Ochratoxin A (OTA) is a significant mycotoxin that contaminates cereal products, coffee, grapes, wine, cheese, and meat. OTA is nephrotoxic, carcinogenic, teratogenic, and immunotoxic. OTA contamination is a serious threat to food safety, endangers human health, and can cause huge economic losses. At present, >20 species of the genera Aspergillus and Penicillium are known to produce OTA. Here we demonstrate that a consensus OTA biosynthetic pathway exists in all OTA-producing fungi and is encoded by a gene cluster containing four highly conserved biosynthetic genes and a bZIP transcription factor.
KEYWORDS: ochratoxin A, biosynthetic pathway, Aspergillus ochraceus, genome, comparative genomics, regulation, mycotoxin
ABSTRACT
Ochratoxin A (OTA) is a toxic secondary metabolite produced by Aspergillus and Penicillium species that widely contaminates food and feed. We sequenced and assembled the complete ∼37-Mb genome of Aspergillus ochraceus fc-1, a well-known producer of OTA. Key genes of the OTA biosynthetic pathway were identified by comparative genomic analyses with five other sequenced OTA-producing fungi: A. carbonarius, A. niger, A. steynii, A. westerdijkiae, and Penicillium nordicum. OTA production was completely inhibited in the deletion mutants (ΔotaA, ΔotaB, ΔotaC, ΔotaD, and ΔotaR1), and OTA biosynthesis was restored by feeding a postblock substrate to the corresponding mutant. The OTA biosynthetic pathway was unblocked in the ΔotaD mutant by the addition of heterologously expressed halogenase. OTA biosynthesis begins with a polyketide synthase (PKS), OtaA, utilizing acetyl coenzyme A (acetyl-CoA) and malonyl-CoA to synthesize 7-methylmellein, which is oxidized to OTβ by cytochrome P450 monooxygenase (OtaC). OTβ and l-β-phenylalanine are combined by a nonribosomal peptide synthetase (NRPS), OtaB, to form an amide bond to synthesize OTB. Finally, OTB is chlorinated by a halogenase (OtaD) to OTA. The otaABCD genes were expressed at low levels in the ΔotaR1 mutant. A second regulator, otaR2, which is adjacent to the biosynthetic gene, could modulate only the expression of otaA, otaB, and otaD. Thus, we have identified a consensus OTA biosynthetic pathway that can be used to prevent and control OTA synthesis and will help us understand the variation and production of the intermediate components in the biosynthetic pathway.
IMPORTANCE Ochratoxin A (OTA) is a significant mycotoxin that contaminates cereal products, coffee, grapes, wine, cheese, and meat. OTA is nephrotoxic, carcinogenic, teratogenic, and immunotoxic. OTA contamination is a serious threat to food safety, endangers human health, and can cause huge economic losses. At present, >20 species of the genera Aspergillus and Penicillium are known to produce OTA. Here we demonstrate that a consensus OTA biosynthetic pathway exists in all OTA-producing fungi and is encoded by a gene cluster containing four highly conserved biosynthetic genes and a bZIP transcription factor.
INTRODUCTION
Ochratoxin A (OTA) is an important mycotoxin that contaminates food and feed, including cereal products, coffee, grapes, beverages, wine, cheese, and ham (1–4). OTA can damage the liver and kidneys in humans following the consumption of contaminated food. Negative health effects may include carcinogenicity, teratogenicity, and immune suppression (5, 6), and OTA has been classified as a possible human carcinogen (group 2B) (7).
OTA was first isolated from Aspergillus ochraceus (8), an important OTA producer (9). To date, more than 20 species of the genera Aspergillus and Penicillium are known to produce OTA (10). Initially, Huff and Hamilton predicted a possible biosynthetic pathway based on the chemical structure of OTA, which consists of a dihydrocoumarin moiety bound to l-β-phenylalanine through an amide linkage (11). Harris and Mantle hypothesized that the dominant OTA biosynthetic process was ochratoxin β → ochratoxin α → ochratoxin A, with ochratoxin β → ochratoxin B → ochratoxin A as an alternative pathway (12). Gallo et al. proposed that ochratoxin β and l-phenylalanine were biotransformed to ochratoxin B (OTB) by a nonribosomal peptide synthetase (NRPS) and that the OTB was transformed to OTA through chlorination in Aspergillus carbonarius (13). Wang et al. characterized two polyketide synthases (PKSs) involved in OTA biosynthesis in A. ochraceus (14). Farber and Geisen demonstrated the potential role of a putative halogenase through the analysis of differentially expressed OTA biosynthesis genes in Penicillium nordicum (15), and Ferrara et al. identified a halogenase involved in OTA biosynthesis in A. carbonarius (16). However, the details of the OTA biosynthetic pathway remained unclear and its regulatory mechanism unknown (10, 17, 18).
The objectives of this study were to elaborate the OTA biosynthetic pathway and its regulatory mechanism. Our working hypothesis proposed that a single biochemical pathway was used to synthesize ochratoxin A, as with other mycotoxins, such as aflatoxin, zearalenone, and fumonisin. This work advances the field by integrating the work of previous investigators. We thus succeed in identifying a single consensus pathway for the biosynthesis of this important mycotoxin in OTA-producing fungi.
RESULTS
Sequencing of the genome of A. ochraceus fc-1.
The genomic sequence of A. ochraceus fc-1 totaled 37.02 Mb. A genome annotation generated by GeneWise software predicted 11,740 genes (Table 1). The predicted protein-coding sequences accounted for 14.4% of the A. ochraceus genome, with an average gene length of 1,680 bp and a GC content of 48.8% (54.9% for exons and 46.1% for introns) (Table 1). The genes were further functionally annotated and classified according to Gene Ontology (GO) terms and the KEGG database (see Fig. S1 in the supplemental material). In the A. ochraceus genome, 53.3% of the genes encoded proteins, with each gene containing an average of 3.18 exons, and 78.5% of the genes containing introns (Table 1). In total, 99 secondary-metabolite biosynthetic gene clusters were predicted in the antiSMASH 3.0 (19) and ClusterBlast analyses, including 23 type I pks, 19 nrps, and 8 hybrid gene clusters (see Table S1 in the supplemental material).
TABLE 1.
Genomic information for A. ochraceus fc-1
| Genetic feature and characteristic | Value |
|---|---|
| Scaffolds | |
| Total length (kb) | 37,020 |
| GC content (%) | 48.8 |
| N50 (kb) | 3,843 |
| No. of protein-coding genes | 11,740 |
| Mean gene length (bp) | 1,680 |
| % of genes encoding proteins | 53.3 |
| % of genes with introns | 78.5 |
| Exons | |
| Avg size (bp) | 475.8 |
| No. | 37,371 |
| Mean no. per gene | 3.2 |
| GC content (%) | 54.9 |
| Total length (bp) | 17,782,101 |
| Introns | |
| Avg size (bp) | 75.9 |
| No. | 25,631 |
| Mean no. per gene | 2.2 |
| GC content (%) | 46.1 |
| Total length (bp) | 1,945,895 |
| RNA | |
| No. of tRNAs | 238 |
| No. of rRNAs | 47 |
Comparative genomic analyses of OTA-producing fungi.
