Abstract
The i-motif represents a paradigmatic example of the wide structural versatility of nucleic acids. In remarkable contrast to duplex DNA, i-motifs are four-stranded DNA structures held together by hemi- protonated and intercalated cytosine base pairs (C:C+). First observed 25 years ago, and considered by many as a mere structural oddity, interest in and discussion on the biological role of i-motifs have grown dramatically in recent years. In this review we focus on structural aspects of i-motif formation, the factors leading to its stabilization and recent studies describing the possible role of i-motifs in fundamental biological processes.
INTRODUCTION
In addition to the B-form DNA double helix (1), DNA can adopt a number of alternative non-B DNA structures (2,3) including G-quadruplex (G4s) (4) and intercalated motif (i-motif) structures (5) (Figure 1). The in vivo existence of these structures in human cells remained uncertain until their recent visualization using antibody fragments that recognize them in a structure-specific manner (6,7). These findings provided key evidence that i-motif structures may be formed in regulatory regions of the human genome, and support the notion that G4 and i-motif structures may play complementary roles in the regulation of gene expression. In this review we focus on structural aspects of i-motif formation and recent studies that describe the possible role of i-motifs in cell biology. We will not extensively review the current uses of i-motif structures in nanotechnological applications, but we will provide a brief summary and refer the reader to excellent reviews on this subject (8–10).
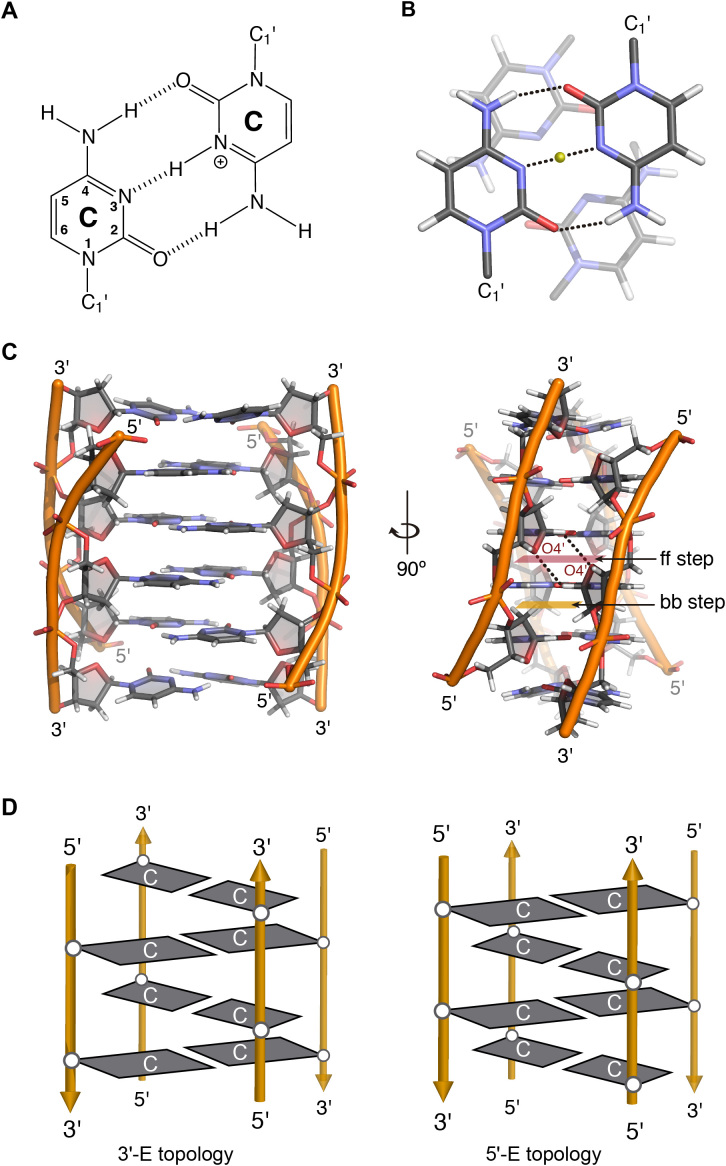
Figure 1.
(A) C:C+ base pair. (B) Stacking between intercalating C:C+ base pairs. (C) Two views of the 3D structure of a tetramolecular i-motif (PDB 1YBR), face-to-face and back-to-back steps are highlighted and two hydrogen bonds between O4′ and H1′ are marked with black dashed lines. (D) Scheme representing the 3′E (left) and 5′E (right) intercalation topologies of an intermolecular i-motif structure.
DNA i-MOTIF-STRUCTURAL FEATURES
The first DNA i-motif was characterized by Gehring et al. for the hexamer sequence d(TCCCCC) forming an intercalated quadruple-helical tetramolecular structure under acidic conditions (5). It consists of two parallel-stranded duplexes intercalated in an antiparallel orientation and held together by hemi-protonated cytosine-cytosine+ (C:C+) base pairs (Figure 1A and B) (5,11). Since this report, a number of i-motif structures has been determined by crystallographic and NMR methods (5,11). As in the case of G4 structures, i-motifs may fold in an intermolecular fashion from the association of two (dimers) or four (tetramers) separate DNA strands, or form an intramolecular structure (monomer) due to the spatial arrangement of four different C-tracts within the same strand.
i-Motifs have other very peculiar structural features. The distance between consecutive base pairs is 3.1 Å, and the right-handed helical twist angle is ∼12–20°, both significantly smaller than those measured in B-DNA (3.4 Å and 36°) (12). The intercalation of base pairs from two parallel duplexes leads to a structure with two major (wide) grooves and two minor (narrow) grooves (Figure 1C). The two minor grooves are extremely narrow (3.1 Å versus ∼5.7 Å for B-DNA), giving rise to a number of inter-strand short distances along the sugar phosphate backbones (12). These parameters result in destabilizing interactions due to close phosphate-phosphate distances, which are partially compensated by favorable inter-strand sugar-sugar contacts. Since the minor grooves are formed between antiparallel strands, sugar-sugar contacts occur in two alternative types: face-to-face (ff) and back-to-back (bb) steps. In the ff steps, the ring oxygens (O4′) face one another (5′-side); however, in the bb steps, the C3′-C4′ edges of the sugar moieties are oriented close to each other (3′-side, Figure 1C).
Due to the spatial arrangement of C:C+ base pairs, i-motif structures can be classified in two different intercalation topologies known as 3′E and 5′E (Figure 1D) (13). When the outmost C:C+ base pair is at the 3′-end, the structure is known as 3′E, while in the 5′E topology, the terminal C:C+ base pair is at the 5′-end (14) (Figure 1D). In absence of additional interactions and for a given number of C:C+ base pairs, the 3′E topology is more stable than the 5′E topology due to the extended sugar-sugar contacts along the narrow grooves (15).
Due to the requirement of hemi-protonated base pairs, it was thought that i-motif structures could only fold at acidic pH values; however, several recent studies have shown that i-motif structures can form at neutral pH depending on the sequence and environmental conditions (16,17). Thus, i-motif structures were observed at neutral pH and low temperatures under molecular crowding conditions (18,19), under negative superhelicity (20), in the presence of silver (21) or copper (I) cations (22), and inside silica nano-channels (23). Chemical modifications such as 2′-deoxy-2′-fluoro-arabinocytidine (2′F-araC, known as 2′F-ANA) also induce formation of DNA i-motif structures under neutral conditions (24,25).
FACTORS AFFECTING THE STABILITY OF i-MOTIF STRUCTURES
Like other nucleic acid structures, i-motif stability depends on many factors, including sequence nature, temperature, and ionic strength. Unlike B-DNA or G-quadruplexes, in which π stacking interactions between sequential nucleobases play an essential role in their stability, the intercalative geometry between consecutive base pairs in i-motif structures gives rise to very little overlap between the six-membered aromatic pyrimidine bases (Figure 1B and C). Although C:C+ base pairs involve the favorable stacking of exocyclic carbonyls and amino groups in an antiparallel fashion, theoretical calculations showed that this and other favorable stacking interactions between consecutive C:C+ base pairs barely compensate the electrostatic repulsion between their charged imino groups (15). Of particular relevance to understand the factors affecting i-motif stability are the multiple studies recently reported on chemically modified i-motif structures, which we discussed in detail below.
The C:C+ base pair
The hemi-protonated C:C+ base pairs are the key interactions for i-motif stability. The three hydrogen bonds of the C:C+ base pair confer a high stability. Computer calculations indicate that the base-pairing energy (BPE) for the C:C+ base pair is 169.7 kJ/mol, higher than BPEs of canonical Watson-Crick G·C (96.6 kJ/mol) and neutral C·C (68.0 kJ/mol) (26). The central hydrogen bonding in a hemiprotonated C:C+ (N3···H···N3) base pair has been described as a double-well potential where the proton delocalizes/oscillates between the two wells (27). Leroy et al. estimated the proton-transfer rate to be 8 × 104 s−1 (28). The NMR structural study of an intramolecular telomeric i-motif (PDB code: 1ELN) showed that the C:C+ base pairs are planar and the N3–N3 distance is around 2.6–2.8 Å (29). Most importantly, protonation at the N3 position produces a positively charged base pair. NMR spectroscopy and computational analyses performed by Lieblein et al. suggested that the N3···H+···N3 bonds possess an asymmetric double-well potential and that the proton in one C:C+ base pair tends to adopt a position that leads to the largest distance with respect to the proton of neighboring C:C+ base pair (27).
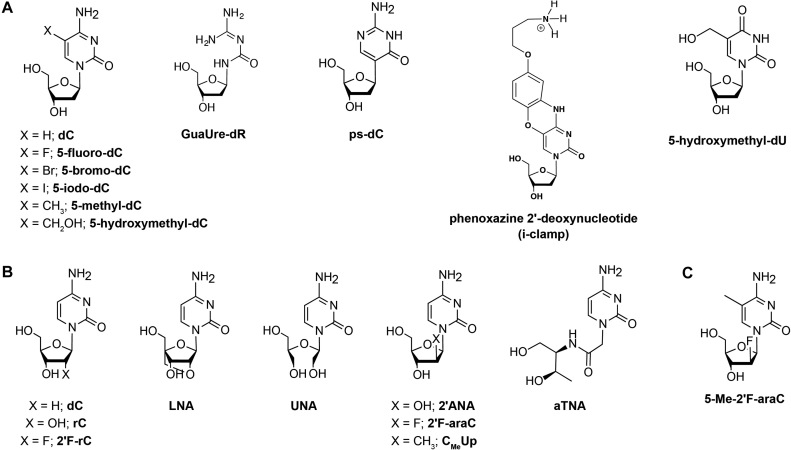
The effect of chemical modifications in the C:C+ base pairs has been investigated in different contexts. Wadkins et al. indicated that a cytosine modification might have different effects on i-motif stability depending on the environmental conditions (30). For instance, substitution of cytosine by its halogenated analogues (Figure 2A), such as 5-fluoro, 5-bromo and 5-iodo derivatives, increases stability of i-motifs at acidic conditions (31). Furthermore, cytosine methylation at position 5 leads to an increase in the pH of mid transition (i.e. pHT) and Tm of i-motif structures (Δ pHT = +0.11 for two sequences containing two 5-methylcytosine substitutions), while hydroxymethylation leads to a decrease in pHT (ΔpHT = –0.2) and Tm (Figure 2A) (32).
Figure 2.
Nucleoside modifications introduced in i-motif structures. (A) Nucleobase modifications. (B) Sugar modifications and (C) A nucleoside analogue possessing both sugar and nucleobase modifications. d: deoxyribose, r: ribose and ara: arabinose sugar.
Recently, Waller's group investigated the effect of 2′-deoxyriboguanylurea (GuaUre-dR) (Figure 2A) on human telomeric i-motif formation (33). GuaUre-dR is a breakdown product of decitabine, a cytidine analogue that acts as an epimutagen and a chemotherapeutic agent. Despite the fact that this analogue can form a base pair with cytosine without protonation, the modified telomeric sequences exhibited a decrease in the pHT (5.8) compared to the unmodified i-motifs (pHT = 6.1) (33). Mir et al. studied the effect of pseudoiso-deoxycytidine (psC) (Figure 2A) on the stability of head-to-head and head-to-tail dimeric i-motif structures (34). An increase of the stability of the studied i-motif structures was observed when the neutral psC:C base pair was located at the end of a C:C+ stack. However, protonated base pairs are required (psC:C+ or psC:psC+) when the psC modifications are located in central positions of the i-motif structures. The results from these two studies underline the importance of the electrostatic interactions in the i-motif stability, where the presence of positive charges in the core of the structures is a key factor for its stability.
Significant i-motif stabilizations have been reported recently by introducing phenoxazine 2′-deoxynucleotides (i-clamps) containing a C8-aminopropynol tether (Figure 2A). These nucleotides are able to form base pairs with protonated cytosines and, simultaneously, interact favorably with the phosphate backbone of the opposite strand. As in the case of psC substitutions, the effect depends on the position of the substitution, where more stabilization is observed when they are located at the 5′-end of the C-stacks (35).
