Abstract
Stem cell maintenance is established by neighboring niche cells that promote stem cell self-renewal. However, it is poorly understood how stem cell activity is regulated by systemic, tissue-extrinsic signals in response to environmental cues and changes in physiological status. Here, we show that neuropeptide F (NPF) signaling plays an important role in the pathway regulating mating-induced germline stem cell (GSC) proliferation in the fruit fly Drosophila melanogaster. NPF expressed in enteroendocrine cells (EECs) of the midgut is released in response to the seminal-fluid protein sex peptide (SP) upon mating. This midgut-derived NPF controls mating-induced GSC proliferation via ovarian NPF receptor (NPFR) activity, which modulates bone morphogenetic protein (BMP) signaling levels in GSCs. Our study provides a molecular mechanism that describes how a gut-derived systemic factor couples stem cell behavior to physiological status, such as mating, through interorgan communication.
Author summary
Communication between different organs is essential to respond quickly to environmental cues or changes in the physiological status of an organism. Recent studies have shown the existence of humoral factors or hormones, which are transported by the circulatory system to different organs and achieve coordination between them. Here, we have analyzed the communication mechanism between organs that regulates proliferation of germline stem cells (GSCs) in the ovary of the fruit fly Drosophila melanogaster. We show that a peptide hormone called neuropeptide F (NPF) is a key player in this process. This peptide is produced in both the brain and the midgut, and, remarkably, we find that only NPF released from the midgut is crucial for controlling post-mating GSC proliferation. Our data suggest that mating stimulates the release of NPF from the endocrine cells of the midgut stimulated by the presence of a seminal peptide. Midgut-derived NPF is then transduced through an NPF-specific G-protein–coupled receptor expressed in the ovary, and this triggers GSC proliferation. Our study identifies an essential interaction between the digestive system and the ovary that regulates the size of stem cell populations in flies depending on mating.
Introduction
Maintenance and regeneration of adult tissues requires a robust stem cell system that balances self-renewal with differentiation [1]. Because abnormalities in stem cell regulation may result in loss of tissue integrity or tumorigenesis, this robust stem cell system is precisely modulated by local and systemic signals [2]. Stem cells reside in a specialized microenvironment, or niche, where they are exposed to local signals required for stem cell function and identity [3,4]. A number of studies have demonstrated the importance of local niche signals in regulating stem cell identity. Less is known, however, regarding how stem cell activity is regulated by systemic, tissue-extrinsic signals in response to environmental cues and changes in physiological status.
The Drosophila ovary is one of the most powerful models for studying adult stem cell behavior in vivo [3,5]. This tissue is composed of many chains of developing egg chambers called ovarioles [6]. The most anterior region of each ovariole, the germarium, contains germline stem cells (GSCs) that give rise to the eggs (Fig 1A). GSCs can divide symmetrically to produce a generative cell population or asymmetrically to produce daughter cells called cystoblasts. Each cystoblast undergoes differentiation into 15 nurse cells and 1 oocyte in each egg chamber, which is surrounded by somatic follicle cells. Therefore, the balance between self-renewal and differentiation of GSCs plays a pivotal role in regulating oogenesis because disruption of this balance may cause germ cell depletion, infertility, or tumorigenesis [1].
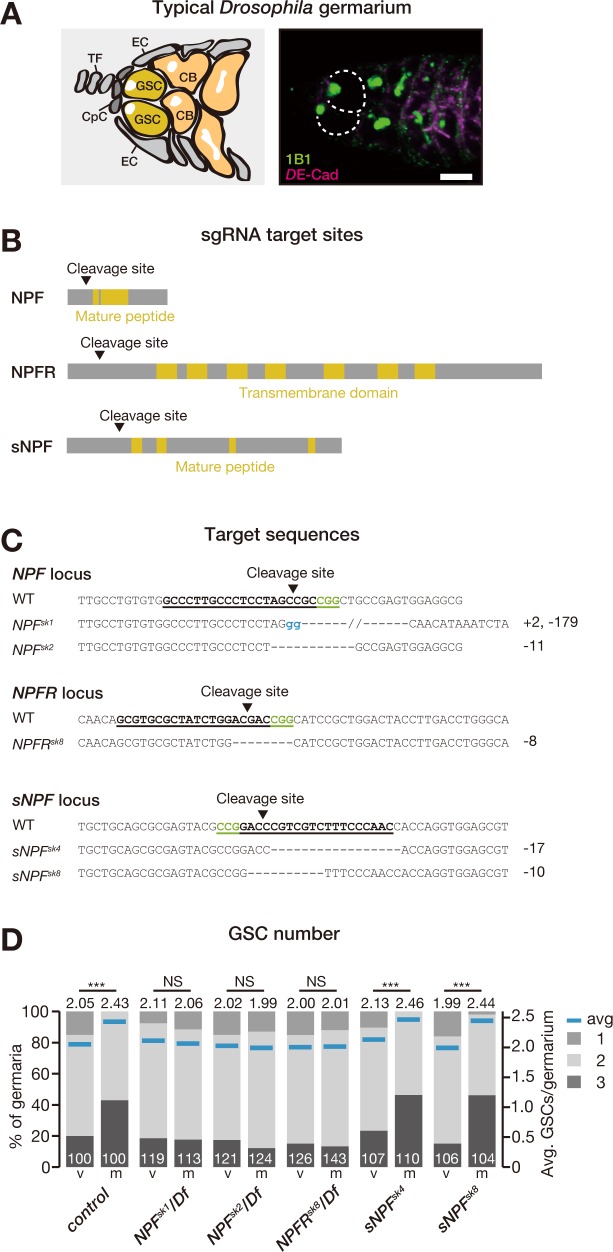
Fig 1. Mating-induced GSC proliferation requires NPF and NPFR.
(A, left) Schematic Drosophila germarium. GSCs reside in a niche formed from somatic cells such as CpCs, TF, and ECs. GSCs are identifiable by a stereotypical spectrosome morphology and location (adjacent to CpCs). GSC division produces 1 self-renewing daughter and 1 CB that differentiates into a germline cyst. (A, right) Representative germarium from a WT fly, containing 2 GSCs. Samples were stained with monoclonal antibody 1B1 (green) and anti-DE-cadherin (magenta), which stain the spectrosome and overall cell membranes, respectively. GSCs are indicated by dotted circles. (B) Schematic representation of sgRNA target sites (cleavage sites) in coding sequences of NPF, NPFR, and sNPF genes. Regions encoding the mature NPF and sNPF peptides and the putative transmembrane domains of NPFR are highlighted in yellow. (C) The target locus in each Cas9-induced mutant was PCR-amplified and sequenced. The WT sequence is shown at the top of each set of sequences as a reference. The Cas9-sgRNA target sequence is underlined with the PAM indicated in green. Deleted nucleotides are shown as dashes. Inserted nucleotides are indicated in blue lowercase letters. The indel size is shown next to each sequence. Each of these indel mutations results in a premature stop codon. (D) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (v) and mated (m) female flies. Loss of NPF or NPFR disrupted the mating-induced increase in GSC number. Flies carrying the CRISPR-induced mutations NPFSK1 or NPFSK2 were crossed with Df(3R)ED10642. Mutant flies carrying NPFRsk8 were crossed with Df(3R)BSC464. The number of germaria analyzed is indicated inside the bars in D. For statistical analysis, a Wilcoxon rank sum test was used for D. ***P ≤ 0.001. Scale bar = 5 μm in panel A. Underlying data can be found in S1 Data. See also S1 Fig. Cas9, CRISPR-associated protein 9; CB, cystoblast; CpC, cap cell; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; EC, escort cell; GSC, germline stem cell; NPF, neuropeptide F; NPFR, neuropeptide F receptor; NS, nonsignificant (P > 0.05); sgRNA, single guide RNA; sNPF, short neuropeptide F precursor; TF, terminal filament; WT, wild-type.
Drosophila female GSCs are anchored to the somatic niche, which comprises cap cells, escort cells, and terminal filaments. The niche produces local signals such as the bone morphogenetic protein (BMP) ligand Decapentaplegic (Dpp), which activates its receptors Saxophone (Sax), Punt (Put), and Thickveins (Tkv) expressed in GSCs to induce GSC division and maintenance [7]. Local signals from the niche have a crucial role in regulating reproduction because impairment of niche signals can cause germ cell depletion, infertility, and tumorigenesis. On the other hand, it is well known that animal reproduction is also coordinated by signals from the external environment [8,9]. One such example involves nutrients, which are important materials for producing eggs. On a protein-poor diet, egg production is restricted through blocking vitellogenesis [8]. A protein-poor diet also results in a reduction of GSC division, which is mediated by neural-derived Drosophila insulin-like peptides (DILPs) [10]. Moreover, in response to nutrients, GSC maintenance is controlled by the adipocyte metabolic pathway [11,12]. Another example of an environmental cue that affects reproduction is mating. Mated females show a dramatic increase in egg production, which is induced by a male-derived peptide from seminal fluid termed sex peptide (SP) [13]. We have previously reported that neural SP signaling also promotes GSC proliferation through its effects on the biosynthesis of ecdysteroids (insect steroid hormones) in the ovary [14,15]. Taken together, these findings suggest that GSC proliferation and maintenance are modulated by tissue-extrinsic signals in response to environmental cues.
Here, we present a series of new findings that reveal a novel and fundamental interorgan communication mechanism controlling GSC proliferation in response to mating. We demonstrate that Drosophila neuropeptide F (NPF), a homolog of mammalian neuropeptide Y (NPY), acts as a key regulator of mating-induced GSC proliferation in Drosophila females. Although NPF is expressed in both the brain and the midgut, we found that only the enteroendocrine-derived peptide—not neuronal NPF—is required for activation of GSCs after mating. The NPF protein is highly accumulated in enteroendocrine cells (EECs) of the middle midgut of virgin female flies and is released in response to SP-dependent signaling upon mating. Through fly injection and ex vivo ovary cultures with synthetic peptide, we show that NPF signaling is sufficient for increasing GSC number in virgin female flies. Notably, after mating, midgut-derived NPF acting on the ovaries through the NPF receptor (NPFR) up-regulates BMP signaling levels in GSCs to induce their proliferation. Our findings describe a mechanism of gut-to-ovary communication that couples stem cell behavior to physiological status by sensing external cues such as mating. Considering NPY’s role in regulating reproduction in many animal species, our study also provides new insights into the role of interorgan communication during animal germline development.
Results
Disruption of NPF function in midgut EECs impairs mating-induced GSC proliferation
We employed a genetic screen using Drosophila fly lines carrying Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-generated mutations in neuropeptide-encoding genes [16] and identified stem cell phenotypes in mutants of NPF (Fig 1B and 1C). In control flies, mated female flies had more GSCs than virgin female flies (Fig 1D), as we reported previously [14]. In contrast, the mating-induced increase in GSC number was suppressed in genetic null mutants for NPF itself or for the gene encoding the NPFR (Fig 1B–1D). We also found that genetic null mutant flies of short neuropeptide F precursor (sNPF), encoding an RxRFamide neuropeptide related to NPF, showed a normal increase in GSC number after mating (Fig 1D), suggesting that the GSC-suppression phenotype is specific to NPF signaling.
Our immunostaining analysis confirmed the presence of anti-NPF signals in the brain [17,18] and EECs of the middle midgut [19,20] in control flies, but not in NPF mutants (S1A Fig). A number of previous studies have already reported that neuronal NPF regulates multiple aspects of physiology and behavior in adult flies, such as circadian rhythm, alcohol sensitivity, male courtship behavior, and food intake [17,18,21–24], whereas the function of EEC-derived NPF remains unclear [20,25]. We first investigated whether neuronal NPF function is required for the normal increase in GSC number induced by mating. However, although RNA interference (RNAi)-mediated knockdown of NPF either pan-neuronally (using nSyb-GAL4) or in neuroendocrine cells (using 386Y-GAL4) resulted in a drastic decrease in NPF protein levels in the brain (S1B Fig), neither manipulation had any effect on post-mating GSC number (S1C Fig). We also confirmed that RNAi driven by neither the nSyb-GAL4 nor 386Y-GAL4 driver resulted in reduced NPF levels in midgut EECs (S1B Fig).
Therefore, we next examined whether the mating-induced increase in GSC number is controlled by midgut-expressed NPF, the other potential source of NPF protein. For this purpose, we utilized the Tk-gut-GAL4 (Tkg-GAL4) driver because this GAL4 driver is known to be active in a restricted population of midgut cells, including NPF-positive EECs [20], but not in the ovary (S2A Fig). Immunostaining analysis with anti-NPF antibody revealed that Tkg-GAL4-mediated transgenic RNAi against NPF (hereafter Tkg>NPFRNAi) dramatically reduced the number of NPF-positive cells in the middle midgut compared with controls (Fig 2A). It should be noted that Tkg-GAL4 is also active in some neuronal cells in the brain and the ventral nerve cord (VNC; S2A and S2B Fig); however, Tkg>NPFRNAi animals did not show a significant reduction in NPF levels in these neuronal cells (S2C Fig). In Tkg>NPFRNAi females, we found that the mating-induced increase in GSC number was severely impaired (Fig 2B). In addition, we performed RNAi-mediated knockdown of NPF driven by several other midgut-GAL4 drivers. Suppression of the increase in GSC number after mating was observed with 4 of these drivers (Fig 2C), which are active in middle midgut EECs [26] but not in ovaries or NPF-positive neurons (S3 Fig). On the other hand, NPF RNAi in enterocytes (using Myo1A-GAL4) or intestinal stem cells and enteroblasts (esg-GAL4) had no effect on the mating-induced increase in GSC number (Fig 2C). We found that the GSC proliferation defect in NPF genetic null mutants was rescued by overexpression of the NPF transgene under the control of the Tkg-GAL4 driver (Fig 2D).