The sequenced A. ochraceus genome was comparable in size to those of five other OTA-producing fungi: A. carbonarius ITEM 5010, Aspergillus niger CBS 513.88 (20), Aspergillus steynii IBT 23096, Aspergillus westerdijkiae CBS 112803 (21), and P. nordicum BFE487 (see Table S2 in the supplemental material). Based on phylogenetic analysis, A. ochraceus was much more similar to A. westerdijkiae, A. steynii, and Aspergillus sclerotiorum, belonging to the Aspergillus section Circumdati (22, 23), than to the other two fungi. Aspergillus section Nigri genes, from A. niger and A. carbonarius, were also close to one another (24), while P. nordicum was more distantly related to the other OTA-producing fungi.
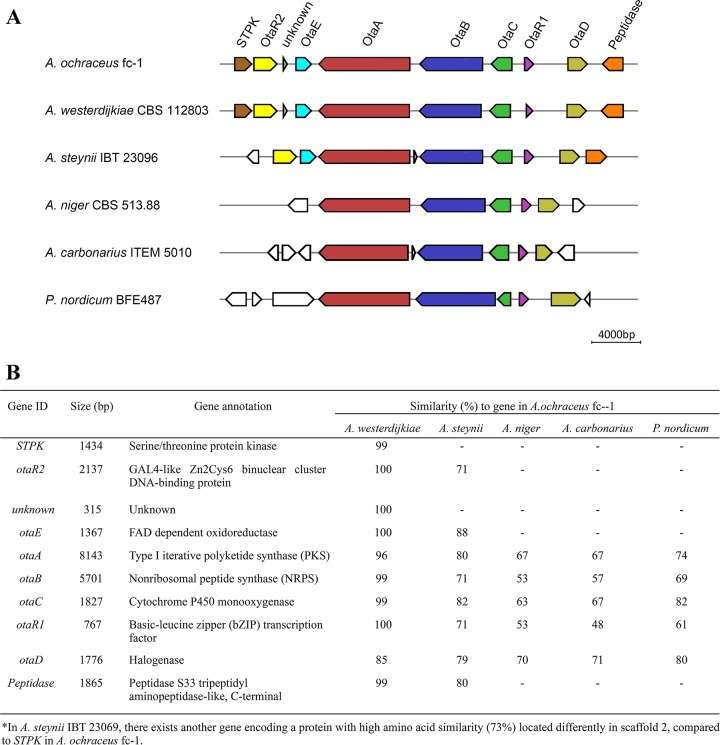
Gene cluster 88 in A. ochraceus displayed high levels of similarity to the corresponding OTA clusters in other OTA-producing fungi. This cluster contained five highly conserved genes, encoding a PKS (otaA), an NRPS (otaB), a cytochrome P450 monooxygenase (otaC), a halogenase (otaD), and a basic leucine zipper (bZIP) transcription factor (otaR1). In addition, genes encoding a flavin adenine dinucleotide (FAD)-dependent oxidoreductase (otaE) and a GAL4-like Zn2Cys6 binuclear DNA-binding protein (otaR2), which were adjacent to the biosynthetic genes, were similar in A. ochraceus, A. westerdijkiae, and A. steynii (Fig. 1A). Similarity analyses of the five proteins encoded by the cluster genes showed high homology levels for this gene cluster among the six strains compared (Fig. 1B). The similarities of the amino acid sequences encoded by these genes to those in A. ochraceus were all >48%, and those of A. westerdijkiae, a new species derived from A. ochraceus (Fig. 1B), were especially high (25, 26).
FIG 1.
Proposed OTA biosynthetic gene cluster. (A) The OTA biosynthetic gene cluster was predicted according to the putative roles of the genes. Genes shown in the same color displayed high similarity with each other. (B) Sequence similarity analyses and predicted functions of the deduced proteins.
Phylogenetic analyses also identified conserved domains in OtaA and OtaB (27), further suggesting their roles in OTA biosynthesis. In addition, OtaA, OtaB, and OtaC were 77%, 72%, and 82% identical to homologous A. steynii 3.53 proteins (NCBI Protein database accession no. AHZ61902.1, AHZ61901.1, and AHZ61900.1) (28). The expression profile of the p450-B03 gene of A. ochraceus HP was similar to that of the pks gene (29), and its encoded protein was 99% identical to OtaC, suggesting that it has a role in OTA biosynthesis. Collectively, these comparative results strengthen the argument that gene cluster 88 is involved in OTA biosynthesis.
Transcriptional analysis of OTA cluster genes.
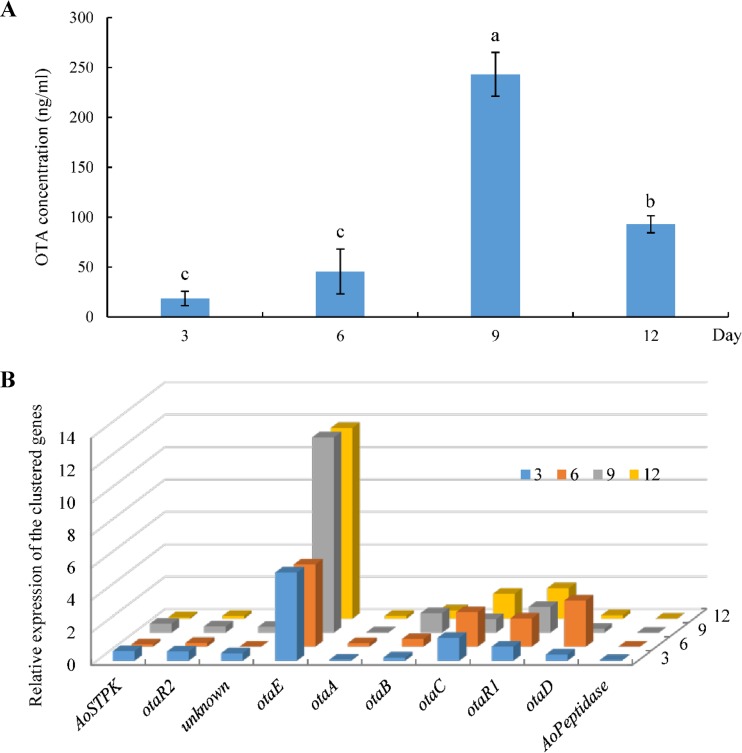
The level of OTA production was highest—243 ng/ml—after 9 days of cultivation (Fig. 2A). Among the cluster genes, otaB expression was especially well correlated with OTA production, although other gene expression patterns displayed few changes (Fig. 2B). Pearson's correlation coefficient (R2) was used to assess the correlation between gene expression and OTA production at various times. A high Pearson correlation coefficient (R2 = 0.836) was found between otaB differential gene expression and OTA production at diverse times, indicating that the expression of this gene was correlated with OTA biosynthesis and regulation. Thus, it was used for further analyses. Expression of otaE, which was identified in only three of the OTA-producing fungi, was also highly correlated with OTA production (R2 = 0.578).
FIG 2.
