A site-specific cross-linking approach reveals the molecular topology of the transit peptide and its nucleotide-dependent movement within the chloroplast protein import channel.
Abstract
Chloroplast protein import is directed by the interaction of the targeting signal (transit peptide) of nucleus-encoded preproteins with translocons at the outer (TOC) and inner (TIC) chloroplast envelope membranes. Studies of the energetics and determinants of transit peptide binding have led to the hypothesis that import occurs through sequential recognition of transit peptides by components of TOC and TIC during protein import. To test this hypothesis, we employed a site-specific cross-linking approach to map transit peptide topology in relation to TOC-TIC components at specific stages of import in Arabidopsis thaliana and pea (Pisum sativum). We demonstrate that the transit peptide is in contact with Tic20 at the inner envelope in addition to TOC complex components at the earliest stages of chloroplast binding. Low levels of ATP hydrolysis catalyze the commitment of the preprotein to import by promoting further penetration across the envelope membranes and stabilizing the association of the preprotein with TOC-TIC. GTP hydrolysis at the TOC receptors serves as a checkpoint to regulate the ATP-dependent commitment of the preprotein to import and is not essential to drive preprotein import. Our results demonstrate the close cooperativity of the TOC and TIC machinery at each stage of transit peptide recognition and membrane translocation during protein import.
INTRODUCTION
The function and diversity of all plastid types relies on the efficient import of thousands of proteins that are encoded in the nucleus and translated in the cytosol. The vast majority of these proteins are synthesized with an N-terminal transit peptide that is recognized by the chloroplast protein import machinery, which facilitates their translocation across the outer and inner chloroplast envelope membranes. Once inside the stroma, proteins may be further targeted to suborganellar compartments, including the inner envelope membrane, thylakoid membrane, or thylakoid lumen (Jarvis and López-Juez, 2013; Shi and Theg, 2013; Paila et al., 2015).
Translocation of the majority of preproteins into chloroplasts occurs via the coordinated activity of translocons at the outer (TOC) and inner (TIC) chloroplast envelope membranes and stromal chaperones, which use ATP to drive import. Studies in both Arabidopsis thaliana and pea (Pisum sativum) support the hypothesis that the Toc34 and Toc159 GTPase import receptor families regulate preprotein recognition and binding to the TOC complex (Perry and Keegstra, 1994; Ma et al., 1996; Kouranov and Schnell, 1997; Oreb et al., 2011). The receptors are associated with Toc75, a β-barrel outer membrane protein, which forms the channel through which preproteins are translocated across the outer membrane (Schnell et al., 1994; Hinnah et al., 2002; Day et al., 2014). Toc75 is also proposed to interact with preproteins in the intermembrane space via its POTRA (polypeptide-transport associated) domains (Chen et al., 2016; Paila et al., 2016; O’Neil et al., 2017), as the POTRAs bind to preproteins and possess molecular chaperone activity in vitro (O’Neil et al., 2017). In addition, the POTRAs interact with two putative chaperones in the intermembrane space, Tic22-III and Tic22-IV, leading to the hypothesis that the Toc75 POTRAs and Tic22 proteins function coordinately to facilitate transit of the preprotein across the intermembrane space (Paila et al., 2016).
Two complexes have been proposed to function as components of the TIC machinery. These include a complex containing the stromal chaperone organizing proteins Tic110 and Tic40, and a small integral membrane protein, Tic20 (Ma et al., 1996; Kouranov and Schnell, 1997; Inaba et al., 2003; Chou et al., 2006). An import associated chaperone network, consisting of Heat-shock protein 93 (Hsp93)/Caseinolytic protease C (ClpC), chloroplast Heat shock protein 70 (cpHsp70), and chloroplast Heat-shock protein 90 (Hsp90C), function as the ATP-dependent import motor to drive membrane translocation and may assist in the suborganellar targeting, folding, or quality control of preproteins in the stroma (Shi and Theg, 2010; Su and Li, 2010; Liu et al., 2014; Sjögren et al., 2014; Paila et al., 2015; Flores-Pérez et al., 2016; Huang et al., 2016; Richardson et al., 2017). In addition, a large 1-MD protein complex, containing Tic214, Tic56, and Tic100, also associates with Tic20 (Kikuchi et al., 2013; Sjögren et al., 2014). The functions of Tic214, Tic56, and Tic100 in protein import have not been defined (Kikuchi et al., 2013; de Vries et al., 2015; Köhler et al., 2015). Tic20 is a common component of both the Tic110-Tic40 and 1-MD complexes, and it exhibits channel activity when reconstituted into proteoliposomes alone or in association with the 1-MD complex (Kovács-Bogdán et al., 2011; Kikuchi et al., 2013), leading to the proposal that it forms a component of the TIC channel.
Several intermediate steps in import have been described based on the energetics of protein import and in vitro binding studies with preproteins and purified import components or isolated chloroplasts. In the first step, preproteins are proposed to interact with TOC in the absence of energy, and transit peptide binding regulates the GTPase cycle of the TOC receptors by controlling receptor-receptor interactions (Ma et al., 1996; Oreb et al., 2011). This has led to the proposal that the TOC GTPase receptors act to gate the TOC channel for translocation of the preprotein across the outer membrane (Kessler and Schnell, 2004; Jarvis, 2008; Oreb et al., 2008; Li and Chiu, 2010; Richardson et al., 2014; Sjuts et al., 2017). This hypothesis is supported by the observation that inhibition of GTP hydrolysis through the use of nonhydrolyzable or slowly hydrolyzable GTP analogs inhibits import (Olsen and Keegstra, 1992; Kessler et al., 1994; Young et al., 1999). Low levels of ATP (∼0.1 mM) promote a second stage, referred to as the early import intermediate (Olsen et al., 1989). At this stage, preprotein binding to chloroplasts is stabilized and the transit peptide is hypothesized to have crossed the outer membrane and associated with TIC components (Kouranov and Schnell, 1997; Kikuchi et al., 2013). Higher levels of ATP promote import into the stroma via the proposed action of the import-associated chaperone network (Shi and Theg, 2010; Su and Li, 2010; Liu et al., 2014; Sjögren et al., 2014; Paila et al., 2015; Flores-Pérez et al., 2016; Huang et al., 2016; Richardson et al., 2017).
The transit peptides of chloroplast preproteins are generally larger and more diverse than the targeting signals for other organellar proteins (Kim and Hwang, 2013), and transit peptides have evolved to mediate tissue-specific or age-dependent import of specific classes of preproteins (Teng et al., 2012; Li and Teng, 2013). Furthermore, a number of TOC complexes with distinct transit peptide selectivities have been identified (Bauer et al., 2000; Ivanova et al., 2004; Kubis et al., 2004; Inoue et al., 2010). These observations have complicated efforts to define the transit peptide determinants that mediate various stages of the import process. Despite these challenges, a number of studies have identified motifs within the transit peptides of specific subclasses of preproteins that are required for efficient import (Lee et al., 2006, 2008; Chotewutmontri et al., 2012; Teng et al., 2012; Holbrook et al., 2016). These studies have led to models in which specific regions of transit peptides mediate interactions of specific TOC-TIC components during import to ensure unidirectional transport of the preprotein from the cytoplasm into the chloroplast stroma (Li and Teng, 2013).
Several major questions regarding the interactions of preproteins with import components and the topology of the preprotein at each stage in translocation remain unanswered. For example, it is clear that Toc34 and Toc159 play key roles in the early stages of import, yet it is still unclear how the GTPase activities of the receptors participate in membrane translocation. Furthermore, it is not clear at what stage the TOC and TIC complexes initiate their cooperative roles in import and which components mediate the initial engagement of the preproteins with TIC. In this study, we aimed to understand how the transit peptide interfaces with the translocation machinery to mediate unidirectional import with high fidelity. Using a site-specific cross-linking approach, we mapped the molecular topology of the transit peptide at energetically defined stages of import and investigated the nucleoside triphosphate (NTP)-dependent transitions between these stages in pea. Our data show that the transit peptide engages both TOC and TIC components at the early stages of binding, in the absence of added NTP. GTP hydrolysis appears to function as a checkpoint to allow preprotein translocation into the chloroplast stroma, and ATP functions to stabilize the interactions of the preprotein with TOC-TIC in addition to providing the energy for preprotein import. This study provides detailed insight into the molecular topology of a transit peptide during translocation across the envelope and provides important information about the molecular mechanism of protein translocation into chloroplasts.