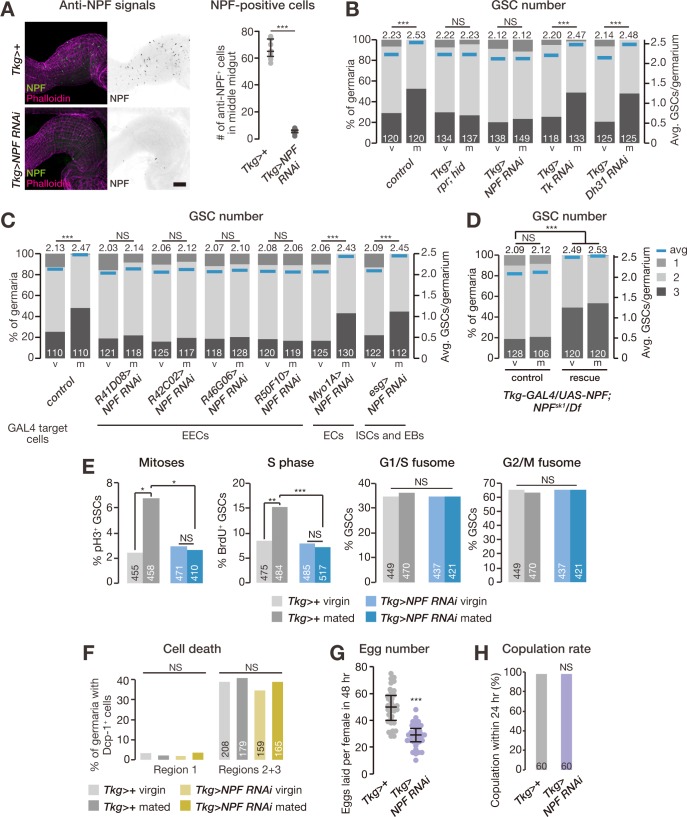
Fig 2. Midgut-derived NPF promotes mating-induced GSC proliferation.
(A) The number of NPF-positive cells was dramatically reduced in the middle midgut of Tkg-GAL4>NPFRNAi animals. (B–D) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. (B) Removal of Tkg-GAL4-positive cells by induced apoptosis or NPF RNAi driven by Tkg-GAL4 suppressed the increase in GSC number after mating. RNAi against Tk or Dh31, which are also expressed in these cells, driven by Tkg-GAL4 had no effect on mating-induced increase in GSC number. (C) NPF RNAi driven by several GAL4 drivers targeting EECs disrupted the increase in GSC number after mating, whereas NPF RNAi driven by esg-GAL4 (ISCs and EBs) or Myo1A-GAL4 (ECs) had no effect on the mating-induced increase in GSC number. (D) The post-mating GSC proliferation defect observed in NPF genetic null mutants was rescued by overexpression of the NPF transgene under the control of Tkg-GAL4. Rescue, Tkg-GAL4/UAS-NPF; NPFsk1/Df(3R)ED10642. Control, Tkg-GAL4/+; NPFsk1/Df(3R)ED10642 or UAS-NPF/+; NPFsk1/Df(3R)ED10642. (E) Mating increased the fraction of GSCs in mitosis (measured by pH3 labeling) and S phase (measured by BrdU incorporation), but not G1/S or G2/M fusome morphologies. The cell cycle promotion observed in GSCs after mating was suppressed in Tkg-GAL4>NPFRNAi animals. (F) Percentages of germaria containing cleaved Dcp-1-positive cells in either region 1 (anterior-most region; containing GSCs, cystoblasts, and dividing cysts) or regions 2 and 3 (containing 16-cell cysts and follicle cells) did not change after mating or NPF RNAi. (G) The total number of eggs produced by females within 48 hours was reduced in Tkg-GAL4>NPFRNAi animals. (H) Receptivity of virgin females was scored as the percentage of females that copulated within 24 hours. The number of germaria analyzed is shown inside the bars in B–F and H. Dots represent the relative intensity of NPF in a single middle midgut (panel A) or the number of eggs produced by a single female fly (panel G); lines represent the median, and whiskers represent the interquartile range. For statistical analysis, a Wilcoxon rank sum test was used for panel B–D. Student t test with Holm’s correction was used for panel A and G. Fisher’s exact test with Holm’s correction was used for panel E, F, and H. ***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05; NS, nonsignificant (P > 0.05). Scale bar = 50 μm in panel A. Underlying data can be found in S1 Data. See also S2, S3 and S4 Figs. BrdU, bromodeoxyuridine; Dcp-1, Death caspase-1; Dh31, diuretic hormone 31; EB, enteroblast; EC, enterocyte; EEC, enteroendocrine cell; GSC, germline stem cell; hid, head involution defective; ISC, intestinal stem cell; NPF, neuropeptide F; pH3, phospho-histone H3; RNAi, RNA interference; rpr, reaper; Tk, Tachykinin; Tkg-GAL4, Tk-gut-GAL4.
Tkg-GAL4-positive cells also express other gut peptide hormone genes, including Tachykinin (Tk) and diuretic hormone 31 (Dh31) [20]. However, transgenic RNAi against either Tk or Dh31 driven by Tkg-GAL4 had no effect on post-mating increase in GSC number (Fig 2B). On the other hand, ablating the Tkg-GAL4-positive cells by expressing the cell death–inducing factor reaper (rpr) and head involution defective (hid) led to suppression of the increase in GSC number after mating (Fig 2B). Taken together, these results suggest that EECs play an important role in regulating the mating-induced increase in GSC number, mainly through the function of NPF.
We next examined whether midgut-derived NPF controls GSC division. For this purpose, we counted the number of GSCs in M phase and S phase by staining with anti-phospho-histone H3 (pH3) and bromodeoxyuridine (BrdU), respectively, in control and Tkg>NPFRNAi adult females. In control female flies, we found that mating increased the frequency of GSCs in both M and S phases (Fig 2E). We also monitored GSC fusome morphology as an indicator of cell cycle phase [27] and did not observe any difference in the frequency of GSCs in G2/M and G1/S phases (Fig 2E). In Tkg>NPFRNAi animals, the increase in the fraction of GSCs in M and S phases was suppressed (Fig 2E), suggesting that midgut-derived NPF promotes GSC progression through both DNA replication and mitosis. We also monitored the fraction of apoptotic cells in the germarium by staining with anti-cleaved Death caspase-1 (Dcp-1), a marker for apoptotic cells [28]. The number of apoptotic cells did not change in Tkg>NPFRNAi female flies compared with controls (Fig 2F), suggesting that the lack of post-mating GSC-number increase seen with NPF RNAi was not caused by increased cell death but mainly by a lack of NPF-induced cell proliferation.
Consistent with their GSC proliferation phenotype, we found that mated Tkg>NPFRNAi female flies laid fewer eggs than mated control females (Fig 2G). Taken together, these data indicate that midgut-derived NPF has a positive impact not only on GSC proliferation but on reproductive fitness after mating as well. To rule out the possibility that the GSC phenotype was due to the absence of mating, we confirmed that the copulation rate of Tkg>NPFRNAi female flies did not change compared with that of control female flies by using males expressing GFP in their sperm (Fig 2H). These findings are all consistent with the idea that NPF from EECs modulates GSC division after mating.
Midgut-derived NPF does not affect gut remodeling after mating
In Drosophila females, mating induces midgut epithelium remodeling, including increases in gut size and in the number of mitotic cells, which is essential for enhancing reproductive output [29,30]. Thus, there may be a possibility that the suppression of mating-induced GSC proliferation in Tkg>NPFRNAi animals is indirectly caused by the dysfunction of midgut remodeling after mating. We therefore examined whether midgut-derived NPF affects gut size or the number of mitotic cells in the midgut after mating; however, Tkg>NPFRNAi females displayed normal increases in mitosis in the midgut epithelium and posterior midgut diameter (S4A and S4B Fig). This suggests that midgut NPF is not involved in tissue remodeling after mating and that the GSC phenotypes in Tkg>NPFRNAi female flies are not due to an indirect effect of defective mating-induced remodeling in the midgut epithelium.
Mating affects NPF accumulation in EECs
To investigate the relationship between mating and NPF in EECs, we performed immunostaining with anti-NPF antibody on the midguts of virgin and mated female flies. Anti-NPF signal was stronger in the EECs of virgin females compared with mated females (Fig 3A). In contrast, mating did not alter NPF mRNA abundance in the middle midgut (Fig 3B), indicating that the observed change in NPF protein levels was not due to transcriptional regulation. This situation was reminiscent of the case of Drosophila insulin-like peptide 2 (Dilp2) because it is well known that increased Dilp2 protein level in insulin-producing cells reflects decreased Dilp2 release into the hemolymph when dilp2 transcription is constant [31]. Similarly, although immunostaining alone cannot completely rule out the contribution of post-transcriptional regulation of NPF, these results do imply that mating promotes NPF release from EECs.
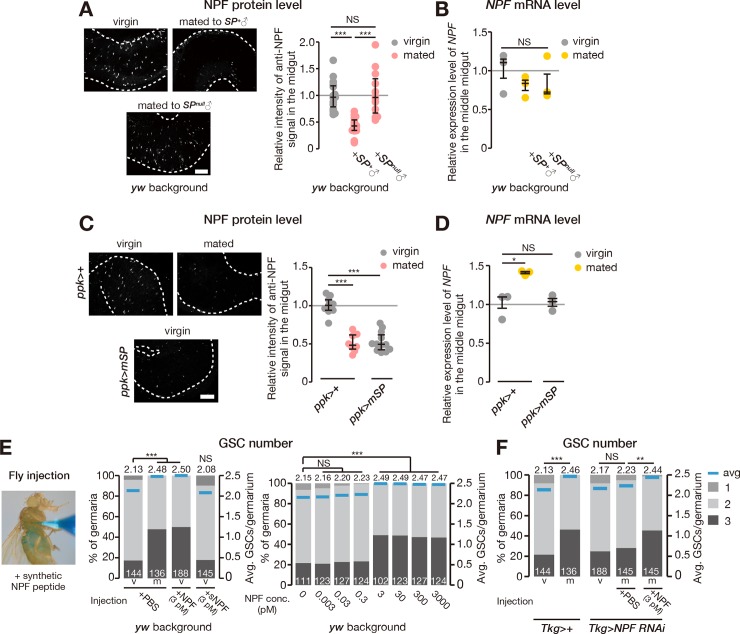
Fig 3. Mating induces NPF accumulation in EECs, and exogenous NPF increases GSC.
(A–D) Levels of NPF protein and NPF transcript in the middle midgut. (A, C) Representative images of anti-NPF immunostaining in the middle midgut are shown on the left. Quantification of anti-NPF signal intensity in the middle midgut is shown on the right graph. (A) Anti-NPF signal accumulated in virgin female flies and females mated with SP-null mutant males (SP0/SPΔ130). (B) Transcript abundance of NPF in the middle midgut did not change after mating with wild-type or SP-null male flies. (C) NPF release was induced without mating by overexpressing mSP in ppk-positive neurons (ppk-GAL4>mSP). (D) Transcript abundance of NPF in the middle midgut was not changed by this manipulation. (E, F) Fraction of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. (E) Microinjection of synthetic NPF peptide, but not sNPF (Bm-sNPF), into virgin female flies induced an increase in GSC number. NPF-dependent increase in GSC number occurred after injecting 3–3,000 pM (but not 0–0.3 pM) NPF. (F) The GSC-number phenotype of NPF RNAi animals was rescued by injecting synthetic NPF peptide. Dots represent the relative signal intensity of anti-NPF in single middle midguts (panel A and C) or the relative expression levels of NPF in the middle midgut (panel B and D); lines represent the median, and whiskers represent the interquartile range. The number of germaria analyzed is shown inside the bars in panel E and F. For statistical analysis, a Wilcoxon rank sum test with Holm’s correction was used for panel A, C, E, and F. Student t test with Holm’s correction was used for panel B and D. ***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05; NS, nonsignificant (P > 0.05). Scale bar = 50 μm in panel A and C. Underlying data can be found in S1 Data. See also S5 Fig. GSC, germline stem cell; mSP, membrane-tethered SP; NPF, neuropeptide F; ppk, pickpocket; RNAi, RNA interfererence; sNPF, short neuropeptide F precursor; SP, sex peptide; Tkg-GAL4, Tk-gut-GAL4.
