Significance
ES cells possess the unique capacity to self-renew as well as differentiate into specialized cell types. It is known that transcription factors and chromatin regulators regulate the cell-fate choices during differentiation. We report unexpectedly that Asf1a, a histone chaperone involved in nucleosome assembly, regulates mouse ES cell differentiation. Mechanistically, we show that Asf1a functions in nucleosome disassembly to resolve the bivalent chromatin domains at lineage-specific genes for gene activation during differentiation. These insights will likely be applicable for understanding human ES cell differentiation and regenerative medicine.
Keywords: embryonic stem cell differentiation, bivalent chromatin domain, histone chaperone, nucleosome disassembly
Abstract
Bivalent chromatin domains containing repressive H3K27me3 and active H3K4me3 modifications are barriers for the expression of lineage-specific genes in ES cells and must be resolved for the transcription induction of these genes during differentiation, a process that remains largely unknown. Here, we show that Asf1a, a histone chaperone involved in nucleosome assembly and disassembly, regulates the resolution of bivalent domains and activation of lineage-specific genes during mouse ES cell differentiation. Deletion of Asf1a does not affect the silencing of pluripotent genes, but compromises the expression of lineage-specific genes during ES cell differentiation. Mechanistically, the Asf1a–histone interaction, but not the role of Asf1a in nucleosome assembly, is required for gene transcription. Asf1a is recruited to several bivalent promoters, partially through association with transcription factors, and mediates nucleosome disassembly during differentiation. We suggest that Asf1a-mediated nucleosome disassembly provides a means for resolution of bivalent domain barriers for induction of lineage-specific genes during differentiation.
ES cells are derived from the inner cell mass of preimplantation blastocysts and can be expanded in culture while maintaining the ability to differentiate into all types of somatic cells (1, 2). Compared with differentiated cells, ES cells maintain a globally “open” chromatin state that is relatively accessible for transcription factors (TFs). At the same time, lineage-specific genes remain silenced. When differentiation has initiated, ES cells exit from this self-renewal state into a state that allows multilineage commitment. As the same cell with its fixed genetic background switches from one state to another, insights into how chromatin dynamics are regulated during this process are of great interest.
At the molecular level, TFs and chromatin regulators play important roles in ES cell pluripotency maintenance as well as lineage-specific gene induction during differentiation (3). It is known that the pluripotent state of ES cells is largely governed by the core TFs, Oct4, Nanog, and Sox2, which sometimes are referred as pluripotent genes (4–6). These three TFs can activate genes necessary to maintain the pluripotent state of ES cells, and, equally importantly, suppress the expression of key lineage-specific genes that drives ES cells into different lineages (7–9). For instance, Oct4 and Sox2 form a complex and silence the transcription of Cdx2, and Nanog suppresses the expression of Gata6 (10, 11). Cdx2 and Gata6 are two key regulators that drive mouse ES cells to differentiate into trophectoderm and endoderm, respectively (12, 13). In addition, Nanog also silences the expression of Brachyury (T), which is important for mesoderm lineage specification (14). Therefore, these three core TFs can function as both activators and suppressors of gene transcription in ES cells.
In addition to TFs, histone modifications also play an important role in ES cell pluripotency and differentiation. For instance, in ES cells, pluripotent genes (Oct4, Nanog, and Sox2) are highly expressed, and the promoters of these genes are enriched with H3K4me3, a mark associated with active gene transcription (15). During differentiation, these pluripotent genes are silenced and their promoters progressively gain repressive marks, including H3K27me3 (16). Moreover, it has been shown that regulatory elements including promoters and enhancers of many lineage-specific and developmentally regulated genes form “bivalent chromatin domain” in ES cells (17). These domains, in general, consist of a large area of nucleosomes modified by H3K27me2/me3 and smaller regions of H3K4me3. Therefore, genes at the bivalent chromatin domains are largely silent or expressed at very low levels, and are poised for activation during differentiation (18). Some pioneer TFs can recognize their target DNA on nucleosomes and can overcome the barrier of repressed chromatin marked by H3K27me3 (19, 20). However, the bivalent domains are largely restrictive for the binding of TFs, and it is underexplored how the bivalent chromatin domains are resolved for the induction of lineage-specific genes.
Asf1 is a histone H3–H4 chaperone conserved from yeast to human cells. Asf1’s role in DNA replication-coupled (RC) nucleosome assembly was first discovered in Drosophila (21). Asf1 binds to the H3 interface involved in the formation of H3–H4 tetramers and forms Asf1–H3–H4 heterotrimeric complex (22). In addition to its role in RC nucleosome assembly, Asf1 is also involved in DNA replication-independent (RI) nucleosome assembly in part through its interactions with downstream chaperone HIRA (23, 24). In mammals, there are two distinct Asf1 isoforms, Asf1a and Asf1b, which are distinguishable by their C-terminal tails. Asf1a and Asf1b bind canonical histone H3.1–H4 dimers and facilitate histone transfer to downstream chaperone CAF-1 in the RC nucleosome assembly pathway. On the contrary, Asf1a also binds to histone H3 variant H3.3 along with H4 and transfers H3.3–H4 dimers to histone chaperone HIRA for nucleosome assembly and histone exchange (25). In addition to its role in nucleosome assembly, Asf1 also has a role in nucleosome disassembly and gene transcription. For instance, in budding yeast, Asf1 mediates nucleosome disassembly at promoter regions and is essential for transcriptional activation of yeast PHO5 and PHO8 genes (26–28). In Drosophila, Asf1 is recruited to specific target promoters, together with other histone modifiers, and regulates gene expression (29, 30). However, it is unknown whether Asf1 also has a role in nucleosome disassembly for gene induction in higher eukaryotic cells (31).
Recently, it has been shown that factors involved in nucleosome assembly pathways play crucial roles in maintaining chromatin states of ES and differentiated cells. For instance, histone chaperone HIRA is required for murine early embryogenesis (32), and HIRA deletion in ES cells alters trophoblast-specific TFs Cdx2 and Hand1 expression in pluripotent states, as well as during differentiation (33). Down-regulation of CAF-1 reverts ES cells to an earlier two-cell–like embryonic stage (34). Furthermore, reduced level of CAF-1 in somatic cells accelerates in vitro cell reprogramming. It is proposed that CAF-1 is required to maintain cell identity of differentiated states (35). On the contrary, histone chaperone Asf1a is necessary for reprogramming of adult dermal fibroblasts into undifferentiated induced pluripotent stem cells (iPSCs) (36). Therefore, it is likely that these histone chaperones play multiple roles in stem cell maintenance and differentiation. Motivated by these studies, we deleted Asf1a and Asf1b from mouse ES cells. Surprisingly, we observed that Asf1a and Asf1b are not required for ES cell proliferation, self-renewal, and silencing of pluripotent genes (Nanog and Oct4) during differentiation. Instead, we found that the expression of lineage-specific genes is compromised during ES cell differentiation upon Asf1a deletion. Mechanistically, we show that Asf1a facilitates nucleosome disassembly at the promoters of these genes. These results reveal a mechanism for the resolution of the repressive bivalent chromatin domains and activation of the lineage-specific genes during ES cell differentiation.
