Abstract
Pif1 family DNA helicases are conserved from bacteria to humans and have critical and diverse functions in vivo that promote genome integrity. Pif1 family helicases share a 23 amino acid region, called the Pif1 signature motif (SM) that is unique to this family. To determine the importance of the SM, we did mutational and functional analysis of the SM from the Saccharomyces cerevisiae Pif1 (ScPif1). The mutations deleted portions of the SM, made one or multiple single amino acid changes in the SM, replaced the SM with its counterpart from a bacterial Pif1 family helicase and substituted an α-helical domain from another helicase for the part of the SM that forms an α helix. Mutants were tested for maintenance of mitochondrial DNA, inhibition of telomerase at telomeres and double strand breaks, and promotion of Okazaki fragment maturation. Although certain single amino acid changes in the SM can be tolerated, the presence and sequence of the ScPif1 SM were essential for all tested in vivo functions. Consistent with the in vivo analyses, in vitro studies showed that the presence and sequence of the ScPif1 SM were critical for ATPase activity but not substrate binding.
INTRODUCTION
RNA and DNA helicases are proteins that perform key functions in all aspects of nucleic acid biology. Bacteria and eukaryotes express multiple helicases, often with non-overlapping functions. For example, Saccharomyces cerevisiae encodes 134 proteins (1) with the sequence motifs of helicases. Helicases were first defined by their ability to use the energy of nucleoside triphosphate (NTP) hydrolysis to catalyze the unwinding of duplex nucleic acids, but it is now realized that helicases can have more diverse activities, such as translocation on single-stranded RNA or DNA and protein displacement from a nucleic acid substrate (2).
Based on amino acid sequence similarity, helicases are divided into six superfamilies (SF1–6) (3). SF1 helicases have a conserved helicase core region consisting of two RecA-like domains (4) and seven helicase motifs (I, Ia, II, III, IV, V and VI) (5). The distance between the motifs is also conserved, although the sequences of these spacers are not (6). Furthermore, each superfamily can be divided into multiple smaller helicase families, named for its first identified member. Helicases in the same family have higher sequence similarity within their helicase motifs and sometimes within the spacer regions between the motifs than they do with helicases in a different family. Pif1-like helicases comprise one of the three families of SF1 helicases (7).
A search of the NCBI protein database (https://www.ncbi.nlm.nih.gov/protein) indicates that Pif1 family helicases are found in virtually all eukaryotes, many eubacteria, and some archaea bacteria (8). Most eukaryotes encode a single Pif1 family helicase (e.g. humans and Schizosaccharomyces pombe) while some single-celled organisms (e.g.S. cerevisiae, Candida albicans and Cryptococcus neoformans) encode two and a few organisms (e.g.Trypanosoma brucei and Arabidopsis thaliana) contain even more, and in these cases some of the sequences are quite divergent (8). The other Pif1 helicase in budding yeast, Rrm3, promotes replication past stable protein complexes at over 1000 genomic sites (9), restricts DNA synthesis during replication stress via its Orc5-binding domain (10) and is important for the repair of replication generated double-strand breaks (DSBs) (11).
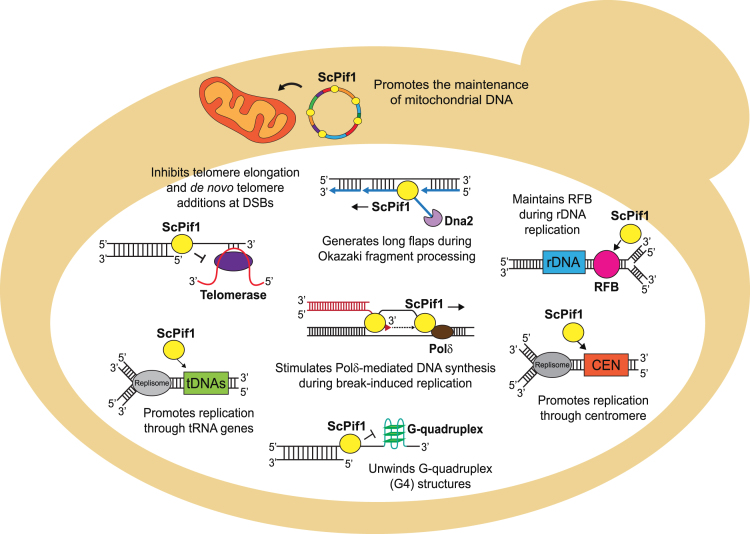
Saccharomyces cerevisiae Pif1 (ScPif1), the prototypical and best-studied helicase in the Pif1 family, has multiple and diverse in vivo functions (Figure 1) [reviewed in (8,12,13)]. ScPif1 was discovered in a screen for genes affecting mitochondrial DNA (mtDNA) recombination (14) and is also critical for the maintenance of mtDNA (15). ScPif1 was re-discovered in a screen for genes affecting telomeres when it was identified as an inhibitor of telomerase-mediated telomere lengthening (16,17). ScPif1 inhibits telomerase processivity at both telomeres and DSBs by removing the enzyme from DNA ends (18,19). Its actions at DSBs is regulated by Mec1-dependent phosphorylation (20). When ScPif1 is unable to remove telomerase from DSBs, the rate of gross chromosomal rearrangements (GCRs) dramatically increases due to the formation of terminal deletions (21).
Figure 1.
Diagram showing the diverse in vivo functions of the ScPif1 DNA helicase. Figure is inspired by an earlier depiction of ScPif1 functions (12). See introduction for references for these functions.
ScPif1’s nuclear functions are not limited to telomeres. ScPif1 contributes to semi-conservative DNA replication in multiple ways. It affects Okazaki fragment maturation by generating long flaps that are cleaved by the nuclease activity of Dna2 (22,23). It promotes fork progression and suppresses DNA damage at several hard to replicate sites, such as tRNA genes (24,25), G-quadruplex (G4) structures (26,27), and centromeres (Chen et al., in preparation). In contrast, ScPif1 inhibits fork progression at the replication fork barrier within ribosomal DNA (rDNA), which ensures that replication and transcription occur in the same direction through rDNA repeats (28). ScPif1 stimulates Pol δ-mediated DNA synthesis and bubble migration during break-induced replication repair of DSBs (29,30). ScPif1 also has multiple in vitro activities. ScPif1 exhibits weak unwinding activity on conventional 5′-tailed duplex DNA, but robustly unwinds forked DNA (31), G4 structures (26,32,33) and RNA/DNA hybrids (R-loops) (33–35). ScPif1 also displaces proteins from single-stranded DNA (ssDNA) (36), enhances the processivity of DNA polymerase δ (29,37) and promotes annealing of complementary DNA single strands (38). In most cases, it is not clear which of its in vitro activities is responsible for a specific in vivo function.
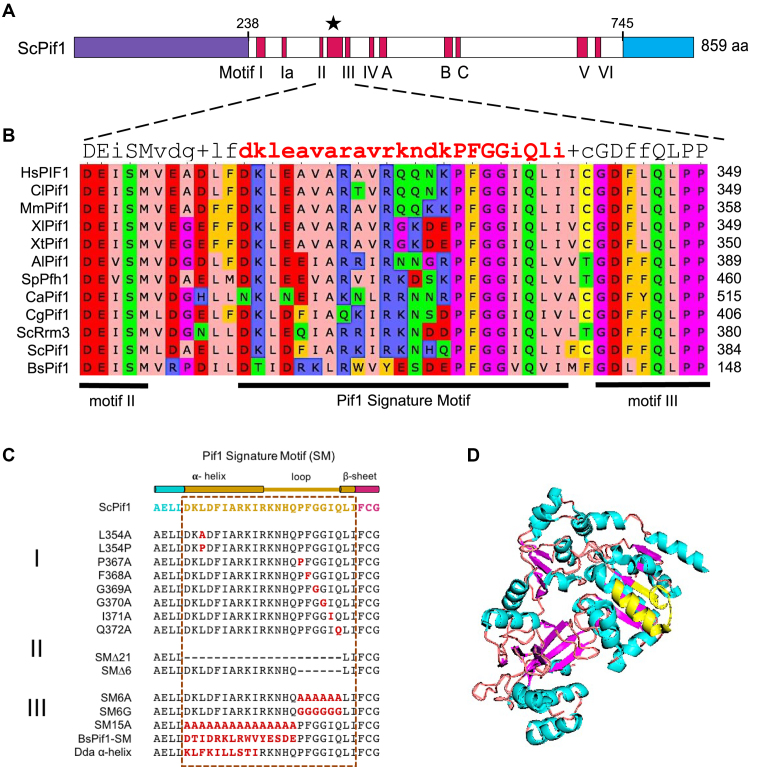
At the structural level, most Pif1 family helicases have three regions: an amino terminus, the ∼400–500 amino acid helicase domain and a carboxyl terminus (Figure 2A). While the helicase core is highly conserved, the amino and carboxyl termini diverge both in size and sequence among Pif1 family members. In addition to the seven helicase motifs, the Pif1 helicase domain contains three motifs (A, B and C) whose functions are unknown which are shared with the Escherichia coli RecD helicase (39,40). Pif1 family helicases also contain the Pif1 signature motif (SM) that is located between motifs II and III (Figure 2A) (8). In our original description of the SM, we said that it was 21 amino acids in length and was absent from plant Pif1 family helicases (8). As more Pif1 family helicases were sequenced, it became clear that the SM is probably better defined as being 23 amino acids in length, as the next two amino acids are also highly conserved (LI or LV) (Figure 2B). Also at least some plant Pif1 family helicases have the SM, although they tend to be more divergent than those in yeasts and multi-cellular organisms (C. L. Geronimo, personal observation).
