Abstract
Background
Cystic fibrosis (CF) is associated with significant morbidity and early mortality due to recurrent acute and chronic lung infections. The chronic use of multiple antibiotics increases the possibility of multidrug resistance (MDR). Antibiotic susceptibility determined by culture-based techniques may not fully represent the resistance profile. The study objective was to detect additional antibiotic resistance using molecular methods and relate the presence of MDR to airway microbiome diversity and pulmonary function.
Methods
Bacterial DNA was extracted from sputum samples and amplified for the V4 region of the 16S rRNA gene. An qPCR array was used to detect antibiotic resistance genes. Clinical culture results and pulmonary function were also noted for each encounter.
Results
Six study participants contributed samples from 19 encounters. Those samples with MDR (n = 7) had significantly lower diversity measured by inverse Simpson's index than those without (n = 12) (2.193 ± 0.427 vs 6.023 ± 1.564, p = 0.035). Differential abundance showed that samples with MDR had more Streptococcus (p = 0.002) and Alcaligenaceae_unclassified (p = 0.002). Pulmonary function was also decreased when MDR was present (FEV1, 51 ± 22.9 vs 77 ± 26.7, p = 0.054; FVC, 64.5 ± 22.7 vs 91.6 ± 27.7, p = 0.047).
Conclusions
The presence of MDR within the CF airway microbiome was associated with decreased microbial diversity, the presence of Alcaligenes, and decreased pulmonary function.
Keywords: Computational biology, Microbiology, Bioinformatics
1. Introduction
Cystic fibrosis (CF) is an autosomal recessive disease associated with significant morbidity and relatively early mortality due to recurrent acute and chronic lung infections [1, 2]. Acute pulmonary exacerbations (APE), which are acute increases in the signs and symptoms of respiratory disease, occur throughout the patients' lives and are treated in part with antibiotics (oral, inhaled, and intravenous (IV)) [3]. Pseudomonas aeruginosa is a very common pathogen in patients, especially after their teenage years; as such, ceftazidime and tobramycin are the most commonly used IV antibiotics for treatment of APE [4, 5, 6, 7]. Antibiotics with a broader range of antimicrobial activity (such as meropenem) are used when antibiotic resistance is presumed or detected, with antibiotic selection being based on both current and past culture results [5]. The chronic use of multiple antibiotics increase with possibility of multidrug resistance (resistance to three or more antibiotic categories) and severely limits the options of antibiotic coverage in patients with advanced disease [8, 9]. However, some studies have shown that CF persons will improve with IV antibiotics despite the in vitro antibiotic susceptibilities of the pathogen grown [10]. One possible explanation for this finding is that these in vitro antibiotic susceptibilities are not representative of the resistance within the microbial community [9].
Antibiotic susceptibility is most often determined by culture-based standard techniques that reveal only the profile of those bacteria grown in the culture. However, this profile of antibiotic susceptibility is not representative of all the organisms colonizing and/or infecting the patient's airways. For example, antibiotic resistance may be the result of the presence of bacteria that are commonly present within the CF airway microbiome and not grown in routine cultures, such as Prevotella species [11]. These beta-lactamases, as well as other transferable antibiotic resistance genes or inducible resistance mechanism, may further influence antibiotic susceptibility of the microbial community beyond what are typically reported with standard culture based techniques [9].
As frequent antibiotic courses are necessary when treating APEs in persons with cystic fibrosis [12], both identifying antibiotic resistance when present and finding ways to limit the development of antibiotic resistance are important to optimize treatment of infections. Additionally, a better understanding of the relationship between microbial diversity and antibiotic resistance may be insightful, as decreasing microbiome diversity within the CF airway has been associated with cumulative antibiotic exposure over time [13, 14] and disease progression [15, 16, 17, 18].
We hypothesized that antibiotic resistance mechanisms are present within the CF airway microbiome that are not detected by standard clinical culture methods and is associated with response to antibiotic therapy. The objective of this study was a) to detect antibiotic resistance genes within respiratory samples with using molecular methods and b) to relate the presence of antibiotic resistance to airway microbiome diversity and pulmonary function.
2. Materials and methods
2.1. Setting and study population
This study cohort is from a larger prospective, longitudinal study of 20 cystic fibrosis patients <21 years of age that was conducted over 18 months at Children's National Health System (CNHS) in Washington DC that collected bronchoalveoloar lavage (BAL), sputum, and oropharyngeal swab respiratory samples. Inclusion criteria for this study included having BAL or sputum respiratory samples collected. Exclusion criteria included not having a corresponding clinical culture obtained and not having enough remaining bacterial DNA with which to perform additional PCR testing. The CNHS Institutional Review Board approved the study (Pro5655, approved 2/20/2015). Written informed consent was obtained from study participants ≥18 years of age and parental permission for those <18 years of age. Assent was obtained from study participants 7–17 years of age.
2.2. Subject encounters
The study was designed to evaluate study participants at four time points. The baseline visit (B) occurred at a routine evaluation when they had not been on IV antibiotics for at least 30 days. The exacerbation visit (E) occurred when they were being hospitalized to begin treatment with IV antibiotics. The treatment visit (T) occurred at the end of their antibiotic treatment course. The recovery visit (R) occurred more than 30 days after completing their IV antibiotic therapy. At every encounter, the study participant provided a respiratory sample, either expectorated sputum (17 samples) or bronchoalveolar lavage fluid (two samples). Study participants who experienced more than one APE requiring IV antibiotics during the study period were asked to participate for each treatment course.
