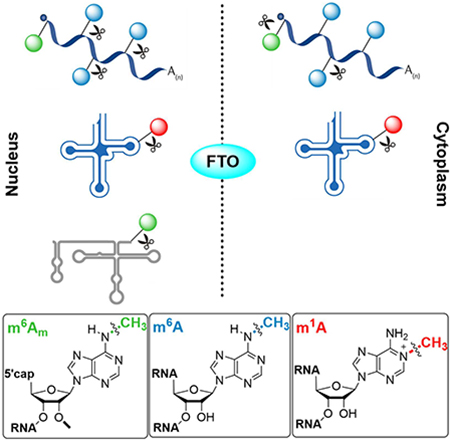
Summary
FTO, the first RNA demethylase discovered, mediates the demethylation of internal N6-methyladenosine (m6A) and N6, 2-O-dimethyladenosine (m6Am) at the +1 position from the 5’ cap in mRNA. Here, we demonstrate that the cellular distribution of FTO is distinct among different cell lines, affecting the access of FTO to different RNA substrates. We find that FTO binds multiple RNA species, including mRNA, snRNA, and tRNA, and can demethylate internal m6A and cap m6A in mRNA, internal m6m A in U6 RNA, internal and cap m6Am in snRNAs, and N1-methyladenosine (m1A) in tRNA. FTO-mediated demethylation shows a greater impact on the transcript levels of mRNAs possessing internal m6A than the ones with cap m6Am in the tested cells. We also show that FTO can directly repress translation by catalyzing m1A tRNA demethylation. Collectively, FTO-mediated RNA demethylation occurs to m6A and m6Am in mRNA and snRNA as well as m1A in tRNA.
In Brief
Wei et al. show that FTO mediates internal m6A and cap m6Am demethylation of polyadenylated RNA with differential substrate preferences in nucleus versus cytoplasm, in which the internal m6A demethylation correlates with transcript level changes. FTO also affects snRNA m6A and m6Am levels and mediates tRNA m1A demethylation.
Introduction
FTO was suggested to be associated with human obesity by GWAS (Fawcett and Barroso, 2010). A genetic variant of FTO has been shown to be associated with increased food intake (Cecil et al., 2008) while loss-of-function mutations in FTO cause severe growth retardation (Boissel et al., 2009) and CNS defects (Ho et al., 2010), which were recapitulated in murine model studies (Church et al., 2010; Fischer et al., 2009; Hess et al., 2013).
Due to these intriguing phenotypes, extensive efforts have been devoted to identify the substrate(s) and to understand the biological function of FTO. FTO was first reported to catalyze the demethylation of 3-methylthymine (3meT) in single-stranded DNA (Gerken et al., 2007) and 3-methyluracil (3meU) in RNA (Jia et al., 2008) as an iron(II)- and αKG-dependent dioxygenase. Later, FTO was identified as the first RNA demethylase that catalyzes reversal of the N6-methyladenosine (m6A) methylation in mRNA in vitro and inside cells (Fu et al., 2013; He, 2010; Jia et al., 2011). m6A is the most abundant internal modification in mammalian mRNAs (Fu et al., 2014). Adjacent to the 5’ cap, the second base in many mRNAs can be 2’-O-methylated (Adams and Cory, 1975; Wei et al., 1975b), with a portion of these bases also bearing m6A methylation to form m6Am (Wei et al., 1975a), deposited by a yet to be identified methyltransferase. This modification was confirmed by transcriptome-wide m6A-seq and exists in considerably lower overall abundance compared with m6A (Linder et al., 2015; Molinie et al., 2016). The m6A portion of m6Am is known to be an in vitro substrate of FTO (Fu, 2012), with a recent study showing that m6Am stabilizes mRNA by preventing DCP2-mediated decapping and microRNA-mediated mRNA degradation (Mauer et al., 2017). However, the functional relevance of m6Am removal by FTO has yet to be fully explored.
Surprisingly, this study also suggested that internal mRNA m6As may not be relevant substrates of FTO (Mauer et al., 2017), despite reports of a range of biological processes affected by the demethylation of internal m6A in the last several years: i) FTO-mediated m6A demethylation is critical in DNA UV damage response, with the m6A methyltransferase complex METTL3/14 exhibiting the opposite function (Xiang et al., 2017); ii) FTO plays noticeable roles in Flaviviridae family viruses infection via demethylating viral RNA m6A in host cells (Gokhale et al., 2016); certain members such as hepatitis C virus do not possess cap nor cap m6Am but still respond to FTO; iii) FTO-catalyzed demethylation significantly affects glioblastoma stem cell differentiation, again with METTL3/14 showing opposing functions (Cui et al., 2017); iv) FTO mediates nuclear demethylation of m6A at the 5’ UTR in heat shock response (Zhou et al., 2015); v) internal m6A demethylation by FTO plays an oncogenic role in a subset of acute myeloid leukemia (Li et al., 2017), and the effect of 2-HG inhibition can be explained through internal m6A methylation changes to oncogenic transcript targets of FTO, such as MYC (Su et al., 2018); vi) using liver-specific Fto-transgenic mice Fto was shown to mediate both m6A and cap m6Am demethylation; however, internal m6A demethylation controls the alternative translation (Zhou et al., 2018); vii) demethylation of internal m6A on a specific mRNA by FTO facilitates the stability and translation of the target transcript in neuronal developments (Yu et al., 2018); and viii) a recent report showing that FTO binds extensively across mRNA and intron regions in premRNA (Bartosovic et al., 2017).
In our biochemical studies, we confirmed that FTO could effectively demethylate both m6A and cap m6Am from purified polyadenylated RNA. We were intrigued by how FTO demethylates m6A and/or m6Am on target polyadenylated RNAs inside cells and the different functional consequences of the demethylation. We found that FTO localization within the nucleus and cytoplasm varies between cell types and that FTO has distinct substrate repertoires inside the cell nucleus and cytoplasm. We further identified additional RNA substrates of FTO, including m1A in tRNA, m6A in U6 RNA, internal and cap m6Am in snRNAs. This study presents the most comprehensive picture of the FTO-mediated RNA demethylation to date. It reveals a previously unappreciated spatial regulation imparted by nuclear versus cytoplasmic demethylation mediated by FTO, which exerts distinct effects on target RNAs.
Results
FTO Mediates Demethylation of Both m6A and m6Am in vitro and inside Cells
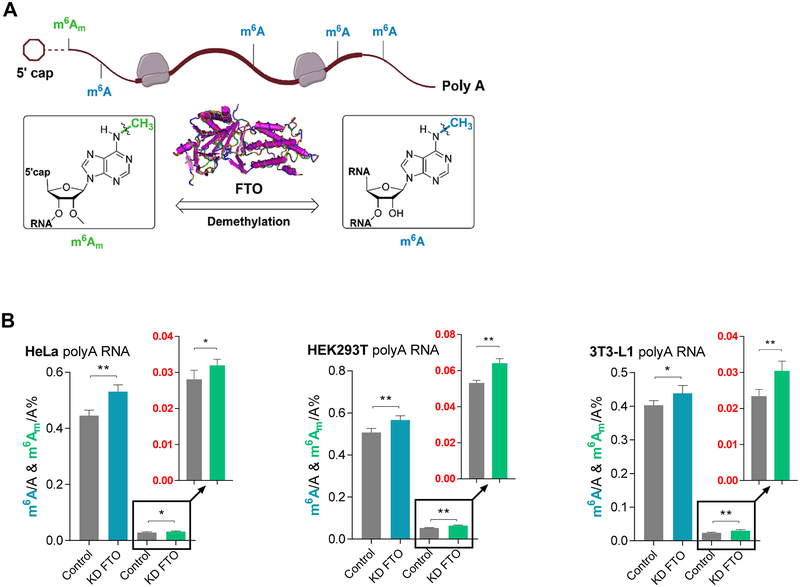
FTO has been shown to demethylate both m6A and m6Am (Fu, 2012; Fu et al., 2013; Jia et al., 2011; Mauer et al., 2017) (Figure 1A). To further validate these previous reports with biologically relevant substrates, we isolated polyadenylated RNAs possessing both naturally occurring m6A and m6Am from HEK293T cells and performed in vitro demethylation using purified FTO (Figure S1A). The substrate polyadenylated RNAs were digested to single nucleosides after FTO treatment. Ultra-High-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) was utilized to quantify levels of both m6A and m6Am from digested RNAs using carefully calibrated standard curves generated using synthetic nucleoside standards (Figure S1B).
Figure 1. FTO Demethylates both m6A and m6Am inside Cells.
See also Figure S1.
(A) A sketch that FTO mediates demethylation of m6A and cap m6Am in polyadenylated RNA.
(B) Quantification of the m6A/A and cap m6Am/A ratio in polyadenylated RNA by LC-MS/MS. In comparison to controls, significant increases in the m6A/A ratio were consistently observed among HeLa, HEK293T, and 3T3-L1 cells upon transient knockdown of FTO (blue bars). Increases of the m6Am/A ratio were consistently observed among HeLa, HEK293T, and 3T3-L1 cells upon transient knockdown of FTO (green bars). P values were determined using Student’s unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n.s. means not significant. Error bars, mean ± s.d. for n = 4 experiments in (B).
As shown in Figure S1C, among HeLa, HEK293T, and 3T3-L1 cells, the levels of m6Am in polyadenylated RNAs are about 1/10 to 1/15 of that of m6A. FTO (2 μM in 20 ul) is capable of almost complete demethylation of all m6Am and 81.4% of m6A in 200 ng polyadenylated RNAs within 1 hour under reported demethylation conditions (Figure S1D) (Jia et al., 2011; Mauer et al., 2017). The use of EDTA in the same reaction, which chelates the essential iron cofactor, serves as a control. We further lowered the FTO concentration to achieve incomplete demethylation of cap m6Am in isolated polyadenylated RNAs and compared the global changes of m6A and m6Am. As shown in Figure S1E, 0.2 μM of FTO (20 ul) could demethylate 81.9% of m6A and 20.3% m6 m A in the purified polyadenylated RNAs. Our results unequivocally show that FTO demethylates both internal m6A and cap m6Am in purified polyadenylated RNAs in vitro. FTO exhibits higher activity towards m6Am; however, it should be noted that because the total amount of m6A is around 10-fold to 15-fold higher than that of m6Am in polyadenylated RNAs (Figure S1C), more m6As are demethylated compared to m6Am in purified polyadenylated RNAs in terms of the number of modifications that can be reversed by FTO in vitro.
