Abstract
Studies have identified abnormalities in the microbiota of patients with arthritis. To evaluate the pathogenicity of human microbiota, we performed fecal microbial transplantation from children with spondyloarthritis and controls to germ-free KRN/B6xNOD mice. Ankle swelling was equivalent in those that received patient vs control microbiota. Principal coordinates analysis revealed incomplete uptake of the human microbiota with over-representation of two genera (Bacteroides and Akkermansia) among the transplanted mice. The microbiota predicted the extent of ankle swelling (R2 = 0.185, p = 0.018). The abundances of Bacteroides (r = −0.510, p = 0.010) inversely and Akkermansia.(r = 0.367, p = 0.078) directly correlated with ankle swelling. Addition of A. muciniphila to Altered Schaedler’s Flora (ASF) resulted in small but statistically significant increased ankle swelling as compared to mice that received ASF alone (4.0 mm, 3.9 – 4.1 versus 3.9 mm, IQR 3.6 – 4.0, p = 0.041), as did addition of A. muciniphila cultures to transplanted human microbiota as compared to mice that received transplanted human microbiota alone (4.5 mm, IQR 4.3 – 5.5 versus 4.1 mm, IQR 3.9 – 4.3, p = 0.019). This study supports previous findings of an association between Akkermansia muciniphila and arthritis.
Introduction
Multiple studies have demonstrated altered microbiota in patients with a variety of categories of arthritis, including juvenile idiopathic arthritis (JIA) [1, 2], rheumatoid arthritis [3], and psoriatic arthritis [4]. Specifically, our previous work demonstrated that a subset of children with juvenile spondyloarthritis had markedly elevated fecal abundance of Akkermansia muciniphila, a commensal organism that is not generally considered to play a role in inflammatory diseases [1]. This organism was isolated based upon its well-characterized ability to degrade intestinal mucin [5], so we hypothesized that degradation of the mucin layer may predispose to arthritis, perhaps by increasing intestinal permeability [1, 6]. A limitation of this line of work is that human cross-sectional studies do not establish causality, and thus animal models of arthritis have been used to evaluate the role of the microbiota. Along those lines, there is evidence in multiple animal models of arthritis that an intact microbiota is necessary for the development of the disease. For example, in the KRN/B6 x NOD (K/BxN) model of RA, the disease is abrogated in the germ-free state but occurs following monocolonization with a single organism, Segmented Filamentous Bacteria [7]. For the current work, we took advantage of the microbiota-dependent nature of the K/BxN model to test the ability of human intestinal microbiota obtained from children with enthesitis-related arthritis (ERA) to induce arthritis. Although we were not able to confirm that the microbiota from patients has greater capacity as compared to that from healthy controls to induce arthritis, we were able to show that a specific organism that our group has previously demonstrated to be associated with arthritis, A. muciniphila [1], was associated with arthritis in the transplanted mice.
Results
Subjects
Clinical characteristics of patients and controls are summarized in Table 1. By design, controls were identical with respect to sex, race, and study site; and ± 1 year of age. None of the controls had HLA-B27 measurements obtained.
Table 1.
Patient population
| Feature | Controls | ERA |
|---|---|---|
| N | 12 | 12 |
| Age at diagnosis (years; median, IQR) | N/A | 13.8 (12.2 – 15.2) |
| Age at collection (years; median, IQR) | 14.1 (11.4 – 15.2) | 13.9 (12.2 – 15.2) |
| Collection site | ||
| CoA | 9, 75% | 9, 75% |
| CHOP | 2, 17% | 2, 17% |
| HUMC | 1, 8.3% | 1, 8.3% |
| Males (n, %) | 7, 58% | 7, 58% |
| Race (n, %) | ||
| Caucasian | 9, 75% | 9, 75% |
| African-American | 3, 25% | 3, 25% |
| HLA-B27+ (n, %) | N/A | 4/12, 33% |
Abbreviations: CHOP = Children’s Hospital of Philadelphia; CoA = Children’s of Alabama; HUMC = Hackensack University Medical Center.
