Summary
The surface of mammalian bodies is colonized by a multitude of microbial organisms, which under normal conditions support the host and are considered beneficial commensals. This requires, however, that the composition of the commensal microbiota is tightly controlled and regulated. The host immune system plays an important role in the maintenance of this microbiota composition. Here we focus on the contribution of one particular immune cell type, invariant Natural Killer T (i NKT) cells, in this process. The iNKT cells are a unique subset of T cells characterized by two main features. First, they express an invariant T‐cell receptor that recognizes glycolipid antigens presented by CD1d, a non‐polymorphic major histocompatibility complex class I‐like molecule. Second, i NKT cells develop as effector/memory cells and swiftly exert effector functions, like cytokine production and cytotoxicity, after activation. We outline the influence that the mucosal microbiota can have on i NKT cells, and how i NKT cells contribute to the maintenance of the microbiota composition.
Keywords: microbiota, mucosal immunology, invariant Natural Killer T cells
Abbreviations
- GF
germ‐free
- HDE
house dust extracts
- IEL
intraepithelial lymphocyte
- iNKT cells
invariant Natural Killer T cells
- RF
restricted flora
- TCR
T‐cell receptor
- αGalCer
α‐galactosyl‐ceramide
Introduction
The bodies of animals are exposed to the environment on the skin and the mucosal surfaces, which have to reconcile two often conflicting aims: to support the exchange of the host with the environment and to protect the host from pathogenic colonization and invasion. Whereas the skin can afford a thick, multilayered barrier, the functional characteristics of the mucosal surfaces, mainly the airways, and the gastrointestinal and urogenital tracts, require at times a single‐cell layer of separation between the host and the environment. These surfaces are colonized by a diverse mixture of microbial organisms, consisting of bacteria, fungi and viruses. Each of the mucosal surfaces hosts a unique microbiome, often with additional niches depending on the particularities of the location. Importantly, the interaction between the host and the microbiota has coevolved in such a manner that it is, under normal conditions, mutually beneficial, and the commensal microbiota contributes to the health of the host, not only at the mucosa, but also systemically. Furthermore, the composition of the commensal microbiota is not random, but is maintained by a complex, location‐specific and bi‐directional communication between the host and its commensals.1, 2, 3 The immune system aims to protect the host against pathogens, and therefore, it is not surprising that it plays an important role in maintaining the truce with the commensal microbiota (Box 1). To this end, the mucosal immune system, composed of cells of haematopoietic origin, interacts closely with various tissue‐resident cell types, such as epithelial cells, microfold cells, and Paneth cells (Box 1).
Box 1. Means of interaction between the immune system and the commensal microbiota .
The commensal microbiota influences the health of the host in many ways, and the host actively shapes the composition of the commensal microbiota. This mutual interaction relies on various means of communication and influence. We outline here shortly those means pertaining the immune system and give examples for invariant Natural Killer T (iNKT) cells were possible.
(A) Means of the microbiota to influence the host immune system:
Three main ways have been described by which the commensal microbiota impacts the host immune system.
(1) Antigens
Microbially derived molecules are detected by the adaptive immune system when they can act as antigens by binding either to the B‐cell receptor (BCR) of B cells (direct binding) or the T‐cell receptor (TCR) of T cells (after processing by antigen‐presenting cells (APCs) and presentation on major histocompatibility complex type molecules). This is the most direct way by which the microbiota can activate the adaptive immune system. The organisms known to carry specific antigens for iNKT cell are discussed in the text and in Table 1. As iNKT cells have some degree of auto‐reactivity,121 there exists another antigen‐dependent method for iNKT cell activation. The microbially derived signal might alter the expression of self‐antigens presented by CD1d on APCs97, 98 or change the expression levels of CD1d itself.97, 122
(2) PAMPs
Microbially derived molecules are sensed similar to pathogen‐associated molecular patterns (PAMPs) by the innate immune system through pattern‐recognition receptors (PRRs). For example, the polysaccharide A from Bacteroides fragilis and the exopolysaccharide from Bifidobacterium longum both can expand regulatory T (Treg) cells in vivo by inducing interleukin‐10 (IL‐10) production in APCs.123, 124 In some cases, receptors for IgA can facilitate the uptake of bacteria.125 Cytokines produced by PAMP‐activated APCs can also stimulate cytokine production by iNKT cells (see text and Figure 1).
(3) Metabolites
Microbially derived molecules that influence metabolic and immunological processes in the host. Short‐chain fatty acids (SCFAs), like acetate, propionate, butyrate; biogenic amines, like taurine and histamine; tryptophan catabolites, are all known to modulate metabolic processes in APCs and thereby the host immune response.123, 124 However, we are not aware of data that link these metabolites to iNKT cells.
(B) Means of the host immune system to influence the microbiota
The host immune system can impact the composition of the commensal microbiota in several ways:
(1) Anti‐microbial peptides
Many anti‐microbial peptides are produced directly by immune cells or are induced by them in, for example, epithelial cells via messenger molecules like cytokines. With regard to iNKT cells, it was shown that iNKT cells, via the production of interferon‐γ, can regulate the production of anti‐microbial peptides by Paneth cells.68
(2) Secretory IgA molecules
The majority of the antibodies produced in mammals are of the secretory IgA (sIgA) type that are transported across the endothelial and epithelial barriers of the mucosal surfaces. For example, around three‐quarters of the commensal bacteria in the mouse gastrointestinal tract are coated with sIgA, which is essential for the maintenance of the bacterial homeostasis in the intestine.125 CD1d‐deficient mice show changes in their IgA‐repertoire compared with wild‐type mice, but it was suggested that this is not due to a direct interaction with iNKT cells, but rather a consequence of the altered microbial flora in the CD1d−/− mice.46 However, it was reported that human iNKT cells could stimulate IgA and IgG secretion by B cells in vitro even in the absence of exogenous iNKT cell antigens.126
(3) Mucus production and glycosylation
The glycosylation of the epithelial cells and the production of mucus by goblet cells and enterocytes are important physical defence mechanisms of the mucosal surfaces.127, 128 For example, the glycosylation of intestinal epithelial cells can be regulated by dendritic cells and innate lymphoid cells (ILC3s). With regard to iNKT cells, it was suggested that iNKT cell‐derived interleukin‐13 (IL‐13) regulates goblet cell homeostasis.129
(4) CD1d‐retrograte signalling
Interaction of the invariant T‐cell receptor (iTCR) of iNKT cells with CD1d not only can activate iNKT cells but can also influence the CD1d‐expressing cells due to CD1d‐retrograde signalling. This has been reported for APC (leading to IL‐12 production);130, 131 epithelial cells (either IL‐12 and IL‐15,94 or IL‐10132, 133); cancer cell lines (IL‐12);134 and ILC3s (IL‐22).135 In the context of the mucosa, pro‐inflammatory responses, IL‐22 by ILC3s,135 and anti‐inflammatory responses, IL‐10 by intestinal epithelial cells,132, 133 have been reported for such CD1d‐retrograte signalling.
Invariant Natural Killer T (iNKT) cells are a unique subset of T lymphocytes that phenotypically and functionally resemble Natural Killer cells as well as memory T cells.4, 5, 6, 7, 8, 9 They are characterized by the expression of a canonical Vα14 to Jα18 T‐cell receptor (TCR) rearrangement (Vα14i) in mice and an orthologous Vα24‐Jα18 TCR chain (Vα24i) in humans. iNKT cells recognize glycolipids, especially glycosphingolipid structures, presented by CD1d, a non‐polymorphic homologue of the major histocompatibility complex class I antigen‐presenting molecules. The first and best‐studied TCR agonist for mouse and human iNKT cells is α‐galactosylceramide (αGalCer), a bacteria‐derived glycolipid chemical optimized to yield its exceptionally strong agonistic potential. The iNKT cells develop as effector/memory cells and, following TCR stimulation, they rapidly produce copious amounts of various cytokines and display strong cytotoxicity.10 Due to their cytokine production iNKT cells can impact a wide variety of different chronic and acute immune processes, ranging from responses to pathogens and tumours, to autoimmune responses.
