Abstract
Alternative splicing (AS) contributes to proteome diversity. As splicing occurs cotranscriptionally, epigenetic determinants such as DNA methylation likely play a part in regulation of AS. Previously, we have shown that DNA methylation marks exons and that a loss of DNA methylation alters splicing patterns in a genome-wide manner. To investigate the influence of DNA methylation on splicing of individual genes, we developed a method to manipulate DNA methylation in vivo in a site-specific manner using the deactivated endonuclease Cas9 fused to enzymes that methylate or demethylate DNA. We used this system to directly change the DNA methylation pattern of selected exons and introns. We demonstrated that changes in the methylation pattern of alternatively spliced exons, but not constitutively spliced exons or introns, altered inclusion levels. This is the first direct demonstration that DNA methylation of exon-encoding regions is directly involved in regulation of AS.
Keywords: alternative splicing, DNA methylation, CRISPR, splicing regulation
INTRODUCTION
Alternative splicing (AS) is an evolutionarily conserved mechanism that increases transcriptomic and proteomic diversity and contributes to organismal complexity by enabling the generation of multiple mRNA products from a single gene (Ast 2004; Barbosa-Morais et al. 2006; Will and Luhrmann 2011; Frenkel-Morgenstern et al. 2012; Kornblihtt et al. 2013). About 95% of human multiexonic genes undergo AS (Pan et al. 2008; Wang et al. 2008). As splicing, or at least the commitment to splicing, occurs cotranscriptionally, epigenetic determinants also play a part in its regulation (Luco et al. 2011; Frenkel-Morgenstern et al. 2012; Kwak et al. 2013). One of these determinants is DNA methylation.
The major site for DNA methylation in mammals is the cytosine at CpG dinucleotide. This epigenetic modification results in 5′ methylation of cytosine (5mC) (Hotchkiss 1948; Li and Zhang 2014). DNA methylation is known to influence gene expression: Hypermethylated CpG islands in promoters may silence transcription of genes (Bird et al. 1985), whereas in gene bodies, hypermethylation may cause elevation in gene expression (Ball et al. 2009; Rauch et al. 2009; Laurent et al. 2010). We recently demonstrated that levels of DNA methylation are higher in genomic regions encoding exons compared to the flanking intron sequences (Gelfman et al. 2013). This suggests a functional role of DNA methylation in splicing regulation. Indeed, knockout of all DNA methylation activity has a profound effect on AS, and manipulation of gene body methylation shows that DNA methylation directly regulates AS (Song et al. 2017). DNA methylation regulates AS by mediating activities of proteins such as CTCF, MeCP2, and HP1 (Shukla et al. 2011; Maunakea et al. 2013; Yearim et al. 2015). Although these data are suggestive, the importance of exonic DNA methylation for regulation of AS has not been directly demonstrated.
DNA methyltransferase 3A (DNMT3A) is a part of the DNMT3 protein family, and it is one of the three conserved enzymes responsible for CpG methylation in mammals (Okano et al. 1998). Its function is essential for normal development (Li et al. 1992; Okano et al. 1999). DNMT3A is responsible for de novo DNA methylation and also error correction (Chedin 2011). After fertilization all DNA methylation is erased, and, at the blastocyst stage, the DNA methylation pattern is reestablished by DNMT3A (Wang et al. 2014). Structurally, DNMT3A has a catalytic domain, a PWWP domain, and a conserved ATRX-DNMT3-DNMT3L domain (ADD domain) (Chen et al. 2004; Chedin 2011). Another member of the DNMT3 protein family is DNMT3-like protein (DNMT3L) (Aapola et al. 2000). DNMT3L lacks catalytic function, as its catalytic domain is truncated, and it is inactive as a DNA methyltransferase. DNMT3L interacts with DNMT3A (Hata et al. 2002) and significantly stimulates its activity (Chedin et al. 2002; Chen et al. 2005).
DNA demethylation is a multistep process in which 5mC is eventually replaced with a cytosine base (Wu and Zhang 2010). In the first and main step of demethylation, 5mC is converted to 5hmC by ten-eleven translocation methylcytosine dioxygenase 1 (TET1) (Guo et al. 2011; Xu et al. 2011), a member of the TET protein family that mostly functions in the demethylation pathway (Ito et al. 2010, 2011). TET1 has a cysteine-rich and DSBH domain (CD), which is the catalytic domain of the enzyme (Tahiliani et al. 2009). This domain is sufficient to ensure demethylation of DNA (Maeder et al. 2013).
To manipulate DNA methylation in vivo in a site-specific manner we established a method that relies on the clustered regularly interspaced short palindromic repeats (CRISPR) system of bacterial DNA editing. This system has previously been modified to allow epigenetic editing (Jinek et al. 2012; Qi et al. 2013). Use of the deactivated endonuclease Cas9 (dCas9) allows targeting of a specific DNA sequence without cleavage; in dCas9 both of its endonuclease domains are mutated, but the protein maintains its ability to bind DNA and single guide RNA (sgRNA) (Gilbert et al. 2013). dCas9 has been used in various biological applications (Mali et al. 2013). Various techniques to change DNA methylation have been reported. Some are quite difficult to execute and have been limited mainly to promoters and transcription start sites (van der Gun et al. 2010; Maeder et al. 2013; Siddique et al. 2013; Bernstein et al. 2015), whereas others use dCas9 as a platform to target the DNA (Choudhury et al. 2016; Liu et al. 2016; Morita et al. 2016; Xu et al. 2016; Okada et al. 2017). None of these studies have examined how DNA methylation of exons and introns affect pre-mRNA splicing and AS in vivo.
Using dCas9 fused to enzymes that methylate or demethylate DNA, we found that DNA methylation occurs upstream of the sgRNA binding site, whereas the DNA demethylation occurs near the sgRNA binding site. We used this method to manipulate DNA methylation at specific sites within exons and introns and investigated how DNA methylation affects exon inclusion. Our results demonstrated that changes in the methylation pattern of alternatively spliced exons, but not introns or constitutively spliced exons, altered inclusion level. This provides the first direct evidence that DNA methylation of exons is involved in regulation of AS.
