Chronic lower airway infection with Pseudomonas aeruginosa is a major contributor to morbidity and mortality in individuals suffering from the genetic disease cystic fibrosis (CF). Whereas it was long presumed that each patient independently acquired unique strains of P. aeruginosa present in their living environment, multiple studies have since demonstrated that shared strains of P. aeruginosa exist among individuals with CF.
KEYWORDS: Pseudomonas aeruginosa, bronchiectasis, cystic fibrosis, epidemic strains, genotyping, infection control, shared strains, strain typing, transmissible strain, Liverpool epidemic strain, clone C
SUMMARY
Chronic lower airway infection with Pseudomonas aeruginosa is a major contributor to morbidity and mortality in individuals suffering from the genetic disease cystic fibrosis (CF). Whereas it was long presumed that each patient independently acquired unique strains of P. aeruginosa present in their living environment, multiple studies have since demonstrated that shared strains of P. aeruginosa exist among individuals with CF. Many of these shared strains, often referred to as clonal or epidemic strains, can be transmitted from one CF individual to another, potentially reaching epidemic status. Numerous epidemic P. aeruginosa strains have been described from different parts of the world and are often associated with an antibiotic-resistant phenotype. Importantly, infection with these strains often portends a worse prognosis than for infection with nonclonal strains, including an increased pulmonary exacerbation rate, exaggerated lung function decline, and progression to end-stage lung disease. This review describes the global epidemiology of clonal P. aeruginosa strains in CF and summarizes the current literature regarding the underlying biology and clinical impact of globally important CF clones. Mechanisms associated with patient-to-patient transmission are discussed, and best-evidence practices to prevent infections are highlighted. Preventing new infections with epidemic P. aeruginosa strains is of paramount importance in mitigating CF disease progression.
INTRODUCTION
Cystic fibrosis (CF) is the most common lethal monogenetic disease affecting the Caucasian population, with an incidence rate of approximately 1 in 3,400 live births (1). CF is caused by the inheritance of two mutant copies (in trans) of the cystic fibrosis transmembrane conductance regulator (CFTR) gene (cftr) found on chromosome 7. While more than 2,000 mutations have been identified, only ∼200 are known to be disease causing (2). Despite the diversity in known mutant alleles, the F508del mutation predominates. The F508del mutation is present in approximately 90% of individuals (where at least one copy can be found), and ∼50% of the CF population are homozygous for this allele. Mutations in the CFTR protein result in impaired cyclic AMP-dependent chloride ion transport across epithelial cell membranes (3). This reduction in chloride ion transport is associated with sodium and water hyperabsorption at epithelial surfaces, resulting in dehydrated mucus (4, 5). There is a strong correlation between clinical phenotype and the degree of CFTR activity (6, 7). Indeed, patients with residual function mutations generally have a milder course and less organ involvement.
While a number of organ systems can be affected, lung disease results in the greatest morbidity and mortality in individuals with CF. The thickened secretions created in CF airways impair mucociliary clearance of inhaled debris and infectious organisms (8). This creates an ideal environment for bacterial colonization and growth and persistence of infection. As CF progresses, repeated episodes of acute and chronic infection and the resultant inflammation lead to airway damage manifesting as bronchiectasis, ultimately resulting in end-stage lung disease (9–11). Despite the progress that has been made in controlling CF airway disease, with the median predicted age of survival increasing to 40 to 50 years of age, respiratory failure remains the most common cause of premature death (12). The microbiology of CF airways has recently been comprehensively reviewed, and the review here focuses specifically on clonal P. aeruginosa infection (13).
The prevalence of P. aeruginosa infection in CF increases as individuals age, and early non-Pseudomonas airway colonizers are often supplanted by P. aeruginosa (13). The natural history of incident P. aeruginosa infections in CF is such that this organism is often intermittently recovered by culture from respiratory secretion samples before chronic infection develops (14). Intermittent isolation of P. aeruginosa from respiratory secretions may be secondary to the fact that the initial nidus of infection is in the sinuses rather than in the lung (15, 16). Whereas in the past, P. aeruginosa chronic airway infection was assumed to be the inevitable consequence of CF, an increasing body of data has demonstrated that the early recognition and implementation of aggressive antibacterial eradication protocols may clear transient infections and at least delay chronic P. aeruginosa infections well into adulthood (17). Early eradication has become the de facto standard of care for incident P. aeruginosa infection (18–23). Once established, P. aeruginosa in the CF airways develops into chronic infections and generally persists indefinitely. Ultimately, 60 to 80% of adults will become chronically infected with P. aeruginosa (24–27). While many definitions of chronic infection have been utilized, “Leeds criteria” (28), by which chronicity is defined as the isolation of P. aeruginosa in >50% of sputum cultures over a 12-month period, have gained widespread acceptance. Patients with chronic P. aeruginosa infections have worse baseline lung function (29–32) and high rates of lung function decline (30, 33, 34) compared to age-matched CF controls. Radiographic scores and symptom scores, including cough, are worse in those with chronic P. aeruginosa infection (35, 36), and nutritional status is similarly worsened compared to those without chronic infection (32). Most importantly, many studies demonstrate that progression to end-stage lung disease and death is more rapid in those with chronic infection than in those without (34, 37–40).
P. aeruginosa is a ubiquitous organism, readily found in soil and aquatic environments (41, 42). As such, CF patients are generally thought to initially acquire P. aeruginosa from their local environments (43). Some studies have found that the closer the patients live to large bodies of water, the greater their risk for having chronic P. aeruginosa infection (44, 45). Furthermore, in CF patients with newly identified lower airway infection with P. aeruginosa, a home environmental reservoir with a genotypically identical strain can sometimes be identified (46). Accordingly, the traditional belief was that, with the exception of siblings, CF patients harbored unique isolates (47–49) and that for transmission of infection to occur, repeated, close, and intimate contact was required. Danish researchers first demonstrated differently in 1986, when they reported the potential spread of a drug-resistant P. aeruginosa clone in a pediatric cohort (50, 51). The potential for the epidemic spread of a clone among a CF patient population was further highlighted by a group from the United Kingdom in 1996, when they used, for the first time, molecular techniques to prove the existence of a shared P. aeruginosa strain, the Liverpool epidemic strain (LES) (52). A precedent for infection transmission between CF patients was well known to exist at that time. Multiple epidemic strains of the Burkholderia cepacia complex (Bcc) (in particular B. cenocepacia strain ET12) were documented in the 1980s and 1990s, and infections with these strains were associated with markedly increased morbidity and mortality (53–55).
It appears that in most instances, infection with these epidemic P. aeruginosa (ePA) strains occurs later in the course of CF lung disease. Early-childhood studies suggest that the majority of first P. aeruginosa infections occur with unique, nonclonal strains (56–58). Shared strains, alternatively, are disproportionally observed in older patient populations (42, 57), likely the product of repeated opportunities for exposure eventually culminating in infection transmission (59–61).
These infection transmission occurrences and their deleterious consequences were responsible for a marked change in infection control practices and the social make-up of CF clinics. However, when finally identified, the prevalence rates of individual clonal strains of P. aeruginosa in clinic populations vastly exceeded those of epidemic Bcc strains, demonstrating the inadequacy of historical infection control attempts. Since then, there have been a large number of epidemiological studies, on the clinic, regional, national, and international levels, investigating CF P. aeruginosa isolates for clonal relatedness; these are detailed in the following sections.
PSEUDOMONAS AERUGINOSA GENETICS
For P. aeruginosa, individual strains generally can be observed to exist within clonal complexes, groupings that have a common ancestor and are therefore notable for shared phenotypic and genotypic features (62). As a result, genome diversity is lower within clonal complexes than between them. There are 5,021 genes that exist across P. aeruginosa genomes, with at least 70% sequence identity between them (62, 63). In fact, 90% of the genes share at least 98% sequence identity (64). The core genome of P. aeruginosa consists of approximately 4,000 genes, common to most P. aeruginosa strains (63). However, the entire genetic complement of P. aeruginosa is suggested to include 10,000 to 40,000 additional genes. Indeed, it has been suggested that the P. aeruginosa pangenome consists of a small number of highly conserved core genes, a larger “shell” of genes with limited conservation, and a vast “cloud” of very rare, poorly conserved genes (62, 63, 65, 66). Additionally, much of the P. aeruginosa genome freely recombines with the genomes of unrelated strains, further increasing diversity.
The P. aeruginosa distribution in the world comprises many rare strains and a few ubiquitous ones (67, 68). Environmental studies generally indicate that most CF strains are nonclonal in nature and are a random sample of the broader P. aeruginosa population within a particular locale (41, 67, 69). The primary focus of this review, however, is those few clones that exist frequently and have been postulated and/or demonstrated to be transmissible among individuals with CF.
Patients with chronic infection are generally infected with a single strain of P. aeruginosa, which persists over prolonged periods of time (70–81). In some instances, several unrelated strains of P. aeruginosa can temporarily, or even permanently, coinfect an established CF P. aeruginosa population. This can lead to total strain replacements or cohabitation, leading to persistent, genetically distinct lineages with different functional characteristics (71, 82, 83). However, even within CF individuals chronically infected with only a single strain, significant diversity within their genetically related colonizing P. aeruginosa strain has been demonstrated. At the genomic level, these organisms undergo point mutations, insertions, and even large-scale deletions, leading to the development of individual clades, persisting to various degrees based on their ability to compete within the complex environment of the CF lower airways (82). Accordingly, phenotypic assessments of serial isolates of the same ancestral strain from an individual patient can demonstrate a large amount of phenotypic heterogeneity (14, 84, 85). This phenotypic heterogeneity in chronically colonizing populations of P. aeruginosa is now well recognized and makes direct comparison of phenotypic properties between specific strains problematic (84, 86–91). This limitation must be considered when comparing traits of individual P. aeruginosa clones (detailed below).
Terminology
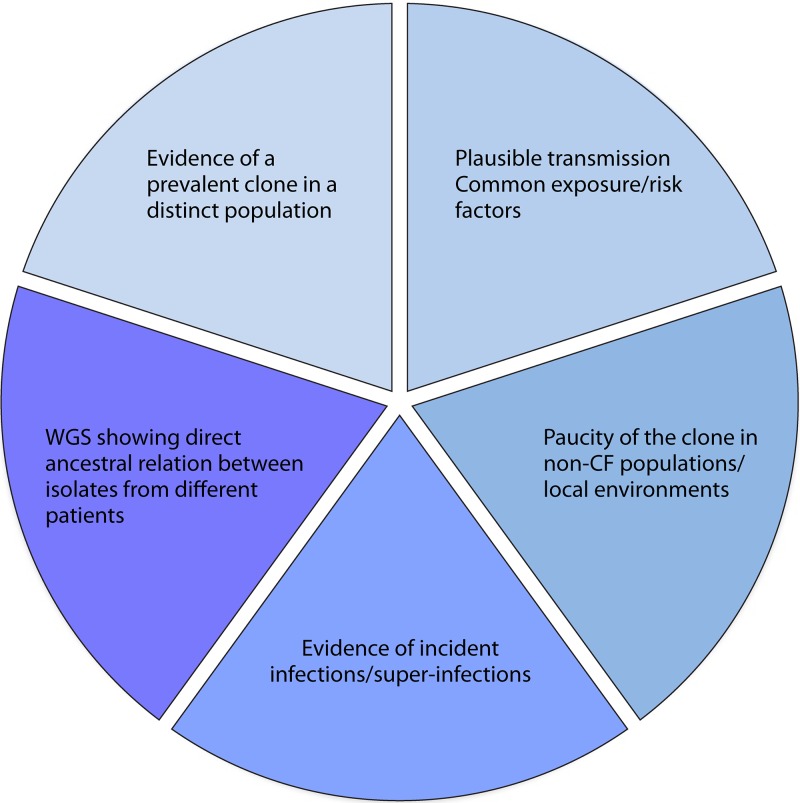
Many terms have been used in CF to describe the potential for a strain to be spread from one individual with CF to another (92, 93). The use of these terms is sometimes inconsistent. Accordingly, we put forward terminology adapted from other recent high-quality reviews (Table 1) (68, 93–95). Any two strains that are identical to a common ancestor can be considered clonal. However, for the strain to be accepted as a potential epidemic P. aeruginosa strain (being readily and frequently spread from one individual to another), several elements must first be documented (illustrated in Fig. 1 and 2). The mere presence of patients infected with the same shared clone of P. aeruginosa is insufficient to label this clone either transmissible or epidemic. Until such time as these elements have been confirmed, the use of terms such as putatively transmissible or potentially epidemic is more appropriate. While some organisms may be transmissible between patients, if they never reach a minimum threshold prevalence, they cannot be considered epidemic. Confounding this is the important distinction that the frequencies of individual clones (even established epidemic clones) may differ from center to center. It is possible that a strain that is epidemic at one site may exist merely infrequently at another site where circumstances do not permit its broad dissemination into a naive population (40, 75, 80). In these other locales, the fact that the numbers of these particular strains have not managed to increase suggests that in addition to having a strain-specific capacity for transmission, environmental and host factors are required as well (Fig. 2 and 3). More recently, the designation “shared strains” has been suggested (96–99). The use of the terminology “shared strain” lacks the potential for negative connotations associated with clonal strains and may therefore facilitate patient recruitment in ongoing epidemiological studies.
TABLE 1.
Proposed terminology with respect to studying the molecular epidemiology of P. aeruginosa in CF
FIG 1.
Features required to fulfill the definition of an epidemic P. aeruginosa strain. For a strain to be considered epidemic, many individual traits should be demonstrated, including the following: (i) a strain must exist at a disproportionately increased prevalence in a defined CF population, be it a clinic, region, or nation; (ii) within a region of increased prevalence, there should be epidemiological linkages showing the potential for spread among patients, including common exposures (camps, fundraising, clinics, or hospitalizations) and increased connectivity among those patients infected with the clone relative to nonclonal isolates; (iii) the clone should either not be found in local non-CF human populations or the natural environment or exist at a greatly increased prevalence in CF populations relative to these non-CF reservoirs; (iv) through the use of prospective sampling, patients who were previously observed either to not be infected with P. aeruginosa or to be infected with unique, nonclonal P. aeruginosa strains should be observed to acquire the putative epidemic strain; and (v) the use of emerging whole-genome sequencing tools enables researchers to demonstrate that the numbers of single nucleotide polymorphisms existing between successive isolates are no higher intrapatient than interpatient, suggesting direct ancestral linkages. Progressive shading indicates those elements with greater weight.
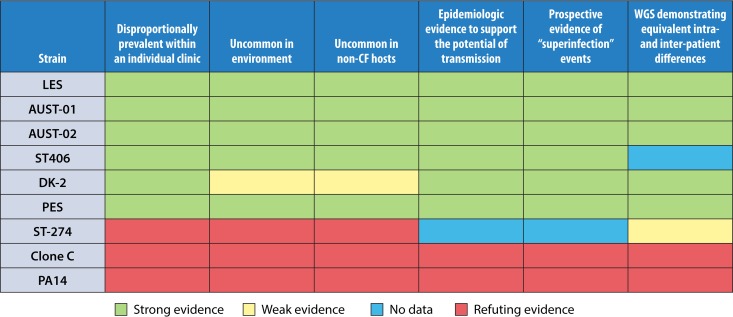
FIG 2.
Evaluating globally important P. aeruginosa CF clones for formal epidemic strain designation.
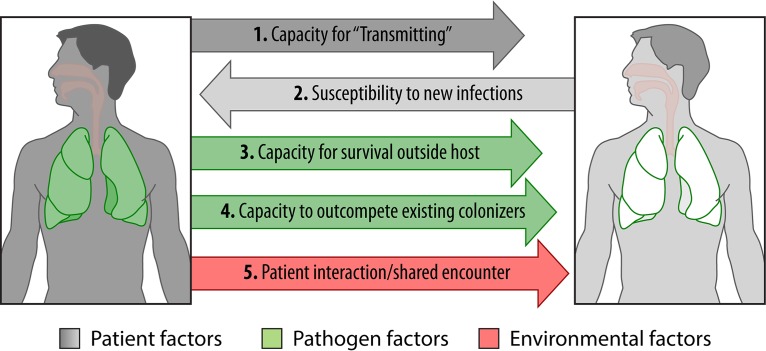
FIG 3.
Events required for P. aeruginosa transmission. A series of events is required for the transmission of P. aeruginosa from one patient to another. The first is patient factors. Both an infected patient capable of transmitting a pathogen and a susceptible host with a lower airway microbiome conducive to invasion with or without superinfection are required. Whether specific demographics increase the risk of either transmission or acquisition is as yet unknown. The second is pathogen factors. While epidemic strains have been shown to differ with respect to several genotypic and phenotypic features, no defining feature of epidemic strains has been identified. An epidemic strain must be able to survive external to the host as a droplet aerosol and resist desiccation. Furthermore, it must be able to outcompete organisms already colonizing the lower airway of the new host. The third is environmental factors. Risk for infection transmission directly correlates with intensity of exposure between the two hosts, which can potentially be minimized with infrastructural controls and/or personal protective equipment.
Genotyping Strategies
The accurate identification of clonal P. aeruginosa strains depends on the genotyping methods used. Multiple genotyping methodologies are available for clinical and research laboratories, and each of them has both advantages and disadvantages. Factors such as discriminatory power, reproducibility, feasibility, turnaround time, cost, and the ability to compare results between laboratories need to be considered when deciding which technique to use in answering specific questions (100).
Untargeted surveillance.
Before genetic-based methodologies were used to type P. aeruginosa, phenotypic typing methods were commonly employed. Lipopolysaccharide (LPS) serotyping relies on the identification of differences in the O-polysaccharide of LPS by using antisera containing polyclonal antibodies derived from rabbits that agglutinate P. aeruginosa in a type-dependent fashion (101, 102). This typing methodology requires a well-developed O-polysaccharide moiety in the bacterial LPS; however, in CF-derived strains, the O-polysaccharide is often deficient, causing agglutination with multiple sera, limiting the ability to discriminate between strains (103). The discriminatory power of this typing methodology is further limited by the fact that there are only 17 antisera, leading to only 17 possible different types that can be identified. Another phenotypic typing method is phage typing, which has similarly been in use since the early 1960s (104, 105). Phage typing relies on distinguishing the patterns of lysis of clinical strains of P. aeruginosa using a defined panel of identified phages (106). As with LPS serotyping, this method is limited by the number of phages available. In general, the phenotypic plasticity of P. aeruginosa strains derived from CF patients makes it challenging to accurately identify genotypically related strains, necessitating genomic-based methodologies in epidemiological studies of CF.
Pulsed-field gel electrophoresis (PFGE) has long been considered to be the gold standard for genotyping clinical isolates, often in the setting of outbreak investigations. Occasionally referred to as “DNA fingerprinting,” PFGE relies on the digestion of bacterial chromosomal DNA with restriction enzymes, resulting in band fragments that are then separated by gel electrophoresis. Because the DNA fragments produced by this macrorestriction enzyme digestion are relatively large, a pulsed electrical field is required to move the DNA efficiently through the agarose gel, hence the name PFGE (107). The most widely used enzyme, SpeI, has approximately 35 cut sites in the P. aeruginosa genome, creating highly individualized fragment sizes (108). Point mutations or insertions and deletions of DNA can alter the restriction profile obtained, leading to distinction between strains. The DNA fragment patterns generated by PFGE are then analyzed, typically based on Tenover criteria (109). According to Tenover guidelines, a banding pattern difference of three fragments can be due to a single genetic event (thus, these isolates are considered highly related), differences of four to six fragments are likely due to two genetic events, and differences of more than seven fragments are due to three or more genetic events (109). In this fashion, PFGE is a highly discriminatory genotyping method, as any random genetic polymorphism that occurs over time during the course of an outbreak can lead to differences in the DNA fragment pattern. PFGE is thus particularly useful in investigating single-center outbreaks. PFGE may not be ideal for comparing strains between centers, as this technique requires considerable skill and results may differ from laboratory to laboratory (110). In addition, the process is time-consuming, labor-intensive, and relatively costly. Another genotyping method that uses the same principles as PFGE is amplified fragment length polymorphism (AFLP) analysis. In AFLP analysis, restriction enzymes are used to digest bacteria, and the fragments are then amplified by PCR before electrophoretic separation of the amplicons on a gel matrix, with visualization of the band pattern. AFLP analysis has been successfully used to identify epidemic strains, such as the LES, Midlands-1 (Md-1), and Manchester (MA) strains, in a fashion comparable to that of PFGE (111).
PCR has also been used to develop typing methods for P. aeruginosa. Random amplified polymorphic DNA (RAPD) analysis is one of the earlier PCR-based techniques using PCR amplification of a random section of the bacterial genome, which is then visualized by using gel electrophoresis, as with PFGE (112). RAPD analysis has the advantage, however, of being simpler and less labor-intensive than PFGE, although it suffers from a lack of reproducibility between different operators and laboratories (102). In addition, repetitive-element-based PCR (Rep-PCR) assays, such as enterobacterial repetitive intergenic consensus (ERIC) sequence PCR (ERIC-PCR) and BOX-PCR, have been used to characterize P. aeruginosa strains isolated from patients with CF (113). Rep-PCR assays utilize primers targeting highly conserved repetitive sequence elements in the bacterial genome. They have the advantage of being reproducible, rapid, and inexpensive (114). Finally, multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) uses PCR to amplify tandem-repeat sections of the bacterial chromosome; polymorphisms in the sequences result in a variable number of tandem repeats, which can be compared between strains (83, 115). The use of a coordinated and centralized nomenclature enables the comparison of a local collection to strains with global isolates and their epidemiologic background.
More recently, sequence-based analysis has been developed as a genotyping approach, focusing on sequencing of single or multiple chromosomal loci. The advantages of sequence-based molecular epidemiology is that it is highly reproducible, has a uniform interpretation, and can be catalogued in databases (https://PubMLST.org/) and, thus, shared between laboratories, as it is based on simple nucleotide sequences (110). One of the most commonly used sequencing-based genotyping techniques is multilocus sequence typing (MLST). MLST typically analyzes seven conserved housekeeping genes, where genetic polymorphisms in sequences at these loci are considered distinct alleles (116). Each isolate has an allelic profile, and isolates with the same allelic profile (7 of 7 alleles) comprise a specific sequence type (ST). Given that MLST examines conserved housekeeping genes, genetic changes in these conserved regions occur slowly over time and are more representative of the evolutionary adaption of a bacterial isolate rather than the more-rapid genetic changes that may occur during the shorter time period of an outbreak (110). Generally, if two strains share 6/7 alleles, they are considered to be likely related, although they have different STs (for example, LES isolates of the same pulsotype have frequently been identified as either ST148 or -683). There are exceptions, however, as P. aeruginosa strains isolated from CF patients with chronic infection can display what is known as a hypermutator phenotype, with rapid mutations occurring in the genes responsible for DNA repair (117). A hypermutable CF P. aeruginosa isolate involving the mutL gene has been described, leading to changes in STs, despite an unchanged PFGE pattern (118).
The most recent technology available to study clonal relatedness and adapted to infer patient-to-patient transmission of organisms is whole-genome sequencing (WGS) (119). WGS has been successfully used to investigate outbreaks of infectious diseases in which patient-to-patient transmission has been well established, such as Clostridium difficile infection (120), methicillin-resistant Staphylococcus aureus (MRSA) infection (121, 122), and tuberculosis (123, 124). Originally considered to be too expensive to be used for routine use, the costs of genome sequencing have deceased considerably in the last several years to the point where it may be considered feasible in certain clinical scenarios. WGS has the advantage of providing a complete view of a bacterial isolate's genome; however, the level of detail renders the analysis complex and time-consuming (125). In fact, the rate-limiting step in whole-genome analysis is the interpretation of the data rather than the sequencing itself. Furthermore, given that WGS detects genetic change down to the level of single nucleotide polymorphisms (SNPs), it can be challenging to determine what degree of genetic relatedness defines clonality between bacterial strains in cross-sectional molecular epidemiological studies. However, when used in longitudinal studies, WGS excels at examining the genetic evolution of bacterial strains over time (126, 127). By using nucleotide substitution frequencies, a molecular clock can be inferred, allowing the establishment of relatedness between strains (128). However, as discussed above, some P. aeruginosa isolates associated with chronic CF infections can have a hypermutator phenotype, which can render the standardization of a molecular clock difficult (117, 129). WGS has been used in epidemiological investigations of patient-to-patient spread of Mycobacterium abscessus (130) and Burkholderia dolosa (131) within CF populations, but it has not yet been used to understand P. aeruginosa transmission dynamics.