2.3. Data collection
REDCap electronic data capture tools were used to collect and store study data [19]. The following data were collected from the electronic medical record: age, gender, race/ethnicity, CFTR genotype (determined by clinical laboratory testing), medications, pulmonary function tests, and clinical laboratory and culture data. Pulmonary function test results were reported using NHANES III reference values [20] and included the percent predicted values for the following: forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC), FEV1/FVC ratio, and forced expiratory flow at 25–75% of the pulmonary volume (FEF25-75). For clinical culture results, the microbiology lab uses MicroScan (BeckmanCoulter, Brea CA) to determine bacterial species identification and drug resistance susceptibility. An internally validated protocol is used for mucoid Pseudomonas aeruginosa isolated from cystic fibrosis samples [21].
2.4. Respiratory sample processing
Research staff collected spontaneously expectorated sputum samples according to standardized procedures. Sputum samples were processed by washing first 1:1 v/v with sterile normal saline, mixing the sample 1:1 v/v with Sputasol (Fisher Healthcare, Houston TX), vortexing for 1 minute, and placing in a 37 °C heated water bath to homogenize the sample. Samples were then pelleted through centrifugation (12,000 g × 10 minutes). Supernatants were removed, and pellets were frozen at −80 °C until they underwent DNA extraction.
2.5. DNA extraction
DNA extraction was performed in the Clinical and Translational Science Institute at CNHS on the QIAsymphony SP (Qiagen, Valencia CA) using the DSP Virus/Pathogen Midi Kit and the Complex800_V6_DSP protocol. Prior to DNA extraction, pellets were thawed, and a chemical lysis step was performed. Specifically, the bacterial pellet was suspended in 500 μl of combination enzyme solution, which included lysozyme (20 mg/mL) and lysostaphin (200 μg/mL) in nuclease free water (Sigma-Aldrich, St. Louis MO). The pellet was then incubated at 37 °C for 30 minutes. The tube was then briefly centrifuged to remove drops from the inside of the lid before placing on the QIAsymphony.
2.6. Next generation sequencing
Extracted DNA was amplified for the V4 region of the 16S rRNA gene and libraries were sequenced on the MiSeq sequencing platform at the University of Michigan (Ann Arbor MI). Raw FASTQ files were processed in mothur (version 1.39.5) [22]. Default settings were used to minimize sequencing errors [23]. Clean sequences were aligned to the SILVA_v123 bacterial reference alignment at http://www.mothur.org. Sequences were clustered into operational taxonomic units (OTUs) at the 0.03 threshold per the Schloss lab MiSeq SOP (version update 4 April 2018) [23]. To remove the effect of sample size bias on community composition, each sample's sequencing file was subsampled to the number of sequences remaining in the smallest file (11,965 sequences).
2.7. Antibiotic resistance gene PCR
The Antibiotic Resistance Genes Microbial DNA qPCR Array (Cat. no. 330261 BAID-1901ZRA, Qiagen, Valencia CA) was used to detect antibiotic resistance genes. This array contains assays for 87 antibiotic resistance genes for aminoglycoside, beta-lactam, erythromycin, fluoroquinolone, macrolide-lincosamide-streptogram B, tetracycline, vancomycin, and multidrug resistance classifications. A total of 1000–3000 ng of extracted DNA was mixed with 1275 μl microbial qPCR Mastermix and microbial DNA-free water as needed to bring the total volume to 2550 μl per sample for each run per the kit's instructions. Using a 96-assay array, 25 μl of sample was added to each well. An array with 25 μl of with no template was added to each well for the control (NTC). The array was tightly sealed with optical thin-wall 8-cap strips and loaded onto an Applied Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City CA). The PCR cycler was programed with an initial PCR activation step at 95 °C for 10 minutes, followed by 2-step cycling of denaturation for 15 seconds at 95 °C and annealing and extension for 2 minutes at 60 °C for 40 cycles. The threshold cycle (CT) for each well was calculated using RQ Manager (Applied Biosystems, Foster City CA), ensuring the CT value for the positive control in each array (PPC) was 22 ± 2. The CT values for each well were imported into the Microbial DNA qPCR Array Template Excel Software for interpretation (Qiagen, Valencia CA). For quality control purposes, in addition to the PPC control described above, six pan-bacteria wells also had to have a CT <29. The ΔCT between the patient sample and the NTC was calculated to determine positivity for a well. Log fold changes >1 were considered positive.
2.8. Statistical analysis
To determine if antibiotic drug resistance is associated with microbial diversity, our cohort was categorized into three groups: 1) presence or absence of methicillin resistant Staphylococcus aureus (MRSA), 2) presence or absence of resistance to two or more beta-lactam antibiotic classes in the gram-negative organisms (e.g., penicillins, cephalosporins, or carbapenems), and 3) presence or absence of multidrug resistant (MDR) bacteria. MDR was defined as resistance to at least one antibiotic within three different antibiotic categories (e.g., beta-lactam, aminoglycoside, and fluoroquinolone) [8]. Participant characteristics were evaluated using Fisher's exact test for categorical variables and two-sided t-test with unequal variance for continuous variables using Stata/IC 15.1 (StataCorp LLC, College Station TX). Microbial diversity was measured using the inverse Simpson's index and the Shannon-Weiner index. The inverse Simpson's index was calculated in Excel (Microsoft, Redmond WA) using the equation 1/Σ[ni*(ni − 1)/(ΣN*(ΣN − 1))]. The Shannon-Weiner index was calculated using the equation −Σ[ln(ni/ΣN)]. OTU and taxonomy tables were imported into Rstudio for subsequent analyses using phyloseq and transformed to DESeq2 files to determine differential abundance between subjects with and without antibiotic resistance [24, 25]. P values used to determine significance when using DESeq2 were adjusted by the Benjamini-Hochberg adjustment [25]. Principle coordinates analysis (PCoA) plots were generated using Bray-Curtis dissimilarity matrices with log transformed counts using DESeq2 to visualize differences in microbial composition between subjects with and without antibiotic resistance. PERMANOVA was also calculated to measure the differences in overall distribution using the adonis function of vegan in Rstudio [26].
