Compared to chemical methods, the biotransformation of glycyrrhizin (GL) into glycyrrhetinic acid 3-O-mono-β-d-glucuronide (GAMG), which has a higher sweetness and stronger pharmacological activity than those of GL, via catalysis by β-glucuronidase is an environmentally friendly approach due to the mild reaction conditions and the high yield of GAMG. However, currently available GUSs show low substrate specificity toward GL and further hydrolyze GAMG to glycyrrhetinic acid (GA) as a by-product, increasing the difficulty of subsequent separation and purification. In the present study, we succeeded in isolating a novel β-glucuronidase (named TpGUS79A) from Talaromyces pinophilus Li-93 that specifically hydrolyzes GL to GAMG without the formation of GA. TpGUS79A also shows higher activity on GL than those of the previously characterized GUSs. Moreover, the gene for TpGUS79A was cloned and its function verified by heterologous expression in P. pastoris. Therefore, TpGUS79A can serve as a powerful biocatalyst for the cost-effective production of GAMG through GL transformation.
KEYWORDS: β-glucuronidase, glycoside hydrolase family 79, glycyrrhetinic acid 3-O-mono-β-d-glucuronide, GAMG, glycyrrhizin, GL, Talaromyces pinophilus Li-93
ABSTRACT
Glycyrrhetinic acid 3-O-mono-β-d-glucuronide (GAMG), which possesses a higher sweetness and stronger pharmacological activity than those of glycyrrhizin (GL), can be obtained by removal of the distal glucuronic acid (GlcA) from GL. In this study, we isolated a β-glucuronidase (TpGUS79A) from the filamentous fungus Talaromyces pinophilus Li-93 that can specifically and precisely convert GL to GAMG without the formation of the by-product glycyrrhetinic acid (GA) from the further hydrolysis of GAMG. First, TpGUS79A was purified and identified through matrix-assisted laser desorption ionization–tandem time of flight mass spectrometry (MALDI-TOF-TOF MS) and deglycosylation, indicating that TpGUS79A is a highly N-glycosylated monomeric protein with a molecular mass of around 85 kDa, including around 25 kDa of glycan moiety. The gene for TpGUS79A was then cloned and verified by heterologous expression in Pichia pastoris. TpGUS79A belonged to glycoside hydrolase family 79 (GH79) but shared low amino acid sequence identity (<35%) with the available GH79 GUS enzymes. TpGUS79A had strict specificity toward the glycan moiety but poor specificity toward the aglycone moiety. Interestingly, TpGUS79A recognized and hydrolyzed the distal glucuronic bond of GL but could not cleave the glucuronic bond in GAMG. TpGUS79A showed a much higher catalytic efficiency on GL (kcat/Km of 11.14 mM−1 s−1) than on the artificial substrate pNP β-glucopyranosiduronic acid (kcat/Km of 0.01 mM−1 s−1), which is different from the case for most GUSs. Homology modeling, substrate docking, and sequence alignment were employed to identify the key residues for substrate recognition. Finally, a fed-batch fermentation in a 150-liter fermentor was established to prepare GAMG through GL hydrolysis by T. pinophilus Li-93. Therefore, TpGUS79A is potentially a powerful biocatalyst for environmentally friendly and cost-effective production of GAMG.
IMPORTANCE Compared to chemical methods, the biotransformation of glycyrrhizin (GL) into glycyrrhetinic acid 3-O-mono-β-d-glucuronide (GAMG), which has a higher sweetness and stronger pharmacological activity than those of GL, via catalysis by β-glucuronidase is an environmentally friendly approach due to the mild reaction conditions and the high yield of GAMG. However, currently available GUSs show low substrate specificity toward GL and further hydrolyze GAMG to glycyrrhetinic acid (GA) as a by-product, increasing the difficulty of subsequent separation and purification. In the present study, we succeeded in isolating a novel β-glucuronidase (named TpGUS79A) from Talaromyces pinophilus Li-93 that specifically hydrolyzes GL to GAMG without the formation of GA. TpGUS79A also shows higher activity on GL than those of the previously characterized GUSs. Moreover, the gene for TpGUS79A was cloned and its function verified by heterologous expression in P. pastoris. Therefore, TpGUS79A can serve as a powerful biocatalyst for the cost-effective production of GAMG through GL transformation.
INTRODUCTION
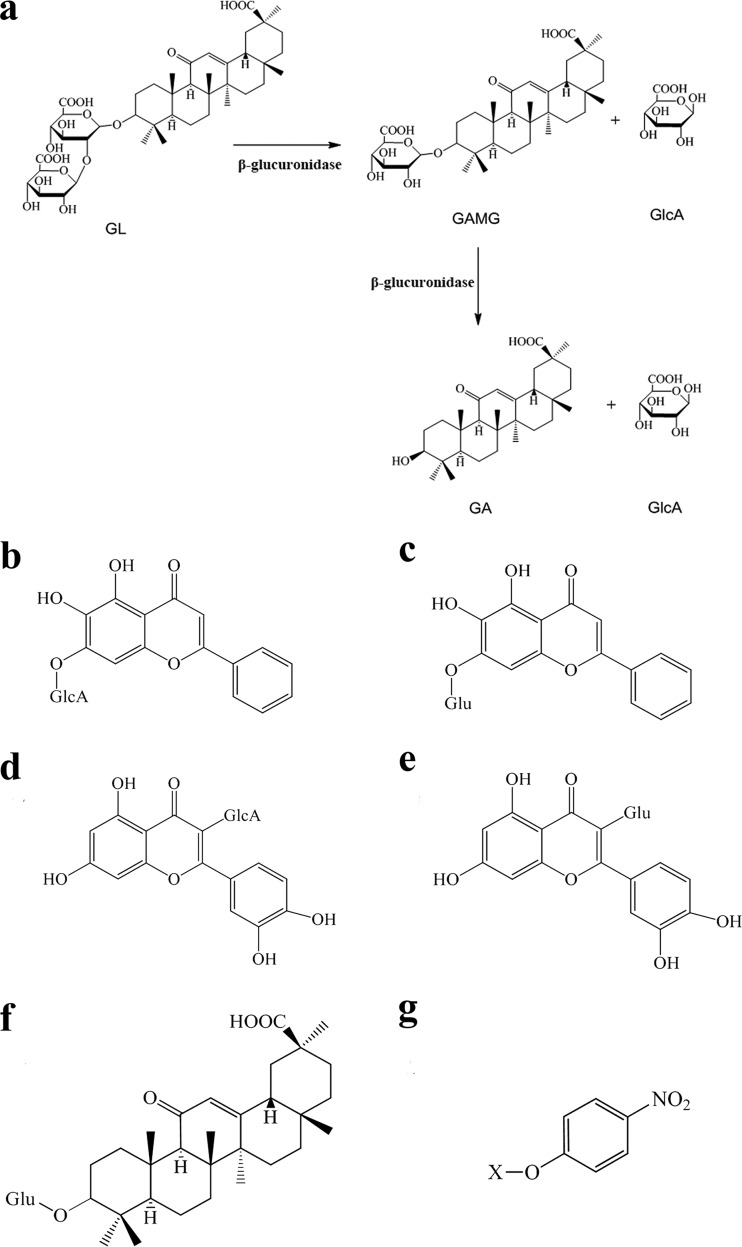
Licorice is one of the most abundantly used medicinal herbs in the world. It is strongly recommended for treatments of pulmonary diseases, deficiencies of the spleen and stomach, and inflammatory disorders (1). Glycyrrhizin (GL), one of the main compounds in licorice, is a triterpenoid saponin consisting of one glycyrrhetinic acid (GA) covalently linked with two glucuronic acids (GlcA) (Fig. 1a). GL has also been used worldwide as a natural sweetener due to its sweet taste. Moreover, various pharmacological activities of GL, such as anti-inflammatory (2), antiallergic (3), anticancer (4), antiviral (5, 6), antibacterial (7), and antioxidative (8) activities, have been discovered. However, the strong polarity of GL causes a poor ability to cross the cell membrane, which results in a low absorption efficiency. Fortunately, elimination of the distal glucuronic acid of GL leads to the formation of glycyrrhetinic acid 3-O-mono-β-d-glucuronide (GAMG), which holds the same biological activities as GL but displays appropriate chemical polarity for efficient absorption (8). Furthermore, GAMG exhibits more efficient physiological functions than those of GL in some cases. For example, GAMG shows a higher activity against Helicobacter pylori, a bacterium which is well known to contribute to gastritis or peptic ulcers (7). Moreover, Park et al. reported that GAMG has more potent antiallergic activity than that of GL (3). In addition, as a sweetener, GAMG is nearly 1,000-fold sweeter than sucrose and 5-fold sweeter than GL (9). Therefore, GAMG has been considered a promising food additive and therapeutic drug with great economic value. However, it is not easy to obtain GAMG directly from licorice because of the very low content. It is also quite difficult to produce GAMG by chemosynthesis. Therefore, the conversion of GL to GAMG is a sensible approach because of the relatively high GL content in licorice (2 to 8% of the dry weight). The transformation of GL into GAMG through chemical methods requires a boiling acidic solution and sophisticated instruments, and the product yield is also quite low (10, 11). In contrast, biotransformation of GL into GAMG via β-glucuronidases (GUSs) or microbes capable of producing GUSs (12–14) is an environmentally friendly approach due to the mild reaction conditions and the higher yield of GAMG.
FIG 1.
(a) Scheme of hydrolysis of glycyrrhizin (GL) into glycyrrhetinic acid 3-O-mono-β-d-glucuronide (GAMG) and glycyrrhetinic acid (GA), catalyzed by β-glucuronidase. (b to g) Structures of typical glucuronides. (b) Baicalin; (c) baicalein 7-O-β-d-glucoside; (d) quercetin 3-O-β-d-glucuronide; (e) quercetin 3-O-β-d-glucoside; (f) GAM-Glc; (g) pNP-sugar. “X” stands for glucopyranose (Glc), galactopyranose (Gal), xylopyranose (Xyl), mannopyranose (M), or glucopyranosiduronic acid (GlcA).