Targeted surveillance.
PCR assays can also be designed to detect specific epidemic clones in CF patients (132, 133). These assays are typically used for the real-time detection of new infections (given their ease and cost-effectiveness) in centers in which particular isolates exist at epidemic rates. Individual assays have been developed for LES (132, 134, 135), AUST-01 (133), the Prairie epidemic strain (PES) (136), MA (137), and Midlands-1 (138). The development of these assays requires a detailed understanding of the genetic background and specificity of genetic elements within each P. aeruginosa strain. These assays have been adapted to directly screen whole sputum as opposed to individual isolates, to ensure better detection of epidemic clones (132). A diagnostic assay developed by using a panel of 188 P. aeruginosa isolates to detect a unique genetic locus in the AUST-01 strain was shown to have 100% sensitivity and 100% specificity when applied directly to whole sputum compared to PFGE of individual isolates (133). In areas where multiple shared strains are hyperendemic, such as the United Kingdom, combined assays may prove more cost-effective. A multiplex PCR assay was reported to have 100% sensitivity and 100% specificity in identifying LES, Midlands-1, and the MA epidemic strain in comparison to PFGE results (139).
There are several factors that limit the broad utilization of specific targeted surveillance methods. Unfortunately, these assays generally rely on the detection of specific genomic islands associated with bacteriophages that can demonstrate genetic instability over time, leading to challenges in the identification of strains. A report from the United Kingdom suggested that certain PCR assays, such as the BOX-PCR assay, cannot accurately identify P. aeruginosa LES isolates, likely due to variations in the carriage of bacteriophages and genomic islands in the genomes of these anomalous LES isolates (137, 140). Furthermore, the approach of looking for specific epidemic strains will not identify novel prevalent strains within distinct CF populations. Logan et al. (141) used real-time PCR probes targeting P. aeruginosa algD and gyrB for the early detection of P. aeruginosa in culture-negative children with CF but also made use of previously reported probes to assess the presence of LES and MA (134, 135, 137, 141). In this work, none of the 400 P. aeruginosa-positive sputum samples were positive for these epidemic strains. However, later, the same group, using a nontargeted protocol for PFGE analysis of P. aeruginosa isolates, identified a clonal pulsotype strain infecting 13 patients, including three sibling pairs, in this cohort (142). The use of probes seeking a limited number of targets is similarly limited by the high frequency of movement of patients from different geographic regions, as patient relocation has the potential to result in the introduction of new epidemic strains into a previously naive population (78, 97). This illustrates an important point: one finds only what one is looking for.
Comparative strategies.
There are few studies that have compared genotyping methodologies head-to-head in their abilities to genotype CF P. aeruginosa isolates (106, 143, 144). In an in vitro genotyping study, 48 well-characterized CF P. aeruginosa isolates were genotyped in a blind fashion in three separate laboratories by using PFGE, RAPD analysis, and MLST (144). The discriminatory powers and congruence between the methods were then compared. The discriminatory powers were comparable between the three genotyping methods, although PFGE and MLST demonstrated the highest congruence. When clonality was defined as agreement between two or more methods, MLST had the highest predictive value (100%) in labeling strains as unique, while PFGE had the highest predictive value (96%) in labeling strains as clonal. In a similar study, Kidd et al. genotyped 104 clinical P. aeruginosa isolates (most of which were from CF patients) using ERIC-PCR, PFGE, and MLST and also found the highest level of agreement between PFGE and MLST, although certain isolates with the same ST had distinct PFGE profiles (143). In molecular typing studies of P. aeruginosa isolates from non-CF patients, PFGE has been shown to have a slightly higher discriminatory power than MLST in comparing isolates, although MLST was better for detecting clonal relatedness (145). When RAPD analysis has been compared to PFGE for genotyping CF P. aeruginosa isolates, results between the two typing methodologies have been comparable, although the reproducibility and interlaboratory variability of RAPD results have been concerns (112, 146).
In a recent study by Johansson et al., MLVA was used to genotype P. aeruginosa strains isolated from CF patients, and the results were compared to those of PFGE (147). In this analysis of 232 P. aeruginosa isolates from 104 CF patients, MLVA results were in agreement with PFGE results in 91% of cases, suggesting that MLVA may represent a rapid and effective genotyping method for routine surveillance in clinics (147). Similarly, a Dutch study comparing genotyping of P. aeruginosa isolates (from various patient populations) using MLVA, PFGE, and MLST found universally high discriminatory abilities for all three methods, with a high degree of congruence (with an adjusted Rand coefficient, which quantifies the global agreement between two methods, of 0.84 for PFGE versus MLVA, 0.91 for PFGE versus MLST, and 0.90 for MLST versus MLVA) (148). Investigators have also used data derived from MLST to define sets of informative SNPs that can be used to infer genetic relationships between isolates (149, 150). In this fashion, complete MLST can be avoided, and a more focused and less costly sequencing-based typing methodology can be utilized. Assays targeting 20 P. aeruginosa SNPs have been shown to have good discriminatory power, being able to recognize more than half of the STs from the P. aeruginosa PubMLST website, including the most common Australian epidemic strains, AUST-01, AUST-02, and AUST-06 (98, 99, 151, 152). A study by Ballarini et al. also used a microarray based on 13 SNPs at conserved loci, the fliC multiallelic locus, and the presence or absence of the exoS-exoU marker genes to genotype P. aeruginosa strains from CF patients as well as from patients with acute infections (153). The congruence between the microarray typing results was higher for comparisons to MLST (adjusted Rand coefficient of 0.559) than for comparisons to PFGE (adjusted Rand coefficient of 0.077). Such molecular approaches should be able to be performed by most clinical microbiology laboratories.
In summary, there are several methods available for genotyping of CF P. aeruginosa isolates based on patterns created by enzymatic digestion, PCR assays, and nucleotide sequencing assays. They each have their advantages and disadvantages, and the choice of a specific method depends on the aim of genotyping. For a clinical laboratory having to acutely investigate potential patient-to-patient transmission, PFGE is likely to be the first testing modality employed, as it is routinely available and requires negligible preparatory time. For a clinical research program evaluating P. aeruginosa epidemiology in a new population, the combined use of MLST as an initial genotype screen and PFGE to confirm strain relatedness is preferable, as it enables both a direct assessment of the relatedness of local isolates and comparison to global ePA strains without significant infrastructural/bioinformatic requirements (144). In clinics where ongoing prospective surveillance for hyperendemic clones such as LES or AUST-1 is required, site-directed PCR assays may be used with reasonable sensitivity and good specificity to monitor specifically for these isolates (133, 139). High-volume reference laboratories with specific clinical typing expertise may use a number of methodologies that vary in cost and output capacity, including RAPD-PCR, Rep-PCR, VNTR analysis, and or MLST. Finally, for those researchers examining the evolutionary adaptation and relatedness of bacterial strains over time, WGS may be the most appropriate genotyping technique (126). Indeed, it is likely that in the next decade, WGS will supplant most other technologies, provided that its costs continue to decline and its capacity to rapidly and easily type strains continues to improve.
GLOBAL DISTRIBUTION OF CLONAL P. AERUGINOSA STRAINS IN CF
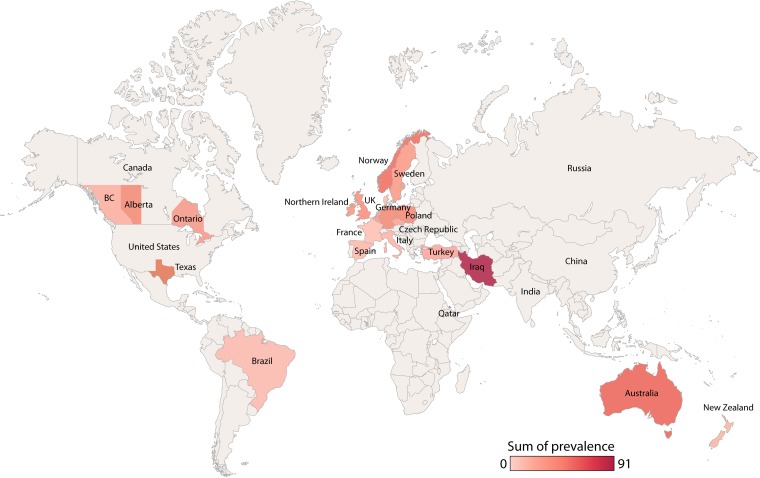
Infection with a clonal P. aeruginosa strain can both significantly impact patient outcomes and serve as a surrogate for the efficacy of infection control policies and procedures. Thus, many studies seeking to understand the local, regional, and national molecular epidemiology of P. aeruginosa have been performed over the last 3 decades. These include a number of single-center (see Table S1 in the supplemental material) and multicenter (Table 2) studies. The estimated prevalence of clonal P. aeruginosa strains in CF populations around the world based on the most recent available data is illustrated on a global map in Fig. 4. What follows is a detailed summary of our knowledge regarding individual named, globally prevalent, and clinically important CF-derived P. aeruginosa strains, listed alphabetically.
TABLE 2.
Multicenter/multiregion CF studies assessing the prevalence of clonal Pseudomonas aeruginosa strainsa
| Country or region | No. of centers | % of population | Focus | Time period (yr) | Testing modality(ies) | Cohort size (no. of subjects) | Main finding(s) | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| Australia | 4 | NS | Peds | 1999–2006 | ERIC-PCR and PFGE | 82 | A subset of the Australasian CF BAL study revealed that all but 3 children (without prior P. aeruginosa infection) experienced new incident infection with novel isolates | 56, 57 |
| 18 | 90 | All | 2007–2010 | ERIC-PCR and PFGE | 983 | 19 clones infecting ≥3 patients were identified in the national ACPinCF study; AUST-01 (22%) and AUST-02 (18%) were most prevalent, found in 89% and 94% of centers, respectively; other common strains were AUST-04 (5%), AUST-05 (4%), AUST-06 (3%), and AUST-07 (2%); only AUST-05 has been observed commonly in the natural environmentb; clonal strains were observed disproportionally in older patients, suggesting that infection control policies have been successful in reducing transmissionc | 96, 97 | |
| Brazil | 4 | NS | All | 2009–2010 | PFGE | 75 | 2 of the 4 clinics had several pairings of unrelated patients with shared strains, representing <20% of each clinic; no shared clones were identified between clinics; no established ePA strains were identified | 289 |
| Belgium | 7 | 100 | All | 2003–2004 | RAPD-PCR and then fAFLP | 276 | 13 clusters of unrelated patients with clonal isolates were identified, including between 2 and 12 patients each; 10 of these clusters could be linked to attendees of the Zeepreventorium; patients in clusters had higher association scores than those with nonclonal isolates; no UK ePA clonal strains were identified | 61 |
| Canada | 8 (Ontario) | NS | Adults (≥16 yr of age) | 2001–2003 | PFGE | 36 | In this study, which sought to identify new strains of P. aeruginosa during pulmonary exacerbations, researchers observed 33% of patients to be infected with a single clone (strain A; LES) | 290 |
| 7 (Ontario) | NS | Adults | 2003–2005 | PFGE and MLST | 446 | Patients were prospectively monitored with quarterly sputum cultures for 3 yr; strain A (LES) and strain B were identified in 22% and 11% of individuals, respectively (each found in 6 of the 7 clinics); 7 new incident infections were observed during the study period | 71, 186, 228 | |
| 7 | NS | All | 2011–2014 | MLST | 483 | High genetic diversity among colonizing strains was observed; rates of ePA were low, with LES and PES observed at 2% and 4% prevalences, respectively; the most common clone was clone C, in 11% of individuals | 281 | |
| France | 10 | NS | All | 2006–2007 | VNTR analysis and MLVA | 156 | In representative samples from multiple clinics, modest clusters of P. aeruginosa spread among the 10 clinics were identified, but no clone exceeded infection in ≥3 patients/center; established ePA strains were not identified | 291 |
| International | 39 | NS | NS | NS | Custom multilocus microarray | 133 | CF isolates were compared against a very large collection of environmental and non-CF clinical isolates; 13 clones represented half of all strains and included environmentally ubiquitous isolates PA14, clone C, and CHA; using a subset of targets within the accessory genome, researchers were able to determine that the vast majority of isolates were indeed different and therefore independently acquired | 222 |
| Italy | 3 | NS | NS | 2006–2008 | AT typing, PFGE, and MLST | 124 | The focus of the study was not on identifying clonal strains, but those researchers identified 7 ST260 isolates from a single center, although these isolates fell into 2 AT types and 5 pulsotypes, suggesting that these isolates were not closely related; indeed, MLST ST260 isolates are commonly found in natural environmentsd | 153 |
| Netherlands | 2 | 46 | All | 2007–2008 | MLST | 313 | 142 different STs were identified as causing infection; in particular, ST406 was found in 15% of subjects, and ST497 was found in 5%; other less common STs identified were ST274 (3%), ST108 and -155 (2% each), ST17, -492, -511, -27, -170, -395, and -517 (2% each), and ST111, -261, -267, -485, -540, and -561 (1% each) | 81 |
| 2 | NS | All | 2007–2011 | MLST | After universal segregation was introduced, no significant changes in prevalences of STs were observed, and 3 cases of ST406 superinfection were observed | 80 | ||
| New Zealand | 7 | 75 | All | 2004–2005 | PFGE | 269 | A heterogeneous P. aeruginosa population was observed, although several small clusters were apparent; the largest cluster involved 9 patients (18% prevalence in one center); patients with this strain were determined to have a higher degree of contact, supporting its potential for transmission; other smaller clusters were present in 6, 5, 4, and 3 individuals; cases of AUST-02 (4 cases), AUST-01 (1 case), and MA (1 case) infections were identified, generally with epidemiological links to zones of endemicity | 60 |
| Norway | 3 | 26 | All | 1994–1998 | PFGE | 60 | A single pulsotype was observed to infect 45% of individuals, and several smaller clusters (2–4 individuals) were identified; infection with the dominant clone was associated with CF camps, health-related CF training camps, and attendance at one particular regional hospital | 59 |
| Spain | 24 | 10 | All | 2013–2014 | MLST, AT marker analysis, and microarray | 75 | A highly heterogeneous P. aeruginosa population was observed; in this limited sampling of individuals from any individual clinic, shared clones were rare; in fact, only one nonkindred pairing was identified; established ePA strains were not observed | 226 |
| United Kingdom (England and Wales) | 31 | 20 | All | 2002–2003 | fAFLP and PFGE | 849 | Importantly, 72% of patients harbored unique isolates; LES was widespread, accounting for 11% of all isolates (found in 48% of centers); less common isolates included Mid-1 (10%), clone C (2%), MA (1%), and Trent (1%); with the exception of LES, most ePA strains were concentrated in a specific center | 111 |
ePA, epidemic P. aeruginosa; Peds, pediatric CF patients; adults, adult CF patients; All, both adult and pediatric CF patients; NS, not stated; fAFLP, fluorescent amplified fragment length polymorphism analysis; AT, ArrayTube.
See reference 41.
FIG 4.
Global prevalence of clonal P. aeruginosa strains. This map is, however, limited. In many cases, a single center's data are extrapolated to an entire country, and admittedly, rates of shared clonal strains have been shown to be highly clinic dependent. Furthermore, rates reported here are from aggregates of the most recently available data. As rates of clonal strain prevalence may change over time, what was once true may no longer be the case. Whereas newer studies have employed technologies such as MLST and VNTR, enabling investigators of individual studies to directly comment on the prevalence of this organism in distant CF populations as well as in non-CF infections and environmental samples, older studies typically used noncomparative genotyping technologies.
AUST-01 (ST649)
AUST-01, originally described as pulsotype 1 or the Australian epidemic strain (AES-1), is hyperendemic among CF patients attending clinics in Southeast Australia (96, 154, 155). While its national prevalence is 22%, it has been found in 94% of CF centers where site-specific prevalences range from 2 to 47% (96). The search that ultimately led to the identification of AUST-01 as an epidemic P. aeruginosa clone was prompted by the death of five children before the age of 5 years shortly after acquiring this clonal isolate (46). However, studies of prospectively collected P. aeruginosa isolates from biobanks have since demonstrated that AUST-01 has been present in the Australian CF population since at least 1991 (34). Importantly, AUST-01 has been observed exclusively in those with CF and has not been identified in environmental reservoirs or non-CF patients (41).
Phenotypic, genotypic, and adaptive characteristics.
Like most CF clonal isolates, AUST-01 lacks a distinctive antibiogram. However, individual isolates of AUST-01 are more likely to be resistant to antibacterials than nonepidemic or unique isolates. In particular, increased levels of antibiotic resistance to beta-lactams, aminoglycosides, and fluoroquinolones are observed (155–157). Furthermore, multidrug resistance (MDR) (typically defined as resistance to all drugs in 2 or more classes of antibiotics) has been identified in as many as 34 to 43% of isolates (156). Limited testing of AUST-01 pathogenicity in animal models has been performed. Relative to PA14, a common reference strain, AUST-01 resulted in increased rates of Caenorhabditis elegans killing. In a mouse model of lung infection, mice infected with AUST-01 exhibited an increased inflammatory response (as measured by tumor necrosis factor alpha [TNF-α] levels) despite a low bacillary load relative to PA14 (158).
Proteomic profiles of AUST-01 show considerable differences compared to the reference strains PAO1 and PA14 (159, 160). In particular, AUST-01 has been shown to overexpress many proteins involved in a range of functions, including virulence (including chitinase, hemolysin, and hydrogen cyanide synthase), siderophore biosynthesis and acquisition (in particular pyochelin), antibiotic resistance (MexX, a multidrug efflux pump component), and LPS biosynthesis (160). In fact, the most abundant protein produced and excreted by AUST-01 is AES_7139, which may be involved in carbohydrate binding, enabling enhanced adherence to epithelial surfaces (160, 161). The gene encoding AES_7139 is highly conserved in AUST-01 isolates.
AUST-01 is also able to produce biofilms of higher biomass than nonepidemic or unique clones (162). Furthermore, AUST-01 is capable of disproportionally invading epithelial cells relative to nonclonal strains; this is particularly true for those isolates adapted to chronic infection (163). In contrast, AUST-01 expresses lower levels of porins (including OprE, OprG, and OprD), which likely contributes to reduced antibiotic permeability conferring resistance compared to nonclonal strains (160). When grown in an artificial medium designed to mimic many conditions existing within the CF airways, AUST-01 had considerable differences in its proteomic profile compared to either PA14 or PAO1. Proteins involved in the biosynthesis of methionine, S-adenosylmethionine, and phenazines and iron acquisition systems were disproportionally expressed in AUST-01 (159). Specifically, pyochelin levels were elevated in early stages of growth, whereas there were no differences in pyoverdin levels. Furthermore, levels of pyocyanin (a known virulence factor in CF) (164) were noted to be increased in AUST-01 relative to PA14 (159, 160).
Genomic differences in AUST-01 compared to the reported reference genome of PAO1 were first assessed by using subtractive hybridization protocols (133). AUST-01-specific genes included those involved in O6 antigen LPS biosynthesis, confirming that AUST-01, like LES and the Midlands-1 strain, belong to the O6 serotype. Other notable differences included differences in phage, restriction endonuclease, DNA repair. and cytosine methyltransferase genes. WGS has since enabled the detailed assessment of the genomic content of AUST-01. AUST-01's genome is 6.254 Mbp and contains 6,957 putative genes (338 unique to it) (165). AUST-01 differs from control strains in the expression of several phage-related gene clusters, in particular the Pf4 prophage gene, which has been implicated in biofilm development and maturation (162, 166). While this prophage is not unique to AUST-01, it is very common within AUST-01 isolates, being found in 100% of screened isolates versus 28% of nonclonal P. aeruginosa isolates (162).
To understand how AUST-01 adapts to the CF lower airway, a study was conducted on serially collected isolates from a CF child who developed chronic infection, where researchers compared the very first infecting isolate with one collected 10 years later (158). WGS demonstrated a loss of function in virulence genes over time (165). However, differential gene expression, as measured by microarray and quantitative PCR (qPCR) analyses, was observed in only 8% of loci. In particular, there was upregulation of adhA, a gene involved in biofilm formation, in the chronic isolate compared to the incident isolate (165). Interestingly, when changing from planktonic to biofilm growth, AUST-01 isolates markedly downregulated genes involved in aerobic respiration (including coxA, coxB, hcnB, hcnC, and hmgA) (158, 162, 167). This same finding was not observed for the control PAO1 and nonclonal strains and may suggest that AUST-01 can more readily adapt to the anaerobic conditions of biofilms and the CF lung (168). In a C. elegans in vivo model, the chronic isolate was not as lethal as the incident isolate. In contrast, in a mouse model of pneumonia, the chronic isolate of AUST-01 resulted in higher levels of inflammation than the incident isolate (158).
Clinical impact.
The impact of AUST-01 was examined by Griffiths et al. (156) in a retrospective cohort study with 3 years of follow-up (Table 3). They demonstrated that children infected with AUST-1 were twice as likely to die as children with nonepidemic strains, although the results did not reach statistical significance (156). A more recent study identified that AUST-01 was present in 38% of the 112 participants and was associated with increased numbers of hospital admission days and pulmonary exacerbation events (169).
TABLE 3.
Clinical outcomes of transmissible P. aeruginosa in individuals with cystic fibrosis by straina
| Strain(s) | Reference | Country | Study design; duration | No. of patients | Study population(s) | Outcome(s) | Main result(s)b |
|---|---|---|---|---|---|---|---|
| Clone C | 178 | UK | Retrospective cohort | 5 (new infections) | Pediatric CF patients | Pre- and post-PEx frequency | No differences |
| AUST-01 | 156 | Australia | Before-and-after cross-sectional cohort, retrospective cohort; 3 yr | 325 (67 with epidemic strain) | CF, consenting patients of all ages | Death | Nonsignificant ↑ OR of death (1.97; 95% CI, 0.57–6.82) |
| 169 | Australia | Prospective cohort; 1 yr | 112 initial/98 monitored (39 with AUST-01) | Adult CF patients, P. aeruginosa infection, 3 yr | PEx, hospital admission days | ↑ PEx frequency, ↑ hospital days | |
| AUST-02 | 169 | Australia | Prospective cohort; 1 yr | 98 (6 with AUST-02) | Adult CF patients, P. aeruginosa infection,c 3 yr | PEx, hospital admission days | No difference in PEx frequency, ↑ hospital days |
| 99e | Australia | Cross-sectional survey at 2 time points + longitudinal cohort (ACPinCF); 3 yr | 166 (70 with AUST-02) | Children + adults with CF and P. aeruginosa infection | Death or transplant, FEV1% decline, BMI decline | ↑ aHR for death/Tx, ↑ hospital days, no significant difference in BMI/FEV1% decline | |
| AUST-03 | 172 | Tasmania | Retrospective cohort; 2 yr | 41 (9 with AUST-03) | CF patients aged ≥15 yr | PEx requiring hospitalization | ↑ PEx hospital admissions and days (in preceding 2 yr) |
| S-1 | 169 | Australia | Prospective cohort; 1 yr | 98 (5 with S-1) | Adult CF patients, P. aeruginosa infection,c 3 yr | PEx, hospital admission days | ↑ PEx frequency, ↑ hospital days |
| S-2 | 169 | Australia | Prospective cohort; 1 yr | 98 (2 with S-2) | Adult CF patients, P. aeruginosa infection,c 3 yr | PEx, hospital admission days | No differences |
| LES | 206 | UK | Retrospective cohort | 157 (18 with LES) | Adult CF patients with sputum sample provided in database | FEV1%, BMI, annual hospital admissions, i.v. antibiotic courses | Nonsignificant trend toward worsened outcomes |
| 207 | UK | Cross-sectional | 157 (93 with LES) | Adult CF patients, PEx free,c 4/52 | HRQoL domains | Worse physical function, Rx burden, respiratory symptoms, health perception | |
| 205 | UK | Prospective cohort; 5 yr | 24 (12 with LES) | CF patients matched for age, sex, spirometry, BMI | FEV1% and BMI annual decline | ↑ FEV1% decline, ↑ BMI decline | |
| MA | 216 | UK | Retrospective cohort; 1 yr | 78 (22 with MA) | Adult CF patients treated for PEx in 2000 | i.v. treatment days, PEx frequency, clinic visits | ↑ PEx frequency, ↑ inpatient episodes and days, ↑ i.v. antibiotic days |
| 217d | UK | Prospective; 8 yr | 80 (28 with MA [21] or LES [7]) | Adult CF patients with chronic P. aeruginosa infection | Survival, FEV1%, BMI, PEx | No survival, FEV1%, BMI difference; ↑ hospitalization days, ↑ i.v. antibiotic days | |
| Midlands-1 | 206 | UK | Retrospective cohort | 157 (47 with Midlands-1) | Adult CF patients with sputum sample provided | FEV1%, BMI, hospital admissions, i.v. abx (preceding) | No differences in disease severity |
| ST406 | 229 | Netherlands | Retrospective cohort | 515 (40 with ST406) | Pediatric and adult CF patients | FEV1%, hospitalization days, BMI | No differences in disease severity or outcomes |
| Irish clones | 294 | Ireland | Retrospective cohort | 32 with clonal strains (23 with pulsotype 1) | Adult CF patients with chronic P. aeruginosa infection | FEV1%, BMI, PEx history | No differences in outcomes pre- and postinfection |
| Italian clones | 295 | Italy | Prospective cohort; 1 yr | 338 (34 with major PFGE clusters) | Pediatric and adult CF patients | FEV1% decline | No outcome differences |
| Polish clones | 296 | Poland | Prospective cohort; 1 yr | 75 (21 with main cluster) | Pediatric CF patients | i.v. antibiotic (hospitalization) days | ↑ i.v. antibiotic/hospital days |
| Strain A/LES | 186 | Canada | Prospective cohort; 3 yr | 446 (67 with LES) | Age ≥18 yr, CF Dx, able to produce sputum | FEV1% decline, time to death/Tx, BMI decline | Similar FEV1 decline, ↑ aHR death/Tx, lower BMI at baseline; no difference over time |
| Strain B | 186 | Canada | Prospective cohort; 3 yr | 446 (32 with strain B) | Age ≥18 yr, CF Dx, able to produce sputum | FEV1% decline, time to death/Tx | No differences in outcomes compared to those with unique P. aeruginosa strains |
| PES | 40c | Canada | Retrospective cohort; 35 yr | 274 (64 with PES) | Age ≥18 yr, 3-mo follow-up | Death or transplant, FEV1% decline, BMI decline | ↑ aHR death/Tx, ↑ FEV1% decline, BMI improvement rate |
PEx, pulmonary exacerbations; Tx, transplant; aHR, adjusted hazard ratio; OR, odds ratio; FEV1%, median forced expiratory volume in 1 s/forced vital capacity; BMI, body mass index; Dx, diagnosis; Rx, treatment; HRQoL, health-related quality of life; i.v., intravenous; CI, confidence interval.