Results
KO of Asf1a or Asf1b in Mouse ES Cells Does Not Affect Cell Growth and Cell Identity.
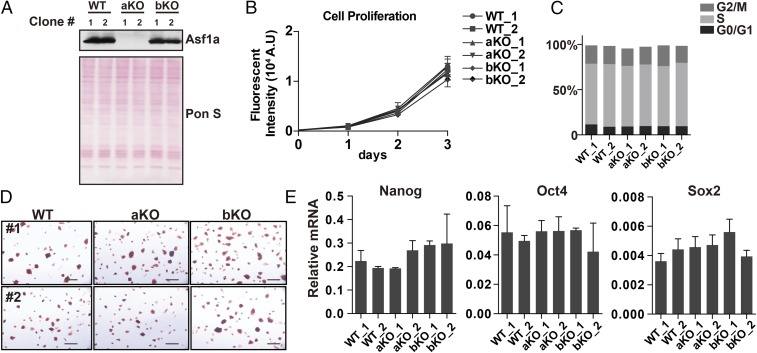
In mammals, there are two isoforms of histone chaperone Asf1, Asf1a and Asf1b, which are involved in RI and RC nucleosome assembly pathways (25). To analyze the function of Asf1a and Asf1b in mouse ES cells, we knocked out Asf1a and Asf1b from these cells by using CRISPR/Cas9 technology (37) and selected two independent clones from different sgRNAs to exclude the potential off-target effects for further analysis. Deletion of both alleles of Asf1a or Asf1b in these clones was confirmed by Sanger sequencing (SI Appendix, Fig. S1A). Western blot analysis showed that Asf1a was not detectable in Asf1a deletion lines, but was expressed normally in Asf1b deletion clones compared with WT cells (Fig. 1A). Western blot analysis of Asf1b was not informative, as our antibodies against Asf1b were not specific. All of the mutant ES cell lines had normal karyotypes (SI Appendix, Fig. S1B). Previous study showed that yeast Asf1 is important for cell viability and cell-cycle progression (21). We therefore tested whether KO of Asf1a or Asf1b affected ES cell growth and cell cycle. Asf1a or Asf1b KO ES cells exhibited similar growth rates (Fig. 1B) and cell-cycle profile (Fig. 1C and SI Appendix, Fig. S1C) as WT cells, suggesting that deletion of either Asf1a or Asf1b has no apparent effect on the proliferation of mouse ES cells. Next, we asked whether Asf1a or Asf1b was required for the self-renewal of mouse ES cells. Asf1a- and Asf1b-KO ES cells were stained positively for alkaline phosphatase (AP; Fig. 1D). Moreover, reverse transcription PCR (RT-PCR) analysis showed that the expression of three core TFs (Nanog, Oct4, and Sox2) was not affected in Asf1a- or Asf1b-KO lines (Fig. 1E). Together, these results show that Asf1a or Asf1b, surprisingly, is dispensable for normal ES cell proliferation and identity.
Fig. 1.
Asf1a or Asf1b KO does not affect mouse ES cell growth and identity. (A) Western blot analysis of Asf1a levels (Top) in two independent WT, Asf1a KO (aKO), and Asf1b KO (bKO) ES cell clones (generated by different sgRNAs). Ponceau S staining (Bottom) was used as a loading control. (B) Growth curves of two independent WT, aKO, and bKO ES cell lines. (C) BrdU/propidium iodide FACS analysis of WT, aKO, and bKO ES cells. The percentage of cells at each stage of the cell cycle was quantified. The original FACS profiles are shown in SI Appendix, Fig. S1C. (D) Representative AP staining images of WT, aKO, and bKO ES cells. (Scale bar: 50 µm.) (E) RT-PCR analysis of the expression of three pluripotent genes (Nanog, Oct4, and Sox2) in two independent WT, aKO, and bKO ES cell lines. The y axis indicates the relative mRNA level to GAPDH. The results are from three independent experiments, and bars represent mean ± SEM.
One possible explanation is that Asf1a and Asf1b are partially redundant with each other for cell growth of mouse ES cells. Consistent with this idea, we failed to generate Asf1a and Asf1b double-KO ES cells despite repeat attempts. To test this idea further, we analyzed the association of Asf1a and Asf1b with histones and downstream chaperones HIRA and CAF-1. We observed a significant increase in the binding of Asf1b with histone chaperone HIRA and histone variant H3.3 (SI Appendix, Fig. S2, lane 4 vs. lane 5) in the Asf1a-KO cells, whereas the interactions of Asf1b with CAF-1 and total H3 were not altered dramatically. This result suggests that, in the absence of Asf1a, Asf1b likely compensates for the role of Asf1a in RI incorporation of H3.3 mediated by HIRA.
Asf1a Is Indispensable for ES Cell Differentiation.
Next, we asked whether Asf1a or Asf1b is required for ES cell differentiation in vitro during the formation of embryoid bodies (EBs) (38), which can recapitulate many aspects of cell differentiation during early embryogenesis. Briefly, WT and mutant ES cells were forced to form multicellular aggregates, called EBs, in hanging drops without leukemia inhibiting factor (LIF) for 3 d (SI Appendix, Fig. S3A). Afterward, these EBs were cultured in suspension without LIF and were collected at different times of the differentiation process (days 3, 5, 7, and 10) for analysis of morphological and molecular changes.
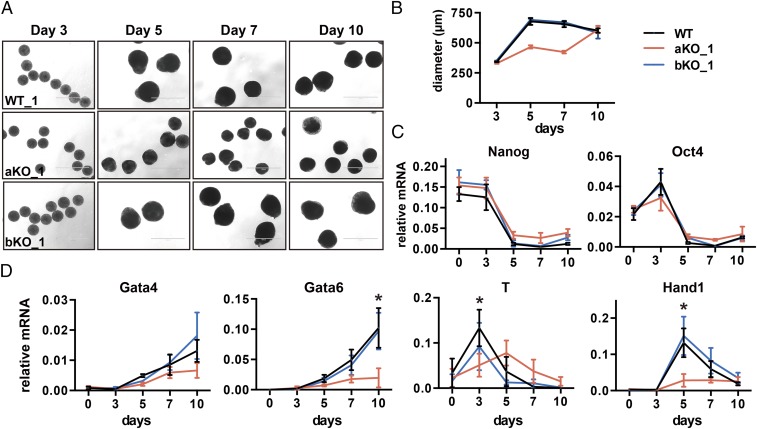
We observed that all of the ES cell lines exhibited similar morphology at day 3. However, starting from day 5, Asf1a-KO cells, but not Asf1b-KO cells, exhibited a significant reduction of EB size compared with WT cells (Fig. 2 A and B), suggesting that Asf1a deletion affects differentiation. Next, we monitored the expression of pluripotent and lineage-specific genes during differentiation by using RT-PCR. We observed no apparent defect in the silencing of two pluripotent genes (Nanog and Oct4) in Asf1a- or Asf1b-KO cells compared with WT cells (Fig. 2C and SI Appendix, Fig. S3). This observation is in contrast with previous reports that depletion of Asf1a by using siRNA affects the expression of pluripotent genes in mouse and human ESCs (36, 39). One possible explanation is that the complete loss of Asf1a in mouse ES cells triggers the complementation by Asf1b. Remarkably, the induction of lineage-specific genes, key for specification of each germ layer, was significantly defective and/or delayed in Asf1a-KO, but not Asf1b-KO, lines (Fig. 2D and SI Appendix, Fig. S3 B and C). These results suggest that Asf1a is not essential for the silencing of pluripotent genes, but is critical for the expression of lineage-specific genes involved in germ-layer specification during mouse ES cell differentiation.
