Abstract
We previously established an in vitro differentiation system based on the inducible expression of the RNA binding protein 6 (RBP6), which initiated differentiation of Trypanosoma brucei non-infectious procyclics to infectious metacyclics (MFs). However, further differentiation to bloodstream forms (BFs) required infection of mice. Here we report the serendipitous isolation of a single point mutation in RBP6 (Q109K), whose expression not only generated MFs, but purified MFs continued the developmental cycle in vitro to BFs expressing variant surface glycoprotein-2 (VSG-2), formerly known as VSG 221. This transition occurred over a period of 11 days and by RNA-Seq, VSG-2 was first measureable on day 1, whereas metacyclic VSGs were detected up to 8 days. We further showed that inducible expression of mutant RBP6 appeared to skip the intermediate epimastigote stage and we highlight the potential involvement of RBP33 in the establishment of metacyclics and in particular in the generation of metacyclics uncharacteristically arrested at the G2/M checkpoint.
Keywords: Trypanosoma brucei, metacyclic trypanosomes, variant surface glycoprotein, RNA binding protein, bloodstream form trypanosomes.
Graphical abstract
1. Introduction
African trypanosomes are the causative agent of sleeping sickness in humans and nagana in animals and remain a substantial public health concern in sub-Saharan Africa. The geographical restriction of the disease is due to the distribution of the tsetse fly vector (Glossina spp.), a blood-feeding dipteran, which transmits the protozoan parasite Trypanosoma brucei. To cope with the changing environment in two host systems, T. brucei transitions through distinct life cycle forms that have evolved to assure parasite survival and successful transmission to the next host [1, 2]. For instance, in the bloodstream of the mammal, the parasites exist mainly as proliferative slender forms, which establish parasitaemia. The differentiation of slender to quiescent, stumpy bloodstream forms (BFs) occurs in response to cell density [3] and these are the forms primed to survive the environmental change associated with the uptake by the tsetse fly [4]. In the midgut of the insect vector stumpy forms differentiate into procyclic forms that have lost their infectivity. In order to regain infectivity, parasites need to go through an intricate developmental program that generates epimastigotes and eventually leads to the establishment of infectious metacyclics in the tsetse salivary glands [5]. Metacyclics, similar to stumpy forms, are quiescent and primed for mammalian invasion [6, 7].
The two-host environments encountered by T. brucei requires a complex cellular differentiation program that includes changes in surface protein expression, metabolism, organelle function, and cytoskeletal architecture. For example, the trypanosome plasma membrane is covered by a distinct, densely packed surface coat consisting of GPI-anchored proteins at different stages of the life cycle. To outmaneuver the immune system when residing in the bloodstream of the mammalian host, T. brucei expresses a variant surface glycoprotein (VSG) coat, which is the paradigm for antigenic variation [8, 9]. In the tsetse midgut, trypanosomes cover their surface with procyclins, a family of EP (Glu/Pro repeat-containing) and GPEET (Gly/Pro/Glu/Glu/Thr repeat-containing) proteins [10, 11], whereas epimastigote forms express a family of proteins known as brucei alanine-rich proteins or BARPs [12]. Metacyclic trypanosomes are covered by a specific subset of VSGs, the metacyclic VSGs (mVSGs), which enables transmission to a vertebrate host [13]. In this context, differential expression of VSGs is one characteristic feature of the T. brucei life cycle and is based on two distinct genetic strategies. The genome of T. brucei has a large repertoire of VSG genes, but bloodstream-form VSG expression occurs predominantly from 1 of about 15 telomeric, polycictronic bloodstream expression sites (BES), and the active BES is sequestered and transcribed in an extranucleolar expression site body [14] by RNA polymerase I (Pol I) [15]. The mechanistic details limiting VSG expression to a single BES are not fully understood [8, 9, 16]. What has emerged is that transcription attenuation and epigenetic silencing play important roles for maintaining monoallelic VSG expression and there is recent evidence for regulation at the level of transcription initiation, involving class I transcription factor A (CITFA), the multi-subunit essential Pol I initiation factor [17].
In contrast to a single dominant VSG being expressed on the surface of bloodstream forms at any one time, metacyclic trypanosomes are heterogeneous, displaying a number of different variants. However, each individual trypanosome expresses just one distinct mVSG, which are located at a telomere and are expressed from a monocistronic Pol I transcription unit [18–20]. Very early studies, based on monoclonal antibodies, have estimated that the repertoire of mVSGs expressed in metacyclics to be between 14 and as many as 27 [21]. Nevertheless, a subsequent survey of VSGs in the T. brucei Lister 427 strain challenged these numbers, since only six mVSGs with a typical metacyclic promoter were found [22]. In addition, metacyclics generated in our in vitro system, based on the inducible expression of the RNA binding protein 6 (RBP6), express five major mVSG transcripts, coding for mVSG-397, −653, −1954, −531 and −639 [23]. Our in vitro results highlighting a more limited repertoire of expressed mVSGs mirror tsetse fly transcriptome data, where four mVSGs were identified [24]. Regardless of the uncertainty about the size of the mVSG repertoire, at present little is known how mVSG gene expression is activated and how the expression is switched to bloodstream-form VSGs, once the parasite enters a mammalian host.
Although inducible expression of RBP6 in T. brucei procyclics initiates a developmental progression to metacyclics [23], multiple attempts to continue the life cycle in vitro to bloodstream forms were unsuccessful. Here, we report the serendipitous isolation of a single point mutation in RBP6 (Q109K), whose expression not only generated MFs, but allowed further differentiation in vitro to BFs expressing VSG-2, formerly known as VSG 221.
