Significance
Gene duplication and divergence are cornerstones of evolution. Genetic redundancy resulting from repeated DNA provides flexibility for transient changes in copy number that may confer selective benefit under new or changing environmental conditions. This work describes a method for creating tandem arrays of specific DNA segments in a bacterium, Acinetobacter baylyi, to accelerate experimental evolution. The induced chromosomal gene amplification mimics a natural process that would otherwise occur more slowly and stochastically. The success of this approach for the evolution of novel protein function was demonstrated with studies of an enzyme that has bioenergy applications in lignin valorization. A protein variant, which emerged from Evolution by Amplification and Synthetic biology, is beneficial in a different bacterium, indicating the broad utility of the method.
Keywords: gene amplification, evolution, P450, guaiacol, Acinetobacter
Abstract
Experimental evolution is a critical tool in many disciplines, including metabolic engineering and synthetic biology. However, current methods rely on the chance occurrence of a key step that can dramatically accelerate evolution in natural systems, namely increased gene dosage. Our studies sought to induce the targeted amplification of chromosomal segments to facilitate rapid evolution. Since increased gene dosage confers novel phenotypes and genetic redundancy, we developed a method, Evolution by Amplification and Synthetic Biology (EASy), to create tandem arrays of chromosomal regions. In Acinetobacter baylyi, EASy was demonstrated on an important bioenergy problem, the catabolism of lignin-derived aromatic compounds. The initial focus on guaiacol (2-methoxyphenol), a common lignin degradation product, led to the discovery of Amycolatopsis genes (gcoAB) encoding a cytochrome P450 enzyme that converts guaiacol to catechol. However, chromosomal integration of gcoAB in Pseudomonas putida or A. baylyi did not enable guaiacol to be used as the sole carbon source despite catechol being a growth substrate. In ∼1,000 generations, EASy yielded alleles that in single chromosomal copy confer growth on guaiacol. Different variants emerged, including fusions between GcoA and CatA (catechol 1,2-dioxygenase). This study illustrates the power of harnessing chromosomal gene amplification to accelerate the evolution of desirable traits.
Evolutionary approaches to improve gene functions have found widespread use and success in many areas of biology including metabolic engineering and synthetic biology. Experimental evolution typically relies on growth or toxicity mitigation to select a desired phenotype (1, 2). Since improved gene function may require many thousands of generations of microbial growth, culturing methods typically employ serial transfers or chemostats (3). The foundation of evolutionary theory rests on the long-accepted principles of duplication and divergence (4). Recently, mounting evidence highlights the importance of a transient period of elevated gene copy number, beyond mere duplication, which facilitates emergence of new functions (5). Increased dosage of an advantageous but weak function may permit survival and facilitate adaptation to new or changing conditions. Moreover, in vivo gene amplification, which is maintained by positive selection, increases the DNA available for mutation. If beneficial mutation(s) arise, a novel allele may be selected with no evidence of this dynamic process, because temporary gene amplification is difficult to investigate and manipulate (6–8). Thus, engineered copy number change has not previously been incorporated in procedures for experimental evolution.
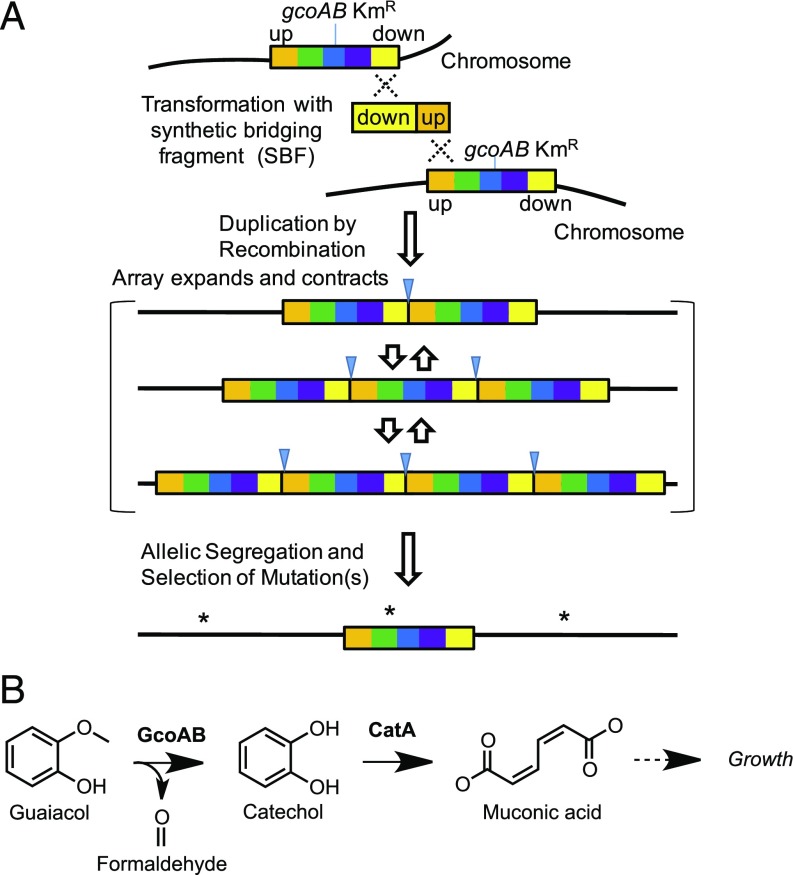
Typical efforts to hasten the isolation of desired mutants include the use of large populations of rapidly growing organisms and, sometimes, the addition of mutagenic agents. Increased gene dosage provides a different and significant benefit by conferring new phenotypes based on the augmentation of a weak or secondary function. The secondary function can then be honed during evolution under selective conditions. Therefore, the creation of precisely amplified chromosomal regions should circumvent the long timeframe typically needed to increase gene dosage by natural and stochastic recombination events. The unique and powerful genetic system of Acinetobacter baylyi ADP1 enables the engineered production of a multicopy chromosomal array of target genes (9–11). This approach was used to develop a method, dubbed Evolution by Amplification and Synthetic biology (EASy; Fig. 1A).
Fig. 1.
