Abstract
Specific interactions between proteins and their molecular partners drive most biological processes, so understanding how these interactions evolve is an important question for biochemists and evolutionary biologists alike. It is often thought that ancestral proteins were systematically more “promiscuous” than modern proteins, and specificity often does evolve by partitioning and refining the activities of multifunctional ancestors after gene duplication. However, recent studies using ancestral protein reconstruction (APR) have found that ligand-specific functions in some modern protein families evolved de novo from ancestors that did not already have those functions. Further, the new specific interactions evolved by simple mechanisms, with just a few mutations changing classically recognized biochemical determinants of specificity, such as steric and electrostatic complementarity. Acquiring new specific interactions during evolution therefore appears to be neither difficult nor rare. Rather, it is likely that proteins continually gain and new lose activities over evolutionary time as mutations cause subtle but consequential changes in the shape and electrostatics of interaction interfaces. Only a lucky few of these, however, are incorporated into the biological processes that contribute to fitness before they are lost to the ravages of further mutation.
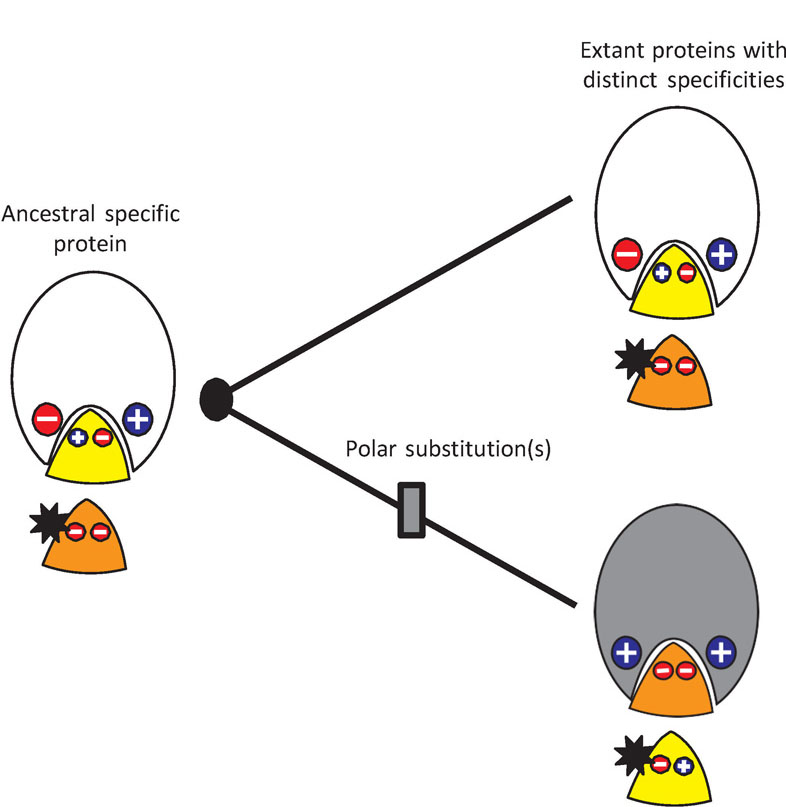
Specific molecular interactions – between enzymes an d substrates, receptors and ligands, and transcription factors and DNA response elements – underlie most cellular processes. How these interactions evolve has long been a source of interest for biochemists and evolutionary biologists [1–5].
Does specificity evolve from multifunctional ancestors?
A widely accepted hypothesis is that ancestral proteins were generalists, recognizing a broad set of ligands, from which modern highly specialized enzymes then evolved by gene duplication, partitioning and refining ancestral activities among the descendants [2, 4–10]. This view holds that the genetic paths to new functions are long, making them unlikely to evolve after a gene duplication but before deleterious mutations eliminate the extra copy [8, 10, 11]. If all the activities of highly specific enzymes were already present in their ancestors, however, evolution could simply partition those functions [12] or refine them under selection after gene duplication [9, 13]. (For definitions of specificity and promiscuity, see Box.)
This idea is appealing for several reasons. First, some present-day enzymes carry out secondary, biologically relevant “moonlighting” activities [14–16], so it is plausible that ancestral proteins might also have had such activities, which subsequently evolved into the primary functions of extant proteins. Second, it has been thought that evolving new biochemical activities should be genetically difficult, requiring many mutations in the context of tight constraints imposed by protein architecture, whereas losing activities already present in an ancestral protein would be much easier. For example, comparative studies of enzyme specificity have shown that homologous enzymes with distinct substrate specificities often differ by many residues, and efforts to transform the specificity of one protein into that of another by swapping a small number of amino acids at structurally important sites often fail [17]. Moreover, directed evolution studies found that new substrate-specificities could evolve more easily and along shorter genetic paths by optimizing low-level side activities present in the starting protein than by acquiring activity on an entirely new substrate [18–20]. This work also showed that some secondary activities could evolve without significantly compromising a protein’s primary function [18, 21], providing a plausible scenario by which ancestral proteins could have slowly acquired the functions that were ultimately partitioned among their descendants.
These observations are consistent with the idea that specific proteins can readily evolve from generalist ancestors, but they are not sufficient to establish that historical evolution always -- or even usually -- occurred this way, or that it is genetically difficult for proteins to evolve specificity de novo. First, the existence of moonlighting activities in many present-day proteins does not necessarily imply that all the functions of extant proteins are derived from secondary functions in their ancestors. Second, directed evolution regimes typically exert very strong selection pressures to produce defined in vitro chemical activities very rapidly, with high mutation rates, very strong selection, few pleiotropic constraints, and little or no opportunity for drift; they are therefore unrepresentative of the long-term historical processes of protein evolution [22]. Finally, comparative studies of homologous proteins are limited in their capacity to identify the minimal causes of functional differences, because long periods of sequence divergence and epistatic interactions among substitutions might obscure relatively simple echanisms by which proteins in the deep past diverged in function. [23, 24].
Reconstructing molecular evolution using ancestral protein reconstruction
Recent developments in ancestral protein reconstruction (APR) make it possible to directly address the historical evolution of specificity. This strategy begins with statistical inference of ancestral protein sequences, followed by gene synthesis, expression of ancestral proteins, and characterization of their physical and functional properties [25]. These techniques allow hypotheses about the specificity or promiscuity of ancestral proteins to be experimentally assessed. Further, amino acid changes from key intervals of phylogenetic history can be re-introduced into reconstructed ancestral proteins, so hypotheses about the mechanisms by which changes in specificity evolved can be directly tested.