Fig. 2.
Asf1a KO affects expression of lineage-specific genes during ES cell differentiation. (A) Representative images of EB morphology during EB differentiation at days 3, 5, 7, and 10 from WT, aKO, and bKO cells. (Scale bar: 1,000 µm.) (B) The diameter of at least 50 EBs at each time point was measured by using ImageJ. (C and D) RT-PCR analysis of pluripotent (C) and lineage-specific gene (D) expression during ES cell differentiation. Data are from three independent experiments. Error bars represent mean ± SEM. The P value was calculated by using a t test between WT and aKO lines (*P < 0.05). The expression of additional germ-layer genes as well as these genes in another independent clone is shown in SI Appendix, Fig. S3 B and C. Note that the effect of Asf1a deletion on the expression of Gata4, although apparent, was not statistically significant for this set of three repeats. The effect of Asf1a deletion on the expression of Gata4 in the rescue experiments shown in Fig. 5F was statistically significant. The difference between these two sets of experiments likely reflects the fact that Asf1a KO on Gata4 expression is small, and therefore some experimental variations during differentiation can mask the difference.
Asf1a Is Required for Histone-Modification Changes During Differentiation.
It has been observed that H3K27me3 was reduced globally during ES cell differentiation (40). We also observed that H3K27me3 levels in WT EBs were significantly lower than in WT ES cells (SI Appendix, Fig. S4A). Compared with WT EBs, Asf1a KO EBs retained markedly higher levels of H3K27me3, suggesting that this mark is not reduced properly during differentiation. The effect is unlikely a result of the impact of Asf1a KO on the expression of H3K27-modifying enzymes, including Ezh2 and Suz12 (two key subunits of H3K27 methyltransferase) and Utx and Jmjd3 (two known H3K27 demethylases), during differentiation (SI Appendix, Fig. S4B). We suggest that the impact of Asf1a KO on H3K27me3 during EB formation likely reflects defects in differentiation (as detailed later).
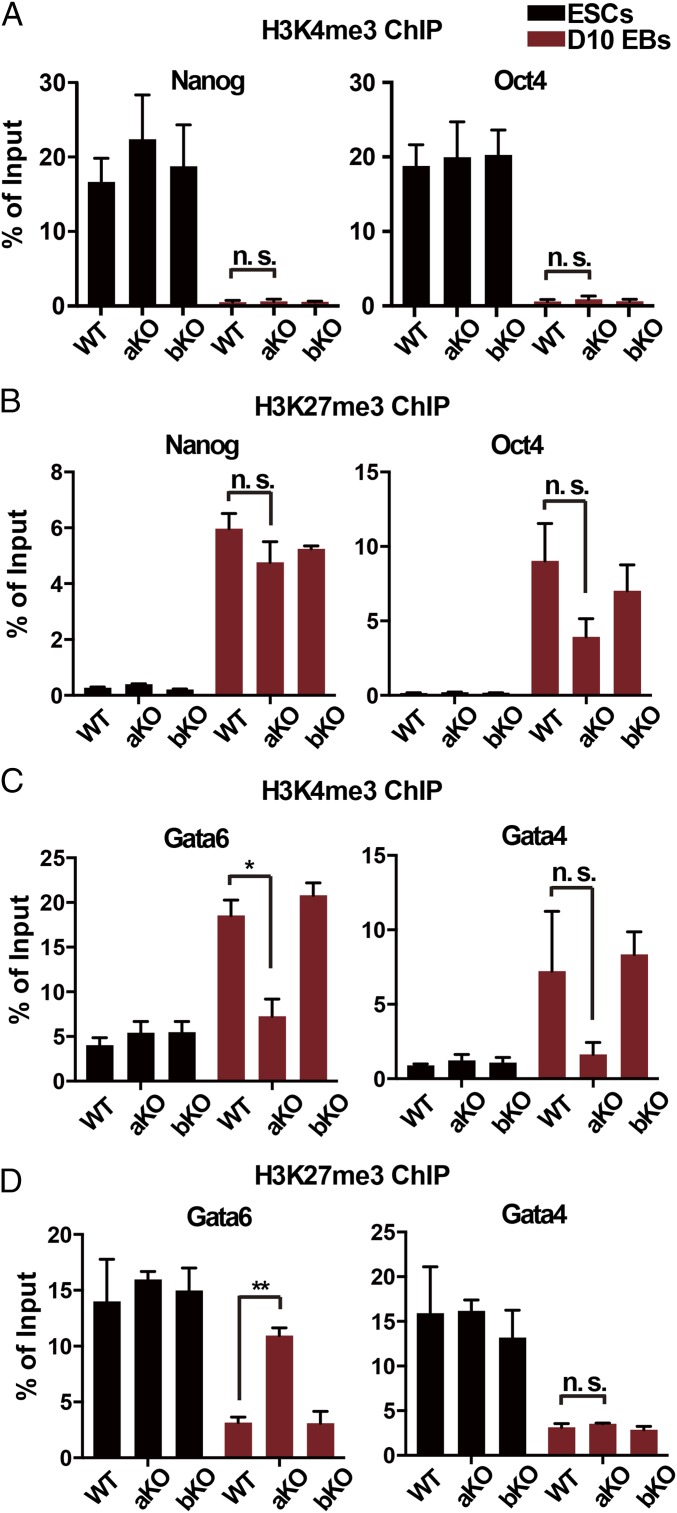
Next, we determined how Asf1a KO affected H3K27me3 and H3K4me3, two marks that coexist at lineage-specific genes (17), at the promoters of Gata4 and Gata6 by using ChIP-PCR. Similar analysis was also performed at two pluripotent genes (Nanog and Oct4). During the differentiation of WT ES cells, the active mark H3K4me3 was dramatically reduced at the promoters of Nanog and Oct4 (Fig. 3A), whereas H3K27me3 dramatically increased (Fig. 3B), consistent with the observations that these genes are silenced in this process. Deletion of Asf1a or Asf1b had no detectable effect on the changes of H3K4me3 and H3K27me3 at the promoters of these two pluripotent genes (Fig. 3 A and B). For lineage-specific genes, H3K4me3 at Gata4 and Gata6 promoters increased dramatically (Fig. 3C), whereas H3K27me3 at these two gene promoters showed a marked reduction upon differentiation of WT ES cells (Fig. 3D), consistent with the induction of these lineage-specific genes in these cells. Depletion of Asf1b had no apparent effect on the dynamic changes of these histone marks at Gata4 and Gata6 promoters. By contrast, deletion of Asf1a compromised the increase in H3K4me3 and the reduction of H3K27me3 at Gata6 promoter (Fig. 3 C and D). At the Gata4 promoter, the increase in H3K4me3 level in Asf1a-KO EBs was slightly compromised, but not significantly so, in three independent experiments compared with WT EBs, whereas the reduction of H3K27me3 was not affected to a detectable degree. The slight impact of Asf1a deletion on changes in H3K4me3 and H3K27me3 levels at the promoters of Gata4 compared with Gata6 during differentiation likely reflects the fact that Asf1a KO affects the induction of Gata4 less than Gata6.
