Abstract
The GTPase elongation factor EF-Tu delivers aminoacyl-tRNAs to the mRNA-programmed ribosome during translation. Cognate codon-anticodon interaction stimulates GTP hydrolysis within EF-Tu. It has been proposed that EF-Tu undergoes a large conformational change subsequent to GTP hydrolysis, which results in the accommodation of aminoacyl-tRNA into the ribosomal A-site. However, this proposal has never been tested directly. Here, we apply single-molecule total internal reflection fluorescence microscopy to study the conformational dynamics of EF-Tu when bound to the ribosome. Our studies show that GTP hydrolysis initiates a partial, comparatively small conformational change of EF-Tu on the ribosome, not directly along the path from the solution ‘GTP’ to the ‘GDP’ structure. The final motion is completed either concomitant with or following dissociation of EF-Tu from the ribosome. The structural transition of EF-Tu on the ribosome is slower when aa-tRNA binds to a cognate versus a near-cognate codon. The resulting longer residence time of EF-Tu on the ribosome may be important for promoting accommodation of the cognate aminoacyl-tRNA into the A-site.
INTRODUCTION
Translation elongation factor Tu (EF-Tu) is a G-protein that binds and delivers aminoacylated tRNAs (aa-tRNAs) to the codon-testing site (A/T site) of the ribosome in a ternary complex (TC) composed of EF-Tu, GTP and aa-tRNA. Upon selection of a cognate aa-tRNA, the ribosome stimulates GTP hydrolysis within EF-Tu, resulting in full accommodation of aa-tRNA into the ribosomal A-site and dissociation of EF-Tu·GDP (1). EF-Tu contains three structural domains, the GTP-binding domain I (or G-domain) and tRNA-binding domains II and III. Cryo-EM and X-ray crystallography studies of various complexes have shown EF-Tu to have two distinct conformations. In the structures of the ‘open’ GDP-bound form of EF-Tu from Escherichia coli and Thermus aquaticus (2,3), the G-domain rotates by ∼90° relative to domains II and III, when compared to the ‘closed’ GDPNP-bound form of EF-Tu from T. thermophilus and T. aquaticus (4,5). More recently, an X-ray crystallography study of the E. coli EF-Tu·GDPNP complex showed it to be in an open conformation, and accompanying single-molecule solution studies indicated that EF-Tu·GTP samples a wide set of conformations between the open and closed extremes (accompanying paper, (6)). On the other hand, there is clear consensus from both structural and solution studies that EF-Tu from either T. thermophilus or E. coli binds to the ribosome in a closed conformation in structures of the TC stalled on the 70S ribosome in the presence of kirromycin (7) or when GDPCP or GDPNP replaces GTP ((8) and (6)).
The mechanism of aa-tRNA selection by the ribosome was first studied by pre-steady state kinetic measurements. Codon-independent initial binding of the TC to the ribosome and codon-recognition steps were monitored by fluorescent tRNA derivatives (9,10), while GTPase activation was followed using fluorescent nucleotides and quenched-flow experiments (11,12). Later, the mechanism of aa-tRNA selection was investigated using single-molecule fluorescence microscopy methods (13). Monitoring of fluorescence resonance energy transfer (FRET) between fluorescently labeled tRNAs enabled detection of the codon-recognition, GTPase activation and aa-tRNA accommodation steps (14). In addition, a possible pseudo-GTPase activated state was postulated in which the TC makes transient contacts with the GTPase associated center and sarcin-ricin loop of the ribosome (15). Motions of aa-tRNA in the A/T-site between the GTPase-activated and the accommodated states were detected even before GTP hydrolysis, implying that aa-tRNA samples positions towards the accommodated state before EF-Tu switches into the GDP conformation (16). Fluorescence polarization studies have also emphasized the dynamic nature of TCs bound to the ribosome (17). Most recently, single-particle cryo-EM studies have resolved near-atomic resolution intermediates of ribosome-bound TCs before (18) and after (19) GTP hydrolysis by E. coli EF-Tu and human eEF1A, respectively.
Together, these studies provided a comprehensive picture of the steps involved in aa-tRNA selection and how the ribosomal GTPase associated center activates GTP hydrolysis by EF-Tu (20), but left unclear the structural dynamics of ribosome-bound EF-Tu, in particular the timing of the transition between the ‘EF-Tu·GTP’ (closed) and ‘EF-Tu·GDP’ (open) conformations.
Previously, we inferred, based on stopped-flow kinetics and single-molecule FRET (smFRET) studies of single-labeled EF-Tu interacting with L11-labeled ribosomes, that the main conformational change of EF-Tu occurs either following or concomitant with EF-Tu dissociation from the ribosome (21). This inference disagrees with earlier proposals of others that the main EF-Tu conformational change occurs, while it is still bound to the ribosome (22,23). Here, using smFRET monitoring of the conformational dynamics of EF-Tu on the ribosome, we provide direct evidence supporting our earlier inference. For this purpose, we employ EF-Tu mutants dual-labeled with Cy3 and Cy5 fluorophores, which allow relative distance measurements between domains I and III of EF-Tu (Figure 1), while it is bound to the ribosome. Our results clearly show that while EF-Tu-dependent GTP hydrolysis induces a partial conformational change of ribosome-bound EF-Tu, the open ‘EF-Tu·GDP’ conformation is not reached until after EF-Tu dissociation. Importantly, the partial conformational change of EF-Tu proceeds more slowly for a cognate TC than for a near-cognate TC, supporting proposals that EF-Tu actively participates in aa-tRNA accommodation (14,16,24–27).
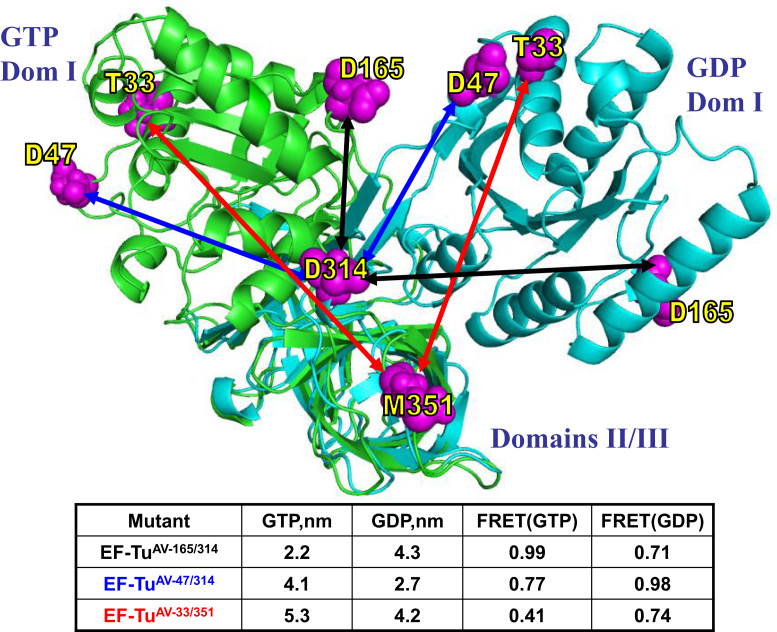
Figure 1.
Design of double-cysteine mutants of EF-Tu for dual-labeling with Cy3 and Cy5. The chosen pairs of cysteines are shown in magenta on the structures of the GDP- and GTP-bound forms of EF-Tu (Protein Data Bank entries 1DG1 and 1OB2, respectively) represented in cyan and green, respectively. Domains II and III of the two structures are superimposed to illustrate the expected distance change between the Cα carbon atoms of the two cysteines in each pair. The arrows show predicted distances for three different double-mutants: EF-TuAV-165/314 (black), EF-TuAV-47/314 (blue) and EF-TuAV-33/351 (red). The table provides the distances in nm along with the corresponding, theoretical FRET values calculated based on Ro = 5.0 nm and the assumption that κ2 = 2/3, which is standard for relative distance measurements (54). The actual distance changes between the Cy dyes could be different due to linker flexibility. The effector region is disordered in 1OB2, rendering the prediction of distances in the GTP-bound form somewhat less trustworthy for EF-TuAV-33/351 and EF-TuAV-47/314. All three dual-labeled EF-Tus are capable of reaching their predicted closed and open structural extremes in solution (6).