β-Glucuronidase (EC 3.2.1.31), a glycosyl hydrolase (GH) widely found in humans, animals, microorganisms, and plants, can hydrolyze the glucuronic bond to remove the glucuronic acid from glycoside. Most GUSs have been classified as GH2 enzymes, and a small number have been categorized as GH1, GH79, and GH30 enzymes (15; http://www.cazy.org). For example, human endogenous GUSs and intestinal bacterial GUSs belonging to GH2 are involved in intracellular redundant mucopolysaccharide degradation to avoid mucopolysaccharide storage disease as well as enterohepatic recirculation of various glucuronides, such as potential toxins, hormones, and drugs (16–20). The GUSs belonging to GH1 and GH79 show limited substrate specificities. For example, Klotho from mammals, belonging to GH1, shows activity on steroid β-glucuronide (21, 22), and GH79 mammalian GUSs (heparanases) specifically degrade the heparan sulfate proteoglycans located on the cell membrane. Similarly, GH79 GUSs from plants and fungi have been reported to act on plant proteoglycans (23, 24). For example, GUSs from Aspergillus niger (AnGlcAase) and Neurospora crassa (NcGlcAase) showed specific activities on the glycan moieties of arabinogalactan-proteins and released both GlcA and 4-O-methyl-GlcA (24). Because of their special physiological and biochemical characteristics, GUSs have been applied conventionally for cancer diagnosis, pharmacotherapy, and genetic manipulation (25–29). Recently, GUSs have attracted great attention due to their ability to modify natural medicinal glycosides by removing the sugar moiety to improve their efficacy and reduce the side effects (30).
Several GUSs derived from animal livers and human intestinal bacteria can hydrolyze GL into GAMG (31, 32), but their low substrate specificities cause the further conversion of GAMG to GA (Fig. 1a). Many fungi, such as Aspergillus terreus (33), A. niger, Penicillium frangnatum, and Penicillium cyclopium (34), can convert GL to GAMG, but they cannot distinguish between GL and GAMG as substrates during hydrolysis. So far, only three GUSs, from the intestinal bacterium Streptococcus LJ-22 (35), the yeast Cryptococcus magnus MG-27 (12), and the filamentous fungus Penicillium purpurogenum Li-3 (13), have been identified to show high substrate specificity for GL. However, their low activities and the lack of knowledge about their catalytic properties and genetic information limit their large-scale application in the production of GAMG through GL hydrolysis.
Recently, we screened a filamentous fungus, Talaromyces pinophilus Li-93, from the soil of a licorice plantation in Xinjiang Province, China, which can convert GL into GAMG without the formation of GA. Furthermore, the genomic sequence of strain Li-93 has been obtained by de novo genome sequencing. In the present study, we aimed to identify and characterize the enzyme selectively hydrolyzing GL into only GAMG from strain Li-93. First, the enzyme (designated TpGUS79A) from strain Li-93 was purified and identified, and then the gene for TpGUS79A was cloned and expressed in Pichia pastoris for functional identification. In addition, the physicochemical properties and substrate specificity of TpGUS79A were disclosed in the study. The substrate recognition mechanism was uncovered by molecular docking and site-directed mutagenesis. A fed-batch fermentation in a 150-liter fermentor was developed to transform GL into GAMG via T. pinophilus Li-93.
RESULTS
Purification and identification of the enzyme from T. pinophilus Li-93 that converts GL into GAMG.
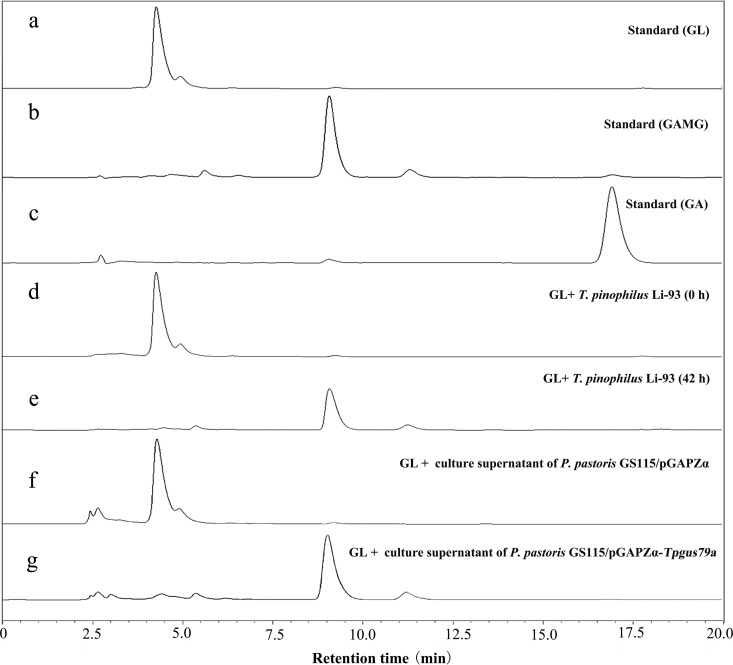
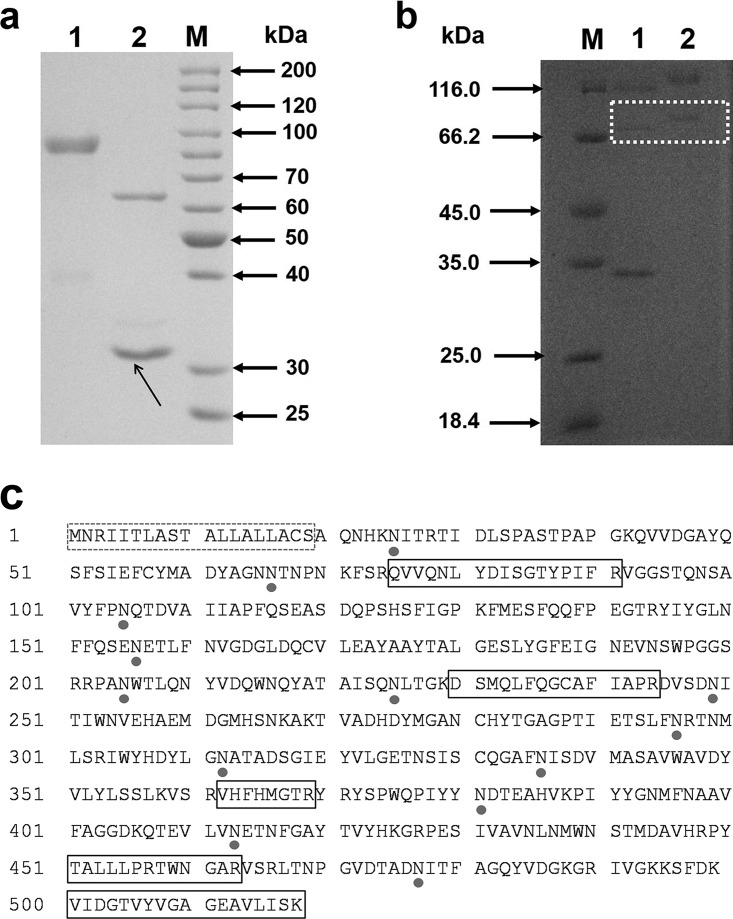
T. pinophilus Li-93, screened from the soil of a licorice plantation by use of GL as the sole carbon source, was found to specifically convert GL to GAMG. As shown in Fig. 2d and e, all the GL was converted to GAMG after 42 h, while no GA was detected. The enzyme (designated TpGUS79A) from strain Li-93 that hydrolyzed GL into GAMG was induced by adding GL to the culture medium. The secreted enzyme was purified through three successive purification steps by monitoring GL hydrolytic activity. The enzyme from the final purification step showed a high purity on SDS-PAGE (Fig. 3a), and its molecular mass was estimated to be 85 kDa. The purification process is summarized in Table 1. Purified TpGUS79A was obtained, with a specific activity of 11.97 U mg protein−1 (the corresponding normalized rate was 11.4 s−1), and the total amount of pure protein was 60 μg from 1,500 ml of culture supernatant.
FIG 2.
HPLC analysis of the transformation of glycyrrhizin (GL) catalyzed by T. pinophilus Li-93 and the engineered strain P. pastoris GS115/pGAPZα-Tpgus79a. (a) GL. (b) GAMG. (c) GA. (d and e) Culture supernatants of T. pinophilus Li-93 after culture for 0 h and 42 h, respectively. (f and g) Products of transformation of GL catalyzed by culture supernatants of the control strain P. pastoris GS115/pGAPZα and the engineered strain P. pastoris GS115/pGAPZα-Tpgus79a, respectively. Note that GlcA has no absorbance at 254 nm, which was the wavelength for HPLC detection, so the peak for GlcA cannot be observed in the HPLC chromatograms.
FIG 3.
(a) SDS-PAGE analysis of purified TpGUS79A from T. pinophilus Li-93. Lane M, molecular size marker; lane 1, purified TpGUS79A; lane 2, N-deglycosylated TpGUS79A. N-Glycosidase F is indicated by an arrow. (b) SDS-PAGE analysis of recombinant TpGUS79A-P expressed in P. pastoris. Lane M, molecular size marker; lane 1, N-deglycosylated TpGUS79A-P; lane 2, purified TpGUS79A-P. Recombinant TpGUS79A-P is emphasized with a white dotted box. (c) Amino acid sequence of TpGUS79A from T. pinophilus Li-93. The putative N-glycosylation sites are shown by closed circles. The dotted box indicates the signal peptide, and solid boxes indicate the peptides identified by MALDI-TOF-TOF MS.
TABLE 1.
Purification of the enzyme converting GL into GAMG from T. pinophilus Li-93
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U mg−1) | Fold purification | Yield (%) |
|---|---|---|---|---|---|
| Culture supernatant | 310.50 | 15.19 | 0.048 | 1.00 | 100 |
| 50% acetone precipitation | 36.63 | 11.43 | 0.31 | 6.46 | 75.25 |
| HiTrap SP FF chromatography | 0.23 | 2.10 | 9.40 | 195.83 | 13.82 |
| HiTrap DEAE HP chromatography | 0.06 | 0.712 | 11.97 | 249.38 | 4.69 |
Peptide mass fingerprinting of TpGUS79A was obtained by matrix-assisted laser desorption ionization–tandem time of flight mass spectrometry (MALDI-TOF-TOF MS), and the results were submitted to Mascot (Matrix Science) for database searching. The results suggested that the hypothetical protein TCE0_044r16781 (accession number GAM42629.1) from Talaromyces cellulolyticus had the highest score for matching the peptide mass fingerprinting of TpGUS79A, but its function was not annotated.
Cloning of putative gene encoding TpGUS79A and functional verification.