Next, we evaluated the effects of FTO perturbation on the cellular m6A and m6Am levels of polyadenylated RNAs. We selected HeLa, HEK293T, and 3T3-L1 cell lines and measured the levels of m6A and m6Am upon transient knockdown of FTO in comparison to the knockdown control by using UHPLC-MS/MS. A 19.2%, 11.8%, and 8.8% increase of the m6A/A ratio was observed in the polyadenylated RNAs in the knockdown samples compared to the knockdown control in HeLa, HEK293T, and 3T3-L1 cell lines, respectively (Figure 1B, blue bars). For m6Am, a 13.7%, 20.6%, and 30.1% increase was consistently observed in the FTO knockdown samples compared to the knockdown control in HeLa, HEK293T, and 3T3-L1 cells, respectively (Figure 1B, green bars). The knockdown efficiency was validated by western blots (Figure S1F). These results revealed that the demethylation percentage of m6A inside cells by FTO is more profound in HeLa cells in comparison to HEK293T and 3T3-L1, while the FTO-mediated demethylation of m6Am is more evident in 3T3-L1 cells compared to the other two cell lines. It again demonstrated both m6A and m6Am demethylation by FTO inside different cell lines. Because the level of internal m6A is 10-fold higher than that of cap m6Am in these cells, 5–10 times more internal m6A are subjected to FTO-mediated demethylation compared with m6Am inside cells.
FTO Preferentially Targets Nuclear m6A and Both Cytoplasmic m6A and m6 m A in mRNA
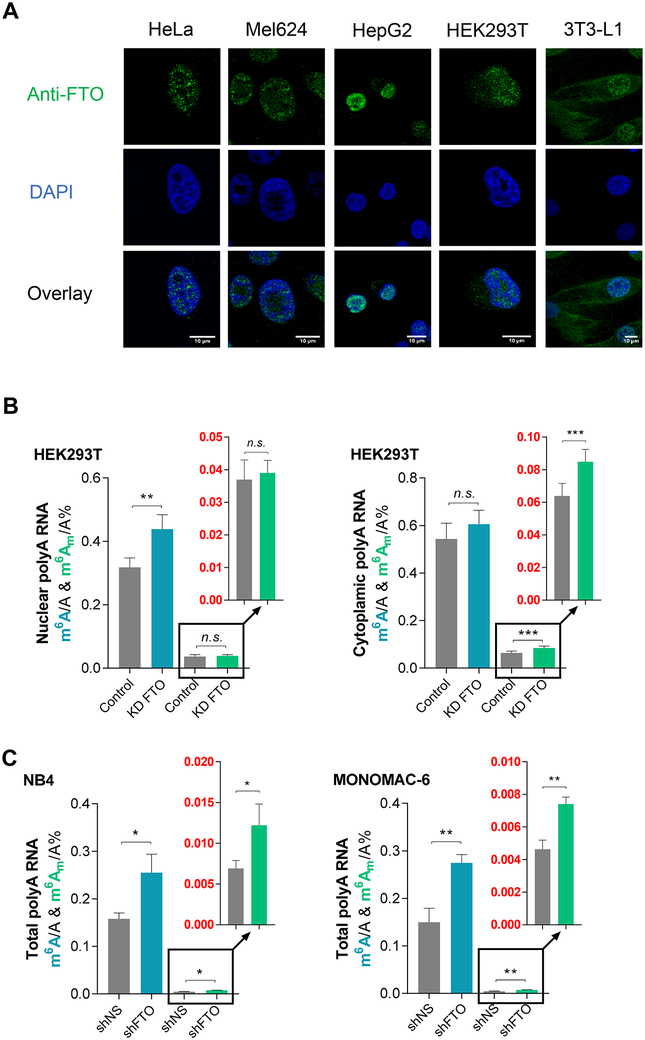
We sought to investigate how FTO mediates differential m6A versus m6Am demethylation in different cell lines and why FTO exhibits much higher m6A demethylation (over ~10-fold) versus m6Am inside certain cells such as HeLa despite m6Am being a preferred in vitro substrate. We reasoned that as almost all mRNA caps are bound by cap-binding proteins inside the cell nucleus (Muller-McNicoll and Neugebauer, 2013), the lack of m6Am demethylation in certain cells might reflect substrate accessibility by FTO. FTO was first reported to be a nuclear protein, but it was later shown to exist in both cell nucleus and cytoplasm in different mammalian cell lines (Aas et al., 2017; Gulati et al., 2014). We asked whether the spatial regulation of FTO plays a crucial role in gaining access to its physiologically relevant substrate(s). We conducted confocal immunofluorescence (IF) imaging with an anti-FTO specific antibody to investigate the cellular localization of FTO in five mammalian cell lines: HeLa, Mel624, HepG2, HEK293T, and 3T3-L1 cell lines. As shown in Figure 2A, the relative proportion of FTO within the nucleus and cytoplasm varies across cell lines. In HeLa, Mel624, and HepG2 cells, FTO predominantly accumulates in the cell nucleus. However, FTO is located in both cell nucleus and cytoplasm in HEK293T cells. The presence of FTO in the cytoplasm is even more notable in 3T3-L1 cells than HEK293T cells. More immunofluorescent images for HeLa, HEK293T, and 3T3-L1 were collected and five representative pictures from each cell line were shown in the Figure S2A. Calculated by ImageJ, the colocalization of the endogenous FTO with the nucleus was 97.7%, 83.5%, and 61.8% in HeLa, HEK293T, and 3T3-L1 cells, respectively. Consistent with our hypothesis, the increasing proportions of cytoplasmic FTO from HeLa, HEK293T, to 3T3-L1 cells, correlate with the corresponding increased cellular demethylation percentage towards m6Am and reduced m6A demethylation percentage in these cell lines (Figure 1B).
Figure 2. Subcellular Distribution of FTO Differs among Cell Lines and the Spatial Localization of FTO Influences its Demethylation of Target Substrates.
See also Figure S2.
(A) Subcellular localization of FTO (green) among HeLa, Mel624, HepG2, HEK293T, and 3T3-L1 cell lines. The nucleus was stained with DAPI (blue). Scale bar = 10 μm. Representative images are selected from three independent experiments.
(B) Quantification of the m6A/A and cap m6Am/A ratios in polyadenylated RNA from HEK293T cells by LC-MS/MS. Transient knockdown of FTO led to a significant increase of nuclear polyadenylated RNA m6A/A ratio but not the cytoplasmic polyadenylated RNA m6A/A ratio (blue bars). Transient knockdown of FTO led to a significant increase of cytoplasmic polyadenylated RNA m6Am/A ratio but no noticeable change of the nuclear polyadenylated RNA m6Am/A ratio (green bars).
(C) Quantification of the m6A/A and cap m6Am/A ratios in polyadenylated RNA from NB4 cells (left) and MONOMAC-6 cells (right) by LC-MS/MS. Stable knockdown of FTO led to both increased m6A/A and cap m6Am/A ratios in total polyadenylated RNA; P values were determined using Student’s unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n.s. means not significant. Error bars, mean ± s.d. for n = 6 experiments in (B); for n = 4 experiments in (C)
We next expressed an FTO mutant lacking the nuclear localization signal (ΔNLS-FTO) (Aas et al., 2017; Sanchez-Pulido and Andrade-Navarro, 2007) in HEK293T cell lines. The LCMS/MS results showed that overexpression of ΔNLS-FTO, which is directed to the cytoplasm, decreases m6Am but not m6A in polyadenylated RNA HEK293T cells. This result contrasts with the effects caused by wild-type FTO overexpression mostly within nucleus (Figure S2B). Next, we separated the cell nuclear and cytoplasmic fractions in HEK293T cells and then measured FTO-mediated demethylation of m6A and m6Am in polyadenylated RNAs in each fraction. As shown in Figure 2B (blue bars) knockdown of FTO led to a 37.9% increase in the m6A level in the cell nucleus but only a slight increase in the m6A level in cytoplasmic polyadenylated RNAs. In contrast, m6Am in cytoplasmic but not nuclear polyadenylated RNAs is significantly increased upon FTO knockdown (Figure 2B, green bars). Furthermore, in HEK293T cells, overexpression of wild-type FTO but not ΔNLS-FTO led to a significant decrease of m6A in polyadenylated RNAs in the cell nucleus, while the nuclear m6Am level in polyadenylated RNAs was not notably affected (Figure S2C). In the cytoplasm, m6A in polyadenylated RNA showed a small percentage change upon FTO overexpression, but m6Am showed a larger decrease upon overexpression of ΔNLS-FTO than wild-type FTO (Figure S2D). The separation of nucleus and cytoplasm for HEK293T cells was validated by western blot (Figure S2E) and qRT-PCR (Figure S2F). The result further confirmed that the FTO-mediated demethylation of m6A is prominent in the cell nucleus, whereas cap m6Am is more a target of FTO in the cytoplasm.
Because the m6A level is at least 10-fold higher than that of cap m6Am in mRNA, even in the cytoplasm FTO still mediates effective m6A and cap m6Am demethylation, in absolute terms. In certain AML cell lines in which we showed proliferation inhibition with FTO knockdown previously (Li et al., 2017), stable knockdown of FTO led to increased m6A in both nucleus and cytoplasm, whereas m6Am only increased in the cytoplasmic but not nuclear fraction (Figure 2C, and Figure S2G-S2J). We found extensive cytoplasmic expression of FTO in these AML cell lines (Figure S2K), and quite dramatic cytoplasmic m6A demethylation (around 40% increase of the m6A level with FTO knockdown), suggesting the critical cytoplasmic m6A demethylation roles of FTO in these cells. It is noteworthy that unlike the ratio of 1/10 to 1/15 between m6Am and m6A in HeLa, HEK293T, and 3T3-L1 cells (Figure S1C), the m6Am to m6A ratio is among 1/25 to 1/30 in the NB4, MONOMAC-6 cells (Figure 2C), and several other AML cells (Li et al., 2017; Su et al., 2018).
