Abstract
Much of our lives are spent in unconstrained rest states, yet cognitive brain processes are primarily investigated using task-constrained states. It may be possible to utilize the insights gained from experimental control of task processes as reference points for investigating unconstrained rest. To facilitate comparison of rest and task functional magnetic resonance imaging data, we focused on activation amplitude patterns, commonly used for task but not rest analyses. During rest, we identified spontaneous changes in temporally extended whole-brain activation-pattern states. This revealed a hierarchical organization of rest states. The top consisted of two competing states consistent with previously identified “task-positive” and “task-negative” activation patterns. These states were composed of more specific states that repeated over time and across individuals. Contrasting with the view that rest consists of only task-negative states, task-positive states occurred 40% of the time while individuals “rested,” suggesting task-focused activity may occur during rest. Together our results suggest that brain activation dynamics form a general hierarchy across task and rest, with a small number of dominant general states reflecting basic functional modes and a variety of specific states potentially reflecting a wide variety of cognitive processes.
Keywords: : community detection, dynamics, fMRI, graph theory, resting-state
Introduction
The brain is a distributed information-processing system with rich spatiotemporal dynamics underlying complex cognitive dynamics. A major goal of cognitive neuroscience is to create a mapping between these two forms of dynamics to better understand the neural basis of cognition. Recent insights in human neuroimaging research have improved this mapping by considering activity in more than one spatial location at a time. These approaches include multivariate pattern analysis (MVPA) of brain activity patterns corresponding to cognitive task events (Haxby et al., 2014; Haynes, 2015), as well as functional connectivity (FC) analysis of brain network dynamics during rest and task (Fox and Raichle, 2007; Hutchison et al., 2013; Smith et al., 2011). Building on these advances, in this study, we use what we term “dynamic MVPA” (dMVPA)—MVPA applied to the temporal evolution of brain processes (Anderson et al., 2012; Betzel et al., 2012; King and Dehaene, 2014; Yuan et al., 2012)—with functional magnetic resonance imaging (fMRI) to characterize the repertoire of brain states across a variety of resting and task cognitive states.
In particular, we used a state space characterization of dynamics previously used to gain insight into other real-world complex systems (Furusawa and Kaneko, 2012; Junejo, 2010). This involves conceptualizing each whole-brain image in time as a single point in a high-dimensional feature space. Brain state changes are thus equivalent to movement through that state space. We primarily utilize distance between these points in state space, using a standard distance metric (spatial pattern correlations) between time points to temporally cluster them into brain states extending through time. More generally, we are proposing the question: how do brain states (whole-brain images) evolve over time? The unique difference here in our state space characterization is the focus on shifts in whole-brain activation patterns across time, in contrast to the sliding-window dynamic FC approaches (Allen et al., 2014; Hutchison et al., 2013), allowing us to observe movement across state space at higher temporal precision (per time point). Here, we map cognitive processes onto brain states in context of a state space to apply methods from dynamical systems to investigate the characteristics of such brain states (Motter, 2015). In this study, we define a brain state as a whole-brain activation pattern at specific point in time and state transitions as changes in activation patterns across time. We can characterize the brain states as “attractor states” based on the cognitive functions and/or behavioral outputs of these states or groups of similar-function brain states as “attractor state clusters/networks” by analyzing the topography of the state space (Motter, 2015). We perform further analyses using graph theoretical formulations. Relative to classical clustering approaches, graph theoretical community detection algorithms can better assign patterns near the edges of a cluster (Newman and Girvan, 2004). This can provide a more comprehensive characterization of a state space's large-scale organization.
We propose the use of the term dMVPA given the similarity of this approach to existing approaches in fMRI FC dynamics (Allen et al., 2014; Hutchison et al., 2013) and in MVPA-based magneto-/electro- encephalogram (M/EEG) methodologies (Khanna et al., 2015; King and Dehaene, 2014). However, whereas standard MVPA uses supervised machine learning (Haynes, 2015) to classify activation patterns during experimentally induced cognitive events, dMVPA can be applied to the moment-by-moment temporal evolution of brain states to characterize spontaneous cognitive events. Furthermore, identifying whole-brain activation pattern states [rather than FC/temporal covariance patterns (Smith et al., 2009, 2012)] facilitates the functional interpretability of those states. This is due to whole-brain fMRI activation patterns being more directly comparable to the vast fMRI task activation literature, which associates cognitive task manipulations with whole-brain spatial activation maps (Laird et al., 2009; Yarkoni et al., 2011). Several recent studies have focused on resting-state activation patterns (Liu and Duyn, 2013; Liu et al., 2013). Specifically, these studies applied k-means clustering to selectively average fMRI time points to identify spatially averaged coactivation patterns. However, these studies focused on relating these patterns to resting-state networks, rather than task activations. In this study, we compare resting-state activation patterns to specific task activation maps—potentially involving coactivation of multiple resting-state networks—to determine whether resting states traverse a similar state space as task states.
We utilize rest data, in part, given the possibility that many brain states are visited in this unconstrained “task-free” context (Fox and Raichle, 2007). This allowed us to obtain a broad sampling of possible brain states across many (N = 97) individuals. We also supplemented this broad repertoire of spontaneous states with experimentally controlled states identified from a variety of tasks involving distinct cognitive functions. We hypothesized that activation-pattern states would overlap between unconstrained rest and task performance, reflecting a common brain activation-pattern state space (He, 2013) between these two highly distinct levels of overt behavior. Conceptualizing activation patterns as brain states puts rest and task in a common conceptual framework, likely facilitating insights into general neural activation-pattern dynamics across a wide variety of mental states.
Materials and Methods
Participants
Data were collected as part of the Washington University-Minnesota Consortium Human Connectome Project (Van Essen et al., 2013). The participants were recruited from the Washington University campus and surrounding area. All participants supplied informed consent. The data were from the “500 Subjects” public data release. We used data from the “100 Unrelated Subjects” set as we wanted a sample representative of the general population (excluding family relations).
We used resting-state fMRI and task-state fMRI data from 100 subjects, with 3 outlier subjects removed for a subset of analyses. The resting-state dataset consisted of four separate runs, each spanning 14.4 min in length. Analyses were performed separately for each rest run. The task data involved seven diverse tasks (Barch et al., 2013). These seven tasks were selected to tap into different cognitive processes as well as the different neural circuitry that supports those functions. The tasks were related to emotion perception, reward learning, language processing, motor responses, relational reasoning, social cognition, and working memory.
MRI parameters
Whole-brain echo-planar scans were acquired with a 32 channel head coil on a modified 3T Siemens Skyra with TR = 720 ms, TE = 33.1 ms, flip angle = 52°, BW = 2290 Hz/Px, in-plane FOV = 208 × 180 mm, 72 slices, 2.0 mm isotropic voxels, with a multiband acceleration factor of 8 (Uğurbil et al., 2013). Data were collected across 2 days. On each day, 28 min of rest (eyes open with fixation) fMRI data were collected across two runs (56 min total), followed by 30 min of task fMRI data collection (60 min total). Each of the seven tasks was completed over two consecutive fMRI runs. Details regarding the resting-state data collection for this dataset can be found elsewhere (Smith et al., 2013), as well as details about the tasks (Barch et al., 2013).
fMRI preprocessing
We used a minimally preprocessed version of the data, which was the result of standard procedures, including spatial normalization to a standard template, motion correction, and intensity normalization. These steps have been described previously (Glasser et al., 2013). We performed analyses on the volume version of these minimally preprocessed data using AFNI (Cox, 1996). We removed variables of no interest from the time series using linear regression, including motion estimates, ventricle and white matter signals, as well as their derivatives. Ventricle, white matter, gray matter, and anatomical structures were identified for each subject using FreeSurfer (Fischl et al., 2002, 2004). A linear trend was removed from the signal of each time series, and the data were spatially smoothed (FWHM = 4 mm). Resting fMRI data are also typically temporally filtered to isolate the low frequency component of the time series. However, we did not apply a temporal filter to the data due to the possibility of relatively rapid brain state transitions. Further data analysis was completed by sampling from a set of 264 regions to capture and explore large-scale regional and system-level questions (Power et al., 2011).
