This is the first report that miR-498 promotes tumor cell proliferation and migration by targeting PTEN. Our data provide new insight into the mechanism of PTEN inactivation in breast tumors and may lead to the development of novel anticancer therapies.
Abstract
Triple negative breast cancer (TNBC) is a subtype of breast cancer with a poor prognosis and high mortality rate. The tumor suppressor phosphatase and tensin homolog deleted on chromosome 10 (PTEN) plays an important role in cell proliferation and cell migration by negatively regulating the PI3K/Akt pathway. PTEN is downregulated by microRNAs in multiple cancers. However, few microRNAs have been reported to directly target PTEN in TNBC. In this study, microRNAs predicted to target PTEN were screened by immunoblotting and luciferase reporter assays. Expression levels of microRNA-498 (miR-498) were measured by TaqMan microRNA assays. We performed clonogenic, cell cycle and scratch wound assays to examine the oncogenic role of miR-498. We demonstrated that miR-498 directly targeted the 3′untranslated region of PTEN mRNA and reduced PTEN protein levels in TNBC cells. Compared with the non-tumorigenic breast epithelial cell line MCF-10A, TNBC cell lines overexpressed miR-498. Moreover, miR-498 promoted cell proliferation and cell cycle progression in TNBC cells in a PTEN-dependent manner. Suppressing miR-498 overexpression impaired the oncogenic effects of miR-498 on cell proliferation and cell migration. This study identified a novel microRNA (miR-498) overexpressed in TNBC cells and its oncogenic role in suppressing PTEN. These results provide new insight into the downregulation of PTEN and indicate a potential therapeutic target for treating TNBC.
Introduction
Breast cancer (BCa) is the most frequently diagnosed cancer in women worldwide (1). As a highly heterogeneous disease, BCa can be classified into intrinsic subtypes depending on the expression of molecular markers, such as the estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER-2)/Neu (2). Triple negative breast cancer (TNBC) is a subtype of BCa in which ER, PR and HER-2 are not expressed (2,3). Although TNBC cases account for only 10–15% of all BCa cases, the prognosis of TNBC patients is poor when compared with that of other BCa subtypes in which ER and PR expression is detected and for which hormone-replacement therapy can often be applied with good outcomes (4). Therefore, despite progress made for these other BCa subtypes, it is critical to identify novel potential therapeutic targets for treating TNBC.
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is a dual phosphatase and acts as a tumor suppressor via its dephosphorylation of phosphatidylinositol 3, 4,5-trisphosphate (PIP3) (5–8). PIP3 is an important second messenger that activates the kinase Akt (formerly protein kinase B) by phosphorylation (7). Activated Akt is a survival factor and is involved in cell proliferation and cell migration through the PI3K/Akt pathway (9–12). PTEN plays a crucial role in suppressing the development of cancer and is one of the most commonly inactivated tumor suppressors in BCa (5,6,13). PTEN inactivity is associated with large tumor sizes, considerable lymph node metastasis and an aggressive triple negative phenotype (13). PTEN expression is much lower in BCa tissue (57.5%, 84/117) than in normal breast tissue (100%, 10/10); however, PTEN mutation rates are approximately 5% in BCa, which indicates that other mechanisms negatively regulate PTEN expression in these tumors (14,15). Recent studies have revealed that microRNAs (miRNAs) can regulate PTEN expression and function (16–18).
miRNAs are a type of small non-coding RNA and are 19–25 nt in length (19). Although they do not encode proteins directly, miRNAs usually bind to the 3′untranslated region (3′UTR) of a protein-encoding mRNA and regulate protein expression by inhibiting target mRNA translation or promoting mRNA degradation (20,21). In prior studies, some miRNAs inactivated PTEN and acted as oncogenic players in multiple cancers (17,18,22,23). For instance, miR-221 and miR-222 were over-expressed in non–small cell lung cancer and hepatocellular carcinoma and promoted tumorigenesis by targeting PTEN (17). However, studies of PTEN regulation by miRNAs in TNBC are limited and provide unclear results.
In this study, to identify novel miRNAs targeting PTEN, we screened 19 miRNAs and discovered that microRNA-498 (miR-498) directly bound to the 3′UTR of PTEN mRNA to negatively regulate PTEN expression. Moreover, miR-498 played an oncogenic role in cell proliferation and migration in TNBC cell lines through suppressing PTEN. These results indicate that miR-498 could be used as a potential target for treating TNBC.
Materials and methods
Cell culture, DNA transfection and DNA transduction
Human HEK293, HEK293T and MCF-7 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). MDA-MB-231, MDA-MB-157, MCF-10A, BT-549, Hs578T and MDA-MB-453 were provided by Drs. David Brindley and Manijeh Pasdar (University of Alberta, Edmonton, AB, Canada). These cell lines were cultured and frozen in liquid nitrogen immediately upon arrival; the cells were routinely tested by PCR for mycoplasma contamination by the following primers: Myco_fw1: 5′-ACACCATGGGAGCTGGTAAT-3′, Myco_rev1: 5′-CTTCATCGACTTTCAGACCCAAGGCA-3′. MDA-MB-231, BT-549, Hs578T, MDA-MB-157 (TNBC model), HEK293, HEK293T and MCF-7 (ER+ and PR+) cells were transfected using the calcium phosphate method or retroviral transduction as previously described (24).
DNA constructs
The PTEN 3′UTR was cloned from HEK293 genomic DNA by PCR and inserted into the pMIR-REPORTTM luciferase expression vector (Ambion). The PTEN open reading frame (ORF) or PTEN ORF plus 3′UTR was cloned into the pBabe-MN-Ires-green fluorescent protein (GFP) expression vector. The sequences of all miRNAs were obtained from the ORIGENE online database (http://www.origene.com) and inserted in the pcDNA3.1 expression vector (Invitrogen). The empty expression vector or scramble-miR was considered as controls (Cont.-miR). For the miR-498 decoy construct, double strands of oligonucleotides containing six repeated sequences that were completely complementary to miR-498 were synthesized (Life Technologies) and inserted into the pMIR-REPORTTM luciferase reporter. The PTEN mutation was synthesized by Life Technologies. The PTEN-shRNA [AGTAGAGGAGCCGTCAAAT, (25)] was cloned into pSUPER-retro-puro vector (Oligoengine) according to the manufacturer’s instructions. All of these constructs were confirmed by DNA sequencing (U. Alberta).
Retroviral transduction
miR-498 was inserted in the pBabe-MN-Ires-GFP vector. The pBabe-MN-Ires-GFP-miR-498 and pCL-Ampho retrovirus packaging vectors were co-transfected into HEK293T cells at a ratio of 1:1, and the viral supernatants were harvested 48 h after transfection. Viral titers were 1 × 106 to 1 × 107 PFU/ml according to plaque assays using HEK293 cells (26). Viral supernatants with equal plaque forming units (PFUs) were added to cell dishes with 8 µg/ml polybrene to enhance viral transduction.
RNA extraction and quantitative reverse transcription PCR
Total RNA was extracted with Trizol (Invitrogen). To quantify mature miR-498, quantitative reverse transcription (qRT)-PCR was performed using the TaqMan microRNA assay (Applied Biosystems, miR-498 assay ID: 001044, U6 snRNA assay ID: 001973). The expression of miR-498 was normalized to the expression of U6 snRNA by the comparative Ct method as previously described (27).