Figure 2.
(A) Schematic of domains of the ScPif1 DNA helicase. The helicase core domain is in white. The red bars within the core domain are the conserved helicase motifs, and the star indicates the position of the Pif1-family Signature Motif (SM), which is located between helicase motifs II and III. The purple and blue domains are the amino and carboxyl regions of ScPif1, respectively, which vary in size and sequence among different Pif1 family helicases. As indicated, the amino terminus of ScPif1 is 237 amino acids, the helix domain is 508 amino acids and the carboxyl region is 114 amino acids. (B) Sequence alignment of Pif1-family SM from different organisms (Al, Aspergillus lentulus; Bs; Bacteroides spp; Ca, Candida albicans; Cg, Candida glabrata; Cl, Canus lupus familiaris; Hs, Homo sapiens; Mm, Mus musculus; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Xl, Xenopus laevis; Xt, Xenopus tropicalis). Protein sequences were obtained from NCBI (htts://www.ncbi.nlm.nih.gov), aligned using Clustal Omega and analyzed using Unipro UGENE. The consensus row at the top indicates the most conserved residue at each position or shows a ‘+’ when two or more residues are equally abundant. Residues are colored in accordance to their physiochemical properties: aliphatic/hydrophobic (pink), aromatic (orange), positive charge (blue), negative charge (red), hydrophilic (green), proline/glycine (magenta) and cysteine (yellow). (C) SM mutations in ScPif1 generated and analyzed in this study. Mutations include alanine substitutions of conserved residues (I), large and small deletions that are represented by dashed lines (II), and successive amino acid substitutions (III). (D) Structure of a truncated ScPif1 helicase (PDB ID: 5O6D, amino acids 237–780) (43) rendered and modified using Pymol (55). Secondary structures are colored as follows: helix (teal), loop (beige) and sheet (pink). The ScPif1 SM, which is shown in yellow, forms an α-helix with an extended loop.
The goal of this study is to determine the functional importance of the SM in Pif1 family helicases, using the budding yeast ScPif1 as a model. Our analysis was informed by the crystal structures of bacterial helicases Bacteroides sp 3-1-23 (BsPif1) (41) and Bacteroides sp 2-1-16 (BaPif1) (42) and the helicase domains of the human Pif1 (hPIF1, amino acids 200–641) and ScPif1 (amino acids 237–780) (43). The structures in these papers show that the Pif1 SM folds into an α-helix with an extended loop (41–43). Specifically, the first 10 amino acids (AELLDKLDFIARKI) of the SM are part of the α-helix (SM residues are in bold and underlined). The next 11 amino acids of the SM form the extended loop with a turn (RKNHQPFGGIQ) (Pro367 in bold represents the first residue in the turn), and the last two amino acids (L373 and I374) in the SM are part of an adjacent β-sheet structure. The SM of BaPif1 is suggested to stabilize the conformation of regions involved in ssDNA-binding (42). Although a leucine to proline mutation in the SM of the hPIF1 helicase (hPIF1-L319P) is associated with an increased risk of breast cancer (44), the contribution of the SM to specific in vivo functions of a Pif1 family helicase has not yet been addressed. By conducting a systematic mutational and functional analysis of the ScPif1 SM, we show that its presence was critical for all tested in vivo functions of ScPif1. Moreover, the SM was essential for ATPase activity in vitro but not for binding to ssDNA. Our in vivo analysis is consistent with in vitro studies in an accompanying paper which shows that the SM in the S. pombe Pif1 helicase is critical for most of its diverse in vitro activities (45).
MATERIALS AND METHODS
S. cerevisiae strains and growth conditions
Unless otherwise indicated, strains were derived from W303 (see Supplementary Table S1 for strains and Supplementary Table S2 for plasmids used herein). To create deletion strains, polymerase chain reaction amplification of selective markers and flanking DNA followed by homologous recombination into the gene of interest was used: PIF1 was deleted with NatMX6 or His3MX6 and DNA2 was deleted with KanMX6. The strain for the GCR (MBY77) assay was derived from YPH500. To generate MBY77, HTX13 on Chr V-L was replaced with the Kluyveromyces lactis URA3 gene (32). Mutant alleles of PIF1 were marked with TRP1 and introduced on the circular centromere plasmid pCG17 via lithium acetate transformation (100 ng of plasmid DNA unless specified otherwise) (46). Depending on the experiment, cells were grown in rich medium YPD (1% yeast extract, 2% peptone and 2% glucose) or synthetic drop-out medium lacking tryptophan (SD–TRP).
Construction of plasmid-borne Pif1 mutant alleles
The PIF1 promoter was excised from plasmid pVS102 (16) by digestion with PspXI and AgeI (New England Biolabs, NEB) and then cloned into plasmid pMB282 (CEN ARS TRP1) containing PIF1 with a C-terminal 3xFLAG tag (32). The resulting plasmid referred to as pCG17 was verified by restriction enzyme digestion and DNA sequencing and used as the target plasmid for mutagenesis. Mutations within the conserved Pif1-family SM were generated in pCG17 using QuikChange Lightning Site-directed Mutagenesis (Agilent Technologies), Q5® Site-directed Mutagenesis (NEB) or cloned in using gBlocks® gene fragments (Integrated DNA Technologies) and the Gibson Assembly® method (NEB). Mutations were verified by NotI and AvaI restriction enzyme digestion (NEB) and DNA sequencing. The SM mutations analyzed in this study are listed in Figure 2C.
Immunoblot analysis
Yeast total protein extracts were prepared using NaOH precipitation (47). Briefly, 1 ml of mid-log phase cells were pelleted, resuspended in 200 μl of 0.1 M NaOH, incubated at room temperature for 10 min, pelleted, resuspended in 60 μl of sample buffer (2X Laemmli buffer with 1M DTT), boiled for 5 min and pelleted again. At least 10 μl of the supernatant was loaded and the proteins were separated on a 5% (37.5:1 polyacrylamide:bis-acrylamide) sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred to a nitrocellulose membrane (GE Healthcare) at 4°C and blocked with 5% non-fat dried milk (Lab Scientific) diluted in Tris-Buffer Saline Tween-20. Detection of FLAG-tagged Pif1 proteins was done using mouse monoclonal anti-FLAG M2 primary antibody (Sigma-Aldrich, 1:1000 dilution) and visualized with horseradish peroxidase-conjugated anti-mouse secondary antibody (Bio-Rad, 1:3000 dilution) and ECL detection kit reagents (GE Healthcare).
Telomere blot analysis
Plasmid pCG17 containing wild-type (WT) or Pif1 mutant alleles or an empty vector control (pMB13) was transformed into a heterozygous PIF1/pif1Δ DNA2/dna2Δ diploid W303 strain. After sporulation and tetrad dissection, spores were genotyped and haploid DNA2+ pif1::NatMX6 spores carrying the plasmids were selected by their KanMXS NatMXR Trp+ phenotype. Genomic DNA was extracted from at least three independent transformants (biological replicates) using MasterPure Yeast DNA Purification kit (Epicentre) ∼100 generations after sporulation. Genomic DNAs were digested with XhoI restriction enzyme (NEB), separated by 1% (w/v) agarose gel electrophoresis in Tris-borate ethylenediaminetetraacetic acid (EDTA) and then transferred to a nylon membrane (GE Healthcare). Telomeres were hybridized with α-32P-labeled Y’-sub-telomeric probe (48), imaged on a Typhoon FLA 9500 Phosphorimager system (GE Healthcare) and quantified using ImageQuant TL software (GE Healthcare).
