Abstract
Background
Selenium, an essential dietary micronutrient, is incorporated into proteins as the amino acid selenocysteine (Sec) in response to in-frame UGA codons. Complex machinery ensures accurate recoding of Sec codons in higher organisms. A specialized elongation factor eEFSec is central to the process.
Scope of review
Selenoprotein synthesis relies on selenocysteinyl-tRNASec (Sec-tRNASec), selenocysteine inserting sequence (SECIS) and other selenoprotein mRNA elements, an in-trans SECIS binding protein 2 (SBP2) protein factor, and eEFSec. The exact mechanisms of discrete steps of the Sec UGA recoding are not well understood. However, recent studies on mammalian model systems have revealed the first insights into these mechanisms. Herein, we summarize the current knowledge about the structure and role of mammalian eEFSec.
Major conclusions
eEFSec folds into a chalice-like structure resembling that of the archaeal and bacterial orthologues SelB and the initiation protein factor IF2/eIF5B. The three N-terminal domains harbor major functional sites and adopt an EF-Tu-like fold. The C-terminal domain 4 binds to Sec-tRNASec and SBP2, senses distinct binding domains, and modulates the GTPase activity. Remarkably, GTP hydrolysis does not induce a canonical conformational change in eEFSec, but instead promotes a slight ratchet of domains 1 and 2 and a lever-like movement of domain 4, which may be critical for the release of Sec-tRNASec on the ribosome.
General significance
Based on current findings, a non-canonical mechanism for elongation of selenoprotein synthesis at the Sec UGA codon is proposed. Although incomplete, our understanding of this fundamental biological process is significantly improved, and it is being harnessed for biomedical and synthetic biology initiatives. This article is part of a Special Issue entitled “Selenium research” in celebration of 200 years of selenium discovery, edited by Dr. Elias Arnér and Dr. Regina Brigelius-Flohe.
Keywords: Selenium, Selenocysteine, Selenoprotein, UGA codon, tRNA, eEFSec
1. Introduction
Intricate machinery is responsible for Sec synthesis and recoding of the Sec UGA codon. Selenium (Se), an essential dietary micronutrient, is found in two dozen human selenoproteins and selenoenzymes [1] that are pivotal for protecting the cell membrane and DNA from oxidative damage, maintaining redox balance and selenium homeostasis, and aiding protein folding and gene expression [2–4]. The biological function of Se is primarily exerted in a form of the amino acid, selenocysteine (Sec), which is required for function and structure of all selenoproteins and selenoenzymes. An intricate process evolved over time that orchestrates extraction of Se from various metabolites, incorporation of Se into Sec, and precise insertion of Sec into the growing selenoprotein chain.
The remarkable ability of Se to alternate between the oxidized and reduced states with ease [5] provides the basis for catalytic superiority of selenoenzymes over the thiol containing counterparts. Whereas thiol-based enzymes are often inactivated through irreversible oxidation and thus require specific enzymes to revert the abysmal situation, selenium-containing enzymes escape such a trap. This resistance to irreversible oxidation makes selenoenzymes advantageous in redox reactions and explains why they are so crucial for the removal of reactive oxygen species. Failure to incorporate Sec diminishes the catalytic prowess of selenoenzymes [6–14] and compromises the structure of selenoproteins, and introduction of Sec improves function of artificial selenoenzymes and confers resistance to inactivation by oxidation (reviewed in [5]). Consistent with its redox and antioxidant function, levels of many selenoprotein genes and selenoproteins are significantly increased under oxidative stress [15,16]. Disruption or deletion of genes encoding selenocysteine tRNA (tRNASec), glutathione peroxidase 4 (GPx4), and thioredoxin reductase 1 (Trx1) and 3 (Trx3) causes embryonically lethal phenotypes in mice, accentuating the significance of maintaining selenoproteome integrity [17–20]. Furthermore, mutations in enzymes facilitating Sec and selenoprotein synthesis cause systemic disorders [3,4,21–27], including severe early childhood degeneration of the human brain [28–30]. Recently, a mutation in tRNASec has been shown to cause deficit in stress-related, but not housekeeping, selenoproteins resulting in a complex clinical profile [31]. Lastly, Se deficiency, low levels of selenoproteins, and mutations in selenoprotein genes cause pathologies of cardiac, muscular, nervous, endocrine, immune, and reproductive systems (reviewed in [2,32–39]). Thus, faithful insertion of Se and Sec into proteins is an essential biological process. Surprisingly, this process is still poorly understood [4,40] and is largely being modeled using the general mechanism of bacterial protein synthesis (reviewed in [41]). While these models permit extrapolations to be drawn, they are often insufficient in their capacity to explain observations and, even worse, may be misleading. The absence of detailed models of selenoprotein synthesis is a consequence of only recent focus on the system and the sheer complexity of the Sec system which presents a number of technical and intellectual challenges.
The Sec UGA recoding process is intimately interlocked with the elaborate cycle of Sec synthesis (Fig. 1). This anabolic cycle provides the obligate substrate for selenoprotein synthesis, but it is so embroidered with particularities that it is prudent to provide a brief mention. In a classical case, proteinogenic amino acids are coupled to cognate tRNAs via specific aminoacyl-tRNA synthetases (aaRSs). However, in case of Sec, indirect aminoacylation is the only pathway to its synthesis and subsequent incorporation into protein. This is because the putative SecRS that would attach Sec onto tRNASec is the only aaRS that never arose during evolution. Also, a cellular pool of free Sec necessary for the direct coupling does not exist due to its high reactivity. Hence, each organism that relies on Sec had to devise a different mechanism for Sec synthesis. The solution involves a multistep process during which Sec is synthesized from a serine (Ser) precursor on the cognate tRNA while utilizing a Se donor molecule [42]. In all organisms, the process begins with tRNASec serylation (Fig. 1), which is catalyzed by a promiscuous SerRS (acts primarily on tRNASer). Subsequently, a single bacterial Sec synthase (SelA) directly converts the seryl moiety to Sec in a reaction mechanism that requires a pyridoxal phosphate (PLP) co-factor and selenophosphate [43] (top panel, Fig. 1). Conversely, in archaea and eukaryotes, the conversion occurs in two steps (bottom panel, Fig. 1). First, a specialized kinase (PSTK) phosphorylates Ser [44,45], and then, the Sec synthase (SepSecS) replaces the phosphoryl group with a selenol moiety using the mechanism analogous to that of the bacterial SelA [44,46,47]. To ensure process fidelity, Sec synthetic enzymes recognize and ‘read’ the distinct fold and structure of tRNASec [43,48–51], the largest and truly remarkable molecule among elongator tRNAs. But where does Se come from and in what form for reactions catalyzed by SelA and SepSecS? Another PLP enzyme, Sec lyase, extracts Se from Sec [52] and releases selenide, which is then used by selenophosphate synthetase (SPS or SPS2) to form selenophosphate [53,54], the major Se donor. In the end, the process achieves its goals: Sec is ‘attached’ to its tRNA while circumventing both the need for free Sec and the absent SecRS.
Fig. 1.
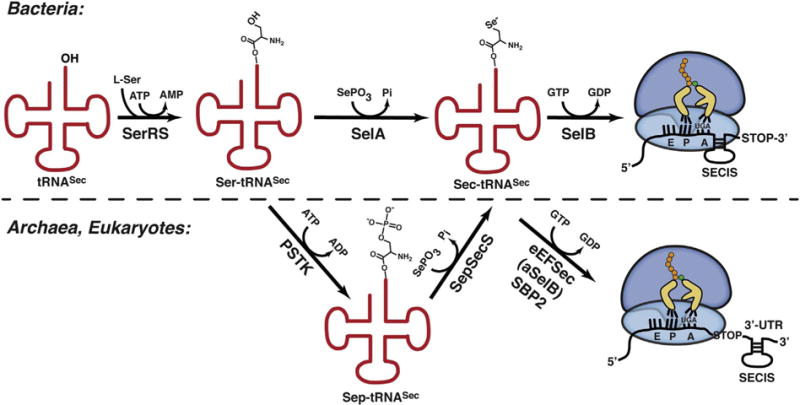
Synthesis and co-translational incorporation of Sec across kingdoms of life. (Top) The bacterial process is initiated with serylation of tRNASec. SelA then directly catalyzes Ser-to-Sec conversion. In the end, SelB delivers Sec-tRNASec to the 70S ribosome in response to an in-frame Sec UGA. The bacterial SECIS, which is in ORF and immediately downstream of the Sec codon, coordinates recoding. (Bottom) As in bacteria, the process begins with tRNASec serylation. However, here the Ser-to-Sec conversion proceeds in two steps: PSTK phosphorylates the Ser group and SepSecS then promotes conversion of Sep-tRNASec to Sec-tRNASec. eEFSec and aSelB promote recoding in eukaryotes and archaea, respectively. In both instances, the 3’-UTR SECIS element coordinates the recoding step. The distinction is that only eukaryotes rely on SBP2 protein factor.