Brain state identification
We conceptualized time in terms of a weighted graph, with each time point as a graph node and edges as the similarities of whole-brain spatial activation patterns at those time points. Activation pattern similarity (similarity of whole-brain activation patterns from one time point to another) was calculated using Pearson correlation. Activation pattern similarity is nearly identical to FC: correlations are performed on the other dimension (FC—correlations between time-series across regions; activation pattern similarity—correlations between spatial pattern across all regions between time points). Pearson correlations were used given that it is a well-established distance measure (Cha, 2007), is invariant to scale changes, and can be conveniently conceptualized as (the square root of) linear variance explained. These correlation-based associations were summarized in a temporal similarity matrix (Fig. 1C), consisting of all pairwise similarities among time points.
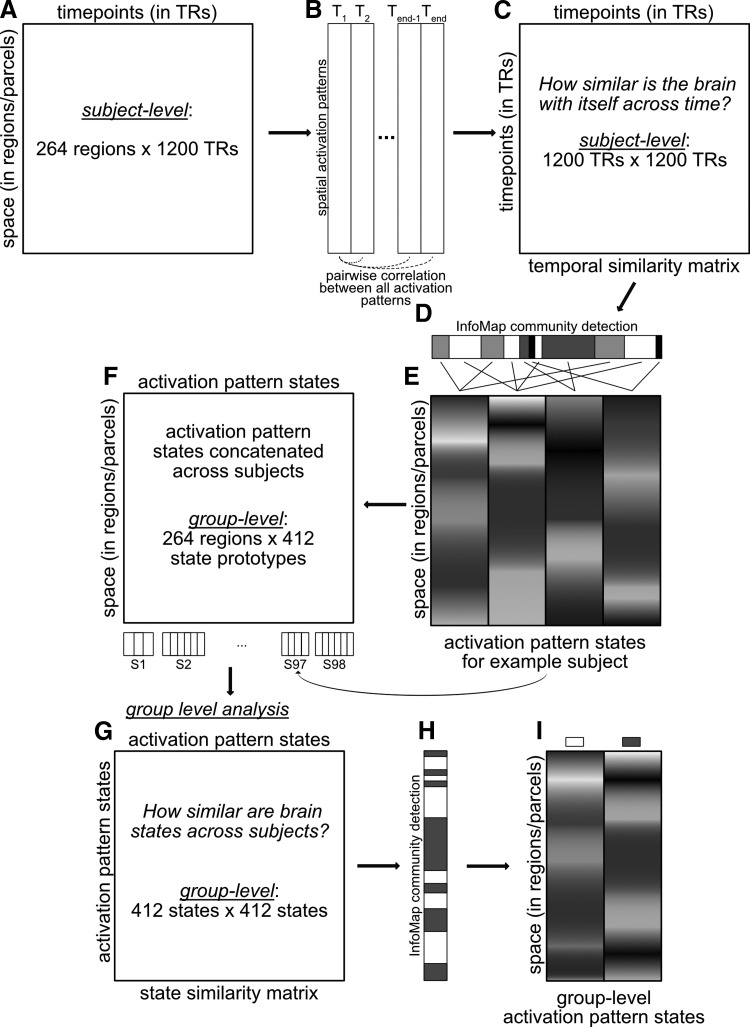
FIG. 1.
Brain state identification methods flowchart. Summary of the analysis pipeline for determining activation pattern states. For each subject's data (A), pairwise correlations were computed between all activation patterns (B), resulting in a temporal similarity matrix (C). Infomap community detection algorithm was applied to this temporal similarity matrix, clustering similar time points into the same unique state (D). State prototypes were calculated by averaging activations across all time points within the same community (E). Group level analyses were conducted by concatenating all individual subject state prototypes into one data matrix (F) and performing the same analysis steps (B–E) this group-level matrix, shown in (G) group-level state similarity matrix, (I) Infomap community detection algorithm solution at the group-level, and (H) group-level activation pattern states.
Time points with strong edges between them were considered as instances of the same brain state. Clusters of similar time points were identified using the Infomap community detection algorithm (Bohlin et al., 2014; Rosvall and Bergstrom, 2011; Rosvall et al., 2009). The Infomap approach is largely based on the Louvain method and optimizes the “map equation” [see www.mapequation.org/code.html (Rosvall et al., 2009) for more details]. Specifically, the algorithm maps the trajectory of a random walker moving through a network to find underlying community structure. In addition, there is an iterative search process step that starts the random walker at different locations in the network to identify the optimal community structure across all iterations (in our study, we set the number of iterations to 100 for computational tractability). Infomap community detection algorithm was first applied to each subject's initial resting-state fMRI run (14.4 min, 1200 time points). Given the likely presence of noise, we thresholded the temporal similarity matrix at thresholds from 1% to 50% connection density (at 1% increments), before the application of Infomap. For each iteration of the connection density thresholding, we applied the Infomap algorithm to assign each time point to unique communities. Modularity was calculated to assess the quality of Infomap's assignment of time points to communities (Blondel et al., 2008; Rubinov and Sporns, 2011). A consensus clustering approach (Lancichinetti and Fortunato, 2012) was applied to derive a final set of community assignment for an individual subject's resting-state run (Fig. 1D). Specifically, consensus clustering converges on a stable community assignment by identifying common community features across each of the 50 Infomap assignments, weighted by their respective modularity scores. This step was performed on a per-subject basis to identify subject-specific community assignments or brain states. This produced on average 4 unique brain states (graph communities) per subject, with a total of 412 brain states across 98 subjects. Note that 2 of the 100 subjects were excluded because the Infomap algorithm returned more than three standard deviations above the average number of states per subject. We reduced the many time points contributing to each brain state to a single “prototype” vector for each brain state via averaging (Fig. 1E).
We also tested two other popular clustering approaches, k-means clustering and hierarchical clustering, on a single subject (to avoid potential overfitting of a specific method on the full dataset). We found that clustering solutions were quite similar to one another, with the Infomap solution performing slightly better based on modularity scores (see Supplementary Fig. S3 for details; Supplementary Data are available online at www.liebertpub.com/brain).
We next conducted a group-level analysis with the goal of identifying shared brain states across subjects (Fig. 1F–I). This involved first computing an adjacency matrix based on the similarities among the brain-state prototypes across all subjects (Fig. 1G). We then applied the Infomap algorithm to that matrix (Fig. 1H) with no thresholding to produce a group-level brain state solution (Fig. 1I). Later, we tested for the presence of a hierarchy of brain states, with the no-threshold (group-level) solution being the top of the hierarchy. We produced lower levels of the hierarchy by rerunning the algorithm (independent of the previous level) using a series of different density thresholds. We began with 100% density (no threshold), going down by 10% increments until the 10% density level. In contrast to the subject-level analysis, no consensus approach was applied across the density thresholds. Each density threshold was analyzed independently, since reducing the density eliminates weaker connections and allows for better isolation and separation of the communities (Power et al., 2011).
Assessing the amount of time spent in each brain state
For resting-state data, each time point of an individual's whole-brain activation was categorized as State A or State B, depending on its similarity to the two-state solution based on the group-level Infomap result. The estimated amount of time spent in each of the two states was then calculated based on the two-state clustering for each individual subject. We then trained a classifier using support vector machines (SVM) to be able to decode whether a subject was in State A or State B at a given time point. Specifically, the SVM classifier was trained on rest runs 2–4 and validated by testing on rest run 1. We then classified every time point for each subject during task sessions. Specifically, the SVM classifier was trained on rest runs 2–4 and validated by testing on rest run 1 before classification of task-state data (for each task separately). SVM training and task-state data classification were all performed within subject (i.e., SVM trained on subject 1's resting-state data before classification of subject 1's task data). Due to this, one additional subject (total N = 97) was removed from the analysis because the subject's resting-state data only exhibited one unique state after group-level clustering remapping. Note that validation with rest run 1 data was based on above-chance classification accuracy, using the group-level Infomap clustering labels as the ground truth. The two-state classification results were used to determine the amount spent in each state for each of the seven tasks.
