Significance
Neurons extend long structures called axons that are highly susceptible to damage and undergo degeneration in many neurological disorders. One strategy for treating these diseases is by elevating the abundance of axon survival factors that suppress axon death. Herein, we describe a protein homeostasis network that regulates axon degeneration by tuning the local levels of axon survival factors. In particular, we find that small-molecule inhibitors targeting a MAPK stress pathway protect axons from pathological degeneration by elevating the local abundance of axon survival factors NMNAT2 and SCG10. Furthermore, we discover that intracellular location imparts sensitivity to distinct protein homeostasis networks. Inactivating multiple nodes in this protein homeostasis network confers maximal therapeutic potential in diseases of axon degeneration.
Keywords: NMNAT2, SCG10, SARM1, axon, DLK
Abstract
Axon degeneration is a prominent event in many neurodegenerative disorders. Axon injury stimulates an intrinsic self-destruction program that culminates in activation of the prodegeneration factor SARM1 and local dismantling of damaged axon segments. In healthy axons, SARM1 activity is restrained by constant delivery of the axon survival factor NMNAT2. Elevating NMNAT2 is neuroprotective, while loss of NMNAT2 evokes SARM1-dependent axon degeneration. As a gatekeeper of axon survival, NMNAT2 abundance is an important regulatory node in neuronal health, highlighting the need to understand the mechanisms behind NMNAT2 protein homeostasis. We demonstrate that pharmacological inhibition of the MAP3Ks dual leucine zipper kinase (DLK) and leucine zipper kinase (LZK) elevates NMNAT2 abundance and strongly protects axons from injury-induced degeneration. We discover that MAPK signaling selectively promotes degradation of palmitoylated NMNAT2, as well as palmitoylated SCG10. Conversely, nonpalmitoylated NMNAT2 is degraded by the Phr1/Skp1a/Fbxo45 ligase complex. Combined inactivation of both pathways leads to synergistic accumulation of NMNAT2 in axons and dramatically enhanced protection against pathological axon degeneration. Hence, the subcellular localization of distinct pools of NMNAT2 enables differential regulation of NMNAT2 abundance to control axon survival.
Neurodegeneration is an omnipresent health concern, with few treatments available to stop or even slow disease progression. Considerable attention is focused on inhibiting neuronal cell death by targeting classical apoptosis and other pathways driving cellular demise. However, functional connectivity in the nervous system is dependent on long projections called axons that undergo degeneration in many neurological disorders, including Alzheimer’s disease, Parkinson’s disease, and peripheral neuropathies associated with chemotherapy or diabetes (1–3). Hence, understanding pathways responsible for protecting axon integrity in response to pathological damage is essential for treatment of these neurodegenerative disorders. Axon transection is a model of traumatic axon injury that stimulates an intrinsic self-destruction pathway known as Wallerian degeneration that resembles pathological axon decay in many neurological disorders (4, 5). Genetic screens for modifiers of Wallerian degeneration identified the Toll/Interleukin-1 receptor domain-containing protein SARM1 as a central executioner of pathological axon degeneration (6, 7). Genetic inactivation of SARM1 confers strong preservation after traumatic nerve injury, vincristine-induced peripheral neuropathy, oxidative stress, and other common neuronal insults, such that inhibiting SARM1 activity has extensive therapeutic potential (6–12).
SARM1-dependent axon degeneration is antagonized by the presence of axon survival factors, including the NAD+ biosynthetic enzyme NMNAT2 and the neuronal stathmin SCG10. Overexpressing NMNAT2 or related NMNAT enzymes suppresses pathological axon degeneration (13), while genetic loss of NMNAT2 stimulates spontaneous SARM1-dependent axon degeneration (14). Elevating or depleting SCG10 evokes the same reciprocal effect on axon health, although to a lesser extent than NMNAT2 (15). Both NMNAT2 and SCG10 are short-lived proteins, and constant delivery to the axon is necessary for axon survival (15, 16). In addition, NMNAT2 and SCG10 are palmitoylated and associated with vesicles that are anterogradely transported through the axon (17–19). Surprisingly, membrane association promotes NMNAT2 degradation in HEK293t cells (20), yet whether this occurs in axons is unknown. Inhibiting pathways responsible for NMNAT2 degradation is neuroprotective, reinforcing the need to understand molecular mechanisms of NMNAT2 protein homeostasis in the axon.
Two pathways are known to regulate NMNAT2 protein turnover in the axon. An atypical SCF E3 ligase complex composed of Phr1/Fbxo45/Skp1a has an evolutionarily conserved role in NMNAT2 degradation, and down-regulating any member of this complex promotes axon survival after injury by elevating NMNAT2 protein levels (21–24). In addition to the Phr1 complex, a MAPK stress-signaling pathway promotes NMNAT2 protein degradation and genetic reduction in the MAP2Ks MKK4 and MKK7 (MKK/4/7) or pharmacological inhibition of the downstream MAPK JNK prolongs the presence of NMNAT2 in injured axons (25). Accordingly, loss of MKK4/7 or all three JNK isoforms confers strong axon protection in vitro and in vivo (26). Whether MAPK signaling and the Phr1 complex work together or independently to promote NMNAT2 degradation is unknown.
Upstream of MKK4/7 and JNK, the MAP3K dual leucine zipper kinase (DLK) is emerging as an appealing therapeutic target in neurological disease. Loss of DLK promotes axon survival in response to axotomy or exposure to vincristine (27). Interestingly, DLK signaling is enhanced in patients with Alzheimer’s disease or ALS (28), and the DLK-MKK4/7-JNK axis promotes apoptosis in response to neuronal injury or trophic factor withdrawal (28–33). Recently developed pharmacological inhibitors of DLK suppress apoptotic cell death and show promise in animal models of neurodegeneration (28, 34), although their role in axon degeneration has not been explored. In the current study, we demonstrate that pharmacological inhibitors of DLK elevate axonal NMNAT2 levels and promote axon survival after injury. We also discover that MAPK signaling selectively targets palmitoylated NMNAT2 and SCG10 for degradation. Conversely, the Phr1 complex targets nonpalmitoylated NMNAT2 for degradation, revealing that distinct pools of NMNAT2 are differentially regulated. Inactivating both pathways leads to a synergistic increase in NMNAT2 within axons and extensive axon protection against multiple forms of injury-induced degeneration. Therefore, the mechanism of NMNAT2 degradation is coupled to its membrane association, and targeting independent pathways responsible for NMNAT2 degradation offers maximal therapeutic potential.
Results
Small-Molecule Inhibition of DLK Protects Axons by Elevating NMNAT2.
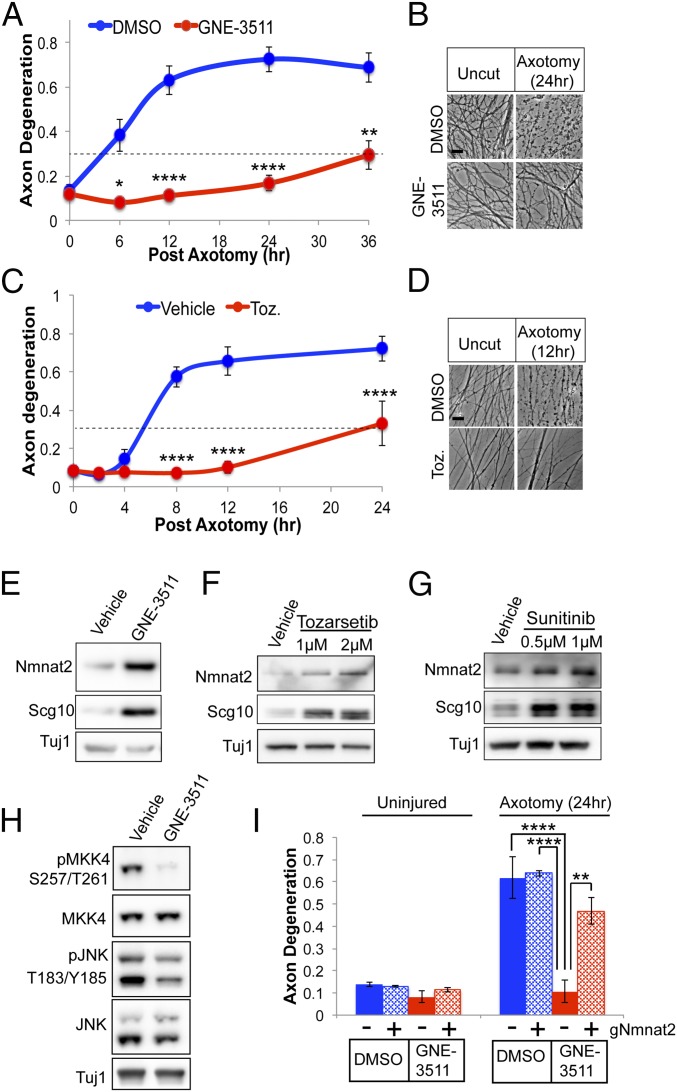
DLK promotes apoptotic cell death in response to excitotoxicity, trophic factor withdrawal, and axonal injury (35). Small-molecule inhibitors targeting DLK reduce apoptotic cell death, yet have not been assessed for neuroprotection in pathological axon degeneration, which utilizes a mechanistically distinct, SARM1-dependent executioner pathway. Since genetic loss of the DLK-MKK4/7-JNK pathway confers protection against Wallerian degeneration we hypothesized that DLK inhibitors will also promote axon survival in models of injury-induced axon degeneration. To test this hypothesis, sensory neurons were treated with the DLK inhibitor GNE-3511 (34), axons were severed with a razor blade, and axon fragmentation was monitored in distal severed axons as previously described (36). In primary sensory neurons, severed axons undergo extensive fragmentation within 6 h after axotomy. However, in the presence of GNE-3511, severed axons are morphologically intact for 36 h following axotomy (Fig. 1 A and B). Furthermore, GNE-3511 suppresses axon degeneration in response to the chemotherapeutic agent vincristine (SI Appendix, Fig. S1). We also examined axon protection in the presence of Tozarsertib, a small molecule originally designed to inhibit aurora kinases but which also shows activity against DLK and prevents death of retinal ganglion cells after injury (31). In the presence of Tozarsertib, severed axons are preserved for almost 24 h after injury (Fig. 1 C and D) suggesting that small-molecule inhibitors of DLK suppress pathological axon degeneration.