Once formed, Sec-tRNASec is delivered to the ribosome in response to a context-dependent in-frame UGA codon (Fig. 1). The delivery and recoding is delegated to a specialized elongation factor, SelB in prokaryotes [55] and eEFSec in eukaryotes [56], which evolved to bind only Sec-tRNASec and no other aa-tRNA. Initially, the name of the bacterial selB gene was born out of a set of pleiotropic, and at the time unmapped, mutations in E. coli that had caused inactivation of formate dehydrogenase [57]. One of these later turned out to be the Sec elongation factor and the name SelB was then used irrespective of the organism. It is now more widely accepted that SelB is used when bacterial and archaeal Sec systems are discussed, and that eEFSec is used in conjunction with the eukaryotic process. Hence, to simplify matters, in the next few sections we shall use SelB, aSelB, and eEFSec for bacterial, archaeal, and eukaryotic Sec elongation factors, respectively.
The deletion of EEFSEC obliterates selenoprotein synthesis in fruit flies [58], further emphasizing the significance of this specialized elongation factor. Intriguingly, eEFSec and SelB cannot recode the Sec UGA on its own. Instead, they require an accessory RNA and/or protein factors. The first such element identified was designated as the SElenoCysteine Insertion Sequence (SECIS). This in-cis element forms a hairpin structure and is present in selenoprotein mRNAs across kingdoms [59–62]. In bacteria, it resides in an open reading frame (ORF) immediately downstream of the Sec UGA, while in archaea and eukaryotes, it is located in the 3′-UTR at variable distances (104–5200 nucleotides) from the Sec codon (Fig. 1). Despite the sequence divergence, SECIS elements share a general structure that is comprised of two RNA helices, an internal loop, and an apical loop. SECIS hairpins can adopt two slightly different forms: (i) Form 1 is composed of a 12–14 base-pair (bp) stem that separates a large apical loop and a single internal loop, and (ii) Form 2 contains an additional 2–7 bp stem, a smaller apical loop, and an additional internal bulge. Both forms harbor a conserved AAR sequence in the apical loop that is essential for Sec incorporation. The internal loop carries unpaired AUGA and UGR sequences at its 5′- and 3′ sides, respectively. The 5′-UGA-3′/5′-GAU-3′ base pairs in the 12–14 bp stem are predicted to adopt a kink turn (K-turn) motif [63,64] that serves as SBP2-binding motif (see below) [65]. It is thought that SECIS aids the differentiation between the Sec and stop UGA codons. Recent findings, however, argue that in some instances SECIS facilitates processive recoding, while in other it prevents mRNA decay [66]. It has also been suggested that certain selenoprotein genes harbor other regulatory sequences within their ORFs in conjunction with SECIS [67,68].
As if this process was already not complex enough, another layer was unearthed in eukaryotes. It was shown that for successful recoding eEFSec requires yet another in-trans factor. The SECIS Binding Protein 2 (SBP2) was identified to be essential for binding to SECIS [69]. Human SBP2 is a protein composed of 854 amino acids that fold into three major domains: (a) the N-Terminal Domain (NTD; residues 1–398), (b) the central Sec-Incorporation Domain (SID; residues 399–584), and (c) the C-terminal RNA-Binding Domain (RBD; residues 585–854) [70,71]. NTD is dispensable for recoding [72] and is completely missing in lower eukaryotes [73]. By contrast, SID and RBD represent the functional end of SBP2. SID is important for Sec insertion and binding to eEFSec, and RBD is essential for binding to the ribosome and SECIS, most likely through interactions with K-turn motifs [70,71]. Importantly, the SID and RBD form a complex with SECIS and support Sec incorporation in-trans [70,74]. This complex is formed only in the presence of SECIS [70] implying that specific interactions and arrangements of these SBP2 domains are required for function. It is also documented that SBP2 and eEFSec might undergo a conformational change upon complex formation [70]. However, the nature of structural rearrangements is not well understood primarily due to the lack of structural information about SBP2 and its complexes. The current consensus states that the SBP2-SECIS complex anchors eEFSec and Sec-tRNASec near the site of translation. Such a recruitment strategy probably serves to avert the looming possibility of premature abortion of selenoprotein synthesis at the Sec UGA codon [75,76], thereby increasing the efficiency of translation. That SBP2 and SECIS are significant has been well-documented by clinical reports. Mutations in SECISBP2 cause disorders impacting a whole spectrum of organ systems including male reproductive organs [21–26] and specific mutations in SECIS elements cause illnesses that resemble disorders caused by deficiencies of specific selenoproteins [77,78]. Lastly, Sec UGA recoding also appears to be modulated by dietary Se levels [79,80].
In spite of process idiosyncrasies across kingdoms, the central role of the Sec elongation factor in Sec UGA recoding is preserved. The current knowledge about the role, structure, and possible mechanism of the mammalian eEFSec is summarized in the following sections.
2. Structure and functional sites of human eEFSec
Prior knowledge about the Sec elongation factors was based on studies on prokaryotic model systems [55,76,81–87]. More recently, crystal structures representing the major functional states of human eEFSec were determined [88]. In practical terms, independent crystal forms of eEFSec complexed with either GDP or non-hydrolyzable GTP analogs (GDPNP and GDPCP) were obtained and then structures determined.
Human eEFSec is composed of four domains (D1–4) organized into a chalice-like structure (Fig. 2A). The height of the chalice is ~100 Å and its width varies from 20 Å at its stem to 60 Å at the cup rim. Domains 1–3 form the cup and resemble general translational elongation factor Tu (EF-Tu). D4 is the base or foot of the chalice and it adopts an oligosaccharide-binding (OB) fold, found in many proteins that interact with nucleic acids. The linker region composed of two β-strands represents the chalice stem that connects the cup and foot. The EF-Tu-like domain carries the main functional crevices, the GTPase site and the Sec-binding pocket (Fig. 2A). The GTPase site completely resides within D1 and it features standard elements found in other GTPases such as the P loop, switch 1, switch 2, and the guanine-binding sequence (Fig. 2B) [88]. Typical interactions between guanine nucleotides and the GTPase site of eEFSec have been observed. The guanine-binding sequence holds the guanine ring in place, while switch 1 serves to anchor the ribose ring and to coordinate waters and Mg2+ (Fig. 2B). On the other hand, the P loop anchors non-bridging oxygens of α- and β-phosphates, and switch 2 coordinates the β- and γ-phosphates and Mg2+ (Fig. 2B). An essential part of switch 2 is the presumed catalytic His96, but this side chain points away from the γ-phosphate in all eEFSec structures (Fig. 2B) [88]. To be fully operational in terms of GTPase activity, eEFSec probably requires additional adjustments that can occur only after binding to the ribosome. It is reasonable to suspect that interactions between switch 2 and 28S rRNA, similar to what was observed in EF-Tu and EF-G complexes [89–91], bring the His96 side chain in proximity of GTP γ-phosphate thus enabling GTP hydrolysis. Interestingly, mutational and activity assays revealed that Thr48, Asp92, and His96 are not essential for GTP/GDP binding, but rather for GTP hydrolysis [88]. Lastly, a minor, but perhaps important, point to take into account is that eEFSec adopts the same structure when in complex with GDPNP and GDPCP [88]. However, this does not necessarily mean that both analogs will be useful for studies that include programmed ribosomes as evidenced by the inability of GDPNP to induce active conformation of RF3 [92] and EF-G [93]. Conversely, EF-Tu and EF-G adopted active conformations on the ribosome in the presence of GDPCP [89–91,94,95].
Fig. 2.
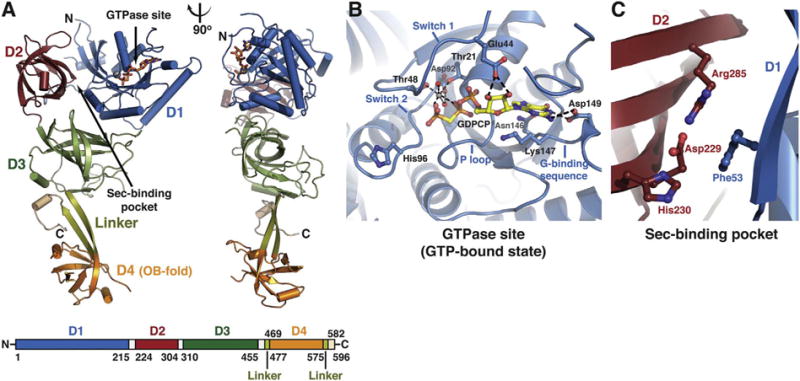
Structure of human eEFSec. (A) eEFSec folds into a chalice-like structure. The structure is shown as cartoon in two orientations that are related by 90° clockwise rotation around the vertical axis. Domains D1 (blue), D2 (red), and D3 (green) form a cup resembling EF-Tu. Flexible linker (light green) connects D3 with an appended D4 (orange), which represents the base of the cup. The C-terminal element (beige) folds below D3 (see: Fig. 3F). The GTPase site and the Sec-binding pocket are indicated with arrows. The domain arrangement and coloring scheme are summarized in a bar diagram below the cartoon diagram. (B) The close-up view of the GTPase site of eEFSec when complexed with a GTP analog. The main elements of the site are labeled and the major interactions between the amino-acid side chains, GTP analog, Mg2+ and water molecules (red spheres) are shown. (C) The close-up view of the Sec-binding pocket of human eEFSec. The pocket resides at the interface of D1 and D2. (For interpretation of the references to colour in this figure legend, the reader is referred to the online version of this chapter.)