MATERIALS AND METHODS
A more detailed description of the applied methods is given in the online Supplementary Data.
Design and preparation of EF-Tu mutants
Site-specific labeling positions (cysteines) within EF-Tu were identified by analyzing crystal structures of the T. thermophilus 70S·EF-Tu·GDP·kirromycin·aa-tRNA complex (PDB codes 1WRN, 1WRO) (7), E. coli EF-Tu·GDPNP·kirromycin·aa-tRNA (1OB2) and EF-Tu·GDP (1DG1) (Figure 1). We used site-directed mutagenesis to separately introduce three pairs of cysteines (C33/C351; C47/C314; C165/C314) into a variant of EF-Tu with two of its three intrinsic cysteines mutated (C81/C137A/C255V, denoted EF-TuAV). This resulted in the three mutants EF-TuAV-33/351, EF-TuAV-47/314 and EF-TuAV-165/314. EF-Tu expression and purification were performed as described (28) with minor modifications described in the Supplementary Materials and Methods.
Labeling of EF-Tu mutants
4-fold molar excesses of both Cy3 and Cy5 maleimides (GE Healthcare) were added to each of EF-TuAV-33/351, EF-TuAV-47/314 and EF-TuAV-165/314, and labeling proceeded for 10 min at room temperature before being quenched with 140 mM β-mercaptoethanol. Excess dye was removed by using PD-10 gel filtration columns (GE Healthcare).
EF-Tu concentrations were determined by the Bio-Rad protein assay (Bio-Rad), while Cy3 and Cy5 concentrations were determined by measuring their absorbances at 550 and 650 nm, respectively (ϵ550 = 150 000 M−1 cm−1, ϵ650 = 250 000 M−1 cm−1).
EF-Tu characterization in ensemble functional assays
The activities of labeled EF-Tu variants in poly(U)-dependent poly(Phe) synthesis, in protecting the labile ester bond in Phe-tRNAPhe against non-enzymatic hydrolysis, and in the formation of an accommodated complex containing Phe-tRNAPhe in the A-site of mRNA-programmed ribosomes were determined as described previously (29).
Total internal reflection fluorescence (TIRF) microscopy-based single-molecule analysis of dual-labeled EF-Tu
The mRNAs used were biotinylated at the 5′ end (Dharmacon, Inc.). Their start codons, marked by a strong Shine-Dalgarno sequence, were followed by A-site codons UUC (cognate, encodes Phe) or CUC (near-cognate, encodes Leu).
The custom-built, objective-type inverted TIRF microscope, preparation of microscope slides and the 70S ribosomal initiation complex (70SIC), containing fMet-tRNAfMet in the P-site and an empty A-site, as well as immobilization of 70SIC to the surface of the microscope slide were described earlier (30). All experiments were conducted under 532 nm laser illumination and using a EMCCD camera (Cascade II, Photometrics). Reducing the imaging area to 128 pixels by 512 pixels permitted 11-ms time resolution. Dual-labeled TC was injected onto the slide at 10 nM concentration in imaging buffer (50 mM Tris–Cl, pH 7.5; 7 mM MgCl2; 30 mM KCl; 70 mM NH4Cl; 1 mM DTT) containing an enzymatic deoxygenation system and triplet state quenchers (30). Intensities of FRET events, which lasted for 33 ms (i.e. three frames) or longer, were extracted and analyzed by using custom-written Matlab (MathWorks) codes and OriginPro (OriginLab Corporation) software. Apparent FRET efficiency was calculated as Eapp = (ICy5)/(ICy5+ICy3), where ICy5 and ICy3 denote the raw fluorescence intensities collected from the Cy5 and Cy3 channels, respectively. Corrected FRET efficiencies were calculated according to the equation: E = (ICy5-χICy3)/(ICy5-χICy3+γICy3), where χ is the leakage of donor emission into the acceptor detection channel, and γ is a correction factor accounting for the ratio of detection efficiencies and quantum yields of the donor and acceptor labels (31). Synchronized apparent FRET traces (32) were calculated from averaged synchronized Cy3 and Cy5 traces before applying these correction factors. The synchronized intensities were subject to global fitting as described in the Supplementary data.
RESULTS
Labeling and characterization of EF-Tu mutants
Wild-type EF-Tu from E. coli contains three Cys residues located at positions 81, 137, and 255. Previously, we have shown that the EF-Tu variant denoted EF-TuAV, in which the Cys residues at positions 137 and 255 are replaced by Ala and Val, respectively, retains substantial activity in protein synthesis. In contrast, Cys81, located between the GTP and tRNA binding sites, is essential (29). In the EF-TuAV background, three different pairs of amino acids, T33/M351, D47/D314 and D165/D314 (Figure 1), were mutated to cysteines for fluorophore labeling. The positions within each pair were selected to enable the analysis of relative distance changes between domains I and III on conversion of the putative closed, GTP-bound form of EF-Tu, as modeled by EF-Tu·GDPNP, to the open, GDP-bound form. In addition, each of the five mutated residues meet the criteria that the native amino acids are neither conserved nor buried, and have no direct contacts with GDP/GTP, aa-tRNA or the ribosome.
The resulting mutants EF-TuAV-33/351, EF-TuAV-47/314 and EF-TuAV-165/314 were labeled with a mixture of Cy3 and Cy5 maleimides, yielding mixtures of EF-Tu labeled with both Cy3 and Cy5 (dual-labeled EF-Tu) as well as EF-Tu labeled with Cy3 or Cy5 only. Nearly equal incorporation of Cy3 and Cy5 was observed. EF-Tu samples labeled with only Cy3 or Cy5 maleimide were prepared similarly. Under these conditions, the labeling (with Cy3 and/or Cy5) of Cys81 was ∼0.1 total dye/protein, while the total labeling of each of the three double mutants was 1.0–1.2 total dye/protein (data not shown).
All labeled EF-Tu mutants showed activities equal to or slightly lower to that of the wild type in poly(Phe) synthesis (Supplementary Figure S1A) and in protection of the ester bond of Phe-tRNAPhe from spontaneous hydrolysis (Supplementary Figure S1B). The hydrolysis protection assay was carried out under conditions (30°C; 30 mM NH4Cl) that do not artificially favor complex formation (see e.g. (33–35)) using 0.4 μM EF-Tu·GTP and 0.1 μM Phe-tRNAPhe. For EF-Tu concentrations ≤2 μM, the poly(Phe) synthesis assay (Supplementary Figure S1A) depended linearly on EF-Tu concentration (Supplementary Figure S1C and D). Rates of A-site accommodation were measured for EF-TuAV mutants labeled with only Cy3 or Cy5 to reduce sample heterogeneity, as well as for EF-TuAV labeled to ∼0.1 dye/protein. All labeled samples showed rates of Phe-tRNAPhe accommodation into the ribosomal A-site comparable to that of wild-type EF-Tu (Supplementary Figure S2).
Single-molecule trajectories and kinetics of dual-labeled EF-TuAV-33/351 on the ribosome
Next, the interactions of TCs containing each of the three dual-labeled EF-Tu variants with the 70S initiation complex (70SIC) were subjected to single-molecule analysis using total internal reflection fluorescence (TIRF) microscopy. As dual-labeled EF-TuAV-33/351 proved to be the best reporter of structural changes, the results obtained for this mutant are described below in most detail.
