Abstract
The constitutive androstane receptor (CAR; NR1I3) is a nuclear receptor orchestrating complex roles in cell and systems biology. Species differences in CAR’s effector pathways remain poorly understood, including its role in regulating liver tumor promotion. We developed transgenic mouse models to assess genome-wide binding of mouse and human CAR, following receptor activation in liver with direct ligands and with phenobarbital, an indirect CAR activator. Genomic interaction profiles were integrated with transcriptional and biological pathway analyses. Newly identified CAR target genes included Gdf15 and Foxo3, important regulators of the carcinogenic process. Approximately 1000 genes exhibited differential binding interactions between mouse and human CAR, including the proto-oncogenes, Myc and Ikbke, which demonstrated preferential binding by mouse CAR as well as mouse CAR-selective transcriptional enhancement. The ChIP-exo analyses also identified distinct binding motifs for the respective mouse and human receptors. Together, the results provide new insights into the important roles that CAR contributes as a key modulator of numerous signaling pathways in mammalian organisms, presenting a genomic context that specifies species variation in biological processes under CAR’s control, including liver cell proliferation and tumor promotion.
INTRODUCTION
The constitutive androstane receptor (CAR) is a nuclear receptor member (NR1I3) of the nuclear receptor superfamily. Typically, nuclear receptors function as transcription factors that are activated by direct ligands (1) and generally share a common structure, an N-terminal A/B domain, a DNA binding domain (DBD), a hinge domain, a ligand binding/ heterodimerization domain (LBD) and an F domain at the C-terminal (2). Upon ligand activation, the LBD would utilize activation function-2 (AF-2) to mediate co-regulator interactions (3). However, CAR is distinguished from other nuclear receptors in that it lacks an A/B domain and is constitutively active in the absence of ligand due to a charge-charge interaction between LBD helixes that mimic an active AF-2 conformation, and therefore capable of co-activator protein interactions in the absence of ligand (4).
CAR was initially characterized as a xenobiotic metabolism modulator. Classified as a Class II nuclear receptor, another unusual property of CAR is its retention in the cytosol in a phosphorylated state, but upon xenobiotic sensing, it undergoes dephosphorylation, translocates into the nucleus, and subsequently binds specific DNA motifs to coordinately regulate transcription of target genes. Targets include genes that encode drug metabolizing functions, such as the Phase I cytochrome P450s, Phase II transferases and Phase III transporters (5–7). With respect to its role in xenobiotic regulation, CAR contributes to regulating the metabolism of approximately 75% of clinically used drugs and is a major determinant in the detoxification of environmental chemicals (6). Consequently, CAR is of toxicological importance in safety evaluations for drug discovery and in the development of industrial chemicals destined for regulatory approval.
More recent evidence identified CAR’s broader roles in regulating energy and lipid metabolism, cell proliferation and carcinogenesis (6). For example, CAR is reported to suppress gluconeogenesis through its targeting of genes such as phosphoenolpyruvate carboxykinase (PEPCK;PCK1) and glucose-6-phosphatase (G6PC), and functions to reverse induced-obesity in rodent models by regulating genes such as sterol regulatory element binding protein 1 (SREBP1;SREBF1) (8). Studies of liver tumorigenesis promoted by phenobarbital (PB) and 1,4-bis[2-(3,5-dichloropyridyloxy)] benzene (TCPOBOP; TCP) revealed that CAR is essential for hepatic tumor progression in initiation-promotion mouse models (9,10). Further studies have demonstrated that CAR induces growth arrest and DNA-damage-inducible beta (GADD45B) and myelocytoblastosis oncogene (MYC), while altering microRNA expression profiles related to hepatocarcinogenesis (11–13). However, detailed roles for CAR’s participation in the carcinogenic process are not well understood and are the subject of some controversy in that chronic human exposures to PB, a known CAR activator, have not been associated with any detectable increase in human liver cancer incidence (14,15). Adding to its overall complexity, another unique feature of CAR among the nuclear receptors is that its activation pathways proceed through both direct ligand interactions and through indirect signaling mechanisms (5,16). The potential receptor conformational alterations inherent in these respective activation pathways may result in differences in CAR genomic targeting, or in biological/ signaling outcomes, areas that have not been well characterized. We hypothesized that differing DNA interaction profiles for the respective species CAR proteins, and perhaps variable chemical activation states, underlie the transcriptional programs driving differences in CAR’s biological function.
Due to technical limitations and poor availability of a chromatin immunoprecipitation (ChIP)-grade antibody, high-resolution genomic mapping for CAR has not been performed in vivo. In this investigation, we deployed ChIP-exo, a modified ChIP-seq method incorporating exonuclease to trim crosslinking ChIP DNA (17), combined with CAR-fusion proteins and a novel delivery system, to conduct high resolution, genome scale profiling for mouse and human CAR (mCAR and hCAR) in vivo, using transgenic mouse models. With a view toward better definition of the role of CAR in carcinogenesis, we identified novel CAR genomic interactions, referenced to published RNA-seq transcriptomic datasets. Further, we assessed CAR’s genomic interactions in the presence of direct vs. indirect activators, performed DNA motif analyses, assessed species differences in CAR genomic binding profiles and identified specific target genes that may contribute to species differences in liver tumor promotion.
MATERIALS AND METHODS
Materials and reagents
TCPOBOP (>99%) was synthesized by the Environmental Health Laboratory in the Department of Environmental and Occupational Health Safety at the University of Washington (Seattle, WA, USA). PB was obtained from the Drug Services Division of the University of Washington. 6-(4-Chlorophenyl: imidazo[2,1-b]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO; CIT) was purchased from BIOMOL Research Laboratories (Plymouth Meeting, MA, USA). Dimethyl sulfoxide (DMSO) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Full length reference/wild type human CAR and mouse CAR cDNAs were sub-cloned into the pEYFP-c1 plasmid (Clontech/Takara, Mountain View, CA, USA) to generate yellow fluorescent protein N-terminal fusion CAR protein constructs (YFP-hCAR and YFP-mCAR). All constructs were validated by DNA sequencing. Adenovirus (AV) constructs containing YFP-hCAR, YFP-mCAR and YFP-empty were produced in the Adeno X Expression System (Clontech/Takara) and amplified to high titer by SignaGen (Rockville, MD, USA).
Animals and treatments
All animal care and experimental procedures complied with protocols approved by the Institutional Animal Care and Use Committee of The Pennsylvania State University. Wild-type (WT) C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA). Breeding pairs of CAR -/- /pregnane X receptor (PXR) -/- double-knockout (DBL KO) mice in the C57BL/6 background were a kind gift of Dr. Wen Xie (University of Pittsburg, Pittsburg, PA), and their derivation was described previously (18). CAR knockout (CAR -/-; CAR KO) mice were generated by crossing WT C57BL/6 mice with DBL KO mice. The mice were maintained under a standard 12 h light, 12 h dark cycle at constant temperature (23 ± 1°C) with 45–65% humidity. A total of six, 8-week-old male CAR -/- mice were infected with YFP-hCAR AV (1 × 1012 virus particles/ml, ∼100 ul diluted injection volume per mouse) through tail-vein injection. At 72 and 94 h, the mice were administrated with two successive doses of PB (75mg/kg, in saline) (n = 3), or CITCO (5 mg/kg, in DMSO) (n = 3), through IP injection. Another six, 8-week-old male CAR -/- mice were infected with YFP-mCAR AV constructs (1 × 1012 virus particles/ml) through tail-vein injection. At 72 and 94 h, two successive doses of PB (75 mg/kg, in saline) (n = 3), or TCPOBOP (2 mg/kg, in DMSO) (n = 3), were administrated. After 96 h, all AV-infected mice were harvested for liver tissues after CO2 asphyxiation-induced euthanasia. For non-adenovirus infected mice, 8-week-old male CAR -/- mice (n = 6) and WT mice (n = 6) were administrated with a single dose of DMSO (4 ml/kg) (n = 3) or TCPOBOP (2 mg/kg, in DMSO) (n = 3), through IP injection. After 24 h, liver tissue extractions were performed following CO2 asphyxiation-induced euthanasia.
Liver extraction and ChIP-exo
Briefly, fresh YFP-hCAR or YFP-mCAR AV infected CAR -/- mouse livers (∼1.0 g) were immediately minced and cross-linked with 1% formaldehyde (diluted from 16% formaldehyde solution methanol-free 1 ml ampules (Thermo Scientific)) for 10 min followed by quenching with 0.125 M glycine for 5 min. Liver tissues were then washed 3× with ice-cold PBS, and subjected to Dounce homogenization followed by 100 nm filtration. Nuclei lysates of liver tissues were prepared by incubating homogenized cells for 15 min in cell lysis buffer (10 mM Tris pH 8.0, 10 mM NaCl, 0.2% IGEPAL) on ice, and then incubating precipitates from previous steps with nuclear lysis buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 0.5% SDS) for 20 min on ice. Nuclei lysates were aliquoted to 700 ul each in 10 ml sonication tubes (Diagenode, Denville, NJ, USA) and sonicated using a Bioruptor 300 instrument (Diagenode, Denville, NJ, USA) for 36 cycles of 30 s on, 30 s off, to achieve an average chromatin fragmentation size of 100–200 bp. Subsequent ChIP-exo procedures and sequencing were performed following previously published protocols (19). Chromatin samples were diluted 2.5-fold with 0.6% (v/v) Triton X-100 and immunoprecipitated using sepharose antibody-conjugated magnetic beads. All YFP-empty, YFP-hCAR and YFP-mCAR construct-infected samples utilized a rabbit anti-GFP antibody Ab290 (Abcam, Cambridge, MA, USA), followed by DNA polishing, A-tailing, Illumina adaptor ligation (ExA2) and subsequent digestion on the beads using lambda and recJ exonuclease. Following single-stranded DNA elution, a primer was annealed to ExA2 and extended with phi29 DNA polymerase, then A-tailed. Exonuclease treated ends were then ligated with a second Illumina sequencing adaptor; the products PCR-amplified and gel-purified.