EASy: Targeted chromosomal amplification. (A) An A. baylyi recipient is transformed with a synthetic bridging fragment (SBF), DNA that promotes homologous recombination between DNA normally upstream (up) and downstream (down) of the target. Selection maintains the tandem array, which can expand and contract via recombination. Here, growth on guaiacol required multiple gcoAB copies. Prolonged selection may result in beneficial mutations (asterisks) that enable growth with one copy of the target region. Blue triangles show a DNA junction introduced by the SBF. (B) The gcoAB-encoded O-demethylase converts guaiacol to catechol, a natural growth substrate for A. baylyi and P. putida that is cleaved by a dioxygenase (CatA) and degraded via the β-ketoadipate pathway (dashed arrow).
As reported here, this method was validated using catabolic studies of guaiacol (2-methoxyphenol), a compound that is readily derived from lignin (12). EASy was used in vivo to improve guaiacol O-demethylation to produce catechol, a key intermediate in aromatic compound catabolism (13). This approach contributes to the emerging use of microbes to transform heterogeneous aromatic compounds derived from lignin to single target products (14–18). The expanded use of aromatics, such as guaiacol, represents an important step in achieving cost-effective microbial conversion of lignin to valuable chemicals (19). The results demonstrate that by inducing targeted gene amplification, EASy enabled selections that might not otherwise be possible.
Results
Identification of gcoAB Genes Predicted to Encode Guaiacol O-Demethylase.
Some bacterial strains of Rhodococcus, Streptomyces, Corynebacterium, and Moraxella can use guaiacol as a carbon source (20–26). This substrate is enzymatically converted to formaldehyde and catechol, which can be further metabolized (Fig. 1B). Although the enzyme responsible for demethylation was partially characterized and used to obtain the N-terminal protein sequence (20–26), the corresponding genes were not identified. With database searches, we discovered gene products in Rhodococcus species that match the 21 N-terminal amino acid sequence previously determined for a guaiacol O-demethylating cytochrome P450 enzyme from Rhodococcus rhodochrous, P450RR1 (21). A 403-amino acid sequence, which we designated GcoA, appears to correspond to the previously studied full-length cytochrome P450 protein. Additional bacterial homologs were identified and, based on convention (27), were classified in the CYP255A family (SI Appendix, Fig. S1 and Table S1).
Each gcoA appears to be cotranscribed with its neighbor, designated gcoB, predicted to encode an oxidoreductase. Most bacterial enzymes in the cytochrome P450 superfamily accept electrons from NAD(P)H using two electron-transfer proteins (28). Sequence analysis of GcoB reveals a type of single-protein partner with three domains. By homology to similar enzymes that function with aromatic ring-hydroxylating dioxygenases (29, 30), we infer that GcoB has an N-terminal 2Fe-2S ferredoxin-like domain, a central flavin-binding domain, and a C-terminal domain that interacts with NAD(P)H (SI Appendix, Fig. S2). In typical cytochrome P450 systems, there is a distinct ferredoxin component.
GcoAB-Mediated Guaiacol O-Demethylase Activity in Vivo.
Putative gcoAB genes from Rhodococcus pyridinivorans AK37, Rhodococcus jostii RHA1, or Amycolatopsis sp. ATCC 39116 were expressed from a plasmid-borne constitutive lac promoter in Pseudomonas putida KT2440. This strain does not naturally metabolize guaiacol, but it can use catechol as a carbon source (13). In minimal medium with glucose and guaiacol, heterologously expressed genes from R. jostii or Amycolatopsis sp. enabled guaiacol to be metabolized (SI Appendix, Fig. S3). However, only gcoAB from Amycolatopsis enabled P. putida to grow on guaiacol as the sole carbon source. In this case, KT2440 expressing gcoAB on pCJ021 completely consumed 6 mM guaiacol (SI Appendix, Fig. S4). Expression of gcoA alone was insufficient for guaiacol metabolism. The requirement for both genes suggests that GcoAB is a two-component cytochrome P450 system with guaiacol O-demethylase activity. Next, to obviate the need for plasmids, Amycolatopsis gcoAB DNA was inserted in the P. putida chromosome. Two strains were made with gcoAB expressed from a constitutive tac promoter. Although integrated in two loci that were used previously to express other genes, neither strain with gcoAB in the chromosome grew on guaiacol as sole carbon source, despite repeated attempts and 200 h of incubation in shake flasks.
Chromosomal Insertion and Increased Dosage of gcoAB in A. baylyi.
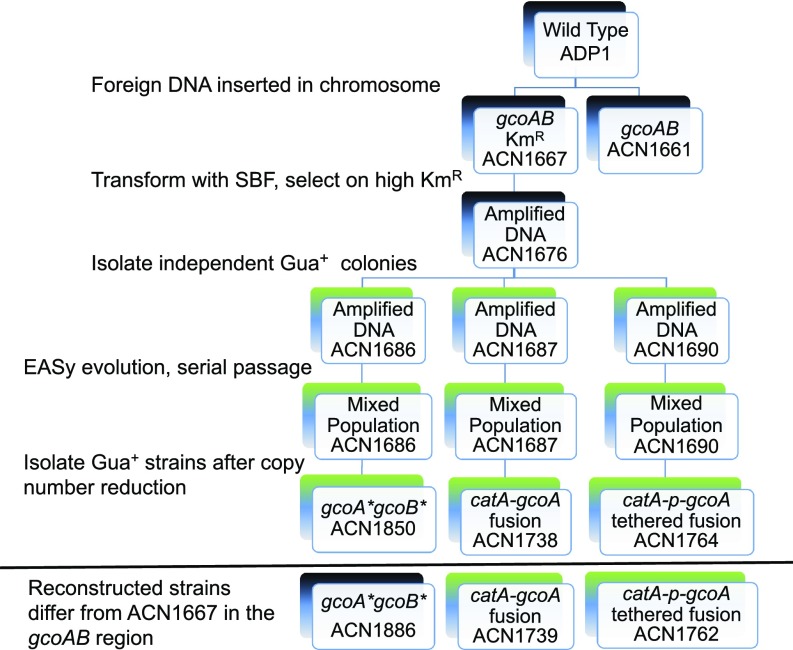
To obtain bacteria that grow on guaiacol without plasmids, we used gene amplification methods in A. baylyi ADP1, a naturally transformable bacterium (9–11, 31). In ADP1, duplication can be induced by transformation with a linear DNA fragment, called a synthetic bridging fragment (SBF), such that the exact endpoints of the duplicated segment (amplicon) are defined (31). Selective growth conditions retain the amplicon and establish its copy number (Fig. 1). In ADP1, the Amycolatopsis gcoAB genes were chromosomally inserted downstream of catA, encoding catechol 1,2-dioxygenase. This insertion, which disrupted an ORF (ACIAD1443) predicted to be cotranscribed with catA, had no discernible effect on growth with carbon sources requiring CatA activity. Insertion of gcoAB yielded ACN1661 and another strain, ACN1667, which has a kanamycin resistance cassette (KmR) downstream of gcoAB to enable selection for increased gene dosage. Strains with one chromosomal copy of gcoAB failed to grow on guaiacol as the carbon source (Gua−). The lineages of these strains and their derivatives are depicted in Fig. 2.
