed in etioplasts of miR-mdh-1, the majority of which lacked normal prolamellar bodies (PLBs; Figure 5) and were deficient in many of the precursors required for forming photosynthetic chloroplasts (protochlorophyllide, PORA, galactolipids, and carotenoids; Figure 6). Thus, the presence of pdNAD-MDH is required, either directly or indirectly, for the proper synthesis of several component classes, which serve as the building blocks of thylakoid membranes. Deficiency of these components, caused by other mutations, is known to lead to aberrant PLBs. For example, etioplasts of Arabidopsis carotenoid and chloroplast regulation mutants have reduced lutein levels and lack PLBs, suggesting that specific carotenoids are essential for PLB formation (Park et al., 2002). PLB formation is absent or aberrant in mutants that cannot make protochlorophyllide (Mascia and Robertson, 1978; Solymosi and Aronsson, 2013) and in the constitutive photomorphogenic1 mutant (Deng et al., 1991; Lebedev et al., 1995), where the lack of PLBs is attributed to a deficiency in PORA or PORB (Sperling et al., 1998).
MGDG and DGDG are the most abundant lipids in thylakoid membranes in chloroplasts, and MGDG is the most dominant lipid in PLBs. The interaction between PORA and MGDG is thought to stabilize the formation of PLBs (Klement et al., 1999; Engdahl et al., 2001; Selstam et al., 2002). There are conflicting data regarding the effects of suppressing MGDG synthase 1 (MGD1) involved in MGDG synthesis (Jarvis et al., 2000; Kobayashi et al., 2007), possibly resulting from differences between the T-DNA alleles. However, recent findings suggest that MGD1 is involved in the initial step of etioplast development by providing a lipid matrix for protochlorophyllide biosynthesis (Fujii et al., 2017). The silencing of MGD1 decreased MGDG levels in etiolated seedlings, as well as total protochlorophyllide levels. Given that these reports show that the deficiency of one component can cause a pleiotropic decrease in the accumulation of another, the finding that the levels of all major PLB components are reduced in miR-mdh-1 does not allow us to pin down a specific metabolic pathway where pdNAD-MDH is required.
In seedlings undergoing photomorphogenesis, proplastids develop into chloroplasts, bypassing the etioplast stage, and the extent to which the etioplast is a good model for this direct route of chloroplast development is debatable (Solymosi and Schoefs, 2010). However, it is important to note that all major PLB components, with the exception of protochlorophyllide and PORA, are also major components of developed chloroplasts, and we observed similar reductions in the levels of these compounds in miR-mdh-1 relative to the wild type in photomorphogenic seedlings. The reduced amounts of these compounds suggest that pdNAD-MDH is required for their proper synthesis in both etioplast-dependent and -independent routes of chloroplast development.
Enzymatically Inactive pdNAD-MDH Complements the pdnad-mdh Mutant
Unexpectedly, we discovered that the embryo-lethal phenotype of the pdnad-mdh mutant is not caused by the loss of plastidial NAD-MDH activity. On one hand, three different mutated forms of the enzyme that were enzymatically inactive and, in two cases, that additionally could not bind NADH, could complement the embryo-lethal phenotype when expressed in pdnad-mdh (Figures 8 and 10). The complemented plants grew normally and were not pale or albino, as were miR-mdh-1 or embryo-complemented pdnad-mdh plants. On the other hand, expressing other NAD-MDH isoforms in chloroplasts failed to complement the pdnad-mdh phenotype (Figure 7). These observations suggest that the phenotypes of pdnad-mdh plants are specifically caused by the absence of the pdNAD-MDH protein itself (Figure 8).
Our data do not rule out the previously proposed role of pdNAD-MDH in balancing redox equivalents via its enzymatic interconversion of malate and oxaloacetate (i.e., the malate valve model; Scheibe, 2004), but we argue that this is not the essential function of the protein. Nevertheless, it is important to note that the pdNAD-MDH protein has MDH activity and that the catalytic residues are conserved among orthologous proteins in other plants (Supplemental Figure 7). Thus, it is likely that the enzymatic activity is important under specific conditions or tissues, such as in pollen tubes during expansion (Selinski et al., 2014). Indeed, in a recent genome-wide association study, pdNAD-MDH was mapped as a quantitative trait locus for malate levels in Arabidopsis (Fusari et al., 2017). Furthermore, mutations in pdNAD-MDH rescued the phenotype of the mosaic death1 (mod1) mutant, which accumulates reactive oxygen species and shows abnormal patterns of programmed cell death (Zhao et al., 2018). Interestingly, the mod1 mutant is proposed to generate reactive oxygen species in the mitochondria in response to a signal from the chloroplast. The fact that mMDH1 mutations could also suppress the mod1 phenotype suggests that a malate valve may facilitate the communication between mitochondria and chloroplasts (Zhao et al., 2018). These findings are consistent with our results, as they suggest that the malate valve is not essential.
Interestingly, we found slightly fewer than the expected 25% of pdnad-mdh plants in the T2 generation (Figure 8C), particularly for the construct that encoded proteins with two amino acid substitutions. This may reflect incomplete complementation by these nonenzymatic proteoforms at certain developmental stage, either because MDH activity is beneficial or because the introduction of amino acid substitutions in the catalytic center may affect the integrity or half-life of the protein itself. Further work will be required to assess the importance of NAD-MDH activity in the chloroplast and the extent to which its role can be compensated by the presence of NADP-MDH. As the loss of NADP-MDH activity alone also has a relatively mild impact on plant growth (Hebbelmann et al., 2012), generating lines expressing the inactive pdNAD-MDH constructs in the pdnad-mdh nadp-mdh double mutant background would be highly valuable for reassessing the importance of MDH activity and the malate valve in the chloroplast.
