Abstract
Post-transcriptional chemical modifications of (t)RNA molecules are crucial in fundamental biological processes, such as translation. Despite their biological importance and accumulating evidence linking them to various human diseases, technical challenges have limited their detection and accurate quantification. Here, we present a sensitive capillary nanoflow liquid chromatography mass spectrometry (nLC-MS) pipeline for quantitative high-resolution analysis of ribonucleoside modifications from complex biological samples. We evaluated two porous graphitic carbon (PGC) materials and one end-capped C18 reference material as stationary phases for reversed-phase separation. We found that these matrices have complementing retention and separation characteristics, including the capability to separate structural isomers. PGC and C18 matrices yielded excellent signal-to-noise ratios in nLC-MS while differing in the separation capability and sensitivity for various nucleosides. This emphasizes the need for tailored LC-MS setups for optimally detecting as many nucleoside modifications as possible. Detection ranges spanning up to six orders of magnitude enable the analysis of individual ribonucleosides down to femtomol concentrations. Furthermore, normalizing the obtained signal intensities to a stable isotope labeled spike-in enabled direct comparison of ribonucleoside levels between different samples. In conclusion, capillary columns coupled to nLC-MS constitute a powerful and sensitive tool for quantitative analysis of modified ribonucleosides in complex biological samples. This setup will be invaluable for further unraveling the intriguing and multifaceted biological roles of RNA modifications.
Keywords: transfer RNA modification, mass spectrometry, porous graphitic carbon, stable isotope labeling, nanoflow liquid chromatography
INTRODUCTION
The translation of the genetic code into functional proteins is fundamental for every living organism. Hence, this process is tightly regulated at multiple layers and integrates many players (Shoemaker and Green 2012) including transfer RNAs (tRNA)—the adapter molecules that link the genetic information to a specific amino acid. These tRNA molecules are decorated by a plethora of evolutionary conserved post-transcriptional chemical modifications (Grosjean 2009; Jackman and Alfonzo 2013; Helm and Alfonzo 2014). These modifications are crucial for fine-tuning translation (Zinshteyn and Gilbert 2013; Grosjean et al. 2014) and thereby maintaining proteome integrity (Nedialkova and Leidel 2015; Thiaville et al. 2016) and can change in response to physiological and chemical stresses (Chan et al. 2010; Alings et al. 2015). Furthermore, a growing number of studies link changes in tRNA modification to various diseases (Sarin and Leidel 2014; Torres et al. 2014), including neoplastic proliferation (Delaunay et al. 2016), type II diabetes (Wei et al. 2011), and developmental disorders (Chen et al. 2009; Suzuki et al. 2011). Despite these advances, our understanding of the mechanisms that underlie tRNA modification is still in its infancy. This is in part because quantitative analyses of precious samples, such as patient material of limited availability, require highly sensitive measurement techniques. Thus, to unravel the biological roles of ribonucleoside modifications, it is essential to improve methods for their accurate, sensitive, and robust quantification (Russell and Limbach 2013; Ross et al. 2016; Thuring et al. 2016).
Albeit conceptually straightforward, technical aspects have limited the broad application of high-throughput analyses of nucleoside modifications. For instance, high-resolution separation of ribonucleosides by liquid chromatography is challenging due to the very polar nature of the analytes (Gehrke and Kuo 1989). Silica-based and end-capped C18 stationary phases have emerged as the standard tool for separation of modified nucleosides (Björk et al. 2007; Alings et al. 2015; Ross et al. 2016; Thuring et al. 2016). This matrix separates many closely related compounds, including positional isomers and nucleoside analogs. However, liquid-chromatography-coupled electrospray ionization mass spectrometry (LC-ESI-MS) detection of certain pyrimidine analogs, such as pseudouridine (Ψ), dihydrouridine (D), and uridine (U), can be challenging using end-capped C18 microbore columns (Alings et al. 2015). This limits an unbiased analysis of complex biological samples, highlighting the need for additional stationary-phase materials with robust separation characteristics and enhanced selectivity.
Porous graphitic carbon (PGC) constitutes an interesting option, as it has been shown to separate highly polar or charged analytes (Pereira 2008; Michel and Buszewski 2009) and structurally closely related species, such as isomers (Gundersen 2001; De Matteis et al. 2012). PGC consists of planar, two-dimensional sheets composed of hexagonally arranged carbon atoms where each sp2 hybridized carbon is joined to three adjacent carbons, creating an extensive polyaromatic scaffold (West et al. 2010). The retention mechanism of polar solutes onto PGC is different from that observed with classical reversed-phase matrices. While, e.g., octadecylsilane-bonded silica phases mainly rely on hydrophobic interactions, the retentive properties of PGC are additionally governed by polar electronic interactions. Therefore, the strength of dispersive interactions is mainly defined by the polarizability of the graphitic surface and the solute. Furthermore, there are various additional interactions, which are summarized by the term “polar retention effect on graphite (PREG)” (Knox and Ross 1997; West et al. 2010). Besides their unique chromatographic characteristics, PGC materials are stable over a wide temperature and pH range (West et al. 2010), making them a popular choice for solid phase extraction (Liu et al. 2011) and in a wide range of analytical applications of biomolecules (Chaimbault et al. 2000; He and Kozak 2012), as well as inorganic (Takeuchi et al. 2000) and organic molecules (Guenu and Hennion 1994).
To explore the possible advantages of PGC materials and to decrease sample input in general, we sought to develop a robust and reproducible high-resolution nanoflow system to quantify ribonucleoside modifications from complex biological samples. Hence, we compared two PGC stationary phases to an end-capped C18 material in a nLC ESI-MS system to evaluate their suitability for ribonucleoside analysis. We obtained excellent chromatographic characteristics with material-specific advantages for selected nucleosides, allowing reliable detection of individual ribonucleosides down to low femtomol concentrations. We showed that PGC materials are able to consistently separate numerous positional isomers, including U, Ψ, and methylated bases of adenosine (A), cytidine (C), and U. However, no material performs optimally for all nucleosides. Finally, we demonstrated the quantitative analysis of ribonucleoside modification levels, both label-free and with a stable isotope labeled spike-in, throughout a linear detection range spanning up to six orders of magnitude. To this end, metabolic labeling of ribonucleosides in Chlamydomonas reinhardtii proves to be a cheap and facile strategy for generating spike-in standards for quantification. Consequently, our methodology is highly suitable for reproducible, quantitative high-resolution analyses of ribonucleoside modifications.
RESULTS
Porous graphitic carbon is well suited for separation of polar compounds
PGC materials efficiently retain polar compounds and are capable of separating structurally related substances (Wan et al. 1995). To test the suitability of PGC columns for reversed-phase separation of chemically modified ribonucleosides, we used a UPLC system coupled with UV-detection and compared the outcome to a C18 material.
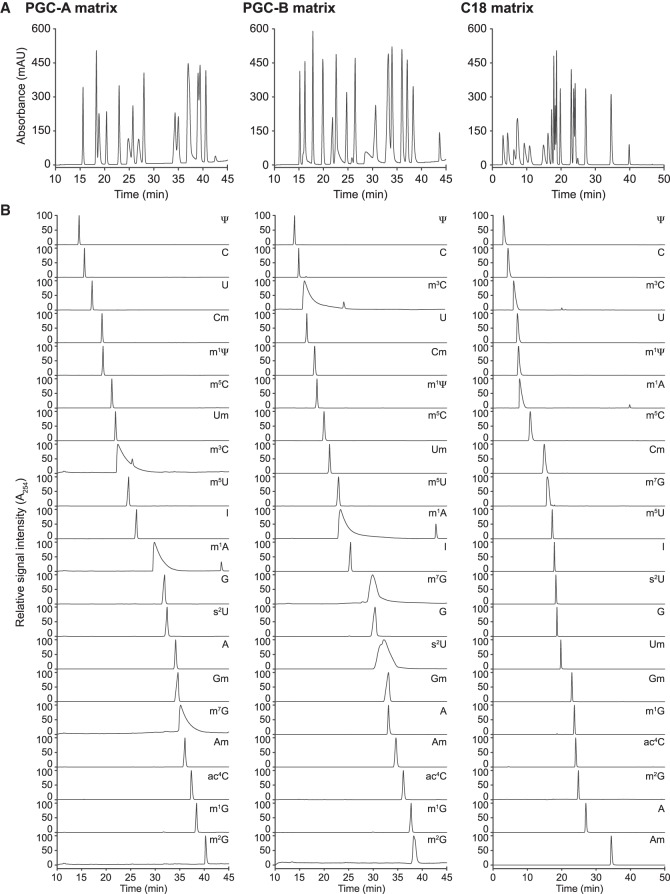
Since the manufacturing of PGC results in a material without precise definition, we used materials from two different suppliers (designated PGC-A and PGC-B; see Materials and Methods). We first established a simple stepwise gradient using microbore columns (I.D. 2.1 mm) to resolve a mix of 20 ribonucleoside standards (Limited Standard Mix, LSM; Supplemental Table 1). Both PGC and C18 materials resolved the individual ribonucleosides, with baseline separation for most analytes (Fig. 1A). In general, peaks were symmetric and signal intensity was high. However, we observed minor variations in resolution and peak symmetry in both PGC materials (Fig. 1A). To evaluate the disparities between the stationary phases, we applied 1 µg of each ribonucleoside standard onto the microbore columns. The materials yielded similar elution profiles with sufficient resolving power to discriminate between positional isomers, such as methylated adenosines (Fig. 1B).
