Abstract
A fundamental as yet incompletely understood feature of Notch signal transduction is a transcriptional shift from repression to activation that depends on chromatin regulation mediated by transcription factor RBP-J and associated cofactors. Incorporation of histone variants alter the functional properties of chromatin and are implicated in the regulation of gene expression. Here, we show that depletion of histone variant H2A.Z leads to upregulation of canonical Notch target genes and that the H2A.Z-chaperone TRRAP/p400/Tip60 complex physically associates with RBP-J at Notch-dependent enhancers. When targeted to RBP-J-bound enhancers, the acetyltransferase Tip60 acetylates H2A.Z and upregulates Notch target gene expression. Importantly, the Drosophila homologs of Tip60, p400 and H2A.Z modulate Notch signaling response and growth in vivo. Together, our data reveal that loading and acetylation of H2A.Z are required to assure tight control of canonical Notch activation.
INTRODUCTION
The Notch signal transduction cascade is highly conserved and is essential for embryonic as well as post-natal development (1–3). Activating mutations in the human NOTCH1 gene are implicated in leukemogenesis (4,5) and loss-of-function of the central transcription factor RBP-J (also known as CSL) promotes tumorigenesis (6). Similarly, deregulation of Notch signaling by the oncogenic protein AML1/ETO promotes acute myeloid leukemia (AML) development (7). Upon ligand binding, the Notch receptor is proteolytically processed and its intracellular domain (NICD) migrates into the nucleus, interacts with RBP-J and activates expression of target genes. The signal is terminated by ubiquitin-dependent proteasomal degradation of the Notch coactivator complex (2,8) and gene repression is re-established by RBP-J that recruits a corepressor complex, waiting for the next wave of Notch activation. At the chromatin level, the activation/repression switch is regulated by dynamic histone acetylation and methylation (9–11). Accordingly, there is an interplay of Notch-associated acetyltransferases (12,13) and HDAC-containing complexes (14) that regulate histone acetylation whereas H3K4 methylation levels are determined by methyltransferase KMT2D (10) and demethylases KDM5A (15) and KDM1A/LSD1 (16–18). Notably, H3K4 methyl modifiers also regulate Notch target genes in vivo as revealed in Drosophila melanogaster (10,15).
The highly conserved histone variant H2A.Z is a regulator of gene expression which is found at promoters in both the non-induced and induced state (19) and its chromatin occupancy negatively correlates with active gene expression (20,21). In mammals, H2A.Z occupancy negatively correlates with the expression of Heparanase, c-Myc, p21WAF1/CIP1, CCND1 and glucocorticoid receptor (GR)-responsive genes (22–26) while it plays a positive role in the expression of estrogen-dependent genes (27). This discrepancy is possibly due to post-translational regulation of H2A.Z: H2A.Z acetylation and ubiquitination have been positively (28–31) and negatively (32,33) linked to gene expression, respectively. First revealed in Saccharomyces cerevisiae, acetylation of H2A.Z is required for gene induction (34) and its acetylation levels correlate genome-wide with gene activity (35). Studies in chicken cells showed that hyperacetylation of H2A.Z results in nucleosome destabilization and an open chromatin conformation (36–38). The vertebrate H2A.Z is mainly acetylated on the lysine residues K4, K7 and K11 (38–40) and genome-wide studies in human cancer cell lines have suggested that H2A.Z acetylation plays a key role in disease-associated gene deregulation (28).
In mammals, the Ep400/p400 (hereafter referred to as p400) subunit of the TRRAP/p400/Tip60 complex loads H2A.Z within chromatin (23) and transcription factors like p53 or the oncogene cMyc recruit p400 to chromatin (23,41,42). Genetic evidence in Drosophila suggests a role for the p400 homolog, called Domino, as a positive regulator of the Notch signaling pathway (43,44); the same holds true for Nipped-A, the Drosophila homolog of the TRRAP subunit (45). The acetyltransferase Kat5/Tip60 (hereafter referred to as Tip60) stimulates H2A.Z exchange in human cells (46).
Here, we show that loss-of-function of H2A.Z leads to the upregulation of canonical Notch target genes. Subsequently, we delineate the role of histone variant H2A.Z occupancy and its acetylation in the context of the Notch signaling response. Using Drosophila as a well-established in vivo model system, we show that the homologs of Tip60, the H2A.Z loader p400 and H2A.Z itself play a key role in Notch-dependent growth control.
MATERIALS AND METHODS
Cell culture, transfection and infection
Murine pre-T lymphoma cell line (Beko) and murine hybridoma mature T cell line (MT) were grown in Iscove's Modified Dulbecco Medium (IMDM, Gibco 21980-065) supplemented with 2% FCS, 0.3 mg/l peptone, 5 mg/l insulin, nonessential aminoacids and penicillin/streptomycin. The MT cells were previously described (18,47). MT NICD-ER cells were induced with (Z)-4-hydroxytamoxifen (4-OHT) at 1 μM final concentration (Sigma-Aldrich H7904–5MG) or ethanol as control. HeLa, 293T and PhoenixTM packaging cells (Orbigen, Inc., San Diego, CA, USA) were cultivated in Dulbecco's modified eagle medium (DMEM, Gibco 61965–059) supplemented with 10% FCS and penicillin/streptomycin. Cells were grown at 37°C with 5% CO2.
Phoenix™ cells were transfected using calcium phosphate as follows: 5 × 106 cells were plated in 72 cm2 flasks and incubated for 16–24 h at 37°C. The day after, 20 μg of plasmid DNA were added to 860 μl of sterile H2O and 120 μl of 2 M CaCl2 were pipetted into the DNA solution and briefly vortexed. To obtain small precipitates, the DNA solution was added dropwise to 1 ml of 2× HBS (50 mM HEPES pH 7.05, 10 mM KCl, 12 mM glucose, 280 mM NaCl, 1.5 mM Na2HPO4) while vortexing and the resulting solution was incubated 20 min at room temperature. In the meantime, Phoenix™ cells were incubated with chloroquine (Sigma-Aldrich C6628-100G) for 10 min at room temperature. The DNA solution was added to the cells and after at least 6 h of incubation at 37°C the medium was changed. For co-immunoprecipitation (CoIP) experiments in Phoenix™ cells, 10 μg of each plasmid DNA were used for transfection as described above.
Retroviral infections of Beko and MT cells were performed as follows: 24 h after transfection of the Phoenix™ cells, the supernatant was filtered and supplemented with polybrene (Sigma-Aldrich H9268) at a final concentration of 2 μg/ml. The retroviral suspension was added to the target cells and the cell/retroviral suspension was centrifuged 45 min at 1800 rpm at 37°C. After centrifugation, the medium was replaced with fresh one. The infection was repeated four times over two days. Cells were grown in culture, analysed by FACS and sorted or selected by using the appropriate selective antibiotic: puromycin (Serva 33835) and/or histidinol (Sigma-Aldrich H6647).
Generation of CRISPR/Cas9 depleted MT cells
CRISPR/Cas9 H2afz/H2afv double depleted MT cells were generated as follows: 3 × 106 293T cells were seeded and, after 24 h, transfected with 2.5 μg psPAX, 1 μg pMD2G and 3.33 μg of the desired lentiCRISPR v2 vector using Lipofectamine 2000 Transfection Reagent (Invitrogen 11668-019) accordingly to manufacturer's instructions. After at least 6 h of incubation at 37°C the medium was replaced with fresh one and 48 h post-transfection the supernatant was filtered, supplemented with polybrene and used for infection of MT cells. Positively infected cells were selected with puromycin and dilutions were performed to establish single cell clones. Individual clones were screened by Western blotting for H2A.Z and the clones, depleted for H2A.Z, were analysed by RT-qPCR to validate the double depletion of H2afz and H2afv at the transcript level. One clone (sgH2afv/H2afz #12) was further analysed by sequencing of the genomic DNA (gDNA). To reach this goal, the gDNA was purified as described below and nested PCR strategies were used as follows: in the case of H2afz, a first PCR was performed using H2afz gDNA1 fw and H2afz gDNA1 rev primers and this product was used as template for a second PCR using H2afz gDNA2 fw and H2afz gDNA2 rev primers; in the case of H2afv, a first PCR was performed using H2afv gDNA1 fw and H2afv gDNA1 rev primers and this product was used as template for a second PCR using H2afv gDNA2 fw and H2afv gDNA2 rev primers. PCR products were cloned in the pSC-A-amp/kan (Agilent Technologies 240205-5) and more colonies were sequenced after purification of the plasmid DNA using standard procedures. Primers are listed in Supplementary Table S5.
Genomic DNA (gDNA) purification
Cells were washed twice in PBS, resuspended in gDNA extraction buffer [10 mM Tris–HCl pH 7.5, 10 mM EDTA pH 8.0, 10 mM NaCl, 0.5% N-Lauroylsarcosine sodium salt, 1 mg/ml proteinase K (Roche 03115852001)] and incubated over night at 37°C. gDNA was precipitated with 100% EtOH in presence of 50 mM NaCl, washed with 70% EtOH, dried and resuspended in TE pH 8.0.
Apoptosis assay
Cells were labeled for 20 min with 5 μl Pacific Blue labeled Annexin V antibody (1:40, BD Biosciences) in 200 μl Annexin V binding buffer (BD Biosciences). The proportion of apoptotic cells was determined at a LSRII flow cytometer (BD Biosciences).
GIEMSA staining
Cells were cytospun in cytofunnels (Thermo Scientific) for 10 min at 1000 rpm in a cytospin4™ cytocentrifuge (Thermo Scientific). Cytospins were then labeled with Giemsa (Merck, 1.09204.0500) following standard procedures.
Cell cycle analysis
Cells were washed twice with PBS and fixed in 70% ethanol on dry ice. After 1 h cells were spun shortly and the pellet was stained for 10 min at 37°C with 50 μg/ml propidium iodide (Merck) in 4 mM sodium citrate containing 0.1 mg/ml RNase in a total volume of 200 μl. Analysis was performed on a FACSCalibur cytometer (BD Biosciences). Cell cycle stages (G1/S/G2M) were determined according to the DNA content. Cells possessing 2n DNA were assigned to be in G1-phase, those having a DNA content between 2n and 4n were defined as S-phase cells and those having approximately 4n DNA content were assumed to be in G2M.
Cell growth analysis
Cells were resuspended in 20 ml IMDM (see above) at a density of 10 000 cells per ml and counted every day for 7 days following standard procedures.
Constructs
All oligonucleotides used in this study are listed in Supplementary Table S5. PCR products were cloned in the pSC-A-amp/kan (Agilent Technologies 240205-5), digested with the selected restriction enzymes (New England Biolabs) and cloned into the destination vectors accordingly to Supplementary Table S6. All plasmids were analysed by sequencing.
The pcDNA 3.1 Flag2 (Invitrogen) and pGEX6P1 (GE Healthcare) were commercially acquired. The lentiCRISPR v2 was a gift from Dr F. Zhang [(48), Addgene plasmid # 52961]. The pcDNA3 mNICD, pcDNA3 mNICD ΔEP and pcDNA3 mNICD+OP were previously described (13). The pGEX4T3-mRBP-J-wt, pcDNA3.1 Flag2 mRBP-J-wt, pGEX6P1-dmSu(H) and pMIGRI Flag-Notch-ER IRES GFP plasmids were previously described (15). The pMY-BioDdx5-IRES-GFP and pMY-BioNICD-IRES-GFP plasmids were previously described (49). The pGEX6P1 mNICD-NT and pGEX6P1 mNICD-CT were previously described (50). The pcDNA3-RBP2N was previously described (51). The GST-NICD and GST-RITA expression plasmids were previously described (13,52). The expression vector pcDNA3 Flag-Tip60β wildtype (wt) and pcDNA3 Flag-Tip60β catalytic-dead mutant (cd) were a generous gift of Dr J. Côté (53). The CβS Flag-p400 was kindly provided by Dr R G. Roeder (54). The pCMV-3Flag-H2A.Z-wt and pCMV-3Flag-H2A.Z-K4R were generously provided by Dr O. Binda.