Fig. 2.
A. baylyi mutants. Growth on guaiacol as sole carbon source is indicated in green (Gua+) or black (Gua−). Changes in gcoAB were point mutations (*) or deletions that encode chimeric proteins. CatA-gcoA encodes a protein with most of CatA connected to most of GcoA. CatA-p-gcoA encodes a protein with most of CatA linked to all of GcoA by a short peptide (p). The mutations were introduced in the chromosomes of strains with wild-type backgrounds (reconstructed strains) to test for Gua+ growth.
The chromosomal region encompassing gcoAB and the KmR gene was duplicated by transforming ACN1667 with linearized pBAC1262, used as the SBF (10, 11, 31). Transformants with multiple copies of a 9.7-kbp amplicon were selected with a high Km concentration, 1 mg/mL. One copy of the drug-resistance marker enables resistance to 25 μg/mL Km, and selections on plates with Km concentrations above 150 μg/mL do not yield colonies in the absence of gene amplification. Quantitative PCR (qPCR) analysis showed that one transformant, ACN1676, had five chromosomal copies of the amplicon. However, despite multiple gcoAB copies, ACN1676 did not grow on guaiacol. Direct selection on plates with 1 mM guaiacol as the sole carbon source, without high Km, resulted in ACN1676-derived Gua+ colonies at a frequency of ∼10−6 within several days. In contrast, the same direct selection method failed to produce any Gua+ colonies from ACN1661 or ACN1667, suggesting that gcoAB amplification facilitates adaptation to Gua+ growth. Copy number for three independent Gua+ isolates (ACN1686, ACN1687, and ACN1690), presumably driven by the demand for GcoAB, was determined to be 7–10 amplicons per chromosome.
EASy Culturing.
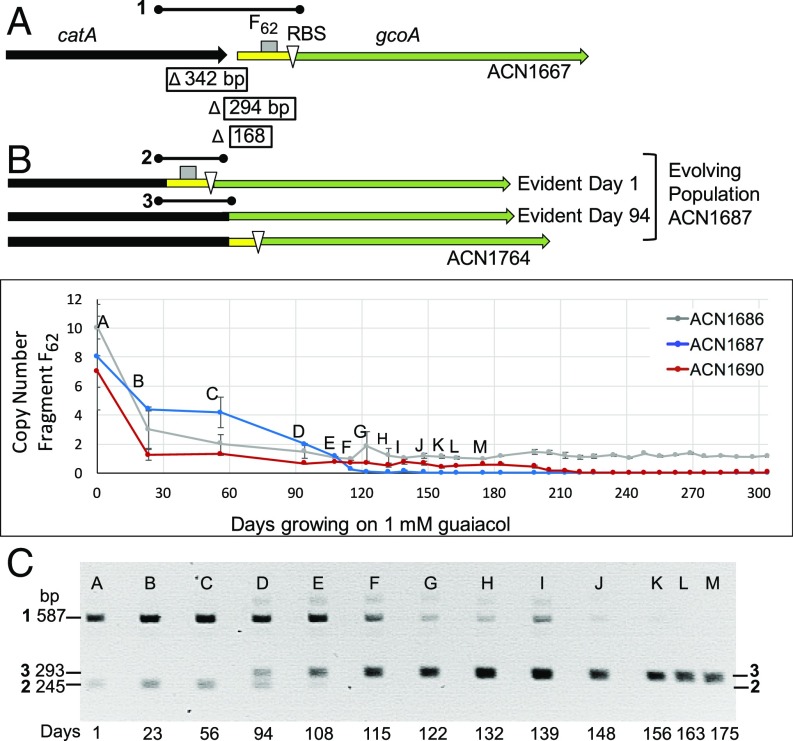
During continued selection for Gua+ growth, gcoAB copies are expected to diverge (5). Beneficial alleles that arise by mutation should reduce or eliminate the demand for multiple copies. To test this model, cultures of ACN1686, ACN1687, and ACN1690 were grown on guaiacol as the only carbon source and diluted (1:50 by volume) each day. Samples from these serial transfers were analyzed by qPCR. Primers were used to detect a 62-bp segment of native DNA in the amplicon adjacent to gcoA. The average copy number of this fragment (F62) decreased as populations of ACN1686, ACN1687, and ACN1690 evolved (Fig. 3). For ACN1686, the average copy number of F62 decreased to one, suggesting that mutation(s) occurred to enable gcoAB-mediated Gua+ growth. For ACN1687 and ACN1690, F62 failed to be detected at later times. This result was unexpected since F62 was intended to serve as a proxy for gcoAB copy number, and at least one copy of these genes should be required for growth on guaiacol. Further assessment, explained below and depicted in SI Appendix, Fig. S5, showed that DNA deletions modified the gco genes and accounted for the failure to detect F62 by qPCR.
Fig. 3.
Evolutionary trajectories. (A) DNA for qPCR analysis (F62) is upstream of gcoA, which was inserted with a ribosome binding site (RBS) to form ACN1667 (SI Appendix, Fig. S5). Deletions of 294 bp or 168 bp arose during EASy to form gene fusions in two evolved strains, ACN1738 and ACN1764, respectively. The deletions remove F62 and prevent detection of this region by our qPCR method. (B) Chromosomal copy number of F62 was monitored by qPCR for samples at different times during serial passage of ACN1686, ACN1687, and ACN1690. (C) PCR was used to amplify genomic DNA of the evolving population of ACN1687. With primers that amplify DNA spanning the 3′ end of catA and the 5′ end of gcoA, a 587-bp fragment 1, corresponding to the parent strain, predominated at the first time point. Letters in B and C correspond to the same samples. The first evidence of a 293-bp fragment was day 94. This fragment 3 reflects a 294-bp deletion in ACN1738. The 245-bp fragment 2 reflects a 342-bp deletion.
