We isolated and determined the complete genome sequence of a KPC-2-producing K. pneumoniae strain from a sampling site in Tokyo Bay, Japan, near a wastewater treatment plant (WWTP). In Japan, the KPC type has been very rarely detected, while IMP is the most predominant type of carbapenemase in clinical carbapenemase-producing Enterobacteriaceae (CPE) isolates. Although laboratory testing thus far suggested that Japan may be virtually free of KPC-producing Enterobacteriaceae, we have detected it from effluent from a WWTP. Antimicrobial resistance (AMR) monitoring of WWTP effluent may contribute to the early detection of future AMR bacterial dissemination in clinical settings and communities; indeed, it will help illuminate the whole picture in which environmental contamination through WWTP effluent plays a part.
KEYWORDS: Klebsiella pneumoniae, blaKPC-2, carbapenemase, effluent, urban sewage
ABSTRACT
Antimicrobial resistance genes (ARGs) and the bacteria that harbor them are widely distributed in the environment, especially in surface water, sewage treatment plant effluent, soil, and animal waste. In this study, we isolated a KPC-2-producing Klebsiella pneumoniae strain (GSU10-3) from a sampling site in Tokyo Bay, Japan, near a wastewater treatment plant (WWTP) and determined its complete genome sequence. Strain GSU10-3 is resistant to most β-lactam antibiotics and other antimicrobial agents (quinolones and aminoglycosides). This strain is classified as sequence type 11 (ST11), and a core genome phylogenetic analysis indicated that strain GSU10-3 is closely related to KPC-2-positive Chinese clinical isolates from 2011 to 2017 and is clearly distinct from strains isolated from the European Union (EU), United States, and other Asian countries. Strain GSU10-3 harbors four plasmids, including a blaKPC-2-positive plasmid, pGSU10-3-3 (66.2 kb), which is smaller than other blaKPC-2-positive plasmids and notably carries dual replicons (IncFII [pHN7A8] and IncN). Such downsizing and the presence of dual replicons may promote its maintenance and stable replication, contributing to its broad host range with low fitness costs. A second plasmid, pGSU10-3-1 (159.0 kb), an IncA/C2 replicon, carries a class 1 integron (containing intI1, dfrA12, aadA2, qacEΔ1, and sul1) with a high degree of similarity to a broad-host-range plasmid present in the family Enterobacteriaceae. The plasmid pGSU10-3-2 (134.8 kb), an IncFII(K) replicon, carries the IS26-mediated ARGs [aac(6')Ib-cr , blaOXA-1, catB4 (truncated), and aac(3)-IId], tet(A), and a copper/arsenate resistance locus. GSU10-3 is the first nonclinical KPC-2-producing environmental Enterobacteriaceae isolate from Japan for which the whole genome has been sequenced.
IMPORTANCE We isolated and determined the complete genome sequence of a KPC-2-producing K. pneumoniae strain from a sampling site in Tokyo Bay, Japan, near a wastewater treatment plant (WWTP). In Japan, the KPC type has been very rarely detected, while IMP is the most predominant type of carbapenemase in clinical carbapenemase-producing Enterobacteriaceae (CPE) isolates. Although laboratory testing thus far suggested that Japan may be virtually free of KPC-producing Enterobacteriaceae, we have detected it from effluent from a WWTP. Antimicrobial resistance (AMR) monitoring of WWTP effluent may contribute to the early detection of future AMR bacterial dissemination in clinical settings and communities; indeed, it will help illuminate the whole picture in which environmental contamination through WWTP effluent plays a part.
INTRODUCTION
Antimicrobial resistance (AMR) is a global health crisis linked to increased and often unrestricted antibiotic use in the clinical and veterinary fields. WHO guidelines have been published for the infection prevention and control (IPC) of carbapenem-resistant Enterobacteriaceae (CRE), carbapenem-resistant Acinetobacter baumannii (CRAB), and carbapenem-resistant Pseudomonas aeruginosa (CRPsA) in health care facilities (1). These bacteria have the potential to facilitate the widespread transmission of AMR via mobile genetic elements through the processes of natural competence, transformation, and plasmid transconjugation that can occur in any environment. For these reasons, it has been concluded that the early recognition of CRE-CRAB-CRPsA should be a high priority to allow evidence-based recommendations to be provided. Furthermore, specific and required IPC practices and procedures should be conducted to effectively prevent the occurrence of these infections and control their spread in acute health care facilities.
In addition to clinical settings, AMR genes (ARGs) and AMR bacteria are widely distributed in the environment, particularly in surface waters (2), sewage treatment plant effluents (3), and soils and animal wastes (4). No direct evidence has yet been uncovered for the transmission of ARGs to humans through the environment, and the extent to which environmental factors contribute to human exposure should be quantified and compared to clinical and veterinary data. A systematic literature review was recently conducted on human exposure to AMR bacteria, including extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in the environment (5). The results indicated that AMR bacteria can be detected at exposure-relevant sites, including recreational areas, and in drinking water, ambient air, shellfish, and fresh produce.