The pMYs-Bio-NCMXH-pSV40-Puro was generated via insertion of the oligo NCMXH (sequence in Supplementary Table S5) into the pMYs-Bio-NCMH-pSV40-Puro predigested with NotI and HindIII which was generated via insertion of the oligo NCMH (sequence in Supplementary Table S5) into the pMYs-Bio-pSV40-Puro predigested with NotI and HindIII. The pcDNA 3.1 Flag-p400–1 was generated by digestion of the CβS Flag-p400 with NotI and ligation of the product into the pcDNA 3.1 Flag2 predigested with NotI.
The pMIGR1 Flag-RBP-J/Tip60-wt or RBP-J/Tip60-cd mutant pSV40-Puro plasmids were generated as follows: first, the RBP-J cDNA was inserted using a PCR cloning strategy to generate the pMIGR1 Flag-RBP-J-wt-pSV40-Puro no STOP which was subsequently used to insert the Tip60-wt or Tip60-cd mutant cDNAs using PCR cloning. The pcDNA3.1 Flag-RBP-J/Tip60-wt and pcDNA3.1 Flag-RBP-J/Tip60-cd mutant plasmids were generated by EcoRI digestion of pMIGR1 Flag-RBP-J/Tip60-wt or RBP-J/Tip60-cd mutant pSV40-Puro plasmids and ligation into the pcDNA3.1 Flag2 plasmid predigested with EcoRI. The Drosophila Domino (AA1557–2352, according to NP_001286676) expression plasmid was generated by PCR aided cloning. The amplified DNA sequence was ligated into the EcoRI and XbaI sites of pcDNA3-Flag1 resulting in pcDNA3-F1-dmDomino (1557–2352).
The pMIGR1–3Flag-H2A.Z-wt and -K4R mutant (lysine in position 4 mutated to arginine) pSV40-Puro plasmids were cloned as follows: the H2A.Z-wt and -K4R mutant were PCR cloned into the pMI-IRES-Berry (generously provided by Drs. E. Surova and H. Jumaa). The pMI-3Flag-H2A.Z-wt and -K4R mutant IRES-Berry plasmids were digested with XhoI and EcoRI and the digestion products were cloned into the pMIGR1-pSV40-Puro predigested with XhoI and EcoRI. The pMIGR1 3Flag-H2A.Z5KR mutant pSV40-Puro was generated by site directed mutagenesis using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies 200521–5) accordingly to manufacturer's instructions with primers listed in Supplementary Table S5 and using the pMIGR1–3Flag-H2A.Z-K4R mutant pSV40-Puro as template.
The CRISPR/Cas9 guides were designed using the online tool available at http://crispr.mit.edu/. The desired 5′ overhangs were added and oligos were phosphorylated, annealed and ligated into the lentiCRISPRv2 predigested with BsmBI.
ShRNA knockdown
For the knockdown (KD) in Beko cells, suitable hairpins were designed with the siRNA Wizard (InvivoGen, http://www.sirnawizard.com/design.php) and cloned into the pSIR Delta U6C Puro vector (kindly provided by Drs D. van Essen and S. Saccani) with HindIII/XhoI. To test knockdown efficiency, the sequence targeted by the siRNA was cloned into the target gene reporter vector pMy-Delta-GFP (kindly provided by Drs D. van Essen and S. Saccani) with EcoRI/NotI. Cells were first infected with the reporter vector and after sorting the GFP positive cells, cells were infected with shRNA-expressing vectors and selected with puromycin. Finally, cells were sorted depending on the GFP shift and the knockdown efficiency was analysed by quantitative real-time PCR (qPCR) and Western blotting. Sequence of the hairpin and primers used for generating the reporter are indicated in Supplementary Table S5.
For the knockdown in MT cells, the pLKO.1 TRC1 shRNA library (SIGMA-ALDRICH) was used. Transfection of 293T cells and infection and selection of MT cells was performed as previously described (10). Sequence of the hairpin is indicated in Supplementary Table S5.
RNA extraction, RT-PCR and qPCR from cell lines
Total RNA was purified using Trizol reagent (Ambion 15596018) accordingly to manufacturer's instructions. 1 μg of RNA was reverse-transcribed into cDNA using random hexamers and M-MuLV reverse transcriptase (New England Biolabs). qPCRs were assembled with Absolute QPCR ROX Mix (Thermo Scientific AB-1139), gene-specific oligonucleotides, double-dye probes (see Supplementary Table S5) and analysed using the StepOnePlus™ Real-Time PCR System (Applied Biosystem). Data were normalized to the housekeeping gene glucuronidase β (GusB).
For RNA-Seq purposes, RNA was purified using the RNeasy Mini Kit (Qiagen #74104) in combination with the QIAshredder (Qiagen #79654) and treatment with DNase I (Qiagen #79254) accordingly to manufacturer′s instructions.
Library preparation and sequencing
Libraries were prepared using the TruSeq® Stranded Total RNA LT-Ribo-Zero Gold kit (Illumina RS-122-2301/2) and sequenced on an Illumina HiSeq 1500 with 50 bases single reads.
RNA-seq analysis
Quality control of RNA-seq reads was performed using the FastQC application available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Filtering and trimming of reads was performed with trimmomatic (55). Reads were aligned using STAR (56) to an index based on mm9 version genome of the mouse genome. FeatureCounts from the Subread package was used for extraction of gene-specific read counts based on mm9 UCSC gene annotations (57). Both genome and gene annotations were downloaded from Illumina's iGenome site (http://support.illumina.com/sequencing/sequencing_software/igenome.html). The results of the FeatureCounts gene expression analysis were imported into R and detection of differentially expressed genes was done using DESeq2 (58). Subsequent data analysis and visualization was done with custom R scripts. Two replicates were analyzed in the case of overexpression of BioNICD and induction of NICD-ER fusion proteins whereas six replicates were analyzed in the case of the CRISPR-mediated depletion of H2A.Z.
For re-analysis of publicly available data on dominant negative Tip60 in Drosophila larval wing discs, the data corresponding to 48 h expression were downloaded from GEO (GSE81159). SRA archives were extracted to fastq format using the fastq-dump program. We used hisat2 (59) in order to align the reads to an index for the dm3 genome (iGenome repository) built by the hisat2-build function. We used UCSC gene models in order to extract splice-sites for the alignment process. Aligned reads were converted to BAM format, sorted and removed for duplicates using samtools. Identification of differentially expressed genes was done after import of BAM-alignment files into R using BioConductor packages (60). Therefore, we used the summarizeOverlaps function of the GenomicAlignments package (61) in order to extract gene-specific read counts. Normalization between experiments and detection of differentially expressed genes was done with the DESeq2 package with default settings (58). In order to identify enriched gene sets based on GO terms, we performed GO enrichment analysis using the enrichGO function and GSEA using the gseGO function of the clusterProfiler package (62). We used the test statistic (stat) of the DESeq2 analysis for ordering the ranked list for GSEA.
Fisher's exact test as implemented in the geneOverlap package (https://www.bioconductor.org/packages/release/bioc/html/GeneOverlap.html) was performed in order to determine whether the observed overlap between two gene lists was significant. Summary statistics on the RNA-Seq data are available in Supplementary Table S2.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed following the Upstate Biotechnology protocol with few modifications. Cells were cross-linked in 1% formaldehyde (FMA) for 10 min at room temperature and the reaction was blocked with 1/8 volume of 1 M glycine pH 8.0. Only in the case of the RBP-J ChIP, cells were pre-fixed with 10 mM dimethyl adipimidate (DMA, Thermo Scientific 20660) in PBS for 1 h at room temperature and cross-linked in 1% FMA for 30 min. Cells were washed twice with PBS and resuspended in SDS Lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.1). The cell suspension was sonicated using the Covaris System S220AFA and, after dilution with ChIP Dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.1, 167 mM NaCl), the chromatin was pre-cleared with pre-saturated protein-A-Sepharose beads (GE Healthcare 17-5280-02) for 30 min at 4°C or 1 h in the case of the Flag ChIP. The chromatin was incubated over night with the proper amount of the desired antibody [RBP-J (Abcam ab25949), p400 (Abcam ab5201), H2A.Z (Abcam ab4174), H2A.Zac (Abcam ab18262), H3 (Abcam ab1791), H3K27ac (Diagenode pA-174-050), H3K4me2 (Diagenode pAb-035-050), H3K18ac (Cell Signaling 9675S)], with IgG (Santa Cruz sc-2027; Diagenode C15410206) as control or 10 μl Flag M2 affinity gel (Sigma-Aldrich A2220). Antibodies were immobilized with pre-saturated protein-A-Sepharose beads for 1 h at 4°C with shaking. Depending on the antibody, different washing conditions were used. Chromatin was eluted from beads with Elution buffer (1% SDS, 0.1 M NaHCO3) and cross-links were reverted at 65°C over night in presence of NaCl. After incubation with Proteinase K for 1 h at 45°C, the DNA was purified by phenol/chloroform extraction and precipitated over night at –20°C in presence of yeast tRNA, glycogen and 2-propanol. After washing with 70% EtOH and drying, the DNA was dissolved in TE pH 8.0 and analysed by qPCR. The BioNICD and BioTip60 ChIP experiments were performed as previously described (50). qPCR analysis were focused at the enhancers of Notch target genes identified by analyzing previously published ChIP-Seq data (63).
qPCRs were assembled with Absolute QPCR ROX Mix (Thermo Scientific AB-1139), gene-specific oligonucleotides, double-dye probes (see Supplementary Table S5) and analysed using the StepOnePlus™ Real-Time PCR System (Applied Biosystem). Where indicated in the figure legend, data where normalized to the positive control (GAPDH 0kb) and eventually to histone H3 or H2A.Z. A region of the chromosome X (chrX: 112357567 ±112357626, indicated as Chrom X) was used as negative control.
ChIP-Seq analysis
Notch-1 and RBP-J ChIP-Seq data [(63), accession # GSE29600] were analysed using MACS14 via the Galaxy Platform. Reads were mapped to the mm9 genome and default settings were used.
GST protein purification and GST pulldown
GST fusion proteins were expressed in Escherichia coli strain BL21 (Stratagene), purified using standard procedures and stored at –80°C. Proteins were in vitro translated in presence of [35S] methionine (Perkin-Elmer NEG709A500UC) using the reticulocyte lysate-coupled transcription/translation system (Promega L4610) accordingly to manufacturer's instructions. Translation and labeling quality were monitored by SDS-PAGE. GST protein and GST fusion proteins were immobilized on Glutathione Sepharose 4 Fast Flow (GE Healthcare 17-5132-01) and incubated together with the in vitro translated proteins in buffer A (40 mM HEPES at pH 7.5, 5 mM MgCl2, 0.2 mM EDTA, 0.5% NP-40, 100 mM KCl) under rotation for 2 h at 4°C. Beads were washed with buffer A, buffer B (40 mM HEPES pH 7.5, 5 mM MgCl2, 0.2 mM EDTA, 0.5% NP-40, 300 mM KCl) and PBS. After washing, beads were resuspended in SDS-PAGE loading buffer and proteins were resolved by SDS-PAGE. Gels were dried and exposed to X-ray films.
Protein extract, CoIP, cell fractionation and Western blotting
Whole Cell Extract (WCE) was prepared as follows: cells were washed twice in PBS, lysed in WCE buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% NP-40, 10% glycerol, 0.5 mM Na3VO4, 10 mM NaF, 1 mM PMSF, 1× protease inhibitor cocktail mix) and incubated 20 min on ice. After centrifuging 15 min at 13 200 rpm at 4°C, the supernatant was recovered and protein concentration measured by Bradford assay (Sigma-Aldrich).
For nuclear extract, cells were washed twice with PBS, resuspended in Hypotonic buffer (20 mM HEPES, 20 mM NaCl, 10% glycerol, 5 mM MgCl2, 0.2 mM PMSF) and incubated 20 min on ice. After vortexing, lysates were centrifuged at 4000 rpm 10 min at 4°C. The supernatant (cytoplasm) was collected and the pellet (nuclei) was washed with PBS. Nuclei were resuspended in Hypertonic buffer (20 mM HEPES pH 7.9, 100 mM NaCl, 0.3% NP-40, 25% glycerol, 1 mM MgCl2, 0.2 mM PMSF, 1× protease inhibitor cocktail mix, 0.3 mM DTT) and Hypertonic buffer 2.2 M NaCl (20 mM HEPES pH 7.9, 2.2 M NaCl, 0.3% NP-40, 25% glycerol, 1 mM MgCl2, 0.2 mM PMSF, 1× protease inhibitor cocktail mix, 0.3 mM DTT) was added dropwise to a final concentration of 0.3 M NaCl. Samples were incubated 20 min on ice in cold room and centrifuged at 40 000 rpm 1 h at 4°C. The supernatant was collected and protein concentration was measured by Bradford assay. In some cases, nuclei were lysed in Hypertonic buffer 300 mM NaCl (20 mM HEPES pH 7.9, 300 mM NaCl, 0.3% NP-40, 25% glycerol, 1 mM MgCl2, 0.2 mM PMSF, 1× protease inhibitor cocktail mix, 0.3 mM DTT).