3. Results
3.1. Participant demographics and antibiotic regimens
Six study participants had corresponding clinical culture data and sufficient bacterial DNA available to complete PCR testing for 19 encounters (Table 1). At the time of study enrollment, the participants had a mean age of 12 years (SD 6.3). Sixty-six percent (n = 4) of the participants were male and 83% (n = 5) were Hispanic. Fifty percent (n = 3) had a homozygous F508del mutation as their CFTR genotype, and 33% (n = 2) had a heterozygous F508del mutation. Fifty percent (n = 3) were in the early stages of disease (FEV1 ≥ 70%), while 17% (n = 1) were considered intermediate (40% ≤ FEV1 < 70%) and 33% (n = 2) were considered advanced (FEV1 < 40%) based on their FEV1 percent predicted at time of enrollment. Additionally, 17% (n = 1) were considered to have mild disease aggressiveness, 50% (n = 3) had moderate disease aggressiveness, and 33% (n = 2) had severe disease aggressiveness based on a combination of age and FEV1 percent predicted at time of enrollment. The majority (n = 4) were receiving inhaled suppressive antibiotics at baseline (B), which included inhaled tobramycin and inhaled aztreonam.
Table 1.
Study participant demographics and clinical characteristics.
| Study ID | Age (years) | Gender | CF Genotype | Baseline FEV1 percent predicted | Common CF bacterial pathogens in past and current cultures |
|---|---|---|---|---|---|
| A | 6 | Female | F508del homozygous | 31 | Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli, Stenotrophomonas maltophilia, Alcaligenes xylosoxidans |
| B | 17 | Male | F508del heterozygous | 65 | Staphylococcus aureus, Pseudomonas aeruginosa, Stenotrophomonas maltophilia |
| C | 7 | Male | Other | 115 | Staphylococcus aureus, Pseudomonas aeruginosa, Haemophilus influenza, Alcaligenes xylosoxidans |
| D | 21 | Female | F508del heterozygous | 34 | Staphylococcus aureus, Pseudomonas aeruginosa |
| E | 7 | Male | F508del homozygous | 82 | Pseudomonas aeruginosa, Stenotrophomonas maltophilia |
| F | 14 | Male | F508del homozygous | 86 | Staphylococcus aureus, Pseudomonas aeruginosa |
The 19 included encounters for these 6 study participants were as follows: 5 baseline (B), 6 exacerbation (E), 2 treatment (T), and 6 recovery (R), which covered a total of 10 hospital admissions for IV antibiotics and subsequent clinic follow up. All antibiotic regimens included a beta-lactam, while 90% also had an aminoglycoside (Table 2). The average duration of antibiotic therapy was 19 days (SD 7.1). The majority of patients also received steroids (80%), and 50% had a concurrent viral pathogen detected at hospital admission (Table 2).
Table 2.
Antibiotic treatment regimens and associated study samples.
| Study ID | Antibiotics received >48 hours | Total days of therapy | Steroids received | Respiratory virus | BETR |
|---|---|---|---|---|---|
| A1 | Piperacillin-tazobactam, imipenem-cilastatin, linezolid | 13 | No | Rhinovirus | E |
| A2 | Meropenem, amikacin | 29 | Yes | Negative | E, T |
| A3 | Meropenem, amikacin, ertapenem | 29 | Yes | Adenovirus | E |
| A4 | Meropenem, amikacin, vancomycin, linezolid | 26 | Yes | Negative | R |
| B | Ceftazidime, tobramycin | 22 | Yes | Not obtained | B, E, R |
| C1 | Ceftazidime, tobramycin | 11 | Yes | Rhinovirus | B, R |
| C2 | Meropenem, tobramycin | 12 | Yes | Rhinovirus | E, R |
| D | Meropenem, tobramycin | 14 | Yes | Not obtained | B, R |
| E | Ceftazidime, tobramycin | 14 | No | Rhinovirus | B, E, T |
| F | Meropenem, tobramycin | 17 | Yes | Not obtained | B, R |
B, baseline; E, exacerbation; T, treatment; R, recovery.
3.2. Determination of antibiotic resistance
The bacteria identified by clinical cultures were methicillin-sensitive Staphylococcus aureus (MSSA, n = 7), Pseudomonas aeruginosa (n = 6), Stenotrophomonas maltophilia (n = 3), Alcaligenes xylosoxidans (n = 2), Escherichia coli (n = 2), MRSA (n = 2), and Haemophilus influenzae (n = 1). While eleven cultures grew multiple bacteria or multiple strains of bacteria, two respiratory cultures only grew normal flora. Using clinical culture reports, antibiotic resistance was noted for each respiratory sample, with resistance being considered present for an antibiotic category if at least one drug in that category was resistant in at least one bacteria grown in culture (Table 3).
Table 3.
