Abstract
Progressive epilepsy with mental retardation (EPMR) is an autosomal recessive disorder discovered recently from an isolated region in Finland. The disorder is characterized by normal early development, generalized tonic-clonic seizures with onset at 5–10 years of age, and progressive mental retardation beginning 2–5 years after the onset of seizures. We recently mapped the EPMR locus to a 7-cM region on chromosome 8p between markers AFM185xb2 and D8S262. A recombination detected with a new microsatellite marker AFMa0S4td9 narrows the region to 4 cM. A yeast artificial chromosome (YAC) contig containing 22 YACs was constructed across the disease gene region. The YAC contig is characterized by a collection of 19 YAC-end sequence-tag sites together with seven microsatellite markers. The entire YAC contig spans a minimum of 3 Mb. Moreover, the distal end of the contig contains a subtelomeric YAC yRM2205 that anchors the contig to the telomere. Construction of a YAC contig across the disease gene region is an essential step toward the isolation of the EPMR gene.
Epilepsy is a common symptom that occurs in ~2%−3% of the population either as an isolated phenomenon or as a cosymptom of a large and diverse set of disease phenotypes (Scheuer and Pedley 1990; Hauser et al. 1993). The most common forms of inherited epilepsies do not display straightforward Mendelian inheritance patterns and are considered to be multifactorial (Gardiner 1990). Among the multifactorial forms, only the locus for juvenile myoclonic epilepsy has been genetically mapped; a subset of these families appear to be linked to DNA markers on chromo-some 6p (Greenberg et al. 1988; Durner et al. 1991; Weissbecker et al. 1991; Delgado-Escueta et al. 1994). In contrast, monogenic inherited epilepsies are rare, but already six loci have been genetically mapped to specific chromosomal re gions: two loci for benign familial neonatal convulsions (BFNC) on chromosomes 20q (EBN1; Leppert et al. 1989) and 8q (EBN2; Lewis et al. 1993); the EPM1 locus for progressive myoclonus epilepsy of Unverricht-Lundborg type on chromosome 21q (Lehesjoki et al. 1991); an EPMR locus for progressive epilepsy with mental retardation locus on 8p (Tahvanainen et al. 1994); a partial epilepsy locus on 10q (Ottman et al. 1995); and an autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) locus on 20q13.2 (Phillips et al. 1995). Recently, a missense mutation in the α4 subunit of the neuronal nicotinic acetylcholine receptor (CHRNA4) gene on chromo-some 20q was reported to be associated with ADNFLE in a large Australian pedigree (Steinlein et al. 1995). It now appears that a putative mutation in the CHRNA4 gene detected in one BFNC family (Beck et al. 1994) may have been an artifact resulting from a DNA sequencing error (Steinlein et al. 1995).
Progressive epilepsy with mental retardation (EPMR), also called Northern epilepsy, has, so far, been found only in a small rural region in northern Finland. To date, 25 patients have been identified. Genealogical records show that, with one exception, all parents of affected children are distant relatives. Children descending from the unrelated parent have a haplotype distinct from the “ common “ haplotype shared by 15 of 19 disease chromosomes. This suggests two different mutations segregating in this Finnish pedigree and may indicate that the disorder is not necessarily as rare as originally anticipated.
The main clinical features of EPMR are normal early development, childhood-onset epilepsy, and mental retardation that starts to develop 2–5 years after the first seizures. All patients have generalized tonic-clonic seizures. One-third of the patients also have complex partial seizures during childhood. Epileptic activity decreases during young adulthood, but complete remission does not occur. Mental retardation is most rapid prior to adulthood, during the period with most frequent seizures. Of the antiepileptic drugs used, only clonazepam has proven effective; it reduces dramatically the number of seizures and can even normalize the EEG pattern when administered early in the disease process (Hirvasniemi et al. 1994, 1995).
To facilitate the isolation and identification of the EPMR gene, we sought to physically map and clone the ~7-cM region encompassing the disease locus. Several microsatellite markers that always segregate with the disease locus were chosen along with markers flanking the EPMR region to screen physical mapping data bases and yeast artificial chromosome (YAC) libraries. A total of 22 YAC clones were identified that span the region between the flanking markers and that are anchored to genomic DNA by seven polymorphic DNA markers. A new microsatellite marker (AFMa054td9), recently positioned on the genetic map by scientists at Genethon, maps to the YAC contig and detects a meiotic recombination in one EPMR family, thereby reducing the minimal genetic region to ~4 cM.
RESULTS
Genetic Linkage Analysis
The EPMR locus has been mapped previously to an approximate 7-cM region defined by the flanking telomeric and centromeric markers, AFM185xb2 and D8S262, respectively (Tah vanainen et al. 1994). We have genotyped a recently discovered microsatellite marker, AFMa054td9, in the EPMR extended pedigree and show that this marker recombines with the disease locus in a single healthy individual who has three affected siblings (Fig. 1). This recombination narrows the EPMR disease gene region to about 4 cM. Because markers D8S504 and D8S264 are uninformative in this branch of the family, the recombination may occur telomeric to AFMaO54td9, leaving the possibility that additional markers will help to narrow the region still further. The maximum two-point logarithm of odds (lod) score for AFMaO54td9 is 5.28 (θ = 0.03) in the extended pedigree. The results of the multipoint linkage analysis are shown in Figure 2. The maximum lod score is 10.70 at a position 0.4 cM distal to D8S264 and 0.6 cM proximal to D8S504. The one-unit lod support interval extends from a point 0.4 cM distal to AFMa054td9 to a point 0.2 cM proximal to AFM185xb2 and covers a total genetic distance of 3.4 cM.