Recent studies have used APR to reconstruct the evolution of specificity (Table 1). Of these, many have found that the specificities of related extant proteins were partitioned [26–30] or enhanced [31–38] from a common ancestor with mult iple functions. These studies establish that multifunctional ancestors sometimes give rise to specific descendants, but they do not reveal the genetic or biochemical mechanisms for how those functions evolved in the first place.
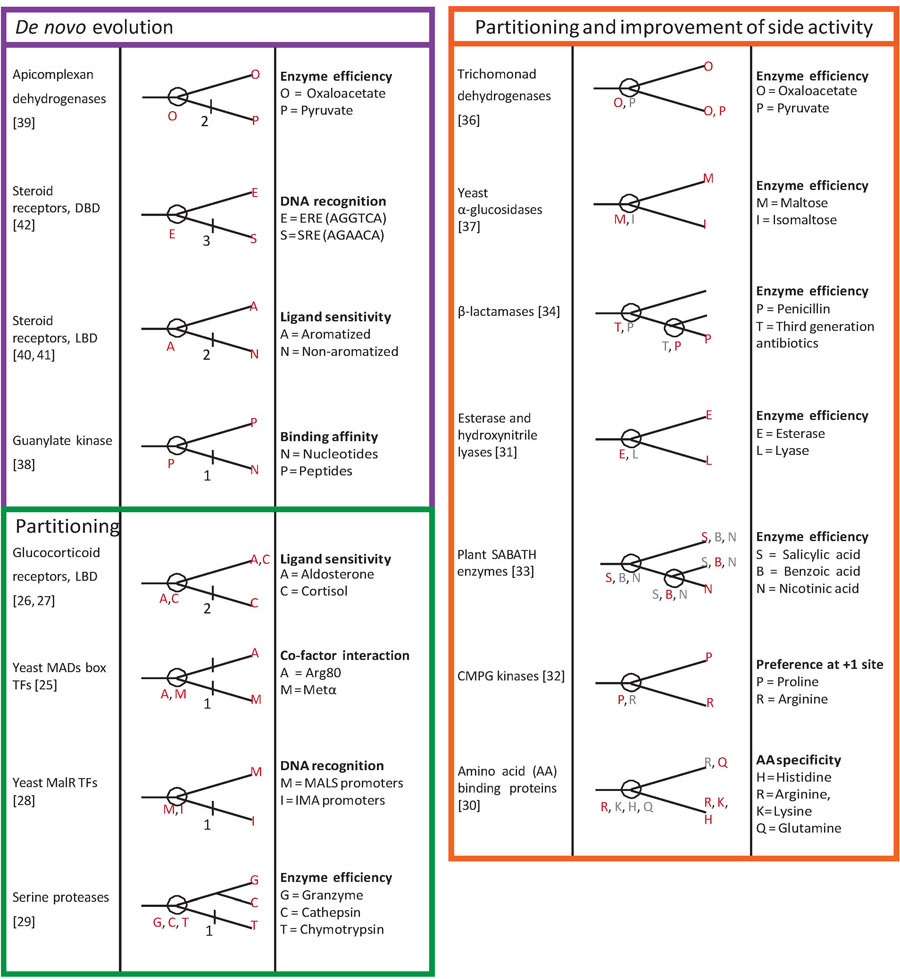
Table 1:
Ancestral reconstruction studies of specific molecular interactions. For each protein family studied, the cladogram shows the specific biological or biochemical property assayed, its distribution among extant and reconstructed ancestral proteins (circles). Protein families are grouped on the basis of their evolutionary history: de novo evolution of a new specific function after gene duplication (purple), partitioning of functions from a multifunctional ancestor (green), or partitioning of functions from a multifunctional ancestor with refinement of an ancestral activity in one or both lineages (orange). For studies that dissect genetic mechanisms, the number of large-effect replacements necessary to recapitulate the shift in specificity is shown on the lineage on which they occurred. Red letters, major function or activity (high affinity, catalytic efficiency, etc.); gray, minor function (lower affinity, etc.).
|
Several recent studies, however, have documented protein families in which the specific biological functions of present-day proteins evolved de novo from ancestral proteins that lacked those functions, and they reveal the mechanisms by which those new functions evolved [39–43]. Here we survey these findings, discussing three case studies in which different kinds of biological specificity evolved de novo: substrate-specificity of an enzyme, ligand-specificity of an allosterically regulated protein, and DNA-specificity of a transcription factor.
Substrate-specificity of metabolic enzymes
Members of evolutionarily related enzyme families typically share similar catalytic chemistry but have diversified in substrate specificity [3, 44, 45]. APR can clarify how this diversity evolved by measuring the activity of ancestral proteins against the substrates of their descendants and then identifying the historical causes, both genetic and structural, that can recapitulate the evolution of the derived specificity.
A recent study examined how highly distinct substrate specificities evolved in a pair of related enzymes – the malate and lactate dehydrogena ses (MDH and LDH) of apicomplexan alveolates, a phylum of single-celled eukaryotes [40]. These two enzymes, which play key metabolic roles, are related by a gene duplication event that occurred 700–900 million years ago. MDH catalyzes reduction of oxaloacetate, whereas LDH catalyzes reduction of pyruvate. Each enzyme is highly specific, with virtually no activity against the substrate of its paralog. The two ligands differ in that oxaloacetate is longer with a C3 hydroxyl group and a charged carboxylate at C4, whereas pyruvate is smaller and ends with a hydrophobic C3 methyl.
The authors reconstructed and characterized the common ancestral protein from which the Apicomplexan MDHs and LDHs arose (AncM/L) by duplication (Figure 1a). They found that AncM/L was not multifunctional; rather, like extant MDHs, it was highly specific for oxaloacetate, with virtually no activity on pyruvate and a preference for oxaloacetate (ratio ofkcat/Km for the two substrates) > 107. In contrast, the subsequent common ancestor of all extant LDHs (AncL) was, like extant LDHs, specific for pyruvate, preferring it over oxaloacetate by >200,000-fold. A discrete switch in specificity – not partitioning of activity from a generalist ancestral protein – therefore occurred during the phylogenetic interval between AncM/L and AncL.