In particular, the widespread detection of carbapenemase-producing Enterobacteriaceae (CPE) in the environment is an emerging environmental issue with potentially serious public health implications. Notably, Xu et al. reported the isolation of Klebsiella pneumoniae carbapenemase (KPC)-producing Citrobacter and Aeromonas isolates from sampling sites near a wastewater treatment plant (WWTP) by the Shifeng River in China (6). An article reviewing antibiotic resistance in China highlighted the issues concerning the enrichment and dissemination of ARGs in the environment in China (7), as well as the need to mitigate the spread of AMR in the environment, particularly under the “One-Health” perspective.
In this study, we isolated and determined the complete genome sequence of a KPC-2-producing K. pneumoniae strain from a sampling site in Tokyo Bay, Japan, near a wastewater treatment plant (WWTP). The first Japanese KPC-2-producing K. pneumoniae clinical isolate, K. pneumoniae strain Kp3018, was classified as sequence type 11 (ST11) and was isolated from a patient treated at a Brazilian hospital in 2012 (8). Since then, there have been no reports documenting additional KPC-2-producing K. pneumoniae in Japan, indicating that the Japanese environment may be almost free of KPC-producing K. pneumoniae. This is the first report on the genomic features of a nonclinical KPC-2-producing Enterobacteriaceae isolate from Japan.
RESULTS
CRE isolate from WWTP effluent.
A large, mucoid colony on CHROMagar ESBL plate was selected for whole-genome sequence (WGS) analysis and was identified as K. pneumoniae and designated GSU10-3. The GSU10-3 isolate carries blaKPC-2, exhibited resistance to meropenem, and had a positive reaction for the Carba NP test. Further antimicrobial susceptibility testing showed that K. pneumoniae GSU10-3 is resistant to most β-lactam antibiotics with Etest (Table 1) and to other antimicrobial agents, including trimethoprim-sulfamethoxazole, ciprofloxacin, kanamycin, gentamicin, streptomycin, minocycline, and tetracycline as determined by disk diffusion test. The string test showed that it had negative stretch and was a low-viscosity colony.
TABLE 1.
Etest results on the K. pneumoniae GSU10-3 isolate
| Antibiotic(s) | MIC (µg/ml) |
|---|---|
| Meropenem | >8 |
| Meropenem-EDTA | >2 |
| Imipenem | >32 |
| Cefotaxime | >256 |
| Cefotaxime-clavulanic acida | >1 |
| Aztreonam | >256 |
| Sulbactam-ampicillin | >256 |
Clavulanic acid present at a fixed concentration of 4 µg/ml.
Whole-genome sequence analysis of K. pneumoniae GSU10-3.
Basic information regarding the complete chromosome and plasmid sequences for K. pneumoniae GSU10-3 is shown in Table 2. An analysis of the complete chromosomal DNA sequence classifies GSU10-3 as ST11 (based on the results of multilocus sequence typing [MLST] analysis). The strain possesses plasmids harboring multiple AMR genes (Table 2). The wzi gene sequence corresponds to the gene in capsular genotype K47 in a homology search against wzi sequence database (9). Strain GSU10-3 does not show a hypermucoviscosity phenotype by the string test, and regulator of mucoid phenotype A (rmpA) (10) and transcriptional activator (rmpA2) (10) genes were not found in both the chromosome and those plasmids.
TABLE 2.
Whole-genome information for K. pneumoniae GSU10-3 from WWTP effluent
| Replicon | % GC | Length (bp) | Inc type | Drug resistance gene(s) | GenBank ID |
|---|---|---|---|---|---|
| Chromosome | 57.4 | 5,478,620 | blaSHV-1, fosA6 | AP018671 | |
| pGSU10-3-1 | 51.6 | 159,072 | IncA/C2 |
dfrA12, aadA2, qacEdelta1, sul1, qnrA1, aac(3)-IId, blaTEM-1B, aph(6)-Id, aph(3'')-Ib, sul2 |
AP018672 |
| pGSU10-3-2 | 52.8 | 134,879 | IncFII | tet(A), aac(6')Ib-cr, blaOXA-1, catB4, aac(3)-IIa | AP018673 |
| pGSU10-3-3 | 54.2 | 66,250 | IncN and IncFII (pHN7A8) | blaKPC-2, fosA3 | AP018674 |
| pGSU10-3-4 | 55.1 | 10,060 | ColRNAI | Not found | AP018675 |
Core genome phylogenetic analysis of K. pneumoniae GSU10-3.
