Abstract
Interest in reconstructing demographic histories has motivated the development of methods to estimate locus-specific pairwise coalescence times from whole-genome sequence data. Here we introduce a powerful new method, ASMC, that can estimate coalescence times using only SNP array data, and is orders of magnitude faster than previous approaches. We applied ASMC to detect recent positive selection in 113,851 phased British samples from the UK Biobank, and detected 12 genome-wide significant signals, including 6 novel loci. We also applied ASMC to sequencing data from 498 Dutch individuals to detect background selection at deeper time scales. We detected strong heritability enrichment in regions of high background selection in an analysis of 20 independent diseases and complex traits using stratified LD score regression, conditioned on a broad set of functional annotations (including other background selection annotations). These results underscore the widespread effects of background selection on the genetic architecture of complex traits.
Introduction
Recently developed methods such as the Pairwise Sequentially Markovian Coalescent (PSMC)1 utilize Hidden Markov Models (HMM) to estimate the coalescence time of two homologous chromosomes at each position in the genome1–6, leveraging previous advances in coalescent theory7–11. These methods have been broadly applied to reconstructing demographic histories of human populations12–20. More generally, methods for inferring ancestral relationships among individuals have potential applications to detecting signatures of natural selection21, genome-wide association studies22–24, and genotype calling and imputation25–28. However, all currently available methods for inferring pairwise coalescence times require whole genome sequencing (WGS) data, and can only be applied to small data sets due to their computational requirements.
Here, we introduce a new method, the Ascertained Sequentially Markovian Coalescent (ASMC), that can efficiently estimate locus-specific coalescence times for pairs of chromosomes using only ascertained SNP array data, which are widely available for hundreds of thousands of samples29. We verified ASMC’s accuracy using coalescent simulations, and determined that it is orders of magnitude faster than other methods when WGS data are available. Leveraging ASMC’s speed, we analyzed SNP array and WGS data sets with the goal of detecting signatures of recent positive selection and background selection using pairwise coalescence times along the human genome. We first analyzed 113,851 British individuals from the UK Biobank data set29, detecting 12 loci with unusually high density of very recent coalescence times as a result of recent positive selection at these sites. These include 6 known loci linked to nutrition, immune response, and pigmentation, as well as 6 novel loci involved in immunity, taste reception, and other aspects of human physiology. We then analyzed 498 unrelated WGS samples from the Genome of the Netherlands data set30 to search for signals of background selection at deeper time scales and finer genomic resolution. We determined that SNPs in regions with low values of average coalescence time are strongly enriched for heritability across 20 independent diseases and complex traits (average N=86k), even when conditioning on a broad set of functional annotations (including other background selection annotations).
Results
Overview of ASMC method
We developed a new method, ASMC, that estimates the coalescence time (which we also refer to as time to most recent common ancestor, TMRCA) for a pair of chromosomes at each site along the genome. ASMC utilizes a Hidden Markov Model (HMM), which is built using the coalescent with recombination process7–11; the hidden states of the HMM correspond to a discretized set of TMRCA intervals, the emissions of the HMM are the observed genotypes, and transitions between states correspond to changes in TMRCA along the genome due to historical recombination events. ASMC shares several key modeling components with previous coalescent-based HMM methods, such as the PSMC1, the MSMC2, and, in particular, the recently developed SMC++3. In contrast with these methods, however, ASMC’s main objective is not to reconstruct the demographic history of a set of analyzed samples. Instead, ASMC is optimized to efficiently compute coalescence times along the genome of pairs of individuals in modern data sets. To this end, the ASMC improves over current coalescent HMM approaches in two key ways. First, by modeling non-random ascertainment of genotyped variants, ASMC enables accurate processing of SNP array data, in addition to WGS data. Second, by introducing a new dynamic programming algorithm, it is orders of magnitude faster than other coalescent HMM approaches, which enables it to process large volumes of data. Details of the method are described in the Online Methods section; we have released open-source software implementing the method (see URLs).
URLs.
The ASMC software is available at http://www.palamaralab.org/software/ and https://github.com/pierpal/ASMC
UK Biobank website: http://www.ukbiobank.ac.uk/
Genome of the Netherlands website: www.nlgenome.nl
UK Biobank Genotyping and QC: http://www.ukbiobank.ac.uk/wp-content/uploads/2014/04/UKBiobank_genotyping_QC_documentation-web.pdf
Human genetic maps: https://mathgen.stats.ox.ac.uk/impute/genetic_maps_b37.tgz
The dbPSHP database of positive selection: ftp://jjwanglab.org/dbPSHP/curation/dbPSHP_20131001.tab
Python’s Scipy library: http://www.scipy.org/
1000 Genomes Project Phase 3 data: ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502
SMC++ program: https://github.com/popgenmethods/smcpp
ARGON simulator: https://github.com/pierpal/ARGON
Simupop software: http://simupop.sourceforge.net/
Selscan software: https://github.com/szpiech/selscan
SLiM simulator: https://messerlab.org/slim/
Simulations
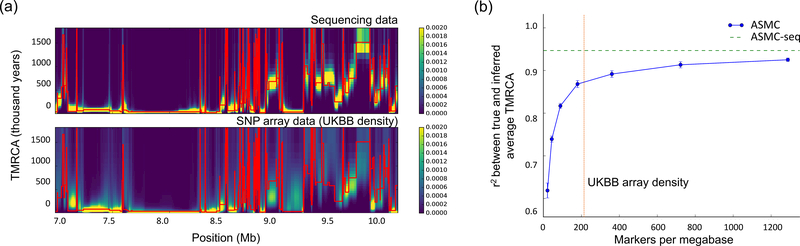
We assessed ASMC’s accuracy in inferring locus-specific pairwise TMRCA from SNP array and WGS data via coalescent simulations using the ARGON software31. Briefly, we measured the correlation between true and inferred average TMRCA for all pairs of 300 individuals simulated using a European demographic model3, for a 30 Mb region with SNP density and allele frequencies matching those of the UK Biobank data set (Figure 1; see Online Methods). As expected, ASMC achieved high accuracy when applied to WGS data (r2=0.95). When sparser SNP array data were analyzed, the correlation remained high (e.g. r2=0.87 at UK Biobank SNP array density), and increased with genotyping density. Similar relative results were obtained when comparing the root mean squared error (RMSE) between true and inferred TMRCA at each site, and the posterior mean estimate of TMRCA attained higher accuracy than the maximum-a-posteriori (MAP) estimate (Supplementary Figure 1). Inferring locus-specific TMRCA is closely related to the task of detecting genomic regions that are identical-by-descent (IBD), which we define as regions for which the true TMRCA is lower than a specified cut-off (other, related definitions have been proposed32). ASMC attained higher IBD detection accuracy (area under the precision-recall curve) than the widely used Beagle IBD detection method33 (Supplementary Table 1).
Figure 1. ASMC accuracy in coalescent simulations.
(a) Sample posterior decoding of TMRCA along a 3 Mb segment for a pair of simulated individuals with ASMC run on WGS data (top) and on SNP array data (bottom). Red lines represent the true TMRCA, while the heat map represents the inferred posterior distribution. Posterior density tends to concentrate more tightly around the true TMRCA when WGS data are analyzed, due to the higher density of polymorphic variants. Posterior estimates using SNP array data are more dispersed for distant TMRCA, but remain highly concentrated for recent TMRCA. (b) Accuracy (r2 between true and inferred average TMRCA) as a function of marker density. TMRCA are inferred using the posterior mean obtained by ASMC at each site. ASMC-seq represents the accuracy obtained using ASMC on WGS data. The red vertical line indicates marker density in the UK Biobank data set. Errors bars represent standard errors. Dots and error bars represent the average and its SE from 10 independent simulations. Numerical results are reported in Supplementary Table 11.