RESULTS
Recombinant, Biotinylated Preprotein Displays Typical Chloroplast Binding/Import Characteristics under Defined Energetic Conditions
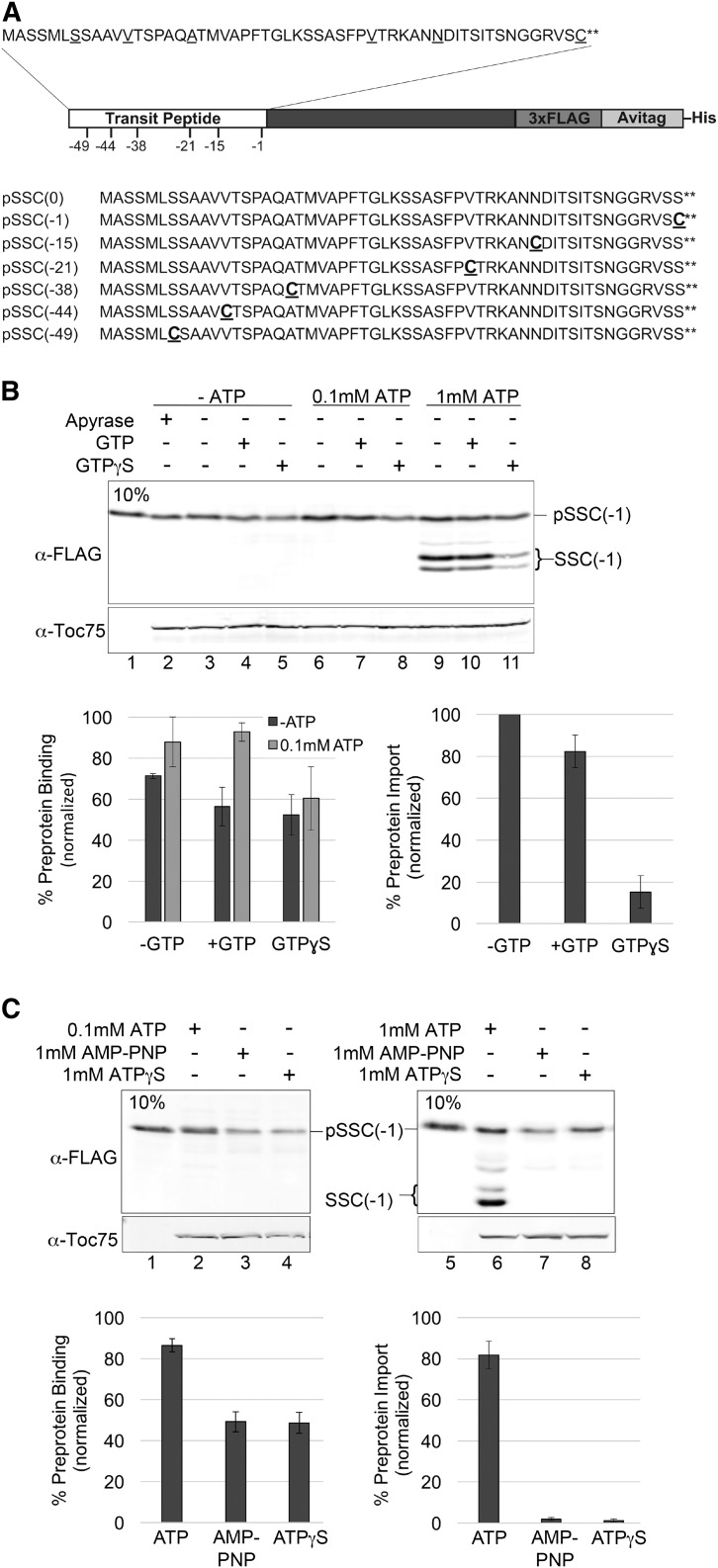
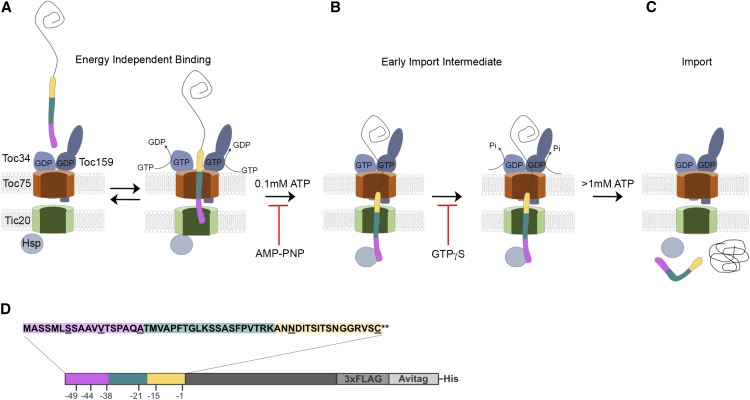
As a first step in mapping the interactions of the transit peptide with import components, we generated versions of the model preprotein, Arabidopsis Rubisco small subunit, in which all cysteine residues were removed from both the transit peptide and mature regions of the protein [pSSC(0)] and replaced with a single cysteine residue at a unique position over the length of the transit peptide. The modified residues are shown in Figure 1A. The single cysteine constructs were fused to a 3x FLAG epitope, an in vivo biotinylation signal (Avitag), and a C-terminal hexahistidine tag. The modified preproteins were expressed and biotinylated in Escherichia coli and purified using nickel affinity chromatography. All of the single cysteine constructs were efficiently imported into isolated pea chloroplasts under standard import conditions at levels comparable to an unmodified Arabidopsis Rubisco small subunit precursor, and the resulting mature processed forms were thermolysin-resistant (Supplemental Figure 1). The energy-dependent stages in import were previously defined with in vitro-translated preproteins; therefore, we tested whether the recombinant, biotinylated preproteins used in this study display typical energy-dependent binding characteristics using pSSC(-1). Chloroplasts isolated from pea were depleted of energy by preincubation with nigericin, an ionophore that dissipates the proton gradient across the thylakoid membrane (Theg et al., 1989), and glycerate, which depletes internal ATP via the ATP-dependent enzymatic activity of glycerate kinase in the stroma (Olsen et al., 1989; Olsen and Keegstra, 1992). In a control reaction, apyrase was added to deplete external NTPs (Figure 1B) (Olsen and Keegstra, 1992). Following incubation with pSSC(-1) for 20 min in the presence or absence of various NTP concentrations, chloroplasts were reisolated, resolved using SDS-PAGE, and subjected to immunoblotting with FLAG epitope antibodies. As shown in Figure 1B, the amount of full-length pSSC(-1) binding to chloroplasts was increased in the presence of 0.1 mM GTP + ATP relative to the absence of added NTP, or those treated with apyrase, consistent with previously observed ATP-stimulated binding (Olsen et al., 1989). In the presence of 1 mM ATP, pSSC(-1) is imported efficiently (Figure 1B). While we did not detect a stimulatory effect of GTP alone on preprotein binding, we did observe that binding and import is inhibited by preincubation with GTPγS, as previously described (Figure 1B) (Olsen and Keegstra, 1992; Young et al., 1999). As expected, we also observed that both AMP-PNP and ATPγS inhibited preprotein binding to chloroplasts and decreased import to barely detectable levels (Figure 1C). These results confirm that the biotinylated, recombinant preproteins used in this study are suitable substrates to investigate energy-dependent transit peptide-translocon cross-linking.
Figure 1.
Chloroplast Binding and Import of Recombinant Biotinylated Preprotein.
(A) Schematic diagram of the single cysteine mutant proteins used in this study. Cys substitutions are shown in bold and underlined.
(B) Isolated pea chloroplasts were incubated with urea-denatured, biotinylated pSSC(-1) in the presence of the indicated amounts of ATP with or without 0.1 mM GTP or 1 mM GTPγS, as indicated. Chloroplasts were reisolated and subjected to SDS-PAGE, and the preprotein [pSSC(-1)] or mature [SSC(-1)] forms were detected by immunoblotting with anti-FLAG and infrared dye-conjugated secondary antibody. Toc75 immunoblots are shown as a loading control. Lane 1 contains 10% of pSSC(-1) added to the reaction. Levels of binding [pSSC(-1)] and import [SSC(-1)] were quantitated as a percentage of either maximal preprotein binding or maximal preprotein import, respectively.
(C) Binding reactions were performed as described in (B), in the presence of 0.1 mM ATP, 1 mM AMP-PNP, or 1 mM ATPγS. Import was performed in the presence of 1 mM ATP, 1 mM AMP-PNP, or 1 mM ATPγS. For all experiments, reactions were performed in at least technical duplicates (same reaction immunoblotted separately) and biological triplicates (using chloroplasts from independent isolations), and the means of biological replicates are shown. Error bars represent the se of the mean between biological replicates.
Site-Specific Cross-Linking to the Early Import Intermediate
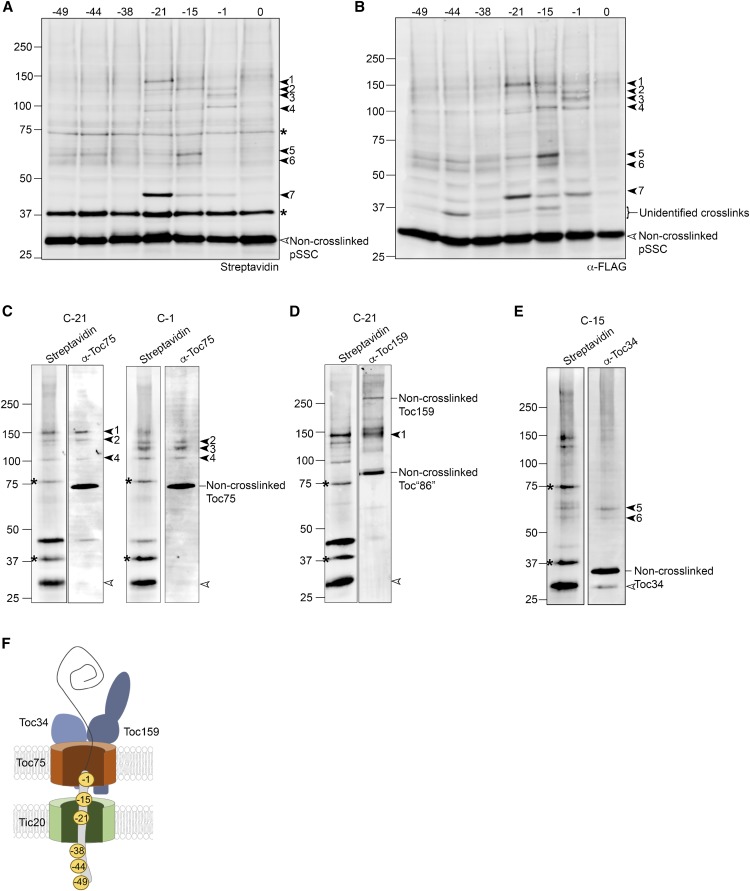
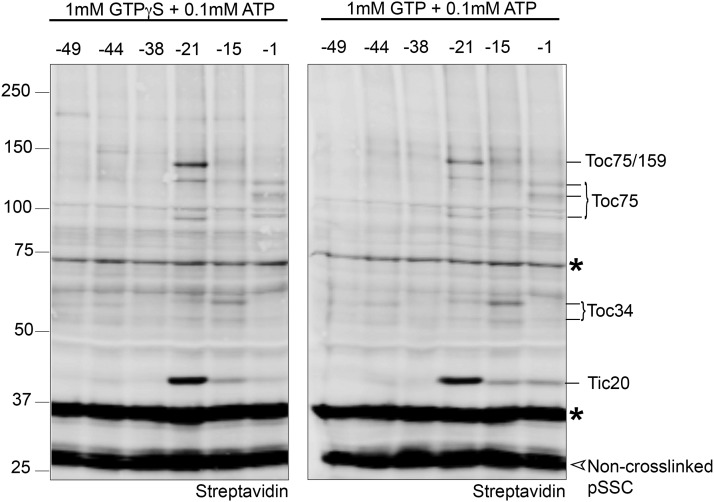
We began by mapping interactions between import components and distinct regions of the transit peptide at the early import intermediate stage. The single cysteine residue within each biotinylated pSSC substrate was chemically modified in vitro with the photoinducible, heterobifunctional cross-linker, 4-n-maleimidobenzophenone (MBP). The photoreactive benzophenone group within this cross-linker is nonselective and generates covalent adducts with a spacer arm length of <5 Å (Corgiat et al., 2014). pSSC cross-linking substrates were incubated with pea chloroplasts under conditions that promote the formation of a stable early import intermediate (0.1 mM GTP + 0.1 mM ATP for 5 min at 26°C in the dark). Chloroplasts were then washed, and cross-linking was induced with UV light. Chloroplast membranes were immunoblotted with streptavidin coupled to an infrared dye to detect pSSC (Figure 2A). Under these early binding conditions, site-specific cross-linking was observed over the length of the transit peptide, as indicated by the higher molecular weight adducts detected with streptavidin (solid arrows in Figure 2A). A consistent pattern of cross-linking was previously shown using a similar approach (Inoue and Akita, 2008).
Figure 2.
Cross-Linking Pattern of the Early Import Intermediate and Identification of Cross-Linked Proteins.
(A) Chloroplasts were incubated with MBP-modified, urea-denatured, biotinylated preprotein in the presence of 0.1 mM GTP + ATP and exposed to UV light for 15 min. Total membranes were isolated and subjected to SDS-PAGE and blotted with streptavidin conjugated to an infrared dye. Non-cross-linked preprotein (pSSC) and endogenous biotinylated chloroplast proteins are indicated by open arrowheads and asterisks, respectively. Site-specific cross-linked products are indicated by closed arrowheads and numbered 1 to 7. The experiment described in (A) is representative of six independent experiments.
(B) Same samples as in a (A) immunoblotted with α-FLAG.
(C) to (E) Cross-linking reactions with the indicated preproteins were scaled up and affinity pull-downs using streptavidin under stringent conditions were performed. Pull-downs were blotted with streptavidin in the 700-nm channel, and antibodies to either Toc75 (α-Toc75) (C), Toc159 (α-Toc159) (D), or Toc34 (α-Toc34) (E) in the 800-nm channel as indicated.