Assay for mitochondrial function
To assay for mitochondrial function, pif1Δ cells carrying plasmid pCG17 bearing either a WT or mutant allele of PIF1 were grown to saturation in 5 ml cultures of SD–TRP medium. From each culture 1 ml of two OD660 yeast cells were pelleted, resuspended in sterile water, and 5 μl of 10-fold serial dilutions were spotted onto SD–TRP medium containing 2% glucose or 3% glycerol and grown at 30°C for at least 3 days.
Suppression of dna2Δ lethality
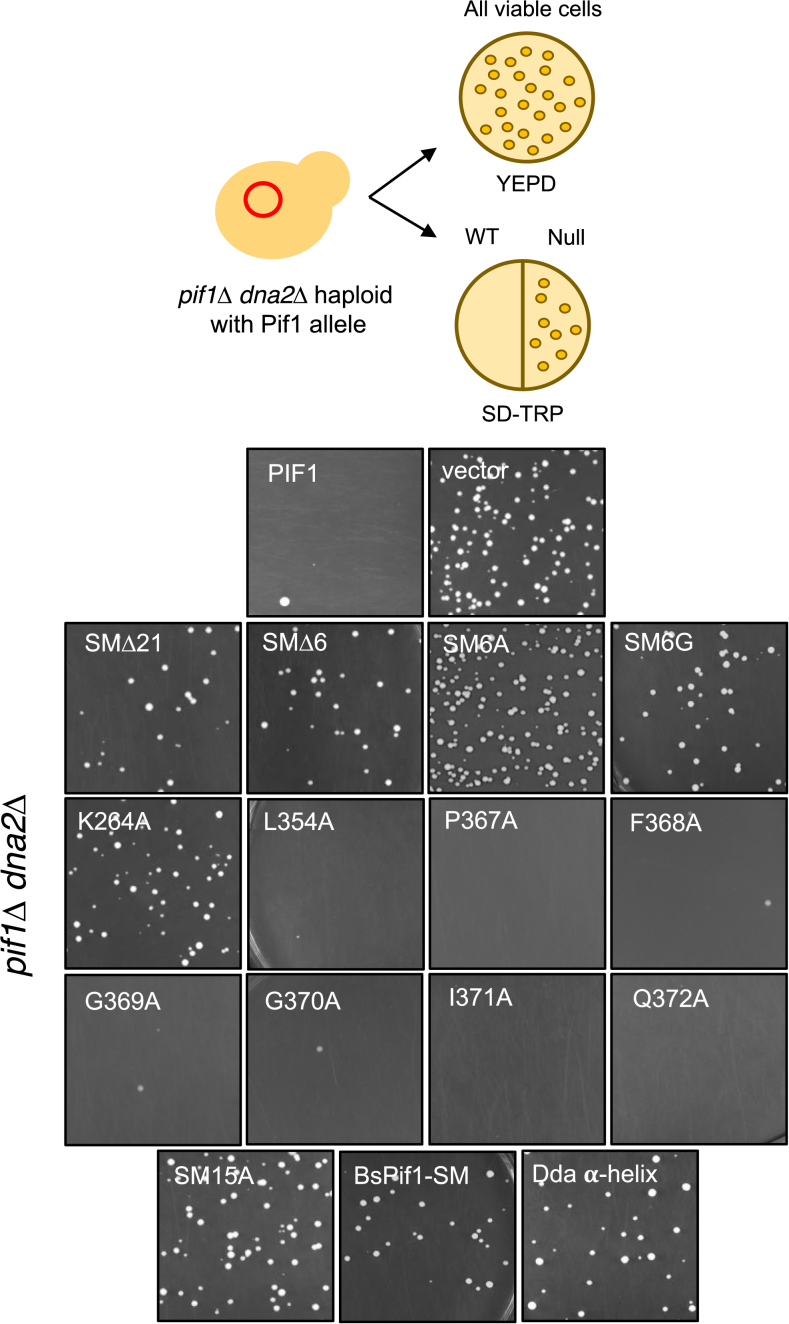
Haploid pif1Δ dna2Δ cells were generated from a doubly heterozygous (PIF1/pif1Δ DNA2/dna2Δ) diploid strain. Using 500 ng pCG17 DNA, Pif1 mutant alleles were transformed into the pif1Δ dna2Δ haploid, plated onto YPD and SD–TRP medium and allowed to grow at 30°C for 3–4 days. Growth on SD–TRP medium indicated that the Pif1 mutant was a null allele in this assay, as it suppressed the dna2Δ lethality. Cells were also plated on YPD to monitor viable cells.
GCR assay
WT or Pif1 mutant alleles were transformed into MBY77 pif1::His3MX6. The GCR assay was performed as previously described (49). Briefly, three to five 5 ml cultures were grown to saturation in SD–TRP medium at 30°C for 72 h. From each culture 100 μl of diluted (10−6) cells were plated onto non-selective medium (SD–TRP), and the plates were incubated at room temperature for 4 days. In addition, 2 ml of cells were pelleted, resuspended in 300 μl of sterile water, plated onto selective medium supplemented with 5-FOA and canavanine sulfate (SD–TRP+FOA+CAN), and the plates were incubated at 30°C for 4–5 days. GCR rates were calculated using the Fluctuation Analysis Calculator (FALCOR) web server and the MMS maximum likelihood method (50).
Purification of ScPif1 mutant variants
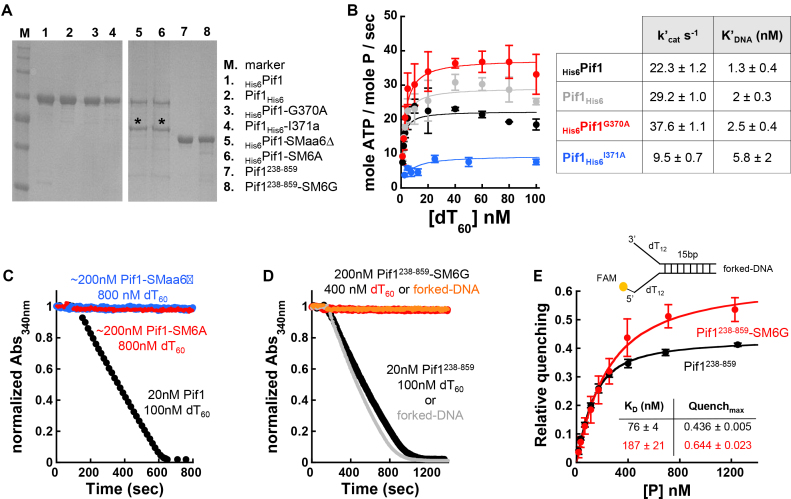
Pif1 variants harboring mutations or deletions within the 23 amino acid SM were generated with standard site-directed mutagenesis protocols. Preliminary expression tests of ScPif1 variants from a pET28b plasmid indicated that large changes within the SM lead to either low expression or poorly soluble proteins. Therefore, ScPif1 and its mutant variants with a His6-tag (N- or C-terminal) were re-cloned into a modified pGEX-6p plasmid into which was introduced a NcoI restriction site after a prescission protease site. After induction with 0.7 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG), Rosetta2 (DE3) pLysS cells were grown overnight at 16°C. Cells were opened in Buffer L (20 mM sodium phosphate buffer, pH 7.4, 600 mM NaCl, 20% glycerol, 1 mM EDTA, 0.5 mM Phenylmethyl sulfonyl fluoride (PMSF) and 1 mM Dithiothreitol (DTT)) and the clarified supernatant incubated with glutathione Sepharose™ 4 Fast Flow (GE Healthcare). Following a high salt wash, the bound proteins were eluted with 25 mM glutathione. After cleavage of the GST-tag during dialysis, the proteins were further purified with anion exchange (HighQ, BioRad), cation exchange (HighS, BioRad) and finally eluted from Ni-NTA (Qiagen), as described previously (51). A ScPif1 construct missing the first 237 amino acids and their variants where the 367PFGGIQ372 motif was substituted for six glycines was overexpressed from a pET28b plasmid and purified as described (51). ScPif1 and its purified mutant variants were quantified spectrophotometrically (51,52); the concentrations of the mutant variants contaminated with the faster migrating band were quantified by gel densitometry using Pif1 WT as a standard.
ATPase and DNA binding assays
The DNA-dependent ATPase activity of ScPif1 and its mutant variants were determined spectrophotometrically using a β-Nicotinamide adenine dinucleotide reduced disodium salt hydrate (NADH) enzyme-coupled assay as previously reported (51,52). ATPase activity was measured at 30°C in Buffer A (10 mM Hepes (pH 7.4), 100 mM NaCl, 8 mM Mg-acetate and 1 mM DTT) as a function of the concentration of ssDNA (dT60) and at a constant concentration of 1 mM ATP. The data were fitted with the equation v = k’cat [DNA]/(K’DNA + [DNA]). DNA binding was measured in Buffer B (50 mM Tris–HCl (pH 8.1), 100 mM NaCl, 1 mM Mg-acetate and 1 mM DTT) by monitoring the fluorescence quenching of a forked-DNA substrate labeled with 6-carboxy fluorescein. DNA binding isotherms were analyzed with a 1:1 binding model, as described in (52). Standard deviations are from three to four independent experiments.