pdNAD-MDH Functions in Complex with AAA-Proteases at the Chloroplast Inner Envelope
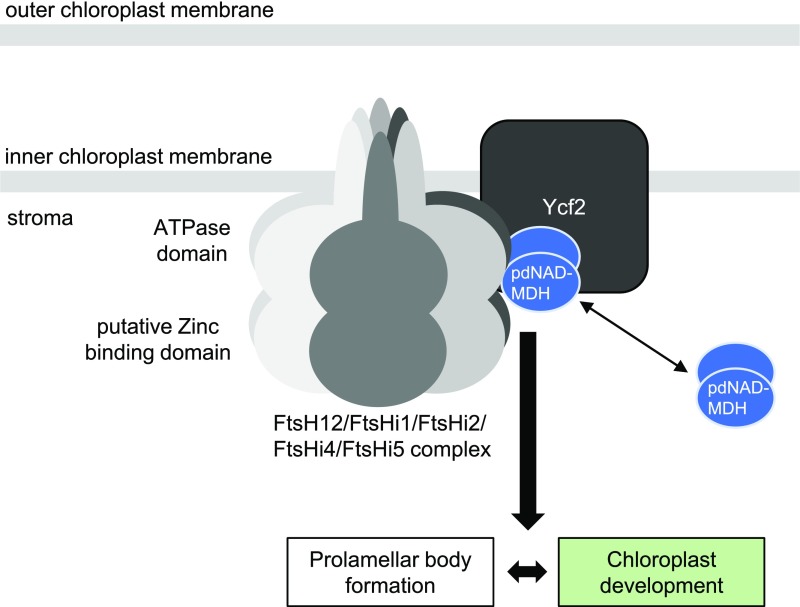
We demonstrate that pdNAD-MDH interacts with members of a proposed large AAA protease complex localized to the chloroplast inner envelope composed of FtsH12 and FtsHi subunits, as well as with Ycf2 (Tables 1 and 2>, Figure 11). Proteomics data indicate that most of the pdNAD-MDH protein is localized to the stroma and chloroplast envelope, and only a minority of the protein associated to the thylakoid membranes (Ferro et al., 2010), suggesting that a fraction of total pdNAD-MDH is stably involved in this interaction. Further, Cvetić et al. (2008) identified pdNAD-MDH in both the stromal and chloroplast envelope fractions of spinach leaf chloroplasts. Plants lacking FtsH12 (emb1047 and emb156), FtsHi2 (emb2083), FtsHi4 (emb3144), or FtsHi5 (emb2458) are all embryo-lethal (Patton et al., 1991, 1998; Franzmann et al., 1995; Sokolenko et al., 2002; Wagner et al., 2012; Lu et al., 2014). Although no Arabidopsis mutant for the chloroplast genome-encoded Ycf2 is currently available, a knockout mutant of the gene could not be generated using plastome transformation in tobacco, suggesting that it is also essential (Drescher et al., 2000). We verified that FtsH12 knockout plants were embryo-lethal and arrested near the globular-to-heart transition stage, similar to pdnad-mdh. Furthermore, Arabidopsis lines with constitutive silencing of FtsH12 expression had a striking resemblance to the miR-mdh-1 line. Thus, the phenotypes observed from the loss of pdNAD-MDH could be explained by the loss of FtsH12 function. Interestingly, in miR-mdh-1, FtsH12 protein levels were strongly reduced compared with the wild type, whereas the converse was not true; in the amiRNA FtsH12 lines, pdNAD-MDH protein levels were similar to the wild type (Figure 9C). Taken together, these data support a hypothesis where pdNAD-MDH plays a role in stabilizing FtsH12, and possibly the entire complex. The exact mechanism by which pdNAD-MDH stabilizes FtsH12 is still unknown, but it does not require pdNAD-MDH catalytic activity or NADH binding, as the inactive pdNAD-MDH proteins could still interact with FtsH12-FtsHi complex members in plants (Table 4). However, other MDH isoforms from different cell compartments could not interact with FtsH12, and no other MDH isoform was identified in the immunoprecipitation experiment with FtsH12, suggesting that the interaction is specific to pdNAD-MDH in chloroplasts (Tables 2 and 3).
Figure 11.
Model for the Interaction of pdNAD-MDH with the Heteromeric FtsH12-FtsHi AAA-ATPase Complex at the Chloroplast Inner Envelope Membrane, Which Plays an Essential Role in Chloroplast Development.
Possible Role of the FtsH12-FtsHi Complex
AAA-type proteins generally form hexamers, where each subunit has an N-terminal transmembrane segment and a C-terminal AAA-ATPase domain expanded in the stroma. The FtsH12-FtsHi complex contains several members of the FtsHi family (FtsHi1, FtsHi2, FtsHi4, and FtsHi5), which lack the zinc binding motif that is considered essential for its metalloprotease activity. In a coexpression network, all members of the FtsH12-FtsHi complex clustered together with genes involved in plastid translation, division, and positioning, as well as amino acid metabolism (Majsec et al., 2017). Nonproteolytic FtsHi proteins were reported to be absent from the cyanobacterium Synechocystis and are thought to have evolved at a later stage of evolution through gene duplication (Sokolenko et al., 2002). However, the activity of the AAA protease complex does not require all six subunits to be active (Martin et al., 2005). AAA-type proteases are also known to generate a pulling force. The mechanism of target protein unfolding by AAA-type proteases includes a conformational change in the AAA domain, which moves conserved, substrate binding hydrophobic residues toward the inner pore of the hexameric complex. This draws the substrate proteins inside the pore and unfolds them (Lee et al., 2001; Langklotz et al., 2012). In yeast mitochondria, the m-AAA protease, described as an ATP-dependent protease that degrades misfolded proteins and mediates protein processing, is proposed to be further involved in the dislocation of imported preproteins from the inner membrane by functioning as an ATP-driven molecular motor (Tatsuta et al., 2007; Botelho et al., 2013). In yeast mitochondria, many nucleus-encoded preproteins are imported into the mitochondrial matrix via the TIM23 translocon (Demishtein-Zohary and Azem, 2017). The FtsH12-FtsHi complex could potentially play a similar role in chloroplasts.
Nakai (2018) recently proposed a novel ATP-driven import motor associated with the TIC (translocon on the inner chloroplast membrane) complex at the inner chloroplast envelope membrane. It is likely that the FtsH12-FtsHi complex, together with pdNAD-MDH, is the proposed ATP-driven motor that imports preproteins across the chloroplast envelopes. Consistent with this hypothesis, we also found large numbers of peptides matching components of the chloroplast protein import machinery (e.g., Ycf1 and Toc159) in our IP with FtsH12 (Table 2). However, further investigations are needed to determine whether the function of FtsH12 in chloroplast development is dependent on its proteolytic activity or solely on its ATPase activity. Further studies will provide exciting new insights into the role of the FtsH12-FtsHi complex in chloroplast development and function.