70SICs were coupled to a streptavidin-coated surface through biotinylated mRNAs containing a strong Shine-Dalgarno sequence upstream of the P-site start codon, AUG (36,37). The P-site of the 70SICs contained initiator fMet-tRNAfMet, while either a phenylalanine (UUC, cognate) or leucine (CUC, near-cognate to Phe-tRNA) codon was positioned in the A-site. In separate experiments for the Phe and Leu codons, dual-labeled EF-TuAV-33/351·GTP in a TC with Phe-tRNAPhe was injected into the flow cell with simultaneous image recording at 11-ms time resolution. Binding of each dual-labeled TC to an immobilized 70SIC resulted in simultaneous increases in Cy3 and Cy5 fluorescence above background levels, which persisted until EF-Tu dissociated. Although one or the other of these signals arose from EF-Tu molecules labeled with Cy3 and Cy5 only, as well as from dual-labeled EF-Tu, only the latter gave rise to sensitized emission from Cy5 due to FRET (Figure 2A–C). Individual, dual-labeled EF-TuAV-33/351 FRET traces obtained with a cognate A-site codon showed a brief low-FRET state (Eapp = 0.45 ± 0.01) at the beginning of the event followed by a rapid increase in FRET signal (Eapp = 0.60 ± 0.01) (Figure 2A). For the near-cognate mRNA, two types of traces were observed in approximately equal amounts. The first showed a single low-FRET state (Eapp = 0.43 ± 0.06, Figure 2B), and the second showed a transition from a lower to a higher FRET state (Eapp 0.43 ± 0.02 to 0.62 ± 0.04, Figure 2C). These FRET signals were clearly ribosome-dependent, as only very few non-specific background interactions were observed in the absence of ribosomes (Figure 2F).
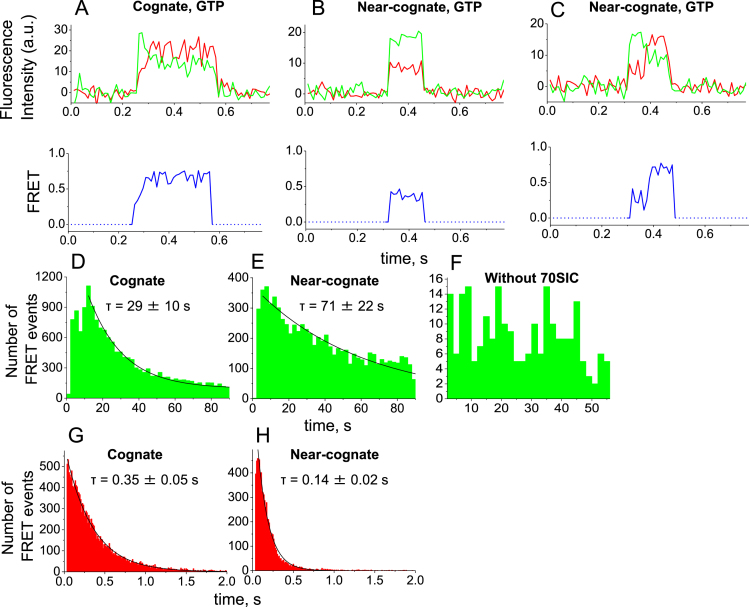
Figure 2.
Single-molecule observations of dual-labeled EF-TuAV-33/351·GTP·Phe-tRNAPhe interacting with 70SIC. (A–C) The upper panels show representative examples of fluorescence trajectories of Cy3 (green) and Cy5 (red) recorded with 532 nm excitation upon interaction of dual-labeled EF-TuAV-33/351·GTP·Phe-tRNAPhe with 70SIC containing a cognate (A) or a near-cognate (B and C) A-site codon. The lower panels show the corresponding apparent FRET efficiencies in blue. (D and E) Decay of available 70SICs displaying a cognate (D) or a near-cognate (E) codon in the ribosomal A-site before dual-labeled EF-TuAV-33/351·GTP·Phe-tRNAPhe binds and occupies the A-site. The histograms show the number of binding events detected in successive 2-second intervals during imaging. (F) Non-specific binding of dual-labeled EF-TuAV-33/351·GTP·Phe-tRNAPhe to the microscope slide, when ribosomes are omitted. (G and H) Dwell-time histograms for the interaction of dual-labeled EF-TuAV-33/351·GTP·Phe-tRNAPhe with the 70SIC when cognate (G) and near-cognate (H) mRNAs are used. The histograms show the number of binding events of specific durations (dwell times). In (D–E) and (G–H), the histograms were fitted to single-exponential decay functions. The resulting decay times of available 70SICs (D-E) and dwell-times on the ribosome (G–H) are shown along with standard deviations of the fits (see also Supplementary Table S1). The histograms show events pooled from 3–4 individual experimental replicates. Approximately 16 800 and 7600 events are included in the analysis for cognate and near-cognate tRNA delivery, respectively.
Fluorescence events lasting <33 ms were excluded from the data analysis to avoid including random noise. Many more such brief events were observed for near-cognate than for cognate mRNA, as expected for non-productive ‘sampling’ interactions between the TCs and the 70SIC during initial selection leading to rapid dissociation. When aa-tRNA was accommodated into the A site and a peptide bond was formed, the ribosome remained in the PRE-translocation state, because EF-G was absent. The appearance rate of FRET events lasting more than 33 ms decayed over time due to the irreversible transformation of 70SICs into pre-translocation complexes having A-sites occupied by fMet-Phe-tRNAPhe (Figure 2D and E). The time constants for the loss of 70SICs were 29 ± 10 s and 61 ± 21 s for the cognate and near-cognate mRNAs, respectively (Supplementary Table S1). We attribute the slower near-cognate decay to the more frequent non-productive sampling of TCs with the near-cognate 70SICs. In contrast, dwell times of dual-labeled EF-TuAV-33/351 on the ribosome, indicated by the duration of the FRET signals, were longer for the cognate versus the near-cognate mRNAs, averaging 0.35 ± 0.05 s and 0.15 ± 0.02 s, respectively (Figure 2G–H, Supplementary Table S1).
Replacement of GTP by GDPNP in the cognate reaction mixture slowed the decay of 70SICs by ∼4-fold to 125 ± 12 s (Supplementary Table S1). This might be caused by one or more of several factors including slower binding, slower accommodation, lower affinity of the GDPNP-containing TCs for the 70SIC (29) and/or more reversible binding events due to the lack of GTPase activity. In contrast, kirromycin decreased the decay time for remaining cognate 70SICs to 17 ± 3 s and had no effect on the decay time of near-cognate 70SICs (Supplementary Table S1). Dwell times of EF-Tu on the ribosome were increased by kirromycin for cognate and near-cognate mRNAs and by GDPNP with cognate mRNA (Supplementary Table S1), in agreement with earlier reports from ensemble experiments (10,11,29). Under these conditions, the duration of the FRET signal was partly limited by photobleaching, leading to an underestimation of the dwell times.
FRET states of dual-labeled EF-TuAV-33/351 on the ribosome
The changes of FRET values of all binding events for both cognate and near-cognate codons can be visualized in a transition density map, which is a contour plot of the final versus the initial apparent FRET efficiencies, Eapp, taken from the last and first frame of each event, respectively. All apparent initial and final FRET values derived from this analysis are listed in Supplementary Table S1. The transition density plot for the binding of a TC formed by dual-labeled EF-TuAV-33/351 to a cognate 70SIC showed a single peak, located above the diagonal, indicating a population of binding events switching from the lower (Eapp = 0.45 ± 0.01) into the higher (Eapp = 0.60 ± 0.01) FRET state (Figure 3A). By contrast, substituting GTP by GDPNP caused the peak to move to the 45° line of identity in the transition density plot (Eapp = 0.51 ± 0.01, Figure 3C), indicating that the FRET change is dependent on GTP hydrolysis. In addition, kirromycin prevented the FRET change (Supplementary Table S1).