Fig. 1.
Small-molecule inhibitors of DLK suppress axon degeneration by elevating NMNAT2/SCG10 protein levels. (A) DRG sensory neurons were pretreated with DMSO or GNE-3511 (500 nM); axons were then severed with a razor blade, and the fragmentation of distal axons was measured over time. An axon degeneration score above 0.3 indicates axons are fragmenting (dotted line). (B) Bright-field images of distal, severed axons 24 h after axotomy, pretreated with vehicle or GNE-3511. (Scale bar, 5 μm.) Pretreating DRG sensory neurons with 1 μM Tozarsertib (Toz.) suppresses injury-induced axon degeneration for ∼24 h after axotomy (C), with bright-field images of distal axons 12 h after axotomy (n = 3) (D). (Scale bar, 5 μm.) Western blots of endogenous NMNAT2 and SCG10 from axon-only extracts derived from DRG axons after treatment with GNE-3511 (E), Tozarsertib (F), or Sunitinib (G) are shown. (H) Western blots of phosphorylated MKK4 and JNK from axon-only extracts after treatment with GNE-3511. Western blots are representative of at least three independent experiments. (I) Cas9-expressing DRGs were transduced with sgRNAs targeting NMNAT2 or a nonspecific scrambled sgRNA control and pretreated with DMSO or GNE-3511 as in A. Loss of NMNAT2 suppresses axon protection afforded by GNE-3511 (n = 3). For A and C, statistical analysis was performed with a repeated-measures, two-way ANOVA, while in I, a standard two-way ANOVA was performed. Post hoc analysis was performed with Bonferroni correction for multiple comparisons, where *P < 0.05, **P < 0.005, and ****P < 0.0001. Error bars represent SEM.
Since downregulating the MAP2Ks MKK4/7 extends protein stability of the axon maintenance factors NMNAT2 and SCG10, we hypothesized that small molecules inhibiting DLK activity would likewise increase axonal levels of these proteins. GNE-3511 treatment for 8 h increases axonal levels of both NMNAT2 and SCG10 (Fig. 1E). Likewise, treating sensory neurons with the DLK inhibitors Tozarsertib or the Food and Drug Administration-approved Sunitinib (31) increases steady-state protein levels of NMNAT2 and SCG10 (Fig. 1 F and G). DLK inhibition decreases steady-state levels of phosphorylated MKK4 and JNK in axons, suggesting this MAP kinase pathway has constitutive function in sensory axons to promote NMNAT2 loss (Fig. 1H).
MAPK signaling functions upstream of SARM1 by tuning NMNAT2/SCG10 levels (25). We previously found that axon protection afforded by loss of MKK4/7 is suppressed by knocking out endogenous NMNAT2, suggesting axon protection is dependent on tuning NMNAT2 protein levels (25). Indeed, axon protection afforded by GNE-3511 is suppressed by targeting the endogenous NMNAT2 gene with single-guide RNAs (sgRNAs) in CAS9-expressing sensory neurons (Fig. 1I and SI Appendix, Fig. S2). Altogether, DLK inactivation increases the local abundance of neuron survival factors and extends axon resistance to pathological axon degeneration.
The MAP3K Leucine Zipper Kinase Promotes Injury-Induced Axon Degeneration.
The extent of axon protection afforded by GNE-3511 was surprising, as genetic loss of DLK does not evoke such a strong effect (27). An enhanced suppressor screen identified the closely related MAP3K leucine zipper kinase (LZK) as another target of DLK small-molecule inhibitors, and combined loss of DLK and LZK leads to additive survival of retinal ganglion cells against injury-induced apoptotic cell death (37). Since nothing is known regarding the contribution of LZK to Wallerian degeneration, we knocked out DLK and LZK individually and in combination from dorsal root ganglion (DRG) sensory neurons using Cas9/CRISPR. While sgRNAs targeting LZK led to a marginal delay in axon degeneration after axotomy, pooling sgRNAs targeting both DLK and LZK results in synergistic axon protection, leaving axons morphologically intact for at least 36 h after axotomy (Fig. 2 A and B). LZK and DLK share the same downstream MAPK partners MKK4/7 and JNK (38), and knocking out these kinases reduced levels of phosphorylated MKK4 and JNK in axons (Fig. 2C); thus, we predicted that double knockout of DLK and LZK would have an additive effect on the downstream targets NMNAT2 and SCG10. Consistent with this hypothesis, combined loss of DLK and LZK leads to enhanced accumulation of NMNAT2 and SCG10 protein levels (Fig. 2 D and E). Therefore, these highly related MAP3Ks share a redundant role in degradation of axon survival factors, and small-molecule inhibitors that target both kinases potently protect axons from injury-induced degeneration.
Fig. 2.
MAP3K LZK promotes axon degeneration by regulating levels of NMNAT2/SCG10. (A) Cas9-expressing DRG sensory neurons were transduced with lentivirus containing sgRNAs to DLK and/or LZK. CRISPR knockout of both DLK and LZK confers additive axon protection in severed distal axons compared with control (Ctrl) sgRNAs or sgRNAs targeting DLK (sgDKL) or LZK (sgLZK). A repeated-measures ANOVA was performed with post hoc Bonferroni tests for multiple comparisons, where ****P < 0.001 (n = 3). (B) Bright-field images of distal axons at the indicated time points after axotomy. (Scale bar, 5 μm.) (C) Western blots of phosphorylated MKK4 and JNK from axon-only extracts in Cas9-expressing DRGs transduced with Ctrl sgRNAs or sgDLK and sgLZK. Western blots are representative of at least three independent experiments. (D) Western blots of NMNAT2 and SCG10 from axon-only extracts derived from DRG sensory neurons transduced with the indicated sgRNAs. (E) Quantification of NMNAT2 and SCG10 protein levels in the presence of the indicated sgRNAs [***P < 0.001, single-factor ANOVA with post hoc t tests (n = 3)]. Error bars represent SEM.
MAPK Signaling Targets Palmitoylated NMNAT2 for Degradation.