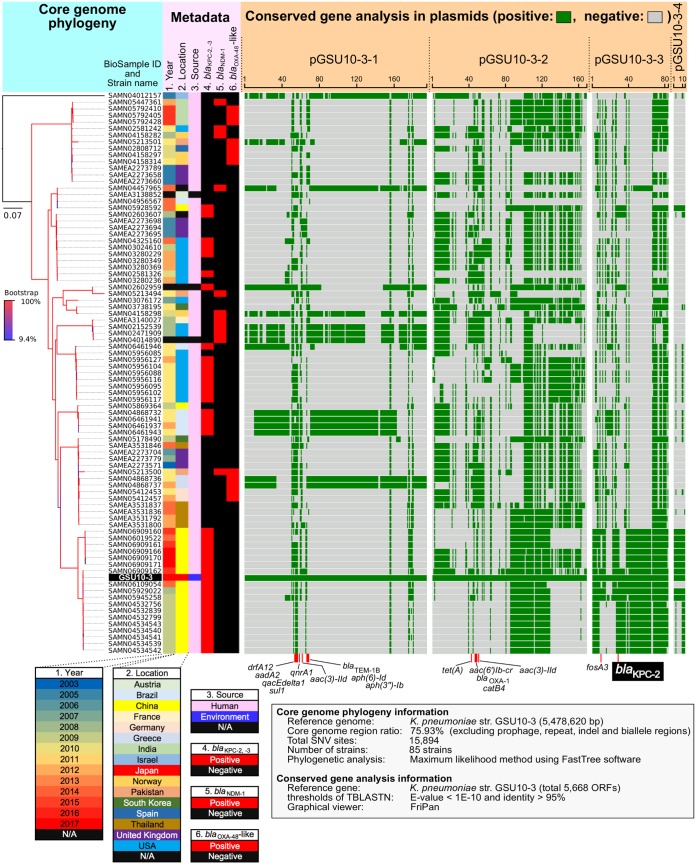
To trace the potential sources of the GSU10-3 strain, we performed core genome phylogenetic analysis using 84 publicly available ST11 K. pneumoniae genome sequences (on 18 April 2018), including draft genomes (see the strain list in Table S1 in the supplemental material). To date, no environmental, ST11 K. pneumoniae genome sequence has been deposited in a public database. Thus, GSU10-3 is the first fully sequenced environmental isolate for which the genome sequence has been characterized. The core genome of those tested K. pneumoniae strains constitutes 75.93% of the genome (4,159,904/5,478,620 bp), and the phylogenetic analysis using 15,894 single-nucleotide variations (SNVs) indicated that strain GSU10-3 shares a common lineage with 19 KPC-2-positive Chinese isolates from human clinical specimens obtained from 2011 to 2017 and is clearly distinct from strains from the European Union (EU) (20 strains), United States (19 strains), and other Asian countries (14 strains) (Fig. 1).
FIG 1.
Representation of a core genome phylogeny and conserved gene analysis of K. pneumoniae strains. A complete picture of the core genome phylogeny of 85 strains of ST11 K. pneumoniae, including GSU10-3, is shown. The core genome phylogeny was constructed by the maximum likelihood method using the core genome region of strain GSU10-3 (75.93% of the genome; 4,159,904/5,478,620 bp), excluding repeat and recombination regions. The isolate year, location, source, and the specific β-lactamases are indicated by the different colors. Conserved genes (≥95% homology) shared with GSU10-3 as a reference genome are highlighted for the chromosome and four plasmids. Antimicrobial resistance genes are indicated below. Information for all tested strains is shown in Table S1 in the supplemental material. N/A, not available; str., strain.
Information on the Klebsiella pneumoniae strains used in whole-genome SNV phylogenetic analysis (Fig. 1). Download Table S1, PDF file, 0.03 MB (29KB, pdf) .
Copyright © 2018 Sekizuka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural comparison of KPC-2 plasmids.
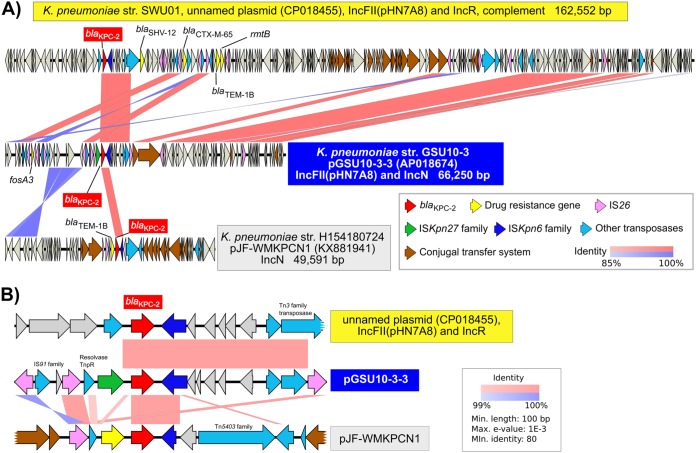
A conserved gene analysis of the four GSU10-3 plasmids indicated that the KPC-2 encoding plasmid (pGSU10-3-3 in strain GSU10-3) shares a number of conserved genes with the above-mentioned 19 KPC-2-positive Chinese isolates (Fig. 1), while the other three plasmids show partial homology to other K. pneumoniae strains. For instance, pGSU10-3-1 (IncA/C replicon plasmid) is similar to plasmids of K. pneumoniae strains isolated in Brazil (four isolates), France (one isolate), Germany (two isolates) and the USA (two isolates). The KPC-2-containing plasmid pGSU10-3-3 (66.2 kb) is on a relatively small IncFII (pHN7A8) replicon compared with other KPC-2-containing plasmids (see Fig. S1), and a pairwise alignment shows that some of the genes involved in the conjugal transfer system (dark brown open reading frames [ORFs] in Fig. 2A) have been removed in pGSU10-3-3. Multiple IS26 might contribute to the loss of conjugation potential by the excision with a possible homologous recombination event. In addition, pGSU10-3-3 has an additional IncN replicon (inverted alignment with dark blue in Fig. 2A) compared to the IncN plasmid pJF-WMKPCN1. Structural comparisons suggested that pGSU10-3-3 carries two Inc replicons (IncFII [pHN7A8] and IncN) and appears to have partially lost conjugational machinery.