Very recently, natural base lesions were introduced in the TAA loops and in the core cytosines of the human telomeric C-rich strand d(CCCTAA)3CCCT (36). This study reveals that i-motifs containing apurinic sites (apA) and 8-oxoadenine (8oxoA) substituting adenine, and 5-hydroxymethyluracil (5hmU) (Figure 2A) substituting thymine exhibit thermal stabilities that depend on the position of these substitutions in the sequence. Thus, in comparison to the unmodified structure, ΔTm values for i-motifs having apA substitutions ranges between −1.7 and +5.0°C, those having 8oxoA vary from −1.4 to +3.5°C, and ΔTm for the ones containing 5hmU is +0.4°C on average. On the contrary, the presence of uracil instead of cytosine (due to the enzymatic or spontaneous deamination of cytosine) substantially reduces i-motif thermal stability. The extent of this destabilization depends on the position of the deaminated cytosines. Whereas the loss of outer cytosines produces a decrease of 11.6–17.0°C in melting temperatures, the destabilization produced by the substitution of inner cytosines is even more pronounced due to the loss of a C:C+ pair intercalated between two inversely oriented pairs (36).
Sugar and phosphate backbone modifications
Sugar modifications
The i-motif folding architecture is destabilized by the close inter-strand phosphorus-phosphorus distances (5.9 Å) along the narrow groove. This destabilizing factor is compensated by an inter-strand favorable interaction network between the deoxyribose sugar moieties along the minor groove (12). Additional stabilizing interactions along the narrow groove results from C-H1′···O4′ interactions within each pair of antiparallel strands (Figure 1C). Moreover, studies have shown intra-nucleoside hydrogen bond between O4′ and H6, and O4′ bonding simultaneously with H1′ and H4′ (12).
Sugar-backbone chemical modifications have shown to be very useful to assess these interactions and modulate i-motif stability. These include cytidines from RNA (37,38), 2′F-RNA (39), locked (LNA) (40) and unlocked (UNA) nucleic acids (41), 2′-arabinonucleic acids (ANA) (42), acyclic threoninol nucleic acids (aTNA) (43), and more recently, 2′-fluoro-arabinonucleic acids (2′F-ANA) (24,25,44) (Figure 2B).
In DNA i-motif structures, the conformation of the glycosidic angles is anti and the deoxyribose sugars mainly adopt C3′-endo (North) puckering. These properties led researchers to investigate the ability of RNA to form i-motifs, since the ribose sugar favors the C3′-endo conformation, in addition to RNA-like modifications favoring the North conformation such as 2′F-RNA, LNA, and 2′-OMe. Reports have shown that oligoribonucleotides form less stable i-motif structures compared to their oligodeoxynucleotide counterparts. Initial studies reported a ΔTm of 29°C between i-motif structures formed by an 18-mer DNA sequence, (d(CCCTCCCTTTTCCCTCCC), 54°C) versus the corresponding 18-mer RNA sequence (r(CCCUCCCUUUUCCCUCCC), 25°C) (38). Since the uracil substituted DNA sequence exhibited a similar melting temperature to the thymine DNA counterpart (56°C), the lower stability of RNA i-motif was ascribed due to the presence of ribose 2′-OH groups (38).
Collin and Gehring studied the effect of replacing one or more DNA residues in dTCCCCC (Tm = 49°C at pH 4.6) by RNA (CR) (37). This study suggested that the destabilization observed due to an RNA residue is almost equal to the destabilization due to the loss of a C:C+ base pair since dTCRCCCC and dTCCRCCC exhibited a melting temperature of 41°C compared to 39°C for dTCCCC. When two hydroxyl groups are juxtaposed in back-to-back step, dTCCRCRCC the distance between them is 0.35 nm compared to 0.65 nm in face-to-face steps, leading to less stable i-motif structures with a Tm = 29.5°C (37).
The r(UC5) sequence led to the formation of two i-motif structures adopting different intercalation topologies. The intercalation topology of the major conformation possesses one less 2′-OH/2′-OH repulsive contact compared to the minor conformation. The free energy of the RNA i-motif per C:C+ base pair (-4 kJ/mol) is almost half that of the DNA i-motif (45). Despite of its modest stability, the existence of RNA i-motifs inside the cells has been suggested in vivo. In a very recent study showing i-motif foci by immunostaining with a structure-specific antibody (iMab) (6), a number of i-motif foci were still observed in the nuclei of HeLa cells upon treatment with DNAse. The authors suggest that those remaining foci might be due to DNA i-motifs somehow inaccessible to DNAse or to the detection of RNA i-motifs, which are not affected by DNAse degradation. In addition, the authors observed a moderate but significant decrease of i-motif foci when treating the cells with RNAse. These observations suggest that RNA i-motif may exist in the cell, and it may have a biological role.
Like RNA, 2′F-RNA (Figure 2B) also adopts a C3′-endo sugar pucker and the fluorine is sterically comparable to a hydroxyl group. Therefore, the main difference between 2′F-RNA and RNA lies in fluorine's lower hydrogen-bonding capability compared to the 2′-OH group. The introduction of a single 2′-fluorine (Cf) enhances the thermal stability of d(TCCCfCC) i-motif (48.8°C) with respect to d(TCCCCC) i-motif (44.8°C) (39). Two consecutive modifications, d(TCCfCfCC), lead to 0.6°C reduction compared to the unmodified strand. On the other hand, ribose modifications in the same positions lead to a destabilization of 20°C. Given the similarity in the size of fluorine and hydroxyl group, the destabilization observed with RNA substitutions is not due to steric clashes but likely due to the solvation of the hydroxyl groups compared to a limited solvent accessibility for the fluorinated minor groove.
LNA is another RNA mimic locked in the C3′-endo conformation with a 2′-OCH2-4′-C methylene bridge (Figure 2B). LNA was also introduced in the hexamer model sequence dTC5 which forms stable intermolecular i-motif structure at pH 4.0 (40). Some of the partially modified sequences exhibit similar (d(TLLCCC), d(TLCLCC), and d(TCLCLC)) or higher (d(TCCLLC)) stability compared to unmodified i-motif, while the fully modified strand does not form an i-motif. The stability in LNA-modified i-motifs depends on the position of LNA in the sequence and is due to the extended hydrogen-bonding network at the back-to-back steps involving the 2′-OCH2-4′-C bridge. Therefore, when LNA is introduced in certain positions, the 2′-OCH2-4′-C bridge creates additional hydrogen bonds that neutralize the unfavorable van der Waals contacts in the minor groove.
Interestingly, substituting the 2′-OH in RNA with its 2′-arabinose epimer leads to stable i-motif structures since the OH group is placed in the wide major groove (42). The different stability observed between riboses and arabinoses confirms the critical significance of sugar–sugar contacts in the minor groove on the stabilization of i-motifs. We recently demonstrated that incorporating 2′F-araC (Figure 2B) modifications in i-motif structures leads to significant stabilization over a wide pH range. 2′F-araC is one of very few chemical modifications that stabilize i-motif structures at neutral conditions not only at the ends of the C-tracts, but also in central positions (24). Furthermore, the 2′F-araC modification stabilized intermolecular centromeric and intramolecular telomeric i-motifs in all the positions tested (24). Despite the fact that the nucleoside 2′F-araC exhibits a lower pKa compared to deoxycytidine (3.9 versus 4.4, respectively), the pHT in the modified i-motif structures was remarkably higher (+0.7 for centromeric sequences and +0.8 for telomeric sequences), allowing for the observation of these structures at neutral pH. NMR structural determination revealed that the 2′F-araC residues adopt a C2′-endo sugar pucker, instead of the C3′-endo conformation that is usually found in unmodified structures, with the fluorine atom oriented in the major groove. Therefore, 2′F-araC modifications do not perturb the hydrogen-bonding network that provides stability to the structure, but instead lead to additional electrostatic interactions that are absent in the unmodified structure. The above results allow the utilization of i-motif structures in several applications, most importantly in biological assays that require physiological temperature and pH conditions. Following these interesting observations, Aviñó et al. investigated the effect of (2′S)-2′-deoxy-2′-C-methyl-cytidine units (CMeUp) (Figure 2B) on telomeric i-motif structures (46). This modification adopts mainly a C3′-endo sugar pucker with the methyl group at C2′ in the ‘arabino’ (or β) orientation. CMeUp was tolerated in i-motif structures; however, stabilization was less pronounced compared to 2′F-araC. This result confirms the role of favorable electrostatic interactions induced by the electronegativity of the fluorine atom in the enhanced stability provoked by 2′F-araC.
With the purpose of finding a modification or combination of modifications that would lead to even higher thermal stability at physiological pH along with higher pHT values compared to the 2′F-araC modification, our groups investigated the effects of 5-Me-2′F-araC (Figure 2C). This nucleoside combines two stabilizing i-motif modifications, 5-methylcytosine nucleobase and 2′-fluoroarabinose sugar in the same nucleotide (44). Interestingly, 5-Me-2′F-araC was found to exhibit a similar stabilization effect to 2′F-araC. However, an i-motif with 2′F-araC:5-Me-dC base pairs exhibited a pHT value of 7.17 compared to 6.53 for a structure containing 2′F-araC:2′F-araC and 5-Me-dC:5-Me-dC base pairs. This suggests that it is possible to tune the pH and thermal stability of i-motif structures by selecting the position and type of modifications and highlights the significance of the nature of base pairs on i-motif stability.
In conclusion, most sugar modifications destabilize i-motif structures, regardless of whether or not they favor the C3′-endo sugar conformation. Substituents oriented toward the compact minor groove cause steric clashes that destabilize the structure. However, chemical modifications that preserve sugar-sugar contacts across the minor groove, such as arabinose sugars, are well tolerated. Of particular relevance is the effect of 2′F-araC substitutions, which lead to stable structures at pH 7.
Phosphate modifications
The arrangement of the sugar-phosphate backbone in i-motif folding gives rise to unusually short distances between adjacent phosphates. In an attempt to suppress the repulsion between the negatively charged phosphate backbones, several backbone modifications have been investigated. Mergny and Lacroix investigated the effect of phosphorothioate, and methylphosphonate, as opposed to the phosphodiester backbone (47). Their studies show that only backbones exhibiting phosphodiester and phosphorothioate bonds allow i-motif formation. They hypothesized that even though the methylphosphonate backbone is neutral, the bulkiness of the methyl group prevents i-motif formation. Additionally, the chirality of the methylphosphonate linkage (presence of both Rp and Sp stereochemistries) might have influenced i-motif formation and stability. The incorporation of phosphorothioates in several DNA C-rich sequences leads to the formation of stable i-motif structures at neutral pH, and they are only a few degrees less stable than the unmodified structures (47). Moreover, the chirality of the phosphorothioate group influences i-motif stability; for instance the Rp-stereochemistry leads to greater stabilization compared to the Sp-stereochemistry (ΔTm = 11°C) (48). Another backbone modification that was investigated involved replacing the negatively-charged sugar-phosphate backbone with a neutral polyamide backbone, i.e. peptide nucleic acid (PNA). Balasubramanian et al. studied the effect of PNA on the model hexanucleotide sequence p(TCCCCC) utilizing nano-electrospray ionization-mass spectrometry (Nano-ESI-MS) and H/D exchange (49). PNA was shown to form stable i-motif structures; however, the i-motif folding occurs at a narrower pH range (4.1–4.5) compared to its DNA counterpart (4.5–6.5) (49). i-Motif formation from a 1:1 mixture of PNA and DNA strands was then studied by Modi et al. (50). FRET studies revealed that structure is stabilized by the intercalation of two parallel DNA-PNA heteroduplexes with the DNA strands occupying one of the minor grooves. The hybrid PNA-DNA i-motifs exhibit an intermediate stability (pH 4.2–5.7) compared to the more stable DNA or less stable PNA i-motifs. This intermediate stability can be attributed to lower electrostatic repulsion compared to the net negatively charged DNA and net positively charged PNA i-motif structures. Another interesting ‘backbone’ modification investigated by Robidoux et al. involves branched oligonucleotides, where dC-rich strands are forced to become parallel by joining them to the vicinal 2′-5′ and 3′-5′ phosphodiester linkages of a branching riboadenosine linker. This study showed that branched oligonucleotides can associate into stable i-motif structures, where certain constructs exhibited a Tm around 25°C at pH 7.0 (51).
C-tract length
In general, under the same experimental conditions, the i-motif structure possessing a higher number of C:C+ base pairs would be more stable (52). Very recently, Waller's group and Burrows’ group investigated the effect of C-tract length on the folding of intramolecular i-motif structures under physiological conditions (16,17).
Waller's group studied several sequences containing at least four C-tracts with different lengths. Their results reveal that, in general, the change in pHT increases as the number of cytosines per tract increases. For instance, pHT increases from 6.1 for C2(T3C2)3 to 6.7 for C3(T3C3)3, to 7.1 for C4(T3C4)3 and 7.2 for C5(T3C7)5. A similar trend was observed for the thermal stability (Tm) where C3(T3C3)3 exhibited a Tm = 7°C while the Tms of C4(T3C4)3 and C5(T3C7)5 were 15.8°C and 26.2°C, respectively. Increasing the C-tract length beyond five enhances the stability; however, it also leads to two melting transitions and to increased hysteresis between the folding and unfolding transitions suggesting the formation of two distinct species at pH 7.4. The sequence with the longest C-tract length tested was C10(T3C10)3 which exhibits a pHT of 7.3. Despite that the tested sequences have tracts of four or more cytosines that fold into i-motif at neutral pH values, more recent studies show that sequences containing shorter C tracts also have the tendency to form stable i-motifs at neutral pH (53,54).