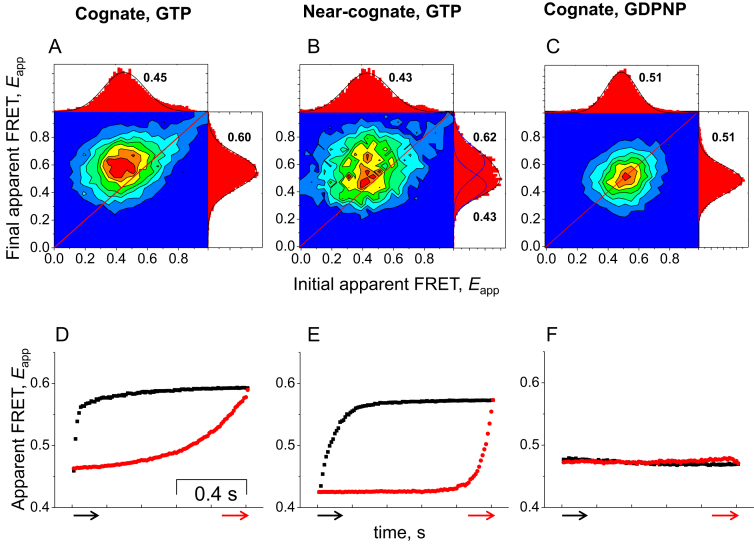
Figure 3.
Analysis of FRET states of dual-labeled EF-TuAV-33/351. Transition density plots (A–C) showing the initial and final apparent FRET values, Eapp, and synchronized apparent FRET trajectories (D–F) of dual-labeled EF-TuAV-33/351·GTP·Phe-tRNAPhe upon interaction with the 70SIC under different conditions. All recorded events are included in these figures. (A and D): cognate mRNA and GTP. (B and E): near-cognate mRNA and GTP. (C and F): cognate mRNA and GDPNP. Each transition density plot has associated FRET histograms showing the distribution of initial (top) and final (right) apparent FRET values. The given initial and final apparent FRET values in (A–C) are the 2D Gaussian fits of the transition density plots (see also Supplementary Table S1). The red and black arrows in (D-F) indicate the trigger points and plotting directions. Event numbers for A, B, D and E are as in Figure 2. 13 000 cognate events were monitored upon replacement of GTP by GDPNP (C and F). In all cases, the events were detected during 3–4 independent experimental replicates.
The transition density plot for the near-cognate mRNA traces showing a transition from a lower to a higher FRET state (Figure 2C) has density above the diagonal (Figure 3B), whereas all of the density for the group of traces maintaining a constant Eapp (Figure 2B) is located near the diagonal line (Figure 3B). Thus, only ∼half of the EF-Tu molecules appear to undergo a conformational change upon binding to the ribosome as part of a near-cognate ternary complex.
Synchronization analysis of dual-labeled EF-TuAV-33/351
Synchronization analysis is used to extract FRET efficiencies and dynamics of the FRET states in the collection of binding events (32,38). Traces of events, which lasted for 33 ms or more were selected for each mutant and aligned at the beginning (pre-synchronization) or at the end (post-synchronization) of the binding events (Figure 4A). The synchronized apparent FRET traces revealed the dynamics of EF-Tu during its interaction with the ribosome. A dynamic change in FRET in the pre-synchronized and the post-synchronized traces was observed for dual-labeled EF-TuAV-33/351 for both the cognate (Figure 3D) and near-cognate binding events (Figure 3E). In contrast, no changes in FRET during EF-Tu binding events were detected for either mRNA in the presence of GDPNP (Figure 3F, cognate), demonstrating that the change in apparent FRET efficiency is dependent on GTP hydrolysis.
Figure 4.
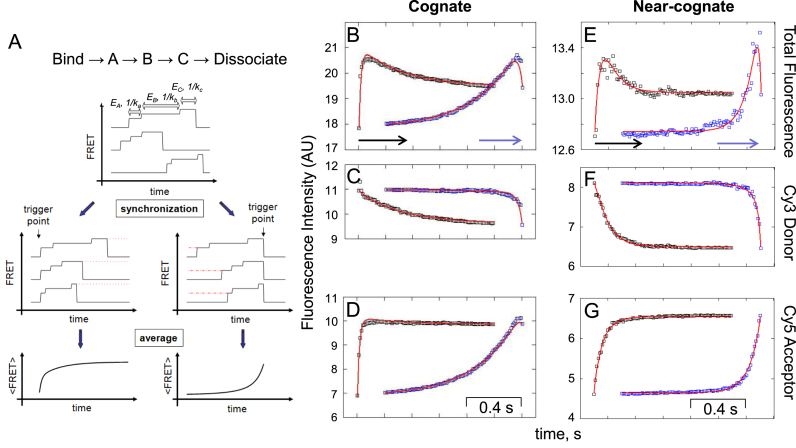
Fitting of synchronized fluorescence intensity traces of dual-labeled EF-TuAV-33/351·GTP·Phe-tRNAPhe interacting with the 70SIC. (A) The fitting model consists of three EF-Tu-containing ribosome complexes A, B and C, which are preceded by EF-Tu binding to the ribosome and terminated by dissociation. The graphics in A illustrate the essentials of the synchronization of events and averaging of traces with the predicted number of steps of TC binding to the 70SIC. The synchronization analysis was applied to traces of events lasting for 33 ms or more. Pre-synchronized (black) and post-synchronized (blue) Cy3 (C, F) and Cy5 (D, G) intensity symbols were summed up to give the total fluorescence intensity (B, E) squares obtained with cognate (B–D) or near-cognate (E–G) mRNA. All of the intensities are absolute intensities in thousands of camera digitizer units. The blue and black arrows indicate the trigger points and plotting directions. The red lines are the global fits according to the model shown in (A). For the near-cognate data, molecules for which EAppfinal – EAppinitial > 0.05 were grouped and synchronized for this analysis. The calculated values are presented in Table 1 (see also Supplementary Figure S3). Event numbers are as in Figures 2 and 3.
With GTP, the averaged synchronized traces for the cognate experiment (Figure 3D) showed a biphasic exponential change in fluorescence with a fast increase of apparent FRET at the beginning of the event followed by a slower apparent FRET increase. The clear two-component approach to the final apparent FRET efficiency in Figure 3D indicates two sequential structural transitions occurring upon progression from an initial complex, A, via an intermediate complex, B, to a final complex, C, before EF-Tu dissociates. Thus, the simplest scheme accounting for the data is:
EF-Tu binding → A → B → C → EF-Tu dissociation (Figure 4A, top), which is similar to one we presented earlier (21). A linear, irreversible scheme is considered, since individual FRET traces did not show clear backward transitions indicating that reversals are relatively rare. In addition, transient binding events due to rapid dissociation from complex A were not included in our data, since only events lasting three frames or longer were analyzed (see above). TC dissociation from intermediate complex B is also considered rare (21). Thus, the simplest scheme above was used for interpreting the data.
Total, Cy3 and Cy5 synchronized fluorescence intensities for traces of cognate binding events showing FRET are displayed in Figure 4B–D. The rapid rise in Cy5 intensity (Figure 4D, black squares), leading to the initial fast increase of Eapp (Figure 3D), is not matched by a corresponding rapid decrease in Cy3 intensity (Figure 4C, black squares). Thus, the resulting initial increase in total fluorescence intensity (Figure 4B) is not a result of FRET but, rather, can be attributed to a local structural change leading to a substantial increase in brightness (quantum yield, QY) of the Cy5 acceptor, which is not surprising as QY of Cy5 is sensitive to environmental changes (39). This QY change was confirmed by repeating the FRET experiment while alternating the excitation wavelength in successive camera frames between 532 nm (Cy3 excitation) and 640 nm (Cy5 excitation) (30,40) (Supplementary Figure S3A–D). As shown in Supplementary Figure S3D, the intensity of Cy5 fluorescence observed upon direct excitation at 640 nm increases rapidly following EF-Tu binding to the ribosome. The intensity traces in Figure 4B–D were therefore fitted by an analytical model of the process (described in Supplementary Materials and Methods) that takes into account the change in QY of Cy5 during the interaction of EF-Tu with the ribosome. The results are summarized in Table 1.