Neurosynth state decoding
Brain states were decoded using the Neurosynth decoder tool (Yarkoni et al., 2011). Neurosynth is a meta-analytical tool that contains a database with brain activation patterns and peak signal coordinates paired with the associated cognitive terms from the fMRI scientific literature. The decoder function takes in our voxelwise representation of brain states, cross-references with the database, and returns a list of cognitive terms each paired with a correlation score indicating how well each brain state is associated with each cognitive term. The decoder returned a list of ∼3406 cognitive terms and their correlation values with the tested brain state. Of the top 50 highest correlated terms, all anatomical terms, redundant terms, nonsensical terms (i.e., “cortexmpfc,” “networkdmn”), and methodological terms were excluded from the list. The remaining terms were visualized as word clouds (Fig. 6) and the relative font sizes of each term were determined by the correlation scores. We also sought to validate the results returned by Neurosynth by running a series of permutation tests for each brain state (Supplementary Fig. S2).
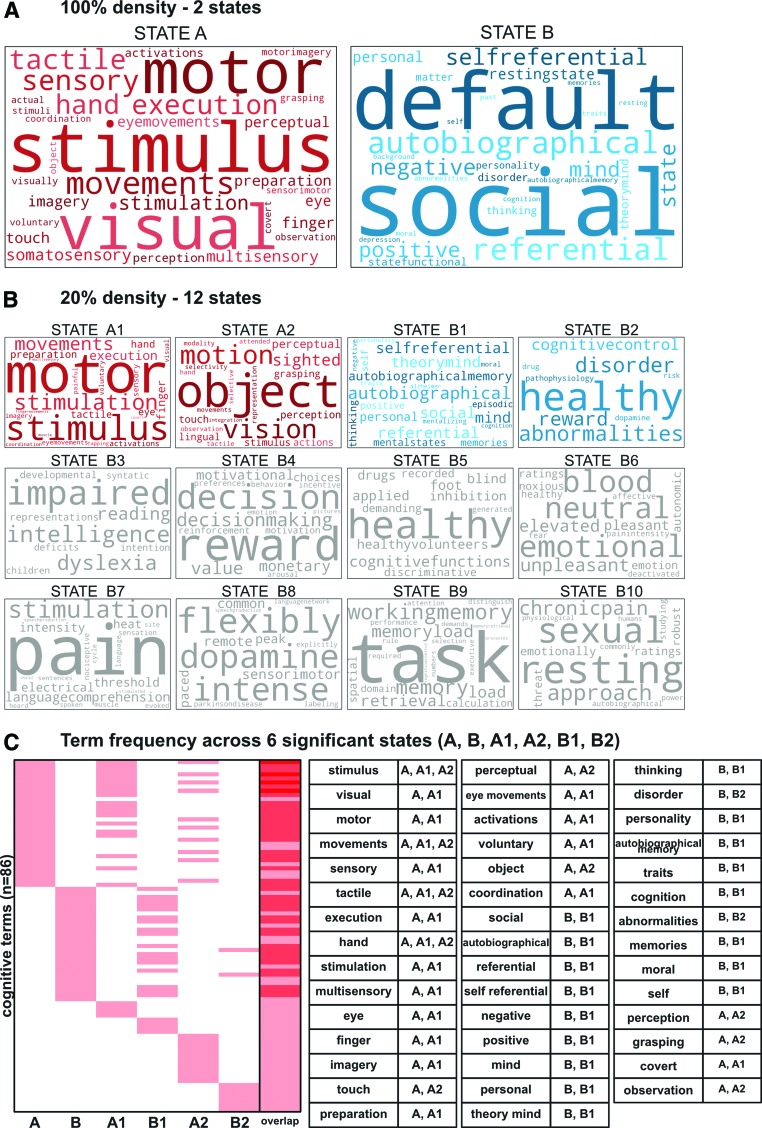
FIG. 6.
Decoding the states using Neurosynth. Each state at the 100% (A) and 20% (B) levels of the hierarchy illustrated in Figure 5 were decoded using Neurosynth. Cognitive terms with the strongest associations with each state are depicted as word clouds (top 50 terms with nonfunctional terms removed; see Materials and Methods section for details on term selection). Terms with higher correlation values with the indicated brain state activation pattern are represented by larger font sizes (with size normalized within each brain state's word cloud). States B3–B10, colored in gray, did not survive the permutation testing (see Supplementary Data for details). (C) Frequency of cognitive terms spread across the top 6 significant states. Only four terms were present in more than two states (“stimulus,” “movements,” “tactile,” “hand”). Terms listed in the table are terms that were present in multiple states (44/86 total terms). Color images available online at www.liebertpub.com/brain
Results
Identifying multivariate brain activation states
We hypothesized that whole-brain fMRI activation patterns would consist of discrete states—configuration patterns that extend (and repeat) through time. To test this possibility, we clustered whole-brain resting-state fMRI activations in time. A standard set of functionally defined regions (Power et al., 2011) was used for computational tractability as well as for the previously-identified functional system assignments (Fig. 2D). Spatial correlations were used as a similarity/distance measure across individual time points, with brain states defined as temporal clusters of similar activation patterns (Fig. 2A).
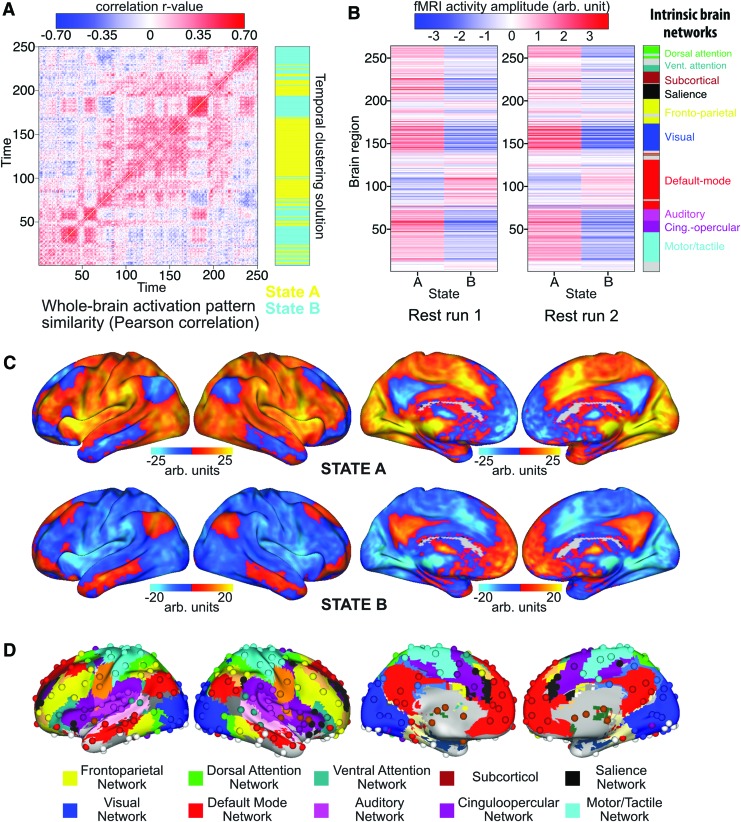
FIG. 2.