As already mentioned, eEFSec harbors the Sec-binding pocket which is required for successful recoding. The Sec-binding pocket sits at the D1-D2 interface and it harbors Asp229, His230, and Arg285 (Fig. 2C) [88]. Positively charged residues are thought to be the main filter that selects Sec over near-cognate Ser, Cys, and perhaps phosphoserine. Interestingly, as long as one positively charged amino acid is present in the pocket, the E. coli SelB is capable of promoting the Sec UGA read through [84]. However, the same is not true for eEFSec. In fact, any change introduced, trimming side chains to Ala or putting in place residues found in EF-Tu, completely diminished recoding of the Sec UGA [88]. This effect was not a consequence of structural problems, as mutants behaved similar to wild-type eEFSec during recombinant expression and purification, but rather suggested that the integrity and composition of the entire site is essential for recoding [88]. What remains unknown is whether mutant eEFSec constructs were completely incapable of binding the Sec moiety within Sec-tRNASec. If so, these mutants could potentially support recoding using misacylated tRNASec species. Lastly, D4 can both sense the nucleotide binding and modulate the GTPase activity of the mammalian eEFSec [96]. It is important to continue studies that analyze the amino-acid discrimination mechanism of eEFSec and the role of D4 as this information would be invaluable for devising a system that supports incorporation of non-standard amino acids.
3. Conservation and divergence of Sec elongation factors
Organisms that rely on Sec and selenoproteins carry the entire Sec-synthetic and Sec-recoding machinery, including the Sec-specific elongation factor. Sec elongation factors exhibit a certain level of conservation and divergence across kingdoms.
Phylogenetic studies suggested a closer evolutionary relationship of eEFSec/aSelB with the translational initiation protein factor IF2/eIF5B than with EF-Tu/EF1A [97,98]. Indeed, the structure of human eEFSec [88] closely resembles aSelB [84] and IF2/eIF5B [99] (Fig. 3A, B, D). In addition, aSelB, just like eEFSec, contains the N-terminal EF-Tu-like domain and an appended C-terminal D4 [84,87]. A relatively good structural agreement exists between D1, D2, and D4 of human eEFSec, archaeal IF2, and yeast eIF5B [88]. This confirmed the earlier proposal that Sec elongation factors are structural chimeras of the general translation elongation and initiation protein factors [84]. It is significant to mention that the general structural organization holds true in the case of SelB, though its D4 is quite distinct (see below; Fig. 3C). Further, eEFSec and aSelB bind GTP and GDP with similar affinities [88,100], which explains why these GTPases do not need a specific guanine nucleotide-exchange factor (GEF) that would cycle their GDP-bound state back into the active, GTP-bound state. This phenomenon suggests that aSelB and eEFSec may not necessarily employ the EF-Tu-based mechanism during decoding and protein elongation (see: “4 The proposed mechanism of action of human eEFSec”).
Fig. 3.
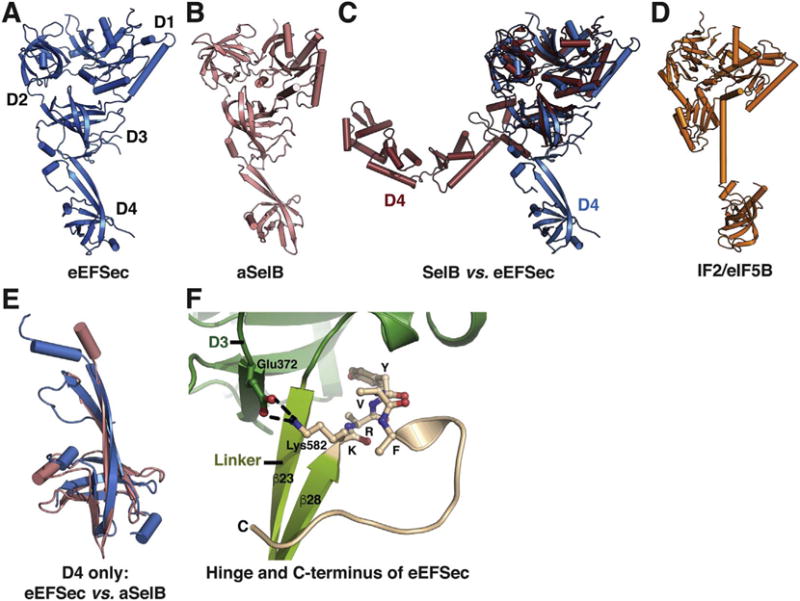
Conservation and divergence of Sec elongation factor structures. The structure of eEFSec (A) resembles that of aSelB (B) in spite of slight divergence of D4. Conversely, analogous comparison with SelB (C) reveals that the structural conservation is preserved within D1–3, whereas D4 adopts both the distinct structures and orientations. A cartoon representation of IF2/eIF5B (D) confirms its phylogenetic relationship with eEFSec and SelB. (E) Structural overlay of D4 from eEFSec (blue) and aSelB (pink) reveals slight differences between the domains. (F) The conformation of the extreme C-terminus (beige) of eEFSec and its interactions with linker (light green) and D3 (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the online version of this chapter.)
The structural homology between eEFSec and SelB [87] is restricted to the EF-Tu-like domain only (Fig. 3C). Minor differences are noted in switch 1 and around the GTPase site in D1, and there are some differences that include size and/or orientation of several solvent-exposed loops in D2 and D3 [88]. The most striking differences are noted in D4. In bacteria, D4 consists of four winged-helix folds and is rotated ~90° around the linker region when compared to the aSelB and eEFSec (Fig. 3C). This structural difference probably reflects divergent constraints posed by the bacterial recoding process that relies on a vicinal SECIS within the reading frame and the absence of SBP2. Naturally, other differences stemming from the ribosome itself should not be discounted.
Analogous comparison between eEFSec and aSelB includes D4 (Fig. 3A, B). Human D4 contains an additional α-helix and a longer C-terminal segment (Fig. 3E). The orientation of human D4 is stabilized by interactions between residues 582-KRYVF-586 in the extreme C-terminus and a segment of D3 (Fig. 3F). Subsequent biochemical, mutagenesis, and activity assays have demonstrated that these interactions have a structural role, but further studies are necessary to assess their potential functional role(s). Taken together, aSelB and eEFSec differ ever so slightly and it is likely that they share the mechanism by which the Sec UGA recoding is accomplished. However, while eukaryotes do rely on SBP2, a corresponding archaeal orthologue remains elusive to this day. Mechanistic differences could be more substantial in the case of SelB, but the core process of tRNA recognition and release may, in fact, be the same.
4. The proposed mechanism of action of human eEFSec
Structures of functional states of eEFSec raised a question whether elongation of selenoprotein synthesis at the Sec UGA codon differs from that of general elongation mechanism that is based on EF-Tu/EF1A. Studies on bacterial systems were particularly useful for delineation of details of the general, canonical mechanism, and the main findings shall be briefly mentioned.
EF-Tu in its GTP-bound state binds and delivers an aminoacyl-tRNA (aa-tRNA) to the ribosome. The establishment of proper codon-anticodon interactions between the mRNA and the anticodon loop of the A-site aa-tRNA in the decoding center of the small ribosomal subunit enables interactions between the GTPase site of EF-Tu and the sarcinricin loop of the 23S rRNA of the large ribosomal subunit. The latter interaction is critical for orienting catalytic residues and a water molecule in the GTPase site of EF-Tu, leading to GTP hydrolysis. What follows the dissolution of GTP at the structural level has enormous functional consequences. Hydrolysis of GTP into GDP and the release of Pi drive D1 to rotate ~90° relative to D2 and D3 [101,102]. This major conformational rearrangement transforms a globular GTP-bound EF-Tu into an elongated GDP-bound EF-Tu. As a consequence, the aminoacyl-binding pocket and tRNA-recognition surfaces are deformed [103], which leads to dissociation of EF-Tu:GDP from the aa-tRNA and the ribosome. The EF-Tu departure releases the acceptor stem of the aa-tRNA, which is significant because the liberated CCA-end can now be properly positioned within the catalytic peptidyl-transferase center (PTC) on the large ribosomal subunit. The accommodation of the CCA-end brings about the aminoacyl moiety into a proper orientation relative to the acyl ester bond in the P-site peptidyl-tRNA and the reaction of peptide bond formation and protein elongation can occur [91,104]. It has been recently suggested that EF1A employs the same mechanism [105]. Hence, in the absence of the GTPase-coupled conformational change of D1 in EF-Tu/EF1A elongation of the nascent protein chain would not take place.