In conclusion, our data define pdNAD-MDH as a moonlighting protein essential for chloroplast development. Moonlighting enzymes perform more than one function, often serving a structural or regulatory function in addition to their known catalytic function (Jeffery, 1999, 2003; Copley, 2003; Moore, 2004). Moonlighting functions occur frequently in highly conserved proteins and are thought to evolve more commonly for soluble, highly abundant proteins that are constitutively expressed (Huberts and van der Klei, 2010; Copley, 2014). Soluble abundant proteins are likely to encounter many more biomolecular interactions within their environment, and advantageous interactions can evolve over time (Copley, 2014). It has also been proposed that acquiring a moonlighting function might be an easier way to expand the functional tool box of an organism without the drawbacks resulting from an expanding genome (Jeffery, 1999). Interestingly, lactate dehydrogenase, a homolog of the MDH family, also plays a moonlighting role as a structural protein in the lenses of bird eyes and was one of the first examples of moonlighting proteins described (Wistow et al., 1987; Hendriks et al., 1988; Huberts and van der Klei, 2010). A well-known example of a moonlighting enzyme in plant metabolism is hexokinase, which is a key enzyme in central metabolism but also acts as a sugar sensor (Jang et al., 1997; Moore et al., 2003). Also, glyceraldehyde-3-phosphate dehydrogenase isoforms, aside from their roles in glycolysis and the Calvin-Benson-Bassham cycle, can regulate DNA stability, control gene expression, function in apoptosis, and act as redox sensors (He et al., 2013; Zaffagnini et al., 2013; Yang and Zhai, 2017). Recently, AROGENATE DEHYDRATASE2 was shown to localize to the chloroplast division machinery in Arabidopsis, suggesting an additional nonenzymatic function besides its enzymatic role in phenylalanine biosynthesis (Bross et al., 2017). However, pdNAD-MDH appears thus far to be a unique example in which the moonlighting function is essential for plant survival.
METHODS
Plant Growth
Arabidopsis thaliana plants were grown in soil in growth cabinets (Percival AR-95 [CLF Plant Climatics]; OR Kälte 3000) fitted with fluorescent lamps and supplemented with red LED panels. Unless stated otherwise, the chambers provided a 12-h-light/12-h-dark cycle, with light intensity of 150 μmol photons m−2 s−1, temperature of 20°C, and relative humidity of 65%.
For experiments with plate-grown seedlings, seeds were surface sterilized and placed on 0.5× Murashige and Skoog medium with vitamins and MES (Duchefa Biochemie) at pH 5.8, solidified with 0.8% (w/v) agar. The seeds were stratified by incubating the plates in the dark at 4°C for 2 d. For etiolated seedlings, germination was stimulated by placing the seeds on plates under white light (150 μmol photons m−2 s−1) for 4 to 6 h, followed by growth for 5 to 6 d in the dark. Light-grown seedlings were placed in either standard growth conditions (as above for plants grown in soil) or in continuous light. For seedlings for lipid, carotenoid, and protochlorophyllide/PORA analysis, seeds were germinated on a 100-μm nylon mesh placed on solid medium to aid harvest. Dark-grown seedlings were harvested under low green light illumination.
To select transformants on plates, the growth medium contained either 15 mg/L BASTA, 15 mg/L hygromycin, or 50 mg/L kanamycin, depending on the resistance marker on the transgene. pdnad-mdh plants transformed with the constructs encoding inactive pdNAD-MDH (R162Q, R234Q, and R162Q R234Q) were selected on soil by spraying them with BASTA (final concentration of 0.018% [w/v] glufosinate; Omya).
The pdnad-mdh T-DNA insertion mutant (ET8629) and constitutive pdNAD-MDH silencing line, miR-mdh-1, were characterized by Beeler et al. (2014). The ftsh12 T-DNA lines (emb1047-1, emb1047-2, ftsh12 1-1, and emb156-1) were ordered from the Nottingham Arabidopsis Stock Centre. The heterozygous pdnad-mdh mutant is in the Landsberg erecta (Ler) background, while miR-mdh-1 and ftsh12 T-DNA insertion lines are in the Columbia (Col-0) background.
Recombinant Protein Expression in Escherichia coli
All sequences of oligonucleotide primers used for cloning the following constructs are listed in Supplemental Table 2. For the recombinant expression of pdNAD-MDH-His and its catalytic inactive variants in E. coli, pdNAD-MDH was cloned into the pET21a+ expression vector. First, the length of the chloroplast transit peptide (cTP) was predicted using the Target P server (http://www.cbs.dtu.dk/services/TargetP/). The full-length coding sequence of pdNAD-MDH without the cTP was amplified with NdeI and NotI restriction sites, using the pdNAD-MDH:pDONR221 vector as a template (Beeler et al., 2014). The PCR product was cloned into the pET21a+ vector (Novagen) using the restriction sites. The point mutations in the catalytic center of pdNAD-MDH were generated in the pdNAD-MDH:pDONR221 vector using a QuikChange site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. The mutated coding sequences were then cloned into pET21a+ as described for the wild-type sequence.
For protein expression, the vectors were transformed into E. coli BL21 (DE3) CodonPlus cells (Agilent Technologies). The cells were cultured in LB medium at 37°C until OD600 of 0.5 to 0.7. Protein expression was induced by adding 1 mM IPTG, and the cultures were incubated overnight at 20°C. Cell lysis and purification of the His-tagged proteins were performed as described (Seung et al., 2013).
NAD-MDH Enzyme Activity Measurements
Enzyme activity of the pdNAD-MDH recombinant proteins was measured spectrophotometrically in an infinite M1000 Pro plate reader (Tecan). Each reaction contained 0.092 M Tris-HCl, pH 7.9, 0.01 M MgCl2, 0.2 mM NADH, and 0.01 μg recombinant protein. The baseline rate at 340 nm was acquired for 5 min at 20°C, measuring absorbance every 20 s. The reaction was started by adding 0.09 mM oxaloacetate. The linear decline in absorbance at 340 nm was monitored and used to calculate the rate of NADH consumption. Activity was determined three times on the same protein preparation to calculate the mean ± se.
Immunoblotting and Native PAGE
To extract soluble proteins, young leaves were homogenized in protein extraction medium (100 mM MOPS, pH 7.2, 1 mM EDTA, 10% [v/v] ethylene glycol, 2 mM dithiothreitol, and 1× Complete Protease Inhibitor cocktail [Roche]). Insoluble material was pelleted at 20,000g. Protein content in the supernatant was determined using the Bradford assay. To extract total protein from leaves (Figure 4), frozen 7-mm leaf discs were ground with two to three glass beads in a mixer mill (Retsch MM 200). The powder was suspended in SDS-PAGE loading buffer (50 mM Tris-HCl, pH 6.8, 100 mM DTT, 2% [w/v] SDS, 30% [v/v] glycerol, and 0.005% [w/v] bromophenol blue) and heated to 95°C for 5 min. Insoluble material was removed via centrifugation, and samples (5 μL) were loaded onto SDS-PAGE gels. To extract total protein from seedlings (Figure 6), frozen seedling samples were extracted with SDS-PAGE loading buffer (at 100 mg/mL) and samples (10 μL) were loaded onto SDS-PAGE gels.
