Abstract
Background: Androgen steroid hormones are key drivers of prostate cancer. Previous work has shown that androgens can drive the expression of alternative mRNA isoforms as well as transcriptional changes in prostate cancer cells. Yet to what extent androgens control alternative mRNA isoforms and how these are expressed and differentially regulated in prostate tumours is unknown.
Methods: Here we have used RNA-Seq data to globally identify alternative mRNA isoform expression under androgen control in prostate cancer cells, and profiled the expression of these mRNA isoforms in clinical tissue.
Results: Our data indicate androgens primarily switch mRNA isoforms through alternative promoter selection. We detected 73 androgen regulated alternative transcription events, including utilisation of 56 androgen-dependent alternative promoters, 13 androgen-regulated alternative splicing events, and selection of 4 androgen-regulated alternative 3′ mRNA ends. 64 of these events are novel to this study, and 26 involve previously unannotated isoforms. We validated androgen dependent regulation of 17 alternative isoforms by quantitative PCR in an independent sample set. Some of the identified mRNA isoforms are in genes already implicated in prostate cancer (including LIG4, FDFT1 and RELAXIN), or in genes important in other cancers (e.g. NUP93 and MAT2A). Importantly, analysis of transcriptome data from 497 tumour samples in the TGCA prostate adenocarcinoma (PRAD) cohort identified 13 mRNA isoforms (including TPD52, TACC2 and NDUFV3) that are differentially regulated in localised prostate cancer relative to normal tissue, and 3 ( OSBPL1A, CLK3 and TSC22D3) which change significantly with Gleason grade and tumour stage.
Conclusions: Our findings dramatically increase the number of known androgen regulated isoforms in prostate cancer, and indicate a highly complex response to androgens in prostate cancer cells that could be clinically important.
Keywords: Androgens, AR, prostate cancer, alternative splicing, alternative promoters, alternative 3' ends, transcription, mRNA isoforms
Introduction
A single human gene can potentially yield a diverse array of alternative mRNA isoforms, thereby expanding both the repertoire of gene products and subsequently the number of alternative proteins produced. mRNAs with different exon combinations are transcribed from most (up to 90%) human genes, and can generate variants that differ in regulatory untranslated regions, or encode proteins with different sub-cellular localisations and functions 1– 5. Altered splicing patterns have been suggested as a new hallmark of cancer cells 6– 8, and in prostate cancer there is emerging evidence that expression of specific mRNA isoforms derived from cancer-relevant genes may contribute to disease progression 9– 11.
Androgen steroid hormones and the androgen receptor (AR) play a key role in the development and progression of prostate cancer, with alternative splicing enabling cancer cells to produce constitutively active ARs 11– 13. The AR belongs to the nuclear receptor superfamily of transcription factors, and is essential for prostate cancer cell survival, proliferation and invasion 14– 16. Classically, androgen binding promotes AR dimerization and its translocation to the nucleus, where it acts as either a transcriptional activator or a transcriptional repressor to dictate prostate specific gene expression patterns 17– 23. The major focus for prostate cancer therapeutics has been to reduce androgen levels through androgen deprivation therapy (ADT), either with inhibitors of androgen synthesis (for example, abiraterone) or with antagonists that prevent androgen binding to the AR (such as bicalutamide or enzalutamide) 24. Although ADT is usually initially effective, most patients ultimately develop lethal castrate resistant disease for which there are limited treatment options 11, 12.
Androgens and other steroid hormones have also been associated with alternative splicing. Recent RNA-sequencing-based analysis of the androgen response of prostate cancer cells grown in vitro and within patients following ADT identified a set of 700 genes whose transcription is regulated by the AR in prostate cancer cells 25. However, in addition to regulating transcriptional levels, steroid hormone receptors can control exon content of mRNA 10, 26– 29. In prostate cancer androgens can modulate the expression of mRNA isoforms via pre-mRNA processing and promoter selection 9, 10, 18, 30. The AR can recruit the RNA binding proteins Sam68 and p68 as cofactors to influence alternative splicing of specific genes, and studies using minigenes driven from steroid responsive promoters indicate that the AR can affect both the transcriptional activity and alternative splicing of a subset of target genes 11, 31, 32. Other steroid hormones also coordinate both transcription and splicing decisions 29. The thyroid hormone receptor (TR) is known to play a role in coordinating the regulation of transcription and alternative splicing 27, and the oestrogen receptor (ER) can both regulate alternative promoter selection and induce alternative splicing of specific gene sets that can influence breast cancer cell behaviour 28, 33– 35.
In previous work we used exon level microarray analysis to identify 7 androgen dependent changes in mRNA isoform expression 10. However, to what extent androgen-regulated mRNA isoforms are expressed in clinical prostate cancer is unclear. To address this, here we have used RNA-Sequencing data to globally profile alternative isoform expression in prostate cancer cells exposed to androgens, and correlated the results with transcriptomic data from clinical tissue. Our findings increase the number of known AR regulated mRNA isoforms by 10 fold and imply that pre-mRNA processing is an important mechanism through which androgens regulate gene expression in prostate cancer.
Methods
Cell culture
Cell culture was as described previously 25, 36. All cells were grown at 37°C in 5% CO 2. LNCaP cells (CRL-1740, ATCC) were maintained in RPMI-1640 with L-Glutamine (PAA Laboratories, R15-802) supplemented with 10% Fetal Bovine Serum (FBS) (PAA Laboratories, A15-101). For androgen treatment of cells, medium was supplemented with 10% dextran charcoal stripped FBS (PAA Laboratories, A15-119) to produce a steroid-deplete medium. Following culture for 72 hours, 10 nM synthetic androgen analogue methyltrienolone (R1881) (Perkin-Elmer, NLP005005MG) was either added (Androgen +) or absent (Steroid deplete) for the times indicated.
RNA-Seq analysis
RNA-seq transcript expression analysis of previously generated data 25 was performed according to the Tuxedo protocol 37. All reads were first mapped to human transcriptome/genome (build hg19) with TopHat 38/Bowtie 39, followed by per-sample transcript assembly with Cufflinks 40. The mapped data was processed with Cuffmerge, Cuffdiff and Cuffcompare, followed by extraction of significantly differentially expressed genes/isoforms; expression changes between cells grown with androgen and cells grown without androgens were assessed. Reference files for the human genome (UCSC build hg19) were downloaded from the Cufflinks pages: ( UCSC-hg19 package from June 2012 was used.). The software versions used for the analysis were: TopHat v1.4.1, SAM tools Version: 0.1.18 (r982:295), bowtie version 0.12.8 (64-bit) and cufflinks v1.3.0 (linked against Boost version 104000). The Tuxedo protocol 37 was carried out as follows: For steps 1–5, no parameters (except for paths to input/output files) were altered. In step 15, additional switches -s, -R, and -C were used when running cuffcompare. Steps 16–18 (extraction of significant results) were performed on the command line.
RNA extraction, RT–PCR and real-time PCR
Cells were harvested and total RNA extracted using TRIzol (Invitrogen, 15596-026) according to manufacturer's instructions. RNA was treated with DNase 1 (Ambion, AM2222) and cDNA was generated by reverse transcription of 500ng of total RNA using the Superscript VILO cDNA synthesis kit (Invitrogen, 11754-050). Alternative events were analysed by either reverse transcriptase PCR or real-time PCR. Exon profiles were monitored and quantified using the Qiaxcel capillary electrophoresis system (Qiagen) and percentage inclusion was calculated as described previously 10. Real time PCR was performed in triplicate on cDNA using SYBR® Green PCR Master Mix (Invitrogen, 4309155) and the QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific). Samples were normalised using the average of three reference genes, GAPDH, β -tubulin and actin. Ct values for each sample were calculated using SDS 2.4 software (Applied Biosystems) and relative mRNA expression was calculated using the 2-ΔΔCt method. All primer sequences are listed in Supplementary Table 1. Raw Ct values are given in Dataset 1 41.
Antibodies
The following commercial antibodies were used in the study: anti-RLN2 rabbit monoclonal (Abcam, ab183505 1:1000 dilution), anti-TACC2 rabbit polyclonal antibody (11407-1-AP, Proteintech 1:500 dilution), anti-NDUFV3 rabbit polyclonal antibody (13430-1-AP, Proteintech 1:500 dilution), anti-actin rabbit polyclonal (A2668, Sigma 1:2000 dilution), anti-α-Tubulin mouse monoclonal (Sigma, T5168 1:2000 dilution), normal rabbit IgG (711-035-152, Jackson labs 1:2000 dilution) and normal mouse IgG (715-036-150, Jackson labs 1:2000 dilution).
Gene ontology analysis
Gene ontology (GO) analysis of RNA-Seq data was carried out as described previously 42. Enrichment of GO terms (with b500 annotations) was calculated using the goseq R package (version 1.18.0). Genes were considered significant at a p-value threshold of 0.05 after adjustment using the Benjamini-Hochberg false discovery rate.
Bioinformatic analysis of patient transcriptome data
Available clinical and processed RNA-Seq data from The Cancer Genome Atlas (TCGA) prostate adenocarcinoma (PRAD) cohort, comprising 497 tumour samples from as many patients with different stages / Gleason grades and 52 matched samples taken from normal prostate tissue (were downloaded from the Broad Institute TCGA Genome Analysis Center (Firehose 16/01/28 run https://doi.org/10.7908/C11G0KM9 43). Transcriptome data from the TCGA PRAD cohort were analysed for alternative isoform expression, with transcript models relying on TCGA GAF2.1, corresponding to the University of California, Santa Cruz (UCSC) genome annotation from June 2011 ( hg19 assembly). This annotation encompassed 42 of the 73 androgen-regulated alternative mRNA isoform pairs identified. These were studied using two types of analysis: 1) differential transcript expression between tumour and normal prostate tissue and 2) correlation between isoform expression in tumour samples and Gleason score or tumour stage.
Differential isoform and gene expression analysis was performed on estimated read counts using the limma software R package (version 3.7) following its RNA-Seq analysis workflow 44. This workflow was also used for differential isoform ratio analysis, relying on logit-transformed ratio (see below). An FDR-adjusted p-value of 0.05 for the moderated t-statistics was used as threshold for significance of differential expression. Individual isoform expression was estimated in TPM (transcripts per million mapped reads). The expression ratio, henceforth called PSI (percent spliced-in), of each annotated androgen-regulated isoform pair in each TCGA sample was calculated as the ratio between the expression of isoform 1 and the total expression of isoforms 1 and 2 combined, i.e. the sum of their expressions. For each isoform pair, ΔPSI is the difference of median PSI between the tumour and the normal groups of samples.
Two-tailed Spearman’s rank correlation tests were used to study the association between isoform expression and both Gleason score and tumour stage (these were used herein as numeric variables). An FDR-adjusted p-value of 0.05 was used as threshold for significance. Isoform expression differences between tumour and normal samples were considered equivalent to those detected in LNCaP cells under androgen stimulation when there was a statistically significant consistent change in the levels of the expected induced or repressed isoform (1 or 2), concomitant with no contradictory change in the PSI. Isoform “switches” were considered equivalent when there was a minimum (ΔPSI > 2.5%) and statistically significant consistent change in the PSI. Equivalent criteria were used to evaluate the equivalence between androgen-dependence and the associations with Gleason score and tumour stage.
Statistical analysis
Statistical analyses were conducted using the GraphPad Prism software (version 5.04/d). PCR quantification of mRNA isoforms was assessed using the unpaired student’s t-test.
Data is presented as the mean of three independent samples ± standard error of the mean (SEM). Statistical significance is denoted as * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 and **** p ≤ 0.0001.
Results
Global identification of androgen-dependent mRNA isoform production in prostate cancer cells predicts a major role for alternative promoter utilisation
We analysed previously published RNAseq data from LNCaP cells 25 to globally profile how frequently androgens drive production of alternative mRNA isoforms in prostate cancer cells. This analysis identified a group of 73 androgen regulated alternative mRNA isoforms, which could be validated by visualisation on the UCSC Genome Browser 45 ( Table 1). 64 AR regulated mRNA isoforms were novel to this study. Experimental validation in an independent RNA sample set using RT-PCR confirmed 17/17 of these alternative events at the mRNA level ( Supplementary Figure 1). 73% of genes (53/73) with identified alternative androgen regulated mRNA isoforms also changed their overall expression levels in response to androgens ( Table 2). Some of the androgen regulated alternative events are in genes are already implicated in in either prostate cancer or other cancer types (summarised in Table 3). However, Gene Ontology analysis of these 73 genes did not identify any significantly enriched biological processes.
Table 1. Details of the 73 androgen regulated mRNA isoforms identified in prostate cancer cells.