On the other hand, the Burrows group was interested in investigating the formation and stability of i-motif structures from several dCn homo-oligonucleotides (n = 10–30) (17). Utilizing different pH-dependent methods an interesting trend was observed between chain length and stability. The highest thermal stabilities and pHT (Tm > 37°C and pHT > 7.2) were observed for dCn strands of length 15, 19, 23 and 27 cytosines (i.e. 4n – 1). This led to identifying 4n – 1 as the ‘sweet spot’ for i-motif folding in deoxycytidine homopolymers. When T nucleotides were introduced to control the length of the i-motif core and to form loops of varying lengths, the most stable structures consisted of an even number of C:C+ base pairs in the core and three loops of only one nucleotide in length, further confirming the results obtained for dCn homo-oligonucleotides. However, it is important to note that these results are not directly applicable to other heteronucleotidic sequences since the presence of loop-to-loop interactions play important roles on the stability of the structures as reviewed in the next section.
Connecting loops and capping interactions
Several systematic studies investigating the number and nature of different nucleobases in the i-motif loops have been undertaken recently in an attempt to comprehend the effect of the interactions between loop nucleobases on i-motif stability (55–57). Capping residues at the end of the i-motif core in addition to the length and nature of their connecting loops are very important factors for i-motif stability (58). Based on the length of the connecting loops, Brooks et al. divided intramolecular i-motif structures into two different classes. i-Motif structures possessing short loops (loop 1 (2-nt): loop 2 (3 to 4-nt): loop 3 (2-nt), i.e. 2:3–4:2) were classified as ‘class I’, while ‘class II’ i-motifs possess longer loops (6–8:2–5:6–7) (59). In general, very short loops, such as one nucleotide, favor the formation of mono and bimolecular i-motifs, whereas longer loops lead preferentially to the formation of intramolecular i-motif structures (60). Since longer loops might allow for extra stabilizing interactions, it has been proposed that class II i-motifs are more stable. However, there is a number of recent studies showing that class I i-motifs are stable at neutral or nearly neutral pH (53,61).
Thymidines are common capping residues in i-motifs since they can form T:T base pairs (Figure 3A) that are isomorphous to C:C+ base pairs and extend the i-motif core (61–63). In fact, T:T base pairs are not only good capping base pairs, but they even are tolerated in the middle of the C:C+ base pair stack (64). Hoogsteen and reverse Watson-Crick A:T base pairs (Figure 3A) in the loops of i-motifs have been found in the crystallographic structure of an oligonucleotide containing a single repeat of the human telomeric [d(TAACCC)] (65) and centromeric sequences (63) (Figure 3A). A:A (Figure 3A) and G:G base pairs have also been observed in several i-motif structures (66–68). This suggests it is not loop size per se but the precise sequence and the resulting interactions of the bases in the loop that are important for stabilization.
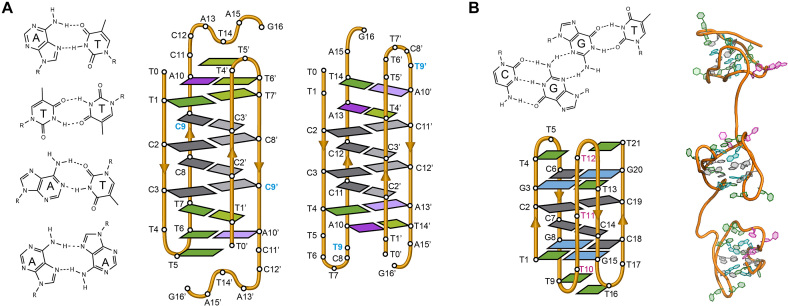
Figure 3.
(A) Several base pairs other than C:C+ that have been observed in i-motif structures (from top to bottom; A:T Hoogsteen, T:T, A:T reverse Watson-Crick and A:A) (Left). Schematic view of the dimeric i-motif structures formed by the two variants of centromeric A-box sequence (63) showing the presence of T:T and A:T reverse Watson–Crick and Hoogsteen base pairs flanking the set of intercalated C:C+ base pairs (Right). The residue in blue denotes the only difference in the primary structure of the two sequences. (B) G:C:G:T minor groove tetrad (Top). Schematic representation of the minimal i-motif structure stabilized by two C:C+ base pairs and two flanking G:C:G:T minor groove tetrads (Bottom) and representation of these structures in tandem (Right) (53).
One of the capping interactions that provokes more significant effects in the i-motif stability is the formation of minor groove tetrads on the ends of the C:C+ tracts. The first example of this kind of interaction was found by Gallego et al. in the structure of the human B-box centromeric sequence, d(TCCCGTTTCCA) (66). This sequence forms a stable head-to-head dimeric i-motif that features a G:T:G:T tetrad between two lateral loops. The two G:T pairs are found in the minor groove side as previously found in other structures (69). Tetrads of the same family have been observed with Watson-Crick G:C base pairs (70) and a combination of G:C and G:T base pairs (69,71). Stabilization of i-motif structures through minor groove tetrads has been observed in other dimeric i-motif structures (72), and more recently in monomeric i-motifs able to form in tandem repeats (53) (Figure 3B). In the latter case, the stabilization provoked by two minor groove tetrads is dramatic, with pHT values close to 8.0.
In cases where a loop connecting two C-tracts is long enough, the formation of other secondary structures, like stem-loop hairpins, is possible. This situation has been observed in a C-rich sequence located near the promoter region of the n-MYC gene (73), although its three-dimensional structure remains to be elucidated.
Impact of ionic strength, molecular crowding and superhelicity
Unlike G4 structures where the nature of the cation leads to significant differences in stability and folding topology, i-motifs are not affected by the nature of the cation but by the ionic strength of the solution. Mergny et al. showed that increasing NaCl concentration from 0 to 100 mM at a pH close to the pKa of cytosine destabilized i-motif structure. Interestingly, higher NaCl concentrations (300 mM) did not cause further destabilization (60). The same trend of decreasing i-motif stability with increasing ionic strength was observed in sequences present in the promoter of n-MYC gene (74).
Molecular crowding agents such as high-molecular weight polyethylene glycols (PEGs) have been widely used to mimic the crowded environment that the nucleic acid would have inside a cell. Crowding conditions preferentially stabilize both i-motif and G4 structures over duplexes and single-stranded DNA (75). For instance, in a 1:1 mixture of G- and C-rich sequences, molecular crowding conditions shift the equilibrium towards G4 and i-motif structures and prevent Watson-Crick duplex formation (76). Dielectric constant effects, such as a shift in the pKa of cytosine by more than 2 units (e.g. 4.8–7.0), or the formation of non-specific PEG/DNA complexes appear to contribute insignificantly to i-motif stabilization (19,75).
Another factor that is associated with i-motif stability is negative superhelicity, which favors DNA double helix unwinding into its component single-stranded sequences. This unwinding relieves negative superhelical stress, facilitating the formation of non-canonical secondary structures in the unwound regions (20). In order to mimic the negative supercoiling induced upstream of a transcription site, Sun and Hurley placed the natural and mutated C- and G-rich sequences of NHE III1 of the c-MYC oncogene promoter in a supercoiled plasmid (20). Using chemical and enzymatic footprinting, they were able to show that i-motif and G4 formation is facilitated by negative superhelicity under physiological conditions. On the contrary, the mutated strands were locally unwound; however, were unable to fold into stable i-motif and G4 structures.
BIOLOGICAL RELEVANCE OF i-MOTIF STRUCTURES
Location of i-motif forming sequences across the genome
The prevalence of G-quadruplex forming sequences along the genome is now well established. In principle, the complementary strand of any G-quadruplex forming sequence is susceptible to forming i-motifs. Thus, Waller's group utilized the search algorithm Quadparser (16), originally designed to find G4-forming sequences, to determine the potential prevalence of i-motif-forming sequences within the human genome. They searched for sequences having four C-tracts of five cytosines separated by tracts ranging between 1 to 19 nucleotides (16). Applying these search criteria they identified 5,125 sequences across the genome with the potential to fold into i-motif structures. Out of these sequences, 637 (i.e. ∼12.4%) were located in gene promoter regions. Through further examination of the gene ontology codes corresponding to the genes under the regulation of those promoters it was found that potential i-motif formation was concentrated in promoters of genes involved in skeletal system development, sequence specific DNA binding, DNA templated transcription and positive regulation of transcription from RNA polymerase II. On the contrary, they did not find sequences fulfilling the search criteria in genes involved in the immune response, G-protein coupled receptor activity and olfactory receptor activity (16).
Using bioinformatics analysis, the Burrows’ group investigated the presence of dCn tracts across the human genome (17). These studies showed the existence of 769 dCn sequences with n between 15 and 81 nucleotides. In addition to promoter regions, this study shows that these C-rich sequences are also present in introns, and 5′- and 3′-UTRs. On the contrary, fewer dCn tracts were observed in the coding and intergenic regions. These two studies highlight the fact that those sequences with higher potential for forming i-motif structures are not randomly located; instead, they are particularly enriched in the promoters of certain genes, which suggests that they may have a role on certain regulatory mechanisms of gene expression; however, these studies are not comprehensive since several sequences, for instance the minimal i-motif structures previously discussed (53), form stable i-motif structures but deviate from the algorithms utilized. Therefore, a better understanding of the sequential requirements for formation of stable i-motifs is necessary to achieve a more accurate mapping of i-motif occurrence along the genome.
Existence of i-motif structures in vivo
The biological relevance of i-motif structures had been largely questioned due to the lack of experimental evidence of their existence in vivo and to the fact that the formation of these structures is favored at pH values more acidic than the intracellular pH. However, several recent studies have changed this paradigm.
A very interesting study on the stability of i-motifs inside cells was performed by Dzatko et al. who applied NMR spectroscopy in living mammalian cells in order to investigate the stability of i-motif structures in the cellular environment (77). Several i-motif forming sequences from different human promoter regions (DAP, HIF-1α, PDGF-A and JAZF1) conveniently labeled with a fluorophore were pre-folded as i-motifs and transfected into HeLa cells. Flow cytometry and confocal microscopy images indicated that the transfected oligonucleotides entered the cells and localized in the nucleus without compromising the cellular viability and the homeostasis of the intracellular pH (pHi). The in-cell NMR spectra for DAP, PDGF-A and JAZF1 exhibited i-motif imino signals up to 35°C, suggesting the presence of folded i-motif structures at physiological conditions. In contrast, no i-motif-specific imino signals were detected for the HIF-1α sequence. Overall, these in-cell NMR spectra indicate that pre-formed i-motif structures introduced into the cells remain stable and persist in the complex intracellular environment of living HeLa cells.
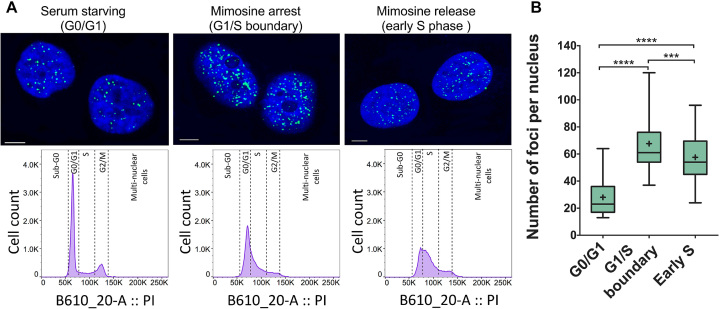
Very recently, Christ's and Dinger's groups developed an antibody, named iMab, able to bind with high affinity and specificity to C-rich DNA sequences forming i-motif structures (6). These groups describe that this antibody is probably the definitive probe for i-motif formation in living human cells. First, the authors demonstrate that iMab antibody binds different well-defined i-motif structures while showing absence of binding to any other DNA structure such as a duplex, hairpin or G-quadruplex. The use of iMab for immunofluorescent staining in three different cell lines (MCF7, U2OS and HeLa) revealed punctuate foci that were attributed to the recognition of i-motifs structures in the nuclei of cells. Very interestingly, the study showed that the number of foci varied along the cell cycle. The authors evaluated the number of foci in cells arrested in three different stages of the cell cycle (early S phase and G0/G1 and G1/S boundaries) showing that the highest number of foci appeared at the G1/S boundary (Figure 4) (6). This observation suggests that i-motif formation can be associated with transcription as late G1 phase is characterized by high transcriptional activity. In contrast, the significant decrease of iMab foci during S phase (Figure 4) suggests that i-motif structures are resolved during DNA replication. Likewise, the number of G4 foci detected in cells by immunofluorescence assays with BG4 antibody also varies along the cell cycle (7,78). In contrast to i-motif, G4 detection was higher during S-phase whose major event is DNA replication. Altogether, these results indicate that i-motif and G4 structures are differently populated in diverse stages of the cell cycle, and suggests that they might play opposing roles in regulating gene expression. This study provides the strongest evidence, so far, for the existence of i-motif structures in vivo and their relevance in key biological processes.
Figure 4.