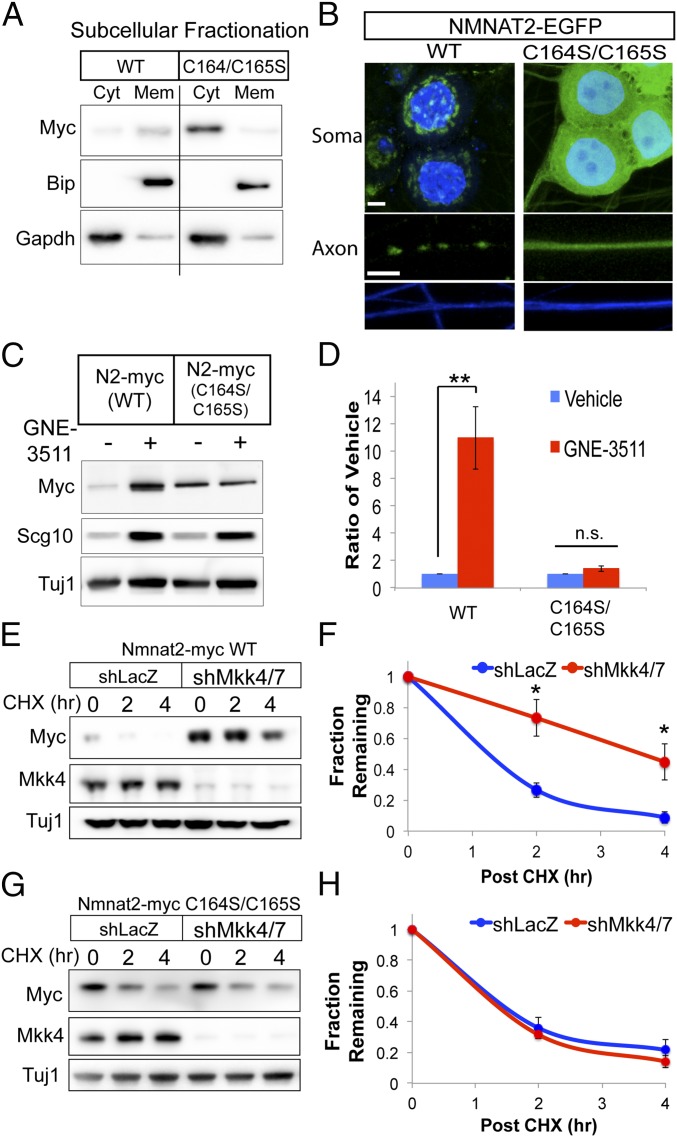
We sought to understand how DLK signaling targets NMNAT2 for degradation. Both NMNAT2 and SCG10 are palmitoylated and attached to vesicles (17, 19). DLK is also palmitoylated, and this modification is required for DLK-dependent activation of MKK4/7 and JNK MAPKs (39). Interestingly, mutagenesis of cysteines in NMNAT2 that are modified by palmitoylation delays NMNAT2 turnover in HEK293t cells (20). Due to these compelling connections, we investigated whether palmitoylation of NMNAT2 is necessary for MAPK-dependent degradation in sensory neurons. We mutated two cysteine residues modified by palmitoylation (C164S/C165S) and expressed this palmitoylation-dead (PD) NMNAT2 protein in primary sensory neurons. Based on subcellular fractionation, a majority of wild-type (WT) NMNAT2 distributes with the membrane fraction, while a smaller pool is soluble, consistent with findings in brain extracts and cell lines (19) (Fig. 3A). In contrast, PD-NMNAT2 distributes predominantly in the soluble fraction. We further examined NMNAT2 localization in sensory neurons using NMNAT2-GFP fusion proteins under fluorescence microscopy. In the soma, WT NMNAT2 is predominantly located at the Golgi and colocalizes with vesicles in the axon (Fig. 3B). However, PD-NMNAT2 is diffuse in both the soma and axon, confirming previous findings that palmitoylation is required for membrane association of NMNAT2 in primary sensory neurons.
Fig. 3.
NMNAT2 palmitoylation is required for MAPK-dependent degradation. (A) Subcellular fractionation of WT or PD NMNAT2 (C164S/C165S) from DRG sensory neurons transduced with Myc-tagged expression constructs. WT NMNAT2 is predominantly found in the membrane (Mem) fraction, while NMNAT2 (C164S/C165S) is detected predominantly in the cytosolic (Cyt) fraction. Successful fractionation was confirmed by immunoblotting for the endoplasmic reticulum resident BiP (Meme) and glycolytic enzyme GAPDH (Cyt). The vertical line indicates samples are from the same Western blot exposure, but they were not on adjacent lanes. This Western blot is representative of three independent experiments. (B) Fluorescence microscopy of NMNAT2-EGFP fusion protein in DRG sensory neurons, where nuclei in soma are labeled with DAPI (blue, Upper) and axons are labeled with TUJ1 (blue, Lower). (Scale bars, 5 μm.) NMNAT2(C164S/C165S)-EGFP is diffusely localized in both soma and axons. (C and D) Steady-state levels of WT and C164S/C165S NMNAT2-myc treated with GNE-3511 for 8 h. Western blots for endogenous SCG10 are shown as positive controls for GNE-3511 treatment. WT and mutant NMNAT2 levels are normalized internally to vehicle control. Statistical comparisons were performed with t tests, where **P < 0.01 (n = 4). n.s., not significant. (E and F) Protein turnover studies using the protein synthesis inhibitor cycloheximide (CHX) show that the half-life of WT NMNAT2-myc is extended in the presence of shRNAs to MKK4/7. (G and H) Turnover of NMNAT2-myc (C164S/C165S) is unaffected in the presence of shRNAs to MKK4/7. In F and H, statistical comparisons were performed with t tests, where *P < 0.05 (n = 3). Error bars represent SEM.
Since Milde et al. (20) evaluated NMNAT2 stability in HEK293t cells, we sought to determine whether abolishing palmitoylation affected protein turnover of NMNAT2 in axons from primary sensory neurons. Notably, the steady-state levels of PD-NMNAT2 are substantially lower in the axon compartment compared with WT NMNAT2; however, the half-life of PD-NMNAT2 in axons was more than doubled (from 1 h to 2 h) (SI Appendix, Fig. S3). In a sensory neuron fraction containing mostly soma, PD-NMNAT2 steady-state levels are elevated compared with WT NMNAT2, while protein degradation is delayed to a similar extent as in axons. These observations demonstrate that palmitoylation promotes NMNAT2 degradation in sensory neurons while facilitating NMNAT2 accumulation in the axon, likely via association with vesicles and anterograde transport.
We next sought to test whether MAPK signaling targets palmitoylated NMNAT2 for degradation. If palmitoylation of NMNAT2 were necessary for MAPK-dependent degradation, then inhibiting DLK activity should have no effect on protein levels of PD-NMNAT2. Indeed, GNE-3511 treatment increases protein levels of WT NMNAT2, while no change is observed in PD-NMNAT2 (Fig. 3 C and D). We made similar observations with a small-molecule JNK inhibitor (SI Appendix, Fig. S4). To confirm that MAPK signaling regulates the degradation of palmitoylated NMNAT2, the turnover rate of WT and PD-NMNAT2 was assessed from sensory neurons after shRNA knockdown of MKK4 and MKK7. As we previously demonstrated (25), knocking down MKK4/7 extends the half-life of WT NMNAT2 (Fig. 3 E and F); however, loss of MAPK signaling has no effect on the turnover rate of PD-NMNAT2 (Fig. 3 G and H). These findings collectively demonstrate that DLK-dependent signaling promotes the selective degradation of palmitoylated NMNAT2.
Palmitoylation Is Required for SCG10 Protein Degradation.
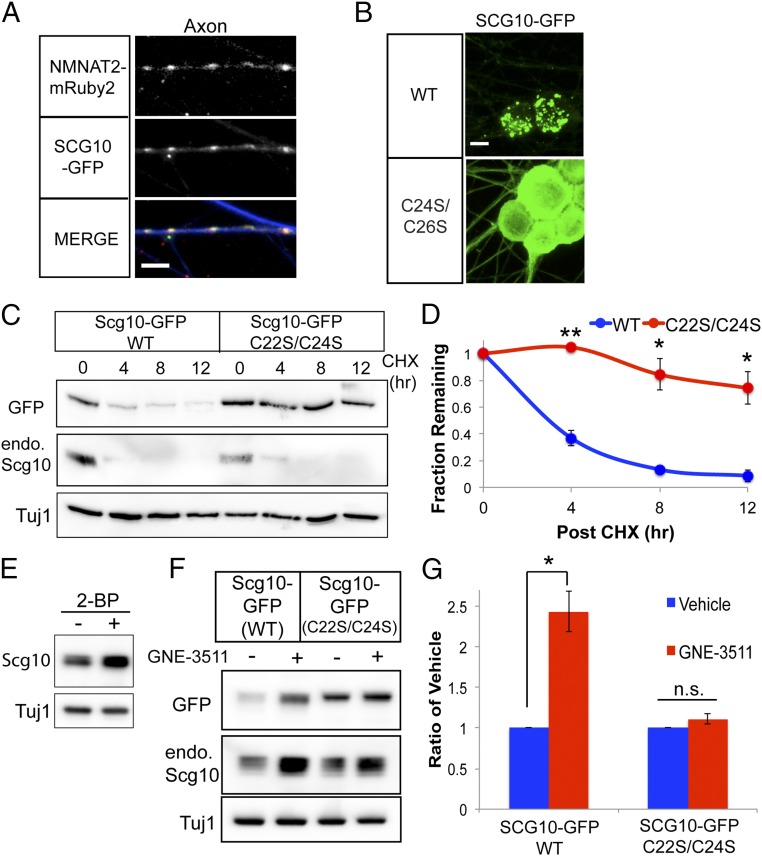
In addition to NMNAT2, DLK-MKK4/7-JNK signaling regulates degradation of the axon maintenance factor and neuronal stathmin SCG10 (15). Since palmitoylation is required for MAPK-dependent degradation of NMNAT2, we examined whether palmitoylation likewise contributes to degradation of SCG10. We first wondered if NMNAT2 and SCG10 reside on overlapping vesicle populations. Indeed, SCG10-EGFP puncta colocalize with NMNAT2-mRuby2 puncta in the axons of sensory neurons (Fig. 4A). SCG10 possesses two cysteines in an N-terminal localization domain that are modified by palmitoylation (17). Mutagenesis of these cysteines (C22S and C24S) redistributes this PD-SCG10 to the cytosol (Fig. 4B), confirming that palmitoylation is necessary for membrane association in sensory neurons.