The repertoire of brain states based on resting-state fMRI. Each individual's prototypical brain states were correlated and clustered using the same Infomap algorithm. (A) An example of individual time point similarity based on spatial correlations. 250 time points from a single subject (HCP subject 100408, time points 100:350) are shown for illustration. The across-subject clustering result (mapped back to this subject's data) is shown in blue and yellow. (B) Brain state prototypes for State A and State B, averaged across all activation vectors across all subjects. The two-state results replicated across all four of the rest runs (results from two runs are shown in B). (C) Voxelwise representations of State A and State B. Note that State B involves activation of the default-mode network as well as the tan/salmon-colored portion of the frontoparietal network in (D). (D) Functionally defined set of 264 regions and the associated functional network assignments. fMRI, functional magnetic resonance imaging. Color images available online at www.liebertpub.com/brain
Brain states were identified first at the individual subject level. Of the 100 subjects, 2 subjects were excluded from subsequent analyses because the clustering algorithm returned more than three standard deviations above the average number of states per subject. On average, 4 unique brain states were identified for each subject for a total of 412 unique brain state prototypes across the remaining 98 subjects. These 412 brain states were reclustered in a group-level analysis. The same processing steps performed at the individual subject level were applied at the group level for determining group-level brain states (see Materials and Methods section for details). Note, while the brain states identified in Figure 2 resemble resting-state FC networks, the brain states were derived from activation pattern correlations, which is orthogonal to FC calculations.
At the group level, two brain states were identified, which we labeled “State A” and “State B” (Fig. 2B, C). The states were summarized by averaging all prototypes with the same clustering label and visualized in 264 regions of interest space (Fig. 2B) and in voxel space (Fig. 2C). An example subject's temporal similarity matrix along with the group-level clustering solution is illustrated in Figure 1A. Periods of highly similar activation patterns can be observed by the blocked structure on the diagonal of the temporal similarity matrix, which are grouped as the same state in the clustering solution. We replicated the current findings using the same approach for the remaining three rest runs and found similar results (second run shown in Fig. 2B). Specifically, State A identified in any given rest run was highly correlated on average with State A identified in other rest runs: rank correlation rho = 0.92 (p < 0.0001 for all pairwise comparisons). This was also the case for State B being correlated with State B identified in other rest runs: average rank correlation rho = 0.91 (p < 0.0001 for all pairwise comparisons). In addition, State A and State B were highly anticorrelated at rho = −0.97 on average (p < 0.0001 for all pairwise comparisons). Note that the clustering approach used to identify distinct activation states prioritizes identification of maximally distinct (e.g., anticorrelated) states. To confirm the statistical significance of the observed anticorrelation despite this prioritization of maximally distinct states, we performed a permutation test that shuffled State A and B labels within subject and found that the observed result was p < 0.0001 relative to chance clustering of time points (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/brain).
Functionally, State A is highly similar to observed “task-positive” activation patterns reported in the literature (Corbetta and Shulman, 2002; Fox et al., 2005; Power et al., 2011). This activation pattern includes active regions in many task-related functional networks, such as the dorsal and ventral attention networks, salience network, and sensory-motor networks (visual, auditory, motor). Conversely, State B is likely to be a “task-negative” state, given the strong activation of the default-mode network (DMN) (Fox et al., 2005; Uddin et al., 2009) (along with a portion of the frontoparietal cognitive control network [FPN]). Note, however, that despite using “task-negative” data (i.e., fMRI data collected during rest), we were able to extract both task-positive and task-negative states.
The relationship between spatial activation patterns (spatial correlations) and FC (temporal correlations)
Using resting-state data, we identified a “task-positive” State A and a “task-negative” State B with patterns observed in both the task activation fMRI literature and the resting-state FC literature. The use of resting-state data both here and in the resting-state FC literature raises the possibility that the effects observed here somehow recapitulate those same resting-state results by different means. Notably, however, the approach used here was quite distinct from FC analyses, as FC analyses used temporal correlations, while the activation-pattern state approach used spatial correlations. Nonetheless, we next sought to verify that the activation-pattern approach produced analytically distinct results from resting-state FC.
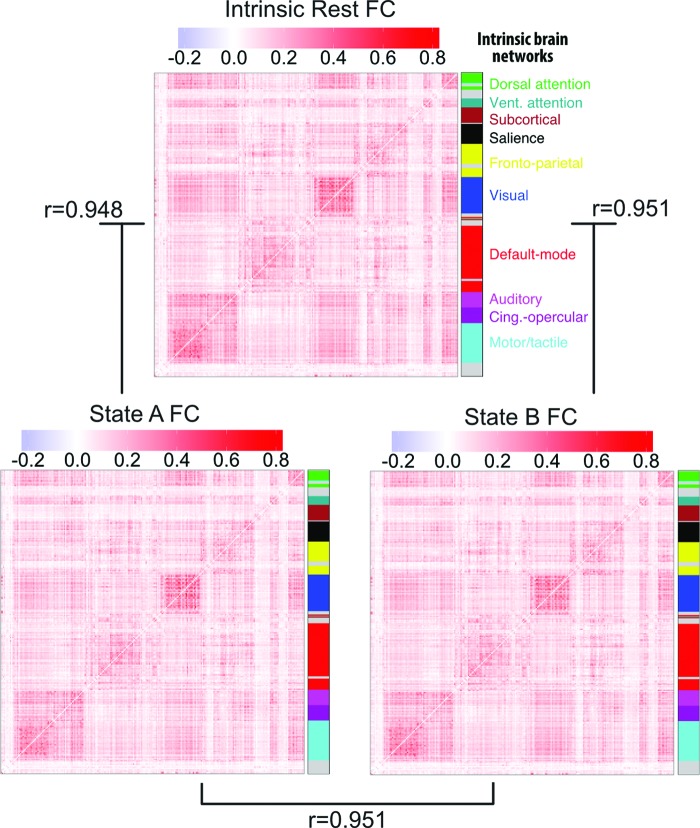
If the FC approach were somehow analytically equivalent to activation-pattern state approach, then we would expect that calculating FC in each of the activation-pattern states would yield distinct FC patterns. We therefore analyzed resting-state FC patterns, including only time points identified as either State A or, separately, State B. Comparing between the FC patterns calculated by averaging across all subjects (Fig. 3), we found that the overall resting-state FC patterns (calculated across the entire rest run, separately for State A time points and State B time points) correlated with State A FC at r = 0.948 and State B FC at r = 0.951. This was inconsistent with patterns identified using resting-state FC (temporal correlations) being mathematically equivalent to activation-pattern states (spatial correlations). In addition, to test whether results were consistent across subjects, FC comparisons were also performed for each subject separately. We then performed random-effects statistical analyses to test for cross-subject consistency. The overall resting-state FC pattern correlated with State A FC on average at r = 0.55 [t(96) = 42.4, p < 0.0001] and State B FC on average at r = 0.54 [t(96) = 49.1, p < 0.0001]. Note that we calculated the overall resting-state FC pattern based on a separate run (e.g., run 2 when comparing to run 1's State A FC) to remove circularity from the analysis (Kriegeskorte et al., 2009). All noncircular combinations (between the first and second resting-state run) were computed and the reported results are based on averages across these comparisons.
FIG. 3.
Independence of activation state dynamics from FC state dynamics. The FC matrices for resting-state (over the entire rest run), State A (just the time points labeled state A), and State B (just the time points labeled state B) are pairwise correlated with each other for each subject. Averaged FC matrices and correlation values across subjects are shown here. These results suggest that the whole-brain multivariate activation states investigated here are independent of FC dynamics. FC, functional connectivity. Color images available online at www.liebertpub.com/brain
The strong correlation between the individual states' FC and overall rest FC suggests that resting-state FC architecture remains consistent across distinct activation brain states. Further supporting this conclusion, cross-subject averaged State A FC correlated with cross-subject averaged State B FC at r = 0.951. This was also the case when computed based on averaging within-subject comparisons: r = 0.49 [t(96) = 48.6, p < 0.0001], suggesting cross-subject consistency. Note that this was calculated by comparing across separate rest runs, as with the overall resting-state FC pattern comparisons above. The resulting average similarity is very high relative to chance, but also in contrast to the strong anticorrelation (rho = −0.97) between State A and State B activation patterns. This suggests that whole-brain activation pattern dynamics are unique and largely independent of FC dynamics.
