Abstract
Fungal pathogens cause devastating infections in millions of individuals each year, representing a huge but underappreciated burden on human health. One of these, the opportunistic fungus Cryptococcus neoformans, kills hundreds of thousands of patients annually, disproportionately affecting people in resource-limited areas. This yeast is distinguished from other pathogenic fungi by a polysaccharide capsule that is displayed on the cell surface. The capsule consists of two complex polysaccharide polymers: a mannan substituted with xylose and glucuronic acid, and a galactan with galactomannan side chains that bear variable amounts of glucuronic acid and xylose. The cell wall, with which the capsule is associated, is a matrix of alpha and beta glucans, chitin, chitosan, and mannoproteins. In this review, we focus on synthesis of the wall and capsule, both of which are critical for the ability of this microbe to cause disease and are distinct from structures found in either model yeasts or the mammals afflicted by this infection. Significant research effort over the last few decades has been applied to defining the synthetic machinery of these two structures, including nucleotide sugar metabolism and transport, glycosyltransferase activities, polysaccharide export, and assembly and association of structural elements. Discoveries in this area have elucidated fundamental biology and may lead to novel targets for antifungal therapy. In this review, we summarize the progress made in this challenging and fascinating area, and outline future research questions.
Keywords: capsule, cell wall, Cryptococcus neoformans, glucuronoxylomannan, glucuronoxylomannogalactan
Introduction
Cryptococcal disease has an enormous impact on AIDS patients and other severely immunocompromised populations worldwide, infecting a million individuals each year and killing close to 20% of them (Kwon-Chung et al. 2014; Denning 2016; Rajasingham et al. 2017). Once inhaled, the infectious particles establish a pulmonary infection that may disseminate to cause lethal meningoenceophalitis (Chayakulkeeree and Perfect 2006; Lin and Heitman 2006). Current therapy for Cryptococcus neoformans is complicated by serious adverse reactions, the need for extended treatment to prevent relapse, and poor access to medications in resource-limited settings. Development of novel therapeutics is complicated by the many metabolic features that are shared between fungi and their human hosts, even while those commonalities have made yeast a favored model system for elucidating glycan biochemistry.
Cryptococcus neoformans is unique among fungal pathogens for surrounding its cell wall with an extensive polysaccharide capsule, which is required for virulence (Fromtling et al. 1982; Chang and Kwon-Chung 1994; Steenbergen and Casadevall 2003; Perfect 2005). The wall and the capsule, both primarily composed of glycoconjugates, serve as essential structural elements of the cell (Figure 1) and as the interface between C. neoformans and the cells of infected hosts. Greater understanding of their biosynthesis may suggest new strategies for fungal drug development.
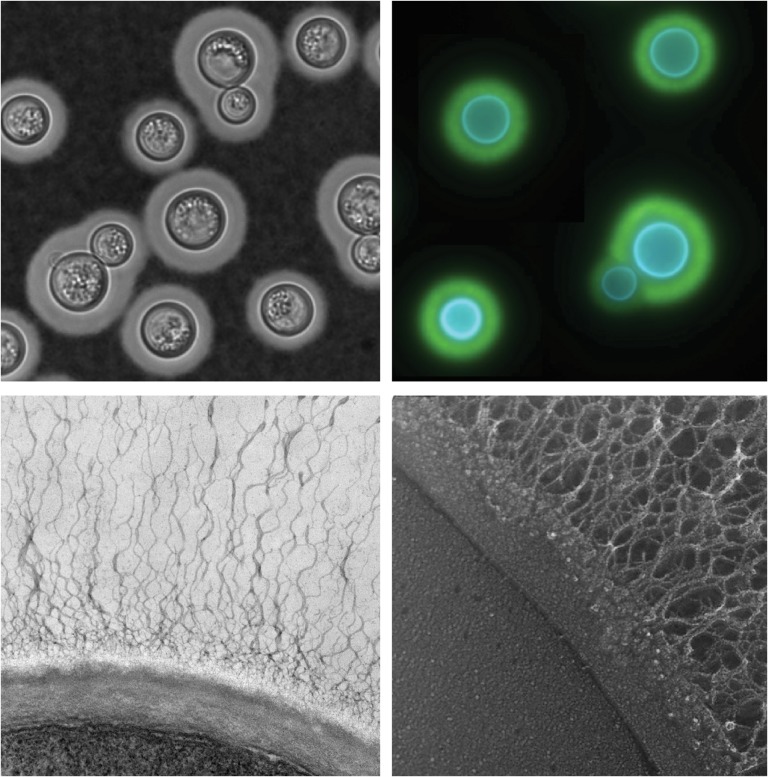
Fig. 1.
Cryptococcus neoformans capsule and cell wall. Clockwise from upper left: Negative stain of cryptococcal cells with India ink; immunofluorescence montage of capsule-induced cells stained with Calcofluor white (blue, stains cell wall) and antibody to the major capsule polysaccharide (green); quick freeze deep-etch electron micrograph (EM) of a portion of the plasma membrane (lower left region of the micrograph), bounded by the two layers of the cell wall and the associated capsule fibers (extending up and to the right); transmission EM of cryptococcal cell edge with capsule fibers extending upwards from the cell wall.
Cell wall
The fungal cell wall is a protective barrier that resists environmental and osmotic stress, while maintaining cellular morphogenesis and regulating membrane permeability (Free 2013). The cryptococcal wall is also critical for the association of capsule polymers with the cell surface (Fonseca et al. 2009; Gilbert et al. 2010; Reese and Doering 2003). Genetic disruption of cell wall synthesis thus reduces cell viability and also diminishes capsule, often yielding avirulent mutants (Gilbert et al. 2011). These features make it an attractive target for development of antifungal therapies (see below), especially because mammalian cells have no equivalent structures.
The cell wall is composed of a matrix of glucose (Glc), N-acetylglucosamine (GlcNAc) and glucosamine (GlcN) polymers (glucan, chitin and chitosan, respectively) with covalently and noncovalently associated glycoproteins. Ultrastructural studies of C. neoformans show that the glycans are arranged in two layers (Figure 1; reviewed in Gilbert et al. 2011). Compositional and imaging studies suggest that the inner layer consists of an alkali-insoluble meshwork of β-glucan and chitin, while the less organized outer layer corresponds to an alkali-soluble fraction containing mainly α- and β-glucans (Reese et al. 2007; Sakaguchi et al. 1993). Studies of mutants with defects in biosynthesis of specific polymers biosynthesis have helped identify the enzymes required for cell wall synthesis and their roles in this process (summarized in Table I).
Table I.
Proteins involved in cell wall synthesis
| Function | Gene | Gene product | Cell wall composition differences | Virulence | Citation |
|---|---|---|---|---|---|
| α-1,3 glucan synthesis | AGS1 | α-1,3 glucan synthase | + | Reduced | Reese and Doering (2003), Reese et al. (2007) |
| β-1,3 glucan synthesisa | FKS1 | β-1,3 glucan synthase | − | Reduced | Thompson et al. (1999) |
| β-1,6 glucan synthesis | KRE5 | β-1,6 glucan synthesis related protein | + | Avirulent | Gilbert et al. (2010) |
| β-1,6 glucan synthesis | KRE6 | β-1,6 glucan synthesis related protein | −b | Avirulent | Gilbert et al. (2010) |
| β-1,6 glucan synthesis | SKN1 | β-1,6 glucan synthesis related protein | −b | Normal | Gilbert et al. (2010) |
| β-1,6 glucan synthesis | KRE61 | β-1,6 glucan synthesis related protein | − | NT | Gilbert et al. (2010) |
| β-1,6 glucan synthesis | KRE62 | β-1,6 glucan synthesis related protein | − | NT | Gilbert et al. (2010) |
| β-1,6 glucan synthesis | KRE63 | β-1,6 glucan synthesis related protein | − | NT | Gilbert et al. (2010) |
| β-1,6 glucan synthesis | KRE64 | β-1,6 glucan synthesis related protein | − | NT | Gilbert et al. (2010) |
| Chitin synthesis | CHS1 | Chitin synthase 1 | − | Normal | Banks et al. (2005) |
| Chitin synthesis | CHS2 | Chitin synthase 2 | − | Normal | Banks et al. (2005) |
| Chitin synthesis | CHS3 | Chitin synthase 3 | + | Avirulent | Baker et al. (2011), Banks et al. (2005) |
| Chitin synthesis | CHS4 | Chitin synthase 4 | + | Normal | Banks et al. (2005) |
| Chitin synthesis | CHS5 | Chitin synthase 5 | + | Normal | Banks et al. (2005) |
| Chitin synthesis | CHS6 | Chitin synthase 6 | − | Normal | Banks et al. (2005) |
| Chitin synthesis | CHS7 | Chitin synthase 7 | − | Normal | Banks et al. (2005) |
| Chitin synthesis | CHS8 | Chitin synthase 8 | − | Normal | Banks et al. (2005) |
| Chitosan synthesis | CDA1 | Chitin deacetylase 1 | −c | NT | Baker et al. (2007), Baker et al. (2011) |
| Chitosan synthesis | CDA2 | Chitin deacetylase 2 | −c | NT | Baker et al. (2007), Baker et al. (2011) |
| Chitosan synthesis | CDA5 | Chitin deacetylase 5 | −c | NT | Baker et al. (2007), Baker et al. (2011) |
| Chitosan synthesis | FPD1 | Putative polysaccharide deacetylase | − | NT | Baker et al. (2007) |
| Cell wall remodeling | PBX1 | Putative glycohydrolase | + | Avirulent | Kumar et al. (2014), Liu et al. (2007) |
| Cell wall remodeling | PBX2 | Putative glycohydrolase | + | Avirulent | Kumar et al. (2014), Liu et al. (2007) |
NT, not tested.
aActivity verified by direct assay.
bAlthough each single mutant wall composition is normal, the kre6Δ skn1Δ double mutant has less cell wall β-1,6-glucan than WT and is reduced in virulence.
cAlthough each single mutant has normal levels of chitosan, its production is abolished in the triple mutant, which is also reduced in virulence.