Figure 1.
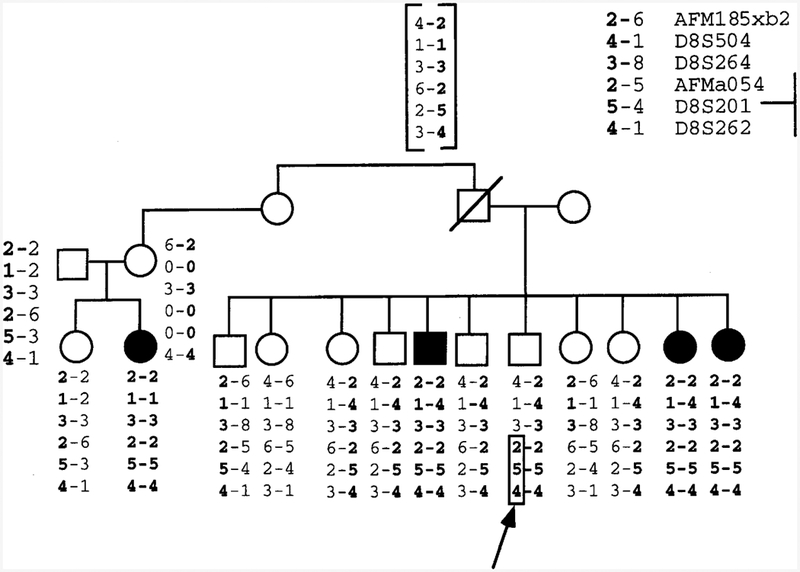
Refinement of the EPMR locus by meiotic breakpoint mapping. DNA marker genotypes across the EPMR locus in an affected family reveal a key recombination event that reduces the EPMR critical region to 4.0 cM. The recombination occurs in the paternal chromosome of a healthy sibling and is marked by an arrow. “Disease homologs” are in boldface type. The boxed region marks the site of chromosomal recombination. (0) Untyped alleles. The inferred hap-Iotype of the deceased father is shown in brackets.
Figure 2.
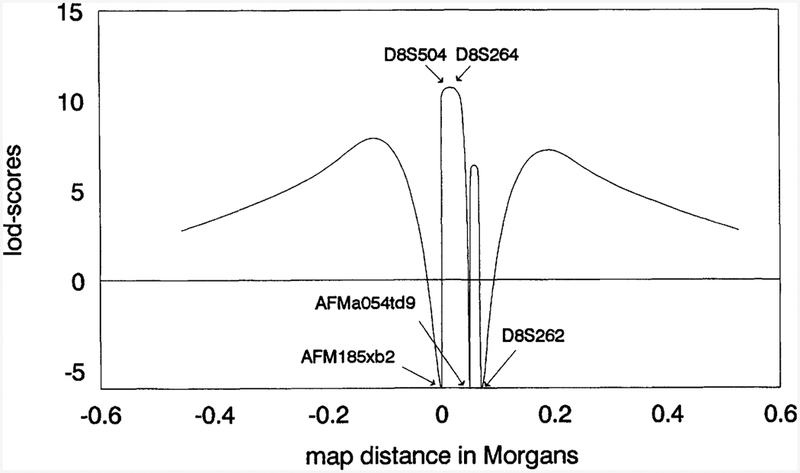
Six-point analysis in EPMR pedigrees assuming a fixed order of five marker loci on chromosome 8 as described in Methods (Linkage). The telomere is to the left.
It has been hypothesized that cathepsin B may play a role in Alzheimer’s disease (Tagawa et al. 1991). Because this gene has been mapped physically to 8p22-p23.1 (Fong et al. 1992), we considered it a candidate gene for EPMR. To exclude cathepsin B as a potential EPMR candidate gene, we analyzed a restriction fragment length polymorphism (RFLP) detected by a cathepsin B intron probe in one of the Finnish families. Several enzymes (EcoRI, PvuII, RsaI, StuI, and TaqI) detected a novel polymorphism in intron 8 (see Methods for details). The TaqI polymorphism revealed recombinations between the disease locus and cathepsin B allowing us to exclude this gene as a candidate for EPMR (data not shown).
Physical Mapping of the EPNR Locus
Library Screening
Fifteen of the YAC clones shown in Figure 2 were ascertained from computerized data bases accessed through the World-Wide Web (see Methods). These are clones characterized as “positive” for markers in the EPMR region or clones that appear to overlap with such clones. To fill a gap between markers D8S264 and D8S504, we screened the Centre d’Etude du Polymorphisme Humain (CEPH) Midi YAC library with both YAC ends from 858E12 (Fig. 3) but found no positive clones. We next screened with D8S264 and the right YAC-end STS from 241d10 and obtained three new YAC clones in both screenings. Probe D8S201 was screened at CEPH against the Mega YAC library (Albertsen et al. 1990; Dausset et al. 1992). Several positive clones were identified (see next sections).
Figure 3.