Based on the gene sequence of the homolog TCE0_044r16781, a putative coding sequence of TpGUS79A, consisting of 1,554 bases (designated Tpgus79a) sharing 98.46% sequence identity with the gene for the protein TCE0_044r16781, was searched against the genome sequence of T. pinophilus Li-93. A coding sequence of 1,554 bases and a genomic sequence of 1,790 bases were amplified from cDNA and genomic DNA of T. pinophilus Li-93, respectively. The sequence alignment indicated that the gene sequence of 1,790 bases included 5 exons and 4 introns. In order to investigate whether the candidate gene encodes TpGUS79A, the candidate gene with 1,554 bases was cloned and transformed into P. pastoris, yielding the engineered strain GS115/pGAPZα-Tpgus79a for the heterologous expression of TpGUS79A (TpGUS79A-P). After cultivation of the strain in yeast extract-peptone-dextrose (YPD) medium for 72 h, the supernatant was harvested for detection of the enzyme expression and activity toward GL. SDS-PAGE analysis indicated that TpGUS79A-P was successfully expressed and secreted into the medium (Fig. 3b). The amount of TpGUS79A-P heterologously expressed in P. pastoris was quite low compared to the amount of TpGUS79A, so the band was weaker than that for TpGUS79A. In addition, a band for an unknown contaminant protein was also observed, which was hard to remove with the current purification protocols. TpGUS79A-P showed activity for hydrolyzing GL into GAMG without GA being detected, indicating that the substrate specificity of TpGUS79A was not changed after heterologous expression in P. pastoris (Fig. 2f and g). Furthermore, the protein band for TpGUS79A-P on the SDS-PAGE gel was identified by MALDI-TOF-TOF MS, and the peptide mass fingerprint acquired was similar to that for TpGUS79A (Fig. 3c). Therefore, we confirmed that the candidate gene Tpgus79a mined from T. pinophilus Li-93 encodes TpGUS79A, which can hydrolyze GL into GAMG with high substrate specificity.
Protein analysis.
Native PAGE and analytical ultracentrifuge analysis suggested that TpGUS79A behaves as a monomeric protein (data not shown). The signal peptide consists of the first 19 amino acid residues, and the N-terminal amino acid sequence was determined to be AQNHK (Fig. 3c). TpGUS79A consists of 517 amino acid residues and has a theoretical molecular weight of 57.2 kDa. However, the molecular weights of both native and recombinant TpGUS79A are much larger than the theoretical value (Fig. 3). Amino acid sequence analysis of TpGUS79A through the NetNGlyc server (http://www.cbs.dtu.dk/services/NetNGlyc/) revealed that there are 13 potential N-glycosylation sites in the enzyme (Fig. 3c). In addition, the molecular weight of TpGUS79A was also sharply reduced after treatment with peptide-N-glycosidase F (PNGase) (Fig. 3a). Therefore, the dramatically higher molecular weight than the theoretical value for TpGUS79A was attributed to its heavy N-glycosylation.
Both domain analysis carried out by Pfam (36) and phylogenetic analysis (see Fig. S1 in the supplemental material) indicated that TpGUS79A belongs to GH79. Interestingly, TpGUS79A showed very low sequence identities with other previously reported GUSs of GH79, such as AnGlcAase from A. niger (34% identity) (24), NcGlcAase from N. crassa (33% identity) (24), and AcGlcA79A from Acidobacterium capsulatum (25% identity) (37).
Physicochemical properties of TpGUS79A.
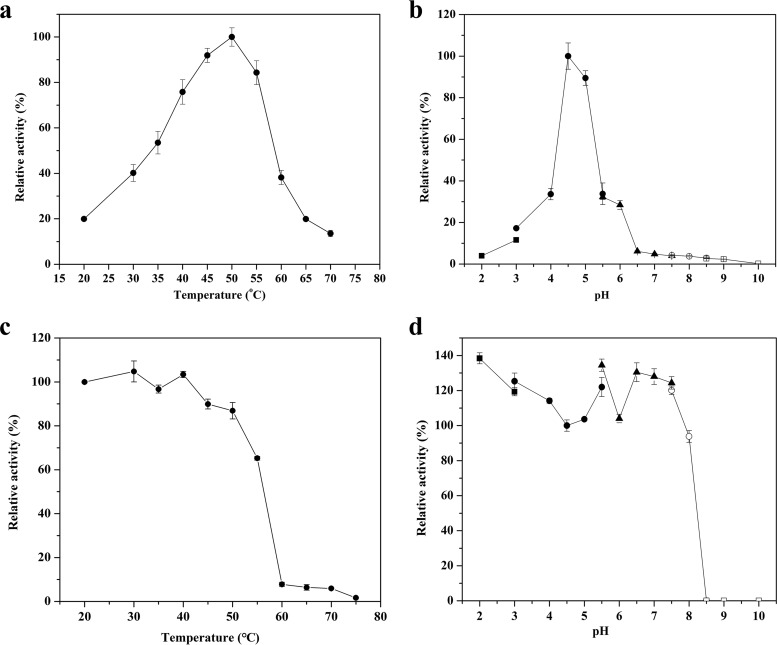
As shown in Fig. 4a and b, the highest activity of TpGUS79A was observed at 50°C and pH 4.5. TpGUS79A was stable after incubation below 40°C for 30 min but lost most of its activity at 60°C (Fig. 4c). On the other hand, TpGUS79A displayed less than 5% relative activity at pH 6.5 and above (Fig. 4b). TpGUS79A was stable in the pH range of 2.0 to 7.5 at 4°C for 12 h, but it lost most of its activity at pHs higher than 8.5 under the same incubation conditions (Fig. 4d). Except for Na+ and K+, all other tested metal ions significantly inhibited TpGUS79A activity (Fig. S2a). More than 90% of TpGUS79A activity was lost in the presence of 10 mM Fe2+, Cu2+, or Al3+. On the other hand, dithiothreitol (DTT), EDTA, glutathione (GSH), β-mercaptoethanol (ME), and urea were not detrimental to TpGUS79A activity, instead stimulating the activity. However, Triton X-100 and SDS significantly inhibited the enzyme activity (Fig. S2b).
FIG 4.
Effects of pH and temperature on TpGUS79A activity. (a) Optimum temperature; (b) optimum pH value; (c) thermostability; (d) pH stability. ■, glycine-HCl buffer (pH 2.0 to 3.0); ●, acetate buffer (pH 3.0 to 5.5); ▲, phosphate buffer (pH 5.5 to 7.5); ○, Tris-HCl buffer (pH 7.5 to 8.5); □, glycine-NaOH (pH 8.5 to 10). The experiments were performed in triplicate, and the error bars show 1 standard deviation.
Substrate specificity of TpGUS79A.
The substrate specificity of TpGUS79A was measured using GL, GAMG, and glycyrrhetinic acid 3-O-mono-β-glucoside (GAM-Glc) as glycyrrhetinic glycosides, quercetin 3-O-β-glucuronide, quercetin 3-O-β-d-glucoside, baicalin, and baicalein 7-O-β-d-glucoside as flavonoid glycosides, and pNP-sugars as artificial substrates (Fig. 1b to g). As shown in Table 2, TpGUS79A showed activity toward the glycyrrhetinic glycoside conjugated with two glucuronic acids (GL) and toward flavonoid substrates conjugated with glucuronic acid but showed no activity against pNP-sugars, except pNP-β-GlcA. This result indicated that TpGUS79A had strict glycan specificity but poor substrate specificity toward aglycone. However, among the tested substrates, TpGUS79A showed the highest activity toward GL (100%), followed by quercetin 3-O-β-d-glucuronide (51.2%), baicalin (13.2%), and pNP-β-GlcA (5.8%). The kinetic parameters for these substrates were determined to further elaborate the substrate specificity of TpGUS79A. The Michaelis-Menten modeling plots of TpGUS79A against these substrates are shown in Fig. S3 in the supplemental material. TpGUS79A showed the highest catalytic efficiency (kcat/Km) on GL, which was 3-fold higher than that on quercetin 3-glucuronide. For flavonoid glucuronides, the kcat/Km value for TpGUS79A toward quercetin 3-glucuronide was 13-fold higher than that for baicalin, mainly due to the Km value for baicalin being 9-fold higher than that for quercetin 3-glucuronide. The Km value for pNP-β-GlcA was 47-fold higher than that for GL, and in addition, the kcat value for pNP-β-GlcA was very low, which resulted in the kcat/Km value toward GL being >1,000-fold higher than that toward pNP-β-GlcA.
TABLE 2.
Substrate specificity of TpGUS79A
| Substrate | Relative activity (%)a | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|
| GL | 100 | 0.54 ± 0.04 | 6.0 ± 0.4 | 11.14 ± 1.0 |
| GAMG | NDb | ND | ND | ND |
| GAM-Glc | ND | ND | ND | ND |
| Quercetin 3-glucuronide | 51.2 | 0.76 ± 0.04 | 2.9 ± 0.3 | 3.83 ± 0.5 |
| Quercetin 3-glucoside | ND | ND | ND | ND |
| Baicalin | 13.2 | 6.88 ± 0.55 | 2.0 ± 0.2 | 0.29 ± 0.01 |
| Baicalein-7-glucoside | ND | ND | ND | ND |
| pNP-β-GlcA | 5.8 | 25.56 ± 1.84 | 0.26 ± 0.01 | 0.01 ± 0.001 |
| pNP-β-Glc | ND | ND | ND | ND |
| pNP-β-Gal | ND | ND | ND | ND |
| pNP-β-Xyl | ND | ND | ND | ND |
| pNP-β-M | ND | ND | ND | ND |
Assays were carried out in 50 mM acetate buffer (pH 4.5) at 45°C. The activity on GL was taken as 100%.
ND, could not be detected.
Homology modeling and substrate recognition analysis.
The homology model of TpGUS79A was first derived from SWISS-MODEL based on the AcGlcA79A structure (Protein Data Bank [PDB] entry 3VNZ; 25% sequence identity). However, the QMEAN Z-score of the model was −7.26, which meant that the structure quality was poor. The structure was further optimized by a 30-ns molecular dynamics (MD) simulation, which showed a QMEAN Z-score of −5.42, indicating that the structure quality was greatly improved. The resulting structure and the structures of two heparanases (PDB entries 5BWI and 5E9B; sequence identities of 27% and 38%, respectively) were employed as templates for homology modeling of TpGUS79A by SWISS-MODEL. The Ramachandran plot indicated that the quality measures were 83.4% in the favored region, 15.9% in the allowed region, and 0.7% in the disallowed region (three residues [Ser154, His386, and Arg427]). A QMEAN score higher than −4 usually indicates a high-quality structure, and our modeled structure showed a QMEAN Z-score of −4.72, indicating that some part of the modeled structure may still be inaccurate. Therefore, we made assumptions for the structural analysis based on the modeled structure and further verified the critical issues in combination with sequence alignment and mutagenesis analysis.