Frozen tissue embedding and immunohistochemistry
YFP-hCAR and YFP-mCAR AV infected CAR -/- mice were perfused with phosphate buffered saline (PBS) (Gibco, Gaithersburg, MD, USA) prior to liver extraction. After overnight incubation in 4% paraformaldehyde, livers were dehydrated with a sucrose solution for 48 h. Then livers were embedded in OCT-filled cryomolds and snap-frozen in liquid nitrogen-cooled 2-methyl butane. Frozen liver tissues were sectioned using a Leica CM1950 Cryostat (Leica Biosystems, Germany). Sectioned liver tissues were immunostained with a chicken anti-GFP antibody (ab13970, Abcam) and an Alexa 488 conjugated antibody (103-545-155, Jackson ImmunoResearch, West Grove, PA, USA), according to the manufacturers’ protocols. Fluorescent imagining was performed using a Nikon Eclipse TE2000-S system equipped with a Nikon Digital Sight DS-Ri1 camera (Nikon, Japan).
Human primary hepatocyte culture and treatments
Human primary hepatocytes (HPH) in six-well plates were procured from the Liver Tissue Cell Distribution System at the University of Pittsburgh, Pittsburg, PA, funded by National Institutes of Health Contract HHSN276201200017C. The HPH culture protocols were detailed previously (20,21). HPH were treated with DMSO or 3 μm CITCO 24 h prior to total RNA extraction.
RNA extraction and quantitative real-time PCR analysis
Mouse liver total RNA was extracted from WT, CAR -/-, YFP-hCAR, YFP-mCAR or YFP-empty AV infected CAR -/- mouse livers using TRIzol (Qiagen, Germantown, MD, USA), following the manufacturer's protocol. Extraction methods for HPH total RNA were described previously (20,21). Concentrations of total RNA were measured using a NanoDrop 1000 spectrophotometer (ThermoScientific, Waltham, MA, USA). Total RNA was converted to cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), following the manufacturer's instructions. Quantitative real-time PCR (qRT-PCR) reactions were performed using a CFX96 Real Time system (Bio-Rad, Hercules, CA, USA) as previously described (22). Each sample was assessed in duplicate. The primer sequences are listed in Supplementary Table S1. mRNA relative expression levels were calculated using the ΔΔCt method as previously described (20), and all target genes were normalized to glyceraldehyde 3-phosphate dehydrogenase (Gapdh) as reference. Primer information is provided in Supplementary Table S1.
Protein extraction and Western blot
Nuclear protein and cytoplasmic protein from YFP-hCAR, YFP-mCAR and YFP-empty AV infected CAR -/- mice livers were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents with Protease inhibitor cocktail set I (Merck Millipore, Billerica, MA, USA) following the manufacturer's instructions. Protein concentrations were measured with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) using a Tecan Infinite M200pro fluorometer absorption function (Tecan, Mannedorf, Switzerland). 30 μM of nuclear or cytoplasmic proteins were loaded onto 10-well precast 10% SDS-PAGE gels (mini-PROTEAN TGX gel, Bio-Rad, Hercules, CA, USA). After denaturing SDS-PAGE separation, proteins were transferred to a 0.2 μm PVDF membrane using the Trans-Blot Turbo Transfer system (Bio-Rad, Hercules, CA, USA) with the Trans-Blot Turbo Transfer Kit (Bio-Rad, Hercules, CA, USA), on the 1 mini TGX preprogrammed setting. Subsequent western blot procedures were performed as previously described (5). Rabbit anti-CYP2B450 was generously provided by Dr Gonzalez from the Laboratory of Metabolism, National Cancer Institute/NIH. Rabbit anti-GFP antibody ab290 was purchased from Abcam. Chicken anti β-actin polyclonal antibody sc-81178 was purchased from Santa Cruz (Santa Cruz Biotechnology, Dallas, TX, USA). All antibodies were diluted 1:1000 prior to incubation.
ChIP sequencing
Sample sequencing was performed on an Illumina NextSeq500 system using a 40 bp paired-end sequencing setting. Reads were mapped to the mouse genome (mm10) using BWA (version 0.7.9a). Each biological replicate contained at least 20 million unique reads.
Genome coverage visualization
For visualization, biological replicates read files were merged into a single file for each sample. Sample reads were separated by forward/reverse strand and respectively converted to genome coverage format bedgraph files; then forward strand and reverse strand bedgraph files were combined and converted using the Integrated Genome Viewer (IGV) tools for IGV browser visualization. Red areas represent forward strand reads coverage, whereas blue area represents reverse strand reads coverage.
Quantitative differential binding analysis
Biological replicate reads files were merged and then peaks were called using the MACS14 default setting (P-value cut off: 1E–5) against the YFP-empty infected control sample (23). MACS14 peaks from all four samples were merged into one peak file using the bedtools merge function (24) and filtered for blacklist regions (25). The merged peak files represent all potential enrichment regions for both mCAR and hCAR. Count matrixes of potential enrichment regions coverage were generated for all biological replicates, then further analyzed using the R corrplot package and the R Bioconductor DESeq2 package. The R corrplot package was used to calculate and plot for Pearson correlation coefficients between two replicates. The DESeq2 utilized a negative binomial-based generalized linear model to examine following differential binding tests: mCAR versus hCAR, mCAR TCPOBOP versus PB, and hCAR CITCO versus PB. The Principal Component Analysis (PCA) data were generated using the DESeq2 package. Hierarchy clustering was performed by submitting the count matrix to ClustVis (26), with average method for both row and column clustering. Enrichment regions with significant differential binding between test groups (fold change ≥ 2, q-value < 0.05) were associated to the closest transcriptional start site (TSS) within 10 kb region and annotated using HOMER (27).
Peak annotation and GO analysis
Based on the highest FRiP score (Fraction of Reads in Peaks), three replicates from mCAR samples and three replicates from hCAR samples were chosen for the stringent peak calling processes. Peak calling processes were performed based on the official specification for the ENCODE TF ChIP-seq processing pipeline, using the Irreproducibility Discovery Rate (IDR) method. Briefly, three replicates from mCAR or hCAR were merged, randomly shuffled into two pseudo replicates, and pseudo replicates were called for peaks using MACS2 and analyzed for IDR consistency R script. Following the IDR framework guideline, the IDR output thresholds, 0.01 for mCAR and 0.0025 for hCAR, were selected. IDR analysis-generated peaks were filtered with blacklist region, then associated with the closest TSS within a 10kb region and annotated by HOMER. Gene ontology (GO) analysis was performed using the HOMER functional enrichment analysis and the DAVID Functional annotation tool (28,29). The mCAR and hCAR top 500 genes ranked by MACS p-value were submitted for GO analysis.
Motif analysis
ChIP-exo peak-pairs for each treatment as well as merged mCAR and hCAR samples were generated following previously published methods (17). Briefly, peak-pairs of 5′ ends of ChIP-exo reads were determined using the Genetrack algorithm (30), with fine grain peak-calling parameters: sigma = 5, exclusion zone = 10. Peak-pairs were merged using the bedtools merge function. The top 500 or 1000 merged peak-pairs overlapping with primary binding sites were used for de novo motif discovery using the MEME algorithm (31). Motifs for mCAR and hCAR differential binding sites were also generated by MEME algorithm using the same settings. Myc, ras-related nuclear protein (Ran), inhibitor of nuclear factor kappa B kinase subunit epsilon (Ikbke), cytochrome P450 (Cyp) family 2, subfamily b, polypeptide 10 (Cyp2b10) and carboxylesterase 2A (Ces2a) were selected for a putative motif location analysis, using the FIMO algorithm (Find Individual Motif Occurences), to scan identified motifs on CAR binding regions near transcription start sites.
RNA-seq re-analysis
To cross-reference our differential genomic species mapping results with transcriptomic data, we utilized a set of published RNA-seq files consisting of C57BL/6 WT mice and humanized CAR-transgenic (hCAR-TG) mice (32). WT_Day60_CornOil (n = 3), WT_Day60_TCPOBOP (n = 3), hCAR-TG_Day60_CornOil (n = 3) and hCAR-TG_Day60_CITCO (n = 3) samples (NCBI Gene Expression Omnibus database GSE98666) were re-analyzed specifically for mCAR and hCAR species difference comparisons. Reads from each sample replicate were mapped to the mm10 mouse genome using HISAT2; then uniquely mapped reads were processed with featureCounts and DESeq2 for differential gene expression analysis. Differential expression analyses were performed between the C57BL/6 WT mouse control and TCPOBOP, and the hCAR-TG mouse control and CITCO (fold change ≥ 2, q-value < 0.05).
RESULTS
Adenovirus transient transgenic mouse characterization
To overcome technical difficulties resulting from the lack of commercial CAR ChIP-grade antibodies, we developed an adenovirus-based system to direct the expression of YFP-CAR fusion constructs in transgenic mouse livers, as depicted in Figure 1A. Successful use of the ChIP-grade anti-GFP Ab290 antibody for YFP-tagged transcription factors in ChIP pull downs has been documented by the supplier (Abcam), including its use in a recent report (33). mCAR and hCAR N-terminal YFP fusion constructs were created and deployed in high titer adenoviral delivery vectors along with YFP-only vehicle constructs serving as controls. YFP-tagged CAR constructs are used frequently for investigating direct and indirect CAR activation, nuclear translocation dynamics and CAR protein-protein interactions (34–37). The Ab290 antibody enabled immunocapture of YFP-CAR fusion proteins bound to their DNA targets with high specificity, allowing genomic mapping of the respective CAR-DNA interactions. The efficacy of the adenovirus-delivered transgenic mouse strategy was verified through immunofluorescence visualization, validation of known CAR target gene mRNA expression and with protein expression analysis. Fluorescence imaging of both YFP-mCAR and YFP-hCAR expression in cyrostat liver sections showed that CAR was highly expressed in mouse liver hepatocytes subsequent to AV delivery (Figure 1B). Although the extent of nuclear translocation appeared to differ between the activated human and mouse constructs, successful transcriptional activation of known target genes for each construct was validated independently. qRT-PCR analyses of liver RNA was conducted in WT and CAR -/- mice, transduced with YFP-empty vector as well as YFP-mCAR and YFP-hCAR constructs. Both YFP-mCAR and YFP-hCAR infected mice exhibited high levels of YFP mRNA expression. Following transduction with the CAR viral vectors, both indirect CAR activator PB and CAR ligand activators TCPOBOP or CITCO, induced Cyp2b10 mRNA expression in CAR -/- mice at comparable levels to WT mice (Figure 1C). Mouse liver western blotting demonstrated the YFP-expressed proteins, exhibiting the anticipated correct size differences between the YFP-empty and YFP fusion vectors, YFP-mCAR and YFP-hCAR. CAR activation by PB induced CYP2B10 protein expression in YFP-mCAR and YFP-hCAR mice, but not in YFP-empty mice (Figure 1D), indicating the functionality of YFP-tagged CAR constructs. These results demonstrated that the AV delivery system effectively expressed the YFP-CAR constructs in mouse liver, and that the resulting fusion proteins retained activity as transcriptional regulators of canonical CAR target genes in the presence of receptor activators.
