Significance
Chromatin comprises the approximately 3 billion bases in the human genome and histone proteins. Histone posttranslational modifications (PTMs) regulate chromatin dynamics and protein transcription to expand the genetic code. Herein we describe the existence of Lys and Arg modifications on histones derived from a glycolytic by-product, methylglyoxal (MGO). These PTMs are abundant modifications, present at similar levels as those of modifications known to modulate chromatin function and leading to altered gene transcription. Using CRISPR-Cas9, we show that the deglycase DJ-1 protects histones from adduction by MGO. These findings demonstrate the existence of a previously undetected histone modification and provide a link between cellular metabolism and the histone code.
Keywords: histone, methylglyoxal, glyoxalase 1, DJ-1, QuARKMod
Abstract
Histone posttranslational modifications (PTMs) regulate chromatin dynamics, DNA accessibility, and transcription to expand the genetic code. Many of these PTMs are produced through cellular metabolism to offer both feedback and feedforward regulation. Herein we describe the existence of Lys and Arg modifications on histones by a glycolytic by-product, methylglyoxal (MGO). Our data demonstrate that adduction of histones by MGO is an abundant modification, present at the same order of magnitude as Arg methylation. These modifications were detected on all four core histones at critical residues involved in both nucleosome stability and reader domain binding. In addition, MGO treatment of cells lacking the major detoxifying enzyme, glyoxalase 1, results in marked disruption of H2B acetylation and ubiquitylation without affecting H2A, H3, and H4 modifications. Using RNA sequencing, we show that MGO is capable of altering gene transcription, most notably in cells lacking GLO1. Finally, we show that the deglycase DJ-1 protects histones from adduction by MGO. Collectively, our findings demonstrate the existence of a previously undetected histone modification derived from glycolysis, which may have far-reaching implications for the control of gene expression and protein transcription linked to metabolism.
Histone posttranslational modifications (PTMs) regulate chromatin structure, DNA accessibility, and transcription to expand the genetic code (1). The side chains of Lys and Arg are subject to numerous PTMs, including acetylation (Lys), methylation (Lys and Arg), citrullination (Arg), ubiquitylation (Lys), and various chain-length acylations (Lys) (2). Many of these PTMs are directly linked to cellular metabolism, mediating feedback mechanisms that maintain homeostasis (3, 4). As a result, histone PTMs are tightly controlled through their enzymatic addition (“writers”) and removal (“erasers”) (3). There is growing interest in electrophilic modifications to histones (5). N-formylation of Lys residues is a nonenzymatic modification stemming from endogenous formaldehyde (6). This PTM is not only abundant (0.04–0.1% of histone Lys), but also resides at numerous epigenetic “hotspots,” including H3K18, H3K23, H4K12, and H4K31 (7). Furthermore, we recently reported the modification of histones by the lipid electrophile 4-oxo-2-nonenal (8). These PTMs are elevated during bouts of inflammation, reside at critical Lys residues (e.g., H3K23, H3K27), and are substrates for the Lys deacylase sirtuin 2 (9, 10).
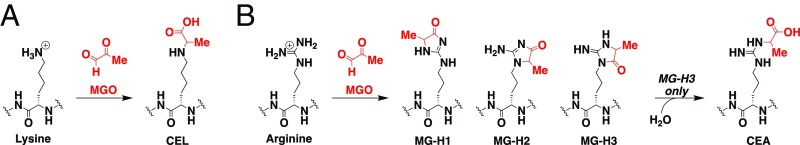
Cellular metabolism produces primary and secondary intermediates capable of reacting with Lys and Arg residues (11, 12). One such metabolite, methylglyoxal (MGO), is generated as a by-product of glycolysis, existing in micromolar quantities in eukaryotic cells (13). This α-oxoaldehyde generates stable protein adducts with the amine-functional groups of Arg and Lys side chains (Fig. 1). To prevent accumulation of MGO, glyoxalase 1 (GLO1) isomerizes the labile, spontaneous hemithioacetal product of the reaction of MGO with GSH to generate lactoylglutathione (14). In a recent study, Qi et al. (15) found significantly elevated MGO levels in patients with diabetic nephropathy, along with an inverse correlation between GLO1 expression and the severity of diabetic nephropathy. Indeed, shRNA knockdown of GLO1 in vivo is known to result in increased MGO-derived protein adducts and inflammatory signaling (16, 17). In addition to diabetes, MGO concentrations are also elevated in cancer, cardiovascular disease, and renal failure (18–20). Although there is a clear link between MGO and MGO-derived PTMs in disease, protein targets for MGO are seldom investigated.
Fig. 1.
MGO generates stable Lys (A) and Arg (B) protein adducts. Three isoforms of MG-H are possible with MG-H3 undergoing hydrolysis to generate CEA.