For CoIP experiments, WCE was pre-cleared with 10 μl protein-A-Sepharose beads (GE Helthcare 17-5280-02) for 1 h at 4°C with rotation. After centrifugation at 3000 rpm for 3 min at 4°C, input was collected and the remaining protein solution was incubated with 10 μl Flag M2 affinity gel (Sigma-Aldrich A2220) for 2 h at 4°C with rotation. Beads were washed three times with WCE buffer and the CoIPs were analysed by Western blotting.
For streptavidin pulldown purposes, 2 mg of nuclear extract and 50 μl of Dynabeads M-280 Streptavidin (Invitrogen, 112.06D) were used for each IP. Dynabeads were washed with 1 ml of ice cold PBS and blocked with 1 ml of 1× TBS and 200 μg of Chicken Egg Albumin (Sigma-Aldrich A5503-1G) for 1 h at 4°C with rotation. In the meantime, the nuclear extract was diluted to have a final concentration of 120 mM NaCl and 10% glycerol, first with buffer without NaCl (20 mM HEPES pH 7.9, 0.2% NP-40, 25% glycerol, 1 mM MgCl2, 0.2 mM PMSF, 1× protease inhibitor cocktail mix, 0.3 mM DTT) and subsequently with buffer without glycerol (20 mM HEPES pH 7.9, 120 mM NaCl, 0.2% NP-40, 1 mM MgCl2, 0.2 mM PMSF, 1× protease inhibitor cocktail mix, 0.3 mM DTT). Input was collected and, after washing the Dynabeads with PBS, pulldown reactions were incubated at 4°C for 3 h with rotation. Pulldowns were washed five times with Washing buffer (20 mM HEPES pH 7.9, 200 mM NaCl, 0.2% NP-40, 1 mM EDTA, 10% glycerol), twice with PBS and finally analysed by Western or streptavidin blotting. For mass-spectrometry purposes, streptavidin pulldown was performed as previously described (49).
For cell fractionation purposes, cells were washed twice in PBS and resuspended in buffer 1 (20 mM HEPES pH 7.4, 20 mM NaCl, 5 mM MgCl2). After incubation on ice for 10 min, cells were lysed with a Dounce pestle and further incubated on ice for 10 min. After centrifugation at 500 g for 5 min at 4°C, the supernatant, containing the cytosolic fraction, was collected and the nuclear pellet was washed once in buffer 1 and finally lysed in buffer 2 (20 mM HEPES pH 7.4, 20 mM NaCl, 5 mM MgCl2, 0.5% NP-40) by vortexing. Samples were incubated on ice for 15 min and centrifuged at 800 g for 5 min at 4°C. The supernatant, containing the nuclear fraction, was collected and the chromatin pellet was resuspended in buffer 3 (20 mM HEPES pH 7.4, 650 mM NaCl, 5 mM MgCl2), incubated on ice for 15 min and sonicated using a Branson sonifier. After centrifugation at 800 g for 5 min at 4°C, the supernatant, containing the chromatin fraction, was collected. Finally, all fractions were supplemented with SDS loading dye, boiled and analysed by Western blotting.
For Western blotting purposes, proteins were resolved by SDS-PAGE and transferred to a Hybond-P PVDF membrane (GE Healthcare 10600023) by wet blotting using the Biorad Mini Trans-Blot system. In the case of H2A.Z (Abcam, ab4174), GAPDH (Abcam, ab8245) and RBP-J (Cosmo Bio Co., Clone T6709) Western blotting, membranes were blocked 1 h at room temperature with 5% nonfat dry milk in 1× TBS, washed with PBS and incubated over night at 4°C with primary antibody (1:1000 antibody in 5% BSA, 1× TBS, 0.3% NP-40). Membranes were washed three times 15 min each with washing buffer (1× TBS, 0.5 M NaCl, 0.5% Triton X-100) and incubated 1 h at room temperature with secondary antibody conjugated to horseradish peroxidase (1:10 000 antibody in 5% BSA, 1× TBS, 0.3% NP-40). Membranes were washed three times 15 min each with washing buffer.
For streptavidin blotting, membranes were blocked 1 h at room temperature with 5% BSA in 1× TBS and incubated with 1 μl of streptavidin–HRP (Perkin-Elmer NEL750001EA) in 5% BSA, 1× TBS for 1 h at room temperature. Finally, membranes were washed three times 15 min each with 1× TBS, 0.5 M NaCl, 0.5% Triton X-100.
In the case of the Flag (Sigma-Aldrich F3165), HA (Roche 11867423001), H3 (Abcam ab1791), H2A.Z (Diagenode C15410201), TBP (Santa Cruz sc-273) and Notch1 (Cell Signaling 2421) Western blotting, membranes were blocked in blocking solution (5% nonfat dry milk, 1× TBS, 0.1% Tween 20) and incubated over night with primary antibody diluted in blocking solution. Membranes were washed in 1× TBS, 0.1% Tween 20 and incubated 1 h at room temperature with the proper secondary antibody diluted 1:5000 in blocking solution. Membranes were washed in 1× TBS, 0.1% Tween 20.
Finally, all membranes were incubated at room temperature with ECL solution and chemiluminescence was detected with a light sensitive film. The following secondary antibody were used: anti-rat IgG HRP (Jackson ImmunoResearch, 112-035-072), anti-mouse IgG HRP (Cell Signaling, 7076S) and anti-rabbit IgG HRP (Cell Signaling #7074S). Western blotting quantification was performed using ImageJ 1.43u.
On bead digestion and nanoLC–MS analysis
Direct endoproteinase digestion of proteins and/or protein complexes immobilized on magnetic bead supports was essentially performed as previously described (64) with modifications indicated in the following. Streptavidin magnetic beads (Dynabeads M-280) from Bio-Streptavidin purifications were finally washed once with wash buffer (lacking NP-40 and glycerol) and resuspended in 50 μl of denaturation buffer (10 mM HEPES–KOH, pH 7.9, 8 M urea). The urea concentration was immediately lowered to 4 M by adding a 50 μl aliquot of 50 mM ammonium bicarbonate. The resulting bead suspension was adjusted to 1 mM DTT and protease digestion was started by the addition of 500 ng Lys-C (Wako). After 1 h incubation at 25°C (Eppendorf Thermomixer, 1200 rpm), beads were separated (magnetic stand) and washed with 200 μl 50 mM ammonium bicarbonate. The supernatant (containing the pre-digested peptides) and the wash were pooled (combined volume = 300 μl) and alkylated with a five-fold excess (over DTT) of iodoacetamide (1.8 mM final concentration) at 25°C (Thermomixer) for 2 h. Over-night digestion (25°C, 10–12 h) was started by addition of 500 ng trypsin (Promega). Proteolysis was stopped by adding TFA to a final concentration of 3%. Tubes were centrifuged at 20 000 g (Eppendorf benchtop, 20°C, 10 min) in order to pellet insoluble material. Finally, the digest was subjected to ‘STAGE Tip′ purification and nanoLC–MS analysis as previously described (50) with the exception that a method employing a 120 min gradient was utilized. The nanoLC column was equilibrated with 2% MS buffer B (0.5% acetic acid/80% MeCN) for 22 min and then the sample was loaded and eluted stepwise with a 24–64% acetonitrile (p.a.) gradient over a period of 95 min (flowrate 0.25 μl/min, 22–25 min: 5%, 25–90 min: 30%, 90–100 min: 50%, 100–109 min: 60%, 109–115 min: 80%, 116–120 min: 2% MS buffer B).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (65) partner repository with the dataset identifier PXD008747.
STRING analysis
STRING analysis was done using the STRING database (66) available at https://string-db.org/ using standard parameters, showing no more than ten interactors and searching in the human database.
Drosophila husbandry
Drosophila RNAi transgenic lines His2Av/H2A.Z-RNAi GD12768, and domino/p400-RNAi GD7787 used in this study were obtained from Vienna Drosophila RNAi Center (VDRC) http://stockcenter.vdrc.at/control/main. Drosophila RNAi transgenic lines His2Av/H2A.Z-RNAi BL28966 and dTip60-RNAi BL28563 were obtained from Bloomington Drosophila Stock Center (BDSC) http://flystocks.bio.indiana.edu. Detailed description of the transgenic flies ey-Gal4, tubulin-Gal4, act>stop>-Gal4, hsp70-FLP, UAS-Dl and UAS-GFP can be found at http://flybase.org/.
As background control the w1118 fly strain was used. All stocks and crosses were reared in standard fly food and kept at 25°C.
Marked FLP-out clones of single and double mutant cells
We generated single and double mutant clones co-expressing the UAS-GFP and UAS-Dl alone or in combination with UAS-RNAi against His2Av/H2A.Z, dTip60 or domino/p400 using the flip-out technique and hs-Flipase (hs-FLP)-mediated recombination. We analysed at least 12 mosaic wing discs of each of the indicated genotypes in Figure 5. Clones were induced by a single heat shock pulse of 10 min at 37°C at 24–48 h after egg laying.
Figure 5.
Depletion of Drosophila His2Av/H2A.Z, dTip60 and domino/p400 leads to an asymmetric response to Delta-dependent Notch signaling and reveals a strong requirement for these factors in Notch responsive cells. (A) Drosophila homologs of RBP-J and p400 proteins, Su(H) and Domino respectively, physically interact with each other. GST pull-down experiments were performed using bacterially purified GST-Su(H) and in vitro transcribed/translated Domino aa 1557–2352 corresponding to the human RBP-J interacting p400–2 fragment (Figure 3D). (B) Scheme of a Drosophila wing disc with the dorsal wing pouch highlighted in green. Notch activation by its ligands Delta/DLL and Serrate/Jagged at the dorsal (d)/ventral (v) boundary results in activation of target genes, such as wingless (wg, in red; a: anterior, p: posterior, N act: activated Notch). Graph shows that the relative area of dorsal wing pouch is reduced in wing discs bearing clones of Delta-expressing cells that are depleted of His2Av/H2A.Z, dTip60 or domino/p400. Data are mean ± SD ([**] P < 0.01, unpaired Student's t-test; n = 12 mosaic wing discs in each case). (C–J) Representative confocal images of mosaic wing imaginal discs. Clones are labelled by GFP (green) and mosaic discs are stained for Wg/WNT (red) and DAPI (blue). (C) Control GFP-labelled clones in a third-instar wing imaginal disc. Endogenous Wg protein (red) is expressed along the d/v boundary (magnified image in C’) in response to Notch activation by its ligands Serrate and Delta and it is also expressed around the wing pouch and in a band across the notum (86). (D) Single Delta overexpressing clones. Note that only dorsal Delta-overexpressing clones activate Notch signaling as denoted by ectopic activation of Wg (red) around the clones. (E) Single His2Av/H2A.Z-RNAi clones and (E’) magnification of clones at the d/v boundary. (F) Double mutant clones overexpressing Delta ligand along with His2Av/H2A.Z-RNAi. (G) dTip60-RNAi clones and (G’) magnification of d/v boundary. (H) Double mutant Delta overexpression and dTip60-RNAi clones. (I) Single domino/p400-RNAi clones and (I’) magnification of clonal cells at d/v boundary. (J) Double mutant clones overexpressing Delta and domino/p400-RNAi transgene. Arrows point to examples of clonal cells where endogenous Wg expression is exclusive (E’ and G’) or downregulated in single RNAi clones (I’). Genotypes: (C) yw hsp70-FLP; actin>stop>Gal4 UAS-GFP (D) yw hsp70-FLP; actin>stop>Gal4 UAS-GFP /+; UAS-Dl/+ (E) yw hsp70-FLP; actin>stop>Gal4 UAS-GFP; +/UAS-His2Av/H2A.Z-RNAiBL28966 (F) yw hsp70-FLP; actin>stop>Gal4 UAS-GFP; UAS-Dl/UAS-His2Av/H2A.Z-RNAiBL28966 (G) yw hsp70-FLP; actin>stop>Gal4 UAS-GFP; +/UAS-dTip60-RNAiBL28563 (H) yw hsp70-FLP; actin>stop>Gal4 UAS-GFP; UAS-Dl/UAS-dTip60-RNAiBL28563 (I) yw hsp70-FLP; actin>stop>Gal4 UAS-GFP/+; +/UAS-domino/p400-RNAiGD7787 (J) yw hsp70-FLP; actin>stop>Gal4 GFP/+; UAS-Dl/UAS-domino/p400-RNAiGD7787. In all wing disc images, dorsal is up and anterior is to the left.
