Twenty-three Klebsiella pneumoniae (blaDHA-1) clinical isolates exhibited a range of susceptibilities to LYS228, with MICs of ≥8 μg/ml for 9 of these. Mutants with decreased susceptibility to LYS228 and upregulated expression of blaDHA-1 were selected from representative isolates.
KEYWORDS: DHA-1, Klebsiella, LYS228, beta-lactamases, peptidoglycan, plasmid
ABSTRACT
Twenty-three Klebsiella pneumoniae (blaDHA-1) clinical isolates exhibited a range of susceptibilities to LYS228, with MICs of ≥8 μg/ml for 9 of these. Mutants with decreased susceptibility to LYS228 and upregulated expression of blaDHA-1 were selected from representative isolates. These had mutations in the chromosomal peptidoglycan recycling gene mpl or ampD. Preexisting mpl mutations were also found in some of the clinical isolates examined, and these had strongly upregulated expression of blaDHA-1.
TEXT
Resistance to β-lactams often results from the expression of ever-evolving serine β-lactamase enzymes (SBLs) that degrade penicillins, cephalosporins, monobactams, and, in some cases, carbapenems (1). Carbapenem-resistant Enterobacteriaceae (CRE) are increasingly regarded as a public health issue (2, 3). β-Lactamase inhibitors (BLIs) counter the effect of SBLs, but metallo-β-lactamases (MBLs) such as New Delhi metallo-β-lactamase-1 (NDM-1), usually expressed together with SBLs, have emerged in Klebsiella pneumoniae and Escherichia coli (4). No inhibitors of MBLs are currently available clinically (5). The monobactam aztreonam (ATM) is stable to MBLs (6, 7) but not all SBLs. To address this, the combination of ATM with the β-lactamase inhibitor avibactam (AVI) is currently in clinical trials (ClinicalTrials registration no. NCT01689207).
We designed the monobactam LYS228, now in phase II clinical trials (ClinicalTrials registration no. NCT03354754), that is stable to both MBLs and SBLs (8). Accordingly, the MIC90 of LYS228 was 1 μg/ml against 271 Enterobacteriaceae clinical isolates, including those expressing a variety of β-lactamases (9). The MIC90 of LYS228 was 1 μg/ml against the 81 K. pneumoniae isolates included in that panel (9). Four of the 81 were annotated as having the class C β-lactamase blaDHA-1. Of these, one was found to have a truncation of the blaDHA-1 gene (data not shown). Of the other three, two were susceptible to LYS228 (NB29263 and NB29289, MIC of 0.125 to 0.5) (Table 1), but one was less susceptible (NB29293, mode MIC of 4 μg/ml) (Table 1). K. pneumoniae lacks a chromosomal ampC but can acquire class C genes (e.g., blaDHA-1) on large plasmids. Isolates with plasmid-borne blaDHA-1 have been reported in several geographic locations (10–24). Plasmid-borne blaDHA-1 likely originated from the chromosome of Morganella morganii and carries with it the associated ampR gene, making it inducible (25, 26). Inducible blaDHA-1 itself may not significantly affect susceptibility to many β-lactams, but combined with other mechanisms, such as porin defects (e.g., OmpK35 and/or OmpK36 in K. pneumoniae), it can become more important in some isolates (27, 28). These factors can complicate susceptibility testing for β-lactams, and a lack of robust methodology for identifying these isolates may result in underestimating their prevalence (19, 29–31). To explore if the variable impact of blaDHA-1 on susceptibility extended to LYS228, we expanded our panel to 23 K. pneumoniae clinical isolates, all having blaDHA-1 (among other enzymes), and determined if this panel trended toward decreased LYS228 susceptibility. The isolates originated in the United States (n = 1), Europe (n = 5), the Middle East (n = 3), and the Asia-Pacific (n = 13). The presence of the ampR-blaDHA-1 region was confirmed for all isolates by PCR and sequencing, although strain NB29381 had a C-to-A mutation 37 bp upstream of blaDHA-1 and NB29390 had an A-to-T mutation 74 bp upstream of blaDHA-1. A range of susceptibilities to LYS228 was observed (0.125 to >64 μg/ml) (Table 1), but the LYS228 MIC was ≥8 μg/ml for 9 of the strains. Therefore, although some isolates were susceptible to LYS228, the panel was overall significantly less susceptible than the set of K. pneumoniae isolates that were not selected based on the presence of blaDHA-1 (9). We partially characterized two strains from this panel that had different susceptibilities to LYS228. K. pneumoniae NB29293 susceptibility was variable, with MICs ranging from 2 to 16 μg/ml, consistent with inducibility of blaDHA-1 (mode MIC, 4 μg/ml) (Table 1). Addition of avibactam (4 μg/ml) improved the LYS228 MIC to 0.25 μg/ml (Table 1). An intact ompK35 porin gene could not be amplified by PCR, and the ompK36 porin gene encoded multiple alterations to the porin protein relative to the reference strain ATCC 43816 (32 and data not shown). These defects may be additive with the effect of the inducible DHA-1, as suggested in previous reports. A second isolate, NB29289, was consistently more susceptible to LYS228 than NB29293 (MIC of 0.125 μg/ml) (Table 1). In contrast to strain NB29293, the genes encoding OmpK35 and OmpK36 only had silent mutations compared to reference strain ATCC 43816 (32). This may explain in part why NB29289 is more sensitive to LYS228 than NB29293, but this remains to be confirmed.