Analysis of Mutations Affecting the Gua+ Phenotype.
A Gua+ colony from each evolved population was isolated: ACN1738 (from ACN1687), ACN1764 (from ACN1690), and ACN1850 (from ACN1686) (Fig. 2). Next, efforts were made to capture their gcoAB region on plasmids using the gap-repair method (32). In two cases these efforts were successful, and the resulting plasmids were then used to construct new strains carrying one chromosomal copy of the gcoAB region from the evolved strain (SI Appendix, Fig. S6). The strains that were constructed in this way (ACN1739 and ACN1762; Fig. 2) grew on guaiacol. In both cases, a deletion fused gcoA to its neighbor, catA. ACN1739 (and ACN1738) have a 294-bp deletion that encodes a chimeric CatA-GcoA protein without nine amino acids at the C terminus of CatA and missing three amino acids at the N terminus of GcoA. This deletion removes F62 (Fig. 3). Similarly, a 168-bp deletion in ACN1762 (and ACN1764) encodes CatA tethered to GcoA by a 34-amino acid linker (SI Appendix, Fig. S5). In this fusion, CatA (missing four C-terminal amino acids) is linked to the complete GcoA protein.
In the third Gua+ mutant (ACN1850), whole genome resequencing revealed two point mutations in gcoAB. One nucleotide change (G → A) replaces glycine with aspartate at position 72 in GcoA. The other nucleotide substitution (also G → A) replaces alanine with threonine at position 4 in GcoB. Reconstructed strains were engineered to differ from ACN1667 by either the gcoA mutation (ACN1881), the gcoB mutation (ACN1863), or both (ACN1886). These strains were Gua−, which suggests that other chromosomal mutations in ACN1850 affect the selected phenotype. Mutations outside the gcoAB region were identified, but their relevance to Gua+ growth is not obvious (SI Appendix, Table S6). Nevertheless, the gcoA mutation may confer some benefit for guaiacol growth, because ACN1886 and ACN1881, which both encode GcoA(G72D), gave rise to spontaneous Gua+ colonies. Comparable strains without this mutation did not (ACN1661, ACN1667, and ACN1863).
Evolution of Faster Gua+ Growth and Lower gcoAB Copy Number.
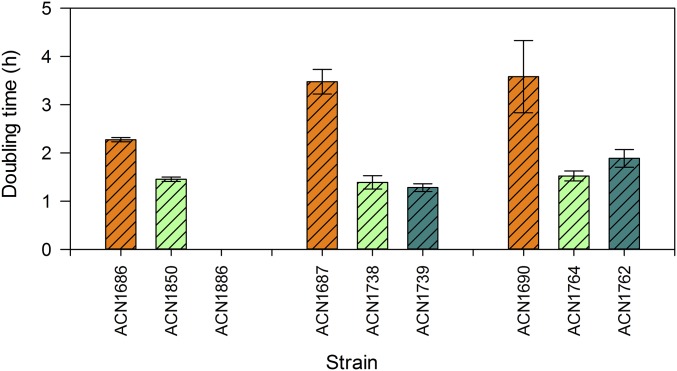
Although ACN1686, ACN1687, and ACN1690 arose from the same Gua− parent (ACN1676; Fig. 2), the populations in initial cultures were not identical. Different Gua+ colonies were chosen and subjected to several rounds of selective restreaking on guaiacol plates before inoculation in liquid culture. Initially, ACN1686 had the highest copy number and grew the fastest on guaiacol, with a doubling time of 2.3 h (Figs. 3 and 4). ACN1687 and ACN1690 grew with 3.5-h doubling times. Evolved isolates (ACN1850, ACN1738, and ACN1764) grew faster than their starting populations with doubling times of ∼1.5 h (Fig. 4). Regardless of other mutations that may be in the evolved strains, each catA-gcoA fusion, in single chromosomal copy, was sufficient for Gua+ growth. Doubling times were 1.3 h and 1.9 h for the reconstructed strains, ACN1739 and ACN1762.
Fig. 4.
Growth of A. baylyi strains on 1 mM guaiacol as the carbon source. Doubling times were determined for populations of ACN1686, ACN1687, and ACN1690 at the start of serial passaging. Each Gua+ isolate is shown adjacent to the parent from which it arose. The rightmost strain in each set is a reconstructed strain (ACN1886, ACN1739, or ACN1762) that has only the changes in catA, gcoA, and gcoB identified in an evolved isolate (ACN1850, ACN1738, and ACN1764). Reconstructed strains do not carry mutations that may have arisen in other chromosomal regions of the evolved Gua+ isolates.
PCR was used to assess the evolution of catA-gcoA fusions (Fig. 3 and SI Appendix, Fig. S7). Primers that generate a 587-bp product for the parent, ACN1667 (fragment 1), generate a 293-bp product (fragment 3) when template DNA carries the catA-gcoA allele of ACN1738 (Fig. 3). PCR analysis of samples from evolving cultures of ACN1687 showed that fragment 1 predominated initially. Over time, its proportion decreased and, concomitantly, the proportion of fragment 3 increased (Fig. 3C). We estimate that fragment 3, first evident at day 94, emerged as the only detectable allele in fewer than 900 generations after the start of serial passaging. A smaller, 245-bp PCR product was observed, purified, and sequenced (fragment 2). This DNA resulted from a 342-bp deletion that removes the 3′ end of catA (286 bp) and generates a catA-p-gcoA tethered fusion. Since this deletion was present on day 1 in liquid culture, it may have been selected during growth of ACN1687 on guaiacol plates. However, this allele alone and in single copy did not confer Gua+ growth in an engineered strain, ACN1887. Once the allele corresponding to fragment 3 emerged, it outcompeted that corresponding to fragment 2.
In similar studies of ACN1690, the first sample contained detectable levels of DNA corresponding to the allele that ultimately emerged in single copy in the evolved isolate, ACN1764 (SI Appendix, Fig. S7). The other evident PCR product corresponded to the parent-strain configuration. In this case, a uniform population where most cells appear to have one chromosomal copy of the gene fusion evolved in ∼1,000 generations.