Disease severity in transplanted mice
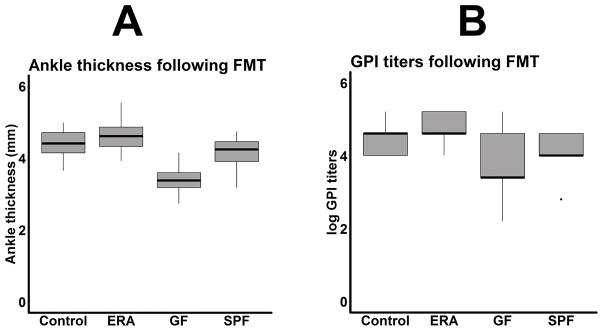
There were no differences in median ankle thickness (4.6 mm, interquartile range (IQR) 4.3 – 4.9 vs 4.4 mm, IQR 4.1 – 4.7, p = 0.242; Figure 1A) or glucose-6 phosphate isomerase (GPI) titers (1:40,960 in both groups; Figure 1B) in mice transplanted with feces derived from enthesitis-related arthritis (ERA) patients as compared to healthy controls. Pooling all the human samples together, mice that received human stool had increased ankle thickness (4.5 mm, IQR 4.3 – 4.8 versus 3.4 mm, IQR 3.1 – 3.6, p < 0.001) and GPI titers (1:40,960 (IQR 1:17,920 – 1:163,840) vs 1:2,560 (IQR 1:2560 – 1:40,960), p = 0.006) as compared to germ-free (GF) mice as well as those moved from the GF to the specific-pathogen free (SPF) facility (ankle thickness: 4.0, IQR 3.7 – 4.3, p = 0.001; GPI titers: 1:10240, IQR 1:10240 – 1:40960, p = 0.020.) The median GPI titers in the GF mice had a bi-modal distribution (Figure 1B). Thus, while there were no obvious differences between mice receiving patient versus control microbiota, mice receiving human microbiota had significantly more severe arthritis as compared to those that remained germ-free.
Figure 1.
Boxplots of ankle thickness (A) and antibody titers against glucose-6 phosphate isomerase (GPI; B) in mice transplanted with fecal material from enthesitis-related arthritis (ERA) patients (n = 12) or healthy controls (n = 12), compared to germ-free (GF) mice (n = 23) as well as those moved from the GF to the specific-pathogen free (SPF) facility (n = 11.)
Analysis of 16S sequence data from transplanted mice
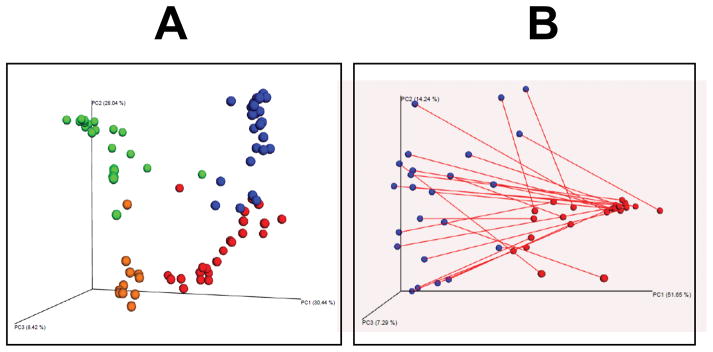
In order to assess the success of the FMT, sequencing of the 16S ribosomal DNA on the transplanted mice was performed, with the results compared to those obtained from the human donors, as well as the GF and SPF mice. As additional controls, 16S sequencing on food pellets obtained from GF mice and on empty tubes was also performed. As shown in Figure 2A, the four groups of subjects – transplanted mice, human donors, GF mice, and SPF mice – all clustered separately, indicating distinct microbiota profiles in each group. Addition of control samples (pellets, empty tubes) to the four groups of subjects demonstrated that these additional controls clustered with the GF mice (Figure S1). Figure 2B depicts the PCoA of the transplanted mice and human donors only, drawn such that each human donor is connected by a line with his or her own murine recipient. It is visually evident that the clustering occurred based upon species, not the donor-recipient dyad. These observations were confirmed with the Permanova test, which showed that species (mouse versus human) was strongly associated with the overall structure of the microbiota (F = 38.9, p = 0.001), while the mouse-human dyad was not (F = 0.42, p = 1.000).
Figure 2.
Principal coordinates of transplanted (n = 24), GF (n = 23), and SPF (n = 11) mice, as well as human donors (n = 24.) 2A shows all four groups (human donors = red, GF mice = green, transplanted mice = blue, SPF mice = orange.) 2B only shows the humans (red) and recipient mice (blue), with each mouse-human dyad connected by a line.
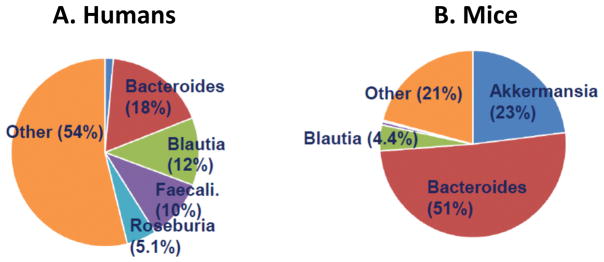
Not surprisingly in light of the observed clustering, substantial taxonomic differences between the human donors and murine recipients were observed at the taxonomic level. At the phylum level, the human samples were dominated by Bacteroidetes (24%) and Firmicutes (64%), which is typical for human fecal microbiota among older children and adults. In contrast, mouse fecal DNA included 23% Verrucomicrobia and 56% Bacteroidetes, while the Firmicutes was only 19%. All three of these differences have p-values well below 0.001. The taxonomic findings at the genus level are shown in Figure 3, and the top 50 OTUs from the transplanted mice are presented in Table S1. Human microbiota had a greater diversity of genera represented as compared to the mice, as indicated both by the chao1 test of richness (929 versus 340, p < 0.001) and the Shannon test of evenness (6.5 versus 4.4, p < 0.001), and as supported by visual inspection of the graphs. Among the murine recipients, the only two genera present at > 5% of the total bacterial abundance were Akkermansia and Bacteroides. Thus, there was incomplete uptake of the transplanted microbiota.