TABLE 1.
Antibiotic susceptibilities of K. pneumoniae clinical isolates harboring blaDHAa
| Strain | Annotated β-lactamase(s)b and relevant mutations | MICc μg/ml |
||||||
|---|---|---|---|---|---|---|---|---|
| LYS228 | LYS228/AVI | ATM | ATM/AVI | CAZ | FEP | MEM | ||
| NB29263 | DHA-1 | 0.5 | ≤0.06 | 2 | 0.25 | 32 | 0.125 | 0.25 |
| NB29289 | SHV-31, DHA-1, VIM-1 | 0.125 | ≤0.06 | 1 | 0.25 | >64 | >16 | 8 |
| NB29289-CDK0033 | NB29289 mpl (1-bp deletion, position 45), selected on LYS228 | 32 | 0.25 | >64 | 8 | >64 | >16 | 8 |
| NB29289-CDK0034 | NB29289 mpl (1-bp deletion, position 275), selected on LYS228 | 8 | ≤0.06 | >64 | 1 | >64 | >16 | >8 |
| NB29293 | SHV-12, CTX-M-15, DHA-1 | 4 | 0.25 | >64 | 1 | >64 | >16 | 0.5 |
| NB29338 | SHV-OSBL(b), DHA-1; mpl (ΔARHVGVLPADAA290–302) | 32 | 0.125 | >64 | 0.25 | >64 | 1 | 2 |
| NB29352 | SHV-OSBL(b), DHA-1; mpl (ΔAEV397–399, LAQKAAAAE*439–448 to RVRLRSLPSA*) | >64 | 1 | >64 | 1 | >64 | 16 | 4 |
| NB29353 | SHV-OSBL(b), TEM-OSBL(b), DHA-1; mpl (G68D) | >64 | ≤0.06 | >64 | ≤0.06 | >64 | 16 | 1 |
| NB29354 | SHV-12(e), DHA-1 | 64 | ≤0.06 | >64 | ≤0.06 | >64 | 32 | 8 |
| NB29379 | SHV-OSBL, TEM-OSBL; DHA-1 | 1 | ≤0.06 | 8 | 0.125 | >64 | 0.125 | 0.06 |
| NB29380 | SHV-OSBL, TEM-OSBL; DHA-1 | 0.25 | 0.125 | 0.5 | 0.25 | 1 | 0.125 | 0.03 |
| NB29381 | SHV-OSBL, DHA-1 | 2 | 0.125 | 16 | 0.25 | 64 | 0.125 | 0.06 |
| NB29382 | SHV-OSBL, DHA-1 | 2 | 0.25 | 8 | 0.125 | >64 | 0.5 | 0.125 |
| NB29383 | SHV-OSBL, TEM-OSBL; DHA-1 | 2 | ≤0.06 | 4 | 0.125 | 32 | 0.25 | 0.06 |
| NB29384 | SHV-OSBL, TEM-OSBL; DHA-1 | 1 | ≤0.06 | 4 | 0.25 | 32 | 0.125 | 0.06 |
| NB29385 | SHV-OSBL, DHA-1 | 8 | 0.25 | 32 | 0.5 | 16 | 4 | 0.25 |
| NB29386 | SHV-OSBL, DHA-1 | 8 | 0.25 | 32 | 0.5 | >64 | 0.25 | 0.06 |
| NB29387 | SHV-OSBL, DHA-1 | 0.5 | 0.125 | 32 | 0.125 | 32 | 0.125 | 0.06 |
| NB29388 | SHV-OSBL, TEM-OSBL; DHA-1; mpl (D63G) | 1 | ≤0.06 | 2 | 0.25 | 64 | 0.125 | 0.125 |
| NB29389 | SHV_OSBL, TEM-OSBL; DHA-1 | 32 | ≤0.06 | 64 | 1 | >64 | 1 | 0.06 |
| NB29390 | SHV-OSBL, DHA-1 | 8 | ≤0.06 | 64 | 0.5 | >64 | 1 | 0.125 |
| NB29391 | SHV-OSBL, DHA-1 | 4 | 0.25 | 8 | 0.25 | 64 | 1 | 0.06 |
| NB29392 | SHV-OSBL, DHA-1 | 1 | ≤0.06 | 4 | 0.125 | 16 | ≤0.06 | 0.06 |
| NB29393 | TEM-OSBL, DHA-1; mpl (G to A, 14 bp upstream of ATG, K131Q, L263F, V432stop) | 32 | 0.125 | >64 | 1 | >64 | 1 | 0.125 |
| NB29293-CDK0021 | NB29293 mpl (D31N), selected on LYS228 | >64 | 0.25 | >64 | 4 | >64 | >16 | 0.5 |
| NB29293-CDK0022 | NB29293 mpl (W247*), selected on LYS228 | >64 | 0.125 | >64 | 4 | >64 | 16 | 0.25 |
| NB29289-CDK0036 | NB29289 ampD (W7G), selected on LYS228 | 16 | ND | >32 | ND | >32 | ND | 4 |
| NB29394 | DHA-1 | 4 | 0.125 | 16 | 0.5 | 64 | 0.25 | 0.125 |
All strains obtained were from International Health Management Associates (IHMA) except for NB29263 (Novartis collection). AVI, avibactam; CAZ, ceftazidime; FEP, cefepime; MEM, meropenem; ND, not determined.