CatA-GcoA Function in Vivo.
The EASy results raise the possibility that genes evolved in A. baylyi may be useful in other organisms or, in terms of synthetic biology, in different chassis. To test this possibility, a catA-gcoA fusion analogous to that of ACN1738 was integrated, with gcoB, into the P. putida KT2440 chromosome. In this location, the native gcoAB configuration does not enable growth on guaiacol. The resulting strain grew on guaiacol as the sole carbon source after a lag (SI Appendix, Fig. S8). Furthermore, there was no effect on Gua+ growth when a gene encoding a second catechol dioxygenase in P. putida KT2440 was deleted. As in A. baylyi, which has a single catA, the catechol-cleaving activity of the fusion protein in P. putida is sufficient for Gua+ growth (SI Appendix, Fig. S8).
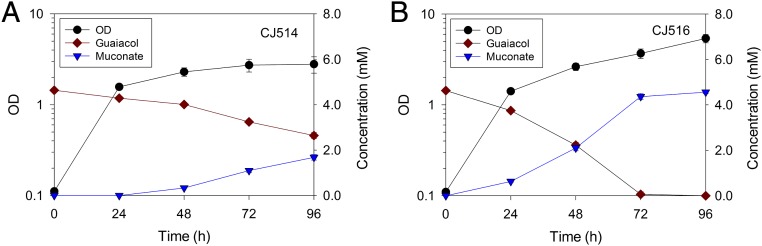
We next sought to compare the rate of guaiacol consumption in vivo mediated by the CatA-GcoA fusion protein relative to that of CatA and GcoA when both proteins are coexpressed but not fused. For this comparison, guaiacol was not used as the sole carbon source, since coexpression of CatA and GcoA, in the absence of their fusion, yield a Gua− phenotype (reported above). Instead, we constructed strains that would accumulate the CatA product, muconate, by deleting the catBC genes required for its further metabolism (33). In these strains, the rates for producing muconate from guaiacol should reflect enzymatic conversion mediated by CatA and GcoA (strain CJ514) or the CatA-GcoA fusion (CJ516) (Fig. 5). During the first 72 h, the strain encoding the CatA-GcoA chimera produced muconate at an average rate ∼3.5 times faster than the strain with separate CatA and GcoA proteins. Thus, combined O-demethylation and ring fission appear to be catalyzed more rapidly by the chimeric enzyme than by the individual proteins.
Fig. 5.
Metabolism of guaiacol (5 mM) by P. putida strains growing on glucose. (A) In CJ514, catA and gcoA are distinct. (B) CJ516 has a catA-gcoA gene fusion analagous to that of A. baylyi ACN1738. Cultures were fed 20 mM glucose at 0, 24, 48, and 72 h. Growth was assessed by OD at 600 nm. Data points represent averages of three biological replicates, and error bars indicate the SDs.
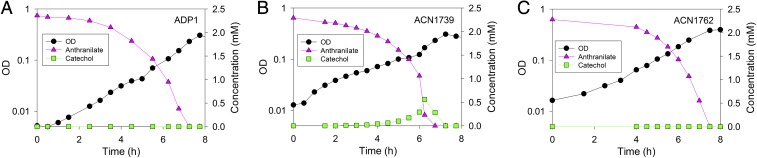
The selective advantage conferred by CatA-GcoA may derive from metabolite channeling. Catechol, generated from guaiacol within this chimeric protein, appears to be cleaved efficiently. We also studied the fate of catechol generated from a compound other than guaiacol. With anthranilate (2-aminobenzoate) as the carbon source, catechol is produced by a dioxygenase encoded by antABC, genes distant from catA on the chromosome (29). Catechol accumulated transiently when ACN1739, but not ADP1, was grown on anthranilate (Fig. 6 A and B). Thus, access of catechol to the active cleavage site in this chimeric enzyme may be improved when catechol is produced from guaiacol but inhibited when produced external to this enzyme.
Fig. 6.
Metabolism of anthranilate as the carbon source by A. baylyi strains. (A) The wild-type (ADP1) has no gco genes. (B) In ACN1739, catechol is cleaved by the CatA moiety of a chimeric protein encoded by a gene fusion (catA-gcoA51738). (C) In ACN1762, a different allele (catA-p-gcoA5164) encodes a tethered fusion protein. Growth was assessed by OD at 600 nm. Graphs display representative results that were obtained in at least three independent experiments.
Although ACN1762 also has a gene fusion, catechol is not detected during growth on anthranilate (Fig. 6C). Thus, catechol accumulation by ACN1739 is not likely due to transcriptional changes resulting merely from the chromosomal insertion of gcoAB. Moreover, different results for ACN1739 and ACN1762 may arise from structural differences in the two fused proteins, one that connects CatA directly to GcoA (in ACN1739) and the other that tethers the two domains (in ACN1762).
Discussion
Evolutionary models highlight naturally dynamic and significant, albeit transient, increases in gene dosage (5, 6). Here, we demonstrate that synthetically engineered increases in regional chromosomal copy number can generate variant enzymes. Using EASy, mutants emerged within ∼1,000 generations. Moreover, the ADP1 genetic system enables variations on the technique in which mutated DNA could be added to growing cultures to effect targeted genomic modification (9).
Traditional evolutionary methods using direct selection on one carbon source were not possible without first increasing gene dosage, since the single-copy gcoAB did not confer growth on guaiacol. Other effective methods (34, 35), which provide alternative carbon sources during adaptation to novel growth substrates, might have yielded Gua+ mutants. However, establishing the conditions to enable mutant isolation by such means can be difficult and may require previous knowledge of metabolic regulation (35). Periods of nonselective growth allow the loss of rare mutations, including potentially beneficial gene duplications (36). Thus, the initial establishment of a multicopy genetic array should prevent some issues that delay the emergence of novel phenotypes during adaptive evolution using other methods.
Whether the similar creation of genetic arrays would enable EASy-type methods to work in other model organisms remains to be tested. Duplicated chromosomal regions in bacteria such as Salmonella enterica or Escherichia coli could be generated by an alternative to SBF-mediated transformation and recombination (e.g., transduction). The possibility that this approach could succeed is supported by studies in S. enterica where rapid evolution occurred when the target gene was placed in a region with the propensity for frequent duplication and amplification (8).