RESULTS
We have previously shown that exons have higher levels of DNA methylation than introns (Gelfman et al. 2013), and we and others also showed that DNA methylation plays a regulatory role in alternative splicing of various exons through mechanisms mediated by different proteins (Shukla et al. 2011; Maunakea et al. 2013; Yearim et al. 2015). Reported experiments do not distinguish whether DNA methylation of exons or introns (or both) is involved in the regulation of these alternative events. In order to examine the importance of DNA methylation of specific gene regions on alternative splicing regulation we developed a method to change DNA methylation in vivo in a site-directed manner. To do so, we utilized the CRISPR technique to target DNA methylation to a specific sequence by fusing the DNMT3A-DNMT3L protein to the deactivated form of Cas9 (dCas9-DNMT3A-3L). We obtained the DNMT3A-DNMT3L from pcDNA3.1-VAZF-3a3lsc(27Linker) plasmid (Siddique et al. 2013) (see Materials and Methods). dCas9 can bind to a specific DNA sequence targeted by sgRNA molecules, but due to the lack of endonuclease activity does not cleave the DNA. Co-transfection of dCas9-DNMT3A-3L with sgRNAs is expected to result in methylation of the targeted genomic region with minimal off-target methylation (Fig. 1). We also designed a reciprocal method to demethylate DNA in vivo site specifically. We fused dCas9 to the core domain of an enzyme necessary for demethylation, TET1. The sequence encoding the catalytic domain (CD domain) of the TET1 protein (Maeder et al. 2013) was cloned downstream from dCas9 to enable synthesis of the fusion protein referred to as dCas9-TET1.
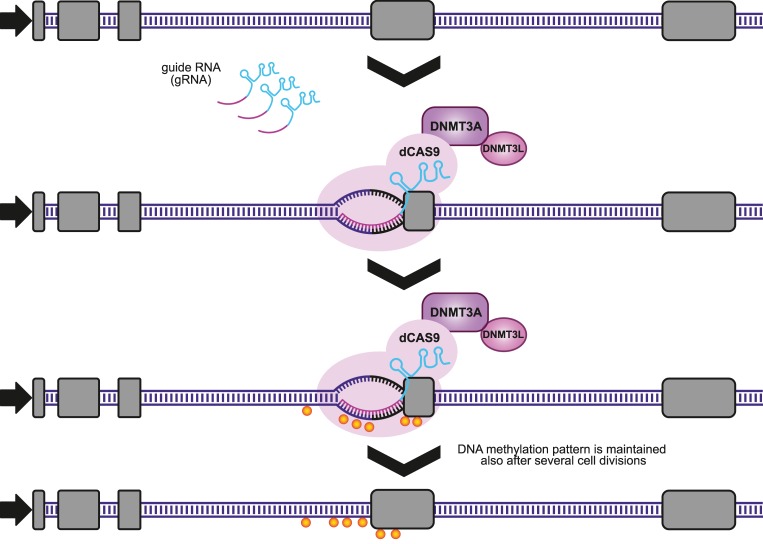
FIGURE 1.
Schematic illustration of region-specific DNA methylation manipulations using the CRISPR system. Cells are co-transfected with a plasmid for expression of dCas9-DNMT3A-3L and the sgRNA expression plasmids. The dCas9 fusion protein is guided to a specific region of the genome by the sgRNAs where it brings DNMT3A-3L close to the DNA, resulting in site-specific methylation. As the transfection is transient, after several cell divisions, the fusion protein expression is lost but the DNA methylation pattern is maintained by the endogenous DNMT1 during genomic DNA replication. A similar method involving the dCas9-TET1 fusion enabled site-specific demethylation of DNA. The yellow spheres indicate methylated CpG.
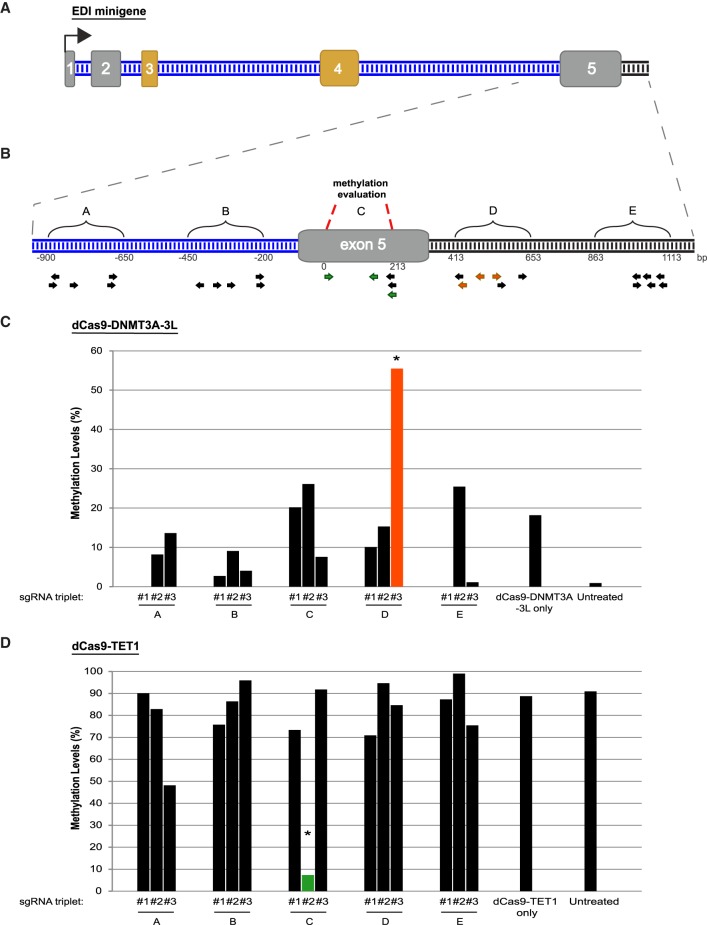
To validate the region-specific DNA methylation manipulation system, we used two different cell lines in which the EDI minigene is integrated into the same genomic position and in the same orientation (Yearim et al. 2015). The precise genomic location of the minigene integration site is not known (this is a proprietary Invitrogen cell line); however, the supplier states that the integration site is intragenic and does not interfere with gene expression of other genes. In one of these Flp-In-HEK293 cell lines all CpG sites in the EDI minigene are methylated (Met+), whereas in the other cell line the EDI minigene is unmethylated (Met−). We previously showed that the original DNA methylation status is maintained during passages (Yearim et al. 2015). The EDI minigene is composed of five exons separated by four introns, and exons 3 and 4 are alternatively spliced (exon skipping). EDI is expressed from a fibronectin promoter that drives expression in a DNA methylation-independent manner (Yearim et al. 2015). EDI minigene was used in previous studies to examine the exon 4 mode of splicing (known as EDI exon [Kadener et al. 2002; de la Mata et al. 2003]), whereas we study the splicing regulation of the whole minigene (Fig. 2A).