Furthermore, iNKT cells are heterogeneous and based on significant biases in cytokine production and the expression of particular transcription factors, several subsets have been described. Some, like NKT1,11 NKT2,11 NKT1012, 13 and NKT1714, 15, 16, 17, 18 cells, develop in the thymus, while others, such as NKTFH 19, 20 and FoxP3+ iNKT21 cells seem to arise or, like NKT10 cells,12, 22, 23 greatly expand after immunization. For some of these subsets a preferential distribution to various organs has been described.12, 24
The TCR‐mediated activation of iNKT cells is called the direct or antigen‐dependent activation.4, 5, 6, 7, 8, 9 Similar to memory T cells, iNKT cells can also be activated in a TCR‐independent manner by cytokines alone. This indirect or antigen‐independent activation can be induced by several pro‐inflammatory cytokines alone or in combination. For some cytokine combinations a preferential activation of a particular iNKT cell subset has been suggested (Fig. 1). However, the direct and indirect activation pathways are not exclusive as cytokines can augment CD1d‐dependent activation too. This synergistic pathway for iNKT cell activation seems particularly critical for stimulation with weak antigens, which, importantly, can be either exogenous (foreign) or host‐derived (self) antigens. Furthermore, surface co‐stimulatory and co‐inhibitory molecules have been suggested to modulate iNKT cell responses.25
Figure 1.
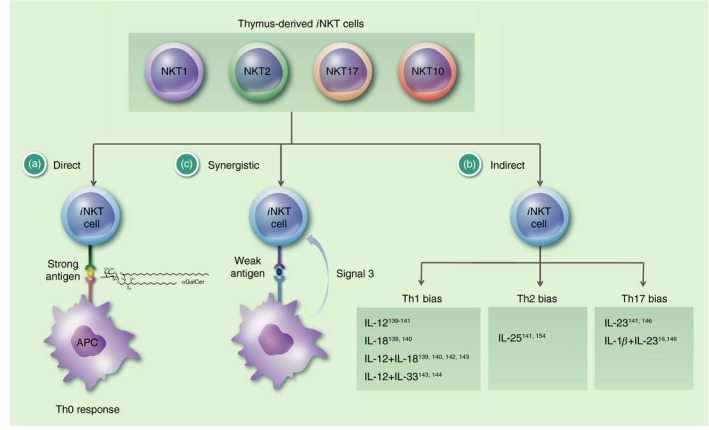
The route of invariant Natural Killer T (i NKT) cell activation impacts their effector function. For i NKT cells in unchallenged control mice, i.e. thymus‐derived i NKT cells, three routes of activation have been described. (a) In the direct or antigen‐dependent activation, a strong antigen, usually of foreign origin, binds to CD1d and activates i NKT cells via their semi‐invariant T‐cell receptor (TCR). αGalCer is depicted as an example. This direct activation leads to the production of both T helper type 1 (Th1) cytokines, like interferon‐γ (IFN‐γ) and tumour necrosis factor, and Th2 cytokines, like interleukin‐4 (IL‐4) and IL‐13, by the i NKT cells, which is sometimes referred to as a Th0 response. (b) The indirect or antigen‐independent activation does not require an engagement of TCR, but rather is achieved by the exposure of the i NKT cells to several pro‐inflammatory cytokines alone or in combination. Some cytokines could induce a preferential production of particular cytokines by the i NKT cells, leading to a Th1‐bias (more IFN‐γ and/or less IL‐4), Th2‐bias (more IL‐4, IL‐13 and/or less IFN‐γ), or Th17‐bias (more IL‐17A). The available data indicate a preferential activation of NKT17 cells in the case of the Th17‐bias. However, in the case of the Th1‐ and Th2‐biases it seems more likely that it is the result of a modulation of the i NKT cell cytokine response. (c) In the synergistic pathway the stimulation of i NKT cells depends both on the TCR and on cytokine receptors. In these cases, the stimulation provided by a weak antigen (signal 1) and sub‐optimal cytokine concentrations (signal 3), which are both too weak on their own to drive i NKT cell activation, can act together to achieve the stimulation of the i NKT cells. The antigens bound to CD1d could be of self or foreign origin. In the direct and synergistic pathways, the signal can also be amplified by the up‐regulation of the expression levels of CD1d97, 122 and/or self‐antigens.97, 98 Additionally, signal two, i.e. co‐stimulation/inhibition, can modulate i NKT cell responses.25
As iNKT cells are positioned in peripheral tissues and display effector functions rapidly after activation, they are part of the first line of defence of the immune system and therefore play an important role at the mucosal surfaces as well. In this review we will outline our current knowledge on the interplay between iNKT cells and the mucosal microbiota, with a special emphasis on the respiratory and gastrointestinal tracts (Fig. 2).
Figure 2.
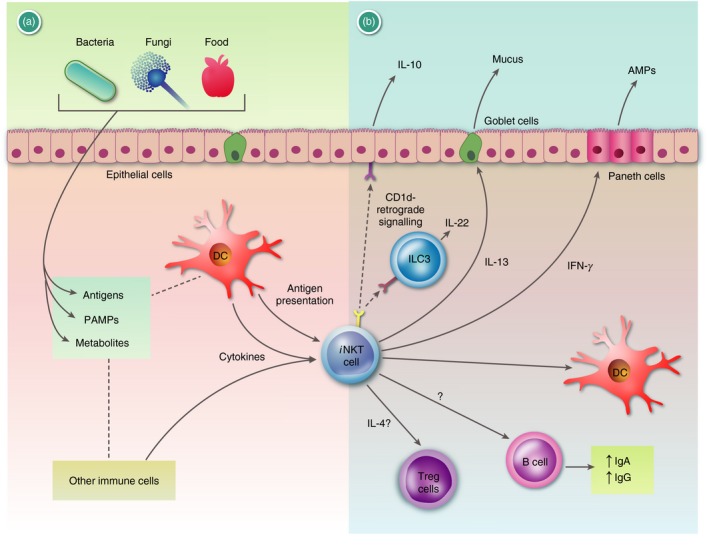
Graphic summary of the functions of invariant Natural Killer T (i NKT) cells at mucosal surfaces: (a) i NKT cells at mucosal surfaces can respond to many signals. Bacterium‐, fungus‐, and food‐derived products can influence the tissue‐resident cells of the mucosa and the local immune cells, via antigens, pathogen‐associated molecular patterns (PAMPs), and metabolites (Box 1). These can alter the frequency and function of various cells (not depicted). Antigens for i NKT cells can be either self‐antigens or antigens derived from the microbiota (Table 1). Some of the bacterially derived glycolipids binding to CD1d can also be inactive, competitive inhibitors. Furthermore, i NKT can respond to local cytokines either directly or synergistically with CD1d‐bound antigens. (b) Invariant NKT cells are known to influence the mucosal microenvironment and the microbial composition via (1) cytokines that they produce and (2) direct cell–cell contact. (1) Invariant NKT cell derived interferon‐γ (IFN‐γ) or interleukin‐13 (IL‐13) has been shown to activate Paneth cells or goblet cells to increase production of anti‐microbial peptides (AMPs) or mucus, respectively. Furthermore, i NKT cells have been shown to boost IgA and IgG production by B cells. Finally, CD1d−/− mice had a lower frequency of CD304+ regulatory T (Treg) cells in the mesenteric lymph nodes and i NKT cell‐derived IL‐4 was implicated.46 (2) Binding of i NKT cells to CD1d can induce, via a CD1d‐retrograde signal, IL‐10 or IL‐22 production by epithelial cells or innate lymphoid cells (ILC3s), respectively, which both support mucosal integrity. Please note that the figure does not attempt to represent a particular mucosal surface, but summarizes available data derived from various tissues and sides. For additional references we refer to the main text and Box 1. AMPs, anti‐microbial peptides; ECs, epithelial cells.