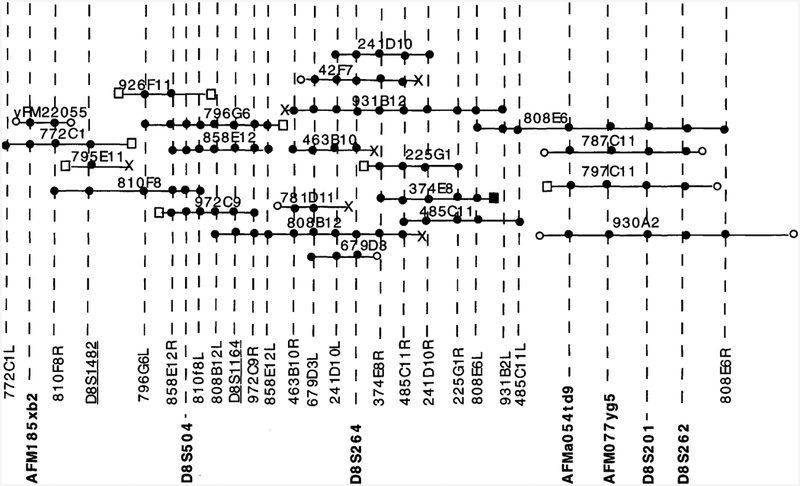
YAC contig of the EPMR region. The contig spans the region between the telomeric marker AFM185xb2 and the centromeric locus D8S262. Microsatellite markers are shown in boldface type. The STSs developed in this study are shown in plain type. STSs derived from computerized data bases (see Methods) are underlined. (●) An STS within a YAC; (□) chimeric YAC ends, with the exception of one (■) that maps to chromosome 8 by PCR analysis but that maps outside the EPMR region. (X) Repetitive YAC ends. (○) Sequence information is not available for these YAC ends. The order of microsatellite markers is determined by genetic and physical mapping.
Characterization of YAC-end Fragments
Ligation-mediated PCR yielded YAC-end DNA sequences in 32 of 40 cases where it was attempted. Because some of these sequences were Alu repeat or nonchromosome 8 sequences, characterization of these YAC ends yielded a total of 17 chromosome 8 STSs. We were not able to generate a PCR product for the YAC ends of clone 810F8 with the ligation-mediated PCR but were successful with the vectorette PCR method (see Methods).
Construction and Characterization of the YAC Contig
Three of the critical microsatellite markers (D8S504, D8S264, and D8S262) from the EPMR region were queried in the CEPH data base for homology to known YAC clones. Preliminary characterization of 12 positive addresses yielded a total of seven YAC clones (796G6, 810F8, 858E12, 931B2, 679D3, 787Cll , and 930A2) that, as shown here, unambiguously map to the disease gene region and contribute to the genomic coverage of the region. Marker D8S201 identified several clones from the Mega YAC library, four of which (787Cll, 797Cll, 808E6, and 930A2) were also positive for DNA markers D8S262, AFMO77yg5, and AFMaO54td9. These four markers span an estimated 3 cM. One of the YAC clones identified by this screening (808E6) overlaps with a D8S264 positive clone (931b12), thus closing the gap between markers D8S264 and the centromeric YACs. Then, YAC-end STSs for three YAC clones (931B2, 858E12, and 796G6) were used to PCR amplify YAC clones obtained from the CEPH data base. Alu-PCR fingerprinting by scientists at CEPH indicated that these clones were members of the same contig with YAC clones known to be positive for markers D8S264 and D8S504. Two YAC clones (972C9 and 926Fll) could be unambiguously mapped to the contig by this procedure and one more, 781Dll, was later shown positive for YAC-end STSs 463B10R and 679D3L. To fill the gap between markers D8S264 and D8S504, we screened the CEPH Midi YAC library (Albertsen et al. 1990; Green and Olson 1990) with DNA marker D8S264 and identified YAC clones 463B10, 241D10, and 42F7. We then screened the library with YAC-end 241D 10R and identified clones 225G1, 374E8, and 485Cll. None of these clones extended to D8S504 and thus did not fill the gap between D8S264 and D8S504. Then, we queried the data base for contigs using YAC 858E12 that contains locus D8S504 and found two, WC-148 and WC-1335 (Center for Genome Research at Whitehead Institute for Biomedical Research, Cambridge, MA). From these contigs we identified three additional overlapping clones, 808B12, 795Ell, and 772C1.
Three unique YAC-end clones (42F7L, 463B10L, and 808B12R) identify similar sets of overlapping YAC clones. Two lines of evidence indicate that these end clones encode low-copy repeat DNA sequences (indicated by “X” in Fig. 3). First, over half of all the CEPH Midi YAC library pools were positive when PCR-amplified with primers specific for 463B10L and 808B12R. Second, Southern blot analysis of TaqI-digested YACs from the contig revealed multiple homologous restriction fragments for the radiolabeled PCR products of 42F7L and 808B12R. Also, hybridization of digested total genomic DNA with radiolabeled PCR product of 42F7L resulted in a smear instead of discrete bands. Unlike YAC clones 42F7 and 808B12, clone 463B10 does not contain the end clones 485CllR and 374E8R, suggesting that it does not extend as far in the centromeric direction as the overlapping clones.