FIG 2.
Structural comparison of the blaKPC-2-positive plasmids. (A) K. pneumoniae SWU01 (BioSample SAMN06109054) was isolated from a clinical blood specimen from a human in China in 2015, K. pneumoniae H154180724 (GenBank KX881941) was isolated from a human in the United Kingdom. (B) Comparison of gene structure around blaKPC-2 is highlighted from panel A. The blaKPC-2 gene in pGSU10-3-3 is flanked by ISKpn27 and ISKpn6. Min., minimum; Max., maximum.
Comparison of genes around blaKPC-2 suggested that multiple insertion sequnces (ISs) could be involved in the acquisition of blaKPC-2, resulting in the gene structure IS26–tnpR–ISKpn27–blaKPC-2–ISKpn6 (Fig. 2B). Part of the gene structure (ISKpn27–blaKPC-2–ISKpn6) is similar to that of a KPC-2-positive IncP-6 plasmid (p121SC21-KPC2, GenBank accession no. or identifier [ID] LT992437) in Citrobacter freundii CF121SC21 isolate obtained from wastewater in 2012 (11), while eight copies of IS26 insertions (pink ORFs in Fig. 2A and B) are a notable genetic feature of pGSU10-3-3 in this study. IS26 is one of the ubiquitous ISs in the family Enterobacteriaceae. IS26 sequences are deposited as follows: 5,176 sequence entries in Klebsiella, 4,255 in Escherichia coli, 1,764 in Acinetobacter, 1,508 in Salmonella, 980 in Enterobacter, and 1,605 in other members of the Enterobacteriaceae family were identified by BLASTp homology search (17 July 2018), suggesting that IS26 could be one of the marked ISs in Klebsiella species.
ARGs.
In addition to blaKPC-2 described above, ARGs are listed in Table 2, and the ARG position in each plasmid is indicated in the linear replicon representation in Fig. 1 and Fig. S1 in the supplemental material.
A whole-plasmid nucleotide sequence search of GSU10-3 plasmids pGSU10-3-1 (A), pGSU10-3-1 (B), pGSU10-3-1 (C), and pGSU10-3-1 (D) against complete plasmid sequence database (13,286 entries as of 5 May 2018). Schematic pairwise alignment of the plasmid is displayed with each significant homologous plasmid as a subject sequence. A red box and a coverage value indicate a region similarity at ≥80% nucleotide identity and shared regions (as a percentage), respectively. Download FIG S1, PPT file, 2.3 MB (2.4MB, ppt) .
Copyright © 2018 Sekizuka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The pGSU10-3-1 plasmid (159,072 bp) is an IncA/C2 replicon that carries a class 1 integron (intI1, dfrA12, aadA2, qacEΔ1, and sul1), qnrA1, aac(3)-IId, blaTEM-1B, aph(6)-Id, aph(3'')-Ib, sul2, and multiple ISs (including an IS91 family member, three IS26 elements, and members of the ISEhe3, IS903D, and IS1182 families). A whole-plasmid search indicates that pGSU10-3-1 shares similarity to the broad-host-range plasmids present in K. pneumoniae and other members of the Enterobacteriaceae, including Aeromonas, Citrobacter, Enterobacter, Escherichia, Proteus, and Vibrio (Fig. S1).
The pGSU10-3-2 plasmid (134,879 bp) is an IncFII(K) replicon that carries IS26-mediated ARGs [aac(6')Ib-cr, blaOXA-1, catB4 (truncated), and aac(3)-IId], tet(A), and a copper/arsenate resistance locus. The copper resistance proteins encoded by the cop operon mediate the sequestration of copper in the periplasm (12), and the ArsAB membrane complex functions in arsenic resistance as an anion-translocating ATPase (13). Such metal resistance genes are likely to increase the persistence, fitness, and propagation of the plasmid in the bacterial host under the conditions of environmental stress present in sewage. A whole-plasmid search indicated that pGSU10-3-2 shares similarity to the only K. pneumoniae-related plasmid, which has a narrow host range.