β-Lactamase annotations are from IHMA. OSBL, original-spectrum β-lactamase. The ampR-blaDHA-1 region was confirmed here by PCR.
AVI used at a fixed concentration of 4 μg/ml. Susceptibility testing was performed using a broth microdilution methodology of the CLSI (41).
Consistent with DHA-1 affecting susceptibility to LYS228, mutants with constitutive upregulation of blaDHA-1 could be selected from strain NB29293 in vitro at a frequency of approximately 10−6 by plating cells on LB agar containing 32 μg/ml of LYS228 (8× the mode MIC). Mutant susceptibility was shifted at least 32-fold (MIC of >64 μg/ml). LYS228 activity was restored by the addition of 4 μg/ml avibactam, confirming involvement of a β-lactamase (NB29293-CDK0022) (Table 1). Genome sequencing of NB29293-CDK0022 using previously described methodology (33) revealed a premature stop codon (encoding W247*) in the chromosomal murein-peptide-ligase gene mpl, which was also present in several other mutants. An NB29293 mutant recovered from a time-kill regrowth experiment, conducted as previously described (9) (NB29293-CDK0021; Table 1), harbored a mutation in mpl encoding D31N. These mutants were constitutively upregulated for expression of blaDHA-1 (Fig. 1). Mutants were also selected from the more sensitive strain NB29289 on agar containing 1 μg/ml LYS228. Of six mutants tested, four had a 1-bp deletion (frameshift) at position 45 and one had a 1-bp deletion at position 275 of mpl. The MIC of LYS228 increased to 8 and 32 μg/ml for these mutants (NB29289-CDK0033 and CDK0034) (Table 1), and again LYS228 susceptibility was restored by the addition of avibactam (Table 1). It should be noted that no mpl mutations were isolated from several non-blaDHA-1 K. pneumoniae isolates during single-step selections described in an accompanying report (33), strengthening the association of mpl mutations with blaDHA-1. Mpl is UDP-N-acetylmuramate:L-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase, involved in peptidoglycan recycling (34, 35), and its mutational loss may induce expression of blaDHA-1 via changes in peptidoglycan intermediates sensed by the plasmid-encoded AmpR regulator. Upregulation of ampC expression caused by mutations in peptidoglycan recycling genes such as ampD or ampG in various bacteria with inducible ampC genes is well studied (36, 37). An in vitro transposon mutagenesis study also found insertions in mpl in Pseudomonas aeruginosa causing AmpR-dependent upregulation of chromosomal ampC and reduced susceptibility to β-lactams (38). A recent report also described the emergence of mpl mutations in P. aeruginosa during serial passaging in the presence of aztreonam (39). The remaining NB29289-derived mutant had an ampD encoding a previously reported W7G substitution shown to upregulate chromosomal ampC expression in some Gram-negative bacteria (40). The MIC of LYS228 increased to 16 for this mutant (Table 1).