The cryptococcal cell wall contains abundant α-glucans, with a larger fraction of α-1,3-linked glucan than model yeast like Saccharomyces cerevisiae (Bose et al. 2003; James et al. 1990). These polymers are made by the α-glucan synthase Ags1, a multimembrane-spanning enzyme with a cytosolic synthase domain that transfers Glc residues from UDP-Glc to the nonreducing end of the growing polysaccharide (Bernard and Latge 2001; Grun et al. 2005; Katayama et al. 1999; Reese and Doering 2003). Studies of Ags1 homologs in other fungi suggest that this protein also acts to assemble and translocate the minor amounts of α-1,4-glucan that are found in the cell wall (Vos et al. 2007). Loss of α-1,3-glucan by deletion of AGS1 decimates the outer layer of the cell wall; although increased chitin synthesis and redistribution of β-glucans partly compensate for this change, the mutant cell walls are still malformed, hypertrophic and fragile. These cells are sensitive to environmental stresses, such as high temperature and SDS, and also exhibit growth defects and avirulence in animal models. Importantly, they do not display surface capsule, although they do shed capsule polysaccharides (Reese and Doering 2003; Reese et al. 2007).
β-1,3-glucan, containing β-1,6 branches (Manners et al. 1973), is also present in cryptococcal cell walls (James et al. 1990). Similar to α-glucan, this polymer is synthesized near the plasma membrane by membrane-bound synthases, which add Glc from UDP-Glc donors to the nonreducing end of the growing glucan. In contrast to the three β-1,3-glucan synthases of S. cerevisiae (Lesage and Bussey 2006), C. neoformans encodes only one, Fks1 (Thompson et al. 1999). This protein associates with the GTP-binding protein Rho1, which controls a signal transduction pathway that regulates growth and cell wall integrity. Association with Rho1 is required for the β-1,3-glucan synthase activity of Fks1, enabling the cell to respond to cell wall stress and fungal growth requirements by modulating β-1,3-glucan production. Compounds of the echinocandin family inhibit β-1,3-glucan synthase activity, and these drugs are used routinely in clinical treatment of Aspergillosis and Candidiasis (Free 2013). Unfortunately, they are ineffective against C. neoformans infections (Abruzzo et al. 1997), which is surprising because Fks1 is essential in C. neoformans and is inhibited by these compounds in vitro (Maligie and Selitrennikoff 2005). The lack of clinical efficacy may be due to poor uptake of the drug, its inactivation by C. neoformans, or some other resistance mechanism.
β-1,6-glucan is another major polysaccharide in the C. neoformans cell wall and is more abundant relative to β-1,3-glucan than in other yeasts (James et al. 1990). In S. cerevisiae, this polymer acts to maintain and organize the cell wall through both covalent and noncovalent interactions with β-1,3-glucan, chitin and GPI-anchored proteins (Free 2013); it is thought to function similarly in C. neoformans. β-1,6-glucan synthesis is mediated by a complex of proteins, although the details of protein function have not been established in C. neoformans. Seven β-1,6-glucan synthesis-related genes have been identified in C. neoformans, and mutant analysis suggests that three of them, KRE5, KRE6 and SKN1, likely play major roles in glucan synthesis (Gilbert et al. 2010). Disruption of KRE5 alone or double deletion of KRE6 and SKN1 alter cell wall integrity and cause sensitivity to high temperature and SDS; not surprisingly, both mutants are avirulent (Gilbert et al. 2010). Interestingly, both mutants also exhibit enlarged capsules compared to wild type under capsule-inducing conditions, although the edge of the capsule does not appear as smooth as that of wild type (Gilbert et al. 2010). These observations suggest that β-1,6-glucan plays a role in capsule polysaccharide organization, either directly through interaction with the capsule polymers or indirectly by influencing other cell wall components. Kre6p and Skn1, in addition, share homology with glycoside hydrolases, and may participate in cell wall remodeling (Kurita et al. 2011; Montijn et al. 1999). The double mutant exhibits altered localization of chitosan in the cell wall and increased shedding of GPI-anchored cell wall proteins compared to wild type (Gilbert et al. 2010).
Chitin is a relatively minor component of yeast cell walls, although it is critical for their resilience and integrity (Minke and Blackwell 1978). This water-insoluble β-1,4-GlcNAc polymer is synthesized by a family of plasma membrane-associated chitin synthases that use UDP-GlcNAc as the sugar donor; it has been postulated that the various enzymes act in different spatial, developmental or environmental scenarios. Cryptococcus neoformans encodes eight putative chitin synthases and three potential chitin regulatory proteins (Banks et al. 2005; Doering 2009). Although no single gene of this group is essential for viability, one synthase (Chs3) and one regulator (Csr2) play dominant roles in cell integrity and cell wall function. Deletion of either gene yields stress-sensitive cells with aberrant morphology and the inability to retain melanin (Banks et al. 2005), a cell wall-associated pigment molecule that is important for resisting environmental stress (Agustinho and Nosanchuk 2017; Eisenman and Casadevall 2012; Nosanchuk et al. 2015). Mislocalization of Chs3 due to loss of post-translational lipid modification is associated with dramatic defects in cell wall integrity and ultrastructure, overall cell morphology and virulence (Santiago-Tirado et al. 2015).
Cell wall chitin may be deacetylated to generate chitosan, a more soluble and flexible polymer of glucosamine (Banks et al. 2005). Cryptococcus neoformans has unusually high levels of chitosan (it may exceed chitin by up to 10-fold) (Banks et al. 2005) even during vegetative growth, while in S. cerevisiae chitosan is only produced during sporulation (Christodoulidou et al. 1996). There are three chitin deacetylases in C. neoformans (Cda1, Cda2 and Cda3); all three of the corresponding genes must be deleted to impair chitosan production (Baker et al. 2007). Additionally, mutants lacking chitin synthase 3 (Chs3) or chitin synthesis regulator 2 (Csr2) do not produce any chitosan; it has been suggested that they form a functional complex with chitin deacetylase (Banks et al. 2005). Cells without chitosan grow slower than wild type, with impaired cell integrity and reduced virulence in animal models (Baker et al. 2011), demonstrating the significance of this cell wall component.
Lastly, glycoproteins are key components of the cell wall in fungi, where they act in critical processes including signal transduction, mating, cell wall synthesis and iron acquisition. These proteins are modified by N- and O-linked oligosaccharides, usually with highly mannosylated structures that are initiated in the ER and extended in the Golgi (Lee et al. 2015; Park et al. 2012; Reilly et al. 2011). Cryptococcal protein-linked glycans are more elaborate than those of the model yeast, S. cerevisiae, and include xylose (Xyl) and Xyl-phosphate moieties (Lee et al. 2015; Park et al. 2012; Reilly et al. 2011). The full spectrum of these glycans has not been defined, and they may include additional components, such as sialic acid (Alviano et al. 1999; Rodrigues et al. 2003, 1997). Over half of cryptococcal wall proteins are also modified by the addition of a GPI-anchor in the ER. As in other eukaryotes, synthesis of these anchors begins with the addition of GlcNAc to phosphatidylinositol, followed by deacetylation of the sugar residue and inositol acylation before further elaboration of the GPI structure, although the specificity of inositol acylation is more relaxed than in S. cerevisiae (Franzot and Doering 1999). Following anchor completion and transfer to protein, the GPI-modified polypeptide may remain membrane-associated or be transferred with part of the anchor to covalent linkage with cell wall glucan (Pittet and Conzelmann 2007; Rodrigues and Djordjevic 2012). Proteomic analysis has identified 29 cryptococcal cell wall proteins as GPI-anchored, including multiple proteases, carbohydrate-active enzymes and phospholipase B1 (Plb1) (Eigenheer et al. 2007). Deletion of PLB1 alters cell morphology and results in increased susceptibility to cell wall stress as well as reduced virulence (Siafakas et al. 2007).
The cell wall is a dynamic structure that undergoes constant remodeling, including modulation of the distribution and cross-linking of its components to accommodate cellular growth and division. The putative glycohydrolase proteins Pbx1 and Pbx2 (Liu et al. 2007), which may participate in this remodeling process (Kumar et al. 2014; Liu et al. 2007), are also required for normal capsule association with the wall and for fungal virulence (Kumar et al. 2014; Liu et al. 2007). The flexibility and adaptability of the cell wall allows fungal cells to maintain integrity through the extraordinary morphologic transformations of growth and development that are part of their life cycle, and to respond to challenges from external stresses including those encountered in mammalian hosts. The key roles of the C. neoformans cell wall in cellular responses to stress, capsule assembly and display of surface antigens, combined with the unfulfilled potential of cryptococcal cell wall glycans as therapeutic targets, highlight the need to pursue the many unanswered questions in this area.
Capsule
The polysaccharide capsule surrounding the cell wall of C. neoformans distinguishes it from other fungal pathogens (Doering 2009; Kumar et al. 2011). Production of capsule polymers is required for cryptococcal disease in animal models (Chang and Kwon-Chung 1994), and mutants exhibiting morphological or structural changes to the capsule generally have reduced virulence (Bulmer et al. 1967; Kwon-Chung and Rhodes 1986). Capsule thickness also changes dramatically with environmental conditions, ranging from undetectable by negative staining to up to 30 μm (Garcia-Hermoso et al. 2004; Guimaraes et al. 2010; Zaragoza and Casadevall 2004)—a remarkable size for cells typically only 3–5 μm in diameter. Upon exposure to a host or host-like conditions, C. neoformans rapidly induces capsule synthesis, although the final capsule thickness depends on the specific environment (Granger et al. 1985). For example, the capsules of cells isolated from the lung and the brain vary greatly in size within the same infected individual (Rivera et al. 1998). The change in capsule thickness is likely mediated at the level of synthesis of individual capsule polysaccharide molecules (Frases et al. 2009; Yoneda and Doering 2008), and the expansion in capsule size can exceed the growth rate of the cell body (Cordero et al. 2013).