SP signaling promotes NPF release from middle midgut EECs upon mating
Our previous study [14] revealed that mating-induced GSC proliferation is mediated by the male seminal-fluid component SP, which plays a central role in triggering dramatic changes in female physiology and behavior after mating [13,32]. SP is received by female neurons expressing the sex peptide receptor (SPR), resulting in the silencing of these neurons [33,34]. We found that female flies mated with male flies lacking SP showed NPF accumulation in middle midgut EECs without significant changes in NPF mRNA levels (Fig 3A and 3B), suggesting that male-derived SP regulates NPF release from EECs upon mating.
The silencing of SPR-positive neurons located on the oviduct, which also express pickpocket (ppk) [35], is particularly important for inducing female GSC proliferation after mating [14]. We found that the expression of a transgene encoding membrane-tethered SP (mSP) in ppk-positive female neurons decreased NPF protein levels in EECs, even in virgin female flies (Fig 3C), while NPF mRNA levels in the middle midgut were not significantly altered (Fig 3D). In contrast, NPF protein levels did not change after expression of mSP with the Tkg-GAL4 driver (S5A and S5B Fig). These results suggest that neuronal—but not midgut—SP signaling is both necessary and sufficient for post-mating NPF release from EECs.
We also examined whether neuronal inactivation of SPR-positive neurons was sufficient to reduce NPF accumulation in EECs in virgin female flies, as the binding of SP to SPR silences SPR-positive neurons [36]. We utilized a mutant of shibire (shits1) that blocks synaptic vesicle release in a temperature-dependent manner [37]. Normal NPF accumulation was observed at the permissive temperature in virgin female flies overexpressing shits1 (S5C Fig). On the other hand, when SPR-positive neurons were silenced in virgin females at the restrictive temperature to mimic mating, NPF protein levels in middle midgut EECs were reduced without any significant changes in NPF mRNA levels (S5C and S5D Fig). These results suggest that NPF release from EECs is induced by silencing the transmitter-release activity of some neurons within the SPR-positive population.
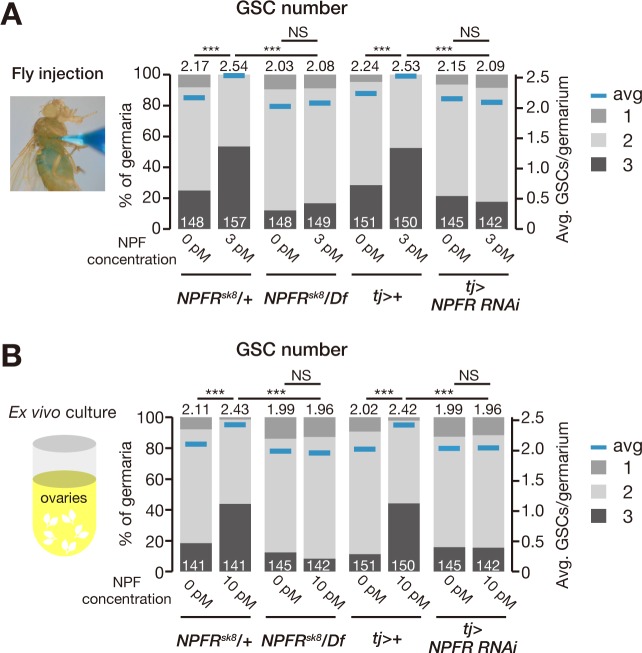
To further examine the necessity and sufficiency of circulating NPF in inducing the increase in GSC number, we manually delivered synthetic NPF peptide by injecting it using glass needles into adult females. NPF injection into wild-type virgin females resulted in a significant increase in GSC number compared with injection of phosphate-buffered saline (PBS) vehicle (Fig 3E). No such increase in GSC number was observed after injection of synthetic sNPF peptide (Fig 3E). Moreover, we observed that the impairment in GSC increase seen in mated Tkg>NPFRNAi animals was restored by NPF injection but not by control PBS injection (Fig 3F). These results suggest that elevation in circulating NPF levels is both necessary and sufficient to trigger mating-induced GSC proliferation.
NPF induces an increase in GSC number by acting on the ovary
The next question to be addressed is whether the NPF signal is directly received by the ovary or is transmitted via other tissues to control mating-induced GSC proliferation. We therefore performed ex vivo ovary culture experiments with synthetic NPF peptide. In this experiment, we dissected ovaries from virgin female flies and cultured them in Schneider’s Drosophila cell culture medium with or without synthetic NPF peptide for 1 day. We found that dissected virgin ovaries cultured with the NPF peptide possessed more GSCs than controls (Fig 4A), indicating that NPF directly affects the ovary in controlling GSC number. This increase was not observed when the ovaries were cultured with synthetic sNPF peptide (Fig 4B), suggesting that the observed response is specific to NPF.
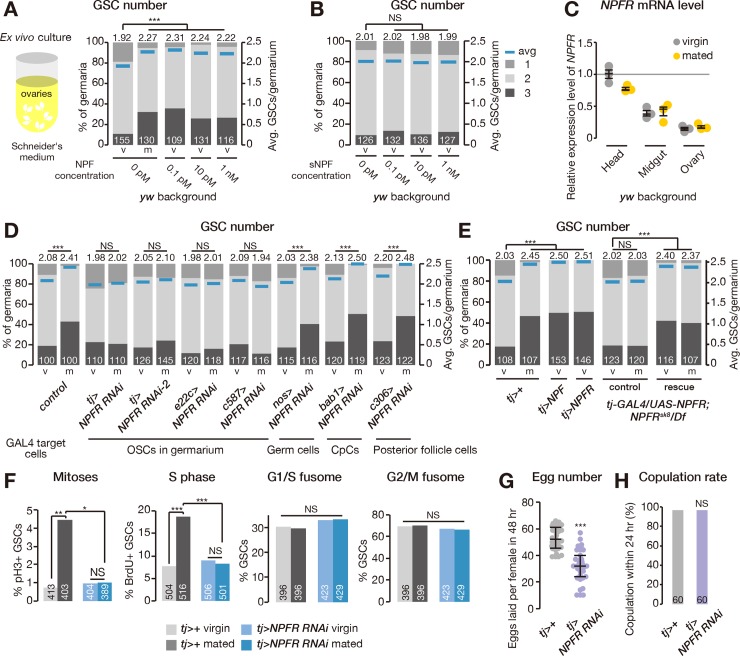
Fig 4. Ovarian NPFR controls mating-induced GSC proliferation.
(A, B, D, E) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. The addition of synthetic NPF peptide (panel A), but not of sNPF (panel B), to the ex vivo ovary culture medium was sufficient to increase GSC number. (C) NPFR transcripts were detected in the ovary, although expression levels were much lower than in the head and midgut. (D) NPFR RNAi driven by tj-GAL4, e22c-GAL4, or c587-GAL4 (targeting germarium somatic cells) inhibited the mating-induced increase in GSC number. The increase in GSC number after mating was not altered by NPFR RNAi driven by nos-GAL4, bab1-GAL4, or c306-GAL4 (germ, cap, and posterior follicle cells, respectively). (E) Overexpressing NPF or NPFR in the somatic cells of the germaria was sufficient to increase GSC number in virgin female flies. The GSC proliferation defect in NPFR genetic null mutants was rescued by overexpressing the NPFR transgene under the control of tj-GAL4. Rescue, tj-GAL4/UAS-NPFR; NPFRsk8/Df(3R)BSC464. Control, tj-GAL4/+; NPFRsk8/Df(3R)BSC464 or UAS-NPFR/+; NPFRsk8/Df(3R)BSC464. (F) Mating increased the frequency of GSCs in M (measured by pH3 labeling) and S (measured by BrdU incorporation) phases, but not G1/S or G2/M fusome morphologies. Cell cycle promotion in GSCs after mating was suppressed in tj-GAL4>NPFRRNAi animals. (G) The total number of eggs produced by females within 48 hours was reduced in tj-GAL4>NPFRRNAi animals. (H) Receptivity of virgin females was scored as the percentage of females that copulated within 24 hours. The number of germaria analyzed is shown inside the bars in panel A, B, D–F, and H. For statistical analysis, a Wilcoxon rank sum test with Holm’s correction was used for A, B, D, and E. Fisher’s exact test with Holm’s correction was used for panel F and H. Student t test was used for panel G. ***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05; NS, nonsignificant (P > 0.05). Underlying data can be found in S1 Data. See also S6 Fig. BrdU, bromodeoxyuridine; CpC, cap cell; GSC, germline stem cell; NPF, neuropeptide F; NPFR, neuropeptide F receptor; OSC, ovarian somatic cell; pH3, phospho-histone H3; RNAi, RNA interference; sNPF, short neuropeptide F precursor.
NPFR activity in the ovarian somatic cells controls mating-induced GSC proliferation
To further understand the role of NPF signaling in the ovary, we focused on the NPFR (CG1147) in Drosophila [38]. We confirmed that NPFR is expressed in the ovary (Fig 4C); however, expression levels in this tissue were much lower than in the head or midgut, the tissues previously reported to express NPFR [17,38]. We also found that the increase in GSC number after mating was suppressed by transgenic NPFR RNAi driven by tj-GAL4, e22c-GAL4, or c587-GAL4 drivers (Fig 4D), which are known to be active in the somatic cells of ovarian germaria, including escort and follicle cells [39–42]. Conversely, we found that overexpression of either NPFR or NPF driven by tj-GAL4 was sufficient to increase GSC number in virgin females (Fig 4E). On the other hand, no suppression of the increase in GSC number was observed when we knocked down NPFR function in germ cells (using nos-GAL4), cap cells (bab1-GAL4), or posterior follicle cells (c306-GAL4; Fig 4D). Although both tj-GAL4 and c587-GAL4 drive expression in the brain and VNC (S6A Fig), pan-neuronal RNAi knockdown of NPFR function did not affect the mating-induced increase in GSC number (S6B Fig). We also confirmed that intestinal RNAi knockdown of NPFR function did not disrupt the mating-induced increase in GSC number (S6C Fig). We found that the GSC proliferation defect in NPFR genetic null mutants was rescued by overexpression of the NPFR transgene under the control of tj-GAL4 (Fig 4E).
We next examined cell cycle progression of GSCs in ovarian NPFR-knockdown females. This manipulation led to phenotypes similar to those seen in Tkg>NPFRNAi female flies—namely, a decrease in GSC frequency in M and S phases after mating without any effects on G1/S or G2/M phase transitions (Fig 4F).
Similar to the case of Tkg>NPFRNAi, mated tj>NPFRRNAi female flies laid fewer eggs than mated control females (Fig 4G). We also found that the copulation rate of tj>NPFRRNAi animals did not change in comparison with control female flies by using males expressing GFP in their sperm (Fig 4H), ruling out the possibility that the GSC phenotype was due to the absence of mating. Taken together, these data suggest that NPFR in ovarian somatic cells is necessary and sufficient for positively controlling mating-induced GSC proliferation and reproductive fitness.
NPF-dependent induction of the increase in GSC number requires NPFR in the ovary
Remarkably, we found that the increase in GSC number induced by in vivo injection or ex vivo ovary culture with synthetic NPF peptide was completely suppressed in NPFR-knockdown animals (Fig 5A and 5B), suggesting that NPFR expressed in the ovary is epistatic to NPF for controlling GSC proliferation. Because the midgut and ovaries are distinct and separate organs, midgut-derived NPF must remotely act on the ovary to control GSC proliferation in response to mating. Therefore, based on our data described above, we hypothesized that a mating stimulus triggers the release of NPF from EECs into the hemolymph, after which the circulating NPF signal can be received by the ovary.
Fig 5. NPF-dependent induction of GSC increase requires NPFR in the ovary.
(A, B) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. Loss of NPFR (NPFRsk8/D(3R)BSC464) or ovarian knockdown of NPFR (tj-GAL4>NPFRRNAi) blocked the NPF-induced increase in GSC number after fly injection (panel A) or ex vivo ovary cultures (panel B). The number of germaria analyzed is shown inside the bars. For statistical analysis, a Wilcoxon rank sum test with Holm’s correction was used. ***P ≤ 0.001. Underlying data can be found in S1 Data. GSC, germline stem cell; NPF, neuropeptide F; NPFR, neuropeptide F receptor; NS, nonsignificant (P > 0.05); RNAi, RNA interference.