For immunodetection of pdNAD-MDH, we used rabbit antisera raised against the Arabidopsis pdNAD-MDH protein (Beeler et al., 2014). For FtsH12, we used rabbit antisera raised against the Arabidopsis FtsH12 protein, which was a gift from Masato Nakai (Osaka University). For the detection of epitope-tagged proteins, we used α-GFP/YFP (ab290; Abcam), α-HA (ab9110; Abcam), α-Flag M2 (F1804; Sigma-Aldrich), and α-PORA (Agrisera). Proteins were detected based on infrared fluorescence using IR800-conjugated secondary antibodies and an Odyssey CLx detection system (Li-Cor). Where actin was detected as a loading control, the rabbit primary antibody was coincubated with a mouse monoclonal antibody against plant actin (A0480; Sigma-Aldrich), which was detected in the same blot using a 680RD-conjugated anti-mouse secondary antibody. Primary antibody dilutions are as follows: α-pdNAD-MDH, 1:2000; α-FtsH12, 1:1000; α-GFP/YFP, 1:10,000; α-HA, 1:7000; α-Flag M2, 1:5000; α-PORA, 1:2000; α-actin, 1:10,000.
Native PAGE observation of NAD-MDH activity was performed as described (Beeler et al., 2014).
Bioinformatic Analyses
For amino acid sequence alignments, all Arabidopsis MDH isoform sequences were retrieved from TAIR, while pdNAD-MDH sequences from different plant species were retrieved from the Phytozome v12 database (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3245001/). Sequences were aligned using Clustal Omega.
For homology-based modeling of the pdNAD-MDH protein structure, the pdNAD-MDH protein sequence without the cTP was modeled using SWISS-MODEL (https://www.ncbi.nlm.nih.gov/pubmed/24782522) onto the human MMDH2 structure as a template (PDB: 2DFD), which was cocrystallized with malate and NAD.
Cloning of Expression Vectors for Plant Transformation
Constructs for Arabidopsis transformation were assembled using Gateway technology (Invitrogen). For multisite Gateway assembly, we prepared the promoter, coding sequence, or tag in appropriate pENTR or pDONR vectors. The ABI3 promoter (2176 bp) was amplified from Arabidopsis genomic DNA, flanked with HindIII restriction sites, and ligated into the pENTR vector between the attL4 and attR1 recombination sites at the 5′ and 3′ ends, respectively. The pdNAD-MDH promoter in pDONR P4-P1r and the pdNAD-MDH coding sequence in pDONR221 were described previously. The coding sequences of the various NAD-MDH isoforms fused to the chloroplast transit peptide of the Rubisco small subunit were synthesized by Biomatik, flanked by attB1 and atttB2 recombination sites. These sequences were recombined directly into the pDONR221 vector. The eYFP and Flag-HA tags were cloned into pDONR P2R-P3 (Beeler et al., 2014; Tschopp et al., 2017). The appropriate promoter, coding sequence, and C-terminal tag were recombined via an LR Clonase reaction into the multisite Gateway binary vector pB7m34GW,0.
To generate overexpressor lines of pdNAD-MDH and FtsH12, the pDONR221 vector containing the corresponding coding sequence was recombined in a LR Clonase reaction into the single Gateway binary vector pB7YWG2,0 (with a 35S promoter and C-terminal YFP tag; Karimi et al., 2002) and pUBC-YFP (Grefen et al., 2010), respectively.
To generate artificial microRNA silencing constructs, we recombined the amiRNA miR-mdh-1 cassette in pDONR221 (described previously; Beeler et al., 2014) into a vector containing an estradiol-inducible cassette (XVE) driven by the UBIQUITIN10 promoter, via the LR Clonase reaction (Lee et al., 2013; Gujas et al., 2017). Two artificial microRNA silencing cassettes for FtsH12 were designed and cloned using the Web MicroRNA Designer tool (WMD3, wmd3.weigelworld.org; Ossowski et al., 2008) according to the provided instructions. The primers used are provided in Supplemental Table 2. The assembled amiRNAs were flanked with attB1 and attB2 recombination sites using the attB1-amiRNA FW and attB2-amiRNA REV primers (Supplemental Table 2) and recombined in pDONR221 vectors using the BP Clonase reaction. Instead of the pART27 vector, we further recombined the insert into the binary pJCV52 vector via an LR Clonase reaction.
To generate stable Arabidopsis lines, the appropriate constructs were transformed into Agrobacterium tumefaciens (strain GV3101). Wild-type Columbia and heterozygous pdnad-mdh plants were transformed using the floral dip method as described previously (Zhang et al., 2006). The T1 generation of transformants was selected using the BASTA or hygromycin selection marker (as described above for plant growth). Up to 10 seedlings per line were screened for transgene expression using immunoblotting (using the antibody against the epitope tag) and genotyped for the pdnad-mdh T-DNA insertion in the case of transformed pdnad-mdh plants. For plants expressing the transgene, the number of T-DNA insertion loci was determined by segregation analysis of the resistance marker gene in the T2 generation. Two independent transformants with single insertions, originating from different T0 parents, were selected for further analysis, either in the T2 generation (BASTA resistant plants that are heterozygous or homozygous for the transgene) or T3 generation (plants that are homozygous for the transgene), as specified.
Transient expression in Nicotiana benthamiana leaves was performed according to Seung et al. (2015).
Immunoprecipitation Experiments
Proteins were extracted from leaf tissue of Arabidopsis plants expressing the epitope tagged proteins by homogenizing in immunoprecipitation medium (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% [v/v] Triton X-100, 1 mM DTT, and Complete Protease Inhibitor cocktail [Roche]). Insoluble material was removed by centrifugation. The supernatant was incubated for 1 h at 4°C with µMACS magnetic beads conjugated to α-YFP or α-HA (Miltenyi Biotec). After incubation, the beads were recovered using a µColumn (Miltenyi Biotec) on a magnetic stand. The beads were washed five times with immunoprecipitation medium before eluting the bound proteins with elution buffer (50 mM Tris-HCl, pH 7.5, and 2% [w/v] SDS). The proteins were precipitated by the addition of 10% (w/v) trichloroacetic acid. After two washes with cold acetone, the protein pellet was dissolved in 10 mM Tris, pH 8.2, and 2 mM CaCl2 and digested with trypsin for 30 min at 60°C. The resulting peptides were dried, dissolved in 0.1% (v/v) formic acid, and analyzed by LC-MS/MS on a nanoAcquity UPLC (Waters) connected to a Q Exactive mass spectrometer (Thermo Scientific) equipped with a Digital PicoView source for electrospray ionization (New Objective). Peptides were trapped on a Symmetry C18 trap column (5 µm, 180 µm × 20 mm; Waters) and separated on a BEH300 C18 column (1.7 µm, 75 µm × 150 mm; Waters) at a flow rate of 250 nL/min using a gradient from 1% solvent B (0.1% formic acid in acetonitrile [Romil])/99% solvent A (0.1% formic acid in water [Romil]) to 40% solvent B/60% solvent A within 90 min. The mass spectrometer settings were as follows: data-dependent analysis; precursor scan range 350 to 1500 m/z, resolution 70,000, maximum injection time 100 ms, threshold 3e6; fragment ion scan range 200 to 2000 m/z, resolution 35,000, maximum injection time 120 ms, and threshold 1e5. Proteins were identified using the Mascot search engine (Matrix Science, version 2.4.1) against the TAIR10 Arabidopsis proteome database, with fragment ion mass tolerance of 0.030 D, parent ion tolerance of 10.0 ppm, and oxidation of methionine was specified in Mascot as a variable modification. Scaffold (Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they achieved a false discovery rate (FDR) of <0.1% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they achieved an FDR of <1.0% and contained at least two identified peptides.