Figure 1.
Adenovirus delivery of YFP-CAR constructs into CAR -/- mice. (A) Scheme of AV delivery system. Each AV YFP-CAR fusion construct was injected into CAR -/- mice 4 days prior to terminal surgery. CAR activator treatments were initiated 24 h and again at 2 h before liver extraction. (B) Fluorescence imaging of liver cryostats in AV infected mice. All samples were treated with PB. DAPI staining was performed for nuclei visualization. Upper panels show anti-YFP immunohistochemistry merged with DAPI in YFP-mCAR infected mice. Lower panels show cryostat imaging of YFP fluorescence merged with DAPI in YFP-hCAR infected mice. (C) qRT-PCR analysis of mice liver mRNA levels. Left: YFP expression levels of YFP-mCAR and YFP-hCAR infected mice were compared to WT mice (3). Student t-tests were performed for each sample versus WT untreated. Right: Expression of Cyp2b10 compared to both untreated WT mice and YFP-empty infected CAR -/-. The data shown are the average from three biological replicates normalized to Gapdh. Student t-tests were performed for each sample versus WT untreated. (D) Nuclear lysate western blot of mice liver. Adenovirus containing YFP-empty, YFP-mCAR and YFP-hCAR constructs infected CAR -/- mice through tail-vein injection. All three samples were treated with PB prior to harvest. In anti-YFP blotting, size differences between YFP-empty, YFP-mCAR and YFP-hCAR constructs could be visualized. All samples were obtained from mice treated with 75 mg/kg PB.
Genome profiling of mCAR and hCAR DNA interactions
Livers from AV infected YFP-mCAR and YFP-hCAR mice with direct and indirect activators treatments (PB treated n = 3, TCPOBOP treated n = 3; PB treated n = 3, CITCO treated n = 3; respectively) were extracted and processed through the ChIP-exo pipeline with paired-ended sequencing. On average, a replicate sample consisted of 27.5 million uniquely mapped reads, with the lowest replicate exhibiting 21 million unique reads. The IGV was used to display ChIP genomic reads coverage for several known CAR-binding genes: glutathione S-transferase 2 and 3 (Gstm2, Gstm3), Cyp2b10 and Ces2a (Figure 2A). The visualization revealed fine structures for CAR binding events. For Cyp2b10, in addition to a well characterized ∼2 kb upstream phenobarbital-responsive enhancer module (PBREM), several CAR binding sites were mapped through the Cyp2b10 promoter region, with the strongest binding localized at ∼10 kb upstream. Initial peak calling analysis across the entire mouse genome was assessed using MACS14 default settings and identified mCAR binding at an average of 18,000 binding loci, whereas hCAR exhibited 48,000 binding loci. Peak distances to TSS were calculated for each hCAR/mCAR treatment and showed that the majority of the detected CAR binding peaks centered on TSSs (Figure 2B).
Figure 2.
Genomic profiling of CAR using ChIP-exo. (A) Visualization of CAR enrichment on selected genes. IGV displayed YFP-CAR proteins with their direct/indirect activator binding locations on selected known CAR binding genes: Cyp2b10 (upper left), Gstm2 and Gstm3 (upper right), and Ces2a (lower left). Mapped reads were separated by strands, with forward strand reads on the upper track (red) and reverse strand reads on the bottom track (blue). From top track to bottom track: hCAR PB, hCAR CITCO, mCAR PB, mCAR TCPOBOP and the Refseq gene track. Track length is 20 kb. (B) Peak distance to TSS plot showing relative distribution of CAR binding peaks to TSS.
Quantitative differential analysis reveals distinct hCAR and mCAR genomic profiles
Previous reports assessing the overall correspondence of transcription factor (TF) binding with gene expression levels concluded positive correlations of TF binding signals to the respective levels of gene expression (38–40). Therefore, delineating gene binding site differences between mCAR and hCAR is likely relevant as a predictor of transcriptional effects and formative in contributing towards our understanding of species variation in downstream CAR signaling.
To quantitatively analyze the difference between genomic binding profile of hCAR and mCAR, we analyzed the genomic coverage for each of the selected ChIP replicates using featureCounts software and used the resulting genomic coverage output for subsequent statistical study. Pearson correlation analysis was carried out for all replicates (Figure 3A). For the hCAR replicates coverage, correlation coefficients approached 1 between any two replicates; for mCAR, five samples exhibited very high correlation coefficients between each other (r > 0.95), with an exception for mCAR_PB_3, which displayed a lower coefficient relative to other mCAR samples (0.42 < r < 0.45). Overall, the results demonstrated marked positive correlations within the hCAR replicates and within the mCAR replicates, indicating that the respective replicate binding profiles were highly similar. However, between the hCAR and mCAR replicates, the coefficients were much lower (0.05 < r < 0.20), suggesting distinctions in the binding profiles between hCAR and mCAR.
Figure 3.
Differential analysis between mCAR and hCAR binding profiles. (A) Pearson correlation analysis for all replicates. Correlation coefficients between mCAR and hCAR replicates were much lower than replicates within the same species, indicating species variances in the respective genomic profiles. (B) PCA analysis for all replicates. Teal dots represent hCAR replicates, orange dots represent mCAR replicates. PCA analysis showed clear separation between mCAR and hCAR replicates. (C) Hierarchy cluster analysis. 1,048 significant mCAR versus hCAR differential binding loci are shown in the clustering. Clustering for all replicates indicated species variation, and consistency within each species. (D) Summary of differential binding loci associated genes. Approximately 1,000 genes exhibit differential mCAR and hCAR binding loci, whereas relatively few genes exhibit variation comparing direct vs indirect activation for hCAR or mCAR replicates. (E) IPA disease and function analysis for mCAR and hCAR differential binding associated genes. The top 10 disease and function terms are shown, ranked by p-value.
DESeq2 package from R Bioconductor uses a negative binomial distribution model to calculate variance-mean dependence in count data. The package is often used for RNA-seq comparative expression analyses, however the same model can be applied to analyze differential genome coverage count data generated from ChIP-seq. In this respect, DESeq2 analysis was performed to compare differential genome coverage, or binding profiles, between mCAR and hCAR replicates. Among 59,508 binding loci examined, DESeq2 revealed 3,511 (binding fold change > 2, adjusted p-value < 0.05) loci with statistically significant differential binding profiles for mCAR versus hCAR, encompassing 5.9% of the total CAR-binding regions. These analyses suggest that overall; mCAR and hCAR binding profiles are similar, although with distinctions. The top 1,000 differential binding loci, ranked by adjusted p-value, were analyzed using PCA (Figure 3B). Each red dot represents a mCAR replicate and blue dots represent hCAR replicates. Separation between the mCAR and hCAR replicate groups is evident, indicating differential DNA binding between mCAR and hCAR.
Hierarchy cluster analysis was also conducted for the mCAR vs hCAR DESeq2 results, using 1,048 significant differential binding loci that were proximal to annotated genes (loci within ±10 kb of TSS, binding fold change ≥ 2, adjusted p-value ≤ 0.05). The red color scale indicates increased locus binding events, compared to blue, indicating decreased binding events. The hierarchy clustering analysis indicates that the hCAR replicates and the mCAR replicates cluster separately, demonstrating differential DNA binding interactions exhibited by the respective species’ receptors. 674 binding loci were associated with stronger mCAR bindings, whereas 331 loci were related to stronger hCAR bindings, as shown in Figure 3D. A comprehensive list of these gene-binding interactions is provided in Supplementary Table S2. An Ingenuity Pathway Analysis (IPA) of differential binding associated genes (Figure 3E) illustrated that the diseases and functional annotations corresponding to the strongest mCAR binding gene regions were largely associated with biological terms: cell proliferation, apoptosis, and cell death; whereas for hCAR, the strongest binding gene regions were annotated with terms that were energy metabolism-related. Together, these results point to interesting species-driven distinctions underlying mCAR vs hCAR DNA binding interactions that likely drive differences in the receptors’ biological functions.
Direct vs indirect CAR activation results in near identical genomic profiles
Applying Pearson correlation analysis for all samples, highly positive correlation coefficients (r > 0.95, except for mCAR_PB_3) were obtained between direct (TCPOBOP / CITCO) and indirect CAR activator (PB) replicates (Figure 3A), indicating remarkably high similarity between the DNA binding profiles resulting from direct vs indirect CAR activator treatments. DESeq2 differential analyses were conducted for TCPOBOP versus PB in mCAR infected CAR -/- mice and CITCO vs PB in hCAR infected CAR -/- mice. In contrast to the differential DNA binding profile results obtained for mCAR versus hCAR, when comparing the profiles for PB versus TCPOBOP treatments (for mouse), or PB versus CITCO (for human), only relatively few genes exhibited differential binding patterns. These results strongly support a conclusion that the genomic CAR binding profiles comparing direct and indirect activators for both mCAR and hCAR are remarkably similar.