Figure 1:
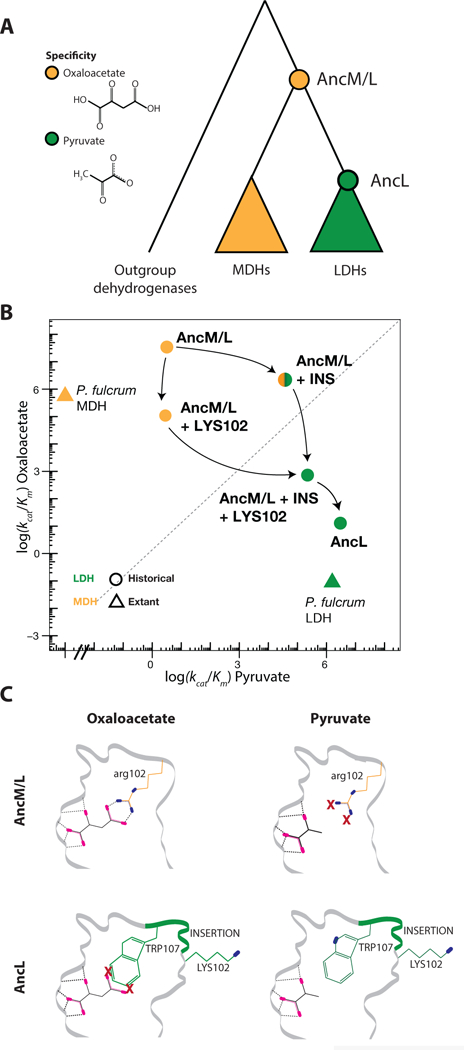
Evolution of lactate and malate dehydrogenase specificity in Apicomplexa [40]. A) Evolutionary relationships of paralogous dehydrogenases and reconstructed ancestral proteins (circles), colored by their substrate-specificity for oxaloacetate (orange) or pyruvate (green). B) Catalytic efficiency of extant and reconstructed dehydrogenases on oxaloacetate and pyruvate. Effect on ancestral activity of the historical amino acid replacement Arg102Lys (LYS102) and an insertion in the active site loop are shown; arrows show possible evolutionary paths from oxaloacetate-to pyruvate-specificity. C) Mechanism for the evolution of pyruvate specificity in LDH. The top row shows a portion of the structure of the ancestor AncM/L with oxaloacetate (left) and pyruvate (right); the bottom row shows the derived ancestor AncL. Sites affected by the key genetic changes are shown, with colors corresponding to panel A. Oxygen atoms, magenta; nitrogen, blue. Dashed lines show hydrogen bonds from enzyme to substrate; X, unsatisfied electrostatic potential.
To identify the best candidates among the 65 substitutions that took place in this interval, Boucher et al. determined the X-ray crystal structures of the two ancestral proteins. Two of the changes were in the active site loop – a six amino acid insertion and an arginine to lysine substitution at a nearby site (Arg102Lys). When introduced together into AncM/L, they recapitulated evolution of the derived specificity, causing a 60,000-fold increase in catalytic efficiency (kcat/Km) on pyruvate and a >40,000-fold reduction on oxaloacetate, thus shifting preference by more than ten orders of magnitude. The loop insertion alone conferred the new function almost entirely, increasing pyruvate efficiency by a factor of >12,000, while only weakly affecting oxaloacetate activity (Figure 1B). Introducing just Arg102Lys moderately reduced catalysis of oxaloacetate activity without affecting pyruvate activity, yielding a weaker enzyme on either substrate. Whichever mutation occurred first, the resulting intermediate form existed only transiently, because AncL and all known descendants possess both sequence features and are pyruvate-specific.
The ancestral crystal structures provided a physical explanation for how the loop insertion conferred the novel activity on pyruvate (Figure 1C). AncM/L was specific for oxaloacetate because the positively charged side chain of Arg102, which paired with oxaloacetate’s negatively charged carboxylate group, was left unsatisfied by pyruvate’s methyl. The insertion placed a Trp residue near this position, which improved hydrophobic packing of the methyl and moved residue 102 out of the active site into a solvent-exposed position, allowing that residue’s electrostatic potential to be satisfied when pyruvate is bound. The structures also showed that the loop and substitution together would likely reduce activity on oxaloacetate because they would leave that substrate’s carboxylate unpaired in the active site.
Thus, LDH appears to have acquired its pyruvate reductase function de novo by a single genetic change that altered a key electrostatic contact between protein and substrate; a second change then abolished the ancestral function by further altering electrostatic contacts. APR was essential to this discovery: extant MDH and LDHs share only ~50% sequence identity, and swapping these two active site changes between paralogs is no longer sufficient to interconvert their functions [40]. This observation indicates that subsequent epistatic substitutions modified the effect that the key sequence changes had when they occurred historically.
Ligand-specificity of allosteric recognition
Many proteins bind and are allosterically regulated by specific interactions with other proteins or small molecules. Our laboratory has studied the evolution of this phenomenon in the vertebrate steroid receptors (SRs), a family of allosterically regulated transcription factors with diverse specificity for steroid hormones [27, 28, 41, 46, 47]. The receptor’s ligand binding domain (LBD) binds a steroid molecule in its hydrophobic core, which causes the transcriptionally active conformation to be favored. There are two phylogenetic classes of SRs, which also correspond to the chemistry of their ligands: the estrogen receptors (ERs) bind steroids with an aromatized A-ring and a 3-hydroxyl, and the non-aromatized steroid receptors (naSRs), which bind androgens (AR), progestogens (PR), glucocorticoids (GR), and mineralocortioids (MR), most of which have a 3-carbonyl (Figure 2a). Eick et al. [41] reconstructed the LBD of AncSR1, the common ancestor of all present-day vertebrate SRs, which duplicated to produce the two classes (Figure 2a). This protein was found to be highly specific for estrogens, activating transcription in response to low doses of vertebrate estrogens but causing no activation in the presence of a large panel of nonaromatic steroids. In contrast, the subsequent common ancestor of all the naSRs (AncSR2) displayed the opposite specificity, with a very sensitive response to a variety of nonaromatized steroids and no activation by any estrogens. A discrete shift in functional specificity therefore occurred on the branch between these two ancestral proteins, with functional sensitivity to a new ligand evolving de novo and the ancestral ligand-sensitivity being abolished.