Capsule production is regulated at the level of transcription, as an integrated response to signaling pathways triggered by stimuli including hypoxia, low iron, low glucose and physiological pH. Interestingly, it is regulated independently of protein glycosylation; expression of most known capsule-related factors is upregulated when capsule is induced, while that of genes encoding known N-glycosylation enzymes is generally down-regulated or unchanged in the same conditions (Maier et al. 2015). We and others have probed the interactions between signaling pathways, transcription factors and downstream synthetic machinery targets that influence capsule synthesis, with the goal of understanding and potentially influencing regulation of this central virulence factor (Chun and Madhani 2010; Gish et al. 2016; Haynes et al. 2013, 2011; Jung et al. 2016, 2015; Kim et al. 2015; Lee et al. 2016; Liu et al. 2008; Maier et al. 2015).
The capsule is composed primarily of two polysaccharides, glucuronoxylomannan (GXM) and glucuronoxylomannogalactan (GXMGal), with trace amounts of mannoproteins (Cherniak et al. 1980; Turner et al. 1984). Both GXM and GXMGal have been implicated in cryptococcal virulence as modulators of the host immune response, inhibiting immune recognition and activation (Kumar et al. 2011; Zaragoza et al. 2009). Hyaluronic acid has also been detected in the capsule, at the interface with the outer cell wall (Chang et al. 2006). This polymer of GlcA and GlcNAc is required for survival at host temperature and for efficient adhesion to endothelial cells, which is in turn a prerequisite for successful infection of the brain by free fungi (Chang et al. 2006; Jong et al. 2007).
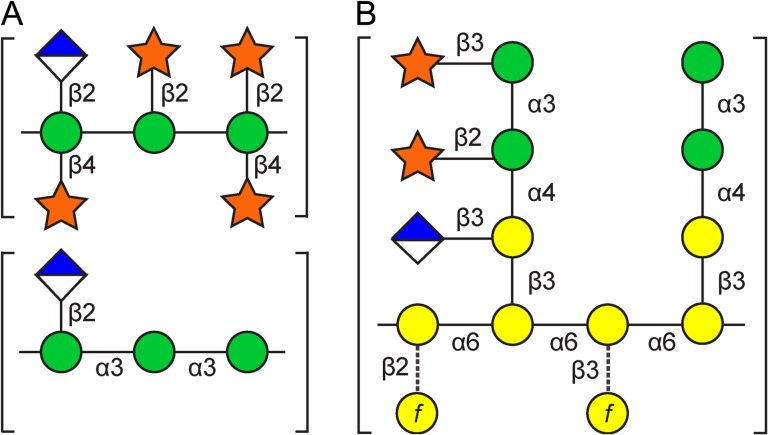
GXM (1700–7000 kDa) accounts for roughly 90% of the capsule mass (Cherniak and Sundstrom 1994; Cherniak et al. 1998; McFadden et al. 2006). It consists of an α-1,3-linked Man backbone substituted with glucuronic acid (GlcA) in β-1,2-linkage and xylose in β-1,2 and β-1,4 linkage (Cherniak et al. 1998; Turner et al. 1992), in a repeating trimer pattern (Figure 2A). NMR analysis of capsule polymers defined six distinct structural reporter groups of GXM that differ in the extent of Xyl and 6-O-acetyl modification of the Man backbone (Cherniak et al. 1998). The differences in the relative proportion of these structural motifs in different serotypes likely result in the distinct antigenic properties of GXM from those strains (Fries et al. 1999).
Fig. 2.
Structures of the capsule polysaccharides. (A) GXM. Schematic of the repeating trimer structure, showing the most and least highly substituted reporter groups. (B) GXMGal. Schematic showing examples of the most and least highly substituted side chains on the galactose backbone. Dashed lines indicate rare modifications. Green circle, mannose; diamond, GlcA; star, Xyl; yellow circle, Gal and Galf. Some mannose residues are also 6-O-acetylated in both capsule polysaccharides and 2-O-acetylated in GXMGal (not shown). See text for details.
GXMGal (Figure 2B) is relatively small (~105 Da) in comparison to GXM and comprises the remainder of the capsule mass. (This capsule polysaccharide was previously termed galactoxylomannan (GalXM) (Cherniak et al. 1982), but was renamed in 2009 to better reflect updated structural studies (Heiss et al. 2009)). GXMGal consists of an α-1,6 linked galactose (Gal) backbone with side chains of galactose and mannose substituted with a variable number of β-linked glucuronic acid (GlcA) and Xyl residues (Cherniak et al. 1982; Heiss et al. 2009; Turner et al. 1984). Some of the galactose backbone is also substituted with galactofuranose (Galf) (De Jesus et al. 2009; Heiss et al. 2013; Previato et al. 2017; Vaishnav et al. 1998).
In contrast to cell wall synthesis, where most of the synthetic enzymes are known (Table I), few of the glycosyltranferases required to generate GXM and GXMGal have been identified. This reflects the challenge of predicting biochemical activity from primary sequence for these enzymes, combined with the fact that many of the capsule polysaccharide linkages are unique, which limits the utility of homology to known proteins in other organisms in predicting their function. The upstream biosynthetic machinery for capsule components is better understood, because much of it also participates in synthesis of other glycoconjugates and is conserved in eukaryotic biology (Table II).
Table II.
Enzymes involved in capsule synthesis
| Function | Gene | Gene product | Protein assayed | Mutant phenotype | Citation | ||
|---|---|---|---|---|---|---|---|
| Altered PS | Capsule size | Virulence | |||||
| UDP-Gal synthesis | UGE1 | UDP-Glc 4-epimerase | − | GXMGal | Reduced | Avirulent | Moyrand et al. (2008) |
| GDP-Man synthesisa | MAN1 | Phosphomannose isomerase | + | GXM, GXMGal | Reduced | Reduced | Wills et al. (2001) |
| UDP-GlcA synthesis | UGD1 | UDP-Glc dehydrogenase | + | GXM, GXMGal | Reduced | Avirulent | Bar-Peled et al. (2004), Moyrand and Janbon (2004) |
| UDP-Xyl synthesis | UXS1 | UDP-GlcA decarboxylase | + | GXM, GXMGal | Reduced | Avirulent | Bar-Peled et al. (2001), Moyrand et al. (2002) |
| UDP-Galf synthesis | UGM | UDP-galactopyranose mutase | + | GXMGal | Normal | Normal | Heiss et al. (2013), Beverley et al. (2005) |
| GDP-Man transport | GMT1 | GDP-Man transporter | + | GXM, GXMGal | Reducedb | Normal | Cottrell et al. (2007), Wang et al. (2014) |
| GDP-Man transport | GMT2 | GDP-Man transporter | + | GXM, GXMGal | Normalb | Normal | Cottrell et al. (2007), Wang et al. (2014) |
| UDP-Galp transport | UGT1 | UDP-Galp transporter | + | GXMGal | Reduced | Avirulent | Li et al. (2017), Moyrand et al. (2007) |
| UDP-GlcA transport | UUT1 | UDP-GlcA transporter | + | GXM, GXMGal | Reduced | Avirulent | Li et al. (2018b) |
| UDP-Xyl/UDP-Galf transport | UXT1 | UDP-Xyl /UDP-Galf transporter | + | GXM, GXMGal | Normalc | Normal | Li et al. (2018a) |
| UDP-Xyl /UDP-Galf transport | UXT2 | UDP-Xyl/UDP-Galf transporter | + | GXM, GXMGal | Normalc | Normal | Li et al. (2018a) |
| Capsule modification | CAS1 | Putative O-acetyltransferase | − | GXM, GXMGal | Reduced | Normal | Janbon et al. (2001) |
| Capsule modification | CXT1 | β-1,2-xylosyltransferase | + | GXM, GXMGal | Normal | NT | Klutts and Doering (2008), Klutts et al. (2007) |
PS, capsular polysaccharide; NT, not tested.
aMan1 catalyzes the first reaction in the pathway for synthesizing GDP-Man (conversion of fructose-6-phosphate to mannose-6-phosphate).
bThe double mutant lacking GMT1 and GMT2 completely lacks capsule and is avirulent.
cuxt1Δ uxt2Δ has a reduced capsule and a virulence defect.
As in many other glycan synthetic pathways, nucleotide sugars are the donor molecules for synthesis of the capsule polysaccharides. Based on the polysaccharide structures (Figure 2), we expect the relevant donors to be GDP-Man, UDP-Galp, UDP-GlcA, UDP-Xyl and UDP-Galf; the synthesis of these compounds is reviewed below and depicted in Figure 3. Sialic acid has also been suggested as a capsule component (Chang et al. 2006; Gahrs et al. 2009; Jong et al. 2007), although analysis of the genome does not reveal homologs of the synthetic machinery required to produce appropriate donors.
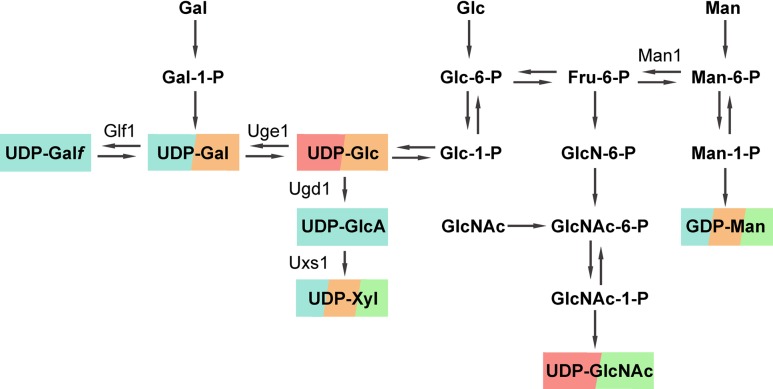
Fig. 3.
Pathways of nucleotide sugar production, with the C. neoformans glycan synthetic pathways they are known to supply. Blue, capsule polysaccharides; orange, glycolipids; green, glycoproteins; pink, cell wall polymers. Bold font, metabolic intermediates and products; regular font, enzymes that have been specifically studied in C. neoformans (other enzymes are not indicated). All sugars are in pyranose forms except for Galf.
GDP-Man, required for synthesis of the GXM backbone and GXMGal side chains, is made through the sequential action of phosphomannose isomerase (Man1 in C. neoformans) (Wills et al. 2001), phosphomannomutase, and GDP-Man pyrophosphorylase on fructose-6-phosphate (Figure 3). As expected for a major precursor of capsule, disrupting GDP-Man synthesis, for example, by deleting MAN1, markedly reduces capsule production and results in an inability to cause disease in mice (Wills et al. 2001).