Due to the Arg- and Lys-rich nature of histones, we hypothesized that these proteins would be susceptible to adduction by MGO. Here we describe the first report of histone modification by a glycolytic by-product in intact chromatin. These PTMs exist basally in cells and in mouse tissues and are elevated during hyperglycemia. Furthermore, MGO-derived PTMs are present at levels comparable to those of other canonical Lys and Arg modifications. Elevations in MGO PTMs are known to decrease H2B acetylation and ubiquitylation, likely a result of the high degree of modification present on H2B. Our findings identify a quantitatively significant histone modification linked to metabolic flux through glycolysis.
Results
MGO Is an Abundant Cellular Metabolite That Modifies Chromatin.
To explore MGO-derived PTMs, we first established a chromatographic method for the detection of methylglyoxal hydroimidazolone (MG-H) 1, MG-H2, MG-H3, and N7-carboxyethyl arginine (CEA) using QuARKMod, a recently described MS-based method for quantifying Arg and Lys PTMs (21). Using stable isotope-labeled standards for each MGO-derived PTM (SI Appendix, Fig. S1), we measured adducts on purified histone H4 treated with 100 µM MGO for 24 h. As shown in SI Appendix, Figs. S1 and S2, we achieved chromatographic separation of each MG-H isomer, although only MG-H1 was detected in treated samples. The bottom trace in SI Appendix, Fig. S2 shows quantifiable levels of MG-H1 and an absence of MG-H2 in chromatin fractions isolated from HEK293 cells. Although MG-H2 is stable, it has not been observed in physiological systems, and our results support this finding (22). MG-H3 was also not observed due to its hydrolysis to yield CEA (SI Appendix, Fig. S1).
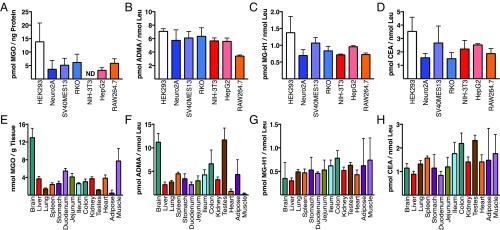
We next quantified cellular MGO in seven distinct cell lines cultured in low-glucose (5 mM) medium for 24 h. As shown in Fig. 2A, MGO is present basally at varying levels. Although no quantifiable MGO was detected in NIH 3T3, this is likely due to the limits of detection with this method. We next measured PTMs in chromatin fractions isolated from each cell line. As shown in Fig. 2 B–D, MG-H1 and CEA are present basally and at levels comparable to those of asymmetric dimethyl Arg (ADMA), which plays a critical role in transcriptional regulation (23). We also measured MGO and chromatin PTMs in tissues isolated from mice (Fig. 2 E–H). In addition to Arg-derived MGO protein adducts, the ε-amine of Lys residues is prone to modification by MGO, generating N-ε-(carboxyethyl)lysine (CEL). The levels of this PTM were found to be at least an order of magnitude lower than those of Arg adducts and often below the limit of detection.
Fig. 2.
(A) Cells were cultured for 24 h in low-glucose medium (5 mM) and cellular MGO was quantified. ND, not detected. (B–D) Chromatin was extracted from the indicated cells and subjected to QuARKMod, demonstrating basal levels of ADMA (B), MG-H1 (C), and CEA (D). (E–H) Basal levels of MGO (E), chromatin ADMA (F), MG-H1 (G), and CEA (H) were also evaluated in tissues isolated from mice. Data are presented as the mean ± SD of three measurements.
CEA Adducts Are Significantly Elevated with Increased Glycolytic Flux.
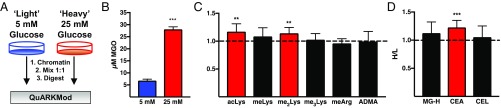
The bulk of MGO is generated through glycolysis (13). We sought to quantify alterations in chromatin PTMs as a result of increased glycolytic flux using stable-isotope labeling of amino acids in cell culture (SILAC). Isotopically labeled light cells (natural abundance isotope amino acids) were cultured in the presence of 5 mM glucose, whereas isotopically labeled heavy cells (13C615N2 Lys, 13C615N4 Arg, and 13C615N Leu) were cultured in the presence of 25 mM glucose for 24 h (Fig. 3A). As shown in Fig. 3B, cells cultured in the presence of 25 mM glucose exhibited an approximate fivefold increase in cellular MGO.
Fig. 3.
SILAC was used to measure global changes in Lys and Arg PTMs following exposure to hyperglycemic conditions. (A) Experimental procedure. (B) Cellular MGO is elevated following hyperglycemia. (C) Changes in canonical histone PTMs following hyperglycemia are shown as an increase in the heavy:light (H/L) ratio. (D) A significant elevation in CEA is observed following hyperglycemia. Red indicates significance from H/L = 1. **P < 0.01; ***P < 0.001. Data are presented as mean ± SD; n > 6.
We next measured the heavy:light ratios (25 mM glucose:5 mM glucose) of both canonical (Fig. 3C) and MGO-derived (Fig. 3D) PTMs on chromatin fractions. A significant elevation in acetylated Lys (acLys) was observed following hyperglycemia, consistent with previous reports (24). In addition, a significant increase in CEA was observed following hyperglycemia; however, no such increase was observed with MG-H1 or CEL adducts (Fig. 3D).