(A) Confocal microscopy images showing fluorescent foci of i-motif recognized by iMab antibody in the nuclei of HeLa cells arrested at different stages of cell cycle. Flow cytometry diagrams show the population of synchronized and propidium iodide stained cells in each stage. (B) Box plot graph showing the quantification of iMab foci in each phase. Boxes represent 25th to 75th percentile. Horizontal line and ‘+’ symbol inside the box represent medians and means, respectively. Whiskers indicate lower and highest values registered in each case. ***P < 0.001, ****P < 0.0001. Reprinted with permission from Zeraati, M., Langley, D.B., Schofield, P., Moye, A.L., Rouet, R., Hughes, W.E., Bryan, T.M., Dinger, M.E. and Christ, D. (2018) I-motif DNA structures are formed in the nuclei of human cells. Nature chemistry, 10, 631–637. Copyright (2018) Springer Nature.
Interaction of DNA i-motifs with ligands and proteins
Compared to the well-documented examples of G4 ligands, the discovery of specific i-motif binding ligands lags far behind. Several compounds such as TMPyP4 (79), bis-acridine (BisA) (80) and phenanthroline derivatives (74) have been evaluated as i-motif ligands. Some of them have a stabilizing effect, but they are not selective since they also bind other DNA structures. Likewise, some complexes with terbium and ruthenium metals have been studied as potential i-motif binders (81,82); however, they lack specificity and lead to a slight destabilization of i-motif structures.
Carboxyl-modified single-walled carbon nanotubes (C-SWNTs) are considered the first selective i-motif ligands. Addition of C-SWNTs leads to a remarkable increase of thermal stability of the intramolecular i-motif formed by the C-rich human telomeric sequence at acidic pH (83). Additionally, it was found that the presence of C-SWNTs induces the formation of this i-motif structure at pH 8.0 and inhibits duplex formation between complementary human telomeric C- and G-rich sequences. Through S1 nuclease assays and fluorescence changes of 2-aminopurine labeled loops, it was proposed that the nanotubes bind to the 5′-end of the major groove of the telomeric i-motif structure. The stabilization effect of the C-SWNTs was explained by the favorable electrostatic interactions between the C:C+ base pairs and the negatively charged C-SWNTs which substantially decrease the pKa of the C:C+ base pairs (83).
Qu et al. investigated the biomedical effect of C-SWNTs on telomerase activity and telomere function (discussed later in this review). This study determined that i-motif structures formed in the presence of C-SWNTs could inhibit telomerase activity, interfere with telomere functions, and lead to senescence and apoptosis in cancer cells in vitro and in vivo (84).
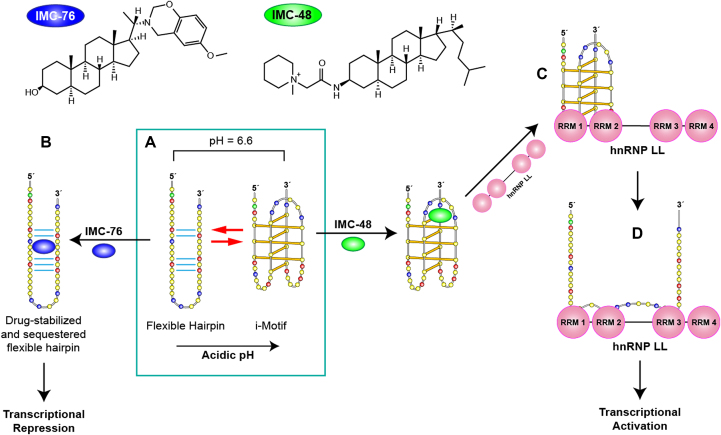
Following the carbon nanotubes, another study has succeeded in identifying a selective i-motif ligand. After screening a library of 1990 compounds, Hurley et al. identified a compound (IMC-48) that binds and stabilizes the i-motif structure formed by the C-rich sequence of BCL2 gene promoter. In parallel, a similar compound (IMC-76) was found to favor an alternative hairpin conformation formed by the same sequence (Figure 5). These molecules can be used to shift the dynamic equilibrium between the two structures formed by this BCL2 promoter sequence, a hairpin and an i-motif structure containing large loops (8:5:7) (85). IMC-48 binds within the central loop of the i-motif (Figure 5A) and is likely stabilized by stacking interactions with thymines. On the other hand, the binding site of IMC-76 was found between the WC hydrogen-bonded tracts in the hairpin structure (Figure 5B). Interestingly, the molecules have opposite effects, while IMC-48 leads to the activation of gene expression, IMC-76 markedly suppresses the levels of BCL2 mRNA.
Figure 5.
(A) Structures of IMC-48 and IMC-76 ligands, specific for BCL2 i-motif and BCL2 hairpin, respectively. (B) BCL2 promoter sequence can adopt two different major conformations which are in a pH-dependent equilibrium. Acidic conditions lead to formation of an i-motif whereas at pH 6.6 there is a mixture of hairpin and i-motif conformations. IMC-76 binds to the hairpin structure shifting the equilibrium towards the hairpin structure which leads to transcriptional repression (A to B) whereas IMC-48 binds to the central loop of the BCL2 i-motif stabilizing the structure that upon binding of hnRNP LL via RRMs 1 and 2 of the protein (A to C) promotes the unfolding of the i-motif and the activation of BCL2 transcription (C to D). Adapted with permission from Kang, H.J., Kendrick, S., Hecht, S.M. and Hurley, L.H. (2014) The Transcriptional Complex Between the BCL2 i-Motif and hnRNP LL Is a Molecular Switch for Control of Gene Expression That Can Be Modulated by Small Molecules. J Am Chem Soc, 136, 4172–4185. Copyright (2014) American Chemical Society.
Several small molecule ligands have been investigated in an attempt to expand the i-motif-specific ligand library such as the type 2 topoisomerase inhibitor mitoxantrone (86), the para-isomer of the peptidomimetic ligand PBP1 (87), which leads to the upregulation of BCL2 gene expression, and thiazole orange (88). Recently, Shu et al. developed a series of acridone derivatives and determined that one of these derivatives, B19, is capable of binding to and stabilizing the i-motif formed in the c-MYC promoter resulting in downregulation of gene expression and eventually to tumor cell death (89). The characterization of more i-motif structures at high resolution as well as its stabilization at neutral pH will contribute to increase the number of ligands that stabilize i-motifs.
Poly-C-binding proteins (PCBP) are proteins that interact with C-rich DNA sequences and play a fundamental role in regulating gene expression. The PCBP family consists of the hnRNP K (heterogeneous nuclear ribonucleoprotein K), αCP1-4, and αCP-KL proteins (90). However, in most cases, it is not clear whether these proteins bind to a given i-motif structure or to the C-rich strand resulting from i-motif unfolding. In an early study, Marsich et al. discovered a highly cytosine-specific protein in human HeLa cells (91). They did not determine the identity of the protein; however, they were able to show that the protein is specific for the human telomeric sequence, d(CCCTAA)n, containing at least four cytosine tracts (92). Later, Lacroix et al. found two proteins that bind telomeric C-rich sequences, hnRNP K and ASF/SF2 (93).
In a recent report, Niu et al. identified BmILF protein of Bombyx mori insect as an i-motif binding protein (94). By pull-down and EMSA assays, the authors demonstrated that BmILF binds to an i-motif structure formed at acidic pH by a C-rich sequence present in the BmPOUM2 gene promoter.
The BCL2 activating transcription factor heterogeneous nuclear ribonucleoprotein LL (hnRNP LL) is one of the very few i-motif binding proteins that have been studied in depth. It was identified through a pull-down assay aimed to find proteins involved in transcription and showing affinity for the i-motif structure formed by the BCL2 promoter oncogene (95). The hnRNP LL protein is a paralog of hnRNP L, which is a pre-mRNA splicing factor capable of binding and stabilizing BCL2 mRNA (95). hnRNP LL was found to bind specifically to i-motif structures as no binding was observed to either BCL2 promoter forming a duplex and to mutated single strand DNA unable to fold into an i-motif. The protein has four RNA recognition motifs (RRMs) and the BCL2 promoter sequence presents two consensus sequences that can be recognized by hnRNP LL RRMs. These consensus sequences correspond to those in the two lateral loops of the i-motif folding. Through EMSA experiments and luciferase reporter assays it was determined that hnRNP LL binds to the two lateral loops of the BCL2 i-motif (Figure 5C) by means of two of its four RRMs. Interestingly, CD and bromine footprinting experiments show that the binding of the protein unfolds the i-motif structures leading to single- stranded sequences. Therefore, the i-motif structure is the most kinetically favorable conformation for protein binding. The unfolding of the i-motif structures upon protein binding provides the more thermodynamically favored single-stranded conformation (Figure 5D). The protein remains bound to the single strand DNA and leads to the activation of BCL2 gene transcription. The discovery of the hnRNP LL as an i-motif binding protein able to activate gene transcription brings i-motif structures into focus as protein recognition sites capable of participating in regulation of gene expression.
Inhibition of telomerase activity
Mammalian telomeric DNA is composed of tandem repeats of the unit 5′-ATTGGG-3′/3′-TAACCC-5′. Importantly, the G-rich strand is a few hundreds of nucleotides longer than its complementary resulting in a G-rich ssDNA 3′-overhang that may form G4 structures. Studies have shown that the stabilization of certain human telomeric G4 topologies with ligands might lead to the inhibition of telomerase activity (96–98); however, the effect of targeting the complementary C-rich strand has not been investigated in depth. Thus, the study conducted by Qu et al. in 2012 was the first to investigate telomerase activity on i-motifs formed by C-rich human telomeric sequences stabilized by C-SWNT (84). As mentioned earlier, C-SWNTs were found to induce duplex dissociation and to stabilize human telomeric i-motifs at physiological pH and temperature under molecular crowding conditions. Likewise, C-SWNTs induce the formation of G4 structures on the complementary G-rich strand (83). In the presence of the C-SWNTs, telomerase activity is inhibited; thereby suggesting that the G4 stabilized on the leading strand can no longer be elongated by telomerase. Further investigations into the effect of C-SWNTs on cell growth and telomere structure and function suggest that the inhibition of cellular growth produced by C-SWNTs was a consequence of an impact on telomere structure rather than on telomerase activity. The persistence of i-motif and G4 structures, leads to telomere uncapping and release of telomere-binding proteins, resulting in telomere dysfunction. Telomere dysfunction induces DNA damage response and activates DNA repair pathways, which in turn trigger cell cycle arrest, senescence, and apoptosis (83).
I-motif in centromeric sequences
The centromere is the chromosomal region on which the kinetochore, a key multiprotein complex for chromosome segregation, assembles during cell division. In most organisms, centromeres contain large arrays of tandemly repeated DNA sequences (DNA satellite). Centromeric DNA sequences substantially vary among species and can also be different in chromosomes of the same organism. In the absence of a shared genetic motif defining the centromere, the presence of nucleosomes containing the centromere-specific histone H3 variant (CENP-A) is recognized as the essential epigenetic feature of centromeric chromatin. However, the possible role of the centromeric DNA sequences in directing the formation of such specialized chromatin has been suggested and is an attractive matter of debate (99,100). Interestingly, Kasinathan and Henikoff proposed in a very recent report that the predominant formation of non-B DNA structures in centromeric DNA sequences might be the basis for the constitution of centromeric chromatin (101).
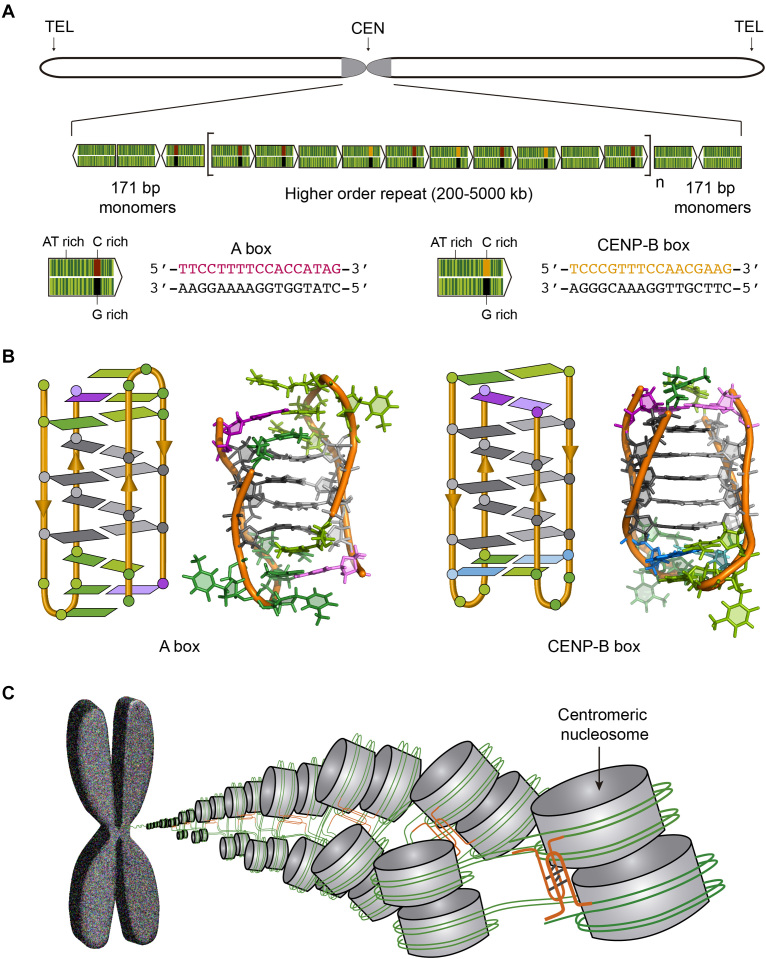
In humans, a 171 bp DNA called alphoid satellite is tandemly repeated along the centromeres (Figure 6A). The alphoid satellite is an AT rich sequence that frequently contains a 17-bp GC rich segment whose sequence has two variants known as CENP-B box and A box (Figure 6A). Interestingly, the CENP-B box, the sequence specifically bound by centromeric protein CENP-B, is absent in lower primates and in human Y chromosome. Gallego et al. found that whereas the G-rich sequence of CENP-B box folds into a structure stabilized by canonical base pairs, its complementary C-rich sequence forms a dimeric i-motif at acidic pH (Figure 6B) (66,102). Likewise, the C-rich sequences of the two variants of A box were reported to fold into dimeric i-motif structures (Figure 3A) (63). The dimeric i-motif structures of truncated versions of both CENP-B box and A box having 11 residues were determined at atomic resolution by NMR spectroscopy (63,66) (Figure 6B). The main difference between both structures is the relative disposition of the loops being at the same side of the structure (face-to-face topology) in CENP-B box i-motif and at opposite sides in A box (head-to-tail topology) (Figure 6B).