We evaluated the robustness of ASMC to various types of model misspecification, including an inaccurate demographic model, inaccurate recombination rate map, and violations of assumptions on allele frequencies in SNP ascertainment. To evaluate the impact of using an inaccurate demographic model, we simulated data under a European demographic history, but assumed a constant effective population size when inferring TMRCA (see Online Methods). As expected, this introduced biases, decreasing the accuracy of inferred TMRCA as measured by the RMSE, but had a negligible effect on the correlation between true and inferred TMRCA (Supplementary Table 2). An inaccurate demographic model is thus likely to result in biased TMRCA estimates, but has little effect on the relative ranking of TMRCA along the genome. Consistent with this observation, IBD detection remained accurate when an incorrect demographic model was used (Supplementary Table 3). We used a similar approach to evaluate the impact of using an inaccurate recombination rate map (see Online Methods), observing only negligible effects on the accuracy of inferred TMRCA (Supplementary Table 4). We next tested the robustness of ASMC to violations of the assumption that observed polymorphisms are ascertained solely based on their frequency, by instead ascertaining more rare variants in certain regions (mimicking genic regions; see Online Methods). We found that the distribution of inferred TMRCA in these “genic” regions did not deviate substantially from other regions (Supplementary Figure 2). Next, we evaluated the impact of varying the number s of discrete TMRCA intervals (i.e. states of the HMM); we observed that increasing s had only a minor impact on posterior mean estimates of TMRCA, although the higher resolution led to noisier MAP estimates (Supplementary Table 5). Finally, we evaluated the effects of ancestry-specific ascertainment of SNPs, mimicking an analysis where ASMC is used to infer coalescence times in individuals that have been genotyped using an array designed for a different, highly diverged population (see Online Methods). Ascertainment of SNPs in a highly diverged population leads to a depletion of informative (high frequency) markers. Consistent with previous simulations with low SNP density (Figure 1B), this leads to reduced accuracy and creates an upward bias in the inferred TMRCAs (Supplementary Figure 3). ASMC should thus be utilized with particular care in the analysis of multi-ethnic cohorts.
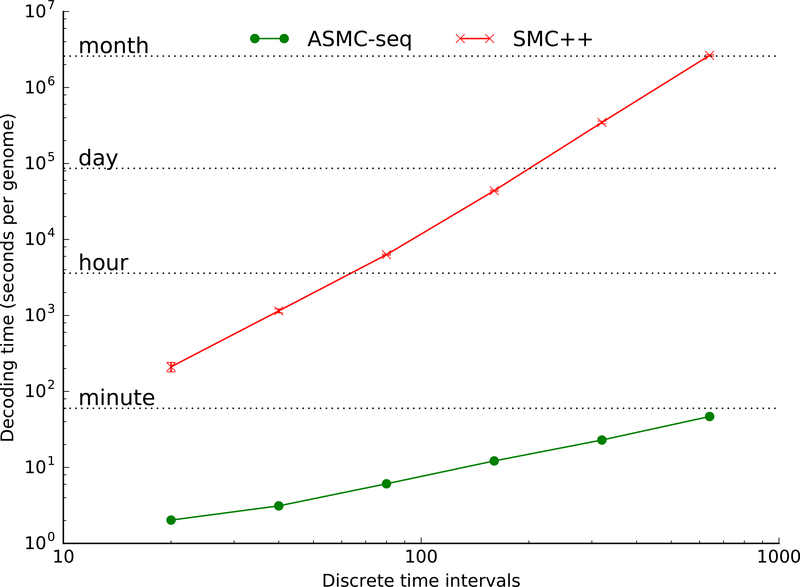
We next evaluated the running time and memory cost of ASMC. ASMC can be run on both SNP array and WGS data. When used to infer coalescence times in WGS data, ASMC is equivalent to the SMC++ method, although it runs considerably faster, making approximations that have only a small impact on accuracy (Figure 1). Letting s be the number of discrete TMRCA intervals (i.e. states of the HMM) and m be the number of observed polymorphic sites, ASMC has asymptotic running time O(sm). In comparison, the SMC++ method, which was shown to be more computationally efficient than other coalescent-based methods3, has asymptotic running time O(s3m). Accordingly, we observed that the running time of ASMC was 2 to 4 orders of magnitude faster than SMC++ when applied to simulated WGS data, depending on the number of discrete TMRCA intervals (Figure 2). For example, analysis of a pair of simulated genomes using 100 discrete time intervals required 7.4 seconds on a single processor for ASMC, compared to 3.3 hours for SMC++. This speedup in the analysis of WGS data leverages approximations that do not result in a significant loss of accuracy (Supplementary Figure 4). The memory cost of ASMC was also efficient compared to SMC++, scaling linearly with s (Supplementary Figure 5).
Figure 2. Running time of ASMC.
We report the running time required to analyze a pair of simulated haploid genomes (extrapolated from running times in 5Mb regions) as a function of the number of discrete TMRCA intervals. Both SMC++ and ASMC-seq were run on WGS data. Numerical results are reported in Supplementary Table 12.
Signals of recent positive selection in the UK Biobank
ASMC’s computational efficiency enables its application to analyses of TMRCA in large data sets. We thus used ASMC to infer locus-specific TMRCA in 113,732 unrelated individuals of British ancestry from the UK Biobank, typed at 678,956 SNPs after QC and phased using Eagle34 (see Online Methods); we note that phasing accuracy in this data set is very high, with average switch error rate on the order of 0.3% (one switch error every ∼10 cM34). We partitioned the data into batches of approximately 10,000 samples each and inferred locus-specific TMRCA for all haploid pairs within each batch, analyzing a total of 2.2 billion pairs of haploid genomes.
We sought to identify genomic regions with an unusually high density of very recent inferred TMRCA events (i.e. within the past several thousand years). Such signals are expected at sites undergoing recent positive selection, since a rapid rise in frequency of a beneficial allele causes all individuals with the beneficial allele to coalesce to a more recent common ancestor than under neutral expectation35; approaches to detect selection based on distortions in inferred coalescence times have been recently applied at different time scales21. We thus computed a statistic, DRCT, reflecting the Density of Recent Coalescence (within the past T generations), averaged within 0.05 cM windows. To compute approximate p-values, we noted that the DRCT statistic under the null is approximately Gamma-distributed. We thus obtained approximate p-values for the DRCT statistic by fitting a Gamma distribution to the null 18% of the genome obtained by conservatively excluding 500Kb windows around regions previously implicated in scans for positive selection (see Online Methods). Using coalescent simulations, we determined that DRC150 is highly sensitive in detecting signals of positive selection within the past ~20,000 years, as compared to other methods36,37 (see Online Methods, Supplementary Figure 6).