Fig. 4.
Protein degradation of SCG10 is dependent on palmitoylation. (A) Colocalization of NMNAT2-mRuby2 with SCG10-GFP in DRG sensory neuron axons. Axons are labeled with TUJ1 (blue) in the merged image. (Scale bar, 5 μm.) (B) Fluorescence microscopy of SCG10-GFP in sensory neurons. WT SCG10-EGFP localizes to puncta, while SCG10(C22S/C24S)-GFP is predominantly diffuse and steady-state levels are much higher. (Scale bar, 10 μm.) (C and D) Turnover of WT or C22S/C24S SCG10-GFP after treatment with cycloheximide (CHX), with quantification. endo., endogenous. (E) Western blot of SCG10 levels from DRGs treated with 50 μM 2-bromopalmitate (2-BP). The Western blot is representative of four independent experiments. (F and G) Treatment with GNE-3511 increases levels of WT SCG10-GFP but does not affect levels of C22S/C24S SCG10-GFP. WT and mutant SCG10 are normalized internally to vehicle control. In D and G, statistical comparisons were performed with a t test, where **P < 0.01 and *P < 0.05 (n = 3). n.s., not significant. Error bars represent SEM.
Although palmitoylation is important for the localization and function of stathmins, no one has assessed whether palmitoylation affects the stability of these proteins. As shown in Fig. 4B, the steady-state levels of PD-SCG10-GFP are much higher compared with WT SCG10-GFP. We next investigated the turnover of this mutant. In sensory neurons, the half-life of WT SCG10-GFP is ∼3 h. However, we observe that the half-life of PD-SCG10-GFP is longer than 12 h (Fig. 4 C and D). As an additional test of this hypothesis, we used 2-bromopalmitate to broadly inhibit DHHC (Asp-His-His-Cys)-palmitoyltransferase activity in sensory neurons (40). We find that 2-bromopalmitate treatment increases the steady-state protein levels of SCG10 (Fig. 4E). These observations support the conclusion that palmitoylation strongly contributes to the degradation of this stathmin. Based on our findings with NMNAT2, we hypothesized PD-SCG10 would not be regulated by MAPK signaling. While GNE-3511 treatment increases the protein levels of WT SCG10-EGFP and endogenous SCG10, GNE-3511 does not affect levels of PD-SCG10-EGFP (Fig. 4 F and G). Hence, similar to NMNAT2, palmitoylation is necessary for MAPK-dependent degradation of SCG10.
The Phr1 E3 Ligase Complex Targets Soluble NMNAT2 for Degradation.
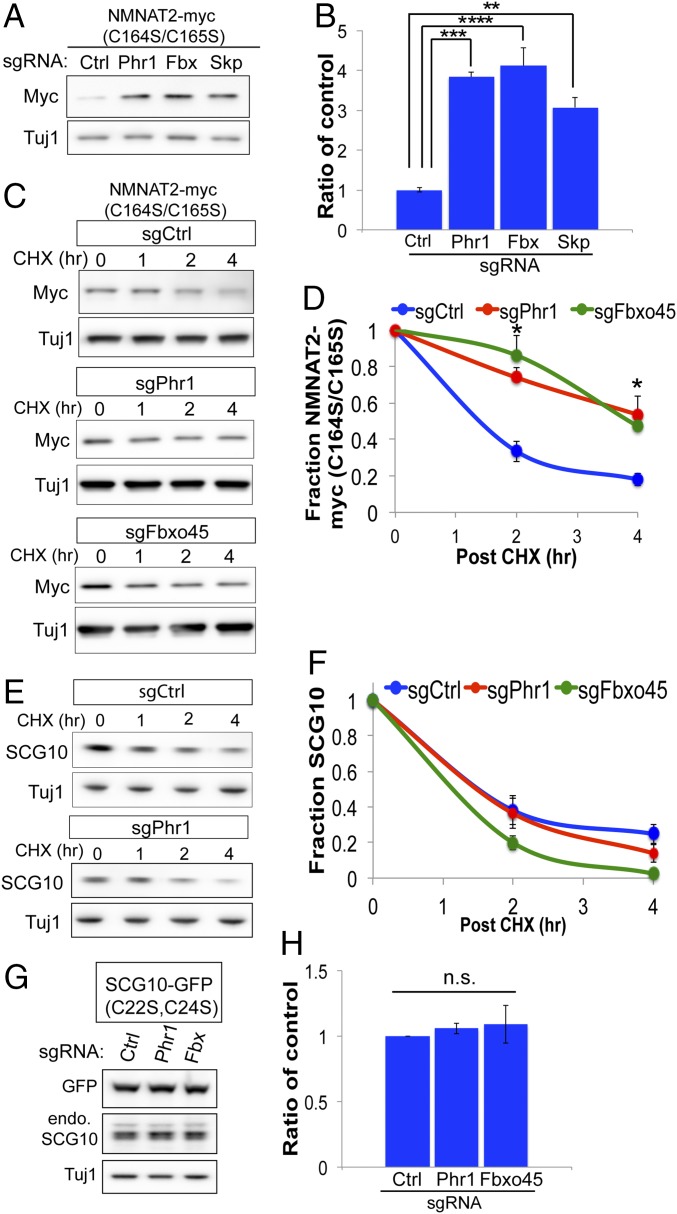
The above findings suggest MAPK signaling selectively promotes degradation of palmitoylated NMNAT2 and SCG10. However, as shown in Fig. 2, abolishing NMNAT2 palmitoylation only partially extends NMNAT2 stability, suggesting additional pathways are responsible for degrading soluble NMNAT2. In addition to DLK signaling, NMNAT2 protein levels are regulated by an atypical SCF ubiquitin ligase complex composed of the E3 ligase Phr1, F-box protein Fbxo45, and Skp1a (21–24). Genetic loss of any component of the ligase complex increases the stability of NMNAT2 in severed axons and extends axon survival in vitro and in vivo. We wondered if this SCF ubiquitin ligase complex targets nonpalmitoylated NMNAT2 protein for degradation. Individual components of the SCF complex were genetically inactivated by sgRNAs in CAS9-expressing sensory neurons. In contrast to inhibiting DLK, steady-state PD-NMNAT2 protein levels increased in neurons lacking Phr1, Fbxo45, or Skp1a (Fig. 5 A and B). CRISPR inactivation of Phr1 or Fbxo45 extended the half-life of PD-NMNAT2 (Fig. 5 C and D). If this SCF ubiquitin ligase complex regulates nonpalmitoylated NMNAT2, we predicted that inhibiting global palmitoylation would render endogenous NMNAT2 more sensitive to perturbations of this E3 complex. To test this hypothesis, CAS9-expressing sensory neurons were treated with 2-bromopalmitate and steady-state levels of NMNAT2 were monitored in the presence of sgRNAs to Fbxo45 or Skp1a. Loss of Fbxo45 or Skp1a has a small effect on basal NMNAT2 protein levels; however, treatment with 2-bromopalmitate in the presence of sgRNAs to Fbxo45 or Skp1a leads to a significant increase in endogenous NMNAT2 protein (SI Appendix, Fig. S5). Hence, preventing NMNAT2 palmitoylation enhances its sensitivity to degradation by the Phr1 E3 ligase complex.
Fig. 5.
Phr1 E3 ligase complex targets cytosolic NMNAT2 for degradation. (A and B) Steady-state levels of NMNAT2(C164S/C165S)-Myc in Cas9-expressing DRG sensory neurons transduced with the indicated sgRNAs, with quantification. A one-way ANOVA was performed with Bonferroni post hoc correction for multiple comparisons, where **P < 0.01, ***P < 0.005, and ****P < 0.0001 for the indicated groups (n = 3). Ctrl, control. (C and D) Turnover of NMNAT2(C164S/C165S)-Myc was assessed in Cas9-expressing DRG sensory neurons treated with cycloheximide (CHX) at the indicated time points, with quantification. Statistical comparisons were performed with t tests, where *P < 0.05 (n = 5). (E and F) Turnover of endogenous SCG10 was assessed in Cas9-expressing DRG sensory neurons, with quantification. No statistically significant differences were observed at any time point (n = 3). (G and H) Steady-state levels of SCG10 (C22S/C24S)-GFP were detected from Cas9-expressing DRG sensory neurons in the presence of the indicated sgRNAs (n = 3). n.s., not significant. Error bars represent SEM.