UDP-Galp is synthesized from UDP-Glc by Uge1, a UDP-glucose 4-epimerase. Cryptococcal mutants lacking Uge1 have no Gal residues in their capsules, consistent with the expected inability to synthesize GXMGal (Moyrand et al. 2007). UDP-Galp may be further modified by UDP-galactopyranose mutase, Glf1, to produce UDP-Galf (Beverley et al. 2005). NMR analysis of capsule glycans isolated from glf1Δ cells confirmed the absence of Galf from GXMGal, although this does not apparently alter capsule size or organization; it also does not alter fungal stress resistance or virulence (Heiss et al. 2013).
UDP-Glc is dehydrogenated by Ugd1 to generate UDP-GlcA; this in turn may be decarboxylated by Uxs1 to form UDP-Xyl (Bar-Peled et al. 2001, 2004). Deleting UGD1 completely abrogates the synthesis of both UDP-GlcA and UDP-Xyl, and disrupts the production of capsule (Griffith et al. 2004), even in the presence of backbone precursors for both GXM and GXMGal. Only trace fibrous material, which may represent abnormally truncated forms of capsule polysaccharides, is observed on the surface of these mutants by electron microscopy (EM). The mutant also demonstrates impaired cell wall integrity and various morphological defects that are not shared by acapsular cells (Griffith et al. 2004; Moyrand and Janbon 2004), which suggests that these two sugar donors are utilized in glycosylation processes other than capsule synthesis. Deletion of UXS1 interrupts UDP-Xyl biosynthesis alone, yielding thin capsules compared to wild type, with deformed fibers (Griffith et al. 2004; Moyrand et al. 2002). Both the ugd1Δ and uxs1Δ mutants are avirulent in mice, consistent with their severe capsule defects (Griffith et al. 2004; Moyrand and Janbon 2004; Moyrand et al. 2002).
While donor molecules such as nucleotide sugars are mainly synthesized in the cytosol, the glycosyltransferase-mediated reactions that generate the majority of C. neoforman’s glycan repertoire, including the capsule polysaccharides, occur in the secretory pathway (Figure 4). The highly charged donors thus require specific transport machinery to cross membranes and gain access to the secretory compartment (Freeze and Elbein 2009); the proteins that serve this function are called nucleotide sugar transporters (NSTs).
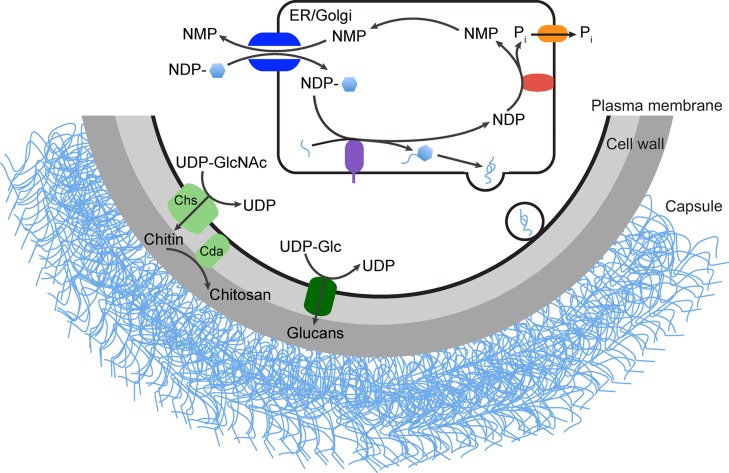
Fig. 4.
Capsule and cell wall biosynthesis. Activated nucleotide sugar donors (shown as NDP-hexagon) are synthesized in the cytosol. Some glycans, including the chitin and glucan polymers of the cell wall, are made directly from these cytosolic precursors by synthetic enzymes (green). Other glycan structures are made in the secretory pathway (ER/Golgi), including protein N- and O-linked glycans and capsule polymers. Nucleotide sugar precursors for these reactions are transported into the secretory compartment by NSTs (dark blue) for use by glycosyltransferase enzymes (purple) located there; this cartoon shows elongation and subsequent export of a capsule fiber (light blue). In many cases glycan synthetic reactions release an NDP moiety that is subsequently cleaved by NDPase (red); the resulting phosphate is exported by the phosphate transporter (orange) while the NMP product serves as an NST antiport substrate.
We used homology to known proteins to identify 10 genes that potentially encode NSTs in the cryptococcal genome. Two of these candidates encode a pair of GDP-Man transporters, Gmt1 and Gmt2 (Cottrell et al. 2007; Wang et al. 2014). Cells lacking either gene still produce capsule although the capsule of gmt1Δ, but not gmt2Δ, is thinner than that of wild type. Deletion of both genes eliminates any detectable capsule synthesis, similar to the phenotype of mutants incapable of synthesizing GDP-Man, although surprisingly the cells remain viable. This is in contrast with S. cerevisiae, where the single GDP-Man transporter is essential (Dean et al. 1997), and suggests that either another transporter can move this precursor or the mannose residues it donates in the secretory pathway are not critical for cell viability.
Our set of 10 potential NSTs also included a UDP-Galp transporter that had been identified by Moyrand et al. (2007), which we confirmed biochemically (Li et al. 2017), two proteins that we have demonstrated to be UDP-Xyl/Galf transporters (Li et al. 2018a), and one that we have shown to transport UDP-GlcA (Li et al. 2018b). Together, the known NSTs account for transport of each activated donor incorporated into the capsule. Completely interrupting the transport of any of these nucleotide sugars, either through single or double knockouts as appropriate, disrupts capsule polysaccharide production and leaves the mutant cells avirulent (Griffith et al. 2004; Li et al. 2017, 2018a, 2018b; Moyrand et al. 2007; Wang et al. 2014). The other four putative NSTs that we identified remain to be investigated; they may transport nucleotide sugars involved in as yet uncharacterized cellular processes or related molecules required for capsule synthesis, such as donors for acetylation reactions.
Glycosyltransferases catalyze the transfer of sugar moieties from activated donors to growing glycan structures, lipids or polypeptides, with specificity for the sugar donor, acceptor and linkage created. The complexity of the cryptococcal capsule polysaccharide structures (Figure 2) suggests that a variety of these enzymes are required for their synthesis. Although roughly 70 cryptococcal genes have been identified as encoding glycosyltransferases (http://www.cazy.org/; (Lombard et al. 2014)), only one has been implicated in capsule synthesis to date (Klutts and Doering 2008). This is partly due to the difficulty of predicting function from sequence, especially when most comparisons are to model yeast with much simpler glycans and no capsule. Confounding the situation is the challenge of assaying these enzymes and the functional redundancy that is common among this family of proteins, especially in fungi.
Cxt1, a β-1,2-xylosyltransferase, is the only glycosyltransferase known to be involved in capsule synthesis (Klutts and Doering 2008; Klutts et al. 2007). In vitro, this protein catalyzes the cation-independent addition of Xyl to the reducing sugar of a Man-α-1,3-Man disaccharide in β-1,2-linkage. cxt1Δ cells lack all β-1,2-linked Xyl in GXMGal and ∼30% of such linkages in GXM, demonstrating a role of this enzyme in capsule synthesis (Klutts and Doering 2008). These mutants also lack β-1,3-linked Xyl in GXMGal, although this may be an indirect effect of loss of the proximal xylose (Klutts and Doering 2008). The defects in xylosylation are not limited to the two capsule glycans but also affect lipid glycosylation (Castle et al. 2008; Reilly et al. 2011), suggesting shared glycan machinery between these biosynthetic pathways.
Xylosyltransferases beyond Cxt1 must exist in C. neoformans, because GXM retains much of its β-1,2-linked Xyl and all β-1,4-linked Xyl despite the loss of this activity. We identified five CXT1 homologs in the cryptococcal genome (Klutts et al. 2007) as candidates for such activity. One of them, Cap10, is already known to be required for capsule formation (Chang and Kwon-Chung 1999; Tefsen et al. 2014), although its activity is not known, while another, Cxt2, also mediates β-1,2-linked Xyl transfer (DP Agustinho, JS Klutts, et al. in preparation). Further biochemical characterization will be required to define the roles of these CXT1 homologs in capsule synthesis and other glycosylation processes.
Studies of other candidates for capsule-implicated glycosyltransferases have proved inconclusive. For example, the GXM backbone is composed of α-1,3-linked Man residues, a linkage also found in the side chains of GXMGal. A protein that catalyzes this Man transfer, Cmt1, was purified from cryptococcal extracts, but deletion of the corresponding gene did not cause any obvious morphologic changes in the capsule (Sommer et al. 2003). It may be that cells compensate for the loss of this glycosyltransferase with functionally redundant enzymes. Supporting this idea, two Cmt1 homologs in C. neoformans, Cap6 and Cap59, are required for capsule formation and virulence in animal models (Chang and Kwon-Chung 1994, 1998), although their biochemical functions are not known. Alternatively, the function of Cmt1 may be unrelated to capsule production. Supporting this hypothesis, Cap59-like protein A in Aspergillus fumigatus (ClpA) is an α-1,3-mannosyltransferase involved in GPI-anchor maturation (Kruger et al. 2016). The biochemical roles of Cmt1 and its relatives thus remain an open question; there may also be unrelated mannosyltransferases that mediate C. neoformans capsule synthesis.
In addition to the monosaccharide components that they have in common, both capsule polymers are O-acetylated. In GXMGal, 80% of the side chain Man residues are acetylated at either the 2-position (75%) or the 6-position (25%), which likely influences the immunoregulatory and immunogenic properties of this polymer (Previato et al. 2017). In GXM, acetylation occurs preferentially on unsubstituted backbone Man residues compared to those that bear GlcA, and does not occur at all on Xyl-substituted Man (Janbon et al. 2001). The overall level of acetylation varies between 20 and 66% (Bacon et al. 1996; Cherniak et al. 1980; Ellerbroek et al. 2004; James et al. 1990; Turner et al. 1992) and contributes to the antigenic properties of GXM, which is the immunodominant epitope of capsule (Cherniak et al. 1980; Kozel et al. 2003; Wozniak and Levitz 2009). The use of O-acetylation-dependent anti-GXM monoclonal antibodies to screen mutants led to the identification of a putative O-acetyltransferase, Cas1 (Janbon et al. 2001). This protein is required for GXM O-acetylation, although its precise biochemical activity has yet to be characterized (Ellerbroek et al. 2004). Double deletion of genes called CAS3 and CAS31 also eliminates O-acetylation and, interestingly, alters xylosylation of GXM (Moyrand et al. 2004).