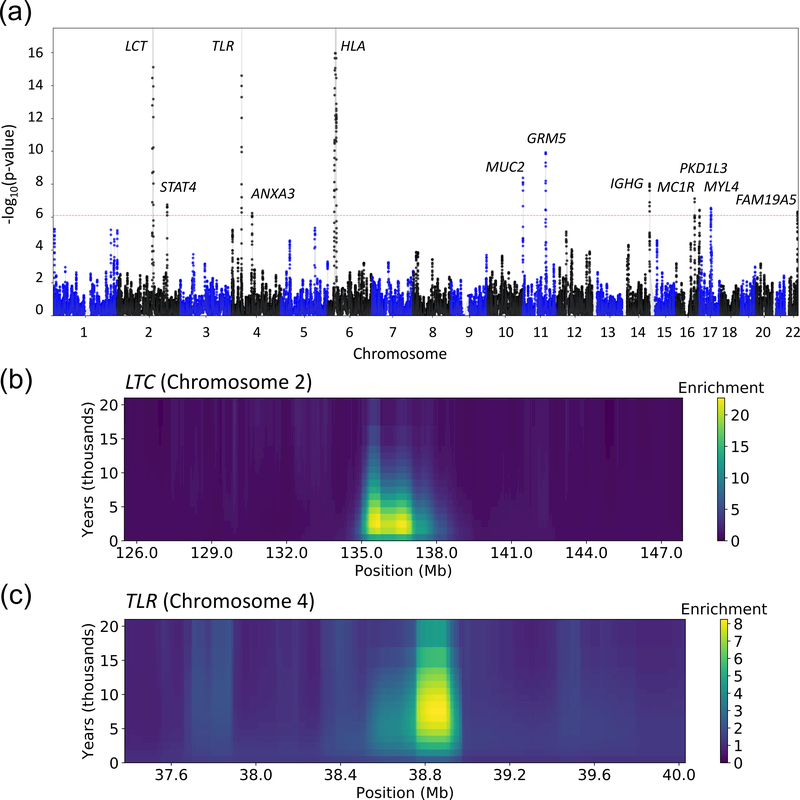
Analyzing 63,103 windows of length 0.05cM in the UK Biobank data set, we detected 12 genome-wide significant loci (p < 0.05 / 63,103 = 7.9 × 10−7; see Figure 3A and Table 1). The loci that we detected exhibited strong enrichment of recent coalescence events spanning up to the past 20,000 years (Figure 3B, 3C and Supplementary Figure 7), consistent with our simulations (Supplementary Figure 6). Of the 12 loci, 6 are loci known to be under recent positive selection, harboring genes linked to nutrition (LCT38), immune response (HLA39, TLR40, IGHG41), eye color (GRM541), and skin pigmentation (MC1R41). We also detected 6 novel loci, harboring genes involved in immune response (STAT442, associated with autoimmune disease43–45); mucus production (MUC5B46 within cluster of mucin genes, involved in protection against infectious disease43, associated with several types of cancer47 and lung disease48); taste reception (PKD1L349, associated with kidney disease50,51); cardiac and fetal muscle (MYL4, associated with atrial fibrillation52); blood coagulation (ANXA353, associated with cancer54 and immune disease55); and brain-specific expression and immune response (FAM19A556). We note that suggestive loci implicated by the DRC150 statistic (p < 10−4; Supplementary Table 6) include known targets of selection linked to eye color (HERC257,58), retinal and cochlear function (PCDH1541), celiac disease (SLC22A458,59) and skin pigmentation (SLC45A258).
Figure 3. Genome-wide scan for recent positive selection in the UK Biobank data set.
(a) Manhattan plot with candidate gene labels for 12 loci detected at genome-wide significance (adjusting for multiple testing, p < 0.05 / 63,103 = 7.9 × 10−7; dashed red line). The DRC150 statistic of recent positive selection was computed using all individuals of British ancestry from the UK Biobank (n=113,851, divided in batches of ~10,000 samples; see Online Methods for details on how p-values were computed). Numerical results for top loci are reported in Table 1; additional suggestive loci are reported in Supplementary Table 6. (b) Enrichment for recent coalescence events at the LCT locus (Chromosome 2). (c) Enrichment for recent coalescence events at the TLR locus (Chromosome 4). y-axis labels assume a 30-year generation time. Analogous plots for other top loci are provided in Supplementary Figure 7.
Table 1. Genome-wide significant signals of recent positive selection.
We report genomic locations, minimum p-value across 0.05cM windows (not adjusted for multiple testing and capped at 10−16), SNP corresponding to signal peak, and candidate gene for the 12 genome-wide significant signals of recent positive selection (adjusting for multiple testing, p < 0.05 / 63,103 = 7.9 × 10−7). Novel loci are denoted in bold font. The The DRC150 statistic of recent positive selection was computed using all individuals of British ancestry from the UK Biobank (n=113,851, divided in batches of ~10,000 samples; see Online Methods for details on how p-values were computed).
| Chromosome | From (Mb) | To (Mb) | Min. p-value | SNP | Candidate gene(s) |
|---|---|---|---|---|---|
| 2 | 134.44 | 139.01 | <10−16 | rs10206673 | LCT38 |
| 2 | 191.73 | 192.07 | 1.81×10−7 | rs7556924 | STAT4 |
| 4 | 38.44 | 38.97 | <10−16 | rs7660745 | TLR gene gamily40 |
| 4 | 79.11 | 79.51 | 5.90×10−7 | rs2867461 | ANXA3 |
| 6 | 25.18 | 33.82 | <10−16 | rs2104362 | HLA39 |
| 11 | 1.08 | 1.23 | 4.21×10−9 | rs11019228 | GRM541 |
| 11 | 88.21 | 90.55 | 1.20×10−10 | rs72636988 | MUC gene family |
| 14 | 106.35 | 107.12 | 9.49×10−9 | rs10142951 | IGHG41 |
| 16 | 70.89 | 71.80 | 7.73×10−8 | rs141399030 | PKD1L3 |
| 16 | 89.12 | 90.14 | 3.78×10−7 | rs62052682 | MC1R41 |
| 17 | 42.64 | 45.18 | 2.87×10−7 | rs75229873 | MYL4 |
| 22 | 48.98 | 49.08 | 4.94×10−7 | rs78014641 | FAM19A5 |
Heritability enrichment in regions under background selection
We next sought to detect signals of natural selection at deeper time scales. To accomplish this, we used ASMC to estimate locus-specific TMRCA for all ~0.5 million pairs of haploid genomes from unrelated individuals in the Genome of the Netherlands (GoNL) WGS data set (498 samples and 19,730,834 variants after QC; see Online Methods); we note that WGS data are required to achieve accurate resolution at deeper time scales (Figure 1A). Motivated by the fact that natural selection modulates the effective population size along the genome35,60, we set out to estimate its strength by measuring average pairwise TMRCA at each site, which is proportional to effective population size61. We refer to this annotation as ASMCavg. Forward-in-time simulations confirmed that the ASMCavg annotation captures the presence of unusual TMRCA variation due to background and positive selection, which leads to lower values of ASMCavg (see Online Methods, Supplementary Figure 8). We expect much or most of the variation in the ASMCavg annotation to be driven by deleterious effects, as supported by several recent studies60,62–66, and thus interpret ASMCavg as an annotation of background selection. We note, however, that in general ASMCavg can be affected by several types of selection that have an impact on effective population size35,60, including background, positive, and balancing selection, and that some authors suggested that positive selection plays an important role in shaping genomic diversity37,67. The genome-wide average of ASMCavg in the GoNL data was 17,399 generations (s.d. = 9,957 generations), consistent with several recent analyses of human effective population size variation1–3,19 , and with an effective size of ~10k commonly assumed in the literature68,69 (we note, however, that our analysis is limited to obtaining posterior TMRCA estimates, which are driven by the demographic model provided in input). We thus expect the ASMCavg annotation to capture background selection occurring within the past several hundred thousand years. As expected, ASMCavg was highly correlated with other measures of background selection, including nucleotide diversity (r=0.50), the McVicker B-statistic60 (r=−0.28), and allele age predicted by ARGWeaver6, quantile-normalized within 10 minor allele frequency bins70 (r=0.26, see Supplementary Table 7).
Analyses using stratified LD score regression (S-LDSC)71 have shown that regions under background selection are enriched for disease and complex trait heritability70; enrichment was observed for the nucleotide diversity, McVicker B-statistic, and ARGWeaver predicted allele age annotations, as well as three other annotations linked to LD and recombination. We evaluated the ASMCavg background selection annotation for heritability enrichment by applying S-LDSC to summary association statistics from 20 independent diseases and complex traits (Supplementary Table 8, average N=86k). We performed both an unconditioned analysis using only the ASMCavg annotation, and a joint analysis conditioned on the 75 annotations from the baselineLD model70 (which includes a broad set of functional annotations, in addition to the six annotations linked to background selection and LD), in order to specifically assess whether our annotation provides additional signal. Focusing on the ASMCavg annotation, we computed the τ* metric70, defined as the proportionate change in per-SNP heritability resulting from a 1 standard deviation increase in the value of the annotation, conditional on other annotations included in the model.