The EcoRI cloning site of the YAC yRM2205 maps within 350–450 kb of the 8p terminus by RecA-assisted restriction endonuclease cleavage mapping (H. Riethman, pers. comm.). The placement of our most distal marker, AFM185xb2, in this YAC clone thereby positions the contig at the very distal region of the short arm of chromosome 8. Somewhat surprisingly, YAC-end STS 772clL appears to map telomeric to yRM2205 (Fig. 3). 772clL does map to chromosome 8 because it amplifies chromosome 8-only cell hybrid DNA, but it does not amplify other YACs in the contig. It seems most likely, therefore, that this STS maps either telomeric to YAC yRM22055, or it maps to a deleted portion of the subtelomeric YAC clone. In Figure 3 we have based the degree of overlap between YAC clones strictly on the number of STSs shared between YACs, and thus the size of the overlaps are not representative of the actual size of the homologous regions. We estimate that the minimum distance between loci AFM185xb2 and AFMa054td9 is the sum of the sizes of the nonover-lapping, nonchimeric YACs 810F8 and 931B2, which is 2.1 Mb. The minimum distance spanned by the entire contig is 3 Mb. Genetic mapping indicates that the relative order of Genethon markers (telomeric to centromeric) is AFMaO54td9, AFMO77yg5, and D8S262. Because D8S201 cosegregates with these three markers in the EPMR pedigree, we are unable to position it within the cluster.
DISCUSSION
The epilepsies are characterized by recurrent convulsive and nonconvulsive seizures resulting from partial or generalized epileptogenic discharges in the cerebrum (Delgado-Escueta et al. 1983). Clinically and etiologically, the epilepsies are a heterogeneous group of diseases. Etiological causes range from predominantly environmental as in the case of posttraumatic epilepsies to predominantly genetic as exemplified by EPMR. In humans there are >140 Mendelian traits (including disorders of amino acids, enzymes, hormones, and vasculature) that include epilepsy as part of a more complex phenotype. These patients, however, account only for a small fraction of all epilepsy patients. Most cases of heritable epilepsy are believed to be multifactorial (Gardiner 1990). Patients with EPMR are distinguished from the majority of epilepsy cases in that they suffer mental deterioration following the onset of seizures. The degree of mental deterioration correlates with the severity of cerebral atrophy. The cause of epilepsy and the generalized atrophic process in the brain are unknown. It is not known whether or to what extent the frequency of seizures or the dose and duration of medication are correlated with cerebral atrophy. In any event, the association between cerebral atrophy and the duration of epilepsy or mental deterioration distinguishes EPMR patients from those with general chronic epilepsy (Hirvasniemi and Karumo 1994).
Recently, a missense mutation in the α4 subunit of the neuronal nicotinic acetylcholine receptor was reported to be associated with the chromosome 20q form of ADNFLE (Steinlein et al. 1995). It is relevant in this regard that neither of the two nicotinic acetylcholine receptor subunits recently mapped to chromosome 8 (Koyama et al. 1994; Spurr et al. 1995) are located near the EPMR locus. We considered the cathepsin B gene to be a reasonable candidate for EPMR based on its physical location and function. Tagawa et al. showed in 1991 that cathepsin B is an intracellular protease with amyloid precursor protein (APP) secretase activity. Because APP secretase cleaves the amyloid β peptide found in senile plaques of A1zheimer patients, we reasoned that a mutant cathepsin B might conceivably lead to neuronal dysfunction in EPMR patients. This possibility was, however, excluded by the identification of recombinations between a cathepsin B RFLP and the EPMR locus. One consequence of the construction of a YAC contig across the EPMR disease locus is that we can now eliminate as candidates any genes that fail to map to YACs in this 3-Mb region. For example, we have already eliminated the squalene synthetase gene that maps to 8p22-p23.1 and is involved in cholesterol synthesis (Shechter et al. 1994) as a possible candidate gene
We describe the physical mapping and cloning of the minimal genetic region encompassing the EPMR locus. A key recombination event has been identified that reduces the minimal genetic region encompassing the disease locus to 4 cM. Haplotype analysis indicates that a single founder effect mutation accounts for 18 of 21 affected individuals within the extended EPMR pedigree, excepting only the 3 affected offspringfected offspring of a married-in father who clearly harbors a disease chromosome with a distinct haplotype (Tahvanainen et al. 1994). Among the DNA markers in the EPMR region, only marker D8S264 is homozygous (allele 3) in the 18 affected individuals who contain identical (15 of 18) or very similar (3 of 18) haplotype and heterozygous for allele 3 in the remaining 3 affected individuals. D8S264 allele 3 occurs with a frequency of ~20% in the general population. Given the strong evidence for a founder effect in this large pedigree, we expect that this marker detects a region of homozygosity across the disease locus and that further delineation of this region will expedite the disease gene search. On the basis of the marked homozygosity of D8S264, we consider the 3-cM chromosomal region bounded by D8S504 and AFM-aO54td9 to be the most likely site for the EPMR gene. The YAC clones spanning this region can now be used as DNA sources for gene isolation and as probes to identify shorter genomic (P1, BAC, or cosmid) clones or cDNA clones. Our immediate goals are to identify physically mapped, polymorphic markers (Petrukhin et al. 1993) that span the minimal genetic region. These markers will be used to define the minimal region of marker homozygosity across the disease locus. We are hopeful that this strategy will continue to narrow the disease locus to a sufficiently small genomic region that gene-coding segments can be systematically identified and characterized as potential disease genes. We have begun to identify cDNA sequences that map to the minimal genetic region using immobilized YAC DNA and selective hybridization conditions as described previously (Bonaldo et al. 1994; Soares et al. 1994)
METHODS
Linkage
Two-point and multipoint analyses were carried out using MLINK and LINKMAP, respectively, of the FASTLINK computer program package (Cottingham et al. 1993; Schäffer et al. 1994). To reduce the computation time required for lod-score calculations, alleles were down-coded. For the multipoint analysis, we used the following fixed order of markers and respective sex-averaged map distances: telomere - AFM185xb2 - D8S504 - D8S264 - AFMa054td9 -D8S262-centromere (1.0, 1.0, 2.0, 3.0 cM). To evaluate linkage between the potential candidate gene, cathepsin B, and the EPMR locus, primers were designed to amplify intronic sequence between exons 8 and 9 (primer sequences: 5’→3’ forward-GGG AGA TAC CCC CAA GTG TAG C, reverse-CCT TCT CGC TAT TGG AGA CGC) (Chan et al. 1986; Gong et al. 1993). Radiolabeled PCR product was used to probe Southern blot filters containing restriction digested DNA (BamHI, BglII, BstI, EcoRI, HaeIII, HindIII, KpnI, MspI, PstI, PvuII, RsaI, StuI, TaqI) from a panel of unrelated individuals to detect RFLPs for linkage analysis.