As described above, the pGSU10-3-3 plasmid (66,250 bp) carries blaKPC-2 and fosA3.
The pGSU10-3-4 plasmid (10,060 bp) is a ColRNAI replicon, and ARGs or virulence determinants have not been found on it.
DISCUSSION
In this study, we isolated a KPC-2-producing K. pneumoniae isolate (GSU10-3) from the effluent of a WWTP in Tokyo Bay, Japan. WWTPs could be a primary source of most AMR bacteria and ARGs in aquatic environments, probably due to the partial purification that occurs during the wastewater treatment processes. A recent study of a Japanese WWTP identified GES- and imipenemase (IMP)-, but not KPC-type, CPE isolates from wastewater (14).
In Japan, IMP is the most predominant type of carbapenemase in clinical CPE isolates (15–17). VIM, OXA48, GES, and NDM carbapenemases are more rarely detected, while the KPC type is very rarely detected, suggesting that Japan may be virtually free of KPC-producing Enterobacteriaceae. In contrast, KPC is predominant in isolates from other East Asian countries. Twenty-four million travelers from Asian countries visited Japan in 2017, with more than a 20% annual rate of increase (as reported by the Japan National Tourism Organization), indicating that in addition to specific, local types of CPE, comprehensive testing should be conducted for every type of imported CPE.
Isolate GSU10-3 is classified as ST11 K. pneumoniae, which is closely related to the dominant clone of KPC-producing K. pneumoniae in China (18). For instance, the KPC-producing K. pneumoniae isolates from central China between 2009 and 2014 were clonally related, with ST11 being the reservoir for the blaKPC-2 and ESBL genes (blaCTX-M-55). These findings demonstrated the high prevalence of carbapenemase- and ESBL-producing K. pneumoniae in central China (19). An outbreak of KPC-2-producing K. pneumoniae (ST11) in a neonatal ward has been reported (20). Hypervirulent and multidrug-resistant K. pneumoniae strains pose a significant threat to public health, and a recent study reported a hospital outbreak in China that involved the dissemination of ST11 with KPC-2-producing K. pneumoniae (21). GSU10-3 in this study showed low-mucoviscosity colony formation and does not carry hypermucoviscosity-related regulator genes (rmpA and rmpA2), suggesting that GSU10-3 might not be the markedly hypervirulent isolate, instead of multiple antimicrobial resistance.
The GSU10-3 isolate possesses the KPC-2-positive plasmid pGSU10-3-3, which contains the Inc replicons IncF and IncN in the same relaxase MOBF family (22). It has been reported that the IncN group can colocalize as a fusion with IncF plasmids (23), suggesting that although pGSU10-3-3 is smaller than other KPC-2-related plasmids, such downsizing could contribute to its stable replication with dual replicons. This scenario would contribute to a broad host range, leading to a rather low fitness cost compared to that of larger KPC-2 plasmids observed in clinical isolates (24) (see Fig. S1 in the supplemental material).
When KPC-producing K. pneumoniae causes an epidemic of carbapenem-resistant Enterobacteriaceae (CRE) in health care settings in developed countries, the spread primarily occurs through patient-to-patient transmission (25). So-called superspreaders are individuals who are likely to have high rectal CRE concentrations, and such CRE carriers are responsible for partial shedding and may play a central role in CRE transmission (26). There have been many reports of CRE in hospital water, and the majority of these reports have described associated clinical outbreaks in intensive care settings, affecting the critically ill and immunocompromised. Drains, sinks, and faucets are the most frequently colonized by CRE, and the most appropriate disinfection method remains unclear. However, it is likely that the replacement of colonized water reservoirs may be required for long-term clearance (27).
Previous studies have sought to establish a possible role for the natural environment in the transmission of clinically relevant AMR bacteria to humans. However, quantitative data analysis from exposure-relevant sites and environmental compartments were not sufficient to determine the abundance of AMR bacteria that pose a significant risk for human exposure, although AMR bacteria have been detected in diverse environments, including wastewater, as shown in this study. The increase in selective pressures due to the overuse of extended-spectrum cephalosporins (widely available as generics) has contributed to the global dissemination of CTX-M-type ESBL strains in all types of environments. Healthy carriers of CTX-M-type ESBL-harboring bacteria are another major public health concern, because carriage rates are increasing, particularly in South East Asia and Eastern Mediterranean regions. Carriers from these regions have the potential to spread these bacteria to other communities (28). Indeed, surfers appear to be at risk of exposure to and colonization by clinically important AMR E. coli in coastal waters (29). Additionally, short-term international travel to Vietnam increases the carriage risk of colistin-resistant, ESBL-producing E. coli (30). Raw vegetables and local foods appear to become contaminated with AMR bacteria due to insufficient hygiene in irrigation water systems, making this a potential means by which such bacteria disseminate to humans via retail vegetables (31).