Let us now consider structural adjustments in eEFSec that take place upon GTP-to-GDP exchange, which is used to mimic the end points of GTP hydrolysis. Somewhat unexpectedly, structures of the GTP- and GDP-bound states of human eEFSec turned out to be remarkably similar (Fig. 4A) [88]. In both states, D1 assumed basically the same orientation relative to D2 and D3 (Fig. 4A). In retrospect, and as already alluded to, the preserved domain arrangement should have been expected because eEFSec (and SelB) bind GTP, GDP, and GTP analogs with almost indistinguishable binding affinity, which stands in sharp contrast to EF-Tu and EF1A. Closer inspection revealed that Sec elongation factors harbor a large solvent-exposed loop (β17-β18) on the dorsal side of D3 that may prevent the canonical rotation of D1. While the movement of D1 was absent, the major change was found elsewhere, in the C-terminal D4 (Fig. 4A). This domain swings ~26° and is translated > 15 Å away from the putative tRNA-binding interface upon GTP-to-GDP exchange (Fig. 4A). The structural results were consistent with an earlier proposal that D4 is somehow capable of ‘sensing’ the nucleotide binding and regulating the GTPase activity of the mammalian eEFSec [96]. The solution-based studies have also confirmed that eEFSec assumes the same conformations in solution as in the crystal [88], thus strongly arguing that the determined structures represent the physiologically relevant states.
Fig. 4.
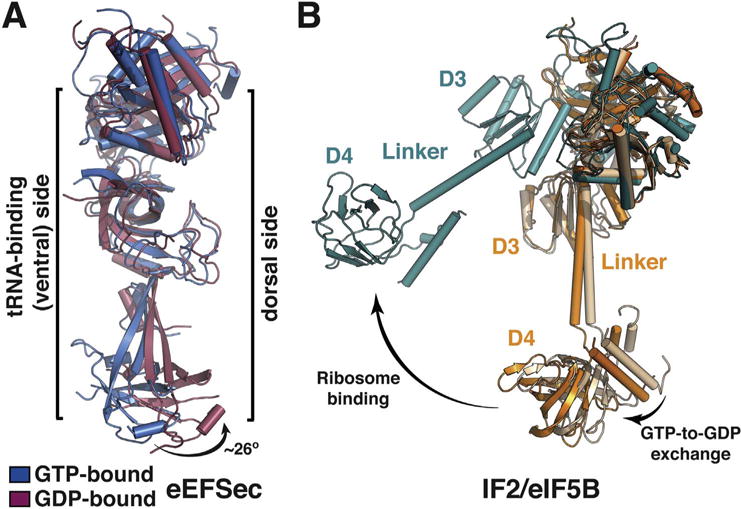
A major, non-canonical conformational change in eEFSec upon GTP-to-GDP exchange. (A) A side view of overlaid GTP- (blue) and GDP-bound (red) states of eEFSec shows that GTP hydrolysis induces a major conformational change in D4, which is distinct from the canonical situation in EF-Tu. In particular, D4 swings ~26° away from the tRNA-binding (or ventral) side of eEFSec and drags with it the linker region. D1 and D2 move towards and away from the tRNA-binding side. (B) The magnitude of the GTPase-coupled conformational change of IF2/eIF5B depends on the ribosome. In the absence of the ribosome, the movement of D3, linker, and D4 is relatively small (beige to orange), whereas the conformational change is quite pronounced when the factor binds to the ribosome (orange, or beige, to teal). (For interpretation of the references to colour in this figure legend, the reader is referred to the online version of this chapter.)
Other, smaller in their magnitude, but perhaps functionally significant changes occur in other domains. Namely, GTP hydrolysis stimulates D1 and D2 to slide in a ratchet-like motion towards and away from the tRNA-binding interface, respectively. These concerted motions have the opposite effect on functional sites as the GTPase site relaxes and the Sec-binding pocket tightens. The relaxation of the GTPase site is manifested by disorder in switch 1 and repositioning of switch 2, as a result of which side chains of Asp92 and His96 are not capable of interacting with the guanine-nucleotide phosphate and Mg2+ ion. On the other hand, the Sec-binding pocket constriction is a result of movements and partial disorder of certain structural and constitutive pocket elements. This observation argues that GTP binding is needed for the Sec-binding pocket to open, which, in turn, would provide a credence for the fact that Sec elongation factors markedly prefer Sec-tRNASec in their GTP-bound state [85].
The structural changes were compared to IF2/eIF5B (Fig. 4B) and used to extrapolate a non-canonical mechanism by which eEFSec aids r (d)ecoding of the Sec UGA codon (Fig. 5). Briefly, a complex between the GTP-bound eEFSec and Sec-tRNASec is anchored to the selenoprotein mRNA by SBP2 and SECIS. Once ribosome stalls at the Sec UGA codon, eEFSec delivers Sec-tRNASec. The binding is such that D4 points in the direction of the central protuberance with its 549KKRAR553 sequence [96]. Just like in the EF-Tu-based mechanism, formation of the codon-anticodon interactions stimulates the GTPase activity of eEFSec. The subsequent concerted constriction of the Sec-binding pocket and the lever-like motion of D4 result in a decreased affinity for Sec-tRNASec. The former motion practically squeezes the Sec moiety out of the Sec-binding pocket, whereas the latter releases its grip on the acceptor-TΨC elbow and the variable arm of tRNASec. These motions eventually lead to eEFSec departure from the recoding assembly. Similar structural rearrangements have been previously observed in aSelB [84] and the translation initiation factor IF2/eIF5B [99,106]. Interestingly, it was demonstrated that the magnitude of D4 movement of IF2/eIF5B is even larger when in complex with the ribosome [106], a scenario that could still be applicable to Sec elongation factors as well. In the end, the mechanism of tRNASec release is probably preserved in bacteria and archaea, but more studies are needed to test this proposition.
Fig. 5.
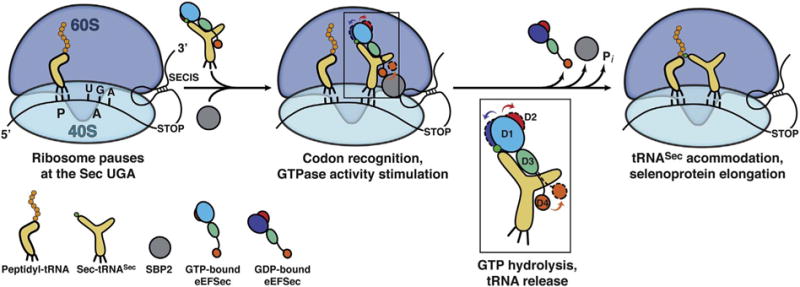
Proposed mechanism of elongation of selenoprotein synthesis as the Sec UGA codon. The 80S ribosome pauses at the SEC UGA codon. The peptidyl-tRNA is bound to the P site and is awaiting the next amino acid. The 3′-UTR SECIS element tethers a large complex composed of SBP2 (grey sphere), the GTP-bound state of eEFSec, and Sec-tRNASec near the ribosome. How eEFSec delivers Sec-tRNASec to the A site and what is the role of SBP2 and ribosomal elements during this step is still not clear. Formation of proper codon-anticodon interactions at the decoding center of the 40S subunit presumably stimulates the GTPase activity of eEFSec. GTP hydrolysis leads to a slight ratchet of D1 (shades of blue) and D2 (red), and a larger swing of D4 (orange). D1 moves towards the ventral (tRNA-binding) side of eEFSec, whereas D2 and D4 move in the opposite direction (see inset). The domain movements, which are different from the EF-Tu-based system, cause dissociation of eEFSec from Sec-tRNASec and the ribosome. This then allows accommodation of Sec-tRNASec within the PTC and the reaction of peptide bond synthesis to occur. The end result is an elongated nascent selenoprotein. (For interpretation of the references to colour in this figure legend, the reader is referred to the online version of this chapter.)
Why is a distinct and peculiar recoding mechanism needed for the Sec UGA codon? Need for Se probably arose during Cambrian “oxygen explosion”, which witnessed subsequent speciation explosion. A peculiar characteristic of Se, an occasionally classified metalloid, is that it can flip between its oxidized and reduced states with ease, something that a close relative from the periodic table of elements, sulfur, is incapable of achieving. Such ability gives advantage to proteins and enzymes that contain Se to avoid permanent inhibition and damage through oxidation. This was then further exploited in combating reactive oxygen species in cells to this day [5]. It could well be then that the evolutionary pressure to have Sec accurately incorporated into the nascent polypeptide yielded a system that took components from a number of already invented tools. For instance, a primordial SerRS was utilized to initiate the anabolic cycle. The absence of editing and anticodon-binding domains in this enzyme eased the process; the former feature meant that the serylation ‘error’ would not be remedied, while the latter signified that the anticodon sequence is not required for catalysis. Hence, enter tRNASec, a UGA codon suppressor, which resembles tRNASer just enough and the cycle can begin. Two distinct anabolic processes, each of which took advantage of the PLP-based mechanism, evolved over time. The one in bacteria relied on a single enzyme, whereas the one in archaea and eukaryotes probably represents a repurposed archaeal machinery originally used for tRNA-dependent Cys synthesis [107]. The Sec UGA codon had to be differentiated from occasional UGA stop codons and a hairpin structure was tagged to the selenoprotein mRNAs to achieve that. With it coevolved an elongation factor which most likely arose from an ancestral initiation factor, perhaps multiple times during evolution as SelB and eEFSec do differ in their requirements. The most recent addition is SBP2, a large multidomain protein that aids only eEFSec during recoding. Naturally, future phylogenetic and biochemical analyses will shed light on this interesting question.