NPF signaling regulates Dpp signaling in GSCs of mated female flies
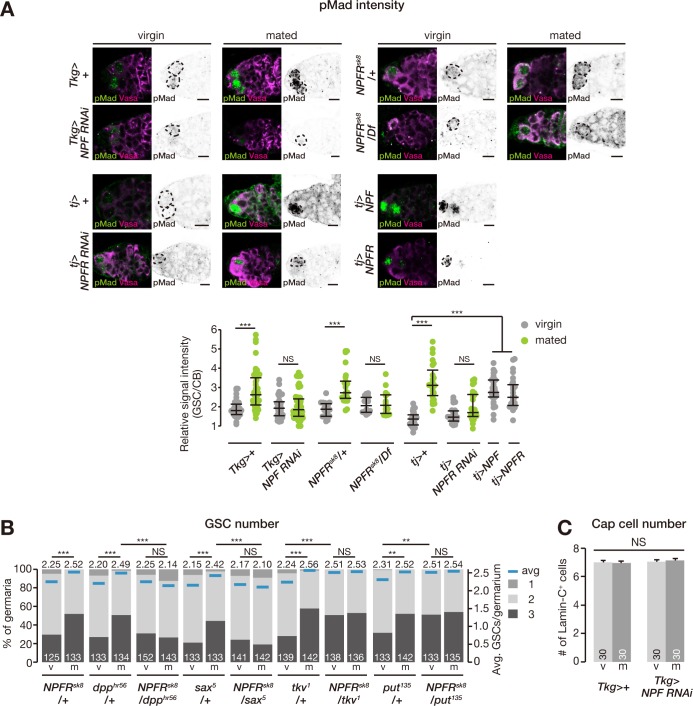
GSC maintenance and proliferation are controlled by signals from the GSC niche, in particular Dpp—the fly counterpart to BMPs [1]. We therefore examined whether down-regulation of NPF signaling affects Dpp signaling by measuring the level of phosphorylated Mad (pMad), a readout of Dpp signaling activation in cells including GSCs [43]. We found that mating increased pMad levels in GSCs of control flies (Fig 6A). On the other hand, Tkg>NPFRNAi led to a reduction in pMad levels in GSCs of mated female flies (Fig 6A). The same phenotype was also observed in ovarian NPFR RNAi female flies and genetic null alleles of NPFR (Fig 6A). Conversely, overexpression of NPF or NPFR in ovarian somatic cells resulted in elevated pMad levels in the GSCs of virgin female flies (Fig 6A).
Fig 6. NPF signaling regulates BMP signaling levels in GSCs and promotes GSC self-renewal towards symmetric GSC divisions.
(A) Representative images of adult female germaria immunostained with anti-pMad antibody (green) and anti-Vasa antibody (germ cell marker; magenta) are shown at the top (GSC, dotted lines). Quantification of relative pMad intensity in GSCs, which was normalized to pMad intensity in CBs, is shown in the bottom graph. For quantification of pMad intensity, the cell boundaries of GSCs and CBs were determined by anti-Vasa staining. Mating-induced increase in pMad expression was suppressed by intestinal NPF RNAi (Tkg-GAL4>NPFRNAi), ovarian NPFR (tj-GAL4>NPFRRNAi), or loss of NPFR function (NPFRsk8/Df(3R)BSC464). Increase in pMad signal intensity in GSCs was induced by ovarian overexpression of NPF or NPFR. (B) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. The mating-induced increase in GSC number was disrupted in the double heterozygous mutant for NPFR and BMP signaling (NPFRsk8/dpphr56 or NPFRsk8/sax5). (C) The number of cap cells did not change after mating or NPF RNAi driven by Tkg-GAL4. Each dot represents the relative intensity of pMad in a single germarium, with lines representing the median and whiskers representing the interquartile range in panel A. The number of germaria analyzed is shown inside the bars in panel B and C. Values are presented as the mean with standard error of the mean in panel C. For statistical analysis, a Wilcoxon rank sum test with Holm’s correction was used for panel A and B. Student t test with Holm’s correction was used for panel C. ***P ≤ 0.001 and **P ≤ 0.01; NS, nonsignificant (P > 0.05). Scale bar = 5 μm. Underlying data can be found in S1 Data. See also S7 Fig. BMP, bone morphogenetic protein; CB, cystoblast; Dpp, Decapentaplegic; GSC, germline stem cell; NPF, neuropeptide F; NPFR, neuropeptide F receptor; pMad, phosphorylated Mad; Put, Punt; Sax, Saxophone; RNAi, RNA interference; Tkg-GAL4, Tk-gut-GAL4; Tkv, Thickveins.
We also tested genetic interactions between NPF and Dpp signaling pathways in controlling the mating-induced increase in GSC number. We counted GSCs in double heterozygous mutant flies carrying NPFRsk8 and one of the Dpp pathway mutations, dpphr56, sax5, tkv1, or put135. The mating-induced increase in GSC number was disrupted in the double heterozygous mutant flies carrying NPFRsk8/dpphr56 and NPFRsk8/sax5 (Fig 6B). On the other hand, the opposite phenotype of increased GSC number in virgin females was observed in NPFRsk8/tkv1 and NPFRsk8/put135 flies (Fig 6B; see Discussion). These results suggest that midgut-derived NPF signaling in ovarian somatic cells affects Dpp signaling in GSCs to control their proliferation after mating.
We also analyzed cap cells, which are critical components of the GSC niche. However, Tkg>NPFRNAi did not change the number of cap cells in virgin or mated female flies (Fig 6C), suggesting that midgut-derived NPF does not affect the overall architecture of the niche. This is consistent with observations that found no mating-induced effects on cap cell numbers [14]. Thus, these findings indicate that the NPF-dependent post-mating increase in GSC proliferation is not dependent on the physical size of the GSC niche but rather is regulated through modulation of the Dpp signaling pathway.
The NPF-dependent increase in GSC number requires ovarian ecdysteroid signaling
Previous studies have revealed that biosynthesis of ecdysone, the major insect steroid hormone, is induced by mating stimuli and that ovarian ecdysteroid transmits its signal directly through the ecdysone receptor (EcR) expressed in the ovarian niche to increase GSC number [14,15,42,44,45]. We therefore examined the relationship between NPF and ecdysteroids in the ovary. Ovarian ecdysteroid levels after mating were not different between control and Tkg>NPFRNAi animals (S7A Fig), indicating that NPF is not essential for mating-induced ecdysteroid biosynthesis. Consistent with this, exogenous application of 20-hydroxyecdysone—the active ecdysteroid—did not rescue the GSC phenotype of Tkg>NPFRNAi animals (S7B Fig). However, ex vivo culture experiments revealed that virgin ovaries dissected from animals with knocked down neverland (nvd), encoding an ecdysteroid biosynthesis enzyme, did not exhibit an increase in GSC number in the presence of synthetic NPF peptide (S7C Fig). In addition, synthetic NPF peptide in ex vivo cultures only induced a minor increase in GSC number in ovaries dissected from EcR RNAi animals (S7C Fig). These results suggest that NPF signaling in this context requires ecdysteroid signaling, which possibly interacts with EcR and/or downstream signaling components (S7D Fig).
Discussion
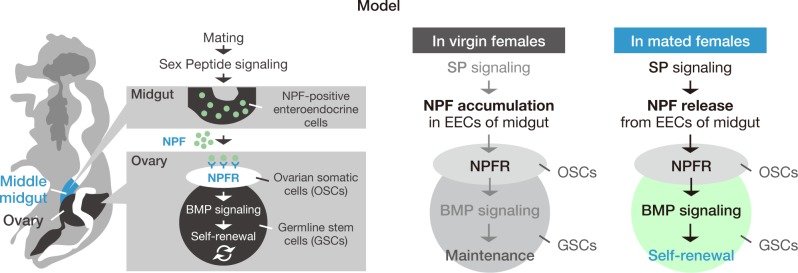
Stem cells are maintained by a specialized microenvironment, or niche, that produces local signals. While the importance of these signals is unquestionable, systemic signals from other tissues are also required for stem cell regulation. However, it remains unclear whether and how crucial systemic signals influence stem cell behavior in response to environmental factors. Our present study demonstrated that midgut-derived NPF modulates GSC proliferation in response to mating stimulus. In mated female flies, SP signaling regulates NPF accumulation levels in EECs in the middle midgut. We showed that NPF acts on the ovary to control GSC proliferation via its receptor NPFR. Furthermore, NPF–NPFR signaling positively modulates Dpp signaling in GSCs to support symmetric GSC divisions. Our results reveal a mechanism of interorgan communication between the gut and the ovary that promotes mating-induced activation of gametogenesis. This is the first study to show that a gut-derived factor modulates GSC activity (Fig 7).
Fig 7. Midgut-derived NPF controls mating-induced GSC proliferation.
A model illustrating the mechanism based on this study. SP signaling induces NPF release from midgut EECs. NPF positively controls pMad expression in GSCs to induce GSC proliferation via ovarian NPFR. BMP, bone morphogenetic protein; EEC, enteroendocrine cell; GSC, germline stem cell; NPF, neuropeptide F; NPFR, neuropeptide F receptor; OSC, ovarian somatic cell; pMad, phosphorylated Mad; SP, sex peptide.
In many animals, reproduction involves significant behavioral and physiological shifts in response to mating. In female Drosophila melanogaster, post-mating responses result from signals, especially SP, delivered by male seminal fluid during mating. Beyond the previously described SP-dependent post-mating changes [13,33,36,46], our data imply that SP induces the post-mating release of NPF from EECs by silencing SPR-positive neurons. However, the molecular and cellular mechanisms by which SP–SPR signaling influences EECs remain unclear. One possibility is that juvenile hormones (JHs) transmit SP-dependent mating signals to the midgut. It has been reported that mating induces JH biosynthesis in an endocrine organ, the corpus allatum, in vivo [29] and that SP stimulates JH biosynthesis in the corpus allatum in vitro [47]. After mating, elevated circulating JH signals are received by the intestinal epithelium, leading to gut remodeling and an increase in gut size, which is required for reproductive success [29]. However, our data indicate that midgut RNAi (in EECs and NPF/Tk/Dh31-positive EECs) against the 2 known genes encoding JH receptors in Drosophila (Methoprene-tolerant and germ cell-expressed bHLH-PAS) does not affect mating-induced GSC proliferation (S4C Fig). Therefore, NPF-dependent control of GSC proliferation appears to be independent of gut JH signaling. It is also unlikely that the midgut directly receives SP because overexpressing SP in EECs did not affect NPF accumulation in EECs (S5A and S5B Fig). Consistent with these results, we did not detect anti-SPR immunostaining signals in EECs. It would be interesting to identify the specific humoral factor that conveys the SP-dependent neuronal signal to EECs.
Another possibility is that nutrient intake after mating is the key in releasing NPF from EECs. Because SP signaling is important for the drastic increase in food intake by females after mating [48], it is possible that SP promotes gut NPF secretion indirectly through some nutrient(s) from food. Notably, the presence of amino acids activates calcium signaling in some EECs in the posterior midgut [49]. In addition, sugar is known to affect a subpopulation of EECs, as activin-β in EECs is up-regulated by chronic high-sugar diets and acts on the fat body [50]. Alternatively, metal ions, such as copper ions, would also be interesting candidates because many NPF-positive cells are located in the stomach-like copper cell region that accumulates copper ions [51,52]. Moreover, NPF has already been identified as a regulatory neuropeptide in discriminating nutritional and food-related conditions [53,54]. Future studies examining whether nutrients can affect NPF release from midgut EECs to control mating-induced GSC proliferation will be worthwhile.
Our findings support the notion that ovarian NPFR enhances BMP signaling in GSCs to promote their self-renewal. Transgenic RNAi and overexpression of NPFR driven by tj-GAL4 revealed that NPFR is necessary and sufficient for induction of female GSC proliferation. More remarkably, ex vivo cultures demonstrated that synthetic NPF peptide is sufficient to induce GSC proliferation in dissected ovaries from virgin females. However, we were unable to address which ovarian cell type expresses NPFR for controlling GSC proliferation. Even though we showed that NPFR transcripts are detected in dissected ovaries (Fig 4C), we failed to observe any clear signals of digoxigenin-labeled NPFR RNA probes by in situ hybridization, whereas the same method was successfully applied for detecting NPFR mRNA in the head and midgut [38]. We speculate that this may be due to lower amounts of transcript in the ovary than in the head and midgut (Fig 4C).
Nevertheless, our analysis using several cell type–specific GAL4 drivers suggests that GSC proliferation requires NPFR acting in some combination of escort or follicle cells but not with germ and cap cells. Although cap cells act as the main niche component by producing the short-range Dpp ligand that ensures GSC self-renewal, escort cells also function in the GSC niche by producing Dpp to repress cystoblast differentiation [55,56]. In escort cells, the Hedgehog (Hh) and Janus Kinase (JAK)-Signal Transducer and Activator of Transcription (STAT) signaling pathways are important for GSC maintenance through activating expression of genes encoding BMP ligands [57–59]. Of these 2 pathways, Hh signaling appears particularly relevant in this context. It is well known that activation of Hh signaling results in stabilization and nuclear localization of the transcription factor Cubitus interruptus (Ci) [60]. Conversely, Hh signaling is negatively regulated by proteolysis of Ci following its phosphorylation by several protein kinases, including cAMP-dependent protein kinase (PKA) [61]. Notably, NPF–NPFR signaling negatively regulates adenylyl cyclase activity, leading to down-regulation of cAMP production [38]. Therefore, it is feasible to hypothesize that NPF–NPFR signaling may result in lower cAMP levels and reduced PKA activity, allowing Ci to persist within the nucleus, leading to enhanced Hh signaling in escort cells. Examining genetic interactions between NPF–NPFR signaling and Hh signaling in ovarian somatic cells is currently underway. It is also of interest to detect in vivo cAMP fluctuation by imaging with fluorescence resonance energy transfer (FRET) probes, such as Epac-based FRET sensors [62], to examine which cells actually respond to NPF and observe whether Hh signaling is involved in NPF–NPFR–dependent GSC proliferation.
