HIV-2 groups A to I resulted from nine independent cross-species transmission events of SIVsmm to humans and differ considerably in their prevalence and geographic spread. Thus, detailed characterization of these viruses offers a valuable means to elucidate immune evasion mechanisms and human-specific adaptations determining viral spread. In a systematic comparison of rare and epidemic HIV-2 groups and their simian SIVsmm counterparts, we found that the ability of Nef to downmodulate the primary viral entry receptor CD4 and the T cell receptor CD3 is conserved, while effects on CD28, CD74, and major histocompatibility complex class I surface expression vary considerably. Furthermore, we show that not only the Env proteins of HIV-2 groups A, AB, F, and I but also those of some SIVsmm isolates antagonize human tetherin. This finding helps to explain why SIVsmm has been able to cross the species barrier to humans on at least nine independent occasions.
KEYWORDS: HIV-2, SIVsmm, tetherin, Env, Nef
ABSTRACT
The host restriction factor tetherin inhibits virion release from infected cells and poses a significant barrier to successful zoonotic transmission of primate lentiviruses to humans. While most simian immunodeficiency viruses (SIV), including the direct precursors of human immunodeficiency virus type 1 (HIV-1) and HIV-2, use their Nef protein to counteract tetherin in their natural hosts, they fail to antagonize the human tetherin ortholog. Pandemic HIV-1 group M and epidemic group O strains overcame this hurdle by adapting their Vpu and Nef proteins, respectively, whereas HIV-2 group A uses its envelope (Env) glycoprotein to counteract human tetherin. Whether or how the remaining eight groups of HIV-2 antagonize this antiviral factor has remained unclear. Here, we show that Nef proteins from diverse groups of HIV-2 do not or only modestly antagonize human tetherin, while their ability to downmodulate CD3 and CD4 is highly conserved. Experiments in transfected cell lines and infected primary cells revealed that not only Env proteins of epidemic HIV-2 group A but also those of a circulating recombinant form (CRF01_AB) and rare groups F and I decrease surface expression of human tetherin and significantly enhance progeny virus release. Intriguingly, we found that many SIVsmm Envs also counteract human as well as smm tetherin. Thus, Env-mediated tetherin antagonism in different groups of HIV-2 presumably stems from a preadaptation of their SIVsmm precursors to humans. In summary, we identified a phenotypic trait of SIVsmm that may have facilitated its successful zoonotic transmission to humans and the emergence of HIV-2.
IMPORTANCE HIV-2 groups A to I resulted from nine independent cross-species transmission events of SIVsmm to humans and differ considerably in their prevalence and geographic spread. Thus, detailed characterization of these viruses offers a valuable means to elucidate immune evasion mechanisms and human-specific adaptations determining viral spread. In a systematic comparison of rare and epidemic HIV-2 groups and their simian SIVsmm counterparts, we found that the ability of Nef to downmodulate the primary viral entry receptor CD4 and the T cell receptor CD3 is conserved, while effects on CD28, CD74, and major histocompatibility complex class I surface expression vary considerably. Furthermore, we show that not only the Env proteins of HIV-2 groups A, AB, F, and I but also those of some SIVsmm isolates antagonize human tetherin. This finding helps to explain why SIVsmm has been able to cross the species barrier to humans on at least nine independent occasions.
INTRODUCTION
In contrast to human immunodeficiency virus type 1 (HIV-1), which is largely responsible for the global AIDS pandemic, HIV-2 has spread less successfully in the human population. While HIV-1 has infected more than 70 million individuals worldwide, fewer than 2 million people live with HIV-2, most of them in West Africa and countries with colonial links to this region (1, 2). These differences in prevalence and spread have been ascribed to lower viral loads in HIV-2-infected individuals, resulting in less efficient sexual and perinatal transmission of HIV-2 than of HIV-1 (3). HIV-2 itself is further subdivided into nine evolutionarily diverse groups (termed A to I), which differ considerably in their spread in the human population (4–11) (Table 1). Each group is the result of an independent cross-species transmission event of simian immunodeficiency viruses (SIVsmm) from sooty mangabeys (Cercocebus atys) to humans (18). While groups A and B have spread epidemically, groups C to I were only identified in one or two individuals and thus may not represent genuine groups but rather dead-end infections in the new human host (4–11) (Table 1). In addition, two recombinant forms derived from HIV-2 groups A and B have been described. The first (CRF01_AB) has been identified in at least seven individuals from different geographic origins and thus represents a bona fide circulating recombinant virus (12, 14). The second (URF01_AB) is a unique recombinant form that has been identified in a single individual only (15, 16).
TABLE 1.
HIV-2 groups A to I
| HIV-2 group | Prevalence | Geographic distribution | Estimated time(s) of the most recent common ancestor | Pathogenicity | Reference |
|---|---|---|---|---|---|
| A | ~92% of all 1 to 2 million HIV-2 infections, prevalence declining | Mainly West Africa (Guinea-Bissau, Gambia, Senegal, Cape Verde Ivory Coast and Sierra Leone, Ghana, Burkina Faso, Mali, Liberia, Guinea), Cameroon, Angola, Mozambique, Brazil, Guiana, Portugal, France, Spain, India, USA | 1940 (1924–1956), 1927 (1879–1964) | CD4+ T cell depletion, AIDS, less pathogenic than HIV-1 | 1, 12, 13 |
| B | ~8% of all 1 to 2 million HIV-2 infections, prevalence declining | Mainly West Africa (Ivory Coast, Ghana, Burkina Faso, Mali) | 1945 (1931–1959), 1934 (1879–1973) | CD4+ T cell depletion, AIDS, less pathogenic than HIV-1 | 1, 12, 13 |
| CRF01_AB | 7 cases known | Ivory Coast, Japan, Nigeria, Guinea-Bissau, Austria, Ghana, Belgium | 1964–1973 | CD4+ T cell depletion, lymphadenopathy, candidiasis, AIDS | 14, 12 |
| URF01_AB | 1 case known | Cameroon | Unknown | 15, 16 | |
| C | 1 case known | Rural Liberia | Isolated from an asymptomatic individual | 4 | |
| D | 1 case known | Rural Liberia | Isolated from an asymptomatic individual | 4 | |
| E | 1 case known | Rural Sierra Leone to USA (Los Angeles, CA) | Isolated from an asymptomatic individual, >500 CD4+ T cells/mm3, probably infected for ∼20 years | 4, 17 | |
| F | 2 cases known | Sierra Leone (Freetown and rural Northern Province), USA (Newark, NJ) | CD4+ T cell depletion | 5, 6 | |
| G | 1 case known | Ivory Coast | Isolated from an asymptomatic blood donor | 7, 8 | |
| H | 2 cases known | Ivory Coast (near Liberian border), France | CD4+ T cell depletion, cerebral neurocysticercosis, AIDS | 9, 10 | |
| I | 1 case known | Rural Ivory Coast (Taï National Park border) | Unknown | 11 |
Successful spread of HIV-1 and -2 in the human population is determined by environmental factors such as male circumcision, commercial sex work, and/or social disruption (19, 20), but it also depends on the adaptation of these zoonotic viruses to their new human host. For example, viruses need to adapt to a variety of so-called host restriction factors that not only suppress viral replication within an infected individual but also may pose significant barriers to successful cross-species transmission events, as they are evolving fast and are often antagonized in a species-specific manner (21, 22). One striking example is the host restriction factor tetherin/BST2, which prevents the release of budding virions from infected cells (23, 24). While the direct simian precursor viruses of HIV-1 and HIV-2 use their accessory protein Nef to counteract this antiviral protein in their natural ape and monkey hosts (25–28), SIV Nef is unable to antagonize human tetherin due to a protective deletion that emerged early during human evolution (29). In the case of HIV-1, only pandemic group M and epidemic group O strains, but not rare group N and P viruses, evolved efficient antagonists of human tetherin (26, 30–34), suggesting that counteraction of this antiviral protein represents a prerequisite for the successful spread of primate lentiviruses in the human population (35). These findings also illustrate that comparison of rare and epidemic groups of HIV represents a valuable means to identify host-virus interactions that might determine efficient spread of HIV and potentially other (zoonotic) viruses in the human population.
While the different groups of HIV-1 are relatively well characterized, most studies analyzing immune evasion mechanisms and human-specific adaptations of HIV-2 focused on the characterization of epidemic group A and (to a lesser extent) group B viruses. For example, it has been shown that HIV-2 group A mastered the tetherin hurdle by switching from Nef to Env to counteract this restriction factor (36–38). However, it has remained unclear whether/how other groups of HIV-2 antagonize human tetherin. Similarly, Nef proteins of HIV-2 group A and B strains have been shown to modulate surface expression of CD3, major histocompatibility complex class I (MHC-I), and other immune receptors, whereas Nef proteins of rare groups C to I were not or only poorly characterized (39, 40).
Here, we systematically analyzed a panel of Nef and Env proteins representing epidemic and rare groups of HIV-2 as well as their simian counterpart, SIVsmm, to determine whether they differ in their immune evasion activities. Experiments in infected primary cells revealed that Nef-mediated downmodulation of CD4 and the T cell receptor CD3 is conserved, whereas the ability of Nef to modulate CD28, CD74, and MHC-I surface levels varies between diverse lineages of SIVsmm and groups of HIV-2. Notably, Env proteins of both epidemic (A and AB) and rare (F and I) groups of HIV-2 efficiently enhance progeny virus release by counteracting human tetherin. This Env-mediated increase of virus release correlated with decreased tetherin levels at the surface of infected cells. While the Nef proteins of SIVsmm generally failed to antagonize human tetherin, we found that some SIVsmm Env proteins increase virus release also in the presence of the human tetherin ortholog. Furthermore, we demonstrate that several SIVsmm lineages encode two tetherin antagonists, as SIVsmm Nef and Env antagonize sooty mangabey tetherin with similar efficiency. Our findings illustrate the enormous flexibility of primate lentiviruses in antagonizing host restriction factors and suggest that preadaptation of some SIVsmm strains to human tetherin facilitates successful zoonotic transmission to humans.
RESULTS
Functional motifs in Nef and Env are conserved between HIV-2 and SIVsmm.
To investigate the evolution of anti-tetherin activity in the SIVsmm/HIV-2 lineage, we selected a panel of evolutionarily diverse Nef and Env proteins representing epidemic (A, B, and CRF01_AB) and rare (F, G, H, and I) groups of HIV-2 as well as different lineages of SIVsmm for functional characterization (Table 2). The Nef and Env proteins of SIVmac 239, a well-characterized macaque-passaged derivative of SIVsmm (55), were included as controls. Unfortunately, Nef and Env proteins of HIV-2 groups C to E could not be analyzed, since neither full-length sequences nor samples from the three respective patients were available.
TABLE 2.