Immunoblotting analysis and antibodies
Cells were lysed in radio-immune precipitation assay (RIPA) lysis buffer supplemented with a protease inhibitor mix (Roche). A total of 50 µg protein was loaded and run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (USA) gels and then transferred to polyvinylidene difluoride membranes for immunoblotting with antibodies. The antibodies used were anti-PTEN (Santa Cruz, SC-7974), anti-β-actin (Sigma, A3854), anti-p-Akt (Santa Cruz, SC-7985-R), anti-Akt (Santa Cruz, SC-5298), anti-GFP (Santa Cruz, SC-9996) and anti-Flag M5 (Sigma, F4042).
Clonogenic assay
Exponentially growing cells were trypsinized and counted. Appropriate numbers of cells were seeded onto 60 mm dishes and incubated for 10–14 days. Colonies with over 50 cells were counted to determine the colony number ratio. The colony number ratio was the average of three independent experiments.
MTT assay
BCa cells were transduced with miR-498 and PTEN ORF or siPTEN, respectively, and 5000 cells/well were then seeded onto 96 well plates. After 48 h, 50 µl MTT (5 mg/ml) was added to the media in each well and incubated in 37°C for 4 h. The media was removed and 150 µl MTT solution was added to each well, and after 30 min the optical density (OD) 550 value was measured.
Luciferase reporter assay
HEK293 cells were transfected with the constructed luciferase reporters, β-galactosidase plasmids and miRNA. After 48 h, the cells were lysed with reporter lysis buffer (Promega). The luciferase activity was measured and normalized to the β-galactosidase activity as previously described (28).
Cell cycle assay
MDA-MB-231 and BT-549 cells were transduced with miR-498. After 48 h, the cells were washed twice, fixed in 70% ethanol, treated with 50 µg/ml RNase A and labeled with 50 µg/ml propidium iodide for 3 h at 4°C. Then the cells were analyzed by flow cytometry (Becton Dickinson). The data were analyzed using FlowJo software (TreeStar).
Scratch wound assay
MDA-MB-231 and BT-549 cells were transduced with miR-498 or the miR-498 decoy. After 48 h, the cells were subjected to scratch wounding in a 24-well plate (28). Two wound tracks were introduced by scraping the cell monolayer with a pipette tip (20 µl), and the two tracks crossed at the center of the well. Phase-contrast images of the wounding areas were acquired at 0, 24 and 48 h after scratching by a charge-coupled device camera. Blank areas of the images were analyzed by the ImageJ MRI Wound Healing Tool.
Statistical analysis
Data are presented as the means ± standard deviation (SD). Statistical comparisons were performed using unpaired two-tailed Student’s t-tests.
Results
Identification of miRNA s regulate PTEN in BCa cell lines
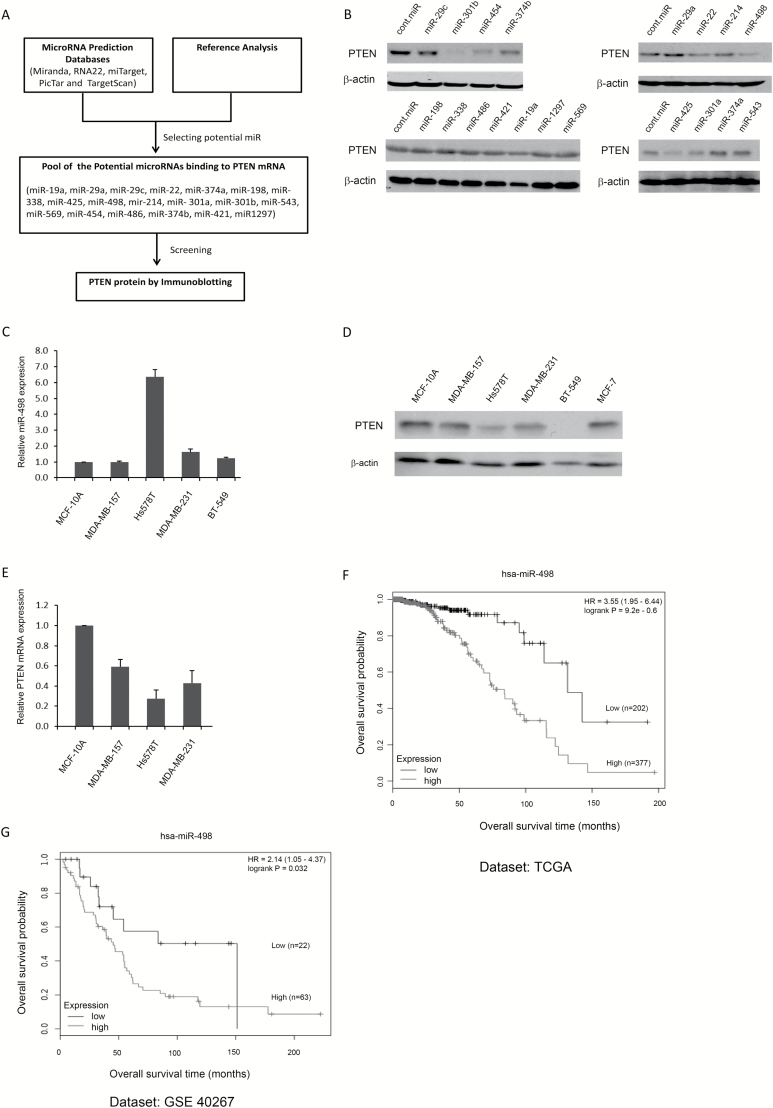
To identify miRNAs that could suppress PTEN protein levels, five miRNA databases (Miranda, RNA22, miTarget, PicTar and TargetScan) were accessed to predict miRNAs that could potentially bind to PTEN mRNA. Nineteen of the predicted miRNAs were selected for further screening (Figure 1A). The precursors of these 19 miRNAs were cloned into an expression vector and then transfected into BCa MCF-7 cells to examine changes in endogenous PTEN protein levels. Of these 19 miRNAs, 6 (miR-22, miR-425, miR-301b, miR-454, miR-374b and miR-498) downregulated PTEN in MCF-7 cells (Figure 1B). Because two miRNAs (miR-22 and miR-425) have been previously reported as PTEN-targeting miRNAs, the other four miRNAs were chosen for further characterization (29,30). The expression levels of miR-498 and the protein levels of PTEN in four TNBC cell lines (MDA-MB157, MDA-MB-231, Hs578T and BT-549), one ER+ and PR+ cell line (MCF-7) and one non-tumorigenic breast epithelial cell line (MCF-10A) were examined by immunoblotting and qRT-PCR. Compared with MCF-10A cells, Hs578T cells highly expressed miR-498 (6-fold increase), and there was a 1.64-fold increase in miR-498 expression in MDA-MB-231 cells (Figure 1C). Consistent with the observed miR-498 expression levels, PTEN levels were significantly lower in Hs578T and MDA-MB-231 cells than breast epithelial MCF-10A cells, but only slightly lower PTEN levels were found in MDA-MB-157 and MCF-7 cells (Figure 1D). The mRNA levels of PTEN in Hs578T and MDA-MB-231 were significantly lower than in MCF-10A (Figure 1E). Moreover, miR-498 expression was negatively correlated with the overall survival of patients with BCa. Kaplan–Meier survival curves revealed that patients with BCa with a high level of miR-498 had a significantly lower overall survival time [Figure 1F and G, kmplot.com, (31)]. Our findings suggested that there was probably an inverse correlation between miR-498 and PTEN expression levels in BCa cell lines.
Figure 1.