Figure 3.
Taxonomy of human donors (n = 24; A) and recipient mice (n = 24; B). Genera with frequency > 4% are shown. Akkermansia is shown in dark blue.
Association of transplanted microbiota with severity of arthritis
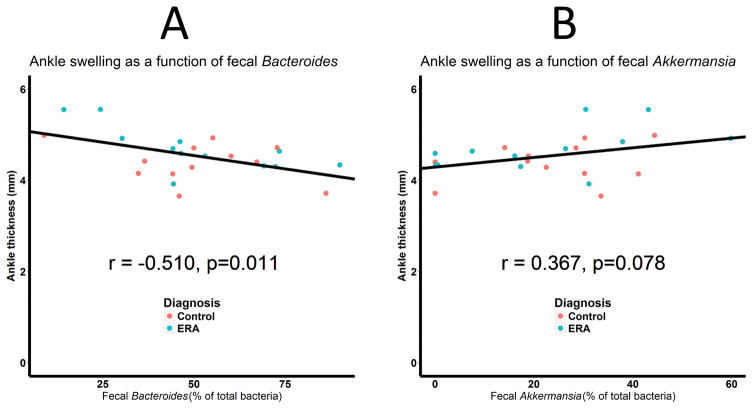
Although the donor’s diagnosis did not predict severity of arthritis (Figure 1), it was nevertheless possible that the makeup of the microbiota among transplanted mice would. Indeed, the Adonis test demonstrated that the contents of the microbiota overall were associated with ankle thickness (R2 = 0.185, p = 0.018). To determine which organisms were driving this association, we measured the correlation between ankle thickness and the two predominant genera, Bacteroides and Akkermansia. As shown in Figure 4, abundance of Bacteroides was inversely associated with ankle thickness (r = −0.510, p = 0.011; Figure 4A), while that of Akkermansia was directly associated with ankle thickness (r = 0.367, p = 0.078; Figure 4B), albeit slightly outside the statistically significant cutoff of 0.05.
Figure 4.
Ankle thickness as a function of the fecal abundance of the Bacteroides (A) and Akkermansia (B) genera. N = 24 for both figures. Microbiota from ERA patients and controls is colored blue and red, respectively.
Monocolonization of Bacteroides fragilis and Akkermansia muciniphila
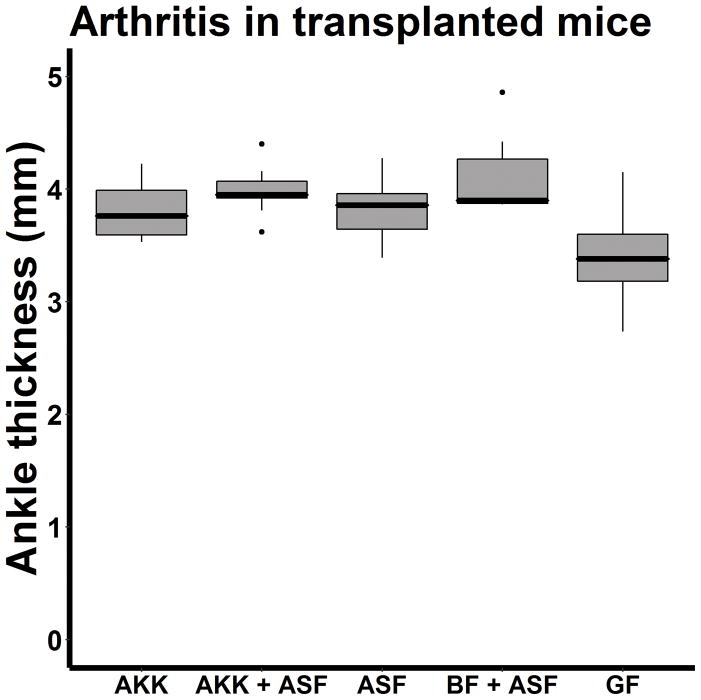
These findings raised the possibility that either Akkermansia contributed towards arthritis, one or more members of the Bacteroides genus were protective, or both. As the abundance of Bacteroides and Akkermansia were inversely associated with each other (r = −0.774, p < 0.001), it was not possible to use statistical testing to choose among these possibilities. Instead, monocolonization of Bacteroides and Akkermansia species was performed. The only known species within the Akkermansia genus is A. muciniphila, so this organism was selected. In contrast, there are multiple species within the Bacteroides genus, and the 16S sequence data was unable to distinguish among them. Bacteroides fragilis was selected for the monocolonization studies, as its polysaccharide tail can induce regulatory T cells [8]. Altered Schaedler’s flora (ASF), a consortium of 8 murine bacterial species that provide gnotobiotic mice with a standardized intestinal microbiota [9, 10], was used as a control for these studies, and was also co-inoculated in most of these experiments. Sequencing of the fecal specimens indicated that the monocolonizations with and without ASF were technically successful (Table 2, S2.) As shown in Figure 5, the extent of ankle swelling among mice transplanted with A. muciniphila with (4.0 mm, 3.9 – 4.1) or without (3.6 mm; IQR 3.8 – 4.1) added ASF, B. fragilis (3.9 mm, IQR 3.9 – 4.4), and ASF alone (3.9 mm, IQR 3.6 – 4.0) were all similar to each other, higher than that observed in GF mice (3.4 mm, IQR 3.1 – 3.6). Despite small differences, the difference in median ankle thickness between mice receiving A. muciniphila + ASF versus those receiving ASF alone was statistically significant (p = 0.041). The abundance of no single species was positively or negatively associated with the extent of arthritis. None of the mice monocolonized with B. fragilis survived the procedure, but there was no indication that B. fragilis when added to ASF was protective. No differences were observed in antibodies against GPI between the mice treated with ASF + A. muciniphila versus those receiving ASF along (median titer 1:40960, IQR 1:25600 – 1:40960 versus 1:40960, IQR 1:10240 – 1:71680, p = 0.905.)