Functional relevance of the two states
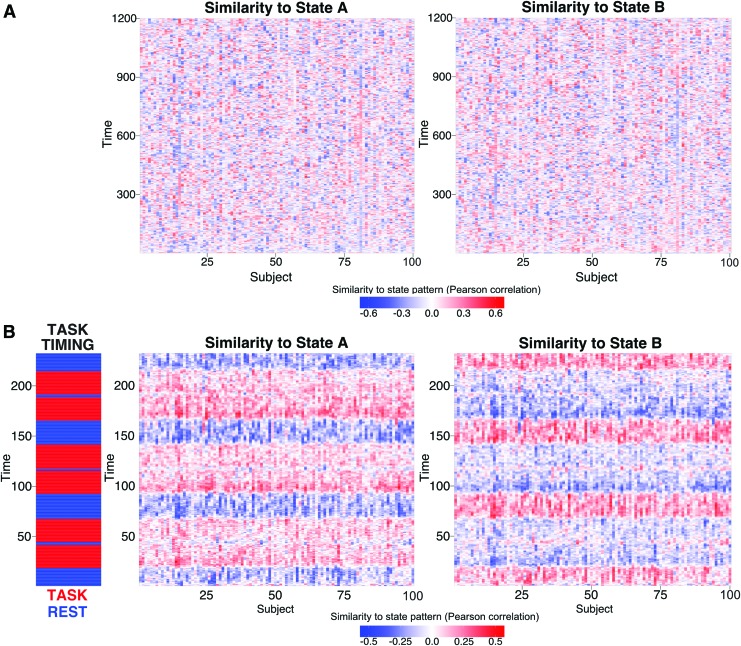
Identifying a task-positive State A when using “task-negative” resting-state data suggests that subjects may have been visiting a common set of states during rest and task. To better test this hypothesis, we directly compared the identified State A and State B activation patterns with rest and task data. This involved testing correlations between the two states' average activation patterns (Fig. 2B) and every individual time point's whole-brain activation pattern. This was done for all subjects individually for resting state and for each of the seven tasks (Fig. 4B shows correlation results for the reasoning task).
FIG. 4.
Functional relevance of the two main states. The two brain states observed at the group level (Fig. 2B) are correlated with each subject's rest and task data (separately for each time point). No discernable temporal organization of State A and State B is observed in resting-state data across all 100 subjects (A). Evidence of State A during rest may suggest subjects are performing covert tasks during rest. Clear structure is seen when correlated with activity during the reasoning task (B). All subjects consistently enter State A when entering task blocks (in red), and enter State B when entering intertask rest periods (in blue). Color images available online at www.liebertpub.com/brain
For resting-state data, no cross-subject temporal patterns were observed (Fig. 4A), as expected. On average, subjects were in State A only 39% of the time, and in State B 61% of the time. In contrast, for each of the seven tasks (Table 1), we found that every subject was in State A most of the time during task blocks and in State B mainly during the intertask rest periods (Fig. 4B), with the exception of the language task. On average, subjects were in State A 54% (State B 46%) of the time during task blocks and in State A 41% (State B 59%) of the time during intertask rest periods. In addition, subjects tended to stay in State A longer for tasks that were likely more challenging or cognitively demanding. For instance, subjects were in State A 59% (State B 41%) for the reasoning task, but only in State A 53% (State B 47%) for the motor task. This suggests that State A is not only relevant to rest data (the data used to derive it) but also likely to be the same high-level state required for performing various active task demands.
Table 1.
Percent Time in Each State for Each Task
| Task blocks | Intertask rest blocks | |||
|---|---|---|---|---|
| Task type | State A, % | State B, % | State A, % | State B, % |
| Reasoning | 59 | 41 | 36 | 64 |
| Emotion | 52 | 48 | 40 | 60 |
| Working memory | 53 | 47 | 39 | 61 |
| Gambling | 57 | 43 | 38 | 62 |
| Language | 49 | 51 | 50 | 50 |
| Social | 55 | 45 | 41 | 59 |
| Motor | 53 | 47 | 40 | 60 |
| Average | 54 | 46 | 41 | 59 |
Percent of time in each state during task blocks and inter-rest rest periods for each task.
Hierarchical organization of brain states
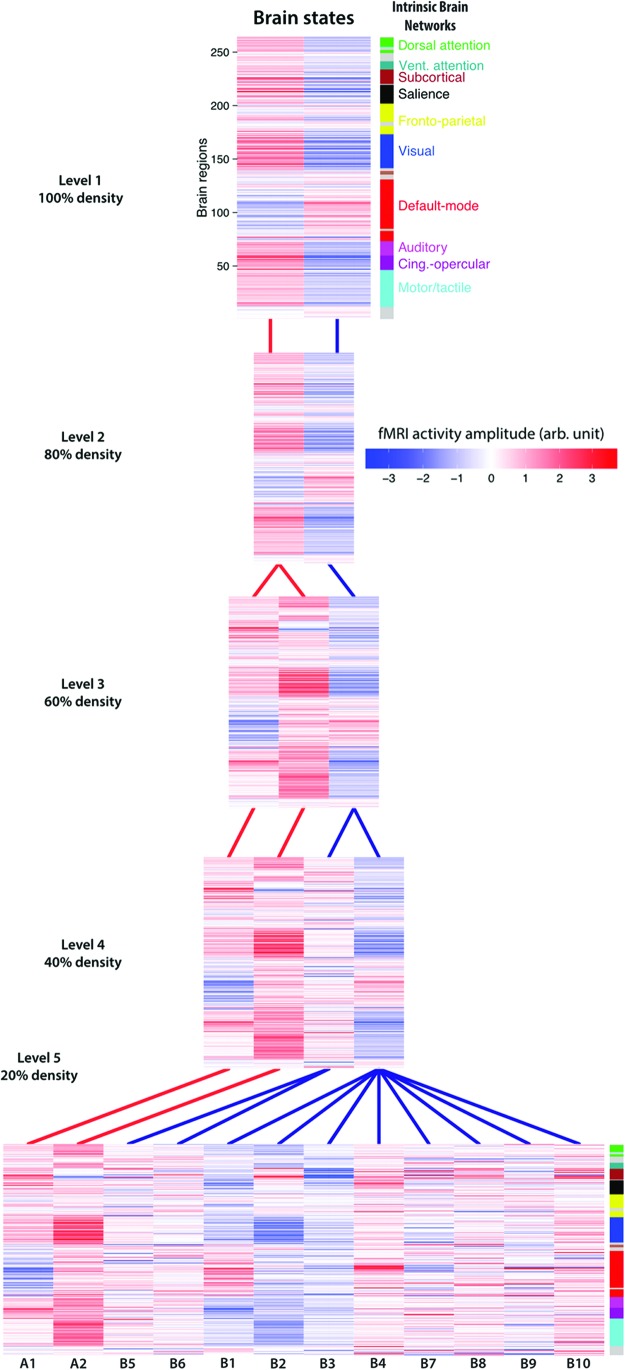
Despite the previous analyses identifying two primary brain activation-pattern states, it is implausible for there to be only two brain states, given the wide variety of possible tasks that brains perform. Furthermore, the vast fMRI literature indicates that each active task (cognitive state) has a unique activation pattern, despite an overarching “task-positive” activation pattern for externally oriented and cognitive control tasks (Corbetta and Shulman, 2002; Fox et al., 2005; Power et al., 2011). We therefore hypothesized that brain states are organized in the form of a hierarchy, with State A and State B at the top level and more specific task states at lower levels. To identify these lower-level task-specific states, the two top-level states were broken down into more states by reducing the connection density of the adjacency matrix before applying Infomap clustering (from 100% to 20% density at decreasing increments of 20%) at the group-level (i.e., after identifying subject-specific brain states). Reducing the connection density of the temporal similarity matrix removes weaker graph edges (lower correlations) between the group state prototypes and increases separation of similar spatial activation patterns. The task-positive State A divides into two states early in the hierarchy (60% density), shown by the red links between the levels of the hierarchy (Fig. 5). The task-negative State B splits into multiple states at the lowest level of the hierarchy (20% density), shown with blue links between the levels of the hierarchy.