GLO1 Knockout Results in Elevated Levels of MGO and MGO-Derived Protein Adducts.
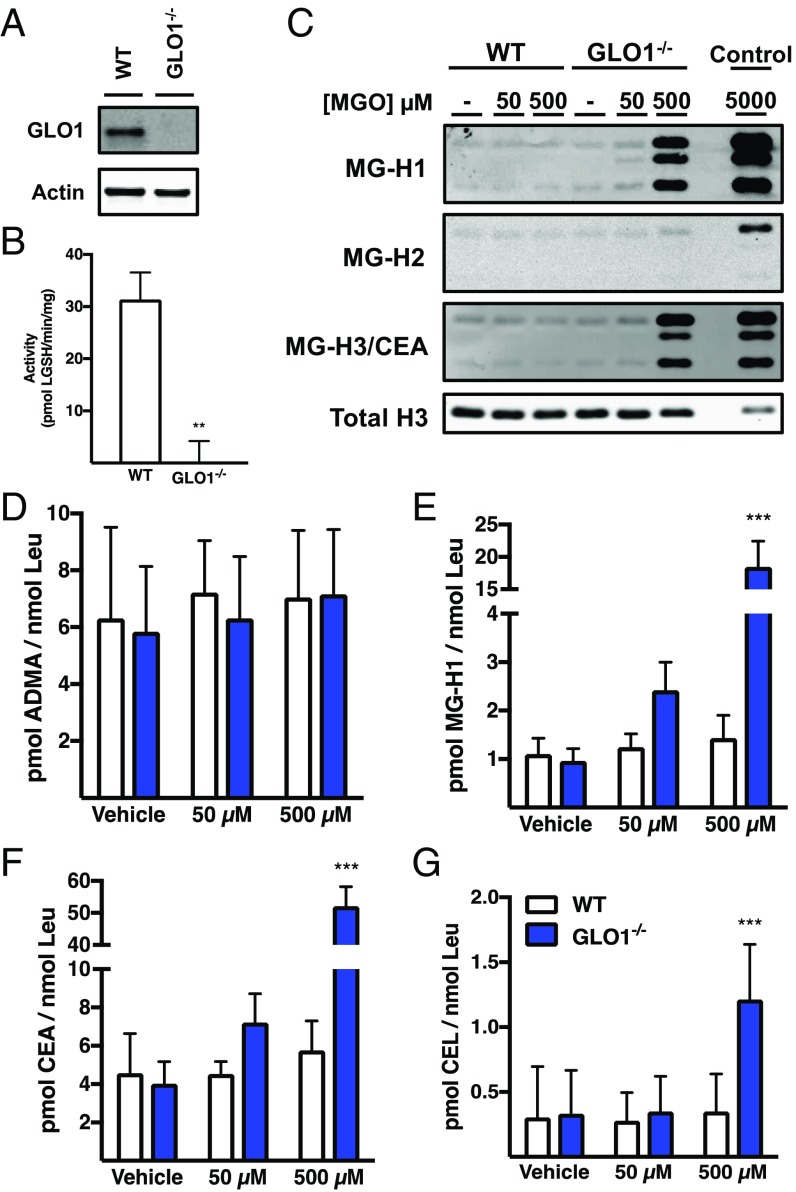
GLO1 is reportedly the major enzyme responsible for the detoxification of MGO, and its expression is significantly decreased in diabetic patients (15). We hypothesized that loss of GLO1 enzymatic activity would increase the levels of MGO-derived PTMs. To test this hypothesis, we developed a cell line using CRISPR-Cas technology lacking the GLO1 gene; transfection protocols are provided in Methods and Dataset S1. These cells (GLO1−/−) lacked any measurable GLO1 expression or activity (Fig. 4 A and B). GLO1−/− cells were more sensitive to MGO toxicity than wild-type (WT) cells, demonstrating a 10-fold decrease in the LC50 (from 367 µM to 46 µM) following a 24-h exposure (SI Appendix, Fig. S3). This is consistent with the finding that following exposure to increasing concentrations of MGO for 1 h, GLO1−/− cells exhibited intracellular MGO concentrations nearly identical to those provided extracellularly, whereas WT cells showed no significant increase (SI Appendix, Fig. S4). When monitored over time, GLO1−/− cells displayed a significant lag-phase in MGO removal following a challenge with 50 µM MGO, and cellular concentrations returned to baseline only after approximately 12 h of exposure (SI Appendix, Fig. S5A). These findings support the accepted role of GLO1 as the primary MGO detoxifying enzyme in the case of exogenously provided MGO and illustrate its high capacity for MGO removal. Interestingly, the basal levels of MGO were not elevated in GLO1−/− cells, implying that alternative mechanisms may exist for the removal of MGO. Nevertheless, GLO1−/− cells provide a useful tool for investigating the chemical biology of MGO-derived protein adducts under conditions of elevated electrophiles, as observed in diabetes.
Fig. 4.