Gene annotations for mCAR and hCAR binding regions
Since the direct- and indirectly-activated CARs exhibited almost identical genomic binding interactions, efforts were then focused toward high confidence peak calling and annotation. Replicates possessing the highest quality parameters for mCAR and hCAR were selected from the sample pool, regardless of activator. Specifically, three replicates possessing the highest FRiP scores were pooled within mCAR and hCAR samples, followed by peak calling using the IDR method. Peaks were mapped to the nearest identified TSS of annotated genes, within ±10 kb, although we realize this is likely a stringent threshold, in that enhancers or other transcriptional regulators may map further distant from a TSS (41). The resulting analysis identified 6,364 unique genes associated with hCAR, and 2,839 unique genes annotated for mCAR, with 2,661 of the genes overlapping (Figure 4A; full gene lists in Supplementary File 2). The genes scored in this manner represent high-confidence CAR-binding genes that specifically passed the strict annotation thresholds, rather than reflecting whole genome binding events. In these respects, the hCAR replicates exhibited higher total reads numbers and higher signal-to-noise ratios compared to the mCAR replicates, which likely allowed more low-affinity hCAR binding sites to pass the annotation thresholds, resulting in higher apparent hCAR total gene counts. Perhaps more pertinent, 94% of mCAR-annotated genes overlapped with hCAR genes. Together with genome browser visualization and DESeq2 quantitative analysis described earlier, the overall results indicate that mCAR and hCAR genomic binding profiles are actually quite similar to one another.
Figure 4.
Gene annotation for mCAR and hCAR binding regions. (A) Venn diagram for total mCAR annotated genes and hCAR annotated genes. Annotated mCAR binding genes (n = 2,839) exhibit 2,661 genes that overlap with hCAR binding genes (n = 6,364). Binding peaks were annotated to the nearest gene TSS within ±10 kb region. (B) GO biological process analysis for the top 500 mCAR and hCAR binding genes. Top 10 GO biological process terms from DAVID GO DIRECT analysis are listed. mCAR and hCAR GO analysis indicated that the top binding genes for CAR are enriched in metabolic pathways, particularly in oxidation-reduction such as cytochrome P450, and lipid metabolism and glucose metabolism.
GO Term analysis was conducted for the top 500 annotated genes using DAVID functional annotation (Figure 4B). The top three enriched GO DIRECT biological process terms for mCAR and hCAR were the same, i.e. lipid metabolic, oxidation-reduction and cholesterol metabolic processes; terms consistent with previously reported roles for CAR as a regulator of phase I cytochrome P450s in drug metabolic pathways and coordinator of energy and lipid metabolism. Glucose metabolic processes were present in both top 10 lists, indicating CAR’s major role in regulating glucose metabolism as well. Drug transmembrane transport processes and glutathione metabolic processes demonstrated the phase II and III regulatory functions of CAR. Gene lists from the top 500 CAR binding genes involved in select biological processes are presented in Table 1. Overall, GO analysis indicated that the most enriched functionalities of CAR lie in drug and energy metabolism, consistent with previous determinations.
Table 1.
The top 500 mCAR and hCAR binding genes as annotated by HOMER. Listed Go terms were selected to represent CAR’s role in drug metabolism, lipid metabolism, glucose metabolism and cell proliferation
| GO term ID | Term | mCAR top 500 binding genes | hCAR top 500 binding genes |
|---|---|---|---|
| GO:0042493 | response to drug | Cyp2b10, Mgst1, Gstm1, Tcf3, Cyp2d9, Cyp2c55, Nfe2l2, Fbp1, Gstm3, Abcb11, Abcc3, Cyp2d22, Nr1i2, Ppm1f | Mgst1, Abcc3, Gstm3, Ppm1f, Nr1i2, Abcc2, Cyp2b10, Tcf3, Abcc4, Gstm1, Abcb4, Abcb11 |
| GO:0006629 | lipid metabolic process | Nr5a2, Thrsp, Dhrs4, G6pc, Scp2, Akr1b7, Hmgcr, Pon1, Erlin1, Ttr, Cyp2b10, Dbi, Por, Cyp2d22, Serpina6, Cyp2c55, C3, Gpd1, Plch2, Cyp26a1, Cyp2d9, Ppard, Decr2, Cps1, Ces1d, Il1rn, Apof, Gpcpd1, Cyp17a1, Stat5a, Qk, Pck1, St6galnac6, Crat, Apoc3, Pla2g12a, Lipe, Ptges, Ptgds, Insig2, Acox2, Agpat2, Cyp4v3, Hsd17b4, Impad1, Apoa2, Dhrs9, Cers2, Cyp27a1, Acsm1, Cln8, Cpt1a, Lrp5, Etnk2 | Cyp2d9, Cyp7a1, Cers2, Cyp17a1, Cyp2b10, Acox2, Pgap2, Cecr5, Cyp2c66, Acot3, Akr1b7, Pon1, Gpat4, Cyp26a1, Apoa2, Acaa1b, Ttr, Cyp2c55, Serpina6, C3, Apof, Fabp6, G6pc, Impad1, Lrp1, Etnk2, Scarb1, Ppard, Ptges, Hsd17b4, Lpin2, Cyp27a1, Erlin1, Cps1, Ptgr2, Ces1d, Tm7sf2, Hacd3, Pck1, Fdx1, Thrsp, Cln8, Gpcpd1, Hmgcr, Por, Abhd5, Cyp2g1, Hsd17b2, Acsm1, Cyp3a13, Gpd1, Decr2, Lrp5, Pcx, Pla2g12a, Scd1, Dhrs9, Stat5a |
| GO:0006006 | glucose metabolic process | Galm, Lrp5, Cpt1a, Car5a, Gapdh, Pdk4, G6pc, Fbp1, Serp1, Pck1, Pgm1, Dlat, Gpd1, Pdk2 | Pck1, Pdhb, G6pc, Pgm1, Serp1, Pdk2, Pcx, Dlat, Akt1, Lrp5, Gpd1 |
| GO:0042127 | regulation of cell proliferation | Lrp5, Cebpb, Lifr, St6gal1, Smad3, Erbb3, Pid1, Nkx2.5, Irs2, Zmiz1, Ptgds, Ptges, Xdh, Birc5, Fgf18, Bcl6, Nr2f2, Hilpda, Klf9, Mlxipl, Hhex, Tcf3, Stat5a, Crip2, Atf5, Ptgfr, Ppard, Cnot8, Cacul1, Cela1, Xirp1, Arg1, Rarb, Nfkbia, Wnt11, Mafg, Lims2, Agt, Por, Slc9a3r1, Grn, Podn, Hmgcr, Ccr7, Shc1, Nr5a2 | Smad6, Serpinf2, Eif4g1, Fosl2, Grn, Bcl6, Abcc4, Agt, Cebpb, Por, Hmgcr, Bak1, Shc1, Cacul1, Rarb, Stat5a, Rnf10, Nfkbia, Eppk1, Lrp5, Slc9a3r1, Hilpda, Sdc4, Morc3, Trp53inp1, Cers2, Zmiz1, Tcf3, Mst1, Mlxipl, Klf9, Akt1, Ern1, Podn, Nr2f2, Jup, Birc5, Pdgfc, Smad3, Tob2, Ppard, Ptges |
Top 500 mCAR and hCAR binding genes categorized by selected GO terms.
Expression analysis on select CAR-binding genes indicates that CAR directly regulates key pathways in hepatocarcinogenesis
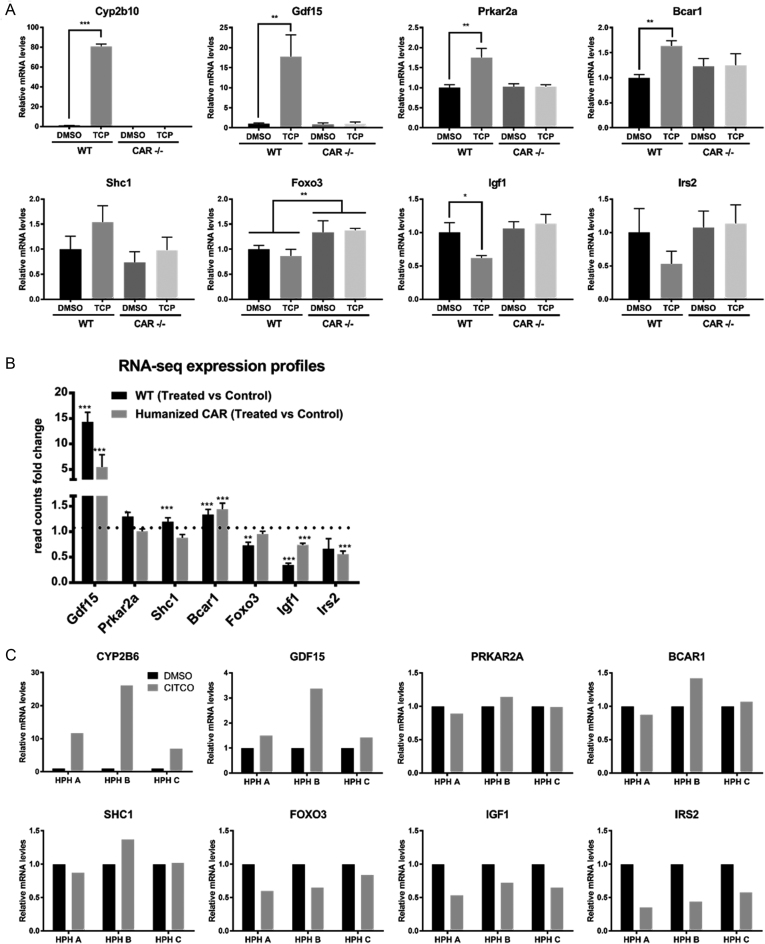
One of the goals of our study was to identify novel gene involvements for CAR in carcinogenesis-related pathways, relationships less well elucidated than other CAR regulatory functions, such as drug and energy metabolism. In these respects, we generated a list of genes from those that overlapped as annotated CAR binding genes with genes associated with common cancer-related pathways, including Wnt/β-catenin, TGF-β pathway, Jak/STAT, PI3K/Akt, p53, apoptosis, etc., and then selected genes whose expression levels were altered in published CAR-related gene expression studies (42). For the selected endpoints, we assessed transcriptomics data (Figure 5), including qRT-PCR mRNA experiments conducted here together with re-analysis of published RNA-seq data (GSE98666). Cyp2b10 or CYP2B6 mRNA served as a positive control for CAR inducibility. Among those CAR-binding genes that were also associated with carcinogenesis pathways, growth/differentiation factor 15 (Gdf15) mRNA expression was remarkable, exhibiting >15-fold induction when comparing TCPOBOP and DMSO treated WT mice; although unchanged in CAR -/- mice. GDF15 is involved in inflammatory and apoptotic pathways (43) and recently associated with liver carcinogenesis through activation of the glycogen synthase kinase 3 beta (GSK-3β) / β-catenin pathway (44). Our ChIP-exo data identified two major CAR binding sites within 5 kb of the Gdf15 gene's TSS. RNA-seq expression analyses also demonstrated that Gdf15 mRNA levels are increased ∼15-fold in WT mice following TCPOBOP treatment, and ∼5-fold with CITCO treatment in humanized CAR mice. Together, these results strongly imply that mCAR directly regulates Gdf15 at the transcriptional level. Although not as remarkable, we conducted mRNA expression assays on three human primary hepatocyte donor cases, and the results were consistent with a trend of increased GDF15 expression with CITCO induction.