As shown in Fig. 5a, the modeled structure of TpGUS79A was very similar to that of AcGlcA79A, and sequence coverage was 82% (residues 40 to 516). Apart from the 19 residues of the signal peptide, 21 residues in the N terminus were missing due to the corresponding missed residues in the template of the AcGlcA79A structure. TpGUS79A is a monomer composed of two domains, namely, a TIM barrel domain (residues 1 to 400) and a β-sandwich domain (residues 401 to 516), similar to the reported GUS structure for GH79. A sequence alignment with other GUSs in GH79 (two fungal GUSs [AnGlcAase and NcGlcAase] and one bacterial GUS [AcGlcAase]) is shown in Fig. 6. Glu192 and Glu325, located in the 4th and 7th β-strands of the TIM barrel, respectively, were identified to be catalytic residues. Two mutants (E192A and E325A) were constructed and showed no activity under the standard activity assay conditions, confirming that these two sites were catalytic residues. Sequence alignment suggested that TpGUS79A adopts a retaining mechanism during catalysis. The specific roles of the two catalytic residues were confirmed by a chemical rescue test (Fig. S4). The activity of only the E325A mutant was partially rescued by adding acetic acid as an exogenous nucleophile, suggesting that E325 acts as a nucleophile.
FIG 5.
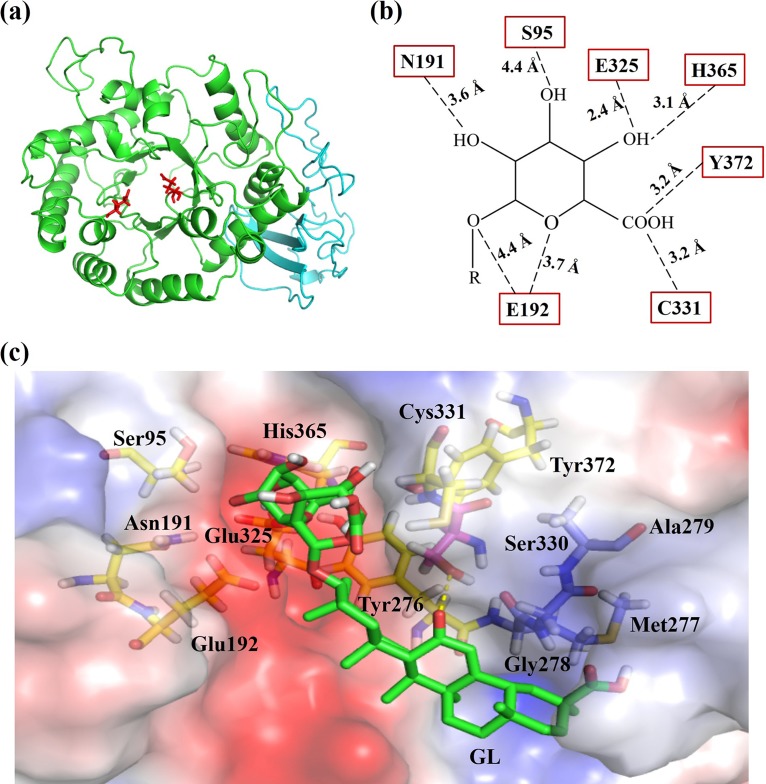
(a) Modeled structure of TpGUS79A based on the structures of AcGlcA79A (PDB entry 3VNZ) and two heparanases (PDB entries 5BWI and 5E9B). The TIM barrel domain is shown in green, and the immunoglobulin-like β-sandwich domain is shown in cyan. The two catalytic residues, Glu192 and Glu325, are shown as red sticks. (b) Residues of TpGUS79A involved in recognizing the substrate GL. Residues interacting with glycan and aglycone moieties are shown as yellow and orange sticks, respectively, and residues forming the hydrophobic region to interact with aglycone are shown as blue sticks. (c) Surface representation of the hydrophobic region of the TpGUS79A catalytic pocket. The hydrophobic area is shown in blue. Ser330 forms a polar contact with aglycone, while three residues, Met277, Gly278, and Ala279, form a hydrophobic region on one side of the substrate channel which may interact with the aglycone of GL. The hydrogen bond is shown by yellow dashed lines.
FIG 6.
Sequence alignment of GH79 GUSs, including AnGlcAase from A. niger, NcGlcAase from N. crassa, and AcGlcA79A from A. capsulatum. The dashed boxes indicate the conserved sequences containing catalytic residues, and stars indicate catalytic residues. Three sequence identity levels (100%, 75%, and 50%) are shown in black, purple, and cyan, respectively.
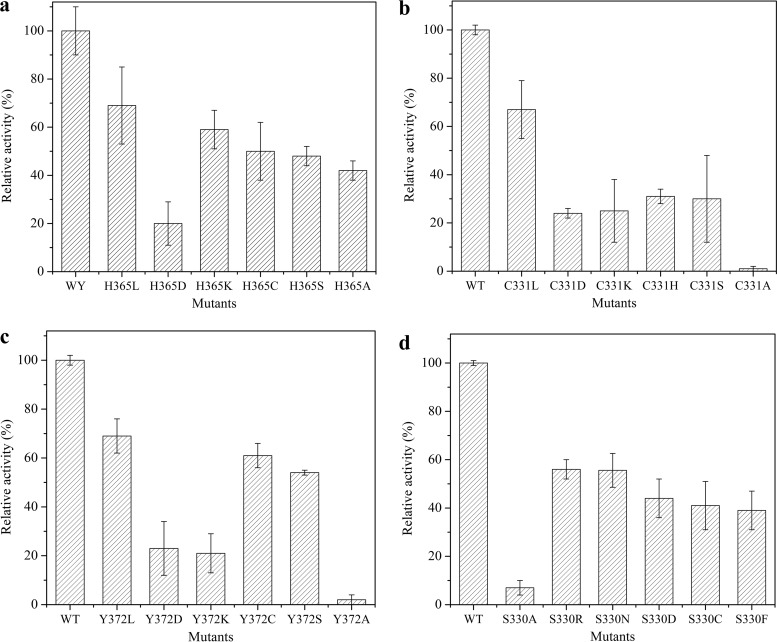
Docking analysis predicted that the following eight residues were involved in recognizing the glycan moiety of GL, through polar interactions and aromatic stacking: Ser95, Asn191, Glu192, Tyr276, Glu325, Tyr372, His365, and Cys331 (Fig. 5b and c). Among these residues, His365, Tyr276, and Asn191 are also conserved in bacterial AcGlcA79A for glycan recognition. Interestingly, seven residues (Ser95, Asn191, Glu192, Glu325, Tyr372, His365, and Cys331) participated in interacting with the first glycan of GL, and little interaction was formed with the second glycan of GL (corresponding to the glycan of GAMG). Therefore, after the first glycan of GL was hydrolyzed, the left glycan could not be recognized due to little interaction, which resulted in TpGUS79A showing no activity toward GAMG. Substantial mutagenesis of these residues was performed to verify their function. The mutants showed similar optimal temperatures for catalysis (data not shown), so the change in activity was not due to the changed temperature profile but to the change of the catalytic pocket caused by the residue mutation. SDS-PAGE analysis of some purified mutants is shown in Fig. S5. As shown in Fig. 7, TpGUS79A lost 80% of its activity if His365 was mutated to aspartic acid, which may be due to the charge change at this site. When Cys331 and Tyr372 were mutated to alanine, TpGUS79A lost more than 95% of its activity, which may be due to the short side chain of alanine not being able to form polar contact with the glycan moiety. When Cys331 and Tyr372 were mutated to aspartic acid or lysine, 75 to 79% activity losses were also observed, perhaps because the charge may not be favorable for interaction with the substrate at these two sites. TpGUS79A lost around 50% of its activity when Ser95 was mutated to alanine, while TpGUS79A was completely deactivated when Asn191 was mutated to alanine. Tyr276 was located parallel to the second glycan moiety of GL to form an aromatic stacking interaction, which may help to stabilize the GL conformation during catalysis. When Tyr276 was mutated to alanine, the enzyme was completely deactivated.
FIG 7.
Relative activities of mutants with substitutions at residues 365 (a), 331 (b), 372 (c), and 330 (d). The activity was tested by GL hydrolysis under standard conditions. The experiment was performed in triplicate, and the error bars show 1 standard deviation.
In contrast to those with glycan, very few polar interactions were seen with the aglycone moiety of GL. As shown in Fig. 5c, only Ser330 formed one hydrogen bond with the O atom of GL. When Ser330 was mutated to alanine, a 93% activity loss was detected due to the short chain of alanine not forming an interaction with aglycone. When Ser330 was mutated to other residues, such as arginine, asparagine, aspartic acid, cystine, phenylalanine, and proline, around 44 to 73% of activity was lost (Fig. 7d). The left side of the aglycone was in close proximity to a negative electrostatic potential, although no interaction was established with the aglycone. This is similar to the results of our previous work, in which the crystal structure of a GH2 fungal β-glucuronidase from Aspergillus oryzae Li-3 in complex with GAMG was solved, and a negative electrostatic potential was also observed in the active pocket of PGUS (Fig. S6). These results indicate that the negative electrostatic potential in the active pocket of TpGUS79A is not out of the ordinary. More importantly, the right side of the aglycone was surrounded by a hydrophobic region composed of three residues, Met277, Gly278, and Ala279, which likely establishes interaction with the hydrophobic aglycone during GL catalysis (Fig. 5c).
Pilot-scale GAMG production by T. pinophilus Li-93.
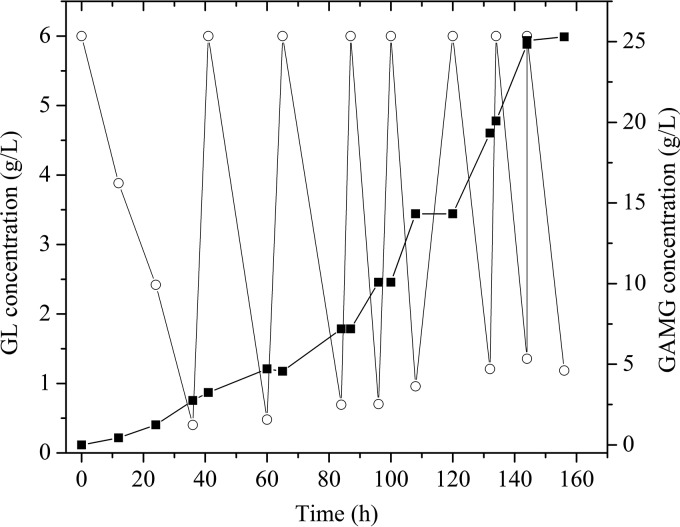
Finally, the potential application of T. pinophilus Li-93 to GAMG preparation from GL hydrolysis was investigated in a 150-liter fermentor in fed-batch mode. At the beginning, the fermentor was filled with 90 liters of potato dextrose broth medium containing 6 g liter−1 ammonium glycyrrhizinate. The biotransformation was initiated by adding T. pinophilus Li-93 seed medium with a 10% inoculum. As shown in Fig. 8, after 36 h, 93% of GL was converted to GAMG. At 41 h, 540 g ammonium glycyrrhizinate was fed into the system every 24 h to maximize the GAMG yield. After 156 h (total of 7 feedings), 25.3 g liter−1 GAMG was obtained, with a GL conversion rate of 95.3%. To our knowledge, this is the largest-scale preparation of GAMG through GL hydrolysis ever reported, indicating that T. pinophilus Li-93 producing TpGUS79A shows great potential for industrial application.