(F) A model illustrating the predominant position of the transit peptide within the translocon at the early import intermediate stage based on the cross-linking pattern in (A). Numbers to the left of the immunoblots indicate the size of molecular weight markers in kilodaltons.
Several site-specific cross-linked products of interest were observed that were absent in the pSSC(0) negative control (numbered 1–7 in Figure 2A). These included higher molecular weight cross-linked products (cross-links #1–4) to the C-terminal half of the transit peptide at positions −1, −15, or −21. We also identified smaller molecular weight cross-links toward the N-terminal region of the transit peptide (−38 and −44 positions) when blotted with anti-FLAG (Figure 2B). These products were not visible using streptavidin as a detection method because their migration is coincident with an abundant biotinylated chloroplast protein that is present in all samples, including the pSSC(0) negative control (Figure 2A, asterisk).
To establish the identity of the cross-linked products, membranes were isolated from pea chloroplasts cross-linked to MBP-modified preproteins and streptavidin affinity pull-downs were performed under stringent solubilization conditions, followed by immunoblotting with antibodies to known TOC/TIC components. As shown in Figure 2C, cross-linked products #1 to 4 identified with pSSC(-1) and (-21) [and presumably pSSC(-15), based on a consistent pattern between these three preproteins] all immunoreacted with anti-Toc75. Cross-link #1 in the pSSC(-21) reaction was also recognized by anti-Toc159 (Figure 2D), corresponding to an adduct with the predominant 86-kD proteolytic fragment of Toc159 generated during the isolation of pea chloroplasts (Bölter et al., 1998). Cross-links #5 and 6, which were observed predominantly with pSSC(-15), reacted with anti-Toc34 (Figure 2E), consistent with its role in the early stages of import. Non-cross-linked TOC components were also observed in streptavidin pull-downs; however, these non-cross-linked species were not found in pull-downs with the pSSC(0) control (Supplemental Figure 2), indicating that they are not due to nonspecific binding to the streptavidin magnetic beads or inadequate dissolution of membranes. These non-cross-linked species likely result from cross-linked adducts that are disrupted during sample processing, residual components of native TOC complexes that were not fully disrupted during membrane solubilization, or are due to the large amount of input needed to enrich cross-linked products for detection using immunoblotting (Supplemental Figure 2). Nevertheless, the identity of the cross-linked products is clear based on their immunoreactivity with specific antibodies and the corresponding shift in molecular weight.
The major cross-link to the early import intermediate occurs with pSSC(-21) and migrates at ∼42 kD (#7). Based on the size of the molecular weight shift, we tested if this adduct corresponded to a cross-link to Tic20. Antibodies of sufficient sensitivity to detect a Tic20 cross-linked product are not available. Consequently, chloroplasts from transgenic Arabidopsis plants expressing Tic20 with an N-terminal myc tag (myc-Tic20) in the tic20-I heterozygous background were used for cross-linking experiments. The tic20-I homozygous mutants display a severe albino phenotype (Kasmati et al., 2011), which is complemented by overexpression of myc-Tic20, demonstrating that it is fully functional (Supplemental Figure 3). Because the myc-Tic20 plants expressed both myc-tagged and non-tagged Tic20, streptavidin detected pSSC(-21) cross-links to both endogenous Tic20 and myc-Tic20 in these pull-downs, with the cross-linked myc-Tic20 showing a slight shift in molecular weight due to the presence of the myc-epitope (Figure 3). Anti-myc detected this higher molecular weight cross-linked product at ∼42 kD in samples from streptavidin pull-downs using chloroplasts isolated from myc-Tic20-expressing Arabidopsis plants, but not in wild-type Arabidopsis chloroplasts (Figure 3). These results show that the abundant ∼42 kD cross-link (#7) corresponds to Tic20.
Figure 3.
Identification of Tic20 as a Major Cross-Link of pSSC(-21).
Chloroplasts isolated from wild-type and transgenic Arabidopsis plants expressing myc-Tic20 were used for cross-linking experiments with pSSC(-21). Cross-linked chloroplast membranes were subjected to streptavidin affinity pull-downs and probed with streptavidin in the 700-nm channel and anti-myc in the 800-nm channel. An asterisk indicates an abundant chloroplast biotinylated protein; open arrowheads indicate non-cross-linked preprotein. Numbers to the left of the immunoblots indicate the size of molecular weight markers in kilodaltons.
The identification of the cross-linked products with individual single Cys mutants is summarized in a model of the topology of the early import intermediate (Figure 2F) based on the cross-linking shown in Figures 2A and 2B, in which residue −21 of the pSSC transit peptide has translocated across the outer envelope and is engaged with Tic20, a component of TIC at the inner membrane. The C-terminal region of the transit peptide, proximal to the cleavage site (residue −1), remains engaged with Toc75. Cross-links to Toc75 and the 86-kD fragment of Toc159 at residue −21 of the transit peptide and Toc34 at position −15 are also observed, albeit at relatively lower abundance relative to Tic20 (Figure 2A). Under high ATP conditions, there is relatively little cross-linking over the length of the transit peptide, with the level of Tic20 cross-link greatly reduced (Supplemental Figure 4). This is consistent with import of pSSC into the stroma and cleavage of the transit peptide.
Repositioning of the Transit Peptide within TOC and TIC during the Transition from Energy-Independent Binding to Formation of the Early Import Intermediate
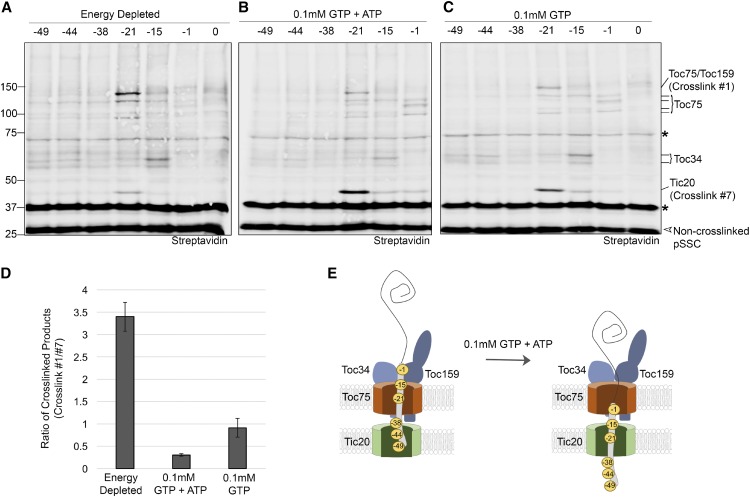
After the identities of the major cross-linked products with the early import intermediate were established, we next examined the topology of the transit peptide during energy-independent binding and the transition to the early import intermediate. To examine energy-independent binding, pea chloroplasts were incubated with nigericin and glycerate to deplete internal ATP and apyrase to deplete external NTPs (Figure 4A). The cross-linking pattern was first compared with the early import intermediate (0.1 mM GTP + ATP; Figure 4B). In energy-depleted chloroplasts, the transit peptide cross-linking pattern was distinct in several respects. First, the levels of cross-links of pSSC(-21) with both Toc75 and Toc159 were increased relative to the early import intermediate, whereas cross-linking of pSSC(-1) to Toc75 was decreased. This is clearly shown when the ratio of the Toc75 or Toc159 cross-link to Tic20 cross-link in the pSSC(-21) sample is compared (Figure 4D). Cross-linking to Toc34 was also more predominant in energy-depleted chloroplasts compared with chloroplasts supplied with GTP and ATP (compare Figures 4A and 4B). These results are consistent with the role for Toc159 and Toc34 in the initial stages of transit peptide recognition. Remarkably, cross-linking of pSSC(-21) to Tic20 was observed even in the absence of NTPs, albeit at reduced levels relative to Toc75 and Toc159 when compared with the early import intermediate formed in the presence of 0.1 mM GTP + ATP (Figure 4D). As summarized in the model in Figure 4E, these data suggest that without added NTP, the mid-region of the transit peptide is engaged with the TOC translocon and can access the TIC complex. Upon addition of 0.1 mM GTP + ATP, the mid-region of the transit peptide is repositioned to make more prevalent contacts with Tic20, and the more C-terminal region of the transit peptide [pSSC(-1)] comes in contact with the Toc75 channel, thereby forming the early import intermediate. The observation that pSSC(-21) cross-links to TOC complex components in the presence of 0.1 mM GTP + ATP suggests that our cross-linking captured a mixed population of import intermediates, with the predominant form corresponding to the early import intermediate and a smaller population that is engaged in initial binding with the translocons.
Figure 4.
Characterization of the Transit Peptide Topology at Defined Energetic Stages of Import.
(A) Pea chloroplasts were depleted of energy by preincubation with apyrase, nigericin, and glycerate in the dark to deplete stromal and external NTP, and cross-linking was assessed as described in Figure 2.
(B) Chloroplasts were preincubated with nigericin and glycerate and then incubated with preprotein in the presence of 0.1 mM GTP and 0.1 mM ATP.
(C) Same as in (B), except chloroplasts were incubated with preprotein in the presence of 0.1 mM GTP only.
(D) The mean ratio between the signal intensity of the pSSC(-21) cross-link to Toc75/Toc159 (cross-link #1) and Tic20 (cross-link #7) was calculated for cross-linking experiments in (A) (energy depleted conditions; n = 5), (B) (0.1 mM GTP+ATP; n = 6), and (C) (0.1 mM GTP; n = 3). Error bars = se.
(E) A model illustrating movement of the transit peptide within the translocon based on the major energy-dependent cross-linking products in (A) to (C). Asterisks indicate abundant chloroplast biotinylated proteins; open arrowheads indicate non-cross-linked preprotein. Energy-depleted, 0.1 mM GTP+ATP, and 0.1 mM GTP cross-linking experiments were performed five, six, and three times, respectively. Numbers to the left of the immunoblots indicate the size of molecular weight markers in kilodaltons.
Roles of GTP and ATP in the Formation of the Early Import Intermediate
In the presence of GTP alone, the overall cross-linking pattern was similar to that of the early import intermediate (Figure 4C). This is most obviously revealed by the increase in cross-linking of pSSC(-21) to Tic20 and pSSC(-1) to Toc75 (compare Figures 4B and 4C). However, quantitation of the ratio of cross-linked Toc75/Toc159 (cross-link #1) to cross-linked Tic20 (cross-link #7) for pSSC(-21) suggested decreased formation of the early import intermediate with GTP alone relative to in the presence of 0.1 mM GTP + ATP (Figure 4D). Quantitative analysis of preprotein binding demonstrates that the level of stable preprotein binding to chloroplasts in the presence of GTP is similar to that observed in the absence of NTPs and lower than that observed in the presence of 0.1 mM ATP (Figure 1B). Together, these observations suggest that GTP alone is sufficient to allow access of the transit peptide to the TIC, but insufficient to support formation of the stably bound early import intermediate. Hydrolysis of GTP appears to be required for the cross-linking pattern observed in the presence of GTP because introducing the slow or nonhydrolyzable analogs of GTP, GTPγS, or GMP-PNP, respectively, resulted in a cross-linking pattern that is largely indistinguishable from that of energy-depleted chloroplasts (Figure 5A; Supplemental Figure 5). It has been suggested previously that the conversion of small amounts of GTP to ATP by adenylate kinases and nucleoside diphosphate kinases at the envelope could mask the specific role for GTP in the early stages of preprotein binding (Soll and Schleiff, 2004). This is consistent with the similarity in cross-linking patterns that we observed with GTP alone and 0.1 mM GTP + ATP (Figures 4B and 4C), a result that is further explored below.