Light and Electron Microscopy
Transmission electron microscopy analyses were conducted as previously described (Beeler et al., 2014). For differential interference contrast images of embryos, seeds were cleared with a glycerin:chloral hydrate:water (1:8:3, w/w/w) solution for 2 to 6 h, depending on the developmental stage of the seeds (Breuninger et al., 2008). Embryos were imaged under an Axio Imager 2 microscope with ZEN pro 2011 software (Zeiss). For imaging of YFP in leaf tissue, confocal laser scanning microscopy was conducted as previously described (Beeler et al., 2014).
Isothermal Titration Calorimetry
Binding of NADH to recombinant pdNAD-MDH proteins (wild type, R162Q, R234Q, and R162Q R234Q) was measured with a VP-ITC instrument at 25°C. Forty injections of cofactor solution (400 µM NADH), each with a volume of 6 µL, were made into a recombinant protein solution (40 µM). An injection spacing of 300 s was selected, and duration was automatically calculated. An initial injection of 2 μL was excluded from data analysis. Default sample cell volume was 1.4644 mL. Stirring speed and reference power were set to 307 RPM and 10 µCal⋅s−1, respectively. Before measurement, samples were degassed for 5 min at 23°C. The proteins were desalted into ITC buffer (20 mM Tris-HCl, pH 7.5, and 10 mM NaCl) with and without 2.5 mM 2-mercaptoethanol (Sigma-Aldrich) using Illustra NAP-5 (Fisher Scientific) prior to the experiment. The ligand tested was NADH (Grade I; Roche) dissolved directly into the ITC buffer. Concentrations of the proteins and NADH were determined spectrophotometrically using following extinction coefficients (Ɛ340nm [NADH] = 6220 M−1 cm−1; Ɛ280nm [pdNAD-MDH] = 8940 M−1 cm−1).
Lipid and Carotenoid Measurements
For targeted lipid profiling, 6-d-old light or dark-grown seedlings were ground into a fine powder in liquid nitrogen using a mortar and pestle. Ground plant material (30–60 mg, precisely weighed for each sample) was extracted with 0.4 mL tetrahydrofuran/methanol (50:50, v/v) including 5 µg/mL hydrogenated MDGD (Matreya) as an internal standard. Ten to fifteen glass beads were added and the samples were homogenized for 3 min at 30 Hz in a tissue lyser (Qiagen). Insoluble material was removed by two rounds of centrifugation at 16,000g for 3 min. Two hundred microliters of supernatant was transferred to an appropriate glass vial for immediate UHPLC-QTOF-MS analysis (Waters) as previously described (Martinis et al., 2011; Kessler and Glauser, 2014). Raw data were processed using MassLynx version 4.1 (waters) for automatic peak analysis. Identification and relative quantification were performed as described by Spicher et al. (2017).
Protochlorophyllide Measurements
Six-day-old etiolated seedlings were harvested and ground into fine powder in liquid nitrogen using a mortar and pestle. Protochlorophyllide was extracted twice in 9:1 mix (v/v) of acetone:0.1 M NH4OH, and insoluble material was removed by pelleting at 13,000g for 10 min after each extraction (Cheminant et al., 2011). The two extractions were combined, and emission spectra were recorded from 500 to 750 nm following excitation at 440 nm using a fluorescence spectrophotometer (Tecan infinite M1000). Protochlorophyllide was quantified at the emission peak of 632 nm and expressed as fluorescence per mg of fresh weight.
Accession Numbers
Sequence data from this article can be found in TAIR (www.arabidopsis.org) under the following accession numbers: pdNAD-MDH (At3g47520), FtsH12 (At1g79560), FtsHi1 (At4g23940), FtsHi2 (At3g16290), FtsHi4 (At5g64580), FtsHi5 (At3g04340), and YCF2 (AtCg00860). TAIR accession numbers of potential interaction partners are provided in Tables 1 to 4.
Supplemental Data
Supplemental Figure 1. Exogenous supply of sucrose did not rescue pdnad-mdh seedlings.
Supplemental Figure 2. Examples of normal, compromised, and absent prolamellar body structures in Col and miR-mdh-1, observed via transmission electron microscopy.
Supplemental Figure 3. Multiple protein sequence alignment of all nine Arabidopsis thaliana malate dehydrogenase isoforms.
Supplemental Figure 4. Native-PAGE gel analysis of heterozygous pdnad-mdh plants expressing various NAD-MDH isoforms.
Supplemental Figure 5. FtsH12 localization and embryo lethality of the ftsh12 mutant.
Supplemental Figure 6. Quantification of FtsH12 in miR-mdh-1.
Supplemental Figure 7. Sequence alignment of pdNAD-MDH isoforms in various plant species, algae, and E. coli.
Supplemental Table 1. Total set of lipids measured in 6-d-old wild-type (Col) and miR-mdh-1 seedlings.
Supplemental Table 2. Oligonucleotide primers used in this study.
Supplemental Data Set 1. Complete list of proteins identified in immunoprecipitates by MS/MS.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was funded by the Swiss National Foundation (Grant 31003A_156987 to O.K. and S.C.Z.) and by ETH Zurich. We thank Claudia Di Césaré from the Neuchâtel Platform of Analytical Chemistry for technical assistance, as well as Peter Hunziker, Yolanda Joho-Auchli, and Simone Wüthrich from the Functional Genomic Centre Zurich for mass spectrometry analysis. We thank Masato Nakai for sharing unpublished work and his thoughts, Enrico Martinoia and Alisdair Fernie for fruitful discussions during the course of this work, and Andrea Ruckle for help in plant culture.
AUTHOR CONTRIBUTIONS
T.B.S., S.C.Z., and O.K. conceived and directed the research. T.B.S., A.C., S.C.Z., and O.K. designed the experiments. T.B.S., A.C., E.D., M.S., F.G., M.S., F.K., S.H., D.A., and B.A.M. performed research and analyzed data. T.B.S. and S.C.Z. wrote the manuscript with input from all of the authors.