FTO-mediated m6A Demethylation in mRNA Influences Transcript Level of Target mRNA
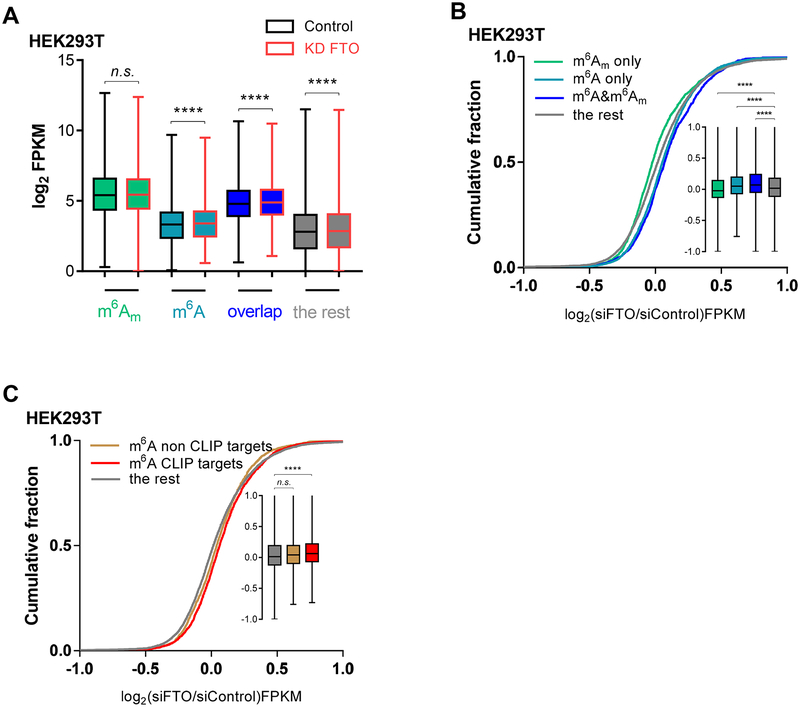
To understand the functional impact of m6A versus m6Am demethylation by FTO in mRNA, we analyzed transcript level changes of m6A and m6Am-containing transcripts upon FTO knockdown in HEK293T cells. The m6A sites were adopted from a previously published meRIP sequencing database (Meyer et al., 2012), and the transcripts starting with m6Am were obtained from the previously published study (Mauer et al., 2017). Almost all of these annotated m6Am sites are found within 500 nt from the transcription start site (TSS) (Figure S3A, top panel). To determine the individual effects of cap m6Am and internal m6A and to avoid cross-effects between these two modifications, we classified transcripts into four groups. The first group of transcripts contains only m6Am but not m6A, the second group of transcripts contains only m6A but not m6Am, the third group of transcripts contains both modifications, and the rest of transcripts were considered as the fourth control group to compare against (Figure S3A bottom panel and Table S1). As shown in Figure 3A, using this classification scheme, we found that the global levels of transcripts containing only m6Am don’t appear to statistically change upon FTO knockdown in HEK293T cells, while the global transcript levels of the other three groups show statistically noticeable increases.
Figure 3. FTO-mediated Demethylation of m6A but not m6Am in Polyadenylated RNA More Affects Transcript Expression Level Changes.
See also Figure S3 and Table S1.
(A) mRNA transcripts are classified into four groups: transcripts containing only m6Am, containing only internal m6A, containing both m6A and m6Am (overlap), and transcripts that contain neither m6A nor m6Am (labeled as the rest). The global expression level of m6Am-alone transcripts did not significantly change upon FTO knockdown while the global transcript level of other three groups increased significantly.
(B) The cumulative fraction of gene expression shows that compared to the transcripts containing only m6A, containing both m6A and m6Am, and containing no m6A or m6Am, the transcripts containing only m6Am but not m6A showed significantly less transcript level change with FTO knockdown compared to the control (in A, P values were determined using the Wilcoxon test; in B, P values were determined using Mann-Whitney test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n.s. means not significant. Error bars, mean ± s.d. for n = 3987, m6A only; n = 2016, m6A only; n = 1307, m6 m A and m6Am; n = 5598, the rest).
(C) The cumulative fraction of gene expression shows that FTO CLIP targets showed significantly increased expression level upon transient knockdown of FTO but not non-targets for transcripts containing only m6A compared to the rest genes. (In C, P values were determined using Mann-Whitney test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n.s. means not significant. Error bars, mean ± s.d. for n = 1895, m6A genes overlapped with FTO targets; n = 2092, non-targets; n = 8896, the rest).
Transient knockdown of FTO led to increased transcript levels of the transcripts harboring only m6A compared to the transcripts without m6A or m6Am in HEK293T cells (Figure 3B). It has previously been observed that increased m6A leads to increased mRNA transcript levels in HepG2 and leukemia cell lines (Huang et al., 2017a; Li et al., 2017). This effect was thought to be mediated by a group of newly identified m6A reader proteins IGF2BP1–3 (Huang et al., 2017a), which bind internal m6A and stabilize methylated mRNAs, or through other yet to be discovered reading mechanisms.
The presence of m6Am adjacent to the 5’ cap was previously reported to stabilize mRNA transcripts by preventing DCP2-mediated cap cleavage and microRNA-mediated mRNA degradation in HEK293T cells (Mauer et al., 2017). However, compared to the other three groups of transcripts (Figure 3B), transcripts containing only m6Am but not m6A showed significantly lower transcript level changes upon FTO knockdown. FTO knockdown should lead to elevated m6Am but this does not appear to correlate with higher levels of these transcripts, despite the proposed transcript-stabilizing role of m6Am at the cap (Mauer et al., 2017). This effect is opposite from those observed for transcripts containing both m6Am and m6A or transcripts containing only m6A. Both of the latter two groups showed significantly larger increases in transcript levels upon FTO knockdown in comparison to the control group. Lastly, grouping just transcripts containing m6A versus non-m6A without considering m6Am, and transcripts containing cap m6Am versus non-m6Am without considering m6A, transcripts containing m6A showed increased transcript levels upon FTO knockdown (Figure S3B), while transcripts containing m6Am showed reduced levels compared with the rest upon FTO knockdown (Figure S3C).
These results suggest that though m6Am could be associated with a higher global transcript level and higher transcript stability, the levels of transcripts bearing cap m6Am dos not appear to change with the knockdown of FTO (Figure 3A). The level change of these transcripts was significantly less than the transcripts containing neither m6Am nor m6A upon FTO knockdown, whereas transcripts containing m6A or both m6A and m6Am had significantly larger increases in their transcript levels than transcripts containing neither or just cap m6Am upon FTO knockdown (Figure 3B). We further cross-compared the list of transcripts contain only m6A but not m6Am with the published FTO CLIP-seq results (Bartosovic et al., 2017) by separating transcripts contains only m6A but not m6Am plus FTO binding into FTO targets and non-targets. Compared to the rest genes (genes except transcripts containing only m6A), we clearly observed in Figure 3C a statistically significant right-shift of the cumulative curve representing the increased transcript levels of the m6A targets (bound by FTO), with no significant change for non-targets possessing m6A but not bound by FTO. qRT-PCR was used to validate the relative transcript levels of three genes containing only m6A or m6Am. The transcript level changes of these genes upon FTO knockdown were consistent with the RNA-seq data (Figure S3D).
FTO Demethylates m6A in U6 RNA and m6Am in snRNAs
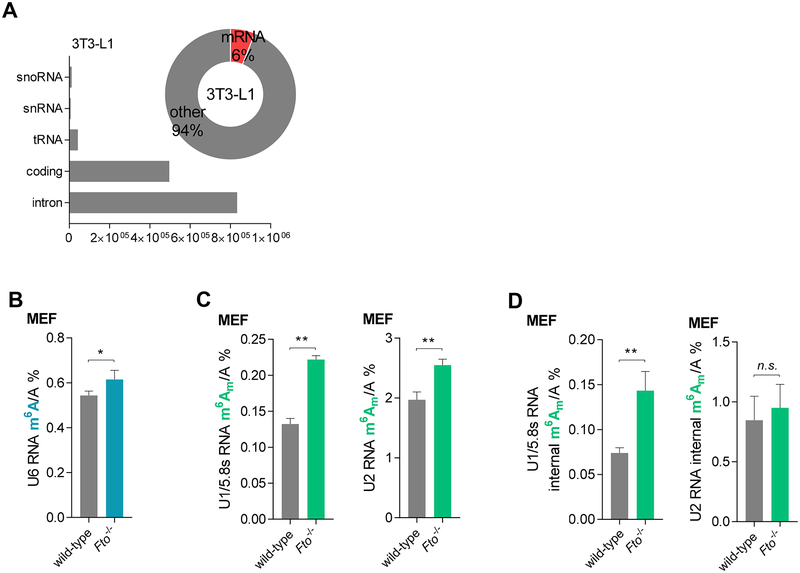
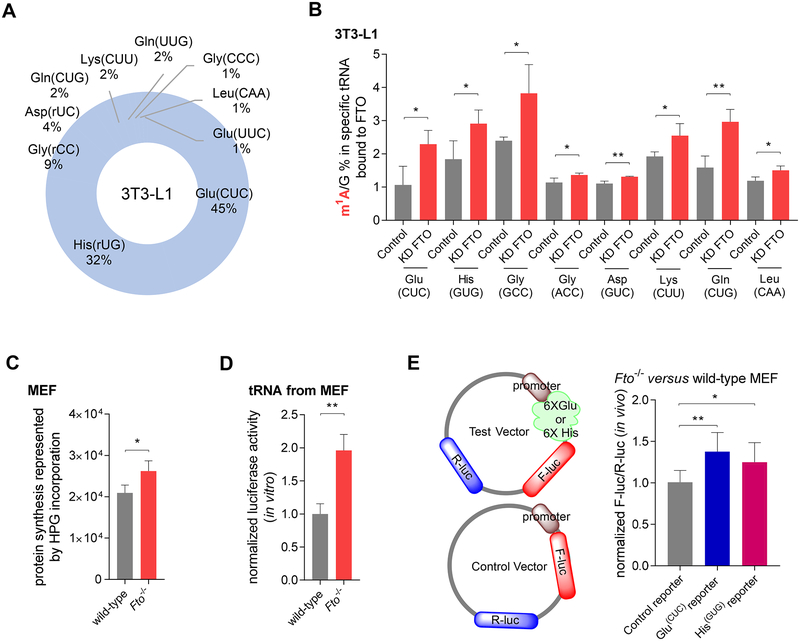
Two recent studies have surveyed FTO-bound RNA species through eCLIP-seq (Van Nostrand et al., 2016) or CLIP-seq of FLAG-tagged FTO (Bartosovic et al., 2017). These studies and our CLIP-seq results in 3T3-L1 cells using FLAG-tagged FTO showed that FTO is able to bind mRNA, tRNA, snRNA, snoRNA (Figure 4A, bar), and rRNA, among which mRNA is enriched (Figure 4A, donut). The purity of anti-FLAG-tagged pulldown was validated in Figure S4A (left) in our studies, and the 32P labeled protein gel of CLIP is shown in Figure S4A (right), showing the formation of covalently linked FTO-nucleic acids complexes. We were interested in non-mRNA CLIP-targeted RNA species and examined the potential activity of FTO on those RNA species. We first assessed the effect of FTO knockdown or overexpression on the total m6A levels in mature rRNA (Figure S4B) in HeLa, HEK293T, and 3T3-L1 cell lines, respectively. No significant changes were observed for m6A in purified mature rRNAs, including 28S rRNA (Figure S4C) and 18S rRNA (Figure S4D). We also compared the m6A levels of top snRNA targets revealed by CLIP-seq, including U1 RNA (a complex band of U1 RNA and 5.8S, Figure S4E), U2 RNA, and U6 RNA, in Fto homozygous knockout (Fto−/−) MEF versus the wild-type MEF cells. Interestingly, we observed increased m6A in the U6 RNA (Figure 4B) but not other snRNAs (Figure S4F) with the depletion of Fto in the Fto−/− compared with the wild-type MEF cells, suggesting that U6 m6A could be demethylated by FTO.