Unless otherwise indicated, the comparator group is patients with unique P. aeruginosa strains.
Compared to patient groups with no P. aeruginosa infection.
The transmissible group comprises those with MA in the majority of cases and thus was not repeated under LES studies.
Disproportionate outcomes were observed exclusively in those with the M3L7 subtype of AUST-02.
AUST-02 (ST775)
AUST-02, originally referred to as pulsotype 2 or AES-2, is particularly common among CF patients attending clinics in Queensland and Western Australia (96). While its national prevalence is 18%, it has been found in 89% of CF centers, where site-specific prevalences range from 2 to 60%. It was first identified in Brisbane, Australia (113, 170), where the earliest-recognized strain of AUST-02 was collected in 1998 (34). Similar to AUST-01, AUST-02 has been observed exclusively in those with CF and has not been identified in environmental reservoirs or non-CF patients (41).
Phenotypic, genotypic, and adaptive characteristics.
AUST-02 is notable for an antibiotic-resistant phenotype. When 63 representative isolates of AUST-02 were compared to 65 nonclonal strains, they were more likely to be resistant to the aminoglycosides and most beta-lactams (imipenem, meropenem, ceftazidime, and piperacillin but not aztreonam) (170). Furthermore, MDR is particularly common, existing disproportionally in AUST-02 relative to nonclonal strains isolated from patients attending the same clinics (range of 28 to 37%) (157, 170).
The genome of AUST-02 is 6.24 Mbp (171). WGS has revealed that the drug-resistant nature of this isolate is secondary to mutations in chromosomal genes rather than the acquisition of novel genetic elements. Indeed, of 11 isolates assessed, all of them had mutations in 49 genes previously established as having the potential to confer antibiotic resistance. Isolates commonly display a hypermutator phenotype (owing to a mutation in mutL), which may be a primary driver of antibiotic resistance. Compared to nonclonal P. aeruginosa isolates, AUST-02 upregulates the expression of the chiC gene (encoding chitinase) and its binding protein (encoded by the cpdD gene) when grown planktonically, similar to what has been observed with LES (167). When AUST-02 is in a biofilm state, it downregulates many genes, including the quorum-sensing regulator rhlR and the effector genes lasB, rhlA, and rhlB. The exceptions are the exsA (transcriptional regulator of ExoS and type III secretion systems [T3SSs]) and prcG (another regulator of T3SSs) genes, both of which are upregulated from its planktonic state.
There appears to be a particularly potent and transmissible sublineage of AUST-02, which thus far is a unique finding among epidemic P. aeruginosa strains. Tai et al. screened mexZ and lasR sequence diversity among a large number of P. aeruginosa clinical strains, including AUST-02 isolates, derived from 70 patients attending the Adult CF Clinic in Brisbane (99). Only two mexZ variants (M2 and M3) and eight lasR variants were detected in the AUST-02 isolates compared to the abundance of variants identified in the non-AUST-02 population. A particular variant, M3L7, was detected exclusively in AUST-02, which those authors proposed represented a distinct subclonal population of AUST-02. This group hypothesized that M3L7 isolates can in fact superinfect patients already infected with the AUST-02 wild-type strain (M2L1). Furthermore, M3L7 AUST-02 demonstrated a particularly drug-resistant phenotype, with 100% of isolates demonstrating resistance to all beta-lactams tested, 94% of isolates demonstrating fluoroquinolone resistance, 80% of isolates showing colistin resistance, and 88% of isolates being tobramycin resistant. Indeed, 100% of isolates had a MDR phenotype. Using longitudinal sampling of prospectively stored isolates, those investigators observed five incident cases of M3L7 AUST-02 infection occurring between 2007 and 2011 among patients previously infected with wild-type AUST-02.
Clinical impact.
In a prospective cohort study identifying epidemic P. aeruginosa clones in CF patients, AUST-02 was identified in 6% of cases and was associated with increased numbers of hospital admission days relative to those associated with nonclonal strains (Table 3) (169). O'Carroll et al. conducted a retrospective cohort study of 100 pediatric and adult patients with CF and noted that those with AUST-02 infection were younger and had lower lung function at baseline than those with unique P. aeruginosa strains (170). With the identification of the M3L7 subtype of AUST-02, clinical outcomes of infection with both this subtype (13 patients) and wild-type AUST-02 (57 patients) were compared to those for patients infected with unique P. aeruginosa strains (96 patients) in a cohort study with longitudinal follow-up. After adjusting for confounders, infection with the M3L7 AUST-02 subtype was associated with an increased hazard ratio for death or lung transplantation, increased numbers of hospital days, and a nonsignificant trend toward declines in lung function and body mass index (BMI) (99). Conversely, infection with the non-M37L AUST-02 subtypes was not associated with an increased risk of death or lung transplantation (99).
AUST-03 (ST242)
AUST-03, originally identified in Tasmania, has been observed most commonly among Australian patients with CF residing in Southern Australia and Tasmania (96). The distribution of AUST-03 is considerably more limited than those of other Australian epidemic strains and was found in only 22% of CF centers, where its prevalence ranged from 1 to 25% (96). Strains of AUST-03 are not known to exist prior to its initial description in 2003 (172, 173). AUST-03 has not been identified in environmental reservoirs or non-CF patients (41).
Phenotypic, genotypic, and adaptive characteristics.
Limited study of AUST-03 has been performed to date. AUST-03 isolates are more likely to be antibiotic resistant (including all beta-lactams, aminoglycosides, and fluoroquinolones) and multidrug resistant, although considerable phenotypic heterogeneity has been documented (172). AUST-03 has not been identified in local hospital environments despite extensive sampling (173).
Clinical impact.
Similarly, there are limited data on the clinical outcomes of AUST-03 infection. A retrospective cohort study of 41 patients aged ≥15 years with CF from multiple regions of Tasmania was conducted. Of these patients, 22% were infected with AUST-03; infection with AUST-03 was associated with a higher frequency of pulmonary exacerbations requiring hospitalizations in the two preceding years than with infection with unique P. aeruginosa strains (Table 3) (172).
Clone C (ST17)
Clone C is thought to be the most abundant P. aeruginosa clone worldwide, having been identified in multiple human and animal infections as well as in freshwater and saltwater environmental reservoirs (67). Clone C has been described as being prevalent to various degrees in multiple individual CF centers, consistent with repeated independent acquisition from environmental reservoirs as opposed to patient-to-patient transmission (79, 82, 174). In a recent survey of P. aeruginosa strains derived from more than 2,200 patients (∼50% of whom had CF) in 143 hospitals in the United Kingdom, clone C was the most common isolate, found in one-third of hospitals, representing 6% of all P. aeruginosa isolates (174). Importantly, a disproportionate abundance in any individual CF center was not observed, suggesting limited potential for patient-to-patient transmission.
Phenotypic, genotypic, and adaptive characteristics.
Clone C has a genome size of 6.902 Mbp with 6,601 open reading frames, which is considerably larger than the reference strain PAO1 (175–177). However, there is wide variation among clone C isolates. In a recent work by Fischer et al., WGS was performed on 57 independently acquired clone C isolates from both human and environmental reservoirs collected over 30 years (62). While they identified significant heterogeneity within the genomes, most sequence diversity was attributed to novel genomic islands and a few regions of specific genome plasticity. In fact, the average strain had 103 unique genes. The accessory genomes of different clone C isolates were as dissimilar as those from unrelated strains from unique backgrounds. Most of the diversity was located in mobile integrative and conjugative elements (ICEs), enabling rapid horizontal spread. Furthermore, intraclonal hot spots for mutations were typically concentrated in phage- or plasmid-related genes as well as in the transcriptional regulator PA2020 and lasR. Within the core genome, however, isolates were different by <100 SNPs. Even within an individual CF host, significant genomic changes and rearrangements are observed (82). Analysis of serial isolates of clone C from an infected patient over 20 years of chronic infection demonstrated both the loss and gain of pathogenicity islands in individual isolates. Those authors observed that clone C exhibited a mutation rate of about 50 SNPs/year of follow-up, which was considerably higher than that observed for an analogous chronic CF infection with PA14, where a mutation rate of 1 SNP/year was observed.
Clinical impact.
There is limited information on the potential impact of clone C. Gopalakaje et al. monitored a cohort of five children with incident clone C infection and did not note a change in the disease trajectory or a predilection for a drug-resistant phenotype (178).
DK-1 (ST387)
DK-1, also known as 6822 or “strain r,” spread rapidly through the Copenhagen, Denmark, CF clinic in 1980 (67). It has persisted in the Danish CF population for over 4 decades. WGS has revealed that DK-1 has a genome size of 6.212 Mbp (179). Relative to its more prevalent comparator, DK-2, our understanding of DK-1 is limited. Analysis of serial isolates from individual patients with chronic infection reveals both high rates of a hypermutator phenotype and mucoidy (secondary to mutations in mucA) (86). While losses of function of the las and rhl quorum-sensing systems occur in 33% of isolates causing chronic infection, this is a lower prevalence than what is observed in DK-2 isolates (180).
DK-2 (ST386)
DK-2 has been referred to by multiple names in the CF literature, including strain 4022 and “strain b.” DK-2 was first recognized in the Danish CF population in 1984, although retrospective analyses of prospectively collected isolates reveal its presence in the CF population since 1972 (67, 129). While considered to be a CF epidemic strain, DK-2 has also been identified to a limited degree in non-CF populations (74).
Phenotypic, genotypic, and adaptive characteristics.
The genome of DK-2 is 6.402 Mbp and shares only 90.7% of its sequence with DK-1, confirming that it is a highly divergent strain acquired from the same locale (179, 181, 182). Yang et al. performed WGS of serial isolates collected from six CF patients from 1973 to 2008 to understand the inter- and intrahost evolution of DK-2 (183). Despite originating from different patients, with a long period between isolate collection, and having significant phenotypic heterogeneity, there was a high level of genome conservation, with only 180 SNPs being identified between the most distantly related clones. Modeling suggests that the common progenitor of the DK-2 epidemic strain entered the Danish CF population immediately before the first isolate from their biobank in 1972. Those researchers proposed that adaptive mutations rapidly arose during the first several years of DK-2's emergence in CF patients. However, thereafter, there was a negative selection bias against adaptive mutations, suggesting that optimal adaptation was achieved early on.
In the C. elegans virulence model, DK-2 strains demonstrated variable lethality (74). In general, isolates of DK-2 from patients with chronic infection demonstrated nonlethality, whereas isolates of DK-2 causing incident infections had lethality comparable to those of the control strains. Marvig et al. similarly studied convergent evolution among 55 DK-2 isolates collected over 34 years from 21 patients (129). Importantly, 20% of these isolates were hypermutators, which were found in 48% of patients. They observed common mutations evolving in loci associated with antibiotic resistance, gene regulation, and cell envelope composition. When those investigators specifically examined these 4,883 intergenic regions, they identified several regions especially prone to mutations (181). The promoter region of the phu system (one of two primary heme uptake systems) was upregulated, suggesting that heme acquisition is an important pathophysiological adaptation of DK-2 during chronic infections. These mutations can confer a growth advantage in in vivo models where hemoglobin is present. Strikingly, this inversely correlated with the production of pyoverdin by DK-2, owing to mutations in pvdS.
The clinical impacts of the Denmark strains have not been reported.
Liverpool Epidemic Strain (ST146 or ST683)
LES represents the most well-known and likely the most prolific epidemic P. aeruginosa strain infecting individuals with CF. It is the only CF P. aeruginosa clone known to be epidemic in individuals with CF in more than one continent (North America and Europe) (184). LES is known to infect 11% of individuals in the United Kingdom, where it is found in almost 50% of clinics (clinic-specific prevalences vary from 5 to 63%) (111). LES infects 15% of CF adults in Ontario, Canada, where it has been found in patients from every part of the province and in over 80% of clinics. Furthermore, sporadic cases have been identified in Australia (96), Spain (185), and other parts of Canada (40), when patients have moved from zones of endemicity to new locales. How LES spread transcontinentally is not clear, but it has been speculated that it transitioned across the Atlantic when children from the United Kingdom attended CF summer camps in Canada (186). The earliest isolate of LES recovered from biobanks was from 1988 (52). LES has been found in the environment only in the immediate vicinity of infected CF patients, such as in air samples and in contaminated quarters, where it is short-lived (187). LES has not been found in non-CF patients without a direct epidemiological link to someone with CF (174). Interestingly, LES has been spread from a child with CF to their non-CF parents as well as to a pet cat, causing symptomatic infection (188, 189).
Phenotypic, genotypic, and adaptive characteristics.
The deleterious effects of LES in CF cases are purported to be due to its extraordinary ability to adapt itself to the CF lung and persist in the airways despite intensive antimicrobial treatment. These adaptations occur in the form of prior acquisition of genes, transcriptional upregulation of gene expression, or a combination of both. Early in vitro studies compared the LESB58 strain (the earliest available isolate and a subtype known to cause chronic infection) to the laboratory reference strains PAO1 and PA14 (190). Although LESB58 had less swimming and twitching motility (acute virulence factors), LESB58 produced more biofilm mass (as measured by a crystal violet assay) than PAO1 and PA14. These findings were confirmed in a rat model of chronic infection. In contrast, when LES431, the isolate associated with the infection of a parent without CF, was examined, it produced less biofilm but caused more death in a mouse model of acute infection than LESB58, reflecting a strain that is more evolved for acute rather than persistent infections (191). Quorum-sensing-regulated exoproducts, such as pyocyanin and LasA protease, are overproduced by clinical LES isolates from CF patients with chronic infection (192). Finally, LES is recognized as a serotype O6 strain, a phenotype associated with more-aggressive disease.
LES is also notorious for its antimicrobial resistance. It was through increased rates of ceftazidime resistance in nonexposed patients that the existence of this transmissible P. aeruginosa strain was first suspected (52). LES is more likely to be resistant to all antibiotics within the classes of aminoglycosides, fluoroquinolones, and beta-lactams than nonclonal unique strains (52, 193–195). Colistin resistance, however, is rarely observed for LES, similar to unique clones (193). Perhaps more concerning is that LES resistance rates have been observed to increase over time in each successive CF cohort (193). Furthermore, pandrug resistance (defined in this particular study as resistance to all agents within the classes of beta-lactams, fluoroquinolones, and aminoglycosides) was observed to increase from 10% in isolates obtained in 2004 to 40% in isolates in 2008. The antimicrobial resistance properties of LES may also permit it to resist clearance, be transmitted, and cause disease. In a transcriptomic analysis of LES subtypes and PAO1, the ampC β-lactamase gene and the mexAB-oprM and mexXY efflux pump genes were all upregulated in isolates LES400 and LES431 compared to strain PAO1 when grown in LB medium as well as in the presence of hydrogen peroxide (196). Expression of these genes confers resistance to β-lactams and aminoglycosides, both mainstays of treatment for P. aeruginosa in CF patients (197). Whole-genome sequencing of a number of LES isolates from individuals with CF from Canada and the United Kingdom identified a number of LES genes involved in antimicrobial resistance, including genes in transporter families that efflux heavy metals and antibiotics (184). These genes were enriched in regions of the accessory genome that were unique to the epidemic strain, a genome measured to be 6.601 Mbp. A hypothesized common pathway for the persistence of LES may be its ability to withstand oxidative stress, be it from activated macrophages, nitric oxide secondary to anaerobic respiration, or antibiotic exposure, thereby avoiding cell death (184, 198). By establishing chronic infection, LES may have more opportunity to spread from patient to patient. Furthermore, LES has demonstrated an enhanced ability to survive on dry surfaces, leading to the potential contamination of clinics and transmission of infection (187).
How do these adaptive changes in LES occur? There is evidence to suggest that these virulence genes occur in genomic islands containing prophages. Whole-genome sequencing of LESB58 revealed the presence of many large genomic islands, including five prophage clusters, one defective (pyocin) prophage cluster, and five nonphage islands (199). Three of these prophages were demonstrated to confer a competitive advantage in a rat model of lung infection. In addition, nonsynonymous polymorphisms frequently occur in the regulatory genes of LES and likely contribute to its phenotypic diversity and variation in virulence (200). In this fashion, LES may evolve in the CF lung through adaptive radiation, responding to key selective pressures. Different antibiotics, for example, have been shown to induce various levels of phage production. Ciprofloxacin can induce high levels of phage, whereas colistin may have an inhibitory effect on phage production (201). Phages are involved in the horizontal transfer of genes, including genes involved in biofilm formation and antimicrobial resistance (202, 203). Thus, the choice of antibiotic therapy could influence phage mobilization, potentially representing an adaptive process. It is therefore not surprising that during periods of pulmonary exacerbation, often characterized by intense antibiotic treatment, variations in phage activity can be detected. In a study of 3 LES-infected patients treated with intravenous (i.v.) antibiotics for a pulmonary exacerbation, there was an increase in the prevalence of the LESGI-5 genomic island, from 10 to 97.5% carriage, detected in the sputum of one patient during a pulmonary exacerbation (73). In addition, LES phages 2 to 4 were detected in the majority of sputum samples tested, indicating widespread phage activity during exacerbations. Finally, in a novel polymicrobial in vitro study of biofilm cocultures of P. aeruginosa LES and various members of the Streptococcus anginosus group (SAG), Whiley et al. demonstrated that in the presence of the SAG, LES produced higher levels of pyocyanin and protease, induced more interleukin-8 (IL-8) production by airway epithelial cells, and caused greater killing of Galleria mellonella larvae than in the absence of the SAG (204). These results illustrate that there are many factors that influence the pathogenesis of LES infection in the CF lung.
Clinical impact.
European and North American studies have independently examined the clinical outcomes of LES infection in CF. A prospective evaluation in the United Kingdom of 12 CF patients with LES matched 1:1 with patients with unique P. aeruginosa strains demonstrated that patients with chronic LES had higher annual rates of decline of FEV1% (median forced expiratory volume in 1 s/forced vital capacity) and BMI (205). When factors associated with LES infection were examined, LES occurred in younger patients who were referred to CF centers at a younger age, and there was a trend toward more-severe disease baseline parameters at screening (206). From a quality-of-life perspective, a cross-sectional survey identified that adults with LES infection scored worse in multiple health-related quality-of-life domains, including physical function, treatment burden, respiratory symptoms, and health perception, than those patients infected with unique P. aeruginosa strains (207). A large prospective multicenter study in Canada by Aaron et al. was undertaken to examine the impact of epidemic P. aeruginosa infection in CF patients on morbidity and mortality over 3 years (186). Of the 446 patients with CF, 67 (15%) had LES, and they were demonstrated to be more than twice as likely to die or undergo lung transplantation as patients with unique strains, but there were no differences noted in lung function or BMI change over time (186). Interestingly, this cohort was subsequently monitored longitudinally to assess differential posttransplant outcomes. Among the 56 lung transplant recipients monitored, no differences in survival following transplantation or rates of acute rejection or occurrence of bronchiolitis obliterans were noted for patients with LES infection compared to patients with infection with unique strains of P. aeruginosa (208).
In addition to accelerated chronic disease progression, LES has been observed to impact CF in many other ways. In patients with incident P. aeruginosa infection with the LES strain, high rates of early eradication treatment failure have been documented (209). Patients infected with LES have also been observed to have disproportionate rates of acute renal failure during pulmonary exacerbations, likely a consequence of the frequent coadministration of nephrotoxic aminoglycosides and colistin in an attempt to treat infection associated with the MDR phenotype (210). LES has been associated with significant respiratory-related morbidity and mortality, causing empyema (211), increased frequencies of pneumothorax (212), and bloodstream infections occurring during acute appendicitis (213).
Manchester Epidemic Strain (ST217)
The Manchester (MA) strain, as its name suggests, was first identified in the Manchester Adult CF Clinic (214). It was first reported in 2001; isolates from an earlier time period have not been reported. Despite existing at epidemic levels within the Manchester CF population, a survey of P. aeruginosa isolates from 31 other CF clinics in England and Wales identified the MA strain in only 3 other clinics (at a prevalence of 2 to 5%). While Manchester isolates have very rarely been isolated outside the United Kingdom, epidemiological links in these cases have always been present (96). The MA strain has been recovered from air samples but not from swabs of solid surfaces in the immediate vicinity of CF patients (215). The MA strain has not been observed in either non-CF populations or environmental surveys (174).
Phenotypic, genotypic, and adaptive characteristics.
The MA strain is notable for its drug-resistant nature, particularly beta-lactams and fluoroquinolones (214). Furthermore, MDR is particularly common, present in up to 91% of patients infected with the MA strain (216). The first genomic studies of MA used subtractive hybridization to assess for differences compared to PAO1 and other epidemic strains (134). Lewis et al. identified a large genomic island of 13.3 kbp (the MA island), which was stable within the genome, containing 18 novel genes and 2 bacteriophage-like regions (137). This bacteriophage gene cluster, Pf4, was unique to the MA strain, and those investigators proposed that it may be involved in its infectivity (137). Those same studies revealed that the MA strain is of serotype O3, unique among other epidemic strains. WGS has since determined that the MA strain's genome is 6.222 Mbp and contains 5,578 putative genes (64). Using comparative genomics, van Mansfeld, however, identified that MA and DK-2 at one point shared a distant ancestor (127).
Clinical impact.
A retrospective cohort study of 78 adult CF patients was performed over a 1-year period, comparing patients with MA infection to those with infection with unique P. aeruginosa strains. Although groups were similar at baseline, patients with MA strain infection had higher pulmonary exacerbation frequencies and higher numbers of inpatient admission episodes, hospitalization days, and intravenous antibiotic days than those with infection with unique strains of P. aeruginosa (216). Jones et al. also conducted a prospective study of 80 patients over 8 years to assess morbidity and mortality of transmissible P. aeruginosa infection compared with infection with unique strains in CF (217). The group infected with transmissible strains consisted of 28 patients, 21 of whom were infected with the MA strain and 7 of whom were infected with the LES strain. Although no survival differences were noted, patients with the transmissible strains had higher hospitalization rates and a higher number of i.v. antibiotic days than patients infected with sporadic strains of P. aeruginosa (217).