GcoAB: A Two-Component Cytochrome P450 O-Demethylase.
The importance of guaiacol metabolism in microbial lignin valorization motivated these studies (19). Database homology searches enabled us to evaluate genes we designated gco (for guaiacol utilization). It is not immediately evident why plasmid-borne Amycolatopsis DNA enabled P. putida to grow on guaiacol as the sole carbon source while the Rhodococcus genes did not, since the corresponding amino acid sequences are highly conserved (SI Appendix, Fig. S1). Differences in enzyme activity, protein stability, and/or heterologous gene expression were not investigated. Members of the large and diverse cytochrome P450 superfamily use molecular oxygen to mediate diverse reactions such as hydroxylation, epoxidation, and dealkylation (37). These enzymes, which are also regioselective and stereospecific, are targets of extensive metabolic engineering efforts (38). The GcoAB sequences, and their EASy-derived variants, make additional applications possible.
GcoB, the reductase for GcoA, is unique among known electron-transfer components of cytochrome P450 systems. GcoB is homologous to three-domain enzymes that transfer electrons from NADH to aromatic-ring hydroxylating dioxygenases, such as benzoate 1,2-dioxygenase (BenC) and anthranilate 1,2-dioxygenase (AntC) (29, 30). GcoB has an N-terminal ferredoxin domain, a central domain that binds FAD, and a C-terminal domain that interacts with NAD(P)H (SI Appendix, Fig. S2). Typically, prokaryotic P450 enzymes interact with two redox partners. Despite many known variations in these components (39), a reductase with the domain structure of GcoB has not previously been reported to interact with a cytochrome P450 enzyme. Characterization of GcoAB, including its atomic-level structure and substrate specificity, are reported in a companion paper (40).
EASy-Derived GcoAB Variants.
The EASy-derived proteins would not have been generated by rational design. The placement of gcoA adjacent to catA was intended to facilitate cotranscription and allow proximal protein synthesis. However, the design of chimeras to improve functionality in guaiacol metabolism is a strategy with no foundation in past discoveries and would not have been attempted. Different fusion proteins appear to vary in catalytic properties, as indicated by catechol accumulation in ACN1739 but not ACN1762 (Fig. 6). Similar fusions emerged several times in additional studies, consistent with the known propensity of amplified DNA to be remodeled by deletion (41). These CatA-GcoA fusions demonstrate the ability of EASy to reveal innovative solutions in response to selective pressure. Furthermore, the successful chromosomal expression of one chimera in P. putida underscores the potential to use this evolutionary method in A. baylyi to generate enzymes for heterologous expression in other organisms.
Point mutations were also isolated. The GcoA(G72D) variant appears to be beneficial although additional mutations are needed for Gua+ growth. Residues at this position in homologs are not highly conserved or known to play a role in catalysis. However, this change provided sufficient benefit to enable spontaneous Gua+ mutants to be isolated from strains with this mutation. One of these arose by spontaneous gene amplification encompassing gcoAB, a result that further emphasizes the importance of gene amplification. An appreciation for this process continues to grow as evolutionary outcomes are documented in diverse contexts (6, 8, 42, 43). The EASy methodology both accelerates evolution in the laboratory and expands options for phenotypic selection.
Methods
Strains, Plasmids, and Growth Conditions.
E. coli hosts DH5α and TOP10 (Invitrogen) were grown in Luria–Bertani (LB) medium (44). NEB 5-alpha F'Iq E. coli (New England Biolabs) was used for hosting plasmids with the tac promoter. A. baylyi strains (SI Appendix, Table S2) (45) were grown, in LB or minimal medium at 37 °C with aeration (11). Unless noted, carbon sources were provided as follows: 20 mM pyruvate, 1 mM guaiacol (Sigma-Aldrich), 2 mM benzoate, or 2.5 mM anthranilate. When used, the final concentration of Km was 25 μg/mL (standard) or 1 mg/mL (high) to select increased gene dosage. When used, the final ampicillin (Ap) concentration was 150 μg/mL. P. putida strains were derived from KT2440 (ATCC 47054) and grown and engineered as previously described (17).
Chromosomal Modification of A. baylyi.
Recipients were transformed with linearized plasmids (10, 46). Transformants were selected in which the linear DNA replaced the homologous chromosomal region. Selection typically relied on drug resistance or loss of the sacB marker in the presence of 10% sucrose. Resulting strains were confirmed by PCR, localized DNA sequencing, and, in some cases, whole genome resequencing. Further information is in SI Appendix, Tables S2–S4 and in SI Appendix, Fig. S5.
Copy Number Analysis in A. baylyi.
An engineered fragment (SBF, linearized pBAC1262) was used as a donor to increase gene dosage, as described (10, 11, 31). A qPCR method was used to assess copy number (11). Primers for qPCR are listed in SI Appendix, Table S3. Standard curves used genomic DNA from ACN1667 for both rpoA and F62 (or other fragments). The ratio of calculated concentration for F62 (or other fragments) relative to rpoA was presumed to equal the copy number of that fragment.
Selection and Evolution of Gua+ A. baylyi Strains.
Growth on guaiacol was defined by colony formation on plates and by ability to reach an OD600 of at least 0.15 in liquid culture after being transferred (1:50) three times with 1 mM guaiacol as carbon source. With guaiacol, cells were grown at 30 °C. Amplification mutants (ACN1686, ACN1687, and ACN1690) were grown with 1 mM guaiacol as the carbon source in batch cultures. After reaching stationary phase, serial transfers were conducted on a daily basis, diluting 1:50 in fresh medium. Weekly, genomic DNA was isolated from evolving populations and analyzed by PCR for gcoAB and the SBF junction. Average growth rates led to ∼5 generations per day for the evolving populations.
DNA Recovery by the Gap-Repair Method (32).
To recover chromosomal regions from Gua+ strains, cells from cultures in midlog phase were mixed with pBAC1282 digested with EcoRV and XbaI (SI Appendix, Table S3) and plated on LB medium. After 6–8 h, transformants with circular plasmids, generated via homologous recombination (SI Appendix, Fig. S6), were selected moving cells scraped from the plate to selective medium with Ap and Km. After overnight growth, drug-resistant colonies were pooled. Plasmid DNA was extracted and introduced into E. coli for maintenance. Plasmids were analyzed and sequenced (Eton Bioscience or Genewiz).