Histones are modified by MGO. (A) Western blot analysis demonstrates complete knockout of GLO1. (B) GLO1−/− cells lack any measurable GLO1 activity. **P < 0.01. (C) Western blot analysis of chromatin fractions using isoform-specific MG-H antibodies reveals histones as targets for modification with markedly increased levels observed in GLO1−/− cells treated with electrophile. Owing to a lack of measurable MG-H2 protein adduction, chromatin was treated with 5 mM MGO for 6 h (control) to serve as a positive control for MGO modification. (D–G) QuARKMod demonstrates the levels of ADMA (D), MG-H1 (E), and CEA (F); the levels of MG-H1 and CEA are comparable to the level of ADMA under basal culture conditions, with marked elevations observed following treatment with MGO. (G) The levels of CEL are an order of magnitude lower than those of Arg-MGO PTMs. Data are presented as the mean ± SD; n = 3. Statistical significance was determined by two-way ANOVA. ***P < 0.001.
We next evaluated the generation of MGO-derived PTMs from total protein using QuARKMod analysis of the cell pellets isolated from the samples used in SI Appendix, Fig. S5A (21). As shown in SI Appendix, Fig. S5 B–D, GLO1−/− cells had elevated levels of MG-H1 and CEA when challenged with MGO, with concentrations peaking after approximately 6 h of exposure. CEL adduct levels were also elevated in GLO1−/− cells, reaching peak concentrations after approximately 6 h, although at approximately 10-fold lower concentrations than those of Arg-MGO adducts.
MGO Protein Adducts Are an Abundant Chromatin PTM.
To explore the effects of elevated intracellular MGO concentrations on MGO-derived chromatin PTMs, we isolated chromatin fractions from WT and GLO1−/− cells treated for 6 h with concentrations mimicking those of the LC50 (50 or 500 µM) (SI Appendix, Fig. S6). We first assessed the presence of MG-H1, MG-H2, and MG-H3/CEA in each condition via immunoblotting with antibodies directed against each isomer (22). As shown in Fig. 4C and SI Appendix, Fig. S7, GLO1−/− cells treated with MGO were found to have marked elevations in both MG-H1 and MG-H3/CEA immunostaining, whereas MG-H2 adducts were absent.
We next applied QuARKMod to chromatin fractions to measure both canonical and MGO-derived PTMs (Fig. 4 D–G and SI Appendix, Fig. S8). Although no alterations were observed in canonical histone PTMs (Fig. 4D and SI Appendix, Fig. S8), the levels of MG-H1, CEA, and CEL were significantly elevated in GLO1−/− cells challenged with MGO (Fig. 4 E–G). As was seen with the basal adduct levels, the CEL levels were an order of magnitude lower than Arg adduct levels.
Identification of Site-Specific MGO Modifications on Histones.
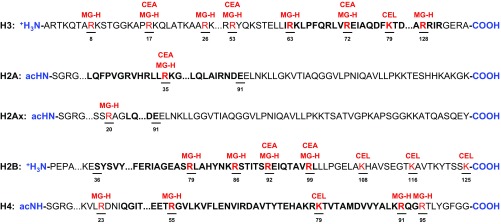
Under basal conditions, MG-H1 and CEA are difficult to detect using standard proteomic approaches, a problem often observed with lower-abundance histone PTMs (25, 26). Attempts were made to immunopurify MGO-modified peptides from untreated samples, but these techniques yielded no enrichment. Thus, exogenous addition of MGO was required to investigate site-specific adducts. GLO1−/− cells were treated with 1 mM MGO for 6 h. This high concentration of MGO was used to generate the broadest possible range of adduction sites. Using a previously described digestion protocol, we mapped MGO adduction sites on each histone, yielding 17 MG-H adducts, 6 CEA adducts, and 5 CEL adducts (Fig. 5; the annotated MS/MS spectra for these adducts are shown in SI Appendix, Figs. S13–S40) (8). Interestingly, all the reported CEA sites were also found to contain an MG-H adduct. These data support previous findings demonstrating that the kinetic product of reaction of MGO with Arg is MG-H3, which is readily hydrolyzed to yield CEA; CEA can then recyclize to yield MG-H1 (27).
Fig. 5.
Identification of 28 site-specific MGO modifications on histones. Proteomic interrogation of site-specific MGO PTMs reveals the N terminus of H3 and the globular domain (bold) of H2B to be heavily susceptible to modification by MGO.
Unlike many histone PTMs, MGO-derived modifications were not restricted to the heavily modified N-terminal tails; many were found on the lateral surface of the nucleosome (SI Appendix, Fig. S9A; adducts are depicted in red). H3R72, H4R23, H4R55, and H2BR92 were located in close proximity to DNA (SI Appendix, Fig. S9B). Furthermore, the presence of MG-H and CEA adducts at H3R53 (SI Appendix, Fig. S9C) may lead to disruption of nucleosomal stability akin to that of H3K56 acetylation (28).
MGO Disrupts Global H2B Modifications.