Immunofluorescence staining
Third instar imaginal discs were fixed and stained by standard procedures using 4D4 anti-Wg antibody (1:100 dilution, Developmental Studies Hybridoma Bank), secondary antibody anti-mouse-AlexaFluor-555 (1:200 Molecular Probes) and DAPI (Sigma Aldrich). Discs were mounted in Fluoromount G (Southern Biotechnology) and the images were captured on a Leica TCS-NT Confocal microscope.
Fluorescence microscopy of transfected HeLa cells was performed essentially as previously described (10,52).
Measurement of mosaic wing areas
Analysis of the area of mosaic wing imaginal discs and of dorsal wing pouch parts in Figure 5 was done using ImageJ software. Data represent mean values of the area of dorsal wing pouch normalized against the total wing disc area.
Drosophila quantitative reverse transcriptase PCR (qRT-PCR)
RNAi knockdown efficiency for His2Av/H2A.Z, dTip60 and for domino/p400 isoforms E, A and D and domino/p400 isoform G was determined by qPCR upon RNAi induction by tubulin-Gal4 promoter. For each genetic condition, total mRNA was isolated from five third instar larvae using RNAeasy-Mini Kit (Qiagen). To remove contaminating DNA, RNA was treated with Turbo DNA-free (Ambion, Life Technologies). cDNA was synthesized with SuperScript® III First-Strand Synthesis System for RT-PCR (Life Technologies) using oligo-dT primers. qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) using gene-specific primers, on an ABI7500 apparatus (Applied Biosystems). Rp49 primers were used for mRNA normalization. Comparative qRT-PCR was performed in triplicates and the relative expression was calculated using the comparative Ct method. Three independent experiments were performed and qPCR data were analysed using two-tailed unpaired t test. Primers used for qPCR are listed in Supplementary Table S5.
RESULTS
Loss-of-function of H2A.Z leads to upregulation of Notch target genes
We have previously defined how the active histone mark H3K4me3 is regulated at the enhancers of Notch target genes (10,15). Given that H3K4me3 and the histone variant H2A.Z colocalize genome-wide (29,30), we hypothesized a role for H2A.Z in the regulation of Notch target gene expression. In order to test this hypothesis, we used the CRISPR/Cas9 technology to deplete H2A.Z in a mature T (MT) cell line characterized by low Notch activity (18,47). In detail, we designed sgRNAs targeting the 5′ untranslated region (5′-UTR) of both H2afz and H2afv genes, encoding for two different H2A.Z isoforms. We could isolate two different clones showing depletion of H2A.Z (Figure 1A and Supplementary Figure S1A and B): the clone sgH2afv/H2afz #12 showed a strong H2A.Z depletion while the clone sgH2afv/H2afz #20 showed only almost 60% reduction in H2A.Z protein levels. Subsequently, we investigated differential expression of Notch target genes upon H2A.Z depletion and we observed that the well-known canonical Notch targets Hes1 and Il2ra (15,63) were significantly upregulated upon H2A.Z depletion in MT cells in both clones (Figure 1B). Given the significant residual H2A.Z protein in clone #20, we focused on the clone #12 (hereafter referred to as sgH2afv/H2afz) and validated the genome editing of the H2A.Z-encoding genes H2afv and H2afz. We observed a different 72 base pairs deletion in the two H2afv alleles whereas the H2afz gene presented with a deletion of 4 base pairs and an insertion of 6 base pairs which were detected in more sequencing reactions, suggesting that both alleles present with the same mutation (Supplementary Figure S1C). Subsequently, we performed RNA-Seq analysis to better define the role of H2A.Z in the Notch-dependent gene expression program. To reach this goal, we first defined Notch target genes as those genes upregulated upon activation of the Notch pathway using two different systems: as first, we developed a Notch-dependent dynamic system by expressing the NICD, fused to the estrogen receptor ligand binding domain (NICD-ER), in our MT cells (scheme in Figure 2A); as a second approach we overexpressed, in MT cells expressing the bacterial biotin ligase BirA, a Bio-tagged NICD wildtype (BioNICD) or an activation-deficient NICD mutant (BioNICD ΔEP) that no longer interacts with the histone acetyltransferase p300 (13). The NICD-ER overexpressing cells were treated with 4-hydroxytamoxifen (4-OHT) to induce the nuclear translocation of the NICD and RNA-Seq analysis were performed in duplicate (Supplementary Figure S2A-B and statistics in Supplementary Table S2) similarly to the BioNICD wildtype and ΔEP mutant overexpressing cells (Supplementary Figure S2C-D and statistics in Supplementary Table S2). By using these two systems we were able to identify 120 genes (log2 Fold Change > 1.5 and Padj < 0.05) that were induced by the Notch pathway [Supplementary Figure S2E (overlapping P-value = 1e–137, odds ratio = 109.1) and Supplementary Table S1]. We compared our set of 120 Notch target genes to a previously published set of Notch-dependent genes observed as downregulated upon inhibition of the Notch signaling pathway in mouse T6E cells (67). Of the 120 Notch target genes identified in our system, seven genes overlapped with the 72 genes of the reference dataset (Ifi202b, Hes1, Pdgfrb, Cd82, Hey1, Isg15 and Dtx1), which was considerably more than expected by chance (odds ratio = 12.4, hypergeometric P-value = 3.9e–06). These data were also validated by qPCR analysis (Supplementary Figure S2F and G). It must be noticed that Lgmn was significantly upregulated in the RNA-Seq analysis of the BioNICD system but not of the NICD-ER system however, it was detected as upregulated in qPCR using both systems. As a next step, we performed RNA-Seq analysis in H2A.Z depleted MT cells (Supplementary Figure S3A and B and statistics in Supplementary Table S2). Using a log2 Fold Change > 1.5 and Padj < 0.05, we observed 21 Notch target genes (e.g. induced in both the NICD-ER and BioNICD system) that are upregulated upon H2A.Z depletion (Figure 1C and Supplementary Figure S3C). The Venn diagram presented in Supplementary Figure S3C (upper) shows a significant overlap between Notch target genes and genes upregulated upon H2A.Z depletion (Overlapping P-value = 6.2e–15, odds ratio = 11.8). There is also an overlap between Notch target genes and genes downregulated upon H2A.Z depletion (bottom, nine genes; overlapping P-value = 8.2e–04, odds ratio = 3.9). Given that well described canonical Notch target genes such as Nrarp, Hes1 and Il2ra are upregulated upon H2A.Z depletion, these data suggest that H2A.Z plays a negative role in the regulation of the canonical Notch signaling pathway. The genes upregulated by Notch signaling but downregulated upon H2A.Z depletion may represent indirect targets. Moreover, Notch target genes show on average a modest but significant tendency of being upregulated in H2A.Z depleted cells (Supplementary Figure S3D, Wilcoxon rank sum test = 0.04384). The downregulation of H2afv and H2afz was also observed in the RNA-Seq experiments (Supplementary Figure S3E) and we further validated the RNA-Seq results by qPCR (Supplementary Figure S3F). We subsequently focused our attention on the enhancers of Notch target genes that were identified making use of previously published ChIP-Seq data versus RBP-J and Notch1 (63). In agreement with our RNA-Seq analysis, we observed increase of H3K4me2 and H3K27ac at the enhancers of Il2ra and Hes1 upon H2A.Z depletion (Figure 1D). Furthermore, H2A.Z depletion in MT cells does not influence cell growth, cell cycle or apoptosis (Supplementary Figure S3G–J). We also validated the RNA-Seq results by using shRNA-mediated knockdown in MT cells, for which we observed upregulation of Hes1 and Il2ra upon loss-of-function of H2A.Z (Supplementary Figure S4A and B). In a constitutively Notch active pre-T cell line (15), Hey1 and Uaca Notch target genes are upregulated upon shRNA-mediated H2A.Z depletion (Supplementary Figure S5A and B). In agreement with previous reports that showed H2A.Z occupancy at enhancers (29,30,68), H2A.Z and its acetylated form (H2A.Zac) are enriched at the enhancers of both Hey1 and Uaca Notch target genes in pre-T cells (Supplementary Figure S5C and D), suggesting a direct role of H2A.Z in the observed regulation. Scanning of the Hey1 locus in pre-T cells showed specific enrichment of H2A.Z, H2A.Zac and of the H2A.Z loader p400 at the Notch/RBP-J-bound enhancers (Supplementary Figure S5D). Altogether, these data support a negative role for H2A.Z in the regulation of Notch target genes.
Figure 1.
Histone variant H2A.Z has a negative impact on the expression of Notch target genes. (A) Histone Variant H2A.Z is efficiently depleted by CRISPR/Cas9 in MT cells. Whole Cell Extract (WCE) was prepared from wildtype (Control) or H2A.Z depleted (clones sgH2afv/H2afz #12 and sgH2afv/H2afz #20) MT cells and analysed by Western blotting. GAPDH was used as loading control. (B) Hes1 and Il2ra Notch target genes are upregulated upon depletion of H2A.Z. Total RNA from wildtype (Control) or H2A.Z depleted (clones sgH2afv/H2afz #12 and sgH2afv/H2afz #20) MT cells was reverse transcribed into cDNA and analysed by qPCR using primers specific for Tbp, Hes1 or Il2ra. Data were normalized to the housekeeping gene GusB (glucuronidase β). Shown is the mean ± SD of five independent experiments ([*] P < 0.05, [***] P < 0.001, unpaired Student's t-test). (C) Heat map showing deregulation of gene expression as log2 Fold Change upon treatment of NICD-ER overexpressing MT cells with (Z)-4-hydroxytamoxifen (4-OHT) versus EtOH control (NICD-ER), overexpression of BioNICD wildtype versus BioNICD ΔEP mutant (BioNICD) in MT cells or H2A.Z depletion (clone sgH2afv/H2afz #12) versus wildtype control (sgH2afv/H2afz) in MT cells for those genes selected as being Notch targets (padj < 0.05 and log2 Fold Change > 1.5). (D) H3K4me2 and H3K27ac increase in MT cells depleted for H2A.Z. Wildtype (Control) or H2A.Z depleted (clone sgH2afv/H2afz #12) MT cells were subjected to ChIP analysis using antibodies against H3K4me2, H3K27ac, H3 or IgG as control. The qPCR analysis was focused at the Notch-dependent enhancers (red squares) represented on the left (Hes1 +0.6 kb and Il2ra -26 kb). Hes5 0 kb was used as negative control (Control). Data were normalized to the positive control (GAPDH 0 kb) and, in the case of H3K4me2 and H3K27ac, further normalized to H3. Shown is the mean ± SD of four independent experiments.
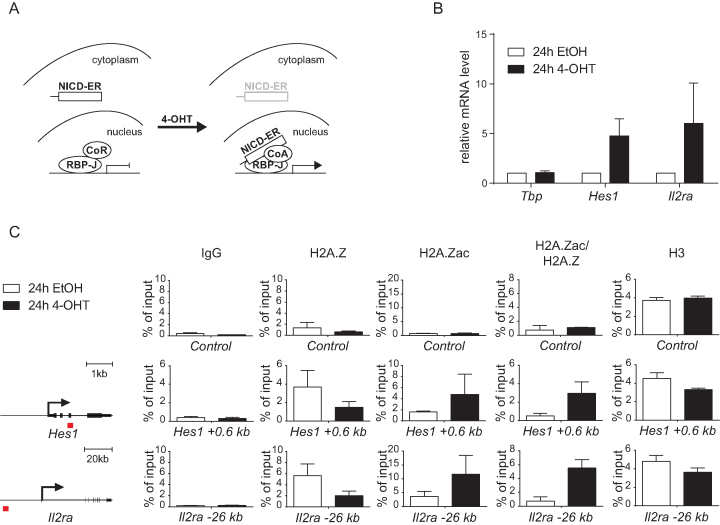
Figure 2.