FIG 1.
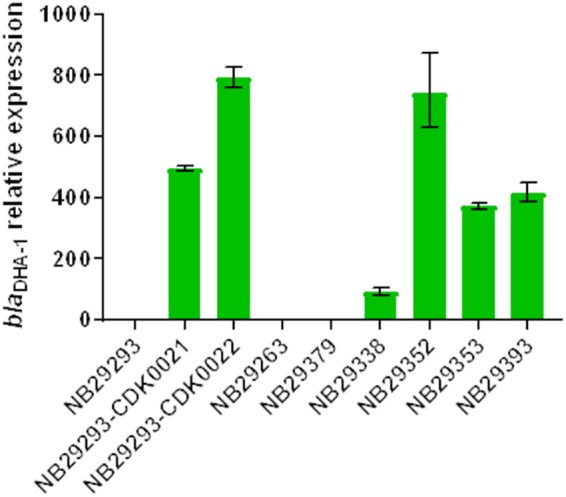
Constitutive blaDHA-1 expression in mpl mutants selected in vitro from K. pneumoniae NB29293 (NB29293-CDK0021 and NB29293-CDK0022) and in clinical isolates harboring alterations in mpl (NB29338, NB29352, NB29353, and NB29393). RT-qPCR was done as previously described (33), and expression analysis was done using the 2−ΔΔCT method (42) relative to uninduced strain NB29293. Two additional clinical isolates harboring wild-type mpl (NB29263 and NB29379) are included for comparison. Data are averages from two biological replicates. Induction of blaDHA-1 transcription (3- to 7-fold) was seen for NB29293 cells exposed to LYS228 at a MIC of 1 μg/ml (data not shown).
The majority of the clinical isolates studied here encoded an Mpl protein identical to that of reference strain ATCC 43816 (32), which does not harbor blaDHA-1. However, isolate NB29353 encoded MplG68D, and mpl from isolates NB29338, NB29352, and NB29393 contained large deletions, frame shifts, and amino acid substitutions (Table 1). Expression of blaDHA-1 was upregulated 90-fold in NB29338 and >300-fold in NB29352, NB29353, and NB29393 relative to strains NB29293, NB29263, and NB29379, which all harbored wild-type mpl (Fig. 1). NB29338, NB29352, NB29353, and NB29393 were also among the least susceptible to LYS228 of the blaDHA-1-containing K. pneumoniae strains tested (Table 1).
In conclusion, this study suggests that inducible blaDHA-1 can decrease susceptibility to LYS228 in some K. pneumoniae clinical isolates, presumably depending on the presence of additional resistance mechanisms in these strains. In cases where β-lactamases may impact the clinical utility of LYS228, pairing with an appropriate β-lactamase inhibitor may restore susceptibility. We also uncovered a novel mechanism of upregulation of plasmid-borne blaDHA-1 expression via chromosomal mpl mutations and show that these mutations occur in clinical isolates. To our knowledge, this is the first report of this mechanism in K. pneumoniae. Delineating the mechanism by which defects in Mpl upregulate DHA-1 expression warrants further study.
ACKNOWLEDGMENTS
We thank Dave Barkan and Peter Skewes-Cox for bioinformatics assistance and Daryl Richie for helpful discussion.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/AAC.01200-18.