2. Material and methods
2.1. T. brucei cell culture, production and purification of MF cells
Inducible expression of an RBP6 (Tb927.3.2930) transgene in the vector pLew100.v5 was carried out in the T. brucei Lister 427(29–13) procyclic strain as described [7, 23]. Different trypanosome cell types (procyclics, epimastigote sand metacyclics) were scored after staining of DNA in live cells with DAPI. Cells were assigned to a type based on their size, shape, position of the kinetoplast relative to the nucleus, and position of the kinetoplast relative to the posterior end of the cell. Metacyclics were purified at room temperature on 0.1 mm diameter zirconia/silica beads columns prepared in Pasteur pipettes in BBSG buffer (50 mM bicine, 50 mM NaCl, 5 mM KCl, 70 mM glucose, pH 8.0) [25]. Purified metacyclics were cultured in BF conditions in HMI-9 media supplemented with 10% FBS at 37°C with 5% CO2. The transformation to BF cells is not very efficient and few cells survive the selection procedure, i.e. around day 5 the culture has less than 103 parasites per ml.
2.2. RNA preparation, RNA-Seq, read processing, data analysis and RT-PCR
Total RNA was prepared from 2 × 107 PF, purified MF, and BF cells. Biological replicates represent experiments performed at least two weeks apart. The RNA was prepared with the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Four independent samples, i.e. biological replicas, were prepared from each life cycle stage and from each time point in a time course and isolation of poly(A)+ mRNA, library preparation and sequencing on an Illumina HiSeq2500 platform were performed at the Yale Center for Genome Analysis. Reads of 75 nt in length were mapped to the T. brucei 11 megabase chromosomes (GeneDB version 5) using the Lasergene 14.1.2 software package from DNASTAR as described [7], except that the minimum aligned length was increased to 70 nt from 35 nt. RPKM (reads assigned per kilobase of target per million mapped reads) was used for normalization and the calculation of RPKM values was restricted to predicted open reading frames. Differential gene expression analysis was done with the Lasergene software package from DNASTAR [7] and differentially expressed genes were analyzed for functional annotation using the Gene Ontology (GO) enrichment tool on the TriTrypDB webserver (http://tritrypdb.org/) and GO terms were condensed by submitting to REVIGO [26].
Since VSG genes share substantial sequence similarities, in particular towards the 3’ end of the gene, read coverage was visually inspected using the DNASTAR SeqMan NGen program, and only genes with a uniform read coverage were included in the data analysis.
Semi-quantitative RT-PCR was performed to confirm RNA-Seq results for selected mRNAs with SuperScript III reverse transcriptase (Invitrogen) and Platinum Pfx DNA polymerase (Invitrogen) following manufacturer’s instructions. First strand cDNA synthesis was performed with random hexamers and the sequences of the PCR primers for the individual transcripts analyzed are available upon request.
RNA-Seq data from this study have been submitted to the NCBI Sequence Read Archive - SRA at http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi with accession numbers: SRP153824, SRP153562 and SRP152737.
3. Results and Discussion
3.1. The RBP6 mutant Q109K promotes an accelerated development to metacyclics
We have previously shown that the T. brucei RNA binding protein RBP6 is a master regulator for the transformation of non-infectious procyclic forms into infectious metacyclic forms [23]. RBP6 is 239 amino acids (aa) long and contains two RNA Recognition Motifs or RRMs, separated by a 42 aa linker containing 11 consecutive glutamine residues (Fig. 1). To begin to understand the mechanism of RBP6 function, we generated N- and C-terminal epitope-tagged versions of the protein. However, this abolished RBP6’s ability to trigger developmental progression, indicating that intact termini are required for its function. Since the T. congolense and T. vivax proteins have a shorter or an absent stretch of glutamines in the linker between the two RRMs, respectively (Fig. 1B), and over-expression of either protein in T. brucei procyclics generated metacyclics with a time course similar to the one induced by the T. brucei protein (unpublished data), we explored the possibility of introducing an epitope tag in place of the stretch of glutamines. This led to the serendipitous isolation of a mutant RBP6 where the first glutamine (Q109) of the stretch of 11 glutamines was changed to a lysine (Fig. 1B).
Fig. 1. Bioinformatics analysis of RBP6.
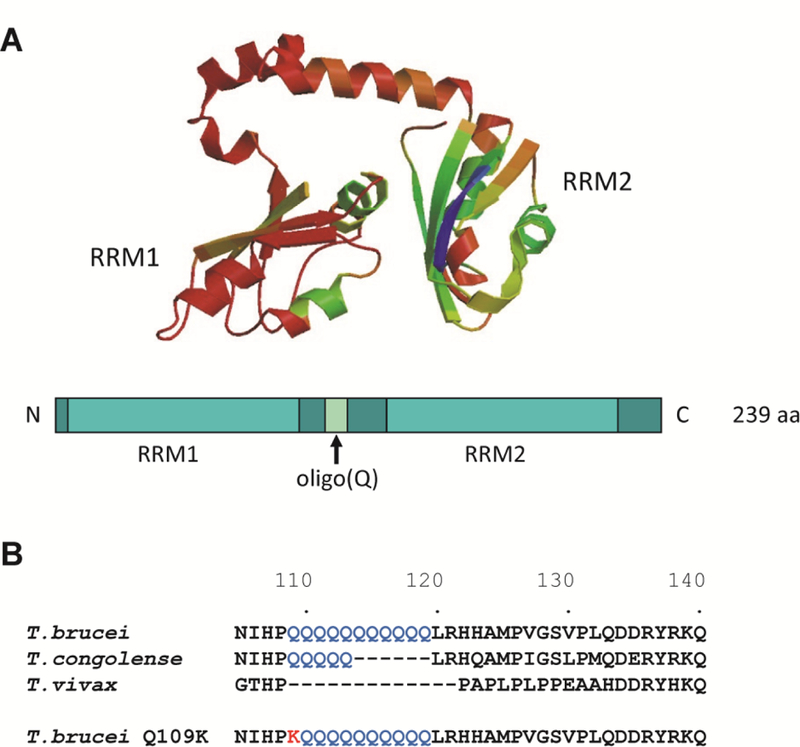
(A) Tertiary structure model of the T. brucei RBP6 (Tb927.3.2930) using the alignment interface of SWISS-MODEL [34] and schematic diagram below. RRM, RNA recognition motif. (B) Sequence alignment of T. brucei, T. congolense (TcIL3000_0_03630), T. vivax (TvY486_0302270) and the mutant T. brucei Q109K RBP6. Note that the alignment was restricted to the oligo(Q) region.
