Abstract
Tooth morphogenesis is initiated by reciprocal interactions between the ectoderm and neural crest–derived mesenchyme. During tooth development, tooth cusps are regulated by precise control of proliferation of cell clusters, termed enamel knots, that are present among dental epithelial cells. The interaction of ectodysplasin-A (EDA) with its receptor, EDAR, plays a critical role in cusp formation by these enamel knots, and mutations of these genes is a cause of ectodermal dysplasia. It has also been reported that deficiency in Nkx2-3, encoding a member of the NK2 homeobox family of transcription factors, leads to cusp absence in affected teeth. However, the molecular role of NKX2-3 in tooth morphogenesis is not clearly understood. Using gene microarray analysis in mouse embryos, we found that Nkx2-3 is highly expressed during tooth development and increased during the tooth morphogenesis, especially during cusp formation. We also demonstrate that NKX2-3 is a target molecule of EDA and critical for expression of the cell cycle regulator p21 in the enamel knot. Moreover, NKX2-3 activated the bone morphogenetic protein (BMP) signaling pathway by up-regulating expression levels of Bmp2 and Bmpr2 in dental epithelium and decreased the expression of the dental epithelial stem cell marker SRY box 2 (SOX2). Together, our results indicate that EDA/NKX2-3 signaling is essential for enamel knot formation during tooth morphogenesis in mice.
Keywords: tooth, development, cell proliferation, tissue-specific transcription factor, transcription regulation, cell differentiation, dental growth, Nkx2–3, odontogenesis
Introduction
Tooth morphogenesis is initiated by reciprocal interactions between the ectoderm and neural crest–derived mesenchyme (1, 2). A good model for understanding the mechanism of ectodermal organ development is tooth development, which features well-defined developmental stages and distinctive cell types. In mice, tooth morphogenesis is initiated by thickening of dental epithelium to form a dental placode, followed by invagination into the surrounding mesenchyme. Thereafter, tooth buds progress into the cap stage, and primary enamel knots are formed in dental epithelium, leading to tooth cusps. In the bell stage, a secondary enamel knot forms at the tip of the cusp-forming area to pattern the cusp and shape the tooth crown (3, 4). In mice, only a single enamel knot is formed in incisors, whereas both primary and secondary enamel knots are formed in molars. An enamel knot is formed by a cluster of cells in dental epithelium and serves as a key signaling center that secretes sonic hedgehog (SHH), 4 bone morphogenetic protein (BMP), fibroblast growth factor (FGF), and members of the MMTV integration site (WNT) family, which are essential for forming the tooth crown shape and related cusps (4–6). These signals are also essential for formation of the apical ectodermal ridge involved in limb development (7, 8), indicating that organ formation is regulated by a common signaling pathway. During tooth growth, the mechanisms of differentiation during the later stages of dental epithelial cell development have been well defined (9–12), whereas regulation and cell fate determination in the early stage of tooth morphogenesis are largely unknown.
Tabby and downless mutant mice have tooth, hair, and sweat gland defects (13). The tabby gene encodes ectodysplasin-A (EDA), a type II membrane protein of the tumor necrosis factor ligand family, whereas the downless gene encodes EDAR, a novel member of the tumor necrosis factor receptor family (14). Edar is expressed in the enamel knot, and mutations of Edar and its ligand the Eda gene lead to ectodermal dysplasia, which includes missing teeth and smaller teeth with reduced cusps, as shown in mutant mice (13, 15). During this process, interactions between EDA and EDAR play a critical role in cusp formation. However, activity downstream of EDA signaling remains unclear.
NK2 homeobox families are tissue-specific transcription factors that feature a common DNA-binding structure and function as homeodomain genes, which critically regulate organ development of evolutionarily distant organisms ranging from the Drosophila genus to humans (16, 17). NKX2-3 is expressed in the pharyngeal floor, as well as oral and branchial arch ectoderm tissues, whereas findings of Nkx2-3 knockout mice have revealed defects in maturation and cellular organization of the salivary glands, as well as largely absent molar cusps (18). Nevertheless, the role of NKX2-3 in tooth development remains largely unknown.
In the present study, we found a high level of Nkx2-3 expression in teeth obtained from embryonic day 14 (E14) mouse embryos using microarray analysis. Subsequently, the expression pattern of NKX2-3 was examined and found localized in the area of the enamel knot, which is closely related to cusp formation. Our findings also showed that Nkx2-3 is induced by EDA and regulates p21 transcription and expression by binding with its promoter region, thus altering cell proliferation. Finally, NKX2-3 induced enhancement of BMP signaling and inhibition of SOX2 expression. These results indicate important roles for NKX2-3 in cusp formation and tooth morphogenesis.
Results
Nkx2-3 highly expressed during tooth morphogenesis
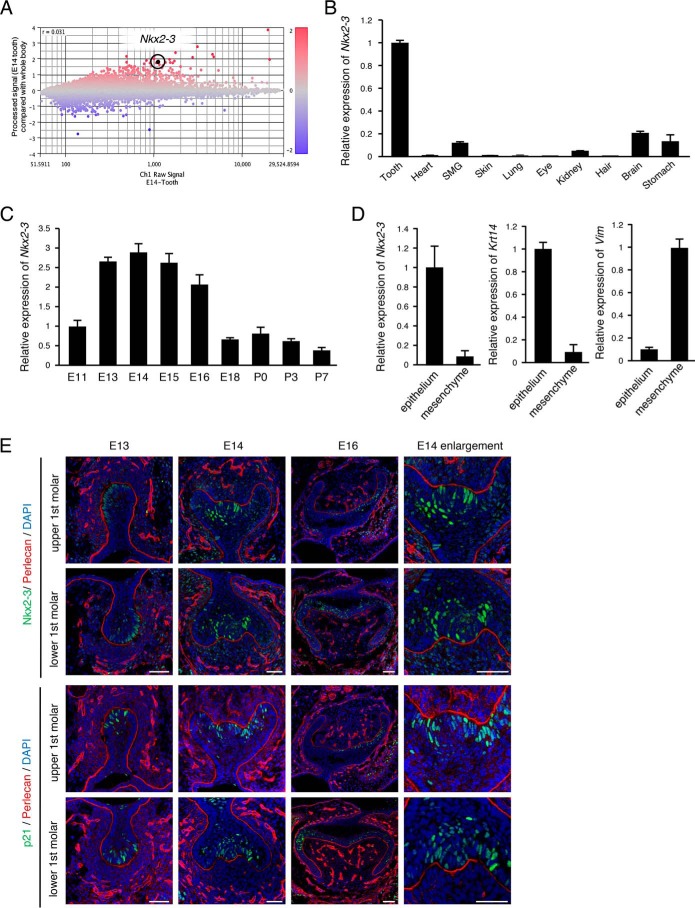
Microarray analysis was performed to identify expression of tooth-specific genes in the tooth morphogenesis stage. Gene expressions in teeth were compared with those in the whole body of mice on E14. Scatter plot data revealed that a large number of genes were either up- or down-regulated in E14 teeth (Fig. 1A). Furthermore, the expression of Nkx2-3 was at a high level in teeth relative to the whole body, which may indicate its important roles during tooth development. To confirm the expression pattern of Nkx2-3 during tooth development, qRT-PCR was performed using total RNA obtained from tooth, heart, submandibular gland, skin, lung, eye, kidney, hair, brain, and stomach samples, as well as during each stage of tooth development (i.e. E11–E18, as well as postnatal day 0 (P0) to P7). Nkx2-3 expression was found to be elevated in teeth as compared with the other examined organs (Fig. 1B) and increased in accordance with tooth morphogenesis stage (E13–E16) (Fig. 1C), indicating that this gene has critical roles in tooth morphogenesis. To confirm whether Nkx2-3 is expressed in dental epithelium or mesenchyme, E14 dental epithelium was separated from mesenchyme under a microscope after treatment with dispase, and expression levels were determined by qRT-PCR. The dental epithelium-specific gene Krt14 was shown to be highly expressed in epithelium, whereas Vim, which encodes the mesenchyme-specific protein vimentin, was highly expressed in mesenchyme, and Nkx2-3 was mainly expressed in dental epithelium (Fig. 1D). Other members of the NK2 family showed different expression patterns as compared with the tooth specificity of Nkx2-3 (Fig. 1B and Fig. S1), indicating that the NK2 family is composed of tissue-specific transcription factors, of which NKX2-3 may play an important role in tooth development. To examine the localization of NKX2-3 during tooth development, we performed immunohistochemistry using first molars obtained from embryo heads on E13, E14, and E16. NKX2-3 expression was shown to begin from the bud stage (E13) and continue in the cap stage (E14), mainly in the primary enamel knot area (Fig. 1E, enlarged right panel). In the bell stage (E16), its expression appeared in the second enamel knot, which is consistent with the enamel knot marker p21 (Fig. 1E, bottom panel). Furthermore, the basement membrane molecule perlecan was stained with NKX2-3 to reveal the border of epithelial and mesenchymal tissues (Fig. 1E). These results indicate that NKX2-3 is highly expressed in dental epithelial cells and localized in enamel knot areas.