Figure 5.
mRNA expression analysis indicates that CAR regulates key genes associated with hepatic carcinogenesis. (A) qRT-PCR assay of selected genes with respect to their mRNA expression levels in mouse liver. qRT-PCR results showed that selected genes exhibit mRNA expression level perturbation in a mCAR dependent manner. Error bars represented standard deviation of biological replicates (n = 3) in each treatment. p-values were calculated by a two-sided Student's t test. (B) RNA-seq examination of selected genes. RNA-seq validated selected genes expression changes in WT mice. Shc1, Prkar2a and Foxo3 expression did not exhibit perturbations in humanized CAR mice. Each column represents the fold-change value calculated by DESeq2, between treated (n = 3) and untreated (n = 3) samples. Asterisks (*) indicate p-values from DESeq2 analysis. (C) Realtime PCR assay of selected genes mRNA level expression in human primary hepatocytes. Realtime PCR showing selected gene mRNA expression changes for three human primary hepatocyte donors. GDF15, IGF1 IRS2 and FOXO3 all showed remarkable trend changes consistent with their mouse ortholog genes.
In addition, cAMP-dependent protein kinase type II-alpha regulatory subunit (Prkar2a) and breast cancer anti-estrogen resistance protein 1, p130cas (Bcar1) exhibited strong CAR binding sites within ±5 kb of each gene's TSS. Expression analyses for the respective genes indicated that both were selectively induced in WT mice but not CAR –/– mice (Figure 5A). SHC-transforming protein 1 (Shc1) showed a similar trend, exhibiting increased CAR dependent expression. These results are likely of biological importance, since cAMP-activated protein kinases (PKA) play many roles, including an interplay in the progression of various tumors (45), and the PKA regulatory subunit RII alpha is specifically reported to block apoptosis in pathological processes (46). Similarly, BCAR1 belongs to the CAS family of adaptor proteins that contribute to signaling pathways involved in cell adhesion, migration and apoptosis (47). As well, SHC1 activates cell proliferation and positively regulates the cell cycle through signaling in the epidermal growth factor (EGF) pathway (48). Together, the ChIP-exo results advance mCAR’s novel role in promoting tumorigenesis through activation of adaptor proteins such as Shc1 and Bcar1, and the PKA kinase subunit (Prkar2a).
Of interest, several genes exhibiting strong CAR-binding sites demonstrated repressed expression levels. Forkhead box protein O3 (Foxo3), functioning in antitumor activities (49), was significantly repressed with respect to mRNA level in qRT-PCR assays conducted with TCPOBOP treated WT mice, but not in CAR -/- mice. RNA-seq expression results indicated that Foxo3 mRNA expression was reduced 30% in TCPOBOP-treated mice compared with controls (Figure 5A, B). These results imply that mCAR may promote tumorigenesis by suppressing Foxo3. Insulin receptor substrate 2 (IRS2) and insulin-like growth factor I (IGF1) have roles in the insulin signaling pathway, promoting cell proliferation (50,51). Here we show that Igf1 mRNA expression levels decreased in TCPOBOP-treated WT mice, and Irs2 displayed a trend toward decreased expression in TCPOBOP-treated WT mice. In sum, these results suggest that CAR may function to repress hepatocarcinogenesis by affecting the insulin signaling pathway, adding to the complexity of understanding CAR’s role in carcinogenesis.
For each of genes alluded to above, we examined expression level of their human ortholog genes in three human primary hepatocytes cases (Figure 5C). GDF15, FOXO3, IRS2 and IGF1 all exhibited similar mRNA expression trends upon exposure to the hCAR-specific activator, CITCO. However, SHC1, PRKAR2A and BCAR1 did not show consistent trend changes in HPH. Potential explanations for these differences include actual species differences in effect between mCAR and hCAR, or the large interindividual differences inherent in humans that the limited donor samples were unable to reflect.
Cross-referencing RNA-seq transcriptomics datasets with identified species differences in CAR-linked oncogenes
Earlier, we characterized genomic profiles of differential binding loci for mCAR and hCAR. Functional annotation of these loci using IPA suggested that mCAR exhibits stronger binding to regions associated with regulation of cell proliferation and apoptosis. Using RNA-seq data for both WT mice and CAR-humanized mice, we cross-examined both genomic and transcriptomic data with the aim of more specifically identifying species differences in the respectively regulated genes.
For the RNA-seq analyses, we set the expression fold-change ≥2 or ≤−2 and adjusted p-value to ≤0.05 as the threshold for screening significantly perturbed genes. Under this threshold, we identified 1,837 mCAR-specifically perturbed genes and 93 hCAR-specifically perturbed genes. Within those gene lists, 48 overlapped with mCAR stronger-binding genes and 3 overlapped with hCAR stronger-binding genes. Detailed gene lists were shown in Supplementary Table S2. Again, we focused on carcinogenesis-related genes. MYC is a well-studied transcription factor regulating cell cycle and apoptosis and reported as a downstream effector of mCAR in liver proliferation (12). Our ChIP-exo data showed that mCAR binds stronger than hCAR on Myc genes (Table S2). Expression analyses with qRT-PCR and RNA-seq demonstrated that Myc mRNA was significantly upregulated in TCPOBOP-treated WT mice by mCAR, in contrast to the modest Myc induction in hCAR-TG mice with CITCO treatment. Given that Myc plays a key role in regulating cell cycle and proliferation, these respective mCAR and hCAR differences in Myc regulation may underpin their differences as biological contributors to hepatocarcinogenesis.
The ChIP-exo data also identified Ikbke and Ran as possessing stronger mCAR binding avidity than hCAR, results that corresponded well with the respective hepatic transcript expression levels obtained in WT but not in CAR -/- mice (Figure 6A). IKBKE was reported as an oncogene and is overexpressed in various tumors (52,53), whereas RAN is suggested to promote tumor cell survival by regulating cell cycle (54). In addition, Bcl2 modifying factor (Bmf), a BCL2 family protein functioning as an apoptotic activator (55), is repressed in TCPOBOP-treated WT mice by 65% based on qRT-PCR assays, and by 70% in corresponding RNA-seq analysis. However, in hCAR-TG mice, CITCO treatment repressed Bmf expression by only 25% (Figure 6B). Although highly donor-dependent, the data in Figure 6C demonstrated reasonable correspondence in primary cultures of human hepatocytes to the inducibility effects noted in mouse liver. Together, these gene targets provide potential insights into the differences between mouse and human with respect to CAR’s regulatory role in carcinogenesis. Specifically, compared to hCAR, mCAR displayed stronger genomic interactions and greater capability of upregulating expression of the proto-oncogenes, Myc, Ikbke and Ran, and repressing tumor suppressor Bmf.
Figure 6.
mRNA expression analysis of CAR-linked oncogenes and tumor repressor genes with species variations. (A) qRT-PCR assay of selected genes mRNA level expression in mouse liver. qRT-PCR mRNA expression analysis showed that selected genes exhibiting species variation in binding were perturbed significantly in WT mice but not in CAR -/- mice. Error bars represent standard deviation of biological replicates (n = 3) in each treatment. p-values were calculated by a two-sided student t test. (B) RNA-seq showing mRNA expression species variations of selected genes. All selected genes show significant expression changes in WT mice with no significant changes in hCAR-TG mice, indicating species variation at the transcription level in the rodent model. Each column represents a fold-change value, calculated by DESeq2 comparing treated (n = 3) and untreated (n = 3) samples. Asterisks (*) indicate p-values from DESeq2. (C) qRT-PCR assay of selected genes mRNA level expression in HPH. qRT-PCR showing selected gene mRNA expression changes within HPH. Due to the limited availability of human donor specimens, statistical evaluation of the data was not conducted.
mCAR prioritizes binding to direct repeat two half-sites motifs whereas hCAR is less stringent in motif recognition
To investigate CAR-binding motifs with higher biological relevance, we used reference transcriptome to filter sites with more functional binding potential. Primary binding sites were defined with peak p-values ranked in the top 500 of binding sites associated with specific genes; as well, we selected only those genes present both in genomic annotations and in the reference transcriptome. We used primary binding sites for the de novo motif analysis. The best characterized binding element for CAR is the PBREM, consisting of two direct repeat 4 (DR4) units serving as CAR-binding sites (56). Each DR4 unit contains two canonical hexamer half-sites AG(T/G)TCA (57). Here, our data showed the preferred mCAR motif, AGGTCANNNNAG(T/G)TCA, displaying the DR4 feature with a slightly weaker hexamer half-site on the left (Figure 7A). In contrast, the preferred hCAR motif, AG(G/T)TCA, showed only a single hexamer half-site, i.e. a degenerate DR4 unit. The single hexamer motif in hCAR’s four-color plot visually formed a single lane in the center, whereas mCAR’s two hexamer direct repeat motif structure shows double lanes (Figure 7A). We also examined motifs generated from weaker binding sites, expanding the input sequence to the top 1,000 sites. At this level, the mCAR motif exhibited only a degenerate single hexamer AG(T/G)TCA, the same as with hCAR (Figure 7B). This disparity of DNA binding sequences was further substantiated by comparing motifs from mCAR and hCAR differential binding regions characterized earlier with DESeq2 (Figure 7C). Motifs from mCAR’s stronger binding sites displayed two strong hexamer half-sites, AGGTCANNNNAG(T/G)TCA, rather than one weak/one strong half-site as seen in the primary binding motif. Once again, the hCAR stronger binding site motif exhibited the single hexamer, AG(T/G)TCA. The hexamer binding sites identified here in the in vivo investigations are consistent with those characterized using in vitro analyses (58). The in vitro studies also demonstrate that hCAR and mCAR bind DNA interchangeably well with human retinoid X receptor (RXR) as the dimer partner, and that CAR is also capable of binding DNA directly as a monomer (57). Combined, the results support the concept that the different hCAR and mCAR binding motif preferences identified here are driven by intrinsic structural differences between the respective CAR proteins, rather than RXR dimerization preferences. The motif analyses suggest that across the genome, mCAR has a stronger tendency to recognize a direct repeat with two half-site structures, but as affinity weakens, mCAR binds to degenerate single half-sites. In contrast, hCAR DNA binding sites were less dependent on the two half-site structure. Recently, several reports demonstrated that DNA binding site sequences can modulate nuclear receptor activities (59,60). Consistent with this view, the motif interactions exhibited by mCAR’s and hCAR’s top binding sites appear to specify the genomic basis for species variation for these receptors, findings that lend new insights into differences in the genomic interactions of rodent vs human CAR.