Footnotes
Articles can be viewed without a subscription.
References
- Bahl J., Francke B., Monéger R. (1976). Lipid composition of envelopes, prolamellar bodies and other plastid membranes in etiolated, green and greening wheat leaves. Planta 129: 193–201. [DOI] [PubMed] [Google Scholar]
- Beeler S., Liu H.-C., Stadler M., Schreier T., Eicke S., Lue W.-L., Truernit E., Zeeman S.C., Chen J., Kötting O. (2014). Plastidial NAD-dependent malate dehydrogenase is critical for embryo development and heterotrophic metabolism in Arabidopsis. Plant Physiol. 164: 1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkemeyer M., Scheibe R., Ocheretina O. (1998). A novel, non-redox-regulated NAD-dependent malate dehydrogenase from chloroplasts of Arabidopsis thaliana L. J. Biol. Chem. 273: 27927–27933. [DOI] [PubMed] [Google Scholar]
- Birktoft J.J., Banaszak L.J. (1983). The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases. X-ray refinement of cytoplasmic malate dehydrogenase. J. Biol. Chem. 258: 472–482. [DOI] [PubMed] [Google Scholar]
- Bodi Z., Zhong S., Mehra S., Song J., Graham N., Li H., May S., Fray R.G. (2012). Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front. Plant Sci. 3: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho S.C., Tatsuta T., von Heijne G., Kim H. (2013). Dislocation by the m-AAA protease increases the threshold hydrophobicity for retention of transmembrane helices in the inner membrane of yeast mitochondria. J. Biol. Chem. 288: 4792–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14: 867–876. [DOI] [PubMed] [Google Scholar]
- Bross C.D., Howes T.R., Abolhassani Rad S., Kljakic O., Kohalmi S.E. (2017). Subcellular localization of Arabidopsis arogenate dehydratases suggests novel and non-enzymatic roles. J. Exp. Bot. 68: 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela H., Pérez-Pérez J.M., Micol J.L. (2011). Uncovering the post-embryonic functions of gametophytic- and embryonic-lethal genes. Trends Plant Sci. 16: 336–345. [DOI] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J.-P., Erhardt M., Hayes S., Terry M.J., Genschik P., Achard P. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A.R., Wigley D.B., Chia W.N., Barstow D., Atkinson T., Holbrook J.J. (1986). Site-directed mutagenesis reveals role of mobile arginine residue in lactate dehydrogenase catalysis. Nature 324: 699–702. [DOI] [PubMed] [Google Scholar]
- Comparot-Moss S., et al. (2010). A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 152: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley S.D. (2003). Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol. 7: 265–272. [DOI] [PubMed] [Google Scholar]
- Copley S.D. (2014). An evolutionary perspective on protein moonlighting. Biochem. Soc. Trans. 42: 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetić T., Veljović-Jovanović S., Vucinić Z. (2008). Characterization of NAD-dependent malate dehydrogenases from spinach leaves. Protoplasma 232: 247–253. [DOI] [PubMed] [Google Scholar]
- Demishtein-Zohary K., Azem A. (2017). The TIM23 mitochondrial protein import complex: function and dysfunction. Cell Tissue Res. 367: 33–41. [DOI] [PubMed] [Google Scholar]
- Deng X.W., Caspar T., Quail P.H. (1991). cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182. [DOI] [PubMed] [Google Scholar]
- Despres B., Delseny M., Devic M. (2001). Partial complementation of embryo defective mutations: a general strategy to elucidate gene function. Plant J. 27: 149–159. [DOI] [PubMed] [Google Scholar]
- Drescher A., Ruf S., Calsa T. Jr., Carrer H., Bock R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22: 97–104. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Engdahl S., Aronsson H., Sundqvist C., Timko M.P., Dahlin C. (2001). Association of the NADPH:protochlorophyllide oxidoreductase (POR) with isolated etioplast inner membranes from wheat. Plant J. 27: 297–304. [DOI] [PubMed] [Google Scholar]
- Feng J., Fan P., Jiang P., Lv S., Chen X., Li Y. (2014). Chloroplast-targeted Hsp90 plays essential roles in plastid development and embryogenesis in Arabidopsis possibly linking with VIPP1. Physiol. Plant. 150: 292–307. [DOI] [PubMed] [Google Scholar]
- Ferro M., et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9: 1063–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann L.H., Yoon E.S., Meinke D.W. (1995). Saturating the genetic-map of Arabidopsis thaliana with embryonic mutations. Plant J. 7: 341–350. [Google Scholar]
- Fujii S., Kobayashi K., Nagata N., Masuda T., Wada H. (2017). Monogalactosyldiacylglycerol facilitates synthesis of photoactive protochlorophyllide in etioplasts. Plant Physiol. 174: 2183–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton D.C., et al. (2008). β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusari C.M., Kooke R., Lauxmann M.A., Annunziata M.G., Encke B., Hoehne M., Krohn N., Becker F.F.M., Schlereth A., Sulpice R., Stitt M., Keurentjes J.J.B. (2017). Genome-wide association mapping reveals that specific and pleiotropic regulatory mechanisms fine-tune central metabolism and growth in Arabidopsis. Plant Cell 29: 2349–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez L.D., Gilday A., Feil R., Lunn J.E., Graham I.A. (2010). AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 64: 1–13. [DOI] [PubMed] [Google Scholar]
- Goward C.R., Nicholls D.J. (1994). Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 3: 1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau U.M., Trommer W.E., Rossmann M.G. (1981). Structure of the active ternary complex of pig heart lactate dehydrogenase with S-lac-NAD at 2.7 A resolution. J. Mol. Biol. 151: 289–307. [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Gujas B., Cruz T.M.D., Kastanaki E., Vermeer J.E.M., Munnik T., Rodriguez-Villalon A. (2017). Perturbing phosphoinositide homeostasis oppositely affects vascular differentiation in Arabidopsis thaliana roots. Development 144: 3578–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M.D., Osmond C.B. (1976). Compartmentation and transport in C4 photosynthesis. In Transport in Plants III, Stocking C.R., Heber U., eds, Volume Vol. 3 (Berlin: Springer; ), pp. 144–184. [Google Scholar]
- He H., Lee M.-C., Zheng L.-L., Zheng L., Luo Y. (2013). Integration of the metabolic/redox state, histone gene switching, DNA replication and S-phase progression by moonlighting metabolic enzymes. Biosci. Rep. 33: e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbelmann I., et al. (2012). Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J. Exp. Bot. 63: 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber U. (1974). Metabolite exchange between chloroplasts and cytoplasm. Annu. Rev. Plant Physiol. 25: 393–421. [Google Scholar]