Due to the relative abundance of MGO-derived PTMs on H3 and H2B, we hypothesized that these adducts may disrupt the formation of canonical PTMs. Consistently, MGO exposure was found to decrease H2B acetylation and ubiquitylation in GLO1−/− cells (SI Appendix, Fig. S10A); this was particularly notable in H2BK120 ubiquitylation (H2BK120ub). H2BK120ub is required for the binding of DOT1L, an H3K79 methyltransferase (29, 30). Despite the loss in H2BK120ub, no alterations in H3K79me2 were observed (SI Appendix, Fig. S10A). In fact, no alterations in any canonical H3, H4, or H2A PTMs were observed (SI Appendix, Fig. S10B). These data are consistent with the hypothesis that MGO disrupts H2B PTMs, likely as a result of the high degree of modification by MGO.
RNA-Seq Reveals Transcripts Regulated by MGO.
Histone PTMs play a critical role in the regulation of gene transcription. We therefore assessed MGO-mediated alterations in transcripts using a global RNA-Seq approach. WT or GLO1−/− cells were treated with vehicle, 50 µM MGO, or 500 µM MGO for 6 h. As shown in SI Appendix, Fig. S11 A and B, treatment of WT cells with either 50 or 500 µM MGO had little impact on total RNA transcripts. This is likely a result of the rapid detoxification of MGO in the presence of GLO1. In GLO1−/− cells, however, a dose-dependent increase in significantly altered transcripts was observed compared with WT vehicle control (SI Appendix, Fig. S11 C–E, log2 fold change >2; P < 0.05).
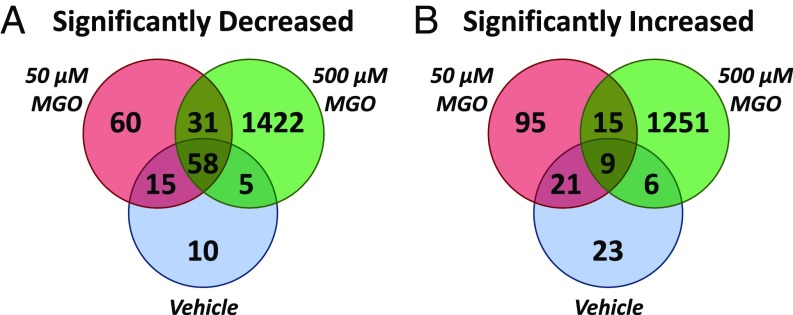
We next evaluated only protein coding transcripts in the GLO1−/− cohorts with and without MGO treatment. As shown in Fig. 6, GLO1−/− cells displayed 88 significantly decreased genes and 59 elevated genes compared with WT vehicle control. Greater changes were observed following treatment with either 50 µM MGO (164 decreased and 140 increased) or 500 µM MGO (1,516 decreased and 1281 increased) (Dataset S2). The large alterations in gene expression observed with the 500 µM MGO treatment are likely attributed to the toxicity observed with these high concentrations (greater than the LC50); therefore, we focused our attention on the 50 µM MGO cohort. Among this group, the most significantly increased genes were ADM2 (log2 fold enrichment, 3.1), POU5F2 (log2 fold enrichment, 2.9), HCLS1 (log2 fold enrichment, 2.7), and SLC6A9 (log2 fold enrichment, 2.7), and the most significantly decreased genes were FZD10 (log2 fold enrichment, −8.8), STS (log2 fold enrichment, −8.5), LTBR (log2 fold enrichment, −7.8), and EVC2 (log2 fold enrichment, −7.8).
Fig. 6.
RNA-Seq reveals transcripts altered by MGO. The Venn diagrams display significantly decreased (A) or increased (B) protein coding transcripts in GLO1−/− cells treated with either vehicle, 50 µM MGO, or 500 µM MGO compared with WT vehicle control. n = 3 for each cohort.
We next performed Gene Ontology analyses using DAVID on vehicle, 50 µM MGO, and 500 µM MGO treatments to attempt to gain insight into possible pathways involved in MGO modifications (31). These analyses yielded little conclusive enrichment (Dataset S3). Notably, no enrichment was observed for antioxidant, endoplasmic reticulum stress, or heat shock responses, consistent with the hypothesis that MGO alters global transcription rather than distinct toxicity-response pathways.
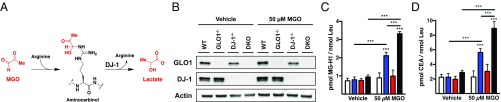
DJ-1 Reduces Histone Arg Modification by MGO.
In a recent study, Richarme et al. (32) found that the deglycase enzyme DJ-1 repaired MGO-derived modifications of guanine. DJ-1 also has been shown to prevent the formation of MGO modifications on Lys and Arg residues by hydrolyzing the aminocarbinol intermediates (Fig. 7A) (33). Therefore, we investigated the effect of DJ-1 on the levels of MGO-derived chromatin adducts. As shown in Fig. 7B, we performed CRISPR-Cas9 to generate DJ-1−/− cells in both WT and GLO1−/− [double KO (DKO)] backgrounds. We then performed QuARKMod on chromatin fractions isolated from cells treated with either vehicle or 50 µM MGO for 6 h (Fig. 7 C and D). MGO treatment of DKO cells resulted in a significant increase in both MG-H1 and CEA compared with treatment of GLO1−/− cells alone. The absence of DJ-1 did not impact the levels of any Lys PTMs or ADMA (SI Appendix, Fig. S12). These data suggest a role for DJ-1 in controlling the levels of MGO-derived Arg modifications on chromatin.