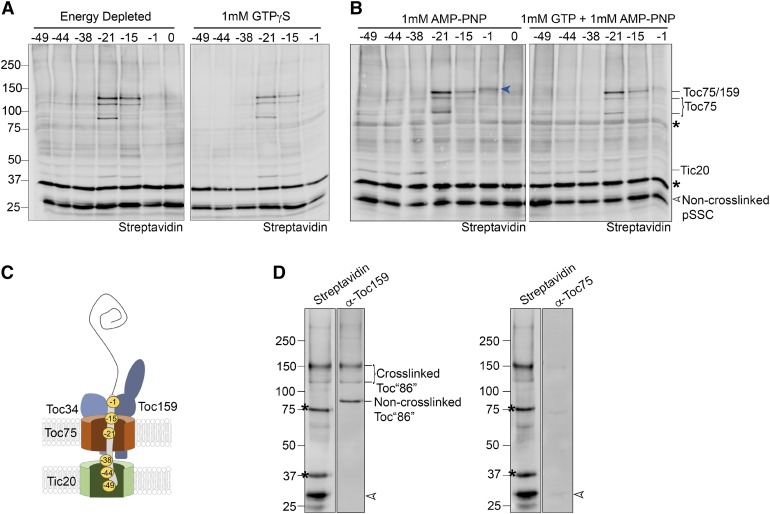
Figure 5.
GTP Does Not Support Transit Peptide Movement in the Presence of AMP-PNP.
(A) Cross-linking was assessed in pea chloroplasts depleted of energy, as described in Figure 4A, and compared with cross-linking in chloroplasts preincubated with 1 mM GTPγS.
(B) Pea chloroplasts were preincubated with nigericin and glycerate in the dark to deplete stromal ATP. Cross-linking in the presence of 1 mM AMP-PNP in the absence or presence of 1 mM GTP was assessed as described in Figure 2. The blue arrow indicates a novel cross-link to pSSC(-1) in the presence of 1mM AMP-PNP.
(C) A model describing the predominant position of the transit peptide within the channel under all conditions tested in (A) and (B). Cross-linking experiments with 1 mM GTPγS, 1 mM AMP-PNP, or 1 mM AMP-PNP + 1 mM GTP were performed twice.
(D) Chloroplast membranes cross-linked with pSSC(-1) in the presence of 1mM AMP-PNP as described in (B) were subjected to streptavidin pull-downs under stringent conditions. Dissolved membranes were divided equally prior to incubation with streptavidin beads, and subsequent pull-downs were probed with either α-Toc159 or α-Toc75. Asterisks indicate abundant chloroplast biotinylated proteins; open arrowheads indicate non-cross-linked preprotein. Numbers to the left of the immunoblots indicate the size of molecular weight markers in kilodaltons.
The observation that GTP alone allows penetration of the transit peptide across the envelope (Figure 4C), as well as the observed inhibitory effect of GTPγS + GMP-PNP on import (Figure 1B), led us to further probe the role of GTP in import. When pea chloroplasts were incubated with 0.1 mM ATP together with GTPγS, pSSC(-21) cross-linking to Tic20 and pSSC(-1) cross-linking to Toc75 were indistinguishable from the early import intermediate (Figure 6). These results show that in the presence of ATP, GTPγS does not prevent the preprotein from accessing the TIC complex. However, GTPγS inhibits protein import at high ATP levels (>1 mM ATP; Figure 1B). Thus, both GTP and ATP hydrolysis are required for the formation of the stable early import intermediate even after the mid-region of the transit peptide has come in contact with Tic20. The observation that ATP alone promotes the formation of the early import intermediate, as well as protein import into the stroma in the absence of added GTP (Figure 1B), suggests that GTP hydrolysis plays a regulatory role rather than an essential energetic role (as shown for ATP) in these processes.
Figure 6.
GTPγS Does Not Inhibit Formation of the Early Import Intermediate.
Cross-linking was performed on chloroplasts depleted of stromal ATP as described in Figure 2, and cross-linking in the presence of 1 mM GTPγS and 0.1 mM ATP was compared with chloroplasts preincubated with 1 mM GTP and 0.1 mM ATP. An asterisk indicates an abundant chloroplast biotinylated protein, and an open arrowhead indicates non-cross-linked preprotein. Cross-linking with 1 mM GTPγS + 0.1 mM ATP was performed twice, with similar results. Numbers to the left of the immunoblots indicate the size of molecular weight markers in kilodaltons.
To further probe the roles of GTP and ATP in the formation of the early import intermediate, we performed pea chloroplast cross-linking experiments with GTP in the presence of 1 mM AMP-PNP, a nonhydrolyzable ATP analog. Incubation with AMP-PNP resulted in a cross-linking pattern with strong similarity to energy-depleted chloroplasts, including minimal cross-linking of pSSC(-21) to Tic20 and increased cross-linking to TOC components relative to the early import intermediate (Figures 5B and 5C). Additionally, a unique pSSC(-1) cross-link was apparent in the presence of AMP-PNP (blue arrow in Figure 5B). Immunoblots of streptavidin pull-downs revealed this to be the 86-kD fragment of Toc159 (Figure 5D). It was also notable that incubation with AMP-PNP resulted in significant cross-linking between the N-terminal region of the transit peptide (positions −49, −44, and −38) and Tic20 (Figure 5B), an effect that was apparent, but not consistently observed in energy-depleted pea chloroplasts (refer to Figures 4A and 5A; Supplemental Figures 6C and 5A). These results show that blocking ATP hydrolysis is more effective at capturing an initial binding stage than simply depleting chloroplasts of internal and external NTPs. In addition, these results clearly demonstrate that the transit peptide has penetrated the TOC complex in the absence of NTP hydrolysis, with the N-terminal region in contact with TIC and the central and C-terminal regions associated with TOC (Figure 5C).
When GTP was added in the presence of AMP-PNP, there was no change in the cross-linking pattern, suggesting that the GTP-dependent effects observed in Figure 4C require a functional ATPase activity. It was also observed that incubation with ATPγS, a slowly hydrolysable analog of ATP, resulted in a cross-linking pattern that was similar to that of the early import intermediate, regardless of whether GTP was present in the reaction (Supplemental Figure 6). ATPγS and AMP-PNP both inhibit preprotein binding and import to the same degree (Figure 1C), but ATPγS can support deeper penetration of the transit peptide into TOC-TIC than occurs when ATP hydrolysis is completely blocked with AMP-PNP.
TOC-TIC Supercomplexes Mediate Preprotein Binding and Import
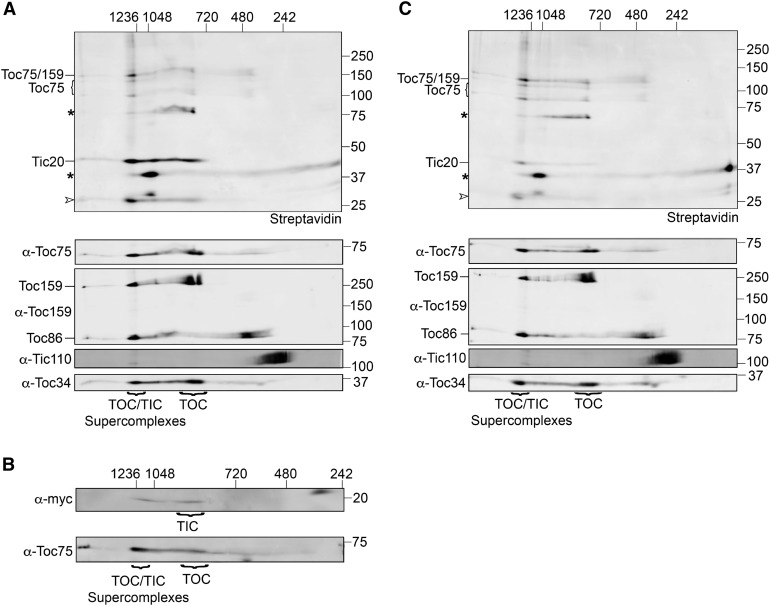
Previous biochemical analyses of isolated import complexes suggest that TOC and TIC complexes exist as both individual complexes and as supercomplexes in which TOC and TIC components are physically associated (Akita et al., 1997; Nielsen et al., 1997; Teng et al., 2006; Köhler et al., 2015; Chen and Li, 2017). Because the transit peptide appears to reposition from a primary contact with TOC components in energy-depleted pea chloroplasts to TIC components at the early import intermediate stage, we next evaluated whether the transit peptide was engaged with either “free” TOC and TIC complexes or TOC/TIC supercomplexes at the stages observed in our cross-linking studies. To this end, pea chloroplasts cross-linked to pSSC(-21) in the absence of energy or in the presence of 0.1 mM GTP + ATP were subjected to 2D Blue Native PAGE (BN-PAGE). As shown in Figure 7A, under early import intermediate conditions, the majority of cross-linked pSSC(-21) was found in high molecular weight complexes of ≥1236 kD, with a small amount of cross-linked Toc75 found in ∼880-kD complexes. Non-cross-linked Toc75 as well as full-length Toc159 and Toc34 were found in both the ∼1236- and ∼880-kD complexes. Small amounts of the inner membrane component Tic110 were found in the supercomplex, with the majority found in a complex between 242 and 480 kD, as previously shown (Kikuchi et al., 2013; Chen and Li, 2017). Based on their respective molecular weights, the ∼1236-kD complex likely corresponds to TOC/TIC supercomplexes, and the 880-kD complex is the previously observed TOC complex (Akita et al., 1997; Kikuchi et al., 2006; Chen and Li, 2007, 2017; Paila et al., 2016). This is supported by detection of myc-Tic20 in two distinct complexes in Arabidopsis plants expressing myc-Tic20: one complex at ∼1200 kD, which comigrates with Toc75, and a second complex that likely represents the 1-MD TIC complex (Figure 7B; Kikuchi et al., 2013; Chen and Li, 2017). The distribution of pSSC(-21) cross-links within these distinct complexes shows no obvious difference in the absence of energy compared with the early import intermediate (other than a lack of Tic20 cross-linking; compare Figures 7A and 7C). These results suggest that the preprotein is associated with TOC/TIC supercomplexes both during binding and at the early import intermediate stage, which demonstrates that supercomplexes represent the functional sites for preprotein recognition and import.
Figure 7.
Cross-Linked Preproteins Are Part of TOC/TIC Supercomplexes.
(A) pSSC(-21) was cross-linked to pea chloroplasts under early import intermediate conditions (0.1 mM GTP + ATP) and chloroplast membranes were isolated, dissolved in 1% digitonin, and subjected to 2D BN-PAGE. Following the second dimension SDS-PAGE, cross-linked proteins were detected using streptavidin conjugated to an infrared dye (Streptavidin, top panel). Non-cross-linked pSSC(-21) and endogenous chloroplast biotinylated proteins are indicated by open arrowheads and asterisks, respectively. Non-cross-linked components of the TOC and TIC complexes are shown using the requisite primary antibodies (bottom panels).