Figure 1.
Expression of NKX2-3 and p21 during tooth morphogenesis. A, in samples obtained on E14, differentially expressed genes were identified by microarray analysis of teeth, and the results were compared with whole embryos. Highlighted plot, Nkx2-3. Red and blue plots, up- and down-regulated genes, respectively. B, qRT-PCR analysis of Nkx2-3 expression in tooth, heart, submandibular gland, skin, lung, eye, kidney, hair, brain, and stomach samples from E14 embryos after normalization to Gapdh mRNA expression. C, qRT-PCR analysis of Nkx2-3 expression in teeth obtained from E11 to P7 after normalization to Gapdh mRNA expression. D, qRT-PCR analysis of Nkx2-3, Krt14, and Vim expressions in tooth epithelium and mesenchyme. E, NKX2-3 (green) expression in E13, E14, and E16 mice, detected by immunocytochemistry. p21 (green) expression in E13, E14, and E16 mice, detected by immunocytochemistry. Perlecan (red) was used to detect the basement membrane. Nuclei were stained using DAPI (blue). Error bars, S.D. Scale bars, 50 μm.
Reduction of Nkx2-3 interrupts cusp formation of molars in an organ culture system
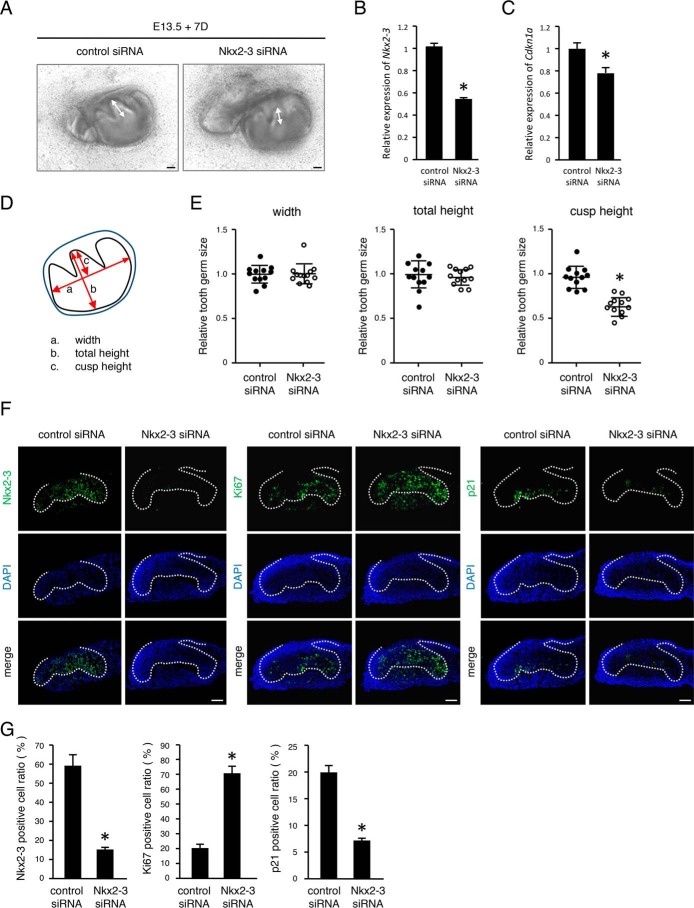
To examine the role of NKX2-3 in enamel knots of developing teeth, E13 mandibular tooth germs were dissected and then transfected with control or Nkx2-3 siRNA (Fig. 2A). Nkx2-3 was specifically down-regulated by 50% by Nkx2-3 siRNA in an ex vivo organ culture system (Fig. 2B). After 1 week of culture, tooth cusp sizes in both groups were measured (Fig. 2D), with no significant differences regarding width or total height between the groups (Fig. 2E). However, cusp height in the presence of Nkx2-3 siRNA was significantly reduced as compared with the control (Fig. 2, A and E). These results suggest that NKX2-3 is critical for molar cusp formation.
Figure 2.
Nkx2-3 siRNA interrupts molar cusp formation and increases dental epithelial cell proliferation. A, 7-day organ cultures of E13 tooth germs transfected with control or Nkx2-3 siRNA. B and C, expression levels of Nkx2-3 mRNA (B) and Cdkn1a mRNA (C) expression levels in 1-day organ cultures of representative E13 tooth germs. D, schematic representation of the method used for measuring cultured tooth germs. E, relative tooth size plot (n = 12), with the average tooth germ size in the control siRNA group set at 1.0. F, 2-day organ cultures of representative E13 tooth germs transfected with control or Nkx2-3 siRNA. NKX2-3, Ki67, and p21 expressions were detected by immunohistochemistry. G, method used for positive cell ratio calculation. Nuclei were stained with DAPI (blue). Broken lines, basement membrane of teeth. *, p < 0.05. Error bars, S.D. Scale bars, 50 μm.
Cusp formation is believed to be regulated by an enamel knot expressing the cyclin-dependent kinase inhibitor p21. To clarify dental epithelial cell proliferation in cultured molars, we examined Ki67, a marker for cell proliferation, as well as p21 using immunohistochemistry. The number of NKX2-3–positive cells was reduced in molars transfected with Nkx2-3 siRNA compared with the control, whereas Ki67-positive cells were increased in the presence of Nkx2-3 siRNA (Fig. 2, F and G). In contrast, p21 was down-regulated in the enamel knot area in Nkx2-3 siRNA–transfected molars (Fig. 2, F and G). In addition, qRT-PCR data revealed that the mRNA level of p21 (Cdkn1a) was also reduced (Fig. 2C), suggesting that NKX2-3 regulates p21 expression in dental epithelial cells. We considered that p21 disappearance in the enamel knot region may be a reason for the inhibition of cusp formation seen in Nkx2-3 siRNA–transfected molars.