Table 1.
Global analysis. Values calculated from global fittings of all pre-synchronized and post-synchronized traces of dual-labeled EF-TuAV-33/351. For the near-cognate data, only molecules for which EAppfinal – EAppinitial > 0.05 were included in this analysis. EA, EB and EC are the actual FRET efficiencies of dual-labeled EF-TuAV-33/351 in complexes A, B and C (Figure 5), and 1/ka, 1/kb and 1/kc are the lifetimes of the three complexes. γA, γB and γC are the values of γ in states A, B and C, respectively, where γ is the combined ratio of QYs and sensitivities in the Cy3 and Cy5 fluorescence detector channels. The FRET values correspond to E = [(1 + χ)Eapp-χ]/[(1+χ – γ)Eapp – χ + γ], with parameters defined and explained in the Supplementary Materials and Methods section ‘Global fitting of synchronized intensities’. The uncertainties are 95% confidence limits calculated from changes of the fitted parameters that increase χ2 four-fold over the global fits or, in the case of ka, from the initial period 3/ka and kc, from the final 3/kb period. 16 800 cognate and 7600 near-cognate events obtained from three to four independent replicates were included in the analysis
| Codon/values | E A | E B | E C | k a, s−1 | k b, s−1 | k c, s−1 | γA | γB | γC |
|---|---|---|---|---|---|---|---|---|---|
| Cognate | 0.41 ± 0.01 | 0.42 ± 0.01 | 0.48 ± 0.01 | 65 ± 14 | 3.2 ± 0.6 | 35 ± 24 | 0.81 ± 0.03 | 1.22 ± 0.05 | 1.03 ± 0.03 |
| Near-cognate | 0.39 ± 0.01 | 0.42 ± 0.02 | 0.51 ± 0.01 | 25 ± 7 | 12 ± 6 | 57 ± 23 | 0.81 ± 0.02 | 1.0 ± 0.07 | 0.92 ± 0.02 |
Similar analysis was carried out for the FRET experiments using near-cognate mRNA (Figure 4E–G). Here, the two populations observed in the near-cognate case (Figure 3B and C) were analyzed separately by splitting the traces into two groups, depending on whether the difference between the final Eapp value and the initial value (before correction for changes in QY) was less than (40% of all near-cognate events) or greater than 0.05 (60% of all near-cognate events). The events coming from either population had decay times of remaining available 70SICs (Eappfinal– Eappinitial ≤ 0.05, 55 ± 19 s; Eappfinal– Eappinitial > 0.05, 61 ± 22 s) and dwell times on the ribosome (Eappfinal– Eappinitial ≤ 0.05, 0.14 ± 0.02 s; Eappfinal– Eappinitial > 0.05, 0.15 ± 0.02 s) that were indistinguishable from one another (data not shown; related to Figure 2E and H showing all FRET events).
Actual FRET efficiencies of complexes A, B and C, corrected for the changes in QY, as well as rate constants determined from the global fitting of cognate and near-cognate, synchronized Cy3 and Cy5 fluorescence intensities, are presented in Table 1 for the group of near-cognate traces in which Eappfinal– Eappinitial > 0.05, while calculated occupancies of the three states are shown in Supplementary Figures S3E and S3F for cognate complexes. Importantly, the actual FRET efficiency does not change substantially between complexes A and B, indicating that this transition is not the major structural change between the GDPNP and GDP forms determined by crystallography of isolated EF-Tu. Rather, the initial change in sensitized emission from Cy5 is due primarily to its change in QY following binding to the ribosome. This change is faster for the cognate codon (ka = 65 ± 14 s−1; fitted value ± 95% confidence interval) than for the near-cognate codon (ka = 25 ± 7 s−1), whereas the subsequent transition step from B to C, corresponding to the decrease in distance between the probes (E changes from 0.42 → 0.48/0.51 for cognate/near-cognate complexes), is slower for the cognate interaction (kb = 3.2 ± 0.6 s−1) than for the near-cognate (kb = 12 ± 6 s−1). The last step, the dissociation of EF-Tu from complex C, has rate constants that are not significantly different for cognate and near-cognate codons (kc ∼35 ± 24 s−1 and 57 ± 23 s−1, respectively). The actual FRET efficiencies in complexes A, B and C are the same for both types of codons. Note that the FRET values in Table 1, corrected for the change in QY (0.41 increasing to 0.48 or 0.39 increasing to 0.51 for cognate or near-cognate complexes, respectively) are somewhat different from the corresponding EF-TuAV-33/351 entries in Supplementary Table S1 (∼0.45 increases to ∼0.60), which are Eapp values, not taking into account the dynamic change of Cy5 QY.
smFRET analysis of dual-labeled EF-TuAV-165/314 and EF-TuAV-47/314
TCs formed by dual-labeled EF-TuAV-165/314 and EF-TuAV-47/314 showed 70SIC-dependent binding events, during which Cy3 and sensitized Cy5 fluorescence intensities simultaneously increased and decreased (Supplementary Figure S4), similar in kind to results reported above for dual-labeled EF-TuAV-33/351 (Figure 2A–C). Despite differences in the parameter values between mutants (Supplementary Table S1), a number of general trends are observable: (i) available ribosomes with empty A-sites (i.e. 70SICs) decay more slowly if programmed with a near-cognate mRNA than with a cognate, due to a higher proportion of reversible sampling events, (ii) replacement of GTP with GDPNP increases the decay time of available 70SICs, (iii) dwell times of EF-Tu on the ribosome are longer in the presence of a cognate codon-anticodon interaction relative to near-cognate, (iv) kirromycin and GDPNP increase the dwell times and (v) kirromycin has no or only minor effects on decay times of available 70SICs. In contrast to EF-TuAV-33/351, EF-TuAV-47/314 and EF-TuAV-165/314 showed very small or no FRET changes, respectively, during their binding events under these various conditions. Their transition density plots displayed peaks centered on the diagonal (Supplementary Figure S6), and their synchronized FRET traces showed only minor changes (Supplementary Figure S7), indicating little distance change.
DISCUSSION
Conformational dynamics of EF-Tu on the ribosome
X-ray crystallographic studies of EF-Tu (1) not bound to the ribosome gave rise to the view that the factor exists in two quite different conformations, denoted ‘closed’ or ‘EF-Tu·GTP’ and ‘open’ or ‘EF-Tu·GDP’ (Figure 1). The GTPase activity of EF-Tu is triggered by the GTPase activating center on the ribosome upon correct codon-anticodon interaction, leading to the hypothesis that the large structural rearrangement on conversion of the closed to the open conformation results in EF-Tu release from both aa-tRNA and the ribosome. However, until recently, capturing a snapshot of EF-Tu·GDP on the ribosome required stalling the factor in a GTP-like structure via addition of the antibiotic kirromycin (41). Thus, information about the structure of EF-Tu on the ribosome post-GTP hydrolysis has been lacking. Using TIRF-based single-molecule FRET microscopy, we have obtained dynamic information about the structural transitions of EF-Tu occurring on the ribosome.
While bound to an mRNA-programmed ribosome, dual-labeled EF-TuAV-33/315 undergoes a single, comparatively minor, FRET change in a relatively slow step (3–12 s−1) (Table 1). Importantly, this change, indicating a decrease in distance between these two sites as expected from the solution crystal structures (Figure 1), is the first direct measurement of a GTP-hydrolysis dependent structural change in EF-Tu bound to a ribosome. This change in FRET efficiency does not occur when ribosome-bound EF-Tu is maintained in a conformation similar to that observed in isolated TCs, either by substituting GDPNP for GTP or in the presence of kirromycin (Supplementary Table S1), as found in cryo-EM and X-ray crystallographic structures of EF-Tu·GDP·kirromycin on the ribosome (7,41). The final FRET value of ribosome-bound EF-Tu, 0.48 ± 0.01 (Table 1), is much lower than that predicted (0.74, Figure 1) or measured for EF-Tu·GDP in solution (0.78 ± 0.1; (6)). These results strongly imply that the full structural change between the closed and open states of EF-Tu occurs not while EF-Tu is bound to the ribosome, but rather is concomitant with or follows dissociation of EF-Tu from the ribosome.