FIGURE 1.
Reliable ribonucleoside separation is achieved by reversed-phase chromatography using porous graphitic carbon (PGC) and C18 materials. (A) Representative chromatograms (A254-trace) obtained with PGC-A (left panel), PGC-B (middle panel), and C18 (right panel) separating 20 ribonucleoside standards (LSM, Supplemental Table 1). (B) Individual chromatograms obtained for each of the 20 ribonucleosides included in the LSM using run conditions as in A. Abbreviations follow the Modomics database convention (Boccaletto et al. 2018).
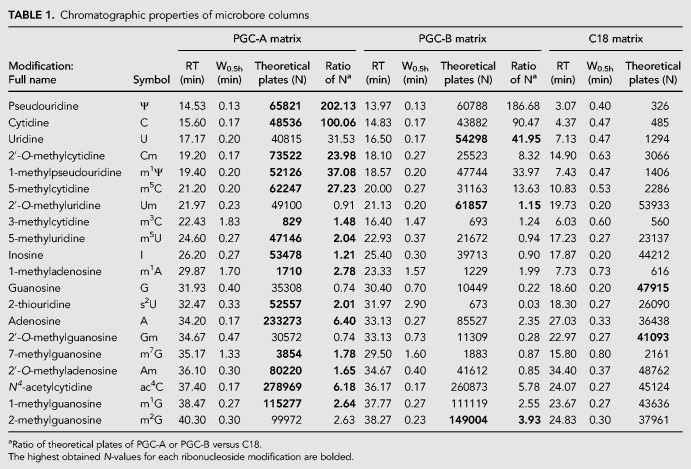
Nonetheless, we observed interesting differences: First, certain ribonucleosides change their elution order, e.g., 3-methylcytidine (m3C) elutes as nucleoside number eight using PGC-A, but as nucleoside number three using PGC-B, coinciding with the position observed for C18 (Fig. 1B). More subtle differences are seen for 2′-O-methylcytidine (Cm), 7-methylguanosine (m7G), and 2-thiouridine (s2U) that change by one or two positions between the PGC materials. In C18, Cm elutes in close proximity to m7G and s2U as nucleoside number eight, placing it three to four nucleosides later than in the PCG materials (Table 1; Fig. 1B). Second, 1-methyladenosine (m1A), m3C, and m7G yield broad peaks in both PGC materials. In general, the peak shape on the C18 column is narrower than for PGC even though ribonucleosides eluting during the first half of the run tend to be broader (Table 1; Fig. 1B). Third, signal intensities differ between the different materials. This is particularly apparent for s2U, but also m7G and, for nucleosides that elute during the first half of the gradient from C18 (Table 1). Fourth, using PGC-A and in particular C18 s2U elutes with a much better peak shape and higher signal intensity than PGC-B. In summary, PGC materials are a powerful strategy to resolve modified ribonucleosides with chromatographic properties that complement C18 materials. Table 1 provides a guideline for choosing the most appropriate material for targeted applications on microbore columns.
TABLE 1.
Chromatographic properties of microbore columns
PGC materials are robust for long-term applications
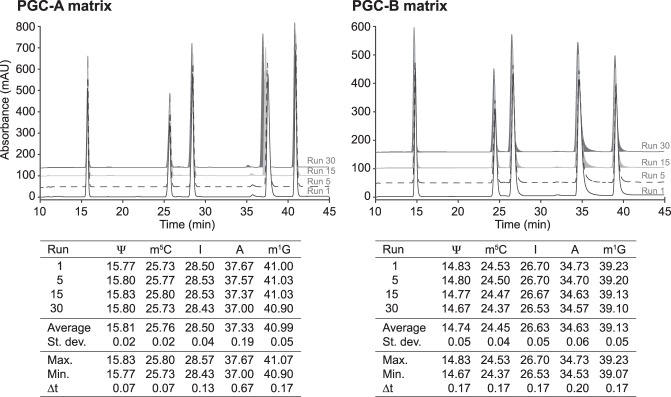
PGC materials are chemically inert, withstanding acidic or basic conditions and high temperatures, but they have been reported to be susceptible to hyper-retention (West et al. 2010). Hyper-retention leads to an irreversible binding of the analyte to the solid phase and can cause a buildup of analytes leading to a loss of resolution and increased backpressure (Reepmeyer et al. 2005; Agrofoglio et al. 2007; Pabst et al. 2010). To assess the longevity of the tested PGC materials, we subjected the columns to successive elution and regeneration cycles loading a mix of five ribonucleosides (Ψ, m5C, I, A, and m1G). These ribonucleosides elute at different time points, allowing us to monitor hyper-retention by analyzing changes in elution time, peak shape, and resolution. Following a series of 30 consecutive runs, we observed only negligible differences for all five analytes (Fig. 2). The largest shift is observed for adenosine in PGC-A, with a difference between the earliest and latest elution time (Δt) of only 0.67 min (Fig. 2). Since the theoretical plate counts remain stable (Table 1), this establishes that the chromatographic performance of the PGC matrices is unaltered throughout the assay.
FIGURE 2.
Ribonucleosides separation is robust and reproducible on PGC materials. Retention time and column durability analysis of 30 consecutive runs with pseudouridine (Ψ), 5-methylcytidine (m5C), inosine (I), adenosine (A), and 1-methylguanosine (m1G) on PGC-A and PGC-B columns. Representative chromatograms from runs 1, 5, 15, and 30 are shown. The table summarizes the retention time parameters for all tested nucleosides. Abbreviations follow the Modomics database convention (Boccaletto et al. 2018).