5. Future directions
Important questions concerning the discrete steps of the elongation phase of selenoprotein synthesis remain unanswered. For instance, the mechanism by which eEFSec selects Sec-tRNASec among other, more abundant aa-tRNAs is not clear. Studies on a bacterial system have suggested that the binding affinity of SelB for Sec-tRNASec is 106-fold higher than that for the unacylated tRNASec [85]. The steep increase may seem too excessive and opens questions as to whether such a protein factor could ever release the tRNA. But, the tight binding is plausible because the cellular concentration of tRNASec is significantly lesser when compared to other elongator tRNAs. Hence, SelB, and by analogy eEFSec, may avoid binding of near- and non-cognate tRNAs by raising the bar of specificity. In other words, Sec elongation factors simply may not interact with anything else to an appreciable extent, which would dramatically decrease the probability of an ‘erroneous’ interaction and mistranslation. Obviously, the alternative could be that eEFSec differs from SelB and that it is not as specific as initially thought. As a consequence, a certain fraction of selenoproteins in each cell type and at all times may be expressed as Ser- and/or Cys-containing variants. The levels of such variants could vary and could be modulated by various exogenous factors, perhaps the most obvious among them being Se dietary intake.
The essential protein factor SBP2 is required for recoding of the Sec UGA codon in eukaryotes, but the mechanism by which this factor achieves its role is not fully resolved. It is believed that SBP2 recruits eEFSec and Sec-tRNASec to the ribosome, which increases the efficiency of selenoprotein synthesis. In one model, SBP2 binds to SECIS and allows eEFSec and Sec-tRNASec to access the ribosomal A site [108,109]. However, it is not clear whether SBP2 remains bound to SECIS and the eEFSec:Sec-tRNASec complex while bound to the ribosome, or whether it dissociates from the ribosome immediately upon ‘cargo’ delivery. It is suggested that after tRNA release, the ribosomal protein eL30 binds to SECIS and displaces SBP2. In another model, after recruitment, SBP2 exchanges for eL30 and induces a conformational change in SECIS, triggering the GTPase activity of eEFSec and the subsequent release of Sec-tRNASec [110]. However, because of its distant location relative to the PTC and the A site, questions may be raised about the exact role of the ribosomal protein eL30 in this process. More recently, the SBP2-dependent structural adjustments in the expansion segment ES7L and helix 89 of the 28S rRNA were observed [111,112], which led to a proposal that SBP2 might aid the Sec-tRNASec accommodation into the PTC and/or stimulate the GTPase activity of eEFSec [112]. Whatever the scenario, it is reasonable to assume that one will have to determine an assemblage of distinct complex structures in order to describe the entire recoding process. A simpler precedent has already been presented in a structural and biochemical study of the bacterial ribosome and SelB [113]. It is important to mention that somewhat mysterious aura of SBP2 is largely due to the absence of structural information about this protein, its domains, and its complexes with SECIS, eEFSec, Sec-tRNASec, and the ribosome. It was suggested that the SBP2 protein factor is inherently unstructured and thus recalcitrant to structural studies [114], and this may indeed be true when this protein factor is without its physiological binding partners. We, therefore, suggest that structural studies of larger Sec recoding assemblies that include programmed human ribosomes should be vigorously pursued.
In that context, we mention additional mechanistic questions about the recoding process itself that require significant scrutiny. For instance, there are two tRNASec isoforms designated as 5-methoxycarbonylmethyluridine (mcm5U) and 5-methoxycarbonylmethyl-2′-O-methyluridine (mcm5Um), in both of which, U34 in the anticodon loop is modified (both isoforms carry N6-isopentenyladenosine, pseudouridine, and 1-methyladenosine at positions 37, 55, and 58, respectively). The only distinction between the two is a single methyl group attached to the 2′-OH of ribose in the ‘hyper methylated’, Um34 isoform. This additional methylation depends on the proper tertiary structure [115] and aminoacylation of tRNASec [116], and higher Se levels [117]. Intriguingly, the Um34 isoform supports expression of stress-related selenoproteins [118,119], whereas the non-Um34 isoform is needed for expression of housekeeping selenoproteins [119–121]. Given these findings, we ask what is the role of 2′-O-methylribosylation of U34 during recoding, how can selective expression be dictated by a seemingly small modification, and what are the impacts of methylribosylation on tRNASec structure, if any? Further, because of its unique fold and long variable arm, we wonder how is tRNASec accommodated in the ribosomal A-site? Does the translocation of the peptidyl-tRNASec from the A site to the P site resemble the canonical process or does it rely on other rearrangements in the PTC and tRNASec? Is the GTPase-coupled conformational change of eEFSec that is bound to the ribosome similar in its magnitude and nature to the one observed in eEFSec in isolation [88] or is it similar to that of IF2/eIF5B [106]? These are just but a few questions illustrating why structural studies of the elongation phase of eukaryotic and human selenoproteins are warranted.
The generality and fundamentality of Sec system is sufficient to explain why one should study this remarkable biological process. But, there are also possibilities for those who wish to venture into the realm of transferring fundamental information into things practical. Studious analyses of all aspects of Sec and selenoprotein synthesis will inevitably provide useful platforms for explaining various human disorders, some of which are of quite vicious disposition. Given significant overlaps with other clinical profiles and the impact of selenoproteins on basic cellular processes that is several orders of magnitude larger than its size, acquired information may lead to defining general principles that govern the development of many maladies. This could be used for the development of novel therapeutic interventions targeting Se homeostasis, redox processes, selenoproteome, proteostasis, or other processes/systems. Lastly, deciphering how Sec UGA codon is recoded will almost certainly find its usefulness in synthetic biology initiatives aiming to take advantage of repurposing of the UGA and other stop codons. This quest will be also helped by most recent discovery that both nonsense and sense codons, other than UGA, encode Sec in various organisms [122].
Acknowledgments
This work was supported by grant from the National Institute of General Medical Sciences (GM097042 to MS), the Provost Deiss Award (to AKP), the CCTS Pre-Doctoral Education for Clinical and Translational Scientists Fellowship (to AKP), and the Chancellor’s Graduate Research Award (to AKP).
Footnotes
Transparency document
The Transparency document associated with this article can be found in online version.
Author contributions
MS and AKP wrote the manuscript.