(B) Total chloroplasts isolated from Arabidopsis plants expressing myc-Tic20 were dissolved in 1% digitonin and subjected to 2D BN-PAGE. The membrane was probed with α-myc to detect myc-Tic20 and α-Toc75.
(C) Cross-linking was performed as in (A) using energy depleted chloroplasts. Numbers to the top and left of the immunoblots indicate the size of molecular weight markers in kilodaltons.
DISCUSSION
Over the past several years, the major components of the chloroplast protein import machinery have been identified; however, we still lack a clear understanding of how translocating proteins interface with the translocation machinery and how this is controlled by NTPs. In this study, we addressed these questions by employing a site-specific cross-linking approach using a heterobifunctional, photoactivatable cross-linker that is uniquely positioned at several sites over the length of the transit peptide of the Rubisco small subunit. This allowed us to assess the position of the transit peptide within the TOC/TIC channels for greater resolution of import intermediates than could be afforded using general cross-linkers. Our results identified major contacts of the transit peptide at all stages and demonstrated repositioning of the transit peptide relative to TOC-TIC components as the preprotein progressed from initial binding to the NTP-dependent early import intermediate. A model integrating the cross-linking results presented here is shown in Figure 8.
Figure 8.
Model of GTP-Dependent Regulation of the Early Import Intermediate.
(A) At the energy-independent stage of import, the TOC GTPases are GDP-loaded.
(B) Interactions of the TOC GTPases with the transit peptide trigger changes in receptor dimerization and promote GDP-to-GTP exchange. This leads to transit peptide insertion into the TOC channel, with its N-terminal region (magenta) in contact the TIC translocon and the mid-region of the transit peptide (teal) inserted into Toc75. The POTRA domains of Toc75 could provide a binding site for the transit peptide in the intermembrane space at this stage.
(C) Low levels of ATP (∼0.1 mM ATP) stimulate repositioning of the transit peptide within the TOC and TIC channels, with the N terminus exposed to the stroma and the mid-region of the transit peptide inserted within the TIC channel. At this stage, the C-terminal region of the transit peptide (yellow) remains in contact with Toc75. Formation of this intermediate is inhibited by preincubation with AMP-PNP and is not promoted by GTP hydrolysis.
(D) GTP hydrolysis at the TOC receptors results in stabilization of the early import intermediate. GTP hydrolysis could act as a checkpoint prior to import, consistent with the observation that GTPγS inhibits the formation of a stable, ATP-dependent early import intermediate.
(E) Import of the preprotein into the stroma requires high levels of ATP (>1 mM) and is facilitated by the ATPase activity of stromal chaperone motors (Hsp).
(F) A schematic diagram indicating the regions referred to as the N-terminal region (magenta), mid-region (teal), and C-terminal region (yellow) of the transit peptide in (A) to (C).
Our studies reveal a previously unknown topological arrangement of the transit peptide in the TOC-TIC machinery at the earliest stage of preprotein binding to the import apparatus. We show that the N-terminal region of the transit peptide (shown in magenta in Figure 8) has penetrated the TOC complex and is in contact with components of the TIC machinery at the inner membrane at the earliest stages of binding (Figure 8B). Energy-independent binding to chloroplasts has been shown to be reversible, and to involve the TOC receptor GTPases and Toc75 (Perry and Keegstra, 1994; Ma et al., 1996). Data presented in this study are consistent with these observations, as we show that the transit peptide cross-links with Toc75 and both of the TOC GTPase receptors in energy-depleted chloroplasts (and chloroplasts treated with AMP-PNP; Figures 4A and 5B). However, our results also show that the mid-region of the transit peptide (shown in teal in Figure 8) is engaged by the Toc75 channel, and the N-terminal region is in contact with Tic20 at the inner membrane (Figures 4A, 5A, and 8B; Supplemental Figures 5C and 6A). This is most apparent in the presence of AMP-PNP (Figure 5B). These results stand in contrast to current models in which initial transit peptide binding is proposed to occur to the cytoplasmic domains of the TOC receptors and that GTP promotes insertion into the Toc75 channel (Jarvis, 2008; Li and Chiu, 2010; Richardson et al., 2014; Sjuts et al., 2017). Therefore, initial recognition of the transit peptide involves a significant interaction with the TOC channel, an interaction that does not require GTP hydrolysis.
The observation that the transit peptide inserts across the TOC channel with its N-terminal region in proximity to Tic20 in the absence of NTP hydrolysis is striking. Although the energetic driving force for insertion of the transit peptide across the TOC channel is unknown, it is possible that an interaction with the receptor GTPases upon arrival at the chloroplast surface induces a conformational change in the TOC complex that promotes transit peptide insertion across the outer membrane. The transit peptide is known to induce changes in the dimerization state of the TOC GTPases and promote GDP for GTP exchange. The changes in dimerization could be sufficient to promote transit peptide insertion even in the absence of nucleotide exchange. It has also been shown that the intermembrane space-localized POTRA domains of Toc75 possess a transit peptide binding site (Paila et al., 2016; O’Neil et al., 2017), which could serve as a docking site for the transit peptide in the intermembrane space and reduce backsliding out of the channel in the absence of energy. In mitochondrial protein import, it has been shown that presequence translocation across the translocon at the mitochondrial outer membrane (TOM) does not require ATP hydrolysis or the membrane potential (Neupert, 1997; Gabriel et al., 2003). The molecular basis for such a mechanism is unknown, but our results suggest that the −21 region of the pre-Rubisco small subunit transit peptide may be important for interaction of the transit peptide with the TOC channel in the absence of energy.
Our results also suggest a modified role for GTP at the TOC receptors compared with that proposed in current models. When incubated with GTP alone, the transit peptide showed a cross-linking pattern resembling the early import intermediate (0.1 mM GTP + ATP) (Figures 4C and 4D). This movement is blocked in the presence of the slowly and nonhydrolyzable GTP analogs, GTPγS and GMP-PNP, respectively (Figures 5A and 8D’ Supplemental Figure 6). This is consistent with previous work by several groups showing that GTP stimulates low levels of preprotein binding to chloroplasts (Olsen and Keegstra, 1992; Kessler et al., 1994; Young et al., 1999). However, we show that AMP-PNP blocks the ability of GTP alone to support low levels of binding and promote insertion of the transit peptide further into the import machinery (Figure 5B). Furthermore, ATP alone is sufficient to support formation of the early import intermediate (Figure 1B). This indicates an essential role for ATP hydrolysis in the transition from energy-independent binding to the early import intermediate (Figure 8C). We conclude that the observed effect of GTP alone may be a result of GTP to ATP conversion by chloroplast adenylate kinases and nucleoside diphosphate kinases (Soll and Schleiff, 2004).
The inhibition of import by GTPγS clearly demonstrates a role for GTP in regulating ATP-dependent formation of the stable early import intermediate (Figures 1B and 8) (Young et al., 1999). The cross-linking pattern observed with GTPγS in the presence of 0.1 mM ATP is very similar to that observed with the early import intermediate (0.1 mM GTP + ATP) (Figure 6), indicating that the transit peptide can penetrate the TOC complex and engage the TIC in the presence of the GTP analog. However, quantitative chloroplast binding studies suggested that this state is less stable than in the presence of ATP alone or GTP + ATP, as indicated by the reduced levels of preprotein binding to chloroplasts observed under these conditions (Figure 1B) (Young et al., 1999). Therefore, we conclude that the cross-linking in the presence of GTPγS + ATP represents a transition state (Figure 8C) between energy-independent binding and the early import intermediate (Figure 8D). In line with these observations, it was previously reported that if GTPγS is added after the formation of the early import intermediate together with >1 mM ATP, import proceeds normally (Wang et al., 2008). This suggests that GTPγS blocks protein import if it is present before the addition of high ATP concentrations (>1 mM) to initiate import (Figure 1B) (Olsen and Keegstra, 1992; Young et al., 1999), but does not block import of the stable early import intermediate into the stroma if it is formed prior to the addition of the analog (Wang et al., 2008). On the basis of these observations, we propose that the role of GTP hydrolysis at the TOC receptors is to regulate the formation of a stable early import intermediate (Figure 8D).
The TOC receptor GTPases directly interact with transit peptides in a transient and GTP-regulated manner (Kouranov and Schnell, 1997; Schleiff et al., 2002; Smith et al., 2004). The transient nature of the interactions is proposed to be important for sequential molecular interactions during import. Transit peptides are important for the GTPase cycle of Toc34 by disrupting receptor dimerization, which promotes GDP to GTP exchange (Oreb et al., 2011; Lumme et al., 2014). The individual role(s) of the TOC GTPase receptors in preprotein recognition and import has been studied through the analysis of mutations that disrupt GTP binding and/or hydrolysis of the major Arabidopsis putative orthologs of pea Toc34 and Toc159, atToc33 and atToc159, respectively. Similar to preincubation with GTPγS, these mutations do not affect preprotein binding to chloroplasts, but they have inhibitory effects on import (Wang et al., 2008; Agne et al., 2009; Lee et al., 2009; Aronsson et al., 2010). In light of our results, these previous findings would support the hypothesis that GTP regulates the transition from reversible binding to formation of a stable, import-competent early import intermediate. As shown in Figure 8, we propose that interactions with the GTPase receptors at the level of TOC promote GDP to GTP exchange and prime the translocon for import (Figures 8A and 8B). Once GTP-loaded, GTP hydrolysis at the TOC GTPase receptors allows stable ATP-dependent interaction of the transit peptide with the translocons in order for subsequent import into the stroma to occur (Figures 8C to 8E). As such, the GTPase activities of the TOC receptors function as a checkpoint in translocation in vivo, rather than contributing to preprotein insertion or as part of the import motor. We envision that such a checkpoint would be important for ensuring that the preprotein is properly positioned within the channel to facilitate unidirectional import, mediated by stromal chaperones. This model is consistent with the observation that import proceeds normally in the absence of added GTP in vitro (Young et al., 1999).
The role for low levels of ATP (∼0.1 mM) in promoting formation of the stably bound early import intermediate is well established and consistent with our cross-linking results (Figures 2 and 4B) (Olsen et al., 1989; Kouranov and Schnell, 1997; Kikuchi et al., 2009). However, we showed that the early import intermediate is inserted into TIC with the mid-region of the transit peptide cross-linking to Tic20 and the C terminus of the transit peptide cross-linking to Toc75, the outer membrane protein translocation channel (Figure 2). These results indicate that the mid-region of the transit peptide repositions from a primary contact with Toc75 to a primary contact with Tic20 at the TIC channel in the presence of low levels of ATP (Figure 8C). At this stage, we hypothesize that the N terminus of the transit peptide is likely protruding into the stroma and engaged with stromal chaperones, consistent with previous studies (Ivey et al., 2000; Shi and Theg, 2010; Su and Li, 2010; Chotewutmontri and Bruce, 2015). Although we did not observe detectable cross-linking to the Hsp70, Hsp93/ClpC, or Hsp90 stromal chaperones at this stage using this method, considerable evidence supports the chaperones’ roles in utilizing ATP to bind the preprotein and stabilize the early import intermediate (Akita et al., 1997; Nielsen et al., 1997; Shi and Theg, 2010; Su and Li, 2010; Inoue et al., 2013; Flores-Pérez et al., 2016; Richardson et al., 2017). We did observe lower molecular weight cross-links of unknown identity (Supplemental Figure 2) that, once identified, may provide additional insight into early stromal events in import.