Inducible expression of wild-type RBP6 (wtRBP6) takes about 6 days to produce 30–40% metacyclics. However, inducible expression of mutant, untagged RBP6 (mutRBP6) generated metacyclic cells in 2–3 days with about 10% of the cells going through this transformation. Visual inspection by microscopy after DNA staining of the culture during the transition from procyclics to metacyclics appeared to suggest that the intermediate epimastigote stage was skipped, since we did not observe epimastigote cells. To confirm this observation, we monitored the levels of a diagnostic marker for short epimastigotes, namely the brucei alanine-rich protein or BARP [12] in cultures after inducibly expressing either the wild-type or mutant RBP6 for two days. As expected, in cultures where wtRBP6 was induced we observed approximately 20% epimastigotes and the levels of both BARP mRNA and protein were highly upregulated as compared to un-induced cells (Fig. 2, lane 2). In contrast, induction of mutRBP6 revealed a minor increase (less than 2-fold) in the level of BARP mRNA and no detectable increase of BARP protein as compared to un-induced procyclic cells (Fig. 2, lane 4). Thus, the mutRBP6 induced a developmental program generating metacyclic trypanosomes in 2–3 days without going through the intermediate epimastigote stage.
Fig. 2. BARP expression.
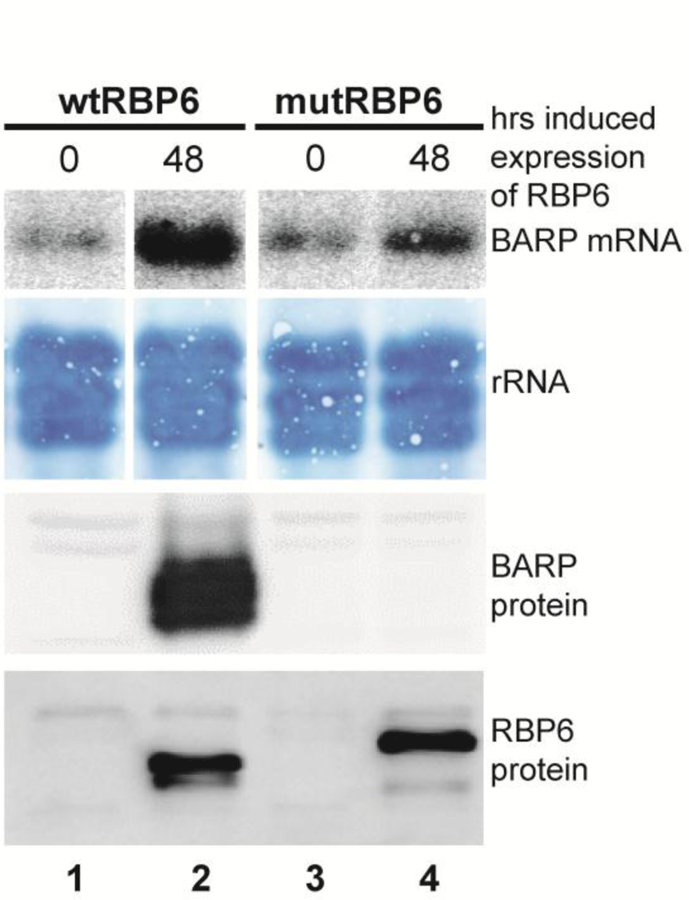
wt and mutRBP6 were induced in procyclic cells for 48 hrs, lanes 2 and 4, and BARP mRNA and protein were monitored by Northern and Western blot, respectively. RBP6 protein expression was monitored by Western blot. Note that mutRBP6 is Flag-tagged.
3.2. Differential mRNA expression during the development from procyclic to metacyclic cells
To begin to understand events triggered by wild-type or mutant RBP6, we first performed RNA-Seq analysis after a 24-hour induction. wtRBP6 changed the level of 184 transcripts, with 152 and 32 transcripts up- and down-regulated 2-fold or more, respectively (Table 1 and suppl. Table S1). Most increased in mRNA abundance was an atypical VSG of unknown function (Tb927.5.3990, 140-fold). In addition, up-regulated transcripts included the mitochondrial alternative oxidase (Tb927.10.7090, 7.1-fold), BARP (Tb927.9.15530, 5.8-fold), six RNA binding proteins (DRBD6A/DRBD6B, Tb927.3.3960/Tb927.3.3930; RBP26, Tb927.7.3730; DRBD12, Tb927.7.5380; PUF5, Tb927.7.4730; and ZC3H45, Tb927.11.8470), two cyclins (Tb927.8.6350 and Tb927.6.5020), as well as several nucleoside and amino acid transporters. Finally, three mRNAs encoding proteins associated with differentiation (PAD, carboxylate-transporter family), implicated as sensors of environmental stimuli and triggers of differentiation [27], were up-regulated, namely PAD4, PAD6 and PAD8 (Tb927.7.5960, 5980 and 6000). Functional categorization by Gene Ontology (GO) analysis of the up-regulated transcripts revealed statistically significant cellular components, including membrane and integral component of the membrane, whereas the down-regulated transcripts were enriched for transporter activity (suppl. Table S2).
Table 1.
Differentially expressed transcripts.
| wtRBP6 | mutRBP6 | |||||
|---|---|---|---|---|---|---|
| 24 h induction |
6 days induction |
Purified metacyclics |
24 h induction |
5 days induction |
Purified metacyclics |
|
|
Significantly
regulated transcripts |
184 | 601 | 1,302 | 22 | 80 | 1,392 |
| Up-regulated | 152 | 504 | 771 | 21 | 71 | 660 |
| Down-regulated | 32 | 97 | 531 | 1 | 9 | 732 |
All changes are compared to un-induced cells.