YACs and YAC Library Screening
YAC clones were isolated by PCR screening of the CEPH Midi and Mega YAC libraries (Albertsen et al. 1990; Dausset et al. 1992), or they were identified from two of several physical mapping data bases available through the World-Wide Web. One YAC clone, yRM2205 [Genome Data Base (GDB) designation D8S596, D8Z5], was isolated from a telomere-enriched library and kindly provided to us by Dr. Harold Riethman (Wistar Institute, Philadelphia, PA). Marker D8S264 and several YAC-end sequence-tagged sites (STSs) (858e12R, 858e12L, and 241d10R) were used to screen the CEPH Midi YAC library by PCR amplification and combinatorial pooling strategies (Albertsen et al. 1990; Green and Olson 1990). Positive YACs were picked from glycerol stocks, colonies were amplified, and the result was verified by PCR with the original marker before YAC-end isolation was attempted. The Mega YACs positive for markers D8S504, D8S264, D8S201, and D8S262, as well as a number of YACs placed in the region by Alu-PCR fingerprinting, were ascertained from the CEPH data base (Cohen et al. 1993). Three YAC clones (808B12, 772C1, 795Ell) were selected from YAC contigs WC-148 and WC-1335 (Center for Genome Research at Whitehead Institute for Biomedical Research, Cambridge, MA). YAC DNA was separated from the yeast chromosomal DNA by pulsed-field gel electrophoresis using a Pharmacia LKB apparatus with a hexagonal electrode and alkali blot filters were prepared. The filters were hybridized with radiolabeled total human genomic DNA to determine the size of each clone.
YAC-end Isolation
YAC ends were isolated by ligation-mediated PCR ampli fication with minor modifications of published protocols (Kere et al. 1992). YAC DNA from single clones was prepared in agarose beads and digested in separate 15-μl reactions with RsaI, AluI, PvuII, and EcoRV. A short linker sequence (2 pmoles), T4 DNA ligase (2 units), and buffer recommended by the enzyme supplier were added, and the reactions were continued for 1–2 hr at 37°C. One microliter of the ligated mixture was amplified by PCR in 20-μl reactions using the linker primer (3 pmoles) and a YAC vector arm-specific primer (30 pmoles). The 5 × PCR buffer contained 20 mM MgC12, 335 mM TRIS-HCL (PH 8.8), 80 MM (NH4)2SO4, 50 mM β-mercaptoethanol, and 0.5 rag/ ml of bovine serum albumin. Temperature cycling conditions were as follows: 94°C for 4 rain followed by 35 cycles of 94°C for I rain, 64°C for 2 rain, and 72°C for 2 rain. One microliter of the reaction mixture was diluted to 500 μl with water and reamplified using the linker primer and an internal YAC vector arm-specific primer in 50-μl reactions under the same concentrations and temperature conditions as above. The primer sequences were as follows: YAC vector arm, left, 5’-CACCCGTTCTCGGAGCACTGTCCGACCGC-3’; internal, left, 5’-TCTCGGTAGCCAAGTTGGTTTAAGG-3’; YAC vector arm, right, 5’-ATATAGGCGCCAGCAACCGCACCTGTGGCG-3’; internal, right, 5’-TCGAACGCCCGATCTCAAGATTAC-3’; linker primer 5’-GCGGTGACCCGGGAGATCTGAATTC-3’; linker 5’-GAATTCAGATC-3’. The reamplification products were run on a 1.5% low-melting-point agarose gel, excised, and sequenced. Both ends of the YAC 810F8 were isolated by the vectorette PCR method (Riley et al. 1990; Wang et al. 1995). The primer pairs specific for the YAC insert end genomic DNA were selected using the PRIMER program (M.J. Daley, S. Lincoln, and E. Lander, Massachusetts Institute of Technology). Each STS was used to amplify the parent clone, DNA from a chromosome 8-only hybrid cell line, and other YACs within the contig. The oligonucleo-tide primers used to detect STSs are presented in Table 1. Some characteristics of the YACs are shown in Table 2.