H2A.Z acetylation (H2A.Zac) but not H2A.Z occupancy positively correlates with activation of Notch target genes. (A) Schematic representation of the NICD-inducible system established in MT cells. The NICD was fused to the estrogen receptor binding domain (NICD-ER) and retrovirally introduced into MT cells. The NICD-ER fusion protein is retained into the cytoplasm unless cells are treated with (Z)-4-hydroxytamoxifen (4-OHT) that induces its nuclear translocation and activation of Notch target genes. (B) Hes1 and Il2ra Notch target genes are induced upon 4-OHT treatment of MT NICD-ER cells. Total RNA from MT NICD-ER cells, treated for 24 h with 4-OHT or EtOH as control, was reverse transcribed into cDNA and analyzed by qPCR using primers specific for Tbp, Hes1 or Il2ra. Data were normalized to the housekeeping gene GusB (glucuronidase β). Shown is the mean ± SD of three independent experiments. (C) H2A.Z acetylation (H2A.Zac) but not H2A.Z occupancy positively correlates with activation of Notch target genes. MT NICD-ER cells were treated for 24 h with 4-OHT or EtOH as control and subjected to ChIP analysis using antibodies against H2A.Z, H2A.Zac, H3 or IgG as control. The qPCR analysis was focused at the Notch-dependent enhancers (red squares) represented on the left (Hes1 +0.6 kb and Il2ra -26 kb). Chrom X was used as negative control (Control). Data were normalized to the positive control (GAPDH 0 kb) and, in the case of H2A.Zac/H2A.Z, the H2A.Zac signals were further normalized to H2A.Z. Shown is the mean ± SD of two independent experiments.
To investigate whether Notch signaling regulates deposition and acetylation of H2A.Z, we used our NICD-ER dynamic system in MT cells (scheme in Figure 2A). Induction of Notch target genes Hes1, Il2ra, Hey1 and Dtx1 was promptly observed upon 4-OHT-induced nuclear translocation of NICD-ER (Figure 2B and Supplementary Figure S6A). By chromatin immunoprecipitation (ChIP) we observed that H2A.Z occupancy negatively correlates with expression of Notch target genes (Figure 2C and Supplementary Figure S6B). Inversely, H2A.Zac positively correlates with expression of Notch target genes (Figure 2C and Supplementary Figure S6B). Nucleosome occupancy was hardly affected, as revealed by pan-H3 ChIP (Figure 2C and Supplementary Figure S6B). Thus, in the active state, total H2A.Z levels are low but levels of acetylated H2A.Z are strongly elevated agreeing with previous reports (28,36).
The TRRAP/p400/Tip60 complex interacts with the RBP-J/Notch complex
Several groups including ours have previously identified components of the RBP-J/NICD coactivator complex (13,17,69,70). Previously, we have purified a Bio-tagged NICD wildtype (NICD) protein and by mass-spectrometry analysis we identified new coactivator components in pre-T cells; among them we have identified and previously described the RNA helicase Ddx5 (49). Here, we also used the activation-deficient BioNICD ΔEP mutant that no longer interacts with the histone acetyltransferase p300 (13). Given that the deletion of the EP domain does not compromise the DNA binding ability of the RBP-J/NICD complex but only its coactivating function (13), we reasoned that the NICD ΔEP mutant may allow us to identify transient NICD interactors. In addition to known coactivator components RBP-J, MAML1, Snw1/SKiP, IKZF1/Ikaros and Ddx5 (49,70–73) (Figure 3A and Supplementary Table S3), we identified several components of the TRRAP/p400/Tip60 complex [(46,53,74–76) and Supplementary Figure S7A], namely p400, TRRAP, Brd8, Vps72/YL-1, Tip60 and several additional subunits. In contrast to the MAML1 coactivator, the TRRAP/p400/Tip60 complex associates more tightly with the NICD ΔEP mutant than with NICD wildtype (Supplementary Table S3), suggesting that this interaction is rather transient under normal conditions. Of note, p400 was previously linked to the loading of H2A.Z within the chromatin (23). suggesting that loading of H2A.Z at Notch target genes is possibly mediated by the TRRAP/p400/Tip60 complex.
Figure 3.
The TRRAP/p400/Tip60 complex interacts with the RBP-J/NICD complex and dynamically localizes at the enhancers of Notch target genes. (A) The TRRAP/p400/Tip60 complex copurifies with NICD. A biotin-tagged hypoactive mutant of the Notch Intracellular Domain (BioNICD ΔEP) was retrovirally introduced in pre-T cells overexpressing the biotin ligase BirA. Upon a single affinity purification with streptavidin beads, mass-spectrometry analysis was performed to identify the interactors of the NICD ΔEP mutant. Notch1 was efficiently recovered as well as the known NICD interactors RBP-J and Maml1. Additionally, Ep400/p400, TRRAP, Brd8 and Kat5/Tip60, subunits of the TRRAP/p400/Tip60 complex, were copurified with NICD ΔEP. Shown is the number of total peptides. The complete list of the BioNICD wildtype (NICD) and ΔEP mutant interactomes is shown in Supplementary Table S3. (B and C) NICD interacts with Tip60 and p400. PhoenixTM cells were transfected with Flag-tagged Tip60 (B) or Flag-tagged p400 (C) together with HA-tagged NICD wildtype or ΔEP mutant. WCE were incubated with Flag antibody and immunoprecipitates were analysed by Western blotting using HA or Flag antibodies. The asterisks indicate unspecific bands. (D) RBP-J directly interacts with p400. GST pull-down experiments were performed using bacterially purified GST-RBP-J and the in vitro transcribed/translated p400 fragments depicted in the schematic representation. Amino acid numbering is accordingly to accession NP_056224.3. p400 domains: HSA, domain in helicases and associated with SANT domains (CDD:214727); DEXDc, DEAD-like helicases superfamily (CDD:238005); HELICc, helicase superfamily C-terminal domain (CDD:238034). (E) p400 dynamically localizes at the enhancers of Hes1 and Il2ra Notch target genes. MT NICD-ER cells were treated for 24h with 4-OHT or EtOH as control and subjected to ChIP analysis using an antibody against p400 or IgG as control. Chrom X was used as negative control (Control). The qPCR analysis was focused at the Notch-dependent enhancers (red squares) represented on the left (Hes1 +0.6 kb and Il2ra -26 kb). Data were normalized to the positive control (GAPDH 0 kb). Shown is the mean ± SD of two independent experiments.
Subsequently, we validated the interaction using co-immunoprecipitation experiments in fibroblasts overexpressing Flag-tagged Tip60 or p400 together with HA-tagged NICD wildtype (NICD) or ΔEP mutant (Figure 3B and C). Flag-affinity purification of Flag-Tip60 (Figure 3B) or Flag-p400 (Figure 3C) co-purified HA-NICD and HA-NICD ΔEP mutant; again the interaction of NICD ΔEP mutant was stronger than NICD wildtype with both Tip60 (Figure 3B, compare lane 3 and 5) and p400 (Figure 3C, compare lanes 3 and 5). Additionally, when we overexpressed a Bio-tagged Tip60 in pre-T cells and performed a streptavidin immunoprecipitation, we were able to copurify the endogenous cleaved Notch (NICD) and RBP-J (Supplementary Figure S7B). In line with the physical interaction between the RBP-J/NICD complex and the TRRAP/p400/Tip60 complex, we observed colocalization of RBP-J, NICD, p400 and Tip60 at the Notch-dependent enhancers of Notch target genes Hey1 and Uaca in pre-T cells (Supplementary Figure S7C). Given the remarkably high number of p400 peptides recovered in our mass-spectrometry analysis, we hypothesized that p400 directly interacts with the RBP-J/NICD complex. To test this hypothesis, we divided the 400 kDa p400 protein in three partially overlapping fragments and used them in GST pulldown assays with a bacterially purified GST-RBP-J protein (Figure 3D). We could observe that fragments p400–1 and p400–2 but not p400–3 directly interact with RBP-J (Figure 3D), suggesting not only that RBP-J and p400 directly interact each other but also that this interaction is mediated by more individual domains within p400. Additionally, we could observe that p400, RBP-J and NICD form a complex in GST PD assays (Supplementary Figure S7D).
In order to investigate the Notch-dependent recruitment of p400, we used ChIP in our Notch-inducible system in MT cells. p400 is found at Notch-dependent enhancers prior to activation and its occupancy increases upon induction of Notch signaling (Figure 3E and Supplementary Figure S8A), suggesting that the NICD stabilizes the RBP-J/p400 interaction. In line with this, we also observed a direct interaction between p400 and NICD (Supplementary Figure S8B). As next, we investigated the RBP-J dependency of the H2A.Z loading by overexpressing Bio-tagged RBP-J in MT cells (Supplementary Figure S8C). We observed that BioRBP-J overexpression leads to reduced H2A.Z occupancy at the enhancers of Notch target genes (Supplementary Figure S8D), suggesting that an excess of RBP-J sequesters p400, inhibiting its capacity to load H2A.Z. Altogether, the biochemical studies on the RBP-J/NICD/p400/Tip60 interaction and their colocalization at Notch-dependent enhancers demonstrate that the TRRAP/p400/Tip60 complex is recruited to Notch target genes prior to Notch induction and suggests p400 involved in the loading of H2A.Z at Notch target genes. The observation that p400 occupancy at enhancers increases upon activation of Notch signaling and that NICD and p400 directly interact with each other suggest that the NICD stabilizes the p400/RBP-J interaction.
H2A.Z acetylation correlates with activation of Notch target genes
To further explore the role of H2A.Z in the regulation of Notch target genes, we focused our attention on its acetylation. In yeast and Drosophila, acetylation of H2A.Z is mediated by the histone acetyltransferase Tip60 (35,77,78), which is a component of the TRRAP/p400/Tip60 complex and is enriched at active enhancer sites (79). In order to evaluate whether Tip60 is responsible for the acetylation of H2A.Z in higher eukaryotes and to evaluate the transcriptional consequences of the Tip60-mediated acetylation of H2A.Z in the context of Notch signaling, we directly fused the transcription factor RBP-J to Tip60 wildtype (wt) or its catalytic-dead mutant [(cd, (53) and Figure 4A]. Wildtype (wt) and catalytic-dead (cd) RBP-J/Tip60 fusion proteins are both expressed in HeLa cells and localize in the nucleus (Supplementary Figure S9A). Furthermore, both RBP-J/Tip60-wt and RBP-J/Tip60-cd mutant interact with NICD and with the RBP-J interactor RITA (52) similarly to RBP-J (Supplementary Figure S9B), demonstrating functionality of both fusion proteins. When RBP-J/Tip60-wt and RBP-J/Tip60-cd mutant were expressed in MT cells (Supplementary Figure S10A), we observed strong upregulation of Notch target genes by RBP-J/Tip60-wt but a milder to absent upregulation by RBP-J/Tip60-cd mutant (Figure 4B and Supplementary Figure S10A). Furthermore, the RBP-J/Tip60 fusion protein leads to increased H2A.Zac at the enhancers of Notch target genes (Figure 4C and Supplementary Figure S10B) but not in the active mark H3K18ac (Supplementary Figure S10C) which is regulated by the histone acetyltransferase p300. This data suggest that Tip60 specifically acetylates H2A.Z and that this acetylation stimulates expression of Notch target genes.
Figure 4.