FIG. 5.
Relationship between levels of the brain state hierarchy. The top level of the hierarchy (i.e., with no thresholding) is depicted at top, with additional states lower in the hierarchy with more stringent thresholding. Each level was defined by removing graph edges between the group prototype states (e.g., 80% density means removing the weakest 20% of edges) before running the community detection algorithm. The lines depict which state in the higher level that the lower-level state was most associated with. Specifically, a line indicates which higher-level state was composed of the largest percentage of the same group-level prototypes as the lower level state. Red links are associated with State A (task-positive) at the top level while blue links are associated with State B (task-negative) at the top level. The state levels below each of the states at 20% density (level 5) corresponds with the state levels in Figure 6. Color images available online at www.liebertpub.com/brain
Decoding brain states using Neurosynth
We next sought to take advantage of the vast fMRI task activation literature to decode the identified resting-state brain states, identifying possible cognitive processes occurring during rest. This meta-analytic decoding approach also allowed us to test the hypothesis that State A and State B are likely to be general states governing basic modes of cognition, while the lower level states (12 states at 20% density) are likely more functionally specific. Note, however, that it will be critical for future research to verify that the mental states predicted from brain activation-pattern states are actually present as predicted.
To examine the specific cognitive functions associated with each state at the 100% and 20% density levels, each of the 14 states (State A, State B, and the 12 states at 20%) were decoded using the Neurosynth decoder function and visualized as word clouds (Poldrack et al., 2009). The word clouds for State A and State B (Fig. 6A) were consistent with our previous interpretation of a “task-positive” state and a “task-negative” state, respectively. In addition, the cognitive terms for each of the lower level states (Fig. 6B, C and Supplementary Fig. S2) are quite distinct from one another, despite certain terms repeating across states (“healthy” for State B2, B5, B6; see Supplementary Fig. S2). State A branched into two lower-level states that shared similar cognitive terms. In contrast, State B is subdivided into 10 lower-level states that include more diverse cognitive terms spanning multiple functional domains not seen in State B word cloud.
We validated that the meta-analytic decoding results were unlikely to have occurred by chance by running a permutation test for each of the brain states (Supplementary Fig. S2). In brief, this involved shuffling terms across activation-pattern states, creating a nonparametric null distribution for each term. We found that all of the cognitive terms of interest depicted in State A and State B word clouds were significantly different when compared against the permutation tests' null distributions (p < 0.001). In addition, all cognitive terms for State A1, A2, B1, and B2 were significantly different (p < 0.05) when compared to the null distributions. Note that the permutation tests controlled for multiple comparisons (family-wise error), since all 3406 term comparisons were included in each permutation. The remaining States B3–B10 (Fig. 6B, in grey) did not survive the permutation tests (at p < 0.05). Together, these results support the effectiveness of Neurosynth decoding of spontaneous activation states and are consistent with the identified brain states corresponding with distinct mental/cognitive states.
Discussion
By characterizing moment-to-moment activation patterns in a high-dimensional state space, we observed a hierarchical organization of functional brain states. Two domain-general states (State A and State B) occupy the highest tier of that hierarchy. These two states can be further subdivided into functionally specific substates. As expected, subjects spent a majority of time in State B during resting-state runs, which matches the activation pattern commonly observed to be more active during rest than during most tasks. Despite using resting-state data for brain state identification, we found that subjects spent a significant portion of time (39%) in State A, which matches the activation pattern commonly observed to be more active during a wide variety of tasks than during rest. The possible functional relevance of these activation patterns was characterized using a variety of distinct task states as well as activation-pattern decoding based on meta-analysis of thousands of fMRI studies. Together, these results suggest that whole-brain activation state configurations correspond between rest and tasks, indicating that the human brain traverses a common activation-pattern state space during rest and a variety of tasks.
Hierarchical organization of brain states
Using the dMVPA approach, we identified a pair of competing anticorrelated states based on spontaneous activation patterns. State A appears to be a “task-positive” state, with high activation amplitudes in common task-active areas such as sensorimotor and cognitive control networks (Cole and Schneider, 2007; Corbetta and Shulman, 2002; Dosenbach et al., 2007; Raichle, 2010). State B appears to be a “task-negative” state, with high activation patterns mainly in the DMN (Buckner et al., 2008; Raichle et al., 2001). Breaking the individual brain states into more clusters, we found 12 distinct brain states that are more functionally specific than State A and B, as indicated by associated terms from the cognitive neuroscience literature. Qualitatively, the activation pattern for each brain state includes highly activated and/or deactivated brain regions that are affiliated with various functional network definitions (Fig. 2, bottom), further supporting the specificity of each brain state. However, detecting the same states across unconstrained rest and task at lower tiers of the hierarchy would be unlikely, given that any two subjects are unlikely to enter the exact same mental state spontaneously. In addition, reverse inferences are more likely to be problematic for lower level (more specific) states, since reverse inferences are often used inaccurately when making overly broad generalizations about specific activations (Poldrack, 2006). Therefore, we focused predominantly on State A and State B, which were both present for 97 of the 98 subjects.
Relationship to resting-state FC
In this study, we emphasized the use of whole-brain activation patterns over alternative FC-based approaches (Allen et al., 2014; Hutchison et al., 2013) for several reasons. Mainly, by focusing on activation amplitudes, we were able to directly compare the identified patterns with the large fMRI activation amplitude literature (Laird et al., 2009; Yarkoni et al., 2011). Also, characterizing activations allowed us to directly test whether subjects were in active task (externally oriented) brain states during “rest” periods, which the results support (Fig. 6). We found that FC architecture remains relatively unchanged across State A and State B, despite significant differences in the underlying activation patterns (Supplementary Fig. S3). This suggests that the activation states identified here are only weakly (if at all) related to FC states (Allen et al., 2014; Hutchison et al., 2013). However, the observed activation states appear to be related to static resting-state FC networks (based on estimating FC across entire resting-state runs). For instance, the State B activation pattern is highly similar to the DMN resting-state network, although a portion of the FPN is also present. Several studies (Chen et al., 2015; Liu et al., 2013) have investigated the relationship between static resting-state networks and coactivation patterns more directly. These studies showed that coactivation patterns can resemble common resting-state networks identified using traditional FC-based approaches, but some differences in the coactivation patterns remained undetected in FC-based analyses. Together, these observations support the use of activation patterns to investigate brain state organization
There have been studies using alternative methodologies for analyzing brain activity in the context of brain states. For example, spectral decomposition has been used to identify unique stimulus-dependent brain state profiles for task and rest and to investigate brain state dynamics for rest and task-active states (Billings et al., 2017). In another study, a hidden Markov model was applied to resting-state data to discover a temporal hierarchy of resting-state networks, in which the networks are naturally organized into two FC metastates (Vidaurre et al. 2017). In contrast, however, the main innovation and strength of our dynamic approach is the ability to detect brain state transitions at per-time-point resolution (as seen in the temporal similarity matrices in Fig. 2) and understanding of the functional context of the activation pattern brain states by relating them to the task activation literature (Fig. 6).
Despite evidence that the present results are independent of FC dynamics, these results appear to be related to observed anticorrelation between DMN and task-positive network time series according to (static) resting-state FC estimates (Fox et al., 2005). That result has been recently questioned, however, given that it is dependent on the global signal regression preprocessing step (Murphy et al., 2009). Importantly, we did not use global signal regression here, such that DMN time series were not anticorrelated with task-positive network time series. This suggests that the observed spatial (not temporal, which is used with FC) anticorrelation between DMN-dominated and task-positive-dominated states are at least somewhat independent of FC-based results. This may have been possible because spatial anticorrelation does not imply temporal anticorrelation (and vice versa). For example, two networks can be activated above baseline (high spatial correlation across time points) while being temporally anticorrelated at higher frequencies. It remains unclear exactly how spatial activation dynamics relate to time series dynamics, however, such that it will be important for future studies to fully characterize the relationship between spatial and temporal brain activity correlations.