Self-packed PGC and C18 capillary columns provide strong advantages for nanoflow ESI-MS
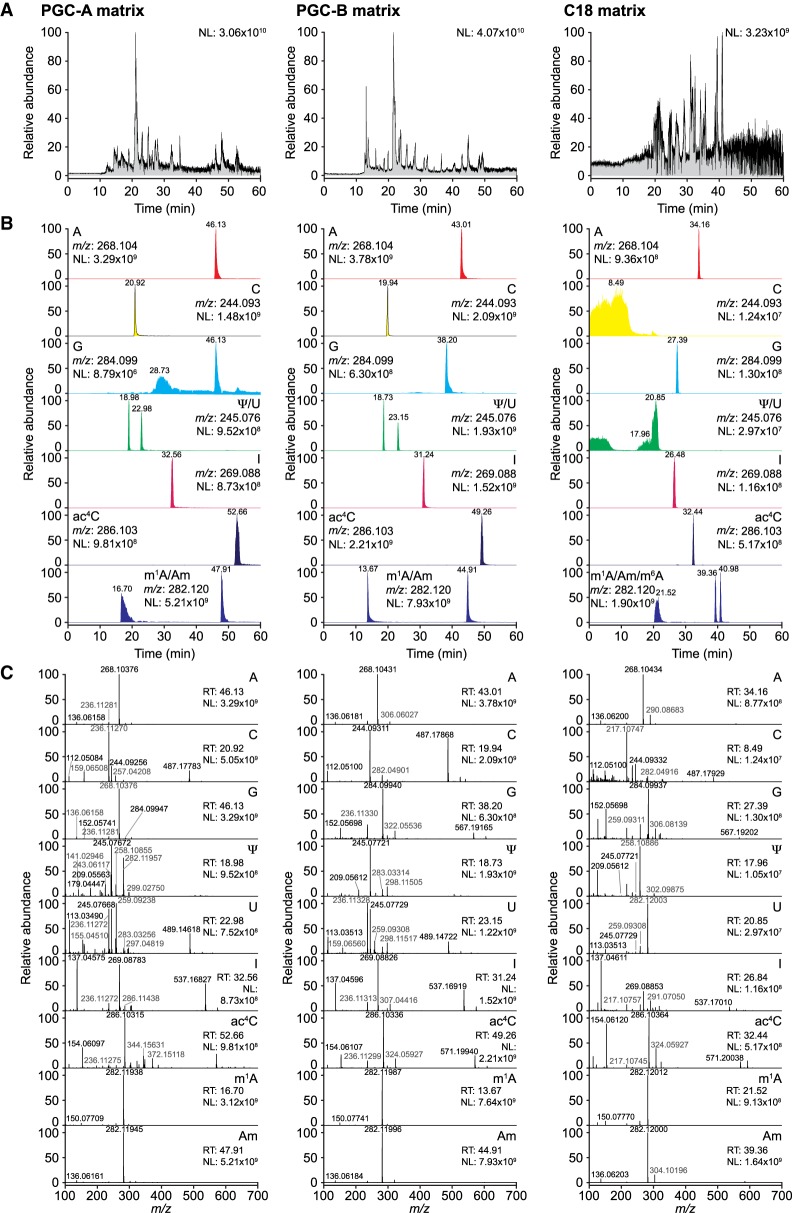
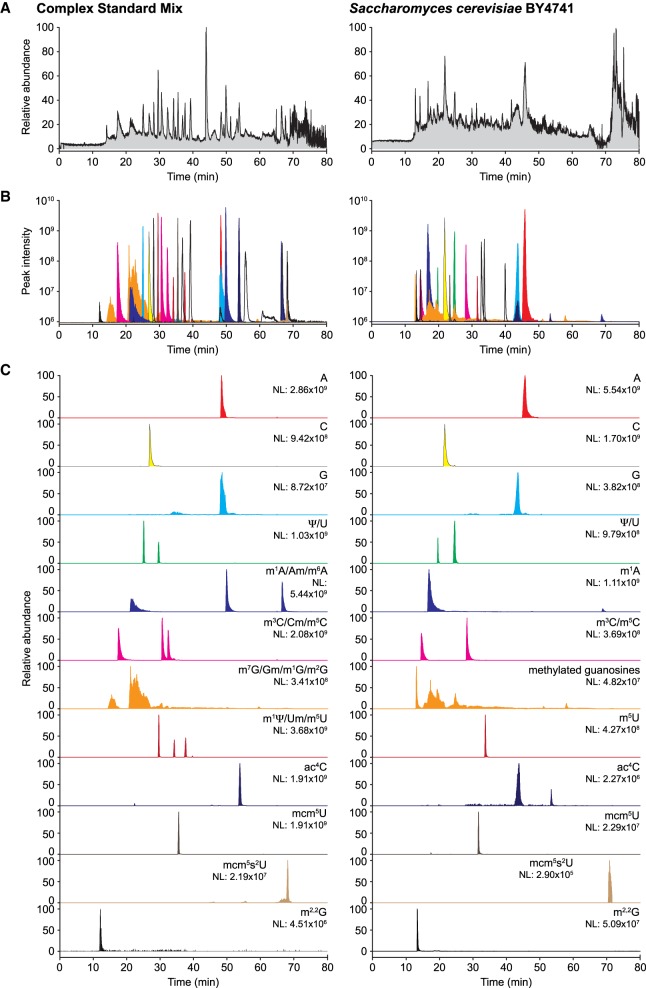
Limiting amounts are often a challenge for quantitative analysis of modified ribonucleosides in biological samples. Even though the sensitivity of a measurement is defined by the instrument, excess material is often required to achieve adequate detection and analysis. Downscaling the chromatographic setup to capillary columns directly coupled with ESI-MS can circumvent this limitation by significantly reducing the amount of input material. However, the microfluidic environment of capillary columns differs from traditional sized columns, preventing direct downscaling. To ensure conformity and comparability, we prepared columns by tightly packing the PGC and C18 materials into silica capillaries (I.D. 75 µm), trimmed to a final length of 500 mm. When loading 100 ng of the LSM (Supplemental Table 1) we achieved good signal-to-noise ratios even though peaks were less distinct than in the UPLC setup (signal intensities 3.06 × 1010, 4.07 × 1010, and 3.23 × 109; Fig. 3A). To evaluate the chromatographic performance of the nLC columns in more detail, we generated extracted ion chromatograms (XIC) for the canonical bases and Ψ, I, N4-acetylcytidine (ac4C), m1A, and 2′-O-methyladenosine (Am) (Fig. 3B) and confirmed the identity of the peaks by MS1 and MS2 spectrum analysis (Fig. 3C). Both PGC materials displayed high peak intensity and acceptable peak symmetry apart from guanosine, which eluted over a broad range (Fig. 3B). Despite the limitations observed for guanosine and its analogs, the PGC materials performed well at separating most ribonucleosides (Supplemental Table 2). For C18 nLC, the signal intensities appeared generally lower and early eluting nucleosides (first ¼ of the run, e.g., C, U, and Ψ ) could not be adequately resolved (Fig. 3B), while later eluting ribonucleosides were well defined (Fig. 3B). In direct comparison, PGC-B yields slightly better resolution and peak symmetry than PGC-A, which we attribute to the smaller particle size and thereby more efficient packaging of the PGC-B material. C18 is on par or slightly exceeds the performance of PGC-B within the 25–50 min elution range (Supplemental Table 2). Since C18 and PGC-B show differences in separation capability of individual nucleosides we evaluated the performance of PGC-B on complex biological samples. We applied 100 ng of either the defined Complete Standard Mix (CSM; Supplemental Table 1) or 250 ng of an enzymatic digest of total yeast tRNA to the PGC-B column and monitored the elution profiles (Fig. 4). As expected, the CSM yields a less complex TIC with readily distinguishable peaks while the yeast tRNA digest leads to a less defined pattern where only a handful of peaks can be distinguished (Fig. 4A). Thus, we generated XICs of all detected ribonucleosides from both analytes (Fig. 4B,C). Regardless of the complexity of the samples, the column separated most ribonucleosides at high resolution despite their strong similarities in structure and chemical properties, as well as significant differences in abundance (Fig. 4B). In our setup, ribonucleosides with overlapping retention times all have different molecular masses, which allows for their accurate identification and quantification. More importantly, most ribonucleosides with identical molecular masses can be readily assigned. In addition to U and Ψ, also the methylated (pseudo)uridines (m1Ψ, Um, and m5U), cytidines (m3C, Cm, and m5C), and adenosines (m1A, Am, m6A) are baseline separated on PGC-B, although the methylated guanosines (m1G, m2G, Gm, m7G) fail to elute as individual peaks (Fig. 4C). Furthermore, the PGC material seems to be capable of separating the acetylated and methylated forms of the same base, as seen in the yeast tRNA digest for ac4C (Fig. 4C).
FIGURE 3.
PGC and C18 capillaries enable the analysis of ribonucleosides in nLC ESI-MS/MS. (A) Representative total ion chromatograms (TIC) of 100 ng of the LSM or CSM (C18 only) analyzed on nanoflow capillary columns packed with PGC-A (left panel), PGC-B (middle panel), or C18 (right panel) material. (B) Extracted ion chromatograms (XIC) for adenosine (A), cytidine (C), guanosine (G), uridine (U), pseudouridine (Ψ), inosine (I), N4-acetylcytidine (ac4C), 1-methyladenosine (m1A), 2′-O-methyladenosine (Am), and N6-methyladenosine (m6A; CSM only). (C) Example MS1 spectra recorded at the retention times corresponding to the nucleosides analyzed in B. (NL) Normalized target level.
FIGURE 4.
Comprehensive analysis of highly complex analytes is achieved using PGC-B. (A) Representative TIC chromatograms of 31 ribonucleoside standards (100 ng of CSM, Supplemental Table 1; left panels) and 250 ng of an enzymatic digest of bulk tRNA isolated from Saccharomyces cerevisiae strain BY4741 (right panels) analyzed on a nano PGC-B capillary column. (B) Overlay of the XICs for all CSM ribonucleosides detected in the respective samples. (C) XICs of selected nucleosides present in the samples. Note the capability of the PGC material to separate, with the exception of methylated guanosine, all positional isomers. (NL) Normalized target level.
Although methylated guanosines are poorly distinguishable from each other, using PCG materials in nLC ESI-MS provides excellent resolution of peaks and high sensitivity for all other ribonucleosides tested. Indeed, weak signals in the range of 104–105 can still be detected, as shown by, e.g., mcm5s2U (Fig. 4C), which implies a dynamic range of at least five orders of magnitude for the accurate detection of ribonucleosides.
