Abstract
Tobacco smoking increases the risk of at least 17 classes of cancer. We analyzed somatic mutations and DNA methylation in 5,243 cancers of types for which tobacco smoking confers an elevated risk. Smoking is associated with increased mutation burdens of multiple distinct mutational signatures, which contribute to different extents in different cancers. One of these signatures, mainly found in cancers derived from tissues directly exposed to tobacco smoke, is attributable to misreplication of DNA damage caused by tobacco carcinogens. Others likely reflect indirect activation of DNA editing by APOBEC cytidine deaminases and of an endogenous clock-like mutational process. Smoking is associated with limited differences in methylation. The results are consistent with the proposition that smoking increases cancer risk by increasing the somatic mutation load, although direct evidence for this mechanism is lacking in some smoking-related cancer types.
Tobacco smoking has been associated with at least 17 types of human cancer (Table 1) and claims the lives of more than six million people every year (1–4). Tobacco smoke is a complex mixture of chemicals among which at least 60 are carcinogens (5). Many of these are thought to cause cancer by inducing DNA damage which, if misreplicated, leads to an increased burden of somatic mutations and hence an elevated chance of acquiring “driver” mutations in cancer genes. Such damage is often in the form of covalent bonding of metabolically activated reactive species of the carcinogen to DNA bases, termed “DNA adducts” (6). Tissues directly exposed to tobacco smoke (e.g. lung) as well as some tissues not directly exposed (e.g. bladder) show elevated levels of DNA adducts in smokers and thus evidence of exposure to carcinogenic components of tobacco smoke (7, 8).
Table 1. Mutational signatures and cancer types associated with tobacco smoking.
Information about the age adjusted odds ratios for current male smokers to develop cancer is taken from refs. (2–4). Odds ratios for small cell lung cancer, lung squamous and lung adenocarcinoma are for an average daily dose of more than 30 cigarettes. Odds ratios for cervix and ovary are for current female smokers. Detailed information about all mutation types, all mutational signatures and DNA methylation is provided in Table S2. Nomenclature for signature IDs is consistent with the COSMIC website, http://cancer.sanger.ac.uk/cosmic/signatures. The patterns of all mutational signatures with elevated mutation burden in smokers are displayed in Fig. 2B. N/A denotes lack of smoking annotation for a given cancer type. * denotes that a signature correlates with pack years smoked in a cancer type.
| Cancer Type | Odds ratios | Non-smokers | Smokers | Total number of mutational signatures found in the cancer type | Signature 4 found in cancer type | Mutational signatures with elevated mutation burden in smokers compared to non-smokers (q-value<0.05) |
|---|---|---|---|---|---|---|
| All Cancer Types | 1,062 | 2,490 | 26 | Y | 4, 5* | |
| Small Cell Lung Cancer | 111.3 | 3 | 145 | 6 | Y | |
| Lung Squamous | 103.5 | 7 | 168 | 8 | Y | 4*, 5 |
| Lung Adenocarcinoma | 21.9 | 120 | 558 | 7 | Y | 2*, 4*, 5*, 13* |
| Larynx | 13.2 | 6 | 117 | 5 | Y | 4*, 5 |
| Pharynx | 6.6 | 27 | 49 | 5 | Y | 5* |
| Oral Cavity | 4.2 | 98 | 265 | 5 | Y | 5* |
| Esophagus Squamous | 3.9 | 99 | 193 | 9 | Y | 5 |
| Esophagus Adenocarcinoma | 3.9 | 67 | 175 | 9 | Y | |
| Bladder | 3.8 | 111 | 288 | 5 | N | 5* |
| Liver | 2.9 | 157 | 235 | 19 | Y | 4*, 5, 16 |
| Stomach | 2.1 | 472 | 13 | N | N/A | |
| Acute Myeloid Leukemia | 2.0 | 202 | 2 | N | N/A | |
| Ovary | 1.9 | 458 | 3 | N | N/A | |
| Cervix | 1.8 | 94 | 74 | 8 | N | |
| Kidney | 1.7 | 154 | 103 | 6 | N | 5 |
| Pancreas | 1.6 | 119 | 120 | 11 | N | |
| Colorectal | 1.3 | 559 | 4 | N | N/A | |
Each biological process causing mutations in somatic cells leaves a mutational signature (9). Many cancers have a somatic mutation in the TP53 gene and catalogues of TP53 mutations compiled two decades ago enabled early exploration of these signatures (10) showing that lung cancers from smokers have more C>A transversions than lung cancers from non-smokers (11–14). To investigate mutational signatures using the thousands of mutation catalogues generated by systematic cancer genome sequencing, we recently described a framework in which each base substitution signature is characterized using a 96 mutation classification that includes the six substitution types together with the bases immediately 5’ and 3’ to the mutated base (15). The analysis extracts mutational signatures from mutation catalogues and estimates the number of mutations contributed by each signature to each cancer genome (15). Using this approach, more than 30 different base substitution signatures have been identified (16–18).