(A) OTA production of A. ochraceus fc-1 in YES liquid medium at different times. Bars labeled with the same letter represent results that are not significantly different from each other. (B) Relative expression of the clustered OTA genes in A. ochraceus fc-1 on days 3, 6, 9, and 12. OtaA is a polyketide synthase; OtaB, a nonribosomal peptide synthetase; OtaC, a cytochrome P450 monooxygenase; OtaD, a halogenase; OtaE, a FAD-dependent oxidoreductase; OtaR1, a bZIP transcription factor; OtaR2, a GAL4-like Zn2Cys6 binuclear cluster DNA-binding protein; STPK, a serine/threonine protein kinase. AoPeptidase, A. ochraceus gene encoding peptidase.
A proposed OTA biosynthetic pathway.
To confirm the involvement of these putative biosynthetic genes in OTA biosynthesis, each of the cluster genes was individually inactivated by a homologous recombination strategy using polyethylene glycol (PEG)-mediated genetic transformation (14). The growth rates of the deletion mutants on peptone-dextrose agar (PDA) and yeast extract and sucrose (YES) medium were not significantly different (P > 0.01) (data not shown). Five genes—otaA, otaB, otaC, otaD, and otaR1—were directly involved in OTA biosynthesis, as evidenced by the fact that OTA production was completely abolished when the disrupted strains grew on YES medium, as well as on maize or on wheat (Table 2). Inactivation of otaE or otaR2 significantly decreased OTA production (P < 0.01) (Table 2). These results confirmed the roles of the five genes listed above in OTA biosynthesis and regulation.
TABLE 2.
OTA production in the mutants compared to that in A. ochraceus fc-1
| Strain or mutation | OTA concna in the following medium: |
||
|---|---|---|---|
| YES (μg/cm2) | Maize (μg/g) | Wheat (μg/g) | |
| WTb | 36.8 ± 3.6 A | 199.3 ± 36.3 A | 28.8 ± 1.6 AB |
| ΔSTPK | 31.7 ± 4.6 A | 277.7 ± 89.0 A | 35.9 ± 4.8 A |
| ΔotaR2 | 13.6 ± 2.0 B | 28.8 ± 9.9 B | 22.0 ± 1.9 BC |
| Δunknown | 41.6 ± 4.6 A | 163.5 ± 69.1 A | 21.9 ± 6.6 BC |
| ΔotaE | 18.3 ± 5.2 B | 14.5 ± 6.5 B | 15.5 ± 6.6 C |
| ΔotaA | 0 C | 0 C | 0 D |
| ΔotaB | 0 C | 0 C | 0 D |
| ΔotaC | 0 C | 0 C | 0 D |
| ΔotaR1 | 0 C | 0 C | 0 D |
| ΔotaD | 0 C | 0 C | 0 D |
| ΔPeptidase | 32.6 ± 1.3 A | 211.4 ± 35.2 A | 19.8 ± 5.9 BC |
The same letter is placed next to concentrations that did not differ significantly from each other.
WT, wild type.
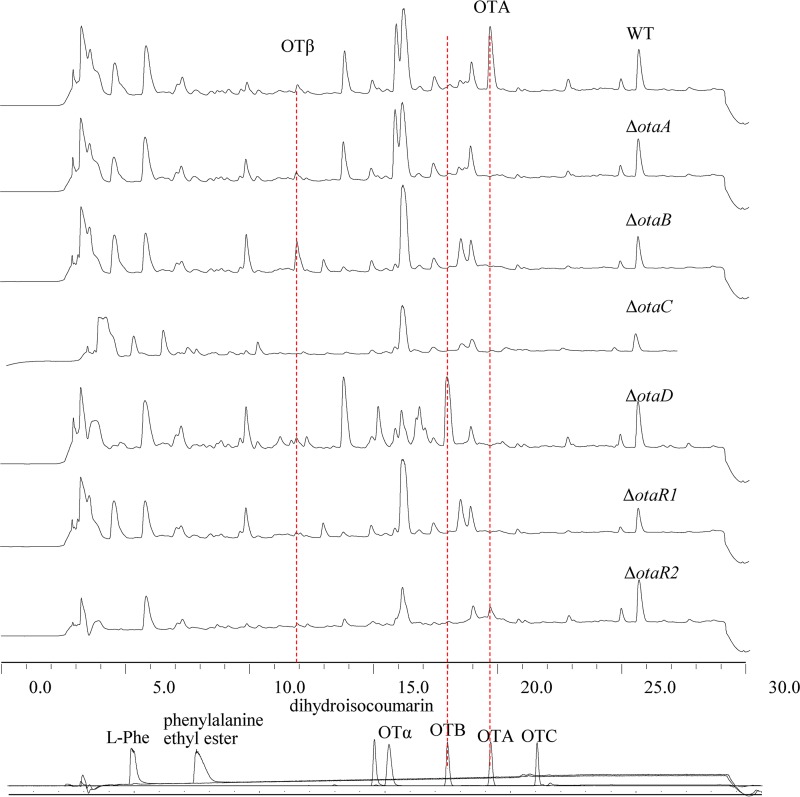
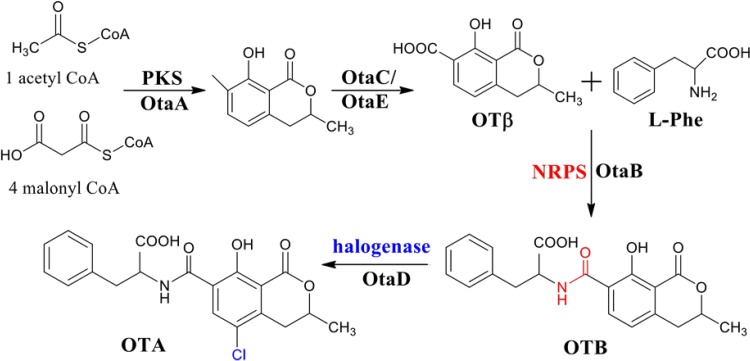
Intermediate metabolites in the OTA biosynthetic pathway were identified by cultivating otaA, otaB, otaC, and otaD deletion mutants and analyzing their metabolite profiles. When the PKS-encoding gene otaA was disrupted, neither OTA nor its precursors were detected, indicating that this step was the first in OTA biosynthesis (Fig. 3). OTA, OTB, and OTβ were not detected in the ΔotaC mutant (Fig. 3). In the ΔotaB mutant, OTβ was the only precursor produced, suggesting that OTβ was a precursor of OTA (Fig. 3). Liquid chromatography-mass spectrometry (LC-MS) analysis of the ΔotaB mutant also detected l-phenylalanine (m/z 166) as an OtaB precursor. OTB (m/z 370) and its analogues accumulated in the ΔotaD mutant (Fig. 3); however, OTβ (m/z 222) did not accumulate, and OTA (m/z 404) was not detected by high-performance liquid chromatography-electrospray ionization (HPLC-ESI) MS, in the ΔotaD mutant.
FIG 3.
HPLC-FLD analysis of mutants with deletions of genes in the cluster involved in OTA biosynthesis. The mutant strains were cultured in maize medium, and the secondary metabolites particularly related to OTA biosynthesis were detected. The last chromatograms show the standards of OTA, OTB, OTC, OTα, l-Phe, phenylalanine ethyl ester, and dihydroisocoumarin. OTB accumulated in the ΔotaD mutant more than in the wild-type (WT) strain. Correspondingly, in the ΔotaB culture, there was a higher peak identified as OTβ.