Nanoflow PGC and C18 capillary columns enable characterization of ribonucleosides at femtomol levels
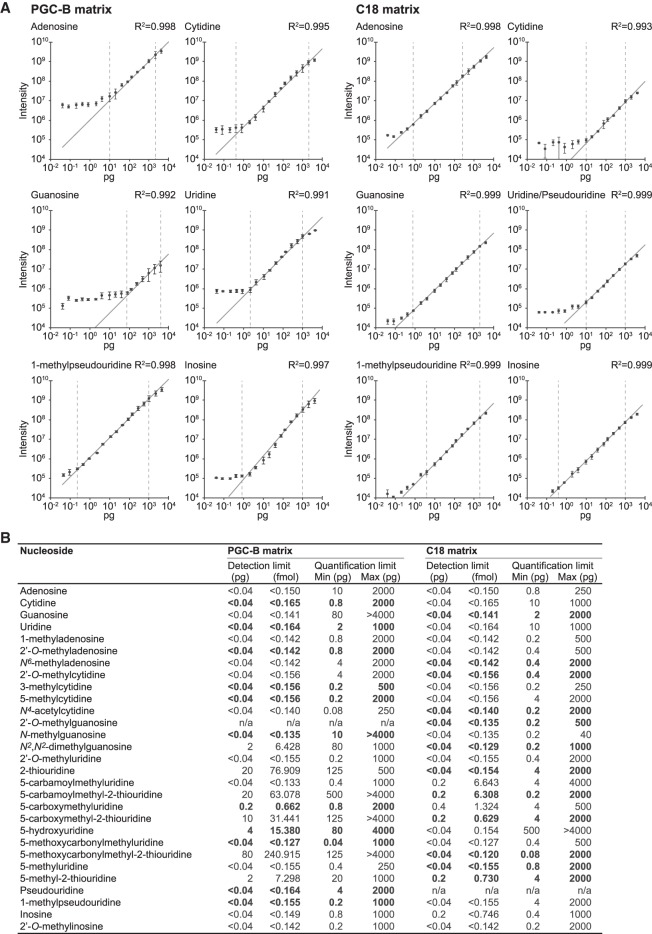
To assess the detection limit and linear range of quantification in our nLC ESI-MS setup using PGC-B and C18 columns, we systematically quantified the maximum peak intensity obtained for each of the 31 ribonucleosides present in the CSM with sample loads spanning six orders of magnitude from 40 fg to 4 ng (Fig. 5). Next, we determined the linear correlation of the maximum peak intensity as a function of the amount of sample loaded. We were able to quantify most of the canonical and modified ribonucleosides within a linear range spanning several orders of magnitude (Fig. 5A,B). As expected for PGC-B, guanosine exhibits only a narrow window of approximately two orders of magnitude, in which it can be reliably quantified, reflecting its suboptimal peak shape on PGC (Fig. 5A,B). In contrast, guanosine displays a much broader quantification window of three orders of magnitude on the C18 matrix, with significant gain at low intensities (Fig. 5A,B). Furthermore, Gm can be separated from the other methylated guanosines, although m1G, m2G, and m7G overlap to form a single peak on the C18 matrix. For thiolated moieties we observe a poor detection limit of 0.1–0.5 ng (∼0.3–1.6 pmol) and a narrow quantification window (Fig. 5B) on PGC-B. However, the C18 matrix consistently yields broader linear quantification windows for these ribonucleosides, ranging lower (0.08–4 pg; 0.24–15.4 fmol) and, most often, higher (2 ng; 6.03–7.69 pmol) than for the PGC-B material. In contrast, remaining ribonucleosides have detection limits lower than 0.04 pg (∼0.1 fmol) on both matrices, although linear correlation is usually achieved at slightly higher loads of 0.2–10 pg (∼0.6–30 fmol) or above (Fig. 5B; Supplemental Table 3). Under optimal conditions, ribonucleosides can be detected and characterized over a very broad range exceeding six orders of magnitude on both materials, whereas the linear range that allows for reliable quantification is significantly smaller.
FIGURE 5.
Absolute quantification can be accomplished over a broad detection range. (A) Calibration curves for representative ribonucleosides analyzed on PGC-B (left panels) and C18 matrices (right panels) showing the observed XIC maximum intensity as a function of sample loaded (0.04–4000 pg). The error bars represent the standard deviation for each data point (n = 3). Linear regression (dark gray line) is used to determine the dynamic range of the instrument for each ribonucleoside on the respective matrix by observing the range at which a linear dependency between input amount and intensity is observed (vertical dashed bars indicate the range). (B) Summary of the linear quantification range determined for all 31 ribonucleosides present in the CSM (linear regression for all moieties is R2 ≥ 0.96). The matrix that provides the best overall performance (detection limit, quantification range, R2-value)—and is the recommended choice for analyzing the selected ribonucleoside—is highlighted in bold.
Metabolic labeling enhances quantitative analysis of ribonucleosides
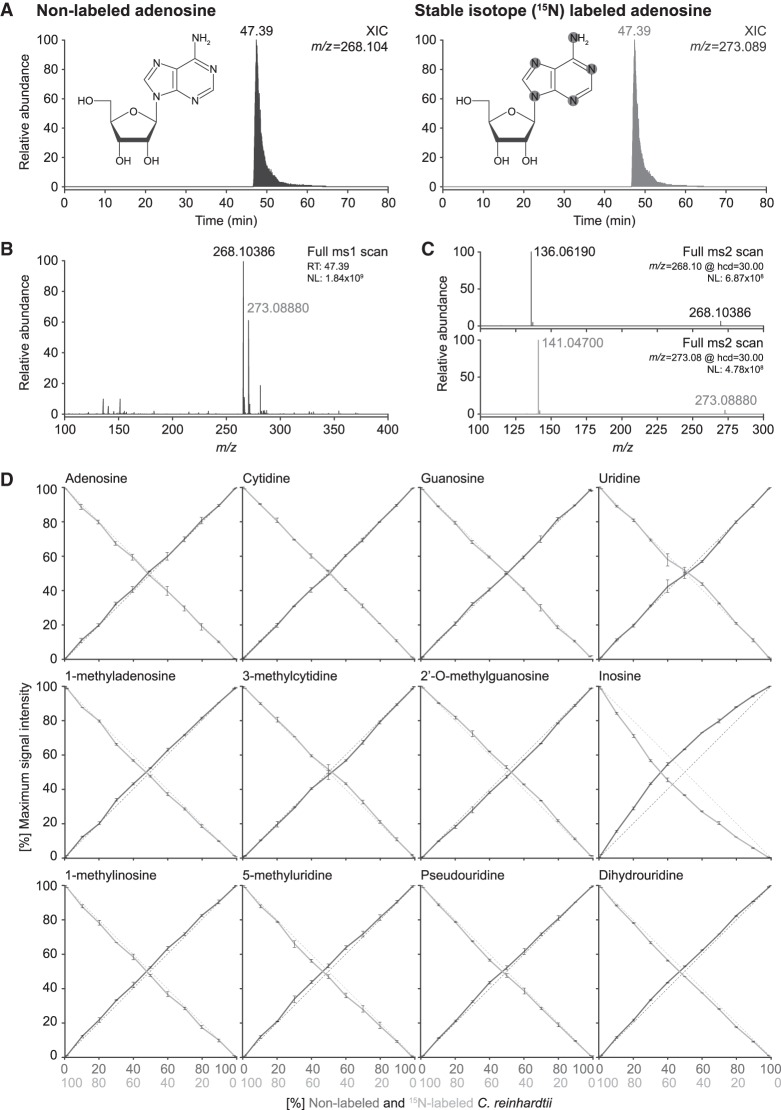
Ribonucleosides can be readily detected and characterized from various biological samples using our nLC ESI-MS setups. Nonetheless, accurate quantitative analysis requires the presence of an invariable internal standard, such as an artificial nucleoside analog (Bruckl et al. 2009), to which the modification levels are normalized. However, differences in structure and charge of the ribonucleosides significantly affect ionization, which might bias results if quantifications are normalized only to a single nucleoside. To circumvent this, stable isotope (15N)-labeled ribonucleosides can be used as internal standards (Fig. 6; Kellner et al. 2014a,b). This allows each modification in the sample to be specifically normalized against its heavier 15N-counterpart, thereby eliminating any variance arising from differences in ionization efficiency (Fig. 6A–C).
FIGURE 6.