Midlands-1 (ST148)
The Midlands-1 (Md-1) strain exists at epidemic levels (27 to 30%) among patients attending the West Midlands Adult CF Centre (111, 174, 206, 218). Importantly, patients infected with Md-1 do not cluster by their postal code, suggesting acquisition from a common CF-related source. Md-1 has been identified in up to 26% of other CF centers in the United Kingdom although relatively infrequently, with a site-specific prevalence of 1 to 5% (111). Md-1 has not been detected in non-CF populations (174). Environmental surveys of Md-1 have not been reported within local afflicted CF populations; however, it has not been reported in general environmental surveys.
Phenotypic, genotypic, and adaptive characteristics.
Antimicrobial resistance does not appear to be a defining feature of Midlands-1, with rates of drug and multidrug resistance similar to those of unique strains (with the exception of colistin). Colistin resistance is present in approximately 13% of Midlands-1 isolates, a 3-fold-higher rate than that observed in nonclonal strains (206).
The use of subtractive hybridization protocols offered the first assessment of the genetic background of Midlands-1 (138). These studies revealed it to be of serotype O6, the most common serotype among epidemic P. aeruginosa strains. Loman et al. reported preliminary data on WGS of over 150 isolates collected over 5 years (219). They reported that this species exhibits a clonal population structure consistent with patient-to-patient spread.
Clinical impact.
Despite its prevalence in the United Kingdom, there are limited data on the outcomes associated with Midlands-1 strain infection in CF. A retrospective study investigating factors associated with P. aeruginosa infection in CF identified that 30% of patients were infected with the Midlands-1 strain. Although these patients were younger at the time of referral to a CF center, there were no other differences noted in disease parameters (including preceding exacerbations or treatments) (206).
PA14 (ST253)
PA14 is one of the most well-studied P. aeruginosa clones; indeed, it is the second most commonly used P. aeruginosa laboratory strain after the standard reference strain PAO1 (ST549) (220). PA14 is ubiquitous in nature and is the second most common (after clone C) isolate recovered from animal, human, and environmental samples (41, 67). In a recent survey of 726 environmental P. aeruginosa strains collected from 47 United Kingdom hospitals, PA14 was the most common strain found, following clone C (174). In the same study of >2,200 patient-derived isolates (∼50% from CF patients), PA14 was found to represent 5.5% of all strains. However, PA14 is relatively uncommon in individuals with CF and is not particularly prevalent in any CF clinic, suggesting that its transmission potential is limited (82).
Phenotypic, genotypic, and adaptive characteristics.
The genome of PA14 was first sequenced in 2006 by Lee et al. and comprises 6.50 Mbp, with 5,973 open reading frames (221). Fischer et al. assessed 42 unrelated P. aeruginosa PA14 isolates collected from multiple environmental and medical sources over 30 years (62). They confirmed that the core genome of PA14 was highly conserved, differing by no more than a few dozen SNPs. However, the PA14 accessory genome was noted to be particularly robust, as the average number of novel genes in each strain was 170 (221, 222). The majority of these genes' closest orthologues were found in other P. aeruginosa clones or non-aeruginosa pseudomonads. Lateral gene transfer seems to be the greatest source of genomic plasticity in PA14 and, in general, all P. aeruginosa strains. Due to its low prevalence in CF patients at any individual CF center, the clinical outcome of PA14 infection in CF has not been assessed.
Prairie Epidemic Strain (ST192)
The Prairie epidemic strain was first identified as clone A097, where it was found in 5% of CF patients attending clinics in Vancouver, Canada. However, it was thought not to represent an epidemic strain at that time, as infection was often transient. However, it has since been identified in 33% of P. aeruginosa-infected patients in the most immediate neighboring CF clinic (Calgary, Canada), approximately 1,000 km away (40, 78, 102, 146). Importantly, patients newly referred to the Calgary adult CF clinic, referred from pediatric and adult CF clinics from other Western Canadian provinces, were identified to be chronically infected with this strain at first assessment, suggesting broad endemicity of PES in the Prairie Provinces of Canada. Longitudinal sampling enabled the observation of multiple PES superinfections, but these occurred exclusively in patients already infected with nonclonal strains of P. aeruginosa (40, 78). Patients infected with PES were more likely to have had a history of attending CF summer camps and being involved in CF fundraising activities (223). By retrospectively assessing prospectively inventoried isolates in a regional CF biobank, investigators identified the first cultured PES isolate from an adult attending a CF clinic in Calgary from 1979 (40). This strain has not been identified in either environmental surveys, studies of the local non-CF bronchiectasis population, or patients experiencing community-acquired P. aeruginosa bloodstream infections (136, 297).
Phenotypic, genotypic, and adaptive characteristics.
Like other epidemic P. aeruginosa strains, PES is notable for increased rates of antimicrobial resistance relative to unique strains (78, 195). In particular, PES isolates are more likely (than unique, nonclonal strains) to be resistant to the following antibiotics: ceftazidime (but not meropenem), tobramycin, and ciprofloxacin. Antimicrobial resistance rates for PES are generally lower than those for comparator epidemic strains from other parts of the world but have been noted to be increasing over time (78). Multidrug resistance, however, remains uncommon in PES.
In vitro studies performed by Duong et al. compared PES to other epidemic strains (including LES and AUST-01 to -04) as well as local nonepidemic P. aeruginosa isolates (195). PES isolates were more likely to exhibit a mucoid phenotype (66% of isolates) than either most nonepidemic isolates (37% of isolates) or other epidemic isolates (14% of isolates). When comparing PES to other epidemic strains, PES produced significantly higher levels of proteases and formed thicker biofilms. Interestingly, superinfecting PES isolates produced less protease and elastase but were more antibiotic resistant than the nonepidemic isolates that they eventually displaced, suggesting that these characteristics may be important in the transmission and establishment of infection by PES (195). To study differential virulence, the Drosophila melanogaster chronic infection fly feeding model was used. PES was observed to kill flies less efficiently than the unique strains that it had replaced (J. Nguyen, J. Duong, M. D. Parkins, and D. G. Storey, presented at the 67th Annual Canadian Society of Microbiology General Conference, Waterloo, ON, Canada, 20 June 2017). Furthermore, in mixed models of infection, where PES was mixed with those same unique strains that it eventually displaced, PES was observed to outcompete the unique, replaced strains, manifesting in reduced fly killing.
In a case-control study comparing the respiratory microbiota of 30 CF patients infected with PES to that of 30 patients infected with unique strains of P. aeruginosa, matched for age, sex, and time period, significant differences were detected (224). Patients infected with PES had microbiota with significantly lower levels of measures of alpha diversity and were more likely to have Pseudomonas as the dominant member of the microflora than patients infected with unique strains. In particular, Prevotella and Veillonella spp. were observed less commonly, suggesting that modulation of resident microflora may be another means by which epidemic P. aeruginosa can influence the disease trajectory. Other works have demonstrated that patients with PES are much less likely than patients with unique P. aeruginosa strains to be coinfected with S. aureus strains (40, 223).
WGS was performed on almost 300 PES isolates from 80 infected CF patients and demonstrated similar levels of inter- and intrapatient diversity (M. G. Surette, personal communication). This work revealed that the genome of PES is 6.35 Mbp. A novel genomic island unique to PES of 40 kb has been well characterized and contains several phage-related genes and hypothetical proteins (136).
Clinical outcome data.
A longitudinal retrospective cohort study was performed to assess the clinical outcomes of PES infection at an adult CF center in Western Canada (40). Of 274 CF patients monitored over a 35-year period, patients with chronic PES infection had a 3-fold-higher hazard ratio for death or lung transplantation than those without P. aeruginosa and a nearly 2-fold-higher hazard ratio for death or lung transplantation than patients with unique P. aeruginosa infection (40). Additionally, PES infection was associated with higher rates of lung function decline and worse nutritional status (as measured by BMI). Similar to LES, when posttransplant outcomes were examined in a cohort of 53 CF patients, no differences in the occurrence of bronchiolitis obliterans syndrome (BOS) or survival were noted between patients infected with PES and those infected with unique strains (225).
CC274 (ST274)
ST274 is common in both CF and non-CF populations, and its role as a transmissible or epidemic P. aeruginosa strain is unknown (41). However, its prevalence in general populations and its predilection for antimicrobial resistance are causes for concern. Occasionally, ST274 has been reported as ST1089 or ST1043 owing to mutations in the acsA or guaA gene, respectively (185, 226). Lopez-Causape et al. performed WGS on 29 ST274 isolates recovered from both Australia and Spain (several isolates were longitudinally recovered from CF patients over a period of up to 18 years) (227). A hypermutator phenotype was found in 31% of isolates and was due to several possible mechanisms, including mutations in the alleles of the mutS and mutL genes. Numbers of SNP differences were significantly higher between patients (51 to 3,256 SNP differences) than within patients (20 to 676 SNP differences). Similar to other common antibiotic-resistant CF strains, antimicrobial resistance in ST274 is generally driven by chromosomal mutations, without evidence of horizontally acquired resistance genes. Clinical-impact studies on ST274-infected CF patients have yet to be performed due to its low prevalence in individual CF clinic populations.
Strain B (ST439)
Strain B was first identified in Ontario, Canada, in 2001 by Aaron et al. (71, 186, 228), where it was present in 8% of CF patients in 86% of CF clinics studied. Since then, it has also been reported in Western Canada following the relocation of several CF patients from Ontario to Alberta (40).
Phenotypic, genotypic, and adaptive characteristics.
Strain B has been shown to overexpress multidrug efflux pump components, specifically mexA and mexX, which are known to contribute to antimicrobial resistance. In addition, in an in vitro susceptibility study, strain B was more resistant to antibiotics (when grown planktonically or as a biofilm) than either laboratory strains or nonclonal P. aeruginosa strains derived from CF patients (194). However, relative to PAO1 and PA14 strains, strain B grew biofilms of low biomass.
Clinical impact.
In the same prospective 3-year study monitoring adult CF patients in Ontario, Canada, performed by Aaron et al., strain B was identified in 7% of participants. Unlike LES, infection with strain B was not associated with an increased hazard ratio for death or lung transplantation and was not associated with increased lung function decline compared with infection with unique P. aeruginosa strains (186).
Dutch Epidemic Strain (ST406)
ST406 has prevalence rates of 20% and 7% in the University Medical Centre Utrecht and Haga Teaching Hospital, respectively (two of the largest CF centers in the Netherlands) (81). The prevalence of ST406 infection was particularly high among those individuals born between 1983 and 1992, with a prevalence exceeding 40%, suggesting potential spread during this time period. Furthermore, having a sibling with CF infected with the same clone was particularly common among those infected with ST406 (229).
ST406 is more likely to be antibiotic resistant and multidrug resistant than nonclonal P. aeruginosa strains (80, 81). van Mansfeld et al. investigated the phenotypic, genotypic, and transcriptomic differences between two isolates of ST406 (the first being the incident isolate) collected 3 years apart from a single patient (127). While considerable phenotypic differences were identified, including a reduced biofilm formation capacity in later isolates, there were relatively few nonsynonymous SNPs in the core genome. Those researchers identified 110 genes with altered transcription levels (more than 2-fold different). The incident isolate was more likely to overexpress genes involved in metabolism and posttranslational modification, whereas the later isolate overexpressed genes encoding cell membrane proteins or proteins involved in cell wall biogenesis or secretion. Interestingly, they observed that the later isolate had 5 genes not present in the initial isolate, possibly representing the incorporation of a prophage.
Using WGS, van Mansfeld et al. demonstrated that the Dutch epidemic strain (ST406) and a second highly prevalent Dutch clone (ST497), both commonly isolated within the same clinics, were not closely related (127).
Clinical impact.
A cohort study of two CF centers in the Netherlands examined clinical outcomes of ST406 infection in 515 CF patients, 265 (52%) of whom were infected with P. aeruginosa (229). Among the P. aeruginosa-infected patients, 18% had ST406 chronic infection. Patients with ST406 were more likely to be receiving chronic inhaled antibiotics than those with nonclonal strains. However, in contrast to many other transmissible strains, CF patients infected with ST406 did not have worse lung function or nutritional status than CF patients infected with unique strains of P. aeruginosa (229).
Other Notable Examples
In addition to large-scale cross-sectional investigations of P. aeruginosa clonality, smaller studies assessing “limited outbreaks” have been performed in the context of unexpected phenotypic patterns. A group in Leeds, United Kingdom, identified the emergence of colistin-resistant P. aeruginosa in six CF patients, four of whom had a clonal strain, as identified by PFGE (230). For these individuals, there were strong epidemiological data to support opportunities for patient-to-patient transmission. Similarly, Johansen et al. reported the emergence of two specific pulsotypes, each causing sequential minioutbreaks, in 1995 and again in 2004, of colistin-resistant P. aeruginosa among a population of Danish CF patients (231). However, as these groups did not report the results of strain typing prior to the emergence of resistance, it is unclear whether these represent events of superinfection by a colistin-resistant clone or the acquisition of adaptive colistin resistance.
The emergence and dissemination of beta-lactamase-producing strains of P. aeruginosa on a global scale are particularly concerning in CF. CF patients with chronic infection with extended-spectrum β-lactamase (ESBL)-producing (232, 233) and metallo-β-lactamase (MBL)-producing (234–236) P. aeruginosa strains have already been documented. A group from Portugal recently reported five patients who were all infected with individual strains of P. aeruginosa, all producing a VIM-2 MBL, epidemic in their local hospital setting (237). In addition to transmission of the strains themselves, transmission of P. aeruginosa mobile genetic elements may be of concern for CF patients.
RISK FACTORS FOR EPIDEMIC P. AERUGINOSA TRANSMISSION AND INFECTION CONTROL STRATEGIES
Potential Mechanisms of Clonal Strain Transmission
While identifying cross-infection events is important for epidemiological purposes, it is even more important to understand the mechanisms and routes by which they have occurred in order to develop evidence-based infection prevention measures in CF. Although a number of routes have been proposed, understanding how the transmission of P. aeruginosa occurs in CF remains poorly understood.
The precise mechanism through which transmission occurs is not clear. Fomites and hands of health care workers (HCWs) have been posed as minor potential sources of transmission. In a 4-week epidemiological study by Doring et al., 81% of sinks at a children's hospital and 42.5% of hands of HCWs were culture positive for P. aeruginosa, suggesting that direct contact may be a source of transmission (238). Panagea et al. also transiently isolated LES from environmental sources (linen, clothing, and respiratory equipment) and HCW hands (187).
Transmission through droplets or droplet nuclei has been suggested as the primary means of cross-infection of transmissible P. aeruginosa in CF by a number of investigators (187, 215, 239–242). When infectious agents are released into the air, they can remain suspended in the form of aerosols, droplets, or droplet nuclei and subsequently infect another individual (243). Aerosols are solid- or liquid-particle suspensions within a gas. Droplets refer to gas-suspended liquid particles that are larger than 10 μm and can be further divided into small (<60-μm) and large (>60-μm) droplets (244). Droplet nuclei are <10 μm in size and represent aerosolized particles with evaporation of much of the water content (245). Critically, droplet nuclei can harbor infectious organisms and have an ability to remain airborne for hours, enabling transmission over further distances (>1 m) (244, 245). As droplet transmission is generally considered short range, to distances of <1 m, transmission in longer ranges will necessitate measures beyond droplet contact precautions (92). The survival of infectious agents varies based on the particle and environmental characteristics, and detection is dependent on the sampling method used; these aspects are described elsewhere (92).
For the transmission of pathogens in persons with CF to occur via the airborne route, an aerosol must be generated (generally following cough), in which infectious particles travel beyond 1 m. In a study of CF patients over a 2-week period, 65% of plated samples (using settle plates) and 20% of air samples were positive for P. aeruginosa following cough (242). Conversely, a mere 5.8% (3/52) of air samples were positive for P. aeruginosa in a study by Speert and Campbell, but the devices used for sampling differed between studies (240). Similarly, an Italian study obtained samples cultured on a sterile sheet or touch plates from 42 CF patients who read aloud a posted text (to re-create normal speaking) or coughing. They identified P. aeruginosa in 0.6% and 1.7% of samples, respectively, again suggesting low transmission potential (246). In a more recent study, Knibbs et al. assessed the viability of P. aeruginosa over distance and time with aerosol sampling systems in 19 patients with CF and in 10 healthy controls (247). All patients with CF had detectable P. aeruginosa, unlike the control patients, and aerosolized colony counts were tightly correlated with sputum bacterial burdens. Viable P. aeruginosa was detected at a distance of up to 4 m (in 17/18 patients), persisting for up to 45 min (in 14/18 patients). The investigators noted that at a ventilation rate of two hourly air changes, the removal of 90% of viable P. aeruginosa bacteria required approximately 50 min after the room had been cleared. Airborne transmission studies at CF centers have rarely cultured P. aeruginosa in the environment up to 3 h following clinical encounters by using spirometry, nebulized therapies, or airway clearance techniques (187, 215). In a study by Ferroni et al. of 22 CF patients in single rooms on a hospital ward, more than 50% of cases had bedroom air samples that were culture positive for strains of P. aeruginosa that were genetically identical to those cultured in their respiratory tract. Notably, the bacterial concentrations were highest after patient waking (154.3 CFU/m3) or airway clearance activities (40.7 CFU/m3) (248).
In addition to clinical studies, a number of laboratory studies have examined the plausibility of airborne transmission of P. aeruginosa. Although it was previously assumed that P. aeruginosa does not survive in droplet nuclei, studies have demonstrated that it can indeed survive in droplet nuclei of artificially generated aerosols for at least 1 min and at a distance of up to 4 m (247, 249, 250). Aerochamber decay models subsequently identified that P. aeruginosa can survive for up to 45 min. Furthermore, mucoid isolates appear to survive longer than nonmucoid isolates (94). In a study of 26 CF patients using cough aerosol sampling, viable P. aeruginosa was identified in the majority of patients (96%), with particle sizes of <3.3 μm in diameter. However, the bacterial quantity was highly variable and was directly related to expiratory flow rates and sputum bacterial loads (241). Similarly, the use of noninvasive ventilation or nebulized therapies in CF patients is associated with aerosol generation, as detected by air sampling surveys of medical wards (251). Importantly, the mere presence of P. aeruginosa in the air does not necessarily mean that transmission events will occur, and it is likely that exposures exceeding a minimum infectious dose are required to institute infection, although work in this area is thus far lacking (252).
Perhaps the most important risk factor for transmission appears to be repeated, close, and prolonged intimate contact. Indeed, coinfected siblings are disproportionally commonly observed among epidemic P. aeruginosa clones (40, 142, 229). In particular, summer camps and, to a lesser extent, winter camps, which were highly prevalent in the late 1970s and 1980s, are thought to have played an important nodal role in infection transmission and dissemination of epidemic P. aeruginosa strains into naive populations at distant CF centers (186, 223). These camps served many purposes, including physical rehabilitation, patient education and motivation, peer support, and peer bonding. Indeed, several studies demonstrated that these camps could produce significant short-term improvements in respiratory health and nutrition (253–255). However, concerns regarding infection transmission quickly became apparent. CF camps are attributed as being a critical node for the spread of Burkholderia cenocepacia (originally referred to as Pseudomonas cepacia) to attendees of multiple CF clinics and subsequent dispersion within individual clinic populations (55, 256) and, accordingly, quickly fell out of favor. As shared strains of P. aeruginosa were more difficult to identify, the potential for clonal P. aeruginosa transmission among campers was not as readily identified. Many CF camp studies have been conducted and assessed the potential transmission of P. aeruginosa both as a new infection in naive patients and as a secondary superinfection event where a previously colonizing isolate could be eliminated by one of these transmissible strains (257). While some studies have not identified transmission (258–260), it may be that the right circumstances to culminate in transmission did not occur in that particular camp. Other prospective studies provide strong observational data in support of P. aeruginosa transmission among camp attendees (257, 261–264). Just like these CF camps, rehabilitation treatment facilities that allow prolonged social contact between patients also exhibited high rates of clonal transmission that likely contributed to the spread of epidemic strains to multiple CF clinics within an individual country (265, 266). Why transmission was noted in many but not all studies is not clear. It may be that a combination of factors are required to come together to facilitate transmission, including a susceptible host, a clone with both improved survival outside the human host and the ability to outcompete and displace existing airway colonizers, a previously infected patient who can spread the infection, and significant opportunities for comingling of patients (Fig. 2). Disturbingly, in a prospective survey of patient beliefs regarding the importance of various infection control practices, patients infected with epidemic P. aeruginosa strains (in particular PES) were more likely to minimize the risk of infection transmission afforded by socialization (223).
Infection Prevention and Control Practices in Cystic Fibrosis
The focus on infection prevention and control (IPC) practices in CF has evolved as our understanding of the potential for pathogen transmission has grown. There has been increasing study of the optimal strategies to prevent infection transmission, but there is much yet to be elucidated. Consensus guidelines have been reported by a number of CF organizations (267–270). Although policies remain variable between continents, guidelines have generally employed a multipronged approach encompassing education, patient separation, hand and cough hygiene, and cleaning and disinfection of equipment. The use of personal protective equipment (PPE) by patients, such as the use of surgical masks, has been recommended in the most recent Cystic Fibrosis Foundation (CFF)-sponsored clinical practice guidelines (268, 270). The use of specific infrastructural controls, incorporating elements of industrial hygiene design and setting (in the development of CF-specific inpatient and outpatient facilities), has alternatively been advocated by others (271).
Separation of CF patients with positive airways cultures into segregated cohorts by pathogen (i.e., epidemic P. aeruginosa) and employing universal segregation (no patient interactions) are longstanding measures to prevent cross-infection and remain widely practiced at European CF centers (268, 272, 273). Retrospective observational studies performed after the introduction of cohort segregation have demonstrated decreases in the numbers of prevalent (156) and incident (80) cases of epidemic P. aeruginosa infections. When infection control measures have been instituted, they have generally been done in conjunction with real-time monitoring for incident cases of new P. aeruginosa strain acquisition. In the year 2000, the Royal Children's Hospital in Melbourne, Australia, introduced a strict cohort segregation policy where patients were separated into specific clinics for P. aeruginosa-negative, unique/nonclonal P. aeruginosa-positive, epidemic AUST-01 P. aeruginosa-positive, Bcc-positive, and methicillin-resistant Staphylococcus aureus (MRSA)-positive infections. Subsequently, prevalence rates of AUST-01 fell from 21% in 1999 to 14% in 2002 and to 6% in 2007, although incident cases continued to occur (46, 155, 156, 274). In the United Kingdom, marked reductions in the prevalence of LES have been observed in successive cohorts of individuals attending the Liverpool adult (275) and pediatric (276) CF clinics after the introduction of cohort segregation. Cohort segregation consisted of seeing CF patients with LES infection in separate, purpose-built outpatient and inpatient facilities based on infection with LES versus unique strains. However, infrequent incident cases continued to be observed despite the intention of absolute segregation of these groups. In Manchester, where patients were initially segregated into P. aeruginosa-positive and -negative groups, continued cases of incident epidemic strain acquisition (either LES or MA) were also observed. However, these cases occurred exclusively among patients in the P. aeruginosa-positive cohort as a result of superinfections (271). No patient who was previously P. aeruginosa naive acquired infection with an epidemic strain. As cohort segregation by pathogen has been associated with ongoing infection transmission, albeit at low levels, universal segregation is increasingly recommended for health care settings. A recent prospective study employing universal segregation and enhanced hand hygiene over a 10-year period identified only one nosocomially acquired case in the study period, thus demonstrating successful prevention of cross-infection of P. aeruginosa in that particular CF center (277). Whether effective infection control measures can be maintained in adult CF clinics, with expanding adult CF populations progressively straining existing infrastructure, remains to be determined.