SIVsmm, SIVmac, and HIV-2 isolates analyzed in this study
| Virus and lineage/group | Isolate | Origin | Isolation method | Reference |
|---|---|---|---|---|
| SIVsmm | ||||
| 1 | PGm5.3 | Pig-tailed macaque, exptl infection, Yerkes Regional Primate Research Center | Virus isolated from lymph nodes was passaged in human PBMCs and CEMx174 cells before genomic viral DNA was isolated | 41 |
| SM2_ FBr12w7 | Sooty mangabey, 3–5 yr old, exptl infection, Yerkes Regional Primate Research Center | Direct isolation of viral RNA from plasma | 42, 43 | |
| 2 | M926 | Sooty mangabey, male, 16 yr old, natural infection, Tulane National Primate Research Center | Viral RNA of SIVsmm lineages 2 to 6 was isolated from plasma or from tissue culture supernatants of short-term sooty mangabey PBMC cultures, in which primary SIVsmm isolates were propagated | 44 |
| 3 | M949 | Sooty mangabey, male, 14 yr old, natural infection, Tulane National Primate Research Center | ||
| 4 | G932 | Sooty mangabey, female, 30 yr old, natural infection, Tulane National Primate Research Center | ||
| 5 | FTq | Sooty mangabey, male, 13 yr old, natural infection, Yerkes National Primate Research Center | ||
| 6 | D215 | Sooty mangabey, female, 24 yr old, natural infection, Tulane National Primate Research Center | ||
| SIVmac | 239 | Rhesus macaque, exptl infection, New England Regional Primate Research Center | Isolation of viral DNA from HUT-78 cells infected with serum | 45, 46 |
| HIV-2 | ||||
| A | ROD10 | Human, male, 32 yr old, Cape Verde, France | Isolation of viral DNA from infected CEM cells | 47, 48, 49 |
| BEN | Human, male, 42 yr old, Mali, Libya, Saudi-Arabia, Germany | Isolation of viral DNA from infected Molt4 clone 8 cells | 50, 51 | |
| B | UC1 | Human, male, Ivory Coast | Isolation of viral DNA from infected SupT1 cells | 52, 53 |
| D205_ ALT | Human, female, Ghana | Isolation of viral DNA from human umbilical cord blood lymphocytes upon coculture with patient PBLs | 54 | |
| CRF01_AB | 7312A | Human, male, 32 yr old, Ivory Coast | Isolation of viral DNA from human PBMCs upon coculture with patient PBMCs | 14 |
| F | NWK08 | Human, male, 68 yr old, Sierra Leone, USA | Isolation of viral DNA from human PBMCs or CEMx174 cells upon coculture with patient PBMCs | 5 |
| G | Abt96 | Human, Ivory Coast | Direct isolation of viral RNA from plasma | 7, 8 |
| H | 12034 | Human, male, 31 yr old, Ivory Coast, France | Direct isolation of proviral DNA from PBMCs | 9 |
| I | 07IC-TNP3 | Human, male, 8 yr old, Ivory Coast | Direct isolation of proviral DNA from blood | 11 |
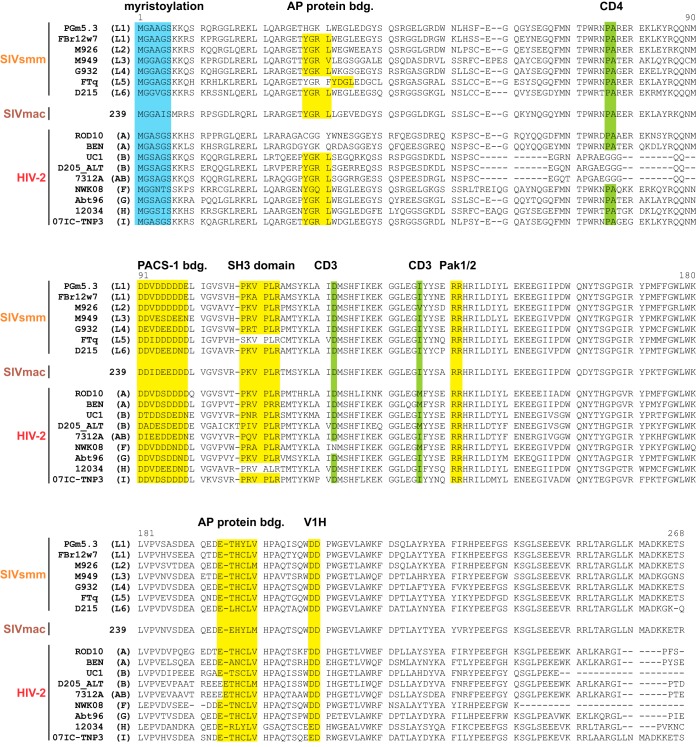
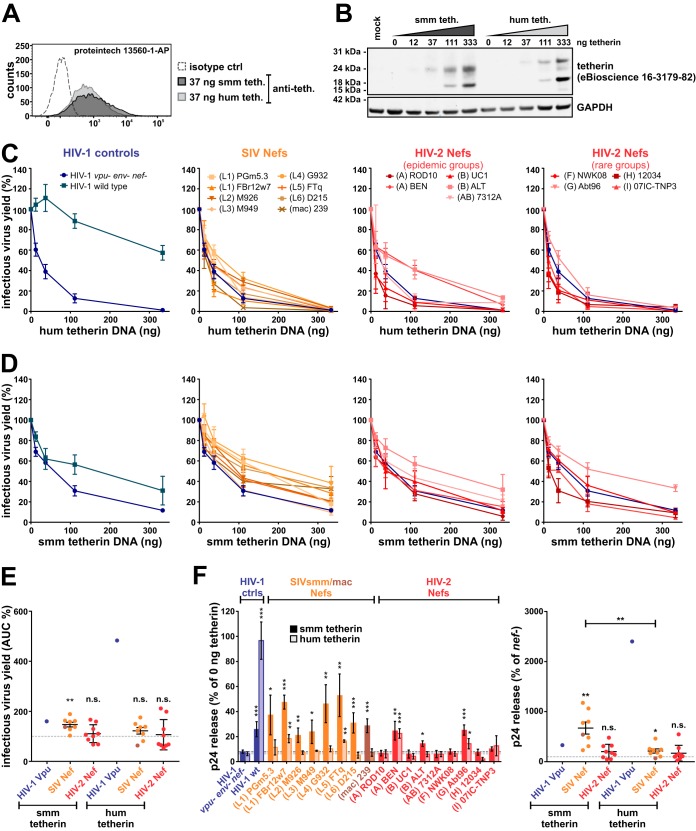
An alignment of the Nef protein sequences revealed that motifs known to be involved in the downmodulation of tetherin, CD4, CD3, and/or MHC-I, are largely conserved (40, 56, 57) (Fig. 1). These include the N-terminal myristoylation site (MGxxxS) (58) as well as protein-protein interaction domains, such as a proline-rich motif (PxxPxR) that binds SH3 domains (59) or an adaptor protein (AP) binding sorting motif (ExxxL[L/V]) in the C-terminal flexible loop (60, 61). Two notable exceptions are the Nef proteins of HIV-2 group B and CRF01-AB, which harbor deletions in their N-terminal domains and lack a proline-alanine motif that has been suggested to be involved in the downmodulation of CD4 by SIVmac Nef (62).
FIG 1.
Nef protein sequence alignment. Alignment of SIVsmm, SIVmac, and HIV-2 Nef protein sequences analyzed in the present study. SIVsmm lineages (L1 to L6) and HIV-2 groups (A, B, AB [CRF01_AB], and F to I) of the respective isolates are indicated in parentheses. The N-terminal Nef myristoylation site is highlighted in blue. Motifs interacting with cellular proteins are shown in yellow (AP, adaptor proteins; PACS-1, phosphofurin acidic cluster sorting protein 1; SH3, Src-homology 3; Pak1/2, p21-activated kinase 1; V1H, subunit H of the vacuolar membrane ATPase; bdg., binding). Selected amino acids involved in the downmodulation of CD3 or CD4 are highlighted in green. Dashes represent gaps that were introduced to improve the alignment.
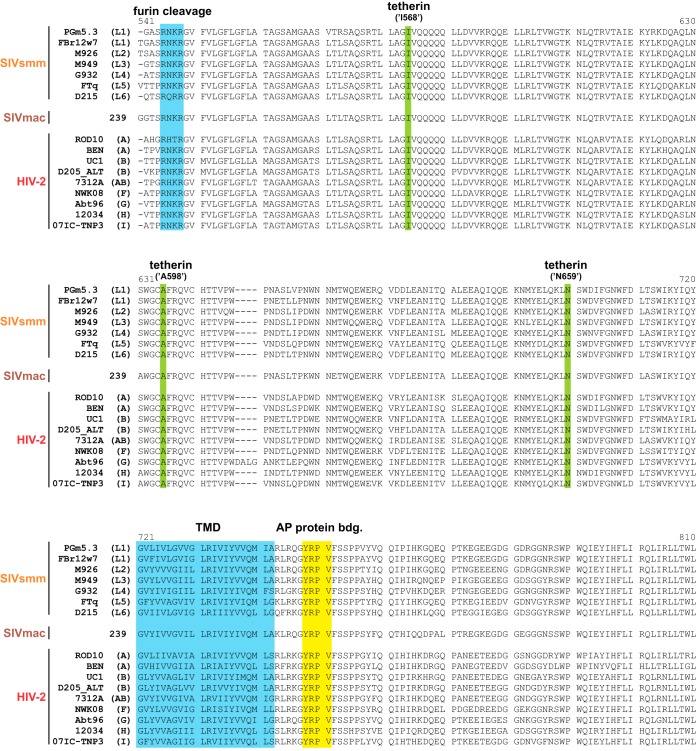
Similar to Nef, known functional domains are also conserved in most HIV-2 and SIVsmm Env proteins analyzed in the present study (Fig. 2). They all harbor the furin cleavage site (Rx[K/R]R) enabling proteolytic processing of gp140 into the mature gp105 and gp36 subunits (63, 64). Moreover, they generally contain an intracellular tyrosine motif (Yxxϕ) that mediates endocytosis via recruitment of AP proteins (65) and is essential for the anti-tetherin activity of HIV-2 group A ROD10 Env (36, 37). In addition to the endocytic sorting motif, residues K422, I568, A598, and N659 are also required for the ability of HIV-2 A Env to antagonize human tetherin (38, 66, 67). While I568, A598, and N659 are conserved in all HIV-2, SIVsmm, and SIVmac strains selected for analysis (Fig. 2), K422 is only present in Env proteins of epidemic HIV-2 groups A, B, and CRF01_AB but not in rare groups F, G, H, and I or SIVsmm.
FIG 2.
Env protein sequence alignment. Alignment of SIVsmm, SIVmac, and HIV-2 gp36 protein sequences analyzed in the present study. SIVsmm lineages (L1 to L6) and HIV-2 groups (A, B, AB [CRF01_AB], and F to I) of the respective isolates are indicated in parentheses. The furin cleavage site and an adaptor protein (AP) binding motif are highlighted in blue and yellow, respectively. Amino acids known to be required for the anti-tetherin activity of HIV-2 A Env are shown in green. Please note that the numbering of the amino acid positions (I568, A598, and N659) used in previous publications (38, 66, 67) is different from the one in this alignment. Dashes represent gaps that were introduced to improve the alignment (TMD, transmembrane domain).
Nef-mediated downmodulation of CD3 and CD4 is conserved among HIV-2 and SIVsmm.
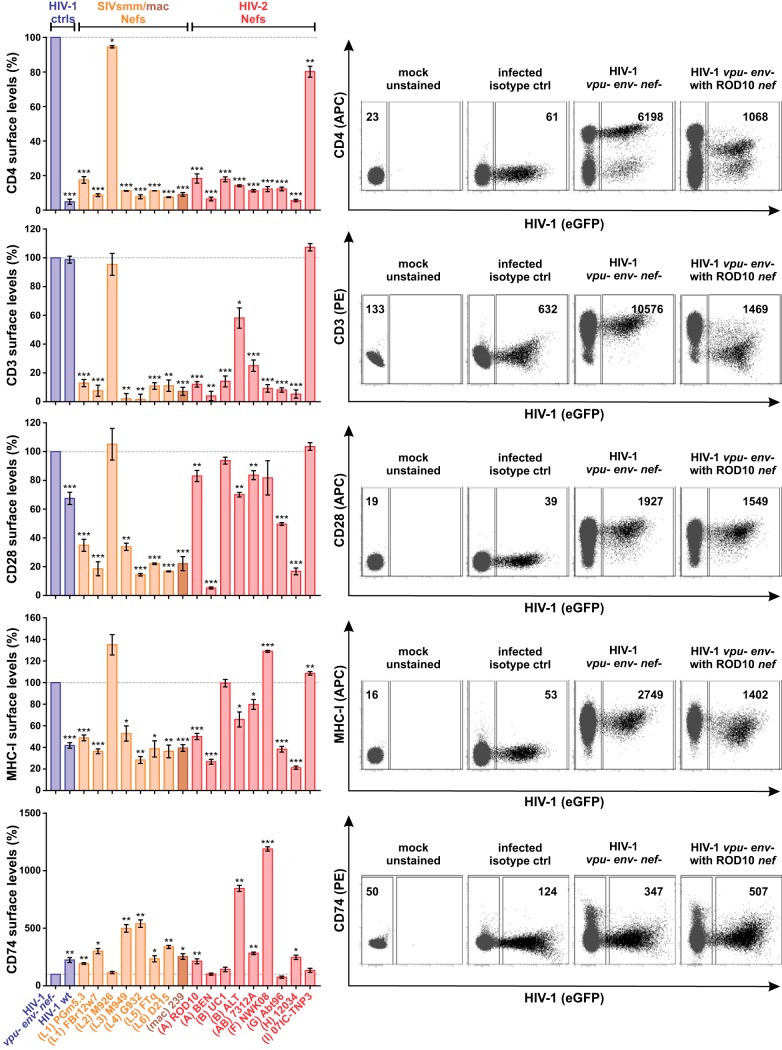
To directly compare Nef functions of different groups of HIV-2, SIVsmm, and SIVmac, human peripheral blood mononuclear cells (PBMCs) were infected with vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped HIV-1 chimeras coexpressing heterologous Nef proteins and enhanced green fluorescent protein (eGFP) from bicistronic mRNA. env and vpu genes were disrupted to abrogate any potential effects of the respective viral proteins on CD4 and tetherin surface expression. Flow cytometric analyses revealed that most SIVsmm, SIVmac, and HIV-2 Nef proteins decreased surface levels of CD4 and CD3 by more than 80% (Fig. 3, top). Notable exceptions were HIV-2 B ALT Nef, which downmodulated CD3 less efficiently, and the SIVsmm M926 and HIV-2 group I Nefs, which were inactive in most assays. Although expression levels differed, all Nef proteins were detected by Western blotting and migrated at the expected molecular weights (Fig. 4A). Furthermore, sequence analyses did not reveal any obvious damaging mutations that would explain the inactivity of SIVsmm M926 and HIV-2 group I Nefs (Fig. 1). As expected, HIV-1 NL4-3 Nef failed to modulate CD3, as this function was lost in HIV-1 and its simian precursor viruses (39, 68). While CD4 downmodulation prevents superinfection and ensures efficient release of fully infectious virus particles by preventing the formation of inactive CD4-Env complexes (69, 70), CD3 downmodulation inhibits hyperactivation and cell death of infected T cells (39). T cell activation is also suppressed by downmodulation of the costimulator CD28. In contrast to CD3, however, the effects of Nef on CD28 varied considerably among the SIV and HIV-2 strains analyzed, and many were only poorly active (Fig. 3, middle). While many HIV-2 Nefs decreased CD28 levels by less than 20%, their SIVsmm counterparts were on average more active (34.71% versus 66.49% downmodulation; P value of 0.061). A similar variability was observed in the effects of HIV-2 Nefs on the surface levels of MHC-I and CD74 (Fig. 3, bottom). Both upregulation of CD74 and downmodulation of MHC-I suppress viral antigen presentation. Thus, the selection pressure to maintain these Nef activities depends on the presence of efficient T cell epitopes and may vary between individual infected hosts.