While bound to the ribosome the other two dual-labeled variants we have studied, EF-TuAV-165/314 and EF-TuAV-47/314, also show no large FRET changes, ruling out formation of the open ‘EF-Tu·GDP’ conformation, but differ from dual-labeled EF-TuAV-33/315 in exhibiting no or only very small FRET changes, respectively (Figure 1 and Supplementary Figure S4). We performed modeling studies demonstrating that this difference can be rationalized if domain 1 of ribosome-bound EF-Tu is subject to rotations and transitions relative to domains 2 and 3 (Supplementary Figure S8 and Supplementary file S1). Our results are thus consistent with a conclusion of Lai et al. (42) that the structural transition of EF-Tu on the ribosome follows an indirect pathway, although they do not support the large domain separation suggested by these authors. We suggest that the differences in dynamic changes observed for EF-TuAV-165/314 and EF-TuAV-47/314versus EF-TuAV-33/315 result from differential mutation and labeling effects on the dynamic equilibrium between the EF-Tu structural extremes. All three EF-Tu mutants in this study have mutations positioned spatially close either to switch I (positions 33 and 47) or to helix C (position 165) and, in the related protein, ras p21, mutations in the corresponding regions are known to affect the balance between two GTP-bound conformations (43,44). Similarly, a mutation in the switch II region causes the EF-Tu·GDP complex to adopt an ‘EF-Tu·GTP’-like behaviour (45).
Our present and earlier results (21), indicate that the full conformational transition of EF-Tu takes place after tRNA has been released from EF-Tu i.e. aa-tRNA release occurs while EF-Tu is still in a conformation closer to the free EF-Tu ‘GTP’ state than the ‘GDP’ one. This is in accord with studies reporting proportionality between the dissociation rate of a given aa-tRNA from EF-Tu·GTP, koff, and the rate of the subsequent peptide-bond formation, kpep (29,46). These results emphasize the apparently optimized affinity of aa-tRNA for EF-Tu·GTP such that koff does not limit kpep. Such optimization would be unnecessary, if aa-tRNA were released from EF-Tu in a GDP-like conformation, since EF-Tu·GDP binds to aa-tRNA at least two orders of magnitude less tightly than does EF-Tu·GTP (47).
Kinetic mechanism and proofreading
Our results permit the construction of a minimal, 4-step kinetic scheme (Steps I–IV, Figure 5), for the formation of the pre-translocation complex upon binding of the ternary complex containing dual-labeled EF-TuAV-33/351 to 70SIC programmed with cognate or near-cognate mRNA. The reaction progresses via complexes A, B and C (equivalent to the model in (21)), which correspond to FRET states A, B and C in the minimal reaction scheme outlined above (Figure 4A, top). This scheme allows evaluation of average FRET efficiencies (E), FRET correction factors including changes in quantum yields (γ), and reaction rate constants (k) (Table 1).
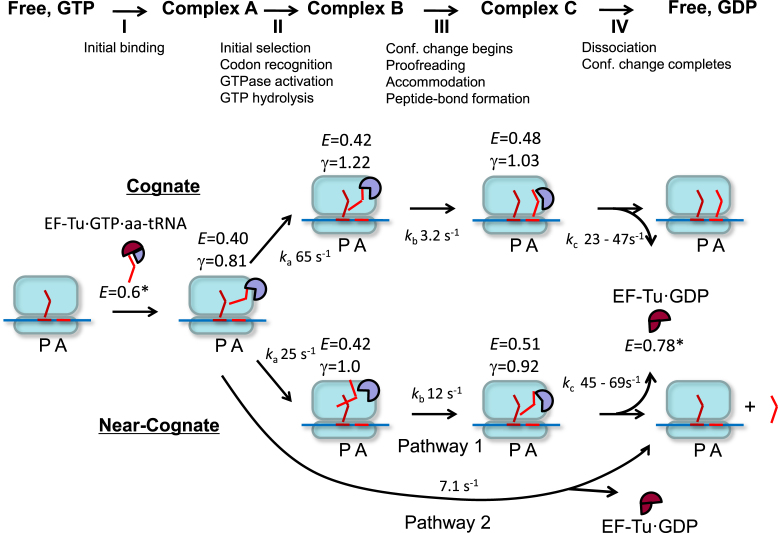
Figure 5.
A kinetic model of interactions between EF-Tu and the ribosome. Actual FRET efficiencies (E), QY ratio correction factors γ, and reaction rate constants ka, kb, kc were determined for dual-labeled EF-TuAV-33/351·GTP·Phe-tRNAPhe interacting with 70SICs programmed with both cognate and near-cognate mRNAs (Table 1). The 70S ribosome is shown in light blue, the mRNA is a blue line, while tRNAs and their corresponding codons are shown in shades of red as bent lines. EF-Tu is shown as a circular sector with different degrees of opening representing different conformations. Classical, GTP-like conformations of EF-Tu are indicated in lavender (FRET states with E in the range from 0.40 to 0.51), while a maroon color is indicative of the classical GDP-like conformation (FRET state E = 0.78*). Thus, the initial FRET state E = 0.6* within the unbound ternary complex, is colored in a combination of maroon and lavender. In the near-cognate case, approximately half of the EF-Tu molecules have no significant FRET or γ change and dissociate with a rate constant of 7.1 s−1 (Pathway 2). kc is given as a range corresponding to ± 1 standard error of the mean. *The FRET values for complexes in solution were determined by (6). These values are not corrected for γ, as the relative QYs and detector sensitivities were not measured in that study. Since the apparent FRET efficiency values for ribosome-bound complexes in (6) (EApp2 values in Figure 3F and G) are similar to the values obtained here, the apparent FRET values for EF-Tu·GTP·aa-tRNA and EF-Tu·GTP in solution are suitable for comparison here.
Upon codon-independent initial binding of the ternary complex to the 70SIC (Step I), complex A is formed and E decreases from 0.61 ± 0.1 in solution (6) to 0.41 (Table 1). For a cognate TC, this is followed by a rapid Step II (65 s−1), which leads to the formation of complex B. Step II is accompanied by a large increase in γ, caused by an increase in the quantum yield (QY) of Cy5 (Figure 4B–D, Supplementary Figure S3 and Equation (3) in Supplementary Materials and Methods), with little change in E. Step II corresponds approximately to the codon recognition and GTPase activation/GTP hydrolysis steps. In accord with this assignment, the change in Cy5 QY is not observed when the non-hydrolysable analog GDPNP replaces GTP (Figure 3F). The individual events occurring during step II cannot be resolved at the present 11-ms time resolution. However, the observation of pronounced repetitive, non-productive binding for near-cognate complexes shows that initial selection does take place in our experimental set-up. Step III, leading to complex C, proceeds more slowly (3.2 s−1). Complex C is characterized by a decrease in γ as well as a significant increase in E (from 0.42 to 0.48), indicative of a decreased distance between these sites in domains I and III (Figure 1). In addition to the conformational change in EF-Tu, step III includes proofreading, aa-tRNA accommodation and peptide-bond formation. Following Step III, EF-Tu·GDP dissociates promptly (23–47 s−1) from the ribosome, so that Step III essentially determines the overall dwell-time of EF-Tu on the ribosome (Supplementary Table S1). A much slower rate for EF-Tu·GDP dissociation (2–3 s−1) was proposed earlier (48), but this probably reflects insufficient resolution of Steps III and IV in that study.