Nkx2-3 inhibits cell proliferation in dental epithelial cells
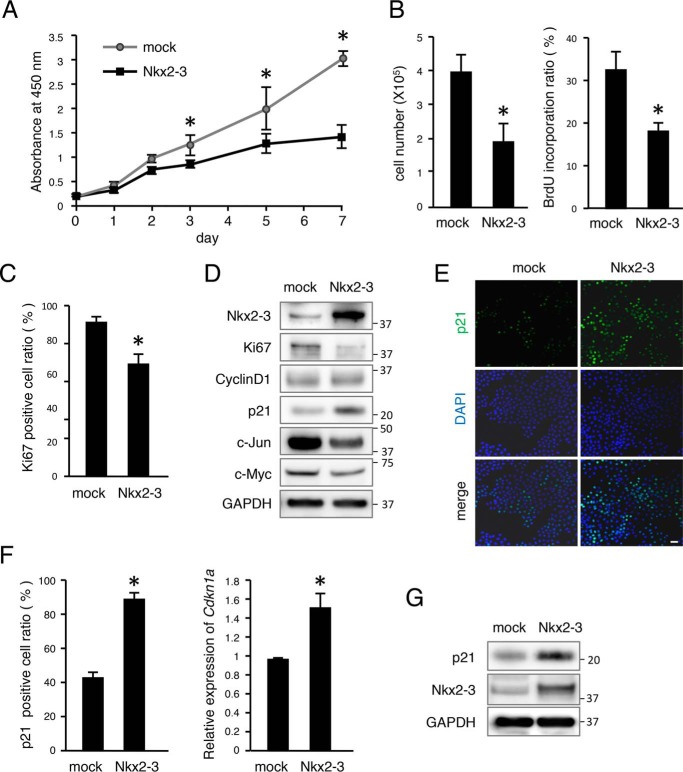
We evaluated cell proliferation using the M3H1 dental epithelial cell line previously established by our group (19). Proliferation was decreased in the Nkx2-3 transfected group as compared with the control (Fig. 3A). Cell count and bromodeoxyuridine (BrdU) incorporation results also showed decreased cell proliferation in the Nkx2-3 transfectant group (Fig. 3B). Furthermore, the ratio of Ki67-positive cells was also reduced (Fig. 3C), which was consistent with our results shown by organ culture immunostaining (Fig. 2, F and G). Together, these findings suggest that NKX2-3 is necessary for regulation of epithelial cell proliferation. We also examined expressions of the cell cycle molecule cyclin D1 and cell proliferation–related markers Ki67, p21, c-Myc, and c-Jun following transfection with Nkx2-3 by Western blot analysis. Ki67, c-Myc, and c-Jun were decreased by Nkx2-3 overexpression, whereas cyclin D1 showed no difference (Fig. 3D). Additionally, p21 was up-regulated in Nkx2-3–overexpressed cells, consistent with findings of the organ cultures. These results suggest that dental epithelial cell proliferation in vitro can be adjusted by NKX2-3.
Figure 3.
NKX2-3 decreased cell proliferation in vitro by inducing p21, a cell cycle regulator. A, cell proliferation was analyzed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay findings after transfection with mock or Nkx2-3 vector. B, the number of cells transfected with Nkx2-3 was determined after 48 h of culture. BrdU incorporation of M3H1 cells was determined after transfection with Nkx2-3 for 48 h, with the ratio calculated as BrdU-positive cells/DAPI-stained nuclei. C, ratio of Ki67-positive cells among M3H1 cells cultured with or without Nkx2-3, calculated as Ki67-positive cells/DAPI-stained nuclei. D, Western blotting results for Ki67, cyclin D1, p21, c-Jun, c-Myc, NKX2-3, and GAPDH in M3H1 cells transfected with mock vector or Nkx2-3 expression vector. GAPDH was used as the internal control. E, p21 (green) expression in M3H1 cells cultured in dishes with or without Nkx2-3 transfection. Nuclei were stained with DAPI (blue). F, ratio of p21-positive cells among M3H1 cells cultured with or without Nkx2-3 transfection, with the ratio calculated as p21-positive cells/DAPI-stained nuclei. qRT-PCR analysis of Cdkn1a expression in M3H1 cells cultured with or without Nkx2-3 transfection. G, Western blotting results of p21, NKX2-3, and GAPDH in M3H1 cells transfected with mock vector or Nkx2-3 expression vector. GAPDH was used as the internal control. *, p < 0.05. Error bars, S.D. Scale bars, 50 μm.
Nkx2-3 regulates cell cycle regulator p21 and binds to its promoter region in dental epithelial cells
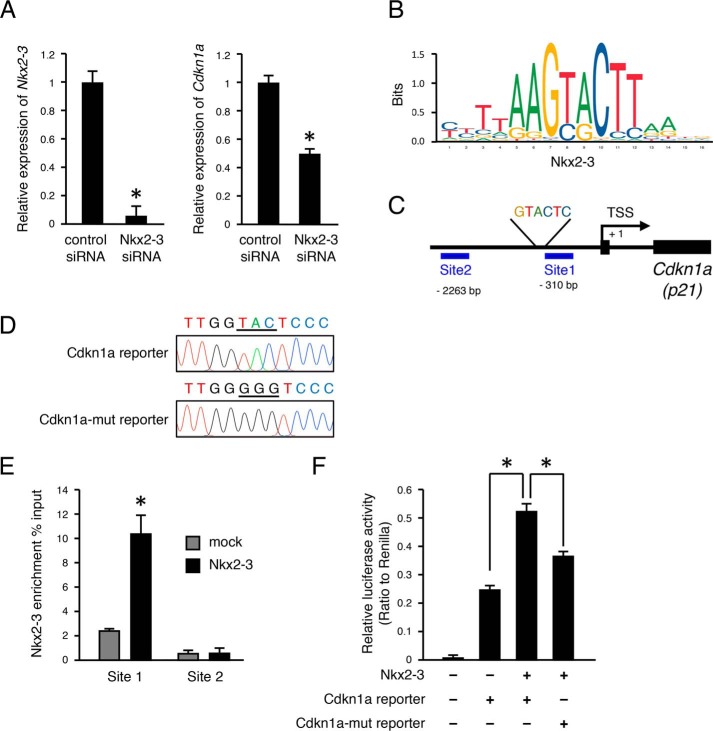
To confirm regulation of p21 expression by NKX2-3, we transfected an Nkx2-3-expressing vector into M3H1 cells and then evaluated the expression level of p21. Our results showed that p21-positive cells were increased at 48 h after transfection with Nkx2-3 (Fig. 3, E and F). qRT-PCR findings also showed that Cdkn1a mRNA expression was increased in the Nkx2-3-transfected group (Fig. 3F), whereas Western blotting revealed that NKX2-3 induced p21 at the protein level (Fig. 3G). These results suggest that NKX2-3 induces p21 expression in mRNA and at the protein level in vitro. The expression level of p21 was also evaluated by qRT-PCR in Nkx2-3 siRNA–transfected M3H1 cells, which showed that Nkx2-3 expression was significantly inhibited and that of Cdkn1a (p21) was also decreased (Fig. 4A). Next, we examined whether NKX2-3 can potentially bind to the promoter region of Cdkn1a using the JASPAR database and found a motif sequence (Fig. 4B). This screening revealed a GTACTC motif at 328 bp upstream of the transcription start site (TSS) of the Cdkna1 gene (Fig. 4C). To identify binding of NKX2-3 in the Cdkn1a promoter region, Nkx2-3–overexpressing M3H1 cells were fixed, and a ChIP assay was performed using antibodies against the V5 tag. DNA segments binding to NKX2-3 protein were detected by qRT-PCR using primer sequences at the proximal promoter region (site 1, bp −310 to −179) containing the consensus DNA-binding sequences as well as the distal region of the promoter (site 2, bp −2263 to −2130), used as a control (Fig. 4C). Expression of NKX2-3 associated with the site 1 region of the Cdkn1a promoter was ∼4.2-fold higher as compared with the mock, whereas it was not significantly different in the control site (Fig. 4E). To further confirm the target sequence of NKX2-3 in the Cdkn1a promoter, we constructed luciferase reporter vectors with insertion of the Cdkn1a promoter or a Cdkn1a promoter mutant by changing the nucleotide sequence GTACTC into GGGGTC (Fig. 4D). A Nkx2-3–expressing vector was transfected into M3H1 cells, and then promoter activities were evaluated using a luciferase reporter assay. Compared with the mock group, the Nkx2-3–overexpressed group had a 2-fold increase in transcriptional activity of the WT sequence Cdkn1a promoter, whereas the use of a triple-base mutation of the Cdkn1a promoter region resulted in a decrease of transcriptional activity as compared with the WT (Fig. 4F). These findings indicated that NKX2-3 directly binds to the promoter region of Cdkn1a and regulates its gene expression in dental epithelial cells.
Figure 4.