Table 2.
Results of monocolonization studies with mouse microbiota.
| Inoculated bacteria | n* | Abundance of Akkermansia (%) | Abundance of Bacteroides (%) | Abundance of ASF (%) |
|---|---|---|---|---|
| AM only | 4 | 88 ± 19 | 0 | 0 |
| ASF + AM | 17 | 56 ± 25 | 0 | 30 ± 25 |
| BF + ASF | 6 | 0 | 11 ± 6.4 | 54 ± 34 |
| ASF alone | 14 | 0 | 0 | 88 ± 19 |
Abbreviations: AM = Akkermansia Muciniphila, ASF = Altered Schaedler’s flora. BF = Bacteroides fragilis.
Due to low-quality samples, sequence data is only available on 14 / 16 of the ASF alone samples and 6/7 of the BF + ASF samples.
Figure 5.
Boxplot of ankle thickness in mice transplanted with A. muciniphila alone (AKK; n = 4), A. muciniphila + Altered Schaedler’s flora (AKK / ASF; n = 16), ASF alone (n = 17), B. fragilis + ASF (BF + ASF; n = 7); or germ-free (GF; n = 23) mice..
Addition of bacterial isolates to human fecal specimens
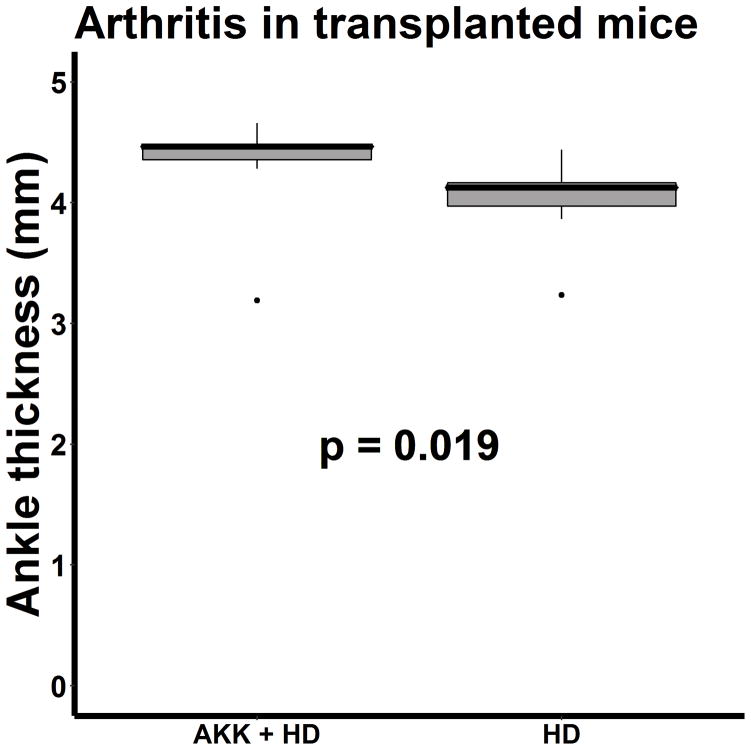
These findings additionally raised the possibility either that bacteria may act in concert in promoting or protecting against disease, or that the immune system of mice colonized with a limited set of bacteria might not be fully developed. These possibilities were further assessed by transplanting GF mice with feces from a single non-arthritic human subject, selected on the basis of low abundance of A. muciniphila (none), with some of the mice receiving supplemental inoculations with A. muciniphila. The technical success of the fecal transplants is shown in Table 3. One of the 9 control mice had a high fecal abundance of A. muciniphila, despite sharing a cage with mice that had no abundance. Supplementation of human feces with A. muciniphila resulted in a small but statistically significant increase in median ankle swelling (4.5 mm, IQR 4.3 – 5.5) compared to that seen in mice that only received human feces (4.1 mm, IQR 3.9 – 4.3, p = 0.019; Figure 6). Overall, there was no association between the fecal abundance of A. muciniphila and ankle swelling (r = 0.325, p = 0.204). However, our previous work indicated that there may be a threshold effect with A. muciniphila, with the phenotype defined by the presence versus absence of abundance ≥ 2% [1], and those mice with an abundance of A. muciniphila ≥ 2% did indeed demonstrate increased ankle thickness as compared to the mice with abundance < 2% (4.5 mm, IQR 4.3 – 4.5, versus 4.1 mm, IQR 3.9 – 4.3, p = 0.036. These differences were not reflected with the ELISA data, which did not identify any differences between the mice receiving human microbiota with or without A. muciniphila (median titer 1:40960, IQR 1:8320 – 1:163840 versus 1:10240, IQR 1:6400 – 1:163840, p = 0.436.) Exclusion of the one control mouse with detectable A. muciniphila did not alter the results: the ankle thickness remained higher in the mice receiving supplemental A. muciniphila (4.5 mm, IQR 4.3 – 4.5 versus 4.1 mm, IQR 3.9 – 4.3, p = 0.027), while the GPI titers remained similar (1:40960, IQR 1:8320 – 1:163840 versus 1:10240, IQR 1:4480 – 1:126560, p = 0.277). Overall, the data with the human microbiota confirm our previous findings that A. muciniphila can promote arthritis in the context of an intact microbiota.