FIGURE 2.
In vivo site-specific changes in DNA methylation induced by dCas9-DNMT3A-3L and dCas9-TET1. (A) Schematic illustration of EDI minigene. Gray and orange boxes indicate constitutive and alternative exons, respectively. Blue and black lines indicate introns and pFRT sequence, respectively. Black arrow represents the transcription start site (TSS). (B) Enlargement of EDI exon 5 region. Regions targeted by sgRNAs are indicated by brackets and letters above the gene schematic. Arrows below the gene schematic represent sgRNA binding sites. The arrow colors represent the different pools of sgRNAs used in panels C and D. Red dashed lines indicate the region analyzed for DNA methylation. (C,D) Methylation levels within exon 5 in (C) Met– cells transfected with dCas9-DNMT3A-3L or (D) Met+ cells transfected with dCas9-TET1 and a combination of three different sgRNAs (from the five or six sgRNAs designed to each region shown in panel B). Different combinations of sgRNAs (marked #1, #2, or #3 below bars) were tested. “dCas9-DNMT3A-3L only” and “dCas9-TET1 only” are controls without sgRNAs and “untreated” indicates cells without transfection. (*) Indicates P-value <0.00503 for data shown in panel C and P-value <1.51 × 10−07 for panel D (t-test).
We first examined where the DNA methylation activity of dCas9-DNMT3A-3L occurs relative to the binding site of sgRNA. sgRNAs were designed (Supplemental Table S1) to regions up- and downstream and within exon 5 of the EDI minigene (Fig. 2B). In design of sgRNAs, we used the web tool (http://crispr.mit.edu/). This tool prepares a list of possible sgRNAs in a given DNA sequence and ranks the sgRNAs based on the presence of a PAM sequence, the number of possible off-target hybridization sites in the genome in question, and the genomic location of the site (exonic, intronic, or intergenic). To address this, in each attempt we used different three sgRNAs combination (sgRNA triplet #1, #2, or #3) that target sites throughout a region of about 250 bp (Fig. 2B).
Met– cells were transfected with dCas9-DNMT3A-3L and different sgRNA plasmid combinations and grown for 4 d. Genomic DNA was extracted and treated with bisulfite (BS), which converts cytosine residues to uracil but leaves 5-methylcytosine residues unaffected. The BS-treated genomic DNA was sequenced with primers specific for exon 5 region of the EDI minigene. Analysis of methylation of exon 5 in cells treated with various combinations of three sgRNAs indicated that only dCas9-DNMT3A-3L in combination with sgRNAs complementary to sequences downstream from exon 5 resulted in methylation in exon 5 (Fig. 2B, orange arrows and Fig. 2C, orange column). Low level of methylation was observed in untreated cells. The region analyzed for methylation in exon 5 was ∼250 base pairs (bp) upstream of the binding site of the sgRNAs, and DNA methylation was elevated to 55%. The level of DNA methylation in cells that expressed dCas9-DNMT3A-3L without sgRNAs was 17%, which is considered the background effect of dCas9-DNMT3A-3L.
We also examined the ability of the dCas9-TET1 construct to demethylate DNA in specific sites (Fig. 2D). Plasmids for expression of dCas9-TET1 and combinations of three sgRNAs were transfected into Met+ cells. After 4 d, CpG methylation in exon 5 was evaluated. The results indicate that dCas9-TET1 indeed demethylate the DNA. We also found that dCas9-TET1 operates close to sgRNA binding site, as the level of methylation in exon 5 was reduced from ∼90% in untreated cells to ∼7% in cells that expressed dCas9-TET1 and a sgRNA combination with complementarity to exon 5 (Fig. 2B, green arrows and Fig. 2D, green column). Thus, with these two fusion proteins and the right combination of sgRNAs, it is possible to manipulate DNA methylation in a desired region of the genome in live cells. To assess possible off-target activity of the sgRNAs, we examined the methylation status for one or two top off-target hybridization sites for each positive sgRNAs co-transfected with dCas9-DNMT3A-3L or dCas9-TET1, used in Figure 2. For most of the sgRNAs, we did not observe any significant changes in the DNA methylation between the different samples. This indicates there was not an off-target binding or activity of dCas9 fused protein with or without any of the sgRNAs (Supplemental Fig. S1).
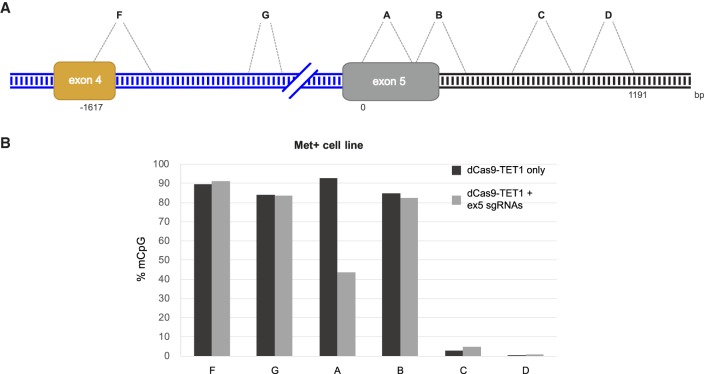
In order to further investigate the functionality of the fusion protein dCas9-TET1, we examined a larger region of the EDI minigene for effects on methylation. We overexpressed dCas9-TET1 with the sgRNA combination targeting exon 5 (Fig. 2B, green arrows). DNA methylation was evaluated in six regions spanning ∼1 kb upstream of exon 5 (Fig. 3A regions F, G), and downstream from exon 5 (Fig. 3A regions B, C, D) and within exon 5 (Fig. 3A, region A). After BS treatment, only these regions were amplified with specific primers and deep sequenced to acquire reads specific for the EDI locus. The reads were mapped to the EDI minigene to identify sites of DNA methylation. The CpG methylation levels of the control sample (dCas9-TET1 without sgRNA) in the regions in the EDI minigene (F, G, A, B) were high as expected for these Met+ cells (Fig. 3B). In cells in which both dCas9-TET1 and sgRNA were expressed, methylation in region A was lower by ∼50% compared to the control sample; methylation was not altered in regions B, F, or G (Fig. 3B). Regions C and D had low levels of CpG methylation in both control and test samples (Fig. 3B). We assume it is related to the fact that they are located within the pFRT integration plasmid and the lacZ promoter, which is part of it, and thus are not uniformly DNA methylated as the EDI minigene. Overall, these results indicate that the range of demethylation by dCas9-TET1 fusion protein is limited to a few hundred base pairs surrounding the sgRNA target zone.