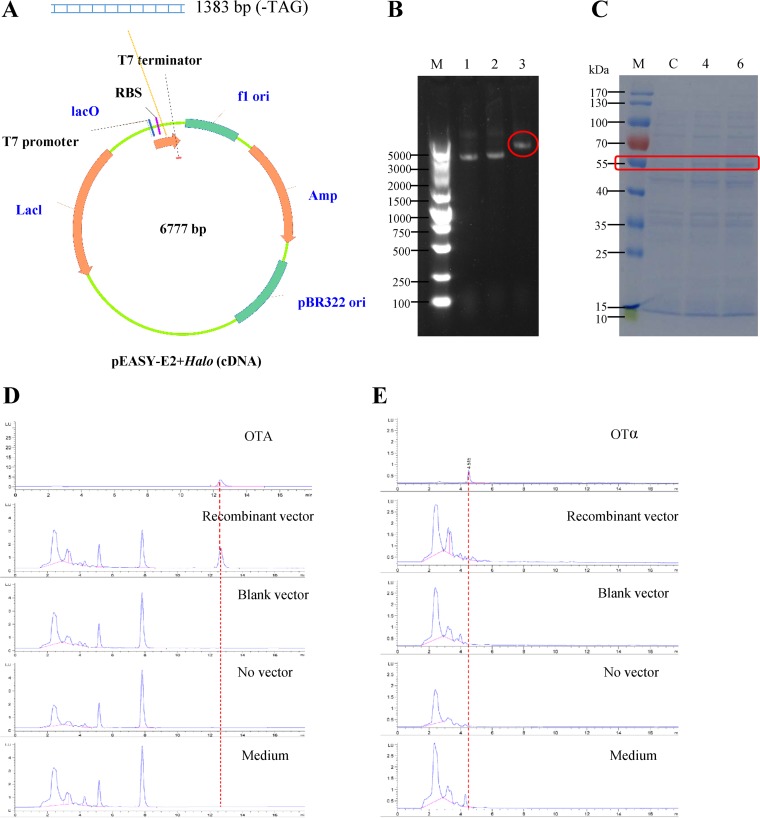
To further confirm the function of the halogenase, the enzyme was expressed heterologously in the pEASY-E2 expression vector in Escherichia coli BL21 (Fig. 4A to C). OTB was added to the culture, which was then incubated for 12 h. The potential enzymatic product, OTA, was detected by HPLC with fluorescence detection (HPLC-FLD) in the transformed strain but not in E. coli BL21 with the empty vector or with no vector (Fig. 4D). When OTβ was mixed with the bacteria, no OTα was synthesized after 12 h of incubation (Fig. 4E). Thus, OtaD transformed OTB to OTA through halogenation, and OTB, rather than OTβ, was the substrate of the halogenase.
FIG 4.
Expression of halogenase in E. coli BL21. (A) Recombinant pEASY vector with halogenase cDNA transformed into E. coli. (B) Confirmation of the recombinant vector by agarose gel electrophoresis. Lanes: 1 and 2, the empty vector; 3, the recombinant vector (pEASY-E2+Halo). (C) Expression of halogenase protein in E. coli BL21. C, 4, and 6 represent induction times of 0, 4, and 6 h, respectively. (D and E) HPLC-FLD analysis of the enzyme activity of heterologously expressed halogenase with OTB (D) or OTβ (E) as the substrate. The chromatograms show (from top to bottom) the OTA (D) or OTα (E) standard and the detection results for strains with a recombinant vector, a blank vector, no vector, or the medium only.
Moreover, inactivation of otaE or otaR2 significantly decreased OTA production (P < 0.01) (Table 2). OtaE may play a role in the biosynthesis of OTA, but it is not essential. Thus, we concluded that four genes, otaABCD, are essential for OTA biosynthesis.
We hypothesize that OTA biosynthesis (Fig. 5) begins with the PKS OtaA, which combines acetyl coenzyme A (acetyl-CoA) and malonyl-CoA to synthesize 7-methylmellein. This compound was also detected during the LC-MS analysis of OtaE mutant cultures. 7-Methylmellein is then oxidized to OTβ by OtaC. OTβ and l-β-phenylalanine are combined by the NRPS (OtaB) to form an amide bond and OTB. OTB is chlorinated by the halogenase OtaD to produce the final product, OTA.
FIG 5.
Proposed OTA biosynthetic pathway, including intermediate metabolites and catalytic enzymes. The process of OTA biosynthesis was confirmed as follows. Starting from acetyl-CoA and malonyl-CoA, the first intermediate was biotransformed by the OtaA enzyme (a polyketide synthase); then it was catalyzed to OTβ (ochratoxin β) by OtaC. OTβ was linked with another substrate, l-phenylalanine, by the nonribosomal peptide synthetase OtaB to form OTB (ochratoxin B) as the penultimate biosynthetic step. Finally, OTB was chlorinated by the halogenase OtaD to produce OTA.
Regulation of OTA biosynthetic genes.
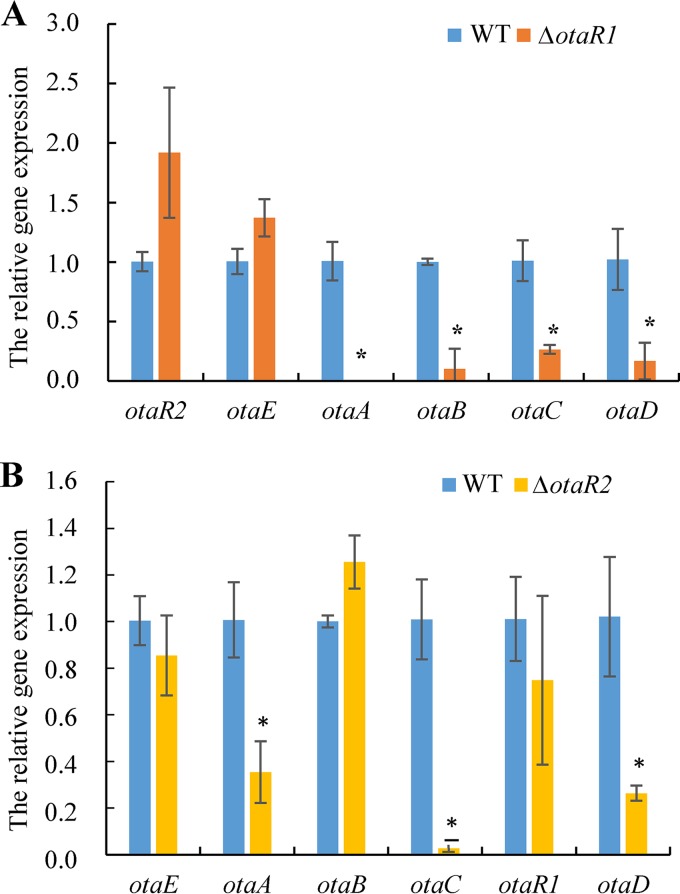
OtaR1, a bZIP transcription factor, is present in the OTA biosynthetic gene cluster. OTA was not detected when the otaR1-inactivated mutant was cultured on YES or maize/wheat medium (Table 2). OtaR1 is probably a pathway-specific regulator that controls OTA production by regulating the four biosynthetic genes. The expression levels of the otaABCD genes, which are directly involved in OTA biosynthesis, decreased significantly in the ΔotaR1 mutant (P < 0.01) (Fig. 6A). otaA expression was completely inhibited, and otaB, otaC, and otaD expression levels were reduced. In contrast, OtaR1 does not regulate either otaR2 or otaE (P > 0.05).