Whole Genome Sequencing.
Approximately 1 µg of genomic DNA from ACN1667 and ACN1850 was fragmented by sonication to an average size of 300–500 bp. End repair, A-tailing, and adapter ligation reactions were performed on the fragmented DNA using the NEBNext Ultra II kit (New England Biolabs). Illumina paired-end sequencing was performed on a NextSeq500 device at the Georgia Genomics Facility (University of Georgia). Sequence analysis and variant calling was done using Geneious version 8.1 with default settings (47). To identify single-nucleotide polymorphisms, the minimum variant frequency was set to 0.8. A reference genome was constructed by mapping ACN1667 whole genome sequencing reads to the ADP1 genome (National Center for Biotechnology Information accession no. NC_005966).
Metabolite Monitoring.
For A. baylyi, 1-mL samples of batch cultures were centrifuged to pellet cells. Supernatant was filtered through a 0.45-μm pore-size nylon syringe filter (Membrane Solutions). A 10-μL sample of the filtrate was analyzed on a 4.6 × 250 mm C18 reverse-phase high-performance liquid chromatography (HPLC) column (Bio-Rad Laboratories). Elution at a rate of 0.7 mL/min isocratic flow was carried out with 30% acetonitrile and 0.1% phosphoric acid. The eluant was monitored by UV detection at 210 nm (Shimadzu LC10A pump with degasser, SIL-20A autosampler, and an SPD-20A detector). The retention times for anthranilate and catechol were 6.5 and 4.8 min, respectively. Peak areas were calculated with the EZ Start Version 7.4 SP1 software package (Shimadzu). Analysis in P. putida cultures was performed as described (48). The retention times for muconate and guaiacol were 4.3 and 12.0 min, respectively.
Supplementary Material
Acknowledgments
We thank David Nelson for assistance in classifying the P450 guaiacol O-demethylases in the CY255A family and Jim C. Spain for discussions of the method. Research was funded by NSF Grants MCB-1361188 and MCB-1615365 (to E.L.N.) and DEB-1556541 (to E.L.N. and M.A.E.). The US Department of Energy (DOE), Energy Efficiency and Renewable Energy, Bioenergy Technologies Office also funded this work via Contract DE-AC36-08GO28308 with the National Renewable Energy Laboratory and its subcontract, XFC-7-70006-01 to the University of Georgia. The US Government retains and the publisher, by accepting the article for publication, acknowledges that the US Government retains a nonexclusive, paid up, irrevocable, worldwide license to publish or reproduce the published form of this work, or allow others to do so, for US Government purposes.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803745115/-/DCSupplemental.
References
- 1.Conrad TM, Lewis NE, Palsson BO. Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol. 2011;7:509. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dragosits M, Mattanovich D. Adaptive laboratory evolution–Principles and applications for biotechnology. Microb Cell Fact. 2013;12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaCroix RA, Palsson BO, Feist AM. A model for designing adaptive laboratory evolution experiments. Appl Environ Microbiol. 2017;83:e03115–e03116. doi: 10.1128/AEM.03115-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno S. Evolution by Gene Duplication. Springer; New York: 1970. [Google Scholar]
- 5.Bergthorsson U, Andersson DI, Roth JR. Ohno’s dilemma: Evolution of new genes under continuous selection. Proc Natl Acad Sci USA. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott KT, Cuff LE, Neidle EL. Copy number change: Evolving views on gene amplification. Future Microbiol. 2013;8:887–899. doi: 10.2217/fmb.13.53. [DOI] [PubMed] [Google Scholar]
- 7.Katju V, Bergthorsson U. Copy-number changes in evolution: Rates, fitness effects and adaptive significance. Front Genet. 2013;4:273. doi: 10.3389/fgene.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Näsvall J, Sun L, Roth JR, Andersson DI. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338:384–387. doi: 10.1126/science.1226521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott KT, Neidle EL. Acinetobacter baylyi ADP1: Transforming the choice of model organism. IUBMB Life. 2011;63:1075–1080. doi: 10.1002/iub.530. [DOI] [PubMed] [Google Scholar]
- 10.Seaton SC, et al. Genome-wide selection for increased copy number in Acinetobacter baylyi ADP1: Locus and context-dependent variation in gene amplification. Mol Microbiol. 2012;83:520–535. doi: 10.1111/j.1365-2958.2011.07945.x. [DOI] [PubMed] [Google Scholar]
- 11.Stoudenmire JL, et al. Malonate degradation in Acinetobacter baylyi ADP1: Operon organization and regulation by MdcR. Microbiology. 2017;163:789–803. doi: 10.1099/mic.0.000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragauskas AJ, et al. Lignin valorization: Improving lignin processing in the biorefinery. Science. 2014;344:1246843. doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs G, Boll M, Heider J. Microbial degradation of aromatic compounds–From one strategy to four. Nat Rev Microbiol. 2011;9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 14.Linger JG, et al. Lignin valorization through integrated biological funneling and chemical catalysis. Proc Natl Acad Sci USA. 2014;111:12013–12018. doi: 10.1073/pnas.1410657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vardon DR, et al. Adipic acid production from lignin. Energy Environ Sci. 2015;8:617–628. [Google Scholar]
- 16.Salvachúa D, Karp EM, Nimlos CT, Vardon DR, Beckham GT. Towards lignin consolidated bioprocessing: Simultaneous lignin depolymerization and product generation by bacteria. Green Chem. 2015;17:4951–4967. [Google Scholar]
- 17.Johnson CW, Beckham GT. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab Eng. 2015;28:240–247. doi: 10.1016/j.ymben.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Mycroft Z, Gomis M, Mines P, Law P, Bugg TD. Biocatalytic conversion of lignin to aromatic dicarboxylic acids in Rhodococcus jostii RHA1 by re-routing aromatic degradation pathways. Green Chem. 2015;17:4974–4979. [Google Scholar]