Figure 7.
Motif analysis for hCAR and mCAR. (A) hCAR and mCAR motifs from the top 500 primary binding sites. The top 500 primary binding sites clearly illustrated two hexamer half-sites DR4 structural motifs for mCAR, whereas hCAR exhibited only one hexamer. Motifs and 2-bits motif logos were generated using MEME suites. Four-color plot represents ±50bp genome sequences, centered on the top 500 primary binding sites. The color scheme for the four-color plot is the same as with motif logos. (B) hCAR and mCAR motifs from the top 1000 primary binding sites. mCAR and hCAR motifs from the top 1000 primary binding sites were very similar, with the mCAR motif degenerated to one hexamer half-site. (C) Motifs characterization of mCAR and hCAR differential binding sites. Differential binding sites analysis revealed that hCAR has high preference for single hexamer motifs, whereas mCAR preferred a complete two half-sites structure. (D) Motif comparison between direct / indirect activators. Binding motifs resulting from direct- or indirect-activated CAR were largely equivalent for both mCAR and hCAR, indicating that different modes of receptor activation do not appear to alter CAR binding profiles.
Subsequently, we compared motifs from direct activator-treated samples with indirect activator-treated samples (Figure 7D). Both hCAR and mCAR exhibited similar motif preferences when compared between direct activators, i.e. CITCO/TCPOBOP, and the indirect activator, PB, suggesting that the DNA sequence recognition of CAR was not influenced by the presence of activator. To validate the characterized motifs, we used FIMO to scan the mCAR motif AGGTCANNNNAG(T/G)TCA for putative binding locations within established CAR binding genes, such as Cyp2b10 and Ces2a (Supplementary Figure S1). We successfully characterized clusters of putative CAR binding sites that were consistent with CAR peaks visualized on the genome browser, particularly within the PBREM of Cyp2b10, a classic CAR-binding element. FIMO motif scans were also conducted for Gdf15, Myc, Ikbke and Ran, and identified putative mCAR motif locations consistent with the differentially mapped binding sites (Figure 8). Therefore, the motif analyses supported the findings presented earlier, depicting differential binding results viewed with respect to binding strength and binding location, strengthening the conclusion that CAR binding to DNA elements is largely unaffected by the presence of ligand.
Figure 8.
Visualization of selected genes and putative motif locations. (A) IGV screenshots showing differential binding peaks and putative motif locations. CAR binding regions on Myc and Ikbke are shown using IGV, indicating preferential binding of mCAR on peaks highlighted by red rectangles. Mapped reads are separated by strands, with forward strand reads on the upper track (green) and reverse strand reads on the bottom track (red). From top to bottom tracks: hCAR, mCAR, Refseq gene and putative mCAR motif locations using FIMO default settings. Track heights are normalized according to mCAR and hCAR total reads number within peaks. Genome tracks are 10k in length. (B) IGV screenshots showing peaks and putative motif locations on Gdf15. Four putative motif locations by FIMO are indicated with red arrows. (C) Putative mCAR motifs and their coordinates. q-values were calculated by FIMO. The mCAR motif logo from top 500 primary binding sites is shown on top of the putative binding sequences.
DISCUSSION
In this study, in vivo genome-wide profiling analyses were conducted to detail binding interactions for the mouse and human CAR transcription factors. These experiments were performed using a novel adenoviral delivery system targeting mouse livers, coupled with high resolution ChIP-exo assays. CAR is unusual in the nuclear receptor superfamily in that although constitutively active, CAR is sequestered in a cytosolic binding complex. Release and subsequent nuclear translocation of the transcriptional regulator occurs either through direct activation with specific ligands, or indirect activation through an epidermal growth factor receptor (EGFR)-mediated signaling pathway (7). Our differential analysis of both mCAR and hCAR genomic binding profiles demonstrated that in terms of binding locations as well as binding signal intensities, direct vs. indirect CAR activation resulted in almost identical DNA binding profiles. Of the 59,508 genomic total binding sites detected in direct/indirect activation differential analysis, only 56 sites (0.09%) showed significant differences for mCAR, whereas only two sites (0.003%) exhibited significant differences for hCAR. Although differing gene expression profiles for CAR were reported previously for direct and indirect activation (61), the motif analysis conducted here revealed that the consensus binding sites for the direct-/indirect-activated hCAR, or for the direct-/indirect-activated mCAR, were indistinguishable (Figure 7D). Along these lines, one consideration is that CAR’s conformation may undergo change in the presence of a direct ligand, potentially affecting the receptor's interaction with transcriptional co-regulators. In these respects, other nuclear receptors are reported to realign and stabilize their helix 12 (H12)/AF2 domain upon ligand binding, thereby directing coactivator recruitment (62). CAR is unique among the nuclear receptors in that its constitutive activity results from a charge-charge interaction between helix 4 and H12/AF2, maintaining an active conformation that mimics ligand docking (63). However, this constitutive activation activity is suggested as somewhat inferior with respect to strength of coactivator recruitment, compared with direct ligand-binding activation (63). Another important consideration is that the indirect activation pathway proceeds through EGFR signaling modulation, which itself may affect other gene expression programs (7,16).
Consistent with CAR’s known regulatory role in drug metabolism and energy homeostasis, gene ontology analysis indicated that most CAR binding genes were enriched in those biological processes. For example, the ChIP-exo results demonstrated strong binding events associated with CAR in genes including Ces2a, ATP binding cassette subfamily C member 3 (Abcc3), cytochrome p450 oxidoreductase (Por), Pepck, G6pc, stearoyl-CoA desaturase-1 (Scd1) and Srebf1. These binding profiles strongly suggest that CAR directly regulates these genes at the transcriptional level, profiles supported by gene expression reports. As one goal of this study was to provide genomic insights into the differential contributions of rodent vs human CAR in cell proliferative and tumorigenic processes, IPA disease and function analysis indicated that regions exhibiting significantly stronger mCAR binding signals were linked to cell proliferation and apoptosis functions, contrasting to hCAR with higher associations with energy / metabolic functions.
Visualization of ChIP-exo data on the genome browser (Figure 2) illustrated that the mCAR and hCAR samples yielded overall matching binding patterns in most scenarios. Further, the hexamer half-site consensus sequences, characterized from the top 1,000 primary binding sites, were AG(G/T)TCA, for both mCAR and hCAR. These results indicated that the mCAR and hCAR interacting genomic profiles are similar in mouse liver. Likely contributing to this result is the highly conserved DBD existing between mCAR and hCAR, demonstrating a similarity of amino acid sequences between the respective species’ of 95.6%, with only 4 non-conservative amino acid replacements occurring in the 91aa length of the DBD (Supplementary Table S3) (74).
Of particular note, differential binding analysis for mCAR and hCAR identified ∼1,000 genes associated with species distinctive binding in mouse liver (Figure 3D). To characterize these binding differences, we used DESeq2 to normalize samples with their signal tag numbers and then quantitatively measured the differences at each of the binding sites. Of all the binding regions examined, 5.9% of the binding peaks exhibited significant mCAR vs. hCAR differences (binding FC > 2, q-value < 0.05). Previous reports suggest significant effector pathway differences programmed through mCAR and hCAR, particularly in cell proliferation and tumor promotion, raising questions for the human relevancy of chemical testing performed in rodent models (64). By cross-referencing the ChIP-exo data with published RNA-seq data, and validating selected results with qRT-PCR mRNA assays, several cancer-related genes were identified both as likely CAR transcriptionally regulated and as exhibiting species differences. With respect to cell proliferation, several genes possessed sites exhibiting both stronger binding for mCAR as well as associated CAR-dependent expression level changes induced by CAR ligands. In these respects, the oncogene Myc is well documented for its roles in tumorigenesis in multiple tissues (65). Overexpression of MYC induces hepatocellular cancer in mouse models (66) and dysregulation of MYC is frequently observed in clinical hepatocellular carcinoma samples (67). Our ChIP-exo data demonstrated that mCAR bound to a site +2 kb downstream of the mouse Myc TSS, whereas hCAR did not. RNA-seq expression levels of Myc also demonstrate similar species variation. Following ligand activation of mCAR in mice, Myc mRNA expression increases by ∼10-fold in WT mice, whereas in mice humanized with hCAR, Myc expression only increases by ∼2-fold (Figure 6). Of related interest, the tumor suppressor FOXO3 is reported to suppress MYC by activating ARF (65). Our data indicate that Foxo3 is repressed by CAR, an event projected to facilitate CAR’s activation of Myc. Given that Myc is reported as a downstream target of CAR in liver proliferation (12), differential regulation of Myc is likely to play a central role in species variability of CAR response in rodent tumorigenesis.