In contrast, inducible expression of mutRBP6 changed abundance levels of 22 transcripts after 24 hours, with 21 and 1 transcripts up- and down-regulated, respectively (Table 1 and suppl. Table S3). 21 of the 22 transcript changes were also observed in the background of wtRBP6 induction, with the transcript for the flagellar calcium-binding 44 kDa protein (Tb927.8.5460) only appearing in the mutant RBP6 induction. However, this transcript increased 2-fold after 4 days of induction of wtRBP6. The low number of detected transcript changes is most likely a reflection of a more heterogeneous population in mutRBP6-induced cells, since only about 10% undergo the differentiation process, as compared to 40–50% in wtRBP6-induced cells.
We next monitored transcript abundance changes during 6 and 5 days of induced expression of wild-type and mutant RBP6, respectively (Table 1 and suppl. Tables S4 and S5). 80 transcripts changed in abundance 2-fold or more during the induction of mutRBP6, as compared to 601 transcripts in wtRBP6-induced expression, and the majority of transcripts changing in mutant RBP6 cells (72 out of 80) were part of the set in wtRBP6 cells. The 8 transcripts changing abundance only in mutRBP6-induced cells are coding for a zinc finger protein (Tb927.10.12760, ZC3H36), an RNA binding protein (Tb927.11.12100, RBP5), a mitochondrial carrier protein (Tb927.10.12840, MCP12), a putative STE/STE11 serine/threonine-protein kinase (Tb927.6.2030), a receptor-type adenylate cyclase (Tb927.6.430, GRESAG 4), a putative protein with a PSP1 C-terminal conserved region (Tb927.9.9370), and two hypothetical proteins (Tb927.4.4940, Tb927.10.4340). Finally, monitoring transcripts encoding VSGs, and in particular the 5 known metacyclic VSGs (VSG-1954, VSG-397, VSG-531, VSG-639, and VSG-653), did not show any significant differences with the 5 known mVSGs becoming the most abundant transcripts around day 2 and 3 in both wild-type and mutant RBP6 inductions.
3.3. Metacyclics arrest outside the G1/G0 checkpoint
In T. brucei the nucleus and kinetoplast (mitochondrial kDNA) have distinct replication cycles, with the kinetoplast beginning replication first [28]. Cells in the G1/G0 phase have 1 inetoplast and 1 nucleus (1K1N), cells with 2 kinetoplasts and 1 nucleus (2K1N) have segregated the kinetoplast and are at the end of S phase, whereas cells with 2 kinetoplasts and 2 nuclei (2K2N) have undergone mitosis but have not yet undergone cytokinesis. As predicted for an asynchronously growing cell population, procyclic parasites are mainly in the 1K1N configuration with around 20–25% of cells representing dividing cells with either a 2K1N or 2K2N configuration [7]. We have shown previously that purified metacyclics (≥ 99% pure) generated by inducibly expressing wtRBP6 are quiescent cells with no evidence of dividing cells and arrested in G1/G0 [7]. In contrast, inducible expression of mutRBP6 resulted in metacyclics, where 75% of the cells were in the 1K1N configuration, and 2K1N and 2K2N configurations represented 6% and 13% of the cells, respectively (Fig. 3). In addition, abnormal cell types were increased to 6% with cells with multiple kinetoplasts and/or nuclei, >2K>2N cells, being the most prevalent. The increased number of 2K2N and >2K>2N cells could indicate a defect in kinetoplast replication/segregation and/or cytokinesis. Since the metacyclics are non-dividing, this result indicated that inducible expression of mutRBP6 not only skipped the epimastigote stage, but also caused cell cycle arrest outside the G1/G0 phase.
Fig. 3. Karyotype of metacyclics following induction of mutRBP6.
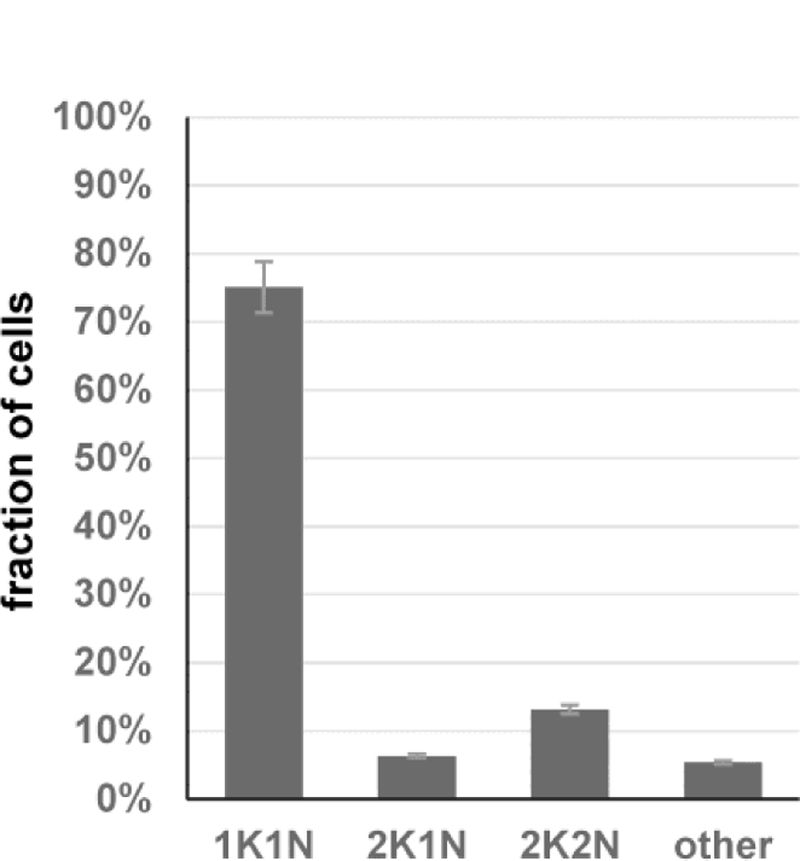
Purified metacyclics were scored for the presence of 1 kinetoplast and 1 nucleus (1K1N), 2 kinetoplasts and 1 nucleus (2K1N), 2 kinetoplasts and 2 nuclei (2K2N) or other configuration (other).