REFERENCES
- 1.Bush K. 2013. Proliferation and significance of clinically relevant beta-lactamases. Ann N Y Acad Sci 1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 2.Watkins RR, Bonomo RA. 2016. Overview: global and local impact of antibiotic resistance. Infect Dis Clin North Am 30:313–322. doi: 10.1016/j.idc.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Shlaes DM, Bradford PA. 2018. Antibiotics-from where to where? How the antibiotic miracle is threatened by resistance and a broken market and what we can do about it. Pathog Immun 13:19–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 5.Drawz SM, Papp-Wallace KM, Bonomo RA. 2014. New beta-lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother 58:1835–1846. doi: 10.1128/AAC.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felici A, Amicosante G. 1995. Kinetic analysis of extension of substrate specificity with Xanthomonas maltophilia, Aeromonas hydrophila, and Bacillus cereus metallo-beta-lactamases. Antimicrob Agents Chemother 39:192–199. doi: 10.1128/AAC.39.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felici A, Amicosante G, Oratore A, Strom R, Ledent P, Joris B, Fanuel L, Frere JM. 1993. An overview of the kinetic parameters of class B beta-lactamases. Biochem J 291(Part 1):151–155. doi: 10.1042/bj2910151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reck F, Bermingham A, Blais J, Capka V, Cariaga T, Casarez A, Colvin R, Dean CR, Fekete A, Gong W, Growcott E, Guo H, Jones AK, Li C, Li F, Lin X, Lindvall M, Lopez S, McKenney D, Metzger L, Moser HE, Prathapam R, Rasper D, Rudewicz P, Sethuraman V, Shen X, Shaul J, Simmons RL, Tashiro K, Tang D, Tjandra M, Turner N, Uehara T, Vitt C, Whitebread S, Yifru A, Zang X, Zhu Q. 2018. Optimization of novel monobactams with activity against carbapenem-resistant Enterobacteriaceae–identification of LYS228. Bioorg Med Chem Lett 28:748–755. doi: 10.1016/j.bmcl.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Blais J, Lopez S, Li C, Ruzin A, Ranjitkar S, Dean CR, Leeds JA, Casarez A, Simmons RL, Reck F. 23 July 2018. In vitro activity of LYS228, a novel monobactam antibiotic, against multidrug-resistant Enterobacteriaceae. Antimicrob Agents Chemother doi: 10.1128/AAC.00552-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compain F, Decre D, Fulgencio JP, Berraho S, Arlet G, Verdet C. 2014. Molecular characterization of DHA-1-producing Klebsiella pneumoniae isolates collected during a 4-year period in an intensive care unit. Diagn Microbiol Infect Dis 80:159–161. doi: 10.1016/j.diagmicrobio.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Voulgari E, Poulou A, Dimitroulia E, Politi L, Ranellou K, Gennimata V, Markou F, Pournaras S, Tsakris A. 2015. Emergence of OXA-162 carbapenemase- and DHA-1 AmpC cephalosporinase-producing sequence type 11 Klebsiella pneumoniae causing community-onset infection in Greece. Antimicrob Agents Chemother 60:1862–1864. doi: 10.1128/AAC.01514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chudackova E, Bergerova T, Fajfrlik K, Cervena D, Urbaskova P, Empel J, Gniadkowski M, Hrabak J. 2010. Carbapenem-nonsusceptible strains of Klebsiella pneumoniae producing SHV-5 and/or DHA-1 beta-lactamases in a Czech hospital. FEMS Microbiol Lett 309:62–70. [DOI] [PubMed] [Google Scholar]
- 13.Empel J, Hrabak J, Kozinska A, Bergerova T, Urbaskova P, Kern-Zdanowicz I, Gniadkowski M. 2010. DHA-1-producing Klebsiella pneumoniae in a teaching hospital in the Czech Republic. Microb Drug Resist 16:291–295. doi: 10.1089/mdr.2010.0030. [DOI] [PubMed] [Google Scholar]