In conclusion, AMR monitoring of WWTP effluent may contribute to the detection of on-going AMR bacterial dissemination in clinical settings and communities. A comprehensive approach will be required to uncover the bigger picture in which environmental contamination through WWTP effluent plays a part.
MATERIALS AND METHODS
Bacterial isolation.
The upper effluent flow of an urban wastewater treatment plant (WWTP) was collected on 23 August 2017 (N35.654861, E139.833358) in Tokyo Bay, Japan. Five hundred milliliters of the effluent was filtered through a polyethersulfone (PES) filter membrane with a pore size of 0.22 µm (Vacuum Filtration "rapid"-Filtermax; TPP Techno Plastic Products, Trasadingen, Switzerland). A quarter of the membrane was incubated with 20 ml of LB broth supplemented with 1 µg/ml meropenem at 37°C for 14 h to select for bacteria with reduced carbapenem susceptibility. The culture (1 to 10 µl) was spread on the CHROMagar ESBL plates (CHROMagar, Paris, France) and incubated at 37°C for 18 h. A colony with a unique morphology, designated K. pneumoniae GSU10-3, was isolated for further molecular genomic analysis. Hypermucoviscosity phenotype K. pneumoniae was checked by the string test (32).
Antimicrobial susceptibility and CPE screening tests.
Antimicrobial susceptibility testing was performed using Etest (bioMérieux, Marcy-l’Étoile, France) and disk diffusion methods under CLSI M100-S28 (33). Carbapenemase production was assessed using a Carba NP test, as described previously (34).
Whole-genome sequence (WGS) analysis.
Genomic DNA from the isolated strain was purified by collecting cells from a 5-ml overnight culture grown in tryptic soy broth (TSB). The cell pellet was resuspended in 500 µl of TE10 (10 mM Tris [pH 8.0] and 10 mM EDTA) supplemented with 500 µl phenol-chloroform, and the cells were subsequently lysed by bead beating for 10 min in ZR BashingBead lysis tubes (Zymo Research, Irvine, CA, USA) attached to a vortex adapter (MO BIO Laboratories, Qiagen, Carlsbad, CA, USA). After centrifugation at 10,000 rpm for 5 min, the upper phase was further purified using a Qiagen DNA purification kit (Qiagen, Germany). Short DNA fragments (approximately 0.5 kb) for paired-end sequencing were generated using an Illumina XT DNA library kit (Illumina). Whole-genome sequencing of the paired-end library was performed using an Illumina NextSeq 500 platform with a 300-cycle NextSeq 500 reagent kit v2 (150-mer paired ends; median coverage, ×96).
The complete genome sequence of the strain was determined using a PacBio Sequel sequencer for long-read sequencing (Sequel SMRT Cell 1M v2 [four/tray]; Sequel sequencing kit v2.1; insert size, approximately 10 kb). Purified genomic DNA (∼200 ng) was used to prepare a SMRTbell library using a SMRTbell template prep kit 1.0 (PacBio, Menlo Park, CA, USA) with barcoding adapters according to the manufacturer’s instructions. Sequencing data were produced with more than 100-fold coverage and assembled using the assembly program SMRT Link v5.
A de novo assembly was performed using Canu version 1.4 (35), minimap version 0.2-r124 (36), racon version 1.1.0 (37), and Circlator version 1.5.3 (38). Error correction of tentative complete circular sequences was performed using Pilon version 1.18 with Illumina short reads (39). Annotation was performed in Prokka version 1.11 (40), InterPro v49.0 (41), and NCBI-BLASTP/BLASTX.
Circular representations of complete genomic sequences were visualized using GView server (42). Antimicrobial resistance (AMR) genes were identified by homology searching against the ResFinder database (43).
Comparative genome sequence analysis.
All publicly available draft genome sequences of K. pneumoniae strains were retrieved (at least ×40 read coverage; see Table S1 in the supplemental material) and compared using bwaMEM read mapping against the K. pneumoniae GSU10-3 complete genome sequence (AP018671) as a reference. After excluding repeat regions and six prophage sequences from the whole-genome sequence, 75.9% of the genome was assigned as the core genome sequence among 85 collected strains, resulting in the identification of 15,894 single-nucleotide variations (SNVs) (Fig. 1). The core genome multilocus sequence typing (cgMLST) was performed using the SNVs described above, and the phylogeny was generated using the maximum likelihood phylogenetic method with FastTree v2.1.10.
Conserved gene sequence analysis among plasmids was performed with BLASTn searches (≥95% nucleotide [nt] identity), followed by visualization using FriPan (http://drpowell.github.io/FriPan/).
Comparative plasmid sequence analysis was performed with BLASTn searches (≥85% nt identity), followed by visualization using Easyfig (44).
Accession number(s).
The complete, annotated genomic sequence of K. pneumoniae GSU10-3 was deposited in a public database (accession numbers AP018671, AP018672, AP018673, AP018674, and AP018675). The short- and long-read sequences for transcriptome sequencing (DNA-Seq) were deposited in the DNA Data Bank of Japan (BioProject PRJDB6962, BioSample SAMD00116246, DRA accession no. DRA006779).