RESULTS
Experimental strategy to determine the in vivo functions of the ScPif1 signature motif (SM)
The SM was identified during sequence alignments of Pif1 proteins as a 21 amino acid stretch that is unique to Pif1 family helicases. When the SM consensus sequence (DKLeXvARaiRKqXkPFGGIQ) was used in a BLAST search, only Pif1 homologues were identified (8). As more Pif1 family helicases were sequenced, it became clear that the SM is probably better defined as being 23 amino acids in length, as the next two amino acids are also highly conserved (LI or LV) (Figure 2B). Although we originally thought the SM was not present in plant Pif1 family helicases, at least some of the plant Pif1 helicases (e.g.Capsella rubella andTrifolium pratense) have this motif, although they are often more divergent than those in yeasts and multi-cellular organisms (C. L. Geronimo, personal observation).
To determine the function of the SM, we designed a series of mutations within the SM, introduced the mutant genes on centromere plasmids into pif1Δ cells and determined the ability of each mutant gene to supply individual Pif1 functions. Our choice of mutations was based in part on structural studies of bacterial and hPIF1 helicases (see ‘Introduction’). As a control, we used the Sc-pif1-K264A allele, where an invariant lysine in the Walker A box is mutated to alanine, which generates a helicase-dead allele (17). Mutant and WT proteins were epitope tagged at their carboxyl termini with three FLAG tags for detection by western blotting.
To determine if the SM is essential for ScPif1 functions, we deleted most of the SM (21 of the 23 amino acids). We also deleted six amino acids (Pro367 to Gln372) that form part of the extended loop (Figure 2C, group II). The first 10 amino acids of the SM form part of an α-helix (41–43). To test the importance of the α-helical portion of the SM, we replaced the first 15 residues (Asp352 to Gln366) with 15 alanines, as poly-alanine cannot form an α-helix, but still provides structural flexibility. To determine if the α-helical structure or simply the sequence of this region was important, we replaced the first 15 SM residues with the SM sequence from the bacterial Bacteroides spp Pif1 (BsPif1), which is different from that of the ScPif1 SM at 12 of the 15 positions (although four of the 12 changes are conservative) (Figure 2B). We also generated a more drastic mutant that replaced the 10 SM amino acids of the α-helix with a completely different sequence that is nonetheless predicted to form an α-helix by the SABLE protein structure prediction server (53). For this allele, we used the Lys123 to Ile132 amino acid region from the T4 bacteriophage Dda helicase (54). We chose the Dda sequence because using the Pymol program (55), it aligned well with the ScPif1 SM α-helix and is located in a similar position as the SM within the Dda protein; i.e. between helicase motifs II and III (Supplementary Figure S1). To test the significance of the SM loop in the carboxyl-terminal region, single alanine substitutions (Figure 2C, group I) or successive amino acid substitutions (Figure 2C, group III) were created. Single amino acid substitutions were also created at specific sites within the SM, including a change that corresponds to the hPIF1-L319P mutation (Sc-pif1-L354P) that is associated with higher breast cancer risk (44) (Figure 2C, group I).The growth rates of cells expressing each TRP1-marked mutant allele were determined for cells growing at 30°C in media lacking tryptophan (Supplementary Figure S2B). Mutations that were functional in the in vivo assays described below had normal growth rates while cells with null or near null alleles grew more slowly, a phenotype that can be explained by their reduced mitochondrial function.
All of the SM alleles except Sc-pif1-L354P produced stable protein in yeast
After generating pif1Δ cells carrying a centromere plasmid borne copy of a mutant ScPIF1 allele, we used western blot analysis to determine if the mutant proteins were expressed in vivo. All of the ScPif1 SM mutants generated for this study were expressed, except for ScPif1-L354P, whose expression was not detectable by western blotting. Therefore, this mutant was not examined in functional assays. Quantification of the expression of key SM mutants indicated some variability in protein levels (Supplementary Figure S2A). However, changes in protein levels did not correlate with loss of ScPif1 function.
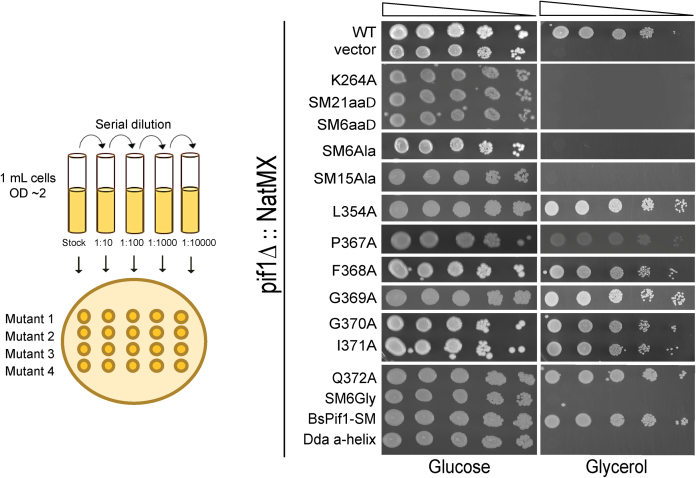
The SM is essential for the mitochondrial functions of ScPif1
ScPif1 is critical for the maintenance of mtDNA as loss of ScPif1 rapidly generates respiratory deficient cells (petites) (15). To test for mitochondrial proficiency we analyzed growth on glycerol, a non-fermentable carbon source. Petites are unable to grow with glycerol as the only carbon source. Plasmids carrying WT or mutant ScPIF1 alleles were transformed into a heterozygous diploid strain (YCG59, pif1Δ/WT), and tetrads were dissected. After spore clones grew up on glucose plates, which allowed for loss of mtDNA, pif1Δ cells carrying either WT or a mutant allele of ScPIF1 were spotted onto selective medium containing either glycerol or glucose. Growth was assessed after ∼3 days at 30°C (Figure 3). Cells expressing single alanine substitutions within the SM exhibited WT mitochondrial function as shown by robust growth on glycerol plates. However, deletion of the entire SM, deletion of the loop region and larger amino acid replacements did not generate respiratory proficient cells. Thus, the SM of ScPif1 is critical for maintenance of mtDNA.
Figure 3.
Analyzing ScPif1 SM mutants for mitochondrial proficiency. As indicated in the cartoon on the left of the figure, cultures were 10-fold serially diluted left-to-right and spotted on selective media containing glucose or glycerol. Respiratory deficient cells grow on media containing glucose but not on media containing glycerol.
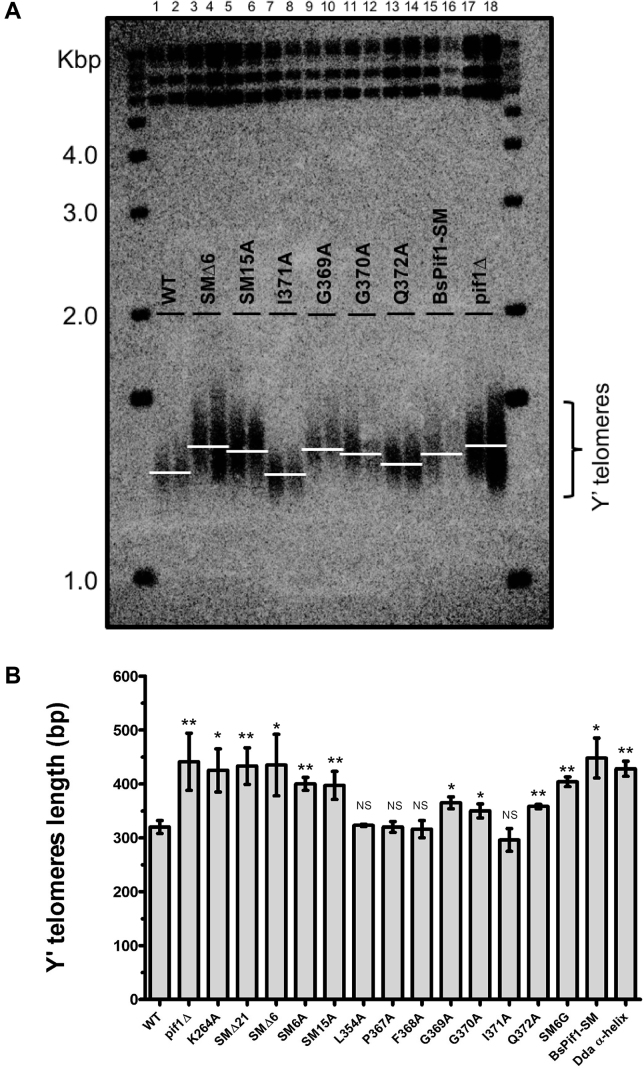
The SM is critical for ScPif1 to remove telomerase from telomeres
ScPif1 catalytically removes telomerase from telomeres in a manner that limits telomere lengthening (18). Thus, compared to WT cells, cells lacking ScPif1 have long telomeres (16,18). We used Southern blot analysis to monitor telomere length in cells expressing each of the mutant alleles (Figure 4). Most of the single alanine substitutions exhibited telomere lengths similar to WT (Figure 4; lanes 1, 2, 7 and 8), indicating that subtle changes in the SM did not disrupt the ability of ScPif1 to regulate telomere length. Cells expressing alleles that deleted the entire SM, the loop region or more substantial amino acid substitutions resulted in long telomeres similar in length to telomeres in pif1Δ cells (Figure 4; lanes 3–6 and 15–18). However, three of the seven single alanine substitutions (G369A, G370A and Q372A) exhibited intermediate telomere phenotypes, suggesting that these residues are required for full inhibition of telomere elongation (Figure 4; lanes 9–14).