| Isoform 1 | Isoform 2 | TCGA PRAD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Event type | Position (hg19) | RefSeq | Position (hg19) | RefSeq | Change with
androgens |
PCR
Validation |
Predicted
to change protein? |
Isoform 1 ID | Isoform 2 ID | Comparable? |
| LIG4 |
Alternative
promoter |
chr13:108859792-
108870716 |
NM_001098268.1 |
chr13:108859792-
108867130 |
NM_002312.3 | Induction of
promoter 2 |
Yes (Qiaxel) | No (5'
UTR) |
uc001vqp.2 | uc001vqn.2 | Yes |
| TACC2 |
Alternative
promoter |
chr10:123748689-
124014060 |
NM_206862.3 |
chr10:123872554-
124014060 |
NM_001291879.1 | Repression of promoter 1 | Yes (Qiaxel) | Yes | uc001lfv.2 | uc001lfx.2 | Yes |
| TPD52 |
Alternative
promoter |
chr8:80947103-
81083894 |
NM_001287144.1 |
chr8:80947103-
80993066 |
NM_001025252.2 | Induction of
promoter 2 |
Yes (Qiaxel) | Yes | uc003ybs.1 | uc003ybr.1 | Yes |
| NUP93 |
Alternative
promoter |
chr16:56764017-
56878861 |
NM_014669.4 |
chr16:56815704-
56878861 |
NM_001242795.1 | Induction of
promoter 1 |
Yes (SYBR) | Yes | uc002eka.2 | uc002ekb.2 | Yes |
| RLN1 |
Alternative
promoter |
chr9:5334932-
5339873 |
NM_006911.3 |
chr9:5335270-
5339396 |
Not annotated |
Repression of
promoter 2 |
Yes (Qiaxel) | Yes
(change from non- coding) |
uc003zjb.1 |
Not
annotated |
No |
| AP2S1 |
Alternative
promoter |
chr19:47341415-
47354252 |
NM_001301078.1 |
chr19:47341415-
47353547 |
NM_001301076.1 | Induction of
promoter 2 |
Yes (SYBR) | Yes | uc002pft.1 |
Not
annotated |
No |
| RLN2 |
Alternative
promoter |
chr9:5299866-
5304611 |
NM_005059.3 |
chr9:5299890-
5304222 |
Not annotated | Induction of
promoter 1 |
Yes (Qiaxel) | Yes
(change from non- coding) |
uc003ziz.1 |
Not
annotated |
No |
| PIK3R1 |
Alternative
promoter |
chr5:67511584-
67597649 |
NM_181523.2 |
chr5:67584252-
67597649 |
NM_181524.1 |
Repression of
promoter 2 |
Yes (SYBR) | Yes | uc003jva.2 | uc003jvc.2 | Yes |
| MAPRE2 |
Alternative
promoter |
chr18:32556892-
32723432 |
NM_001143826.2 |
chr18:32621324-
32723432 |
NM_014268.3 | Switch to
promoter 2 |
Yes (Qiaxel) | Yes | uc010xcb.1 | uc002kyf.2 | Yes |
| NDUFAF4 |
Alternative
promoter |
chr6:97337187-
97345767 |
NM_014165.3 |
chr6:97337227-
97345368 |
Not annotated |
Repression of
promoter 2 |
Yes (Qiaxel) | Yes
(change from non- coding) |
uc003pov.2 |
Not
annotated |
No |
| DCXR |
Alternative
promoter |
chr17:79993757-
79995573 |
NM_016286.3 |
chr17:79993765-
79995217 |
Not annotated |
Repression of
promoter 2 |
Yes (Qiaxel) | Yes | uc002kdg.2 |
Not
annotated |
No |
| PEX10 |
Alternative
promoter |
chr1:2336241-
2344010 |
NM_002617.3 | Not annotated | Switch to
promoter 2 |
Yes (Qiaxel) | Yes | uc001ajh.2 |
Not
annotated |
No | |
| SNAPC2 |
Alternative
promoter |
chr19:7985194-
7988136 |
NM_003083.3 |
chr19:7985867-
7988136 |
NR_030717.1 | Switch to
promoter 2 |
Yes (SYBR) | Yes
(change to non- coding) |
uc002miw.1 | uc002mix.1 | Yes |
| ATP6V0D1 |
Alternative
promoter |
chr16:67471917-
67515089 |
NM_004691.4 |
chr16:67471931-
67475338 |
Not annotated |
Repression of
promoter 2 |
Yes | uc002ete.1 |
Not
annotated |
No | |
| ARRDC1 |
Alternative
promoter |
chr9:140500092-
140509812 |
NM_001317968.1 |
chr9:140506874-
140509793 |
Not annotated | Induction of
promoter 2 |
Yes
(change to non- coding) |
uc004cnp.1 |
Not
annotated |
No | |
| DENND1A |
Alternative
promoter |
chr9:126141933-
126692417 |
NM_020946.1 |
chr9:126143408-
126586780 |
Not annotated |
Repression of
promoter 2 |
Yes | uc004bnz.1 |
Not
annotated |
No | |
| KLHL36 |
Alternative
promoter |
chr16:84682117-
84701292 |
NM_024731.3 |
chr16:84684274-
84701134 |
Not annotated | Induction of
promoter 2 |
Yes | uc002fig.2 |
Not
annotated |
No | |
| RAB3IL1 |
Alternative
promoter |
chr11:61664768-
61687741 |
NM_001271686.1 |
chr11:61664768-
61685081 |
NM_013401.3 |
Repression of
promoter 2 |
Yes | uc001nsp.2 | uc001nso.2 | Yes | |
| ACER3 |
Alternative
promoter |
chr11:76571917-
76737841 |
NM_018367.6 |
chr11:76631206-
76737818 |
Not annotated |
Repression of
promoter 2 |
Yes | uc009yum.1 |
Not
annotated |
No | |
| OSBPL1A |
Alternative
promoter |
chr18:21742011-
21977833 |
NM_080597.3 |
chr18:21742011-
21852196 |
NM_018030.4 | Induction of
promoter 2 |
Yes | uc002kve.2 | uc002kvd.2 | Yes | |
| TRIM16 |
Alternative
promoter |
chr17:15531280-
15586193 |
NM_006470.3 |
chr17:15530970-
15555735 |
Not annotated | Induction of
promoter 2 |
Yes | uc002gow.2 |
Not
annotated |
No | |
| VSIG10L |
Alternative
promoter |
chr19:51834795-
51845378 |
NM_001163922.1 |
chr19:51834795-
51843009 |
Not annotated | Induction of
promoter 1 |
Yes | uc002pwf.2 |
Not
annotated |
No | |
| SEPT5 |
Alternative
promoter |
chr22:19701987-
19710845 |
NM_002688.5 |
chr22:19705958-
19710845 |
NM_001009939.2 |
Repression of
promoter 2 |
Yes | uc002zpv.1 | uc002zpw.1 | Yes | |
| HMGCR |
Alternative
promoter |
chr5:74632154-
74657926 |
NM_000859 |
chr5:74632993-
74657926 |
NM_000859.2 |
Repression of
promoter 1 |
Yes | uc011cst.1 | uc003kdp.2 | Yes | |
| RDH13 |
Alternative
promoter |
chr19:55555692-
55580914 |
NM_138412.3 |
chr19:55555692-
55574585 |
NM_001145971.1 | Induction of
promoter 1 |
Yes | uc002qip.2 | uc010esr.1 | Yes | |
| GPRIN2 |
Alternative
promoter |
chr10:46993001-
47000677 |
Not annotated |
chr10:46993546-
47000568 |
NM_014696.3 |
Repression of
promoter 2 |
No (5' UTR) | Not annotated | uc001jec.2 | No | |
| CLK3 |
Alternative
promoter |
chr15:74900713-
74922542 |
NM_003992.4 |
chr15:74,908,246-
74,922,542 |
NM_003992 |
Repression of
promoter 1 |
Yes | uc002ayg.3 | uc002ayj.3 | Yes | |
| RNH1 |
Alternative
promoter |
chr11:494512-
507283 |
NM_203387.2 |
chr11:494512-
506821 |
NM_002939.3 | Induction of
promoter 1 |
No (5' UTR) | uc001lpp.1 | uc001lpl.1 | Yes | |
| ZFAND6 |
Alternative
promoter |
chr15:80351910-
80430735 |
NM_001242911.1 |
chr15:80364903-
80430735 |
NM_001242916.1 |
Repression of
promoter 2 |
No (5' UTR) | uc002bff.1 | uc002bfh.1 | Yes | |
| CDIP1 |
Alternative
promoter |
chr16:4560677-
4588816 |
NM_013399.2 |
chr16:4560677-
4588471 |
NM_001199054.1 |
Repression of
promoter 2 |
No (5' UTR) | uc002cwu.2 | uc002cwv.2 | Yes | |
| YIF1B |
Alternative
promoter |
chr19:38794200-
38806606 |
NM_001039672.2 |
chr19:38794200-
38806445 |
NM_001145461.1 | Switch to
promoter 2 |
Yes | uc002ohz.2 | uc002ohx.2 | Yes | |
| LIMK2 |
Alternative
promoter |
chr22:31608250-
31676066 |
NM_005569.3 |
chr22:31644348-
31676066 |
NM_016733.2 | Switch to
promoter 2 |
Yes | uc003akh.2 | uc003aki.2 | Yes | |
| TSC22D3 |
Alternative
promoter |
chrX:106956452-
106959711 |
NM_001015881.1 |
chrX:106956452-
106960291 |
NM_004089.3 |
Repression of
promoter 1 |
Yes | uc004enf.2 | uc004eng.2 | Yes | |
| ALDH1A3 |
Alternative
promoter |
chr15:101419897-
101456830 |
NM_000693.3 |
chr15:101438281-
101457072 |
Not annotated |
Repression of
promoter 1 |
Yes | uc002bwn.3 |
Not
annotated |
No | |
| TRABD |
Alternative
promoter |
chr22:50624341-
50638028 |
NM_001320485.1 |
chr22:50628979-
50638028 |
NM_001320487.1 | Switch to
promoter 2 |
No (5' UTR) | uc003bjq.1 | uc003bjs.1 | Yes | |
| LIMCH1 |
Alternative
promoter |
chr4:41361624-
41702061 |
NM_001289124.1 |
chr4:41362648-
41702061 |
NM_001289122.2 |
Repression of
promoter 2 |
Yes | uc003gvu.3 |
Not
annotated |
No | |
| GMFB |
Alternative
promoter |
chr14:54941209-
54955744 |
NM_004124.2 |
chr14:54941314-
54955637 |
Not annotated | Induction of
promoter 2 |
Yes
(change to non- coding) |
uc010tqz.1 |
Not
annotated |
No | |
| MLST8 |
Alternative
promoter |
chr16:2255178-
2259418 |
NM_022372.4 |
chr16:2255732-
2259418 |
NM_001199174.1 | Switch to
promoter 1 |
No (5' UTR) | uc010uvy.1 | uc002cpf.2 | Yes | |
| TLE3 |
Alternative
promoter |
chr15:70340130-
70390256 |
NM_020908.2 |
chr15:70340130-
70387124 |
NM_001282982.1 | Induction of
promoter 2 |
Yes | uc002asn.2 | uc002ask.2 | Yes | |
| UBA1 |
Alternative
promoter |
chrX:47050199-
47074527 |
NM_153280.2 |
chrX:47053201-
47074527 |
NM_003334.3 |
Repression of
promoter 1 |
No (5' UTR) | uc004dhj.3 | uc004dhk.3 | Yes | |
| TNRC6B |
Alternative
promoter |
chr22:40440821-
40731812 |
NM_001024843.1 |
chr22:40573929-
40731812 |
NM_001162501.1 |
Repression of
promoter 2 |
Yes | uc003aym.2 | uc011aor.1 | Yes | |
| FDFT1 |
Alternative
promoter |
chr8:11660120-
11696818 |
NM_004462.4 |
chr8:11665926-
11696818 |
NM_001287750.1 |
Repression of
promoter 2 |
Yes | uc003wui.2 | uc010lsb.2 | Yes | |
| GREB1 |
Alternative
promoter |
chr2:11674242-
11782912 |
NM_014668.3 |
chr2:11680080-
11728355 |
NM_148903.2 | Induction of
promoter 2 |
Yes | uc002rbo.1 | uc002rbl.2 | Yes | |
| NCAPD3 |
Alternative
promoter |
chr11:134022337-
134094426 |
NM_015261.2 |
chr11:134022772-
134093593 |
Not annotated | Induction of
promoter 2 |
Yes | uc001qhd.1 |
Not
annotated |
No | |
| SLC36A4 |
Alternative
promoter |
chr11:92877337-
92931141 |
NM_152313.3 |
chr11:92877337-
92930621 |
NM_001286139.1 | Induction of
promoter 2 |
Yes | uc001pdn.2 |
Not
annotated |
No | |
| KLC2 |
Alternative
promoter |
chr11:66024765-
66035331 |
NM_001134775.1 |
chr11:66025174-
66035331 |
NM_022822.2 |
Repression of
promoter 1 |
No (5' UTR) | uc010rov.1 | uc001ohb.2 | Yes | |
| RAP1GAP |
Alternative
promoter |
chr1:21922708-
21978348 |
NM_001145658.1 |
chr1:21922533-
21946950 |
Not annotated |
Repression of
promoter 1 |
Yes | uc001bez.1 |
Not
annotated |
No | |
| TMEM79 |
Alternative
promoter |
chr1:156252704-
156262234 |
NR_026678.1 |
chr1:156254070-
156262234 |
NM_032323.2 |
Repression of
promoter 1 |
No (5' UTR) | uc001fod.2 | uc010phi.1 | Yes | |
| NR4A1 |
Alternative
promoter |
chr12:52416616-
52453291 |
NM_001202233.1 |
chr12:52445186-
52453291 |
NM_173157.2 | Induction of
promoter 2 |
Yes | uc010sno.1 | uc001rzr.2 | Yes | |
| ZNF32 |
Alternative
promoter |
chr10:44139307-
44144326 |
NM_001324166.1 |
chr10:44139307-
44144326 |
NM_001324167.1 |
Repression of
promoter 2 |
No (5' UTR) | uc001jbc.2 | uc001jbb.2 | Yes | |
| C1QTNF3 |
Alternative
promoter |
chr5:34017963-
34043371 |
NM_181435.5 |
chr5:34018571-
34035881 |
Not annotated | Induction of
promoter 1 |
Yes | uc003jio.2 |
Not
annotated |
No | |
| UBE2D3 |
Alternative
promoter |
chr4:103715540-
103748710 |
NM_181887.2 |
chr4:103715540-
103749105 |
NM_181886.3 | Switch to
promoter 2 |
No (5' UTR) | uc003hwk.2 | uc011cet.1 | Yes | |
| KRT8 |
Alternative
promoter |
chr12:53290971-
53343650 |
NM_001256293.1 |
chr12:53,290,971-
53,298,868 |
NM_002273 |
Repression of
promoter 1 |
No (5' UTR) | uc009zml.1 | uc001sbd.2 | Yes | |
| ELOVL1 |
Alternative
promoter |
chr1:43829068-
43833745 |
NM_022821.3 |
chr1:43829093-
43832057 |
Not annotated | Induction of
promoter 2 |
Yes
(change to non- coding) |
uc001cjb.2 |
Not
annotated |
No | |
| RCAN1 |
Alternative
promoter |
chr21:35888740-
35987441 |
NM_004414.6 |
chr21:35888740-
35899308 |
NM_203418.2 | Induction of
promoter 2 |
Yes | uc002yue.2 | uc002yub.2 | Yes | |
| SORBS3 |
Alternative
promoter |
chr8:22409251-
22433008 |
NM_005775.4 |
chr8:22422332-
22433100 |
Not annotated | Induction of
promoter 2 |
Yes | uc003xbv.2 |
Not
annotated |
No | |
| MAT2A |
Alternative
3' end |
chr2:85766101-
85772403 |
NM_005911.5 |
chr2:85,766,101-
85,770,775 |
NM_005911 |
Repression of
isoform 2 |
Yes (Qiaxel) | Yes | uc002spr.2 | uc010ysr.1 | Yes |
| CNNM2 |
Alternative
3' end |
chr10:104678075-
104687375 |
NM_199077.2 |
chr10:104678075-
104838344 |
NM_017649.4 | Induction of
isoform 1 |
Yes (SYBR) | Yes | uc001kwl.2 | uc001kwm.2 | Yes |
| TMEM125 |
Alternative
3' end |
chr1:43735698-
43736343 |
Not annotated |
chr1:43735665-
43739673 |
NM_144626.2 | Induction of
isoform 1 |
Yes
(change to non- coding) |
Not
annotated |
uc001cir.2 | No | |
| CBWD2 |
Alternative
3' end |
chr2:114195268-
114253781 |
NM_172003.3 |
chr2:114195169-
114199073 |
Not annotated | Induction of
isoform 2 |
Yes | uc002tju.2 |
Not
annotated |
No | |
| NDUFV3 |
Alternative
exon |
chr21:44313378-
44329773 |
NM_021075.3 |
chr21:44313378-
44329773 |
NM_001001503.1 | Switch to isoform 2 (exon
excluded) |
Yes | uc002zcm.2 | uc002zcn.2 | Yes | |
| ZNF678 |
Alternative
exon |
chr1:227751220-
227850164 |
NM_178549.3 | Not annotated | Switch to isoform 2 (exon
excluded) |
Yes
(change to non- coding) |
uc009xet.1 |
Not
annotated |
No | ||
| ZNF121 |
Alternative
exon |
chr19:9676404-
9695209 |
NM_001308269.1 |
chr19:9676404-
9695209 |
NM_001008727.3 | Switch to isoform 2 (exon
excluded) |
Yes | uc010xkq.1 | uc010xkp.1 | Yes | |
| SPATC1L |
Alternative
exon |
chr21:47581062-
47604373 |
NM_032261.4 | Not annotated | Induction of isoform 2 (exon
included) |
Yes | uc002zii.2 |
Not
annotated |
No | ||
| MOCOS |
Alternative
exon |
chr18:33767480-
33848685 |
NM_017947.2 | Not annotated | Switch to isoform 2 (exon
excluded) |
Yes | uc002kzq.3 |
Not
annotated |
No | ||
| RBM45 |
Alternative
exon |
chr2:178977151-
178994382 |
NM_152945.3 | Not annotated | Switch to isoform 2 (exon
included) |
Yes | uc002ulv.2 |
Not
annotated |
No | ||
| MIPEP |
Alternative
exon |
chr13:24304328-
24463587 |
NM_005932.3 | Not annotated | Repression of isoform 2 (exon
excluded) |
Yes | uc001uox.3 |
Not
annotated |
No | ||
| BBS4 |
Alternative
exon |
chr15:72978520-
73030817 |
NM_001320665.1 | Not annotated | Induction of isoform 2 (exon
included) |
Yes | uc002avb.2 |
Not
annotated |
No | ||
| FAM195A |
Alternative
exon |
chr16:691804-
698474 |
NM_138418.3 |
chr16:691804-
698474 |
NR_138607.1 | Switch to isoform 1 (exon
exluded) |
Yes
(change from non- coding) |
uc002cic.1 | uc002cie.2 | Yes | |
| LINC01133 |
Alternative
exon |
chr1:159931008-
159948851 |
ENST00000443364.6 |
chr1:159931014-
159948876 |
NR_038849.1 | Induction of isoform 1 (exon
excluded) |
Both non-coding |
Not
annotated |
uc001fuu.2 | No | |
| SS18 |
Alternative
exon |
chr18:23596217-
23670611 |
NM_001007559.2 |
chr18:23596217-
23670611 |
NM_005637.3 | Switch to isoform 2 (exon
excluded) |
Yes | uc002kvm.2 | uc002kvn.2 | Yes | |
| RHOC |
Alternative
exon |
chr1:113243897-
113249757 |
ENST00000369638.6 |
chr1:113243947-
113249742 |
ENST00000369636.6 | Switch to isoform 2 (exon
excluded) |
No (5' UTR) | uc009wgk.1 | uc001ecr.1 | Yes | |
| ZNF226 |
Retained
intron |
chr19:44669215-
44681838 |
NM_001319088.1 |
chr19:44669249-
44679582 |
NM_015919.3 | Switch to isoform 1 (intron
included) |
Yes | uc002oyo.2 | uc002oyn.2 | Yes | |
Table 2. Quantitative changes in gene expression in response to androgens for the 73 genes with AR regulated alternative mRNA isoforms.