Here, we studied 5,243 cancer genome sequences (4,633 exomes and 610 whole genomes) of cancer classes for which smoking increases risk to identify mutational signatures and methylation changes associated with tobacco smoking (Table S1). 2,490 samples were reported to be from tobacco smokers and 1,063 from never smokers (Table 1) enabling investigation of the mutational consequences of smoking by comparing somatic mutations and methylation in smokers with non-smokers for lung, larynx, pharynx, oral cavity, esophagus, bladder, liver, cervix, kidney and pancreas cancers (Fig. 1 and Table S2).
Fig. 1. Comparison between tobacco smokers and lifelong non-smokers.
Bars are used to display average values for numbers of somatic substitutions per megabase, numbers of indels per megabase, numbers of dinucleotide mutations per megabase, numbers of breakpoints per megabase, fraction of the genome that shows copy number changes and numbers of mutations per megabase attributed to mutational signatures found in multiple cancer types associated with tobacco smoking. Light gray bars are non-smokers, while dark gray bars are smokers. Comparisons between smokers and non-smokers for all features, including mutational signatures specific for a cancer type and overall DNA methylation are provided in Table S2. Error bars correspond to 95% confidence intervals for each feature. Each q-value is based on a two-sample Kolmogorov-Smirnov test corrected for multiple hypothesis testing for all features in a cancer type. Cancer types are ordered based on their age adjusted odds ratios for smoking as provided in Table 1. Data for numbers of breakpoints per megabase and fraction of the genome that shows copy number changes were not available for liver cancer and small cell lung cancer. Adeno stands for Adenocarcinoma; Esophag. stands for Esophagus. Note that the presented data include only a few cases (<10) of nonsmokers for lung small cell, lung squamous and cancer of the larynx.
We first compared total numbers of base substitutions, small insertions and deletions (indels) and genomic rearrangements. Total base substitutions were higher in smokers compared to non-smokers for all cancer types together (q-value<0.05) and, for individual cancer types, in lung adenocarcinoma, larynx, liver and kidney cancers (Table S2). Total numbers of indels were higher in smokers compared to non-smokers in lung adenocarcinoma and liver cancer (Table S2). The whole genome sequenced cases allowed comparison of genome rearrangements between smokers and non-smokers in pancreatic and liver cancer, where no differences were found (Table S2). However, sub-chromosomal copy number changes entail genomic rearrangement and can serve as surrogates for rearrangements. Lung adenocarcinomas from smokers exhibited more copy number aberrations than from non-smokers (Table S2).