FIG 8.
Fed-batch fermentation to prepare GAMG through GL hydrolysis by T. pinophilus Li-93 in a 150-liter fermentor. ○, GL; ■, GAMG.
DISCUSSION
In this study, a unique β-glucuronidase (designated TpGUS79A) capable of selectively hydrolyzing GL into GAMG without the production of GA as a by-product was identified from the filamentous fungus T. pinophilus Li-93. TpGUS79A is in monomeric form in solution, with a molecular weight of 85 kDa. In addition, TpGUS79A is heavily N-glycosylated, since its molecular weight was decreased by 20 kDa after deglycosylation in vitro. These results suggest that TpGUS79A is very different from β-glucuronidases I and II in selectively converting GL into GAMG (35). For example, GUS from Streptococcus LJ-22 is a homodimer with a molecular weight of 140 kDa (35), and GUS purified from P. purpurogenum Li-3 belongs to GH2 and is a homotetramer, each subunit of which has a molecular weight of 69.72 kDa (38). GUS from C. magnus MG-27 also showed high substrate selectivity for GL, but it was not isolated and characterized, so no comparison can be made with TpGUS79A.
This study reports a GUS gene coding for an enzyme (TpGUS79A) with the capability of selectively hydrolyzing GL into GAMG only, without the production of GA. TpGUS79A has been identified to belong to GH79, which is quite different from the families for previously reported GUSs for converting GL, such as AtGUS (from A. terreus) (33), PGUS (from A. oryzae; GenBank accession number ABU68712), and AuGUS (from Aspergillus ustus) (39), which belong to GH2. Though TpGUS79A is classified as a GH79 enzyme, it shares very low sequence identity (less than 35%) with other GUSs from GH79. Phylogenetic analysis (see Fig. S1 in the supplemental material) indicated that TpGUS79A is not in the same clade with GH79 GUSs from other fungi and is even further away from GUSs from bacteria and plants in terms of genetic distance.
Compared to other GUSs with low substrate specificity for GL (33, 40), TpGUS79A was more sensitive to heat and alkaline pH, similar to other GH79 GUSs (23, 24). Except for monovalent metal ions and Mg2+, all other tested metal ions were detrimental to the activity of TpGUS79A (Fig. 5a). In contrast, the activities of PGUS-E (40), PGUS-P (40, 41), AtGUS-E (33), and SvGUS (from Scutellaria viscidula) (42) were not inhibited by the common metal ions and were inhibited only by heavy metal ions. Our previous study reported that N-glycosylated GUS was more sensitive to metal ions than nonglycosylated GUS (40). We found that TpGUS79A entirely formed inclusion bodies without activity when its gene was expressed in Escherichia coli. In addition, TpGUS79A lost most catalytic activity after its deglycosylation by PNGase F, indicating that N-glycosylation is important for TpGUS79A to maintain activity (Fig. S7). These phenomena may indicate that N-linked glycans assist TpGUS79A to fold correctly and maintain its structure. Surprisingly, in contrast to metal ions, most inhibitors (except Triton X-100 and SDS) did not inhibit the enzyme activity; rather, they slightly activated TpGUS79A activity. The presence of N-linked glycans has generally been thought to protect enzymes from inhibitors (40, 43). Therefore, it may be reasonable that the presence of inhibitors did not significantly affect enzyme activity. While the phenomenon that enzyme activity can be activated by inhibitors is rarely reported, the reason for it still remains to be explored.
Most of the GUSs used for biotransformation of natural glucuronides showed much higher binding affinities and catalytic activities on artificial substrates than on the natural glucuronides. For instance, the Km value of LcGUS30 (from Lactobacillus brevis) for baicalin is 11-fold higher than that for pNP-β-GlcA, and its catalytic efficiency (kcat/Km) on pNP-β-GlcA is 19-fold higher than that on baicalin (44). Both PGUS-E and PGUS-P showed much lower Km values and higher kcat/Km values on pNP-β-GlcA than on GL (40). However, TpGUS79A showed the lowest binding affinity for pNP-β-GlcA (Km value of 25.56 mM) and the lowest activity on pNP-β-GlcA, which was only 5.8% of that on GL. This implies that the structure of TpGUS79A may be quite different from those of other GUSs used for GL transformation; perhaps it harbors a unique substrate binding domain that renders its high substrate specificity for GL.
According to our results, TpGUS79A demonstrates a higher specific activity (11.97 U mg−1) and catalytic efficiency (kcat/Km value of 11.14 mM−1 s−1) on GL than those of the other available GUSs (Table S1), such as β-glucuronidase I (35), β-glucuronidase II (35), PGUS-E (40), PGUS-P (40), and AtGUS-E (33). Therefore, TpGUS79A is a highly competitive biocatalyst ready to be applied in the production of GAMG through GL hydrolysis, not only because of its high substrate selectivity but also because of its high activity on GL. Moreover, TpGUS79A also displays activity on flavonoid glucuronides, such as quercetin 3-glucuronide and baicalin. However, its activity on baicalin (kcat/Km value of 0.29 mM−1s−1; specific activity of 0.565 U mg−1) is much lower than those of the GUSs with high specificity for baicalin (Table S2), such as LcGUS30 (44) and SvGUS (42). So far, the GUSs specifically hydrolyzing quercetin 3-glucuronide have rarely been reported. In the present study, TpGUS79A showed a higher activity on quercetin 3-glucuronide than on baicalin, and this activity was even more than half of its activity on GL (Table 2). These results indicate that TpGUS79A does not show strict substrate specificity toward aglycones but displays significantly different affinities and activities on them.
Similar to other GUSs in GH79, TpGUS79A adopts a retaining mechanism, with Glu325 and Glu192 acting as a nucleophile and an acid/base, respectively, as demonstrated by the mutational analysis and activity rescue test. The acid/base residues of GH79 GUSs from bacteria, fungi, and plants are located in the conserved sequence (W/F)XXXGNE (45), which is similar to that (WXXXNE) of GH2 GUSs and heparanases (Fig. 6). Nonetheless, the nucleophilic residues of GH79 GUSs from bacteria and fungi are located in the downstream conserved sequence L(F)G(T)ETNS, which is quite different from those of plant GH79 GUSs and mammal heparanases (PGKKV-WLGE) and of GH2 GUSs.
Conclusions.
In this study, a novel GUS (TpGUS79A) from the filamentous fungus T. pinophilus Li-93 was isolated, purified, and characterized. TpGUS79A displays high substrate specificity and precisely hydrolyzes GL into GAMG without the formation of GA as a by-product. Additionally, the gene for TpGUS79A was cloned and verified by heterologous expression in P. pastoris. TpGUS79A belongs to GH79 and shows a higher GL hydrolytic activity (11.97 U mg−1) and catalytic efficiency (kcat/Km value of 11.14 mM−1 s−1) than those of the previously characterized GUSs. A fed-batch fermentation in a 150-liter fermentor was established to prepare GAMG through GL hydrolysis by T. pinophilus Li-93, with a GL conversion rate of 95.3% after 7 feedings.
MATERIALS AND METHODS
Plasmids, hosts, and chemicals.
The vector PMD-19T (TaKaRa, Dalian, China) and E. coli Top10 were used for gene cloning and plasmid preparation. The vector pGAPZαA and P. pastoris GS115 (Invitrogen, Carlsbad, CA, USA) were used for gene expression. Restriction enzymes and T4 DNA ligase were supplied by Thermo Scientific (Lithuania). Ammonium glycyrrhizinate (70%) was purchased from Xinjiang Tianshan Pharmaceutical Co. (China). Baicalin (baicalein 7-O-β-d-glucuronide), quercetin 3-O-β-d-glucuronide, and baicalein 7-O-β-d-glucoside were purchased from ChromaBio (Chengdu, China). Quercetin 3-O-β-d-glucoside, p-nitrophenyl β-d-glucopyranoside (pNP-β-Glc), pNP β-d-galactopyranoside (pNP-β-Gal), pNP β-xylopyranoside (pNP-β-Xyl), pNP β-glucopyranosiduronic acid (pNP-β-GlcA), trypsin, bovine serum albumin, and glycyrrhizin (GL) were purchased from Sigma-Aldrich (San Diego, CA, USA). All other chemicals used in this study were of the highest analytical grade commercially available.
Strain screening and enzyme production conditions.
Microorganisms from licorice plantation soil in Xinjiang, China, were cultured on plates with screening medium containing glycyrrhizin as the sole carbon source (3 g liter−1 ammonium glycyrrhizinate, 3 g liter−1 NaNO3, 1 g liter−1 K2HPO4, 1 g liter−1 KCl, 0.5 g liter−1 MgSO4·7H2O, 30 g liter−1 agar). The microorganisms growing on screening plates were continually cultured in liquid screening medium, and their culture supernatants were detected by high-pressure liquid chromatography (HPLC) (corresponding parameters are mentioned below). Except for GL, only GAMG could be detected in the culture supernatant of a filamentous fungus named T. pinophilus Li-93 during the culture process. To obtain and isolate the enzyme converting GL to GAMG secreted by strain Li-93, the strain was cultured in potato dextrose broth at 30°C for 48 h in a shaking incubator (200 rpm), and then the 48-h culture was transferred into seed medium with a 10% inoculum and shaken for 24 h at 30°C. Lastly, the seed culture was transferred into enzyme-producing medium with a 10% inoculum and incubated with shaking for 36 to 48 h at 30°C. The preparation method for seed medium was as follows: 200 g scrubbed and diced potatoes was added to 1 liter water, boiled for 30 min, and then filtered by use of a fine sieve; 40 g glucose was added to the potato extract and boiled until dissolved; the pH was adjusted to 5.0; and the medium was diluted to 5 liters with water. The preparation method for enzyme-producing medium was the same as that for seed medium, but 25 g ammonium glycyrrhizinate (5 g liter−1) was added to the medium instead of 40 g glucose.
Purification of the native enzyme.