| LNCaP RNA-Seq (+/- androgens for 24 hours) | Reciprocal RNA-Seq (also change in 7
patients following ADT) |
||||
|---|---|---|---|---|---|
| No change | Upregulated | Downregulated | No change | Upregulated | Downregulated |
| RLN2 | LIG4 | NUP93 | LIG4 | TPD52 | None |
| DENND1A | TACC2 | PIK3R1 | TACC2 | AP2S1 | |
| RAB3IL1 | RLN1 | MAPRE2 | NUP93 | DCXR | |
| OSBPL1A | AP2S1 | NDUFAF4 | RLN1 | PEX10 | |
| TRIM16 | DCXR | ACER3 | RLN2 | HMGCR | |
| Sep-05 | PEX10 | GPRIN2 | PIK3R1 | ALDH1A3 | |
| RDH13 | SNAPC2 | TLE3 | MAPRE2 | FDFT1 | |
| ZFAND6 | ATP6V0D1 | TNRC6B | NDUFAF4 | GREB1 | |
| CDIP1 | ARRDC1 | SORBS3 | SNAPC2 | NCAPD3 | |
| LIMK2 | KLHL36 | ZNF121 | ATP6V0D1 | RAP1GAP | |
| TSC22D3 | VSIG10L | LINC01133 | ARRDC1 | TMEM79 | |
| GMFB | HMGCR | DENND1A | KRT8 | ||
| MLST8 | CLK3 | KLHL36 | ELOVL1 | ||
| znf32 | RNH1 | RAB3IL1 | TMEM125 | ||
| C1QTNF3 | YIF1B | ACER3 | |||
| UBE2D3 | PAK1IP1 | OSBPL1A | |||
| MAT2A | ALDH1A3 | TRIM16 | |||
| CBWD2 | TRABD | VSIG10L | |||
| ZNF678 | LIMCH1 | SEPT5 | |||
| MOCOS | UBA1 | RDH13 | |||
| FDFT1 | GPRIN2 | ||||
| GREB1 | CLK3 | ||||
| NCAPD3 | RNH1 | ||||
| SLC36A4 | ZFAND6 | ||||
| KLC2 | CDIP1 | ||||
| RAP1GAP | YIF1B | ||||
| TMEM79 | LIMK2 | ||||
| NR4A1 | TSC22D3 | ||||
| KRT8 | TRABD | ||||
| ELOVL1 | LIMCH1 | ||||
| RCAN1 | GMFB | ||||
| CNNM2 | MLST8 | ||||
| TMEM125 | TLE3 | ||||
| NDUFV3 | UBA1 | ||||
| SPATC1L | TNRC6B | ||||
| RBM45 | SLC36A4 | ||||
| MIPEP | KLC2 | ||||
| BBS4 | NR4A1 | ||||
| FAM195A | znf32 | ||||
| SS18 | C1QTNF3 | ||||
| RHOC | UBE2D3 | ||||
| ZNF226 | RCAN1 | ||||
| TPD52 | SORBS3 | ||||
| MAT2A | |||||
| CNNM2 | |||||
| CBWD2 | |||||
| NDUFV3 | |||||
| ZNF678 | |||||
| ZNF121 | |||||
| SPATC1L | |||||
| MOCOS | |||||
| RBM45 | |||||
| MIPEP | |||||
| BBS4 | |||||
| FAM195A | |||||
| LINC01133 | |||||
| SS18 | |||||
| RHOC | |||||
| ICAM3 | |||||
| ZNF226 | |||||
Table 3. Alternative events in genes previously linked to cancer.
| Gene name | Function | Clinical importance and
roles in other cancer types |
Clinical importance and roles in prostate
cancer |
|---|---|---|---|
|
TACC2
Transforming Acidic Coiled- Coil Containing Protein 2 |
centrosome- and
microtubule-interacting protein |
Growth and prognosis of
breast cancer 56 |
castration-resistant growth of prostate
cancer 57 |
| LIG4 | DNA ligase with role in DNA
repair |
Prognostic marker in
nasopharyngeal cancer 58 Upregulated in colorectal cancer with role in wnt signalling 59 |
Predictor of poor prognosis 60 |
|
RLN1 and RLN2
(Relaxin1 and 2) |
Endocrine hormones (part of
insulin gene superfamily) |
Breast cancer
invasiveness 61, 62 metastasis of human osteosarcoma 63 Thyroid cancer oncogenesis 64, 65 |
Well characterised role in the development
and progression of prostate cancer 5, 50– 55. |
|
TPD52
(Tumor Protein D52) |
Role in proliferation and exo-
and endocytic pathways |
Well characterised role
in numerous cancer types 46, 66– 69 |
Known AR target, overexpressed and
amplified in prostate cancer 70 Oncogene in prostate cancer 71 Neuroendocrine transdifferentiation of prostate cancer 72 Isoform produced by alternative promoter known as PrLZ and already linked to prostate cancer 47– 49, 73, 74 |
|
FDFT1
(Farnesyl-Diphosphate Farnesyltransferase 1) |
squalene synthase | Role in lung cancer
metastasis 75 |
Linked to prostate cancer risk and
aggressiveness 76 |
|
TLE3
(Transducin Like Enhancer Of Split 3) |
Negative regulator of Wnt/β-
catenin signaling |
Predictive marker for
response to therapy in ovarian and breast cancer 77, 78 Represses colon cancer proliferation 79 |
Upregulated in prostate tumours
80 and
linked to wnt signalling in castrate resistant disease 81 |
|
CNNM2
(Cyclin & CBS Domain Divalent Metal Cation Transport Mediator 2) |
Magnesium transporter | Proposed oncogenic role
via increasing magnesium uptake 82 |
Unknown |
| NUP93 | Nucleoporin protein – role in
apoptosis |
Driver mutation linked to
breast cancer 83 |
Unknown |
|
MAT2A
Methionine adenosyltransferase II |
Biosynthesis of
S-adenosylmethionine, the principal biological methyl donor and precursor of polyamines and glutathione. |
Upregulated in liver and
colon cancer, potential drug target 84, 85 Tumour suppressor in kidney carcinogenesis 86 Role in other cancer types 87 |
Upregulated in prostate cancer and linked
to cell migration via miR-34a and miR- 34b 87, 88 |
| PIK3R1 | PI3K regulatory subunit | Underexpressed in breast
cancer 89 High mutation frequency in endometrial cancer 90 |
Controlled by androgens and repressed in
prostate cancer cells 21 |
|
SNAPC2
(Small Nuclear RNA Activating Complex Polypeptide 2) |
Subunit of the snRNA-
activating protein complex. Necessary for RNA polymerase II and III dependent small-nuclear RNA gene transcription |
Epigenetic silencing
is prognostic in glioblastoma 91 |
Unknown |
|
ZNF678
(Zinc Finger Protein 678) |
Potential role in
transcriptional regulation |
Unknown | Unknown |
|
NDUFV3
(NADH:Ubiquinone Oxidoreductase Subunit V3) |
Subunit of part of the
mitochondrial respiratory chain |
Unknown | Androgen regulated alternative splice
isoform previously identified by our exon array study 10 |
|
OSBPL1A
(Oxysterol Binding Protein Like 1A) |
Intracellular lipid receptor | Alternative promoter use in
colorectal cancer 92 |
Unknown |
|
RDH13
(Retinol Dehydrogenase 13) |
Role in retinoic acid
production and protection against oxidative stress |
Unknown | Unknown |
|
ZNF121
(Zinc Finger Protein 121) |
Potential role in
transcriptional regulation |
Interacts with MYC.
Upregulated in breast cancer 93 |
Unknown |
|
SLC36A4.1
(Solute Carrier Family 36 Member 4) |
amino acid transporter | Unknown | Unknown |
|
RCAN1
(Regulator of Calcineurin 1) |
Inhibits calcineurin-
dependent signaling pathways |
Inhibits NF-κB and
suppresses lymphoma growth in mice 94. Role in cancer cell migration 95 |
Unknown |
|
DCXR
(Dicarbonyl & l-xylulose reductase) |
Role in the uronate cycle of
glucose metabolism |
Low expression
indicates poor prognosis for hepatocellular carcinoma 96. Role in cell adhesion 97, 98 |
Upregulated and potential biomarker in
prostate cancer 99 |
|
NDUFAF4
(NADH:Ubiquinone Oxidoreductase Complex Assembly Factor 4) |
Role in the mitochondrial
respiratory chain |
Unknown | Unknown |
|
MAPRE2
(Microtubule Associated Protein RP/EB Family Member 2) |
Microtubule-associated
protein that is necessary for spindle symmetry during mitosis |
Role in the invasion of
pancreatic cancer cells 100 |
Unknown |
|
PEX10
(Peroxisomal Biogenesis Factor 10) |
Involved in import of
peroxisomal matrix proteins |
Unknown | Unknown |
|
AP2S1
(Adaptor Related Protein Complex 2 Sigma 1 Subunit) |
Function in protein transport
across membranes |
Unknown | Unknown |
|
LINC01133
(long non-coding RNA) |
Long non-coding RNA | Poor prognosis in
colorectal cancer 101 Upregulated and linked to poor prognosis in lung cancer 102 |
Unknown |
|
ZNF226
(Zinc Finger Protein 226) |
Potential role in
transcriptional regulation |
Unknown | Unknown |
|
CDIP1
(Cell death inducing p53 target 1) |
p53 apoptotic effector
Regulates TNF-alpha- mediated apoptosis |
sensitivity to TNFα-
induced apoptosis in cancer cells 103 |
Unknown |
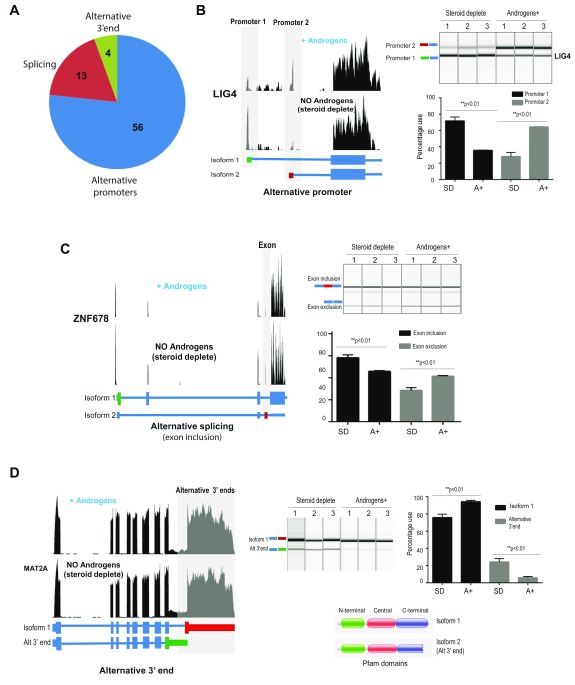
The 73 identified mRNA isoforms were generated via androgen-regulated utilisation of 56 alternative promoters, 4 alternative 3′ ends and 13 alternative splicing events ( Figure 1A). Of the 56 androgen regulated alternative promoters that were identified, 23 alternative promoters were induced by androgens (including LIG4, Figure 1B), 26 promoters were repressed by androgens, and for 7 genes there was a switch in usage from one promoter to another ( Table 1). The alternative splicing events that were under androgen control included 12 alternative exons and one androgen-regulated intron retention ( Table 1). 10 of these are novel to this study, including exclusion of an alternative exon in ZNF678 ( Figure 1C). Of the alternative exons, six genes contained switches in previously unannotated protein-coding exons in response to androgen-exposure. We also identified four androgen regulated alternative mRNA 3' end isoform switches, including a switch in the 3’ end of the mRNA transcript for the MAT2A gene ( Figure 1D).
Figure 1. Global identification of androgen-dependent mRNA isoform production in prostate cancer cells predicts a major role for alternative promoter utilisation.
(A) Analysis of RNAseq data from LNCaP cells grown with (A+) or without androgens (R1881) (steroid deplete, SD) for 24 hours identified 73 androgen regulated alternative mRNA isoforms. The 73 alternative events were generated via androgen-regulated utilisation of 56 alternative promoters, 4 alternative 3' ends and 13 alternative splicing events. (B) Androgens drive a promoter switch in the LIG4 gene, which produces an mRNA isoform with an alternative 5’UTR. Visualisation of our LNCaP cell RNA-seq reads for the LIG4 gene on the UCSC genome browser identified a switch from promoter 1 to alternative promoter 2 in cells grown in the presence of androgens. Promoter 2 is predicted to produce a different 5’UTR without influencing the protein sequence (left panel). Quantitative PCR using primers specific to each promoter indicate that in response to androgens there is repression of promoter 1 and induction of promoter 2 (right panel). (C) Androgens drive alternative splicing of the ZNF678 gene. Visualisation of our LNCaP cell RNA-seq reads for the ZNF678 gene on the UCSC genome browser identified a switch to inclusion of a cassette exon in the presence of androgens. Inclusion of the alternative cassette exon in the ZNF678 gene is predicted to induce a switch to an alternative non-coding mRNA isoform (left panel). Quantitative PCR using primers in flanking exons confirmed increased inclusion of the alternative exon in LNCaP cells exposed to androgens (right panel). (D) Androgens promote selection of an alternative 3’ end for the MAT2A gene. Visualisation of our LNCaP cell RNA-seq reads for the MAT2A gene on the UCSC genome browser indicates a switch to reduced usage of an alternative 3’ end in the presence of androgens (left panel). Quantitative PCR using primers specific to each isoform confirmed down-regulation of an alternative 3’ end (p<0.01). Alternative 3’ ends for the MAT2A gene are predicted to produce proteins with different amino acid sequences and to influence a known Pfam domain (right panel).
Androgen regulated events control the production of alternative protein isoforms, non-coding RNAs and alternative 5' UTRs
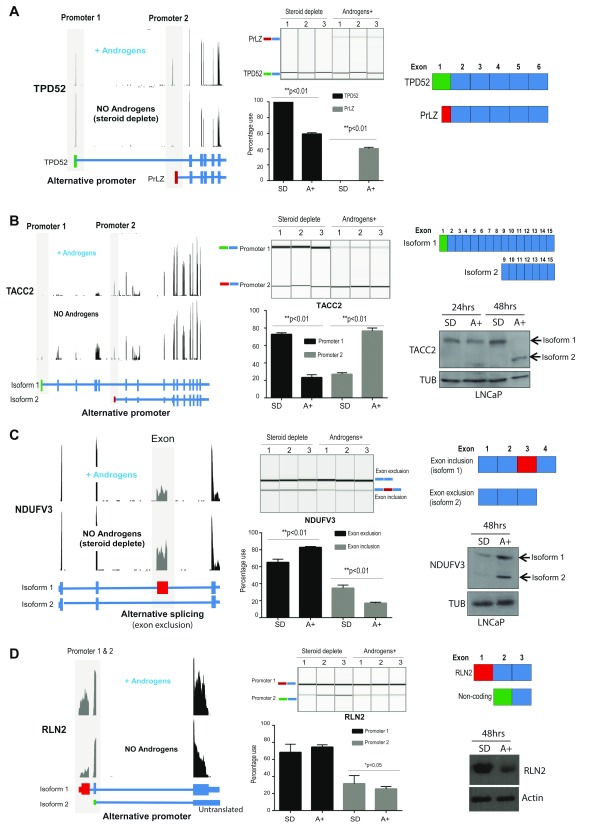
48/73 (66%) of the androgen regulated alternative events detected in response to androgen stimulation are predicted to change the amino acid sequence of the resulting protein ( Table 1). Some of these are already known to have a well characterised role in prostate cancer progression, including an alternative promoter in the oncogene TPD52 that produces a protein isoform called PrLZ ( Figure 2A) 46– 49. Others are not so well characterised. Using western blotting we could detect a novel shorter protein isoform corresponding to androgen-driven selection of an alternative promoter in the TACC2 gene ( Figure 2B); and exclusion of a cassette exon in the NDUFV3 gene, which we show also produces a novel shorter protein isoform ( Figure 2C). We also detected a switch in the 3' end of the mRNA transcript for the MAT2A gene, which is predicted to produce a protein isoform with a shorter C-terminal domain ( Figure 1D); and induction of an alternative 3' isoform of CNNM2, which is predicted to be missing a conserved CBS domain ( Table 1 and Supplementary Figure 1).
Figure 2. Androgen regulated mRNA isoform switches control alternative protein isoforms and non-coding RNAs.
( A) Androgens induce an alternative promoter in the oncogene TPD52 that produces an isoform called PrLZ. Visualisation of our LNCaP cell RNA-seq reads for the TPD52 gene on the UCSC genome browser identified a switch from promoter 1 to alternative promoter 2 in cells grown in the presence of androgens. Promoter 2 is known to produce an alternative protein isoform of TPD52 known as PrLZ (left panel). Quantitative PCR using primers specific to each promoter indicate an induction of the PrLZ isoform in response to androgens (middle panel). PrLZ has an alternative N-terminal amino acid sequence which results in an alternative protein isoform and disrupts a known Pfam domain (right panel). ( B) Androgens induce an alternative promoter in the TACC2 gene that produces a novel alternative protein isoform. Visualisation of our LNCaP cell RNA-seq reads for the TACC2 gene on the UCSC genome browser identified a switch from promoter 1 to alternative promoter 2 in cells grown in the presence of androgens. Promoter 2 is predicted to produce an alternative shorter protein isoform of TACC2 (isoform 2) (left panel). Quantitative PCR using primers specific to each promoter indicate a switch from isoform 1 to isoform 2 in response to androgens (middle panel). Detection of TACC2 protein in LNCaP by western blotting (cells were grown with or without androgens for 24 or 48 hours). Tubulin was used as a loading control. Exposure to androgens for 48 hours induces expression of the alternative TACC2 protein isoform (right panel). ( C) Androgens drive alternative splicing of the NDUFV3 gene. Visualisation of our LNCaP cell RNA-seq reads for the NDUFV3 gene on the UCSC genome browser identified a switch to exclusion of a cassette exon in the presence of androgens (left panel). Quantitative PCR using primers in flanking exons confirmed less inclusion of the alternative exon in LNCaP cells exposed to androgens (middle panel). Exclusion of the alternative cassette exon is predicted to produce an alternative protein isoform. Detection of NDUFV3 protein in LNCaP cells using western blotting (right panel). ( D) Androgens suppress an alternative promoter in the RLN2 gene, which produces a shorter non-coding mRNA isoform. Visualisation of our LNCaP cell RNA-seq reads for the RLN2 gene on the UCSC genome browser identified a switch from promoter 1 to alternative promoter 2 in cells grown in the presence of androgens. Promoter 2 is predicted to produce an untranslated non-coding mRNA isoform (left panel). Quantitative PCR using primers specific to each promoter indicated a significant switch in promoter usage in response to androgens (middle panel). Detection of RLN2 protein in LNCaP by western blotting (cells were grown with or without androgens for 48 hours). Actin was used as a loading control. As seen previously 55, androgens suppress RLN2 protein levels.
