ABSTRACT
Transcription by RNA polymerase II (Pol II) is accomplished with the aid of numerous accessory factors specific to each transcriptional stage. The structure of the Pol II elongation complex (EC) bound with Spt4/5, Elf1, and TFIIS unveiled the sophisticated basal EC architecture essential for transcription elongation and other transcription-related events.
KEYWORDS: RNA polymerase II, transcription elongation, elongation complex, Spt4/5, Elf1, TFIIS, X-ray crystallography, cryo-EM
Introduction
RNA polymerase II (Pol II) governs the intricate process of eukaryotic gene transcription through three major stages: initiation, elongation, and termination. While Pol II itself is a huge protein complex composed of 12 subunits, it associates with many other protein factors to form higher-order complexes, which are prerequisite for nuclear transcription. Many of these accessory factors are recruited to the transcription complex in a stage-specific fashion, and play the essential roles required for each particular stage. Recently, with the rapid evolution of cryo-electron microscopy (cryo-EM) technology, several near-atomic structures of such higher-order transcription complexes of Pol II have been reported, especially those involved in transcription initiation [1]. In contrast, the structural information of higher-order complexes formed during the elongation stage is still limited. Recently, we elucidated the structure of the Pol II elongation complex (EC) from the yeast Komagataella pastoris, in the complex with “basal” elongation factors [2]. The structure presented the first basic architecture of an EC operating within cells, and especially shedded new light on Spt4/5 and Elf1, which extend the Pol II structure and capability as integral EC components for processive transcription, regulation, and various transcription-coupled events.
The “basal” elongation factors
In the elongation stage, numerous proteins and protein complexes with diverse functions are associated with the transcribing Pol II. Among these, three elongation factors, Spt4/5, Elf1, and TFIIS, are conserved not only in eukaryotes, but also in archaea (Spt5 even in bacteria). Therefore, they were considered to play the most general and basic roles in transcription elongation. Spt4/5 is a heterodimer of Spt4 and Spt5, and supports processive RNA elongation by suppressing transcriptional pause/arrest [3,4]. In higher eukaryotes, it is also known as DRB sensitivity-inducing factor (DSIF), and is involved in the promoter-proximal pausing together with negative elongation factor (NELF) [3,5]. Spt5 is composed of multiple, tandemly connected domains. The NusG N-terminal homology (NGN) domain is conserved in all three domains of life, and forms a tight complex with the zinc-finger protein Spt4 in eukaryotes and archaea [6]. In contrast, the Kyrpides-Ouzounis-Woese (KOW) domains are diverse: the bacterial NusG and archaeal Spt5 proteins typically have only one KOW domain, whereas the eukaryotic Spt5 has multiple KOW repeats, and additionally contains an intrinsically disordered C-terminal repeat region (CTR). Depending on its phosphorylation state, the CTR recruits various factors involved in mRNA processing, chromatin remodeling, and DNA repair [7–9].
Elf1 (elongation factor 1; Elof1 in humans) is a small zinc-finger protein with a molecular mass of ∼10 kDa, and is conserved in eukaryotes and several archaea. Although Elf1's function in transcription had been enigmatic due to limited studies, several lines of evidence implied its participation in the Pol II elongation complex (EC). In yeast, Elf1 showed synthetic-lethality with several elongation factors, including Spt4/5 and TFIIS, and the knockdown of Elf1 impaired the chromatin stability in actively transcribed genes [10]. In addition, the presence of Elf1 in some archaeal species raised the speculation that Elf1 could participate in functions independent from chromatin maintenance [11]. While the direct binding with Pol II was not confirmed in the initial study [10], chromatin immunoprecipitation (ChIP) analyses suggested that Elf1 is associated with transcribing Pol II [10,12].