Fig. 7.
DJ-1 protects chromatin from modification by MGO. (A) DJ-1 hydrolyzes the intermediate aminocarbinol product of MGO and Arg. (B) Western blot analysis demonstrates complete knockout of DJ-1 in both WT and GLO1−/− cell lines (DKO). (C and D) QuARKMod was performed on chromatin fractions isolated from each cohort, demonstrating significant increases in MG-H1 and CEA in DKO cells compared with GLO1−/− alone. Treatments were provided for 6 h. Data are presented as mean ± SD; n = 3. Statistical significance was determined via two-way ANOVA. ***P < 0.001.
Discussion
In this study, we identify histones as targets for modification by the glycolytic by-product MGO in intact chromatin. We report MGO-derived Arg modifications as abundant histone PTMs that are present basally and at the same order of magnitude as ADMA, a PTM involved in transcriptional regulation (23). Under conditions of high intracellular MGO, we identify 28 site-specific modifications on histones, four of which reside on the PTM-rich N-terminal tail of H3. These modifications disrupt basal canonical histone PTMs, most notably on H2B, where a global loss of acetylation and ubiquitylation is observed. We also demonstrate that the deglycase protein, DJ-1, may play a role in limiting the accumulation of MG-H1 and CEA on chromatin. Although the functional role of these PTMs is currently under investigation, we hypothesize that MGO may serve as a key regulatory signaling molecule involved in global responses due to altered glycolytic flux. This is of particular importance in the context of diabetic nephropathy, where GLO1 activity is significantly decreased and levels of MGO-modified proteins are markedly elevated (15).
Cellular metabolism intricately controls gene expression and transcription through histone PTMs. This is perhaps most notable through the acetylation of histone Lys residues via acetyl-CoA (34, 35). Many of these modifications regulate gene expression by altering chromatin structure and architecture (36). For example, acetylation at H3K56 disrupts the stability of the nucleosome, leading to an “unwrapping” of DNA (28). The presence of MG-H or CEA adducts at H3R53, which lies adjacent to H3K56 in the entry-exit site of the nucleosome, would lead to a similar loosening of the DNA around the nucleosome. In addition to H3R53, MG-H modifications present at H3R8 and H3R26 may have a profound impact on chromatin function. These residues are critical for the binding of the epigenetic modulators AF9 and KDM4A, respectively (37, 38), and their adduction by MGO will alter their ability to form critical H-bonding interactions (37, 38). Thus, the presence of MGO adducts, notably under basal conditions and across multiple cell lines, may suggest a mechanism by which cellular metabolism controls chromatin dynamics. Indeed, MGO was found to significantly alter the expression of numerous genes, notably in GLO1−/− cells treated with 50 µM MGO, where 164 transcripts were decreased and 140 transcripts were increased. Nonetheless, despite these global alterations in gene expression, no clear pathway enrichment was observed. Although nonhistone proteins are also known to be modified by MGO (24, 39), these data support the idea that MGO forms PTMs that play an active role in the regulation of gene expression, rather than simply serving as a reactive molecule responsible for the induction of electrophile-response pathways (40).
The modification of proteins by MGO is reportedly the result of nonenzymatic reactions with Arg and Lys residues; however, our data suggest that these modifications are regulated by eraser proteins. In support of Richarme et al. (33), we demonstrate that DJ-1 “protects” Arg residues from modification by MGO, while not impacting other canonical PTMs. Although no increases MG-H1 or CEA were observed in the DJ-1−/− cells, this is likely due to the rapid metabolism of MGO in cells expressing GLO1. Furthermore, these studies support a recent report that defined a critical role for DJ-1 in maintaining DNA integrity via the removal of intermediates that lead to MGO modifications (32). Although much work is still needed to fully understand the role of MGO-derived PTMs under both physiological and pathophysiological situations, the present study sheds light on a key link between glycolysis and the regulation of chromatin.
Methods
More detailed information on the study methodology is provided in SI Appendix, Methods.
Cell Culture.
HEK293 cells were cultured in low-glucose DMEM (1 g/L glucose) supplemented with 10% FBS. Cells were incubated at 37 °C and under 5% CO2. SILAC cells were cultured in low-glucose SILAC medium supplemented with 0.1 g/L 13C615N2 Lys, 0.1 g/L 13C615N4 Arg, 0.1 g/L 13C615N Leu for “heavy” cells or natural abundance isotopes for “light” cells, and 10% dialyzed FBS (Fisher Scientific). Isotope incorporation was monitored at each passage via QuARKMod and labeling was considered complete when incorporation exceeded 95%.