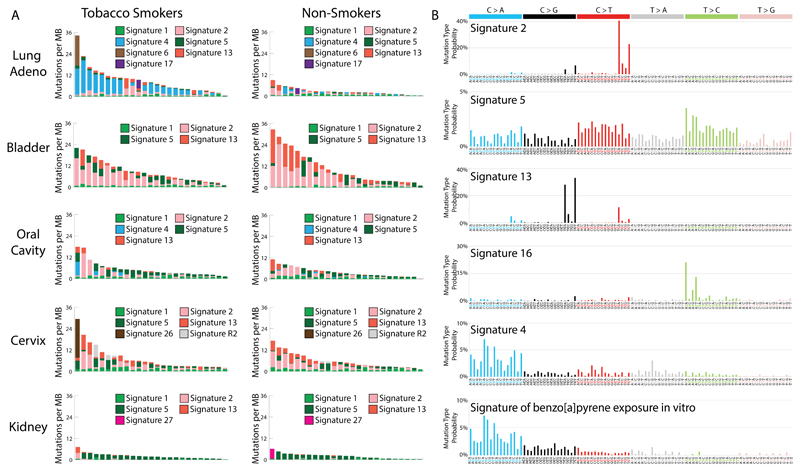
We then extracted mutational signatures, estimated the contributions of each signature to each cancer and compared the numbers of mutations attributable to each signature in smokers and non-smokers. Increases in smokers compared to non-smokers were seen for signatures 2, 4, 5, 13 and 16 (the mutational signature nomenclature is that used in COSMIC and references (16–18)). There was sufficient statistical power to show that these increases were of clonal mutations (mutations present in all cells of each cancer) for signatures 4 and 5 (q-value<0.05) as expected if they are due to cigarette smoke exposure prior to neoplastic change (Supplementary Text).
Signature 4 is characterized mainly by C>A mutations with smaller contributions from other base substitution classes (Fig. 2B and Fig. S1). It was found only in cancer types in which tobacco smoking increases risk and mainly in those derived from epithelia directly exposed to tobacco smoke (Fig. S2 and S3). Signature 4 is very similar to the mutational signature induced in vitro by exposing cells to benzo[a]pyrene (cosine similarity=0.94; Figs. 2B and S3), a tobacco smoke carcinogen (19). The similarity extends to the presence of a transcriptional strand bias indicative of transcription-coupled nucleotide excision repair (NER) of bulky DNA adducts on guanine (Fig. S1), the proposed mechanism of DNA damage by benzo[a]pyrene. Thus, signature 4 is likely the direct mutational consequence of misreplication of DNA damage induced by tobacco carcinogens.
Fig. 2. Mutational signatures associated with tobacco smoking.
(A) Each panel contains 25 randomly selected cancer genomes (represented by individual bars) from either smokers or non-smokers in a given cancer type. The y-axes reflect numbers of somatic mutations per megabase. Each bar is colored proportionately to the numbers of mutations per megabase attributed to the mutational signatures found in that sample. Naming of mutational signatures is consistent with previous reports (16–18). (B) Each panel contains the pattern of a mutational signature associated with tobacco smoking. Signatures are depicted using a 96 substitution classification defined by the substitution type and sequence context immediately 5’ and 3’ to the mutated base. Different colors are used to display different types of substitutions. The percentages of mutations attributed to specific substitution types are on the vertical axes, while the horizontal axes display different types of substitutions. Mutational signatures are depicted based on the trinucleotide frequency of the whole human genome. Signatures 2, 4, 5, 13 and 16 are extracted from cancers associated with tobacco smoking. The signature of benzo[a]pyrene is based on in vitro experimental data (19). Numerical values for these mutational signatures are provided in Table S6.