Fig. 3.
Asf1a KO affects dynamic changes of histone modifications at lineage-specific genes during differentiation. (A and B) Analysis of H3K4me3 (A) and H3K27me3 (B) at the promoters of two pluripotent genes (Nanog and Oct4) by ChIP-PCR. (C and D) Analysis of H3K4me3 (C) and H3K27me3 (D) at the promoters of two lineage-specific genes (Gata4 and Gata6). Black and red bars represent ES cells and day 10 EBs, respectively. Data were from three independent experiments. Error bars represent mean ± SEM (*P < 0.05 and **P < 0.01).
Asf1a Is also Required for Induction of Lineage-Specific Genes During Differentiation to Neural Precursors.
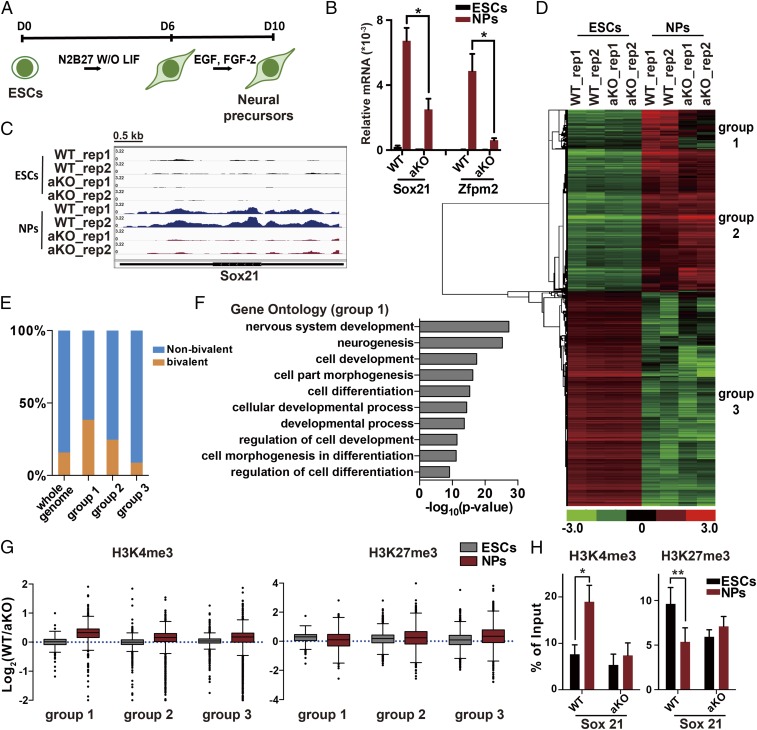
To gain additional insight into the role of Asf1a in ES differentiation, we differentiated the ES cells along a neural pathway in adherent serum-free culture as described previously (41, 42). Briefly, ES cells were cultured in serum-free medium without LIF for 6 d and then replated and maintained in FGF-2– and EGF–containing medium. The multipotent neural precursors (NPs) were collected at day 10 (Fig. 4A). We then analyzed the expression levels of two bivalent genes (Sox21 and Zfpm2), which are induced during this differentiation process (17). Consistent with the results we observed during EB differentiation, cells without Asf1a showed defects in the induction of Sox21 and Zfpm2 compared with WT cells (Fig. 4B). As NPs are relatively homogenous, we then employed this system to analyze the transcriptome of WT and Asf1a-KO ES cells and NPs by using RNA sequencing (RNA-seq). As expected, Sox21 level is affected in Asf1a-KO NPs in both experimental replicates (Fig. 4C). To analyze the impact of Asf1a deletion on gene expression during differentiation, we first identified genes whose expression were altered during the differentiation of WT and Asf1a-KO ES cells. We found that the expression of 2,742 genes increased, whereas that of 3,247 genes decreased, in WT ES cells during differentiation. Interestingly, Asf1a KO did not affect expression of genes that were silenced (group 3; Fig. 4D and SI Appendix, Fig. S5A). In contrast, Asf1a KO affected the expression of a subgroup of genes (group 1) that were induced during differentiation (Fig. 4D), whereas it had little effect on the induction of the second group of genes (group 2). The group 1 genes were enriched with bivalent genes, which contain both H3K27me3 and H3K4me3 at gene promoters, compared with the whole genome as well as genes in group 2 and group 3 (Fig. 4E). Gene Ontology (GO) analysis revealed that the group 1 genes are involved in cell differentiation and neural development, whereas group 2 genes are mainly involved in metabolic process and protein transport (Fig. 4F and SI Appendix, Fig. S5B). These results strongly support the idea that Asf1a is important for the induction of “bivalent” genes during lineage specification.
Fig. 4.
Asf1a is required for induction of lineage-specific genes during neural differentiation. (A) An outline of the monolayer ES cell in vitro neural differentiation. (B) Gene-expression analysis of Sox21 and Zfpm2 in Asf1a-KO cells expressing EV (aKO) or WT Asf1a (WT) during neural differentiation using RT-PCR. Data are from three independent experiments. Error bars represent mean ± SEM. The P value was calculated by using a t test between WT and aKO NPs (*P < 0.05). (C) Snapshot of RNA-seq results at the Sox21 locus. Data are from two independent experiments (rep1 and rep2). (D) The hierarchical clustering analysis of the differentially expressed genes during neural differentiation of WT and aKO cells identified by RNA-seq. (E) The percentage of bivalent genes in each of the three subgroups of genes identified in D. (F) GO analysis of the group 1 genes. The top 10 significant GO terms and P value are displayed. (G) Relative levels of H3K4me3 and H3K27me3 between WT and aKO at the promoters of three subgroups of genes identified in D. The y axis represents the log2 ratio of ChIP-seq reads between WT and aKO lines. (H) Analysis of changes in H3K4me3 and H3K27me3 at the gene promoter of Sox21 in WT and aKO lines by ChIP-PCR. The P value was calculated by using a t test between ES cells and NPs (*P < 0.05 and **P < 0.01). Note that Asf1a-KO and WT clones were the same as used in Fig. 5.