FIG 3.
Nef-mediated modulation of CD4, CD3, CD28, MHC-I, and CD74. Human PBMCs were infected with VSV-G-pseudotyped HIV-1 vpu- env- IRES eGFP reporter viruses expressing the indicated Nef proteins. Three days postinfection, surface levels of CD4, CD3, CD28, MHC-I, and CD74 were determined by flow cytometry. The mean fluorescence intensity of the respective surface receptor on infected (i.e., eGFP positive) cells was normalized to that of uninfected cells, and the control lacking nef (vpu- env- nef-) was set to 100%. The panels on the left show mean values from 3 to 11 independent experiments ± standard errors of the means (SEM). Asterisks indicate statistically significant differences compared to the negative control (vpu- env- nef-) (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). Examples of primary data are shown on the right.
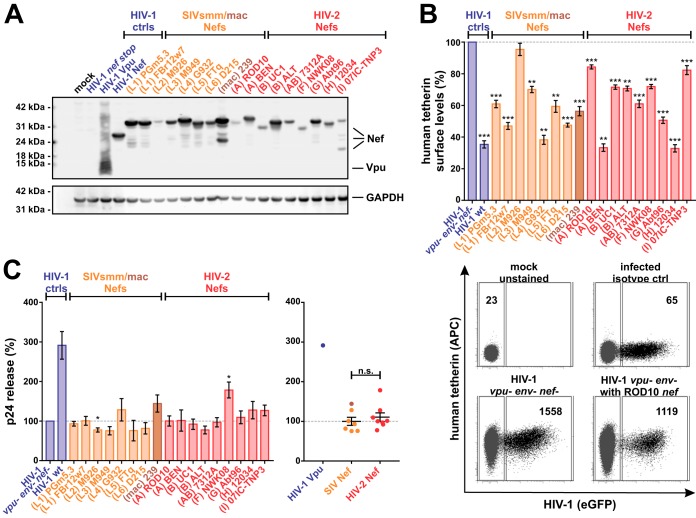
FIG 4.
Expression of Nef and Nef-mediated counteraction of human tetherin in primary cells. (A) HEK293T cells were transfected with expression plasmids for the indicated C-terminally AU1-tagged Vpu and Nef proteins. Two days posttransfection, cells were lysed and viral protein expression was monitored by Western blotting using an anti-AU1 antibody. GAPDH served as a loading control. (B) Human PBMCs were infected with HIV-1 NL4-3-derived viruses expressing the indicated Nef proteins and analyzed by flow cytometry as described for Fig. 3. The top panel shows mean values from 3 to 12 independent experiments ± SEM. Examples of primary data are shown in the bottom panel. (C) In parallel, p24 contents in the cells and cell culture supernatants were determined by ELISA 3 days postinfection. Relative p24 release from PBMCs was calculated by normalizing the amount of cell-free p24 to that of cell-associated p24. The values of the negative control (vpu- env- nef-) were set to 100%. Mean values from 4 to 12 independent experiments ± SEM are shown on the left. On the right, samples were grouped into SIVsmm/mac and HIV-2 Nefs. n.s., not significant. Asterisks in panels B and C indicate statistically significant differences compared to the negative control (vpu- env- nef-) (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Nef proteins of different groups of HIV-2 fail to efficiently antagonize human tetherin.
To elucidate the role of tetherin in the evolution of HIV-2, we tested the ability of SIVsmm and HIV-2 Nefs to decrease the surface expression of this innate antiviral protein (Fig. 4B). Surprisingly, both SIVsmm and HIV-2 Nefs decreased tetherin surface levels of infected PBMCs by about 20 to 60%, although human tetherin has previously been shown to be resistant to SIV Nefs (25, 26, 28). However, Nef may also indirectly affect tetherin expression, e.g., by suppressing T cell activation via downmodulation of CD3/CD28 (31) or by exerting general effects on cellular trafficking. Furthermore, tetherin surface levels do not always correlate with progeny virus release (71). We therefore also determined relative virus release by quantifying p24 contents in the cells and culture supernatants (Fig. 4C, left). While HIV-1 M Vpu increased virion release by more than 3-fold, SIV and HIV-2 Nefs had no or only marginal effects. On average, there was no significant difference between SIVsmm/mac and HIV-2 Nefs (Fig. 4C, right), arguing against a specific adaptation of Nef to human tetherin.
To directly compare the counteraction of human and sooty mangabey tetherin, we monitored infectious virus yield in HEK293T cells transfected with increasing amounts of the respective tetherin expression plasmid. Both human and sooty mangabey tetherin were expressed to similar levels (Fig. 5A and B). In agreement with the data obtained in primary cells, HIV-1 M Vpu efficiently rescued infectious virus production in the presence of human tetherin, whereas SIVsmm/mac and HIV-2 Nefs had no or (in the case of HIV-2 BEN and ALT) modest effects (Fig. 5C). Surprisingly, the effects of SIV and HIV-2 Nefs on infectious virus production in the presence of sooty mangabey tetherin were generally weak (Fig. 5D). On average, however, SIVsmm/mac Nefs significantly increased infectious virus yield by counteracting the tetherin ortholog from their original sooty mangabey host, while this was not the case for HIV-2 Nefs (Fig. 5E). To exclude a possible bias by effects of Nef on virion infectivity, virus release was also quantified by enzyme-linked immunosorbent assay (ELISA). Normalization of p24 amounts in the cell culture supernatant to the amount of cell-associated p24 confirmed that SIVsmm Nefs significantly increase virion release in the presence of sooty mangabey tetherin but were not or only poorly active against human tetherin (Fig. 5F). Again, most HIV-2 Nefs did not antagonize the human ortholog of tetherin and had little, if any, effect on the release of newly formed virions. Only HIV-2 BEN and Abt96 Nef significantly enhanced viral egress from transfected HEK293T cells. However, these two Nef proteins had no effect on virus release in infected primary cells (Fig. 4C and 5F). In summary, these results confirm that human tetherin is largely resistant to SIV Nef proteins (26, 29, 35, 72) and show that HIV-2 Nefs have not evolved efficient activity against human tetherin.
FIG 5.
Nef-mediated counteraction of human and sooty mangabey tetherin in HEK293T cells. (A and B) HEK293T cells were transfected with expression plasmids for human or sooty mangabey tetherin. Two days posttransfection, cell surface levels (A) and total protein levels (B) of tetherin were determined by flow cytometry and Western blotting, respectively. (C to E) HEK293T cells were cotransfected with HIV-1 vpu- env- IRES eGFP reporter constructs expressing the indicated Nef proteins and increasing amounts of expression plasmids for human (C) or sooty mangabey (D) tetherin. Two days posttransfection, infectious virus yield was determined by infection of TZM-bl reporter cells and normalized to the control sample without tetherin. Mean values from 3 to 11 independent experiments performed in triplicate ± SEM are shown. (E) The area under the curve (AUC) of the titrations shown in panels C and D was calculated, and samples were grouped into SIVsmm/mac and HIV-2 Nefs. (F) HEK293T cells were transfected as described for panels C and D. However, a fixed dose of 37 ng was used for the tetherin expression plasmid. Two days posttransfection, p24 concentrations in the cells and cell culture supernatants were determined by ELISA. Relative p24 release from HEK293T cells was calculated by normalizing the amount of cell-free p24 to that of cell-associated p24. Mean values from 4 to 11 independent experiments ± SEM are shown on the left. On the right, samples were grouped into SIVsmm/mac and HIV-2 Nefs. Asterisks in panels E and F indicate statistically significant differences compared to the negative control (vpu- env- nef-) (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
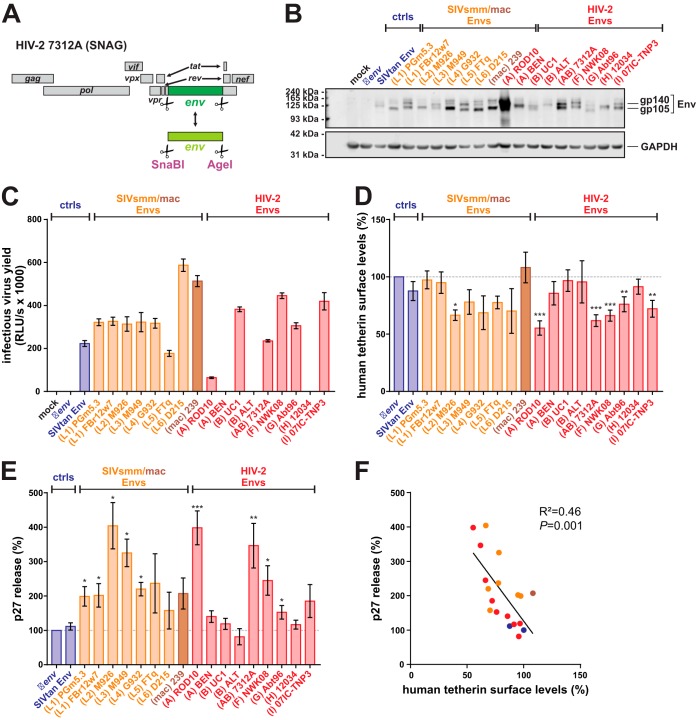
SIVsmm and HIV-2 group A, F, I, and CRF01_AB Env counteract human tetherin.
Since the Env proteins of some HIV-2 group A strains exert anti-tetherin activity in human cells (36–38), we tested whether other groups of HIV-2 are also capable of Env-mediated tetherin counteraction. To analyze the effects of Env on tetherin restriction in infected cells, we inserted heterologous HIV-2 and SIV env sequences into the proviral HIV-2 clone 7312A (Fig. 6A). The env gene of SIVtan, infecting tantalus monkeys (Chlorocebus tantalus), was included as a positive control, as it efficiently antagonizes human and sooty mangabey tetherin (73). To prevent concomitant alterations of tat, rev, and nef genes, only the sequences encoding the transmembrane and extracellular domains of Env were exchanged. The latter has previously been shown to mediate tetherin binding and, thus, counteraction of this restriction factor (74). All chimeric viruses expressed Env proteins of the expected size (Fig. 6B), and most of them mediated infection of TZM-bl reporter cells (Fig. 6C). Exceptions were the constructs expressing HIV-2 A BEN, B ALT, and H 12034 Env, which required VSV-G pseudotyping for infection (Fig. 6C and data not shown). Flow cytometric analysis of PBMCs infected with VSV-G-pseudotyped HIV-2 chimeras revealed that not only HIV-2 A Env but also the envelope proteins of HIV-2 groups F, G, and I, the CRF01_AB recombinant, and SIVsmm M926 significantly decreased tetherin surface levels by 25 to 45% (Fig. 6D). Unexpectedly, the SIVtan Env chimera had no significant effect on tetherin surface levels, possibly because HIV-2 and SIVtan Env are too diverse to form a chimeric protein maintaining its anti-tetherin activity. To investigate whether the observed reductions of tetherin surface levels by some Env proteins translate into increased virus release, we determined relative p27 release from PBMCs by ELISA. Here, the chimeras involving Env proteins of HIV-2 groups A, F, G, and I and the CRF01_AB recombinant increased virion release by about 2- to 4-fold (Fig. 6E). Intriguingly, SIVsmm Envs representing lineages 1 to 5 showed similar enhancing effects (Fig. 6E). This comprises SIVsmm alleles never passaged in human cells, suggesting that some SIVsmm Envs counteract human tetherin in primary target cells without prior adaptation. Overall, Env-mediated enhancement of virion release correlated well with the reduction of tetherin surface levels (Fig. 6F).
FIG 6.