Most lung and larynx cancers from smokers had many signature 4 mutations. There were more signature 4 mutations in cancers from smokers compared to non-smokers in all cancer types together (Table S2) and in lung squamous, lung adenocarcinoma and larynx cancers (Table S2) accounting, in large part, for differences in total numbers of base substitutions (Table 1). 13.8% of lung cancers in non-smokers showed many signature 4 mutations (Fig. 2A;>1 mutation per MB) which may be due to passive smoking, misreporting of smoking habits or annotation errors. Signature 4 mutations were also found in oral cavity, pharynx and esophagus cancers, albeit in much smaller numbers than in lung and larynx cancers perhaps due to less exposure to tobacco smoke or more efficient clearance. Differences in mutation burden attributed to signature 4 between smokers and non-smokers were not observed in these cancer types (Fig. 1). Signature 4 mutations were found at low levels in cancers of the liver, an organ not directly exposed to tobacco smoke, and were elevated in smokers compared to non-smokers (Fig. 1).
Signature 4 was not extracted from bladder, cervix, kidney or pancreas cancers, despite the known risks conferred by smoking and the presence of many smokers in these series. It was also not extracted from cancers of the stomach, colorectum, ovary and acute myeloid leukemia for which the smoking status in the analyzed series was unknown but among which many are likely to have been smokers. The tissues from which all these cancer types are derived are not directly exposed to tobacco smoke. Simulations indicate that the lack of signature 4 is not due to statistical limitations (Supplementary Text and Fig. S4). The absence of signature 4 suggests that misreplication of direct DNA damage due to tobacco smoke constituents does not contribute substantially to mutation burden in these cancers even though DNA adducts indicative of tobacco-induced DNA damage are present in the tissues from which they arise (7).
Signatures 2 and 13 are characterized by C>T and C>G mutations respectively at TpC dinucleotides and have been attributed to overactive DNA editing by APOBEC deaminases (20, 21). The cause of the over-activity in most cancers has not been established although APOBECs are implicated in the cellular response to entrance of foreign DNA, retrotransposon movement and local inflammation (22). Signatures 2 and 13 showed more mutations in smokers than non-smokers in lung adenocarcinoma (Table S2). Since they are found in many other cancer types, where they are apparently unrelated to tobacco smoking, it seems unlikely that the signature 2 and 13 mutations associated with smoking in lung adenocarcinoma are direct consequences of misreplication of DNA damage induced by tobacco smoke. More plausibly, the cellular machinery underlying signatures 2 and 13 is activated by tobacco smoke, perhaps as a result of inflammation arising from deposition of particulate matter or by indirect consequences of DNA damage.
Signature 5 is characterized by mutations distributed across all 96 subtypes of base substitution, with predominance of T>C and C>T mutations (Fig. 2B) and evidence of transcriptional strand bias for T>C mutations (18). Signature 5 is found in all cancer types, including those unrelated to tobacco smoking, and in most cancer samples. It is “clock-like” in that the number of mutations attributable to this signature correlates with age of diagnosis in many cancer types (17). Signature 5, together with signature 1, is thought to contribute to mutation accumulation in most normal somatic cells and in the germline (17, 23). The mechanisms underlying signature 5 are not well understood, although an enrichment of signature 5 mutations was found in bladder cancers harboring inactivating mutations in ERCC2 which encodes a component of NER (24).
Signature 5 (or a similar signature that is difficult to differentiate from it because of their relatively flat profiles) was increased between 1.3-fold and 5.1-fold (q-value<0.05; Table S2) in smokers compared to non-smokers in all cancer types together and in lung squamous, lung adenocarcinoma, larynx, pharynx, oral cavity, esophageal squamous, bladder, liver and kidney cancers. The association of smoking with signature 5 mutations across these nine cancer types therefore includes some for which the risks conferred by smoking are modest and for which normal progenitor cells are not directly exposed to cigarette smoke (Table 1). Given the clock-like nature of signature 5 (17), its presence in the human germline (23), its ubiquity in cancer types unrelated to tobacco smoking (18) and its widespread occurrence in non-smokers, it seems unlikely that signature 5 mutations associated with tobacco smoking are direct consequences of misreplication of DNA damaged by tobacco carcinogens. It is more plausible that smoking affects the machinery generating signature 5 mutations (24). Presumably as a consequence of the effects of smoking, signature 5 mutations correlated with age of diagnosis in non-smokers (p-value:0.001) but not in smokers (p-value:0.59).