- Hendriks W., Mulders J.W., Bibby M.A., Slingsby C., Bloemendal H., de Jong W.W. (1988). Duck lens epsilon-crystallin and lactate dehydrogenase B4 are identical: a single-copy gene product with two distinct functions. Proc. Natl. Acad. Sci. USA 85: 7114–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyno E., Innocenti G., Lemaire S.D., Issakidis-Bourguet E., Krieger-Liszkay A. (2014). Putative role of the malate valve enzyme NADP-malate dehydrogenase in H2O2 signalling in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrer D., Flütsch S., Pazmino D., Matthews J.S.A., Thalmann M., Nigro A., Leonhardt N., Lawson T., Santelia D. (2016). Blue light induces a distinct starch degradation pathway in guard cells for stomatal opening. Curr. Biol. 26: 362–370. [DOI] [PubMed] [Google Scholar]
- Huberts D.H.E.W., van der Klei I.J. (2010). Moonlighting proteins: an intriguing mode of multitasking. Biochim. Biophys. Acta 1803: 520–525. [DOI] [PubMed] [Google Scholar]
- Jang J.C., León P., Zhou L., Sheen J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P., Dörmann P., Peto C.A., Lutes J., Benning C., Chory J. (2000). Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. USA 97: 8175–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery C.J. (1999). Moonlighting proteins. Trends Biochem. Sci. 24: 8–11. [DOI] [PubMed] [Google Scholar]
- Jeffery C.J. (2003). Moonlighting proteins: old proteins learning new tricks. Trends Genet. 19: 415–417. [DOI] [PubMed] [Google Scholar]
- Journet E.P., Neuburger M., Douce R. (1981). Role of glutamate-oxaloacetate transaminase and malate dehydrogenase in the regeneration of NAD for glycine oxidation by spinach leaf mitochondria. Plant Physiol. 67: 467–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kessler F., Glauser G. (2014). Prenylquinone profiling in whole leaves and chloroplast subfractions. Methods Mol. Biol. 1153: 213–226. [DOI] [PubMed] [Google Scholar]
- Kinoshita H., Nagasaki J., Yoshikawa N., Yamamoto A., Takito S., Kawasaki M., Sugiyama T., Miyake H., Weber A.P.M., Taniguchi M. (2011). The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J. 65: 15–26. [DOI] [PubMed] [Google Scholar]
- Klement H., Helfrich M., Oster U., Schoch S., Rüdiger W. (1999). Pigment-free NADPH:protochlorophyllide oxidoreductase from Avena sativa L. Purification and substrate specificity. Eur. J. Biochem. 265: 862–874. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Kondo M., Fukuda H., Nishimura M., Ohta H. (2007). Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc. Natl. Acad. Sci. USA 104: 17216–17221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langklotz S., Baumann U., Narberhaus F. (2012). Structure and function of the bacterial AAA protease FtsH. Biochim. Biophys. Acta 1823: 40–48. [DOI] [PubMed] [Google Scholar]
- Lebedev N., Van Cleve B., Armstrong G., Apel K. (1995). Chlorophyll synthesis in a deetiolated (det340) mutant of Arabidopsis without NADPH-protochlorophyllide (PChlide) oxidoreductase (POR) A and photoactive PChlide-F655. Plant Cell 7: 2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Schwartz M.P., Prakash S., Iwakura M., Matouschek A. (2001). ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell 7: 627–637. [DOI] [PubMed] [Google Scholar]
- Lee Y., Rubio M.C., Alassimone J., Geldner N. (2013). A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412. [DOI] [PubMed] [Google Scholar]
- Lu X., Zhang D., Li S., Su Y., Liang Q., Meng H., Shen S., Fan Y., Liu C., Zhang C. (2014). FtsHi4 is essential for embryogenesis due to its influence on chloroplast development in Arabidopsis. PLoS One 9: e99741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majsec K., Bhuiyan N.H., Sun Q., Kumari S., Kumar V., Ware D., van Wijk K.J. (2017). The plastid and mitochondrial peptidase network in Arabidopsis thaliana: a foundation for testing genetic interactions and functions in organellar proteostasis. Plant Cell 29: 2687–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield S.G., Briarty L.G. (1991). Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can. J. Bot. 69: 461–476. [Google Scholar]
- Martin A., Baker T.A., Sauer R.T. (2005). Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature 437: 1115–1120. [DOI] [PubMed] [Google Scholar]
- Martinis J., Kessler F., Glauser G. (2011). A novel method for prenylquinone profiling in plant tissues by ultra-high pressure liquid chromatography-mass spectrometry. Plant Methods 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascia P.N., Robertson D.S. (1978). Studies of chloroplast development in four maize mutants defective in chlorophyll biosynthesis. Planta 143: 207–211. [DOI] [PubMed] [Google Scholar]
- Minárik P., Tomásková N., Kollárová M., Antalík M. (2002). Malate dehydrogenases--structure and function. Gen. Physiol. Biophys. 21: 257–265. [PubMed] [Google Scholar]
- Moore Bd. (2004). Bifunctional and moonlighting enzymes: lighting the way to regulatory control. Trends Plant Sci. 9: 221–228. [DOI] [PubMed] [Google Scholar]
- Moore B., Zhou L., Rolland F., Hall Q., Cheng W.H., Liu Y.X., Hwang I., Jones T., Sheen J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336. [DOI] [PubMed] [Google Scholar]
- Musrati R.A., Kollárová M., Mernik N., Mikulásová D. (1998). Malate dehydrogenase: distribution, function and properties. Gen. Physiol. Biophys. 17: 193–210. [PubMed] [Google Scholar]
- Nakai M. (2018). New perspectives on chloroplast protein import. Plant Cell Physiol. 59: 1111–1119. [DOI] [PubMed] [Google Scholar]
- Nicholls D.J., Miller J., Scawen M.D., Clarke A.R., Holbrook J.J., Atkinson T., Goward C.R. (1992). The importance of arginine 102 for the substrate specificity of Escherichia coli malate dehydrogenase. Biochem. Biophys. Res. Commun. 189: 1057–1062. [DOI] [PubMed] [Google Scholar]
- Nickelsen J. (2005). The green alga Chlamydomonas reinhardtii: A genetic model organism. In Progress in Botany, Esser K., Lüttge U., Beyschlag W., Murata J., eds (Heidelberg, Germany: Springer-Verlag; ), pp. 68–89. [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690. [DOI] [PubMed] [Google Scholar]
- Park H., Kreunen S.S., Cuttriss A.J., DellaPenna D., Pogson B.J. (2002). Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton D.A., Franzmann L.H., Meinke D.W. (1991). Mapping genes essential for embryo development in Arabidopsis thaliana. Mol. Gen. Genet. 227: 337–347. [DOI] [PubMed] [Google Scholar]