Antibiotic resistance detected by clinical culture and PCR.
| Study ID | Clinical culture |
Antibiotic resistance gene PCR |
Inverse Simpson index | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MRSA | 1 BL | 2 BL | AG | FQ | MRSA | BL | AG | FQ | ||
| A1-E | + | + | + | + | + | + | 1.436 | |||
| A2-E | + | + | + | + | + | + | 4.325 | |||
| A2-T | + | + | + | + | + | + | + | + | + | 1.690 |
| A3-E | + | + | 1.240 | |||||||
| A4-R | + | + | + | + | 1.919 | |||||
| B-B | + | + | + | 14.333 | ||||||
| B-E | + | + | + | + | + | + | 12.257 | |||
| B-R | + | + | 14.533 | |||||||
| C1-B | + | + | + | + | + | 1.411 | ||||
| C1-R | 1.881 | |||||||||
| C2-E | 2.126 | |||||||||
| C2-R | + | 2.718 | ||||||||
| D-B | + | + | + | 3.049 | ||||||
| D-R | + | + | + | + | 3.585 | |||||
| E-B | + | + | + | + | + | 1.017 | ||||
| E-E | + | + | + | + | + | + | + | 1.915 | ||
| E-T | + | + | + | + | + | + | 1.014 | |||
| F-B | 10.438 | |||||||||
| F-R | 7.275 | |||||||||
1 BL, one beta-lactam; 2 BL, two or more beta-lactam; AG, aminoglycoside; FQ, fluoroquinolone.
Next, antibiotic resistance was determined using PCR. An antibiotic resistance gene was considered present if it showed at least a one log fold change above control based on the ΔCT between the patient sample and the control. As expected, antibiotic resistance genes were detected for additional antibiotic categories beyond those detected in culture in 32% (n = 6) of the samples. However, some antibiotic resistance detected in culture was not identified by the PCR (Table 3).
Partial congruence of at least one antibiotic category between the resistance detected by clinical culture and that detected by PCR was 68 percent. Full congruence was only 42 percent. However, when evaluating the congruence by each antibiotic category using Fisher's exact test, no statistically significant differences were noted (MRSA p = 0.298, beta-lactam p = 0.350, aminoglycoside p = 0.350, fluoroquinolone p = 0.272, and MDR p = 0.155). For all further analysis, antibiotic resistance for each respiratory sample was defined as the combination of resistance detected by either standard clinical culture or PCR.
3.3. Antibiotic multidrug resistance is associated with microbiome diversity and differential abundance
When evaluating the presence/absence of MDR bacteria, those samples having resistance detected (n = 7) had significantly lower diversity measured by inverse Simpson's index than those without (n = 12) (2.193 ± 0.427 vs 6.023 ± 1.564, p = 0.035).
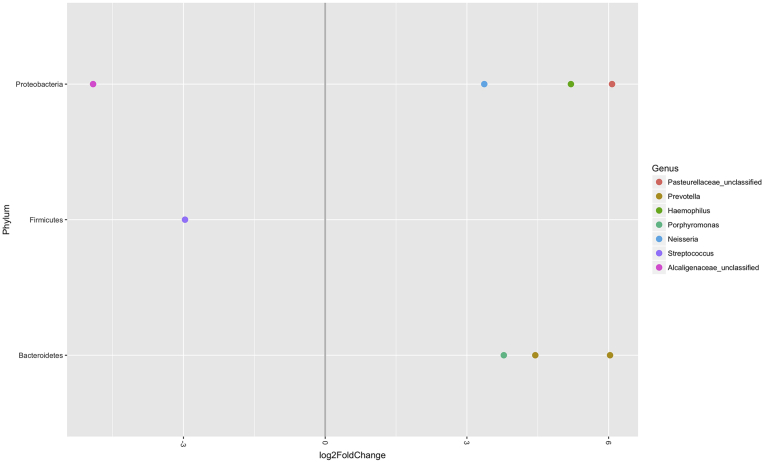
Next, differential abundance of OTUs was measured between those respiratory samples with and without MDR bacteria. Those samples with MDR bacteria present were more likely to have Streptococcus (p = 0.002) and Alcaligenaceae_unclassified (p = 0.002) within the bacterial community (Fig. 1).
Fig. 1.
Differential abundance of OTUs present in respiratory samples with and without MDR. The bacterial genera on the left side of the graph were more abundant in the samples with MDR present, while those on the right side of the graph were more abundant in the samples with MDR absent. Log2 fold changes are shown on the x-axis. All fold-changes are significant at p < 0.05.
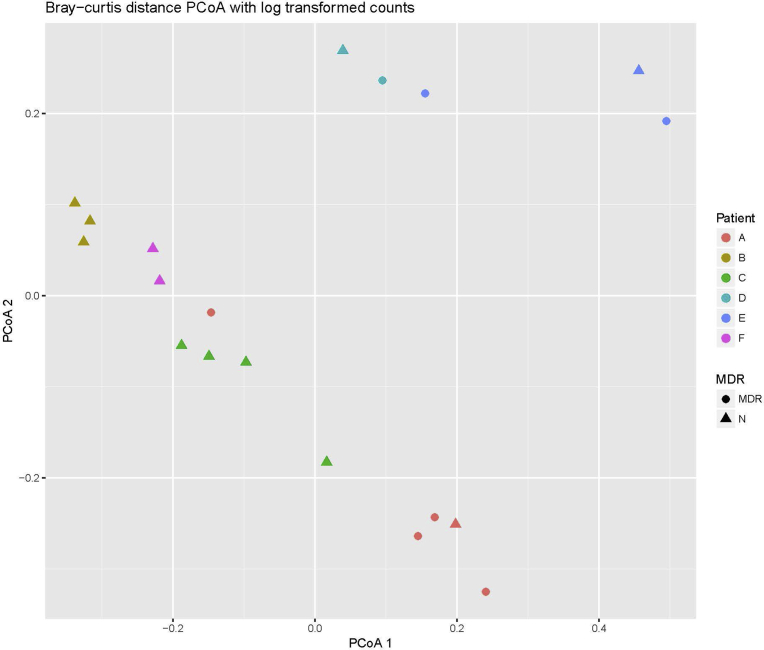
We also used Bray-Curtis dissimilarity matrices and PERMANOVA to measure differences between bacterial distributions in the respiratory samples with and without MDR bacteria. No difference in bacterial distribution was detected based on MDR (p = 0.289); however, there was a significant difference detected based on patient (p < 0.001). The permutation test for homogeneity of multivariate dispersions based on patient was also significant (p = 0.016). The corresponding Bray-Curtis PCoA plot using log transformed counts demonstrates some clustering based on patient, but none based on presence or absence of MDR bacteria (Fig. 2).