The culture supernatant of T. pinophilus Li-93 was obtained by centrifugation (13,000 × g for 10 min) after incubation for 42 h. The cell-free culture supernatant was mixed with ice-cold acetone at a 1:1 (vol/vol) ratio and centrifuged at 15,000 × g for 15 min. The precipitate was suspended in buffer A (30 mM NaAc-HAc (sodium acetate-acetic acid), pH 4.5) and dissolved by continuous stirring for 30 min. The insoluble materials from the supernatant were removed by centrifugation at 13,000 × g for 20 min. Subsequently, the supernatant was loaded onto a HiTrap SP FF column (GE Healthcare) equilibrated with buffer A. The column was washed with 5 column volumes (CVs) of buffer A at a flow rate of 2 ml min−1, and the protein was eluted by use of an increasing gradient of elution buffer (30 mM NaAc-HAc, pH 4.5, 500 mM NaCl) at concentrations ranging from 0 to 100% at a flow rate of 1 ml min−1. The active fractions were combined, and the buffer was exchanged with buffer B (30 mM Tris-HCl, pH 7.3) by use of an Amicon Ultra 30K device (Merck Millipore Corp., MA, USA) and loaded onto a HiTrap DEAE FF column (GE Healthcare) equilibrated with buffer B. Afterward, the column was washed with 5 CVs of buffer B at a flow rate of 1 ml min−1. The protein was eluted by use of an increasing gradient of elution buffer (30 mM Tris-HCl, pH 7.3, 500 mM NaCl) at concentrations ranging from 10 to 100% at flow rate of 1 ml min−1. The fractions with high specific activity were combined and used for further investigations. The whole purification process for the native enzyme was conducted at 4°C. Enzyme purity was assessed by SDS-PAGE, and the protein concentration was determined by use of a bicinchoninic acid (BCA) protein assay kit (Bio-Rad Co. Ltd., CA, USA), using bovine serum albumin as a standard.
Protein identification.
The active fraction from the final purification step was identified by MALDI-TOF-TOF MS. The bands for candidate proteins were excised from the SDS-PAGE gel, diced into small pieces, and subsequently subjected to in-gel digestion according to a standard procedure. Briefly, the samples were destained with acetonitrile (ACN)/NH4HCO3, reduced with DTT, alkylated with iodoacetamide, and digested with trypsin at 37°C for 16 h. The tryptic peptide mixtures were extracted, lyophilized, and redissolved in 0.1% trifluoroacetic acid for mass spectrometric analysis. Subsequently, the samples were analyzed by use of a model 4700 proteomics analyzer (MALDI-TOF-TOF; Applied Biosystems, USA). The scope of peptide mass fingerprinting (PMF) mass scanning was from 700 Da to 3,500 Da. The dominant peptides were selected for further analysis by secondary ion mass spectrometry. The data were searched using Mascot (Matrix Science). The parameters for searching in the database were set as follows: the fixed modification was set as carbamidomethyl (C), the variable modification was set as oxidation (M), the database was the NCBI nr database, retrieval taxonomy was set as fungi, the maximum missing cleavage was set as 1, the peptide mass tolerance error scope was ±100 ppm, and the fragment mass tolerance quality error scope was ±0.6 Da.
N-terminal amino acid sequencing and deglycosylation of the enzyme.
The active fraction from the final purification step was loaded onto a polyvinylidene difluoride (PVDF) membrane (Merck Millipore Corp., MA, USA) for N-terminal amino acid sequencing. N-terminal amino acid sequencing of the enzyme was carried out on a model PPSQ-31A protein sequencer (Shimadzu Co. Ltd., Kyoto, Japan).
TpGUS79A was deglycosylated separately under denaturing and nondenaturing reaction conditions by use of peptide-N-glycosidase F (PNGase F) according to the manufacturer's procedure (New England BioLabs, Evry, France).
(i) Denaturing reaction conditions.
Nine microliters of enzyme solution (0.1 to 0.2 mg/ml) mixed with 1 μl glycoprotein denaturing buffer (10×) was denatured by heating at 100°C for 10 min and then was chilled on ice and centrifuged for 10 s. Two microliters of GlycoBuffer 2 (10×), 2 μl 10% NP-40, and 6 μl double-distilled water (ddH2O) were added to the denatured enzyme solution, for a total reaction volume of 20 μl. PNGase F (1 μl) was added to the 20-μl mixture, followed by incubation at 37°C for 1 to 3 h. The deglycosylation results were analyzed by SDS-PAGE.
(ii) Nondenaturing reaction conditions.
PNGase F (2 μl) was added to 10 μl reaction mixture containing 9 μl TpGUS79A solution (0.1 to 0.2 mg/ml) and 1 μl GlycoBuffer 2 (10×) to remove the glycan at 37°C for 1 to 6 h. For nondenatured deglycosylation, an aliquot of TpGUS79A solution mixed with GlycoBuffer 2 and storage buffers of PNGase F after incubation at 37°C for 1 to 6 h was prepared as a negative control, and TpGUS79A subjected to denaturing deglycosylation was used as a positive control for the completely deglycosylated protein. An aliquot of reaction solution was sampled for enzyme activity and SDS-PAGE analysis every 1 h.
Total RNA extraction and cDNA library construction.
T. pinophilus Li-93 was cultivated at 30°C for 3 days in modified Czapek medium (3 g liter−1 NaNO3, 1 g liter−1 K2HPO4, 1 g liter−1 KCl, 0.5 g liter−1 MgSO4·7H2O, 0.01 g liter−1 FeSO4·7H2O, 3 g liter−1 GL, pH 5.0) with shaking (200 rpm). For extraction of total RNA, the hyphae were harvested by centrifugation at 13,000 × g for 10 min, washed two times with sterile water, and ground into a fine powder by use of a pestle in liquid nitrogen. Total RNA was extracted using a fungal RNA kit (Omega Bio-Tek, GA, USA) according to the manufacturer's instructions. The cDNA was synthesized using PrimeScript RT master mix (TaKaRa, China).
Gene cloning of the native enzyme.
A forward primer (5′-ATGAATCGCATCATCACCCTCGCCT-3′) and a reverse primer (5′-TTAAACCCAGCCCTGTGCAAGTCTC-3′) were designed and synthesized for cloning of the Tpgus79a gene based on the results of protein identification. The PCR mixture (25 μl) consisted of 1 μl of cDNA as the template, 0.2 μM (each) primers, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 0.5 U of rTaq polymerase (TaKaRa Bio Co., Ltd., Dalian, China), and 10 μl of 10× PCR buffer. The PCR conditions were as follows: 94°C for 5 min and 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min. The amplified DNA fragment was sequenced and then cloned into the pMD19-T vector, resulting in the plasmid pMD19T-Tpgus79a.
Expression of the recombinant enzyme in P. pastoris.
The Tpgus79a gene was amplified from plasmid pMD19T-Tpgus79a. PCR was carried out using the following primers: 5′-CGGGGTACCATGAATCGCATCATCACCCTCGCCTC-3′ and 5′-AAGGAAAAAAGCGGCCGCTTAAACCCAGCCCTGTGCAAGTCTC-3′ (KpnI and NotI restriction sites are underlined). The PCR mixture (50 μl) consisted of 50 ng of pMD19T-Tpgus79a plasmid DNA, 0.2 μM (each) primers, 0.5 U of Pfu DNA polymerase (TaKaRa Bio, Co., Ltd., Dalian, China), a 200 μM concentration of each dNTP, and 5 μl of DNA polymerase buffer. Two-step PCR was performed with 30 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 90 s. The PCR product was purified by use of a gel purification kit (Tiangen, Beijing, China), digested with KpnI and NotI, and ligated into the vector pGAPZα by use of T4 DNA ligase, resulting in the recombinant expression plasmid pGAPZα-Tpgus79a. After linearization with AvrII, the expression plasmid was transformed into P. pastoris GS115. The transformants were screened on YPD plates containing Zeocin. For high-level expression of the target protein, a multicopy recombinant was selected from plates and cultured in YPD broth with shaking at 30°C for 2 to 4 days. The supernatant of the expression culture was used for the measurement of enzyme activity and for SDS-PAGE analysis.
Enzyme activity assay.
Enzymatic activity was measured using ammonium glycyrrhizinate, GAMG, GAM-Glc (glycyrrhetinic acid 3-O-mono-β-glucoside), quercetin 3-O-β-glucuronide, quercetin 3-O-β-glucoside, baicalin, baicalein-7-O-β-glucoside, pNP-β-GlcA, and other pNP-linked monosaccharides as substrates. The assay was performed in 50 mM acetate buffer (NaAc-HAc) (pH 4.5) at 45°C, and the reaction was stopped by adding 150 μl 0.4 M Na2CO3 (for pNP-linked monosaccharides) or by heating at 100°C for 5 min (for flavonoid and terpenoid glucuronides). The concentration of p-nitrophenol produced by the enzymatic reaction was determined by measuring the absorbance at 405 nm, based on an extinction coefficient of 22,412 M−1 cm−1 for p-nitrophenol, by use of a microplate spectrophotometer (BioTek, USA). When flavonoid or terpenoid glucuronides were used as substrates, the concentration of the product was measured by HPLC using a Shimadzu LC-10A system (Shimadzu, Japan) equipped with a 2.5C18-MS-II column (3.0-mm internal diameter × 100 mm) (Cosmosil; Nacalai Tesque, Inc.). When GL, GAMG, or GAM-Glc was the substrate, the mobile phase of HPLC consisted of 0.6% acetic acid-methanol (18:82 [vol/vol]) and the eluate was monitored at a 254-nm excitation wavelength. On the other hand, when quercetin 3-O-β-glucuronide or quercetin 3-O-β-glucoside was the substrate, the mobile phase consisted of 0.38% phosphoric acid-methanol (55:45 [vol/vol]), and the eluate was monitored at a 280-nm excitation wavelength. Similarly, when the substrate was baicalin or baicalein-7-O-β-glucoside, the mobile phase consisted of 0.4% phosphoric acid-methanol (47:53 [vol/vol]), and the eluate was monitored at a 360-nm excitation wavelength. One unit of enzyme activity was defined as the amount of enzyme required to produce 1 μmol of product/min.
Kinetic characterization.
The kinetic parameters Km and kcat of TpGUS79A on different substrates were measured in 50 mM NaAc-HAc buffer (pH 4.5) at 45°C. The initial velocity of GL hydrolysis at 5 min was measured by variation of the ammonium glycyrrhizinate concentration from 0.22 to 1.86 mM, with an enzyme concentration of 0.013 mg ml−1. The initial velocity of quercetin 3-glucuronide hydrolysis at 8 min was detected by variation of the quercetin 3-glucuronide concentration from 0.25 to 2.51 mM, with an enzyme concentration of 0.034 mg ml−1. The initial velocity of baicalin hydrolysis at 20 min was measured by variation of the baicalin concentration from 0.81 to 8.07 mM, with an enzyme concentration of 0.084 mg ml−1. The initial velocity of pNP-β-GlcA hydrolysis at 20 min was measured by variation of the pNP-β-GlcA concentration from 3 to 60 mM, with an enzyme concentration of 0.095 mg ml−1. The kinetic parameters were obtained by fitting the data to the Michaelis-Menten equation through nonlinear regression, using Origin 8.0. All assays were performed in duplicate, and reported kinetic constants are the mean values derived from triplicate independent experiments.