Figure 4. FTO-mediated Internal m6A, Internal and Cap m6Am Demethylation in snRNAs.
See also Figure S4.
(A) Bar graph summarizing the reads distribution of the FTO target RNA species in the 3T3-L1 cells; donut graph showing the percentage of the FTO-bound mRNA in all FTO CLIP-seq targets in the 3T3-L1 cell line.
(B) An increased m6A/A ration in the U6 RNA was observed by LC-MS/MS in Fto−/− MEF cells compared to the control wild-type MEF cells.
(C) Quantification of the m6Am/A ratio of the U1/5.8s RNA and U2 RNA by LC-MS/MS after decapping, showing increased m6Am/A ratios in the Fto−/− MEF cells compared to the wild-type control.
(D) Quantification of the m6Am/A ratio in U1/5.8s RNA and U2 RNA by LC-MS/MS without decapping, showing increased internal m6Am/A ratios in Fto−/− MEF cells compared to the wild-type control. P values were determined using Student’s unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n.s. means not significant. Error bars, mean ± s.d. for n = 6 experiments in (B) to (D).
Besides m6A, we also tested FTO demethylation of m6Am in these U RNAs. After decapping, increased m6Am was observed in both U1/5.8s RNA and U2 RNA (Figure 4C). The quantification results showed a reasonable level of m6Am in U2 RNA, with close to one m6Am per U2 RNA molecule (187 bp long). To determine if the m6Am is near the cap or internal, we measured the internal m6Am level without decapping; decapping is required to remove the cap structure and expose cap m6Am before nuclease P1 and alkaline phosphatase digestion. Internal m6Am was also observed in both U2 and U1/5.8S RNA (Figure 4D), with increased internal m6Am observed in U1/5.8s RNA in the Fto−/− compared to the wild-type MEF cells. However, considering the average length of mammalian U1 snRNA and 5.8s rRNA (around 160 bp), the 0.1% ratio of internal m6Am/A indicates less than 0.05 m6Am per molecule of U1 snRNA. In U2 RNA both the cap and internal m6Am modifications are more abundant, approaching to 0.5 per molecule of U2 RNA, respectively. The Fto knockout led to a significant increase of the total m6Am level, most likely at the cap. Given the role of snRNPs in the splicing of mRNA (Maniatis and Reed, 1987), and previous work suggesting that FTO deficiency promotes splicing (Zhao et al., 2014), it will be interesting to investigate this potential link between FTO-induced modification changes in snRNAs and mRNA splicing in the future.
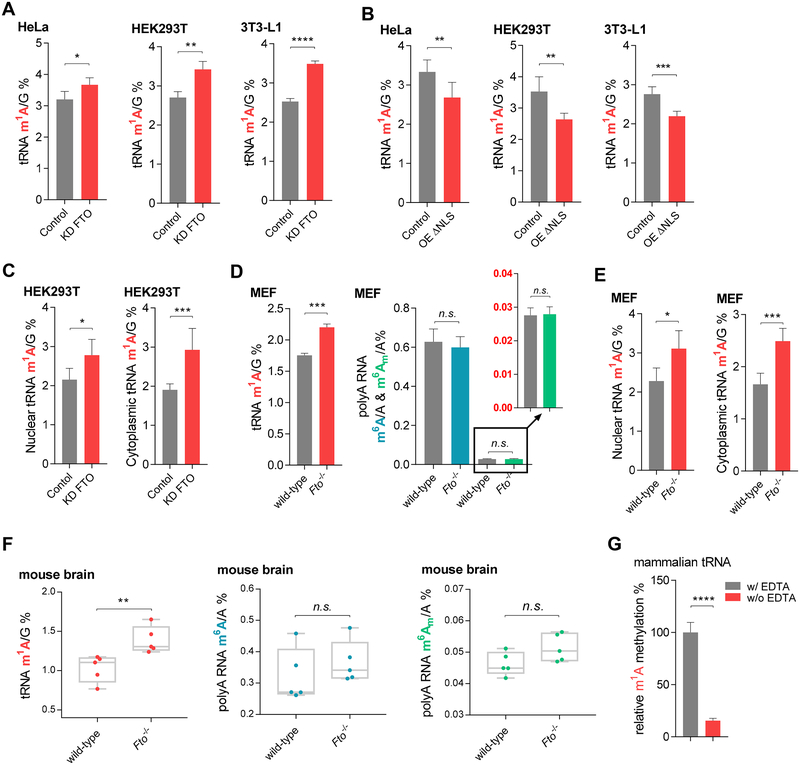
FTO Mediates m1A Demethylation in tRNA
In the CLIP-seq results, we observed that FTO associates with tRNA and the distribution of CLIP reads in tRNA is significantly enriched compared to input (Table S2 and Table S3). We found that the knockdown of FTO led to the increase of 14.7%, 26.5%, and 38.1% of the total m1A level in tRNA (Figure 5A) compared with the knockdown control in HeLa, HEK293T, and 3T3-L1 cells, respectively. The transcript levels of ALKBH1 and ALKBH3, which were previously shown to mediate tRNA m1A demethylation (Liu et al., 2016; Ueda et al., 2017), remain unchanged upon the knockdown of FTO (Figure S5A) (Mauer et al., 2017), The overexpression of ΔNLS-FTO led to the opposite effects with about 19.6%, 23.1%, and 20.4% decrease of m1A in tRNA observed (Figure 5B) when compared with controls in HeLa, HEK293T, and 3T3-L1 cells, respectively. We then separated the nuclear and cytoplasmic fractions of HEK293T cells with FTO knocked down versus the control. Both nuclear tRNA m1A and cytoplasmic tRNA m1A increased upon FTO knockdown, with cytoplasmic tRNA m1A showing a larger increase (Figure 5C).
Figure 5. FTO-mediated tRNA m1A Demethylation in vitro, inside Cells, and in Brain Tissues.
See also Figure S5, Table S2, and Table S3.
(A) Quantification of the m1A/G ratio in total tRNA purified from HeLa, HEK293T, and 3T3-L1 cells by LC-MS/MS. The transient knockdown led to an increase of at least ~15% of the m1A/G ratio in total tRNA compared to control cells.
(B) A consistent decrease (~20%) of the total tRNA m1A/G ratio was observed upon overexpression of ΔNLS-FTO compared to the control.
(C) Transient knockdown of FTO led to a significant increase of both nuclear and cytoplasmic total tRNA m1A/G ratio in HEK293T cells.
(D) A significant increase of m1A in tRNA but not m6A or m6Am in the polyadenylated RNA was observed in the Fto−/− MEF cells compared to the wild-type MEF cells.
(E) Knockout of Fto led to significant increases of both nuclear and cytoplasmic tRNA m1A levels in tRNA in MEF cells.
(F) A significant increase of the total tRNA m1A but not polyadenylated RNA m6A or m6Am could be observed in Fto−/− mouse brain tissues compared to wild-type mouse brains.
(G) Demethylation of m1A in tRNA isolated from HEK293T cells by recombinant FTO. EDTA chelates cofactor iron and inactivates FTO. The ratio of m1A/G was shown. P values were determined using Student’s unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n.s. means not significant. Error bars, mean ± s.d. for n = 4 experiments in (A) to (E); for n = 5 experiments in (F); for n = 3 experiments in (G).
Interestingly, the m1A level in tRNA is dramatically affected by FTO-mediated demethylation in comparison to m6A and cap m6Am in polyadenylated RNAs in Fto knockout mouse cells and tissues. In Fto−/− MEF cells, we observed a significant increase of the total m1A in tRNA with no noticeable differences of m6A or m6Am in polyadenylated RNAs observed (Figure 5D). Both nuclear tRNA m1A and cytoplasmic tRNA m1A showed noticeable increases in Fto−/− MEF cells compared with the wild-type MEF cells (Figure 5E). The separation of nucleus and cytoplasm for MEF cells was validated by western blot (Figure S5B) and qRT-PCR (Figure S5C). We further tested the changes of the m1A levels in tRNA as well as m6A and cap m6Am in polyadenylated RNAs in the brain tissue of Fto−/− mice (Figure S5D). While the total m6A and m6Am showed slight increases in five pairs of Fto−/− versus the wild-type mouse brains, the total m1A level in tRNA showed the largest increases in the brain of the Fto−/− mice (Figure 5F). It is possible that the surviving Fto knockout mice have adapted to the loss of Fto with compensated changes of the total m6A or cap m6Am levels in polyadenylated RNA. Many of these knockout mice still die early (several weeks after birth) in our hands, suggesting the lack of demethylation of individual mRNA or other RNA species still exhibits a noticeable effect on these mice. We also tested the level changes of m1A in tRNA in two cell lines related to human leukemia. Stable knockdown of FTO leads to significantly increased tRNA m1A in NB4 and MONOMAC-6 cells (Figure S5E).
We examined whether FTO exhibits biochemical demethylation activity towards m1A in tRNA by treating the purified total tRNA from HEK293T cells with the FTO protein under reported demethylation conditions. The levels of abundant tRNA methylations, including m1A, m1G, m7G, and m5C, m2G, m22G, Am, and m3C, were measured in the presence or absence of EDTA. Incubation of purified tRNA with FTO led to a dramatic decrease of the m1A level (Figure 5G), and a slight decrease of m3C, but not levels of other modifications (Figure S5F), in comparison to the control samples, confirming that m1A in tRNA is a biochemically viable substrate of FTO in addition to m6A and m6Am in the polyadenylated RNAs. We then compared the demethylation activity of FTO towards tRNA m1A and mRNA m6A by incubating similar numbers of m1A and m6A together with FTO in the presence or absence of EDTA. The demethylation activity of FTO towards tRNA m1A is lower than that of polyadenylated RNA m6A (Figure S5G). However, considering the abundances of tRNA and polyadenylated RNA inside mammalian cells, the demethylation towards tRNA m1A could still be functionally important.
m1A58 is the most common m1A site in tRNA, residing in the loop region of a TΨC stem-loop motif of tRNA (Hopper and Phizicky, 2003; Phizicky and Hopper, 2010), and can be reversed through demethylation in HeLa and MEF cells (Liu et al., 2016). We reasoned that FTO may preferentially mediate m1A demethylation in tRNAs with a selectivity towards a stem-loop structure, as previous studies suggested low activities of FTO towards m1A in single-stranded nucleic acid substrates (Jia et al., 2008). We synthesized two ssRNA probes, one with m1A in a stem-loop sequence mimicking the TΨC loops in tRNAHis(GUG) (contains m1A58) and the other one with m1A in an unstructured sequence (Figure S5H). Indeed, FTO exhibited low activity towards m1A in the unstructured probe but a high preference for the stem-loop probe (Figure S5I). This biochemical result, combined with the quantifications of modification changes in various cells and tissues, support m1A in tRNA as another substrate of FTO.