iNKT cells and the microbiota in the gastrointestinal tract
The gastrointestinal tract, reaching from the mouth to the rectum, is usually divided into the upper gastrointestinal tract (pharynx, oesophagus, stomach), the small intestine (duodenum, jejunum, ileum), and the large intestine (caecum, colon, rectum). Its main function is to extract and absorb nutrients from the food via its large surface of approximately 200 m2, made up largely of small intestinal villi and microvilli. This digestive process is supported by the commensal microbiota of the intestine. The gastrointestinal tract harbours diverse sets of bacteria, fungi, Archaea, and viruses with varying densities, although the vast majority of the microbiome are bacteria. Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria are the dominant bacterial phyla in the human intestine, with estimates suggesting over 1000 distinct species.26 The bacterial density increases along the gastrointestinal tract, spanning from 103 to 104 bacteria/ml at the beginning of the small intestine up to 1011 bacteria/ml in the colon.27 Besides digestion, the composition of the gut microbiota can influence many aspects of human health, including neural, gastrointestinal, metabolic and skeletal systems, as well as the immune system.1, 2, 3 Holding such large numbers of bacteria, separated from the body by only one cell layer, at bay requires several defence mechanisms, with the immune system in a prominent position. The closest are the intraepithelial lymphocytes (IELs), which are sited between the mucosal epithelial cells. There are approximately 10–15 IELs for every 100 epithelial cells in the small intestine with fewer cells in the large intestine.28 Over 90% of these IELs are T cells with the large majority expressing CD8α. Furthermore, basically all IEL T cells express an effector/memory phenotype, bear a TCR with limited diversity, and many have unusual developmental requirements unique to this location.28 In contrast, the lamina propria lymphocytes, which are scattered throughout the lamina propria underneath the epithelial layer, are more diverse in their cellular composition and resemble more those of other lymphatic organs.29
iNKT cells can be detected in the mouse intestine at frequencies only slightly lower than in other peripheral organs, like the spleen and lymph nodes (stomach,30 small intestine,31, 32, 33 large intestine33, 34, 35, 36, 37). Approximately 65% (BALB/c mice) to 85% (C57BL/6 mice) of intestinal iNKT cells are NKT1 cells, with the rest largely being NKT2 cells, and no differences in this distribution were noted for IELs versus lamina propria lymphocytes.24 The distribution of intestinal iNKT cells correlated indirectly with the bacterial density, i.e. more iNKT cells in the small intestine than the large intestine, and the proximity of the bacteria, i.e. more iNKT cells in the lamina propria lymphocytes than in the IELs.33 Invariant NKT cells can also be found in the human intestine, however, less is known about their distribution.38, 39, 40
Although iNKT cells do not require the commensal microbiota to develop an activated/memory phenotype and the ability to produce cytokines,33, 35, 41, 42, 43 the microbiota nonetheless impacts their functionality. Indeed, iNKT cells from germ‐free mice (GF) differed in their TCR Vβ7 frequency and expressed lower levels of activation markers33 compared with the control specific pathogen‐free mice. Furthermore, these iNKT cells were hypo‐responsive and performed weaker effector functions (cytokine production, cytotoxicity) after antigenic stimulation.33 Reconstitution of the GF animals with bacteria that contain iNKT cell antigens (Sphingobium yanoikuyae), but not with antigen‐negative bacteria (Escherichia coli), could fully establish phenotypic and functional maturity of the iNKT cells.33 Interestingly, the microbiota affected the distribution of iNKT cells as well. Whereas the frequency of iNKT cells in GF animals was lower in spleen and liver,35, 42 it was increased in the small and large intestines33 and the colon.35, 37 This distribution was established within the first 5 weeks of life, as reconstitution of the GF mice with a normal microbiota at a later time did not change the frequency of iNKT cells.35 Importantly, the increased frequency of iNKT cells in the intestine of GF mice has also functional consequences for intestinal immune responses. Oxazolone‐induced colitis is a mouse model of ulcerative colitis in which iNKT cells are known to be pathogenic.44 As a result of the higher numbers of effector iNKT cells in the mucosa at the onset of the disease, these GF animals were also more sensitive to oxazolone‐induced colitis.35 CXCL16 production by epithelial cells was implicated in the recruitment of iNKT cells to the intestine;35 however, as the authors themselves published conflicting data,37 the role of CXCL16 appears unclear. Data similar to the GF mice were obtained with mice with a highly restricted intestinal flora (RF mice), which is devoid of Sphingobium but enriched for Firmicutes.42 The frequency of iNKT cells in RF mice was reduced in the spleen and liver, they displayed an altered TCR Vβ7 frequency, lower expression of activation markers, and produced smaller amounts of cytokines following activation with αGalCer.33, 42 Moreover, in the RF mice the intra‐gastric administration of S. yanoikuyae bacteria increased the frequency of spleen and liver iNKT cells and their expression of activation markers within a day.42 The data in the GF and RF animals demonstrate that it is not the microbiota per se that affects iNKT cells, but the composition of the microbiota and the presence of particular iNKT cell antigens. When we compared control specific pathogen‐free mice obtained from different vendors, which are known to differ in their gut flora,45 we again noticed differences in the frequency, Vβ7‐usage, and tumour necrosis factor production of iNKT cells.33 These differences were abolished by co‐housing the offspring, which served to equalize the microbial flora.33
Several lines of data support the idea that specific iNKT cell antigens can be provided by the intestinal microbiota. First, the effects observed in the GF animals were independent of interleukin‐12 or MyD88.33, 35 Second, using Nur77 expression as an indication for TCR‐mediated activation, a CD1d‐dependent stimulation of iNKT cells in the intestine was shown in the presence of the intestinal microbiota.46 Third, intra‐gastric administration of GF mice with a mixture of heat‐killed bacteria leads to the expression of CD1d–antigen complexes, detected with the antibody clone L363,47 on liver dendritic cells.48 Finally, and most directly, iNKT cell antigens have been discovered in several bacteria present in the intestinal microbiota.
Besides the above mentioned α‐Proteobacteria Sphingobium yanoikuyae,49, 50 iNKT cell antigens have been described in the closely related Sphingomonas paucimobilis,49, 50, 51 Sphingomonas wittichii,52 Sphingomonas capsulata,49 Novosphingobium aromaticivorans,53 and Ehrichia muris.49 The Proteobacterium Heliobacter pylori is a common colonizer of the stomach, present in about half of the world population,54 that can cause gastric ulcers. About 25% of H. pylori's lipids are cholesteryl α‐glucosides55 that contain several related antigens for iNKT cells.30, 56 The intestinal microbiota in humans and mice is dominated by members of the phyla Firmicutes and Bacteriodetes (human,57 mouse58). For one common member of these phyla, Bacteroides fragilis, an α‐galactosylceramide antigen (α‐GalCer(Bf)) reminiscent of αGalCer has been described that could stimulate human and mouse iNKT cells.59 Surprisingly, a subsequent study described a B. fragilis‐derived α‐galactosylceramide (GSL‐Bf717) that could bind CD1d but was not stimulatory.37 This suggests that some commensal‐derived glycolipids can act as competitive inhibitors for other stimulatory iNKT cell antigens. Another example for this was the inhibition of αGalCer‐induced iNKT cell activation in vitro by a lipid extract of Bifidobacterium infantis.60 In addition to B. fragilis, Prevotella copri and Bacteriodes vulgatus, two other commensals belonging to the Bacteriodetes phylum, express iNKT cell antigens, although at approximately 100‐fold lower concentrations than B. fragilis.61 Within the Firmicutes phylum so far only the commensal strain Lactobacillus casei has been described to express an iNKT cell antigen.62 Furthermore, a few other reports reported iNKT cell antigens from commensal bacteria, without clearly defining the source48 or the exact structure of the antigen (e.g. B. infantis 60). Besides bacteria, an iNKT cell antigen (ChAcMan) has been reported for the fungus Candida albicans, a gut commensal and opportunistic pathogen.63 Finally, iNKT cell antigens have been described for several mucosal pathogens (Table 1). In summary, in the last decade iNKT cell antigens were discovered in many bacteria, fungi and protozoa, indicating that such antigens are widely distributed in the environment.64
Table 1.