TFIIS is a well-known elongation factor conserved in eukaryotes and archaea. It reactivates an arrested EC by stimulating the transcript cleavage activity of Pol II, similar to its bacterial functional counterparts GreA and GreB [13]. Several structures of Pol II bound with TFIIS have been reported. TFIIS inserts its C-terminal domain into the secondary channel of Pol II, to remodel the active site [14,15].
The architecture of the Pol II EC
By combining X-ray crystallography and cryo-EM single particle reconstruction, we elucidated the structure of the Pol II EC bound with the three basal elongation factors, Spt4/5, Elf1, and TFIIS (Figure 1(A)). The elongation factors bind and modify a wide area of the Pol II surface, establishing a functional EC as a whole. In particular, Elf1 and Spt4/5 bind near the DNA/RNA binding sites of Pol II, reinforcing or newly establishing the paths for DNA and RNA. Within the Pol II main cleft, Elf1, the NGN domain of Spt5, and Spt4 are aligned along the DNA, covering it from the downstream to the upstream region through the transcription bubble. At the downstream part of the DNA, Elf1 seals the Pol II cleft, completing the “DNA-entry tunnel”. Meanwhile, Spt4 and the NGN domain of Spt5, together with the KOW1 domain of Spt5, surround the upstream part of the DNA, establishing the “DNA-exit tunnel”. These interactions establish a continuous tunnel covering ∼30 bp of the DNA (positions –15 to +16), defining the size of the transcription bubble. Spt5 also modifies and remodels the RNA exit tunnel of Pol II by its KOW domains. The KOW5 domain of Spt5 binds at the RNA exit site, and directs the nascent RNA together with the KOW4 domain. The Spt5 KOW1 domain physically separates the DNA and RNA exits, preventing possible DNA-RNA interactions, such as R-loop formation. These features represent the basic architecture essential for accomplishing processive transcription, and counteracting pauses or premature termination.
Figure 1.
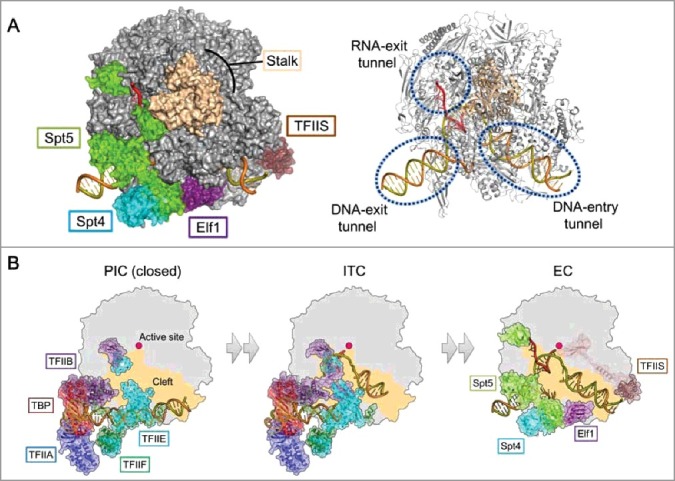
(A) Overall structure of the Pol II-EC [2]. Transcription factors are removed in the right panel to show the internal nucleic acids. (B) Structural transition from the preinitiation complex to the elongation complex. Left: Preinitiation complex (closed form), based on PDB 5FZ5 [17]. Middle: Initial transcribing complex, based on PDB 4V1N [16]. Right: Pol II EC [2]. The TFIIF dimerization domain and Spt5 KOW4 are removed for clarity.
Structural change during transition from initiation to elongation
The architecture of the Pol II EC drastically differs from those of the reported Pol II initiation complexes [16,17]. A structural comparison revealed that Spt4, Spt5, and Elf1 in the elongation complex share overlapping locations with general transcription factors (GTFs) and DNA in the initiation complexes (Figure 1(B)). This indicates that the binding of the elongation factors and GTFs is mutually exclusive, and they must be exchanged during the transition from the initiation to the early elongation stages of transcription. The exchange of protein factors rearranges the Pol II complex architecture, switching its functions from those required for transcription initiation to those for transcription elongation. The elongation factors should stabilize the established EC, preventing possible bubble collapse and dissociation of DNA/RNA, while providing the basic framework for regulation and transcription-coupled events. The details underlying this rearrangement remain as important questions to be answered.