3.4. RNA-Seq analysis of metacyclics
Since the RNA-Seq time course analysis described above monitored a heterogeneous population with only about 10% of cells becoming metacyclics after mutRBP6 induction, we next interrogated the transcriptome of purified metacyclics. This revealed 1,392 differentially expressed transcripts in metacyclics when compared to un-induced procyclic trypanosomes, with 660 and 732 transcripts up- and down-regulated, respectively (Table 1 and suppl. Table S6). Overall, metacyclics derived from inducible expression of mutRBP6 had a predominantly bloodstream-form type transcriptome with the largest changes occurring in genes encoding cell surface components. This result mirrored our previous transcriptome analysis of metacyclics obtained after induction of wtRBP6 [7] and thus, the majority of changes (1,097, 79%) were in common. Nevertheless, there were noteworthy differences, in particular in the set of transcripts down-regulated only in mutant metacyclics (231 transcripts). 13 subunits of eukaryotic translation initiation factors 2, 3 and 4 were less abundant in metacyclics than in procyclics, with 8 transcripts encoding subunits of eukaryotic initiation factor 3 (eIF3), the largest and most complex initiation factor. Furthermore, 8 aminoacyl-tRNA synthetases were down-regulated, thus highlighting translation initiation and accurate mRNA translation. GO analysis also identified components of the mitochondrial membrane as significantly down-regulated in this data set (suppl. Table S2). Conversely, the 30 transcripts only down-regulated in metacyclics generated by wtRBP6 induction were enriched for ribosomal proteins (7 out of 30). Thus, although both metacyclic transcriptomes clearly highlight a down-regulation of transcripts encoding components of the translation machinery, there appears to be a subtle difference in what components are affected in the respective transcriptomes, which will require further investigations. Finally, among the 63 transcripts only up-regulated in metacyclics following mutRBP6 induction was RBP33 (suppl. Table S6) and this transcript was also one of the most highly expressed mRNAs in the metacyclic transcriptome (number 112; suppl. Table S7). This is an intriguing result, since it was previously shown that overexpression of RBP33 leads to a rapid growth arrest in G2/M, i.e. accumulation of 2K2N cells [29], which is one of the configurations enriched in metacyclics (13%; Fig. 3). Overexpression of RBP33 also resulted in a decrease of total mRNA [29], an observation we noted previously in metacyclics when compared to procyclics [7]. Thus, RBP33 could play a role in the establishment of metacyclics and in particular in the generation of metacyclics uncharacteristically arrested at the G2/M checkpoint.
The similarity of the transcriptomes of the two metacyclic populations was also true for the mRNAs encoding VSGs. Although using 0.1% of VSG mRNA as a cutoff, there were 23 VSG transcripts in the wild-type and 9 VSGs in the mutant RBP6 metacyclics, the overall representation of the 5 know metacyclic VSGs was comparable (Fig. 4). As shown previously [7], VSG-397 transcript was the most abundant representing about 50% of the VSG mRNAs in both populations, and for the transcripts derived from the 5 known metacyclic VSG expression sites comprised between 86 and 90 % of VSG mRNAs.
Fig. 4. Metacyclic VSG expression.
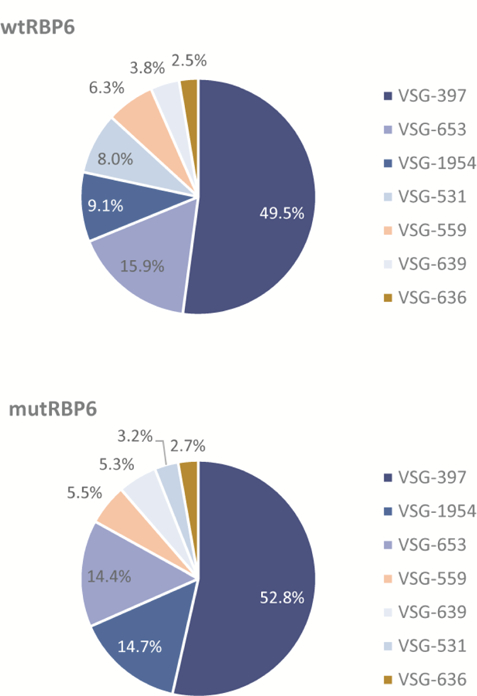
Presented are the percentages for the seven most abundant VSGs. Metacyclic VSGs are shown in shades of blue.