FIG 6.
Relative expression of the biosynthetic genes in the ΔotaR1 (A) and ΔotaR2 (B) mutants. The expression levels of the key biosynthetic genes in the mutants were separately compared to those for the wild-type (WT) strain in 6-day cultures by one-way analysis of variance (P < 0.01). Asterisks indicate significant differences from expression in A. ochraceus fc-1.
The zinc finger DNA-binding protein OtaR2 is located adjacent to the biosynthetic genes. OTA production in the ΔotaR2 mutant was reduced on all three test media (P < 0.01) (Table 2). OtaR2 can modulate otaA, otaC, and otaD expression levels. The expression of all three of these genes was significantly reduced in the ΔotaR2 mutant (P < 0.01) (Fig. 6B). In contrast, OtaR2 does not regulate the expression of otaB, otaE, or otaR1 (P > 0.05) (Fig. 6B).
DISCUSSION
OTA is one of the most significant mycotoxins, produced mainly by Aspergillus and Penicillium species (2), widely contaminating food and feed and seriously harming human health. Several biosynthetic pathways have been proposed for OTA. Its biosynthesis was originally predicted by its structural composition; it consists of a dihydrocoumarin moiety linked to l-β-phenylalanine. According to Huff and Hamilton (11), the polyketide mellein is initially synthesized by acetic acid condensation. Mellein is transformed to OTβ, which is subsequently chlorinated to form OTα. OTα is transformed to OTC by binding with phenylalanine ethyl ester to protect the intermediate. OTC is then transformed to OTA by de-esterification. However, Harris and Mantle (12) proposed a different pathway for OTA production based on precursor feeding experiments. They proposed biotransformation from OTβ to OTα and finally to OTA by direct linking with phenylalanine in a potentially dominant pathway. In this proposed biosynthetic pathway, OTC was not a precursor in OTA biosynthesis. Harris and Mantle (12) also proposed an alternative pathway in which OTβ was linked to phenylalanine and then transformed to OTB, with a final chlorination step to produce OTA. In this pathway, OTB was a significant intermediate during OTA biosynthesis. Gallo et al. (13) suggested a third potential biosynthetic pathway. They hypothesized an OTA biosynthetic pathway similar to the alternative pathway proposed by Harris and Mantle, except that they believed that OTα was a byproduct of OTA hydrolysis.
In the past decade, the molecular mechanisms underlying OTA biosynthesis have been increasingly studied. The biosynthesis of most polyketides is catalyzed by a multifunctional enzyme, PKS, in a manner similar to that of the chain elongation step of classical fatty acid biosynthesis. Homologues of the pks gene have been characterized, and their roles in OTA biosynthesis have been identified in OTA-producing fungi by use of gene inactivation (14, 30–35). In addition, the role of an nrps gene in A. carbonarius has been confirmed by gene disruption (13). A P450 monooxygenase that oxidized the polyketide moiety during OTA biosynthesis was discovered through differential expression analysis of OTA-producing and non-OTA-producing strains of A. westerdijkiae (36). Three genes encoding a cytochrome P450 monooxygenase, an NRPS, and a PKS were determined to be involved in OTA biosynthesis by use of 5′ rapid amplification of cDNA ends (5′ RACE) and the genome-walking method in A. steynii (28). A partial, putative OTA gene cluster was cloned, and pks, nrps, and transporter genes were identified, in P. nordicum (33, 37). The halogenase gene was associated with OTA biosynthesis by use of differential-display reverse transcriptase PCR (RT-PCR) with P. nordicum grown on minimal media either suitable or inadequate for OTA production (15). Transcriptome sequencing (RNA-Seq) and reverse transcriptase quantitative PCR (RT-qPCR) analysis were used to associate a PKS, an NRPS, and a chloroperoxidase with the OTA biosynthetic process (38).
Previously, the OTA biosynthetic pathway and its molecular basis have been characterized separately. In this study, we provide a detailed correlation between the biochemical synthesis of OTA and its genetic basis. Here we used comparative genomic analysis of six OTA-producing fungi to identify a conserved OTA biosynthetic gene cluster. Subsequent gene knockout mutants identified four structural genes (otaABCD) and one regulatory gene (otaR1) that were conserved in all six species and were required for OTA production. Potential pathway intermediates consistent with the proposed biosynthetic pathway were identified chemically and were used in cross-feeding experiments of deletion mutants. OTC and OTα were not intermediates in OTA biosynthesis in our biosynthetic pathway. OTβ and OTB are proposed as the intermediates in OTA biosynthesis. From these structures, OTB is a dechlorinated analogue of OTA, while OTβ differs from OTα in the presence/absence of a chlorine in the dihydrocoumarin moiety of OTA.
Most of the analogues of OTA (e.g., OTB, OTα, and OTβ) are much less toxic than OTA. For example, OTB does not cause microscopically obvious damage in rat kidneys as OTA does (39). OTB also reduces the toxic effects of OTA if the compounds are administered together. OTA is highly nephrotoxic, with a 50% infective concentration (IC50) of 0.5 μM for immortalized human kidney epithelial (IHKE) cells. OTα, the dihydrocoumarin moiety of OTA, was not cytotoxic even at levels as high as 50 μM (40). Considering the toxicity of OTA and its analogues, our proposal for the OTA biosynthesis pathway can potentially assist in controlling and detoxifying OTA.
Regarding regulation, usually only one transcription factor exists per gene cluster, and this transcription factor controls the expression of all the genes in the cluster. For example, in Aspergillus flavus, AflR, a 47-kDa specific zinc finger DNA-binding protein, regulates the expression of all the genes in the aflatoxin cluster except for aflJ, which modulates the expression of structural genes and serves as a coactivator with aflR during aflatoxin biosynthesis (39–41). In Aspergillus oryzae, AflR is affected by environmental factors, such as carbon sources and temperature, in a manner similar to that in Aspergillus parasiticus (42). In the OTA cluster, deletion of otaR1 prevents OTA production, while inactivation of otaR2 merely reduces production. In transcriptional analyses, OtaR1 could alter the expression of all four of the biosynthetic genes, particularly otaA, but did not alter otaR2 expression. OtaR2 does not regulate otaB or otaR1 but does regulate the remaining three biosynthetic genes. Thus, OtaR1 probably functions as an OTA pathway-specific regulator. An otaR1 deletion had no effect on otaR2 transcription, and vice versa. Therefore, the mechanisms underlying the way in which OTA biosynthesis regulation responds to different environmental cues remain to be elucidated.
In summary, we have determined the steps of OTA biosynthesis in OTA-producing fungi and have provided a framework for its regulation. OtaR1 and OtaR2 both regulate OTA biosynthesis, with OtaR1 acting as the pathway-specific regulator and controlling the expression of the four biosynthetic genes otaABCD. Our results provide a consensus OTA biosynthetic pathway that is anchored to a gene cluster, in which particular genes are associated with particular enzymes and metabolic intermediates. These results should settle the points of uncertainty regarding the biosynthetic mechanism for OTA that have arisen from studies less comprehensive than the current one.