Table 3.
Results of monocolonization studies with human microbiota
| Inoculated bacteria | n | Abundance of A. Muciniphila (%) |
|---|---|---|
| Human microbiota only | 9 | 8.6 ± 4.8 |
| Human microbiota + A. muciniphila | 9 | 2.0 ± 0.1 |
Due to low-quality samples, sequence data is only available on 8 / 9 of the stool samples from mice receiving supplemental A. muciniphila.
Figure 6.
Boxplot of ankle thickness in mice receiving feces from a healthy human donor (HD) with (n = 9) or without (n = 9) supplementation with A. muciniphila.
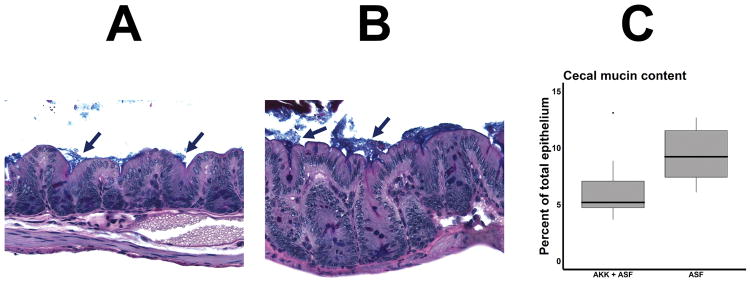
To evaluate a potential mechanism by which A. muciniphila might be permissive towards arthritis, cecal sections were stained for mucin deposition in mice transplanted with A. muciniphila plus ASF versus ASF alone, using Alcian Blue pH 2.5 + PAS to stain the mucin, and measuring the extent of the fraction of the total epithelium that was stained with mucin. Not unexpectedly, the mice colonized with A. muciniphila + ASF had median of 7.1% (IQR 5.2 – 11) mucin coverage as compared to 9.5% (7.1 – 12), p = 0.013 in the mice inoculated with ASF (Figure 7.) There was no association between mucin coverage and arthritis in these mice (r = −0.002), with a weak and non-significant association between abundance of A. muciniphila and mucin content (r = −0.374, p = 0.115.)
Figure 7.
Mucin thickness of transplanted mice. Cecal sections were stained with the Alcian Blue pH 2.5 + Periodic acid–Schiff (PAS) stain, and the area of mucus and epithelium were determined by manual tracing using Image Pro Plus v7.0. 7A and 7B depict representative sections of mice receiving ASF with (7A) or without (7B) supplemental A. muciniphila. Mucin stain appears as a blue layer (arrows) overlying the epithelium. A Boxplot summarizing the results is shown in 7C (n = 10 for A. muciniphila + ASF, n = 9 for ASF alone.)
Discussion
This study provides additional evidence linking the human intestinal microbiota with the pathogenesis of arthritis. We did not demonstrate differences in the extent of arthritis between mice colonized with patient versus control microbiota, findings that could be attributed to the failure of human-mouse FMT to recapitulate the precise microbial contents of the human donors, as observed herein (Figure 3) and previously [11, 12]. Our prior work demonstrated that a subset of children with ERA demonstrated a high intestinal abundance of A. muciniphila [1]. This study was subsequently corroborated by work in the HLA-B27 animal model of SpA, which revealed an expansion of A. muciniphila early in the disease process [13]. Data opposing a pathogenic role of A. muciniphila in arthritis was published by Scher et al., which showed that adult patients with psoriatic arthritis had decreased intestinal abundance of A muciniphila as compared to healthy adult controls [4].