3.5. Metacyclics generated by inducibly expressing mutRBP6 can progress to bloodstream forms in vitro
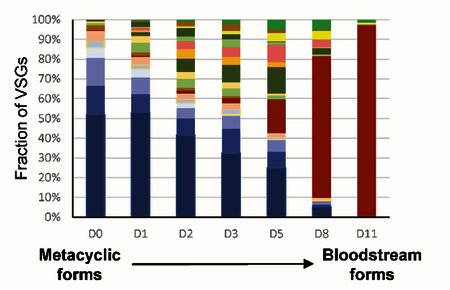
Although inducible expression of wtRBP6 triggered differentiation of procyclics to metacyclics in culture, further differentiation to bloodstream forms required infection of mice, despite numerous attempts to continue the development in vitro. The unusual characteristics of mutRBP6, especially the apparent arrest of some cells at a different checkpoint, as compared to metacyclics generated by wtRBP6, led us to test whether this cell population is able to differentiate into bloodstream forms in vitro. We therefore incubated enriched metacyclics in bloodstream form media at 37°C and within two weeks an exponentially growing culture was obtained, which by visual inspection had characteristics of bloodstream form (BF) trypanosomes. We next investigated the identity of the expressed VSG(s) in the established BF culture, i.e. did the cells switch expression from metacyclic VSGs to a BF VSG? We performed RT-PCR with common primers and following cloning and sequencing the prominent VSG in the in vitro generated BF cells was VSG-2. The appearance of VSG-2 can be explained by the observation that BF trypanosomes expressing VSG-2 have a shorter doubling time in vitro, compared to cells expressing other VSGs [30]. This result was confirmed with gene-specific primers, which detected VSG-2 mRNA at day 2 after transfer to BF conditions and on day 11 only VSG-2, but not VSG-397 was detected (Fig. 5), providing strong evidence that the metacyclic VSGs were silenced and the BF VSG-2 was activated during this transformation process. In order to more accurately monitor the VSG switching event, we performed high-throughput mRNA sequencing for cells collected at similar time points. Our RNA-Seq data revealed that multiple VSGs were being expressed during this transition. Using 0.1% of VSG mRNA as an arbitrary cutoff, 23 VSGs were expressed on day 0 with the known five mVSGs being the most prominent VSGs (>80%; Fig. 6). The number of different expressed VSGs increased to 67 at day 3 and then decreased to 38 at day 11. Many of these VSGs, e.g. VSG-2, −21, −3, −8, −17, −11 and −13 are known to be present in BESs in Lister 427 [31]. Whereas the mVSG gradually decreased, VSG-2 was above the threshold on day 1 (0.4%) and steadily increased till it represented more than 90% of VSG mRNAs on day 11, but it was only at day 8 that VSG-2 became the most prominent VSG.
Fig. 5. RT-PCR using primer specific for VSG-397, VSG-2 and a-tubulin.
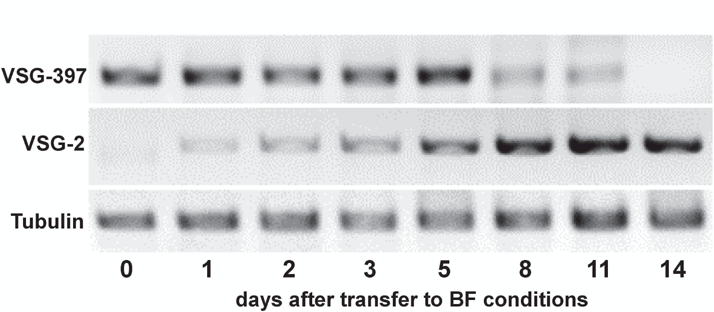
Fig. 6. Dynamics of VSG expression during transformation from metacyclics (D0) to bloodstream forms (D11).
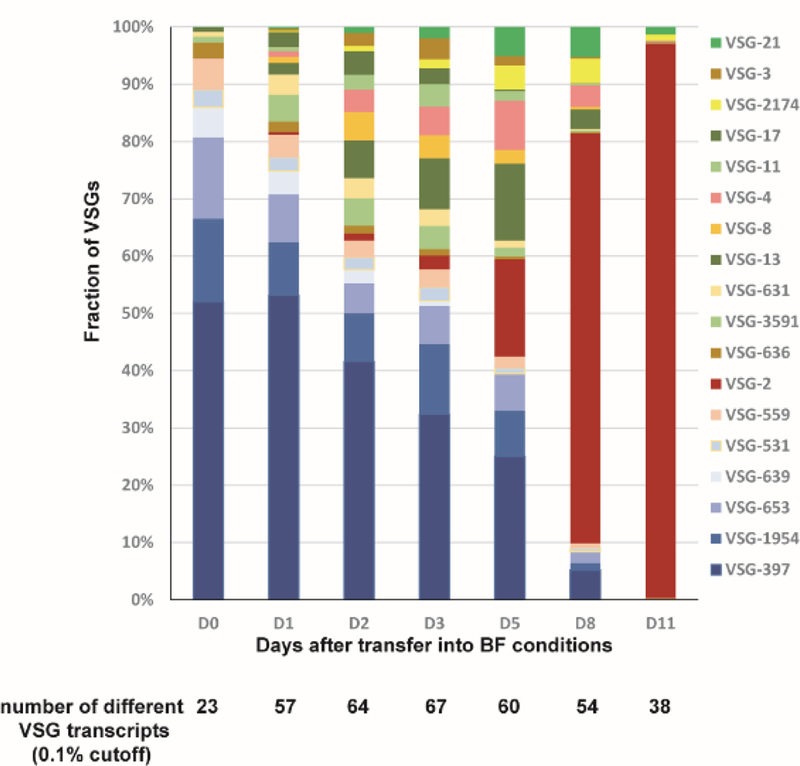
Each colored box represents an individual VSG’s presence in the population. Only variants present at greater than 0.1% of the population at each time point are shown. Metacyclic VSGs are shown in shades of blue.
Monitoring the ESAGs revealed that the mRNAs for ESAG7 and ESAG6, the two subunits of the transferrin receptor and the first two genes in a BSF expression site, were upregulated from the first day of transition, reached a peak at day 3 and then decreased (Fig. 7). It is intriguing to note that the ESAG7/6 mRNA accumulation mirrors the increased diversity of VSGs expressed, suggesting that multiple expression sites were activated in the population initially, but this will require further investigations.
Fig. 7.