Mucosal commensals and pathogens with known invariant Natural Killer T (iNKT) cell antigens
| Organism | Phylum | Pathogenicity | Antigen | References |
|---|---|---|---|---|
| Bacteria | ||||
| Mycobacterium tuberculosis | Actinobacteria | Pathogen | Phosphatidylinositol mannoside (PIM) | 78 |
| Saccharopolyspora | Actinobacteria | Environmental, opportunistic pathogen | M‐AcM‐MAG | 63 |
| Rothia dentocariosa | Actinobacteria | Commensal, opportunistic pathogen | M‐AcM‐MAG | 63 |
| Arthrobacter | Actinobacteria | Commensal, opportunistic pathogen | M‐AcM‐MAG | 63 |
| Bacteroides fragilis | Bacteriodetes | Commensal, opportunistic pathogen | αGalCer(Bf) | 59 |
| Prevotella copri | Bacteroidetes | Commensal | αGalCer (~ 100 × lower concentration than B. fragilis) | 61 |
| Bacteroides vulgatus | Bacteriodetes | Commensal, opportunistic pathogen | αGalCer (~ 100 × lower concentration than B. fragilis) | 61 |
| Streptococcus pneumoniae and Group B streptococcus | Firmicutes | Commensal, opportunistic pathogens |
SPN‐Glc‐DAG SPN‐Gal‐Glc‐DAG |
62 |
| Lactobacillus casei | Firmicutes | Commensal | Glc‐DAG | 62 |
| Sphingomonas paucimobilis | Proteobacteria | Commensal, opportunistic pathogen | α‐glucuronosyl ceramide (GSL‐1/aGlcUCer) | 50, 51 |
| Sphingomonas yanoikuyae | Proteobacteria | Environmental, commensal, opportunistic pathogen | α‐galacturonosyl‐ceramides | 50 |
| Ehrlichia muris | Proteobacteria | Pathogen in rodents, but not in humans | Antigen not defined | 49 |
| Helicobacter pylori | Proteobacteria | Commensal, opportunistic pathogen | Cholestoryl‐α‐glucosides, especially monoacyl α‐CPG | 30, 56 |
| Sphingomonas wittichii | Proteobacteria | No pathogenicity reported | α‐galacturonosyl‐ceramides | 52 |
| Borrelia burgdorferi | Spirochaetae | Pathogen | BbGL‐II (1,2‐di‐O‐acyl‐ 3‐O‐α‐d‐galactopyranosyl‐sn‐glycerol, 6) | 52 |
| Fungi | ||||
| Aspergillus fumigatus and Aspergillus niger (latter with lower antigenic content) | Ascomycota | Opportunistic pathogen | Asperamide B | 136 |
| Candida albicans | Ascomycota | Commensal, opportunistic pathogen | ChAcMan | 63 |
| Protozoa | ||||
| Entamoeba histolytica | Amoebozoa | Opportunistic pathogen (often asymptomatic) | lipopeptidophosphoglycan (EhLPPG) | 137 |
| Leishmania donovani | Euglenozoa | Opportunistic pathogen (often asymptomatic) | Lipophosphoglycan (LPG) | 138 |
Given the impact of the intestinal microbiota on iNKT cells it might not be too surprising that the elimination of the gut flora with antibiotics likewise influences iNKT cell responses. After 2 weeks of antibiotic treatment, the frequency of iNKT cells increased in the colon of wild‐type C57BL/6 mice.65 Interestingly, this increase disappeared within 1–2 weeks following the bacterial reconstitution.65 Changes were also noted beyond the intestine. For example, the iNKT cell frequency in liver increased following antibiotic‐mediated commensal depletion.66 Furthermore, for several models the changes observed after the depletion of the commensal microbiota by antibiotics were dependent on iNKT cells: a delayed liver regeneration after partial hepatectomy,66 an amelioration of experimental autoimmune encephalomyelitis,67 but also an augmented concanavalin A‐induced liver injury.48
Importantly, the interaction between the commensal microbiota and iNKT cells is mutual, as iNKT cells can influence the composition of the intestinal microflora. CD1d‐deficient mice were found to host an altered gut microbiota,46, 68, 69 which was pro‐inflammatory upon transfer into wild‐type animals.69 Furthermore, CD1d‐deficient mice were more susceptible to intestinal colonization by pathogenic bacteria as well.68 Whereas in control mice the intestinal bacteria were largely separated from the intestinal epithelial cells by a mucus layer, this layer was impaired in the CD1d‐deficient animals, leading to a direct contact of the bacteria and the epithelial cells.46 The impact of iNKT cells on the intestinal microbiota was stronger in the small than the large intestine,46, 69 in line with the higher frequency of iNKT cells in the small intestine.33 This might also explain why an analysis of faeces from CD1d‐deficient pigs did not reveal any differences in the bacterial composition.70 The changes in the intestinal microbiota seen in the CD1d‐deficient animals could be replicated in mice where CD1d was selectively missing on CD11c+ cells.46 In contrast, in mice with a CD11c‐specific deletion of CD1d, the separation between the intestinal bacteria and the epithelial cells was intact, as in the control animals.46 This demonstrates that iNKT cells influence the intestinal microbiota in at least two independent ways involving CD11c+ cells, presumably dendritic cells, and other CD1d+ cells in the intestine. Finally, the intestinal microbiota could be influenced by antigen‐specific activation of iNKT cells with αGalCer. On the one hand, oral challenge of control mice with αGalCer led to an increase in Bacteriodetes and Proteobacteria but a decrease in Firmicutes species.46 On the other hand, injection of αGalCer into GF mice delayed their reconstitution with bacteria given orally.68 However, information is still limited on how iNKT cells could achieve this influence on the microbiota (Box 1).
iNKT cells and the microbiota in the respiratory tract
The mammalian respiratory system is generally divided into the upper and lower respiratory tract. The upper respiratory tract is composed of the nose, nasal cavity and sinuses, pharynx (throat) and larynx (voice box) and is the first mucosal site where the body encounters air‐borne microorganisms. The lower respiratory tract includes the conducting airways (trachea, bronchi, bronchioles) and the alveoli, in which the gas exchange occurs. The upper respiratory tract retains larger particles; however, particles smaller than 1–3 μm, such as microorganisms, pollen and smoke, can pass to the lower respiratory tract. There, particles or microorganisms can be trapped in the mucous that is secreted by submucosal mucous glands and lines the lower airways. The rhythmic pulse‐like movements by the cilia of the epithelial cells then transport the mucus and its trapped particles to the upper respiratory tract where they are eliminated either via the digestive tract or through the sneeze and cough reflexes. Furthermore, this mucus and the liquid layer on the airway surfaces of the lower respiratory tracts contain various antimicrobial peptides and antigen‐specific secretory IgA to protect the lung.71, 72 Despite these mechanical and chemical defence mechanisms, the lower airways are not sterile, but host a unique commensal microbiota.
The airways are colonized immediately after birth and a stable commensal microbiota develops within the first years of life.73 This commensal microbiota contributes to lung development and function, and to host defence; a disordered microbiota (dysbiosis) is a common feature of chronic lung inflammations, like asthma.71, 73 The core microbiota of the healthy lung contains five bacteria phyla: Bacteroidetes, Firmicutes, Proteobacteria, Fusobacteria and Actinobacteria.71, 73
iNKT cells are more frequent in the lung than in the secondary lymphatics,24 with a strong influx from the blood following lung inflammation.74 There is strong evidence for a major role for iNKT cells in regulating immune and inflammatory responses in the respiratory system. They are important contributors for most asthma models and have been implicated in the pathogenesis of human asthma.75, 76 Furthermore, iNKT cells are important for a protective immune response in the lung to many infectious agents.77 It is clear now that several of these airway pathogens contain iNKT cell antigens.
Besides the Sphingobium bacteria mentioned above, iNKT cell antigens have been described for Mycobacterium tuberculosis, the causative bacteria of tuberculosis,78 Streptococcus pneumoniae, the leading cause of community‐acquired pneumonia,62 group B streptococcus,62 Saccharopolyspora spp., the leading cause for Farmer's lung,63 and the fungus Aspergillus fumigatus.79 In contrast to M. tuberculosis, which is considered an obligate pathogen, the other organisms on this list are opportunistic pathogens as they can be found in the human microbiota to varying extents and are usually asymptomatic in healthy individuals. Nonetheless, the presence of particular commensals or the composition of the lung microbiota can probably impact inflammatory responses. For example, patients with poorly controlled asthma have a higher bacterial count in the lung, and some of the taxa (e.g. Sphingobium) are known to bear iNKT cell antigens.80
Additionally, several of the organisms mentioned above can readily be detected in the environment, like Sphingobium spp., Saccharopolyspora spp., or Aspergillus fumigatus, indicating that the inhaled air is another source for iNKT cell antigens.64 Indeed, the majority of sterile house dust extracts (HDEs) that we tested contained antigens for mouse and human iNKT cells.81 HDEs are simple aqueous preparations from house dust that provide a relatively complete sampling of the environment without the need for a priori assumptions imposed by the nature of the purification.82 The HDEs displayed adjuvant‐like properties in an iNKT‐cell‐dependent allergen‐induced mouse asthma model.81 When different HDEs were tested, we noted a large variability in the antigenic strength as well as in the chemical nature, indicating that different HDEs could contain several distinct iNKT cell antigens.81 Furthermore, as our experiments indicated that dust mites are probably not the source of the antigenic activity found in HDE,81 we consider bacteria as the most likely source of these environmental iNKT cell antigens.64 However, it should be noted that some preliminary data suggest that plants and their pollen might contain iNKT cell antigens as well.83, 84, 85 Altogether, recent findings indicate that antigens for iNKT cells are almost ubiquitous indoors and in the environment. As they will reach the airways with the inhaled air, it seems likely that iNKT cell antigens are present under normal conditions in the lung at low levels and could under certain conditions contribute to airway inflammation.