Figure 6.
(A) Human chromosome showing the centromere in grey and the structure of the human centromeric alpha satellite DNA. The 171 bp monomers (green arrows) are AT-rich sequences that can contain either A box (red and black) or CENP-B box (orange and black). The monomers are tandemly repeated and form a higher order repeat, which in turn appears repeated along the centromere. (B) Schematic and cartoon structures of the dimeric i-motifs formed by truncated A box (PDB ID: 2MRZ) and CENP-B box (PDB ID: 1C11). Cytosines are represented in grey, thymines in green, adenines in magenta and guanines in blue. (C) Proposed model for nucleosome organization at the centromere. The i-motifs formed by C-rich sequences of A and CENP-B boxes (orange) would maintain the structural organization of the centromere.
The centromeric region of chromosome 3 of Droshophila melanogaster contains the dodeca satellite DNA, which is composed by tandem repeats of 11/12 bp (CCCGTACTGGT/CCCGTACTCGGT). The capability of these sequences to fold into i-motif structures has been also evaluated. Sequences derived from both 11 bp and 12 bp repeat units were able to fold into dimeric i-motif structures in vitro at acidic pH (103).
These observations lead to propose a possible role for i-motif structures as a structural element providing long-range interactions between laterally associated centromeric nucleosomes (63) (Figure 6C). The presence of the above mentioned i-motif forming sequences at the entrance and exit of the nucleosome would facilitate their participation in dimeric i-motifs formation (Figure 6C). Thus, not only non-B DNA structures may have a role in directing centromere location (101) but also in providing particular architectural features to the centromere.
Transcriptional regulation of gene expression
A substantial amount of data suggests that i-motif structures may be involved in regulation of transcription. A recent report showing i-motif foci in vivo determined that number of i-motif foci were higher during G1/S phase of the cell cycle (Figure 4) in which the transcription activity is higher (6). This fact, together with the evidence described below suggest that these structures may have been selected as recognition motifs for proteins involved in the activation of the transcriptional machinery.
The BCL2 oncogene promotes cell survival and proliferation through an anti-apoptotic mechanism. It is overexpressed in some cancer cells, while its under-expression leads to neurodegenerative diseases (104). As discussed earlier, the C-rich sequence of the BCL2 promoter folds into a hairpin and an i-motif structures that are in dynamic equilibrium (105). Two back-to-back studies by Hurley's group showcase two compounds (Figure 5) and a protein capable of modulating BCL2 transcription in vitro and in vivo by specifically targeting either the i-motif or the hairpin form in dynamic equilibrium (85,95). Apparent stabilization of the i-motif structure via IMC-48 compound leads to significant upregulation of BCL2. On the contrary, apparent stabilization of the flexible hairpin species via IMC-76 compound leads to transcriptional repression in lymphoma cell lines (Figure 5B) (85).
In a consecutive study, the Hurley's group determined the effect of the hnRNP LL protein on BCL2 transcription. Two of the four RNA recognition motifs (RRMs) in hnRNP LL are required for stable binding to a single-stranded RNA or DNA. The lateral loops of BCL2 i-motif possess sequences capable of binding to the RRMs in hnRNP LL (Figure 5C). These studies demonstrate that hnRNP LL activates transcription by recognizing and unfolding the BCL2 i-motif (Figure 5). The presence of IMC-48 shifts the equilibrium in favor of the i-motif and therefore increases the i-motif population available for binding to hnRNP LL. The research group demonstrated that IMC-48 binds to the central loop of the BCL2 i-motif, followed by recognition and binding of hnRNP LL to the two lateral loops. Consequently, hnRNP LL binding leads to i-motif unfolding which, in turn, leads to transcriptional activation. In conclusion, these studies demonstrate the effect of two small molecules (IMC-76 and IMC-48) and a transcriptional factor (hnRNP LL) on the relative population of i-motif and its impact on gene expression.
Recently, a study conducted by Muniyappa and co-authors demonstrated the potential of the C-rich sequences of the PI and PII promoters of human acetyl-CoA carboxylase 1 (ACC1) gene to fold into intramolecular i-motif structures at neutral pH under molecular crowding conditions (106). The authors state that, experiments in HeLa cells including i-motif-forming sequences in a promoter region regulating luciferase transcription lead to a decrease in protein expression and suggest a significant role of i-motif-forming sequences in the regulation of ACC1 gene expression. However, the authors also claim that G4 and i-motif structures in plasmids containing wild type sequences exhibit a cooperative effect leading to a decrease in luciferase expression. Therefore, it is unclear whether i-motif or G4 structures are responsible for transcriptional repression. This ambiguity is addressed in a recent study on the PDGFR-β promoter region (107) where point mutations of the G4 structures led to upregulation of gene expression. This upregulation is due to the effect introduced by the G4 mutations on the i-motif-forming strand.
In a similar study conducted by Niu et al., it was reported that the binding of BmILF protein to an i-motif activates the transcription of BmPOUM2, a Bombyx mori gene involved in developmental processes during metamorphosis. Luciferase expression assays demonstrated that vectors lacking the i-motif-forming sequence in the promoter of the gene resulted in significant decreasing of luciferase activity. Additionally, by using oligonucleotides complementary to the i-motif and G4 forming sequences present in the promoter, it was determined that only the hybridization of i-motif forming sequence with complementary oligonucleotides produced significant decrease in promoter activity. The authors propose that during transcription, the dsDNA at the promoter is melted and the i-motif is formed and bound by BmILF, which may recruit other factors to activate BmPOUM2 expression.
Burrows et al. investigated the effect that modified G4 and i-motif-forming sequences from VEGF gene promoter have on luciferase and renilla expression levels (108). In addition, this study reveals that sequences containing nucleobases produced during oxidative stress such as 8-oxo-7,8-dihydroguanine lead to the up- or down-regulation of transcription depending on whether the modified nucleobases are located on the coding or on the template strand of the promoter, respectively.
Kendrick et al. used IMC-76 in combination with ellipticine (GQC-05) in order to simultaneously target BCL2 and MYC oncogene promoters in diffuse large B-cell lymphoma (DLBCL) (109). GQC-05 stabilizes the G4 formed by G-rich MYC promoter sequence, supposedly acting as an ‘off-switch’ to impede gene expression (110). The simultaneous stabilization of the hairpin over the i-motif (via IMC-76) in the BCL2 promoter and the G4 (via GQC-05) in the MYC promoter decreased the mRNA levels of both genes and enhanced the sensitivity of DLBCL cells towards cyclophosphamide (a chemotherapeutic drug). This study was the first to simultaneously target two different DNA secondary structures, i.e. i-motifs and G4s, leading to dual transcriptional repression and providing an effective approach to treat aggressive malignancies (109).
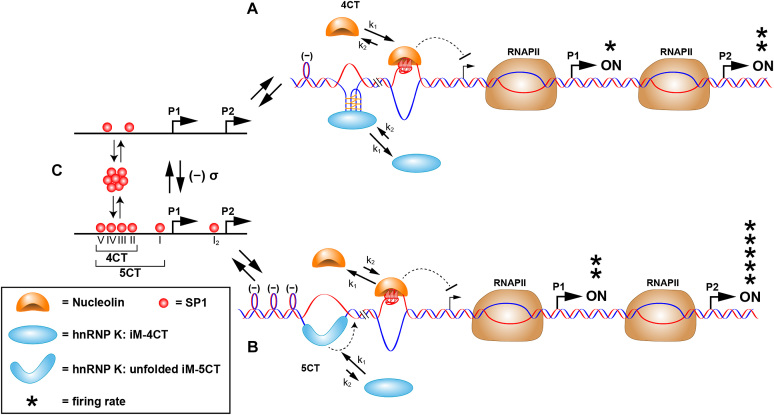
Following the above mentioned reports, several studies demonstrate how G4s and i-motifs act as on/off molecular switches for the regulation of tyrosine hydroxylase (Th) (111), MYC (112), platelet-derived growth factor receptor β (PDGFR-β) (107), and KRAS promoters (113). For instance, in the case of the MYC promoter it is the extent of transcriptionally induced negative superhelicity that determines whether MYC expression can be turned on. The authors propose that binding of SP1 to its promoter binding sites induces negative superhelicity, which in turn drives duplex melting at 4CT or 5CT GC-rich elements. The mechanism by which the protein contributes to increase negative superhelicity is not known. At low levels of SP1 there is only sufficient unwinding of the duplex to expose the 4CT element which contains six C-tracts that provides the two hnRNP K binding sites in the lateral loops of the i-motif (Figure 7C to A). However, this provides insufficient binding strength for the hnRNP K–i-motif complex to compete with nucleolin, which binds to the G4 structure to repress gene transcription (i.e. acting as an OFF switch). However, with increased concentrations of SP1 available to bind to the duplex DNA, this results in increased transcriptionally induced negative superhelicity and the two additional C-tracts become available (Figure 7C to B). This provides an additional CT binding site for hnRNP K uniquely found in the 5CT elements. Ultimately, two steps are needed to activate transcription, the first being recognition by hnRNP K of the two CT elements displayed in the lateral loops of the i-motif. In the second step, unfolding of the i-motif occurs, providing access to the third CT element, which is uniquely found in the 5CT elements. Now with three KH domains from the hnRNP K bound to the 3CT elements, this thermodynamically stable complex is able to compete with nucleolin–G4 complex and MYC transcription is turned on (112).
Figure 7.
Proposed scheme for the molecular mechanosensor mechanism for differential control of MYC expression through the NHE III1. (A) Low SP1 occupancy of duplex promoter binding sites (as shown in (C), upper) results in low negative supercoiling and basal levels of transcription from the P1 and P2 promoters. In this scenario, hnRNP K can only access the central and lateral loops in the i-motif that bind to two of the KH domains; as such only the 4CT element is accessed, and nucleolin binding predominates over hnRNP K binding to the equilibrating G-quadruplex and i-motif, which are mutually exclusive. (B) At higher occupancy levels of SP1 to the duplex promoter binding sites (as shown in (C), lower), which results in enhanced negative supercoiling, the 5CT element is now fully melted, and hnRNP K forms a thermodynamically stable complex binding through the addition of the CT element. The binding affinity of hnRNP K to the unfolded i-motif and the additional CT element involving three KH domains now exceeds that of nucleolin to the G-quadruplex, and MYC expression from P1 and P2 is at peak levels. Reprinted with permission from Sutherland, C., Cui, Y.X., Mao, H.B. and Hurley, L.H. (2016) A Mechanosensor Mechanism Controls the G-Quadruplex/i-Motif Molecular Switch in the MYC Promoter NHE III1. J Am Chem Soc, 138, 14138–14151. Copyright (2016) American Chemical Society.
Regulation of DNA biosynthesis
Very recently, Sugimoto and co-authors investigated the effect of several non-canonical DNA structures on DNA replication by the Klenow fragment (KF) of DNA polymerase (114). Different i-motif-forming sequences inserted in the template strand of the replication reaction were found to stall DNA polymerase and thus impede DNA replication or repair. The stalling effect produced by i-motifs on replication was higher than that of other structures with similar thermodynamic stability such us hairpins or mixed-type G4s. This is justified by the unique intercalating topology of base pairs in i-motif structures, which complicate its unwinding and subsequent replication by DNA polymerase. As mentioned earlier, the base pairs in an i-motif structure are intercalated between two parallel duplexes and lack the base stacking interactions like in antiparallel duplexes (Figure 1B). Therefore, unzipping is particularly disfavored since consecutive base pairs belong to different duplexes. In addition, the arrangement of the loops in the i-motif structure might cause steric hindrance for polymerase binding. These two factors may contribute to the fact that i-motif unwinding by KF requires higher activation energy than that of other non-canonical structures. These data suggest that i-motifs could modulate DNA replication in vivo and pose a greater impact compared to other secondary structures.