The present empirical results of an anticorrelation between two brain states raise important theoretical implications involving neural synchrony and oscillations. Specifically, neural synchrony and oscillations, commonly observed in electrophysiological and EEG studies (Laumann et al., 2017; Wang, 2010), have been utilized to help facilitate understanding of spontaneous neural activity during resting-state fMRI (Deco et al., 2011; Fox and Raichle, 2007; Zuo et al., 2010). Furthermore, it has been suggested that anticorrelation patterns, similar to the present results, may have an equally important role in understanding neural synchrony (Fox et al., 2005). One might conceptualize State A and State B as a pair of anticorrelated sinusoidal oscillations, in which neural activity spontaneously flips back and forth during resting state. However, modeling neural oscillations as sinusoidal functions can be misleading, considering that resting-state activity has been known to be both nonstationary and nonsinusoidal (Cole and Voytek, 2018; Jones et al., 2012). Nonetheless, it will be important for future studies to fully characterize the functional relevance of anticorrelations in understanding neural synchrony and oscillations.
Brain activation states common across task and rest
By using a data-driven approach across dozens of subjects, we were able to obtain a comprehensive characterization (at the spatiotemporal scale of multiband fMRI) of brain states across rest and task. The results confirmed our hypothesis that “task-positive” brain states occur regularly during rest periods. We specifically found that subjects spent the majority of rest in State B (61% of the time) but that a substantial portion of rest was also spent in State A (39% of the time). This ratio is reversed (as expected) during the seven tasks: subjects were in State A more often (54% of the time on average) during task periods. The only exception was the language task, where subjects were in State A only 49% of the time during task periods. One potential explanation is that during the “story” condition of the language task subjects enter a self-reflective cognitive state consistent with State B. This could be either due to the introspective nature of stories or due to a lack of active task demands, which may bias the percentages in favor of State B over State A during task periods.
A popular account of resting periods is that mind wandering is the primary mental phenomenon occurring during those times. Past studies have linked increased activity in the DMN, present in the State B activation pattern, with mind wandering (Mason et al., 2007; Mittner et al., 2016). However, it is likely that increased DMN activity is not the only neural mechanism underlying mind wandering. The significant presence of State A suggests that the brain could possibly be performing active tasks, even during “rest.” Several studies have suggested personally relevant planning as one such task, which involves activity in the autobiographical memory system (Baird et al., 2011) and executive control systems (Christoff et al., 2009; Smallwood et al., 2012). Alternatively, common self-reported experiences of engaging in problem solving during rest (Smallwood and Schooler, 2006) may also require increased activity in areas such as the FPN. In addition, one might expect periodic activations in motor and sensory systems that are largely involved with passive attending to sensorimotor events (Fox et al., 2006). These “task-positive” networks are present in the State A activation pattern, suggesting that State A might be important in a full explanation of mind wandering. However, future work will be required to assess the specific tasks that are performed during unconstrained resting-state mind wandering, determining whether task-positive-like brain states during rest actually correspond with task-positive mental states. Also of note, State B was present for a substantial portion of time during task performance. These results suggest that there is a balance between these two states—with only moderate shifting from this homeostatic baseline—regardless of outward behavioral state.
Limitations
The present study involves several limitations that will be important for future studies to address. For instance, with fMRI, the choice of a representative “baseline” for activation analyses is a complex issue. For standard task analyses, this is often circumvented using intertask rest periods as the baseline to compare across task conditions. This choice has several issues, especially given that some regions are more active during rest than task (Stark and Squire, 2001). Yet, even this imperfect baseline choice is unavailable when investigating activations during resting states. Building on common practice in the fMRI FC literature, we used each region's (or voxel's) time series mean as baseline. However, differences in the choice of baseline may influence the amplitude of the activations we observe, which in turn may affect the interpretability of activation patterns. Nonetheless, the majority of our analyses and findings are rooted in correlation-based methods, which are designed to remove the influence of baseline (or linear scaling) shifts in signals. Thus, while the choice of a valid baseline remains an important problem to explore in fMRI research, its effect is likely minimized by the analytical approaches used in this study.
In the proposed dMVPA approach, we correlated spatial patterns between each individual timepoints to identify unique brain states that occur during resting-state fMRI. However, we did not measure the influence of temporal autocorrelations induced by the hemodynamic response on the timepoint-by-timepoint comparisons, which may be potentially confounding. Based on this, we avoid exploring state characteristics such as state dwell times and state transitions as these measures are more likely to be influenced by hemodynamic-induced temporal autocorrelations. It will be important for future research to assess the impact of hemodynamic-induced temporal autocorrelations on our proposed approach. For instance, future work could apply preprocessing steps to minimize the effects of temporal autocorrelations such as blind deconvolution (Havlicek et al., 2011), which can account for hemodynamics to infer underlying neural processes in time series.
Conclusion
We used a novel dMVPA approach combining insights from multivariate activation methods as well as graph theoretical methods to identify and characterize activation-pattern brain states during resting-state and task-state fMRI. This provided three primary benefits. First, relative to related M/EEG dMVPA approaches, the increased spatial accuracy of fMRI facilitated identification of activation patterns. Second, use of both resting-state and task-state data across 98 subjects allowed for an especially comprehensive identification of human brain activation states. Third, a focus on fMRI activation amplitude patterns allowed us to begin to interpret even spontaneous activations in terms of specific functions, given the vast task fMRI activation literature and associated cognitive functions. Together, these benefits revealed a hierarchical organization of brain states with shared features across rest periods and task performance. It will be important for future studies to build on these results, improving our understanding of the many brain states that are entered spontaneously and as a result of cognitive task instructions along with their relationship to ongoing brain network dynamics.
Supplementary Material
Acknowledgments
We thank Patrick Shafto and Bart Krekelberg for helpful conversations during preparation of this article. The authors acknowledge support from NIH awards K99-R00 MH096801 (M.W.C.), R01 R01MH109520 (M.W.C.), and R01 R01AG055556 (M.W.C.). Data were provided by the Human Connectome Project, Washington University-Minnesota Consortium (principal investigators David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH institutes and centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author Disclosure Statement
No competing financial interests exist.