Acetylation of H2A.Z is required for upregulation of Hes1 Notch target gene. (A) Schematic representation of the Flag-RBP-J/Tip60 fusion proteins used in Figure 4B and C and in Supplementary Figures S9 and S10. Amino acid numbering is accordingly to accession NP_033061.3 for RBP-J and NP_874368.1 for Tip60. RBP-J domain: LAG1-DNAbind, LAG1 DNA binding (CDD:255260); Tip60 domain: MOZ/SAS, MOZ/SAS family (CDD:250916). (B) RBP-J/Tip60 wildtype (wt) but not its catalytic dead (cd) mutant upregulates Hes1 expression in MT cells. MT cells were infected with retroviral particles delivering plasmids encoding Flag-tagged RBP-J/Tip60-wt, cd mutant or empty vector (Control). Total RNA was reverse transcribed into cDNA and analysed by qPCR using primers specific for Tbp or Hes1. Data were normalized to the housekeeping gene GusB (glucuronidase β). Shown is the mean ± SD ([**] P < 0.01, [***] P < 0.001, unpaired Student's t-test). (C) RBP-J/Tip60 fusion protein leads to increased acetylation of H2A.Z. MT RBP-J/Tip60−wt or control cells were subjected to ChIP analysis versus H2A.Zac, H3 or IgG as control. The qPCR analysis was focused at the Notch-dependent enhancer within the Hes1 gene (red square) represented on the left (Hes1 +0.6 kb). Hes5 0 kb was used as negative control (Control). Shown is the mean ± SD of raw data from three independent experiments. (D) Schematic representation of the lysine residues that are known to be acetylated within the histone variant H2A.Z. (E) H2A.Z wildtype (wt) and acetylation defective mutant (5KR) are expressed in MT cells and enriched in the chromatin-bound fraction. MT cells were infected with retroviral particles delivering plasmids encoding Flag-tagged H2A.Z-wt, 5KR mutant or empty vector (Control). Cells were subjected to fractionation to purify the cytosolic, nuclear or chromatin-bound proteins and analysed by Western blotting using antibodies against Flag, H3, TBP or GAPDH. (F) H2A.Z-wt but not the 5KR mutant leads to upregulation of Hes1 in MT cells. Total RNA from Flag-H2A.Z-wt, 5KR mutant or control MT cells was reverse transcribed into cDNA and analysed by qPCR using primers specific for Tbp or Hes1. Data were normalized to the housekeeping gene GusB (glucuronidase β). Shown is the mean ± SD of two independent experiments measured twice each ([***] P < 0.001, unpaired Student's t-test).
In order to further define the requirement of H2A.Zac for the regulation of Notch target genes, we overexpressed in MT cells H2A.Z wildtype (H2A.Z-wt) or an acetylation-defective mutant where the five lysine residues known to be acetylated [(31,38–40) and Figure 4D] have been mutated to arginine residues (H2A.Z-5KR). Both proteins are efficiently expressed and enriched in the chromatin fraction (Figure 4E). While H2A.Z-wt leads to upregulation of Hes1, the upregulation given by H2A.Z-5KR is only minor (Figure 4F), suggesting again that acetylation of H2A.Z has a stimulatory function in the expression of Notch target gene Hes1. Additionally, when we investigated the chromatin occupancy of H2A.Z-wt and H2A.Z-5KR by Flag ChIP, we noticed that H2A.Z-wt is more enriched in the chromatin compared to the H2A.Z-5KR mutant (Supplementary Figure S10D), suggesting that acetylation of H2A.Z may influence its chromatin incorporation and/or stability.
Altogether, our data support a role for Tip60 in regulating the acetylation of histone variant H2A.Z and link H2A.Zac to active Notch target gene expression.
Depletion of the Drosophila melanogaster homologs of Tip60, p400 and H2A.Z enhance Notch-mediated tissue growth in vivo
Next, we investigated whether the function of the Drosophila melanogaster homologs of p400, Tip60 and H2A.Z within the Notch pathway, is conserved. The homolog of p400 in D. melanogaster is called Domino and we observed that the region corresponding to the RBP-J-interacting p400–2 fragment is well conserved within the Domino protein (Supplementary Figure S11). Thus, we tested whether this region of Domino interacts with the Drosophila homolog of RBP-J, Suppressor of Hairless [Su(H)]. In GST PD assays, we observed a physical interaction between Su(H) and Domino (Figure 5A). We next tested genetic interactions of Notch with domino/p400, histone variant His2Av/H2A.Z and dTip60 in vivo using the growing eye imaginal disc of Drosophila, whose growth critically depends on the Notch signaling pathway (80–82). Changes in eye size are induced by spatial and temporal activation or repression of Notch signaling using the eye-specific driver eyeless-Gal4 [hereafter referred to as ey-Gal4, (83)] in combination with overexpression of the Notch ligand Delta (Dl) by ey-Gal4 (hereafter referred to as ey-Gal4 UAS-Dl), which results in mild overgrowth of the ventral region of the eye [(81) and Supplementary Figure S12] and has been used as a sensitive genetic background to identify genes that cooperate with hyperactivated Notch signaling activity in converting Delta-mediated mild tissue overgrowth into tumor growth (84,85). RNAi-mediated depletion of dTip60, domino/p400 or His2Av/H2A.Z (qPCR control of RNAi efficiency in Supplementary Figure S13) induced an overall smaller eye size and necrosis, indicating a requirement for these factors in eye cell proliferation and/or viability (Supplementary Figure S12B). Surprisingly, combination of Delta overexpression with depletion of any of these factors resulted in marked reduction in the ventral region, where cells are highly responsive to Delta signals, and hyperplasia or tumor in the dorsal region (50%, 80% and 48.6% penetrance for His2Av/H2A.Z, dTip60 and domino/p400, respectively) where cells are normally insensitive to Delta overexpression (Supplementary Figure S12A and C and statistics in Supplementary Figure S12D). To substantiate these results, we next assessed the effect on Delta-dependent Notch signaling in marked clones in another developmental paradigm, the wing disc. In this disc, Delta is a ventral to dorsal signal [(86) and scheme in Figure 5B left]. Delta and Serrate activate Notch signaling and target the gene wingless (wg/Wnt) in a dorsal/ventral (d/v) stripe of cells along the prospective wing margin. We found that clones with depleted expression of any of His2Av/H2A.Z, dTip60 or domino/p400 could be recovered in both dorsal and ventral regions; however, clones were less prominent along the d/v boundary, the region with high Notch activity. Mutant cells in this domain lacked or had reduced expression of Wg, resulting, in the case of dTip60-RNAi clones, in a thinner stripe of Wg (compare Figure 5C’ with 5G’), indicating a requirement for this factor in Notch receiving cells. Double clones overexpressing Delta along with RNAi-based depletion of any of these factors revealed absolute requirement of them for wing cell proliferation or viability as clones could not be recovered or very infrequently in the dorsal wing pouch, a region responsive to Delta signaling, while ventral clones were recovered and displayed overgrowth compared with single Delta overexpressing clones (Figure 5F, H and J compared to Figure 5E, G and I, respectively). Likewise, we found that size of the dorsal wing pouch in respect to the overall wing disc area is much reduced in the double mosaic discs (Figure 5B right): 0.1518 ± 0.009 (n = 12), 0.1553 ± 0.007 (n = 12) and 0.1515 ± 0.006 (n = 12) for His2Av/H2A.Z-RNAi, dTip60-RNAi and domino/p400-RNAi respectively, compared to 0.2014 ± 0.011 (n = 12) of single Delta-overexpressing clones (arbitrary units).
In view of these interactions, we revisited the publicly available RNA-Seq data that investigated differential gene expression in Drosophila dissected late L3 wings expressing Tip60 wildtype (wt) or Tip60 dominant negative (dn) mutant (87). Gene Set Enrichment Analysis (GSEA) of this RNA-Seq study unveiled downregulation by Tip60-dn of two different GO terms associated with Notch signaling (Supplementary Figure S14 and Supplementary Table S4; GO:0007219, P = 0.002058 and Padj = 0.0052; GO:0008593, P = 0.00209205 and Padj = 0.0052). Among the genes associated with these GO terms, we found Hairless (H), which is the main negative regulator of Notch signaling in Drosophila (88–90) and smoothened (smo), encoding for an essential component of the Hedgehog signaling pathway and which we recently found to negatively regulate Notch signaling response (91). These observations suggest that dTip60 might negatively regulate Notch signaling response in part by transcriptionally regulate the expression of negative regulators of Delta-Notch signaling. Additionally, Tip60-dn leads to downregulation of CoREST that was previously described as another negative regulator of Notch signaling (92,93) as well as dishevelled (dsh), which encodes for a component of the Wnt signaling pathway that downregulates Notch signaling in some cellular contexts (94–96). To note, also krz and dx, which protein products form a complex involved in the degradation of the Notch receptor (97), are downregulated in the Tip60-dn. Altogether with our functional analysis in vivo these data support that the TRRAP/p400/Tip60 complex might regulate cell proliferation through regulation of Notch signaling at multiple nodal points (98). Depletion of dTip60 and domino/p400 in the wing discs also supports a role for these factors in physiological high Notch signaling for target gene expression and/or viability, which is consistent with our results in mouse T cells. Thus, this strongly suggests a conserved role for H2A.Z/His2Av in Notch signaling and Notch-mediated growth control in vivo.
DISCUSSION
Here we show that the CRISPR/Cas9 system can be efficiently used to deplete the histone variant H2A.Z in murine mature T cells. Either CRISPR/Cas9-mediated depletion or knockdown of H2A.Z increase expression of Notch target genes, suggesting that H2A.Z acts as a negative regulator that buffers transcriptional activation. Previously, H2A.Z was shown to be either a positive (26,27) or a negative regulator (23) of gene expression but how this works mechanistically remained elusive. Our data strongly suggest that post-translational modifications of H2A.Z are at least predictive or might even direct the transcriptional response. H2A.Z per se prevents expression of Notch target genes but, once acetylated, it supports gene expression. These conclusions are supported by the increased acetylation of H2A.Z upon Notch activation and by the RBP-J/Tip60 fusion protein that leads to increased H2A.Zac and gene expression. The upregulation of Hes1 observed upon overexpression of H2A.Z-wt but, to a lesser extent by the H2A.Z-5KR acetylation-defective mutant, is also in line with this hypothesis. In summary, our data support a model where the non-acetylated H2A.Z negatively correlates with Notch target gene expression, while H2A.Zac positively correlates with expression of Notch-dependent genes (Figure 6). This is in line with data previously reported in different systems showing that H2A.Zac correlates with active gene expression (28–30). Our observation that H2A.Z is reduced upon Notch activation is in line with data in D. melanogaster showing a reduced occupancy of His2Av, the homolog of H2A.Z, upon Notch activation (99), marking the evolutionary conservation of the H2A.Z function in regard to Notch signaling. It still remains unclear how H2A.Z is evicted from the chromatin upon gene activation. However, two hypotheses could explain this: (a) a passive, charge-repulsion mechanism could take place since the acetylation of H2A.Z has a destabilizing effect; alternatively (b) it might require an active mechanism that involves enzymatic activities aimed to remove or replace the H2A.Z/H2B dimer. Of note, nucleosome occupancy does not change upon Notch activation, suggesting that the H2A.Z eviction most likely does not involve the whole nucleosome but just the H2A.Z/H2B dimer. The observation that the H2A.Z-5KR acetylation-defective mutant is still able to upregulate gene expression, though to a lower extent compared to the H2A.Z-wt, suggests that additional lysine residues are acetylated and these might have a minor impact on transcription compared to the acetylation of the lysines in position 4, 7, 11, 13 and 15 which are mutated in our H2A.Z-5KR construct. Alternatively, additional post-translational modifications, not yet identified, of H2A.Z might be responsible for this phenotype. For example, ubiquitination and methylation of H2A.Z have been linked to gene repression (33,100). We cannot exclude that the occurrence of these modifications at different residues may be involved in gene activation. The observation that H2A.Z-wt is more enriched on the chromatin compared to the H2A.Z-5KR mutant suggests that H2A.Z acetylation may be required for proper chromatin incorporation of this histone variant and/or that H2A.Z acetylation may regulate its stability.
Figure 6.