Mutual exclusivity of i-motifs and G-quadruplexes
Most of the earlier studies focused on investigating the biological effects of G4s and i-motifs separately in single-stranded DNA fragments. However, in the genome and with the exception of chromosome ends, the formation of G4 and i-motif structures is compromised by hybridization between complementary strands to form a duplex DNA. Several factors have been found to affect this competence. The first G4/i-motif-duplex interconversion studies were carried out by Phan and Mergny. They inferred that 1:1 mixtures of the C-rich d((C3TA2)3C3T) and G-rich d(AG3(T2AG3)3) telomeric sequences at acidic pH (<5) and in the presence of KCl produced predominantly i-motif and G4, respectively (115). However, at pH 7.0 and 100 mM NaCl the duplex formed by the hybridization of both sequences was the predominant species. Several groups have investigated the factors influencing interconversion kinetics and determined that the sequence, the experimental conditions (i.e. ionic strength, temperature, and pH) (116,117), and the incorporation of chemical modifications all play a significant role in favoring the tetraplex structures over duplexes and vice versa.
Mao et al. reported single-molecule concentration jump experiments showing that in the absence of any force, human telomeric DNA i-motif structures exhibit a half-life of 2.60 s at neutral pH. This half-life is sufficient for the interaction of i-motifs with proteins or small molecules leading to the modulation of several biological processes (118). i-Motifs half-life in the presence of their complementary strands can be dramatically extended by introducing 2′F-araC modifications (Figure 2B) in telomeric sequences (25). When 2′F-araC modified i-motif and/or 2′F-araG modified G4 structures were pre-folded, their unfolding poses a significant barrier to duplex formation. In this scenario i-motifs and G4s located at the termini of a duplex co-exist at neutral pH conditions for >30 days (25).
Using magneto-optical tweezers, Mao's group recently quantitatively evaluated for the first time how chemical (ions and pH) and mechanical (superhelicity and molecular crowding) factors influence the population dynamics of G4s and i-motifs formed by the insulin-linked polymorphic region (ILPR) (119). By applying 32 different sets of chemical and mechanical experimental conditions, it was found that chemical factors, especially concentration of potassium ions and acidic pH, have the most substantial effect on the formation and stabilization of G4s and i-motifs, respectively. Among the mechanical factors, superhelicity was found to have more significant impact than molecular crowding. Negative superhelicity reduces the stability of DNA duplexes consequently favoring the formation of G4s in the G-rich strand and i-motifs in the complementary C-rich strand.
In the last few years, several researchers have begun to investigate on the co-existence of G4 and i-motif structures. The question as whether i-motif and G4 structures can appear simultaneously in complementary single strands or are mutually exclusive was a matter of debate due to contradictory reports. One of the earlier reports by Sun and Hurley studied the influence of negative superhelicity on the formation of G4 and i-motif structures in the promoter regions of cMYC and their impact on gene expression (20). By means of enzymatic and chemical footprinting assay, they showed that both the G4 and i-motif structures are present at the same time in opposite strands and with slight displacement relative to each other. Three out of the four required GC tracts were shared between the two tetraplexes.
However, in a later study in collaboration with Mao's group they showed that the G-quadruplex and the i-motif in the MYC promoter are mutually exclusive in the NHE III1 (112). This finding of mutual exclusivity has also been demonstrated in ILPR region by the Mao group (120). Using chemical footprinting and single molecule techniques, they were able to show that the G-rich sequence folds into a G4 structure at pH 7.4 and 100 mM K+ while the C-rich strand folds into an i-motif structure at pH 5.5 and 100 mM Li+. However, under conditions that favor the formation of both structures (pH 5.5 and 100 mM K+), either the G4 or the i-motif structure forms, but not both, which the authors attribute to mutual steric hindrance. In a more recent work, it was observed that that both tetraplexes can form simultaneously when the G4 and i-motif forming sequences are offset with respect to each other in complementary strands (121). This study further suggests that mutual exclusivity is governed by steric hindrance between the structures arising from the complementary strands. The mutually exclusivity phenomenon also suggests that G4 and i-motif structures may play opposing biological roles at the same location of the genome. As described above, several studies strongly suggest that G4 formation suppresses transcription while i-motif formation lead to the activation of gene expression (59). The observation that the highest number of i-motif (6) and G4 (7) foci in the nuclei of cells is observed at different stages of the cell cycle also supports that both structures might not coexist in the cellular context.
SOME APPLICATIONS OF i-MOTIF STRUCTURES
The particular features of i-motif structures have inspired the design of nanotechnological systems for analytical and biomedical purposes (8–10). Many of these devices are based on the structural transitions that i-motif-forming oligonucleotides experience as a result of pH variations. Taking advantage of this structural response to pH, several nanodevices based on i-motif folding/unfolding were devised to monitor pH changes inside cellular context. Measuring the intracellular pH is a fundamental goal in biosciences given the crucial influence of pH in cellular processes and the implications of dysregulated intracellular pH in certain diseases such as cancer (122). The development of the so-called ‘I-switch’ by the Krishnan group (123) was the first example of an i-motif-based nanomachine able to sense and report pH changes along endosomal maturation both inside living cells in culture (123) and in multicellular organism (124). Interestingly, by adding a tag to the device it is possible to target any biotinylated protein and measure the pH associated to its function (123,125).
The use of gold nanoparticles (AuNPs) functionalized in their surface with C-rich sequences able to form i-motifs has also been successfully applied for monitoring changes of pH inside living cells. The particular spectroscopic properties of AuNPs and their facility to be functionalized and internalized inside cells make them very attractive alternatives for the development of efficient pH sensors of the cellular environment. Thus, several designs based on AuNPs modified with i-motif-forming sequences have been used to monitor pH changes inside endocytic vesicles (126–130).
These examples provide evidence of the compatibility of i-motif structures with the intracellular environment and highlight their potential as building blocks for nanobiotechnological applications. Indeed, some other i-motif-based nanodevices such as biosensors or drug release platforms have already been developed and serve as proof of principle for their application in vivo (8,9,131).
CONCLUSIONS/PERSPECTIVE
Despite the tremendous advances in the field of i-motif structural biology in the recent years, many aspects still require further investigation. The present data strongly suggest that i-motifs form transiently in the cell. However, more in vivo studies are definitively needed to confirm i-motif formation in different phases of the cell cycle. In addition, further research on i-motif recognition by proteins and small ligands in vitro and in vivo are essential to elucidate the role of i-motifs in different biological processes. Due to the dynamic nature of i-motifs, these studies are hampered, particularly in vivo, by the difficulty of discriminating between recognition of C-rich sequences or recognition of the actual i-motif structure. Synthetic i-motif constructions stabilized by chemical modifications (e.g. 2′F-ANA) can facilitate these investigations by ‘freezing’ these intrinsically dynamic sequences in the potentially active i-motif conformation. Chemically modified i-motifs, stable over a wide range of conditions compared to their unmodified counterparts, can be also used for screening experiments to identify new proteins and small ligands that specifically recognize this structure.
There is still much to know about i-motif structures. Compared to G4s, there are only a few i-motif structures determined by NMR or crystallographic methods. At present, it is not possible to predict the stability of an i-motif based on its sequence. Therefore, more structural information is needed to fully understand the effect of capping interactions and the loops connecting the C-tracts on i-motif stability. In spite of the recent findings of i-motif ligands, the number of known i-motif-specific binders is very limited in comparison with G4 ligands. No three-dimensional structure of an i-motif/ligand complex has been determined yet. This will be a major achievement for the development of potential drugs based on i-motif recognition.
Since the first years of the century, several aspects of G4s have gained considerable research interest mainly due to their thermodynamic stability at physiological conditions. However, the i-motif structure has been the ugly duckling in the family of non-canonical DNA structures for many years. The recent results shed a new light on the i-motif field, which will blossom in the coming years.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Professor Daniel Christ and Professor Laurence Hurley for providing the original copy of Figures 4, 5 and 7 to be used in this review.
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERCC); MINECO grant [BFU2017-89707-P]; Juan de la Cierva postdoctoral Fellowship [FJCI-2016-28474 to M.G.]. Funding for open access charge: NSERCC Discovery grant ( to M.J.D.); MINECO grant [BFU2017-89707-P to C.G.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Watson J.D., Crick F.H.C.. Molecular structure of nucleic acids - a structure for deoxyribose nucleic acid. Nature. 1953; 171:737–738. [DOI] [PubMed] [Google Scholar]
- 2. Du Y., Zhou X.. Targeting non-B-form DNA in living cells. Chem. Rec. 2013; 13:371–384. [DOI] [PubMed] [Google Scholar]
- 3. Choi J., Majima T.. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011; 40:5893–5909. [DOI] [PubMed] [Google Scholar]
- 4. Sen D., Gilbert W.. Formation of parallel 4-Stranded complexes by Guanine-Rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 5. Gehring K., Leroy J.L., Gueron M.. A tetrameric DNA-structure with protonated cytosine.cytosine Base-Pairs. Nature. 1993; 363:561–565. [DOI] [PubMed] [Google Scholar]
- 6. Zeraati M., Langley D.B., Schofield P., Moye A.L., Rouet R., Hughes W.E., Bryan T.M., Dinger M.E., Christ D.. I-motif DNA structures are formed in the nuclei of human cells. Nat Chem. 2018; 10:631–637. [DOI] [PubMed] [Google Scholar]
- 7. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alba J.J., Sadurni A., Gargallo R.. Nucleic acid i-Motif structures in analytical chemistry. Crit. Rev. Anal. Chem. 2016; 46:443–454. [DOI] [PubMed] [Google Scholar]
- 9. Dembska A. The analytical and biomedical potential of cytosine-rich oligonucleotides: A review. Anal. Chim. Acta. 2016; 930:1–12. [DOI] [PubMed] [Google Scholar]
- 10. Yatsunyk L.A., Mendoza O., Mergny J.L.. “Nano-oddities”: Unusual nucleic acid assemblies for DNA-Based nanostructures and nanodevices. Acc. Chem. Res. 2014; 47:1836–1844. [DOI] [PubMed] [Google Scholar]
- 11. Kang C.H., Berger I., Lockshin C., Ratliff R., Moyzis R., Rich A.. Crystal-Structure of intercalated 4-Stranded D(C3t) at 1.4 angstrom resolution. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:11636–11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berger I., Egli M., Rich A.. Inter-strand C-H...O hydrogen bonds stabilizing four-stranded intercalated molecules: Stereoelectronic effects of 04′ in cytosine-rich DNA. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:12116–12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Phan A.T., Leroy J.L.. Intramolecular i-motif structures of telomeric DNA. J. Biomol. Struct. Dyn. 2000; 245–251. [DOI] [PubMed] [Google Scholar]
- 14. Gueron M., Leroy J.L.. The i-motif in nucleic acids. Curr. Opin. Struct. Biol. 2000; 10:326–331. [DOI] [PubMed] [Google Scholar]