Interestingly, however, the age at the time of the antigen exposure seems to be important as well. Whereas the iNKT cell antigens in the HDEs greatly augmented symptoms in an allergen‐induced asthma model in adult mice,81 they seemed to have an opposite effect early in life. The ‘Urban Environment and Childhood Asthma’ (URECA) project is a longitudinal study that follows infants born to parents with asthma from birth through to age 14–16 years.86 As part of the URECA study we correlated the iNKT cell–antigen content of the house dust from children when they were 3 months old with the clinical symptoms of those children at age 3–7 years.87 Our data indicate that infants growing up in homes containing more iNKT cell antigens were less likely to develop asthma.87 A child's house that is rich in antigens for iNKT cells probably reflects an environment with increased microbe exposure. According to the ‘hygiene hypothesis’, increased microbial exposure in the first years of life protects children from asthma.88, 89 In line with this hypothesis are also the data on lung iNKT cells in GF mice. Such GF mice had a higher frequency of iNKT cells in the lung than specific pathogen‐free control mice.35 Similar to the intestine, this frequency remained high in the lung if the mice were not colonized with bacteria within the first 3 weeks of life.35 Due to this increased frequency of effector iNKT cells in the lung, such mice were more susceptible to allergen‐induced asthma as well.35 Similarly, the deliberate activation of iNKT cells in 2‐week‐old mice with an H. pylori‐derived antigen or the T helper type 1‐biasing antigen C‐GalCer protected mice against asthma 6 weeks later.56 Surprisingly, a Sphingobium‐derived antigen was not able to induce this protection.56 These data demonstrate that the influence of the commensal microbiota on iNKT cells can have important roles in the regulation of the immune response in the airways and suggest that this impact is influenced by the age of the individual. In contrast to the intestine, no information is currently available as to whether iNKT cells can influence the lung microbiota or can regulate the production of anti‐microbial peptides in the lung under steady‐state conditions.
iNKT cells and microbiota in other mucosal tissues
Much less is known about the role of iNKT cells at other mucosal surfaces and of their interaction there with the local commensals than for the intestine or the lung. Although, the microbial composition of the urogenital tract differs from the intestine, several of the bacteria known to bear iNKT cell antigens can be found in the urogenital tract of healthy individuals.90, 91 iNKT cells are functional in the bladder, as αGalCer activation was shown to support bacterial clearance in urinary tract infections.92, 93 Furthermore, urethral epithelial cells were shown to be receptive to retrograde signalling downstream of CD1d.94 Therefore, it seems likely that iNKT cells play a role in the protection of the mucosal surface of the urogenital tract as well.
Concluding remarks
It is becoming increasingly clear how intricate and far‐reaching the interaction between the host and its commensal microbiota is, where each individual player seems to have its own role at maintaining and fostering this mutually beneficial relationship. These data also establish a mutual interaction between the commensal microbiota and iNKT cells, where the microbiota is required for iNKT cells to gain function, and where iNKT cells can influence the composition of the mucosal flora. Moreover, they suggest that iNKT cells play an important role in supporting the health and homeostasis of the mucosal tissues by acting as an earlier sensor for tissue damage and bacterial translocation.
The microbiota is a major source for iNKT cell antigens. These include bacteria, fungi, protozoa and metazoa that either colonize the mucosa (true or facultative commensals, opportunistic pathogens), or temporarily present themselves at the mucosa following ingestion or inhalation (commensals, pathogens, environmental antigens) (Table 1). Besides microbes, environmental antigens64 could be derived from food components too, like milk lipids,95, 96 and from airborne pollen,83 either directly or after processing by the commensals. Furthermore, microbe‐induced changes in the presentation of host‐derived self‐lipids or the expression levels of CD1d itself could conceivably modulate mucosal iNKT cell responses as well.97, 98 Finally, although the GF data indicated that pathogen‐associated molecular pattern‐induced signals are not necessary for full iNKT cell maturation,33, 35 it is likely that such signals are involved in the regulation of iNKT cell functions in the mucosa (Box 1). Such indirect signals could explain the long‐lasting effects on iNKT cell frequency and functionality following virus infection.56, 99 In this context, it is notable that iNKT cells can also be activated by bacterially derived superantigens.100, 101
Moreover, anything that changes the composition of the commensal microbiota could also potentially impact iNKT cells. Host‐derived variables, like gender and genetics, are known to impact the microbiota.102, 103 The list is longer for environmental variables, inducing the diet (e.g. fibre content, antibiotics/probiotics), housing (e.g. urban/rural, pets), and the presence of chronic infections (e.g. helminths) or other ‘pathobionts’.104, 105, 106, 107 Consequently, the microbiota is dynamic.108
An intriguing finding is the far‐reaching impact that the microbiota can have on the host. On the one hand, changes in one mucosal surface can lead to changes in other mucosal sides. For example, using αGalCer as adjuvant for an sublingual vaccination led to increases in antigen‐specific antibody levels also in vaginal washes.109 Furthermore, changes in the microbiota of one mucosal tissue can impact the frequency and function of immune cells of other mucosal tissues.110 On the other hand, the impact of the microbiota is not limited to the mucosal surfaces and can influence apparently any part of the body, including seemingly remote areas, such as the brain.111 With regard to iNKT cells, the above‐mentioned systemic effects of antibiotic treatment and the role of iNKT cells are relevant in sepsis,112 which is often caused by translocated intestinal bacteria.113 Furthermore, one member of the commensal Sphingobium species (Novosphingobium aromaticivorans) has been linked to iNKT cell‐dependent autoimmune responses against the bile duct in mice53 and humans.114, 115
However, many open questions remain with regard to the details of the mutual regulation of iNKT cells and the commensal microbiota. For many of the observed influences the mechanistic understanding is still rudimentary, and many new microbial mediators will probably be discovered, adding to the complexity. It seems likely that different commensals provide at times complementary or opposing influences, as reported for example for B. fragilis.37, 59 Furthermore, the response of iNKT cells towards microbial‐derived signals can be pro‐inflammatory or anti‐inflammatory and the decisive factors governing this outcome are largely unclear. Whereas the nature of the antigen‐presenting cell probably plays a role,116 the potential involvement of different iNKT cell subsets is currently unexplored. Finally, much needs to be learned about the mechanisms of the systemic impact on iNKT cells and the extent to which the microbiota impacts iNKT cell functions all over the body.
Invariant NKT cells are of great therapeutic potential as the lock‐and‐key principle of CD1d/iTCR is basically shared by every human being. Consequently, iNKT cell antigens are already in clinical trials for cancer therapy and for several vaccination approaches,117, 118 and we expect many new applications, in particular for mucosal vaccinations, in the near future. The data reviewed here also suggest that iNKT cells could be a promising therapeutic target to address microbial dysbiosis, which is linked to many mucosal diseases.119, 120 Furthermore, the finding that neonatal changes can have life‐long impacts on the frequency of mucosal iNKT cells is intriguing, as it suggests an option for preventive approaches to treat, for example, asthma.
Disclosures
The authors declare that they have no competing interests.
Acknowledgements
The authors would like to thank Dr Duygu Sag for critically reading the manuscript. This work was supported by grants to GW by TÜBITAK (no. 116Z272, no. 117Z216); the European Molecular Biology Organization (EMBO Installation Grant no. 3073); and the Dokuz Eylul University (no. 2017.KB.SAG.029). The funders had no role in the preparation of the manuscript.