Thus, the precise role of A. muciniphila in arthritis remains to be elucidated. The data in this study demonstrated that it was sufficient to cause arthritis in animals, but that it could do so only in the context of an intact microbiota. The mechanism by which A. muciniphila may contribute to arthritis may pertain to its ability to degrade intestinal mucins; indeed, it was given its name in 2004 due to its ability to thrive on intestinal mucins [5]. Intestinal mucins serve a barrier function between intestinal epithelial cells and luminal bacteria [14]. This barrier helps maintain a tolerogenic state between the mucosal immune system and the intestinal microbiota, while invasive bacteria typically engender an inflammatory immune response (reviewed by [15].) Thus, our data support the possibility that abundance of A. muciniphila early in the disease process may permit some other species of bacteria to become invasive, with this other strain triggering mucosal autoimmunity. This possibility was suggested as well by a study of infection with Salmonella Typhimurium, in which co-inoculation of A. muciniphila and S. Typhimurium resulted in increased intestinal damage and S. typhimurium invasiveness, compared to mice that did not receive A. muciniphila [16]; as with our findings, there was also increased mucin loss in mice receiving A. muciniphila. The decreased abundance of A. muciniphila previously reported in adults with psoriatic arthritis [4], as well as in studies of patients with IBD (e.g. [17, 18]), may reflect declines in the population of this species over time due to loss of its primary substrate, intestinal mucin, as previously suggested [19]. The findings of decreased intestinal mucin content reported herein are consistent with reports in children with JIA [20] as well as in adults with ankylosing spondylitis [21] of increased intestinal permeability. Although we did not see an association between cecal mucin content and arthritis among mice transplanted with ASF, other mechanisms may have been involved, or other regions of the intestine may need to be evaluated for mucin content. Notably, all interventions performed in this study, including ASF alone, resulted in increased ankle thickness compared to that observed in GF mice (Figures 1, 5). This likely reflects non-specific effects on host immunity, as even single organisms can impact the development of the murine immune system [22]. However, even in this context, the presence of A. muciniphila with other organisms resulted in increased arthritis as compared to ASF or human microbiota without A. muciniphila.
This study has limitations. Although the subjects had ERA, the animal model used in the study is not a model of SpA. Nevertheless, the mechanism by which A. muciniphila may predispose to arthritis that was demonstrated in this study may likely apply to human subjects, and the previously published findings of decreased intestinal permeability in children with JIA were not limited to those with SpA [20]. In addition, the absolute differences between the groups were small, albeit statistically significant, and the ELISA findings were minimal. Nevertheless, the findings were internally consistent and corroborate our previous data implicating A. muciniphila in the pathogenesis of arthritis [1], indicating that more research on the role of this organism is warranted. Finally, although the mice that received human microbiota had a more complex microbiota than that obtained from monocolonizations alone, the transplants did not fully recapitulate the contents of the human intestinal microbiota.
In summary, our data has provided further evidence implicating A. muciniphila in the pathogenesis of arthritis. Additional work may focus on the nature of the barrier layer in children with arthritis and means of protecting it from further damage.
Materials and Methods
Animals
KRN/B6 and NOD mice were obtained from Jackson labs (Bar Harbor, ME); the KRN/B6 mice were obtained with the kind permission of Dr. Christophe Benoist. Derivation into the germ-free state was performed separately on both parental strains using SWR foster mothers following C-Section delivery of the offspring, and the mice were maintained in the gnotobiotic facility in isolators. Gastric gavage of feces or pure cultures of bacteria (Akkermansia muciniphila BAA-835 (ATCC) or Bacteroides fragilis strain 086-5443-2-2 (kindly donated by Cindy Sears) was performed using standard mouse gavage technique, with manual restraint, using sterile 20 ga x 30 mm disposable plastic rodent feeding tubes (Instech FTP-20-30-50, Plymouth Meeting, PA), and 1 ml sterile disposable syringes. For the monocolonizations, the cultures were grown to visual turbidity. 0.2 ml of each fecal or pure bacterial suspension was administered. Following gavage, each mouse was maintained in its own isolator to prevent horizontal transfer of microbiota. Inoculation of ASF was performed via co-housing of GF mice with mice colonized exclusively with ASF, permitting horizontal transfer of ASF to the previously GF mice. Ankle thickness was measured with a Mitituyo thickness gager (Middleburg Heights, OH), which is an accepted measure of disease activity in this model; assessments were performed by one investigator (MLS) [7]. Fecal transplants were generally performed at age 2 months, with the mice evaluated approximately 3 weeks thereafter; male and female mice were used interchangeably. All experiments were conducted under protocols approved by the UAB IACUC animal-review committee.