- 15. Malliavin T.E., Gau J., Snoussi K., Leroy J.L.. Stability of the I-motif structure is related to the interactions between phosphodiester backbones. Biophys. J. 2003; 84:3838–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wright E.P., Huppert J.L., Waller Z.A.E.. Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 2017; 45:2951–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fleming A.M., Ding Y., Rogers R.A., Zhu J., Zhu J., Burton A.D., Carlisle C.B., Burrows C.J.. 4n-1 is a “Sweet Spot” in DNA i-Motif folding of 2′-Deoxycytidine homopolymers. J. Am. Chem. Soc. 2017; 139:4682–4689. [DOI] [PubMed] [Google Scholar]
- 18. Zhou J., Wei C., Jia G., Wang X., Feng Z., Li C.. Formation of i-motif structure at neutral and slightly alkaline pH. Mol. Biosyst. 2010; 6:580–586. [DOI] [PubMed] [Google Scholar]
- 19. Rajendran A., Nakano S., Sugimoto N.. Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. 2010; 46:1299–1301. [DOI] [PubMed] [Google Scholar]
- 20. Sun D., Hurley L.H.. The importance of negative superhelicity in inducing the formation of G-Quadruplex and i-Motif structures in the c-Myc Promoter: Implications for drug targeting and control of gene expression. J. Med. Chem. 2009; 52:2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Day H.A., Huguin C., Waller Z.A.E.. Silver cations fold i-motif at neutral pH. Chem. Commun. 2013; 49:7696–7698. [DOI] [PubMed] [Google Scholar]
- 22. Abdelhamid M.A., Fabian L., MacDonald C.J., Cheesman M.R., Gates A.J., Waller Z.A.. Redox-dependent control of i-Motif DNA structure using copper cations. Nucleic Acids Res. 2018; 46:5886–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Satpathi S., Das K., Hazra P.. Silica nano-channel induced i-motif formation and stabilization at neutral and alkaline pH. Chem. Commun. 2018; 54:7054–7057. [DOI] [PubMed] [Google Scholar]
- 24. Abou Assi H., Harkness R.W.V, Martin-Pintado N., Wilds C.J., Campos-Olivas R., Mittermaier A.K., González C., Damha M.J.. Stabilization of i-motif structures by 2′-beta-fluorination of DNA. Nucleic Acids Res. 2016; 44:4998–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abou Assi H., El-Khoury R., González C., Damha M.J.. 2′-Fluoroarabinonucleic acid modification traps G-quadruplex and i-motif structures in human telomeric DNA. Nucleic Acids Res. 2017; 45:11535–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang B., Rodgers M.T.. Base-pairing energies of proton-bound heterodimers of cytosine and modified cytosines: implications for the stability of DNA i-motif conformations. J. Am. Chem. Soc. 2014; 136:282–290. [DOI] [PubMed] [Google Scholar]
- 27. Lieblein A.L., Kramer M., Dreuw A., Furtig B., Schwalbe H.. The nature of hydrogen bonds in cytidine…H+…cytidine DNA base pairs. Angew. Chem. Int. Ed. Engl. 2012; 51:4067–4070. [DOI] [PubMed] [Google Scholar]
- 28. Leroy J.L., Gehring K., Kettani A., Gueron M.. Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization, and proton exchange properties. Biochemistry. 1993; 32:6019–6031. [DOI] [PubMed] [Google Scholar]
- 29. Phan A.T., Gueron M., Leroy J.L.. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000; 299:123–144. [DOI] [PubMed] [Google Scholar]
- 30. Bhaysar-Jog Y.P., Van Dornshuld E., Brooks T.A., Tschumper G.S., Wadkins R.M.. Epigenetic modification, dehydration, and molecular crowding effects on the thermodynamics of i-Motif structure formation from C-Rich DNA. Biochemistry. 2014; 53:1586–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lannes L., Halder S., Krishnan Y., Schwalbe H.. Tuning the pH Response of i-Motif DNA Oligonucleotides. ChemBioChem. 2015; 16:1647–1656. [DOI] [PubMed] [Google Scholar]
- 32. Xu B.C., Devi G., Shao F.W.. Regulation of telomeric i-motif stability by 5-methylcytosine and 5-hydroxymethylcytosine modification. Org. Biomol. Chem. 2015; 13:5646–5651. [DOI] [PubMed] [Google Scholar]
- 33. Wright E.P., Lamparska K., Smith S.S., Waller Z.A.E.. Substitution of cytosine with guanylurea decreases the stability of i-Motif DNA. Biochemistry. 2017; 56:4879–4883. [DOI] [PubMed] [Google Scholar]
- 34. Mir B., Soles X., González C., Escaja N.. The effect of the neutral cytidine protonated analogue pseudoisocytidine on the stability of i-motif structures. Sci. Rep. 2017; 7:2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsvetkov V.B., Zatsepin T.S., Belyaev E.S., Kostyukevich Y.I., Shpakovski G.V., Podgorsky V.V., Pozmogova G.E., Varizhuk A.M., Aralov A.V.. i-Clamp phenoxazine for the fine tuning of DNA i-motif stability. Nucleic Acids Res. 2018; 46:2751–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dvorakova Z., Renciuk D., Kejnovska I., Skolakova P., Bednarova K., Sagi J., Vorlickova M.. i-Motif of cytosine-rich human telomere DNA fragments containing natural base lesions. Nucleic Acids Res. 2018; 46:1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Collin D., Gehring K.. Stability of chimeric DNA/RNA cytosine tetrads: Implications for i-motif formation by RNA. J. Am. Chem. Soc. 1998; 120:4069–4072. [Google Scholar]
- 38. Lacroix L., Mergny J.L., Leroy J.L., Helene C.. Inability of RNA to form the i-motif: Implications for triplex formation. Biochemistry. 1996; 35:8715–8722. [DOI] [PubMed] [Google Scholar]
- 39. Fenna C.P., Wilkinson V.J., Arnold J.R., Cosstick R., Fisher J.. The effect of 2′-fluorine substitutions on DNA i-motif conformation and stability. Chem. Commun. 2008; 3567–3569. [DOI] [PubMed] [Google Scholar]
- 40. Kumar N., Nielsen J.T., Maiti S., Petersen M.. i-Motif formation with locked nucleic acid (LNA). Angew. Chem. Int. Ed. Engl. 2007; 46:9220–9222. [DOI] [PubMed] [Google Scholar]
- 41. Pasternak A., Wengel J.. Modulation of i-motif thermodynamic stability by the introduction of UNA (unlocked nucleic acid) monomers. Bioorg. Med. Chem. Lett. 2011; 21:752–755. [DOI] [PubMed] [Google Scholar]
- 42. Robidoux S., Damha M.J.. D-2-deoxyribose and D-arabinose, but not D-ribose, stabilize the cytosine tetrad (i-DNA) structure. J. Biomol. Struct. Dyn. 1997; 15:529–535. [DOI] [PubMed] [Google Scholar]
- 43. Perez-Rentero S., Gargallo R., González C., Eritja R.. Modulation of the stability of i-motif structures using an acyclic threoninol cytidine derivative. RSC Adv. 2015; 5:63278–63281. [Google Scholar]
- 44. Abou Assi H., Lin Y.C., Serrano I., González C., Damha M.J.. Probing synergistic effects of DNA methylation and 2′-beta-Fluorination on i-Motif stability. Chem-Eur J. 2018; 24:471–477. [DOI] [PubMed] [Google Scholar]
- 45. Snoussi K., Nonin-Lecomte S., Leroy J.L.. The RNA i-motif. J. Mol. Biol. 2001; 309:139–153. [DOI] [PubMed] [Google Scholar]
- 46. Aviñó A., Dellafiore M., Gargallo R., González C., Iribarren A.M., Montserrat J., Eritja R.. Stabilization of telomeric I-Motif structures by (2′S)-2′-Deoxy-2′-C-Methylcytidine residues. ChemBioChem. 2017; 18:1123–1128. [DOI] [PubMed] [Google Scholar]
- 47. Mergny J.L., Lacroix L.. Kinetics and thermodynamics of i-DNA formation: phosphodiester versus modified oligodeoxynucleotides. Nucleic Acids Res. 1998; 26:4797–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kanaori K., Sakamoto S., Yoshida H., Guga P., Stec W., Tajima K., Makino K.. Effect of phosphorothioate chirality on i-motif structure and stability. Biochemistry. 2004; 43:5672–5679. [DOI] [PubMed] [Google Scholar]
- 49. Krishnan-Ghosh Y., Stephens E., Balasubramanian S.. PNA forms an i-motif. Chem. Commun. 2005; 5278–5280. [DOI] [PubMed] [Google Scholar]
- 50. Modi S., Wani A.H., Krishnan Y.. The PNA-DNA hybrid I-motif: implications for sugar-sugar contacts in i-motif tetramerization. Nucleic Acids Res. 2006; 34:4354–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robidoux S., Klinck R., Gehring K., Damha M.J.. Association of branched oligonucleotides into the i-motif. J. Biomol. Struct. Dyn. 1997; 15:517–527. [DOI] [PubMed] [Google Scholar]
- 52. Fojtik P., Vorlickova M.. The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH. Nucleic Acids Res. 2001; 29:4684–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mir B., Serrano I., Buitrago D., Orozco M., Escaja N., González C.. Prevalent sequences in the human genome can form Mini i-Motif structures at physiological pH. J. Am. Chem. Soc. 2017; 139:13985–13988. [DOI] [PubMed] [Google Scholar]
- 54. Brazier J.A., Shah A., Brown G.D.. I-motif formation in gene promoters: unusually stable formation in sequences complementary to known G-quadruplexes. Chem. Commun. 2012; 48:10739–10741. [DOI] [PubMed] [Google Scholar]
- 55. Benabou S., Garavís M., Lyonnais S., Eritja R., González C., Gargallo R.. Understanding the effect of the nature of the nucleobase in the loops on the stability of the i-motif structure. Phys. Chem. Chem. Phys. 2016; 18:7997–8004. [DOI] [PubMed] [Google Scholar]
- 56. Fujii T., Sugimoto N.. Loop nucleotides impact the stability of intrastrand i-motif structures at neutral pH. Phys. Chem. Chem. Phys. 2015; 17:16719–16722. [DOI] [PubMed] [Google Scholar]
- 57. Fleming A.M., Stewart K.M., Eyring G.M., Ball T.E., Burrows C.J.. Unraveling the 4n - 1 rule for DNA i-motif stability: base pairs vs. loop lengths. Org. Biomol. Chem. 2018; 16:4537–4546. [DOI] [PubMed] [Google Scholar]
- 58. Dai J., Hatzakis E., Hurley L.H., Yang D.. I-motif structures formed in the human c-MYC promoter are highly dynamic–insights into sequence redundancy and I-motif stability. PLoS One. 2010; 5:e11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brooks T.A., Kendrick S., Hurley L.. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010; 277:3459–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mergny J.L., Lacroix L., Han X.G., Leroy J.L., Helene C.. Intramolecular folding of pyrimidine oligodeoxynucleotides into an I-DNA Motif. J. Am. Chem. Soc. 1995; 117:8887–8898. [Google Scholar]
- 61. Lieblein A.L., Furtig B., Schwalbe H.. Optimizing the kinetics and thermodynamics of DNA i-motif folding. ChemBioChem. 2013; 14:1226–1230. [DOI] [PubMed] [Google Scholar]
- 62. Canalia M., Leroy J.L.. Structure, internal motions and association-dissociation kinetics of the i-motif dimer of d(5mCCTCACTCC). Nucleic Acids Res. 2005; 33:5471–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garavís M., Escaja N., Gabelica V., Villasante A., González C.. Centromeric Alpha-Satellite DNA adopts dimeric i-Motif structures capped by AT hoogsteen base Pairs. Chem-Eur J. 2015; 21:9816–9824. [DOI] [PubMed] [Google Scholar]
- 64. Canalia M., Leroy J.L.. [5mCCTCTCTCC]4: an i-motif tetramer with intercalated T*T pairs. J. Am. Chem. Soc. 2009; 131:12870–12871. [DOI] [PubMed] [Google Scholar]
- 65. Kang C.H., Berger I., Lockshin C., Ratliff R., Moyzis R., Rich A.. Stable loop in the crystal-sructure of the intercalated 4-stranded cytosine-rich metazoan telomere. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:3874–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gallego J., Chou S.H., Reid B.R.. Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimer. J. Mol. Biol. 1997; 273:840–856. [DOI] [PubMed] [Google Scholar]
- 67. Esmaili N., Leroy J.L.. i-motif solution structure and dynamics of the d(AACCCC) and d(CCCCAA) tetrahymena telomeric repeats. Nucleic Acids Res. 2005; 33:213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen Y.W., Jhan C.R., Neidle S., Hou M.H.. Structural basis for the identification of an i-motif tetraplex core with a parallel-duplex junction as a structural motif in CCG triplet repeats. Angew. Chem. Int. Ed. Engl. 2014; 53:10682–10686. [DOI] [PubMed] [Google Scholar]
- 69. Viladoms J., Escaja N., Pedroso E., González C.. Self-association of cyclic oligonucleotides through G:T:G:T minor groove tetrads. Bioorg.Med. Chem. 2010; 18:4067–4073. [DOI] [PubMed] [Google Scholar]
- 70. Escaja N., Gomez-Pinto I., Pedroso E., González C.. Four-stranded DNA structures can be stabilized by two different types of minor groove G:C:G:C tetrads. J. Am. Chem. Soc. 2007; 129:2004–2014. [DOI] [PubMed] [Google Scholar]
- 71. Escaja N., Gelpi J.L., Orozco M., Rico M., Pedroso E., González C.. Four-stranded DNA structure stabilized by a novel G:C:A:T tetrad. J. Am. Chem. Soc. 2003; 125:5654–5662. [DOI] [PubMed] [Google Scholar]
- 72. Escaja N., Viladoms J., Garavís M., Villasante A., Pedroso E., González C.. A minimal i-motif stabilized by minor groove G:T:G:T tetrads. Nucleic Acids Res. 2012; 40:11737–11747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Benabou S., Ferreira R., Aviñó A., González C., Lyonnais S., Sola M., Eritja R., Jaumot J., Gargallo R.. Solution equilibria of cytosine- and guanine-rich sequences near the promoter region of the n-myc gene that contain stable hairpins within lateral loops. Biochim. Biophys. Acta. 2014; 1840:41–52. [DOI] [PubMed] [Google Scholar]
- 74. Day H.A., Pavlou P., Waller Z.A.. i-Motif DNA: structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014; 22:4407–4418. [DOI] [PubMed] [Google Scholar]
- 75. Cui J., Waltman P., Le V.H., Lewis E.A.. The effect of molecular crowding on the stability of human c-MYC promoter sequence I-motif at neutral pH. Molecules. 2013; 18:12751–12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Miyoshi D., Matsumura S., Nakano S., Sugimoto N.. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004; 126:165–169. [DOI] [PubMed] [Google Scholar]