Signature 16 is predominantly characterized by T>C mutations at ApT dinucleotides (Fig. 2B), exhibits a strong transcriptional strand bias consistent with almost all damage occurring on adenine (Fig. S5) and has only been found thus far in liver cancer. The underlying mutational process is currently unknown. Signature 16 exhibited a higher mutation burden in smokers compared to non-smokers in liver cancer (Table S2).
For smokers with lung, larynx, pharynx, oral cavity, esophageal, bladder, liver, cervix, kidney and pancreas cancers quantitative data on cumulative exposure to tobacco smoke were available (Table S1). Total numbers of base substitution mutations positively correlated with pack years smoked for all cancer types together (q-value<0.05) and for lung adenocarcinoma (Table S3). For individual mutational signatures, correlations with pack years smoked were found in multiple cancer types for signatures 4 and 5 (Table S3). Signature 4 correlated with pack years in lung squamous, lung adenocarcinoma, larynx and liver cancers. Signature 5 correlated with pack years in all cancers together, in lung adenocarcinoma, pharynx, oral cavity and bladder cancers (Table S3). In lung adenocarcinoma, correlations with pack years smoked were also observed for signatures 2 and 13. The rates of these correlations allow estimation of the approximate numbers of mutations accumulated in a normal cell of each tissue due to smoking a pack of cigarettes a day for a year: lung, 150 mutations; larynx, 97; pharynx, 39; oral cavity 23; bladder, 18; liver, 6 (Table S3).
Consistent with our results, previous studies have reported the higher numbers in smokers compared to non-smokers of total base substitutions in lung adenocarcinoma (mainly due to C>A substitutions) (25, 26), of signatures 4 and 5 in lung adenocarcinoma (18), of signature 4 in liver cancer (27) and of signature 5 in bladder cancer (24).
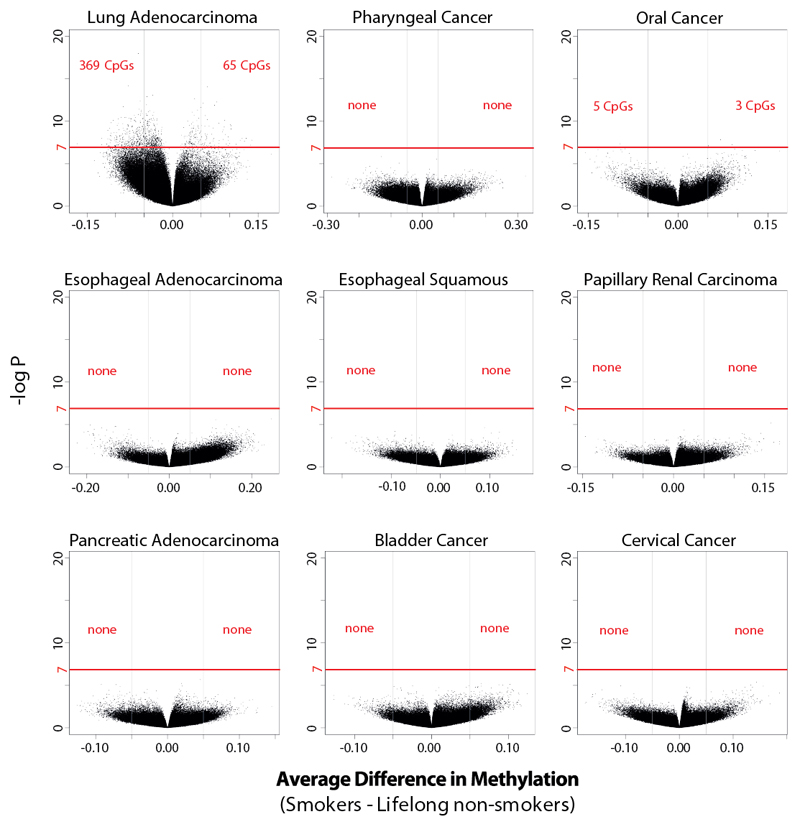
Differential methylation of the DNA of normal cells of smokers compared to non-smokers has been reported (28). Using data from methylation arrays, each containing ~470,000 of the ~28 million CpG sites in the human genome, we evaluated whether differences in methylation are found in cancers. Overall levels of CpG methylation in DNA from cancers were similar in smokers and non-smokers for all cancer types (Fig. S6). Individual CpGs were differentially methylated (>5% difference) only in two cancer types: 369 CpGs were hypo- and 65 were hyper-methylated in lung adenocarcinoma, with 5 hypo- and 3 hyper-methylated in oral cancer (Fig. 3 and S7). CpGs exhibiting differences in methylation clustered in certain genes but were neither associated with known cancer genes more than expected by chance nor with genes hypo-methylated in normal blood or buccal cells of tobacco smokers (Fig. S8; Tables S4 and S5) (28). Therefore, with the exception of lung cancer, CpG methylation showed limited differences between the cancers of smokers and non-smokers (Fig. 3).
Fig. 3. Differentially methylated individual CpGs in tobacco smokers across cancers associated with tobacco smoking.