Env-mediated counteraction of human tetherin in primary PBMCs. (A) Genome organization of the SNAG variant of HIV-2 7312A, containing unique SnaBI/AgeI sites that enable the exchange of a large fragment of env. (B) HEK293T cells were transfected with proviral constructs expressing the indicated Env chimeras. Two days posttransfection, cells were lysed and viral protein expression was monitored by Western blotting using an anti-HIV-2 Env antiserum. GAPDH served as a loading control. (C) Infectivity of chimeric HIV-2 7312A constructs expressing the indicated Env proteins was determined by infecting TZM-bl reporter cells with the supernatants of transfected HEK293T cells. One representative experiment, measured in triplicates ± SEM, is shown. (D and E) Tetherin surface levels (D) and relative p27 release (E) in infected human PBMCs were determined 3 days postinfection, as described for Fig. 4B and C. Mean values from 3 to 11 independent experiments ± SEM are shown. (F) Correlation of tetherin surface levels shown in panel D and relative p27 release shown in panel E. Asterisks indicate statistically significant differences compared to the Δenv negative control (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
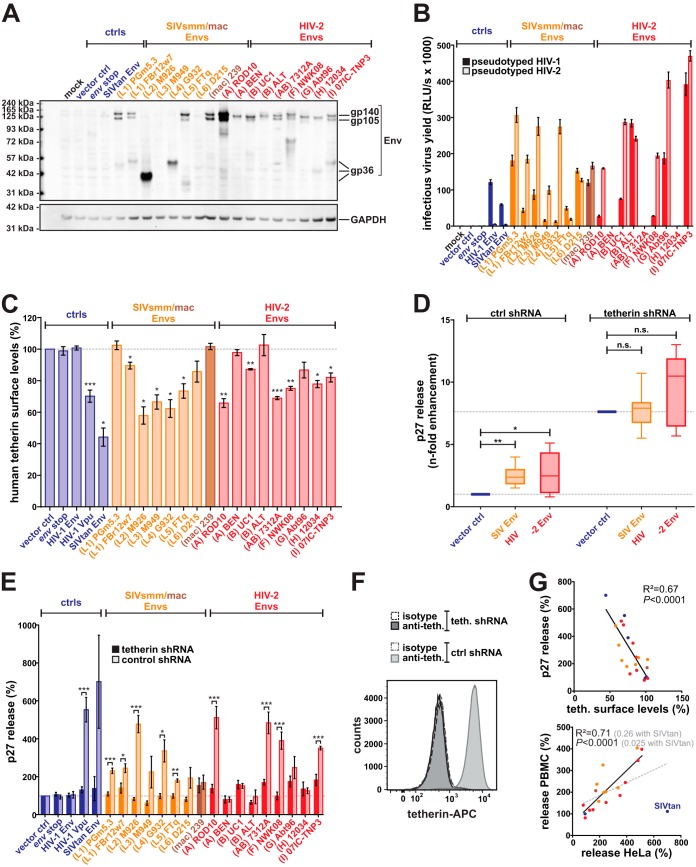
To test whether exchange of the N and C termini altered the anti-tetherin activities of SIV and HIV-2 Envs in our chimeric viruses, we also analyzed the effects of the respective full-length Env proteins on tetherin surface levels and virus release. Most full-length SIV and HIV-2 Env proteins expressed from pCAGGS vectors were detectable by Western blotting (Fig. 7A). Notably, differences in detection do not necessarily reflect different expression levels but may also be attributable to the HIV-2 gp105-specific antiserum used for detection. With the exception of HIV-2 A BEN, B ALT, and H 12034 Env, all envelope proteins mediated infection of env-deficient HIV-1 and/or HIV-2 (Fig. 7B). Experiments in transfected HeLa cells endogenously expressing tetherin revealed that not only Env proteins of rare and epidemic groups of HIV-2 but also those of SIVsmm lineages 1 to 5 downmodulate human tetherin about as efficiently as HIV-1 M Vpu (Fig. 7C). In agreement with a study by Towers and colleagues (73), SIVtan Env also efficiently decreased tetherin protein levels in this experimental setup. This supports the hypothesis that fusion to an only distantly related HIV-2 Env abrogated anti-tetherin activity of SIVtan Env in the chimeric viruses analyzed in infected PBMCs. The observed Env-mediated reductions in tetherin surface levels were accompanied by a 2- to 5-fold increase in virion release (Fig. 7D and E). Notably, increased virion release was a direct result of Env-mediated tetherin counteraction, as the enhancing effect was largely lost upon knockdown of tetherin (Fig. 7D and E). Env-mediated enhancement of virion release in HeLa cells correlated well with the reduction of tetherin surface levels (Fig. 7G, upper) and enhancement of virion release in primary cells (Fig. 7G, lower). In summary, these findings demonstrate that not only Env proteins of epidemic HIV-2 group A viruses but also those of rare groups F and I, the circulating recombinant form CRF01_AB, as well as several primary SIVsmm isolates counteract restriction by human tetherin.
FIG 7.
Counteraction of human tetherin by full-length SIVsmm/mac and HIV-2 Env proteins. (A) HEK293T cells were transfected with pCAGGS expression plasmids for the indicated Env proteins. Two days posttransfection, cells were lysed and Env expression was monitored by Western blotting using an anti-HIV-2 Env antiserum. GAPDH served as a loading control. (B) Env functionality was determined by infection of TZM-bl reporter cells with pseudotyped env-deficient mutants of HIV-1 M NL4-3 or HIV-2 AB 7312A. One representative experiment, measured in triplicates ± SEM, is shown. (C) Tetherin surface levels of HeLa cells transiently expressing the indicated Env proteins and an env-deficient mutant of HIV-2 7312A. Flow cytometry was performed 2 days posttransfection, and the mean fluorescence intensity of the tetherin staining in Env-expressing cells was normalized to that of the empty vector control. Mean values from 3 to 6 independent experiments ± SEM are shown. (D and E) Relative p27 release from tetherin knockdown HeLa cells and the respective control cell line expressing scrambled short hairpin RNA (shRNA). HeLa cells were cotransfected with an env-deficient mutant of HIV-2 7312A and expression plasmids for the indicated Env proteins. Two days posttransfection, p27 concentrations were determined by ELISA, and relative p27 release was calculated by normalizing the amount of cell-free p27 to that of cell-associated p27. Mean values from 3 to 8 independent experiments ± SEM are shown. In panel D, values were normalized to control cells expressing tetherin but no antagonist. In panel E, the vector controls of both tetherin-expressing and tetherin knockdown cells was set to 100%. Asterisks in panels C, D, and E indicate statistically significant differences compared to the vector control (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). (F) Flow cytometric analysis of HeLa cells stably expressing scrambled (ctrl.) or tetherin-specific (teth.) shRNA. Cells were either stained with a tetherin-specific APC-conjugated antibody (anti-teth.) or the respective isotype control. (G) The top panel illustrates the correlation of tetherin surface levels shown in panel C and relative p27 release shown in panel E. In the lower panel, p27 release from HeLa cells was correlated with p27 release from PBMCs (Fig. 6E). As fusion of SIVtan Env to HIV-2 7312A resulted in a loss of anti-tetherin activity, correlation with (gray) and without (black) SIVtan Env is shown.
Determinants of anti-tetherin activity map to the central part of SIVsmm Env.
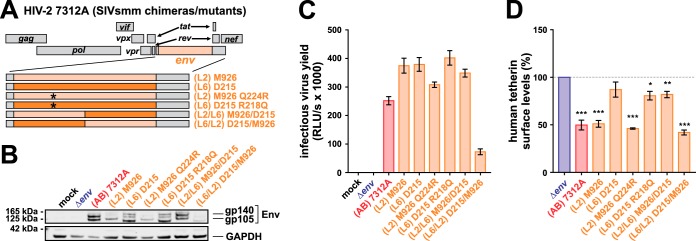
Comparing Env protein sequences, we found that SIVsmm D215 Env, which failed to significantly antagonize tetherin (Fig. 6E and 7C and E), harbors an arginine residue at position 218, whereas all active SIVsmm Envs harbor a conserved glutamine residue at this site. Intriguingly, an arginine-to-glutamine exchange at this position also emerged as an adaptive mutation of SIVsmm to human cells during in vivo passaging in mice (75). We therefore hypothesized that Q218 is involved in Env-mediated counteraction of human tetherin and introduced this residue in the inactive (L6) D215 Env (Fig. 8A). Although the respective mutant was expressed from the HIV-2 7312A backbone and able to mediate infection (Fig. 8B and C), the R218Q exchange did not increase the ability of SIVsmm D215 Env to downmodulate human tetherin (Fig. 8D). Furthermore, the inverse mutation (Q224R) did not result in a loss of tetherin antagonism in SIVsmm M926 Env (Fig. 8D), demonstrating that an R/Q change at this position is not associated with increased activity against human tetherin. To narrow down the determinants of tetherin counteraction in SIVsmm Env, we generated chimeras comprising domains of the active M926 and inactive D215 isolates (Fig. 8A). While an M926 chimera harboring the N-terminal half of the D215 ectodomain was still as active as wild-type M926, the exchange of the central part of Env abrogated the ability of M926 Env to downmodulate human tetherin from the surface of transfected HeLa cells (Fig. 8D). Vice versa, introduction of the central domain of M926 Env conferred full anti-tetherin activity to the inactive D215 isolate. This demonstrates that residues in the central part of Env, comprising the membrane-proximal ectodomain and the transmembrane domain, are important determinants of tetherin downmodulation by SIVsmm Env.
FIG 8.
Mapping of the anti-tetherin activity in SIVsmm Env. (A) Genome organization of HIV-2 7312A constructs encoding mutated or chimeric Env proteins. Asterisks illustrate point mutations that were introduced in the N-terminal half of Env. (B) HEK293T cells were transfected with proviral constructs expressing the indicated Env chimeras/mutants. Two days posttransfection, cells were lysed and viral protein expression was monitored by Western blotting using an anti-HIV-2 Env antiserum. GAPDH served as a loading control. (C) Infectivity of chimeric HIV-2 7312A constructs expressing the indicated Env proteins was determined by infecting TZM-bl reporter cells with the supernatants of transfected HEK293T cells. One representative experiment, measured in triplicates ± SEM, is shown. (D) Tetherin surface levels of HeLa cells transfected with proviral constructs expressing the indicated Env chimeras/mutants. Flow cytometry was performed 2 days posttransfection, and the mean fluorescence intensity of the tetherin staining in Env-expressing cells was normalized to that of the empty vector control. Mean values from 5 independent experiments ± SEM are shown. Asterisks indicate statistically significant differences compared to mock-transfected cells (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
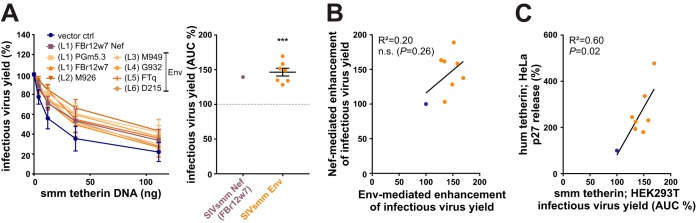
SIVsmm uses both Nef and Env to counteract sooty mangabey tetherin.
Quantification of infectious virus yield in the presence of sooty mangabey tetherin revealed that the anti-tetherin activity of SIVsmm Envs is not limited to the human ortholog, as all SIVsmm Env proteins also counteracted smm tetherin (Fig. 9A, left). On average, SIVsmm Envs increased infectious HIV-2 production by about 50% (Fig. 9A, right). This enhancing effect is similar to that observed in the presence of SIVsmm Nefs (Fig. 5E). Although some SIVsmm Nefs [e.g., (L3) M949] were rather poor antagonists of sooty mangabey tetherin (Fig. 5D to F), we found no evidence for compensation via Env when correlating Nef- and Env-mediated counteraction of smm tetherin (Fig. 9B). However, SIVsmm Env-mediated counteraction of sooty mangabey tetherin correlated well with its ability to antagonize human tetherin (Fig. 9C). Overall, these findings demonstrate that many primary SIVsmm isolates use both Nef and Env to antagonize tetherin in their natural monkey host.
FIG 9.
Counteraction of sooty mangabey tetherin by SIVsmm Env proteins. (A) HEK293T cells were cotransfected with HIV-2 (AB) 7312A Δenv, expression plasmids for VSV-G, the indicated SIVsmm Nef or Env proteins, and increasing amounts of vectors expressing sooty mangabey tetherin. Two days posttransfection, infectious virus yield was determined by infection of TZM-bl reporter cells and normalized to the control sample without tetherin. Mean values from 4 independent experiments performed in triplicate ± SEM are shown on the left. On the right, the area under the curve (AUC) was calculated and normalized to the vector control. Asterisks indicate statistically significant differences compared to the vector control (***, P ≤ 0.001). (B) Correlation analysis of SIVsmm Nef- and Env-mediated enhancement of infectious virus release in the presence of sooty mangabey tetherin (AUC %), as shown in Fig. 5E and panel A of this figure, respectively. (C) Correlation analysis of SIVsmm-mediated counteraction of human and sooty mangabey tetherin as described in Fig. 7E and panel A of this figure, respectively. In panels B and C, the empty vector control and SIVsmm Env/Nef samples are shown in blue and orange, respectively.
DISCUSSION
Although SIVs have been identified in more than 40 African primate species (76), only Central chimpanzees (Pan troglodytes troglodytes), Western lowland gorillas (Gorilla gorilla gorilla), and sooty mangabeys (Cercocebus atys) successfully transmitted their viruses to humans (77). One hurdle that SIVs have to overcome in order to spread efficiently in the human population is the host restriction factor tetherin, which is often antagonized in a species-specific manner (35). Here, we show that SIVsmm, the direct precursor of HIV-2, has a selection advantage upon transmission to humans, as it is able to antagonize the human tetherin ortholog in the absence of human-specific adaptive changes. Our data demonstrate that SIVsmm encodes two tetherin antagonists, i.e., Nef and Env. While the human tetherin ortholog is resistant against SIVsmm Nef (25, 28), it is susceptible to counteraction by SIVsmm Env. Thus, Env-mediated anti-tetherin activity in SIVsmm may have facilitated zoonotic transmission of this virus and help to explain why it crossed the monkey-human barrier on at least nine independent occasions, giving rise to HIV-2 groups A to I (11). This is in stark contrast to SIVcpz and SIVgor, which lack Env-mediated anti-tetherin activity and had to adapt their Vpu and Nef proteins to enable efficient spread of HIV-1 in the human population (26, 31, 35).