11 of the remaining identified androgen-regulated alternative events change the expression of mRNAs from coding to non-coding or untranslated (not predicted to produce a protein) ( Table 1). These included promoter switches for the RLN1 and RLN2 genes which encode peptide hormones that may be important in prostate cancer 5, 50– 55. Androgens drive a promoter switch in both RLN1 and RLN2 to produce predicted non-coding or untranslated mRNA isoforms, reducing expression of protein-coding RLN1 and RLN2 mRNA isoforms. To test whether prostate cancer cells turn off gene expression by switching between utilisation of promoters that generate coding and noncoding mRNAs, we analysed RLN2 protein levels. Consistent with our hypothesis and a previous study 55, RLN2 protein production was negatively regulated by androgens in parallel to the switch to the non-coding mRNA isoform ( Figure 2D).
14 of the identified androgen-dependent mRNA isoforms lead to/result in coding mRNAs with altered 5’ untranslated regions (5′ UTR) with no impact on the coding sequence. These include a promoter switch in the LIG4 gene ( Figure 1B).
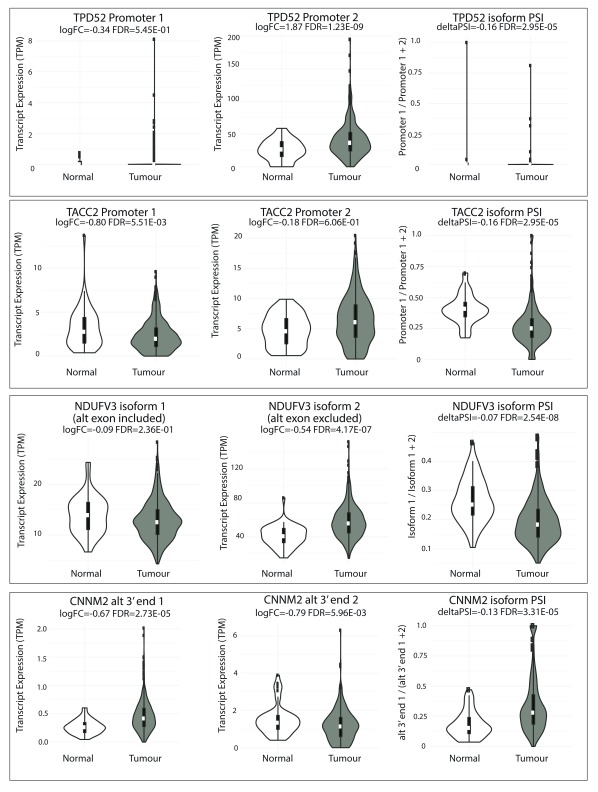
Differential expression of androgen-dependent mRNA isoforms in prostate adenocarcinoma versus normal tissue
To investigate potential links between androgen-dependent mRNA isoforms and tumourigenesis, we analysed the expression of 41 androgen-regulated mRNA isoform pairs in clinical prostate adenocarcinoma and normal prostate tissues. This analysis utilised transcriptomic data from 497 tumour samples and 52 normal samples in the PRAD TCGA cohort 104. The remaining isoform pairs identified within our dataset have not been previously annotated by UCSC, therefore it was not possible to include them in our comparison. A description of the cohort used is summarised in Table 4.
Table 4. Description of the TCGA PRAD cohort.
| Features | Total Cases |
|---|---|
| Cohort | 497 patients |
| Tumour | 497 |
| Normal | 52 (w/tumour matched
sample available) |
| Gleason grade | |
| 6 | 50 |
| 7 | 287 |
| 8 | 67 |
| 9 | 140 |
| 10 | 4 |
| Tumour stage | |
| T2a | 14 |
| T2b | 10 |
| T2c | 192 |
| T3a | 173 |
| T3b | 140 |
| T4 | 12 |
| Gleason grade (alternative gleason grade
grouping) | |
| 1 (primary +
secondary score ≤ 6) |
50 |
| 2 (3 + 4) | 171 |
| 3 (4 + 3) | 123 |
| 4 (4 + 4) | 93 |
| 5 (primary +
secondary score ≥ 9) |
111 |
All tumours were hormone naive (not subject to ADT) at the time of sample collection
33 of the 42 mRNA isoform pairs exhibited significant differences in the expression of at least one of the isoforms, or in the isoform expression ratio between tumour and normal tissues ( Table 5). 13 of those tumour-specific alterations mimicked the effect of androgen stimulation in LNCaP cells: the changes were in form of alternative promoters for TACC2, TPD52, NUP93, PIK3R1, RDH13, ZFAND6, CDIP1, YIF1B, LIMK2, and FDFT1; an alternative 3´ end in CNNM2; and alternative exons in NDUFV3 and SS18 ( Figure 3, Table 5 & Supplementary Figure 2). Two of the alternative promoters ( ZFAND6 and CDIP1) are predicted to introduce a change in the 5′UTR, whereas all the others are predicted to alter the resulting protein isoform. A number of mRNA isoforms that were androgen responsive in LNCaP cells showed tumour specific alterations opposite to the effect of androgen stimulation. These were LIG4, MAPRE2, OSBPL1A, SEPT5, NR4A1, and RCAN1 (all predicted to alter the resulting protein isoform except LIG4). For the remaining 14 mRNA isoform pairs, the data was inconclusive according to the consistency conditions listed in the methods section ( Table 5).
Table 5. Summarised results of the differential expression analysis of androgen-regulated isoforms between tumour and normal tissue samples in the TCGA PRAD cohort.
| Isoform 1 | Isoform 2 | PSI | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Event type | Change with androgens (LNCap) | log2FC | Av.Expr.
(TPM) |
FDR | log2FC | Av.Expr.
(TPM) |
FDR | Delta
PSI |
Av. PSI | FDR | Consistency
of change in tumours |
| LIG4 | Alternative promoter | Induction of promoter 2 | -0.81 | 1.77 | 4.31E-02 | -1.53 | 1.28 | 4.48E-05 | 0.06 | 0.597300667 | 9.85E-02 | Opposite |
| TACC2 | Alternative promoter | Repression of promoter 1 | -0.80 | 2.42 | 5.51E-03 | 0.18 | 6.22 | 6.06E-01 | -0.16 | 0.284239843 | 2.95E-05 | Consistent |
| TPD52 | Alternative promoter | Induction of promoter 2 | -0.34 | 0.17 | 5.45E-01 | 1.87 | 39.20 | 1.23E-09 | 0.00 | 0.011365308 | 8.11E-06 | Consistent |
| NUP93 | Alternative promoter | Induction of promoter 1 | 0.25 | 25.52 | 6.45E-04 | 0.31 | 7.20 | 6.08E-01 | 0.01 | 0.828738669 | 7.52E-01 | Consistent |
| RLN1 | Alternative promoter | Repression of promoter 2 | -0.45 | 133.50 | 4.97E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| AP2S1 | Alternative promoter | Induction of promoter 2 | 0.48 | 191.44 | 2.24E-05 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| RLN2 | Alternative promoter | Induction of promoter 1 | 0.48 | 5.07 | 2.41E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| PIK3R1 | Alternative promoter | Repression of promoter 2 | -1.79 | 7.15 | 3.26E-12 | -1.79 | 1.26 | 8.20E-06 | -0.02 | 0.820282185 | 7.52E-01 | Consistent |
| MAPRE2 | Alternative promoter | Switch to promoter 2 | 1.17 | 1.52 | 1.22E-01 | -0.34 | 0.07 | 1.96E-01 | 0.09 | 0.730349729 | 4.67E-02 | Opposite |
| NDUFAF4 | Alternative promoter | Repression of promoter 2 | 0.55 | 0.06 | 5.86E-02 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| DCXR | Alternative promoter | Repression of promoter 2 | 0.68 | 623.07 | 2.05E-05 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| PEX10 | Alternative promoter | Switch to promoter 2 | 0.92 | 75.55 | 7.84E-06 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| SNAPC2 | Alternative promoter | Switch to promoter 2 | 0.38 | 5.42 | 1.23E-01 | 0.22 | 37.58 | 3.20E-02 | -0.01 | 0.130583106 | 8.29E-01 | Inconclusive |
| ATP6V0D1 | Alternative promoter | Repression of promoter 2 | -0.12 | 109.86 | 1.42E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| ARRDC1 | Alternative promoter | Induction of promoter 2 | 0.46 | 12.78 | 2.34E-05 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| DENND1A | Alternative promoter | Repression of promoter 2 | 0.04 | 7.09 | 9.11E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| KLHL36 | Alternative promoter | Induction of promoter 2 | -0.38 | 10.58 | 4.61E-06 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| RAB3IL1 | Alternative promoter | Repression of promoter 2 | 0.34 | 0.28 | 5.07E-01 | 0.05 | 4.68 | 6.91E-01 | 0.01 | 0.062673984 | 4.28E-01 | Inconclusive |
| ACER3 | Alternative promoter | Repression of promoter 2 | 0.13 | 6.32 | 8.52E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| OSBPL1A | Alternative promoter | Induction of promoter 2 | 0.14 | 4.11 | 5.75E-01 | -1.06 | 3.56 | 3.44E-09 | 0.17 | 0.522207286 | 1.03E-08 | Opposite |
| TRIM16 | Alternative promoter | Induction of promoter 2 | -0.65 | 6.87 | 1.03E-14 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| VSIG10L | Alternative promoter | Induction of promoter 1 | -1.01 | 1.91 | 5.49E-04 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| SEPT5 | Alternative promoter | Repression of promoter 2 | 0.80 | 11.47 | 1.79E-09 | 1.09 | 3.86 | 1.82E-06 | -0.03 | 0.749615358 | 1.90E-01 | Opposite |
| HMGCR | Alternative promoter | Repression of promoter 1 | -0.86 | 0.59 | 1.07E-01 | -0.55 | 17.41 | 1.09E-02 | 0.00 | 0.029105295 | 9.62E-01 | Inconclusive |
| RDH13 | Alternative promoter | Induction of promoter 1 | 1.67 | 2.10 | 1.31E-08 | 0.72 | 0.05 | 5.88E-03 | 0.00 | 0.962155441 | 9.33E-02 | Consistent |
| GPRIN2 | Alternative promoter | Repression of promoter 2 | -- | -- | -- | -0.48 | 3.31 | 3.98E-02 | -- | -- | -- |
Not
assessed |
| CLK3 | Alternative promoter | Repression of promoter 1 | 0.10 | 31.34 | 1.07E-01 | -- | 0.04 | -- | 0.00 | 0.998537929 | 6.18E-01 | Inconclusive |
| RNH1 | Alternative promoter | Induction of promoter 1 | -0.16 | 4.38 | 7.95E-01 | -0.19 | 6.56 | 5.74E-01 | 0.00 | 0.375368151 | 7.52E-01 | Inconclusive |
| ZFAND6 | Alternative promoter | Repression of promoter 2 | -0.10 | 37.63 | 6.33E-01 | -1.51 | 2.29 | 5.59E-03 | 0.03 | 0.935657481 | 3.73E-02 | Consistent |
| CDIP1 | Alternative promoter | Repression of promoter 2 | 0.77 | 0.35 | 1.16E-01 | -1.83 | 3.70 | 2.77E-11 | 0.06 | 0.142411928 | 1.46E-03 | Consistent |
| YIF1B | Alternative promoter | Switch to promoter 2 | 0.50 | 2.52 | 3.18E-01 | 2.83 | 3.08 | 1.60E-04 | -0.32 | 0.497841217 | 1.64E-02 | Consistent |
| LIMK2 | Alternative promoter | Switch to promoter 2 | -0.90 | 6.80 | 1.50E-03 | 0.58 | 10.99 | 1.10E-05 | -0.19 | 0.382613244 | 2.85E-06 | Consistent |
| TSC22D3 | Alternative promoter | Repression of promoter 1 | -- | 35.48 | -- | -1.08 | 173.59 | 8.13E-15 | 0.01 | 0.203019277 | 2.97E-01 | Inconclusive |
| ALDH1A3 | Alternative promoter | Repression of promoter 1 | 0.71 | 279.09 | 7.51E-03 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| TRABD | Alternative promoter | Switch to promoter 2 | 1.57 | 21.80 | 3.42E-02 | 0.87 | 0.54 | 1.18E-01 | 0.00 | 0.958501941 | 5.17E-01 | Inconclusive |
| LIMCH1 | Alternative promoter | Repression of promoter 2 | -- | 0.01 | -- | -- | -- | -- | -- | -- | -- |
Not
assessed |
| GMFB | Alternative promoter | Induction of promoter 2 | -0.11 | 11.91 | 7.54E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| MLST8 | Alternative promoter | Switch to promoter 1 | 0.87 | 0.19 | 9.88E-04 | 1.51 | 4.90 | 9.60E-03 | 0.02 | 0.121241399 | 5.81E-01 | Inconclusive |
| TLE3 | Alternative promoter | Induction of promoter 2 | 0.10 | 0.10 | 8.70E-01 | -0.20 | 5.14 | 4.28E-01 | 0.00 | 0.02562604 | 6.14E-01 | Inconclusive |
| UBA1 | Alternative promoter | Repression of promoter 1 | 0.21 | 23.51 | 1.39E-01 | 0.01 | 131.71 | 9.46E-01 | 0.01 | 0.190009964 | 2.99E-01 | Inconclusive |
| TNRC6B | Alternative promoter | Repression of promoter 2 | 0.18 | 2.27 | 3.34E-02 | -0.43 | 0.03 | 4.15E-01 | 0.00 | 0.988593061 | 3.56E-02 | Inconclusive |
| FDFT1 | Alternative promoter | Repression of promoter 2 | -0.57 | 94.14 | 1.13E-07 | -1.07 | 1.05 | 5.62E-12 | 0.00 | 0.986642757 | 2.13E-02 | Consistent |
| GREB1 | Alternative promoter | Induction of promoter 2 | 1.45 | 1.01 | 6.45E-04 | 0.28 | 1.48 | 3.21E-01 | 0.14 | 0.378280864 | 3.40E-02 | Inconclusive |
| NCAPD3 | Alternative promoter | Induction of promoter 2 | 0.16 | 75.75 | 6.55E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| SLC36A4 | Alternative promoter | Induction of promoter 2 | -0.91 | 2.15 | 1.60E-03 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| KLC2 | Alternative promoter | Repression of promoter 1 | 0.47 | 0.27 | 4.16E-01 | -0.76 | 3.64 | 8.12E-02 | 0.00 | 0.1048405 | 4.53E-01 | Inconclusive |
| RAP1GAP | Alternative promoter | Repression of promoter 1 | 1.94 | 3.42 | 3.45E-08 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| TMEM79 | Alternative promoter | Repression of promoter 1 | 0.21 | 3.77 | 7.91E-01 | -1.40 | 1.67 | 2.05E-05 | 0.19 | 0.399443544 | 5.07E-02 | Inconclusive |
| NR4A1 | Alternative promoter | Induction of promoter 2 | -0.40 | 1.86 | 2.34E-01 | -0.74 | 5.81 | 7.87E-03 | 0.06 | 0.292753045 | 2.53E-01 | Opposite |
| ZNF32 | Alternative promoter | Repression of promoter 2 | 0.03 | 67.26 | 7.14E-01 | 0.03 | 4.12 | 7.14E-01 | 0.00 | 0.942446541 | 1.00E+00 | Inconclusive |
| C1QTNF3 | Alternative promoter | Induction of promoter 1 | -0.30 | 3.41 | 4.67E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| UBE2D3 | Alternative promoter | Switch to promoter 2 | -0.50 | 8.00 | 5.09E-04 | -0.13 | 0.32 | 8.18E-01 | -0.01 | 0.953413055 | 5.49E-01 | Inconclusive |
| KRT8 | Alternative promoter | Repression of promoter 1 | -0.08 | 2.08 | 8.55E-01 | 0.48 | 697.27 | 1.26E-05 | 0.00 | 0.003455479 | 9.85E-02 | Inconclusive |
| ELOVL1 | Alternative promoter | Induction of promoter 2 | -0.10 | 100.07 | 1.38E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| RCAN1 | Alternative promoter | Induction of promoter 2 | -0.31 | 1.39 | 4.66E-01 | -1.40 | 6.90 | 4.40E-07 | 0.09 | 0.2372612 | 1.64E-02 | Opposite |
| SORBS3 | Alternative promoter | Induction of promoter 2 | 0.21 | 6.33 | 6.20E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| MAT2A | Alternative 3' end | Repression of isoform 2 | -0.36 | 102.47 | 6.63E-02 | 0.27 | 13.41 | 2.87E-01 | -0.03 | 0.888519015 | 5.32E-03 | Inconclusive |
| CNNM2 | Alternative 3' end | Induction of isoform 1 | 0.67 | 0.44 | 2.73E-05 | -0.79 | 1.22 | 5.96E-03 | 0.13 | 0.331082656 | 3.31E-05 | Consistent |
| TMEM125 | Alternative 3' end | Induction of isoform 1 | -- | -- | -- | 0.45 | 40.70 | 9.40E-04 | -- | -- | -- |
Not
assessed |
| CBWD2 | Alternative 3' end | Induction of isoform 2 | 0.00 | 16.56 | 9.88E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| NDUFV3 | Alternative exon | Switch to isoform 2 (exon excluded) | -0.09 | 12.98 | 2.36E-01 | 0.54 | 56.19 | 4.17E-07 | -0.07 | 0.201011 | 2.54E-08 | Consistent |
| ZNF678 | Alternative exon | Switch to isoform 2 (exon excluded) | 0.32 | 0.97 | 2.23E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| ZNF121 | Alternative exon | Switch to isoform 2 (exon excluded) | 0.90 | 0.08 | 5.97E-03 | 0.02 | 3.09 | 9.28E-01 | 0.00 | 0.037899858 | 9.85E-02 | Inconclusive |
| SPATC1L | Alternative exon | Induction of isoform 2 (exon included) | 0.35 | 36.98 | 4.71E-02 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| MOCOS | Alternative exon | Switch to isoform 2 (exon excluded) | -0.82 | 2.24 | 1.14E-09 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| RBM45 | Alternative exon | Switch to isoform 2 (exon included) | 0.25 | 7.85 | 9.96E-07 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| MIPEP | Alternative exon | Repression of isoform 2 (exon excluded) | 0.87 | 49.00 | 9.53E-04 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| BBS4 | Alternative exon | Induction of isoform 2 (exon included) | 0.02 | 21.63 | 9.71E-01 | -- | -- | -- | -- | -- | -- |
Not
assessed |
| FAM195A | Alternative exon | Switch to isoform 1 (exon exluded) | 0.87 | 43.81 | 4.03E-08 | 0.99 | 5.57 | 1.01E-08 | -0.01 | 0.884563881 | 2.50E-01 | Inconclusive |
| LINC01133 | Alternative exon | Induction of isoform 1 (exon excluded) | -- | -- | -- | -1.58 | 2.77 | 1.39E-08 | 0.00 | -- | -- |
Not
assessed |
| SS18 | Alternative exon | Switch to isoform 2 (exon excluded) | -1.47 | 3.70 | 1.97E-02 | -0.14 | 33.31 | 1.18E-02 | -0.07 | 0.087763421 | 2.88E-02 | Consistent |
| RHOC | Alternative exon | Switch to isoform 2 (exon excluded) | 0.62 | 1.48 | 3.71E-06 | 0.13 | 153.20 | 1.96E-01 | 0.00 | 0.009830219 | 1.46E-03 | Inconclusive |
| ZNF226 | Retained intron | Switch to isoform 1 (intron included) | -0.13 | 2.48 | 5.37E-01 | -0.08 | 13.49 | 7.40E-01 | -0.01 | 0.184522223 | 8.77E-01 | Inconclusive |
Figure 3. Differential expression of androgen dependent mRNA isoforms in prostate cancer versus normal tissue within the PRAD TCGA cohort for TPD52, TACC2, NDUFV3 and CNNM2.