Effects of pH and temperature on enzyme activity.
The physicochemical properties of the enzyme were determined using ammonium glycyrrhizinate as a substrate. The optimum temperature of the enzyme was measured in 50 mM acetate buffer (pH 4.5) at various temperatures. The thermal stability of the enzyme was assessed via determining the residual activities at 40°C after incubating the enzyme in 50 mM acetate buffer (pH 4.5) at each temperature for 30 min. The optimal pH value was measured at 45°C, using the following 50 mM buffer solutions: glycine-HCl buffer of pH 2.0 to 3.0, acetate buffer of pH 3.0 to 5.5, phosphate buffer of pH 6.0 to 7.5, Tris-HCl buffer of pH 7.5 to 8.5, and glycine-NaOH buffer of pH 8.5 to 10. The pH stability was assessed by measuring the residual activities in 50 mM acetate buffer (pH 4.5) at 45°C after incubating the enzyme in various 50 mM buffer solutions at 4°C for 12 h.
Effects of metal ions and inhibitors on enzyme activity.
The effects of metal ions and inhibitors on enzyme activity were determined by ammonium glycyrrhizinate hydrolysis at the optimum temperature and pH. The enzyme activity was assayed with different metal ions at final concentrations of 1 and 10 mM or with reductants and inhibitors at final concentrations of 5 and 10 mM.
Homology modeling of TpGUS79A structure.
TpGUS79A belongs to the GH79 family according to sequence analysis. So far, only the structural features of AcGlcA79A (GUS from A. capsulatum) belonging to GH79 have been described (37). Therefore, homology modeling of TpGUS79A was performed by use of SWISS-MODEL software (https://www.swissmodel.expasy.org/), based on the crystal structure of AcGlcA79A (PDB entry 3VNZ). The modeled structure of TpGUS79A was further optimized by a 30-ns all-atom MD simulation. The MD simulation was performed with NAMD 2.9. The model system was solvated in TIP2P25 with a 15-Å water box. The periodic boundary conditions were applied for energy minimization and equilibration. The solvated protein was ionized using 0.15 M NaCl to neutralize the charge. The temperature was kept constant at 310 K in the NVT ensemble. The trajectory was recorded every 5,000 steps (1 step means 2 fs), and the par_all27_prot_lipid parameter was applied to all calculations in the CHARMM force field. The resulting structure and the structures of two heparanases (PDB entries 5BWI and 5E9B) were employed as templates for homology modeling of TpGUS79A by SWISS-MODEL. The quality of the optimized structure of TpGUS79A was evaluated by checking the Ramachandran plot parameters and the QMEAN (composite scoring of the geometric aspects of protein structures). The structure of GL was first optimized by a 30-ns MD simulation (the conditions were the same as those for TpGUS79A) until the root mean square deviation (RMSD) became stable to minimize the energy. The optimized GL structure was then used for docking with TpGUS79A, using AutoDock Vina (version 4.2.6). The structure was visualized and analyzed by use of PyMOL.
Site-directed mutagenesis of TpGUS79A.
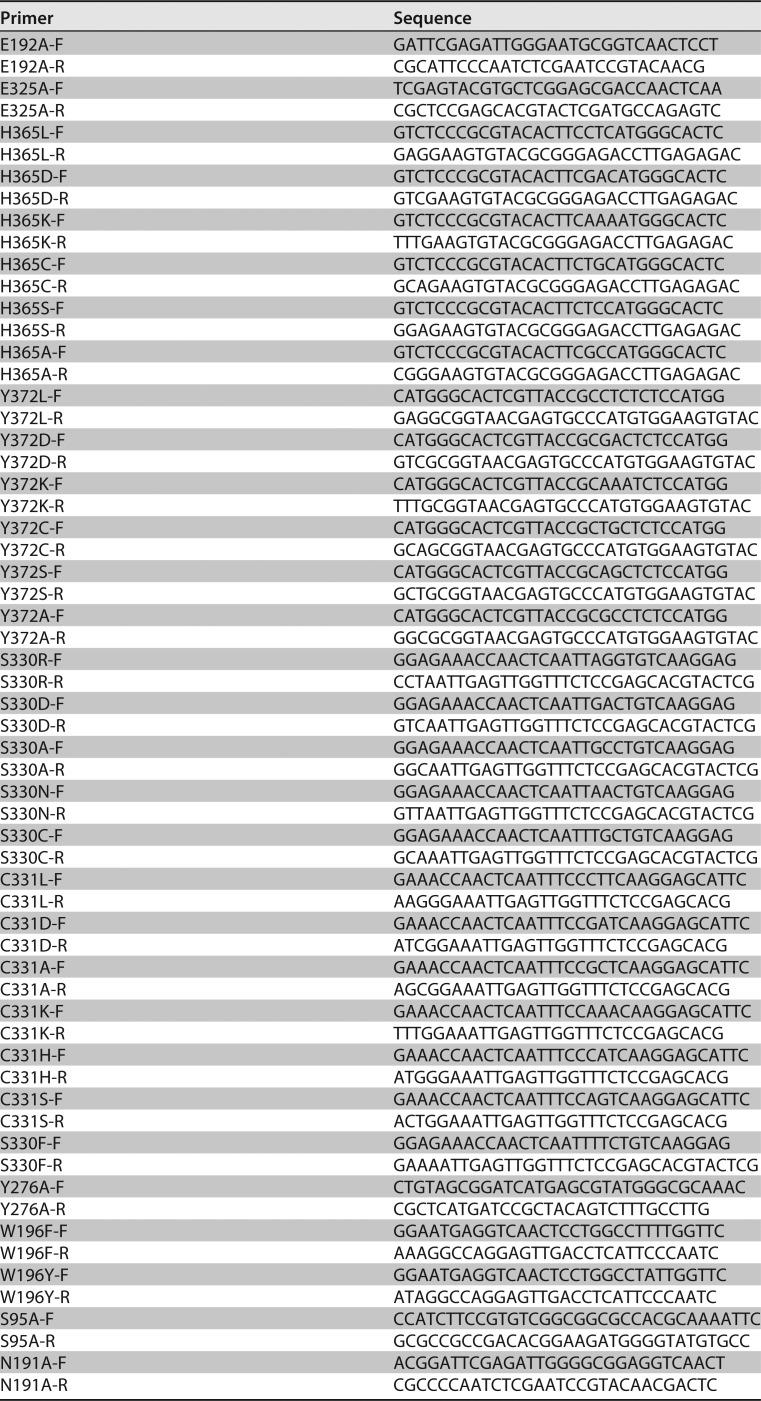
The residues predicted to play a catalytic role in the active center of TpGUS79A were mutated by site-directed mutagenesis through inverse-PCR amplification. The expression plasmids for the mutants were amplified using TpGAPZα-Tpgus79a as the template and the primers shown in Table 3. The mutant enzymes were produced extracellularly by transformed P. pastoris GS115. Each variant protein was purified from 72-h culture supernatant by acetone precipitation and cation-exchange chromatography steps (refer to the native enzyme purification process described above). The mutant enzyme activity was assayed by using ammonium glycyrrhizinate as the substrate and the methods described above.
TABLE 3.
Primers used for site-directed mutagenesis of TpGUS79A
Preparation of GAMG by T. pinophilus Li-93 in a 150-liter fermentor.
To inspect the real application of T. pinophilus Li-93 for GAMG production through GL hydrolysis, a pilot-scale fed-batch fermentation was performed in a 150-liter fermentor (Baoxin, Shanghai, China). The medium, seed preparation, inoculum amount, and culture temperature were the same as those used for the enzyme production process mentioned above. The rate of agitation was 200 rpm, and the fermentation pH was held at pH 5.0 throughout the fermentation process. The first feed was performed after 41 h, and then 540 g substrate was fed every 24 h. The GL conversion rate (CGL) was calculated as follows: CGL (%) = (S0 − St)/S0 × 100, where S0 stands for the total substrate concentration provided and St stands for the substrate concentration at time t.
Accession number(s).
The nucleotide sequence of the Tpgus79a gene has been deposited in GenBank under accession number KY609919.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 21506011, 21425624, and 21606019).
We thank Shutao Sun from the Institute of Microbiology, Chinese Academy of Sciences, for her technical assistance and guidance on MALDI-TOF-TOF MS analysis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00755-18.