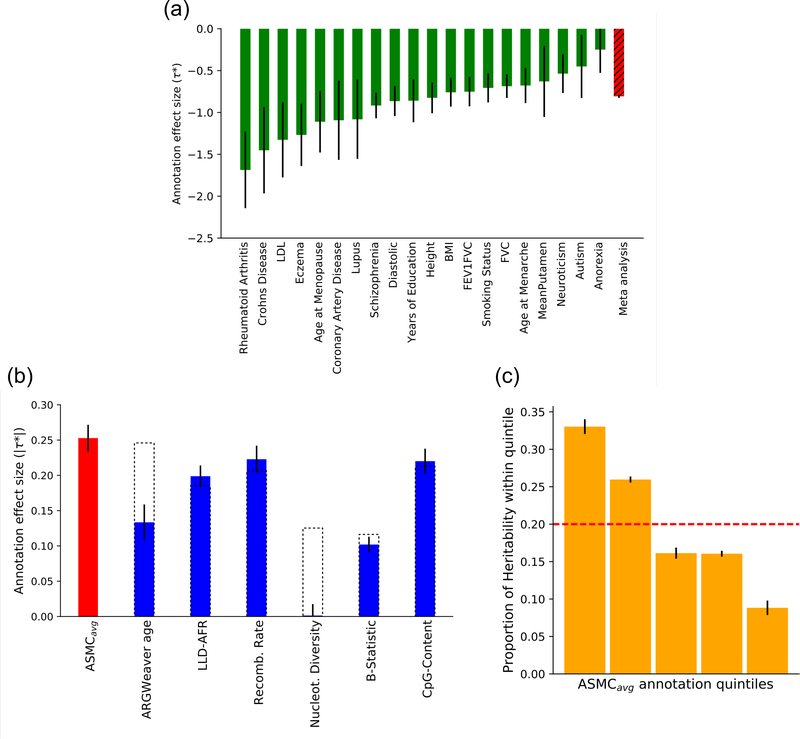
In the unconditioned analysis, lower ASMCavg was associated with higher per-SNP heritability for all 20 traits analyzed (Figure 4A), confirming that regions under background selection are enriched for disease heritability. Meta-analyzed across the 20 traits, the τ* for ASMCavg had a value of −0.81 (s.e. = 0.01; Z-test p < 10−300). After conditioning on the baselineLD model, the τ* for ASMCavg remained strongly significant at −0.25 (s.e. = 0.01; Z-test p = 7×10−153), implying that ASMCavg remains informative for disease heritability after conditioning on other annotations linked to background selection as well as a broad set of functional annotations. Furthermore, ASMCavg attained a larger value of τ* than each of the other six annotations linked to background selection (Figure 4B), implying that it was the most disease-informative background selection annotation in this analysis; we note that adding ASMCavg to the baselineLD model reduced the |τ*| of the nucleotide diversity annotation from 0.13 to 0.00 and reduced the |τ*| of the ARGWeaver6 predicted allele age annotation from 0.25 to 0.13, indicating that ASMCavg subsumes signals from these annotations. We computed the proportion of heritability explained by each quintile of the ASMCavg annotation, which provides a more intuitive interpretation of the strength of the annotation’s effect (Figure 4C). We observed that SNPs in the smallest quintile of the annotation explained 33.1% (s.e. 0.5%) of heritability, 3.8x more than SNPs in the highest quintile (8.7%, s.e. 0.5%), the largest ratio among annotations linked to background selection (Supplementary Table 9) (tied with the nucleotide diversity annotation, whose effect was however subsumed by the ASMCavg annotation; Figure 4B). Annotations constructed based on average pairwise TMRCA conditional on the allele present on each chromosome were further informative for disease heritability (Supplementary Figure 9 and Supplementary Figure 10; see Online Methods).
Figure 4. S-LDSC analysis of ASMCavg background selection annotation and disease heritability.
(a) τ* value of the ASMCavg annotation for 20 independent diseases and complex traits (sample sizes in Supplementary Table 8). Error bars represent SE of the τ* estimate. (b) Absolute values of τ* estimates (meta-analyzed across 20 independent diseases and complex traits, sample sizes in Supplementary Table 8) in joint analysis conditioned on baselineLD annotations. Error bars represent SE of the meta-analyzed τ* estimate. Dashed bars reflect values for six baselineLD annotations linked to background selection before the introduction of the ASMCavg annotation. (c) Proportion of heritability explained by SNPs within different quintiles of ASMCavg annotation (in joint analysis conditioned on baselineLD annotations). Error bars represent SE of the estimated proportions. Numerical results are reported in Supplementary Table 13.
Discussion
We have introduced a new method for inferring pairwise coalescence times, ASMC, that can be applied to either SNP array or WGS data and is highly computationally efficient. Exploiting ASMC’s speed, we analyzed ~2.2 billion pairs of haploid chromosomes from 113,851 British samples within the UK Biobank data set, and detected strong evidence of recent positive selection at 6 known loci and 6 novel loci linked to immune response and other biological functions. We further used ASMC to detect background selection at deeper time scales, estimating the average TMRCA at each position along the genome of 498 WGS phased samples from the Netherlands. Using this annotation in a stratified LD score regression analysis of 20 diseases and complex traits, we detected a strong enrichment for heritability in regions predicted to be under background selection; our annotation had the largest effect among available annotations quantifying background selection.
High-throughput inference of ancestral relationships has a number of applications beyond those related to recent positive selection and disease heritability that we have pursued in this work. Genotype calling and imputation methods25–28, for instance, infer unobserved genotypes relying on ancestral relationships, which are usually estimated using computationally efficient approximations of the coalescent model (e.g. the copying model72). Related ideas have been applied to detect phenotypic associations22–24. The processing speed achieved by the ASMC approach, on the other hand, enables making minimal simplifications to the full coalescent process, while retaining high computational scalability. In addition, accurate detection of very recent common ancestors (IBD regions) across samples is a key component of several other types of analysis, including long-range phasing34,73,74, estimation of recombination rates using haplotype boundaries75–77, haplotype-based association studies78, estimation of mutation and gene conversion rates79. In addition, ASMC’s linear-time forward-backward algorithm can be leveraged to scale up demographic inference in both WGS and SNP array data. The use of this approach in large SNP data sets, in particular, will allow to accurately infer fine-scale demographic history within the past tens of generations, improving on methods that focus on summary statistics of shared long-range haplotypes80–82, rather than directly estimating recent coalescence rates.