Violin-boxplots of expression in transcripts per million mapped reads (TPM) of Isoforms 1 (left panel) and 2 (central panel), and of their expression ratio in PSI (right panel) in normal and tumour samples. The mean log2 fold-change (logFC) in expression between tumour and normal samples and the associated FDR-adjusted p-value for the moderated t-statistic of differential expression are shown for both isoforms (left and central panels). The mean difference in PSI (deltaPSI) between tumour and normal samples and the associated FDR-adjusted p-value for the Mann-Whitney U test of differential splicing are shown (right panel).
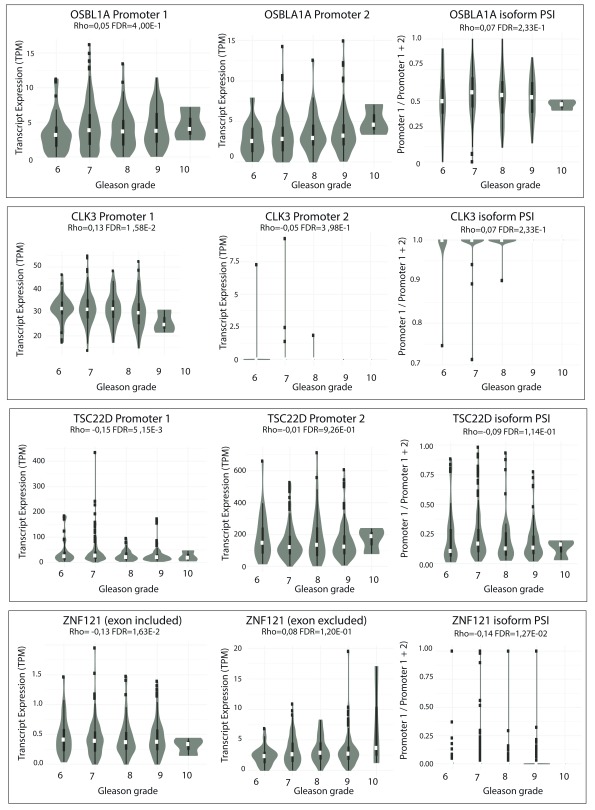
Changes in androgen-dependent mRNA isoform expression during tumour progression
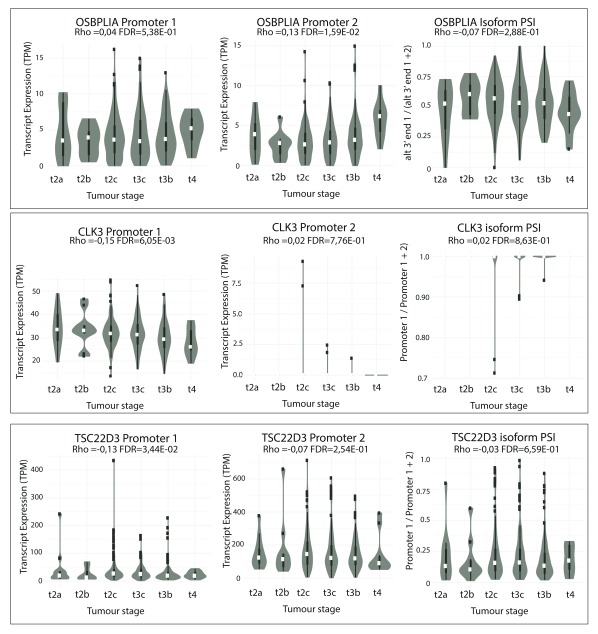
We next investigated whether the identified androgen-dependent mRNA isoforms are differentially expressed during prostate cancer progression by correlating the expression levels of each isoform with Gleason scores and prostate tumour grades within the PRAD TCGA cohort ( Figure 4 & Figure 5, Table 6 & Table 7 and Supplementary Figure 3 & Supplementary Figure 4). For 6 of the alternative mRNA isoforms responsive to androgens (made from alternative promoters in LIG4, OSBPL1A, CLK3, TSC22D3 & ZNF32 and utilising an alternative exon in ZNF121), the expression changed significantly with Gleason score and showed specific alterations consistent with the effect of androgen stimulation. Conversely, 9 alternative isoforms (which were androgen responsive in LNCaP cells) showed tumour specific alterations opposite to the effect of androgen stimulation (including an alternative promoters in NUP93 and the alternative 3´end of MAT2A). 3 androgen regulated mRNA isoforms ( OSBPL1A, CLK3 and TSC22D3) change significantly with both Gleason grade and tumour stage.
Figure 4. Differential alternative mRNA isoform expression in the TGCA PRAD cohort across different Gleason grades for OSBPL1A, CLK3, TSC22D and ZNF121.
Violin-boxplots of expression in transcripts per million mapped reads (TPM) of Isoforms 1 (left panel) and 2 (central panel), and of their expression ratio (right panel) by Gleason grade. Their respective Spearman’s correlation coefficient (Rho) with grade and associated FDR-adjusted p-value are shown.
Figure 5. Differential alternative mRNA isoform expression in the TGCA PRAD cohort across different tumour stages for OSBPL1A, CLK3 and TSC22D3.
Violin-boxplots of expression in transcripts per million mapped reads (TPM) of Isoforms 1 (left panel) and 2 (central panel), and of their expression ratio (right panel) by tumour stage. Their respective Spearman’s correlation coefficient (Rho) with stage and associated FDR-adjusted p-value are shown.
Table 6. Summarised results of the correlation analysis of androgen-regulated isoforms expression with Gleason score in the TCGA PRAD cohort.
| Isoform 1 | Isoform 2 | PSI | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Event type | Change with androgens
(LNCap) |
Rho | FDR | Rho | FDR | Rho | FDR | Consistency
of change with Gleason |
| LIG4 | Alternative promoter | Induction of promoter 2 | -0.07 | 1.92E-01 | 0.09 | 1.07E-01 | -0.18 | 4.21E-04 | Consistent - |
| TACC2 | Alternative promoter | Repression of promoter 1 | -0.08 | 1.55E-01 | 0.01 | 9.26E-01 | -0.08 | 1.88E-01 | Inconclusive |
| TPD52 | Alternative promoter | Induction of promoter 2 | 0.00 | 9.51E-01 | 0.02 | 7.73E-01 | 0.00 | 9.46E-01 | Inconclusive |
| NUP93 | Alternative promoter | Induction of promoter 1 | -0.18 | 7.92E-04 | -0.07 | 1.81E-01 | 0.04 | 4.75E-01 | Opposite |
| RLN1 | Alternative promoter | Repression of promoter 2 | -0.16 | 1.98E-03 | -- | -- | -- | -- | Not assessed |
| AP2S1 | Alternative promoter | Induction of promoter 2 | -0.01 | 8.72E-01 | -- | -- | -- | -- | Not assessed |
| RLN2 | Alternative promoter | Induction of promoter 1 | -0.10 | 6.03E-02 | -- | -- | -- | -- | Not assessed |
| PIK3R1 | Alternative promoter | Repression of promoter 2 | -0.07 | 2.51E-01 | 0.09 | 1.20E-01 | -0.17 | 1.29E-03 | Inconclusive |
| MAPRE2 | Alternative promoter | Switch to promoter 2 | -0.07 | 1.92E-01 | -0.06 | 2.73E-01 | 0.06 | 3.23E-01 | Inconclusive |
| NDUFAF4 | Alternative promoter | Repression of promoter 2 | 0.00 | 9.79E-01 | -- | -- | -- | -- | Not assessed |
| DCXR | Alternative promoter | Repression of promoter 2 | -0.29 | 4.07E-09 | -- | -- | -- | -- | Not assessed |
| PEX10 | Alternative promoter | Switch to promoter 2 | 0.08 | 1.50E-01 | -- | -- | -- | -- | Not assessed |
| SNAPC2 | Alternative promoter | Switch to promoter 2 | 0.15 | 5.48E-03 | -0.18 | 3.55E-04 | 0.21 | 5.13E-05 | Opposite |
| ATP6V0D1 | Alternative promoter | Repression of promoter 2 | -0.11 | 3.43E-02 | -- | -- | -- | -- | Not assessed |
| ARRDC1 | Alternative promoter | Induction of promoter 2 | 0.12 | 2.00E-02 | -- | -- | -- | -- | Not assessed |
| DENND1A | Alternative promoter | Repression of promoter 2 | -0.02 | 8.10E-01 | -- | -- | -- | -- | Not assessed |
| KLHL36 | Alternative promoter | Induction of promoter 2 | -0.13 | 1.67E-02 | -- | -- | -- | -- | Not assessed |
| RAB3IL1 | Alternative promoter | Repression of promoter 2 | 0.06 | 3.17E-01 | 0.32 | 9.13E-12 | -0.02 | 7.15E-01 | Opposite |
| ACER3 | Alternative promoter | Repression of promoter 2 | 0.16 | 3.79E-03 | -- | -- | -- | -- | Not assessed |
| OSBPL1A | Alternative promoter | Induction of promoter 2 | 0.05 | 4.00E-01 | 0.13 | 1.58E-02 | -0.07 | 2.33E-01 | Consistent |
| TRIM16 | Alternative promoter | Induction of promoter 2 | 0.10 | 6.06E-02 | -- | -- | -- | -- | Not assessed |
| VSIG10L | Alternative promoter | Induction of promoter 1 | -0.16 | 1.98E-03 | -- | -- | -- | -- | Not assessed |
| SEPT5 | Alternative promoter | Repression of promoter 2 | 0.17 | 1.12E-03 | 0.12 | 1.93E-02 | -0.04 | 4.91E-01 | Opposite |
| HMGCR | Alternative promoter | Repression of promoter 1 | 0.03 | 6.56E-01 | -0.05 | 4.54E-01 | 0.07 | 2.33E-01 | Inconclusive |
| RDH13 | Alternative promoter | Induction of promoter 1 | 0.03 | 7.01E-01 | 0.08 | 1.20E-01 | -0.10 | 1.00E-01 | Inconclusive |
| GPRIN2 | Alternative promoter | Repression of promoter 2 | -- | -- | -0.01 | 8.93E-01 | -- | -- | Not assessed |
| CLK3 | Alternative promoter | Repression of promoter 1 | -0.13 | 1.58E-02 | -0.05 | 3.98E-01 | 0.07 | 2.33E-01 | Consistent |
| RNH1 | Alternative promoter | Induction of promoter 1 | 0.05 | 4.41E-01 | 0.07 | 1.83E-01 | -0.01 | 9.23E-01 | Inconclusive |
| ZFAND6 | Alternative promoter | Repression of promoter 2 | 0.07 | 1.87E-01 | 0.05 | 3.82E-01 | -0.03 | 6.36E-01 | Inconclusive |
| CDIP1 | Alternative promoter | Repression of promoter 2 | 0.02 | 8.10E-01 | 0.03 | 6.81E-01 | -0.01 | 9.23E-01 | Inconclusive |
| YIF1B | Alternative promoter | Switch to promoter 2 | 0.02 | 8.10E-01 | -0.04 | 5.42E-01 | 0.05 | 4.39E-01 | Inconclusive |
| LIMK2 | Alternative promoter | Switch to promoter 2 | -0.02 | 8.10E-01 | -0.03 | 6.30E-01 | 0.00 | 9.49E-01 | Inconclusive |
| TSC22D3 | Alternative promoter | Repression of promoter 1 | -0.15 | 5.15E-03 | -0.01 | 9.26E-01 | -0.09 | 1.14E-01 | Consistent |
| ALDH1A3 | Alternative promoter | Repression of promoter 1 | -0.12 | 2.00E-02 | -- | -- | -- | -- | Not assessed |
| TRABD | Alternative promoter | Switch to promoter 2 | 0.14 | 8.04E-03 | -0.04 | 5.43E-01 | 0.05 | 4.39E-01 | Inconclusive |
| LIMCH1 | Alternative promoter | Repression of promoter 2 | 0.05 | 4.34E-01 | -- | -- | -- | -- | Not assessed |
| GMFB | Alternative promoter | Induction of promoter 2 | 0.08 | 1.55E-01 | -- | -- | -- | -- | Not assessed |
| MLST8 | Alternative promoter | Switch to promoter 1 | 0.19 | 5.32E-04 | 0.19 | 2.05E-04 | 0.07 | 2.14E-01 | Inconclusive |
| TLE3 | Alternative promoter | Induction of promoter 2 | 0.05 | 4.28E-01 | -0.10 | 7.19E-02 | 0.07 | 2.33E-01 | Inconclusive |
| UBA1 | Alternative promoter | Repression of promoter 1 | 0.09 | 8.99E-02 | 0.03 | 5.95E-01 | 0.01 | 8.68E-01 | Inconclusive |
| TNRC6B | Alternative promoter | Repression of promoter 2 | -0.05 | 4.00E-01 | -0.09 | 1.19E-01 | 0.09 | 1.11E-01 | Inconclusive |
| FDFT1 | Alternative promoter | Repression of promoter 2 | -0.02 | 7.41E-01 | 0.07 | 2.07E-01 | -0.07 | 2.14E-01 | Inconclusive |
| GREB1 | Alternative promoter | Induction of promoter 2 | -0.05 | 4.41E-01 | -0.14 | 5.45E-03 | 0.04 | 4.60E-01 | Opposite |
| NCAPD3 | Alternative promoter | Induction of promoter 2 | -0.23 | 3.61E-06 | -- | -- | -- | -- | Not assessed |
| SLC36A4 | Alternative promoter | Induction of promoter 2 | 0.12 | 1.88E-02 | -- | -- | -- | -- | Not assessed |
| KLC2 | Alternative promoter | Repression of promoter 1 | -0.02 | 8.10E-01 | 0.13 | 1.58E-02 | -0.04 | 4.60E-01 | Inconclusive |
| RAP1GAP | Alternative promoter | Repression of promoter 1 | 0.01 | 8.79E-01 | -- | -- | -- | -- | Not assessed |
| TMEM79 | Alternative promoter | Repression of promoter 1 | -0.04 | 4.70E-01 | 0.15 | 3.46E-03 | -0.09 | 1.11E-01 | Inconclusive |
| NR4A1 | Alternative promoter | Induction of promoter 2 | 0.10 | 5.44E-02 | 0.00 | 9.79E-01 | 0.10 | 7.40E-02 | Inconclusive |
| ZNF32 | Alternative promoter | Repression of promoter 2 | -0.22 | 1.32E-05 | -0.22 | 1.11E-05 | -0.09 | 1.31E-01 | Consistent - |
| C1QTNF3 | Alternative promoter | Induction of promoter 1 | 0.08 | 1.58E-01 | -- | -- | -- | -- | Not assessed |
| UBE2D3 | Alternative promoter | Switch to promoter 2 | 0.18 | 7.24E-04 | 0.08 | 1.27E-01 | -0.02 | 7.15E-01 | Inconclusive |
| KRT8 | Alternative promoter | Repression of promoter 1 | -0.05 | 3.81E-01 | -0.16 | 2.07E-03 | 0.01 | 8.68E-01 | Inconclusive |
| ELOVL1 | Alternative promoter | Induction of promoter 2 | 0.18 | 7.24E-04 | -- | -- | -- | -- | Not assessed |
| RCAN1 | Alternative promoter | Induction of promoter 2 | 0.10 | 5.13E-02 | -0.01 | 8.70E-01 | 0.12 | 3.69E-02 | Inconclusive |
| SORBS3 | Alternative promoter | Induction of promoter 2 | 0.12 | 2.21E-02 | -- | -- | -- | -- | Not assessed |
| MAT2A | Alternative 3' end | Repression of isoform 2 | 0.04 | 5.39E-01 | 0.27 | 3.68E-08 | -0.33 | 8.82E-13 | Opposite |
| CNNM2 | Alternative 3' end | Induction of isoform 1 | -0.06 | 3.30E-01 | 0.03 | 5.87E-01 | -0.08 | 2.04E-01 | Inconclusive |
| TMEM125 | Alternative 3' end | Induction of isoform 1 | -- | -- | -0.19 | 2.05E-04 | -- | -- | Not assessed |
| CBWD2 | Alternative 3' end | Induction of isoform 2 | 0.13 | 1.37E-02 | -- | -- | -- | -- | Not assessed |
| NDUFV3 | Alternative exon | Switch to isoform 2 (exon
excluded) |
0.14 | 8.04E-03 | -0.07 | 2.48E-01 | 0.13 | 2.23E-02 | Opposite |
| ZNF678 | Alternative exon | Switch to isoform 2 (exon
excluded) |
-0.07 | 1.87E-01 | -- | -- | -- | -- | Not assessed |
| ZNF121 | Alternative exon | Switch to isoform 2 (exon
excluded) |
-0.13 | 1.63E-02 | 0.08 | 1.20E-01 | -0.14 | 1.27E-02 | Consistent |
| SPATC1L | Alternative exon | Induction of isoform 2
(exon included) |
-0.13 | 1.58E-02 | -- | -- | -- | -- | Not assessed |
| MOCOS | Alternative exon | Switch to isoform 2 (exon
excluded) |
-0.01 | 8.72E-01 | -- | -- | -- | -- | Not assessed |
| RBM45 | Alternative exon | Switch to isoform 2 (exon
included) |
0.12 | 2.45E-02 | -- | -- | -- | -- | Not assessed |
| MIPEP | Alternative exon | Repression of isoform 2
(exon excluded) |
-0.14 | 9.92E-03 | -- | -- | -- | -- | Not assessed |
| BBS4 | Alternative exon | Induction of isoform 2
(exon included) |
-0.08 | 1.87E-01 | -- | -- | -- | -- | Not assessed |
| FAM195A | Alternative exon | Switch to isoform 1 (exon
exluded) |
0.04 | 5.43E-01 | 0.14 | 5.35E-03 | -0.18 | 4.65E-04 | Opposite |
| LINC01133 | Alternative exon | Induction of isoform 1
(exon excluded) |
-- | -- | -0.02 | 7.51E-01 | -- | -- | Not assessed |
| SS18 | Alternative exon | Switch to isoform 2 (exon
excluded) |
0.04 | 4.86E-01 | -0.06 | 2.51E-01 | 0.07 | 2.33E-01 | Inconclusive |
| RHOC | Alternative exon | Switch to isoform 2 (exon
excluded) |
0.29 | 4.07E-09 | 0.15 | 4.24E-03 | 0.21 | 3.63E-05 | Opposite |
| ZNF226 | Retained intron | Switch to isoform 1 (intron
included) |
0.01 | 8.67E-01 | -0.10 | 7.49E-02 | 0.11 | 6.74E-02 | Inconclusive |
Table 7. Summarised results of the correlation analysis of androgen-regulated isoforms expression with tumour stage in the TCGA PRAD cohort (related to Figure 4 and Supplementary Figure 5).