Figure 4.
(A) Analysis of telomere lengths in cells expressing different PIF1 alleles. Genomic DNA was isolated from three or more independent isolates from each of the 14 SM mutant strains as well as from WT, pif1Δ and catalytically dead pif1-K264A cells (in this figure two representative examples are shown for 9 of the 17 strains examined). DNA was digested with XhoI, separated on 1% agarose gels and analyzed by Southern hybridization using a radiolabeled sub-telomeric Y’ probe (48). The lanes contain DNA from WT (lanes 1 and 2), SMΔ6 (lanes 3 and 4), SM15A (lanes 5 and 6), I371A (lanes 7 and 8), G369A (lanes 9 and 10), G370A (lanes 11 and 12), Q372A (lanes 13 and 14), BsPif1-SM (lanes 15–16) and pif1Δ (lanes 17 and 18). (B) Quantitation of telomere lengths. To obtain the number of base pairs of TG1–3 telomeric DNA, 875 bp, the amount of Y’ DNA in the terminal XhoI restriction fragment (48) was subtracted from the size of the terminal fragments. The average telomere length was determined for at least three biological isolates per strain. The white lines represent the peak DNA signal that indicates the average length of the Y’ telomeres. Telomeres were classified in Table 1 as having WT (320 ± 12 bp), intermediate (357 ± 11 bp) or null (441 ± 53 bp) telomere lengths. Error bars are standard deviations. Given that loss of ScPif1 function results in longer telomeres, a one-tailed student t-test was used to compare the mean telomere lengths between WT and ScPif1 mutants to determine if the increase in telomere lengths observed in the SM mutants was statistically significant; NS, not significant, *P<0.05, **P<0.005.
The SM is critical for ScPif1’s role in Okazaki fragment processing
Deletion of PIF1 suppresses the lethality caused by deletion of DNA2, an essential gene whose protein product has both helicase and nuclease activities (22,23). The basis for this suppression is that Pif1 is required to generate Okazaki fragments with long 5′ flaps whose resolution is Dna2-dependent. To assess the importance of the SM for ScPif1’s role during Okazaki fragment processing, the viability of pif1Δ dna2Δ cells expressing a plasmid borne allele with a SM mutant was assessed (Figure 5). Seven mutants each with a single alanine substitution (e.g. L354A and P367A) failed to suppress the lethality of a dna2Δ mutant indicating that these alleles were able to generate long-flap Okazaki fragments. However, deletions and more substantial amino acid substitutions (e.g. successive alanine changes or α-helix sequence replacements) generated viable cells, indicating that the mutant ScPif1 suppressed the dna2Δ lethality. Therefore, the SM was essential for Pif1 function during Okazaki fragment processing. Replacement of the SM with the bacterial BsPif1 SM sequence also suppressed the dna2Δ lethality. Thus, as with telomere length regulation (Figure 4), the bacterial SM was unable to provide the Okazaki fragment processing activity of WT ScPif1, even though it did provide its mitochondrial function (Figure 3). Of the 15 SM mutants tested here, the bacterial SM substitution mutant (BsPif1-SM allele) was the only separation of function allele we obtained.
Figure 5.
Analysis of the ability of ScPif1 SM mutants to suppress the lethality of a dna2Δ cells. Plasmid-borne WT and SM alleles were introduced into pif1Δ dna2Δ cells and analyzed for growth on selective media as indicated in the cartoon. WT alleles are unable to suppress the lethality of dna2Δ cells. Images shown are cells plated on selective media lacking tryptophan; growth indicates null ScPif1 function.
The SM is critical for ScPif1 to remove telomerase from double strand breaks
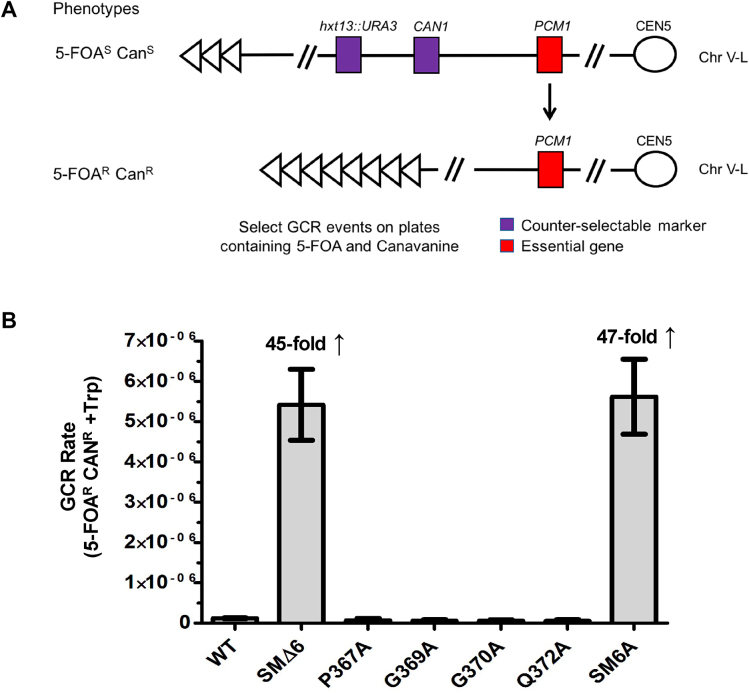
In addition to regulating the lengths of existing telomeres, Pif1 removes telomerase from DSBs, thus inhibiting de novo telomere addition (19). De novo telomere additions were detected by a GCR assay that selects for the simultaneous loss of two markers on the left arm of chromosome V (Figure 6A) (21). To determine if the SM affects de novo telomere addition, the rate at which GCR events occurred in strain MBY77 carrying plasmid borne WT or mutant alleles of ScPIF1 was calculated (Figure 6B). The GCR rate for cells expressing WT ScPif1 was 1.20 × 10−7 events per cell division. The rates for cells expressing pif1-P367A as well as those for the three alleles that had intermediate telomere lengths (pif1-G369A, G370A and Q372A) were similar to WT cells. In contrast, the GCR rates in cells expressing an allele lacking part of the SM (SMΔ6) or with multiple tandem alanine substitutions (SM6A) were almost 50-fold higher than in WT cells. Thus, the SM is also essential to inhibit de novo telomere additions.
Figure 6.
(A) Schematic of gross chromosomal rearrangement (GCR) assay (49). (B) Bar graph shows GCR rates of WT ScPif1, P367A, SMΔ6, G369A, G370A, Q372A and SM6A mutants. The average rate in cells expressing WT ScPif1 was 1.20 × 10−7 events per cell division and the average rates in mutant cells expressing single alanine substitutions were similarly low. The GCR rate for cells expressing P367A, an allele that was WT in all in vivo assays, was 6.68 × 10−8. The GCR rate for cells expressing G369A, G370A and Q372A, alleles that exhibited an intermediate telomere phenotype, were 5.85 × 10−8, 5.79 × 10−8 and 5.36 × 10−8, respectively. In contrast, the average rates in mutant strains SMΔ6 and SM6A were almost 50-fold higher compared to WT. Experiments were done on at least three biological replicates. Error bars show standard deviations.