Accurate relative quantification of ribonucleosides can be achieved using stable isotope labeled internal spike-in standards. (A) XICs of adenosine in its native nonlabeled (left panel) form and with stable isotope (15N)-labeling (right panel, spike-in standard), as well as a schematic representation of the chemical structure of adenosine showing the 15N incorporation sites highlighted in gray. (B) MS1 spectrum for adenosine at RT 39.30 min. Note the increase in mass (∼5 Da) resulting from the incorporation of 15N in the base (15N-labeled mass in gray, nonlabeled mass in black). (C) MS2 fragmentation spectra of nonlabeled (top panel; m/z = 268.10) and 15N-labeled (bottom panel; m/z = 273.09) adenosine. Note the appearance of the base at m/z = 136.06 and m/z = 141.05, respectively, corresponding to the expected neutral loss of ribose (−132.04). (D) Cross-dilution series of nonlabeled and 15N-labeled enzymatic digests of bulk tRNA isolated from Chlamydomonas reinhardtii. Quantification of the maximum peak intensity of nonlabeled (solid dark gray line) versus 15N-labeled (solid light gray line) XICs of the canonical bases and representative ribonucleoside modifications. Shown is the abundance ratio of nonlabeled (N) and 15N-labeled (15N) maximum signal intensities (MaxI); Abundance = MaxI15N/(MaxI15N + MaxIN) or vice versa. The dotted lines represent the expected ratio of 15N-labeled (light gray) versus nonlabeled (dark gray) material present in the samples. (NL) Normalized target level.
Since metabolic labeling of yeast requires a set of expensive reagents, we devised a cheap and simple strategy to generate metabolically labeled spike-in standards. To this end, we grew the single-celled green algae C. reinhardtii in a 15N-containing growth media (Barth et al. 2014), achieving full labeling within a few days. To test the suitability of stable isotope labeled spike-in standards for normalization and to simulate a data set that corresponds to SILAC labeling of ribonucleosides, we devised a cross-dilution series where nonlabeled and 15N-labeled ribonucleosides isolated from C. reinhardtii are combined in specific ratios ranging from 100:0 to 0:100 (Fig. 6D). The 15N-labeled ribonucleosides yielded a strong signal throughout the cross-dilution series. As little as 2.5 ng (99:1 ratio) of spike-in standards were sufficient for reliable detection and quantification when mixing with 99-fold more concentrated nonlabeled ribonucleosides (247.5 ng load). Moreover, the intensities observed throughout the cross-dilution series (Fig. 6) were within the linear range of the instrument (Fig. 5).
Hence, we analyzed a total of 19 ribonucleosides semi-automatically using pyQms (Leufken et al. 2017) and manually using QualBrowser, yielding identical outcomes (Supplemental Fig. 2; Supplemental Table 4). The ratio of nonlabeled and labeled ribonucleosides was calculated to determine the relative proportion of each isoform in the cross-dilution series. Importantly, the measured relative abundance of the isoforms accurately reflects the theoretical cross-dilution ratio for almost all ribonucleosides (Fig. 6D; Supplemental Fig. 2; Supplemental Table 4). Interestingly, for m5C, Cm, and I, the point of equal intensity between the nonlabeled and 15N-labeled isoform shifted significantly toward a higher ratio of the nonlabeled isoform (Fig. 6D; Supplemental Fig. 2). As the reproducibility of the measurements (n = 3) following normalization to the 15N-labeled isoform is very high (m5C ±0.62%; Cm ±0.83%; I ±0.42%), it is highly unlikely that technical issues caused this difference. This suggests that the observed shift stems from marginally different amounts of m5C, Cm and I in the two samples. Since the C. reinhardtii cultures yielding the metabolically 15N-labeled and the nonlabeled tRNA samples were generated six months apart of each other, they may have encountered slightly different growth conditions that resulted in the observed differences. Furthermore, sample preparation procedures involve the enzymatic digestion and dephosphorylation of ribonucleosides, which might have introduced trace amounts of deaminases (Singhal et al. 2013), potentially altering the amount of I in the sample.
This shows that SILAC-type experiments are feasible for ribonucleoside analysis. Furthermore, we are able to reliably detect minute changes in modification levels from complex biological samples, highlighting the need of spike-in standards as a powerful tool for normalization.
DISCUSSION
PGC and C18 materials are well suited as column matrices for reversed-phase chromatography of post-transcriptionally modified ribonucleosides. Due to their particular retention mechanisms, both materials offer unique advantages that nicely complement each other. In this study, using different PGC stationary phases we demonstrate that many modified ribonucleosides can be baseline separated at high peak capacity in microbore (Fig. 1) and capillary nanoflow (Fig. 3) column format, achieving equal or slightly better performance than with an end-capped C18 material (Table 1; Supplemental Table 2). It is straightforward to establish separation as even a simple linear gradient and different buffer systems yield satisfying results (Supplemental Fig. 3). Although the two PGC materials are very similar in their chemical and physical properties, slight differences in retention time, peak resolution, peak shape, and peak symmetry are apparent (Figs. 1B, 3B). This can most likely be attributed to differences in physical parameters, e.g., different particle size of the PGC matrices (3 µm versus 2.1 µm) and different column lengths in the microbore setup (150 mm versus 100 mm). Importantly, we did not observe a buildup of analytes due to hyper-retention, which would lead to a gradual decrease in chromatographic performance (Pabst et al. 2010; West et al. 2010). Both PGC matrices remained stable throughout thirty consecutive runs, showing only minute changes in retention time and peak resolution, as well as only slight fluctuations in peak intensity (Fig. 2). This shows that both PGC materials maintain uniform long-term chromatographic performance.
In the nLC setup, the smaller particle size of PGC-B (2.1 µm) shows greater peak capacity compared to PGC-A (3.0 µm) albeit at the cost of increased backpressure. However, the strong binding characteristic of both PGC materials allows challenging uridine analogs to be resolved in highly symmetric peaks. In contrast, the C18 reference material fails to retain pseudouridine in the nLC setup (Fig. 3B). Overall, both PGC materials bind ribonucleosides more strongly than C18, as reflected by the need for significantly higher acetonitrile concentrations to elute the analytes. We successfully baseline separated more than 25 ribonucleosides from both synthetic and complex biological samples on PGC capillary columns, achieving good signal-to-noise ratios in a detection range that spans up to six orders of magnitude (Figs. 4, 5). However, the four methylated guanosines (m/z = 298.118) included in the CSM (m1G, m2G, m7G, Gm) remain poorly resolved on both PGC matrices (Fig. 4C), although guanosine itself does not exhibit a well defined peak (Figs. 3B, 4C). Conversely, N2,N2-dimethylguanosine (m2,2G), a double methylated form of guanosine, was detected without difficulty at high signal intensity in the biological sample (Fig. 4C). On the other hand, the C18 material poorly resolves C (Fig. 3B) and other ribonucleosides that elute during the first ⅓ of the run (Supplemental Table 2). Another good example are N6-threonylcarbamoyladenosine (t6A) and cyclic N6-threonylcarbamoyladenosine (ct6A). While t6A is exceptionally well separated on C18 material, it cannot be reliably detected on PGC material. The opposite is true for ct6A (data not shown). However, with few exceptions, the tested columns separate ribonucleosides with identical molecular weights using simple mobile phase gradients. This enables reliable quantification without the need for MS2 or MS3 fragmentation strategies. Nevertheless, it is crucial to use ribonucleoside standards to determine the elution order as it may vary slightly between different PGC materials.