- 77. Dzatko S., Krafcikova M., Hansel-Hertsch R., Fessl T., Fiala R., Loja T., Krafcik D., Mergny J.L., Foldynova-Trantirkova S., Trantirek L.. Evaluation of the stability of DNA i-Motifs in the nuclei of living mammalian cells. Angew. Chem. Int. Ed. Engl. 2018; 57:2165–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Biffi G., Di Antonio M., Tannahill D., Balasubramanian S.. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014; 6:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Martino L., Pagano B., Fotticchia I., Neidle S., Giancola C.. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B. 2009; 113:14779–14786. [DOI] [PubMed] [Google Scholar]
- 80. Alberti P., Ren J., Teulade-Fichou M.P., Guittat L., Riou J.F., Chaires J., Helene C., Vigneron J.P., Lehn J.M., Mergny J.L.. Interaction of an acridine dimer with DNA quadruplex structures. J. Biomol. Struct. Dyn. 2001; 19:505–513. [DOI] [PubMed] [Google Scholar]
- 81. Xu H., Zhang H., Qu X.. Interactions of the human telomeric DNA with terbium-amino acid complexes. J. Inorg. Biochem. 2006; 100:1646–1652. [DOI] [PubMed] [Google Scholar]
- 82. Shi S., Geng X., Zhao J., Yao T., Wang C., Yang D., Zheng L., Ji L.. Interaction of [Ru(bpy)2(dppz)]2+ with human telomeric DNA: preferential binding to G-quadruplexes over i-motif. Biochimie. 2010; 92:370–377. [DOI] [PubMed] [Google Scholar]
- 83. Li X., Peng Y., Ren J., Qu X.. Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:19658–19663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen Y., Qu K., Zhao C., Wu L., Ren J., Wang J., Qu X.. Insights into the biomedical effects of carboxylated single-wall carbon nanotubes on telomerase and telomeres. Nat. Commun. 2012; 3:1074. [DOI] [PubMed] [Google Scholar]
- 85. Kendrick S., Kang H.J., Alam M.P., Madathil M.M., Agrawal P., Gokhale V., Yang D.Z., Hecht S.M., Hurley L.H.. The dynamic character of the BCL2 promoter i-Motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J. Am. Chem. Soc. 2014; 136:4161–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wright E.P., Day H.A., Ibrahim A.M., Kumar J., Boswell L.J., Huguin C., Stevenson C.E., Pors K., Waller Z.A.. Mitoxantrone and analogues bind and stabilize i-Motif forming DNA sequences. Sci. Rep. 2016; 6:39456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Debnath M., Ghosh S., Chauhan A., Paul R., Bhattacharyya K., Dash J.. Preferential targeting of i-motifs and G-quadruplexes by small molecules. Chem. Sci. 2017; 8:7448–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sheng Q., Neaverson J.C., Mahmoud T., Stevenson C.E.M., Matthews S.E., Waller Z.A.E.. Identification of new DNA i-motif binding ligands through a fluorescent intercalator displacement assay. Org. Biomol. Chem. 2017; 15:5669–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shu B., Cao J., Kuang G., Qiu J., Zhang M., Zhang Y., Wang M., Li X., Kang S., Ou T.M. et al. Syntheses and evaluation of new acridone derivatives for selective binding of oncogene c-myc promoter i-motifs in gene transcriptional regulation. Chem. Commun. 2018; 54:2036–2039. [DOI] [PubMed] [Google Scholar]
- 90. Yoga Y.M., Traore D.A., Sidiqi M., Szeto C., Pendini N.R., Barker A., Leedman P.J., Wilce J.A., Wilce M.C.. Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides. Nucleic Acids Res. 2012; 40:5101–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Marsich E., Piccini A., Xodo L.E., Manzini G.. Evidence for a HeLa nuclear protein that binds specifically to the single-stranded d(CCCTAA)(n) telomeric motif. Nucleic Acids Res. 1996; 24:4029–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Marsich E., Xodo L.E., Manzini G.. Widespread presence in mammals and high binding specificity of a nuclear protein that recognises the single-stranded telomeric motif (CCCTAA)n. Eur. J. Biochem. 1998; 258:93–99. [DOI] [PubMed] [Google Scholar]
- 93. Lacroix L., Lienard H., Labourier E., Djavaheri-Mergny M., Lacoste J., Leffers H., Tazi J., Helene C., Mergny J.L.. Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 2000; 28:1564–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Niu K., Zhang X., Deng H., Wu F., Ren Y., Xiang H., Zheng S., Liu L., Huang L., Zeng B. et al. BmILF and i-motif structure are involved in transcriptional regulation of BmPOUM2 in Bombyx mori. Nucleic Acids Res. 2018; 46:1710–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kang H.J., Kendrick S., Hecht S.M., Hurley L.H.. The transcriptional complex between the BCL2 i-Motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014; 136:4172–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Burger A.M., Dai F., Schultes C.M., Reszka A.P., Moore M.J., Double J.A., Neidle S.. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005; 65:1489–1496. [DOI] [PubMed] [Google Scholar]
- 97. Phatak P., Cookson J.C., Dai F., Smith V., Gartenhaus R.B., Stevens M.F., Burger A.M.. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br. J. Cancer. 2007; 96:1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tauchi T., Shin-ya K., Sashida G., Sumi M., Okabe S., Ohyashiki J.H., Ohyashiki K.. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene. 2006; 25:5719–5725. [DOI] [PubMed] [Google Scholar]
- 99. Villasante A., Abad J.P., Mendez-Lago M.. Centromeres were derived from telomeres during the evolution of the eukaryotic chromosome. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:10542–10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Aldrup-MacDonald M.E., Kuo M.E., Sullivan L.L., Chew K., Sullivan B.A.. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 2016; 26:1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Henikoff S., Thakur J., Kasinathan S., Talbert P.B.. Remarkable evolutionary plasticity of centromeric chromatin. Cold Spring Harb. Symp. Quant. Biol. 2017; 82:71–82. [DOI] [PubMed] [Google Scholar]
- 102. Gallego J., Golden E.B., Stanley D.E., Reid B.R.. The folding of centromeric DNA strands into intercalated structures: a physicochemical and computational study. J. Mol. Biol. 1999; 285:1039–1052. [DOI] [PubMed] [Google Scholar]
- 103. Garavís M., Mendez-Lago M., Gabelica V., Whitehead S.L., González C., Villasante A.. The structure of an endogenous Drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci. Rep. 2015; 5:13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bar-Am O., Weinreb O., Amit T., Youdim M.B.. Regulation of Bcl-2 family proteins, neurotrophic factors, and APP processing in the neurorescue activity of propargylamine. FASEB J. 2005; 19:1899–1901. [DOI] [PubMed] [Google Scholar]
- 105. Kendrick S., Akiyama Y., Hecht S.M., Hurley L.H.. The i-Motif in the bcl-2 P1 promoter forms an unexpectedly stable structure with a unique 8:5:7 loop folding pattern. J. Am. Chem. Soc. 2009; 131:17667–17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Muniyappa K., Kaulage M.H., Bhattacharya S.. Structural characterization of i-Motif structure in the human Acetyl-CoA carboxylase 1 gene promoters and their role in the regulation of gene expression. ChemBioChem. 2018; 19:1078–1087. [DOI] [PubMed] [Google Scholar]
- 107. Brown R.V., Wang T., Chappeta V.R., Wu G., Onel B., Chawla R., Quijada H., Camp S.M., Chiang E.T., Lassiter Q.R. et al. The consequences of overlapping G-Quadruplexes and i-Motifs in the Platelet-Derived growth factor receptor beta core promoter nuclease hypersensitive element can explain the unexpected effects of mutations and provide opportunities for selective targeting of both structures by small molecules to downregulate gene expression. J. Am. Chem. Soc. 2017; 139:7456–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Fleming A.M., Zhu J., Ding Y., Burrows C.J.. 8-Oxo-7,8-dihydroguanine in the context of a gene promoter G-Quadruplex is an On-Off switch for transcription. ACS Chem. Biol. 2017; 12:2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kendrick S., Muranyi A., Gokhale V., Hurley L.H., Rimsza L.M.. Simultaneous drug targeting of the promoter MYC G-Quadruplex and BCL2 i-Motif in diffuse large B-Cell lymphoma delays tumor growth. J. Med. Chem. 2017; 60:6587–6597. [DOI] [PubMed] [Google Scholar]
- 110. Brown R.V., Danford F.L., Gokhale V., Hurley L.H., Brooks T.A.. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J. Biol. Chem. 2011; 286:41018–41027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Banerjee R., Russo N., Liu M., Basrur V., Bellile E., Palanisamy N., Scanlon C.S., van Tubergen E., Inglehart R.C., Metwally T. et al. TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat. Commun. 2014; 5:4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sutherland C., Cui Y.X., Mao H.B., Hurley L.H.. A Mechanosensor mechanism controls the G-Quadruplex/i-Motif molecular switch in the MYC promoter NHE III1. J. Am. Chem. Soc. 2016; 138:14138–14151. [DOI] [PubMed] [Google Scholar]
- 113. Kaiser C.E., Van Ert N.A., Agrawal P., Chawla R., Yang D., Hurley L.H.. Insight into the complexity of the i-Motif and G-Quadruplex DNA structures formed in the KRAS promoter and subsequent Drug-Induced gene repression. J. Am. Chem. Soc. 2017; 139:8522–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Takahashi S., Brazier J.A., Sugimoto N.. Topological impact of noncanonical DNA structures on Klenow fragment of DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:9605–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Phan A.T., Mergny J.L.. Human telomeric DNA: G-quadruplex, i-motif and watson-crick double helix. Nucleic Acids Res. 2002; 30:4618–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bucek P., Jaumot J., Aviñó A., Eritja R., Gargallo R.. pH-modulated Watson-Crick duplex-quadruplex equilibria of guanine-rich and cytosine-rich DNA sequences 140 base pairs upstream of the c-kit transcription initiation site. Chem-Eur. J. 2009; 15:12663–12671. [DOI] [PubMed] [Google Scholar]
- 117. Kumar N., Sahoo B., Varun K.A., Maiti S., Maiti S.. Effect of loop length variation on quadruplex-Watson Crick duplex competition. Nucleic Acids Res. 2008; 36:4433–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jonchhe S., Shrestha P., Ascencio K., Mao H.. A new concentration jump strategy reveals the lifetime of i-Motif at physiological pH without force. Anal. Chem. 2018; 90:3205–3210. [DOI] [PubMed] [Google Scholar]
- 119. Selvam S., Mandal S., Mao H.. Quantification of chemical and mechanical effects on the formation of the G-Quadruplex and i-Motif in duplex DNA. Biochemistry. 2017; 56:4616–4625. [DOI] [PubMed] [Google Scholar]
- 120. Dhakal S., Yu Z.B., Konik R., Cui Y.X., Koirala D., Mao H.B.. G-quadruplex and i-motif are mutually exclusive in ILPR double-stranded DNA. Biophys. J. 2012; 102:2575–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Cui Y.X., Kong D.M., Ghimire C., Xu C.X., Mao H.B.. Mutually exclusive formation of G-Quadruplex and i-Motif is a general phenomenon governed by steric hindrance in duplex DNA. Biochemistry. 2016; 55:2291–2299. [DOI] [PubMed] [Google Scholar]
- 122. Webb B.A., Chimenti M., Jacobson M.P., Barber D.L.. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer. 2011; 11:671–677. [DOI] [PubMed] [Google Scholar]
- 123. Modi S., M G.S., Goswami D., Gupta G.D., Mayor S., Krishnan Y.. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 2009; 4:325–330. [DOI] [PubMed] [Google Scholar]
- 124. Surana S., Bhat J.M., Koushika S.P., Krishnan Y.. An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nat. Commun. 2011; 2:340. [DOI] [PubMed] [Google Scholar]
- 125. Modi S., Nizak C., Surana S., Halder S., Krishnan Y.. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 2013; 8:459–467. [DOI] [PubMed] [Google Scholar]
- 126. Wang C., Du Y., Wu Q., Xuan S., Zhou J., Song J., Shao F., Duan H.. Stimuli-responsive plasmonic core-satellite assemblies: i-motif DNA linker enabled intracellular pH sensing. Chem. Commun. 2013; 49:5739–5741. [DOI] [PubMed] [Google Scholar]
- 127. Huang J., He Y., Yang X., Wang K., Ying L., Quan K., Yang Y., Yin B.. I-motif-based nano-flares for sensing pH changes in live cells. Chem. Commun. 2014; 50:15768–15771. [DOI] [PubMed] [Google Scholar]
- 128. Peng J., He X., Wang K., Tan W., Wang Y., Liu Y.. Noninvasive monitoring of intracellular pH change induced by drug stimulation using silica nanoparticle sensors. Anal. Bioanal. Chem. 2007; 388:645–654. [DOI] [PubMed] [Google Scholar]
- 129. Huang J., Ying L., Yang X., Yang Y., Quan K., Wang H., Xie N., Ou M., Zhou Q., Wang K.. Ratiometric fluorescent sensing of pH values in living cells by dual-fluorophore-labeled i-motif nanoprobes. Anal. Chem. 2015; 87:8724–8731. [DOI] [PubMed] [Google Scholar]
- 130. Cao Y., Qian R.C., Li D.W., Long Y.T.. Raman/fluorescence dual-sensing and imaging of intracellular pH distribution. Chem. Commun. 2015; 51:17584–17587. [DOI] [PubMed] [Google Scholar]
- 131. Sellner S., Kocabey S., Zhang T., Nekolla K., Hutten S., Krombach F., Liedl T., Rehberg M.. Dexamethasone-conjugated DNA nanotubes as anti-inflammatory agents in vivo. Biomaterials. 2017; 134:78–90. [DOI] [PubMed] [Google Scholar]