Although the ASMC offers new opportunities for inference of pairwise coalescence times, we note several limitations. First, the ASMC can operate either on pairs of unphased chromosomes within a single diploid individual, or on pairs of phased chromosomes across individuals. To prevent biases3, the latter application requires haplotypes phased with extremely high accuracy, which may be difficult to obtain. In this work, extremely accurate phasing was possible in the UK Biobank data set due to the very large sample size paired with the Eagle phasing algorithm34 (on the order of one switch error every ~10cM; also see ref. 72,82), and also possible in the GoNL data set due to leveraging trio information. Second, ASMC assumes a demographic model that includes a single panmictic population, and does not allow for the presence of samples from multiple ethnic backgrounds. Analyses of multi-ethnic samples will require extending the current approach so that it can accommodate demographic models involving multiple populations. Furthermore, ancestry-specific SNP ascertainment may lead to a depletion of high-frequency markers, creating an upward bias (Supplementary Figure 3). Third, ASMC is not currently applicable to imputed data. We have shown that higher genotyping density leads to higher accuracy in the inference of coalescence times. However, our preliminary tests involving the use of ASMC on imputed data where only markers with high-quality imputation accuracy are retained (e.g. imputation r2>0.99) resulted in substantial upward biases of the inferred coalescence times, which are due to spurious genotype calls. Effectively extending the ASMC to handle imputed data will thus require additional modeling of imputation accuracy. Fourth, our approach to assess the statistical significance of loci under recent positive selection is based on approximate p-value calculations. The use of approximate p-values has previously been adopted in detecting signals of positive selection37, and is more conservative than the widespread approach of simply ranking top loci36; nonetheless, the construction of an improved null model is a desirable direction of future development83. Finally, we note that although ASMC’s speed enables the analysis of large data sets, the computational costs of inferring pairwise coalescence times scale quadratically with the number of analyzed individuals. It may be possible to improve on this quadratic scaling given that at each location in the genome the ancestral relationships of a set of n samples can be efficiently represented using a tree-shaped genealogy containing n-1 nodes. The task of efficiently reconstructing a samples’ ancestral recombination graph (ARG)6,24,84, however, is substantially more complex than that of estimating pairwise TMRCA, and remains an exciting direction of future research. Despite these limitations and avenues for further improvement, we expect that ASMC will be a valuable tool for computationally efficient inference of pairwise coalescence times using SNP array or WGS data.
Online Methods
We provide an overview of the main components of the ASMC approach. An extended description can be found in the Supplementary Note.
ASMC model overview.
The ASMC is a coalescent-based HMM1-4 (see Supplementary Note for background on related methods). At each site along the genome, hidden states represent the time at which a pair of analyzed haploid individuals coalesce, which we also refer to as their time to most recent common ancestor (TMRCA). In this model, time is discretized using a set of s user-specified time intervals, each representing a possible hidden state. The TMRCA may change between adjacent sites whenever a recombination event occurs along the lineages connecting the two individuals to their MRCA. The transition probability between states is modeled using a Markovian approximation5 of the full coalescent process. Observations are obtained using genotypes of the pair of analyzed samples, as well as a set of additional samples, as detailed below, and emission probabilities reflect the chance of observing a specific genotypic configuration, conditional on the pair’s TMRCA at a site. Calculations of the initial state distribution, the transition, and the emission probabilities consider the demographic history of the analyzed sample, which is separately estimated (e.g. using other coalescent HMMs run on available WGS data for the analyzed population) and provided as input. The main goal of the ASMC is to perform high-throughput inference of posterior TMRCA probabilities along the genome for many pairs of haploid individuals genotyped using either WGS or SNP array platforms.
Emission model.
ASMC’s emission probability calculations rely on the recently developed Conditioned Sample Frequency Spectrum (CSFS)4, which is extended to handle non-randomly ascertained genotype observations (e.g. SNP array data). Consider a sample of n individuals, and define 2 of them as distinguished, (n-2) of them as undistinguished. We are interested in estimating posterior TMRCA probabilities at a set of observed sites in the genome of the 2 distinguished samples. At each site, the CSFS model4 allows computing the HMM emission probability P(d,u|τ), i.e. the probability that d ∈{0,1,2} derived alleles are carried by the distinguished pair of samples, while u ∈[0, n-2] derived alleles are observed in the (n-2) undistinguished samples, conditioned on the fact that the distinguished pair’s TMRCA (the HMM’s hidden state) is τ. Intuitively, this approach enables exploiting the relationship between an allele’s frequency and its age, which is modeled using the set of undistinguished samples and used to improve the inference of TMRCA for the distinguished pairs4. Because the set of undistinguished samples is solely used to obtain allele frequencies, their ancestral relationships need not be tracked, leading to a substantially simplified and tractable model. In the ASMC, this approach is extended to accommodate the fact that the observed sites may not be a randomly ascertained subset of polymorphic variants in the sample. To this end, we write the emission probability as P(obs|d+u)×P(d,u|τ), where the additional term P(obs|d+u) represents the probability that a site with (d+u)∈[0, n] carriers of the derived allele is observed in the ascertained data. In the ASMC, this probability is estimated using the ratio between the empirical allele frequency spectrum obtained from the analyzed data and the allele frequency spectrum that is expected under neutrality for the demographic model provided in input. Details are provided in the Supplementary Note. The emission model enables handling both major/minor and ancestral/derived genotype data encoding. We verified using coalescent simulation (see Simulations), that the number of individuals used when computing the CSFS model does not have a substantial impact on accuracy (Supplementary Table 10).
Transition model.
The transition model describes the probability of transitioning along the genome between any pair of the s possible time intervals for the TMRCA of the two analyzed samples (which we referred to as distinguished individuals in the emission model). These transition probabilities are computed using the conditional Simonsen-Churchill model (CSC)5,6. In contrast to previously proposed Markovian approximations of the coalescent process, such as the SMC7 and the SMC’8, the CSC model remains accurate even if the observed genotypes are distant from one another5. This is an important requirement in the analysis of SNP array data, as markers in this type of data are separated by substantially larger genetic distances than in the case of WGS data. Details on the calculation of transition probabilities can be found in the Supplementary Note. ASMC supports variable recombination rates along the genome through a genetic map provided in input.
Inference.
The standard HMM forward-backward algorithm to perform posterior inference has computational cost O(s2m) for analysis using s hidden states in a sequence of length m9. Current analyses making use of coalescent HMMs to infer demographic histories utilize a number of hidden states in the order of 102. When human WGS data is analyzed, the number of observed sites is in the order of 109. Thus, the computational cost of applying the standard HMM approach is very high, and a number of solutions to speed up the inference have been proposed (see Supplementary Note for an overview). Here, we devise a new approach that uses dynamic programming to reduce the computational dependence on the number s of hidden states from quadratic to linear, resulting in a gain of 2 orders of magnitude for the average analysis compared to the standard algorithm. A related procedure exists for the SMC transition model10, but cannot be applied to the more accurate and more complex CSC approach used in this work. The speed-up in the HMM forward algorithm is obtained by simplifying the key operation of computing an updated α′ vector of forward probabilities using the current forward vector, α, and the transition matrix, T, which is obtained from the CDC model. Computing the i-th entry of this vector normally requires performing the summation , which has computational cost O(s). This operation, however, can be rewritten as a linear combination of three terms, each of which can be recursively computed in time O(1), reducing the cost of computing the entire forward vector from O(s2) to O(s) (see the Supplementary Note for a detailed derivation). An equivalent speed-up can be obtained for the backward algorithm. Furthermore, to reduce the dependence of ASMC’s running time on sequence length when WGS data are analyzed, we make the following approximation. Consider two polymorphic sites separated by a stretch of n monomorphic sites. Computing an updated forward probability vector α′ using the standard approach would require performing the operation α′ = α(TE0)nTEp, where E0 is a diagonal matrix with emission probabilities for a monomorphic site in its diagonal entries and Ep is the equivalent matrix for the emission at the next polymorphic site in the sequence. For short genetic distances that are observed between polymorphic sites, the matrix T is close to diagonal, and we can thus effectively approximate this product as αTnEn0TEp (see Supplementary Figure 4). Using the previously described dynamic programming approach, this operation can be computed in time O(s), and only needs to be performed at a subset of polymorphic sites, resulting in a further speedup of 2-3 orders of magnitude compared to the standard forward/backward approach operating on all sites. This approximation is not needed when SNP array data are analyzed, as we need not integrate over large stretches of monomorphic sites, treating instead all sites between a pair of genotyped SNPs as unobserved. In addition to this, most quantities involved in the O(ms) forward/backward operations can be precomputed and stored in a cache, substantially reducing constant terms in the computation.