In complex biological samples, detecting the ribonucleoside of interest is merely the first step, as crucial information about its function and regulation can only be deduced from its abundance. This necessitates an approach whereby absolute or relative abundances of the analytes can be precisely quantified by the use of calibration curves, spike-in standards, and a fine-tuned instrument with a broad dynamic range. By determining the limit of detection and linear range for each ribonucleoside, it is possible to set up the mass spectrometer to measure absolute abundances. Indeed, we found that most of the ribonucleosides studied have a linear range spanning three to four orders of magnitude (Fig. 5). Within this range, we can determine absolute quantities of ribonucleosides present in any given sample by correlating maximum peak intensity to sample amounts. The major caveat of this approach is the need for nucleoside standards, i.e., each ribonucleoside of interest can only be quantified once the detection limits of the instrument have been determined using a pure ribonucleoside standard (Fig. 5). Even though the number of nucleosides that are commercially available is increasing, absolute quantification of many modifications requires expensive custom-synthesis. However, for most biological applications the need for nucleoside standards can be circumvented, as the change in relative abundance between samples is more informative than absolute quantification. Here, stable isotope (15N-)labeled ribonucleosides from C. reinhardtii provide an invariable internal reference point, to which different samples can be compared and normalized (Fig. 6; Supplemental Fig. 2). Using C. reinhardtii provides a simple source of 15N-labeled ribonucleosides that are approximately 10–15 times cheaper than those isolated from 15N-labeled yeast. This circumvents metabolic labeling of biological samples in cases where it is not straightforward, e.g., due to the high costs associated with defined isotope-labeled growth media, or in the cases where it is simply not possible, such as clinical samples. As we have successfully demonstrated, even low amounts of metabolically 15N-labeled spike-in standards yield sufficiently strong signals for reliable normalization and quantification (Fig. 6D). Moreover, the technical reproducibility of measurements from the same biological sample can be significantly improved by internal normalization. For example, the maximum peak intensity recorded for A, C, G, and U in three technical repeats yielded 10.6%, 10.0%, 12.2%, and 9.4% standard deviation prior to normalization, but merely 0.89%, 0.46%, 0.57%, and 0.78% following normalization to the 15N-isoform. This constitutes a 12- to 22-fold increase in precision. Furthermore, subtle changes in culture conditions or unwarranted enzymatic activity in sample preparation may lead to changes in individual tRNA modifications, which can be detected by the use of labeled ribonucleosides in SILAC-like experiments. Finally, we established that pyQms (Leufken et al. 2017) is a robust tool for semi-automated MS based ribonucleoside quantification, achieving identical results to manual quantification in a fraction of the time required for analysis.
In conclusion, we have shown that a nLC ESI-MS setup coupled to capillary PGC or C18 nLC columns can be successfully used to quantitatively analyze chemically modified ribonucleosides. The excellent resolution and strong signal-to-noise ratio obtained for, in particular, pyrimidine-based ribonucleosides makes PGC an excellent chromatographic matrix for the study of complex biological samples. Furthermore, combining this chromatographic setup to a MS detector with improved sensitivity could further lower the detection boundary past attomol amounts and expand the linear range for quantification.
MATERIALS AND METHODS
Preparation of synthetic ribonucleoside standards
Mass spectrometry grade ribonucleosides (Carbosynth Ltd.; apart from 5-methoxycarbonylmethyluridine [mcm5U], 5-methoxycarbonylmethyl-2-thiouridine [mcm5s2U], 5-carbamoylmethyluridine [ncm5U], 5-carbamoylmethyl-2-thiouridine [ncm5s2U], N6-threonylcarbamoyladenosine [t6A], and cyclic N6-threonylcarbamoyladenosine [ct6A] that were synthesized by A.D., K.D., and E.S.) were individually dissolved in 5 mM NH4HCO2 pH 5.3 at a concentration of 50 ng/µL. All four canonical bases and 27 ribonucleoside modifications (Supplemental Table 1) were combined in equal amounts to yield the CSM at a concentration of 1.6 µg/µL. A LSM consisting of 20 bases (Supplemental Table 1) with a concentration of 1.0 µg/µL was also prepared for UPLC analysis.
Cell culture and metabolic labeling of ribonucleosides
The haploid Saccharomyces cerevisiae strain S288C BY4741 was grown in rich growth media (YPD; Formedium) for tRNA isolation. In brief, overnight starter cultures were grown at 30°C, 200 rpm. The starter cultures were inoculated into prewarmed medium to yield an OD600 = 0.2, and the yeast were grown at 30°C to the logarithmic growth phase (OD600 = 0.8–1.0) and harvested by centrifugation for 3 min at 5000g.
The cell wall deficient CW15 strain of the green algae Chlamydomonas reinhardtii was grown heterotrophically at 22°C with a light intensity of 20 µE/m2/sec and continuous aeration in a rotary shaker at 120 rpm for several generations in tris-acetate-phosphate (TAP) medium (Harris 2009) containing 15NH4Cl (99.4% 15N, Cambridge Isotope Laboratories, Inc.) as the sole nitrogen source. The cells were harvested by centrifugation at 2500g for 5 min (Barth et al. 2014). Nonlabeled ribonucleosides were obtained as described above, apart from using NH4Cl with a natural isotope distribution.
tRNA isolation and preparation for LC-MS analysis
Isolation and purification of tRNA from C. reinhardtii and S. cerevisiae was performed as previously described (Alings et al. 2015). C. reinhardtii tRNA was additionally subjected to gel extraction from a denaturing 8 M urea 8% polyacrylamide gel (Lecanda et al. 2016). Subsequently, tRNA was enzymatically digested and dephosphorylated to yield monoribonucleosides (Alings et al. 2015). Briefly, 10 µg of tRNA was combined with 40 mU of Nuclease P1 from Penicillium citrinum (Sigma-Aldrich Biochemie GmbH) resuspended in 0.2 M CH3CO2Na (sodium acetate) pH 5.3 and 0.1 U of Shrimp Alkaline Phosphatase (Thermo Scientific) in 30 µL reactions at 37°C containing 2 mg/mL ZnCl2 and 1×NEB3 buffer (New England Biolabs GmbH) for near neutral reaction conditions, or 20 mM CH3CO2Na pH 5.3 for acidic conditions. After 1.5 h the reaction mixture was supplemented with 15 µL 0.5 M NH4HCO3 and incubation at 37°C was resumed for 1 h. The reaction was terminated by adding 5.0% C2HF3O2 (trifluoroacetic acid; TFA; Sigma-Aldrich Chemie GmbH) in water to a final concentration of 1.0%. The ribonucleosides were purified with HyperSep Hypercarb SPE Spin Tips (Thermo Fisher Scientific GmbH), dried to completion in a Savant SpeedVac concentrator (Thermo Fisher Scientific GmbH), and finally resuspended in 5 mM NH4HCO2 pH 5.3.
Reversed-phase high performance liquid chromatography of ribonucleosides
Nucleoside analysis was performed on two PGC matrices; hereafter referred to as PGC-A (Hypercarb 3 µm; pore size 250 Å; Thermo Fisher Scientific GmbH) and PGC-B (Prototype PGC 2.1 µm; pore size 250 Å; MilliporeSigma); and a C18 reference matrix (Synergi 4 µm Hydro-RP 80 Å; Phenomenex Ltd.). The PGC-A (2.1 × 150 mm), PGC-B (2.1 × 100 mm), and C18 (2.0 × 150 mm) columns were connected to a Knauer PLATINblue UPLC system equipped with an Autosampler 3950, a PLATINBlue T-1 column thermostat, and a MW-1 UV detector (KNAUER Wissenschaftliche Geräte GmbH). The solvent system consisted of 5 mM NH4HCO2 pH 5.3 (solvent A) and 100% C2H3N (acetonitrile; ACN; solvent B) for PGC and 20% C2H3N for C18, respectively. Either 25 µg of the LSM or 2 µg of each individual synthetic ribonucleoside was loaded onto the columns. For all runs, a flow rate of 0.45 mL/min was applied and absorption at 254 nm was recorded.
The PGC columns were equilibrated in solvent A with 2% B until a stable baseline was achieved. After sample loading, the PGC columns were washed for 5 min using solvent A with 2% B. Separation was obtained by applying a linear gradient from 2% to 98% for 40 min, followed by a wash at 98% B for 5 min and regeneration in solvent A with 2% B for 10 min, respectively. Conversely, a complex gradient of 2%–10% B for 1 min, 10% B for 2 min, 10%–25% B for 25 min, 25%–50% B for 10 min, and 50%–98% B for 1 min, followed by a wash at 98% B and regeneration in solvent A with 2% B for 5 min and 10 min, respectively, was also used. The on-column temperature was maintained at 55°C throughout the PGC runs, with post-column cooling set to 25°C.
The C18 column was equilibrated in solvent A at a flow rate of 0.45 mL/min for 12 min. A linear gradient from 0%–50% B for 40 min, followed by a wash of 50%–80% B for 9 min and of 80%B for 4 min. Column regeneration was achieved by lowering solvent B concentration from 80%–0% within 5 min followed by 12 min of 100% solvent A. Also used was a multistep gradient of 100% A for 10 min, 0%–13% B for 3 min, 13%–19% B for 13 min, 19%–32% for 9 min, 32%–50% for 5 min followed by two wash steps of 50%–80% B for 9 min and 80% B for 4 min. Column regeneration was achieved by lowering solvent B concentration from 80%–0% within 5 min followed by 12 min of 100% solvent A. The column was thermostatically controlled at 15°C and post-column cooling at 25°C.