Immunoblotting for miRNA that regulate PTEN protein levels and miR-498 expression in BCa cell lines. (A) Summary of the screen for miRNA that regulate PTEN protein levels. (B) Endogenous PTEN protein levels were screened by immunoblotting after miRNA transfections in MCF-7 cells. (C) miR-498 expression profiles were determined by TaqMan probe qRT-PCR in the non-tumorigenic breast epithelial cell line MCF-10A and four TNBC cell lines as indicated. (D) Levels of endogenous PTEN protein expression were detected by western blotting in MCF-10A, MCF-7 and four TNBC cell lines. An antibody against β-actin was used as a loading control. (E) The levels of PTEN mRNA expression were detected by qRT-PCR in the indicated cell lines. (F and G) Kaplan–Meier survival curves generated from two datasets [TCGA (F) and GSE 40267 (G)] revealed that patients with BCa with high levels of miR-498 had a significantly lower overall survival (kmplot.com, 30 31). C and E are results obtained from three independent repeats.
miR-498 negatively regulates PTEN protein levels in TNBC cell lines
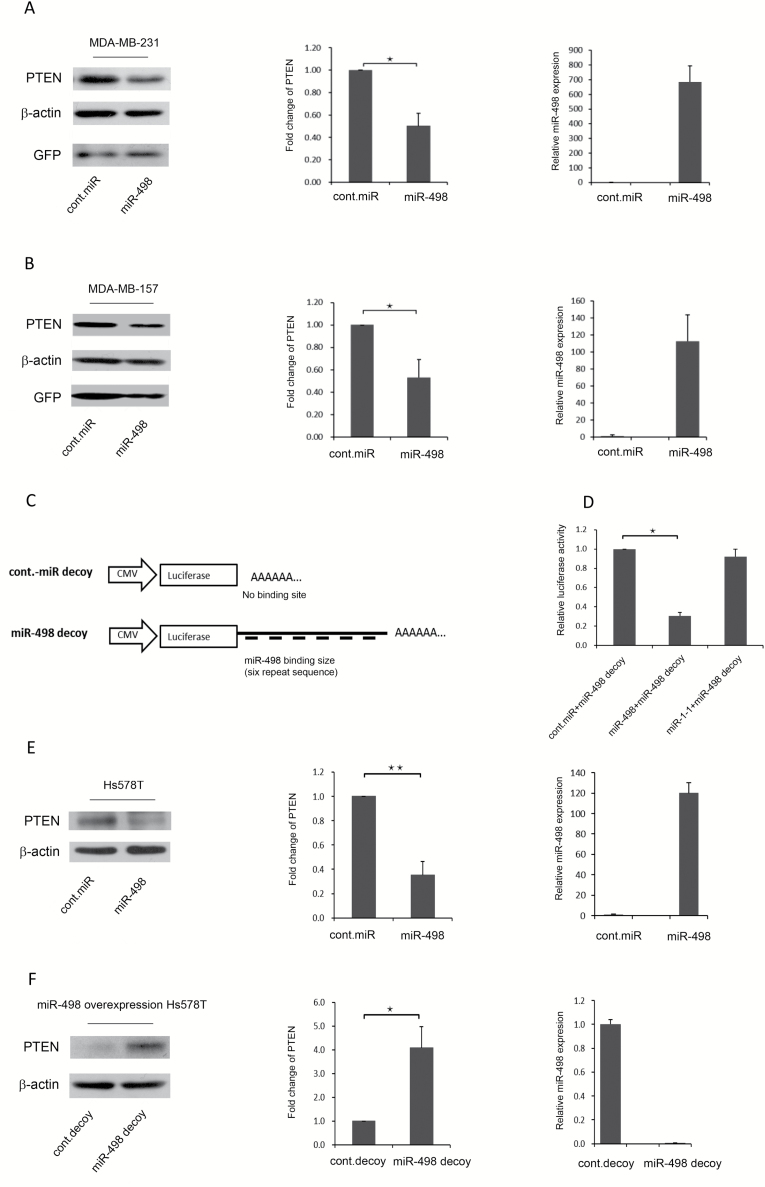
Because TNBC is the subtype of BCa with the highest rate of mortality, we investigated whether miR-498 overexpression reduced PTEN protein levels in MDA-MB-231 and MDA-MB-157 cells. miR-498 was transduced into MDA-MB-231 and MDA-MB-157 cells, and endogenous PTEN was examined by immunoblotting. Compared with control miRNA transduction, miR-498 overexpression led to a significant downregulation of endogenous PTEN protein and mRNA levels in MDA-MB-231 (Figure 2A, left and middle panels) and MDA-MB-157 cells (Figure 2B, left and middle panels). GFP was used as a transduction efficiency marker. In addition, miR-498 expression levels in transduced MDA-MB-231 and MDA-MB-157 cells were measured by qRT-PCR, and the results confirmed the overexpression of miR-498 [Figure 2A (right panel) and B (right panel)]. Together, these results suggested that miR-498 overexpression can downregulate PTEN and that miR-498 levels are negatively correlated with PTEN protein levels in TNBC cell lines.
Figure 2.
miR-498 negatively regulates PTEN protein levels in TNBC cell lines. (A) MDA-MB-231 cells were transduced with a retrovirus encoding control-miRNA or miR-498 as indicated. Levels of PTEN were detected by western blotting. GFP was used as a marker for retroviral transduction efficiency (left panel). Densitometry was performed with ImageJ software (NIH), and relative PTEN band intensities were normalized to β-actin. Relative fold changes in PTEN levels induced by miR-498 are shown (middle panel). miR-498 expression levels were measured by qRT-PCR (right panel). (B) Similar to A, except that MDA-MB-157 cells were used. (C) Structure of the miR-498 decoy. (D) miR-498 bound exclusively to the miR-498 decoy and suppressed target luciferase translation. (E) Hs578T cells were transfected with the miR-498 expression plasmid, and miR-498-overexpressing Hs578T cells were selected by hygromycin for 3 weeks. PTEN protein levels were detected by immunoblotting (left panel). Densitometry was performed with ImageJ software and relative PTEN band intensity was normalized to β-actin. The corresponding graph is a schematic of a densitometry analysis of a western blot (middle). Levels of miR-498 expression were measured by TaqMan qRT-PCR (right panel). (F) The miR-498 decoy was transduced into miR-498-overexpressing Hs578T cells. Endogenous miR-498 expression and PTEN protein levels were measured by immunoblotting (left panel). The fold change in PTEN protein levels was compared with the control decoy and the miR-498 decoy (middle). Levels of miR-498 expression were detected by qRT-PCR in the indicated cells (right panel). Error bars indicate the SD, *P < 0.01, ** P < 0.001 (n = 3); results are from three independent experiments.
To determine the functional roles of endogenous miR-498, a miR-498 decoy was constructed in which six tandem miR-498 sequences complementary to miR-498 were linked to a luciferase reporter gene [Figure 2C, (28)]. The luciferase activity of the miR-498 decoy was significantly reduced by miR-498 when miR-498 and the miR-498 decoy were co-transfected into HEK293 cells; however, the luciferase activity of the miR-498 decoy was not affected by either the control miRNA or miR-1-1 (Figure 2D). Our findings indicated that the miR-498 decoy was highly specific for miR-498 and was capable of binding to miR-498.