Generation of GLO1−/− and DJ-1−/− Cells Using CRISPR-Cas9.
gRNA oligonucleotides (Dataset S1) were designed to target restriction enzyme recognition sites in the initial exons of the GLO1 or DJ-1 locus and ligated into the pSpCas9(BB)-2A-Puro plasmid as described by Cong et al. (41). To generate cells lacking GLO1 or DJ-1, 2 × 105 HEK293 cells were plated in 2 mL of DMEM supplemented with 10% FBS in six-well plates. The next day, 5 µg of each construct was combined with 10 µL of Lipofectamine 2000 reagent (Life Technologies) in 1 mL of Opti-MEM and incubated at room temperature for 30 min. The DMEM was replaced with the plasmid-Lipofectamine 2000 solution, and the cells were incubated at 37 °C for 24 h. The medium was replaced, after which the cells were allowed to recover for 24 h at 37 °C. The medium was then replaced with serum-containing DMEM with 0.75 µg/mL puromycin, and the cells were incubated at 37 °C for 48 h. The medium was then replaced with puromycin-free medium, and the cells were incubated for another 24 h, after which the medium was replaced to remove any traces of puromycin. Cultures were then pelleted, resuspended in sorting buffer (PBS + 4% FBS), and strained. Solutions were sorted by flow cytometry using a BD FACSAria III cell sorter to isolate single cell cultures in 3 × 96-well plates for each cell line, yielding approximately 100 viable clones.
To generate DKO cells, the transfection procedure was performed using the DJ-1 gRNA in GLO1−/− cells rather than WT cells. Flow cytometry experiments were performed at the Vanderbilt University Medical Center Flow Cytometry Shared Resource. Clones were maintained to approximately 80% confluency and passaged until a sufficient number of cells could be harvested for indel analysis. A restriction-fragment length polymorphism (RFLP) assay (SI Appendix, Table S1) was used to assess for indel mutations; target genes were PCR-amplified and subjected to digestion by restriction enzymes specific to the WT sequence at the gRNA target site. The absence of restriction enzyme activity was indicative of mutations in both alleles at the gRNA target site, and clones with homozygous mutations of GLO1 or DJ-1 were validated as genetic knockouts. PCR products from RFLP assays were purified using the Nucleospin PCR Clean-Up Kit and sequenced with the forward primer (Dataset S1). Sequencing samples were analyzed by GenHunter Corporation.
Supplementary Material
Acknowledgments
We thank William P. Tansey and Carol A. Rouzer for their thoughtful discussions and input. Financial support was provided by National Institutes of Health Grants CA87819 and S10 OD017997 (to L.J.M.) and the SENS Foundation (to D.A.S.). The Vanderbilt University Medical Center Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (Grant P30 CA068485) and the Vanderbilt Digestive Disease Research Center (Grant DK058404). Vanderbilt Technologies for Advanced Genomics (VANTAGE) is supported in part by the Clinical and Translational Science Awards Program (Grant 5UL1 RR024975-03), the Vanderbilt Ingram Cancer Center (Grant P30 CA68485), the Vanderbilt Vision Center (Grant P30 EY08126), and National Center for Research Resources (Grant G20 RR030956). Supplemental funding to VANTAGE and Vanderbilt Technologies for Advanced Genomics Analysis and Research Design is provided by National Institutes of Health Grants P50 GM115305 and U19 HL065962.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802901115/-/DCSupplemental.
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: Histone modifications. Cell. 2014;159:458–458.e1. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem Biol. 2015;10:95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol. 2017;18:90–101. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galligan JJ, Marnett LJ. Histone adduction and its functional impact on epigenetics. Chem Res Toxicol. 2017;30:376–387. doi: 10.1021/acs.chemrestox.6b00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci USA. 2007;104:60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wisniewski JR, Zougman A, Mann M. N-epsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008;36:570–577. doi: 10.1093/nar/gkm1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galligan JJ, et al. Stable histone adduction by 4-oxo-2-nonenal: A potential link between oxidative stress and epigenetics. J Am Chem Soc. 2014;136:11864–11866. doi: 10.1021/ja503604t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui Y, Li X, Lin J, Hao Q, Li XD. Histone ketoamide adduction by 4-oxo-2-nonenal is a reversible posttranslational modification regulated by Sirt2. ACS Chem Biol. 2017;12:47–51. doi: 10.1021/acschembio.6b00713. [DOI] [PubMed] [Google Scholar]
- 10.Jin J, He B, Zhang X, Lin H, Wang Y. SIRT2 reverses 4-oxononanoyl lysine modification on histones. J Am Chem Soc. 2016;138:12304–12307. doi: 10.1021/jacs.6b04977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sola-Penna M, Da Silva D, Coelho WS, Marinho-Carvalho MM, Zancan P. Regulation of mammalian muscle type 6-phosphofructo-1-kinase and its implication for the control of the metabolism. IUBMB Life. 2010;62:791–796. doi: 10.1002/iub.393. [DOI] [PubMed] [Google Scholar]