ACKNOWLEDGMENTS
This work was supported by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development, AMED (grants JP17fk0108121 and JP18fk0108048). This work was also supported by a grant for Research on Emerging and Re-emerging Infectious Diseases and Immunization (H30 Shinkogyosei-Ippan-002) from the Ministry of Health, Labour and Welfare, Japan.
The funding agencies had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation.
T. Sekizuka, K. Yatsu, and M. Kuroda contributed to the isolation of the KPC-2-positive strains. T. Segawa performed antimicrobial susceptibility testing. Y. Inamine, M. Nishio, and N. Kishi performed the genome sequencing. T. Sekizuka and M. Kuroda performed the comparative genome analysis. M. Kuroda wrote the manuscript.
REFERENCES
- 1.World Health Organization. 2017. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 2.Ng C, Tay M, Tan B, Le TH, Haller L, Chen H, Koh TH, Barkham TMS, Gin KY. 2017. Characterization of metagenomes in urban aquatic compartments reveals high prevalence of clinically relevant antibiotic resistance genes in wastewaters. Front Microbiol 8:2200. doi: 10.3389/fmicb.2017.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karkman A, Do TT, Walsh F, Virta MPJ. 2018. Antibiotic-resistance genes in waste water. Trends Microbiol 26:220–228. doi: 10.1016/j.tim.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Sharma M, Reynnells R. 2016. Importance of soil amendments: survival of bacterial pathogens in manure and compost used as organic fertilizers. Microbiol Spectr 4(4) doi: 10.1128/microbiolspec.PFS-0010-2015. [DOI] [PubMed] [Google Scholar]
- 5.Huijbers PM, Blaak H, de Jong MC, Graat EA, Vandenbroucke-Grauls CM, de Roda Husman AM. 2015. Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ Sci Technol 49:11993–12004. doi: 10.1021/acs.est.5b02566. [DOI] [PubMed] [Google Scholar]
- 6.Xu H, Wang X, Yu X, Zhang J, Guo L, Huang C, Jiang X, Li X, Feng Y, Zheng B. 2018. First detection and genomics analysis of KPC-2-producing Citrobacter isolates from river sediments. Environ Pollut 235:931–937. doi: 10.1016/j.envpol.2017.12.084. [DOI] [PubMed] [Google Scholar]
- 7.Qiao M, Ying GG, Singer AC, Zhu YG. 2018. Review of antibiotic resistance in China and its environment. Environ Int 110:160–172. doi: 10.1016/j.envint.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Saito R, Takahashi R, Sawabe E, Koyano S, Takahashi Y, Shima M, Ushizawa H, Fujie T, Tosaka N, Kato Y, Moriya K, Tohda S, Tojo N, Koike R, Kubota T. 2014. First report of KPC-2 carbapenemase-producing Klebsiella pneumoniae in Japan. Antimicrob Agents Chemother 58:2961–2963. doi: 10.1128/AAC.02072-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decré D. 2013. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol 51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Yao Y, Lazaro-Perona F, Falgenhauer L, Valverde A, Imirzalioglu C, Dominguez L, Canton R, Mingorance J, Chakraborty T. 2017. Insights into a novel blaKPC-2-encoding IncP-6 plasmid reveal carbapenem-resistance circulation in several Enterobacteriaceae species from wastewater and a hospital source in Spain. Front Microbiol 8:1143. doi: 10.3389/fmicb.2017.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cha JS, Cooksey DA. 1991. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci U S A 88:8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diorio C, Cai J, Marmor J, Shinder R, DuBow MS. 1995. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in Gram-negative bacteria. J Bacteriol 177:2050–2056. doi: 10.1128/jb.177.8.2050-2056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomi R, Matsuda T, Yamamoto M, Chou PH, Tanaka M, Ichiyama S, Yoneda M, Matsumura Y. 2018. Characteristics of carbapenemase-producing Enterobacteriaceae in wastewater revealed by genomic analysis. Antimicrob Agents Chemother 62:e02501-17. doi: 10.1128/AAC.02501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koyano S, Saito R, Nagai R, Tatsuno K, Okugawa S, Okamura N, Moriya K. 2013. Molecular characterization of carbapenemase-producing clinical isolates of Enterobacteriaceae in a teaching hospital, Japan. J Med Microbiol 62:446–450. doi: 10.1099/jmm.0.050708-0. [DOI] [PubMed] [Google Scholar]