To test whether the miR-498 decoy could suppress endogenous miR-498 and restore PTEN expression, a stable miR-498 overexpressing Hs578T cell line was generated. The plasmid expressing miR-498 was transfected into Hs578T cells, and hygromycin (200 ng/ml) was used to select cells stably overexpressing miR-498; cells were also transfected with control miRNA and treated with the same concentration of hygromycin. After 2 weeks of selection, stable miR-498-overexpressing cells were obtained. PTEN protein levels were then measured by immunoblotting, and miR-498 overexpression was confirmed by qRT-PCR. Endogenous PTEN levels were significantly lower in the miR-498-overexpressing cells than in the stable control miRNA cells (Figure 2E, left and middle panels). miR-498 expression was confirmed by qRT-PCR (Figure 2E, right panel). Next, the miR-498 decoy was transduced into the stable miR-498-overexpressing Hs578T cells to investigate its role in suppressing miR-498 and restoring PTEN. As shown in Figure 2F (left and middle panels), compared with the control miRNA decoy, the miR-498 decoy significantly restored PTEN protein levels. miR-498 expression was also suppressed by the miR-498 decoy and reached only 0.25% of the miR-498 expression level in the control miRNA decoy-transduced cells (Figure 2F, right panel). These results indicated that the miR-498 decoy suppressed miR-498 and restored PTEN protein levels and that the miR-498 decoy could be used as an inhibitor of miR-498 overexpression.
PTEN is a direct target of miR-498
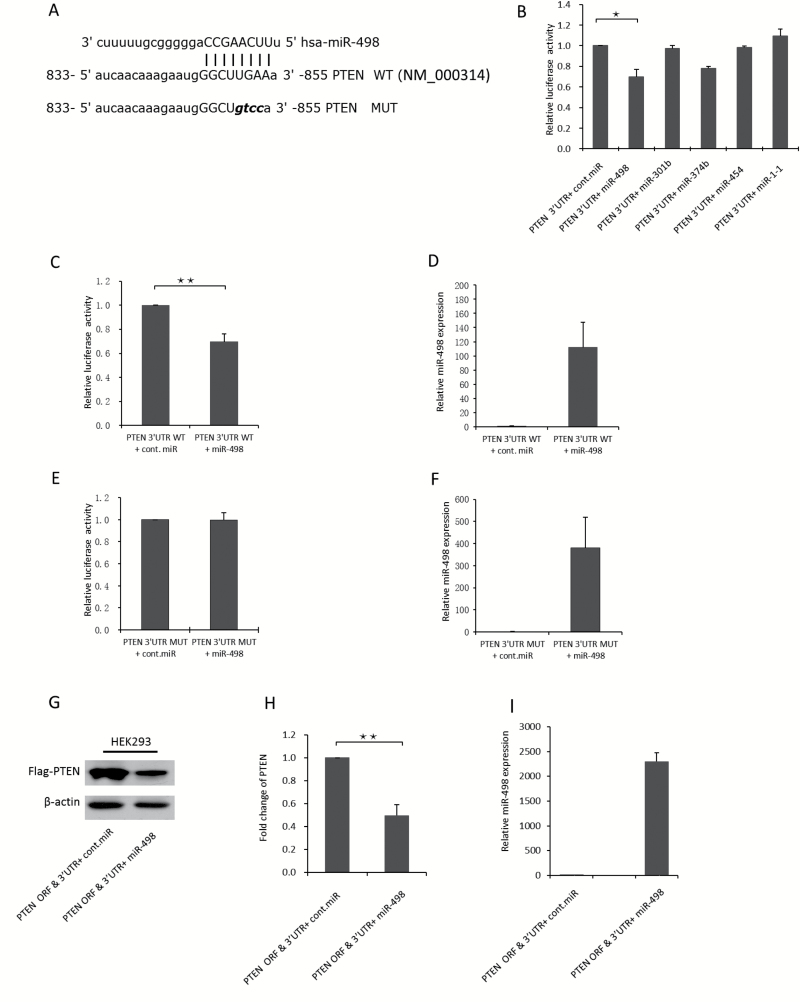
To identify the miR-498 binding site in the 3′UTR of PTEN mRNA, we used the microRNA.org database to search for the putative binding sites. Then, we cloned the putative binding sites into a luciferase reporter construct to examine their binding with miR-498 in vitro. The luciferase reporter and miR-498 were co-transfected into HEK293 cells, which have a high transfection efficiency and are widely used in validation of miRNA targets (32). After 48 h, the cells were lysed, and luciferase activity was measured. The results of the luciferase reporter assays revealed that the putative binding site at 833 to 855 bp on the 3′UTR of PTEN mRNA was the most dominant binding site (Figure 3A). Four miRNAs as indicated were co-transfected with the PTEN mRNA 3′UTR luciferase reporter into HEK293 cells to confirm their binding to the 3′UTR of PTEN mRNA. Of the four miRNAs, compared with the control miRNA, miR-498 significantly decreased luciferase reporter activity, which was consistent with miR-498 binding to the 3′UTR of PTEN mRNA (Figure 3B).
Figure 3.
PTEN is directly targeted by miR-498. (A) Predicted binding sites of miR-498 in the 3′UTR of PTEN. The mutated sequence (MUT) was changed by four DNA bases. (B) The binding of four potential miRNAs as indicated to the 3′UTR of PTEN mRNA was examined by luciferase assays. miR-1-1 was used as a negative control miRNA. Plasmids expressing each miRNA or control miRNA were co-transfected with a luciferase reporter containing the PTEN 3′UTR into HEK293 cells, and luciferase activities were measured. Error bars indicate the SD, * P < 0.05 (n = 2). (C) miR-498 binding to the wild-type 3′UTR of PTEN mRNA was determined by luciferase assay. Plasmids expressing miR-498 or control miRNA were co-transfected into HEK293 cells with a luciferase reporter containing the WT-PTEN 3′UTR; after 48 h, the cells were lysed, and luciferase activity was measured. (D) Levels of miR-498 expression were determined by TaqMan probe qRT-PCR. (E) miR-498 binding to the mutated 3′UTR of PTEN mRNA was determined by luciferase assay. Plasmids expressing miR-498 or control miRNA were co-transfected into HEK293 cells with a luciferase reporter containing the MUT PTEN 3′UTR; after 48 h, the cells were lysed, and luciferase activity was measured. (F) Levels of miR-498 expression were detected by TaqMan probe qRT-PCR. (G) miR-498 overexpression decreased the exogenous PTEN protein levels in HEK293 cells. ORF and 3′UTR PTEN constructs and miR-498 were co-transfected into HEK293 cells, and exogenous PTEN was examined by immunoblotting. (H) Exogenous PTEN levels were normalized to those of the loading control β-actin and compared with the control miRNA. (I) miR-498 overexpression was examined by TaqMan probe qRT-PCR. ** P < 0.001 (n = 3); the results are from three independent experiments.
Furthermore, the overexpression of miR-498 reduced luciferase activity by more than 30% compared with the control miRNA (Figure 3C and D). These results suggest that miR-498 can bind to this site to inhibit PTEN expression. Moreover, mutations to this binding site abolished the reduced luciferase activity induced by miR-498 overexpression (Figure 3E and F), which confirmed that miR-498 was capable of binding to this site on the 3′UTR of PTEN mRNA.