The elution order and average retention time for all ribonucleoside modifications was determined from three technical repetitions using automated peak analysis in OpenLAB CDS EZChrom Edition (KNAUER Wissenschaftliche Geräte GmbH). The signal-to-noise ratio is calculated as intensities of the signal/background noise, where signal is the maximum peak intensity for the analyte and background noise is the average intensity for the base areas adjacent to the peak.
Lifetime performance of porous graphitic carbon in reversed-phase chromatography
The performance of the PGC-A (2.1 × 150 mm) and PGC-B (2.1 × 100 mm) columns were monitored over 30 consecutive runs using 2 µg each of pseudouridine (Ψ), 5-methylcytidine (m5C), inosine (I), adenosine (A), and 1-methylguanosine (m1G). The same chromatographic conditions were applied as described above for the complex gradient. Column performance (e.g., backpressure, signal-to-noise ratio, etc.) was monitored from the run parameters and changes in ribonucleoside separation (retention time, peak symmetry, and peak resolution) were determined using automated peak analysis in OpenLAB CDS EZChrom Edition (KNAUER Wissenschaftliche Geräte GmbH).
Packing of PCG and C18 into fused silica capillaries
PGC-A, PGC-B, and C18 were packed into 75 µm inner diameter uncoated fused silica capillaries (Z-FSS-075365; Postnova Analytics GmbH), to which frits were prepared (Meiring et al. 2002). In essence, 3 parts of potassium silicate 28/30° solution (Kremer Pigmente) and 1 part of formamide were mixed thoroughly and the resulting precipitate was spun down at 14,600g for 20 sec. One end of the silica capillary was immersed for ∼10 sec in the top layer of the polymerization solution, after which the capillary was baked at 100°C for 4 h. The newly polymerized frit was conditioned with 100% ACN for 5 min.
Next, ∼50 mg of stationary phase was suspended in a 750 µL of a 1:1 C3H8O (isopropanol; IPA):C2H6O (ethanol) solution and ultrasonicated for 5 min in 2 mL flat-bottomed glass vial (MilliporeSigma). The vial was tightly connected to the loading bomb and the slurry was injected into the fritted capillary at an initial pressure of ∼50 bar, followed by a gradual increase to ∼100 bar. Once the desired bed length was reached (it takes ∼8 h to reach 550 mm), the column frit was trimmed to a final length of about 4–5 mm, the capillary was connected to a Proxeon EASY nLC-1000 (Thermo Fisher Scientific GmbH), and the matrix was compacted to its final bed length (500 mm) with a flow of 0.1% formic acid at a constant pressure of 500 bar.
Liquid chromatography mass spectrometry of ribonucleosides
LC-MS analysis was performed using a Proxeon EASY nLC (Thermo Fisher Scientific GmbH) online coupled via a self-packed PGC or C18 capillary column and an electropolished stainless steel emitter (Proxeon ES542, 30 µm × 40 mm), connected via a MicroTight True ZDV union (IDEX Europe GmbH), to a Q Exactive mass spectrometer (Thermo Finnigan LLC). The column was attached to a Proxeon nano ESI source (Thermo Fisher Scientific GmbH). Unless otherwise stated, the total load was 100 ng for the synthetic ribonucleosides and 250 ng for ribonucleosides derived from biological samples. The solvent system used for separation on PGC consisted of 5 mM ammonium formate pH 5.3 (solvent A) and 2:1 IPA:ACN (solvent B). Separation on PGC was achieved using a multistep gradient (0%–12% B in 1 min; 12%–60% B in 60 min; 60%–100% B in 1 min; hold at 100% B for 25 min; 100%–0% B in 1 min; hold at 0% B for 12 min) at a flow rate of 250 nL/min, with the on-column temperature set to 55°C. The solvent system used for the C18 capillary consisted of 5 mM ammonium formate pH 5.3 (solvent A) and 40% ACN (solvent B). Separation on C18 was achieved using a multistep gradient (0% B for 2 min; 0%–80% B in 50 min; 80%–100% B in 8 min; hold at 100% B for 5 min; 100%–0% B in 5 min; hold at 0% B for 12 min) at a flow rate of 250 nL/min, with the on-column temperature set to 20°C. The mass spectrometer was run in the positive mode at a resolution of 70,000. Full MS spectra were recorded in profile mode in the scan range from m/z = 100–700, with the AGC target value set to 3 × 106 and the maximum fill time to 50 msec. The five most intense ions were selected for fragmentation. MS2 scans were recorded at a resolution of 35.000 in an isolation window of 4.0 m/z with the ACG target value set to 2 × 105 and the maximum fill time to 120 msec. Only single charged ions were allowed (M + H+). All measurements were carried out in three technical replicates.
Linear response calibration curves
Dilution of the CSM yields the following concentrations: 1000, 500, 250, 125, 62.5, 31.25, 20, 10, 5, 2.5, 1, 0.5, 0.2, 0.1, 0.05, 0.02, and 0.01 pg/µL. Four microliters of each dilution was subjected to LC-MS analysis in three technical replicates.
Quantitative analysis of LC-MS data
Manual identification and quantification of ribonucleosides was performed using Qual Browser in the Xcalibur suite (Thermo Fisher Scientific GmbH). A lookup list containing the chemical formulae of the expected modifications was compiled (ribonucleoside data from MODOMICS) (Boccaletto et al. 2018), and the corresponding m/z values (Nuc + H+) were generated using the isotope-simulation function in Qual Browser. Next, XIC were created within a tolerance range of ±0.002 Da of the theoretical m/z value, and the maximum intensity of the peak (MaxI) was determined using manual peak annotation (Add Peaks). The identity of the ribonucleoside was verified by its retention time and fragmentation pattern in the MS1 and MS2 scans. The MaxI values were normalized against their 15N-labeled isoforms in samples spiked with a stable isotope labeled standard.
Semi-automated quantitative data analysis was carried out using pymzML (version 2.0.0) (Bald et al. 2012; Kösters et al. 2018) and pyQms (version 0.5.0) (Barth et al. 2014; Leufken et al. 2017). Prior to quantification, Thermo RAW files were converted to the mzML format (Martens et al. 2011) using msconvert as part of Proteowizard (version 3.0.10738) (Kessner et al. 2008). Using a manually curated lookup list of all known ribonucleosides (data from MODOMICS) (Boccaletto et al. 2018), pyQms calculates high accuracy isotopologue patterns derived from their chemical formulas and matches those onto MS1 spectra. Quantification is based on the maximum intensity of the matched isotope pattern chromatogram (MIC) of each ribonucleoside (Leufken et al. 2017). In addition, pyQms generates a weighted similar match score (mScore) to assess the quality of all quantifications (Gower 1971).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Dr. Daniel Shollenberger and Dr. David Bell at MilliporeSigma for kindly providing PGC material, Professor Michael Hippler for Chlamydomonas material and media, Annalen Nolte and Karin Scharmann for technical support, and members of the Leidel laboratory and SPP1784 for critical discussions. R.L.R. thanks the University of Cincinnati RITE program for supporting his international travel and research experience. We thank Dr. Ulrich Hennecke for suggestions on the manuscript. This work is dedicated to the memory of Professor Andrzej Małkiewicz. This work was supported by grants from the Max Planck Society, the North Rhine-Westphalian Ministry for Innovation, Science and Research (314-400 010 09 to S.A.L.), the European Research Council (ERC-2012-StG 310489-tRNAmodi to S.A.L.); the Sigrid Jusélius Foundation (Sigrid Jusélius Fellowship to L.P.S.); the National Science Centre, Poland (UMO-2017/25/B/ST5/00971 to E.S.) and the National Institutes of Health (NIH GM58843 to P.A.L.). Funding for open access charge: The Max Planck Society. S.A.L. is a member of SPP1784: “Chemical Biology of Native Nucleic Acid Modifications.”
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.065482.117.
Freely available online through the RNA Open Access option.