Table 1.
YAC-end STSs Listed by the YAC Address
| STS NAME | PRIMERS 5′−3′ |
|---|---|
| 225G1R | 1 ATT CCT TTG CCC TGT ACA CG 2 GTG CAG ATA AAC CAG TGT CTG C |
| 241D10L | 1 GGG TAA GGA AAG ATG CCA CA 2 AAA CAG GAC GGA CTC CAG G |
| 241D10R | 1 ATT CCG CTT TGG TTT CCC 2 CCA ACA CAT TCC CAG TGC TA |
| 374E8R | 1 ATT CGC TCT GTA CTG AAA GGT G 2 TCA CAG GCC CCA ATT CTC |
| 463B10R | 1 CAC TTC CCA GCA GAC ATT CC 2 TAT GAG CAA GCC TGG AGT CC |
| 485C11L | 1 AAA GGA TGC TGG GCC TTA TT 2 CCA GGT CCC TGG TGT TTT AA |
| 485C11R | 1 TTC AGT TGC TTC TGT ATT TCC A 2 TGG CTG CAT CAG TAG CTC TG |
| 679D3L | 1 CTT TGA CTT CCC GTA CTG GG 2 CTG GAA TTT GTA TTT AGG GCA |
| 772C1L | 1 TCT CAC CTG ACC CAT GAT GA 2 TTT TAC TCA GCC TGG CGT G |
| 796G6L | 1 TTT CAC TCT CTC TGT TTT C 2 TGT TCG ATC ACA TAG CTC |
| 808B12L | 1 CAC AGG TGT GTA GAG CTC AGA A 2 CTC CCA GAG ACA GAA CCT GG |
| 808E6L | 1 TCA TCT CGG TCC TTC TCA GA 2 CAT TTT GCT ATT CCT TTT AGC C |
| 808E6R | 1 TTC GTC GAT TTT CCT GCT CT 2 AGA AAT GGG GGA AAG GAG AA |
| 810F8L | 1 AGA AAC TTC CGA GTC CAG TCC 2 TTT CTG TGA GTT CCC GAG CT |
| 810F8R | 1 TGA GGT GCT TCT GTT TGT GG 2 GAA TTC TGG GAA GGT GGT AGC |
| 858E12L | 1 ATG AAA CGC AGC GGA AAC 2 GGC AGC AAT TCC ATC AAG |
| 858E12R | 1 CCA GTC TAC ACA ATC AGA AAA C 2 ACC TGT GTC GCT TAG ATG |
| 931B2L | 1 ACT TGA AGT TCC GAT GCC C 2 TTC TGC TGA GTG GGA GCC |
| 972C9R | 1 AGG GTC AGT TTC TGG AGA TTT G 2 TGG GAA GGG AAA GAA TTG G |
(R) Right and (L) left arm of the vector. Primer sequences are shown.
Table 2.
Characteristics of YAC Clones in the EPMR Contig
| YAC address | YAC sizea | Right endb | Left endb |
|---|---|---|---|
| 225G1 | 280 | + | Chi |
| 241D1O | 470 | + | + |
| 374E8 | 360 | + | Chi |
| 42F7 | 350 | N.D. | Rep |
| 463B10 | 270 | + | Rep |
| 485C11 | 360 | + | + |
| 679D3 | 740*, 360/280 | N.D. | + |
| 772C1 | 1770*, 1300 | Chi | + |
| 781D11 | 580*, 650 | N.D. | Rep |
| 787C11 | 1740*, 1600 | N.D. | N.D. |
| 795E11 | 1800 | Rep | Chi |
| 796G6 | 1280*, 950 | Chi | + |
| 797C11 | 1670*, 1250/950 | Chi | N.D. |
| 808B12 | 1230*, 900 | Rep | + |
| 808E6 | 1780*, 1200 | + | + |
| 810F8 | 1180*, 1000 | + | + |
| 858E12 | 810*, 800 | + | + |
| 926F11 | 950 | Chi | Chi |
| 930A2 | 1480* | N.D | N.D. |
| 931B2 | 1230*, 1100 | Rep | + |
| 972C9 | 850 | + | Chi |
| yRM2205 | 250* | N.D. | N.D. |
YAC sizes were determined in our laboratory, except those marked by an asterisk (*), which were derived from computer data bases (see Methods). In cases where the YAC contained more than one insert, the sizes of each insert are separated by a / symbol.
(+) Unique sequence DNA mapping to chromosome 8; (Chi) chimeric, indicating that the YAC end does not map to chromosome 8; (Rep) repetitive DNA sequence; (N.D.) not determined.