To evaluate the effect of miR-498 on PTEN protein levels in HEK293 cells, we generated a Flag-tagged PTEN construct that contained the ORF and 3′UTR of PTEN mRNA to mimic PTEN mRNA. We then co-transfected this PTEN construct with miR-498 into HEK293 cells. Flag-PTEN levels were examined by immunoblotting, and exogenous Flag-PTEN was significantly decreased by approximately 50% by the overexpression of miR-498 compared with control miRNA (Figure 3G and H). miR-498 expression levels were also measured by qRT-PCR (Figure 3I). Notably, the levels of PTEN protein did not change when cells were transfected with plasmids expressing the PTEN-coding sequence (without the 3′UTR of PTEN) along with miR-498 (data not shown). Similar results were obtained from H1299 and MCF-7 cells (data not shown). These results are consistent with miR-498 directly bound to the 3′UTR of PTEN mRNA to negatively regulate PTEN protein levels.
miR-498 plays an oncogenic role dependent on PTEN suppression in TNBC cells
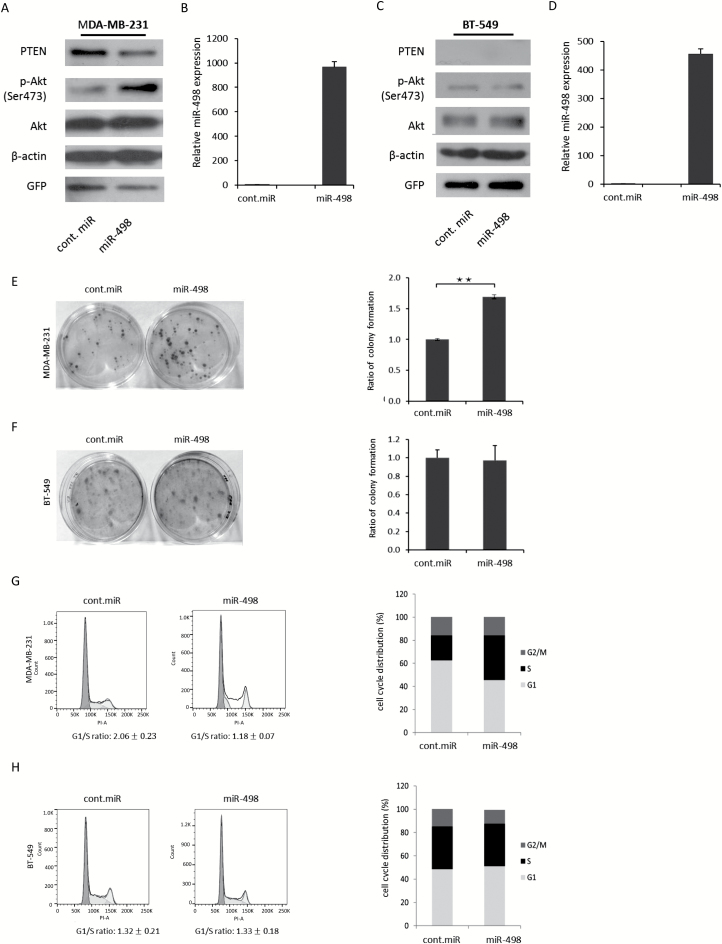
It has been reported that PTEN acts as a tumor suppressor and suppresses cell proliferation and cell cycle progression by negatively regulating the p-Akt signaling pathway (7,8,33,34). On the basis of the results described earlier, miR-498 overexpression suppressed PTEN in the TNBC cell lines MDA-MB-231 and MDA-MB-157 (Figure 2A and B) to exert an oncogenic effect in vitro. To determine whether the oncogenic effect of miR-498 is dependent on PTEN suppression and p-Akt activation, miR-498 was transduced into wild-type PTEN MDA-MB-231 cells and PTEN null BT-549 cells, and the levels of PTEN and p-Akt were measured by immunoblotting. Compared with the control miRNA cells, miR-498 suppressed PTEN and increased p-Akt levels in MDA-MB-231 cells (Figure 4A); the expression of miR-498 was determined by qRT-PCR (Figure 4B). However, no obvious changes in p-Akt protein levels were observed in PTEN null BT-549 cells overexpressing miR-498 (Figure 4C and D). To determine the oncogenic effects of miR-498 overexpression, cell clonogenic assays were performed. As shown in Figure 4E, miR-498 significantly increased the colony formation ratio in MDA-MB-231 cells. However, no significant changes in the colony formation ratio were found in BT-549 (PTEN-null) cells (Figure 4F). These results support that miR-498 promoted cell proliferation through suppressing PTEN and activating p-Akt signaling. PTEN has been reported to induce cell cycle arrest through negative regulation of the PI3K/Akt pathway (33,34). To determine the effects of miR-498 on promoting cell cycle progression in TNBC cells by suppressing PTEN, MDA-MB-231 and BT-549 cells were transduced with miR-498. The overexpression of miR-498 promoted cell cycle progression in MDA-MB-231 cells, and their G1/S ratio was 1.18; the G1/S ratio of cells transduced with the control miRNA was 2.06 (Figure 4G). However, no differences in the G1/S ratio were shown in PTEN null BT-549 cells transduced with miR-498 or control miRNA (Figure 4H), which suggested that miR-498-overexpression-induced cell cycle arrest was dependent on PTEN repression. Together, these results demonstrated that miR-498 promoted cell proliferation and cell cycle progression in a PTEN-dependent manner.
Figure 4.
miR-498 promotes cell proliferation and cell cycle arrest by suppressing PTEN and increasing p-Akt levels in TNBC cells. (A) MDA-MB-231 cells were transduced with the indicated expression plasmids and analyzed by immunoblotting with specific antibodies. (B) Levels of miR-498 expression were measured by qRT-PCR. (C) Similar to A, except that BT-549 (PTEN null) cells were used. (D) Levels of miR-498 expression were measured by qRT-PCR. (E) MDA-MB-231 cells were transduced with the indicated expression plasmids. Colony-forming assays were performed. Ratio of colony formation is shown in the right panel as indicated. (F) Similar to E, except that BT-549 cells were used. All experiments were performed in triplicate. *P < 0.05. **P < 0.01. (G) MDA-MB-231 cells were transduced with plasmids expressing miR-498 or control-miRNA. The cell cycle profile was determined by propidium iodide staining and flow cytometry (left panel). The cell cycle distribution was shown in the right panel. (H) Similar to G, except that BT-549 cells were used. **P < 0.001 (n = 3); the results are from three independent experiments.