FIGURE 3.
dCas9-TET1 induces DNA demethylation in a limited region. (A) Schematic illustration of EDI minigene exons 4 and 5. Exon 4 is alternatively spliced, whereas exon 5 is constitutively spliced. Blue and black lines indicate EDI introns and pFRT sequence, respectively. The six regions analyzed for methylation are indicated with letters. (B) The level of methylated CpGs (in percent) within the six regions denoted in panel A after transfection of dCas9-TET1 plasmid into Met+ cells with or without sgRNA expression (black and gray bars, respectively).
DNA methylation of exons affects AS
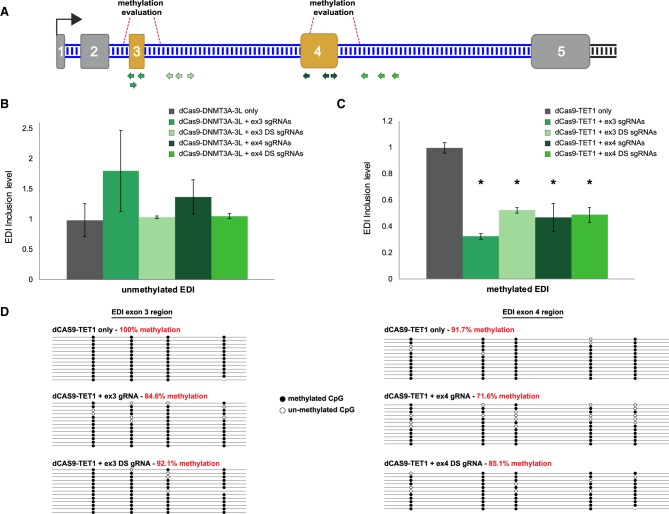
Next, we manipulated DNA methylation within the EDI minigene and examined how this influenced AS. sgRNAs were designed to the alternative exons in the EDI minigene, exon 3 and exon 4, and intronic regions located 250 bp downstream from these exons (Fig. 4A). We overexpressed dCas9-DNMT3A-3L or dCas9-TET1 with the various sgRNA combinations in Met− or Met+ cell lines, respectively. Each region was targeted with three sgRNAs to target the fusion proteins to an ∼200-bp region. After 4 d, total RNA was extracted, and the EDI splicing pattern (the inclusion level of both alternative exons 3 and 4) was examined by reverse transcription followed by qualitative PCR (RT-qPCR) analysis.
FIGURE 4.
Changes of DNA methylation level in alternative exons affect alternative splicing. (A) Schematic illustration of EDI minigene. Small arrows in various green shades indicate sgRNA binding sites. Red dashed lines indicate regions where methylation was evaluated. (B,C) RT-qPCR analysis from (B) Met– cells transfected with dCas9-3A3L and the indicated sgRNAs and (C) Met+ cells transfected with dCas9-TET1 and the indicated sgRNAs. Values were normalized to levels in untreated cells. Plotted are means ± SEM, (*) indicates P-value <0.003 (t-test), n = 3. (D) Lollipop maps of bisulfite sequencing analysis of exon 3 (left) and exon 4 (right) from conditions indicated. In red are the methylation percentages. Methylated CpG and unmethylated CpG are shown in black and white circles, respectively.
dCas9-DNMT3A-3L and dCas9-TET1 exerted different effects on the splicing of the EDI pre-mRNA. In Met− cells, dCas9-DNMT3A-3L activity caused an elevation in the inclusion level of the alternatively spliced exons when the sgRNAs targeted the methylation activity to the exons themselves (Fig. 4B). Although these alterations in the inclusion levels were not statistically significant, the direction in the shift of the splicing pattern is visible. In contrast, in Met+ cells, when sgRNAs targeted dCas9-TET1 to exons or to introns, the inclusion levels decreased significantly (Fig. 4C). This indicates that DNA methylation has a positive effect on the AS regulation of the EDI minigene, since the lack of DNA methylation hinders the recognition of exon 3 & 4 as exons and their inclusion in the mature mRNA. This effect on the EDI inclusion levels was seen only when the alternative exons were targeted, as the inclusion level did not change when a constitutive exon (exon 5, for DNA methylation levels see Fig. 2) or the middle of an intron were targeted (Supplemental Fig. S2).
DNA methylation levels for each of the two alternatively spliced exons were examined in the Met+ cell line after overexpressing dCas9-TET1 with or without the sgRNA triplets. When dCas9-TET1 with sgRNAs targeting alternatively spliced exons or introns, there was a decrease in the methylation levels compared to cells in which only dCas9-TET1 was expressed (Fig. 4D). Although the change in the methylation was not large (between 15% and 30%), it was enough to cause a shift in the inclusion level of the alternative exons. Moreover, no significant change in the transcription level of the EDI minigene was observed in the Met+ cell line after dCas9-TET1 OE with or without sgRNAs. In the Met− cell line, a slight down-regulation was observed when the dCas9-DNMT3A-3L was targeted downstream from one of the alternative exons. However, this down-regulation did not correspond to the decrease in the inclusion level of the alternative exons of the EDI minigene (Supplemental Fig. S3).