Although NPF–NPFR signaling positively affects pMad levels in GSCs, it should be noted that there are both positive and negative roles for BMP receptors in NPF–NPFR–dependent regulation of mating-induced GSC proliferation. Our genetic data suggest that the BMP type-I receptor Sax positively regulates mating-induced GSC proliferation, while the other type-I receptor Tkv and the type-II receptor Put negatively regulate it. This complex situation is reminiscent of a recent finding that Tkv plays both positive and negative roles in female GSC proliferation in Drosophila [63]. In the latter case, Tkv proteins in escort cells sequester excess cap cell–produced Dpp, thereby reducing Dpp activity. Thus, it is possible that each subtype of BMP receptor may have a unique function in mediating NPF–NPFR signaling in different cell types of the germarium.
In addition to ecdysteroids [44,64] and insulin [10,65], we have identified NPF as a new essential humoral regulator for GSC proliferation and self-renewal. It will be important to investigate whether and how these endocrine signals reciprocally work in GSCs and the GSC niche. While previous studies demonstrated parallel regulation by ecdysone and insulin for GSC proliferation [14,44], our study suggests that NPF–NPFR signaling requires ecdysteroid signaling to control GSC proliferation (S7D Fig). Ecdysteroid signaling is known to be crucial for GSC maintenance that is dependent on intrinsic epigenetic machinery [64,66]. Thus, NPF–NPFR signaling may affect chromatin remodeling in GSCs to control mating-induced GSC proliferation.
The question as to the effective dose of synthetic NPF peptide in adult female flies must be addressed. As shown in Fig 3E, the threshold concentration to induce GSC increase by injection is 3 pM of synthetic NPF peptide, and we estimate a single-fly injection amount of 100 nL. Assuming that the total amount of hemolymph per single female fly is approximately 1 μL, the injected NPF peptide should increase hemolymph NPF titers by approximately 0.3 pM. On the other hand, binding assays with isotope-labeled NPF in mammalian cells shows a half maximal inhibitory concentration (IC50) of 65 nM NPF on NPFR [38]. Based on the pharmacological profiles of NPFR, in conjunction with the assumption that the peptide is rapidly degraded in the hemolymph [67], the minimal effective amount of injected synthetic NPF peptide (0.3 pM) seems to be very low. Coupled with a lack of published evidence on the actual chemical characteristics of NPF peptide in hemolymph, the uncertainty as to the exact fate of injected NPF and why our results were reproducible even at concentrations well below the theoretical threshold require further examination. Future studies must address how exogenous NPF peptide behaves in the animal after injection (e.g., which tissues NPF accumulates in or how long it is stable in the hemolymph), whether and how NPF in vivo is biochemically modified or complexed before interaction with the ovary, and the measurement of the actual amount of circulating NPF in virgin and mated females.
In many animal species, NPY has a role in regulating reproduction. In planarians, neuronal neuropeptide Y-8 (NPY-8) and neuropeptide Y receptor Y1 (NPYR-1) signaling regulate germline development, including GSC differentiation [68]. In mammals, administration of NPY results in various effects on luteinizing hormone (LH) and gonadotropin-releasing hormone (GnRH) secretion, either stimulatory or inhibitory. Injection of NPY into ovariectomized and sex steroid-treated rats stimulates secretion of LH and GnRH [69,70]. Conversely, NPY suppresses the gonadotropic axis and delays sexual maturation in intact rats [71–73]. NPY also has a role in coordinating mammalian reproductive function and energy balance [74]. Although several studies have described the role of neuronal NPY signaling on reproduction, the role of intestinal NPY is poorly understood. Therefore, it would be of interest to explore the role of intestinal NPY on reproduction in the context of mammalian nutritional status.
Materials and methods
Drosophila strains
Flies were raised on cornmeal-yeast-agar medium at 25°C; temperature-sensitive mutants were cultured at 29°C for 1 day prior to performing the assays. yw was used as the control strain. The mutant alleles NPFsk1, NPFsk2, sNPFsk4, sNPFsk8, and NPFRsk8 were created in a yw background using CRISPR/Cas9 as previously described [16]. The following guide RNA (gRNA) sequences were used: NPF, 5ʹ-GCCCTTGCCCTCCTAGCCGC-3ʹ; sNPF, 5ʹ-GTTGGGAAAGACGACGGGTC-3ʹ; NPFR, 5ʹ-GCGTGCGCTATCTGGACGAC-3ʹ. Breakpoint details of NPFsk1, NPFsk2, and NPFRsk8 are described in Fig 1. The following transgenic and mutant stocks were used: nSyb-GAL4 (Bloomington 51941), elav-GAL4 (Bloomington 8765), 386Y-GAL4 (Bloomington 25410), Tk-gut-GAL4 [20] (gift from Masayuki Miura, the University of Tokyo, Japan), Myo1A-GAL4 [75] (gift from Kazutaka Akagi, National Center for Geriatrics and Gerontology, Japan), esg-GAL4 (Bloomington 26816), SPR-GAL4::VP16 (see the section “Establishing the SPR-GAL4::VP16 strain” below in Materials and methods for details), tj-GAL4 (Kyoto 104055), e22c-GAL4 (Kyoto 106609), c587-GAL4 [76] (gift from Hiroko Sano, Kurume University, Japan), bab1-GAL4 [77] (gift from Satoru Kobayashi, University of Tsukuba, Japan), c306-GAL4 (Bloomington 3743), nos-GAL4 (Kyoto 107748), UAS-NPF [17] (gift from Ping Shen, University of Georgia, USA), UAS-NPFR (gift from Ping Shen), UAS-rpr; UAS-hid (gift from Katja Brückner, University of California San Francisco), UAS-shits1 (Bloomington 44222), dj-GFP/CyO [78] (Bloomington 5417), Df(3R)ED10642 (Kyoto 150266), Df(3R)BSC464 (Bloomington 24968), dpphr56 [7] (Bloomington 36528), sax5 (Bloomington 8785), tkv1 (Bloomington 427), put135 [7] (Bloomington 3100), and SP0 [79] and SPΔ130 [79] (gifts from Nobuaki Tanaka, Hokkaido University, Japan). Janelia GAL4 stocks [80] were obtained from the Bloomington Drosophila Stock Center: R41D08-GAL4 (45279), R42C03-GAL4 (50148), R46G06-GAL4 (41271), and R50F10-GAL4 (45998). RNAi constructs targeting NPFR (KK107663), gce (KK101814), and Met (KK100638) were obtained from the Vienna Drosophila Resource Center (VDRC) [81] and lines targeting NPF (27237), NPFR (NPFR RNAi-2; 25939), Tk (25800), and Dh31 (41957) were obtained from the Bloomington TRiP collection [82].
Behavioral assays
Flies were reared at 25°C and aged for 5 to 7 days. Virgin female flies were mated overnight to yw male flies at 25°C (10 males and 5–10 females per vial). For egg-laying assays, individual female flies were transferred to a fresh chamber with cornmeal-yeast-agar medium, after which the eggs were counted manually.
Immunohistochemistry
Ovaries and midguts were dissected in Grace’s supplemented insect medium (Gibco) and fixed in 4% paraformaldehyde in Grace’s medium for 30 to 60 minutes at room temperature (RT). Fixed samples were washed 3 times in PBS supplemented with 0.1% Triton X-100. After washing, the samples were blocked in blocking solution (PBS with 0.1% Triton X-100 and 0.2% bovine serum albumin [BSA]) for 1 hour at RT and then incubated with a primary antibody in blocking solution at 4°C overnight. Primary antibodies used in this study were mouse anti-Hts 1B1 [83] (1:50; Developmental Studies Hybridoma Bank [DSHB]), rat anti-DE-cadherin DCAD2 [84] (1:50; DSHB), rabbit anti-pH3 (1:1000; Merck Millipore), rabbit monoclonal anti-pMad (1:1000; Abcam), mouse anti-Lamin-C LC28.26 [85] (1:10; DSHB), rat anti-BrdU (1:50; Abcam), rabbit cleaved Dcp-1 (1:100; Cell Signaling Technology), rabbit anti-NPF (1:2000; provided by Ping Shen), rat anti-Vasa (1:1000; DSHB), and Alexa Fluor 546 phalloidin (1:200; Invitrogen). When anti-pMad antibody was used, the immunofluorescent signals were enhanced by Can Get Signal Solution B (ToYoBo). After washing, fluorophore (Alexa Fluor 488 or 546)-conjugated secondary antibodies (Invitrogen) were used at a 1:200 dilution, and the samples were incubated for 2 hours at RT in blocking solution. After another washing step, all samples were mounted in FluorSave reagent (Merck Millipore). For BrdU incorporation, dissected ovaries were incubated in Grace’s medium containing 10 μM BrdU (Sigma-Aldrich) for 1 hour at RT, washed, and then fixed with 4% paraformaldehyde in Grace’s medium for 1 hour. Ovaries were denatured in 2 N HCl for 30 minutes, neutralized in 100 mM borax for 2 minutes, and then immunostained using mouse anti-BrdU (1:50; Abcam). GSC number was determined based on morphology and positioning of their anteriorly anchored spherical spectrosome [44]. Samples were visualized using a Zeiss LSM 700 confocal microscope or Zeiss Axioplan 2. Images were processed using ImageJ software (NIH).
Quantitative reverse transcription PCR
To quantify mating-induced changes in gene expression, the middle midguts from 5 to 10 adult female flies were dissected. Total RNA was extracted using RNAiso Plus reagent (TaKaRa). cDNA was prepared with ReverTra Ace qPCR RT Master Mix with gDNA Remover (ToYoBo). Quantitative reverse transcription PCR (qRT-PCR) was performed using the Universal SYBR Select Master Mix (Applied Biosystems) with a Thermal Cycler Dice TP800 system (TaKaRa). Serial dilutions of a plasmid containing the open reading frame of each gene were used as standard. The amount of target RNA was normalized to ribosomal protein 49 (rp49) and then relative fold changes were calculated. The following primer pairs were used to measure transcript level: rp49 forward, 5ʹ-CGGATCGATATGCTAAGCTGT-3ʹ and reverse, 5ʹ-GCGCTTGTTCGATCCGTA-3ʹ; NPF forward, 5ʹ-CTCCGCGAAAGAACGATGTCAACAC-3ʹ and reverse, 5ʹ-CCTCAGGATATCCATCAGCGATCCG-3ʹ; NPFR forward, 5´-GATCCTGTCCAAGTACTGGCCCTAC-3´ and reverse, 5´-ACGATCACCTGATATCTGTCGAAGGC-3´. All qRT-PCR runs were performed in triplicate.
Establishing the SPR-GAL4::VP16 strain
To prepare the SPR-GAL4::VP16 line, we used a recombineering approach based on previously described methods [86]. To prepare a landing-site cassette, 5′ and 3′ homology arms were amplified from the GAL4/terminator gene of pBPGUw [87] and were used to flank the positive/negative selectable marker RpsL-kana [88], conferring kanamycin resistance and streptomycin sensitivity. SPR-specific arms were added to this landing-site cassette by PCR using the following primers: SPR-F, 5ʹ-gaattaaggcagcgccaggggaatccgctcgagaaacccacgtccacgagATGAAGCTACTGTCTTCTATCGAACAAGC-3ʹ and SPR-R, 5ʹ-ttggtgtgcacactaaattatcgatataaacaacaagccatttaacttacGATCTAAACGAGTTTTTAAGCAAACTCACTCCC-3ʹ. These primers contained 50 bases matching the SPR locus (in lower case) and sequences corresponding to the GAL4 and terminator arms that were previously added (in upper case); note the underlined ATG of both SPR and GAL4. This cassette was recombined into the bacterial artificial chromosome CH321-69P02 [89] (obtained from the Children’s Hospital Oakland Research Institute), containing the SPR locus within 88 kb of genomic DNA sequence; recombinants were then selected on kanamycin medium. In a second round of recombination, the landing pad was replaced with full-length GAL4::VP16+terminators amplified from pBPGAL4.2::VP16Uw [90]; recombinants were identified by streptomycin resistance. Potential regulatory elements in flanking regions, upstream and downstream untranslated regions, and introns remained intact, although regions downstream of the target region are presumably no longer transcribed. The recombined regions were sequenced, and the finished bacterial artificial chromosome was integrated into the attP40 site by Rainbow Transgenic Flies (Camarillo, CA).
Fly injection
Fly injection was performed using a previously described technique [91]. NPF peptide amidated at its C-terminus (SNSRPPRKNDVNTMADAYKFLQDLDTYYGDRARVRF-NH2) was synthesized by Eurofins Genomics. The synthetic NPF peptide was diluted in PBS to 3 pM. The NPF peptide solution was injected into the thoraces of virgin female flies chilled on ice. Although we could not control the exact amount of peptide solution for fly injection because of limitations imposed by our injection apparatus, we can roughly estimate the amount, which should be 100 nL. Injected flies were transferred into vials with standard fly food. Flies were cultured at RT in vials with or without males for 16 hours. Afterwards, female flies were dissected and immunostained to count GSC number. Synthetic Bm-sNPF peptide (SPSRRLRF-NH2) [92] was a gift from Yoshiaki Tanaka (National Agricultural and Food Research Organization, Japan).