Defining the order of the synthetic events that generate GXM and GXMGal will require further identification and elucidation of capsule biosynthetic machinery. Some hints as to reaction order, however, may be gleaned by compositional and linkage analysis of glycoconjugates from mutants in the known synthetic pathways. For example, the abrogation of UDP-GlcA transport alone prevents production of capsule and/or a recognizable precursor molecule (Li et al. 2018b) although GlcA occurs only as a monomeric side chain in both capsule polymers (Cherniak et al. 1998; Heiss et al. 2009). This suggests that GlcA is required for capsule polysaccharide synthesis, although this may be either direct (e.g. as part of the substrate necessary for the action of xylosyl- or mannosyltransferases) or indirect (e.g. to prevent degradation of nascent capsule polymers). Alternatively, the UDP-GlcA transporter may be required to assemble a functional protein complex for capsule synthesis, or to provide precursor for some protein modification that is required for capsule synthesis. In contrast, the absence of Xyl or O-acetyl modifications of Man backbone residues, which are mutually exclusive in GXM, does not impact the addition of additional Xyl or GlcA (Griffith et al. 2004; Janbon et al. 2001; Li et al. 2018a, 2018b, Moyrand et al. 2002). These findings suggest that while the addition of Xyl may be influenced by the acetylation status of backbone Man, or vice versa, both are relatively late steps in the capsule synthetic process.
Following synthesis, capsule glycans must travel to the cell surface for display or release (Figure 4). This likely occurs through the classical secretory pathway, since immuno-EM studies show that mutants in this pathway accumulate secretory vesicles containing material recognized by anti-GXM monoclonal antibodies (Yoneda and Doering 2009). We do not yet know, however, whether this capsule material consists of short segments (e.g. the repeating trimer unit of GXM) that are subsequently fused when they are released into the extracellular space, or long polysaccharide chains that are synthesized in the luminal space before being packaged (Doering 2009). The pattern of antibody deposition, on both the cell surface and in vesicles, and the appearance of some micrographs are consistent with the export of large polysaccharides in secretory vesicles, rather than subunit assembly at the cell surface, although it has been challenging to show this directly (Doering 2009).
Exported capsule fibers associate with the cell surface in a process that is independent of capsule polysaccharide production (Bulmer and Sans 1968; Kozel 1977; Kozel and Hermerath 1984); many acapsular mutants bind capsule material shed from wild-type cells even though they shed none themselves (Kumar et al. 2014). α-Glucanase treatment of acapsular cells eliminates this ability, and mutants lacking α-1,3-glucan display no surface capsule (although capsule polysaccharide shedding is unaffected). These observations suggest that this cell wall polysaccharide participates in capsule polymer association (Reese and Doering 2003; Reese et al. 2007), potentially interacting with capsule material directly or via some glycan-binding protein. α-1,3-Glucan has also been reported to occur within the capsule, in a scattered pattern (Cordero, Pontes et al. 2011), which may be evidence of cell wall components being shed from the cell surface or of a role for this material in capsule assembly. Treatment of cells with chitinase or chitin synthesis inhibitors after capsule induction yields thinner capsules and an increase in shed capsule polysaccharides (Fonseca et al. 2013, 2009; Gilbert et al. 2010; Ramos et al. 2012; Rodrigues, Alvarez et al. 2008). This implicates chitin in capsule organization and/or attachment, although this again could be via direct interactions or by indirectly influencing other aspects of cell wall structure.
The integration of new capsule polysaccharide into the existing structure has been controversial. One model is that capsule enlargement occurs by the addition of new material at the distal edge (Zaragoza et al. 2006). Another hypothesis is that polysaccharide material is added at the inner portion of the capsule near the cell wall, and then gradually displaced towards the outer edge by subsequent polymer incorporation. This is supported by pulse-chase labeling of capsule using fluorophore-tagged antibodies or radiolabelled sugars (Pierini and Doering 2001) and by live imaging (Cordero et al. 2013).
How capsule fibers interact is also incompletely understood, although they may self-associate (Evans 1960; Kozel and Hermerath 1984; McFadden et al. 2006) and their assembly on the cell surface is dependent on the concentration of divalent cations (Nimrichter et al. 2007). Depletion of Ca2+ by EDTA, for instance, increases GXM shedding, and decreases capsule size and the viscosity of GXM. It has been postulated that the calcium ions form salt bridges between the negatively charged GlcA residues of neighboring GXM fibers to facilitate the extension and stability of capsule fibers on the cell (Nimrichter et al. 2007).
Although GXM constitutes the majority of the capsule material, GXMGal may play a key role in capsule organization, since several mutants defective in GXMGal production actually have thicker capsules (Li et al. 2017; Moyrand et al. 2007). It has also been observed that capsule material is progressively cross-linked over time, as suggested by quick-freeze deep-etch EM studies during capsule maturation (Pierini and Doering 2001) and by the greater susceptibility of newly incorporated radiolabelled Man in the capsule to gamma radiation-induced release compared to older material (Zaragoza et al. 2006). Capsule polymer organization may be further modulated by β-1,6 glucan or chitin, as discussed above.
In addition to cross-linking of distinct capsule fibers, individual polysaccharide polymers may be branched, an idea supported by viscosity, light scattering and morphological measurements (Cordero, Frases et al. 2011). Whether and how branching is regulated remains largely unexplored. One advance in this area is the discovery that a putative secreted lactonohydrolase, Lhc1, decreases the structural complexity of capsule polysaccharides, although the mechanism is not known. Deletion of LHC1 also yields a larger capsule (Park et al. 2014).
Capsule polymers are released into the extracellular space, where they modulate host responses and facilitate traversal of the blood–brain barrier (Vecchiarelli et al. 2011, 2013). This shed material may be liberated in some way from the existing cell wall-associated structure. Alternatively, free polysaccharide could derive from newly synthesized polysaccharide that exits the cell via classical secretion, as suggested by the significant reduction in soluble secreted capsular material observed following loss of function mutations in that export pathway. Capsule polysaccharide may also be contained in vesicles that are shed from cryptococcal cells (Rodrigues, Nakayasu et al. 2008; Rodrigues et al. 2007); this is supported by the observation that RNAi suppression of extracellular vesicle formation reduces capsule shedding (Panepinto et al. 2009). Cells in this study still exhibited capsules of normal thickness, which suggests that glycans destined to be incorporated into the capsule and those that are released into the extracellular millieu might exit the cell through independent pathways. A similar pattern, of reduced capsule shedding with normal capsule thickness, is seen upon elimination of a putative flippase (Rizzo et al. 2013). If there are two distinct pathways, they may compete for capsular material; disruption of the ESCRT-I (endosomal sorting complex required for transport) protein Vps23 decreased capsule size and increased shedding of capsule polysaccharides (Hu et al. 2013). This combination of phenotypes may also result if any component of the mechanisms for capsule assembly or association with the cell is perturbed, as with the cells lacking Ags1 discussed above (Reese and Doering 2003; Reese et al. 2007). Further examples of dissociation of capsule thickness and shedding are provided by studies of regulatory mutants. In these strains capsule enlargement and shedding can vary either coordinately or in opposite directions (Gish et al. 2016; Kmetzsch et al. 2011; O’Meara et al. 2014), perhaps reflecting global regulation of capsule polysaccharide synthesis versus specific regulation of capsule association with the cell surface or of individual export pathways.
Finally, it is important to consider that most investigations of capsule composition and function use the readily available material that accumulates in culture supernatant fractions, assuming that shed polysaccharides are derived from the capsule (or at least the same intracellular pool) and that the biochemical properties and structural characteristics are representative of those of capsule polysaccharides. However, capsule glycans released from cells by dimethylsulfoxide treatment or gamma radiation exhibit distinct physical, chemical and immunogenic properties as compared to those isolated either by precipitation or filtration (Frases et al. 2008). Although some of the differences (e.g. elevated glucose content) may be due to cell wall disruption by these treatments or to the method of isolation (Cordero, Pontes et al. 2011; Reese and Doering 2003), the relationship between intact capsule polysaccharides and shed materials will require further investigation.
Conclusion
Cryptococcal capsule and cell wall synthesis are fascinating and highly dynamic processes that are extremely responsive to the metabolic state of the cell and environmental stimuli. Studies in these areas have progressed, so that we now have a good understanding of upstream events, such as nucleotide sugar synthesis and transport. Most of the enzymes required for cell wall synthesis have also been identified, along with a few that act to build capsule components.
Despite significant recent progress, multiple compelling questions about cell wall and capsule synthesis remain to be answered. For the cell wall, outstanding questions include the details of its maturation and response to environmental cues, the role of chitosan, the inability of echinocandins to inhibit synthesis in vivo, and the mechanisms of capsule polysaccharide binding. Many more gaps remain in our understanding of capsule synthesis. Most of the glycosyltransferases that synthesize capsule polysaccharides remain to be discovered, along with their specificity and how they are regulated. We do not know whether these proteins form complexes or associate with other components of the capsule synthetic machinery such as NSTs, or where they occur in the secretory pathway. The enzymes that mediate capsule acetylation and the substrates of unassigned NST-like proteins that influence capsule synthesis also still remain a mystery. Finally, although we are beginning to infer the order of synthetic events from the study of mutants, this still remains to be defined, along with the temporal and spatial dynamics of capsule synthesis and organization.
Despite the daunting list of questions, new tools suggest that progress towards answering them will accelerate in the coming years. Recent experimental advances, including computational modeling and network analysis, biochemical tools for glycoconjugate analysis and discrimination, and the availability of genome-wide resources for study of C. neoformans, will undoubtedly propel this field forward. The fundamental understanding that will result from these investigations will advance the field of glycobiology, inform the study of other pathogenic microbes, and move us closer to influencing these synthetic processes to improve human health.