As mentioned above, 60% of the events for dual-labeled EF-TuAV-33/315 binding to 70SIC as part of a near-cognate ternary complex exhibited large enough FRET changes so they could be analyzed similarly and compared with the cognate ternary complex, as summarized in Table 1. Although the E values for the various complexes are little affected, there are substantial differences in the rate constants ka (Step II), which decreases from 65 s−1 to 25 s−1, and kb (Step III), which increases from 3.2 s−1 to 12 s−1. The magnitude of ka for the cognate mRNA and the fact that it decreases for the near-cognate case is compatible with GTP hydrolysis occurring during Step II, and the 2.5-fold decrease we measure is in line with the low efficiency of initial selection observed earlier by Rodnina and coworkers using low-fidelity conditions similar to ours (49,50). In (49), a subsequent kinetic step (termed k4 occurring at 50–60 s−1) that accounted for an additional lag in the recorded signals was assigned to the GTP-to-GDP conformational change. Our result that the FRET change is much slower indicates that the k4 of Pape et al. is not the rate constant for the main EF-Tu structural change. This is in agreement with later work by Rodnina and coworkers showing that the previously assigned structural change is more likely to be attributed to Pi dissociation (51).
The γ change observed in step II is codon-dependent, and significantly lower γ values are observed for complexes B and C for the near-cognate versus the cognate complexes. This indicates that the QY change in Cy5 is not dye or position dependent, but reflects subtle differences in the interactions between the ribosome, aa-tRNA and EF-Tu in these states (for a description of relevant structural differences between cognate and near-cognate complexes see (18,52)), providing a rationale for the larger value of kb in the near-cognate case, and the resulting 2.5–3-fold decrease in EF-Tu dwell time.
Previously, EF-Tu was thought to be passively released from aa-tRNA and the ribosome in its open, GDP-bound conformation without playing a role during proofreading and accommodation. Instead, structural transitions in the tRNA via a ribosomal corridor lined by conserved bases have been considered a major driving force during these two processes (53). The differences in EF-Tu dwell time that we observe suggest, to the contrary, that EF-Tu improves proofreading by favoring near-cognate dissociation prior to accommodation and contributes to cognate tRNA accommodation by increasing its residence time. Notably, the relative decreases in EF-Tu residence time for near-cognate versus cognate tRNA is observed for all three dual-labeled EF-Tus (Supplementary Table S1), which argues against the effect reflecting a mutation and/or labeling artifact. Earlier indications supporting the view that EF-Tu affects proofreading can be found in the literature (14,16,25). Interestingly, a mutation in EF-Tu located at the domain I/III interface (Ala375-to-Thr) was found to lower the rejection rate of near-cognate tRNAs during both initial binding and proofreading (24) in support of a possible role of EF-Tu during proofreading. More recently, proofreading has been suggested to occur in at least two discrete steps (26,27), only the first of which involves EF-Tu. In this first step, which occurs following GTP hydrolysis and Pi release, near-cognate aa-tRNA in complex with EF-Tu·GDP can dissociate from the ribosome prior to accommodation, while accommodation is favored for cognate aa-tRNA in complex with EF-Tu·GDP (26). Our results indicate that such accommodation takes place with EF-Tu in a relatively closed conformation. This can be understood from an energetic point of view, since proofreading accuracy decreases linearly with decreasing affinity between EF-Tu and aa-tRNA (26,27) and the closed EF-Tu conformation has a 100-fold higher affinity for aa-tRNA than the open conformation (47).
Conclusion
Our smFRET studies reveal a small conformational change of ribosome-bound EF-Tu following GTP hydrolysis, while the full conformational change of EF-Tu, as expected based on X-ray crystallographic studies, thus occurs concomitant with or after its release from the ribosome. The structural change of EF-Tu on the ribosome, which occurs via a path different from the direct route between the ‘GTP’ and ‘GDP’ structures, is slower for ternary complexes bound to a cognate versus a near-cognate programmed 70SIC, suggesting that this transition, in concert with transitions in the tRNA, can play an active role in the partitioning of aa-tRNA between a first, EF-Tu-dependent part of proofreading and accommodation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Karen Margrethe Nielsen and Xiaonan Cui for skilled technical assistance. Recombinant TEV protease was kindly donated by Dr Ditlev E. Brodersen.
Author Contributions: Y.E.G., B.S.C. and C.R.K. conceived the project and advised on experimental design. D.K. and W.L. designed, produced, labeled and characterized mutants of EF-Tu. D.K. and C.C. performed single-molecule studies. D.K., C.C., B.S.C., Y.E.G. and C.R.K. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Leo Pharma Research Foundation (to D.K. and C.R.K.); Novo Nordisk Foundation (to C.R.K.); Danish Council for Independent Research (Natural Sciences) (to C.R.K.); National Institutes of Health [R01-GM080376 and R35-GM118139 to Y.E.G., B.S.C. and W.L.]; American Heart Association Postdoctoral Fellowship [12POST8910014 to C.C.]. Funding for open access charge: Private Danish foundations.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kavaliauskas D., Nissen P., Knudsen C.R.. The busiest of all ribosomal assistants: elongation factor tu. Biochemistry. 2012; 51:2642–2651. [DOI] [PubMed] [Google Scholar]
- 2. Kjeldgaard M., Nyborg J.. Refined structure of elongation factor EF-Tu from Escherichia coli. J. Mol. Biol. 1992; 223:721–742. [DOI] [PubMed] [Google Scholar]
- 3. Polekhina G., Thirup S., Kjeldgaard M., Nissen P., Lippmann C., Nyborg J.. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure. 1996; 4:1141–1151. [DOI] [PubMed] [Google Scholar]
- 4. Berchtold H., Reshetnikova L., Reiser C.O., Schirmer N.K., Sprinzl M., Hilgenfeld R.. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993; 365:126–132. [DOI] [PubMed] [Google Scholar]
- 5. Kjeldgaard M., Nissen P., Thirup S., Nyborg J.. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993; 1:35–50. [DOI] [PubMed] [Google Scholar]
- 6. Johansen J.S., Kavaliauskas D., Pfeil S.H., Blaise M., Cooperman B.S., Goldman Y.E., Thirup S.S., Knudsen C.R.. E. coli elongation factor Tu bound to a GTP analogue displays an open conformation equivalent to the GDP-bound form. Nucleic Acids Res. 2018; doi:10.1093/nar/gky697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmeing T.M., Voorhees R.M., Kelley A.C., Gao Y.G., Murphy F.V.T., Weir J.R., Ramakrishnan V.. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009; 326:688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Voorhees R.M., Schmeing T.M., Kelley A.C., Ramakrishnan V.. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010; 330:835–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodnina M.V., Pape T., Fricke R., Kuhn L., Wintermeyer W.. Initial binding of the elongation factor Tu.GTP.aminoacyl-tRNA complex preceding codon recognition on the ribosome. J. Biol. Chem. 1996; 271:646–652. [DOI] [PubMed] [Google Scholar]
- 10. Rodnina M.V., Fricke R., Wintermeyer W.. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry. 1994; 33:12267–12275. [DOI] [PubMed] [Google Scholar]
- 11. Rodnina M.V., Fricke R., Kuhn L., Wintermeyer W.. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 1995; 14:2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pape T., Wintermeyer W., Rodnina M.V.. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998; 17:7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blanchard S.C. Single-molecule observations of ribosome function. Curr. Opin. Struct. Biol. 2009; 19:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blanchard S.C., Gonzalez R.L., Kim H.D., Chu S., Puglisi J.D.. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004; 11:1008–1014. [DOI] [PubMed] [Google Scholar]