- Patton D.A., Schetter A.L., Franzmann L.H., Nelson K., Ward E.R., Meinke D.W. (1998). An embryo-defective mutant of arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 116: 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J.E., Smith S.M. (2007). Arabidopsis peroxisomal malate dehydrogenase functions in beta-oxidation but not in the glyoxylate cycle. Plant J. 50: 381–390. [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Zhou W., Smith S.M. (2010). Fatty acid beta-oxidation in germinating Arabidopsis seeds is supported by peroxisomal hydroxypyruvate reductase when malate dehydrogenase is absent. Plant Mol. Biol. 72: 101–109. [DOI] [PubMed] [Google Scholar]
- Ruppel N.J., Hangarter R.P. (2007). Mutations in a plastid-localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana. BMC Plant Biol. 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryberg M., Sundqvist C. (1982). Characterization of prolamellar bodies and prothylakoids fractionated from wheat etioplasts. Physiol. Plant. 56: 125–132. [Google Scholar]
- Ryberg M., Sundqvist C. (1988). The regular ultrastructure of isolated prolamellar bodies depends on the presence of membrane‐bound NADPH‐protochlorophyllide oxidoreductase. Physiol. Plant. 73: 218–226. [Google Scholar]
- Scheibe R. (1987). NADP+‐malate dehydrogenase in C3‐plants: Regulation and role of a light‐activated enzyme. Physiol. Plant. 71: 393–400. [Google Scholar]
- Scheibe R. (2004). Malate valves to balance cellular energy supply. Physiol. Plant. 120: 21–26. [DOI] [PubMed] [Google Scholar]
- Selinski J., König N., Wellmeyer B., Hanke G.T., Linke V., Neuhaus H.E., Scheibe R. (2014). The plastid-localized NAD-dependent malate dehydrogenase is crucial for energy homeostasis in developing Arabidopsis thaliana seeds. Mol. Plant 7: 170–186. [DOI] [PubMed] [Google Scholar]
- Selstam E., Schelin J., Brain T., Williams W.P. (2002). The effects of low pH on the properties of protochlorophyllide oxidoreductase and the organization of prolamellar bodies of maize (Zea mays). Eur. J. Biochem. 269: 2336–2346. [DOI] [PubMed] [Google Scholar]
- Seung D., Thalmann M., Sparla F., Abou Hachem M., Lee S.K., Issakidis-Bourguet E., Svensson B., Zeeman S.C., Santelia D. (2013). Arabidopsis thaliana AMY3 is a unique redox-regulated chloroplastic α-amylase. J. Biol. Chem. 288: 33620–33633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Soyk S., Coiro M., Maier B.A., Eicke S., Zeeman S.C. (2015). PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 13: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sew Y.S., Ströher E., Fenske R., Millar A.H. (2016). Loss of mitochondrial malate dehydrogenase activity alters seed metabolism impairing seed maturation and post-germination growth in Arabidopsis. Plant Physiol. 171: 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein E., Sulebele G. (1969). Catalytic mechanism of pig heart mitochondrial malate dehydrogenase studied by kinetics at equilibrium. Biochemistry 8: 2543–2550. [DOI] [PubMed] [Google Scholar]
- Sokolenko A., Pojidaeva E., Zinchenko V., Panichkin V., Glaser V.M., Herrmann R.G., Shestakov S.V. (2002). The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41: 291–310. [DOI] [PubMed] [Google Scholar]
- Solymosi K., Aronsson H. (2013). Plastid development in leaves during growth and senescence. Advances in Photosynthesis and Respiration, Biswal B., Krupinska K., Biswal U.C., eds, Volume 36 (Dordrecht, The Netherlands: Springer Science+Business Media; ). [Google Scholar]
- Solymosi K., Schoefs B. (2010). Etioplast and etio-chloroplast formation under natural conditions: the dark side of chlorophyll biosynthesis in angiosperms. Photosynth. Res. 105: 143–166. [DOI] [PubMed] [Google Scholar]
- Sperling U., Franck F., van Cleve B., Frick G., Apel K., Armstrong G.A. (1998). Etioplast differentiation in arabidopsis: both PORA and PORB restore the prolamellar body and photoactive protochlorophyllide-F655 to the cop1 photomorphogenic mutant. Plant Cell 10: 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicher L., Almeida J., Gutbrod K., Pipitone R., Dörmann P., Glauser G., Rossi M., Kessler F. (2017). Essential role for phytol kinase and tocopherol in tolerance to combined light and temperature stress in tomato. J. Exp. Bot. 68: 5845–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M., Miyake H. (2012). Redox-shuttling between chloroplast and cytosol: integration of intra-chloroplast and extra-chloroplast metabolism. Curr. Opin. Plant Biol. 15: 252–260. [DOI] [PubMed] [Google Scholar]
- Tatsuta T., Augustin S., Nolden M., Friedrichs B., Langer T. (2007). m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 26: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaz T., Bagard M., Pracharoenwattana I., Lindén P., Lee C.P., Carroll A.J., Ströher E., Smith S.M., Gardeström P., Millar A.H. (2010). Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol. 154: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp M.-A., Iki T., Brosnan C.A., Jullien P.E., Pumplin N. (2017). A complex of Arabidopsis DRB proteins can impair dsRNA processing. RNA 23: 782–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Aigner H., Funk C. (2012). FtsH proteases located in the plant chloroplast. Physiol. Plant. 145: 203–214. [DOI] [PubMed] [Google Scholar]
- Wigley D.B., Gamblin S.J., Turkenburg J.P., Dodson E.J., Piontek K., Muirhead H., Holbrook J.J. (1992). Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 A resolution. J. Mol. Biol. 223: 317–335. [DOI] [PubMed] [Google Scholar]
- Wistow G.J., Mulders J.W., de Jong W.W. (1987). The enzyme lactate dehydrogenase as a structural protein in avian and crocodilian lenses. Nature 326: 622–624. [DOI] [PubMed] [Google Scholar]
- Yang S.S., Zhai Q.H. (2017). Cytosolic GAPDH: a key mediator in redox signal transduction in plants. Biol. Plant. 61: 417–426. [Google Scholar]
- Yu B., Wakao S., Fan J., Benning C. (2004). Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol. 45: 503–510. [DOI] [PubMed] [Google Scholar]
- Zaffagnini M., Fermani S., Costa A., Lemaire S.D., Trost P. (2013). Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front. Plant Sci. 4: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zhao Y., et al. (2018). Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res. 28: 448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]