Fig. 2.
Two-dimensional PCoA plot of respiratory samples with and without MDR bacteria. The PCoA was created using Bray-Curtis dissimilarity matrices based on log transformed counts. Differences in MDR are shown by shape, while differences in patient are shown by different colors. MDR = multidrug resistant bacteria present, N = multidrug resistant bacteria absent.
3.4. Antibiotic multidrug resistance is associated with decreased pulmonary function
Lastly, study participant demographic and clinical features were examined for differences between the respiratory samples with MDR bacteria and those without. No significant differences were noted regarding gender, race/ethnicity, CFTR genotype, or disease stage of the patient (Table 4). However, disease aggressiveness trended toward significance (p = 0.099), with those with MDR present having more aggressive disease.
Table 4.
Study participant demographics based on presence of MDR bacteria.
| MDR bacteria absent from all samples (n = 3) | MDR bacteria present in at least one sample (n = 3) | P value | |
|---|---|---|---|
| Gender (n, %)a | 0.400 | ||
| Female | 0 (0%) | 2 (67%) | |
| Male | 3 (100%) | 1 (33%) | |
| Race/Ethnicity (n, %)a | >0.999 | ||
| White, non-Hispanic | 1 (33%) | 0 (0%) | |
| White, Hispanic | 2 (67%) | 3 (100%) | |
| CFTR genotype (n, %)a | >0.999 | ||
| F508del/F508del | 1 (33%) | 2 (67%) | |
| F508del/other | 1 (33%) | 1 (33%) | |
| Other | 1 (33%) | 0 (0%) | |
| Disease stage (n, %)b | 0.239 | ||
| Early | 2 (67%) | 1 (33%) | |
| Intermediate | 1 (33%) | 0 (0%) | |
| Advanced | 0 (0%) | 2 (67%) | |
| Disease aggressiveness (n, %)b | 0.099 | ||
| Mild | 1 (33%) | 0 (0%) | |
| Moderate | 2 (67%) | 1 (33%) | |
| Severe | 0 (0%) | 2 (67%) |
Two-sided Fisher's exact test.
Two-sample Wilcoxon rank-sum test.
Additionally, at the time of the sample collection, there was no significant difference in percent predicted FEV1/FVC or FEF25-75 (Table 5). Other measures of pulmonary function approached significance (percent predicted FEV1, p = 0.054) or were significant (percent predicted FVC, p = 0.047), showing decreased pulmonary function when MDR bacteria was present in the corresponding respiratory sample. The BETR timing of the sample collected was not significant (p = 0.094) but trended toward more resistance at the time of exacerbation (E) and when on antibiotic therapy (T).
Table 5.
Clinical features associated with the presence of MDR bacteria at time of sample collection.
| MDR bacteria absent (n = 12) | MDR bacteria present (n = 7) | P value | |
|---|---|---|---|
| Timing of sample collectiona (n, %) | 0.094 | ||
| Baseline | 5 (42%) | 0 (0%) | |
| Exacerbation | 3 (25%) | 3 (43%) | |
| Treatment | 0 (0%) | 2 (28.5%) | |
| Recovery | 4 (33%) | 2 (28.5%) | |
| Pulmonary functionb (n, SD) | |||
| FEV1 | 77 (26.7) | 51 (22.9) | 0.054 |
| FVC | 91.6 (27.7) | 64.5 (22.7) | 0.047 |
| FEV1/FVC | 73.3 (9.7) | 72 (12.0) | 0.830 |
| FEF25-75 | 51.7 (28.3) | 33.3 (28.8) | 0.229 |
Bold represents p value < 0.05.
Two-sided Fisher's exact test.
Two-sided t test.
Lastly, we evaluated the relationship between pulmonary function and the relative abundance of Alcaligenaceae_unclassified and Streptococcus using linear regression. For Alcaligenaceae_unclassified, the relationship with FEV1 was a modest negative correlation (r = -0.325, p = 0.189, adjusted R2 = 0.050). This was also true for FVC (r = -0.381, p = 0.119, adjusted R2 = 0.092), while FEV1/FVC and FEF25-75 showed no statistical significance (p = 0.576 and p = 0.389 respectively). For Streptococcus, the relationship with FEV1/FVC was a modest positive correlation (r = 0.368, p = 0.133, adjusted R2 = 0.082). No statistical significance was noted for other measures of pulmonary function and the relative abundance of Streptococcus (FEV1 p = 0.765, FVC p = 0.369, and FEF25-75 p = 0.544).