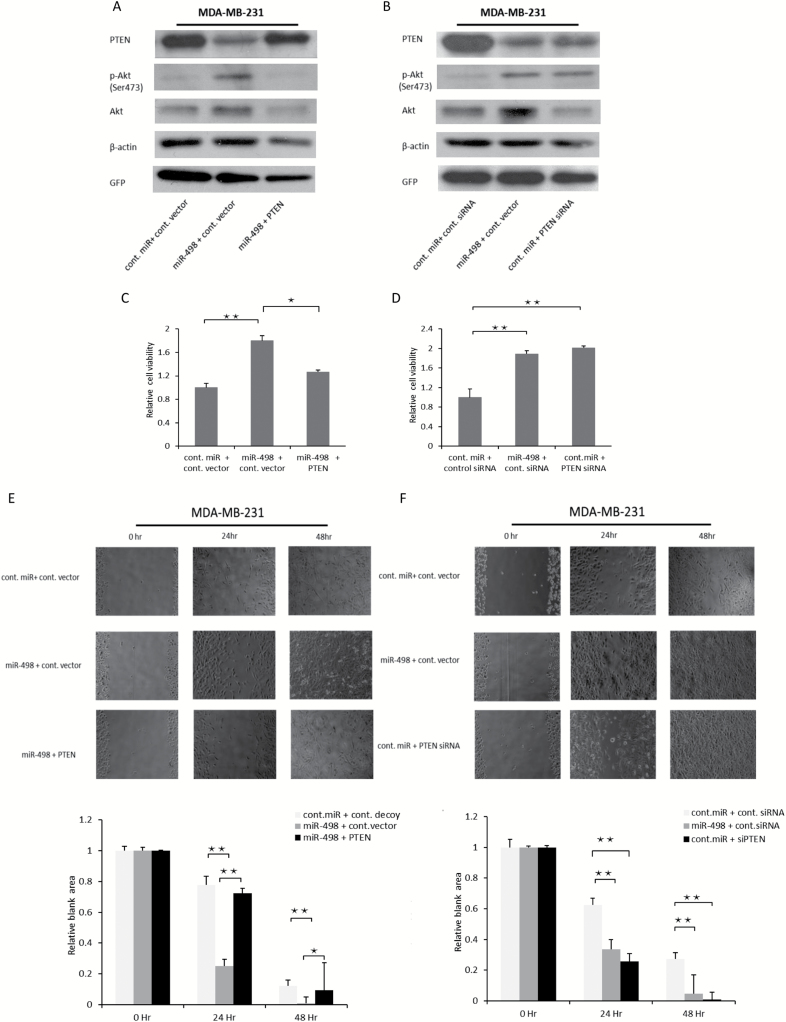
Introduction of the PTEN ORF rescues the phenotypes associated with miR-498 overexpression
As described earlier, overexpression of miR-498 exhibited an oncogenic effect. To determine whether the oncogenic effect of miR-498 overexpression was dependent on PTEN suppression, we introduced PTEN ORF to rescue the suppression of PTEN by miR-498 overexpression. As shown in Figure 5A, the decreased protein levels of PTEN by miR-498 overexpression were largely rescued by introducing PTEN ORF and the enhanced p-Akt levels were also inhibited by PTEN ORF in MDA-MB-231 cells. Furthermore, we compared the effect of miR-498 overexpression and siRNA (siPTEN) depletion of PTEN on cell proliferation and cell migration. Similar to the PTEN depletion, overexpression of miR-498 decreased the levels of PTEN protein and increased p-Akt levels (Figure 5B). As expected, cell viability was significantly enhanced by miR-498 overexpression and inhibited by the introduced PTEN (Figure 5C). Moreover, cell migration was significantly promoted by overexpression of miR-498 and inhibited by restored PTEN (Figure 5E). In terms of cell proliferation and cell migration, miR-498 overexpression was comparable to siPTEN in MDA-MB-231 cells (Figure 5D and F). Taken together, our results indicate that the introduced PTEN ORF rescues the phenotype of miR-498 overexpressing cells.
Figure 5.
Introduction of PTEN rescues miR-498-overexpression-induced phenotypes. (A) MDA-MB-231 cells were transfected with the miR-498 and PTEN ORF or control vector and analyzed by immunoblotting with indicated antibodies. (B) Similar to A, except that plasmids expressing miR-498 and PTEN-siRNA or control-siRNA were co-transfected into MDA-MB-231 cells. (C) MDA-MB-231 cells were transfected with the indicated expression plasmids and seeded on 96 well plates and the value of OD 550 nm was measured and normalized to the value of the control group after 48 h. (D) Similar to C, the indicated expression plasmids were transfected into MDA-MB-231 cells, and cell viability was examined by the MTT assay. Error bars indicated the SD, *P < 0.05, **P < 0.01 (n = 3) and results were the mean value of three independent experiments. (E) MDA-MB-231 cells were trypsinized and counted 2 days after transfection with the indicated plasmids. The scratch assay was carried out in a 24-well plate. The wound areas were tracked continuously for 48 h on the stage of a phase-contrast/fluorescence microscope (Zeiss Axiovert 100M) coupled to a charge-coupled device camera. The data were analyzed with Metamorph software (Molecular Devices); phase-contrast images of three to five selected fields were acquired at 24 and 48 h. Quantitative analyses of the open wound areas were measured using the ImageJ MRI Wound Healing Tool. The relative blank areas of the scratches were measured to evaluate cell migration, and statistical analyses of the cell migration are shown in the lower panel as indicated. (F) Similar to E, except that MDA-MB-231 cells were transfected with the different expression plasmids as indicated. Error bars indicated the SD, *P < 0.01, **P < 0.001 (n = 3) and results were the mean value of three independent experiments.
In addition, we compared miR-498 with miR-425, which is an oncogenic PTEN-targeted miRNA, as a positive control. We observed that both miR-498 and miR-425 decreased endogenous PTEN levels and increased p-Akt levels in MDA-MB-231 and MCF-7 (Supplementary Figure 1A and B, available at Carcinogenesis Online). In terms of cell proliferation and cell migration, miR-498 overexpression was on par with miR-425 in MDA-MB-231 and MCF-7 (Supplementary Figure 1C–F, available at Carcinogenesis Online).
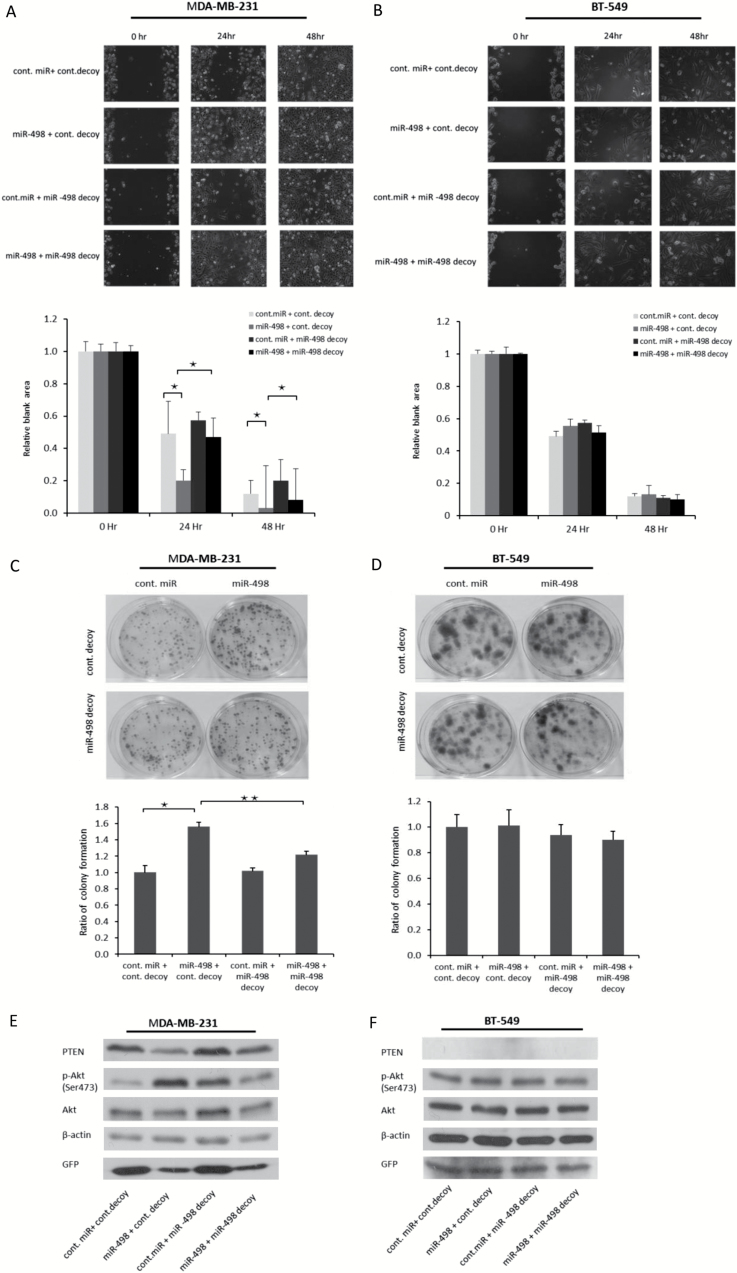
The PTEN-dependent oncogenic effect of miR-498 overexpression is inhibited by the miR-498 decoy
It has been established that PTEN suppresses cell migration (35,36). To determine whether miR-498 overexpression affects cell migration through suppressing PTEN, we used wound-healing assays to visualize the migration of TNBC MDA-MB-231 and BT-549 cells transduced with miR-498 or the miR-498 decoy. As shown in Figure 6A, 24 h after wounding, miR-498 overexpression significantly enhanced the migration of MDA-MB-231 cells (PTEN wt) compared with that of cells transduced with control miRNA or the control miRNA decoy. However, the miR-498 decoy blocked the enhanced cell migration induced by miR-498 overexpression. These results revealed that miR-498 overexpression promoted cell migration and that the miR-498 decoy could be used as an anti-miR-498 agent to inhibit the function of miR-498. The relative blank areas of the wounds were measured to evaluate cell migration. Statistical analyses of the cell migration are shown in the lower panel of Figure 6A. Furthermore, in BT-549 cells (PTEN null) at 24 h, miR-498 overexpression did not enhance cell migration, which suggested that the enhanced cell migration was dependent on PTEN. In addition, no significant differences in cell migration were found between miR-498-overexpressing cells and miR-498 decoy-expressing cells (Figure 6B).