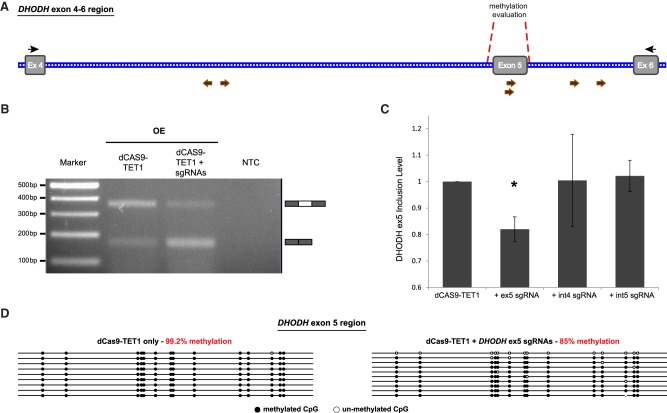
To further examine the importance of DNA methylation of exon on the regulation of exon selection and splicing, we used this technique to demethylate an exon in an endogenous gene and examined its splicing pattern in HCT 116 cells, a colorectal carcinoma cell line. We chose exon 5 of the DHODH gene as it inclusion levels were down-regulated in DNMT1 and DNMT3B double knockout HCT 116 cells compared to the wild-type HCT 116 cells (Maunakea et al. 2013). Various sgRNAs were designed to target either DHODH exon 5 or its flanking introns. Intron 4 and intron 5 targeted sgRNAs were designed to hybridize approximately at the middle of the introns and away from any splice sites (Fig. 5A). These sgRNAs were then cloned into separate expression vectors. The sgRNAs (two per region) were co-transfected with the dCas9-TET1 expression plasmid into HCT 116 cells. As a control, cells were transfected with only the dCas9-TET1 plasmid. Following an incubation period of 4 d, cells were harvested and total RNA was extracted. Initially, to examine the splicing pattern of the exon, reverse transcription PCR (RT-PCR) was performed using primers designed to evaluate splicing of exon 5. In the presence of the DHODH exon 5 targeted sgRNAs, the splicing pattern shifts toward the skipped isoform as compared to the control sample (Fig. 5B). To further this, RT-qPCR analysis indicated a significant decrease of ∼20% in the inclusion level of DHODH exon 5 when exon 5 was targeted for demethylation, whereas no significant change in splicing was observed when sgRNAs targeting the surrounding introns were used (Fig. 5C). The DNA methylation levels of DHODH exon 5 were also evaluated by extracting genomic DNA and performing BS conversion and sequencing with primers specific for the exon 5 region. The DHODH exon 5 is fully methylated when dCas9-TET1 is not targeted to exon 5 (Fig. 5D, left). The DNA methylation levels decreased by 15% when exon 5 targeting sgRNAs were used (Fig. 5D, right). This decrease in methylation levels is similar to that observed when the EDI minigene alternative exons were targeted for demethylation (Fig. 4D). This further indicates that DNA methylation of exon is more important for splicing regulation than introns.
FIGURE 5.
dCas9-TET1 targeting of DHODH exon 5 affects alternative splicing. (A) Schematic illustration of genomic region of DHODH from exon 4 to exon 6. Gray boxes and blue lines indicate exons and introns, respectively. Brown and black arrows represent sgRNA binding sites and RT-PCR primer binding sites, respectively. Red dashed lines indicate the region analyzed for DNA methylation. (B) Gel electrophoresis of RT-PCR products of the DHODH exon 5 region after overexpression of dCas9-TET1 with or without sgRNAs targeting exon 5 of DHODH gene in HCT116 cells. RNA was extracted, reverse transcribed, and primers were used to amplify the included (370 bp) or the skipped (172 bp) isoforms. NTC is non-template control. (C) HCT116 cells were either transfected with dCas9-TET1 only (control) or co-transfected with dCas9-TET1 and sgRNAs targeting different regions: DHODH exon 5 itself (+ex5 gRNA), DHODH intron 4 (+int4 gRNA), or DHODH intron 5 (+int5 gRNA). For each sample, RT-qPCR was performed to analyze inclusion levels (for primer sequences see Supplemental Table S1). Plotted are means ± SEM, (*) indicates P-value <7.70539 × 10−05 (t-test), n = 3. (D) Lollipop maps of BS sequencing analysis of DHODH exon 5 from indicated treatments. In red are the methylation percentages. Methylated CpG and unmethylated CpG are shown in black and white circles, respectively.
DISCUSSION
Here we demonstrate a method, based on the CRISPR technology (Mali et al. 2013; Choudhury et al. 2016; Liu et al. 2016; Morita et al. 2016; Okada et al. 2017), for in vivo site-specific manipulation of DNA methylation in a targeted manner. By transient expression a dCas9 fused to either a DNA methyltransferase (DNMT3A-3L) or a component of the demethylation system (TET1) and sgRNAs, we demonstrated alterations in CpG methylation in proximity of the sgRNA binding sites. The biochemical pathway of DNA methylation in the nucleus is a simple and straight forward addition of a methyl group to the cytosine base and that involves one enzyme, a DNA methyltransferase (Jones and Liang 2009). The opposite process of demethylation is complex and involves multiple enzymes and replacement of the 5-methyl C base with a C (Wu and Zhang 2010). The TET1 enzyme is the first to act and triggers the rest of the process (Guo et al. 2011; Xu et al. 2011). So, it is interesting, but not surprising, that dCas9-DNMT3A-3L and dCas9-TET1's direction of function is not the same; one functions upstream of its DNA binding site, whereas the other functions on a range of a few hundreds of bases at the DNA binding site itself, respectively. We attribute this difference to the location of the active site in the fused protein. Perhaps these results can imply to the function of the endogenous enzymes in vivo. This suggests that dCas9 might be a useful platform for analysis of functions of various DNA and chromatin binding proteins.
How exons, with an average length of 150 bp, which are engulfed by introns of thousands of base pairs (Gelfman et al. 2012), are recognized is a question that researchers have only partially been able to answer. We and others have previously shown that chromatin organization and epigenetic markers play a major role in the process. There is higher nucleosome occupancy on exons compared to introns (Schwartz et al. 2009; Tilgner et al. 2009), and there is more DNA methylation of exons than flanking introns (Gelfman et al. 2013). We have shown that DNA methylation directly regulates AS of exons in a genome-wide manner and specifically regulates splicing of the EDI minigene (Yearim et al. 2015). These experiments could not distinguish effects of DNA methylation levels in exons and introns. Here, we demonstrated that DNA methylation of exons has a greater effect on the inclusion level of alternatively spliced exons than does the level of DNA methylation of introns.
In this work, we presented two examples, in different cell types, of how manipulating the DNA methylation level of exons affects their splicing. Significant changes in the AS of exons of the EDI minigene were observed when a small reduction in the percentage of CpG methylation of the alternatively spliced exon was induced using dCas9-TET1 (Fig. 4C,D). When DHODH exon 5 was targeted for demethylation, there were small but significant changes in the inclusion level of the exon and decrease in its DNA methylation (Fig. 5C,D). That DNA methylation had a stronger effect on the EDI minigene may be due to the tight regulation of DHODH exon 5 splicing because DHODH enzyme has an essential role in the cell. DHODH gene encodes dihydroorotate dehydrogenase which is involved in pyrimidine biosynthesis (Munier-Lehmann et al. 2013) and associated with the electron transfer chain in the mitochondria (Fang et al. 2013). Inhibitors of this enzyme have been tested as treatments for various types of cancer such as melanoma (White et al. 2011; Dorasamy et al. 2017) and leukemia (Lewis et al. 2016) as well for autoimmune diseases such as rheumatoid arthritis (Munier-Lehmann et al. 2013).