ASMC simulations.
We performed extensive coalescent simulations to assess the accuracy of the ASMC method. Unless otherwise specified, all simulations use the setup described in this section (standard setup). We used the ARGON simulator11 (v0.1.160415), incorporating a human recombination rate map (see URLs) and a recent demographic model for European individuals4. We simulated 300 haploid individuals and a region of 30Mb. To simulate SNP array data, we subsampled polymorphic variants to match the genotype density and allele frequency spectrum observed in the UK Biobank data set (described below). We used recombination rates from the first 30Mb of Chromosome 2, whose average rate of 1.66 cM/Mb well represents the recombination rates observed along the genome (mean 1.45 cM/Mb, s.d. 0.33 cM/Mb across autosomes). The demographic model and genetic map used to simulate the data were used when running ASMC, unless otherwise specified.
Time discretization.
We ran ASMC using different numbers of discrete time intervals, which were chosen to correspond to quantiles of the pairwise coalescence distribution induced by the demographic model. To achieve increased resolution into the recent past, some simulations utilized 160 discretization intervals chosen as follows: 40 intervals of length 10 between generations 0 and 400, 80 intervals of length 20 between generations 400 and 2,000, and 40 intervals corresponding to quantiles of the coalescence distribution, starting at generation 2,000. While using a larger number of time intervals provides increased resolution, the choice of time discretization should take into account that a larger number of time intervals typically results in noisier MAP estimates of TMRCA (see Supplementary Table 5).
Accuracy evaluation.
ASMC’s inference accuracy was evaluated using two metrics. For a given region, and for all pairs of samples in a simulated data set, we computed the squared correlation (r2) between the true and inferred sum of TMRCA at each site within the region. This metric captures the accuracy of inferred genetic kinship, but is unchanged by potential scaling factors and possible systematic biases in the TMRCA estimates. We thus also measured the root mean square error (RMSE) between true and inferred TMRCA at individuals sites, which we usually report as a percent difference compared to analysis of WGS data for improved readability. For our analysis of IBD detection accuracy, we defined as true IBD regions all sites for which pairwise TMRCA were lower than a specified time threshold (note that several definitions exist for IBD sharing among unrelated individuals12, and that IBD is also sometimes defined as the set of sufficiently long genomic regions where two chromosomes share a common ancestor uninterrupted by recombination13,14). We ran Beagle15 (v4.1) providing the true genetic map and using default parameters, and used threshold values for the output LOD score (ibdlod) to select the set of inferred IBD sites. To detect IBD using ASMC, we obtained MAP estimates of TMRCA at all sites using 160 discretization intervals (see Time discretization), and used thresholds on the inferred TMRCA values to select the set of inferred IBD sites. For both methods, we computed accuracy using the precision-recall curve. Neither Beagle nor ASMC enable obtaining recall values in the full [0,1] range, due to the presence of a lower bound for Beagle’s admissible LOD threshold values, and ASMC’s time discretization. To compare the two methods’ accuracies in each simulation, we computed the area under the precision-recall curve (auPRC) only within the range in which the accuracy of both methods could be measured, and reported the percent difference between the two methods’ auPRC (see Supplementary Figure 11 for an illustration). The PRC curve between observed points was interpolated using the method of ref 16.
Model misspecifications.
To mimic inaccuracies in the genetic map we simulated data using a human recombination map for the simulated region, but run ASMC using a map with added noise. To introduce noise, the recombination rate between each pair of contiguous markers in the map was altered by randomly adding or subtracting a fraction of its true value (see Supplementary Table 4). To test whether deviations from the assumption of frequency-based ascertainment introduce significant biases, we mimicked over-ascertainment of rare variants in genic regions of the genome. To this end, we randomly sampled ~25% of the markers from 10Kb-long genes placed every 200Kb, while the remaining variants were sampled to match the UK Biobank frequency spectrum as in standard simulations, and compared the distribution of coalescent times within over-ascertained regions and the rest of the genome (Supplementary Figure 2). To test ASMC’s robustness to an accurate demographic model we simulated data under a European demographic history, but ran ASMC assuming a constant population size of 10,000 diploid individuals (see Supplementary Table 2). To test the effects of ancestry-specific SNP ascertainment, we simulated an analysis where a group of individuals is genotyped using an array that has been designed using a different, highly diverged population. To this end, we simulated two populations that split 2,000 generations (or ~60,000 years) in the past. The two populations have identical, European-like effective population size histories after the split, and a symmetric migration rate of 0.0, 3×10−5 or 1×10−2 per generation. We simulated ancestry-specific marker ascertainment by selecting SNPs based on frequencies from only one of the two populations, matching the spectrum observed in the UK Biobank. We then inferred coalescence times in both populations independently as described in previous experiments involving a single population. Results are reported in Supplementary Figure 3.
UK Biobank (UKBB) data set.
The UK Biobank interim release data comprise 152,729 samples, from which we extracted 113,851 individuals of British ancestry (as described in ref. 17). 95 trio parents were excluded and used to assess phasing quality with the Eagle18 software, leaving a total of 113,756 samples. From these, we created 11 batches with 10,000 samples and 1 batch with the remaining 3,756 samples, which we analyzed using ASMC. Out of the original ~800k variants (for basic quality control details see URLs: UK Biobank Genotyping and QC), we analyzed a total of 678,956 SNPs that were autosomal, polymorphic in the set of analyzed samples, biallelic, with missingness ≤10%, and not included in a set of 65 variants with significantly different allele frequencies between the UK BiLEVE array and the UK Biobank array. We divided the genome in 39 autosomal regions from different chromosomes or separated by centromeres.
Detection of recent positive selection.
To detect the occurrence of recent positive selection, we computed a statistic related to the Density of Recent Coalescence events within the past T generations (DRCT statistic). The DRCT statistic was measured as follows. At a given site along the genome, we first averaged all posterior TMRCA estimates obtained from all analyzed pairs of samples and renormalized these averages to obtain an average pairwise coalescence distribution at the site. The DRCT statistic was then obtained by integrating this distribution between generations 0 and T. The statistic was measured in windows of 0.05 cM, reporting an average of all SNPs within each window. We tested the sensitivity of the DRCT statistic in detecting recent positive selection using extensive simulation. Details for these simulations can be found in the Supplementary Note.
Null model calibration.
Given n samples from a population of recent effective size N, the DRCT statistic is approximately Gamma-distributed under the null for sufficiently small values of T and n⟨⟨N. The rationale of this approximation is that for n⟨⟨N, a small number of coalescence events will have occurred within the short time span of T generations. In this regime, the coalescence time of each pair of lineages may be modeled as independent and exponentially distributed, which allows approximating the total number of early coalescence events as a Gamma-distributed random variable. Similar approximations have been recently used elsewhere19,20. We thus computed approximate p-values for our selection scan in the UKBB data set using the following approach. We first extracted a subset of “neutral” genomic regions, spanning a total of 18% of the genome, and defined as any genotyped site at a distance greater than 500Kb from regions contained in a recent database of positive selection21 (see URLs: database of positive selection). We then built an empirical null model by fitting a Gamma distribution (using Python’s Scipy library, see URLs) to these putatively neutral regions, and used this model to obtain approximate p-values throughout the genome. We analyzed 63,103 windows, using a Bonferroni significance threshold of 0.05 / 63,103 = 7.9 x 10-7. One of the genome-wide significant signals that we detected (PKD1L3 locus, chr16:70.89-71.80Mb) fell within the putatively neutral portion of the genome. We thus iterated this procedure, excluding this locus from the set of putatively neutral loci.
Genome of the Netherlands (GoNL) data set.