We asked why FTO could have distinct RNA species as demethylation substrates especially the preference for the loop structure. We examined the published protein structures and realized the unique structural features of FTO that allow it to gain access to the modification sites in multiple RNA species. We noticed that the overall structure of FTO has similarities with NSUN6, a tRNA m5C methyltransferase (Haag et al., 2015). In the overlapped structures of FTO and NSUN6 with bound tRNA, we observed that the FTO active site could gain access to tRNA m1A58 (Figure S5J, left: overlapped structures of FTO (raspberry) with the complex of NSUN6 (purple) and tRNA (orange), with the iron center depicted in a yellow ball). However, another known mRNA m6A demethylase, ALKBH5, does not carry the similar structural fold as FTO does, which may explain the more constrained substrate specificity of ALKBH5 on mRNA m6A (Figure S5J, right: overlapped structures of ALKBH5 (green) with the complex of NSUN6 (blue) and tRNA (orange)).
FTO-mediated tRNA Demethylation Affects Translation
We further validated the m1A demethylation activity of FTO on selected target tRNAs (Figure 6A). Biotin-labeled DNA probes complementary to tRNA species were used to specifically isolate the corresponding tRNA from 3T3-L1 cells. The m1A level of the purified specific tRNA was then measured using LC-MS/MS. The results demonstrated that the knockdown of FTO leads to consistent increases of the m1A levels in most top confident tRNAs targets bound to FTO, including tRNAGlu(CUC), tRNAHis(GUG), tRNAGly(GCC), tRNAGly(ACC), tRNAAsp(GUC), tRNALys(CUU), tRNAGln(CUG), and tRNALeu(CAA) (Figure 6B). In addition, we also tested several tRNAs that were not FTO targets according to our FTO CLIP-seq data, including tRNAAla(AGC), tRNAAsn(GUU), tRNAGly(UCC), and tRNAPhe(GAA). Results revealed that FTO does not exhibit enzymatic activity on these non-targeted tRNAs (Figure S6A). Therefore, the majority of tRNAs identified by CLIP-seq are substrates of FTO.
Figure 6. FTO Mediates Specific tRNA m1A Demethylation and Suppresses Translation.
See also Figure S6.
(A) Pie chart showing the top FTO-bound tRNAs identified by CLIP-seq with FLAG-tagged FTO in 3T3-L1 cells.
(B) Quantification of the m1A/G ratio in tRNA targets bound to FTO by LC-MS/MS. In comparison to the control, m1A/G ratios of tRNAGlu(CUC), tRNAHis(GUG), tRNAGly(GCC), tRNAGly(ACC), tRNAAsp(GUC), tRNALys(CUU), tRNAGln(CUG), and tRNALeu(CAA) noticeably increased upon transient knockdown of FTO in 3T3-L1 cell.
(C) Quantification of total protein synthesis in MEF cells by flow cytometry. HPG incorporation into wild-type MEF cell was recorded in gray and the Fto−/− MEF cells in red.
(D) Supplementation of total tRNA purified from Fto−/− MEF cell leads to increased translation efficiency in vitro compared to using the total tRNA from wild-type MEF cells.
(E) Cartoon illustration of the reporter assays was shown: firefly luciferase (F-luc) was used as the reporter and Renilla luciferase (R-luc) on the same plasmid was used as the internal transfection control. 6 × GUG(Glu)-coding sequences (recognized by tRNAGlu(CAC)) and 6 × CAC(His)-coding sequences (recognized by tRNAHis(GUG)) were inserted after the PLK promoter region of F-luc as the positive reporter, respectively. The effects of tRNAGlu(CUC) and tRNAHis(GUG) were revealed by the reporter assay. The reporter assay showed significant increases of protein synthesis in the Fto−/− MEF cell compared to the wild-type MEF cell when the control reporter and test reporters (with inserted Glu(CUC)-coding sequence or His(GUG)-coding sequence) are transiently expressed in the wild-type and Fto−/− MEF cell, respectively. P value was determined using Student’s unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n.s. means not significant. Error bars, mean ± s.d. for n = 3 experiments in (B) and (C); n = 8 experiments in (D) and (E).
We then used an alkyne-modified glycine analog, L-homopropargylglycine (HPG), to metabolically label newly synthesized proteins in Fto−/− and wild-type MEF cells. The HPG-labeled cells were then fluorescently labeled and analyzed by flow cytometry. The results showed a notable increase in the protein synthesis in the Fto−/− cells compared to the wild-type cells (Figure 6C and Figure S6B).
To separate the effects of the FTO-mediated tRNA m1A demethylation from the effects of the FTO-mediated adenosine demethylation in mRNA on translation, we performed an in vitro translation assay by using a rabbit reticulocyte-based luciferase reporter system (Promega). tRNAs extracted from Fto−/− and wild-type MEF cells were subjected to the in vitro luciferase mRNA translation assay using rabbit reticulocyte lysate, respectively. The activity of the translated luciferase protein was measured in the incubation mixture by a luminometer. As shown in Figure 6D, the incubation of tRNAs extracted from Fto−/− cells induced significantly higher luciferase signals than tRNAs extracted from the wild-type MEF cells, indicating that the FTO-mediated tRNA demethylation led to increased protein translation efficiency.
We further designed a reporter assay with six repeated codon sequences (6×GAG(Glu) for tRNAGlu(CUC) target and 6×CAC(His) for tRNAHis(GUG) target) added to the 5’ of firefly luciferase (F-luc) (Figure 6E, left); a second Renilla luciferase (R-luc) encoded in the same plasmid was used to normalize the expression efficiency. For both reporters, the significantly elevated translation was observed in the Fto−/− MEF cells compared to the wild-type MEF cells (Figure 6E, right). We also applied a reporter assay with six repeated codon sequences (6×GGA(Gly)) for non-target tRNAGly(UCC) to the same reporter system mentioned above as the negative control. As expected, there was no noticeable change of translation between Fto−/− MEF cells and the wild-type MEF cells (Figure S6C). These results further indicate that the FTO-mediated tRNA m1A demethylation negatively affects translation, which is consistent with the previous discovery that m1A-methylated tRNAs are preferentially recognized and delivered to translation-active polysomes (Liu et al., 2016).
Discussion
FTO harbors two structural domains, an N- and a C-terminal domain. The active site of FTO resides in its N-terminal domain and is exposed to the protein surface (Han et al., 2010). Unlike ALKBH1, which possesses a distinct tRNA-binding domain (Liu et al., 2016), FTO contains an L1-loop surrounding its active site across to the Jelly-roll motif on the other side of the active site. The L1-loop facilitates FTO’s selectivity towards single-stranded RNA substrates. The additional C-terminal domain, which is absent in the other AlkB family proteins (Yang et al., 2008; Yu et al., 2006), may endow FTO with the ability to associate with potential protein partners. These structural features may allow FTO to accomplish demethylation of different RNA substrates via interacting with diverse partner proteins depending on the cellular context.
Demethylation of m6A and Cap m6Am in mRNA by FTO in the Nucleus and Cytoplasm
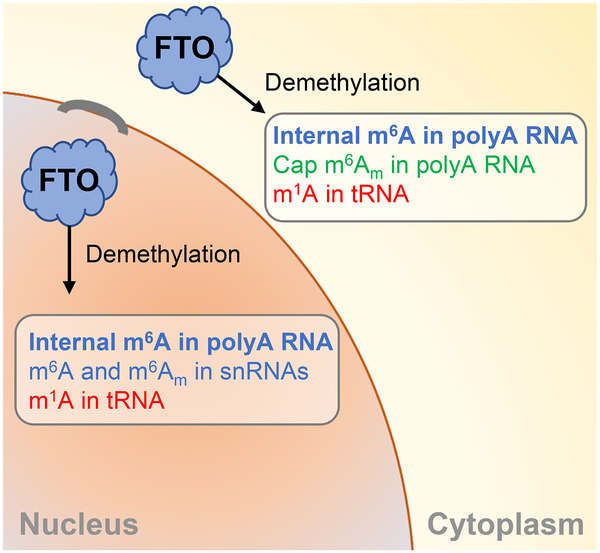
Both internal m6A and cap m6Am exist in polyadenylated RNA in mammalian cells. The levels of internal m6A are about 10–20 folds higher than those of cap m6Am in different cell lines (Wei et al., 1975a; Wei et al., 1975b). Our cellular results and biochemical assays using FTO and purified mRNA unequivocally show demethylation of both internal m6A and cap m6Am by FTO. While FTO catalyzes more potent demethylation on cap m6Am over m6A in vitro, the extent of demethylation inside cells is opposite with 5–10 folds more m6A demethylated by FTO compared with m6Am in cell lines tested, in absolute terms. The primary polyadenylated RNA substrate of FTO in the cell nucleus appears to be m6A, with cap m6Am more a target of FTO in the cytoplasm (Figure 7).
Figure 7. A Model for FTO-mediated RNA Demethylation.
Nuclear m6A is the main substrate of FTO in the cell nucleus. FTO also mediates m1A tRNA demethylation as well as m6A (U6) and m6Am (U2 in particular but also U1) snRNA demethylation in the nucleus. FTO can localize to the cytoplasm and mediate mRNA m6A and cap m6Am demethylation as well as tRNA m1A demethylation in the cytoplasm.
Nuclear mRNAs consist of only a portion of total cellular mRNAs. A notable fraction of nuclear mRNAs or other polyadenylated RNAs are substrates of FTO-mediated demethylation. In HeLa cells, although FTO could impact a significant fraction of nuclear polyadenylated m6A, this effect is “diluted” when examining the total mRNA pool. It should be noted that when FTO shuttles to the cytoplasm, it may still demethylate a portion of m6A in cytoplasmic mRNAs as it could gain access to both internal m6A and cap m6Am on mRNA (Figure 7). In certain AML cells, the cytoplasmic m6A demethylation by FTO can affect almost 40% of the total m6A (Figure S2H and Figure S2J). Because the total m6A level is considerably higher than that of m6Am in mRNA, a change of the similar percentage of m6A versus m6Am manifests as a much larger number of m6A changes in mRNA.