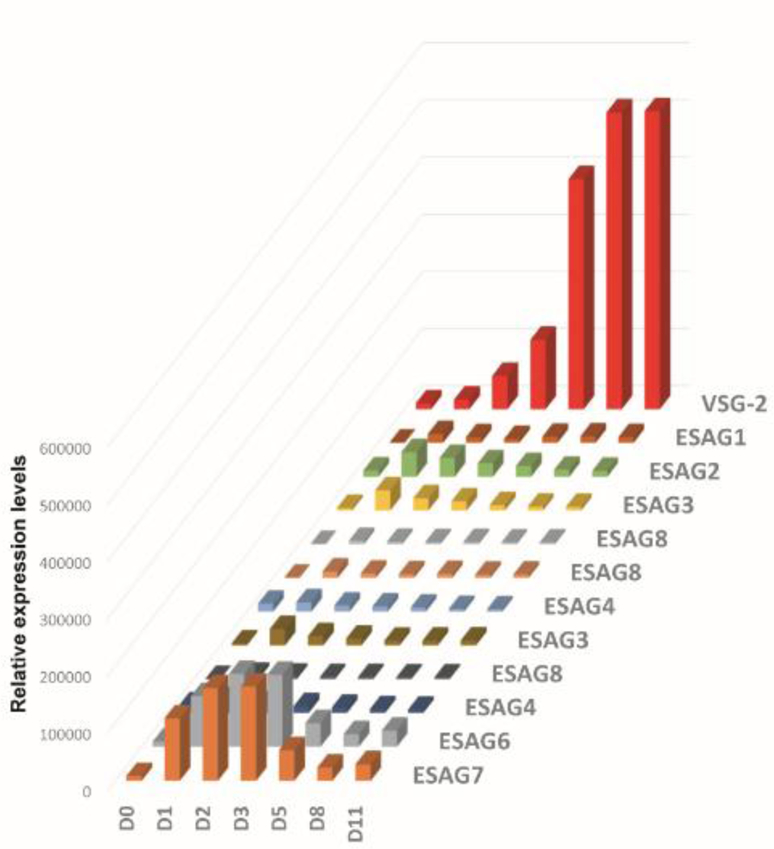
Dynamics of ESAG and VSG-2 expression during transformation from metacyclics (D0) to bloodstream forms (D11).
Including the changes in VSG and ESAG transcript abundance, 560 and 73 transcripts were up- and down-regulated, respectively, during the day 0 to day 11 time course (suppl. Table S8). GO analysis revealed a significant enrichment of transporter activity and membrane components in the up-regulated transcripts. A similar representation emerged when the metacyclic transcriptome (day 0) was compared to the bloodstream form one (day 11), with 449 and 61 transcripts up- and down-regulated, respectively (suppl. Table S9). Consistent with our previous observation that the transcriptome of metacyclics was similar to that of bloodstream forms, the two transcriptomes showed a very good correlation (R2=0.97), and a majority of the up-regulated transcripts (71 out of 449) encoded putative VSG- and ESAG-related proteins. Notable highly up-regulated transcripts include chromosomal passenger complex 2 (11.5-fold; Tb927.11.14840), two procyclic-enriched flagellar receptor adenylate cyclases (7-fold; ACP1, Tb927.11.17040 and ACP2, Tb927.10.16190), PUF nine target 1 (4.9-fold; Tb927.11.6550), and a family of 5 almost identical proteins (Tb927.5.2160, Tb927.5.2170, Tb927.5.2200, Tb927.5.2230, and Tb927.5.2260), up-regulated between 7- and 10-fold. The 5 transcripts encode a 116 aa-long protein with a predicted META domain, a small domain of about 100 aa found in proteins of unknown function. Nevertheless, in Leishmania major the syntenic protein meta-1 localized to the flagellar pocket [32], was mainly expressed in infective metacyclics [33] and overexpression in L. amazonensis generated a more virulent strain [33]. Intriguingly, the T. brucei transcripts encoding a META domain-containing protein were significantly down-regulated in metacyclics as compared to procyclics (between 8.6- and 11-fold; suppl. Tables S6 an S9) in both the wt and mutRBP6 background, which was also evident at the protein level in wtRBP6 metacyclics [7]. Thus, further experiments are warranted to elucidate the function of these META domain-containing proteins in the T. brucei life cycle.
In conclusion, inducible expression of a mutant RBP6 where the first glutamine (Q109) of a stretch of 11 glutamines between the two RRMs was changed to a lysine not only offers an experimental system to investigate how trypanosomes receive instructions to begin synthesizing the mVSG coat, and how each cell expresses a single mVSG, but more importantly will also provide insights how mVSG expression is repressed and switched to the expression of bloodstream-form VSGs. The effect of this single amino acid change is quite remarkable, considering that removing the entire stretch of 11 glutamines did not impact RBP6 function. The next important question to be tackled is the mechanism of action of this mutant RBP6 protein.
Supplementary Material
Acknowledgements
We thank Isabel Roditi for anti-BARP antibodies, Megan Ericson and Francesca Tomaino for performing the T. vivax and T. congolense RBP6 experiments, and Nikolay Kolev for discussions and comments on the manuscript. This work was supported by the National Institutes of Health (http://www.nih.gov) [grant numbers AI028798 and AI110325]. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- BARP
brucei alanine-rich protein
- BES
bloodstream expression site
- BF
bloodstream forms
- GO
Gene Ontology
- MF
metacyclic forms
- mVSG
metacyclic variant surface glycoprotein
- Pol I
RNA polymerase I
- PF
procyclic forms
- RBP6
RNA binding protein 6
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Overexpression of a single point mutation in the Trypanosoma brucei RBP6 (Q109K) generates metacyclics that further differentiate in vitro to bloodstream forms, expressing VSG-2.
The authors declare no financial conflict of interest.
Contributor Information
Huafang Shi, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, 60 College Street, New Haven, Connecticut 06520, USA..
Kiantra Butler, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, 60 College Street, New Haven, Connecticut 06520, USA..
Christian Tschudi, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, 60 College Street, New Haven, Connecticut 06520, USA..