MATERIALS AND METHODS
Strains and DNA/RNA preparation.
Total DNA was extracted from Aspergillus ochraceus fc-1 cultured in yeast extract and sucrose (YES) medium for 6 days at 28°C according to the instructions in the E.Z.N.A. Fungal DNA kit (Omega Bio-tek, Norcross, GA, USA). RNA was extracted from A. ochraceus fc-1 cultured in YES liquid medium for 3, 6, 9, or 12 days by use of an RNeasy Plant minikit (Qiagen, Hilden, Germany).
Genome sequencing, assembly, and RNA-Seq analysis.
The genome of A. ochraceus was sequenced using Roche 454 GS-FLX Plus pyrosequencing technology and paired-end linker libraries. Sequence reads were assembled by ALLPATHS-LG software (Broad Institute of MIT and Harvard, Cambridge, MA, USA), and hybrid data sets were assembled by the HaploMerger pipeline (43). High-quality data were obtained for the assembly of scaffolds with high coverage using 454 GS-FLX technology, and paired-end linker sequences were assembled and merged with the original 454 whole-genome shotgun sequence data. Improved scaffold coverage and scaffold lengths spanning longer gene intervals resulted.
Four transcriptome libraries were constructed and sequenced with an Illumina HiSeq 2000 platform. To estimate the gene expression levels in A. ochraceus, Illumina HiSeq 2000 reads from each library were mapped to assembled isotigs (nonsingleton transcripts of the PUT set) from all the genomic reads. Each transcriptome library comprised the means of results from three biological replicates. The expression of the OTA biosynthetic genes was analyzed by use of the RNA-Seq data.
Gene prediction and functional annotation.
PASA (44) and EVidenceModeler (45) were used to generate consensus gene models based on predictions from several types of gene finders, including AUGUSTUS (46), FGENESH, geneid, GeneMark-ES (47), GlimmerHMM (48), and GeneWise (49).
Functional annotation of the genome was carried out by Blast2GO, and the GO annotations were summarized by Aspergillus-specific GO Slim. GO enrichment analysis (Fisher's exact test) was used (P < 0.05) to determine the functional categories of the overrepresented cellular components, biological processes, and molecular functions with statistical significance (false discovery rate [FDR], <0.05). GO terms were mapped to a GO annotation content overview using the CateGOrizer program (50).
The same gene annotation protocol was performed to annotate the predicted genes from five sequenced OTA-producing fungi. The strains used (with National Center for Biotechnology Information [NCBI] and Joint Genome Institute [JGI; U.S. Department of Energy] genome assembly accession numbers given in parentheses) were as follows: A. westerdijkiae CBS 112803 (ASM130734v1, GCA_001307345.1), A. steynii IBT23069, A. niger CBS 513.88 (ASM285v2, GCA_000002855.2), A. carbonarius ITEM 5010, and P. nordicum BFE487 (PnBFE487-1.0, GCA_000733025.2).
Phylogenetic analysis of genomes from Aspergillus and Penicillium species.
Twenty-five sequenced Aspergillus and Penicillium species were selected for phylogenetic analysis. Using hidden Markov models (HMM) (51), putative orthologs were identified with the BUSCO (Benchmarking Universal Single-Copy Orthologs) database (52) as having the highest full-sequence HMM bit scores with a minimum E value of 1E−50, and the gene family of each ortholog was aligned using MAFFT (53). Ambiguous alignment regions were masked by using Gblocks with default settings (54). Data sets with one or more complete sequences missing and those retaining <20% of the original amino acids were omitted from further analysis. The remaining protein alignment sequences were concatenated into a single superalignment using a customized Perl script. ProtTest, version 3.2, was applied to select the best-fit protein evolution model (55), and trees were reconstructed using RAxML with the best-fit model PROTGAMMAIJTTF and 100 rapid bootstraps (56).
In silico analyses of secondary-metabolite clusters and the OTA cluster.
The antiSMASH 3.0 platform (19) was used to predict gene clusters of secondary metabolites in the masked scaffolds and to perform ClusterBlast analysis on available fungal genome sequences. The same protocol was used to predict secondary-metabolite clusters from six additional OTA-producing fungi. Genetic similarity analysis of the predicted OTA biosynthetic cluster among these OTA-producing fungi was performed. The potential OTA cluster was predicted by analyzing the amino acid similarities between the reported PKS protein and the proteins related to OTA biosynthesis in the sequenced OTA-producing fungi.
Construction of gene deletion mutants.
To disrupt the cluster genes, deletion mutants were designed and constructed via homologous recombination with DNA fragments based on the primers listed in Table 3. Genetic transformation of A. ochraceus protoplasts was performed as described previously (14), and mutants were selected on a medium containing 100 μg/ml hygromycin B. The replacements were confirmed by diagnostic PCR with appropriate primers based on the insertion site (Table 3).
TABLE 3.
Primers used for the construction of knockout mutants in A. ochraceus fc-1
OTA detection and intermediate analysis of cultured strains.
Metabolites from strains cultured on a maize or wheat medium for 6 days were analyzed. Cultures were ground, and 5 g of the powdered samples was resolved in 25 ml of methanol. A 10-ml volume of the supernatant was evaporated, and the metabolites were resolved with 2 ml of methanol and were filtered. OTA was measured in the filtrate by HPLC-FLD (14). Metabolites produced by mutants of key structural genes were analyzed by LC-MS on an Agilent Accurate-Mass-QTOF LC/MS 6520 instrument.
Biosynthetic gene expression in regulatory gene knockout mutants.
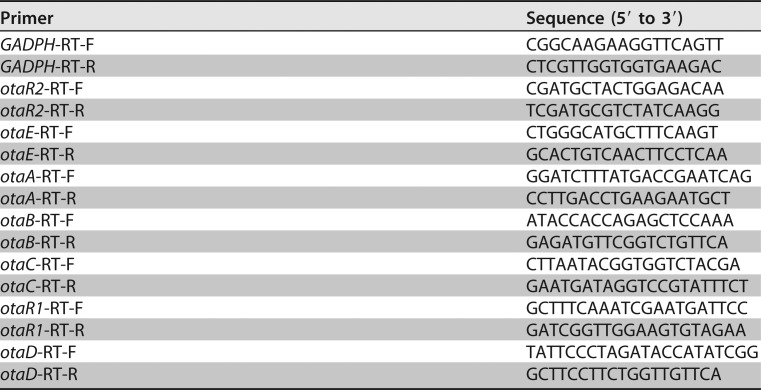
Both the wild-type (WT) strain and regulatory gene mutants were cultured for 6 days. RNA was extracted with an RNeasy Plant minikit (Qiagen, Hilden, Germany). cDNA was obtained by using a PrimeScript RT Reagent kit with gDNA Eraser (TaKaRa, Kyoto, Japan). Quantitative PCR analysis of the OTA biosynthetic genes was performed with Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA, USA) on an Applied Biosystems 7500 real-time PCR system (Life Technologies, Waltham, MA, USA). The expression of OTA biosynthetic genes was quantified by the comparative 2−ΔΔCT method using their respective primers in triplicate (Table 4).
TABLE 4.
Primers used for the expression of biosynthetic genes
Heterologous expression of the halogenase gene in E. coli.