Functional Relevance of Cap m6Am and Internal m6 A Demethylation by FTO
A previous study showed that the presence of m6 Am adjacent to the 5’ cap prevents decapping and stabilizes mRNA transcripts in HEK293T cells (Mauer et al., 2017). In order to further probe the effect of m6Am, we categorized the previously identified transcripts (Mauer et al., 2017) into four classes: cap m6 Am -only transcripts, m6 Am-only transcripts, transcripts containing both modifications, and transcripts with neither modification. In HEK293T cells, m6A-only transcripts showed increased transcript levels upon FTO knockdown (Figure 3A and 3B), suggesting the presence of mechanisms that stabilize m6A-modification transcripts. The YTHDF2-mediated decay of methylated transcripts operates in HeLa cells (Ke et al., 2017; Zhao et al., 2014), but may not be the predominant pathway affecting the transcript levels of m6A-containing transcripts in HEK293T and AML cells (Li et al., 2017).
To our surprise, when we analyzed cap m6Am-only transcripts, we found that FTO knockdown does not noticeably affect mRNA transcript levels of cap m6Am-only genes in reported HEK293T cells. A careful analysis indicated that the previous analysis combined cap m6Am-only transcripts with transcripts that contain both cap m6Am and m6A (Mauer et al., 2017).
The transcript stabilizing effect is most likely derived from the internal m6As that co-exist with cap m6Am in transcripts that contain both modifications. So far, all functional relevant reports of FTO have been ascribed to internal m6A in mRNA, which include a wide range of different systems (Cui et al., 2017; Gokhale et al., 2016; Su et al., 2018; Xiang et al., 2017; Yu et al., 2018; Zhou et al., 2015; Zhou et al., 2018; Zou et al., 2016). A recently published FTO CLIP-seq result revealed a peak of FTO binding closely around known m6A sites and in intronic regions in HEK cells, suggesting that FTO mediates intronic m6A demethylation (Bartosovic et al., 2017).
The discoveries of the cap m6Am in stabilizing mRNA and its demethylation by FTO are interesting and could potentially open new research directions (Mauer et al., 2017); however, the effects of FTO on cap m6Am of these mRNA transcripts require future investigations. The methyltransferase will need to be identified and the phenotypes of the methyltransferase knockout cells or mice need to be investigated and compared to severe phenotypes observed in Fto knockout mice. The role of cap m6Am demethylation in RNA species other than mRNA merits consideration. Appropriate and sensitive detection method and procedures are critical to investigate the FTO-mediate demethylation in the future research of the community.
tRNA m1A and Small Nuclear RNA Demethylation by FTO
CLIP-seq results of FTO followed by LC-MS/MS tests revealed additional substrates of FTO, including m6A in U6 RNA (Figure 4B) as well as internal and cap m6Am in snRNA (Figure 4C and 4D). The demethylation towards modifications in these U RNAs may affect mRNA splicing. However, cautions must be taken as the ratio of m6A/A or m6Am/A for these small nuclear RNAs ranges from 0.05–2%. For short RNA species such as U1 and U6, the very low modified A/A ratio (0.05–0.5%) indicated far below stoichiometric methylation with 0.02–0.2 m6Am or m6A per snRNA molecule. Close to one m6Am, either at the cap or internal, exists per U2 RNA molecule, and the cap U2 m6Am demethylation by FTO can have functional consequences in splicing, as previously suggested (Maniatis and Reed, 1987; Zhao et al., 2014).
When comparing the modification levels in the Fto−/− mouse MEF cells and brain tissues with the wild-type control we observed a large change of m1A in tRNA. m1A methylation in tRNA is important for the cell viability and fitness (Oerum et al., 2017), with m1A/A ratio ranging around 1–3%. Previous studies have shown that m1A58 is installed by the methyltransferase complex Trmt6/61 (Anderson et al., 1998) and can be reversed by ALKBH1 (Liu et al., 2016). The dynamic regulation of tRNA m1A58 could impact translation initiation and elongation. In the current study, our CLIP sequencing data showed that FTO binds m1A58-containing tRNAs. Further biochemical characterization revealed that FTO possesses effective tRNA m1A demethylation activity in vitro and inside cells (both nucleus and cytoplasm). In fact, FTO was originally shown to be able to reverse DNA 1mA damage in 2007 (Gerken et al., 2007), most likely on ssDNA (Jia et al., 2008). The reporter assay confirmed that the FTO-mediated tRNA demethylation affects translation, which is reminiscent of the mechanism used by ALKBH1 in regulating translation elongation. FTO mediates both nuclear and cytoplasmic m1A demethylation in tRNA. The mRNA cap m6Am demethylation (Mauer et al., 2017) and tRNA m1A demethylation by FTO could collectively repress translation, with cap demethylation perhaps destabilizing mRNA in certain cells or growth conditions and tRNA demethylation reducing translation. The m1A tRNA demethylation by FTO may also impact tRNA fragments during certain development processes, which merits further investigations.
In summary, we uncover a previously unrecognized spatial regulation of RNA demethylation through FTO with distinct roles in the cell nucleus versus cytoplasm. Internal m6A is the primary target of nuclear FTO in most cells. FTO can be localized to the cytoplasm and mediate both m6A and cap m6Am demethylation in polyadenylated RNA (Figure 7). In certain AML cells, we observed dramatic (up to 40%) m6A mRNA demethylation by FTO in the cytoplasm. FTO also mediates demethylation of m1A in tRNA in both cell nucleus and cytoplasm, which may tune translation together with cytoplasmic cap m6Am mRNA demethylation. Some of the phenotypes observed for FTO in brain tissues may be partially derived from tuning translation through tRNA demethylation. We also identified additional RNA substrates including m6A and internal and cap m6Am in snRNAs (Figure 7). The snRNA demethylation, in particular, m6Am in U2 RNA, may contribute to effects on splicing (Zhao et al., 2014). Lastly, the nuclear-centralized m6A demethylation and the ability of FTO to mediate m1A demethylation suggest potential nuclear roles of the m6A or m1A demethylation, on both polyadenylated RNA and other nuclear RNA species, in affecting gene expression, which will be investigated in the future.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests may be directed to and will be fulfilled by the lead contact corresponding author Chuan He (chuanhe@uchicago.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
Wild-type MEF (mouse embryonic fibroblasts) and Fto−/− MEF cell lines were obtained from Prof. Arne Klungland. Spontaneously immortalized clonal MEF cell lines were established from individual E13.5-E15.5 days-old embryos obtained from heterozygous Fto+/− crossings. Limbs were removed and remaining tissue chopped into small pieces and homogenized into single-cell suspension using a pipette. MEFs are grown in DMEM with 10% serum, 2 mM glutamine and penicillin/streptomycin. Cells grown for 4–7 days were trypsinized and frozen in complete medium with 10% DMSO as stock. Cells are grown in complete medium for 2–4 days before using them for following analysis. Human HeLa, HepG2, HEK293T, Mel624, and 3T3-L1 cell lines were purchased from ATCC. The aforementioned cells were maintained in DMEM (Gibco, #11965) with 10% fetal bovine serum (FBS, Gibco, 10438–026) and 1× Pen/Strep (Gibco, 15140) at 37°C with 5% CO2. FTO stable knockdown and the knockdown control AML cells were obtained from Prof. Jianjun Chen and cultured as previously reported (Li et al., 2017).
Tissues
Wild-type and Fto−/− mouse brains were obtained from Prof. Arne Klungland. Littermate animals were used as Fto+/+ controls (wild-type). The mice were bred and housed in a 12-hour light/dark cycle at the Comparative Medicine, Oslo University Hospital, Rikshospitalet, Norway, with a diet of pellets and water ad libitum. Brains were carefully removed from decapitated mice and quickly frozen on dry ice and stored at −80°C.
METHOD DETAILS
Plasmid construction
FLAG-tagged human FTO in the pcDNA 3.0 vector was obtained as previously reported (Jia et al., 2011). FLAG-tagged mouse FTO in the pCMV6-Entry vector was purchased from OriGene (MR208064). FLAG-tagged human ΔNLS-FTO mutant in pCMV6-Entry was generated by the deletion of 17 amino acids from the N-terminal of FTO full-length cDNA. High-purity plasmids used for mammalian cell transfection were prepared using HiSpeed Plasmid Maxi Kit (QIAGEN).
siRNA knockdown, and plasmid transfection
AllStars negative control siRNA (Qiagen, 1027281) was used as control siRNA in knockdown experiments. siRNA directed against human FTO (used in HEK293T and HeLa cells) was ordered from Qiagen (SI04293625, Hs_FTO_7 FlexiTube siRNA). Silencer® Select against mouse FTO (used in 3T3-L1 cells) was ordered from ThermoFisher (assay ID 166879). Cells were resuspended 16 hours prior to the transfection and maintained at 50% confluency for siRNA transfection or 80% confluency for plasmid transfection. Transfection was achieved by using Lipofectamine RNAiMAX (Invitrogen) for siRNA and Lipofectamine 2000 (Invitrogen) for DNA plasmids, following the manufacturer’s protocols, respectively. The cells were harvested after 48 hours post siRNA transfection or 24 hours post plasmid transfection for subsequent experiments.
Cytoplasm and nucleus separation
Cytoplasm and nucleus were separated using NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, 78833) following the manufacturer’s protocol. The capability of this kit to efficiently separate cytoplasm and nucleus for HEK293T cell and MEFs cell has been reported previously (Huang et al., 2017b; McKenna and Wright, 2015). According to the published protocol (Conrad and Ørom, 2017), the separation efficiency was validated using qRT-PCR for total RNA purified from different cellular fractions as well as western blot.
Immunofluorescent imaging sample preparation
Immunofluorescent imaging was performed as reported previously (Zhao et al., 2014). In brief, the cells were cultured in an 8-well chamber (Lab-Tek) after treatment indicated in each experiment. The cells were washed once in phosphate buffered saline (PBS) and then fixed in 4% paraformaldehyde in PBST (PBS with 0.05% Tween-20) at the room temperature for 15 min. The fixing solution was removed, and −20 °C chilled methanol was immediately added to each chamber. After incubated for 10 min at the room temperature, the cells were rinsed once with PBS and blocked with blocking solution (1% BSA with PBST) for 1 hour at the room temperature. After that, the blocking solution was replaced with the primary antibody (ab126605 for FTO, 1:1000 dilution in the blocking solution, identical to the previous report (Aas et al., 2017)) and incubated for 1 hour at room temperature. After being washed 4 times with PBST (300 μl, 5–10 min for each wash), secondary antibody (1:300 dilution in PBST) was added to the mixture and incubated at room temperature for 1 hour. After washing 4 times with PBST (300 μl, 5–10 min for each wash), anti-fade reagent (Slowfade, Invitrogen) was added to mount the slides.