Figure 2:
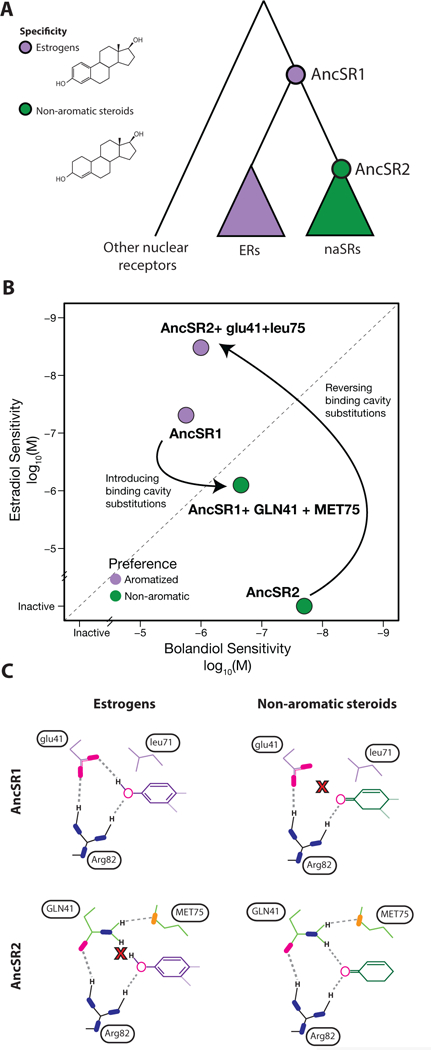
Evolution of ligand specificity in vertebrate steroid receptors [41, 46]. A) Evolutionary relationship of estrogen receptors (ERs) and non-aromatized steroid receptors (naSRs). Circles represent reconstructed ancestral proteins (AncSR1 and AncSR2), colored by their ligand specificity for estrogens (purple) or steroids with a nonaromatized A-ring (green). B) Effect of two historical substitutions on preference for aromatized or non-aromatized steroids. Each protein is plotted according to the ligand concentration at which half-maximal activation of a luciferase reporter was achieved (EC50), for the estrogen estradiol and the otherwise identical but nonaromatized androgen bolandiol. Effect of ancestral and derived states (lower and upper case, respectively) of historical mutations glu41GLN and leu75MET are shown when introduced into the reconstructed ancestral proteins. C) Mechanism for large-effect historical mutations. Top row shows AncSR1 with ancestral states in complex with the aromatized ring of estrogens (left) and nonaromatized steroids (right). Bottom row shows AncSR2 with derived states. Key residues are labeled; Arg82 is conserved among all steroid receptors. Oxygen atoms, magenta; nitrogen, blue; sulfur, orange. Dashed lines show hydrogen bonds from ligand to receptor; X, unsatisfied electrostatic potential.
Extant ERs and naSRs differ at about 70 percent of sequence sites, and even AncSR1 and AncSR2 differ by 171 substitutions, suggesting that the mechanism for the difference in specificity might be very complex. But APR, along with an X-ray crystal structure of the ancestral protein, allowed the authors to identify two substitutions that occurred during the interval between AncSR1 and AncSR2 that can account for most of the evolutionary change in specificity [46]. Reversing these two sites in AncSR2 to their ancestral states conferred a >100,000-fold shift in preference for estrogens vs. non-aromatized steroids. Introducing the derived states into AncSR1 also conferred activation by nonaromatized steroids and dramatically shifted the receptor’s preference from estrogens to these hormones (Figure 2b).
The two key mutations altered the electrostatics of the pocket and its complementarity to polar groups on the two ligands (Figure 2c). AncSR1 coordinated the hormone’s A-ring using a network of hydrogen-bond acceptors that are fully satisfied with estrogens, including a Glu that accepts a hydrogen bond from the 3-hydroxyl of estrogens. This network effectively excluded nonaromatized steroids, most of which contain a 3-carbonyl, because that complex would contain an excess of unsatisfied hydrogen-bond acceptors in the pocket; even nonaromatized steroids that contain a potential donor at this position present it at the wrong angle to pair with the Glu [47]. The two key substitutions replaced this Glu with Gln and introduced a second polar residue into the pocket, which created a new arrangement of donors and acceptors that could pair with nonaromatized steroids but left the 3-hydroxyl of estrogens unpaired. Molecular dynamics and hydrogen-deuterium exchange experiments both showed that the derived protein’s hydrogen bond network is consistently satisfied in the active conformation when nonaromatized steroids are bound. But with estrogen in the pocket, AncSR2 explores numerous suboptimal conformations in which the hydrogen bonding potential of the pocket is unfulfilled, unless water molecules are drawn into the protein’s interior, disrupting the active conformation. Thus, as in MDH/LDH evolution, just two historical mutations are sufficient to in the ancestral background to remodel the binding site, changing specific electrostatic complementarity to the ancestral ligand into specific complementarity to the derived ligand.
DNA specificity of transcription factors
Regulation of cell state and activity depends on specific interactions between transcription factors and DNA response elements (REs) in the vicinity of target genes. Evolution of DNA specificity has been studied using APR in several families of TFs, including the DNA-binding domain of SRs. All SRs bind as dimers to palindromes of a 6 base-pair “half-site.” ERs bind preferentially to EREs -- palindromes of AGGTCA – whereas naSRs bind to SREs (palindromes of AGAACA and variants) [48, 49]; ERs activate reporter transcription effectively from EREs but not SREs, and the converse is true of naSRs (Figure 3a).