Schematic summary representing the link between H2A.Z and Notch signaling. In absence of Notch signaling (OFF, left side), H2A.Z occupancy increases while its acetylation (H2A.Zac) is low. Upon Notch activation (ON, right side), H2A.Z occupancy is reduced while its acetylation increases.
p400, the homolog of the yeast Eaf1 and Swr1 (53,74) is responsible of the H2A.Z loading (23,101,102). Thus, the identification of the TRRAP/p400/Tip60 complex as an interactor of the RBP-J/NICD complex suggests p400 as the H2A.Z loader at Notch target genes. Our ChIP results looking at the H2A.Z loading machinery, TRRAP/p400/Tip60 complex, show that its occupancy increases upon Notch activation. So far, we can only speculate how this stabilization occurs. It is possible that the NICD, together with other coactivators, form an additional interaction surface for p400, as suggested by our biochemical data. However, we cannot exclude that the RBP-J/NICD complex interacts with other complex components. It is also possible that additional transcription factors, such as Ets factors (63), are needed to form an enhanceosome structure. Strikingly, we observe increased binding of this complex to a mutant NICD complex that recruits coactivators MAML and p300 less efficiently (13). We speculate that the TRRAP/p400/Tip60 interaction with the RBP-J/NICD complex is rather transient and relies on cumulative protein–protein interactions. Such transient intermediates are important for amplitude and duration of the Notch response and as consequence for the biological output. Our data using RNAi-mediated knockdown of Drosophila His2Av/H2A.Z, dTip60, and domino/p400 targeted to the developing eye and wing with or without Delta-driven Notch hyperactivation underline the in vivo genetic interaction between His2Av/H2A.Z, dTip60 and domino/p400 in Notch-dependent signaling. In addition, activation of target gene wg and overgrowth response support the biological relevance of the physical interaction between Domino/p400 and Su(H). The domino/p400- and His2Av/H2A.Z-RNAi data are in line with the mammalian data and they further stress a synthetic lethality between a mild gain of Notch signaling and depletion of components of the TRRAP/p400/Tip60 complex. This lethality suggests that wing cells expressing Delta cannot tolerate the simultaneous depletion of dTip60, which would be expected to lead to extreme Delta-Notch signaling activation and target upregulation. The TRRAP/p400/Tip60 complex can also regulate other processes and other signaling pathways, but the synthetic lethality provides strong argument for functional requirements of these components in the regulation of Delta-Notch signaling. It is also possible that this complex regulates Notch at different steps and also via feedback loops. This last hypothesis is supported by the different spatio-temporal recruitment of Tip60 and p400 observed at the p21 locus (23) and is further supported by the observation that negative Notch modulators, such as H, smo, CoREST, dsh, krz and dx are downregulated by the Tip60-dn mutant in Drosophila late L3 wings (87). As consequence, Tip60 may play several roles in the regulation of the Notch pathway: it might regulate Notch modulators and in addition directly impact on Notch/RBP-J-dependent genes. Our tethering approach using the RBP-J/Tip60 fusion allows to discriminate between these two functions of Tip60 focusing exclusively on the role of Tip60 in the regulation of Notch/RBP-J-dependent genes. Additionally, domino/p400, dTip60 as well as Nipped-A, the Drosophila homolog encoding for TRRAP, were previously identified as Notch regulators (44,98).
We found that murine p400 directly interacts with RBP-J leading to H2A.Z loading prior to Notch activation and that the acetylation of H2A.Z is mediated by the acetyltransferase Tip60, in line with previous studies in Drosophila (77) and yeast (35,78). To increase the local concentration of Tip60 directly at Notch-dependent enhancers, we fused the transcription factor RBP-J with the acetyltransferase Tip60. Tip60-mediated acetylation of H2A.Z supports gene activation which is in line with our model that H2A.Z acetylation is stimulatory. So far, we have not addressed the role of additional subunits of the TRRAP/p400/Tip60 complex. Since genome-wide occupancy of Brd2 was previously linked to H2A.Z occupancy (103,104) and BRD inhibitor JQ1 has effects on Notch-dependent leukemia (105), it remains to be investigated whether the Brd8 subunit of the TRRAP/p400/Tip60 complex is involved in the regulation of Notch target genes.
In summary, our data reveal that the histone variant H2A.Z, thought to be a generic factor, is not essential for cell viability. While repression of Notch target genes is associated with increased occupancy of H2A.Z but reduced H2A.Zac (Figure 6, left), activation is accompanied by reduced H2A.Z occupancy and increased H2A.Zac (Figure 6, right).
DATA AVAILABILITY
RNA-Seq data have been deposited at GEO under the accession number GSE111029. Mass-spectrometry data are available via ProteomeXchange with identifier PXD008747.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to M. Krötschel, S. Fietzeck, C. Grubisic, P. Käse and T. Schmidt-Wöll for excellent technical assistance. We thank Drs D. van Essen and S. Saccani for providing us with protocols and plasmids, Dr J. Côté, Dr R. Roeder, Dr O. Binda, Dr J. Meier-Soelch, Dr M. Kracht, Dr E. Surova and Dr H. Jumaa for providing us with plasmids. We want to thank Drs V. Di Cerbo and R. Schneider for providing us with protocols. We want to thank J. Bespalowa, S. Jarmer, A. Würch and S. Hobitz for excellent FACS sorting.
B.D.G., F.F., D.M.V., K.H., I.G.P., P.S. and F.O. performed experiments. B.D.G. and T.B. designed experiments, supervised the work and wrote the manuscript with contributions from other authors. D.M.V. and M.D. designed the Drosophila in vivo experiments. A.N. and T.S. performed deep-sequencing. G.M. performed the mass-spectrometry. T.Z. and M.B. performed the bioinformatics analysis. S.H. performed FACS sorting.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the collaborative research grant TRR81 and the Heisenberg program (BO 1639/5–1) by the DFG (German Research Foundation), the Max Planck Society and the EXC 294 in Freiburg and the Excellence Cluster for Cardio Pulmonary System (ECCPS) in Giessen to T.B and S.H. This work was further supported by the DFG through collaborative research grants (SFB 1074/A3 and KFO309 service project Z1) and by the BMBF (Federal Ministry of Education and Research, research nucleus SyStAR) to F.O. Spanish Grants [BFU2015–64239-R co-financed by the European Regional Development Fund (ERDF), SEV-2013–0317, PROMETEO/2017/146]; Botin Foundation and AECC Foundation (CICP16001DOMI) to M.D. Funding for open access charge: DFG collaborative research [TRR81]. This study was supported in part by the Excellence Initiative of the German Federal and State Governments [GSC-4, Spemann Graduate School].
Conflict of interest statement. None declared.
REFERENCES
- 1. Artavanis-Tsakonas S., Rand M.D., Lake R.J.. Notch signaling: cell fate control and signal integration in development. Science. 1999; 284:770–776. [DOI] [PubMed] [Google Scholar]
- 2. Kopan R., Ilagan M.X.. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009; 137:216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bray S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016; 17:722–735. [DOI] [PubMed] [Google Scholar]
- 4. Weng A.P., Ferrando A.A., Lee W., Morris J.P. 4th, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C.. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004; 306:269–271. [DOI] [PubMed] [Google Scholar]
- 5. Puente X.S., Pinyol M., Quesada V., Conde L., Ordonez G.R., Villamor N., Escaramis G., Jares P., Bea S., Gonzalez-Diaz M. et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011; 475:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kulic I., Robertson G., Chang L., Baker J.H., Lockwood W.W., Mok W., Fuller M., Fournier M., Wong N., Chou V. et al. Loss of the Notch effector RBPJ promotes tumorigenesis. J. Exp. Med. 2015; 212:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thiel V.N., Giaimo B.D., Schwarz P., Soller K., Vas V., Bartkuhn M., Blatte T.J., Dohner K., Bullinger L., Borggrefe T. et al. Heterodimerization of AML1/ETO with CBFbeta is required for leukemogenesis but not for myeloproliferation. Leukemia. 2017; 31:2491–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borggrefe T., Lauth M., Zwijsen A., Huylebroeck D., Oswald F., Giaimo B.D.. The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFbeta/BMP and hypoxia pathways. Biochim. Biophys. Acta. 2016; 1863:303–313. [DOI] [PubMed] [Google Scholar]
- 9. Bray S.J. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006; 7:678–689. [DOI] [PubMed] [Google Scholar]
- 10. Oswald F., Rodriguez P., Giaimo B.D., Antonello Z.A., Mira L., Mittler G., Thiel V.N., Collins K.J., Tabaja N., Cizelsky W. et al. A phospho-dependent mechanism involving NCoR and KMT2D controls a permissive chromatin state at Notch target genes. Nucleic Acids Res. 2016; 44:4703–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giaimo B.D., Oswald F., Borggrefe T.. Dynamic chromatin regulation at Notch target genes. Transcription. 2017; 8:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurooka H., Honjo T.. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 2000; 275:17211–17220. [DOI] [PubMed] [Google Scholar]
- 13. Oswald F., Tauber B., Dobner T., Bourteele S., Kostezka U., Adler G., Liptay S., Schmid R.M.. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 2001; 21:7761–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oswald F., Winkler M., Cao Y., Astrahantseff K., Bourteele S., Knochel W., Borggrefe T.. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 2005; 25:10379–10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liefke R., Oswald F., Alvarado C., Ferres-Marco D., Mittler G., Rodriguez P., Dominguez M., Borggrefe T.. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010; 24:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mulligan P., Yang F., Di Stefano L., Ji J.Y., Ouyang J., Nishikawa J.L., Toiber D., Kulkarni M., Wang Q., Najafi-Shoushtari S.H. et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell. 2011; 42:689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yatim A., Benne C., Sobhian B., Laurent-Chabalier S., Deas O., Judde J.G., Lelievre J.D., Levy Y., Benkirane M.. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol. Cell. 2012; 48:445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu T., Park S.S., Giaimo B.D., Hall D., Ferrante F., Ho D.M., Hori K., Anhezini L., Ertl I., Bartkuhn M. et al. RBPJ/CBF1 interacts with L3MBTL3/MBT1 to promote repression of Notch signaling via histone demethylase KDM1A/LSD1. EMBO J. 2017; 36:3232–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D.. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005; 123:233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li B., Pattenden S.G., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L.. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. PNAS. 2005; 102:18385–18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang H., Roberts D.N., Cairns B.R.. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005; 123:219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farris S.D., Rubio E.D., Moon J.J., Gombert W.M., Nelson B.H., Krumm A.. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J. Biol. Chem. 2005; 280:25298–25303. [DOI] [PubMed] [Google Scholar]
- 23. Gevry N., Chan H.M., Laflamme L., Livingston D.M., Gaudreau L.. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007; 21:1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. John S., Sabo P.J., Johnson T.A., Sung M.H., Biddie S.C., Lightman S.L., Voss T.C., Davis S.R., Meltzer P.S., Stamatoyannopoulos J.A. et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell. 2008; 29:611–624. [DOI] [PubMed] [Google Scholar]
- 25. Sutcliffe E.L., Parish I.A., He Y.Q., Juelich T., Tierney M.L., Rangasamy D., Milburn P.J., Parish C.R., Tremethick D.J., Rao S.. Dynamic histone variant exchange accompanies gene induction in T cells. Mol. Cell. Biol. 2009; 29:1972–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dalvai M., Fleury L., Bellucci L., Kocanova S., Bystricky K.. TIP48/Reptin and H2A.Z requirement for initiating chromatin remodeling in estrogen-activated transcription. PLos Genet. 2013; 9:e1003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gevry N., Hardy S., Jacques P.E., Laflamme L., Svotelis A., Robert F., Gaudreau L.. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009; 23:1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Valdes-Mora F., Song J.Z., Statham A.L., Strbenac D., Robinson M.D., Nair S.S., Patterson K.I., Tremethick D.J., Stirzaker C., Clark S.J.. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 2012; 22:307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu G., Cui K., Northrup D., Liu C., Wang C., Tang Q., Ge K., Levens D., Crane-Robinson C., Zhao K.. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013; 12:180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ku M., Jaffe J.D., Koche R.P., Rheinbay E., Endoh M., Koseki H., Carr S.A., Bernstein B.E.. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 2012; 13:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Law C., Cheung P.. Expression of non-acetylatable H2A.Z in myoblast cells blocks myoblast differentiation through disruption of MyoD expression. J. Biol. Chem. 2015; 290:13234–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sarcinella E., Zuzarte P.C., Lau P.N., Draker R., Cheung P.. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell. Biol. 2007; 27:6457–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Draker R., Sarcinella E., Cheung P.. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 2011; 39:3529–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Halley J.E., Kaplan T., Wang A.Y., Kobor M.S., Rine J.. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 2010; 8:e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Millar C.B., Xu F., Zhang K., Grunstein M.. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006; 20:711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bruce K., Myers F.A., Mantouvalou E., Lefevre P., Greaves I., Bonifer C., Tremethick D.J., Thorne A.W., Crane-Robinson C.. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005; 33:5633–5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Myers F.A., Lefevre P., Mantouvalou E., Bruce K., Lacroix C., Bonifer C., Thorne A.W., Crane-Robinson C.. Developmental activation of the lysozyme gene in chicken macrophage cells is linked to core histone acetylation at its enhancer elements. Nucleic Acids Res. 2006; 34:4025–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ishibashi T., Dryhurst D., Rose K.L., Shabanowitz J., Hunt D.F., Ausio J.. Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry. 2009; 48:5007–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beck H.C., Nielsen E.C., Matthiesen R., Jensen L.H., Sehested M., Finn P., Grauslund M., Hansen A.M., Jensen O.N.. Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteomics: MCP. 2006; 5:1314–1325. [DOI] [PubMed] [Google Scholar]