RNA purification
Total RNA was purified with TRIzol® reagents (Thermofisher Scientific, #15596018) from cell lysate. Polyadenylated RNA was purified from total RNA with two rounds of polyA tail purification using Dynabeads® mRNA DIRECT™ kit (Thermofisher Scientific, #61006). The rRNAs was further removed using RiboMinus™ Eukaryote kit (Thermofisher Scientific, A1083708). Small RNAs (< 200 nt) were separated from total RNA and then subjected to size selection using 15% TBE-urea gel (Thermofisher Scientific, EC6885BOX). tRNA was then sliced and recovered from the gel.
For small nuclear RNAs, the nuclei were first isolated using aforementioned NE-PER kit and the total nuclear RNA was purified using TRIzol® reagents. Small nuclear RNAs (< 200 nt) were separated from total nuclear RNA and then subjected to size selection using 6% TBE-urea gel (Thermofisher Scientific, EC6865BOX). Individual small nuclear RNA was then sliced based on molecular weight and recovered from the gel.
Biotinylated single-stranded DNA probes:
As previously reported (Liu et al., 2016), DNA probes were designed to complement with the 3’ gene sequences of tRNAGlu(CUC), tRNAHis(GUG), tRNAGly(GCC), tRNAGly(ACC), tRNAAsp(GUC), tRNALys(CUU), tRNAGln(CUG), and tRNALeu(CAA), tRNAAla(rGC), tRNAAsn(GUU), tRNAGly(UCC), and tRNAPhe(rAA). The probes used for tRNA selection are listed in Table S4.
Individual tRNA isolation
As previously reported (Liu et al., 2016), Streptavidin-conjugated C1 magnetic Dynabeads (Invitrogen) were used for individual tRNA target isolation. 20 μl of RNase-free beads were washed once with buffer A (10 mM Tris-HCl, pH 7.5, 2 mM EDTA, 2 M NaCl), and finally resuspended in 20 μl of buffer A. Subsequently, 200 μM of biotinylated oligonucleotides in 20 μl of water were mixed with resuspended Dynabeads in buffer A and incubated at room temperature for 30 min. After the incubation, the oligonucleotide-coated Dynabeads were then washed for four times with buffer B (5 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 M NaCl) and equilibrated in 6× SSC solution (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0). Total tRNA in 6× SSC solutions and the oligonucleotide-coated Dynabeads and were heated for 10 min at 75°C separately. The tRNA and beads were mixed and incubated together at 75°C for another 10 min. Thereafter, the suspension was gently rotated at room temperature for 3 hours before washed, in succession, three times with 3× SSC, twice with 1× SSC, and several times with 0.1× SSC until the absorbance of the wash solution at 260 nm closed to zero. tRNA retained on the beads was eluted three times using RNase-free water.
Quantitative analysis of RNA modification levels
For the quantification of the m6A and cap m6Am in polyadenylated RNA, purified polyadenylated RNA (described in the RNA purification section above) was further treated with RppH (NEB, M0356S) in NEB Thermopol buffer for 2 hours to remove the cap. 50 ng polyadenylated RNA after the removal of the cap, purified tRNA and purified snRNAs were digested by nuclease P1 (Sigma, N8630), respectively, in 20 l of buffer containing 25 mM NaCl and 2.5 mM ZnCl2 for 1 h at 42 °C. Subsequently, 1 unit of alkaline phosphatase (Sigma, P5931) and NH4HCO3 (100 mM) were added and the sample was incubated for another 1 h at 37 °C. The samples were then filtered (0.22 m, Millipore) and injected into a C18 reverse phase column coupled online to Agilent 6460 LC-MS/MS spectrometer in positive electrospray ionization mode. The nucleosides were quantified by using retention time and the nucleoside to base ion mass transitions (268-to-136 for A; 296-to-150 for m6Am, 282-to-150 for m6A and m1A (retention times of 2.5 and 0.9 min, respectively)). Quantification was performed by comparing with the standard curve obtained from pure nucleoside standards running with the same batch of samples. The m6A and m6Am levels were calculated as the ratio of m6A and m6Am to A, respectively. The tRNA m1A level was calculated as the ratio of m1A to G.
Biochemistry assay of FTO activity in vitro
Similar to a previous report (Liu et al., 2016), the demethylation activity assay was performed in standard 20 μl of reaction buffer containing KCl (100 mM), MgCl2 (2 mM), SUPERNase In (0.2 U/μl, life technology), L-ascorbic acid (2 mM), α-ketoglutarate (300 M), (NH4)2Fe(SO4)2·6H2O (150 μM), and 50 mM of HEPES buffer (pH 6.5). For m6A and m6Am in polyadenylated RNA, 200 ng polyadenylated RNA purified from HEK293T cells was incubated with 2 M FTO purified in mammalian cells (High concentration) or 0.2 M FTO (low concentration) in the above reaction buffer for 3 hours and then quenched by the addition of 5 mM of EDTA, respectively. For tRNA m1A, 200 ng tRNA purified from HEK293T cell was incubated with 2 M FTO purified in mammalian cells in the aforementioned reaction buffer for 3 hours and then quenched by the addition of 1 mM of EDTA. Excessive amount of EDTA was added to control samples. To incubate similar amounts of m1A and m6A with FTO, 100 ng purified mammalian polyadenylated RNA and 20 ng tRNA were pooled together and reacted under the aforementioned demethylation conditions; tRNA m1A/A ratio is around 2–4% while polyadenylated RNA m6AA ratio is around 0.4–0.8%. The RNAs were then isolated with 100 μl TRIzol® reagents (Thermofisher Scientific, #15596018) using standard protocol and subjected to RNA digestion prior to LC-MS/MS analysis.
Biochemical demethylation assays of FTO towards m1A in single-stranded RNA (ssRNA) were conducted in 20 μl of reaction mixture together with 2 M FTO purified from the mammalian cells and 100 ng probes. The reaction was incubated at 37°C for 1 h and quenched by addition of 5 mM of EDTA. Excessive amount of EDTA was added to control samples. The samples were then centrifuged at 16000 g for 30 min at 4°C. The supernatant was collected for RNA digestion prior to LC-MS/MS analysis. The sequences of the probes were listed in the STAR Method.
Luciferase reporter assay
The luciferase activity was measured by the luminometer (SYNERGY|HTX) following the manufacturing settings of luciferase reporter assay.
Similar to a previous report (Ueda et al., 2017), in vitro translation reaction was performed using the Flexi® Rabbit Reticulocyte Lysate System (Promega, L4540). Total tRNA purified from wild-type MEF cell and Fto−/− MEF cell was incubated for 30 min at 30°C with the reaction mixture following the manufacturing protocol.
For luciferase reporter inside cells, tRNAHis(GUG) reporter was obtained as previously reported (Liu et al., 2016). Similarly, F-luc-6 × Glu(CAC) and F-luc-6 × Gly(UCC) reporter plasmid were obtained by inserting the sequence of GAGGAGGAGGAGGAGGAG and GGAGGAGGAGGAGGAGGA, respectively, before the F-luc coding region. To normalize translation differences between Fto−/− and wild-type MEF cells introduced by any other factors including initiation (i.e. variations of cellular levels of tRNAiMet and other tRNAs), a control reporter (F-luc plus R-luc) devoid of these 6× sequences was also transfected. The normalization factor from this control reporter was applied to signals obtained from the 6×GAG(Glu)-reporter, 6×CAC(His)-reporter, and 6×UCC(Gly)-reporter. General protocol: The wild-type and Fto−/− MEF cells were maintained in the 6-well plates at 80% confluency. 500 ng of reporter plasmids (pmirGlo empty vector as control or pmirGlo-specific tRNA anti-codon inserted vector) were transfected into wild-type and Fto−/− MEF cells following the aforementioned plasmid transfection protocol. After 6 hours, each well was each well was trypsin-digested, extensively washed with PBS, and re-seeded into a 96-well plate. After 18 hours from re-seeding, the cells in the 96-well plate were assayed using Dual-Glo® Luciferase Assay System (Promega, E2920). Renilla Luciferase (R-luc) was used to normalize firefly luciferase (F-luc) activity to evaluate the translation efficiency of the reporter.
Measurement of protein synthesis rate
HPG (Life Technologies; 50 nM final concentration) was added to the culture medium and incubated for 1 h. Cells were then lifted from plates and washed twice with phosphate buffered saline (PBS) and then fixed in 0.5 ml of 1% paraformaldehyde (Affymetrix) in PBS for 15 min on ice followed by permeabilizing in 200 μl PBS supplemented with 3% FBS (Sigma) and 0.1% saponin (Sigma) for 5 min at room temperature. The azide-alkyne reaction was performed with the Click-iT Cell Reaction Buffer Kit (Life Technologies) and azide-modified Alexa Fluor 488 (Life Technologies) at 5 mM final concentration. After a 30-minute reaction, the cells were washed twice with 3% BSA. All cells were filtered through a 40-μm cell strainer to obtain single cell suspensions. For flow cytometric analysis, all sorted fractions were double sorted to ensure high purity. Data were analyzed by FlowJo (Tree Star) software.
CLIP-seq
Similar to a previous report (Liu et al., 2016), in brief, for each cell line, 5 plates of cells in 15 cm style dish were grown until 80% confluency (~107 cells). The medium was aspirated and the cells were washed with PBS once and irradiated once with 400 mJ/cm2 at 254 nm in Stratalinker on ice. After irradiation, cells were harvested with a scraper, transferred to a microtube, pelleted at 4000 rpm for 3 min at 4°C, and then lysed in RIPA lysis buffer (Sigma, R2078) with cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail and SUPERNase In at 4°C for 4 hours. Subsequently, the lysis mixture was centrifuged at 17,000 g at 4°C for 30 min and the supernatant was carefully collected. Meanwhile, add 100 μl Anti-FLAG® M2 Magnetic Beads to a fresh microtube, wash beads twice with 1 ml lysis buffer. Change to a new tube, leave the beads in last wash until IP experiment. 100 μl of Anti-FLAG® M2 Magnetic Beads was added into the supernatant and mixed gently at 4°C for 4 hours. The supernatant was then discarded and the beads were washed for 3 times with 1 ml of high salt buffer and another 8 times with 1 ml of wash buffer (Wang et al., 2009). The RNAs were then eluted by proteinase K digestion following the manufacturer’s protocol. Subsequently, RNAs were recovered by phenol/chloroform extraction and then subjected to library preparation using NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, E7300S).