Figure 3:
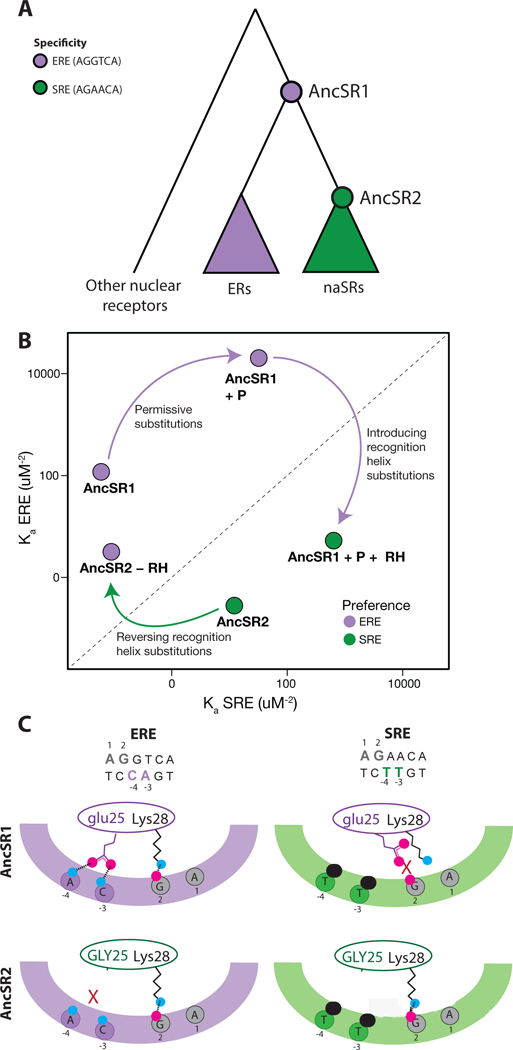
Evolution of DNA specificity in vertebrate steroid receptors [43]. A) Evolutionary relationship of DNA binding domains (DBDs) of ERs and naSRs. Circles represent the ancestral proteins, AncSR1 and AncSR2. Colors show preference for estrogen or steroid response elements (ERE, purple; SRE, green). B) Effect of historical amino acid replacements on DNA affinity for ERE and SRE, shown as the association constant (Ka) for each motif in a fluorescence anisotropy binding assay. Purple arrow shows the preference-switching effect of introducing the derived states at three residues in the recognition helix (+RH) and the non-specific effect of permissive changes (+P) on AncSR1. Green arrows show the effect of reverting to the ancestral residues at the RH sites (-RH). C) Mechanism for the effect of historical RH substitution glu25GLY on specificity. The top and bottom rows shows AncSR1 and AncSR2, respectively, in complex with ERE (left) and SRE (right). The protein’s RH is shown as an oval, and the DNA major groove as a colored arc; bases that participate in specific interactions are shown, numbered according to their strand. Bases that differ between the REs are colored. Side chains at site 25 and the conserved residue Lys28 are shown. Magenta, oxygen atoms; blue, nitrogen; black, methyl group. Hydrogen bonds are shown with dashed lines. X, unsatisfied hydrogen bonding potential. A steric clash between methyl groups on the SRE and the ancestral glu25 repositions that side chain, disrupting two ancestral hydrogen bonds; substitution of GLY25 relieves this clash and allows SRE binding.
Reconstruction and characterization of the DBDs of AncSR1 and AncSR2 illuminated the evolutionary trajectory and mechanisms by which these specificities diversified [43]. AncSR1’s DBD, like its LBD, was highly specific, activating reporter expression from EREs but showing no activation on SREs. AncSR2’s specificity was inverted, activating from SREs but not at all on EREs. Thus, SRE-mediated transcriptional activity evolved de novo from an ancestor that did not already possess that function. Follow-up biochemical studies showed that this change in biological specificity was mediated by massive changes in affinity between AncSR1 and AncSR2 – an increase in affinity for SRE by a factor of ~200 and a ~400-fold reduction in ERE affinity, together yielding an 80,000-fold shift in relative affinity. Underscoring the point that low-level chemical activities are not necessarily relevant in biological terms, AncSR1 did have weak affinity for SREs, and AncSR2 had weak affinity for EREs, but in both cases those affinities were too low to mediate transcriptional activation.
The shift in specificity was caused in large part by a few genetic changes. The authors focused on three key substitutions that occurred during the AncSR1-AncSR2 interval, were conserved in different states between the two classes of SR, and are located on the proteins “recognition helix” (RH), which inserts into the DNA major groove and contacts the variable base pairs in the half-site. Experiments showed that these were indeed large-effect historical substitutions: reversing them in AncSR2 to the ancestral state was sufficient to switch the protein’s DNA specificity in reporter assays from SRE back to ERE and to confer a ~1,500-fold increase in relative affinity for EREs (Figure 3b). Conversely, when the derived states were introduced into AncSR1, they increased specificity for SREs by four to five orders of magnitude by both increasing SRE affinity and reducing ERE affinity. When introduced together with a group of permissive mutations that nonspecifically affected DNA affinity, these three substitutions yielded a DBD that mediated robust transcription on SRE.
In this case, too, the mutations reversed the protein’s specificity by changing electrostatic complementarity to the ligands. X-ray crystal structures and MD simulations of the ancestral proteins showed that the three substitutions did not establish any new positive interactions to SREs; rather, they affected exclusionary interactions, removing steric clashes and unsatisfied polar/charged groups that had reduced the AncSR1’s affinity for SRE, while introducing new incompatibilities with ERE. For example, AncSR1 bound SRE poorly because an ancestral glutamate on AncSR1’s recognition helix sterically clashed with two methyl groups on the DNA that are unique to SRE, leaving the glutamate and several other charged or polar moieties in the interface unsatisfied. In AncSR2, this glutamate is replaced with a glycine, which both removes the charged side chain and its unsatistfied potential and relieves the steric clash, increasing SRE affinity without introducing any new positive interactions. In turn, the derived side chains also left several polar groups on the ERE unpaired. Thus, specificity was shifted not by generating new positive interactions with the derived RE but by changing the negative determinants of specificity to exclude the ancestral binding site and allow binding to the derived site.
Evolution and the biochemical mechanisms of specificity
These studies used APR to investigate different kinds of molecular specificity – between enzymes and substrates, receptors and ligands, and transcription factors and DNA – but they reveal similarities in the genetic and physical mechanisms of functional evolution. Although the number of cases is relatively small (Table 1), three major lessons emerge. First, the specific biological functions of extant proteins have in some cases emerged de novo following gene duplications; although such de novo trajectories are not the most frequent kind of history observed, they can and do occur. Second, when new specific interactions with molecular partners evolved, they did so through a small number of large-effect substitutions, with other historical sequences changes exerting permissive, fine-tuning, or inconsequential effects [50–52]. Third, the mechanisms by which these large-effect substitutions conferred new functions were relatively simple, involving steric clashes and changes in polar interactions between protein and ligand.
These evolutionary observations are consistent with fundamental knowledge concerning the biochemical causes of protein specificity span [53, 54]. Although a satisfied hydrogen bond makes only a small contribution to affinity in solvent -- because the system forms the same number of hydrogen bonds whether or not the complex is bound -- an unsatisfied polar or charged group on a hydrophobic interface can strongly reduce affinity by causing the bound complex to make fewer total hydrogen bonds than the unbound state. [54, 55]. A polar or charged group can therefore discriminate strongly against ligands that provide no complementary group to fulfill the residue’s electrostatic potential. Steric clashes can also have dramatic effects on specificity, because Pauli repulsion effects are associated with very high energies; a protein structure may adjust to minimize such clashes, but these changes often disrupt other interactions in the complex.