Ex vivo ovary culture
Adult females were cultured on standard medium and dissected in Schneider’s insect medium (Gibco). Approximately 6 ovaries were transferred to a microcentrifuge tube containing 20 μL Schneider’s medium supplemented with 15% fetal calf serum and 0.6% penicillin-streptomycin with the addition of NPF peptide, sNPF peptide, or PBS. Cultures were incubated at RT for 16 hours, and then samples were immunostained to count GSC number.
Statistical analysis
All experiments were performed independently at least twice. Fluorescence intensity in confocal sections was measured via ImageJ. For NPF quantification, an average of 10 cells were examined for each midgut. For pMad quantification, signal intensity was calculated by measuring the fluorescence intensity in GSCs and CBs, which were costained with anti-Vasa antibody to visualize their cell boundaries. Size of the posterior midgut in confocal sections was measured via ImageJ. Sample sizes were chosen based on the number of independent experiments required for statistical significance and technical feasibility. The experiments were not randomized, and the investigators were not blinded. All statistical analyses were carried out in the “R” software environment [93]. The P value is provided in comparison with control and indicated as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and “NS” for nonsignificant (P > 0.05).
Supporting information
(A, B) Representative images of adult female brains and midguts immunostained with anti-NPF antibody (green) and monoclonal nc82 (neuropil marker; magenta) or phalloidin (magenta). Anti-NPF signals in neuroendocrine cells (arrowhead) were dramatically reduced by loss of NPF (panel A) or neuronal knockdown of NPF (panel B). Anti-NPF signals in midgut EEC were also reduced by loss of NPF (panel A) but not after neuronal knockdown of NPF (panel B). (C) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. NPF RNAi driven by nSyb-GAL4 (pan-neuronal cells) or 386Y-GAL4 (neuroendocrine cells) had no effect on the mating-induced increase in GSC number. The number of germaria analyzed is shown inside the bars in panel C. For statistical analysis, a Wilcoxon rank sum test was used for panel C. ***P ≤ 0.001. Scale bar = 50 μm in panel A and B. Underlying data can be found in S1 Data. GSC, germline stem cell; NPF, neuropeptide F.
(TIF)
(A–C) Representative images of adult female brains and midguts immunostained with anti-NPF antibody (green) or anti-GFP antibody (green) and monoclonal nc82 (neuropil marker; magenta) or phalloidin (magenta). (A, B) Tkg-GAL4 driver expressed in the brain and VNC but not in the ovary. (C) NPF RNAi driven by the Tkg-GAL4 driver did not reduce anti-NPF levels in the brain and VNC. Scale bar = 50 μm (brain and VNC) and 100 μm (ovary). NPF, neuropeptide F; Tkg-GAL4, Tk-gut-GAL4; VNC, ventral nerve cord.
(TIF)
Representative images of adult female brains and midguts immunostained with anti-GFP antibody (green), anti-NPF antibody (magenta), or phalloidin (magenta). These GAL4 drivers were expressed in NPF-positive EECs. Anti-GFP signals were also detected in the brain, VNC, and oviduct, but not the ovary. Scale bar = 50 μm (brain and VNC) and 100 μm (ovary and midgut). AG, accessory gland; EEC, enteroendocrine cell; NPF, neuropeptide F; OV, oviduct; SP, spermatheca; VNC, ventral nerve cord.
(TIF)
(A, B) The number of mitotic cells (panel A) or size (panel B) of the posterior midgut in virgin or mated female flies was not affected by NPF RNAi driven by Tkg-GAL4 (NPF-positive EECs). (C) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. Met RNAi or gce RNAi driven by Myo1A-GAL4 (ECs) or Tkg-GAL4 (NPF/Tk/Dh31-positive EECs) had no effect on the mating-induced increase in GSC number. Dots represent the number of mitotic cells in a single middle midgut (panel A) or the diameter of a single posterior midgut (panel B); lines represent the median, and whiskers represent the interquartile range. For statistical analysis, a Wilcoxon rank sum test was used in panel A and C. Student t test was used in panel B. ***P ≤ 0.001 and **P ≤ 0.01. Underlying data can be found in S1 Data. Dh31, diuretic hormone 31; EC, enterocyte; EEC, enteroendocrine cell; gce, germ cell-expressed bHLH-PAS; GSC, germline stem cell; Met, Methoprene tolerant; NPF, neuropeptide F; pH3, phospho-histone H3; Tk, Tachykinin; Tkg-GAL4, Tk-gut-GAL4.
(TIF)
(A, C) Representative images of anti-NPF antibody immunostaining in the middle midgut are shown on the left. Quantification of anti-NPF signal intensity in the middle midgut is shown on the right graph. Anti-NPF signal intensity did not change after overexpressing membrane-thethered SP (mSP) in EECs (Tkg-GAL4>mSP). (B) NPF mRNA level did not change in Tkg-GAL4>mSP animals. (C) NPF accumulation was reduced by silencing SPR-positive neurons (SPR-GAL4>shits1), mimicking SP binding to SPR at the restrictive temperature, without mating. (D) Transcript abundance of NPF in the middle midgut did not change by this manipulation. Dots represent the relative signal intensity of anti-NPF in a single middle midgut (panel A and C) or relative expression levels of NPF in the middle midgut (panel B and D); lines represent the median, and whiskers represent the interquartile range. For statistical analysis, a Wilcoxon rank sum test with Holm’s correction was used for panel A and C. Student t test with Holm’s correction was used for panel B and D. ***P ≤ 0.001 and **P ≤ 0.01; NS, nonsignificant (P > 0.05). Scale bar = 50 μm in panel A and C. Underlying data can be found in S1 Data. EEC, enteroendocrine cell; mSP, membrane-tethered SP; NPF, neuropeptide F; shi, shibire; SP, sex peptide; SPR, sex peptide receptor; Tkg-GAL4, Tk-gut-GAL4; ts, temperature-sensitive.
(TIF)
(A) Representative images of adult female brains, VNCs, and midguts immunostained with anti-GFP antibody (green) and monoclonal nc82 (neuropil marker; magenta) or phalloidin (magenta) in c587-GAL4>mCD::GFP or tj-GAL4>mCD8::GFP females. Both GAL4 drivers are expressed in the CNS but not the midgut. (B, C) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. (B) NPFR RNAi driven by nSyb-GAL4 or elav-GAL4 (pan-neuronal) did not affect the mating-induced increase in GSC number. (C) NPFR RNAi driven by esg-GAL4 (ISCs and EBs), Myo1A-GAL4 (ECs), or Tkg-GAL4 (NPF/Tk/Dh31-positive EECs) had no effect on GSC number after mating. The number of germaria analyzed is shown inside the bars in panel B and C. For statistical analysis, a Wilcoxon rank sum test was used for panel B and C. ***P ≤ 0.001 and **P ≤ 0.01. Scale bar = 50 μm (brain and VNC) or 100 μm (midgut) in panel A. Underlying data can be found in S1 Data. Dh31, diuretic hormone 31; EB, enteroblast; EC, enterocyte; EEC, enteroendocrine cell; GSC, germline stem cell; ISC, intestinal stem cell; NPF, neuropeptide F; NPFR, neuropeptide F receptor; Tk, Tachykinin; VNC, ventral nerve cord.
(TIF)
(A) Ecdysteroid levels in mated ovaries did not change in Tkg-GAL4>NPFRNAi animals. (B, C) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. (B) GSC phenotype in NPF RNAi animals was not rescued by feeding with the active form of ecdysteroid, 20E. Flies were fed on standard cornmeal-yeast-agar yeast medium mixed with a solution of 20E in ethanol, resulting in a final concentration of 0.1 mM 20E. (C) Ovarian knockdown of nvd (c587-GAL4>nvdRNAi) or EcR (c587-GAL4>EcRRNAi) blocked the NPF-induced increase in GSC number in ex vivo ovary cultures. (D) A model illustrating GSC regulation by NPF and ecdysteroid. NPF-dependent increase in GSC number requires ovarian ecdysteroid signaling. Dots represent ovarian ecdysteroid levels of female flies (panel A); lines represent the median, and whiskers represent the interquartile range. The number of germaria analyzed is shown inside the bars in panel B and C. For statistical analysis, Student t test was used for panel A, and a Wilcoxon rank sum test with Holm’s correction was used for panel B and C. ***P ≤ 0.001 and *P ≤ 0.05; NS, nonsignificant (P > 0.05). Underlying data can be found in S1 Data. 20E, 20-hydroxyecdysone; BMP, bone morphogenetic protein; EcR, ecdysone receptor; GSC, germline stem cell; NPF, neuropeptide F; NPFR, neuropeptide F receptor; nvd, neverland; Tkg-GAL4, Tk-gut-GAL4.
(TIF)
In this file, separate worksheets contain the data used in each figure panel as indicated.
(XLSX)
Acknowledgments
We thank Toshiro Aigaki, Kazutaka Akagi, Katja Brückner, Yoshiki Hayashi, Ken-ichi Kimura, Satoru Kobayashi, Masayuki Miura, Norbert Perrimon, Hiroko Sano, Ping Shen, Nobuaki Tanaka, Yoshiaki Tanaka, Bloomington Stock Center, Kyoto Stock Center (DGRC), the National Institute of Genetics, the Vienna Drosophila RNAi Center, Children’s Hospital Oakland Research Institute, and the Developmental Studies Hybridoma Bank for stocks and reagents; Reiko Kise for her technical assistance; and Makoto Hayashi, Satoru Kobayashi, Masanao Sato, and Irene Miguel-Aliaga for helpful discussion and critical reading of the manuscript.
Abbreviations
- BMP
bone morphogenetic protein
- BrdU
bromodeoxyuridine
- BSA
bovine serum albumin
- Cas9
CRISPR-associated protein 9
- Ci
Cubitus interruptus
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- Dcp-1
Death caspase-1
- Dh31
diuretic hormone 31
- Dilp2
Drosophila insulin-like peptide 2
- Dpp
Decapentaplegic
- DSHB
Developmental Studies Hybridoma Bank
- EcR
ecdysone receptor
- EEC
enteroendocrine cell
- FRET
fluorescence resonance energy transfer
- GnRH
gonadotropin-releasing hormone
- gRNA
guide RNA
- GSC
germline stem cell
- Hh
Hedgehog
- hid
head involution defective
- IC50
the half maximal inhibitory concentration
- JAK-STAT
Janus Kinase-Signal Transducer and Activator of Transcription
- JH
juvenile hormone
- LH
luteinizing hormone
- mSP
membrane-tethered SP
- NPF
neuropeptide
- NPFR
neuropeptide F receptor
- NPY
neuropeptide
- NPYR-1
neuropeptide Y receptor Y1
- nvd
neverland
- PBS
phosphate-buffered saline
- pH3
phospho-histone H3
- PKA
cAMP-dependent protein kinase
- pMad
phosphorylated Mad
- ppk
pickpocket
- Put
Punt
- qRT-PCR
quantitative reverse transcription PCR
- RNAi
RNA interference
- rp49
ribosomal protein 49
- rpr
reaper
- RT
room temperature
- Sax
Saxphone
- sNPF
short neuropeptide F precursor
- SP
sex peptide
- SPR
sex peptide receptor
- Tk
Tachykinin
- Tkv
Thickveins
Data Availability
All relevant data are within the paper and its Supporting Information files. In particular, all numerical data for Figures are described in S1 Data in Supporting Information.