References
- [1].Vickerman K, Tetley L, Hendry KA, Turner CM. Biology of African trypanosomes in the tsetse fly. Biol Cell 1988;64:109–19. [DOI] [PubMed] [Google Scholar]
- [2].Gadelha C, Holden JM, Allison HC, Field MC. Specializations in a successful parasite: what makes the bloodstream-form African trypanosome so deadly? Mol Biochem Parasitol 2011;179:51–8. [DOI] [PubMed] [Google Scholar]
- [3].Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci 1997;110 ( Pt 21): 2661–71. [DOI] [PubMed] [Google Scholar]
- [4].Silvester E, McWilliam KR, Matthews KR. The Cytological Events and Molecular Control of Life Cycle Development of Trypanosoma brucei in the Mammalian Bloodstream. Pathogens 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dyer NA, Rose C, Ejeh NO, Acosta-Serrano A. Flying tryps: survival and maturation of trypanosomes in tsetse flies. Trends Parasitol 2013;29:188–96. [DOI] [PubMed] [Google Scholar]
- [6].Shapiro SZ, Naessens J, Liesegang B, Moloo SK, Magondu J. Analysis by flow cytometry of DNA synthesis during the life cycle of African trypanosomes. Acta Trop 1984;41:313–23. [PubMed] [Google Scholar]
- [7].Christiano R, Kolev NG, Shi H, Ullu E, Walther TC, Tschudi C. The proteome and transcriptome of the infectious metacyclic form of Trypanosoma brucei define quiescent cells primed for mammalian invasion. Mol Microbiol 2017;106:74–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Horn D Antigenic variation in African trypanosomes. Mol Biochem Parasitol 2014;195:123– 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mugnier MR, Stebbins CE, Papavasiliou FN. Masters of Disguise: Antigenic Variation and the VSG Coat in Trypanosoma brucei. PLoS Pathog 2016;12:e1005784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Richardson JP, Beecroft RP, Tolson DL, Liu MK, Pearson TW. Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol Biochem Parasitol 1988;31:203–16. [DOI] [PubMed] [Google Scholar]
- [11].Roditi I, Schwarz H, Pearson TW, Beecroft RP, Liu MK, Richardson JP, et al. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol 1989;108:737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Urwyler S, Studer E, Renggli CK, Roditi I. A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol Microbiol 2007;63:218–28. [DOI] [PubMed] [Google Scholar]
- [13].Tetley L, Turner CM, Barry JD, Crowe JS, Vickerman K. Onset of expression of the variant surface glycoproteins of Trypanosoma brucei in the tsetse fly studied using immunoelectron microscopy. J Cell Sci 1987;87:363–72. [DOI] [PubMed] [Google Scholar]
- [14].Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 2001;414:759–63. [DOI] [PubMed] [Google Scholar]
- [15].Gunzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, et al. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell 2003;2:542–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cestari I, Stuart K. Transcriptional Regulation of Telomeric Expression Sites and Antigenic Variation in Trypanosomes. Curr Genomics 2018;19:119–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nguyen TN, Muller LS, Park SH, Siegel TN, Günzl A. Promoter occupancy of the basal class I transcription factor A differs strongly between active and silent VSG expression sites in Trypanosoma brucei. Nucleic Acids Res 2014;42:3164–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Graham SV, Barry JD. Transcriptional regulation of metacyclic variant surface glycoprotein gene expression during the life cycle of Trypanosoma brucei. Mol Cell Biol 1995;15:5945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Alarcon CM, Son HJ, Hall T, Donelson JE. A monocistronic transcript for a trypanosome variant surface glycoprotein. Mol Cell Biol 1994;14:5579–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ginger ML, Blundell PA, Lewis AM, Browitt A, Gunzl A, Barry JD. Ex vivo and in vitro identification of a consensus promoter for VSG genes expressed by metacyclic-stage trypanosomes in the tsetse fly. Eukaryot Cell 2002;1:1000–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Turner CM, Barry JD, Maudlin I, Vickerman K. An estimate of the size of the metacyclic variable antigen repertoire of Trypanosoma brucei rhodesiense. Parasitology 1988;97:269–76. [DOI] [PubMed] [Google Scholar]
- [22].Cross GA, Kim HS, Wickstead B. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol Biochem Parasitol 2014;195:59–73. [DOI] [PubMed] [Google Scholar]
- [23].Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science 2012;338:1352–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Savage AF, Kolev NG, Franklin JB, Vigneron A, Aksoy S, Tschudi C. Transcriptome profiling of Trypanosoma brucei development in the tsetse fly vector Glossina morsitans. PLoS One 2016;11:e0168877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ramey-Butler K, Ullu E, Kolev NG, Tschudi C. Synchronous expression of individual metacyclic variant surface glycoprotein genes in Trypanosoma brucei. Mol Biochem Parasitol 2015;200:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 2011;6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dean S, Marchetti R, Kirk K, Matthews KR. A surface transporter family conveys the trypanosome differentiation signal. Nature 2009;459:213–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci 1990;95:49–57. [DOI] [PubMed] [Google Scholar]
- [29].Cirovic O, Trikin R, Hoffmann A, Doiron N, Jakob M, Ochsenreiter T. The nuclear RNA binding protein RBP33 influences mRNA and spliced leader RNA abundance in Trypanosoma brucei. Mol Biochem Parasitol 2017;212:16–20. [DOI] [PubMed] [Google Scholar]
- [30].Doyle JJ, Hirumi H, Hirumi K, Lupton EN, Cross GA. Antigenic variation in clones of animal-infective Trypanosoma brucei derived and maintained in vitro. Parasitology 1980;80:359–69. [DOI] [PubMed] [Google Scholar]
- [31].Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, Bason N, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One 2008;3:e3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nourbakhsh F, Uliana SR, Smith DF. Characterisation and expression of a stage-regulated gene of Leishmania major. Mol Biochem Parasitol 1996;76:201–13. [DOI] [PubMed] [Google Scholar]
- [33].Uliana SR, Goyal N, Freymuller E, Smith DF. Leishmania: overexpression and comparative structural analysis of the stage-regulated meta 1 gene. Exp Parasitol 1999;92:183–91. [DOI] [PubMed] [Google Scholar]
- [34].Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. , SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res 46 (2018) W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