Total RNA was extracted and was converted to cDNA. The halogenase gene without introns was amplified with I-5 2× Hi-Fi PCR master mix (MCLAB, South San Francisco, CA). The DNA product was resolved on a 2% agarose gel and was extracted with a SanPrep Column DNA gel extraction kit (Sangon Biotech, Shanghai, China). The recombinant vector was constructed in a pEASY-Blunt E2 expression system (Transgen, Beijing, China). The transformed E. coli strains were spread onto LB medium containing 100 μg/ml ampicillin. Transformants were identified and were cultured in LB medium with 100 μg/ml ampicillin for 12 to 16 h. The plasmid was extracted by using a SanPrep Column Plasmid Mini-Preps kit (Sangon Biotech, Shanghai, China). The plasmid was transformed into E. coli BL21 and its presence confirmed by growth on selective media and amplification of the inserted DNA.
Protein production in E. coli was optimized based on the concentration (0.1, 0.25, 0.5, or 1 mM) of isopropyl-β-d-thiogalactopyranoside (IPTG) and the induction time (4, 6, or 8 h). After optimization, the bacterial cells were broken ultrasonically at 300 W for 20 min (5-s sonication; 5-s interval). Cell debris was pelleted by centrifugation at 11,000 × g for 10 min at 4°C. A 40-μl volume of the protein sample was resolved via SDS-PAGE with a 5% stacking gel and a 10% separation gel. The gel was first stained with Coomassie brilliant blue R-250 for 4 h and then washed in 40% (vol/vol) methanol with 10% (vol/vol) acetic acid for 12 h. The desired protein was ∼53 kDa, and optimization conditions were selected based on the presence and intensity of this band.
Functional analysis of the expressed halogenase.
Different strains of E. coli BL21 (with a recombinant vector, a control vector, or no vector) were cultured separately and were induced under optimized conditions (0.5 mM IPTG and induction for 6 h). Strains were grown on LB medium supplemented with 20 μl OTB or OTβ. OTA and related metabolites were extracted from cultures growing on supplemented media with 3 volumes of methanol, and the metabolites present—especially the putative biosynthesized product OTA or OTα—were identified by HPLC-FLD.
Statistical analysis.
The statistical analysis of data was performed by IBM SPSS Statistics, version 21.0.
Accession number(s).
The genomic sequence data obtained in this study have been submitted to NCBI under BioProject accession no. PRJNA264608 and BioSample accession no. SAMN03140103.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the editorial input of Joan W. Bennett and the genomic data analyses provided by the Institute of Microbiology, Chinese Academy of Sciences.
This work was supported by the National Key Research and Development Program of China (grant 2017YFC1600900), the National Science Foundation of China (grant 31601577), and the National Basic Research Program of China (973 program) (grant 2013CB127800).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01009-18.
REFERENCES
- 1.Jørgensen K. 2005. Occurrence of ochratoxin A in commodities and processed food—a review of EU occurrence data. Food Addit Contam 22(Suppl 1):26–30. doi: 10.1080/02652030500344811. [DOI] [PubMed] [Google Scholar]
- 2.Zhihong L, Kunlun H, Yunbo L. 2015. Ochratoxin A and ochratoxin-producing fungi on cereal grain in China: a review. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 32:461–470. doi: 10.1080/19440049.2014.981763. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Li Y, Wang H, Gu X, Zheng X, Wang Y, Diao J, Peng Y, Zhang H. 2016. Screening and identification of novel ochratoxin A-producing fungi from grapes. Toxins (Basel) 8:E333. doi: 10.3390/toxins8110333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez A, Medina A, Cordoba JJ, Magan N. 2014. The influence of salt (NaCl) on ochratoxin A biosynthetic genes, growth and ochratoxin A production by three strains of Penicillium nordicum on a dry-cured ham-based medium. Int J Food Microbiol 178:113–119. doi: 10.1016/j.ijfoodmicro.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Joint FAO/WHO Expert Committee on Food Additives. 2001. Safety evaluation of certain mycotoxins in food. WHO Food Additives Series 47. FAO Food and Nutrition Paper 74. International Programme on Chemical Safety, World Health Organization, Geneva, Switzerland: http://www.fao.org/3/a-bc528e.pdf. [Google Scholar]
- 6.Petzinger E, Ziegler K. 2000. Ochratoxin A from a toxicological perspective. J Vet Pharmacol Ther 23:91–98. doi: 10.1046/j.1365-2885.2000.00244.x. [DOI] [PubMed] [Google Scholar]
- 7.IARC. 1993. Ochratoxin A. IARC Monogr Eval Carcinog Risks Hum 56:489–521. [PMC free article] [PubMed] [Google Scholar]
- 8.van der Merwe KJ, Steyn PS, Fourie L, Scott DB, Theron JJ. 1965. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh. Nature 205:1112–1113. doi: 10.1038/2051112a0. [DOI] [PubMed] [Google Scholar]
- 9.Wilson DM, Mubatanhema W, Jurjevic Z. 2002. Biology and ecology of mycotoxigenic Aspergillus species as related to economic and health concerns. Adv Exp Med Biol 504:3–17. doi: 10.1007/978-1-4615-0629-4_2. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Wang L, Liu F, Wang Q, Selvaraj JN, Xing F, Zhao Y, Liu Y. 2016. Ochratoxin A producing fungi, biosynthetic pathway and regulatory mechanisms. Toxins (Basel) 8:E83. doi: 10.3390/toxins8030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huff WE, Hamilton PB. 1979. Mycotoxins—their biosynthesis in fungi: ochratoxins—metabolites of combined pathways. J Food Prot 42:815–820. doi: 10.4315/0362-028X-42.10.815. [DOI] [PubMed] [Google Scholar]
- 12.Harris JP, Mantle PG. 2001. Biosynthesis of ochratoxins by Aspergillus ochraceus. Phytochemistry 58:709–716. doi: 10.1016/S0031-9422(01)00316-8. [DOI] [PubMed] [Google Scholar]
- 13.Gallo A, Bruno KS, Solfrizzo M, Perrone G, Mulè G, Visconti A, Baker SE. 2012. New insight into the ochratoxin A biosynthetic pathway through deletion of a nonribosomal peptide synthetase gene in Aspergillus carbonarius. Appl Environ Microbiol 78:8208–8218. doi: 10.1128/AEM.02508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Wang Y, Wang Q, Liu F, Selvaraj JN, Liu L, Xing F, Zhao Y, Zhou L, Liu Y. 2015. Functional characterization of new polyketide synthase genes involved in ochratoxin A biosynthesis in Aspergillus ochraceus fc-1. Toxins (Basel) 7:2723–2738. doi: 10.3390/toxins7082723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farber P, Geisen R. 2004. Analysis of differentially-expressed ochratoxin A biosynthesis genes of Penicillium nordicum. Eur J Plant Pathol 110:661–669. doi: 10.1023/B:EJPP.0000032405.21833.89. [DOI] [Google Scholar]
- 16.Ferrara M, Perrone G, Gambacorta L, Epifani F, Solfrizzo M, Gallo A. 2016. Identification of a halogenase involved in the biosynthesis of ochratoxin A in Aspergillus carbonarius. Appl Environ Microbiol 82:5631–5641. doi: 10.1128/AEM.01209-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallo A, Ferrara M, Perrone G. 2017. Recent advances on the molecular aspects of ochratoxin A biosynthesis. Curr Opin Food Sci 17:49–56. doi: 10.1016/j.cofs.2017.09.011. [DOI] [Google Scholar]