References
- 1.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 2.Rayman MP. Selenoproteins and human health: insights from epidemiological data. Biochim Biophys Acta. 2009;1790:1533–1540. doi: 10.1016/j.bbagen.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Dumitrescu AM, Refetoff S. Inherited defects of thyroid hormone metabolism. Ann Endocrinol (Paris) 2011;72:95–98. doi: 10.1016/j.ando.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 2014;94:739–777. doi: 10.1152/physrev.00039.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reich HJ, Hondal RJ. Why nature chose selenium. ACS Chem Biol. 2016;11:821–841. doi: 10.1021/acschembio.6b00031. [DOI] [PubMed] [Google Scholar]
- 6.Rocher C, Lalanne JL, Chaudiere J. Purification and properties of a recombinant sulfur analog of murine selenium-glutathione peroxidase. Eur J Biochem. 1992;205:955–960. doi: 10.1111/j.1432-1033.1992.tb16862.x. [DOI] [PubMed] [Google Scholar]
- 7.Maiorino M, Aumann KD, Brigelius-Flohe R, Doria D, van den Heuvel J, McCarthy J, Roveri A, Ursini F, Flohe L. Probing the presumed catalytic triad of selenium-containing peroxidases by mutational analysis of phospholipid hydro-peroxide glutathione peroxidase (PHGPx) Biol Chem Hoppe Seyler. 1995;376:651–660. doi: 10.1515/bchm3.1995.376.11.651. [DOI] [PubMed] [Google Scholar]
- 8.Gladyshev VN, Jeang KT, Stadtman TC. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc Natl Acad Sci U S A. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura T, Stadtman TC. A new selenoprotein from human lung adenocarcinoma cells: purification, properties, and thioredoxin reductase activity. Proc Natl Acad Sci U S A. 1996;93:1006–1011. doi: 10.1073/pnas.93.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuiper GG, Klootwijk W, Visser TJ. Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase reduces catalytic efficiency and alters substrate preference. Endocrinology. 2003;144:2505–2513. doi: 10.1210/en.2003-0084. [DOI] [PubMed] [Google Scholar]
- 11.Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckett GJ, Arthur JR. Selenium and endocrine systems. J Endocrinol. 2005;184:455–465. doi: 10.1677/joe.1.05971. [DOI] [PubMed] [Google Scholar]
- 13.Toppo S, Vanin S, Bosello V, Tosatto SC. Evolutionary and structural insights into the multifaceted glutathione peroxidase (Gpx) superfamily. Antioxid Redox Signal. 2008;10:1501–1514. doi: 10.1089/ars.2008.2057. [DOI] [PubMed] [Google Scholar]
- 14.Lee BC, Dikiy A, Kim HY, Gladyshev VN. Functions and evolution of selenoprotein methionine sulfoxide reductases. Biochim Biophys Acta. 2009;1790:1471–1477. doi: 10.1016/j.bbagen.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Touat-Hamici Z, Legrain Y, Bulteau AL, Chavatte L. Selective up-regulation of human selenoproteins in response to oxidative stress. J Biol Chem. 2014;289:14750–14761. doi: 10.1074/jbc.M114.551994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zahia TH, Yona L, Anne-Laure B, Laurent C. Selective up-regulation of human selenoproteins in response to oxidative stress. Free Radic Biol Med. 2014;75(Suppl. 1):S25. doi: 10.1016/j.freeradbiomed.2014.10.745. [DOI] [PubMed] [Google Scholar]
- 17.Bösl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci U S A. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweizer U, Schomburg L, Savaskan NE. The neurobiology of selenium: lessons from transgenic mice. J Nutr. 2004;134:707–710. doi: 10.1093/jn/134.4.707. [DOI] [PubMed] [Google Scholar]
- 19.Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakupoglu C, Przemeck GK, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, de Angelis MH, Wurst W, Bornkamm GW, Brielmeier M, Conrad M. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol. 2005;25:1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, Moeller LC, Boran G, Schomburg L, Weiss RE, Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat Genet. 2005;37:1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- 22.Dumitrescu AM, Di Cosmo C, Liao XH, Weiss RE, Refetoff S. The syndrome of inherited partial SBP2 deficiency in humans. Antioxid Redox Signal. 2009;12:905–920. doi: 10.1089/ars.2009.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Cosmo C, McLellan N, Liao XH, Khanna KK, Weiss RE, Papp L, Refetoff S. Clinical and molecular characterization of a novel selenocysteine insertion sequence-binding protein 2 (SBP2) gene mutation (R128X) J Clin Endocrinol Metab. 2009;94:4003–4009. doi: 10.1210/jc.2009-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azevedo MF, Barra GB, Naves LA, Ribeiro Velasco LF, Godoy Garcia Castro P, de Castro LC, Amato AA, Miniard A, Driscoll D, Schomburg L, de Assis Rocha Neves F. Selenoprotein-related disease in a young girl caused by nonsense mutations in the SBP2 gene. J Clin Endocrinol Metab. 2010;95:4066–4071. doi: 10.1210/jc.2009-2611. [DOI] [PubMed] [Google Scholar]
- 25.Hamajima T, Mushimoto Y, Kobayashi H, Saito Y, Onigata K. Novel compound heterozygous mutations in the SBP2 gene: characteristic clinical manifestations and the implications of GH and triiodothyronine in longitudinal bone growth and maturation. Eur J Endocrinol. 2012;166:757–764. doi: 10.1530/EJE-11-0812. [DOI] [PubMed] [Google Scholar]
- 26.Schoenmakers E, Agostini M, Mitchell C, Schoenmakers N, Papp L, Rajanayagam O, Padidela R, Ceron-Gutierrez L, Doffinger R, Prevosto C, Luan J, Montano S, Lu J, Castanet M, Clemons N, Groeneveld M, Castets P, Karbaschi M, Aitken S, Dixon A, Williams J, Campi I, Blount M, Burton H, Muntoni F, O’Donovan D, Dean A, Warren A, Brierley C, Baguley D, Guicheney P, Fitzgerald R, Coles A, Gaston H, Todd P, Holmgren A, Khanna KK, Cooke M, Semple R, Halsall D, Wareham N, Schwabe J, Grasso L, Beck-Peccoz P, Ogunko A, Dattani M, Gurnell M, Chatterjee K. Mutations in the selenocysteine insertion sequence-binding protein 2 gene lead to a multisystem selenoprotein deficiency disorder in humans. J Clin Invest. 2010;120:4220–4235. doi: 10.1172/JCI43653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito Y, Shichiri M, Hamajima T, Ishida N, Mita Y, Nakao S, Hagihara Y, Yoshida Y, Takahashi K, Niki E, Noguchi N. Enhancement of lipid peroxidation and its amelioration by vitamin E in a subject with mutations in the SBP2 gene. J Lipid Res. 2015;56:2172–2182. doi: 10.1194/jlr.M059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agamy O, Ben Zeev B, Lev D, Marcus B, Fine D, Su D, Narkis G, Ofir R, Hoffmann C, Leshinsky-Silver E, Flusser H, Sivan S, Söll D, Lerman-Sagie T, Birk OS. Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am J Hum Genet. 2010;87:538–544. doi: 10.1016/j.ajhg.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makrythanasis P, Nelis M, Santoni FA, Guipponi M, Vannier A, Bena F, Gimelli S, Stathaki E, Temtamy S, Megarbane A, Masri A, Aglan MS, Zaki MS, Bottani A, Fokstuen S, Gwanmesia L, Aliferis K, Bustamante Eduardo M, Stamoulis G, Psoni S, Kitsiou-Tzeli S, Fryssira H, Kanavakis E, Al-Allawi N, Sefiani A, Al Hait S, Elalaoui SC, Jalkh N, Al-Gazali L, Al-Jasmi F, Bouhamed HC, Abdalla E, Cooper DN, Hamamy H, Antonarakis SE. Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum Mutat. 2014;35:1203–1210. doi: 10.1002/humu.22617. [DOI] [PubMed] [Google Scholar]
- 30.Anttonen AK, Hilander T, Linnankivi T, Isohanni P, French RL, Liu Y, Simonović M, Söll D, Somer M, Muth-Pawlak D, Corthals GL, Laari A, Ylikallio E, Lahde M, Valanne L, Lonnqvist T, Pihko H, Paetau A, Lehesjoki AE, Suomalainen A, Tyynismaa H. Selenoprotein biosynthesis defect causes progressive encephalopathy with elevated lactate. Neurology. 2015;85:306–315. doi: 10.1212/WNL.0000000000001787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoenmakers E, Carlson B, Agostini M, Moran C, Rajanayagam O, Bochukova E, Tobe R, Peat R, Gevers E, Muntoni F, Guicheney P, Schoenmakers N, Farooqi S, Lyons G, Hatfield D, Chatterjee K. Mutation in human selenocysteine transfer RNA selectively disrupts selenoprotein synthesis. J Clin Invest. 2016;126:992–996. doi: 10.1172/JCI84747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bellinger FP, Raman AV, Reeves MA, Berry MJ. Regulation and function of selenoproteins in human disease. Biochem J. 2009;422:11–22. doi: 10.1042/BJ20090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt RL, Simonović M. Synthesis and decoding of selenocysteine and human health. Croat Med J. 2012;53:535–550. doi: 10.3325/cmj.2012.53.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweizer U, Dehina N, Schomburg L. Disorders of selenium metabolism and selenoprotein function. Curr Opin Pediatr. 2011;23:429–435. doi: 10.1097/MOP.0b013e32834877da. [DOI] [PubMed] [Google Scholar]
- 35.Hatfield DL, Tsuji PA, Carlson BA, Gladyshev VN. Selenium and selenocysteine: roles in cancer, health, and development. Trends Biochem Sci. 2014;39:112–120. doi: 10.1016/j.tibs.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohrle J, Gartner R. Selenium and thyroid. Best Pract Res Clin Endocrinol Metab. 2009;23:815–827. doi: 10.1016/j.beem.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Lescure A, Rederstorff M, Krol A, Guicheney P, Allamand V. Selenoprotein function and muscle disease. Biochim Biophys Acta. 2009;1790:1569–1574. doi: 10.1016/j.bbagen.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Duntas LH, Benvenga S. Selenium: an element for life. Endocrine. 2015;48:756–775. doi: 10.1007/s12020-014-0477-6. [DOI] [PubMed] [Google Scholar]