Elf1 and the DNA-entry tunnel
Elf1 intervenes between the Pol II clamp and lobe, to keep the downstream DNA held within the DNA-entry tunnel. Although it was disordered in the structure, the Elf1 N-terminal tail, which is rich in basic amino-acid residues, could interact with the downstream DNA to gain additional DNA affinity. It is notable that Elf1 binds to a basic patch of the Pol II lobe, which serves as the DNA-binding site (“outer” DNA-binding site) in the pre-initiation complex (closed form) [17]. Elf1 binds through its C-terminal helix, which is rich in acidic residues (the “Pol II-binding helix”). Thus, Elf1 blocks the outer DNA-binding site required for transcription initiation, while it directs and holds the DNA within the “inner” binding site, which is essential for transcription elongation. Except for its role as an integral component of the elongation complex, the biological functions of Elf1 remain enigmatic. As Elf1 constitutes the downstream edge of the EC and genetically interacts with Spt4, Spt5, Spt6, TFIIS, and the Paf1 complex [10], it might be implicated in chromatin-mediated transcription or chromatin remodeling.
Spt4/5 and the DNA-exit tunnel
The Spt5 NGN domain forms a tight complex with Spt4, and mediates the Rpb1 clamp and Rpb2 protrusion domains. This binding site is conserved in the archaeal Spt5 and bacterial NusG [18–21]. Many basic residues in the Spt5 NGN domain interact with the upstream DNA duplex and the non-template DNA strand within the transcription bubble. As in Elf1, the locations of the Spt5 NGN and Spt4 overlap with the duplex DNA in the pre-initiation complex (closed form) [17]. Therefore, the presence of the Spt5 NGN, Spt4, and Elf1 should be significant for maintaining the transcription bubble, while preventing the DNA from rewinding into a duplex. The Spt5 KOW1 domain lies adjacent to the Spt5 NGN domain and Spt4, and they completely encircle the upstream DNA duplex to form the DNA-exit tunnel for the upstream DNA. The newly established DNA-exit tunnel defines the upstream DNA orientation and the size of the transcription bubble. The DNA-exit tunnel should be another important site for coordination with transcription-coupled phenomena. In fact, the DNA repair factor Rad26 binds to the upstream DNA of the Pol II EC in a mutually exclusive manner with Spt4 and the Spt5 NGN and KOW1 domains, to bend the DNA and expand the transcription bubble [22].
The Spt5 KOW domains and the RNA-exit tunnel
The Spt5 KOW5 domain is tightly docked within a complementary pocket formed by five Pol II subunits, next to the RNA exit site. The Spt5 KOW4 domain is located at the stalk subunit Rpb7, and part of the KOW4-KOW5 linker contacts the exiting RNA. Thus, the Spt5 KOW4-KOW5 domains augment the RNA-exit tunnel, probably supporting the efficient exit of the growing RNA. The Spt5 KOW5 domain also seems to adjust the EC conformation favorable for RNA elongation, by restricting inter-subunit movements within Pol II [23]. Spt4/5 is implicated in many co-transcriptional events, including mRNA processing. Spt5 has an intrinsically disordered CTR following the KOW5 domain. Similar to the C-terminal domain (CTD) of the Rpb1 of Pol II, the CTR is a phosphorylation target, and serves as an interface to recruit various factors [7–9]. The CTR should extrude from KOW5 near the RNA exit, thus facilitating the transfer of the growing nascent RNA from Pol II to mRNA processing machineries.