The cross-links observed between the transit peptide and Tic20 at all stages of import (Figure 8) strongly supports a central role for this component in TIC function. Tic20 has been shown to associate with both the Tic110-Tic40 complexes, which are involved in assembling import associated chaperone complexes in the stroma, and with the 1-MD complexes, which contain Tic56, Tic100, and Tic214. Although the roles of these two complexes have been a subject of considerable debate (de Vries et al., 2015; Köhler et al., 2015, 2016; Nakai, 2015; Bölter and Soll, 2017), our results provide strong support for Tic20 as a central component of the translocation channel at the inner membrane. Tic20 has been shown to possess membrane channel activity (Kovács-Bogdán et al., 2011), consistent with this hypothesis. Our data do not provide insights into the relative contributions of the other TIC components to protein import because we did not observe detectable cross-links other than those to Tic20 and low molecular weight adducts of unknown identity (Supplemental Figure 2).
Our data also demonstrate close coordination between TOC and TIC at all stages of protein import. In addition to contact of the transit peptide with Tic20 at the earliest stages of preprotein binding, we show (using 2D BN-PAGE) that the cross-linked preprotein is mainly associated with large (>1.2 MD) TOC/TIC supercomplexes, both in the absence of energy and at the early import intermediate stage (Figure 7). Significant proportions of Toc34, Toc75, Toc159, and Tic20 are associated with these supercomplexes (Figure 7). This observation disfavors a model in which initial recognition of preproteins at the TOC complex triggers an association with TIC. Instead, these results indicate that TOC/TIC supercomplexes represent active import sites and are consistent with a recent study showing an association of importing preprotein with high molecular weight complexes containing both TOC and TIC components (Chen and Li, 2017).
Together, the site-specific cross-linking results presented in this study provide important mechanistic insight into how the transit peptide interfaces with the translocation channel during import. One unexplored implication of our findings is the identification of conserved motifs within the transit peptide that interact with specific translocation components at distinct stages in import. Generally, there is little sequence conservation among transit peptides, although they are enriched in uncharged residues toward the N terminus, where they are proposed to interact with Hsp70 (Chotewutmontri et al., 2012; Li and Teng, 2013), and many contain a degenerate FGLK motif proposed to be important for an interaction with the TOC receptors (Lee et al., 2006; Chotewutmontri et al., 2012; Holbrook et al., 2016). Additionally, when transit peptide subgroups are analyzed separately, some common motifs can be identified that appear to be important for import based on import efficiency analysis of members of each of these subgroups (Lee et al., 2008). Transit peptides can also define tissue or age-dependent import, which raises the question of which core motifs are needed for translocation through TOC and TIC, and which motifs are responsible for specific recognition of different substrates in different tissues (Li and Teng, 2013). The site-specific cross-linking approach used in this study is a powerful tool to begin to investigate regions of transit peptides that function as core motifs for translocation versus substrate-specific recognition. Furthermore, with clarification of the intermediate stages of import, the preprotein binding step can be the subject of more focused investigation and separated from the early import intermediate step wherein the transit peptide is already inserted into the TIC channel. As an example, our results show that the FGLK motif and a LKSSA motifs, which are known to be critical for import (Lee et al., 2006; Chotewutmontri et al., 2012), both lie adjacent to a region that shows strong cross-linking to TOC in the absence of energy and cross-linking to Tic20 at the early binding stage [pSSC(-21)] (Figures 2, 4A, and 4B). These data are consistent with the proposal that these motifs interact with both the outer and inner translocation complexes and are thus core regions necessary for interactions with the TOC and TIC channels (Lee et al., 2006; Chotewutmontri et al., 2012). Expanding this approach to examine representatives from diverse groups of transit peptides may lend insight into the substrate specificity observed in different tissues and developmental stages (Teng et al., 2012; Li and Teng, 2013).
In conclusion, this work helps resolve models of translocation at the molecular level by providing fundamental insight into the NTP-dependent mechanism of protein translocation, particularly the role of GTP and ATP hydrolysis, and the molecular interactions of the transit peptide with the translocation machinery during import. These results have implications for future studies aimed at identifying critical motifs within transit peptides that are broadly necessary for import and identifying the molecular mechanism of substrate specificity of plastid protein import.
METHODS
Cloning and Generation of Transgenic Arabidopsis Plants
The cysteine-less pSSU skeleton clone [pET21a:pSSC(0)3xFLAG-His] used to generate single Cys pSSU constructs was generated in several cloning steps. First, pET8C-pS/ProtA-1 (Kouranov and Schnell, 1997), which encodes pea (Pisum sativum) SSU with a Cys-to-Ser mutation at the −1 position, was subcloned into pET21b to generate pET21b:pS-1-His. The 5′ portion of pS-1 was removed and replaced with the sequence encoding residues 1 to 95 of Arabidopsis thaliana pSSU via compatible restriction enzyme sites to generate pET21b:pSSC(0). This plasmid consists of the first 95 residues of Arabidopsis pSSU, and residues 99 to 169 of pea SSU (subunit 3C), as this portion of pea SSU does not contain any Cys residues. The resulting pSSC(0) insert was subcloned into pET21a:pre-Tic40-3xFLAG (Inoue et al., 2013) following removal of the pre-Tic40 insert by digestion. The resulting plasmid, pET21a:pSSC(0)3xFLAG-His, was used as a template for PCR-based site-directed mutagenesis to mutate single residues to Cys at indicated positions within the transit peptide, as described by Inoue and Akita (2008). Single Cys mutants were further modified to incorporate an Avitag biotinylation signal (GLNDIFEAQKIEWHE) by subcloning via PCR into each single Cys mutant using an XhoI site between the last residue of the 3xFLAG epitope and first residue of the His tag (Inoue et al., 2013). Primers used are listed in Supplemental Table 1.
For overexpression of myc-Tic20 in Arabidopsis, pEG100-TPmycTic20 was generated, which corresponds to full-length TIC20-I with a myc-tag positioned after the first two residues of the mature region of the protein (i.e., between residues 102 and 103 of Tic20-I), driven by the 35S CaMV promoter. To generate pEG100-TPmycTic20, the TIC20-I CDS was first subcloned into pENTRD-TOPO to generate pENTRD-Tic20. The sequence encoding the myc-tag was amplified by PCR with overhangs that overlap flanking sequences in TIC20-I. This amplified fragment was then used for overlap extension PCR using pENTRD-Tic20 as the template, digested with DpnI, and transformed into Escherichia coli. Positive colonies were sequenced by the MSU Genomics Facility to verify proper insertion of the myc-tag. pENTRD-TPmycTic20 was then used in a LR recombination reaction with pEG100 (Earley et al., 2006) to generate pEG100-TPmycTic20. Primers used for cloning are listed in Supplemental Table 1. Agrobacterium tumefaciens strain GV3101 was transformed with pEG100-TPmycTic20 using the floral-dip method (Clough and Bent, 1998) and used to transform the heterozygous Arabidopsis tic20-I mutant (SALK_39676). Transformants were selected with BASTA and genotyped for the tic20-I T-DNA insertion by PCR. Expression of myc-Tic20 was confirmed in T1 plants via immunoblotting. T2 seeds were pooled and plants selected on MS medium supplemented with 1% sucrose, 100 μg/mL timentin (PhytoTechnology Laboratories), and 20 μg/mL phosphinothricin (PhytoTechnology Laboratories). Plants were grown at 22°C under long-day conditions (16 h of light/8 h of dark), 100 μmol m−2 s−1 under fluorescent grow lights. Thirteen-day-old T2 plants were used for chloroplast isolation and cross-linking experiments. The mycTic20 protein was confirmed to be functional by complementation of the tic20-I mutant in the T3 generation.
Expression, Purification, and MBP Modification of Single-Cys Mutants
Plasmids encoding each single Cys mutant were transformed into BL21 (DE3) already containing pBirA (a plasmid carrying the biotin ligase gene; Avidity) and selected on LB with 100 µg/mL ampicillin and 10 µg/mL chloramphenicol. Cultures for protein expression were grown in LB with 100 µg/mL ampicillin, at 37°C to an OD600 of 0.6 to 0.8, and protein expression and biotinylation induced by addition of 1 mM IPTG and 50 μM biotin from a 1 M d-biotin (Sigma-Aldrich) stock dissolved in 10 mM Bicine pH 8.3. Cultures were grown at 37°C for 3 to 4 h and chilled for 10 min on ice, and cells were collected by centrifugation at 8000g for 15 min at 4°C. All proteins were expressed in inclusion bodies. Cells were lysed in 25 mM Tris-HCl, pH 8, 10 mM EDTA, and 50 mM glucose (TGE) with 200 µg/mL lysozyme followed by sonication. Inclusion bodies were collected by centrifugation (11,000 rpm for 20 min, 4°C) and washed three times with TGE + 1% Triton X-100, and once with TGE, then dissolved in binding buffer (BB; 20 mM Tris-HCl pH 8, 0.5 M NaCl, 10 mM imidazole) with 6 M urea and bound to His-bind resin (Novagen). The column was washed with 10 column volumes of BB with 6 M urea, 6 column volumes of BB with 20 mM imidazole and 6 M urea, and eluted in BB with 0.5 M imidazole and 6 M urea. Protein was precipitated with 10% trichloroacetic acid (TCA), washed with ice-cold 0.5% TCA, followed by ice-cold 100% acetone. The protein pellet was dissolved in S-buffer (25 mM HEPES-KOH, pH 7.0, 50 mM KCl, 2 mM MgCl2, and 6 M urea) to 100 μM and stored at −80°C. Prior to import reactions, protein aliquots were thawed on ice, and MBP (Sigma-Aldrich) dissolved in DMSO was added to a final concentration of 2.5 mM and incubated in the dark for ∼4 h. The reaction was quenched with the addition of 5 mM DTT. Modified preprotein was TCA precipitated overnight and washed with 0.5% TCA, followed by 100% ice-cold acetone. The protein pellet was resuspended in S-buffer, quantitated via Bradford assay, and diluted to a final concentration of 10 μM in S-buffer prior to cross-linking.
Chloroplast Isolation
Chloroplasts were isolated from pea plants essentially as described by Smith et al. (2003). Approximately 25 g of tissue from 7-d-old pea (cv Green Arrow; Jung Seed Co.) plants grown under a 24°C 16-h-light/ 22°C 8-h-dark cycle at 100 μmol m−2 s−1 under fluorescent grow lights were used. Aerial tissue was harvested and homogenized in ∼300 mL of grinding buffer (GB; 25 mM HEPES-KOH, pH 7.5, 330 mM sorbitol, 2 mM EDTA, 1 mM MgCl2, 1 mM MnCl2, 0.25% [w/v] BSA, and 5 mM sodium ascorbate) using a rotary homogenizer at 40 rpm. Homogenate was centrifuged at 1500g for 4 min at 4°C. The chloroplast pellet was resuspended in ∼12 mL of GB and overlaid onto three PBF-Percoll 40%/85% step gradients. Gradients were centrifuged for 10 min at 1500g, 4°C, with no brake, and the upper layer of broken chloroplasts (40%:GB interface) was removed and discarded. The lower layer of intact chloroplasts at the 40%:85% interface was collected and diluted into two tubes with ∼30 mL of ice-cold HS buffer (25 mM HEPES-KOH, pH 7.5, and 330 mM sorbitol) each. Intact chloroplasts were collected by centrifugation at 4000g for 2 min and resuspended in ∼200 μL of cold HS buffer. Chlorophyll content was measured as described previously (Smith et al., 2003). For isolation of Arabidopsis chloroplasts, the protocol has been previously described in detail elsewhere (Inoue et al., 2011).