- 12.Moellering RE, Cravatt BF. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science. 2013;341:549–553. doi: 10.1126/science.1238327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabbani N, Thornalley PJ. Measurement of methylglyoxal by stable isotopic dilution analysis LC-MS/MS with corroborative prediction in physiological samples. Nat Protoc. 2014;9:1969–1979. doi: 10.1038/nprot.2014.129. [DOI] [PubMed] [Google Scholar]
- 14.Rabbani N, Xue M, Thornalley PJ. Activity, regulation, copy number and function in the glyoxalase system. Biochem Soc Trans. 2014;42:419–424. doi: 10.1042/BST20140008. [DOI] [PubMed] [Google Scholar]
- 15.Qi W, et al. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat Med. 2017;23:753–762. doi: 10.1038/nm.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giacco F, et al. Knockdown of glyoxalase 1 mimics diabetic nephropathy in nondiabetic mice. Diabetes. 2014;63:291–299. doi: 10.2337/db13-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Osta A, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thornalley PJ. Pharmacology of methylglyoxal: Formation, modification of proteins and nucleic acids, and enzymatic detoxification—A role in pathogenesis and antiproliferative chemotherapy. Gen Pharmacol. 1996;27:565–573. doi: 10.1016/0306-3623(95)02054-3. [DOI] [PubMed] [Google Scholar]
- 19.Kilhovd BK, et al. Increased serum levels of methylglyoxal-derived hydroimidazolone-AGE are associated with increased cardiovascular disease mortality in nondiabetic women. Atherosclerosis. 2009;205:590–594. doi: 10.1016/j.atherosclerosis.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 20.Agalou S, Ahmed N, Babaei-Jadidi R, Dawnay A, Thornalley PJ. Profound mishandling of protein glycation degradation products in uremia and dialysis. J Am Soc Nephrol. 2005;16:1471–1485. doi: 10.1681/ASN.2004080635. [DOI] [PubMed] [Google Scholar]
- 21.Galligan JJ, et al. Quantitative analysis and discovery of lysine and arginine modifications. Anal Chem. 2016;89:1299–1306. doi: 10.1021/acs.analchem.6b04105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T, Streeter MD, Spiegel DA. Generation and characterization of antibodies against arginine-derived advanced glycation endproducts. Bioorg Med Chem Lett. 2015;25:4881–4886. doi: 10.1016/j.bmcl.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Fuhrmann J, Thompson PR. Protein arginine methylation and citrullination in epigenetic regulation. ACS Chem Biol. 2016;11:654–668. doi: 10.1021/acschembio.5b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nokin MJ, et al. Hormetic potential of methylglyoxal, a side-product of glycolysis, in switching tumours from growth to death. Sci Rep. 2017;7:11722. doi: 10.1038/s41598-017-12119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H, Lin S, Garcia BA, Zhao Y. Quantitative proteomic analysis of histone modifications. Chem Rev. 2015;115:2376–2418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen SC, et al. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci Signal. 2016;9:rs9. doi: 10.1126/scisignal.aaf7329. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, Kartika R, Spiegel DA. Exploring post-translational arginine modification using chemically synthesized methylglyoxal hydroimidazolones. J Am Chem Soc. 2012;134:8958–8967. doi: 10.1021/ja301994d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 29.Zhou L, et al. Evidence that ubiquitylated H2B corrals hDot1L on the nucleosomal surface to induce H3K79 methylation. Nat Commun. 2016;7:10589. doi: 10.1038/ncomms10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitcomb SJ, et al. Histone monoubiquitylation position determines specificity and direction of enzymatic cross-talk with histone methyltransferases Dot1L and PRC2. J Biol Chem. 2012;287:23718–23725. doi: 10.1074/jbc.M112.361824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Richarme G, et al. Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science. 2017;357:208–211. doi: 10.1126/science.aag1095. [DOI] [PubMed] [Google Scholar]
- 33.Richarme G, et al. Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J Biol Chem. 2015;290:1885–1897. doi: 10.1074/jbc.M114.597815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y, Thomas PM, Kelleher NL. Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat Commun. 2013;4:2203. doi: 10.1038/ncomms3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galdieri L, Vancura A. Acetyl-CoA carboxylase regulates global histone acetylation. J Biol Chem. 2012;287:23865–23876. doi: 10.1074/jbc.M112.380519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 37.Su Z, et al. Reader domain specificity and lysine demethylase-4 family function. Nat Commun. 2016;7:13387. doi: 10.1038/ncomms13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 2014;159:558–571. doi: 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oya T, et al. Methylglyoxal modification of protein: Chemical and immunochemical characterization of methylglyoxal-arginine adducts. J Biol Chem. 1999;274:18492–18502. doi: 10.1074/jbc.274.26.18492. [DOI] [PubMed] [Google Scholar]
- 40.Liu Q, et al. RNA-seq data analysis at the gene and CDS levels provides a comprehensive view of transcriptome responses induced by 4-hydroxynonenal. Mol Biosyst. 2013;9:3036–3046. doi: 10.1039/c3mb70114j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cong L, Zhang F. Genome engineering using CRISPR-Cas9 system. Methods Mol Biol. 2015;1239:197–217. doi: 10.1007/978-1-4939-1862-1_10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.