In both examples we targeted for demethylation, sgRNAs were designed to hybridize to internal exons and thus affect their AS. There may be specific CpG methylation positions that are more important than the others, but at the resolution of the current experiments, this cannot be determined. Mechanistically, DNA methylation may influence AS by altering the level of tri-methylation of histone 3 at lysine 9 (H3K9me3). The H3K9me3 recruits HP1 proteins, which bind several AS regulatory proteins, to generate a high concentration of these regulatory proteins on alternative exons at the chromatin level (Yearim et al. 2015). We assume that during transcription, the splicing factors move from the chromatin onto the alternative exons at the RNA level and thus regulate their splicing in a site-specific manner.
Processes other than splicing are also likely to be influenced by DNA methylation. For example, it has been shown by others that methylation of promoters can cause gene silencing (Choudhury et al. 2016; Liu et al. 2016; Morita et al. 2016; Xu et al. 2016; Okada et al. 2017). The method we describe here has potential to be a powerful tool to investigate many different regulatory processes. This method can be used to examine whether epigenetic features such as GC content, chromatin modifications, or genomic location (inter or intra-genic) impact the activity of TET1 or DNMT3A enzymes and will promote our understanding of how DNA methylation regulates various cellular activities.
MATERIALS AND METHODS
dCas9-DNMT3A-3L cloning
The cloning of the pcDNA-dCas9-DNMT3A-DNMT3L-T2A-Puromycin (dCas9-3A3L) was done using Gibson assembly as described in Gibson et al. (2009). Briefly, 1 µg pcDNA3.1-dCas9 plasmid was restricted with AflII (NEB), downstream from dCas9 CDS, according to manufacturer's instructions. DNMT3A-DNMT3L and T2A-Puromycin insert were amplified from pcDNA3.1-VAZF-3a3lsc(27Linker) (Siddique et al. 2013) and pX459 (Ran et al. 2013), respectively using Q5 High Fidelity DNA Polymerase (NEB) according to the manufacturer's instructions. Primers were designed according to the following guidelines: The binding section (bold and underlined) should have an annealing temperature of ∼60°C and be no longer than 28 bp; the homology section of the primer was 40 bp. The primers were synthesized at IDT, and sequences are as follows:
DNMT3A-DNMT3L insert
GIB dCas9_3A Fw:
5′-gtgaccccaagaagaagaggaaggtgggcGgatctagacAcggcccaaaaaagaagagaaagg-3′
GIB 3L Rev 5′-caccgcatgttagcagacttcctctgccctctccactgccatggtgatggtgatgatgaccg-3′
T2A-Puromycin insert
GIB 3L_pur: 5′-ggatctgaatatgcataccggtcatcatcaccatcaccatggcagtggagagggcaga-3′
GIB pur_dCAS Rev: 5′-caactagaaggcacagtcgaggctgatcagcggtttaaactcaggcaccgggcttgc-3′
Special considerations in the design were disruption of a stop codon (represented by capital G in the GIB dCas9_3A Fw primer) and the maintenance of a continuous open reading frame by adding 1 nucleotide (nt) as spacer between inserts (represented by capital A in GIB dCas9_3A Fw primer). A single product was validated on 1% agarose gel. The inserts were size selected from the gel and purified using the Wizard SV Gel and PCR Clean-Up System (Promega). Gibson assembly was done in 15-µL reaction mix as previously described (Gibson et al. 2009) containing ISO buffer X1, T5 exonuclease, Phusion polymerase, and Taq ligase, supplemented with 3.1 µL linear pcDNA3.1-dCas9 plasmid (100 ng), 0.5 µL each insert, and 0.9 µL nuclease free water. The reaction was incubated at 50°C for 1 h. A 10-µL aliquot of this reaction was used to transform competent 10XL Gold E. coli after heat shock (ampicillin resistance selection was used). Plasmids were extracted from 16 colonies using Wizard Plus SV Miniprep Kit (Promega). Sequences were validated using a standard colony PCR protocol with several primer pairs tailing the construct. Final validation was performed by Sanger sequencing at the Inter-Departmental Research Facility Unit.
dCas9-TET1 cloning
The cloning of the pcDNA-dCas9-TET1_CD-T2A-Puromycin (dCas9-TET1) was done using Gibson assembly essentially as described above using the following primers:
TET1_CD insert
TET1CD_C7 GIB FWD: 5′-tgaccccaagaagaagaggaaggtgggcGgatctagacAAagcattgtggcccagctg-3′
TET1CD_C7 GIB REV: 5′-ctcttctgagatgagtttttgttcgaagggccctctagagacccaatggttatagggccc-3′
T2A-Puromycin insert
Puro_TET1 GIB FWD: 5′-tgctctcacacacgttgcggggccctataaccattgggtctctagagggcccttcgaaca-3′
GIB pur_dCAS Rev: 5′-caactagaaggcacagtcgaggctgatcagcggtttaaactcaggcaccgggcttgc -3′
A stop codon (represented by capital G in TET1CD_C7 GIB FWD primer) was disrupted, and the open reading frame was maintained by adding 2 nt as spacer between inserts (represented by AA at TET1CD_C7 GIB FWD primer). The TET1_CD insert was amplified from pJFA344C7 (Maeder et al. 2013).
sgRNA design and cloning
sgRNA expression plasmid pX552 (Swiech et al. 2015) was cut with SapI (NEB) according to manufacturer's protocol, verified by agarose gel electrophoresis, and purified using Wizard Plus SV Miniprep DNA Purification System (Promega). All sgRNA sequences were designed using the CRISPR Design Tool (http://crispr.mit.edu/). Sequences with the highest scores were chosen. Each sgRNA was designed together with its complement sequence (sgRNA_comp), and 3 nt, ACC or CAA, were added to the 5′ end of each, respectively, to complement the SapI restriction site. Each sgRNA oligonucleotide and complementary oligonucleotide was phosphorylated at the 5′ end using T4 Polynucleotide Kinase (NEB) in T4 ligase buffer (NEB) according to manufacturer's protocol. sgRNA and sgRNA_comp oligonucleotides were annealed (thermo-cycling program: 30 min at 37°C PNK reaction, 20 min at 65°C heat inactivation, 5 min at 95°C denaturation and ramp to 4°C at 0.5°C per sec to anneal). For each ligation reaction, sgRNAs were diluted 1:25 dilution. Ligations of sgRNA inserts with linearized pX552 were done using T4 ligase (NEB) according to manufacturer's instructions (50 ng vector with 1 µL diluted sgRNA in a 20-µL reaction volume). A 2-µL aliquot of the ligation reaction was used to transform competent 10XL Gold E. coli after heat shock (ampicillin selection was used). Plasmids extracted from three colonies were purified using the Wizard Plus SV Miniprep Kit (Promega), and positive colonies were confirmed by Sanger sequencing at the Inter-Departmental Research Facility Unit.