In Vitro Chloroplast Binding/Import Assay and UV Cross-Linking
Isolated chloroplasts from both pea and Arabidopsis were incubated with 0.4 μM Nigericin (Calbiochem), 10 mM d-glycerate (Sigma-Aldrich), and with or without apyrase (2 units per 100 μL of chloroplasts) (Sigma-Aldrich) in the dark for ∼30 min (Inoue et al., 2011). Chloroplasts at 1 mg chlorophyll/mL were incubated in import buffer (50 mM HEPES-KOH, pH 7.5, 330 mM sorbitol, 25 mM KOAc, and 5 mM MgOAC) with the indicated concentrations of NTP for 5 min at 26°C in the dark before the addition of 200 nM preprotein. For experiments with GTPγS, chloroplasts were preincubated with all components except ATP and preprotein for 10 min, followed by incubation with ATP for 5 min before the addition of 200 nM preprotein. Reactions were incubated for a further 5 min at 26°C for chloroplast binding or for 20 min at 26°C for import analysis. Reactions were stopped by the addition of 500 μL ice-cold HS buffer. Chloroplasts were collected by centrifugation at 1800g for 3 min, 4°C, and resuspended in 100 μL HS buffer. Chloroplast suspension was layered on top of ∼800 μL 40% Percoll and centrifuged at 6000g for 5 min. Intact chloroplasts were washed once with 500 μL HS buffer, resuspended in 25 μL 2× SDS-PAGE sample buffer (350 mM Tris base, 5% SDS, 160 mM DTT, 7.5% glycerol, and 0.1% bromophenol blue), and subjected to SDS-PAGE and immunoblotting with monoclonal anti-FLAG antibodies (1:5000) (Sigma-Aldrich; catalog no. F1804, lot 124K6106). Binding and import efficiency were quantitated by band densitometry using ImageStudio software (Li-Cor). The experiment was repeated three times with independent chloroplast isolations, each with two technical replicates (quantitation of two immunoblots from the same sample), and binding/import efficiencies averaged between biological replicates (independent chloroplast isolations). As a loading control, membranes were immunoblotted with Toc75 antibodies, and binding/import efficiencies normalized based on relative Toc75 signal.
For UV cross-linking, following incubation at 26°C for 5 min, chloroplasts were washed with ∼4× volume of HS buffer and resuspended in the original reaction volume of HS (100–500 μL). Reactions were transferred to culture plates (12- or 24-well) prechilled on ice and exposed to UV light at 302 nm for 15 min at a distance of ∼2 cm. Chloroplasts were then transferred to microcentrifuge tubes and stored at −80°C. To analyze the cross-linking profile, membranes equivalent to 20 µg of chlorophyll were isolated by dilution in ice-cold 2 mM EDTA with 200 mM NaCl and centrifugation at max speed for 30 min. Membrane pellets were resuspended in 2× SDS-PAGE sample buffer and resolved on a 5 to 12% gradient SDS-PAGE gel. Experiments were performed at least in duplicate (the number of replicates is indicated in the figure legends).
Streptavidin Pull-Downs and Immunoblotting
Cross-linked chloroplast samples from pea or Arabidopsis (0.5–1 mg chlorophyll) in HS buffer were thawed on ice and diluted ∼10× with ice-cold resuspension buffer (RB) (50 mM Tricine-KOH, pH 7.5, 150 mM NaCl, and 2 mM EDTA) with 1% plant protease inhibitor cocktail (Sigma-Aldrich). Total membranes were collected by ultracentrifugation at 35,000 rpm in a TH-641 swinging bucket rotor (Sorvall). The membrane pellet was resuspended in RB (final [chlorophyll] of ∼0.5 mg/mL), diluted 1:1 with RB with 2% Triton X-100, 2% SDS, and EDTA-free protease inhibitor (Roche), and incubated at room temperature for ∼1 h while gently rocking until the membrane pellet dissolved. Samples were centrifuged at 21,100g for 30 min, 4°C, to collect any undissolved material, and the supernatant was incubated with prewashed streptavidin magnetic beads (Dynabeads Streptavidin MyOne C1; Invitrogen) overnight at 4°C while gently rocking. Beads were collected and washed in RB with 1% Triton X-100 and 1% SDS, five times for 5 min while gently rocking. Bound proteins were eluted in 35 μL 4× LDS-SB (Invitrogen) supplemented with 160 mM DTT and ∼10 μM d-biotin pH 8.0 (Sigma-Aldrich), and incubated at 70°C for 15 min. Eluted protein was transferred to a clean tube, resolved on a 5 to 12% gradient SDS-PAGE gel and blotted with the desired primary antibody, followed by goat anti-rabbit 800CW secondary antibody and Streptavidin 680-LT (Li-Cor Biosciences). Immunoblots were imaged on a Li-Cor Odyssey CLx imaging system. Antisera raised against pea Toc86 and Toc34 were used for immunodetection of Toc159 (1:2500) and Toc34 (1:2500), respectively, and have been previously described (Kouranov et al., 1998). Pea Toc75 antibodies (1:2500) were affinity purified from antisera raised against pea Toc75 that has been previously described (Kouranov et al., 1998). Monoclonal Myc antibody (9B11; 1:2500) was purchased from Cell Signaling Technology (catalog no. 2276S, lot 19). Li-Cor secondary antibodies were used at 1:15,000 (P/N 925-32210, 925-32211). Biotinylated proteins were detected with Li-Cor streptavidin conjugated to an infrared dye fluorescing in either the 700- or 800-nm channels (P/N 925-68031 and 925-32230, respectively), at a 1:5000 dilution.
Two-Dimensional BN-PAGE
2D BN-PAGE was performed essentially as previously described (Kikuchi et al., 2006; Hsu et al., 2012; Paila et al., 2016). Chloroplasts isolated from pea were resuspended in a hypotonic lysis buffer (10 mM HEPES-KOH, pH 7.5, 1 mM MgCl2, and 10 μL/mL plant protease inhibitor cocktail [Sigma-Aldrich]) for 10 min on ice and centrifuged at 18,000g, 4°C, for 30 min. The membrane pellet was solubilized in BN-PAGE buffer (50 mM Bis-Tris-HCl, pH 7, and 0.5 M 6-aminocaproic acid) with 10% glycerol and 10 μL/mL plant protease inhibitor cocktail containing 1% (w/v) digitonin (Sigma-Aldrich) at 0.5 mg chlorophyll/mL for 20 min on ice. Insoluble material was removed by centrifugation at 18,000g for 20 min at 4°C. One hundred microliters of supernatant was mixed with 3.25 μL of 5% SERVA Blue G Dye solution (50 mM Bis-Tris-HCl, pH 7.0, 5% [w/v] Serva Blue G Dye, and 0.5 M 6-aminocaproic acid). The sample was loaded onto a 3.5 to 10% gradient native SDS-PAGE gel (50 mM Bis-Tris-HCl, pH 7, and 0.5 M 6-aminocaproic acid, with a 0–16% glycerol gradient). Electrophoresis was performed in blue cathode buffer (50 mM Tricine, 15 mM Bis-Tris-KOH, pH 7, and 0.02% [w/v] Serva Blue G) and anode buffer (50 mM Bis-Tris, pH 7) at 30 V for ∼16 h at 4°C. Following BN-PAGE, sample lanes were excised and incubated at 37°C in 3.3% SDS, 4% 2-mercaptoethanol, 65 mM Tris-HCl, pH 6.8, for 1 h. The gel strip was layered on top of a 5 to 12% gradient SDS-PAGE gel and run according to standard procedures.
Statistical Analysis
To facilitate quantitative comparison of gel images, means and se of the mean were calculated using Microsoft Excel.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers:Arabidopsis TIC20-I (AT1G04940), Arabidopsis RUBISCO SMALL SUBUNIT 1B (AT5G38430), Arabidopsis Toc75-I protein sequence (AT1G35860), pea Toc159 protein sequence (AAF75761), pea Toc34 protein sequence (Q41009), pea Toc75 protein sequence (Q43715), pea RUBISCO SMALL SUBUNIT 3A (KY283547), and pea Tic110 protein sequence (O24303).
Supplemental Data
Supplemental Figure 1. Import of single Cys pSSC mutants.
Supplemental Figure 2. Control immunoblots of streptavidin pull-downs.
Supplemental Figure 3. Complementation of tic20-I Arabidopsis mutants with expression of myc-Tic20.
Supplemental Figure 4. Cross-linking pattern under import (1 mM ATP) conditions.
Supplemental Figure 5. Cross-linking in the presence of GMP-PNP.
Supplemental Figure 6. Cross-linking profile with ATPγS.
Supplemental Table 1. Primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by National Institutes of Health Grant 2RO1-GM061893 to D.J.S. L.G.L.R. is a recipient of a Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada. We thank Mitchell Kimber and Matthew Bennett for technical assistance.
AUTHOR CONTRIBUTIONS
L.G.L.R., D.J.S., and H.I. contributed to conceptualization and design of the research. L.G.L.R. performed the research, and L.G.L.R. and D.J.S. wrote and revised the manuscript. E.L.S. contributed to data acquisition and revision of the manuscript.