- 19.Beckham GT, Johnson CW, Karp EM, Salvachúa D, Vardon DR. Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol. 2016;42:40–53. doi: 10.1016/j.copbio.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 20.Dardas A, et al. The demethylation of guaiacol by a new bacterial cytochrome P-450. Arch Biochem Biophys. 1985;236:585–592. doi: 10.1016/0003-9861(85)90662-9. [DOI] [PubMed] [Google Scholar]
- 21.Eltis LD, Karlson U, Timmis KN. Purification and characterization of cytochrome P450RR1 from Rhodococcus rhodochrous. Eur J Biochem. 1993;213:211–216. doi: 10.1111/j.1432-1033.1993.tb17750.x. [DOI] [PubMed] [Google Scholar]
- 22.Karlson U, et al. Two independently regulated cytochromes P-450 in a Rhodococcus rhodochrous strain that degrades 2-ethoxyphenol and 4-methoxybenzoate. J Bacteriol. 1993;175:1467–1474. doi: 10.1128/jb.175.5.1467-1474.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawahara N, et al. Purification and characterization of 2-ethoxyphenol-induced cytochrome P450 from Corynebacterium sp. strain EP1. Can J Microbiol. 1999;45:833–839. [PubMed] [Google Scholar]
- 24.Sauret-Ignazi G, Dardas A, Pelmont J. Purification and properties of cytochrome P-450 from Moraxella sp. Biochimie. 1988;70:1385–1395. [PubMed] [Google Scholar]
- 25.Sutherland JB. Demethylation of veratrole by cytochrome P-450 in Streptomyces setonii. Appl Environ Microbiol. 1986;52:98–100. doi: 10.1128/aem.52.1.98-100.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YH, Engesser KH, Kim SJ. Physiological, numerical and molecular characterization of alkyl ether-utilizing rhodococci. Environ Microbiol. 2007;9:1497–1510. doi: 10.1111/j.1462-2920.2007.01269.x. [DOI] [PubMed] [Google Scholar]
- 27.Nelson DR. Cytochrome P450 nomenclature, 2004. Methods Mol Biol. 2006;320:1–10. doi: 10.1385/1-59259-998-2:1. [DOI] [PubMed] [Google Scholar]
- 28.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems–Biological variations of electron transport chains. Biochim Biophys Acta. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Eby DM, Beharry ZM, Coulter ED, Kurtz DM, Jr, Neidle EL. Characterization and evolution of anthranilate 1,2-dioxygenase from Acinetobacter sp. strain ADP1. J Bacteriol. 2001;183:109–118. doi: 10.1128/JB.183-1.109-118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlsson A, et al. X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1. J Mol Biol. 2002;318:261–272. doi: 10.1016/S0022-2836(02)00039-6. [DOI] [PubMed] [Google Scholar]
- 31.Reams AB, Neidle EL. Genome plasticity in Acinetobacter: New degradative capabilities acquired by the spontaneous amplification of large chromosomal segments. Mol Microbiol. 2003;47:1291–1304. doi: 10.1046/j.1365-2958.2003.03342.x. [DOI] [PubMed] [Google Scholar]
- 32.Gregg-Jolly LA, Ornston LN. Recovery of DNA from the Acinetobacter calcoaceticus chromosome by gap repair. J Bacteriol. 1990;172:6169–6172. doi: 10.1128/jb.172.10.6169-6172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanks BH, Keeling PL. Bioprivileged molecules: Creating value from biomass. Green Chem. 2017;19:3177–3185. [Google Scholar]
- 34.Lee DH, Palsson BO. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a Nonnative carbon source, L-1,2-propanediol. Appl Environ Microbiol. 2010;76:4158–4168. doi: 10.1128/AEM.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Hofwegen DJ, Hovde CJ, Minnich SA. Rapid evolution of citrate utilization by Escherichia coli by direct selection requires citT and dctA. J Bacteriol. 2016;198:1022–1034. doi: 10.1128/JB.00831-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth JR, Maisnier-Patin S. Reinterpreting long-term evolution experiments: Is delayed adaptation an example of historical contingency or a consequence of intermittent selection? J Bacteriol. 2016;198:1009–1012. doi: 10.1128/JB.00110-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLean JK, Leys D, Munro WA. Microbial cytochromes P450. In: Ortiz de Montellano RP, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. Springer International Publishing; Cham, Switzerland: 2015. pp. 261–407. [Google Scholar]
- 38.McIntosh JA, Farwell CC, Arnold FH. Expanding P450 catalytic reaction space through evolution and engineering. Curr Opin Chem Biol. 2014;19:126–134. doi: 10.1016/j.cbpa.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLean KJ, Luciakova D, Belcher J, Tee KL, Munro AW. Monooxygenase, Peroxidase and Peroxygenase Properties and Mechanisms of Cytochrome P450. Springer; Cham, Switzerland: 2015. Biological diversity of cytochrome P450 redox partner systems; pp. 299–317. [DOI] [PubMed] [Google Scholar]
- 40.Mallinson SJB, et al. A promiscuous cytochrome P450 aromatic O-demethylase for lignin conversion. Nat Commun. 2018 doi: 10.1038/s41467-018-04878-2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reams AB, Roth JR. Mechanisms of gene duplication and amplification. Cold Spring Harb Perspect Biol. 2015;7:a016592. doi: 10.1101/cshperspect.a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toussaint JP, et al. Gene duplication in Pseudomonas aeruginosa improves growth on adenosine. J Bacteriol. 2017;199:e00261-17. doi: 10.1128/JB.00261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dormeyer M, et al. Hierarchical mutational events compensate for glutamate auxotrophy of a Bacillus subtilis gltC mutant. Environ Microbiol Rep. 2017;9:279–289. doi: 10.1111/1758-2229.12531. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Lab Press; New York: 1989. [Google Scholar]
- 45.Vaneechoutte M, et al. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl Environ Microbiol. 2006;72:932–936. doi: 10.1128/AEM.72.1.932-936.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClung DJ, et al. Novel heterologous bacterial system reveals enhanced susceptibility to DNA damage mediated by yqgF, a nearly ubiquitous and often essential gene. Microbiology. 2016;162:1808–1821. doi: 10.1099/mic.0.000355. [DOI] [PubMed] [Google Scholar]
- 47.Kearse M, et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson CW, et al. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab Eng Commun. 2016;3:111–119. doi: 10.1016/j.meteno.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.