Patients
Patients were treatment-naïve children with the ERA category of JIA with active peripheral or axial arthritis [23]. Controls were children either recruited from the community (n = 9) or were derived from children referred to rheumatology to evaluate for arthritis but found to have non-inflammatory causes of joint pain or irrelevant laboratory markers (n = 3.) Each control was matched to a patient by sex, race, site, and age ± 1 year. Subjects with recent (6-month) exposure to antibiotics were excluded. Patients and controls were recruited at three sites: Children’s Hospital of Alabama / University of Alabama at Birmingham (UAB), The Children’s Hospital of Philadelphia (CHOP), and Hackensack University Medical Center (HUMC.) These studies were approved by the IRB at UAB (coordinating center), as well as at CHOP and HUMC. Written consent was obtained from the guardians of all subjects, and assent and consent were obtained from the subjects themselves, as appropriate.
Collection of fecal specimens
Fecal specimens were placed immediately into Cary-Blair media, which permits transport of live bacteria [24] and shipped by commercial carrier overnight to UAB. The feces were frozen in 10% glycerol as a cryopreservative at −80°C until transfer.
ELISAs
Antibodies directed against GPI, the hallmark antibody of the model, were measured as described [25]. E. coli containing plasmid expressing mouse GPI as a fusion protein was obtained as a kind gift from Dr. Altan Ercan from Harvard University, with GP-GST obtained via glutathione sepharose column purification per protocol [25]. Product was dialyzed 3x with PBS. EIA/RIA 96 well high binding plates (Costar 3590) plates (Fisher, Pittsburgh, PA) were coated with recombinant mouse GPI at 5 ug/ml overnight at 4° C. Plates were blocked with BSA (1%) for one hour. After blocking, diluted mouse serum was added in at serial four-fold dilutions starting at 1:160, for one hour at room temperature (RT). Subsequently, alkaline-phosphatase (AP)-conjugated anti-mouse IgG (Jackson ImmunoResearch; West Grove, PA) was added at 1:5000 for one hour at RT, followed by AP substrate (Sigma-Aldrich, St. Louis, MO). After substrate development, the reaction was stopped with the addition of 3N NaOH. The antibody titer was obtained from a Bio-Tek Synergy HT ELISA reader (Winoosky, VT). NOD serum was used as a negative control.
Mucin staining
Cecal tissue was removed at time of euthanasia and immediately placed in Carnoy’s solution for four hours. Material was then removed and trimmed into transversed sections and placed in 80% ethanol. Sections were stained with the Alcian Blue pH 2.5 + Periodic acid–Schiff (PAS) stain. Color images were made using a 40x objective lens. For each image, the area of mucus on the mucosal surface and the area of mucosal epithelium were determined by manual tracing using Image Pro Plus v7.0. Results were expressed as the ratio of the area of mucus to the area of epithelium, and values for individual images were averaged for each mouse. Assessments were made by one investigator blinded to treatment (TRS).
Sequencing of 16S ribosomal DNA
This was done as previously described [1]. Briefly, fecal DNA was purified with the Zymo Research kit (Irvine, CA), and the V4 16S rDNA region was amplified and sequenced on the MiSeq apparatus (Illumina, San Diego, CA). Following quality filtering, generation of operational taxonomic units and assessments of alpha and beta diversity were performed with the quantitative insight into microbial ecology (QIIME) pipeline [26].
Statistical analysis
Assessments of association between metadata and the distance matrix generated by the QIIME beta_diversity.py script were evaluated with the Permanova test for dichotomous variables and the Adnois test for continuous variables, using the QIIME compare_categories.py script. Continuous variables such as comparisons of ankle swelling and GPI titers, were performed with the Mann-Whitney U-test, and correlation analysis was performed with the Pearson test. All analyses were conducted with SPSS (Chicago, IL) Version 22.
Supplementary Material
Figure S1. Principal coordinates of transplanted (n = 24; blue), GF (n = 23; green), and SPF (n = 11; orange) mice, as well as human donors (n = 24; red.) Also included are microbiota obtained from food pellets used in the GF facility (n = 3; yellow) and that obtained from blank tubes (n = 3; purple.)
Table S1. Top 50 OTUs among murine recipients of human fecal material (Excel file).
Table S2. Abundance of genera corresponding to ASF, Akkermansia muciniphila, and Bacteroides fragilis in mice receiving monocolonizations with or without ASF (Excel file).
Acknowledgments
This work was supported by funding from the NIH / NIEHS (1R21ES024413-01; PI Dr. Stoll), NIH / NIAMS (P60 AR064172), the American College of Rheumatology (PI Dr. Stoll), and the Childhood Arthritis Rheumatology Research Alliance (PI Dr. Stoll.)
The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: Comprehensive Cancer Center (P30AR050948), Center for Clinical Translational Science (UL1TR001417), University Wide Institutional Core and Heflin Center for Genomic Sciences.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare.
Supplemental information is available on Genes and Immunity’s website.