With growing evidence of the potential for P. aeruginosa (and other pathogens) in CF to be transmitted by droplets or droplet nuclei, IPC guidelines have added measures relating to diagnostics (i.e., pulmonary function testing [PFT]) and the use of PPE, including gowns and gloves for providers and surgical masks for patients (270). PFT recommendations include allowing time for clearance of potential aerosols, using designated laboratories with HEPA filters, or using negative-pressure rooms to decrease transmission risks (270). A study of 11 CF patients by Driessche et al. was conducted to assess the effectiveness of surgical masks in reducing the amount of aerosolized P. aeruginosa (278). They identified that the quantity of P. aeruginosa bacteria recovered by culture was reduced by 88% when a surgical mask was donned compared to no mask use. Similarly, Wood et al. studied 25 adults with CF and chronic P. aeruginosa infection using an aerosol sampling device to measure viable aerosolized P. aeruginosa bacteria following talking or coughing activities (279). The use of surgical face masks was associated with a lower burden of aerosolized P. aeruginosa than uncovered coughing maneuvers, in which 76% generated viable aerosols. Cough etiquette was also beneficial compared to uncovered coughing but provided only ∼50% of the reduction seen with face mask use (279). In contrast, Zuckerman et al. also conducted a randomized controlled study assessing the impact of mask use on air contamination (280). A total of 303 patients were enrolled, with 149 and 154 being randomly assigned to the mask and no-mask groups, respectively. The overall contamination rate was low at 4.6%, with higher rates in spirometry rooms (4.0%) than in exam rooms (1.0%), but rates were similar between the two patient groups. The investigators also noted that increased frequency of cough was associated with increased rates of air contamination (280).
THE FUTURE OF EPIDEMIC STRAINS OF P. AERUGINOSA
Evidence suggests that the current incidence of epidemic P. aeruginosa infections has been markedly reduced from its peak levels. In many instances, these reductions took place even prior to the introduction of more-stringent infection control policies, with the realization that infections can be transmitted between CF patients who are in contact with one another (40, 186). The development of cohort segregation and universal segregation has led to a further reduction in rates of incident infection, although new infections still occasionally occur. Whether the adoption of CFF-sponsored infection control guidelines will lead to further reductions in the rates of incident cases is as yet unproven (270). Furthermore, it is likely that future studies assessing the clinical impact of infection with epidemic P. aeruginosa clones will be less likely to demonstrate a negative clinical outcome. With infrequent incident cases of epidemic P. aeruginosa infection, the overall health of this cohort will be skewed by the survivor effect. Indeed, those few patients continuing to survive with chronic epidemic P. aeruginosa infection will have a milder phenotype and lower rate of disease progression than those who had already succumbed to disease (281). This may be particularly relevant for future studies coming from countries such as the United States, which to date remains notably underrepresented in terms of studies of shared P. aeruginosa strains.
This does not mean, however, that the study of clonal P. aeruginosa in CF should be abandoned. The advent of next-generation sequencing holds great promise in the quest to understand the biology of epidemic P. aeruginosa strains. Recently, an international Pseudomonas aeruginosa consortium has been developed and has worked to develop an international reference panel, including environmentally prevalent strains, epidemic strains, and CF phenotypically adapted strains (282, 283). Ultimately, this group intends to sequence and compare the genomes of 1,000 P. aeruginosa isolates and develop a comprehensive user-friendly genomic pipeline to aid in the rapid assessment of the P. aeruginosa strain background. Furthermore, understanding the biology of superinfection and strain replacement is required in order to ensure that our strategies to prevent new infections are optimized.
Understanding who is at risk of propagating infection is particularly important. All studies to date assessing the presence of clonal P. aeruginosa strains have focused on those patients with active CF lung disease and have excluded those who have received lung transplantation. However, the proportion of individuals with CF having received life-saving lung transplantation continues to increase. These individuals continue to have CF chronic sinus disease, where they continue to harbor those same organisms that chronically colonized their diseased lower airways (284–286). Presently, 10.5% of Canadians living with CF have received transplantation, the majority of which are lung transplantations (24). With improving transplant survival, this number is likely to increase. Indeed, a number of epidemic P. aeruginosa strains have been recovered from patients following lung transplantation in either bronchoalveolar lavage fluid or expectorated sputum, including LES (208), PES (287), and DK-2 (74, 288). Whether individuals following transplantation are capable of transmitting this infection to others is as yet unknown. Similarly, whether patients are capable of acquiring epidemic P. aeruginosa strain superinfections after transplantation is unclear.
CONCLUSIONS
In summary, epidemic strains of P. aeruginosa in CF excel in their ability to be transmitted and cause disease due to adaptive mechanisms that contribute to chronic infection, such as enhanced biofilm formation, antimicrobial resistance, and the downregulation of acute virulence factors, thereby contributing to immune evasion. Epidemic P. aeruginosa was a particular concern for CF populations in the 1980s and 1990s, when socialization among CF patients was prioritized for peer support. Fortunately, with the introduction of increasingly stringent infection control measures, the incidence of epidemic P. aeruginosa strains in CF patients continues to fall. However, continued genotypic surveillance is recommended in order to monitor the emergence and spread of potential shared strains. Furthermore, novel treatment strategies specifically targeting the adapted epidemic P. aeruginosa phenotype are required to improve the clinical outcome of those patients living with chronic infection.
Supplementary Material
Biographies

Michael D. Parkins completed an M.Sc. in Cystic Fibrosis (CF) microbiology and M.D. at the University of Calgary in 2000 and 2003, respectively. He became a Fellow of the Royal College of Physicians and Surgeons of Canada in Internal Medicine and Infectious Diseases in 2008 before entering into a CF Fellowship at Queen's University of Belfast, United Kingdom, in 2009. He is currently an Associate Professor in the Department of Medicine and the Department of Microbiology and Infectious Disease in the Cumming School of Medicine at the University of Calgary. He is the Clinic Director of the Southern Alberta Adult Cystic Fibrosis Clinic. His research interests focus on CF airway disease, in particular harnessing the CF microbiome to predict clinical outcomes and treatment response, and the study of infection transmission risk among those with suppurative lung disease.

Ranjani Somayaji completed her M.D. in 2010 followed by residency training in Internal Medicine and Infectious Disease from the University of Calgary and an M.P.H. from Johns Hopkins University. She subsequently completed a one-year clinical CF fellowship and is currently completing a two-year research CF fellowship at the University of Washington. She is currently a clinical lecturer with the Department of Medicine in the Cumming School of Medicine at the University of Calgary. Her research interests include understanding the epidemiology and management of airway infections and their effects on clinical outcomes in individuals with CF.

Valerie J. Waters is an Associate Professor of Pediatrics at the University of Toronto. She has been a full-time staff physician in the Division of Infectious Diseases and is the infectious diseases consultant for the cystic fibrosis (CF) clinic at the Hospital for Sick Children, Toronto, since 2005. She is also Senior Associate Scientist with the Research Institute at the Hospital for Sick Children. Dr. Waters received her M.D. from McGill University, Montreal, and did her pediatric residency at the Hospital for Sick Children and her infectious diseases fellowship at Columbia Presbyterian Medical Center, New York, NY. She also obtained a M.Sc. in biostatistics from Columbia University, New York, NY. Dr. Waters' primary research focus is the diagnosis and management of multidrug-resistant pathogens in CF and biofilm antimicrobial susceptibility testing. She also studies the clinical impact of pulmonary exacerbations in CF and the role of the pulmonary microbiome in CF.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/CMR.00019-18.
REFERENCES
- 1.O'Sullivan BP, Freedman SD. 2009. Cystic fibrosis. Lancet 373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 2.Ferec C, Cutting GR. 2012. Assessing the disease-liability of mutations in CFTR. Cold Spring Harb Perspect Med 2:a009480. doi: 10.1101/cshperspect.a009480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalemans W, Barbry P, Champigny G, Jallat S, Dott K, Dreyer D, Crystal RG, Pavirani A, Lecocq JP, Lazdunski M. 1991. Altered chloride ion channel kinetics associated with the delta F508 cystic fibrosis mutation. Nature 354:526–528. doi: 10.1038/354526a0. [DOI] [PubMed] [Google Scholar]
- 4.Boucher RC. 2007. Cystic fibrosis: a disease of vulnerability to airway surface dehydration. Trends Mol Med 13:231–240. doi: 10.1016/j.molmed.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Fahy JV, Dickey BF. 2010. Airway mucus function and dysfunction. N Engl J Med 363:2233–2247. doi: 10.1056/NEJMra0910061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Gracia J, Mata F, Alvarez A, Casals T, Gatner S, Vendrell M, de la Rosa D, Guarner L, Hermosilla E. 2005. Genotype-phenotype correlation for pulmonary function in cystic fibrosis. Thorax 60:558–563. doi: 10.1136/thx.2004.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirtz S, Gonska T, Seydewitz HH, Thomas J, Greiner P, Kuehr J, Brandis M, Eichler I, Rocha H, Lopes AI, Barreto C, Ramalho A, Amaral MD, Kunzelmann K, Mall M. 2004. CFTR Cl-channel function in native human colon correlates with the genotype and phenotype in cystic fibrosis. Gastroenterology 127:1085–1095. doi: 10.1053/j.gastro.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Hoegger MJ, Fischer AJ, McMenimen JD, Ostedgaard LS, Tucker AJ, Awadalla MA, Moninger TO, Michalski AS, Hoffman EA, Zabner J, Stoltz DA, Welsh MJ. 2014. Impaired mucus detachment disrupts mucociliary transport in a piglet model of cystic fibrosis. Science 345:818–822. doi: 10.1126/science.1255825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ratjen F. 2008. Recent advances in cystic fibrosis. Paediatr Respir Rev 9:144–148. doi: 10.1016/j.prrv.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Ratjen F. 2009. Update in cystic fibrosis 2008. Am J Respir Crit Care Med 179:445–448. doi: 10.1164/rccm.200812-1927UP. [DOI] [PubMed] [Google Scholar]
- 11.Ratjen F, Doring G. 2003. Cystic fibrosis. Lancet 361:681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- 12.Stephenson AL, Sykes J, Stanojevic S, Quon BS, Marshall BC, Petren K, Ostrenga J, Fink AK, Elbert A, Goss CH. 2017. Survival comparison of patients with cystic fibrosis in Canada and the United States: a population-based cohort study. Ann Intern Med 166:537–546. doi: 10.7326/M16-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LiPuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 15.Aanaes K. 2012. Bacterial sinusitis can be a focus for initial lung colonisation and chronic lung infection in patients with cystic fibrosis. J Cyst Fibros 12(Suppl 2):S1–S20. doi: 10.1016/S1569-1993(13)00150-1. [DOI] [PubMed] [Google Scholar]
- 16.Whiteson KL, Bailey B, Bergkessel M, Conrad D, Delhaes L, Felts B, Harris JK, Hunter R, Lim YW, Maughan H, Quinn R, Salamon P, Sullivan J, Wagner BD, Rainey PB. 2014. The upper respiratory tract as a microbial source for pulmonary infections in cystic fibrosis. Parallels from island biogeography. Am J Respir Crit Care Med 189:1309–1315. doi: 10.1164/rccm.201312-2129PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones AM. 2005. Eradication therapy for early Pseudomonas aeruginosa infection in CF: many questions still unanswered. Eur Respir J 26:373–375. doi: 10.1183/09031936.05.00069705. [DOI] [PubMed] [Google Scholar]
- 18.Blanchard AC, Horton E, Stanojevic S, Taylor L, Waters V, Ratjen F. 2017. Effectiveness of a stepwise Pseudomonas aeruginosa eradication protocol in children with cystic fibrosis. J Cyst Fibros 16:395–400. doi: 10.1016/j.jcf.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Hansen CR, Pressler T, Hoiby N. 2008. Early aggressive eradication therapy for intermittent Pseudomonas aeruginosa airway colonization in cystic fibrosis patients: 15 years experience. J Cyst Fibros 7:523–530. doi: 10.1016/j.jcf.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Langton Hewer SC, Smyth AR. 2014. Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis. Cochrane Database Syst Rev 2014:CD004197. doi: 10.1002/14651858.CD004197.pub4. [DOI] [PubMed] [Google Scholar]
- 21.Ratjen F, Munck A, Kho P, Angyalosi G, ELITE Study Group. 2010. Treatment of early Pseudomonas aeruginosa infection in patients with cystic fibrosis: the ELITE trial. Thorax 65:286–291. doi: 10.1136/thx.2009.121657. [DOI] [PubMed] [Google Scholar]
- 22.Stanojevic S, Waters V, Mathew JL, Taylor L, Ratjen F. 2014. Effectiveness of inhaled tobramycin in eradicating Pseudomonas aeruginosa in children with cystic fibrosis. J Cyst Fibros 13:172–178. doi: 10.1016/j.jcf.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Tramper-Stranders GA, Wolfs TF, van Haren Noman S, van Aalderen WM, Nagelkerke AF, Nuijsink M, Kimpen JL, van der Ent CK. 2010. Controlled trial of cycled antibiotic prophylaxis to prevent initial Pseudomonas aeruginosa infection in children with cystic fibrosis. Thorax 65:915–920. doi: 10.1136/thx.2009.126128. [DOI] [PubMed] [Google Scholar]
- 24.Cystic Fibrosis Canada. 2017. The Canadian Cystic Fibrosis Patient Registry: 2016 annual data report. Cystic Fibrosis Canada, Toronto, Ontario, Canada. [Google Scholar]
- 25.Hauser AR, Jain M, Bar-Meir M, McColley SA. 2011. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin Microbiol Rev 24:29–70. doi: 10.1128/CMR.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambiase A, Raia V, Del Pezzo M, Sepe A, Carnovale V, Rossano F. 2006. Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect Dis 6:4. doi: 10.1186/1471-2334-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razvi S, Quittell L, Sewall A, Quinton H, Marshall B, Saiman L. 2009. Respiratory microbiology of patients with cystic fibrosis in the United States, 1995 to 2005. Chest 136:1554–1560. doi: 10.1378/chest.09-0132. [DOI] [PubMed] [Google Scholar]
- 28.Lee TW, Brownlee KG, Conway SP, Denton M, Littlewood JM. 2003. Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. J Cyst Fibros 2:29–34. doi: 10.1016/S1569-1993(02)00141-8. [DOI] [PubMed] [Google Scholar]
- 29.Kerem E, Corey M, Gold R, Levison H. 1990. Pulmonary function and clinical course in patients with cystic fibrosis after pulmonary colonization with Pseudomonas aeruginosa. J Pediatr 116:714–719. doi: 10.1016/S0022-3476(05)82653-8. [DOI] [PubMed] [Google Scholar]
- 30.Konstan MW, Morgan WJ, Butler SM, Pasta DJ, Craib ML, Silva SJ, Stokes DC, Wohl ME, Wagener JS, Regelmann WE, Johnson CA, Scientific Advisory Group and the Investigators and Coordinators of the Epidemiologic Study of Cystic Fibrosis. 2007. Risk factors for rate of decline in forced expiratory volume in one second in children and adolescents with cystic fibrosis. J Pediatr 151:134.e1–139.e1. doi: 10.1016/j.jpeds.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Navarro J, Rainisio M, Harms HK, Hodson ME, Koch C, Mastella G, Strandvik B, McKenzie SG. 2001. Factors associated with poor pulmonary function: cross-sectional analysis of data from the ERCF. European Epidemiologic Registry of Cystic Fibrosis. Eur Respir J 18:298–305. doi: 10.1183/09031936.01.00068901. [DOI] [PubMed] [Google Scholar]
- 32.Pamukcu A, Bush A, Buchdahl R. 1995. Effects of Pseudomonas aeruginosa colonization on lung function and anthropometric variables in children with cystic fibrosis. Pediatr Pulmonol 19:10–15. doi: 10.1002/ppul.1950190103. [DOI] [PubMed] [Google Scholar]
- 33.Kosorok MR, Zeng L, West SE, Rock MJ, Splaingard ML, Laxova A, Green CG, Collins J, Farrell PM. 2001. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol 32:277–287. [DOI] [PubMed] [Google Scholar]
- 34.Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, Grimwood K. 2001. Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J Pediatr 138:699–704. doi: 10.1067/mpd.2001.112897. [DOI] [PubMed] [Google Scholar]
- 35.Sawicki GS, Rasouliyan L, McMullen AH, Wagener JS, McColley SA, Pasta DJ, Quittner AL. 2011. Longitudinal assessment of health-related quality of life in an observational cohort of patients with cystic fibrosis. Pediatr Pulmonol 46:36–44. doi: 10.1002/ppul.21325. [DOI] [PubMed] [Google Scholar]
- 36.West SE, Zeng L, Lee BL, Kosorok MR, Laxova A, Rock MJ, Splaingard MJ, Farrell PM. 2002. Respiratory infections with Pseudomonas aeruginosa in children with cystic fibrosis: early detection by serology and assessment of risk factors. JAMA 287:2958–2967. doi: 10.1001/jama.287.22.2958. [DOI] [PubMed] [Google Scholar]
- 37.Courtney JM, Bradley J, McCaughan J, O'Connor TM, Shortt C, Bredin CP, Bradbury I, Elborn JS. 2007. Predictors of mortality in adults with cystic fibrosis. Pediatr Pulmonol 42:525–532. doi: 10.1002/ppul.20619. [DOI] [PubMed] [Google Scholar]
- 38.Demko CA, Byard PJ, Davis PB. 1995. Gender differences in cystic fibrosis: Pseudomonas aeruginosa infection. J Clin Epidemiol 48:1041–1049. doi: 10.1016/0895-4356(94)00230-N. [DOI] [PubMed] [Google Scholar]
- 39.Hudson VL, Wielinski CL, Regelmann WE. 1993. Prognostic implications of initial oropharyngeal bacterial flora in patients with cystic fibrosis diagnosed before the age of two years. J Pediatr 122:854–860. doi: 10.1016/S0022-3476(09)90007-5. [DOI] [PubMed] [Google Scholar]
- 40.Somayaji R, Lam JC, Surette MG, Waddell B, Rabin HR, Sibley CD, Purighalla S, Parkins MD. 2017. Long-term clinical outcomes of ‘Prairie epidemic strain’ Pseudomonas aeruginosa infection in adults with cystic fibrosis. Thorax 72:333–339. doi: 10.1136/thoraxjnl-2015-208083. [DOI] [PubMed] [Google Scholar]
- 41.Kidd TJ, Ritchie SR, Ramsay KA, Grimwood K, Bell SC, Rainey PB. 2012. Pseudomonas aeruginosa exhibits frequent recombination, but only a limited association between genotype and ecological setting. PLoS One 7:e44199. doi: 10.1371/journal.pone.0044199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranganathan SC, Skoric B, Ramsay KA, Carzino R, Gibson AM, Hart E, Harrison J, Bell SC, Kidd TJ, Australian Respiratory Early Surveillance Team for Cystic Fibrosis. 2013. Geographical differences in first acquisition of Pseudomonas aeruginosa in cystic fibrosis. Ann Am Thorac Soc 10:108–114. doi: 10.1513/AnnalsATS.201209-077OC. [DOI] [PubMed] [Google Scholar]
- 43.Kelly NM, Fitzgerald MX, Tempany E, O'Boyle C, Falkiner FR, Keane CT. 1982. Does Pseudomonas cross-infection occur between cystic-fibrosis patients. Lancet ii:688–690. [DOI] [PubMed] [Google Scholar]
- 44.Goeminne PC, Nawrot TS, De Boeck K, Nemery B, Dupont LJ. 2015. Proximity to blue spaces and risk of infection with Pseudomonas aeruginosa in cystic fibrosis: a case-control analysis. J Cyst Fibros 14:741–747. doi: 10.1016/j.jcf.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Psoter KJ, De Roos AJ, Wakefield J, Mayer JD, Bryan M, Rosenfeld M. 2016. Association of meteorological and geographical factors and risk of initial Pseudomonas aeruginosa acquisition in young children with cystic fibrosis. Epidemiol Infect 144:1075–1083. doi: 10.1017/S0950268815002411. [DOI] [PubMed] [Google Scholar]
- 46.Armstrong DS, Grimwood K, Carlin JB, Carzino R, Gutierrez JP, Hull J, Olinsky A, Phelan EM, Robertson CF, Phelan PD. 1997. Lower airway inflammation in infants and young children with cystic fibrosis. Am J Respir Crit Care Med 156:1197–1204. doi: 10.1164/ajrccm.156.4.96-11058. [DOI] [PubMed] [Google Scholar]
- 47.Grothues D, Koopmann U, von der Hardt H, Tummler B. 1988. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol 26:1973–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomassen MJ, Demko CA, Doershuk CF, Root JM. 1985. Pseudomonas aeruginosa isolates: comparisons of isolates from campers and from sibling pairs with cystic fibrosis. Pediatr Pulmonol 1:40–45. doi: 10.1002/ppul.1950010110. [DOI] [PubMed] [Google Scholar]
- 49.Wolz C, Kiosz G, Ogle JW, Vasil ML, Schaad U, Botzenhart K, Doring G. 1989. Pseudomonas aeruginosa cross-colonization and persistence in patients with cystic fibrosis. Use of a DNA probe. Epidemiol Infect 102:205–214. doi: 10.1017/S0950268800029873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedersen SS, Jensen T, Pressler T, Hoiby N, Rosendal K. 1986. Does centralized treatment of cystic fibrosis increase the risk of Pseudomonas aeruginosa infection? Acta Paediatr Scand 75:840–845. doi: 10.1111/j.1651-2227.1986.tb10299.x. [DOI] [PubMed] [Google Scholar]
- 51.Pedersen SS, Koch C, Hoiby N, Rosendal K. 1986. An epidemic spread of multiresistant Pseudomonas aeruginosa in a cystic fibrosis centre. J Antimicrob Chemother 17:505–516. doi: 10.1093/jac/17.4.505. [DOI] [PubMed] [Google Scholar]
- 52.Cheng K, Smyth RL, Govan JR, Doherty C, Winstanley C, Denning N, Heaf DP, van Saene H, Hart CA. 1996. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 348:639–642. doi: 10.1016/S0140-6736(96)05169-0. [DOI] [PubMed] [Google Scholar]
- 53.Ledson MJ, Gallagher MJ, Corkill JE, Hart CA, Walshaw MJ. 1998. Cross infection between cystic fibrosis patients colonised with Burkholderia cepacia. Thorax 53:432–436. doi: 10.1136/thx.53.5.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Millar-Jones L, Paull A, Saunders Z, Goodchild MC. 1992. Transmission of Pseudomonas cepacia among cystic fibrosis patients. Lancet 340:491. doi: 10.1016/0140-6736(92)91817-R. [DOI] [PubMed] [Google Scholar]