These findings imply that specificity should be evolvable via one or a very few mutations. A substitution that introduces a donor or acceptor onto a hydrophobic interface can increase the free energy of binding by up to 5 kcal/mol, decreasing affinity by 5,000-fold [55]. Conversely, a substitution that removes an existing polar or charged group – or satisfies an existing unpaired group – will increase affinity by the same amount. A single substitution that accomplishes both of these changes, leaving a donor or acceptor unsatisfied with one ligand and satisfying a previously unpaired group – can therefore shift specificity by up to 107-fold. Substitutions that induce ligand-specific steric clashes or introduce multiple unsatisfied donors or acceptors into a binding site can have even larger effects. These are precisely the kinds of mechanisms – and magnitude of effects – that APR studies show drove de novo acquisition of specific functions during historical protein evolution.
Why are trajectories from generalist ancestors so common?
If specific functions have evolved de novo in some cases by such simple mechanisms, then why does there appear to be a preponderance of protein families in which specific paralogs evolved by partitioning from a multifunctional ancestor (Table 1)? There are three requirements for paralogs with distinct specific functions to evolve. Without specifying an order in which they must occur, those requirements are: 1) a gene duplication must take place; 2) the distinct biochemical activities found in the extant paralogs must be differentially gained and/or lost among the paralogs in a way that produces distinct specificities; and 3) the genes and their relevant functions must be preserved to the present. We propose that the third requirement often represents the rate-limiting factor in long-term protein evolution and is the primary cause of differences in the observed frequency of trajectories in which specific paralogs evolve from multifunctional ancestors vs. those in which new specificity evolves de novo.
Both de novo and partitioning histories require a gene duplication, so the first requirement per se cannot account for the preponderance of trajectories in the former category. What about the second requirement: is it difficult or unlikely for biochemical activities to originate de novo? The answer is almost certainly no. Virtually all extant proteins have a large number of chemical side-activities – sometimes high-efficiency ones -- that do not contribute to the organism’s function or fitness [reviewed in 16, 44]. Further, as the studies we have discussed make clear, new specific activities can often evolve via just one or a few substitutions. It is therefore likely that proteins are continually gaining and losing secondary biochemical activities during evolution because of the stochastic processes of mutation and drift. The purported difficulty of evolving new biochemical activities is thus not a plausible explanation for the preponderance of histories in which the ancestor already had the functions of its descendants. Indeed, new activities must have originated during evolution at some point to have been present in the ancestor and/or its descendants; partitioning histories merely push these events further back in time.
We propose that the key difference between the two kinds of trajectories is how they affect the third requirement – that duplicated genes and biochemical activities be retained over evolutionary time. Unless genes are conserved by purifying selection, mutation and drift erode their sequences quickly, leading to loss of function, expression, or the reading frame itself. To be preserved over long periods of evolutionary time, then, a duplicate gene and its biochemical activity must contribute to fitness. In some cases, such as during adaptation to strong selection pressures like novel antibiotics or pesticides, a new chemical activity might be immediately advantageous and become selectively maintained [57–61]. But in most cases, the divergent specificities of proteins involve endogenous ligands, and these activities are more slowly integrated into the organism’s biology – its development, physiology, or metabolism, for example. This suggests that subsequent genetic changes at other loci, affecting such processes as production of a substrate or ligand, utilization of the product, or regulation of the gene’s expression, were likely required to incorporate the new activity into the organism’s biological processes and fitness. Because additional genetic changes across multiple genes are involved, this process is likely to be a major limiting factor in the long-term evolution of most new protein functions.
A key difference between de novo and partitioning evolutionary histories is that the latter leave more time for a new biochemical activity to become biologically significant and subject to purifying selection. In de novo evolution, a biochemical activity must originate after gene duplication and then be incorporated into the biological functions of the organism before the activity or the gene is lost. In contrast, partitioning from a multifunctional ancestor requires the new activity to evolve before the duplication, and it can become biologically significant either during this pre-duplication period, when the single copy of the gene is protected from degenerative mutation by purifying selection, or after. Partitioning histories therefore leave more time than de novo histories do for this rate-limiting step to occur and are for that reason—not because evolving a new function is genetically difficult—more likely to produce pairs of paralogs with distinct specificities. A partitioning trajectory also requires the ancestral activities to be partially lost in a complementary fashion among the paralogs, but this step is unlikely to be rate-limiting because it can occur by degenerative mutation and neutral drift [12].
A corollary of this argument is that there is no reason to believe that ancient proteins were in general more multifunctional or promiscuous than extant ones. When we say that an extant protein evolved some specific function de novo, this means only that the ancestral protein from which it evolved did not have that function, not that it did not have any – or many – other side activities. Today’s proteins will eventually be the ancestors of tomorrow’s. Between now and then, many promiscuous activities will be gained and lost; only those that both become biological functions constrained by natural selection and undergo gene duplication will ultimately produce paralogs with distinct specificities. Of the innumerable biochemical activities that extant proteins now possess or will acquire, the only ones that scientists of the future will observe – and the only ones for which we will demand an account of their ancestors – are these happy few.
Box 1. Defining specificity (and evolutionary paths to it).
A ligand-specific protein carries out biologically relevant functions only when it physically interacts with certain other molecules. Specificity evolves de novo when a descendant protein possesses a ligand-specific function that was absent in its ancestor. A multifunctional or generalist ancestor possesses several such biological functions, which can be partitioned (and potentially amplified or refined) among its descendants. A ligand-specific biological function is distinct from a promiscuous chemical side-activity, which does not affect cells or organisms [14, 16]. Biologically insignificant chemical activities are present in virtually all molecules [56]: even the lowest-affinity interactions still form complexes sometimes, and the least efficient enzymes turn over substrate at some rate. Side-activities may also be insignificant because the relevant substrates or ligands do not naturally occur or because the products do not contribute to the organism’s function or fitness. It is therefore trivial to observe that a protein with ligand-specific function descends from an ancestral protein that promiscuously interacted with the ligand to some degree. Only biologically significant functions are relevant to understanding the evolutionary dynamics that drive the evolution of specificity; unless they affect the organism’s biology, low-level promiscuous activities are invisible to natural selection and have no effect on the protein’s evolutionary fate.
Highlights:
Ancestral protein reconstruction has revealed how molecular interactions evolved.
Not all paralogs with distinct functions evolved from promiscuous ancestors.
Some specific interactions evolved de novo by very simple genetic mechanisms.
Changes in electrostatic complementarity have driven the evolution of specificity.
Ancient proteins were not systematically more promiscuous than modern ones.