NKX2-3 binds to Cdkn1a promoter region. A, qRT-PCR analysis of Nkx2-3 expression in M3H1 cells cultured with control siRNA or Nkx2-3 siRNA transfection. qRT-PCR analysis of Cdkn1a expression in M3H1 cells cultured with control siRNA or Nkx2-3 siRNA transfection. B, NKX2-3 binding motif in mice from the JASPAR database. C, diagram of Cdkn1a promoter region. Genomic PCR was performed using primer sets for sites 1 (−310 to −179 bp) and 2 (−2263 to −2130 bp) in the Cdkn1a promoter. D, sequences of WT Cdkn1a promoter and mutant promoter region. E, ChIP analysis of NKX2-3 enrichment (percentage input of V5/original sample) of Cdkn1a promoter in M3H1 cells cultured with mock or Nkx2-3 transfection. F, Cdkn1a promoter reporter activities were evaluated in M3H1 cells transfected with the Nkx2-3 expression vector. *, p < 0.05. Error bars, S.D.
EDA/EDAR signaling regulates Nkx2-3 expression in dental epithelial cells
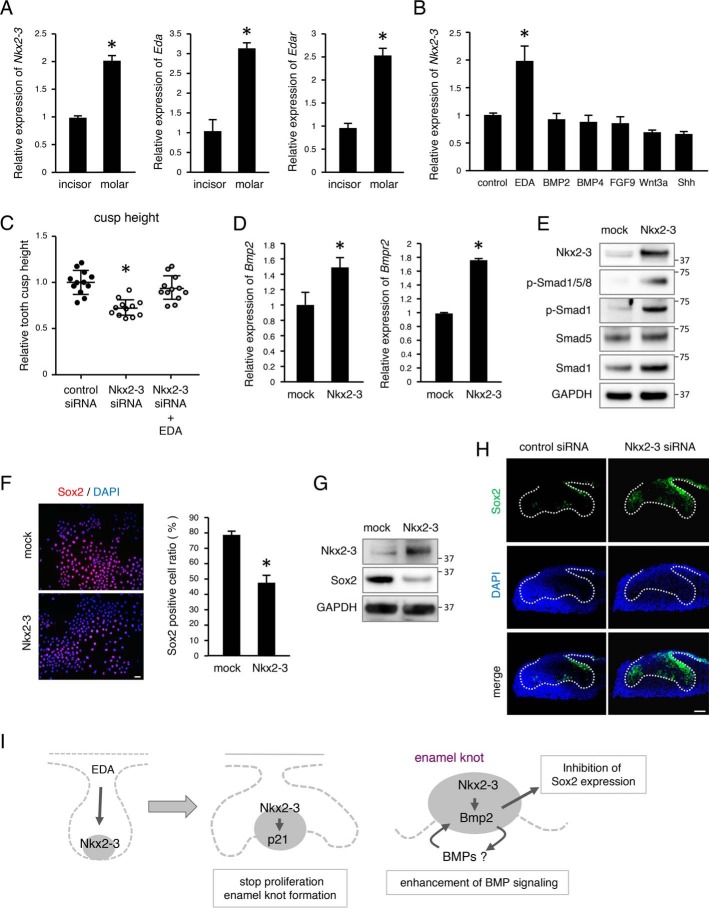
EDA/EDAR signaling is important for enamel knot formation and tooth morphogenesis. In mice, only a single enamel knot is formed in incisors, whereas both primary and secondary enamel knots are formed in molars. qRT-PCR results revealed that Nkx2-3 as well as Eda and Edar were expressed at higher levels in molars as compared with incisors (Fig. 5A), thus suggesting that NKX2-3 is a key regulator for enamel knot formation. To examine whether EDA/EDAR regulates Nkx2-3 expression, M3H1 cells were stimulated with Eda-a1 as well as BMPs, FGF9, Wnt3a, and SHH. Nkx2-3 was shown to be significantly induced by Eda-a1 (Fig. 5B). Furthermore, we added Eda-a1 to organ culture medium used for culturing teeth transfected with Nkx2-3 siRNA, which apparently abrogated the decrease in cusp height (Fig. 5C), suggesting that NKX2-3 is a target transcription factor regulated by the EDA/EDAR signaling pathway in the enamel knot (Fig. 5I). Other molecules, including BMPs, did not have effects on cusp formation in Nkx2-3 siRNA-treated organ cultures (data not shown).
Figure 5.
EDA/EDAR signaling regulates Nkx2-3 expression in dental epithelial cells. A, expressions of Nkx2-3, Eda, and Edar in mandibular incisors and molars of E14 embryos. B, qRT-PCR analysis of Nkx2-3 expression in M3H1 cells cultured with the addition of different growth factors. C, relative tooth cusp height plot (n = 12), with the average tooth germ size in the control siRNA group set at 1.0. D, qRT-PCR analysis of Bmp2 and Bmpr2 expressions in M3H1 cells cultured with mock or Nkx2-3 transfection. E, Western blotting results of p-Smad1/5/8, p-Smad1, Smad5, Smad1, NKX2-3, and GAPDH in M3H1 cells transfected with mock vector or Nkx2-3 expression vector. GAPDH was used as the internal control. F, SOX2 (red) expression in M3H1 cells cultured in dishes with or without Nkx2-3 transfection. Nuclei were stained with DAPI (blue). The ratio of SOX2-positive cells among M3H1 cells cultured with or without Nkx2-3 transfection was calculated as SOX2-positive cells/DAPI-stained nuclei. G, Western blotting results of SOX2, NKX2-3, and GAPDH in M3H1 cells transfected with mock vector or NKX2-3 expression vector. GAPDH was used as the internal control. H, 2-day organ cultures of representative E13 tooth germs transfected with control or Nkx2-3 siRNA. SOX2 (green) expression was detected by immunohistochemistry. I, schematic diagram showing roles of Nkx2-3 during tooth development. *, p < 0.05. Error bars, S.D. Scale bars, 50 μm.
BMP signaling is also an inducer of p21 and associated with epithelial–mesenchymal interaction in the enamel knot region (20). In an examination of the ability of BMPs to induce Nkx2-3, we found no change in expression level of Nkx2-3 in M3H1 cells after adding BMP2 or BMP4 (Fig. 5B). However, it was also noted that transfected Nkx2-3 induced Bmp2 and Bmpr2 mRNA expressions (Fig. 5D) as well as phosphorylation of Smad1/5/8 and Smad1 (Fig. 5E) in M3H1 cells. Thus, NKX2-3 may directly or indirectly regulate BMP signaling in the enamel knot (Fig. 5I).
Nkx2-3 decreases Sox2+ cells in the buccal side of dental epithelial cells
Recently, a cell lineage tracing method was used to show that mouse molar anterior buccal secondary enamel knots are derived from primary enamel knots, which were originally Sox2+ cells (21). To elucidate whether NKX2-3 can drive stem cells into differentiation, we examined expression of the dental epithelial stem cell marker SOX2 in vitro. We cultured M3H1 cells transfected with Nkx2-3 or a mock vector for 48 h and noted that the number of Sox2+ cells among the Nkx2-3–transfected cells was decreased (Fig. 5F). Western blot analysis also showed that SOX2 expression was decreased in Nkx2-3–overexpressed M3H1 cells (Fig. 5G). We also used an ex vivo organ culture system with or without Nkx2-3 siRNA, and found that Sox2+ cells were increased on the buccal side of the tooth germ and in the enamel knot in the Nkx2-3 siRNA–transfected group (Fig. 5H). Together, these results suggest that NKX2-3 contributes to cell fate determination of the enamel knot for cusp formation (Fig. 5I).
Discussion
Hypodontia, congenital absence of teeth, can occur without or with association with certain genetic syndromes. Several genes are known to be involved with isolated or nonsyndromic hypodontia, such as PAX9 (22), MSX1 (23), AXIN2 (24), and EDA (25), whereas other reports have described isolated tooth agenesis or microdontia in families with mutations of EDA (26–28) and EDAR (29–31), suggesting that EDA/EDAR signaling is a critical component of tooth development. Furthermore, these molecules associated with tooth anomalies also regulate enamel knot formation and function. In the present study, we demonstrated that NKX2-3 is a potential target gene of EDA/EDAR signaling, as it regulates cell proliferation through p21 transcription in the molar enamel knot.