Notably, most SIVsmm Env proteins analyzed in this study represent primary isolates from evolutionarily diverse lineages (78, 79) (Table 1), demonstrating that Env-mediated tetherin counteraction is not an artifact of virus passaging in vitro but widespread in SIVsmm in vivo. In agreement with a preadaptation of SIVsmm Env to human tetherin, we also show that not only epidemic HIV-2 group A but also rare groups F and I use their Env protein to antagonize human tetherin. Furthermore, Env-mediated anti-tetherin activity was also detected in a circulating recombinant form (CRF01_AB), which expresses a chimeric envelope protein derived from HIV-2 groups A and B (12). Interestingly, we found no evidence for further adaption of Env to human tetherin, as SIVsmm and HIV-2 Envs were, on average, equally active in downmodulating human tetherin from the cell surface and enhancing virion release. In fact, the Env protein of the primary SIVsmm isolate M926 antagonized human tetherin as efficiently as the most potent HIV-2 Envs (i.e., ROD10 and 7312A). Thus, SIVsmm may already optimally counteract human tetherin, and/or further adaptation may not be possible due to functional and structural constraints in the viral envelope protein.
Several studies have mapped residues in HIV-2 group A Env proteins that are essential for efficient counteraction of human tetherin. These comprise a tyrosine-based endocytosis motif in the cytoplasmic domain, as well as several residues in the extracellular domain (i.e., K422, I568, A598, and N659) (36, 38, 66, 67, 80, 81). Most of them, including N659, which is required for direct interaction of Env with human tetherin (67), are already present in SIVsmm Envs and conserved among different groups of HIV-2. The only exception is K422, which is absent from rare HIV-2 groups F, G, H, and I as well as from SIVsmm. Notably, however, some Env proteins efficiently antagonize tetherin in the absence of K422, suggesting that this particular amino acid is only important in the context of certain Env backbones.
In addition to mutational analyses, passaging experiments were performed to identify adaptive changes that may confer anti-tetherin activity to SIVsmm, SIVmac, or HIV-2 Env. For example, a nef-deficient mutant of SIVmac and the neurotropic SIVsmm clone E543 accumulated mutations in the Env cytoplasmic domain during in vivo passage in macaques that enabled the virus to efficiently counteract macaque tetherin (82, 83). Similarly, Exline and colleagues showed that the inactive HIV-2 ROD14 Env acquired the ability to antagonize tetherin via two mutations (K796R and D830G) in its intracellular tail upon replication in JLTRG reporter cells (84). While G830 is only found in ROD10 Env, R796 is present in almost all SIVsmm and HIV-2 Env isolates analyzed in the present study. Only the envelope protein of HIV-2 H 12034, which was inactive in all assays, harbors a histidine at this position. To recapitulate the adaptive changes during the emergence of HIV-2, Schmitt and colleagues recently passaged an SIVsmm clone in humanized mice (75). After five serial passages, the virus replicated more efficiently in human cells than the parental SIVsmm strain, and the authors identified several fixed mutations in the nef and env genes. Here, we show that the Env R222Q exchange identified in this study is not associated with increased counteraction of human tetherin. In agreement with this, chimeric Env constructs revealed that important determinants of SIVsmm Env-mediated tetherin counteraction are located further downstream, in the central domain of Env. However, further mutational analyses are required to map the exact residues in the transmembrane domain and/or membrane-proximal ectodomain that confer anti-tetherin activity to SIVsmm Env.
Efficient tetherin antagonism may increase viral replicative fitness by several means. First, removal of tetherin from the sites of virion budding enables efficient progeny virus release and spread of infection within and between individuals (23, 24). Second, it prevents the accumulation of tethered virions at the cell surface that may render infected cells more susceptible to antibody-dependent cell-mediated cytotoxicity (ADCC) (85, 86). Finally, tetherin counteraction limits the induction of an antiviral immune response, since tetherin also acts as a pattern recognition receptor that activates NF-κB-dependent gene expression upon sensing of budding virions (87). In agreement with this multilayered antiviral activity, we have recently shown that tetherin is a major component of the interferon (IFN)-induced immune response against viruses and that its counteraction is especially important during acute infection, when IFN levels are particularly high (88, 89). The selection advantage of an efficient tetherin antagonist is also supported by the finding that Vpu-mediated tetherin counteraction is highly conserved among primary HIV-1 group M isolates throughout the course of infection (90). Thus, it may come as a surprise that several HIV-2 Envs analyzed in the present study (including both isolates of epidemic group B) showed no or only marginal activity against human tetherin. This is even more surprising as inactive Env proteins may act in a transdominant manner and prevent tetherin counteraction by otherwise active Env proteins (66). Notably, however, a previous study showed that only about 50% of all primary HIV-2 group A isolates encode an Env protein that antagonizes this restriction factor (38). Thus, although Env-mediated anti-tetherin activity can be found in different groups of HIV-2, it may be less conserved than Vpu-mediated tetherin counteraction in HIV-1 group M. One likely explanation is the accessibility and antigenicity of the viral envelope protein. Env is a highly variable protein that acquires mutations to evade recognition by humoral and cellular immune responses. In some cases, these evasion mutations may come at the cost of reduced anti-tetherin activity. Nevertheless, many other viruses, including Ebola, Marburg, influenza, equine infectious anemia, herpes simplex, feline immunodeficiency, and human endogenous retroviruses, also use their envelope glycoproteins to counteract tetherin-mediated restriction (91–97).
It has been suggested that efficient tetherin counteraction is crucial for (cell-free) virus transmission but less important for viral spread within an infected individual when tetherin restriction can be overcome by direct cell-to-cell spread (98, 99). Thus, the poor conservation of Env-mediated anti-tetherin activity in HIV-2 may also explain why this virus is, on average, transmitted less efficiently than HIV-1 (100). In agreement with a key role of tetherin in viral transmission, previous studies suggested that differences in anti-tetherin activity contribute to the differential spread and prevalence of HIV-1 groups M, N, O, and P. While pandemic group M and epidemic group O viruses evolved potent anti-tetherin activity, rare group N and P strains fail to efficiently antagonize the human tetherin ortholog (26, 30–33, 35). In contrast, we show here that lack of anti-tetherin activity does not explain the differential spread of the various groups of HIV-2 in the human population, since the Env proteins of rare HIV-2 groups F and I efficiently counteract human tetherin.
Besides tetherin counteraction, Nef-mediated antagonism of the host restriction factor SERINC5 has also been shown to correlate with the prevalence of primate lentiviruses (101). However, epidemic and rare groups of HIV-2 counteracted SERINC5 with similar efficiencies (101), suggesting that other viral or environmental factors determined the poor spread of rare HIV-2 groups. For example, the time point of zoonotic spillover is not known for most groups of HIV-2 (Table 1), and low prevalence rates may simply stem from later emergence in the human population.
Here, we further show that Nef-mediated downmodulation of the T cell receptor (TCR) CD3 and the primary receptor CD4 is highly conserved among different lineages of SIVsmm and different groups of HIV-2. In contrast, Nef-mediated downmodulation of the CD3 costimulator CD28 was less well conserved, probably because efficient downmodulation of CD3 already potently blocks TCR signaling, thereby reducing the selection pressure to also downmodulate CD28. Interestingly, HIV-2 Nefs decreased CD28 less efficiently on average than their SIVsmm counterparts, a finding that is in agreement with a previous study (39). Similar to CD28, the ability to modulate surface levels of MHC-I and the MHC-II-associated invariant chain (CD74) varied substantially among different SIVsmm and HIV-2 isolates. These variations may reflect specific adaptations to an infected individual, since the selection pressure to modulate antigen presentation strongly depends on the MHC genotype and the viral epitopes presented.
In summary, our findings show that SIVsmm Env and Nef proteins cooperate to counteract tetherin. Notably, this is not the only retrovirus encoding two tetherin antagonists. We have previously identified an HIV-1 group O strain that uses both Vpu and Nef to antagonize tetherin (34). Similarly, a chimpanzee-adapted HIV-1 strain evolved Vpu- and Nef-mediated anti-tetherin activity, with both proteins exerting additive effects (102). The expression of two tetherin antagonists may not only increase the efficacy of tetherin counteraction but also facilitate transmission to a new host species, as different antagonists target different domains of tetherin. If one viral protein (e.g., Nef) is inactive, the other one may still perform its function. In the case of SIVsmm, the evolution of two tetherin antagonists resulted in a preadaptation to humans that may have facilitated the emergence of different groups of HIV-2.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney 293T (HEK293T) and HeLa cells (obtained from the American Type Culture Collection [ATCC]) were first described by DuBridge et al. and Scherer et al., respectively (103, 104). The generation of tetherin knockdown HeLa cells (T3) and a corresponding control cell line (TR) has been previously described (105). TZM-bl cells are a HeLa-derived reporter cell line and were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from John C. Kappes, Xiaoyun Wu, and Tranzyme, Inc. (106). HEK293T, HeLa, and TZM-bl cells were authenticated by the ATCC or NIH and cultured in Dulbecco's modified Eagle medium (DMEM) containing 10% heat-inactivated fetal calf serum (FCS) plus 2 mM glutamine, 100 μg/ml streptomycin, and 100 U/ml penicillin. Human PBMCs were isolated using lymphocyte separation solution (Biochrom) and cultured in RPMI 1640 medium with 10% FCS, 2 mM glutamine, 100 μg/ml streptomycin, 100 U/ml penicillin, and 10 ng/ml interleukin 2 (IL-2). Three days before infection, PBMCs were activated using 1 μg/ml phytohemagglutinin (PHA).
Expression vectors and infectious molecular clones.
Using standard cloning techniques, lentiviral nef and vpu genes were C-terminally AU1 tagged and inserted into the cytomegalovirus immediate-early promoter-based pCG expression vector via XbaI/MluI. The pCG_nef and pCG_vpu vectors coexpress eGFP via an internal ribosome entry site (IRES). While HIV-2 NWK08 nef was directly amplified from patient DNA (5), SIVsmm FBr12w7, M926, M949, and G932 nef genes had been amplified by reverse transcription-PCR (RT-PCR) from the plasma of naturally infected sooty mangabeys, and FTq nef was isolated from the supernatant of an SIVsmm-infected sooty mangabey PBMC culture in previous studies (107, 108). HIV-2 ROD10, BEN, 7312A, SIVsmm PGm5.3, and SIVmac 239 nef genes, as well as HIV-1 NL4-3 nef and vpu, were amplified from previously described proviral constructs (41, 47, 50, 55, 109). All remaining nef genes (SIVsmm D215, HIV-2 UC1, ALT, Abt96, 12034, and 07IC-TNP3) were synthesized by GenScript (Piscataway), Biomatik (Cambridge, Canada), or BaseClear (Leiden) according to published sequences. Untagged nef genes were inserted in an HIV-1 NL4-3 proviral construct coexpressing eGFP via an IRES (110). Most env genes were synthesized according to published sequences (SIVsmm M926, M949, G932, FTq, D215, HIV-2 BEN, ALT, Abt96, 12034, and 07IC-TNP3). Ambiguous sites in the sequences of HIV-2 Abt96 and 07IC-TNP3 env and nef were replaced by the respective nucleotides found in the SIVsmm/HIV-2 consensus sequence. During synthesis, SnaBI and AgeI restriction sites were introduced in the env genes of SIVsmm M926, M949, G932, FTq, D215, HIV-2 BEN, ALT, and 07IC-TNP3 to facilitate insertion into the proviral HIV-2 7312A backbone. The remaining env genes were amplified from patient DNA (HIV-2 NWK08) (5), monkey plasma (SIVsmm FBr12w7) (42, 43), proviral constructs (HIV-2 ROD10, 7312A [SNAG variant], SIVsmm PGm5.3, and SIVmac 239) (41, 47, 55, 109), or expression plasmids (HIV-2 UC1 and SIVtan) (73, 111, 112). All of them were cloned into the pCAGGS expression vector via EcoRI/KpnI. An env stop control mutant was generated by removing a KpnI fragment from HIV-2 H 12034 env and reinserting it in the reverse orientation. env fragments encoding the entire envelope ectodomain and transmembrane domain (but not the N-terminal signal peptide and the cytoplasmic tail) were inserted into a previously described variant of pJK7312A (109). This variant, termed SNAG, facilitates the exchange of a large env fragment, as it contains unique SnaBI and AgeI restriction sites 69 nucleotides downstream of the env start codon and 36 nucleotides upstream of tat2-rev2, respectively. An env-defective version (termed NSMF) was generated by deleting small fragments in the env gene using NsiI and MfeI (113). Like the original pJK7312A construct, SNAG and NSMF were inserted into pBlueScript II KS+ and contain cellular sequences flanking the viral long terminal repeats (LTR). To facilitate cloning, internal KpnI, AgeI, EcoRI, and XbaI restriction sites were disrupted by introducing silent mutations in SIVsmm M949 env, G932 env, FTq env, and HIV-2 UC1 nef, respectively. Chimeras of SIVsmm M926 and D215 env were generated by overlap extension PCR (OE-PCR) and also inserted into SNAG via SnaBI and AgeI. Point mutations in SIVsmm M926 and D215 env were introduced by Q5 site-directed mutagenesis. Human and sooty mangabey tetherin-bst2 genes were expressed from pCG expression vectors (114). Of two known sooty mangabey tetherin variants, we used the sm1 clone (115). The pHIT/G vector expressing vesicular stomatitis virus glycoprotein (VSV-G) has been previously described (116). None of the cellular or viral genes was codon optimized.