Since our findings with MAPK signaling suggest there are parallel mechanisms of regulation between NMNAT2 and SCG10, we next wondered if SCG10 is similarly regulated by the Phr1 E3 complex. However, CRISPR inactivation of Phr1 or Fbxo45 has no effect on the turnover rate of SCG10 (Fig. 5 E and F). Since nonpalmitoylated NMNAT2 was strongly regulated by this E3 ligase complex, we wondered if nonpalmitoylated SCG10 would be affected by loss of these factors. However, CRISPR inactivation of Phr1 or Fbxo45 does not affect levels of PD-SCG10-GFP (Fig. 5 G and H). Consequently, the Phr1 E3 complex regulates axon degeneration by selectively tuning levels of the axon survival factor NMNAT2, and not SCG10.
Dual Inhibition of MAPK Signaling and Phr1 Enhances Axon Survival Against Pathological Axon Degeneration.
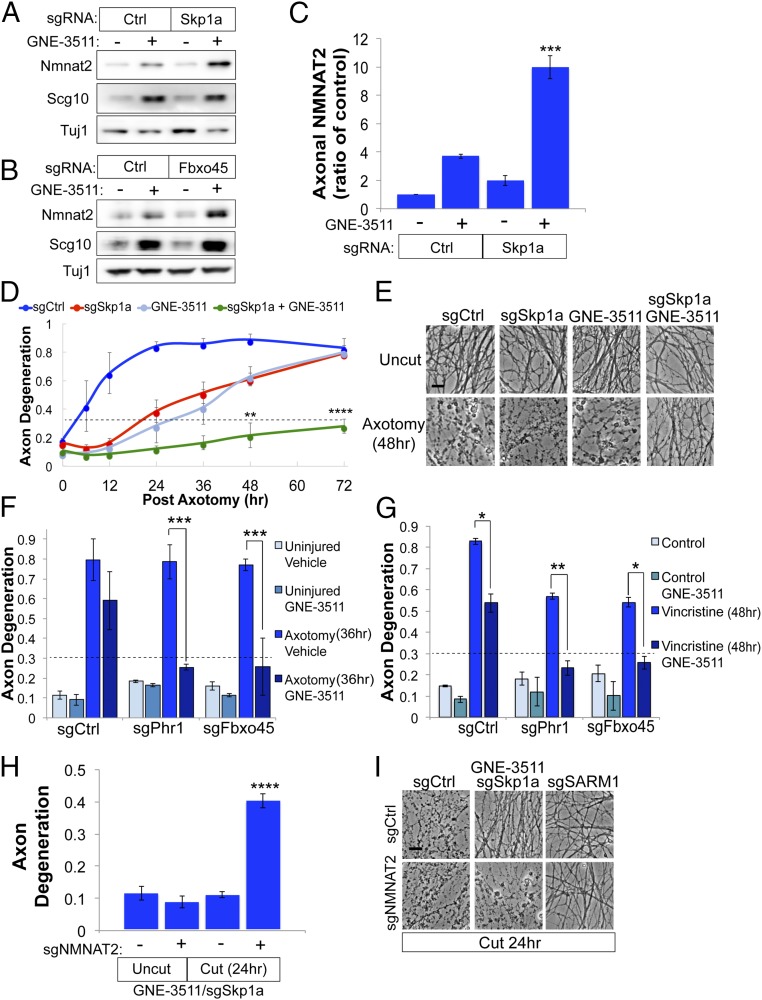
The studies described above show that MAPK signaling and the Phr1 E3 ligase complex regulate different NMNAT2 subpopulations and function as parallel, independent pathways driving NMNAT2 protein degradation. This finding implies that combined inactivation of both pathways will lead to additive accumulation of NMNAT2 in axons. To test this hypothesis, we pretreated sensory neurons with GNE-3511 to inhibit DLK/LZK and inactivated components of the Phr1 E3 complex with CRISPR. Knocking out Skp1a or Fbxo45 in combination with GNE-3511 treatment results in a synergistic increase in axonal NMNAT2 protein (Fig. 6 A–C). If axon survival is linked to NMNAT2 abundance, then combined inhibition of DLK signaling and the Phr1 E3 ligase complex should confer enhanced axon protection. Indeed, GNE-3511 treatment in the presence of sgRNAs targeting Skp1a results in dramatically enhanced axon protection against axotomy-induced degeneration, delaying fragmentation for 72 h after axotomy (Fig. 6 D and E). GNE-3511 treatment also confers profound protection in combination with sgRNAs to Phr1 or Fbxo45 (Fig. 6F). Since JNK MAPKs act downstream of DLK/LZK, we predicted that pharmacological inhibition of JNK should also evoke enhanced axon preservation with sgRNAs targeting the E3 ligase complex. Indeed, treating DRG sensory neurons with a small-molecule JNK inhibitor does enhance axon protection in the presence of sgRNAs to Phr1, Fbxo45, or Skp1a after axotomy (SI Appendix, Fig. S6).
Fig. 6.
Combined inactivation of MAPK signaling and E3 ligase complex confers additive axon protection. Cas9-expressing DRG sensory neurons transduced with control (Ctrl) sgRNAs or sgRNAs targeting Skp1a (A) or Fbxo45 (B) were treated with GNE-3511 for 14 h, and levels of endogenous NMNAT2 were assessed from axon-only extracts by Western blot. (C) Quantification of axonal NMNAT2 levels from DRGs treated as in A [***P < 0.001, single-factor ANOVA with Bonferroni post hoc test for multiple comparisons (n = 3)]. (D and E) Time course of axon degeneration after axotomy from DRGs treated with vehicle or GNE-3511 and Ctrl sgRNAs or sgRNAs targeting Skp1a. Axons are preserved for 72 h after axotomy when pretreated with GNE-3511 and sgRNAs to Skp1a, with representative images shown in E. (Scale bar, 5 μm.) A repeated-measures ANOVA was performed with Bonferroni post hoc tests for multiple comparisons, where ****P < 0.0001 and **P < 0.005. (F) Axon degeneration in distal axons 36 h after axotomy from DRGs treated with DMSO or GNE-3511 and sgRNAs targeting Phr1 or Fbxo5. (G) Axon degeneration in distal axons from DRG sensory neurons treated with 40 nM vincristine transduced with the indicated sgRNAs and GNE-3511. For F and G, a two-way ANOVA with post hoc Bonferroni test for multiple comparisons was performed, where *P < 0.01, **P < 0.005, and ***P < 0.001 (n = 3). (H) Cas9-expressing DRGs were treated as described in D, except cells were transduced with sgRNAs to NMNAT2 on DIV4. Axon degeneration was assessed in severed axons 24 h after axotomy (two-way ANOVA with Bonferroni correction for multiple comparisons, where ****P < 0.0001). Error bars represent SEM. (I) Representative bright-field images of distal axons from H with sgRNA to SARM1 included as a control. (Scale bar, 5 μm.)
We next examined whether combined inhibition of the DLK/Phr1 pathways will enhance axon survival against the chemotherapeutic agent vincristine, another model of pathological axon degeneration. GNE-3511 treatment protects axons from vincristine-induced degeneration for 24 h (SI Appendix, Fig. S1); however, this effect is lost after 48 h of exposure. The same effect is true for sgRNAs targeting Fbxo45 or Skp1a. However, combining GNE-3511 with sgRNAs to either Fbxo45 or Skp1a strongly suppresses axon degeneration for 48 h in the presence of vincristine (Fig. 6G). Finally, we confirmed that axon protection in this model is dependent on NMNAT2 by using sgRNAs to knock out endogenous NMNAT2. As predicted, axon protection afforded by loss of Skp1a and MAPK signaling is suppressed by the addition of sgRNAs targeting NMNAT2 (Fig. 6 H and I). In contrast, axons are still protected in a SARM1-deficient background even in the presence of sgRNAs targeting NMNAT2 (Fig. 6I), consistent with previous findings that SARM1 acts downstream of NMNAT2 to promote degeneration (14). Collectively, these findings reveal that NMNAT2 subpopulations are regulated by distinct cellular pathways that tune local abundance of this neuron survival factor to control SARM1-dependent axon degeneration.