The NK2 homeobox family members are known to function as tissue-specific homeobox transcription factors that regulate various fundamental cellular processes, including head patterning, cardiac and lung development, and neural cell specification (32, 33). Nkx2-1 encodes thyroid transcription factor-1 (TTF-1), which is a protein that regulates transcription of genes specific for the thyroid, lungs, and diencephalon. Nkx2-1 mutations lead to surfactant protein dysregulation, which causes interstitial lung disease in patients with brain-lung-thyroid syndrome (34, 35). Our results showed that Nkx2-1 was highly expressed in lung and brain tissues obtained from E14 mouse embryos (see Fig. S1), suggesting that the specific expression pattern of the NK2 homeobox family is critical for tissue-specific growth and development. NKX2-3 controls development of small intestine splenic morphology as well as shaping of lymphocyte dynamics and vasculature (36–41) and also salivary gland and tooth development (18). Absence of Nkx2-3 in mouse molars was reported to result in no cusp formation, whereas there were no phenotypical changes in the incisors (18), suggesting that NKX2-3 is a critical regulator for molar cusp formation. However, the molecular mechanism of NKX2-3 in regard to tooth development remains unclear.
An important event during tooth development is formation of the enamel knot and its expression of several signaling molecules. During knot formation, a group of proliferating cells exit the cell cycle and are regulated by the cyclin-dependent kinase inhibitor p21 (20). In the present study, we found NKX2-3 to be highly expressed in the enamel knot region of molars and involved in regulation of p21 expression in both primary and secondary enamel knots as a homeobox transcription factor. BMP2 and BMP4, as well as BMP7 are expressed in early dental epithelium and regulate tooth morphogenesis (42–46). The expression of Bmp4 shifts to the mesenchyme at the bud stage (47), indicating that BMPs are critical regulators for epithelial–mesenchymal interaction during early tooth development. BMP signaling is an inducer of p21 and associated with apoptosis in the enamel knot (20). Together, these findings suggest that BMP2 in the enamel knot may induce BMP4 in mesenchyme through an epithelial–mesenchymal interaction at the bud stage, and then BMP signaling is accelerated by stimulation of BMP2 and BMP4 in the enamel knot area in an autocrine or paracrine manner (Fig. 5I). BMP signaling may also be critically involved in cusp formation and tooth germ development by regulating cell proliferation and differentiation in the enamel knot. The present findings revealed that NKX2-3 regulates Bmp2 and Bmpr2 expression, indicating NKX2-3 as a regulator of the BMP signaling pathway in the enamel knot. Furthermore, we found that BMP2 had no effects on cusp formation in an Nkx2-3 siRNA–treated organ culture (data not shown), which may be related to down-regulation of Bmp2 and Bmpr2 expression in Nkx2-3-silencing cells.
SOX2 is a member of the family of SRY-related HMG transcription factors that are important for cell fate and differentiation in developmental processes and are also essential for embryonic development (48–50). During tooth development, Sox2+ cells contribute to all epithelial cell lineages and function as dental epithelial stem cells in incisor and molar development (51, 52). Using a lineage tracing method, a recent study revealed the cell lineage of enamel knots (21), which demonstrated that the mouse incisor enamel knot is derived from Sox2+ cells, some of which differentiate into ameloblasts but not into stem cells in the cervical loop. On the other hand, in molars, primary enamel knot cells give rise to a buccal secondary enamel knot. Thus, cusp formation is controlled by construction of signaling centers that were originally SOX2-expressing stem cells. The present findings showed that NKX2-3 regulates SOX2 expression in M3H1 cells and ex vivo in the tooth germ, suggesting a crucial role for NKX2-3 in enamel knot formation and cell fate determination. Furthermore, differences in tooth shape between incisors and molars are due to the level of expression of NKX2-3 during odontogenesis.
We also identified NKX2-3 bound to the promoter region of Cdkn1a (p21) in ChIP assay findings. An NKX2-3 binding motif exists near the TSS of Cdkn1a, indicating that NKX2-3 directly regulates Cdkn1a expression by binding at the promoter region. NKX2-3 has been shown to bind with a variety of different genes, suggesting that other genes may be targeted for control of cell proliferation during tooth development.
In summary, we found that NKX2-3 is highly expressed in teeth, especially during the early developmental stage. Nkx2-3 is induced by EDA as a target molecule of the EDA/EDAR pathway in dental epithelial cells. NKX2-3 regulates cell proliferation by controlling p21 gene expression by binding directly to its promoter or through the BMP signaling pathway. We concluded that NKX2-3 is a critical regulator of cusp formation and has an important role in regulating the fate of enamel knot cells.
Experimental procedures
Microarray analysis
All animal experiments were approved by the ethics committee of the Kyushu University Animal Experiment Center (protocol A26-208-0). Total RNA samples from lower first molars and whole embryos from E14 mice were isolated using TRIzol reagent (Life Technologies, Inc.) and purified with an RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. RNA quality was verified using an Experion automated electrophoresis system (Bio-Rad), with the results showing RQI (RNA quality indicator) values of 10.0 for tooth RNA and 9.9 for whole-embryo RNA. Labeling and array hybridization were performed using standard protocols at the Research Support Center of the Research Center for Human Disease Modeling of Kyushu University. Gene expression profiles were analyzed using a chip-based gene array (MouseWG-6 version 2, Illumina, Santa Clara, CA). Data were normalized using Genome Studio (Illumina). Gene expression analysis was performed using Subio Platform version 1.18 (Subio, Amami, Japan).
Tissue preparation
Pregnant mice were euthanized by anesthesia, and mouse embryos were immediately dissected. Embryo heads were fixed with 4% paraformaldehyde in PBS for 5 h at 4 °C, and samples were embedded in O.C.T. compound (Sakura Finetek, Tokyo, Japan). Frozen sections of mandibular molars and incisors were obtained from mouse heads at each stage (E13, E14, and E16) and cut into 10-μm-thick sections.
RNA isolation and qRT-PCR analysis
Total RNA was isolated from E14 mouse tissues (tooth, skin, lung, liver, kidney, heart, eye, and brain) and from molar tooth buds at each developmental stage (E11, E13, E14, E15, and E18) using TRIzol reagent (Life Technologies) and then purified using an RNeasy Mini kit (Qiagen). cDNA was synthesized using SuperScript III reverse transcriptase reagent (Life Technologies). The specific forward and reverse primers used for qRT-PCR were as follows: Nkx2-3, 5′-tggccctgatgatgttacca-3′ and 5′-ggaaaactgcgtcccttcag-3′; Cdkn1a, 5′-tcccgactcttgacattgct-3′ and 5′-tccaaaatagaggggcagct-3′; Krt14, 5′-gtacgagaagatggcggaga-3′ and 5′-ctttcatgctgagctgggac-3′; Vim, 5′-cagcagtatgaaagcgtgg-3′ and 5′-ggaagaaaaggttggcagag-3′; and glyceraldehyde 3-phosphate dehydrogenase (Gapdh), 5′-ggagcgagaccccactaacatc-3′ and 5′-ctcgtggttcacacccatcac-3′. Expression of each gene was normalized to that of Gapdh. qRT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad) with a CFX Connect Real-Time PCR detection system (Bio-Rad).
Organ cultures
Tooth germs from mandibular molars dissected from E13 mice embryos were seeded into cell culture inserts (BD Falcon, BD Biosciences) and grown using an air–liquid interface culture technique in Dulbecco's modified Eagle's medium (DMEM)/F-12, supplemented with 20% fetal bovine serum (Gibco/Life Technologies), 180 μg/ml ascorbic acid, 2 mm l-glutamine, and 50 units/ml penicillin/streptomycin at 37 °C in a humidified atmosphere of 5% CO2 for 7 days, as described previously (19, 53). For siRNA-mediated knockdown, tooth germs were transfected with siRNA for Nkx2-3 (ON-TARGET Plus L-057189-01–0002, Dharmacon) or control siRNA (ON-TARGET Plus Nontargeting Control Pool D-001810-05, Dharmacon) at a concentration of 500 nm using Lipofectamine 3000 reagent, according to the manufacturer's protocol. EDA-A1 (200 ng/ml; R&D Systems) was added to organ culture medium. Organs were cultured for 2 days and embedded into frozen blocks, and then the sections were immunostained. For evaluation of tooth size, E13 tooth germs were cultured for 7 days, and tooth size was determined using ImageJ software (Wayne Rasband, National Institutes of Health).