- 16.Ohno Y, Nakamura A, Hashimoto E, Matsutani H, Abe N, Fukuda S, Hisashi K, Komatsu M, Nakamura F. 2017. Molecular epidemiology of carbapenemase-producing Enterobacteriaceae in a primary care hospital in Japan, 2010-2013. J Infect Chemother 23:224–229. doi: 10.1016/j.jiac.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto N, Asada R, Kawahara R, Hagiya H, Akeda Y, Shanmugakani RK, Yoshida H, Yukawa S, Yamamoto K, Takayama Y, Ohnishi H, Taniguchi T, Matsuoka T, Matsunami K, Nishi I, Kase T, Hamada S, Tomono K. 2017. Prevalence of, and risk factors for, carriage of carbapenem-resistant Enterobacteriaceae among hospitalized patients in Japan. J Hosp Infect 97:212–217. doi: 10.1016/j.jhin.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66:307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 19.Guo X, Cao Z, Dai Z, Li Y, He X, Hu X, Tian F, Ren Y. 2017. Antimicrobial susceptibility and molecular epidemiology of multidrug-resistant Klebsiella pneumoniae in Central China. Jpn J Infect Dis 70:229–234. doi: 10.7883/yoken.JJID.2016.049. [DOI] [PubMed] [Google Scholar]
- 20.Yu J, Tan K, Rong Z, Wang Y, Chen Z, Zhu X, Wu L, Tan L, Xiong W, Sun Z, Chen L. 2016. Nosocomial outbreak of KPC-2- and NDM-1-producing Klebsiella pneumoniae in a neonatal ward: a retrospective study. BMC Infect Dis 16:563. doi: 10.1186/s12879-016-1870-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan L, Wang S, Guo Y, Jin Y, Duan J, Hao Z, Lv J, Qi X, Hu L, Chen L, Kreiswirth BN, Zhang R, Pan J, Wang L, Yu F. 2017. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol 7:182. doi: 10.3389/fcimb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 23.Deng Y, He L, Chen S, Zheng H, Zeng Z, Liu Y, Sun Y, Ma J, Chen Z, Liu JH. 2011. F33:A-:B- and F2:A-:B- plasmids mediate dissemination of rmtB-blaCTX-M-9 group genes and rmtB-qepA in Enterobacteriaceae isolates from pets in China. Antimicrob Agents Chemother 55:4926–4929. doi: 10.1128/AAC.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He S, Chandler M, Varani AM, Hickman AB, Dekker JP, Dyda F. 2016. Mechanisms of evolution in high-consequence drug resistance plasmids. mBio 7:e01987-16. doi: 10.1128/mBio.01987-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerner A, Adler A, Abu-Hanna J, Cohen Percia S, Kazma Matalon M, Carmeli Y. 2015. Spread of KPC-producing carbapenem-resistant Enterobacteriaceae: the importance of super-spreaders and rectal KPC concentration. Clin Microbiol Infect 21:470.e1–470.e7. doi: 10.1016/j.cmi.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 27.Kizny Gordon AE, Mathers AJ, Cheong EYL, Gottlieb T, Kotay S, Walker AS, Peto TEA, Crook DW, Stoesser N. 2017. The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections—a systematic review of the literature. Clin Infect Dis 64:1435–1444. doi: 10.1093/cid/cix132. [DOI] [PubMed] [Google Scholar]
- 28.Woerther PL, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leonard AFC, Zhang L, Balfour AJ, Garside R, Hawkey PM, Murray AK, Ukoumunne OC, Gaze WH. 2018. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey). Environ Int 114:326–333. doi: 10.1016/j.envint.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama T, Kumeda Y, Kawahara R, Yamaguchi T, Yamamoto Y. 2018. Carriage of colistin-resistant, extended-spectrum beta-lactamase-producing Escherichia coli harboring the mcr-1 resistance gene after short-term international travel to Vietnam. Infect Drug Resist 11:391–395. doi: 10.2147/IDR.S153178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Hoek AH, Veenman C, van Overbeek WM, Lynch G, de Roda Husman AM, Blaak H. 2015. Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int J Food Microbiol 204:1–8. doi: 10.1016/j.ijfoodmicro.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; twenty-eighth informational supplement M100–S28. Clinical Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaser R, Sovic I, Nagarajan N, Sikic M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 41.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SC, Wu CH, Xenarios I, Yeh LS, Young SY, Mitchell AL. 2017. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. 2010. Interactive microbial genome visualization with GView. Bioinformatics 26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information on the Klebsiella pneumoniae strains used in whole-genome SNV phylogenetic analysis (Fig. 1). Download Table S1, PDF file, 0.03 MB (29KB, pdf) .
Copyright © 2018 Sekizuka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A whole-plasmid nucleotide sequence search of GSU10-3 plasmids pGSU10-3-1 (A), pGSU10-3-1 (B), pGSU10-3-1 (C), and pGSU10-3-1 (D) against complete plasmid sequence database (13,286 entries as of 5 May 2018). Schematic pairwise alignment of the plasmid is displayed with each significant homologous plasmid as a subject sequence. A red box and a coverage value indicate a region similarity at ≥80% nucleotide identity and shared regions (as a percentage), respectively. Download FIG S1, PPT file, 2.3 MB (2.4MB, ppt) .
Copyright © 2018 Sekizuka et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.