The PFGGIQ region within the ScPif1 SM is required for ATPase activity
Analysis of the in vivo phenotypes of SM mutants (Table 1) indicated that while no single amino acid was required for function, the entire 23 amino acid region as well as subsets of the region were necessary. Within the 23 amino acid SM, deletion or substitution of the conserved 367PFGGIQ372 motif was sufficient to lead to a null phenotype in vivo indistinguishable from that of pif1Δ or the pif1-K264A allele. This observation suggests that the change of this six amino acid stretch affects one or more of ScPif1’s biochemical functions, e.g. DNA binding, the DNA-dependent ATPase activity or the ATPase-coupled unwinding activity. To distinguish among these possibilities, we purified ScPif1 variants containing selected SM mutations. Consistent with a WT phenotype in vivo, the ScPif1 constructs harboring the single-point mutations G370A or I371A retained DNA-dependent ATPase activity in vitro, the major catalytic activity that drives ScPif1 functions (Figure 7B). The differences in the DNA-dependent ATPase activity (both in the apparent K’DNA and k’cat; see table associated with Figure 7B) between WT ScPif1 and these single-point mutant variants did not result in dramatic differences in ScPif1 functions in vivo (Table 1). In contrast, the full-length ScPif1 variants harboring a deletion of the 367PFGGIQ372 motif (SMΔ6) or a substitution of this region with six alanines (SM6A) did not show ssDNA-dependent ATPase activity, even at a 10-fold higher concentration of enzyme (Figure 7C). However, both variants co-purified with a contaminating band that could not be removed, even with size-exclusion chromatography (Figure 7A, asterisks). To test whether the loss of ATPase activity in these mutants was due to the presence of the contaminating band, we generated a substitution of the 367PFGGIQ372 motif with six glycines (SM6G) within a Pif1 construct that lacked the N-terminal region of the helicase (Pif1238-859-SM6G). The purified protein did not contain detectable amounts of the contaminating band (Figure 7A), yet, neither ssDNA nor a forked-DNA substrate (i.e. an unwinding substrate) stimulated ATPase activity, even at an enzyme concentration that was 10-fold higher than that of its WT counterpart (Figure 7D). However, the SM6G variant of ScPif1238-859 retained DNA binding activity, as monitored by quenching of fluorescence intensity of a forked-DNA substrate labeled with 6-carboxyfluorescein (Figure 7E). Thus, loss of DNA-dependent ATPase activity in this mutant was not due to loss of DNA binding activity. Taken together, the in vitro results indicate that while large changes within the 367PFGGIQ372 motif of the SM did not lead to loss of DNA binding activity of ScPif1, they eliminated its ATP binding and/or hydrolysis activities.
Table 1.
Summary table showing effects of SM mutations on individual ScPif1 functions in vivo. (A) All the SM mutants were tested for effects on mitochondrial function, telomere length and Okazaki fragment processing. Telomeres were classified as having WT (320 ± 12 bp), intermediate (357 ± 11 bp) or null (441 ± 53 bp) telomere lengths. (B) A subset of the SM mutants exhibiting WT and partial loss of function phenotypes was further tested for effects on de novo telomere addition. The GCR rate for the SM mutants tested was classified as WT or null. Dash in Table 1A indicates that the GCR rate for that SM mutant was not determined
| (A) | ||||
|---|---|---|---|---|
| ScPif1 allele | Mitochondrial function | Telomere length | De novo telomere addition | Okazaki fragment processing |
| WT | WT | WT | WT | WT |
| pif1Δ | Null | Null | – | Null |
| K264A | Null | Null | – | Null |
| SM amino terminal mutants | ||||
| SMΔ21 | Null | Null | – | Null |
| L354A | WT | WT | – | WT |
| SM15A | Null | Null | – | Null |
| BsPif1-SM | WT | Null | – | Null |
| Dda α-helix | Null | Null | – | Null |
| SM carboxyl terminal mutants | ||||
| SMΔ6 | Null | Null | Null | Null |
| P367A | WT | WT | WT | WT |
| F368A | WT | WT | – | WT |
| G369A | WT | Intermediate | WT | WT |
| G370A | WT | Intermediate | WT | WT |
| I371A | WT | WT | – | WT |
| Q372A | WT | Intermediate | WT | WT |
| SM6A | Null | Null | Null | Null |
| SM6G | Null | Null | – | Null |
| (B) | ||||
| ScPif1 allele | Average GCR Rate (5-FOAR CANR + TRP) | |||
| WT | 1.20 × 10−7 | |||
| SMΔ6 | 5.42 × 10−6 | |||
| P367A | 6.68 × 10−8 | |||
| G369A | 5.85 × 10−8 | |||
| G370A | 5.79 × 10−8 | |||
| Q372A | 5.36 × 10−8 | |||
| SM6A | 5.62 × 10−6 | |||
Figure 7.
The SM of ScPif1 is required for DNA dependent ATPase activity in vitro. (A) SDS-PAGE analysis of the purified ScPif1 WT and mutant ScPif1 proteins prepared in Escherichia coli. The asterisk indicates a contaminating band that co-purifies with full-length Pif1 variants with mutations within the SM. (B) DNA-dependent ATPase activity of Pif1 (N- and C-terminal His6-tagged) and the single-point mutant variants G370A and I371A. (C) Time course of ATP hydrolysis, monitoring the absorbance of NADH, of Pif1 and its SMaa6Δ and SM6A variants. (D) Time courses of ATP hydrolysis of Pif1238-859 and its SM6G variant. (E) Forked-DNA binding activity of Pif1238-859 and its SM6G variant.
DISCUSSION
The SM is a unique feature of Pif1 family helicases, yet its importance for the diverse functions of these proteins was unknown. By making a series of mutations in the SM of ScPif1, we showed that the SM is critical for ATPase activity in vitro and for multiple in vivo functions of this multi-functional DNA helicase. These conclusions agree with and complement those in an accompanying paper that shows that the SM in the fission yeast Pfh1, another member of the Pif1 family of DNA helicases, is essential for all of its in vitro activities, except DNA annealing, including its preferential unwinding of G4 and RNA/DNA hybrids (see accompanying paper by Mohammad et al. (45)).
Previous studies have addressed the in vitro functions of the SM in bacterial helicases (41,42). Mutation of the equivalent amino acid identified in the cancer-variant of hPIF1 resulted in more than 90% reduction of ssDNA binding, double-stranded DNA (dsDNA) and G4-DNA unwinding activities of BaPif1 (42). Also, a deletion of the conserved four amino acids PFGG within the loop region of BsPif1 abolishes its unwinding activity (41). In our SM study, we tested mutations of almost all the residues within the SM for effects on ScPif1 functions in vivo as well as several of these mutations for their effects in vitro. Although substitution of an alanine residue at seven different highly conserved residues throughout the SM yielded active protein in vivo (and for two of these substitutions, in vitro as well), deletion of most of the 23 amino acid SM (SMΔ21) was as defective in vivo as a complete deletion of the PIF1 gene or the catalytically dead K264A allele (Table 1). As the SMΔ21 allele produced a stable protein, its lack of in vivo function was not due to its producing an unstable product (Supplemental Figure S2A). Likewise, deletion of six amino acids near the carboxyl end of the SM (SMΔ6), one of the most highly conserved regions within the motif (Figure 2B), was a null allele in vivo, as was the SM6A and SM6G alleles where the same six amino acids were replaced with consecutive alanines or glycines. The null phenotypes of the SMΔ6 and SM6A alleles in vivo can be explained by our in vitro data showing their lack of ATPase activity, but not DNA binding. Thus, although single alanine substitutions at these positions were WT for several in vivo functions, their consecutive replacement generated non-functional protein. In the crystal structure, starting with the proline residue, which is proposed to create a kink, these six amino acids form part of an extended loop (41–43). Thus, not only the presence but also the sequence of this highly conserved six amino acid portion of the loop (367PFGGIQ372) were essential for ScPif1 functions in vivo. The Pif1238-859 SM6G protein bound a forked-DNA substrate in vitro, although its affinity was ∼2.5x lower than the WT ScPif1 (Figure 7E). Thus, the SM is not essential for substrate binding, but it may enhance binding as suggested in (42). The SM is situated between helicase motifs II (Walker B box) and III, and these canonical motifs are known to be involved in NTP hydrolysis and DNA binding, respectively (4). Given the SM’s location, we speculate that the conserved loop region may provide the appropriate spatial configuration required for the residues from motifs II and III to coordinate and couple the energy of ATP binding/hydrolysis with DNA unwinding.