Discussion
NMNAT2 is a major gatekeeper for axon health and resistance to pathological axon degeneration. The studies described herein identify converging regulatory pathways controlling local NMNAT2 abundance and axon survival after injury. In Wallerian degeneration, axon transection deprives the distal axon segment of NMNAT2 protein and evokes an intrinsic axon dismantling program dependent on the central executioner SARM1. SARM1 is an NADase that rapidly consumes NAD+ from severed axons (41, 42). Elevating NMNAT2 or related NMNAT enzymes suppresses this NADase activity and preserves axon integrity (43); however, translating this neuroprotective mechanism to human disease is challenging. In contrast to other NMNAT enzymes, NMNAT2 protein is short-lived and subject to constitutive, regulated degradation (16). This unique aspect of NMNAT2 biology offers an unexpected opportunity for therapeutic intervention by targeting proteostasis networks responsible for NMNAT2 degradation to elevate NMNAT2 abundance and suppress SARM1 activity. Our studies highlight the neuroprotective function of NMNAT2 in SARM1-dependent axon degeneration; however, NMNAT2 also displays chaperone activity (44), so elevating local NMNAT2 levels could provide additional therapeutic benefits in other neurological disorders.
One major regulatory node in NMNAT2 proteostasis is MAPK signaling via the MAP3K DLK. This stress kinase is particularly relevant to neurodegeneration, as Le Pichon et al. (28) report that DLK signaling is enhanced in neurological diseases, including Alzheimer’s disease and ALS. Inactivating DLK signaling via genetic loss or small-molecule inhibitors suppresses apoptotic cell death and ameliorates neurodegeneration in several models. In addition to preventing cell death, we find that kinase inhibitors targeting DLK have a second neuroprotective role in axons, blocking SARM1-dependent axon degeneration by elevating levels of NMNAT2 and SCG10. Our studies suggest the axoprotective potency of these drugs is a consequence of targeting DLK and the related MAP3K LZK, which shares 86% homology in the kinase domain with DLK (37). Knocking out both DLK and LZK leads to synergistic axon protection, revealing a previously unknown function for LZK in pathological axon degeneration. Consistent with our pharmacological studies, genetic loss of DLK/LZK promotes axon protection by elevating NMNAT2 abundance in the axon. Consequently, targeting the DLK/LZK MAP kinase pathway is a very appealing option in neurodegeneration by suppressing apoptotic death of the neuronal cell body as well as degeneration of a damaged axon via distinct mechanisms. Yang et al. (26) reported strong additive axon protection when DLK is deleted in combination with two other MAP3Ks, MEKK4 and MLK2. These kinases might cooperate with DLK to promote NMNAT2 degradation or activate axon degeneration through an independent pathway. Redundancy in the MAP3K network may explain why loss of DLK alone does not protect axons from degeneration in some injury models (33). Future studies will resolve how this MAPK network influences axon health and degeneration in disease.
Axon survival is dependent on constant delivery of NMNAT2 even in the absence of damage or disease. To facilitate axon transport, NMNAT2 is modified by palmitoylation at the trans-Golgi network and associated with vesicles to promote anterograde transport through the axon. Our data suggest membrane association protects NMNAT2 from recognition by the Phr1 E3 ligase pathway. In primary neurons and brain extracts, a majority of the NMNAT2 is associated with membranes (ref. 19 and this study), although membrane association might be dynamic and vary depending on the cell type, context, or neuronal compartment. Based on our findings, relocalizing NMNAT2 between cytosolic and membrane compartments would control whether NMNAT2 is more or less susceptible to MAPK-dependent turnover and endow axons with another stratum of regulatory control over degradation of this survival factor.
How palmitoylated NMNAT2 evades degradation by MAPK signaling in the axon requires further study. While it would be attractive to hypothesize that the MAPKs are only activated upon stress, our findings instead demonstrate that constitutive DLK/LZK signaling regulates NMNAT2 degradation. This is consistent with our prior finding that axotomy does not accelerate degradation of SCG10 (15). However, constitutive MAPK signaling and stress-activated MAPK signaling are not mutually exclusive, and a role for chronic stress activation of MAPK-dependent NMNAT2 degradation could have broad implications for neurodegeneration.
Palmitoylation is required for MAPK-dependent degradation of both NMNAT2 and SCG10, suggesting that this lipid modification is a component of a broader strategy for regulating protein degradation of neuronal proteins. The profound stability observed in a PD-SCG10 mutant indicates membrane association is critical for regulated degradation of this stathmin. We find that the Phr1 E3 ligase complex does not contribute to SCG10 degradation, suggesting that NMNAT2 is a specific substrate for this complex. Furthermore, our finding that NMNAT2 and SCG10 are colocalized raises the possibility that MAPK signaling regulates protein turnover of specific vesicle populations. DLK is also palmitoylated (39) and might be targeted to NMNAT2/SCG10-containing vesicles in response to local stimuli. For example, the DLK-MKK4/7-JNK axis is stimulated in response to perturbations to the cytoskeleton and to kinesins (45–47), suggesting that NMNAT2/SCG10 turnover might be tuned by changes in cytoskeletal dynamics or vesicular transport. Whether this vesicle population represents a “neuron survival vesicle” and is subject to specialized regulation remains to determined. Identifying the other residents of this vesicle and the fate of this vesicle population will reveal important insights into axon biology and regulation of axon survival factors in health and disease.
In addition to MAPK signaling, the Phr1 E3 ligase complex represents another regulatory layer in NMNAT2 proteostasis. Our biochemical and epistasis studies suggest that MAPK signaling and the Phr1 E3 ligase complex function independently to promote degradation of distinct NMNAT2 subpopulations. Inhibiting both pathways leads to a dramatic increase in axonal NMNAT2 and synergistic axon survival in response to traumatic injury or vincristine-induced degeneration. The network of NMNAT2 proteostasis is likely very rich and will include additional unidentified factors. For example, a small-molecule screen identified several candidate signaling pathways that modulate NMNAT2 mRNA and protein levels in cortical neurons (48). Furthermore, how NMNAT2 and other short-lived factors traverse long distances in a nerve and evade protein degradation is unknown. Whether supporting glia influence MAPK signaling or other protein homeostasis networks to regulate NMNAT2 axon delivery deserves future attention. Connecting these pathways in axon development and disease will reveal additional regulatory nodes in NMNAT2 biology and other therapeutic targets for diseases of axon loss.
Methods
Reagents and Plasmids.
The following chemicals were obtained from Sigma–Aldrich: vincristine sulfate salt (catalog no. V8879) poly-d-lysine hydrobromide (catalog no. P0899), cycloheximide (catalog no. C7698), digitonin (catalog no. D141), and 2-bromopalmitate (catalog no. 238422). For culture of primary DRG sensory neurons, Neurobasal Medium (catalog no. 21103-049; Life Technologies) was prepared with B27 supplement (catalog no. 17504-044; Life Technologies), nerve growth factor (catalog no. B.5017; Envigo Bioproducts), 5-fluoro-2′-deoxyuridine (catalog no. F0503; Sigma), and uridine (catalog no. U3003; Sigma). Laminin mouse protein was obtained from Thermo Fisher Scientific (catalog no. 23017015). Protein kinase inhibitors were as follows: Tozarsertib (catalog no. MK-0457; Apexbt), Sunitinib (catalog no. S-8803; LC Laboratories), GNE-3511 (catalog no. 19174; Cayman Chemicals), and JNK inhibitor VIII (catalog no. 15946; Cayman Chemicals). For all pharmacology studies, dimethyl sulfoxide (DMSO) was used as the vehicle control. Primary and secondary antibodies used in this study are shown in SI Appendix, Table S1. For expression plasmids, NMNAT2 was subcloned in FUGW lentiviral backbone. Details on the SCG10 construct are provided elsewhere (15). For EGFP/mRuby2 fusion constructs, the internal ribosome entry site-Venus was removed. For knockdown of MKK4 and MKK7, we used shRNAs from the RNAi consortium available at Sigma (MKK4: TRCN0000345130, MKK7: TRCN000012609). For biochemical studies, NMNAT2 was tagged at the C terminus with a myc tag and 6× HIS moiety. Mutations were generated with the Infusion (Clontech) procedure and confirmed by sequencing. For Cas9/CRISPR studies, sgRNAs were cloned in Lentiguide plasmid backbone after restriction digest of a BsmBI site. CRISPR targeting sequences for genes of interest were designed in consultation with the Genome Engineering and iPSC Core at Washington University School of Medicine. Sequences for individual sgRNAs used in this study are shown in SI Appendix, Table S2.
Isolation and Culture of DRG Sensory Neurons.