References
- 1. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity 2017; 46:562–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol 2017; 18:851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brandtzaeg P. Role of the intestinal immune system in health In: Baumgart DC, ed. Crohn's Disease and Ulcerative Colitis: From Epidemiology and Immunobiology to a Rational Diagnostic and Therapeutic Approach. Cham: Springer International Publishing, 2017: 23–56. [Google Scholar]
- 4. Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol 2007; 25:297–336. [DOI] [PubMed] [Google Scholar]
- 5. Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1‐ and MR1‐restricted T cells. Annu Rev Immunol 2014; 32:323–66. [DOI] [PubMed] [Google Scholar]
- 6. Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d‐restricted antigens by natural killer T cells. Nat Rev Immunol 2012; 12:845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol 2005; 23:877–900. [DOI] [PubMed] [Google Scholar]
- 8. Wingender G, Kronenberg M. Role of NKT cells in the digestive system. IV. The role of canonical natural killer T cells in mucosal immunity and inflammation. Am J Physiol Gastrointest Liver Physiol 2008; 294:G1–8. [DOI] [PubMed] [Google Scholar]
- 9. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol 2013; 13:101–17. [DOI] [PubMed] [Google Scholar]
- 10. Wingender G, Krebs P, Beutler B, Kronenberg M. Antigen‐specific cytotoxicity by invariant NKT cells in vivo is CD95/CD178‐dependent and is correlated with antigenic potency. J Immunol 2010; 185:2721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady‐state production of IL‐4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol 2013; 14:1146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL‐10‐producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest 2014; 124:3725–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wingender G, Sag D, Kronenberg M. NKT10 cells: a novel iNKT cell subset. Oncotarget 2015; 6:26552–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB et al Identification of an IL‐17‐producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med 2007; 204:995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Michel ML, Mendes‐da‐Cruz D, Keller AC, Lochner M, Schneider E, Dy M et al Critical role of ROR‐γt in a new thymic pathway leading to IL‐17‐producing invariant NKT cell differentiation. Proc Natl Acad Sci USA 2008; 105:19845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doisne JM, Soulard V, Bécourt C, Amniai L, Henrot P, Havenar‐Daughton C et al Cutting edge: crucial role of IL‐1 and IL‐23 in the innate IL‐17 response of peripheral lymph node NK1.1‐ invariant NKT cells to bacteria. J Immunol 2011; 186:662–6. [DOI] [PubMed] [Google Scholar]
- 17. Doisne JM, Becourt C, Amniai L, Duarte N, Le Luduec JB, Eberl G et al Skin and peripheral lymph node invariant NKT cells are mainly retinoic acid receptor‐related orphan receptor γt+ and respond preferentially under inflammatory conditions. J Immunol 2009; 183:2142–9. [DOI] [PubMed] [Google Scholar]
- 18. Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS et al Diverse cytokine production by NKT cell subsets and identification of an IL‐17‐producing CD4–NK1.1– NKT cell population. Proc Natl Acad Sci USA 2008; 105:11287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A et al Identification of Bcl‐6‐dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol 2011; 13:35–43. [DOI] [PubMed] [Google Scholar]
- 20. King IL, Fortier A, Tighe M, Dibble J, Watts GF, Veerapen N et al Invariant natural killer T cells direct B cell responses to cognate lipid antigen in an IL‐21‐dependent manner. Nat Immunol 2011; 13:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monteiro M, Almeida CF, Caridade M, Ribot JC, Duarte J, Agua‐Doce A, Wollenberg I, Silva‐Santos B, Graca L. Identification of regulatory Foxp3+ invariant NKT cells induced by TGF‐beta. J Immunol 2010; 185:2157–63. [DOI] [PubMed] [Google Scholar]
- 22. Wingender G, Birkholz AM, Sag D, Farber E, Chitale S, Howell AR et al Selective conditions are required for the induction of invariant NKT cell hyporesponsiveness by antigenic stimulation. J Immunol 2015; 195:3838–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Birkholz AM, Girardi E, Wingender G, Khurana A, Wang J, Zhao M et al A novel glycolipid antigen for NKT cells that preferentially induces IFN‐γ production. J Immunol 2015; 195:924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee YJ, Wang H, Starrett GJ, Phuong V, Jameson SC, Hogquist KA. Tissue‐specific distribution of iNKT cells impacts their cytokine response. Immunity 2015; 43:566–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shissler SC, Lee MS, Webb TJ. Mixed signals: co‐stimulation in invariant natural killer T cell‐mediated cancer immunotherapy. Front Immunol 2017; 8:1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016; 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lambolez FMS, Cheroutre H. Lymphocytes: Intraepithelial in eLS (Ed). 2013. 10.1002/9780470015902.a0001197.pub3 [DOI] [Google Scholar]
- 29. López MC. Mucosal immunity * A2 In: McQueen CA, ed. Comprehensive Toxicology, 2nd edn Oxford: Elsevier, 2010: 203–15. [Google Scholar]
- 30. Ito Y, Vela JL, Matsumura F, Hoshino H, Tyznik A, Lee H et al Helicobacter pylori cholesteryl α‐glucosides contribute to its pathogenicity and immune response by natural killer T cells. PLoS ONE 2013; 8:e78191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR et al Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med 2000; 192:741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang JH, Lee JM, Youn HJ, Lee KA, Chung Y, Lee AY et al Functional maturation of lamina propria dendritic cells by activation of NKT cells mediates the abrogation of oral tolerance. Eur J Immunol 2008; 38:2727–39. [DOI] [PubMed] [Google Scholar]
- 33. Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B et al Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology 2012; 143:418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ronet C, Darche S, Leite de Moraes M, Miyake S, Yamamura T, Louis JA et al NKT cells are critical for the initiation of an inflammatory bowel response against Toxoplasma gondii. J Immunol 2005; 175:899–908. [DOI] [PubMed] [Google Scholar]
- 35. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A et al Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Montbarbon M, Pichavant M, Langlois A, Erdual E, Maggiotto F, Neut C et al Colonic inflammation in mice is improved by cigarette smoke through iNKT cells recruitment. PLoS ONE 2013; 8:e62208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk‐Hasdemir D et al Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 2014; 156:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Keeffe J, Doherty DG, Kenna T, Sheahan K, O'Donoghue DP, Hyland JM et al Diverse populations of T cells with NK cell receptors accumulate in the human intestine in health and in colorectal cancer. Eur J Immunol 2004; 34:2110–9. [DOI] [PubMed] [Google Scholar]
- 39. Fuss IJ, Heller F, Boirivant M, Leon F, Yoshida M, Fichtner‐Feigl S et al Nonclassical CD1d‐restricted NK T cells that produce IL‐13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest 2004; 113:1490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grose RH, Cummins AG, Thompson FM. Deficiency of invariant natural killer T cells in coeliac disease. Gut 2007; 56:790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park AY, Hondowicz BD, Scott P. IL‐12 is required to maintain a Th1 response during Leishmania major infection. J Immunol 2000; 165:896–902. [DOI] [PubMed] [Google Scholar]
- 42. Wei B, Wingender G, Fujiwara D, Chen DY, McPherson M, Brewer S et al Commensal microbiota and CD8+ T cells shape the formation of invariant NKT cells. J Immunol 2010; 184:1218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeng J, Howard JC. Spontaneous focal activation of invariant natural killer T (iNKT) cells in mouse liver and kidney. BMC Biol 2010; 8:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL‐13‐producing NK‐T cells. Immunity 2002; 17:629–38. [DOI] [PubMed] [Google Scholar]
- 45. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saez de Guinoa J, Jimeno R, Gaya M, Kipling D, Garzón MJ, Dunn‐Walters D et al CD1d‐mediated lipid presentation by CD11c+ cells regulates intestinal homeostasis. EMBO J, 2018; 37:pii: e97537 https://doi.org/10.15252/embj.201797537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Im JS, Arora P, Bricard G, Molano A, Venkataswamy MM, Baine I et al Kinetics and cellular site of glycolipid loading control the outcome of natural killer T cell activation. Immunity 2009; 30:888–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wei Y, Zeng B, Chen J, Cui G, Lu C, Wu W et al Enterogenous bacterial glycolipids are required for the generation of natural killer T cells mediated liver injury. Sci Rep 2016; 6:36365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mattner J, Debord KL, Ismail N, Goff RD, Cantu C 3rd, Zhou D et al Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 2005; 434:525–9. [DOI] [PubMed] [Google Scholar]
- 50. Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD et al Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 2005; 434:520–5. [DOI] [PubMed] [Google Scholar]
- 51. Sriram V, Du W, Gervay‐Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d‐specific ligands for NKT cells. Eur J Immunol 2005; 35:1692–701. [DOI] [PubMed] [Google Scholar]