REFERENCES
- Agrofoglio LA, Bezy V, Chaimbault P, Delépée R, Rhourri B, Morin P. 2007. Mass spectrometry based methods for analysis of nucleosides as antiviral drugs and potential tumor biomarkers. Nucleosides Nucleotides Nucleic Acids 26: 1523–1527. [DOI] [PubMed] [Google Scholar]
- Alings F, Sarin LP, Fufezan C, Drexler HC, Leidel SA. 2015. An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 21: 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bald T, Barth J, Niehues A, Specht M, Hippler M, Fufezan C. 2012. pymzML—Python module for high-throughput bioinformatics on mass spectrometry data. Bioinformatics 28: 1052–1053. [DOI] [PubMed] [Google Scholar]
- Barth J, Bergner SV, Jaeger D, Niehues A, Schulze S, Scholz M, Fufezan C. 2014. The interplay of light and oxygen in the reactive oxygen stress response of Chlamydomonas reinhardtii dissected by quantitative mass spectrometry. Mol Cell Proteomics 13: 969–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR, Huang B, Persson OP, Byström AS. 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13: 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckl T, Globisch D, Wagner M, Müller M, Carell T. 2009. Parallel isotope-based quantification of modified tRNA nucleosides. Angew Chem Int Ed Engl 48: 7932–7934. [DOI] [PubMed] [Google Scholar]
- Chaimbault P, Petritis K, Elfakir C, Dreux M. 2000. Ion-pair chromatography on a porous graphitic carbon stationary phase for the analysis of twenty underivatized protein amino acids. J Chromatogr A 870: 245–254. [DOI] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. 2010. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6: e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Tuck S, Byström AS. 2009. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 5: e1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay S, Rapino F, Tharun L, Zhou Z, Heukamp L, Termathe M, Shostak K, Klevernic I, Florin A, Desmecht H, et al. 2016. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med 213: 2503–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis CI, Simpson DA, Euerby MR, Shaw PN, Barrett DA. 2012. Chromatographic retention behaviour of monosubstituted benzene derivatives on porous graphitic carbon and octadecyl-bonded silica studied using molecular modelling and quantitative structure-retention relationships. J Chromatogr A 1229: 95–106. [DOI] [PubMed] [Google Scholar]
- Gehrke CW, Kuo KC. 1989. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J Chromatogr 471: 3–36. [DOI] [PubMed] [Google Scholar]
- Gower JC. 1971. A general coefficient of similarity and some of its properties. Biometrics 27: 857–871. [Google Scholar]
- Grosjean H. 2009. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. In DNA and RNA modification enzymes: structure, mechanism, function and evolution (ed. Grosjean H), pp. 1–18. Landes Bioscience, Austin, TX. [Google Scholar]
- Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barre A, Yoshizawa S, Fourmy D, de Crecy-Lagard V, et al. 2014. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet 10: e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenu S, Hennion MC. 1994. On-line sample handling of water-soluble organic pollutants in aqueous samples using porous graphitic carbon. J Chromatogr A 665: 243–251. [Google Scholar]
- Gundersen JL. 2001. Separation of isomers of nonylphenol and select nonylphenol polyethoxylates by high-performance liquid chromatography on a graphitic carbon column. J Chromatogr A 914: 161–166. [DOI] [PubMed] [Google Scholar]
- Harris EH. 2009. Appendix: resources for the investigator. In The chlamydomonas sourcebook, 2nd ed., (ed. Witman EH et al. ), pp. 303–308. Academic Press, London. [Google Scholar]
- He X, Kozak M. 2012. Development of a liquid chromatography–tandem mass spectrometry method for plasma-free metanephrines with ion-pairing turbulent flow online extraction. Anal Bioanal Chem 402: 3003–3010. [DOI] [PubMed] [Google Scholar]
- Helm M, Alfonzo JD. 2014. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol 21: 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Alfonzo JD. 2013. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscipl Rev RNA 4: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. 2014a. Profiling of RNA modifications by multiplexed stable isotope labelling. Chem Commun (Camb) 50: 3516–3518. [DOI] [PubMed] [Google Scholar]
- Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M. 2014b. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res 42: e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P. 2008. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24: 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JH, Ross P. 1997. Carbon-based packing materials for liquid chromatography. Adv Chromatogr 74: 162. [PubMed] [Google Scholar]
- Kösters M, Leufken J, Schulze S, Sugimoto K, Klein J, Zahedi RP, Hippler M, Leidel SA, Fufezan C. 2018. pymzML v2.0: introducing a highly compressed and seekable gzip format. Bioinformatics 34: 2513–2514. [DOI] [PubMed] [Google Scholar]
- Lecanda A, Nilges BS, Sharma P, Nedialkova DD, Schwarz J, Vaquerizas JM, Leidel SA. 2016. Dual randomization of oligonucleotides to reduce the bias in ribosome-profiling libraries. Methods 107: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leufken J, Niehues A, Sarin LP, Wessel F, Hippler M, Leidel SA, Fufezan C. 2017. pyQms enables universal and accurate quantification of mass spectrometry data. Mol Cell Proteomics 16: 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Shi J, Zeng L, Wang T, Cai Y, Jiang G. 2011. Evaluation of graphene as an advantageous adsorbent for solid-phase extraction with chlorophenols as model analytes. J Chromatogr A 1218: 197–204. [DOI] [PubMed] [Google Scholar]
- Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Römpp A, Neumann S, Pizarro AD, et al. 2011. mzML—a community standard for mass spectrometry data. Mol Cell Proteomics 10: R110 000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiring HD, van der Heeft E, ten Hove GJ, de Jong APJM. 2002. Nanoscale LC–MS(n): technical design and applications to peptide and protein analysis. J Sep Sci 25: 557–568. [Google Scholar]
- Michel M, Buszewski B. 2009. Porous graphitic carbon sorbents in biomedical and environmental applications. Adsorption 15: 193–202. [Google Scholar]
- Nedialkova DD, Leidel SA. 2015. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161: 1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst M, Grass J, Fischl R, Leonard R, Jin C, Hinterkorner G, Borth N, Altmann F. 2010. Nucleotide and nucleotide sugar analysis by liquid chromatography-electrospray ionization-mass spectrometry on surface-conditioned porous graphitic carbon. Anal Chem 82: 9782–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L. 2008. Porous graphitic carbon as a stationary phase in HPLC: theory and applications. J Liquid Chromatogr Relat Technol 31: 1687–1731. [Google Scholar]
- Reepmeyer JC, Brower JF, Ye H. 2005. Separation and detection of the isomeric equine conjugated estrogens, equilin sulfate and Δ8,9-dehydroestrone sulfate, by liquid chromatography–electrospray-mass spectrometry using carbon-coated zirconia and porous graphitic carbon stationary phases. J Chromatogr A 1083: 42–51. [DOI] [PubMed] [Google Scholar]
- Ross R, Cao X, Yu N, Limbach PA. 2016. Sequence mapping of transfer RNA chemical modifications by liquid chromatography tandem mass spectrometry. Methods 107: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SP, Limbach PA. 2013. Evaluating the reproducibility of quantifying modified nucleosides from ribonucleic acids by LC-UV-MS. J Chromatogr B Analyt Technol Biomed Life Sci 923–924: 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin LP, Leidel SA. 2014. Modify or die? - RNA modification defects in metazoans. RNA Biol 11: 1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. 2012. Translation drives mRNA quality control. Nat Struct Mol Biol 19: 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal A, Arora G, Sajid A, Maji A, Bhat A, Virmani R, Upadhyay S, Nandicoori VK, Sengupta S, Singh Y. 2013. Regulation of homocysteine metabolism by Mycobacterium tuberculosis S-adenosylhomocysteine hydrolase. Sci Rep 3: 2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Nagao A, Suzuki T. 2011. Human mitochondrial diseases caused by lack of taurine modification in mitochondrial tRNAs. Wiley Interdiscip Rev RNA 2: 376–386. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Kojima T, Miwa T. 2000. Ion chromatography of inorganic anions on graphitic carbon as the stationary phase. J High Resolut Chromatogr 23: 590–594. [Google Scholar]
- Thiaville PC, Legendre R, Rojas-Benitez D, Baudin-Baillieu A, Hatin I, Chalancon G, Glavic A, Namy O, de Crecy-Lagard V. 2016. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb Cell 3: 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuring K, Schmid K, Keller P, Helm M. 2016. Analysis of RNA modifications by liquid chromatography-tandem mass spectrometry. Methods 107: 48–56. [DOI] [PubMed] [Google Scholar]
- Torres AG, Batlle E, Ribas de Pouplana L. 2014. Role of tRNA modifications in human diseases. Trends Mol Med 20: 306–314. [DOI] [PubMed] [Google Scholar]
- Wan QH, Shaw N, Davies MC, Barrett DA. 1995. Chromatographic behaviour of positional isomers on porous graphitic carbon. J Chromatogr A 697: 219–227. [Google Scholar]
- Wei FY, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A, Matsui H, Atta M, Michiue H, Fontecave M, et al. 2011. Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest 121: 3598–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C, Elfakir C, Lafosse M. 2010. Porous graphitic carbon: a versatile stationary phase for liquid chromatography. J Chromatogr A 1217: 3201–3216. [DOI] [PubMed] [Google Scholar]
- Zinshteyn B, Gilbert WV. 2013. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet 9: e1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.