Funding Statement
JSPS https://kaken.nii.ac.jp/en/grant/KAKENHI-PROJECT-16H04792/ (grant number KAKENHI 16H04792). Received by RN. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. JST PRESTO http://www.jst.go.jp/kisoken/presto/en/index.html (grant number JPMJPR12M1). Received by RN. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. AMED-CREST, AMED https://www.amed.go.jp/koubo/010720170310_kettei_kadai01.html (grant number JP18gm1110001). Received by RN. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The Naito Foundation https://www.naito-f.or.jp/en/. Received by RN. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. JSPS https://kaken.nii.ac.jp/en/grant/KAKENHI-PROJECT-15J00652/ (grant number KAKENHI 15J00652). Received by TA. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. MEXT https://kaken.nii.ac.jp/en/grant/KAKENHI-PLANNED-25114005/ (grant number KAKENHI 25114005). Received by GY; Grant-in-Aid for Scientific Research on Innovative Areas 'Mechanisms regulating gamete formation in animals’. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The Inoue Foundation for Science https://www.inoue-zaidan.or.jp. Received by YSN. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Spradling A, Fuller MT, Braun RE, Yoshida S. Germline stem cells. Cold Spring Harb Perspect Biol. 2011;3: a002642 10.1101/cshperspect.a002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drummond-Barbosa D. Stem cells, their niches and the systemic environment: an aging network. Genetics. 2008;180: 1787–1797. 10.1534/genetics.108.098244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirilly D, Xie T. The Drosophila ovary: an active stem cell community. Cell Res. 2007;17: 15–25. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17199109. 10.1038/sj.cr.7310123 [DOI] [PubMed] [Google Scholar]
- 4.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414: 98–104. 10.1038/35102160 [DOI] [PubMed] [Google Scholar]
- 5.Wong MD, Jin Z, Xie T. Molecular mechanisms of germline stem cell regulation. Annu Rev Genet. 2005;39: 173–195. 10.1146/annurev.genet.39.073003.105855 [DOI] [PubMed] [Google Scholar]
- 6.Spradling AC. Developmental genetics of oogenesis In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, Cold Spring Harbor Laboratory Press; 1993. p. 1–70. [Google Scholar]
- 7.Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94: 251–60. 10.1016/S0092-8674(00)81424-5 [DOI] [PubMed] [Google Scholar]
- 8.Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231: 265–278. 10.1006/dbio.2000.0135 [DOI] [PubMed] [Google Scholar]
- 9.Soller M, Bownes M, Kubli E. Mating and sex peptide stimulate the accumulation of yolk in oocytes of Drosophila melanogaster. Eur J Biochem. 1997;243: 732–738. 10.1111/j.1432-1033.1997.00732.x [DOI] [PubMed] [Google Scholar]
- 10.LaFever L, Drummond-Barbosa D. Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science 2005;309: 1071–1073. 10.1126/science.1111410 [DOI] [PubMed] [Google Scholar]
- 11.Armstrong AR, Laws KM, Drummond-Barbosa D. Adipocyte amino acid sensing controls adult germline stem cell number via the amino acid response pathway and independently of Target of Rapamycin signaling in Drosophila. Development. 2014;141: 4479–4488. 10.1242/dev.116467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuoka S, Armstrong AR, Sampson LL, Laws KM, Drummond-Barbosa D. Adipocyte metabolic pathways regulated by diet control the female germline stem cell lineage in Drosophila melanogaster. Genetics. 2017;206: 953–971. 10.1534/genetics.117.201921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubli E. Sex-peptides: seminal peptides of the Drosophila male. Cell Mol Life Sci. 2003;60: 1689–1704. 10.1007/s00018-003-3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ameku T, Niwa R. Mating-Induced Increase in Germline Stem Cells via the Neuroendocrine System in Female Drosophila. PLoS Genet. 2016;12(6): e1006123 10.1371/journal.pgen.1006123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ameku T, Yoshinari Y, Fukuda R, Niwa R. Ovarian ecdysteroid biosynthesis and female germline stem cells. Fly. 2017;11: 185–193. 10.1080/19336934.2017.1291472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo S, Ueda R. Highly Improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195: 715–721. 10.1534/genetics.113.156737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci. 2005;102: 2141–2146. 10.1073/pnas.0406814102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee G, Bahn JH, Park JH. Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc Natl Acad Sci U S A. 2006;103: 12580–12585. 10.1073/pnas.0601171103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veenstra JA, Agricola HJ, Sellami A. Regulatory peptides in fruit fly midgut. Cell Tissue Res. 2008;334: 499–516. 10.1007/s00441-008-0708-3 [DOI] [PubMed] [Google Scholar]
- 20.Song W, Veenstra JA, Perrimon N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9: 40–47. 10.1016/j.celrep.2014.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erion R, King AN, Wu G, Hogenesch JB, Sehgal A. Neural clocks and neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue. Elife. 2016;5: e13552 10.7554/eLife.13552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shohat-Ophir G, Kaun KR, Azanchi R, Mohammed H, Heberlein U. Sexual Deprivation Increases Ethanol Intake in Drosophila. Science 2012;335: 1351–1355. 10.1126/science.1215932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang QP, Lin YQ, Zhang L, Wilson YA, Oyston LJ, Cotterell J, et al. Sucralose Promotes Food Intake through NPY and a Neuronal Fasting Response. Cell Metab. 2016;24: 75–90. 10.1016/j.cmet.2016.06.010 [DOI] [PubMed] [Google Scholar]
- 24.Beshel J, Dubnau J, Zhong Y. A Leptin Analog Locally Produced in the Brain Acts via a Conserved Neural Circuit to Modulate Obesity-Linked Behaviors in Drosophila. Cell Metab. 2017;25: 208–217. 10.1016/j.cmet.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown MR, Crim JW, Arata RC, Cai HN, Chun C, Shen P. Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides. 1999;20: 1035–42. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10499420. [DOI] [PubMed] [Google Scholar]
- 26.Marianes A, Spradling AC. Physiological and stem cell compartmentalization within the Drosophila midgut. Elife. 2013;2: e00886 10.7554/eLife.00886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu H-J, LaFever L, Drummond-Barbosa D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev Biol. 2008;313: 700–712. 10.1016/j.ydbio.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Z, McCall K, Steller H. DCP-1, a Drosophila cell death protease essential for development. Science. 1997;275: 536–540. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8999799. [DOI] [PubMed] [Google Scholar]
- 29.Reiff T, Jacobson J, Cognigni P, Antonello Z, Ballesta E, Tan KJ, et al. Endocrine remodelling of the adult intestine sustains reproduction in Drosophila. Elife. 2015;4: e06930 10.7554/eLife.06930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shingleton AW. Physiology: Female flies have the guts for reproduction. Curr Biol. 2015;25: R716–R718. 10.1016/j.cub.2015.06.073 [DOI] [PubMed] [Google Scholar]
- 31.Géminard C, Rulifson EJ, Léopold P. Remote Control of Insulin Secretion by Fat Cells in Drosophila. Cell Metab. 2009; 10.1016/j.cmet.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 32.Chapman T, Bangham J, Vinti G, Seifried B, Lung O, Wolfner MF, et al. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc Natl Acad Sci U S A. 2003;100: 9923–9928. 10.1073/pnas.1631635100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yapici N, Kim Y-J, Ribeiro C, Dickson BJ. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature. 2008;451: 33–37. 10.1038/nature06483 [DOI] [PubMed] [Google Scholar]
- 34.Häsemeyer M, Yapici N, Heberlein U, Dickson BJ. Sensory neurons in the Drosophila genital tract regulate female reproductive behavior. Neuron. 2009;61: 511–518. 10.1016/j.neuron.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 35.Rezával C, Pavlou HJ, Dornan AJ, Chan Y-B, Kravitz EA, Goodwin SF. Neural circuitry underlying Drosophila female postmating behavioral responses. Curr Biol. 2012;22: 1155–1165. 10.1016/j.cub.2012.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng K, Palfreyman MT, Häsemeyer M, Talsma A, Dickson BJ. Ascending SAG neurons control sexual receptivity of Drosophila females. Neuron. 2014;83: 135–148. 10.1016/j.neuron.2014.05.017 [DOI] [PubMed] [Google Scholar]
- 37.White B, Osterwalder T, Keshishian H. Molecular genetic approaches to the targeted suppression of neuronal activity. Curr Biol. 2001;11: R1041–R1053. 10.1016/S0960-9822(01)00621-2 [DOI] [PubMed] [Google Scholar]
- 38.Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides. 2002;23: 773–780. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11897397. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, et al. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 34: 58–61. 10.1002/gene.10137 [DOI] [PubMed] [Google Scholar]
- 40.Morris LX, Spradling AC. Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development. 2011;138: 2207–2215. 10.1242/dev.065508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barth JMI, Hafen E, Köhler K. The lack of autophagy triggers precocious activation of Notch signaling during Drosophila oogenesis. BMC Dev Biol. 2012;12: 35 10.1186/1471-213X-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris LX, Spradling AC. Steroid signaling within Drosophila ovarian epithelial cells sex-specifically modulates early germ cell development and meiotic entry. PLoS ONE. 2012;7(10): e46109 10.1371/journal.pone.0046109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kai T, Spradling A. An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc Natl Acad Sci U S A. 2003;100: 4633–4638. 10.1073/pnas.0830856100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ables ET, Drummond-Barbosa D. The steroid hormone ecdysone functions with intrinsic chromatin remodeling factors to control female germline stem cells in Drosophila. Cell Stem Cell. 2010;7: 581–592. 10.1016/j.stem.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.König A, Yatsenko AS, Weiss M, Shcherbata HR. Ecdysteroids affect Drosophila ovarian stem cell niche formation and early germline differentiation. EMBO J. 2011;30: 1549–1562. 10.1038/emboj.2011.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. Insect seminal fluid proteins: Identification and function. Annu Rev Entomol 2011;56: 21–40. 10.1146/annurev-ento-120709-144823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moshitzky P, Fleischmann I, Chaimov N, Saudan P, Klauser S, Kubli E, et al. Sex-peptide activates juvenile hormone biosynthesis in the Drosophila melanogaster corpus allatum. Arch Insect Biochem Physiol. 1996;32: 363–374. [DOI] [PubMed] [Google Scholar]
- 48.Carvalho GB, Kapahi P, Anderson DJ, Benzer S. Allocrine Modulation of Feeding Behavior by the Sex Peptide of Drosophila. Curr Biol. 2006;16: 692–696. 10.1016/j.cub.2006.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park JH, Chen J, Jang S, Ahn TJ, Kang KJ, Choi MS, et al. A subset of enteroendocrine cells is activated by amino acids in the Drosophila midgut. FEBS Lett. 2016;590: 493–500. 10.1002/1873-3468.12073 [DOI] [PubMed] [Google Scholar]
- 50.Song W, Cheng D, Hong S, Sappe B, Hu Y, Wei N, et al. Midgut-Derived Activin Regulates Glucagon-like Action in the Fat Body and Glycemic Control. Cell Metab. 2017;25: 386–399. 10.1016/j.cmet.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubreuil RR. Copper cells and stomach acid secretion in the Drosophila midgut. Int J Biochem Cell Biol. 2004;36: 745–752. 10.1016/j.biocel.2003.07.004 [DOI] [PubMed] [Google Scholar]
- 52.Li H, Qi Y, Jasper H. Preventing Age-Related Decline of Gut Compartmentalization Limits Microbiota Dysbiosis and Extends Lifespan. Cell Host Microbe. 2016;19: 240–253. 10.1016/j.chom.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beshel J, Zhong Y. Graded Encoding of Food Odor Value in the Drosophila Brain. J Neurosci. 2013;33: 15693–15704. 10.1523/JNEUROSCI.2605-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slade JD, Staveley BE. Manipulation of components that control feeding behavior in Drosophila melanogaster increases sensitivity to amino acid starvation. Genet Mol Res. 2016;15 10.4238/gmr.15017489 [DOI] [PubMed] [Google Scholar]
- 55.Song X, Wong MD, Kawase E, Xi R, Ding BC, McCarthy JJ, et al. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131: 1353–1364. 10.1242/dev.01026 [DOI] [PubMed] [Google Scholar]
- 56.Kirilly D, Wang S, Xie T. Self-maintained escort cells form a germline stem cell differentiation niche. Development. 2011;138: 5087–5097. 10.1242/dev.067850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: Similar somatic stem cells and signals. Dev Cell. 2005;9: 501–510. 10.1016/j.devcel.2005.08.012 [DOI] [PubMed] [Google Scholar]
- 58.Wang L, Li Z, Cai Y. The JAK/STAT pathway positively regulates DPP signaling in the Drosophila germline stem cell niche. J Cell Biol. 2008;180: 721–728. 10.1083/jcb.200711022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rojas-Ríos P, Guerrero I, González-Reyes A. Cytoneme-mediated delivery of Hedgehog regulates the expression of bone morphogenetic proteins to maintain germline stem cells in Drosophila. PLoS Biol. 2012;10(4): e1001298 10.1371/journal.pbio.1001298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee RT, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143: 367–372. 10.1242/dev.120154 [DOI] [PubMed] [Google Scholar]