References
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. 2014. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex 24:663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Fincham JM, Schneider DW, Yang J. 2012. Using brain imaging to track problem solving in a complex state space. Neuroimage 60:633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird B, Smallwood J, Schooler JW. 2011. Back to the future: autobiographical planning and the functionality of mind-wandering. Conscious Cogn 20:1604–1611 [DOI] [PubMed] [Google Scholar]
- Barch DM, Burgess GC, Harms MP, Petersen SE, Schlaggar BL, Corbetta M, et al. . 2013. Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage 80:169–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Erickson MA, Abell M, O'Donnell BF, Hetrick WP, Sporns O. 2012. Synchronization dynamics and evidence for a repertoire of network states in resting EEG. Front Comput Neurosci 6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings JCW, Medda A, Shakil S, Shen X, Kashyap A, Chen S, et al. . 2017. Instantaneous brain dynamics mapped to a continuous state space. Neuroimage 162:344–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008:10008 [Google Scholar]
- Bohlin L, Edler D, Lancichinetti A, Rosvall M. 2014. Community detection and visualization of networks with the map equation framework. In: Ding Y, Rousseau R, and Wolfram D (eds.) Measuring Scholarly Impact. New York, NY: Springer; p. 3–34 [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. 2008. The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci 1124:1–38 [DOI] [PubMed] [Google Scholar]
- Cha S-H. 2007. Comprehensive survey on distance/similarity measures between probability density functions. Int J Math Model Methods Appl Sci 1:300–307 [Google Scholar]
- Chen JE, Chang C, Greicius MD, Glover GH. 2015. Introducing co-activation pattern metrics to quantify spontaneous brain network dynamics. Neuroimage 111:476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoff K, Gordon AM, Smallwood J, Smith R, Schooler JW. 2009. Experience sampling during fMRI reveals default network and executive system contributions to mind wandering. Proc Natl Acad Sci U S A 106:8719–8724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Schneider W. 2007. The cognitive control network: integrated cortical regions with dissociable functions. Neuroimage 37:343–360 [DOI] [PubMed] [Google Scholar]
- Cole SR, Voytek B. 2018. Cycle-by-cycle analysis of neural oscillations. bioRxiv 302000. [DOI] [PubMed] [Google Scholar]
- Corbetta M, Shulman GL. 2002. Control of goal-directed and stimulus-driven attention in the brain. Nat Rev Neurosci 3:201–215 [DOI] [PubMed] [Google Scholar]
- Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29:162–173 [DOI] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, McIntosh AR. 2011. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat Rev Neurosci 12:43–56 [DOI] [PubMed] [Google Scholar]
- Dosenbach NUF, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RAT, et al. . 2007. Distinct brain networks for adaptive and stable task control in humans. Proc Natl Acad Sci U S A 104:11073–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. . 2002. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33:341–355 [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Ségonne F, Quinn BT, Dale AM. 2004. Sequence-independent segmentation of magnetic resonance images. Neuroimage 23:S69–S84 [DOI] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. 2006. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc Natl Acad Sci U S A 103:10046–10051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci 8:700–711 [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. 2005. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A 102:9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa C, Kaneko K. 2012. A dynamical-systems view of stem cell biology. Science 338:215–217 [DOI] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, et al. . 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80:105–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlicek M, Friston KJ, Jan J, Brazdil M, Calhoun VD. 2011. Dynamic modeling of neuronal responses in fMRI using cubature Kalman filtering. Neuroimage 56:2109–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxby JV, Connolly AC, Guntupalli JS. 2014. Decoding neural representational spaces using multivariate pattern analysis. Annu Rev Neurosci 37:435–456 [DOI] [PubMed] [Google Scholar]
- Haynes J-D. 2015. A primer on pattern-based approaches to fMRI: principles, pitfalls, and perspectives. Neuron 87:257–270 [DOI] [PubMed] [Google Scholar]
- He BJ. 2013. Spontaneous and task-evoked brain activity negatively interact. J Neurosci 33:4672–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, et al. . 2013. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage 80:360–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Vemuri P, Murphy MC, Gunter JL, Senjem ML, Machulda MM, et al. . 2012. Non-stationarity in the “resting brain's” modular architecture. PLoS One 7:13949–13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junejo IM. (2010). Learning self-similarities for action recognition using conditional random fields. In: Rebai A. (ed.) Bayesian Network, Rijeka, Croatia: IntechOpen; Available from: www.intechopen.com/books/bayesian-network/learning-self-similarities-for-action-recognition-using-conditional-random-fields [Google Scholar]
- Khanna A, Pascual-Leone A, Michel CM, Farzan F. 2015. Microstates in resting-state EEG: current status and future directions. Neurosci Biobehav Rev 49:105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JR, Dehaene S. 2014. Characterizing the dynamics of mental representations: the temporal generalization method. Trends Cogn Sci 18:203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Simmons WK, Bellgowan PSF, Baker CI. 2009. Circular analysis in systems neuroscience: the dangers of double dipping. Nat Neurosci 12:535–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Eickhoff SB, Kurth F, Fox PM, Uecker AM, Turner JA, et al. . 2009. ALE meta-analysis workflows via the BrainMap database: progress towards a probabilistic functional brain atlas. Front Neuroinform 3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancichinetti A, Fortunato S. 2012. Consensus clustering in complex networks. Sci Rep 2:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Snyder AZ, Mitra A, Gordon EM, Gratton C, Adeyemo B, et al. . 2017. On the stability of BOLD fMRI correlations. Cereb Cortex 27:4719–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chang C, Duyn JH. 2013. Decomposition of spontaneous brain activity into distinct fMRI co-activation patterns. Front Syst Neurosci 7:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Duyn JH. 2013. Time-varying functional network information extracted from brief instances of spontaneous brain activity. Proc Natl Acad Sci U S A 110:4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MF, Norton MI, Van Horn JD, Wegner DM, Grafton ST, Macrae CN. 2007. Wandering minds: the default network and stimulus-independent thought. Science 315:393–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittner M, Hawkins GE, Boekel W, Forstmann BU. 2016. A neural model of mind wandering. Trends Cogn Sci 20:570–578 [DOI] [PubMed] [Google Scholar]
- Motter AE. 2015. Networkcontrology. Chaos 25:97621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. 2009. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage 44:893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ, Girvan M. 2004. Finding and evaluating community structure in networks. Phys Rev E Stat Nonlin Soft Matter Phys 69:1–15 [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Halchenko YO, Hanson SJ. 2009. Decoding the large-scale structure of brain function by classifying mental states across individuals. Psychol Sci 20:1364–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA. 2006. Can cognitive processes be inferred from neuroimaging data? Trends Cogn Sci 10:59–63 [DOI] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. . 2011. Functional network organization of the human brain. Neuron 72:665–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME. 2010. Two views of brain function. Trends Cogn Sci 14:180–190 [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. 2001. A default mode of brain function. Proc Natl Acad Sci U S A 98:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall M, Axelsson D, Bergstrom CT. 2009. The map equation. Eur Phys J Spec Top 178:13–23 [Google Scholar]
- Rosvall M, Bergstrom CT. 2011. Multilevel compression of random walks on networks reveals hierarchical organization in large integrated systems. PLoS One 6:e18209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2011. Weight-conserving characterization of complex functional brain networks. Neuroimage 56:2068–2079 [DOI] [PubMed] [Google Scholar]
- Smallwood J, Brown K, Baird B, Schooler JW. 2012. Cooperation between the default mode network and the frontal–parietal network in the production of an internal train of thought. Brain Res 1428:60–70 [DOI] [PubMed] [Google Scholar]
- Smallwood J, Schooler JW. 2006. The restless mind. Psychol Bull 132:946–958 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, et al. . 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A 106:13040–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, et al. . 2012. Temporally-independent functional modes of spontaneous brain activity. Proc Natl Acad Sci U S A 109:3131–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, et al. . 2011. Network modelling methods for FMRI. Neuroimage 54:875–891 [DOI] [PubMed] [Google Scholar]
- Smith SM, Vidaurre D, Beckmann CF, Glasser MF, Jenkinson M, Miller KL, et al. . 2013. Functional connectomics from resting-state fMRI. Trends Cogn Sci 17:666–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark CE, Squire LR. 2001. When zero is not zero: the problem of ambiguous baseline conditions in fMRI. Proc Natl Acad Sci U S A 98:12760–12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Kelly AM, Biswal BB, Castellanos FX, Milham MP. 2009. Functional connectivity of default mode network components: correlation, anticorrelation, and causality. Hum Brain Mapp 30:625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K. 2013. The WU-Minn Human Connectome Project: an overview. Neuroimage 80:62–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre D, Smith SM, Woolrich MW. 2017. Brain network dynamics are hierarchically organized in time. Proc Natl Acad Sci 35:201705120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-J. 2010. Neurophysiological and computational principles of cortical rhythms in cognition. Physiol Rev 90:1195–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. 2011. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods 8:665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Zotev V, Phillips R, Drevets WC, Bodurka J. 2012. Spatiotemporal dynamics of the brain at rest—exploring EEG microstates as electrophysiological signatures of BOLD resting state networks. Neuroimage 60:2062–2072 [DOI] [PubMed] [Google Scholar]
- Zuo XN, Di Martino A, Kelly C, Shehzad ZE, Gee DG, Klein DF, et al. . 2010. The oscillating brain: complex and reliable. Neuroimage 49:1432–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.