Recently, the cryo-EM structure of bovine Pol II EC bound with Spt4/5 was reported [24]. This structure revealed the tandem array of four KOW domains (KOW2–3–X–4), which were sandwiched between the Pol II body and the stalk. In contrast, our yeast EC structure revealed a density of a single KOW domain (KOW4), which contacts the Pol II stalk, but does not reach the Pol II body [2]. The tandem array of the KOW domains in the bovine structure seems to occur because the stalk is more tilted toward the Pol II body than those in the yeast EC [2] and the bovine EC without Spt4/5 (PDB:5FLM) [25]. Therefore, the observed structural difference may reflect various EC states, which could have originated from distinct sample preparation procedures. For example, the bovine sample contained NELF, although its density was not observed [24].
EC models of archaeal RNAP and eukaryotic Pol I
The Pol II EC structure allowed us to build plausible EC models of other RNA polymerases. Based on the Sulfolobus solfataricus RNAP structure (PDB: 3HKZ) [26], we created a structural model of an archaeal EC containing Elf1 and Spt4/5 (Figure 2(A)). As in the Pol II EC, the archaeal Elf1, the NGN domain of Spt5, and Spt4 are likely to seal the RNAP cleft to form a long DNA tunnel. As the KOW1 domain of archaeal Spt5 is smaller than its eukaryotic counterpart, due to the lack of a eukaryote-specific insertion, a completely closed DNA-exit tunnel should not be formed. Since archaeal Spt5 has only a single KOW domain (KOW1), the expansion of the RNA-exit tunnel by extra KOW domains and the CTR seems to be a eukaryote-specific feature required for more elaborate transcription-coupled mechanisms.
Figure 2.
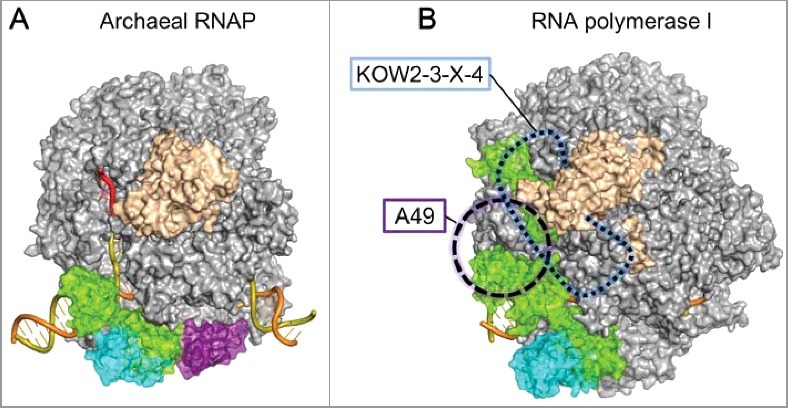
(A) Model of the archaeal EC bound with Elf1 and Spt4/5. (B) Superimposition of Pol I EC with Spt4/5 in the Pol II ECs. The position of Spt5 KOW2-3-X-4 is indicated by a blue dashed line. The position of the A49 tandem winged-helix domain is indicated as a violet circle.
Spt4/5 is reportedly implicated in transcription by eukaryotic RNA polymerase I (Pol I), which is specialized in ribosomal RNA synthesis [27,28]. By using the Saccharomyces cerevisiae Pol I EC structure (PDB: 5M5X) [29], we created a hypothetical model of the Pol I EC with Spt4/5 (Figure 2(B)). This model is consistent with the previous biochemical observation that the central region of Spt5, including NGN to KOW5, is important for binding both Pol I and Pol II [27]. However, the model creation required the adjustments of the Spt4/5 and Pol I domains, reflecting a few critical differences between the Pol I and Pol II ECs. First, since the clamp coiled-coil domain of Pol I is much longer than that of Pol II, the binding manner of the NGN domain to the coiled-coil domain may differ from that in the Pol II EC. Second, Pol I has a Pol I-specific insertion between the clamp and stalk (“Clamp-insertion” [30]), which could perturb or interfere with the KOW2–3–X–4 binding to Pol I. However, the stalk is tilted in the Pol I EC (PDB: 5M5X), as in the bovine Pol II EC bound with Spt4/5 (PDB: 5OIK), and this might be favorable for the KOW2–3–X–4 binding. Third, the Spt5 KOW1 domain in the model clashes with the tandem winged-helix domain of the A49 subunit in one of the Pol I EC structures (PDB: 5M64) [29]. Therefore, the KOW1 domain must replace the tandem winged-helix domain for the Spt4/5 binding to the Pol I EC. Alternatively, as a genetic interaction has been reported between A49 and Spt5 [27], they might work in a compensatory fashion.