Acknowledgements
We thank Liza C. Miller and Daniel Agustinho for their constructive comments on the article.
Abbreviations
- EM
electron microscopy
- ER
endoplasmic reticulum
- ESCRT
endosomal sorting complex required for transport
- Gal
galactose
- Galf
galactofuranose
- Galp
galactopyranose
- Glc
glucose
- GlcA
glucuronic acid
- GlcN
glucosamine
- GlcNAc
N-acetylglucosamine
- GXM
glucuronoxylomannan
- GXMGal
glucuronoxylomannogalactan
- GPI
glycophosphatidylinositol
- Man
mannose
- NST
nucleotide sugar transporter
- Xyl
xylose
Funding
Unpublished work on cryptococcal glycan synthesis that is cited here was supported by funding from the National Institutes of Health to T.L.D. (grants R21 AI109623 and R01 GM 071007). Z.A.W. was partly supported by an Alexander and Gertrude Berg Departmental Fellowship from the Department of Molecular Microbiology, Washington University School of Medicine. L.X.L. received funding from a National Institutes of Health training grant (T32 GM007200), a Sondra Schlesinger Graduate Fellowship from the Department of Molecular Microbiology, Washington University School of Medicine, and a National Institute of Health Predoctoral Fellowship Award (F30 AI120339).
Conflict of interest statement
None declared.
References
- Abruzzo GK, Flattery AM, Gill CJ, Kong L, Smith JG, Pikounis VB, Balkovec JM, Bouffard AF, Dropinski JF, Rosen H et al. . 1997. Evaluation of the echinocandin antifungal MK-0991 (L-743,872): Efficacies in mouse models of disseminated aspergillosis, candidiasis, and cryptococcosis. Antimicrob Agents Chemother. 41:2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agustinho DP, Nosanchuk JD. 2017. Functions of Fungal Melanins. Reference Module in Life Sciences. Amsterdam: Elsevier; Available from: http://www.sciencedirect.com/science/article/pii/B9780128096338120916. [Google Scholar]
- Alviano CS, Travassos LR, Schauer R. 1999. Sialic acids in fungi: A minireview. Glycoconjugate J. 16:545–554. [DOI] [PubMed] [Google Scholar]
- Bacon BE, Cherniak R, Kwon-Chung KJ, Jacobson ES. 1996. Structure of the O-deacetylated glucuronoxylomannan from Cryptococcus neoformans Cap70 as determined by 2D NMR spectroscopy. Carbohydr Res. 283:95–110. [DOI] [PubMed] [Google Scholar]
- Baker LG, Specht CA, Donlin MJ, Lodge JK. 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell. 6:855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LG, Specht CA, Lodge JK. 2011. Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot Cell. 10:1264–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, Lodge JK. 2005. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 4:1902–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M, Griffith CL, Doering TL. 2001. Functional cloning and characterization of a UDP- glucuronic acid decarboxylase: The pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc Natl Acad Sci USA. 98:12003–12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M, Griffith CL, Ory JJ, Doering TL. 2004. Biosynthesis of UDP-GlcA, a key metabolite for capsular polysaccharide synthesis in the pathogenic fungus Cryptococcus neoformans. Biochem J. 381:131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard M, Latge JP. 2001. Aspergillus fumigatus cell wall: Composition and biosynthesis. Med Mycol. 39(Suppl 1):9–17. [PubMed] [Google Scholar]
- Beverley SM, Owens KL, Showalter M, Griffith CL, Doering TL, Jones VC, McNeil MR. 2005. Eukaryotic UDP-galactopyranose mutase (GLF gene) in microbial and metazoal pathogens. Eukaryot Cell. 4:1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I, Reese AJ, Ory JJ, Janbon G, Doering TL. 2003. A yeast under cover: The capsule of Cryptococcus neoformans. Eukaryot Cell. 2:655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer GS, Sans MD. 1968. Cryptococcus neoformans. 3. Inhibition of phagocytosis. J Bacteriol. 95:5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer GS, Sans MD, Gunn CM. 1967. Cryptococcus neoformans. I. Nonencapsulated mutants. J Bacteriol. 94:1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle SA, Owuor EA, Thompson SH, Garnsey MR, Klutts JS, Doering TL, Levery SB. 2008. Beta1,2-xylosyltransferase Cxt1p is solely responsible for xylose incorporation into Cryptococcus neoformans glycosphingolipids. Eukaryot Cell. 7:1611–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Jong A, Huang S, Zerfas P, Kwon-Chung KJ. 2006. CPS1, a homolog of the Streptococcus pneumoniae type 3 polysaccharide synthase gene, is important for the pathobiology of Cryptococcus neoformans. Infect Immun. 74:3930–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 14:4912–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. 1998. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect Immun. 66:2230–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J Bacteriol. 181:5636–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayakulkeeree M, Perfect JR. 2006. Cryptococcosis. Infect Dis Clin North Am. 20:507–544, v–vi. [DOI] [PubMed] [Google Scholar]
- Cherniak R, Reiss E, Slodki ME, Plattner RD, Blumer SO. 1980. Structure and antigenic activity of the capsular polysaccharide of Cryptococcus neoformans serotype A. Mol Immunol. 17:1025–1032. [DOI] [PubMed] [Google Scholar]
- Cherniak R, Reiss E, Turner SH. 1982. A galactoxylomannan antigen of Cryptococcus neoformans serotype A. Carbohydr Res. 103:239–250. [Google Scholar]
- Cherniak R, Sundstrom JB. 1994. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect Immun. 62:1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherniak R, Valafar H, Morris LC, Valafar F. 1998. Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin Diagn Lab Immunol. 5:146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulidou A, Bouriotis V, Thireos G. 1996. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J Biol Chem. 271:31420–31425. [DOI] [PubMed] [Google Scholar]
- Chun CD, Madhani HD. 2010. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans. PLoS One. 5(9):e12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero RJ, Bergman A, Casadevall A. 2013. Temporal behavior of capsule enlargement for Cryptococcus neoformans. Eukaryot Cell. 12(10):1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero RJ, Frases S, Guimaraes AJ, Rivera J, Casadevall A. 2011. Evidence for branching in cryptococcal capsular polysaccharides and consequences on its biological activity. Mol Microbiol. 79:1101–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero RJ, Pontes B, Guimaraes AJ, Martinez LR, Rivera J, Fries BC, Nimrichter L, Rodrigues ML, Viana NB, Casadevall A. 2011. Chronological aging is associated with biophysical and chemical changes in the capsule of Cryptococcus neoformans. Infect Immun. 79:4990–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell TR, Griffith CL, Liu H, Nenninger AA, Doering TL. 2007. The pathogenic fungus Cryptococcus neoformans expresses two functional GDP-mannose transporters with distinct expression patterns and roles in capsule synthesis. Eukaryot Cell. 6:776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jesus M, Nicola AM, Rodrigues ML, Janbon G, Casadevall A. 2009. Capsular localization of the Cryptococcus neoformans polysaccharide component galactoxylomannan. Eukaryot Cell. 8:96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N, Zhang YB, Poster JB. 1997. The VRG4 gene is required for GDP-mannose transport into the lumen of the Golgi in the yeast, Saccharomyces cerevisiae. J Biol Chem. 272:31908–31914. [DOI] [PubMed] [Google Scholar]
- Denning DW. 2016. Minimizing fungal disease deaths will allow the UNAIDS target of reducing annual AIDS deaths below 500 000 by 2020 to be realized. Philos Trans R Soc Lond B Biol Sci. 371:1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TL. 2009. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu Rev Microbiol. 63:223–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenheer RA, Jin Lee Y, Blumwald E, Phinney BS, Gelli A. 2007. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res. 7:499–510. [DOI] [PubMed] [Google Scholar]
- Eisenman HC, Casadevall A. 2012. Synthesis and assembly of fungal melanin. Appl Microbiol Biotechnol. 93:931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerbroek PM, Lefeber DJ, van Veghel R, Scharringa J, Brouwer E, Gerwig GJ, Janbon G, Hoepelman AI, Coenjaerts FE. 2004. O-acetylation of cryptococcal capsular glucuronoxylomannan is essential for interference with neutrophil migration. J Immunol. 173:7513–7520. [DOI] [PubMed] [Google Scholar]
- Evans EE. 1960. Capsular reactions of Cryptococcus neoformans. Ann NY Acad Sci. 89:184–192. [Google Scholar]
- Fonseca FL, Guimaraes AJ, Kmetzsch L, Dutra FF, Silva FD, Taborda CP, Araujo GD, Frases S, Staats CC, Bozza MT et al. . 2013. Binding of the wheat germ lectin to Cryptococcus neoformans chitooligomers affects multiple mechanisms required for fungal pathogenesis. Fungal Genet Biol. 60:64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca FL, Nimrichter L, Cordero RJ, Frases S, Rodrigues J, Goldman DL, Andruszkiewicz R, Milewski S, Travassos LR, Casadevall A et al. . 2009. Role for chitin and chitooligomers in the capsular architecture of Cryptococcus neoformans. Eukaryot Cell. 8:1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzot SP, Doering TL. 1999. Inositol acylation of glycosylphosphatidylinositols in the pathogenic fungus Cryptococcus neoformans and the model yeast Saccharomyces cerevisiae. Biochem J. 340(Pt 1):25–32. [PMC free article] [PubMed] [Google Scholar]
- Frases S, Nimrichter L, Viana NB, Nakouzi A, Casadevall A. 2008. Cryptococcus neoformans capsular polysaccharide and exopolysaccharide fractions manifest physical, chemical, and antigenic differences. Eukaryot Cell. 7:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frases S, Pontes B, Nimrichter L, Viana NB, Rodrigues ML, Casadevall A. 2009. Capsule of Cryptococcus neoformans grows by enlargement of polysaccharide molecules. Proc Natl Acad Sci USA. 106:1228–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free SJ. 2013. Fungal cell wall organization and biosynthesis. Adv Genet. 81:33–82. [DOI] [PubMed] [Google Scholar]