dCas9-DNMT3A-3L or dCas9-TET1 and sgRNA overexpression
HEK293 cells with stably integrated unmethylated or methylated EDI (Yearim et al. 2015) were seeded into 12-well plates (140,000 cells in each well). For dCas9-DNMT3A-3L overexpression, cells were co-transfected 24 h after plating with either with 400 ng dCas9-3A3L plasmid and 177 ng of each of three different sgRNA expression plasmids or 931 ng dCas9-3A3L plasmid or were untreated. For dCas9-TET1 overexpression, at 24 h after plating, cells were co-transfected with 400 ng of dCas9-TET1 plasmid and 166 ng of each of three sgRNA expression plasmids or 898 ng of dCas9-TET1 plasmid or were untreated. Transfection was done using TransIT-LT1 (Mirus Bio), according to the manufacturer's recommended protocol. A similar protocol was used in experiments with the HCT 116 cells with the following changes: approximately 300,000 cells were seeded in 6-well plate, and after 24 h cells were co-transfected with 1 µg of dCas9-TET1 plasmid and 415 ng of each of two sgRNA expression plasmids or 1.83 µg of dCas9-TET1 plasmid or were untreated. Following transfection, cells were incubated for 4 d and then genomic DNA was extracted using High Pure PCR Template Preparation Kit (Roche) or using the following protocol: Cells from each well were pelleted by centrifugation and washed with PBS. Pellets were resuspended in DNA Lysis Buffer (50 mM Tris-HCl, 60 mM NaCl, 0.65% SDS, 0.8 mg/mL Proteinase K [NEB]) and incubated at 37°C overnight or at 55°C for 3 h. Samples were treated with RNase A. DNA was extracted using Phase Lock Gel tubes (5Prime) and phenol:chloroform:isoamyl alcohol (25:24:1) saturated with 100 mM Tris, pH 8.0 (Sigma) according to the protocol provided by 5Prime. One-tenth volume of 3 M NaOAc and 2.5 volumes of cold ethanol were added. Samples were incubated either in −80°C for 1 h or at −20°C overnight and then centrifuged at 10000 rpm for 15 min at 4°C. Then samples were washed with cold 70% ethanol, centrifuged for 5 min at 10,000 rpm at 4°C, and supernatant removed. Pellets were air dried for 15 min and resuspended in nuclease-free water.
Bisulfite DNA methylation analysis
Genomic DNA samples (500 ng) were bisulfate converted using EZ DNA Methylation Kit (Zymo Research). The specific regions of interest in the EDI minigene were amplified by BS-PCR using FastStart High Fidelity PCR System, dNTPack (Roche) according to manufacturer's instructions (thermo-cycling program: 35 cycles where each cycle was consisted of 30 sec at 95°C, 30 sec at 60°C, and 35 sec at 72°C). Special primers designed for bisulfate converted genomic DNA were used. An aliquot of 5 µL of each sample was run on a 1.5% agarose gel to verify that there was a single PCR product. The samples were cloned using TOPO TA Cloning (Invitrogen) according to manufacturer's instructions and transformed into competent XL-10 Gold E. coli after heat shock (kanamycin or ampicillin selection). Positive colonies for the BS-PCR insert were selected, and plasmid extraction was performed using Presto Mini Plasmid Kit (Geneaid). DNA sequencing was performed by the Inter-Departmental Research Facility Unit. The sequences were analyzed using BiQ Analyzer software (Bock et al. 2005) to give CpG methylation maps. BS specific primers are listed in Supplemental Table S1.
Analysis of splicing patterns by RT-PCR and quantitative RT-PCR
Total RNA was extracted from the cells with TRI reagent (Sigma) according to the manufacturers’ instructions. Approximately 1 µg of total RNA was reverse transcribed with SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) using an oligo(dT)20 primer. For RT-PCR of DHODH exon 5 alternatively spliced products, PCR was performed using Biotools DNA Polymerase (Biotools) on a ProFlex PCR System (Applied BioSystems) using the following thermocycling parameters: 2 min at 94°C followed by 30 cycles of 30 sec at 94°C, 45 sec at 59°C, and 30 sec at 72°C, ending with a final extension of 10 min at 72°C. Qualitative PCR was performed using KAPA SYBR FAST Universal qPCR Kit (KAPA Biosystems) using two exon–exon junction primer pairs designed to detect the inclusion isoform and the skipping isoform. To calculate EDI inclusion levels of methylated relative to unmethylated cells from qPCR we used the following equation:
qPCR reactions were performed on an ABI StepOnePlus Real-Time PCR System (Applied Biosystems) using the following thermocycling parameters: 3 min at 95°C followed by 40 cycles of 3 sec at 95°C and 30 sec at 60°C, ending with a dissociation curve. Primer sequences are listed in Supplemental Table S1.
Cell culture
Flp-In-HEK293 cells stably transformed with the methylated or unmethylated EDI minigene were cultured in complete DMEM medium with high glucose (Sigma), 10% fetal bovine serum (Sigma), 2 mg/mL l-alanyl-l-glutamine (Biological Industries Israel), 100 U/mL penicillin, 0.1 mg/mL streptomycin (Biological Industries Israel) supplemented with 100 µg/mL hygromycin B (InvivoGen). HCT 116 human colon carcinoma cells were cultured in complete RPMI medium with high glucose (Sigma), 10% fetal bovine serum (Sigma), 2 mg/mL l-alanyl-l-glutamine (Biological Industries Israel), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Biological Industries Israel). All cells were grown at 37°C in a humidified atmosphere with 5% CO2.