DRGs were dissected from embryonic day (E) 13.5 mouse embryos (equal number of male and female embryos). After dissociation in trypsin, the DRGs were seeded as spot cultures in plastic plates precoated with poly-d-lysine and laminin. DRGs were cultured in Neurobasal Medium supplemented with 2% B27, 50 ng/mL nerve growth factor, and 1 μM 5-fluoro-2′-deoxyuridine/1 μM uridine to suppress proliferation of nonneuronal cells. Fresh culture media were added every 2–3 d. For CRISPR studies, Cas9 knock-in (49) mice were crossed to mice expressing Cre downstream of the Actin promoter to induce ubiquitous Cas9 expression. E13.5 DRGs were isolated from the progeny of this cross and cultured as described above. For all other studies, pregnant CD1 mice were from Charles River Laboratories. For studies employing shRNAs, we infect sensory neurons 2 days after plating on days in vitro 2 (DIV2) with lentivirus expressing Bcl-xL to prevent nonspecific toxicity. Bcl-xL is not applied to any other experiments in this study as we do not observe nonspecific toxicity related to sgRNAs or Cas9 expression. All experimental protocols in this study using mice were reviewed and approved by the Washington University School of Medicine Institutional Animal Care and Use Committee.
Measurement of Axon Degeneration.
Axotomy was performed with a razor blade by manually severing sensory neuron axons seeded in plastic 96-well plates on DIV7 or DIV8. Bright-field images from severed distal axons were collected with an Operetta automated imaging system (PerkinElmer). Axon degeneration was quantified from bright-field images using an ImageJ (NIH)-based macro previously described (36). Axon degeneration is calculated as a ratio of fragmentated axons to total axon area in each image. Fragmentated axons are defined in each binarized image based on pixel circularity of an axon particle. Within each well, six fields were imaged that were distal to the cut site. For each experimental replicate, at least three wells were examined per condition. Independent sensory neuron cultures were used for each experimental replicate.
Production of Lentiviral Particles and Transduction of Primary Sensory Neurons.
Lentivirus was generated as previously described (50). In brief, lentiviral plasmids were cotransfected in HEK293t cells with vesicular stomatis virus glycoprotein and psPAX2 lentiviral packing vector. Two days later, the culture supernatant was collected and centrifuged for 1 min at 500 × g and the supernatant containing lentivirus was collected and stored at −80 °C. Lentivirus was applied to DRGs on DIV2 except in CRISPR studies, which were performed as follows. For all sgRNAs except those targeting NMNAT2, Cas9-expressing DRGs were transduced with lentivirus containing sgRNAs several hours after seeding on poly-d-lysine/laminin-coated plates. For NMNAT2 suppression studies, sgRNAs targeting NMNAT2 were added on DIV4 (SI Appendix, Fig. S2). Validation of CRISPR efficiency was performed by Western immunoblotting for endogenous protein levels of the target (SI Appendix, Fig. S7). For each experimental replicate, independent viruses were produced.
Subcellular Fractionation of DRG Neurons.
Methods were modified based on the subcellular fractionation protocol described by Milde and Coleman (51). On DIV8, DRGs were gently lifted from the plate with cold PBS and transferred to a plastic microcentrifuge tube on ice. DRGs were collected by centrifugation (500 × g for 5 min at 4 °C) and gently resuspended in fractionation buffer [150 mM NaCl, 50 mM Hepes (pH 7.4), 100 μg/mL digitonin prepared fresh in sterile H2O, 1 mM PMSF, and protease inhibitor mixture (Roche)]. DRGs were gently aspirated with a plastic pipette ∼10 times to disperse, but not lyse, the cells. DRGs were incubated in fractionation buffer for 45 min, constantly rotating at 4 °C. Digitonin-permeabilized DRGs were collected by centrifugation at 5,000 × g for 1 min at 4 °C. The supernatant represents the cytosolic fraction and was saved. The pellet was gently resuspended in cold fractionation buffer, and digitonin-permeabilized DRGs were collected again by centrifugation at 5,000 × g for 1 min at 4 °C. The supernatant was discarded, and digitonin-permeabilized DRGs were resuspended in cold fractionation with 1% Triton-X. The DRGs were vortexed for incubation at 4 °C for 30 min with intermittent vortexing to ensure complete membrane permeabilization. Cell debris, nuclei, and unpermeabilized cells were collected by centrifugation at 5,000 × g for 1 min at 4 °C. The supernatant (membrane fraction) was saved. Laemmli buffer was added to the cytosolic and membrane fractions, and extracts were analyzed by SDS/PAGE and Western immunoblotting. Western immunoblotting for GAPDH and BiP was used as a control for the cytosolic and membrane fractions, respectively.
Cycloheximide Chase Analysis of Protein Turnover and Western Immunoblotting.
On DIV7, neurons were treated with 25 μg/mL cycloheximide to inhibit protein synthesis and lysed as described below at the indicated time points after cycloheximide addition. To generate cell extracts, neurons were washed with cold PBS and then lysed in cold radioimmunoprecipitation assay buffer [150 mM NaCl, 50 mM Hepes (pH 7.4), 0.5% sodium deoxycholate, 0.1% Triton-X, 0.5% SDS, 1 mM PMSF, 1× protease inhibitor mixture (Roche), and 1× phosphatase inhibitor mixture (Sigma)]. Cell extracts were precleared of debris by centrifugation at 4,000 × g for 4 min at 4 °C. Clarified extracts were then mixed with Laemmli buffer and analyzed by SDS/PAGE and Western immunoblotting for the indicated protein. All quantification from Western immunoblotting was performed with ImageJ. For GNE-3511 studies of exogenously tagged protein levels, intensity levels were normalized to the DMSO control.
Confocal Microscopy of NMNAT2/SCG10 Localization.
DRGs were seeded in chamber slides coated as described above with poly-d-lysine and laminin. DRGs were transduced with lentivirus expressing NMNAT2-mRuby2 and SCG10-EGFP on DIV3. On DIV8, DRGs were fixed with 4% paraformaldehyde (in PBS) for 1 h. For TUJ1 immunostaining, cells were permeabilized for 30 min in 0.1% Triton-X/PBS solution with 3% goat serum. Permeabilized cells were incubated with anti-TUJ1 (1:1,000) overnight at 4 °C. Cells were washed three times with PBS and then incubated with goat anti-mouse antisera conjugated with Cy5 for 1 h. Cells were washed three times in PBS and then mounted in Vectashield (with DAPI) for visualization with a Leica DMI4000 B microscope under confocal settings. Axons and soma were visualized under oil immersion with a 60× objective. Images were generated from projection of confocal Z-stacks using a Leica software package, and final images were assembled with ImageJ.
Supplementary Material
Acknowledgments
We thank members of the A.D. and J.M. laboratories for helpful conversations and support. The Genome Engineering and iPSC Core at Washington University School of Medicine helped with design of sgRNAs for CRISPR studies. This work was supported by funds from NIH Grants R01-NS65053 (to A.D.), RF1-AG013730 (to J.M.), R01-NS087632 (to J.M. and A.D.), and R01-CA219866 (to A.D. and J.M.). D.W.S. is supported by Muscular Dystrophy Association Development Grant MDA344513, and A.D. is supported by Muscular Dystrophy Association Research Grant MDA349925.
Footnotes
Conflict of interest statement: J.M. and A.D. are cofounders of Disarm Therapeutics and members of its scientific advisory board.
This article is a PNAS Direct Submission. Y.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806933115/-/DCSupplemental.