- 55.Pegues DA, Carson LA, Tablan OC, FitzSimmons SC, Roman SB, Miller JM, Jarvis WR. 1994. Acquisition of Pseudomonas cepacia at summer camps for patients with cystic fibrosis. Summer Camp Study Group. J Pediatr 124:694–702. doi: 10.1016/S0022-3476(05)81357-5. [DOI] [PubMed] [Google Scholar]
- 56.Hu H, Harmer C, Anuj S, Wainwright CE, Manos J, Cheney J, Harbour C, Zablotska I, Turnbull L, Whitchurch CB, Grimwood K, Rose B, FBAL Study Investigators. 2013. Type 3 secretion system effector genotype and secretion phenotype of longitudinally collected Pseudomonas aeruginosa isolates from young children diagnosed with cystic fibrosis following newborn screening. Clin Microbiol Infect 19:266–272. doi: 10.1111/j.1469-0691.2012.03770.x. [DOI] [PubMed] [Google Scholar]
- 57.Kidd TJ, Ramsay KA, Vidmar S, Carlin JB, Bell SC, Wainwright CE, Grimwood K, ACFBAL Study Investigators. 2015. Pseudomonas aeruginosa genotypes acquired by children with cystic fibrosis by age 5-years. J Cyst Fibros 14:361–369. doi: 10.1016/j.jcf.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 58.Stapleton PJ, Izydorczyk C, Wang P, Diaz Caballero J, Yau Y, Beaudoin T, Waters VJ, Guttman D. 2017. Abstract 408: Whole genome evolutionary analysis of new onset Pseudomonas aeruginosa infection in children with cystic fibrosis. Pediatr Pulmonol 52(S47):371. [Google Scholar]
- 59.Fluge G, Ojeniyi B, Hoiby N, Digranes A, Ciofu O, Hunstad E, Haanaes OC, Storrosten OT. 2001. Typing of Pseudomonas aeruginosa strains in Norwegian cystic fibrosis patients. Clin Microbiol Infect 7:238–243. doi: 10.1046/j.1469-0691.2001.00247.x. [DOI] [PubMed] [Google Scholar]
- 60.Schmid J, Ling LJ, Leung JL, Zhang N, Kolbe J, Wesley AW, Mills GD, Brown PJ, Jones DT, Laing RT, Pattemore PK, Taylor DR, Grimwood K. 2008. Pseudomonas aeruginosa transmission is infrequent in New Zealand cystic fibrosis clinics. Eur Respir J 32:1583–1590. doi: 10.1183/09031936.00099508. [DOI] [PubMed] [Google Scholar]
- 61.Van Daele S, Vaneechoutte M, De Boeck K, Knoop C, Malfroot A, Lebecque P, Leclercq-Foucart J, Van Schil L, Desager K, De Baets F. 2006. Survey of Pseudomonas aeruginosa genotypes in colonised cystic fibrosis patients. Eur Respir J 28:740–747. doi: 10.1183/09031936.06.00002706. [DOI] [PubMed] [Google Scholar]
- 62.Fischer S, Klockgether J, Moran Losada P, Chouvarine P, Cramer N, Davenport CF, Dethlefsen S, Dorda M, Goesmann A, Hilker R, Mielke S, Schonfelder T, Suerbaum S, Turk O, Woltemate S, Wiehlmann L, Tummler B. 2016. Intraclonal genome diversity of the major Pseudomonas aeruginosa clones C and PA14. Environ Microbiol Rep 8:227–234. doi: 10.1111/1758-2229.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hilker R, Munder A, Klockgether J, Losada PM, Chouvarine P, Cramer N, Davenport CF, Dethlefsen S, Fischer S, Peng H, Schonfelder T, Turk O, Wiehlmann L, Wolbeling F, Gulbins E, Goesmann A, Tummler B. 2015. Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ Microbiol 17:29–46. doi: 10.1111/1462-2920.12606. [DOI] [PubMed] [Google Scholar]
- 64.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, Olavarietta R, Doud M, Smith RS, Montgomery P, White JR, Godfrey PA, Kodira C, Birren B, Galagan JE, Lory S. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A 105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koonin EV, Wolf YI. 2012. Evolution of microbes and viruses: a paradigm shift in evolutionary biology? Front Cell Infect Microbiol 2:119. doi: 10.3389/fcimb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mosquera-Rendon J, Rada-Bravo AM, Cardenas-Brito S, Corredor M, Restrepo-Pineda E, Benitez-Paez A. 2016. Pangenome-wide and molecular evolution analyses of the Pseudomonas aeruginosa species. BMC Genomics 17:45. doi: 10.1186/s12864-016-2364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cramer N, Wiehlmann L, Ciofu O, Tamm S, Hoiby N, Tummler B. 2012. Molecular epidemiology of chronic Pseudomonas aeruginosa airway infections in cystic fibrosis. PLoS One 7:e50731. doi: 10.1371/journal.pone.0050731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cramer N, Wiehlmann L, Tummler B. 2010. Clonal epidemiology of Pseudomonas aeruginosa in cystic fibrosis. Int J Med Microbiol 300:526–533. doi: 10.1016/j.ijmm.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 69.Pirnay JP, Bilocq F, Pot B, Cornelis P, Zizi M, Van Eldere J, Deschaght P, Vaneechoutte M, Jennes S, Pitt T, De Vos D. 2009. Pseudomonas aeruginosa population structure revisited. PLoS One 4:e7740. doi: 10.1371/journal.pone.0007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Breitenstein S, Walter S, Bosshammer J, Romling U, Tummler B. 1997. Direct sputum analysis of Pseudomonas aeruginosa macrorestriction fragment genotypes in patients with cystic fibrosis. Med Microbiol Immunol 186:93–99. doi: 10.1007/s004300050050. [DOI] [PubMed] [Google Scholar]
- 71.Burkett A, Vandemheen KL, Giesbrecht-Lewis T, Ramotar K, Ferris W, Chan F, Doucette S, Fergusson D, Aaron SD. 2012. Persistency of Pseudomonas aeruginosa in sputum cultures and clinical outcomes in adult patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis 31:1603–1610. doi: 10.1007/s10096-011-1483-8. [DOI] [PubMed] [Google Scholar]
- 72.Fernandez-Olmos A, Garcia-Castillo M, Alba JM, Morosini MI, Lamas A, Romero B, Galan JC, del Campo R, Canton R. 2013. Population structure and antimicrobial susceptibility of both nonpersistent and persistent Pseudomonas aeruginosa isolates recovered from cystic fibrosis patients. J Clin Microbiol 51:2761–2765. doi: 10.1128/JCM.00802-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fothergill JL, Mowat E, Ledson MJ, Walshaw MJ, Winstanley C. 2010. Fluctuations in phenotypes and genotypes within populations of Pseudomonas aeruginosa in the cystic fibrosis lung during pulmonary exacerbations. J Med Microbiol 59:472–481. doi: 10.1099/jmm.0.015875-0. [DOI] [PubMed] [Google Scholar]
- 74.Jelsbak L, Johansen HK, Frost AL, Thogersen R, Thomsen LE, Ciofu O, Yang L, Haagensen JA, Hoiby N, Molin S. 2007. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect Immun 75:2214–2224. doi: 10.1128/IAI.01282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kalferstova L, Vilimovska Dedeckova K, Antuskova M, Melter O, Drevinek P. 2016. How and why to monitor Pseudomonas aeruginosa infections in the long term at a cystic fibrosis centre. J Hosp Infect 92:54–60. doi: 10.1016/j.jhin.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 76.Marvig RL, Dolce D, Sommer LM, Petersen B, Ciofu O, Campana S, Molin S, Taccetti G, Johansen HK. 2015. Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients. BMC Microbiol 15:218. doi: 10.1186/s12866-015-0563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ, Brockhurst MA, Winstanley C. 2011. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med 183:1674–1679. doi: 10.1164/rccm.201009-1430OC. [DOI] [PubMed] [Google Scholar]
- 78.Parkins MD, Glezerson BA, Sibley CD, Sibley KA, Duong J, Purighalla S, Mody CH, Workentine ML, Storey DG, Surette MG, Rabin HR. 2014. Twenty-five-year outbreak of Pseudomonas aeruginosa infecting individuals with cystic fibrosis: identification of the prairie epidemic strain. J Clin Microbiol 52:1127–1135. doi: 10.1128/JCM.03218-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Romling U, Fiedler B, Bosshammer J, Grothues D, Greipel J, von der Hardt H, Tummler B. 1994. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J Infect Dis 170:1616–1621. doi: 10.1093/infdis/170.6.1616. [DOI] [PubMed] [Google Scholar]
- 80.van Mansfeld R, de Vrankrijker A, Brimicombe R, Heijerman H, Teding van Berkhout F, Spitoni C, Grave S, van der Ent C, Wolfs T, Willems R, Bonten M. 2016. The effect of strict segregation on Pseudomonas aeruginosa in cystic fibrosis patients. PLoS One 11:e0157189. doi: 10.1371/journal.pone.0157189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Mansfeld R, Willems R, Brimicombe R, Heijerman H, van Berkhout FT, Wolfs T, van der Ent C, Bonten M. 2009. Pseudomonas aeruginosa genotype prevalence in Dutch cystic fibrosis patients and age dependency of colonization by various P. aeruginosa sequence types. J Clin Microbiol 47:4096–4101. doi: 10.1128/JCM.01462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cramer N, Klockgether J, Wrasman K, Schmidt M, Davenport CF, Tummler B. 2011. Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ Microbiol 13:1690–1704. doi: 10.1111/j.1462-2920.2011.02483.x. [DOI] [PubMed] [Google Scholar]
- 83.Vu-Thien H, Corbineau G, Hormigos K, Fauroux B, Corvol H, Clement A, Vergnaud G, Pourcel C. 2007. Multiple-locus variable-number tandem-repeat analysis for longitudinal survey of sources of Pseudomonas aeruginosa infection in cystic fibrosis patients. J Clin Microbiol 45:3175–3183. doi: 10.1128/JCM.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ashish A, Paterson S, Mowat E, Fothergill JL, Walshaw MJ, Winstanley C. 2013. Extensive diversification is a common feature of Pseudomonas aeruginosa populations during respiratory infections in cystic fibrosis. J Cyst Fibros 12:790–793. doi: 10.1016/j.jcf.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tingpej P, Smith L, Rose B, Zhu H, Conibear T, Al Nassafi K, Manos J, Elkins M, Bye P, Willcox M, Bell S, Wainwright C, Harbour C. 2007. Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J Clin Microbiol 45:1697–1704. doi: 10.1128/JCM.02364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Hoiby N. 2010. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 156:1108–1119. doi: 10.1099/mic.0.033993-0. [DOI] [PubMed] [Google Scholar]
- 87.Foweraker JE, Laughton CR, Brown DF, Bilton D. 2009. Comparison of methods to test antibiotic combinations against heterogeneous populations of multiresistant Pseudomonas aeruginosa from patients with acute infective exacerbations in cystic fibrosis. Antimicrob Agents Chemother 53:4809–4815. doi: 10.1128/AAC.00269-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Foweraker JE, Laughton CR, Brown DF, Bilton D. 2005. Phenotypic variability of Pseudomonas aeruginosa in sputa from patients with acute infective exacerbation of cystic fibrosis and its impact on the validity of antimicrobial susceptibility testing. J Antimicrob Chemother 55:921–927. doi: 10.1093/jac/dki146. [DOI] [PubMed] [Google Scholar]
- 89.Gillham MI, Sundaram S, Laughton CR, Haworth CS, Bilton D, Foweraker JE. 2009. Variable antibiotic susceptibility in populations of Pseudomonas aeruginosa infecting patients with bronchiectasis. J Antimicrob Chemother 63:728–732. doi: 10.1093/jac/dkp007. [DOI] [PubMed] [Google Scholar]
- 90.Winstanley C, O'Brien S, Brockhurst MA. 2016. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol 24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Workentine ML, Sibley CD, Glezerson B, Purighalla S, Norgaard-Gron JC, Parkins MD, Rabin HR, Surette MG. 2013. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One 8:e60225. doi: 10.1371/journal.pone.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clifton IJ, Peckham DG. 2010. Defining routes of airborne transmission of Pseudomonas aeruginosa in people with cystic fibrosis. Expert Rev Respir Med 4:519–529. doi: 10.1586/ers.10.42. [DOI] [PubMed] [Google Scholar]
- 93.Fothergill JL, Walshaw MJ, Winstanley C. 2012. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J 40:227–238. doi: 10.1183/09031936.00204411. [DOI] [PubMed] [Google Scholar]
- 94.Clifton IJ, Fletcher LA, Beggs CB, Denton M, Conway SP, Peckham DG. 2010. An aerobiological model of aerosol survival of different strains of Pseudomonas aeruginosa isolated from people with cystic fibrosis. J Cyst Fibros 9:64–68. doi: 10.1016/j.jcf.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 95.Oliver A, Mulet X, Lopez-Causape C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 96.Kidd TJ, Ramsay KA, Hu H, Marks GB, Wainwright CE, Bye PT, Elkins MR, Robinson PJ, Rose BR, Wilson JW, Grimwood K, Bell SC, ACPinCF Investigator Group. 2013. Shared Pseudomonas aeruginosa genotypes are common in Australian cystic fibrosis centres. Eur Respir J 41:1091–1100. doi: 10.1183/09031936.00060512. [DOI] [PubMed] [Google Scholar]
- 97.Kidd TJ, Soares Magalhaes RJ, Paynter S, Bell SC, ACPinCF Investigator Group. 2015. The social network of cystic fibrosis centre care and shared Pseudomonas aeruginosa strain infection: a cross-sectional analysis. Lancet Respir Med 3:640–650. doi: 10.1016/S2213-2600(15)00228-3. [DOI] [PubMed] [Google Scholar]
- 98.Syrmis MW, Moser RJ, Kidd TJ, Hunt P, Ramsay KA, Bell SC, Wainwright CE, Grimwood K, Nissen MD, Sloots TP, Whiley DM. 2013. High-throughput single-nucleotide polymorphism-based typing of shared Pseudomonas aeruginosa strains in cystic fibrosis patients using the Sequenom iPLEX platform. J Med Microbiol 62:734–740. doi: 10.1099/jmm.0.055905-0. [DOI] [PubMed] [Google Scholar]
- 99.Tai AS, Bell SC, Kidd TJ, Trembizki E, Buckley C, Ramsay KA, David M, Wainwright CE, Grimwood K, Whiley DM. 2015. Genotypic diversity within a single Pseudomonas aeruginosa strain commonly shared by Australian patients with cystic fibrosis. PLoS One 10:e0144022. doi: 10.1371/journal.pone.0144022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tenover FC, Arbeit RD, Goering RV, Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect Control Hosp Epidemiol 18:426–439. doi: 10.2307/30141252. [DOI] [PubMed] [Google Scholar]
- 101.Homma JY, Kim KS, Yamada H, Ito M, Shionoya H. 1970. Serological typing of Pseudomonas aeruginosa and its cross-infection. Jpn J Exp Med 40:347–359. [PubMed] [Google Scholar]
- 102.Speert DP. 2002. Molecular epidemiology of Pseudomonas aeruginosa. Front Biosci 7:e354–e361. doi: 10.2741/A929. [DOI] [PubMed] [Google Scholar]
- 103.Hancock RE, Mutharia LM, Chan L, Darveau RP, Speert DP, Pier GB. 1983. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun 42:170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Clokie MR, Millard AD, Letarov AV, Heaphy S. 2011. Phages in nature. Bacteriophage 1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Postic B, Finland M. 1961. Observations on bacteriophage typing of Pseudomonas aeruginosa. J Clin Invest 40:2064–2075. doi: 10.1172/JCI104432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.International Pseudomonas aeruginosa Typing Study Group. 1994. A multicenter comparison of methods for typing strains of Pseudomonas aeruginosa predominantly from patients with cystic fibrosis. J Infect Dis 169:134–142. doi: 10.1093/infdis/169.1.134. [DOI] [PubMed] [Google Scholar]
- 107.Carle GF, Frank M, Olson MV. 1986. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science 232:65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- 108.Romling U, Grothues D, Bautsch W, Tummler B. 1989. A physical genome map of Pseudomonas aeruginosa PAO. EMBO J 8:4081–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Singh A, Goering RV, Simjee S, Foley SL, Zervos MJ. 2006. Application of molecular techniques to the study of hospital infection. Clin Microbiol Rev 19:512–530. doi: 10.1128/CMR.00025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Scott FW, Pitt TL. 2004. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J Med Microbiol 53:609–615. doi: 10.1099/jmm.0.45620-0. [DOI] [PubMed] [Google Scholar]
- 112.Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol 34:1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Syrmis MW, O'Carroll MR, Sloots TP, Coulter C, Wainwright CE, Bell SC, Nissen MD. 2004. Rapid genotyping of Pseudomonas aeruginosa isolates harboured by adult and paediatric patients with cystic fibrosis using repetitive-element-based PCR assays. J Med Microbiol 53:1089–1096. doi: 10.1099/jmm.0.45611-0. [DOI] [PubMed] [Google Scholar]
- 114.Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Onteniente L, Brisse S, Tassios PT, Vergnaud G. 2003. Evaluation of the polymorphisms associated with tandem repeats for Pseudomonas aeruginosa strain typing. J Clin Microbiol 41:4991–4997. doi: 10.1128/JCM.41.11.4991-4997.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Waine DJ, Honeybourne D, Smith EG, Whitehouse JL, Dowson CG. 2008. Association between hypermutator phenotype, clinical variables, mucoid phenotype, and antimicrobial resistance in Pseudomonas aeruginosa. J Clin Microbiol 46:3491–3493. doi: 10.1128/JCM.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Garcia-Castillo M, Maiz L, Morosini MI, Rodriguez-Banos M, Suarez L, Fernandez-Olmos A, Baquero F, Canton R, del Campo R. 2012. Emergence of a mutL mutation causing multilocus sequence typing–pulsed-field gel electrophoresis discrepancy among Pseudomonas aeruginosa isolates from a cystic fibrosis patient. J Clin Microbiol 50:1777–1778. doi: 10.1128/JCM.05478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 120.Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O'Connor L, Ip CL, Golubchik T, Batty EM, Finney JM, Wyllie DH, Didelot X, Piazza P, Bowden R, Dingle KE, Harding RM, Crook DW, Wilcox MH, Peto TE, Walker AS. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Harris SR, Cartwright EJ, Torok ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kong Z, Zhao P, Liu H, Yu X, Qin Y, Su Z, Wang S, Xu H, Chen J. 2016. Whole-genome sequencing for the investigation of a hospital outbreak of MRSA in China. PLoS One 11:e0149844. doi: 10.1371/journal.pone.0149844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJ, Brinkman FS, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 124.Torok ME, Reuter S, Bryant J, Koser CU, Stinchcombe SV, Nazareth B, Ellington MJ, Bentley SD, Smith GP, Parkhill J, Peacock SJ. 2013. Rapid whole-genome sequencing for investigation of a suspected tuberculosis outbreak. J Clin Microbiol 51:611–614. doi: 10.1128/JCM.02279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marvig RL, Sommer LM, Jelsbak L, Molin S, Johansen HK. 2015. Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. Future Microbiol 10:599–611. doi: 10.2217/fmb.15.3. [DOI] [PubMed] [Google Scholar]
- 126.Diaz Caballero J, Clark ST, Coburn B, Zhang Y, Wang PW, Donaldson SL, Tullis DE, Yau YC, Waters VJ, Hwang DM, Guttman DS. 2015. Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung. mBio 6:e00981-15. doi: 10.1128/mBio.00981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van Mansfeld R, de Been M, Paganelli F, Yang L, Bonten M, Willems R. 2016. Within-host evolution of the Dutch high-prevalent Pseudomonas aeruginosa clone ST406 during chronic colonization of a patient with cystic fibrosis. PLoS One 11:e0158106. doi: 10.1371/journal.pone.0158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Marvig RL, Sommer LM, Molin S, Johansen HK. 2015. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 129.Marvig RL, Johansen HK, Molin S, Jelsbak L. 2013. Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet 9:e1003741. doi: 10.1371/journal.pgen.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR Jr, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, McAdam AJ, Priebe GP, Kishony R. 2011. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet 43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Panagea S, Winstanley C, Parsons YN, Walshaw MJ, Ledson MJ, Hart CA. 2003. PCR-based detection of a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. Mol Diagn 7:195–200. doi: 10.1007/BF03260038. [DOI] [PubMed] [Google Scholar]
- 133.Williams HL, Turnbull L, Thomas SJ, Murphy A, Stinear T, Armstrong DS, Whitchurch CB. 2010. A diagnostic PCR assay for the detection of an Australian epidemic strain of Pseudomonas aeruginosa. Ann Clin Microbiol Antimicrob 9:18. doi: 10.1186/1476-0711-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Parsons YN, Panagea S, Smart CH, Walshaw MJ, Hart CA, Winstanley C. 2002. Use of subtractive hybridization to identify a diagnostic probe for a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. J Clin Microbiol 40:4607–4611. doi: 10.1128/JCM.40.12.4607-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smart CH, Walshaw MJ, Hart CA, Winstanley C. 2006. Use of suppression subtractive hybridization to examine the accessory genome of the Liverpool cystic fibrosis epidemic strain of Pseudomonas aeruginosa. J Med Microbiol 55:677–688. doi: 10.1099/jmm.0.46461-0. [DOI] [PubMed] [Google Scholar]
- 136.Workentine M, Poonja A, Waddell B, Duong J, Storey DG, Gregson D, Somayaji R, Rabin HR, Surette MG, Parkins MD. 2016. Development and validation of a PCR assay to detect the prairie epidemic strain of Pseudomonas aeruginosa from patients with cystic fibrosis. J Clin Microbiol 54:489–491. doi: 10.1128/JCM.02603-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lewis DA, Jones A, Parkhill J, Speert DP, Govan JR, Lipuma JJ, Lory S, Webb AK, Mahenthiralingam E. 2005. Identification of DNA markers for a transmissible Pseudomonas aeruginosa cystic fibrosis strain. Am J Respir Cell Mol Biol 33:56–64. doi: 10.1165/rcmb.2004-0352OC. [DOI] [PubMed] [Google Scholar]
- 138.Smart CH, Scott FW, Wright EA, Walshaw MJ, Hart CA, Pitt TL, Winstanley C. 2006. Development of a diagnostic test for the Midlands 1 cystic fibrosis epidemic strain of Pseudomonas aeruginosa. J Med Microbiol 55:1085–1091. doi: 10.1099/jmm.0.46604-0. [DOI] [PubMed] [Google Scholar]
- 139.Fothergill JL, Upton AL, Pitt TL, Hart CA, Winstanley C. 2008. Diagnostic multiplex PCR assay for the identification of the Liverpool, Midlands 1 and Manchester CF epidemic strains of Pseudomonas aeruginosa. J Cyst Fibros 7:258–261. doi: 10.1016/j.jcf.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 140.Fothergill JL, White J, Foweraker JE, Walshaw MJ, Ledson MJ, Mahenthiralingam E, Winstanley C. 2010. Impact of Pseudomonas aeruginosa genomic instability on the application of typing methods for chronic cystic fibrosis infections. J Clin Microbiol 48:2053–2059. doi: 10.1128/JCM.00019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Logan C, Habington A, Lennon G, Cronin F, O'Sullivan N. 2010. Evaluation of the efficacy of real-time polymerase chain reaction for the routine early detection of Pseudomonas aeruginosa in cystic fibrosis sputum and throat swab specimens. Diagn Microbiol Infect Dis 68:358–365. doi: 10.1016/j.diagmicrobio.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 142.Logan C, Habington A, Lennon G, Grogan J, Byrne M, O'Leary J, O'Sullivan N. 2012. Genetic relatedness of Pseudomonas aeruginosa isolates among a paediatric cystic fibrosis patient cohort in Ireland. J Med Microbiol 61:64–70. doi: 10.1099/jmm.0.035642-0. [DOI] [PubMed] [Google Scholar]
- 143.Kidd TJ, Grimwood K, Ramsay KA, Rainey PB, Bell SC. 2011. Comparison of three molecular techniques for typing Pseudomonas aeruginosa isolates in sputum samples from patients with cystic fibrosis. J Clin Microbiol 49:263–268. doi: 10.1128/JCM.01421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Waters V, Zlosnik JE, Yau YC, Speert DP, Aaron SD, Guttman DS. 2012. Comparison of three typing methods for Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis 31:3341–3350. doi: 10.1007/s10096-012-1701-z. [DOI] [PubMed] [Google Scholar]