Acknowledgments:
We thank members of the Thornton Lab for helpful discussions and comments. This work was supported by NIH grant R01-GM104397 (JWT), NIH grant R01-GM121931, National Science Foundation grant DEB-1501877 (JWT/MAS), NSF graduate research fellowship (MAS), a Chicago Research Fellowship (GKAH), and NIH training grant T32-GM007197 (MAS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Financial Interests. The authors declare no competing financial interests.
Works cited
- 1.Horowitz NH (1945). On the Evolution of Biochemical Syntheses. Proc Natl Acad Sci U S A 31, 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen RA (1976). Enzyme recruitment in evolution of new function. Annu Rev Microbiol 30, 409–425. [DOI] [PubMed] [Google Scholar]
- 3.Petsko GA, Kenyon GL, Gerlt JA, Ringe D, and Kozarich JW (1993). On the origin of enzymatic species. Trends Biochem Sci 18, 372–376. [DOI] [PubMed] [Google Scholar]
- 4.Waley SG (1969). Some aspects of the evolution of metabolic pathways. Comp Biochem Physiol 30, 1–11. [DOI] [PubMed] [Google Scholar]
- 5.Ycas M (1974). On earlier states of the biochemical system. J Theor Biol 44, 145–160. [DOI] [PubMed] [Google Scholar]
- 6.Weng JK, Philippe RN, and Noel JP (2012). The rise of chemodiversity in plants. Science 336, 1667–1670. [DOI] [PubMed] [Google Scholar]
- 7.Pandya C, Farelli JD, Dunaway-Mariano D, and Allen KN (2014). Enzyme promiscuity: engine of evolutionary innovation. Journal of Biological Chemistry 289, 30229–30236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Brien PJ, and Herschlag D (1999). Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol 6, R91–R105. [DOI] [PubMed] [Google Scholar]
- 9.Khersonsky O, and Tawfik DS (2010). Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem 79, 471–505. [DOI] [PubMed] [Google Scholar]
- 10.James LC, and Tawfik DS (2003). Conformational diversity and protein evolution--a 60-year-old hypothesis revisited. Trends Biochem Sci 28, 361–368. [DOI] [PubMed] [Google Scholar]
- 11.Tokuriki N, Jackson CJ, Afriat-Jurnou L, Wyganowski KT, Tang R, and Tawfik DS (2012). Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat Commun 3, 1257. [DOI] [PubMed] [Google Scholar]
- 12.Force A, Lynch M, Pickett FB, Amores A, Yan YL, and Postlethwait J (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conant GC, and Wolfe KH (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9, 938–950. [DOI] [PubMed] [Google Scholar]
- 14.Copley SD (2012). Moonlighting is mainstream: paradigm adjustment required. Bioessays 34, 578–588. [DOI] [PubMed] [Google Scholar]
- 15.Jeffery CJ (1999). Moonlighting proteins. Trends Biochem Sci 24, 8–11. [DOI] [PubMed] [Google Scholar]
- •• 16.Copley SD (2015). An evolutionary biochemist’s perspective on promiscuity. Trends Biochem Sci 40, 72–78. An insightful review of protein side-activities and their evolution, with consideration of the distinction between biologically consequential and inconsequential activities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlt JA, and Babbitt PC (2009). Enzyme (re)design: lessons from natural evolution and computation. Curr Opin Chem Biol 13, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aharoni A, Gaidukov L, Khersonsky O, McQ Gould S, Roodveldt C, and Tawfik DS (2005). The ‘evolvability’ of promiscuous protein functions. Nat Genet 37, 73–76. [DOI] [PubMed] [Google Scholar]
- 19.O’Loughlin TL, Patrick WM, and Matsumura I (2006). Natural history as a predictor of protein evolvability. Protein Eng Des Sel 19, 439–442. [DOI] [PubMed] [Google Scholar]
- 20.Renata H, Wang ZJ, and Arnold FH (2015). Expanding the enzyme universe: accessing non-natural reactions by mechanism-guided directed evolution. Angew Chem Int Ed Engl 54, 3351–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokuriki N, Jackson CJ, Afriat-Jurnou L, Wyganowski KT, Tang R, and Tawfik DS (2012). Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat Commun 3, 1257. [DOI] [PubMed] [Google Scholar]
- 22.Bloom JD, and Arnold FH (2009). In the light of directed evolution: pathways of adaptive protein evolution. Proc Natl Acad Sci U S A 106 Suppl 1, 9995–10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harms MJ, and Thornton JW (2010). Analyzing protein structure and function using ancestral gene reconstruction. Current opinion in structural biology 20, 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochberg GKA, and Thornton JW (2017). Reconstructing Ancient Proteins to Understand the Causes of Structure and Function. Annual Review of Biophysics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thornton JW (2004). Resurrecting ancient genes: experimental analysis of extinct molecules. Nature Reviews Genetics 5, 366–375. [DOI] [PubMed] [Google Scholar]
- • 26.Baker CR, Hanson-Smith V, and Johnson AD (2013). Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science 342, 104–108. Using ancestral protein reconstruction, this study shows how paralogous transcription factors that interact with different co-factors and regulate different target genes evolved from a multifunctional ancestor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bridgham JT, Carroll SM, and Thornton JW (2006). Evolution of hormone-receptor complexity by molecular exploitation. Science 312, 97–101. [DOI] [PubMed] [Google Scholar]
- 28.Ortlund EA, Bridgham JT, Redinbo MR, and Thornton JW (2007). Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317, 1544–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pougach K, Voet A, Kondrashov FA, Voordeckers K, Christiaens JF, Baying B, Benes V, Sakai R, Aerts J, Zhu B et al. (2014). Duplication of a promiscuous transcription factor drives the emergence of a new regulatory network. Nat Commun 5, 4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wouters MA, Liu K, Riek P, and Husain A (2003). A despecialization step underlying evolution of a family of serine proteases. Mol Cell 12, 343–354. [DOI] [PubMed] [Google Scholar]
- • 31.Clifton BE, and Jackson CJ (2016). Ancestral Protein Reconstruction Yields Insights into Adaptive Evolution of Binding Specificity in Solute-Binding Proteins. Cell Chem Biol 23, 236–245. Strong specificity for glutamine evolved from an ancestral protein with preference for arginine but secondary binding to glutamine; the ancestral promiscuity arises from the protein’s capacity to occupy two different conformations. [DOI] [PubMed] [Google Scholar]