We also performed H3K4me3 and H3K27me3 ChIP-deep sequencing (ChIP-seq) by using ES cells and NPs and analyzed the effect of Asf1a KO on changes of H3K4me3 and H3K27me3 at gene promoters. As expected, at the promoters of group 1 genes, the level of H3K4me3 in WT cells increased more than in Asf1a-KO cells when ES cells differentiated into NPs. H3K27me3 levels were reduced more in Asf1a WT than in Asf1a-KO cells (Fig. 4G). ChIP-PCR analysis confirmed that, at the Sox21 promoter, Asf1a KO affected the increase of H3K4me3 and reduction of H3K27me3 during differentiation of ES cells into NPs (Fig. 4H). In contrast, the effect of Asf1a KO on H3K4me3 and H3K27me3 at the promoters of group 2 and group 3 genes was less pronounced than in group 1 genes (Fig. 4G). These results are consistent with the idea that the induction of group 1 genes is compromised in Asf1a-KO cells.
The Asf1a–Histone H3 Interaction Is Important for Gene Transcription During ES Cell Differentiation.
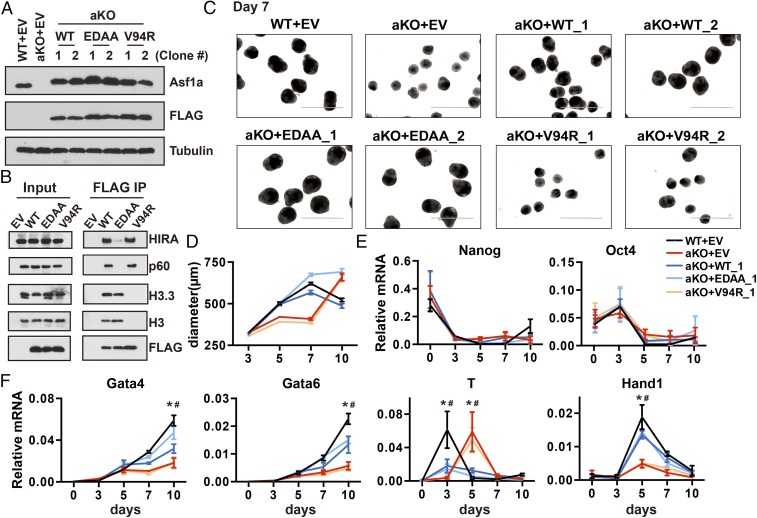
Asf1 in yeast has nucleosome assembly and disassembly activity. To understand how Asf1a regulates the expression of lineage-specific genes, we ectopically expressed FLAG-tagged WT Asf1a, Asf1a mutant that cannot bind HIRA and CAF-1 [Asf1a (E36A, D37A, marked as EDAA)] and is defective in nucleosome assembly, or Asf1a mutant deficient for H3–H4 binding [Asf1a (V94R)] that is defective in nucleosome assembly and disassembly in Asf1a-KO ES cells and analyzed the effect of these Asf1a mutations on the expression of lineage-specific and pluripotent genes (i.e., control) by using EB differentiation assays. As shown in Fig. 5A, WT, Asf1a (EDAA), and Asf1a (V94R) mutants were expressed at similar levels. Moreover, Asf1a (EDAA) mutant exhibited defects in its association with HIRA and CAF-1 (p60) but not histone. In contrast, Asf1a (V94R) mutant was defective in binding to histone H3 but not histone chaperones (Fig. 5B). These results confirm the differential effects of two Asf1a mutants on histone and chaperone binding and are consistent with previous findings that HIRA and CAF-1 bind to Asf1a through the same domain and compete for the association with Asf1a in vivo (43).
Fig. 5.
The Asf1a–histone interaction is important for the induction of gene transcription during differentiation. (A) Analysis of the exogenously expressed Asf1a and mutants in Asf1a-KO ES cells. “WT” indicates FLAG-tagged WT Asf1a; “EDAA” indicates Asf1a (E36A, D37A, indicated as EDAA), and “V94R” indicates Asf1a (V94R). Two independent lines were analyzed by using Western blot and compared with endogenous Asf1a level. (B) Analysis of interactions of Asf1a and Asf1a mutants with histone H3 and chaperones HIRA and CAF-1 (p60). Asf1a-KO cells expressing FLAG-tagged WT or Asf1a mutants were immunoprecipitated with M2 beads, and coimmunoprecipitated proteins were analyzed by Western blot. (C and D) Representative images of day 7 EBs (C) and the average size of EBs formed from ES cells expressing WT Asf1a and Asf1a mutants (D). Two independent rescue lines were analyzed, and the results of another line are shown in SI Appendix, Fig. S5A. (Scale bar: 1,000 µm.) (E and F) RT-PCR analysis of pluripotent (E) and four lineage-specific genes (F) of ES clones expressing WT or mutant Asf1a during differentiation. The expression of additional germ-layer marks and the expression results obtained from another independent line are shown in SI Appendix, Fig. S5 B and C [*P < 0.05, (WT+EV) vs. (aKO+EV); #P < 0.05 (WT+EV) vs. (aKO+V94R)].
We then performed an EB differentiation assay by using these rescue cell lines. Upon differentiation, we observed that the size of EBs expressing WT and mutant Asf1a (EDAA) was similar to that of EBs formed by WT ES cells infected with empty vector (EV; Fig. 5 C and D and SI Appendix, Fig. S6A). RT-PCR analysis showed that expression of WT or Asf1a (EDAA) mutant had no apparent effect on the silencing of Nanog and Oct4 (Fig. 5E) and at least partially rescued the defects in the expression of lineage-specific genes caused by Asf1a KO (Fig. 5F and SI Appendix, Fig. S6 B and C). By contrast, expression of Asf1a (V94R) mutant failed to rescue the defects in EB size (Fig. 5 C and D and SI Appendix, Fig. S6A) and the induction of lineage-specific genes (Gata4, Gata6, T, and Hand1) caused by Asf1a deletion (Fig. 5F and SI Appendix, Fig. S6 B and C). These results indicate that the Asf1a–histone H3 interaction is important for the role of Asf1a in regulating lineage-specific genes, whereas the interactions between Asf1a and its downstream chaperones (HIRA and CAF-1) are dispensable. These results imply that Asf1a’s role in nucleosome disassembly, but not in nucleosome assembly, mediates its function in gene regulation during differentiation.
Asf1a Binds to Lineage-Specific Gene Promoters.