In addition, Gdf15, a gene associated with cell proliferation, was identified as bound by both species’ CAR and transcriptionally activated. Gdf15 is overexpressed in clinical liver cancer tissues, inducing proliferation of liver cancer stem cells through activating AKT/GSK-3β/β-catenin pathways (44). The Wnt/β-catenin pathway has been extensively studied with respect to hepatic carcinogenesis (68,69). In the current investigation, ChIP-exo data revealed several peaks with strong binding signals in the Gdf15 promoter region, and putative binding motifs were characterized using local analysis of consensus sequences. Both RNA-seq results and qRT-PCR assays (Figure 5) demonstrate that the expression level of Gdf15 is increased markedly, up to ∼15-fold, with CAR ligand treatment. The qRT-PCR assays further illustrated that induction of Gdf15 was CAR-dependent, as CAR -/- mice show no induction. These results suggest that regulation of Gdf15 is of high relevance with respect to CAR’s role in promoting mouse liver carcinogenesis. In addition to Gdf15, other novel cancer-related genes that were identified as CAR-regulated include Shc1, Prkar2a, and Bcar1; and, Foxo3, a gene with apoptotic-repressing function. These interactions reveal apparently complex mechanisms of CAR that underscore its importance as a regulator of the carcinogenic process.
Adding further complexity, our ChIP-exo investigation identified apparent intensive genomic cross-talk of CAR with other nuclear and soluble receptors. For examples, CAR exhibited significant binding to various receptors’ promoter regions, including hepatocyte nuclear factor 4 alpha (Hnf4a), retinoic acid recetor beta ( Rarb), retinoid X receptor beta ( Rxrb), PXR (Nr1i2), peroxisome proliferator activated receptor delta (Ppard), estrogen receptor alpha (Esr1), COUP transcription factor 2 (Nr2f2), small heterodimer partner (Nr0b2; Shp1), as well as aryl hydrocarbon receptor (Ahr;AHR), and CAR (Nr1i3) itself. Although genomic binding does not necessarily denote a direct link to biological function, the concept of nuclear receptor cross-talk at the genomic level is fascinating, in particular for cases where CAR’s genomic binding events are further correlated as directly regulating expression. In these respects, several of the aforementioned CAR-binding genes; specifically, Rarb, Shp1 and Ahr, demonstrate RNA-seq derived expression level responsiveness to CAR activators (70). In this context, expression of both mouse and human CAR is reportedly modulated through AHR interaction (71). Consistent with these concepts, recent studies examining functional attributes of nuclear receptors suggest important roles for their recognition as dynamic scaffolding proteins, capable of fine-tuning transcriptional signaling outcomes through modification by ligands, and DNA sequence interactions that themselves can alter co-regulator binding (72).
Increasingly, humanized CAR/PXR rodent models are being deployed to facilitate hepatocarcinogenesis studies and to inform toxicology investigations in industry. In these respects, results from the present investigation, comparatively assessing mCAR and hCAR genomic profiling across the mouse genome, are of increasing relevance. Although extrapolation of hCAR genomic profiling from rodent systems to human should be viewed with some caution, the DBD structural conservation between the receptors suggests that a high degree of functional conservation likely exists among the receptors with respect to target gene interactions and subsequent regulation (73). Combined with further transcriptomics and biological pathway validation, studies such as those described here should serve to guide further investigations of CAR’s regulatory roles in the human biological interface.
CONCLUSIONS
Using in vivo high-resolution in vivo profiling of mCAR and hCAR genomic interactions in transgenic liver models, integrated with transcriptomic data analyses, this investigation identified several novel CAR target genes established as key regulators of cell proliferation and carcinogenesis. Comparative analysis of mCAR and hCAR genomic binding revealed species differences in their respective interaction profiles and regulation of several oncogenes that serve to underlie biological variation in xenobiotic response. These findings substantiate the importance of CAR as a general transcription activator in the hepatic environment and its role as a critical regulator not only of xenobiotic and energy metabolism, but also general coordinator of cell proliferation, apoptosis and liver cancer development.
DATA AVAILABILITY
The datasets containing sequencing reads are available in the NCBI Gene Expression Omnibus repository (Accession GE112199). The scripts and command lines used to process the bioinformatics data reported in this manuscript are available in the github repository, through the following link: https://github.com/omielab-car/omiecinski_car_chip_exo. A UCSC browser session for the sample reads bigWig files are available through the following link: https://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=nxb939&hgS_otherUserSessionName=ChIP_exo_CAR.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Wen Xie (University of Pittsburgh School of Pharmacy) for providing the CAR/PXR double-KO mice that were used to generate the CARKO mice in this study. The authors also gratefully acknowledge the efforts of Dr Aswathy Sebastian for general bioinformatics counseling and differential binding analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH grants from the National Institute of General Medical Sciences [GM066411 to C.J.O.]; National Environmental Health Sciences Institute [ES013768 to B.F.P.]; National Institute of Food and Agriculture, U.S. Department of Agriculture [2014-06624 to C.J.O.]. Funding for open access charge: NIH and endowment funds.
Conflict of interest statement. B.F.P. has a financial interest in Peconic, LLC, which utilizes the ChIP-exo technology implemented in this study and could potentially benefit from the outcomes of this research.
REFERENCES
- 1. Krasowski M.D., Yasuda K., Hagey L.R., Schuetz E.G.. Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors). Nucl. Recept. 2005; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steinmetz A.C., Renaud J.P., Moras D.. Binding of ligands and activation of transcription by nuclear receptors. Annu. Rev. Biophys. Biomol. Struct. 2001; 30:329–359. [DOI] [PubMed] [Google Scholar]
- 3. Kojetin D.J., Matta-Camacho E., Hughes T.S., Srinivasan S., Nwachukwu J.C., Cavett V., Nowak J., Chalmers M.J., Marciano D.P., Kamenecka T.M. et al. Structural mechanism for signal transduction in RXR nuclear receptor heterodimers. Nat. Commun. 2015; 6:8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dussault I., Lin M., Hollister K., Fan M., Termini J., Sherman M.A., Forman B.M.. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol. Cell. Biol. 2002; 22:5270–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Omiecinski C.J., Coslo D.M., Chen T., Laurenzana E.M., Peffer R.C.. Multi-species analyses of direct activators of the constitutive androstane receptor. Toxicol. Sci. 2011; 123:550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang H., Wang H.. Signaling control of the constitutive androstane receptor (CAR). Protein Cell. 2014; 5:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mutoh S., Sobhany M., Moore R., Perera L., Pedersen L., Sueyoshi T., Negishi M.. Phenobarbital indirectly activates the constitutive active androstane receptor (CAR) by inhibition of epidermal growth factor receptor signaling. Sci. Signal. 2013; 6:ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao J., Xie W.. Targeting xenobiotic receptors PXR and CAR for metabolic diseases. Trends Pharmacol. Sci. 2012; 33:552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamamoto Y., Moore R., Goldsworthy T.L., Negishi M., Maronpot R.R.. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004; 64:7197–7200. [DOI] [PubMed] [Google Scholar]
- 10. Huang W., Zhang J., Washington M., Liu J., Parant J.M., Lozano G., Moore D.D.. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol. Endocrinol. 2005; 19:1646–1653. [DOI] [PubMed] [Google Scholar]
- 11. Columbano A., Ledda-Columbano G.M., Pibiri M., Cossu C., Menegazzi M., Moore D.D., Huang W., Tian J., Locker J.. Gadd45beta is induced through a CAR-dependent, TNF-independent pathway in murine liver hyperplasia. Hepatology. 2005; 42:1118–1126. [DOI] [PubMed] [Google Scholar]
- 12. Blanco-Bose W.E., Murphy M.J., Ehninger A., Offner S., Dubey C., Huang W., Moore D.D., Trumpp A.. C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology. 2008; 48:1302–1311. [DOI] [PubMed] [Google Scholar]
- 13. Shizu R., Shindo S., Yoshida T., Numazawa S.. MicroRNA-122 down-regulation is involved in phenobarbital-mediated activation of the constitutive androstane receptor. PLoS One. 2012; 7:e41291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Braeuning A., Schwarz M.. Is the question of phenobarbital as potential liver cancer risk factor for humans really resolved. Arch. Toxicol. 2016; 90:1525–1526. [DOI] [PubMed] [Google Scholar]
- 15. La Vecchia C., Negri E.. A review of epidemiological data on epilepsy, phenobarbital, and risk of liver cancer. Eur. J. Cancer Prev. 2014; 23:1–7. [DOI] [PubMed] [Google Scholar]
- 16. Negishi M. Phenobarbital meets phosphorylation of nuclear receptors. Drug Metab. Dispos. 2017; 45:532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rhee H.S., Pugh B.F.. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011; 147:1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saini S.P., Sonoda J., Xu L., Toma D., Uppal H., Mu Y., Ren S., Moore D.D., Evans R.M., Xie W.. A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol. Pharmacol. 2004; 65:292–300. [DOI] [PubMed] [Google Scholar]