Each dot represents an individual CpG. The horizontal axes reflect differences in methylation between lifelong non-smokers and smokers, where positive values correspond to hyper-methylation and negative values to hypo-methylation. The vertical axes depicted levels of statistical significance. Results satisfying a Bonferroni threshold of 10-7 (above the red line) are considered statistically significant.
The genomes of smoking-associated cancers permit reassessment of our understanding of how tobacco smoke causes cancer. Consistent with the proposition that an increased mutation load caused by tobacco smoke contributes to increased cancer risk, the total mutation burden is elevated in smokers compared to non-smokers in lung adenocarcinoma, larynx, liver and kidney cancers. However, differences in total mutation burden were not observed in the other smoking-associated cancer types and in some there were no statistically significant smoking-associated differences in mutation load, signatures or DNA methylation. Caution should be exercised in the interpretation of the latter observations. In addition to limitations of statistical power, multiple rounds of clonal expansion over many years are often required for development of a symptomatic cancer. It is thus conceivable that, in the normal tissues from which smoking associated cancer types originate, there are more somatic mutations in smokers than in non-smokers (or differences in methylation) but that these differences become obscured during the intervening clonal evolution. Moreover, some theoretical models predict that relatively small differences in mutation burden caused by smoking in pre-neoplastic cells could account for the observed increases in cancer risks (29) and others that differences in mutation burden between smokers and non-smokers need not be observed in the final cancers (Supplementary Text and Fig. S6). Thus, increased somatic mutation loads in precancerous tissues may still explain the smoking-induced risks of most cancers, although other mechanisms have been proposed (30, 31).
The generation of the increased somatic mutation burden by tobacco smoking, however, appears to be mechanistically complex. Smoking correlates with increases in base substitutions of multiple mutational signatures, together with increases in indels and copy number changes. The extent to which these distinct mutational processes operate differs between tissue types, at least in part depending on the degree of direct exposure to tobacco smoke, and their mechanisms range from misreplication of DNA damage caused by tobacco smoke constituents to activation of more generally operative mutational processes. Although we cannot exclude roles for covariate behaviours of smokers or differences in the biology of cancers arising in smokers compared to non-smokers, smoking itself is most plausibly the cause of these differences.
Supplementary Material
One Sentence Summary.
Multiple distinct mutational processes associated with tobacco smoking in cancer reflect direct and indirect effects of tobacco smoke.