- 14.Livermore DM, Mushtaq S, Meunier D, Hopkins KL, Hill R, Adkin R, Chaudhry A, Pike R, Staves P, Woodford N, Committee BRSS. 2017. Activity of ceftolozane/tazobactam against surveillance and “problem” Enterobacteriaceae, Pseudomonas aeruginosa and non-fermenters from the British Isles. J Antimicrob Chemother 72:2278–2289. doi: 10.1093/jac/dkx136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingti B, Paul D, Maurya AP, Bora D, Chanda DD, Chakravarty A, Bhattacharjee A. 2017. Occurrence of bla DHA-1 mediated cephalosporin resistance in Escherichia coli and their transcriptional response against cephalosporin stress: a report from India. Ann Clin Microbiol Antimicrob 16:13. doi: 10.1186/s12941-017-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdet C, Benzerara Y, Gautier V, Adam O, Ould-Hocine Z, Arlet G. 2006. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob Agents Chemother 50:607–617. doi: 10.1128/AAC.50.2.607-617.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennequin C, Robin F, Cabrolier N, Bonnet R, Forestier C. 2012. Characterization of a DHA-1-producing Klebsiella pneumoniae strain involved in an outbreak and role of the AmpR regulator in virulence. Antimicrob Agents Chemother 56:288–294. doi: 10.1128/AAC.00164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdalhamid B, Albunayan S, Shaikh A, Elhadi N, Aljindan R. 2017. Prevalence study of plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae lacking inducible ampC from Saudi hospitals. J Med Microbiol 66:1286–1290. doi: 10.1099/jmm.0.000504. [DOI] [PubMed] [Google Scholar]
- 19.Vanwynsberghe T, Verhamme K, Raymaekers M, Cartuyvels R, Van Vaerenbergh K, Boel A, De Beenhouwer H. 2009. A large outbreak of Klebsiella pneumoniae (DHA-1 and SHV-11 positive): importance of detection and treatment of ampC B-lactamases. Open Infect Dis J 3:55–60. doi: 10.2174/1874279300903010055. [DOI] [Google Scholar]
- 20.Jean SS, Hsueh PR, SMART Asia-Pacific Group. 2017. Distribution of ESBLs, AmpC beta-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008–14: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). J Antimicrob Chemother 72:166–171. doi: 10.1093/jac/dkw398. [DOI] [PubMed] [Google Scholar]
- 21.Lin WP, Wang JT, Chang SC, Chang FY, Fung CP, Chuang YC, Chen YS, Shiau YR, Tan MC, Wang HY, Lai JF, Huang IW, Lauderdale TL. 2016. The antimicrobial susceptibility of Klebsiella pneumoniae from community settings in Taiwan, a trend analysis. Sci Rep 6:36280. doi: 10.1038/srep36280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh WS, Wang NY, Feng JA, Weng LC, Wu HH. 2015. Identification of DHA-23, a novel plasmid-mediated and inducible AmpC beta-lactamase from Enterobacteriaceae in Northern Taiwan. Front Microbiol 6:436. doi: 10.3389/fmicb.2015.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoo JS, Byeon J, Yang J, Yoo JI, Chung GT, Lee YS. 2010. High prevalence of extended-spectrum beta-lactamases and plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae isolated from long-term care facilities in Korea. Diagn Microbiol Infect Dis 67:261–265. doi: 10.1016/j.diagmicrobio.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Liu XQ, Liu YR. 2016. Detection and genotype analysis of AmpC beta-lactamase in Klebsiella pneumoniae from tertiary hospitals. Exp Ther Med 12:480–484. doi: 10.3892/etm.2016.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnaud G, Arlet G, Verdet C, Gaillot O, Lagrange PH, Philippon A. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible beta-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob Agents Chemother 42:2352–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philippon A, Arlet G, Jacoby GA. 2002. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother 46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin SY, Bae IK, Kim J, Jeong SH, Yong D, Kim JM, Lee K. 2012. Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA-1 and loss of OmpK35 and/or OmpK36. J Med Microbiol 61:239–245. doi: 10.1099/jmm.0.037036-0. [DOI] [PubMed] [Google Scholar]