- 15. Lee T.H., Blanchard S.C., Kim H.D., Puglisi J.D., Chu S.. The role of fluctuations in tRNA selection by the ribosome. PNAS. 2007; 104:13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geggier P., Dave R., Feldman M.B., Terry D.S., Altman R.B., Munro J.B., Blanchard S.C.. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 2010; 399:576–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mishra P.P., Qureshi M.T., Ren W., Lee T.H.. Codon-dependent tRNA fluctuations monitored with fluorescence polarization. Biophys. J. 2010; 99:3849–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loveland A.B., Demo G., Grigorieff N., Korostelev A.A.. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature. 2017; 546:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Behrmann E., Loerke J., Budkevich T.V., Yamamoto K., Schmidt A., Penczek P.A., Vos M.R., Burger J., Mielke T., Scheerer P. et al. . Structural snapshots of actively translating human ribosomes. Cell. 2015; 161:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodnina M.V., Fischer N., Maracci C., Stark H.. Ribosome dynamics during decoding. Philos. Trans. R. Soc. London B, Biol. Sci. 2017; 372:20160182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu W., Chen C., Kavaliauskas D., Knudsen C.R., Goldman Y.E., Cooperman B.S.. EF-Tu dynamics during pre-translocation complex formation: EF-Tu·GDP exits the ribosome via two different pathways. Nucleic Acids Res. 2015; 43:9519–9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodnina M. Quality control of mRNA decoding on the bacterial ribosome. Adv. Protein Chem. Struct. Biol. 2012; 86:95–128. [DOI] [PubMed] [Google Scholar]
- 23. Voorhees R.M., Ramakrishnan V.. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013; 82:203–236. [DOI] [PubMed] [Google Scholar]
- 24. Tapio S., Kurland C.G.. Mutant EF-Tu increases missense error in vitro. Mol. Gen. Genet. 1986; 205:186–188. [DOI] [PubMed] [Google Scholar]
- 25. Polikanov Y., Starosta A., Juette M., Altman R.B., Terry D.S., Lu W., Burnett B., Dinos G., Reynolds K., Blanchard S.C. et al. . Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics hygromycin A and A201A. Mol. Cell. 2015; 58:832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Noel J.K., Whitford P.C.. How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome. Nat. Commun. 2016; 7:13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ieong K.W., Uzun U., Selmer M., Ehrenberg M.. Two proofreading steps amplify the accuracy of genetic code translation. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:13744–13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Knudsen C., Clark B., Degn B., Wiborg O.. One-step purification of E. coli elongation factor Tu. Biochem. Int. 1992; 28:353–362. [PubMed] [Google Scholar]
- 29. Liu W., Kavaliauskas D., Schrader J.M., Poruri K., Birkedal V., Goldman E., Jakubowski H., Mandecki W., Uhlenbeck O.C., Knudsen C.R. et al. . Labeled EF-Tus for rapid kinetic studies of pretranslocation complex formation. ACS Chem. Biol. 2014; 9:2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen C., Stevens B., Kaur J., Cabral D., Liu H., Wang Y., Zhang H., Rosenblum G., Smilansky Z., Goldman Y.E. et al. . Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol. Cell. 2011; 42:367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ha T., Ting A.Y., Liang J., Caldwell W.B., Deniz A.A., Chemla D.S., Schultz P.G., Weiss S.. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen C., Greenberg M.J., Laakso J.M., Ostap E.M., Goldman Y.E., Shuman H.. Kinetic schemes for post-synchronized single molecule dynamics. Biophys. J. 2012; 102:L23–L25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vorstenbosch E.L., Potapov A.P., de Graaf J.M., Kraal B.. The effect of mutations in EF-Tu on its affinity for tRNA as measured by two novel and independent methods of general applicability. J. Biochem. Biophys. Methods. 2000; 42:1–14. [DOI] [PubMed] [Google Scholar]
- 34. Antonsson B., Leberman R.. Stabilization of the ternary complex EF-Tu.GTP.valyl-tRNAval by ammonium salts. Biochimie. 1982; 64:1035–1040. [DOI] [PubMed] [Google Scholar]
- 35. Delaria K., Guillen M., Louie A., Jurnak F.. Stabilization of the Escherichia coli elongation factor Tu-GTP-aminoacyl-tRNA complex. Arch. Biochem. Biophys. 1991; 286:207–211. [DOI] [PubMed] [Google Scholar]
- 36. Roy R., Hohng S., Ha T.. A practical guide to single-molecule FRET. Nat Methods. 2008; 5:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen C., Stevens B., Kaur J., Smilansky Z., Cooperman B.S., Goldman Y.E.. Allosteric versus spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:16980–16985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Veigel C., Wang F., Bartoo M.L., Sellers J.R., Molloy J.E.. The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 2002; 4:59–65. [DOI] [PubMed] [Google Scholar]
- 39. Chen A.K., Cheng Z., Behlke M.A., Tsourkas A.. Assessing the sensitivity of commercially available fluorophores to the intracellular environment. Anal. Chem. 2008; 80:7437–7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kapanidis A.N., Laurence T.A., Lee N.K., Margeat E., Kong X., Weiss S.. Alternating-laser excitation of single molecules. Acc. Chem. Res. 2005; 38:523–533. [DOI] [PubMed] [Google Scholar]
- 41. Fischer N., Neumann P., Konevega A.L., Bock L.V., Ficner R., Rodnina M.V., Stark H.. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature. 2015; 520:567–570. [DOI] [PubMed] [Google Scholar]
- 42. Lai J., Ghaemi Z., Luthey-Schulten Z.. Conformational change in Elongation factor-Tu involves separation of its domains. Biochemistry. 2017; 56:5972–5979. [DOI] [PubMed] [Google Scholar]
- 43. Shima F., Ijiri Y., Muraoka S., Liao J., Ye M., Araki M., Matsumoto K., Yamamoto N., Sugimoto T., Yoshikawa Y. et al. . Structural basis for conformational dynamics of GTP-bound Ras protein. J. Biol. Chem. 2010; 285:22696–22705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsumoto K., Shima F., Muraoka S., Araki M., Hu L., Ijiri Y., Hirai R., Liao J., Yoshioka T., Kumasaka T. et al. . Critical roles of interactions among switch I-preceding residues and between switch II and its neighboring alpha-helix in conformational dynamics of the GTP-bound Ras family small GTPases. J. Biol. Chem. 2011; 286:15403–15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Knudsen C.R., Wieden H.J., Rodnina M.. The importance of structural transitions of the switch II region for the functions of elongation factor Tu on the ribosome. J. Biol. Chem. 2001; 276:22183–22190. [DOI] [PubMed] [Google Scholar]
- 46. Schrader J.M., Chapman S.J., Uhlenbeck O.C.. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. PNAS. 2011; 108:5215–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pingoud A., Block W., Wittinghofer A., Wolf H., Fischer E.. The elongation factor Tu binds aminoacyl-tRNA in the presence of GDP. J. Biol. Chem. 1982; 257:11261–11267. [PubMed] [Google Scholar]
- 48. Wohlgemuth I., Pohl C., Mittelstaet J., Konevega A., Rodnina M.. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Phil. Trans. R. Soc. B. 2011; 366:2979–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pape T., Wintermeyer W., Rodnina M.. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999; 18:3800–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gromadski K.B., Rodnina M.V.. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004; 13:191–200. [DOI] [PubMed] [Google Scholar]
- 51. Kothe U., Rodnina M.V.. Delayed release of inorganic phosphate from elongation factor Tu following GTP hydrolysis on the ribosome. Biochemistry. 2006; 45:12767–12774. [DOI] [PubMed] [Google Scholar]
- 52. Agirrezabala X., Schreiner E., Trabuco L.G., Lei J., Ortiz-Meoz R.F., Schulten K., Green R., Frank J.. Structural insights into cognate versus near-cognate discrimination during decoding. EMBO J. 2011; 30:1497–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sanbonmatsu K.Y., Joseph S., Tung C.S.. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15854–15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dale R., Eisinger J., Blumberg W.. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys. J. 1979; 26:161–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.