CLIP samples were pooled and sequenced with Hiseq-2000. Raw reads from CLIP samples were first trimmed according to recommended settings (Bolger et al., 2014). Gene structure annotations were downloaded from UCSC mm10 RefSeq/Repeatmasker. For CLIP-seq, we used a modified version of PARalyzer (Corcoran et al., 2011) to allow for all mutations, rather than just T to C mutations. For tRNA, the reads in each tRNA gene were counted. Similar to the previous report (Bartosovic et al., 2017), RNA-seq with rRNA depletion for 3T3-L1 was reanalyzed and used as the input for CLIP-seq (Schick et al., 2015). The reads distribution in tRNA of input (RNA-seq) in 3T3-L1 cells were shown in Table S2. The CLIP-seq in HEK293T cells was reanalyzed (Bartosovic et al., 2017). The reads distribution in tRNA of CLIP-seq and input (RNA-seq) in HEK293T cells were shown in Table S3.
Classification of the m6A and m6Am only genes
The m6Am list was obtained from supplementary table 1 in the previous study (Mauer et al., 2017). The m6A list was obtained from table S6 in the previous report (Meyer et al., 2012). In the reported m6A list, the peak site was annotated. The genes containing both m6A and m6Am were first classified by the overlap of two lists followed by deducting the genes that have m6A in 5’UTR (though there must be some over deduction in this step as definitely not all the 5’UTR m6As are m6Am, but just to be on the safe side). m6A only genes were classified by deducting genes containing both m6A and m6Am mentioned above from the genes in 2012 list. Similarly, cap m6Am only genes were classified by deducting genes containing both m6A and m6Am mentioned above from the genes in 2017 list. The list of three groups of genes was provided in Table S1.
Replication
For the quantification of modification changes using LC-MS-MS: 6 replicates (RNAs were purified from three plates of cells each time and two rounds of experiments were performed) were tested for most samples of transient knockdown or overexpression and controls; 4 replicates (RNAs were purified from two plates of cells each time and two rounds of experiments were performed) were tested for samples with Fto knockout, wild-type or FTO stable knockdown and controls; 5 replicates (RNAs from five Fto−/− and wild type brains, respectively) were tested for the mouse brain samples of Fto knockout and wild-type; and 3 replicates were tested for the individual tRNAs purified from Fto−/− and wild-type MEF cells. For the reporter assay experiments: in vitro, 8 replicates were performed (tRNAs were purified from four plates of cells each time and two rounds of experiments were performed); inside cells, 8 replicates were performed (cells were seeded in eight wells parallelly in one 96-well plate). For the HPG incorporation experiment, 3 replicates (three plates of cells) were tested.
QUANTIFICATION AND STATISTICAL ANALYSIS
P values were determined using the two-tailed Student’s unpaired t-test for unpaired samples in LC-MS/MS results, protein synthesis results, and reporter results. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent mean ± s.d.. n.s. means not significant, n = 3 for experiments in the Figure 5G, 6B, 6C, S4C, S4D, S5F, S5G, S5I, and S6A; n = 4 for experiments in the Figure 1B, 2C, 5A to 5E, S2G to J, and S5E; n = 5 for experiments in Figure 5F; n = 6 for experiments in Figure 2B, 4B to 4D, S1C to S1E, S2B to S2D, S3D, and S4F; n = 8 for experiments in Figure 6D, 6E and S6C. P values were determined using the Wilcoxon test in expression level analysis and using Mann-Whitney test in cumulative fraction results. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent mean ± s.d.. n.s. means not significant, for n = transcript numbers in Figure 3 and Figure S3.
DATA AND SOFTWARE AVAILABILITY
Data resources
Sequencing data have been deposited into the Gene Expression Omnibus (GEO) under the accession number GSE106395.
Supplementary Material
Highlights.
FTO mediates both internal m6A and cap m6Am demethylation of polyadenylated RNA
FTO exhibits differential substrate preferences in nucleus versus cytoplasm
The internal mRNA m6A demethylation by FTO correlates with transcript level changes
FTO affects snRNA m6A and m6Am levels and mediates tRNA m1A demethylation
ACKNOWLEDGEMENTS
This work is supported by the National Institute of Health (GM071440 and HG008935 to C.H., and CA214965 to J.C.) and the National Basic Research Program of China (2014CB964900). The Mass Spectrometry Facility of the University of Chicago is funded by National Science Foundation (CHE-1048528). The University of Chicago Cancer Center is supported by National Institute of Health CA014599. C.H. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information including six figures and four tables can be found with this article online.
DECLARATION OF INTERESTS
Chuan He is a scientific founder of Accent Therapeutics, Inc. and a member of its scientific advisory board.
REFERENCE
- Aas A, Isakson P, Bindesboll C, Alemu EA, Klungland A, and Simonsen A (2017). Nucleocytoplasmic Shuttling of FTO Does Not Affect Starvation-Induced Autophagy. PLoS One 12, e0168182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM, and Cory S (1975). Modified nucleosides and bizarre 5[prime]-termini in mouse myeloma mRNA. Nature 255, 28–33. [DOI] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, and Hinnebusch AG (1998). The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & Development 12, 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, and Vanacova S (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res 45, 11356–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, Meyre D, Golzio C, Molinari F, Kadhom N, et al. (2009). Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet 85, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil JE, Tavendale R, Watt P, Hetherington MM, and Palmer CN (2008). An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med 359, 2558–2566. [DOI] [PubMed] [Google Scholar]
- Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, Wells S, Bruning JC, Nolan PM, Ashcroft FM, et al. (2010). Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet 42, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, and Ørom UA (2017). Cellular Fractionation and Isolation of Chromatin-Associated RNA In Enhancer RNAs: Methods and Protocols, Ørom UA, ed. (New York, NY: Springer New York; ), pp. 1–9. [DOI] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, and Ohler U (2011). PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 12, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. (2017). m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 18, 2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett KA, and Barroso I (2010). The genetics of obesity: FTO leads the way. Trends Genet 26, 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Bruning JC, and Ruther U (2009). Inactivation of the Fto gene protects from obesity. Nature 458, 894–898. [DOI] [PubMed] [Google Scholar]
- Fu Y (2012). Dynamic Regulation of Rna Modifications by AlkB Family Dioxygenases In Chemistry Department (Proquest, Order No. 3548228: The University of Chicago; ). [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, and He C (2014). Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 15, 293–306. [DOI] [PubMed] [Google Scholar]
- Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun 4, 1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, et al. (2016). N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 20, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati P, Avezov E, Ma M, Antrobus R, Lehner P, O’Rahilly S, and Yeo GS (2014). Fat mass and obesity-related (FTO) shuttles between the nucleus and cytoplasm. Biosci Rep 34, e00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag S, Warda AS, Kretschmer J, Gunnigmann MA, Hobartner C, and Bohnsack MT (2015). NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 21, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Niu T, Chang J, Lei X, Zhao M, Wang Q, Cheng W, Wang J, Feng Y, and Chai J (2010). Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 464, 1205–1209. [DOI] [PubMed] [Google Scholar]
- He C (2010). Grand challenge commentary: RNA epigenetics? Nat Chem Biol 6, 863–865. [DOI] [PubMed] [Google Scholar]
- Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Bronneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento O, et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci 16, 1042–1048. [DOI] [PubMed] [Google Scholar]
- Ho AJ, Stein JL, Hua X, Lee S, Hibar DP, Leow AD, Dinov ID, Toga AW, Saykin AJ, Shen L, et al. (2010). A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc Natl Acad Sci U S A 107, 8404–8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, and Phizicky EM (2003). tRNA transfers to the limelight. Genes Dev 17, 162–180. [DOI] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. (2017a). Recognition of RNA N6-methyladenosine by IGF2BP Proteins Enhances mRNA Stability. Nat Chem Biol under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JH, Ku WC, Chen YC, Chang YL, and Chu CY (2017b). Dual mechanisms regulate the nucleocytoplasmic localization of human DDX6. Sci Rep 7, 42853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang Y-G, et al. (2011). N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, and He C (2008). Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett 582, 3313–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S, Hanna JH, Black DL, Darnell JE Jr., and Darnell RB (2017). m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31, 990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. (2017). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, and Jaffrey SR (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, et al. (2016). ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 167, 816–828 e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, and Reed R (1987). The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature 325, 673–678. [DOI] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. (2017). Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna S, and Wright CJ (2015). Inhibiting IκBβ–NFκB signaling attenuates the expression of select pro-inflammatory genes. J Cell Sci 128, 2143–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, and Jaffrey SR (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinie B, Wang J, Lim KS, Hillebrand R, Lu Z. x., Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P, et al. (2016). m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat Meth 13, 692–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-McNicoll M, and Neugebauer KM (2013). How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 14, 275–287. [DOI] [PubMed] [Google Scholar]
- Oerum S, Degut C, Barraud P, and Tisne C (2017). m1A Post-Transcriptional Modification in tRNAs. Biomolecules 7, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, and Hopper AK (2010). tRNA biology charges to the front. Genes Dev 24, 1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, and Andrade-Navarro MA (2007). The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick S, Fournier D, Thakurela S, Sahu SK, Garding A, and Tiwari VK (2015). Dynamics of chromatin accessibility and epigenetic state in response to UV damage. J. Cell Sci. 128, 4380–4394. [DOI] [PubMed] [Google Scholar]
- Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 172, 90–105 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K, and Tsujikawa K (2017). AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep 7, 42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. (2016). Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tollervey J, Briese M, Turner D, and Ule J (2009). CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods 48, 287–293. [DOI] [PubMed] [Google Scholar]
- Wei C-M, Gershowitz A, and Moss B (1975a). N6, O2[prime]-dimethyladenosine a novel methylated ribonucleoside next to the 5[prime] terminal of animal cell and virus mRNAs. Nature 257, 251–253. [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, and Moss B (1975b). Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell 4, 379–386. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. (2017). RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, and He C (2008). Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452, 961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, and Hunt JF (2006). Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature 439, 879–884. [DOI] [PubMed] [Google Scholar]
- Yu J, Chen M, Huang H, Zhu J, Song H, Zhu J, Park J, and Ji SJ (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res 46, 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 24, 1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, and Qian S-B (2015). Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Shu XE, Mao Y, Liu X-M, Yuan X, Zhang X, Hess ME, Brüning JC, and Qian S-B (2018). N6-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol Cell 69, 636–647.e637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Toh JDW, Wong KHQ, Gao Y-G, Hong W, and Woon ECY (2016). N6-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci Rep 6, 25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.