4. Discussion
In our study, we found an association of multidrug resistance with decreased diversity, an increased relative abundance of Alcaligenaceae_unclassified, and decreased pulmonary function. Decreasing diversity has been associated with both prolonged antibiotic use [13, 14]; however, no prior studies have tried to associate the presence of antibiotic resistance within the microbial community of the airway to community diversity. Of the small number of prior studies generally evaluating antibiotic resistance within the microbial community of CF sputa, the majority have done so using a metagenomic approach [27, 28, 29]. Lim et al. evaluated ten sputum samples from three adult CF patients [27]. Similar to our study, their study participants were culture positive for ESBL producing Escherichia coli, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia. They found the antibiotic resistance gene profiles of the patients were dynamic over time, and that a previously underappreciated diversity of antibiotic resistance genes was detected using their methods. Feigelman et al. evaluated the sputa of 6 CF patients, which they compared to sputa from patients with COPD, healthy subjects, and smokers [28]. They found high variability in microbial diversity amongst the CF patients compared to the other groups, and were able to predict the antibiotic resistance found in culture by screening their sequencing results to a database of annotated antibiotic resistance genes. Lastly, Bacci et al. studied the sputa of 12 CF patients at baseline conditions, of whom 6 had mild lung disease and 6 had severe lung disease [29]. Those with severe disease had a significantly higher percentage of antibiotic resistance genes detected, with multidrug efflux pumps being the most abundant. Similar to our study, antibiotic resistance genes were related to the presence of Alcaligenes xylosoxidans.
Alcaligenes (also called Achromobacter) xylosoxidans is intrinsically resistant to aminoglycosides, cephalosporins (except ceftazidime) and aztreonam [30, 31]. It can also acquire resistance to other beta-lactams through the acquisition of mobile genetic elements with beta-lactamases, and can acquire efflux pump systems that provide resistance to additional antimicrobials, including fluoroquinolones [30, 31]. Alcaligenes xylosoxidans is often considered an opportunistic pathogen in CF, as patients with respiratory cultures positive for Alcaligenes xylosoxidans are often older with more advanced disease and lower lung function values [32]. A small retrospective case control study found that those patients chronically infected with Alcaligenes xylosoxidans did not seem to do worse than negative controls [33]. However, case reports have demonstrated that Alcaligenes xylosoxidans can cause rapidly progressive lung disease [34]. In our study, there was a modest negative correlation between the relationship between FEV1 and FVC and the relative abundance of Alcaligenaceae_unclassified. It requires more study to know if Alcaligenes xylosoxidans may be an underappreciated driver of declining pulmonary function.
The finding of Streptococcus being more abundant in MDR samples is less easy to explain. A few studies have demonstrated the potential role of the Streptococcus milleri group being associated with both acute and chronic lung infections [35, 36]. However, more recent studies have shown Streptococcus to be more abundant in early lung disease and associated with the presence of less inflammation [37], or to be associated with clinical stability [38]. Furthermore, an association between the relative abundance of Streptococcus and FEV1, FVC, or FEF25-75 was not found in our study, while FEV1/FVC had a modest positive correlation.
We did identify an association between the presence of MDR bacteria and decreased pulmonary function. However, it should be noted that repeated episodes of APE drive down lung function [3], and that each APE is usually treated with antibiotics [3, 39]. Furthermore, it is well established that chronic antibiotic use drives the development of antibiotic resistance [9]. Lastly, opportunistic bacteria such as Alcaligenes xylosoxidans, Stenotrophomonas maltophilia, and Burkholderia cepacia are more often present in advanced CF disease [32, 37], and these organisms have high levels of antibiotic resistance [30, 31].
We found a fair amount of discrepancy between antibiotic resistance identified by culture and antibiotic resistance identified by PCR. While we expected the cultures to not fully represent the resistance of the microbial community, we had expected the PCR assay to be more thorough in its detection. This discrepancy may have been the result of PCR bias based on primers or relative abundance of template DNA limiting the ability of the PCR to detect the resistance gene. Prior studies using quantitative real-time PCR for bacteria detection in CF sputa specimens found a congruence rate of 54–90%, depending on the bacteria being identified [40]. Our study had a partial congruence rate of 68% and a full congruence rate of only 42%. Another possible explanation for the discordance is that PCR is limited by the number of assays run. While our PCR method allowed for the detection of 87 antibiotic resistance genes, there may have been additional antibiotic resistance mechanisms present that we were not set up to detect [9].
A possible solution to these two limitations would be using a metagenomic sequencing approach, as done in other studies [27, 28, 29]. This could allow for simultaneous detection of bacterial species present and antibiotic resistance by cross-referencing sequences against a larger database of known antibiotic resistance genes [41, 42, 43]. Metagenomic sequencing may also provide an opportunity to identify additional emerging and opportunistic pathogens, such as Bordetella bronchiseptica, which have not been as well studied in the CF population [44, 45].
Other limitations of this study include the small sample size, not having corresponding PCR results for each study time point, the reliance on clinical culture results, and the limitations of PCR as described above. Another limitation is that the management of APEs requires more than just antibiotic treatment, and the effects of airway clearance and other therapies such as steroids, which are not accounted for in this study [4, 39]. Lastly, our study population had lower than average FEV1 percent predicted based on the 2016 CF Patient Registry report, which limits the generalizability to the general CF population [46]. However, the findings from this study can inform future studies in larger cohorts, as the association between antibiotic resistance, microbial diversity, and lung function is important to establish.