| Isoform 1 | Isoform 2 | PSI | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Event type | Change with androgens
(LNCap) |
Rho | FDR | Rho | FDR | Rho | FDR | Consistency
of change with stage |
| LIG4 | Alternative promoter | Induction of promoter 2 | -0.04 | 6.05E-01 | 0.02 | 6.82E-01 | -0.09 | 1.82E-01 | Inconclusive |
| TACC2 | Alternative promoter | Repression of promoter 1 | -0.08 | 1.74E-01 | -0.05 | 4.47E-01 | -0.04 | 5.65E-01 | Inconclusive |
| TPD52 | Alternative promoter | Induction of promoter 2 | -0.02 | 7.85E-01 | -0.02 | 6.82E-01 | -0.02 | 7.95E-01 | Inconclusive |
| NUP93 | Alternative promoter | Induction of promoter 1 | -0.12 | 3.95E-02 | 0.03 | 6.65E-01 | -0.05 | 4.43E-01 | Opposite |
| RLN1 | Alternative promoter | Repression of promoter 2 | -0.22 | 1.82E-05 | -- | -- | -- | -- | Not assessed |
| AP2S1 | Alternative promoter | Induction of promoter 2 | -0.04 | 5.51E-01 | -- | -- | -- | -- | Not assessed |
| RLN2 | Alternative promoter | Induction of promoter 1 | -0.16 | 5.68E-03 | -- | -- | -- | -- | Not assessed |
| PIK3R1 | Alternative promoter | Repression of promoter 2 | -0.02 | 7.92E-01 | 0.11 | 5.92E-02 | -0.14 | 3.27E-02 | Opposite - |
| MAPRE2 | Alternative promoter | Switch to promoter 2 | -0.02 | 7.56E-01 | -0.02 | 6.82E-01 | 0.03 | 1.00E+00 | Inconclusive |
| NDUFAF4 | Alternative promoter | Repression of promoter 2 | 0.08 | 1.89E-01 | -- | -- | -- | -- | Not assessed |
| DCXR | Alternative promoter | Repression of promoter 2 | -0.30 | 6.32E-10 | -- | -- | -- | -- | Not assessed |
| PEX10 | Alternative promoter | Switch to promoter 2 | 0.10 | 9.95E-02 | -- | -- | -- | -- | Not assessed |
| SNAPC2 | Alternative promoter | Switch to promoter 2 | 0.13 | 2.87E-02 | -0.23 | 5.57E-06 | 0.20 | 2.40E-04 | Opposite |
| ATP6V0D1 | Alternative promoter | Repression of promoter 2 | -0.11 | 5.43E-02 | -- | -- | -- | -- | Not assessed |
| ARRDC1 | Alternative promoter | Induction of promoter 2 | 0.08 | 2.06E-01 | -- | -- | -- | -- | Not assessed |
| DENND1A | Alternative promoter | Repression of promoter 2 | -0.01 | 8.49E-01 | -- | -- | -- | -- | Not assessed |
| KLHL36 | Alternative promoter | Induction of promoter 2 | -0.10 | 1.04E-01 | -- | -- | -- | -- | Not assessed |
| RAB3IL1 | Alternative promoter | Repression of promoter 2 | 0.08 | 1.71E-01 | 0.33 | 4.58E-12 | 0.00 | 9.75E-01 | Opposite |
| ACER3 | Alternative promoter | Repression of promoter 2 | 0.16 | 4.77E-03 | -- | -- | -- | -- | Not assessed |
| OSBPL1A | Alternative promoter | Induction of promoter 2 | 0.04 | 5.38E-01 | 0.13 | 1.59E-02 | -0.07 | 2.88E-01 | Consistent |
| TRIM16 | Alternative promoter | Induction of promoter 2 | 0.06 | 3.95E-01 | -- | -- | -- | -- | Not assessed |
| VSIG10L | Alternative promoter | Induction of promoter 1 | -0.12 | 5.43E-02 | -- | -- | -- | -- | Not assessed |
| SEPT5 | Alternative promoter | Repression of promoter 2 | 0.11 | 7.96E-02 | 0.07 | 2.54E-01 | -0.01 | 8.89E-01 | Inconclusive |
| HMGCR | Alternative promoter | Repression of promoter 1 | 0.00 | 9.91E-01 | -0.04 | 5.77E-01 | 0.04 | 6.25E-01 | Inconclusive |
| RDH13 | Alternative promoter | Induction of promoter 1 | -0.03 | 7.33E-01 | 0.10 | 7.19E-02 | -0.12 | 9.32E-02 | Inconclusive |
| GPRIN2 | Alternative promoter | Repression of promoter 2 | -- | -- | 0.03 | 6.48E-01 | -- | -- | Not assessed |
| CLK3 | Alternative promoter | Repression of promoter 1 | -0.15 | 6.05E-03 | 0.02 | 7.76E-01 | 0.02 | 8.63E-01 | Consistent |
| RNH1 | Alternative promoter | Induction of promoter 1 | -0.02 | 7.92E-01 | 0.10 | 6.12E-02 | -0.08 | 2.28E-01 | Inconclusive |
| ZFAND6 | Alternative promoter | Repression of promoter 2 | 0.03 | 6.50E-01 | 0.04 | 5.78E-01 | -0.04 | 6.05E-01 | Inconclusive |
| CDIP1 | Alternative promoter | Repression of promoter 2 | 0.10 | 1.04E-01 | 0.02 | 7.82E-01 | 0.06 | 3.78E-01 | Inconclusive |
| YIF1B | Alternative promoter | Switch to promoter 2 | -0.01 | 8.87E-01 | -0.10 | 6.71E-02 | 0.06 | 3.97E-01 | Inconclusive |
| LIMK2 | Alternative promoter | Switch to promoter 2 | 0.00 | 9.67E-01 | -0.05 | 4.72E-01 | 0.00 | 9.75E-01 | Inconclusive |
| TSC22D3 | Alternative promoter | Repression of promoter 1 | -0.13 | 3.44E-02 | -0.07 | 2.54E-01 | -0.03 | 6.59E-01 | Consistent |
| ALDH1A3 | Alternative promoter | Repression of promoter 1 | -0.18 | 7.69E-04 | -- | -- | -- | -- | Not assessed |
| TRABD | Alternative promoter | Switch to promoter 2 | 0.06 | 3.95E-01 | -0.03 | 6.48E-01 | 0.03 | 7.83E-01 | Inconclusive |
| LIMCH1 | Alternative promoter | Repression of promoter 2 | 0.02 | 7.85E-01 | -- | -- | -- | -- | Not assessed |
| GMFB | Alternative promoter | Induction of promoter 2 | 0.07 | 2.57E-01 | -- | -- | -- | -- | Not assessed |
| MLST8 | Alternative promoter | Switch to promoter 1 | 0.10 | 8.19E-02 | 0.15 | 6.14E-03 | 0.02 | 7.83E-01 | Inconclusive |
| TLE3 | Alternative promoter | Induction of promoter 2 | 0.03 | 6.38E-01 | -0.11 | 3.84E-02 | 0.04 | 5.65E-01 | Opposite |
| UBA1 | Alternative promoter | Repression of promoter 1 | 0.12 | 5.43E-02 | 0.00 | 9.72E-01 | 0.06 | 3.99E-01 | Inconclusive |
| TNRC6B | Alternative promoter | Repression of promoter 2 | -0.04 | 6.31E-01 | -0.03 | 6.48E-01 | 0.02 | 7.83E-01 | Inconclusive |
| FDFT1 | Alternative promoter | Repression of promoter 2 | -0.05 | 4.82E-01 | 0.04 | 5.46E-01 | -0.08 | 2.28E-01 | Inconclusive |
| GREB1 | Alternative promoter | Induction of promoter 2 | -0.11 | 7.48E-02 | -0.18 | 7.01E-04 | 0.01 | 8.96E-01 | Opposite |
| NCAPD3 | Alternative promoter | Induction of promoter 2 | -0.23 | 1.82E-05 | -- | -- | -- | -- | Not assessed |
| SLC36A4 | Alternative promoter | Induction of promoter 2 | 0.07 | 2.59E-01 | -- | -- | -- | -- | Not assessed |
| KLC2 | Alternative promoter | Repression of promoter 1 | -0.03 | 6.33E-01 | 0.13 | 1.81E-02 | -0.08 | 2.78E-01 | Inconclusive |
| RAP1GAP | Alternative promoter | Repression of promoter 1 | 0.02 | 7.85E-01 | -- | -- | -- | -- | Not assessed |
| TMEM79 | Alternative promoter | Repression of promoter 1 | -0.08 | 1.71E-01 | 0.16 | 1.97E-03 | -0.10 | 1.20E-01 | Inconclusive |
| NR4A1 | Alternative promoter | Induction of promoter 2 | 0.01 | 8.49E-01 | -0.06 | 3.69E-01 | 0.08 | 2.62E-01 | Inconclusive |
| ZNF32 | Alternative promoter | Repression of promoter 2 | -0.15 | 6.70E-03 | 0.02 | 7.34E-01 | -0.08 | 2.33E-01 | Inconclusive |
| C1QTNF3 | Alternative promoter | Induction of promoter 1 | 0.03 | 6.74E-01 | -- | -- | -- | -- | Not assessed |
| UBE2D3 | Alternative promoter | Switch to promoter 2 | 0.20 | 2.96E-04 | 0.07 | 2.17E-01 | -0.02 | 7.83E-01 | Inconclusive |
| KRT8 | Alternative promoter | Repression of promoter 1 | -0.04 | 6.05E-01 | -0.24 | 2.72E-06 | 0.04 | 6.05E-01 | Inconclusive |
| ELOVL1 | Alternative promoter | Induction of promoter 2 | 0.13 | 2.87E-02 | -- | -- | -- | -- | Not assessed |
| RCAN1 | Alternative promoter | Induction of promoter 2 | 0.09 | 1.26E-01 | -0.01 | 8.69E-01 | 0.10 | 1.20E-01 | Inconclusive |
| SORBS3 | Alternative promoter | Induction of promoter 2 | 0.11 | 7.96E-02 | -- | -- | -- | -- | Not assessed |
| MAT2A | Alternative 3' end | Repression of isoform 2 | 0.01 | 9.35E-01 | 0.18 | 7.83E-04 | -0.21 | 8.42E-05 | Opposite |
| CNNM2 | Alternative 3' end | Induction of isoform 1 | 0.05 | 3.95E-01 | 0.05 | 4.47E-01 | -0.04 | 6.05E-01 | Inconclusive |
| TMEM125 | Alternative 3' end | Induction of isoform 1 | -- | -- | -0.16 | 2.80E-03 | -- | -- | Not assessed |
| CBWD2 | Alternative 3' end | Induction of isoform 2 | 0.08 | 1.74E-01 | -- | -- | -- | -- | Not assessed |
| NDUFV3 | Alternative exon | Switch to isoform 2 (exon
excluded) |
0.11 | 7.48E-02 | -0.05 | 4.72E-01 | 0.11 | 1.00E-01 | Inconclusive |
| ZNF678 | Alternative exon | Switch to isoform 2 (exon
excluded) |
-0.02 | 7.43E-01 | -- | -- | -- | -- | Not assessed |
| ZNF121 | Alternative exon | Switch to isoform 2 (exon
excluded) |
-0.08 | 1.80E-01 | 0.03 | 6.48E-01 | -0.09 | 1.82E-01 | Inconclusive |
| SPATC1L | Alternative exon | Induction of isoform 2
(exon included) |
-0.10 | 9.95E-02 | -- | -- | -- | -- | Not assessed |
| MOCOS | Alternative exon | Switch to isoform 2 (exon
excluded) |
0.03 | 6.33E-01 | -- | -- | -- | -- | Not assessed |
| RBM45 | Alternative exon | Switch to isoform 2 (exon
included) |
0.08 | 1.71E-01 | -- | -- | -- | -- | Not assessed |
| MIPEP | Alternative exon | Repression of isoform 2
(exon excluded) |
-0.16 | 4.48E-03 | -- | -- | -- | -- | Not assessed |
| BBS4 | Alternative exon | Induction of isoform 2
(exon included) |
-0.06 | 3.85E-01 | -- | -- | -- | -- | Not assessed |
| FAM195A | Alternative exon | Switch to isoform 1 (exon
exluded) |
0.06 | 3.37E-01 | 0.10 | 6.85E-02 | -0.10 | 1.20E-01 | Inconclusive |
| LINC01133 | Alternative exon | Induction of isoform 1
(exon excluded) |
-- | -- | 0.00 | 9.72E-01 | -- | -- | Not assessed |
| SS18 | Alternative exon | Switch to isoform 2 (exon
excluded) |
0.04 | 5.68E-01 | -0.04 | 5.46E-01 | 0.06 | 3.97E-01 | Inconclusive |
| RHOC | Alternative exon | Switch to isoform 2 (exon
excluded) |
0.15 | 6.05E-03 | 0.11 | 3.84E-02 | 0.11 | 1.00E-01 | Inconclusive |
| ZNF226 | Retained intron | Switch to isoform 1 (intron
included) |
-0.03 | 6.64E-01 | -0.09 | 1.23E-01 | 0.07 | 3.35E-01 | Inconclusive |
Copyright: © 2018 Munkley J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Copyright: © 2018 Munkley J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Discussion
The main function of the androgen receptor (AR) is as a DNA binding transcription factor that regulates gene expression. Here we show the AR can couple hormone induced gene transcription to alternative mRNA isoform expression in prostate cancer. In response to androgens, the AR can induce the use of alternative promoters, induce the expression of alternatively spliced mRNA isoforms, regulate the expression of non-coding mRNA transcripts, and promote the transcription of mRNA isoforms encoding different protein isoforms. Importantly, we also find that some of these alternative mRNA isoforms are differentially regulated in prostate cancer versus normal tissue and also significantly change expression during tumour progression. Our data suggest that most androgen regulated alternative mRNA isoforms are generated through alternative promoter selection rather than through internal alternative exon splicing mechanisms. This suggests expression of alternative isoforms of specific genes can be a consequence of RNA polymerase being recruited to different promoters in response to androgen stimulation. Alternative promoter usage has been observed for many genes and is believed to play a significant role in the control of gene expression 4, 105, 106. Alternative promoter use can also generate mRNA isoforms with distinct functional activities from the same gene, sometimes having opposing functions 11.