Deep sequencing and bioinformatics analysis of DNA methylation
After genomic DNA extraction and bisulfite conversion, regions of interest were amplified using primers designed for bisulfite DNA sequencing that have a 3′ tail of eight random nucleotides (N). This tail was added in order to remove redundant reads created by PCR bias. The BS-PCR samples were purified using Presto Gel/PCR DNA Fragments Extraction Kit (Geneaid) according to manufacturer's instructions. As a part of the preparation for deep-sequencing, samples amplified from different regions were mixed together to a final concentration of 10 nM. The sequencing libraries were prepared with INCPM ChIP-Seq Kits by the INCPM at the Weizmann Institute (Israel). Deep sequencing of the libraries was done in one lane on a MiSeq Nano V2 (300 cycles) for 150-base paired-end reads by the INCPM. As PCR samples are of low complexity, a PhiX library was added to the run at a concentration of 20%. For each PCR mix, reads (N = 78,000, 95% CI [77000, 79000]) were produced. PCR duplicates were removed and adapters were trimmed using FASTX-Toolkit (hannonlab.cshl.edu/fastx_toolkit/). Unique reads (N = 560, 95% CI [445, 677]) were mapped to EDI minigene sequence. For methylation analysis, Bismark (www.bioinformatics.bbsrc.ac.uk/projects/bismark/) (Krueger and Andrews 2011) and Bowtie 2 (bowtie-bio.sourceforge.net/bowtie2/index.shtml) (Langmead and Salzberg 2012) software were applied to perform read alignment, analyze methylated DNA signaling and output cytosine methylation sites in the EDI minigene. All parameters were set at default values.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
G.A. is supported by grants from the Israel Science Foundation (ISF 61/09 and ISF 142/13). I.K. is partly supported by the Safra Bioinformatics Center at Tel Aviv University.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.064865.117.
REFERENCES
- Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, Shintani A, Minoshima S, Krohn K, Antonarakis SE, et al. 2000. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 65: 293–298. [DOI] [PubMed] [Google Scholar]
- Ast G. 2004. How did alternative splicing evolve? Nat Rev Genet 5: 773–782. [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Carmo-Fonseca M, Aparicio S. 2006. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res 16: 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DL, Le Lay JE, Ruano EG, Kaestner KH. 2015. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J Clin Investig 125: 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. 1985. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell 40: 91–99. [DOI] [PubMed] [Google Scholar]
- Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. 2005. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21: 4067–4068. [DOI] [PubMed] [Google Scholar]
- Chedin F. 2011. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog Mol Biol Transl Sci 101: 255–285. [DOI] [PubMed] [Google Scholar]
- Chedin F, Lieber MR, Hsieh CL. 2002. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci 99: 16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Tsujimoto N, Li E. 2004. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol 24: 9048–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. 2005. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J Cell Biochem 95: 902–917. [DOI] [PubMed] [Google Scholar]
- Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. 2016. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 7: 46545–46556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532. [DOI] [PubMed] [Google Scholar]
- Dorasamy MS, Choudhary B, Nellore K, Subramanya H, Wong PF. 2017. Dihydroorotate dehydrogenase inhibitors target c-Myc and arrest melanoma, myeloma and lymphoma cells at S-phase. J Cancer 8: 3086–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Uchiumi T, Yagi M, Matsumoto S, Amamoto R, Takazaki S, Yamaza H, Nonaka K, Kang D. 2013. Dihydro-orotate dehydrogenase is physically associated with the respiratory complex and its loss leads to mitochondrial dysfunction. Biosci Rep 33: e00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel-Morgenstern M, Lacroix V, Ezkurdia I, Levin Y, Gabashvili A, Prilusky J, Del Pozo A, Tress M, Johnson R, Guigo R, et al. 2012. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res 22: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfman S, Burstein D, Penn O, Savchenko A, Amit M, Schwartz S, Pupko T, Ast G. 2012. Changes in exon-intron structure during vertebrate evolution affect the splicing pattern of exons. Genome Res 22: 35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfman S, Cohen N, Yearim A, Ast G. 2013. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res 23: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. 2011. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129: 1983–1993. [DOI] [PubMed] [Google Scholar]
- Hotchkiss RD. 1948. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem 175: 315–332. [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu sc, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Liang G. 2009. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 10: 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Fededa JP, Rosbash M, Kornblihtt AR. 2002. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc Natl Acad Sci 99: 8185–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. 2013. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14: 153–165. [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis TA, Sykes DB, Law JM, Munoz B, Rustiguel JK, Nonato MC, Scadden DT, Schreiber SL. 2016. Development of ML390: a human DHODH inhibitor that induces differentiation in acute myeloid leukemia. ACS Med Chem Lett 7: 1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Zhang Y. 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926. [DOI] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. 2016. Editing DNA methylation in the mammalian genome. Cell 167: 233–247.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. 2013. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol 31: 1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. 2013. Cas9 as a versatile tool for engineering biology. Nat Methods 10: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Chepelev I, Cui K, Zhao K. 2013. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res 23: 1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, Sakai A, Nakashima H, Hata K, Nakashima K, et al. 2016. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol 34: 1060–1065. [DOI] [PubMed] [Google Scholar]
- Munier-Lehmann H, Vidalain PO, Tangy F, Janin YL. 2013. On dihydroorotate dehydrogenases and their inhibitors and uses. J Med Chem 56: 3148–3167. [DOI] [PubMed] [Google Scholar]
- Okada M, Kanamori M, Someya K, Nakatsukasa H, Yoshimura A. 2017. Stabilization of Foxp3 expression by CRISPR-dCas9-based epigenome editing in mouse primary T cells. Epigenetics Chromatin 10: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. 2009. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci 106: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. 2009. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol 16: 990–995. [DOI] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, Jeltsch A. 2013. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol 425: 479–491. [DOI] [PubMed] [Google Scholar]
- Song K, Li L, Zhang G. 2017. The association between DNA methylation and exon expression in the Pacific oyster Crassostrea gigas. PLoS One 12: e0185224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. 2015. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33: 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001. [DOI] [PubMed] [Google Scholar]
- van der Gun BT, Maluszynska-Hoffman M, Kiss A, Arendzen AJ, Ruiters MH, McLaughlin PM, Weinhold E, Rots MG. 2010. Targeted DNA methylation by a DNA methyltransferase coupled to a triple helix forming oligonucleotide to down-regulate the epithelial cell adhesion molecule. Bioconjug Chem 21: 1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Li G, Ci W, Li W, et al. 2014. Programming and inheritance of parental DNA methylomes in mammals. Cell 157: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. 2011. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu sc, Zhang Y. 2010. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. 2011. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42: 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, Ruan K, Wang F, Xu GL, Hu R. 2016. A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2: 16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm JP, Nissim-Rafinia M, Cohen AH, Rippe K, Meshorer E, et al. 2015. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep 10: 1122–1134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.