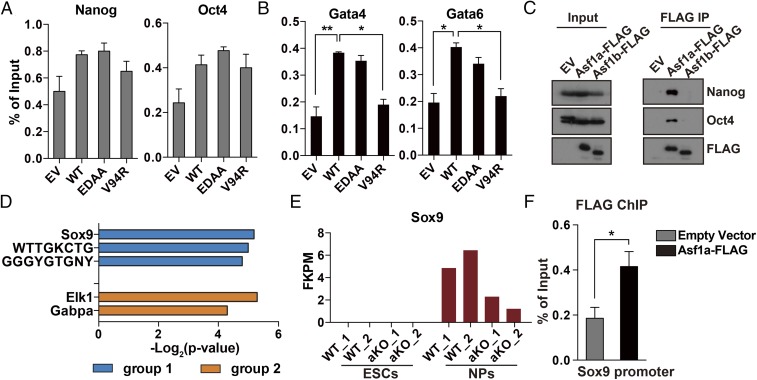
To understand how Asf1a regulates the expression of a subset of genes during differentiation, we first asked whether Asf1a localized at some of these gene promoters. We performed Asf1a ChIP by using antibodies against the FLAG epitope in Asf1a-KO ES cells stably expressing FLAG-tagged WT or mutant Asf1a. As shown in Fig. 6A, we did not detect significant enrichment of Asf1a at the promoters of Nanog and Oct4 compared with controls. In contrast, Asf1a was enriched at the promoters of Gata4 and Gata6, two tested lineage-specific gene promoters (Fig. 6B). We also noticed that Asf1a (V94R), but not Asf1a (EDAA), mutant showed defects in binding to Gata4 and Gata6 promoters (Fig. 6B). This result suggests that Asf1a chromatin binding depends on its ability to bind its cargo, H3–H4, an observation consistent with the chromatin binding of other histone chaperones, including CAF-1 and TONSL-MMS22L (44, 45). Together, these results show that Asf1a can bind to the promoters of lineage-specific genes and that this binding depends on the presence of its cargo, H3–H4.
Fig. 6.
Asf1a binds to the promoters of lineage-specific genes. (A and B) Asf1a does not bind to the promoters of Nanog and Oct4 (A) but binds to the promoters of Gata4 and Gata6 (B). Asf1a ChIP-PCR was performed by using ES cells expressing WT Asf1a and Asf1a mutants described in Fig. 5A. Results are from three different experiments, and error bars represent mean ± SEM (*P < 0.05 and **P < 0.01). (C) Asf1a, but not Asf1b, was coimmunoprecipitated with Nanog and Oct4. (D) Identification of TF binding sites (TFBSs) that are enriched in group 1 but not group 2 genes or vice-versa. Top hits were listed together with the −log2 (P value). (E) Expression of Sox9 in ES cells and NPs during neural differentiation based on RNA-seq reads. (F) Asf1a binds to the promoter of Sox9. FLAG ChIP was performed in aKO ES cells expressing EV or WT FLAG-Asf1a, and ChIP DNA was analyzed by quantitative PCR at the Sox9 promoter (*P < 0.05).
Second, we asked whether Asf1a, like Drosophila Asf1, could interact with TFs. We found that Asf1a, but not Asf1b, interacted with TFs Nanog and Oct4 (Fig. 6C). Third, we identified which TF binding sites are enriched at the promoters of group 1 genes but not at those of group 2 genes identified in Fig. 4D and vice-versa. We found that promoters of group 1 genes are enriched for the Sox9 binding sites, whereas the promoters of group 2 genes are enriched with Elk1 (Fig. 6D). Sox9 is a bivalent and lineage-specific gene important for neural differentiation (46). Inspection of RNA-seq results indicates that Asf1a KO affected the induction of Sox9, but not Elk1, during the differentiation of ES cells into NPs (Fig. 6E and SI Appendix, Fig. S7). Moreover, ChIP-PCR analysis indicates that Asf1a binds to the promoter of Sox9 (Fig. 6F). These results suggest that Asf1a is recruited to lineage gene promoters in part through its interactions with Nanog and Oct4, and regulates expression key genes such as Sox9 during in vitro neural differentiation.
Asf1a Mediates Nucleosome Disassembly at Bivalent Gene Promoters During Differentiation.
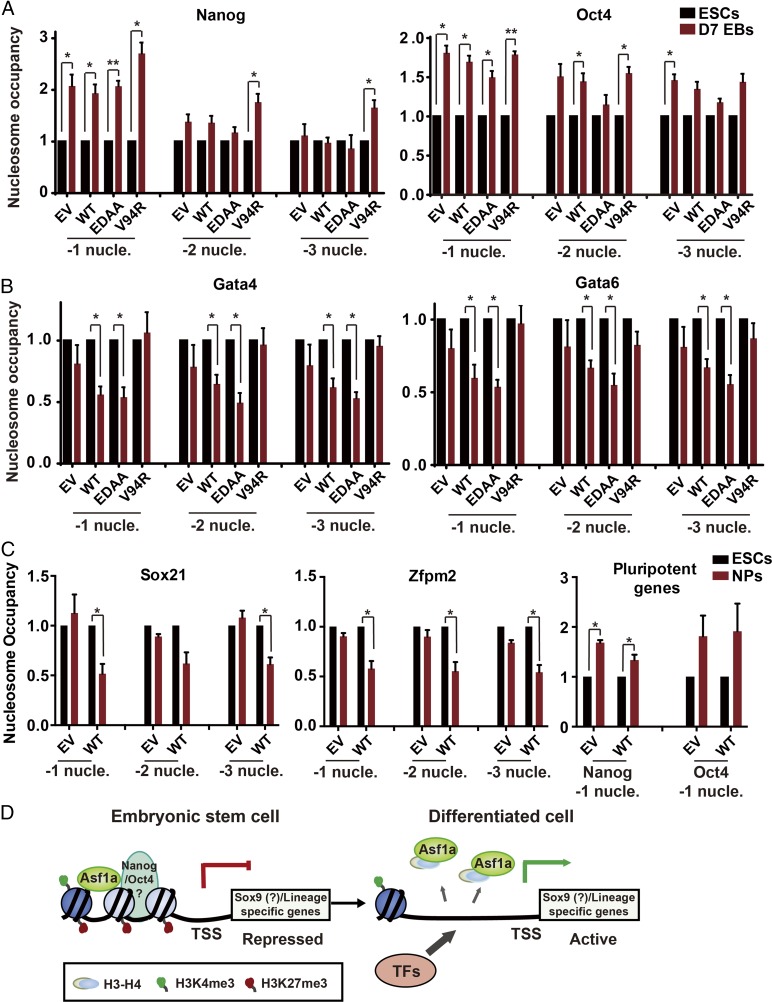
Our results presented here indicate that Asf1a regulates gene expression through its role in nucleosome disassembly. To test this idea experimentally, we asked whether deletion of Asf1a affects nucleosome occupancy at the promoters of Gata4 and Gata6 in ES cells and EBs first. Briefly, chromatin from Asf1a-KO ES cells and EBs expressing EV, WT, or mutant Asf1a were digested to mononucleosomes with micrococcal nuclease. Nucleosomes were then precipitated by using antibodies against H3, and ChIP DNAs were analyzed by using PCR primers targeting three individual nucleosomes (−1 to −3) upstream of the transcription start site (TSS) of Nanog, Oct4, Gata4, and Gata6 (SI Appendix, Fig. S8A). The nucleosome positions at these genes were based on a recently published study (47). We observed a significant increase in nucleosome occupancy at the −1 nucleosomes upstream of the TSSs of Nanog and Oct4 genes in WT EBs compared with corresponding ES cells (Fig. 7A). Moreover, deletion of Asf1a or expression of WT or mutant Asf1a in Asf1a-KO cells had no apparent effects on the nucleosomal changes at the −1 nucleosomes of these two pluripotent genes (Fig. 7A). At the −2 and −3 nucleosomes of Nanog and Oct4, we did not detect consistent changes in nucleosome occupancy in any of the samples analyzed. These results support the idea that a more compact chromatin state at the promoter of these two genes is formed during silencing of Nanog and Oct4, and that Asf1a deletion has no apparent effect on chromatin compaction at these two genes during EB formation. In contrast, at gene promoters of Gata4 and Gata6, nucleosome occupancy at three nucleosomes (−1 to −3) upstream of the TSS of Gata4 or Gata6 was reduced in WT or Asf1a (EDAA) EBs compared with their corresponding ES cells (Fig. 7B), consistent with an open chromatin state at these genes. In contrast, the reduction in nucleosome occupancy at these three positions of these two genes upon differentiation was not detectable in Asf1a-deleted cells (i.e., EV) or in cells expressing Asf1a (V94R) mutant. However, deletion of Asf1a had no apparent effect on nucleosome occupancy at promoter of housekeeping gene GAPDH in ES and EBs (SI Appendix, Fig. S8B), consistent with the idea that this gene is highly expressed in ES cells and EBs. Finally, inhibition of transcription had no apparent effect on nucleosomal