- Freeze HH, Elbein AD. 2009. Glycosylation precursors In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Fries BC, Goldman DL, Cherniak R, Ju R, Casadevall A. 1999. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect Immun. 67:6076–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromtling RA, Shadomy HJ, Jacobson ES. 1982. Decreased virulence in stable, acapsular mutants of Cryptococcus neoformans. Mycopathologia. 79:23–29. [DOI] [PubMed] [Google Scholar]
- Gahrs W, Tigyi Z, Emody L, Makovitzky J. 2009. Polarization optical analysis of the surface structures of various fungi. Acta Histochem. 111:308–315. [DOI] [PubMed] [Google Scholar]
- Garcia-Hermoso D, Dromer F, Janbon G. 2004. Cryptococcus neoformans capsule structure evolution in vitro and during murine infection. Infect Immun. 72:3359–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert NM, Donlin MJ, Gerik KJ, Specht CA, Djordjevic JT, Wilson CF, Sorrell TC, Lodge JK. 2010. KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol Microbiol. 76:517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert NM, Lodge JK, Specht CA. 2011. The cell wall of Cryptococcus In: Heitman J, Kozel TR, Kwon-Chung KJ, Perfect J, Casadevall A, editors. Cryptococcus From Human Pathogen to Model Yeast. Washington: ASM Press; p. 67–79. [Google Scholar]
- Gish SR, Maier EJ, Haynes BC, Santiago-Tirado FH, Srikanta DL, Ma CZ, Li LX, Williams M, Crouch EC, Khader SA et al. . 2016. Computational analysis reveals a key regulator of Cryptococcal virulence and determinant of host response. mBio. 7:e00313–e00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger DL, Perfect JR, Durack DT. 1985. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Invest. 76:508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith CL, Klutts JS, Zhang L, Levery SB, Doering TL. 2004. UDP-glucose dehydrogenase plays multiple roles in the biology of the pathogenic fungus Cryptococcus neoformans. J Biol Chem. 279:51669–51676. [DOI] [PubMed] [Google Scholar]
- Grun CH, Hochstenbach F, Humbel BM, Verkleij AJ, Sietsma JH, Klis FM, Kamerling JP, Vliegenthart JF. 2005. The structure of cell wall alpha-glucan from fission yeast. Glycobiology. 15:245–257. [DOI] [PubMed] [Google Scholar]
- Guimaraes AJ, Frases S, Cordero RJ, Nimrichter L, Casadevall A, Nosanchuk JD. 2010. Cryptococcus neoformans responds to mannitol by increasing capsule size in vitro and in vivo. Cell Microbiol. 12:740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BC, Maier EJ, Kramer MH, Wang PI, Brown H, Brent MR. 2013. Mapping functional transcription factor networks from gene expression data. Genome Res. 23:1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BC, Skowyra ML, Spencer SJ, Gish SR, Williams M, Held EP, Brent MR, Doering TL. 2011. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS Pathog. 7:e1002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss C, Klutts JS, Wang Z, Doering TL, Azadi P. 2009. The structure of Cryptococcus neoformans galactoxylomannan contains beta-D-glucuronic acid. Carbohydr Res. 344:915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss C, Skowyra ML, Liu H, Klutts JS, Wang Z, Williams M, Srikanta D, Beverley SM, Azadi P, Doering TL. 2013. Unusual galactofuranose modification of a capsule polysaccharide in the pathogenic yeast Cryptococcus neoformans. J Biol Chem. 288:10994–11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B, Chan V, Liu V, Kronstad J. 2013. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect Immun. 81:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James PG, Cherniak R, Jones RG, Stortz CA, Reiss E. 1990. Cell-wall glucans of Cryptococcus neoformans Cap 67. Carbohydr Res. 198:23–38. [DOI] [PubMed] [Google Scholar]
- Janbon G, Himmelreich U, Moyrand F, Improvisi L, Dromer F. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol Microbiol. 42:453–467. [DOI] [PubMed] [Google Scholar]
- Jong A, Wu CH, Chen HM, Luo F, Kwon-Chung KJ, Chang YC, Lamunyon CW, Plaas A, Huang SH. 2007. Identification and characterization of CPS1 as a hyaluronic acid synthase contributing to the pathogenesis of Cryptococcus neoformans infection. Eukaryot Cell. 6:1486–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KW, So YS, Bahn YS. 2016. Unique roles of the unfolded protein response pathway in fungal development and differentiation. Sci Rep. 6:33413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KW, Yang DH, Maeng S, Lee KT, So YS, Hong J, Choi J, Byun HJ, Kim H, Bang S et al. . 2015. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Commun. 6:6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Hirata D, Arellano M, Perez P, Toda T. 1999. Fission yeast alpha-glucan synthase Mok1 requires the actin cytoskeleton to localize the sites of growth and plays an essential role in cell morphogenesis downstream of protein kinase C function. J Cell Biol. 144:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Jung KW, Maeng S, Chen YL, Shin J, Shim JE, Hwang S, Janbon G, Kim T, Heitman J et al. . 2015. Network-assisted genetic dissection of pathogenicity and drug resistance in the opportunistic human pathogenic fungus Cryptococcus neoformans. Sci Rep. 5:8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutts JS, Doering TL. 2008. Cryptococcal xylosyltransferase 1 (Cxt1p) from Cryptococcus neoformans plays a direct role in the synthesis of capsule polysaccharides. J Biol Chem. 283:14327–14334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutts JS, Levery SB, Doering TL. 2007. A beta-1,2-xylosyltransferase from Cryptococcus neoformans defines a new family of glycosyltransferases. J Biol Chem. 282:17890–17899. [DOI] [PubMed] [Google Scholar]
- Kmetzsch L, Joffe LS, Staats CC, de Oliveira DL, Fonseca FL, Cordero RJ, Casadevall A, Nimrichter L, Schrank A, Vainstein MH et al. . 2011. Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol Microbiol. 81:206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel TR. 1977. Non-encapsulated variant of Cryptococcus neoformans. II. Surface receptors for cryptococcal polysaccharide and their role in inhibition of phagocytosis by polysaccharide. Infect Immun. 16:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel TR, Hermerath CA. 1984. Binding of cryptococcal polysaccharide to Cryptococcus neoformans. Infect Immun. 43:879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel TR, Levitz SM, Dromer F, Gates MA, Thorkildson P, Janbon G. 2003. Antigenic and biological characteristics of mutant strains of Cryptococcus neoformans lacking capsular O-acetylation or xylosyl side chains. Infect Immun. 71:2868–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger AT, Engel J, Buettner FF, Routier FH. 2016. Aspergillus fumigatus Cap59-like protein A is involved in alpha1,3-mannosylation of GPI-anchors. Glycobiology. 26:30–38. [DOI] [PubMed] [Google Scholar]
- Kumar P, Heiss C, Santiago-Tirado FH, Black I, Azadi P, Doering TL. 2014. Pbx proteins in Cryptococcus neoformans cell wall remodeling and capsule assembly. Eukaryot Cell. 13:560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Yang M, Haynes BC, Skowyra ML, Doering TL. 2011. Emerging themes in cryptococcal capsule synthesis. Curr Opin Struct Biol. 21:597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita T, Noda Y, Takagi T, Osumi M, Yoda K. 2011. Kre6 protein essential for yeast cell wall beta-1,6-glucan synthesis accumulates at sites of polarized growth. J Biol Chem. 286:7429–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Fraser JA, Doering TL, Wang Z, Janbon G, Idnurm A, Bahn YS. 2014. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 4:a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Rhodes JC. 1986. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 51:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Bahn YS, Kim HJ, Chung SY, Kang HA. 2015. Unraveling the novel structure and biosynthetic pathway of O-linked glycans in the Golgi apparatus of the human pathogenic yeast Cryptococcus neoformans. J Biol Chem. 290:1861–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KT, So YS, Yang DH, Jung KW, Choi J, Lee DG, Kwon H, Jang J, Wang LL, Cha S et al. . 2016. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat Commun. 7:12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G, Bussey H. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 70:317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LX, Ashikov A, Liu H, Griffith CL, Bakker H, Doering TL. 2017. Cryptococcus neoformans UGT1 encodes a UDP-Galactose/UDP-GalNAc transporter. Glycobiology. 27:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LX, Rautengarten C, Heazlewood JL, Doering TL. 2018. a. Xylose donor transport is critical for fungal virulence. PLoS Pathog. 14(1):e1006765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LX, Rautengarten C, Heazlewood JL, Doering TL. 2018. b. UDP-glucuronic acid transport is required for virulence of Cryptococcus neoformans. mBio. 9:e02319-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Heitman J. 2006. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 60:69–105. [DOI] [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 135:174–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, Kelly MJ, Chow ED, Madhani HD. 2007. Parallel beta-helix proteins required for accurate capsule polysaccharide synthesis and virulence in the yeast Cryptococcus neoformans. Eukaryot cell. 6:630–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier EJ, Haynes BC, Gish SR, Wang ZA, Skowyra ML, Marulli AL, Doering TL, Brent MR. 2015. Model-driven mapping of transcriptional networks reveals the circuitry and dynamics of virulence regulation. Genome Res. 25:690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maligie MA, Selitrennikoff CP. 2005. Cryptococcus neoformans resistance to echinocandins: (1,3)Beta-glucan synthase activity is sensitive to echinocandins. Antimicrob Agents Chemother. 49:2851–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners DJ, Masson AJ, Patterson JC, Bjorndal H, Lindberg B. 1973. The structure of a beta-(1–6)-D-glucan from yeast cell walls. Biochem J. 135:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DC, De Jesus M, Casadevall A. 2006. The physical properties of the capsular polysaccharides from Cryptococcus neoformans suggest features for capsule construction. J Biol Chem. 281:1868–1875. [DOI] [PubMed] [Google Scholar]
- Minke R, Blackwell J. 1978. The structure of alpha-chitin. J Mol Biol. 120:167–181. [DOI] [PubMed] [Google Scholar]