Conclusion
Structural analyses have unveiled the architecture of the Pol II EC with basal factors. The structure not only revealed the sophisticated architecture required for processive transcription, but also provided insights into the stage-specific rearrangements of the transcription machineries, transcription regulation, and various transcription-coupled phenomena. Further studies are needed to enhance our understanding of these processes, with comparisons to other polymerase systems.
Funding Statement
This work was supported by Japan Society for the Promotion of Science KAKENHI [grant number JP15H04344].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Nogales E, Louder RK, He Y. Structural insights into the eukaryotic transcription initiation machinery. Annu Rev Biophys. 2017 May;46:59–83. doi: 10.1146/annurev-biophys-070816-033751. PMID:28532216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ehara H, Yokoyama T, Shigematsu H, et al. . Structure of the complete elongation complex of RNA polymerase II with basal factors. Science. 2017 Sep;357(6354):921–924. doi: 10.1126/science.aan8552. PMID:28775211. [DOI] [PubMed] [Google Scholar]
- [3].Wada T, Takagi T, Yamaguchi Y, et al. . DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998 Feb;12(3):343–356. PMID:9450929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hirtreiter A, Damsma GE, Cheung AC, et al. . Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010 Jul;38(12):4040–4051. doi: 10.1093/nar/gkq135. PMID:20197319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li J, Gilmour DS. Promoter proximal pausing and the control of gene expression. Curr Opin Genet Dev. 2011 Apr;21(2):231–235. doi: 10.1016/j.gde.2011.01.010. PMID:21324670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Smollett K, Blombach F, Reichelt R, et al. . A global analysis of transcription reveals two modes of Spt4/5 recruitment to archaeal RNA polymerase. Nat Microbiol. 2017 Mar;2:17021. doi: 10.1038/nmicrobiol.2017.21. PMID:28248297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wier AD, Mayekar MK, Héroux A, et al. . Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci USA. 2013 Oct;110(43):17290–17295. doi: 10.1073/pnas.1314754110. PMID:24101474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mayer A, Schreieck A, Lidschreiber M, et al. . The spt5 C-terminal region recruits yeast 3' RNA cleavage factor I. Mol Cell Biol. 2012 Apr;32(7):1321–31. doi: 10.1128/MCB.06310-11. PMID:22290438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ding B, LeJeune D, Li S. The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. J Biol Chem. 2010 Feb;285(8):5317–526. doi: 10.1074/jbc.M109.082818. PMID:20042611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Prather D, Krogan NJ, Emili A, et al. . Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae. Mol Cell Biol. 2005 Nov;25(22):10122–10135. doi: 10.1128/MCB.25.22.10122-10135.2005. PMID:16260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Daniels JP, Kelly S, Wickstead B, et al. . Identification of a crenarchaeal orthologue of Elf1: implications for chromatin and transcription in Archaea. Biol Direct. 2009 Jul;4:24. PMID:19640276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mayer A, Lidschreiber M, Siebert M, et al. . Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010 Oct;17(10):1272–1278. doi: 10.1038/nsmb.1903. PMID:20818391. [DOI] [PubMed] [Google Scholar]
- [13].Hogan BP, Hartsch T, Erie DA. Transcript cleavage by Thermus thermophilus RNA polymerase. Effects of GreA and anti-GreA factors. J Biol Chem. 2002 Jan;277(2):967–975. doi: 10.1074/jbc.M108737200. PMID:11606592. [DOI] [PubMed] [Google Scholar]