Figure 6.
The oncogenic effect of miR-498 overexpression was inhibited by the miR-498 decoy. (A) MDA-MB-231 and (B) BT-549 cells were trypsinized and counted 2 days after transfection with indicated plasmids. The scratch assay was carried out as described in Figure 5E. The relative blank areas of the scratches were measured to evaluate cell migration, and statistical analyses of the cell migration are shown in the lower panel of A and B. *P < 0.05. (C). MDA-MB-231 cells were transfected with the indicated expression plasmids. Colony-forming assays were performed. Ratio of colony formation is shown in the lower panel as indicated. All experiments were performed in triplicate. *P < 0.05. **P < 0.01. (D) Similar to C, except that BT-549 cells were used. (E and F) miR-498 and the miR-498 decoy were transduced into MDA-MB-231 (E) and BT-549 (F) cells. The levels of PTEN and p-Akt were determined by immunoblotting as indicated. *P < 0.01 (n = 3).
To confirm that the oncogenic effect of miR-498 overexpression on cell proliferation was inhibited by the miR-498 decoy, miR-498 and the miR-498 decoy were transduced into the TNBC cell lines MDA-MB-231 and BT-549. As shown in Figure 6C, miR-498 increased the colony numbers in MDA-MB-231 cells, whereas the miR-498 decoy significantly reduced the colony numbers. These results support that miR-498 overexpression promoted proliferation in MDA-MB-231 cells, and this oncogenic effect was inhibited by the miR-498 decoy. In contrast, in BT-549 cells (PTEN null), no significant changes in the colony numbers were found after transducing miR-498 or the miR-498 decoy (Figure 6D). These results indicated that the oncogenic role of miR-498 was PTEN-dependent in vitro. In addition, the protein levels of PTEN and p-Akt in MDA-MB-231 and BT-549 cells transduced with miR-498 or the miR-498 decoy were measured. Consistently, p-Akt was activated by miR-498 overexpression and suppressed by the miR-498 decoy in PTEN wild-type MDA-MB-231 cells (Figure 6E). However, in PTEN null BT-549 cells, p-Akt levels were not changed by miR-498 overexpression (Figure 6F). On the basis of these data, we concluded that the oncogenic role of miR-498 was PTEN-dependent and can be inhibited by the miR-498 decoy.
Discussion
The tumor suppressor PTEN is a negative regulator of the PI3K/Akt pathway and is involved in cell proliferation, cell migration and cell cycle arrest (7,8,27,33–37). PTEN has been reported to be absent or expressed at low levels in multiple cancers, e.g. prostate cancer, gastric cancer, colorectal cancer, non–small cell lung cancer and BCa (38–41). Recent studies have shown that reduced PTEN expression is more common in BCa tissues than normal breast tissues and is positively associated with tumor size, lymph node metastasis and an aggressive triple-negative phenotype. Taken together, these findings support a role that the tumor suppressor PTEN plays in the development of TNBC (13). Due to its poor prognosis and the lack of effective therapies, TNBC is the most lethal subtype of BCa (3). The median survival of metastatic TNBC patients is only 13 months (42). Therefore, it is urgent and necessary to find effective targets to prevent the progression of TNBC. In this study, we aimed to investigate the relation of TNBC and oncogenic miRNAs that can suppress PTEN.
miRNAs are one of the mechanisms through which PTEN can be inactivated. In fact, PTEN is suppressed by miRNAs in multiple cancers. Meng et al. (43) revealed that miR-21 downregulated PTEN in hepatocellular cancer, and Nip et al. (44) identified that overexpressing the oncogenic miR-4534 negatively regulated PTEN in prostate cancer. However, in TNBC, studies of PTEN regulation by miRNAs are limited. In this study, we screened predicted PTEN-targeting miRNAs and identified that miR-498 is overexpressed in TNBC cell lines and suppresses PTEN by directly binding to the 3′UTR of PTEN mRNA.
The role of miR-498 in cancers appears to be unclear. miR-498 is expressed at low levels in non–small cell lung cancer and ovarian cancer (45,46). However, Matamala et al.(47) reported miR-498 overexpression in TNBC tissues according to microarray analyses, and we detected miR-498 overexpression in TNBC cell lines according to the qRT-PCR analyses in our study. In addition, miR-498 is significantly higher in metastatic medullary thyroid carcinoma than in primary medullary thyroid carcinoma (48). These results indicate the diversity of miR-498 expression profiles in different tissues. Furthermore, in addition to PTEN, other targets of miR-498 have been reported. For instance, telomerase reverse transcriptase enhances cell growth in an ER-α-dependent manner. Vitamin D–induced miR-498 overexpression targets telomerase reverse transcriptase mRNA and suppresses telomerase reverse transcriptase levels in ovarian cancer cells, and miR-498 functions as a tumor suppressor (49). However, in TNBC, which has no ER-α expression, miR-498 is highly expressed in tissues and cell lines and negatively regulates the tumor suppressor PTEN, which contributes to oncogenesis. Therefore, the diversity of miR-498 targets and expression levels might explain the conflicting reports of the effects of miR-498.
In conclusion, our data revealed that miR-498 is overexpressed in TNBC cells and acts as an oncogenic factor by negatively regulating the tumor suppressor PTEN. Our findings demonstrated that the oncogenic effect of miR-498 is dependent on PTEN suppression. This study expands our understanding of the function of miR-498 and provides new information for the development of TNBC therapies.
Supplementary material
Supplementary Figure 1 can be found Carcinogenesis online.
Funding
This work was supported by grants from the Women & Children’s Health Research Institute (WCHRI), the Alberta Heritage Foundation for Medical Research (AHFMR), the Canadian Breast Cancer Foundation and the Canadian Institutes of Health Research (CIHR) to R.P.L. C.C was supported by a Scholarship from China Scholarship Council. D.D.E. holds the Muriel & Ada Hole Kids with Cancer Society Chair in Pediatric Oncology, University of Alberta.
Supplementary Material
Acknowledgements
We thank Dr. Wei Wu and Dr. Abou Zeinab in our laboratory for their technical assistance. We thank Dr. David Brindley and Dr. Manijeh Pasdar for generously providing us several breast cancer cell lines (BT 549, MDA-MB157, Hs578T). Conception and design: R.P.L. and C.C.; Development of methodology: C.C., H.W., B.W. and R.P.L; Analysis and interpretation of the data: C.C., H.W., B.W., D.D.E. and R.P.L.; Writing, review and revision of the manuscript: C.C., D.D.E. and R.P.L. All authors have read the final version.