- 40. Bonenfant D., Coulot M., Towbin H., Schindler P., van Oostrum J.. Characterization of histone H2A and H2B variants and their post-translational modifications by mass spectrometry. Mol. Cell. Proteomics: MCP. 2006; 5:541–552. [DOI] [PubMed] [Google Scholar]
- 41. Chan H.M., Narita M., Lowe S.W., Livingston D.M.. The p400 E1A-associated protein is a novel component of the p53 → p21 senescence pathway. Genes Dev. 2005; 19:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fuchs M., Gerber J., Drapkin R., Sif S., Ikura T., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M.. The p400 complex is an essential E1A transformation target. Cell. 2001; 106:297–307. [DOI] [PubMed] [Google Scholar]
- 43. Eissenberg J.C., Wong M., Chrivia J.C.. Human SRCAP and Drosophila melanogaster DOM are homologs that function in the notch signaling pathway. Mol. Cell. Biol. 2005; 25:6559–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saj A., Arziman Z., Stempfle D., van Belle W., Sauder U., Horn T., Durrenberger M., Paro R., Boutros M., Merdes G.. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev. Cell. 2010; 18:862–876. [DOI] [PubMed] [Google Scholar]
- 45. Gause M., Eissenberg J.C., Macrae A.F., Dorsett M., Misulovin Z., Dorsett D.. Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in Notch signaling during wing development. Mol. Cell. Biol. 2006; 26:2347–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Choi J., Heo K., An W.. Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic Acids Res. 2009; 37:5993–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Giaimo B.D., Ferrante F., Borggrefe T.. Chromatin immunoprecipitation (ChIP) in mouse T-cell lines. J. Visual. Exp: JoVE. 2017; 124:e55907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sanjana N.E., Shalem O., Zhang F.. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014; 11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jung C., Mittler G., Oswald F., Borggrefe T.. RNA helicase Ddx5 and the noncoding RNA SRA act as coactivators in the Notch signaling pathway. Biochim. Biophys. Acta. 2013; 1833:1180–1189. [DOI] [PubMed] [Google Scholar]
- 50. Hein K., Mittler G., Cizelsky W., Kuhl M., Ferrante F., Liefke R., Berger I.M., Just S., Strang J.E., Kestler H.A. et al. Site-specific methylation of Notch1 controls the amplitude and duration of the Notch1 response. Sci.Signal. 2015; 8:ra30. [DOI] [PubMed] [Google Scholar]
- 51. Oswald F., Kostezka U., Astrahantseff K., Bourteele S., Dillinger K., Zechner U., Ludwig L., Wilda M., Hameister H., Knöchel W. et al. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 2002; 21:5417–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wacker S.A., Alvarado C., von Wichert G., Knippschild U., Wiedenmann J., Clauss K., Nienhaus G.U., Hameister H., Baumann B., Borggrefe T. et al. RITA, a novel modulator of Notch signalling, acts via nuclear export of RBP-J. EMBO J. 2011; 30:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doyon Y., Selleck W., Lane W.S., Tan S., Cote J.. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004; 24:1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Park J.H., Sun X.J., Roeder R.G.. The SANT domain of p400 ATPase represses acetyltransferase activity and coactivator function of TIP60 in basal p21 gene expression. Mol. Cell. Biol. 2010; 30:2750–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liao Y., Smyth G.K., Shi W.. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013; 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang H., Zou J., Zhao B., Johannsen E., Ashworth T., Wong H., Pear W.S., Schug J., Blacklow S.C., Arnett K.L. et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. PNAS. 2011; 108:14908–14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Aktas T., Avsar Ilik I., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A.. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017; 544:115–119. [DOI] [PubMed] [Google Scholar]
- 65. Vizcaino J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Weng A.P., Millholland J.M., Yashiro-Ohtani Y., Arcangeli M.L., Lau A., Wai C., Del Bianco C., Rodriguez C.G., Sai H., Tobias J. et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006; 20:2096–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brunelle M., Nordell Markovits A., Rodrigue S., Lupien M., Jacques P.E., Gevry N.. The histone variant H2A.Z is an important regulator of enhancer activity. Nucleic Acids Res. 2015; 43:9742–9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wu L., Sun T., Kobayashi K., Gao P., Griffin J.D.. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol. Cell. Biol. 2002; 22:7688–7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhou S., Fujimuro M., Hsieh J.J., Chen L., Miyamoto A., Weinmaster G., Hayward S.D.. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function. Mol. Cell. Biol. 2000; 20:2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu L., Aster J.C., Blacklow S.C., Lake R., Artavanis-Tsakonas S., Griffin J.D.. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000; 26:484–489. [DOI] [PubMed] [Google Scholar]
- 72. Wilson J.J., Kovall R.A.. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006; 124:985–996. [DOI] [PubMed] [Google Scholar]
- 73. Geimer Le Lay A.S., Oravecz A., Mastio J., Jung C., Marchal P., Ebel C., Dembele D., Jost B., Le Gras S., Thibault C. et al. The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci. Signal. 2014; 7:ra28. [DOI] [PubMed] [Google Scholar]
- 74. Doyon Y., Cote J.. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004; 14:147–154. [DOI] [PubMed] [Google Scholar]
- 75. Cai Y., Jin J., Florens L., Swanson S.K., Kusch T., Li B., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W.. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 2005; 280:13665–13670. [DOI] [PubMed] [Google Scholar]
- 76. Robert F., Hardy S., Nagy Z., Baldeyron C., Murr R., Dery U., Masson J.Y., Papadopoulo D., Herceg Z., Tora L.. The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol. Cell. Biol. 2006; 26:402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kusch T., Florens L., Macdonald W.H., Swanson S.K., Glaser R.L., Yates J.R. 3rd, Abmayr S.M., Washburn M.P., Workman J.L.. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004; 306:2084–2087. [DOI] [PubMed] [Google Scholar]
- 78. Keogh M.C., Mennella T.A., Sawa C., Berthelet S., Krogan N.J., Wolek A., Podolny V., Carpenter L.R., Greenblatt J.F., Baetz K. et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006; 20:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ravens S., Yu C., Ye T., Stierle M., Tora L.. Tip60 complex binds to active Pol II promoters and a subset of enhancers and co-regulates the c-Myc network in mouse embryonic stem cells. Epigenet. Chromatin. 2015; 8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cho K.O., Choi K.W.. Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature. 1998; 396:272–276. [DOI] [PubMed] [Google Scholar]
- 81. Dominguez M., de Celis J.F.. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998; 396:276–278. [DOI] [PubMed] [Google Scholar]
- 82. Papayannopoulos V., Tomlinson A., Panin V.M., Rauskolb C., Irvine K.D.. Dorsal-ventral signaling in the Drosophila eye. Science. 1998; 281:2031–2034. [DOI] [PubMed] [Google Scholar]
- 83. Hazelett D.J., Bourouis M., Walldorf U., Treisman J.E.. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development. 1998; 125:3741–3751. [DOI] [PubMed] [Google Scholar]
- 84. Ferres-Marco D., Gutierrez-Garcia I., Vallejo D.M., Bolivar J., Gutierrez-Avino F.J., Dominguez M.. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006; 439:430–436. [DOI] [PubMed] [Google Scholar]
- 85. Palomero T., Sulis M.L., Cortina M., Real P.J., Barnes K., Ciofani M., Caparros E., Buteau J., Brown K., Perkins S.L. et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 2007; 13:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Doherty D., Feger G., Younger-Shepherd S., Jan L.Y., Jan Y.N.. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev. 1996; 10:421–434. [DOI] [PubMed] [Google Scholar]
- 87. Flegel K., Grushko O., Bolin K., Griggs E., Buttitta L.. Roles for the histone modifying and exchange complex NuA4 in cell cycle progression in Drosophila melanogaster. Genetics. 2016; 203:1265–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Morel V., Lecourtois M., Massiani O., Maier D., Preiss A., Schweisguth F.. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr. Biol.: CB. 2001; 11:789–792. [DOI] [PubMed] [Google Scholar]
- 89. Nagel A.C., Krejci A., Tenin G., Bravo-Patino A., Bray S., Maier D., Preiss A.. Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell. Biol. 2005; 25:10433–10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Barolo S., Stone T., Bang A.G., Posakony J.W.. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002; 16:1964–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Da Ros V.G., Gutierrez-Perez I., Ferres-Marco D., Dominguez M.. Dampening the signals transduced through hedgehog via microRNA miR-7 facilitates notch-induced tumourigenesis. PLoS Biol. 2013; 11:e1001554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jarriault S., Greenwald I.. Suppressors of the egg-laying defective phenotype of sel-12 presenilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans. Genes Dev. 2002; 16:2713–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lopez C.I., Saud K.E., Aguilar R., Berndt F.A., Canovas J., Montecino M., Kukuljan M.. The chromatin modifying complex CoREST/LSD1 negatively regulates notch pathway during cerebral cortex development. Dev. Neurobiol. 2016; 76:1360–1373. [DOI] [PubMed] [Google Scholar]
- 94. Collu G.M., Hidalgo-Sastre A., Acar A., Bayston L., Gildea C., Leverentz M.K., Mills C.G., Owens T.W., Meurette O., Dorey K. et al. Dishevelled limits Notch signalling through inhibition of CSL. Development. 2012; 139:4405–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Munoz-Descalzo S., Sanders P.G., Montagne C., Johnson R.I., Balayo T., Arias A.M.. Wingless modulates the ligand independent traffic of Notch through Dishevelled. Fly. 2010; 4:182–193. [DOI] [PubMed] [Google Scholar]
- 96. Axelrod J.D., Matsuno K., Artavanis-Tsakonas S., Perrimon N.. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science. 1996; 271:1826–1832. [DOI] [PubMed] [Google Scholar]
- 97. Mukherjee A., Veraksa A., Bauer A., Rosse C., Camonis J., Artavanis-Tsakonas S.. Regulation of Notch signalling by non-visual beta-arrestin. Nat. Cell Biol. 2005; 7:1191–1201. [DOI] [PubMed] [Google Scholar]
- 98. Kwon M.H., Callaway H., Zhong J., Yedvobnick B.. A targeted genetic modifier screen links the SWI2/SNF2 protein domino to growth and autophagy genes in Drosophila melanogaster. G3 (Bethesda). 2013; 3:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Krejci A., Bray S.. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007; 21:1322–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Binda O., Sevilla A., LeRoy G., Lemischka I.R., Garcia B.A., Richard S.. SETD6 monomethylates H2AZ on lysine 7 and is required for the maintenance of embryonic stem cell self-renewal. Epigenetics. 2013; 8:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J.. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004; 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C.. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004; 303:343–348. [DOI] [PubMed] [Google Scholar]
- 103. Surface L.E., Fields P.A., Subramanian V., Behmer R., Udeshi N., Peach S.E., Carr S.A., Jaffe J.D., Boyer L.A.. H2A.Z.1 monoubiquitylation antagonizes BRD2 to maintain poised chromatin in ESCs. Cell Rep. 2016; 14:1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Draker R., Ng M.K., Sarcinella E., Ignatchenko V., Kislinger T., Cheung P.. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLos Genet. 2012; 8:e1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Knoechel B., Roderick J.E., Williamson K.E., Zhu J., Lohr J.G., Cotton M.J., Gillespie S.M., Fernandez D., Ku M., Wang H. et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat. Genet. 2014; 46:364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data have been deposited at GEO under the accession number GSE111029. Mass-spectrometry data are available via ProteomeXchange with identifier PXD008747.