- 18.Geisen R, Schmidt-Heydt M, Touhami N, Himmelsbach A. 2018. New aspects of ochratoxin A and citrinin biosynthesis in Penicillium. Curr Opin Food Sci 23:23–31. doi: 10.1016/j.cofs.2018.04.001. [DOI] [Google Scholar]
- 19.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH. 2015. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43(W1):W237–W243. doi: 10.1093/nar/gku1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d'Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, et al. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 21.Han X, Chakrabortti A, Zhu J, Liang ZX, Li J. 2016. Sequencing and functional annotation of the whole genome of the filamentous fungus Aspergillus westerdijkiae. BMC Genomics 17:633. doi: 10.1186/s12864-016-2974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khalesi M, Khatib N. 2011. The effects of different ecophysiological factors on ochratoxin A production. Environ Toxicol Pharmacol 32:113–121. doi: 10.1016/j.etap.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Visagie CM, Varga J, Houbraken J, Meijer M, Kocsubé S, Yilmaz N, Fotedar R, Seifert KA, Frisvad JC, Samson RA. 2014. Ochratoxin production and taxonomy of the yellow aspergilli (Aspergillus section Circumdati). Stud Mycol 78:281–289. doi: 10.1016/j.simyco.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garmendia G, Vero S. 2016. Occurrence and biodiversity of Aspergillus section Nigri on ‘Tannat’ grapes in Uruguay. Int J Food Microbiol 216:31–39. doi: 10.1016/j.ijfoodmicro.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Frisvad JC, Frank JM, Houbraken JAMP, Kuijpers AFA, Samson RA. 2004. New ochratoxin A producing species of Aspergillus section Circumdati. Stud Mycol 50:23–43. [Google Scholar]
- 26.Taniwaki MH. 2006. An update on ochratoxigenic fungi and ochratoxin A in coffee. Adv Exp Med Biol 571:189–202. doi: 10.1007/0-387-28391-9_12. [DOI] [PubMed] [Google Scholar]
- 27.Gallo A, Ferrara M, Perrone G. 2013. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins (Basel) 5:717–742. doi: 10.3390/toxins5040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gil-Serna J, Vázquez C, González-Jaén MT, Patiño B. 2015. Clustered array of ochratoxin A biosynthetic genes in Aspergillus steynii and their expression patterns in permissive conditions. Int J Food Microbiol 214:102–108. doi: 10.1016/j.ijfoodmicro.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 29.O'Callaghan J, Stapleton PC, Dobson ADW. 2006. Ochratoxin A biosynthetic genes in Aspergillus ochraceus are differentially regulated by pH and nutritional stimuli. Fungal Genet Biol 43:213–221. doi: 10.1016/j.fgb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Abbas A, Coghlan A, O'Callaghan J, Garcia-Estrada C, Martin J-F, Dobson ADW. 2013. Functional characterization of the polyketide synthase gene required for ochratoxin A biosynthesis in Penicillium verrucosum. Int J Food Microbiol 161:172–181. doi: 10.1016/j.ijfoodmicro.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Bacha N, Atoui A, Mathieu F, Liboz T, Lebrihi A. 2009. Aspergillus westerdijkiae polyketide synthase gene “aoks1” is involved in the biosynthesis of ochratoxin A. Fungal Genet Biol 46:77–84. doi: 10.1016/j.fgb.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 32.Gallo A, Knox BP, Bruno KS, Solfrizzo M, Baker SE, Perrone G. 2014. Identification and characterization of the polyketide synthase involved in ochratoxin A biosynthesis in Aspergillus carbonarius. Int J Food Microbiol 179:10–17. doi: 10.1016/j.ijfoodmicro.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Karolewiez A, Geisen R. 2005. Cloning a part of the ochratoxin A biosynthetic gene cluster of Penicillium nordicum and characterization of the ochratoxin polyketide synthase gene. Syst Appl Microbiol 28:588–595. doi: 10.1016/j.syapm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 34.O'Callaghan J, Caddick MX, Dobson ADW. 2003. A polyketide synthase gene required for ochratoxin A biosynthesis in Aspergillus ochraceus. Microbiology 149:3485–3491. doi: 10.1099/mic.0.26619-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Zhu L, Chen H, Li M, Zhu X, Gao Q, Wang D, Zhang Y. 2016. A polyketide synthase encoded by the gene An15g07920 is involved in the biosynthesis of ochratoxin A in Aspergillus niger. J Agric Food Chem 64:9680. doi: 10.1021/acs.jafc.6b03907. [DOI] [PubMed] [Google Scholar]
- 36.Sartori D, Massi FP, Ferranti LS, Fungaro MHP. 2014. Identification of genes differentially expressed between ochratoxin-producing and non-producing strains of Aspergillus westerdijkiae. Indian J Microbiol 54:41–45. doi: 10.1007/s12088-013-0408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geisen R, Schmidt-Heydt M, Karolewiez A. 2006. A gene cluster of the ochratoxin A biosynthetic genes in Penicillium. Mycotoxin Res 22:134–141. doi: 10.1007/BF02956777. [DOI] [PubMed] [Google Scholar]
- 38.Gerin D, De Miccolis Angelini RM, Pollastro S, Faretra F. 2016. RNA-Seq reveals OTA-related gene transcriptional changes in Aspergillus carbonarius. PLoS One 11:e0147089. doi: 10.1371/journal.pone.0147089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandes M, Keller NP, Adams TH. 1998. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol Microbiol 28:1355–1365. doi: 10.1046/j.1365-2958.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- 40.Payne GA, Nystrom GJ, Bhatnagar D, Cleveland TE, Woloshuk CP. 1993. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl Environ Microbiol 59:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang P-K. 2003. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol Genet Genomics 268:711–719. doi: 10.1007/s00438-003-0809-3. [DOI] [PubMed] [Google Scholar]
- 42.Liu B-H, Chu FS. 1998. Regulation of aflR and its product, AflR, associated with aflatoxin biosynthesis. Appl Environ Microbiol 64:3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang S, Chen Z, Huang G, Yu T, Yang P, Li J, Fu Y, Yuan S, Chen S, Xu A. 2012. HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res 22:1581–1588. doi: 10.1101/gr.133652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, Salzberg SL, White O. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol 9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33(Web Server issue):W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res 18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majoros WH, Pertea M, Salzberg SL. 2004. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 49.Birney E, Clamp M, Durbin R. 2004. GeneWise and Genomewise. Genome Res 14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Z-L, Bao J, Reecy JM. 2008. CateGOrizer: a Web-based program to batch analyze Gene Ontology classification categories. Online J Bioinform 9:108–112. [Google Scholar]
- 51.Collyda C, Diplaris S, Mitkas P, Maglaveras N, Pappas C. 2007. Enhancing the quality of phylogenetic analysis using fuzzy hidden Markov model alignments. Stud Health Technol Inform 129(Part 2):1245–1249. [PubMed] [Google Scholar]
- 52.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. 2013. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res 41(Database issue):D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 54.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 55.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.