To test if the six highly conserved amino acids in the loop region are sufficient for SM function, we replaced the amino terminal 15 amino acids of the SM with fifteen alanines (SM15A). In all tested in vivo functions, SM15A was a null allele, even though it produced stable protein in vivo. Thus, the conserved six amino acid portion of the loop region was necessary but not sufficient for SM function. The amino terminal 10 amino acids are part of an α-helix (41–43). Given that SM15A was a null allele and contained a poly-alanine sequence that is unable to form an α-helix, these results suggest that the α-helical structure of this region was also critical for function. To determine if the presence of an α-helix plus the loop region were sufficient to supply SM activity, we replaced the 10 amino acid helix-forming region with 10 amino acids from the bacteriophage dda helicase (Dda α-helix), which is predicted to form an α-helix. We also replaced this region with the comparable part of the Bacteriodes spp Pif1 family helicase (BsPif1-SM), which also forms an α-helix (41,42), but whose sequence is fairly divergent from that of the equivalent region of ScPif1 (Figure 2B). Although expressed normally, both of these mutant proteins lacked all (Dda) or some (BsPif1-SM) of the in vivo functions of ScPif1 (Supplemental Figure S2A, Table 1) Therefore, not only the structure but also the sequences of the α-helix and the loop regions in the SM are essential for in vivo function.
ScPif1 and the S. pombe Pfh1 DNA helicases are multi-functional in vivo (see introduction and (56)). Unlike Rrm3 (57) or the S. pombe Pfh1 (58), ScPif1 does not move with the replisome but is probably recruited to its diverse targets during or after replication of the sequence (27) (Chen et al., in preparation). A long standing interest in our lab is to identify regions of ScPif1 that might target it to its diverse in vivo substrates. However, with few exceptions, a given SM allele was either functional or non-functional for all tested in vivo functions. The exceptions include three of the single alanine substitutions (G369A, G370A and Q372A) in the loop region of the SM. All three had intermediate telomere lengths, longer than WT cells but shorter than telomeres in pif1Δ cells (Figure 4). Small differences in average telomere lengths are probably easier to detect than similarly small differences in the other in vivo functions examined in this paper. However, of the 15 SM mutants, these three were the only ones to have intermediate effects on telomere length. By both in vivo and in vitro assays, ScPif1 uses its ATPase activity to remove telomerase from DNA ends, resulting in a less stable telomerase–telomeric DNA interaction and hence a less processive telomerase (18,19). In contrast, single-point mutations of residues G370 and I371 resulted in different effects on the ATPase activity in vitro, with I371A having a negative impact and G370A being slightly more active than WT ScPif1 (Figure 7). The molecular origin of these differences remain to be determined, as is the quantitative relationship between the degree of ATPase activity in vitro and telomere function in vivo. Because the DNA-dependent ATPase activity of Pif1-G370A was similar, if not slightly better, to that of the WT enzyme (Figure 7B), the defect in these telomere-specific partial loss of function alleles may be manifest only in vivo. For example, the intermediate telomere length phenotypes could be due to partial disruption of a protein–protein interaction that recruits ScPif1 to telomeres.
The BsPif1-SM allele is our best example of a separation of function allele. In this allele, the 15 amino acid stretch that forms part of the α-helix and the first part of the loop in the ScPif1 SM was replaced with the corresponding sequence from Bacteroides spp Pif1. The SM sequence in this allele also contains the six highly conserved amino acids in the loop region (367PFGGIQ372). The BsPif1-SM allele had WT mitochondrial function but was a null for three nuclear functions (Table 1). Because all tested nuclear functions were defective in this mutant, its phenotype is hard to explain by loss of a protein–protein interaction that targets ScPif1 to a specific substrate. However, loss of nuclear functions could be explained if the BsPif1-SM protein is deficient in nuclear localization. Translation of ScPif1 can begin at either the first or second methionine in the open reading frame (16,17). Between the first and second methionine is a mitochondrial localization signal. Thus, if translation begins at the first methionine, as it commonly does (59), the translated protein is targeted to mitochondria. If translation begins at the second methionine, the protein is targeted to nuclei. From its sequence, the ScPif1 SM is unlikely to encode a nuclear localization signal (NLS). However, the protein made by the BsPif1-SM allele might change the presentation of the as yet unidentified NLS, as in the crystal structures, the BsPif1 SM interacts with the 1B domain, which folds into an unstructured loop (41), while the 1B domain of ScPif1 forms an α-helix (43).
In both ScPif1 (Figure 7) and the fission yeast Pfh1 (see accompanying paper by Mohammad et al., (45)) the SM was not needed to bind to ssDNA and/or other DNA substrates in vitro. Moreover, the ScPif1 SM was not required for protein stability as ScPif1 and Pfh1 variants lacking almost the entire SM were stable in vitro (Figure 7) (see accompanying paper by Mohammad et al. (45)). Only one of the 15 ScPif1 SM mutants was unstable in vivo, ScPif1-L354P, the equivalent of the cancer-associated hPIF1 variant, hPIF1-L319P. In contrast, the equivalent S. pombe SpPfh1-L430P mutant was stable in vivo and in vitro but inactive for in vivo and in vitro ATP-dependent activities (44) (see accompanying paper by Mohammad et al. (45)). Similar to the S. pombe Pfh1, analysis of the equivalent Bacteriodes spp BaPif1 I118P mutant showed a significant reduction in ssDNA binding and dsDNA and G4-unwinding activities (42).
The analyses here and in the accompanying paper argue that the SM is essential for all functions that require the ATPase activity of Pif1 family DNA helicases. By extension, ScPif1 must act enzymatically for the tested in vivo functions. In contrast, the ability of ScPif1 (and Pfh1) to anneal complementary DNA strands does not require ATP hydrolysis (38,45). By using helicase dead alleles, such as pif1-K264A, all of the tested in vivo activities of ScPif1 are ATPase dependent [see, for examples (17,29,60), Chen et al., in preparation]. Thus, as yet, there is no in vivo evidence for a function for ScPif1- (or Pfh1-) mediated strand annealing. However, the role of ScPif1 in fortifying the replication fork barrier in the rDNA (28), which has not been tested for ATPase-dependence, is a candidate for being mediated by strand annealing. Finally, if ScPif1 is recruited to its in vivo targets by specific protein–protein interactions, these interactions usually do not involve the SM region of ScPif1.
In summary, the combined data here and in the accompanying paper show that the SM regions of ScPif1 and fission yeast Pfh1 were essential for ATPase activity and for all tested in vivo functions but not for substrate binding or DNA annealing in vitro. Because we found little evidence for SM alleles that support only a subset of ScPif1’s in vivo functions, sequences that define ScPif1 targeting to a specific genome region likely lie mainly outside the SM. The demonstration in the accompanying paper that Pfh1, like ScPif1 (34,38), has single strand annealing activity and preferentially unwinds RNA/DNA hybrids provides additional support that members of the Pif1 family of DNA helicases share not only sequence similarity but also common biochemical properties.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Y. Kim for help with some of the experiments. We thank A. Tracyzk for useful discussions on protein structure, which helped us with the design of certain mutations and T. Pohl for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [1R35GM118279 to V.A.Z., 2R01GM098509 to R.G]; National Science Foundation [DGE1656466 to C.L.G.]. Funding for open access charge: NIH [1R35GM118279 to V.A.Z.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Shiratori A., Shibata T., Arisawa M., Hanaoka F., Murakami Y., Eki T.. Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast. 1999; 11:60–65. [DOI] [PubMed] [Google Scholar]
- 2. Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23–50. [DOI] [PubMed] [Google Scholar]
- 3. Gorbalenya A.E., Koonin E.V.. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993; 3:419–429. [Google Scholar]
- 4. Raney K.D., Byrd A.K., Aarattuthodiyil S.. Structure and mechanism of SF1 DNA helicases. Advances in Experimental Medicine and Biology. 2013; 767:17–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M.. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989; 17:4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tuteja N., Tuteja R.. Unraveling DNA helicases Motif, structure, mechanism and function. Eur. J. Biochem. 2004; 271:1849–1863. [DOI] [PubMed] [Google Scholar]
- 7. Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bochman M.L., Sabouri N., Zakian V.A.. Unwinding the functions of the Pif1 family helicases. DNA Repair. 2010; 9:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ivessa A.S., Lenzmeier B.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A.. The saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone Protein-DNA complexes. Mol. Cell. 2003; 12:1525–1536. [DOI] [PubMed] [Google Scholar]