ACKNOWLEDGMENTS
We thank the EPMR families for their excellent cooperation. We would also like to thank Dr. Harold Riethman for providing a key telomeric YAC clone, Dr. Juha Kere for helpful discussions, and the CEPH for their assistance in screening YAC libraries. This work was supported by the Academy of Finland (A.E.L., A.C.), the Ulla Hjelt Foundation (S.R.), Deutsche Forschungsgemeinschaft (Klifo H6rforschung; Zel 49/6–1) (S.M.L.), and HG00008 from the National Center for Human Genome Research (Jurg Ott/ S.M.L.). Dr. Ott is a professor of Genetics and Development at Columbia University. Part of this study was performed at the Folkhälsan Institute of Genetics.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Albertsen HM, Abderrahim H, Cann HM, Dausset J, Le Paslier D, and Cohen D. 1990. Construction and characterization of a yeast artificial chromosome library containing seven haploid human genome equivalents. Proc. Natl. Acad. Sci 87: 4256–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C et al. 1994. A nonsense mutation in the α4 subunit of the nicotinergic acetylcholine receptor (CHRNA4) cosegregates with 20q-linked benign neonatal familial convulsions (EBN1). Neurobiol. Dis l: 95–99. [DOI] [PubMed] [Google Scholar]
- Bonaldo MF, Yu M−T, Jelenc P, Brown S, Su L, Lawton L, Deaven L, Efstratiadis A, Warburton D, and Soares MB. 1994. Selection of cDNAs using chromosome-specific genomic clones: Application to human chromosome 13. Hum. Mol. Genet 3: 1663–1673. [DOI] [PubMed] [Google Scholar]
- Chan SJ, San Segundo B, McCormick MB, and Steiner DF. 1986. Nucleotide and predicted amino acid sequences of cloned human and mouse preprocathepsin B cDNAs. Proc. Natl. Acad. Sci 83: 7721–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Chumakov I, and Weissenbach J. 1993. A first-generation physical map of the human genome. Nature 336: 698–701. [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr., Idury RM, and Schaffer AA. 1993. Faster sequential genetic linkage computations. Am. J. Hum. Genet 53: 252–263. [PMC free article] [PubMed] [Google Scholar]
- Dausset J, Ougen P, Abderrahim H, Billault A, Sambucy J-L, Cohen D, and Le Paslier D. 1992. The CEPH YAC library. Behring Inst. Mitt 91: 13–20. [PubMed] [Google Scholar]
- Delgado-Escueta AV, Treiman DM, and Walsh GO. 1983. The treatable epilepsies. N. Engl. J. Med 308: 1508–1514. [DOI] [PubMed] [Google Scholar]
- Delgado-Escueta AV, Serratosa JM, Liu AW, Medina MT, and Sparkes RS. 1994. A juvenile myoclonic epilepsy locus proximal to HLA: The value of studying a large family. Epilepsia 35: S7, 8. [Google Scholar]
- Durner M, Sander T, Greenberg DA, Johnson K, Beck-Mannagetta G, and Janz D. 1991. Localization of idiopathic generalized epilepsy on chromosome 6p in families of juvenile rnyoclonic epilepsy patients. Neurology 41: 1651–1655. [DOI] [PubMed] [Google Scholar]
- Fong D, Chan MM-Y, Hsieh W-T, Menninger JC, and Ward DC. 1992. Confirmation of the human cathepsin B gene (CTSB) assignment to chromosome 8. Hum. Genet 89: 10–12. [DOI] [PubMed] [Google Scholar]
- Gardiner RM 1990. Genes and epilepsy. J. Med. Genet 27: 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Chan SJ, Bajkowski AS, Steiner DF, and Frankfater A. 1993. Characterization of the cathepsin B gene and multiple mRNAs in human tissue: Evidence for alternative splicing of cathepsin B pre-mRNA. DNA Cell Biol 12: 299–309. [DOI] [PubMed] [Google Scholar]
- Green ED and Olson MV. 1990. Systematic screening of yeast artificial-chromosome libraries by use of the polymerase chain reaction. Proc. Natl. Acad. Sci 87: 1213–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Delgado-Escueta AV, Widelitz H, Sparkes RS, Treiman L, Maldonado HM, Park MS, and Terasaki PI. 1988. Juvenile myoclonic epilepsy (]ME) may be linked to the BF and HLA loci on human chromosome 6. Am. J. Med. Genet 31: 185–192. [DOI] [PubMed] [Google Scholar]
- Hauser WA, Annegers JF, and Kurland LT. 1993. Incidence of epilepsy and unprovoked seizures in Rochester, Minnesota: 1935–1984. Epilepsia 34: 453–468. [DOI] [PubMed] [Google Scholar]
- Hirvasniemi A and Karumo J. 1994. Neuroradiological findings in the northern epilepsy syndrome. Acta Neurol. Scand 90: 388–393. [DOI] [PubMed] [Google Scholar]
- Hirvasniemi A, Lang H, Lehesjoki A-E, and Leisti J. 1994. Northern epilepsy syndrome: An inherited childhood onset epilepsy with associated mental deterioration. J. Med. Genet 31: 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvasniemi A, Herrala P, and Leisti J. 1995. Northern epilepsy syndrome: Clinical course and the effect of medication on seizures. Epilepsia 36: 792–797. [DOI] [PubMed] [Google Scholar]
- Kere J, Nagaraja R, Mumm S, Ciccodicola A, D’Urso M, and Schlessinger D. 1992. Mapping human chromosomes by walking with sequence-tagged sites from end fragments of yeast artificial chromosome inserts. Genomics 14: 241–248. [DOI] [PubMed] [Google Scholar]