Acknowledgements
This work was supported by the Wellcome Trust (grant number 098051). S.N.-Z. is a Wellcome-Beit Prize Fellow and is supported through a Wellcome Trust Intermediate Fellowship (grant WT100183MA). P.J.C. is personally funded through a Wellcome Trust Senior Clinical Research Fellowship (grant WT088340MA). M.R.S. is a paid advisor for GRAIL, a company developing technologies for sequencing of circulating tumor DNA for the purpose of early detection of cancer. L.B.A. is personally supported through a J. Robert Oppenheimer Fellowship at Los Alamos National Laboratory. This research used resources provided by the Los Alamos National Laboratory Institutional Computing Program, which is supported by the U.S. Department of Energy National Nuclear Security Administration under Contract No. DE-AC52-06NA25396. Research performed at Los Alamos National Laboratory was carried out under the auspices of the National Nuclear Security Administration of the United States Department of Energy. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001202), the UK Medical Research Council (FC001202), and the Wellcome Trust (FC001202). D.H.P. is funded by Cancer Research UK (grant number C313/A14329), the Wellcome Trust (Grants 101126/Z/13/Z and 101126/B/13/Z), the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Health Impact of Environmental Hazards at King’s College London in partnership with Public Health England (PHE) and by the project EXPOSOMICS, grant agreement 308610-FP7 (European Commission). P.V. was partially supported by the project EXPOSOMICS, grant agreement 308610-FP7 (European Commission). Y.T. and T.S. are supported by the Practical Research for Innovative Cancer Control from Japan Agency for Medical Research and Development (15ck0106094h0002), and National Cancer Center Research and Development Funds (26-A-5). We would like to thank The Cancer Genome Atlas (TCGA), the International Cancer Genome Consortium (ICGC), and the authors of all studies cited in Tables S1 for providing free access to their somatic mutational data.
References
- 1.Secretan B, et al. A review of human carcinogens--Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 2009;10:1033–1034. doi: 10.1016/s1470-2045(09)70326-2. [DOI] [PubMed] [Google Scholar]
- 2.Lim SS, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pesch B, et al. Cigarette smoking and lung cancer--relative risk estimates for the major histological types from a pooled analysis of case-control studies. International journal of cancer. 2012;131:1210–1219. doi: 10.1002/ijc.27339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agudo A, et al. Impact of cigarette smoking on cancer risk in the European prospective investigation into cancer and nutrition study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:4550–4557. doi: 10.1200/JCO.2011.41.0183. [DOI] [PubMed] [Google Scholar]
- 5.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nature reviews. Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 6.Phillips DH. In: The Cancer Handbook. Allison MR, editor. Macmillan; London: 2002. pp. 293–306. [Google Scholar]
- 7.Phillips DH. Smoking-related DNA and protein adducts in human tissues. Carcinogenesis. 2002;23:1979–2004. doi: 10.1093/carcin/23.12.1979. [DOI] [PubMed] [Google Scholar]
- 8.Phillips DH, Venitt S. DNA and protein adducts in human tissues resulting from exposure to tobacco smoke. International journal of cancer. 2012;131:2733–2753. doi: 10.1002/ijc.27827. [DOI] [PubMed] [Google Scholar]
- 9.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Current opinion in genetics & development. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 11.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 12.Pfeifer GP, Denissenko MF. Formation and repair of DNA lesions in the p53 gene: relation to cancer mutations? Environ Mol Mutagen. 1998;31:197–205. doi: 10.1002/(sici)1098-2280(1998)31:3<197::aid-em1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 13.Smith LE, et al. Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. J Natl Cancer Inst. 2000;92:803–811. doi: 10.1093/jnci/92.10.803. [DOI] [PubMed] [Google Scholar]
- 14.Le Calvez F, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res. 2005;65:5076–5083. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
- 15.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell reports. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrov LB. Understanding the origins of human cancer. Science. 2015;350:1175. doi: 10.1126/science.aad7363. [DOI] [PubMed] [Google Scholar]