changes at the promoters of Gata4 and Gata6 (SI Appendix, Fig. S8C). These results indicate that the impact of Asf1a deletion on nucleosomal changes at Gata4 and Gata6 during EB formation is unlikely to be caused by a global change in nucleosome occupancy and/or a consequence of active antisense transcription. Second, we also analyzed nucleosomal changes at −1 to −3 nucleosomes surrounding TSS of Sox21 and Zfpm2 during ES cell neural differentiation. We found that nucleosome occupancy was also reduced at the −1 nucleosome of Sox21 and from −1 to −3 nucleosomes of Zfpm2 during neural differentiation of WT ES cells. Importantly, this reduction was not detectable in Asf1a-KO cells in which changes in nucleosome occupancy of Nanog and Oct4 were not affected (Fig. 7C). These results strongly suggest that Asf1a is directly or indirectly involved in nucleosome disassembly at the promoters of at least some of the lineage-specific genes for their induction during differentiation.
Fig. 7.
Asf1a mediates nucleosome disassembly at lineage-specific gene promoter. (A and B) Nucleosome occupancy at two pluripotent genes (Nanog and Oct4, A) and two lineage-specific genes (Gata4 and Gata6, B) during EB formation. H3 ChIP was performed by using chromatin from Asf1a-KO ES cells and day 7 (D7) EBs expressing EV, WT, or mutant Asf1a. Quantitative PCR was performed targeting each of three nucleosomes upstream of TSSs of two pluripotent genes (A) and two lineage-specific genes (B). (C) Nucleosome occupancy at two bivalent gene (Sox21 and Zfpm2) promoters and two pluripotent gene (Nanog and Oct4) promoters during neural differentiation. The H3 ChIP enrichment was normalized against its enrichment at the gene body of GAPDH, and the nucleosome occupancy of ES cells in each line was set as 1. Results are from three different experiments, and error bars represent mean ± SEM (*P < 0.05 and **P < 0.01). (D) A working model depicting that Asf1a mediates nucleosome disassembly at lineage-specific gene promoters and facilitates subsequent association of TFs.
Discussion
How the repressive bivalent chromatin states at lineage-specific genes are resolved for the activation of these genes during mouse ES cell differentiation is largely unknown. Here we show that Asf1a, one of the two Asf1 isoforms in mammalian cells, is important for nucleosome disassembly at lineage-specific genes and the activation of these genes during differentiation, uncovering an unexpected role of Asf1a in mouse ES cell differentiation.
Asf1 is best known for its role in nucleosome assembly in yeast and human cells (48). Budding yeast cells lacking Asf1 grow poorly and accumulate in the G2/M phase of cell cycle (21). In Schizosaccharomyces pombe, Asf1 is essential for cell viability (49). In human cell lines, depletion of Asf1b, an isoform of Asf1 in mammalian cells, impairs continued cell proliferation (50). Therefore, it is surprising that mouse ES cells lacking Asf1a or Asf1b do not show any obvious defects in cell proliferation. We suggest that Asf1a and Asf1b are partially redundant in mouse ES cells despite the fact that Asf1a and Asf1b have distinct functions. Consistent with this idea, we observed that, in Asf1a-deleted mouse ES cells, Asf1b, which is not known to bind H3.3 or HIRA, could bind H3.3 and HIRA (SI Appendix, Fig. S2). Moreover, we could not generate mouse ES cells lacking Asf1a and Asf1b despite repeated attempts, suggesting that mouse ES cells lacking Asf1a and Asf1b are likely not viable.
Unexpectedly, we made several observations supporting the idea that Asf1a is important for mouse ES cell differentiation. First, we observed that EBs formed in vitro, which contain three germ layers, grow significantly smaller in size after deletion of Asf1a, but not Asf1b. Gene-expression analysis revealed that Asf1a deletion has no apparent effect on the silencing of pluripotent genes (Nanog and Oct4). Instead, the induction of genes critical for germ-layer formation was defective and/or dramatically delayed. These results suggest that Asf1a is important for the regulation of lineage-specific genes during the in vitro random differentiation of ES cells. Second, Asf1a is also indispensable during ES neural differentiation. In this in vitro differentiation process, Asf1a regulates the expression of a group of genes that are involved in the neural development process, and this group of genes is enriched with bivalent genes. Together, these studies support our conclusion that Asf1a is dispensable for the silencing of pluripotent genes but important for the induction of lineage-specific genes in these two different in vitro differentiation processes.
How does Asf1a manage to regulate a subgroup of genes during differentiation? Similar as in Drosophila, Asf1a could be recruited to target gene promoters through the associations with TFs. Supporting this idea, we show that Asf1a interacts with Nanog and Oct4. During the in vitro neural differentiation, we found that genes regulated by Asf1a are enriched with the Sox9 binding motif. Sox9 is a bivalent gene that is important for the induction and maintenance of multipotent NPs (46). Moreover, we also found that Asf1a binds to the Sox9 promoter and is required for the induction of Sox9 during neural differentiation. These findings support a model in which Asf1a is recruited to target promoters through interactions with TFs, such as Nanog and Oct4, facilitates the induction of Sox9 and other key lineage-specific genes, thereby regulating a subgroup of genes involved in neural differentiation (Fig. 7D).