- 52. Wu D, Xing GW, Poles MA, Horowitz A, Kinjo Y, Sullivan B et al Bacterial glycolipids and analogs as antigens for CD1d‐restricted NKT cells. Proc Natl Acad Sci USA 2005; 102:1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mattner J, Savage PB, Leung P, Oertelt SS, Wang V, Trivedi O et al Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe 2008; 3:304–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lin D, Koskella B. Friend and foe: factors influencing the movement of the bacterium Helicobacter pylori along the parasitism–mutualism continuum. Evol Appl 2015; 8:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hirai Y, Haque M, Yoshida T, Yokota K, Yasuda T, Oguma K. Unique cholesteryl glucosides in Helicobacter pylori: composition and structural analysis. J Bacteriol 1995; 177:5327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chang YJ, Kim HY, Albacker LA, Lee HH, Baumgarth N, Akira S et al Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest 2011; 121:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M et al Diversity of the human intestinal microbial flora. Science 2005; 308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier‐Kolthoff JP, Kumar N et al The Mouse Intestinal Bacterial Collection (miBC) provides host‐specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol 2016; 1:16131. [DOI] [PubMed] [Google Scholar]
- 59. Wieland Brown LC, Pukall R, Abt B, Foesel BU, Meier‐Kolthoff JP, Kumar N et al Production of α‐galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol 2013; 11:e1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liang S, Webb T, Li Z. Probiotic antigens stimulate hepatic natural killer T cells. Immunology 2014; 141:203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. von Gerichten J, Schlosser K, Lamprecht D, Morace I, Eckhardt M, Wachten D et al Diastereomer‐specific quantification of bioactive hexosylceramides from bacteria and mammals. J Lipid Res 2017; 58:1247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X et al Invariant natural killer T cells recognize glycolipids from pathogenic Gram‐positive bacteria. Nat Immunol 2011; 12:966–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shimamura M, Yamamura M, Nabeshima T, Kitano N, van den Elzen P, Yesilkaya H et al Activation of invariant natural killer T cells stimulated with microbial alpha‐mannosyl glycolipids. Sci Rep 2017; 7:9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wingender G. From the deep sea to everywhere: environmental antigens for iNKT cells. Arch Immunol Ther Exp (Warsz) 2016; 64:291–8. [DOI] [PubMed] [Google Scholar]
- 65. Burrello C, Garavaglia F, Cribiù FM, Ercoli G, Bosari S, Caprioli F et al Short‐term oral antibiotics treatment promotes inflammatory activation of colonic invariant natural killer T and conventional CD4+ T cells. Front Med (Lausanne) 2018; 5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu X, Sun R, Chen Y, Zheng X, Bai L, Lian Z et al Oral ampicillin inhibits liver regeneration by breaking hepatic innate immune tolerance normally maintained by gut commensal bacteria. Hepatology 2015; 62:253–64. [DOI] [PubMed] [Google Scholar]
- 67. Yokote H, Miyake S, Croxford JL, Oki S, Mizusawa H, Yamamura T. NKT cell‐dependent amelioration of a mouse model of multiple sclerosis by altering gut flora. Am J Pathol 2008; 173:1714–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nieuwenhuis EE, Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, Simons‐Oosterhuis Y et al Cd1d‐dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest 2009; 119:1241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Selvanantham T, Lin Q, Guo CX, Surendra A, Fieve S, Escalante NK et al NKT cell‐deficient mice harbor an altered microbiota that fuels intestinal inflammation during chemically induced colitis. J Immunol 2016; 197:4464–72. [DOI] [PubMed] [Google Scholar]
- 70. Yang G, Artiaga BL, Hackmann TJ, Samuel MS, Walters EM, Salek‐Ardakani S et al Targeted disruption of CD1d prevents NKT cell development in pigs. Mamm Genome 2015; 26:264–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 2017; 15:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu BG, Segal LN. Lung microbiota and its impact on the mucosal immune phenotype. Microbiol Spectr 2017; 5 10.1128/microbiolspec.bad-0005-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Torow N, Marsland BJ, Hornef MW, Gollwitzer ES. Neonatal mucosal immunology. Mucosal Immunol 2017; 10:5–17. [DOI] [PubMed] [Google Scholar]
- 74. Thanabalasuriar A, Neupane AS, Wang J, Krummel MF, Kubes P. iNKT cell emigration out of the lung vasculature requires neutrophils and monocyte‐derived dendritic cells in inflammation. Cell Rep 2016; 16:3260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zeissig S, Blumberg RS. Commensal microbial regulation of natural killer T cells at the frontiers of the mucosal immune system. FEBS Lett 2014; 588:4188–94. [DOI] [PubMed] [Google Scholar]
- 76. Umetsu DT, Dekruyff RH. Natural killer T cells are important in the pathogenesis of asthma: the many pathways to asthma. J Allergy Clin Immunol 2010; 125:975–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Paget C, Trottein F. Role of type 1 natural killer T cells in pulmonary immunity. Mucosal Immunol 2013; 6:1054–67. [DOI] [PubMed] [Google Scholar]
- 78. Fischer K, Scotet E, Niemeyer M, Koebernick H, Zerrahn J, Maillet S et al Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d‐restricted T cells. Proc Natl Acad Sci USA 2004; 101:10685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 1999; 12:310–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J et al Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol 2011; 127:372–81. e1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wingender G, Rogers P, Batzer G, Lee MS, Bai D, Pei B et al Invariant NKT cells are required for airway inflammation induced by environmental antigens. J Exp Med 2011; 208:1151–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Horner AA. Regulation of aeroallergen immunity by the innate immune system: laboratory evidence for a new paradigm. J Innate Immun 2010; 2:107–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Agea E, Russano A, Bistoni O, Mannucci R, Nicoletti I, Corazzi L et al Human CD1‐restricted T cell recognition of lipids from pollens. J Exp Med 2005; 202:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mirotti L, Florsheim E, Rundqvist L, Larsson G, Spinozzi F, Leite‐de‐Moraes M et al Lipids are required for the development of Brazil nut allergy: the role of mouse and human iNKT cells. Allergy 2013; 68:74–83. [DOI] [PubMed] [Google Scholar]
- 85. Abos‐Gracia B, del Moral MG, López‐Relaño J, Viana‐Huete V, Castro L, Villalba M et al Olea europaea pollen lipids activate invariant natural killer T cells by upregulating CD1d expression on dendritic cells. J Allergy Clin Immunol 2013; 131:1393–9. e5. [DOI] [PubMed] [Google Scholar]
- 86. Gern JE, Visness CM, Gergen PJ, Wood RA, Bloomberg GR, O'Connor GT et al The Urban Environment and Childhood Asthma (URECA) birth cohort study: design, methods, and study population. BMC Pulm Med 2009; 9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chandra S, Wingender G, Greenbaum JA, Khurana A, Gholami AM, Ganesan AP et al Development of asthma in inner‐city children: possible roles of MAIT cells and variation in the home environment. J Immunol 2018; 200:1995–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bach JF. The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat Rev Immunol 2018; 18:105–20. [DOI] [PubMed] [Google Scholar]
- 89. Lambrecht BN, Hammad H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat Immunol 2017; 18:1076–83. [DOI] [PubMed] [Google Scholar]
- 90. Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ et al Integrated next‐generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med 2012; 10:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Park YJ, Lee HK. The role of skin and orogenital microbiota in protective immunity and chronic immune‐mediated inflammatory disease. Front Immunol 2017; 8:1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hsieh YJ, Fu CL, Hsieh MH. Helminth‐induced interleukin‐4 abrogates invariant natural killer T cell activation‐associated clearance of bacterial infection. Infect Immun 2014; 82:2087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Minagawa S, Ohyama C, Hatakeyama S, Tsuchiya N, Kato T, Habuchi T. Activation of natural killer T cells by α‐galactosylceramide mediates clearance of bacteria in murine urinary tract infection. J Urol 2005; 173:2171–4. [DOI] [PubMed] [Google Scholar]
- 94. Kawana K, Matsumoto J, Miura S, Shen L, Kawana Y, Nagamatsu T et al Expression of CD1d and ligand‐induced cytokine production are tissue specific in mucosal epithelia of the human lower reproductive tract. Infect Immun 2008; 76:3011–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jyonouchi S, Abraham V, Orange JS, Spergel JM, Gober L, Dudek E et al Invariant natural killer T cells from children with versus without food allergy exhibit differential responsiveness to milk‐derived sphingomyelin. J Allergy Clin Immunol 2011; 128:102–9. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Brennan PJ, Tatituri RV, Heiss C, Watts GF, Hsu FF, Veerapen N et al Activation of iNKT cells by a distinct constituent of the endogenous glucosylceramide fraction. Proc Natl Acad Sci USA 2014; 111:13433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. De Libero G, Moran AP, Gober HJ, Rossy E, Shamshiev A, Chelnokova O et al Bacterial infections promote T cell recognition of self‐glycolipids. Immunity 2005; 22:763–72. [DOI] [PubMed] [Google Scholar]
- 98. Darmoise A, Teneberg S, Bouzonville L, Brady RO, Beck M, Kaufmann SH et al Lysosomal alpha‐galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity 2010; 33:216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Barthelemy A, Ivanov S, Fontaine J, Soulard D, Bouabe H, Paget C et al Influenza A virus‐induced release of interleukin‐10 inhibits the anti‐microbial activities of invariant natural killer T cells during invasive pneumococcal superinfection. Mucosal Immunol 2017; 10:460–9. [DOI] [PubMed] [Google Scholar]