- 17.Alexandrov LB, et al. Clock-like mutational processes in human somatic cells. Nature genetics. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nik-Zainal S, et al. The genome as a record of environmental exposure. Mutagenesis. 2015;30:763–770. doi: 10.1093/mutage/gev073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer discovery. 2015;5:704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahbari R, et al. Timing, rates and spectra of human germline mutation. Nature genetics. 2016;48:126–133. doi: 10.1038/ng.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nature genetics. 2016 doi: 10.1038/ng.3557. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govindan R, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimoto A, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nature genetics. 2016;48:500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 28.Teschendorff AE, et al. Correlation of Smoking-Associated DNA Methylation Changes in Buccal Cells With DNA Methylation Changes in Epithelial Cancer. JAMA oncology. 2015;1:476–485. doi: 10.1001/jamaoncol.2015.1053. [DOI] [PubMed] [Google Scholar]
- 29.Tomasetti C, Marchionni L, Nowak MA, Parmigiani G, Vogelstein B. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:118–123. doi: 10.1073/pnas.1421839112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sopori M. Effects of cigarette smoke on the immune system. Nature reviews. Immunology. 2002;2:372–377. doi: 10.1038/nri803. [DOI] [PubMed] [Google Scholar]
- 31.Rubin H. Selective clonal expansion and microenvironmental permissiveness in tobacco carcinogenesis. Oncogene. 2002;21:7392–7411. doi: 10.1038/sj.onc.1205800. [DOI] [PubMed] [Google Scholar]
- 32.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandrov LB, Nik-Zainal S, Siu HC, Leung SY, Stratton MR. A mutational signature in gastric cancer suggests therapeutic strategies. Nature communications. 2015;6 doi: 10.1038/ncomms9683. 8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulze K, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nature genetics. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behjati S, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolli N, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature communications. 2014;5 doi: 10.1038/ncomms3997. 2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju YS, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife. 2014;3 doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murchison EP, et al. Transmissible [corrected] dog cancer genome reveals the origin and history of an ancient cell lineage. Science. 2014;343:437–440. doi: 10.1126/science.1247167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nature reviews. Genetics. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nik-Zainal S, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nature genetics. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature genetics. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nature medicine. 2015;21:751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagener R, et al. Analysis of mutational signatures in exomes from B-cell lymphoma cell lines suggest APOBEC3 family members to be involved in the pathogenesis of primary effusion lymphoma. Leukemia. 2015;29:1612–1615. doi: 10.1038/leu.2015.22. [DOI] [PubMed] [Google Scholar]
- 46.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnett V, Lewis T. Outliers in statistical data. ed. 3rd. Wiley; Chichester ; New York: 1994. Wiley series in probability and mathematical statistics Applied probability and statistics; p. xvii. 584. [Google Scholar]
- 48.Holland PW, Welsch RE. Robust Regression Using Iteratively Reweighted Least-Squares. Communications in Statistics: Theory and Methods. 1977;A6:813–827. [Google Scholar]
- 49.Huber PJ, Ronchetti E. Robust statistics. 2nd. Wiley; Hoboken, N.J.: 2009. Wiley series in probability and statistics; p. xvi. 354 p. [Google Scholar]
- 50.Street J, Carroll R, Ruppert D. A Note on Computing Robust Regression Estimates Via Iteratively Reweighted Least Squares. The American Statistician. 1988;42 [Google Scholar]
- 51.Abdullah MB. On a Robust Correlation Coefficient. The Statistician. 1990;39:455–460. [Google Scholar]
- 52.Nordling CO. A new theory on cancer-inducing mechanism. British journal of cancer. 1953;7:68–72. doi: 10.1038/bjc.1953.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. British journal of cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silverman DT, Hartge P, Morrison AS, Devesa SS. Epidemiology of bladder cancer. Hematol Oncol Clin North Am. 1992;6:1–30. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.