Transfection.
HEK293T cells were transfected using a standard calcium phosphate method (5 to 6 μg DNA and 13 μl CaCl2 per well of a 6-well plate), and HeLa cells were transfected using linear polyethylenimine (3 μg DNA and 9 μg polyethylenimine per well of a 6-well plate) (117). Two days posttransfection, cells and supernatants were harvested for further analysis.
Western blotting.
To monitor expression of Vpu, Nef, and Env, HEK293T cells were transfected with pCG or pCAGGS expression plasmids or chimeric pBlueScript II KS+_7312A constructs. To compare expression of human and smm tetherin, HEK293T cells were transfected with increasing amounts of pCG expression plasmids (0, 12, 37, 111, and 333 ng). Two days posttransfection, cells were lysed in Western blot lysis buffer (150 mM NaCl, 50 mM HEPES, 5 mM EDTA, 0.1% [vol/vol] NP-40, 0.5 mM Na3VO4, 0.5 mM NaF, pH 7.5). After addition of 22.5% protein-loading dye (LI-COR) and 2.5% β-mercaptoethanol, samples were incubated at 95°C for 5 min, separated on a NuPAGE Bis-Tris 4 to 12% gradient gel (Invitrogen), and blotted onto an Immobilon-FL polyvinylidene difluoride (PVDF) membrane (Merck Millipore). The membrane was blocked in 5% bovine serum albumin (BSA) or 5% milk, and proteins were stained using primary antibodies directed against the AU1-tag (dilution, 1:1,000; NB600-453; Novus Biologicals), Env (dilution, 1:500; ARP418; J Cordell, CFAR), tetherin (dilution, 1:500; 16-3179-82; eBioscience), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; dilution, 1:500; sc-365062; Santa Cruz). After incubation with secondary antibodies conjugated with infrared dyes (dilution, 1:20,000; 925-32210 or 925-68071; LI-COR), proteins were detected using the infrared imager Odyssey 9120 (LI-COR) and Image Studio Lite, version 4.0 (LI-COR).
Flow cytometry.
To compare expression of human and smm tetherin, HEK293T cells were cotransfected with the respective pCG expression plasmids (37 ng) and an expression construct for eGFP (1 μg). HeLa cells endogenously expressing tetherin were transfected with proviral DNA (2 μg), pHIT/G (0.5 μg), and env expression vectors (0.5 μg). Two days posttransfection, cells were harvested and analyzed by flow cytometry. Surface levels of human and sooty mangabey tetherin in HEK293T cells were determined using a primary antibody against tetherin (13560-1-AP; Proteintech) and a secondary allophycocyanin (APC)-conjugated antibody (A-21245; Life Technologies). Surface expression of endogenous human tetherin in HeLa cells was determined using an APC-conjugated antibody (348410; BioLegend). Transfected cells were identified by eGFP reporter gene expression or by staining of p27 with a fluorescein isothiocyanate (FITC)-conjugated antibody against p24 Gag (6604665; Beckman Coulter). The mean fluorescence intensity (MFI) of the APC signal in transfected HeLa cells was normalized to that of the untransfected cells of the same sample. Further, the APC MFI of cells expressing Nef and/or Env was normalized to that of cells transfected with control plasmids.
PHA-activated PBMCs were infected with VSV-G-pseudotyped HIV-1 NL4-3 or HIV-2 7312A particles expressing heterologous Nef and Env proteins, respectively. Three days postinfection, cells were analyzed by flow cytometry. To this end, cells were stained for surface expression of CD4 (APC conjugated; MHCD0405; Invitrogen; or phycoerythrin [PE] conjugated; 555347; BD), CD3 (PE conjugated; 555333; BD), CD28 (APC conjugated; 559770; BD), MHC-I (APC conjugated; 311410; BioLegend), CD74 (PE conjugated; 226-050; Ancell), and tetherin (APC conjugated; 348410; BioLegend). HIV-1- and HIV-2-infected cells were identified by eGFP reporter gene expression and intracellular p27 staining (FITC conjugated; 6604665; Beckman Coulter). The MFI ratios of infected and uninfected cells were used to determine changes in protein surface levels. In all experiments, appropriate isotype control antibodies IgG1κ-APC (400122, MOPC-21; BioLegend), IgG1κ-PE (556027_51-35405X; MOPC-21; BD), IgG2α-APC (MG2A05; Invitrogen), IgG2α-PE (281-050; Ancell), and IgG2α-APC (400222; MOPC-173; BioLegend) were used to determine background fluorescence and unspecific antibody binding.
Infectivity assay.
To test functional expression of HIV and SIV Envs from the pCAGGS expression plasmids, a pseudotyping assay was performed. To this end, HEK293T cells were cotransfected with 1 μg of pCAGGS expression vector and 5 μg proviral DNA encoding env-defective mutants of HIV-1 NL4-3 or HIV-2 7312A. Infectivity of chimeric HIV-2 SNAG constructs was determined by transfecting HEK293T cells with 5 μg proviral DNA. Two days posttransfection, supernatants were harvested and used to infect TZM-bl reporter cells. Three days later, β-galactosidase activity was measured.
Virus release assay.
HEK293T cells were cotransfected with 4 μg proviral HIV-1 constructs encoding heterologous nef genes, 1 μg pHIT/VSV-G and increasing amounts of a tetherin expression construct (0, 12, 37, 111, and 333 ng) or 4 μg proviral HIV-2 constructs, 1 μg env expression plasmid, 0.5 μg pHIT/VSV-G, and increasing amounts of an smm tetherin expression construct (0, 4, 12, 37, and 111 ng). HeLa cells were transfected with 2 μg proviral DNA, 0.5 μg pHIT/VSV-G, and 0.5 μg env expression plasmid or 2.5 μg proviral HIV-2 constructs expressing heterologous env ectodomains. Two days posttransfection, supernatants and cells were lysed and a home-made p24 sandwich ELISA was performed. The antibodies used (anti-HIV-1 p24, clone 183-H12-5C; NIH; polyclonal anti-HIV-1 p24 rabbit antiserum, home-made; anti-rabbit, HRP-conjugated; 111-035-008; Dianova) detect both HIV-1 p24 and HIV-2 p27. Relative virus release was determined by normalizing the amount of p24/p27 in the culture supernatant to the amount of cell-associated p24/p27. Furthermore, infectious virus production was determined via infection of TZM-bl reporter cells and measurement of the β-galactosidase activity 3 days later. The home-made ELISA was also used to determine relative p24/p27 release from PBMCs 3 days postinfection.
Statistical analyses.
All statistical calculations were performed with a two-tailed unpaired Student's t test or a one-sample t test using GraphPad Prism 7. P values of ≤0.05 were considered significant. Correlations were calculated with the linear regression module.
Alignments.
Nef and Env protein sequence alignments were prepared using MultAlin (http://multalin.toulouse.inra.fr/multalin/) (118).
Ethical statement.
Experiments involving human peripheral blood mononuclear cells were reviewed and approved by the Institutional Review Board (i.e., the Ethics Committee of Ulm University), and individuals and/or their legal guardians provided written informed consent prior to donating blood. All blood samples were anonymized before use. No nonhuman primates were involved, harmed, sampled, or kept for this study.
Accession number(s).
The GenBank accession numbers of the nef and env sequences used in this study are FJ400519.1 (SIVsmm PGm5.3 nef), AF077017.1 (SIVsmm PGm5.3 env), KF477993 (SIVsmm FBr12w7 nef), KF477993 (SIVsmm FBr12w7 env), JX860420 (SIVsmm M926 env), JX860426 (SIVsmm M949 env), JX860416 (SIVsmm G932 env), JX860414 (SIVsmm FTq env), JX860413 (SIVsmm D215 nef), JX860413 (SIVsmm D215 env), FJ400519.1 (SIVmac 239 nef), AF077017.1 (SIVmac 239 env), KY272752 (HIV-2 ROD10 nef), M30502.1 (HIV-2 BEN env), L07625.1 (HIV-2 UC1 nef), L07625.1 (HIV-2 UC1 env), X61240.1 (HIV-2 ALT nef), X61240.1 (HIV-2 ALT env), L36874.1 (HIV-2 7312A nef), L36874.1 (HIV-2 7312A env), AF208027.1 (HIV-2 Abt96 nef), AF208027.1 (HIV-2 Abt96 env), AY530889.1 (HIV-2 96FR12034 nef), AY530889.1 (HIV-2 96FR12034 env), KC693505.1 (HIV-2 07IC-TNP3 nef), KC693505.1 (HIV-2 07IC-TNP3 env), U58991.1 (SIVtan env), KM390026.1 (HIV-1 NL4-3 nef), KM390026.1 (HIV-1 NL4-3 env), and KM390026.1 (HIV-1 NL4-3 vpu). The nef and env sequences determined in this study are available from GenBank under accession numbers MH541052 to MH541058.
ACKNOWLEDGMENTS
We thank Susanne Engelhart, Daniela Krnavek, Regina Linsenmeyer, Martha Mayer, and Kerstin Regensburger for excellent technical assistance. Beatrice H. Hahn kindly provided a pSM expression plasmid for HIV-2 B UC1 Env and the infectious molecular clones of HIV-2 7312A SNAG and NSMF (111–113). We also thank Todd A. Reinhart for providing a pGEX13 expression plasmid of HIV-2 B UC1 Nef (119) and Nicoletta Casartelli for providing tetherin knockdown HeLa cells (T3) and a corresponding control cell line (TR) (105). TZM-bl cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from John C. Kappes, Xiaoyun Wu, and Tranzyme, Inc. (106). Bruce Chesebro and Kathy Wehrly provided the HIV-1 p24 monoclonal antibody (183-H12-5C) through the NIH AIDS Reagent Program (120).
F.K. is supported by the DFG and the ERC. D.S. is supported by the Junior Professorship Programme of the State of Baden-Wuerttemberg, Germany, and the Baustein Programme of the Medical Faculty of Ulm University. E.H. is supported by the International Graduate School in Molecular Medicine Ulm. C.A. was supported by RO1 AI065325 (NIH-NIAID). P.S. is supported by RO1 AI119346 (NIH-NIAID).
The HIV-2 F NWK08 clone used in this study is cited in a U.S. patent (121).