- 145.Johnson JK, Arduino SM, Stine OC, Johnson JA, Harris AD. 2007. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J Clin Microbiol 45:3707–3712. doi: 10.1128/JCM.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Speert DP, Campbell ME, Henry DA, Milner R, Taha F, Gravelle A, Davidson AG, Wong LT, Mahenthiralingam E. 2002. Epidemiology of Pseudomonas aeruginosa in cystic fibrosis in British Columbia, Canada. Am J Respir Crit Care Med 166:988–993. doi: 10.1164/rccm.2203011. [DOI] [PubMed] [Google Scholar]
- 147.Johansson E, Welinder-Olsson C, Gilljam M, Pourcel C, Lindblad A. 2015. Genotyping of Pseudomonas aeruginosa reveals high diversity, stability over time and good outcome of eradication. J Cyst Fibros 14:353–360. doi: 10.1016/j.jcf.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 148.van Mansfeld R, Jongerden I, Bootsma M, Buiting A, Bonten M, Willems R. 2010. The population genetics of Pseudomonas aeruginosa isolates from different patient populations exhibits high-level host specificity. PLoS One 5:e13482. doi: 10.1371/journal.pone.0013482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Eusebio N, Pinheiro T, Amorim AA, Gamboa F, Saraiva L, Gusmao L, Amorim A, Araujo R. 2013. SNaPaer: a practical single nucleotide polymorphism multiplex assay for genotyping of Pseudomonas aeruginosa. PLoS One 8:e66083. doi: 10.1371/journal.pone.0066083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Robertson GA, Thiruvenkataswamy V, Shilling H, Price EP, Huygens F, Henskens FA, Giffard PM. 2004. Identification and interrogation of highly informative single nucleotide polymorphism sets defined by bacterial multilocus sequence typing databases. J Med Microbiol 53:35–45. doi: 10.1099/jmm.0.05365-0. [DOI] [PubMed] [Google Scholar]
- 151.Anuj SN, Whiley DM, Kidd TJ, Ramsay KA, Bell SC, Syrmis MW, Grimwood K, Wainwright CE, Nissen MD, Sloots TP. 2011. Rapid single-nucleotide polymorphism-based identification of clonal Pseudomonas aeruginosa isolates from patients with cystic fibrosis by the use of real-time PCR and high-resolution melting curve analysis. Clin Microbiol Infect 17:1403–1408. doi: 10.1111/j.1469-0691.2010.03439.x. [DOI] [PubMed] [Google Scholar]
- 152.Syrmis MW, Kidd TJ, Moser RJ, Ramsay KA, Gibson KM, Anuj S, Bell SC, Wainwright CE, Grimwood K, Nissen M, Sloots TP, Whiley DM. 2014. A comparison of two informative SNP-based strategies for typing Pseudomonas aeruginosa isolates from patients with cystic fibrosis. BMC Infect Dis 14:307. doi: 10.1186/1471-2334-14-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ballarini A, Scalet G, Kos M, Cramer N, Wiehlmann L, Jousson O. 2012. Molecular typing and epidemiological investigation of clinical populations of Pseudomonas aeruginosa using an oligonucleotide-microarray. BMC Microbiol 12:152. doi: 10.1186/1471-2180-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Anthony M, Rose B, Pegler MB, Elkins M, Service H, Thamotharampillai K, Watson J, Robinson M, Bye P, Merlino J, Harbour C. 2002. Genetic analysis of Pseudomonas aeruginosa isolates from the sputa of Australian adult cystic fibrosis patients. J Clin Microbiol 40:2772–2778. doi: 10.1128/JCM.40.8.2772-2778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Armstrong DS, Nixon GM, Carzino R, Bigham A, Carlin JB, Robins-Browne RM, Grimwood K. 2002. Detection of a widespread clone of Pseudomonas aeruginosa in a pediatric cystic fibrosis clinic. Am J Respir Crit Care Med 166:983–987. doi: 10.1164/rccm.200204-269OC. [DOI] [PubMed] [Google Scholar]
- 156.Griffiths AL, Jamsen K, Carlin JB, Grimwood K, Carzino R, Robinson PJ, Massie J, Armstrong DS. 2005. Effects of segregation on an epidemic Pseudomonas aeruginosa strain in a cystic fibrosis clinic. Am J Respir Crit Care Med 171:1020–1025. doi: 10.1164/rccm.200409-1194OC. [DOI] [PubMed] [Google Scholar]
- 157.Smith DJ, Ramsay KA, Yerkovich ST, Reid DW, Wainwright CE, Grimwood K, Bell SC, Kidd TJ. 2016. Pseudomonas aeruginosa antibiotic resistance in Australian cystic fibrosis centres. Respirology 21:329–337. doi: 10.1111/resp.12714. [DOI] [PubMed] [Google Scholar]
- 158.Harmer CJ, Wynn M, Pinto R, Cordwell S, Rose BR, Harbour C, Triccas JA, Manos J. 2015. Homogentisate 1-2-dioxygenase downregulation in the chronic persistence of Pseudomonas aeruginosa Australian epidemic strain-1 in the CF lung. PLoS One 10:e0134229. doi: 10.1371/journal.pone.0134229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hare NJ, Soe CZ, Rose B, Harbour C, Codd R, Manos J, Cordwell SJ. 2012. Proteomics of Pseudomonas aeruginosa Australian epidemic strain 1 (AES-1) cultured under conditions mimicking the cystic fibrosis lung reveals increased iron acquisition via the siderophore pyochelin. J Proteome Res 11:776–795. doi: 10.1021/pr200659h. [DOI] [PubMed] [Google Scholar]
- 160.Hare NJ, Solis N, Harmer C, Marzook NB, Rose B, Harbour C, Crossett B, Manos J, Cordwell SJ. 2012. Proteomic profiling of Pseudomonas aeruginosa AES-1R, PAO1 and PA14 reveals potential virulence determinants associated with a transmissible cystic fibrosis-associated strain. BMC Microbiol 12:16. doi: 10.1186/1471-2180-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Scott NE, Hare NJ, White MY, Manos J, Cordwell SJ. 2013. Secretome of transmissible Pseudomonas aeruginosa AES-1R grown in a cystic fibrosis lung-like environment. J Proteome Res 12:5357–5369. doi: 10.1021/pr4007365. [DOI] [PubMed] [Google Scholar]
- 162.Manos J, Arthur J, Rose B, Tingpej P, Fung C, Curtis M, Webb JS, Hu H, Kjelleberg S, Gorrell MD, Bye P, Harbour C. 2008. Transcriptome analyses and biofilm-forming characteristics of a clonal Pseudomonas aeruginosa from the cystic fibrosis lung. J Med Microbiol 57:1454–1465. doi: 10.1099/jmm.0.2008/005009-0. [DOI] [PubMed] [Google Scholar]
- 163.Harmer CJ, Triccas JA, Hu H, Rose B, Bye P, Elkins M, Manos J. 2012. Pseudomonas aeruginosa strains from the chronically infected cystic fibrosis lung display increased invasiveness of A549 epithelial cells over time. Microb Pathog 53:37–43. doi: 10.1016/j.micpath.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 164.Caldwell CC, Chen Y, Goetzmann HS, Hao Y, Borchers MT, Hassett DJ, Young LR, Mavrodi D, Thomashow L, Lau GW. 2009. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am J Pathol 175:2473–2488. doi: 10.2353/ajpath.2009.090166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Naughton S, Parker D, Seemann T, Thomas T, Turnbull L, Rose B, Bye P, Cordwell S, Whitchurch C, Manos J. 2011. Pseudomonas aeruginosa AES-1 exhibits increased virulence gene expression during chronic infection of cystic fibrosis lung. PLoS One 6:e24526. doi: 10.1371/journal.pone.0024526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Webb JS, Lau M, Kjelleberg S. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol 186:8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Manos J, Arthur J, Rose B, Bell S, Tingpej P, Hu H, Webb J, Kjelleberg S, Gorrell MD, Bye P, Harbour C. 2009. Gene expression characteristics of a cystic fibrosis epidemic strain of Pseudomonas aeruginosa during biofilm and planktonic growth. FEMS Microbiol Lett 292:107–114. doi: 10.1111/j.1574-6968.2008.01472.x. [DOI] [PubMed] [Google Scholar]
- 168.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI0213870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tingpej P, Elkins M, Rose B, Hu H, Moriarty C, Manos J, Barras B, Bye P, Harbour C. 2010. Clinical profile of adult cystic fibrosis patients with frequent epidemic clones of Pseudomonas aeruginosa. Respirology 15:923–929. doi: 10.1111/j.1440-1843.2010.01792.x. [DOI] [PubMed] [Google Scholar]
- 170.O'Carroll MR, Syrmis MW, Wainwright CE, Greer RM, Mitchell P, Coulter C, Sloots TP, Nissen MD, Bell SC. 2004. Clonal strains of Pseudomonas aeruginosa in paediatric and adult cystic fibrosis units. Eur Respir J 24:101–106. doi: 10.1183/09031936.04.00122903. [DOI] [PubMed] [Google Scholar]
- 171.Sherrard LJ, Tai AS, Wee BA, Ramsay KA, Kidd TJ, Ben Zakour NL, Whiley DM, Beatson SA, Bell SC. 2017. Within-host whole genome analysis of an antibiotic resistant Pseudomonas aeruginosa strain sub-type in cystic fibrosis. PLoS One 12:e0172179. doi: 10.1371/journal.pone.0172179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bradbury R, Champion A, Reid DW. 2008. Poor clinical outcomes associated with a multi-drug resistant clonal strain of Pseudomonas aeruginosa in the Tasmanian cystic fibrosis population. Respirology 13:886–892. doi: 10.1111/j.1440-1843.2008.01383.x. [DOI] [PubMed] [Google Scholar]
- 173.Bradbury RS, Champion AC, Reid DW. 2009. Epidemiology of Pseudomonas aeruginosa in a tertiary referral teaching hospital. J Hosp Infect 73:151–156. doi: 10.1016/j.jhin.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 174.Martin K, Baddal B, Mustafa N, Perry C, Underwood A, Constantidou C, Loman N, Kenna DT, Turton JF. 2013. Clusters of genetically similar isolates of Pseudomonas aeruginosa from multiple hospitals in the UK. J Med Microbiol 62:988–1000. doi: 10.1099/jmm.0.054841-0. [DOI] [PubMed] [Google Scholar]
- 175.Kresse AU, Dinesh SD, Larbig K, Romling U. 2003. Impact of large chromosomal inversions on the adaptation and evolution of Pseudomonas aeruginosa chronically colonizing cystic fibrosis lungs. Mol Microbiol 47:145–158. doi: 10.1046/j.1365-2958.2003.03261.x. [DOI] [PubMed] [Google Scholar]
- 176.Larbig KD, Christmann A, Johann A, Klockgether J, Hartsch T, Merkl R, Wiehlmann L, Fritz HJ, Tummler B. 2002. Gene islands integrated into tRNA(Gly) genes confer genome diversity on a Pseudomonas aeruginosa clone. J Bacteriol 184:6665–6680. doi: 10.1128/JB.184.23.6665-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schmidt KD, Tummler B, Romling U. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J Bacteriol 178:85–93. doi: 10.1128/jb.178.1.85-93.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gopalakaje S, O'Brien CJ, Robb A, Spencer DA. 2013. Clinical profile of Pseudomonas aeruginosa clone C infection in CF children. J Cyst Fibros 12:S75. [Google Scholar]
- 179.Norman A, Ciofu O, Amador CI, Hoiby N, Jelsbak L. 2016. Genome sequence of Pseudomonas aeruginosa strain DK1-NH57388A, a stable mucoid cystic fibrosis isolate. Genome Announc 4:e00008-16. doi: 10.1128/genomeA.00008-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bjarnsholt T, Jensen PO, Jakobsen TH, Phipps R, Nielsen AK, Rybtke MT, Tolker-Nielsen T, Givskov M, Hoiby N, Ciofu O, Scandinavian Cystic Fibrosis Study Consortium. 2010. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One 5:e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Marvig RL, Damkiaer S, Khademi SM, Markussen TM, Molin S, Jelsbak L. 2014. Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. mBio 5:e00966-14. doi: 10.1128/mBio.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rau MH, Marvig RL, Ehrlich GD, Molin S, Jelsbak L. 2012. Deletion and acquisition of genomic content during early stage adaptation of Pseudomonas aeruginosa to a human host environment. Environ Microbiol 14:2200–2211. doi: 10.1111/j.1462-2920.2012.02795.x. [DOI] [PubMed] [Google Scholar]
- 183.Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Hoiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A 108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dettman JR, Rodrigue N, Aaron SD, Kassen R. 2013. Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 110:21065–21070. doi: 10.1073/pnas.1307862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lopez-Causape C, Rojo-Molinero E, Mulet X, Cabot G, Moya B, Figuerola J, Togores B, Perez JL, Oliver A. 2013. Clonal dissemination, emergence of mutator lineages and antibiotic resistance evolution in Pseudomonas aeruginosa cystic fibrosis chronic lung infection. PLoS One 8:e71001. doi: 10.1371/journal.pone.0071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Aaron SD, Vandemheen KL, Ramotar K, Giesbrecht-Lewis T, Tullis E, Freitag A, Paterson N, Jackson M, Lougheed MD, Dowson C, Kumar V, Ferris W, Chan F, Doucette S, Fergusson D. 2010. Infection with transmissible strains of Pseudomonas aeruginosa and clinical outcomes in adults with cystic fibrosis. JAMA 304:2145–2153. doi: 10.1001/jama.2010.1665. [DOI] [PubMed] [Google Scholar]
- 187.Panagea S, Winstanley C, Walshaw MJ, Ledson MJ, Hart CA. 2005. Environmental contamination with an epidemic strain of Pseudomonas aeruginosa in a Liverpool cystic fibrosis centre, and study of its survival on dry surfaces. J Hosp Infect 59:102–107. doi: 10.1016/j.jhin.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 188.McCallum SJ, Gallagher MJ, Corkill JE, Hart CA, Ledson MJ, Walshaw MJ. 2002. Spread of an epidemic Pseudomonas aeruginosa strain from a patient with cystic fibrosis (CF) to non-CF relatives. Thorax 57:559–560. doi: 10.1136/thorax.57.6.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mohan K, Fothergill JL, Storrar J, Ledson MJ, Winstanley C, Walshaw MJ. 2008. Transmission of Pseudomonas aeruginosa epidemic strain from a patient with cystic fibrosis to a pet cat. Thorax 63:839–840. doi: 10.1136/thx.2007.092486. [DOI] [PubMed] [Google Scholar]
- 190.Kukavica-Ibrulj I, Bragonzi A, Paroni M, Winstanley C, Sanschagrin F, O'Toole GA, Levesque RC. 2008. In vivo growth of Pseudomonas aeruginosa strains PAO1 and PA14 and the hypervirulent strain LESB58 in a rat model of chronic lung infection. J Bacteriol 190:2804–2813. doi: 10.1128/JB.01572-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Carter ME, Fothergill JL, Walshaw MJ, Rajakumar K, Kadioglu A, Winstanley C. 2010. A subtype of a Pseudomonas aeruginosa cystic fibrosis epidemic strain exhibits enhanced virulence in a murine model of acute respiratory infection. J Infect Dis 202:935–942. doi: 10.1086/655781. [DOI] [PubMed] [Google Scholar]
- 192.Fothergill JL, Panagea S, Hart CA, Walshaw MJ, Pitt TL, Winstanley C. 2007. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol 7:45. doi: 10.1186/1471-2180-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ashish A, Shaw M, Winstanley C, Ledson MJ, Walshaw MJ. 2012. Increasing resistance of the Liverpool epidemic strain (LES) of Pseudomonas aeruginosa (Psa) to antibiotics in cystic fibrosis (CF)—a cause for concern? J Cyst Fibros 11:173–179. doi: 10.1016/j.jcf.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 194.Beaudoin T, Aaron SD, Giesbrecht-Lewis T, Vandemheen K, Mah TF. 2010. Characterization of clonal strains of Pseudomonas aeruginosa isolated from cystic fibrosis patients in Ontario, Canada. Can J Microbiol 56:548–557. doi: 10.1139/W10-043. [DOI] [PubMed] [Google Scholar]
- 195.Duong J, Booth SC, McCartney NK, Rabin HR, Parkins MD, Storey DG. 2015. Phenotypic and genotypic comparison of epidemic and non-epidemic strains of Pseudomonas aeruginosa from individuals with cystic fibrosis. PLoS One 10:e0143466. doi: 10.1371/journal.pone.0143466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Salunkhe P, Smart CH, Morgan JA, Panagea S, Walshaw MJ, Hart CA, Geffers R, Tummler B, Winstanley C. 2005. A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J Bacteriol 187:4908–4920. doi: 10.1128/JB.187.14.4908-4920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Waters V, Ratjen F. 2006. Multidrug-resistant organisms in cystic fibrosis: management and infection-control issues. Expert Rev Anti Infect Ther 4:807–819. doi: 10.1586/14787210.4.5.807. [DOI] [PubMed] [Google Scholar]
- 198.Jani M, Mathee K, Azad RK. 2016. Identification of novel genomic islands in Liverpool epidemic strain of Pseudomonas aeruginosa using segmentation and clustering. Front Microbiol 7:1210. doi: 10.3389/fmicb.2016.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Winstanley C, Langille MG, Fothergill JL, Kukavica-Ibrulj I, Paradis-Bleau C, Sanschagrin F, Thomson NR, Winsor GL, Quail MA, Lennard N, Bignell A, Clarke L, Seeger K, Saunders D, Harris D, Parkhill J, Hancock RE, Brinkman FS, Levesque RC. 2009. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res 19:12–23. doi: 10.1101/gr.086082.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jeukens J, Boyle B, Kukavica-Ibrulj I, Ouellet MM, Aaron SD, Charette SJ, Fothergill JL, Tucker NP, Winstanley C, Levesque RC. 2014. Comparative genomics of isolates of a Pseudomonas aeruginosa epidemic strain associated with chronic lung infections of cystic fibrosis patients. PLoS One 9:e87611. doi: 10.1371/journal.pone.0087611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fothergill JL, Mowat E, Walshaw MJ, Ledson MJ, James CE, Winstanley C. 2011. Effect of antibiotic treatment on bacteriophage production by a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:426–428. doi: 10.1128/AAC.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Blahova J, Hupkova M, Krcmery V Sr. 1994. Phage F-116 transduction of antibiotic resistance from a clinical isolate of Pseudomonas aeruginosa. J Chemother 6:184–188. doi: 10.1080/1120009X.1994.11741150. [DOI] [PubMed] [Google Scholar]
- 203.Goerke C, Matias y Papenberg S, Dasbach S, Dietz K, Ziebach R, Kahl BC, Wolz C. 2004. Increased frequency of genomic alterations in Staphylococcus aureus during chronic infection is in part due to phage mobilization. J Infect Dis 189:724–734. doi: 10.1086/381502. [DOI] [PubMed] [Google Scholar]
- 204.Whiley RA, Sheikh NP, Mushtaq N, Hagi-Pavli E, Personne Y, Javaid D, Waite RD. 2014. Differential potentiation of the virulence of the Pseudomonas aeruginosa cystic fibrosis Liverpool epidemic strain by oral commensal streptococci. J Infect Dis 209:769–780. doi: 10.1093/infdis/jit568. [DOI] [PubMed] [Google Scholar]
- 205.Al-Aloul M, Crawley J, Winstanley C, Hart CA, Ledson MJ, Walshaw MJ. 2004. Increased morbidity associated with chronic infection by an epidemic Pseudomonas aeruginosa strain in CF patients. Thorax 59:334–336. doi: 10.1136/thx.2003.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chambers D, Scott F, Bangur R, Davies R, Lim A, Walters S, Smith G, Pitt T, Stableforth D, Honeybourne D. 2005. Factors associated with infection by Pseudomonas aeruginosa in adult cystic fibrosis. Eur Respir J 26:651–656. doi: 10.1183/09031936.05.00126704. [DOI] [PubMed] [Google Scholar]
- 207.Ashish A, Shaw M, McShane J, Ledson MJ, Walshaw MJ. 2012. Health-related quality of life in cystic fibrosis patients infected with transmissible Pseudomonas aeruginosa strains: cohort study. JRSM Short Rep 3:12. doi: 10.1258/shorts.2011.011119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Srour N, Chaparro C, Vandemheen K, Singer LG, Keshavjee S, Aaron SD. 2015. Effect of infection with transmissible strains of Pseudomonas aeruginosa on lung transplantation outcomes in patients with cystic fibrosis. J Heart Lung Transplant 34:588–593. doi: 10.1016/j.healun.2014.09.040. [DOI] [PubMed] [Google Scholar]
- 209.France M, Dodd ME, Govan JR, Doherty CJ, Webb AK, Jones AM. 2006. First isolation of Pseudomonas aeruginosa: failure of eradication treatment associated with a clonal strain. Thorax 91:ii34. [Google Scholar]
- 210.Al-Aloul M, Miller H, Stockton P, Ledson MJ, Walshaw MJ. 2005. Acute renal failure in CF patients chronically infected by the Liverpool epidemic Pseudomonas aeruginosa strain (LES). J Cyst Fibros 4:197–201. doi: 10.1016/j.jcf.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 211.Mohan K, Lakshman V, Fothergill JL, Ledson MJ, Winstanley C, Walshaw MJ. 2010. Empyema due to a highly transmissible Pseudomonas aeruginosa strain in an adult cystic fibrosis patient. J Med Microbiol 59:614–616. doi: 10.1099/jmm.0.014696-0. [DOI] [PubMed] [Google Scholar]
- 212.Nazareth D, Frost F, Mohan K, Greenwood J, Cullen D, Walshaw M. 2017. Abstract 455: Is transmissible Pseudomonas aeruginosa a risk factor for pneumothorax in CF? Pediatr Pulmonol 52(S47):389. [Google Scholar]
- 213.Gilchrist FJ, Doherty CJ, Govan JR, Webb AK, Jones AM. 2011. Pseudomonas aeruginosa bacteraemia in an adult with cystic fibrosis and acute appendicitis. J Cyst Fibros 10:477–478. doi: 10.1016/j.jcf.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 214.Jones AM, Govan JR, Doherty CJ, Dodd ME, Isalska BJ, Stanbridge TN, Webb AK. 2001. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet 358:557–558. doi: 10.1016/S0140-6736(01)05714-2. [DOI] [PubMed] [Google Scholar]
- 215.Jones AM, Govan JR, Doherty CJ, Dodd ME, Isalska BJ, Stanbridge TN, Webb AK. 2003. Identification of airborne dissemination of epidemic multiresistant strains of Pseudomonas aeruginosa at a CF centre during a cross infection outbreak. Thorax 58:525–527. doi: 10.1136/thorax.58.6.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jones AM, Dodd ME, Doherty CJ, Govan JR, Webb AK. 2002. Increased treatment requirements of patients with cystic fibrosis who harbour a highly transmissible strain of Pseudomonas aeruginosa. Thorax 57:924–925. doi: 10.1136/thorax.57.11.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jones AM, Dodd ME, Morris J, Doherty C, Govan JR, Webb AK. 2010. Clinical outcome for cystic fibrosis patients infected with transmissible Pseudomonas aeruginosa: an 8-year prospective study. Chest 137:1405–1409. doi: 10.1378/chest.09-2406. [DOI] [PubMed] [Google Scholar]
- 218.Waine DJ, Honeybourne D, Smith EG, Whitehouse JL, Dowson CG. 2009. Cross-sectional and longitudinal multilocus sequence typing of Pseudomonas aeruginosa in cystic fibrosis sputum samples. J Clin Microbiol 47:3444–3448. doi: 10.1128/JCM.00459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Loman N, Manzoor S, Quick J, Fothergill JL, Gossain S, Kenna D, Turton J, Walshaw M, Whitehouse JL, Pallen M, Winstanley C, Hawkey P, Nash EF. 2014. Use of whole-genome sequencing to identify transmission of Pseudomonas aeruginosa between cystic fibrosis patients. J Cyst Fibros 13:148.24440167 [Google Scholar]
- 220.Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, Larbig KD, Heeb S, Schock U, Pohl TM, Wiehlmann L, Tummler B. 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol 192:1113–1121. doi: 10.1128/JB.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol 7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wiehlmann L, Wagner G, Cramer N, Siebert B, Gudowius P, Morales G, Kohler T, van Delden C, Weinel C, Slickers P, Tummler B. 2007. Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 104:8101–8106. doi: 10.1073/pnas.0609213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Somayaji R, Waddell B, Workentine ML, Surette MG, Brager NP, Rabin HR, Parkins MD. 2015. Infection control knowledge, beliefs and behaviours amongst cystic fibrosis patients with epidemic Pseudomonas aeruginosa. BMC Pulm Med 15:138. doi: 10.1186/s12890-015-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Acosta Amador PN, Heirali A, Somayaji R, Waddell B, Surette M, Rabin H, Parkins M. 2016. Abstract 361: Epidemic Pseudomonas aeruginosa: a driving force in the cystic fibrosis (CF) microbiome. Pediatr Pulmonol 51(S45):330. [Google Scholar]