- • 32.Devamani T, Rauwerdink AM, Lunzer M, Jones BJ, Mooney JL, Tan MA, Zhang ZJ, Xu JH, Dean AM, and Kazlauskas RJ (2016). Catalytic Promiscuity of Ancestral Esterases and Hydroxynitrile Lyases. J Am Chem Soc 138, 1046–1056. Ancestral protein reconstruction of a rare case in which homologs evolved distinct reaction chemistries. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howard CJ, Hanson-Smith V, Kennedy KJ, Miller CJ, Lou HJ, Johnson AD, Turk BE, and Holt LJ (2014). Ancestral resurrection reveals evolutionary mechanisms of kinase plasticity. Elife 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang R, Hippauf F, Rohrbeck D, Haustein M, Wenke K, Feike J, Sorrelle N, Piechulla B, and Barkman TJ (2012). Enzyme functional evolution through improved catalysis of ancestrally nonpreferred substrates. Proc Natl Acad Sci U S A 109, 2966–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, and Sanchez-Ruiz JM (2013). Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian beta-lactamases. J Am Chem Soc 135, 2899–2902. [DOI] [PubMed] [Google Scholar]
- 36.Shih PM, Occhialini A, Cameron JC, Andralojc PJ, Parry MA, and Kerfeld CA (2016). Biochemical characterization of predicted Precambrian RuBisCO. Nat Commun 7, 10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steindel PA, Chen EH, Wirth JD, and Theobald DL (2016). Gradual neofunctionalization in the convergent evolution of trichomonad lactate and malate dehydrogenases. Protein Science 25, 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voordeckers K, Brown CA, Vanneste K, van der Zande E, Voet A, Maere S, and Verstrepen KJ (2012). Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol 10, e1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 39.Anderson DP, Whitney DS, Hanson-Smith V, Woznica A, Campodonico-Burnett W, Volkman BF, King N, Thornton JW, and Prehoda KE (2016). Evolution of an ancient protein function involved in organized multicellularity in animals. Elife 5, e10147 A single substitution was sufficient to confer on an ancestral kinase moderate affinity for a peptide because of similarities in the two ligands’ surface properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boucher JI, Jacobowitz JR, Beckett BC, Classen S, and Theobald DL (2014). An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases. Elife 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eick GN, Colucci JK, Harms MJ, Ortlund EA, and Thornton JW (2012). Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genetics 8, e1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harms MJ, and Thornton JW (2014). Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature 512, 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKeown AN, Bridgham JT, Anderson DW, Murphy MN, Ortlund EA, and Thornton JW (2014). Evolution of DNA specificity in a transcription factor family produced a new gene regulatory module. Cell 159, 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nobeli I, Spriggs RV, George RA, and Thornton JM (2005). A ligand-centric analysis of the diversity and evolution of protein-ligand relationships in E.coli. J Mol Biol 347, 415–436. [DOI] [PubMed] [Google Scholar]
- 45.Todd AE, Orengo CA, and Thornton JM (2001). Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol 307, 1113–1143. [DOI] [PubMed] [Google Scholar]
- 46.Harms MJ, Eick GN, Goswami D, Colucci JK, Griffin PR, Ortlund EA, and Thornton JW (2013). Biophysical mechanisms for large-effect mutations in the evolution of steroid hormone receptors. Proc Natl Acad Sci U S A 110, 11475–11480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thornton JW, Need E, and Crews D (2003). Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science 301, 1714–1717. [DOI] [PubMed] [Google Scholar]
- 48.So AY, Chaivorapol C, Bolton EC, Li H, and Yamamoto KR (2007). Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet 3, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welboren WJ, Sweep FC, Span PN, and Stunnenberg HG (2009). Genomic actions of estrogen receptor alpha: what are the targets and how are they regulated. Endocr Relat Cancer 16, 1073–1089. [DOI] [PubMed] [Google Scholar]
- 50.Bloom JD, Gong LI, and Baltimore D (2010). Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328, 1272–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong LI, Suchard MA, and Bloom JD (2013). Stability-mediated epistasis constrains the evolution of an influenza protein. Elife 2, e00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Starr TN, and Thornton JW (2016). Epistasis in protein evolution. Protein Sci 25, 1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 53.Fersht AR, Shi JP, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MM, and Winter G (1985). Hydrogen bonding and biological specificity analysed by protein engineering. Nature 314, 235–238. Classic paper establishing that hydrogen bonds make large contributions to specificity but not affinity. [DOI] [PubMed] [Google Scholar]
- 54.Fersht AR (1987). The Hydrogen-Bond in Molecular Recognition. Trends in Biochemical Sciences 12, 301–304. [Google Scholar]
- •• 55.von Hippel PH, and Berg OG (1986). On the specificity of DNA-protein interactions. Proc Natl Acad Sci USA 83, 1608–1612. Seminal paper showing that polar residues drive specificity because unsatisfied electrostatic potential reduces affinity, not because hydrogen bonds increase affinity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nobeli I, Favia AD, and Thornton JM (2009). Protein promiscuity and its implications for biotechnology. Nat Biotechnol 27, 157–167. [DOI] [PubMed] [Google Scholar]
- 57.Davies Julian, and Davies Dorothy. (2010). Origins and evolution of antibiotic resistance. Microbiology and molecular biology reviews 74, 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wootton John C., Feng Xiaorong, Ferdig Michael T., Cooper Roland A., Mu Jianbing, Baruch Dror I., Magill Alan J., and Su Xin-zhuan. (2002). Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418, 320–323. [DOI] [PubMed] [Google Scholar]
- 59.Martin Rowena E., Marchetti Rosa V., Cowan Anna I., Howitt Susan M., Bröer Stefan, and Kirk Kiaran. (2009). Chloroquine transport via the malaria parasite’s chloroquine resistance transporter. Science 325, 1680–1682. [DOI] [PubMed] [Google Scholar]
- 60.Powles Stephen B., and Yu Qin. (2010). Evolution in action: plants resistant to herbicides. Annual review of plant biology 61, 317–347. [DOI] [PubMed] [Google Scholar]
- 61.Broser Matthias, Glöckner Carina, Gabdulkhakov Azat, Guskov Albert, Buchta Joachim, Kern Jan, Frank Müh Holger Dau, Saenger Wolfram, and Zouni Athina. (2011). Structural basis of cyanobacterial photosystem II inhibition by the herbicide terbutryn. Journal of Biological Chemistry 286, 15964–15972. [DOI] [PMC free article] [PubMed] [Google Scholar]