In conclusion, the presence of multidrug resistance to antibiotics within the CF airway microbiome was associated with decreased microbial diversity, the presence of Alcaligenes, and decreased pulmonary function. Future studies should incorporate shotgun sequencing with screening against antimicrobial resistance databases to more fully define the mechanisms of resistance within the CF airway microbiome. Longitudinal studies of a larger cohort would also be able to explore the impact of short courses of antibiotics on changes in antibiotic resistance over time and the relationship of antibiotic resistance to pulmonary function.
Declarations
Author contribution statement
Andrea Hahn: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Aszia Burrell, Hollis Chaney, Geovanny F. Perez, Iman Sami, Anastassios C. Koumbourlis Performed the experiments.
Hani Fanous Performed the experiments; Contributed reagents, materials, analysis tools or data.
Robert J. Freishtat, Keith A. Crandall Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by Award Number UL1TR000075 from the NIH National Center for Advancing Translational Sciences. Andrea Hahn was supported by K12 Career Development Program K12HL119994 award from the National Heart, Lung, and Blood Institute.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at NCBI SRA repository under the accession number PRJNA437613. The corresponding manuscript sample description and BioProject descriptions are matched as follows: A1-E (AHCF01b2), A2-E (AHCF01b3), A2-T (AHCF01c3), A3-E (AHCF01b4), A4-R (AHCF01d5), B-B (AHCF03a), B-E (AHCF03b), B-R (AHCF03d), C1-B (AHCF04a), C1-R (AHCF04d), C2-E (AHCF04b2), C2-R (AHCF04d2), D-B (AHCF20a), D-R (AHCF20d), E-B (AHCF25a), E-E (AHCF25b), E-T (AHCF25c), F-B (AHCF27a), and F-R (AHCF27d).
References
- 1.MacKenzie T., Gifford A.H., Sabadosa K.A. Longevity of patients with cystic fibrosis in 2000 to 2010 and beyond: survival analysis of the cystic fibrosis foundation patient registry. Ann. Intern. Med. 2014;161(4):233–241. doi: 10.7326/M13-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramsey B.W. Management of pulmonary disease in patients with cystic fibrosis. N. Engl. J. Med. 1996;335(3):179–188. doi: 10.1056/NEJM199607183350307. [DOI] [PubMed] [Google Scholar]
- 3.Wagener J.S., Rasouliyan L., Vandevanter D.R. Oral, inhaled, and intravenous antibiotic choice for treating pulmonary exacerbations in cystic fibrosis. Pediatr. Pulmonol. 2013;48(7):666–673. doi: 10.1002/ppul.22652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flume P.A., Mogayzel P.J., Robinson K.A. Cystic fibrosis pulmonary guidelines: treatment of pulmonary exacerbations. Am. J. Respir. Crit. Care Med. 2009;180(9):802–808. doi: 10.1164/rccm.200812-1845PP. [DOI] [PubMed] [Google Scholar]
- 5.Doring G., Conway S.P., Heijerman H.G.M. Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis: a European consensus. Eur. Respir. J. 2000;16(4):749–767. doi: 10.1034/j.1399-3003.2000.16d30.x. [DOI] [PubMed] [Google Scholar]
- 6.Zobell J.T., Young D.C., Waters C.D. A survey of the utilization of anti-pseudomonal beta-lactam therapy in cystic fibrosis patients. Pediatr. Pulmonol. 2011;46(10):987–990. doi: 10.1002/ppul.21467. [DOI] [PubMed] [Google Scholar]
- 7.Van Meter D.J., Corriveau M., Ahern J.W., Lahiri T. A survey of once-daily dosage tobramycin therapy in patients with cystic fibrosis. Pediatr. Pulmonol. 2009;44(4):325–329. doi: 10.1002/ppul.20985. [DOI] [PubMed] [Google Scholar]
- 8.Magiorakos A.P., Srinivasan A., Carey R.B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 9.Sherrard L.J., Tunney M.M., Elborn J.S. Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet (London, England) 2014;384(9944):703–713. doi: 10.1016/S0140-6736(14)61137-5. [DOI] [PubMed] [Google Scholar]
- 10.Smith A.L., Fiel S.B., Mayer-Hamblett N., Ramsey B., Burns J.L. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: lack of association in cystic fibrosis. Chest. 2003;123(5):1495–1502. doi: 10.1378/chest.123.5.1495. [DOI] [PubMed] [Google Scholar]
- 11.Sherrard L.J., McGrath S.J., McIlreavey L. Production of extended-spectrum β-lactamases and the potential indirect pathogenic role of Prevotella isolates from the cystic fibrosis respiratory microbiota. Int. J. Antimicrob. Agents. 2016;47(2):140–145. doi: 10.1016/j.ijantimicag.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.VanDevanter D.R., LiPuma J.J. Microbial diversity in the cystic fibrosis airways: where is thy sting? Future Microbiol. 2012;7(7):801–803. doi: 10.2217/fmb.12.55. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J., Schloss P.D., Kalikin L.M. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc. Natl. Acad. Sci. 2012;109(15):5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klepac-Ceraj V., Lemon K.P., Martin T.R. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ. Microbiol. 2010;12(5):1293–1303. doi: 10.1111/j.1462-2920.2010.02173.x. [DOI] [PubMed] [Google Scholar]
- 15.Zemanick E.T., Harris J.K., Wagner B.D. Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0062917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coburn B., Wang P.W., Diaz Caballero J. Lung microbiota across age and disease stage in cystic fibrosis. Sci. Rep. 2015;5(1):10241. doi: 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox M.J., Allgaier M., Taylor B. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010;5(6) doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flight W.G., Smith A., Paisey C. Rapid detection of emerging pathogens and loss of microbial diversity associated with severe lung disease in cystic fibrosis. J. Clin. Microbiol. 2015;53(7):2022–2029. doi: 10.1128/JCM.00432-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)-A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inf. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.NHAHNES III . 1988–1994. Centers for Disease Control and Prevention.https://wwwn.cdc.gov/nchs/nhanes3/default.aspx [Google Scholar]