REFERENCES
- 1.Visseaux B, Damond F, Matheron S, Descamps D, Charpentier C. 2016. Hiv-2 molecular epidemiology. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis 46:233–240. [DOI] [PubMed] [Google Scholar]
- 2.Ariën KK, Abraha A, Quiñones-Mateu ME, Kestens L, Vanham G, Arts EJ. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J Virol 79:8979–8990. doi: 10.1128/JVI.79.14.8979-8990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Cock KM, Adjorlolo G, Ekpini E, Sibailly T, Kouadio J, Maran M, Brattegaard K, Vetter KM, Doorly R, Gayle HD. 1993. Epidemiology and transmission of HIV-2: why there is no HIV-2 pandemic. JAMA 270:2083–2086. [DOI] [PubMed] [Google Scholar]
- 4.Gao F, Yue L, Robertson DL, Hill SC, Hui H, Biggar RJ, Neequaye AE, Whelan TM, Ho DD, Shaw GM. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol 68:7433–7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith SM, Christian D, de Lame V, Shah U, Austin L, Gautam R, Gautam A, Apetrei C, Marx PA. 2008. Isolation of a new HIV-2 group in the US. Retrovirology 5:103. doi: 10.1186/1742-4690-5-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Luckay A, Sodora DL, Telfer P, Reed P, Gettie A, Kanu JM, Sadek RF, Yee J, Ho DD, Zhang L, Marx PA. 1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J Virol 71:3953–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan CA, Yamaguchi J, Vallari AS, Hickman RK, Devare SG. 1997. Genetic variation in human immunodeficiency virus type 2: identification of a unique variant from human plasma. AIDS Res Hum Retrovir 13:401–404. doi: 10.1089/aid.1997.13.401. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi J, Devare SG, Brennan CA. 2000. Identification of a new HIV-2 subtype based on phylogenetic analysis of full-length genomic sequence. AIDS Res Hum Retrovir 16:925–930. doi: 10.1089/08892220050042864. [DOI] [PubMed] [Google Scholar]
- 9.Damond F, Worobey M, Campa P, Farfara I, Colin G, Matheron S, Brun-Vézinet F, Robertson DL, Simon F. 2004. Identification of a highly divergent HIV type 2 and proposal for a change in HIV type 2 classification. AIDS Res Hum Retrovir 20:666–672. doi: 10.1089/0889222041217392. [DOI] [PubMed] [Google Scholar]
- 10.Visseaux B, Hurtado-Nedelec M, Charpentier C, Collin G, Storto A, Matheron S, Larrouy L, Damond F, Brun-Vézinet F, Descamps D, ANRS CO 05 HIV-2 Cohort . 2012. Molecular determinants of HIV-2 R5-X4 tropism in the V3 loop: development of a new genotypic tool. J Infect Dis 205:111–120. doi: 10.1093/infdis/jir698. [DOI] [PubMed] [Google Scholar]
- 11.Ayouba A, Akoua-Koffi C, Calvignac-Spencer S, Esteban A, Locatelli S, Li H, Li Y, Hahn BH, Delaporte E, Leendertz FH, Peeters M. 2013. Evidence for continuing cross-species transmission of SIVsmm to humans: characterization of a new HIV-2 lineage in rural Côte d'Ivoire. AIDS 27:2488–2491. doi: 10.1097/01.aids.0000432443.22684.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibe S, Yokomaku Y, Shiino T, Tanaka R, Hattori J, Fujisaki S, Iwatani Y, Mamiya N, Utsumi M, Kato S, Hamaguchi M, Sugiura W. 2010. HIV-2 CRF01_AB: first circulating recombinant form of HIV-2. J Acquir Immune Defic Syndr 54:241–247. doi: 10.1097/QAI.0b013e3181dc98c1. [DOI] [PubMed] [Google Scholar]
- 13.Lemey P, Pybus OG, Wang B, Saksena NK, Salemi M, Vandamme A-M. 2003. Tracing the origin and history of the HIV-2 epidemic. Proc Natl Acad Sci U S A 100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao F, Yue L, White AT, Pappas PG, Barchue J, Hanson AP, Greene BM, Sharp PM, Shaw GM, Hahn BH. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature 358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi J, Vallari A, Ndembi N, Coffey R, Ngansop C, Mbanya D, Kaptué L, Gürtler LG, Devare SG, Brennan CA. 2008. HIV type 2 intergroup recombinant identified in Cameroon. AIDS Res Hum Retrovir 24:86–91. doi: 10.1089/aid.2007.0190. [DOI] [PubMed] [Google Scholar]
- 16.Ndembi N, Abraha A, Pilch H, Ichimura H, Mbanya D, Kaptue L, Salata R, Arts EJ. 2008. Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J Clin Microbiol 46:177–184. doi: 10.1128/JCM.00428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho DD, Lee S, Moudgil T, Kerndt PR. 1990. HIV-2 in Los Angeles. AIDS 4:1301–1302. doi: 10.1097/00002030-199012000-00028. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 19.Sousa JD, Temudo MP, Hewlett BS, Camacho RJ, Müller V, Vandamme A-M. 2016. Male circumcision and the epidemic emergence of HIV-2 in West Africa. PLoS One 11:e0166805. doi: 10.1371/journal.pone.0166805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Sousa JD, Alvarez C, Vandamme A-M, Müller V. 2012. Enhanced heterosexual transmission hypothesis for the origin of pandemic HIV-1. Viruses 4:1950–1983. doi: 10.3390/v4101950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluge SF, Sauter D, Kirchhoff F. 2015. SnapShot: antiviral restriction factors. Cell 163:774–774.e1. doi: 10.1016/j.cell.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Colomer-Lluch M, Gollahon LS, Serra-Moreno R. 2016. Anti-HIV factors: targeting each step of HIV's replication cycle. Curr HIV Res 14:175–182. doi: 10.2174/1570162X14999160224094621. [DOI] [PubMed] [Google Scholar]
- 23.Neil SJD, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 24.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauter D, Schindler M, Specht A, Landford WN, Münch J, Kim K-A, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang SJ, Lopez LA, Hauser H, Exline CM, Haworth KG, Cannon PM. 2010. Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 7:13. doi: 10.1186/1742-4690-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog 5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauter D, Vogl M, Kirchhoff F. 2011. Ancient origin of a deletion in human BST2/Tetherin that confers protection against viral zoonoses. Hum Mutat 32:1243–1245. doi: 10.1002/humu.21571. [DOI] [PubMed] [Google Scholar]
- 30.Yang SJ, Lopez LA, Exline CM, Haworth KG, Cannon PM. 2011. Lack of adaptation to human tetherin in HIV-1 group O and P. Retrovirology 8:78. doi: 10.1186/1742-4690-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kluge SF, Mack K, Iyer SS, Pujol FM, Heigele A, Learn GH, Usmani SM, Sauter D, Joas S, Hotter D, Bibollet-Ruche F, Plenderleith LJ, Peeters M, Geyer M, Sharp PM, Fackler OT, Hahn BH, Kirchhoff F. 2014. Nef proteins of epidemic HIV-1 group O strains antagonize human tetherin. Cell Host Microbe 16:639–650. doi: 10.1016/j.chom.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauter D, Unterweger D, Vogl M, Usmani SM, Heigele A, Kluge SF, Hermkes E, Moll M, Barker E, Peeters M, Learn GH, Bibollet-Ruche F, Fritz JV, Fackler OT, Hahn BH, Kirchhoff F. 2012. Human tetherin exerts strong selection pressure on the HIV-1 group N Vpu protein. PLoS Pathog 8:e1003093. doi: 10.1371/journal.ppat.1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauter D, Hué S, Petit SJ, Plantier J-C, Towers GJ, Kirchhoff F, Gupta RK. 2011. HIV-1 group P is unable to antagonize human tetherin by Vpu, Env or Nef. Retrovirology 8:103. doi: 10.1186/1742-4690-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mack K, Starz K, Sauter D, Langer S, Bibollet-Ruche F, Learn GH, Stürzel CM, Leoz M, Plantier J-C, Geyer M, Hahn BH, Kirchhoff F. 2017. Efficient Vpu-mediated tetherin antagonism by an HIV-1 group O strain. J Virol 91:e02177-16. doi: 10.1128/JVI.02177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauter D, Specht A, Kirchhoff F. 2010. Tetherin: holding on and letting go. Cell 141:392–398. doi: 10.1016/j.cell.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 36.Le Tortorec A, Neil SJD. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83:11966–11978. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauser H, Lopez LA, Yang SJ, Oldenburg JE, Exline CM, Guatelli JC, Cannon PM. 2010. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 7:51. doi: 10.1186/1742-4690-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C-Y, Shingai M, Welbourn S, Martin MA, Borrego P, Taveira N, Strebel K. 2016. Antagonism of BST-2/tetherin is a conserved function of the Env glycoprotein of primary HIV-2 isolates. J Virol 90:11062–11074. doi: 10.1128/JVI.01451-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindler M, Münch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Müller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, Bailes E, Roques P, Sodora DL, Silvestri G, Sharp PM, Hahn BH, Kirchhoff F. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 40.Khalid M, Yu H, Sauter D, Usmani SM, Schmokel J, Feldman J, Gruters RA, van der Ende ME, Geyer M, Rowland-Jones S, Osterhaus AD, Kirchhoff F. 2012. Efficient Nef-mediated downmodulation of TCR-CD3 and CD28 is associated with high CD4+ T cell counts in viremic HIV-2 infection. J Virol 86:4906–4920. doi: 10.1128/JVI.06856-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novembre FJ, De Rosayro J, O'Neil SP, Anderson DC, Klumpp SA, McClure HM. 1998. Isolation and characterization of a neuropathogenic simian immunodeficiency virus derived from a sooty mangabey. J Virol 72:8841–8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmökel J, Li H, Shabir A, Yu H, Geyer M, Silvestri G, Sodora DL, Hahn BH, Kirchhoff F. 2013. Link between primate lentiviral coreceptor usage and Nef function. Cell Rep 5:997–1009. doi: 10.1016/j.celrep.2013.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milush JM, Reeves JD, Gordon SN, Zhou D, Muthukumar A, Kosub DA, Chacko E, Giavedoni LD, Ibegbu CC, Cole KS, Miamidian JL, Paiardini M, Barry AP, Staprans SI, Silvestri G, Sodora DL. 2007. Virally induced CD4+ T cell depletion is not sufficient to induce AIDS in a natural host. J Immunol 179:3047–3056. [DOI] [PubMed] [Google Scholar]
- 44.Fischer W, Apetrei C, Santiago ML, Li Y, Gautam R, Pandrea I, Shaw GM, Hahn BH, Letvin NL, Nabel GJ, Korber BT. 2012. Distinct evolutionary pressures underlie diversity in simian immunodeficiency virus and human immunodeficiency virus lineages. J Virol 86:13217–13231. doi: 10.1128/JVI.01862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansfield KG, Lerch NW, Gardner MB, Lackner AA. 1995. Origins of simian immunodeficiency virus infection in macaques at the New England Regional Primate Research Center. J Med Primatol 24:116–122. doi: 10.1111/j.1600-0684.1995.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 46.Daniel MD, Letvin NL, King NW, Kannagi M, Sehgal PK, Hunt RD, Kanki PJ, Essex M, Desrosiers RC. 1985. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science 228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 47.Clavel F, Guyader M, Guétard D, Sallé M, Montagnier L, Alizon M. 1986. Molecular cloning and polymorphism of the human immune deficiency virus type 2. Nature 324:691–695. doi: 10.1038/324691a0. [DOI] [PubMed] [Google Scholar]
- 48.Ryan-Graham MA, Peden KW. 1995. Both virus and host components are important for the manifestation of a Nef- phenotype in HIV-1 and HIV-2. Virology 213:158–168. doi: 10.1006/viro.1995.1556. [DOI] [PubMed] [Google Scholar]
- 49.Clavel F, Guétard D, Brun-Vézinet F, Chamaret S, Rey MA, Santos-Ferreira MO, Laurent AG, Dauguet C, Katlama C, Rouzioux C. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233:343–346. doi: 10.1126/science.2425430. [DOI] [PubMed] [Google Scholar]
- 50.Kirchhoff F, Jentsch KD, Bachmann B, Stuke A, Laloux C, Lüke W, Stahl-Hennig C, Schneider J, Nieselt K, Eigen M. 1990. A novel proviral clone of HIV-2: biological and phylogenetic relationship to other primate immunodeficiency viruses. Virology 177:305–311. doi: 10.1016/0042-6822(90)90484-9. [DOI] [PubMed] [Google Scholar]
- 51.Klemm E, Schneweis KE, Horn R, Tackmann W, Schulze G, Schneider J. 1988. HIV-II infection with initial neurological manifestation. J Neurol 235:304–307. doi: 10.1007/BF00314179. [DOI] [PubMed] [Google Scholar]
- 52.Barnett SW, Quiroga M, Werner A, Dina D, Levy JA. 1993. Distinguishing features of an infectious molecular clone of the highly divergent and noncytopathic human immunodeficiency virus type 2 UC1 strain. J Virol 67:1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans LA, Moreau J, Odehouri K, Legg H, Barboza A, Cheng-Mayer C, Levy JA. 1988. Characterization of a noncytopathic HIV-2 strain with unusual effects on CD4 expression. Science 240:1522–1525. [DOI] [PubMed] [Google Scholar]
- 54.Kühnel H, Briesen H von, Dietrich U, Adamski M, Mix D, Biesert L, Kreutz R, Immelmann A, Henco K, Meichsner C. 1989. Molecular cloning of two west African human immunodeficiency virus type 2 isolates that replicate well in macrophages: a Gambian isolate, from a patient with neurologic acquired immunodeficiency syndrome, and a highly divergent Ghanian isolate. Proc Natl Acad Sci U S A 86:2383–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naidu YM, Kestler HW, Li Y, Butler CV, Silva DP, Schmidt DK, Troup CD, Sehgal PK, Sonigo P, Daniel MD. 1988. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol 62:4691–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geyer M, Fackler OT, Peterlin BM. 2001. Structure–function relationships in HIV-1 Nef. EMBO Rep 2:580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manrique S, Sauter D, Horenkamp FA, Lülf S, Yu H, Hotter D, Anand K, Kirchhoff F, Geyer M. 2017. Endocytic sorting motif interactions involved in Nef-mediated downmodulation of CD4 and CD3. Nat Commun 8:442. doi: 10.1038/s41467-017-00481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geyer M, Munte CE, Schorr J, Kellner R, Kalbitzer HR. 1999. Structure of the anchor-domain of myristoylated and non-myristoylated HIV-1 Nef protein. J Mol Biol 289:123–138. doi: 10.1006/jmbi.1999.2740. [DOI] [PubMed] [Google Scholar]
- 59.Saksela K, Cheng G, Baltimore D. 1995. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J 14:484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bresnahan PA, Yonemoto W, Ferrell S, Williams-Herman D, Geleziunas R, Greene WC. 1998. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr Biol 8:1235–1238. doi: 10.1016/S0960-9822(07)00517-9. [DOI] [PubMed] [Google Scholar]