Construction of expression vectors
Nkx2-3 expression vectors were constructed using a Gateway cloning system (Life Technologies), according to the manufacturer's protocol. Briefly, the coding sequence of mouse Nkx2-3 cDNA without a stop codon was cloned into a pENTR/D-TOPO vector, and then the following forward and reverse primers were used: 5′-cacccaattaagtggccctgatgatgttaccaagcccggtcacc-3′ and 5′-caaaggatccaccgccggt-3′. Expression vectors were constructed using an LR recombination reaction (pcDNA-DEST40; Life Technologies) tagged with V5-His.
Cell cultures and transfection
Dental epithelial stem cells (M3H1 cell line) were previously isolated from the cervical loop of the mandible incisor of 4-month-old mice (19). Cells were cultured in low-Ca2+ DMEM, which included DMEM (21068028, no calcium, Gibco/Life Technologies) supplemented with 3.0 mm CaCl2, 1% l-glutamine, 1% sodium pyruvate, 1% penicillin/streptomycin, and 10% Ca2+-free fetal bovine serum (10270, Gibco/Life Technologies) at 37 °C in a humidified incubator in an atmosphere containing 5% CO2. Cells were cultured for 48 h in low-Ca2+ DMEM with additional recombinant protein growth factors, such as BMP2 (100 ng/ml; Wako), BMP4 (100 ng/ml; Wako), EDA-A1 (200 ng/ml; R&D Systems), SHH (100 ng/ml; R&D Systems), FGF9 (2.5 ng/ml; R&D Systems), and Wnt3a (5 ng/ml; R&D Systems).
For transfection, cells were cultured in 12-well plates at a density of 2 × 105 cells/well in low-Ca2+ DMEM and then transfected with an expression vector using Lipofectamine 3000 with Plus reagent (Life Technologies), according to the manufacturer's protocol.
Cell proliferation and BrdU incorporation
M3H1 cells were seeded into 96-well plates and then transfected with an Nkx2-3 expression vector. Proliferation was determined after culturing for 1, 2, 3, 5, and 7 days using a Cell Counting Kit (CCK)-8 (Dojindo Laboratories), according to the manufacturer's protocol. BrdU incorporation was assayed using a BrdU labeling kit (Roche Diagnostics). Cells transfected with Nkx2-3 were cultured for 48 h, and then BrdU was applied to the plates for 30 min, and incorporated BrdU was detected after washing with PBS, according to the manufacturer's protocol. BrdU-positive cells were counted under a fluorescent microscope.
Immunohistochemistry
Immunostaining of frozen sections and M3H1 cells was performed using primary antibodies against NKX2-3 (1:500; Abcam), p21 (1:500, Abcam), perlecan (1:500, Chemicon International), Ki67 (1:500, Cell Signaling Technology), and SOX2 (1:500, Abcam) for 16 h at 4 °C. Cells and tissue sections were then incubated with species-specific secondary antibodies conjugated with Alexa 488 or Alexa 594 fluorescent dye (Life Technologies) for 1 h at room temperature. Nuclei were visualized by DAPI. Sections were mounted with Vectashield mounting medium (Mountant, PermaFluor, TA-030-FM, Thermo Scientific). Images were captured using a C2 confocal microscope (Nikon, Tokyo, Japan) and analyzed with NIS-Elements AR software, version 4.00 (Nikon). Sox2+ cells were counted under a fluorescent microscope.
Western blotting
Cells were washed twice with 1 mm ice-cold sodium orthovanadate (Sigma-Aldrich) in PBS and lysed with CelLytic M (Sigma-Aldrich) supplemented with a 1% protease inhibitor mixture (Sigma-Aldrich) and 1 mm phenylmethylsulfonyl fluoride (Sigma-Aldrich), and then 10 μg of protein from each sample was separated using a 4–12% SDS-polyacrylamide gel (NuPAGE, Invitrogen) and analyzed by Western blotting. The blotted membranes were incubated with antibodies to NKX2-3 (1:500; Abcam), p21 (1:500; Abcam), c-Myc (1:500; Cell Signaling Technology), c-Jun (1:500; Cell Signaling Technology), cyclin D1 (1:500; Cell Signaling Technology), SOX2 (1:500; Abcam), Ki67 (1:500; Cell Signaling Technology), p-Smad1/5/8 (1:500; Cell Signaling Technology), p-Smad1 (1:500; Cell Signaling Technology), Smad1 (1:500; Cell Signaling Technology), Smad5 (1:500; Cell Signaling Technology), and GAPDH (1:500; Cell Signaling Technology), and then signals were detected using an ECL kit (Amersham Biosciences) and visualized with an ImageQuant LAS 4000 system (GE Healthcare).
ChIP assay
ChIP assays were performed using M3H1 cells transfected with an Nkx2-3 vector or mock vector with a ChIP-IT express kit (Active Motif), according to the manufacturer's protocol. Cross-linked chromatin lysates were sonicated and incubated with protein G–magnet beads with a V5 antibody (M215-11, MBL) for 6 h at 4 °C. Cross-linking was reversed using a reverse cross-linking buffer, and then DNA fragments were analyzed by qRT-PCR using the following primer pairs: Cdkn1a site 1, 5′-tccttttctgggaagtggtg-3′ and 5′-tcctctggggctgatagaaa-3′; Cdkn1a site 2, 5′-cctagaaagcaagcctgtgg-3′ and 5′-agcagcccataaacatccat-3′.
Luciferase assay
A Cdkn1a reporter plasmid was constructed by inserting a −435 to +81 promoter sequence into a pGL4.15 vector (Promega). The following forward and reverse primers were used: 5′-tctatcctgaccctcgtgct-3′ and 5′-tggggtctctgtctccattc-3′. A Cdkn1a mutant reporter plasmid was constructed using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies), and the following forward and reverse primers were used: 5′-ctgacagtccctctttgggggtcccctgtccttttctgg-3′ and 5′-gactgtcagggagaaacccccaggggacaggaaaagacc-3′. These two reporter vectors were transfected into M3H1 cells either with a mock vector or the Nkx2-3 expression vector. A pRL-TK vector that encodes Renilla luciferase was used as an internal control to co-transfect M3H1 cells. Activity was determined 48 h later using a Dual-Luciferase reporter assay system (Promega) with a luminometer (LB942, Berthold). Firefly luciferase activity was normalized for Renilla luciferase activity as an internal control.
Statistical analysis
All experiments were repeated at least three times to confirm reproducibility. Statistical significance was determined using a two-tailed unpaired Student's t test with Prism version 6 (GraphPad Software, La Jolla, CA). One-way analysis of variance and Tukey's multiple-comparison test were utilized for quantification between multiple groups. Differences with a p value < 0.05 were considered to be statistically significant.
Author contributions
X. H., K. Y., and S. F. designed the study, performed experiments, analyzed data, and wrote the paper. K. M., C. A., K. F., T. Y., Y. C., K. S., T. I., A. Y., and T. T. participated in performing the experiments and analysis of data. S. F. and I. T. edited the paper. All authors have read and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We thank Megumi Kiyota (Research Support Center, Graduate School of Medical Sciences, Kyushu University) for assistance with the microarray analysis.
This work was supported in part by Grants-in-Aid for Scientific Research 18H03012, 17K19765, and 15H05688 (to K. Y.); 16H07065 (to K. M.); 15K11359 (to K. S.); 16H05548 (to A. Y.); and 17H01606 (to S. F.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1.