- 10. Syed S., Desler C., Rasmussen L.J., Schmidt K.H.. A novel Rrm3 function in restricting DNA replication via an Orc5-Binding domain is genetically separable from Rrm3 function as an ATPase/Helicase in facilitating fork progression. PLoS Genet. 2016; 12:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muñoz-Galván S., García-Rubio M., Ortega P., Ruiz J.F., Jimeno S., Pardo B., Gómez-González B., Aguilera A.. A new role for Rrm3 in repair of replication-born DNA breakage by sister chromatid recombination. PLoS Genet. 2017; 13:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chung W.-H. To peep into Pif1 helicase: multifaceted all the way from genome stability to repair-associated DNA synthesis. J. Microbiol. 2014; 52:89–98. [DOI] [PubMed] [Google Scholar]
- 13. Geronimo C.L., Zakian V.A.. Getting it done at the ends: Pif1 family DNA helicases and telomeres. DNA Repair. 2016; 44:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Foury F., Kolodynski J.. pif mutation blocks recombination between mitochondrial rho+ and rho- genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lahaye A., Stahl H., Thines-Sempoux D., Foury F.. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991; 10:997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schulz V.P., Zakian V.A.. The saccharomyces Pif1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994; 76:145–155. [DOI] [PubMed] [Google Scholar]
- 17. Zhou J.-Q., Monson E.K., Teng S.-C., Schulz V.P., Zakian V.A.. Pif1p helicase, a catalytic inhibitor of telomerase in Yeast. Science. 2000; 289:771–774. [DOI] [PubMed] [Google Scholar]
- 18. Boulé J.-B., Vega L.R., Zakian V.A.. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005; 438:57–61. [DOI] [PubMed] [Google Scholar]
- 19. Phillips J.A., Chan A., Paeschke K., Zakian V.A.. The Pif1 helicase, a negative regulator of telomerase, acts preferentially at long telomeres. PLOS Genet. 2015; 11:e1005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Makovets S., Blackburn E.H.. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat. Cell Biol. 2009; 11:1383–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Myung K., Chen C., Kolodner R.D.. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001; 411:1073–1076. [DOI] [PubMed] [Google Scholar]
- 22. Budd M.E., Reis C.C., Smith S., Campbell J.L., Myung K.. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 Helicase / Nuclease and DNA polymerase δ. Mol. Cell. Biol. 2006; 26:2490–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pike J.E., Burgers P.M.J., Campbell J.L., Bambara R.A.. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J. Biol. Chem. 2009; 284:25170–25180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tran P.L.T., Pohl T.J., Chen C.-F., Chan A., Pott S., Zakian V.A.. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat. Commun. 2017; 8:15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Osmundson J.S., Kumar J., Yeung R., Smith D.J.. Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat. Struct. Mol. Biol. 2016; 24:162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ribeyre C., Lopes J., Boulé J.-B.B., Piazza A., Guédin A., Zakian V.A., Mergny J.-L.L., Nicolas A.. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009; 5:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paeschke K., Capra J.A., Zakian V.A.. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011; 145:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ivessa A.S., Zhou J.Q., Zakian V.A.. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000; 100:479–489. [DOI] [PubMed] [Google Scholar]
- 29. Wilson M.A., Kwon Y., Xu Y., Chung W.-H., Chi P., Niu H., Mayle R., Chen X., Malkova A., Sung P. et al. . Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013; 502:393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saini N., Ramakrishnan S., Elango R., Ayyar S., Zhang Y., Deem A., Ira G., Haber J.E., Lobachev K.S., Malkova A.. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013; 502:389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lahaye A., Leterme S., Foury F.. PIF 1 DNA Helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J. Biol. Chem. 1993; 268:26155–26161. [PubMed] [Google Scholar]
- 32. Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A.. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013; 497:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou R., Zhang J., Bochman M.L., Zakian V.A.. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife. 2014; 3:e02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boulé J.B., Zakian V.A.. The yeast Pif1p DNA helicase preferentially unwinds RNA-DNA substrates. Nucleic Acids Res. 2007; 35:5809–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chib S., Byrd A.K., Raney K.D.. Yeast helicase Pif1 unwinds RNA:DNA hybrids with higher processivity than DNA:DNA duplexes. J. Biol. Chem. 2016; 291:5889–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galletto R., Tomko E.J.. Translocation of Saccharomyces cerevisiae Pif1 helicase monomers on single-stranded DNA. Nucleic Acids Res. 2013; 41:4613–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koc K.N., Singh S.P., Stodola J.L., Burgers P.M., Galletto R.. Pif1 removes a Rap1-dependent barrier to the strand displacement activity of DNA polymerase δ. Nucleic Acids Res. 2016; 44:3811–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramanagoudr-Bhojappa R., Byrd A.K., Dahl C., Raney K.D.. Yeast Pif1 accelerates annealing of complementary DNA strands. Biochemistry. 2014; 53:7659–7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang D.H., Zhou B., Huang Y., Xu L.X., Zhou J.Q.. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006; 34:1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmidt K.H., Kolodner R.D.. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 2004; 24:3213–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen W.-F., Dai Y.-X., Duan X.-L., Liu N.-N., Shi W., Li N., Li M., Dou S.-X., Dong Y.-H., Rety S. et al. . Crystal structures of the BsPif1 helicase reveal that a major movement of the 2B SH3 domain is required for DNA unwinding. Nucleic Acids Res. 2016; 44:2949–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou X., Ren W., Bharath S.R., Tang X., He Y., Chen C., Liu Z., Li D., Song H.. Structural and functional insights into the unwinding mechanism of bacteroides sp Pif1. Cell Rep. 2016; 14:2030–2039. [DOI] [PubMed] [Google Scholar]
- 43. Lu K.-Y., Chen W.-F., Rety S., Liu N.-N., Wu W.-Q., Dai Y.-X., Li D., Ma H.-Y., Dou S.-X., Xi X.-G.. Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase. Nucleic Acids Res. 2017; 46:1486–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chisholm K.M., Aubert S.D., Freese K.P., Zakian V.A., King M.C., Welcsh P.L.. A genomewide screen for suppressors of Alu-Mediated rearrangements reveals a role for PIF1. PLoS One. 2012; 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mohammad J.B., Wallgren M., Sabouri N.. The Pif1 signature motif of Pfh1 is necessary for both protein displacement and helicase unwinding activities, but is dispensable for strand-annealing activity. Nucleic Acids Res. 2018; doi:10.1093/nar/gky654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gietz R.D., Schiestl R.H.. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007; 2:5–8. [DOI] [PubMed] [Google Scholar]
- 47. Kushnirov V.V. Rapid and reliable protein extraction from yeast. Yeast. 2000; 16:857–860. [DOI] [PubMed] [Google Scholar]
- 48. Conrad M.N., Wright J.H., Wolf A.J., Zakian V.A.. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990; 63:739–750. [DOI] [PubMed] [Google Scholar]
- 49. Putnam C.D., Kolodner R.D.. Determination of gross chromosomal rearrangement rates. Cold Spring Harb. Protoc. 2010; 5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hall B.M., Ma C.-X., Liang P., Singh K.K.. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009; 25:1564–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Singh S.P., Koc K.N., Stodola J.L., Galletto R.. A monomer of Pif1 unwinds Double-Stranded DNA and it is regulated by the nature of the Non-translocating strand at the 3′-End. J. Mol. Biol. 2016; 428:1053–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barranco-Medina S., Galletto R.. DNA binding induces dimerization of saccharomyces cerevisiae Pif1. Biochemistry. 2010; 49:8445–8454. [DOI] [PubMed] [Google Scholar]
- 53. Adamczak R., Porollo A., Meller J.. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins Struct. Funct. Genet. 2005; 59:467–475. [DOI] [PubMed] [Google Scholar]
- 54. He X., Byrd A.K., Yun M.-K., Pemble C.W., Harrison D., Yeruva L., Dahl C., Kreuzer K.N., Raney K.D., White S.W.. The T4 phage SF1B helicase Dda is structurally optimized to perform DNA strand separation. Structure. 2012; 20:1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. The PyMOL Molecular Graphics System. Version 2.0 Schrödinger, LLC. [Google Scholar]
- 56. Sabouri N. The functions of the multi-tasking Pfh1Pif1 helicase. Curr. Genet. 2017; 63:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Azvolinsky A., Dunaway S., Torres J.Z., Bessler J.B., Zakian V.A.. The S. cerevisiae Rrm3p DNA helicase moves with the replication forkand affects replication of allyeast chromosomes. Genes Dev. 2006; 20:3104–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McDonald K.R., Guise A.J., Pourbozorgi-Langroudi P., Cristea I.M., Zakian V.A., Capra J.A., Sabouri N.. Pfh1 is an accessory replicative helicase that interacts with the replisome to facilitate fork progression and preserve genome integrity. PLoS Genet. 2016; 12:e1006238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kozak M. How do eukaryotic ribosomes select imitation regions in messenger RNA. Cell. 1978; 15:1109–1123. [DOI] [PubMed] [Google Scholar]
- 60. Pike J.E., Henry R.A., Burgers P.M.J., Campbell J.L., Bambara R.A.. An alternative pathway for okazaki fragment processing. J. Biol. Chem. 2010; 285:41712–41723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.