The data set consists of 748 individuals who passed quality control and were sequenced at an average of ~13x (quality control details for the Release 4 data are described elsewhere22). We analyzed 19,730,834 sequenced variants for 498 trio-phased unrelated parents, excluding centromeres and dividing the genome in the same 39 autosomal regions used for analysis of the UKBB data set.
ASMCavg annotation.
We set out to estimate the strength of background selection by measuring variation in local effective population size along the genome23. We used ASMC to estimate the posterior mean TMRCA at all sites and for all pairs of haploid individuals in the GoNL data set. We averaged these estimates at each site to obtain an annotation of background selection, which we refer to as ASMCavg. We similarly computed other annotations, conditioning on whether either or both individuals at a site carried a mutated allele. The ASMChet annotation (Supplementary Figure 9), was obtained by averaging at each site the posterior mean TMRCA estimates for all pairs of individuals that were found to be heterozygous at each site. Other annotations were similarly computed using only pairs carrying e.g. minor/major alleles at each site (see Supplementary Figure 10). We verified that the ASMChet annotation captures the effects of natural selection using forward simulation. Details for these simulations can be found in the Supplementary Note.
Stratified LD Score (S-LDSC) analysis
We investigated whether large values of our annotations related to background selection corresponded to an enrichment in heritability for 20 complex traits and diseases listed in Supplementary Table 8. The S-LDSC analysis was run on data sets containing European individuals using standard guidelines24. The sets of LD-score, regression, and heritability SNPs were defined as follows. LD score SNPs were set to be 9,997,231 biallelic SNPs with at least 5 minor alleles observed in 489 European samples from the 1000 Genomes Phase 3 data set25 (see URLs); regression SNPs were set as 1,217,312 HapMap Project Phase 3 SNPs; and Heritability SNPs, used to compute trait heritability, were chosen as the 5,961,159 reference SNPs with MAF ≥ 0.05. The MHC region (2Mb 25-34 on Chromosome 6) and SNPs with χ2>80 or 0.0001N were excluded from the analysis. Annotations contained in the baselineLD model, which we included in our joint analyses, can be found in Supplementary Table S9 of ref. 26. To avoid minor allele frequency (MAF)-mediated effects, all ASMC-related annotations used in the S-LDSC analysis were quantile-normalized with respect to MAF of regression SNPs. Specifically, we used 10 MAF ranges specified in the baselineLD model, corresponding to 10 frequency quantiles for the regression SNPs. For each range, we ranked values of an annotation for the corresponding SNPs, and mapped them to quantiles of a Standard Normal distribution. Annotation effects, τ*, were obtained from the output of S-LDSC, as described in ref. 26. Independent traits were selected on the basis of low genetic correlation, as previously described24. Meta-analysis of τ* values across independent traits was performed computing a weighted average of individual estimates of τ*, weighted using 1/(hi2εi2), where hi2 represents heritability for the i-th trait, and εi represents the standard error of the trait’s τ* estimate.
Data and code availability
The ASMC program and source code, as well as genomic annotations of positive and background selection can be downloaded at http://www.palamaralab.org/software/ and https://github.com/pierpal/ASMC.
Supplementary Material
Acknowledgements
We thank Po-Ru Loh for suggesting several coding improvements for the ASMC software, and for support with the phasing and processing of the UK Biobank data; Steven Gazal for support with the S-LDSC analysis and the baselineLD model; Ilya Shlyakhter for support with the COSI2 simulator; Yair Field for support with the simulation setup in the analysis of recent positive selection; David Reich for providing computational resources; Hilary Finucane, Yakir Reshef, and Sasha Gusev for helpful discussions. This research was conducted using publicly available data sets (see URLs): the UK Biobank Resource under Application #16549, and the Genome of the Netherlands resource under Application #2017149. We thank the participants of the UK Biobank and the Genome of the Netherlands projects. P.P. and A.L.P. were supported by NIH grants R01 MH101244, R01 HG006399 and R01 GM105857; J.T. and Y.S.S. were supported in part by an NIH grant R01 GM094402, and a Packard Fellowship for Science and Engineering; Y.S.S. is a Chan Zuckerberg Biohub investigator.
Footnotes
Competing interests
The authors declare no competing financial interests.
References
- 1.Li H & Durbin R Inference of human population history from individual whole-genome sequences. Nature 475, 493–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiffels S & Durbin R Inferring human population size and separation history from multiple genome sequences. Nat Genet 46, 919–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terhorst J, Kamm JA & Song YS Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet 49, 303–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hobolth A, Christensen OF, Mailund T & Schierup MH Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet 3, e7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheehan S, Harris K & Song YS Estimating variable effective population sizes from multiple genomes: a sequentially markov conditional sampling distribution approach. Genetics 194, 647–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen MD, Hubisz MJ, Gronau I & Siepel A Genome-wide inference of ancestral recombination graphs. PLoS Genet 10, e1004342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudson RR & Kaplan NL The coalescent process in models with selection and recombination. Genetics 120, 831–40 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiuf C & Hein J Recombination as a point process along sequences. Theor Popul Biol 55, 248–59 (1999). [DOI] [PubMed] [Google Scholar]
- 9.McVean GA & Cardin NJ Approximating the coalescent with recombination. Philos Trans R Soc Lond B Biol Sci 360, 1387–93 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marjoram P & Wall JD Fast “coalescent” simulation. BMC Genet 7, 16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobolth A & Jensen JL Markovian approximation to the finite loci coalescent with recombination along multiple sequences. Theor Popul Biol 98, 48–58 (2014). [DOI] [PubMed] [Google Scholar]
- 12.1000 Genomes Project, C. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skoglund P et al. Genetic evidence for two founding populations of the Americas. Nature 525, 104–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghavan M et al. POPULATION GENETICS. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science 349, aab3884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green RE et al. A draft sequence of the Neandertal genome. Science 328, 710–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prufer K et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sankararaman S et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vernot B & Akey JM Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–21 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Tennessen JA et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart JR & Stringer CB Human evolution out of Africa: the role of refugia and climate change. Science 335, 1317–21 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Hunter-Zinck H & Clark AG Aberrant Time to Most Recent Common Ancestor as a Signature of Natural Selection. Mol Biol Evol 32, 2784–97 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris AP, Whittaker JC & Balding DJ Fine-scale mapping of disease loci via shattered coalescent modeling of genealogies. Am J Hum Genet 70, 686–707 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zollner S & Pritchard JK Coalescent-based association mapping and fine mapping of complex trait loci. Genetics 169, 1071–92 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minichiello MJ & Durbin R Mapping trait loci by use of inferred ancestral recombination graphs. Am J Hum Genet 79, 910–22 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howie BN, Donnelly P & Marchini J A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchsberger C, Abecasis GR & Hinds DA minimac2: faster genotype imputation. Bioinformatics 31, 782–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchini J, Howie B, Myers S, McVean G & Donnelly P A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39, 906–13 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Le SQ & Durbin R SNP detection and genotyping from low-coverage sequencing data on multiple diploid samples. Genome Res 21, 952–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudlow C et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genome of the Netherlands, C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 46, 818–25 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Palamara PF ARGON: fast, whole-genome simulation of the discrete time Wright-fisher process. Bioinformatics 32, 3032–4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakeley J & Wilton P Coalescent and models of identity by descent in Encyclopedia of Evolutionary Biology Vol. 1 (ed. Kliman RM) 287–292 (Oxford Academic Press, 2016). [Google Scholar]