- 100. Hayworth JL, Mazzuca DM, Maleki Vareki S, Welch I, McCormick JK, Haeryfar SM. CD1d‐independent activation of mouse and human iNKT cells by bacterial superantigens. Immunol Cell Biol 2012; 90:699–709. [DOI] [PubMed] [Google Scholar]
- 101. Hammamieh R, Chakraborty N, Lin Y, Shupp JW, Miller SA, Morris S et al Characterization of the interaction of staphylococcal enterotoxin B with CD1d expressed in human renal proximal tubule epithelial cells. BMC Microbiol 2015; 15:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chen L, Zhang YH, Huang T, Cai YD. Gene expression profiling gut microbiota in different races of humans. Sci Rep 2016; 6:23075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Haro C, Rangel‐Zúñiga OA, Alcalá‐Díaz JF, Gómez‐Delgado F, Pérez‐Martínez P, Delgado‐Lista J et al Intestinal microbiota is influenced by gender and body mass index. PLoS ONE 2016; 11:e0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nagai M, Obata Y, Takahashi D, Hase K. Fine‐tuning of the mucosal barrier and metabolic systems using the diet‐microbial metabolite axis. Int Immunopharmacol 2016; 37:79–86. [DOI] [PubMed] [Google Scholar]
- 105. Brosschot TP, Reynolds LA. The impact of a helminth‐modified microbiome on host immunity. Mucosal Immunol 2018. 10.1038/s41385-018-0008-5 [DOI] [PubMed] [Google Scholar]
- 106. Tao L, Reese TA. Making mouse models that reflect human immune responses. Trends Immunol 2017; 38:181–93. [DOI] [PubMed] [Google Scholar]
- 107. Zengler K, Zaramela LS. The social network of microorganisms – how auxotrophies shape complex communities. Nat Rev Microbiol 2018; 16:383–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sinha R, Goedert JJ, Vogtmann E, Hua X, Porras C, Hayes R et al Quantification of human microbiome stability over six months: implications for epidemiologic studies. Am J Epidemiol 2018; 187:1282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Singh S, Yang G, Byrareddy SN, Barry MA, Sastry KJ. Natural killer T cell and TLR9 agonists as mucosal adjuvants for sublingual vaccination with clade C HIV‐1 envelope protein. Vaccine 2014; 32:6934–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gray J, Oehrle K, Worthen G, Alenghat T, Whitsett J, Deshmukh H. Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci Transl Med 2017; 9 10.1126/scitranslmed.aaf9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Mu C, Yang Y, Zhu W. Gut microbiota: the brain peacekeeper. Front Microbiol 2016; 7:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Szabo PA, Anantha RV, Shaler CR, McCormick JK, Haeryfar SM. CD1d‐ and MR1‐restricted t cells in sepsis. Front Immunol 2015; 6:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Schwandt T, Schumak B, Gielen GH, Jüngerkes F, Schmidbauer P, Klocke K et al Expression of type I interferon by splenic macrophages suppresses adaptive immunity during sepsis. EMBO J 2012; 31:201–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Selmi C, Balkwill DL, Invernizzi P, Ansari AA, Coppel RL, Podda M et al Patients with primary biliary cirrhosis react against a ubiquitous xenobiotic‐metabolizing bacterium. Hepatology 2003; 38:1250–7. [DOI] [PubMed] [Google Scholar]
- 115. Olafsson S, Gudjonsson H, Selmi C, Amano K, Invernizzi P, Podda M et al Antimitochondrial antibodies and reactivity to N. aromaticivorans proteins in Icelandic patients with primary biliary cirrhosis and their relatives. Am J Gastroenterol 2004; 99:2143–6. [DOI] [PubMed] [Google Scholar]
- 116. De Libero G, Mori L. Professional differences in antigen presentation to iNKT cells. Immunity 2014; 40:5–7. [DOI] [PubMed] [Google Scholar]
- 117. Fujii S, Shimizu K, Okamoto Y, Kunii N, Nakayama T, Motohashi S et al NKT cells as an ideal anti‐tumor immunotherapeutic. Front Immunol 2013; 4:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Tefit JN, Crabé S, Orlandini B, Nell H, Bendelac A, Deng S et al Efficacy of ABX196, a new NKT agonist, in prophylactic human vaccination. Vaccine 2014; 32:6138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang B, Yao M, Lv L. The human microbiota in health and disease. Engineering 2017; 3:71–82. [Google Scholar]
- 120. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 2015; 26:26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gapin L. iNKT cell autoreactivity: what is ‘self’ and how is it recognized? Nat Rev Immunol 2010; 10:272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kawana K, Quayle AJ, Ficarra M, Ibana JA, Shen L, Kawana Y et al CD1d degradation in Chlamydia trachomatis‐infected epithelial cells is the result of both cellular and chlamydial proteasomal activity. J Biol Chem 2007; 282:7368–75. [DOI] [PubMed] [Google Scholar]
- 123. Sokolowska M, Frei R, Lunjani N, Akdis CA, O'Mahony L. Microbiome and asthma. Asthma Res Pract 2018; 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tomkovich S, Jobin C. Microbiota and host immune responses: a love–hate relationship. Immunology 2016; 147:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Mathias A, Pais B, Favre L, Benyacoub J, Corthésy B. Role of secretory IgA in the mucosal sensing of commensal bacteria. Gut Microbes 2014; 5:688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zeng SG, Ghnewa YG, O'Reilly VP, Lyons VG, Atzberger A, Hogan AE et al Human invariant NKT cell subsets differentially promote differentiation, antibody production, and T cell stimulation by B cells in vitro . J Immunol 2013; 191:1666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 2017; 279:70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol 2016; 16:639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. De Paiva CS, Raince JK, McClellan AJ, Shanmugam KP, Pangelinan SB, Volpe EA et al Homeostatic control of conjunctival mucosal goblet cells by NKT‐derived IL‐13. Mucosal Immunol 2011; 4:397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yue SC, Shaulov A, Wang R, Balk SP, Exley MA. CD1d ligation on human monocytes directly signals rapid NF‐κB activation and production of bioactive IL‐12. Proc Natl Acad Sci USA 2005; 102:11811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Yue SC, Nowak M, Shaulov‐Kask A, Wang R, Yue D, Balk SP et al Direct CD1d‐mediated stimulation of APC IL‐12 production and protective immune response to virus infection in vivo . J Immunol 2010; 184:268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Olszak T, Neves JF, Dowds CM, Baker K, Glickman J, Davidson NO et al Protective mucosal immunity mediated by epithelial CD1d and IL‐10. Nature 2014; 509:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Colgan SP, Hershberg RM, Furuta GT, Blumberg RS. Ligation of intestinal epithelial CD1d induces bioactive IL‐10: critical role of the cytoplasmic tail in autocrine signaling. Proc Natl Acad Sci USA 1999; 96:13938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Miura S, Kawana K, Schust DJ, Fujii T, Yokoyama T, Iwasawa Y et al CD1d, a sentinel molecule bridging innate and adaptive immunity, is downregulated by the human papillomavirus (HPV) E5 protein: a possible mechanism for immune evasion by HPV. J Virol 2010; 84:11614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Saez de Guinoa J, Jimeno R, Farhadi N, Jervis PJ, Cox LR, Besra GS et al CD1d‐mediated activation of group 3 innate lymphoid cells drives IL‐22 production. EMBO Rep, 2017; 18:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Albacker LA. Invariant natural killer T cells recognize a fungal glycosphingolipid that can induce airway hyperreactivity. Nat Med 2013; 19:1297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Lotter H, González‐Roldán N, Lindner B, Winau F, Isibasi A, Moreno‐Lafont M et al Natural killer T cells activated by a lipopeptidophosphoglycan from Entamoeba histolytica are critically important to control amebic liver abscess. PLoS Pathog 2009; 5:e1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Amprey JL, Im JS, Turco SJ, Murray HW, Illarionov PA, Besra GS et al A subset of liver NK T cells is activated during Leishmania donovani infection by CD1d‐bound lipophosphoglycan. J Exp Med 2004; 200:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol 2007; 178:2706–13. [DOI] [PubMed] [Google Scholar]
- 140. Tyznik AJ, Tupin E, Nagarajan NA, Her MJ, Benedict CA, Kronenberg M. Cutting edge: the mechanism of invariant NKT cell responses to viral danger signals. J Immunol 2008; 181:4452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Watarai H, Sekine‐Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R et al Development and function of invariant natural killer T cells producing Th2‐ and Th17‐cytokines. PLoS Biol 2012; 10:e1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Fujibayashi Y, Fujimori Y, Kasumoto I, Kai S, Hara H, Okamura H et al Interleukin‐18 regulates T helper 1 or 2 immune responses of human cord blood CD4+ Vα24+Vβ11+ natural killer T cells. Int J Mol Med 2007; 20:241–5. [PubMed] [Google Scholar]
- 143. Smithgall MD, Comeau MR, Yoon BR, Kaufman D, Armitage R, Smith DE. IL‐33 amplifies both Th1‐ and Th2‐type responses through its activity on human basophils, allergen‐reactive Th2 cells, iNKT and NK cells. Int Immunol 2008; 20:1019–30. [DOI] [PubMed] [Google Scholar]
- 144. Bourgeois E, Van LP, Samson M, Diem S, Barra A, Roga S et al The pro‐Th2 cytokine IL‐33 directly interacts with invariant NKT and NK cells to induce IFN‐γ production. Eur J Immunol 2009; 39:1046–55. [DOI] [PubMed] [Google Scholar]
- 145. Terashima A, Watarai H, Inoue S, Sekine E, Nakagawa R, Hase K et al A novel subset of mouse NKT cells bearing the IL‐17 receptor B responds to IL‐25 and contributes to airway hyperreactivity. J Exp Med 2008; 205:2727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Price AE, Reinhardt RL, Liang HE, Locksley RM. Marking and quantifying IL‐17A‐producing cells in vivo . PLoS ONE 2012; 7:e39750. [DOI] [PMC free article] [PubMed] [Google Scholar]