Conflict of Interest Statement: None declared. No potential competing interests are disclosed.
Abbreviations
- 3′UTR
3′untranslated region
- BCa
breast cancer
- ER
estrogen receptor
- GFP
green fluorescent protein
- miR-498
microRNA-498
- miRNA
microRNA
- ORF
open reading frame
- PR
progesterone receptor
- PTEN
phosphatase and tensin homolog deleted on chromosome 10
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- TNBC
triple negative breast cancer
References
- 1. Garcia M., et al. (2007)Global Cancer Facts & Figures 2007. American Cancer Society, Atlanta, GA: 1, p. 52. [Google Scholar]
- 2. Brenton J.D., et al. (2005)Molecular classification and molecular forecasting of breast cancer: ready for clinical application?J. Clin. Oncol., 23, 7350–7360. [DOI] [PubMed] [Google Scholar]
- 3. Dent R., et al. (2007)Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res., 13, 4429–4434. [DOI] [PubMed] [Google Scholar]
- 4. Anders C.K., et al. (2009)Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin. Breast Cancer, 9 (suppl. 2, S73–S81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Steck P.A., et al. (1997)Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet., 15, 356–362. [DOI] [PubMed] [Google Scholar]
- 6. Li J., et al. (1997)PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science, 275, 1943–1947. [DOI] [PubMed] [Google Scholar]
- 7. Maehama T., et al. (1998)The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem., 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- 8. Stambolic V., et al. (1998)Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell, 95, 29–39. [DOI] [PubMed] [Google Scholar]
- 9. Datta S.R., et al. (1999)Cellular survival: a play in three Akts. Genes Dev., 13, 2905–2927. [DOI] [PubMed] [Google Scholar]
- 10. Romashkova J.A., et al. (1999)NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature, 401, 86–90. [DOI] [PubMed] [Google Scholar]
- 11. Zhou B.P., et al. (2001)HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol., 3, 973–982. [DOI] [PubMed] [Google Scholar]
- 12. Ogawara Y., et al. (2002)Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J. Biol. Chem., 277, 21843–21850. [DOI] [PubMed] [Google Scholar]
- 13. Li S., et al. (2017)Loss of PTEN expression in breast cancer: association with clinicopathological characteristics and prognosis. Oncotarget, 8, 32043–32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang H.Y., et al. (2013)PTEN mutation, methylation and expression in breast cancer patients. Oncol. Lett., 6, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rhei E., et al. (1997)Mutation analysis of the putative tumor suppressor gene PTEN/MMAC1 in primary breast carcinomas. Cancer Res., 57, 3657–3659. [PubMed] [Google Scholar]
- 16. Pezzolesi M.G., et al. (2008)Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in Cowden syndrome. The Ame. J. Human Genetics, 82, 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garofalo M., et al. (2009)miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell, 16, 498–509. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Huse J.T., et al. (2009)The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev., 23, 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pang K.C., et al. (2007)RNAdb 2.0–an expanded database of mammalian non-coding RNAs. Nucleic Acids Res., 35(Database issue), D178–D182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pillai R.S., et al. (2005)Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science, 309, 1573–1576. [DOI] [PubMed] [Google Scholar]
- 21. Olsen P.H., et al. (1999)The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol., 216, 671–680. [DOI] [PubMed] [Google Scholar]
- 22. Darido C., et al. (2011)Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell, 20, 635–648. [DOI] [PubMed] [Google Scholar]
- 23. Yang H., et al. (2008)MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res., 68, 425–433. [DOI] [PubMed] [Google Scholar]
- 24. Wu H., et al. (2011)UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat. Med., 17, 347–355. [DOI] [PubMed] [Google Scholar]
- 25. Bononi A., et al. (2013)Identification of PTEN at the ER and MAMs and its regulation of Ca(2+) signaling and apoptosis in a protein phosphatase-dependent manner. Cell Death Differ., 20, 1631–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rowe W.P., et al. (1970)Plaque assay techniques for murine leukemia viruses. Virology, 42, 1136–1139. [DOI] [PubMed] [Google Scholar]
- 27. Livak K.J., et al. (2001)Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 28. Wang B., et al. (2017)MicroRNA-1301 suppresses tumor cell migration and invasion by targeting the p53/UBE4B pathway in multiple human cancer cells. Cancer Lett., 401, 20–32. [DOI] [PubMed] [Google Scholar]
- 29. Ma J., et al. (2014)NF-kappaB-dependent microRNA-425 upregulation promotes gastric cancer cell growth by targeting PTEN upon IL-1β induction. Mol. Cancer, 13, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bar N., et al. (2010)miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PLoS One, 5, e10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lánczky A., et al. (2016)miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat., 160, 439–446. [DOI] [PubMed] [Google Scholar]
- 32. Hansen T.B., et al. (2013)Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388. [DOI] [PubMed] [Google Scholar]
- 33. Li D.M., et al. (1998)PTEN/MMAC1/TEP1 suppresses the tumorigenicity and induces G1 cell cycle arrest in human glioblastoma cells. Proc. Natl. Acad. Sci. USA, 95, 15406–15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun H., et al. (1999)PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl. Acad. Sci. USA, 96, 6199–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suzuki A., et al. (2003)Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J. Exp. Med., 197, 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liliental J., et al. (2000)Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr. Biol., 10, 401–404. [DOI] [PubMed] [Google Scholar]
- 37. Groszer M., et al. (2001)Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science, 294, 2186–2189. [DOI] [PubMed] [Google Scholar]
- 38. Whang Y.E., et al. (1998)Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc. Natl. Acad. Sci. USA, 95, 5246–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soria J.C., et al. (2002)Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin. Cancer Res., 8, 1178–1184. [PubMed] [Google Scholar]
- 40. Kang Y.H., et al. (2002)Promoter methylation and silencing of PTEN in gastric carcinoma. Lab. Invest., 82, 285–291. [DOI] [PubMed] [Google Scholar]
- 41. Goel A., et al. (2004)Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res., 64, 3014–3021. [DOI] [PubMed] [Google Scholar]
- 42. Kassam F., et al. (2009)Survival outcomes for patients with metastatic triple-negative breast cancer: implications for clinical practice and trial design. Clin. Breast Cancer, 9, 29–33. [DOI] [PubMed] [Google Scholar]
- 43. Meng F., et al. (2007)MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology, 133, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nip H., et al. (2016)Oncogenic microRNA-4534 regulates PTEN pathway in prostate cancer. Oncotarget, 7, 68371–68384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cong J., et al. (2015)Low miR-498 expression levels are associated with poor prognosis in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci., 19, 4762–4765. [PubMed] [Google Scholar]
- 46. Wang M., et al. (2015)MicroRNA-498 is downregulated in non-small cell lung cancer and correlates with tumor progression. J. Cancer Res. Ther., 11 (suppl 1, C107–C111. [DOI] [PubMed] [Google Scholar]
- 47. Matamala N., et al. (2016)MicroRNA deregulation in triple negative breast cancer reveals a role of miR-498 in regulating BRCA1 expression. Oncotarget, 7, 20068–20079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Santarpia L., et al. (2013)A miRNA signature associated with human metastatic medullary thyroid carcinoma. Endocr. Relat. Cancer, 20, 809–823. [DOI] [PubMed] [Google Scholar]
- 49. Kasiappan R., et al. (2012)1,25-Dihydroxyvitamin D3 suppresses telomerase expression and human cancer growth through microRNA-498. J. Biol. Chem., 287, 41297–41309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.