- Montijn RC, Vink E, Muller WH, Verkleij AJ, Van Den Ende H, Henrissat B, Klis FM. 1999. Localization of synthesis of beta1,6-glucan in Saccharomyces cerevisiae. J Bacteriol. 181:7414–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyrand F, Chang YC, Himmelreich U, Kwon-Chung KJ, Janbon G. 2004. Cas3p belongs to a seven-member family of capsule structure designer proteins. Eukaryot Cell. 3:1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyrand F, Fontaine T, Janbon G. 2007. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol Microbiol. 64:771–781. [DOI] [PubMed] [Google Scholar]
- Moyrand F, Janbon G. 2004. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37 degrees C and for capsule biosynthesis. Eukaryot cell. 3:1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyrand F, Klaproth B, Himmelreich U, Dromer F, Janbon G. 2002. Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol Microbiol. 45:837–849. [DOI] [PubMed] [Google Scholar]
- Moyrand F, Lafontaine I, Fontaine T, Janbon G. 2008. UGE1 and UGE2 regulate the UDP-glucose/UDP-galactose equilibrium in Cryptococcus neoformans. Eukaryot Cell. 7:2069–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimrichter L, Frases S, Cinelli LP, Viana NB, Nakouzi A, Travassos LR, Casadevall A, Rodrigues ML. 2007. Self-aggregation of Cryptococcus neoformans capsular glucuronoxylomannan is dependent on divalent cations. Eukaryot Cell. 6:1400–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosanchuk JD, Stark RE, Casadevall A. 2015. Fungal melanin: What do we know about structure? Front Microbiol. 6:1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Xu W, Selvig KM, O’Meara MJ, Mitchell AP, Alspaugh JA. 2014. The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host. Mol Cell Biol. 34:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepinto J, Komperda K, Frases S, Park YD, Djordjevic JT, Casadevall A, Williamson PR. 2009. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol Microbiol. 71:1165–1176. [DOI] [PubMed] [Google Scholar]
- Park JN, Lee DJ, Kwon O, Oh DB, Bahn YS, Kang HA. 2012. Unraveling unique structure and biosynthesis pathway of N-linked glycans in human fungal pathogen Cryptococcus neoformans by glycomics analysis. J Biol Chem. 287:19501–19515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YD, Shin S, Panepinto J, Ramos J, Qiu J, Frases S, Albuquerque P, Cordero RJ, Zhang N, Himmelreich U et al. . 2014. A role for LHC1 in higher order structure and complement binding of the Cryptococcus neoformans capsule. PLoS Pathog. 10:e1004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR. 2005. Cryptococcus neoformans: A sugar-coated killer with designer genes. FEMS Immunol Med Microbiol. 45:395–404. [DOI] [PubMed] [Google Scholar]
- Pierini LM, Doering TL. 2001. Spatial and temporal sequence of capsule construction in Cryptococcus neoformans. Mol Microbiol. 41:105–115. [DOI] [PubMed] [Google Scholar]
- Pittet M, Conzelmann A. 2007. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1771:405–420. [DOI] [PubMed] [Google Scholar]
- Previato JO, Vinogradov E, Maes E, Fonseca LM, Guerardel Y, Oliveira PAV, Mendonca-Previato L. 2017. Distribution of the O-acetyl groups and beta-galactofuranose units in galactoxylomannans of the opportunistic fungus Cryptococcus neoformans. Glycobiology. 27:582–592. [DOI] [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect Dis. 17(8):873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos CL, Fonseca FL, Rodrigues J, Guimaraes AJ, Cinelli LP, Miranda K, Nimrichter L, Casadevall A, Travassos LR, Rodrigues ML. 2012. Chitin-like molecules associate with Cryptococcus neoformans glucuronoxylomannan to form a glycan complex with previously unknown properties. Eukaryot Cell. 11:1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese AJ, Doering TL. 2003. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol. 50:1401–1409. [DOI] [PubMed] [Google Scholar]
- Reese AJ, Yoneda A, Breger JA, Beauvais A, Liu H, Griffith CL, Bose I, Kim MJ, Skau C, Yang S et al. . 2007. Loss of cell wall alpha(1–3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol Microbiol. 63:1385–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MC, Aoki K, Wang ZA, Skowyra ML, Williams M, Tiemeyer M, Doering TL. 2011. A xylosylphosphotransferase of Cryptococcus neoformans acts in protein O-glycan synthesis. J Biol Chem. 286:26888–26899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Feldmesser M, Cammer M, Casadevall A. 1998. Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect Immun. 66:5027–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo J, Oliveira DL, Joffe LS, Hu G, Gazos-Lopes F, Fonseca FL, Almeida IC, Frases S, Kronstad JW, Rodrigues ML. 2013. Role of the Apt1 protein in polysaccharide secretion by Cryptococcus neoformans. Eukaryot Cell. 13(6):715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Alvarez M, Fonseca FL, Casadevall A. 2008. Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot Cell. 7:602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Djordjevic JT. 2012. Unravelling secretion in Cryptococcus neoformans: More than one way to skin a cat. Mycopathologia. 173:407–418. [DOI] [PubMed] [Google Scholar]
- Rodrigues ML, Dobroff AS, Couceiro JN, Alviano CS, Schauer R, Travassos LR. 2003. Sialylglycoconjugates and sialyltransferase activity in the fungus Cryptococcus neoformans. Glycoconj J. 19:165–173. [DOI] [PubMed] [Google Scholar]
- Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell. 7:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 6:48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Rozental S, Couceiro JN, Angluster J, Alviano CS, Travassos LR. 1997. Identification of N-acetylneuraminic acid and its 9-O-acetylated derivative on the cell surface of Cryptococcus neoformans: Influence on fungal phagocytosis. Infect Immun. 65:4937–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi N, Baba T, Fukuzawa M, Ohno S. 1993. Ultrastructural study of Cryptococcus neoformans by quick-freezing and deep-etching method. Mycopathologia. 121:133–141. [DOI] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Peng T, Yang M, Hang HC, Doering TL. 2015. A single protein S-acyl transferase acts through diverse substrates to determine cryptococcal morphology, stress tolerance, and pathogenic outcome. PLoS Pathog. 11:e1004908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siafakas AR, Sorrell TC, Wright LC, Wilson C, Larsen M, Boadle R, Williamson PR, Djordjevic JT. 2007. Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J Biol Chem. 282:37508–37514. [DOI] [PubMed] [Google Scholar]
- Sommer U, Liu H, Doering TL. 2003. An alpha-1,3-mannosyltransferase of Cryptococcus neoformans. J Biol Chem. 278:47724–47730. [DOI] [PubMed] [Google Scholar]
- Steenbergen JN, Casadevall A. 2003. The origin and maintenance of virulence for the human pathogenic fungus Cryptococcus neoformans. Microb Infect. 5:667–675. [DOI] [PubMed] [Google Scholar]
- Tefsen B, Grijpstra J, Ordonez S, Lammers M, van Die I, de Cock H. 2014. Deletion of the CAP10 gene of Cryptococcus neoformans results in a pleiotropic phenotype with changes in expression of virulence factors. Res Microbiol. 165(6):399–410. [DOI] [PubMed] [Google Scholar]
- Thompson JR, Douglas CM, Li W, Jue CK, Pramanik B, Yuan X, Rude TH, Toffaletti DL, Perfect JR, Kurtz M. 1999. A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J Bacteriol. 181:444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SH, Cherniak R, Reiss E. 1984. Fractionation and characterization of galactoxylomannan from Cryptococcus neoformans. Carbohydr Res. 125:343–349. [DOI] [PubMed] [Google Scholar]
- Turner SH, Cherniak R, Reiss E, Kwon-Chung KJ. 1992. Structural variability in the glucuronoxylomannan of Cryptococcus neoformans serotype A isolates determined by 13C NMR spectroscopy. Carbohydr Res. 233:205–218. [DOI] [PubMed] [Google Scholar]
- Vaishnav VV, Bacon BE, O’Neill M, Cherniak R. 1998. Structural characterization of the galactoxylomannan of Cryptococcus neoformans Cap67. Carbohydr Res. 306:315–330. [DOI] [PubMed] [Google Scholar]
- Vecchiarelli A, Pericolini E, Gabrielli E, Chow SK, Bistoni F, Cenci E, Casadevall A. 2011. Cryptococcus neoformans galactoxylomannan is a potent negative immunomodulator, inspiring new approaches in anti-inflammatory immunotherapy. Immunotherapy. 3:997–1005. [DOI] [PubMed] [Google Scholar]
- Vecchiarelli A, Pericolini E, Gabrielli E, Kenno S, Perito S, Cenci E, Monari C. 2013. Elucidating the immunological function of the Cryptococcus neoformans capsule. Future Microbiol. 8:1107–1116. [DOI] [PubMed] [Google Scholar]
- Vos A, Dekker N, Distel B, Leunissen JA, Hochstenbach F. 2007. Role of the synthase domain of Ags1p in cell wall alpha-glucan biosynthesis in fission yeast. J Biol Chem. 282:18969–18979. [DOI] [PubMed] [Google Scholar]
- Wang ZA, Griffith CL, Skowyra ML, Salinas N, Williams M, Maier EJ, Gish SR, Liu H, Brent MR, Doering TL. 2014. Cryptococcus neoformans dual GDP-mannose transporters and their role in biology and virulence. Eukaryot Cell. 13:832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills EA, Roberts IS, Del Poeta M, Rivera J, Casadevall A, Cox GM, Perfect JR. 2001. Identification and characterization of the Cryptococcus neoformans phosphomannose isomerase-encoding gene, MAN1, and its impact on pathogenicity. Mol Microbiol. 40:610–620. [DOI] [PubMed] [Google Scholar]
- Wozniak KL, Levitz SM. 2009. Isolation and purification of antigenic components of Cryptococcus. Methods Mol Biol. 470:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. 2008. Regulation of Cryptococcus neoformans capsule size is mediated at the polymer level. Eukaryot Cell. 7:546–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. 2009. An unusual organelle in Cryptococcus neoformans links luminal pH and capsule biosynthesis. Fungal Genet Biol. 46:682–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Casadevall A. 2004. Experimental modulation of capsule size in Cryptococcus neoformans. Biol Proced Online. 6:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol. 68:133–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Telzak A, Bryan RA, Dadachova E, Casadevall A. 2006. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol Microbiol. 59:67–83. [DOI] [PubMed] [Google Scholar]