Androgen exposure further drives a smaller number of alternative splicing events suggesting that the AR could contribute to altered patterns of splicing in prostate cancer cells. Tumour progression is believed to be associated with a coordinated change in splicing patterns which is regulated by several factors including signalling molecules 7. We also identified 4 AR regulated alternative mRNA 3′ end isoform switches. This is the first time that regulation of 3′ mRNA end processing has been shown to be controlled by androgens. The selection of alternative 3′ ends can produce mRNA isoforms differing in the length of their 3′ UTRs (which can lead to the inclusion or exclusion of regulatory elements and influence gene expression), or in their C-terminal coding region (which can contribute to proteome diversity) 107– 114. Defective 3′ mRNA processing of numerous genes has been linked to an oncogenic phenotype 115– 119, and the 3′ mRNA end profiles of samples from multiple cancer types significantly differ from those of healthy tissue samples 115, 119– 121.
Based on the findings presented in this study, we propose that activated AR has the ability to coordinate both transcriptional activity and mRNA isoform decisions through the recruitment of co-regulators to specific promoters. The genomic action of the AR is influenced by a large number of collaborating transcription factors 122– 124. Specifically, Sam68 and p68 have been shown to modulate AR dependent alternative splicing of specific genes and are significantly overexpressed in prostate cancer 31, 32. In future work it will be important to define the role of specific AR co-regulators in AR mediated isoform selection.
Some of the androgen dependent mRNA isoforms identified here are predicted to yield protein isoforms that may be clinically important, or to switch off protein production via generation of noncoding mRNA isoforms. Although the functional significance of the alternative mRNA isoforms identified in this study is yet largely unexplored, as is their role in the cellular response to androgens, the presented results emphasize the importance of analysing gene regulation and function at the mRNA isoform level.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2018 Munkley J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
The RNASeq data from LNCaP cells has been published previously https://doi.org/10.1016/j.ebiom.2016.04.018 25
The RNAseq custom tracks are available in Supplementary File 1. To view these files please load them onto the UCSC website using the ‘My data’ tab and ‘custom tracks’. Then ‘Paste URLs or data’. The data is aligned to Feb 2009 (GRCh37/hg19).
Prostate adenocarcinoma cohort RNA-Seq data was downloaded from the Broad Institute TCGA Genome Analysis Center: Firehose 16/01/28 run https://doi.org/10.7908/C11G0KM9 43
Dataset 1: Real-time PCR raw Ct values 10.5256/f1000research.15604.d212873 41
Dataset 2: Raw unedited western blot images 10.5256/f1000research.15604.d212874 125
Funding Statement
This work was funded by Prostate Cancer UK [PG12-34, S13-020 and RIA16-ST2-011].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 3 approved]
Supplementary material
Supplementary Table 1: Details of primer sequences used.
Supplementary File 1: RNA-Seq reads custom tracks for visualisation on UCSC genome browser
Supplementary Figure 1: PCR validation of 17 androgen regulated alternative events.
Supplementary Figure 2: Differential alternative mRNA isoform expression in theTGCA PRAD cohort. Normal vs. tumour (unpaired samples) analysis. Violin-boxplots of expression in transcripts per million mapped reads (TPM) of Isoforms 1 (left panel) and 2 (central panel), and of their expression ratio in PSI (right panel) in normal and tumour samples. The mean log2 fold-change (logFC) in expression between tumour and normal samples and the associated FDR-adjusted p-value for the moderated t-statistic of differential expression are shown for both isoforms (left and central panels). The mean difference in PSI (deltaPSI) between tumour and normal samples and the associated FDR-adjusted p-value for the Mann-Whitney U test of differential splicing are shown (right panel).
Supplementary Figure 3: Differential alternative mRNA isoform expression in the TGCA PRAD cohort across different Gleason grades. Violin-boxplots of expression in transcripts per million mapped reads (TPM) of Isoforms 1 (left panel) and 2 (central panel), and of their expression ratio (right panel) by Gleason grade. Their respective Spearman’s correlation coefficient (Rho) with grade and associated FDR-adjusted p-value are shown.
Supplementary Figure 4: Differential alternative mRNA isoform expression in the TGCA PRAD cohort across different tumour stages. Violin-boxplots of expression in transcripts per million mapped reads (TPM) of Isoforms 1 (left panel) and 2 (central panel), and of their expression ratio (right panel) by tumour stage. Their respective Spearman’s correlation coefficient (Rho) with stage and associated FDR-adjusted p-value are shown.
References
- 1. Johnson JM, Castle J, Garrett-Engele P, et al. : Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302(5653):2141–4. 10.1126/science.1090100 [DOI] [PubMed] [Google Scholar]
- 2. Pan Q, Shai O, Lee LJ, et al. : Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–5. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- 3. Wang ET, Sandberg R, Luo S, et al. : Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–6. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davuluri RV, Suzuki Y, Sugano S, et al. : The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24(4):167–77. 10.1016/j.tig.2008.01.008 [DOI] [PubMed] [Google Scholar]
- 5. Trapnell C, Williams BA, Pertea G, et al. : Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hanahan D, Weinberg RA: Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 7. Oltean S, Bates DO: Hallmarks of alternative splicing in cancer. Oncogene. 2014;33(46):5311–8. 10.1038/onc.2013.533 [DOI] [PubMed] [Google Scholar]
- 8. Ladomery M: Aberrant alternative splicing is another hallmark of cancer. Int J Cell Biol. 2013;2013: 463786. 10.1155/2013/463786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rajan P, Elliott DJ, Robson CN, et al. : Alternative splicing and biological heterogeneity in prostate cancer. Nat Rev Urol. 2009;6(8):454–60. 10.1038/nrurol.2009.125 [DOI] [PubMed] [Google Scholar]
- 10. Rajan P, Dalgliesh C, Carling PJ, et al. : Identification of novel androgen-regulated pathways and mRNA isoforms through genome-wide exon-specific profiling of the LNCaP transcriptome. PLoS One. 2011;6(12):e29088. 10.1371/journal.pone.0029088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munkley J, Livermore K, Rajan P, et al. : RNA splicing and splicing regulator changes in prostate cancer pathology. Hum Genet. 2017;136(9):1143–54. 10.1007/s00439-017-1792-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Livermore KE, Munkley J, Elliott DJ: Androgen receptor and prostate cancer. AIMS Mol Sci. 2016;3(2):280–99. 10.3934/molsci.2016.2.280 [DOI] [Google Scholar]
- 13. Mills IG: Maintaining and reprogramming genomic androgen receptor activity in prostate cancer. Nat Rev Cancer. 2014;14(3):187–98. 10.1038/nrc3678 [DOI] [PubMed] [Google Scholar]
- 14. Snoek R, Cheng H, Margiotti K, et al. : In vivo knockdown of the androgen receptor results in growth inhibition and regression of well-established, castration-resistant prostate tumors. Clin Cancer Res. 2009;15(1):39–47. 10.1158/1078-0432.CCR-08-1726 [DOI] [PubMed] [Google Scholar]
- 15. Hara T, Miyazaki H, Lee A, et al. : Androgen receptor and invasion in prostate cancer. Cancer Res. 2008;68(4):1128–35. 10.1158/0008-5472.CAN-07-1929 [DOI] [PubMed] [Google Scholar]
- 16. Hååg P, Bektic J, Bartsch G, et al. : Androgen receptor down regulation by small interference RNA induces cell growth inhibition in androgen sensitive as well as in androgen independent prostate cancer cells. J Steroid Biochem Mol Biol. 2005;96(3–4):251–8. 10.1016/j.jsbmb.2005.04.029 [DOI] [PubMed] [Google Scholar]
- 17. Munkley J, Oltean S, Vodák D, et al. : The androgen receptor controls expression of the cancer-associated sTn antigen and cell adhesion through induction of ST6GalNAc1 in prostate cancer. Oncotarget. 2015;6(33):34358–74. 10.18632/oncotarget.6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Munkley J, Rajan P, Lafferty NP, et al. : A novel androgen-regulated isoform of the TSC2 tumour suppressor gene increases cell proliferation. Oncotarget. 2014;5(1):131–9. 10.18632/oncotarget.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Massie CE, Lynch A, Ramos-Montoya A, et al. : The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30(13):2719–33. 10.1038/emboj.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Munkley J, Lafferty NP, Kalna G, et al. : Androgen-regulation of the protein tyrosine phosphatase PTPRR activates ERK1/2 signalling in prostate cancer cells. BMC Cancer. 2015;15:9. 10.1186/s12885-015-1012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Munkley J, Livermore KE, McClurg UL, et al. : The PI3K regulatory subunit gene PIK3R1 is under direct control of androgens and repressed in prostate cancer cells. Oncoscience. 2015;2(9):755–64. 10.18632/oncoscience.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karantanos T, Corn PG, Thompson TC: Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32(49):5501–11. 10.1038/onc.2013.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cai C, He HH, Chen S, et al. : Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20(4):457–71. 10.1016/j.ccr.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yuan X, Cai C, Chen S, et al. : Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33(22):2815–25. 10.1038/onc.2013.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Munkley J, Vodak D, Livermore KE, et al. : Glycosylation is an Androgen-Regulated Process Essential for Prostate Cancer Cell Viability. EBioMedicine. 2016;8:103–16. 10.1016/j.ebiom.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Auboeuf D, Hönig A, Berget SM, et al. : Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298(5592):416–9. 10.1126/science.1073734 [DOI] [PubMed] [Google Scholar]
- 27. Satoh T, Katano-Toki A, Tomaru T, et al. : Coordinated regulation of transcription and alternative splicing by the thyroid hormone receptor and its associating coregulators. Biochem Biophys Res Commun. 2014;451(1):24–9. 10.1016/j.bbrc.2014.07.029 [DOI] [PubMed] [Google Scholar]
- 28. Dutertre M, Gratadou L, Dardenne E, et al. : Estrogen regulation and physiopathologic significance of alternative promoters in breast cancer. Cancer Res. 2010;70(9):3760–70. 10.1158/0008-5472.CAN-09-3988 [DOI] [PubMed] [Google Scholar]
- 29. Dowhan DH, Hong EP, Auboeuf D, et al. : Steroid hormone receptor coactivation and alternative RNA splicing by U2AF 65-related proteins CAPERalpha and CAPERbeta. Mol Cell. 2005;17(3):429–39. 10.1016/j.molcel.2004.12.025 [DOI] [PubMed] [Google Scholar]
- 30. Cochrane DR, Wang Z, Muramaki M, et al. : Differential regulation of clusterin and its isoforms by androgens in prostate cells. J Biol Chem. 2007;282(4):2278–87. 10.1074/jbc.M608162200 [DOI] [PubMed] [Google Scholar]
- 31. Rajan P, Gaughan L, Dalgliesh C, et al. : The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J Pathol. 2008;215(1):67–77. 10.1002/path.2324 [DOI] [PubMed] [Google Scholar]
- 32. Clark EL, Coulson A, Dalgliesh C, et al. : The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68(19):7938–46. 10.1158/0008-5472.CAN-08-0932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salama SA, Mohammad MA, Diaz-Arrastia CR, et al. : Estradiol-17β upregulates pyruvate kinase M2 expression to coactivate estrogen receptor-α and to integrate metabolic reprogramming with the mitogenic response in endometrial cells. J Clin Endocrinol Metab. 2014;99(10):3790–9. 10.1210/jc.2013-2639 [DOI] [PubMed] [Google Scholar]
- 34. Lal S, Allan A, Markovic D, et al. : Estrogen alters the splicing of type 1 corticotropin-releasing hormone receptor in breast cancer cells. Sci Signal. 2013;6(282):ra53. 10.1126/scisignal.2003926 [DOI] [PubMed] [Google Scholar]
- 35. Bhat-Nakshatri P, Song EK, Collins NR, et al. : Interplay between estrogen receptor and AKT in estradiol-induced alternative splicing. BMC Med Genomics. 2013;6:21. 10.1186/1755-8794-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Munkley J, McClurg UL, Livermore KE, et al. : The cancer-associated cell migration protein TSPAN1 is under control of androgens and its upregulation increases prostate cancer cell migration. Sci Rep. 2017;7(1): 5249. 10.1038/s41598-017-05489-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trapnell C, Roberts A, Goff L, et al. : Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim D, Pertea G, Trapnell C, et al. : TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Langmead B, Salzberg SL: Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trapnell C, Hendrickson DG, Sauvageau M, et al. : Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnol. 2013;31(1):46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Munkley J, Maia TM, Ibarluzea N, et al. : Dataset 1 in: Androgen-dependent alternative mRNA isoform expression in prostate cancer cells. F1000Research. 2018. 10.5256/f1000research.15604.d212873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Young MD, Wakefield MJ, Smyth GK, et al. : Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Center BITGDA: Analysis-ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. Broad Institute of MIT and Harvard Dataset. 2016. 10.7908/C11G0KM9 [DOI] [Google Scholar]
- 44. Ritchie ME, Phipson B, Wu D, et al. : limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tyner C, Barber GP, Casper J, et al. : The UCSC Genome Browser database: 2017 update. Nucleic Acids Res. 2017;45(D1):D626–D34. 10.1093/nar/gkw1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roslan N, Bieche I, Bright RK, et al. : TPD52 represents a survival factor in ERBB2-amplified breast cancer cells. Mol Carcinog. 2014;53(10):807–19. 10.1002/mc.22038 [DOI] [PubMed] [Google Scholar]
- 47. Zhang H, Wang J, Pang B, et al. : PC-1/PrLZ contributes to malignant progression in prostate cancer. Cancer Res. 2007;67(18):8906–13. 10.1158/0008-5472.CAN-06-4214 [DOI] [PubMed] [Google Scholar]
- 48. Zhang D, He D, Xue Y, et al. : PrLZ protects prostate cancer cells from apoptosis induced by androgen deprivation via the activation of Stat3/Bcl-2 pathway. Cancer Res. 2011;71(6):2193–202. 10.1158/0008-5472.CAN-10-1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu L, Shang ZF, Wang J, et al. : PC-1/PrLZ confers resistance to rapamycin in prostate cancer cells through increased 4E-BP1 stability. Oncotarget. 2015;6(24):20356–69. 10.18632/oncotarget.3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Feng S, Agoulnik IU, Bogatcheva NV, et al. : Relaxin promotes prostate cancer progression. Clin Cancer Res. 2007;13(6):1695–702. 10.1158/1078-0432.CCR-06-2492 [DOI] [PubMed] [Google Scholar]
- 51. Feng S, Agoulnik IU, Li Z, et al. : Relaxin/RXFP1 signaling in prostate cancer progression. Ann N Y Acad Sci. 2009;1160:379–80. 10.1111/j.1749-6632.2008.03793.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Neschadim A, Summerlee AJ, Silvertown JD: Targeting the relaxin hormonal pathway in prostate cancer. Int J Cancer. 2015;137(10):2287–95. 10.1002/ijc.29079 [DOI] [PubMed] [Google Scholar]
- 53. Silvertown JD, Ng J, Sato T, et al. : H2 relaxin overexpression increases in vivo prostate xenograft tumor growth and angiogenesis. Int J Cancer. 2006;118(1):62–73. 10.1002/ijc.21288 [DOI] [PubMed] [Google Scholar]
- 54. Silvertown JD, Symes JC, Neschadim A, et al. : Analog of H2 relaxin exhibits antagonistic properties and impairs prostate tumor growth. FASEB J. 2007;21(3):754–65. 10.1096/fj.06-6847com [DOI] [PubMed] [Google Scholar]
- 55. Thompson VC, Morris TG, Cochrane DR, et al. : Relaxin becomes upregulated during prostate cancer progression to androgen independence and is negatively regulated by androgens. Prostate. 2006;66(16):1698–709. 10.1002/pros.20423 [DOI] [PubMed] [Google Scholar]