References
- 1.Stoll ML, Kumar R, Morrow CD, Lefkowitz EJ, Cui X, Genin A, et al. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis Res Ther. 2014;16(6):486. doi: 10.1186/s13075-014-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tejesvi MV, Arvonen M, Kangas SM, Keskitalo PL, Pirttila AM, Karttunen TJ, et al. Faecal microbiome in new-onset juvenile idiopathic arthritis. Eur J Clin Microbiol Infect Dis. 2016;35(3):363–70. doi: 10.1007/s10096-015-2548-x. [DOI] [PubMed] [Google Scholar]
- 3.Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016 doi: 10.1002/art.39783. [DOI] [PubMed] [Google Scholar]
- 4.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67(1):128–39. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(Pt 5):1469–76. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 6.Stoll ML, Cron RQ. The microbiota in pediatric rheumatic disease: epiphenomenon or therapeutic target? Curr Opin Rheumatol. 2016 doi: 10.1097/BOR.0000000000000312. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu HJ, Ivanov, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–7. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65(8):3287–92. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes-Neto JC, Mantz S, Held K, Sinha R, Segura Munoz RR, Schmaltz R, et al. A real-time PCR assay for accurate quantification of the individual members of the Altered Schaedler Flora microbiota in gnotobiotic mice. J Microbiol Methods. 2017;135:52–62. doi: 10.1016/j.mimet.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar R, Maynard CL, Eipers P, Goldsmith KT, Ptacek T, Grubbs JA, et al. Colonization potential to reconstitute a microbe community in patients detected early after fecal microbe transplant for recurrent C. difficile. BMC Microbiol. 2016;16:5. doi: 10.1186/s12866-015-0622-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arrieta MC, Walter J, Finlay BB. Human Microbiota-Associated Mice: A Model with Challenges. Cell Host Microbe. 2016;19(5):575–8. doi: 10.1016/j.chom.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Asquith MJ, Stauffer P, Davin S, Mitchell C, Lin P, Rosenbaum JT. Perturbed Mucosal Immunity and Dysbiosis Accompany Clinical Disease in a Rat Model of Spondyloarthritis. Arthritis Rheumatol. 2016;68(9):2151–62. doi: 10.1002/art.39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson ME. Mucus layers in inflammatory bowel disease. Inflamm Bowel Dis. 2014;20(11):2124–31. doi: 10.1097/MIB.0000000000000117. [DOI] [PubMed] [Google Scholar]
- 15.Grigg JB, Sonnenberg GF. Host-Microbiota Interactions Shape Local and Systemic Inflammatory Diseases. J Immunol. 2017;198(2):564–571. doi: 10.4049/jimmunol.1601621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One. 2013;8(9):e74963. doi: 10.1371/journal.pone.0074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michail S, Durbin M, Turner D, Griffiths AM, Mack DR, Hyams J, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. 2012;18(10):1799–808. doi: 10.1002/ibd.22860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7(6):e39242. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah R, Cope JL, Nagy-Szakal D, Dowd S, Versalovic J, Hollister EB, et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes. 2016;7(5):384–96. doi: 10.1080/19490976.2016.1190073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picco P, Gattorno M, Marchese N, Vignola S, Sormani MP, Barabino A, et al. Increased gut permeability in juvenile chronic arthritides. A multivariate analysis of the diagnostic parameters. Clin Exp Rheumatol. 2000;18(6):773–8. [PubMed] [Google Scholar]
- 21.Martinez-Gonzalez O, Cantero-Hinojosa J, Paule-Sastre P, Gomez-Magan JC, Salvatierra-Rios D. Intestinal permeability in patients with ankylosing spondylitis and their healthy relatives. Br J Rheumatol. 1994;33(7):644–7. doi: 10.1093/rheumatology/33.7.644. [DOI] [PubMed] [Google Scholar]
- 22.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6(220):220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31(2):390–2. [PubMed] [Google Scholar]
- 24.Cary SG, Blair EB. New Transport Medium for Shipment of Clinical Specimens. I. Fecal Specimens. J Bacteriol. 1964;88:96–8. doi: 10.1128/jb.88.1.96-98.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monach PA, Mathis D, Benoist C. The K/BxN arthritis model. Curr Protoc Immunol. 2008;Chapter 15(Unit 15):22. doi: 10.1002/0471142735.im1522s81. [DOI] [PubMed] [Google Scholar]
- 26.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Principal coordinates of transplanted (n = 24; blue), GF (n = 23; green), and SPF (n = 11; orange) mice, as well as human donors (n = 24; red.) Also included are microbiota obtained from food pellets used in the GF facility (n = 3; yellow) and that obtained from blank tubes (n = 3; purple.)
Table S1. Top 50 OTUs among murine recipients of human fecal material (Excel file).
Table S2. Abundance of genera corresponding to ASF, Akkermansia muciniphila, and Bacteroides fragilis in mice receiving monocolonizations with or without ASF (Excel file).