- 39.Hatfield DL, Schweizer U, Tsuji PA, Gladyshev VN. Selenium: Its Molecular Biology and Role in Human Health. 4th. Springer Nature; New York: 2016. [Google Scholar]
- 40.Shetty SP, Copeland PR. Selenocysteine incorporation: a trump card in the game of mRNA decay. Biochimie. 2015;114:97–101. doi: 10.1016/j.biochi.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 42.Su D, Hohn MJ, Palioura S, Sherrer RL, Yuan J, Söll D, O’Donoghue P. How an obscure archaeal gene inspired the discovery of selenocysteine biosynthesis in humans. IUBMB Life. 2009;61:35–39. doi: 10.1002/iub.136. [DOI] [PubMed] [Google Scholar]
- 43.Itoh Y, Brocker MJ, Sekine S, Hammond G, Suetsugu S, Söll D, Yokoyama S. Decameric SelA*tRNA(Sec) ring structure reveals mechanism of bacterial selenocysteine formation. Science. 2013;340:75–78. doi: 10.1126/science.1229521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherrer RL, O’Donoghue P, Söll D. Characterization and evolutionary history of an archaeal kinase involved in selenocysteinyl-tRNA formation. Nucleic Acids Res. 2008;36:1247–1259. doi: 10.1093/nar/gkm1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan J, Palioura S, Salazar JC, Su D, O’Donoghue P, Hohn MJ, Cardoso AM, Whitman WB, Söll D. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc Natl Acad Sci U S A. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobosz-Bartoszek M, Simonović M. Structure and mechanism of selenocysteine synthases. In: Hatfield DL, Schweizer U, Tsuji PA, Gladyshev VN, editors. Selenium: Its Molecular Biology and Role in Human Health. Springer Science +Business Media; New York: 2016. pp. 101–112. [Google Scholar]
- 48.Palioura S, Sherrer RL, Steitz TA, Söll D, Simonović M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science. 2009;325:321–325. doi: 10.1126/science.1173755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiba S, Itoh Y, Sekine S, Yokoyama S. Structural basis for the major role of O-phosphoseryl-tRNA kinase in the UGA-specific encoding of selenocysteine. Mol Cell. 2010;39:410–420. doi: 10.1016/j.molcel.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 50.Wang C, Guo Y, Tian Q, Jia Q, Gao Y, Zhang Q, Zhou C, Xie W. SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Res. 2015;43:10534–10545. doi: 10.1093/nar/gkv996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holman KM, Puppala AK, Lee JW, Lee H, Simonović M. Insights into substrate promiscuity of human seryl-tRNA synthetase. RNA. 2017;23:1685–1699. doi: 10.1261/rna.061069.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esaki N, Nakamura T, Tanaka H, Soda K. Selenocysteine lyase, a novel enzyme that specifically acts on selenocysteine. Mammalian distribution and purification and properties of pig liver enzyme. J Biol Chem. 1982;257:4386–4391. [PubMed] [Google Scholar]
- 53.Veres Z, Kim IY, Scholz TD, Stadtman TC. Selenophosphate synthetase. Enzyme properties and catalytic reaction. J Biol Chem. 1994;269:10597–10603. [PubMed] [Google Scholar]
- 54.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J. 2007;404:115–120. doi: 10.1042/BJ20070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forchhammer K, Leinfelder W, Böck A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- 56.Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leinfelder W, Forchhammer K, Zinoni F, Sawers G, Mandrand-Berthelot MA, Böck A. Escherichia coli genes whose products are involved in selenium metabolism. J Bacteriol. 1988;170:540–546. doi: 10.1128/jb.170.2.540-546.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirosawa-Takamori M, Chung HR, Jäckle H. Conserved selenoprotein synthesis is not critical for oxidative stress defence and the lifespan of Drosophila. EMBO Rep. 2004;5:317–322. doi: 10.1038/sj.embor.7400097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berry MJ, Banu L, Harney JW, Larsen PR. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ringquist S, Schneider D, Gibson T, Baron C, Böck A, Gold L. Recognition of the mRNA selenocysteine insertion sequence by the specialized translational elongation factor SELB. Genes Dev. 1994;8:376–385. doi: 10.1101/gad.8.3.376. [DOI] [PubMed] [Google Scholar]
- 61.Liu Z, Reches M, Groisman I, Engelberg-Kulka H. The nature of the minimal ‘selenocysteine insertion sequence’ (SECIS) in Escherichia coli. Nucleic Acids Res. 1998;26:896–902. doi: 10.1093/nar/26.4.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84:765–774. doi: 10.1016/s0300-9084(02)01405-0. [DOI] [PubMed] [Google Scholar]
- 63.Walczak R, Carbon P, Krol A. An essential non-Watson-Crick base pair motif in 3’UTR to mediate selenoprotein translation. RNA. 1998;4:74–84. [PMC free article] [PubMed] [Google Scholar]
- 64.Walczak R, Westhof E, Carbon P, Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 65.Fletcher JE, Copeland PR, Driscoll DM, Krol A. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA. 2001;7:1442–1453. [PMC free article] [PubMed] [Google Scholar]
- 66.Fradejas-Villar N, Seeher S, Anderson CB, Doengi M, Carlson BA, Hatfield DL, Schweizer U, Howard MT. The RNA-binding protein Secisbp2 differentially modulates UGA codon reassignment and RNA decay. Nucleic Acids Res. 2017;45:4094–4107. doi: 10.1093/nar/gkw1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howard MT, Aggarwal G, Anderson CB, Khatri S, Flanigan KM, Atkins JF. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 2005;24:1596–1607. doi: 10.1038/sj.emboj.7600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Howard MT, Moyle MW, Aggarwal G, Carlson BA, Anderson CB. A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. RNA. 2007;13:912–920. doi: 10.1261/rna.473907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Donovan J, Caban K, Ranaweera R, Gonzalez-Flores JN, Copeland PR. A novel protein domain induces high affinity selenocysteine insertion sequence binding and elongation factor recruitment. J Biol Chem. 2008;283:35129–35139. doi: 10.1074/jbc.M806008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caban K, Kinzy SA, Copeland PR. The L7Ae RNA binding motif is a multi-functional domain required for the ribosome-dependent Sec incorporation activity of Sec insertion sequence binding protein 2. Mol Cell Biol. 2007;27:6350–6360. doi: 10.1128/MCB.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bubenik JL, Miniard AC, Driscoll DM. Characterization of the UGA-recoding and SECIS-binding activities of SECIS-binding protein 2. RNA Biol. 2014;11:1402–1413. doi: 10.1080/15476286.2014.996472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takeuchi A, Schmitt D, Chapple C, Babaylova E, Karpova G, Guigo R, Krol A, Allmang C. A short motif in Drosophila SECIS binding protein 2 provides differential binding affinity to SECIS RNA hairpins. Nucleic Acids Res. 2009;37:2126–2141. doi: 10.1093/nar/gkp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mehta A, Rebsch CM, Kinzy SA, Fletcher JE, Copeland PR. Efficiency of mammalian selenocysteine incorporation. J Biol Chem. 2004;279:37852–37859. doi: 10.1074/jbc.M404639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mansell JB, Guevremont D, Poole ES, Tate WP. A dynamic competition between release factor 2 and the tRNA(Sec) decoding UGA at the recoding site of jt>Escherichia coli formate dehydrogenase H. EMBO J. 2001;20:7284–7293. doi: 10.1093/emboj/20.24.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kotini SB, Peske F, Rodnina MV. Partitioning between recoding and termination at a stop codon-selenocysteine insertion sequence. Nucleic Acids Res. 2015;43:6426–6438. doi: 10.1093/nar/gkv558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Allamand V, Richard P, Lescure A, Ledeuil C, Desjardin D, Petit N, Gartioux C, Ferreiro A, Krol A, Pellegrini N, Urtizberea JA, Guicheney P. A single homozygous point mutation in a 3′-untranslated region motif of selenoprotein N mRNA causes SEPN1-related myopathy. EMBO Rep. 2006;7:450–454. doi: 10.1038/sj.embor.7400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maiti B, Arbogast S, Allamand V, Moyle MW, Anderson CB, Richard P, Guicheney P, Ferreiro A, Flanigan KM, Howard MT. A mutation in the SEPN1 selenocysteine redefinition element (SRE) reduces selenocysteine incorporation and leads to SEPN1-related myopathy. Hum Mutat. 2009;30:411–416. doi: 10.1002/humu.20879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsuji PA, Carlson BA, Anderson CB, Seifried HE, Hatfield DL, Howard MT. Dietary selenium levels affect Selenoprotein expression and support the interferon-gamma and IL-6 immune response pathways in mice. Nutrients. 2015;7:6529–6549. doi: 10.3390/nu7085297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Howard MT, Carlson BA, Anderson CB, Hatfield DL. Translational redefinition of UGA codons is regulated by selenium availability. J Biol Chem. 2013;288:19401–19413. doi: 10.1074/jbc.M113.481051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Förster C, Ott G, Forchhammer K, Sprinzl M. Interaction of a selenocysteine-incorporating tRNA with elongation factor Tu from E coli. Nucleic Acids Res. 1990;18:487–491. doi: 10.1093/nar/18.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baron C, Böck A. The length of the aminoacyl-acceptor stem of the selenocysteine-specific tRNA(Sec) of Escherichia coli is the determinant for binding to elongation factors SELB or Tu. J Biol Chem. 1991;266:20375–20379. [PubMed] [Google Scholar]