- 56. Onodera Y, Takagi K, Miki Y, et al. : TACC2 (transforming acidic coiled-coil protein 2) in breast carcinoma as a potent prognostic predictor associated with cell proliferation. Cancer Med. 2016;5(8):1973–82. 10.1002/cam4.736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Takayama K, Horie-Inoue K, Suzuki T, et al. : TACC2 is an androgen-responsive cell cycle regulator promoting androgen-mediated and castration-resistant growth of prostate cancer. Mol Endocrinol. 2012;26(5):748–61. 10.1210/me.2011-1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim DH, Oh SY, Kim SY, et al. : DNA ligase4 as a prognostic marker in nasopharyngeal cancer patients treated with radiotherapy. Asian Pac J Cancer Prev. 2014;15(24):10985–9. 10.7314/APJCP.2014.15.24.10985 [DOI] [PubMed] [Google Scholar]
- 59. Jun S, Jung YS, Suh HN, et al. : LIG4 mediates Wnt signalling-induced radioresistance. Nat Commun. 2016;7:10994. 10.1038/ncomms10994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grupp K, Roettger L, Kluth M, et al. : Expression of DNA ligase IV is linked to poor prognosis and characterizes a subset of prostate cancers harboring TMPRSS2:ERG fusion and PTEN deletion. Oncol Rep. 2015;34(3):1211–20. 10.3892/or.2015.4080 [DOI] [PubMed] [Google Scholar]
- 61. Cao WH, Liu HM, Liu X, et al. : Relaxin enhances in-vitro invasiveness of breast cancer cell lines by upregulation of S100A4/MMPs signaling. Eur Rev Med Pharmacol Sci. 2013;17(5):609–17. [PubMed] [Google Scholar]
- 62. Binder C, Hagemann T, Husen B, et al. : Relaxin enhances in-vitro invasiveness of breast cancer cell lines by up-regulation of matrix metalloproteases. Mol Hum Reprod. 2002;8(9):789–96. 10.1093/molehr/8.9.789 [DOI] [PubMed] [Google Scholar]
- 63. Ma J, Niu M, Yang W, et al. : Role of relaxin-2 in human primary osteosarcoma. Cancer Cell Int. 2013;13(1):59. 10.1186/1475-2867-13-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hombach-Klonisch S, Bialek J, Trojanowicz B, et al. : Relaxin enhances the oncogenic potential of human thyroid carcinoma cells. Am J Pathol. 2006;169(2):617–32. 10.2353/ajpath.2006.050876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Radestock Y, Willing C, Kehlen A, et al. : Relaxin enhances S100A4 and promotes growth of human thyroid carcinoma cell xenografts. Mol Cancer Res. 2010;8(4):494–506. 10.1158/1541-7786.MCR-09-0307 [DOI] [PubMed] [Google Scholar]
- 66. Byrne JA, Frost S, Chen Y, et al. : Tumor protein D52 (TPD52) and cancer-oncogene understudy or understudied oncogene? Tumour Biol. 2014;35(8):7369–82. 10.1007/s13277-014-2006-x [DOI] [PubMed] [Google Scholar]
- 67. Byrne JA, Balleine RL, Schoenberg Fejzo M, et al. : Tumor protein D52 (TPD52) is overexpressed and a gene amplification target in ovarian cancer. Int J Cancer. 2005;117(6):1049–54. 10.1002/ijc.21250 [DOI] [PubMed] [Google Scholar]
- 68. Zhao Z, Liu H, Hou J, et al. : Tumor Protein D52 (TPD52) Inhibits Growth and Metastasis in Renal Cell Carcinoma Cells Through the PI3K/Akt Signaling Pathway. Oncol Res. 2017;25(5):773–9. 10.3727/096504016X14774889687280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li J, Li Y, Liu H, et al. : The four-transmembrane protein MAL2 and tumor protein D52 (TPD52) are highly expressed in colorectal cancer and correlated with poor prognosis. PLoS One. 2017;12(5):e0178515. 10.1371/journal.pone.0178515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rubin MA, Varambally S, Beroukhim R, et al. : Overexpression, amplification, and androgen regulation of TPD52 in prostate cancer. Cancer Res. 2004;64(11):3814–22. 10.1158/0008-5472.CAN-03-3881 [DOI] [PubMed] [Google Scholar]
- 71. Goto Y, Nishikawa R, Kojima S, et al. : Tumour-suppressive microRNA-224 inhibits cancer cell migration and invasion via targeting oncogenic TPD52 in prostate cancer. FEBS Lett. 2014;588(10):1973–82. 10.1016/j.febslet.2014.04.020 [DOI] [PubMed] [Google Scholar]
- 72. Moritz T, Venz S, Junker H, et al. : Isoform 1 of TPD52 (PC-1) promotes neuroendocrine transdifferentiation in prostate cancer cells. Tumour Biol. 2016;37(8):10435–46. 10.1007/s13277-016-4925-1 [DOI] [PubMed] [Google Scholar]
- 73. Shang ZF, Wei Q, Yu L, et al. : Suppression of PC-1/PrLZ sensitizes prostate cancer cells to ionizing radiation by attenuating DNA damage repair and inducing autophagic cell death. Oncotarget. 2016;7(38):62340–51. 10.18632/oncotarget.11470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li L, Xie H, Liang L, et al. : Increased PrLZ-mediated androgen receptor transactivation promotes prostate cancer growth at castration-resistant stage. Carcinogenesis. 2013;34(2):257–67. 10.1093/carcin/bgs337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang YF, Jan YH, Liu YP, et al. : Squalene synthase induces tumor necrosis factor receptor 1 enrichment in lipid rafts to promote lung cancer metastasis. Am J Respir Crit Care Med. 2014;190(6):675–87. 10.1164/rccm.201404-0714OC [DOI] [PubMed] [Google Scholar]
- 76. Fukuma Y, Matsui H, Koike H, et al. : Role of squalene synthase in prostate cancer risk and the biological aggressiveness of human prostate cancer. Prostate Cancer Prostatic Dis. 2012;15(4):339–45. 10.1038/pcan.2012.14 [DOI] [PubMed] [Google Scholar]
- 77. Susini T, Berti B, Carriero C, et al. : Topoisomerase II alpha and TLE3 as predictive markers of response to anthracycline and taxane-containing regimens for neoadjuvant chemotherapy in breast cancer. Onco Targets Ther. 2014;7:2111–20. 10.2147/OTT.S71646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Samimi G, Ring BZ, Ross DT, et al. : TLE3 expression is associated with sensitivity to taxane treatment in ovarian carcinoma. Cancer Epidemiol Biomarkers Prev. 2012;21(2):273–9. 10.1158/1055-9965.EPI-11-0917 [DOI] [PubMed] [Google Scholar]
- 79. Yang RW, Zeng YY, Wei WT, et al. : TLE3 represses colorectal cancer proliferation by inhibiting MAPK and AKT signaling pathways. J Exp Clin Cancer Res. 2016;35(1):152. 10.1186/s13046-016-0426-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80. Nakaya HI, Beckedorff FC, Baldini ML, et al. : Splice variants of TLE family genes and up-regulation of a TLE3 isoform in prostate tumors. Biochem Biophys Res Commun. 2007;364(4):918–23. 10.1016/j.bbrc.2007.10.097 [DOI] [PubMed] [Google Scholar]
- 81. Guan H, Liu C, Fang F, et al. : MicroRNA-744 promotes prostate cancer progression through aberrantly activating Wnt/β-catenin signaling. Oncotarget. 2017;8(9):14693–707. 10.18632/oncotarget.14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Giménez-Mascarell P, Oyenarte I, Hardy S, et al. : Structural Basis of the Oncogenic Interaction of Phosphatase PRL-1 with the Magnesium Transporter CNNM2. J Biol Chem. 2017;292(3):786–801. 10.1074/jbc.M116.759944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee JH, Zhao XM, Yoon I, et al. : Integrative analysis of mutational and transcriptional profiles reveals driver mutations of metastatic breast cancers. Cell Discov. 2016;2:16025. 10.1038/celldisc.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang W, Sviripa V, Chen X, et al. : Fluorinated N, N-dialkylaminostilbenes repress colon cancer by targeting methionine S-adenosyltransferase 2A. ACS Chem Biol. 2013;8(4):796–803. 10.1021/cb3005353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Frau M, Feo F, Pascale RM: Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J Hepatol. 2013;59(4):830–41. 10.1016/j.jhep.2013.04.031 [DOI] [PubMed] [Google Scholar]
- 86. Wang X, Guo X, Yu W, et al. : Expression of methionine adenosyltransferase 2A in renal cell carcinomas and potential mechanism for kidney carcinogenesis. BMC Cancer. 2014;14:196. 10.1186/1471-2407-14-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Maldonado LY, Arsene D, Mato JM, et al. : Methionine adenosyltransferases in cancers: Mechanisms of dysregulation and implications for therapy. Exp Biol Med (Maywood). 2018;243(2):107–17. 10.1177/1535370217740860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tomasi ML, Cossu C, Spissu Y, et al. : S-adenosylmethionine and methylthioadenosine inhibit cancer metastasis by targeting microRNA 34a/b-methionine adenosyltransferase 2A/2B axis. Oncotarget. 2017;8(45):78851–69. 10.18632/oncotarget.20234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cizkova M, Vacher S, Meseure D, et al. : PIK3R1 underexpression is an independent prognostic marker in breast cancer. BMC Cancer. 2013;13:545. 10.1186/1471-2407-13-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cheung LW, Hennessy BT, Li J, et al. : High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 2011;1(2):170–85. 10.1158/2159-8290.CD-11-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ma J, Hou X, Li M, et al. : Genome-wide methylation profiling reveals new biomarkers for prognosis prediction of glioblastoma. J Cancer Res Ther. 2015;11 Suppl 2:C212–5. 10.4103/0973-1482.168188 [DOI] [PubMed] [Google Scholar]
- 92. Thorsen K, Schepeler T, Øster B, et al. : Tumor-specific usage of alternative transcription start sites in colorectal cancer identified by genome-wide exon array analysis. BMC Genomics. 2011;12:505. 10.1186/1471-2164-12-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Luo A, Zhang X, Fu L, et al. : Zinc finger factor ZNF121 is a MYC-interacting protein functionally affecting MYC and cell proliferation in epithelial cells. J Genet Genomics. 2016;43(12):677–85. 10.1016/j.jgg.2016.05.006 [DOI] [PubMed] [Google Scholar]
- 94. Liu C, Zheng L, Wang H, et al. : The RCAN1 inhibits NF- κB and suppresses lymphoma growth in mice. Cell Death Dis. 2015;6(10):e1929. 10.1038/cddis.2015.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Espinosa AV, Shinohara M, Porchia LM, et al. : Regulator of calcineurin 1 modulates cancer cell migration in vitro. Clin Exp Metastasis. 2009;26(6):517–26. 10.1007/s10585-009-9251-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hang X, Wu Z, Chu K, et al. : Low expression of DCXR protein indicates a poor prognosis for hepatocellular carcinoma patients. Tumour Biol. 2016;37(11):15079–85. 10.1007/s13277-016-5302-9 [DOI] [PubMed] [Google Scholar]
- 97. Ebert B, Kisiela M, Maser E: Human DCXR - another 'moonlighting protein' involved in sugar metabolism, carbonyl detoxification, cell adhesion and male fertility?. Biol Rev Camb Philos Soc. 2015;90(1):254–78. 10.1111/brv.12108 [DOI] [PubMed] [Google Scholar]
- 98. Cho-Vega JH, Vega F, Schwartz MR, et al. : Expression of dicarbonyl/L-xylulose reductase (DCXR) in human skin and melanocytic lesions: morphological studies supporting cell adhesion function of DCXR. J Cutan Pathol. 2007;34(7):535–42. 10.1111/j.1600-0560.2006.00661.x [DOI] [PubMed] [Google Scholar]
- 99. Cho-Vega JH, Tsavachidis S, Do KA, et al. : Dicarbonyl/L-xylulose reductase: a potential biomarker identified by laser-capture microdissection-micro serial analysis of gene expression of human prostate adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2615–22. 10.1158/1055-9965.EPI-07-0684 [DOI] [PubMed] [Google Scholar]
- 100. Abiatari I, Gillen S, DeOliveira T, et al. : The microtubule-associated protein MAPRE2 is involved in perineural invasion of pancreatic cancer cells. Int J Oncol. 2009;35(5):1111–6. 10.3892/ijo_00000426 [DOI] [PubMed] [Google Scholar]
- 101. Zhang JH, Li AY, Wei N: Downregulation of long non-coding RNA LINC01133 is predictive of poor prognosis in colorectal cancer patients. Eur Rev Med Pharmacol Sci. 2017;21(9):2103–7. [PubMed] [Google Scholar]
- 102. Zhang J, Zhu N, Chen X: A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol. 2015;36(10):7465–71. 10.1007/s13277-015-3460-9 [DOI] [PubMed] [Google Scholar]
- 103. Brown-Endres L, Schoenfeld D, Tian F, et al. : Expression of the p53 target CDIP correlates with sensitivity to TNFα-induced apoptosis in cancer cells. Cancer Res. 2012;72(9):2373–82. 10.1158/0008-5472.CAN-11-3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Fraser M, Sabelnykova VY, Yamaguchi TN, et al. : Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017;541(7637):359–64. 10.1038/nature20788 [DOI] [PubMed] [Google Scholar]
- 105. Ayoubi TA, Van De Ven WJ: Regulation of gene expression by alternative promoters. FASEB J. 1996;10(4):453–60. 10.1096/fasebj.10.4.8647344 [DOI] [PubMed] [Google Scholar]
- 106. Baek D, Davis C, Ewing B, et al. : Characterization and predictive discovery of evolutionarily conserved mammalian alternative promoters. Genome Res. 2007;17(2):145–55. 10.1101/gr.5872707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Millevoi S, Vagner S: Molecular mechanisms of eukaryotic pre-mRNA 3' end processing regulation. Nucleic Acids Res. 2010;38(9):2757–74. 10.1093/nar/gkp1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hollerer I, Grund K, Hentze MW, et al. : mRNA 3'end processing: A tale of the tail reaches the clinic. EMBO Mol Med. 2014;6(1):16–26. 10.1002/emmm.201303300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tian B, Manley JL: Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38(6):312–20. 10.1016/j.tibs.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Moore MJ: From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–8. 10.1126/science.1111443 [DOI] [PubMed] [Google Scholar]
- 111. Miyamoto S, Chiorini JA, Urcelay E, et al. : Regulation of gene expression for translation initiation factor eIF-2 alpha: importance of the 3' untranslated region. Biochem J. 1996;315(Pt 3):791–8. 10.1042/bj3150791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Takagaki Y, Seipelt RL, Peterson ML, et al. : The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87(5):941–52. 10.1016/S0092-8674(00)82000-0 [DOI] [PubMed] [Google Scholar]
- 113. Edwalds-Gilbert G, Veraldi KL, Milcarek C: Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25(13):2547–61. 10.1093/nar/25.13.2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lutz CS: Alternative polyadenylation: a twist on mRNA 3' end formation. ACS Chem Biol. 2008;3(10):609–17. 10.1021/cb800138w [DOI] [PubMed] [Google Scholar]
- 115. Mayr C, Bartel DP: Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–84. 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Enciso-Mora V, Hosking FJ, Di Stefano AL, et al. : Low penetrance susceptibility to glioma is caused by the TP53 variant rs78378222. Br J Cancer. 2013;108(10):2178–85. 10.1038/bjc.2013.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Stacey SN, Sulem P, Jonasdottir A, et al. : A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43(11):1098–103. 10.1038/ng.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wiestner A, Tehrani M, Chiorazzi M, et al. : Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109(11):4599–606. 10.1182/blood-2006-08-039859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lin Y, Li Z, Ozsolak F, et al. : An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40(17):8460–71. 10.1093/nar/gks637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Morris AR, Bos A, Diosdado B, et al. : Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res. 2012;18(19):5256–66. 10.1158/1078-0432.CCR-12-0543 [DOI] [PubMed] [Google Scholar]
- 121. Fu Y, Sun Y, Li Y, et al. : Differential genome-wide profiling of tandem 3' UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21(5):741–7. 10.1101/gr.115295.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wang Q, Li W, Liu XS, et al. : A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27(3):380–92. 10.1016/j.molcel.2007.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Andreu-Vieyra C, Lai J, Berman BP, et al. : Dynamic nucleosome-depleted regions at androgen receptor enhancers in the absence of ligand in prostate cancer cells. Mol Cell Biol. 2011;31(23):4648–62. 10.1128/MCB.05934-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhao JC, Fong KW, Jin HJ: FOXA1 acts upstream of GATA2 and AR in hormonal regulation of gene expression. Oncogene. 2016;35(33):4335–44. 10.1038/onc.2015.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Munkley J, Maia TM, Ibarluzea N, et al. : Dataset 2 in: Androgen-dependent alternative mRNA isoform expression in prostate cancer cells. F1000Research. 2018. 10.5256/f1000research.15604.d212874 [DOI] [PMC free article] [PubMed] [Google Scholar]