REFERENCES
- 1.Pharmacopeia Commission. 2015. Pharmacopeia of People's Republic of China, vol 1 China Medical Science Press, Beijing, China. [Google Scholar]
- 2.Matsui S, Matsumoto H, Sonoda Y, Ando K, Aizu-Yokota E, Sato T, Kasahara T. 2004. Glycyrrhizin and related compounds down-regulate production of inflammatory chemokines IL-8 and eotaxin 1 in a human lung fibroblast cell line. Int Immunopharmacol 4:1633–1644. doi: 10.1016/j.intimp.2004.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park HY, Park SH, Yoon HK, Han MJ, Kim DH. 2004. Anti-allergic activity of 18β-glycyrrhetinic acid-3-O-β-d-glucuronide. Arch Pharm Res 27:57–60. doi: 10.1007/BF02980047. [DOI] [PubMed] [Google Scholar]
- 4.Huang RY, Chu YL, Jiang ZB, Chen XM, Zhang X, Zeng X. 2014. Glycyrrhizin suppresses lung adenocarcinoma cell growth through inhibition of thromboxane synthase. Cell Physiol Biochem 33:375–388. doi: 10.1159/000356677. [DOI] [PubMed] [Google Scholar]
- 5.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. 2003. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harada S. 2005. The broad anti-viral agent glycyrrhizin directly modulates the fluidity of plasma membrane and HIV-1 envelope. Biochem J 392:191–199. doi: 10.1042/BJ20051069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krausse R, Bielenberg J, Blaschek W, Ullmann U. 2004. In vitro anti-Helicobacter pylori activity of extractum liquiritiae, glycyrrhizin and its metabolites. J Antimicrob Chemother 54:243–246. doi: 10.1093/jac/dkh287. [DOI] [PubMed] [Google Scholar]
- 8.Asl MN, Hosseinzadeh H. 2008. Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother Res 22:709–724. doi: 10.1002/ptr.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizutani K, Kuramoto T, Tamura Y, Ohtake N, Doi S, Nakaura M, Tanaka O. 1994. Sweetness of glycyrrhetic acid 3-O-β-d-monoglucuronide and the related glycosides. Biosci Biotechnol Biochem 58:554–555. doi: 10.1271/bbb.58.554. [DOI] [PubMed] [Google Scholar]
- 10.Chaturvedula VSP, Yu O, Mao G. 2014. Isolation and NMR spectral assignments of 18-glycyrrhetinic acid-3-O-d-glucuronide and 18-glycyrrhetinic acid. IOSR J Pharm 4:5. [Google Scholar]
- 11.Wang R, Lin C, Liu J, Yu F, Gao J, Pan X. 2012. Pressured microwave-assisted hydrolysis of crude glycyrrhizic acid for preparation of glycyrrhetinic acid. Chin J Chem Eng 20:152–157. doi: 10.1016/S1004-9541(12)60375-9. [DOI] [Google Scholar]
- 12.Kuramoto T, Ito Y, Oda M, Tamura Y, Kitahata S. 1994. Microbial production of glycyrrhetic acid 3-O-mono-β-d-glucuronide from glycyrrhizin by Cryptococcus magnus MG-27. Biosci Biotechnol Biochem 58:3. doi: 10.1271/bbb.58.455. [DOI] [Google Scholar]
- 13.Feng S, Li C, Xu X, Wang X. 2006. Screening strains for directed biosynthesis of β-d-mono-glucuronide-glycyrrhizin and kinetics of enzyme production. J Mol Catal B Enzym 43:63–67. doi: 10.1016/j.molcatb.2006.06.016. [DOI] [Google Scholar]
- 14.Amin HAS, El-Menoufy HA, El-Mehalawy AA, Mostafa ES. 2011. Biosynthesis of glycyrrhetinic acid 3-O-mono-beta-d-glucuronide by free and immobilized Aspergillus terreus beta-d-glucuronidase. J Mol Catal B Enzym 69:54–59. doi: 10.1016/j.molcatb.2010.12.010. [DOI] [Google Scholar]
- 15.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schollhammer I, Poll DS, Bickel MH. 1975. Liver microsomal beta-glucuronidase and UDP-glucuronyltransferase. Enzyme 20:269–276. doi: 10.1159/000458949. [DOI] [PubMed] [Google Scholar]
- 17.Paigen K. 1989. Mammalian beta-glucuronidase—genetics, molecular-biology, and cell biology. Prog Nucleic Acid Res Mol Biol 37:155–205. doi: 10.1016/S0079-6603(08)60698-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhu BT, Evaristus EN, Antoniak SK, Sarabia SF, Ricci MJ, Liehr JG. 1996. Metabolic deglucuronidation and demethylation of estrogen conjugates as a source of parent estrogens and catecholestrogen metabolites in Syrian hamster kidney, a target organ of estrogen-induced tumorigenesis. Toxicol Appl Pharmacol 136:186–193. doi: 10.1006/taap.1996.0023. [DOI] [PubMed] [Google Scholar]
- 19.Hazenberg MP, Deherder WW, Visser TJ. 1988. Hydrolysis of iodothyronine conjugates by intestinal bacteria. FEMS Microbiol Lett 54:9–16. doi: 10.1111/j.1574-6968.1988.tb02707.x. [DOI] [PubMed] [Google Scholar]
- 20.Wallace BD, Wang HW, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, Redinbo MR. 2010. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tohyama O, Imura A, Iwano A, Freund JN, Henrissat B, Fujimori T, Nabeshima Y. 2004. Klotho is a novel beta-glucuronidase capable of hydrolyzing steroid beta-glucuronides. J Biol Chem 279:9777–9784. doi: 10.1074/jbc.M312392200. [DOI] [PubMed] [Google Scholar]
- 22.Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, Iwasaki H, Iida A, Shiraki-Iida T, Nishikawa S, Nagai R, Nabeshima Y. 1997. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 23.Eudes A, Mouille G, Thevenin J, Goyallon A, Minic Z, Jouanin L. 2008. Purification, cloning and functional characterization of an endogenous beta-glucuronidase in Arabidopsis thaliana. Plant Cell Physiol 49:1331–1341. doi: 10.1093/pcp/pcn108. [DOI] [PubMed] [Google Scholar]
- 24.Konishi T, Kotake T, Soraya D, Matsuoka K, Koyama T, Kaneko S, Igarashi K, Samejima M, Tsumuraya Y. 2008. Properties of family 79 beta-glucuronidases that hydrolyze beta-glucuronosyl and 4-O-methyl-beta-glucuronosyl residues of arabinogalactan-protein. Carbohydr Res 343:1191–1201. doi: 10.1016/j.carres.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Shuttleworth E, Allen N. 1980. Csf beta-glucuronidase assay in the diagnosis of neoplastic meningitis. Arch Neurol 37:684–687. doi: 10.1001/archneur.1980.00500600032003. [DOI] [PubMed] [Google Scholar]
- 26.Sly WS, Vogler C. 1997. Gene therapy for lysosomal storage disease: a no-brainer? Transplants of fibroblasts secreting high levels of beta-glucuronidase decrease lesions in the brains of mice with Sly syndrome, a lysosomal storage disease. Nat Med 3:719–720. [DOI] [PubMed] [Google Scholar]
- 27.Papot S, Combaud D, Bosslet K, Gerken M, Czech J, Gesson JP. 2000. Synthesis and cytotoxic activity of a glucuronylated prodrug of nornitrogen mustard. Bioorg Med Chem Lett 10:1835–1837. doi: 10.1016/S0960-894X(00)00353-X. [DOI] [PubMed] [Google Scholar]
- 28.Jefferson RA, Kavanagh TA, Bevan MW. 1987. Gus fusions—beta-glucuronidase as a sensitive and versatile gene fusion marker in higher-plants. EMBO J 6:3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, Wang X. 2015. Klotho: a novel biomarker for cancer. J Cancer Res Clin Oncol 141:961–969. doi: 10.1007/s00432-014-1788-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lv B, Sun H, Huang S, Feng X, Jiang T, Li C. 2018. Structure-guided engineering of the substrate specificity of a fungal β-glucuronidase toward triterpenoid saponins. J Biol Chem 293:433–443. doi: 10.1074/jbc.M117.801910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DH, Jang IS, Lee HK, Jung EA, Lee KY. 1996. Metabolism of glycyrrhizin and baicalin by human intestinal bacteria. Arch Pharm Res 19:5. [Google Scholar]
- 32.Akao TA, Akao T, Hattori M, Kanaoka M, Yamamoto K, Namba T, Kobashi K. 1991. Hydrolysis of glycyrrhizin to 18β-glycyrrhetyl monoglucuronide by lysosomal β-d-glucuronidase of animal livers. Biochem Pharmacol 41:5. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Huangfu J, Qi F, Kaleem I, E W, Li C. 2012. Effects of a non-conservative sequence on the properties of beta-glucuronidase from Aspergillus terreus Li-20. PLoS One 7:e30998. doi: 10.1371/journal.pone.0030998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amin HA, El-Menoufy HA, El-Mehalawy AA, Mostafa EE. 2010. Microbial production of glycyrrhetic acid 3-O-mono-β-d-glucuronide from glycyrrhizin by Aspergillus terreus. Malays J Microbiol 6:8. [Google Scholar]
- 35.Hye-Young P, Kim NY, Han MJ, Bae EA, Kim DH. 2005. Purification and characterization of two novel beta-d-glucuronidases converting glycyrrhizin to 18β-glycyrrhetinic acid-3-O-beta-d-glucuronide from Streptococcus LJ-22. J Microbiol Biotechnol 15:792–799. [Google Scholar]
- 36.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michikawa M, Ichinose H, Momma M, Biely P, Jongkees S, Yoshida M, Kotake T, Tsumuraya Y, Withers SG, Fujimoto Z, Kaneko S. 2012. Structural and biochemical characterization of glycoside hydrolase family 79 beta-glucuronidase from Acidobacterium capsulatum. J Biol Chem 287:14069–14077. doi: 10.1074/jbc.M112.346288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou SP, Liu GY, Kaleem I, Li C. 2013. Purification and characterization of a highly selective glycyrrhizin-hydrolyzing beta-glucuronidase from Penicillium purpurogenum Li-3. Process Biochem 48:358–363. doi: 10.1016/j.procbio.2012.12.008. [DOI] [Google Scholar]
- 39.Wang X, Liu Y, Wang C, Feng X, Li C. 2015. Properties and structures of β-glucuronidases with different transformation types of glycyrrhizin. RSC Adv 5:68345–68350. doi: 10.1039/C5RA11484E. [DOI] [Google Scholar]
- 40.Zou S, Guo S, Kaleem I, Li C. 2013. Purification, characterization and comparison of Penicillium purpurogenum β-glucuronidases expressed in Escherichia coli and Pichia pastoris. J Chem Technol Biotechnol 88:1913–1919. doi: 10.1002/jctb.4050. [DOI] [Google Scholar]
- 41.Zou SP, Xie LP, Liu YL, Kaleem I, Zhang GF, Li C. 2012. N-linked glycosylation influences on the catalytic and biochemical properties of Penicillium purpurogenum beta-d-glucuronidase. J Biotechnol 157:399–404. doi: 10.1016/j.jbiotec.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 42.Zhang CZ, Zhang YF, Chen JP, Liang XM. 2005. Purification and characterization of baicalin-beta-d-glucuronidase hydrolyzing baicalin to baicalein from fresh roots of Scutellaria viscidula Bge. Process Biochem 40:1911–1915. doi: 10.1016/j.procbio.2004.07.003. [DOI] [Google Scholar]
- 43.Nuylert A, Ishida Y, Asano Y. 2017. Effect of glycosylation on the biocatalytic properties of hydroxynitrile lyase from the passion fruit, Passiflora edulis: a comparison of natural and recombinant enzymes. Chembiochem 18:257–265. doi: 10.1002/cbic.201600447. [DOI] [PubMed] [Google Scholar]
- 44.Sakurama H, Kishino S, Uchibori Y, Yonejima Y, Ashida H, Kita K, Takahashi S, Ogawa J. 2014. β-Glucuronidase from Lactobacillus brevis useful for baicalin hydrolysis belongs to glycoside hydrolase family 30. Appl Microbiol Biotechnol 98:4021–4032. doi: 10.1007/s00253-013-5325-8. [DOI] [PubMed] [Google Scholar]
- 45.Sasaki K, Taura F, Shoyama Y, Morimoto S. 2000. Molecular characterization of a novel beta-glucuronidase from Scutellaria baicalensis georgi. J Biol Chem 275:27466–27472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.