- 28.Tsai YK, Liou CH, Fung CP, Lin JC, Siu LK. 2013. Single or in combination antimicrobial resistance mechanisms of Klebsiella pneumoniae contribute to varied susceptibility to different carbapenems. PLoS One 8:e79640. doi: 10.1371/journal.pone.0079640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanwynsberghe T, Verhamme K, Raymaekers M, Cartuyvels R, Boel A, De Beenhouwer H. 2007. Outbreak of Klebsiella pneumoniae strain harbouring an AmpC (DHA-1) and a blaSHV-11 in a Belgian hospital, August-December 2006. Euro Surveill 12:E070201–E070203. [DOI] [PubMed] [Google Scholar]
- 30.Reuland EA, Hays JP, de Jongh DM, Abdelrehim E, Willemsen I, Kluytmans JA, Savelkoul PH, Vandenbroucke-Grauls CM, al Naiemi N. 2014. Detection and occurrence of plasmid-mediated AmpC in highly resistant gram-negative rods. PLoS One 9:e91396. doi: 10.1371/journal.pone.0091396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moland ES, Hanson ND, Black JA, Hossain A, Song W, Thomson KS. 2006. Prevalence of newer beta-lactamases in gram-negative clinical isolates collected in the United States from 2001 to 2002. J Clin Microbiol 44:3318–3324. doi: 10.1128/JCM.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broberg CA, Wu W, Cavalcoli JD, Miller VL, Bachman MA. 2014. Complete genome sequence of Klebsiella pneumoniae strain ATCC 43816 KPPR1, a rifampin-resistant mutant commonly used in animal, genetic, and molecular biology studies. Genome Announc 22:e00924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dean CR, Barkan DT, Bermingham A, Blais J, Casey F, Casarez A, Colvin R, Fuller J, Jones AK, Li C, Lopez S, Metzger LE IV, Mostafavi M, Prathapam R, Rasper D, Reck F, Ruzin A, Shaul J, Shen X, Simmons RL, Skewes-Cox P, Takeoka KT, Tamrakar P, Uehara T, Wei J-R. 2018. Mode of action of the monobactam LYS228 and mechanisms decreasing in vitro susceptibility in Escherichia coli and Klebsiella pneumoniae. Antimicrob Agents Chemother 62:e01200-18. doi: 10.1128/AAC.01200-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mengin-Lecreulx D, van Heijenoort J, Park JT. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate: l-alanyl-gamma-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J Bacteriol 178:5347–5352. doi: 10.1128/jb.178.18.5347-5352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uehara T, Park JT. 15 October 2008, posting date. Peptidoglycan recycling. EcoSal Plus 2008 doi: 10.1128/ecosalplus.4.7.1.5. [DOI] [PubMed] [Google Scholar]
- 36.Jones RN, Baquero F, Privitera G, Inoue M, Wiedermann B. 1997. Inducible B-lactamase-mediated resistance to third-generation cephalosporins. Clin Microbiol Infect 3:S7–S18. doi: 10.1111/j.1469-0691.1997.tb00643.x. [DOI] [Google Scholar]
- 37.Luan Y, Li GL, Duo LB, Wang WP, Wang CY, Zhang HG, He F, He X, Chen SJ, Luo DT. 2015. DHA-1 plasmid-mediated AmpC beta-lactamase expression and regulation of Klebsiella pneumoniae isolates. Mol Med Rep 11:3069–3077. doi: 10.3892/mmr.2014.3054. [DOI] [PubMed] [Google Scholar]
- 38.Tsutsumi Y, Tomita H, Tanimoto K. 2013. Identification of novel genes responsible for overexpression of ampC in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 57:5987–5993. doi: 10.1128/AAC.01291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorth P, McLean K, Ratjen A, Secor PR, Bautista GE, Ravishankar S, Rezayat A, Garudathri J, Harrison JJ, Harwood RA, Penewit K, Waalkes A, Singh PK, Salipante SJ. 2017. Evolved aztreonam resistance is multifactorial and can produce hypervirulence in Pseudomonas aeruginosa. mBio 8:e00517-17. doi: 10.1128/mBio.00517-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrosino JF, Pendleton AR, Weiner JH, Rosenberg SM. 2002. Chromosomal system for studying AmpC-mediated β-lactam resistance mutation in Escherichia coli. Antimicrob Agents Chemother 46:1535–1539. doi: 10.1128/AAC.46.5.1535-1539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacterial that grow aerobically. Approved Standard, 10th ed, supplement M07-A10 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 42.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]