Asf1 is involved in nucleosome assembly and disassembly. We present two lines of evidence supporting the idea that Asf1a’s role in nucleosome disassembly or histone eviction is important for the induction of lineage-specific genes. First, we observed that the ability of Asf1a to bind H3–H4 is required for the induction of lineage-specific genes, but its ability to bind CAF-1 and HIRA is dispensable for this process. Because Asf1 functions in nucleosome assembly by transferring H3–H4 to downstream chaperones, CAF-1 and HIRA, these results suggest that Asf1a’s role in nucleosome disassembly, but not nucleosome assembly, is critical for the induction of lineage-specific genes. Second, we show that nucleosome occupancy decreases at four bivalent promoters tested in WT cells in two different differentiation processes, suggesting that nucleosomes at the promoters of these genes are disassembled. Importantly, Asf1a deletion compromised the reduction in nucleosome occupancy at these four genes. We noted that Asf1a deletion compromised the establishment of H3K4me3 and removal of H3K27me3 at some lineage-specific promoters during differentiation (Fig. 4G). It is possible that these effects represent a combination of nucleosome occupancy and histone modification changes. Nonetheless, these results are consistent with the idea that Asf1a helps evict nucleosomes at these gene promoters, aiding the establishment of proper histone modifications for gene transcription during differentiation.
Why is nucleosome disassembly important for the induction of these lineage-specific genes? These genes contain bivalent chromatin domains, consisting of a large area of nucleosomes modified by H3K27me3 and smaller regions of H3K4me3. These domains create a barrier for the association of TFs with their binding sites on the DNA (19). Therefore, this bivalent chromatin barrier must be resolved for gene activation. A recent report indicates that early postreplicative chromatin lacks H3K27me3 mark after the induction of ES cell differentiation (51). It is proposed that a delayed nucleosome assembly of this mark provides a more accessible chromatin for the recruitment of TFs. Thus, we suggest that nucleosome disassembly by Asf1a at bivalent domains provides another means to resolve the bivalent chromatin domains at lineage-specific genes, which in turn facilitates the recruitment of TFs for activation of lineage-specific genes (Fig. 7D).
Materials and Methods
Cell Culture.
Mouse E14 ES cells (provided by Tom Fazzio, University of Massachusetts Medical School, Worcester, MA) were cultured in standard ES medium with LIF. Detailed procedures of cell culture and CRISPR/Cas9 targeting are described in SI Appendix, Supplementary Materials and Methods.
In Vitro Differentiation Assays and RT-PCR Analysis.
Detailed information of EB formation and neural differentiation is provided in SI Appendix, Supplementary Materials and Methods.
ChIP-Seq and ChIP–Real-Time PCR.
A detailed protocol is provided in SI Appendix, Supplementary Materials and Methods, and primer sets are listed in SI Appendix, Table S1.
RNA-Seq and ChIP-Seq Data Analysis.
Information about data analysis is provided in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Tom Fazzio for E14 stem cell lines, LIF-expression plasmids, and protocols and Drs. Dong Fang, Xiangdong Lv, and Minjie Zhang for reading this manuscript. This work was supported by National Institute of Health Grant GM119015 (to Z.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.L. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE114424).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801909115/-/DCSupplemental.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 6.Yeo J-C, Ng H-H. The transcriptional regulation of pluripotency. Cell Res. 2013;23:20–32. doi: 10.1038/cr.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 8.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TI, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 11.Yuan P, et al. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrisey EE, et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strumpf D, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 14.Mora-Castilla S, et al. Transient downregulation of Nanog and Oct4 induced by DETA/NO exposure in mouse embryonic stem cells leads to mesodermal/endodermal lineage differentiation. Stem Cells Int. 2014;2014:379678. doi: 10.1155/2014/379678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Voigt P, Tee W-W, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwafuchi-Doi M, Zaret KS. Cell fate control by pioneer transcription factors. Development. 2016;143:1833–1837. doi: 10.1242/dev.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soufi A, et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyler JK, et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 22.English CM, Maluf NK, Tripet B, Churchill MEA, Tyler JK. ASF1 binds to a heterodimer of histones H3 and H4: A two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 24.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 25.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: An escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 26.Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Adkins MW, Williams SK, Linger J, Tyler JK. Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Mol Cell Biol. 2007;27:6372–6382. doi: 10.1128/MCB.00981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Goodfellow H, et al. Gene-specific targeting of the histone chaperone asf1 to mediate silencing. Dev Cell. 2007;13:593–600. doi: 10.1016/j.devcel.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 30.Moshkin YM, et al. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35:782–793. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Pal S, et al. The commercial antibodies widely used to measure H3 K56 acetylation are non-specific in human and Drosophila cells. PLoS One. 2016;11:e0155409. doi: 10.1371/journal.pone.0155409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts C, et al. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol Cell Biol. 2002;22:2318–2328. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banaszynski LA, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishiuchi T, et al. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol. 2015;22:662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- 35.Cheloufi S, et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature. 2015;528:218–224. doi: 10.1038/nature15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez-Muñoz E, Arboleda-Estudillo Y, Otu HH, Cibelli JB. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345:822–825. doi: 10.1126/science.1254745. [DOI] [PubMed] [Google Scholar]
- 37.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurosawa H. Methods for inducing embryoid body formation: In vitro differentiation system of embryonic stem cells. J Biosci Bioeng. 2007;103:389–398. doi: 10.1263/jbb.103.389. [DOI] [PubMed] [Google Scholar]
- 39.Tan Y, Xue Y, Song C, Grunstein M. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc Natl Acad Sci USA. 2013;110:11493–11498. doi: 10.1073/pnas.1309914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 41.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 43.Tang Y, et al. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol. 2006;13:921–929. doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saredi G, et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex. Nature. 2016;534:714–718. doi: 10.1038/nature18312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott CE, et al. SOX9 induces and maintains neural stem cells. Nat Neurosci. 2010;13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- 47.Voong LN, et al. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell. 2016;167:1555–1570.e15. doi: 10.1016/j.cell.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mousson F, Ochsenbein F, Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116:79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- 49.Umehara T, Chimura T, Ichikawa N, Horikoshi M. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes Cells. 2002;7:59–73. doi: 10.1046/j.1356-9597.2001.00493.x. [DOI] [PubMed] [Google Scholar]
- 50.Corpet A, et al. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011;30:480–493. doi: 10.1038/emboj.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petruk S, et al. Delayed accumulation of H3K27me3 on nascent DNA is essential for recruitment of transcription factors at early stages of stem cell differentiation. Mol Cell. 2017;66:247–257.e5. doi: 10.1016/j.molcel.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.