- SHH
- sonic hedgehog
- qRT-PCR
- quantitative real-time PCR
- BMP
- bone morphogenetic protein
- FGF
- fibroblast growth factor
- E
- embryonic day
- TSS
- transcription start site
- DMEM
- Dulbecco's modified Eagle's medium
- BrdU
- bromodeoxyuridine
- DAPI
- 4′,6-diamidino-2-phenylindole
- p-
- phosphorylated
- MMTV
- mouse mammary tumor virus
- EDA
- ectodysplasin-A
- EDAR
- ectodysplasin-A receptor.
References
- 1. Lumsden A. G. (1988) Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development 103, 155–169 [DOI] [PubMed] [Google Scholar]
- 2. Thesleff I., and Hurmerinta K. (1981) Tissue interactions in tooth development. Differentiation 18, 75–88 10.1111/j.1432-0436.1981.tb01107.x [DOI] [PubMed] [Google Scholar]
- 3. MacKenzie A., Ferguson M. W., and Sharpe P. T. (1992) Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development 115, 403–420 [DOI] [PubMed] [Google Scholar]
- 4. Jernvall J., and Thesleff I. (2000) Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 92, 19–29 10.1016/S0925-4773(99)00322-6 [DOI] [PubMed] [Google Scholar]
- 5. Catón J., and Tucker A. S. (2009) Current knowledge of tooth development: patterning and mineralization of the murine dentition. J. Anat. 214, 502–515 10.1111/j.1469-7580.2008.01014.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thesleff I., Keränen S., and Jernvall J. (2001) Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation. Adv. Dent. Res. 15, 14–18 10.1177/08959374010150010401 [DOI] [PubMed] [Google Scholar]
- 7. Tickle C. (2015) How the embryo makes a limb: determination, polarity and identity. J. Anat. 227, 418–430 10.1111/joa.12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bénazet J. D., and Zeller R. (2009) Vertebrate limb development: moving from classical morphogen gradients to an integrated 4-dimensional patterning system. Cold Spring Harb. Perspect. Biol. 1, a001339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukumoto S., Kiba T., Hall B., Iehara N., Nakamura T., Longenecker G., Krebsbach P. H., Nanci A., Kulkarni A. B., and Yamada Y. (2004) Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 167, 973–983 10.1083/jcb.200409077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshizaki K., Yamamoto S., Yamada A., Yuasa K., Iwamoto T., Fukumoto E., Harada H., Saito M., Nakasima A., Nonaka K., Yamada Y., and Fukumoto S. (2008) Neurotrophic factor neurotrophin-4 regulates ameloblastin expression via full-length TrkB. J. Biol. Chem. 283, 3385–3391 10.1074/jbc.M704913200 [DOI] [PubMed] [Google Scholar]
- 11. Arakaki M., Ishikawa M., Nakamura T., Iwamoto T., Yamada A., Fukumoto E., Saito M., Otsu K., Harada H., Yamada Y., and Fukumoto S. (2012) Role of epithelial-stem cell interactions during dental cell differentiation. J. Biol. Chem. 287, 10590–10601 10.1074/jbc.M111.285874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshizaki K., Hu L., Nguyen T., Sakai K., Ishikawa M., Takahashi I., Fukumoto S., DenBesten P. K., Bikle D. D., Oda Y., and Yamada Y. (2017) Mediator 1 contributes to enamel mineralization as a coactivator for Notch1 signaling and stimulates transcription of the alkaline phosphatase gene. J. Biol. Chem. 292, 13531–13540 10.1074/jbc.M117.780866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tucker A. S., Headon D. J., Courtney J. M., Overbeek P., and Sharpe P. T. (2004) The activation level of the TNF family receptor, Edar, determines cusp number and tooth number during tooth development. Dev. Biol. 268, 185–194 10.1016/j.ydbio.2003.12.019 [DOI] [PubMed] [Google Scholar]
- 14. Ferguson B. M., Brockdorff N., Formstone E., Ngyuen T., Kronmiller J. E., and Zonana J. (1997) Cloning of Tabby, the murine homolog of the human EDA gene: evidence for a membrane-associated protein with a short collagenous domain. Hum. Mol. Genet. 6, 1589–1594 10.1093/hmg/6.9.1589 [DOI] [PubMed] [Google Scholar]
- 15. Pispa J., Jung H. S., Jernvall J., Kettunen P., Mustonen T., Tabata M. J., Kere J., and Thesleff I. (1999) Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev. Biol. 216, 521–534 10.1006/dbio.1999.9514 [DOI] [PubMed] [Google Scholar]
- 16. Harvey R. P. (1996) NK-2 homeobox genes and heart development. Dev. Biol. 178, 203–216 10.1006/dbio.1996.0212 [DOI] [PubMed] [Google Scholar]
- 17. Stanfel M. N., Moses K. A., Schwartz R. J., and Zimmer W. E. (2005) Regulation of organ development by the NKX-homeodomain factors: an NKX code. Cell. Mol. Biol. (Noisy-le-grand) Suppl 51, OL785–OL799 [PubMed] [Google Scholar]
- 18. Biben C., Wang C. C., and Harvey R. P. (2002) NK-2 class homeobox genes and pharyngeal/oral patterning: Nkx2-3 is required for salivary gland and tooth morphogenesis. Int. J. Dev. Biol. 46, 415–422 [PubMed] [Google Scholar]
- 19. Arai C., Yoshizaki K., Miyazaki K., Saito K., Yamada A., Han X., Funada K., Fukumoto E., Haruyama N., Iwamoto T., Takahashi I., and Fukumoto S. (2017) Nephronectin plays critical roles in Sox2 expression and proliferation in dental epithelial stem cells via EGF-like repeat domains. Sci. Rep. 7, 45181 10.1038/srep45181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jernvall J., Aberg T., Kettunen P., Keränen S., and Thesleff I. (1998) The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development 125, 161–169 [DOI] [PubMed] [Google Scholar]
- 21. Du W., Hu J. K., Du W., and Klein O. D. (2017) Lineage tracing of epithelial cells in developing teeth reveals two strategies for building signaling centers. J. Biol. Chem. 292, 15062–15069 10.1074/jbc.M117.785923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stockton D. W., Das P., Goldenberg M., D'Souza R. N., and Patel P. I. (2000) Mutation of PAX9 is associated with oligodontia. Nat. Genet. 24, 18–19 10.1038/71634 [DOI] [PubMed] [Google Scholar]
- 23. Vastardis H., Karimbux N., Guthua S. W., Seidman J. G., and Seidman C. E. (1996) A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 13, 417–421 10.1038/ng0896-417 [DOI] [PubMed] [Google Scholar]
- 24. Mostowska A., Biedziak B., and Jagodzinski P. P. (2006) Axis inhibition protein 2 (AXIN2) polymorphisms may be a risk factor for selective tooth agenesis. J. Hum. Genet. 51, 262–266 10.1007/s10038-005-0353-6 [DOI] [PubMed] [Google Scholar]
- 25. Tao R., Jin B., Guo S. Z., Qing W., Feng G. Y., Brooks D. G., Liu L., Xu J., Li T., Yan Y., and He L. (2006) A novel missense mutation of the EDA gene in a Mongolian family with congenital hypodontia. J. Hum. Genet. 51, 498–502 10.1007/s10038-006-0389-2 [DOI] [PubMed] [Google Scholar]
- 26. Mues G., Tardivel A., Willen L., Kapadia H., Seaman R., Frazier-Bowers S., Schneider P., and D'Souza R. N. (2010) Functional analysis of Ectodysplasin-A mutations causing selective tooth agenesis. Eur. J. Hum. Genet. 18, 19–25 10.1038/ejhg.2009.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rasool M., Schuster J., Aslam M., Tariq M., Ahmad I., Ali A., Entesarian M., Dahl N., and Baig S. M. (2008) A novel missense mutation in the EDA gene associated with X-linked recessive isolated hypodontia. J. Hum. Genet. 53, 894–898 10.1007/s10038-008-0323-x [DOI] [PubMed] [Google Scholar]