- 21.Zimmer B.L., Bacsafra M., Churc E.A., Mattes T.M., Mendoza-Morales G.V.P.L. 2004. Antimicrobial Susceptibility Testing of Cystic Fibrosis Isolates of Pseudomonas aeruginosa: Evaluation of the MicroScan Dried Overnight Gram Negative Panel and Instrument Systems, Frozen Broth Microdilution Panels, and Disk Diffusion. Abstract C-138. [Google Scholar]
- 22.Schloss P.D., Westcott S.L., Ryabin T. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMurdie P.J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12) doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oksanen J., Blanchet F.G., Friendly M. 2017. Vegan: Community Ecology Package.https://CRAN.R-project.org/package=veganhttps://github.com/vegandevs/vegan/issues%0Ahttps://github.com/vegandevs/vegan/issues%0Ahttps://github.com/vegandevs/vegan R Packag version 24-4. [Google Scholar]
- 27.Lim Y.W., Evangelista J.S., Schmieder R. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J. Clin. Microbiol. 2014;52(2):425–437. doi: 10.1128/JCM.02204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feigelman R., Kahlert C.R., Baty F. Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details. Microbiome. 2017;5(1) doi: 10.1186/s40168-017-0234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bacci G., Mengoni A., Fiscarelli E. A different microbiome gene repertoire in the airways of cystic fibrosis patients with severe lung disease. Int. J. Mol. Sci. 2017;18(8) doi: 10.3390/ijms18081654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbott I.J., Peleg A.Y. Stenotrophomonas, achromobacter, and nonmelioid burkholderia species: antimicrobial resistance and therapeutic strategies. Semin. Respir. Crit. Care Med. 2015;36(1):99–110. doi: 10.1055/s-0034-1396929. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y., Zhu Y., Ma Y. Genomic insights into intrinsic and acquired drug resistance mechanisms in Achromobacter xylosoxidans. Antimicrob. Agents Chemother. 2015;59(2):1152–1161. doi: 10.1128/AAC.04260-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Baets F., Schelstraete P., Van Daele S., Haerynck F., Vaneechoutte M. Achromobacter xylosoxidans in cystic fibrosis: prevalence and clinical relevance. J. Cyst. Fibros. 2007;6(1):75–78. doi: 10.1016/j.jcf.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Tan K., Conway S.P., Brownlee K.G., Etherington C., Peckham D.G. Alcaligenes infection in cystic fibrosis. Pediatr. Pulmonol. 2002;34(2):101–104. doi: 10.1002/ppul.10143. [DOI] [PubMed] [Google Scholar]
- 34.De Baets F., Schelstraete P., Haerynck F. Achromobacter xylosoxidans induced bronchiolitis obliterans in cystic fibrosis. Pediatr. Pulmonol. 2014;49(4):414–416. doi: 10.1002/ppul.22864. [DOI] [PubMed] [Google Scholar]
- 35.Sibley C.D., Parkins M.D., Rabin H.R., Duan K., Norgaard J.C., Surette M.G. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc. Natl. Acad. Sci. 2008;105(39):15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkins M.D., Sibley C.D., Surette M.G., Rabin H.R. The Streptococcus milleri group-an unrecognized cause of disease in cystic fibrosis: a case series and literature review. Pediatr. Pulmonol. 2008;43(5):490–497. doi: 10.1002/ppul.20809. [DOI] [PubMed] [Google Scholar]
- 37.Zemanick E.T., Wagner B.D., Robertson C.E. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur. Respir. J. 2017;50(5) doi: 10.1183/13993003.00832-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filkins L.M., Hampton T.H., Gifford A.H. Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J. Bacteriol. 2012;194(17):4709–4717. doi: 10.1128/JB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth A., Elborn J.S. Exacerbations in cystic fibrosis: 3-Management. Thorax. 2008;63(2):180–184. doi: 10.1136/thx.2006.060905. [DOI] [PubMed] [Google Scholar]
- 40.Zemanick E.T., Wagner B.D., Sagel S.D., Stevens M.J., Accurso F.J., Kirk Harris J. Reliability of quantitative real-time PCR for bacterial detection in cystic fibrosis airway specimens. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahn A., Warnken S., Pérez-Losada M., Freishtat R.J., Crandall K.A. Microbial diversity within the airway microbiome in chronic pediatric lung diseases. Infect. Genet. Evol. 2017 doi: 10.1016/j.meegid.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McArthur A.G., Waglechner N., Nizam F. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57(7):3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia B., Raphenya A.R., Alcock B. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallet F., Perez T., Armand S., Wallaert B., Courcol R.J. Pneumonia due to Bordetella bronchiseptica in a cystic fibrosis patient: 16S rRNA sequencing for diagnosis confirmation. J. Clin. Microbiol. 2002 doi: 10.1128/JCM.40.6.2300-2301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brady C., Ackerman P., Johnson M., McNamara J. Bordetella bronchiseptica in a pediatric Cystic Fibrosis center. J. Cyst. Fibros. 2014 doi: 10.1016/j.jcf.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Barley M., Mcnally J., Marshall B. 2016. Annual Data Report 2016 Cystic Fibrosis Foundation Patient Registry. Cyst Fibros Found Patient Regist. [Google Scholar]