- 61.Craig HM, Pandori MW, Guatelli JC. 1998. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc Natl Acad Sci U S A 95:11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iafrate AJ, Carl S, Bronson S, Stahl-Hennig C, Swigut T, Skowronski J, Kirchhoff F. 2000. Disrupting surfaces of nef required for downregulation of CD4 and for enhancement of virion infectivity attenuates simian immunodeficiency virus replication in vivo. J Virol 74:9836–9844. doi: 10.1128/JVI.74.21.9836-9844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 64.Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G. 1992. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem 267:16396–16402. [PubMed] [Google Scholar]
- 65.Rowell JF, Stanhope PE, Siliciano RF. 1995. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol 155:473–488. [PubMed] [Google Scholar]
- 66.Bour S, Akari H, Miyagi E, Strebel K. 2003. Naturally occurring amino acid substitutions in the HIV-2 ROD envelope glycoprotein regulate its ability to augment viral particle release. Virology 309:85–98. doi: 10.1016/S0042-6822(02)00128-9. [DOI] [PubMed] [Google Scholar]
- 67.Dufrasne FE, Lombard C, Goubau P, Ruelle J. 2016. Single amino acid substitution N659D in HIV-2 envelope glycoprotein (Env) impairs viral release and hampers BST-2 antagonism. Viruses 8:E285. doi: 10.3390/v8100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Howe AY, Jung JU, Desrosiers RC. 1998. Zeta chain of the T-cell receptor interacts with nef of simian immunodeficiency virus and human immunodeficiency virus type 2. J Virol 72:9827–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lama J, Mangasarian A, Trono D. 1999. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol 9:622–631. doi: 10.1016/S0960-9822(99)80284-X. [DOI] [PubMed] [Google Scholar]
- 70.Benson RE, Sanfridson A, Ottinger JS, Doyle C, Cullen BR. 1993. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J Exp Med 177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyagi E, Andrew AJ, Kao S, Strebel K. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface downmodulation and intracellular depletion. Proc Natl Acad Sci U S A 106:2868–2873. doi: 10.1073/pnas.0813223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmökel J, Sauter D, Schindler M, Leendertz FH, Bailes E, Dazza M-C, Saragosti S, Bibollet-Ruche F, Peeters M, Hahn BH, Kirchhoff F. 2011. The presence of a vpu gene and the lack of Nef-mediated downmodulation of T cell receptor-CD3 are not always linked in primate lentiviruses. J Virol 85:742–752. doi: 10.1128/JVI.02087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gupta RK, Mlcochova P, Pelchen-Matthews A, Petit SJ, Mattiuzzo G, Pillay D, Takeuchi Y, Marsh M, Towers GJ. 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc Natl Acad Sci U S A 106:20889–20894. doi: 10.1073/pnas.0907075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, Oldenburg J, Cannon PM. 2010. Ebola virus glycoprotein counteracts BST-2/tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J Virol 84:7243–7255. doi: 10.1128/JVI.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmitt K, Mohan Kumar D, Curlin J, Remling-Mulder L, Stenglein M, O'Connor S, Marx P, Akkina R. 2017. Modeling the evolution of SIV sooty mangabey progenitor virus towards HIV-2 using humanized mice. Virology 510:175–184. doi: 10.1016/j.virol.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aghokeng AF, Peeters M. 2005. Simian immunodeficiency viruses (SIVs) in Africa. J Neurovirol 11(Suppl 1):S27–S32. [PubMed] [Google Scholar]
- 77.Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, Falkenstein S, Bohm R, Koehler J, Traina-Dorge V, Williams T, Staprans S, Plauche G, Veazey RS, McClure H, Lackner AA, Gormus B, Robertson DL, Marx PA. 2005. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J Virol 79:8991–9005. doi: 10.1128/JVI.79.14.8991-9005.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Apetrei C, Gautam R, Sumpter B, Carter AC, Gaufin T, Staprans SI, Else J, Barnes M, Cao R, Garg S, Milush JM, Sodora DL, Pandrea I, Silvestri G. 2007. Virus subtype-specific features of natural simian immunodeficiency virus SIVsmm infection in sooty mangabeys. J Virol 81:7913–7923. doi: 10.1128/JVI.00281-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abada P, Noble B, Cannon PM. 2005. Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production. J Virol 79:3627–3638. doi: 10.1128/JVI.79.6.3627-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Noble B, Abada P, Nunez-Iglesias J, Cannon PM. 2006. Recruitment of the adaptor protein 2 complex by the human immunodeficiency virus type 2 envelope protein is necessary for high levels of virus release. J Virol 80:2924–2932. doi: 10.1128/JVI.80.6.2924-2932.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Serra-Moreno R, Jia B, Breed M, Alvarez X, Evans DT. 2011. Compensatory changes in the cytoplasmic tail of gp41 confer resistance to tetherin/BST-2 in a pathogenic nef-deleted SIV. Cell Host Microbe 9:46–57. doi: 10.1016/j.chom.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsuda K, Chen C-Y, Whitted S, Chertova E, Roser DJ, Wu F, Plishka RJ, Ourmanov I, Buckler-White A, Lifson JD, Strebel K, Hirsch VM. 2016. Enhanced antagonism of BST-2 by a neurovirulent SIV envelope. J Clin Investig 126:2295–2307. doi: 10.1172/JCI83725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Exline CM, Yang SJ, Haworth KG, Rengarajan S, Lopez LA, Droniou ME, Seclen E, Cannon PM. 2015. Determinants in HIV-2 Env and tetherin required for functional interaction. Retrovirology 12:67. doi: 10.1186/s12977-015-0194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arias JF, Heyer LN, von Bredow B, Weisgrau KL, Moldt B, Burton DR, Rakasz EG, Evans DT. 2014. Tetherin antagonism by Vpu protects HIV-infected cells from antibody-dependent cell-mediated cytotoxicity. Proc Natl Acad Sci U S A 111:6425–6430. doi: 10.1073/pnas.1321507111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alvarez RA, Hamlin RE, Monroe A, Moldt B, Hotta MT, Rodriguez Caprio G, Fierer DS, Simon V, Chen BK. 2014. HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J Virol 88:6031–6046. doi: 10.1128/JVI.00449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJD. 2012. Innate sensing of HIV-1 assembly by tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kmiec D, Iyer SS, Stürzel CM, Sauter D, Hahn BH, Kirchhoff F. 2016. Vpu-mediated counteraction of tetherin is a major determinant of HIV-1 interferon resistance. mBio 7:e00934-16. doi: 10.1128/mBio.00934-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yamada E, Nakaoka S, Klein L, Reith E, Langer S, Hopfensperger K, Iwami S, Schreiber G, Kirchhoff F, Koyanagi Y, Sauter D, Sato K. 2018. Human-specific adaptations in Vpu conferring anti-tetherin activity are critical for efficient early HIV-1 replication in vivo. Cell Host Microbe 23:110–120. doi: 10.1016/j.chom.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 90.Pickering S, Hué S, Kim E-Y, Reddy S, Wolinsky SM, Neil SJD. 2014. Preservation of tetherin and CD4 counter-activities in circulating Vpu alleles despite extensive sequence variation within HIV-1 infected individuals. PLoS Pathog 10:e1003895. doi: 10.1371/journal.ppat.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. 2009. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A 106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kühl A, Banning C, Marzi A, Votteler J, Steffen I, Bertram S, Glowacka I, Konrad A, Stürzl M, Guo J-T, Schubert U, Feldmann H, Behrens G, Schindler M, Pöhlmann S. 2011. The Ebola virus glycoprotein and HIV-1 Vpu employ different strategies to counteract the antiviral factor tetherin. J Infect Dis 204(Suppl 3):S850–S860. doi: 10.1093/infdis/jir378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gnirß K, Zmora P, Blazejewska P, Winkler M, Lins A, Nehlmeier I, Gärtner S, Moldenhauer A-S, Hofmann-Winkler H, Wolff T, Schindler M, Pöhlmann S. 2015. Tetherin sensitivity of influenza A viruses is strain specific: role of hemagglutinin and neuraminidase. J Virol 89:9178–9188. doi: 10.1128/JVI.00615-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morrison JH, Guevara RB, Marcano AC, Saenz DT, Fadel HJ, Rogstad DK, Poeschla EM. 2014. Feline immunodeficiency virus envelope glycoproteins antagonize tetherin through a distinctive mechanism that requires virion incorporation. J Virol 88:3255–3272. doi: 10.1128/JVI.03814-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lemaître C, Harper F, Pierron G, Heidmann T, Dewannieux M. 2014. The HERV-K human endogenous retrovirus envelope protein antagonizes Tetherin antiviral activity. J Virol 88:13626–13637. doi: 10.1128/JVI.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Y, Luo S, He S, Zhang M, Wang P, Li C, Huang W, Hu B, Griffin GE, Shattock RJ, Hu Q. 2015. Tetherin restricts HSV-2 release and is counteracted by multiple viral glycoproteins. Virology 475:96–109. doi: 10.1016/j.virol.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 97.Yin X, Hu Z, Gu Q, Wu X, Zheng Y-H, Wei P, Wang X. 2014. Equine tetherin blocks retrovirus release and its activity is antagonized by equine infectious anemia virus envelope protein. J Virol 88:1259–1270. doi: 10.1128/JVI.03148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jolly C, Booth NJ, Neil SJD. 2010. Cell-cell spread of human immunodeficiency virus type 1 overcomes tetherin/BST-2-mediated restriction in T cells. J Virol 84:12185–12199. doi: 10.1128/JVI.01447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liang Z, Zhang Y, Song J, Zhang H, Zhang S, Li Y, Tan J, Qiao W. 2017. The effect of bovine BST2A1 on the release and cell-to-cell transmission of retroviruses. Virol J 14:173. doi: 10.1186/s12985-017-0835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kanki PJ, Travers KU, MBoup S, Hsieh CC, Marlink RG, Gueye-NDiaye A, Siby T, Thior I, Hernandez-Avila M, Sankalé JL. 1994. Slower heterosexual spread of HIV-2 than HIV-1. Lancet 343:943–946. doi: 10.1016/S0140-6736(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 101.Heigele A, Kmiec D, Regensburger K, Langer S, Peiffer L, Stürzel CM, Sauter D, Peeters M, Pizzato M, Learn GH, Hahn BH, Kirchhoff F. 2016. The potency of Nef-mediated SERINC5 antagonism correlates with the prevalence of primate lentiviruses in the wild. Cell Host Microbe 20:381–391. doi: 10.1016/j.chom.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Götz N, Sauter D, Usmani SM, Fritz JV, Goffinet C, Heigele A, Geyer M, Bibollet-Ruche F, Learn GH, Fackler OT, Hahn BH, Kirchhoff F. 2012. Reacquisition of Nef-mediated tetherin antagonism in a single in vivo passage of HIV-1 through its original chimpanzee host. Cell Host Microbe 12:373–380. doi: 10.1016/j.chom.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol 7:379–387. doi: 10.1128/MCB.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Scherer WF, Syverton JT, Gey GO. 1953. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med 97:695–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Casartelli N, Sourisseau M, Feldmann J, Guivel-Benhassine F, Mallet A, Marcelin A-G, Guatelli J, Schwartz O. 2010. Tetherin restricts productive HIV-1 cell-to-cell transmission. PLoS Pathog 6:e1000955. doi: 10.1371/journal.ppat.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schmökel J, Li H, Bailes E, Schindler M, Silvestri G, Hahn BH, Apetrei C, Kirchhoff F. 2009. Conservation of Nef function across highly diverse lineages of SIVsmm. Retrovirology 6:36. doi: 10.1186/1742-4690-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schindler M, Schmökel J, Specht A, Li H, Münch J, Khalid M, Sodora DL, Hahn BH, Silvestri G, Kirchhoff F. 2008. Inefficient Nef-mediated downmodulation of CD3 and MHC-I correlates with loss of CD4+ T cells in natural SIV infection. PLoS Pathog 4:e1000107. doi: 10.1371/journal.ppat.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Silva TI, Aasa-Chapman M, Cotten M, Hué S, Robinson J, Bibollet-Ruche F, Sarge-Njie R, Berry N, Jaye A, Aaby P, Whittle H, Rowland-Jones S, Weiss R. 2012. Potent autologous and heterologous neutralizing antibody responses occur in HIV-2 infection across a broad range of infection outcomes. J Virol 86:930–946. doi: 10.1128/JVI.06126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schindler M, Münch J, Kirchhoff F. 2005. Human immunodeficiency virus type 1 inhibits DNA damage-triggered apoptosis by a Nef-independent mechanism. J Virol 79:5489–5498. doi: 10.1128/JVI.79.9.5489-5498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kong R, Li H, Georgiev I, Changela A, Bibollet-Ruche F, Decker JM, Rowland-Jones SL, Jaye A, Guan Y, Lewis GK, Langedijk JPM, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2012. Epitope mapping of broadly neutralizing HIV-2 human monoclonal antibodies. J Virol 86:12115–12128. doi: 10.1128/JVI.01632-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kong R, Li H, Bibollet-Ruche F, Decker JM, Zheng NN, Gottlieb GS, Kiviat NB, Sow PS, Georgiev I, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2012. Broad and potent neutralizing antibody responses elicited in natural HIV-2 infection. J Virol 86:947–960. doi: 10.1128/JVI.06155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tanaka M, Herr W. 1990. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 115.McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJD, Bieniasz PD. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog 5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fouchier RA, Meyer BE, Simon JH, Fischer U, Malim MH. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J 16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ehrhardt C, Schmolke M, Matzke A, Knoblauch A, Will C, Wixler V, Ludwig S. 2006. Polyethylenimine, a cost-effective transfection reagent. Signal Transduct 6:179–184. doi: 10.1002/sita.200500073. [DOI] [Google Scholar]
- 118.Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schaefer TM, Bell I, Pfeifer ME, Ghosh M, Trible RP, Fuller CL, Ashman C, Reinhart TA. 2002. The conserved process of TCR/CD3 complex downmodulation by SIV Nef is mediated by the central core, not endocytic motifs. Virology 302:106–122. doi: 10.1006/viro.2002.1628. [DOI] [PubMed] [Google Scholar]
- 120.Chesebro B, Wehrly K, Nishio J, Perryman S. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol 66:6547–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Smith SM, Marx PA. November 2009. Pathogenic west african human immunodeficiency virus type 2 (HIV-2) group F isolate. US patent 8,747,862.