- 61.Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 1999;13: 2828–2837. 10.1101/gad.13.21.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patel N, Gold MG. The genetically encoded tool set for investigating cAMP: More than the sum of its parts. Front Pharmacol. 2015. 10.3389/fphar.2015.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo L, Wang H, Fan C, Liu S, Cai Y. Wnt ligands regulate Tkv expression to constrain Dpp activity in the Drosophila ovarian stem cell niche. J Cell Biol. 2015;209: 595–608. 10.1083/jcb.201409142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ables ET, Bois KE, Garcia CA, Drummond-Barbosa D. Ecdysone response gene E78 controls ovarian germline stem cell niche formation and follicle survival in Drosophila. Dev Biol. 2015;400: 33–42. 10.1016/j.ydbio.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu H-J, Drummond-Barbosa D. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc Natl Acad Sci U S A. 2009;106: 1117–1121. 10.1073/pnas.0809144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ables ET, Drummond-Barbosa D. Steroid Hormones and the Physiological Regulation of Tissue-Resident Stem Cells: Lessons from the Drosophila Ovary. Curr Stem Cell Reports. 2017;3: 9–18. 10.1007/s40778-017-0070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isaac RE, Bland ND, Shirras AD. Neuropeptidases and the metabolic inactivation of insect neuropeptides. Gen Comp Endocrinol. 2009; 10.1016/j.ygcen.2008.12.011 [DOI] [PubMed] [Google Scholar]
- 68.Saberi A, Jamal A, Beets I, Schoofs L, Newmark PA. GPCRs Direct Germline Development and Somatic Gonad Function in Planarians. PLoS Biol. 2016;14(5): e1002457 10.1371/journal.pbio.1002457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Urban JH, Das I, Levine JE. Steroid modulation of neuropeptide Y-induced luteinizing hormone releasing hormone release from median eminence fragments from male rats. Neuroendocrinology. 1996;63: 112–119. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9053775. 10.1159/000126947 [DOI] [PubMed] [Google Scholar]
- 70.Sabatino FD, Collins P, McDonald JK. Investigation of the effects of progesterone on neuropeptide Y-stimulated luteinizing hormone-releasing hormone secretion from the median eminence of ovariectomized and estrogen-treated rats. Neuroendocrinology. 1990;52: 600–607. Available from: http://www.ncbi.nlm.nih.gov/pubmed/2126610. 10.1159/000125651 [DOI] [PubMed] [Google Scholar]
- 71.Rettori V, Milenkovic L, Aguila MC, McCann SM. Physiologically significant effect of neuropeptide Y to suppress growth hormone release by stimulating somatostatin discharge. Endocrinology. 1990;126: 2296–301. 10.1210/endo-126-5-2296 [DOI] [PubMed] [Google Scholar]
- 72.Kerkerian L, Guy J, Lefèvre G, Pelletier G. Effects of neuropeptide Y (NPY) on the release of anterior pituitary hormones in the rat. Peptides. 1985;6: 1201–1204. Available from: http://www.ncbi.nlm.nih.gov/pubmed/3914636. [DOI] [PubMed] [Google Scholar]
- 73.Pierroz DD, Catzeflis C, Aebi AC, Rivier JE, Aubert ML. Chronic administration of neuropeptide Y into the lateral ventricle inhibits both the pituitary-testicular axis and growth hormone and insulin-like growth factor I secretion in intact adult male rats. Endocrinology. 1996;137: 3–12. 10.1210/endo.137.1.8536627 [DOI] [PubMed] [Google Scholar]
- 74.Aubert ML, Pierroz DD, Gruaz NM, D’Allèves V, Vuagnat B a, Pralong FP, et al. Metabolic control of sexual function and growth: role of neuropeptide Y and leptin. Mol Cell Endocrinol. 1998;140: 107–113. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9722177. [DOI] [PubMed] [Google Scholar]
- 75.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat Signaling Mediates Regeneration and Homeostasis in the Drosophila Midgut. Cell. 2009;137: 1343–1355. 10.1016/j.cell.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu C-H, Xie T. Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development. 2003;130: 2579–2588. 10.1242/dev.00499 [DOI] [PubMed] [Google Scholar]
- 77.Bolívar J, Pearson J, López-Onieva L, González-Reyes A. Genetic dissection of a stem cell niche: The case of the Drosophila ovary. Dev Dyn. 2006;235: 2969–2979. 10.1002/dvdy.20967 [DOI] [PubMed] [Google Scholar]
- 78.Santel A, Winhauer T, Blümer N, Renkawitz-Pohl R. The Drosophila don juan (dj) gene encodes a novel sperm specific protein component characterized by an unusual domain of a repetitive amino acid motif. Mech Dev. 1997;64: 19–30. 10.1016/S0925-4773(97)00031-2 [DOI] [PubMed] [Google Scholar]
- 79.Liu H, Kubli E. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2003;100: 9929–9933. 10.1073/pnas.1631700100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jenett A, Rubin GM, Ngo TTB, Shepherd D, Murphy C, Dionne H, et al. A GAL4-Driver Line Resource for Drosophila Neurobiology. Cell Rep. 2012;2: 991–1001. 10.1016/j.celrep.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448: 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- 82.Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, et al. The transgenic RNAi project at Harvard medical school: Resources and validation. Genetics. 2015;201: 843–852. 10.1534/genetics.115.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ding D, Parkhurst SM, Lipshitz HD. Different genetic requirements for anterior RNA localization revealed by the distribution of Adducin-like transcripts during Drosophila oogenesis. Proc Natl Acad Sci U S A. 1993;90: 2512–2516. 10.1073/pnas.90.6.2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev Biol. 1994;165: 716–726. 10.1006/dbio.1994.1287 [DOI] [PubMed] [Google Scholar]
- 85.Riemer D, Stuurman N, Berrios M, Hunter C, Fisher PA, Weber K. Expression of Drosophila lamin C is developmentally regulated: analogies with vertebrate A-type lamins. J Cell Sci. 1995;108: 3189–3198. [DOI] [PubMed] [Google Scholar]
- 86.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33: 1–12. 10.1093/nar/gki140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pfeiffer BD, Jenett A, Hammonds AS, Ngo T-TB, Misra S, Murphy C, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci. 2008;105: 9715–9720. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang S, Zhao Y, Leiby M, Zhu J. A new positive/negative selection scheme for precise BAC recombineering. Mol Biotechnol. 2009;42: 110–116. 10.1007/s12033-009-9142-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Venken KJT, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6: 431–434. 10.1038/nmeth.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pfeiffer BD, Ngo TTB, Hibbard KL, Murphy C, Jenett A, Truman JW, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186: 735–755. 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cochemé HM, Logan A, Prime TA, Abakumova I, Quin C, McQuaker SJ, et al. Using the mitochondria-targeted ratiometric mass spectrometry probe MitoB to measure H2O2 in living Drosophila. Nat Protoc. 2012;7: 946–958. 10.1038/nprot.2012.035 [DOI] [PubMed] [Google Scholar]
- 92.Yamanaka N, Yamamoto S, Zitan D, Watanabe K, Kawada T, Satake H, et al. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS ONE. 2008;3(8): e3048 10.1371/journal.pone.0003048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.R Development Core Team. R: A Language and Environment for Statistical Computing. In: R Foundation for Statistical Computing; Vienna Austria: [Internet]. 2011. 10.1038/sj.hdy.6800737 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A, B) Representative images of adult female brains and midguts immunostained with anti-NPF antibody (green) and monoclonal nc82 (neuropil marker; magenta) or phalloidin (magenta). Anti-NPF signals in neuroendocrine cells (arrowhead) were dramatically reduced by loss of NPF (panel A) or neuronal knockdown of NPF (panel B). Anti-NPF signals in midgut EEC were also reduced by loss of NPF (panel A) but not after neuronal knockdown of NPF (panel B). (C) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. NPF RNAi driven by nSyb-GAL4 (pan-neuronal cells) or 386Y-GAL4 (neuroendocrine cells) had no effect on the mating-induced increase in GSC number. The number of germaria analyzed is shown inside the bars in panel C. For statistical analysis, a Wilcoxon rank sum test was used for panel C. ***P ≤ 0.001. Scale bar = 50 μm in panel A and B. Underlying data can be found in S1 Data. GSC, germline stem cell; NPF, neuropeptide F.
(TIF)
(A–C) Representative images of adult female brains and midguts immunostained with anti-NPF antibody (green) or anti-GFP antibody (green) and monoclonal nc82 (neuropil marker; magenta) or phalloidin (magenta). (A, B) Tkg-GAL4 driver expressed in the brain and VNC but not in the ovary. (C) NPF RNAi driven by the Tkg-GAL4 driver did not reduce anti-NPF levels in the brain and VNC. Scale bar = 50 μm (brain and VNC) and 100 μm (ovary). NPF, neuropeptide F; Tkg-GAL4, Tk-gut-GAL4; VNC, ventral nerve cord.
(TIF)
Representative images of adult female brains and midguts immunostained with anti-GFP antibody (green), anti-NPF antibody (magenta), or phalloidin (magenta). These GAL4 drivers were expressed in NPF-positive EECs. Anti-GFP signals were also detected in the brain, VNC, and oviduct, but not the ovary. Scale bar = 50 μm (brain and VNC) and 100 μm (ovary and midgut). AG, accessory gland; EEC, enteroendocrine cell; NPF, neuropeptide F; OV, oviduct; SP, spermatheca; VNC, ventral nerve cord.
(TIF)
(A, B) The number of mitotic cells (panel A) or size (panel B) of the posterior midgut in virgin or mated female flies was not affected by NPF RNAi driven by Tkg-GAL4 (NPF-positive EECs). (C) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. Met RNAi or gce RNAi driven by Myo1A-GAL4 (ECs) or Tkg-GAL4 (NPF/Tk/Dh31-positive EECs) had no effect on the mating-induced increase in GSC number. Dots represent the number of mitotic cells in a single middle midgut (panel A) or the diameter of a single posterior midgut (panel B); lines represent the median, and whiskers represent the interquartile range. For statistical analysis, a Wilcoxon rank sum test was used in panel A and C. Student t test was used in panel B. ***P ≤ 0.001 and **P ≤ 0.01. Underlying data can be found in S1 Data. Dh31, diuretic hormone 31; EC, enterocyte; EEC, enteroendocrine cell; gce, germ cell-expressed bHLH-PAS; GSC, germline stem cell; Met, Methoprene tolerant; NPF, neuropeptide F; pH3, phospho-histone H3; Tk, Tachykinin; Tkg-GAL4, Tk-gut-GAL4.
(TIF)
(A, C) Representative images of anti-NPF antibody immunostaining in the middle midgut are shown on the left. Quantification of anti-NPF signal intensity in the middle midgut is shown on the right graph. Anti-NPF signal intensity did not change after overexpressing membrane-thethered SP (mSP) in EECs (Tkg-GAL4>mSP). (B) NPF mRNA level did not change in Tkg-GAL4>mSP animals. (C) NPF accumulation was reduced by silencing SPR-positive neurons (SPR-GAL4>shits1), mimicking SP binding to SPR at the restrictive temperature, without mating. (D) Transcript abundance of NPF in the middle midgut did not change by this manipulation. Dots represent the relative signal intensity of anti-NPF in a single middle midgut (panel A and C) or relative expression levels of NPF in the middle midgut (panel B and D); lines represent the median, and whiskers represent the interquartile range. For statistical analysis, a Wilcoxon rank sum test with Holm’s correction was used for panel A and C. Student t test with Holm’s correction was used for panel B and D. ***P ≤ 0.001 and **P ≤ 0.01; NS, nonsignificant (P > 0.05). Scale bar = 50 μm in panel A and C. Underlying data can be found in S1 Data. EEC, enteroendocrine cell; mSP, membrane-tethered SP; NPF, neuropeptide F; shi, shibire; SP, sex peptide; SPR, sex peptide receptor; Tkg-GAL4, Tk-gut-GAL4; ts, temperature-sensitive.
(TIF)
(A) Representative images of adult female brains, VNCs, and midguts immunostained with anti-GFP antibody (green) and monoclonal nc82 (neuropil marker; magenta) or phalloidin (magenta) in c587-GAL4>mCD::GFP or tj-GAL4>mCD8::GFP females. Both GAL4 drivers are expressed in the CNS but not the midgut. (B, C) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. (B) NPFR RNAi driven by nSyb-GAL4 or elav-GAL4 (pan-neuronal) did not affect the mating-induced increase in GSC number. (C) NPFR RNAi driven by esg-GAL4 (ISCs and EBs), Myo1A-GAL4 (ECs), or Tkg-GAL4 (NPF/Tk/Dh31-positive EECs) had no effect on GSC number after mating. The number of germaria analyzed is shown inside the bars in panel B and C. For statistical analysis, a Wilcoxon rank sum test was used for panel B and C. ***P ≤ 0.001 and **P ≤ 0.01. Scale bar = 50 μm (brain and VNC) or 100 μm (midgut) in panel A. Underlying data can be found in S1 Data. Dh31, diuretic hormone 31; EB, enteroblast; EC, enterocyte; EEC, enteroendocrine cell; GSC, germline stem cell; ISC, intestinal stem cell; NPF, neuropeptide F; NPFR, neuropeptide F receptor; Tk, Tachykinin; VNC, ventral nerve cord.
(TIF)
(A) Ecdysteroid levels in mated ovaries did not change in Tkg-GAL4>NPFRNAi animals. (B, C) Frequency of germaria containing 1, 2, and 3 GSCs (left axis) and the average number of GSCs per germarium (right axis) in virgin (v) and mated (m) female flies. (B) GSC phenotype in NPF RNAi animals was not rescued by feeding with the active form of ecdysteroid, 20E. Flies were fed on standard cornmeal-yeast-agar yeast medium mixed with a solution of 20E in ethanol, resulting in a final concentration of 0.1 mM 20E. (C) Ovarian knockdown of nvd (c587-GAL4>nvdRNAi) or EcR (c587-GAL4>EcRRNAi) blocked the NPF-induced increase in GSC number in ex vivo ovary cultures. (D) A model illustrating GSC regulation by NPF and ecdysteroid. NPF-dependent increase in GSC number requires ovarian ecdysteroid signaling. Dots represent ovarian ecdysteroid levels of female flies (panel A); lines represent the median, and whiskers represent the interquartile range. The number of germaria analyzed is shown inside the bars in panel B and C. For statistical analysis, Student t test was used for panel A, and a Wilcoxon rank sum test with Holm’s correction was used for panel B and C. ***P ≤ 0.001 and *P ≤ 0.05; NS, nonsignificant (P > 0.05). Underlying data can be found in S1 Data. 20E, 20-hydroxyecdysone; BMP, bone morphogenetic protein; EcR, ecdysone receptor; GSC, germline stem cell; NPF, neuropeptide F; NPFR, neuropeptide F receptor; nvd, neverland; Tkg-GAL4, Tk-gut-GAL4.
(TIF)
In this file, separate worksheets contain the data used in each figure panel as indicated.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. In particular, all numerical data for Figures are described in S1 Data in Supporting Information.