- 33.Browning BL & Browning SR Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loh PR, Palamara PF & Price AL Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet 48, 811–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bamshad M & Wooding SP Signatures of natural selection in the human genome. Nat Rev Genet 4, 99–111 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Voight BF, Kudaravalli S, Wen X & Pritchard JK A map of recent positive selection in the human genome. PLoS Biol 4, e72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Field Y et al. Detection of human adaptation during the past 2000 years. Science 354, 760–764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bersaglieri T et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet 74, 1111–20 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barreiro LB & Quintana-Murci L From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet 11, 17–30 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Wellcome Trust Case Control, C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabeti PC et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thierfelder WE et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature 382, 171–4 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Liang YL et al. Association of STAT4 rs7574865 polymorphism with autoimmune diseases: a meta-analysis. Mol Biol Rep 39, 8873–82 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi S et al. Association of STAT4 with susceptibility to rheumatoid arthritis and systemic lupus erythematosus in the Japanese population. Arthritis Rheum 58, 1940–6 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Korman BD, Kastner DL, Gregersen PK & Remmers EF STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep 8, 398–403 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gendler SJ & Spicer AP Epithelial mucin genes. Annu Rev Physiol 57, 607–34 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Kufe DW Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer 9, 874–85 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seibold MA et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med 364, 1503–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishimaru Y et al. Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci U S A 103, 12569–74 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li A, Tian X, Sung SW & Somlo S Identification of two novel polycystic kidney disease-1-like genes in human and mouse genomes. Genomics 81, 596–608 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Ishimaru Y et al. Interaction between PKD1L3 and PKD2L1 through their transmembrane domains is required for localization of PKD2L1 at taste pores in taste cells of circumvallate and foliate papillae. FASEB J 24, 4058–67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gudbjartsson DF et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet 47, 435–44 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Raynal P & Pollard HB Annexins: the problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim Biophys Acta 1197, 63–93 (1994). [DOI] [PubMed] [Google Scholar]
- 54.Wu N, Liu S, Guo C, Hou Z & Sun MZ The role of annexin A3 playing in cancers. Clin Transl Oncol 15, 106–10 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Okada Y et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet 44, 511–6 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Tom Tang Y et al. TAFA: a novel secreted family with conserved cysteine residues and restricted expression in the brain. Genomics 83, 727–34 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Sturm RA et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet 82, 424–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathieson I et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huff CD et al. Crohn’s disease and genetic hitchhiking at IBD5. Mol Biol Evol 29, 101–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McVicker G, Gordon D, Davis C & Green P Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet 5, e1000471 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wakeley J Coalescent theory : an introduction, xii, 326 p. (Roberts & Co. Publishers, Greenwood Village, Colo, 2009). [Google Scholar]
- 62.Hernandez RD et al. Classic selective sweeps were rare in recent human evolution. Science 331, 920–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charlesworth B Background selection 20 years on: the Wilhelmine E. Key 2012 invitational lecture. J Hered 104, 161–71 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Comeron JM Background selection as null hypothesis in population genomics: insights and challenges from Drosophila studies. Philos Trans R Soc Lond B Biol Sci 372(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gazal S et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nature genetics (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torres R, Szpiech ZA & Hernandez RD Human demographic history has amplified the effects background selection across the genome. bioRxiv, 181859 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Enard D, Messer PW & Petrov DA Genome-wide signals of positive selection in human evolution. Genome Res 24, 885–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serre D et al. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol 2, E57 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pritchard JK, Pickrell JK & Coop G The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol 20, R208–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gazal S et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat Genet (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li N & Stephens M Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–33 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loh PR et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kong A et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet 40, 1068–75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kong A et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467, 1099–103 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Hinch AG et al. The landscape of recombination in African Americans. Nature 476, 170–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wegmann D et al. Recombination rates in admixed individuals identified by ancestry-based inference. Nat Genet 43, 847–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gusev A et al. DASH: a method for identical-by-descent haplotype mapping uncovers association with recent variation. Am J Hum Genet 88, 706–717 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palamara PF et al. Leveraging Distant Relatedness to Quantify Human Mutation and Gene-Conversion Rates. Am J Hum Genet 97, 775–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palamara PF, Lencz T, Darvasi A & Pe’er I Length distributions of identity by descent reveal fine-scale demographic history. Am J Hum Genet 91, 809–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ralph P & Coop G The geography of recent genetic ancestry across Europe. PLoS Biol 11, e1001555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Browning SR & Browning BL Accurate Non-parametric Estimation of Recent Effective Population Size from Segments of Identity by Descent. Am J Hum Genet 97, 404–18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nei M, Suzuki Y & Nozawa M The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet 11, 265–89 (2010). [DOI] [PubMed] [Google Scholar]
- 84.Griffiths RC & Marjoram P An ancestral recombination graph. Institute for Mathematics and its Applications 87, 257 (1997). [Google Scholar]
References for Online Methods
- 1.Li H & Durbin R Inference of human population history from individual whole-genome sequences. Nature 475, 493–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheehan S, Harris K & Song YS Estimating variable effective population sizes from multiple genomes: a sequentially markov conditional sampling distribution approach. Genetics 194, 647–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiffels S & Durbin R Inferring human population size and separation history from multiple genome sequences. Nat Genet 46, 919–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terhorst J, Kamm JA & Song YS Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet 49, 303–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hobolth A & Jensen JL Markovian approximation to the finite loci coalescent with recombination along multiple sequences. Theor Popul Biol 98, 48–58 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Simonsen KL & Churchill GA A Markov Chain Model of Coalescence with Recombination. Theor Popul Biol 52, 43–59 (1997). [DOI] [PubMed] [Google Scholar]
- 7.McVean GA & Cardin NJ Approximating the coalescent with recombination. Philos Trans R Soc Lond B Biol Sci 360, 1387–93 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marjoram P & Wall JD Fast "coalescent" simulation. BMC Genet 7, 16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabiner LR & Juang B-H An introduction to hidden Markov models. ASSP Magazine, IEEE 3, 4-16 %@ 0740-7467 (1986). [Google Scholar]
- 10.Harris K, Sheehan S, Kamm JA & Song YS Decoding coalescent hidden Markov models in linear time. Res Comput Mol Biol 8394, 100–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palamara PF ARGON: fast, whole-genome simulation of the discrete time Wright-fisher process. Bioinformatics 32, 3032–4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakeley J & Wilton P Coalescent and models of identity by descent. in Encyclopedia of Evolutionary Biology Vol. 1 (ed. Kliman RM) 287–292 (Oxford Academic Press, 2016). [Google Scholar]
- 13.Palamara PF, Lencz T, Darvasi A & Pe'er I Length distributions of identity by descent reveal fine-scale demographic history. Am J Hum Genet 91, 809–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browning BL & Browning SR Detecting identity by descent and estimating genotype error rates in sequence data. Am J Hum Genet 93, 840–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Browning BL & Browning SR Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis J & Goadrich M The relationship between Precision-Recall and ROC curves. 233–240 %@ 1595933832 (ACM, 2006).
- 17.Galinsky KJ, Loh PR, Mallick S, Patterson NJ & Price AL Population Structure of UK Biobank and Ancient Eurasians Reveals Adaptation at Genes Influencing Blood Pressure. Am J Hum Genet 99, 1130–1139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh PR, Palamara PF & Price AL Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet 48, 811–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathieson I & McVean G Demography and the age of rare variants. PLoS Genet 10, e1004528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field Y et al. Detection of human adaptation during the past 2000 years. Science 354, 760–764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li MJ et al. dbPSHP: a database of recent positive selection across human populations. Nucleic Acids Res 42, D910–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genome of the Netherlands, C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 46, 818–25 (2014). [DOI] [PubMed] [Google Scholar]
- 23.McVicker G, Gordon D, Davis C & Green P Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet 5, e1000471 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.1000 Genomes Project, C. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gazal S et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat Genet (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ASMC program and source code, as well as genomic annotations of positive and background selection can be downloaded at http://www.palamaralab.org/software/ and https://github.com/pierpal/ASMC.