- 225.Pritchard J, Thakrar MV, Somayaji R, Surette MG, Rabin HR, Helmersen D, Lien D, Purighalla S, Waddell B, Parkins MD. 2016. Epidemic Pseudomonas aeruginosa infection in patients with cystic fibrosis is not a risk factor for poor clinical outcomes following lung transplantation. J Cyst Fibros 15:392–399. doi: 10.1016/j.jcf.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 226.Lopez-Causape C, de Dios-Caballero J, Cobo M, Escribano A, Asensio O, Oliver A, Del Campo R, Canton R, Sole A, Cortell I, Asensio O, Garcia G, Martinez MT, Cols M, Salcedo A, Vazquez C, Baranda F, Giron R, Quintana E, Delgado I, de Miguel MA, Garcia M, Oliva C, Prados MC, Barrio MI, Pastor MD, Olveira C, de Gracia J, Alvarez A, Escribano A, Castillo S, Figuerola J, Togores B, Oliver A, Lopez C, de Dios Caballero J, Tato M, Maiz L, Suarez L, Canton R. 2017. Antibiotic resistance and population structure of cystic fibrosis Pseudomonas aeruginosa isolates from a Spanish multi-centre study. Int J Antimicrob Agents 50:334–341. doi: 10.1016/j.ijantimicag.2017.03.034. [DOI] [PubMed] [Google Scholar]
- 227.Lopez-Causape C, Sommer LM, Cabot G, Rubio R, Ocampo-Sosa AA, Johansen HK, Figuerola J, Canton R, Kidd TJ, Molin S, Oliver A. 2017. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci Rep 7:5555. doi: 10.1038/s41598-017-05621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Altman E, Wang Z, Aaron SD, Liu X, Vandemheen KL, Ferris W, Giesbrecht T, Li J. 2009. Epidemiological investigation and glycotyping of clinical Pseudomonas aeruginosa isolates from patients with cystic fibrosis by mass spectrometry: association with multiple drug resistance. J Microbiol Methods 76:204–208. doi: 10.1016/j.mimet.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 229.de Vrankrijker AM, Brimicombe RW, Wolfs TF, Heijerman HG, van Mansfeld R, van Berkhout FT, Willems RJ, Bonten MJ, van der Ent CK. 2011. Clinical impact of a highly prevalent Pseudomonas aeruginosa clone in Dutch cystic fibrosis patients. Clin Microbiol Infect 17:382–385. doi: 10.1111/j.1469-0691.2010.03295.x. [DOI] [PubMed] [Google Scholar]
- 230.Denton M, Kerr K, Mooney L, Keer V, Rajgopal A, Brownlee K, Arundel P, Conway S. 2002. Transmission of colistin-resistant Pseudomonas aeruginosa between patients attending a pediatric cystic fibrosis center. Pediatr Pulmonol 34:257–261. doi: 10.1002/ppul.10166. [DOI] [PubMed] [Google Scholar]
- 231.Johansen HK, Moskowitz SM, Ciofu O, Pressler T, Hoiby N. 2008. Spread of colistin resistant non-mucoid Pseudomonas aeruginosa among chronically infected Danish cystic fibrosis patients. J Cyst Fibros 7:391–397. doi: 10.1016/j.jcf.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 232.Gonzalez-Villa M, Ribas-Aparicio RM, Coria-Jimenez R, Donis-Rocandio JE, Aparicio-Ozores G. 2012. Detection of extended spectrum beta-lactamase OXA-141 in Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis. Enferm Infecc Microbiol Clin 30:535–541. (In Spanish.) doi: 10.1016/j.eimc.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 233.Vali P, Shahcheraghi F, Seyfipour M, Zamani MA, Allahyar MR, Feizabadi MM. 2014. Phenotypic and genetic characterization of carbapenemase and ESBLs producing Gram-negative bacteria (GNB) isolated from patients with cystic fibrosis (CF) in Tehran hospitals. J Clin Diagn Res 8:26–30. doi: 10.7860/JCDR/2014/6877.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Li Y, Zhang X, Wang C, Hu Y, Niu X, Pei D, He Z, Bi Y. 2015. Characterization by phenotypic and genotypic methods of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolated from patients with cystic fibrosis. Mol Med Rep 11:494–498. doi: 10.3892/mmr.2014.2685. [DOI] [PubMed] [Google Scholar]
- 235.Pollini S, Fiscarelli E, Mugnaioli C, Di Pilato V, Ricciotti G, Neri AS, Rossolini GM. 2011. Pseudomonas aeruginosa infection in cystic fibrosis caused by an epidemic metallo-beta-lactamase-producing clone with a heterogeneous carbapenem resistance phenotype. Clin Microbiol Infect 17:1272–1275. doi: 10.1111/j.1469-0691.2011.03466.x. [DOI] [PubMed] [Google Scholar]
- 236.Tarhani M, Hakemi-Vala M, Hashemi A, Nowroozi J, Khanbababee G. 2016. Detection of metallo-β-lactamases and Klebsiella pneumoniae carbapenemases in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Arch Pediatr Infect Dis 4:e35905. [Google Scholar]
- 237.Cardoso O, Alves AF, Leitao R. 2008. Metallo-beta-lactamase VIM-2 in Pseudomonas aeruginosa isolates from a cystic fibrosis patient. Int J Antimicrob Agents 31:375–379. doi: 10.1016/j.ijantimicag.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 238.Doring G, Ulrich M, Muller W, Bitzer J, Schmidt-Koenig L, Münst L, Grupp H, Wolz C, Stern M, Botzenhart K. 1991. Generation of Pseudomonas aeruginosa aerosols during handwashing from contaminated sink drains, transmission to hands of hospital personnel, and its prevention by use of a new heating device. Zentrabl Hyg Umweltmed 191:494–505. [PubMed] [Google Scholar]
- 239.Humphreys H, Peckham D, Patel P, Knox A. 1994. Airborne dissemination of Burkholderia (Pseudomonas) cepacia from adult patients with cystic fibrosis. Thorax 49:1157–1159. doi: 10.1136/thx.49.11.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Speert D, Campbell ME. 1987. Hospital epidemiology of Pseudomonas aeruginosa from patients with cystic fibrosis. J Hosp Infect 9:11–21. doi: 10.1016/0195-6701(87)90089-2. [DOI] [PubMed] [Google Scholar]
- 241.Wainwright C, France MW, O'Rourke P, Anuj S, Kidd TJ, Nissen MD, Sloots TP, Coulter C, Ristovski Z, Hargreaves M, Rose BR, Harbour C, Bell SC, Fennelly KP. 2009. Cough-generated aerosols of Pseudomonas aeruginosa and other bacteria from cystic fibrosis patients. Thorax 64:926–931. doi: 10.1136/thx.2008.112466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zimakoff J, Hoiby N, Rosendal K, Guilbert JP. 1983. Epidemiology of Pseudomonas aeruginosa infection and the role of contamination of the environment in a cystic fibrosis clinic. J Hosp Infect 4:31–40. doi: 10.1016/0195-6701(83)90062-2. [DOI] [PubMed] [Google Scholar]
- 243.Cox C. 1995. Physical aspects of bioaerosol particles, p 15–25. In Cox CS, Wathes CS (ed), Bioaerosols handbook. Lewis Publishers, Boca Raton, FL. [Google Scholar]
- 244.Tang J, Li Y, Eames I, Chan PKS, Ridgway GL. 2006. Factors involved in the aerosol transmission of infection and control of ventilation in healthcare premises. J Hosp Infect 64:100–114. doi: 10.1016/j.jhin.2006.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wells W. 1934. On air-borne infection. II. Droplets and droplet nuclei. Am J Hyg 20:611–618. [DOI] [PubMed] [Google Scholar]
- 246.Festini F, Taccetti G, Galici V, Neri S, Bisogni S, Ciofi D, Braggion C. 2010. A 1-m distance is not safe for children with cystic fibrosis at risk for cross-infection with Pseudomonas aeruginosa. Am J Infect Control 38:244–245. doi: 10.1016/j.ajic.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 247.Knibbs LD, Johnson GR, Kidd TJ, Cheney J, Grimwood K, Kattenbelt JA, O'Rourke PK, Ramsay KA, Sly PD, Wainwright CE, Wood ME, Morawska L, Bell SC. 2014. Viability of Pseudomonas aeruginosa in cough aerosols generated by persons with cystic fibrosis. Thorax 69:740–745. doi: 10.1136/thoraxjnl-2014-205213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ferroni A, Werkhauser-Bertrand A, Le Bourgeois M, Beauvais R, Vrielynck S, Durand C, Lenoir G, Berche P, Sermet-Gaudelus I. 2008. Bacterial contamination in the environment of hospitalised children with cystic fibrosis. J Cyst Fibros 7:477–482. doi: 10.1016/j.jcf.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 249.Clifton I, Fletcher LA, Beggs CB, Denton M, Peckham DG. 2008. A laminar flow model of aerosol survival of epidemic and non-epidemic strains of Pseudomonas aeruginosa isolated from people with cystic fibrosis. BMC Microbiol 8:105. doi: 10.1186/1471-2180-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Schaal K. 1991. Medical and microbiological problems arising from airborne infection in hospitals. J Hosp Infect 18(Suppl A):451–459. doi: 10.1016/0195-6701(91)90056-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Roberts K, Hathway A, Fletcher LA, Beggs CB, Elliott MW, Sleigh PA. 2006. Bioaerosol production on a respiratory ward. Indoor Built Environ 15:35–40. doi: 10.1177/1420326X06062562. [DOI] [Google Scholar]
- 252.Leggett HC, Cornwallis CK, West SA. 2012. Mechanisms of pathogenesis, infective dose and virulence in human parasites. PLoS Pathog 8:e1002512. doi: 10.1371/journal.ppat.1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Blau H, Mussaffi-Georgy H, Fink G, Kaye C, Szeinberg A, Spitzer SA, Yahav J. 2002. Effects of an intensive 4-week summer camp on cystic fibrosis: pulmonary function, exercise tolerance, and nutrition. Chest 121:1117–1122. doi: 10.1378/chest.121.4.1117. [DOI] [PubMed] [Google Scholar]
- 254.Goldbart AD, Cohen AD, Weitzman D, Tal A. 2007. Effects of rehabilitation winter camps at the Dead Sea on European cystic fibrosis patients. Isr Med Assoc J 9:806–809. [PubMed] [Google Scholar]
- 255.Kaplan TA, McKey RM Jr, Toraya N, Moccia G. 1992. Impact of CF summer camp. Clin Pediatr (Phila) 31:161–167. doi: 10.1177/000992289203100307. [DOI] [PubMed] [Google Scholar]
- 256.Centers for Disease Control and Prevention. 1993. Pseudomonas cepacia at summer camps for persons with cystic fibrosis. MMWR Morb Mortal Wkly Rep 42:456–459. [PubMed] [Google Scholar]
- 257.Brimicombe RW, Dijkshoorn L, van der Reijden TJ, Kardoes I, Pitt TL, van den Broek PJ, Heijerman HG. 2008. Transmission of Pseudomonas aeruginosa in children with cystic fibrosis attending summer camps in The Netherlands. J Cyst Fibros 7:30–36. doi: 10.1016/j.jcf.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 258.Greenberg D, Yagupsky P, Peled N, Goldbart A, Porat N, Tal A. 2004. Lack of evidence of transmission of Pseudomonas aeruginosa among cystic fibrosis patients attending health camps at the Dead Sea, Israel. Isr Med Assoc J 6:531–534. [PubMed] [Google Scholar]
- 259.Ridderberg W, Andersen C, Vaeth M, Bregnballe V, Norskov-Lauritsen N, Schiotz PO. 2016. Lack of evidence of increased risk of bacterial transmission during cystic fibrosis educational programmes. J Cyst Fibros 15:109–115. doi: 10.1016/j.jcf.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 260.Smith DL, Smith EG, Gumery LB, Stableforth DE, Dalla Costa LM, Pitt TL. 1993. Epidemiology of Pseudomonas aeruginosa infection in cystic fibrosis and the use of strain genotyping. J Infect 26:325–331. doi: 10.1016/0163-4453(93)95709-R. [DOI] [PubMed] [Google Scholar]
- 261.Hoogkamp-Korstanje JA, Meis JF, Kissing J, van der Laag J, Melchers WJ. 1995. Risk of cross-colonization and infection by Pseudomonas aeruginosa in a holiday camp for cystic fibrosis patients. J Clin Microbiol 33:572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Hoogkamp-Korstanje JA, van der Laag J. 1980. Incidence and risk of cross colonization in cystic fibrosis holiday camps. Antonie Van Leeuwenhoek 46:100–101. doi: 10.1007/BF00422237. [DOI] [PubMed] [Google Scholar]
- 263.Hunfeld KP, Schmidt C, Krackhardt B, Posselt HG, Bargon J, Yahaf Y, Schafer V, Brade V, Wichelhaus TA. 2000. Risk of Pseudomonas aeruginosa cross-colonisation in patients with cystic fibrosis within a holiday camp—a molecular-epidemiological study. Wien Klin Wochenschr 112:329–333. [PubMed] [Google Scholar]
- 264.Ojeniyi B, Frederiksen B, Hoiby N. 2000. Pseudomonas aeruginosa cross-infection among patients with cystic fibrosis during a winter camp. Pediatr Pulmonol 29:177–181. doi:. [DOI] [PubMed] [Google Scholar]
- 265.Tummler B, Koopmann U, Grothues D, Weissbrodt H, Steinkamp G, von der Hardt H. 1991. Nosocomial acquisition of Pseudomonas aeruginosa by cystic fibrosis patients. J Clin Microbiol 29:1265–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Van Daele SG, Franckx H, Verhelst R, Schelstraete P, Haerynck F, Van Simaey L, Claeys G, Vaneechoutte M, de Baets F. 2005. Epidemiology of Pseudomonas aeruginosa in a cystic fibrosis rehabilitation centre. Eur Respir J 25:474–481. doi: 10.1183/09031936.05.00050304. [DOI] [PubMed] [Google Scholar]
- 267.Cystic Fibrosis Trust Clinical Standards and Accreditation Group. 2001. Standards for the clinical care of children and adults with cystic fibrosis in the UK. Cystic Fibrosis Trust, London, United Kingdom: https://www.cysticfibrosis.org.uk/~/media/documents/the-work-we-do/care/consensus-docs-with-new-address/cystic-fibrosis-trust-standards-of-care.ashx?la=en Accessed 15 December 2017. [Google Scholar]
- 268.Kerem E, Conway S, Elborn S, Heijerman H. 2005. Standards of care for patients with cystic fibrosis: a European consensus. J Cyst Fibros 4:7–26. doi: 10.1016/j.jcf.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 269.Saiman L, Siegel J, Cystic Fibrosis Foundation. 2003. Infection control recommendations for patients with cystic fibrosis: microbiology, important pathogens, and infection control practices to prevent patient-to-patient transmission. Infect Control Hosp Epidemiol 24:S6–S52. doi: 10.1086/503485. [DOI] [PubMed] [Google Scholar]
- 270.Saiman L, Siegel JD, LiPuma JJ, Brown RF, Bryson EA, Chambers MJ, Downer VS, Fliege J, Hazle LA, Jain M, Marshall BC, O'Malley C, Pattee SR, Potter-Bynoe G, Reid S, Robinson KA, Sabadosa KA, Schmidt HJ, Tullis E, Webber J, Weber DJ, Cystic Fibrosis Foundation, Society for Healthcare Epidemiology of America. 2014. Infection prevention and control guideline for cystic fibrosis: 2013 update. Infect Control Hosp Epidemiol 35(Suppl 1):S1–S67. doi: 10.1086/676882. [DOI] [PubMed] [Google Scholar]
- 271.Jones AM, Dodd ME, Govan JR, Doherty CJ, Smith CM, Isalska BJ, Webb AK. 2005. Prospective surveillance for Pseudomonas aeruginosa cross-infection at a cystic fibrosis center. Am J Respir Crit Care Med 171:257–260. doi: 10.1164/rccm.200404-513OC. [DOI] [PubMed] [Google Scholar]
- 272.Elborn J, Hodson M, Bertram C. 2009. Implementation of European standards of care for cystic fibrosis—control and treatment of infection. J Cyst Fibros 8:211–217. doi: 10.1016/j.jcf.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 273.Frederiksen B, Koch C, Hoiby N. 1999. Changing epidemiology of Pseudomonas aeruginosa infection in Danish cystic fibrosis patients (1974-1995). Pediatr Pulmonol 28:159–166. doi:. [DOI] [PubMed] [Google Scholar]
- 274.Griffiths AL, Wurzel DF, Robinson PJ, Carzino R, Massie J. 2012. Australian epidemic strain Pseudomonas (AES-1) declines further in a cohort segregated cystic fibrosis clinic. J Cyst Fibros 11:49–52. doi: 10.1016/j.jcf.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 275.Ashish A, Shaw M, Winstanley C, Humphreys L, Walshaw MJ. 2013. Halting the spread of epidemic Pseudomonas aeruginosa in an adult cystic fibrosis centre: a prospective cohort study. JRSM Short Rep 4:1. doi: 10.1258/shorts.2012.012018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Morris D, Fallon L, Heaf L, Burrows EF, Wallace H, McNamara PS, Winstanley C, Southern KW. 2009. Abstract 324: A reducing prevalence of the Liverpool epidemic strain of Pseudomonas aeruginosa in children attending the index paediatric clinic. Pediatr Pulmonol 44(S32):325.19330772 [Google Scholar]
- 277.Wiehlmann L, Cramer N, Ulrich J, Hedtfeld S, Weissbrodt H, Tummler B. 2012. Effective prevention of Pseudomonas aeruginosa cross-infection at a cystic fibrosis centre—results of a 10-year prospective study. Int J Med Microbiol 302:69–77. doi: 10.1016/j.ijmm.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 278.Driessche KV, Hens N, Tilley P, Quon BS, Chilvers MA, de Groot R, Cotton MF, Marais BJ, Speert DP, Zlosnik JE. 2015. Surgical masks reduce airborne spread of Pseudomonas aeruginosa in colonized patients with cystic fibrosis. Am J Respir Crit Care Med 192:897–899. doi: 10.1164/rccm.201503-0481LE. [DOI] [PubMed] [Google Scholar]
- 279.Wood ME, Stockwell RE, Johnson GR, Ramsay KA, Sherrard LJ, Jabbour N, Ballard E, O'Rourke P, Kidd TJ, Wainwright CE, Knibbs LD, Sly PD, Morawska L, Bell SC. 2017. Face masks and cough etiquette reduce the cough aerosol concentration of Pseudomonas aeruginosa in people with cystic fibrosis. Am J Respir Crit Care Med 197:348–355. doi: 10.1164/rccm.201707-1457OC. [DOI] [PubMed] [Google Scholar]
- 280.Zuckerman J, Clock SA, Prato SB, McDevitt JJ, Zhou JJ, Leclair LW, Lucas FL, Saiman L. 2015. Air contamination with bacteria in cystic fibrosis clinics: implications for prevention strategies. Am J Respir Crit Care Med 191:598–601. doi: 10.1164/rccm.201410-1877LE. [DOI] [PubMed] [Google Scholar]
- 281.Middleton ML, Layeghifard M, Klingel M, Stanojevic S, Yau Y, Zlosnik J, Speert D, Coriati A, Ratjen F, Tullis DE, Stephenson A, Wilcox P, Freitag A, Chilvers M, McKinney ML, Lavoie A, Wang P, Guttman D, Waters VJ. 2017. Abstract 328: Distribution and clinical impact of clonal Pseudomonas aeruginosa infection in the Canadian cystic fibrosis population. Pediatr Pulmonol 52(S47):339. [Google Scholar]
- 282.De Soyza A, Hall AJ, Mahenthiralingam E, Drevinek P, Kaca W, Drulis-Kawa Z, Stoitsova SR, Toth V, Coenye T, Zlosnik JE, Burns JL, Sa-Correia I, De Vos D, Pirnay JP, Kidd TJ, Reid D, Manos J, Klockgether J, Wiehlmann L, Tummler B, McClean S, Winstanley C, EU FP7 Funded COST Action BM1003 Cell Surface Virulence Determinants of Cystic Fibrosis Pathogens. 2013. Developing an international Pseudomonas aeruginosa reference panel. Microbiologyopen 2:1010–1023. doi: 10.1002/mbo3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Freschi L, Jeukens J, Kukavica-Ibrulj I, Boyle B, Dupont MJ, Laroche J, Larose S, Maaroufi H, Fothergill JL, Moore M, Winsor GL, Aaron SD, Barbeau J, Bell SC, Burns JL, Camara M, Cantin A, Charette SJ, Dewar K, Deziel E, Grimwood K, Hancock RE, Harrison JJ, Heeb S, Jelsbak L, Jia B, Kenna DT, Kidd TJ, Klockgether J, Lam JS, Lamont IL, Lewenza S, Loman N, Malouin F, Manos J, McArthur AG, McKeown J, Milot J, Naghra H, Nguyen D, Pereira SK, Perron GG, Pirnay JP, Rainey PB, Rousseau S, Santos PM, Stephenson A, Taylor V, Turton JF, Waglechner N, et al. 2015. Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front Microbiol 6:1036. doi: 10.3389/fmicb.2015.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Ciofu O, Johansen HK, Aanaes K, Wassermann T, Alhede M, von Buchwald C, Hoiby N. 2013. P. aeruginosa in the paranasal sinuses and transplanted lungs have similar adaptive mutations as isolates from chronically infected CF lungs. J Cyst Fibros 12:729–736. doi: 10.1016/j.jcf.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 285.Fothergill JL, Neill DR, Loman N, Winstanley C, Kadioglu A. 2014. Pseudomonas aeruginosa adaptation in the nasopharyngeal reservoir leads to migration and persistence in the lungs. Nat Commun 5:4780. doi: 10.1038/ncomms5780. [DOI] [PubMed] [Google Scholar]
- 286.Mainz JG, Hentschel J, Schien C, Cramer N, Pfister W, Beck JF, Tummler B. 2012. Sinonasal persistence of Pseudomonas aeruginosa after lung transplantation. J Cyst Fibros 11:158–161. doi: 10.1016/j.jcf.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 287.Syed SA, Whelan FJ, Waddell B, Rabin HR, Parkins MD, Surette MG. 2016. Reemergence of lower-airway microbiota in lung transplant patients with cystic fibrosis. Ann Am Thorac Soc 13:2132–2142. doi: 10.1513/AnnalsATS.201606-431OC. [DOI] [PubMed] [Google Scholar]
- 288.Lee B, Haagensen JA, Ciofu O, Andersen JB, Hoiby N, Molin S. 2005. Heterogeneity of biofilms formed by nonmucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J Clin Microbiol 43:5247–5255. doi: 10.1128/JCM.43.10.5247-5255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ferreira AG, Leao RS, Carvalho-Assef AP, da Silva EA, Firmida MDC, Folescu TW, Paixao VA, Santana MA, de Abreu e Silva FA, Barth AL, Marques EA. 2015. Low-level resistance and clonal diversity of Pseudomonas aeruginosa among chronically colonized cystic fibrosis patients. APMIS 123:1061–1068. doi: 10.1111/apm.12463. [DOI] [PubMed] [Google Scholar]
- 290.Aaron SD, Ramotar K, Ferris W, Vandemheen K, Saginur R, Tullis E, Haase D, Kottachchi D, St Denis M, Chan F. 2004. Adult cystic fibrosis exacerbations and new strains of Pseudomonas aeruginosa. Am J Respir Crit Care Med 169:811–815. doi: 10.1164/rccm.200309-1306OC. [DOI] [PubMed] [Google Scholar]
- 291.Llanes C, Pourcel C, Richardot C, Plesiat P, Fichant G, Cavallo JD, Merens A, GERPA Study Group. 2013. Diversity of beta-lactam resistance mechanisms in cystic fibrosis isolates of Pseudomonas aeruginosa: a French multicentre study. J Antimicrob Chemother 68:1763–1771. doi: 10.1093/jac/dkt115. [DOI] [PubMed] [Google Scholar]
- 292.Chen SH, Chen RY, Xu XL, Chen HT. 2014. Multilocus sequencing typing of Pseudomonas aeruginosa isolates and analysis of potential pathogenicity of typical genotype strains from occupational oxyhelium saturation divers. Undersea Hyperb Med 41:135–141. [PubMed] [Google Scholar]
- 293.Delic-Attree I, Toussaint B, Froger A, Willison JC, Vignais PM. 1996. Isolation of an IHF-deficient mutant of a Pseudomonas aeruginosa mucoid isolate and evaluation of the role of IHF in algD gene expression. Microbiology 142(Part 10):2785–2793. doi: 10.1099/13500872-142-10-2785. [DOI] [PubMed] [Google Scholar]
- 294.Keating D, Crowe MJ, Kennedy B, Salmon A, Britton D, Gallagher CG, McKone EF, Schaffer K. 2013. Molecular detection of an atypical, highly resistant, clonal Pseudomonas aeruginosa isolate in cystic fibrosis patients. J Cyst Fibros 12:141–146. doi: 10.1016/j.jcf.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 295.Cigana C, Melotti P, Baldan R, Pedretti E, Pintani E, Iansa P, De Fino I, Favari F, Bergamini G, Tridello G, Cirillo DM, Assael BM, Bragonzi A. 2016. Genotypic and phenotypic relatedness of Pseudomonas aeruginosa isolates among the major cystic fibrosis patient cohort in Italy. BMC Microbiol 16:142. doi: 10.1186/s12866-016-0760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Milczewska J, Wolkowicz T, Zacharczuk K, Kwiatkowska M. 2015. Cross-infections with Pseudomonas aeruginosa in patients with cystic fibrosis attending the Warsaw Centre. Dev Period Med 19:60–65. [PubMed] [Google Scholar]
- 297.Woo TE, Lim R, Surette MG, Waddell B, Bowron JC, Somayaji R, Duong J, Mody CH, Rabin HR, Storey DG, Parkins MD. Epidemiology and natural history of Pseudomonas aeruginosa airway infections in non-cystic fibrosis bronchiectasis. ERJ Open Res 4:00162-2017. doi: 10.1183/23120541.00162-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.