- 83.Rudinger J, Hillenbrandt R, Sprinzl M, Giegé R. Antideterminants present in minihelix(Sec) hinder its recognition by prokaryotic elongation factor Tu. EMBO J. 1996;15:650–657. [PMC free article] [PubMed] [Google Scholar]
- 84.Leibundgut M, Frick C, Thanbichler M, Böck A, Ban N. Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 2005;24:11–22. doi: 10.1038/sj.emboj.7600505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paleskava A, Konevega AL, Rodnina MV. Thermodynamic and kinetic frame-work of selenocysteyl-tRNASec recognition by elongation factor SelB. J Biol Chem. 2010;285:3014–3020. doi: 10.1074/jbc.M109.081380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paleskava A, Konevega AL, Rodnina MV. Thermodynamics of the GTP-GDP-operated conformational switch of selenocysteine-specific translation factor SelB. J Biol Chem. 2012;287:27906–27912. doi: 10.1074/jbc.M112.366120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Itoh Y, Sekine S, Yokoyama S. Crystal structure of the full-length bacterial selenocysteine-specific elongation factor SelB. Nucleic Acids Res. 2015;43:9028–9038. doi: 10.1093/nar/gkv833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dobosz-Bartoszek M, Pinkerton MH, Otwinowski Z, Chakravarthy S, Söll D, Copeland PR, Simonović M. Crystal structures of the human elongation factor eEFSec suggest a non-canonical mechanism for selenocysteine incorporation. Nat Commun. 2016;7:12941. doi: 10.1038/ncomms12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin J, Gagnon MG, Bulkley D, Steitz TA. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell. 2015;160:219–227. doi: 10.1016/j.cell.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA. 2012;18:230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou XL, Ruan ZR, Huang Q, Tan M, Wang ED. Translational fidelity maintenance preventing Ser mis-incorporation at Thr codon in protein from eukaryote. Nucleic Acids Res. 2013;41:302–314. doi: 10.1093/nar/gks982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Y, Feng S, Kumar V, Ero R, Gao YG. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol. 2013;20:1077–1084. doi: 10.1038/nsmb.2645. [DOI] [PubMed] [Google Scholar]
- 95.Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gonzalez-Flores JN, Gupta N, DeMong LW, Copeland PR. The selenocysteine-specific elongation factor contains a novel and multi-functional domain. J Biol Chem. 2012;287:38936–38945. doi: 10.1074/jbc.M112.415463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sacerdot C, Dessen P, Hershey JW, Plumbridge JA, Grunberg-Manago M. Sequence of the initiation factor IF2 gene: unusual protein features and homologies with elongation factors. Proc Natl Acad Sci U S A. 1984;81:7787–7791. doi: 10.1073/pnas.81.24.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Keeling PJ, Fast NM, McFadden GI. Evolutionary relationship between translation initiation factor eIF-2gamma and selenocysteine-specific elongation factor SELB: change of function in translation factors. J Mol Evol. 1998;47:649–655. doi: 10.1007/pl00006422. [DOI] [PubMed] [Google Scholar]
- 99.Roll-Mecak A, Cao C, Dever TE, Burley SK. X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell. 2000;103:781–792. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 100.Rother M, Wilting R, Commans S, Böck A. Identification and characterisation of the selenocysteine-specific translation factor SelB from the archaeon Methanococcus jannaschii. J Mol Biol. 2000;299:351–358. doi: 10.1006/jmbi.2000.3756. [DOI] [PubMed] [Google Scholar]
- 101.Kjeldgaard M, Nissen P, Thirup S, Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993;1:35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- 102.Polekhina G, Thirup S, Kjeldgaard M, Nissen P, Lippmann C, Nyborg J. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure. 1996;4:1141–1151. doi: 10.1016/s0969-2126(96)00122-0. [DOI] [PubMed] [Google Scholar]
- 103.Jurnak F. Structure of the GDP domain of EF-Tu and location of the amino acids homologous to ras oncogene proteins. Science. 1985;230:32–36. doi: 10.1126/science.3898365. [DOI] [PubMed] [Google Scholar]
- 104.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVT, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crepin T, Shalak VF, Yaremchuk AD, Vlasenko DO, McCarthy A, Negrutskii BS, Tukalo MA, El’skaya AV. Mammalian translation elongation factor eEF1A2: X-ray structure and new features of GDP/GTP exchange mechanism in higher eukaryotes. Nucleic Acids Res. 2014;42:12939–12948. doi: 10.1093/nar/gku974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fernandez IS, Bai XC, Hussain T, Kelley AC, Lorsch JR, Ramakrishnan V, Scheres SH. Molecular architecture of a eukaryotic translational initiation complex. Science. 2013;342:1240585. doi: 10.1126/science.1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR, 3rd, Ibba M, Söll D. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 108.Kinzy SA, Caban K, Copeland PR. Characterization of the SECIS binding protein 2 complex required for the co-translational insertion of selenocysteine in mammals. Nucleic Acids Res. 2005;33:5172–5180. doi: 10.1093/nar/gki826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kossinova O, Malygin A, Krol A, Karpova G. A novel insight into the mechanism of mammalian selenoprotein synthesis. RNA. 2013;19:1147–1158. doi: 10.1261/rna.036871.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chavatte L, Brown BA, Driscoll DM. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat Struct Mol Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 111.Kossinova O, Malygin A, Krol A, Karpova G. The SBP2 protein central to selenoprotein synthesis contacts the human ribosome at expansion segment 7L of the 28S rRNA. RNA. 2014;20:1046–1056. doi: 10.1261/rna.044917.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Caban K, Copeland PR. Selenocysteine insertion sequence (SECIS)-binding protein 2 alters conformational dynamics of residues involved in tRNA accommodation in 80 S ribosomes. J Biol Chem. 2012;287:10664–10673. doi: 10.1074/jbc.M111.320929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fischer N, Neumann P, Bock LV, Maracci C, Wang Z, Paleskava A, Konevega AL, Schröder GF, Grubmüller H, Ficner R, Rodnina MV, Stark H. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature. 2016;540:80–85. doi: 10.1038/nature20560. [DOI] [PubMed] [Google Scholar]
- 114.Olieric V, Wolff P, Takeuchi A, Bec G, Birck C, Vitorino M, Kieffer B, Beniaminov A, Cavigiolio G, Theil E, Allmang C, Krol A, Dumas P. SECIS-binding protein 2, a key player in selenoprotein synthesis, is an intrinsically disordered protein. Biochimie. 2009;91:1003–1009. doi: 10.1016/j.biochi.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 115.Kim LK, Matsufuji T, Matsufuji S, Carlson BA, Kim SS, Hatfield DL, Lee BJ. Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure. RNA. 2000;6:1306–1315. doi: 10.1017/s1355838200000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim JY, Carlson BA, Xu XM, Zeng Y, Chen S, Gladyshev VN, Lee BJ, Hatfield DL. Inhibition of selenocysteine tRNA[Ser]Sec aminoacylation provides evidence that aminoacylation is required for regulatory methylation of this tRNA. Biochem Biophys Res Commun. 2011;409:814–819. doi: 10.1016/j.bbrc.2011.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hatfield D, Lee BJ, Hampton L, Diamond AM. Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res. 1991;19:939–943. doi: 10.1093/nar/19.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chittum HS, Baek HJ, Diamond AM, Fernandez-Salguero P, Gonzalez F, Ohama T, Hatfield DL, Kuehn M, Lee BJ. Selenocysteine tRNA[Ser]Sec levels and selenium-dependent glutathione peroxidase activity in mouse embryonic stem cells heterozygous for a targeted mutation in the tRNA[Ser]Sec gene. Biochemistry. 1997;36:8634–8639. doi: 10.1021/bi970608t. [DOI] [PubMed] [Google Scholar]
- 119.Moustafa ME, Carlson BA, El-Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, Mansur DB, Burk RF, Berry MJ, Diamond AM, Lee BJ, Gladyshev VN, Hatfield DL. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol Cell Biol. 2001;21:3840–3852. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Carlson BA, Moustafa ME, Sengupta A, Schweizer U, Shrimali R, Rao M, Zhong N, Wang S, Feigenbaum L, Lee BJ, Gladyshev VN, Hatfield DL. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J Biol Chem. 2007;282:32591–32602. doi: 10.1074/jbc.M707036200. [DOI] [PubMed] [Google Scholar]
- 121.Carlson BA, Xu XM, Gladyshev VN, Hatfield DL. Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J Biol Chem. 2005;280:5542–5548. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- 122.Mukai T, Englert M, Tripp HJ, Miller C, Ivanova NN, Rubin EM, Kyrpides NC, Söll D. Facile recoding of selenocysteine in nature. Angew Chem Int Ed Eng. 2016;55:5337–5341. doi: 10.1002/anie.201511657. [DOI] [PMC free article] [PubMed] [Google Scholar]