- Koyama K, Sudo K, and Nakamura Y. 1994. Mapping of the human nicotinic acetylcholine receptor β3 gene (CHRNB3) within chromosome 8p11.2. Genomics 21: 460–461. [DOI] [PubMed] [Google Scholar]
- Lehesjoki A-E, Koskiniemi M, Sistonen P, Miao J, Hastbacka J, Norio R, and de la Chapelle A. 1991. Localization of a gene for progressive myoclonus epilepsy to chromosome 21q22. Proc. Natl. Acad. Sci 88: 3696–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppert M, Anderson VE, Quattlebaum T, Stauffer D, O’Connell P, Nakamura Y, Lalouel J-M, and White R. 1989. Benign familial neonatal convulsions linked to genetic markers on chromosome 20. Nature 337: 647–648. [DOI] [PubMed] [Google Scholar]
- Lewis TB, Leach RJ, Ward K, O’Connell P, and Ryan SG. 1993. Genetic heterogeneity in Benign familial neonatal convulsions: Identification of a new locus on chromosome 8q. Am. J. Hum. Genet 53: 670–675. [PMC free article] [PubMed] [Google Scholar]
- Ottman R, Risch N, Hauser WA, Pedley TA, Lee JH, Barker-Cummings C, Lustenberger A, Nagle KJ, Lee KS, Scheuer ML, Neystat M, Susser M, and Wilhelmsen KC. 1995. Localization of a gene for partial epilepsy to chromosome 10q. Nature Genet 10: 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrukhin K, Fischer SG, Pirastu M, Tanzi RE, Chernov I, Devoto M, Brzustowicz LM, Cayanis E, Vitale E, Russo JJ, Matseoane D, Boukhgalter B, Wasco W, Figus AL, Loudianos J, Cao A, Sternlieb I, Evgrafov O, Parano E, Pavone L, Warburton D, Ott J, Penchaszadeh GK, Scheinberg IH, and Gilliam TC. 1993. Mapping, cloning and genetic characterization of the region containing the Wilson disease gene. Nature Genet 5: 338–343. [DOI] [PubMed] [Google Scholar]
- Phillips HA, Scheffer IE, Berkovic SF, Hollway GE, Sutherland GR, and Mulley JC. 1995. Localization of a gene for autosomal dominant nocturnal frontal lobe epilepsy to chromosome 20q13.2. Nature Genet 10: 117–118. [DOI] [PubMed] [Google Scholar]
- Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, and Markham AF. 1990. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res 18: 2887–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, and Cottingham RW Jr. 1994. Avoiding recomputation in linkage analysis. Hum. Hered 44: 225–237. [DOI] [PubMed] [Google Scholar]
- Scheuer ML and Pedley TA. 1990. The evaluation and treatment of seizures. N. Engl. J. Med 323: 1468–1474. [DOI] [PubMed] [Google Scholar]
- Shechter I, Conrad DG, Hart I, Berger R, McKenzie TL, Bleskan J, and Patterson D. 1994. Localization of the Squalene synthase gene (FDFT1) to human chromosome 8p22-p23.1. Genomics 20: 116–118. [DOI] [PubMed] [Google Scholar]
- Soares MB, Bonaldo MF, Jelene P, Su L, Lawton L, and Efstratiadis A. 1994. Construction and characterization of a normalized cDNA library. Proc. Natl. Acad. Sci 91: 9228–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr NK, Blanton R, Bookstein R, Clarke R, Cottingham R, Daiger S, Drayna D, Faber P, Horrigan S, Kas K, et al. 1995. Report and abstracts of the second international workshop on human chromosome 8 mapping. Oxford, United Kingdom, September 16–18, 1994. Cytogenet. Cell Genet 68:– 147–164. [DOI] [PubMed] [Google Scholar]
- Steinlein OK, Mulley JC, Propping P, Wallace RH, Phillips HA, Sutherland GR, Scheffer IE, and Berkovic SF. 1995. A missense mutation in the neuronal nicotinic acetylcholine receptor α4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nature Genet 11: 201–203. [DOI] [PubMed] [Google Scholar]
- Tagawa K, Kunishita T, Maruyama K, Yoshikawa K, Kominami E, Tsuchiya T, Suzuki K, Tabira T, Sugita H, and Ishiura S. 1991. Alzheimer’s disease amyloid β-clipping enzyme (APP secretase): Identification, purification and characterization of the enzyme. Biochem. Biophys. Res. Commun 177: 377–387. [DOI] [PubMed] [Google Scholar]
- Tahvanainen E, Ranta S, Hirvasniemi A, Karila E, Leisti J, Sistonen P, Weissenbach J, Lehesjoki A-E, and de la Chapelle A. 1994. The gene for a recessively inherited human childhood progressive epilepsy with mental retardation maps to the distal short arm of chromosome 8. Proc. Natl. Acad. Sci 91: 7267–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Kleyn PW, Vitale E, Ross BM, Lien L, Xu J, Carter TA, Brzustowicz LM, Obici S, Selig S, Pavone L, Parono E, Penchaszadeh GK, Munsat T, Kunkel LM, and Gilliam TC. 1995. Refinement of the Spinal muscular atropy locus by genetic and physical mapping. Am. J. Hum. Genet 56: 202–209. [PMC free article] [PubMed] [Google Scholar]
- Weissbecker KA, Durner M, Janz D, Scaramelli A, Sparkes RS, and Spence MA. 1991. Confirmation of linkage between juvenile myoclonic epilepsy locus and the HLA region of chromosome 6. Am. J. Med. Genet 38: 32–36. [DOI] [PubMed] [Google Scholar]