References
- 1.Salvadores N, Sanhueza M, Manque P, Court FA. Axonal degeneration during aging and its functional role in neurodegenerative disorders. Front Neurosci. 2017;11:451. doi: 10.3389/fnins.2017.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cashman CR, Höke A. Mechanisms of distal axonal degeneration in peripheral neuropathies. Neurosci Lett. 2015;596:33–50. doi: 10.1016/j.neulet.2015.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kneynsberg A, Combs B, Christensen K, Morfini G, Kanaan NM. Axonal degeneration in tauopathies: Disease relevance and underlying mechanisms. Front Neurosci. 2017;11:572. doi: 10.3389/fnins.2017.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conforti L, Gilley J, Coleman MP. Wallerian degeneration: An emerging axon death pathway linking injury and disease. Nat Rev Neurosci. 2014;15:394–409. doi: 10.1038/nrn3680. [DOI] [PubMed] [Google Scholar]
- 5.Gerdts J, Summers DW, Milbrandt J, DiAntonio A. Axon self-destruction: New links among SARM1, MAPKs, and NAD+ metabolism. Neuron. 2016;89:449–460. doi: 10.1016/j.neuron.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osterloh JM, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J Neurosci. 2013;33:13569–13580. doi: 10.1523/JNEUROSCI.1197-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y, et al. MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J Exp Med. 2007;204:2063–2074. doi: 10.1084/jem.20070868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henninger N, et al. Attenuated traumatic axonal injury and improved functional outcome after traumatic brain injury in mice lacking Sarm1. Brain. 2016;139:1094–1105. doi: 10.1093/brain/aww001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisler S, et al. Prevention of vincristine-induced peripheral neuropathy by genetic deletion of SARM1 in mice. Brain. 2016;139:3092–3108. doi: 10.1093/brain/aww251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turkiew E, Falconer D, Reed N, Höke A. Deletion of Sarm1 gene is neuroprotective in two models of peripheral neuropathy. J Peripher Nerv Syst. 2017;22:162–171. doi: 10.1111/jns.12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziogas NK, Koliatsos VE. Primary traumatic axonopathy in mice subjected to impact acceleration: A reappraisal of pathology and mechanisms with high-resolution anatomical methods. J Neurosci. 2018;38:4031–4047. doi: 10.1523/JNEUROSCI.2343-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman MP, Freeman MR. Wallerian degeneration, wld(s), and nmnat. Annu Rev Neurosci. 2010;33:245–267. doi: 10.1146/annurev-neuro-060909-153248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilley J, Orsomando G, Nascimento-Ferreira I, Coleman MP. Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell Rep. 2015;10:1974–1981. doi: 10.1016/j.celrep.2015.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin JE, et al. SCG10 is a JNK target in the axonal degeneration pathway. Proc Natl Acad Sci USA. 2012;109:E3696–E3705. doi: 10.1073/pnas.1216204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilley J, Coleman MP. Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 2010;8:e1000300. doi: 10.1371/journal.pbio.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Paolo G, et al. Targeting of SCG10 to the area of the Golgi complex is mediated by its NH2-terminal region. J Biol Chem. 1997;272:5175–5182. doi: 10.1074/jbc.272.8.5175. [DOI] [PubMed] [Google Scholar]
- 18.Lutjens R, et al. Localization and targeting of SCG10 to the trans-Golgi apparatus and growth cone vesicles. Eur J Neurosci. 2000;12:2224–2234. doi: 10.1046/j.1460-9568.2000.00112.x. [DOI] [PubMed] [Google Scholar]
- 19.Mayer PR, et al. Expression, localization, and biochemical characterization of nicotinamide mononucleotide adenylyltransferase 2. J Biol Chem. 2010;285:40387–40396. doi: 10.1074/jbc.M110.178913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milde S, Gilley J, Coleman MP. Subcellular localization determines the stability and axon protective capacity of axon survival factor Nmnat2. PLoS Biol. 2013;11:e1001539. doi: 10.1371/journal.pbio.1001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong X, et al. The Highwire ubiquitin ligase promotes axonal degeneration by tuning levels of Nmnat protein. PLoS Biol. 2012;10:e1001440. doi: 10.1371/journal.pbio.1001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babetto E, Beirowski B, Russler EV, Milbrandt J, DiAntonio A. The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep. 2013;3:1422–1429. doi: 10.1016/j.celrep.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brace EJ, Wu C, Valakh V, DiAntonio A. SkpA restrains synaptic terminal growth during development and promotes axonal degeneration following injury. J Neurosci. 2014;34:8398–8410. doi: 10.1523/JNEUROSCI.4715-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamagishi Y, Tessier-Lavigne M. An atypical SCF-like ubiquitin ligase complex promotes wallerian degeneration through regulation of axonal Nmnat2. Cell Rep. 2016;17:774–782. doi: 10.1016/j.celrep.2016.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker LJ, et al. MAPK signaling promotes axonal degeneration by speeding the turnover of the axonal maintenance factor NMNAT2. eLife. 2017;6:e22540. doi: 10.7554/eLife.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, et al. Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell. 2015;160:161–176. doi: 10.1016/j.cell.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller BR, et al. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat Neurosci. 2009;12:387–389. doi: 10.1038/nn.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Pichon CE, et al. Loss of dual leucine zipper kinase signaling is protective in animal models of neurodegenerative disease. Sci Transl Med. 2017;9:eaag0394. doi: 10.1126/scitranslmed.aag0394. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh AS, et al. DLK induces developmental neuronal degeneration via selective regulation of proapoptotic JNK activity. J Cell Biol. 2011;194:751–764. doi: 10.1083/jcb.201103153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pozniak CD, et al. Dual leucine zipper kinase is required for excitotoxicity-induced neuronal degeneration. J Exp Med. 2013;210:2553–2567. doi: 10.1084/jem.20122832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welsbie DS, et al. Functional genomic screening identifies dual leucine zipper kinase as a key mediator of retinal ganglion cell death. Proc Natl Acad Sci USA. 2013;110:4045–4050. doi: 10.1073/pnas.1211284110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watkins TA, et al. DLK initiates a transcriptional program that couples apoptotic and regenerative responses to axonal injury. Proc Natl Acad Sci USA. 2013;110:4039–4044. doi: 10.1073/pnas.1211074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes KA, Harder JM, John SW, Shrager P, Libby RT. DLK-dependent signaling is important for somal but not axonal degeneration of retinal ganglion cells following axonal injury. Neurobiol Dis. 2014;69:108–116. doi: 10.1016/j.nbd.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel S, et al. Scaffold-hopping and structure-based discovery of potent, selective, and brain penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine inhibitors of dual leucine zipper kinase (DLK, MAP3K12) J Med Chem. 2015;58:8182–8199. doi: 10.1021/acs.jmedchem.5b01072. [DOI] [PubMed] [Google Scholar]
- 35.Tedeschi A, Bradke F. The DLK signalling pathway–A double-edged sword in neural development and regeneration. EMBO Rep. 2013;14:605–614. doi: 10.1038/embor.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerdts J, Sasaki Y, Vohra B, Marasa J, Milbrandt J. Image-based screening identifies novel roles for IkappaB kinase and glycogen synthase kinase 3 in axonal degeneration. J Biol Chem. 2011;286:28011–28018. doi: 10.1074/jbc.M111.250472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welsbie DS, et al. Enhanced functional genomic screening identifies novel mediators of dual leucine zipper kinase-dependent injury signaling in neurons. Neuron. 2017;94:1142–1154.e6. doi: 10.1016/j.neuron.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen M, et al. Leucine zipper-bearing kinase promotes axon growth in mammalian central nervous system neurons. Sci Rep. 2016;6:31482. doi: 10.1038/srep31482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holland SM, et al. Palmitoylation controls DLK localization, interactions and activity to ensure effective axonal injury signaling. Proc Natl Acad Sci USA. 2016;113:763–768. doi: 10.1073/pnas.1514123113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jennings BC, et al. 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J Lipid Res. 2009;50:233–242. doi: 10.1194/jlr.M800270-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, Milbrandt J. SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science. 2015;348:453–457. doi: 10.1126/science.1258366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Essuman K, et al. The SARM1 Toll/Interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron. 2017;93:1334–1343.e5. doi: 10.1016/j.neuron.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasaki Y, Nakagawa T, Mao X, DiAntonio A, Milbrandt J. NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD+ depletion. eLife. 2016;5:e19749. doi: 10.7554/eLife.19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ali YO, et al. NMNAT2:HSP90 complex mediates proteostasis in proteinopathies. PLoS Biol. 2016;14:e1002472. doi: 10.1371/journal.pbio.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valakh V, Walker LJ, Skeath JB, DiAntonio A. Loss of the spectraplakin short stop activates the DLK injury response pathway in Drosophila. J Neurosci. 2013;33:17863–17873. doi: 10.1523/JNEUROSCI.2196-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valakh V, Frey E, Babetto E, Walker LJ, DiAntonio A. Cytoskeletal disruption activates the DLK/JNK pathway, which promotes axonal regeneration and mimics a preconditioning injury. Neurobiol Dis. 2015;77:13–25. doi: 10.1016/j.nbd.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, et al. Restraint of presynaptic protein levels by Wnd/DLK signaling mediates synaptic defects associated with the kinesin-3 motor Unc-104. eLife. 2017;6:e24271. doi: 10.7554/eLife.24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali YO, Bradley G, Lu HC. Screening with an NMNAT2-MSD platform identifies small molecules that modulate NMNAT2 levels in cortical neurons. Sci Rep. 2017;7:43846. doi: 10.1038/srep43846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasaki Y, Vohra BPS, Lund FE, Milbrandt J. Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J Neurosci. 2009;29:5525–5535. doi: 10.1523/JNEUROSCI.5469-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milde S, Coleman MP. Identification of palmitoyltransferase and thioesterase enzymes that control the subcellular localization of axon survival factor nicotinamide mononucleotide adenylyltransferase 2 (NMNAT2) J Biol Chem. 2014;289:32858–32870. doi: 10.1074/jbc.M114.582338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.