- 28. Song S., Han D., Qu H., Gong Y., Wu H., Zhang X., Zhong N., and Feng H. (2009) EDA gene mutations underlie non-syndromic oligodontia. J. Dent. Res. 88, 126–131 10.1177/0022034508328627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zeng B., Zhao Q., Li S., Lu H., Lu J., Ma L., Zhao W., and Yu D. (2017) Novel EDA or EDAR mutations identified in patients with X-linked hypohidrotic ectodermal dysplasia or non-syndromic tooth agenesis. Genes (Basel) 8, 259 10.3390/genes8100259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kieri C. F., Bergendal B., Lind L. K., Schmitt-Egenolf M., and Stecksén-Blicks C. (2014) EDAR-induced hypohidrotic ectodermal dysplasia: a clinical study on signs and symptoms in individuals with a heterozygous c.1072C > T mutation. BMC Med. Genet. 15, 57 10.1186/1471-2350-15-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kimura R., Yamaguchi T., Takeda M., Kondo O., Toma T., Haneji K., Hanihara T., Matsukusa H., Kawamura S., Maki K., Osawa M., Ishida H., and Oota H. (2009) A common variation in EDAR is a genetic determinant of shovel-shaped incisors. Am. J. Hum. Genet. 85, 528–535 10.1016/j.ajhg.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McGinnis W., and Krumlauf R. (1992) Homeobox genes and axial patterning. Cell 68, 283–302 10.1016/0092-8674(92)90471-N [DOI] [PubMed] [Google Scholar]
- 33. Garcia-Fernàndez J. (2005) The genesis and evolution of homeobox gene clusters. Nat. Rev. Genet. 6, 881–892 10.1038/nrg1723 [DOI] [PubMed] [Google Scholar]
- 34. Guillot L., Carré A., Szinnai G., Castanet M., Tron E., Jaubert F., Broutin I., Counil F., Feldmann D., Clement A., Polak M., and Epaud R. (2010) NKX2-1 mutations leading to surfactant protein promoter dysregulation cause interstitial lung disease in “Brain-Lung-Thyroid Syndrome”. Hum. Mutat. 31, E1146–E1162 10.1002/humu.21183 [DOI] [PubMed] [Google Scholar]
- 35. Carré A., Szinnai G., Castanet M., Sura-Trueba S., Tron E., Broutin-L'Hermite I., Barat P., Goizet C., Lacombe D., Moutard M. L., Raybaud C., Raynaud-Ravni C., Romana S., Ythier H., Léger J., and Polak M. (2009) Five new TTF1/NKX2.1 mutations in brain-lung-thyroid syndrome: rescue by PAX8 synergism in one case. Hum. Mol. Genet. 18, 2266–2276 10.1093/hmg/ddp162 [DOI] [PubMed] [Google Scholar]
- 36. Pabst O., Förster R., Lipp M., Engel H., and Arnold H. H. (2000) NKX2.3 is required for MAdCAM-1 expression and homing of lymphocytes in spleen and mucosa-associated lymphoid tissue. EMBO J. 19, 2015–2023 10.1093/emboj/19.9.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang C. C., Biben C., Robb L., Nassir F., Barnett L., Davidson N. O., Koentgen F., Tarlinton D., and Harvey R. P. (2000) Homeodomain factor Nkx2-3 controls regional expression of leukocyte homing coreceptor MAdCAM-1 in specialized endothelial cells of the viscera. Dev. Biol. 224, 152–167 10.1006/dbio.2000.9749 [DOI] [PubMed] [Google Scholar]
- 38. Tarlinton D., Light A., Metcalf D., Harvey R. P., and Robb L. (2003) Architectural defects in the spleens of Nkx2-3-deficient mice are intrinsic and associated with defects in both B cell maturation and T cell-dependent immune responses. J. Immunol. 170, 4002–4010 10.4049/jimmunol.170.8.4002 [DOI] [PubMed] [Google Scholar]
- 39. Pabst O., Zweigerdt R., and Arnold H. H. (1999) Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development 126, 2215–2225 [DOI] [PubMed] [Google Scholar]
- 40. Czömpöly T., Lábadi A., Kellermayer Z., Olasz K., Arnold H. H., and Balogh P. (2011) Transcription factor Nkx2-3 controls the vascular identity and lymphocyte homing in the spleen. J. Immunol. 186, 6981–6989 10.4049/jimmunol.1003770 [DOI] [PubMed] [Google Scholar]
- 41. Robles E. F., Mena-Varas M., Barrio L., Merino-Cortes S. V., Balogh P., Du M. Q., Akasaka T., Parker A., Roa S., Panizo C., Martin-Guerrero I., Siebert R., Segura V., Agirre X., Macri-Pellizeri L., et al. (2016) Homeobox NKX2-3 promotes marginal-zone lymphomagenesis by activating B-cell receptor signalling and shaping lymphocyte dynamics. Nat. Commun. 7, 11889 10.1038/ncomms11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thesleff I., and Sharpe P. (1997) Signalling networks regulating dental development. Mech. Dev. 67, 111–123 10.1016/S0925-4773(97)00115-9 [DOI] [PubMed] [Google Scholar]
- 43. Feng J., Yang G., Yuan G., Gluhak-Heinrich J., Yang W., Wang L., Chen Z., Schulze McDaniel J., Donly K. J., Harris S. E., Macdougall M., and Chen S. (2011) Abnormalities in the enamel in bmp2-deficient mice. Cells Tissues Organs 194, 216–221 10.1159/000324644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Laurikkala J., Kassai Y., Pakkasjärvi L., Thesleff I., and Itoh N. (2003) Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev. Biol. 264, 91–105 10.1016/j.ydbio.2003.08.011 [DOI] [PubMed] [Google Scholar]
- 45. Jia S., Kwon H. E., Lan Y., Zhou J., Liu H., and Jiang R. (2016) Bmp4-Msx1 signaling and Osr2 control tooth organogenesis through antagonistic regulation of secreted Wnt antagonists. Dev. Biol. 420, 110–119 10.1016/j.ydbio.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zouvelou V., Luder H. U., Mitsiadis T. A., and Graf D. (2009) Deletion of BMP7 affects the development of bones, teeth, and other ectodermal appendages of the orofacial complex. J. Exp. Zool. B Mol. Dev. Evol. 312, 361–374 10.1002/jez.b.21262 [DOI] [PubMed] [Google Scholar]
- 47. Vainio S., Karavanova I., Jowett A., and Thesleff I. (1993) Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell 75, 45–58 10.1016/S0092-8674(05)80083-2 [DOI] [PubMed] [Google Scholar]
- 48. Kamachi Y., Uchikawa M., and Kondoh H. (2000) Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 16, 182–187 10.1016/S0168-9525(99)01955-1 [DOI] [PubMed] [Google Scholar]
- 49. Juuri E., Jussila M., Seidel K., Holmes S., Wu P., Richman J., Heikinheimo K., Chuong C. M., Arnold K., Hochedlinger K., Klein O., Michon F., and Thesleff I. (2013) Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development 140, 1424–1432 10.1242/dev.089599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Juuri E., Isaksson S., Jussila M., Heikinheimo K., and Thesleff I. (2013) Expression of the stem cell marker, SOX2, in ameloblastoma and dental epithelium. Eur. J. Oral Sci. 121, 509–516 10.1111/eos.12095 [DOI] [PubMed] [Google Scholar]
- 51. Juuri E., Saito K., Ahtiainen L., Seidel K., Tummers M., Hochedlinger K., Klein O. D., Thesleff I., and Michon F. (2012) Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev. Cell 23, 317–328 10.1016/j.devcel.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li J., Feng J., Liu Y., Ho T. V., Grimes W., Ho H. A., Park S., Wang S., and Chai Y. (2015) BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev. Cell 33, 125–135 10.1016/j.devcel.2015.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miyazaki K., Yoshizaki K., Arai C., Yamada A., Saito K., Ishikawa M., Xue H., Funada K., Haruyama N., Yamada Y., Fukumoto S., and Takahashi I. (2016) Plakophilin-1, a novel Wnt signaling regulator, is critical for tooth development and ameloblast differentiation. PLoS One 11, e0152206 10.1371/journal.pone.0152206 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.