References
- Agne B., Infanger S., Wang F., Hofstetter V., Rahim G., Martin M., Lee D.W., Hwang I., Schnell D., Kessler F. (2009). A toc159 import receptor mutant, defective in hydrolysis of GTP, supports preprotein import into chloroplasts. J. Biol. Chem. 284: 8670–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita M., Nielsen E., Keegstra K. (1997). Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136: 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H., Combe J., Patel R., Agne B., Martin M., Kessler F., Jarvis P. (2010). Nucleotide binding and dimerization at the chloroplast pre-protein import receptor, atToc33, are not essential in vivo but do increase import efficiency. Plant J. 63: 297–311. [DOI] [PubMed] [Google Scholar]
- Bauer J., Chen K., Hiltbunner A., Wehrli E., Eugster M., Schnell D., Kessler F. (2000). The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403: 203–207. [DOI] [PubMed] [Google Scholar]
- Bölter B., Soll J. (2017). Ycf1/Tic214 is not essential for the accumulation of plastid proteins. Mol. Plant 10: 219–221. [DOI] [PubMed] [Google Scholar]
- Bölter B., May T., Soll J. (1998). A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide. FEBS Lett. 441: 59–62. [DOI] [PubMed] [Google Scholar]
- Chen K.Y., Li H.M. (2007). Precursor binding to an 880-kDa Toc complex as an early step during active import of protein into chloroplasts. Plant J. 49: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.J., Li H.M. (2017). Stable megadalton TOC-TIC supercomplexes as major mediators of protein import into chloroplasts. Plant J. 92: 178–188. [DOI] [PubMed] [Google Scholar]
- Chen Y.L., Chen L.J., Li H.M. (2016). Polypeptide transport-associated domains of the Toc75 channel protein are located in the intermembrane space of chloroplasts. Plant Physiol. 172: 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P., Bruce B.D. (2015). Non-native, N-terminal Hsp70 molecular motor recognition elements in transit peptides support plastid protein translocation. J. Biol. Chem. 290: 7602–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P., Reddick L.E., McWilliams D.R., Campbell I.M., Bruce B.D. (2012). Differential transit peptide recognition during preprotein binding and translocation into flowering plant plastids. Plant Cell 24: 3040–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.L., Chu C.C., Chen L.J., Akita M., Li H.M. (2006). Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 175: 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Corgiat B.A., Nordman J.C., Kabbani N. (2014). Chemical crosslinkers enhance detection of receptor interactomes. Front. Pharmacol. 4: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day P.M., Potter D., Inoue K. (2014). Evolution and targeting of Omp85 homologs in the chloroplast outer envelope membrane. Front. Plant Sci. 5: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J., Sousa F.L., Bölter B., Soll J., Gould S.B. (2015). YCF1: A Green TIC? Plant Cell 27: 1827–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú., Bédard J., Tanabe N., Lymperopoulos P., Clarke A.K., Jarvis P. (2016). Functional analysis of the Hsp93/ClpC chaperone at the chloroplast envelope. Plant Physiol. 170: 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K., Egan B., Lithgow T. (2003). Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J. 22: 2380–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah S.C., Wagner R., Sveshnikova N., Harrer R., Soll J. (2002). The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys. J. 83: 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook K., Subramanian C., Chotewutmontri P., Reddick L.E., Wright S., Zhang H., Moncrief L., Bruce B.D. (2016). Functional analysis of semi-conserved transit peptide motifs and mechanistic implications in precursor targeting and recognition. Mol. Plant 9: 1286–1301. [DOI] [PubMed] [Google Scholar]
- Hsu S.C., Nafati M., Inoue K. (2012). OEP80, an essential protein paralogous to the chloroplast protein translocation channel Toc75, exists as a 70-kD protein in the Arabidopsis thaliana chloroplast outer envelope. Plant Mol. Biol. 78: 147–158. [DOI] [PubMed] [Google Scholar]
- Huang P.K., Chan P.T., Su P.H., Chen L.J., Li H.M. (2016). Chloroplast Hsp93 directly binds to transit peptides at an early stage of the preprotein import process. Plant Physiol. 170: 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T., Li M., Alvarez-Huerta M., Kessler F., Schnell D.J. (2003). atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278: 38617–38627. [DOI] [PubMed] [Google Scholar]
- Inoue H., Akita M. (2008). The transition of early translocation intermediates in chloroplasts is accompanied by the movement of the targeting signal on the precursor protein. Arch. Biochem. Biophys. 477: 232–238. [DOI] [PubMed] [Google Scholar]
- Inoue H., Rounds C., Schnell D.J. (2010). The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. Plant Cell 22: 1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Wang F., Inaba T., Schnell D.J. (2011). Energetic manipulation of chloroplast protein import and the use of chemical cross-linkers to map protein-protein interactions. Methods Mol. Biol. 774: 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Li M., Schnell D.J. (2013). An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. USA 110: 3173–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova Y., Smith M.D., Chen K., Schnell D.J. (2004). Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 15: 3379–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey R.A. III, Subramanian C., Bruce B.D. (2000). Identification of a Hsp70 recognition domain within the rubisco small subunit transit peptide. Plant Physiol. 122: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008). Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179: 257–285. [DOI] [PubMed] [Google Scholar]
- Jarvis P., López-Juez E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14: 787–802. [DOI] [PubMed] [Google Scholar]
- Kasmati A.R., Töpel M., Patel R., Murtaza G., Jarvis P. (2011). Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J. 66: 877–889. [DOI] [PubMed] [Google Scholar]
- Kessler F., Schnell D.J. (2004). Chloroplast protein import: solve the GTPase riddle for entry. Trends Cell Biol. 14: 334–338. [DOI] [PubMed] [Google Scholar]
- Kessler F., Blobel G., Patel H.A., Schnell D.J. (1994). Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 266: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Kikuchi S., Hirohashi T., Nakai M. (2006). Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant Cell Physiol. 47: 363–371. [DOI] [PubMed] [Google Scholar]
- Kikuchi S., Oishi M., Hirabayashi Y., Lee D.W., Hwang I., Nakai M. (2009). A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell 21: 1781–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M., Takase M., Ide T., Nakai M. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Hwang I. (2013). Direct targeting of proteins from the cytosol to organelles: the ER versus endosymbiotic organelles. Traffic 14: 613–621. [DOI] [PubMed] [Google Scholar]
- Köhler D., Montandon C., Hause G., Majovsky P., Kessler F., Baginsky S., Agne B. (2015). Characterization of chloroplast protein import without Tic56, a component of the 1-megadalton translocon at the inner envelope membrane of chloroplasts. Plant Physiol. 167: 972–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler D., Helm S., Agne B., Baginsky S. (2016). Importance of translocon subunit Tic56 for rRNA processing and chloroplast ribosome assembly. Plant Physiol. 172: 2429–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Schnell D.J. (1997). Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139: 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Chen X., Fuks B., Schnell D.J. (1998). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143: 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács-Bogdán E., Benz J.P., Soll J., Bölter B. (2011). Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 11: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S., Patel R., Combe J., Bédard J., Kovacheva S., Lilley K., Biehl A., Leister D., Ríos G., Koncz C., Jarvis P. (2004). Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Lee G.J., Lee K.H., Kim S., Cheong G.W., Hwang I. (2006). Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol. 140: 466–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Kim J.K., Lee S., Choi S., Kim S., Hwang I. (2008). Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 20: 1603–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Wang F., Schnell D.J. (2009). Toc receptor dimerization participates in the initiation of membrane translocation during protein import into chloroplasts. J. Biol. Chem. 284: 31130–31141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.M., Chiu C.C. (2010). Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61: 157–180. [DOI] [PubMed] [Google Scholar]
- Li H.M., Teng Y.S. (2013). Transit peptide design and plastid import regulation. Trends Plant Sci. 18: 360–366. [DOI] [PubMed] [Google Scholar]
- Liu L., McNeilage R.T., Shi L.X., Theg S.M. (2014). ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens. Plant Cell 26: 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumme C., et al. (2014). Nucleotides and substrates trigger the dynamics of the Toc34 GTPase homodimer involved in chloroplast preprotein translocation. Structure 22: 526–538. [DOI] [PubMed] [Google Scholar]
- Ma Y., Kouranov A., LaSala S.E., Schnell D.J. (1996). Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 134: 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M. (2015). The TIC complex uncovered: The alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim. Biophys. Acta 1847: 957–967. [DOI] [PubMed] [Google Scholar]
- Neupert W. (1997). Protein import into mitochondria. Annu. Rev. Biochem. 66: 863–917. [DOI] [PubMed] [Google Scholar]
- Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16: 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil P.K., Richardson L.G.L., Paila Y.D., Piszczek G., Chakravarthy S., Noinaj N., Schnell D. (2017). The POTRA domains of Toc75 exhibit chaperone-like function to facilitate import into chloroplasts. Proc. Natl. Acad. Sci. USA 114: E4868–E4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L.J., Keegstra K. (1992). The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J. Biol. Chem. 267: 433–439. [PubMed] [Google Scholar]
- Olsen L.J., Theg S.M., Selman B.R., Keegstra K. (1989). ATP is required for the binding of precursor proteins to chloroplasts. J. Biol. Chem. 264: 6724–6729. [PubMed] [Google Scholar]
- Oreb M., Tews I., Schleiff E. (2008). Policing Tic ‘n’ Toc, the doorway to chloroplasts. Trends Cell Biol. 18: 19–27. [DOI] [PubMed] [Google Scholar]
- Oreb M., Höfle A., Koenig P., Sommer M.S., Sinning I., Wang F., Tews I., Schnell D.J., Schleiff E. (2011). Substrate binding disrupts dimerization and induces nucleotide exchange of the chloroplast GTPase Toc33. Biochem. J. 436: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila Y.D., Richardson L.G.L., Schnell D.J. (2015). New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 427: 1038–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila Y.D., Richardson L.G., Inoue H., Parks E.S., McMahon J., Inoue K., Schnell D.J. (2016). Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. eLife 5: e12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S.E., Keegstra K. (1994). Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson L.G., Paila Y.D., Siman S.R., Chen Y., Smith M.D., Schnell D.J. (2014). Targeting and assembly of components of the TOC protein import complex at the chloroplast outer envelope membrane. Front. Plant Sci. 5: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson L.G.L., Singhal R., Schnell D.J. (2017). The integration of chloroplast protein targeting with plant developmental and stress responses. BMC Biol. 15: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Soll J., Sveshnikova N., Tien R., Wright S., Dabney-Smith C., Subramanian C., Bruce B.D. (2002). Structural and guanosine triphosphate/diphosphate requirements for transit peptide recognition by the cytosolic domain of the chloroplast outer envelope receptor, Toc34. Biochemistry 41: 1934–1946. [DOI] [PubMed] [Google Scholar]
- Schnell D.J., Kessler F., Blobel G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012. [DOI] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2010). A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2013). The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta 1833: 314–331. [DOI] [PubMed] [Google Scholar]
- Sjögren L.L., Tanabe N., Lymperopoulos P., Khan N.Z., Rodermel S.R., Aronsson H., Clarke A.K. (2014). Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J. Biol. Chem. 289: 11318–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjuts I., Soll J., Bölter B. (2017). Import of soluble proteins into chloroplasts and potential regulatory mechanisms. Front. Plant Sci. 8: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.D., Schnell D.J., Fitzpatrick L., Keegstra K. (2003). In vitro analysis of chloroplast protein import. Curr. Protoc. Cell Biol. 11: 16. [DOI] [PubMed] [Google Scholar]
- Smith M.D., Rounds C.M., Wang F., Chen K., Afitlhile M., Schnell D.J. (2004). atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 165: 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J., Schleiff E. (2004). Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 5: 198–208. [DOI] [PubMed] [Google Scholar]
- Su P.H., Li H.M. (2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y.S., Su Y.S., Chen L.J., Lee Y.J., Hwang I., Li H.M. (2006). Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. Plant Cell 18: 2247–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y.S., Chan P.T., Li H.M. (2012). Differential age-dependent import regulation by signal peptides. PLoS Biol. 10: e1001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theg S.M., Bauerle C., Olsen L.J., Selman B.R., Keegstra K. (1989). Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 264: 6730–6736. [PubMed] [Google Scholar]
- Wang F., Agne B., Kessler F., Schnell D.J. (2008). The role of GTP binding and hydrolysis at the atToc159 preprotein receptor during protein import into chloroplasts. J. Cell Biol. 183: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.E., Keegstra K., Froehlich J.E. (1999). GTP promotes the formation of early-import intermediates but is not required during the translocation step of protein import into chloroplasts. Plant Physiol. 121: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]