- 19. Rhee H.S., Pugh B.F.. ChIP-exo Method for Identifying Genomic Location of DNA-Binding Proteins with Near-Single-Nucleotide Accuracy. Curr. Protoc. Mol. Biol. 2012; doi:10.1002/0471142727.mb2124s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Page J.L., Johnson M.C., Olsavsky K.M., Strom S.C., Zarbl H., Omiecinski C.J.. Gene expression profiling of extracellular matrix as an effector of human hepatocyte phenotype in primary cell culture. Toxicol. Sci. 2007; 97:384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olsavsky K.M., Page J.L., Johnson M.C., Zarbl H., Strom S.C., Omiecinski C.J.. Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues. Toxicol. Appl. Pharmacol. 2007; 222:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hao R., Su S., Wan Y., Shen F., Niu B., Coslo D.M., Albert I., Han X., Omiecinski C.J.. Bioinformatic analysis of microRNA networks following the activation of the constitutive androstane receptor (CAR) in mouse liver. Biochim. Biophys. Acta. 2016; 1859:1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Metsalu T., Vilo J.. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic. Acids. Res. 2015; 43:W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 29. Huang da W., Sherman B.T., Lempicki R.A.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic. Acids. Res. 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Albert I., Wachi S., Jiang C., Pugh B.F.. GeneTrack–a genomic data processing and visualization framework. Bioinformatics. 2008; 24:1305–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng S.L., Bammler T.K., Cui J.Y.. RNA sequencing reveals age and species differences of constitutive androstane receptor-targeted drug-processing genes in the liver. Drug Metab. Dispos. 2017; 45:867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boogerd C.J., Aneas I., Sakabe N., Dirschinger R.J., Cheng Q.J., Zhou B., Chen J., Nobrega M.A., Evans S.M.. Probing chromatin landscape reveals roles of endocardial TBX20 in septation. J. Clin. Invest. 2016; 126:3023–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mutoh S., Osabe M., Inoue K., Moore R., Pedersen L., Perera L., Rebolloso Y., Sueyoshi T., Negishi M.. Dephosphorylation of threonine 38 is required for nuclear translocation and activation of human xenobiotic receptor CAR (NR1I3). J. Biol. Chem. 2009; 284:34785–34792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H., Chen T., Cottrell J., Wang H.. Nuclear translocation of adenoviral-enhanced yellow fluorescent protein-tagged-human constitutive androstane receptor (hCAR): a novel tool for screening hCAR activators in human primary hepatocytes. Drug Metab. Dispos. 2009; 37:1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sueyoshi T., Moore R., Sugatani J., Matsumura Y., Negishi M.. PPP1R16A, the membrane subunit of protein phosphatase 1beta, signals nuclear translocation of the nuclear receptor constitutive active/androstane receptor. Mol. Pharmacol. 2008; 73:1113–1121. [DOI] [PubMed] [Google Scholar]
- 37. Chen T., Laurenzana E.M., Coslo D.M., Chen F., Omiecinski C.J.. Proteasomal interaction as a critical activity modulator of the human constitutive androstane receptor. Biochem. J. 2014; 458:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheng C., Alexander R., Min R., Leng J., Yip K.Y., Rozowsky J., Yan K.K., Dong X., Djebali S., Ruan Y. et al. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. 2012; 22:1658–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ouyang Z., Zhou Q., Wong W.H.. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:21521–21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng C., Gerstein M.. Modeling the relative relationship of transcription factor binding and histone modifications to gene expression levels in mouse embryonic stem cells. Nucleic Acids Res. 2012; 40:553–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012; 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ochsner S.A., Tsimelzon A., Dong J., Coarfa C., McKenna N.J.. Research resource: a reference transcriptome for constitutive androstane receptor and pregnane X receptor xenobiotic signaling. Mol. Endocrinol. 2016; 30:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zimmers T.A., Jin X., Hsiao E.C., McGrath S.A., Esquela A.F., Koniaris L.G.. Growth differentiation factor-15/macrophage inhibitory cytokine-1 induction after kidney and lung injury. Shock. 2005; 23:543–548. [PubMed] [Google Scholar]
- 44. Xu Q., Xu H.X., Li J.P., Wang S., Fu Z., Jia J., Wang L., Zhu Z.F., Lu R., Yao Z.. Growth differentiation factor 15 induces growth and metastasis of human liver cancer stem-like cells via AKT/GSK-3beta/beta-catenin signaling. Oncotarget. 2017; 8:16972–16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caretta A., Mucignat-Caretta C.. Protein kinase a in cancer. Cancers (Basel). 2011; 3:913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benetti L., Roizman B.. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9411–9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cabodi S., del Pilar Camacho-Leal M., Di Stefano P., Defilippi P.. Integrin signalling adaptors: not only figurants in the cancer story. Nat. Rev. Cancer. 2010; 10:858–870. [DOI] [PubMed] [Google Scholar]
- 48. Zheng Y., Zhang C., Croucher D.R., Soliman M.A., St-Denis N., Pasculescu A., Taylor L., Tate S.A., Hardy W.R., Colwill K. et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013; 499:166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deng Y., Wang F., Hughes T., Yu J.. FOXOs in Cancer Immunity: Knowns and Unknowns. Semin. Cancer Biol. 2018; 50:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reiss K., Del Valle L., Lassak A., Trojanek J.. Nuclear IRS-1 and cancer. J. Cell. Physiol. 2012; 227:2992–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. White M.F. IRS2 integrates insulin/IGF1 signalling with metabolism, neurodegeneration and longevity. Diabetes Obes. Metab. 2014; 16(Suppl 1):4–15. [DOI] [PubMed] [Google Scholar]
- 52. Hutti J.E., Shen R.R., Abbott D.W., Zhou A.Y., Sprott K.M., Asara J.M., Hahn W.C., Cantley L.C.. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol. Cell. 2009; 34:461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zubair H., Azim S., Srivastava S.K., Ahmad A., Bhardwaj A., Khan M.A., Patel G.K., Arora S., Carter J.E., Singh S. et al. Glucose metabolism reprogrammed by overexpression of IKKepsilon promotes pancreatic tumor growth. Cancer Res. 2016; 76:7254–7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barres V., Ouellet V., Lafontaine J., Tonin P.N., Provencher D.M., Mes-Masson A.M.. An essential role for Ran GTPase in epithelial ovarian cancer cell survival. Mol. Cancer. 2010; 9:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pinon J.D., Labi V., Egle A., Villunger A.. Bim and Bmf in tissue homeostasis and malignant disease. Oncogene. 2008; 27(Suppl 1):S41–S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sueyoshi T., Kawamoto T., Zelko I., Honkakoski P., Negishi M.. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J. Biol. Chem. 1999; 274:6043–6046. [DOI] [PubMed] [Google Scholar]
- 57. Handschin C., Meyer U.A.. A conserved nuclear receptor consensus sequence (DR-4) mediates transcriptional activation of the chicken CYP2H1 gene by phenobarbital in a hepatoma cell line. J. Biol. Chem. 2000; 275:13362–13369. [DOI] [PubMed] [Google Scholar]
- 58. Frank C., Gonzalez M.M., Oinonen C., Dunlop T.W., Carlberg C.. Characterization of DNA complexes formed by the nuclear receptor constitutive androstane receptor. J. Biol. Chem. 2003; 278:43299–43310. [DOI] [PubMed] [Google Scholar]
- 59. Meijsing S.H., Pufall M.A., So A.Y., Bates D.L., Chen L., Yamamoto K.R.. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009; 324:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang J., Chalmers M.J., Stayrook K.R., Burris L.L., Wang Y., Busby S.A., Pascal B.D., Garcia-Ordonez R.D., Bruning J.B., Istrate M.A. et al. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat. Struct. Mol. Biol. 2011; 18:556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li D., Mackowiak B., Brayman T.G., Mitchell M., Zhang L., Huang S.M., Wang H.. Genome-wide analysis of human constitutive androstane receptor (CAR) transcriptome in wild-type and CAR-knockout HepaRG cells. Biochem. Pharmacol. 2015; 98:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shan L., Vincent J., Brunzelle J.S., Dussault I., Lin M., Ianculescu I., Sherman M.A., Forman B.M., Fernandez E.J.. Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol. Cell. 2004; 16:907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dussault I., Lin M., Hollister K., Fan M., Termini J., Sherman M.A., Forman B.M.. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol. Cell. Biol. 2002; 22:5270–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ross J., Plummer S.M., Rode A., Scheer N., Bower C.C., Vogel O., Henderson C.J., Wolf C.R., Elcombe C.R.. Human constitutive androstane receptor (CAR) and pregnane X receptor (PXR) support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogens phenobarbital and chlordane in vivo. Toxicol. Sci. 2010; 116:452–466. [DOI] [PubMed] [Google Scholar]
- 65. Stine Z.E., Walton Z.E., Altman B.J., Hsieh A.L., Dang C.V.. MYC, metabolism, and cancer. Cancer Discov. 2015; 5:1024–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shachaf C.M., Kopelman A.M., Arvanitis C., Karlsson A., Beer S., Mandl S., Bachmann M.H., Borowsky A.D., Ruebner B., Cardiff R.D. et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004; 431:1112–1117. [DOI] [PubMed] [Google Scholar]
- 67. Wang L., Zhang X., Jia L.T., Hu S.J., Zhao J., Yang J.D., Wen W.H., Wang Z., Wang T., Zhao J. et al. c-Myc-mediated epigenetic silencing of MicroRNA-101 contributes to dysregulation of multiple pathways in hepatocellular carcinoma. Hepatology. 2014; 59:1850–1863. [DOI] [PubMed] [Google Scholar]
- 68. Cui J., Zhou X., Liu Y., Tang Z.. Mutation and overexpression of the beta-catenin gene may play an important role in primary hepatocellular carcinoma among Chinese people. J. Cancer Res. Clin. Oncol. 2001; 127:577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wong C.M., Fan S.T., Ng I.O.. beta-Catenin mutation and overexpression in hepatocellular carcinoma: clinicopathologic and prognostic significance. Cancer. 2001; 92:136–145. [DOI] [PubMed] [Google Scholar]
- 70. Cui J.Y., Klaassen C.D.. RNA-Seq reveals common and unique PXR- and CAR-target gene signatures in the mouse liver transcriptome. Biochim. Biophys. Acta. 2016; 1859:1198–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Patel R.D., Hollingshead B.D., Omiecinski C.J., Perdew G.H.. Aryl-hydrocarbon receptor activation regulates constitutive androstane receptor levels in murine and human liver. Hepatology. 2007; 46:209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nwachukwu J.C., Nettles K.W.. The nuclear receptor signalling scaffold: insights from full-length structures. EMBO J. 2012; 31:251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hemberg M., Kreiman G.. Conservation of transcription factor binding events predicts gene expression across species. Nucleic Acids Res. 2011; 39:7092–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. di Masi A., De Marinis E., Ascenzi P., Marino M.. Nuclear receptors CAR and PXR: Molecular, functional, and biomedical aspects. Mol. Aspects Med. 2009; 30:297–343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets containing sequencing reads are available in the NCBI Gene Expression Omnibus repository (Accession GE112199). The scripts and command lines used to process the bioinformatics data reported in this manuscript are available in the github repository, through the following link: https://github.com/omielab-car/omiecinski_car_chip_exo. A UCSC browser session for the sample reads bigWig files are available through the following link: https://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=nxb939&hgS_otherUserSessionName=ChIP_exo_CAR.