- [14].Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003 Aug;114(3):347–357. PMID:12914699. [DOI] [PubMed] [Google Scholar]
- [15].Wang D, Bushnell DA, Huang X, et al. . Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009 May;324(5931):1203–1206. doi: 10.1126/science.1168729. PMID:19478184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Plaschka C, Larivière L, Wenzeck L, et al. . Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015 Feb;518(7539):376–380. doi: 10.1038/nature14229. PMID:25652824. [DOI] [PubMed] [Google Scholar]
- [17].Plaschka C, Hantsche M, Dienemann C, et al. . Transcription initiation complex structures elucidate DNA opening. Nature. 2016 May;533(7603):353–358. doi: 10.1038/nature17990. PMID:27193681. [DOI] [PubMed] [Google Scholar]
- [18].Klein BJ, Bose D, Baker KJ, et al. . RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci USA. 2011 Jan;108(2):546–550. doi: 10.1073/pnas.1013828108. PMID:21187417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu B, Steitz TA. Structural insights into NusG regulating transcription elongation. Nucleic Acids Res. 2017 Jan;45(2):968–974. doi: 10.1093/nar/gkw1159. PMID:27899640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Martinez-Rucobo FW, Sainsbury S, Cheung AC, et al. . Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011 Apr;30(7):1302–1310. doi: 10.1038/emboj.2011.64. PMID:21386817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Said N, Krupp F, Anedchenko E, et al. . Structural basis for λN-dependent processive transcription antitermination. Nat Microbiol. 2017 Apr;2:17062. doi: 10.1038/nmicrobiol.2017.62. PMID:28452979. [DOI] [PubMed] [Google Scholar]
- [22].Xu J, Lahiri I, Wang W, et al. . Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017 Nov;551(7682):653–657. doi: 10.1038/nature24658. PMID:29168508; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sekine S, Murayama Y, Svetlov V, et al. . The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015 Feb;57(3):408–421. doi: 10.1016/j.molcel.2014.12.014. PMID:25601758. [DOI] [PubMed] [Google Scholar]
- [24].Bernecky C, Plitzko JM, Cramer P. Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat Struct Mol Biol. 2017 Oct;24(10):809–815. doi: 10.1038/nsmb.3465. PMID:28892040. [DOI] [PubMed] [Google Scholar]
- [25].Bernecky C, Herzog F, Baumeister W, et al. . Structure of transcribing mammalian RNA polymerase II. Nature. 2016 Jan;529(7587):551–554. doi: 10.1038/nature16482. PMID:26789250. [DOI] [PubMed] [Google Scholar]
- [26].Hirata A, Klein BJ, Murakami KS. The X-ray crystal structure of RNA polymerase from Archaea. Nature. 2008 Feb;451(7180):851–854. doi: 10.1038/nature06530. PMID:18235446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Viktorovskaya OV, Appling FD, Schneider DA. Yeast transcription elongation factor Spt5 associates with RNA polymerase I and RNA polymerase II directly. J Biol Chem. 2011 May;286(21):18825–18833. doi: 10.1074/jbc.M110.202119. PMID:21467036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Anderson SJ, Sikes ML, Zhang Y, et al. . The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J Biol Chem. 2011 May;286(21):18816–18824. doi: 10.1074/jbc.M110.202101. PMID:21467039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tafur L, Sadian Y, Hoffmann NA, et al. . Molecular Structures of Transcribing RNA Polymerase I. Mol Cell. 2016 Dec;64(6):1135–1143. doi: 10.1016/j.molcel.2016.11.013. PMID:27867008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Engel C, Sainsbury S, Cheung AC, et al. . RNA polymerase I structure and transcription regulation. Nature. 2013 Oct;502(7473):650–655. doi: 10.1038/nature12712. PMID:24153182. [DOI] [PubMed] [Google Scholar]


